- 1School of Life Science, South China Normal University, Guangzhou, China
- 2Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Sciences, Guangzhou, China
- 3College of Life Science and Technology, Jinan University, Guangzhou, China
The yellow pond turtle (Mauremys mutica) is widely cultured for food, medicine and for keeping as pets in China. The high risk of disease outbreaks and mortality caused by sudden sharp temperature drop events has significant negative effects on the aquaculture industry of M. mutica. However, the mechanism underlying the damage caused by cold stress is still unclear. To fill this gap, we performed transcriptome sequencing of M. mutica samples collected at three sampling time points (3h, 24h and 48h) during cold treatment, and at a recovery time point (96 h after the end of cold stress). The results showed that immunity and metabolism-related pathways (neuroactive ligand-receptor interactions, cytokine-cytokine receptor interactions and NOD-like receptor signaling pathways) were significantly enriched at both 24h and 48h. The expression of immunity and metabolism-related genes (BS2A1, PIWIL2 and Fads1) significantly decreased at all-time points compared to the control group. These results suggested that impaired immunity and depressed metabolism under cold stress may be the main cause of the massive cold-stress mortality of M. mutica. This study provides novel insights into the molecular mechanisms underlying mass disease and mortality caused by sudden sharp temperature drop events in M. mutica.
1 Introduction
As global climate change becomes increasingly unpredictable, extreme climate events would increase in frequency and intensity, and may lead to sub-lethal physiological and behavioral influences on many species (Rahmstorf and Coumou, 2011). Among these species, aquatic organisms are more vulnerable to extreme climate events, which have been posing significant challenges to aquaculture industry (Ahmed et al., 2019). Temperature is one of the most important environmental factors in aquaculture industry, as this factor can influence the growth, feeding, metabolism and immune function of aquaculture organisms (Yang et al., 2016; Liu et al., 2022). Extreme high or low temperature events can result in illness, death and even extinction of species in some cases (Bucciarelli et al., 2020). Hence, it is urgent to explore the mechanisms underlying the damages to aquaculture organisms, aiming to provide novel insights into the prevention and control of illness and death of aquaculture species.
Recent studies on aquatic species showed that extreme temperatures have a significant influence on immune and metabolic levels, which may be responsible for the illness and even death in aquaculture species. For heat stress, transcriptome analysis showed that acute high temperatures significantly reduced the metabolic levels of Palaemon gravieri (Shi et al., 2022). Another case study on Mauremys mutica highlighted impaired immunity under heat stress following observation of down-regulation of all the differently expressed genes in the immune-related categories of complement and coagulation cascade pathways (Gao et al., 2021). For cold stress, studies on Oreochromis niloticus showed that renal metabolism and immune-related genes were suppressed under cold stress, indicating that differently expressed genes in immune-related pathways of the phagosome and cell adhesion molecules were significantly down-regulated at low temperatures (Zhou et al., 2019). Moreover, ectothermic animals can tolerate low temperatures, but cold stress has been shown to have a significant influence on physiological processes, such as circadian rhythms and steroid and fatty acid biosynthesis in Puntius tetrazona (Liu et al., 2020). Although the molecular mechanism of response to cold stress has been investigated in traditional aquaculture organisms such as fish, the molecular mechanism underlying illness and death in aquaculture reptiles is still unclear.
The yellow pond turtle (Mauremys mutica), is widely cultured for food, medicine and for keeping as pets in China (Yuan et al., 2021). In recent years, the industrial production of M. mutica has been hampered by high risks of disease outbreak and mortality caused by extreme temperature events in autumn. A previous study found that the expression of immune-related genes of transferrin was down-regulated in M. mutica under heat stress at 32°C(Wei et al., 2020). Although dysfunction of immune-related complement and coagulation cascade pathways has been identified as the major cause of illness outbreak and death rates under heat stress in M. mutica (Gao et al., 2021), it is possible that the mechanisms of damage in M. mutica under cold stress are also related to immune-related genes and pathways, but need to be further explored.
High-throughput sequencing-based transcriptome sequencing is a useful tool for identification of differently expressed genes (DEGs), which have potential roles in response to environmental stress (Wang et al., 2009). In this study, we performed transcriptome sequencing of M. mutica samples collected at three sampling time points (3h, 24h and 48h) during the cold treatment and a recovery time point (96 h after the end of cold stress). The following questions were addressed (1) identify DEGs between three cold stress time points, a recovery time point and the control group, and describe the functional categories of these DEGs; (2) explore how pathways related to immunity and metabolism play a regulatory role. The findings of this study facilitate reveal the mechanism of physiological damage to M. mutica caused by cold stress.
2 Materials and methods
2.1 Sample collection
Living samples of M. mutica (body weight = 84 ± 6 g, n = 20) were purchased from a turtle farm in Guangzhou, China in May 2020. The samples were acclimated at a water temperature of 28°C for one week, replicating the culture conditions on the farm. During this period of acclimatization, all M. mutica were fed once daily with a commercial turtle feed (Shenzhen Inch-Gold Fish Food Co., Ltd., Guangdong, China).
2.2 Cold stress
At the end of acclimatization, four individuals were randomly selected as a control group (28°C). A total of 16 individuals were randomly selected as the cold stress group (20°C) and the recovery group. Within half an hour, we reduced the culture temperature to 20°C in an incubator, imitating a sudden temperature drop in autumn. 20°C is the temperature level, at which M. mutica usually shows low survival rates in hatchlings and high abnormality rates based on our previous laboratory observations. At 20°C, M. mutica were able to feed, but their activity was reduced. The intestine is an important immune and digestive organ, and easily vulnerable to environmental turbulence (Zhou et al., 2020). Therefore, intestinal tissues were collected at 3h, 24h and 48h of treatment and 96 h after the end of cold stress, intestinal tissues had been removed from the intestinal contents and rinsed with saline and stored in liquid nitrogen until RNA extraction.
