- 1Department of Family Medicine, Shengjing Hospital of China Medical University, Shenyang, China
- 2Science and Technology Research Center of China Customs, Beijing, China
- 3Department of Developmental Cell Biology, Key Laboratory of Cell Biology, Ministry of Public Health, China Medical University, Shenyang, China
- 4College of Metrology and Measurement Engineering, China Jiliang University, Hangzhou, China
- 5Department of General Practice, The First Hospital of China Medical University, Shenyang, China
- 6Department of Infection Diseases, The First Affiliated Hospital of China Medical University, Shenyang, China
Background/objective: Hypervirulent Klebsiella pneumoniae (hvKP) is an emerging global health threat, exhibiting increased virulence and multidrug resistance compared to classic K. pneumoniae. Understanding the research landscape surrounding hvKP is crucial for developing effective control strategies. This study aimed to comprehensively analyze the global research trends in hvKP from 2013 to 2024 using bibliometric and topic modeling techniques.
Methods: Data from 1,014 articles on hvKP, retrieved from the Web of Science Core Collection, were analyzed using Bibliometrix, CiteSpace, and VOSviewer to assess publication trends, collaborations, geographical distribution, and keyword co-occurrence. Latent Dirichlet Allocation (LDA) topic modeling was employed to identify key research themes.
Results: The analysis revealed a steadily increasing volume of hvKP research, with China and the United States as major contributors. Four primary research themes emerged: high virulence phenotypes and mechanisms; drug resistance and treatment strategies; genetic and molecular mechanisms; and epidemiological and transmission characteristics. Research hotspots included virulence mechanisms, drug resistance, genomic detection approaches, and epidemiological features.
Conclusion: This bibliometric analysis provides a comprehensive overview of hvKP research, highlighting key trends and research gaps. The identified research hotspots inform future research directions and contribute to the development of effective strategies for combating hvKP infections. The increasing research volume underscores the urgent need for continued investigation into this significant public health threat.
1 Introduction
Hypervirulent Klebsiella pneumoniae (hvKP) is a hypervirulent (hypermucoviscous) variant of K. pneumoniae, distinct from classic K. pneumoniae (cKP). It represents an evolving pathotype characterized by enhanced virulence (1). Early reports emerged from Taiwan in the mid-1980s and 1990s, describing a Klebsiella species associated with a unique clinical syndrome, causing liver abscesses as a sole pathogen (2). Subsequently, many reports referred to it as hypermucoviscous K. pneumoniae due to the hypermucoviscous colony morphology observed on specific agar plates (3). In 2004, Taiwanese researchers identified a novel gene, magA (“mucoviscosity-associated gene A”), in hypermucoviscous K. pneumoniae strains (3), initially considered a hallmark gene for hvKP (4–7), leading to initial ambiguity in hvKP definition and research. Subsequent studies revealed that not all hypermucoviscous strains carry magA (8), and many hvKP strains exhibit non-K1/K2 capsular serotypes (9, 10). In 2013, Alyssa et al. comprehensively defined hvKP based on its characteristic features (11), formally establishing the term hypervirulent K. pneumoniae (hvKP). However, the precise definition of hvKP remains a subject of ongoing refinement due to its diverse genetic background and complex virulence mechanisms (12).
HvKP is predominantly prevalent in the Asia-Pacific region, with colonization rates in healthy adults ranging from 2.7% in Thailand to 16.7% in Japan (13). While the Asia-Pacific remains the epicenter of hvKP infections, increasing case reports from Europe and North America highlight its expanding global footprint (14, 15). Large-scale genomic analyses have identified multiple concurrent antibiotic-resistant clusters of hvKP strains worldwide, underscoring its accelerating global spread (16).
Clinically, hvKP is associated with severe infections, including pneumonia, bloodstream infections, and even brain abscesses, leading to high morbidity and mortality (17). For instance, a case report from Japan documented a diabetic patient who developed emphysematous cholecystitis and disseminated infection due to hvKP K2-ST65, ultimately resulting in fatal multi-organ failure (18). Similarly, another study reported a case of community-acquired pneumonia caused by hvKP K2-ST86, where the patient rapidly deteriorated and died (19).
The convergence of hypervirulence and antimicrobial resistance has emerged as a defining evolutionary trajectory of K. pneumoniae, posing a significant and emerging public health threat (20). Retrospective studies have shown a marked increase in hvKP infections over recent years, closely linked to patterns of antimicrobial resistance (21). In China, carbapenem-resistant hvKP (CR-hvKP) strains have been widely reported across multiple regions (22), with regional dissemination patterns exemplified by the ST25 CR-hvKP strains isolated in central-southern China (23). Hospital outbreaks of multidrug-resistant hvKP, such as the ST11 strain, further highlight the dual threats of virulence and resistance evolution (24, 25).
Despite three decades of extensive basic and clinical research since its discovery, our understanding of hvKP remains incomplete. Key challenges include the lack of objective diagnostic tests, which hampers accurate prevalence estimation; unclear genotype/phenotype markers, leaving the mechanisms of infection partially understood; and insufficient data to determine optimal antimicrobial therapies for hvKP infections (26, 27). Current research predominantly consists of scattered case reports and small-scale studies, lacking systematic integration and comprehensive analysis.
This study employs bibliometric analysis and topic modeling approaches to comprehensively examine the research domain of hvKP. With advancements in bibliometric software and scientific databases (28), researchers have increasingly utilized relational bibliometrics for network analysis (29), which offers robust analytical capabilities for uncovering macro-level research trends and knowledge structures. Here, we analyzed literature metadata (e.g., authors, journals, and keywords) to construct knowledge maps, revealing the overall structure and key nodes in hvKP research. Data were sourced from the Web of Science Core Collection and analyzed using tools such as Bibliometrix, CiteSpace, and VOSviewer for visualization. However, bibliometric analysis primarily focuses on quantitative relationships and co-occurrence patterns, limiting its ability to explore thematic content and research directions in depth. To address this, we integrated topic modeling methods, employing the Latent Dirichlet Allocation (LDA) model from the Python Gensim library to perform topic mining on literature abstracts. As an unsupervised machine learning technique, the LDA model identifies latent themes within texts and represents each topic using word probability distributions (30, 31). During preprocessing, we applied stopword removal, punctuation removal, and stemming to enhance model accuracy and efficiency. After evaluating models with varying numbers of topics, the optimal theme model was selected.
2 Materials and methods
This study selected the Web of Science Core Collection (WoSCC) as the data source, given its comprehensive coverage of high-quality research publications. The search covered all WoSCC editions, including the Science Citation Index Expanded (SCI-EXPANDED), Social Sciences Citation Index (SSCI), Current Chemical Reactions (CCR-EXPANDED), and Index Chemicus (IC). The search query was “((((TS = (hypervirulent Klebsiella pneumoniae)) OR TS = (hvKP)) OR TS = (hypermucoviscous Klebsiella pneumoniae)) OR TS = (hypervirulent Klebsiella pneumoniae)) OR TS = (hypermucoviscous Klebsiella pneumoniae))),” with a time span from 2013 to 2024, resulting in 1,147 records.
2.1 Inclusion and exclusion criteria
2.1.1 Inclusion criteria
1. Document types include research articles and review articles.
2. Documents are published in English.
3. The content directly relates to research on hypervirulent Klebsiella pneumoniae (hvKP).
4. The publication date is between 2013 and 2024.
2.1.2 Exclusion criteria
1. Document types such as book reviews, editorials, news articles or conference papers.
2. Documents published in languages other than English.
3. Documents with irrelevant or low relevance to hvKP.
4. Duplicate publications or records in the database.
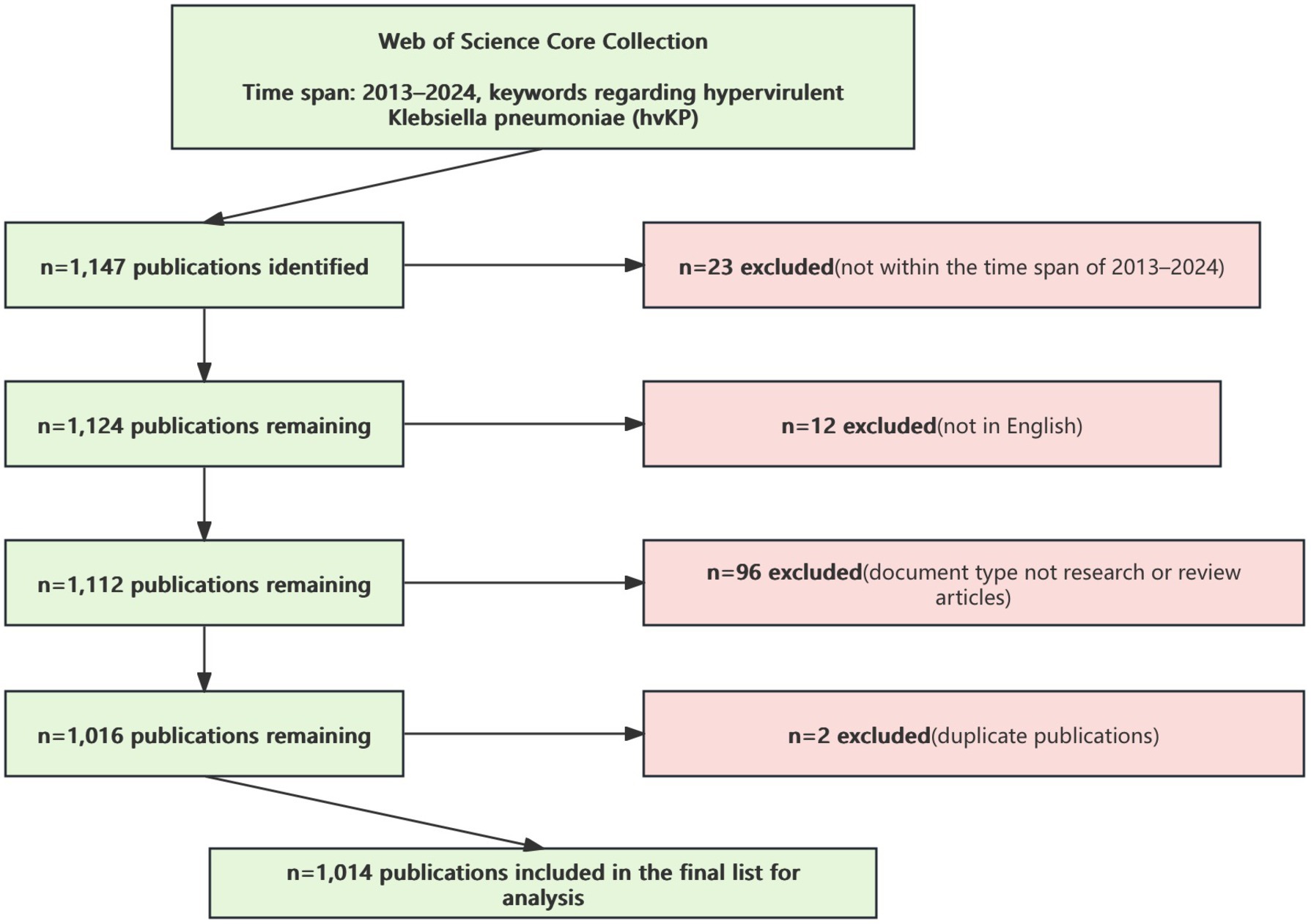
After excluding irrelevant document types, languages, and duplicates, a total of 1,014 publications were included (Figure 1).
2.2 Data export and processing
The data were exported as plain text files, including full records and cited references. The cleaned data were imported into the following tools for analysis and visualization.
• Bibliometrix 4.3.1: Used for analyzing annual publication output, citation counts, authors’ h-index, publication trends over time, three-field plots, and document co-citation analysis.
• CiteSpace 6.1.R6 (64-bit) Advanced: Used for generating structural co-occurrence maps, journal dual-map overlays, and keyword analysis.
• VOSviewer 1.6.19: Used for author co-authorship analysis and country network visualization.
