- 1Instituto de Estudos Costeiros (IECOS), Universidade Federal do Pará (UFPA), Bragança, Brazil
- 2Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio)/Resex Ipaú Anilzinho, Tucuruí, Brazil
Introduction: All Sphyrna species occurring along the Brazilian coast are threatened with extinction, largely due to intense harvesting to supply the shark fin and meat trade. This scenario is worsened by the high vulnerability of these species to fishing pressure, resulting from their morphological, behavioral, and physiological characteristics. The lack of effective management and enforcement enables the landing and commercialization of hammerhead sharks without morphological identification, as they are often sold headless and finless. In this context, the use of alternative approaches such as forensic genetics becomes essential for species identification.
Methods: We developed a multiplex PCR protocol using the mitochondrial markers NADH dehydrogenase subunit 2 (NADH2) and tRNA-Trp for the rapid identification of S. mokarran, S. lewini, S. tudes, S. alleni and S. media. All primers were tested in silico and experimentally validated for the target species included in this study.
Results: The species-specific primers produced distinct band profiles: S. mokarran (1,500 bp), S. lewini (1,500 and 400 bp), S. media (3,000, 700, and 400 bp), S. tudes (400 bp), and S. alleni (400 and 300 bp). The target species showed consistent band patterns across most tested concentrations, with 100 ng/µL yielding the best performance for all species. In cross-amplification tests, the protocol also allowed for the distinction of C. limbatus from Sphyrna species.
Discussion: Therefore, the developed multiplex protocol represents a viable and effective tool, and based on its reproducibility, sensitivity, validation, and specificity, we recommend its application as support for enforcement actions and to help combat the illegal trade of threatened hammerhead shark species.
1 Introduction
The family Sphyrnidae comprises two genera: Eusphyra, with a single species, and Sphyrna, currently with nine recognized species (Gonzalez et al., 2024; Jabado et al., 2024). Among the hammerhead sharks of the genus Sphyrna, six occur in the Atlantic Ocean and have already been recorded along the Brazilian coast: Sphyrna mokarran, Sphyrna lewini, Sphyrna tudes, Sphyrna alleni, Sphyrna zygaena, and Sphyrna media (Feitosa et al., 2018; Cruz et al., 2021a; Gonzalez et al., 2024). According to national legislation and international agreements, all these species are classified under some threat category (MMA, 2022; IUCN, 2025; CITES, 2025), except S. alleni, a recently described species (Gonzalez et al., 2024) that has not yet undergone a formal risk assessment. Nevertheless, they are heavily captured by both industrial and artisanal fisheries, either as target species or as bycatch (Stevens et al., 2000; Feitosa et al., 2018; Marceniuk et al., 2019).
This pressure is exacerbated by species-specific traits. For example, the distinctive cephalofoil (hammer-shaped head) makes these sharks more susceptible to entanglement in fishing nets compared to other species (Compagno, 1984). Additionally, their gregarious behavior can lead to mass captures during a single fishing event, while their heightened sensory sensitivity contributes to higher bycatch rates in longline fisheries (Compagno, 1984). These vulnerabilities are further intensified by their low tolerance to post-capture stress (Morgan and Burgess, 2007; Gallagher et al., 2014).
The high demand for fins in the international market and the growing consumption of shark meat, especially in developing countries such as Brazil, further drive their capture (Dent and Clarke, 2015; Martins et al., 2021). In the Brazilian context, the absence of an official monitoring and enforcement system worsens the situation, hindering the control of elasmobranch capture, landing, and trade (Barreto et al., 2016). Existing data are mostly derived from academic research, which indicates the recurrent commercialization of sharks, including Sphyrna, in ports, fairs, and markets along the coast (Feitosa et al., 2018; Guimarães-Costa et al., 2020; Cruz et al., 2021b; Martins et al., 2021; Souza-Araujo et al., 2021).
These animals are often sold under generic names such as “cação”, “panã”, or “cação panã”, and in a discharacterized state—without head and fins—which makes morphological identification unfeasible, as specimens are commercialized as steaks, fillets, or “charutos” (Feitosa et al., 2018; Martins et al., 2021). This discharacterization compromises fishery monitoring and hinders the development of public policies and conservation measures (Feitosa et al., 2018; Martins et al., 2021).
Given this context, morphological identification proves ineffective, and alternative methods such as forensic genetics become essential (Feitosa et al., 2018; Martins et al., 2021). Molecular tools based on mitochondrial gene sequencing have been widely used for elasmobranch identification (Ward et al., 2005; Steinke et al., 2017; Feitosa et al., 2018). Although the COI gene (Cytochrome C oxidase subunit 1) and the ITS (Internal Transcribed Spacer) region are broadly applied in forensic studies and multiplex PCR protocols (Caballero et al., 2012; Nachtigall et al., 2017; Cardeñosa et al., 2018), other markers may offer specific advantages. The NADH2 gene (NADH dehydrogenase subunit 2) shows conserved intraspecific regions and high interspecific variability, making it suitable for studies involving sharks of the order Carcharhiniformes (Díaz-Jaimes et al., 2016). Its higher mutation rate provides greater phylogenetic resolution, with intraspecific divergences comparable to those of COI, but with improved discrimination among species of the same genus (Naylor et al., 2005, 2012).
More recently, rapid and low-cost approaches such as multiplex PCR protocols have gained prominence for enabling species-specific identification without the need for sequencing (Barbosa et al., 2021; Lutz et al., 2023). This technique allows the simultaneous amplification of different DNA targets in a single reaction, with identification based on band patterns obtained through agarose gel electrophoresis (Ali et al., 2014). Among its advantages are lower cost, speed, and applicability in enforcement contexts (Lutz et al., 2023).
Multiplex PCR has also proven highly effective in detecting commercial fraud across various fish families, particularly those with high market value (Barbosa et al., 2021; Lutz et al., 2023). In the case of hammerhead sharks, it has been used to identify threatened species present in landings and trade (Abercrombie et al., 2005; Sodré et al., 2024).
In this context, the present study developed a multiplex PCR protocol aimed at the rapid and specific identification of the five hammerhead shark species recorded along the Brazilian coast: S. mokarran, S. lewini, S. tudes, S. alleni and S. media (Gonzalez et al., 2024; Jabado et al., 2024). The implementation of this protocol may strengthen enforcement and conservation efforts by promoting greater control over the trade and landing of threatened species and facilitating the application of current environmental legislation.
2 Materials and methods
2.1 Ethics statement
The samples used for the development of the present study were collected from landing ports, markets, and fish fairs, with all individuals acquired already dead. Therefore, no permits were required for the collection of live organisms, nor were ethical procedures for euthanasia or prior authorization from the Animal Ethics Committee necessary.
2.2 Sampling, DNA isolation, amplification, and sequencing
A total of 77 samples were used. Of these, 50 were previously identified using the COI gene by Martins et al. (2021) (Figure 1A), and 27 were identified using the NADH2 gene in the present study (Figure 1B). Among the total, 32 Sphyrna samples were used in the replicability test; 26 shark and ray samples in the cross-amplification test; and 21 samples in the validation test. The complete composition of the tests — including sample codes, common names, species, tests, and origin - is detailed in Supplementary Material 1. Despite sampling efforts, only two individuals of S. media were collected, which explains the limited number of samples used in the tests.
