- 1Laboratory of Microbiology, Faculty of Fisheries Sciences, Hokkaido University, Hakodate, Japan
- 2Hakodate Fisheries Research, Hokkaido Research Organization, Local Independent Administrative Agency, Hakodate, Japan
The overexploitation of the wild populations of Apostichopus japonicus has caused a significant decline in their numbers, prompting the establishment of a sea cucumber aquaculture industry that has become a major player in East Asia. Recent research has focused on identifying the various factors that may affect sea cucumber aquaculture, including water quality, diet, and the microbiome associated with their hosts. Notably, studies on host-associated microbiomes have emerged as a prominent area of research, particularly in the context of investigating the relationship of the microbiome with diseases that have caused significant economic losses. Intestinal atrophy of A. japonicus larvae has been reported at a number of sea cucumber farms in Japan, but there have been no factor screenings, and there is currently no efficient mitigation. In this study, we applied metagenomic analyses for the first time to an intestinal atrophic case of the sea cucumber A. japonicus larvae, which occurred in an aquaculture facility in Hokkaido, Japan, elucidating potential causative microbial factors and comparing the diseased microbiome against the healthy pan-microbiome. Through our metagenome comparisons, we have found significant increases in Oceanicoccus, Ruegeria, Nioella, Labrenzia, Tenacibaculum, and Muricauda in the diseased larvae. Among those, Tenacibaculum was estimated to potentially be the most pathogenic bacterium in the development of intestinal atrophic disease due to previous outbreaks of sea urchin diseases in the same facility. Our metagenomic approach demonstrates the further need of microbial screening of such causative agents and identification of the pathogenesis of such diseases, which could contribute to establishing more sustainable aquaculture in the sea cucumber industry.
Introduction
The sea cucumber Apostichopus japonicus (Selenka, 1867), belonging to Echinodermata: Holothuroidea, is one of the most important and valuable aquaculture resources in Asia, and it has been consumed as both a delicacy and a traditional medicine for over 100 years (Chen and Chang, 2015). Due to the growing commercial value of sea cucumber, wild populations of A. japonicus were overfished in the 20th century, leading to declines in wild populations and an increase in the price of the product, and it has been listed as a threatened species in the IUCN due to this high demand and the lack of regulations, (Hamel and Mercier, 2013). To address the decline in wild populations, aquaculture of A. japonicus was first developed in the 1950s (Yang et al., 2015). Since the first successful artificial cultivation of A. japonicus, aquaculture bloomed and rapidly developed into a major industry in East Asia, with China being the largest producer of cultured A. japonicus (Han et al., 2016). A study used genetic markers to investigate the genetic diversity and structure of wild and cultured A. japonicus populations in China, indicating that wild populations exhibited higher genetic diversity than cultured populations, and there was a genetic gap in different geographic regions, highlighting the importance of preserving genetic diversity in both wild and cultured populations for sustainable management of the industry (Chen et al., 2008).
In recent decades, research efforts have been devoted to identifying factors that impact the aquaculture of the sea cucumber A. japonicus, such as investigating areas such as water quality, stock density, genetic factors, and diet (Li and Li, 2010; Gao et al., 2017; Feng et al., 2022). In recent years, the study of host-associated microbiomes has achieved a prominent position in research, with a growing body of literature highlighting its importance in elucidating the biology of host organisms and enabling a range of practical applications (Adair and Douglas, 2017). Expanding research into sea cucumber has accumulated knowledge on the microbiomes associated with the host organism, in particular, by providing valuable insights into host growth performance and disease prevention and treatment and encouraging the development of novel aquaculture applications in sea cucumber (Yamazaki et al., 2016; Chen et al., 2022a; Zhao et al., 2022). Our own previous research has also demonstrated a relationship between the gut microbiota of sea cucumber and growth performance and also during gut regeneration and larval development (Yamazaki et al., 2016; Yamazaki et al., 2020; Yu et al., 2022). These findings suggest the potential for interventions targeting the host-associated microbiome to help enhance the aquaculture of A. japonicus by promoting host health, development, and growth performance.
The rapid expansion of sea cucumber aquaculture has given rise to various diseases, such as rotting edges syndrome and skin ulceration disease, which have resulted in significant economic losses and hindered sustainable development of the industry (Wang et al., 2004; Yang et al., 2015). Skin ulceration syndrome (SUS) is one of the most severe diseases affecting adult sea cucumbers and has been reported in multiple sea cucumber species worldwide, primarily caused by Vibrio splendidus, Vibrio alginolyticus, Vibrio cyclitrophicus, and Shewanella marisflavi bacteria (Deng et al., 2009; Li and Li, 2010; Zhang et al., 2015). SUS causes skin ulcers that lead to a decline in sea cucumber health and eventual death (Zhang, 2006). In addition, sea cucumber larval development is affected by diseases such as rotting edges syndrome, caused by Vibrio lentus during the auricularia stage, and off-plate syndrome, caused by pathogenic Vibrio sp. during the doliolaria and pentactula settlement stages (Yu et al., 2010). The impact of these diseases has prompted extensive research to better understand their underlying causes and to develop effective prevention and control measures (Yasoda et al., 2006). For instance, recent studies have explored the use of probiotics, such as Bacillus, and dietary supplements, including β-glucan, to regulate the growth, immune response, intestinal microbial homeostasis, and pathogen resistance of A. japonicus (Zhao et al., 2012; Yang et al., 2015; Yang et al., 2017). Moreover, ecofriendly agents such as immunostimulants are being investigated to enhance immune response in A. japonicus and to aid resistance against bacterial diseases in aquaculture (Zhang et al., 2021). During the study of a healthy sea cucumber microbiome in its early life stages from fertilized eggs to benthic-settled juveniles under laboratory aquarium conditions in our laboratory, a disease characterized by an atrophic digestive system occurred, and it halted metamorphosis at the auricularia stage at a local sea cucumber farm on site. Outbreaks of such a disease, named as intestinal atrophic disease, have occurred sporadically in various facilities in Hokkaido, Japan. Despite these sporadic occurrences, we have been unable to find causative agents related to this disease in sea cucumber larvae. This knowledge gap places an obstacle to the sustainable seed production of sea cucumber in related aquaculture industries. Addressing these challenges requires further investigation of the mechanisms and pathogenesis of intestinal atrophic disease in sea cucumber larvae.
To our knowledge, this is the first study to assess the structure, function, and dynamics of the microbiome on intestinal atrophic diseased larvae using a metagenomic approach. We also used a unique metagenome data set of the pan-microbiome of healthy larvae as a baseline control to identify the microbial factors causing the structural and functional differences in larvae affected by intestinal atrophic disease. The broad assessment of the atrophic-associated microbiomes performed in this study identified significantly increased numbers of potential pathogenic microorganisms, such as Tenacibaculum, which is known to be a fish and shellfish pathogen. Furthermore, this study highlights the relative abundance of specific microbial metabolisms, including succinate dehydrogenase, cytochrome c oxidases, and pathogenic islands, in larvae with internal atrophic disease. Additionally, the influence of environmental rearing seawater on the intestinal microbiome was also evaluated. These findings offer novel insights into the disturbance of the microbiome and the potential involvement of pathogenic bacteria in the development of intestinal atrophic disease during the larval stage of sea cucumber, thereby advancing our understanding of host–microbe interactions and aiding in disease prevention in sea cucumber aquaculture.
Materials and methods
Sample collection of healthy and atrophic larvae
Due to the limitations in collecting healthy samples from the farm during the disease outbreaks, we decided to utilize a pan-microbiome from healthy larvae reared in our laboratory as a baseline control. For healthy samples, fertilized egg or gastrula samples of the sea cucumber A. japonicus were collected on three occasions in 2021. Fertilized eggs or gastrula were prepared at 18.7°C at a farm in Hokkaido Aquaculture Promotion Corporation Kumaishi Branch, Japan (42.12574, 139.99966), at 11:00 am on 30th June 2021 and 12th July 2021. Other fertilized egg samples were prepared at 18.7°C at a farm in Hokkaido Aquaculture Promotion Corporation Shiriuchi Branch, Japan (41.61463, 140.38335), at 11:00 am on 30th July 2021. Fertilized egg or gastrula samples were further reared in our laboratory breeding system for sample collection at different developmental stages, and the sub-sampling procedures were performed as described in a previous study (Yu et al., 2022). In brief, the final density of fertilized eggs in this study reached 7,500 eggs per liter in an 8-liter volume aquarium. The aquarium was prepared using a sterilized 8-liter glass bottle (Ishizuka Glass Co. Ltd., Aichi, Japan). Each bottle was filled with 7.5 liters of sterilized artificial seawater (Sea Life, MARINETECH, Japanese Seawater Co. Ltd., Japan). Additionally, 0.22 μm Sterivex filters were installed at the exit of the aeration system to prevent contamination. A commercially available diatom, Chaetoceros gracilis (Hakodate Fisheries Research, Japan), was fed to the sea cucumber larvae daily. Larvae samples with intestinal atrophic were collected directly from a farm in the Shiriuchi Branch, Japan (42.12574, 139.99966), on the day disease occurred on August 12th, and the larvae were starved for 3 days from August 15th, and feeding was restarted on August 16th 2021. As described above, the fertilized eggs collected from the Shiriuchi farm were subsequently cultured in our laboratory system to establish a healthy pan-microbiome profile for early-stage sea cucumber, A. japonicus. This approach was employed as a control due to the limited availability of healthy samples from the farm during the atrophic sample collection. We acknowledge the limitations in obtaining an adequate number of healthy controls and have addressed this concern by using the laboratory-raised larvae as a comparative reference (Figure S1).
Microbial DNA extraction and 16S rRNA gene sequencing
Microbial DNA extraction from sea cucumber was performed using the NucleoSpin Soil Kit (MACHEREY-NAGEL, Düren, Germany) according to the manufacturer’s protocol. Microbial DNA extraction from seawater was performed using the NucleoSpin Tissue kit (MACHEREY-NAGEL) according to the modified manufacturer’s protocol. In brief, seawater samples were heated at 55°C for 1 hour to add an active cell lysis process in the TE buffer (10 mM Tris-HCl, 1 mM EDTA) containing 20% SDS and proteinase K (20 mg mL-1) instead of buffer T1. In the third step, for the lyse sample, 1 mL of buffer B3 was used instead of 200 μL.
The hypervariable V1-V2 region of the 16S rRNA gene was amplified by PCR with barcoded 27Fmod and 338R primers with Illumina adaptor sequences (Yamazaki et al., 2019; Yu et al., 2022). PCR amplicons were purified using AMPure XP magnetic purification beads (Beckman Coulter, Brea, CA, USA), and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies Japan). Equal amounts of each PCR amplicon were mixed and then sequenced using MiSeq Reagent Kit v3 (600-cycles) with the MiSeq Illumina platform. Based on sample-specific barcodes, obtained reads were assigned to each sample.