2.3 Total RNA extraction and transcriptome sequencing
Total RNA from intestinal tissues was extracted using Trizol RNA Isolation Reagent (Sangon, Shanghai, China) according to the manufacturer’s instructions. Genomic DNA contaminants were removed by digestion of the total RNA samples with RNase-free DNase I (Sangon, Shanghai, China). Concentration and purity were assessed using a Nanodrop One spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Qualified RNA samples were used for the construction of an RNA-seq library. In brief, mRNA was enriched with magnetic beads with Oligo (dT) and subsequently randomly fragmented with Fragmentation Buffer. The first cDNA strand was synthesized by random hexamers using mRNA as a template. Then the second cDNA strand was synthesized by adding buffer solution, dNTPs, RNase H and DNA polymerase I. The cDNA strand was purified using AMPure XP beads. AMPure XP beads were used for segment size selection. Transcriptome sequencing libraries were constructed using Hieff NGSf MaxUp Dual-mode mRNA Library Prep Kits for Illumina® (Yeasen, Shanghai, China) and sequenced on an Illumina novaseq 6000 platform with PE 150 bp.
2.4 Data processing and de novo assembly
Raw reads were first quality filtered using Trimmomatic v 0.36 with default parameters to remove adaptors, contaminants and low-quality reads (average Phred quality score (Q) <20 per read). The obtained clean reads were de novo assembled using Trinity v 2.4.0 with default settings. After removal of redundant sequences from the assembled transcripts, the resultant unigenes (transcripts) were used as reference sequences for subsequent mapping. For functional annotation, the unigene sequences were annotated by searching against NR (ftp://ftp.ncbi.nih.gov/blast/db/nr), Swiss-Prot (http://www.uniprot.org/), GO (http://www.geneontology.org/), COG (http://www.ncbi.nlm.nih.gov/COG/), CDD (https://www.ncbi.nlm.nih.gov/cdd/), TrEMBL (http://www.ensembl.org/), and KEGG (http://www.genome.jp/kegg/) databases using BLASTX programs, the BLAST parameter E-value is not greater than 1e-5 and the HMMER parameter E-value is not greater than 1e-10.
2.5 The identification of DEGs and enrichment analysis
Gene expression levels were measured as fragment values per kilobase transcript per mapped read per million (FPKM). DESeq2 was used to identify the differential expressed genes (DEGs) between the treatment group and control group. Genes with fold changes >2 and adjusted p-values (padj) < 0.05 were considered as DEGs (Wang et al., 2010). These DEGs were annotated by Gene Ontology (GO) functional enrichment and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analysis. Multiple hypothesis tests were performed on the p-values (≤ 0.05) from the enrichment analysis.
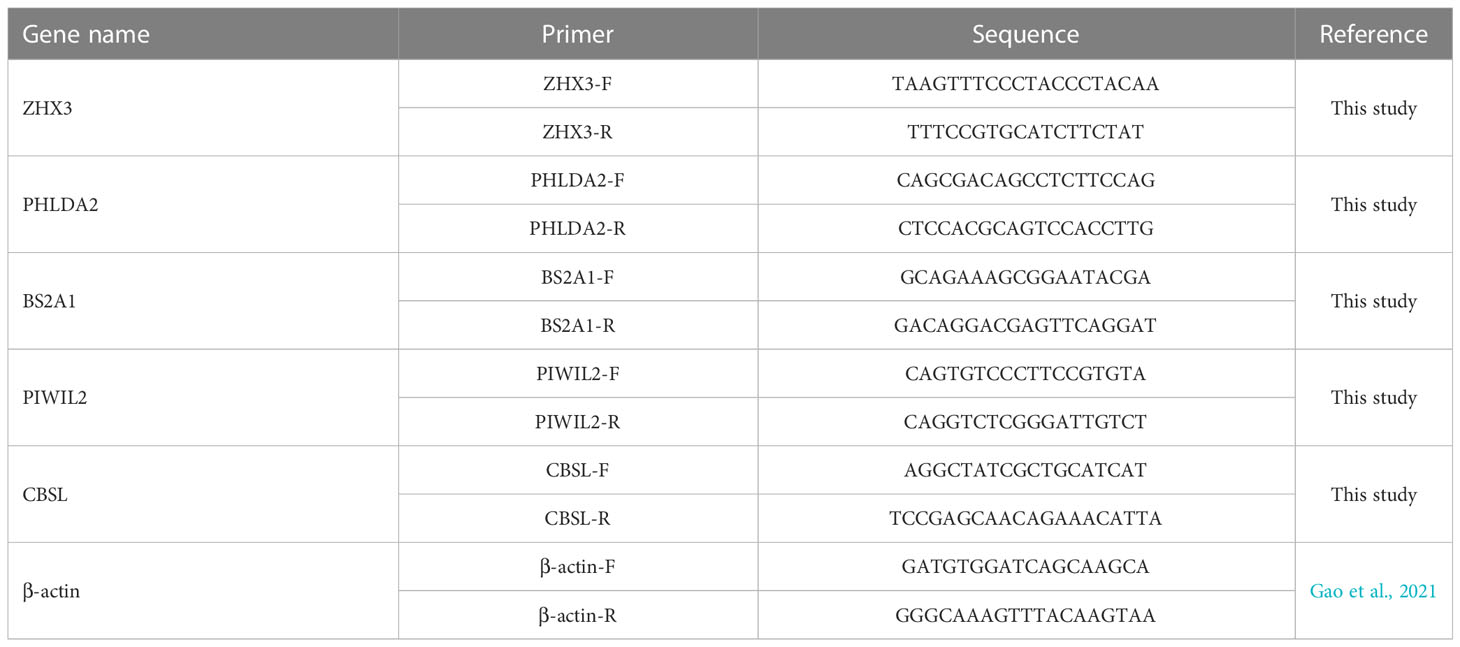
2.6 Real-time PCR validation of genes