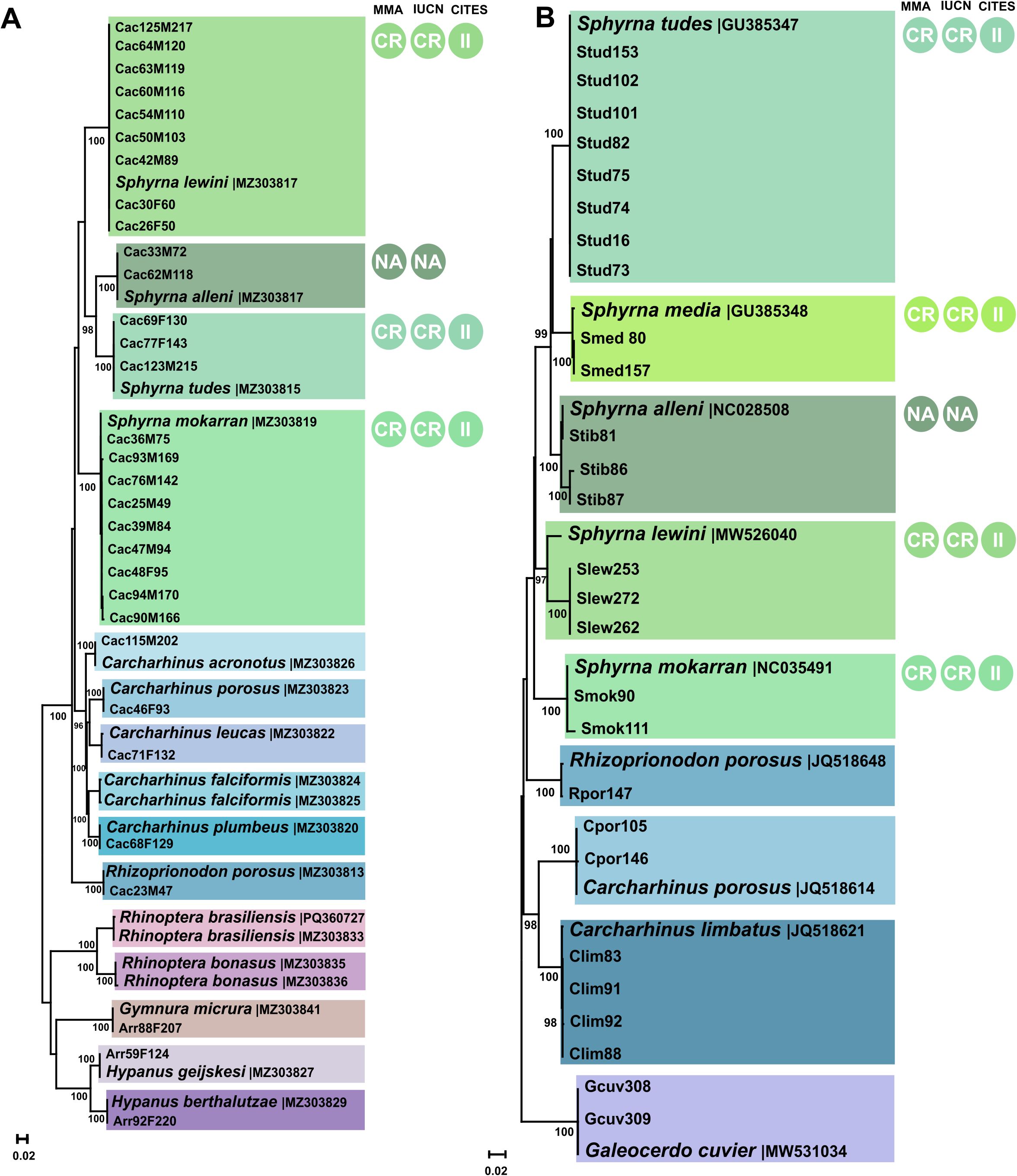
Figure 1. Neighbor-Joining trees constructed using the K2P model. Bootstrap support values are shown on branches, based on 1,000 pseudoreplicates. The resulting clusters indicate the species used in the multiplex protocol tests. (A) Samples and sequences from Martins et al. (2021). (B) Samples and sequences obtained in the present study for validation testing. Circles indicate the conservation status of each species according to the MMA (2022) and IUCN (2025), with the following categories: NA – Not Assessed; CR – Critically Endangered. Species listed in the CITES (2025) appendices are also indicated, with II – Appendix II.
The fillets collected in this study were sanitized with 98% alcohol, and muscle tissue portions were stored in microtubes with 70% alcohol at −18°C. DNA extraction followed the saline (NaCl) protocol of Aljanabi and Martinez (1997), adapted to the laboratory. DNA concentration and purity were assessed using a NanoDrop Lite Plus (Thermo Scientific™), and integrity was verified on a 1% agarose gel.
Amplification of the mitochondrial NADH2 gene was performed using the primers Ilem-Mustelus and Asn-Mustelus (Naylor et al., 2005), in a final reaction volume of 15 µL: 2.4 µL of dNTPs (1.25 mM); 1.5 µL of buffer (10×); 0.6 µL of MgCl2 (50 mM); 0.4 µL of each primer (10 ng); 0.15 µL of BSA (5 mg/mL); 0.3 µL of DNA; 0.2 µL of Taq polymerase (5 U/µL); and ultrapure water. PCR conditions were: initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 30 s, annealing at 48°C for 45 s, extension at 72°C for 1 min; and a final extension at 72°C for 7 min.
Positive PCR products were purified with PEG according to Paithankar and Prasad (1991) and subjected to electrophoresis using an ABI 3500 capillary sequencer (Thermo Fisher), with unidirectional sequencing of the forward strand (5’) using the Ilem-Mustelus primer.
All samples are available at the Laboratório de Genética Aplicada (LAGA), Instituto de Estudos Costeiros (IECOS), Universidade Federal do Pará – Campus Bragança.
2.3 Dataset and analyses
NADH2 sequences obtained in this study were edited using BioEdit v. 7.1.3.0 (Hall, 1999) and automatically aligned with the CLUSTAL W application (Thompson et al., 1994). NADH2 haplotypes were identified using DnaSP (Librado and Rozas, 2009) to guide sample identification.
Species identification was based on genetic similarity with reference sequences from GenBank (NCBI, http://www.ncbi.nlm.nih.gov), adopting a threshold of 98%–100% similarity (Hebert et al., 2003).
COI and NADH2 sequences were organized into separate databases according to the marker. The COI database, previously identified by Martins et al. (2021), was considered the reference. Reference sequences from NCBI were added to the NADH2 database. For both databases, Neighbor-Joining (NJ) trees were constructed using the Kimura 2-parameter substitution model (Kimura, 1980) in MEGA 11 software (Tamura et al., 2021), with statistical support estimated by bootstrap (1000 pseudoreplicates; Felsenstein (1985). The absence of stop codons was verified using the same software.
The conservation status of hammerhead shark species was consulted according to national legislation (MMA Ordinance No. 148/2022), the International Union for Conservation of Nature Red List (IUCN), and the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) agreement.
2.4 Primer development
The NADH2 gene was selected due to the presence of forensic informative sites for Carcharhiniformes (Díaz-Jaimes et al., 2016), with the target species of the protocol being S. mokarran, S. lewini, S. tudes, S. alleni and S. media. For primer design, sequences available in GenBank were used, prioritizing distinct haplotypes within each species to adequately represent intraspecific variability. This approach allowed the identification of conserved intraspecific and divergent interspecific regions, which are essential for the development of species-specific primers.
The sequences used were: S. mokarran (NC035491, JQ518692, MT881534, MT881535, MT881536), S. lewini (NC022679, MW526040, MW526041, OM165180, OM165184, OM165181), S. tudes (GU385347, JQ518690), S. alleni (NC028508, OM165196, OM165197, JQ518693) and S. media (GU385348, OM165195).
