- 1University Key Laboratory for Integrated Chinese Traditional and Western Veterinary Medicine and Animal Healthcare in Fujian Province, Fujian Agriculture and Forestry University, Fuzhou, Fujian Province, China
- 2Fujian Key Laboratory of Traditional Chinese Veterinary Medicine and Animal Health, Fujian Agriculture and Forestry University, Fuzhou, Fujian Province, China
- 3Wuhan Animal Disease Control Center, Wuhan, Hubei Province, China
Introduction: Matrine (MT) is a potential resistance reversal agent. However, it remains unclear whether MT can reverse the resistance of Haemophilus parasuis (H. parasuis) to β-lactams, and, if so, by what mechanism MT works.
Methods: We screened one cefaclor (CEC)-resistant strain (clinical strain C7) from eight clinical (H. parasuis) strains and determined the underlying resistance mechanism. Then, we investigated the reversal effect of MTon the resistance of this strain to CEC.
Results and Discussion: The production of β-lactamase, overexpression of AcrAB-TolC system, and formation of biofilm might not be responsible for the resistance of clinical strain C7 to CEC. Fourteen mutation sites were found in four PBP genes (ftsI, pbp1B, mrcA, and prcS) of clinical strain C7, among which the mutation sites located in ftsI (Y103D and L517R) and mrcA (A639V) genes triggered the resistance to CEC. The minimum inhibitory concentration (MIC) of CEC against clinical strain C7 was reduced by two to eight folds after MT treatment, accompanied by the significant down-regulated expression of mutated ftsI and mrcA genes. Based on such results, we believed that MT could reverse the resistance of H. parasuis to CEC by inhibiting the mutations in ftsI and mrcA genes. Our research would provide useful information for restoring the antimicrobial activity of β-lactams and improving the therapeutic efficacy of Glässer’s disease.
Introduction
Glässer’s disease is a bacterial infectious disease caused by Haemophilus parasuis (H. parasuis). It is usually clinically manifested as serositis, arthritis, bronchitis, meningitis, and septicemia, and has caused huge economic losses to the pig industry (Costa-Hurtado et al., 2020). β-lactams may be the current drugs of choice for the treatment of Glässer’s disease because of their powerful antibacterial activity and good safety. However, the surveys on the prevalence and antimicrobial susceptibilities of H. parasuis from 2013 to 2017 in China revealed high resistance rates and increasing resistance to β-lactams (Zhao et al., 2018; Zhang et al., 2019), which posed a serious challenge to the effectiveness of β-lactams in the treatment of Glässer’s disease. Thus, there is an urgent need to explore new and more effective therapies.
Screening of resistance-reversal agents from phytochemicals and combining them with antimicrobials is expected to address the challenge of antimicrobial resistance (Ayaz et al., 2019; Khameneh et al., 2019). Some phytochemicals show little or no antimicrobial activity. Nonetheless, they can neutralize the resistance mechanism, enabling the antimicrobials to still be effective against resistant microbes (Zhou et al., 2012; Cai et al., 2016; Barbieri et al., 2017; Pourahmad and Mohammadi, 2018; Itzia Azucena et al., 2019; Qu et al., 2019; Wei et al., 2020). Matrine (MT) is one of the main quinolizidine alkaloids extracted from plants such as Sophora flavescens, Sophora alopecuroide, and Sophora subprostrata (Pourahmad and Mohammadi, 2018). Previous in vitro experiments showed that MT reversed the resistance of Escherichia coli (E. coli) to a variety of antimicrobials by inhibiting the AcrAB-TolC efflux pump (Zhou et al., 2012; Pourahmad and Mohammadi, 2018). Another study revealed the inhibitory effect of MT on the biofilm formation of antimicrobial-resistant E. coli, which helped to reduce the resistance to antimicrobials from the aspect of interbacterial population communication (Sun et al., 2019). In addition to the predictable resistance reversal activity, MT also exhibits good pharmacokinetic properties and safety (You et al., 2020), which makes it a promising resistance-reversal agent for veterinary clinical use. However, it remains unclear whether MT can also reverse the resistance of H. parasuis to β-lactams, and, if so, by what mechanism MT works.
The objectives of this study were to investigate the reversal effect of MT on the resistance of H. parasuis strains to β-lactams and the underlying mechanism. The results would provide a theoretical basis for the further application of MT in veterinary practice.
Materials and methods
Chemicals
The standards of MT, ampicillin (AMP), amoxicillin (AMX), oxacillin (OXA), penicillin (PEN), ceftiofur (CEF), cefaclor (CEC), cefepime (CPE) and cefotaxime (CTX) were purchased from Shanghai Yuanye Bio-Technology Co. Ltd. (Shanghai, China) with a purity ≥95%. Stock standard solutions of MT and 8 β-lactams were prepared separately by dissolving each compound in sterile distilled water at a concentration of 5,120 μg/mL and then stored at −20°C. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), as an efflux pump inhibitor, was purchased from Macklin Inc. (Shanghai, China) with a purity ≥98%. Stock standard solution of CCCP was prepared in dimethylsulfoxide (DMSO) at a concentration of 200 mg/mL and then stored at −20°C. Other chemical reagents were of analytical grade and purchased from the Sinopharm Chemical Reagent (Shanghai, China).
Bacterial strains and culture conditions
A total of 8 clinical strains of H. parasuis were used in this study. Six strains were kindly provided by National Reference Laboratory of Veterinary Drug Residues at Huazhong Agricultural University (Wuhan, China). The other two strains were obtained from Fujian Academy of Agricultural Sciences (Fuzhou, China). These strains were isolated from the diseased pigs suffering fibrinous polyserositis, polyarthritis, and meningitis diagnosed in China during March 2014 to May 2018, and were confirmed by polymerase chain reaction (PCR). All of the clinical strains were stored at −80°C in milk. Prior to use, each strain was sub-cultured at least three times on modified tryptone soya agar (TSA) (BD Biosciences, Franklin Lakes, NJ, United States) supplemented with 5 μg/mL of nicotinamide adenine dinucleotide (NAD) (Sigma-Aldrich, St. Louis, MO, United States) and 5% (v/v) new-born calf serum (Tianhang Biotechnology Co. Ltd., Huzhou, Zhejiang Province, China) to ensure viability and purity. Tryptone soya broth (TSB) (BD Biosciences, Franklin Lakes, NJ, United States) and modified TSB supplemented with 5 μg/mL of NAD and 5% (v/v) new-born calf serum were used for screening of β-lactam-resistant strains, determining possible resistance mechanism, and investigating the reversal effect of MT on the resistance of H. parasuis strains to β-lactams. E. coli (ATCC25922) and Staphylococcus aureus (S. aureus, ATCC25923 and ATCC29213) were kept in our laboratory.
