- 1Jeju Bio Research Center, Korea Institute of Ocean Science & Technology (KIOST), Jeju, Republic of Korea
- 2South Sea Environment Research Center, Korea Institute of Ocean Science & Technology (KIOST), Geoje, Republic of Korea
- 3Department of Life Science, Woosuk University, Jincheon, Republic of Korea
- 4Tropical & Subtropical Research Center, Korea Institute of Ocean Science & Technology (KIOST), Jeju, Republic of Korea
- 5KIOST School, Marine Technology and Convergence Engineering,University of Science and Technology (UST), Daejeon, Republic of Korea
- 6Marine Biotechnology & Bioresource Research Department, Korea Institute of Ocean Science & Technology (KIOST), Busan, Republic of Korea
Introduction: Global ocean warming is known to disrupt interactions between corals and their symbiotic microbiota; however, the temporal sequence of structural and functional changes within microbial communities under thermal stress remains poorly understood. In this study, we investigated the microbial response of the soft coral Eleutherobia rubra following short-term heat exposure.
Methods: Colonies of E. rubra were collected near Eoyudo, Korea, exposed to short-term thermal stress (26 °C for 24 hours) alongside controls (16 °C). 16S rRNA gene sequencing was conducted to assess taxonomic shifts. Predicted functional profiles were inferred using PICRUSt2, and microbial community changes were evaluated through alpha and beta diversity analyses, LEfSe, and correlation analyses.
Results: The overall taxonomic composition showed minimal change, with noticeable variations restricted to rare taxa. In contrast, functional predictions revealed consistent and pronounced reorganization of metabolic potential, characterized by increased core metabolic activities and a decline in several stress associated pathways. Beta diversity and LEfSe analyses indicated limited taxonomic shifts, but more distinct functional differentiation. Correlation analyses further suggested that specific microbial taxa may be driving these functional changes.
Discussion: Our findings suggest that in E. rubra, functional restructuring of the microbiome precedes taxonomic shifts during 24h of heat stress exposure. These observation represent microbial responses under the experimental conditions and provide a basis for future studies on longer-term dynamics.
1 Introduction
Since 1970, the global ocean has warmed unabated, absorbing over 90% of the excess heat in the climate system, with the rate of ocean warming more than doubling since 1993 (Bindoff et al., 2022; Cheng et al., 2019). This rapid increase in ocean temperature has significantly affected marine ecosystems, leading to more frequent and intense marine heatwaves, widespread coral bleaching, and disruptions in marine biodiversity (Hoegh-Guldberg et al., 2007; Hughes et al., 2017; Oliver et al., 2018; Smale et al., 2019). Coral reef ecosystems are highly sensitive indicators of such environmental changes (Hughes et al., 2017). The Korean Peninsula, located at the northwestern boundary of the Pacific Ocean, is strongly influenced by the Kuroshio Current, which flows northward along the western edge of the North Pacific (Lee et al., 2021; Nan et al., 2015; Wu et al., 2012; Yeh and Kim, 2010). This current plays a critical role in regional biogeochemical processes, including the transport of nutrients and sediments, as well as local climate regulation (Das et al., 2021). Persistent warming of the Kuroshio Current has been associated with the northward shift of distribution ranges of corals, fishes, and other marine organisms (Lu and Lee, 2014; Venegas et al., 2023). Indeed, the sea surface temperature (SST) in the waters surrounding South Korea, including Jeju Island, has shown a gradual increase, with the annual mean SST increasing at a rate approximately 2.6 times that of the global average over the past five decades (Han et al., 2023).
Coral reef ecosystems, known for their exceptional biodiversity, provide essential habitat and are particularly vulnerable to stressors such as warming and pollution due to the sessile nature of corals (Sobha et al., 2023; Sun et al., 2016). Elevated ocean temperatures contribute significantly to coral bleaching and microbial dysbiosis, ultimately leading to increased coral mortality (Ainsworth et al., 2016; Hoegh-Guldberg et al., 2007). Corals maintain complex interactions not only with symbiotic dinoflagellates but also with diverse microbial consortia, including bacteria, archaea, and fungi. These microbial communities, collectively referred to as the coral microbiome, are critical for coral immunity, metabolism, and environmental adaptation, and are considered key regulators of coral health and disease resistance (Pollock et al., 2018; Sun et al., 2016). However, most studies on coral microbiomes and bleaching have focused on tropical hermatypic corals, with relatively little attention given to soft corals lacking symbiotic dinoflagellates (Li et al., 2022). This research gap is particularly notable in the temperate coastal regions of Korea, where soft corals dominate local benthic communities. Despite their ecological importance, the structural and functional dynamics of microbial communities associated with soft corals under environmental stress remain poorly understood. In particular, the research on soft corals in highlatitude regions is very rare compared to the study on tropical hard corals. A few studies were reported about the soft coral distribution due to the changes in the benthic ecosystem in high-latitude regions (Ribas-Deulofeu et al., 2023), the analysis of symbiotic algae in soft corals in high-latitude regions (De Palmas et al., 2015), and the composition changes in bacterial community according to latitude in soft coral Scleronephthya gracillimum (Woo et al., 2017).
In this study, we investigated the microbial community and predicted functional profiles associated with a soft coral species, Eleutherobia rubra. It is distributed primarily in temperate waters of Korea, Japan, and the United States (California, Monterey), as well as in tropical waters off Western Australia at depths deeper than 80 m, with Korean populations predominantly inhabiting areas within 20 m depth along the western and southern coasts (Imahara et al., 2014; Song, 1976; Verseveldt and Bayer, 1988). The East Asian marginal seas, including the waters surrounding the Korean Peninsula, are experiencing accelerated warming driven by climate change, a trend further intensified by the influence of the Kuroshio Current (Sasaki and Umeda, 2021; Wang and Wu, 2022). Over the past decade, sea surface temperatures along the southern coast of Korea and around Jeju Island have increased markedly, and transcriptomic analyses of E. rubra indicate that this thermal stress may significantly threaten the survival of this representative coral species in the region (Ribas-Deulofeu et al., 2023; Ryu et al., 2019). We selected this coral species because it has no zooxanthellae, the effect of symbiotic algae on microbiome change can be avoided in response to the elevated seawater temperature. And also because of its distribution, this temperate coral species data can be useful to trace changes in coral ecosystems across the Pacific Northwest region.
Our findings provide foundational insights into the effects of climate-driven ocean warming on the symbiotic microbiota of soft corals. Unlike most prior studies that focus on tropical reef-building corals, this study uniquely explores the structural and functional shifts in the microbiome of a temperate soft coral, contributing to the broader understanding of coral holobiont resilience in rapidly warming seas.
2 Materials and methods
2.1 Sample collection and stress exposure
Colonies of the soft coral E. rubra were collected at a depth of approximately 15 to 20 m near Eoyudo, located off the southern coast of Korea (34°39′14.00′′N, 128°34′19.00′′E), in November, 2023 using standard SCUBA techniques, and the seawater temperature during collection was 16–17°C (Figure 1). The specimens were transported to a laboratory aquaria facility and acclimated for three days in filtered seawater maintained at 16°C and 33 ppt salinity, under a 14-hour light and 10-hour dark cycle at the ambient light intensity (800–900 lux). Seawater, pumped from a depth of 10 m in proximity to the institute (300 m from the coast) and filtered serially through 100, 10, and 1 µm filters (Filtertech, South Korea), was continuously supplied to four 100 L tanks at a minimal rate of 50 L h−1 for the coral acclimation, and seawater finally filtered through a 0.1 µm membrane was used for the thermal stress experiment. After acclimation, three intact colonies of corals were assigned to 26°C, 28°C, and 30°C experimental tanks respectively for 24 hours. The control group for the thermal experiment was kept in seawater at 16°C, and the experimental conditions were controlled by AquaController III Pro (Neptune Systems, Morgan Hill, CA, USA). Upon completion of the exposure, coral samples were immediately stored in an ultra-low temperature freezer at –80°C until DNA extraction.
