- 1Laboratory of Marine Biology, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
- 2Advanced Genomics Center, National Institute of Genetics, Shizuoka, Japan
- 3School and Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Yokohama, Japan
- 4Aqua-Bioresource Innovation Center, Kyushu University, Fukuoka, Japan
- 5Laboratory of Aquatic Molecular Developmental Biology, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
- 6Center for Integrative Genetics, Faculty of Biosciences, Norwegian University of Life Sciences, Oslo, Norway
Sexual development and reproduction are largely linked to epigenetic changes in many fish species. However, understanding of epigenetic regulation in scombrid species, such as tunas and mackerels, is limited. This study investigates DNA methylation’s impact on cyp19a expression, crucial for estrogen synthesis, focusing on chub mackerel. Given the commercial significance of scombrids and susceptibility of marine fish to environmental changes, elucidating epigenetic mechanisms, particularly in the context of global warming, becomes imperative. We aimed to generalize observations from chub mackerel to other scombrids. Additionally, we studied DNA methylation patterns across fish with different sexual systems to understand aromatase regulation’s phenotypic plasticity. Our in silico analysis revealed highly conserved promoter sequences within scombrids, sharing TFBS like Foxl2, FOS::JUN, ESRR, and Sox3, while CpG content varies. This indicates a conserved regulatory network controlling gene expression. We found sexual dimorphism in DNA methylation, with males hypermethylated and aromatase expression downregulated. Despite similar dnmt1 expression, tet1, tet2, and tet3 were higher expressed in females, suggesting that the observed DNA methylation patterns are maintained through active demethylation rather than differential methylation. Gonochoristic Japanese anchovy and protogynous bamboo leaf wrasse displayed similar trends, but species-specific methylation patterns highlight DNA methylation’s complex role in gonadal changes. In vitro assays confirmed methylation’s regulatory role and identified an SF-1 binding site relevant for promoter activation in chub mackerel. Another studied SF-1 site, present in both chub mackerel and bamboo leaf wrasse, showed regulatory effects, indicating potential similar regulatory mechanisms for cyp19a expression. Overall, our findings suggest that while global methylation affects cyp19a transcription, the variation in CpG density and location could be introducing nuances in its epigenetic regulation. This study contributes to our understanding of the cyp19a regulation in fish gonad maturation.
1 Introduction
Environmental changes, including disturbances to fish breeding grounds, pose a threat to successful breeding, embryo/larval survival, and sexual differentiation, impacting viability in aquaculture and natural environments (Barange and Perry, 2009). Epigenetic modifications at the genome level, triggered by global changes, drive acclimatization and adaptation across generations (Ryu et al., 2018). Specifically, DNA methylation plays a pivotal role in fine-tuning gene expression during reproductive maturation. Reproduction is finely controlled by the hypothalamic-pituitary-gonadal (HPG) axis and subsequent modulation of sex steroid production. Some studies have shown that epigenetic marks are disrupted by environmental factors, triggering changes in fish phenotypes. For example, in the article by Anastasiadi et al., 2017 larvae of European sea bass exposed to an increase in temperature experienced changes in global methylation and subsequent expression of genes linked to stress response, growth and differentiation.
Among the genes regulated by DNA methylation, Cytochrome P450 Family 19 Subfamily A (Cyp19a), stands out for its essential role in estrogen synthesis across vertebrates. Highly expressed in ovarian tissue, Cyp19a catalyzes the conversion of androgens into 17β-Estradiol (E2) in granulosa cells, which then acts over the liver to produce vitellogenin (Kime, 1993) during the reproductive season. Cyp19a is modulated by a plethora of transcription factors (TF), including the Forkhead box L2 (Foxl2), which is essential for sex differentiation and maintenance (Huang et al., 2017), and can activate aromatase transcription by binding to its promoter. Additionally, in the presence of steroidogenic factor 1 (SF-1), Foxl2 synergistically upregulates Cyp19a expression (Wang et al., 2007). Other TF have been found to influence aromatase expression as well. For instance, the estrogen-related receptor (ESRR) gamma stimulates aromatase synthesis in human placental cells and, when knockdown, drastically inhibits its expression (Kumar and Mendelson, 2011), while FOS::JUN (also known as AP-1) motif is implicated in aromatase expression up-regulation induced by leptin in MCF-7 cell line (Catalano et al., 2003).
The epigenetic regulation of cyp19a, has been extensively investigated in commonly consumed fish species such as the Chinese sea perch (Chen et al., 2018), the Japanese flounder (Wen et al., 2014) and the European sea bass (Navarro-Martín et al., 2011). However, little is known about the epigenetic regulation of cyp19a in the Scombridae family, which encompasses the most commercially valuable and widely consumed fish, including tunas and mackerels (Supplementary Figure S1). There is a significant gap in our knowledge regarding the epigenetic mechanisms affecting these important food fishes. It is said that pelagic fish are more susceptible to environment changes (Petrik et al., 2020). Given that cyp19a modulation has proven to be influenced in early developmental stages by environmental factors, such as temperature, inducing masculinization in fish (Piferrer and Blázquez, 2005), studying cyp19a modulation in the context of global warming is imperative. Furthermore, understanding how DNA methylation influences cyp19a expression in species with varied sexual systems, such as gonochorists and sequential hermaphrodites, could provide crucial information about the flexibility and adaptability of reproductive strategies in different ecological contexts.
In our study, we aim to investigate the conservation level of the cyp19a promoter sequence within the Scombridae family, shedding light on its evolutionary significance and the regulatory mechanisms that operate across related species. Using chub mackerel (Scomber japonicus) as a model, we aimed to unravel the modulation of cyp19a expression by DNA methylation and identify potential epigenetically influenced regulatory elements within this region. To achieve this, we conducted in silico comparative analysis, assessed methylation levels in the ovaries and testes of mature chub mackerel and performed in vitro assays to further study potential transcription factors within the promoter. Additionally, we compared its DNA methylation patterns with those of Engraulis japonicus and Pseudolabrus sieboldi to gain insights into the similarities and differences among species with different sexual systems. This study revealed the evolutionary and ecological drivers behind reproductive diversity, enhancing our knowledge of the complexity of epigenetic regulation in marine fish species.
2 Materials and methods
2.1 In silico comparative study of scombrids’ cyp19a promoter
Sequences from scombrids, including southern bluefin tuna (Thunnus maccoyii), yellowtail tuna (T. albacares), and chub mackerel, along with the non-scombrid teleosts Japanese medaka (Oryzias latipes), Nile tilapia (Oreochromis niloticus) and Japanese flounder (Paralichthys olivaceus), were obtained from the NCBI database (Supplementary Table S1). Two additional sequences from our laboratory, chub mackerel (here after referred as Q-saba strain) and the bamboo leaf wrasse (P. sieboldi) were also aligned. As a reference, we constructed a phylogenetic tree of the Scombridae family using the online tool TimeTree 5 (Kumar et al., 2022), which is included in the Supplementary Materials. A sequence spanning approximately 1500bp upstream of the start codon and covering both the first exon and intron, was selected for the analysis. The alignment was conducted with Clustal Omega software (Madeira et al., 2022) and transcription factor binding sites (TFBSs) search was done using JASPAR (Castro-Mondragon et al., 2022) and AnimalTFDB (Shen W et al., 2023) databases. The search was conducted using default parameters, and a minimum relative score of 85% was applied. A bibliographic search was conducted to pre-select and identify relevant TFBSs in the database.
2.2 Fish rearing and handling
50 Q-saba brooders were reared in a 5-ton semi-circular FRP tank, with continuous water exchange. Fish were maintained at 18~20°C with a constant photoperiod of 16:8 (light: dark) hours for a period of 3 months. Food was fed twice a day at 2% of fish body weight using commercial pelleted food. After checking the maturity status, fish were injected with human chorionic gonadotrophin (400IU/kg fish) and artificial fertilization was conducted. 3 days after hatching (dah), fish were fed with live feed, followed by artificial food from 21dah. Later, at around, 50dah (Fork length ~8cm), fish were transferred to 10mX10m in open sea cages in Ao-suisan’s fish farm, at Karatsu bay, Japan. Fish were fed with commercial pelleted feed (Marubeni Nissin, Japan) and regularly examined for reproductive status. 11-month-old fish were euthanized and sampled for further analysis. All experiments were conducted in accordance with the Kyushu University Animal Husbandry Guidelines, and all protocols were approved by the competent committee.