• LDA topic modeling: For LDA topic modeling, the LdaModel class in the Gensim library was employed, supporting online, constant memory, and distributed training for efficient large-scale corpus processing. The core estimation code is based on the onlineldavb.py script by Matthew D. Hoffman, David M. Blei, and Francis Bach: “Online Learning for Latent Di-richlet Allocation,” NIPS 2010. Preprocessing involved text cleaning, including synonym merging and phrase recognition, and Porter Stemming for stemming. The model was trained using the preprocessed document vectors. The optimal number of topics (k) was determined to be 4 by evaluating model coherence and perplexity across a range of 2–15 topics. Model parameters were set as follows: num_topics = 4; other parameters used default values (alpha = ‘symmetric’, eta = None, decay = 0.5, offset = 1.0, etc.). The model was trained incrementally using the LdaModel’s update() method, saved and loaded using save() and load() methods, respectively, and new document topic distributions were inferred using the getitem method.
3 Results
3.1 Publication and citation trends
Figure 2 illustrates the trends in the number of publications and citations related to hypervirulent Klebsiella pneumoniae (hvKP) from 2013 to 2024 (data up to November 2024). The results demonstrate a consistent yearly increase in both publication output and citation counts for hvKP-related research. To project future trends, we fitted quadratic polynomial regression models to the publication and citation data, yielding the following predictive equations: Publication count: y = 84.5 + 219.26x + 31.7x2; Citation count: y = 2288.33 + 7627.95x + 2304.3x2 (where x represents the year, with 2013 as year 0). These equations can be used to estimate future publication and citation counts for hvKP research. Before 2013, the annual publication count was consistently ≤7. The sharp increase from 2013 onwards is likely attributable to the expanding geographical distribution of hvKP. During this period, hvKP emerged as an important pathogen in various regions, including the United States, Canada, Europe, Israel, South Africa, and Australia (32), with infections no longer confined to individuals of Asian descent or those with recent travel history to Asia (33). This broadened recognition of hvKP as a globally significant cause of lethal pneumonia fueled increased research interest. The quadratic polynomial regression models predict the continued robust growth of hvKP research. Sustained attention and investment in hvKP research are crucial for safeguarding global public health.
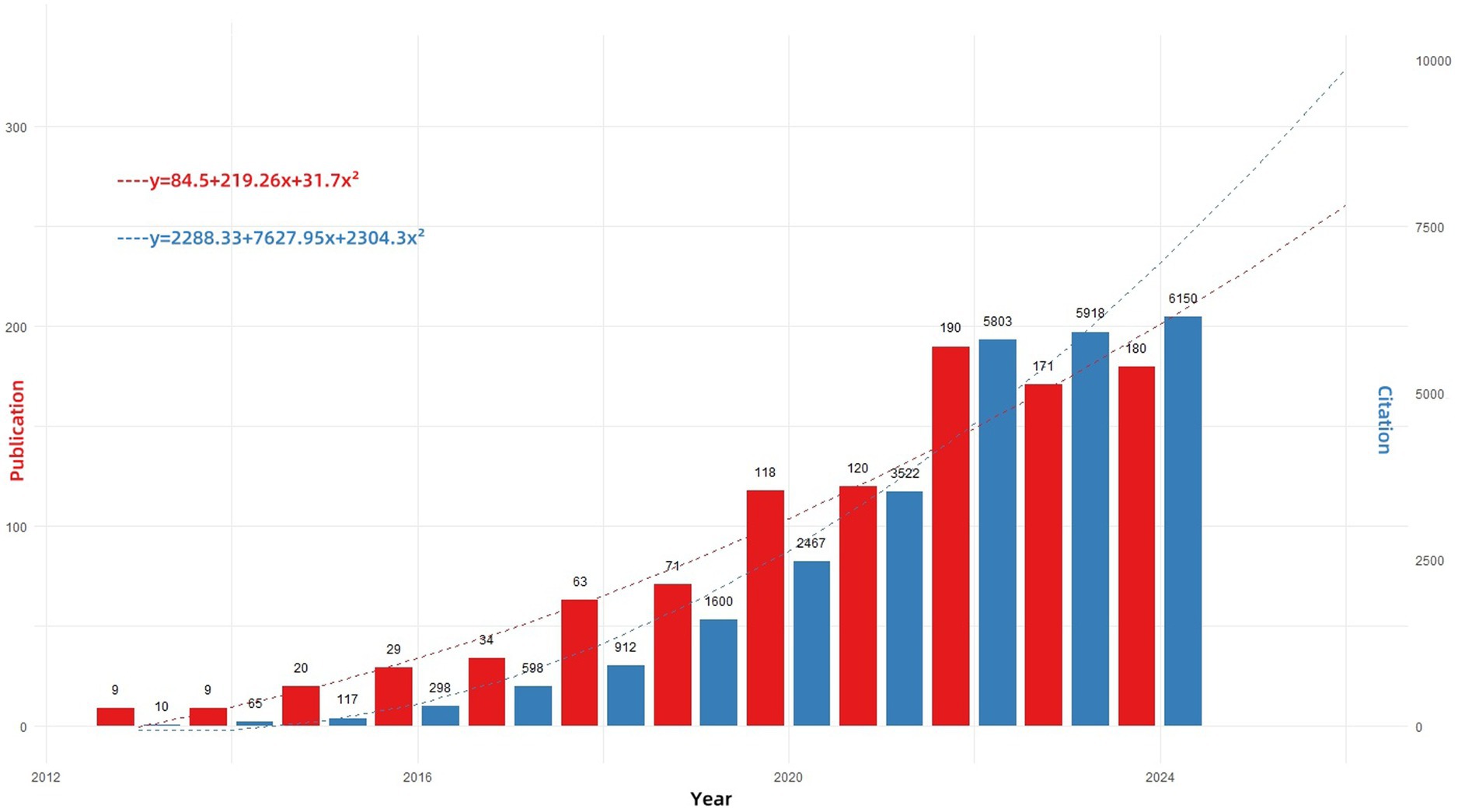
Figure 2. Annual publications and MeanTCperYear in the research field from 2012 to 2024. “Publications (red)” indicates the annual number of papers published; “MeanTCperYear (blue)” (mean total citations per paper divided by the number of citable years) is a useful metric to assess the yearly impact of this research field.
3.2 Author analysis
Figure 3A presents key metrics for the top five most prolific authors in the hvKP research field, including publication count, average citation count, and h-index (34). Liu, Yang and Sylvain Brisse stand out as the most prolific and highly cited authors, respectively, demonstrating significant influence in the field. Liu, Yang from Nanchang University focuses on rapid molecular detection of hvKP (35), antimicrobial resistance mechanisms [e.g., carbapenem resistance (36) and ceftazidime/avibactam resistance (37)], and infection pathogenesis (38). Sylvain Brisse from the Institut Pasteur, Paris, was recognized as a “Highly Cited Researcher in the field of Microbiology” in 2020, and his research primarily centers on hvKP genomics (39, 40). Figure 3B displays the publication timelines for the top 10 most prolific authors. Zhang, Rong (Zhejiang University), Edward Wai-Chi (City University of Hong Kong), and Chen, Sheng exhibited a peak in publications in 2018, indicating a significant surge in their influence and recognition within the hvKP research community that year. This may be linked to their collaborative work on three publications in 2018 focusing on carbapenem-resistant hypervirulent K. pneumoniae ST11 (25, 41), facilitated by an outbreak at an affiliated hospital of Zhejiang University in China, allowing for detailed epidemiological and microevolutionary analyses. These three authors also form the most densely connected research group (warmest color tones in Figure 3D), with Chen, Sheng playing a crucial role in information dissemination and collaboration (the center of the green collaboration module in Figure 3C). Their collaborative work centers on the epidemiology and transmission patterns of hvKP (42) and the study of hvKP virulence plasmids (43, 44).
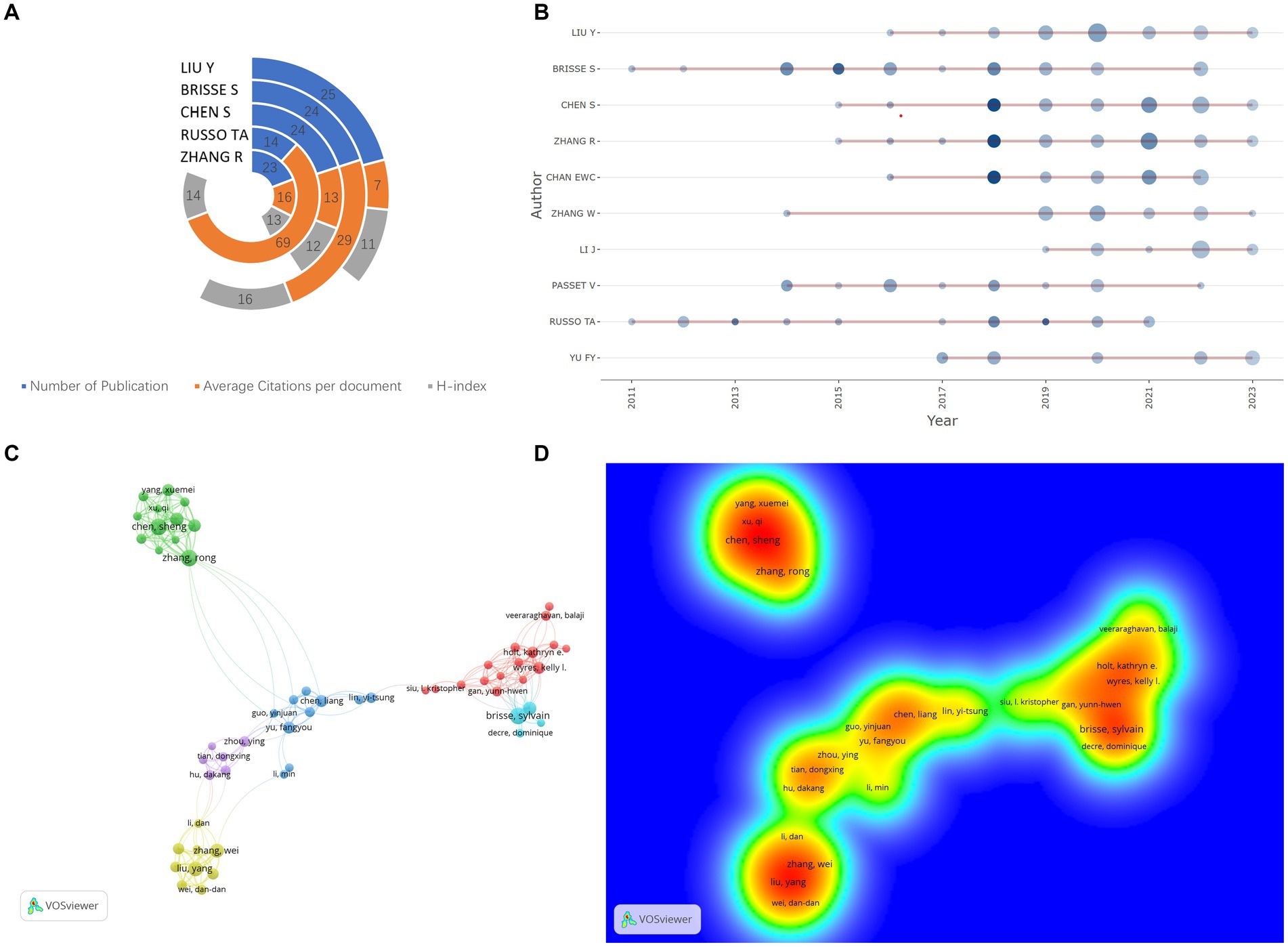
Figure 3. (A) Academic impact metrics of top 5 authors: Total publications, average citations per document, and H-index [Quantifying scholar influence by balancing publication count and citation impact (114)]. (B) Authors’ temporal productivity [Dot size reflects annual article count, shading intensity denotes Total Citations per Year (TCpY)]. (C) Co-authorship network analysis; (D) Co-authorship density visualization.
3.3 Institutional analysis
Zhejiang University is the leading contributor, with 54 publications. Other contributing institutions include Wenzhou Medical University (23), Shanghai Jiao Tong University (20) (Figure 4A). Figure 4B depicts the institutional collaboration network for hvKP research. A total of 345 institutions participated (N = 345), with 721 collaborations (E = 721) and a network density of 0.0122, close to zero, indicating a relatively low overall network density. However, several tightly knit sub-networks exist, notably those centered around Zhejiang University (Count = 54, BC = 0.20) and the Institut Pasteur (Count = 20, BC = 0.12). These institutions not only exhibit high publication output but also high BC, signifying the formation of strong collaborative groups within the hvKP research field.