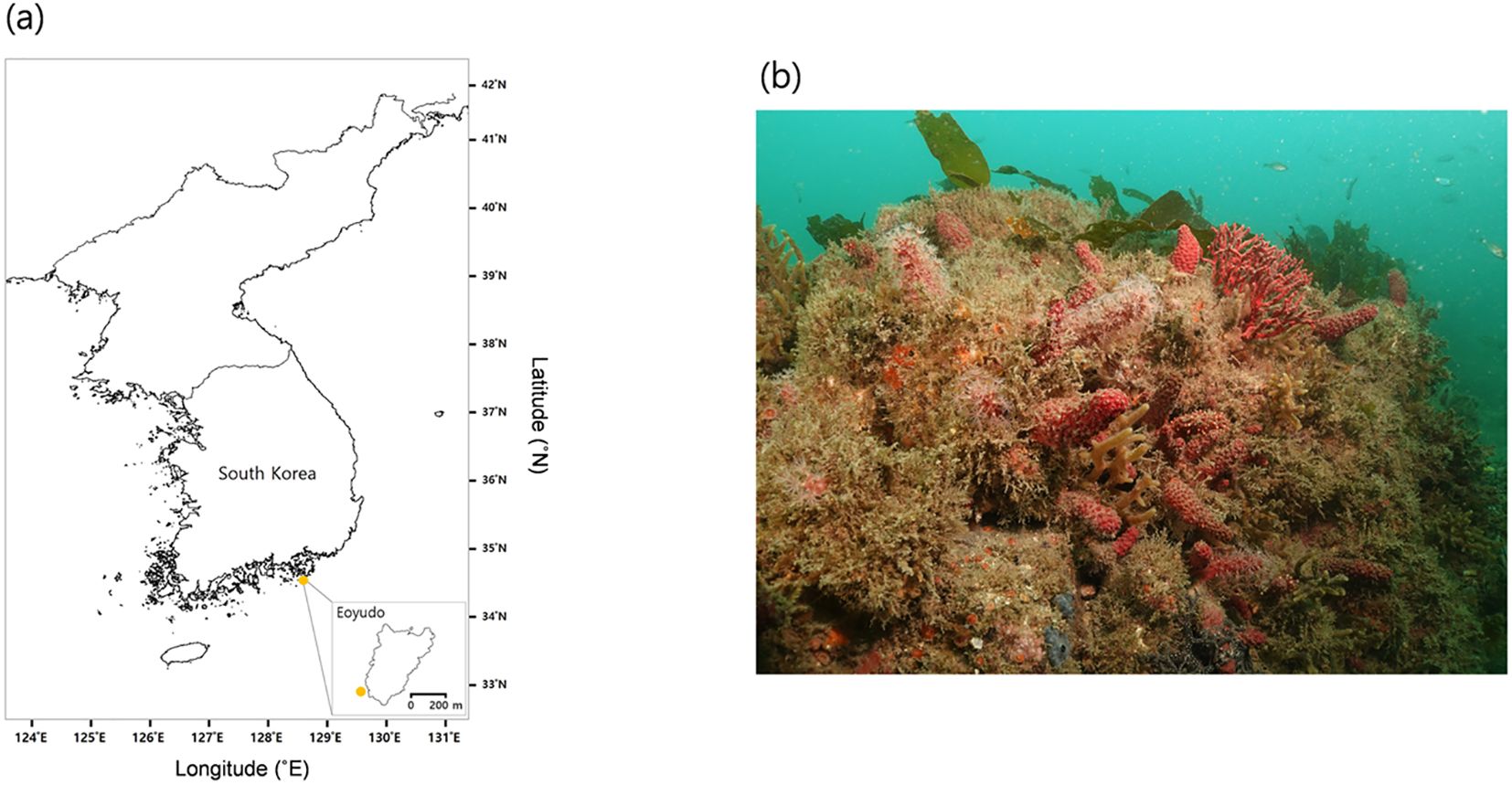
Figure 1. Sampling location and habitat overview of E. rubra. (a) Map showing the sampling site around Eoyudo, located off the southern coast of Korea. The inset map highlights the exact collection point near the island (orange dot). (b) Underwater photograph of the natural habitat of E. rubra.
2.2 DNA extraction
Frozen tissue samples were ground in a mortar with liquid nitrogen. Approximately 200 mg of powdered tissue was homogenized in 1 mL of CTAB lysis buffer containing 3% CTAB, 5.0 M NaCl, 3% PVP, 0.5 M EDTA (pH 8.0), 1 M Tris-HCl (pH 8.0), and 0.2% β-mercaptoethanol. The homogenate was incubated at 65°C for 1 hour. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was then added to denature proteins, followed by centrifugation at 10,000 ×g for 15 minutes at room temperature. The aqueous phase was collected, and RNase A (30 mg/mL) was added, followed by a 1-hour incubation at 37°C. DNA was further extracted with phenol:chloroform:isoamyl alcohol (25:24:1), then purified again using chloroform:isoamyl alcohol (24:1). After centrifugation at 10,000 ×g for 15 minutes at room temperature, the aqueous phase was recovered. DNA was precipitated by adding one-tenth volume of 3 M sodium acetate (pH 5.2) and an equal volume of 100% ethanol. The precipitated DNA was washed with 70% ethanol and resuspended in ion-exchanged ultrapure water. DNA concentration was measured using the Picogreen method with a Victor 3 fluorometer (Perkin Elmer Inc., Waltham, MA, USA), and quality was confirmed by agarose gel electrophoresis.
2.3 Library preparation and sequencing
Sequencing libraries were prepared according to the Illumina 16S Metagenomic Sequencing Library protocol to amplify the V3–V4 region. A total of 2 ng of genomic DNA was amplified in a PCR reaction containing 5× reaction buffer, 1 mM dNTP mix, 500 nM of each universal forward and reverse primer, and Herculase II fusion DNA polymerase (Agilent Technologies, Santa Clara, CA, USA). The first PCR was conducted with an initial denaturation at 95°C for 3 minutes, followed by 25 cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C, and a final extension at 72°C for 5 minutes. The primers used in the first amplification included Illumina adapter overhangs: V3-F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and V4-R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′). The resulting PCR product was purified using AMPure beads (Agencourt Bioscience, Beverly, MA, USA), and 2 µL of the purified product was used as the template for a second PCR using NexteraXT indexed primers. The second PCR followed the same thermal cycling conditions as the first but was run for 10 cycles. The final PCR products were again purified using AMPure beads. Quantification was performed using qPCR (KAPA Library Quantification Kit for Illumina platforms), and quality was assessed using the TapeStation D1000 ScreenTape system (Agilent Technologies, Waldbronn, Germany). Paired-end sequencing (2 × 300 bp) was conducted on the Illumina MiSeq platform by Macrogen (Daejeon, South Korea).
2.4 OTU analysis
After sequencing, the raw MiSeq data were demultiplexed based on index sequences, and fastq files were generated for each sample (Chen et al., 2018). Adapter sequences were removed using the fastp program, and sequencing errors in the overlapping region of paired-end reads were corrected. Paired-end reads were merged into single sequences using FLASH (v1.2.11) (Magoč and Salzberg, 2011). Assembled reads shorter than 400 bp or longer than 500 bp were removed. Low-quality sequences, ambiguous bases, and chimeric reads were filtered out using CD-HIT-OUT (Fu et al., 2012), and the remaining reads were clustered into species-level operational taxonomic units (OTUs). Representative sequences for each OTU were identified using BLAST+ (v2.9.0) (Zhang et al., 2000) against the NCBI 16S Microbial Reference Database. Taxonomic assignments were based on the best hit, but OTUs were left unclassified if the query coverage or identity of the best hit was below 85%. The resulting OTU table was used for comparative analyses of microbial communities using QIIME (v1.9) (Caporaso et al., 2010).
2.5 Taxonomic and functional profiling
Microbial diversity within samples was evaluated using alpha diversity indices, including Shannon, Inverse Simpson, and Chao1, calculated based on rarefied OTU tables. Comparisons between the control and heat stress groups were performed using Student’s t-test and visualized as boxplots with Plotly. To assess beta diversity, principal coordinate analysis (PCoA) was performed using both weighted and unweighted UniFrac distances. To ensure direct comparability with the functional data, we also calculated Bray–Curtis dissimilarities on the rarefied OTU relative abundance table (square root transformed). The resulting distance matrix was subjected to principal coordinates analysis (PCoA) and to PERMANOVA and PERMDISP with 999 permutations using scikit-bio (Supplementary Table S4). Group distribution trends were visualized with 95% confidence ellipses using the EllipticEnvelope method. To verify sequencing depth sufficiency, rarefaction curves were generated using Chao1 and Observed OTUs. All samples showed curves approaching saturation, indicating adequate coverage for diversity estimates (Supplementary Figure S1).
Taxonomic composition was compared at the genus and phylum levels based on relative abundance and visualized as stacked bar plots. Genus-level composition was further evaluated using rank–abundance curves to simultaneously assess dominant and rare taxa. To statistically compare phylum-level relative abundances between control and 26°C-treated (heat stress) groups, both Welch’s t-test and the Mann–Whitney U test were applied to account for variance and non-normal distribution. The average difference (Δ) was calculated as the mean relative abundance difference between groups (Supplementary Tables S1, S2). Functional analysis was performed using PICRUSt2 to predict KEGG and MetaCyc pathways. Significant changes were evaluated based on log2 fold change and the Mann–Whitney U test. Pathways with marked increases or decreases were visualized using bar plots, and the top 50 most abundant functions were shown as a log-scaled heatmap. Functional PCoA based on Bray–Curtis distances was also conducted and visualized with Seaborn and Matplotlib. LEfSe analysis was used to identify significantly enriched species-level taxa, and the results were visualized using a bidirectional bar plot based on log-transformed LDA scores. Lastly, to evaluate correlations between genus-level taxa and predicted metabolic functions, Spearman correlation analysis was performed, and relationships among the top 20 genera and functions were visualized as a heatmap. All analyses were conducted using various Python libraries, including Pandas, Scipy, Seaborn,Matplotlib, andPlotly (Barrett et al., 2005; McKinney, 2011; Sievert, 2020; Virtanen et al., 2020; Waskom, 2021).
