- 1School of Marine Sciences, Ningbo University, Ningbo, Zhejiang, China
- 2Ningbo Institute of Oceanography, Ningbo, Zhejiang, China
- 3Ningbo Yifeng Aquaculture Company, Ningbo, Zhejiang, China
Ammonia nitrogen is highly toxic to crustaceans; however, studies investigating the molecular mechanisms underlying its metabolism and excretion in these organisms remain scarce. The present study aims to identify key genes and investigate the molecular mechanisms in response to ammonia nitrogen using RNA sequencing in Scylla paramamosain, a key economic crab species in Asia that is subjected to ammonia nitrogen stress in aquaculture. The median lethal concentrations (LC50) of ammonia nitrogen exposure were determined as 325.4 mg/L for 24 hours, 253.7 mg/L for 48 hours, and 198.2 mg/L for 72 hours. Subsequently, crabs were exposed to 325.13 mg/L NH4Cl for durations of 24, 48, and 96 hours, followed by transcriptome sequencing to identify differentially expressed genes (DEGs) related to ammonia nitrogen metabolism in the anterior and posterior gills of Scylla paramamosain. Ammonia nitrogen stress caused significant damage to the morphology and structure of the gills, with prolonged exposure leading to further damage, including cellular vacuolization and narrowing of the gill hemolymph chambers. A total of 184.66 Gb of clean data and 36,439 DEGs were obtained, including 7,880 DEGs in the anterior gill and 28,559 DEGs in the posterior gill, which were implicated in the regulation of ammonia nitrogen metabolism-related pathways. KEGG enrichment analysis revealed that ammonia nitrogen stress induced changes in the expression of metabolism- and immune-related genes. Following ammonia nitrogen stress, the expression of ammonia nitrogen metabolism-related genes (punA, XDH, rocF, allB) in the anterior gill was upregulated, whereas the expression of genes in the posterior gill (GART, rocF, alc) was downregulated. These results indicate that ammonia nitrogen stress enhances ammonia nitrogen metabolism in the anterior gill, while inhibiting this process in the posterior gill. The expression of immune-related genes (Hsp10, Hsp70, Hsp90, CTL) in both the anterior and posterior gills was downregulated following ammonia nitrogen stress, suggesting that ammonia nitrogen stress diminished the organism’s immune capacity. These findings provide a theoretical foundation for understanding the molecular mechanisms regulating ammonia nitrogen metabolism and for improving the artificial culture of Scylla paramamosain.
1 Introduction
Global production of mud crabs reached 152,000 tons in 2021 (FAO, 2024). This species is an important aquaculture organism in Asia due to its rapid growth, desirable taste, and high economic value (Zheng et al., 2020). By 2023, China’s mariculture output of mud crabs increased to 157,012 tons, reflecting a 1.52% growth from the previous year, with overall aquaculture production continuing to rise annually (Wang, 2024). Despite the continuous increase in production, the yield of mud crabs in artificial breeding is constrained by various environmental factors in culture ponds. Crustaceans are widely distributed in aquatic ecosystems and play an important role in the food chain. However, crustaceans are extremely sensitive to environmental changes, so they are often used for toxicological and ecological risk assessment (Lin et al., 2017). Notably, high concentrations of ammonia nitrogen are toxic to mud crabs, leading to significant reductions in production and substantial economic losses in aquaculture operations (Kır et al., 2004; Preena et al., 2021). However, the adaptive regulatory mechanisms of mud crabs in response to ammonia nitrogen stress remain poorly understood. This study aims to elucidate the underlying molecular mechanisms governing their adaptation to ammonia nitrogen stress.
Ammonia is a common pollutant in aquaculture, primarily derived from aquatic animal excreta and food residues (Lin et al., 2023). In water bodies, ammonia primarily exists in the form of ionized ammonia (NH4+) and unionized ammonia (NH3). NH4Cl dissociates in water to generate NH4+ and Cl-, while NH4+ and NH3 can undergo mutual conversion in water bodies, maintaining a dynamic equilibrium (Cheng et al., 2019). Previous studies have shown that high levels of ammonia nitrogen are toxic and lethal to crustaceans (Tang et al., 2022), studies have shown that ammonia nitrogen stress can increase the severity of pathogen infections in aquatic organisms, leading to disease occurrence, and both the prevalence rate and severity of the disease correspondingly escalate with rising concentrations of ammonia nitrogen in water bodies (Liu and Chen, 2004). Ammonia nitrogen stress has brought huge losses to the artificial culture of mud crab, therefore, it is essential to investigate ammonia nitrogen stress in mud crabs. There are three primary forms of ammonia nitrogen toxicity in crustaceans. First, ammonia nitrogen enters the bodies of crustaceans through the gill epithelium and accumulates in the hemolymph, where it competes with Na+ for passive cation diffusion (Eddy, 2005). Ammonia stress also leads to the collapse of gill lamellae, hindering gas exchange (Romano and Zeng, 2013). Third, ammonia stress damages cell membranes and entire cells, leading to apoptosis (Liang et al., 2016). Recent research shows that high ammonia levels can disrupt the integrity of the gill epithelium, subsequently affecting the ion transport capacity of the posterior gill-associated epithelium (Li et al., 2024). However, the molecular mechanisms regulating ammonia nitrogen metabolism in mud crabs remain understudied and unclear. Gills are in direct contact with the external environment and are considered the primary barrier against external invasions. They are highly vulnerable to environmental changes, including ammonia stress (Negro and Collins, 2017). Therefore, studying the molecular mechanisms of gill function under ammonia nitrogen stress is crucial for the healthy and efficient breeding of crustaceans, provide more targeted prevention and control measures for the artificial breeding of mud crab.
Mud crab gills consist of eight structures, with the first four classified as anterior gills (1-4) and the last four as posterior gills (5-8) (Yuan et al., 2024). The anterior and posterior gills differ in both structure and function. The anterior gills primarily contain a thin respiratory epithelium (1-2 μm thick), which facilitates gas exchange (Mo et al., 2024b). In contrast, the posterior gills consist of a thicker ion transport epithelium (10-20 μm thick), which plays a crucial role in ion regulation and transport (Henry et al., 2012; Mo et al., 2024a; Rahi et al., 2018; Shen et al., 2024). Recent studies have focused on the tissue structure and molecular regulation of crustacean gills under environmental stressors. For instance, dissolved oxygen is a crucial environmental factor for aquatic animals, and acute hypoxia significantly damages gill structure, affects respiratory metabolism, increases oxygen consumption, and reduces ammonia nitrogen metabolism rates (Ning et al., 2023). In low-dissolved oxygen environments, the HIF-1 signaling pathway is activated, reducing oxygen consumption (Yang et al., 2021). Moreover, previous studies have indicated that gill filaments and gill structure are damaged under pH stress (Zhu et al., 2022). The down-regulation of letm1 expression in low pH environments reduces cellular damage, while the up-regulation of NHE3 expression in high pH environments mitigates damage. However, studies on the molecular regulatory mechanisms of the anterior and posterior gills of mud crabs in response to ammonia nitrogen stress are limited. Previous studies on crustaceans under ammonia nitrogen stress have primarily focused on the metabolic pathways of ammonia nitrogen in the body. Ammonia nitrogen metabolism in crustaceans occurs through three primary pathways: the synthesis of glutamic acid followed by glutamine release into the hemolymph (Pan et al., 2018), the synthesis of arginine and the subsequent urea excretion (Dong et al., 2019), and the synthesis of xanthine with subsequent urea excretion (Zhang et al., 2020). Building on this, the present study preliminarily explores the molecular mechanisms regulating ammonia nitrogen metabolism in mud crabs following ammonia nitrogen exposure.
The aim of this study was to investigate the effects of ammonia nitrogen stress on the gill morphology and structure of mud crabs, as well as to identify differentially expressed genes (DEGs) associated with this stress using transcriptome sequencing. In order to explore the adaptation and regulation mechanism of mud crab to ammonia nitrogen stress, reduce the loss of ammonia nitrogen toxicity to mud crab breeding, and provide a theoretical basis for the future mud crab breeding. Initially, we conducted ammonia nitrogen exposure experiments on groups of mud crabs to determine the half-lethal concentration for different exposure durations, followed by an assessment of ammonia nitrogen toxicity in mud crabs. Next, we analyzed DEGs in the anterior and posterior gills of mud crabs exposed to different durations of ammonia nitrogen stress using transcriptome sequencing, and examined changes in tissue morphology through histological sections. Furthermore, the study aimed to investigate the molecular mechanisms underlying ammonia nitrogen metabolism in mud crabs, providing a foundation for future research on ammonia nitrogen stress in this species.
2 Materials and methods
2.1 Animals and samples
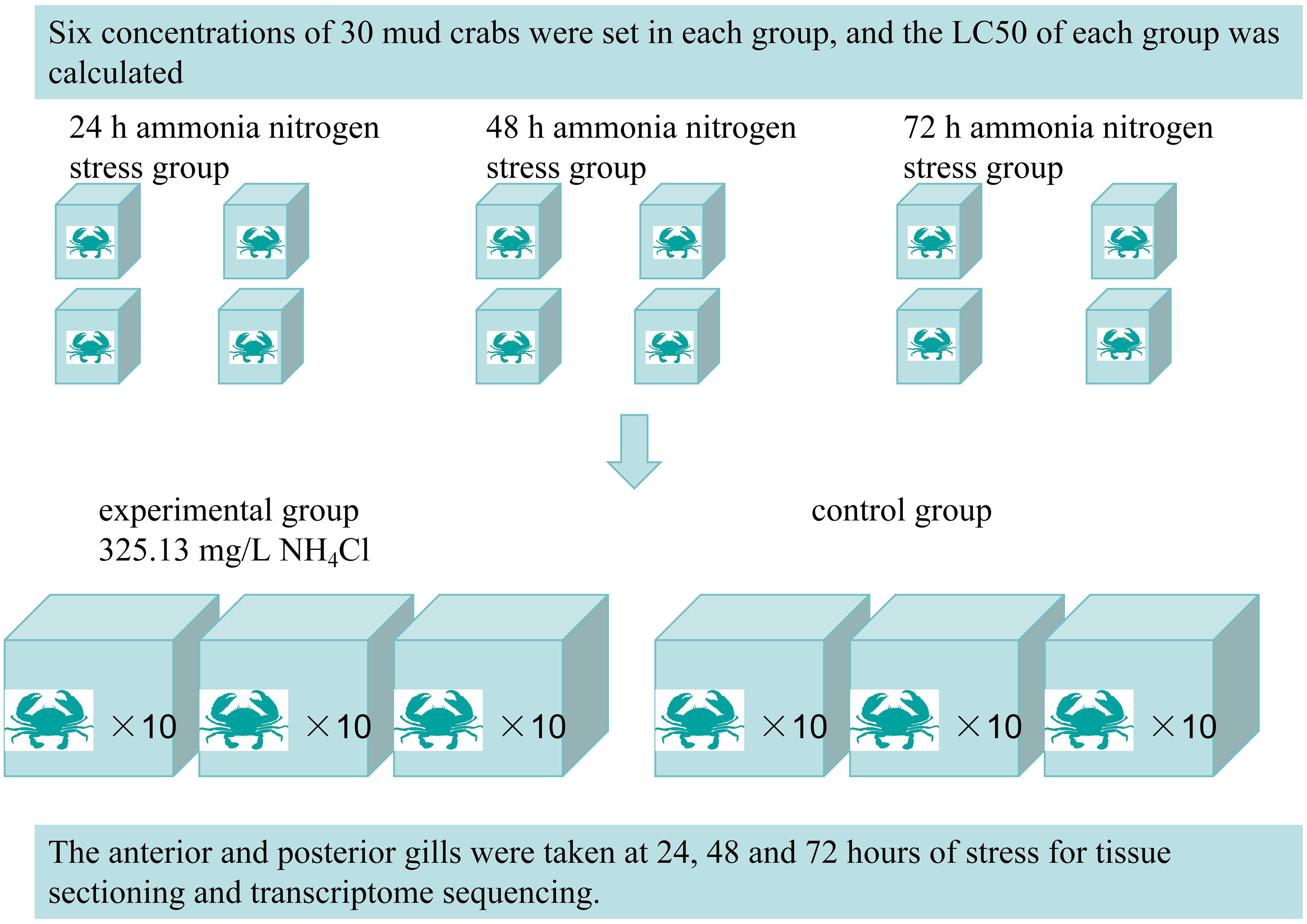
Mud crabs were obtained from the Ningbo Yifeng Aquaculture Company in Ninghai, China (29°10’45.5”N, 121°31’08.5”E) and acclimated in a cement tank for one week prior to the experiment. Throughout the experiment, environmental conditions were stable, with water temperature maintained at 28.37 ± 0.11°C, dissolved oxygen at 5.68 ± 0.46 mg/L, salinity at 21.26 ± 0.1 ppt, and pH at 8.17 ± 0.1. After acclimation, The mud crabs were divided into three exposure duration groups: 24 hour, 48 hour, and 72 hour. For the 24 hour group, ammonia nitrogen concentrations were set at 280 mg/L, 310 mg/L, 340 mg/L, and 370 mg/L. Both the 48 hour and 72 hour groups were exposed to ammonia nitrogen concentrations of 160 mg/L, 210 mg/L, 260 mg/L, and 310 mg/L. Each concentration level within all groups contained 30 crabs. The median lethal concentration (LC50) for each exposure duration was calculated based on mortality data under the respective experimental conditions. Then 60 crabs were randomly assigned to two groups: the ammonia nitrogen stress group (325.13 mg/L) and the control group, with both groups maintained in six water tanks (102 cm × 69 cm × 61 cm). Each treatment group was replicated three times (Figure 1).
