- 1Department of Marine Life Science (BK21 FOUR) and Marine Life Research Institute, Jeju National University, Jeju, Republic of Korea
- 2Department of Aquatic Life Medicine, College of Ocean Science and Technology, Kunsan National University, Gunsan, Republic of Korea
- 3Department of Marine Biology and Aquaculture, Gyeongsang National University, Tongyeong, Republic of Korea
- 4Department of Applied Aquabiology, National Fisheries University, Shimonoseki, Yamaguchi, Japan
- 5Biodiversity Research and Cooperation Division, National Institute of Biological Resources, Incheon, Republic of Korea
Pufferfish from the genus Takifugu are vital commercial resources in East Asia. Within the genus, the taxonomic status of two commercially important species, T. rubripes and T. chinensis, remains ambiguous, especially given their morphological variability. Recent observations of suspected hybrids between T. rubripes and T. chinensis on Jeju Island, South Korea, displaying intermediate phenotypes, have further confused their classification. In this study, we analyzed 73 pufferfish, including wild-caught T. rubripes, T. chinensis, suspected hybrids, and farm-bred T. rubripes, using 16 microsatellite loci to explore their population structure and evolutionary relationships. The Bayesian clustering and principal coordinate analysis showed minimal genetic differentiation among the wild populations, regardless of phenotype. This finding suggests that T. rubripes and T. chinensis might represent a single species with considerable morphological diversity. In contrast, farm-bred T. rubripes exhibited significant genetic differentiation from wild populations, likely due to domestication-induced genetic drift. These results challenge the existing taxonomic distinctions between T. rubripes and T. chinensis and highlight the profound impact of aquaculture on the genetics of captive populations. This study underscores the necessity for ongoing research into the taxonomy and population genetics of the T. rubripes-chinensis complex to guide conservation and management strategies and stresses the importance of genetic monitoring in pufferfish aquaculture to counteract inbreeding and genetic drift.
1 Introduction
Pufferfish species of the genus Takifugu are widely consumed as a culinary delicacy in South Korea, Japan, and China, holding a significant position in the gastronomic culture of these countries. In particular, Takifugu rubripes is highly esteemed for its exquisite texture and flavor, making it a species of considerable economic value and a widely cultivated target in aquaculture. As of 2022, approximately 16,000 tons of farmed T. rubripes were produced in China and around 3,000 tons in Japan (FAO, 2024). The global aquaculture production of this species is on the rise, reflecting the increasing market demand for pufferfish. However, the aquaculture production of T. rubripes in South Korea remains limited, with annual production of less than 70 tons, primarily confined to specific regions such as the southern part of Jeju Island (FAO, 2024). Consequently, the majority of pufferfish consumed domestically relies on annual wild catches of 3,000 to 5,000 tons, supplemented by imports of approximately 3,000 to 5,000 tons of pufferfish from China.
Pufferfish contain tetrodotoxin (TTX), a potent neurotoxin, and TTX-related food poisoning incidents have primarily resulted from improper consumption of pufferfish (Guardone et al., 2020). Such incidents have been consistently reported in countries where pufferfish are consumed (Kim et al., 2003; Watari et al., 2021). The distribution of this toxin within tissues varies among different pufferfish species (Noguchi et al., 2006b; Noguchi and Arakawa, 2008); therefore, accurate species identification is essential for preventing TTX poisoning by precisely determining the toxin distribution, and consumption is restricted to specific pufferfish species with verified edible parts by the Ministry of Food and Drug Safety of Korea. While the accumulation of TTX in wild pufferfish poses a threat to food safety, farmed pufferfish, which can be systematically managed, may serve as a viable alternative for the safe supply of seafood. Since pufferfish acquire TTX through exogenous sources, primarily from their diet, they do not accumulate TTX in their bodies when reared in environments isolated from TTX sources, such as aquaculture facilities (Noguchi et al., 2006a). Therefore, continuous management of the pufferfish aquaculture industry is essential to ensure the supply of pufferfish as a safe seafood product free from the risk of TTX poisoning. However, despite the commercial value of farmed pufferfish, the genetic impacts of selective breeding on farmed populations remain understudied. Assessing genetic diversity in both wild and farmed pufferfish is crucial to identify potential issues affecting aquaculture sustainability and productivity. Population structure is influenced by genetic drift and natural selection. It typically develops over generations but can form rapidly through founder effects or genetic bottlenecks (Peacock et al., 2009). In aquaculture, genetic drift is inevitable due to selective breeding, yet reports on genetic divergence from wild populations are rare (Barría et al., 2023). Severe genetic drift is well-documented in livestock but less reported in fish. However, population structure is evident in ornamental fish such as goldfish (Chen et al., 2020) and Betta fish (Zhang et al., 2022), which show significant genetic divergence from their wild counterparts due to domestication.
Two pufferfish species, T. rubripes and its closely related species T. chinensis, which are distributed in the coastal waters of South Korea, are morphologically distinguishable: T. rubripes features a white anal fin and irregular black spots on dorsal half of posterior part of the body, whereas T. chinensis does not exhibit these markings (Baek et al., 2018). Additionally, T. rubripes is known to spawn at depths of 20–50 m, whereas T. chinensis spawns at depths below 100 m, indicating distinct ecological differences between the two species. Furthermore, T. rubripes is generally recognized as a larger species compared to T. chinensis in terms of maximum body size (Matsuura, 2017). However, advances in molecular phylogenetics have prompted a reevaluation of the taxonomic classification within the genus Takifugu over the past several decades. The research consistently supports the notion that T. rubripes and T. chinensis, previously considered distinct species based on morphological differences, may be a single species. This hypothesis is backed by studies utilizing mitochondrial haplotypes, microsatellites, and genome-wide SNPs (Reza et al., 2011; Baek et al., 2018; Park et al., 2020; Takahashi et al., 2023; Liu et al., 2024). In addition, a noticeable increase in pufferfish displaying intermediate morphological traits between T. rubripes and T. chinensis has been observed in Korea, suggesting possible natural hybridization between the two species. Intermediate forms of this combination have also been observed in Japan (Reza et al., 2008), and since the edible parts (muscle, skin, and testes) of both species are identical, consumption is allowed only for pufferfish exhibiting these intermediate traits among hybrid pufferfish (Ministry of Health, Labour and Welfare of Japan, 2020). Despite the increasing occurrence of intermediate forms, information on their occurrence and genetic population structure remains insufficient. These potential hybrids, referred to here as “hybrid-suspected pufferfish” (HSP), may spark debate over the accuracy of existing species classifications and the broader ecological and evolutionary implications of interspecific hybridization. To address these issues, it is critical to explore the genetic population structure and evolutionary relationships between T. rubripes, T. chinensis, and the HSP. Such studies are crucial for clarifying taxonomic ambiguities and devising appropriate conservation and management strategies. Furthermore, examining the genetic underpinnings of observed morphological variations within this group could offer deeper insights into the drivers of phenotypic variation and adaptation in marine fishes.