Screening of β-lactam-resistant strains
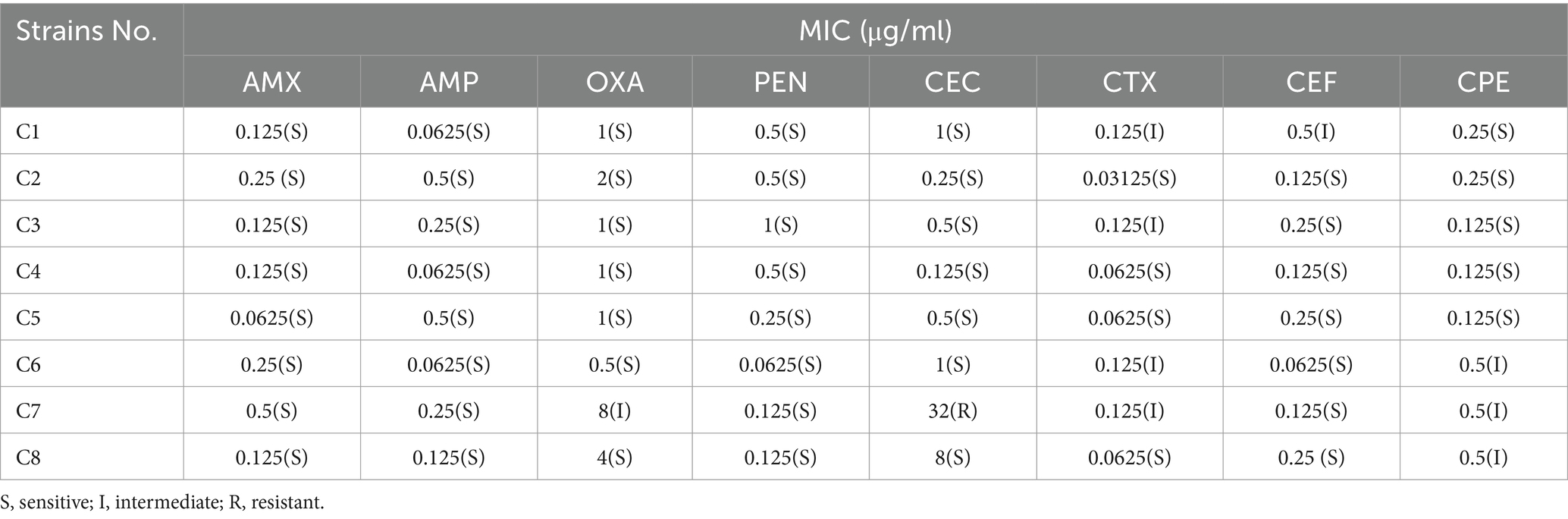
The susceptibility of 8 clinical H. parasuis strains to β-lactams was determined using the microdilution method according to the Clinical and Laboratory Standards Institute (CLSI, 2020). Eight β-lactams were tested, included AMX, AMP, OXA, PEN, CEC, CTX, CEF, and CPE. Aliquots of modified TSB supplemented with β-lactams (100 μL) at concentrations ranging from 0.015625 to 256 μg/mL were prepared in a 96-well plate. An exponentially growing bacterial culture was diluted with modified TSB to 1.0 × 106 cfu/mL, and then 100 μL of diluted bacteria was added to each treatment. The mixture was incubated at 37°C for 24–48 h. E. coli (ATCC25922) and S. aureus (ATCC29213) were used for quality control. All experiments were repeated three times. The minimum inhibitory concentration (MIC) was defined as the lowest concentration at which the wells remained clear. β-lactam-resistant strain was identified according to the determined MICs and the breakpoints established by Chen et al. (2021). The MIC of MT against β-lactam-resistant strain was also determined as described above.
Determination of possible resistance mechanism
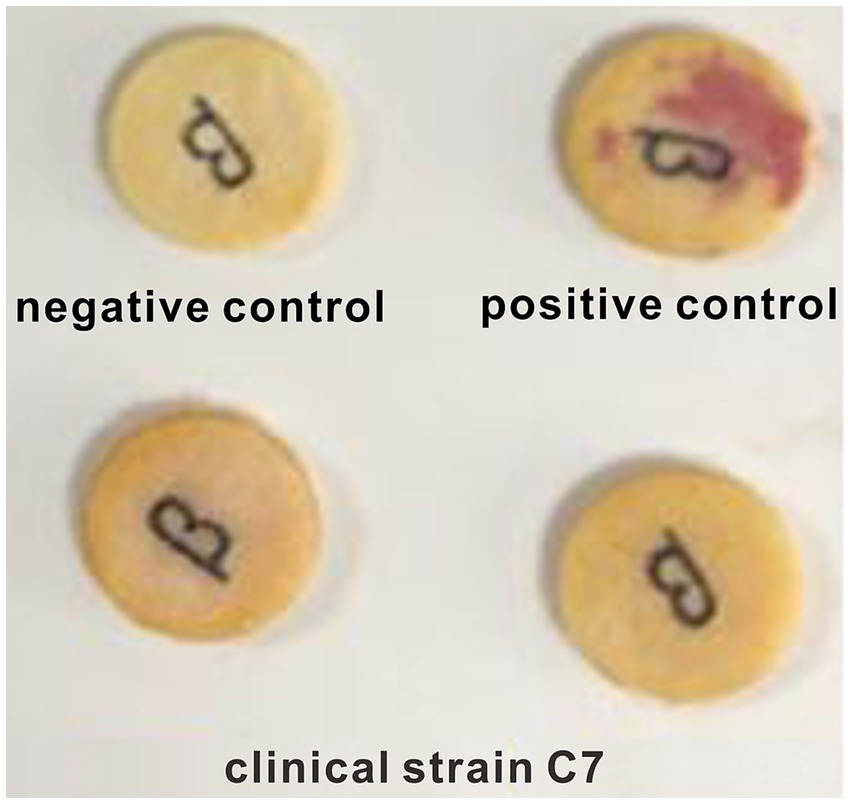
β-lactamase activity in β-lactam-resistant strain was assessed by nitrocefin test and enzyme linked immunosorbent assay (ELISA). The tested strain was sub-cultured twice, then a single colony was selected and applied to nitrocefin paper (Wenzhou Kangtai Biotechnology Co. Ltd., Wenzhou, China). Visual observation of the colour change of the nitrocefin paper was performed after standing at room temperature for 5 min. S. aureus (ATCC29213) and S. aureus (ATCC25923) were used as negative and positive controls, respective. ELISA was performed according to the manufacturer’s instructions of β-lactamase ELISA kit (Shanghai Zhuochai Biology Co. Ltd., Shanghai, China). All experiments were repeated three times. β-lactamase-producing strain was identified according to the colour change (from yellow to red) of the nitrocefin paper and the results of ELISA.