2.3 DNA isolation and bisulfite conversion treatment
gDNA and total RNA were isolated from gonads of 11-month-old mature fish (4 females and 4 males), which were stored in DNA/RNA shield (Zymo Research, USA), using the Quick-DNA/RNA Magbead kit (Zymo Research, USA) and following manufacturer’s instructions. Then, around 200ng of gDNA was subjected to bisulfite conversion treatment employing EZ-96 DNA Methylation-Gold MagPrep (Zymo Research, USA). Bisulfite-converted DNA was stored at -20°C until use.
2.4 Cyp19a regulatory regions PCR and DNA methylation pattern scoping
Bisulfite sequencing (BS-seq) was conducted to study the DNA methylation patterns of cyp19a promoter in chub mackerel. Primers were designed targeting the regulatory region surrounding cyp19a start codon, with amplicons with average 250bp in length (Supplementary Table S2). Primer design criteria mainly focused on including all in silico detected CpGs in the analyzed region. Bisulfite-converted DNA was amplified with KOD -Multi & Epi enzyme (Toyobo, Japan). Reaction conditions were as follows: pre-denaturation at 94°C for 2 min, followed by 45 cycles of 98°C for 10 s, 59°C for 30 s and extension at 68°C for 15 s. PCR products were then purified with the NucleoSpin Gel and PCR Clean-up kit (Takara, Japan). Since the fragments obtained after PCR were blunt-ended, an A-tailing step was added, using dA-overhang reaction Mix (Nippongene, Japan). The fragments were then ligated into a pGEM-T Easy Vector (Promega, USA) following manufacturer’s instructions. After incubating the ligation reactions overnight at -4°C, bacteria were transformed, insert-positive colonies cultured and plasmid then purified, with the FastGene Plasmid Mini Kit (NIPPON Genetics, Japan) and sent to sequence, using Sanger platform. Relative methylation levels per position were calculated as the percentage of unmethylated cytosines over the total coverage for that position.
2.5 Expression measurement of cyp19a and genes involved in maintenance of DNA methylation
To study gene expression of cyp19a, dnmt1, tet1, tet2 and tet3 in ovaries and testes, RT-qPCR was conducted. Used primers can be found in Supplementary Table S2. Around 1 µg of total RNA was reverse transcribed into cDNA with Superscript IV (Invitrogen, USA). RT-qPCR was carried out using SSoFast EvaGreen Supermix (Bio-Rad, USA) in a qTOWER3 G touch machine (Analytik jena, Germany). Samples were run in duplicate and a final melting curve step was added to confirm single amplicon amplification. The housekeeping rpl8 gene was also measured for expression normalization.
2.6 Inter-species DNA methylation analysis
DNA methylation patterns were also compared across species with different sexual systems. Data on cyp19a promoter methylation in the gonochoristic fish Japanese anchovy (E. japonicus) and the protogynous fish bamboo leaf wrasse was collected. Using a similar method to the one described above, bisulfite-converted gonad DNA was PCR amplified and then sequenced either by Sanger sequencing or Amplicon sequencing. A comparative analysis was conducted by aligning the target sequences, identifying common TFBSs, and comparing both global and CpG position-wise methylation levels.
2.7 Cyp19a vector construction
Specific primers were designed to amplify the proximal promoter region of cyp19a, including the sequence previously studied for DNA methylation patterns (Supplementary Table S2) and with an overhang sequence overlapping pGL3-TK vector. PCR was carried out with PrimeSTAR Max DNA Polymerase (Takara, Japan), under the following conditions: 45 cycles of 98°C for 10 s, 56°C for 15 s and extension at 72°C for 15 s. The purified amplicon was then inserted into the Xhol and Bgl II restriction enzyme cutting sites of the pGL3-TK plasmid, using the In-Fusion® Snap Assembly kit (Takara, Japan).
To study whereas methylation of cyp19a promoter was actually affecting its activity, whole methylation of the constructed plasmid was conducted with CpG Methyltransferase (M.SssI) enzyme (NEB, USA), as specified by manufacturer’s instructions. Verification of successful methylation was achieved through digestion with the SmaI enzyme and subsequent gel electrophoresis.
To further investigate regulatory elements and their potential involvement in cyp19a promoter activation, some of the previously found TFBSs during the in silico analysis were selected for deletion. The process included inverse PCR of the cyp19a/pGL3-TK plasmid using PrimeSTAR Max DNA Polymerase (Takara, Japan) and primers designed to be oriented in opposite directions (Supplementary Table S2), overlapping the target sequences to be deleted. Three 11bp deletions were introduced in positions -821/-810 (ATGGCCTTCAG), -707/-696 (TTGTCCTTGTG), and -217/-207 (TCAAGGTCGC). The nucleotide positions relative to the ATG start codon site were considered as +1.
2.8 Cell culture and dual luciferase reporter assay
Chinese hamster ovary (CHO) cells were cultured in Ham’s F-12 (Wako, Japan) and supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. The cells were seeded in 96 well/plate at an amount 10 000 cells per well and co-transfected the next day with 100ng of a mix of reporter plasmids: cyp19a/pGL3-TK (either methylated, control or promoter with deletion) and the inner control pRL-SV40 (Promega, USA), in triplicates. Cells were harvested 24 hours after transfection, and the Dual-Luciferase Reporter (DLR) assay was performed according to the manual (Promega, USA). Measurement was carried out using the luminometer function of the Thermo Scientific Varioskan® Flash.
All statistical analysis were done with RStudio (version 2023.06.1 + 524). The data didn’t follow a normal distribution, so comparisons were made with no parametric Kruskal test and a posteriori contrast with Dunn’s test. UpSet plots (Lex et al., 2014) for visualization of TFBSs per fish group were created with ComplexUpset package (Krassowski et al., 2022).
3 Results
3.1 Surrounding region of cyp19a gene is highly conserved within Scombridae group
To investigate how conserved cyp19a regulatory region was, and identify common regulatory elements and CpG sites, an in silico study was conducted (Figure 1). Chub mackerel sequences (Wild and Q-saba strain) were 95.8% identical, which interestingly is slightly lower than that observed for the two species of tunas (98.6%), even though the former correspond to the same species. When comparing chub mackerel sequences with those of other species, the similarity percentage varies, ranging from 44.7% in Japanese medaka to 55% in Japanese flounder (Supplementary Figure S2). The TATA box and ATG were consistently aligned in all examined sequences. In most instances, a CpG site is detected within an 11bp proximity to the TATA box. However, in tuna species, the closest CpG is approximately 50bp away. Similarly, for all species except tunas, a CpG site is typically located around 25–29bp downstream from the ATG codon.

Figure 1 Comparative analysis of cyp19a promoter sequences across several scombrids and non-scombrids. Putative TFBSs as well as TATA box and start codon are highlighted in some way (e.g. colored, highlighted, underlined, etc.).
Within the nucleotide discrepancies found, three potential TFBSs are present in the Q-saba strain mackerel: Sox3 (-919/-910, TCTTTCTTTT), also present in the tunas’ sequence, Dmrt1 (240/248, TACCAAGTA) and AR (-34/-13, TTCAGAACAACCTGTACACACC) (Figure 2, indicated in red). Conversely, two novel Sox3 were absent in the Q-saba strain mackerel, TTTTTGTCCC (-1033/-1024) and ATACAATGAAA (-1165/-1155) (Figure 2, indicated in yellow). We found that the studied sequence is highly conserved within the Scombridae family, with a similarity of 72.8% in average. Both tuna species, southern bluefin and yellowtail tuna, share all 24 TFBSs discovered, with 12 of them also common to chub mackerel. These TFBSs include SF-1, Nr5A2, Sox3, FOS::JUN, Foxl2, and ESRR. TFBSs conservation varies across species. One Foxl2 site, consistently identified around position -170, is present across all examined species. Additionally, two regions harboring multiple putative TFBSs (ESRR, SF-1, and Nr5A2), located relatively close upstream of this Foxl2 site, are commonly observed in most sequences. Moreover, unique TFBSs were discovered for each fish species. Notably, the Bamboo leaf wrasse distinguishes itself with 20 out of 25 (80%) exclusive TFBSs, predominantly featuring FOS::JUN, Foxl2, and Sox3. Furthermore, this species boasts the highest count of cAMP-response element binding protein (CREB), Foxl2, and Dmrt1 sites identified (Figure 2).