While genomic techniques are invaluable for identifying signs of domestication and pinpointing genes linked to local adaptation, more accessible genetic markers, such as microsatellites, remain crucial. These markers are effective in deciphering population structures, detecting genetic drift impacts, and assessing inbreeding within domesticated populations, especially when they show polymorphism (Prunier et al., 2023). In this study, we utilized a suite of 16 microsatellite markers to examine the genetic relationships among wild-caught T. rubripes, T. chinensis, and HSP from the waters around Jeju Island, alongside farm-bred T. rubripes from local aquaculture operations. Our analysis of genetic diversity and population structure aimed to (1) clarify the taxonomic distinctions between T. rubripes and T. chinensis, (2) provide genetic evidence of natural hybridization between these species, and (3) assess the genetic impacts of selective breeding on farmed T. rubripes populations. Our findings offer fresh insights into the evolutionary history and current genetic landscape of the T. rubripes-chinensis species complex, which hold significant implications for their taxonomy, conservation, and sustainable management.
2 Materials and methods
2.1 Sampling and DNA extraction
From 2020 to 2022, a total of 73 pufferfishes were purchased from the market on the east, south and Jeju Island and an aquafarm in Jeju Island (Figure 1, Table 1). Based on phenotypic features, obtained pufferfishes were identified as 4 groups; T. rubripes, T. chinensis, and HSP type 1 and 2. T. rubripes with white anal fin and irregular black marks on the side of the body can be distinguished by T. chinensis (Figures 1, 2, Supplementary Table S1). HSP have phenotypic features of both species. HSP type1 are characterized by a white anal fin (T. rubripes) and no irregular black marks on the side of the body (T. chinensis), and the HSP type2 have a black anal fin (T. chinensis) and irregular black marks (T. rubripes, Figure 2, Supplementary Table S1). Genomic DNA used for microsatellite analysis was extracted from the pectoral fin using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocols.
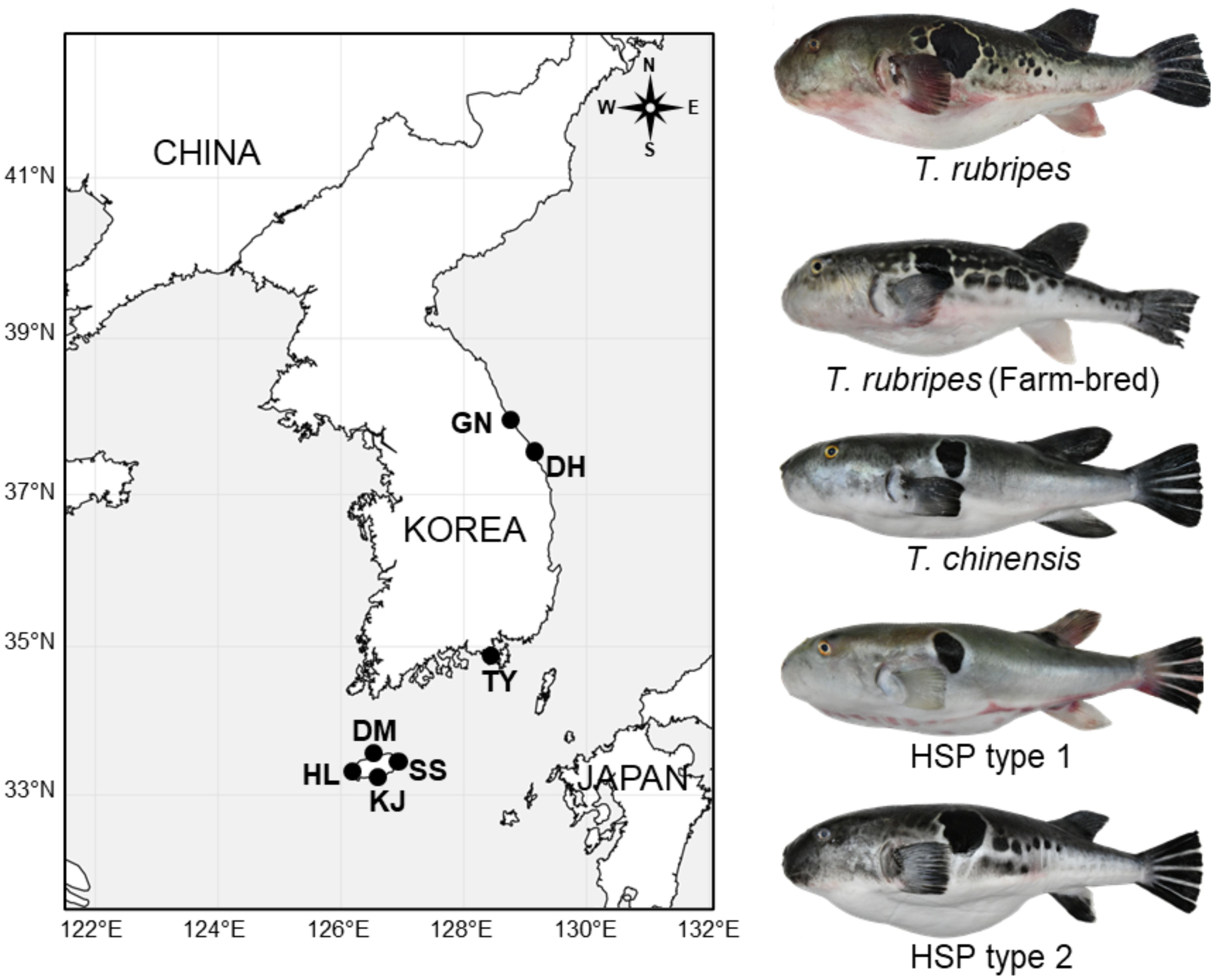
Figure 1. Sampling locations of Takifugu rubripes and T. chiensis in South Korea. T. rubripes (wild type), T. rubripes from fish farm, T. chinensis, and two types of hybrid suspect pufferfish (HSP) are shown with representative images. Other abbreviations on the map refer to locality of South Korea where these fish species have been collected: HL (Hallim market, Jeju), KJ (Kangjeong market, Jeju), DM (Dongmun market, Jeju), SS (Seongsan farm, Jeju), TY (Seoho market and offshore collection, Tongyeong), DH (Mukhwo market, Donghae), and GN (Jumunjin market, Gangneung).