Multidrug efflux pump activity in β-lactam-resistant strain was assessed by comparison the MICs to β-lactams before and after treatment with CCCP. For the treatment group, each test well contained CCCP at a final concentration of 10 μg/mL. The MICs were determined as described above. Efflux pump-positive phenotype was identified when the ratio of MIC to MIC_CCCP treatment was equal to or greater than 4.
Biofilm-forming ability of β-lactam-resistant strain was assessed by crystal violet staining. Briefly, 10 μL of exponentially growing bacterial culture (1.0 × 108 cfu/mL) was added into individual well of a 96-well plate containing 100 μL of modified TSB. The well containing only modified TSB was used as control. Then, the 96-well plate was incubated at 37°C for 48 h. After incubation, the plate was washed three times with 200 μL of PBS (pH = 7.2) and fixed with 100 μL of methanol for 15 min. The excess methanol was discarded and the plate was left to dry at ambient temperature. The plate was then strained with 1% (w/v) crystal violet for 5 min. Unbound dye was wash away with PBS (pH = 7.2) and the plate was air-dried again. Finally, 100 μL of 33% (v/v) glacial acetic acid was added to each well and the optical density (OD) was measured at 630 nm using a microplate reader (Bio-Rad Laboratories, Hercules, CA, United States). All experiments were repeated three times. Strain which has strong ability to form biofilm was identified when the ratio of OD_test to OD_control was equal to or greater than 2.
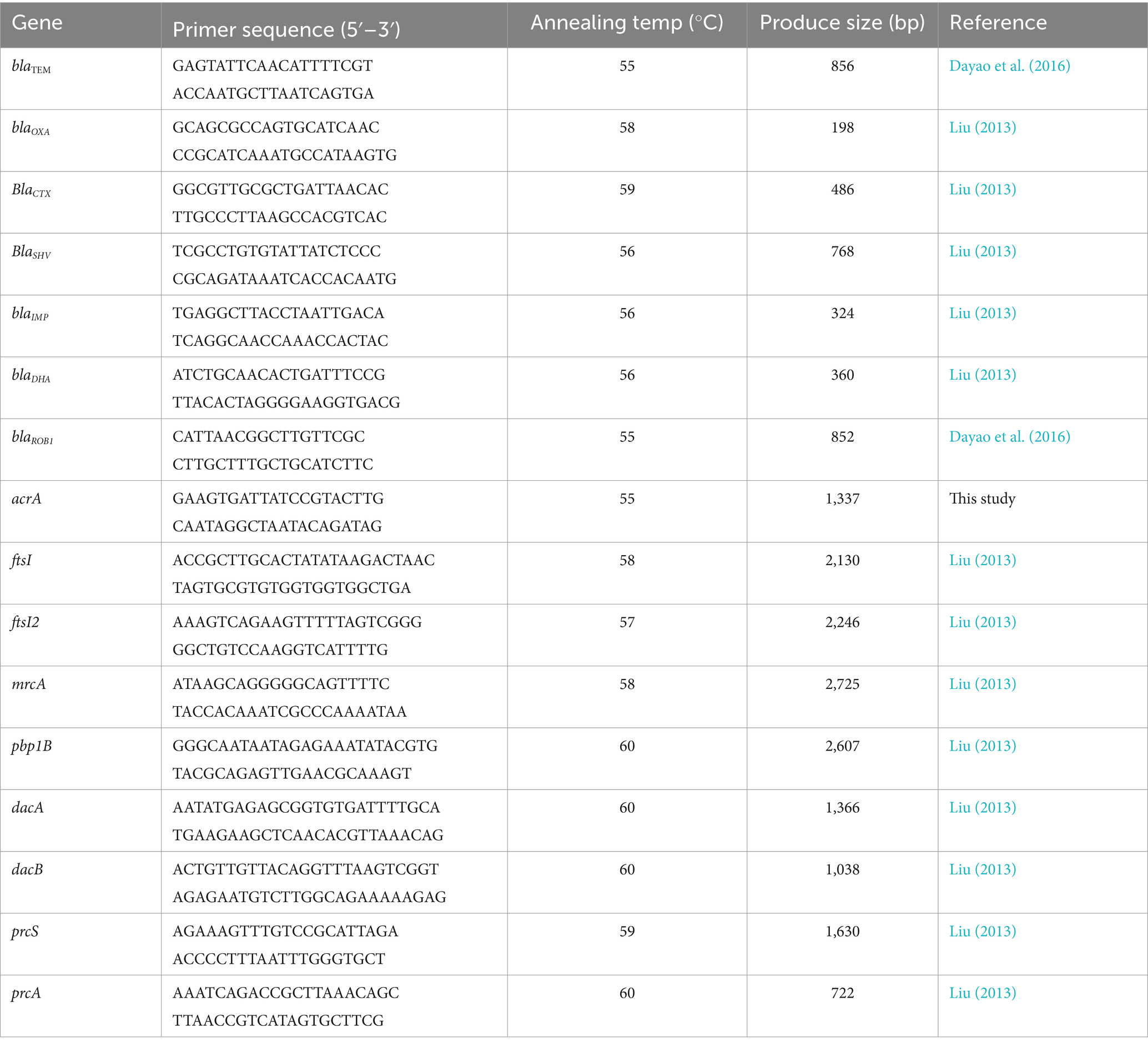
The presence of antimicrobial-resistant genes (ARGs) was determined by PCR in β-lactam-resistant strain. These ARGs included blaTEM, blaOXA, blaSHV, blaCTX, blaIMP, blaDHA, blaROB, blaROB1, and acrA. The genomic DNA was extracted using a TIANamp Bacteria DNA Kit (Tiangen Biotech Co. Ltd., Shanghai, China), according to the manufacture’s instruction. The primers for PCR (Table 1) were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China). The PCR system contained 12.5 μL of 2 × CataAmp TaqPCR Mix (Beijing Catascis Biotech Co. Ltd., Beijing, China), 2 μL of DNA template, 1 μL of forward primer, 1 μL of reverse primer, and 8.5 μL of RNase-free water. Amplification conditions began with an initial denaturation at 98°C for 3 min, followed by 30 cycles of 98°C for 10 s, 52–59°C for 20 s, and 72°C for 20 s, and terminated at 72°C for 10 min.
