- 1Institute of Life Sciences, Nalco Square, Bhubaneswar, India
- 2School of Biotechnology, KIIT University, Bhubaneswar, India
Circular RNAs (circRNAs) are a newly discovered family of regulatory RNAs generated through backsplicing. Genome-wide profiling of circRNAs found that circRNAs are ubiquitously expressed and regulate gene expression by acting as a sponge for RNA-binding proteins (RBPs) and microRNAs (miRNAs). To identify circRNAs expressed in mouse skeletal muscle, we performed high-throughput RNA-sequencing of circRNA-enriched gastrocnemius muscle RNA samples, which identified more than 1,200 circRNAs. In addition, we have identified more than 14,000 and 15,000 circRNAs in aging human skeletal muscle tissue and satellite cells, respectively. A subset of abundant circRNAs was analyzed by RT-PCR, Sanger sequencing, and RNase R digestion assays to validate their expression in mouse skeletal muscle tissues. Analysis of the circRNA-miRNA-mRNA regulatory network revealed that conserved circNfix might associate with miR-204-5p, a suppressor of myocyte enhancer factor 2c (Mef2c) expression. To support the hypothesis that circNfix might regulate myogenesis by controlling Mef2c expression, silencing circNfix moderately reduced Mef2c mRNA expression and inhibited C2C12 differentiation. We propose that circNfix promotes MEF2C expression during muscle cell differentiation in part by acting as a sponge for miR-204-5p.
Introduction
Skeletal muscle is the largest organ contributing to one-third of the human body weight. Skeletal muscle is responsible for voluntary movement and has a high degree of regeneration ability. The fusion of the myoblasts generates the multinucleated long muscle fibers with contractile properties, and the process is known as myogenesis (Okazaki and Holtzer, 1966). The expression of muscle-specific transcription factors such as MyoD, Myog, Myf5, and Mef2a-d at various points collaboratively regulate multiple muscle formation steps, including myoblast proliferation, cell cycle exit, muscle-specific gene expression, myoblast fusion, and sarcomere assembly (Buckingham and Rigby, 2014). In addition to the well-known myogenic transcription factors, noncoding RNAs such as microRNAs, lncRNAs, and the recently discovered circular RNAs (circRNAs) were also involved in muscle cell differentiation and muscle development (Zhao et al., 2019; Das et al., 2020).
CircRNAs are one class of closed-loop single-stranded RNA molecules generated by the head-to-tail splicing of pre-mRNA (Chen and Yang, 2015). Due to the closed circular nature, circRNAs are resistant to exonucleases and very stable compared to linear RNAs. circRNAs have been reported to regulate gene expression by acting as sponges for RNA-binding proteins (RBPs) and miRNAs (Panda, 2018; Das et al., 2021). Also, a few circRNAs are translated into polypeptides with the cap-independent translation mechanisms, and some circRNAs regulate transcription by associating with snRNA and RNA pol II (Bose and Ain, 2018; Sinha et al., 2021). CircRNAs are dynamically expressed and involved in various physiological processes, including development, differentiation, and aging (Abdelmohsen et al., 2015; Di Timoteo et al., 2020). Interestingly, global upregulation of circRNAs was observed in the aged brain compared to young mice and drosophila (Westholm et al., 2014; Gruner et al., 2016). Although recent studies underscored the importance of circRNAs in muscle physiology, the expression and functions of circRNAs in muscle regeneration are still not understood completely.
This study sought to characterize circRNAs expressed in mouse skeletal muscle. Here, we identified circRNAs expressed in young and aged skeletal muscles. We also predicted their association with muscle miRNAs and downstream target genes. The circRNA-miRNA-mRNA regulatory network highlighted their possible role in muscle physiology. Also, we analyzed the previously published RNA-sequencing data from young and old human skeletal muscle and satellite cells to identify age-associated circRNAs. Together, our study provides new information to better understand the age-associated decline in muscle function and develop a new therapeutic strategy for age-associated muscle diseases.
Methods
Animals and Sampling
Young (6-weeks old) and aged (17-months old) male BALB/c mice were acquired from the Institute of Life Sciences breeding colonies. All animals were raised under standard conditions. The gastrocnemius muscle was collected from each mouse, washed in ice-cold PBS, and lysed in TRIzol for RNA isolation immediately or stored in −80°C for further use. All experimental procedures were conducted according to the approved guidelines of the institutional animal ethics committee of the Institute of Life Sciences.