2.2 Molecular identification
For the molecular identification of the species, we performed DNA barcoding using the primer that targeted the mitochondrial COI gene (Fish F1: 5’ TCA ACC AAC CAC AAA GAC ATT GGC AC 3’ and R1: 5’ TAG ACT TCT GGG TGG CCA AAG AAT CA 3’ (Ward et al., 2005). We employed primers commonly used in teleosts for the DNA barcoding analysis and followed the PCR conditions described by (Ward et al., 2005). The PCR products were verified through 1% agarose gel electrophoresis, and the positive products were submitted to Macrogen for sequencing. The obtained sequencing results were used to identify the species based on the most similar species to the query using BLAST. Maximum likelihood tree was reconstructed to address the phylogenetic position among closely related species using the software IQ-TREE2 with default parameters (-b 1000) (Minh et al., 2020).
2.3 Microsatellite analysis
The amplification efficiency and the polymorphism for some of the samples were examined in 31 primer sets developed from T. rubripes and T. pseudommus using electrophoresis, and a total of 16 primer sets were finally selected. We performed the polymerase chain reaction (PCR) to amplify selected microsatellite loci, and the information of each microsatellite marker were summarized in Table 2. Forward primers of selected 16 microsatellite markers were 5´-fluorescently labeled using 6-FAM, HEX, or TAMRA to distinguish different alleles on the sequence analyzer. The PCR was conducted in a total volume of 50 µL, containing 100 ng of template DNA, PCR Master Mix 2X (ThermoFisher), and 10 µM of each primer. For Cst-1 and Cst-4, The PCR conditions were as follows: initial denaturation at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 58°C (Cst-1) or 60°C (Cst-4) for 1 min, extension at 72°C for 2 min, and final elongation at 72°C for 10 min. For the other markers, The conditions were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 50°C (Tru-7, Tru-8) or 60°C (Tru-9, Tru18, Fms13, F160, F112, F178, F86, F61, Tru-17, F65, F160, and F204) for 30 sec, extension at 72°C for 30 sec, and final elongation at 72°C for 1 min or 2 min. The amplification of fragments were confirmed by electrophoresis in 2.5% agarose gels. Fragment analysis of amplified products was performed by Macrogen Inc. (Seoul, Korea) using ABI 3730xl analyzer (Applied Biosystems, Foster City, CA, USA), and allele size was determined using the Peak Scanner Software v.1.0 (Applied Biosystems). The scored allele size data were evaluated for the existence of null alleles, allele dropout and stutter, and other scoring errors using MICRO-CHECKER v.2.2.3 (Van Oosterhout et al., 2004).
2.4 Genetic diversity and population structure analysis
The total number of alleles per locus (A), allelic richness (Ar), and observed and expected heterozygosity (HO and HE) were calculated using GenAlex v.6.51 (Peakall and Smouse, 2012) and FSTAT v.2.9.4 (Goudet, 2001). Arlequin v.3.5.2.2 was used to calculate the pairwise FST values between populations based on microsatellite loci with 1,000 permutations (Excoffier and Lischer, 2010). The genetic structure of populations was estimated using the Bayesian model-based clustering method implemented in the software STRUCTURE v.2.3.4 (Pritchard et al., 2000). The burn-in period was set to 5,000 and the number of Markov chain Monte Carlo (MCMC) repetitions to 50,000. Twenty iterations for each K-value were carried out assuming cluster K from 2 to 9 to examine population structure. The optimal number of genetic clusters was estimated using STRUCTURE harvester v.0.6.94 (Earl and vonHoldt, 2012), and CLUMPAK (Cluster Markov Packager Across K) was used to visualize the STRUCTURE output (Kopelman et al., 2015). Principal Coordinates Analysis (PCoA) based on tri distance matrix were used to visualize the genetic relationships among individuals and populations using covariance-standardized method as implemented in GenAlex v.6.51 (Peakall and Smouse, 2012). Contemporary effective population size was measured using NeEstimator to assess the effect of domestication on the bottleneck. Hybrid index for the putative hybrids between T. rubripes and T. chiensis was estimated using Genodive (Meirmans, 2020) with default parameters. We used the software POPTREE2 (Takezaki et al., 2014) to reconstruct the phylogenetic relationship of the five groups. Trees were generated using the neighbor-joining (NJ) method with 1,000 bootstrap replicates.