The mutation of penicillin-binding proteins (PBPs) genes in β-lactam-resistant strain was detected by PCR amplification and conventional DNA sequencing. These PBP genes included ftsI, ftsI2, mrcA, pbp1B, dacA, dacB, prcA, and prcS. The primes for PCR (Table 1) were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China). The PCR system contained 25 μL of 2 × CataAmp High Fidelity PCR Mix (Fuzhou Yihe Gene Technology Co. Ltd., Fuzhou, China), 1 μL of DNA template, 2 μL of forward primer, 2 μL of reverse primer, and 20 μL of RNase-free water. The DNA extraction and PCR amplification were performed as described above. The amplified products were sequenced by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China). The mutations in PBP genes were identified by comparing the sequencing results with the corresponding sequences in H. parasuis SH0165. To identify the association between the mutations in PBP genes and the resistance to β-lactams, the mutated PBP genes were cloned into PLB-T vector (Tiangen Biotech Co. Ltd., Beijing, China), and transformed into E. coil DH5α (Tiangen Biotech Co. Ltd., Beijing, China), according to the manufacturer’s instructions. Then, the susceptibility of these transformants to β-lactams was determined according to the microdilution method recommended by the CLSI (2020). Transformants containing wild-type PBP genes were used as controls.
Investigation into the reversal effect of MT on the resistance to β-lactams
The changes in the susceptibility of β-lactam-resistant H. parasuis to CEC were determined after treatment with various concentrations of MT. Briefly, 100 μL of exponentially growing bacterial culture (1.0 × 108 cfu/mL) was incubated with an equal volume of modified TSB contained various concentrations of MT (0, 0.25, 0.5, 4, 16, and 32 μg/mL) for 22 h at 37°C. Aliquots (100 μL) of the bacterial culture were taken at 1, 4, 7, 10, and 13 h and subsequently centrifugated at 3000 rpm for 5 min at 4°C. The pellets were washed with sterile PBS (pH = 7.2) to remove residual MT. Then, the bacteria were sub-cultured twice and cultured in modified TSB to the exponential growth phase (1.0 × 108 cfu/mL). The MICs of CEC against the exponentially growing bacteria were determined as described above. All experiments were repeated three times.
The changes in the biofilm-forming ability of β-lactam-resistant H. parasuis were determined after treatment with various concentrations of MT (0, 0.125, 0.25, 2, 8, and 16 μg/mL). The incubation of β-lactam-resistant strain with MT and the assessment of biofilm formation were performed as described above. All experiments were repeated three times.
The morphological change of β-lactam-resistant H. parasuis was observed using scanning electron microscopy (SEM, Hitachi Higher Technologies Co., Tokyo, Japan) after treatment with MT at 16 μg/mL for 7 h. The incubation procedure was the same as described above.
The changes in the relative expression level of mutated PBP genes were determined by real-time fluorescence quantitative PCR (qRT-PCR), after the β-lactam-resistant H. parasuis was treated with various concentrations of MT. The incubation procedure was the same as described above. The total RNA was extracted using a RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech Co. Ltd., Shanghai, China), according to the manufacture’s instruction. Then, the cDNA was synthesized according to the manufacture’s instruction of FastKing gDNA Dispelling RT Super-Mix kit (Tiangen Biotech Co. Ltd., Shanghai, China). QRT-PCR was performed with Taq SYBR Green qPCR Premix (Yeasen Biotechnology Co. Ltd., Shanghai, China) using the Roche LightCycler® 384 RT-PCR system (Roche, Basel, Switzerland). Relative quantification was performed by the 2−ΔΔCt method, and ppiB was used as internal control. The primers for qRT-PCR (Table 2) were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China). Each sample was measured with three technical replicates per target gene per independent experiment.
Statistical analysis
All values were expressed as means ± standard deviation (SD). Statistical analysis was performed using two-tailed t-test with SPSS version 21 (IBM Co., Armonk, NY, United States). A p-value of <0.05 was considered statistically significant.
Results
Susceptibility of clinical Haemophilus parasuis strains to β-lactams
The MICs of eight β-lactams against eight clinical H. parasuis strains are shown in Table 3. Most strains were susceptive or intermediate to eight β-lactams. Only one strain (clinical strain C7) showed resistance to CEC. This strain was used for subsequent experiments. The MIC of MT against the clinical strain C7 was 512 μg/mL.
Mechanism of resistance to CEC in clinical strain C7
A negative result was observed in nitrocefin test (Figure 1). Similar result was obtained by ELISA assay. The β-lactamase content in clinical strain C7 was much lower than the limit of detection (0.1 U/L) of ELISA method. The consistency was also observed between phenotype and genotype. None of β-lactamase-encoding genes was found in clinical strain C7. These results indicated that the clinical strain C7 was not a β-lactamase-producing H. parasuis.
Efflux pump inhibition test showed that the ratio of MIC to MIC_CCCP treatment was equal to 2, which suggested the resistance of clinical strain C7 to CEC might not be associated with the overexpression of AcrAB-TolC efflux pump. This speculation was supported by the fact that acrA gene was not detected in clinical strain C7.
Crystal violet staining showed that the ratio of OD_test to OD_control was equal to 1.15, which suggested the clinical strain C7 had a weak ability to form biofilm.
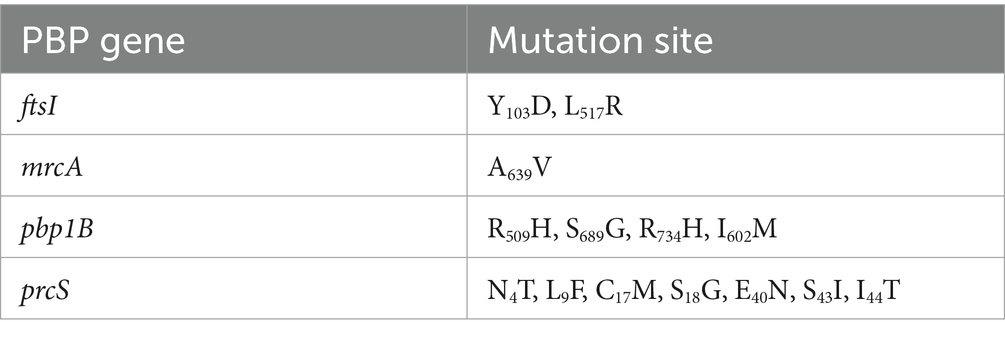
As shown in Table 4, a total of fourteen mutation sites were found in four PBP genes (ftsI, pbp1B, mrcA, and prcS). The presence of positive bands at 2607, 2725, 1,630, and 2,130 bp (Figure 2), coupled with subsequent sequencing results indicated that the transformants containing the PLB-T vector with mutated PBP genes had been successfully constructed. The MICs of transformants carrying mutated ftsI and mrcA genes to CEC were increased 4 and 2 times respectively, when compared with those of transformants carrying wild-type PBP genes. No changes in susceptibility to CEC were observed when the other mutated PBP genes (pbp1B and prcS) were cloned into PLB-T vector and then transformed into E. coil DH5α.