Primer design was performed in the portion of the NADH dehydrogenase 2 gene and the translation of leader peptide (tRNA-Trp) (Supplementary Figure 1). The FASTA alignment of these sequences was analyzed using BioEdit 7.1.3.0 software (Hall, 1999), focusing on the identification of species-specific polymorphisms at the 3’ end of the primers.
Seven primers were developed: Smok1, Slew1, Slew2, Stud1, Stib1, Smed1 and Reverse1. Primer quality was evaluated using the Multiple Primer Analyzer software (Thermo Fisher Scientific), considering melting temperature (Tm), self-complementarity, GC content at the 3’ end, and dimer formation. The primers that showed the best performance and were used in the multiplex PCR are available in Table 1 and Supplementary Figure 1. The protocol is currently undergoing the patenting process, and detailed information on reagent concentrations will be made available upon legal authorization. The method is intended to be applied in collaboration with enforcement agencies.
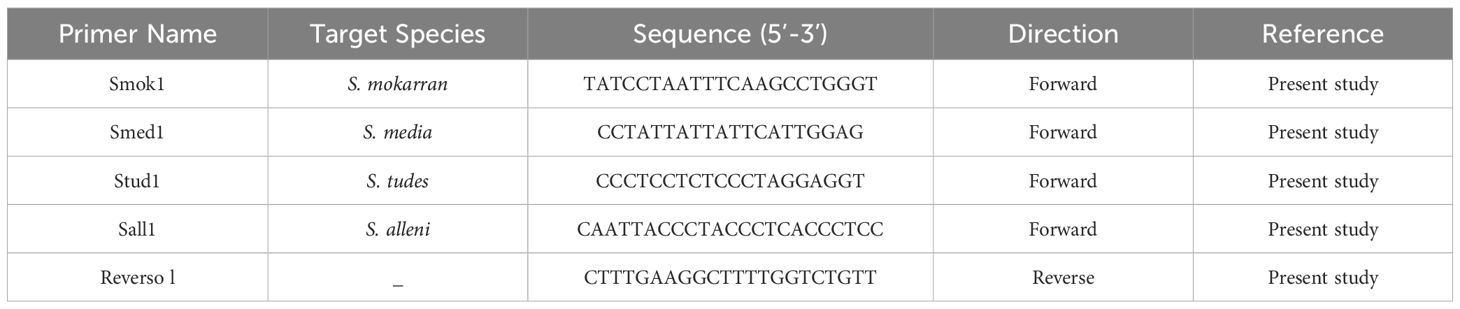
Table 1. Details of the primers developed in the present study for the identification of hammerhead shark species (genus Sphyrna).
2.5 Specificity and replicability tests
With primers validated in silico and assessed for quality, experimental tests of specificity and replicability were conducted as described below.
A database was assembled with 32 tissue samples from genetically identified sharks using the COI and NADH2 genes, representing the following species: S. mokarran (N = 10), S. lewini (N = 10), S. tudes (N = 5), S. alleni (N = 5), and S. media (N = 2).
For the specificity test, two individuals of each species were selected. Each species-specific primer was initially tested on its target species and then on the other species of the genus to verify the absence of cross-amplification.
Replicability tests included a larger number of individuals per species, limited by sample availability in the Applied Genetics Laboratory database. S. mokarran and S. lewini had greater representation (N = 10) due to the higher availability of tissue samples. Samples of S. tudes and S. alleni were supplemented with individuals collected in this study, totaling five individuals each. The species S. media was only recorded in the current sampling, with a maximum of two individuals. Since standardizing the number of samples would require reducing all species to the lowest available N (i.e., S. media, with N = 2), the tests were performed using the total available samples per species.
After the initial tests, a temperature gradient test was conducted to determine the optimal hybridization temperature. Each primer pair (species-specific forward + universal reverse) was tested at temperatures ranging from 48°C to 70°C. Based on the results, the hybridization temperature was standardized at 52.4°C, as it provided the best amplification results for all species.
2.6 Multiplex PCR
After determining the optimal annealing temperature for all primers, multiplex PCR amplification was performed using only the primers previously validated as positive, maintaining the parameters used in the single PCR. Two individuals of each species were used for the tests. After several assay rounds, the most efficient reaction setup was selected. Primers 16S-L2949 and 16S-H3058 were included as positive controls for the samples (Kartavtsev et al., 2007). The multiplex PCR reaction was standardized in a final volume of 15 µL, containing: 2.4 µL of dNTPs (1.25 mM); 1.5 µL of buffer (10×); 0.6 µL of MgCl2 (50 mM); 0.4 to 0.15 µL (10 pmol) of each of the five species-specific primers; 0.12 µL (10 pmol) of each positive control primer; 0.15 µL of BSA (5 mg/mL); 0.4 µL of DNA (100 ng/µL); 0.3 µL of Taq DNA polymerase (5 U/µL); and ultrapure water to complete the volume. Amplification conditions were: initial denaturation at 94°C for 3 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 52.4°C for 45 seconds, and extension at 72°C for 1 minute; with a final extension at 72°C for 10 minutes. Band profiles were visualized by electrophoretic migration during 1 hour and 40 minutes on a 2.5% agarose gel, using 5 µL of the PCR product mixed with 5 µL of loading buffer (Blue Juice) containing GelRed.
2.7 Sensitivity test
The sensitivity of the multiplex PCR protocol was evaluated using different DNA concentrations, as described by Lutz et al. (2023). Concentrations of 100 ng/μL, 50 ng/μL, 20 ng/μL, 10 ng/μL, and 1 ng/μL were tested for five species from the family Sphyrnidae and one species from the family Carcharhinidae, aiming to assess the detection capability and the minimum threshold required to obtain species-specific banding profiles.
2.8 Multiplex PCR validation and cross-amplification
To validate the specificity of the banding profiles obtained, 10 individuals of S. mokarran and S. lewini, five individuals of S. tudes and S. alleni, and two individuals of S. media were used. To test the protocol’s specificity and the absence of cross-amplification, 26 samples belonging to the following species were included: Rhizoprionodon porosus, Carcharhinus porosus, Carcharhinus acronotus, Carcharhinus leucas, Carcharhinus falciformis, Carcharhinus plumbeus, Carcharhinus limbatus, Galeocerdo cuvier, Gymnura micrura, Rhinoptera brasiliensis, Rhinoptera bonasus, Hypanus geijskesi, and Hypanus berthalutzae, with two individuals per species. In these assays, a DNA concentration of 100 ng/μL, previously determined as optimal, was used, maintaining the amplification parameters.
3 Results
3.1 Multiplex protocol for hammerhead sharks (Sphyrna)
Following the single PCR tests, all primers that yielded consistent banding patterns were selected for the multiplex PCR. Among them, the primer Slew2, although it amplified in single PCR, did not produce a banding profile in the multiplex format and was therefore excluded from the final stage of the protocol. The remaining primers performed well, generating species-specific profiles with single or multiple bands, which enabled the unequivocal identification of the analyzed species. All individuals amplified the 200 base pair (bp) control band, corresponding to the 16S rRNA gene.
The fragments amplified with the primers designed for the NADH2 gene allowed for species differentiation based on the following banding profiles: S. mokarran, a single 1,500 bp band; S. lewini, two bands of 1,500 bp and 400 bp; S. media, three bands of 3,000 bp, 700 bp, and 400 bp; S. tudes, a single 400 bp band; and S. alleni, two bands of 400 bp and 300 bp (Figure 2).