3 Results
3.1 Species identification
The COI barcode sequences obtained in this study showed 100% similarity between T. rubripes and T. chinensis, making it impossible to distinguish between the two species using the barcode sequence. All samples used in this study matched the sequence of T. rubripes-chinensis species complex. The ML phylogeny showed that the individuals used in this study clustered within the T. rubripes-chinensis-pseudommus complex (Supplementary Figure S1). No significant differences in the color of the anal fin were observed between wild and farmed T. rubripes (Figures 1, 2), while an inter-specific level difference was observed between T. rubripes and T. chinensis. A substantial difference in the distribution of black spots on the body was observed between the wild and farmed populations of T. rubripes. The farmed individuals showed larger spots on the body, while the wild population showed smaller spots that were similar in size to the diameter of the eye. Two hybrid-suspected groups (HSP type 1 and 2) exhibited the mosaic pattern in their spots on the body and color of the anal fin.
3.2 Genetic diversity of pufferfish
Sixteen microsatellite markers were used to measure the genetic diversity of 73 individuals, including T. rubripes, T. chinensis, and HSP type 1 and 2. The results showed that the species-level genetic diversity varied in five groups. The number of alleles varies considerably, from 3.438 in HSP type 1 to 15.563 in T. chinensis (Table 3). Observed and expected heterozygosity were relatively consistent across populations, ranging from 0.724 to 0.875 for observed heterozygosity and 0.664 to 0.878 for expected heterozygosity. Overall, the descriptive index suggests the domesticated group exhibits lower genetic diversity than wild populations (Table 3). No significant differences were observed in HSP types. Hybrid index analysis revealed that two HSP type 1 individuals showed affinity with T. chinensis (h = 0.000 - 0.211, zero indicates affinity with T. chinensis). For the HSP type 2 individuals, hybrid indexes were varied in samples (h = 0.000 - 0.614). The estimated effective population size (Ne) for each population was infinite for all populations except for the farmed T. rubripes population (Ne = 10.9, 95% CI = 8.4 - 14.3).
3.3 Intraspecific population structure
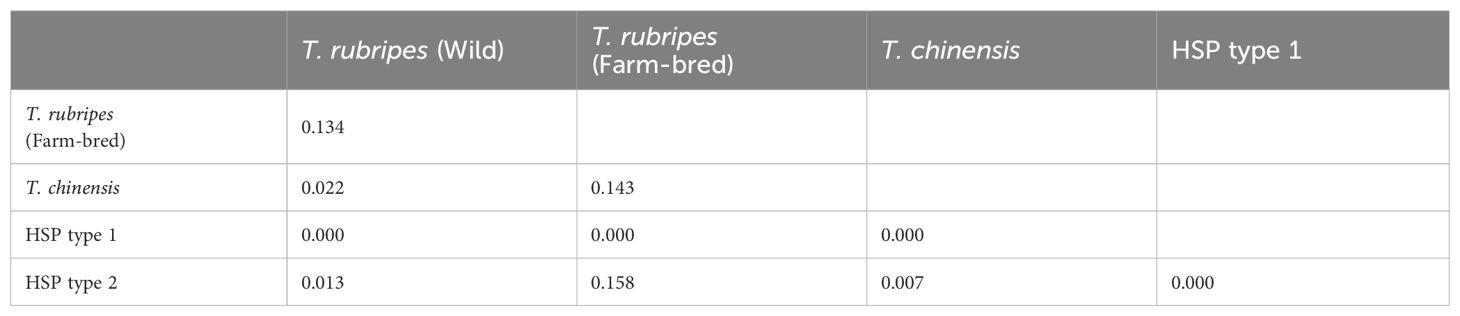
The highest FST is observed between farm-bred T. rubripes and others, ranging from 0.134 to 0.158, while other comparisons were below 0.03 (Table 4). The wild T. rubripes and T. chinensis exhibit no significant differentiations (FST = 0.022, p < 0.05). According to Frankham et al. (2002), FST value greater than 0.15 can be considered significant in differentiating populations. Our STRUCTURE analysis indicated that the significant population structure was observed between farm T. rubripes and the others (Best K = 2), while no significant structure was observed among wild T. rubripes, wild T. chinensis, and the moderate groups (HSP1 and HSP2) (Figure 3A). The PCoA plot revealed the distinct clustering patterns, with two wild pufferfish populations and the moderate groups forming a cluster together, while T. rubripes from the farm showed a distinct cluster from others (Figure 3B). The first two principal coordinates account for 12.18% and 3.85% of the total genetic variation, respectively. Interestingly, farmed T. rubripes shows the genetic distinctiveness within the group. The genetic distance-based tree indicated that farmed T. rubripes showed the sister relationship with wild T. rubripes although moderate bootstrap support value (Supplementary Figure S2).