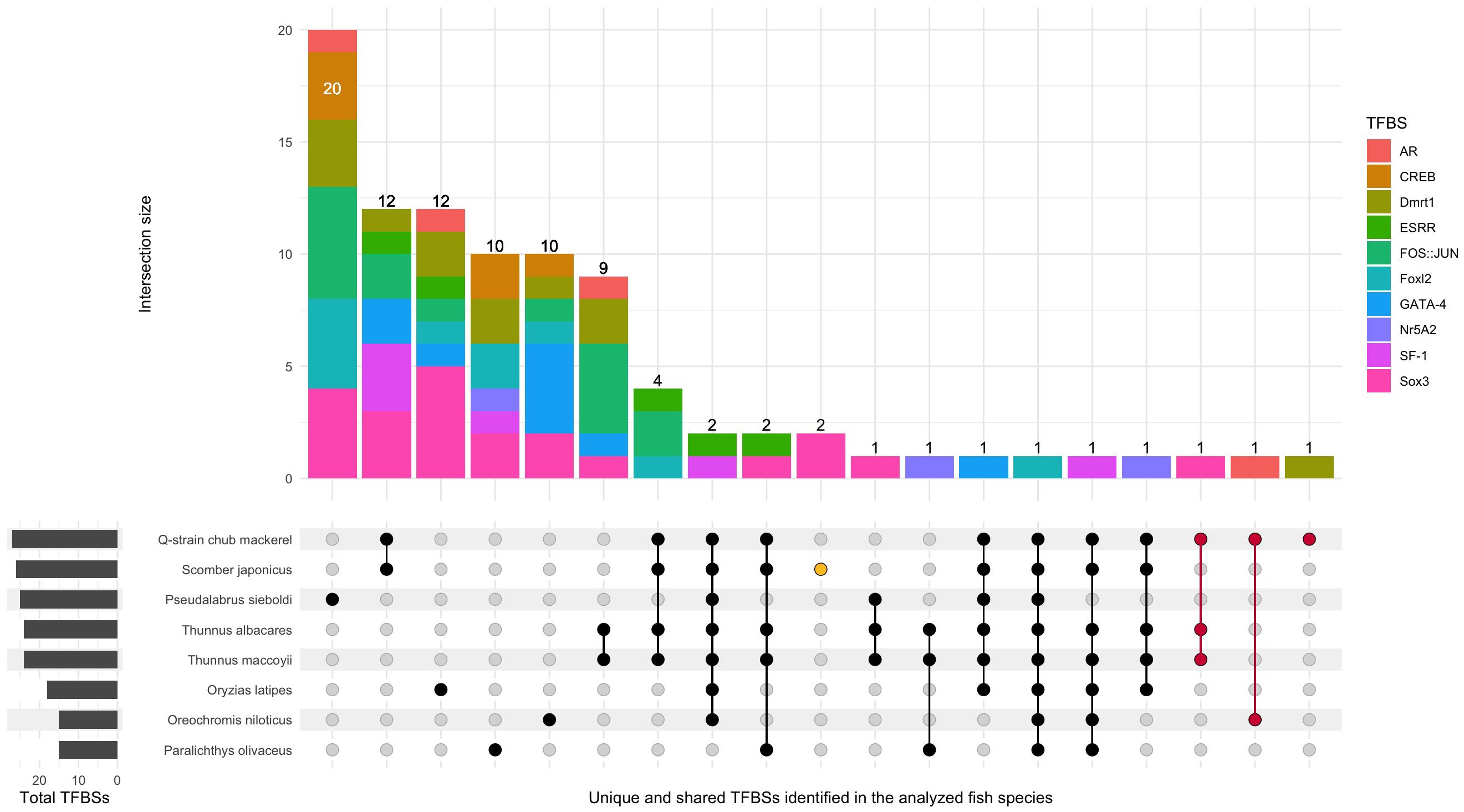
Figure 2 UpSet plot illustrating unique and shared TFBSs among the analyzed fish species. The stacked bar plot indicates the frequency of each interaction, with each color representing a different TFBS. The gray bar plot on the left shows the total number of TFBSs found for each fish species. Black dots connected by lines indicate TFBSs that overlap in different species, while unconnected dots represent TFBSs unique to a particular species. Interactions highlighted in red denote TFBSs that are novel to the Q-saba strain, while those in yellow indicate TFBSs unique to wild chub mackerel.
Exploring CpG content reveals CpG islands exclusively in Japanese flounder (within the gene body, 108bp long) and Nile tilapia (-1321/-1209 upstream of the start codon, 113bp long), with no overlap. CpG site content is highest in flounder and tilapia (34 and 28 sites, respectively), followed by chub mackerel with 20 sites. In contrast, southern bluefin tuna and yellowtail tuna exhibit the lowest CpG site counts (11 and 12 sites, respectively). Notably, sequences with the highest CpG content, namely Japanese flounder and Nile tilapia, also display the lowest amount of TFBSs (15 TFBSs in both species). Interestingly, additional CpG sites found in chub mackerel but absent in tunas are predominantly within the gene body (Figure 1). Despite high sequence conservation between tunas and chub mackerel, their CpG sites differ significantly in distribution and abundance.
3.2 Sexual dimorphism in cyp19a DNA methylation patterns and gene expression
Next, to explore the cyp19a promoter further and gain insights into the molecular dynamics within a member of the Scombridae family, we conducted BS-seq to examine DNA methylation patterns in chub mackerel. To do so, we first analyzed gonadal DNA methylation levels of cyp19a promoter and first exon. The Kruskal-Wallis test revealed significant differences in DNA methylation patterns between females and males (p-value < 0.001). Consistency in CpG methylation levels was observed among individuals of the same sex. Specifically, females exhibited an average DNA methylation of 30.92% ± 3.42%, while males showed 96.19% ± 1.12% (expressed as mean ± S.E.M).
Furthermore, post-hoc comparisons using Dunn’s test revealed sex-related differences for almost all CpGs included in this study, with DNA methylation levels significantly higher in males (Figure 3). Notably, the most significant differences in methylation were observed in the distal part of the promoter, whereas differences within the 5’-UTR and the first exon were less pronounced. CpG sites downstream the start codon exhibit an increased in methylation levels in females (58~60%), compared to the sites located further upstream (9~21%). It’s noteworthy that the majority of these CpGs are exclusive to S. japonicus and not found in tunas.

Figure 3 Relative DNA methylation levels in the promoter region of gonad cyp19a for males (blue) and females (pink). Average methylation was computed for each CpG position. Error bars indicate standard error and * denotates significant sexual differences for each CpG position (adjusted p value <0.05). N=8 fish.
When analyzing the link between DNA methylation levels and cyp19a expression, a statistically significant negative correlation of -0.52 was identified (p-value < 0.001). This finding suggests that fish with higher gonad DNA methylation levels tend to exhibit lower expression of aromatase. Additionally, the strength of this correlation varies across different CpG positions analyzed, ranging from a robust -0.70 correlation at CpG position 25 to a milder -0.27 correlation at CpG position 107 (Figure 4).