3 Results
3.1 Changes in microbial diversity under heat stress
To investigate changes in the symbiotic microbial community of corals under elevated seawater temperatures, colonies of E. rubra (a soft coral species) were collected from coastal waters near Eoyudo, located off the southern coast of Korea. The collected corals were transferred to experimental aquaria and acclimated for three days in filtered seawater maintained at 16°C. Based on the sea surface temperature data around South Sea, Korea (Ribas-Deulofeu et al., 2023) and the other soft coral, S. gracillimum, study on heat stress in our laboratory (Woo and Yum, 2022), the heat stress experiment of E. rubra was designed originally at 26°C, 28°C, and 30°C but the groups of 28°C and 30°C were melted down within 24 hr and only 26°C group was compared with the control group to investigate its microbial community. Total DNA was extracted from each sample, followed by 16S rRNA-based metagenomic sequencing.
The impact of thermal exposure on the microbial community of E. rubra was assessed through alpha and beta diversity analyses. All alpha diversity indices (Shannon, Chao1, and Inverse Simpson) showed a modest increase under the heat stress condition, indicating a potential expansion of microbial diversity in response to heat stress (Figure 2). The similar alpha diversity values suggest that the overall microbial diversity remained relatively stable. However, high inter-individual variability precluded statistical significance. To evaluate whether this non-significance was simply due to small sample size (n = 3 per group), we performed a post-hoc power analysis using Cohen’s d for each index. Moderate-to-large effect sizes were observed for Shannon (d =0.79) and Inverse Simpson (d =0.93), yet the resulting statistical power was low across all metrics (Shannon ≈11.7%, Inverse Simpson ≈14.4%, Chao1 ≈5.2%; see Supplementary Table S3).
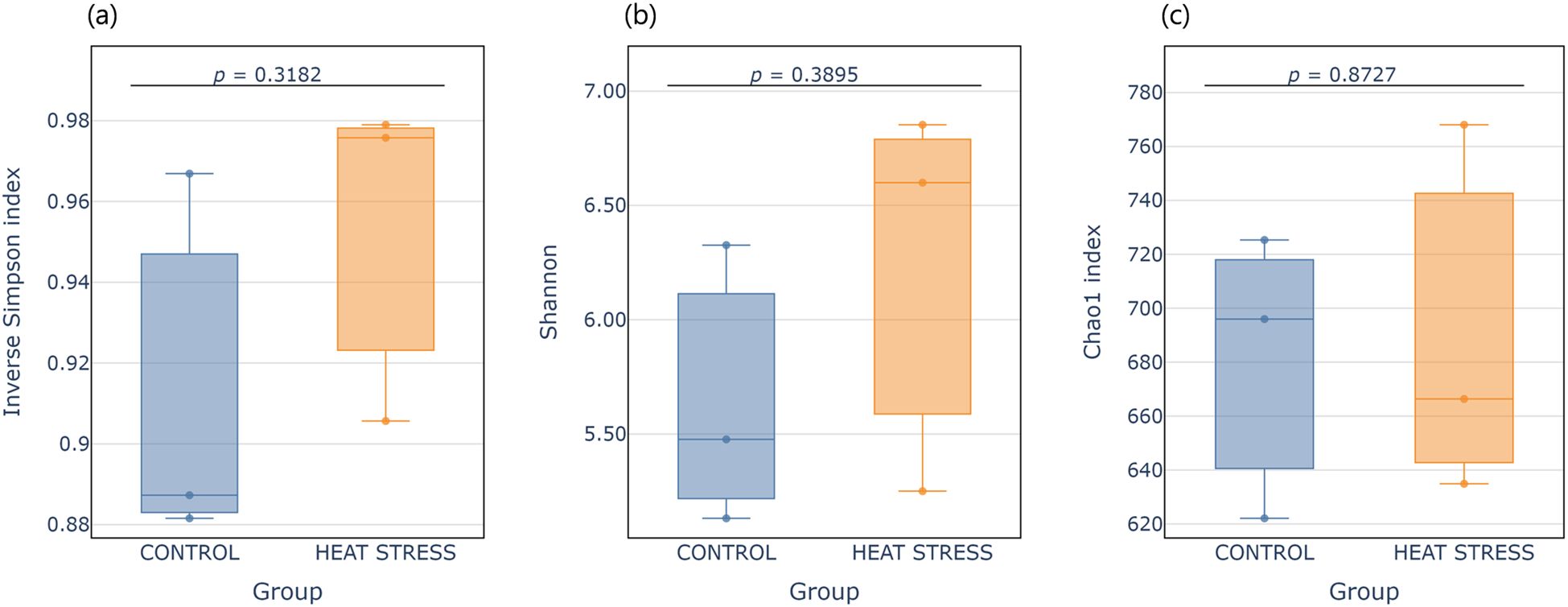
Figure 2. Boxplots showing alpha diversity indices for the control and heat treatment groups. (a) Inverse Simpson index, (b) Shannon index, and (c) Chao1 index. Each box represents the interquartile range (IQR), with the median shown as a horizontal line. Individual samples are overlaid as vertical jittered dots. p-values were calculated using Student’s t-test.
For beta diversity, principal coordinate analysis (PCoA) was performed using both weighted and unweighted UniFrac distances (Figures 3a, b). In the weighted UniFrac analysis, PC1 explained 72.8% of the total variance, and the centroids of the two groups were located in close proximity, indicating that the composition and relative proportions of dominant taxa remained largely unchanged after heat stress. In contrast, the unweighted UniFrac analysis showed greater separation between groups, with PC1 and PC2 explaining 25.1% and 21.9% of the variance, respectively. In addition to the UniFrac-based comparisons, we conducted a complementary Bray–Curtis β-diversity analysis on the same OTU relative-abundance table. Using Bray–Curtis distances, the first two PCoA axes accounted for 45.1% and 21.8% of the variance, respectively (Figure 3). Although PERMANOVA (pseudo-F =1.08, p =0.205) and PERMDISP (F =0.099, p =0.786) did not indicate statistically significant group separation (Supplementary Table S4), the ordination nonetheless displayed a visibly clearer segregation between CONTROL and HEAT-STRESS samples than the UniFrac plots.
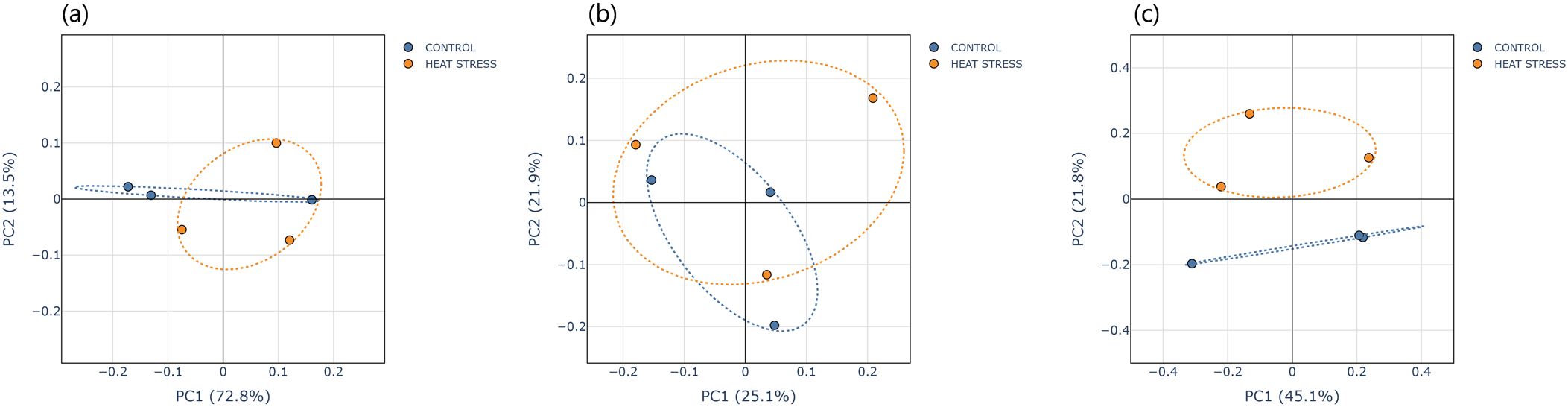
Figure 3. Principal Coordinates Analysis (PCoA) plots based on (a) Weighted UniFrac, (b) Unweighted UniFrac, and (c) Bray-Curtis distances. Each point represents an individual sample, and dotted ellipses indicate the 95% confidence region for each group.
This pattern was also reflected in the rank-abundance curve analysis (Figure 4b). Compared to the control group, the heat stress group exhibited higher overall abundance, and the curve was more gradual in the latter half (after rank 150), indicating increased evenness and a relative enrichment of rare taxa. These results are consistent with the alpha and beta diversity findings, supporting the interpretation that heat stress may induce community restructuring through increasing rare microbial populations.
