- 1Fisheries College, Jimei University, Xiamen, China
- 2Mangrove Conservation Foundation, Shenzhen, China
- 3Quanzhou Bay Estuary Wetland Nature Reserve Development Center, Quanzhou Forestry Bureau, Quanzhou, China
- 4Hunan Shuitiandi Environmental Technology Co., Ltd., Changsha, China
- 5Sustainable Ocean Governance Centre, National Sun Yat-sen University, Kaohsiung, Taiwan
Background: The mangrove ecosystem has been severely threatened by the invasive species Spartina alterniflora for a long time. The macrobenthos diversity, as a biological indicator for assessing ecosystem sensitivity, can respond sensitively to the removal of the invasive S. alterniflora. Traditional morphological method has been commonly used to study macrobenthos diversity. However, it is difficult to comprehensively and accurately assess using traditional morphological method alone. Few studies have systematically assessed the effectiveness of environmental DNA in monitoring the diversity of macrobenthos in mangrove ecosystems.
Methods: Traditional morphological method and eDNA metabarcoding (using COI primers) were combined to analyze changes in macrobenthos diversity in September (before S. alterniflora removal), October (after S. alterniflora removal but before mangrove planting), and December (after S. alterniflora removal and mangrove planting) of 2022 at the Quanzhou Bay Mangrove Wetland Nature Reserve, Fujian Province, China.
Results: The results showed that 26 species were identified using traditional morphological method, while 212 species were identified through eDNA metabarcoding, especially more Arthropod were detected. However, no species were found to be common between the two methods at the species level. In terms of abundance, Arthropods and Annelids were the dominant groups for two methods, both 28.38% in the traditional morphological method and 38.38% and 32.94% in the eDNA metabarcoding, respectively. Additionally, the traditional morphological method indicated that following the removal of S. alterniflora, species richness, density, biomass, and diversity indices of macrobenthos initially declined, but subsequently showed a consistent recovery trend.
Discussion: Our study suggests that eDNA metabarcoding has tremendous potential to monitor macrobenthos diversity, but shows low consistency in species with traditional method. For broader application in macrobenthos diversity using eDNA metabarcoding, further optimization is needed. Additionally, the results suggested that after the removal of S. alterniflora, macrobenthos diversity initially declined but gradually recovered, and long-term monitoring is warranted in order to better understand the changes in macrobenthos diversity with planting of mangroves.
1 Introduction
Macrobenthos refers to invertebrate taxa that inhabit the bottom of water bodies or sediments, with an individual size typically greater than 500 μm (Nalepa and Robertson, 1981; Buss and Borges, 2008; Ji et al., 2022), and these organisms primarily include members of Annelida, Mollusca, Arthropoda, Cnidaria, Chordata, and Sipuncula, among others (Tagliapietra and Sigovini, 2010). As an important component of the coastal salt marsh wetland ecosystem, macrobenthos serve as a key nutrient hub and a key link in the food web (Herman et al., 1999; Zou et al., 2018; Talley and Levin, 2001). Due to the strong regional specificity, limited migration ability, and high sensitivity to environmental changes, macrobenthos are widely regarded as biological indicators for assessing ecosystem sensitivity. Their presence and abundance can serve as credible indicators of the overall health of the wetland ecosystem (Lv et al., 2018). Traditional morphological method has been commonly used to study macrobenthos diversity. However, due to the neglect errors, difficulties in classification, and some flaws in survey techniques, it is challenging to conduct a comprehensive and complete assessment (Thomsen and Willerslev, 2015; Elbrecht and Leese, 2017; Wang K. et al., 2023). The eDNA metabarcoding is a technique for rapidly and accurately identification of multiple species by extracting DNA directly from environmental samples, like sediments and aquatic samples, and it has been widely applied to groups like microorganisms, plankton, and fish (Pawlowski et al., 2018; Duarte et al., 2021). However, the monitoring of macrobenthos diversity using eDNA started relatively late (Wang T. et al., 2023; Ji et al., 2021b; Pawlowski et al., 2022; Meng et al., 2019), particularly in mangrove habitats, with research mainly focusing on certain groups of macrobenthos rather than the community. For example, Bravo et al. (2021) assessed the diversity of gastropods and crabs in the mangroves of Hong Kong, China, and the Greater Bay Area, identifying 44 species of gastropods and 58 species of brachyurans in Hong Kong’s mangroves, and 5 new records of gastropods and 7 new records of crabs in the Greater Bay Area (Bravo et al., 2021). Liu et al. (2018) utilized eDNA to investigate the insect fauna within the mangrove ecosystems located on Hainan Island, China, and successfully identified pests (Liu et al., 2018). Accordingly, it is worth exploring the applicability of eDNA metabarcoding to systematically assess the diversity of macrobenthos communities in mangrove ecosystems merits exploration.
Nearly half of mangroves have been destroyed in the world, and the protection and restoration of mangroves has become an important task for global environmental conservation (Polidoro et al., 2010; Zheng et al., 2023). S. alterniflora, a herbaceous perennial native to the east coast of North America and the Gulf of Mexico, can tolerate high salinity and varying temperatures. With the strong adaptability, S. alterniflora rapidly proliferates in new environments (Cao et al., 2021; An et al., 2007). Its rapid growth has invaded the living space of mangroves, resulting in a sharp decline in mangrove biodiversity and ecosystem functions (Jiang et al., 2021). S. alterniflora has become rampant along the coastal areas and is a serious ecological threat to the security of coastal wetlands. Since 2022, the removal of S. alterniflora has been comprehensively carried out along coastal zones of China (Ge et al., 2020). Generally, the removal of S. alterniflora includes three main methods: physical, chemical, and biological control (Zhou et al., 2015). Chemical control, such as herbicides, is simple and easy to implement but may lead to environmental pollution. Biological control methods are not yet sufficiently developed and may carry a potential risk of new invasive species invading the ecology (Xie et al., 2018; Zhao et al., 2019). Therefore, physical control methods, such as burning, mowing, plowing, and flooding, are most commonly practiced in China (Yuan et al., 2011). The “mowing + plowing” is one of the main and most widely applied methods. Therefore, studying the changes in macrobenthos after the “mowing + plowing” of S. alterniflora will provide important references for the restoration and management of macrobenthos resources (Patten and O’Casey, 2007).
The Quanzhou Bay Mangrove Nature Reserve is located in Quanzhou, Fujian Province, at the estuaries of the Jinjiang and Luoyang Rivers. It covers a total of 7,065.31 hectares, primarily consists of mangroves, mudflats, wetlands, water bodies, and some artificial environments, serves as a critical role to protect mangrove estuarine wetlands and typifies the subtropical estuarine wetlands in China (Ji et al., 2015). Historically, mangroves flourished in the reserve, establishing it as the largest artificial mangrove restoration area on the southeast coast of China. However, the mangrove area has been increasingly affected by human disturbances, including land reclamation, aquaculture, and the introduction of S. alterniflora (Lu et al., 2018). S. alterniflora was introduced to the region in 1982, and by 2005, its coverage had expanded to 563 hectares (Ji et al., 2015). To safeguard the ecological security of this wetland system, the Quanzhou Bay Reserve initiated physical removal of S. alterniflora using “mowing + plowing” in October 2022, followed by the planting of mangrove species. The planting density was 4 seedlings per square meter, with the primary species being Kandelia obovata, Avicennia marina, Aegiceras corniculatum, Bruguiera gymnorhiza, and Rhizophora stylosa (Hu et al., 2025). Therefore, the traditional morphological method and eDNA metabarcoding were applied to detect macrobenthos diversity changes in September (before S. alterniflora removal), October (after S. alterniflora removal but before mangrove planting), and December (after S. alterniflora removal and mangrove planting), and analyze the changes in macrobenthos diversity before and after S. alterniflora removal and mangrove planting in the nature reserve. The objectives of this study are: (1) to assess the effectiveness of eDNA metabarcoding technology in monitoring macrobenthos diversity in mangrove, and (2) to monitor the short-term changes in macrobenthos communities during the S. alterniflora removal and mangrove restoration process.
2 Materials and methods
2.1 Sampling station setting and field sampling methods
This study included three sampling events: before the removal of S. alterniflora (September 2022), after its removal but before mangrove planting (October 2022), and after mangrove planting (December 2022). In the area where S. alterniflora was removed, six sampling stations (A, B, C, D, E, F) were set up based on the varying tidal zone adaptability of different mangrove species. Station A and station B were located at high tide, stations C and F were located at medium tide, and stations D and E were located at low tide. The diagram of the sampling stations is shown in Figure 1.