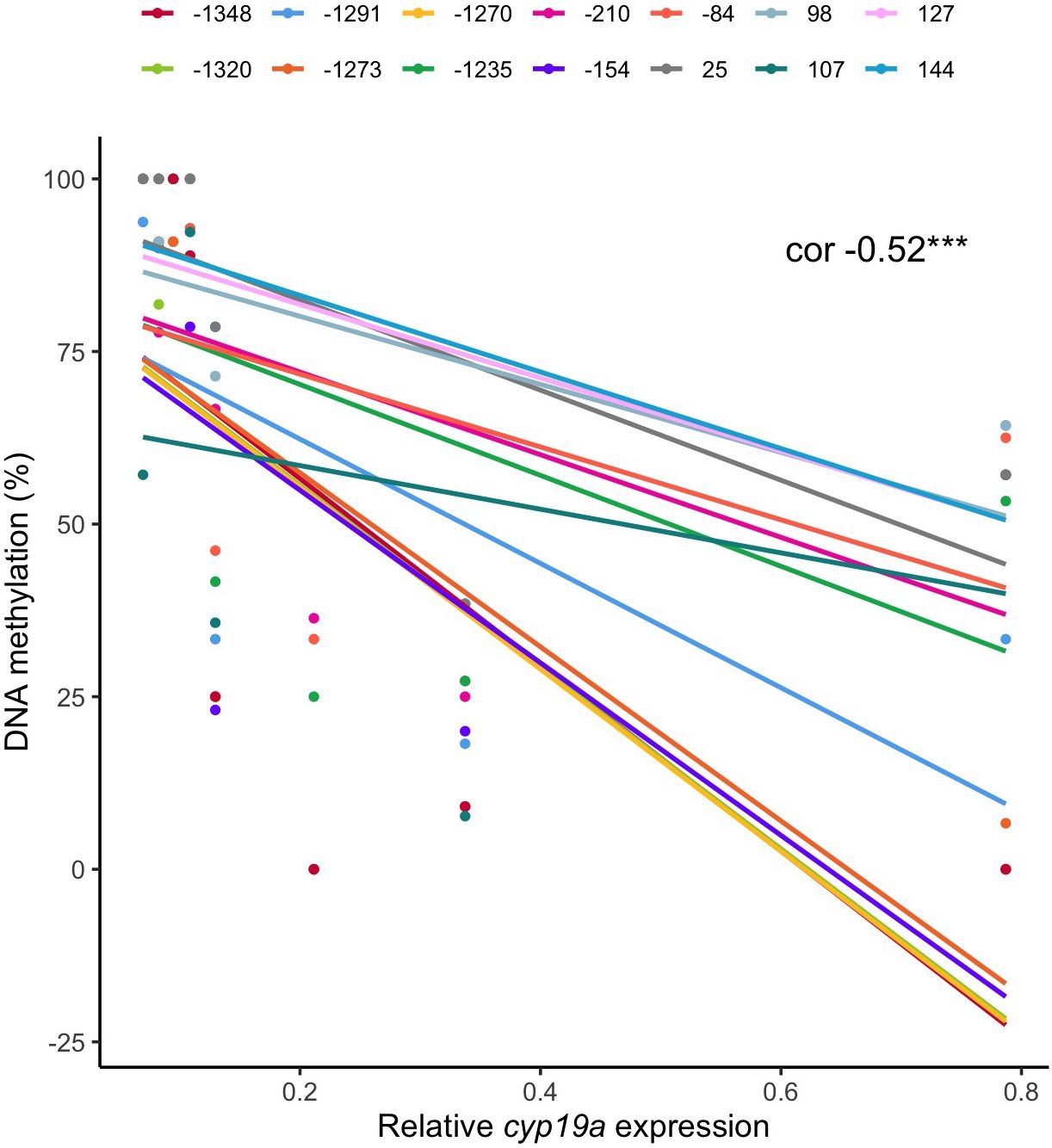
Figure 4 Correlation between cyp19a expression and DNA methylation in gonad, for each CpG studied (represented in different colors). The global correlation coefficient is -0.52 and this inversely proportional relationship prevails for most CpG positions.
We also measured gene expression of enzymes responsible for maintaining DNA methylation dynamics. While no significant differences were observed in dnmt1 expression, all three Tet enzymes exhibited significantly higher expression levels in mature ovaries compared to testes. Notably, tet1, tet2, and tet3 were expressed approximately 5, 12, and 9 times more in females, respectively (Figure 5). This gender-specific variation in the expression of these enzymes aligns with the overall lower methylation levels observed in ovaries.
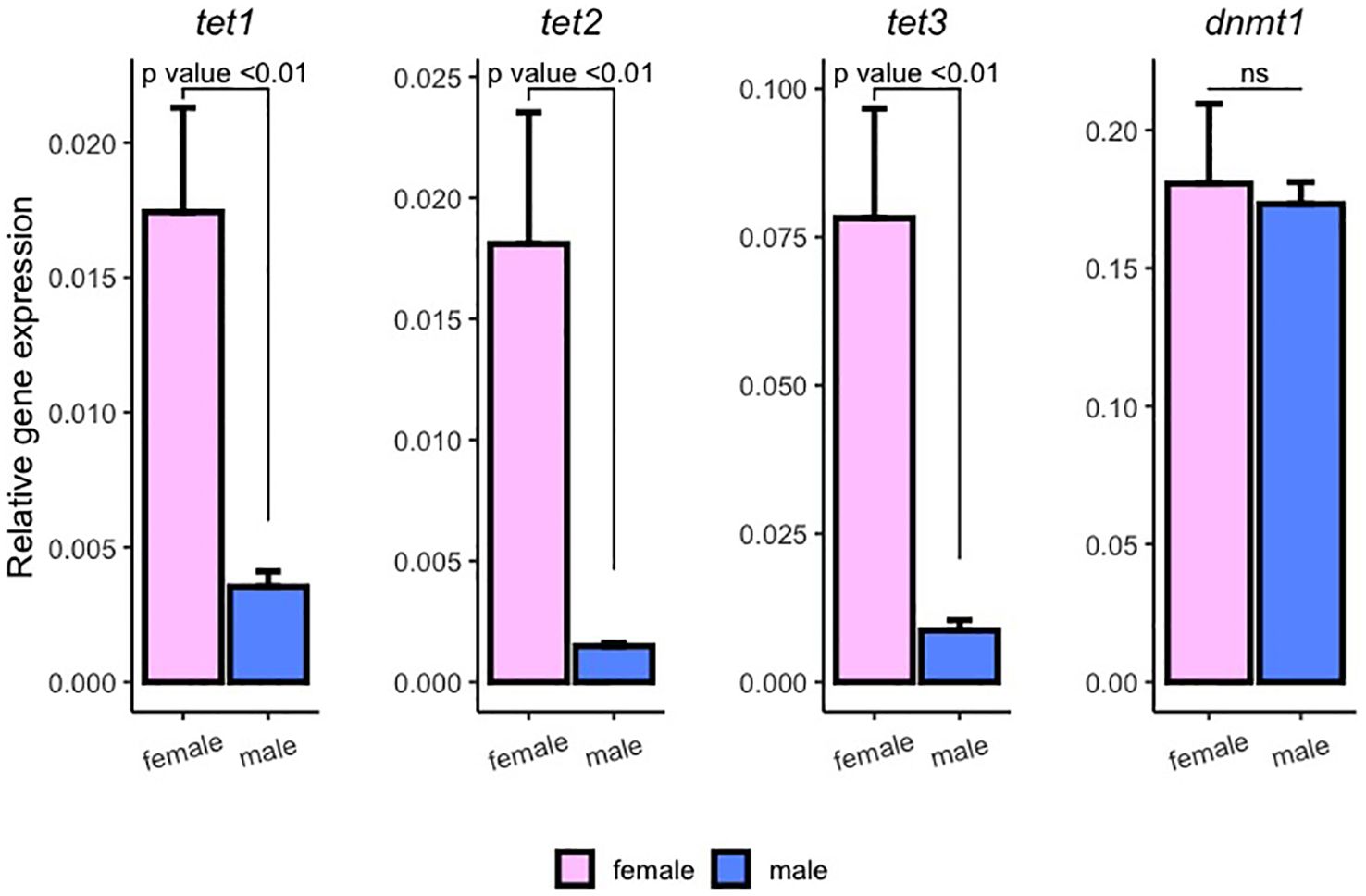
Figure 5 Mean relative expression of tet1, tet2, tet3 and dnmt1 in ovaries (pink) and testes (blue). Error bars indicate standard error and significant differences are denoted by their p-values (Dun’s test). N=15~16 fish.