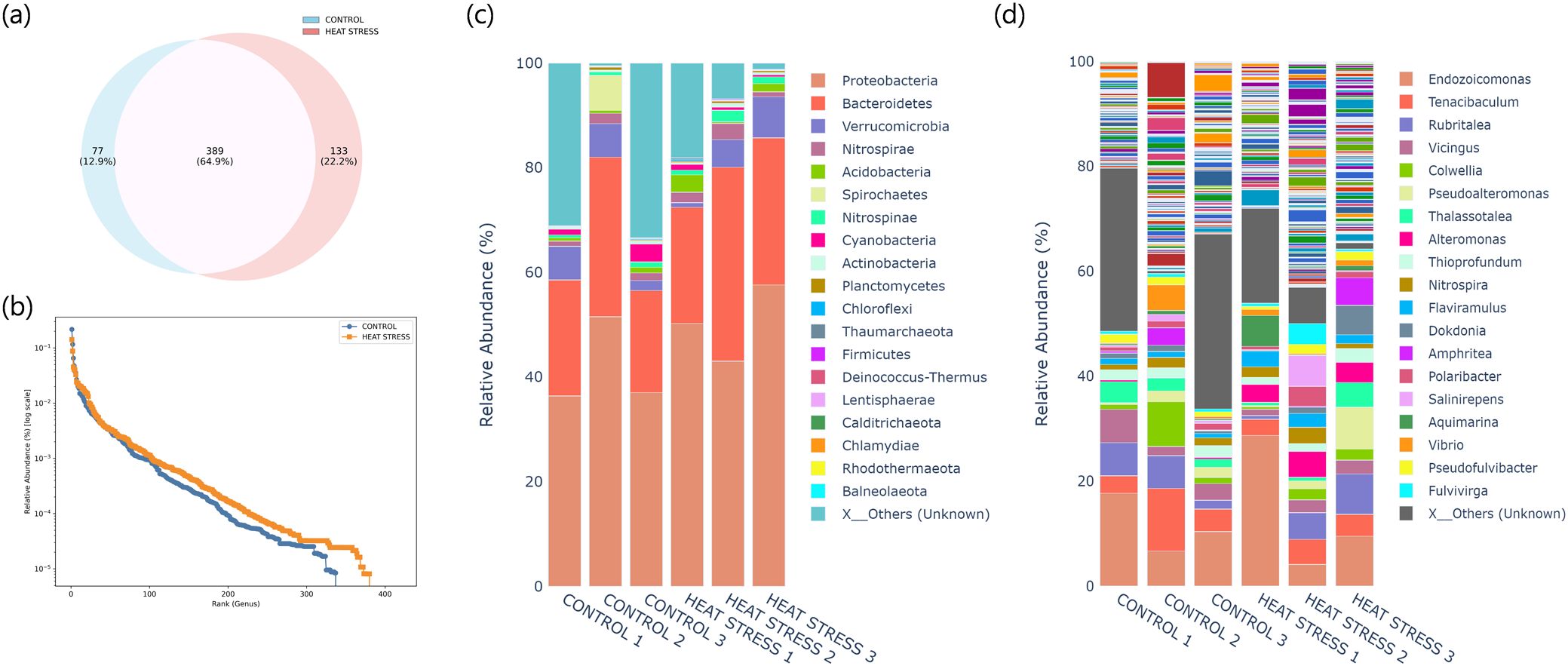
Figure 4. Taxonomic composition of microbial communities under control and heat stress conditions. (a) Venn diagram showing the number of shared and unique OTUs between the two groups at the species level. (b) Rank-abundance curves based on genus-level relative abundance (log scale). (c) Stacked barplot of relative abundance at the phylum level. (d) Stacked barplot of relative abundance at the genus level. “X_Others” indicates low-abundance or unclassified taxa grouped for clarity.
3.2 Structural shifts in bacterial communities under heat stress
To evaluate the effects of heat stress on the symbiotic microbial community structure of E. rubra in more detail, we analyzed the distribution and relative abundance of bacterial taxa. Among a total of 599 species identified across both groups, 389 species (64.9%) were shared between the control and heat stress samples, while 77 species (12.9%) were unique to the control group and 133 species (22.2%) were detected only under heat stress conditions (Figure 4a). Taxonomic composition was further examined at both the phylum and genus levels (Figures 4c, d; Supplementary Tables S1, S2). At the phylum level, the dominant group Proteobacteria increased from an average of 41.7 ± 8.6% in the control group to 50.3 ± 7.3% in the heat stress group. Bacteroidetes also showed an increasing trend, from 24.0 ± 5.7% to 29.1 ± 7.5%, although these changes were not statistically significant (p > 0.05). Several minor phyla, including Acidobacteria, Firmicutes, and Nitrospinae, also showed increasing trends under heat stress, but without statistical significance. At the genus level, Alteromonas, Phaeobacter, Pseudomaricurvus, Oleibacter, and Pseudoruegeria were significantly enriched in the heat-stress group (p < 0.05), while Thiohalophilus, along with flavobacterial genera such as Zobellia and Paracoccus, showed significant decreases in relative abundance (p < 0.05).
Given the high variation among samples, group-level comparisons based solely on average relative abundance can be misleading. To better quantify taxonomic shifts associated with heat stress, LEfSe (Linear Discriminant Analysis Effect Size) analysis was conducted (Figure 5; Supplementary Table S5). Using a threshold of LDA score > 2.0 and p < 0.05, a total of 43 species showed significant differences between the groups. The top five species enriched under heat stress were Colwellia aquamaris, Wenzhouxiangella marina, Microbulbifer hydrolyticus, Poseidonibacter lekithochrous, and Pelagimonas phthalicica. In contrast, species that were less abundant in the heat stress group included Endozoicomonas elysicola, Thiohalocapsa marina, and Dulcicalothrix necridiiformans, some of which are known for their photosynthetic capacity or benthic adaptations. For completeness, we also repeated the LEfSe analysis using a stricter LDA cut-off of 3.0. As a result, only 15 discriminatory species were retained, all of which are a subset of those shown in Figure 5. This result is provided in Supplementary Figure S2.
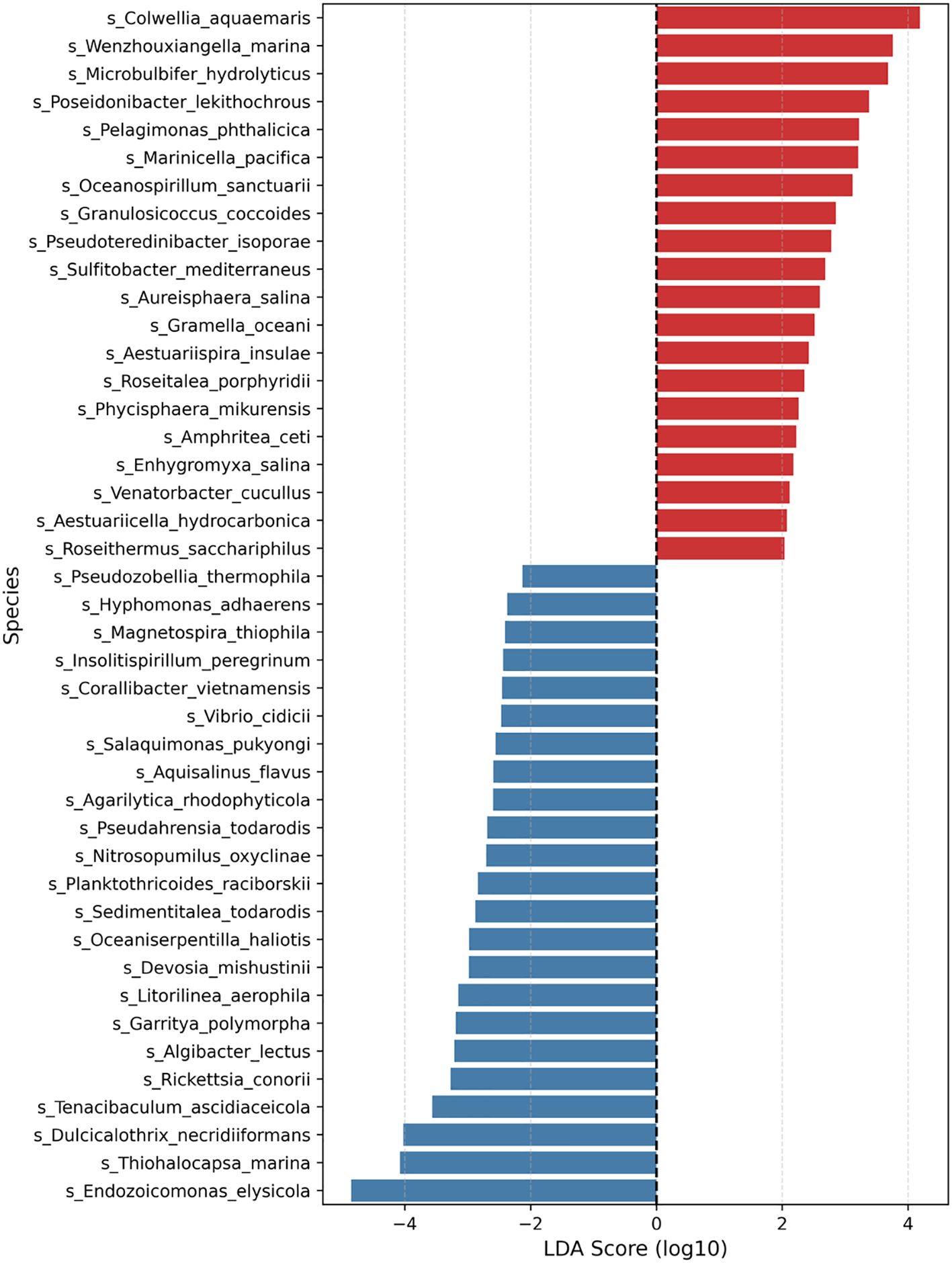
Figure 5. Differentially abundant species between the heat stress and control groups were identified by LEfSe analysis. The plot shows species with LDA scores (log10) > 2.0 and p < 0.05. Red bars indicate species enriched in the heat stress group, while blue bars represent species enriched in the control group.