2.2 Sample collection
In this experiment, samples were collected at 24, 48, and 72 hours following ammonia nitrogen stress exposure. The sample groups included the anterior gill control group (AG), anterior gill 24 hour group (AG24), anterior gill 48 hour group (AG48), anterior gill 72 hour group (AG72), posterior gill control group (PG), posterior gill 24 hour group (PG24), posterior gill 48 hour group (PG48), and posterior gill 72 hour group (PG72). The collected samples were immediately frozen in liquid nitrogen and stored at -80°C for subsequent RNA extraction. The samples designated for tissue observation were stored in a fixative solution for subsequent histological analysis.
2.3 HE staining
The gills were exposed and fixed in a cryotube containing tissue fixative according to the method described by Ozawa et al (Ozawa and Sakaue, 2020). The gills were embedded in paraffin and sectioned at a thickness of 5 μm. The sections were dewaxed in xylene and rehydrated through an ethanol series. The gill morphology and ultrastructure were observed using light microscopy after hematoxylin-eosin (HE) staining, purchased from Invitrogen (Carlsbad, CA, USA).
2.4 Transcriptome analysis
2.4.1 Total RNA extraction, library construction for sequencing
Total RNA was extracted from anterior and posterior gill samples using TRI reagent (Invitrogen, USA). RNA quality was assessed using a microplate reader at A260/280, and RNA integrity was evaluated with the Agilent 2100 Bioanalyzer system (Agilent Technologies, USA). Samples with an RNA Integrity Number (RIN) greater than 7.0 were selected for library preparation. First-strand cDNA synthesis was carried out using M-MLV reverse transcriptase (Promega, USA), with the addition of a 3’ RACE adaptor and Oligo (dT) to the cDNA tail. Following PCR amplification, the PCR products were purified using AMPure XP beads to obtain the final library. Following library construction, preliminary quantification was performed using the Qubit 2.0, and the library was diluted to 1.5 ng/μL. The Agilent 2100 Bioanalyzer system was used to assess the insert size of the library. A total of 18 sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA), following the manufacturer’s instructions. Index codes were added to assign sequences to each sample. The libraries were subsequently sequenced on an Illumina HiSeq 4000 platform, generating 150 bp paired-end reads. Raw sequencing data from the Illumina Genome Analyzer were processed using SeqQC V2.1 for quality control, including filtering high-quality reads based on score values, removal of reads containing primer/adaptor sequences, and trimming of read lengths.
2.4.2 Data analysis
Raw reads in FASTQ format were initially processed using fastp (Chen et al., 2018), and low-quality reads were removed to generate clean reads. The clean reads were then aligned to the reference genome using HISAT2 (Kim et al., 2015). The FPKM of each gene was calculated, and the read counts were obtained using HTSeq-count (Anders et al., 2014). Principal component analysis (PCA) was conducted using R (v 3.2.0) to assess the biological replicates of the samples. Differential expression analysis was carried out using DESeq2 (Love et al., 2014). A q-value < 0.05 and a fold change > 2 or < 0.5 were set as thresholds for identifying significantly differentially expressed genes (DEGs). Hierarchical clustering of DEGs was performed using R (v 3.2.0) to visualize gene expression patterns across different groups and samples. A radar plot of the top 10 genes was generated to display the expression of upregulated and downregulated DEGs using the R package ggradar.
2.5 Quantitative real-time reverse transcription PCR
To validate the accuracy of the transcriptome sequencing results, eight DEGs were selected for RT-PCR analysis. Primers for the target genes were designed using Primer 5, and total RNA was extracted from fresh anterior and posterior gills. First-strand cDNA was synthesized using the PrimeScript RT Kit (Takara) containing gDNA Eraser, and SYBR Green RT-PCR was conducted using the ABI 7500 Sequence Detection System (Applied Biosystems) to validate the transcriptome data. The 18S rRNA of mud crab (GenBank accession number: fj646616.1) was used as an internal control to normalize gene expression levels, and all experiments were conducted in triplicate. Gene expression levels were analyzed using the comparative CT method (2 −ΔΔCT).
2.6 Statistical analysis
Statistical analysis in this experiment was conducted using SPSS v22.0 (IBM, USA) and GraphPad Prism v10.0 (GraphPad Software Inc., San Diego, CA, USA). Differences between the control and treatment groups were compared using a t-test, and χ² was employed to assess the homogeneity of variance between the two groups. The significance level was set at a p-value < 0.05.
3 Results
3.1 Exploration of the semi lethal concentration of ammonia nitrogen
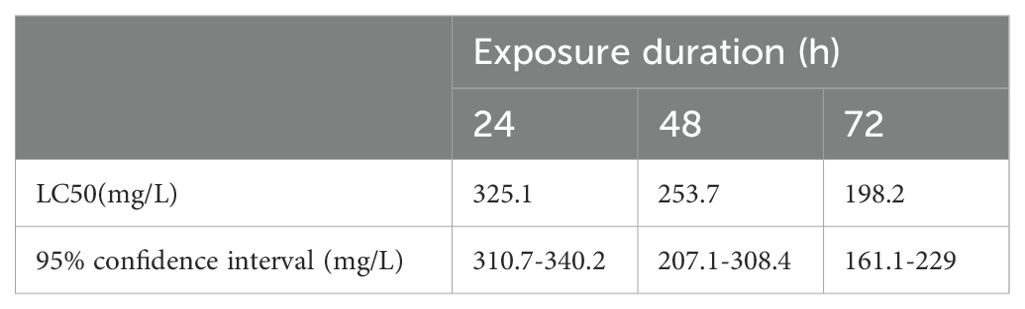
This study utilized NH4Cl to simulate external ammonia nitrogen stress and investigate its toxicity in mud crabs. The 24 hour LC50 for ammonia nitrogen stress was 325.4 mg/L, with a 95% confidence interval ranging from 310.7 mg/L to 340.2 mg/L. After 48 hours, the LC50 was 253.7 mg/L, with a 95% confidence interval ranging from 207.1 mg/L to 308.4 mg/L. After 72 hours, the LC50 was 198.2 mg/L, with a 95% confidence interval ranging from 161.1 mg/L to 229 mg/L (Table 1). The LC50 of 24 h was selected as the experimental concentration in the subsequent ammonia nitrogen stress experiments of this experiment.
3.2 Histology analysis
This study examined the structural changes in gill filaments following ammonia nitrogen stress. In the anterior gill, cells appeared vacuolated, accompanied by morphological changes in the epithelial cells. Under severe stress, the stratum corneum breaks and detaches from the epithelial cells, which subsequently slough off (Figures 2A–D). Meanwhile, the hemolymph cavity shrinks and collapses, compromising the integrity of the gill structure. In the posterior gill, cells showed vacuolization following ammonia nitrogen stress. Additionally, the epithelial cells were separated from the stratum corneum, and the hemolymph cavity shrank (Figures 2E–H).