3.3 The sex-biased methylation of cyp19a promoter is well conserved, but methylation patterns are species-specific
We compared the DNA methylation patters found in chub mackerel with two other species. The target regions, even though variable in length, were all established around the cyp19a start codon (Supplementary Data). We found that Japanese anchovy also shares a putative Foxl2 binding site with the other fish previously analyzed, located at position -226/-218. This site is situated relatively close to a well-conserved SF-1 site (position -247/-236), with a CpG site in-between. The SF-1 site is also present in chub mackerel and wrasse and overlaps with an ESRR site in these two species (Supplementary Figure S3). When compared, the methylation levels at that CpG site followed the expected trend, with females displaying lower methylation levels (38% and 46%) than males (94% in both cases) for Japanese anchovy and chub mackerel, respectively. In the case of bamboo leaf wrasse, while the trend persists, the difference is much smaller (92% for females and 100% for males). Across all three species, we noted higher cyp19a promoter methylation levels in testes compared to ovaries, consistent with the downregulation of aromatase expression (Figure 6). However, these differences in methylation were notably smaller in the hermaphrodite fish. While the males of the two gonochoristic fish were between 38 to 69% more methylated than females, this contrast was only 10% in the bamboo leaf wrasse. Furthermore, this pattern completely inverts at the CpG site nearest to the start codon, with hypermethylation observed in females (85%) rather than males (29%). We found several putative TFBSs overlapping that CpG, including CREBL1, CREB3L1, HES2, HES5 and Hey2, which make that region an interesting focus for future research.
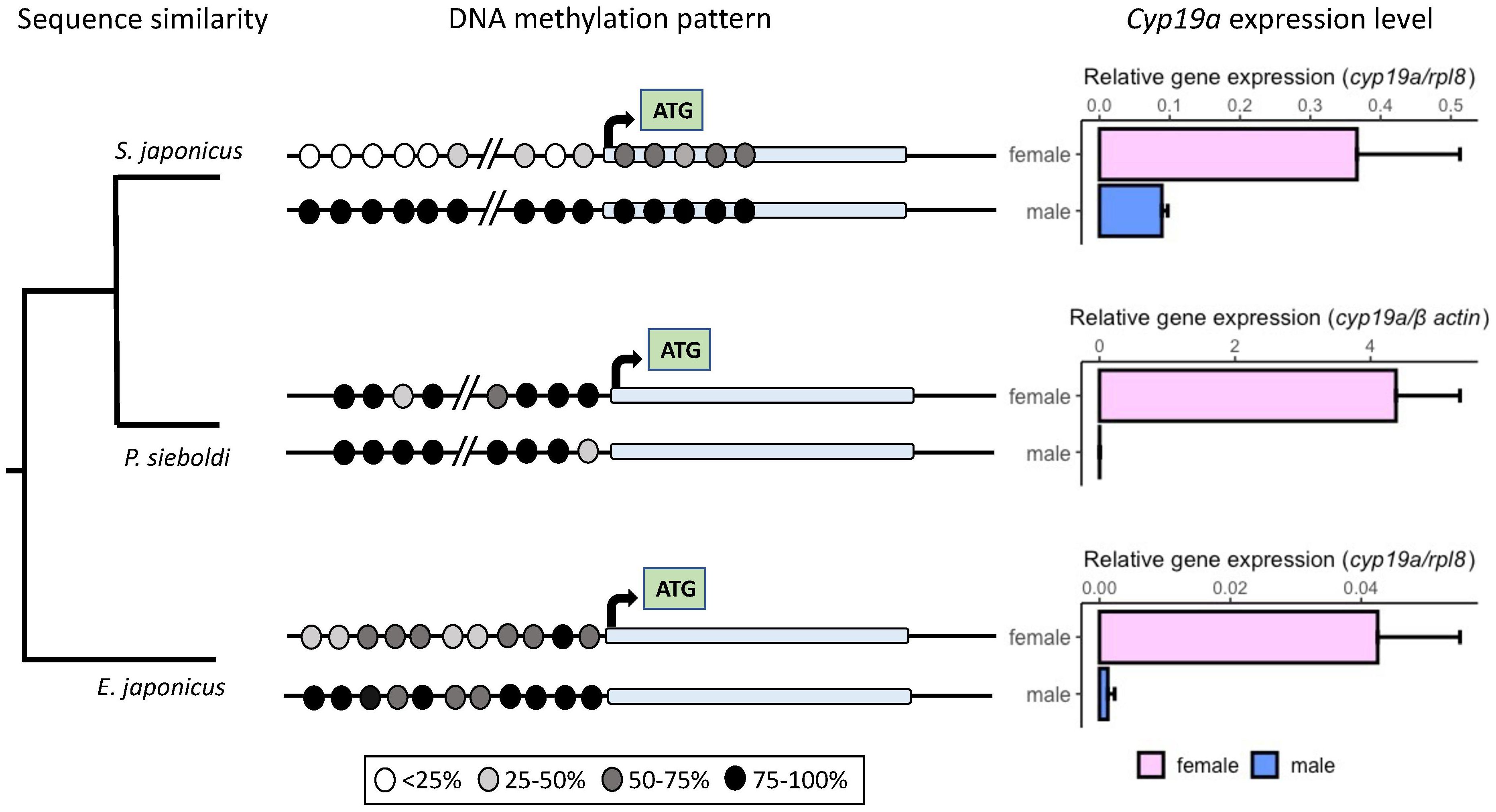
Figure 6 Comparative analysis of the methylation patterns within the cyp19a promoter region in two gonochoristic fish (S. japonicus and E. japonicus) and a protogynous hermaphrodite fish (P. sieboldin). Methylation range percentages are depicted in grayscale. On the right, cyp19a expression is presented with standard error values in ovaries (pink) and testes (blue) for all three species.
3.4 Cyp19a promoter activation in vitro
Finally, to investigate the impact of cyp19a promoter methylation on gene expression in chub mackerel, we conducted an in vitro experiment involving the artificial methylation of the studied sequence. A significant reduction in the activation of the methylated promoter was observed (adjusted p-value < 0.001). These findings align with our previous observations of decreased cyp19a expression under conditions of increased methylation, further strengthening the link between DNA methylation and cyp19a downregulation.
Additionally, we investigated specific regulatory elements on the cyp19a promoter through deletion analysis. Specifically, we investigated three putative binding sites for SF-1. Two of these sites (positions -821/-810 and -707/-696) were unique to chub mackerel and one (-217/-207) was shared by most species, also overlapping with ESRR and Nr5A2. Interestingly, only the deletion introduced at position -821/-810 upstream of the start codon significantly repressed promoter activity (Figure 7). These findings suggest that this deleted region, present in chub mackerel but absent in other scombrids, play a role in the upregulation of cyp19a. Moreover, while the other two introduced deletions did not exhibit significant differences, a trend of reduced luciferase activity compared to the control was observed, suggesting potential effects on cyp19a regulation. Specifically, the deletion introduced at position -217/-207, shared with bamboo leaf wrasse, contains a differentially methylated CpG (mCpG) site in both species. The sexual dimorphism at this position is evident in both species, but it is more pronounced in chub mackerel, with methylation levels at 46% for females and 94% for males, whereas in wrasse, it stands at 92% for females and 100% for males. These results position the deleted sequence as a TFBS candidate, shared across species.