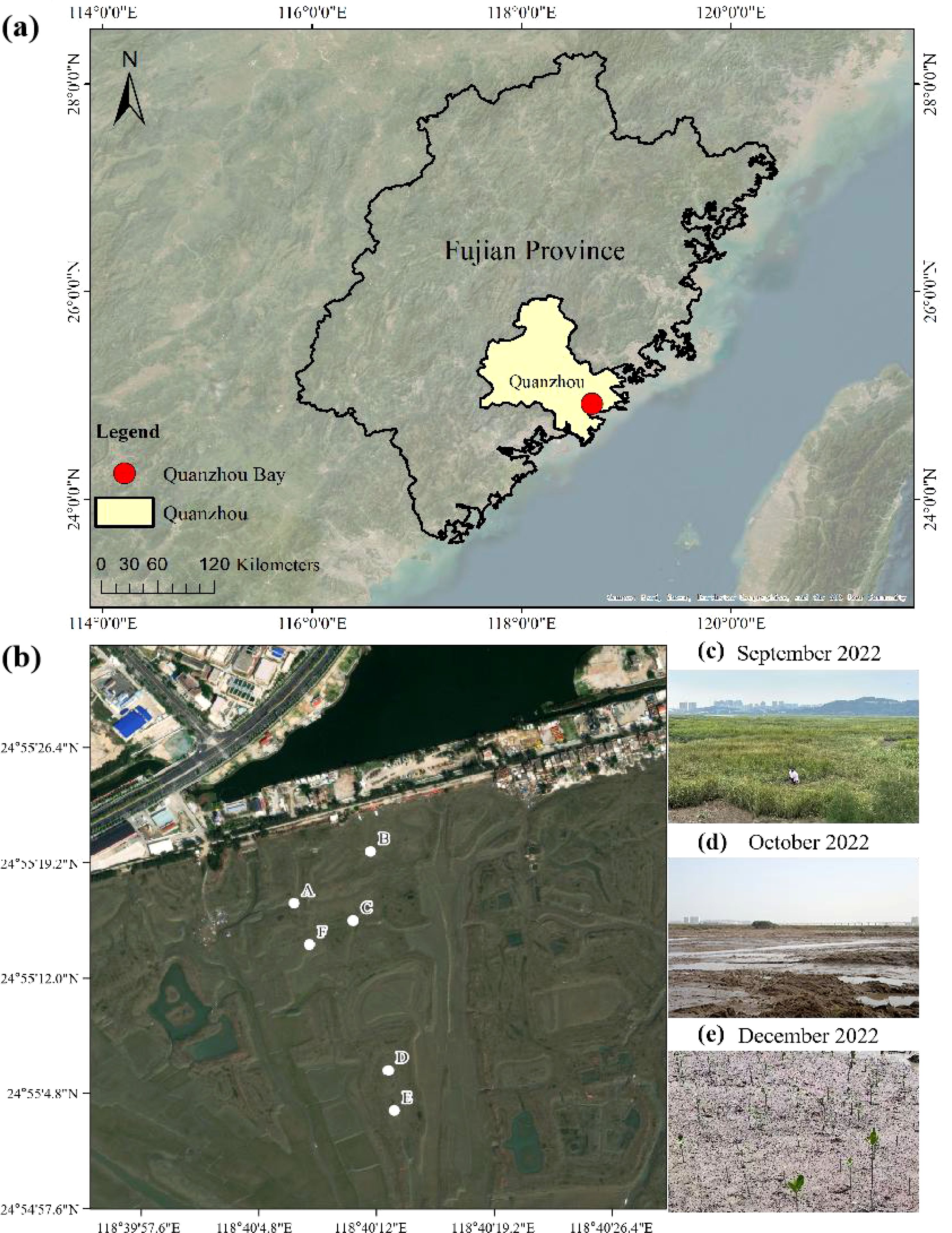
Figure 1. Map of mangrove wetland study area (a) and sampling station (b) in Quanzhou Bay. (c-e) represents the sampling station overview in September, October and December respectively in 2022.
2.2 Traditional morphological method
Macrobenthos samples were collected using a frame with dimensions of 30 cm × 30 cm × 30 cm, and sorted with a sieve of 0.5 mm mesh size. The sorted biological samples were immediately fixed in 95% ethanol. After being brought back to the laboratory, different species were identified, counted, and weighed. All species were identified to the species or genus level whenever possible. The biomass was measured with an accuracy of 0.001 g. Sample processing, preservation, and weighing were conducted following the Marine Biological Survey Specification (GB/T 12763.6-2007) (China Marine Standardization Technical Committee, 2007).
2.3 eDNA metabarcoding
2.3.1 eDNA sampling
Surface sediments (0 - 1 cm) were collected from each site and immediately placed into 5 ml sterile centrifuge tubes, and then stored in liquid nitrogen. All samples were stored at - 80°C until DNA extraction (DiBattista et al., 2020).
2.3.2 DNA extraction, PCR amplification and Illumina Miseq sequencing
The FastDNA® Spin Kit for soil was used to extract genomic DNA from sediment samples 1.5% agarose gel electrophoresis was applied to detect the quality of DNA extraction, and the Qubit® dsDNA HS Assay Kit was used to detect DNA concentration and purity. The DNA samples used for PCR amplification were uniformly diluted to a concentration of 20 ng/μL. The COI gene was amplified by PCR for the mitochondrial gene variable region using the primer pairs mlCOIintF (5’-GGWACWGGWTGAACWGTWTAYCCYCC-3’) and jgHCO2198 (5’- TANACYTCNGGRTGNCCRAARAAYCA -3’) (Leray et al., 2013). The following is the principle of PCR amplification: initial denaturation at 94°C for 3 minutes, followed by 25 cycles of denaturing at 94°C for 15 seconds, annealing at 46°C for 15 seconds and extension at 72°C for 10 seconds, and single extension at 72°C for 3 minutes. The PCR amplification system was as follows:TransStart FastPfu buffer solution 2.5μL, dNTPs 2μL, forward primer (10uM) 0.5μL, reverse primer (10uM) 0.5μL, TransStart Taq DNA polymerase 0.5μL, template eDNA 1uL, nuclease-free water supplementation to 25μL. The above PCR products were purified using magnetic beads. Use 1.5% agarose gel electrophoresis for detection, and the recovered products were detected and quantified by the multifunctional enzyme - labeler INFINITE F NANO+ of Tecan Company, followed by the construction of the library, which was sequenced using Illumina’s Miseq PE300 platform.
2.3.3 Sequence processing
Trimmomatic software was used for original sequencing sequences for quality control, and FLASH software was used for splicing. UPARSE software (version 7.1 http://drive5.com/uparse/) was applied for OTU clustering of sequences with 97% similarity and remove chimeras. RDP classifier (http://rdp.cme.msu.edu/) was used to categorize and annotate each sequence, and compare it with NT database. Non-target species that are typically not classified as benthos (such as bacteria, archaea and plankton) were removed. Additionally, micro- and meiobenthos, such as nematodes, rotifers, and platyhelminthes, were also removed (Han et al., 2018; Ji et al., 2021a). Target species include macrobenthos identified in previous studies or known to exist in adjacent waters, as well as species that were not previously recorded but are capable of surviving in the habitat. The species-filtering steps to identify target species for eDNA metabarcoding are detailed in Supporting information: Supplementary Figure S1. The raw reads of samples for September and October were deposited into the NCBI Sequence Read Archive (SRA) database BioProject ID: PRJNA1220310 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1220310). And the raw reads of samples for December were from PRJNA1171633 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1171633; Hu et al., 2025).
2.4 Data processing
2.4.1 Number of species
Count the number of macrobenthos species at each taxonomic level. The percentage of species richness for each group is determined by dividing the number of species in that group by the total number of species (Sun et al., 2022; Lejzerowicz et al., 2015).
2.4.2 Density and biomass
Calculation formula for the species density of macrobenthos:
Where Ni denotes the number of individuals of the ith species, and A is the sampling area.
Calculation formula for the biomass of macrobenthos:
Where Bi denotes the weight of the ith species, and A is the sampling area.
2.4.3 Relative abundance
The estimation of relative abundance in eDNA technology is based on the OTU (Operational Taxonomic Unit) sequence abundance table, while traditional method is based on species count calculation, that is, the number of species in this taxonomic group divided by the total number of species (Cilleros et al., 2019; Li et al., 2019).
2.4.4 Species diversity
Simpson Index:
Margalef Index:
Pielou Index:
Shannon Index:
Where Ni denotes the number of individuals of the ith species, N denotes a total number of individuals for all species, and S denotes species number.
2.5 Data analysis
The data were processed and analyzed utilizing ArcGIS Pro 2.8, Origin 2024, Primer 6.0. The discrepancy in environmental factors, including TN, TP, and total organic carbon (TOC), at each site across the three months was analyzed using the Kruskal-Wallis test in SPSS 26.0. The correlation analysis between the biomass and density from the traditional morphological method and the sequence abundance obtained using eDNA metabarcoding at each site across the three months was also conducted using SPSS 26.0. The sampling station map was generated using ArcGIS Pro 2.8. The species relative abundance bar chart was drawn with Origin 2024. The diversity index data were analyzed with DIVERSE in Primer 6.0.
3 Result
3.1 Community species composition of macrobenthos
A total of 26 species belonging to 5 phyla, 6 classes, 6 orders and 11 families were identified by traditional morphological method in September, October and December 2022 (Table 1). In September 2022, a total of 11 species of macrobenthos were recorded, belonging to 4 phyla, 4 classes, 3 orders and 4 families. In October 2022, a total of 6 species of macrobenthos were recorded, belonging to 4 phyla, 4 classes, 4 orders and 6 families. In September, 82% of the species were eliminated, and 4 new species were recorded. Including Rhinogobius brevirostris, Alpheus japonicus, Neanthes glandicincta and Glycera chirori. 16 species belonging to 5 phyla, 6 classes, 6 orders and 10 families were recorded in December 2022. Compared with October, 12 new recorded species accounted for 75% of the species in December, and the new recorded species included Perinereis aibuhitensis, L. melanostoma, Mugilogobius abei, Metaplax longipes, Periophthalmus magnuspinnatus, Macrophthalmus erato, Perinereis nuntia Savigny, Phascolosoma esculenta, Nephthys oligobranchia, Bivalvia sp, Mediomastus sp. and Neanthes sp.
