- 1Key Laboratory of Mariculture & Stock Enhancement in North China’s Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University, Dalian, China
- 2College of Biological Science and Technology, Shenyang Agricultural University, Shenyang, China
- 3College of Life Science, Liaoning Normal University, Dalian, China
Comparative microRNA (miRNA) and mRNA transcriptome analyses were performed on Strongylocentrotus intermedius of the same sex with significant gonadal color differences. The results indicated that 1) the color of female gonads was generally superior to that of males. 2) Comparative and integrated miRNA and mRNA transcriptome analyses identified differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) in female and male individuals with significant gonadal color differences. Common and sex-specific DEGs and “DEM-DEG” pairs involved in carotenoid absorption, accumulation, and transformation were identified as candidates correlated with gonad coloration in S. intermedius. Collectively, the results from this study have enriched our knowledge of the process of sea urchin gonad coloration and should provide additional clues for increasing the gonad quality of commercial sea urchins from molecular and metabolic aspects.
1 Introduction
Sea urchins are representative invertebrates inhabiting shallow coastal areas. At present, there are nearly a thousand sea urchin species distributed in various sea regions worldwide. Among the existing sea urchin species, only 30 species are edible and are of economic value (Zhao, 2005). Only about 10 species of edible sea urchins have been extensively studied and utilized in large-scale aquaculture (Kelly, 2005).
The gonad (also named as the “uni” or “roe” in the market) is the major edible part of sea urchins. With the increasing consumption of sea urchin gonads, the international trade in sea urchins has seen increasing activity in recent years. In the market, gonad color is considered as one of the most important metrics determining the quality and the presentation degree of sea urchin products, and this consequently influences the price of sea urchin gonads. Typically, gonads with bright mango or yellow color are the most desirable, and those that are pale or dark brown are the least desirable. Therefore, developing suitable color in the gonads of farmed sea urchins has been an important concern in sea urchin aquaculture (Mcbride et al., 2004). It has been acknowledged that gonad coloration in sea urchins is the result of dietary carotenoid accumulation (Kelly and Symonds, 2013). Based on this context, efforts have been made to improve or enhance the gonad color of sea urchins through diet adjustment. For example, kelp is considered as the best diet for enhancing the color of sea urchin gonads compared to other algae or prepared foods (Qiu et al., 2005). The addition of β-carotene to the diet can enable the sea urchins Strongylocentrotus droebachiensis and Psammechinus miliaris to obtain a better gonadal color (Suckling et al., 2011). In addition, it has recently been reported that sea urchin lineages with desirable gonad color can be obtained by distant hybridization or selective breeding (Liu et al., 2020). However, knowledge of the molecular mechanism of gonad coloration is still limited at present, and strategies for gonad color enhancement from a molecular aspect are lacking.
The temperate sea urchin Strongylocentrotus intermedius is the edible species with the highest quality gonads. This species was introduced to northern China from Japan in 1989 (Chang, 2004). Currently, S. intermedius is the predominant commercially valuable sea urchin species farmed in large-scale aquaculture systems along the coast of Liaoning and Shandong provinces in China (Gao and Chang, 1999). Consistent with the studies on gonad coloration mentioned above, we found that diet improvement, hybridization, and selective breeding could enhance the gonad color of S. intermedius (Tang et al., 2022). In addition, during the farming process, we found that gonadal color differences existed even in sea urchins of the same sex reared under identical culture and feeding conditions and a consistent genetic background. We therefore hypothesized that gene expression and regulation may also play important roles in gonad coloration in S. intermedius.
To examine our hypothesis, we first determined the gonad colors of S. intermedius individuals with consistent genetic backgrounds and identical breeding conditions and without morphological trait differences, and then, S. intermedius individuals of the same sex and with significant gonad color differences were identified and selected. Finally, an integrated microRNA (miRNA)-mRNA analysis was performed to identify candidate genes, miRNAs, and miRNA-mRNA modules associated with gonad coloration in S. intermedius. The results of this study will not only deepen our understanding of the molecular mechanism of gonad coloration in sea urchins but will also provide additional biomarkers for gonad color improvement and enhancement in sea urchin aquaculture.
2 Materials and methods
2.1 Specimen collection
A total of 100 S. intermedius individuals [average shell diameter: (3.84 ± 0.16) cm; average height: (2.17 ± 0.17) cm; average body weight: (22.08 ± 2.65) g] were randomly selected from a family broodstock cultured at the Key Laboratory of Mariculture & Stock Enhancement in North China’s Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University. All specimens were at the same age and were cultured in 500 L recirculating seawater tanks under identical conditions [room temperature; natural light cycle; pH: (8.10 ± 0.03); salinity: (30.15 ± 0.20); diet: kelp (Saccharina japonica); continuous aeration; seawater changed every 2 days].
2.2 Trait measurements, gonad color determination, and sex identification
Phenotypic traits including test diameter, test height, live body weight, and gonad weight were measured initially. Test diameter and test height were measured by using a digital display electronic Vernier caliper (precision: 0.01 mm; 16EW; Mahr, Germany). For measurement of live body weight, the specimens were dried with paper towels and then weighed individually using a digital balance with a sensitivity of 0.01 g (JJ300, Changshu Shuangjie Testing Instrument Factory, Jiangsu, China). Specimens with no significant differences in phenotypic traits were then selected. The gonad tissues were dissected on ice and washed with sterile seawater to remove the intestines and other tissues on the surface as much as possible. Then, each gonad was divided into three parts: the first part for gonadal color determination, the second part for sex determination, and the third part for further RNA sequencing (RNA-seq).
Gonadal color was determined with a Color Cue 2 colorimeter (PANTONE, USA) in D65 mode. Based on the standards of light orange-yellow (L1* = 68.9, a1* = 28.7, b1* = 60.4) and light yellow (L2* = 74.6, a2* = 28.7, b2* = 66.1), the total chromatic aberration between individual gonadal color and standard color was defined as ΔE. The chromatic aberration between individual gonadal color and the light orange-yellow standard color was defined as ΔE1, while the chromatic aberration between individual gonadal color and the light yellow standard color was defined as ΔE2. The ΔE (both ΔE1 and ΔE2) values were calculated via the method described in the International Commission on Illumination LAB (CIELAB) 1976 standard (Mcbride et al., 2004). The calculation formula was as follows:
where L* is the lightness of the gonad; a* is the redness of the gonad, and b* is the yellowness of the gonad; ΔL1* (or ΔL2*) is the difference between gonad lightness and the light orange-yellow (or light yellow) standard color; Δa1* (or Δa2*) is the difference between gonad redness and the light orange-yellow (or light yellow) standard color; and Δb1* (or Δb2*) is the difference between gonad yellowness and the light orange-yellow (or light yellow) standard color.
EBR1 and Bindin genes were selected for sex determination of each individual according to the method proposed by Cui et al. (Cui et al., 2022). The reaction system and reaction conditions followed the methods described by Zhan et al. (Zhan et al., 2020), and the relative expression level of each gene was determined using the 2−ΔΔCT method (Feng et al., 2023). Primers for sex determination are listed in Supplementary Table 2.
2.3 RNA-seq sample preparation, library construction, sequencing and data processing
To avoid having phenotypic trait differences affect our results, the specimens with no significant differences in phenotypic traits were selected. For each sex, specimens with a ΔE (both ΔE1 and ΔE2) in the range of 20.00–35.00 were considered as having delicate color gonads, and specimens with ΔE (both ΔE1 and ΔE2) values less than 10 were considered as having lovely color gonads (designated as the L group). Detailed specimen information is presented in Supplementary Table 1.
A total of six specimens of each sex were selected, including three specimens with delicate color gonads (designated as the D group) and three specimens having lovely color gonads (designated as the L group). The gonad tissues from each specimen were divided into three parts: one part for mRNA library construction, another other part for small RNA library construction, and the last part for quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) validation. For the D group, the mRNA library of females was designated as C_D, with three replicates (C_D_1, C_D_2, and C_D_3); the mRNA library of males was designated as X_D, with three replicates (X_D_1, X_D_2, and X_D_3); the small RNA library of females was designated as sC_D, with three replicates (sC_D_1, sC_D_2, and sC_D_3); and the small RNA library of males was designated as sX_D, with three replicates (sX_D_1, sX_D_2, and sX_D_3). For the L group, the mRNA library of females was designated as C_L, with three replicates (C_L_1, C_L_2, and C_L_3); the mRNA library of males was designated as X_L, with three replicates (X_L_1, X_L_2, and X_L_3); the small RNA library of females was designated as sC_L, with three replicates (sC_L_1, sC_L_2, and sC_L_3); and the small RNA library of males was designated as sX_L, with three replicates (sX_L_1, sX_L_2, and sX_L_3). All samples were stored at −80°C until RNA extraction.