3.3 Functional shifts in the microbial community under heat stress
Changes at the phylogenetic level alone do not provide direct insight into how microbial community restructuring may influence the physiological functions of E. rubra. In marine microbial ecosystems, taxonomically distinct organisms can share similar metabolic pathways due to functional redundancy. Therefore, an increase or decrease in the abundance of a specific taxon does not necessarily translate into changes in overall metabolic capacity. Conversely, even minor changes in taxonomic composition may lead to substantial shifts in host–microbe interactions if the expression of key metabolic pathways is altered. To address this, we reconstructed functional profiles based on predicted metabolic potential using PICRUSt2 and examined how temperature changes affected the overall metabolic spectrum of the community. We also investigated how these functional shifts correlated with specific bacterial taxa. A total of 366 metabolic pathways were predicted and quantified using the MetaCyc database in PICRUSt2, and Bray–Curtis distance-based PCoA was conducted. PC1 explained 68% of the total variance, and samples from the control and heat stress groups were separated (Figure 6a). A heatmap based on the log10-transformed median abundance of predicted metabolic pathways also revealed overall differences in functional profiles between the groups (Figure 6b), which is consistent with the separation pattern observed in the PCoA analysis. Based on the median abundance of each pathway, log2 fold changes were calculated. Among the 366 predicted pathways, 47 (12.8%) showed at least a twofold increase (log2FC > 1), and 25 (6.8%) exhibited at least a twofold decrease (log2FC < –1) under heat stress (Supplementary Table S6). The syringate degradation pathway showed the largest increase, followed by elevated levels of pathways related to carbohydrate and cofactor metabolism, including enterobacterial common antigen biosynthesis, methylaspartate cycle, adenosylcobalamin biosynthesis, and isoprene biosynthesis. In contrast, pathways involved in membrane components and fermentation, such as vitamin E biosynthesis, chondroitin sulfate degradation, and pyruvate fermentation to acetone, were reduced. These results suggest that metabolic pathways may be reorganized in response to thermal stress (Figures 7a, b). To evaluate how these functional changes were linked to taxonomic composition, we calculated Spearman correlation coefficients between the top 20 most abundant genera and the predicted metabolic functions, and visualized the results in a heatmap (Figure 8). Genera such as Thalassotalea, Amphritea, and Rubritalea exhibited positive correlations with most major metabolic pathways, especially those involved in amino acid and nucleotide biosynthesis. In contrast, genera such as Alteromonas and Nitrospira showed negative correlations with the same pathways, indicating that different genera contributed differentially to the functional profiles under heat stress. Interestingly, we observed cases where changes in genus-level abundance did not align with changes in functional output. This suggests that the taxa responsible for particular metabolic functions may have shifted under heat stress, even when overall abundance patterns remained relatively stable.
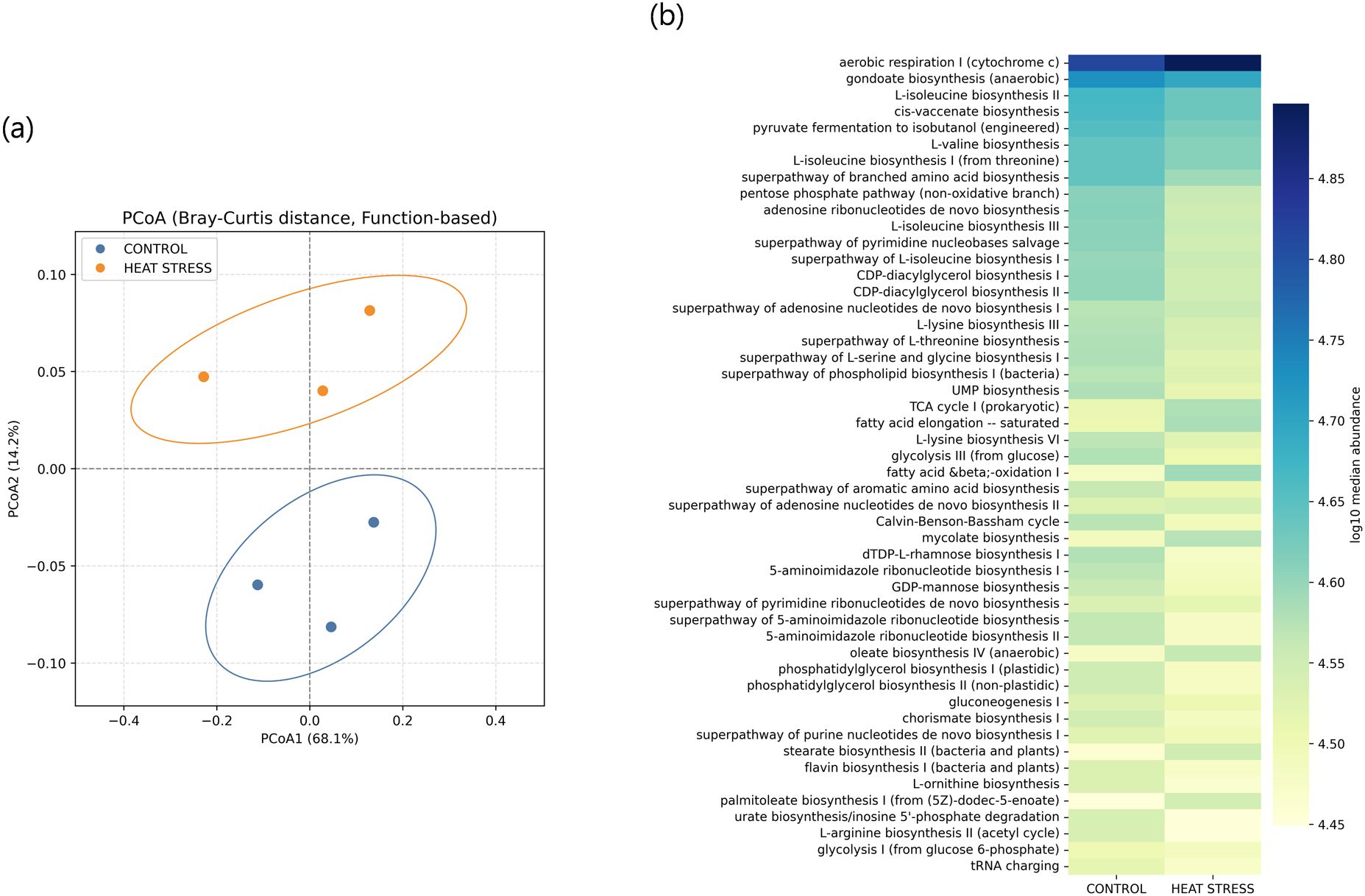
Figure 6. Functional analysis of microbial communities based on PICRUSt2-predicted MetaCyc pathways. (a) PCoA based on Bray–Curtis distances of functional profiles. Each point represents a sample, with ellipses indicating the 95% confidence intervals. (b) Heatmap displaying the log10-transformed median abundance of the top 50 predicted pathways in each group.
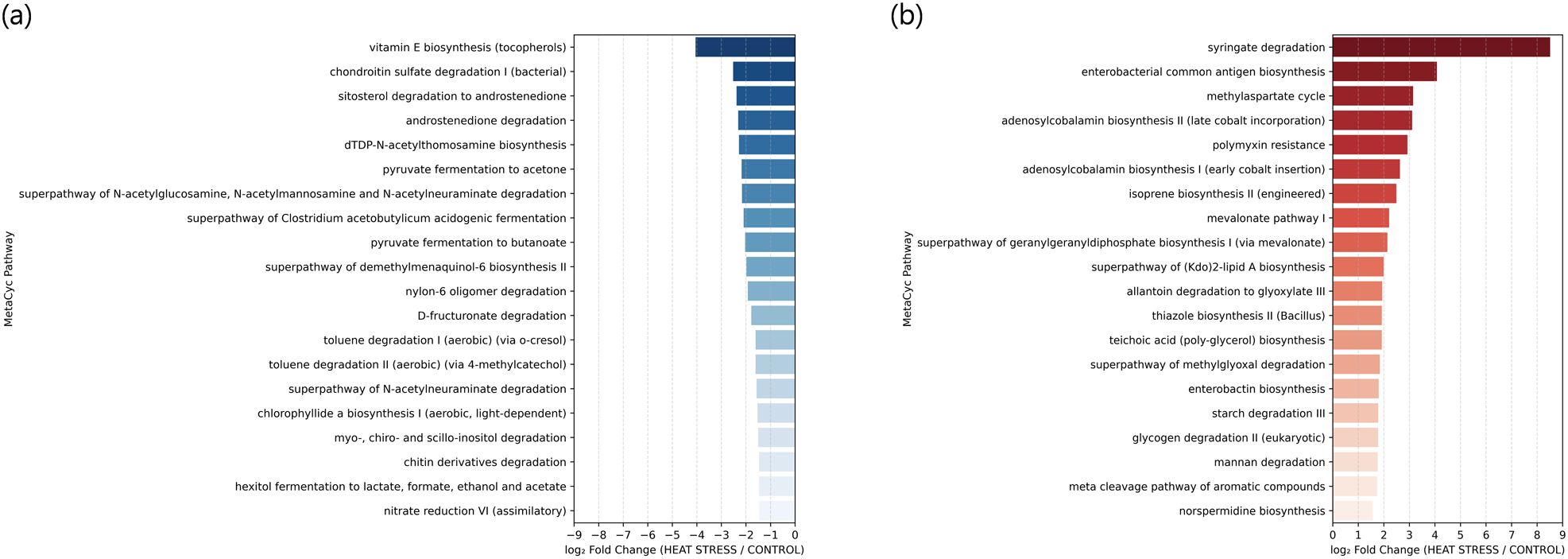
Figure 7. Top 20 predicted metabolic pathways showing the most significant changes in relative abundance between the heat stress and control groups based on log2 fold change of median values. (a) Pathways with decreased abundance in the heat stress group (log2FC < 0, blue). (b) Pathways with increased abundance in the heat stress group (log2FC > 0, red).