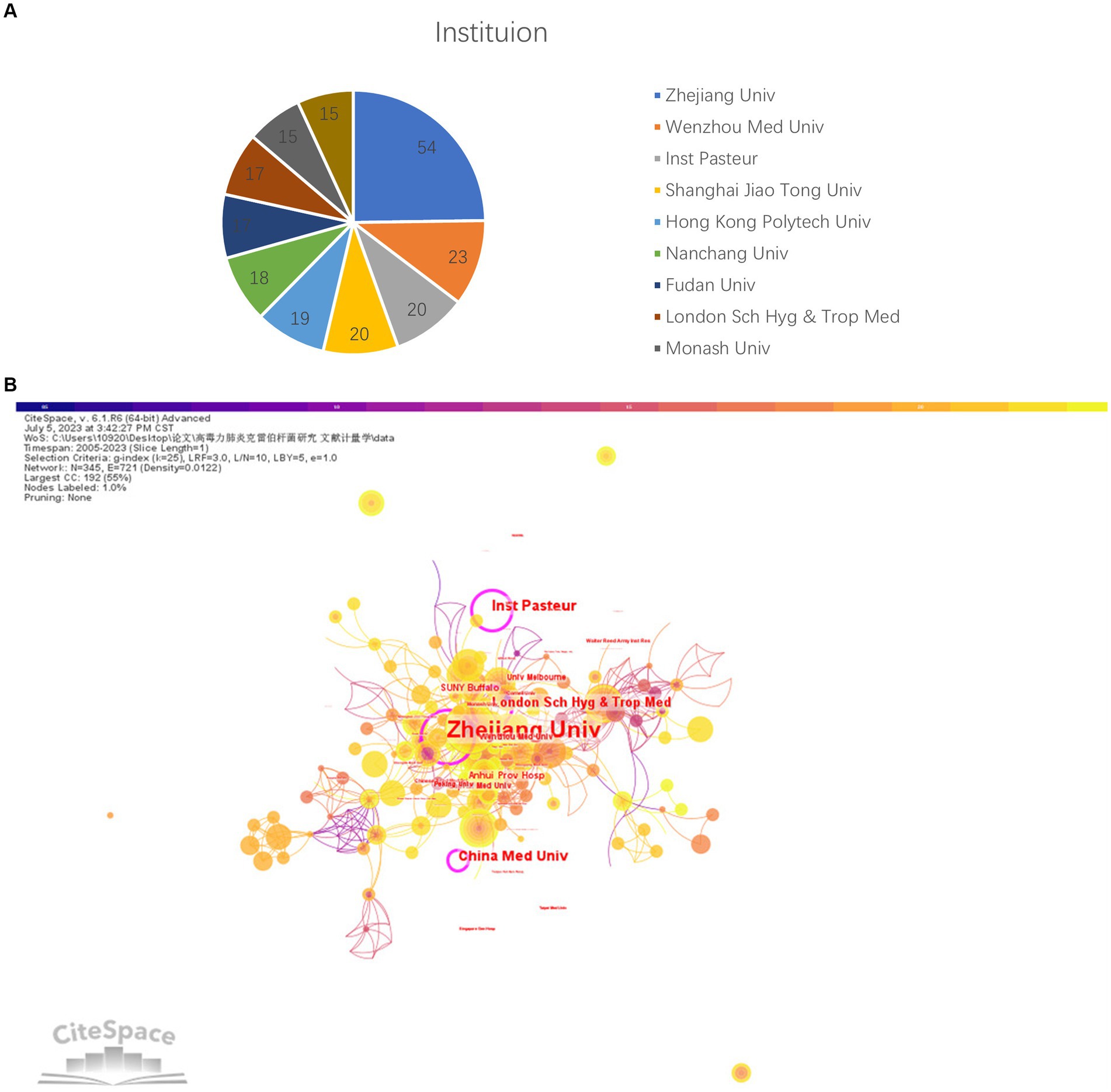
Figure 4. (A) Top 10 institutions with the highest number of papers. (B) Inter-institutional Collaboration Network [Purple node periphery denotes high betweenness centrality (BC ≥ 0.1), reflecting bridging frequency in shortest-path connections (115)].
3.4 Country analysis
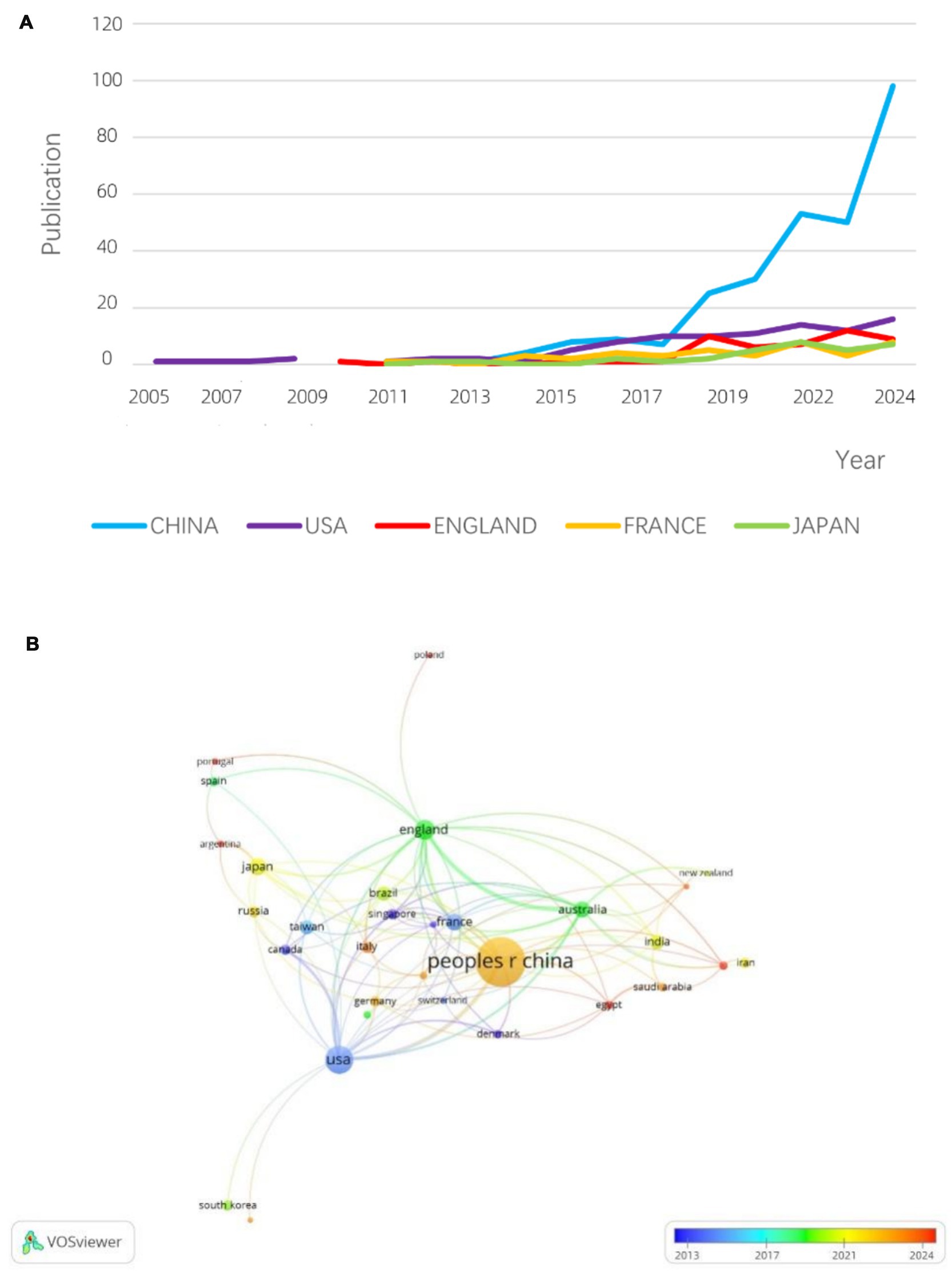
Figure 5A presents the publication timelines for the top five countries/regions. Before 2011, the United States and the United Kingdom dominated hvKP research. However, after 2017, China experienced exponential growth in hvKP-related publications, surpassing other countries/regions. This may be attributed to China’s large population and its longstanding status as a high-prevalence region for hvKP infections. A 2016 study in China reported that 30.9% of K. pneumoniae isolates tested positive using the string test (45). Figure 5B shows the inter-country/region collaboration network. The United States and China exhibit the most frequent collaborations, although the collaborative output is relatively low compared to their individual outputs, suggesting significant potential for further collaboration. The UK and Australia show a strong collaborative relationship, with 16 joint publications representing 47% of Australia’s total output, providing a model for other countries.
3.5 Journal analysis
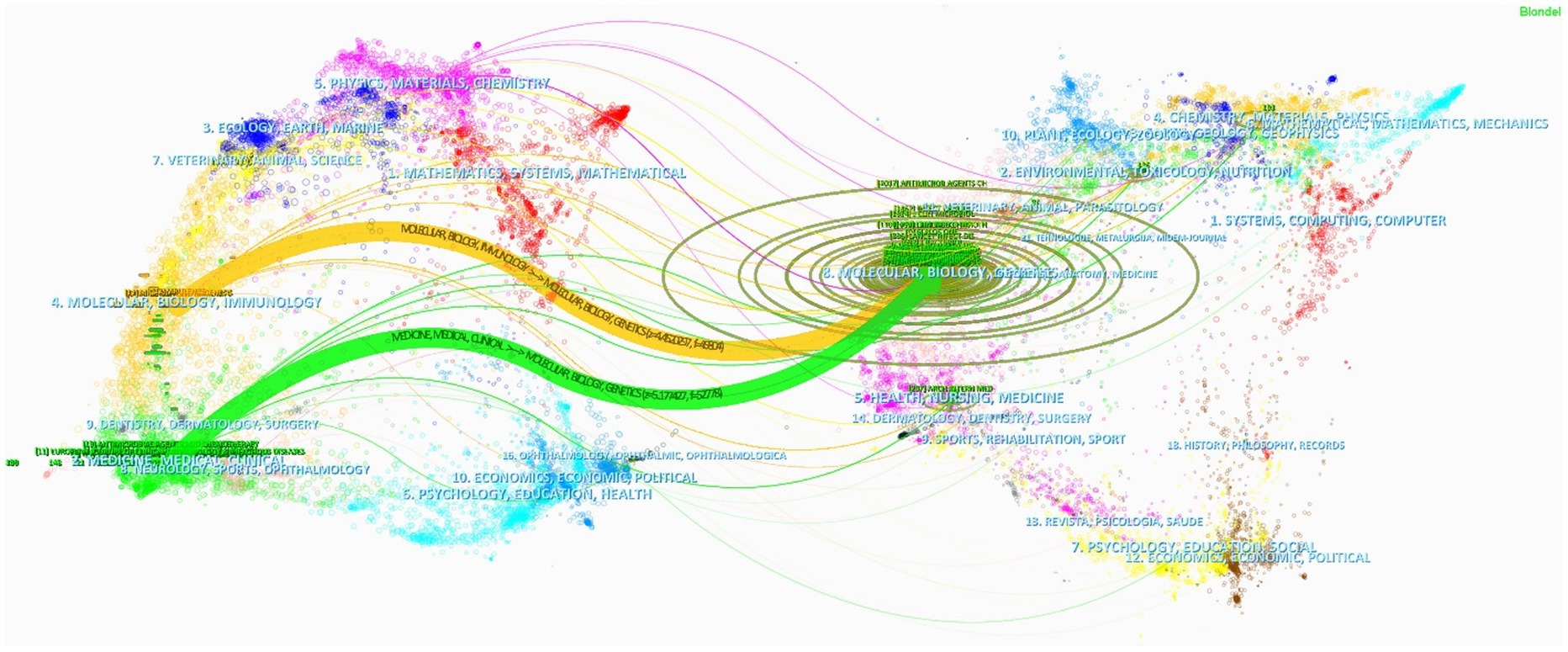
Supplementary Table S1 reveals that the driving force behind hvKP research is distributed across numerous Q2 journals. While Frontiers in Cellular and Infection Microbiology is the only Q1 journal among the top 10, its substantial publication count (third place) indicates significant research output. This suggests that the field encompasses both high-impact research and a broad base of in-depth investigations, providing a solid foundation for future advancements. Figure 6 displays a dual-map overlay of journals, showing that journals in immunology and clinical medicine are predominantly cited by journals focused on molecular biology, genetics, and microbiology. This highlights the importance of interdisciplinary integration in advancing research, exemplified by studies exploring the impact of genetic variations on hvKP-host interactions and pathogenesis (46), and the role of hvKP’s molecular mechanisms of high mortality in informing effective clinical treatments (47).