To check the reliability of the RNA-seq, five genes were randomly selected from DEGs and used for validation using quantitative real-time RT-PCR (qRT-PCR): Zinc fingers and homeoboxes protein 3 (ZHX3), PREDICTED: pleckstrin homology-like domain family A member 2 (PHLDA2), Butyrophilin subfamily 2 member A1 (BS2A1), piwi-like protein 2 (PIWIL2), cystathionine beta-synthase-like (CBSL). β-actin was used as a reference gene. The primers (Table 1) were designed using Primer Express 3.0 and synthesized by Sangon Biotech (Shanghai) Co., Ltd. A total of 2 μg RNA from each sample were reversely transcribed to cDNA using an RT-PCR kit (PrimeScript™ RT kit, Takara). qPCR was performed in a CFX96 Touch™ Real Time PCR Detection System. The qPCR mixture consisted of 0.5 μL of primer (10 μM), 1 μL of first-strand cDNA as the template, 12.5 μL of Roche FastStart Universal SYBR Green Master (Rox) and 10.5 μL of water. The qPCR reactions were exposed to initial denaturation (95°C for 4 min) followed by 35 cycles (95°C for 30 s, annealing at 57°C for 20 s) and extension at 72°C for 30 s in 25 μL of the reaction mixture. Relative expression ratios of target genes to β-actin genes were calculated using the 2 - ΔΔCT method and all data are presented as relative mRNA expression.
3 Results
3.1 Transcriptome sequencing and de novo assembly
The RNA-seq generated a total of 564,354,538 raw reads (NCBI BioProject accession:PRJNA949287, SRA accession:SRR24172447-SRR24172458). After technical filter, we obtained 466,900,447 clean reads, which generated a total of 100,351 unigenes (Table 2). Of these unigenes, a total of 23,879 unigenes were successfully annotated. When clean reads from each sample were mapped to the reference transcriptome, the average mapping rate was 73.16% (17,080,052), ranging from 71.62% (15,465,566) to 74.73% (15,509,747).
Table 2 Summary of sequencing and assembly of the transcriptome data (C01~C04: control groups; L-3-1~L-3-4: 3 h cold stress groups; L-24-1~L-24-4: 24 h cold stress groups; L-48-1~L-48-4: 48 h cold stress groups; L-R-1~L-R-4: recovery groups).
3.2 Differential expressed genes (DEGs)
DESeq2 analysis showed that a total of 151, 2418, 2383 and 827 DEGs were identified at 3 h, 24 h, 48 h and the recovery group, respectively. With increasing time of cold stress, the number of DEGs increased rapidly within 24 hours and stabilized within 48 hours. The number of up- and down-regulated DEGs showed a continuous increase at the first three time points (3 h, 24 h and 48 h) and then decreased in the recovery group. Of these DEGs, 38, 1050, 915 and 411 genes were up-regulated at 3 h, 24 h, 48 h and in the recovery group, respectively. In contrast, 118, 1368, 1468 and 416 genes showed down-regulation in the 3 h, 24 h, 48 h and recovery groups, respectively (Figure 1).
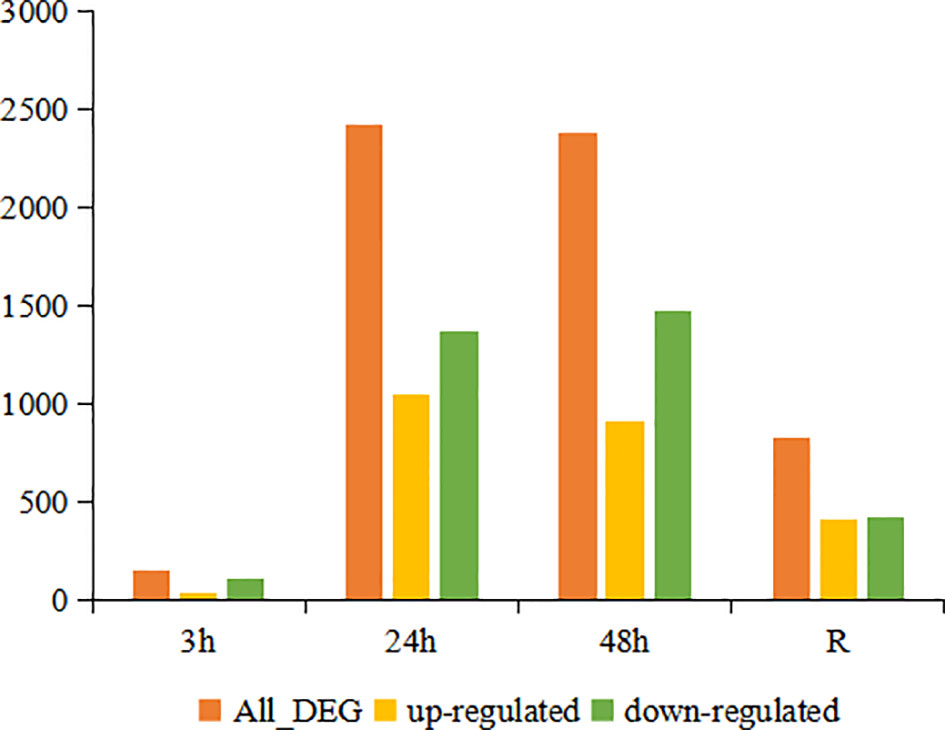
Figure 1 Number of DEGs in the continuous cold stress groups (3 h, 24 h and 48 h) and recovery group (R) compared to the control group.