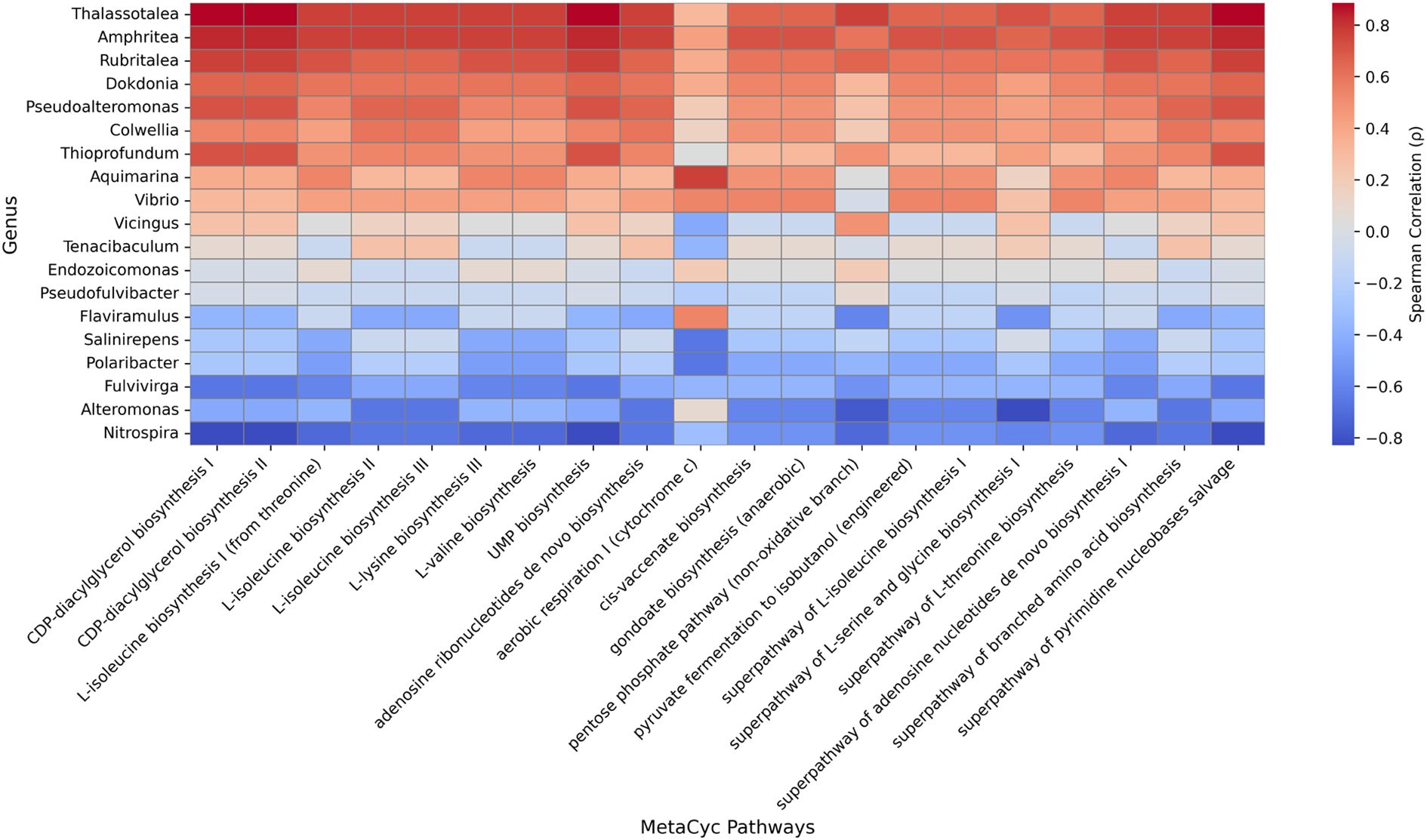
Figure 8. Spearman correlation heatmap between the top 19 genera and selected MetaCyc pathways. Each cell represents the Spearman correlation coefficient (p) between genus-level relative abundance and pathway abundance predicted by PICRUSt2. Red and blue colors indicate positive and negative correlations, respectively. Pathways and genera are ordered by mean correlation values to highlight overall patterns.
4 Discussion
In this study, we examined how short-term thermal stress influences the structure and function of the symbiotic microbial community associated with the temperate soft coral E. rubra. While the dominant microbial taxa remained relatively stable, alpha and beta diversity patterns, alongside shifts in lowabundance taxa, indicate early microbial community destabilization. These changes were not uniform across samples, instead exhibiting greater inter-individual variation under heat stress, a phenomenon that aligns with the Anna Karenina Principle (AKP) (Zaneveld et al., 2017). According to this model, microbiomes under stress become more individualized and stochastic in their responses, leading to increased dispersion without a consistent directional shift. This interpretation is supported by unweighted UniFrac and rank-abundance analyses, which revealed greater community variability and rare taxon enrichment in heat stress samples, even in the absence of major taxonomic turnover. Nevertheless, our PERMDISP analyses did not reveal statistically significant differences in β-diversity dispersion between the CONTROL and HEAT-STRESS groups (unweighted UniFrac: F =0.838, p =0.275; weighted UniFrac: F =0.003, p =0.790) (Supplementary Table S4). This suggests that the statistical power may have been insufficient to detect the dispersion differences predicted by the Anna Karenina principle, likely due to the small sample size (n =3 per group) and substantial within-group variability. This limitation in statistical power is further supported by our post hoc analysis of alpha diversity indices. Despite observing moderate to large effect sizes for the Shannon (d =0.79) and Inverse Simpson (d =0.93) indices, the corresponding statistical power remained below 15%. This result reflects the impact of the limited sample size and pronounced inter-individual variation. These post hoc power estimates (Supplementary Table S3) indicate that genuine biological shifts may have gone undetected. Nevertheless, consistent trends in functional profiles were still observed, suggesting that biologically meaningful changes were present. Future studies with larger sample sizes and longer exposure periods will be necessary to detect community-wide structural shifts with greater statistical confidence.
In contrast to the relatively subtle taxonomic shifts, functional changes were more pronounced. According to PICRUSt2-based predictions, a total of 47 metabolic pathways showed more than a twofold increase, while 25 pathways significantly decreased in response to heat stress. The Bray–Curtis distance-based PCoA also showed a clear separation between the two groups. These results indicate that functional responses may occur earlier and more sensitively than structural reorganization at the community level during the initial phase of induced extreme temperature. This may be attributed to the functional redundancy of marine microbial communities, where functional roles can be redistributed without substantial taxonomic turnover (Louca et al., 2016). Species- and genus-level analyses revealed that specific taxa changed significantly in relative abundance following heat stress. For example, among the enriched species identified by LEfSe analysis, Colwellia aquamaris and Microbulbifer hydrolyticus are known to be involved in polysaccharide degradation and fatty acid metabolism (Liu et al., 2014; Zhong and Agarwal, 2024). These species may play central roles in organic matter turnover and energy metabolism in marine ecosystems and may have been selectively enriched due to their high adaptability to stress. In contrast, taxa such as Endozoicomonas elysicola and Thiohalocapsa marina, which decreased under heat stress, are known to establish stable mutualistic relationships with marine invertebrates such as corals and sponges. Some of these taxa are functionally linked to sulfur metabolism and photosynthesis (Anil Kumar et al., 2009; Hochart et al., 2023; Neave et al., 2017; Reyes et al., 2025). Their decline may reflect a breakdown in symbiotic associations or the loss of functional specialists under heat stress, potentially weakening host–microbe interactions. In particular, Endozoicomonas has been recognized as a core microbiome member in various coral species, contributing to pathogen resistance and metabolic homeostasis (Neave et al., 2017; Ziegler et al., 2019). Therefore, its reduction may pose physiological risks to E. rubra under thermal stress. These findings suggest that the compositional shifts observed in response to heat are not merely structural but may involve the replacement of functionally critical taxa at early stages. Zhu et al. (2023) reported significant increases in bacterial α-diversity, β-dispersion, and core microbiome restructuring in the tropical stony coral Porites cylindrica under both acute and chronic thermal stress. Notably, beneficial taxa like Delftia decreased, while opportunistic pathogens such as Nautella emerged. In contrast, our findings in the temperate soft coral E. rubra under short-term heat stress revealed a stable taxonomic composition but pronounced shifts in functional pathways. Given the differences in experimental conditions, it remains unclear whether both tropical stony and temperate soft corals would exhibit similar functional shifts under short-term stress or undergo taxonomic changes with prolonged exposure. Future studies under identical conditions are needed to clarify this.