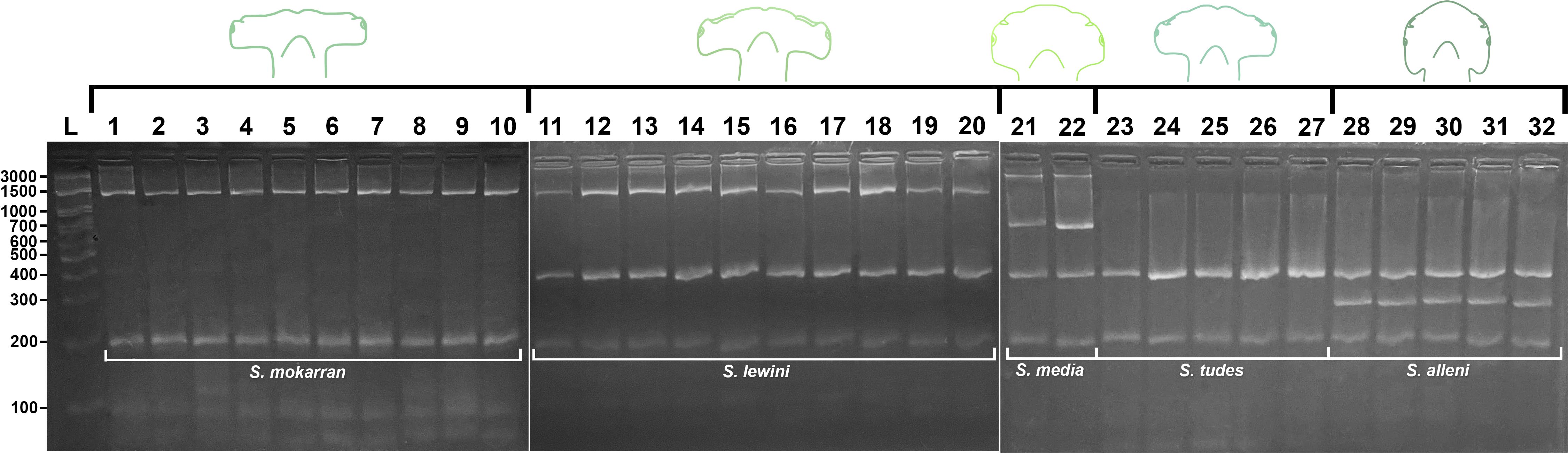
Figure 2. Species-specific band profiles obtained by multiplex PCR for Sphyrna species distributed along the Brazilian coast. L: 100 bp DNA ladder; lanes 1–10: S. mokarran (1,500 bp); lanes 11–20: S. lewini (1,500 and 400 bp); lanes 21–22: S. media (3,000, 700, and 400 bp); lanes 23–27: S. tudes (400 bp); lanes 28–32: S. alleni (400 and 300 bp). A 200 bp control band is present in all species.
3.2 Influence of DNA concentration on specificity and band visualization
In the sensitivity test, all target species exhibited the expected specific banding profiles at all concentrations, except for S. media, which at 50 ng/µL showed a pattern identical to that of C. limbatus at 100 ng/µL. It was also observed that C. limbatus, between 50 and 10 ng/µL, presented a profile similar to that of S. tudes, becoming indistinguishable from the latter at a concentration of 1 ng/µL (Figure 3).
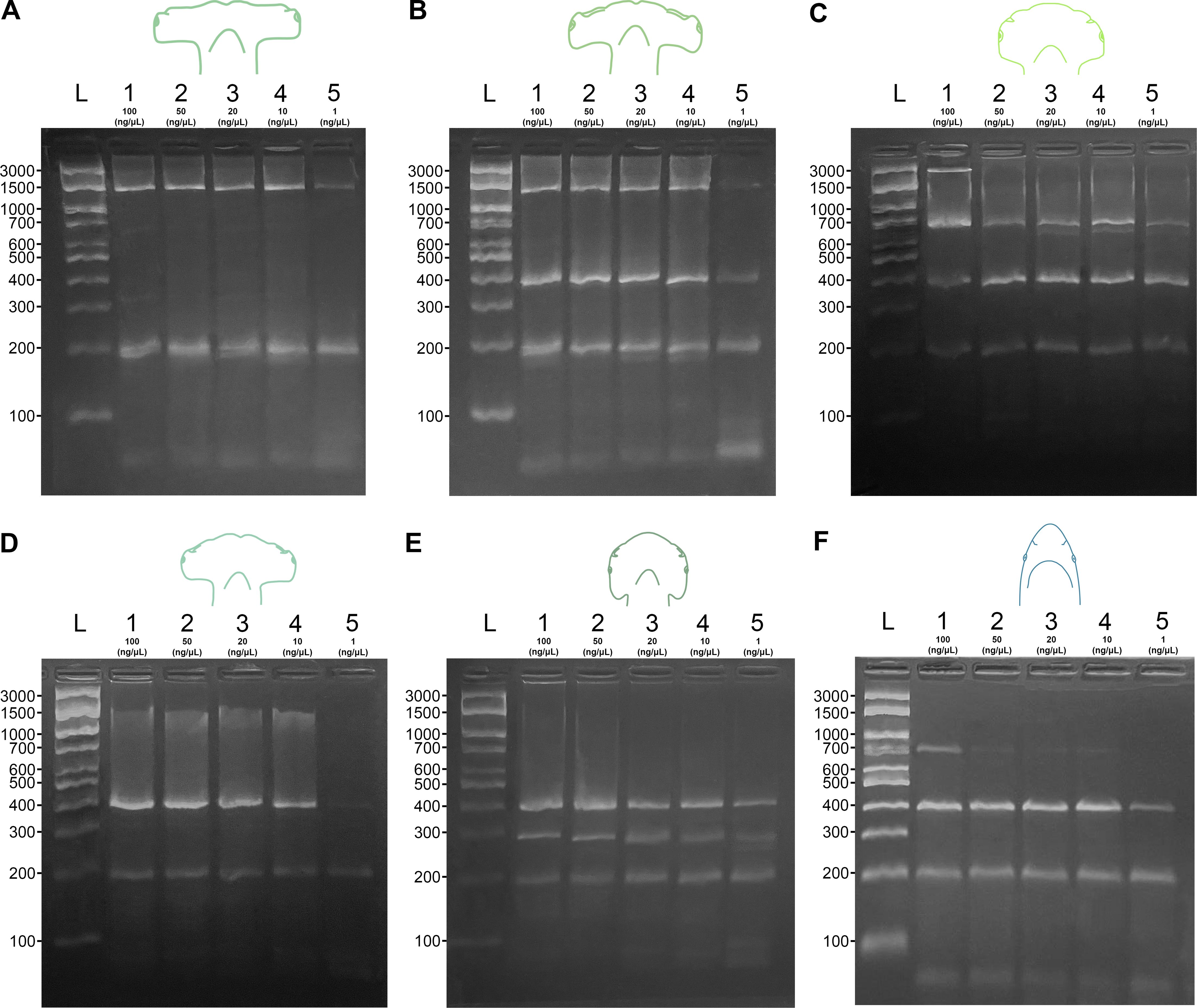
Figure 3. Sensitivity test of the multiplex PCR protocol using Sphyrna DNA at concentrations of 100 ng/µL, 50 ng/µL, 20 ng/µL, 10 ng/µL, and 1 ng/µL. The test was applied to all target species: (A) S. mokarran, (B) S. lewini, (C) S. media, (D) S. tudes, (E) S. alleni, and to the non-target species (F) C limbatus. L: 100 bp DNA ladder.