3.3 GO annotations
To understand the gene function of these DEGs, we performed GO annotation analysis: the results revealed a similar pattern of GO category distribution across the four time points. A large proportion of DEGs were associated with biological processes, followed by cellular components and molecular functions (Figure 2). Within the biological process category, most DEGs were identified as cellular processes, single organism processes, metabolic processes and biological regulation. In the cellular components category, cells, cell parts, cell membranes and organelles were the most represented. In addition, most of the DEGs associated with molecular functions were related to catalytic activity, binding, signal transduction activity, and molecular transduction activity.
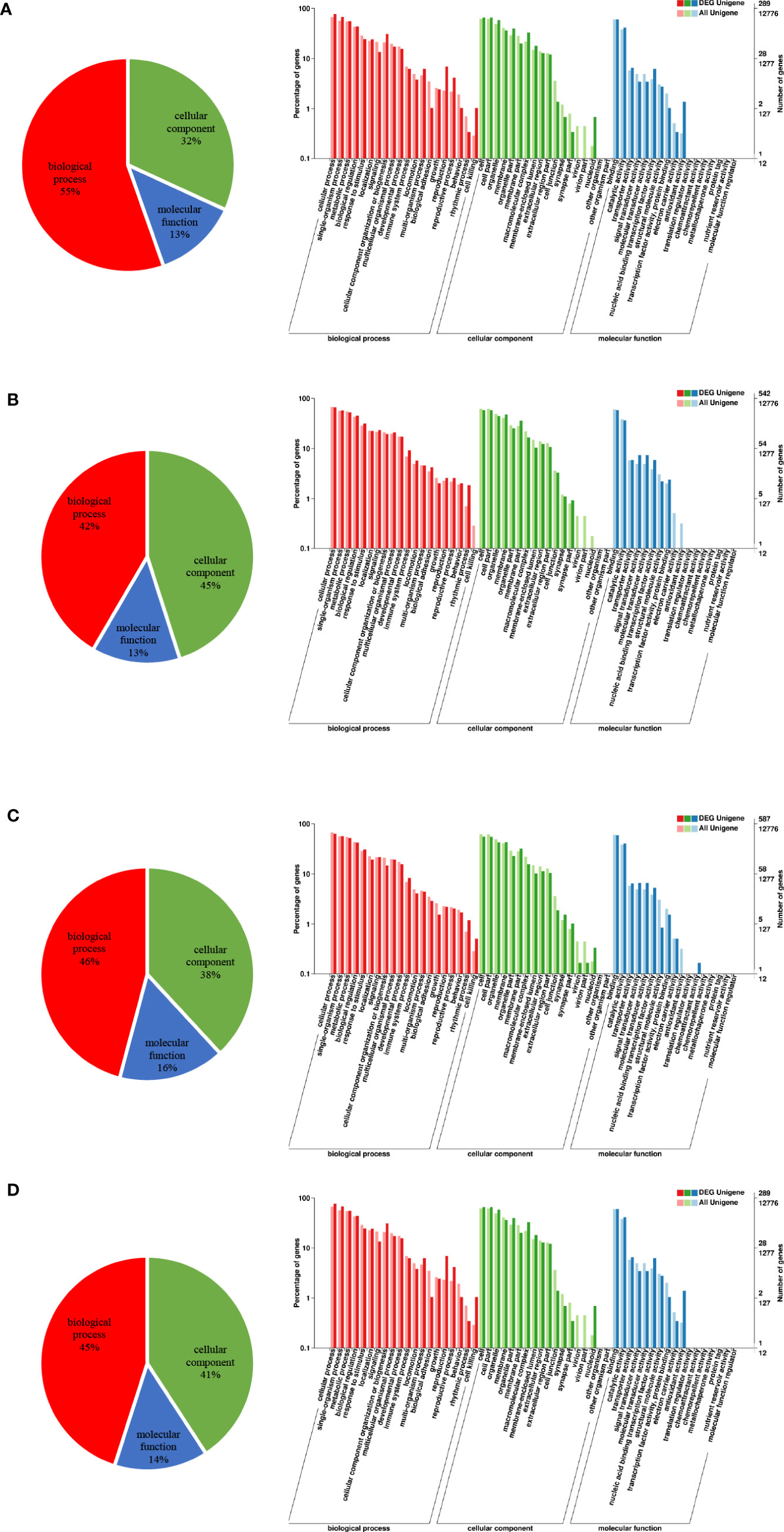
Figure 2 Distribution of GO second-level functional annotations. (A) GO classifications of DEGs between the control and 3 h cold stress groups. (B) GO classifications of DEGs between the control and 24 h cold stress groups. (C) GO classifications of DEGs between the control and 48 h cold stress groups. (D) GO classifications of DEGs between the control and recovery groups.