Meta16S analysis and plot construction
The paired-end sequence data with quality scores were analyzed using Quantitative Insights Into Microbial Ecology 2 (QIIME 2, version 2022.2) (Bolyen et al., 2019). Quality controls and merging paired-end sequences were performed using DADA2 (Callahan et al., 2016). Reads with 100% similarity constituted an amplicon sequence variance (ASV). ASVs were assigned to taxonomy using the naïve Bayes classifier and Greengenes database. Using subsampled reads, unweighted UniFrac distances as beta-diversity were calculated and visualized in PCoA plots (Lozupone et al., 2011). Significant differences of unweighted UniFrac distance were tested by permutational multivariate analysis of variance (PERMANOVA) (FDR-corrected p<0.05). The phylogenic tree was generated by FastTree (Price et al., 2010). The Z-score was calculated using the genefilter package in R (Gentleman et al., 2023). Pheatmap, ggplot, and the DESeq2 package in R (Version 4.0.5) were used for heatmap construction and statistical analysis (McMurdie and Holmes, 2013; Love et al., 2014). Venn diagrams of shared ASV between sea cucumber and seawater were generated by Venny 2.1 (Oliveros, 2007-2015). Re-analysis of individual parameters using linear-constrained ordination redundancy analysis (RDA) was performed by vegan-2.6-4 (Oksanen et al., 2015). Spearman correlations were used for correlation network construction by R package Hmisc and psych (Revelle, 2017; Harrell and Dupont, 2019). LDA Effect Size (LEfSe) was used for assessing the significantly different taxa and microbial functions (Segata et al., 2011). EZR package was used for the t-test, F-test, and Shapiro–Wilk test to compare the size of digestive systems between diseased and healthy individuals (Kim, 2020).
Metagenomic sequencing and functional profile
A total of 20 templates with ≥ 300 ng DNA material extracted from sea cucumber larvae were used for paired-end shotgun metagenomic sequencing on the HiSeq platform. For analysis of sea cucumber larval metagenome samples, the metagenomics RAST server (MG-RAST) was used for eukaryotic taxonomic classification and microbial functional annotation (Meyer et al., 2008). The low-quality regions were trimmed by using SolexaQA, followed by dereplication using the k-mer approach (Cox et al., 2010). Duplicate Read Inferred Sequencing Error Estimation (DRISEE) was used to analyze the sets of Artificial Duplicate Reads (ADRs) (Gomez-Alvarez et al., 2009; Keegan et al., 2012). Reads with 97% identity were clustered, and the longest sequences were picked as the cluster representatives. The cluster representatives were further assigned to taxonomy using a BLAT similarity search, which integrates SILVA, Greengenes, and RDP (Cole et al., 2003; DeSantis et al., 2006; Pruesse et al., 2007). After the abundance profile was generated, the SEED subsystem was used for the functional profiles (Overbeek et al., 2005).
Results
Descriptions of signs of intestinal atrophic disease
Microscopic observation of sea cucumber with internal atrophic indicated that the digestive system, including the intestines and stomach, were clearly atrophic (Figures 1A, B). The size of the intestine and stomach in diseased larvae was significantly smaller than in healthy larvae, showing a digestive system with a width of 57.1-107.1 μm and length of 128.5-199.9 μm. Compared to the healthy larvae, the width and length of intestine and stomach was significantly narrower and shorter (t-test, p < 0.01, Table S1). No parasitic eukaryotes-like cells were observed in those atrophic samples. In healthy larvae, it was very easy to define the intestine and stomach under light microscopy (Figure 1C), but in atrophic larvae, the digestive system was atrophied, and it was very difficult to distinguish the different digestive organs, such as the intestines and stomach. An additional sign of diseased larvae was the halt in metamorphosis when the disease occurred.
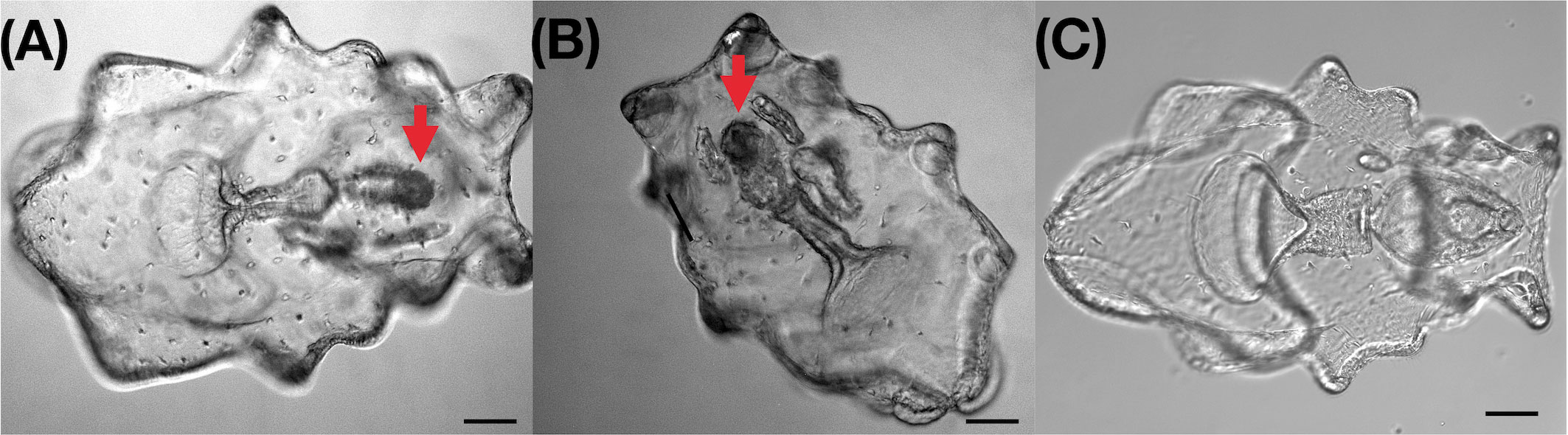
Figure 1 Light microscopy image of healthy larvae and larvae with intestinal atrophic disease of Apostichopus japonicus. (A, B) Larvae with intestinal atrophic disease. Size of digestive system (stomach and intestine) is obviously smaller, width is between 50-70μm, length is between 150-170μm; (C) Healthy larvae. Width of digestive system is between 170-200μm, length is between 250 and 270μm; bar:100μm.
The significantly different bacterial diversity and composition in larvae with intestinal atrophic disease compared to healthy pan-microbiome
In order to understand whether the microbial community varied in larvae with intestinal atrophic disease and which taxa showed increases in atrophic individuals, we compared the atrophic Meta16S data against the healthy pan-Meta16S data set. A total of 165,675 and 256,140 Meta16S sequence reads were obtained to create a “healthy pan-Meta16S data set” from 17 sea cucumber and 14 seawater samples collected from healthy larvae, respectively. A total of 32,759 and 41,608 Meta16S sequence reads were obtained from 3 sea cucumber and 3 seawater samples collected from larvae with intestinal atrophic disease, respectively. Alpha and beta diversity of larval microbiota were assessed and compared (Figure 2). Alpha diversity based on Chao1 and the Shannon index indicated a relatively higher alpha diversity in atrophic larvae, but beta diversity based on the Unweighted UniFrac distance showed a significantly decreased beta diversity in atrophic larvae (PERMANOVA, p<0.05), indicating a more complex but conservative microbiota in larvae with intestinal atrophic disease (Figures 2A, B). The PCoA plot based on the Unweighted UniFrac distance also illustrated a clustering microbiota in atrophic larvae, which separated the healthy larvae at same developmental stage as the atrophic samples, indicating a distinct microbiota in atrophic larvae (Figure 2C).
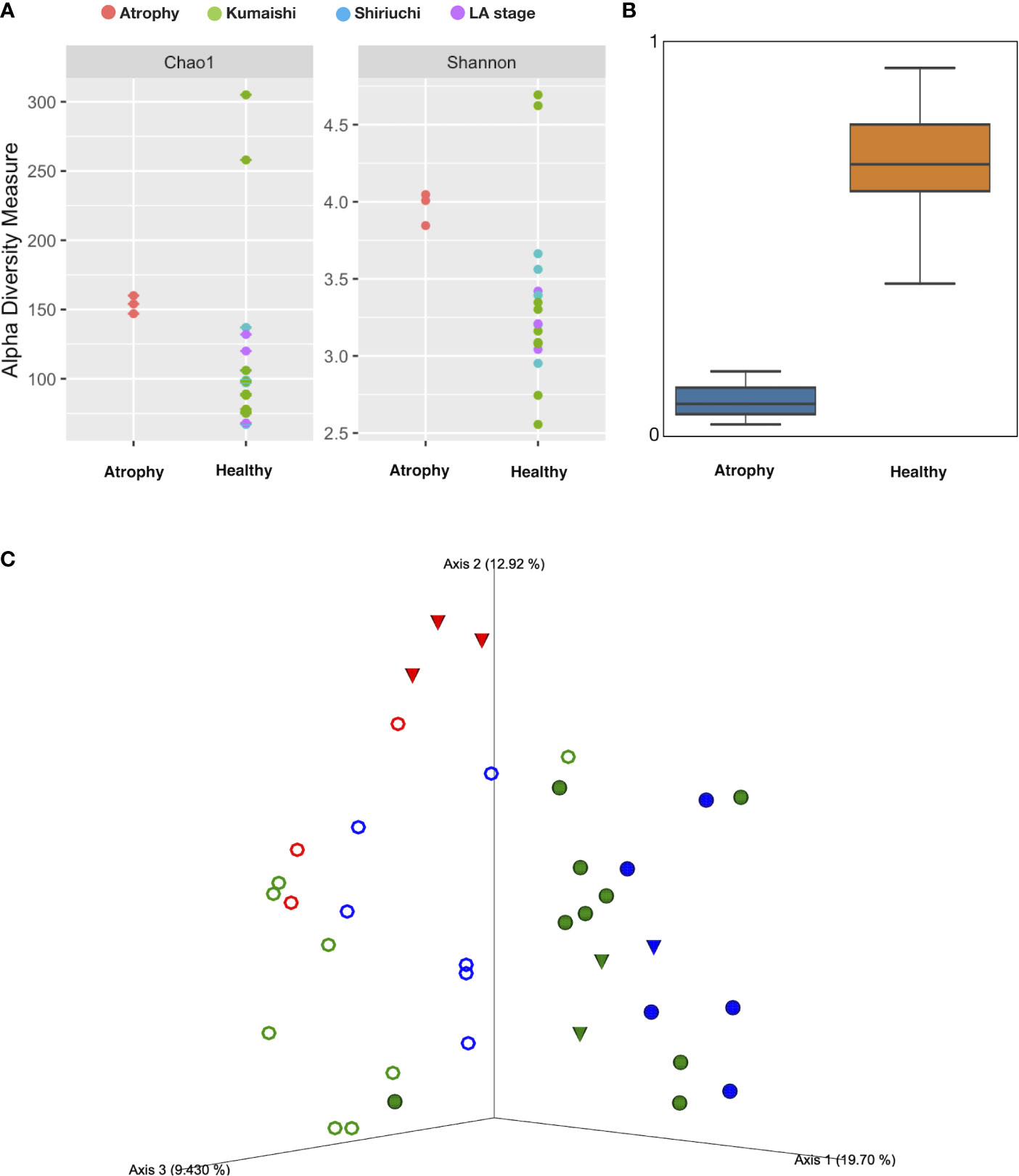
Figure 2 Alpha and beta diversity among sea cucumber samples. (A) Alpha diversity based on Chao1 and Shannon index. Red: atrophic larvae samples; green: healthy larvae collected from Kumaishi; blue: healthy larvae collected from Shiriuchi; purple: healthy larvae at late auricularia stage. (B) Boxplot based on unweighted UniFrac distance (p<0.05, q<0.05). Scale represents similarity within samples. (C) PCoA plot based on unweighted UniFrac distances obtained by comparison of microbiotas of sea cucumber larvae at different health status. Red: atrophic larvae and their rearing seawater; blue: healthy larvae collected from Shiriuchi; green: healthy larvae collected from Kumaishi; closed circle: healthy larvae; open circle: larvae’s rearing seawater; triangle: larvae at late auricularia stage.