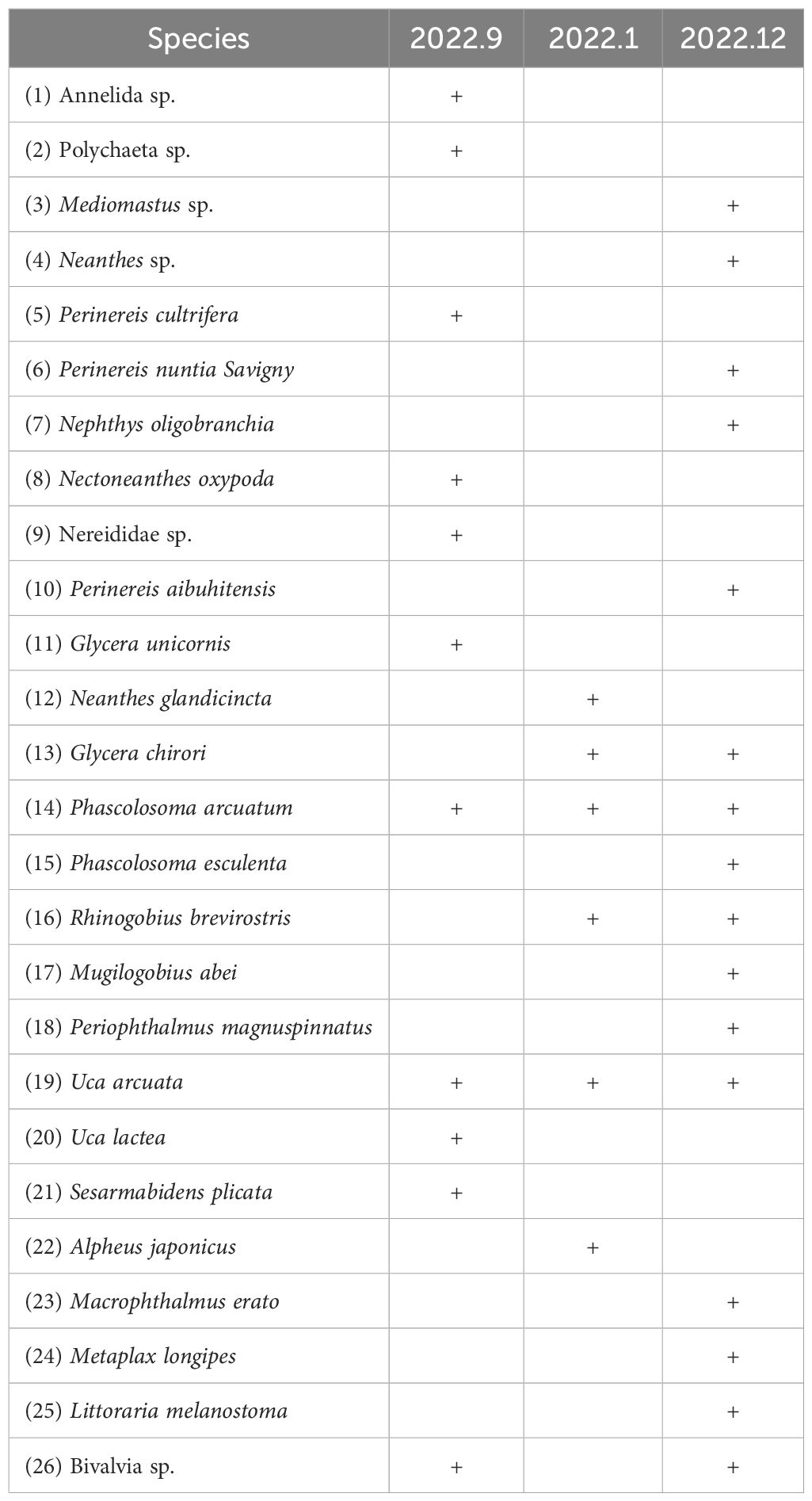
Table 1. Macrobenthos detected by traditional morphological method in Quanzhou Bay Mangrove Wetland from September to December 2022.
The eDNA metabarcoding obtained a total of 152,160 sequences over three months, with 57,886, 37,229, and 56,975 sequences recorded in September, October, and December of 2022, respectively (Supplementary Table S1). A total of 212 species were identified over the three months, with 25, 44, and 184 species identified in September, October, and December, respectively. These species belong to 6 phyla, including Sipuncula, Chordata, Annelida, Arthropoda, Mollusca, and Cnidaria. Among them, Mollusca and Arthropoda accounted for a relatively large proportion in the identification over the three months, accounting for 41.04% and 42.45%, respectively. The eDNA metabarcoding identified more Cnidaria taxa than traditional morphological method (Figure 2).
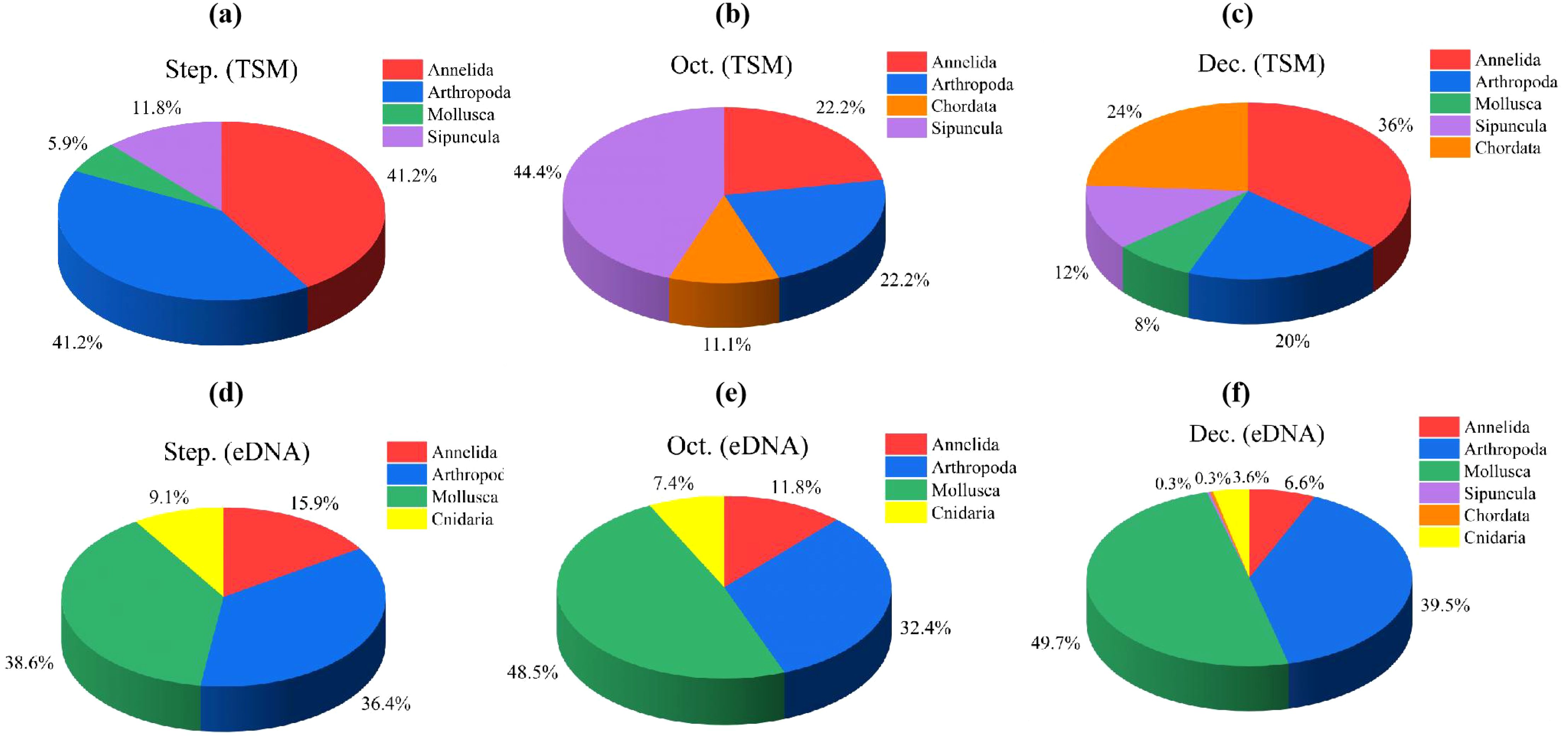
Figure 2. Species composition of macrobenthos at the phylum level detected by eDNA metabarcoding (a-c) and traditional morphological method (d-f) in September, October and December.
At the species level, the two methods did not identify the same species. At the generic level, the two methods did not co-identify taxa in September and October, and 4 genera were co-identified by the two methods in December, which were Macrophthalmus, Nephtys, Phascolosoma and Neanthes. At the family level, the two methods were used together to identify Nereididae in September, Alpheidae in October, and Gobiidae, Ocypodidae, Grapsidae, Phascolosomatidae, Nereididae and Nephtyidae in December (Figure 3).