3.4 KEGG pathway
To identify the biological pathways that play a key role in the response to cold stress, we enriched these DEGs against the KEGG database, but there were no shared KEGG pathways among the four time points. Neuroactive ligand receptor interactions, cytokine-cytokine receptor interactions and NOD-like receptor signaling pathways were shared at 24h and 48 h. Specifically, the expression of most DEGs in the NOD-like receptor signaling pathway were down-regulated, two key DEGs (CXCR3 and CXCL10) showed down-regulated expression in the cytokine-cytokine receptor interaction pathway (Supplementary Figure 1). The ether lipid metabolism pathway was shared in the 24 h and recovery groups. Steroid hormone biosynthesis, biosynthesis of unsaturated fatty acid, and glycine serine and threonine metabolism were shared between the 48 h and recovery groups (Figure 3). Interestingly, most DEGs in steroid hormone biosynthesis pathways showed down-regulated expression at 48 h, while most DEGs showed up-regulated expression in the recovery group (Supplementary Figure 1).
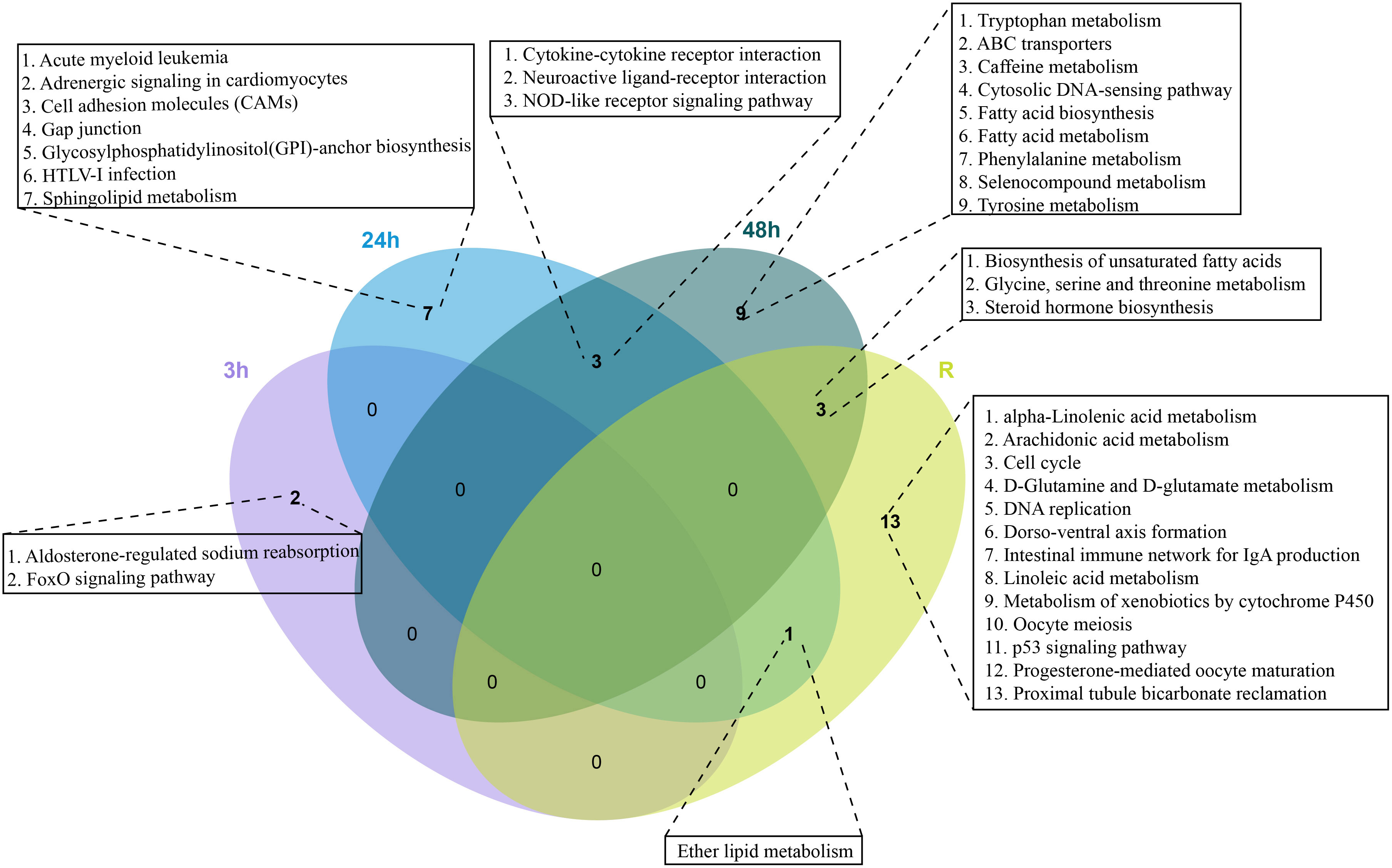
Figure 3 Venn diagram of enriched KEGG pathways in the continuous cold stress groups (3h, 24 h, 48 h and Recovery).
3.5 Validation by real-time PCR
To validate the expression profiles, the relative mRNA expression levels of five genes with random selection from DEGs were measured using qPCR. For most time points and genes, the expression trends determined by RNA-seq and qPCR showed good consistency (Figure 4).