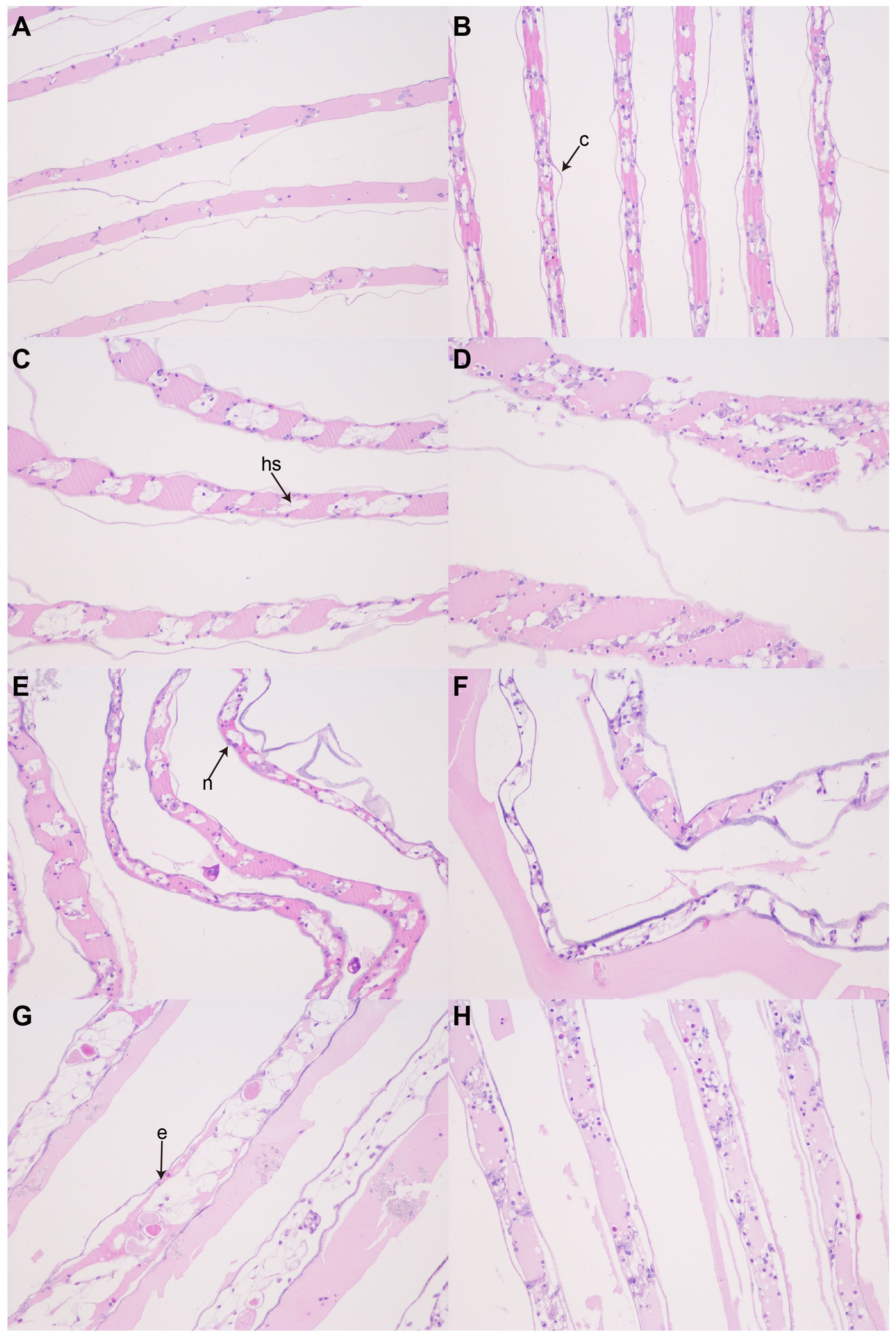
Figure 2. Microscopic images of histological sections of S. Paramamosains gills (200×). (A) AG; (B) AG24; (C) AG48; (D) AG72; (E)PG; (F) PG24; (G) PG48; (H) PG72. c, cuticle; hs, hemolymph space; e, epithelial cell; n, nucleus.
3.3 Analysis of differentially expressed genes
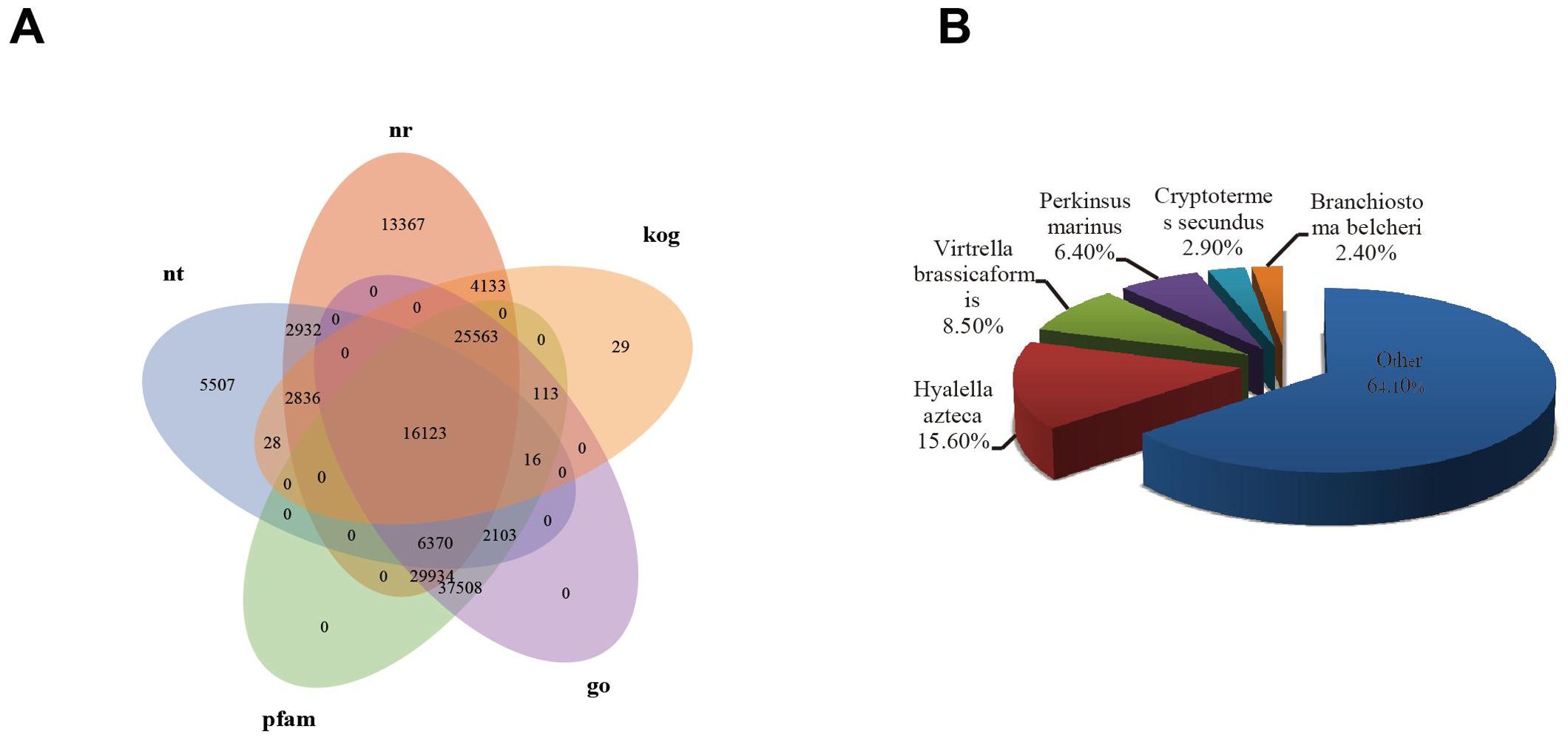
We annotated all 250,975 unigenes using the NR, NT, Swiss-Prot, PFAM, GO, KOG, and KEGG databases. Of these unigenes, 101,258 (40.34%) had significant BLAST matches with known proteins in NR, 35,915 (14.31%) in NT, 81,611 (32.51%) in Swiss-Prot, 117,730 (46.9%) in PFAM, 117,730 (46.9%) in KOG, 48,841 (19.46%) in GO, and 43,263 (17.23%) in KEGG, respectively. A total of 146,696 (58.45%) unigenes had the best BLAST matches in at least one of the seven databases, while 13,266 (5.63%) showed the best BLAST matches in all seven databases. Regarding the main species distribution matched against the NR database, 15.6% of the matched unigenes showed similarities to Hyalella azteca, followed by Vitrella brassicaformis (8.5%), Perkinsus marinus (6.4%), Cryptotermes secundus (2.9%), and Branchiostoma belcheri (2.4%).
We identified genes whose expression was significantly altered by ammonia stress, using log2 fold change > 1 and p-value < 0.005 as the thresholds. Among the anterior gills, the highest number of differentially expressed genes (DEGs) was observed in the comparison between AG24 and AG (5,336 DEGs), and between PG24 and PG (20,525 DEGs) in the posterior gills. Notably, the number of DEGs in the posterior gills was higher than in the anterior gills under ammonia stress. Temporal sequence analysis of the DEGs revealed 46 differentially expressed genes in the anterior gill across three time points, and 36 differentially expressed genes in the posterior gill across the same time points. NR annotation was performed on the shared differentially expressed genes between the anterior and posterior gills. Among the 46 DEGs in the anterior gill, 9 genes were not annotated, and 5 were classified as hypothetical proteins. In the posterior gill, 6 of the 36 DEGs were not annotated, and 8 were hypothetical proteins (Figures 3–5).
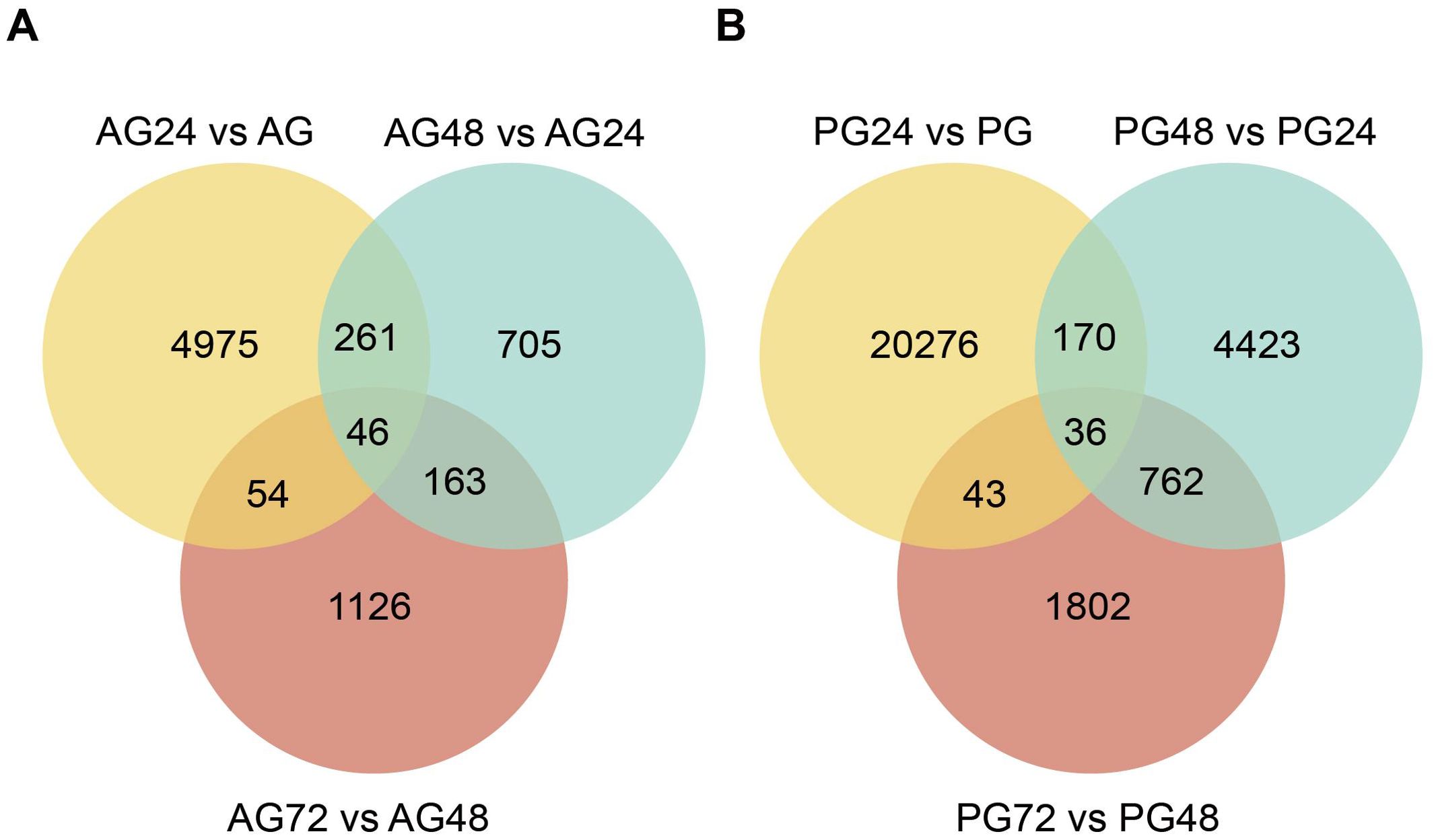
Figure 4. Wayne diagram of differential expressed genes between anterior and posterior gills. (A) Number of differentially expressed genes in the anterior gill at different ammonia nitrogen stress times; (B) Number of differentially expressed genes in the posterior gill under different ammonia nitrogen stress times.
