- 1Department of Botany, School of Biological Sciences, North-West University, Mmabatho, South Africa
- 2Department of Chemistry, Biology and Biotechnology, University of Perugia, Perugia, Italy
- 3Department of Biology, College of Science and Technology, Wenzhou-Kean University, Wenzhou, China
It is increasingly clear that plant genomes encode numerous complex multidomain proteins that harbor functional adenylyl cyclase (AC) centers. These AC containing proteins have well-documented roles in development and responses to the environment. However, it is only for a few of these proteins that we are beginning to understand the intramolecular mechanisms that govern their cellular and biological functions, as detailed characterizations are biochemically and structurally challenging given that these poorly conserved AC centers typically constitute only a small fraction (<10%) of complex plant proteins. Here, we offer fresh perspectives on their seemingly cryptic activities specifically showing evidence for the presence of multiple functional AC centers in a single protein and linking their catalytic strengths to the Mg2+/Mn2+-binding amino acids. We used a previously described computational approach to identify candidate multidomain proteins from Arabidopsis thaliana that contain multiple AC centers and show, using an Arabidopsis leucine-rich repeat containing protein (TAIR ID: At3g14460; AtLRRAC1) as example, biochemical evidence for multienzymatic activities. Importantly, all AC-containing fragments of this protein can complement the AC-deficient mutant cyaA in Escherichia coli, while structural modeling coupled with molecular docking simulations supports catalytic feasibility albeit to varying degrees as determined by the frequency of suitable substrate binding poses predicted for the AC sites. This statistic correlates well with the enzymatic assays, which implied that the greatly reduced AC activities is due to the absence of the negatively charged [DE] amino acids previously assigned to cation-, in particular Mg2+/Mn2+-binding roles in ACs.
Introduction
It has been established that cyclic nucleotide monophosphates, cyclic guanosine monophosphate, and cyclic adenosine monophosphate (cAMP), and their generating enzymes, guanylyl cyclases (GCs) and adenylyl cyclases (ACs), play critical roles in many diverse biological processes of living organisms ranging from prokaryotes (e.g., Escherichia coli) to the complex multicellular Homo sapiens (Moutinho et al., 2001; Newton and Smith, 2004; Schaap, 2005; Meier and Gehring, 2006; Lomovatskaya et al., 2008). It has also become clear that plant GCs (e.g., Meier et al., 2010; Qi et al., 2010; Kwezi et al., 2011; Mulaudzi et al., 2011; Irving et al., 2012; Turek and Gehring, 2016) have distinct and varied domain architectures and can be part of multifunctional enzymes or “moonlighting” proteins with two or more distinct functions (Jeffery, 2003; Irving et al., 2012; Muleya et al., 2014; Kwezi et al., 2018; Su et al., 2019). This is likely to be similar in plant ACs, whereby AC domains coexist and cofunction with other domains. Few ACs known for this include a class III AC protein (MpCAPE) in the liverwort, Marchantia polymorpha, with phosphodiesterase activity at its N-terminus end (Kasahara et al., 2016) and two Arabidopsis proteins, AtKUP5 and AtKUP7, with K+-uptake permease activity (Al-Younis et al., 2015, 2018).
Despite the recent discoveries of plant ACs, the mechanism of actions, cellular regulations, and molecular dynamics of such catalytic centers remain largely unclear. These poorly conserved ACs retain only key residues of their catalytic centers that have been incorporated into larger multidomain proteins likely through divergent evolution (Zhang and Ma, 2012; Guo and Fang, 2014). ACs and other poorly conserved enzymes of such nature have consistently recorded low catalytic activities (Lomovatskaya et al., 2011; Hartwig et al., 2014; Gehring and Turek, 2017; Wong et al., 2018), and concerns on their biological significance were previously raised and discussed (Ashton, 2011; Berkowitz et al., 2011). Typically constituting only a small fraction of a complex plant protein (<10%), detailed characterization of AC centers becomes more challenging both biochemically and structurally (Irving et al., 2012, 2018; Wong and Gehring, 2013a; Kwezi et al., 2018). Initial efforts to identify ACs in higher plants were based on the construction of a 14-amino-acid search motif derived from annotated and experimentally tested GCs and ACs catalytic centers whereby the amino acid at position 1 forms hydrogen bonds with purine, amino acid at position 3 confers substrate specificity, and amino acid in position 14 stabilizes the transition from ATP to cAMP. The [DE] amino acids at one to three residues downstream of position 14 participates in Mg2+/Mn2+ binding (Figure 1A; Gehring, 2010). This approach has successfully identified several ACs in Arabidopsis many of which have since been experimentally validated, while the catalytic roles of key amino acids at the catalytic centers have also been verified by mutagenesis experiments (Gehring and Turek, 2017; Wong et al., 2018). However, the role of [DE] in regulating catalytic efficiency has never been experimentally assessed nor verified. Here, we offer fresh perspectives on the regulation of these seemingly cryptic activities specifically presenting evidence for the presence of multiple functional AC centers in a single protein and linking their catalytic strengths to the (Mg2+/Mn2+)-binding [DE] amino acids.