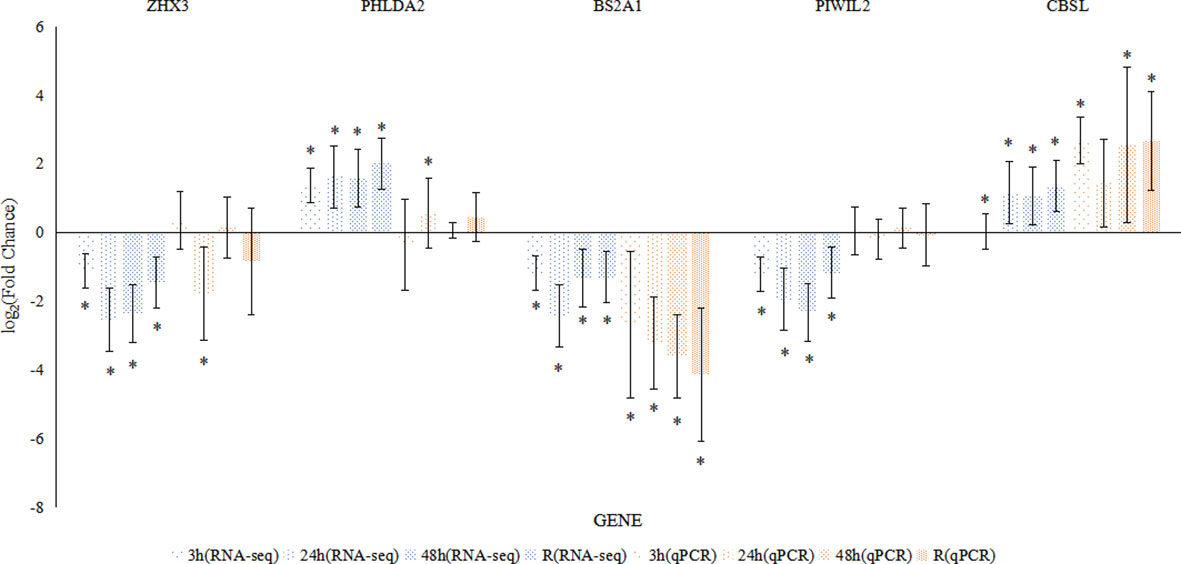
Figure 4 Comparison of the expression profiles of five DEGs determined by RNA-seq and qRT-PCR in the intestine. Each bar indicates the log2 (fold change) of a gene’s expression compared to that in the control group, and each asterisk represents the values (mean ± SD) that were significantly different from those in the control group.
4 Discussion
Cold stress caused by sudden temperature drops is one of the most severe threats to aquaculture organisms. Previous studies highlighted the importance of innate cellular and humoral parameters (phagocytosis, respiratory burst (RB), IgM levels, lysozyme and complement activity) in response to cold stress in tilapia (Chen et al., 2002). Our study revealed that immunity and metabolism-related pathways (NOD-like receptor signaling pathways, cytokine-cytokine receptor interactions and neuroactive ligand-receptor interactions) were significantly enriched at both 24h and 48h with the down-regulated expression of BS2A1, PIWIL2 and Fads1 at all-time points, indicating that impaired immunity and depressed metabolism under cold stress may be the main cause of the massive mortality in M. mutica. The finding was consistent with previous studies on Nile tilapia following exposure to hypothermia, which revealed renal dysfunction and down-regulated gene expression in immune-related pathways (Zhou et al., 2019).
The NOD-like receptor is an important component of the immune signaling pathway (NOD-like receptor signaling pathways), which can protect the host from pathogen invasion (Zhang et al., 2021). In the present study, the expression of most DEGs in the NOD-like receptor signaling pathway were down-regulated, suggesting that cold stress could depress immunity by impeding this immunity-related pathway. Similar findings were also found in other aquatic organisms, such as Larimichthys polyactis, many genes (e.g. NOD2,TRIP6, Nemo, NFKB and JNK) in the NOD-like receptor signaling pathway are down-regulated following cold stress (Chu et al., 2020).
Cytokine-cytokine receptor interaction is one of the crucial pathways in immunity, and plays an important role in the inflammatory and defense process of the host (Cheng et al., 2017). In the present study, two key DEGs (CXCR3 and CXCL10) showed down-regulated expression in the cytokine-cytokine receptor interaction pathway. CXCR3, a G protein-coupled receptor of the CXC chemokine receptor family, is expressed mainly in activated T lymphocytes and NK cells, as well as some epithelial cells (Karin and Razon, 2018). CXCL10 is a key driver of chemokines and a potent target for the treatment of autoimmune diseases such as inflammatory bowel disease, multiple sclerosis and rheumatoid arthritis (Muller et al., 2010). The down-regulated expressions of both DEGs suggested that cold stress may inhibit the expression of both DEGs and further disrupt the function of the cytokine-cytokine receptor interaction pathway. Therefore, we inferred that the down-regulated expression of DEGs caused by cold stress could lead to impaired immunity in M. mutica. However, our results showed no significant enrichment of this pathway in the recovery group, indicating that immunity recovered when the cold stress was removed, which may be a mechanism of resistance to temperature stress. In summary, impaired immunity may be an important mechanism leading to disease and mortality in M. mutica under cold stress.
