- 1Department of Microbiology, Oregon State University, Corvallis, OR, United States
- 2Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA, United States
- 3Marine Science Institute, University of California, Santa Barbara, Santa Barbara, CA, United States
- 4U.S. Geological Survey, Eastern Ecological Science Center at Patuxent Research Refuge, Laurel, MD, United States
- 5BioSciences Department, Rice University, Houston, TX, United States
- 6Department of Biology, Boston University, Boston, MA, United States
- 7Department of Biological Sciences, Florida Atlantic University, Davie, FL, United States
- 8Minderoo Foundation, Perth, WA, Australia
- 9Center for Global Discovery and Conservation Science, Arizona State University, Hilo, HI, United States
- 10Friday Harbor Laboratories, University of Washington, Friday Harbor, WA, United States
- 11Rutgers New Jersey Medical School Molecular and Genomics Core, Rutgers University, NJ, United States
- 12National Institute of Water and Atmospheric Research, Christchurch, New Zealand
- 13Antarctic Research Centre, Victoria University of Wellington, Wellington, New Zealand
- 14Department of Environmental Science, Policy, and Management, University of California, Berkeley, Berkeley, CA, United States
Coral reefs play vital roles in global marine systems and are currently facing increased threats of bleaching. Coral bleaching is heavily influenced by the host-associated microeukaryote community – most notably the dinoflagellate family Symbiodiniaceae. The apicomplexan family Corallicolidae, is the second most abundant member of the microeukaryote community, yet their role in coral health is largely unknown. To explore the role that this apicomplexan and the greater non-metazoan microeukaryotic community play in coral health, samples of a thermally sensitive scleractinian coral, Acropora hyacinthus, were collected over the course of a severe coral bleaching event and its aftermath. Through 18S rRNA gene sequencing analysis, we found that taxa within the family Corallicolidae were relatively enriched in corals during, and immediately after, the severe bleaching event as compared to before or one year after. Although utilizing 18S rRNA gene sequencing methods is not the standard for Symbiodiniaceae community profiling, we were able to observe symbiont shuffling among the Symbiodiniaceae communities, as the dominant algal symbiont shifted from the genus Cladocopium to the genus Symbiodinium following the bleaching event. Furthermore, the non-metazoan microeukaryote community displayed a general shift towards a state of dysbiosis; evidenced by substantial changes in both microeukaryote community composition and dispersion. These results offer insight into the dynamics of apicomplexans throughout the course of an increasingly common global coral reef stressor.
1 Introduction
Globally, coral reefs face a plethora of challenges (Riegl et al., 2009), with concerns such as marine heatwaves and bleaching events becoming more frequent and intense (Hughes et al., 2018) as a result of increased atmospheric CO2 (Coles and Brown, 2003). The health of both reefs and individual corals is intimately linked to the composition and ecological dynamics within the holobiont, as disruptions to community structure may lead to reduced coral health and fitness due to dysbiosis (Reshef et al., 2006; Zaneveld et al., 2017). The holobiont is comprised of the coral host, and the collection of microorganisms from all domains of life as well as viruses (Rohwer et al., 2002; Knowlton and Rohwer, 2003; van Oppen et al., 2009; Correa et al., 2021). Despite this diversity, much of the focus in coral holobiont studies centers around bacteria, viruses, and the photosynthetic, microeukaryote symbiont, Symbiodiniaceae (Rohwer et al., 2002; Vega Thurber et al., 2017). However, recent initiatives have been made to uncover the broader diversity of the microeukaryotes inhabiting the coral holobiont, beyond Symbiodiniaceae (Clerissi et al., 2018; Dennis et al., 2020; Galindo-Martínez et al., 2022; Iha et al., 2021; Kwong et al., 2019; Mathur et al., 2018; Paulino et al., 2020; Qiu et al., 2021; Sweet et al., 2019; Tandon et al., 2023; Vohsen et al., 2020).
Given their likely role in coral health, a recently identified microeukaryote from the phylum Apicomplexa has garnered interest in the microeukaryotic fraction of the coral holobiont (Kwong et al., 2019). The phylum Apicomplexa falls under the eukaryotic supergroup SAR (Stramenopile, Alveolata, Rhizaria) and is comprised mainly of single-celled obligate intracellular parasites (Gräf et al., 2015) that evolved from free-living phototrophic ancestors and have retained a non-photosynthetic plastid. Within Apicomplexa, there exist several other pathogenic parasites known to cause disease; well-known examples include Plasmodium and Toxoplasma which, in humans, cause malaria and toxoplasmosis, respectively (Weiss and Kim, 2013; Snow et al., 2005). These are just two in over 6,000 named species within Apicomplexa – although this is likely a vast underestimation of the group’s diversity (Adl et al., 2007). The apicomplexan Corallicolida was recently identified as the second most abundant coral-associated microeukaryote after Symbiodiniaceae in most coral genera (Kwong et al., 2019; Keeling et al., 2021). Apicomplexan DNA has been found in corals from a variety of locations and hosts, such as in low latitudes (Janouškovec et al., 2015; Upton and Peters, 1986; Toller et al., 2002; Slapeta and Linares, 2013; Janouškovec et al., 2012; Kwong et al., 2019; Kirk et al., 2013; Janouškovec et al., 2013), in cold and deep-sea corals, and in diverse hosts including both octocorals (Octocorallia) and hexacorals (Hexacorallia) (Mathur et al., 2018; Vohsen et al., 2020). This range has even recently been described outside of the coral holobiont within the fireworm species Hermodice carunculata, a native of the Atlantic Ocean and Mediterranean Sea. The circumstances in which Corallicolida exist within this non-coral host indicate that this is a fireworm-specific infection and raises the possibility that H. carunculata is a vector for Corallicolida. If H. carunculata is indeed a vector, it is unlikely that they are the only species filling this role, thereby suggesting the ecological effects of Corallicolida reach beyond corals alone (Bonacolta et al., 2025). Despite their range and abundance in coral microbiomes, Apicomplexa remain relatively understudied, thereby leaving our understanding of the effects of Corallicolida on their coral hosts quite limited.
The phylogenetic placement of Corallicolida within the anthozoan-infecting phylum Apicomplexa suggests a potentially parasitic relationship with corals (Keeling et al., 2021). At present, however, there is no evidence suggesting that Corallicolida are detrimental to coral health. In fact, these microeukaryotes are largely found in apparently healthy corals (Kwong et al., 2019). This association with apparently healthy corals does not rule out the potential for members of Corallicolida to act as opportunistic parasites shifting from a seemingly commensal relationship with their host to a parasitic relationship in the presence of a stressor, as has been shown in corals infected with the bacteria Aquarickettsia (Klinges et al., 2019). A parasitic relationship has been previously suggested, as apicomplexans identified in Mussismila braziliensis corals were more abundant in diseased colonies (Garcia et al., 2013). Additionally, the Corallicolidae family were found to be significantly enriched in the thermally sensitive octocoral Paramuricea clavata, whereas the dinoflagellate order Syndiniales was significantly enriched in thermally resistant individuals (Bonacolta et al., 2024). Most of these studies into the relationship between climate stress and apicomplexans have focused on soft octocorals, and this research has yet to extend to hexacorals (which include the reef-building scleractinian corals). The connection between members of Corallicolida and thermal sensitivity in corals is not yet understood, though based on prior observations of the Corallicolida-host dynamic, potential connections include a context-dependent relationship between Corallicolida and the host in which Corallicolida act as opportunistic pathogens in the presence of environmental stressors. Therefore, as shifting global conditions continue to threaten the health and resilience of coral reef ecosystems, an understanding of Corallicolida and broader microeukaryote community dynamics within the coral holobiont is needed.
To address this, we utilized 18S rRNA gene sequencing to assess the diversity and abundance of Corallicolidae and other non-metazoan microeukaryotes within the thermosensitive, scleractinian coral Acropora hyacinthus. Our study spans a two-year period that includes an extreme marine heatwave and associated coral bleaching event on the island of Mo’orea, French Polynesia. Based on previous reports of Corallicolida enrichment in thermally sensitive octocorals (Bonacolta et al., 2024) and corals in a state of stress following disease (Garcia et al., 2013), we hypothesize that A. hyacinthus corals facing extreme thermal stress would exhibit heightened abundances of Corallicolida populations.
2 Materials and methods
2.1 Experimental design
The corals sampled in the present study were taken across several sites around the lagoon (back reef) and north fore reef of the island of Mo’orea, French Polynesia over several time points from August 2018 to March 2020. This sampling regime captured a severe marine heatwave and related bleaching event in March of 2019 (Speare et al., 2022). The coral species sampled, Acropora hyacinthus, belongs to a family of stony corals known as acroporids, which are a group particularly susceptible to bleaching and mortality in the face of bleaching (Marshall and Baird, 2000; Jones et al., 2008; Obura 2001; Mccowan et al., 2012). Overall, high mortality among all corals, specifically acroporids, occurred during the 2019 bleaching event (Winslow et al., 2024). We analyzed corals sampled before (August 2018), during (March 2019), and after (August 2019, March 2020) the bleaching event.
Samples were collected from sites 1, 2, 3, 4, and 5 from established Mo’orea Coral Reef Long-Term Ecological Research (MCR-LTER) sites (Leichter et al., 2013). Additional sampling sites (0, 3.5, and 5.5) were added to ensure each side of the island (north, east, and west) had three replicate sites. A map of Mo’orea with annotated LTER sites and coral samples is available in Supplementary Figure 1. Specimens from replicate coral colonies were taken from both fore reef and back reef habitats, though very few samples (n=3) were taken from the fore reef due to high mortality at those locations. It is important to note that these sites represent highly distinct reef habitats (the fore reef being offshore oceanside of the lagoon and typically ~10 meters deeper than the back reef), which has been shown to lead to differences in microbial community makeup within corals (Nelson et al., 2011). However, for this analysis, patterns remained consistent when analyses were performed with and without the fore reef samples; therefore, they were retained and grouped together for all analyses. At each of the original LTER sites, thermistors located ten feet deep record temperature data every twenty minutes. Sites included in this study were chosen based on the presence of individual corals that survived through August 2019, which was true for all sites but LTER 6. At this site, complete mortality of all corals was observed by August 2019. Because of this disparity in survival rates of coral between sites, there are an uneven number of samples taken from each site, presenting a potential limitation in the statistical power of our analyses, particularly as it pertains to comparisons between shores of the island. Total sample numbers for each site are presented in Supplementary Table 1.
2.2 Sample collection
Approximately 2-3 g of target corals (mucus, tissue and skeleton) were sampled on fore (FOR) and back (BAK) reefs using sterile bone cutters and placed in individual 15 mL sterile tubes containing 6 mL of DNA/RNA Shield, five 6.35-mm ceramic beads, and 1.35 g of garnet bead matrix (MP Biomedicals, Ohio, USA). Samples were placed on ice and returned to Gump Marine Station for initial processing. Bead tubes were then vortexed at maximum speed (3,000 rpm) for 25 minutes to disrupt coral cells and skeleton completely. The sample was allowed to settle at room temperature and 1.5 mL aliquots of coral slurry were placed in 2 mL sterile tubes for storage at -40°C. Frozen slurries were transported to Oregon State University on Techni Ice (Frankston, Vic, AU).