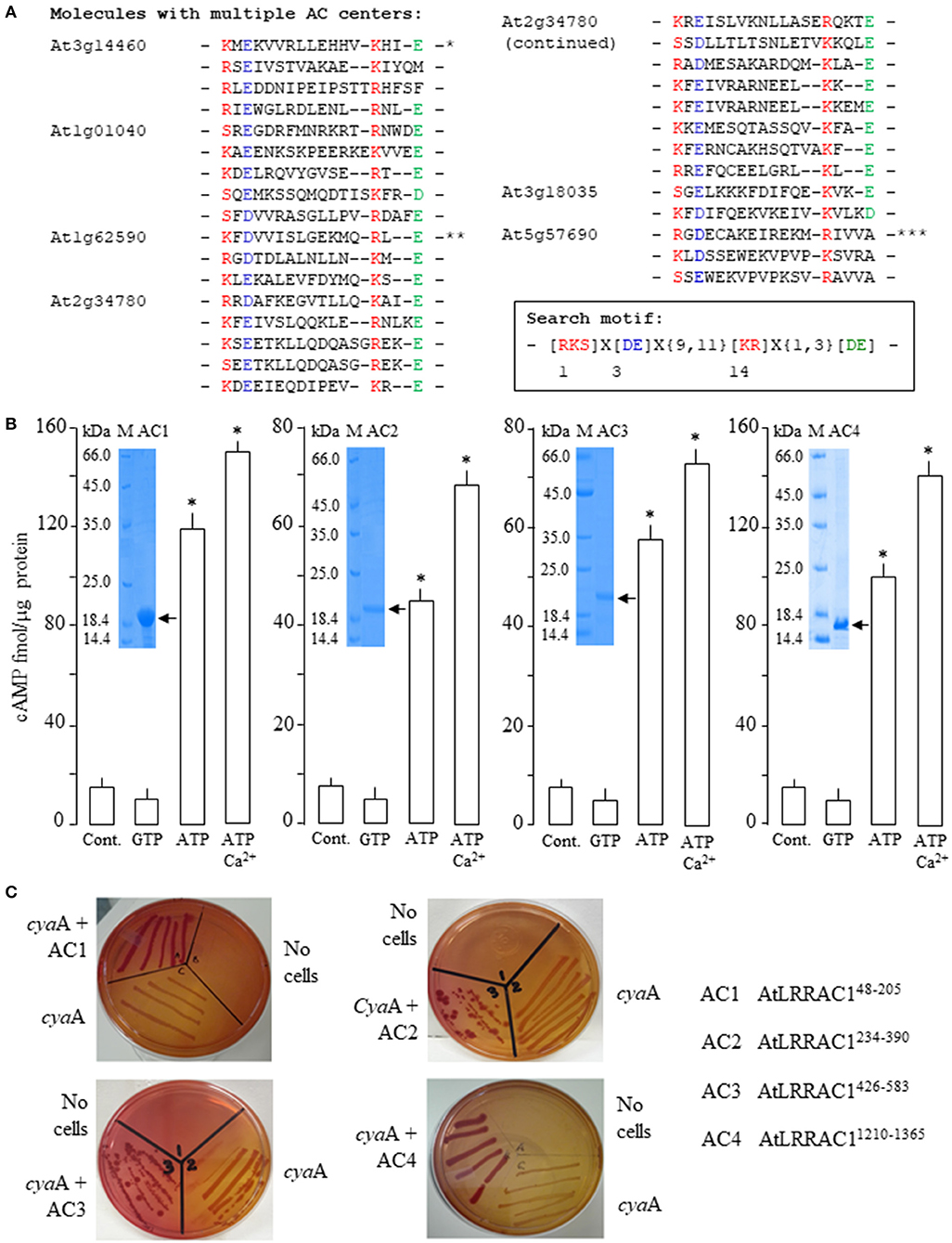
Figure 1. Computational identification of proteins with multiple adenylyl cyclase (AC) centers in Arabidopsis thaliana and functional characterizations of AtLRRAC1 AC catalytic centers. (A) Alignment of the AC catalytic centers of proteins with multiple AC centers in Arabidopsis thaliana. Asterisks denote AC centers previously confirmed to be catalytically active; *AtLRRAC1 (Ruzvidzo et al., 2013), **AtPPR-AC (Bianchet et al., 2019), and ***AtDGK4 albeit with extraordinarily high recombinant proteins used in the enzymatic assay (Dias et al., 2019). Inset: The 14-amino-acid AC search motif derived from annotated and experimentally tested guanylyl cyclases (GCs) and ACs catalytic centers. The residue forming hydrogen bonding with the purine at position 1 is highlighted in red, the residue conferring substrate specificity in position 3 is highlighted in blue, while the amino acid in position 14 that stabilizes the transition state from ATP to cyclic adenosine monophosphate (cAMP) is highlighted in red. The [DE] amino acid at one to three residues downstream from position 14 participates in Mg2+/Mn2+ binding and is colored green (Gehring, 2010). (B) Cyclic AMP generated by 5 μg of the AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365 recombinant proteins in the presence (at final concentrations) of 1 mM ATP or GTP, or 1 mM ATP and 250 μM Ca2+ when 5 mM Mn2+ ion is the cofactor (control reaction contained all other components except the protein and Ca2+). Insets: Coomassie brilliant blue-stained gels after resolution of the affinity-purified His-tagged recombinant AtLRRAC1 proteins (arrows) by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Data are mean values (n = 3), and error bars show SE of the mean. Asterisks denote values significantly different from those of the control (P < 0.05) as is determined by ANOVA and the post hoc Student–Newman–Keuls multiple range tests. (C) Complementation of cyaA mutation by the AC centers of AtLRRAC1. Recombinant AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365 proteins harboring the four AC centers of AtLRRAC1 complemented the cyaA SP850 mutant E. coli in lactose metabolism (Shah and Peterkofsky, 1991) as indicated by the growth of deep red colonies on MacConkey agar compared to the noncomplemented cyaA mutants that yielded yellowish colonies.