Total RNA isolation and qualification were performed following the method proposed by Zhan et al. (Zhan et al., 2019).
For UMI-mRNA transcriptome sequencing, total RNA purity and concentration were examined using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), and total RNA integrity and quantity were finally measured using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system. High-quality RNAs (3 μg/sample) were used to construct sequencing libraries using a NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s instructions. Illumina sequencing of mRNA library preparations was performed on an Illumina Novaseq 6000 platform.
For small RNA transcriptome sequencing, small RNA libraries based on 2 μg high-quality RNA per sample were constructed using a NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system with DNA High Sensitivity Chips. Small RNA library preparations were finally sequenced on a Novaseq 6000 platform.
The original sequencing reads from both mRNA and small RNA transcriptomes that contained low-quality reads with adapters were preliminarily processed to generate clean reads. After trimming using standard methods (Zhan et al., 2019), all RNA clean data were submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) Sequence Database (Accession No. PRJNA949413). Q20 (percentage of bases with a Phred value > 20), Q30 (percentage of bases with a Phred value > 30), and GC-content were also quantified to evaluate the quality of the clean data.
The high-quality clean data (reads without adapters, reads with N bases less than 10%, and reads with a base number of qphred ≤ 20 accounted for less than 50% of the total data) were used for subsequent analyses. The reference library was generated by assembling clean data into transcriptomes following the method previously described by Grabherr et al. (Grabherr et al., 2011). Gene expression levels were then calculated with RSEM (v. 1.2.15). The completeness of the gene assembly was evaluated by BUSCO v. 3.0.2.
Transcriptome annotation was performed using the Basic Local Alignment Search Tool (BLAST). Databases employed were the NCBI non-redundant (Nr) database, NCBI nucleotide sequences (Nt), Swiss-Prot, Protein family, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups (COG). Blast2GO with NR annotation and InterProScan5 were employed for Gene Ontology (GO) and InterPro annotation, respectively.
DESeq2 v. 1.6.3 was used to analyze differentially expressed genes (DEGs) in the mRNA libraries. A |log2 fold change| > 0.5 was set as the threshold for significant differential expression. DEGs were classified based on GO (GOseq 1.10.0 and 2.10.0 method) and KEGG (KOBAS 2.0.12 method).
Small RNA clean data were trimmed from the raw data of small RNA libraries by removing adapter dimers, junk (reads with 5’ pollution or without a 3’ sequence), and low-complexity reads (i.e., reads with a qphred base number ≤ 20 that accounted for more than 30% of the total data, reads with more than 10% N bases, and reads with polyA/T/G/C). To remove tags originating from protein-coding genes, repeat sequences, rRNAs, tRNAs, snRNAs, snoRNAs, and small RNA tags were mapped to RepeatMasker and Rfam databases as much as possible. All clean data were then submitted to the NCBI SRA Sequence Database (Accession No. PRJNA949420). Small RNAs were annotated by the Rfam (http://rfam.sanger.ac.uk/) database, and rRNAs, tRNAs, snRNAs, and snoRNAs were discarded. Novel miRNA prediction and analyses were performed using miREvo and mirdeep 2.
Differentially expressed miRNAs (DEMs) were analyzed using the DESeq R package (1.10.1) with the threshold of |log2 fold change| > 0.75 for significant differential expression by default. The DEMs were cluster analyzed with a value of log10 (TPM+1).
GO terms and KEGG pathways of DEGs and candidate target genes of DEMs were analyzed. A corrected P-value < 0.05 indicated significant enrichment (Young et al., 2010), and a rich factor indicated enrichment degree. P-values and rich factors were calculated using the following equations:
where M is the number of genes annotated as a specific pathway among all genes; N is the number of all genes with pathway annotation; n is the number of differentially expressed genes in N; x is the number of differentially expressed genes annotated as in a specific pathway; and j is a constant ranging from 0 to x − 1.
where K is the number of DEGs annotated to a specific pathway, and M is the number of genes annotated to a specific pathway among all genes.
2.4 miRNA target gene prediction by RNA-seq
The miRNAs obtained from small RNA-seq analysis in this study were mapped to reference sequences using Bowtie. Moreover, we also employed the whole genome sequencing data from Strongylocentrotus purpuratus (NCBI Taxonomy ID: 7668; the same family as S. intermedius) as another reference to enhance the accuracy of the prediction of miRNA target genes in this study.
MiRanda was used to identify sequence conservation, binding sites, and the UTR base distribution of each microRNA (score = 140.0, energy = −1.0). Because miRNA regulation could be both positive and negative, we therefore focused on those miRNA-mRNA pairs with the same and opposite relative expression trends in this study.
2.5 qRT-PCR validation
qRT-PCR was used to validate candidate DEGs and DEMs. Total RNA was extracted using the Trizol reagent. A PrimeScript™ RT reagent kit (TaKaRa, Japan) and an miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN, China) were employed to generate the first strand cDNA of DEGs and DEMs, respectively.
A total of 12 DEGs (up: 9; down: 3) and three DEMs (up: 1; down: 2) were selected from RNA-seq and small RNA-seq of females, and 12 DEGs (up: 10; down: 2) and 10 DEMs (up: 6; down: 4) were selected from RNA-seq and small RNA-seq of males. Primers were designed by primer premier 5.0 (http://www.premierbiosoft.com/) (Supplementary Table 2). β-actin and RUN6B (U6) were used as internal controls (Zhou et al., 2018). The qRT-PCR was performed in a 20 μL reaction volume containing 2 μL of cDNA, 10 μL of 2×TB Green Premix Ex Taq II (Tli RNaseH Plus), 6.4 μL of double-distilled H2O water, and 0.8 μL of each primer. The cycling program was as follows: 95°C for 600 s; 95°C for 15 s, 60°C for 60 s, followed by 40 cycles; 95°C for 10 s, 65°C for 60 s, 97°C for 1 s, and 37°C for 30 s. Single-PCR product confirmation was analyzed using melting curve analysis at the end of the amplification. The relative expression level of each candidate DEG or DEM was determined by the 2−ΔΔCT method (Feng et al., 2023).
2.6 Data analysis
All data are expressed as the mean value ± standard deviation (mean ± S.D.). Statistical analysis was performed using IBM SPSS Statistics (v. 26). Normal distributions and homogeneity of variance were tested by using SPSS v26.0 (IBM, Shanghai, China).
The measured variables were normally distributed in this analysis. One-way ANOVA was used to analyze the test diameter, test height, live body weight, gonad weight, and chromatic aberration. In addition, gene and miRNA relative expression levels were quantified according to the results of statistical analyses of three independent experiments (n = 3). One-way ANOVA was used to analyze the differences. Statistical significance was set as P < 0.05; highly statistically significance was set as P < 0.01, and extremely statistically significance was set as P < 0.001.
3 Results
3.1 Gonad color differences between female and male S. intermedius
The chromatic aberration data showed that the ΔE1 range of the specimen selected for RNA-seq was from 2.34 to 26.24, and the ΔE2 range of the specimen selected for RNA-seq was from 2.80 to 34.21. The average ΔE1 of gonads of all examined females was 10.54 ± 4.46, which was less than that of gonads of all examined males (13.77 ± 7.60). The average ΔE2 of gonads of all examined females was 13.67 ± 7.50, which was also less than that of gonads of all examined males (18.65 ± 9.96) (Supplementary Table 1). Further analysis showed that 1) the proportion of females (46.43%) was higher than that of males (34.00%) when ΔE (both ΔE1 and ΔE2) was less than 10; 2) when 10 ≤ ΔE (both ΔE1 and ΔE2) ≤ 20, the proportion of females (42.86%) was higher than that of males (12.00%); 3) the proportion of females (10.71%) was lower than that of males (54.00%) when ΔE (both ΔE1 and ΔE2) was greater than 20 (Figure 1).