Additionally, the banding profile of S. media varied according to concentration. At 100 ng/µL, bands of approximately 3,000 bp, 700 bp, and 400 bp were observed; at concentrations of 20, 10, and 1 ng/µL, the pattern changed to 700 bp, ~690 bp, and 400 bp. The 3,000 bp and ~690 bp bands were attributed to nonspecific amplification (Figure 3).
At a concentration of 1 ng/µL, some bands became difficult to visualize, whereas at 100 ng/µL the results were more consistent for all species. The control band was present in all species and concentrations (Figure 3).
3.3 Cross-amplification tests for non-target elasmobranchs
In the cross-amplification tests, only the control band (200 bp) was generally observed in the non-target species. Exceptions were recorded for C. limbatus, which exhibited bands of 700 bp and 400 bp, and for R. brasiliensis, which showed two distinct profiles: ~600 bp, ~630 bp, 400 bp and ~600 bp, 400 bp, 300 bp (Figure 4). Despite the presence of nonspecific bands in these species, the observed patterns clearly differed from those of the target Sphyrna species (Figure 2).
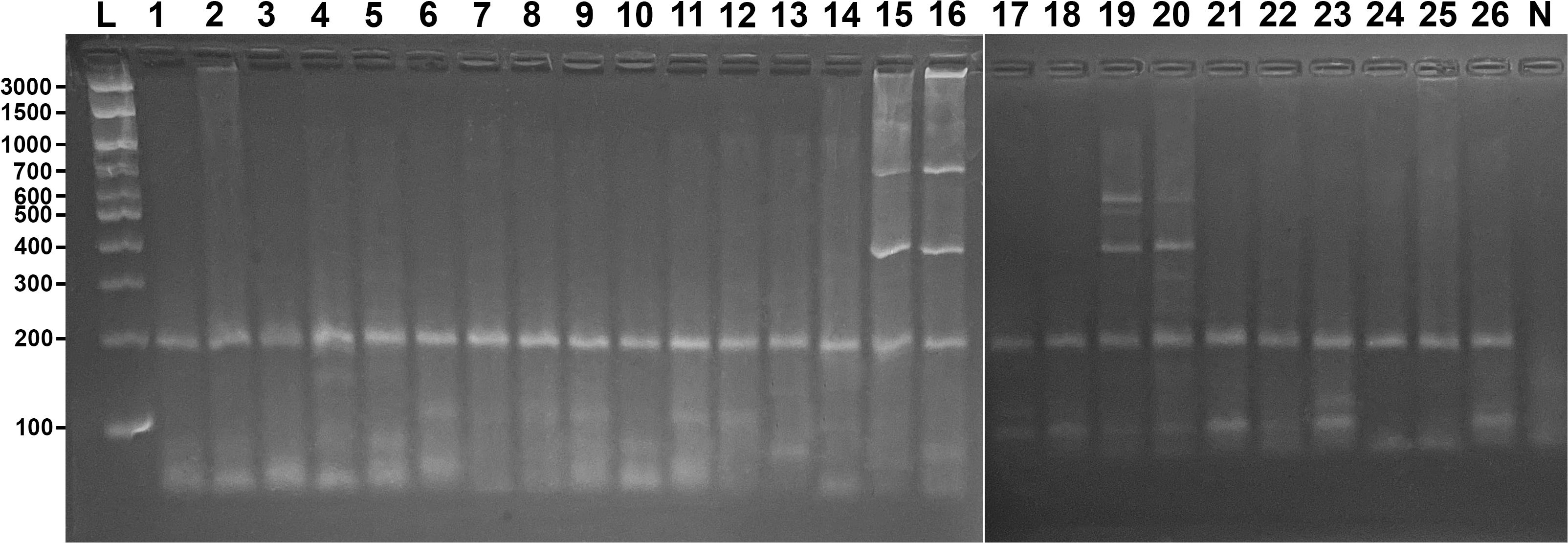
Figure 4. Cross-amplification test using non-target species to assess the specificity of the multiplex PCR protocol developed for Sphyrna species. L: 100 bp DNA ladder; lanes 1–2: R. porosus; 3–4: C. porosus; 5–6: C. acronotus; 7–8: C. leucas; 9–10: C. falciformis; 11–12: C. plumbeus; 13–14: G. cuvier; 15–16: C. limbatus; 17–18: G. micrura; 19–20: R. brasiliensis; 21–22: R. bonasus; 23–24: H. geijskesi; 25–26: H. berthalutzae; lane 27: negative control. A 200 bp control band was observed in all species.
A total of 26 non-target elasmobranch samples were tested, including eight from shark species and five from ray species (Figure 4).
3.4 Multiplex validation for Sphyrna identification
The validation of the multiplex protocol was carried out using samples from individuals marketed under generic names that could correspond to species of the genus Sphyrna, such as “cação”, “panã”, “cação panã”, “cação panã roxa”, “cação panã branca”, “panã amarela”, “cação sacuri”, and “sacuri”. All analyzed samples exhibited species-specific banding profiles consistent with the expected patterns for the target species: S. mokarran (1,500 bp), S. lewini (1,500 and 400 bp), S. media (3,000, 700, and 400 bp), S. tudes (400 bp), and S. alleni (400 and 300 bp). A reproducible pattern was also observed for the non-target species C. limbatus (700 and 400 bp). Other non-target species amplified only the control band, as observed for R. porosus and C. porosus (200 bp) (Figure 5).
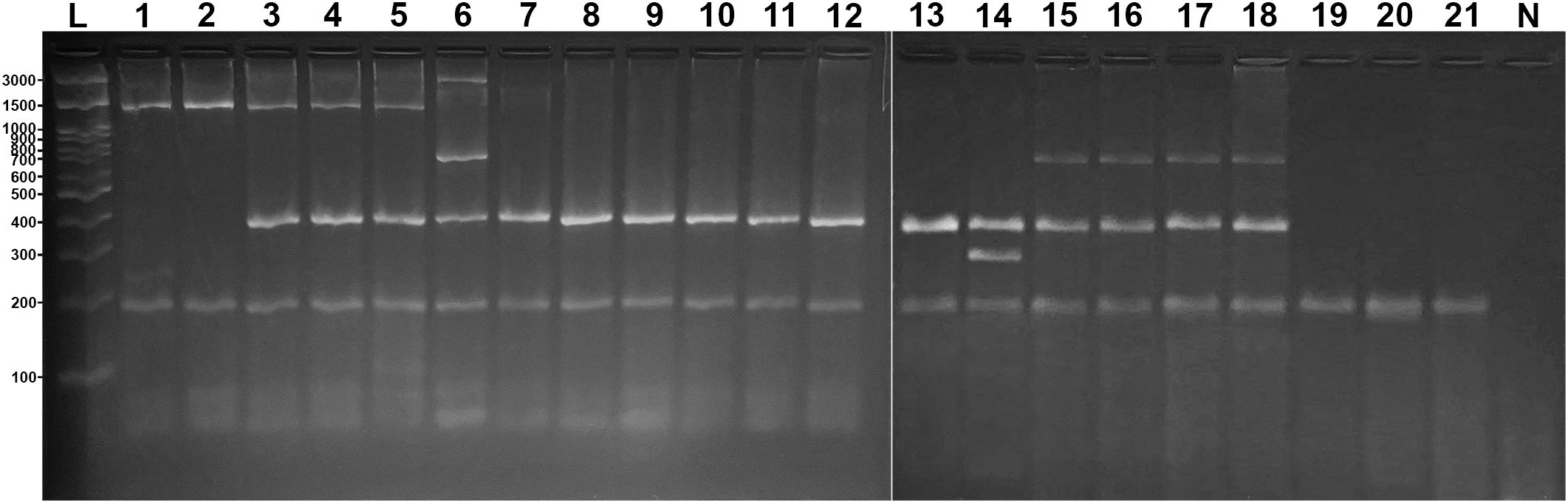
Figure 5. Validation test using samples from sharks identified by commercial names corresponding to Sphyrna species. Lanes 1–2: S. mokarran; 3–5: S. lewini; 6: S. media; 7–13: S. tudes; 14: S. alleni; 15–18: C. limbatus; 19: R. porosus; 20–21: C. porosus; lane 22: negative control. A 200 bp control band was present in all samples.