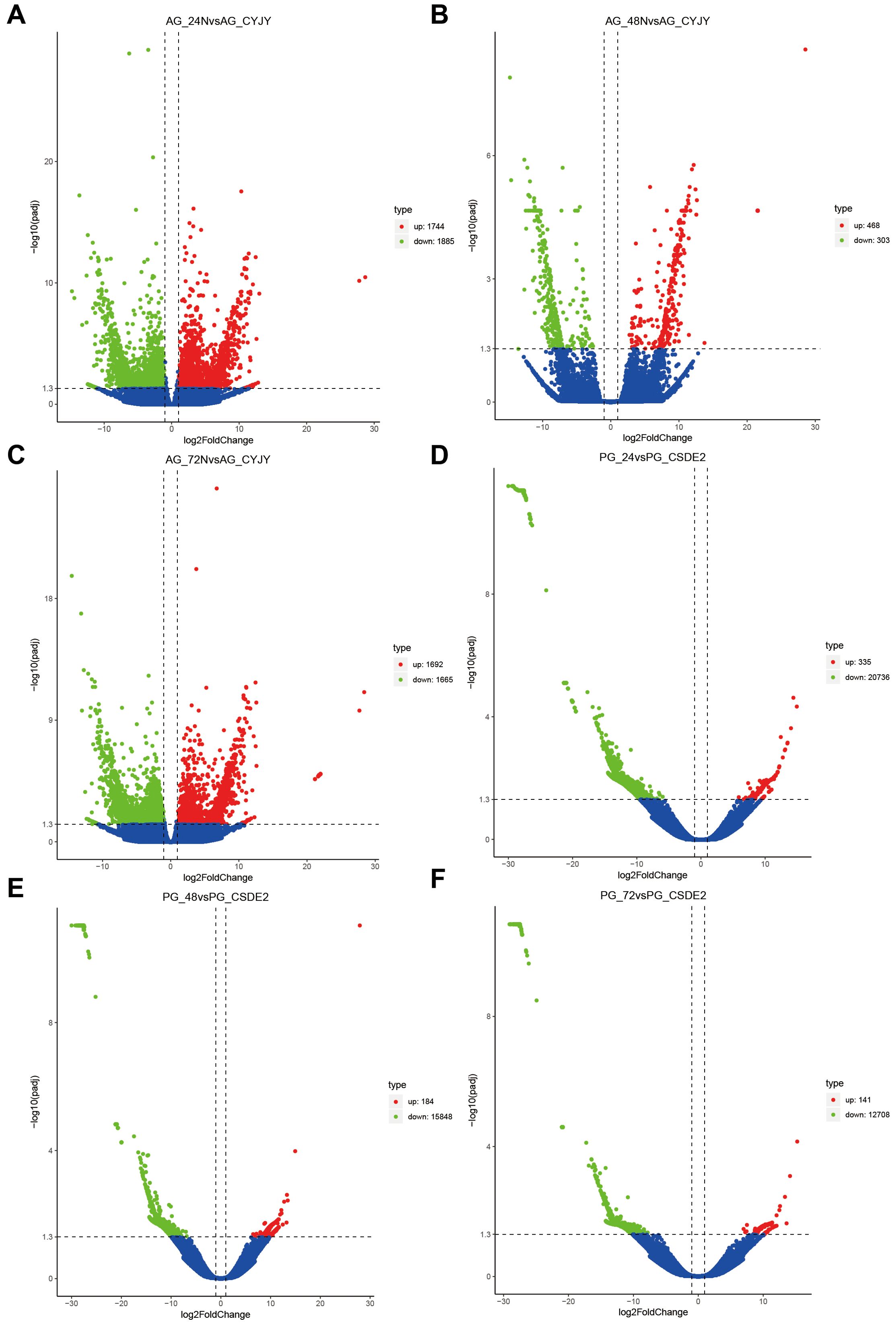
Figure 5. Volcano plot of the DEGs. (A) AG24 VS AG; (B) AG48 VS AG; (C) AG72 VS AG; (D) PG24 VS PG; (E) PG48 VS PG; (F) PG72 VS PG.
3.4 GO enrichment analysis of the DEGs
Gene Ontology (GO) enrichment analysis is a valuable tool for evaluating gene functions and biological processes. In this study, GO analysis was employed to assess the impact of ammonia nitrogen stress on gene expression, growth, and development in mud crabs, with the goal of further elucidating the molecular mechanisms underlying their resistance to ammonia nitrogen stress.
The differentially expressed genes (DEGs) in both anterior and posterior gills were enriched in the Gene Ontology (GO) database. In AG24 vs. AG, the DEGs were predominantly enriched in the biological process category, particularly in cell process regulation and biological regulation. In the molecular function group, they were mainly enriched in transcription factor activity, sequence-specific DNA binding, and nucleic acid binding transcription factor activity. In the cellular component group, the DEGs were mostly enriched in viral envelope and viral membrane (Figure 6A). In AG48 vs. AG, the DEGs were enriched in regeneration, regeneration process, and multi-organ regeneration process within the biological process group. The cellular component group was primarily enriched in the Golgi membrane and the intrinsic components of the Golgi membrane. The molecular function group was mainly enriched in protein binding and mannosyltransferase activity (Figure 6B). In AG72 vs. AG, the DEGs showed significant enrichment in the organic matter metabolic process, macromolecular metabolic process, and primary metabolic process within the biological process group. In the cellular component group, they were significantly enriched in ribonucleoprotein complex and ribosome. The molecular function group was significantly enriched in structural molecular activity and structural components of ribosome (Figure 6C). In PG24 vs. PG, the DEGs were significantly enriched in amide biosynthesis, ribonucleoprotein complex biogenesis, and ribosome biogenesis within the biological process group. In the cellular component group, they were significantly enriched in ribosomes, and in the molecular function group, they were significantly enriched in catalytic activity, heterocyclic compound binding, and organic ring compound binding (Figure 6D). In PG48 vs. PG, the DEGs were significantly enriched in amide biosynthesis, peptide biosynthesis, and ribosome biogenesis within the biological process group. In the cellular component group, they were significantly enriched in ribonucleoprotein complex and ribosome, while the molecular function group was significantly enriched in catalytic activity, heterocyclic compound binding, and organic cyclic compound binding (Figure 6E). In PG72 vs. PG, the DEGs were significantly enriched in cellular amide metabolism, amide biosynthesis, and peptide biosynthesis within the biological process group. In the cellular component group, they were significantly enriched in ribonucleoprotein complex and ribosome, while in the molecular function group, they were significantly enriched in catalytic activity, heterocyclic compound binding, and organic cyclic compound binding (Figure 6F).
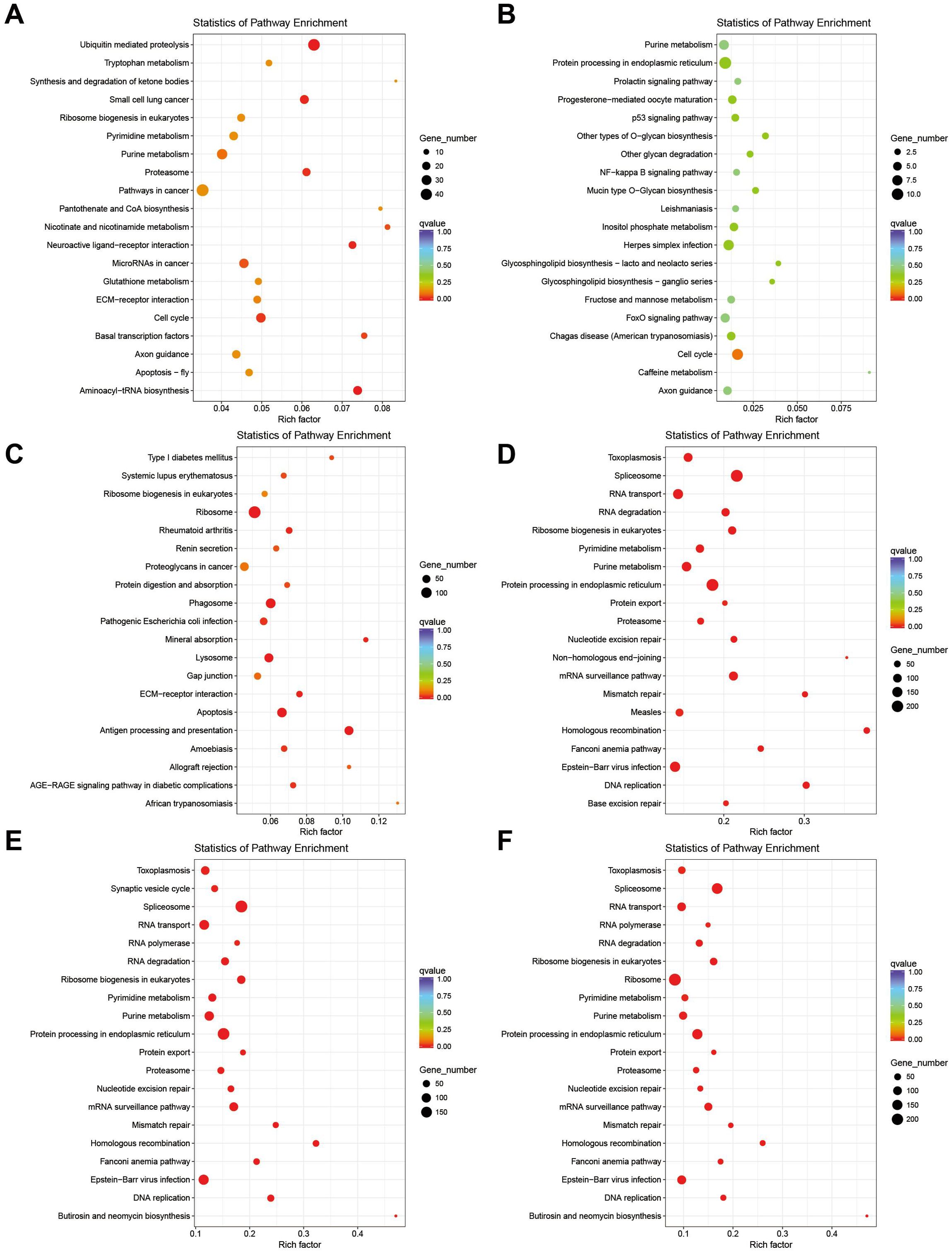
Figure 6. KEGG enrichment pathway plots for DEGs in the anterior and posterior gills. (A) AG24 VS AG; (B) AG48 VS AG; (C) AG72 VS AG; (D) PG24 VS PG; (E) PG48 VS PG; (F) PG72 VS PG.