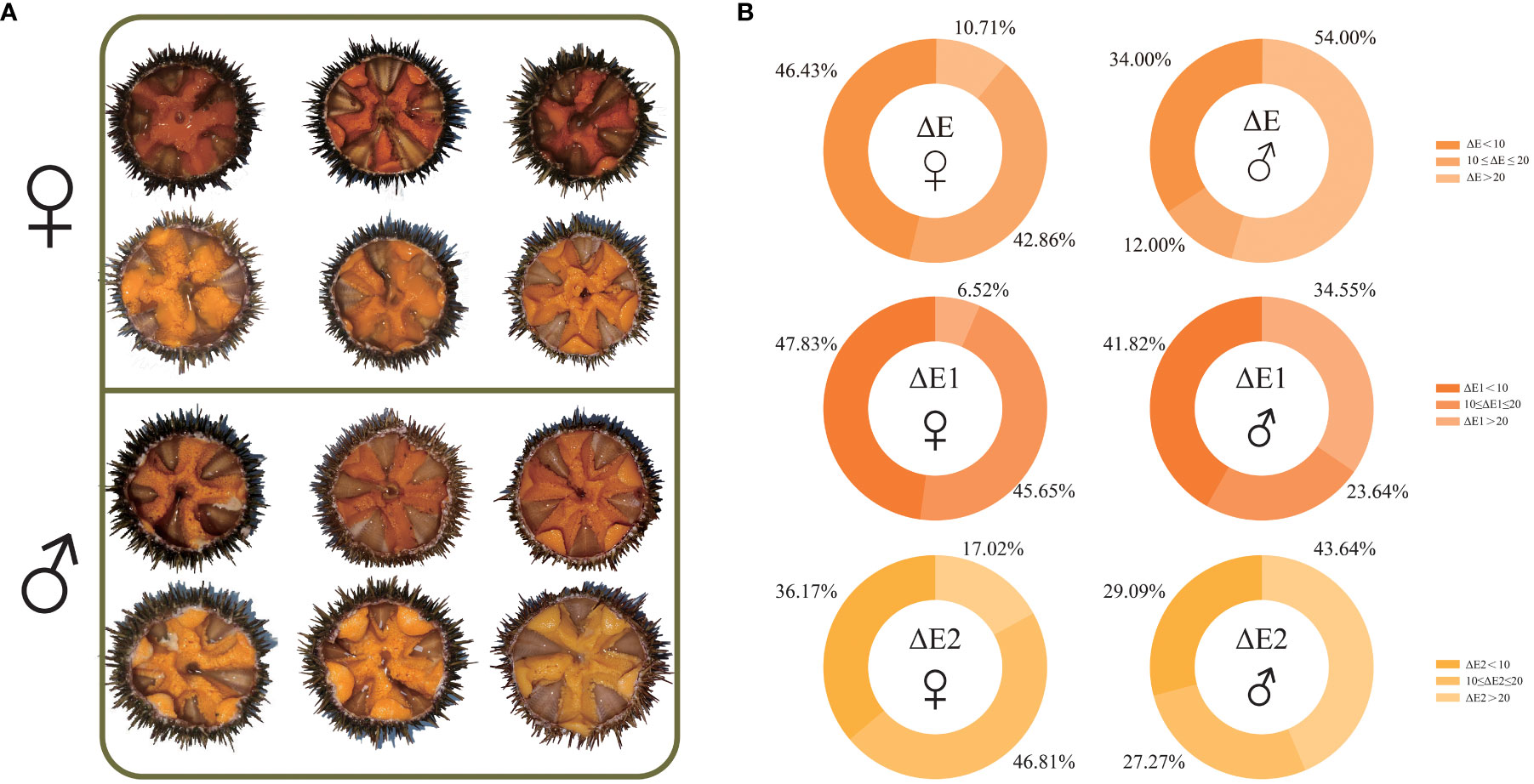
Figure 1 Anatomical images of gonads of selected Strongylocentrotus intermedius for RNA sequencing (A) and gonad color determination summary of all examined S. intermedius (B). ΔE: total chromatic aberration between individual gonadal color and standard color; ΔE1: chromatic aberration between individual gonadal color and light orange-yellow standard color; ΔE2: chromatic aberration between individual gonadal color and light yellow standard color.
3.2 UMI-mRNA and small RNA sequencing
For UMI-mRNA sequencing, about 45.43–60.28 million clean reads were obtained from each sample after trimming. The Q20 of clean reads ranged from 96.89% to 97.37%, and the Q30 range was 91.84%–92.65%. The GC content range of the samples was 39.14%–44.77%. The completeness assessment data showed that the assembled unigenes were deemed 87.8% complete by BUSCO using a eukaryotic database of 978 genes, of which 42.8% were single genes and 45.0% were duplicated genes. A total of 145,207 transcripts were obtained with an average length of 1091 bp, and the N50 and N90 length averages of all transcripts were 2163 bp and 395 bp, respectively. Approximately 132,621 unigenes with a mean length of 1171 bp and an N50 length of 2212 bp were also generated. After BLAST, 97,693 unigenes were annotated in at least one database, and 6960 unigenes could be annotated in all databases. The lengths of the unigenes with the largest numbers of bases ranged from 300 bp to 500 bp (Supplementary Figures 1 and 2).
Based on the results of sRNA sequencing, a total of 10,639,960–14,784,908 clean reads were observed as candidates and were included in the subsequent comparative miRNA analysis. A total of 41 precursor miRNAs and 35 mature miRNAs were identified, of which mature miRNAs comprised 22 known miRNAs and 13 novel miRNAs (Supplementary Figures 1 and 2).
3.3 Comparative mRNA and miRNA transcriptome analyses and verification
A total of 78,025 DEGs (up: 41,181; down: 36,844) between the C_L group and the C_D group and 69,147 DEGs (up: 35,198; down: 33,949) between the X_L group and the X_D group were identified by mRNA transcriptome comparisons (Figure 2).
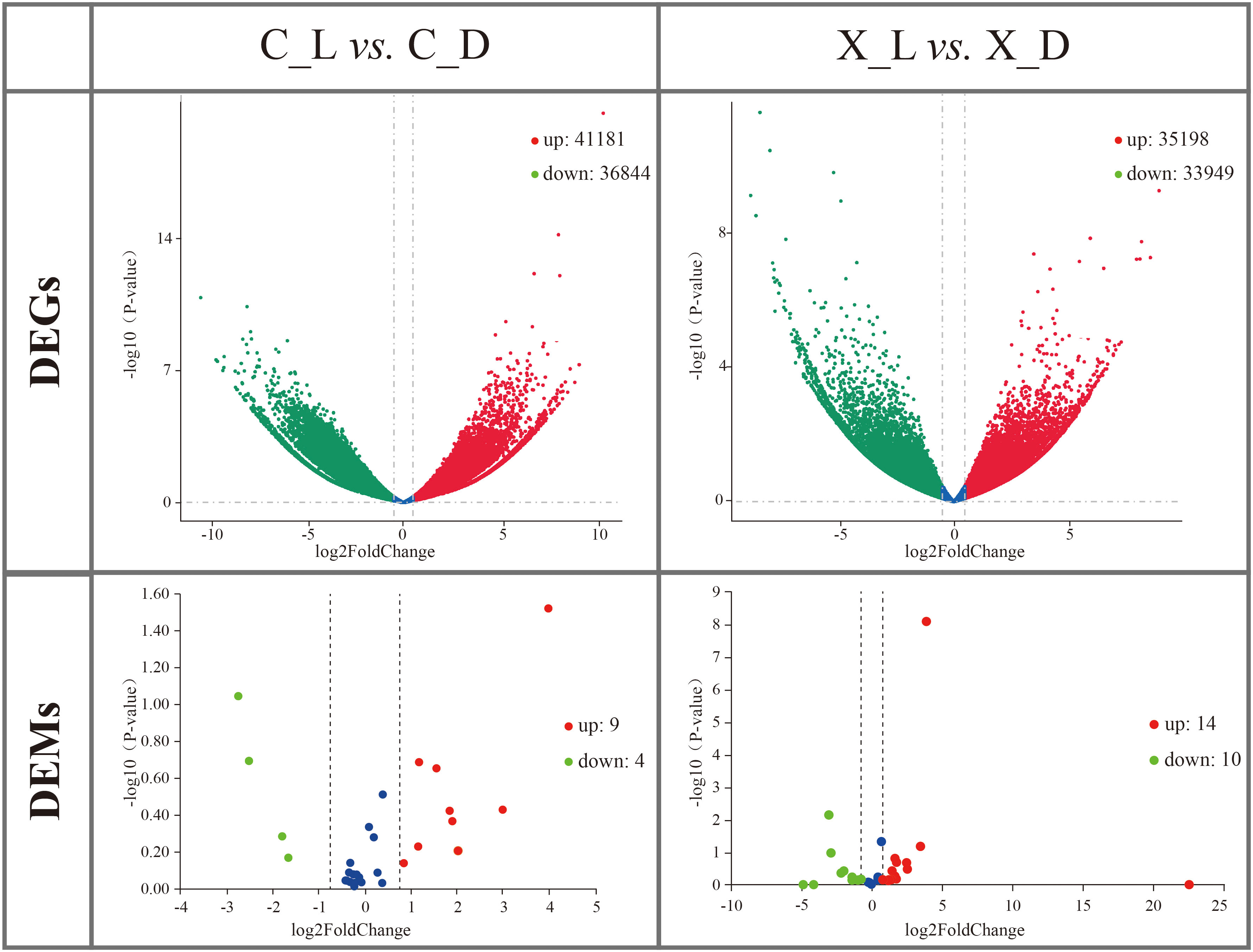
Figure 2 Volcano plots of differentially expressed genes (DEGs) and differentially expressed microRNAs (DEMs). Red spots represent upregulated DEGs or DEMs; green spots represent downregulated DEGs or DEMs.