4 Discussion
4.1 Molecular diagnosis in the monitoring of elasmobranch trade
Investigating the trade of threatened sharks is essential, and in this context, the use of molecular techniques has proven to be more effective than morphological identification, especially considering that sharks are often marketed in a visually altered form — in cuts such as “charutos” (cigar-shaped), fillets, or steaks (Feitosa et al., 2018). Although Interministerial Normative Instruction No. 14 (ICMBio, 2012) prohibits the landing of sharks without fins attached to the body, it is common for individuals to be landed without heads and fins (Personal observation), which makes species-level morphological identification unfeasible (Feitosa et al., 2018; Martins et al., 2021).
The use of generic names such as “cação” or “panã” further increases taxonomic uncertainty, reinforcing the importance of genetic methodologies as an essential tool for accurate identification (Feitosa et al., 2018; Cruz et al., 2021b; Martins et al., 2021).
Several studies have already employed molecular tools to identify elasmobranchs at the species level, many of which are threatened with extinction. Feitosa et al. (2018), using the COI and NADH2 markers, identified 17 species at landing ports, 13 of which are threatened. Among them, S. tudes, S. tiburo, S. mokarran, and S. lewini stood out, with the latter two being the most frequently recorded. A similar result was reported by Martins et al. (2021), who identified 20 taxa being traded. Of these, 17 are under some degree of threat, with S. mokarran and S. lewini again among the most frequently recorded. This scenario is also observed in southeastern Brazil, where Cruz et al. (2021a) identified, using the COI gene, nine traded species, more than half of which are threatened with extinction, including S. zygaena, which was recorded as the second most abundant species in the trade.
These studies highlight the effectiveness of genetic tools in elucidating the species-level identity of sharks, especially when morphological methods are unfeasible. However, most of these analyses rely on DNA sequencing—a robust technique, but one that is more costly, time-consuming, and dependent on advanced laboratory infrastructure (Lutz et al., 2023). In this context, alternative approaches such as PCR emerge as faster, more cost-effective, and accessible solutions (Barbosa et al., 2021; Lutz et al., 2023), particularly for large-scale applications such as environmental enforcement actions.
Based on this premise, the multiplex PCR protocol developed in this study proved to be an efficient tool for the molecular identification of five Sphyrnidae species (S. mokarran, S. lewini, S. tudes, S. alleni, and S. media) and one Carcharhinidae species (C. limbatus), through species-specific amplification profiles. Each taxon exhibited a unique gel banding pattern, enabling direct visual distinction, in addition to consistent amplification of the control band (16S gene, ~200 bp) in all individuals tested. This approach has already been successfully applied to several taxonomic groups (Abercrombie et al., 2005; Castigliego et al., 2015; Barbosa et al., 2021; Lutz et al., 2023).
The functionality of the protocol depends on the careful optimization of PCR parameters, such as DNA and reagent concentrations, as well as the characteristics of the agarose gel and the composition of the running buffer (Castigliego et al., 2015). The sensitivity of the protocol was evaluated, and a DNA concentration of 100 ng/µL proved to be the most effective for ensuring specific band amplification for all tested species, particularly C. limbatus, S. tudes, and S. media, whose banding patterns changed at lower concentrations and could even overlap.
Cross-amplification tests revealed unexpected patterns in C. limbatus and R. brasiliensis. For the latter, two individuals exhibited distinct profiles, which may be related to primer non-specificity, hybridization temperature, or inherent characteristics of the multiplex reaction itself, as reported by Castigliego et al. (2015) and Lutz et al. (2023).
In the case of S. media, although the banding pattern was consistent between the two individuals analyzed, the small sample size limits the assessment of the protocol’s replicability for this species, highlighting the need for future studies with a larger sample size to ensure more robust validation.
These findings reinforce the importance of strict standardization and continuous validation of the protocol, especially considering its practical application in the monitoring and control of elasmobranch trade.
4.2 Practical applications and distinctive features of the developed protocol
The primers developed exhibited species-specific profiles: S. mokarran and S. tudes showed single-band patterns, S. media presented a triple pattern, and S. lewini and S. alleni exhibited double patterns. This variation can be explained by the complexity of multiplex PCR reactions, which often generate profiles different from those observed in single PCRs (Lutz et al., 2023), as well as the difficulty in designing highly specific primers for phylogenetically close species (Lim et al., 2010).
Previous studies, such as that of Sodré et al. (2024), also reported specific profiles for S. mokarran and S. lewini using the ITS2 region of nuclear rDNA. However, in that case, it was not possible to differentiate S. tudes from S. tiburo, as both exhibited the same amplification pattern. Protocols developed for use in Asian markets (Abercrombie et al., 2005) and along the Mexican coast (Aguilar-Rendón et al., 2020) were focused on a limited number of species, such as S. lewini, S. mokarran, and S. zygaena.
The protocol developed in this study represents a significant advancement by enabling the simultaneous identification of five Sphyrna species—a higher number than previously reported—along with one Carcharhinus species. These results indicate broader applicability and adaptability to the conditions of the Brazilian coastline.
Based on replicability, sensitivity, cross-amplification, and validation tests, the protocol demonstrated robustness and reliability, proving feasible for implementation by environmental and fisheries enforcement agencies such as Brazilian Institute of Environment and Renewable Natural Resources (IBAMA), the Special Secretariat for Aquaculture and Fisheries (SAP), Chico Mendes Institute for Biodiversity Conservation (ICMBio), and state environmental agencies. Its application can support efforts to combat the illegal trade of threatened species and contribute to fisheries monitoring and management actions.
4.3 Diagnostic tools and conservation of threatened Sphyrna species
The global decline of elasmobranch populations has been widely attributed to overfishing, as evidenced by numerous studies. Pacoureau et al. (2021) estimated a 70.1% reduction in the global abundance of sharks and rays over the past five decades, primarily driven by an 18-fold increase in fishing effort, which has contributed to the listing of more than half of elasmobranch species in threatened categories. According to the IUCN (2025), 37% of these species are threatened with extinction, a proportion also reflected in Brazil’s National List of Threatened Species (MMA, 2022) and in the CITES (2025).
Within the hammerhead shark group (family Sphyrnidae), the situation is particularly critical (IUCN, 2025; Jabado et al., 2024). All assessed species of the genus Sphyrna are categorized as threatened on both national and international lists (MMA, 2022; IUCN, 2025), due to widespread targeted and incidental capture linked to the trade in meat and fins (Stevens et al., 2000; Dent and Clarke, 2015; Feitosa et al., 2018; Marceniuk et al., 2019). Brazil is among the main shark-catching countries, standing out for both meat imports and local fishing activity (Dent and Clarke, 2015).
Despite the severity of the situation, the lack of updated statistical data and effective tracking systems hinders the implementation of more precise management measures (Barreto et al., 2016). Regional reports indicate that shark meat remains widely available in local markets and fairs, suggesting that exploitation continues at a large scale (Bornatowski and Abilhoa, 2012; Guimarães-Costa et al., 2020; Souza-Araujo et al., 2021).