3.6 Highly cited publications
This study identified the top 10 most frequently cited articles to analyze the current research trends in hvKP (Supplementary Table S2). The two most frequently cited articles, both authored by Russo, Thomas A., are review papers on hvKP. Russo’s 2013 review (11) was the first to define this emerging strain as hypervirulent K. pneumoniae (hvKP), focusing on its pathogenic mechanisms, epidemiological characteristics, and diagnostic and therapeutic approaches. His 2019 review (1) further explored the pathogenesis, colonization, and infection processes of hvKP, emphasizing the role of virulence factors. The third most cited article, a 2016 review by Paczosa and Mecsas (48), primarily discussed the K-antigen of hvKP. A 2012 review by Siu et al. (49) highlighted the unique invasive syndrome caused by hvKP, particularly its association with liver abscesses.
Three research articles—by Holt, Kathryn E; Bialek-Davenet, Suzanne; and Struve, Carsten—focused on genomic sequencing of hvKP. Holt et al. conducted whole-genome sequencing of Klebsiella species, including high-virulence clones (40). Bialek-Davenet et al. developed an openly accessible database, BIGSdb-Kp, to extract medically and epidemiologically relevant information from Klebsiella pneumoniae genome sequences (39). Struve et al.’s work emphasized the CC23 clone (50). Li et al.’s retrospective study analyzed the epidemiology, risk factors, and drug resistance of hvKP (51). A review by Carlos Catalan-Najera et al. (52) distinguished between the high-mucosity and high-virulence phenotypes. Zhang et al.’s study examined the geographic distribution, clinical characteristics, and antimicrobial resistance of hvKP in China (45).
In summary, the top 10 most cited articles on hvKP primarily focus on reviews and genomic studies. The high proportion of review articles suggests that hvKP is a research hotspot, with many investigators contributing to the field and achieving significant research depth and breadth. The prominence of genomic studies may be attributed to the critical role of genetics in understanding hvKP’s virulence, drug resistance, and transmissibility. By investigating the genetic makeup of hvKP, scientists can gain deeper insights into its biological characteristics, providing a theoretical foundation for the prevention and treatment of hvKP infections.
3.7 Keyword analysis
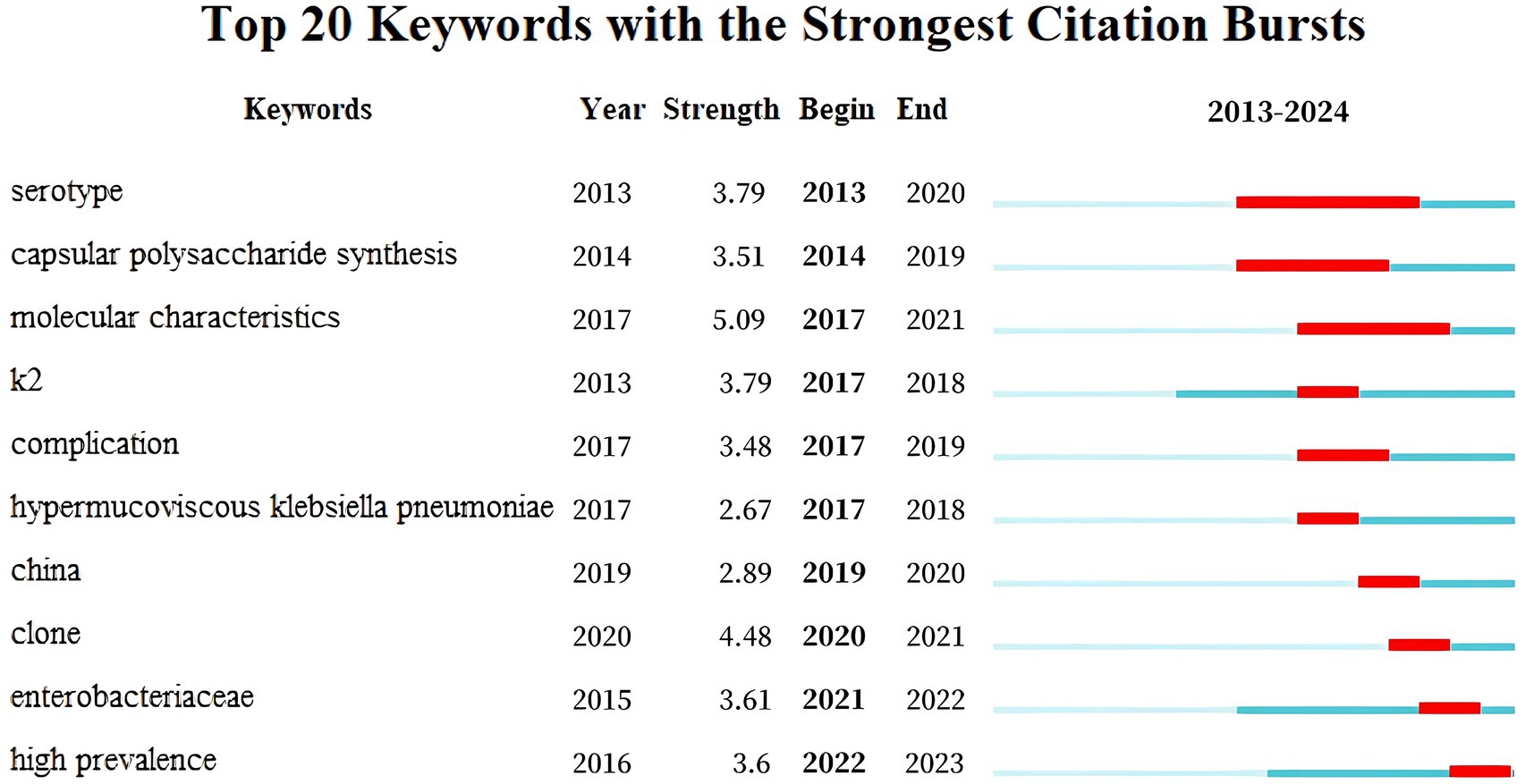
Supplementary Tables S3a,b list the top 15 keywords by centrality and frequency, respectively. “Pyogenic liver abscess” exhibits a betweenness centrality (BC) of 0.11 > 0.1, highlighting its importance in hvKP research. Figure 7 illustrates keyword burst detection, enabling the division of hvKP research trends into four phases: (2013–2016) Basic Research Phase: focusing on basic biological characteristics (serotype, capsular polysaccharide synthesis, molecular features) and early clinical complications, with initial studies on the K2 gene; (2017–2018) Rise of Hypermucoviscous hvKP: research shifted toward the pathogenesis of hypermucoviscous hvKP, with peak interest in the K2 gene; (2019–2020) Chinese Outbreak and Clone Studies: focus on outbreaks and clones in China, investigating epidemiological characteristics; (2021–2023) Enterobacteriaceae and High Prevalence: research expanded to the entire Enterobacteriaceae family, focusing on the public health impact and control strategies of hvKP.
3.8 Key research themes
Preprocessing involved removing stop words and punctuation, and stemming to ensure data consistency and cleanliness. The model was trained using the LdaModel class in Gensim, implementing an online LDA algorithm with the following parameter settings: alpha = “symmetric” (symmetric prior) and eta = None (default prior). These parameters control the prior beliefs regarding the document-topic and topic-word distributions. Topic optimization involved training LDA models with varying numbers of topics (2 to 15). Lower perplexity indicates better model fit (Figure 8A), while higher coherence suggests a more coherent topic structure (Figure 8B). The scatter plot in Figure 8C shows that models with 4, 5, and 7 topics balance perplexity and coherence. Further manual analysis indicated that 5 and 7 topics resulted in overly fine-grained and fragmented classifications. A 4-topic model was ultimately selected. Representative keywords for each topic are presented in Supplementary Table S4.
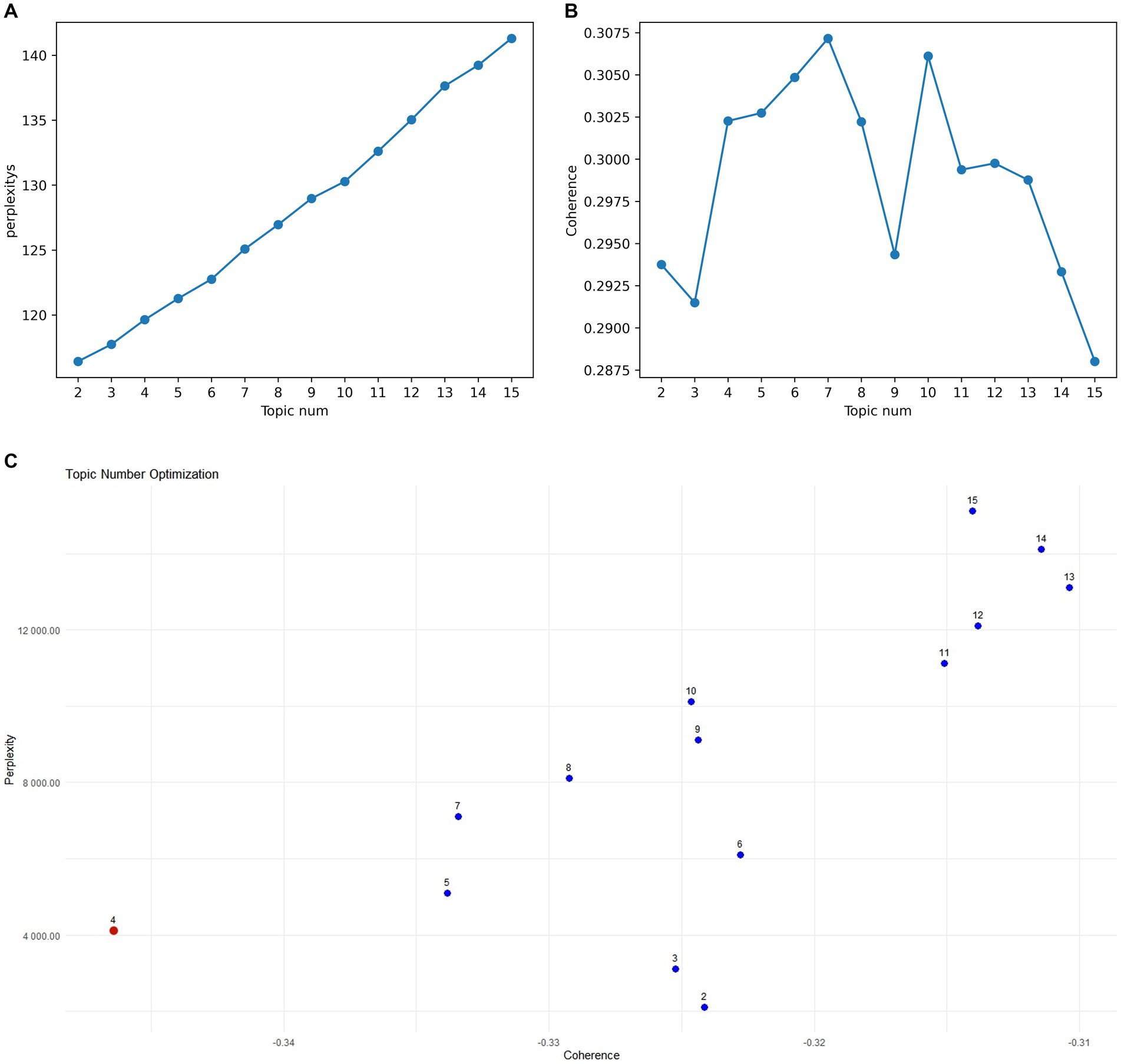
Figure 8. (A) Perplexity for topics 2–15. (B) Coherence for topics 2–15. (C) Topic model optimal parameter selection diagram.