Since the significantly different microbiota in atrophic larvae was revealed, we wondered how microbial compositions were different in the atrophic larvae. A total of 1,915 ASVs from sea cucumber and seawater samples were obtained and further assigned to 210 families and 457 genera. Flavobacteriaceae (23.0 ± 5.1%) and Rhodobacteraceae (22.8 ± 8.3%) were the most dominant bacteria families in the microbiota of atrophic larvae, followed by Spongiibacteraceae (7.5 ± 4.0%), Desulfuromonadia PB19 (5.3 ± 2.6%), Stappiaceae (4.7 ± 4.0%), Alphaproteobacteria NRL2 (3.5 ± 1.7%), Halieaceae (3.4 ± 0.9%), Crocinitomicaceae (2.8 ± 2.2%), Hyphomonadaceae (2.5 ± 0.9%), and Bdellovibrionaceae (1.7 ± 1.7%), consisting of over 77% microbiota in atrophic larvae (Figure S2). Comparing the microbial composition between atrophic and healthy larvae, there were several bacterial families that were significantly different in the atrophic microbiome found by LEfSe analysis (Figure S3). Flavobacteriaceae was significantly enriched in atrophic larvae but relatively rare in healthy larvae (5.6 ± 6.3%). Other families dominant in atrophic larvae, including Spongiibacteraceae, Desulfuromonadia PB19, Stappiaceae, Alphaproteobacteria NRL2, Halieaceae, and Crocinitomicaceae, were barely or not detected in healthy larvae (Figures S3A, B, Table S2). On the other hand, Alteromonadaceae, Methylophagaceae, and Lachnospiraceae were significantly abundant in healthy samples but dramatically rare in atrophic larvae, with a relative abundance of 0.7 ± 0.6% and below the undetectable limit (BUL), respectively. Since a significantly different microbial structure and composition was observed in atrophic larvae, we assumed these significantly varied bacteria in atrophic larvae were related to intestinal atrophic disease.
In order to further understand how bacteria were different in atrophic larval microbiota and whether there were pathogenetic bacteria resulting in intestinal atrophic disease, we also assessed the significantly different bacteria at the genera level (Figure 3; Figure S4). In atrophic larvae, Oceanicoccus (7.1± 3.6%) and Ruegeria (5.6 ± 3.0%) were the most dominant genera, followed by Nioella (4.4 ± 3.0%), Labrenzia (4.2 ± 3.9%), Tenacibaculum (3.6 ± 3.1%), and Muricauda (3.2 ± 1.3%). These dominant bacteria significantly increased in atrophic larvae but were barely or not detected at all in healthy larvae (Figure 3). The abundances of Nioella, Labrenzia, Tenacibaculum, and Muricauda in healthy sea cucumber were 0.6 ± 1.0%, 0.0% ± 0.1%, 0.2 ± 0.4%, and 0.3 ± 0.4%, respectively. Sulfitobacter, which includes probiotic candidate strains, belonging to Rhodobacteraceae was significantly dominant in healthy samples (3.5 ± 7.2%) but not detected in atrophic larvae (Figure 3; Table S3). Neptunibacter and Alteromonas were also significantly abundant in healthy larvae compared to atrophic larvae, indicating these bacteria may be responsible for different functions associated with host biology in healthy larvae.
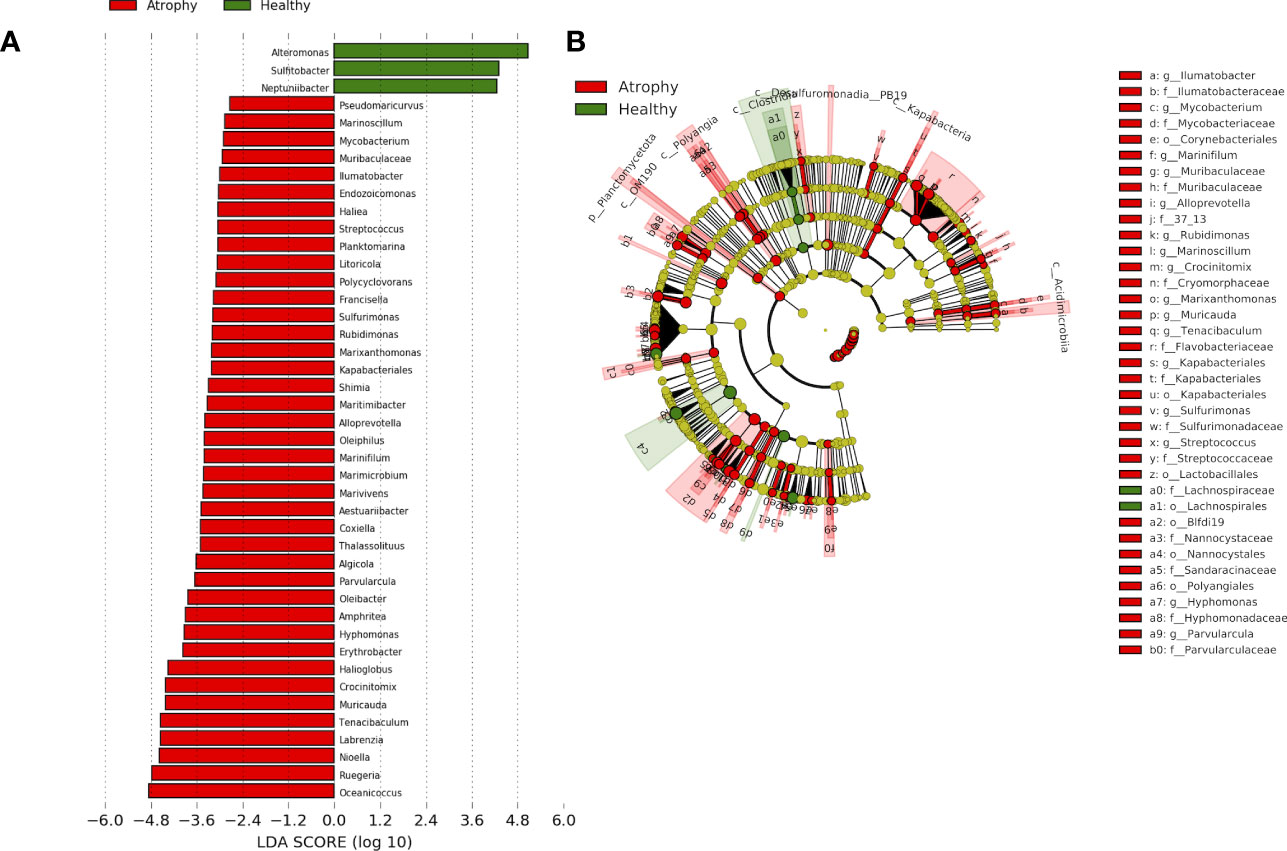
Figure 3 The linear discriminant analysis effect size (LEfSe) analysis of microbial abundance among sea cucumber larvae samples with different health status. (A) Only genera-level taxa with significant difference in atrophic and healthy larvae were detected by LEfSe analysis with an LDA threshold score of 3.5 and a significance of 0.05. Red: atrophic larvae; green: healthy larvae. (B) The cladogram of detected prokaryotic taxa for atrophic and healthy larval microbial community. Red: atrophic larvae; green: healthy larvae.
Further linear-constrained ordination redundancy analysis (RDA) with forward selection revealed that health status significantly impacts on microbial community composition (ANOVA, p<0.05) (Figure 4). Four families, namely, Alteromonadaceae, Methylophagaceae, Myxococcaceae, and Lachnospiraceae, showed positive correlations with healthy larval microbiota, whereas Flavobacteriaceae, Spongiibacteraceae, Cellvibrionaceae, Stappiaceae, Crocinitomicaceae, Halieaceae, Alphaproteobacteria NRL2, and Desulfuromonadia PB19 were positively correlated with atrophic larval microbiota (Figure 4A). At the genera level, our analysis revealed a positive correlation between healthy microbiota and the presence of Alteromonas, Sulfitobacter, and Neptunibacter. Conversely, we observed a positive correlation between atrophic microbiota and the presence of Oceannicoccus, Tenacibaculum, Labranzia, Ruegeria, Nioella, and Muricauda (Figure 4B). These findings highlight the potential association between specific bacterial groups and the health status of sea cucumber larvae, providing valuable insights into the microbial dynamics and potential contributors to the development of intestinal atrophic disease.