Given the intense pressure on threatened species and the lack of effective mechanisms for tracking traded specimens, investing in diagnostic technologies that combine accuracy, speed, and applicability becomes strategic. Molecular methods that do not require sequencing, such as the multiplex PCR protocol developed in this study, represent a significant advancement in this regard. In addition to lower costs and shorter analysis times (Barbosa et al., 2021; Lutz et al., 2023), this approach enables the rapid screening of protected species and is particularly useful in contexts of illegal trade enforcement (Abercrombie et al., 2005; Aguilar-Rendón et al., 2020). By enabling the reliable differentiation of five Sphyrna species, the protocol developed here adds a valuable molecular tool to the arsenal of regulatory and environmental control bodies. Its cost-effectiveness and diagnostic precision make it especially suitable for adoption by laboratories linked to environmental and fisheries oversight, including IBAMA, SAP/MAPA, and state agencies. In this way, it offers concrete potential to enhance species identification workflows, strengthen enforcement against illegal trade, and improve conservation outcomes for threatened elasmobranchs.
Finally, we recommend the inclusion of the species-specific primer for S. zygaena in regions where this species has already been recorded, such as the southeastern coast of Brazil (Cruz et al., 2021a).
5 Conclusion
A multiplex PCR protocol was developed for the identification of Sphyrna species exploited along the Brazilian coast. The method enables reliable detection of five hammerhead shark species (S. mokarran, S. lewini, S. tudes, S. alleni, and S. media) and the species C. limbatus, each displaying distinct amplification profiles. This technique is efficient, rapid, and highly accurate, making it suitable for implementation by environmental enforcement agencies. Its use may support efforts to curb the illegal trade of threatened species and facilitate more effective fisheries monitoring and management actions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PQ522203–PQ522229.
Ethics statement
The requirement of ethical approval was waived by Ethics Committee CEUA Of the Federal University of Pará for the studies involving animals because It was not necessary to request approval from the Ethics Committee because all specimens were obtained from the trade. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
TM: Formal analysis, Methodology, Conceptualization, Writing – original draft, Writing – review & editing. ÍL: Data curation, Formal analysis, Writing – original draft, Conceptualization, Methodology. RD: Writing – original draft, Formal analysis, Methodology. GM: Writing – review & editing, Methodology, Software. PS: Writing – review & editing, Formal analysis, Methodology. NS: Methodology, Formal analysis, Writing – review & editing. MV: Writing – original draft, Funding acquisition, Resources. IS: Supervision, Project administration, Writing – original draft, Resources. GE-G: Supervision, Conceptualization, Writing – review & editing, Validation, Writing – original draft, Investigation, Visualization, Funding acquisition, Resources, Formal analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), (CAPES, grant number: 88887.494609/2020-00), to CNPq for financing this study (Universal Project #439113/2018-0).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1638479/full#supplementary-material
References
Abercrombie D. L., Clarke S. C., and Shivji M. S. (2005). Global-scale genetic identification of hammerhead sharks: Application to assessment of the international fin trade and law enforcement. Conserv. Genet. 6, 775–788. doi: 10.1007/s10592-005-9036-2
Aguilar-Rendón T. A., Rendón-Herrera J. J., Osuna-González V., Oñate González E. C., Domínguez-Domínguez O., and Saavedra-Sotelo N. C. (2020). Molecular identification of hammerhead shark trunks from the southern Gulf of California using multiplex PCR. J. Fisheries Sci. 2, 22–29. doi: 10.30564/jfsr.v2i1.1685
Ali M., Razzak A., and Hamid S. B. A. (2014). Multiplex PCR in species authentication: probability and prospects—A review. Food Anal. Methods 7, 1933–1949. doi: 10.1007/s12161-014-9844-4
Aljanabi S. and Martinez I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Barbosa A. J., Sampaio I., and Santos S. (2021). Re-visiting the occurrence of mislabeling in frozen “pescada-branca” (Cynoscion leiarchus and Plagioscion squamosissimus – Sciaenidae) sold in Brazil using DNA barcoding and octaplex PCR assay. Food Res. Int. 143, 110308. doi: 10.1016/j.foodres.2021.110308
Barreto R., Ferretti F., Flemming J. M., Amorim A., Andrade H., Worm B., et al. (2016). Trends in the exploitation of South Atlantic shark populations. Conserv. Biol. 30, 792–804. doi: 10.1111/cobi.12663
Bornatowski H. and Abilhoa V. (2012). Tubarões e raias capturados pela pesca artesanal no Paraná: guia de identificação (Curitiba: Hori Consultoria Ambiental).
Caballero S., Cardenosa D., Soler G., and Hyde J. (2012). Application of multiplex PCR approaches for shark molecular identification: feasibility and applications for fisheries management and conservation in the Eastern Tropical Pacific. Mol. Ecol. Resour 12, 233–237. doi: 10.1111/j.1755-0998.2011.03089.x
Cardeñosa D., Quinlan J., Shea K. H., and Chapman D. D. (2018). Multiplex real-time PCR assay to detect illegal trade of CITES-listed shark species. Sci. Rep. 8, 16313. doi: 10.1038/s41598-018-34663-6
Castigliego L., Armani A., Tinacci L., Gianfaldoni D., and Guidi A. (2015). Two alternative multiplex PCRs for the identification of the seven species of anglerfish (Lophius spp.) using an end-point or a melting curve analysis real-time protocol. Food Chem. 166, 1–9. doi: 10.1016/j.foodchem.2014.06.014
CITES. (2025). Checklist of CITES species. Available online at: http://checklist.cites.org/ (Accessed May 25, 2025).
Compagno L. J. V. (1984). FAO species catalogue. Vol. 4. Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 2 (Rome: FAO Fisheries Synopsis No. 125).
Cruz V. P. da, Adachi A. M. C. d., Ribeiro G. da S., Oliveira P. H. de, Oliveira C., Oriano Junior R., et al. (2021a). A shot in the dark for conservation: Evidence of illegal commerce in endemic and threatened species of elasmobranch at a public fish market in southern Brazil. Aquat Conserv. 31, 1650–1659. doi: 10.1002/aqc.3572
Cruz M. M., Szynwelski B. E., and Ochotorena de Freitas T. R. (2021b). Biodiversity on sale: The shark meat market threatens elasmobranchs in Brazil. Aquat Conserv. 31, 3437–3450. doi: 10.1002/aqc.3710
Dent F. and Clarke S. (2015). State of the global market for shark products. FAO Fisheries Aquaculture Tech. paper. 590, 1–196.
Díaz-Jaimes P., Bayona-Vásquez N. J., Adams D. H., and Uribe-Alcocer M. (2016). Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta Gene 7, 48–55. doi: 10.1016/j.mgene.2015.11.005
Feitosa L. M., Martins A. P. B., Giarrizzo T., Macedo W., Monteiro I. L., Gemaque R., et al. (2018). DNA-based identification reveals illegal trade of threatened shark species in a global elasmobranch conservation hotspot. Sci. Rep. 8, 3347. doi: 10.1038/s41598-018-21683-5
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evol. (N Y) 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gallagher A., Serafy J., Cooke S., and Hammerschlag N. (2014). Physiological stress response, reflex impairment, and survival of five sympatric shark species following experimental capture and release. Mar. Ecol. Prog. Ser. 496, 207–218. doi: 10.3354/meps10490
Gonzalez C., Postaire B., Driggers W., Caballero S., and Chapman D. (2024). Sphyrna alleni sp. nov., a new hammerhead shark (Carcharhiniformes, Sphyrnidae) from the Caribbean and the Southwest Atlantic. Zootaxa 5512, 491–511. doi: 10.11646/zootaxa.5512.4.2
Guimarães-Costa A., MaChado F. S., Reis-Filho J. A., Andrade M., Araújo R. G., Corrêa E. M. R., et al. (2020). DNA barcoding for the assessment of the taxonomy and conservation status of the fish bycatch of the northern Brazilian shrimp trawl fishery. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.566021
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids symposium Ser. 41, 95–98.