AtLRRAC1 Harbors Multiple Catalytically Active AC Centers that Complemented AC-Deficient E. coli
Applying a previously detailed computational approach to identify candidate AC domains in proteins from Arabidopsis thaliana (Ludidi and Gehring, 2003; Gehring, 2010; Wong and Gehring, 2013a; Wong et al., 2018), the At3g14460 gene was shown to encode a leucine-rich repeat (LRR) containing protein that harbors four distinct AC centers (Figure 1A), which are spatially distributed throughout the entire protein (Supplementary Figure 1). The AC centers are referred to as AC1, AC2, AC3, and AC4, respectively, and were tested individually for catalytic activity. Besides At3g14460, A. thaliana also harbors five other proteins with multiple AC centers: At1g01040 with 5, At1g62590 with 3, At2g34780 with 13, At3g18035 with 2, and At5g57690 with 3 centers (Figure 1A). Notably, At3g14460 (AtLRRAC1), At1g62590 (AtPPR-AC), and At5g57690 (AtDGK4) (Figure 1A) have been experimentally shown to be functional ACs (Ruzvidzo et al., 2013; Bianchet et al., 2019; Dias et al., 2019).
After the identification of the four AC centers in AtLRRAC1, each center was then separately cloned, expressed, and affinity purified to produce four respective truncated recombinant proteins: AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365 (Figure 1B, insets). When these truncated protein fractions were tested in vitro for their ability to convert ATP to cAMP, they all demonstrated significant Mn2+-dependent AC activities that could be enhanced by Ca2+ (Figure 1B). Incidentally, Ca2+-dependent activity increases in plant GCs and ACs have been observed previously (Muleya et al., 2014; Wheeler et al., 2017; Chatukuta et al., 2018), and in GCs, Ca2+ can act as switch between the kinase and GC activities of the dual-functioning phytosulfokine receptor (AtPSKR1) (Muleya et al., 2014). However, when comparing the in vitro AC activities of the four AC centers of AtLRRAC1, the activities of AtLRRAC1234−390 and AtLRRAC1426−583 were approximately half those of AtLRRAC148−205 and AtLRRAC11210−1365 (Figure 1B). We speculate that this reduction in activity is due to the absence of the negatively charged [DE] amino acids downstream of the AC catalytic centers of AtLRRAC1234−390 and AtLRRAC1426−583 (marked green in Figure 1A) responsible for Mg2+/Mn2+ binding (Ludidi and Gehring, 2003; Gehring, 2010).
To further support our in vitro AC activity findings, each of the four AC centers of AtLRRAC1 was tested to see if it could rescue the AC-deficient E. coli mutant strain SP850 with a cyaA mutation essential for lactose fermentation. When each of the AC centers was cloned and expressed in the SP850 mutant followed by growth on MacConkey agar, cells turned deep red, indicating a rescue to the wild-type phenotype (Figure 1C). This result corresponds well with the biochemical assay, which also showed catalytic activity in all four AC-containing recombinant proteins (Figure 1B).
AC Centers Harboring Negatively Charged Mg2+/Mn2+-Binding [DE] Amino Acids Docked with ATP at Higher Correct Binding Pose Frequencies
We then examined the structures of the four AC-containing fragments of AtLRRAC1 by iterative threading assembly (Zhang, 2008) followed by molecular docking of ATP to their AC centers using AutoDock Vina (Trott and Olson, 2010) and evaluated the binding poses and their free energies. We observed that ATP docked to AtLRRAC148−205 and AtLRRAC11210−1365 with its adenine and phosphate ends situated spatially close to the key amino acids of the AC centers (Figure 2A). However, in AtLRRAC1234−390 and AtLRRAC1426−583, ATP docked in a manner that its adenine or phosphate appears to be too distant for interactions with one of the key amino acids at the AC centers (see black arrows in Figure 2A). In addition to this spatial consideration, we also analyzed the orientations of docked ATP for “correct binding pose” according to previously ascertained deductions and rationales (Wong and Gehring, 2013b; Wong et al., 2015). Here, we define a “correct binding pose” as ATP orientation with its adenine head pointing toward the amino acid at position 1 and its phosphate tail pointing toward the positively charged amino acid at position 14 of the AC motif (Figure 1A inset; Supplementary Figure 2)–a definition that is consistent with that of previous works on plant ACs and GCs (Wong and Gehring, 2013b; Wong et al., 2015). This orientation of nucleotides has been deemed favorable for catalysis in previously characterized ACs and GCs (Al-Younis et al., 2015, 2018; Wheeler et al., 2017; Chatukuta et al., 2018; Bianchet et al., 2019) where, notably, mutagenesis of these residues have yielded reduced catalytic activities (Wheeler et al., 2017; Al-Younis et al., 2018). To evaluate the likelihood of the “correct ATP binding pose” at the AC centers of the AtLRRAC1 protein, a total of 18 solutions generated by AutoDock Vina were evaluated and expressed as percentage. We discovered that docking of ATP to the AC1 center yielded the highest “correct binding pose” frequency of 38.9% compared to the other AC centers. The “correct binding pose” frequencies for the other AC centers were 33.3% for the AC4 center, 16.7% for the AC3 center, and 11.1% for the AC2 center (Figure 2B). In AC2 and AC3 centers, even in instances where the “correct binding pose” was obtained, part of the ATP substrate (adenine or phosphate) seems too far from the key amino acids at the AC centers as indicated by black arrows in the surface models (Figure 2A; Supplementary Figure 2). Once again, this finding is consistent with our in vitro biochemical results that showed that the AtLRRAC148−205 and AtLRRAC11210−1365 recombinants generate approximately twice as much cAMP than the AtLRRAC1234−390 and AtLRRAC1426−583 recombinants (Figure 1B). All clusters, docking data and interpretation of docking solutions are provided in Supplementary Figure 2. Modeling and docking experiments have provided visualizations of how the respective catalytic centers could accommodate ATP. Furthermore, docking statistics not only provided valuable support to our biochemical data but also increased the degree of confidence of both the experimental results and our proposed roles for the [DE] residues in the motif. However, at this point, we are unsure how the absence of [DE] resulted in poorer ATP docking poses in AC2 and AC3, but we speculate that absence of [DE] resulted in loosened substrate binding pockets.