2.3 DNA extraction and rRNA metabarcoding
To increase microbial nucleic acid yield, DNA extraction of individual coral samples was performed using a modified protocol followed by a Zymo-BIOMICS ™ DNA/RNA Miniprep Kit (Zymo Research, Irvine, CA, USA; RRID: SCR_008968). Coral slurry (300 µL) was enzymatically digested with 30 µL Chicken Egg White Lysozyme (10 mg/µL), 1.8 µL mutanolysin (50 KU/mL), 1.8 µL lysostaphin (4 KU/mL). These samples were then incubated at 37°C for 1 hour, followed by the addition of 15 µL of proteinase K (20 mg/µL) and 30 µL of Proteinase K digestion buffer (0.1M NaCl, 10 mM Tris, 1mM EDTA, 0.5% SDS, RNA/DNAse-Free water) (Kearney et al., 2018; Yang et al., 2021; Belser et al., 2023). These samples were once again incubated, this time for 1 hour at 50°C. One volume of DNA/RNA Lysis Buffer (ZymoBIOMICS™ DNA/RNA Miniprep) was then added to each sample, and the rest of the extraction was performed according to the ZymoBIOMICS™ protocol. Eluted DNA from each sample was then stored at -80 °C.
Amplicons were prepared using a modified nested polymerase chain reaction (PCR) approach validated in 2019 (del Campo et al., 2019). Step 1 of this approach was a 25 µL PCR reaction consisting of 12.5 µL of Platinum II Hot-Start PCR Master Mix, 1 µL of forward and 1 µL of reverse UnonMet-PCR metazoan excluding primers (10 µM) targeting the V4 region of 18S eukaryotic rRNA gene (UnonMetaF 5’-GTGCCAGCAGCCGCG-3’, UnonMetaR 5’-TTTAAGTTTCAGCCTTGCG-3’) (Bower et al., 2004), and 1 µL of template DNA. PCR was performed using the following thermocycler protocol: 30 s at 98°C, 35 cycles each consisting of 10 s at 98°C, 30 s at 51.1°C, 1 min at 72°C, and finishing with 5 min at 72°C. PCR products were visualized on a 1.5% agarose gel. Successfully amplified PCR products were then purified using Agencourt AMPure XP beads (Beckman Coulter Inc. Brea, California, USA) following the manufacturer’s guidelines. However, 80% ethanol, rather than the recommended 70%, was used for the washing steps, and a 5-minute drying step was included after the final ethanol wash to evaporate excess ethanol. One µL of the purified DNA from the first step was used as the template for a second amplification using the eukaryotic-specific primer set from Comeau et al. (2011) targeting the V4 region of the 18S eukaryotic rRNA gene, E572F+E1009R (E572F: 5’-CYGCGGTAATTCCAGCTC-3’, E1009R: 5’-AYGGTATCTRATCRTCTTYG-3’) (Comeau et al., 2011). These 18S rRNA primers, though open to amplification bias present in many PCR methods, perform as well as or sometimes better than other commonly used universal 18S rRNA primers in their ability to amplify the microeukaryotic V4 region, with the exception of metazoans, which the primers are designed to exclude (del Campo et al., 2019). Primers from this step were already attached to unique identifying barcodes that were specific to each sample (Holt et al., 2022). These PCR products were once again cleaned using the method above and quantified with a high-sensitivity (HS) dsDNA assay kit and Qubit 4.0 fluorometer (Invitrogen, Waltham, Massachusetts, USA). Three PCR negative controls were also prepared following the same protocol. Appropriate equimolar volumes of each sample were then calculated and pooled for high throughput sequencing at the Center for Quantitative Life Sciences (CQLS) at Oregon State University using a 2x300 paired-end sequencing approach on the Illumina MiSeq (Illumina, San Diego, California, USA) platform with v3 chemistry. Prior to sequencing, DNA quality was assessed using TapeStation HS-D5000 (Agilent, Santa Clara, California, USA).
2.4 18S rRNA gene analysis
2.4.1 Sequence read preprocessing
Individual quality control reports for each sequencing read were created using FastQC (Andrews, 2010). Aggregated quality reports were then generated using MultiQC (Ewels et al., 2016) to visualize the mean quality scores for the forward and reverse sequencing reads individually. Forward, reverse, and the reverse complement of both primer sequences were removed from all reads using a two-step cutadapt (Martin, 2011) approach. Further quality control was performed using R and RStudio (v. 4.3.0; R Core Team, 2022) following the standard DADA2 (v. 1.28.0; Callahan et al., 2016) pipeline. Low-quality base regions identified in the MultiQC report were removed by truncating the forward and reverse reads at 270 and 220 base pairs, respectively. Aside from increasing the truncQ value from 0 to 2, default parameters were used for filterAndTrim with FastQC. Following the filtering steps, forward and reverse reads were merged and only those ranging from 396 – 406 bp were kept. Chimeric sequences were then removed.
Taxonomy was then assigned to the ASV (amplicon sequence variant) level using default parameters and the PR2 (Protist Ribosomal Reference) database (v. 5.0.0; Guillou et al., 2013). Sequence taxonomy was also assessed via BLASTn (Supplementary File 1). Off-target sequences identified as metazoan (206,484 reads corresponding to animal reads from the phyla Cnidaria, Porifera, Arthropoda, and Annelida) and bacterial (49 reads), which accounted for 1% of the total input reads, were removed from the analysis. A phyloseq object was then created using the phyloseq package in R Studio (v.1.44.0 R Core Team, 2022; McMurdie and Holmes, 2013). Using the ‘combined detection method’ in the decontam package (v. 1.20.0; Davis et al., 2018), four contaminant ASVs identified by both prevalence and quantification thresholds (threshold = 0.4) were removed from all samples. Furthermore, a single ASV not annotated to the domain level was removed. After all contaminant ASVs were removed, negative controls were excluded from downstream analysis. The number of reads removed per sample at each preprocessing step can be found in Supplementary File 1.
To then account for differences in library sizes across samples, the rrarefy function from the vegan package (v. 2.6-8; Oksanen et al., 2012) was used to randomly subsample sample counts to 14,205 reads because this depth captured a majority of the observed diversity (Supplementary Figure 2). This approach resulted in the loss of two samples that had 1,084 and 1,865 reads, respectively, resulting in a total of 83 samples remaining. The rarefied phyloseq object was subsequently used for the alpha diversity and relative abundance analyses, and the non-rarefied phyloseq object was used for the beta diversity and differential abundance analyses. After all filtering and quality control steps were performed, 1,205 ASVs corresponding to 10,319,623 reads across 83 samples remained in the non-rarefied phyloseq object for subsequent analysis. In the non-rarefied phyloseq object, the minimum, maximum, and average read depth was 14,205, 592,613 and 124,332.8 reads, respectively. Final sample numbers, read counts per sample group, and the observed number of non-metazoan eukaryotic taxa are reported in Supplementary Table 2 and Supplementary File 1.
2.4.2 Microbiome analysis
Both Shannon diversity (richness and evenness) and Chao1 richness indices were calculated to assess within-sample alpha diversity. This was done using the estimate_richness function in the phyloseq R package (McMurdie and Holmes, 2013). Chao1 diversity data were then log-transformed to obtain normally distributed model residuals. To evaluate the effects of time and site on Shannon diversity and Chao1 diversity, a linear mixed effect model was created using the lme4 R package (v. 1.1.35.1; Bates et al., 2015). In this model, timepoint, LTER site, and their interaction were set as fixed effects, and coral sample ID was set as the random effect. Pairwise comparisons were made using the emmeans package (v. 1.10.0; Lenth, 2017) to calculate estimated marginal means (EMM), with the Tukey-Kramer p-value adjustment method. Taxon counts were transformed to reflect their relative abundance per sample, and the top 100 most abundant taxa were included in the plot. Data were also subset to only include the taxa within the phylum Apicomplexa, transformed to relative abundance values, and then pruned to only plot the top 100 most abundant taxa. Taxa that were not annotated to the species level were reported at the next best taxonomic rank. Significant differences in Corallicolidae mean relative abundance by location within time points were assessed using the Kruskal-Wallis test, followed by the Dunn post hoc test with false discovery rate p-value correction. Beta diversity analysis was performed using unrarefied data that were robust centered log-ratio (rCLR) transformed using the microViz package (v. 0.12.5; Martino et al., 2019; Barnett, 2021). Transformed data were ordinated using a principal component analysis (PCA). Differences in beta diversity were identified by a permutational analysis of variance (PERMANOVA) using the adonis2 functions in the vegan package (v. 2.6-8; Oksanen et al., 2012) and pairwise.adonis (Martinez Arbizu, 2020) for pairwise comparisons. Dispersion, as distance-to-centroid, was calculated using the vegan::betadisper function, and statistical significance was determined using an ANOVA. Any significant results were then reanalyzed using a permutation test to identify significant pairwise comparisons. Finally, differential abundance analysis was performed using ANCOM-BC2 (v. 2.6.0; Lin and Peddada, 2024) on unrarefied data to identify differentially abundant taxa by shore type within single time points. Multiple pairwise comparisons were made, therefore we incorporated the mixed directional false discovery rate (mdFDR) into the ANCOM-BC model. P-values were adjusted using the FDR method, and only those taxa also passing a sensitivity threshold were considered differentially abundant. For alpha diversity, beta diversity, dispersion, and differential abundance, the following number of samples were used for each analysis: 9 samples from the north and east, and 4 samples from the west in March 2019, 12 samples from the north, 9 samples from the east, and 4 samples from the west in August 2019, and 5 samples from the north, 4 samples from the east, and 3 samples from the west in March 2020.
3 Results
3.1 The health of Acropora hyacinthus corals across Mo’orea was significantly affected by a severe thermal stress event in 2019
During early 2019, the island of Mo’orea, French Polynesia, experienced a severe marine heatwave that lasted from approximately December 2018 to July 2019, with peak temperatures above 30°C occurring in March, April, and May 2019 (Speare et al., 2022). Bottom thermistor temperatures for this period were largely consistent across sites (Figure 1A, Pearson correlation test, p < 0.01), despite a data gap at LTER 2 (green line) and LTER 3 (brown line) from July 2018 to September 2019.