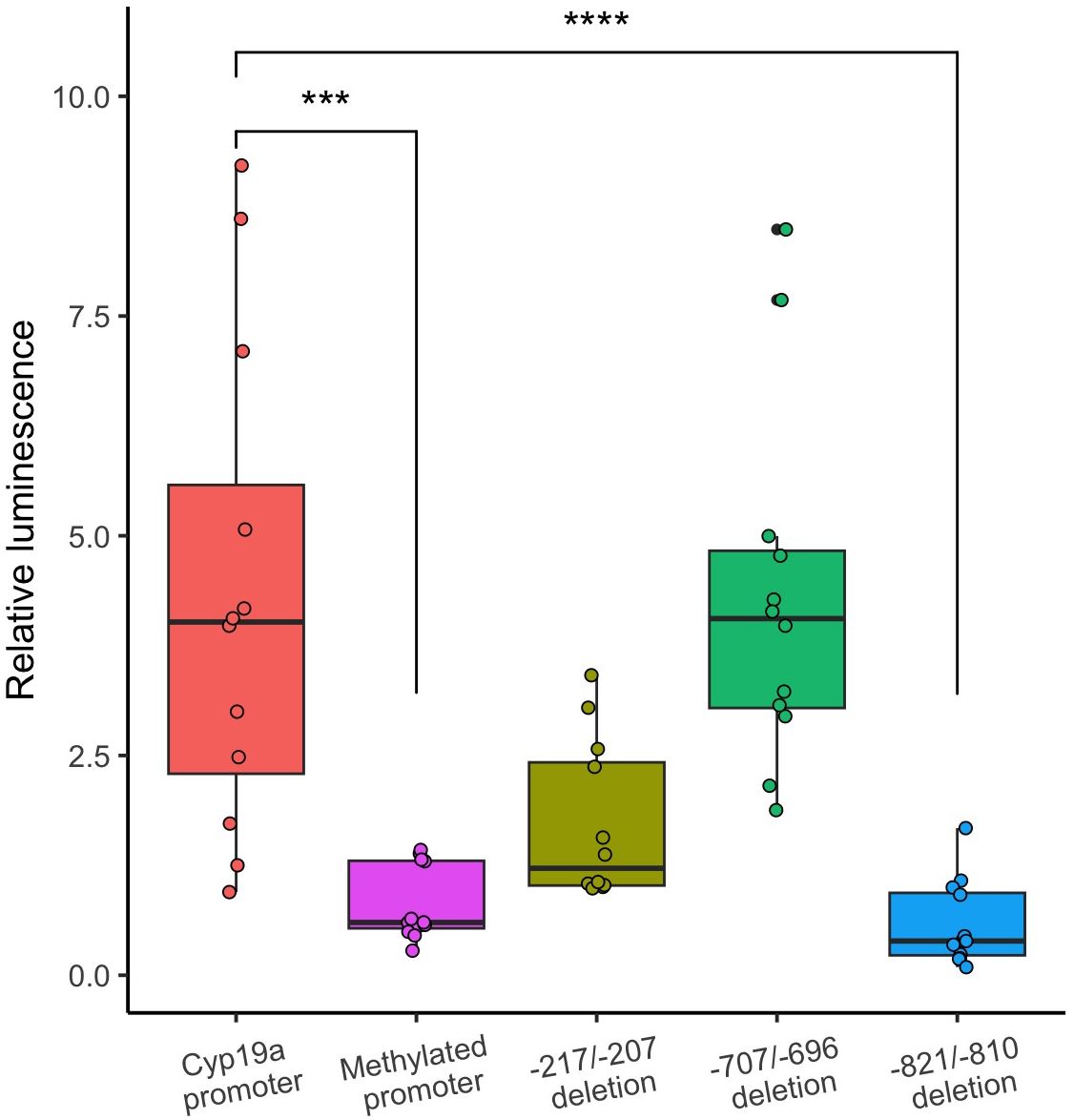
Figure 7 In vitro assay of cyp19a promoter. Luciferase activity in the methylated cyp19a promoter group (purple) was significantly repressed, compared to the unmethylated group (***, adjusted p-value < 0.001). All introduced deletions exhibited lower luciferase activity, (measured as relative luminescence) than the control, but only the -821/-810 deletion was significantly different (****, adj. p-value < 0.0001).
4 Discussion
4.1 The regulatory elements of the cyp19a promoter are highly conserved across scombrid species, though their CpG content varies
The comparative in silico examination of the cyp19a promoter reveals several remarkably conserved elements among tunas and chub mackerel, which are absent in other species. Southern bluefin and yellowtail tuna share all 24 TFBSs discovered, half of them also common in chub mackerel. These shared putative TFBSs include Foxl2, Sox3, SF-1, and ESRR, indicating potential roles in gene expression regulation. For instance, Foxl2 has been found to upregulate cyp19a gene expression by directly binding to the promoter in Nile tilapia (Wang et al., 2007), consistent with other studies reporting a female-biased expression of Foxl2 in gonad (Li et al., 2020; Shu et al., 2022; Shen X. et al., 2023). Similarly, we also found that foxl2 gene is highly expressed in ovaries of mature chub mackerel (data not shown). Furthermore, the co-transfection of Foxl2 with SF-1 was shown to have a synergetic influence in the in vitro activation of cyp19a promoter, effect lost when the promoter is hypermethylated (Navarro-Martín et al., 2011). Likewise, Sox3 has also been found to be differentially higher expressed in females (Li et al., 2020; Shen X. et al., 2023) and is suggested to affect cyp19a transcription by directly binding to the promoter in a frog species (Oshima et al., 2009). The presence of these common regulatory elements across scombrids suggests a conserved regulatory network governing gene expression. The modulation of cyp19a expression, may exhibit similar regulatory mechanisms across these species. Moreover, the methylation status of the promoter could influence the binding of a combination of these TFs and consequently modulate aromatase expression in the gonad. To this date, there are several articles discussing the relationship of DNA methylation with transcription. In a global analysis of the human TF and CpG methylation, for example, researchers found that most of the 542 TFBSs analyzed were affected by mCpG, with the great majority being inhibited while some TF preferred to bind to methylated DNA (Yin et al., 2017). The location and content of CpGs also hold importance in gene regulation. While the majority of CpG sites located in the distal part of the promoter were conserved in both tunas and chub mackerel, those closer to the start codon were largely absent in tunas. Despite the high conservation of the cyp19a promoter region within the Scombridae family, the same level of conservation does not extend to individual CpG sites. On a broader scale, we also observed a variation in CpG content across species, with some exhibiting CpG islands (notably in Japanese flounder and Nile tilapia), and higher CpG content. We identified conserved SF-1 and Foxl2 sites, along with a CpG site in-between, across all species examined (with the CpG site absent only in the medaka sequence). Some TFBSs were present in most species, but shifted location, like the androgen receptor (AR). We found CpG sites surrounding the TATA box at varying distances across different species. In a study conducted in 2007, Kitazawa and Kitazawa observed that a single-CpG site adjacent to the TATA box of the mouse RANKL gene exhibited a tissue-specific methylation pattern and, when methylated, influenced TBP binding subsequently affecting gene expression. It is likely that certain CpG sites play varying roles in the regulation of aromatase expression, with some contributing more significantly while others exerting a lesser impact.
4.2 Differential DNA methylation patterns of cyp19a promoter were found in adult chub mackerel gonad, consistent with sex-biased cyp19a expression
We found a clear difference in methylation patterns between adult female and male chub mackerel. We detected significantly higher methylation levels in male fish, matching the lower observed aromatase expression, which suggests a potential downregulation effect on cyp19a expression. Methylation of the promoter may influence the activation of cyp19a transcription by disrupting the binding of TF to the promoter. Conversely, while still displaying sexual dimorphism, the observed differences in methylation levels were considerably smaller for most mCpG sites located downstream of the start codon. Interestingly, almost all CpG sites located within cyp19a gene body were exclusively present in chub mackerel. Whether methylation of these intragenic CpGs is relevant or not on expression modulation is yet to be confirmed. There is limited information regarding potential explanations in fish models, but some studies in other vertebrates suggest that methylation within gene bodies may not exert the same regulatory influence as methylation within promoters. For instance, gene body methylation was found not to be associated with gene repression on the human X chromosome (Hellman and Chess, 2007). Moreover, in mammalian genes, active expression has been observed in presence of extensive methylation of intragenic CpG islands (Larsen et al., 1993; Unoki and Nakamura, 2003). These findings collectively emphasize that DNA methylation has context-dependent effects on gene expression regulation. Intragenic CpG sites, absent on other scombrids, could be affecting the overall modulation of cyp19a to a lesser extent, affecting transcription elongation or alternative splicing (Jones, 2012). It is also worth mentioning that, due to experimental limitations of the bisulfite treatment, commonly included in most DNA methylation related studies, it is not possible to differentiate between mCpG and hydroxymethylated CpGs. Cytosine hydroxymethylation is an epigenetic modification of DNA which effects on gene regulation may vary from those of DNA methylation. It was found to be enriched on tissue-specific gene bodies and associated with active transcription (Cui et al., 2020), in contrast with the common association of DNA methylation with gene repression. There is a possibility that the higher methylation levels observed within actively transcribed gene bodies correspond to hydroxymethylated cytosines.