Notably, structural analyses alone may underestimate the true extent of microbiome reorganization under environmental stress. The most prominent responses under heat stress were observed at the level of metabolic function, providing insights into how microbial communities deploy adaptive strategies under environmental change (Figures 7a, b). Among the most highly increased pathways was syringate degradation, which is involved in the breakdown of aromatic compounds. This increase may reflect metabolic adaptation to oxidative stress or the accumulation of plant-derived compounds under thermal stress. A similar pathway enrichment has been reported in Arctic Ocean microbiomes as part of their metabolic response to stress and the influx of terrestrial organic matter (Grevesse et al., 2022). Other upregulated pathways, including enterobacterial common antigen biosynthesis, methylaspartate cycle, adenosylcobalamin (vitamin B12) biosynthesis, and isoprene biosynthesis, are associated with cofactor metabolism and membrane fluidity. Enterobacterial common antigen, for example, plays a key role in outer membrane integrity and permeability control, enhancing microbial survival under stress (Rai and Mitchell, 2020). Vitamin B12 accumulation has also been linked to osmoregulatory stress tolerance and improved microbial survival (Wang et al., 2024). These observations suggest that metabolic pathways enriched under heat stress may contribute to maintaining cellular homeostasis, adjusting membrane fluidity, and synthesizing key cofactors to enhance microbial resilience. In contrast, pathways such as vitamin E biosynthesis, chondroitin sulfate degradation, and pyruvate fermentation to acetone were reduced under heat stress (Figure 7b). This suggests that some physiological functions may be downregulated under stress. However, the decreased pathways generally represent non-core metabolic functions and are biologically less interpretable compared to the upregulated core functions. Therefore, the reduction in these pathways may be incidental, resulting from community restructuring rather than targeted suppression by environmental cues.
The Spearman correlation heatmap between metabolic functions and genus-level taxa revealed strong positive correlations between functional pathways and genera such as Thalassotalea, Rubritalea, and Amphritea, whereas Alteromonas and Nitrospira were negatively correlated with those functions (Figure 8). These findings suggest that specific genera may play central roles in driving or suppressing key metabolic processes under changing environmental conditions. Overall, our analysis highlights that microbial functional responses can occur more rapidly and sensitively than taxonomic changes and may offer valuable insight into the physiological adaptation and resilience of host organisms. Even within the 24 h exposure window, genus- and species-level LEfSe analysis detected several statistically significant shifts. Only a handful of taxa, Colwellia aquaemaris, Wenzhouxiangella marina, and Endozoicomonas elysicola, combined statistical significance with ecologically meaningful abundance, whereas most discriminant species occurred at < 0.1% relative abundance. Such low-abundance, functionally specialised microbes may therefore act as early biomarkers of microbiome destabilisation. To visualise these subtle patterns, we provide a species-level stacked-bar plot (Supplementary Figure S3) and an accompanying table of per-sample species abundances (Supplementary Table S7). In this context, we define early destabilization as a phase in which statistically discernible shifts in predicted functional pathways emerge before significant changes are detectable in taxonomic composition, β-diversity dispersion, or α-diversity indices. Such functional adjustments, particularly in stress-response and metabolic pathways, may therefore signal the onset of holobiont stress adaptation.
Taken together, our findings highlight that functional responses of the microbiome may serve as earlier and more sensitive indicators of coral stress than structural shifts alone. This underscores the importance of incorporating functional profiling in future coral microbiome research, particularly under climate change scenarios (Kim et al., 2025). Similar integrative approaches combining isolation, metabolic screening, and genome-guided analysis have been proposed to explore the biosynthetic and ecological potential of symbiotic microbes in sponge systems (Kim et al., 2025), and may serve as a useful framework for future coral holobiont research. While our study focused on short-term responses, longer-term experiments with larger sample sizes will be critical to determine whether the observed functional plasticity leads to adaptive resilience or cumulative dysbiosis. Moreover, targeted validation of predicted functions, e.g., transcriptomic or metabolomic analyses, will help clarify the ecological roles of different microbial taxa in coral health and stress recovery.
5 Conclusion
This study demonstrates that short-term heat stress may elicit early functional responses in the microbial community associated with the soft coral E. rubra, even in the absence of marked taxonomic shifts. While the composition of dominant microbial taxa remained relatively stable, predictive functional profiling suggested detectable metabolic changes. These findings imply that microbial communities may begin to adjust their functional potential in response to thermal stress before large-scale structural reorganization becomes apparent.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/, https://doi.org/10.6084/m9.figshare.29148449.v1.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
JK: Investigation, Writing – review & editing, Writing – original draft, Data curation. YJ: Investigation, Writing – review & editing. SH: Writing – review & editing, Data curation, Investigation. CO: Writing – review & editing, Funding acquisition, Data curation. HY: Writing – review & editing, Investigation. JL: Investigation, Writing – review & editing, Writing – original draft, Data curation. SW: Conceptualization, Writing – review & editing, Writing – original draft, Data curation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Korea Institute of Ocean Science and Technology Research grants (PEA0314 and PEA0302), and “Marine Biotics project (20210469)” funded by Ministry of Ocean and Fisheries, Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1635356/full#supplementary-material
References
Ainsworth T. D., Heron S. F., Ortiz J. C., Mumby P. J., Grech A., Ogawa D., et al. (2016). Climate change disables coral bleaching protection on the great barrier reef. Science 352, 338–342. doi: 10.1126/science.aac7125
Anil Kumar P., Srinivas T. N. R., Thiel V., Tank M., Sasikala C., Ramana C. V., et al. (2009). Thiohalocapsa marina sp. nov., from an Indian marine aquaculture pond. Int. J. Syst. Evol. Microbiol. 59, 2333–2338. doi: 10.1099/ijs.0.003053-0
Barrett P., Hunter J., Miller J. T., Hsu J.-C., and Greenfield P. (2005). “matplotlib–a portable python plotting package,” in Astronomical data analysis software and systems XIV. eds. P. L. Shopbell, M. C. Britton & R. Ebert, Astronomical Society of the Pacific (San Francisco, USA), 347. , 91.
Bindoff N. L., Cheung W. W. L., Kairo J. G., Ar´ıstegui J., Guinder V. A., Hallberg R., et al. (2022). Changing Ocean, Marine Ecosystems, and Dependent Communities (United Kingdom: Cambridge University Press), 447–587. doi: 10.1017/9781009157964
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen S., Zhou Y., Chen Y., and Gu J. (2018). fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng L., Abraham J., Hausfather Z., and Trenberth K. E. (2019). How fast are the oceans warming? Science 363, 128–129. doi: 10.1126/science.aav7619
Das P., Lin A. T.-S., Chen M.-P. P., Miramontes E., Liu C.-S., Huang N.-W., et al. (2021). Deep-sea submarine erosion by the kuroshio current in the manila accretionary prism, offshore southern Taiwan. Tectonophysics 807, 228813. doi: 10.1016/j.tecto.2021.228813
De Palmas S., Denis V., Ribas-Deulofeu L., Loubeyres M., Woo S., Hwang S. J., et al. (2015). Symbiodinium spp. associated with high-latitude scleractinian corals from jeju island, South Korea. Coral Reefs 34, 919–925. doi: 10.1007/s00338-015-1286-y
Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012). Cd-hit: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Grevesse T., Guéguen C., Onana V. E., and Walsh D. A. (2022). Degradation pathways for organic matter of terrestrial origin are widespread and expressed in arctic ocean microbiomes. Microbiome 10, 237. doi: 10.1186/s40168-022-01417-6
Han I.-S., Lee J.-S., and Jung H.-K. (2023). Long-term pattern changes of sea surface temperature during summer and winter due to climate change in the korea waters. Fisheries Aquat. Sci. 26, 639–648. doi: 10.47853/fas.2023.e56
Hochart C., Paoli L., Ruscheweyh H.-J., Salazar G., Boissin E., Romac S., et al. (2023). Ecology of endozoicomonadaceae in three coral genera across the pacific ocean. Nat. Commun. 14, 3037. doi: 10.1038/s41467-023-38502-9
Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Hughes T. P., Kerry J. T., Alvarez´ Noriega M., Alvarez´ Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Imahara Y., Iwase F., and Namikawa H. (2014). The Octocorals of Sagami Bay. Tokai University Press, Hadano City, Kanagawa, Japan
Kim J.-A., Choi S.-S., Lim J. K., and Kim E.-S. (2025). Unlocking marine treasures: isolation and mining strategies of natural products from sponge-associated bacteria. Natural Product Rep. 42, 1195–1225. doi: 10.1039/d5np00013k
Lee S.-H., Hwang J.-S., Kim K.-Y., and Molinero J. C. (2021). Contrasting effects of regional and local climate on the interannual variability and phenology of the scyphozoan, aurelia coerulea and nemopilema nomurai in the korean peninsula. Diversity 13, 214. doi: 10.3390/d13050214
Li T., Huang J., Du H., Liu X., Zhong C., and Lin S. (2022). Coral bleaching from a nutrient perspective is understudied: A bibliometric survey. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.926783
Liu Y., Liu L.-Z., Zhong Z.-P., Zhou Y.-G., Liu Y., and Liu Z.-P. (2014). Colwellia aquaemaris sp. nov., isolated from the cynoglossus semilaevis culture tank in a recirculating mariculture system. Int. J. Syst. Evol. Microbiol. 64, 3926–3930. doi: 10.1099/ijs.0.063305-0
Louca S., Parfrey L. W., and Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lu H.-J. and Lee H.-L. (2014). Changes in the fish species composition in the coastal zones of the kuroshio current and China coastal current during periods of climate change: Observations from the set-net fishery, (1993–2011. Fish Res. 155, 103–113. doi: 10.1016/j.fishres.2014.02.032
Magoč T. and Salzberg S. L. (2011). Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
McKinney W. (2011). “pandas: a foundational python library for data analysis and statistics,” in Python for high performance and scientific computing, 14. , 1–9.