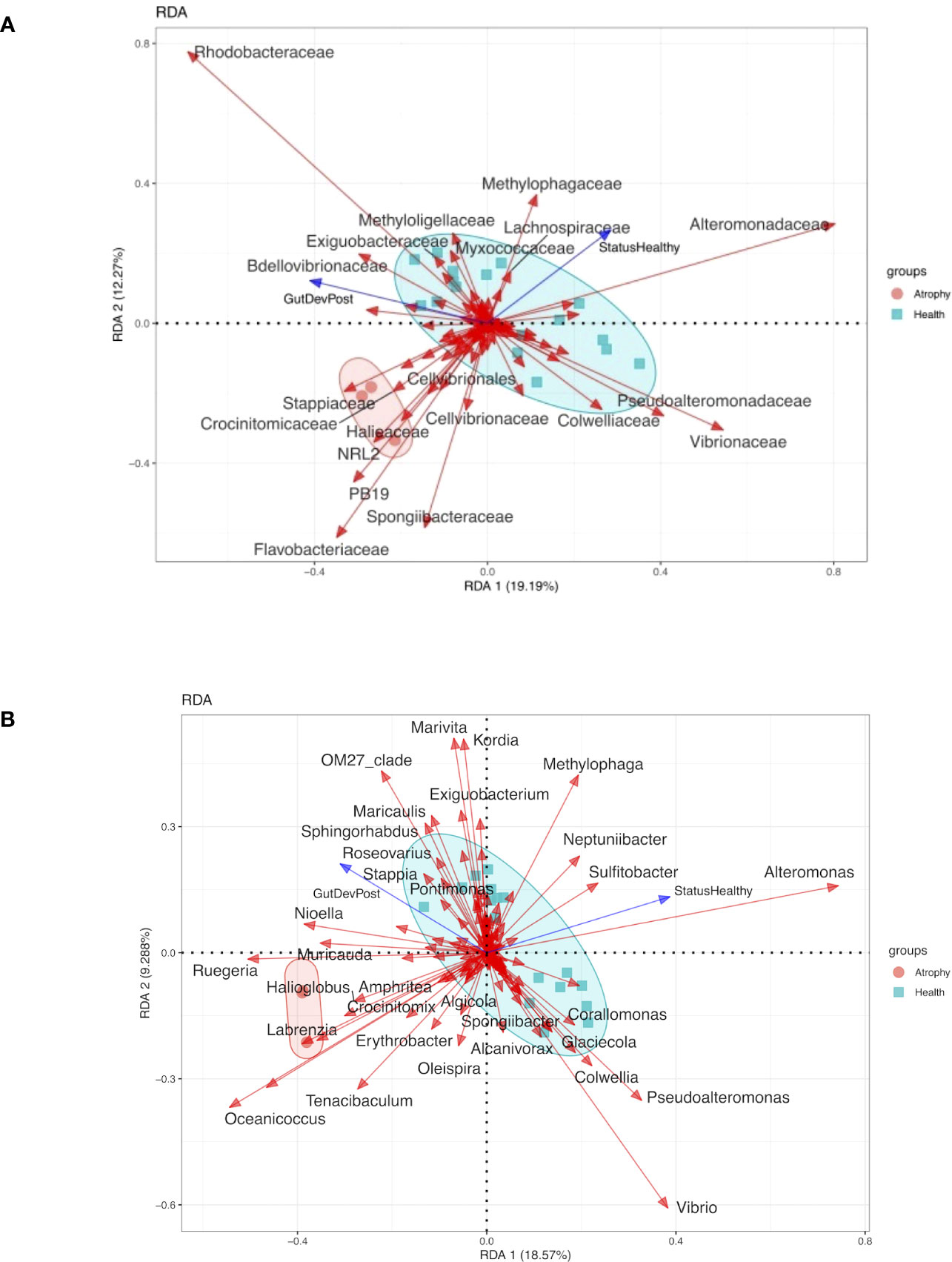
Figure 4 Redundancy analysis (RDA) of the relationship between the health status and the relative abundance of bacterial group of atrophy and healthy larvae samples. Circles represent atrophy samples, cubes represent healthy samples. The abbreviation of samples is shown in Table S1. Arrows indicated the direction and magnitude of variables (p<0.05). (A) RDA of the relationship between the health status and the relative abundance of bacterial group at family level. (B) RDA f the relationship between the health status and the relative abundance of bacterial group at genera level.
Since the bacterial structure and composition were significantly different in atrophic larvae from those of healthy larvae, and no parasitic eukaryotes-like cells were observed under a microscope, we also performed meta16S comparison to determine whether the eukaryote community also varied and was associated with intestinal atrophic disease using mitochondrial reads. Although alpha diversity based on the Shannon index was not significantly different between atrophic and healthy larvae (Figure S5), Onchocercidae and Perkinsidae were significantly increased in atrophic larvae after LEfSe analysis, with relative abundances of 1.1± 0.1% and 0.2± 0.2%, respectively (Figure S5).
Impact of environmental rearing seawater on microbiome of atrophic larvae
In order to understand the rearing seawater impact on the microbial differences in atrophic larvae and its relationship with intestinal atrophic disease, the microbial structure and composition of rearing seawater were also assessed. The microbial diversity of alpha diversity based on the Shannon index and beta diversity based on the Unweighted UniFrac distance showed significant differences between atrophic larvae and their rearing seawater (p<0.05) (Figures 5A, B). A total of 118 ASVs (33.5%) were shared within atrophic larvae and the rearing seawater, indicating the seawater also influenced the atrophic microbiome moderately (Figure 5C). Rhodobacteraceae, Flavobacteriaceae, Alteromonadaceae, Pseudohongiellaceae, OM182 Clade (Gammaproteobacteria), Microbacteriaceae, Legionellaceae, Hyphomonadaceae, Stappiaceae, and Saccharospirillaceae were the dominant families in the rearing seawater of atrophic larvae, comprising over 86% of microbiota in the rearing seawater (Table S2). The significantly abundant bacteria in atrophic larvae Spongiibacteraceae (7.5± 4.0%), Desulfuromonadia PB19 (5.3 ± 2.5%), and Halieaceae (3.4 ± 0.9%) were only enriched in atrophic larvae but significantly decreased in their rearing seawater, with relative abundances of 0.4 ± 0.2%, 0.1 ± 0.1%, and 0.1 ± 0.2%, respectively (Figure S3C, Table S2). Other dominant bacteria in atrophic larvae including Flavobacteriaceae, Stappiaceae, Alphaproteobacteria NRL2, and Crocinitomicaceae that might relate with intestinal atrophic disease were also abundant in their rearing seawater (Figure S3C, Table S2). Furthermore, a heatmap based on significantly different genera between atrophic and healthy larvae was generated to better understand how bacteria varied in atrophic larvae, their rearing seawater, and healthy larvae (Figure 6). At the genus level, Planktomarina, the NS3a marine group (Flavobacteriaceae), Pseudohongiella, Nioella, Aestuariibacter, Jejudonia, Pontimonad, Marivita, Alteromonas, and Labrenzia were the most dominant bacterial taxa in the rearing seawater of atrophic larvae, consisting of over 60% of the microbiota in rearing seawater (Table S3). Among these dominant bacteria in the rearing seawater of atrophic larvae, Nioella and Labrenzia were also significantly abundant in atrophic larvae and might be affected by their rearing seawater. Whereas Oceanicoccus, Ruegeria, Crocinitomix, and Muricauda were significantly increased in atrophic larvae compared to healthy larvae, they were significantly reduced in their rearing seawater. The same tendency could also be observed in Halioglobus and Amphritea. Moreover, while Tenacibaculum was dominant in atrophic samples and potentially associated with intestinal atrophic disease, it was only abundant in seawater collected on the first day when disease occurred and not detected in later seawater samples.
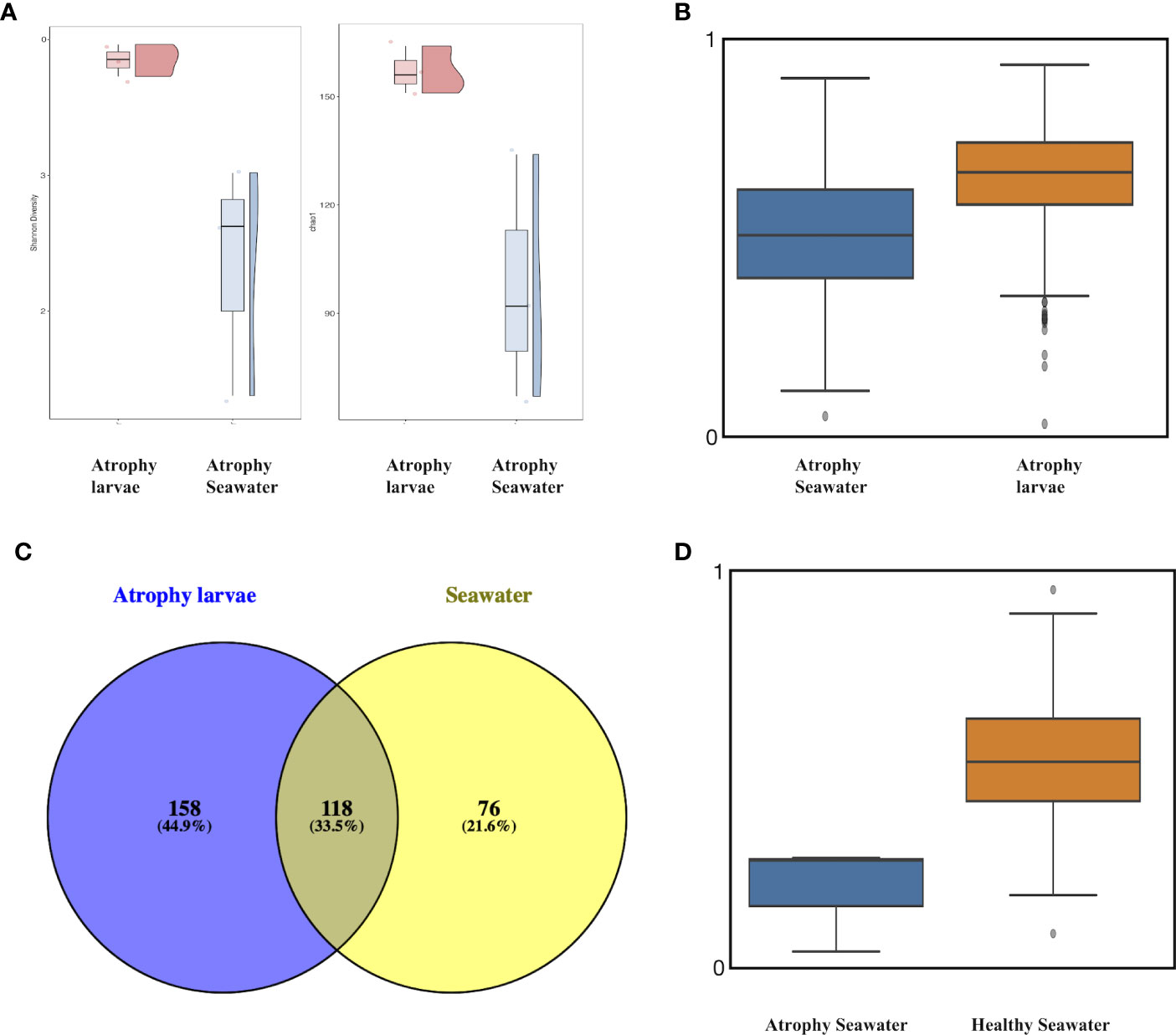
Figure 5 Alpha and beta diversity among atrophic sea cucumber and seawater samples. (A) Alpha diversity based on Chao1 and Shannon index. Red: atrophic larvae samples; blue: atrophic larvae rearing seawater. (B) Boxplot based on unweighted UniFrac distance of atrophic larvae and their rearing seawater (p<0.05, q<0.05). Scale represents similarity within samples. (C) Venn diagram depicting unique and shared OTUs among atrophic larvae and their rearing seawater. (D) Boxplot based on unweighted UniFrac distance of atrophic larvae’s rearing seawater and healthy larvae’s rearing seawater (p<0.05, q<0.05). Scale represents similarity within samples.