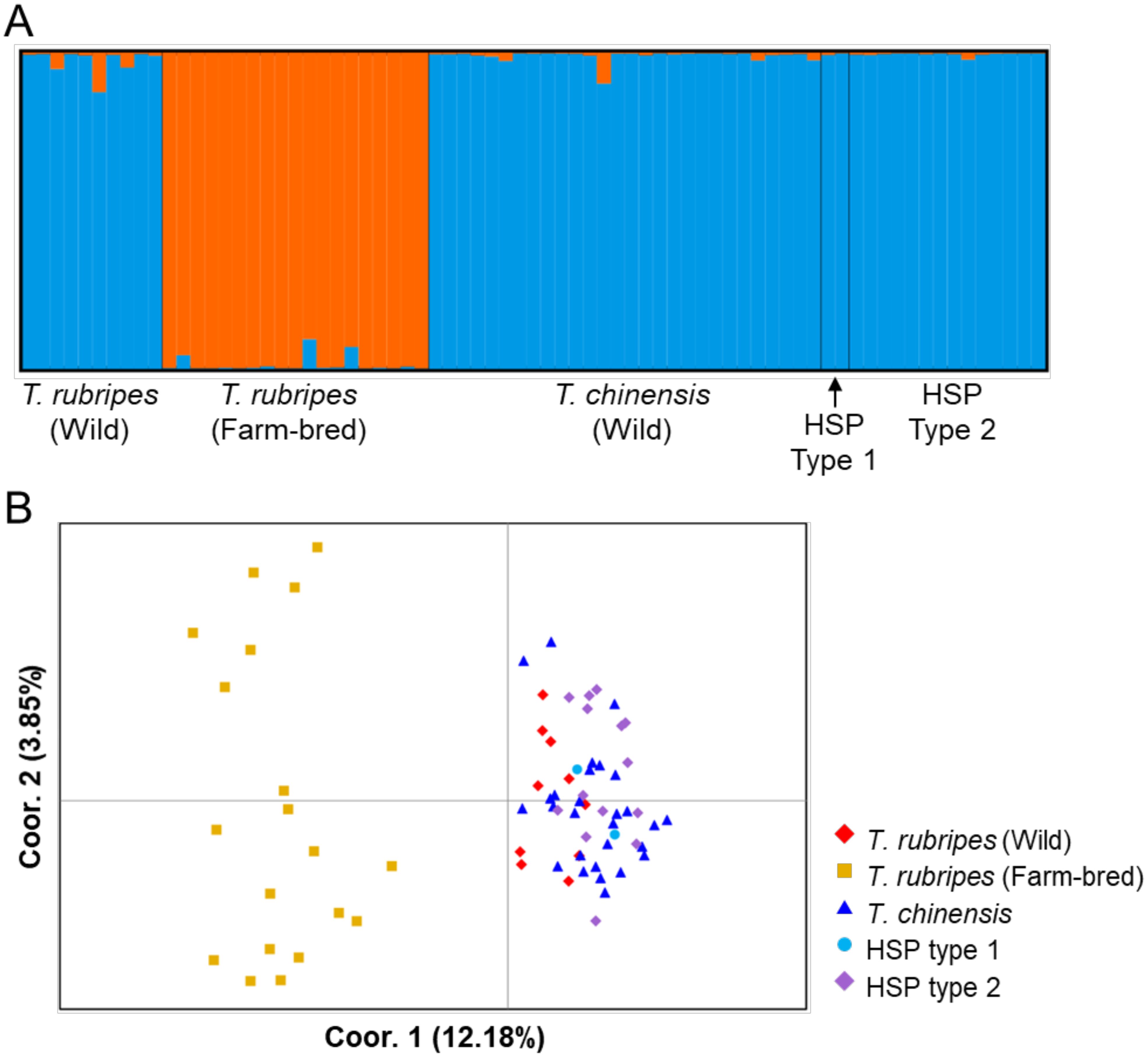
Figure 3. STRUCTURE plot (Best K = 2) of Takifugu rubripes-chinensis complex showing the proportion of genetic clusters (Q-values) on the Y-axis (A). PCoA plot of the T. rubripes-chinensis complex (B).
4 Discussion
In this study, we performed the genetic analysis using 16 microsatellite markers to determine the population structure and evolutionary relationships of T. rubripes and T. chinensis throughout Korean waters. Our results revealed no significant population structure throughout the individuals except for the farmed T. rubripes population. No clear genetic differentiation was observed in the two species, which was consistent with the previous studies using mitochondrial markers to genomic SNP markers (Baek et al., 2018; Park et al., 2020; Takahashi et al., 2023; Liu et al., 2024). The individuals with moderate morphological characteristics between species were also indistinguishable in terms of allele frequency. However, the estimated genetic diversity for the HSP group should be interpreted with caution due to the limited sample size. This implies that the species identity of T. rubripes and T. chinensis should be reconsidered given the fact that no genetic and genomic divergences between the species (Reza et al., 2011; Baek et al., 2018; Park et al., 2020; Takahashi et al., 2023). The fact that the species complex possesses diverse phenotypic characteristics despite having no genetic divergence suggests that the phenotypic traits (i.e., the coloration of the anal fin and lateral bands on the body) of this species complex exhibit intraspecific morphological variation. Our study highlights that the severe genetic drift during domestication is responsible for the rapid change of allele frequency, which may have resulted in the population structure.
Due to its economic significance, species identification of Takifugu species using genetic and genomic markers has been studied for decades (Reza et al., 2011; Baek et al., 2018; Park et al., 2020; Takahashi et al., 2023; Liu et al., 2024). Yet, there was no evidence to support the genetic isolation between T. rubripes and T. chinensis. We used the microsatellite markers developed by five independent research groups (Takagi et al., 2001, 2003; Furukawa et al., 2004; Cui et al., 2005; Kai et al., 2005) and found no differences in allele frequencies between the two species. Given that different species become reproductively isolated, thereby limiting gene flow, and consequently possess different allele frequencies across their genomes, this can be detected through allele frequency among the randomly selected markers. Previous studies that investigated the differences in allele frequencies of SNPs in genomic regions also indicated that the lack of clear differences in allele frequencies suggests that these two species are genetically undifferentiated and belong to the same species. Furthermore, the sharing of mitochondrial haplotypes, considering the characteristics of mitochondrial loci reflecting historical differentiation, suggests the possibility that the two species have not historically differentiated and may be the same species. Indeed, our result of the hybrid index clearly indicate that no signal of gene flow between species due to the lack of genetic divergence between the species. The distinct morphological characteristics observed in pufferfish may be attributed to the influence of environmental variables, including water temperature and salinity, on their developmental processes. Further investigation is necessary to ascertain the extent to which environmental factors shape morphological diversity.