DEMs were identified by miRNA transcriptome comparison. A total of 13 DEMs (up: 9; down: 4) in sC_L vs. sC_D and 24 DEMs (up: 14; down: 10) in sX_L vs. sX_D were identified (Figure 2).
The qRT-PCR results indicated that the expression trends of selected DEGs and DEMs were well correlated with those obtained in RNA-seq and small RNA-seq analyses, indicating the reliability and accuracy of our RNA-seq and small RNA-seq data, respectively (Figure 3).
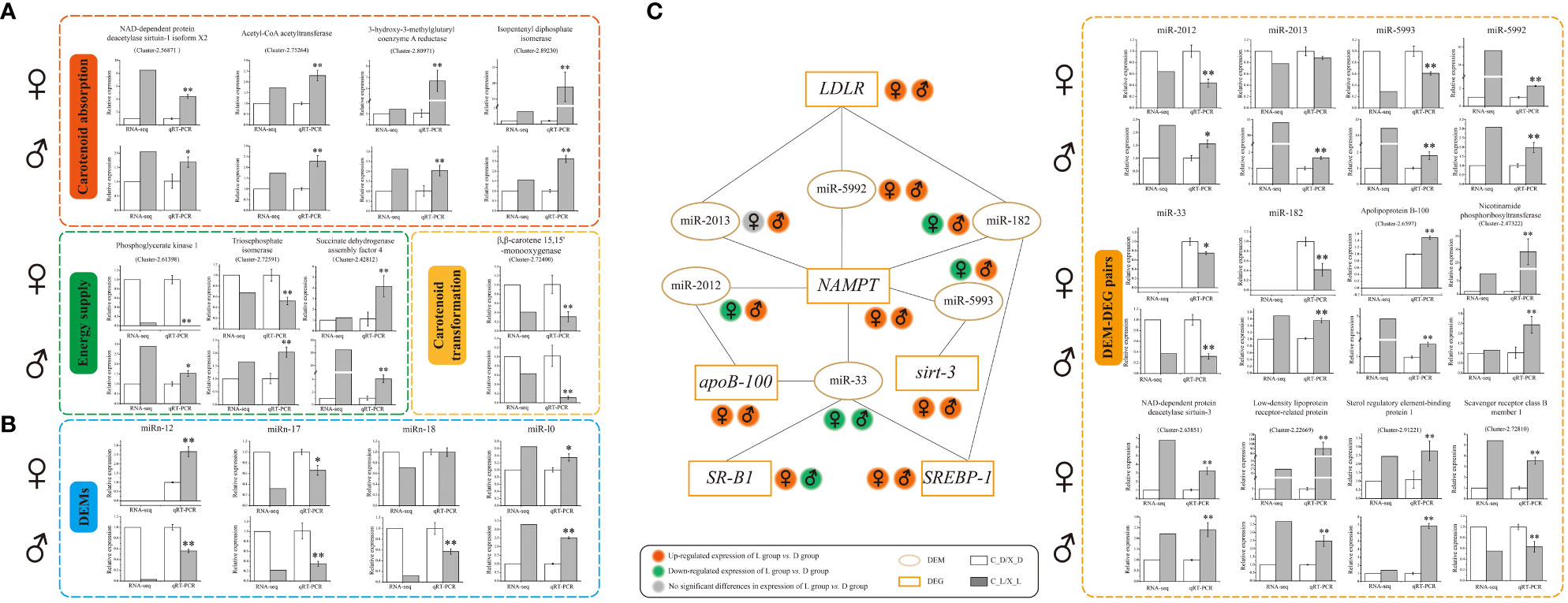
Figure 3 Verification of differentially expressed genes (DEGs), differentially expressed microRNAs (DEMs), and DEM-DEG pairs by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). (A) qRT-PCR verification of DEGs; (B) qRT-PCR verification of DEMs; (C) interaction schematic diagram and qRT-PCR verification of DEM-DEG pairs. Values are expressed as the mean ± S.D. (n = 3). *: P < 0.05 vs. control (C_D group or X_D group). **: P < 0.01 vs. control (C_D group or X_D group).
3.4 GO and KEGG enrichment analysis of DEGs and potential DEM target genes
Annotation of the identified DEGs and DEMs was performed using KEGG and GO terms. In females, there were 219 identified DEGs enriched in 170 pathways in C_L vs. C_D. KEGG analysis showed that the nicotine addiction pathway was the most enriched pathway, followed by the Glycosaminoglycan biosynthesis-CS/DS pathway and the Pantothenate and CoA biosynthesis pathway. GO term analysis revealed that the U2-type spliceosomal complex pathway and the U2-type post-mRNA release spliceosomal complex pathway were the most significant enriched pathways. About 41.38% of the predicted target genes of DEMs were enriched in the inflammatory bowel disease (IBD) pathway. A total of 541 DEM target genes were clustered in categories related to the intrinsic components of membranes in the cellular component category, and 536 DEM target genes were clustered in categories related to the integral components of membranes in the cellular component category (Figure 4).
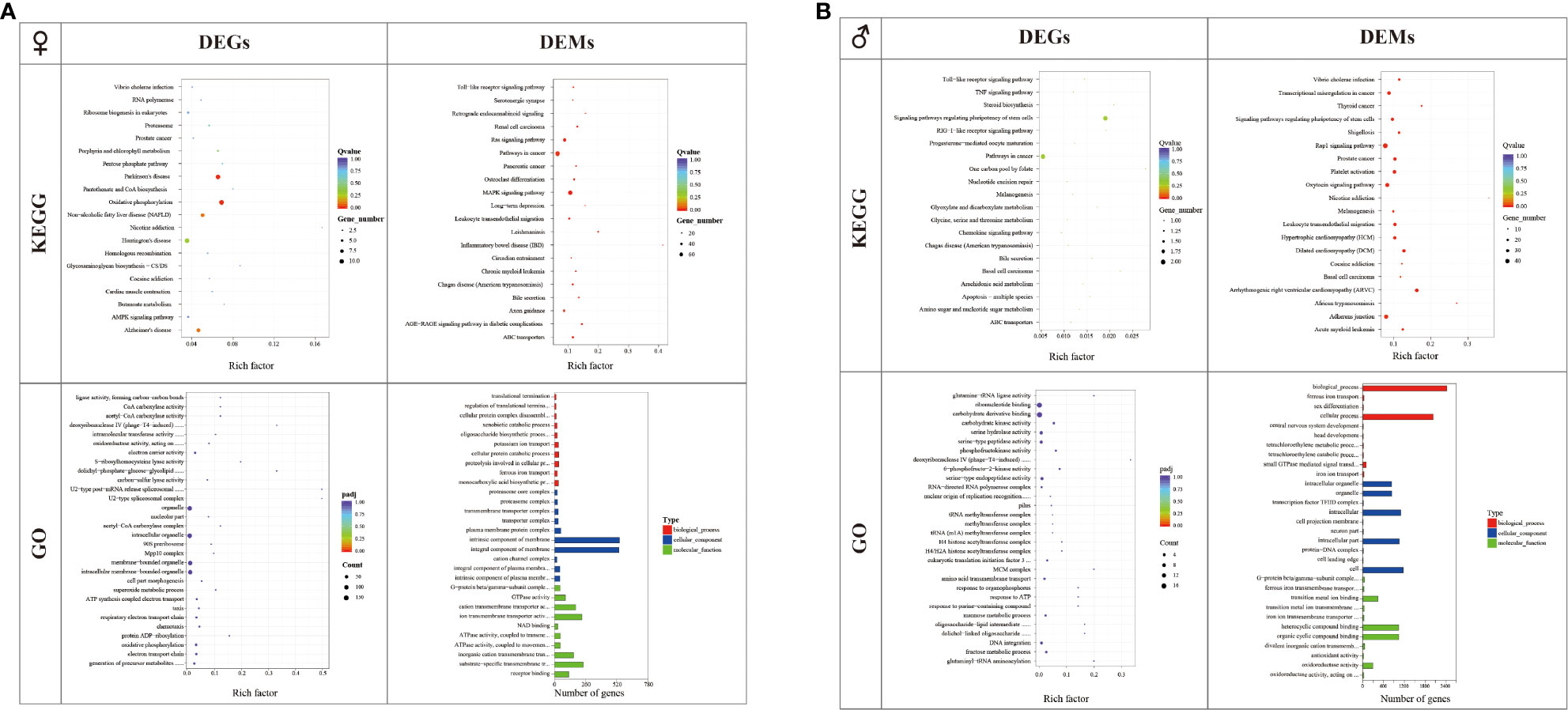
Figure 4 GO and KEGG analyses of differentially expressed genes (DEGs) and the target genes of differentially expressed microRNAs (DEMs) in the C_L group and C_D group (A) and X_L group and X_D group (B).