Continuing our analysis, we performed in vitro experiments to evaluate the impact of promoter methylation on transcriptional activation. Our results indicated a significant repression of luciferase activity in the methylated cyp19a promoter group compared to the unmethylated control, confirming the regulatory role of methylation in gene expression. To elucidate the underlying mechanisms, we then explored the involvement of three previously identified SF-1 binding sites in gene regulation. We successfully identified a putative SF-1 binding site that, when deleted, led to a remarkable reduction in promoter activity, comparable with the methylated promoter group, suggesting an essential role of this motif in aromatase regulation. In addition, the other two deletions, even though not significant, negatively affected luciferase activity. Remarkably, the deleted sequence from position -217 to -207 contains three TFBSs (SF-1, ESRR, and Nr5A2) and is shared by most of the analyzed fish species. Additionally, it overlaps with a CpG site present in scombrids and bamboo leaf wrasse. Our previous analysis of sf-1 gene expression in gonads revealed comparable levels between females and males (data not shown). Given SF-1 is present in both ovaries and testes, methylation of the promoter may serve to obstruct the binding of available SF-1, which is partially responsible for the downregulation of cyp19a.
When analyzing gene expression of enzymes involved in methylation dynamics, we found that dnmt1 expression levels were similar in all fish, whereas Tet enzymes expression exhibited a significant increase in the female group. Dnmt1 plays a crucial role in maintaining methylation patterns within the genome (Bestor, 2000), while Tet enzymes are responsible for catalyzing the oxidation of methyl groups (Tahiliani et al., 2009). Tet enzymes, due to facilitating active DNA demethylation, offer a dynamic mechanism for modulating gene expression in response to environmental cues. Interestingly, despite similar levels of dnmt1 mRNA abundance, the notably elevated synthesis of oxidases could be indicating active mCpG oxidation and consequently activation of epigenetically modulated genes. Our findings suggest that the distinct sexual dimorphic methylation patterns observed in chub mackerel cyp19a promoter are due to active demethylation by tet enzymes, rather than male-biased dnmt1 activity.
4.3 DNA methylation patterns are species-specific and may differ depending on the sexual system
The link between promoter methylation and cyp19a expression has been previously studied in other species of fish. Sex differences in methylation levels were also found in the cyp19a promoter of Chinese sea perch (Chen et al., 2018) and Japanese flounder (Wen et al., 2014), among others. In our laboratory, we also conducted similar experiments with bamboo leaf wrasse, a hermaphrodite fish, and Japanese anchovy. We found conserved binding sites for Foxl2 and SF-1, both of which have been shown to enhance the in vitro activation of the cyp19a promoter (Nakamoto et al., 2007; Navarro-Martín et al., 2011), suggesting that these two closely located TFBSs jointly play a role in cyp19a regulation.
In this study, we observed an increased methylation level in the testes’ aromatase promoter, consistent with the previously mentioned studies in other species. We also identified sexually and species-variable DNA methylation patterns. The overall sexual difference in methylation levels is much more distinct in the gonochoristic species analyzed, whereas for the bamboo leaf wrasse, the differences were much smaller. In a specific case, we examined the methylation levels of a single CpG position (-210 in chub mackerel and -131 in bamboo leaf wrasse), which coincides with one of the deletions introduced in the experiment mentioned above (Figure 7, position -217/-207). This deleted sequence encompasses two shared TFBSs by these fish, SF-1 and ESRR. At this position, the sexual bias in methylation is much more pronounced in chub mackerel. We observed a trend of repression when deleting the sequence in chub mackerel, suggesting that this site may have a mild effect on promoter activation. However, further investigation is needed to determine the extent to which the methylation of this common site is relevant.
Sex-change is a complex process that usually involves gonadal transformation. Previously, a sharp downregulation of ovarian aromatase was found to precede sex-change in three species of wrasse (Thomas et al., 2019), supporting the idea of Cyp19a as a proximal regulator of gonadal change in protogynous species. Hypermethylation of the aromatase promoter during sex-change has also been investigated before. In one study, scientists observed an overall rise in methylation levels following masculinization of bluehead wrasse, which is also evident in the hypermethylation of a CpG island associated with the transcription start site (TSS) (Todd et al., 2019). This aligns with the overall increase in methylation discovered in the bamboo leaf wrasse but contrasts with the specific decrease in methylation observed at the CpG site nearest to the TSS. An overall hypermethylated promoter with a punctual hypomethylated CpG site was also observed in the protogynous orange-spotted grouper after sex-change (Guo et al., 2021). As mentioned earlier, even differential methylation at a single CpG site can sufficiently modulate gene expression (Kitazawa and Kitazawa, 2007). Given their capacity for sex change, we anticipate that hermaphrodite fish will display greater variability and plasticity in their methylation patterns. Furthermore, this fish presented the highest unique TFBS count among the rest of fish analyzed earlier, which highlights how distinct its regulatory region is from those of gonochoristic fishes. DNA methylation shifts during sex change seem to globally increase, while DNA methylation patterns exhibit unique species-specific characteristics. This implies that while the global increase in DNA methylation is a commonality, the nuanced patterns are intricately tailored to the specific genetic makeup of each species, contributing to the complexity of the observed phenomenon.
This study contributes to our understanding of Cyp19a regulation and gonad maturation in fish. Our research confirms that DNA methylation of cyp19a exhibits sex-specific patterns and is prevalent among adult fish. Notably, our findings reveal that the cyp19a promoter is highly conserved in scombrids, suggesting that the results obtained from chub mackerel could provide valuable insights into the same process in other members of the Scombridae family. Moreover, we discovered that, while overall methylation affects cyp19a activity, differences in CpG content may introduce subtle distinctions in its regulation. These findings are pivotal in advancing our comprehension of sexual development and reproductive processes in fish, shedding light on the intricate mechanisms underlying these biological phenomena.
Data availability statement
The data presented in the study are deposited in the NCBI repository (https://www.ncbi.nlm.nih.gov/), accession numbers PP825915 and PP825916.
Ethics statement
The animal study was approved by Kyushu University Animal Husbandry Guidelines. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MG: Writing – original draft, Methodology, Investigation, Formal analysis, Conceptualization. AD: Writing – review & editing, Methodology. YE: Writing – review & editing, Methodology. AT: Writing – review & editing. TI: Writing – review & editing. SM: Writing – review & editing. YO: Writing – review & editing. MS: Writing – review & editing. MM: Writing – review & editing. TC: Writing – review & editing. KO: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.This research was financially supported by JSPS KAKENHI grant numbers: JP16H04981, JP16H06279 (PAGS), JP19H03049, JP22H00386, JP22K05832,and JP22K19211.
Acknowledgments
The authors thank all the staff of Aqua-BioResource Innovation Center (ABRIC) Karatsu satellite, Kyushu University, for fish maintenance and sampling support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1408561/full#supplementary-material
References
Anastasiadi D., Díaz N., Piferrer F. (2017). Small ocean temperature increases elicit stage-dependent changes in DNA methylation and gene expression in a fish, the European sea bass. Sci. Rep. 7, 12401. doi: 10.1038/s41598–017-10861–6
Barange M., Perry R. I. (2009). “Physical and ecological impacts of climate change relevant to marine and inland capture fisheries and aquaculture,” in Climate change implications for fisheries and aquaculture: Overview of current scientific knowledge (FAO Fisheries and Aquaculture Technical Paper No. 530. Eds. Cochrane K., De Young C., Soto D., Bahri T. (FAO, Rome), 7–106).