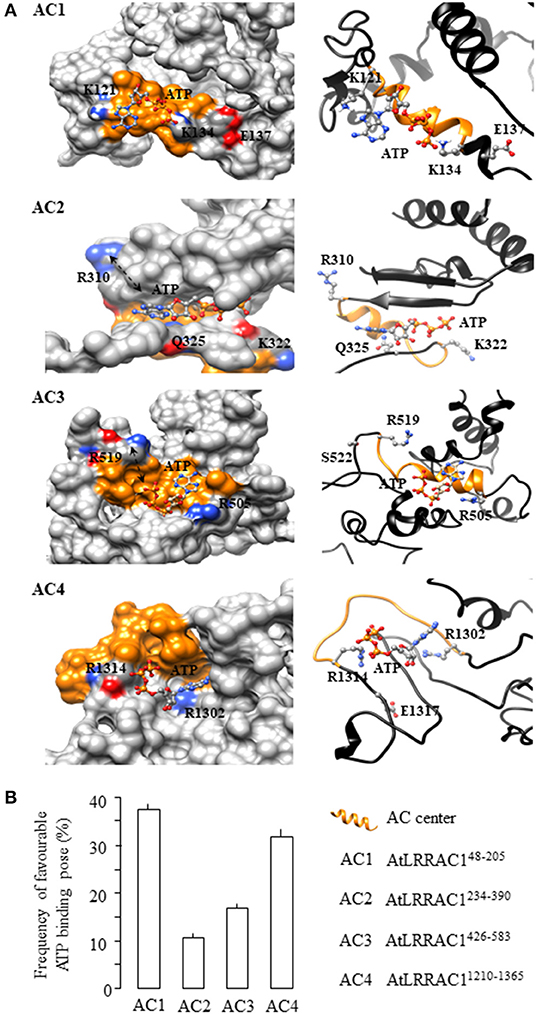
Figure 2. Computational assessment of the adenylyl cyclase (AC) centers of AtLRRAC1. (A) Representative images of the four AC-containing fragments of AtLRRAC1 (AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365) docked with ATP are shown as AC1, AC2, AC3, and AC4, respectively. The interaction of ATP with key residues at the AC catalytic centers of each fragment is shown as surface (left panel) and ribbon models (right panel), respectively. The amino acid residues at positions 1 and 14 of the AC centers, which are implicated in interactions with ATP, are colored according to their charges in the surface models and shown as individual atoms in the ribbon models. AC centers are colored orange, and black arrows in the surface models of AtLRRAC1234−390 and AtLRRAC1426−583 show the far distance between ATP and the key amino acids in the AC centers than in AtLRRAC148−205 and AtLRRAC11210−1365. Full-length AtLRRAC1 model was generated using the iterative threading assembly refinement (I-TASSER) method on the online server: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ (Zhang, 2008), and ATP docking simulations were performed using AutoDock Vina (ver. 1.1.2) (Trott and Olson, 2010). Molecular graphics and analyses were performed with the UCSF Chimera package (Pettersen et al., 2004). (B) Frequency of favorable ATP binding pose at the AC centers of AtLRRAC1 as estimated by molecular docking. All four AC-containing fragments of the AtLRRAC1 (AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365) were docked with ATP at their AC centers, and a total of 18 solutions generated by AutoDock Vina (ver. 1.1.2) (Trott and Olson, 2010) for each fragment were then evaluated and expressed as percentage. All clusters, docking data, and interpretation of docking solutions are provided in Supplementary Figure 2. Orientations and binding poses were analyzed with the UCSF Chimera package (Pettersen et al., 2004). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Taken together, structural assessments of the AC centers in AtLRRAC1 showed that the AtLRRAC1234−390 and AtLRRAC1426−583 fragments accommodated ATP less favorably (both spatially and in terms of frequency of the “correct binding pose”) compared to the AtLRRAC148−205 and AtLRRAC11210−1365 fragments. Again, it is tempting to speculate that this might be due to the absence of the negatively charged [DE] amino acids downstream of the AC catalytic centers of the AtLRRAC1234−390 and AtLRRAC1426−583 fragments (marked green in Figure 1). These amino acids appear at two amino acids downstream of the AC centers of the AtLRRAC148−205 and AtLRRAC11210−1365 fragments, and they normally appear at one to three amino acids downstream of the AC centers and have been previously assigned with the role of cation binding and in particular Mg2+ or Mn2+ (e.g., Gehring, 2010). Previous AC searches and characterizations have excluded hits without [DE] (Wong and Gehring, 2013b; Gehring and Turek, 2017; Wong et al., 2018), although the more recently created AC/GC prediction tools, ACPred and GCPred, offer the option for excluding [DE] in their servers (Xu et al., 2018a,b) to provide greater flexibility. Divalent cation (Mg2+/Mn2+) binding residues [DE] are known to coordinate the phosphates of ATP/ADP or GTP/GDP to the catalytic site of various enzymes including but not limited to kinases, ATP-/GTP-ases and phosphodiesterases. Divalent cation (Mg2+/Mn2+) binds to conserved [DE] amino acids in bacterial- and animal-soluble GCs typically at one to three residues downstream of the catalytic centers. Our AC and GC motifs were initially constructed by extracting key functional amino acids which are conserved at catalytic sites of canonical ACs and GCs across species, including the [DE] amino acids. Our motifs have since identified several ACs and GCs, many of which have been experimentally validated, but the role of [DE] in regulating their catalytic efficiency has never been assessed experimentally. Here, we showed for the first time that AC centers missing the [DE] residues have significantly reduced activities, and these preliminary data could explain past results, e.g., AtDGK4 by Dias et al. (2019) and/or guide future characterization works.