In addition to the DEGs in the above enriched pathways, we also identified three key DEGs in the continuous cold stress group (3 h, 24 h and 48 h) and in the recovery group (R): Butyrophilin subfamily 2 member A1, PIWIL-like protein 2, and Fatty acid desaturase 1, these DEGs showed decreased expression at all four time points. Butyrophilin subfamily 2 member A1 is a cell surface transmembrane glycoprotein, a member of the lactophilic casein superfamily, which has been shown to modulate immune function and polymorphisms in protein-coding sequences associated with susceptibility to inflammatory diseases (Murakata et al., 2014). PIWIL2 is a member of the Piwi subfamily and is important in stem cell development, self-renewal, RNA transcriptional regulatory mechanisms and induced silencing (Feng et al., 2021). PIWIL2 can bind directly to IKK and promote its phosphorylation, leading to phosphorylation of IκB and subsequent nuclear translocation of NF-κB to inhibit apoptosis (Zhao et al., 2021). Fatty acid desaturase is a key enzyme in the synthesis pathway of polyunsaturated fatty acids (PUFAs), which can mediate the development of related diseases by regulating the transcriptional levels or expression of some nuclear and enzymatic genes (Lee et al., 2016). In this study, the decreased expression of these genes suggested that M. mutica has a weak ability to regulate immunity under cold stress, thus potentially leading to an increased risk of infection.
Moreover, three metabolism-related pathways were significantly enriched in both 48h and recovery groups: steroid hormone biosynthesis, biosynthesis of unsaturated fatty acid, and glycine serine and threonine metabolism. Previous studies showed that the expression of key genes encoding proteins in steroid hormone biosynthesis and lipid metabolism are significantly down-regulated in fish, indicating a redistribution of lipid reserves and cholesterol metabolism, highlighting the sensitivity of steroid metabolism to cold stress in fish (Qian and Xue, 2016). In the present study, most DEGs in steroid hormone biosynthesis pathways showed down-regulated expression at 48 h, while most DEGs showed up-regulated expression in the recovery group, indicating that cold stress inhibited steroid hormone biosynthesis in M. mutica, but these hormones could recover to normal levels after the remove of cold stress. Similarly, lipid metabolism involves lipolysis and synthesis, and it has been established that Oreochromis niloticus may alter energy metabolism through carbohydrate-rich and lipid metabolism to resist cold stress (Zhou et al., 2019). This phenomenon was similar to our findings: the biosynthetic pathway of unsaturated fatty acids was significantly enriched in both the 48 h and recovery groups, suggesting that M. mutica could produce more unsaturated fatty acids to resist cold stress. Glycine can be synthesized from choline, serine, hydroxyproline and threonine, and plays key roles in maintaining health by inhibiting inflammation and macrophage activation (Razak et al., 2017). Here, most DEGs in the glycine, serine and threonine metabolic pathways showed up-regulated expression at 48 h and in the recovery group, suggesting that M. mutica may respond to cold stress by increasing metabolic levels. In general, metabolic imbalance may be another important mechanism leading to illness and mortality in M. mutica under cold stress.
In conclusion, our results showed that cold stress has a significant effect on the expressions of DEGs in immunity and metabolism-related pathways in M. mutica, indicating that impaired immunity and imbalanced metabolism caused by cold stress may be a major cause of high morbidity and mortality in M. mutica. This study provides novel insights into the molecular mechanisms underlying mass disease and mortality under cold stress in M. mutica.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR24172447-SRR24172458 (NCBI BioProject accession: PRJNA949287, SRA accession: SRR24172447-SRR24172458).
Ethics statement
The animal study was reviewed and approved by Animal Experimental Ethical Inspection Form of Guangdong Institute of Applied Biological Resources GZABR20190501.
Author contributions
Research idea: SG, JZ, and YCG. Methodology: YCG and YW. Writing—original draft: JO and YCG. Writing—review & editing: JO, YCG, YW, HH, YG, JZ, and SG. All authors read and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Thousand PhD Program of Guangdong Academy of Sciences (2020GDASYL-20200103099), the Guangdong Basic and Applied Basic Research Foundation (2019A1515110455), the Scientific and Technological Program of Guangzhou (202103000082), the National Natural Science Foundation of China (32270542), GDAS Special Project of Science and Technology Development (2022GDASZH-2022010106).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1224166/full#supplementary-material
References
Ahmed N., Thompson S., Glaser M. (2019). Global aquaculture productivity, environmental sustainability, and climate change adaptability. Environ. Manage. 63, 159–172. doi: 10.1007/s00267-018-1117-3
Bucciarelli G. M., Clark M. A., Delaney K. S., Riley S. P. D., Shaffer H. B., Fisher R. N., et al. (2020). Amphibian responses in the aftermath of extreme climate events. Sci. Rep. 10, 3409. doi: 10.1038/s41598-020-60122-2