Pufferfish have adapted to various aquatic ecosystems (Yamanoue et al., 2009; Santini et al., 2013). Understanding the functional and evolutionary significance of their diverse phenotypic characteristics can contribute to the effective management and conservation of wild populations. By exploring the adaptive mechanisms that have enabled pufferfish to thrive in different environments, we can gain valuable insights into their ecological roles and develop strategies to protect their habitats and maintain their genetic diversity. Furthermore, the study of phenotypic variation in pufferfish can shed light on the underlying molecular and developmental processes that give rise to phenotypic variation. This knowledge can be applied to other species facing similar environmental challenges, advancing our understanding of how organisms respond to changing conditions and adapt to new niches. Consequently, recognizing the significance of phenotypic variation in pufferfish and investigating its evolutionary and functional implications are crucial for effective conservation and management.
The effects of strong natural selection and genetic drift resulting from reduced effective population size and long-term inbreeding are the most prominent features of domestication (Nei and Tajima, 1981). This study observed a marked differentiation in allele frequencies in the cultured population. PCA and STRUCTURE also strongly supported the differentiation between the cultured population and the wild T. rubripes-chinensis complex. Furthermore, extremely low effective population size (Ne ≈ 10) and heterozygosity indicate reduced genetic diversity. When the effective population size is lower than 50, it indicates that the population is at a high risk of genetic drift and inbreeding (Jamieson and Allendorf, 2012; Ralls et al., 2018).
The changes in allele frequencies across 16 unlinked loci distributed throughout the genome could be attributed to strong selection and genetic drift. Although our study could not determine the specific types of selective pressures acting on these loci, the absence of intentional selection on parental individuals in the fish farm suggests that the observed changes may be due to genetic drift. To further investigate whether selection-driven differentiation has contributed to the major genomic differentiation, future research should employ genome-wide deep sequencing data and GWAS (genomewide association study) to detect genomic regions displaying genetic differentiation between wild and cultured populations. By examining whether these regions are the primary contributors to the overall genomic differentiation, it will be possible to elucidate the role of selection-driven differentiation in promoting genome-wide differentiation, which in turn leads to phylogenetic divergence.
Decreased genetic diversity can lead to problems, such as inbreeding depression, increased mortality rates, and reduced productivity in the aquaculture industry (Ye et al., 2022). Inbreeding depression is a phenomenon where the offspring of genetically related individuals exhibit reduced fitness, which can manifest as lower survival rates, slower growth, and decreased resistance to diseases (Smallbone et al., 2016). In aquaculture, this can result in significant economic losses due to reduced yields and increased susceptibility to diseases (Doyle, 2016). Furthermore, the loss of genetic diversity can limit the ability of a population to adapt to changing environmental conditions, such as climate change or emerging pathogens. This lack of adaptability can threaten the long-term sustainability of the aquaculture industry (Regan et al., 2021; Li, 2022). Therefore, to ensure the sustainable maintenance and growth of the aquaculture industry, further research is required to investigate the negative effects of reduced genetic diversity and develop strategies to mitigate these impacts (Robinson et al., 2023). This may include implementing breeding programs (i.e., genetic rescue strategy) that prioritize the maintenance of genetic diversity, exploring the use of wild broodstock to introduce new genetic material, and developing tools for monitoring and managing genetic diversity in aquaculture populations (Li, 2022; Robinson et al., 2023).
In conclusion, our microsatellite analysis of the T. rubripes-chinensis species complex challenges the current taxonomic distinction between T. rubripes and T. chinensis. The lack of genetic differentiation among wild populations, despite the substantial morphological variation, suggests that these nominal species may represent a single species expressing high phenotypic variation. This finding has important implications for the management and conservation of wild Takifugu populations, as well as for the regulation of the pufferfish food trade, given the differences in toxicity between species. In stark contrast, the significant genetic divergence observed in farm-bred T. rubripes highlights the profound impact of aquaculture practices on the genetic makeup of captive populations. The rapid genetic drift and potential inbreeding associated with selective breeding underscore the need for genetic monitoring in pufferfish aquaculture to maintain the health and productivity of farmed stocks. Future research should focus on resolving the taxonomic status of the T. rubripes-chinensis complex using high-resolution genomic markers and investigating the genetic basis of toxicity variation within this group. Furthermore, the development of sustainable breeding programs that prioritize the maintenance of genetic diversity is essential to ensure the long-term viability of the Takifugu aquaculture industry. Our study emphasizes the importance of integrating genetic data into the management of both wild and captive populations to promote the conservation and sustainable exploitation of these economically and culturally significant pufferfishes. Given the lack of genetic differentiation between what were previously considered distinct species, conservation efforts should focus on preserving the overall genetic diversity of this species complex rather than managing them as separate taxa. Conservation strategies should include establishing genetic monitoring protocols, creating a management plan that minimize inbreeding, and potentially implementing genetic rescue approaches by periodically introducing wild genetic material. These conservation measures would not only help preserve this economically important species complex but also maintain its ecological functions in marine ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because We analyzed commercially available fishes.
Author contributions
NK: Formal analysis, Investigation, Writing – original draft. HK: Investigation, Writing – review & editing. HH: Investigation, Writing – review & editing. WL: Writing – review & editing. HT: Writing – review & editing. KC: Writing – review & editing, Funding acquisition, Supervision. HJ: Conceptualization, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by a grant (20163MFDS641) from the Ministry of Food and Drug Safety of Korea in 2024. This study was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2019-NR040078).
Acknowledgments
We are grateful to the staff of the Shellfish Aquaculture and Research Laboratory of Jeju National University for their help in the sampling and analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1437097/full#supplementary-material
Supplementary Figure 1 | Maximum likelihood tree of Takifugu species based on mitochondrial COI sequences. Bootstrap values shown at branch nodes. Accession numbers and species name are provided for each terminal branch.
Supplementary Figure 2 | PopTree plot showing the relationship among five groups. The numbers in the node represent the bootstrap values.
Supplementary Table 1 | Morphological characteristics of two Takifugu species and HSP types.