In males, there were 23 identified DEGs enriched in 37 pathways in X_L vs. X_D. KEGG analysis showed that the one carbon pool by the folate pathway was the most enriched pathway, followed by the basal cell carcinoma pathway and the steroid biosynthesis pathway. GO term analysis revealed that the deoxyribonuclease IV (phage-T4-induced) activity pathway was the most enriched pathway. For small RNA libraries, 35.71% of the predicted target genes of DEMs were enriched in the nicotine addiction pathway. A total of 2415 DEM target genes were clustered in categories related to biological processes, and 2025 DEM target genes were clustered in categories related to cellular processes in the biological process category (Figure 4).
3.5 Prediction of interactions between DEMs and DEGs
A total of six DEMs were identified as possibly having a targeted regulatory relationship with six DEGs (Figure 3 and Supplementary Table 3). Among these, there were 15 “DEM-DEG” pairs (Figure 3 and Supplementary Table 3).
The qRT-PCR verification data showed that the expression trends of 15 paired selected DEM-DEGs (“miR-2012 - Apolipoprotein B-100 (apoB-100),” “miR-2012 - Nicotinamide phosphoribosyltransferase (NAMPT),” “miR-2013 - Low-density lipoprotein receptor-related protein (LDLR),” “miR-2013 - NAMPT,” “miR-5993 - NAD-dependent protein deacetylase sirtuin-3 (Sirt-3),” “miR-5993 - NAMPT,” “miR-5992 - LDLR,” “miR-5992 - NAMPT,” “miR-33 - apoB-100,” “miR-33 - Sterol regulatory element-binding protein 1 (SREBP-1),” “miR-33 - Scavenger receptor class B member 1 (SR-B1 or SCARB1),” “miR-33 - NAMPT,” “miR-182 - NAMPT,” “miR-182 - LDLR,” and “miR-182 - SREBP-1”) were well correlated with the prediction of correlations in expression between DEMs and DEGs (Figure 3 and Supplementary Table 3).
4 Discussion
Economical trait improvement has always been a core pursuit in aquaculture. As one of the important quality factors affecting the marketability of sea urchin products, gonad color improvement and enhancement have been the focus of the sea urchin aquaculture industry. Carotenoids are the main pigments present in sea urchin gonads, and their concentrations directly determine the gonad color (Symonds et al., 2009). In terms of adult sea urchins, variation in carotenoid concentration between female and male individuals has been documented in Heliocidaris erythrogramma and H. tuberculatus (Tsushima, 2007). Similar to previous reports (Phillips et al., 2009), our data showed that the color of female gonads was generally superior to that of males. It has been reported that the gonadal color of female sea urchins is better than that of males due to carotenoids being deposited in eggs rather than in sperm (Borisovets et al., 2002). We thus hypothesize that there may be variations in carotenoid metabolism between female and male individuals of S. intermedius, and further in-depth research is required. In addition, we also noticed that some previous studies reported that the differential expression of genes related to the amino acid metabolism of S. intermedius in different sexes was one of the factors impacting the gonadal flavor of S. intermedius (Gou et al., 2022), and there was no clear difference in gonadal carotenoid concentration between female and male individuals of Strongylocentrotus franciscanus, Lytechinus variegatus, and Evechinus chloroticus (Suckling et al., 2020). There have been no further reports indicating gonadal color differences between females and males in these sea urchin species. Therefore, further extensive investigation is needed to determine whether there is sex-driven gonadal color variation within all sea urchins.
How gene expression and regulation affect carotenoid absorption, accumulation, and transformation and consequently lead to differences in gonad color was our core concern in this study. We therefore first focused on the differential expression of genes associated closely with carotenoid absorption, accumulation, and carotenoid metabolism.
It has been well documented that carotenoid absorption is associated with all types of lipoproteins to varying degrees in human and animal models (Von Lintig et al., 2020). Among lipoproteins, the low-density lipoproteins (LDLs), a type of lipoproteins rich in cholesterol, are considered as the main carriers involved in the absorption and transport of 60–70% of the β-carotene to different sites within the organism (Yang et al., 1994). It is acknowledged that the amounts of apoB-100 and cholesterol are two major factors determining the abundance of LDLs (Cisse et al., 2021). The apoB-100 is necessary for LDL assembly; it not only comprises the core structural component of LDLs but also provides a ligand for LDL receptors. Moreover, apoB-100 can also mediate the conversion from very low-density lipoproteins (VLDLs) to LDLs, the main pathway responsible for LDL production. In this study, the qRT-PCR validation results indicated that upregulated relative expression of apoB-100 was observed in S. intermedius with the lovely gonad color compared with those with delicate gonad color in both females and males. Therefore, combining our data with the results of previous studies, we speculate that upregulated apoB-100 expression on the one hand promotes the transformation of VLDLs into LDLs, while on the other hand, it enhances LDL assembly, subsequently leading to an increase in dietary carotenoid absorption (Figures 3 and 5). In addition to the upregulation of apoB-100, we also found several clues from cholesterol metabolism concerning the increase of LDL levels in S. intermedius with lovely gonadal color compared with those with delicate gonadal color in both females and males (Figure 3). A sufficient acetyl-CoA supply is the basal condition for cholesterol synthesis. Glucose, fatty acid, and amino acid catabolism is responsible for acetyl-CoA production. Based on our RNA-seq data analysis and qRT-PCR validation results, we noted the upregulation of both nicotinamide adenine dinucleotide (NAD+)–dependent protein deacetylase related genes (sirt-3 and sirt-1) and NAMPT in S. intermedius with lovely gonadal color compared to those with delicate gonadal color in both females and males. The sirtuins are a family of highly conserved NAD+-dependent enzymes with deacetylase activity responsible for removing the acetyl groups from acetylated lysine residues of numerous target proteins, including histones and transcription factors. Since NAD+ is the core substrate of the sirtuins, it is therefore thought that the NAD+ level within an organism determines the activities of the sirtuins. NAMPT is the acknowledged regulator of the intracellular nicotinamide adenine dinucleotide (NAD) pool, with the role of enhancing the biosynthesis of NAD and thus enhancing the activities of NAD-dependent enzymes. Combining our results and the known functions of both the sirtuins and NAMPT, we therefore speculate that the lovely gonadal color in S. intermedius may be due to the enhanced protein deacetylation that provides more acetyl-CoA to produce LDL and thus the ability to absorb more carotenoids. Of course, an increase of acetyl-CoA cannot fully demonstrate the enhancement of cholesterol anabolism. Increased expression levels of genes associated with cholesterol anabolism such as Acetyl-CoA acetyltransferase (ACAT), 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (HMGCR), and Isopentenyl diphosphate isomerase (IDI) were also observed in this study. ACAT is the first enzyme involved in terpenoid synthesis during endogenous cholesterol synthesis, as it catalyzes the formation of acetoacetyl-CoA from acetyl-CoA by transferring an acetyl group from one acetyl-CoA molecule to another. HMGCR is the rate-limiting enzyme catalyzing the step of conversion of HMG-CoA into mevalonate (MVA) during endogenous cholesterol synthesis. IDI performs the regulatory isomerization of isopentenyl diphosphate (IPP) into dimethylallyl diphosphate, a key rate-limiting step in terpenoid biosynthesis during endogenous cholesterol synthesis. Since cholesterol is also a major factor that determines the synthesis of LDL, we therefore speculate that active cholesterol synthesis enhances the production of LDL and thus the absorption of more carotenoids. Taken together, the above results suggest that enhanced absorption of carotenoids induced by elevated LDL production is a common mechanism contributing to gonadal color variation in both sexes of sea urchins (Figures 3 and 5).