Chen W. H., Sun L. T., Tsai C. L., Song Y. L., Chang C. F. (2002). Cold-stress induced the modulation of catecholamines, cortisol, immunoglobulin m, and leukocyte phagocytosis in tilapia. Gen. Comp. Endocrinol. 126, 90–100. doi: 10.1006/gcen.2001.7772
Cheng Q., Wang Y. X., Yu J., Yi S. (2017). Critical signaling pathways during wallerian degeneration of peripheral nerve. Neural Regener. Res. 12, 995–1002. doi: 10.4103/1673-5374.208596
Chu T., Liu F., Qin G., Zhan W., Wang M., Lou B. (2020). Transcriptome analysis of the Larimichthys polyactis under heat and cold stress. Cryobiology 96, 175–183. doi: 10.1016/j.cryobiol.2020.06.014
Feng D., Yan K., Liang H., Liang J., Wang W., Yu H., et al. (2021). CBP-mediated Wnt3a/beta-catenin signaling promotes cervical oncogenesis initiated by Piwil2. Neoplasia 23, 1–11. doi: 10.1016/j.neo.2020.10.013
Gao Y., Wei Y., Cao D., Ge Y., Gong S. (2021). Transcriptome analysis reveals decreased immunity under heat stress in Mauremys mutica. Aquaculture 531. doi: 10.1016/j.aquaculture.2020.735894
Karin N., Razon H. (2018). Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine 109, 24–28. doi: 10.1016/j.cyto.2018.02.012
Lee J. M., Lee H., Kang S., Park W. J. (2016). Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients 8. doi: 10.3390/nu8010023
Liu Y., Lv H., Xu L., Zhang K., Mei Y., Chen J., et al. (2022). The effect of dietary lactic acid bacteria on intestinal microbiota and immune responses of crucian carp (Carassius auratus) under water temperature decrease. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.847167
Liu L., Zhang R., Wang X., Zhu H., Tian Z. (2020). Transcriptome analysis reveals molecular mechanisms responsive to acute cold stress in the tropical stenothermal fish tiger barb (Puntius tetrazona). BMC Genomics 21, 737. doi: 10.1186/s12864-020-07139-z
Muller M., Carter S., Hofer M. J., Campbell I. L. (2010). Review: the chemokine receptor CXCR3 and its ligands CXCL9, CXCL10 and CXCL11 in neuroimmunity–a tale of conflict and conundrum. Neuropathol. Appl. Neurobiol. 36, 368–387. doi: 10.1111/j.1365-2990.2010.01089.x
Murakata Y., Fujimaki T., Yamada Y. (2014). Association of a butyrophilin, subfamily 2, member A1 gene polymorphism with hypertension. BioMed. Rep. 2, 818–822. doi: 10.3892/br.2014.340
-Qian B., Xue L. (2016). Liver transcriptome sequencing and de novo annotation of the large yellow croaker (Larimichthy crocea) under heat and cold stress. Mar. Genomics 25, 95–102. doi: 10.1016/j.margen.2015.12.001
Rahmstorf S., Coumou D. (2011). Increase of extreme events in a warming world. Proc. Natl. Acad. Sci. U. S. A. 108, 17905–17909. doi: 10.1073/pnas.1101766108
Razak M. A., Begum P. S., Viswanath B., Rajagopal S. (2017). Multifarious beneficial effect of nonessential amino acid, glycine: a review. Oxid. Med. Cell Longev. 2017, 1716701. doi: 10.1155/2017/1716701
Shi W., Hu R., Wang P., Zhao R., Shen H., Li H., et al. (2022). Transcriptome analysis of acute high temperature-responsive genes and pathways in Palaemon gravieri. Comp. Biochem. Physiol. Part D Genomics Proteomics. 41, 100958. doi: 10.1016/j.cbd.2021.100958
Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wei Y., Gao Y., Cao D., Ge Y., Shi H., Gong S. (2020). Effects of acute temperature stress on mRNA expression of transferrin in the yellow pond turtle Mauremys mutica. Asian Herpetol. Res. 11, 124–131. doi: 10.16373/j.cnki.ahr.200006
Yang Y., Yu H., Li H., Wang A., Yu H. Y. (2016). Effect of high temperature on immune response of grass carp (Ctenopharyngodon idellus) by transcriptome analysis. Fish Shellfish Immunol. 58, 89–95. doi: 10.1016/j.fsi.2016.09.014
Yuan J., Wang Y., Liu F., Li W., Hong X., Chen C., et al. (2021). Comparative transcriptomic analysis reveals the gonadal development-related gene response to environmental temperature in Mauremys mutica. Comp. Biochem. Physiol. Part D Genomics Proteomics 40, 100925. doi: 10.1016/j.cbd.2021.100925
Zhang J., Wu Z., He Y., Li X., Li J. (2021). Transcriptome analysis reveals impaired fertility and immunity under salinity exposure in juvenile grass carp. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.697813
Zhao X., Huang L., Lu Y., Jiang W., Song Y., Qiu B., et al. (2021). PIWIL2 interacting with IKK to regulate autophagy and apoptosis in esophageal squamous cell carcinoma. Cell Death Differ. 28, 1941–1954. doi: 10.1038/s41418-020-00725-4
Zhou T., Gui L., Liu M., Li W., Hu P., Duarte D. F. C., et al. (2019). Transcriptomic responses to low temperature stress in the Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 84, 1145–1156. doi: 10.1016/j.fsi.2018.10.023
Keywords: Mauremys mutica, cold stress, transcriptomics, immunity, metabolism
Citation: OuYang J, Gao Y, Wei Y, Huang H, Ge Y, Zhao J and Gong S (2023) Transcriptome analysis reveals reduced immunity and metabolic level under cold stress in Mauremys mutica. Front. Mar. Sci. 10:1224166. doi: 10.3389/fmars.2023.1224166
Received: 17 May 2023; Accepted: 23 June 2023;
Published: 10 July 2023.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaReviewed by:
Xidong Mu, Chinese Academy of Fishery Sciences, ChinaMin Yang, South China Agricultural University, China
Copyright © 2023 OuYang, Gao, Wei, Huang, Ge, Zhao and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiping Gong, Z3NwNjIxQDE2My5jb20=; Jun Zhao, YmlvempAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jiahui OuYang
Jiahui OuYang Yangchun Gao2†
Yangchun Gao2† Shiping Gong
Shiping Gong