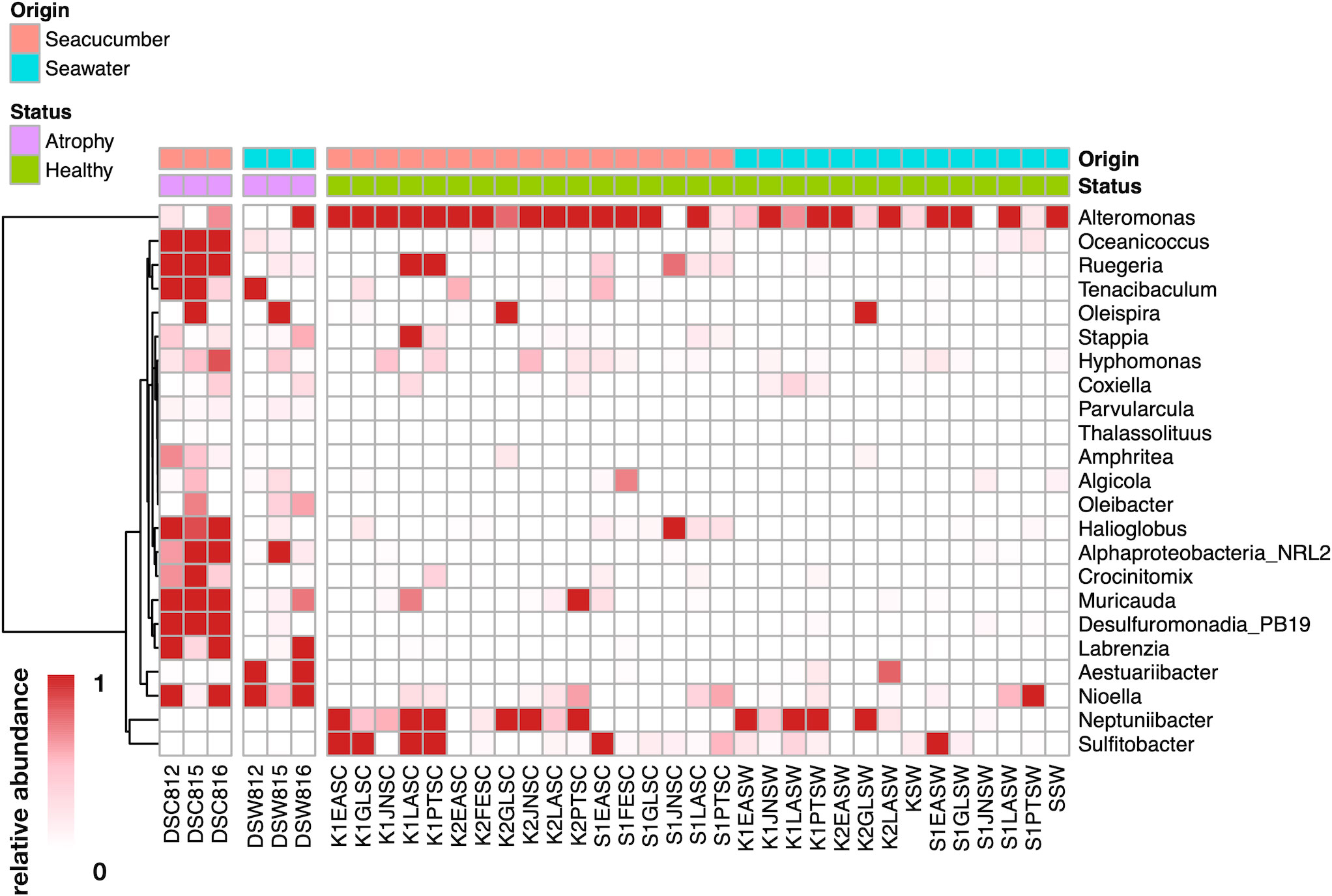
Figure 6 Dynamics of significantly different genera between atrophic larvae, atrophic larvae’s rearing seawater, healthy larvae, and healthy larvae’s rearing seawater. Scale represents relative abundance. DSC812-816 represent atrophic larvae samples; DSW812-816 represent atrophic larvae’s rearing seawater; K*SC and S*SC represent healthy larvae; K*SW and S*SW represent healthy larvae’s rearing seawater. Legend bar shows the sample types and health status. Origin shows the original samples collected from sea cucumber or seawater, red represents sea cucumber, blue represents seawater, Status shows the healthy status of samples, purple represents atrophic samples, and green represents healthy samples.
In addition, we compared the microbiota between atrophic larvae rearing seawater and healthy larvae rearing seawater to further investigate whether seawater impacted the alteration of the larval microbiome and induced potential pathogenetic bacteria. Although alpha diversity based on the Shannon index was not significantly different between diseased larval seawater and healthy larval seawater, beta diversity based on the unweighted UniFrac distance was significantly different between environmental seawater in atrophic and healthy larvae (Figure 5D). At the family level, Alphaproteobacteria NRL2, Micrococcaceae, Parvularculaceae, Porticoccaceae, and Thalassobaculaceae significantly increased in atrophic larval rearing seawater, and Alphaproteobacteria NRL2 was significantly enriched in atrophic larvae, which could be induced by their rearing seawater (Table S2, Figure S6A). At the genus level, Planktomarina, the NS3a marine group (Flavobacteriaceae), Pseudohongiella, Nioella, Aestuariibacter, Jejudonia, Pontimonad, Marivita, Alteromonas, and Labrenzia were the most dominant bacterial taxa in the rearing seawater of atrophic larvae, consisting of over 60% of the microbiota (Table S3). Among these bacteria, Nioella, Oleibacter, Muricauda, and Algicola were found to significantly increase in the seawater used for rearing atrophic larvae but decrease in the seawater used for rearing healthy larvae. These bacteria were also found to be significantly abundant in atrophic larvae, suggesting a potential influence of the rearing seawater on their presence and abundance in atrophic larval microbiome (Figure 6; Figure S6B, Table S3). In addition, the proportion of Tenacibaculum (BUL) in healthy larval rearing seawater was very small compared to atrophic larval rearing seawater (1.1% ± 1.6%), indicating Tenacibaculum was likely to be introduced in the rearing seawater of atrophic larvae. These results suggested rearing seawater impacted on atrophic larval microbiota in a moderate way, causing the differences in microbiome of atrophic larvae.
Microbial function analysis of healthy and atrophic larvae
In order to further understand how microbial functions varied in larvae with intestinal atrophic disease, microbial function profile was also performed by MG-RAST, and it revealed the significantly different microbial functions in atrophic larvae. In general, the most abundant microbial functions were Serine-glyoxylate cycle, methionine biosynthesis, ribosome LSU bacterial, cobalt–zinc–cadmium resistance, purine biosynthesis, Ton and Tol transport systems, respiratory complex I, phosphate metabolism, and RNA polymerase bacterial and DNA replication in atrophic samples (Figure 7A). Heatmaps based on LEfSe analysis revealed the significantly different functions in atrophic larvae (Figure 7B). Compared to healthy larvae, there were more microbial functions enriched in atrophic larvae related to vitamin B (Thiamin) biosynthesis, metabolism of aromatic compounds (phenylalkanoic acid degradation), carbohydrates metabolism (glycolysis and gluconeogenesis), amino acid metabolism (lysine biosynthesis), and protein metabolism (metallocenter biosynthesis). In addition, microbial functions related to respiration including biogenesis of c-type cytochromes, succinate dehydrogenase, and ATP synthase increased in atrophic larvae. These functions may contribute to bacterial pathogens regulating their metabolism for growth in atrophic larvae, enabling them to colonize larval microbiota and induce pathogenic damage. In addition, the function of pathogenicity islands, detected in large numbers of bacterial pathogens, was also enhanced in atrophic larvae, probably in relation to intestinal atrophic disease.
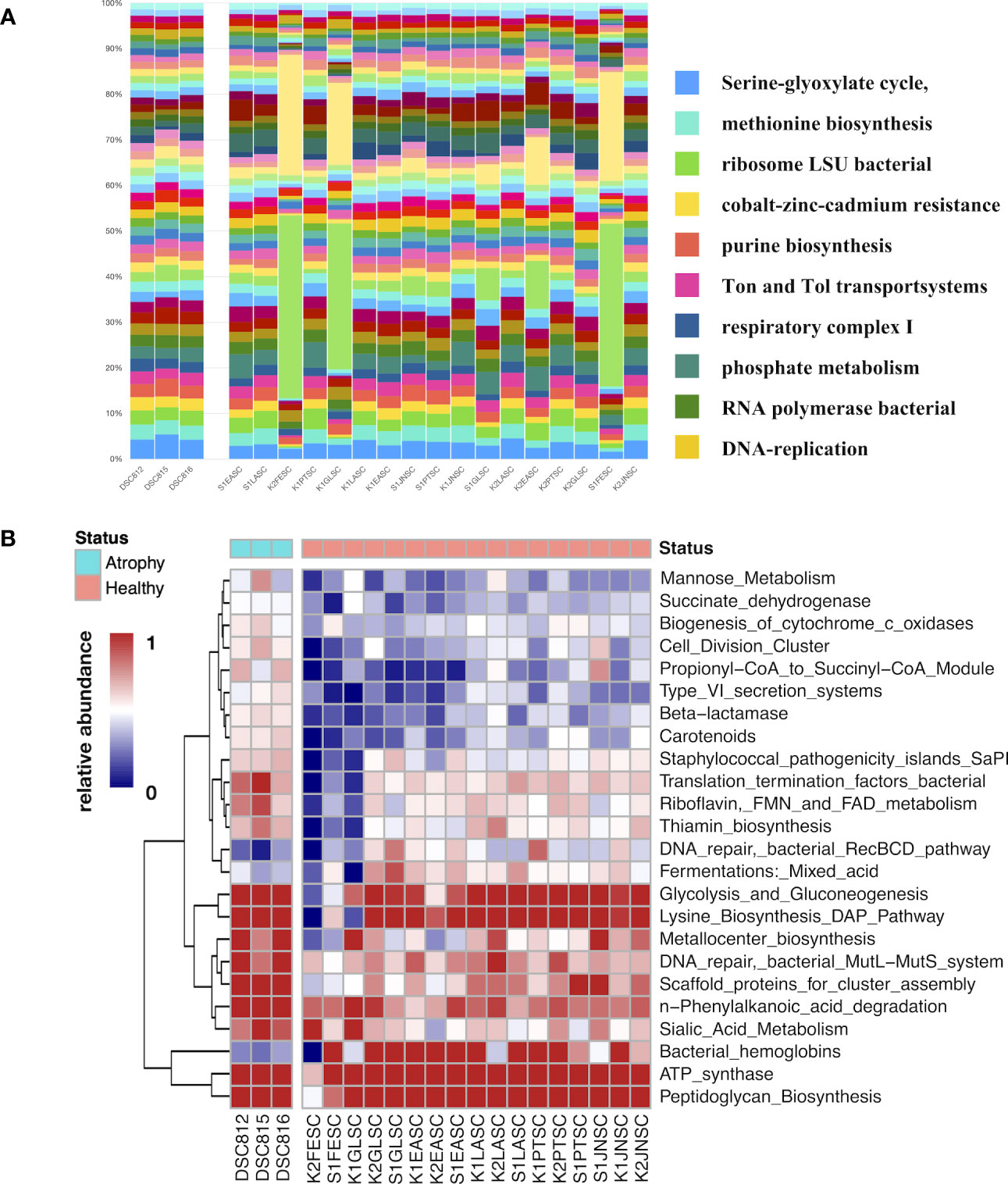
Figure 7 Microbial function analysis of atrophic and healthy larvae. (A) Distribution of microbial functions among atrophic and healthy larvae samples. Bars represent the relative percentage of each microbial functions. DSC812-816 represent diseased sea cucumber larvae and others represent healthy sea cucumber larvae. (B) Dynamics of significantly different microbial functions between atrophic larvae and healthy larvae. Scale represents relative abundance. DSC812-816 represent atrophic larvae samples; K*SC and S*SC represent healthy larvae. Legend bar shows health status. Purple represents atrophic samples, and green represent healthy samples.