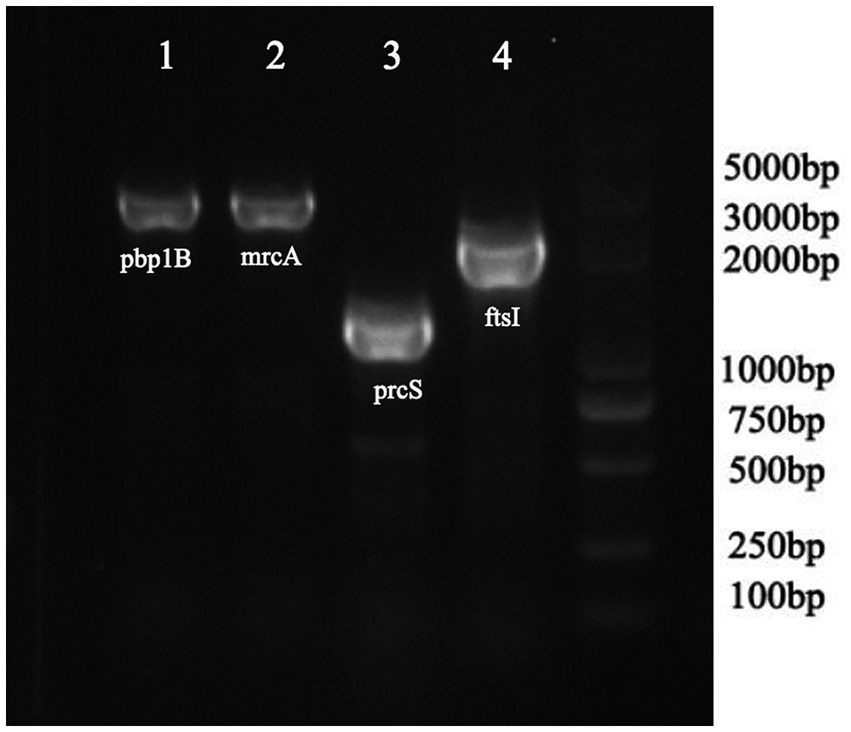
Figure 2. PCR analysis confirming the transformants containing the PLB-T vector with mutated PBP gens.
Reversal effect of MT on the resistance to CEC in clinical strain C7
As shown in Figure 3, a 2-to 8-fold decrease in MIC was observed after the clinical strain C7 was treatment with various concentrations of MT for 13 h. However, such reversal effect did not appear to be concentration-or time-dependent.
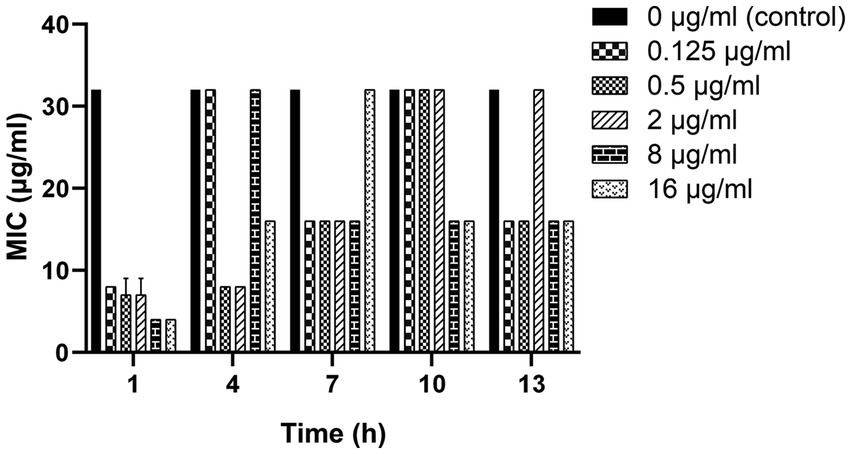
Figure 3. Changes in the susceptibility of clinical strain C7 to CEC after treatment with MT. Each bar represented the mean ± SD of the three independent experiments.
As shown in Figure 4, the biofilm-forming ability of clinical strain C7 was inhibited in the presence of low concentration of MT. However, such inhibitory effect was not significantly different (p > 0.05) from that of the control group (clinical strain C7 without treatment by MT).
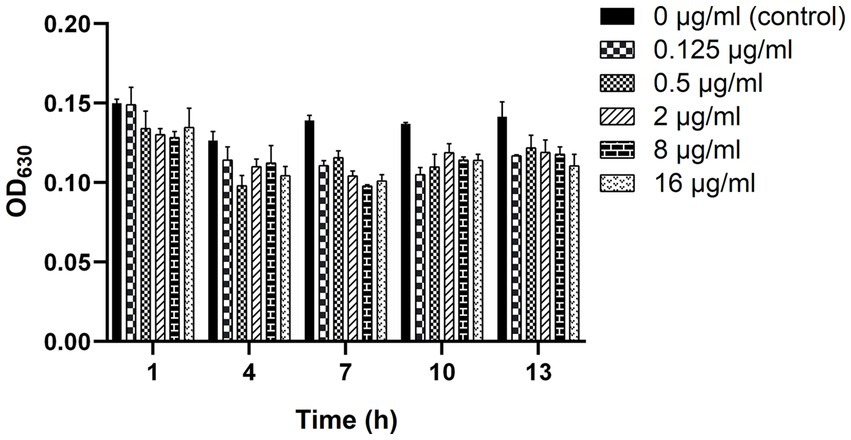
Figure 4. Changes in the biofilm-forming ability of clinical strain C7 after treatment with MT. Each bar represented the mean ± SD of the three independent experiments.
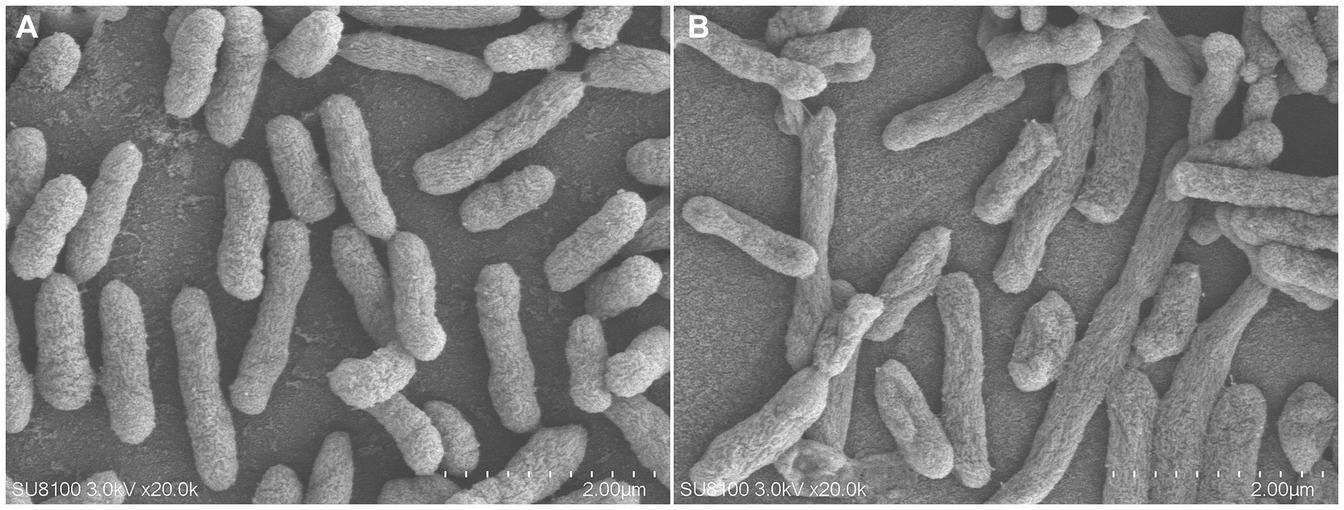
As shown in Figure 5, compared with the control group, the bacterial membrane of clinical strain C7 became wrinkled and concaved after treatment with MT at 16 μg/mL for 7 h.