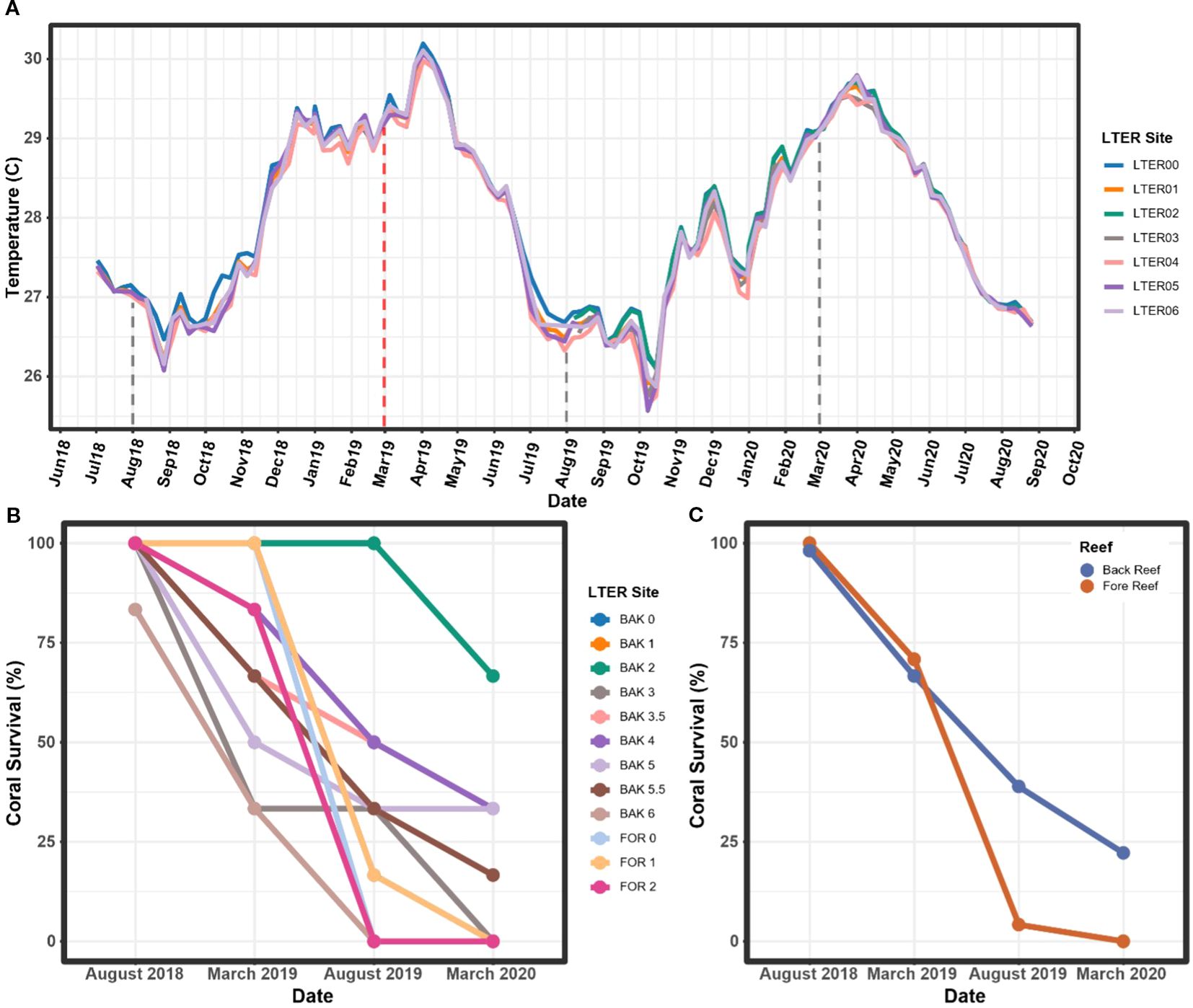
Figure 1. (A) Mean daily temperature, in degrees Celsius (°C), by week at each site located around Mo’orea, French Polynesia, from July 2018 to September 2020. A data gap for LTER 2 and 3 is located between July 2018 and September 2019, though temperature at these sites is strongly correlated with each of the other sites (p < 0.01) and temperatures from this period in LTER 3 are expected to follow trends seen in the other sites. (B) A. hyacinthus survival rates by site. (C) A. hyacinthus survival rates by habitat type (fore and back reefs). Sampling time points are denoted by the dashed lines, and the heat wave sampling timepoint is denoted by the red dashed line. For the purposes of this study, “BAK” denotes that the sample was taken from the back reef, “FOR” denotes that the sample was taken from the fore reef, and numbers designate which site the sample was taken from.
Acropora hyacinthus experienced extensive mortality during the marine heatwave. Eleven out of the twelve sites began August 2018 with 100% of target coral individuals alive (n = 6/6 colonies), yet between August 2018 and March 2020, each site saw some degree of mortality (Figure 1B). Sites BAK 6 and FOR 2 underwent a complete loss of all six target individuals by August 2019, followed by complete losses at BAK 3 and FOR 1 in March 2020 (Figure 1B). The site with the highest coral survivorship was BAK 2, which saw 66.7% (n = 4/6) of corals surviving at the March 2020 time point (Figure 1B). Every other site had an overall mortality of greater than 60% (two or fewer colonies remaining) at the March 2020 time point (Figure 1B). In terms of habitat, the back reefs had higher coral survival by percentage than the fore reef, with a final average percentage of 22% of corals alive as opposed to 0% on the fore reef in March 2020 (Figure 1C). However, it is important to note that there were more sites and coral individuals analyzed in the back reef than the fore reef, leading to an unbalanced design when considering habitat as a variable. Coral survival was significantly linked to time point in our linear mixed model (LMM, p < 0.001), but site and habitat were not found to impact the percentage of corals alive.
3.2 Time and space impact relative abundance of non-metazoan microeukaryotes within the coral holobiont
To understand the makeup of the non-metazoan microeukaryote community within A. hyacinthus samples, relative abundances of the top 100 taxa were evaluated (Figure 2A). Taxa within the family Corallicolidae were observed at generally higher relative abundances on the east (sites 3, 3.5, and 4) and west (sites 5 and 5.5) shores of the island compared to the north shore (Figure 2A). Specifically, during the heatwave in March 2019, the east shore had a significantly higher Corallicolidae mean relative abundance (9.63% ± 7.05%) compared to the north shore (3.94% ± 3.34%, p = 0.037, z = -2.5, Dunn Test). After the heatwave in August 2019, the west shore had a significantly higher member relative abundance (35.07% ± 30.74%) compared to the north shore (7.7% ± 17.7%, p = 0.032, z = 2.5, Dunn Test). In both cases, the significant increase in mean relative abundance was supported by a large effect size (d = 1.1 and 1.5, respectively). Additionally, although we were focused on the Apicomplexa, when evaluating all non-metazoan microeukaryotes, we found that two dinoflagellate genera within the family Symbiodiniaceae, Cladocopium and Symbiodinium, had the highest relative abundances across sites (Figure 2A). Symbiodinium was found in higher abundances on the north shore of the island (sites 0, 1, and 2), increasingly in the time points occurring after the bleaching event. Other less common microeukaryotic taxa also became apparent on the north shore in August 2019 following the bleaching event. These include a diatom, Cylindrotheca closterium, found only at LTER 4 and several red algae varieties, Hydrolithon onkodes, a member of the genus Polysiphonia, and a taxon from the Rhodomelaceae family at LTER 3.
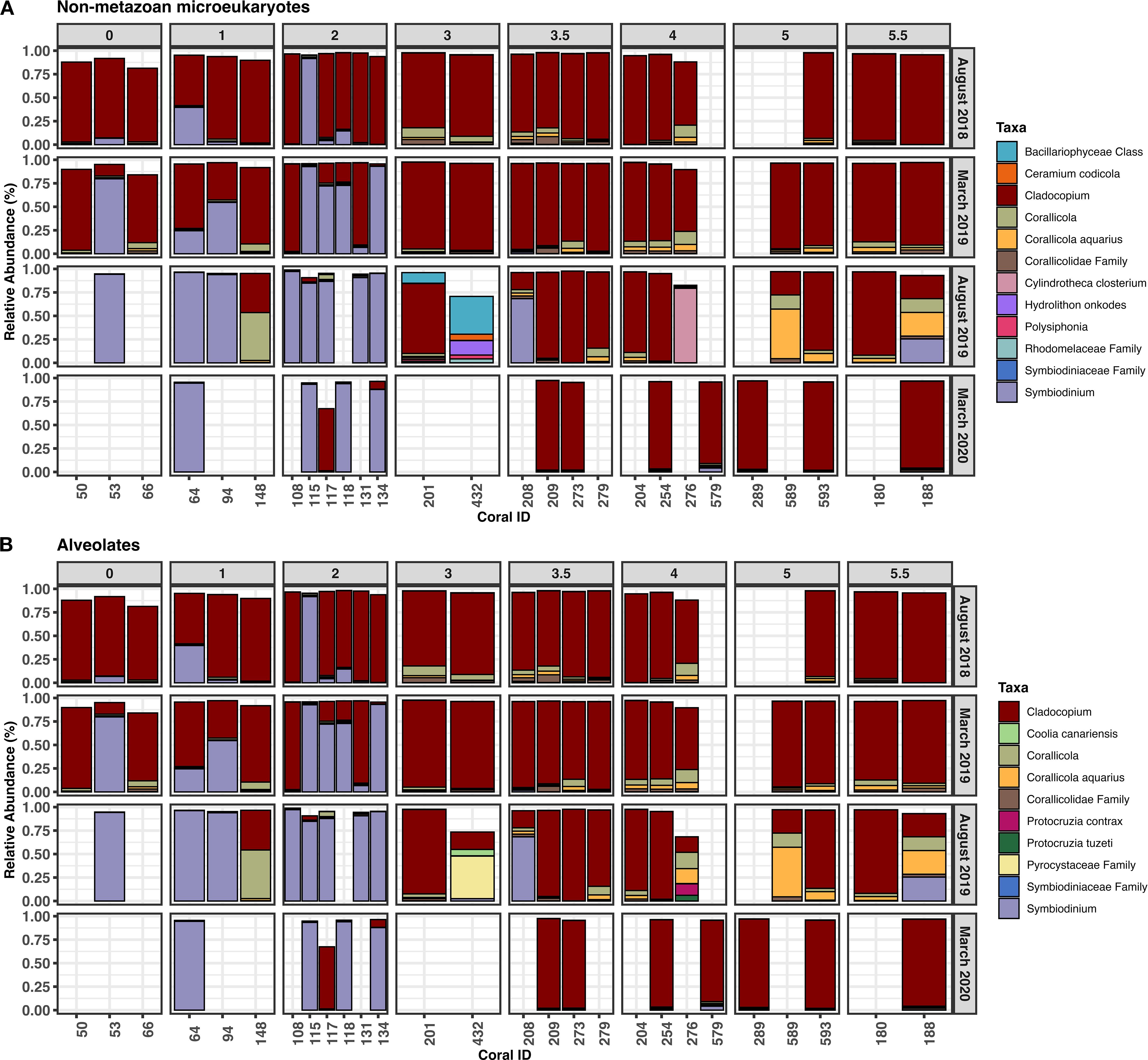
Figure 2. Relative abundances of the top 100 taxa observed over time within (A) all non-metazoan microeukaryotes and (B) alveolates only, grouped by site. Taxa that were not annotated to the species level were reported at the next highest taxonomic rank. Abundances do not total to 100% because only the top 100 most abundant taxa across all samples were plotted. Blank columns represent samples lost during collection or coral death. Sample descriptions are in Supplementary File 1.
We observed similar trends in overall microeukaryote abundance, when looking exclusively at alveolates (Figure 2B). Notably, another photosynthetic dinoflagellate, Coolia canariensis was detected in LTER 3 corals in August 2019 and two species of ciliate, Protocruzia contrax and Protocruzia tuzeti, were found exclusively after the bleaching event at LTER 4 in August 2019 (Figure 2B).
3.3 Time and space impact alpha diversity of non-metazoan microeukaryotes
In order to understand community richness and evenness of the non-metazoan microeukaryotic populations in A. hyacinthus corals around Mo’orea, we explored both Shannon and Chao1 diversity. Timepoint was a significant driver of Shannon diversity (linear mixed effect model (LMEM); p = 0.012, F = 3.53, F(3, 71)), whereas location alone and the interaction of location and timepoint did not significantly explain differences in Shannon diversity. We found that at the August 2019 time point, Shannon diversity was significantly higher in the east and west shores compared to the north (p = 0.01 and 0.03, t = -2.95 and -2.54, and df = 71 and 71, respectively; Figure 3A; Supplementary File 1). For Chao1 diversity, only the interaction between timepoint and location was found to be a significant driver of alpha diversity (LMEM, p = 0.048, F = 2.28, F(6, 57.1)), whereas timepoint or location alone did not significantly affect alpha diversity. In accordance with Shannon diversity, Chao1 diversity was higher in the east shore compared to the north shore (p = 0.002, t = -3.56, df = 70.76) but also compared to the west shore (p = 0.02, t = 2.73, df = 69.83; Figure 3B; Supplementary File 1). Additionally, within the east shore, higher Chao1 diversity was observed in August 2019 (after) compared to March 2019 (during) (p = 0.01, t = 3.24, df = 48.23; Figure 3C). These differences in both Shannon and Chao1 diversity appear to be largely influenced by LTER site 3, because this site displayed much higher alpha diversity values than other sites (Supplementary Figure 4).