Correlation network analysis revealed a more complex microbiome in atrophic larvae
To further understand the species–species association in the microbiome of atrophic larvae, a correlation network of larval microbiota based on Spearman correlation was generated (Figure S7). In healthy larval microbiome, Spongiibacter and Nitrosomonas were the most important bacteria linked to seven other genera, followed by Reinekea and Enrythrobacter related to six other genera (Figure S7A). Compared to healthy larval microbiota, there were more microbial connections observed in atrophic larvae (Figure S7B). Nioella and Ruegeria were in the center of the correlations, related to most other bacteria in atrophic larvae. Algicola was negatively correlated with Nioella and Oceanicaulis, and Aestuariibacter was negatively correlated with unclassified Pseudomonadaceae. The dominant taxon in healthy larval Alteromonas was correlated with abundant taxa in atrophic larvae Stappia. The potential pathogen Tenacibaculum was correlated with Amphritea and unclassified Cryomorphaceae, which was rare in healthy larvae. The microbial correlations in atrophic larvae revealed a more complex microbial correlation in atrophic larval microbiota, which is probably related to intestinal atrophic disease.
Discussion
Disease in sea cucumber A. japonicus has been exclusively studied in recent decades to reduce the mortality of sea cucumber and contribute to the development of its aquaculture, including skin ulceration disease, viscera ejection syndrome, and acute peristome edema disease (Wang et al., 2007; Deng et al., 2009). However, most studies only concentrated on the disease of adult sea cucumber, and there has been a lack of knowledge of the disease that occurs during larval ontogenesis in A. japonicus, which has impacted seed production and resulted in enormous economic losses (Chen et al., 2022b). This study represents the first report of intestinal atrophic disease occurring at the auricularia stage of sea cucumber larval ontogenesis, highlighting it as a novel warning sign during larvae production in sea cucumber aquaculture. These findings have important implications for the industry and emphasize the need for further research and preventive measures to ensure the sustainable development of sea cucumber aquaculture. We revealed the significantly varied microbiome and their correlations in atrophic larvae, in which significantly abundant Tenacibaculum, Oceanicoccus, and Labrenzia were detected. In addition, it was demonstrated that significantly different microbial functions in atrophic larvae related to adaptations in the metabolism contributed to the colonization of bacterial pathogens in the host. Environmental rearing seawater impacted on host microbiome in atrophic larvae moderately, probably associated with the introduction of potential pathogens, highlighting the importance of standardized aquaculture management.
Host-associated microbiomes in marine invertebrates play an important role in host health maintenance, immunity development, and disease resistance (O’Brien et al., 2019). The disturbance of microbial composition and function is directly related to outbreaks of diseases in marine organisms (Hou et al., 2018; Rubio-Portillo et al., 2021). Our study indicated the significantly different microbial structure and function in larvae with intestinal atrophic disease, indicating the microbial alteration in atrophic larvae is probably related to such diseases. In the sea cucumber A. japonicus, diseases caused by bacterial pathogens such as Vibrio spp. and Pseudoalteromonas spp. have been well studied to understand the symptoms, histopathology, and possible treatment for those diseases (Zhang et al., 2021). A study on skin ulceration disease revealed the significantly changed microbiota in diseased sea cucumber and indicated a potential pathogen, Lactococcus garviaeae, related to disease outbreaks (Zhang et al., 2018). V. lentus can cause rotting edges syndrome in sea cucumber larvae at the auricularia stage, which results in the autolysis of larvae and significantly increased mortality (Yang et al., 2015). V. splendidus and V. alginolyticus are also reported to be related to several diseases in A. japonicus, including skin ulceration disease and viscera ejection syndrome (Deng et al., 2009; Zhang et al., 2014). Pseudoalteromonas spp. cause skin ulceration disease in A. japonicus, indicating different bacterial pathogens that could be related to the same disease in sea cucumber (Liu et al., 2010). According to our study, neither Vibrio spp. nor Pseudoalteromonas spp. were enriched in atrophic larval microbiota, showing that these two common pathogens in A. japonicus may not be related to intestinal atrophic disease.
The significantly increased bacterial taxa in atrophic larvae, including Muricauda, Oceanicoccus, Ruegeria, Tenacibaculum, Labrenzia, and Nioella, have been identified as potential causative microbial factors related to disease. While Muricauda spp. have been reported as relatively abundant in the microbiome of diseased sea grape, it has not been directly implicated as a pathogenic bacterium causing disease outbreaks (Kopprio et al., 2021). Bacteria in the genus Ruegeria are associated with coral diseases such as Montipora white syndrome (MWS), but they can also act as commensal bacteria in healthy corals (Casey et al., 2015). The genus Nioella has been detected in shrimp larvae with low survival rates and in the gut microbiome of shrimp with white feces syndrome caused by Vibrio spp. (Lu et al., 2020; Callac et al., 2023). However, apart from Tenacibaculum, there have been no studies reporting these bacteria as pathogenic bacteria associated with diseases or syndromes in marine invertebrates. The increased abundance of these bacteria may be associated with disturbances in the host microbiome and can serve as indicators of diseased status. On the other hand, significantly decreased Sulfitobactor, Alteromonas, and Neptunibacter in atrophic larvae are common core microbes in the pan-microbiome of healthy larvae. The significantly increased bacteria in healthy larval microbiome such as Alteromonas spp. and Sulfitoacter spp. have been used as probiotics in marine mollusks and fish against pathogens, which could probably be applied to investigating probiotics to modulate the host microbiome and prevent intestinal atrophic disease (Emilia Noor and Eguchi, 2012; Chi et al., 2014).
Although there have been no studies reporting significantly increased bacteria Tenacibaculum in atrophic larvae and related diseases in sea cucumber, Tenacibaculum was shown to have been able to cause spotting disease in sea urchin in the Shiriuch farm in 2000 (Masuda et al., 2004). Sea urchins that were exposed to Tenacibaculum F2 exhibited 100% mortality within 24 hours of cultivation and displayed detachment of spines and the development of dark violet spotting lesions (Tajima et al., 1997). Considering the high occurrence of intestinal atrophic disease and the cessation of metamorphosis in sea cucumber larvae cultivated in the same farm at Shiriuchi, we hypothesize that Tenacibaculum spp. present a strong candidate as the potential pathogenic bacterium responsible for causing atrophic disease in sea cucumbers. However, we did not isolate any Tenacibaculum strains from the atrophic larvae in this study, so further investigations are needed to confirm their role and elucidate the underlying mechanisms of pathogenesis. Tenacibaculum spp. are also reported to be a fish pathogenetic bacterium causing “tenacibaculosis” in different fish species, resulting in tissue loss, exposure of jaw bones, and frayed caudal fins (Spilsberg et al., 2022). In one study, Atlantic salmon and rainbow trout were exposed to Tenacibaculum dicentrarchi, resulting in the development of tail and peduncle lesions. The mortality rates significantly increased up to 65% and 93% within one hour, respectively (Avendaño-Herrera et al., 2006). The genomic study on Tenacibaculum revealed virulence-associated genes, including putative virulence, antimicrobial resistance genes, pathogenicity islands, toxins, and hemolysins, being present in most Tenacibaculum species (Nowlan et al., 2023). A metagenomic functional profile revealed that pathogenicity islands were also enriched in atrophic larvae, probably related to the high abundance of Tenacibaculum. Although Tenacibaculum spp. show their virulence factors and potential pathogenicity in marine organisms, the isolation and in situ detection of Tenacibaculum spp. in intestinal atrophic disease during sea cucumber larval ontogenesis also need to be further examined.
Our study also revealed parasitic disease-associated protozoans Perkinsidae belonging to Perkinsea, which is a microeukaryotic parasitic related to varieties of mollusk infection disease (Villalba et al., 2004). A study of sea cucumber with skin ulceration disease also indicated a significantly increased Perkinsidae detected in its eukaryotes community (Delroisse et al., 2020). The histology of Perkinsus species belonging to Perkinsidae exposed to clams indicated trophozoites (15 μm in diameter) of Perkinsus, which could be observed under microscopy at five days after infection (Waki et al., 2012). Since we did not observe any trophozoites nor protozoans in diseased larvae, the significantly increased Perkinsidae may not be the factor causing intestinal atrophic disease but is probably related to alteration of host physiology.
Microbial functions of succinate dehydrogenase, pathogenicity islands, and biosynthesis of cytochrome c oxidases enriched in atrophic larvae were related to bacterial pathogenicity. The metabolism of c-type cytochromes is essential for several pathogens as it enables respiration under both external and host environments. These cytochromes play a crucial role in the pathogen’s virulence by contributing to various mechanisms (Wilson, 2015). Succinate dehydrogenase activity increased in sea cucumber A. japonicus during pathogen V. splendidus infection, involved in innate immunity through its participation in mitochondrial ROS generation (Sun et al., 2020). Such studies provide evidence that significantly increased succinate dehydrogenase in atrophic larvae might also be related to pathogen infection. In particular, studies of Tenacibaculum maritimum detected gene encoding with succinate dehydrogenase and cytochrome c oxidases in its genome, further explaining that Tenacibaculum is the potential pathogen, resulting in intestinal atrophic (Perez-Muñoz et al., 2017). The alteration of the microbial functions allowed the bacterial pathogens to adjust their metabolism for survival and growth in atrophic larvae and further cause pathogenetic infection.