Hebert P. D. N., Cywinska A., Ball S. L., and deWaard J. R. (2003). Biological identifications through DNA barcodes. Proc. R Soc. Lond B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
ICMBio. (2012). Instrução Normativa Interministerial MPA/MMA N° 14, de 26 de novembro de 2012. Available online at: https://www.icmbio.gov.br/cepsul/images/stories/legislacao/Instrucao_normativa/2012/in_inter_mpa_mma_14_2012_normasprocedimentoscapturatubaroes_raias.pdf (Accessed February 20, 2025).
IUCN. (2025). International union for conservation of nature, red list of threatened species 2025. Available online at: https://www.iucnredlist.org (Accessed January 13, 2025).
Jabado R. W., Morata A. Z. A., Bennett R. H., Finucci B., Ellis J. R., Fowler S., et al. (2024). The global status of sharks, rays, and chimaeras (Switzerland: Gland).
Kartavtsev Y. P., Jung S.-O., Lee Y.-M., Byeon H.-K., and Lee J.-S. (2007). Complete mitochondrial genome of the bullhead torrent catfish, Liobagrus obesus (Siluriformes, Amblycipididae): Genome description and phylogenetic considerations inferred from the Cyt b and 16S rRNA genes. Gene 396, 13–27. doi: 10.1016/j.gene.2007.01.027
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Librado P. and Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lim D. D., Motta P., Mara K., and Martin A. P. (2010). Phylogeny of hammerhead sharks (Family Sphyrnidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenet Evol. 55, 572–579. doi: 10.1016/j.ympev.2010.01.037
Lutz Í., Miranda J., Martins T., Santana P., Ferreira C., Muhala V., et al. (2023). A multiplex PCR forensic protocol for the molecular certification of sea catfishes (Ariidae – Siluriformes) from coastal Amazon, Brazil. Microchemical J. 195, 109417. doi: 10.1016/j.microc.2023.109417
Marceniuk A. P., Barthem R. B., Wosiacki W. B., Klautau M. A. G. C., Junior T. V., Rotundo M. M., et al. (2019). Sharks and batoids (Subclass Elasmobranchii) caught in the industrial fisheries off the Brazilian North coast. Rev. Nordestina Biol. 27, 120–142. doi: 10.22478/ufpb.2236-1480.2019v27n1.47112
Martins T., Santana P., Lutz Í., da Silva R., Guimarães-Costa A., Vallinoto M., et al. (2021). Intensive commercialization of endangered sharks and rays (Elasmobranchii) along the coastal amazon as revealed by DNA barcode. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.769908
MMA. (2022). Ministério do Meio Ambiente, Portaria MMA n° 148, de 7 de junho de 2022 (Brazil: Portaria MMA n° 148). Available online at: https://www.in.gov.br/en/web/dou/-/portaria-mma-n-148-de-7-de-junho-de-2022-407293037 (Accessed November 25, 2024).
Morgan A. and Burgess G. H. (2007). At-vessel fishing mortality for six species of sharks caught in the northwest atlantic and gulf of Mexico. Gulf Caribb Res. 19. doi: 10.18785/gcr.1902.15
Nachtigall P., Rodrigues-Filho L., Sodré D., Vallinoto M., and Pinhal D. (2017). A multiplex PCR approach for the molecular identification and conservation of the Critically Endangered daggernose shark. Endanger Species Res. 32, 169–175. doi: 10.3354/esr00798
Naylor G. J. P., Caira J. N., Jensen K., Rosana K. A. M., White W. T., and Last P. R. (2012). A DNA sequence–based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull. Am. Mus Nat. Hist 367, 1–262. doi: 10.1206/754.1
Naylor G. J. P., Ryburn J. A., Fedrigo O., and López J. A. (2005). “Phylogenetic relationships among the major lineages of modern elasmobranchs,” in Reproductive biology and phylogeny of chondrichthyes sharks, batoids and chimaeras. Ed. Hamlett W. C. (Boca Raton: Science Publishers), 1–25.
Pacoureau N., Rigby C. L., Kyne P. M., Sherley R. B., Winker H., Carlson J. K., et al. (2021). Half a century of global decline in oceanic sharks and rays. Nature 589, 567–571. doi: 10.1038/s41586-020-03173-9
Paithankar K. R. and Prasad K. S. N. (1991). Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 19, 1346–1346. doi: 10.1093/nar/19.6.1346
Sodré C. F. L., Macedo W., Feitosa L. M., Sousa N. S. M., Carvalho-Neta R. N. F., Carvalho Costa L. F., et al. (2024). Molecular identification of sharks from the genus Sphyrna (Elasmobranchii: Chondrichthyes) in Maranhão Coast (Brazil). Braz. J. Biol. 84. doi: 10.1590/1519-6984.274862
Souza-Araujo J., Souza-Junior O. G., Guimarães-Costa A., Hussey N. E., Lima M. O., and Giarrizzo T. (2021). The consumption of shark meat in the Amazon region and its implications for human health and the marine ecosystem. Chemosphere 265, 129132. doi: 10.1016/j.chemosphere.2020.129132
Steinke D., Bernard A. M., Horn R. L., Hilton P., Hanner R., and Shivji M. S. (2017). DNA analysis of traded shark fins and mobulid gill plates reveals a high proportion of species of conservation concern. Sci. Rep. 7, 9505. doi: 10.1038/s41598-017-10123-5
Stevens J. D., Bonfil R., Dulvy N. K., and Walker P. A. (2000). The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. ICES J. Mar. Sci. 57, 476–494. doi: 10.1006/jmsc.2000.0724
Tamura K., Stecher G., and Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson J. D., Higgins D. G., and Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Keywords: elasmobranchs, hammerhead shark, NADH2, Sphyrnidae, threatened species
Citation: Martins T, Lutz Í, Da Silva R, Monteiro De Lima G, Santana P, Brígida NS, Vallinoto M, Sampaio I and Evangelista-Gomes G (2025) Forensic genetics for monitoring the illegal trade of hammerhead sharks (Sphyrna spp.) using a multiplex PCR protocol. Front. Mar. Sci. 12:1638479. doi: 10.3389/fmars.2025.1638479
Received: 30 May 2025; Accepted: 31 July 2025;
Published: 26 August 2025.
Edited by:
David Seth Portnoy, Texas A&M University Corpus Christi, United StatesReviewed by:
Khaled Mohammed Geba, Menoufia University, EgyptIoannis A. Giantsis, Aristotle University of Thessaloniki, Greece
Copyright © 2025 Martins, Lutz, Da Silva, Monteiro De Lima, Santana, Brígida, Vallinoto, Sampaio and Evangelista-Gomes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Grazielle Evangelista-Gomes, Z3JhemllbGxlQHVmcGEuYnI=
 Thais Martins
Thais Martins Ítalo Lutz
Ítalo Lutz Raimundo Da Silva
Raimundo Da Silva Gabriel Monteiro De Lima
Gabriel Monteiro De Lima Paula Santana
Paula Santana Nicolly Santa Brígida
Nicolly Santa Brígida Marcelo Vallinoto
Marcelo Vallinoto Iracilda Sampaio
Iracilda Sampaio Grazielle Evangelista-Gomes
Grazielle Evangelista-Gomes