Bestor T. H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402. doi: 10.1093/hmg/9.16.2395
Castro-Mondragon J. A., Riudavets-Puig R., Rauluseviciute I., Berhanu Lemma R., Turchi L., Blanc-Mathieu R., et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. doi: 10.1093/nar/gkab1113
Catalano S., Marsico S., Giordano C., Mauro L., Rizza P., Panno M. L., et al. (2003). Leptin enhances, via AP-1, expression of aromatase in the MCF-7 cell line. J. Biol. Chem. 278, 28668–28676. doi: 10.1074/jbc.M301695200
Chen X., He Y., Wang Z., Li J. (2018). Expression and DNA methylation analysis of cyp19a1a in Chinese sea perch Lateolabrax maculatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 226, 85–90. doi: 10.1016/j.cbpb.2018.07.008
Cui X. L., Nie J., Ku J., Dougherty U., West-Szymanski D. C., Collin F., et al. (2020). A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation. Nat. Commun. 11, 6161. doi: 10.1038/s41467-020-20001-w
Guo C.-Y., Tseng P.-W., Hwang J.-S., Wu G.-C., Chang C.-F. (2021). Potential role of DNA methylation of cyp19a1a promoter during sex change in protogynous orange-spotted grouper, Epinephelus coioides. Gen. Comp. Endocrinol. 311, 113840. doi: 10.1016/j.ygcen.2021.113840
Hellman A., Chess A. (2007). Gene body-specific methylation on the active X chromosome. Science 80). 315, 1141–1143. doi: 10.1126/science.1136352
Huang S., Ye L., Chen H. (2017). Sex determination and maintenance: the role of DMRT1 and FOXL2. Asian J. Androl. 19:619–624. doi: 10.4103/1008-682X.194420
Jones P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/nrg3230
Kime D. E. (1993). Classical and non-clssical reproductive steroids in fish. Rev. Fish Biol. Fish. 3, 160–180. doi: 10.1007/BF00045230
Kitazawa R., Kitazawa S. (2007). Methylation status of a single cpG locus 3 bases upstream of TATA-box of receptor activator of nuclear factor-κB ligand (RANKL) gene promoter modulates cell- and tissue-specific RANKL expression and osteoclastogenesis. Mol. Endocrinol. 21, 148–158. doi: 10.1210/me.2006-0205
Krassowski M., Arts M., Lagger C., Max (2022). krassowski/complex-upset: v1.3.5. Zenodo. doi: 10.5281/zenodo.7314197
Kumar P., Mendelson C. R. (2011). Estrogen-Related receptor γ (ERRγ) mediates oxygen-Dependent induction of aromatase (CYP19) gene expression during human trophoblast differentiation. Mol. Endocrinol. 25, 1513–1526. doi: 10.1210/me.2011-1012
Kumar S., Suleski M., Craig J. M., Kasprowicz A. E., Sanderford M., Li M., et al. (2022). TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174. doi: 10.1093/molbev/msac174
Larsen F., Solheim J., Prydz H. (1993). A methylated CpG island 3’ in the apolipoprotein-E gene does not repress its transcription. Hum. Mol. Genet. 2, 775–780. doi: 10.1093/hmg/2.6.775
Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H. (2014). UpSet: visualization of intersecting sets. IEEE Trans. Visualization Comput. Graphics 20, 1983–1992. doi: 10.1109/TVCG.2014.2346248
Li S., Lin G., Fang W., Huang P., Gao D., Huang J., et al. (2020). Gonadal transcriptome analysis of sex-related genes in the protandrous yellowfin seabream (Acanthopagrus latus). Front. Genet. 11. doi: 10.3389/fgene.2020.00709
Madeira F., Pearce M., Tivey A. R. N., Basutkar P., Lee J., Edbali O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276—W279. doi: 10.1093/nar/gkac240
Nakamoto M., Wang D.-S., Suzuki A., Matsuda M., Nagahama Y., Shibata N. (2007). Dax1 suppresses P450arom expression in medaka ovarian follicles. Mol. Reprod. Dev. 74, 1239–1246. doi: 10.1002/mrd.20689
Navarro-Martín L., Viñas J., Ribas L., Díaz N., Gutiérrez A., Di Croce L., et al. (2011). DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PloS Genet. 7. doi: 10.1371/journal.pgen.1002447
Oshima Y., Naruse K., Nakamura Y., Nakamura M. (2009). Sox3: A transcription factor for Cyp19 expression in the frog Rana rugosa. Gene 445, 38–48. doi: 10.1016/j.gene.2009.05.011
Petrik C. M., Stock C. A., Andersen K. H., van Denderen P., Daniël W. J. R. (2020). Large pelagic fish are most sensitive to climate change despite pelagification of ocean food webs. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.588482
Piferrer F., Blázquez M. (2005). Aromatase distribution and regulation in fish. Fish Physiol. Biochem. 31, 215–226. doi: 10.1007/s10695-006-0027-0
Ryu T., Veilleux H. D., Donelson J. M., Munday P. L., Ravasi T. (2018). The epigenetic landscape of transgenerational acclimation to ocean warming. Nat. Clim. Change 8, 504–509. doi: 10.1038/s41558-018-0159-0
Shen W.-K., Chen S.-Y., Gan Z.-Q., Zhang Y.-Z., Yue T., Chen M.-M., et al. (2023). AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 51, D39–D45. doi: 10.1093/nar/gkac907
Shen X., Yáñez J. M., Bastos Gomes G., Poon Z. W. J., Foster D., Alarcon J. F., et al. (2023). Comparative gonad transcriptome analysis in cobia (Rachycentron canadum). Front. Genet. 14. doi: 10.3389/fgene.2023.1128943
Shu C., Wang L., Zou C., Tan X., Zou Y., Kong L., et al. (2022). Function of Foxl2 and Dmrt1 proteins during gonadal differentiation in the olive flounder Paralichthys olivaceus. Int. J. Biol. Macromol. 215, 141–154. doi: 10.1016/j.ijbiomac.2022.06.098
Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Thomas J. T., Todd E. V., Muncaster S., Lokman P. M., Damsteegt E. L., Liu H., et al. (2019). Conservation and diversity in expression of candidate genes regulating socially-induced female-male sex change in wrasses. PeerJ 7, e7032. doi: 10.7717/peerj.7032
Todd E. V., Ortega-Recalde O., Liu H., Lamm M. S., Rutherford K. M., Cross H., et al. (2019). Stress, novel sex genes, and epigenetic reprogramming orchestrate socially controlled sex change. Sci. Adv. 5, 1–14. doi: 10.1126/sciadv.aaw7006
Unoki M., Nakamura Y. (2003). Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett. 554, 67–72. doi: 10.1016/S0014-5793(03)01092-5
Wang D.-S., Kobayashi T., Zhou L.-Y., Paul-Prasanth B., Ijiri S., Sakai F., et al. (2007). Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21, 712–725. doi: 10.1210/me.2006-0248
Wen A. Y., You F., Sun P., Li J., Xu D. D., Wu Z. H., et al. (2014). CpG methylation of dmrt1 and cyp19a promoters in relation to their sexual dimorphic expression in the Japanese flounder Paralichthys olivaceus. J. Fish Biol. 84, 193–205. doi: 10.1111/jfb.12277
Keywords: DNA methylation, cyp19a expression, scombrid, aromatase, epigenetics, sexual plasticity
Citation: Galotta M, Dam AT, Eto Y, Toyoda A, Itoh T, Mohapatra S, Ogino Y, Saitou M, Matsuyama M, Chakraborty T and Ohta K (2024) Insights into epigenetic regulation of cyp19a via comparative analysis using the scombrid chub mackerel as model. Front. Mar. Sci. 11:1408561. doi: 10.3389/fmars.2024.1408561
Received: 28 March 2024; Accepted: 03 May 2024;
Published: 07 June 2024.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Amit Kumar Sinha, University of Arkansas at Pine Bluff, United StatesAparna Chaudhari, Central Institute of Fisheries Education (ICAR), India
Biswajit Maiti, Nitte University, India
Copyright © 2024 Galotta, Dam, Eto, Toyoda, Itoh, Mohapatra, Ogino, Saitou, Matsuyama, Chakraborty and Ohta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tapas Chakraborty, dGFwYXNfY2hAYWdyLmt5dXNodS11LmFjLmpw; Kohei Ohta, a19vaHRhQGFnci5reXVzaHUtdS5hYy5qcA==
 Mariel Galotta
Mariel Galotta Anh Tuan Dam
Anh Tuan Dam Yuhei Eto1
Yuhei Eto1 Atsushi Toyoda
Atsushi Toyoda Takehiko Itoh
Takehiko Itoh Yukiko Ogino
Yukiko Ogino Michiya Matsuyama
Michiya Matsuyama Tapas Chakraborty
Tapas Chakraborty Kohei Ohta
Kohei Ohta