In considering possible factors shaping the microbiome and inducing the bacterial pathogen, environmental factors including rearing water and diets are the most common source of contamination (Chen et al., 2022b). Free-living microbiota within aquatic ecosystems influence the microbiome of aquatic vertebrates such as aquatic mammals and fish, indicating that a shift of environment could lead to ecological disruptions that impact the microbiota and health of aquatic vertebrates (Sehnal et al., 2021). Our results revealed that 33.5% of ASVs were shared with the microbiome in atrophic larvae and their rearing seawater, including the significantly changed microbes and potential pathogen Tenacibaculum. In particular, on the first day we observed the occurrence of intestinal atrophic disease in sea cucumber larvae at the farm, the abundance of Tenacibaculum increased in both atrophic larvae and rearing seawater in the farm (Figure 6, Table S3). These results indicate that the rearing seawater impacted the atrophic larval microbiota, especially related to the natural presence of potential pathogens Tenacibaculum. Similar to our results, the microbiome of sea cucumber with Body Vesicular Syndrome (BVS) was significantly altered and shaped by rearing pond water, resulting in changes of the microbial metabolism and BVS occurrence (Zhao et al., 2022). Studies of environmental factors on skin ulceration syndrome (SUS) outbreaks in A. japonicus also demonstrated that the microbiome significantly changed in SUS-diseased sea cucumber under environmental stressors and pathogen V. splendidus exposure (Zhang et al., 2019). The environmental stresses, including high temperature, mechanical damage, nitrite-N, and ammonium, promoted pathogenetic infection by shaping the gut microbiome and increasing the possibility of pathogenic invasion (Zhang et al., 2019). Another study also emphasized the influence of high temperature on rearing water and sediments of A. japonicus and its microbiome, showing the microbiota was significantly changed with the increasing proportion of potential pathogens (Wang et al., 2021). When we considered the possible side factors accelerating intestinal atrophic disease during larval development, it was found that higher humidity in the breeding center during the summer season might cause the aerosol contamination in the rearing environmental seawater via the aeration system. Formation of dew was observed in the tubes of the aeration system, possibly affecting water quality and providing the possibility for bacteria to colonize the small tube or other components in the aeration system under humid conditions. We observed a significant increase in Tenacibaculum on the first day of disease occurrence and a high abundance of Nioella and Labrenzia in rearing seawater, indicating a contamination event that potentially altered the microbiome of the environmental seawater. This contamination could have induced the potential pathogens, leading to the disturbance of the larval microbiome and resulting in the development of intestinal atrophic disease in the sea cucumber larvae (Herpertz-Dahlmann et al., 2017). In addition, diet should be also considered as a possible reason causing the alteration of the microbiome in the rearing seawater and sea cucumber larvae. Unfortunately, since the diets fed for both healthy and atrophic larvae were originally collected from the same research center (Hakodate Fisheries Research, Japan) and stored under the same conditions (4°C), the microbial community of the diet was not investigated in this study. Our findings on the impact of environmental factors on the atrophic larval microbiome highlight the importance of aquaculture management, in particular, the breeding conditions including temperature, humidity, and salinity, which could be key factors influencing the seed production of sea cucumber. The disturbance of the microbiome in intestinal atrophic larvae also gives us new insights into microbial homeostasis contributing to host disease resistance and health maintenance. Further approaches to isolating potential pathogens and exposing them to sea cucumber larvae could help us better understand the mechanism of intestinal atrophic disease in sea cucumber larvae and contribute to strengthening its aquaculture industry.
Conclusion
In this study, we report for the first time the occurrence of intestinal atrophic disease during sea cucumber larval production in a Japanese farm and investigate the variation in the associated microbiome. Our analysis reveals a significant alteration in the composition of the microbiome in atrophic larvae compared to the healthy pan-microbiome, characterized by increased abundance of keystone bacteria, including Oceanicoccus, Ruegeria, Nioella, Labrenzia, and Tenacibaculum. Our findings suggest that the environmental rearing seawater was the factor impacting the atrophic larval microbiome moderately, emphasizing the need for standardized aquaculture management, particularly with regards to culture conditions. Additionally, this study infers that a potential pathogen, Tenacibaculum, is associated with intestinal disease, underscoring the need for further investigation into the role of potential pathogens in sea cucumber development. Such investigations provide new insights into host–microbe interactions and ultimately benefit the seed production and aquaculture of sea cucumber.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval were not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
TS and JY designed the study. JY, RY, SK, and YS performed the experiment. JY, TS, and SM obtained the data and performed the data analysis. JY and TS drafted the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank the Hokkaido University DX Doctoral Fellowship for the PhD scholarship. This study was supported by Kaken 19K22262. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1225318/full#supplementary-material
References
Adair K. L., Douglas A. E. (2017). Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 35, 23–29. doi: 10.1016/j.mib.2016.11.002
Avendaño-Herrera R., Toranzo A. E., Magariños B. (2006). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis. Aquat Organ 71 (3), 255–266. doi: 10.3354/dao071255
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Callac N., Giraud C., Boulo V., Wabete N., Pham D. (2023). Microbial biomarker detection in shrimp larvae rearing water as putative bio-surveillance proxies in shrimp aquaculture. PeerJ 11, e15201. doi: 10.7717/peerj.15201
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Casey J. M., Connolly S. R., Ainsworth T. D. (2015). Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5 (1), 11903. doi: 10.1038/srep11903
Chen J., Chang Y. (2015). Sea cucumber aquaculture in China. Echinoderm Aquacult., 317–330. doi: 10.1002/9781119005810.ch14
Chen C.-Z., Li P., Liu L., Li Z.-H. (2022a). Exploring the interactions between the gut microbiome and the shifting surrounding aquatic environment in fisheries and aquaculture: A review. Environ. Res. 214, 114202. doi: 10.1016/j.envres.2022.114202
Chen L., Li Q., Yang J. (2008). Microsatellite genetic variation in wild and hatchery populations of the sea cucumber (Apostichopus japonicus Selenka) from northern China. Aquacult. Res. 39 (14), 1541–1549. doi: 10.1111/j.1365-2109.2008.02027.x
Chen J., Lv Z., Guo M. (2022b). Research advancement of Apostichopus japonicus from 2000 to 2021. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.931903
Chi C., Liu J.-Y., Fei S.-Z., Zhang C., Chang Y.-Q., Liu X.-L., et al. (2014). Effect of intestinal autochthonous probiotics isolated from the gut of sea cucumber (Apostichopus japonicus) on immune response and growth of A. japonicus. Fish Shellfish Immunol. 38 (2), 367–373. doi: 10.1016/j.fsi.2014.04.001
Cole J. R., Chai B., Marsh T. L., Farris R. J., Wang Q., Kulam S. A., et al. (2003). The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31 (1), 442–443. doi: 10.1093/nar/gkg039
Cox M. P., Peterson D. A., Biggs P. J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinf. 11 (1), 485. doi: 10.1186/1471-2105-11-485
Delroisse J., Van Wayneberghe K., Flammang P., Gillan D., Gerbaux P., Opina N., et al. (2020). Epidemiology of a SKin Ulceration Disease (SKUD) in the sea cucumber Holothuria scabra with a review on the SKUDs in Holothuroidea (Echinodermata). Sci. Rep. 10 (1), 1–19. doi: 10.1038/s41598-020-78876-0
Deng H., He C., Zhou Z., Liu C., Tan K., Wang N., et al. (2009). Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture 287 (1), 18–27. doi: 10.1016/j.aquaculture.2008.10.015
DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72 (7), 5069–5072. doi: 10.1128/AEM.03006-05
Emilia Noor S., Eguchi M. (2012). Mixed Cultures of the Phytoplankton Nannochloropsis oculata and the Marine Bacterium Sulfitobacter sp. RO3 Inhibit the Growth of Virulent Strains of the Major Fish Pathogen Vibrio Anguillarum. Aquacult. Sci. 60 (1), 39–45. doi: 10.11233/aquaculturesci.60.39
Feng Q.-M., Ru X.-S., Zhang L.-B., Zhang S.-Y., Yang H.-S. (2022). Differences in feeding behavior and intestinal microbiota may relate to different growth rates of sea cucumbers (Apostichopus japonicus). Aquaculture 559, 738368. doi: 10.1016/j.aquaculture.2022.738368
Gao L., He C., Bao X., Tian M., Ma Z. (2017). Transcriptome analysis of the sea cucumber (Apostichopus japonicus) with variation in individual growth. PloS One 12 (7), e0181471. doi: 10.1371/journal.pone.0181471
Gentleman R., Carey V., Huber W., Hahne F. (2023). genefilter: genefilter: methods for filtering genes from high-throughput experiments. R package version 1.82.1. Available at: https://bioconductor.org/packages/release/bioc/html/genefilter.html
Gomez-Alvarez V., Teal T. K., Schmidt T. M. (2009). Systematic artifacts in metagenomes from complex microbial communities. Isme J. 3 (11), 1314–1317. doi: 10.1038/ismej.2009.72
Hamel J. F., Mercier A. (2013). “Apostichopus japonicus. The IUCN red list of threatened species 2013,” Gland: IUCN. doi: 10.2305/IUCN.UK.2013-1.RLTS.T180424A1629389.en
Han Q., Keesing J. K., Liu D. (2016). A review of sea cucumber aquaculture, ranching, and stock enhancement in China. Rev. Fish. Sci. Aquacult. Taylor Francis 24 (4), 326–341. doi: 10.1080/23308249.2016.1193472
Harrell J. F., Dupont Ch. (2019). Hmisc: Harrell Miscellaneous R Package Version 4.2-0. Available at: https://CRAN.R-project.org/package=Hmisc
Herpertz-Dahlmann B., Seitz J., Baines J. (2017). Food matters: how the microbiome and gut–brain interaction might impact the development and course of anorexia nervosa. Eur. Child Adolesc. Psychiatry 26 (9), 1031–1041. doi: 10.1007/s00787-017-0945-7
Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2018). Intestinal bacterial signatures of white feces syndrome in shrimp. Appl. Microbiol. Biotechnol. 102, 3701–3709. doi: 10.1007/s00253-018-8855-2
Keegan K. P., Trimble W. L., Wilkening J., Wilke A., Harrison T., D’Souza M., et al. (2012). A platform-independent method for detecting errors in metagenomic sequencing data: DRISEE. PloS Comput. Biol. Public Library Sci. 8 (6), e1002541. doi: 10.1371/journal.pcbi.1002541
Kim J. (2020) , R package version 0.1.4. Available at: https://github.com/jinkim3/ezr.