As shown in Figure 6, significant decreases in the relative expressions of mutated ftsI and mrcA genes were observed after the clinical strain C7 was treated with various concentrations of MT for 1, 10 and 13 h. However, such changes did not appear to be concentration-or time-dependent too.
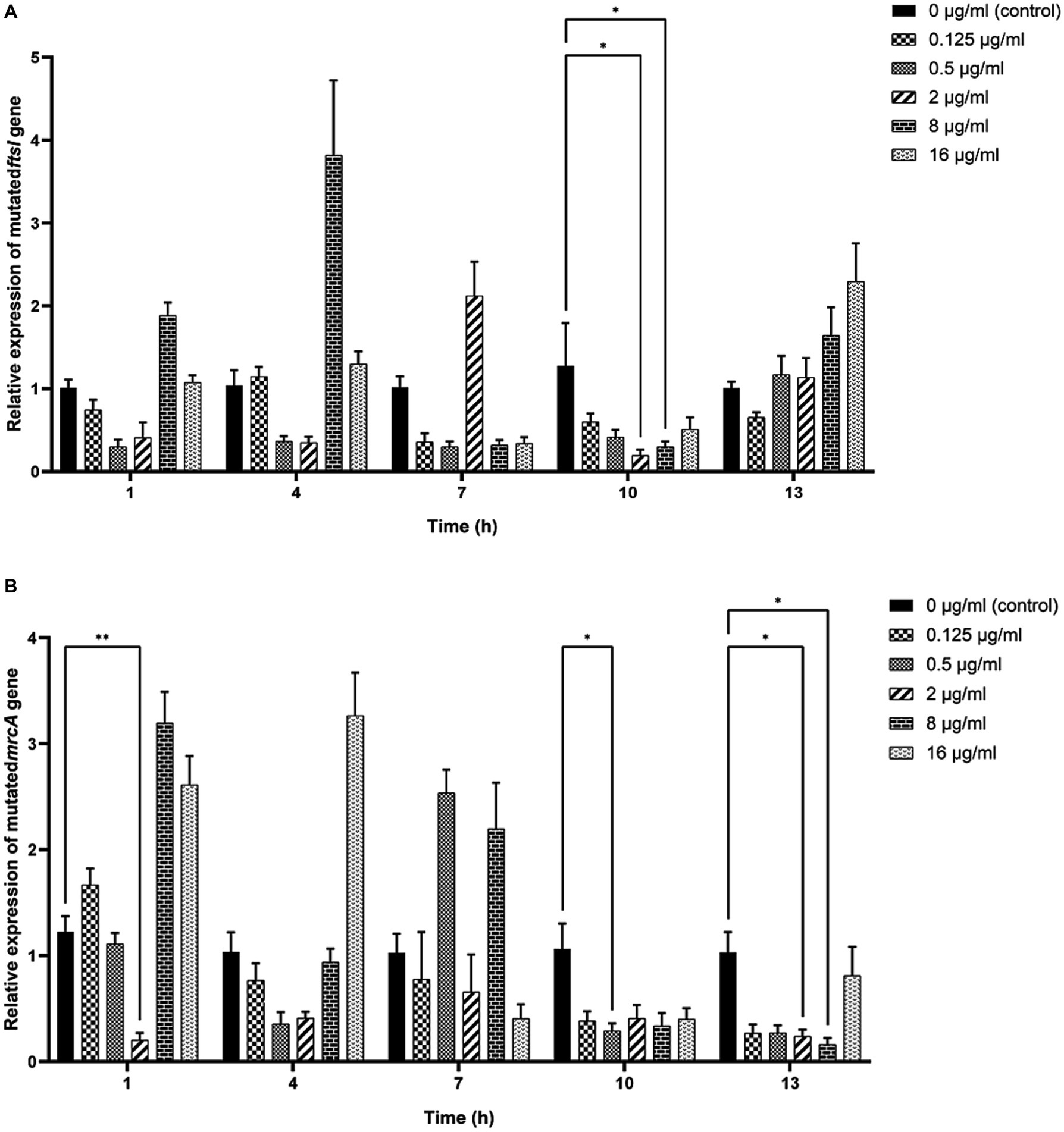
Figure 6. Effects of MT on the relative expression of mutated ftsI (A) and mrcA (B) genes after treatment with MT (* indicated a significant difference at p < 0.05; ** indicated a significant difference at p < 0.01). Each bar represented the mean ± SD of the three independent experiments.
Discussion
The widespread use of antimicrobials in livestock husbandry can induce antimicrobial resistance, which leads to the shortened service lifespan of antimicrobials and failure of anti-infective therapy. Screening of resistance-reversal agents and investigating their mechanism of action may be a promising strategy to address the challenge of antimicrobial resistance. In this study, we screened one CEC-resistant H. parasuis (clinical strain C7) from eight clinical H. parasuis strains and determined the underlying resistance mechanism. Then, we investigated the reversal effect of MT on the resistance of this strain to CEC. We believed that our research would provide useful information for restoring the antimicrobial activity of β-lactams and improving the therapeutic efficacy of Glässer’s disease.
Four possible mechanisms may contribute to β-lactam resistance in H. parasuis. First, the production of β-lactamase encoded by blaTEM, blaOXA, blaSHV, blaCTX and so no, which can inactivate β-lactams by hydrolyzing the β-lactam ring (San Millan et al., 2007; Yang et al., 2013; Moleres et al., 2015; An et al., 2023). Second, the overexpression of efflux pump such as AcrAB-TolC system, which can actively expel a wide range of toxic compounds, including β-lactams, from the cell (Feng et al., 2014). Third, the formation of biofilm, which was proved to be positive correlation with resistance to β-lactams (Zhang et al., 2014). Fourth, the mutations in PBP genes, which may reduce the affinity of PBP transpeptidases for β-lactams. To our knowledge, for H. parasuis, the association between the mutations in PBP genes and the resistance to β-lactams has not been identified. The results of our study showed that (i) the production of β-lactamase, overexpression of AcrAB-TolC system, and formation of biofilm might not be responsible for the resistance of clinical strain C7 to CEC; and (ii) fourteen mutation sites were found in four PBP genes (ftsI, pbp1B, mrcA, and prcS) of clinical strain C7, among which the mutation sites located in ftsI (Y103D and L517R) and mrcA (A639V) genes triggered the resistance to CEC, which was basically consistent with Liu’s prediction (Liu, 2013). This conclusion was also supported indirectly by the facts that the MIC of CEC against clinical strain C7 was reduced by two to eight folds after MT treatment, accompanied by the significant down-regulated expression of mutated ftsI and mrcA genes. Nevertheless, it should be noted that there might be some other drug efflux pumps besides AcrAB-TolC system on the surface of H. parasuis. Overexpression of these efflux pumps might still induce resistance of H. parasuis to CEC.