Discussion and Outlook
The possibility of proteins harboring multiple catalytically active AC centers is intriguing as it adds yet another layer of complexity to the regulation of signaling pathways in plant cells (Wong et al., 2015; Marondedze et al., 2017; Irving et al., 2018; Kwezi et al., 2018; Swiezawska et al., 2018; Su et al., 2019). Multiple functional AC centers in a single protein may also offer an additive effect which can accumulatively increase the catalytic strength. If these catalytic centers are proven to be functional in vivo, questions such as “are the centers catalytically active at the same time?” “how are their activities regulated?” and “do they cross-talk?” will require further investigations in vivo and in planta. Interestingly, a recent work by Dias et al. (2019) showed that the Arabidopsis thaliana DGK4 protein has GC but not AC activity in vitro with 10 or 40 μg recombinant protein. AC activity was only detected when the recombinant protein was raised to an unphysiologically high amount of 400 μg. Notably, this protein has a nucleotide cyclase center with the [DE] Mg2+/Mn2+-binding residue, and although it has three AC centers (Figure 1A), they all lacked the [DE] amino acids which might account for the weakened AC activity. From physiological studies, the authors have assigned this protein to various signaling roles regulating lipid composition, cytoskeletal dynamics, and pollen tube growth that affected fertilization in planta. Since the AC centers in DGK4 satisfied our AC motif (Figure 1A) and was also predicted by ACPred (Xu et al., 2018b), these could be indirect evidence for the molecular and biological roles of [DE] in GC/AC centers of such nature. Further mutagenesis experiments that add or remove the [DE] amino acids from the respective AC centers of AtLRRAC1 will validate our current data. We also hypothesize that the multidomain ACs might be capable of forming intramolecular dimers that in turn affect catalytic activities and, consequently, cAMP-dependent downstream signaling. In conclusion, our evidence for the presence of multiple functional AC centers in a single protein and linking catalytic strength to the Mg2+/Mn2+-binding [DE] amino acids has offered fresh perspectives that will contribute to the eventual goal of elucidating the intricate molecular regulations of such poorly conserved catalytic centers.
Materials and Methods
Generation of the AtLRRAC1 Recombinant Proteins
Total RNA was extracted from 6-week-old A. thaliana ecotype Columbia-0 (Col-0) seedlings using the RNeasy plant mini kit, in combination with DNase 1 treatment, as instructed by the manufacturer (Qiagen, Crawley, UK). Copy DNA (cDNA) sequence of AtLRRAC1 or At3g14460 was retrieved from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org) and checked for presence of multiple AC catalytic centers using the PROSITE database located within the Expert Protein Analysis System (ExPASy) proteomics server (https://www.expasy.org). cDNA synthesis from the total RNA and subsequent amplification of each of the identified multiple AC catalytic centers from the cDNA were simultaneously performed in the presence of a set of each of the respective sequence-specific primer pairs in Supplementary Table 1, using a Verso 1-Step RT-PCR kit and in accordance with the manufacturer's instructions (Thermo Scientific, Rockford, USA). The PCR products were then cloned into a pTrcHis2-TOPO expression vector via the TA cloning system (Invitrogen Corp., Carlsbad, USA) to make pTrcHis2-TOPO:AtLRRAC1 fusion expression constructs with C-terminus His purification tags. Expression, purification, and refolding processes of the recombinant AtLRRAC1 proteins were undertaken as is detailed elsewhere (Meier et al., 2010; Kwezi et al., 2011; Chatukuta et al., 2018) and their relative molecular masses estimated using the ProtParam tool on the ExPasy Proteomics Server (http://au.expasy.org/tool/.protpatram.html). The purified proteins were then used for in vitro enzymatic assays.