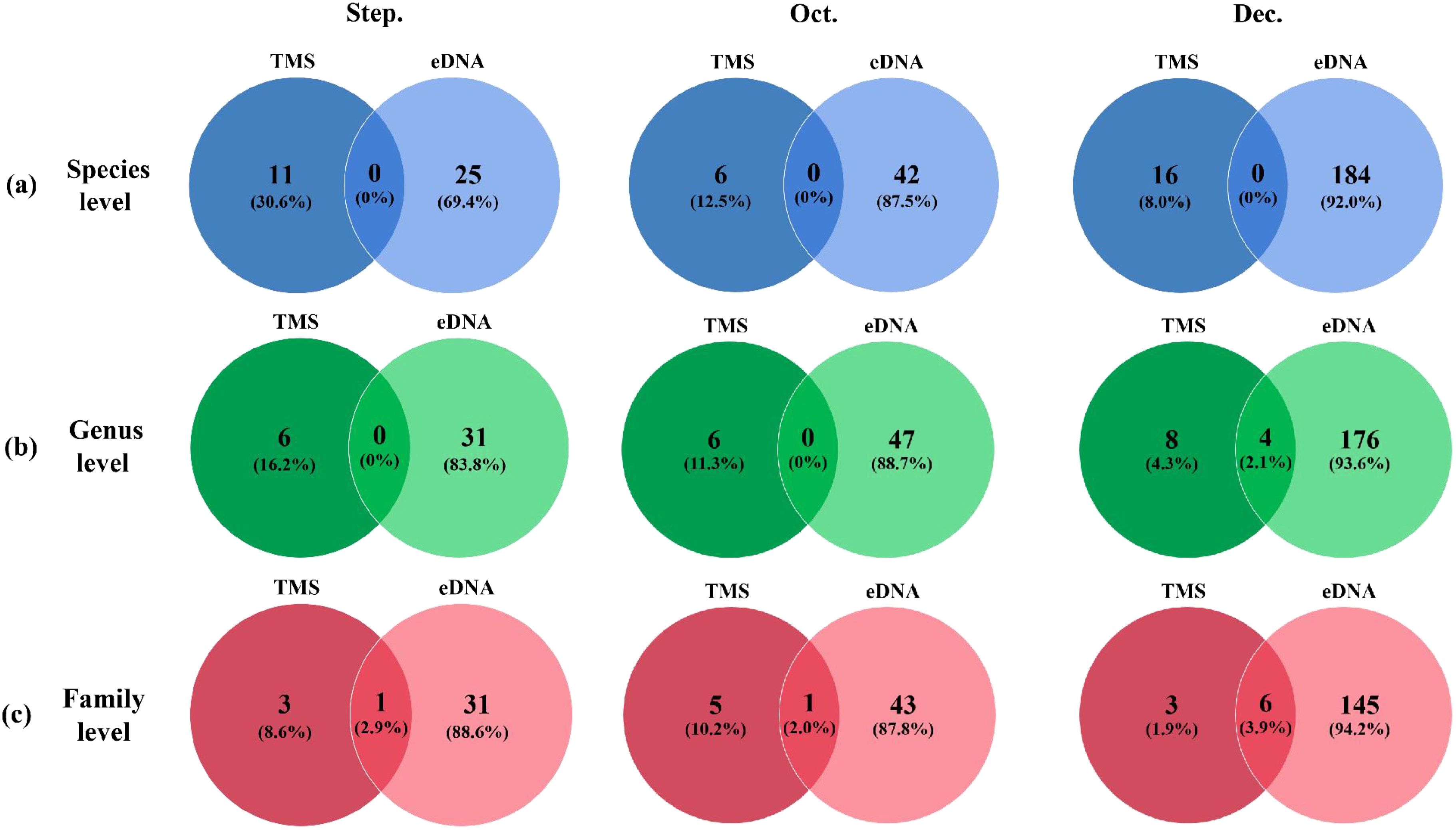
Figure 3. Venn diagrams of species composition at the species (a), genus (b) and family (c) levels by eDNA metabarcoding and traditional method from September to December in 2022.
3.2 Abundance of macrobenthos
The average density of macrobenthos was 68.55 ind./m2. The average density in September was 25.94 ind./m2, the average density in October was 14.82 ind./m2, and the average density in December was 27.79 ind./m2. The highest average density appeared in December 2022, and the lowest appeared in October 2022. The average density of macrobenthos in different months showed the following trends: December 2022 > September 2022 > October 2022 (Figure 4).
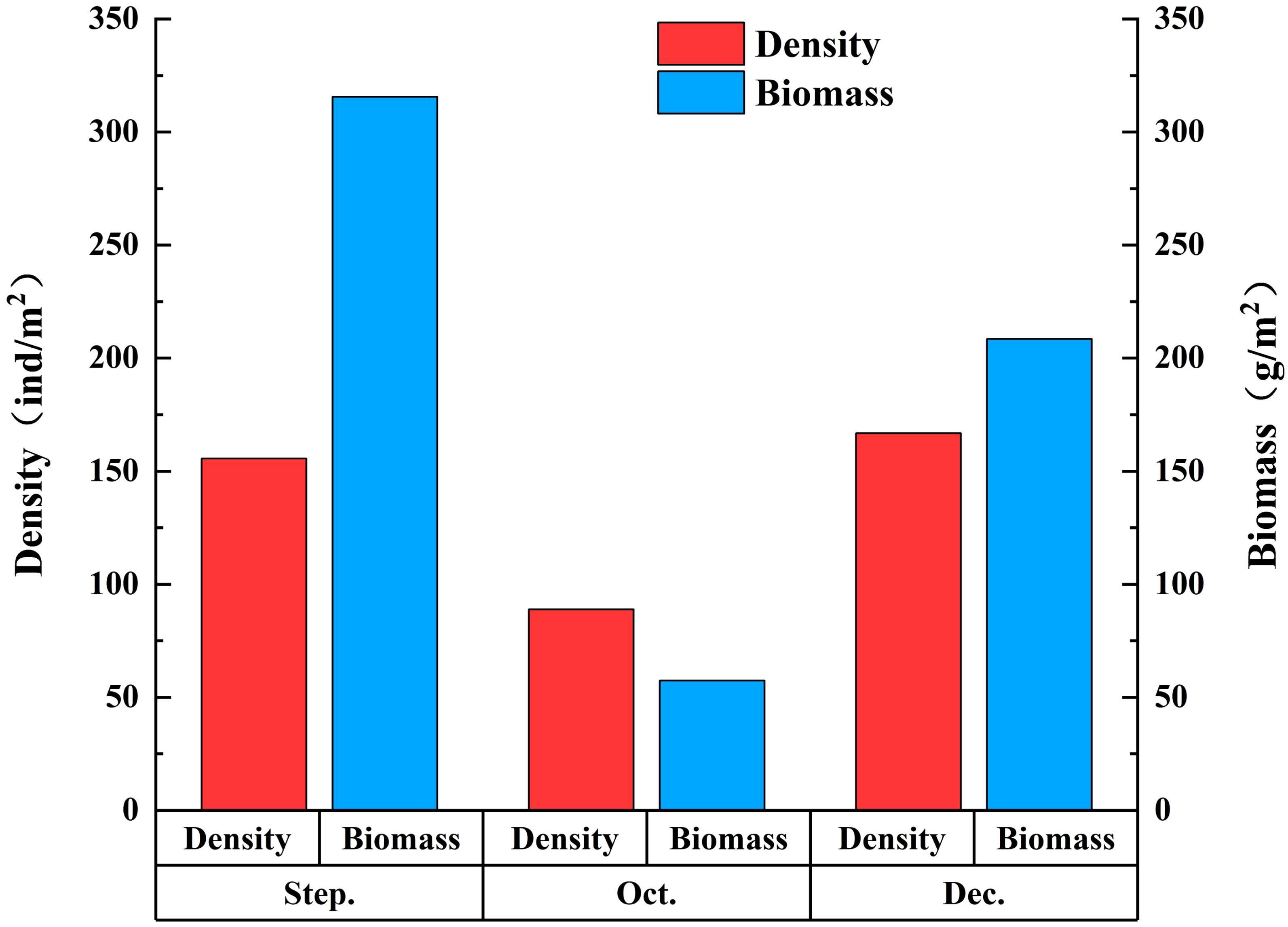
Figure 4. Density and biomass of macrobenthos detected by traditional morphological method from September to December in 2022.
The average biomass of macrobenthos was 96.90 g/m2. The average biomass in September was 52.59 g/m2, the average biomass in October was 9.57 g/m2, and the average biomass in December was 34.74 g/m2. The highest value of biomass appeared in September 2022, and the lowest appeared in October 2022. The change trends of macrobenthos biomass in different months were successively as follows: September 2022 > December 2022 > October 2022 (Figure 4).
In terms of relative abundance, the results of traditional morphological method showed that: at the phylum level, September Arthropoda were the dominant group, accounting for 46.43% of the total abundance. In October, the dominant group was Sipuncula, accounting for 68.75%. In December, the relative abundance of Annelia was the highest, accounting for 36.67%, followed by Chordate and Arthropoda, accounting for 26.67% and 20.00%, respectively (Figure 5). At the species level, the individual abundances of U. arcuata and P. arcuatum were the highest in September, accounting for 39.29% and 21.43%, respectively. P. arcuatum in October was the dominant species with 68.75% abundance. In December, the main dominant groups were R. brevirostris and N. oligobranchia, with an abundance ratio of 13.33%, followed by M. erato and P. magnuspinnatus, with an abundance ratio of 10.00% (Figure 6).
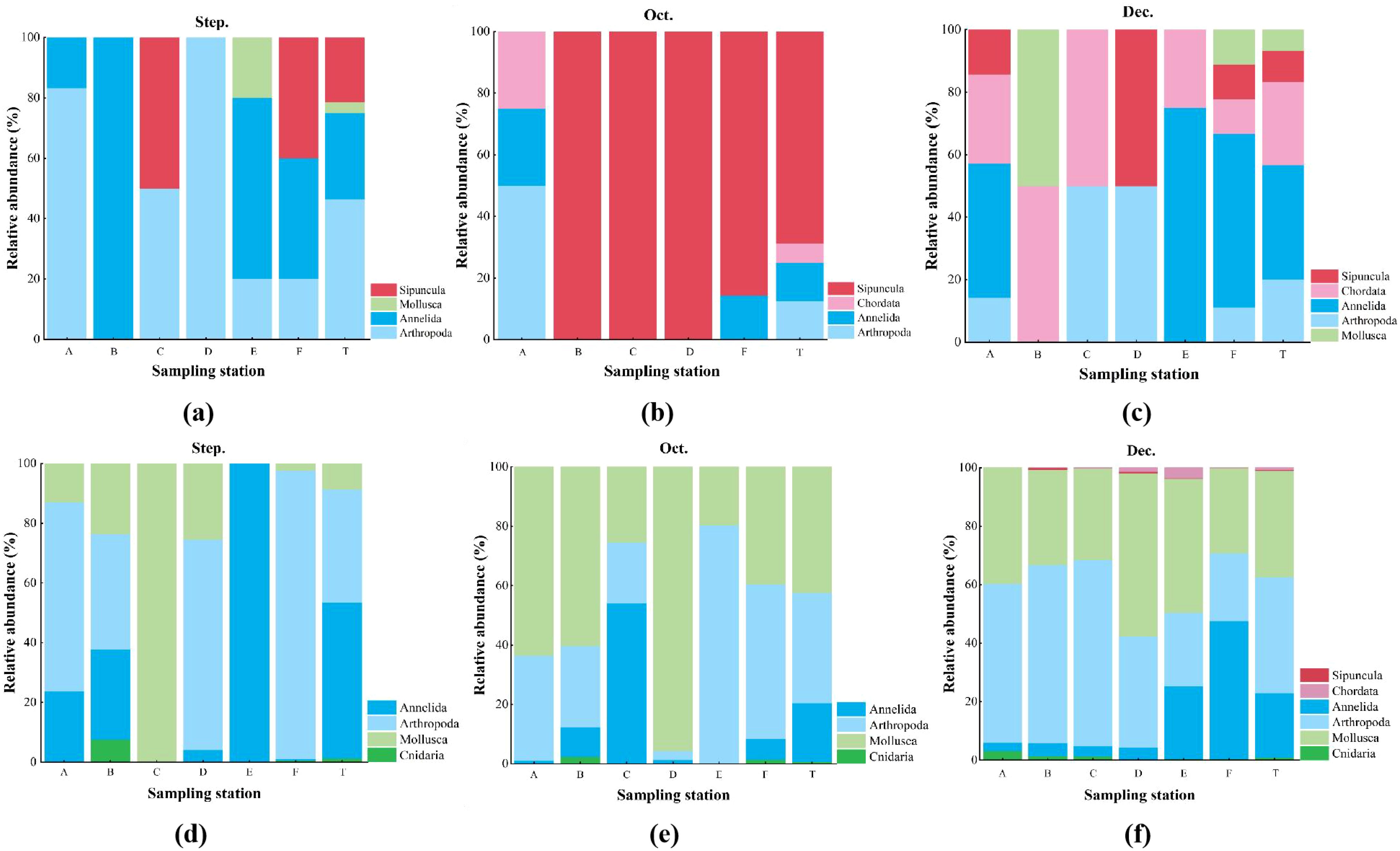
Figure 5. Relative abundance (%) of macrobenthos at the phylum level in every sampling station using traditional method (a-c) and eDNA metabarcoding (d-f) from September to December in 2022.