To investigate the antibiotic-resistance reversal effect of MT, the clinical strain C7 was treated with various concentrations of MT (0, 0.125, 0.25, 2, 8, and 16 μg/mL) for predetermined periods of time (1, 4, 7, 10, and 13 h). The concentrations and incubation time were chosen according to the plasma pharmacokinetic (PK) profiles of MT in rat (Jiang et al., 2015) and human (Zhang et al., 2009). When rat was administered orally at a dose of 10 mg/kg, the peak plasma concentration was 1.44 ± 0.14 μg/mL, and the elimination half-life was 7.80 ± 2.25 h. The corresponding PK parameters were 2.38 ± 0.72 μg/mL and 7.80 ± 0.70 h after the volunteers received a single oral dose of MT 400 mg soft gelatin capsule. For clinical strain C7, the concentration and time of exposure to MT were comparable to the plasma PK profiles of MT in rat and human. Although H. parasuis was commonly found in the upper respiratory tract of pigs (Little, 1970), the plasma PK profiles of MT could still be used as an alternative standard for evaluating the therapeutic efficacy because of the high tissue permeability of MT (Yang et al., 2010) and the correlation between plasma and tissue PK profile. From a pharmacokinetic/pharmacodynamic perspective, we believed that the antibiotic-resistance reversal effect of MT observed in vitro might also be present in vivo.
The antibiotic-resistance reversal effect of MT was proved by the 2-to 8-fold decrease in MIC of CEC against clinical strain C7 and significant down-regulated expression of mutated ftsI and mrcA genes after MT treatment. However, such changes did not appear to be concentration-or time-dependent. It was interesting that prolonged exposure to high concentration of MT (exposure to 16 μg/mL of MT for 13 h) caused the up-regulated expression of mutated ftsI gene (Figure 6), which would allow the clinical strain C7 to regain resistance to CEC. We speculated that this may be due to the sensitive perception of non-lethal effect of MT by H. parasuis. In fact, the bacterial membrane of clinical strain C7 became wrinkled and concaved after treatment with MT at 16 μg/mL (although much lower than the MIC) for 7 h (Figure 5). Such significant morphological changes might be sufficient for H. parasuis to perceive and respond to the non-lethal effect of MT. The mechanism of H. parasuis perceiving and responding to the resistance-reversal agent should be further investigated to achieve a significant and long-lasting antibiotic-resistance reversal effect.
Conclusion
In summary, the mutations in PBP genes (ftsI and mrcA) could increase the resistance of clinical strain C7 to CEC; MT could reverse the resistance of H. parasuis to CEC by inhibiting the mutations in ftsI and mrcA genes. A further study on the mechanism of H. parasuis perceiving and responding to the resistance-reversal agent should be undertaken.
Data availability statement
The datasets presented in this study can be found in online repositories at the NCBI under accession numbers: 391471, 391472, 391473, 391474.
Author contributions
JZ: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft. WY: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft. HD: Data curation, Methodology, Writing – review & editing. DoL: Data curation, Resources, Software, Writing – original draft. QW: Data curation, Formal analysis, Writing – original draft. LY: Methodology, Writing – review & editing. QK: Methodology, Validation, Writing – original draft. RX: Data curation, Validation, Writing – original draft. DiL: Validation, Visualization, Writing – original draft. RL: Validation, Visualization, Writing – original draft. DY: Supervision, Writing – original draft, Writing – review & editing. BY: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the grants from the Natural Science Foundation of Fujian Province of China (Grant no. 2023J01076) and the National Natural Science Foundation of China (Grant no. 32172906).
Acknowledgments
The clinical strains of H. parasuis were kindly donated by National Reference Laboratory of Veterinary Drug Residues at Huazhong Agricultural University (Wuhan, China) and Fujian Academy of Agricultural Sciences (Fuzhou, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, J., Zhang, C., and Li, Y. (2023). Outer membrane vesicles associated with the delivery of amoxicillin resistance in Glaesserella parasuis. Microb. Drug Resist. 29, 263–270. doi: 10.1089/mdr.2022.0184
Ayaz, M., Ullah, F., Sadiq, A., Ullah, F., Ovais, M., Ahmed, J., et al. (2019). Synergistic interactions of phytochemicals with antimicrobial agents: potential strategy to counteract drug resistance. Chem. Biol. Interact. 308, 294–303. doi: 10.1016/j.cbi.2019.05.050
Barbieri, R., Coppo, E., Marchese, A., Daglia, M., Sobarzo-Sánchez, E., Nabavi, S. F., et al. (2017). Phytochemicals for human disease: An update on plant-derived compounds antibacterial activity. Microbiol. Res. 196, 44–68. doi: 10.1016/j.micres.2016.12.003
Cai, W., Fu, Y., Zhang, W., Chen, X., Zhao, J., Song, W., et al. (2016). Synergistic effects of baicalein with cefotaxime against Klebsiella pneumoniae through inhibiting CTX-M-1 gene expression. BMC Microbiol. 16:181. doi: 10.1186/s12866-016-0797-1
Chen, C., Chen, J., Zhou, X., Wu, X., Wang, J., Huang, A., et al. (2021). Establishment of epidemiological cut-off values and determination of drug resistance of Haemophilus parasuis with β-lactam drugs. Acta Vet. Zootech. Sin. 52, 3234–3235.
Clinical and Laboratory Standards Institute (CLSI) (2020). “Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals” in CLSI supplement VET01SEd5E. 5th. eds. J. L. Watts, T. R. Shryock, M. Apley, D. J. Bade, S. D. Brown, J. T. Gray, et al. (Wayne, PA: CLSI).