In vitro AC Enzymatic Assays and Detection of cAMP
The AC activity of each of the purified recombinant AtLRRAC1 proteins were determined in vitro by incubating 5 μg of the protein in 50 mM Tris–Cl buffer (pH 8.0), containing at, final concentration, 5 mM Mn2+, 1 mM ATP or 1 mM GTP, and 2 mM 3-isobutyl-1-methylxanthine, followed by measurement of the generated cAMP. Since Ca2+ has been shown to enhance the activity of ACs (Chatukuta et al., 2018) or GCs and indeed acting as a cellular switch between the GC and kinase activities of AtPSKR1 (Muleya et al., 2014), it was also tested against the AtLRRAC1 recombinants at a final concentration of 250 μM and in the presence of 5 mM Mn2+, 1 mM ATP, and 2 mM 3-isobutyl-1-methylxanthine. Levels of the generated cAMP were determined by enzyme immunoassaying following its acetylation protocol as described by the supplier's manual (Sigma-Aldrich Corp., Missouri, USA; code: CA201). The methods are detailed elsewhere (Ruzvidzo et al., 2013).
Complementation of cyaA Mutation in the E. coli AC-deficient Strain
The E. coli cyaA mutant SP850 strain [lam-, el4-, relA1, spoT1, cyaA1400 (:kan), thi-1] (Shah and Peterkofsky, 1991), deficient in the adenylyl cyclase (cyaA) gene, was obtained from the E. coli Genetic Stock Centre (Yale University, New Haven, USA) (accession no. 7200). The strain was prepared to be chemically competent followed by its transformation with the different pTrcHis2-TOPO:AtLRRAC1 fusion constructs (through heat shock at 42°C for 2 min). The transformed bacteria were then grown at 37°C for 18–40 h on MacConkey agar supplemented with 1% lactose, 15 μg/ml kanamycin, and 0.5 mM of the transgene inducer, isopropyl-β-D-thiogalactopyranoside (Sigma-Aldrich Corp., Missouri, USA) (Moutinho et al., 2001; Ruzvidzo et al., 2013; Swiezawska et al., 2015).
Computational Assessment of the AtLRRAC1 Multiple AC Catalytic Centers
An AtLRRAC1 model was generated using the iterative threading assembly refinement (I-TASSER) method (Zhang, 2008). The full-length AtLRRAC1 amino acid sequence (1,401 aa) was submitted to the I-TASSER server available online at http://zhanglab.ccmb.med.umich.edu/I-TASSER/, and a model with the highest C-score was downloaded from the server and used for the subsequent molecular docking experiments. Docking of ATP to AC centers of the various AtLRRAC1 fragments, AtLRRAC148−205, AtLRRAC1234−390, AtLRRAC1426−583, and AtLRRAC11210−1365 was performed using AutoDock Vina (ver. 1.1.2) (Trott and Olson, 2010). The AC centers of the various AtLRRAC1 fragments and ATP docking poses were analyzed and all images created by UCSF Chimera (ver. 1.10.1) (Pettersen et al., 2004). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). To evaluate the frequency of the “correct ATP binding pose” at the AC centers, a total of 18 solutions generated by AutoDock Vina were evaluated and expressed as percentage.
Statistical Analysis
All data of immunoassay in this work was subjected in triplicates (n = 3) to analysis of variance (ANOVA) (Super-Anova, Statgraphics Version 7, Statgraphics Corporation, USA). Where ANOVA revealed significant differences between treatments, means were separated by post hoc Student–Newman–Keuls multiple range test (P < 0.05).
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
OR, CG, and AW conceived and designed the study. OR performed the functional characterizations. AW undertook the structural works. All authors wrote the manuscript and read and approved its final version.