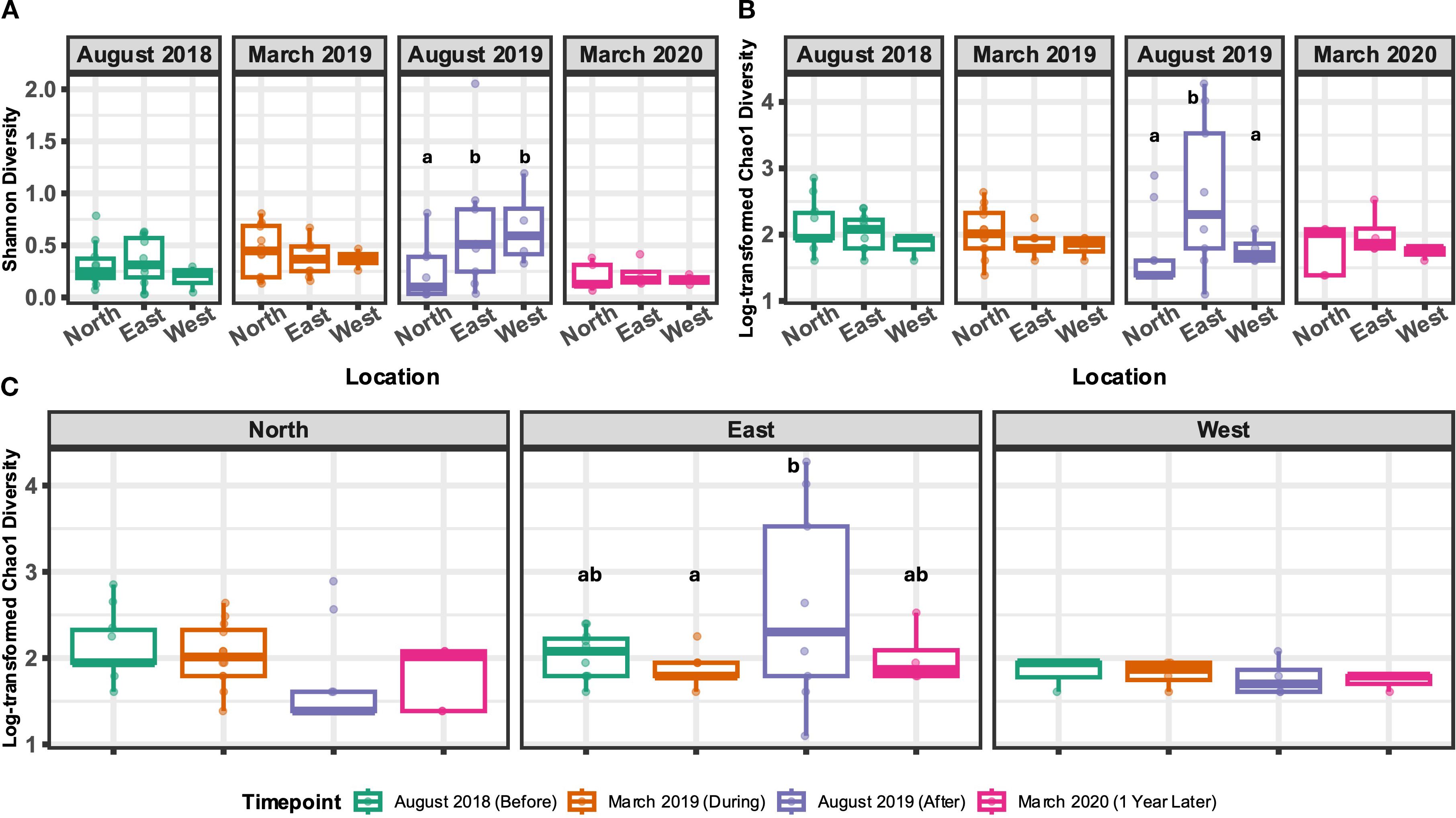
Figure 3. Alpha diversity metrics, Shannon diversity (A) and Chao1 diversity (B, C), for non-metazoan microeukaryotes calculated at the genus level. Boxplots that do not share a letter indicate a statistically significant difference between the groups. A and B display alpha diversity metrics between shore types within single timepoints, whereas C displays Chao1 diversity across timepoints within specific shores. Included in analyses are 22 total samples from March 2019 from both the north and east (n=9, n=9) and west (n=4); 25 total in August 2019 from the north (n=12), east (n=9), and west (n=4); and 12 total from March 2020 from north (n=5), east (n=4), and west (n=3).
3.4 Non-metazoan microeukaryote community composition shifts significantly as a result of space and time
To explore variation within the non-metazoan microeukaryotic communities residing in A. hyacinthus samples, we calculated beta diversity over time and space. In August 2018, prior to the extreme marine heat wave and bleaching event, baseline microeukaryote microbiome composition significantly differed by location around the island (north, east, and west shores) (p = 0.027, pseudo-F = 1.6, R2 = 0.13). This appeared to be driven largely by the north shore, because communities between the east and west shores displayed no significant differences at this, or any, time point (Supplementary File 1). Unlike the east and west shores, the north shore also contained fore reef sites (n = 5). When these sites were removed from the analysis, however, the spatial beta diversity patterns remained consistent; therefore, these samples were retained throughout the analysis.
Without the confounding effect of location, we subset the data by shore type in order to investigate how the heat wave affected community structure over time. After parsing, no significant differences between sites within locations were observed at any time point (Supplementary File 1). On the north shore, we observed significant differences in microeukaryotic community composition over time between August 2018 and August 2019 (p = 0.01, pseudo-F = 4.89, R2 = 0.22), between March 2019 and August 2019 (p = 0.02, pseudo-F = 2.79, R2 = 0.13), and between August 2018 and March 2020 (p = 0.02, pseudo-F = 2.83, R2 = 0.18). However, no significant differences in community composition were identified across time in the east or west shores – although the east shore displayed significantly higher beta dispersion (variance) in August 2019, compared to both August 2018 and March 2019 (Figure 4).
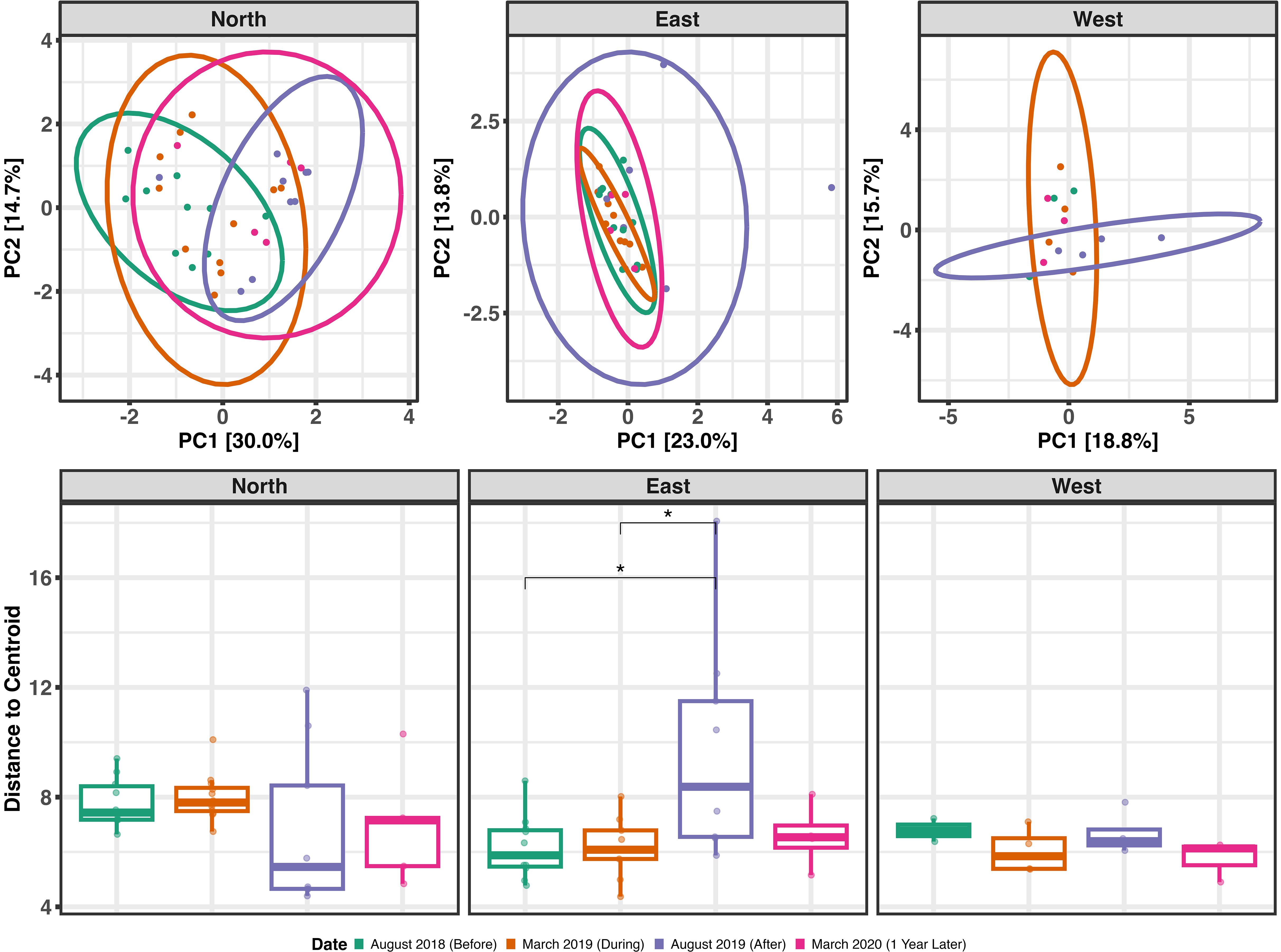
Figure 4. Top: Principal components analysis ordination of beta diversity over time based on Euclidean distances, separated by location around the island (north, east, and west). Time points are designated by the color of points and ellipses. Ellipses represent the 95% confidence interval. Bottom: Beta dispersion (within-group variation) over time as distance-to-centroid, separated by location around the island. Boxplots that do not share a letter indicate a statistically significant difference between the groups. Included in analyses are 22 total samples from March 2019 from both the north and east (n=9, n=9) and west (n=4); 25 total in August 2019 from the north (n=12), east (n=9), and west (n=4); and 12 total from March 2020 from north (n=5), east (n=4), and west (n=3).
3.5 Heat stress drove changes in the relative abundance of specific microeukaryotic taxa
To understand island-wide patterns, we identified taxa that were differentially abundant across shore types within each time point. Prior to the bleaching event, no taxa were identified as differentially abundant between any of the shores (Supplementary File 1). During the bleaching event in March 2019, a single Corallicola aquarius ASV (ASV 9) had significantly higher relative abundance in the west shore relative to the north (Figure 5), although the log fold change (LFC) was quite small (LFC = 1.53; Supplementary File 1). This trend was sustained, and amplified, in the August 2019 time point (LFC = 4.31; Supplementary File 1), but was not detected in March 2020 (Figure 5). Compared to the north shore, several Symbiodinium ASVs were significantly reduced in relative abundance in the east shore at all time points – but most notably in March 2019 (Figure 5). In the west shore, however, only a single Symbiodinium ASV (ASV 2) had significantly lower relative abundance compared to the north shore in March 2019 and March 2020. In August 2019 and March 2020, several Cladocopium ASVs significantly increased in relative abundance in both the east and west shores relative to the north shore (Figure 5). Across all time points, no taxa were identified as significantly differentially abundant between the west and east shores (Figure 5; Supplementary File 1). Importantly it is notable that 18S rRNA copy number can vary across Symbiodiniaceae genera and lineages (Gong and Marchetti, 2019; Davies et al., 2023), but data are not currently available to correct for this potential variation in this study.