Figure 9 uses word clouds to visually represent the interrelationships between different research themes, with each cluster marked by a different color. Clusters are interconnected through shared keywords (e.g., red nodes for Topic 0), with node size reflecting keyword frequency and connection thickness representing the strength of word distribution within a specific topic. The analysis reveals significant cross-cluster associations. For example, Cluster 1 (Core Mechanisms and Antimicrobial Resistance) shows a strong association with Cluster 3 (Treatment and Prevention) because the type of resistance genes directly influences antibiotic selection and efficacy. For instance, hvKp strains carrying the NDM-1 gene exhibit high resistance to carbapenem antibiotics (e.g., imipenem). A study on hvKp infections in India found that strains carrying the NDM-1 gene showed resistance rates exceeding 90% against imipenem, leading to a significant increase in treatment failure rates (53). Additionally, Cluster 4 (Epidemiology and Transmission) correlates with Cluster 2 (Clinical Manifestations and Diagnosis) as epidemiological data aids in predicting and preventing infections. For example, patients with a travel history to East Asia should be vigilant about the possibility of hvKP infection (54).
The following sections (3.8.1–3.8.4) delve into these four major topics in depth, exploring their significance and the latest advances in current research.
3.8.1 Phenotype and virulence determinants
Klebsiella pneumoniae (hvKP) exhibits hypervirulence primarily due to its virulence factors and phenotypic traits, such as hypermucoviscosity, capsular serotypes (e.g., K1 and K2), virulence genes (e.g., rmpA, rmpA2, iucA, magA), high serum resistance, and strong invasiveness (55–57).
3.8.1.1 Hypermucoviscosity
The hypermucoviscous phenotype, a key feature of hvKP, results from increased capsular polysaccharide synthesis (58, 59). This phenotype enhances bacterial virulence by promoting immune evasion, particularly resistance to neutrophil-mediated killing (60). The thickened and sticky capsular polysaccharides form a physical barrier, hindering immune cell recognition and phagocytosis, thereby promoting immune evasion and persistent infection (58, 59). Additionally, hypermucoviscosity may further augment virulence by influencing biofilm formation and bacterial invasiveness. In experimental studies, the hypermucoviscous phenotype is typically detected using the “string test,” where a string length ≥ 0.5 mm indicates positivity (61). This method is simple and widely used in laboratory settings. Furthermore, whole-genome sequencing (WGS) and PCR techniques can identify genes associated with hypermucoviscosity (62). For instance, rmpA and rmpA2 are key regulatory genes for hypermucoviscosity, often located on large plasmids and upregulated to enhance capsular polysaccharide synthesis (58, 59). However, some hvKP strains exhibit hypermucoviscosity in the absence of rmpA/rmpA2, suggesting alternative regulatory mechanisms (63). These mechanisms may involve other genes or regulators in the capsular polysaccharide synthesis pathway, warranting further investigation.
3.8.1.2 Capsular serotypes
The capsular serotypes of hvKP, including K1, K2, K5, K20, K54, and K57, are often co-present with virulence genes (e.g., rmpA, rmpA2, iucA), further enhancing their virulence (64, 65). Among these, K1 and K2 are the most extensively studied and prevalent serotypes (66). Khairuddin et al. found that K1 and K2 serotypes accounted for 11.1 and 6.1% of hvKP isolates, respectively (56). These serotypes are frequently associated with severe community-acquired infections, such as liver abscesses, pneumonia, and meningitis (64). In virulence assessments, K1 serotype hvKP demonstrated 100% lethality in the Galleria mellonella infection model, while K2 serotype exhibited high serum resistance (67, 68). Whole-genome sequencing has revealed complex relationships between hvKP serotypes and virulence genes. For example, magA is a unique virulence gene in the K1 serotype (56), while rmpA and rmpA2 are detected in 99.4 and 98.6% of K1 and K2 serotypes, respectively (69). Additionally, K1 serotype hvKP is often associated with the ST23 sequence type (70, 71). Although K54 and K57 serotypes are less frequent, they have also been reported as virulence markers in some studies (65, 72). In experimental research, PCR is commonly used to detect capsular serotypes and virulence genes; however, variations in detection standards across laboratories may lead to inconsistent results (65).
3.8.1.3 Virulence genes
The virulence genes of hvKP, including rmpA, rmpA2, iucA, aerobactin, iroB, and peg-344, significantly enhance pathogenicity by regulating mucoid phenotypes, iron acquisition systems, and other virulence factors.
• rmpA and rmpA2: Studies have shown that rmpA and rmpA2 are highly conserved in hvKP and strongly associated with hypervirulence (73–75). For instance, a study in Malaysia reported that all hvKP strains carried rmpA and rmpA2 (56). These genes act as key regulators of capsular synthesis by targeting the promoter of the capsular gene cluster (cps), promoting high capsular production and enhancing bacterial adhesion and invasiveness (57, 66, 76). Further research has revealed that the promoter activity of rmpA is closely linked to virulence. Strong promoter activity (e.g., P11T and P12T) correlates with high capsular production and invasive virulence, while weak activity (e.g., P9T and P10T) may enhance bacterial colonization. Additionally, mutations (e.g., insertions/deletions) in rmpA and rmpA2 can shift hvKP from a hypermucoid to a hypomucoid phenotype, reducing invasive virulence but enhancing colonization. This adaptive change allows hvKP to persist and spread within hosts, increasing its epidemiological significance (77).
• iucA and aerobactin: The presence of iucA and aerobactin in hvKP is strongly associated with hypervirulence, particularly in severe infections such as liver abscesses (78, 79). iucA encodes aerobactin synthetase, and aerobactin serves as the primary siderophore for hvKP, facilitating iron acquisition and significantly enhancing bacterial survival and pathogenicity (80). Compared to the unstable RmpA2, aerobactin exhibits higher stability in carbapenem-resistant hvKP (CR-hvKP) and has been proposed as a reliable marker for hvKP identification (81, 82).
• iroB: The iroB gene, encoding the salmochelin siderophore system, plays a critical role in iron acquisition, significantly enhancing hvKP survival and pathogenicity (67, 77). Gene knockout studies have demonstrated that iroB deletion markedly reduces virulence, confirming its importance in hvKP pathogenicity (83). iroB is prevalent in hvKP, a study in Sudan reported a 57.9% detection rate of iroB in hvKP clinical isolates (74). Notably, iroB often coexists with other virulence genes (e.g., iucA, rmpA2), which encode siderophores and mucoid phenotype regulators, further augmenting hvKP virulence (84, 85).
• peg-344: The presence of peg-344 is strongly correlated with hypervirulence in hvKP. For example, peg-344-positive hvKP strains exhibit enhanced serum resistance, biofilm formation, and invasiveness, as well as increased pathogenicity in animal models (35, 84, 86).
3.8.2 Antibiotic resistance mechanisms
In recent years, the issue of antibiotic resistance in hypervirulent Klebsiella pneumoniae (hvKP) has become increasingly severe, particularly with the emergence of multidrug-resistant (MDR) and carbapenem-resistant (CRKP) strains, posing significant challenges to clinical treatment (55, 87). The development of novel antibiotics and vaccines targeting hvKP, as well as the use of gene-editing technologies such as CRISPR-Cas9 to knockout resistance genes, are likely to be future research hotspots (88, 89). Below, we systematically elucidate the resistance mechanisms of hvKP from two perspectives: antibiotic classification and molecular mechanisms.
Resistance mechanisms in hvKP involve outer membrane porin mutations, plasmid-mediated gene transfer, efflux pumps, and biofilm formation. Mutations in outer membrane porins (such as ompK35 and ompK36) reduce the ability of antibiotics to enter bacterial cells, thereby enhancing resistance. Zhao et al. (90) found that the L359R mutation in ompK36 is associated with hvKP’s resistance to ceftazidime/avibactam. Additionally, hvKP carries multiple resistance genes (such as blaCTX-M, blaTEM, blaSHV, etc.) via plasmids and spreads resistance among strains through conjugation (90–92). The efflux pump system (e.g., acrAB and tolC) actively expels antibiotics from the cell, reducing their intracellular concentration and thereby enhancing hvKP’s resistance to multiple antibiotics (93). Alharbi et al. (94) noted that hvKP protects itself by forming biofilms, increasing its tolerance to antibiotics.
The antibiotic classification-based resistance mechanisms involve carbapenems, colistin, fosfomycin, and multidrug resistance mechanisms. Carbapenem resistance is mainly mediated by the spread of carbapenemase genes (such as blaKPC and blaNDM), which are transferred via plasmids or chromosomes, conferring hvKP resistance to carbapenems (95–97). Liu et al. (95) demonstrated that the blaKPC gene spreads via IncFII plasmids in ST11-type hvKP, while Tang et al. (97) showed that the blaNDM-1 gene can be horizontally transferred through outer membrane vesicles (OMVs). For instance, Liu et al. (95) found that the blaKPC gene spreads through IncFII plasmids in ST11-type hvKP, while Tang et al. (97) confirmed that the blaNDM-1 gene can be horizontally transferred in hvKP via outer membrane vesicles (OMVs). Colistin resistance is primarily associated with mgrB gene mutations and overexpression of the phoPQ system, which modify the lipopolysaccharide (LPS) structure of the bacterial outer membrane, reducing colistin binding capacity and thereby inducing resistance (87). Intrinsic mechanisms of fosfomycin resistance include the UhpTE350Q mutation and the presence of fosA6/5 genes, which affect fosfomycin uptake or metabolism, leading to hvKP resistance to fosfomycin. Furthermore, hvKP achieves multidrug resistance and high virulence by carrying multiple resistance genes (such as blaKPC, blaNDM, blaOXA, etc.) and virulence genes (such as rmpA, iucA, etc.) (98, 99). For example, Zhang et al. (100) reported a hybrid plasmid in ST11-KL64 type hvKP carrying blaKPC-2 and rmpA2 genes, indicating the co-evolution of resistance and virulence genes.
3.8.3 Genomic detection approaches
Traditional culture and biochemical identification methods, while reliable, are limited in terms of timeliness and sensitivity. With the rapid development of molecular biology techniques, genome-based detection methods have gradually become mainstream, leading to significant progress in the genomic research of hvKp. This has unveiled its virulence mechanisms, evolutionary pathways, and drug resistance characteristics (101, 102).
3.8.3.1 Research methods
• Whole genome sequencing: WGS is a core technology for the classification of hvKP. By sequencing the entire genome of hvKP strains, researchers can identify virulence-associated genes and mutation sites, providing comprehensive genomic information. For example, by comparing the genomes of different hvKP strains, high-virulence-associated gene clusters such as rmpA and rmpA2 can be identified. This method offers crucial insights for the classification and evolutionary studies of hvKP (103, 104).
• Single nucleotide polymorphism analysis: SNP analysis is another commonly used genomic detection method. By comparing SNP sites in hvKP strains, phylogenetic trees can be constructed, enabling strain classification and evolutionary analysis. This approach is particularly valuable for tracing the transmission routes and evolutionary relationships of hvKP (105, 106).
• Machine learning algorithms: In recent years, machine learning algorithms have seen increasing application in genomic data analysis. By training deep learning models, researchers can automatically identify hvKP features and perform classification from high-throughput genomic data. For instance, models based on convolutional neural networks (CNN) and long short-term memory networks (LSTM) have demonstrated high accuracy in hvKP classification (107, 108).
3.8.3.2 Application directions
• Virulence gene detection: The detection of virulence genes is a critical basis for hvKP classification. Using PCR or high-throughput sequencing, researchers can identify virulence-associated genes such as rmpA, aerobactin, and iroN. The presence or absence of these genes directly reflects the virulence level of the strains (103, 104).
• Genomic evolution and diversity: Through whole-genome sequencing and comparative genomic analysis, hvKp has been classified into multiple clonal lineages, such as ST23, ST65, and ST86, with ST23-K1 and ST86-K2 being the predominant hvKp clonal lineages (101, 102).
• Drug resistance mechanism research: hvKp has increasingly exhibited resistance to multiple antibiotics, particularly carbapenems, which is closely associated with the presence of resistance genes such as blaKPC and blaNDM (101, 109).
3.8.4 Clinical epidemiology
The clinical epidemiology of hypervirulent Klebsiella pneumoniae (hvKP) is characterized by its high-risk populations, infection types, low resistance but high virulence, and transmission patterns. hvKP primarily affects individuals with diabetes, long-term hospitalized patients, and immunocompromised populations, leading to severe infections such as pyogenic liver abscess (PLA) and ventilator-associated pneumonia (VAP) (110). Although hvKP exhibits lower antibiotic resistance compared to carbapenem-resistant Klebsiella pneumoniae (CRKP), its virulence is significantly higher, particularly in strains carrying virulence genes such as iucA and rmpA and those of the ST23 lineage, which are associated with a hypermucoid phenotype and severe infections (111).