Funding
This project was funded by the National Research Foundation (NRF) of South Africa (Grant Nos. CSUR78843 and CSUR93635) awarded to OR and the National Natural Science Foundation of China (Grant No. 31850410470) and the Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ19C130001) awarded to AW.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2019.00136/full#supplementary-material
References
Al-Younis, I., Wong, A., and Gehring, C. (2015). The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic center. FEBS Lett. 589, 3848–3852. doi: 10.1016/j.febslet.2015.11.038
Al-Younis, I., Wong, A., Lemtiri-Chlieh, F., Schmöckel, S., Tester, M., Gehring, C., et al. (2018). The Arabidopsis thaliana K+-uptake permease 5 (AtKUP5) contains a functional cytosolic adenylate cyclase essential for K+ transport. Front. Plant Sci. 9:1645. doi: 10.3389/fpls.2018.01645
Ashton, A. R. (2011). Guanylyl cyclase activity in plants? Proc. Natl. Acad. Sci. U.S.A. 108:E96. doi: 10.1073/pnas.1101007108
Berkowitz, G. A., Gehring, C., Irving, H. R., and Kwezi, L. (2011). Reply to Ashton: the putative guanylyl cyclase domain of AtPepR1 and similar plant receptors. Proc. Natl. Acad. Sci. U.S.A. 108, E97–E98. doi: 10.1073/pnas.1103313108
Bianchet, C., Wong, A., Quaglia, M., Alqurashi, M., Gehring, C., Ntoukakis, V., et al. (2019). An Arabidopsis thaliana leucine-rich repeat protein harbors an adenylyl cyclase catalytic center and affects responses to pathogens. J. Plant Physiol. 232, 12–22. doi: 10.1016/j.jplph.2018.10.025
Chatukuta, P., Dikobe, T. B., Kawadza, D. T., Sehlabane, K. S., Takundwa, M. M., Wong, A., et al. (2018). An Arabidopsis clathrin assembly protein with a predicted role in plant defense can function as an adenylate cyclase. Biomolecules 8:E15. doi: 10.3390/biom8020015
Dias, F. V., Serrazina, S., Vitorino, M., Marchese, D., Heilmann, I., Godinho, M., et al. (2019). A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tube growth. New Phytol. 222, 1434–1446. doi: 10.1111/nph.15674
Gehring, C. (2010). Adenyl cyclases and cAMP in plant signaling - past and present. Cell Commun. Signal. 8:15. doi: 10.1186/1478-811X-8-15
Gehring, C., and Turek, I. S. (2017). Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant Sci. 8:1704. doi: 10.3389/fpls.2017.01704
Guo, T., and Fang, Y. (2014). Functional organization and dynamics of the cell nucleus. Front. Plant Sci. 5:378. doi: 10.3389/fpls.2014.00378
Hartwig, C., Bähre, H., Wolter, S., Beckert, U., Kaever, V., and Seifert, R. (2014). cAMP, cGMP, cCMP and cUMP concentrations across the tree of life: high cCMP and cUMP levels in astrocytes. Neurosci. Lett. 5, 183–187. doi: 10.1016/j.neulet.2014.07.019
Irving, H. R., Cahill, D. M., and Gehring, C. (2018). Moonlighting proteins and their role in the control of signaling microenvironments, as exemplified by cGMP and phytosulfokine receptor 1 (PSKR1). Front. Plant Sci. 9:415. doi: 10.3389/fpls.2018.00415
Irving, H. R., Kwezi, L., Wheeler, J., and Gehring, C. (2012). Moonlighting kinases with guanylate cyclase activity can tune regulatory signal networks. Plant Signal. Behav. 7, 201–204. doi: 10.4161/psb.18891
Jeffery, C. J. (2003). Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19, 415–417. doi: 10.1016/S0168-9525(03)00167-7
Kasahara, M., Suetsugu, N., Urano, Y., Yamamoto, C., Ohmori, M., Takada, Y., et al. (2016). An adenylyl cyclase with a phosphodiesterase domain in basal plants with a motile sperm system. Sci. Rep. 6:39232. doi: 10.1038/srep39232
Kwezi, L., Ruzvidzo, O., Wheeler, J. I., Govender, K., Iacuone, S., Thompson, P. E., et al. (2011). The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 286, 22580–22588. doi: 10.1074/jbc.M110.168823
Kwezi, L., Wheeler, J. I., Marondedze, C., Gehring, C., and Irving, H. R. (2018). Intramolecular crosstalk between catalytic activities of receptor kinases. Plant Signal. Behav. 13:e1430544. doi: 10.1080/15592324.2018.1430544
Lomovatskaya, L. A., Romanenko, A. S., and Filinova, N. V. (2008). Plant adenylate cyclases. J. Recept. Signal. Transduct. Res. 28, 531–542. doi: 10.1080/10799890802602308
Lomovatskaya, L. A., Romanenko, A. S., Filinova, N. V., and Dudareva, L. V. (2011). Determination of cAMP in plant cells by a modified enzyme immunoassay method. Plant Cell Rep. 30, 125–132. doi: 10.1007/s00299-010-0950-5
Ludidi, N., and Gehring, C. (2003). Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 278, 6490–6494. doi: 10.1074/jbc.M210983200
Marondedze, C., Wong, A., Thomas, L., Irving, H., and Gehring, C. (2017). “Cyclic nucleotide monophosphates in plants and plant signaling,” in Non-canonical Cyclic Nucleotides, ed R. Seifert (Cham: Springer International Publishing), 87–103.
Meier, S., and Gehring, C. (2006). Emerging roles in plant biotechnology for the second messenger cGMP-guanosine 3', 5'-cyclic monophosphate. Afr. J. Biotechnol. 5, 1687–1692. doi: 10.5897/AJB06.416
Meier, S., Ruzvidzo, O., Morse, M., Donaldson, L., Kwezi, L., and Gehring, C. (2010). The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE 5:e8904. doi: 10.1371/journal.pone.0008904
Moutinho, A., Hussey, P. J., Trewavas, A. J., and Malho, R. (2001). cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. U.S.A. 98, 10481–10486. doi: 10.1073/pnas.171104598
Mulaudzi, T., Ludidi, N., Ruzvidzo, O., Morse, M., Hendricks, N., Iwuoha, E., et al. (2011). Identification of a novel Arabidopsis thaliana nitric oxide-binding molecule with guanylate cyclase activity in vitro. FEBS Lett. 585, 2693–2697. doi: 10.1016/j.febslet.2011.07.023
Muleya, V., Wheeler, J. I., Ruzvidzo, O., Freihat, L., Manallack, D. T., Gehring, C., et al. (2014). Calcium is the switch in the moonlighting dual function of the ligand-activated receptor kinase phytosulfokine receptor 1. Cell Commun. Signal. 12:60. doi: 10.1186/s12964-014-0060-z
Newton, R. P., and Smith, C. J. (2004). Cyclic nucleotides. Phytochemistry 65, 2423–2437. doi: 10.1016/j.phytochem.2004.07.026
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Qi, Z., Verma, R., Gehring, C., Yamaguchi, Y., Zhao, Y. C., Ryan, C. A., et al. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. U.S.A. 107, 21193–21198. doi: 10.1073/pnas.1000191107
Ruzvidzo, O., Dikobe, B. T., Kawadza, D. T., Mabadahanye, G. H., Chatukuta, P., and Kwezi, L. (2013). “Recombinant expression and functional testing of candidate adenylate cyclase domains,” in Cyclic Nucleotide Signaling in Plants: Methods and Protocols, ed C. Gehring (Totowa, NJ: Humana Press), 13–25.