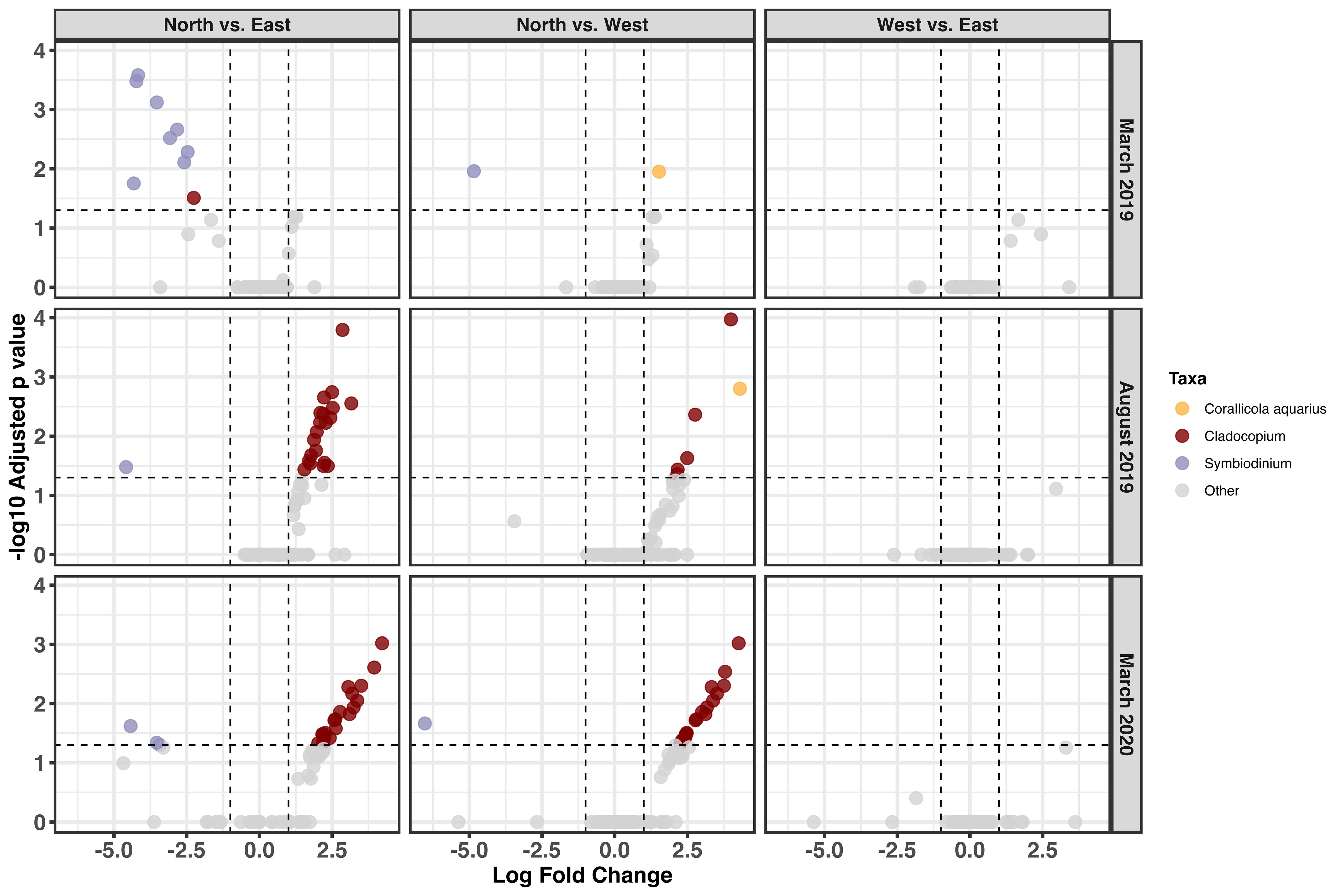
Figure 5. Volcano plot depicting differentially abundant non-metazoan microeukaryotic taxa in Acropora hyacinthus in March 2019 and August 2019. Taxa below the horizontal dotted line are determined to be non-significantly different. Each dot represents a single ASV, and dots are colored by genus or species. Log fold change values are for the second group in the comparison. Sampling numbers are as follows: in March 2019 north and east (n=9, n=9), west (n=4); in August 2019 north (n=12), east (n=9), and west (n=4); and in March 2020 north (n=5), east (n=4), and west (n=3) were used.
4 Discussion
4.1 Apicomplexan abundances in coral host may be influenced by marine heat waves
Species from the phylum Apicomplexa appeared in A. hyacinthus samples on all shores in Mo’orea throughout the course of the study (Supplementary Figure 3). Within the phylum, the genus Corallicola was the singular representative. This genus lies within the order Corallicolida, a deep-branching clade that is known to be associated with corals (Kwong et al., 2021). Corallicola aquarius was the only taxon that was identified as a named species present in our data, though several other taxa only annotated to the genus level were also identified (Supplementary File 1).
Apicomplexans have been identified in several, often healthy, coral species (Kwong et al., 2019; Toller et al., 2002; Kirk et al., 2013; Janouškovec et al., 2015; Upton and Peters, 1986; Slapeta and Linares, 2013; Janouškovec et al., 2013; Mathur et al., 2018; Vohsen et al., 2020). In some cases, however, it is thought that this association may have negative implications because apicomplexans have been shown to proliferate in corals under disease and thermal stressors (Garcia et al., 2013; Bonacolta et al., 2024). In octocorals, Corallicola differential abundance has been shown to predict coral thermal susceptibility and mortality (Bonacolta et al., 2024), and our findings of enrichment during periods of thermal stress may suggest a similar connection between Corallicola and hexacorals.
Both Corallicola aquarius and taxa from the genus Corallicola appeared in healthy corals around the island at low relative abundances, which indicates that the relationship between A. hyacinthus and Corallicola is fairly common, widespread, and robust around the island (Supplementary Figure 3). Therefore, our results support that under normal conditions, apicomplexans (namely of the genus Corallicola) can exist as a neutral or commensal member of the A. hyacinthus microeukaryotic community – as shown by low relative abundance prior to, and one year after, a severe bleaching event. Despite its consistent low relative abundance, significant increases in relative abundance were documented – namely in the east and west shores during and immediately after an environmental disturbance. This apparent increase may also be due, in part, to an overall reduction in photosymbiont density given that photosynthetic dinoflagellates are known to decrease in coral hosts in response to heat stress events (Nielsen et al., 2018); therefore, the decrease in Symbiodiniaceae may result in a perceived increase in Apicomplexa relative abundance. Alternatively, the increased relative abundance may highlight the possible shift from a commensal apicomplexan-host relationship to one of opportunistic parasitism that further weakens host health. However, parasitism in an ecological framework can also provide possible benefits, with parasite-mediated effects on population dynamics, interspecific competition, and energy flow possibly resulting from a system rich in parasitic individuals (Hudson et al., 2006). When put in the context of the coral holobiont, as suggested in Bonacolta et al. (2024), parasitism complicates our understanding of Corallicola’s role within the coral holobiont, even if a parasitic relationship between Corallicola and its coral host can be more substantially defined by previous findings (Bonacolta et al., 2024).
Yet, due to the limited knowledge about how these organisms interact with their coral hosts, it remains difficult to speculate whether their role is beneficial or harmful. Additionally, given the compositional nature of sequencing data and 18S rRNA gene copy number variation (Countway et al., 2005), without true quantitative data informing total apicomplexan abundance, it is difficult to directly infer the impacts that these apparent changes in relative abundance have on coral health. Furthermore, without other quantitative molecular methods such as qPCR, we are unable to definitively determine if the total abundance of Corallicola increased, or if the perceived increase in relative abundance was simply due to the reduction in other taxa. However, from this study, it was found that Corallicola exist in A. hyacinthus populations around Mo’orea, both in healthy populations as well as corals experiencing extreme thermal stress. This establishes Corallicola as a likely common member of the A. hyacinthus holobiont, as well as provides insight into the dynamics of members of Corallicola within A. hyacinthus corals during a common stress event for stony corals.
4.2 Marine heat waves may promote shifting dynamics within the dominant dinoflagellate populations in A. hyacinthus hosts
As expected, given that they are obligate symbionts of coral, dinoflagellates in the family Symbiodiniaceae were the most abundant non-metazoan microeukaryote recovered from the A. hyacinthus holobiont and were detected from every sample across all sites and timepoints. Under normal conditions before the bleaching event, Cladocopium was the most abundant genus. However, on the north shore, apparent shuffling from Cladocopium to Symbiodinium occurred after the bleaching event – most notably in the August 2019 timepoint. This shift is supported visually (Figure 2) and statistically (Figure 5), because several ASVs within the Cladocopium genus were enriched following the marine heatwave. The dominance of the genus Cladocopium in A. hyacinthus corals is consistent with compositions found in studies conducted on A. hyacinthus corals prior to the 2019 bleaching event in Mo’orea (Putnam et al., 2012; Epstein et al., 2019; Kriefall et al., 2022). In other studies that observed symbiont composition following this bleaching event, Durusdinium and Symbiodinium were identified as more abundant than Cladocopium (Grupstra et al., 2021; Leinbach et al., 2022). Leinbach et al. in particular observed a dominance of the genus Symbiodinium in A. hyacinthus samples post-bleaching event, where corals sampled pre-bleaching event from Mo’orea saw a dominance of the genus Cladocopium (Kriefall et al., 2022), which is similar to what we observed in this study – particularly on the north shore.
Symbiont shuffling, as we observed in the north shore, is an adaptive strategy by which corals can resist heat stress by exchanging their current algal symbionts for more thermally tolerant ones (Baker et al., 2004). As with many resistance and resilience strategies, there are often tradeoffs associated with symbiont shuffling such as weakened resilience against other stressors due to reduced photosynthetic efficiency and energy allocation (Jones and Berkelmans, 2010, 2011). Additionally, not all individuals will utilize this method during heat stress – as we observed in the east and west shores. One possibility that may explain the differences in symbiont shuffling across shores could be due to environmental heterogeneity across the island – particularly in terms of nutrient enrichment – because nutrient ratios often drive host-symbiont interactions (Shantz and Burkepile, 2014; Baker et al., 2018; Morris et al., 2019). In the present study, we observed that the north shore displayed a higher degree of symbiont shuffling compared to the east and west shores (Figures 2, 5). This north shore is also known to be nutrient rich due to anthropogenic activity. The geography of the north shore includes two large valleys in which much of the human population lives. When land was cleared for agricultural use and urban development (Walker et al., 2014), waste from animal livestock, runoff from agricultural and landscaping activities, and treated and untreated human waste made the north shore particularly susceptible to periods of high nutrient enrichment (Haßler et al., 2019; Adam et al., 2021). In previous studies, elevated nutrients – particularly nitrogen – were shown to increase bacterial and archaeal microbiome diversity (McConnell, 2019). Further, excess nutrients can have direct effects on the coral microbiome, causing increased coral microbiome dispersion and dissimilarity (Messyasz et al., 2021).
Alternatively, the compositional change in the algal symbiont community may reflect a more negative development, rather than a beneficial adaptation. Under various climate change scenarios, Symbiodinium species are known to transition from a mutualistic to parasitic relationship within the coral host (Lesser et al., 2013; Baker et al., 2018) – especially under nutrient enrichment conditions. Given that this study did not directly measure nutrients at the LTER sites, further work could enhance our understanding of whether or not this transition is potentially beneficial or harmful to the host. It should also be noted that understanding Symbiodiniaceae dynamics within these samples was not the original intent of the study, and as such, the marker gene that was used (18S rRNA) is not the typical marker gene used to study these dinoflagellates. The 18S rRNA method used here does not allow us to resolve lineage and instead allows for comparison at the clade level (Sampayo et al., 2009). As a result, we cannot definitively resolve the functional shifts associated with the changes in the dominant symbiont observed in this study.