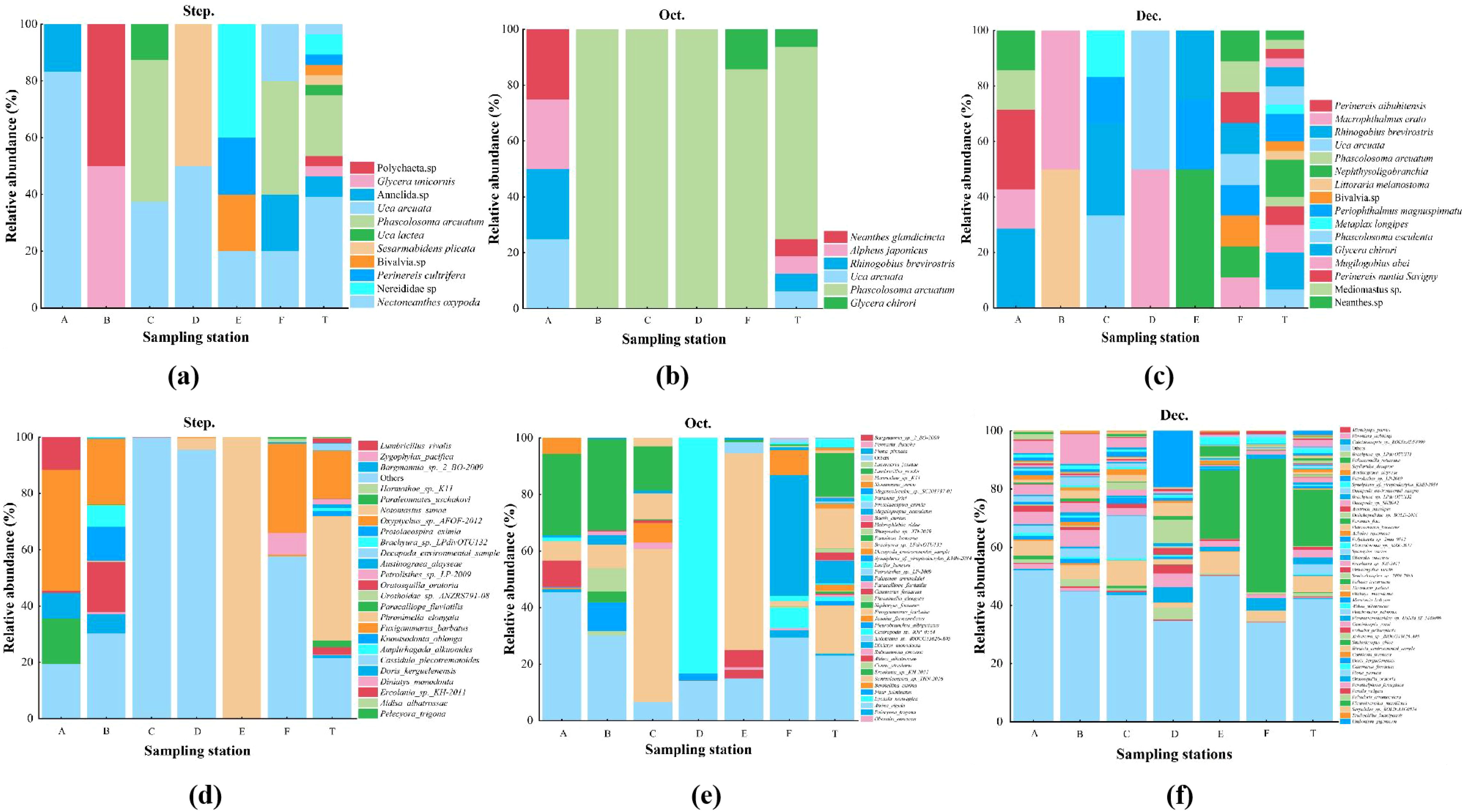
Figure 6. Relative abundance (%) of macrobenthos at the phylum level in every sampling station using traditional method (a-c) and eDNA metabarcoding (d-f) from September. Species with relative abundance lower than 2% were contained in “others” at the overall level.
The results of eDNA metabarcoding showed that the relative abundances of Annelida and Arthropoda in September were the highest at the phylum level, which were 52.18% and 37.88% respectively. The largest relative abundances in October were Mollusca and Arthropoda, with 42.36% and 37.14%, respectively. In December, the relative abundances of Arthropoda and Mollusca were the highest, 39.71% and 36.41%, respectively (Figure 5). At the species level, in September, N. sunae was the dominant species with a relative abundance of 44.09%. The dominant species in October was one species of the genus Harmothoe, which was 16.57%. The Paranai frici was the dominant species in December, which was 19.42% (Figure 6).
The correlation analysis between density and biomass from traditional method and sequence abundance from the eDNA metabarcoding indicates that, there was a significant correlation between density and biomass (P < 0.01), but no correlation between abundance and density or biomass (P > 0.05).
3.3 Diversity of macrobenthos
The Margalef index (D) in September, October and December 2022 ranged from 0 to 3.64, with the maximum value appearing at station F in December, which was 3.6. And the lowest value appearing at stations B, C, D and E in October, which was 0. Pielou index (J’) was between 0 and 1.00, with the maximum value of 1.00 at stations B and D in September and stations B, D and F in December, and the lowest value of 0 at stations B, C, D and E in October. Shannon-Wiener index (H’) was between 0 and 2.20. The maximum value was 2.20 at station F in December, and the lowest value was 0 at station B, C, D and E in October. Simpson index (1-λ) ranges from 0 to 0.89, with the maximum value appearing at station F in December, which was 0.89. And the lowest value appearing at station B, C, D, and E in October, which was 0.
In September, the maximum values of Margalef index, Shannon-Wiener index and Simpson index appeared at stations E and F, and the maximum values of Pielou index appeared at stations B and D. In October, the maximum values of the four indexes all appeared at station A, but the four indexes of station B, C, D and E were all 0. In December, the maximum values of the four indexes all appeared at station F, and the maximum values of Pielou index also appeared at station B and D. In general, the four exponential curve trends of the six stations all decreased first and then increased (Figure 7).