References
Baek J. I., Han K. H., Lee S. H., Kim J. K. (2018). Morphological and molecular comparison among three species and one unidentified Takifugu species. Korean J. Fish. Aquat. Sci. 51, 404–410. doi: 10.5657/KFAS.2018.0404 (in Korean with English abstract)
Barría A., Peñaloza C., Papadopoulou A., Mahmuddin M., Doeschl-Wilson A., Benzie J. A. H., et al. (2023). Genetic differentiation following recent domestication events: A study of farmed Nile tilapia (Oreochromis niloticus) populations. Evol. Appl. 16, 1220–1235. doi: 10.1111/eva.13560
Chen D., Zhang Q., Tang W., Huang Z., Wang G., Wang Y., et al. (2020). The evolutionary origin and domestication history of goldfish (Carassius auratus). Proc. Natl. Acad. Sci. 117, 29775–29785. doi: 10.1073/pnas.2005545117
Cui J. Z., Shen X. Y., Yang G. P., Gong Q. L., Gu Q. Q. (2005). Characterization of microsatellite DNAs in Takifugu rubripes genome and their utilization in the genetic diversity analysis of T. rubripes and T. pseudommus. Aquaculture 250, 129–137. doi: 10.1016/j.aquaculture.2005.04.041
Doyle R. W. (2016). Inbreeding and disease in tropical shrimp aquaculture: a reappraisal and caution. Aquac. Res. 47, 21–35. doi: 10.1111/are.12472
Earl D. A., vonHoldt B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Excoffier L., Lischer H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
FAO (2024). “Fishery and aquaculture statistics,” in Global aquaculture production Quantity. Available at: https://www.fao.org/fishery/statistics-query/en/aquaculture/aquaculture_quantity (Accessed December 1, 2024).
Frankham R., Briscoe D. A., Ballou J. D. (2002). Introduction to conservation genetics (United Kingdom: Cambridge University Press).
Furukawa S., Takeshima H., Otaka T., Mitsuboshi T., Shirasu K., Ikeda D., et al. (2004). Isolation of microsatellite markers by in silico screening implicated for genetic linkage mapping in Japanese pufferfish Takifugu rubripes. Fish. Sci. 70, 620–628. doi: 10.1111/j.1444-2906.2004.00849.x
Goudet J. (2001).FSTAT, a program to estimate and test gene diversity and fixation indices (version 2.9.3). Available online at: https://cir.nii.ac.jp/crid/1572824500657842432 (Accessed August 3, 2022). http://www2.unil.ch/popgen/softwares/fstat.htm.
Guardone L., Maneschi A., Meucci V., Gasperetti L., Nucera D., Armani A. (2020). A global retrospective study on human cases of tetrodotoxin (TTX) poisoning after seafood consumption. Food Rev. Int. 36, 645–667. doi: 10.1080/87559129.2019.1669162
Jamieson I. G., Allendorf F. W. (2012). How does the 50/500 rule apply to MVPs? Trends Ecol. Evol. 27, 578–584. doi: 10.1016/j.tree.2012.07.001
Kai W., Kikuchi K., Fujita M., Suetake H., Fujiwara A., Yoshiura Y., et al. (2005). A genetic linkage map for the tiger pufferfish, Takifugu rubripes. Genetics 171, 227–238. doi: 10.1534/genetics.105.042051
Kim J.-H., Gong Q.-L., Mok J.-S., Min J.-G., Lee T.-S., Park J.-H. (2003). Characteristics of puffer fish poisoning outbreaks in Korea, (1991-2002). J. Food Hyg. Saf. 18, 133–138 (in Korean with English abstract).
Kopelman N. M., Mayzel J., Jakobsson M., Rosenberg N. A., Mayrose I. (2015). CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191. doi: 10.1111/1755-0998.12387
Li C. (2022). Conservation of genetic resources for sustainable aquaculture. J. World Aquac. Soc 53, 4–7. doi: 10.1111/jwas.12875
Liu K., Sun H., Zhao X., Wang C., An C., Li A., et al. (2024). DNA barcoding, identification, and validation of the pufferfish (Order: Tetraodontiformes) in China coastal waters. Ecol. Evol. 14, e10944. doi: 10.1002/ece3.10944
Matsuura K. (2017). Pufferfishes and their allies of Japan (Hiratsuka, Japan: Tokai University Press).
Meirmans P. G. (2020). GENODIVE version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20, 1126–1131. doi: 10.1111/1755-0998.13145
Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Ministry of Health, Labour and Welfare of Japan (2020). Available online at: https://www.mhlw.go.jp/content/11130500/000682445.pdf (in Japanese) (Accessed December 1, 2024).