4.3 Other non-metazoan microeukaryote populations are potentially diversified by a marine heat wave
Though in small abundances compared to members of Symbiodiniaceae and Corallicola, several non-metazoan microeukaryotes from other taxonomic groups were found to inhabit A. hyacinthus. This was largely true after the bleaching event, at the August 2019 time point which could signal a tendency for these coral microbial communities to shift towards a more diverse state during the bleaching event. In the August 2019 timepoint, we observed higher Shannon diversity and dispersion in the east shore. This may indicate that, in terms of composition, the community may be relatively stable but the relative abundance of taxa may drive within-group heterogeneity. It is also possible, as discussed previously with the Apicomplexa, that with the loss of Symbiodiniaceae that commonly accompanies bleaching events, these low abundance taxa became more apparent during sequencing. Not much is understood about the community dynamics among microeukaryotic communities within coral during thermal stress events outside of Symbiodiniaceae dynamics, though similar results were seen in bacterial communities in corals around Mo’orea and in the Florida Keys during various thermal bleaching events (Maher et al., 2020; Wang et al., 2018). Here, highly diverse bacterial communities were observed immediately after the bleaching event – several months after peak temperatures had been reached. This is thought to occur due to a delayed establishment of opportunistic taxa until temperatures subsided, thereby increasing in richness as the event continues (Maher et al., 2020; Wang et al., 2018). It has also been suggested that external stressors on hosts can induce “Anna Karenina Principle” effects on host microbiomes (including corals), in which beta diversity increases during periods of above average temperatures in coral surface microbiomes (Zaneveld et al., 2017). We saw the addition of several microeukaryotes to the microbiome of A. hyacinthus over the course of the bleaching event, including members of Rhodophyta, Cylindrotheca (diatoms), and Protocruzia (ciliates). This signals a shift toward dysbiosis in the microeukaryotic community of A. hyacinthus corals, potentially in response to the stressful conditions experienced.
It is likely that red turf-forming algae such as Ceramium codicola (and to a lesser extent family Rhodomelaceae and genus Polysiphonia) are detected as the result of potential blooms or overgrowth in these coral individuals. Although Rhodomelaceae and Polysiphonia are typical members of a reef ecosystem (Guiry and Guiry 2020), C. codicola is more unusual, because it typically inhabits the colder waters on the Pacific coast of the United States (Cho et al., 2019). Even though it is unusual, members of this family are commonly listed as introduced or cryptic species globally – most often introduced through the shipping industry (Williams and Smith, 2007). Depending on the species and environment, turf-forming algae can compete with corals, potentially weakening or overgrowing their competition (Jompa and McCook, 2003). This competitive relationship suggests that under stress such as that faced by corals during a thermal stress event, it is possible that red turf-forming algae growth increased but the coral was unable to compete at its normal capacity. Similarly, shifts in diatom abundance such as Cylindrotheca closterium may also be indicative of dysbiosis in the community, because C. closterium abundances in Montipora corals have been shown to shift in response to thermal stress – although in cold temperatures (Yamashiro et al., 2012). Diatoms are not uncommon inhabitants of coral mucus (Cavada et al., 2011), but extreme thermal stress may destabilize the relationship, thereby leading to a potential bloom. In this present study, Cylindrotheca closterium was only found in one coral individual at LTER 4 during the August 2019 time point, though it dominated the microeukaryotic community at that time. It is possible that in response to the thermal stress this coral individual was facing during the spring of 2019, C. closterium and other members of the class Bacillariophyceae were able to establish an infection and dominate the microeukaryotic community.
Other microeukaryotic members identified during this period of stress include Hydrolithon onkodes and the ciliates Protocruzia contrax and Protocruzia tuzeti. Hydrolithon onkodes is a coralline algae typical of the reefs surrounding Mo’orea (Tribollet and Payri, 2001) within Rhodophyta. This species often plays a fundamental role in coral larvae recruitment (Birrell et al., 2008) and is therefore considered to be beneficial. Despite this, overgrowth of crustose coralline algae has been documented on scleractinian corals (Eckrich et al., 2011), which may indicate that H. onkodes is not necessarily beneficial if physically overgrowing the coral. The coral individual in which H. onkodes was mainly identified, in LTER 3, is also where most other Rhodophyta members were identified, indicating that H. onkodes growth may have been part of the potential algal overgrowth on that coral individual. Ciliates within the genus Protocruzia have been described to perform a wide range of ecological functions within coral reef ecosystems, including bacterivory (Sweet et al., 2019), nutrient acquisition during stress events (Kramarsky-Winter et al., 2006), and protection against pathogens through antibiotic secretion (Rohwer et al., 2002). These ciliates were most notably present during the August 2019 timepoint after the severe bleaching event. Their presence may serve a supporting role to corals during this stress period through nutrient acquisition or pathogen protection, or a more opportunistic role because ciliates are often attracted to diseased or decaying coral tissue (Ravindran et al., 2022). Both ciliates and H. onkodes may be less detrimental to the coral holobiont compared to diatom and turf algae overgrowth but may still be circumstantially harmful.
With all of the microeukaryotes observed during the August 2019 timepoint however, it is important to note that no widespread patterns across space or time were observed. Most microeukaryotes not belonging to Corallicolida or Symbiodiniaceae were only found in specific individuals at specific time points, making it difficult to speculate on any overarching roles or causation regarding these organisms as they relate to thermal stress or bleaching events within the coral holobiont.
5 Conclusion
In this study, we found that in A. hyacinthus, apicomplexans from the genus Corallicola increased in relative abundance during and after a severe bleaching event. During the bleaching event, Cladocopium and Symbiodinium (photosynthetic dinoflagellates) abundances were dynamic especially on the north shore of the island, where we saw shuffling from Cladocopium dominance to Symbiodinium dominance. We also showed that, overall, the non-metazoan microeukaryotic community became more diverse immediately after the bleaching event, with taxa such as Rhodophyta, diatoms, and ciliates populating corals during a period of stress. Through this work, we identified patterns of Corallicolida population dynamics in response to a severe marine heat wave and bleaching event and gained insight into apicomplexan dynamics within the coral holobiont during this increasingly frequent stress event. At present, sequencing data such as in this study rely on relative abundances, however implementing the use of qPCR for Corallicolida genomic signatures in future bleaching event experiments could provide quantitative information about true fluctuations in species abundance. Further, including more frequent and widespread sampling, explicit monitoring of other environmental parameters such as nutrients and salinity and developing microscopy methods to understand localization of these microorganisms could enhance understanding of apicomplexan dynamics within the coral holobiont during stress events.
Data availability statement
Scripts used for bioinformatic and statistical analyses can be found at: https://github.com/pattonsunni/apicomplexan. The 18S rRNA gene dataset supporting the conclusions of this article is available in the NCBI Sequence Read Archive (SRA) repository under the BioProject accession number PRJNA1194564.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
AP: Conceptualization, Investigation, Writing - original draft, Visualization. SP: Formal Analysis, Validation, Writing – review & editing, Investigation, Visualization. ERS: Conceptualization, Investigation, Writing – review & editing. CG: Investigation, Writing – review & editing. LH-K: Writing – review & editing, Investigation. JK: Investigation, Writing – review & editing. RM: Investigation, Writing – review & editing. AM: Investigation, Writing – review & editing. SS: Writing – review & editing, Investigation. ART: Investigation, Writing – review & editing, Funding acquisition. AMSC: Funding acquisition, Investigation, Writing – review & editing. RVT: Conceptualization, Writing – review & editing, Supervision, Investigation, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by funding from the National Science Foundation OCE-1635913 to RVT and ART, OCE -1635798 and OCE-2145472 to AMSC, and OCE-1933165 to ART. Additionally, this work was supported by the Sheila van Zandt Student Research Experience Scholarship from the Oregon State University Department of Microbiology awarded to AP and ERS.
Acknowledgments
This manuscript uses data collected by the U.S. National Science Foundation’s (NSF) Mo’orea Coral Reef Long Term Ecological (MCR LTER) research site under NSF Award OCE 1637396. Additional financial support to the MCR LTER site was provided through a generous gift from the Gordon and Betty Moore Foundation. We would like to acknowledge Dr. Nathan Kirk for his assistance in the conceptualization of this project. We would also like to acknowledge Dr. Alex Vompe for his contribution and assistance in the visualization of data. Additionally, we would like to thank the members of the MCR LTER research site and the members of Gump Research Station for their contributions in the care and maintenance of the field site from which samples were collected. Research was completed under permits issued by the French Polynesian Government (Délégation à la Recherche) and the Haut-commissariat de la République en Polynésie Française (DTRT) (Protocole d’Accueil 2005-2021). Finally, we would like to thank the Oregon State University Center for Quantitative Life Sciences for their next-generation sequencing services. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1626071/full#supplementary-material
Supplementary Table 1 | The number of coral individuals analyzed at each site for August 2018 - August 2019, as well as March 2020.
Supplementary Table 2 | Number of specimens, number of reads, and number of non-metazoan microeukaryotic taxa observed according to the side of the island (Mo’orea, French Polynesia) in which specimens were collected (N = north, E = east, W = west) at each LTER site and time point. These data were obtained from the non-rarefied phyloseq object.
Supplementary Figure 1 | Map of Mo’orea, French Polynesia, with the sampling scheme for the original Mo’orea Virus Project. Labels denoting Long Term Ecological Research (LTER) sites 1-6 as well as the added 0, 3.5, and 5.5 for the Mo’orea Virus Project are marked with black stars and red dots indicate where coral samples were collected. Only a subset of the red dots corresponds to samples collected and analyzed for this study. Adapted from an unpublished figure by Dr. Emily Schmeltzer. Note that corals at site LTER 6 did not survive through August 2019 so data from this site are not included in this study. The sample numbers reflect total samples at all timepoints (resampling) – not the number of coral individuals.
Supplementary Figure 2 | Rarefaction curve for the outputs of number of species and number of individuals found per sample. Samples were rarefied to 14,205 reads, losing two samples with reads of 1,084 and 1,865.
Supplementary Figure 3 | Corallicolidae total abundance across shore types over time for both nonrarefied and rarefied data. Taxa within Corallicolidae were identified in all 83 samples in the nonrarefied data and in 82 samples in the rarefied data.
Supplementary Figure 4 | Alpha diversity metrics, Shannon diversity (A) and Chao1 diversity (B), for non-metazoan microeukaryotes calculated at the genus level. Asterisks with bars denote statistical significance between groups.
References
Adam T. C., Burkepile D. E., Holbrook S. J., Carpenter R. C., Claudet J., Loiseau C., et al. (2021). Landscape-scale patterns of nutrient enrichment in a coral reef ecosystem: Implications for coral to algae phase shifts. Ecological Applications: A Publication of the Ecological Society of America, 31 (1), e2227.
Adl S. M., Leander B. S., Simpson A. G. B., Archibald J. M., Anderson O. R., Bass D., et al. (2007). Diversity, nomenclature, and taxonomy of protists. Systematic Biol. 56, 684–689. doi: 10.1080/10635150701494127
Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Accessed April 15 2024).
Baker D. M., Freeman C. J., Wong J. C. Y., Fogel M. L., and Knowlton N. (2018). Climate change promotes parasitism in a coral symbiosis. ISME J. 12, 921–930. doi: 10.1038/s41396-018-0046-8
Baker A. C., Starger C. J., McClanahan T. R., and Glynn P. W. (2004). Corals’ adaptive response to climate change. Nature 430, 741–741. doi: 10.1038/430741a
Barnett (2021). microViz: an R package for microbiome data visualization and statistics. J. Open Source Software 6, 3201. doi: 10.21105/joss.032012
Bates D., Mächler M., Bolker B., and Walker S. (2015). Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software67 (1). https://doi.org/10.18637/jss.v067.i01.