Kopprio G. A., Luyen N. D., Cuong L. H., Fricke A., Kunzmann A., Huong L. M., et al. (2021). Bacterial community composition of the sea grape Caulerpa lentillifera: a comparison between healthy and diseased states. BioRxiv 2021, 450479. doi: 10.1101/2021.06.30.450479
Li L., Li Q. (2010). Effects of stocking density, temperature, and salinity on larval survival and growth of the red race of the sea cucumber Apostichopus japonicus (Selenka). Aquacult. Int. 18 (3), 447–460. doi: 10.1007/s10499-009-9256-4
Liu H., Zheng F., Sun X., Hong X., Dong S., Wang B., et al. (2010). Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka). J. Invertebrate Pathol. 105 (3), 236–242. doi: 10.1016/j.jip.2010.05.016
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5 (2), 169–172. doi: 10.1038/ismej.2010.133
Lu J., Zhang X., Qiu Q., Chen J., Xiong J. (2020). Identifying potential polymicrobial pathogens: moving beyond differential abundance to driver taxa. Microbial. Ecol. 80 (2), 447–458. doi: 10.1007/s00248-020-01511-y
Masuda Y., Tajima K., Ezuru Y. (2004). Resuscitation of Tenacibaculum sp., the causative bacterium of spotting disease of sea urchin Strongylocentroutus intermedius, from the viable but non-culturable state. Fish. Sci. 70 (2), 277–284. doi: 10.1111/j.1444-2906.2003.00801.x
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE Public Library Sci. 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 9 (1), 386. doi: 10.1186/1471-2105-9-386
Nowlan J. P., Sies A. N., Britney S. R., Cameron A. D. S., Siah A., Lumsden J. S., et al. (2023). Genomics of Tenacibaculum species in British Columbia, Canada. Pathogens 12 (1), 101. doi: 10.3390/pathogens12010101
O’Brien P. A., Webster N. S., Miller D. J., Bourne D. G. (2019). Host-microbe coevolution: applying evidence from model systems to complex marine invertebrate holobionts. MBio 10 (1), e02241–e02218. doi: 10.1128/mBio.02241-18
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2015). “vegan: community ecology package. 2019” R Package Version 2, 10. Available at: https://cran.r-project.org/web/packages/vegan/vegan.pdf
Oliveros J. C. (2007-2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Overbeek R., Begley T., Butler R. M., Choudhuri J. V., Chuang H.-Y., Cohoon M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33 (17), 5691–5702. doi: 10.1093/nar/gki866
Perez-Muñoz M. E., Arrieta M. C., Ramer-Tait A. E., Walter J. (2017). A critical assessment of the ‘sterile womb’ and ‘in utero colonization’ hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 5 (1), 48. doi: 10.1186/s40168-017-0268-4
Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2 – approximately Maximum-likelihood trees for large alignments. PloS ONE Public Library Sci. 5 (3), e9490. doi: 10.1371/journal.pone.0009490
Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35 (21), 7188–7196. doi: 10.1093/nar/gkm864
Revelle W. (2017). psych: Procedures for Personality and Psychological Research Software. Available at: https://CRAN.R-project.org/package=psych
Rubio-Portillo E., Ramos-Esplá A. A., Antón J. (2021). Shifts in marine invertebrate bacterial assemblages associated with tissue necrosis during a heat wave. Coral Reefs 40 (2), 395–404. doi: 10.1007/s00338-021-02075-0
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60
Sehnal L., Brammer-Robbins E., Wormington A. M., Blaha L., Bisesi J., Larkin I., et al. (2021). Microbiome composition and function in aquatic vertebrates: small organisms making big impacts on aquatic animal health. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.567408
Selenka E. (1867). Beiträge zur Anatomie und Systematik der Holothurien. Zeitschrift für wissenschaftliche Zoologie 17 (2), 291–374. pls. XVII-XX., Available online at: https://www.biodiversitylibrary.org/page/45007590#page/321/mode/1up.
Spilsberg B., Nilsen H. K., Tavornpanich S., Gulla S., Jansen M. D., Lagesen K., et al. (2022). Tenacibaculosis in Norwegian Atlantic salmon (Salmo salar) cage-farmed in cold sea water is primarily associated with Tenacibaculum finnmarkense genomovar finnmarkense. J. Fish Dis. 45 (4), 523–534. doi: 10.1111/jfd.13577
Sun L., Zhou F., Shao Y., Lv Z., Li C. (2020). The iron–sulfur protein subunit of succinate dehydrogenase is critical in driving mitochondrial reactive oxygen species generation in Apostichopus japonicus. Fish Shellfish Immunol. 102, 350–360. doi: 10.1016/j.fsi.2020.04.060
Tajima K., HIrano T., Shimizu M., Ezura Y. (1997). Isolation and pathogenicity of the causative bacterium of spotting disease of sea urchin strongylocentrotus intermedius. Fish. Sci. 63, 249–252. doi: 10.2331/fishsci.63.249
Villalba A., Reece K. S., Ordás M. C., Casas S. M., Figueras A. (2004). Perkinsosis in molluscs: a review. Aquat. Living Resour. 17 (4), 411–432. doi: 10.1051/alr:2004050
Waki T., Shimokawa J., Watanabe S., Yoshinaga T., Ogawa K. (2012). Experimental challenges of wild Manila clams with Perkinsus species isolated from naturally infected wild Manila clams. J. Invertebrate Pathol. 111(1), 50–55. doi: 10.1016/j.jip.2012.05.009
Wang P., Chang Y., Yu J., Li C., Xu G. (2007). Acute peristome edema disease in juvenile and adult sea cucumbers Apostichopus japonicus (Selenka) reared in North China. J. Invertebrate Pathol. 96 (1), 11–17. doi: 10.1016/j.jip.2007.03.001
Wang L., Wei C., Chang Y., Ding J. (2021). Response of bacterial community in sea cucumber Apostichopus japonicus intestine, surrounding water and sediment subjected to high-temperature stress. Aquaculture 535, 736353. doi: 10.1016/j.aquaculture.2021.736353
Wang Y.-G., Zhang C.-Y., Rong X.-J., Chen J.-J., Shi C.-Y. (2004). Diseases of cultured sea cucumber, Apostichopus japonicus, in China. FAO Fish Tech Pap 46, 297–310.
Wilson A. (2015). Bacterial c-type cytochromes and pathogenicity. Eur. J. BioMed. Res. 1 (1), 17–21. doi: 10.18088/ejbmr.1.1.2015.pp17-21
Yamazaki Y., Meirelles P. M., Mino S., Suda W., Oshima K., Hattori M., et al. (2016). Individual Apostichopus japonicus fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci. Rep. 6 (6), 21631. doi: 10.1038/srep21631
Yamazaki Y., Sakai Y., Mino S. (2019). Repeated selective enrichment process of sediment microbiota occurred in sea cucumber guts. Environ. Microbiol. Rep. 11, 797–807. doi: 10.1111/1758-2229.12791
Yamazaki Y., Sakai Y., Yu J. W., Mino S., Sawabe T. (2020). Tracking the dynamics of individual gut microbiome of sea cucumber Apostichopus japonicus during gut regeneration. PeerJ 8, e10260. doi: 10.7717/peerj.10260
Yang H., Hamel J.-F., Mercier A. (2015). The Sea Cucumber Apostichopus japonicus: History, Biology and Aquaculture (China: Academic Press).
Yang G., Peng M., Tian X., Dong S. (2017). Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: An example of sea cucumber. Sci. Rep. 7 (1), 4778. doi: 10.1038/s41598-017-05312-1
Yang G., Xu Z., Tian X., Dong S., Peng M. (2015). Intestinal microbiota and immune related genes in sea cucumber (Apostichopus japonicus) response to dietary β-glucan supplementation. Biochem. Biophys. Res. Commun. (China: Academic Press) 458 (1), 98–103. doi: 10.1016/j.bbrc.2015.01.074
Yasoda H. N., Chi Z., Zhu K. (2006). Probiotics and sea cucumber farming. SPC Bechedemer Inf. Bull. 24 (5), 45–48.
Yu J. W., Sakai Y., Mino S., Sawabe T. (2022). Characterization of microbiomes associated with the early life stages of sea cucumber Apostichopus japonicus selenka. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.801344
Yu D. X., Sun H. L., Chen S. Q., Zhang Y., Liu C. L., Tan J. (2010). Healthy Aquaculture Technology of Sea cucumber (Beijing: Ocean Press).
Zhang B. (2006). Isolation and identification of causative pathogen for skin ulcerative syndrome in Apostichopus japonicus. Journal of fisheries of China 30 (1). https://www.china-fishery.com/scxuebao/article/abstract/20060119?st=search
Zhang J., Cao Z., Li Z., Wang L., Li H., Wu F., et al. (2015). Effect of Bacteriophages on Vibrio alginolyticus Infection in the Sea Cucumber, Apostichopus japonicus (Selenka). J. World Aquacult. Soc. 46 (2), 149–158. doi: 10.1111/jwas.12177
Zhang W., Li C., Guo M. (2021). Use of ecofriendly alternatives for the control of bacterial infection in aquaculture of sea cucumber Apostichopus japonicus. Aquaculture 545, 737185. doi: 10.1016/j.aquaculture.2021.737185
Zhang P., Li C., Zhang P., Jin C., Pan D., Bao Y. (2014). iTRAQ-based proteomics reveals novel members involved in pathogen challenge in sea cucumber Apostichopus japonicus. PloS ONE Public Library Sci. 9 (6), e100492. doi: 10.1371/journal.pone.0100492
Zhang Z., Xing R., Lv Z., Shao Y., Zhang W., Zhao X., et al. (2018). Analysis of gut microbiota revealed Lactococcus garviaeae could be an indicative of skin ulceration syndrome in farmed sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 80, 148–154. doi: 10.1016/j.fsi.2018.06.001
Zhang Z., Zhang W., Hu Z., Li C., Shao Y., Zhao X., et al. (2019). Environmental factors promote pathogen-induced skin ulceration syndrome outbreak by readjusting the hindgut microbiome of Apostichopus japonicus. Aquaculture 507, 155–163. doi: 10.1016/j.aquaculture.2019.03.054
Zhao Z., Jiang J., Pan Y., Dong Y., Wang B., Gao S., et al. (2022). Sea cucumber body vesicular syndrome is driven by the pond water microbiome via an altered gut microbiota. Msystems 7 (3), e01357–e01321. doi: 10.1128/msystems.01357-21
Zhao Y., Zhang W., Xu W., Mai K., Zhang Y., Liufu Z. (2012). Effects of potential probiotic Bacillus subtilis T13 on growth, immunity and disease resistance against Vibrio splendidus infection in juvenile sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 32 (5), 750–755. doi: 10.1016/j.fsi.2012.01.027
Keywords: Apostichopus japonicus, host-microbe interaction, larval development, disease susceptibility, metagenomics
Citation: Yu J, Sawabe T, Yamano R, Koike S, Sakai Y and Mino S (2023) Inferring potential causative microbial factors of intestinal atrophic disease in the sea cucumber Apostichopus japonicus. Front. Mar. Sci. 10:1225318. doi: 10.3389/fmars.2023.1225318
Received: 19 May 2023; Accepted: 11 August 2023;
Published: 30 August 2023.
Edited by:
Chris Michael Pearce, Fisheries and Oceans Canada (DFO), CanadaReviewed by:
Amy Long, Fisheries and Oceans Canada (DFO), CanadaOjas Natarajan, University of South Florida, United States
Copyright © 2023 Yu, Sawabe, Yamano, Koike, Sakai and Mino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juanwen Yu, anVhbndlbi55dUBmaXNoLmhva3VkYWkuYWMuanA=; Tomoo Sawabe, c2F3YWJlQGZpc2guaG9rdWRhaS5hYy5qcA==
 Juanwen Yu
Juanwen Yu Tomoo Sawabe
Tomoo Sawabe Ryota Yamano1
Ryota Yamano1 Yuichi Sakai
Yuichi Sakai Sayaka Mino
Sayaka Mino