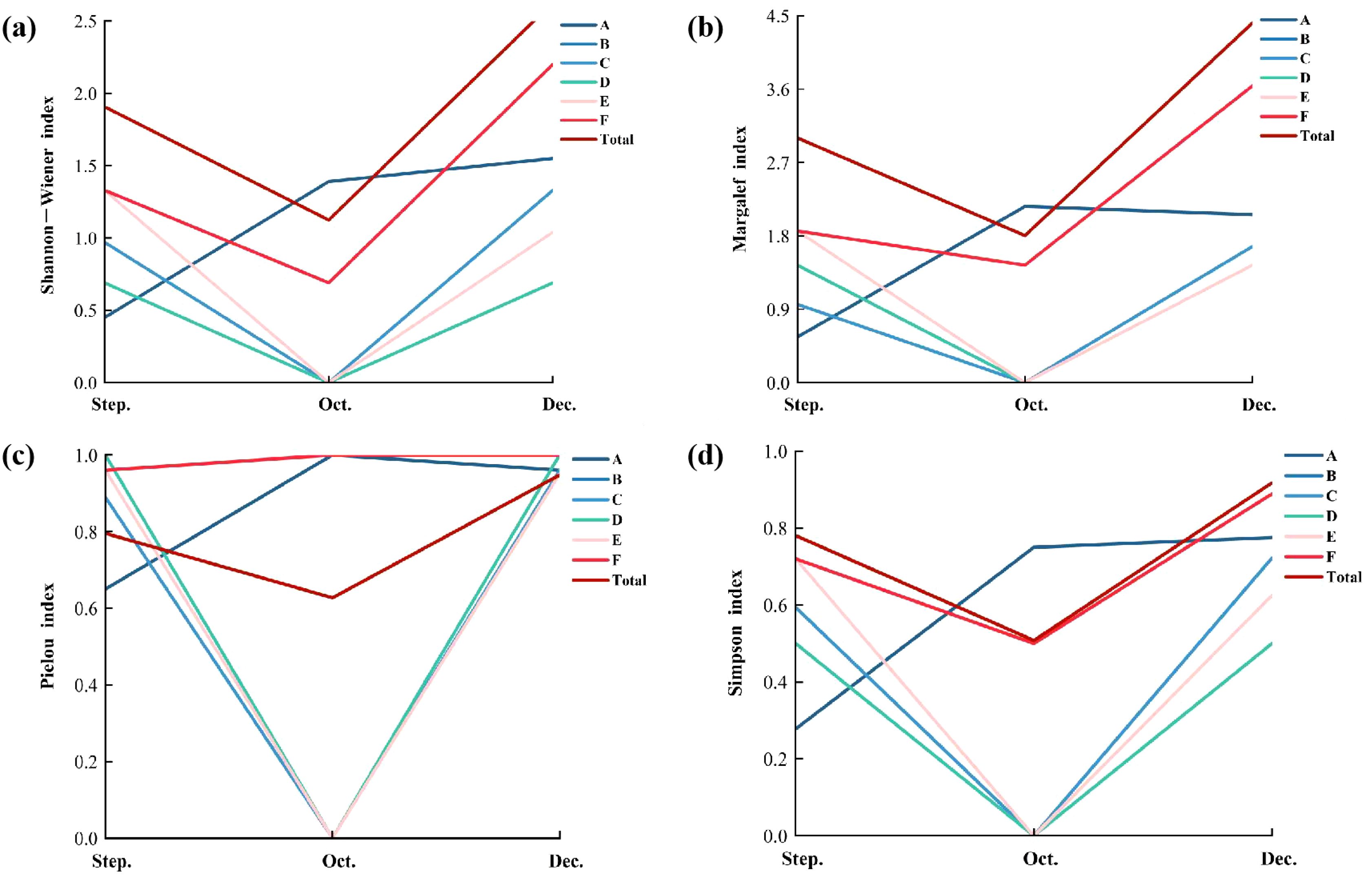
Figure 7. Macrobenthos biodiversity index using traditional morphological method, include Shannon−Wiener index (a), Margalef index (b), Pielou index (c) and Simpon index (d).
4 Discussion
4.1 eDNA metabarcoding for monitoring macrobenthos diversity in mangrove ecosystems
In this study, traditional morphological method and eDNA metabarcoding were used to identify macrobenthos diversity. In three months, 212 macrobenthos were identified using eDNA metabarcoding, significantly more than 26 species identified using traditional morphological method. In terms of species composition, the traditional morphological method was dominated by Annelida and Arthropoda, accounting for 50.00% and 23.08%, respectively. The species composition of the eDNA metabarcoding was mainly Arthropoda (42.45%) and Mollusca (41.04%). Marshall and Stepien (2020) used the eDNA metabarcoding with Coi as primer to monitor the benthos of river ecosystems, and found that eDNA metabarcoding could detect more benthos than traditional method, especially significantly more insects. Ji (2022) monitored and evaluated macrobenthos diversity in Chaobai River using eDNA metabarcoding with Coi primer, that compared with the results of traditional method, eDNA metabarcoding detected significantly more species, especially the Arthropoda species that were difficult to identify and whose life history was partly in water. In general, our study also suggests that the eDNA metabarcoding has higher species resolution in monitoring macrobenthos taxa, especially Arthropoda. This may be related to the Coi primers used in this study, which are biased towards Arthropoda (Ratnasingham and Hebert, 2007) and may overestimate the abundance or species number of Arthropoda. Compared with COI, 18S can capture a wider range of species information in a more balanced manner (Sawaya et al., 2019), therefore, the use of multi-primers for co-monitoring the macrobenthos. In addition, the same species could only be identified at the family level by the two methods in September and October, and the common groups identified were only Sericidae and Pannidae, respectively. In December, both methods co-identified species at both genus and family levels (Figure 3). Schwesig et al. (2024) used eDNA metabarcoding and the traditional method detected Odonata insects in lakes and ponds and found that only 10 of the 29 species detected by the two methods were identified together. Doloiras-Laraño et al. (2023) used eDNA metabarcoding, bulk-sample metabarcoding, and morphological approaches to compare benthic macroinvertebrate diversity from Shikoku Island, Japan. Of the 114 species detected, 90 were identified by eDNA, 29 by morphology, and only 5 species were common to both methods. According to the results of this study, the two methods have low commonality in detecting the species composition. Apart from the differences caused by the selection of primers, the accuracy of species annotation using eDNA metabarcoding is largely dependent on the integrity and quality of the reference database (Goldberg et al., 2016). Cahill et al. (2018) argued that the reference database for identifying mOTU is limited, and only species with COI sequences in the database can be assigned to a taxon. Because the reference database is incomplete, sequenced mOTU may actually come from organisms that are not present in the database. Many mOTUs cannot be assigned beyond the gate level, and even within the gate, their data sets may be assigned to the class level. Many studies have also shown that, poor species representativeness in databases may lead to misidentification and affect ecological quality assessments (Deiner et al., 2017; Hering et al., 2018; Ruppert et al., 2019). Therefore, our study shows that eDNA metabarcoding can detect more species, especially Arthropoda groups, but compared with traditional morphological method, too few species are monitored detected jointly. Multi-primers co-monitoring and the improvement of database may be the key to more comprehensive and effective monitoring of macrobenthos by using eDNA metabarcoding.
In addition, no mollusks could be collected by the traditional morphological method in October, and only partial remains of mollusks could be collected during the sampling process, but the highest species proportion and abundance detected by eDNA metabarcoding in October were Mollusca. The differences in sampling principles between the two methods may partially explain the discrepancies in the results. eDNA metabarcoding can rapidly monitor the community structure and diversity from a large number of species hybrid samples, animal residues and environmental samples with extremely high classification resolution. Following the “mowing + plowing” of the study area, a large number of snail residues were exposed, which increased the release of mollusk DNA. However, the traditional morphological method relies on manual collection, and the residual body is ignored in the process of analysis because it is difficult to collect or identify (Taberlet et al., 2012). Gong et al. (2024) analyzed the impact of ecological restoration project on macrobenthos communities structure in Nanhui Beach in Shanghai and found that many Assiminea latericea residues were collected after the project was carried out. The impact of engineering is more obvious for mollusks with poor mobility. Therefore, the increased difficulty in collecting mollusks and the challenge of identifying their remains after tillage may be key reasons for their low proportion in the traditional morphological method. Moreover, the correlation analysis results showed that the abundance obtained from eDNA metabarcoding had no correlation with density or biomass based on the traditional morphological method. Firstly, in terms of abundance, the abundance obtained using traditional morphological method is based on the number of individuals identified for each species, while the abundance obtained using eDNA metabarcoding is based on the number of sequences identified for each species, and the relationship between the number of individuals and the sequence is still unclear (Lejzerowicz et al., 2015), so it may be difficult for eDNA abundance to accurately reflect individual density or biomass. Deiner et al. (2017) reported that eDNA may detect higher species richness at a certain site, not because of the presence of the species itself, but because their DNA is released by a large amount of DNA caused by water flow or external interference of organisms, thus producing a bias in the results. Furthermore, eDNA reflects the accumulation of DNA signals in the environment and cannot distinguish between living and dead organisms, whereas traditional morphological method primarily collects living organisms (Ruppert et al., 2019). Therefore, sampling processing and analysis data may also partly explain the differences in the evaluation results of the two methods. In summary, our study suggested that eDNA metabarcoding can detect more species, but compared with traditional morphological method, the number of species commonly detected is lower, and the reliability of detection results is restricted by primer deviation, database integrity, species representativeness standardization and sampling principal defect. Therefore, the application of COI primers in the evaluation of mangrove macrobenthos has potential but needs further optimization.
4.2 Short-term effects of S. alterniflora management projects on macrobenthos
The “mowing + plowing” of S. alterniflora will obviously change the habitat environment and have obvious effects on macrobenthos (Gong et al., 2024). In this study, the diversity of S. alterniflora decreased significantly one month after removal. The traditional morphological method showed that there were fewer six annelids in October than in September, including one species of the genus Glycera and one species of the genus Nectoneanthes, however, R. brevirostris and A. japonicus were newly detected. The Nereididae (belong to burrowing animals (Gong et al., 2016), previous studies have shown that infestations of S. alterniflora can increase the number of burrowing animal groups (Lana and Guiss, 1992; Netto and Lana, 1997; Chen et al., 2017). With the removal of S. alterniflora, the effect of promoting sedimentation and making land decreased, and the frequency and time of tidal inundation of the habitat increased (Gong et al., 2016), which provided living conditions for aquatic benthic invertebrates. Therefore, R. brevirostris and A. japonicus were collected in October. Judging from the results of biological density and biomass, S. alterniflora was removed in October due to strong human interference, and its density and biomass were the lowest (Figure 7). After the planting of mangrove plants in December, the two indexes gradually increased, and more Nereididae and crabs were added than in October, and a typical species of mangrove wetland, L. melanostoma, was collected. As the developed root system of mangrove plants changed the sediment conditions in the area and provided different food sources, snails and crabs gradually joined the project site. Meanwhile, the special morphological structure of mangrove plants can provide a variety of habitat microhabitats for mollusks, and these conditions provide favorable habitat conditions for mollusks such as Nereididae (Su et al., 2020). Therefore, changes in species richness and composition respond well to habitat changes.