The transmission of hvKP is closely linked to healthcare settings, especially in long-term care facilities and intensive care units, where it poses a significant threat to vulnerable populations Additionally, the increasing incidence of community-acquired hvKP infections highlights its ability to spread beyond hospital environments, further complicating its control and management (112). Studies have shown that diabetic patients and those with gallstones are particularly susceptible to hvKP-related PLA, while elderly patients and those with pressure ulcers in long-term care facilities are at higher risk of hvKP infections (110, 113).
Research methodologies such as retrospective analyses, genomic studies, and phenotypic experiments have been instrumental in understanding hvKP’s epidemiology. For instance, Guo et al. used genomic analysis to differentiate between PLA- and VAP-associated strains (110), while Alfaifi et al. conducted retrospective studies to examine resistance patterns in long-term care settings. However, limitations such as small sample sizes and regional focus in some studies may affect the generalizability of findings (113). Further research is needed to elucidate hvKP’s transmission mechanisms, its interactions with host immunity, and the development of novel therapeutic strategies, particularly in the context of its increasing prevalence in Asia and its potential crossover with CRKP (110, 112, 113).
4 Study limitations
This study has the following limitations.
1. Database selection: This study exclusively used the Web of Science (WoS) database as the data source and did not include other important databases such as PubMed/MEDLINE, Cochrane, or Embase/SCOPUS. This may have resulted in the omission of some relevant literature, particularly clinical and regional studies. Future research should consider incorporating more databases to ensure comprehensive coverage of the literature.
2. Limitations of topic modeling: Although topic modeling can reveal the latent structure of research themes, its results depend on text preprocessing and model parameter selection. The Latent Dirichlet Allocation (LDA) model used in this study, while effective in identifying topics, may have been influenced by text cleaning, stop-word removal, and stemming processes. Additionally, the choice of the number of topics is somewhat subjective and may affect the final topic classification.
Despite these limitations, this study provides a systematic analytical framework for understanding the knowledge structure and research trends in the field of hvKP through bibliometric and topic modeling methods, offering valuable insights for future research.
5 Conclusion
This study comprehensively reviewed the progress in the field of hypervirulent Klebsiella pneumoniae (hvKP) research through bibliometric analysis and topic modeling methods. The results indicate that hvKP research has shown significant growth over the past decade, particularly in Asia, with China emerging as the primary contributor in this field. Research hotspots primarily focus on hvKP’s phenotypic and virulence determinants, antibiotic resistance mechanisms, genomic detection methods, and clinical epidemiology. Through bibliometric analysis, we identified the formation of collaborative networks among core authors, institutions, and countries, with particularly strong collaborations between China, the United States, and France. Additionally, topic modeling revealed major research directions, including the identification of virulence genes, the elucidation of resistance mechanisms, and the application of genomics in hvKP classification and evolutionary studies.
Despite significant advancements, hvKP research still faces numerous challenges. Firstly, the precise definition and diagnostic criteria for hvKP remain inconsistent, making it difficult to directly compare results across different studies. Secondly, the mechanisms of hvKP virulence and the evolution of resistance are complex, requiring further in-depth research to uncover their molecular basis. Moreover, the global transmission trends and epidemiological characteristics of hvKP still need to be validated through large-scale, multicenter studies. Future research should focus on the genomic evolution of hvKP, the co-evolution of resistance and virulence, and the development of novel therapeutic strategies, particularly for multidrug-resistant hvKP strains.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
TT: Data curation, Software, Visualization, Validation, Writing – original draft, Writing – review & editing. HH: Writing – original draft, Writing – review & editing. Z-HG: Writing – original draft, Writing – review & editing. KZ: Conceptualization, Writing – review & editing. XH: Data curation, Methodology, Software, Writing – original draft. WW: Formal analysis, Writing – review & editing. XuZ: Project administration, Writing – review & editing. FZ: Software, Writing – original draft. LW: Writing – review & editing. XiZ: Writing – review & editing. J-HW: Supervision, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Key Research and Development Program of China (2022YFC2602204). Hui Han, a co-first author of this article, is the provider of this funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1545678/full#supplementary-material
References
1. Russo, TA, and Marr, CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. (2019) 32:e00001–19. doi: 10.1128/Cmr.00001-19
2. Wang, JH, Liu, YC, Lee, SS, Yen, MY, Chen, YS, Wang, JH, et al. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis. (1998) 26:1434–8. doi: 10.1086/516369
3. Fang, CT, Chuang, YP, Shun, CT, Chang, SC, and Wang, JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. (2004) 199:697–705. doi: 10.1084/jem.20030857
4. Fang, FC, Sandler, N, and Libby, SJ. Liver abscess caused by magA plus Klebsiella pneumoniae in North America. J Clin Microbiol. (2005) 43:991–2. doi: 10.1128/Jcm.43.2.991-992.2005
5. Fang, C-T. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. (2007) 45:405–6. doi: 10.1086/519262
6. Fung, CP, Chang, FY, Lee, SC, Hu, BS, Kuo, BI, Liu, CY, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut. (2002) 50:420–4. doi: 10.1136/gut.50.3.420
7. Yeh, KM, Kurup, A, Siu, LK, Koh, YL, Fung, CP, Lin, JC, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. (2007) 45:466–71. doi: 10.1128/jcm.01150-06
8. Turton, JF, Englender, H, Gabriel, SN, Turton, SE, Kaufmann, ME, and Pitt, TL. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol. (2007) 56:593–7. doi: 10.1099/jmm.0.46964-0
9. Brisse, S, Fevre, C, Passet, V, Issenhuth-Jeanjean, S, Tournebize, R, Diancourt, L, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. (2023) 4:e4982. doi: 10.1371/journal.pone.0004982
10. Yu, WL, Ko, WC, Cheng, KC, Lee, CC, Lai, CC, and Chuang, YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. (2008) 62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
11. Shon, AS, Bajwa, RPS, and Russo, TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. (2013) 4:107–18. doi: 10.4161/viru.22718
12. Lan, P, Jiang, Y, Zhou, J, and Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. (2021) 25:26–34. doi: 10.1016/j.jgar.2021.02.020
13. Chen, Y, and Chen, Y. Clinical challenges with hypervirulent Klebsiella pneumoniae (hvkp) in China. J Transl Intern Med. (2021) 9:71–5. doi: 10.2478/jtim-2021-0004
14. Sturm, E, Tai, A, Lin, B, Kwong, J, Athan, E, Howden, BP, et al. Bilateral osteomyelitis and liver abscess caused by hypervirulent Klebsiella pneumoniae-a rare clinical manifestation (case report). BMC Infect Dis. (2018) 18:380. doi: 10.1186/s12879-018-3277-4
15. Bilal, S, Volz, MS, Fiedler, T, Podschun, R, and Schneider, T. Klebsiella pneumoniae–induced liver abscesses, Germany. Emerg Infect Dis. (2014) 20:1939–40. doi: 10.3201/eid2011.140149
16. Spadar, A, Perdigão, J, Campino, S, and Clark, TG. Large-scale genomic analysis of global Klebsiella pneumoniae plasmids reveals multiple simultaneous clusters of carbapenem-resistant hypervirulent strains. Genome Med. (2025) 15:3. doi: 10.1186/s13073-023-01153-y
17. Lee, CR, Lee, JH, Park, KS, Jeon, JH, Kim, YB, Cha, CJ, et al. Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. (2017) 7:483. doi: 10.3389/fcimb.2017.00483
18. Kubota, Y, Ishioka, H, Harada, S, Suzuki, M, Shiotsuka, J, Lefor, AK, et al. Septic shock with emphysematous cholecystitis and disseminated infection caused by hypervirulent Klebsiella pneumoniae capsular genotype K2-St65 in a Japanese man with diabetes mellitus: a case report. J Infect Chemother. (2021) 27:350–3. doi: 10.1016/j.jiac.2020.09.017
19. Yamamoto, H, Iijima, A, Kawamura, K, Matsuzawa, Y, Suzuki, M, and Arakawa, Y. Fatal fulminant community-acquired pneumonia caused by hypervirulent Klebsiella pneumoniae K2-ST86. Medicine. (2020) 90:e20360. doi: 10.1097/MD.0000000000020360
20. Marr, CM, and Russo, TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti-Infect Ther. (2019) 17:71–3. doi: 10.1080/14787210.2019.1555470
21. Liu, C, and Guo, J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. (2019) 18:4. doi: 10.1186/s12941-018-0302-9
22. Shi, Q, Quan, J, Lan, P, Huang, D, Zhou, J, Jiang, Y, et al. Prevalence and characteristics of pks gene cluster harbouring Klebsiella pneumoniae from bloodstream infection in China. Epidemiol Infect. (2020) 148:e69. doi: 10.1017/S0950268820000655
23. Li, J, Huang, ZY, Yu, T, Tao, XY, Hu, YM, Wang, HC, et al. Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. (2019) 19:219. doi: 10.1186/s12866-019-1593-5
24. He, Z, Xu, W, Zhao, H, Li, W, Dai, Y, and Lu, H. Epidemiological characteristics an outbreak of ST11 multidrug-resistant and hypervirulent Klebsiella pneumoniae in Anhui, China. Front Microbiol. (2022) 13:996752. doi: 10.3389/fmicb.2022.996753/full
25. Gu, D, Dong, N, Zheng, Z, Lin, D, Huang, M, Wang, L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. doi: 10.1016/S1473-3099(17)30489-9
26. Sanikhani, R, Moeinirad, M, Shahcheraghi, F, Lari, A, Fereshteh, S, Sepehr, A, et al. Molecular epidemiology of hypervirulent Klebsiella pneumoniae: a systematic review and meta-analysis. Iran J Microbiol. (2021) 13:257–65. doi: 10.18502/ijm.v13i3.6384
27. Namikawa, H, Oinuma, KI, Yamada, K, Kaneko, Y, Kakeya, H, and Shuto, T. Predictors of hypervirulent Klebsiella pneumoniae infections: a systematic review and meta-analysis. J Hosp Infect. (2023) 134:153–60. doi: 10.1016/j.jhin.2023.02.005
28. Donthu, N, Kumar, S, Mukherjee, D, Pandey, N, and Lim, WM. How to conduct a bibliometric analysis: an overview and guidelines. J Bus Res. (2021) 133:285–96. doi: 10.1016/j.jbusres.2021.04.070
29. Stuart, D. Open bibliometrics and undiscovered public knowledge. Online Information Rev. (2018) 42:209. doi: 10.1108/Oir-07-2017-0209
30. Chauhanuttam, S. Topic modeling using latent Dirichlet allocation. ACM Comput Surveys. (2021) 54:1–35. doi: 10.1145/3462478
31. Yang, Y, Ngai, EWT, and Wang, L. Resistance to artificial intelligence in health care: literature review, conceptual framework, and research agenda. Inf Manag. (2024) 61:103961. doi: 10.1016/j.im.2024.103961
32. Pomakova, DK, Hsiao, CB, Beanan, JM, Olson, R, MacDonald, U, Keynan, Y, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. (2012) 31:981–9. doi: 10.1007/s10096-011-1396-6
33. Rafat, C, Messika, J, Barnaud, G, Dufour, N, Magdoud, F, Billard-Pomarès, T, et al. Hypervirulent Klebsiella pneumoniae, a 5-year study in a French Icu. J Med Microbiol. (2018) 67:1083–9. doi: 10.1099/jmm.0.000788
34. Hirsch, JE. An index to quantify an individual’s scientific research output. Proc Natl Acad Sci USA. (2005) 102:16569. doi: 10.1073/pnas.0507655102