In the anterior gills, 31 unigenes were annotated in the GO database, which were categorized into 15 biological processes. Notably, “oxidation-reduction reactions” and “single-organism metabolic processes” were the most highly represented. Other relevant processes, such as “tRNA metabolic processes,” “response to oxidative stress,” “lipid transport,” and “amino acid activation,” were also well represented. A total of 15 functional groups were assigned to molecular functions, with the most significant being oxidoreductase activity.
In the posterior gills, most differentially expressed genes (DEGs) were enriched in three major categories. Among the biological processes, the most highly represented terms included “biological regulation,” “regulation of cellular processes,” “cellular response to stimulus,” “response to stimulus,” and “cell communication.” In terms of cellular components, DEGs were predominantly associated with the membrane, including “integral components of the membrane,” “intrinsic components,” and “membrane parts.” For molecular functions, the DEGs were most notably enriched in “signaling receptor activity” and “hydroxymethyl, formyl, and related transferase activity”.
3.5 KEGG enrichment analysis of DEGs
The KEGG pathway database was categorized into six major groups: environmental information processing, genetic information processing, cellular processes, human diseases, metabolism, and organismal systems. By analyzing the DEGs of the anterior and posterior gills of mud crabs exposed to ammonia nitrogen stress at different time points, this study aimed to explore the molecular mechanisms underlying the mud crabs’ resistance to ammonia nitrogen stress. The pathways identified from the KEGG analysis provide valuable insights into the biological processes and molecular pathways involved in the crabs’ adaptive responses to environmental stress (Figure 6).
The aim of this study was to identify the biological pathways activated or inactivated during the response of mud crabs to ammonia stress. The results of this experiment indicated that the differentially expressed genes (DEGs) in AG24 vs AG were predominantly enriched in metabolic pathways, including ubiquitin-mediated proteolysis (ko04120), purine metabolism (ko00230), glutathione metabolism (ko00480), nicotinate and nicotinamide metabolism (ko00760), and pyrimidine metabolism (ko00240) (Figure 7A). In AG48 vs AG, the DEGs were primarily enriched in the cell cycle (ko04110), protein processing in the endoplasmic reticulum (ko04141), and purine metabolism (ko00230) (Figure 7B). In AG72 vs AG, the DEGs were primarily enriched in immune-related pathways, such as the ribosome (ko03010), phagosome (ko04145), apoptosis - multiple species (ko04215), and antigen processing and presentation (ko04612) (Figure 7C). In PG24 vs PG, the DEGs were predominantly enriched in the ribosome (ko03010), protein processing in the endoplasmic reticulum (ko04141), endocytosis (ko04144), pyrimidine metabolism (ko00240), and purine metabolism (ko00230) (Figure 7D). In PG48 vs PG, the DEGs were predominantly enriched in the ribosome (ko03010), protein processing in the endoplasmic reticulum (ko04141), endocytosis (ko04144), phagosome (ko04145), purine metabolism (ko00230), and pyrimidine metabolism (ko00240) (Figure 7E). In PG72 vs PG, the DEGs were predominantly enriched in the ribosome (ko03010), protein processing in the endoplasmic reticulum (ko04141), endocytosis (ko04144), phagosome (ko04145), purine metabolism (ko00230), and pyrimidine metabolism (ko00240) (Figure 7F).
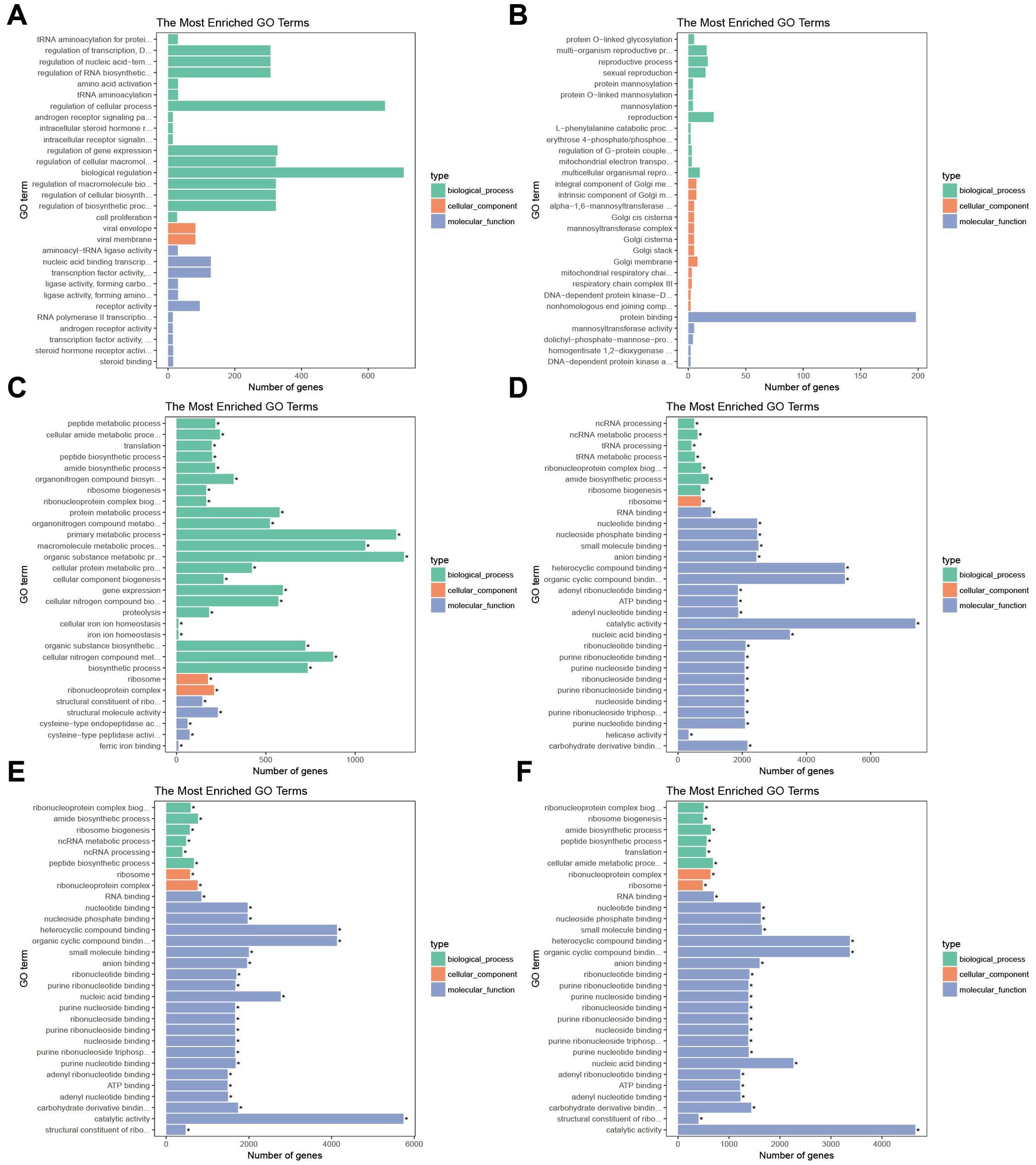
Figure 7. Top 30 of GO enrichment pathways of DEGs between anterior and posterior gills. Green is biological process, orange is cellular component, blue is molecular function. (A) AG24 VS AG; (B) AG48 VS AG; (C) AG72 VS AG; (D) PG24 VS PG; (E) PG48 VS PG; (F) PG72 VS PG.
3.6 Validation of RNA-seq by RT-qPCR
Eight genes were randomly selected for quantitative RT-qPCR to validate the accuracy of the transcriptome sequencing results. The results demonstrated that the gene expression patterns were consistent with those observed in the transcriptome sequencing data (Figure 8).
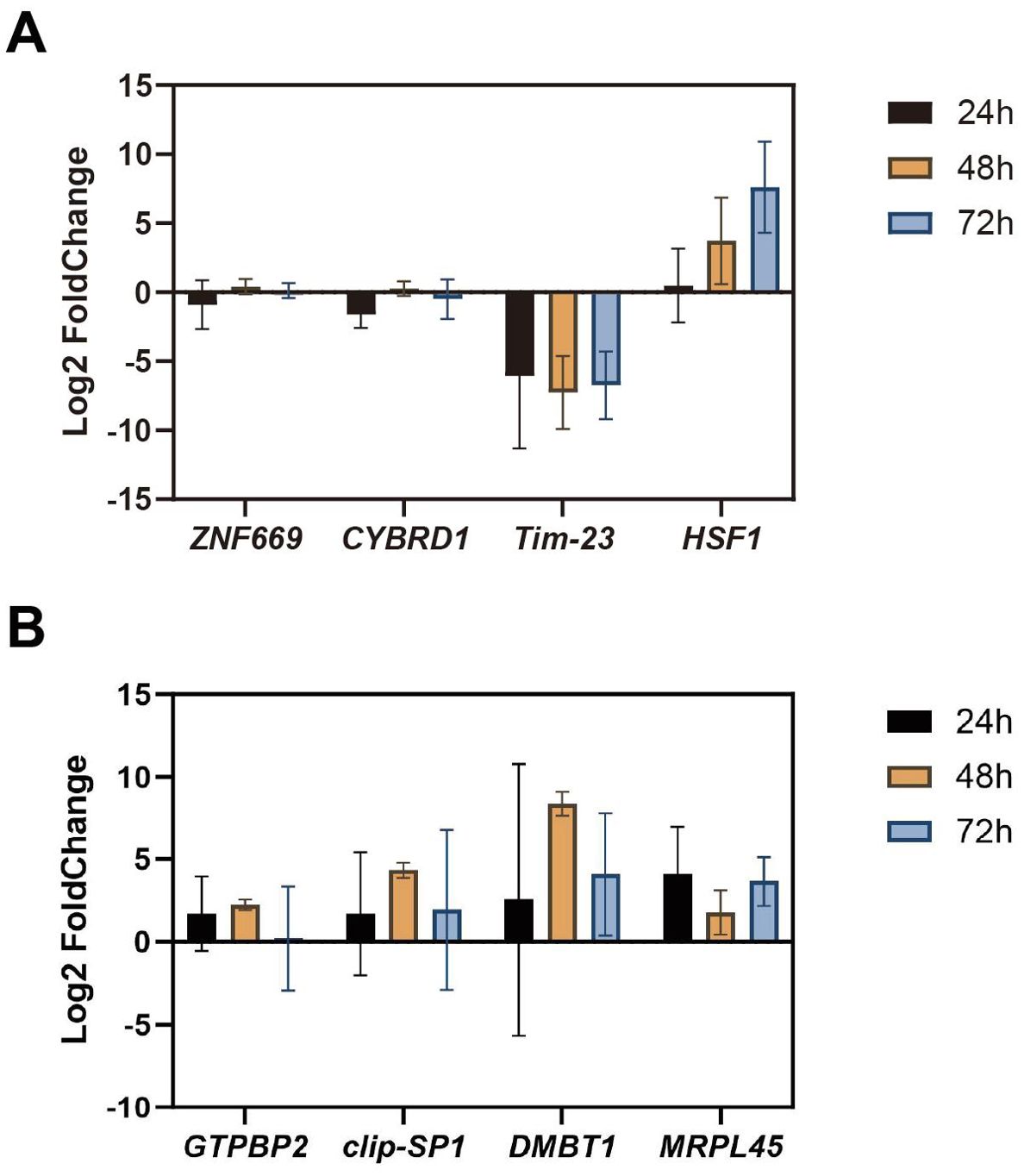
Figure 8. Validation of RNA-seq data. (A) Quantitative validation of anterior gill RNA-Seq results by RT-qPCR; (B) Quantitative validation of the posterior gill RNA-Seq results by RT-qPCR.