Belser C., Poulain J., Labadie K., Gavory F., Alberti A., Guy J., et al. (2023). Integrative omics framework for characterization of coral reef ecosystems from the tara pacific expedition. Sci. Data 10, 326. doi: 10.1038/s41597-023-02204-0
Birrell C. L., McCook L. J., Willis B. L., and Harrington L. (2008). Chemical effects of macroalgae on larval settlement of the broadcast spawning coral acropora millepora. Mar. Ecol. Prog. Ser. 362, 129–137. doi: 10.3354/meps07524
Bonacolta A. M., Miravall J., Gómez-Gras D., Ledoux J.-B., López-Sendino P., Garrabou J., et al. (2024). Differential apicomplexan presence predicts thermal stress mortality in the mediterranean coral paramuricea clavata. Environ. Microbiol. 26, e165485. doi: 10.1111/1462-2920.16548
Bonacolta A. M., Weiler B. A., Grimes C. J., Trznadel M., Vermeij M. J.A., Keeling P. J., et al. (2025). Fireworms are a reservoir and potential vector for coral-infecting apicomplexans. ISME J. 19, wraf078. doi: 10.1093/ismejo/wraf078
Bower S. M., Carnegie R. B., Goh B., Jones S. R., Lowe G. J., and Mak M. W. (2004). Preferential PCR amplification of parasitic protistan small subunit rDNA from metazoan tissues. J. Eukaryotic Microbiol. 51, 325–325. doi: 10.1111/j.1550-7408.2004.tb00574.x
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. Jo A., and Holmes S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–835. doi: 10.1038/nmeth.3869
Cavada F., Ayala R., Troccoli L., and Cruz-Motta J. J. (2011). Microalgae from the mucus layer of two massive corals: More than sunken plankton. Mar. Biol. 158 (11), 2495–2504.
Cho T. Oh, Boo S. M., and Hansen G. I. (2019). Structure and reproduction of the genus ceramium (Ceramiales, rhodophyta) from oregon, USA. Phycologia 40, 547–571. doi: 10.2216/i0031-8884-40-6-547.1
Clerissi C., Brunet Sébastien, Vidal-Dupiol J., Adjeroud M., Lepage P., Guillou L., et al. (2018). Protists within corals: the hidden diversity. Front. Microbiol. 9, 407707. doi: 10.3389/fmicb.2018.02043
Coles S. L. and Brown B. E. (2003). Coral bleaching–capacity for acclimatization and adaptation. Adv. Mar. Biol. 46, 183–223. doi: 10.1016/S0065-2881(03)46004-5
Comeau AndréM., Li W. K.W., Tremblay Jean-Éric, Carmack E. C., and Lovejoy C. (2011). Arctic Ocean Microbial Community Structure before and after the 2007 Record Sea Ice Minimum. PloS One 6, e274925. doi: 10.1371/journal.pone.0027492
Correa A. M.S., Howard-Varona C., Coy S. R., Buchan A., Sullivan M. B., and Weitz J. S. (2021). Revisiting the rules of life for viruses of microorganisms. Nat. Rev. Microbiol. 19, 501–135. doi: 10.1038/s41579-021-00530-x
Countway P. D., Gast R. J., Savai P., and Caron D. A. (2005). Protistan diversity estimates based on 18S rDNA from seawater incubations in the western north atlantic. J. Eukaryotic Microbiol. 52, 95–106. doi: 10.1111/j.1550-\7408.2005.05202006.x
Davies S. W., Gamache M. H., Howe-Kerr L. I., Kriefall N. G., Baker A. C., Banaszak A. T., et al. (2023). Building consensus around the assessment and interpretation of symbiodiniaceae diversity. PeerJ 11, e15023. doi: 10.7717/peerj.15023
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., and Callahan B. J. (2018). 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226. doi: 10.1186/s40168-018-0605-2
del Campo J., Pons M. J., Herranz M., Wakeman K. C., Valle J. D., Vermeij M. J.A., et al. (2019). Validation of a universal set of primers to study animal-associated microeukaryotic communities. Environ. Microbiol. 21, 3855–3615. doi: 10.1111/1462-2920.14733
Dennis M. M., Becker A. A.M.J., and Freeman M. A. (2020). Pathology of multifocal purple spots, a nonspecific lesion morphology of caribbean sea fans gorgonia spp. Dis. Aquat. Organisms 141, 79–89. doi: 10.3354/dao03523
Eckrich C. E., Engel M. S., and Peachey R. B. J. (2011). Crustose, calcareous algal bloom. (Ramicrusta sp.) overgrowing scleractinian corals, gorgonians, a hydrocoral, sponges, and other algae in lac bay, bonaire, dutch caribbean. Coral Reefs 30, 131–131. doi: 10.1007/s00338-010-0683-5
Epstein H. E., Smith H. A., Cantin N. E., Mocellin V. J.L., Torda G., and van Oppen M. J.H. (2019). Temporal variation in the microbiome of acropora coral species does not reflect seasonality. Front. Microbiol. 10, 460849. doi: 10.3389/fmicb.2019.01775
Ewels P., Magnusson M., Lundin S., and Käller M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Galindo-Martínez C. T., Weber M., Avila-Magaña V., Enríquez S., Kitano H., Medina Mónica, et al. (2022). The role of the endolithic alga ostreobium spp. during coral bleaching recovery. Sci. Rep. 12, 1–125. doi: 10.1038/s41598-022-07017-6
Garcia G. D., Gregoracci G. B., Santos E. de O., Meirelles P. M., Silva G. G.Z., Edwards R., et al. (2013). Metagenomic analysis of healthy and white plague-affected mussismilia Braziliensis corals. Microbial Ecol. 65, 1076–1086. doi: 10.1007/s00248-012-0161-4
Gong W. and Marchetti A. (2019). Estimation of 18S gene copy number in marine eukaryotic plankton using a next-generation sequencing approach. Front. Mar. Sci. 6, 453814. doi: 10.3389/fmars.2019.00219
Gräf R., Batsios P., and Meyer I. (2015). Evolution of Centrosomes and the Nuclear Lamina: Amoebozoan Assets. European Journal of Cell Biology94 (6), 249–256.
Grupstra C. G.B., Rabbitt K. M., Howe-Kerr L. I., and Correa A. M.S. (2021). Fish predation on corals promotes the dispersal of coral symbionts. Anim. Microbiome 3, 1–125. doi: 10.1186/s42523-021-00086-4
Guillou L., Bachar D., Audic Stéphane, Bass D., Berney Cédric, Bittner L., et al. (2013). The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Guiry M. and Guiry W. (2020). AlgaeBase, World-wide electronic publication (Galway: National University of Ireland). Available online at: http://www.algaebase.org (Accessed April 20 2024).
Haßler K., Dähnke K., Kölling M., Sichoix L., Nickl A.-L., and Moosdorf N. (2019). Provenance of Nutrients in Submarine Fresh Groundwater Discharge on Tahiti and Moorea, French Polynesia. Applied Geochemistry100 (January), 181–189.
Holt C. C., Boscaro V., Van Steenkiste N. W., Herranz M., Mathur V., Irwin N. A., et al. (2022). Microscopic marine invertebrates are reservoirs for cryptic and diverse protists and fungi. Microbiome 10, 10. doi: 10.1186/s40168-022-01363-3
Hudson P. J., Dobson A. P., and Lafferty K. D. (2006). Is a Healthy Ecosystem One That Is Rich in Parasites?. Trends in Ecology & Evolution21 (7), 381–385.
Hughes T. P., Anderson K. D., Connolly S. R., Heron S. F., Kerry J. T., Lough J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Iha C., Dougan K. E., Varela J. A., Avila V., Jackson C. J., Bogaert K. A., et al. (2021). Genomic adaptations to an endolithic lifestyle in the coral-associated alga ostreobium. Curr. Biology: CB 31, 1393–1402.e5. doi: 10.1016/j.cub.2021.01.018
Janouškovec J., Horák Aleš, Barott K. L., Rohwer F. L., and Keeling P. J. (2012). Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr. Biology: CB 22, R518–R519. doi: 10.1016/j.cub.2012.04.047
Janouškovec J., Horák Aleš, Barott K. L., Rohwer F. L., and Keeling P. J. (2013). Environmental distribution of coral-associated relatives of apicomplexan parasites. ISME J. 7, 444–475. doi: 10.1038/ismej.2012.129
Janouškovec J., Tikhonenkov D. V., Burki F., Howe A. T., Kolísko M., Mylnikov A. P., et al. (2015). Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. United States America 112, 10200–12075. doi: 10.1073/pnas.1423790112
Jompa J. and McCook L. J. (2003). Coral-algal competition: macroalgae with different properties have different effects on corals. Mar. Ecol. Prog. Ser. 258, 87–95. doi: 10.3354/meps258087
Jones A. and Berkelmans R. (2010). Potential costs of acclimatization to a warmer climate: growth of a reef coral with heat tolerant vs. Sensitive symbiont types. PloS One 5, e10437. doi: 10.1371/journal.pone.0010437
Jones A. M. and Berkelmans R. (2011). Tradeoffs to thermal acclimation: energetics and reproduction of a reef coral with heat tolerant symbiodinium type-D. J. Mar. Sci. 2011, 185890. doi: 10.1155/2011/185890
Jones A. M., Berkelmans R., van Oppen M. J. H., Mieog J. C., and Sinclair W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc. R. Soc. B: Biol. Sci. 275, 1359–1365. doi: 10.1098/rspb.2008.0069
Kearney S. M., Gibbons S. M., Poyet M., Gurry T., Bullock K., Allegretti J. R., et al. (2018). Endospores and other lysis-resistant bacteria comprise a widely shared core community within the human microbiota. ISME J. 12, 2403–2165. doi: 10.1038/s41396-018-0192-z
Keeling P. J., Mathur V., and Kwong W. K. (2021). Corallicolids: The elusive coral-infecting apicomplexans. PloS Pathog. 17), e1009845. doi: 10.1371/journal.ppat.1009845
Kirk N. L., Ritson-Williams R., Coffroth M. A., Miller M. W., Fogarty N. D., and Santos S. R. (2013). Tracking transmission of apicomplexan symbionts in diverse caribbean corals. PloS One 8, e806185. doi: 10.1371/journal.pone.0080618
Klinges J. G., Rosales S. M., McMinds R., Shaver E. C., Shantz A. A., Peters E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13), 2938–2953. doi: 10.1038/s41396-019-0482-0
Knowlton N. and Rohwer F. (2003). Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 162, S51–S62. doi: 10.1086/378684
Kramarsky-Winter E., Harel M., Siboni N., Dov E. B., Brickner I., Loya Y., et al. (2006). Identification of a protist-coral association and its possible ecological role. Mar. Ecol. Prog. Ser. 317, 67–73. doi: 10.3354/meps317067
Kriefall N. G., Kanke M. R., Aglyamova G. V., and Davies S. W. (2022). Reef Environments Shape Microbial Partners in a Highly Connected Coral Population. Proceedings of the Royal Society B 289, 20212459. January. https://doi.org/10.1098/rspb.2021.2459.
Kwong W. K., Campo J. D., Mathur V., Vermeij M. J.A., and Keeling P. J. (2019). A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature 568, 103–107. doi: 10.1038/s41586-019-1072-z
Kwong W. K., Irwin N. A.T., Mathur V., Na I., Okamoto N., Vermeij M. J.A., et al. (2021). Taxonomy of the apicomplexan symbionts of coral, including corallicolida ord. Nov., reassignment of the genus gemmocystis, and description of new species corallicola aquarius gen. Nov. Sp. Nov. and anthozoaphila gnarlus gen. Nov. Sp. Nov. J. Eukaryotic Microbiol. 68, e12852. doi: 10.1111/jeu.12852
Leichter J. J., Alldredge A. L., Bernardi G., Brooks A. J., Carlson C. A., Carpenter R. C., et al. (2013). “Biological and physical interactions on a tropical island coral reef: transport and retention processes on moorea, French Polynesia. Oceanography 26, 52–63. doi: 10.5670/oceanog.2013.45
Leinbach S. E., Speare K. E., and Strader M. E. (2022). Reef habitats structure symbiotic microalgal assemblages in corals and contribute to differential heat stress responses. Coral Reefs 42, 205–217. doi: 10.1007/s00338-022-02316-w
Lenth R. V. (2017). Emmeans: Estimated Marginal Means, Aka Least-Squares Means. CRAN: Contributed Packages. The R Foundation. https://doi.org/10.32614/cran.package.emmeans.