Costa-Hurtado, M., Barba-Vidal, E., Maldonado, J., and Aragon, V. (2020). Update on Glässer's disease: how to control the disease under restrictive use of antimicrobials. Vet. Microbiol. 242:108595. doi: 10.1016/j.vetmic.2020.108595
Dayao, D., Gibson, J. S., Blackall, P. J., and Turni, C. (2016). Antimicrobial resistance genes in Actinobacillus pleuropneumoniae, Haemophilus parasuis and Pasteurella multocida isolated from Australian pigs. Aust. Vet. J. 94, 227–231. doi: 10.1111/avj.12458
Feng, S., Xu, L., Xu, C., Fan, H., Liao, M., and Ren, T. (2014). Role of acrAB in antibiotic resistance of Haemophilus parasuis serovar 4. Vet. J. 202, 191–194. doi: 10.1016/j.tvjl.2014.05.045
Itzia Azucena, R. C., José Roberto, C. L., Martin, Z. R., Rafael, C. Z., Leonardo, H. H., Gabriela, T. P., et al. (2019). Drug susceptibility testing and synergistic antibacterial activity of curcumin with antibiotics against Enterotoxigenic Escherichia coli. Antibiotics (Basel) 8:43. doi: 10.3390/antibiotics8020043
Jiang, M., Wang, L., Jiang, W., and Huang, S. (2015). Simultaneous determination of 14-thienyl methylene matrine and matrine in rat plasma by high-performance liquid chromatography-tandem mass spectrometry and its application in a pharmacokinetic study. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 974, 126–130. doi: 10.1016/j.jchromb.2014.10.041
Khameneh, B., Iranshahy, M., Soheili, V., and Fazly Bazzaz, B. S. (2019). Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob. Resist. Infect. Control 8:118. doi: 10.1186/s13756-019-0559-6
Liu, Y. Y. (2013) Investigation and mechanism of drug resistance in H. parasuis. [dissertation/Docter's thesis]. [Wuhan (Hubei Province)]: Huazhong Agricultural University.
Moleres, J., Santos-López, A., Lázaro, I., Labairu, J., Prat, C., Ardanuy, C., et al. (2015). Novel blaROB-1-bearing plasmid conferring resistance to β-lactams in Haemophilus parasuis isolates from healthy weaning pigs. Appl. Environ. Microbiol. 81, 3255–3267. doi: 10.1128/AEM.03865-14
Pourahmad, R. J., and Mohammadi, P. (2018). The effect of total alkaloid extract of local Sophora alopecuroides on minimum inhibitory concentration and intracellular accumulation of ciprofloxacin, and acrA expression in highly resistant Escherichia coli clones. J. Glob. Antimicrob. Resist. 12, 55–60. doi: 10.1016/j.jgar.2017.09.005
Qu, S., Dai, C., Shen, Z., Tang, Q., Wang, H., Zhai, B., et al. (2019). Mechanism of synergy between tetracycline and quercetin against antibiotic resistant Escherichia coli. Front. Microbiol. 10:2536. doi: 10.3389/fmicb.2019.02536
San Millan, A., Escudero, J. A., Catalan, A., Nieto, S., Farelo, F., Gibert, M., et al. (2007). Beta-lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob. Agents Chemother. 51, 2260–2264. doi: 10.1128/AAC.00242-07
Sun, T., Li, X. D., Hong, J., Liu, C., Zhang, X. L., Zheng, J. P., et al. (2019). Inhibitory effect of two traditional Chinese medicine monomers, Berberine and Matrine, on the quorum sensing system of antimicrobial-resistant Escherichia coli. Front. Microbiol. 10:2584. doi: 10.3389/fmicb.2019.02584
Wei, S., Yang, Y., Tian, W., Liu, M., Yin, S., and Li, J. (2020). Synergistic activity of fluoroquinolones combining with Artesunate against multidrug-resistant Escherichia coli. Microb. Drug Resist. 26, 81–88. doi: 10.1089/mdr.2018.0463
Yang, Z., Gao, S., Yin, T., Kulkarni, K. H., Teng, Y., You, M., et al. (2010). Biopharmaceutical and pharmacokinetic characterization of matrine as determined by a sensitive and robust UPLC-MS/MS method. J. Pharm. Biomed. Anal. 51, 1120–1127. doi: 10.1016/j.jpba.2009.11.020
Yang, S. S., Sun, J., Liao, X. P., Liu, B. T., Li, L. L., Li, L., et al. (2013). Co-location of the erm(T) gene and blaROB-1 gene on a small plasmid in Haemophilus parasuis of pig origin. J. Antimicrob. Chemother. 68, 1930–1932. doi: 10.1093/jac/dkt112
You, L., Yang, C., Du, Y., Wang, W., Sun, M., Liu, J., et al. (2020). A systematic review of the pharmacology, toxicology and pharmacokinetics of Matrine. Front. Pharmacol. 11:01067. doi: 10.3389/fphar.2020.01067
Zhang, B., Ku, X., Yu, X., Sun, Q., Wu, H., Chen, F., et al. (2019). Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci. Rep. 9:9908. doi: 10.1038/s41598-019-45482-8
Zhang, X. L., Xu, H. R., Chen, W. L., Chu, N. N., Li, X. N., Liu, G. Y., et al. (2009). Matrine determination and pharmacokinetics in human plasma using LC/MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 3253–3256. doi: 10.1016/j.jchromb.2009.08.026
Zhang, J., Xu, C., Shen, H., Li, J., Guo, L., Cao, G., et al. (2014). Biofilm formation in Haemophilus parasuis: relationship with antibiotic resistance, serotype and genetic typing. Res. Vet. Sci. 97, 171–175. doi: 10.1016/j.rvsc.2014.04.014
Zhao, Y., Wang, Q., Li, J., Lin, X., Huang, X., and Fang, B. (2018). Epidemiology of Haemophilus parasuis isolates from pigs in China using serotyping, antimicrobial susceptibility, biofilm formation and ERIC-PCR genotyping. PeerJ 6:e5040. doi: 10.7717/peerj.5040
Keywords: Haemophilus parasuis, matrine, cefaclor, resistance, penicillin-binding protein, ftsI, mrcA
Citation: Zhao J, Yang W, Deng H, Li D, Wang Q, Yi L, Kuang Q, Xu R, Li D, Li R, Yu D and Yang B (2024) Matrine reverses the resistance of Haemophilus parasuis to cefaclor by inhibiting the mutations in penicillin-binding protein genes (ftsI and mrcA). Front. Microbiol. 15:1364339. doi: 10.3389/fmicb.2024.1364339
Edited by:
Zhigang Qiu, Tianjin Institute of Environmental and Operational Medicine, ChinaCopyright © 2024 Zhao, Yang, Deng, Li, Wang, Yi, Kuang, Xu, Li, Li, Yu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: DaoJin Yu, yudaojin@yeah.net; Bo Yang, ybwhsw@sina.com
†These authors have contributed equally to this work and share first authorship
 JingChao Zhao
JingChao Zhao Wen Yang1,2†
Wen Yang1,2† Hui Deng
Hui Deng Di Li
Di Li Bo Yang
Bo Yang