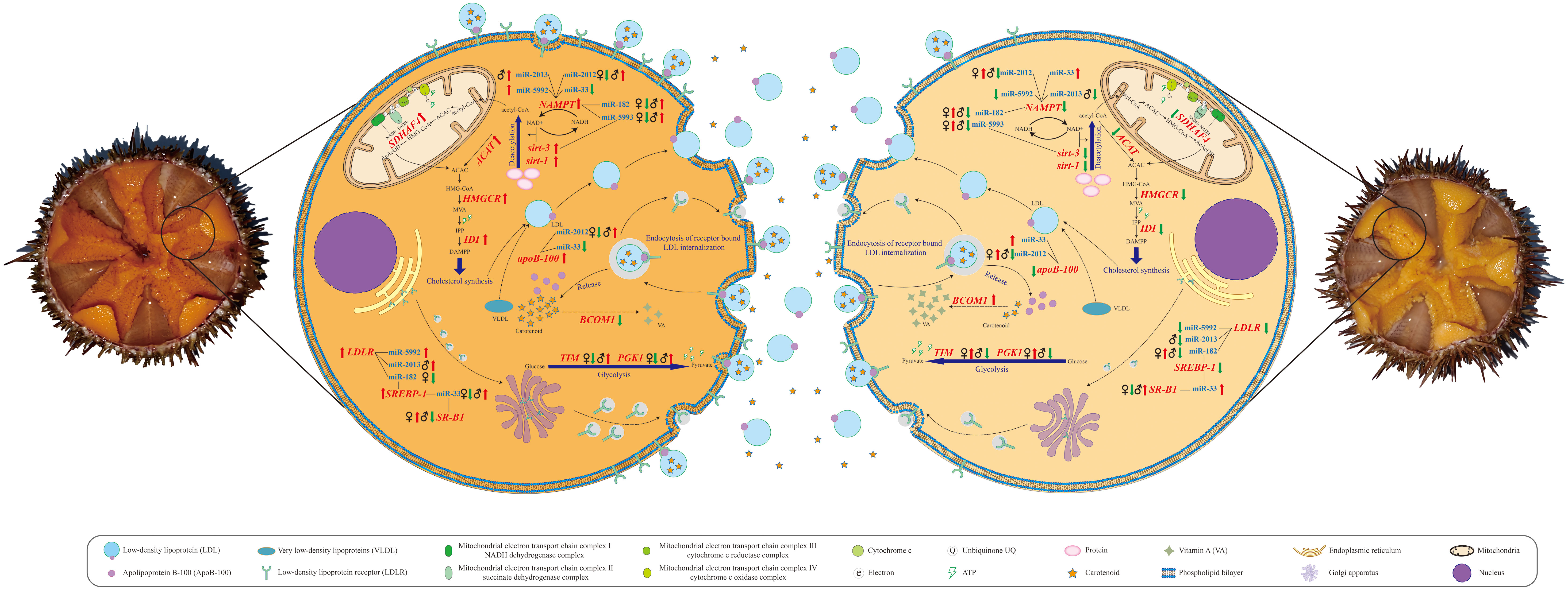
Figure 5 Schematic diagrams of metabolic pathways correlated with gonad coloration in Strongylocentrotus intermedius.
Concerning carotenoid accumulation, it is accepted that endocytosis of receptor-bound LDL is the main mechanism responsible for cellular accumulation of carotenoids (Berthelot et al., 2012). As might be expected, upregulated relative expression of LDLR was observed in sea urchins with lovely gonadal color compared to those with delicate gonadal color in both females and males. This observation suggests to some extent that more LDL-carotenoid packages might be endocytosed by the LDLR to further contribute to the gonads having a superior color in S. intermedius. Moreover, the upregulation of SREBP-1 in both females and males with lovely gonadal color also attracted our attention. SREBPs are a type of unique membrane-bound transcription factors that control lipid metabolism (Shimano, 2001). Emerging evidence indicates that SREBP-1 can enhance the expression of LDLR on the surface of cell membranes and further promote the transport of LDL from the plasma to the cells (Guo et al., 2014). Another interesting observation is the sex-specific expression alteration of SR-B1 that was upregulated in females with lovely gonadal color compared with females with delicate gonadal color and was downregulated in males with lovely gonadal color compared with males with delicate gonadal color. It has been well documented that the SR-B1 gene encodes a multi-ligand cell surface receptor for native and modified lipoproteins (including LDL, HDL, and VLDL) that mediates the absorption of carotenoids in both vertebrates and invertebrates (Toomey et al., 2017). Importantly, the relative expression level of the SR-B1 gene has been demonstrated to be highly correlated with muscle carotenoid accumulation, leading to a red muscle color in farmed Atlantic salmon (Salmo salar) and shellfish (Patinopecten yessoensis) (Ren et al., 2012). Combining the results of previous studies mentioned above with our data, we therefore suppose that the gonadal color variation in both sexes of sea urchins is determined by the expression level of LDLR that is responsible for LDL-carotenoid internalization and that SR-B1-induced enhancement of LDL-carotenoid internalization is a female-specific pathway contributing to cellular accumulation of carotenoids and resulting in gonadal color variation in S. intermedius (Figure 3 and Figure 5).
Obviously, both the absorption and the accumulation processes of carotenoids require a supply of energy (ATP). Is the supply of energy (or ATP) correlated with the gonadal color variation? and is there a sex-specific difference in energy supply associated with gonadal color variation? We observed the upregulation of Succinate dehydrogenase (SDH) assembly factor 4 (SDHAF4) in S. intermedius (in both females and males) with lovely gonadal color compared with those with delicate gonadal color. SDHAF4 serves as a chaperone for SDHA (the catalytic flavoprotein subunit of SDH)-flavin adenine dinucleotide (FAD) and promotes the assembly of SDH (Moosavi et al., 2019). It has been reported that loss of SDHAF4 function can suppress the activity of SDH and the assembly of mitochondrial respiratory complex II with a subsequent reduction of energy production in organisms (Wang et al., 2022). Based on this context, we suggest that the upregulation of SDHAF4 in the gonads of S. intermedius could provide more energy to promote the anabolism of key factors associated with absorption and the accumulation of carotenoids and therefore enable S. intermedius individuals to produce a better gonadal color. Interestingly, we also found that the relative expression level changes in Phosphoglycerate kinase 1 (PGK1) and Triosephosphate isomerase (TIM) differed in both females and males with lovely gonadal color compared with those having delicate gonadal color. Specifically, downregulation of PGK1 and TIM was observed in female S. intermedius with lovely gonadal color, whereas upregulation of PGK1 and TIM was observed in male S. intermedius with lovely gonadal color. PGK1 is the first enzyme involved in the production of ATP via glycolysis by catalyzing the reversible transfer of a phosphate group from 1, 3-bisphosphoglycerate (1, 3-BPG) to adenosine 5′-diphosphate (ADP) (Liu et al., 2022). TIM interconverts dihydroxyacetone phosphate and D-glyceraldehyde-3-phosphate, the fifth step of glycolysis, and it plays an important role in energy production (Wierenga et al., 2010). Key enzymes are conserved mechanisms in the direction and efficiency regulation of metabolic pathways. Our observations suggest that sex-specific energy production efficiency of glycolysis may be correlated with gonadal color variation in sea urchins, i.e., male individuals produce better gonadal color through upregulating PGK1 and TIM expression to elevate energy production via glycolysis (Figure 3 and Figure 5).
Transformation into retinoids (vitamin A and its derivatives) is an important part of carotene metabolism, and the conversion efficiency determines the content of carotenoids to some extent. The enzyme β,β-carotene 15,15’-monooxygenase 1 (BCMO1) is currently considered the key enzyme for retinoid metabolism, as it can oxidatively cleave β-carotene into two molecules of all-trans-retinal (Harrison and Kopec, 2020). As expected, downregulation of BCMO1 was observed in both female and male S. intermedius with lovely gonadal color compared with those with delicate gonadal color. This observation indicated that inhibition of the transformation of carotene to retinoids is a common strategy for both female and male sea urchins to obtain better gonadal coloration (Figures 3 and 5).