Lesser M. P., Stat M., and Gates R. D. (2013). The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs 32, 603–611. doi: 10.1007/s00338-013-1051-z
Mccowan D. M., Pratchett M. S., and Baird A. (2012). Bleaching Susceptibility and Mortality among Corals with Differing Growth Forms.Proceedings of the 12th International Coral Reef Symposium,9–13.
Maher R. L., Schmeltzer E. R., Meiling S., McMinds R., Ezzat Leïla, Shantz A. A., et al. (2020). Coral microbiomes demonstrate flexibility and resilience through a reduction in community diversity following a thermal stress event. Front. Ecol. Evol. 8, 555698. doi: 10.3389/fevo.2020.555698
Marshall P. A. and Baird A. H. (2000). Bleaching of corals on the great barrier reef: differential susceptibilities among taxa. Coral Reefs 19(2), 155–163. doi: 10.1007/s003380000086
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads”. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Martinez Arbizu P. (2020). pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.4.
Martino C., Morton J. T., Marotz C. A., Thompson L. R., Tripathi A., Knight R., et al. (2019). A novel sparse compositional technique reveals microbial perturbations. mSystems 4. doi: 10.1128/mSystems.00016-19
Mathur V., Del Campo J., Kolisko M., and Keeling P. J. (2018). Global diversity and distribution of close relatives of apicomplexan parasites. Environ. Microbiol. 20, 2824–2335. doi: 10.1111/1462-2920.14134
McConnell K. M. (2019). “Reef microbiomes of mo’orea, French Polynesia, a socio-ecological island system,” in Moorea, îles du vent, French Polynesia, (Hypsographic) (Corvallis, OR USA: Oregon State University).
McMurdie P. J. and Holmes S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e612175. doi: 10.1371/journal.pone.0061217
Messyasz A., Maher R. L., Meiling S. S., and Thurber R. V. (2021). Nutrient enrichment predominantly affects low diversity microbiomes in a marine trophic symbiosis between algal farming fish and corals. Microorganisms 9, 1873. doi: 10.3390/microorganisms9091873
Morris L. A., Voolstra C. R., Quigley K. M., Bourne D. G., and Bay L. K. (2019). Nutrient availability and metabolism affect the stability of coral–symbiodiniaceae symbioses. Trends Microbiol. 27, 678–689. doi: 10.1016/j.tim.2019.03.004
Nielsen D. A., Petrou K., and Gates R. D. (2018). Coral Bleaching from a Single Cell Perspective. The ISME Journal12 (6), 1558–1567.
Nelson C. E., Alldredge A. L., McCliment E. A., Amaral-Zettler L. A., and Carlson C. A. (2011). Depleted dissolved organic carbon and distinct bacterial communities in the water column of a rapid-flushing coral reef ecosystem. ISME J. 5, 1374–1875. doi: 10.1038/ismej.2011.12
Obura D. (2001). Can Differential Bleaching and Mortality among Coral Species Offer Useful Indicators for Assessment and Management of Reefs under Stress. Bulletin of Marine Science69 (2), 421–442.
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2012). vegan: Community Ecology Package. Software http://CRAN.R-project.org/package=vegan (Accessed April 30 2024).
Lin H. and Peddada S. D. (2024). Multigroup Analysis of Compositions of Microbiomes with Covariate Adjustments and Repeated Measures. Nature Methods 21 (1), 83–91.
Paulino G. V.B., Félix C. R., and Landell M. F. (2020). Diversity of filamentous fungi associated with coral and sponges in coastal reefs of northeast Brazil. J. Basic Microbiol. 60, 103–115. doi: 10.1002/jobm.201900394
Putnam H. M., Stat M., Pochon X., and Gates R. D. (2012). Endosymbiotic flexibility associates with environmental sensitivity in scleractinian corals. Proc. R. Soc. B: Biol. Sci. 2728–2743. doi: 10.1098/rspb.2012.1454
Qiu D., Huang L., Zhuang Y., Zhong Yu, Tan Y., Li X., et al. (2021). Dinoflagellate-targeted PCR reveals highly abundant and diverse communities of parasitic dinoflagellates in and near zhubi reef, south China sea. Coral Reefs 40, 1931–1395. doi: 10.1007/s00338-021-02168-w
Ravindran C., Raveendran H. P., and Irudayarajan L. (2022). Ciliated protozoan occurrence and association in the pathogenesis of coral disease. Microbial Pathogenesis 162, 105211. doi: 10.1016/j.micpath.2021.105211
R Core Team (2022). “R: A language and environment for statistical computing,” in R foundation for statistical computing, vienna. Available online at: https://www.R-project.org.
Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., and Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2735. doi: 10.1111/j.1462-2920.2006.01148.x
Riegl B., Bruckner A., Coles S. L., Renaud P., and Dodge R. E. (2009). Coral reefs. Ann. New York Acad. Sci. 1162, 136–186. doi: 10.1111/j.1749-6632.2009.04493.x
Rohwer F., Seguritan V., Azam F., and Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Sampayo E. M., Dove S., and Lajeunesse T. C. (2009). Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus symbiodinium. Mol. Ecol. 18, 500–519. doi: 10.1111/j.1365-294X.2008.04037.x
Shantz A. A. and Burkepile D. E. (2014). Context-dependent effects of nutrient loading on the coral–algal mutualism. Ecology 95, 1995–2005. doi: 10.1890/13-1407.1
Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., and Hay S. I. (2005). The global distribution of clinical episodes of plasmodium falciparum malaria. Nature 434, 214–217. doi: 10.1038/nature03342
Šlapeta J. and Linares M. C. (2013). Combined Amplicon Pyrosequencing Assays Reveal Presence of the Apicomplexan ‘type-N’ (cf. Gemmocystis Cylindrus) and Chromera Velia on the Great Barrier Reef, Australia. PLOS ONE8 (9),e76095.
Speare K. E., Adam T. C., Winslow E. M., Lenihan H. S., and Burkepile D. E. (2022). Size-dependent mortality of corals during marine heatwave erodes recovery capacity of a coral reef. Global Change Biol. 28, 1342–1585. doi: 10.1111/gcb.16000
Sweet M., Burian A., Fifer J., Bulling M., Elliott D., and Raymundo L. (2019). Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7, 1–145. doi: 10.1186/s40168-019-0759-6
Tandon K., Pasella M. M., Iha C., Ricci F., Hu J., O’Kelly C. J., et al. (2023). Every refuge has its price: ostreobium as a model for understanding how algae can live in rock and stay in business. Semin. Cell Dev. Biol. 134, 27–36. doi: 10.1016/j.semcdb.2022.03.010
Thurber R. V., Payet JérômeP., Thurber A. R., and Correa A. M.S. (2017). Virus-host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 15, 205–165. doi: 10.1038/nrmicro.2016.176
Toller W., Rowan R., and Knowlton N. (2002). Genetic evidence for a protozoan (phylum apicomplexa) associated with corals of the montastraea annularis species complex. Coral Reefs 21, 143–146. doi: 10.1007/s00338-002-0220-2
Tribollet A. and Payri C. (2001). Bioerosion of the coralline alga hydrolithon onkodes by microborers in the coral reefs of moorea, French Polynesia. Oceanologica Acta 24, 329–425. doi: 10.1016/S0399-1784(01)01150-1
Upton S. J. and Peters E. C. (1986). A New and Unusual Species of Coccidium (Apicomplexa: Agamococcidiorida) from Caribbean Scleractinian Corals. Journal of Invertebrate Pathology47 (2), 184–193.
van Oppen M. J.H., Leong J.-A., and Gates. R. D. (2009). Coral-virus interactions: A double-edged sword? Symbiosis 47, 1–8. doi: 10.1007/BF03179964
Vohsen S. A., Anderson K. E., Gade A. M., Gruber-Vodicka H. R., Dannenberg R. P., Osman E. O., et al. (2020). Deep-sea corals provide new insight into the ecology, evolution, and the role of plastids in widespread apicomplexan symbionts of anthozoans. Microbiome 8, 345. doi: 10.1186/s40168-020-00798-w
Walker B. L. E., López-Carr D., Chen C., and Currier K. (2014). Perceptions of Environmental Change in Moorea, French Polynesia: The Importance of Temporal, Spatial, and Scalar Contexts. GeoJournal79 (6), 705–719.
Wang Lu, Shantz A. A., Payet JérômeP., Sharpton T. J., Foster A., Burkepile D. E., et al. (2018). Corals and their microbiomes are differentially affected by exposure to elevated nutrients and a natural thermal anomaly. Front. Mar. Sci. 5, 353060. doi: 10.3389/fmars.2018.00101
Weiss L. M. and Kim K. (2013). Toxoplasma gondii: the model apicomplexan -perspectives and methods (Amsterdam, Netherlands: Academic Press).
Williams S. L. and Smith J. E. (2007). A global review of the distribution, taxonomy, and impacts of introduced seaweeds 38, 327–359. doi: 10.1146/annurev.ecolsys.38.091206.095543
Winslow E. M., Speare K. E., Adam T. C., Burkepile D. E., Hench J. L., and Lenihan H. S. (2024). Corals survive severe bleaching event in refuges related to taxa, colony size, and water depth. Sci. Rep. 14, 1–115. doi: 10.1038/s41598-024-58980-1
Yamashiro H., Mikame Y., and Suzuki H. (2012). Localized outbreak of attached diatoms on the coral Montipora due to low-temperature stress. Sci. Rep. 2, 552.
Yang Y., Zhou Q., Zahr K., Harding M. W., Feindel D., and Feng J. (2021). Impact of DNA extraction efficiency on the sensitivity of PCR-based plant disease diagnosis and pathogen quantification. Eur. J. Plant Pathology/European Foundation Plant Pathol. 159, 583–915. doi: 10.1007/s10658-020-02189-1
Keywords: apicomplexan, coral, Corallicolidae, microeukaryote, Acropora hyacinthus, 18S rRNA
Citation: Peterson A, Patton S, Schmeltzer ER, Grupstra CGB, Howe-Kerr LI, Klinges JG, Maher RL, Messyasz A, Seabrook S, Thurber AR, Correa AMS and Vega Thurber RL (2025) Apicomplexan and non-metazoan microeukaryotes in the thermosensitive reef-building coral Acropora hyacinthus shift in abundance throughout an extreme coral bleaching event. Front. Mar. Sci. 12:1626071. doi: 10.3389/fmars.2025.1626071
Received: 09 May 2025; Accepted: 29 August 2025;
Published: 22 September 2025.
Edited by:
Cole G Easson, Middle Tennessee State University, United StatesReviewed by:
Guoxin Cui, King Abdullah University of Science and Technology, Saudi ArabiaXiaopeng Yu, Guangxi University, China
Zhu Wentao, Hainan University, China
Copyright © 2025 Peterson, Patton, Schmeltzer, Grupstra, Howe-Kerr, Klinges, Maher, Messyasz, Seabrook, Thurber, Correa and Vega Thurber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca L. Vega Thurber, cnZlZ2F0aHVyYmVyQHVjc2IuZWR1
 Athena Peterson
Athena Peterson Sunni Patton
Sunni Patton Emily R. Schmeltzer4,1
Emily R. Schmeltzer4,1 Carsten G. B. Grupstra
Carsten G. B. Grupstra Rebecca L. Maher
Rebecca L. Maher Sarah Seabrook
Sarah Seabrook