In this study, the density, biomass, and diversity index of macrobenthos reached their lowest levels after the removal but gradually increased following the planting of mangrove species (Figure 7). Additionally, sediment environmental factors, including TN, TP, and total organic carbon (TOC), exhibited an overall decreasing trend across the three periods (Supplementary Figure S2), with TN and TOC showing a highly significant decline (P<0.05). During the removal of S. alterniflora, highly mobile macrobenthos likely escaped due to physical disturbances, while less mobile species struggled to adapt to the changing habitat and could be unable to survive, resulting in the lowest observed density, biomass, and diversity of macrobenthos. Following mangrove planting, the environmental factors gradually stabilized and improved (Supplementary Figure S2). The rich food resources and favorable habitat provided by mangroves may be key factors in the recovery of macrobenthos density, biomass, and diversity. Wei et al. (2012) by comparing the light flat, S. alterniflora flat and mangrove, found that the density, species and individual density of macrobenthos in mangrove were higher, and the composition also changed greatly (Wei et al., 2012). Chen et al. (2007) reported although there was no significant difference in the density of macrobenthos between mangroves flat and the light beach, the number of species was substantially higher in mangroves than that on adjacent light beach at Jiulong River Estuary in China. These studies show that mangrove plants effectively improve the mudflat environment and restore biodiversity. Therefore, our study showed that the species richness, abundance and diversity of macrobenthos would initially decrease but subsequently increase following the tillage and removal of S. alterniflora. Meanwhile, we recommend conducting long-term monitoring of macrobenthos communities integrating traditional morphological method with eDNA metabarcoding to capture the dynamic changes in macrobenthos diversity following mangrove restoration.
5 Conclusions
In this study, the traditional morphological method and eDNA metabarcoding were applied to detect macrobenthos communities diversity changes in September (before S. alterniflora removal), October (after S. alterniflora removal but before mangrove planting), and December (after S. alterniflora removal and mangrove planting) in Quanzhou Bay mangrove Reserve. Twenty-six species were detected by the traditional method and 212 species by eDNA metabarcoding. Compared with traditional results, eDNA metabarcoding can discover more species, especially Arthropoda. However, the two methods did not identify the same species, so we suggest that the broad application of eDNA metabarcoding in macrobenthos diversity monitoring needs to be optimized from the aspects of database, primers and normalizing the relationship between individual counts and sequences. Therefore, it is currently recommended to monitor macrobenthos diversity using jointly traditional morphological method and environmental eDNA metabarcoding technology in the mangrove. In addition, after the removal of S. alterniflora, macrobenthos diversity in coastal wetlands initially declined but gradually recovered with the planting of mangrove plants. In summary, we believe that the “mowing + plowing” treatment of S. alterniflora followed by mangrove planting is a process of restoring the macrobenthos communities. Long-term monitoring is recommended to assess the interannual variations of macrobenthos communities, providing valuable insights for the effective conservation and management of macrobenthos diversity in mangrove ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
LL: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. YG: Conceptualization, Formal analysis, Methodology, Writing – review & editing. GW: Investigation, Methodology, Writing – review & editing. SF: Investigation, Methodology, Writing – review & editing. KL: Investigation, Methodology, Writing – review & editing. MH: Formal analysis, Methodology, Writing – review & editing. MY: Formal analysis, Methodology, Writing – review & editing. CC: Formal analysis, Methodology, Writing – review & editing. RC: Formal analysis, Methodology, Writing – review & editing. SD: Formal analysis, Methodology, Writing – review & editing. ZP: Formal analysis, Methodology, Writing – review & editing. FJ: Conceptualization, Funding acquisition, Supervision, Validation, Writing – review & editing. YS: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by Natural Science Foundation of Fujian Province, China (No. 2023J05156), Fujian Provincial Department of Education Science and Technology projects (No. JAT220179) and The Green Home Carbon Reduction “Red” Power Plan Project.
Acknowledgments
We are grateful to Shenzhen Mangrove Conservation Foundation and Quanzhou Bay estuary wetland nature reserve development center for the help while sampling and the sharing of background information.
Conflict of interest
Author ZP was employed by the company Hunan Shuitiandi Environmental Technology Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1581442/full#supplementary-material
References
An S. Q., Gu B. H., Zhou C. F., Wang Z. S., Deng Z. F., Zhi Y. B., et al. (2007). Spartina invasion in China: implications for invasive species management and future research. Weed Res. 47, 183–191. doi: 10.1111/j.1365-3180.2007.00559.x
Bravo H., Cheng C. L., Iannucci A., Natali C., Quadros A., Rhodes M., et al. (2021). A DNA barcode library for mangrove gastropods and crabs of Hong Kong and the Greater Bay Area reveals an unexpected faunal diversity associated with the intertidal forests of Southern China. BMC Ecol. Evol. 21, 1–15. doi: 10.1186/s12862-021-01914-6
Buss D. F., Borges E. L. (2008). Application of rapid bioassessment protocols (RBP) for benthic macroinvertebrates in Brazil: comparison between sampling techniques and mesh sizes. Neotrop. Entomol 37, 288–295. doi: 10.1590/S1519-566X2008000300007
Cahill A. E., Pearman J. K., Borja A., Carugati L., Carvalho S., Danovaro R., et al. (2018). A comparative analysis of metabarcoding and morphology-based identification of benthic communities across different regional seas. Ecol. Evol. 8, 8908–8920. doi: 10.1002/ece3.4283
Cao M., Cui L., Sun H., Zhang X., Zheng X., Jiang J. (2021). Effects of Spartina alterniflora invasion on soil microbial community structure and ecological functions. Microorganisms 9, 138. doi: 10.3390/microorganisms9010138
Chen G. C., Ye Y., Lu C. Y. (2007). Changes of macro-benthic faunal community with stand age of rehabilitated Kandelia candel mangrove in Jiulongjiang Estuary, China. Ecol. Eng. 31, 215–224. doi: 10.1016/j.ecoleng.2007.07.002
Chen S. Y., Chen B., Liao J. J., Chen G. L., Huang Y., Chen G. C. (2017). Composition and distribution pattern of Littorinid snails in young rehabilitated mangroves (in Chinese). Ecol. J. 36, 460. doi: 10.13292/j.1000-4890.201702.022
China Marine Standardization Technical Committee (2007). The specification for marine monitoring—Part 6: Marine biological survey (Accessed 18 May 2007).
Cilleros K., Valentini A., Allard L., Dejean T., Etienne R., Grenouillet G., et al. (2019). Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Res. 19, 27–46. doi: 10.1111/1755-0998.12900
Deiner K., Bik H. M., Mächler E., Seymour M., Lacoursière-Roussel A., Altermatt F., et al. (2017). Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
DiBattista J. D., Reimer J. D., Stat M., Masucci G. D., Biondi P., De Brauwer M., et al. (2020). Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Sci. Rep. 10, 8365. doi: 10.1038/s41598-020-64858-9
Doloiras-Laraño A. D., Yaegashi S., Serrana J. M., Ishitani N., Watanabe K. (2023). Comparison of eDNA, bulk-sample metabarcoding, and morphological approaches: A case study of riverine benthic macroinvertebrate communities. bioRxiv. doi: 10.1101/2023.05.30.542510
Duarte S., Leite B. R., Feio M. J., Costa F. O., Filipe A. F. (2021). Integration of DNA-based approaches in aquatic ecological assessment using benthic macroinvertebrates. Water 13, 331. doi: 10.3390/w13030331
Elbrecht V., Leese F. (2017). Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Front. Environ. Sci. 5. doi: 10.3389/fenvs.2017.00011
Ge B., Jiang S., Yang L., Zhang H., Tang B. (2020). Succession of macrofaunal communities and environmental properties along a gradient of smooth cordgrass Spartina alterniflora invasion stages. Mar. Environ. Res. 156, 104862. doi: 10.1016/j.marenvres.2019.104862
Goldberg C. S., Turner C. R., Deiner K., Klymus K. E., Thomsen P. F., Murphy M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Gong K., Bao Y. X., Ren P., Fang F., Hai H., Wang H. (2016). Macrobenthic functional groups in different habitats in the reclaimed wetland Xuanmen Bay (in Chinese). Acta Ecol. Sin. 36, 8214–8223. doi: 10.5846/stxb201504260860
Gong L., Hu Y., Li B., Song Y. H., Li Z. Y., Li T. Y., et al. (2024). Effects of Spartina alterniflora eradication project on macrobenthos community structure: A case study of Nanhui tidal flat in Shanghai. Chin. J. Ecol. 43, 2892–2900. doi: 10.13292/j.1000-4890.202409.006
Han Z., Deng M., Yuan A., Wang J., Li H., Ma J. (2018). Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 625, 106–113. doi: 10.1016/j.scitotenv.2017.12.209
Hering D., Borja A., Jones J. I., Pont D., Boets P., Bouchez A., et al. (2018). Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res. 138, 192–205. doi: 10.1016/j.watres.2018.03.003
Herman P. M., Middelburg J. J., Van de Koppel J., Heip C. H. (1999). Ecology of estuarine macrobenthos. Adv. Ecol. Res. 29, 195–240. doi: 10.1016/S0065-2504(08)60194-4
Hu M., Guo Y., Ji F., Shih Y., Liu K., Yi X., et al. (2025). Environmental DNA metabarcoding shows potential for monitoring meiofauna and marine nematodes diversity in mangrove ecosystems in China. Ecol. Indic. 170, 113134. doi: 10.1016/j.ecolind.2025.113134
Ji F. F. (2022). Construction of eDNA technology for three types of aquatic organisms and its application in biodiversity assessment of typical lakes and reservoirs. Huazhong Agricultural University, China (Wuhan.