Since we mined and validated gene candidates correlated with gonadal coloration, we subsequently focused on finding and clarifying the molecular driving factors that led to differential expression of these gene candidates. Gene expression is regulated by multiple factors, among which the regulation of non-coding RNAs (ncRNAs) has garnered much attention in recent years (Panni et al., 2020). Extensive studies have demonstrated that miRNAs are a class of short endogenous ncRNAs that are vital regulators of multiple physiological and biochemical processes in eukaryotes (O'Brien et al., 2018). Our comparative miRNA transcriptome data showed that the number of DEMs correlated with gonadal color variation in males was greater than that in females. This observation indicates that more transcriptional regulation occurred in gonadal coloration in males, suggesting that the color of male gonads is more sensitive to changes than the color of female gonads. This could also partly explain why male gonads are generally less colored than female gonads. Among DEMs, we first focused on two known miRNAs (miR-33 and miR-182) that were predicted to have potential targeting relationships with gene candidates associated with carotenoid absorption, accumulation, and transformation (Figures 3 and 5). MiR-33 is an intronic miRNA located within SREBP genes and is a transcriptional regulator of cholesterol synthesis; it modulates the expression of genes involved in cellular lipid metabolism in mammals and marine teleosts (Aryal et al., 2017). It has been reported that miR-33 negatively regulates SREBP-1 expression in mice (Horie et al., 2013). Similarly, upregulation of SREBP-1 expression was detected in both females and males with lovely gonadal color, while downregulation of miR-33 expression was observed in both sexes of S. intermedius by qRT-PCR verification, and a negative regulatory relationship between miR-33 and SREBP-1 was predicted by integrated DEM-DEG analysis. We thus speculate that decreased miR-33 expression may contribute to the upregulation of SREBP-1 and in turn promote the expression of LDLR, leading to an enhancement of carotenoid cellular accumulation. Unlike miR-33, the relative expression changes of miR-182 varied in a sex-specific manner in this study. Specifically, the relative expression of miR-182 was decreased in female S. intermedius with lovely gonadal color compared with female S. intermedius with delicate gonadal color. It has been demonstrated that miR-182 knockdown can increase the levels of SREBP-1 in rats (Zhang et al., 2020); based on the upregulation of SREBP-1 observed in this study, we suggest that miR-182 may negatively regulate gonadal color variation through targeting SREBP-1 in female S. intermedius. However, increased relative expression of miR-182 was observed in male S. intermedius with lovely gonadal color compared with males with delicate gonadal color. Since our knowledge of the complexity of posttranscriptional regulation of small RNA is still limited, whether miR-182 regulates gonadal color variation through other genes rather than SREBP-1 in male S. intermedius and whether miR-182 positively regulates gonadal color variation in male S. intermedius through targeting SREBP-1 remain to be investigated in the future. In addition to miR-33 and miR-182, we also obtained some known miRNAs (miR-10, miR-2012, miR-2013, miR-5992, and miR-5993) and some novel miRNAs (miRn-12, miRn-17, and miRn-18) that might be correlated closely with gonad coloration in S. intermedius. Potential interactions between these candidate miRNAs and their target genes were also predicted; however, as there are too few references concerning these miRNAs, more efforts should be made to investigate and clarify the roles of these candidate miRNAs in sea urchin gonad coloration. Moreover, we also noticed that some known miRNAs might have multiple functions; for example, miR-2013 was identified as one of the miRNA candidates responsible for gonad coloration in S. intermedius, but it has also exhibited potential regulatory functions against infection and heat stress in S. intermedius coelomocytes (Sacks et al., 2018; Han et al., 2022). This observation supports the hypothesis that the regulatory functions of miRNAs are complicated, and also suggests that the function of a given miRNA might be tissue specific.
Undoubtedly, all of the above identified DEGs, DEMs, and “DEM-DEG” pairs that were closely correlated with gonad coloration in S. intermedius have strong potential to be used as molecular markers or indicators for high-quality sea urchin selective breeding or in the assessment of sea urchin gonad products in the market. Therefore, it is necessary to further verify the stability of these molecular markers in the future.
5 Conclusion
In this study, we analyzed the molecular mechanism of gonad color differentiation in farmed sea urchins from the aspect of differential expression of miRNAs and mRNAs. Common and sex-specific molecular markers closely related to sea urchin gonad coloration were identified. Common and sex-specific transcriptional regulation mechanisms associated with sea urchin gonad coloration were preliminarily characterized. The observations from this study will deepen and enrich our knowledge of the process of sea urchin gonad coloration, and the results provide further clues for increasing the gonad quality of commercial sea urchins from molecular and metabolic aspects.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA949413 https://www.ncbi.nlm.nih.gov/, PRJNA949420.
Ethics statement
The animal study was approved by Office of Ethics Committee of Dalian Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YZ and YC conceived and designed the experiments. BW, RJ, DC, and TZ performed the experiments. YZ, BW, JS, and YC analyzed the data. YZ and BW drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Liaoning Province Innovative Talent Support Program of Colleges and Universities (LR2020065) and the Liaoning Revitalization Talents Program (XLYC2002107).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1247470/full#supplementary-material
References
Aryal B., Singh A. K., Rotllan N., Price N., Fernández-Hernando C. (2017). MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 28, 273–280. doi: 10.1097/MOL.0000000000000420
Berthelot K., Estevez Y., Deffieux A., Peruch F. (2012). Isopentenyl diphosphate isomerase: A checkpoint to isoprenoid biosynthesis. Biochimie 94, 1621–1634. doi: 10.1016/j.biochi.2012.03.021
Borisovets E. E., Zadorozhny P. A., Kalinina M. V., Lepskaya N. V., Yakush E. V. (2002). Changes of major carotenoids in gonads of sea urchins (Strongylocentrotus intermedius and S. nudus) at maturation. Comp. Biochem. Phys. B 132, 779–790. doi: 10.1016/s1096-4959(02)00099-4
Chang Y. (2004). “Biological characteristics of main economic species of sea urchins in China,” in Sea cucumbers, sea urchins biology research and breeding. Ed. Chang Y. (Beijing, CN: China Ocean Press), 216–224.
Cisse A., Schachner-Nedherer A. L., Appel M., Beck C., Ollivier J., Leitinger G., et al. (2021). Dynamics of apolipoprotein B-100 in interaction with detergent probed by incoherent neutron scattering. J. Phys. Chem. Lett. 12, 12402–12410. doi: 10.1021/acs.jpclett.1c03141
Cui D., Liu L., Zhao T., Song J., Zhang W., Yin D., et al. (2022). Responses of sea urchins (Strongylocentrotus intermedius) with different sexes to CO2-induced seawater acidification: histology, physiology, and metabolomics. Mar. pollut. Bull. 178, 113606. doi: 10.1016/j.marpolbul.2022.113606
Feng C., Zhang J., Bao J., Luan D., Jiang N., Chen Q. (2023). Transcriptome analysis of germ cell changes in male Chinese mitten crabs (Eriocheir sinensis) induced by rhizocephalan parasite, Polyascus gregaria. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1144448
Gao X., Chang Y. (1999). “Main economic categories in China,” in China's economic sea urchin and its aquaculture. Eds. Gao X., Chang Y. (Beijing, CN: China Agriculture Press), 4–9.
Gou P., Wang Z., Yang J., Wang X., Qiu X. (2022). Comparative transcriptome analysis of differentially expressed genes in the testis and ovary of sea urchin (Strongylocentrotus intermedius). Fishes 7, 152. doi: 10.3390/FISHES7040152
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo D., Bell E. H., Mischel P., Chakravarti A. (2014). Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Design 20, 2619–2626. doi: 10.2174/13816128113199990486
Han L., Wu Y., Hao P., Ding B., Li Y., Wang W., et al. (2022). Corrigendum: Sea urchins in acute high temperature and low oxygen environments: The regulatory role of microRNAs in response to environmental stress. Front. Mar. Sci 9. doi: 10.3389/FMARS.2022.1027240
Harrison E. H., Kopec R. E. (2020). Enzymology of vertebrate carotenoid oxygenases. BBA-Mol. Cell Biol. L. 1865, 158653. doi: 10.1016/j.bbalip.2020.158653
Horie T., Nishino T., Baba O., Kuwabara Y., Nakao T., Nishiga M., et al. (2013). MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 4, 2883. doi: 10.1038/ncomms3883
Kelly M. S. (2005). Echinoderms: their culture and bioactive compounds. Prog. Mol. Subcell. Biol. 39, 139–165. doi: 10.1007/3-540-27683-1_7
Kelly M. S., Symonds R. (2013). “Carotenoids in sea urchins,” in Sea urchins: biology and ecology. Ed. Lawrence J. M. (London, UK: Academic Press), 171–178.