4 Discussion
The gill of crustaceans is a crucial organ primarily involved in physiological processes such as respiration, immunity, and osmotic regulation (Xing et al., 2025). Gills are directly exposed to the external environment and are highly sensitive to environmental factors, making them commonly used as a model organ for studying environmental impacts (Zhou et al., 2025). Ammonia nitrogen is a major environmental stressor and pollutant in the artificial breeding of crustaceans (Liu et al., 2024), potentially causing stress to cultured animals by damaging the gills and inducing a range of toxic effects. In this study, we employed RNA-seq analysis to investigate the transcriptome of mud crabs under ammonia stress, aiming to gain insights into the molecular mechanisms underlying the detrimental effects of ammonia. The results revealed a decrease in the number of gill epithelial cells, an increase in intercellular space, and narrowing and collapse of the hemolymph cavity following ammonia nitrogen stress. These structural changes are likely to impair the gas exchange capacity of the anterior gills, consistent with previous reports on the toxic effects of ammonia nitrogen in crustaceans. Previous studies have shown that hypoxia can exacerbate the influx of ammonia nitrogen. Furthermore, other research has demonstrated that the accumulation of ammonia nitrogen within the organism can significantly reduce the oxygen-carrying capacity of the blood, further diminishing respiratory efficiency (Liang et al., 2016). Finally, oxidative stress can lead to cellular damage.
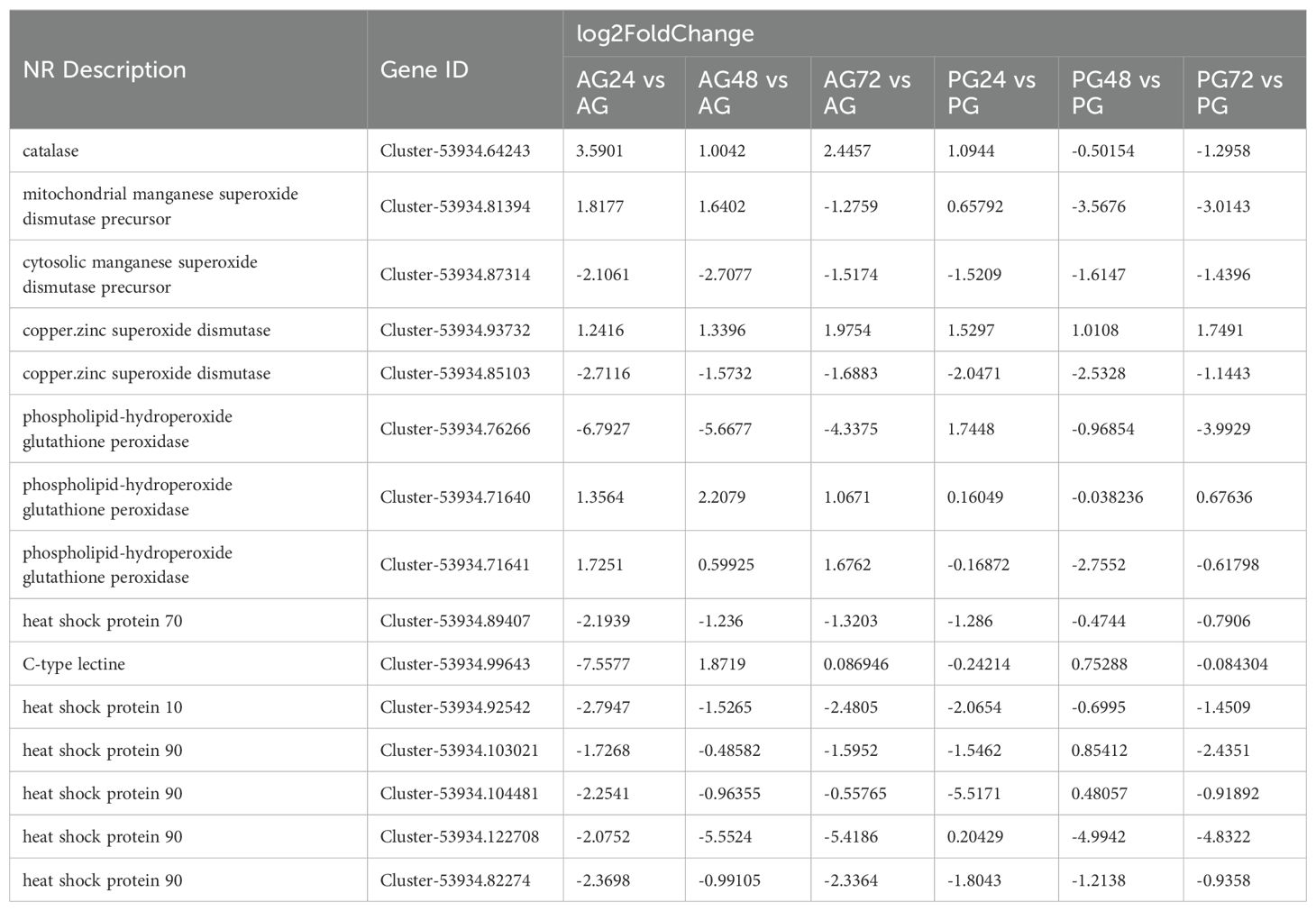
In this experiment, NH4Cl with a 24 h LC50 was used to simulate the ammonia nitrogen stress environment. The study showed that ammonia in the water mainly exists in ionic and non-ionic forms, and the two forms can be converted to each other (Cheng et al., 2019). However, this experiment mainly focused on the ammonia nitrogen stress of mud crab under this concentration. This concentration was obtained from the previous ammonia nitrogen lethality study. There are certain limitations in setting a single concentration, and a single concentration cannot reflect the regulation mechanism of mud crab under different ammonia nitrogen stress intensity. Excessive ammonia nitrogen stress reduces the immune capacity of crustaceans and increases their susceptibility to invasion by external pathogenic organisms (Cheng and Chen, 2002; Lin et al., 2023; Tang et al., 2022). Previous studies have shown that ammonia reduces the immune capacity, phagocytic activity, and antibacterial enzyme activity in white shrimp (Litopenaeus vannamei) (Yeh et al., 2004). Recent studies have demonstrated that ammonia stress decreases the expression of immune-related genes (Xiao et al., 2019). Exposure to ammonia nitrogen in mollusks has been shown to reduce lysosomal integrity, survival, cellular and immune responses, and disrupt energy allocation (Cong et al., 2017). In this study, the expression of immune-related genes, including HSP10, HSP70, HSP90, and CLEC, was downregulated in both the anterior and posterior gills after exposure to ammonia nitrogen stress, which weakened the immune capacity of the mud crab, consistent with previous studies. However, CLEC expression was downregulated after 24 hours of ammonia nitrogen stress but upregulated after 48 and 72 hours. This pattern was also observed in the posterior gill. This pattern may be associated with the activation of the immune response due to bacterial invasion (Table 2).
Previous studies have shown that ammonia nitrogen stress results in oxidative stress, characterized by increased oxygen consumption and decreased osmotic adjustment capacity, which affects the molting frequency of crustaceans and damages the hepatopancreas and gill tissues (Liang et al., 2019). It is noteworthy that aquatic animals possess an antioxidant stress system that involves a variety of enzymes, including catalase (CAT), superoxide dismutase (SOD), and glutathione peroxidase (GPx), which play a role in mitigating oxidative stress (Sun et al., 2024). In this experiment, we observed differences in the expression of related genes in the anterior and posterior gills. The expression of CAT was upregulated in the anterior gill, and upregulated in the posterior gill after 24 hours of ammonia nitrogen exposure, but downregulated at 48 and 72 hours. Furthermore, it has been demonstrated that organisms produce reactive oxygen species (ROS) in response to external stressors, which can cause cellular damage. SOD reduces superoxide radicals to form hydrogen peroxide and oxygen, while CAT and GPx further reduce hydrogen peroxide, effectively eliminating the harmful effects of reactive oxygen species (Liu et al., 2010). We observed differences in the expression of these genes between the anterior and posterior gills, as well as across different time points of stress, indicating distinct expression patterns of the antioxidant stress system in the gills of the mud crab under oxidative stress. This result not only demonstrates that the regulatory pathways of ammonia nitrogen metabolism in the gills of mud crabs differ before and after ammonia nitrogen stress, but also that the antioxidant stress pathways differ.
The results indicate that a total of 46 differentially expressed genes (DEGs) were identified in the anterior gills, while 36 DEGs were identified in the posterior gills across the different groups. These DEGs identified in the gills of mud crabs will aid in the discovery of genes related to ammonia resistance. Further analysis of the DEGs in the anterior gill revealed that upregulated DEGs were primarily enriched in receptor activity and signal receptor activity after short-term ammonia nitrogen stress. In contrast, downregulated DEGs were mainly enriched in nitrogen compound metabolism and cellular components, suggesting that short-term ammonia nitrogen stress leads to the downregulation of nitrogen metabolism-related genes and the inhibition of nitrogen metabolism pathways. After prolonged ammonia nitrogen stress, upregulated DEGs were primarily enriched in metabolic processes, hydrolase activity, and the cytoplasm, while downregulated DEGs were predominantly enriched in nitrogen compound metabolism, protein binding, and cellular macromolecule metabolism. This suggests that prolonged ammonia nitrogen stress continues to block nitrogen metabolism pathways, and the accumulation of ammonia nitrogen in the body begins to impair protein binding. To mitigate the adverse effects of ammonia nitrogen stress, the organism upregulated the expression of relevant hydrolase genes and enhanced the metabolism of other substances.
Further transcriptome analysis revealed distinct gene expression patterns between the anterior and posterior gills under ammonia nitrogen stress. The expression of ubiquitin-mediated proteolysis genes in AG24 and AG72 was significantly downregulated, whereas DEGs downregulated in PG24 and PG72 were primarily enriched in metabolism-related pathways, including pyrimidine and purine metabolism. Ubiquitin-mediated proteins are involved in various biochemical processes, playing a particularly important role in immunity (Wang et al., 2012). Purine and pyrimidine metabolism are key pathways in ammonia nitrogen metabolism (Liu et al., 2014). These findings suggest that short-term ammonia nitrogen stress leads to a decline in immune capacity in the anterior gills and the blockage of ammonia nitrogen metabolism in the posterior gills. Upregulated DEGs in AG24 were primarily enriched in ECM-receptor interaction pathways, which are crucial in various biological processes, particularly in wound healing (Farhadi et al., 2024). The upregulation of this pathway may be linked to tissue damage caused by ammonia nitrogen stress, with increased expression potentially aiding in tissue repair. DEGs upregulated in PG24 were primarily enriched in the biosynthesis of unsaturated fatty acids, which are essential nutrients involved in key processes such as biofilm function and immune response (Sun et al., 2020). In contrast, DEGs upregulated in AG72 were significantly enriched in nitrogen metabolism, while DEGs upregulated in PG72 were significantly enriched in immune-related pathways, particularly lysosome activity. The differential enrichment of DEGs between the anterior and posterior gills may be attributed to their distinct functional roles.