Nan F., Xue H., and Yu F. (2015). Kuroshio intrusion into the south China sea: A review. Prog. Oceanography 137, 314–333. doi: 10.1016/j.pocean.2014.05.012
Neave M. J., Michell C. T., Apprill A., and Voolstra C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7, 40579. doi: 10.1038/srep40579
Oliver E. C. J., Donat M. G., Burrows M. T., Moore P. J., Smale D. A., Alexander L. V., et al. (2018). Longer and more frequent marine heatwaves over the past century. Nat. Commun. 9, 1324. doi: 10.1038/s41467-018-03732-9
Pollock F. J., McMinds R., Smith S., Bourne D. G., Willis B. L., Medina M., et al. (2018). Coralassociated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 4921. doi: 10.1038/s41467-018-07275-x
Rai A. K. and Mitchell A. M. (2020). Enterobacterial common antigen: Synthesis and function of an enigmatic molecule. mBio 11, 01914–01920. doi: 10.1128/mbio.01914-20
Reyes G., Betancourt I., Andrade B., Pozo Y., Sorroza L., Trujillo L. E., et al. (2025). Genomic sequence data of thiohalocapsa marina: a sulfur-oxidizing bacterium prevalent in treated municipal wastewater and commercial shrimp hatchery effluents. BMC Res. Notes 18, 97. doi: 10.1186/s13104-025-07162-x
Ribas-Deulofeu L., Loubeyres M., Denis V., De Palmas S., Hwang S.-J., Woo S., et al. (2023). Jeju island: a sentinel for tracking ocean warming impacts on high-latitude benthic communities. Coral Reefs 42, 1097–1112. doi: 10.1007/s00338-023-02400-9
Ryu T., Cho I.-Y., Hwang S.-J., Yum S., Kim M.-S., and Woo S. (2019). First transcriptome assembly of the temperate azooxanthellate octocoral eleutherobia rubra. Mar. Genomics 48, 100682. doi: 10.1016/j.margen.2019.04.007
Sasaki Y. N. and Umeda C. (2021). Rapid warming of sea surface temperature along the kuroshio and the China coast in the east China sea during the twentieth century. J. Climate 34, 4803–4815. doi: 10.1175/JCLI-D-20-0421.1
Sievert C. (2020). Interactive web-based data visualization with R, plotly, and shiny (New York, USA: Chapman and Hall/CRC).
Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Climate Change 9, 306–312. doi: 10.1038/s41558-019-0412-1
Sobha T. R., Vibija C. P., and Fahima P. (2023). Coral Reef: A Hot Spot of Marine Biodiversity (Singapore: Springer Nature Singapore), 171–194. doi: 10.1007/978-981-19-5841-08
Song J.-I. (1976). A study on the classification of the korean anthozoa: 2. alcyonacea. Korean J. zoology 19, 51–62.
Sun W., Anbuchezhian R., and Li Z. (2016). Association of Coral-Microbes, and the Ecological Roles of Microbial Symbionts in Corals (Cham: Springer International Publishing), 347–357. doi: 10.1007/978-3-319-31305-422
Venegas R. M., Acevedo J., and Treml E. A. (2023). Three decades of ocean warming impacts on marine ecosystems: A review and perspective. Deep Sea Res. Part II Top. Stud. Oceanogr 212, 105318. doi: 10.1016/j.dsr2.2023.105318
Verseveldt J. and Bayer F. M. (1988). Revision of the genera bellonella, eleutherobia, nidalia and nidaliopsis (octocorallia: Alcyoniidae and nidalliidae), with descriptions of two new genera. Zoologische Verhandelingen 245, 1–131.
Virtanen P., Gommers R., Oliphant T. E., Haberland M., Reddy T., Cournapeau D., et al. (2020). Scipy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods 17, 261–272. doi: 10.1038/s41592-019-0686-2
Wang Q., Wang Z., Guan J., and Song J. (2024). Transcriptome analysis reveals the important role of vitamin b12 in the response of natronorubrum daqingense to salt stress. Int. J. Mol. Sci. 25, 4168. doi: 10.3390/ijms25084168
Wang Y.-L. and Wu C.-R. (2022). Rapid surface warming of the pacific asian marginal seas since the late 1990s. J. Geophysical Research: Oceans 127, e2022JC018744. doi: 10.1029/2022JC018744
Waskom M. L. (2021). Seaborn: statistical data visualization. J. Open Source Softw 6, 3021. doi: 10.21105/joss.03021
Woo S., Yang S.-H., Chen H.-J., Tseng Y.-F., Hwang S.-J., De Palmas S., et al. (2017). Geographical variations in bacterial communities associated with soft coral scleronephthya gracillimum. PloS One 12, e0183663. doi: 10.1371/journal.pone.0183663
Woo S. and Yum S. (2022). Transcriptional response of the azooxanthellate octocoral scleronephthya gracillimum to seawater acidification and thermal stress. Comp. Biochem. Physiol. Part D: Genomics Proteomics 42, 100978. doi: 10.1016/j.cbd.2022.100978
Wu L., Cai W., Zhang L., Nakamura H., Timmermann A., Joyce T., et al. (2012). Enhanced warming over the global subtropical western boundary currents. Nat. Climate Change 2, 161–166. doi: 10.1038/nclimate1353
Yeh S.-W. and Kim C.-H. (2010). Recent warming in the yellow/east China sea during winter and the associated atmospheric circulation. Continental Shelf Res. 30, 1428–1434. doi: 10.1016/j.csr.2010.05.002
Zaneveld J. R., McMinds R., and Vega Thurber R. (2017). Stress and stability: applying the anna karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121. doi: 10.1038/nmicrobiol.2017.121
Zhang Z., Schwartz S., Wagner L., and Miller W. (2000). A greedy algorithm for aligning dna sequences. J. Comput. Biol. 7, 203–214. doi: 10.1089/10665270050081478
Zhong W. and Agarwal V. (2024). Polymer degrading marine microbulbifer bacteria: an un(der)utilized source of chemical and biocatalytic novelty. Beilstein J. Org Chem. 20, 1635–1651. doi: 10.3762/bjoc.20.146
Zhu W., Wang H., Li X., Liu X., Zhu M., Wang A., et al. (2023). Consistent responses of coral microbiome to acute and chronic heat stress exposures. Mar. Environ. Res. 185, 105900. doi: 10.1016/j.marenvres.2023.105900
Keywords: Eleutherobia rubra, thermal stress response, taxonomic and functional profiling, coral microbiome, coral-microbe interaction
Citation: Kim J-A, Jo Y, Hwang S-J, Oh C, Yang H-S, Lim JK and Woo S (2025) Functional reorganization and taxonomic shifts in the symbiotic microbiota of the temperate soft coral Eleutherobia rubra under heat stress. Front. Mar. Sci. 12:1635356. doi: 10.3389/fmars.2025.1635356
Received: 26 May 2025; Accepted: 05 August 2025;
Published: 29 August 2025.
Edited by:
Xiubao Li, Hainan University, ChinaReviewed by:
Xiaopeng Yu, Guangxi University, ChinaLivia Villela, Federal University of Rio de Janeiro, Brazil
Copyright © 2025 Kim, Jo, Hwang, Oh, Yang, Lim and Woo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae Kyu Lim, ai5rLmxpbUBraW9zdC5hYy5rcg==; Seonock Woo, Y3dvb0BraW9zdC5hYy5rcg==
 Jeong-A Kim
Jeong-A Kim Yejin Jo2
Yejin Jo2 Chulhong Oh
Chulhong Oh Jae Kyu Lim
Jae Kyu Lim Seonock Woo
Seonock Woo