Liu L., Sun J., Zhan Y., Zhao T., Zou Y., Yan H., et al. (2020). Gonadal traits and nutrient compositions of novel sea urchin hybrids of Hemicentrotus pulcherrimus (♀) and Strongylocentrotus intermedius (♂). Aquacult. Rep. 18, 100439. doi: 10.1016/j.aqrep.2020.100439
Liu H., Wang X., Shen P., Ni Y., Han X. (2022). The basic functions of phosphoglycerate kinase 1 and its roles in cancer and other diseases. Eur. J. Pharmacol. 920, 174835. doi: 10.1016/j.ejphar.2022.174835
Mcbride S. C., Price R. J., Tom P. D., Lawrence J. M., Lawrence A. L. (2004). Comparison of gonad quality factors: color, hardness and resilience, of Strongylocentrotus franciscanus between sea urchins fed prepared feed or algal diets and sea urchins harvested from the Northern California fishery. Aquaculture 233, 405–422. doi: 10.1016/j.aquaculture.2003.10.014
Moosavi B., Berry E. A., Zhu X. L., Yang W. C., Yang G. F. (2019). The assembly of succinate dehydrogenase: a key enzyme in bioenergetics. Cell. Mol. Life Sci. 76, 4023–4042. doi: 10.1007/s00018-019-03200-7
O'Brien J., Hayder H., Zayed Y., Peng C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9. doi: 10.3389/fendo.2018.00402
Panni S., Lovering R. C., Porras P., Orchard S. (2020). Non-coding RNA regulatory networks. BBA-Gene Regul. Mech. 1863, 194417. doi: 10.1016/j.bbagrm.2019.194417
Phillips K., Bremer P., Silcock P., Hamid N., Delahunty C., Barker M., et al. (2009). Effect of gender, diet and storage time on the physical properties and sensory quality of sea urchin (Evechinus chloroticus) gonads. Aquaculture 288, 205–215. doi: 10.1016/j.aquaculture.2008.11.0260
Qiu D., Kong Y., Wang Q., Chen S., Cheng Z., Zhong W. (2005). Influence of feedstuffs on qualities of sea urchins (Strongylocentrotus intermedius). Fisheries Sci. 24, 32–34. doi: 10.16378/j.cnki.1003-1111.2005.07.010
Ren X., Hou R., Wang S., Zhan Y., Huang X., Bao Z. (2012). Identification of genes relating to carotenoids accumulation in adductor muscles of Yesso scallops (Patinopecten yessoensis). Periodical Ocean Univ. China 42, 41–47. doi: 10.16441/j.cnki.hdxb.2012.09.007
Sacks D., Baxter B., Campbell B. C. V., Carpenter J. S., Cognard C., Dippel D., et al. (2018). Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int. J. Stroke 13 (6), 612–632. doi: 10.1177/1747493018778713
Shimano H. (2001). Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 40, 439–452. doi: 10.1016/s0163-7827(01)00010-8
Suckling C., Kelly M., Symonds R. (2020). “Chapter 11 - Carotenoids in sea urchins,” in Developments in aquaculture and fisheries science. Ed. Lawrence J. M. (London, UK: Academic Press), 209–217.
Suckling C. C., Symonds R. C., Kelly M. S., Young A. J. (2011). The effect of artificial diets on gonad colour and biomass in the edible sea urchin Psammechinus miliaris. Aquaculture 318, 335–342. doi: 10.1016/j.aquaculture.2011.05.042
Symonds R. C., Kelly M. S., Suckling C. C., Young A. J. (2009). Carotenoids in the gonad and gut of the edible sea urchin Psammechinus miliaris. Aquaculture 288, 120–125. doi: 10.1016/j.aquaculture.2008.11.018
Tang L., Heqiu Y., Chi X., Zhang F., Ning Y., Ding J., et al. (2022). Effects of two phospholipids addition in plam oil-based diet on growth, gonad development and nutritional composition of sea urchins (Strongylocentrotus intermedius). Chin. J. Anim. Nutr. 34, 4674–4688. doi: 10.3969/j.issn.1006-267x.2022.07.054
Toomey M. B., Lopes R. J., Araújo P. M., Johnson J. D., Gazda M. A., Afonso S., et al. (2017). High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. P. Natl. Acad. Sci. U.S.A. 114, 5219–5224. doi: 10.1073/pnas.1700751114
Tsushima M. (2007). “Carotenoids in sea urchins,” in Edible sea urchins: biology and encology, 2nd ed. Ed. Lawrence J. M. (Boston, USA: Elsevier), 159–166.
Von Lintig J., Moon J., Lee J., Ramkumar S. (2020). Carotenoid metabolism at the intestinal barrier. BBA-Mol. Cell Biol. L. 1865, 158580. doi: 10.1016/j.bbalip.2019.158580
Wang X., Zhang X., Cao K., Zeng M., Fu X., Zheng A., et al. (2022). Cardiac disruption of SDHAF4-mediated mitochondrial complex II assembly promotes dilated cardiomyopathy. Nat. Commun. 13, 3947. doi: 10.1038/s41467-022-31548-1
Wierenga R. K., Kapetaniou E. G., Venkatesan R. (2010). Triosephosphate isomerase: a highly evolved biocatalyst. Cell. Mol. Life Sci. 67, 961–982. doi: 10.1007/s00018-010-0473-9
Yang C., Gu Z. W., Yang M., Gotto A. M. Jr (1994). Primary structure of apoB-100. Chem. Phys. Lipids 67-68, 99–104. doi: 10.1016/0009-3084(94)90128-7
Young M. D., Wakefield M. J., Smyth G. K., Oshlack A.. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhan Y., Cui D., Xing D., Zhang J., Zhang W., Li Y., et al. (2020). CO2-driven ocean acidification repressed the growth of adult sea urchin Strongylocentrotus intermedius by impairing intestine function. Mar. pollut. Bull. 153, 110944. doi: 10.1016/j.marpolbul.2020.110944
Zhan Y., Li J., Sun J., Zhang W., Li Y., Cui D., et al. (2019). The impact of chronic heat stress on the growth, survival, feeding, and differential gene expression in the sea urchin Strongylocentrotus intermedius. Front. Genet. 10. doi: 10.3389/fgene.2019.00301
Zhang H., Yang L., Wang Y., Huang W., Li Y., Chen S., et al. (2020). Oxymatrine alleviated hepatic lipid metabolism via regulating miR-182 in non-alcoholic fatty liver disease. Life Sci. 257, 118090. doi: 10.1016/j.lfs.2020.118090
Zhao R. X. (2005). Outline of main producing places of sea urchin (Echinoidea) in the world. Modern Fisheries Inf. 20, 18–21.
Keywords: Strongylocentrotus intermedius, gonad color, different sexes, mRNA transcriptome, microRNA transcriptome, microRNA-mRNA integrated analysis
Citation: Wu B, Jiao R, Cui D, Zhao T, Song J, Zhan Y and Chang Y (2023) Integrated microRNA-mRNA analysis provides new insights into gonad coloration in the sea urchin Strongylocentrotus intermedius. Front. Mar. Sci. 10:1247470. doi: 10.3389/fmars.2023.1247470
Received: 26 June 2023; Accepted: 21 August 2023;
Published: 07 September 2023.
Edited by:
Giovanna Romano, Anton Dohrn Zoological Station, ItalyReviewed by:
Sabrina Carrella, Anton Dohrn Zoological Station Naples, ItalyLei Wei, Ludong University, China
Chunde Wang, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Wu, Jiao, Cui, Zhao, Song, Zhan and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyao Zhan, enl5ZGxvdUBob3RtYWlsLmNvbQ==; Yaqing Chang, eXFrZXlsYWJAaG90bWFpbC5jb20=
 Boqiong Wu1
Boqiong Wu1 Tanjun Zhao
Tanjun Zhao Yaoyao Zhan
Yaoyao Zhan Yaqing Chang
Yaqing Chang