A pathway diagram of ammonia nitrogen metabolism was constructed based on the KEGG enrichment results (Figure 9). The figure shows that both the anterior and posterior gills share the same ammonia nitrogen metabolism pathways, but differ in the regulatory genes involved. In this study, NH4Cl was used to simulate external ammonia nitrogen stress to investigate the molecular mechanisms underlying ammonia nitrogen metabolism in the anterior and posterior gills of the mud crab. Previous studies have shown that ammonia nitrogen metabolism is a complex process, with crustacean nitrogen metabolism primarily involving three pathways (Liu et al., 2014). This study focused on the process of urea synthesis and excretion following NH4+ stress, as indicated by the expression of related enzyme genes. The experiment revealed that after 24 hours of ammonia nitrogen stress, the expression of glnA in the anterior gill was inhibited, which weakened the conversion of NH4+ into L-Glutamine, blocked the NH4+ metabolic pathway, and maintained a prolonged stress state, thereby impairing ammonia nitrogen metabolism and exacerbating the toxic effects of ammonia nitrogen on the organism. However, the upregulation of Puna and XDH genes in this pathway accelerated subsequent material conversion, ultimately leading to the transformation of ammonia nitrogen into urea for excretion. Increased expression of ROCF in another pathway enhanced the conversion of arginine to urea and boosted ammonia nitrogen metabolism in this pathway. After 72 hours of ammonia nitrogen stress, the expression of the glnA gene increased compared to 24 hours, which accelerated the conversion of NH4+ to L-Glutamine. Expression of additional genes in this pathway also increased, further enhancing ammonia nitrogen metabolism. In contrast to the 24 hour stress condition, the expression of argH in another pathway decreased, resulting in reduced ammonia nitrogen metabolism in this pathway. After 24 hours of ammonia nitrogen stress in the posterior gill, the expression of GART and ROCF in both pathways decreased, leading to reduced ammonia nitrogen metabolism in the posterior gill. Expression of ALC decreased after prolonged ammonia nitrogen stress compared to 24 hours, diminishing the response of both pathways in converting ammonia nitrogen into urea.
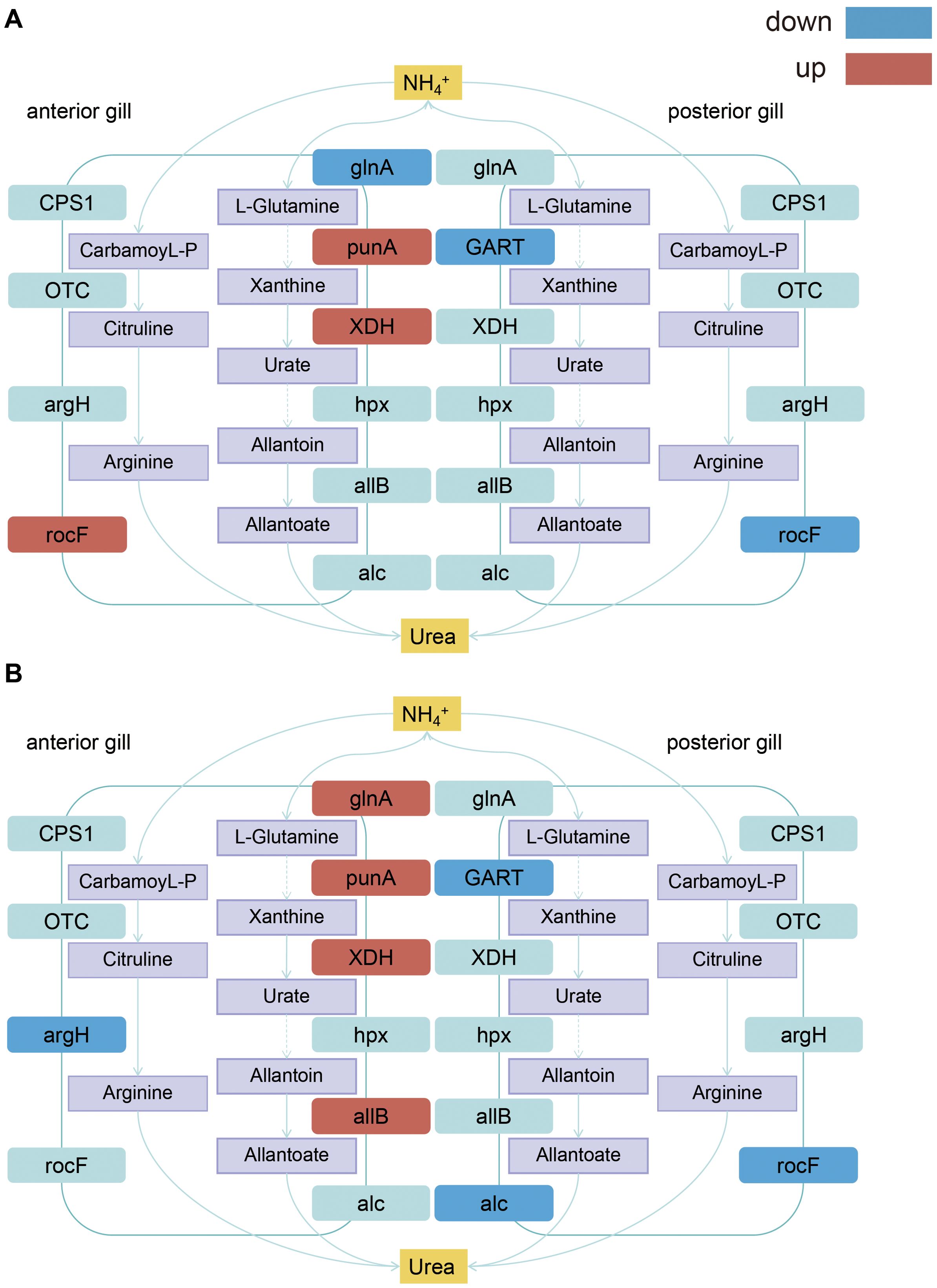
Figure 9. Schematic mechanisms of signaling pathways of anterior and posterior gills nitrogen metabolism in S. paramamosain. (A) Ammonia nitrogen metabolic pathway under 24 hour ammonia nitrogen stress; (B) Ammonia nitrogen metabolic pathway under 72 hour ammonia nitrogen stress. Purple is the corresponding metabolite, red is the up-regulated differentially expressed gene, blue is the down regulated differentially expressed gene, and light blue is the non differentially expressed gene on the metabolic pathway.
5 Conclusion
In summary, this study employed 325.13 mg/L NH4Cl to simulate external ammonia nitrogen stress and investigated the differential gene expression between the anterior and posterior gills, along with the molecular mechanisms of ammonia nitrogen metabolism. Tissue section observations revealed that ammonia nitrogen stress causes the cuticle to break, epithelial cells in the gill to die and detach, and the gill hemolymph cavity to shrink and collapse. These changes result in a reduction in the gill’s gas exchange capacity. This study explored the molecular mechanisms underlying ammonia nitrogen metabolism in mud crabs and examined the associated metabolic pathways. Through the analysis of ammonia nitrogen metabolism related pathways, it was found that there were differences in ammonia nitrogen metabolism pathways between the anterior gills and posterior gills, and the expression of ammonia nitrogen metabolism related genes in the anterior gill was relatively up-regulated, such as punA and XDH The expression of ammonia nitrogen metabolism related genes in the gill was down regulated, such as GART and rocf. The results showed that after ammonia nitrogen stress, the immune function of mud crab decreased, and the expression of HSP10, HSP70, Hsp90 and other immune genes decreased. These findings provide a solid foundation for future research on ammonia nitrogen stress. The results of this study provide new insights into the toxicological mechanisms of ammonia nitrogen on crustaceans.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors upon reasonable request, without undue reservation.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
CYW: Data curation, Formal analysis, Writing – review & editing, Investigation, Methodology. XL: Data curation, Methodology, Writing – original draft, Investigation. JZ: Writing – review & editing, Data curation, Formal analysis. SXW: Writing – review & editing, Data curation, Formal analysis. XZ: Writing – review & editing, Data curation, Formal analysis. ZHL: Writing – review & editing, Data curation, Formal analysis. YYF: Conceptualization, Methodology, Project administration, Resources, Writing – review & editing, Data curation. ZWJ: Conceptualization, Methodology, Project administration, Resources, Writing – review & editing, Investigation. LL: Conceptualization, Methodology, Project administration, Resources, Writing – review & editing, Formal analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key R & D Program of China (grant number 2023YFD2401005), the Dongying Science and Technology Innovation Major Project (grant number 2023ZDJH30), the Ningbo Municipal Public Welfare Research Plan (grant number 2023S128), and the K.C. Wong Magna Fund of Ningbo University.
Conflict of interest
Author ZWJ was employed by the company Ningbo Yifeng Aquaculture Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1587303/full#supplementary-material
Supplementary Table 1 | List of genes with their respective forward and reverse primers used for RT-PCR analysis.
Supplementary Table 2 | Summary of annotation results for transcripts of S. paramamosain.
Supplementary Table 3 | KOG classification of the unigenes.
Supplementary Table 4 | GO classification of all unigenes.
Supplementary Table 5 | KEGG classification of the unigenes.
Supplementary Table 6 | Summary of SNPs in the transcripts of S. paramamosain.
Supplementary Table 7 | List of the differentially expressed genes by AG24 vs AG.
Supplementary Table 8 | List of the differentially expressed genes by AG48 vs AG.
Supplementary Table 9 | List of the differentially expressed genes by AG72 vs AG.
Supplementary Table 10 | List of the differentially expressed genes by PG24 vs PG.
Supplementary Table 11 | List of the differentially expressed genes by PG48 vs PG.
Supplementary Table 12 | List of the differentially expressed genes by PG72 vs PG.
References
Anders S., Pyl P. T., Huber W. (2014). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng C.-H., Ma H.-L., Su Y.-L., Deng Y.-Q., Feng J., Xie J.-W., et al. (2019). Ammonia toxicity in the mud crab (Scylla paramamosain): The mechanistic insight from physiology to transcriptome analysis. Ecotoxicol. Environ. Saf. 179, 9–16. doi: 10.1016/j.ecoenv.2019.04.033
Cheng W., Chen J.-C. (2002). The virulence of Enterococcus to freshwater prawn Macrobrachium rosenbergii and its immune resistance under ammonia stress. Fish Shellfish Immunol. 12, 97–109. doi: 10.1006/fsim.2001.0363
Cong M., Wu H., Yang H., Zhao J., Lv J. (2017). Gill damage and neurotoxicity of ammonia nitrogen on the clam Ruditapes philippinarum. Ecotoxicology 26, 459–469. doi: 10.1007/s10646-017-1777-4
Dong Z., Mao S., Chen Y., Ge H., Li X., Wu X., et al. (2019). Effects of air-exposure stress on the survival rate and physiology of the swimming crab Portunus trituberculatus. Aquaculture 500, 429–434. doi: 10.1016/j.aquaculture.2018.10.049
Eddy F. B. (2005). Ammonia in estuaries and effects on fish. J. Fish Biol. 67, 1495–1513. doi: 10.1111/j.1095-8649.2005.00930.x
FAO (2024). Fishery and aquaculture statistics – yearbook 2021 (Rome, Italy: Food & Agriculture Org). Available at: https://books.google.it/books?id=mLn7EAAAQBAJ (Accessed February 2024).