35. Liao, W, Long, D, Huang, Q, Wei, D, Liu, X, Wan, L, et al. Rapid detection to differentiate hypervirulent Klebsiella pneumoniae (hvKp) from classical K. pneumoniae by Identifying peg-344 with loop-mediated isothermal amplication (LAMP). Front Microbiol. (2020) 11:1189. doi: 10.3389/fmicb.2020.01189
36. Liao, W, Wang, LD, Li, D, Du, FL, Long, D, Liu, Y, et al. High prevalence of 16s rrna Methylase genes among Carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Microb Drug Resist. (2021) 27:44–52. doi: 10.1089/mdr.2019.0482
37. Li, D, Liao, W, Huang, H, Du, FL, Wei, DD, Mei, YF, et al. Emergence of hypervirulent ceftazidime/avibactam-resistant Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Infect Drug Resist. (2020) 13:2673–80. doi: 10.2147/Idr.S257477
38. Li, P, Luo, W, Xiang, TX, Jiang, Y, Liu, P, Wei, DD, et al. Horizontal gene transfer via Omvs co-carrying virulence and antimicrobial-resistant genes is a novel way for the dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. (2022) 13:945972. doi: 10.3389/fmicb.2022.945972
39. Bialek-Davenet, S, Criscuolo, A, Ailloud, F, Passet, V, Jones, L, Delannoy-Vieillard, AS, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. (2014) 20:1812–20. doi: 10.3201/eid2011.140206
40. Holt, KE, Wertheim, H, Zadoks, RN, Baker, S, Whitehouse, CA, Dance, D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. (2015) 112:E3574–81. doi: 10.1073/pnas.1501049112
41. Dong, N, Yang, X, Zhang, R, Chan, EW, and Chen, S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microb Infect. (2018) 7:146. doi: 10.1038/s41426-018-0146-6
42. Shu, L, Dong, N, Lu, J, Zheng, Z, Hu, J, Zeng, W, et al. Emergence of OXA-232 Carbapenemase-producing Klebsiella pneumoniae that carries a plvpk-like virulence plasmid among elderly patients in China. Antimicrob Agents Chemother. (2019) 63:e02246. doi: 10.1128/Aac.02246-18
43. Yang, X, Dong, N, Chan, EWC, Zhang, R, and Chen, S. Carbapenem resistance-encodinc and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. (2021) 29:65–83. doi: 10.1016/j.tim.2020.04.012
44. Dong, N, Lin, D, Zhang, R, Chan, EWC, and Chen, S. Carriage of bla(KPC-2) by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother. (2018) 73:3317–21. doi: 10.1093/jac/dky358
45. Zhang, Y, Zhao, C, Wang, Q, Wang, X, Chen, H, Li, H, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. (2016) 60:6115–20. doi: 10.1128/Aac.01127-16
46. Raj, S, Sharma, T, Pradhan, D, Tyagi, S, Gautam, H, Singh, H, et al. Comparative analysis of clinical and genomic characteristics of Hypervirulent Klebsiella pneumoniae from hospital and community settings: experience from a tertiary healthcare center in India. Microbiol Spectr. (2022) 10:e0037622–2. doi: 10.1128/spectrum.00376-22
47. Xu, Q, Xie, M, Liu, X, Heng, H, Wang, H, Yang, C, et al. Molecular mechanisms underlying the high mortality of hypervirulent Klebsiella pneumoniae and its effective therapy development. Signal Transduct Target Ther. (2023) 8:221. doi: 10.1038/s41392-023-01490-9
48. Paczosa, MK, and Mecsas, J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/Mmbr.00078-15
49. Siu, LK, Yeh, KM, Lin, JC, Fung, CP, and Chang, FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. (2012) 12:881–7. doi: 10.1016/S1473-3099(12)70205-0
50. Struve, C, Roe, CC, Stegger, M, Stahlhut, SG, Hansen, DS, Engelthaler, DM, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. Mbio. (2015) 6:e00630. doi: 10.1128/mbio.00630-15
51. Li, W, Sun, G, Yu, Y, Li, N, Chen, M, Jin, R, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. (2014) 58:225–32. doi: 10.1093/cid/cit675
52. Catalán-Nájera, JC, Garza-Ramos, U, and Barrios-Camacho, H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. (2017) 8:1111–23. doi: 10.1080/21505594.2017.1317412
53. Pandhare, A. Bacterial resistance in India: studying plasma antibiotic levels. Indian J Crit Care Med. (2015) 19:574–5. doi: 10.4103/0972-5229.167032
54. Nakamura, K, Nomoto, H, Harada, S, Suzuki, M, Yomono, K, Yokochi, R, et al. Infection with capsular genotype K1-St23 hypervirulent Klebsiella pneumoniae isolates in Japan after a stay in east asia: two cases and a literature review. J Infect Chemother. (2021) 27:1508. doi: 10.1016/j.jiac.2021.05.011
55. Hefzy, EM, Taha, R, Abd El Salam, S, Abdelmoktader, A, and Khalil, MAF. Hypervirulent Klebsiella pneumoniae: epidemiology, virulence factors, and antibiotic resistance. Novel Res Microbiol J. (2023) 7:1857–72. doi: 10.21608/nrmj.2023.287177
56. Khairuddin, A, Zuraina, N M N N, Nasir, N S M, Chuan, CW, Halim, HS, Yusof, W, et al. Microbiological and clinical characteristics of Hypervirulent 3 Klebsiella pneumoniae isolated from patients in tertiary centers[a/Ol]. medRxiv [Preprint]. medRxiv: 2023.11.17.23298703[2025-02-26] (2023). Available online at: https://www.medrxiv.org/content/10.1101/2023.11.17.23298703v1
57. Emam, SM, Abdelrahman, S, Hasan, AA, and Melouk, MS. Hypervirulent Klebsiella pneumoniae at benha university hospitals. Egypt J Hosp Med. (2023) 90:3592–7. doi: 10.21608/ejhm.2023.292752
58. Xu, Q, Yang, X, Chan, EWC, and Chen, S. The hypermucoviscosity of hypervirulent K. pneumoniae confers the ability to evade neutrophil-mediated phagocytosis. Virulence. (2021) 12:2050–9. doi: 10.1080/21505594.2021.1960101
59. Choby, JE, Howard-Anderson, J, and Weiss, DS. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med. (2020) 287:283–300. doi: 10.1111/joim.13007
60. He, Z, Xu, W, Zhao, H, Li, W, Dai, Y, Lu, H, et al. Epidemiological characteristics an outbreak of ST11 multidrug-resistant and hypervirulent Klebsiella pneumoniae in Anhui, China. Front Microbiol. (2022) 13:6753. doi: 10.3389/fmicb.2022.996753
61. Neumann, B, Stürhof, C, Rath, A, Kieninger, B, Eger, E, Müller, JU, et al. Detection and characterization of putative hypervirulent Klebsiella pneumoniae isolates in microbiological diagnostics. Sci Rep. (2023) 13:19025. doi: 10.1038/s41598-023-46221-w
62. Altayb, HN, Elbadawi, HS, Baothman, O, Kazmi, I, Alzahrani, FA, Nadeem, MS, et al. Genomic analysis of multidrug-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae strain lacking the hypermucoviscous regulators (rmpA/rmpA2). Antibiotics. (2022) 11:596. doi: 10.3390/antibiotics11050596
63. Dey, T, Chakrabortty, A, Kapoor, A, Warrier, A, Nag, VL, Sivashanmugam, K, et al. Unusual hypermucoviscous clinical isolate of Klebsiella pneumoniae with no known determinants of hypermucoviscosity. Microbiol Spectr. (2022) 10:e00393–22. doi: 10.1128/spectrum.00393-22
64. Zhao, D, Tang, M, Ma, Z, Hu, P, Fu, Q, Yao, Z, et al. Synergy of bacteriophage depolymerase with host immunity rescues sepsis mice infected with hypervirulent Klebsiella pneumoniae of capsule type K2. Virulence. (2024) 15:2415945. doi: 10.1080/21505594.2024.2415945
65. Lee, J, Hwang, JH, Yeom, JH, Lee, S, and Hwang, JH. Analysis of virulence profiles in clinical isolates of Klebsiella pneumoniae from renal abscesses: clinical significance of hypervirulent isolates. Front Cell Infect Microbiol. (2024) 14:111. doi: 10.3389/fcimb.2024.1367111
66. Chae, KJ, Lee, J, Hwang, JH, and Hwang, JH. Invasive hypervirulent Klebsiella pneumoniae syndrome originating from an anorectal abscess as opposed to a pyogenic liver abscess. Medicina. (2022) 58:1450. doi: 10.3390/medicina58101450
67. Sohrabi, M, Alizade Naini, M, Rasekhi, A, Oloomi, M, Moradhaseli, F, Ayoub, A, et al. Emergence of K1 St23 and K2 St65 hypervirulent Klebsiella pneumoniae as true pathogens with specific virulence genes in cryptogenic pyogenic liver abscesses shiraz Iran. Front Cell Infect Microbiol. (2022) 12:290. doi: 10.3389/fcimb.2022.964290
68. Al-Busaidi, B, Al-Muzahmi, M, Al-Shabibi, Z, Rizvi, M, Al-Rashdi, A, Al-Jardani, A, et al. Hypervirulent capsular serotypes K1 and K2 Klebsiella pneumoniae strains demonstrate resistance to serum bactericidal activity and galleria mellonella lethality. Int J Mol Sci. (2024) 25:1944. doi: 10.3390/ijms25031944
69. Liao, CH, Huang, YT, and Hsueh, PR. Multicenter surveillance of capsular serotypes, virulence genes, and antimicrobial susceptibilities of Klebsiella pneumoniae causing bacteremia in Taiwan, 2017–2019. Front Microbiol. (2022) 13:523. doi: 10.3389/fmicb.2022.783523
70. Wang, S, Ding, Q, Zhang, Y, Zhang, A, Wang, Q, Wang, R, et al. Evolution of virulence, fitness, and carbapenem resistance transmission in St23 hypervirulent Klebsiella pneumoniae with the capsular polysaccharide synthesis gene wcaJ inserted via insertion sequence elements. Microbiol Spectr. (2022) 10:e0240022. doi: 10.1128/spectrum.02400-22
71. Morales-León, F, Matus-Köhler, M, Araya-Vega, P, Aguilera, F, Torres, I, Vera, R, et al. Molecular characterization of the convergent carbapenem-resistant and hypervirulent Klebsiella pneumoniae strain K1-St23, collected in Chile during the Covid-19 pandemic. Microbiol Spectr. (2023) 11:e00540–23. doi: 10.1128/spectrum.00540-23
72. Shao, C, Xin, L, Mi, P, Jiang, M, and Wu, H. Phenotypic and molecular characterization of K54-St29 hypervirulent Klebsiella pneumoniae causing multi-system infection in a patient with diabetes. Front Microbiol. (2022) 13:140. doi: 10.3389/fmicb.2022.872140
73. Ali, MR, Yang, Y, Dai, Y, Lu, H, He, Z, Li, Y, et al. Prevalence of multidrug-resistant hypervirulent Klebsiella pneumoniae without defined hypervirulent biomarkers in Anhui, China: a new dimension of hypervirulence. Front Microbiol. (2023) 14:91. doi: 10.3389/fmicb.2023.1247091
74. Mohammed, KAM, Elhag, SAA, and Ahmed, STES. Molecular detection of virulence genes (rmpA2, iuc & iroB) of hypervirulent Klebsiella pneumoniae in clinical isolates from patients in Khartoum state, Sudan. Asian J Res Infect Dis. (2022) 9:23–31. doi: 10.9734/ajrid/2022/v9i430275
75. Shanthini, T, Manohar, P, Hua, X, Leptihn, S, and Nachimuthu, R. Detection of hypervirulent Klebsiella pneumoniae from clinical samples in Tamil Nadu[a/Ol]. Infectious diseases (except Hiv/aids). medRxiv [Preprint] (2023). Available online at: http://medrxiv.org/lookup/doi/10.1101/2023.02.19.23286158
76. Goh, KJ, Altuvia, Y, Argaman, L, Raz, Y, Bar, A, Lithgow, T, et al. Ril-seq reveals extensive involvement of small Rnas in virulence and capsule regulation in hypervirulent Klebsiella pneumoniae. Nucleic Acids Res. (2024) 52:9119–38. doi: 10.1093/nar/gkae440
77. Teng, G, Zhang, M, Fu, Y, Yang, X, Kang, Y, Qin, Q, et al. Adaptive attenuation of virulence in hypervirulent carbapenem-resistant Klebsiella pneumoniae. mSystems. (2024) 9:e01363–23. doi: 10.1128/msystems.01363-23
78. Liu, S, Huang, Z, Kong, J, Zhao, Y, Xu, M, Zhou, B, et al. Effects of aerobactin-encoding gene iucB and regulator of mucoid phenotype rmpA on the virulence of Klebsiella pneumoniae causing liver abscess. Front Cell Infect Microbiol. (2022) 12:955. doi: 10.3389/fcimb.2022.968955
79. Russo, TA, Lebreton, F, and Mcgann, PT. A step forward in hypervirulent Klebsiella pneumoniae diagnostics. Emerg Infect Dis. (2025) 31:516. doi: 10.3201/eid3101.241516
80. Russo, TA, Olson, R, Macdonald, U, Beanan, J, and Davidson, BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. (2015) 83:3325–33. doi: 10.1128/iai.00430-15
81. Parrott, AM, Shi, J, Aaron, J, Green, DA, Whittier, S, and Wu, F. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a new York City hospital through screening of virulence genes. Clin Microbiol Infect. (2021) 27:583–9. doi: 10.1016/j.cmi.2020.05.012
82. Shankar, C, Basu, S, Lal, B, Shanmugam, S, Vasudevan, K, Mathur, P, et al. Aerobactin seems to be a promising marker compared with unstable RmpA2 for the identification of hypervirulent carbapenem-resistant Klebsiella pneumoniae: in Silico and in vitro evidence. Front Cell Infect Microbiol. (2021) 11:681. doi: 10.3389/fcimb.2021.709681
83. Zi, C, Yang, S, Fu, X, Wang, W, Luo, Y, Zhang, J, et al. An efficient method for knocking out genes on the virulence plasmid of hypervirulent Klebsiella pneumoniae. New Microbiol. (2023) 46:186–95.