Nei M., Tajima F. (1981). Genetic drift and estimation of effective population size. Genetics 98, 625–640. doi: 10.1093/genetics/98.3.625
Noguchi T., Arakawa O. (2008). Tetrodotoxin – Distribution and accumulation in aquatic organisms, and cases of human intoxication. Mar. Drugs 6, 220–242. doi: 10.3390/md6020220
Noguchi T., Arakawa O., Takatani T. (2006a). Toxicity of pufferfish Takifugu rubripes cultured in netcages at sea or aquaria on land. Comp. Biochem. Physiol. Part D Genomics Proteomics 1, 153–157. doi: 10.1016/j.cbd.2005.11.003
Noguchi T., Arakawa O., Takatani T. (2006b). TTX accumulation in pufferfish. Comp. Biochem. Physiol. Part D Genomics Proteomics 1, 145–152. doi: 10.1016/j.cbd.2005.10.006
Park Y. J., Lee M. N., Noh J. K., Noh E. S., Kang J. H., Park J. Y., et al. (2020). Classification of Takifugu rubripes, T. chinensis and T. pseudommus by genotyping-by-sequencing. PLoS One 15, e0236483. doi: 10.1371/journal.pone.0236483
Peacock M. M., Beard K. H., O’neill E. M., Kirchoff V. S., Peters M. B. (2009). Strong founder effects and low genetic diversity in introduced populations of Coqui frogs. Mol. Ecol. 18, 3603–3615. doi: 10.1111/j.1365-294X.2009.04308.x
Peakall R., Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Prunier J. G., Veyssière C., Loot G., Blanchet S. (2023). Comparing the utility of microsatellites and single nucleotide polymorphisms in conservation genetics: Insights from a study on two freshwater fish species in France. Diversity 15, 681. doi: 10.3390/d15050681
Ralls K., Ballou J. D., Dudash M. R., Eldridge M. D. B., Fenster C. B., Lacy R. C., et al. (2018). Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 11, e12412. doi: 10.1111/conl.12412
Regan T., Bean T. P., Ellis T., Davie A., Carboni S., Migaud H., et al. (2021). Genetic improvement technologies to support the sustainable growth of UK aquaculture. Rev. Aquac. 13, 1958–1985. doi: 10.1111/raq.12553
Reza M., Kinoshita S., Furukawa S., Mochizuki T., Watabe S. (2011). Microsatellite and mitochondrial DNA analyses reveal no genetic difference between two pufferfish species torafugu Takifugu rubripes and karasu T. chinensis. Fish. Sci. 77, 59–67. doi: 10.1007/s12562-010-0310-y
Reza S., Furukawa S., Mochizuki T., Matsumura H., Watabe S. (2008). Genetic comparison between torafugu Takifugu rubripes and its closely related species karasu Takifugu chinensis. Fish. Sci. 74, 743–754. doi: 10.1111/j.1444-2906.2008.01585.x
Robinson N. A., Robledo D., Sveen L., Daniels R. R., Krasnov A., Coates A., et al. (2023). Applying genetic technologies to combat infectious diseases in aquaculture. Rev. Aquac. 15, 491–535. doi: 10.1111/raq.12733
Santini F., Nguyen M. T. T., Sorenson L., Waltzek T. B., Lynch Alfaro J. W., Eastman J. M., et al. (2013). Do habitat shifts drive diversification in teleost fishes? An example from the pufferfishes (Tetraodontidae). J. Evol. Biol. 26, 1003–1018. doi: 10.1111/jeb.12112
Smallbone W., van Oosterhout C., Cable J. (2016). The effects of inbreeding on disease susceptibility: Gyrodactylus turnbulli infection of guppies, Poecilia reticulata. Exp. Parasitol. 167, 32–37. doi: 10.1016/j.exppara.2016.04.018
Takagi M., Sakai H., Takei T., Kaneko Y. (2001). Mechanism of inbreeding and genetic diversity of the tiger puffer Takifugu rubripes. Jpn. Fish. Resour. Conserv. Assoc. Rep. 41–47 (in Japanese).
Takagi M., Sato J., Monbayashi C., Aoki K., Tsuji T., Hatanaka H., et al. (2003). Evaluation of microsatellites identified in the tiger puffer Takifugu rubripes DNA database. Fish. Sci. 69, 1085–1095. doi: 10.1111/j.0919-9268.2003.00732.x
Takahashi H., Kakioka R., Nagano A. J. (2023). Development and validation of novel SNP markers for the rapid identification of natural hybrids of the 11 closely related pufferfish species (Takifugu spp.) distributed in Japan. Aquac. Rep. 31, 101650. doi: 10.1016/j.aqrep.2023.101650
Takezaki N., Nei M., Tamura K. (2014). POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 31, 1622–1624. doi: 10.1093/molbev/msu093
Van Oosterhout C., Hutchinson W. F., Wills D. P. M., Shipley P. (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. N. (2005). DNA barcoding Australia’s fish species. Philos. Trans. R. Soc B Biol. Sci. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Watari T., Tachibana T., Okada A., Nishikawa K., Otsuki K., Nagai N., et al. (2021). A review of food poisoning caused by local food in Japan. J. Gen. Fam. Med. 22, 15–23. doi: 10.1002/jgf2.384
Yamanoue Y., Miya M., Matsuura K., Miyazawa S., Tsukamoto N., Doi H., et al. (2009). Explosive speciation of Takifugu: another use of Fugu as a model system for evolutionary biology. Mol. Biol. Evol. 26, 623–629. doi: 10.1093/molbev/msn283
Ye Y., Ren W., Zhang S., Zhao L., Tang J., Hu L., et al. (2022). Genetic diversity of fish in aquaculture and of common carp (Cyprinus carpio) in traditional rice–fish coculture. Agriculture 12, 997. doi: 10.3390/agriculture12070997
Keywords: Takifugu, microsatellites, population structure, genetic drift, domestication
Citation: Kajino N, Kim H-J, Hong H-K, Lee W-O, Takahashi H, Choi K-S and Jeon H-B (2025) Genetic differentiation between captive and wild populations induced by selective breeding in morphologically diverse Takifugu pufferfish. Front. Mar. Sci. 12:1437097. doi: 10.3389/fmars.2025.1437097
Received: 23 May 2024; Accepted: 04 April 2025;
Published: 29 April 2025.
Edited by:
Mhd Ikhwanuddin, University of Malaysia Terengganu, MalaysiaReviewed by:
Khaled Mohammed Geba, Menoufia University, EgyptIoannis A. Giantsis, Aristotle University of Thessaloniki, Greece
Seunghyun Kang, Korea Polar Research Institute, Republic of Korea
Copyright © 2025 Kajino, Kim, Hong, Lee, Takahashi, Choi and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwang-Sik Choi, c2tjaG9pQGplanVudS5hYy5rcg==; Hyung-Bae Jeon, amhiNTY5NkBnbWFpbC5jb20=
 Nobuhisa Kajino
Nobuhisa Kajino Hyoun-Joong Kim
Hyoun-Joong Kim Hyun-Ki Hong
Hyun-Ki Hong Wan-Ok Lee1
Wan-Ok Lee1 Kwang-Sik Choi
Kwang-Sik Choi Hyung-Bae Jeon
Hyung-Bae Jeon