Ji J. F., Cai L. Z., Chen X. W., Rao Y. Y., Li X., Yan L. (2015). Current status of benthic fauna resources in Quanzhou Bay mangrove wetland and its management strategies (in Chinese). Wetl. Sci. Manage. 2), 4. doi: 10.3969/j.issn.1673-3290.2015.02.10
Ji F. F., Han D. Y., Yan L., Yan S. H., Zha J. M., Shen J. Z. (2021a). Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method. Sci. Total Environ. 806, 150587. doi: 10.1016/j.scitotenv.2021.150587
Ji F. F., Han D. Y., Yan L., Yan S. H., Zha J. M., Shen J. Z. (2022). Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method. Sci. Total Environ. 806, 150587. doi: 10.1016/j.scitotenv.2021.150587
Ji F. F., Yan L., Yan S. H., Qin T. L., Shen J. Z., Zha J. M. (2021b). Estimating aquatic plant diversity and distribution in rivers from Jingjinji Region, China, using environmental DNA metabarcoding and a traditional survey method. Environ. Res. 199, 111348. doi: 10.1016/j.envres.2021.111348
Jiang S. Y., Chen L. L., Yan L., Liu C. Y., Peng Z. R., Zhang C. X., et al. (2021). Impacts of Spartina alterniflora invasion on the benthic food web in the Yellow River Delta during autumn. J. Appl. Ecol. 32, 4499–4507. doi: 10.13287/j.1001-9332.202112.009
Lana P. C., Guiss C. (1992). Macrofauna–plant-biomass interactions in a euhaline salt marsh in Paranaguá Bay (SE Brazil). Mar. Ecol. Prog. Ser. 80, 57–64. doi: 10.3354/meps080057
Lejzerowicz F., Esling P., Pillet L., Wilding T. A., Black K. D., Pawlowski J. (2015). High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 5, 13932. doi: 10.1038/srep13932
Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Ranwez V., et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 1–14. doi: 10.1186/1742-9994-10-34
Li Z. F., Jiang X. M., Wang J., Meng X. L., Heino J., Xie Z. C. (2019). Multiple facets of stream macroinvertebrate alpha diversity are driven by different ecological factors across an extensive altitudinal gradient. Ecol. Evol. 9, 1306–1322. doi: 10.1002/ece3.4841
Liu L., Guo Z., Zhong C., Shi S. (2018). DNA barcoding reveals insect diversity in the mangrove ecosystems of Hainan Island, China. Genome 61, 797–806. doi: 10.1139/gen-2018-0062
Lu C., Liu J., Jia M., Liu M., Man W., Fu W., et al. (2018). Dynamic analysis of mangrove forests based on an optimal segmentation scale model and multi-seasonal images in Quanzhou Bay, China. Remote Sens 10, 2020. doi: 10.3390/rs10122020
Lv W., Zhou W., Zhao Y. (2018). Macrobenthos functional groups as indicators of ecological restoration in reclaimed intertidal wetlands of China’s Yangtze Estuary. Reg. Stud. Mar. Sci. 22, 93–100. doi: 10.1016/j.rsma.2018.06.003
Marshall N. T., Stepien C. A. (2020). Macroinvertebrate community diversity and habitat quality relationships along a large river from targeted eDNA metabarcode assays. Environ. DNA 2, 572–586. doi: 10.1002/edn3.90
Meng L., Tingting W., Boyang S., Xiyang H., Haigen X., Hongying S. (2019). Biodiversity monitoring of freshwater benthic macroinvertebrates using environmental DNA. Bio. Sci. 27, 480. doi: 10.17520/biods.2018227
Nalepa T. F., Robertson A. (1981). Screen mesh size affects estimates of macro-and meio-benthos abundance and biomass in the Great Lakes. Can. J. Fish. Aquat. Sci. 38, 1027–1034. doi: 10.1139/f81-141
Netto S. A., Lana P. C. (1997). Influence of Spartina alterniflora on superficial sediment characteristics of tidal flats in paranaguá bay (South-Eastern Brazil). Estuar. Coast. Shelf Sci. 44, 641–648. doi: 10.1006/ecss.1996.0154
Patten K., O’Casey C. (2007). Use of Willapa Bay, Washington, by shorebirds and waterfowl after Spartina control efforts. J. Field Ornithol. 78, 395–400. doi: 10.1111/j.1557-9263.2007.00128.x
Pawlowski J., Bruce K., Panksep K., Aguirre F. I., Amalfitano S., Apothéloz-Perret-Gentil L., et al. (2022). Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods. Sci. Total Environ. 818, 151783. doi: 10.1016/j.scitotenv.2021.151783
Pawlowski J., Kelly-Quinn M., Altermatt F., Apothéloz-Perret-Gentil L., Beja P., Boggero A., et al. (2018). The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 637, 1295–1310. doi: 10.1016/j.scitotenv.2018.05.002
Polidoro B. A., Carpenter K. E., Collins L., Duke N. C., Ellison A. M., Ellison J. C., et al. (2010). The loss of species: mangrove extinction risk and geographic areas of global concern. PloS One 5, e10095. doi: 10.1371/journal.pone.0010095
Ratnasingham S., Hebert P. D. (2007). BOLD: the barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Ruppert K. M., Kline R. J., Rahman M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547. doi: 10.1016/j.gecco.2019.e00547
Sawaya N. A., Djurhuus A., Closek C. J., Hepner M., Olesin E., Visser L., et al. (2019). Assessing eukaryotic biodiversity in the Florida Keys National Marine Sanctuary through environmental DNA metabarcoding. Ecol. Evol. 9, 1029–1040. doi: 10.1002/ece3.4742
Schwesig K., Zizka V., Scherber C., Hölzel N. (2024). Comparing eDNA and transect methods for aquatic biodiversity assessment in lakes and ponds. Mol. Ecol. Res. 25, e14060. doi: 10.1111/1755-0998.14060
Su M., Zhang C., Zheng Y. T., Wang W. Q., Wang M., Chen G. G. (2020). Distribution and community survey of mollusks in the mangrove nature reserve of Huizhou District (in Chinese). Cent. South For. Investig. Plan. 39, 43–46. doi: CNKI:SUN:ZLDF.0.2020-01-010
Sun J., Song Y., Shi Y., Zhai J., Yan W. (2022). Progress of marine biodiversity studies in China seas in the past decade. Bio. Sci. 30, 22526. doi: 10.17520/biods.2022526
Taberlet P., Coissac E., Pompanon F., Brochmann C., Willerslev E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x
Tagliapietra D., Sigovini M. (2010). Benthic fauna: collection and identification of macrobenthic invertebrates. Terre Environ. 88, 253–261.
Talley T. S., Levin L. A. (2001). Modification of sediments and macrofauna by an invasive marsh plant. Biol. invasions 3, 51–68. doi: 10.1023/A:1011453003168
Thomsen P. F., Willerslev E. (2015). Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Wang T. T., Wang X. D., Wang D. Y., Fan S. D., Wang S., Chen Z. B., et al. (2023). Aquatic invertebrate diversity profiling in heterogeneous wetland habitats by environmental DNA metabarcoding. Ecol. Indic. 150, 110126. doi: 10.1016/j.ecolind.2023.110126
Wang K., Wang S., Zhang X., Wang W., Li F., Dong L., et al. (2023). Potential ecological impacts of physical control on Spartina alterniflora in coastal wetland: Migration and transformation of nutrients and the response of bacterial community structure. J. Cleaner Prod. 398, 136556. doi: 10.1016/j.jclepro.2023.136556
Wei D. Z., Xiang C. Y., Bao Y. X., Li L. F., Zhang S. S. (2012). Effect of mangrove planting on macrobenthic community structure and functional group. J. Zhejiang Norm. Univ. 35, 195–202. doi: 10.3969/j.issn.1001-5051.2012.02.014
Xie B., Wang A., Zhao Y., Zhu S., Song J., Han G., et al. (2018). Effects of mowing plus waterlogging on germination and seedling growth of Spartina alterniflora (in Chinese). Chin. J. Ecol. 37, 417–423. doi: CNKI:SUN:STXZ.0.2018-02-017
Yuan L., Zhang L., Xiao D., Huang H. (2011). The application of cutting plus waterlogging to control Spartina alterniflora on saltmarshes in the Yangtze Estuary, China. Estuarine Coastal Shelf Sci. 92, 103–110. doi: 10.1016/j.ecss.2010.12.019
Zhao C., Li J., Zhao X. (2019). Mowing plus shading as an effective method to control the invasive plant Spartina alterniflora. Flora 257, 151408. doi: 10.1016/j.flora.2019.05.007
Zheng J., Wei H., Chen R., Liu J., Wang L., Gu W. (2023). Invasive trends of Spartina Alterniflora in the southeastern Coast of China and potential distributional impacts on mangrove forests. Plants 12, 1923. doi: 10.3390/plants12101923
Zhou T., Liu S., Feng Z., Liu G., Gan Q., Peng S. (2015). Use of exotic plants to control Spartina alterniflora invasion and promote mangrove restoration. Sci. Rep. 5, 12980. doi: 10.1038/srep12980
Keywords: macrobenthos, mangrove, environmental DNA metabarcoding, biodiversity, Quanzhou Bay
Citation: Lin L, Guo Y, Wang G, Feng S, Liu K, Hu M, Ye M, Cao C, Chen R, Ding S, Peng Z, Ji F and Shih Y-J (2025) Changes in macrobenthos communities during the invasive Spartina alterniflora removal and mangrove restoration. Front. Mar. Sci. 12:1581442. doi: 10.3389/fmars.2025.1581442
Received: 22 February 2025; Accepted: 03 April 2025;
Published: 29 April 2025.
Edited by:
Jinhyun Kim, Korea Polar Research Institute, Republic of KoreaReviewed by:
Arnelyn D. Doloiras-Laraño, Ehime University, JapanTa-Jen Chu, Taiwan Wetland Society, Taiwan
Copyright © 2025 Lin, Guo, Wang, Feng, Liu, Hu, Ye, Cao, Chen, Ding, Peng, Ji and Shih. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenfen Ji, ZmZqaTEyMTMyNjYyMjZAMTYzLmNvbQ==; Yi-Jia Shih, ZWphMDMxM0BtYWlsLm5zeXN1LmVkdS50dw==
 Lin Lin1
Lin Lin1 Kai Liu
Kai Liu Chunxi Cao
Chunxi Cao Zenghui Peng
Zenghui Peng Fenfen Ji
Fenfen Ji