Schaap, P. (2005). Guanylyl cyclases across the tree of life. Front. Biosci. 10, 1485–1498. doi: 10.2741/1633
Shah, S., and Peterkofsky, A. (1991). Characterization and generation of Escherichia coli adenylate cyclase deletion mutants. J. Bacteriol. 173, 3238–3242. doi: 10.1128/jb.173.10.3238-3242.1991
Su, B., Qian, Z., Li, T., Zhou, Y., and Wong, A. (2019). PlantMP: a database for moonlighting plant proteins. Database 2019:baz050. doi: 10.1093/database/baz050
Swiezawska, B., Duszyn, M., Jaworski, K., and Szmidt-Jaworska, A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9:1428. doi: 10.3389/fpls.2018.01428
Swiezawska, B., Jaworski, K., Szewczuk, P., Pawelek, A., and Szmidt-Jaworska, A. (2015). Identification of a Hippeastrum hybridum guanylyl cyclase responsive to wounding and pathogen infection. J. Plant Physiol. 189, 77–86. doi: 10.1016/j.jplph.2015.09.014
Trott, O., and Olson, A. J. (2010). Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Turek, I., and Gehring, C. (2016). The plant natriuretic peptide receptor is a guanylyl cyclase and enables cGMP-dependent signaling. Plant Mol. Biol. 91, 275–286. doi: 10.1007/s11103-016-0465-8
Wheeler, J. I., Wong, A., Marondedze, C., Groen, A. J., Kwezi, L., Freihat, L., et al. (2017). The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J. 91, 590–600. doi: 10.1111/tpj.13589
Wong, A., and Gehring, C. (2013a). Computational identification of candidate nucleotide cyclases in higher plants. Methods Mol. Biol. 1016, 195–205. doi: 10.1007/978-1-62703-441-8_13
Wong, A., and Gehring, C. (2013b). The Arabidopsis thaliana proteome harbors undiscovered multi-domain molecules with functional guanylyl cyclase catalytic centers. Cell Commun. Signal. 11:48. doi: 10.1186/1478-811X-11-48
Wong, A., Gehring, C., and Irving, H. R. (2015). Conserved functional motifs and homology modelling to predict hidden moonlighting functional sites. Front. Bioeng. Biotechnol. 3:82. doi: 10.3389/fbioe.2015.00082
Wong, A., Tian, X., Gehring, C., and Marondedze, C. (2018). Discovery of novel functional centers with rationally designed amino acid motifs. Comput. Struct. Biotechnol. J. 16, 70–76. doi: 10.1016/j.csbj.2018.02.007
Xu, N., Fu, D., Li, S., Wang, Y., and Wong, A. (2018a). GCPred: a web tool for guanylyl cyclase functional center prediction from amino acid sequence. Bioinformatics 34, 2134–2135. doi: 10.1093/bioinformatics/bty067
Xu, N., Zhang, C., Lim, L. L., and Wong, A. (2018b). “Bioinformatic analysis of nucleotide cyclase functional centers and development of ACPred webserver,” in Proceedings of the ACM-BCB'18: 9th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics 2018 (Washington, DC), 122–129.
Zhang, L., and Ma, H. (2012). Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 195, 248–263. doi: 10.1111/j.1469-8137.2012.04143.x
Keywords: Arabidopsis thaliana, leucine-rich repeat, adenylyl cyclase, cAMP, multidomain proteins, multiple AC centers
Citation: Ruzvidzo O, Gehring C and Wong A (2019) New Perspectives on Plant Adenylyl Cyclases. Front. Mol. Biosci. 6:136. doi: 10.3389/fmolb.2019.00136
Received: 10 September 2019; Accepted: 13 November 2019;
Published: 03 December 2019.
Edited by:
Barbara Cellini, University of Perugia, ItalyReviewed by:
Song Xiang, Tianjin Medical University, ChinaChristopher Berndsen, James Madison University, United States
Copyright © 2019 Ruzvidzo, Gehring and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oziniel Ruzvidzo, b3ppbmllbC5ydXp2aWR6b0Bud3UuYWMuemE=; Aloysius Wong, QWx3b25nQGtlYW4uZWR1
 Oziniel Ruzvidzo
Oziniel Ruzvidzo Chris Gehring
Chris Gehring Aloysius Wong
Aloysius Wong