Farhadi A., Xue L., Zhao Q., Han F., Xu C., Chen H., et al. (2024). Identification of key genes and molecular pathways associated with claw regeneration in mud crab (Scylla paramamosain). Comp. Biochem. Physiol. Part D: Genomics Proteomics 49, 101184. doi: 10.1016/j.cbd.2023.101184
Henry R. P., Lucu C., Onken H., Weihrauch D. (2012). Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals [Review. Front. Physiol. 3. doi: 10.3389/fphys.2012.00431
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kır M., Kumlu M., Eroldoğan O. T. (2004). Effects of temperature on acute toxicity of ammonia to Penaeus semisulcatus juveniles. Aquaculture 241, 479–489. doi: 10.1016/j.aquaculture.2004.05.003
Li Y., Zhang X., Tong R., Xu Q., Zhang N., Liao Q., et al. (2024). Mechanisms of ammonotelism, epithelium damage, cellular apoptosis, and proliferation in gill of Litopenaeus vannamei under NH4Cl exposure. Environ. Sci. Pollution Res. 31, 15153–15171. doi: 10.1007/s11356-024-32111-9
Liang C., Liu J., Cao F., Li Z., Chen T. (2019). Transcriptomic analyses of the acute ammonia stress response in the hepatopancreas of the kuruma shrimp (Marsupenaeus japonicus). Aquaculture 513, 734328. doi: 10.1016/j.aquaculture.2019.734328
Liang Z., Liu R., Zhao D., Wang L., Sun M., Wang M., et al. (2016). Ammonia exposure induces oxidative stress, endoplasmic reticulum stress and apoptosis in hepatopancreas of pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 54, 523–528. doi: 10.1016/j.fsi.2016.05.009
Lin Y., Huang J.-j., Dahms H.-U., Zhen J.-j., Ying X.-p. (2017). Cell damage and apoptosis in the hepatopancreas of Eriocheir sinensis induced by cadmium. Aquat. Toxicol. 190, 190–198. doi: 10.1016/j.aquatox.2017.07.008
Lin W., Luo H., Wu J., Hung T.-C., Cao B., Liu X., et al. (2023). A review of the emerging risks of acute ammonia nitrogen toxicity to aquatic decapod crustaceans. Water 15, 27. Available at: https://www.mdpi.com/2073-4441/15/1/27 (Accessed December 2022).
Liu C.-H., Chen J.-C. (2004). Effect of ammonia on the immune response of white shrimpLitopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol. 16, 321–334. doi: 10.1016/S1050-4648(03)00113-X
Liu H.-P., Chen F.-Y., Gopalakrishnan S., Qiao K., Bo J., Wang K.-J. (2010). Antioxidant enzymes from the crab Scylla paramamosain: Gene cloning and gene/protein expression profiles against LPS challenge. Fish Shellfish Immunol. 28, 862–871. doi: 10.1016/j.fsi.2010.02.008
Liu S., Pan L., Liu M., Yang L. (2014). Effects of ammonia exposure on nitrogen metabolism in gills and hemolymph of the swimming crab Portunus trituberculatus. Aquaculture 432, 351–359. doi: 10.1016/j.aquaculture.2014.05.029
Liu Z., Shi A., Zhao M., Zheng C., Chen J., Niu C., et al. (2024). Dietary N-carbamylglutamate (NCG) supplementation improves the survival, digestive and immunological parameters by promoting ammonia excretion in Chinese mitten crab (Eriocheir sinensis), under ammonia nitrogen stress. Aquaculture Fisheries. doi: 10.1016/j.aaf.2023.12.008
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mo N., Feng T., Zhu D., Liu J., Shao S., Han R., et al. (2024a). Analysis of adaptive molecular mechanisms in response to low salinity in antennal gland of mud crab, Scylla paramamosain. Heliyon 10. doi: 10.1016/j.heliyon.2024.e25556
Mo N., Shao S., Yang Y., Bao C., Cui Z. (2024b). Identifying low salinity adaptation gene expression in the anterior and posterior gills of the mud crab (Scylla paramamosain) by transcriptomic analysis. Comp. Biochem. Physiol. Part D: Genomics Proteomics 49, 101166. doi: 10.1016/j.cbd.2023.101166
Negro C. L., Collins P. (2017). Histopathological effects of chlorpyrifos on the gills, hepatopancreas and gonads of the freshwater crab Zilchiopsis collastinensis. Persistent effects after exposure. Ecotoxicol. Environ. Saf. 140, 116–122. doi: 10.1016/j.ecoenv.2017.02.030
Ning X., Han B., Shi Y., Qian X., Zhang K., Yin S. (2023). Hypoxia stress induces complicated miRNA responses in the gill of Chinese mitten crab (Eriocheir sinensis). Aquat. Toxicol. 261, 106619. doi: 10.1016/j.aquatox.2023.106619
Ozawa A., Sakaue M. (2020). New decolorization method produces more information from tissue sections stained with hematoxylin and eosin stain and masson-trichrome stain. Ann. Anat. - Anatomischer. Anzeiger 227, 151431. doi: 10.1016/j.aanat.2019.151431
Pan L., Si L., Liu S., Liu M., Wang G. (2018). Levels of metabolic enzymes and nitrogenous compounds in the swimming crab portunus trituberculatus exposed to elevated ambient ammonia-N. J. Ocean Univ. China 17, 957–966. doi: 10.1007/s11802-018-3574-y
Preena P. G., Rejish Kumar V. J., Singh I. S. B. (2021). Nitrification and denitrification in recirculating aquaculture systems: the processes and players. Rev. Aquaculture 13, 2053–2075. doi: 10.1111/raq.12558
Rahi M. L., Moshtaghi A., Mather P. B., Hurwood D. A. (2018). Osmoregulation in decapod crustaceans: physiological and genomic perspectives. Hydrobiologia 825, 177–188. doi: 10.1007/s10750-018-3690-0
Romano N., Zeng C. (2013). Toxic effects of ammonia, nitrite, and nitrate to decapod crustaceans: A review on factors influencing their toxicity, physiological consequences, and coping mechanisms. Rev. Fisheries Sci. 21, 1–21. doi: 10.1080/10641262.2012.753404
Shen M., Wang Y., Tang Y., Zhu F., Jiang J., Zhou J., et al. (2024). Effects of different salinity reduction intervals on osmoregulation, anti-oxidation and apoptosis of Eriocheir sinensis megalopa. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 291, 111593. doi: 10.1016/j.cbpa.2024.111593
Sun Y., Fu Z., Liu X., Ma Z. (2024). The Impact of Acute Ammonia Nitrogen Stress on the Gill Tissue Structure and Antioxidant Ability of Gills and Red and White Muscle in Juvenile Yellowfin Tuna (Thunnus albacares). Antioxidants 13, 1357. Available at: https://www.mdpi.com/2076-3921/13/11/1357 (Accessed November 2024).
Sun P., Zhou Q., Monroig Ó., Navarro J. C., Jin M., Yuan Y., et al. (2020). Cloning and functional characterization of an elovl4-like gene involved in the biosynthesis of long-chain polyunsaturated fatty acids in the swimming crab Portunus trituberculatus. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 242, 110408. doi: 10.1016/j.cbpb.2020.110408
Tang D., Wu Y., Wu L., Bai Y., Zhou Y., Wang Z. (2022). The effects of ammonia stress exposure on protein degradation, immune response, degradation of nitrogen-containing compounds and energy metabolism of Chinese mitten crab. Mol. Biol. Rep. 49, 6053–6061. doi: 10.1007/s11033-022-07393-2
Wang Q., Chen L., Wang Y., Li W., He L., Jiang H. (2012). Expression characteristics of two ubiquitin/ribosomal fusion protein genes in the developing testis, accessory gonad and ovary of Chinese mitten crab, Eriocheir sinensis. Mol. Biol. Rep. 39, 6683–6692. doi: 10.1007/s11033-012-1474-6
Xiao J., Li Q.-Y., Tu J.-P., Chen X.-L., Chen X.-H., Liu Q.-Y., et al. (2019). Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotoxicol. Environ. Saf. 180, 491–500. doi: 10.1016/j.ecoenv.2019.05.029
Xing Y.-f., Feng Y.-f., Tian J., Zhang Y., Han C., Liu X.-j., et al. (2025). Nitrite and sulfide stress affects physiological and metabolic functions of gills in crab Eriocheir sinensis. Aquaculture 594, 741437. doi: 10.1016/j.aquaculture.2024.741437
Yang C., Zhong Z.-F., Wang S.-P., Vong C.-T., Yu B., Wang Y.-T. (2021). HIF-1: structure, biology and natural modulators. Chin. J. Natural Medicines 19, 521–527. doi: 10.1016/S1875-5364(21)60051-1
Yeh S.-T., Liu C.-H., Chen J.-C. (2004). Effect of copper sulfate on the immune response and susceptibility to Vibrio alginolyticus in the white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 17, 437–446. doi: 10.1016/j.fsi.2004.04.016
Yuan Z., Niu J., Qin K., Wang C., Mu C., Wang H. (2024). Metabolic mechanism of Scylla paramamosain gill mitochondria in response to acute low salinity stress. Aquaculture Rep. 37, 102234. doi: 10.1016/j.aqrep.2024.102234
Zhang X., Pan L., Wei C., Tong R., Li Y., Ding M., et al. (2020). Crustacean hyperglycemic hormone (CHH) regulates the ammonia excretion and metabolism in white shrimp, Litopenaeus vannamei under ammonia-N stress. Sci. Total Environ. 723, 138128. doi: 10.1016/j.scitotenv.2020.138128
Zheng P., Han T., Li X., Wang J., Su H., Xu H., et al. (2020). Dietary protein requirement of juvenile mud crab Scylla paramamosain. Aquaculture 518, 734852. doi: 10.1016/j.aquaculture.2019.734852
Zhou L., Zhao F., Shi Y., Liu H., Zhang K., Zhang C., et al. (2025). Comprehensive whole transcriptome analysis reveals specific lncRNA- and circRNA-ceRNA networks of Chinese mitten crab (Eriocheir sinensis) in response to hypoxia stress. Aquaculture 598, 742017. doi: 10.1016/j.aquaculture.2024.742017
Keywords: Scylla paramamosain, ammonia nitrogen, gills, transcriptome, differentially expressed genes
Citation: Wang C, Liu X, Zhou J, Wang S, Zheng X, Li Z, Fu Y, Jin Z and Liu L (2025) Transcriptome analysis of gills reveal the key metabolism-related genes and pathways response to ammonia nitrogen stress in Scylla paramamosain. Front. Mar. Sci. 12:1587303. doi: 10.3389/fmars.2025.1587303
Received: 04 March 2025; Accepted: 07 April 2025;
Published: 09 May 2025.
Edited by:
Yong-Jun Chen, Southwest University, ChinaReviewed by:
D. K. Meena, Central Inland Fisheries Research Institute (ICAR), IndiaBaoquan Gao, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2025 Wang, Liu, Zhou, Wang, Zheng, Li, Fu, Jin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Liu, bGl1bGVpMUBuYnUuZWR1LmNu
 Chaoyue Wang
Chaoyue Wang Xiao Liu1
Xiao Liu1 Lei Liu
Lei Liu