84. Yan, Q, Zhou, M, Zou, M, and Liu, WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. (2016) 35:387–96. doi: 10.1007/s10096-015-2551-2
85. Piazza, A, Perini, M, Mauri, C, Comandatore, F, Meroni, E, Luzzaro, F, et al. Antimicrobial susceptibility, virulence, and genomic features of a hypervirulent serotype K2, St65 Klebsiella pneumoniae causing meningitis in Italy. Antibiotics. (2022) 11:261. doi: 10.3390/antibiotics11020261
86. Sattler, J, Ernst, CM, Zweigner, J, and Hamprecht, A. High frequency of acquired virulence factors in carbapenemase-producing Klebsiella pneumoniae isolates from a large german university hospital, 2013–2021. Antimicrob Agents Chemother. (2024) 68:e0060224–4. doi: 10.1128/aac.00602-24
87. Liu, X, Wu, Y, Zhu, Y, Jia, P, Li, X, Jia, X, et al. Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerg Microbes Infect. (2022) 11:648–61. doi: 10.1080/22221751.2022.2036078
88. Hao, M, Shi, X, Lv, J, Niu, S, Cheng, S, du, H, et al. In vitro activity of apramycin against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Front Microbiol. (2020) 11:425. doi: 10.3389/fmicb.2020.00425
89. Huang, J, Yi, M, Yuan, Y, Xia, P, Yang, B, Liao, J, et al. Emergence of a fatal ST11-Kl64 tigecycline-resistant hypervirulent Klebsiella pneumoniae clone cocarrying blaNDM and blaKPC in plasmids. Microbiol Spectr. (2022) 10:e02539–22. doi: 10.1128/spectrum.02539-22
90. Zhao, J, Pu, D, Li, Z, Zhang, Y, Liu, X, Zhuo, X, et al. Loss and gain of ceftazidime-avibactam susceptibility in a non-carbapenemase-producing K1-St23 hypervirulent Klebsiella pneumoniae. Virulence. (2024) 15:2348251. doi: 10.1080/21505594.2024.2348251
91. Han, B, Feng, C, Jiang, Y, Ye, C, Wei, Y, Liu, J, et al. Mobile genetic elements encoding antibiotic resistance genes and virulence genes in Klebsiella pneumoniae: important pathways for the acquisition of virulence and resistance. Front Microbiol. (2025) 16:157. doi: 10.3389/fmicb.2025.1529157
92. Shankar, C, Vasudevan, K, Jacob, JJ, Baker, S, Isaac, BJ, Neeravi, AR, et al. Hybrid plasmids encoding antimicrobial resistance and virulence traits among hypervirulent Klebsiella pneumoniae St2096 in India. Front Cell Infect Microbiol. (2022) 12:116. doi: 10.3389/fcimb.2022.875116
93. Suresh, K, and Pillai, D. Prevalence of antimicrobial resistance, biofilm formation, efflux pump activity, and virulence capabilities in multi-drug-resistant Klebsiella pneumoniae isolated from freshwater fish farms. J Water Health. (2024) 22:721–34. doi: 10.2166/wh.2024.382
94. Alharbi, MT, Almuhayawi, MS, Nagshabandi, MK, Tarabulsi, MK, Alruhaili, MH, Gattan, HS, et al. Antimicrobial resistance pattern, pathogenicity and molecular properties of hypervirulent klebsiella pneumonia (hvKp) among hospital-acquired infections in the intensive care unit (ICU). Microorganisms. (2023) 11:661. doi: 10.3390/microorganisms11030661
95. Liu, Z, Guan, J, Chen, Z, Tai, C, Deng, Z, Chao, Y, et al. CpxR promotes the carbapenem antibiotic resistance of Klebsiella pneumoniae by directly regulating the expression and the dissemination of blaKPC on the Incfii conjugative plasmid. Emerg Microbes Infect. (2023) 12:2256427. doi: 10.1080/22221751.2023.2256427
96. Wang, Q, Liu, Y, Chen, R, Zhang, M, Si, Z, Wang, Y, et al. Genomic insights into the evolution and mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae co-harboring blaKPC and blaNDM: implications for public health threat mitigation. Ann Clin Microbiol Antimicrob. (2024) 23:27. doi: 10.1186/s12941-024-00686-3
97. Tang, B, Yang, A, Liu, P, Wang, Z, Jian, Z, Chen, X, et al. Outer membrane vesicles transmitting blaNDM-1 mediate the emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Antimicrob Agents Chemother. (2023) 67:e0144422–2. doi: 10.1128/aac.01444-22
98. Chen, L, Zhou, Y, Wang, S, Wu, C, Zhou, P, Wang, B, et al. Genomic analysis of carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese tertiary hospital. Infection Drug Resist. (2023) 16:6385–94. doi: 10.2147/Idr.S425949
99. Mukherjee, S, Bhadury, P, Mitra, S, Naha, S, Saha, B, Dutta, S, et al. Hypervirulent Klebsiella pneumoniae causing neonatal bloodstream infections: emergence of NDM-1-producing hypervirulent ST11-K2 and ST15-K54 strains possessing plvpk-associated markers. Microbiol Spectr. (2023) 11:e04121–2. doi: 10.1128/spectrum.04121-22
100. Zhang, F, Li, L, Zhao, Y, Dong, H, Zhao, B, Zhao, X, et al. Molecular characterization of hybrid virulence plasmids in ST11-KL64 KPC-2-producing multidrug-resistant hypervirulent Klebsiella pneumoniae from China. Front Microbiol. (2024) 15:849. doi: 10.3389/fmicb.2024.1353849
101. Miliotis, G, Mcdonagh, F, Singh, NK, O'Connor, L, Tuohy, A, Morris, D, et al. Genomic analysis reveals the presence of emerging pathogenic klebsiella lineages aboard the international space station. Microbiol Spectr. (2023) 11:e01897–23. doi: 10.1128/spectrum.01897-23
102. Kamau, E, Ranson, EL, Tsan, AT, Bergmann-Leitner, ES, Garner, OB, and Yang, S. Clinical and genomic characterization of hypervirulent Klebsiella pneumoniae (hvKp) infections via passive surveillance in Southern California, 2020–2022. Front Microbiol. (2022) 13:1169. doi: 10.3389/fmicb.2022.1001169
103. Rezvannejad, E, Asadollahpour Nanaei, H, and Esmailizadeh, A. Detection of candidate genes affecting milk production traits in sheep using whole-genome sequencing analysis. Vet Med Sci. (2022) 8:1197–204. doi: 10.1002/vms3.731
104. Ahmad, A, Imran, M, and Ahsan, H. Biomarkers as biomedical bioindicators: approaches and techniques for the detection, analysis, and validation of novel biomarkers of diseases. Pharmaceutics. (2023) 15:1630. doi: 10.3390/pharmaceutics15061630
105. Chiara, M, Horner, DS, Ferrandi, E, Gissi, C, and Pesole, G. HaploCoV: unsupervised classification and rapid detection of novel emerging variants of Sars-CoV-2. Commun Biol. (2023) 6:1–15. doi: 10.1038/s42003-023-04784-4
106. Medvedev, A, Lebedev, M, Ponomarev, A, Kosaretskiy, M, Osipenko, D, Tischenko, A, et al. Grape: genomic relatedness detection pipeline[a/Ol]. F1000Res. (2023) 11:589. doi: 10.12688/f1000research.111658.2
107. Kumar, R, Rao, T S, Walid Md, A A, Seeniappan, K, Maranan, R, and Saratha, M. Integrating diverse omics data using graph convolutional networks: advancing comprehensive analysis and classification in colorectal cancer[C/Ol]. In: 4th International Conference on Smart Electronics and Communication (Icosec), pp. 1329–1335; (2023).
108. Park, H, Lim, SJ, Cosme, J, O'Connell, K, Sandeep, J, Gayanilo, F, et al. Investigation of machine learning algorithms for taxonomic classification of marine metagenomes. Microbiol Spectr. (2023) 11:e0523722–2. doi: 10.1128/spectrum.05237-22
109. Nishiyama, N, Arakaki, W, Uechi, K, Utsumi, D, Sato, Y, Nakamatsu, M, et al. Prevalence and genomic analysis of hypermucoviscous Klebsiella pneumoniae in a single center in Japan: insights into virulence and potential clinical implications. Open Forum Infect Dis. (2023) 10:ofad500.369. doi: 10.1093/ofid/ofad500.369
110. Guo, M, Gao, B, Su, J, Zeng, Y, Cui, Z, Liu, H, et al. Phenotypic and genetic characterization of hypervirulent Klebsiella pneumoniae in patients with liver abscess and ventilator-associated pneumonia. BMC Microbiol. (2023) 23:338. doi: 10.1186/s12866-023-03022-5
111. Chang, D, Sharma, L, Dela Cruz, CS, and Zhang, D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front Microbiol. (2021) 12:662. doi: 10.3389/fmicb.2021.750662
112. Rahmat Ullah, S, Jamal, M, Rahman, A, and Andleeb, S. Comprehensive insights into Klebsiella pneumoniae: unravelling clinical impact, epidemiological trends and antibiotic-resistance challenges. J Antimicrob Chemother. (2024) 79:1484–92. doi: 10.1093/jac/dkae184
113. Alfaifi, BA, Alkhaldi, SA, Alanazi, MD, Shuraim, WA, Aldossry, MA, Alzahrani, HS, et al. Epidemiology and antibiotic resistance patterns of Klebsiella pneumoniae infections among male patients in a long-term care hospital in Riyadh, Saudi Arabia: a retrospective study. Cureus. (2024) 16:101. doi: 10.7759/cureus.72101
114. Bihari, A, Tripathi, S, and Deepak, A. A review on h-index and its alternative indices. J Inf Sci. (2023) 49:624–65. doi: 10.1177/01655515211014478
Keywords: hypervirulent Klebsiella pneumoniae, Klebsiella pneumoniae, hypervirulence, antimicrobial resistance, bibliometrics, topic modeling, epidemiological characteristics
Citation: Tian T, Han H, Guan Z-H, Zhang K, Huang X, Wang W, Zhang X, Zhang F, Wei L, Zhang X and Wang J-H (2025) A systematic review of hypervirulent Klebsiella pneumoniae research: bibliometric and topic modeling perspectives. Front. Med. 12:1545678. doi: 10.3389/fmed.2025.1545678
Edited by:
Zisis Kozlakidis, International Agency for Research on Cancer (IARC), FranceReviewed by:
Amira Awad Moawad, Friedrich Loeffler Institut, GermanyPriyavardhan Mishra, Padmashree Dr. D.Y. Patil University, India
Copyright © 2025 Tian, Han, Guan, Zhang, Huang, Wang, Zhang, Zhang, Wei, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Han, aGFuaHVpMjAwMkAxNjMuY29t; Xin Zhang, emhhbmd4aW44MDA3MDVAMTYzLmNvbQ==; Jia-He Wang, d2FuZ2poMUBzai1ob3NwaXRhbC5vcmc=
†These authors have contributed equally to this work
 Tian Tian
Tian Tian Hui Han2*†
Hui Han2*† Ke Zhang
Ke Zhang Fei Zhang
Fei Zhang Leijia Wei
Leijia Wei Xin Zhang
Xin Zhang Jia-He Wang
Jia-He Wang