Muscle Cell Differentiation, Circular RNA Silencing, and Immunostaining
High glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% FBS and antibiotics was used to culture the C2C12 mouse myoblast cell line in 5% CO2 at 37°C (Pandey et al., 2020). Human skeletal muscle myoblasts (HSMM) cells (Lonza) were cultured in Skeletal Muscle Cell Growth Medium-2 Bullet Kit (Lonza, CC-3245) and subcultured using subculture reagents (Lonza, CC-5034) following the instructions. The growth media of the sub-confluent C2C12 and HSMM cells were replaced with the differentiation media (DMEM supplemented with 2% horse serum), and the cells were differentiated into myotubes for up to 4 days. The C2C12 and HSMM differentiation was monitored with a phase-contrast microscope to check the formation of elongated multinucleated myotubes. The differentiated HSMM myotubes were immunostained with Anti-heavy chain Myosin/MYH3 antibody (Abcam# ab124205) followed by Goat Anti-rabbit IgG H&L (Alexa Fluor® 488) fluorescent secondary antibody against MYH3 antibody (Abcam #ab150077) and DAPI (Sigma #D9542) for nuclear staining. For circNfix silencing experiments, the C2C12 cells were transfected with control GapmeR (ctrl GapmeR) or circNfix GapmeR twice (36 and 12 h before inducing differentiation) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) following manufacturer’s instructions. The transfected cells were allowed to differentiate for 4 days before using them for differentiation analysis. The 4-day differentiated cells were stained with Jenner–Giemsa stains to analyze the differentiation efficiency as described previously (Velica and Bunce, 2011).
RNA Sequencing and Circular RNA Analysis
The total RNA from two young and two aged gastrocnemius muscles was prepared using TRIzol (Thermo). Seven μg of total RNA was treated with RNase R to enrich the circRNA population. A total of 500 ng RNase R treated RNA was fragmented, and the cDNA library was prepared using the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina, following the manufacturer’s instructions. The cDNA library quantity and quality was analyzed using Qubit and TapeStation, respectively. The libraries were sequenced for 75 bp paired-end reads on the Illumina NextSeq 550 platform using NextSeq 550 High Output Kit v2 (Illumina TG-160–2002); data are deposited in ENA (accession number PRJEB46548). The two young samples were sequenced with 53 and 62 million reads, while the aged samples were sequenced with 40 and 55 million reads. The adapter contamination was removed from the raw fastq files, then aligned to the mouse genome (mm10) using the STAR aligner using ChimSegmentMin-10 parameter. CIRCexplorer2 (v2.3.6) pipeline was used to identify the circRNAs (Zhang et al., 2016). Since circRNA read numbers were low for most circRNAs, we used transcripts per million (TPM) as normalized circRNA expression levels using the formula (circRNA read number/total reads in the sample x 1,000,000). The circRNA backsplice junction read numbers from two young and two aged muscle samples are provided in Supplementary Table S1.
Previously published total RNA-seq data of human skeletal muscle samples from young (22–35 years) and aged (70–82) were downloaded from NCBI GEO (Tumasian et al., 2021) (GEO accession: GSE164471; SRA ID: SRP300916) (Supplementary Table S2). In addition, total RNA-seq data of human muscle satellite cells from young and aged donors were obtained from NCBI GEO (GEO Accession No. GSE78611) using the SRA toolkit (v2.10.0). SRA data was converted into fastq data for further analysis. FastQC software (v0.11.2) was used to assess the sequencing quality of raw data in fastq format. The circRNA expression in these human muscle tissue and satellite cells was identified using CIRCexplorer2 (Zhang et al., 2016). The normalized circRNA expression in each samples of human muscle tissue and satellite cells were obtained using TPM calculation as described above (Supplementary Table S3, S4). A subset of abundant circRNAs with higher read numbers in the RNA-seq analysis and with length <2 Kb was selected for further analysis.
RT-PCR, Sanger Sequencing, RNase R Treatment, and QuantitativePCR
Total RNA from gastrocnemius muscle were isolated using TRIzol or Total RNA isolation kit (HiMedia) followed by cDNA synthesis using High Capacity cDNA Reverse Transcription kit following the manufacturer’s protocol (Thermo Fisher Scientific). The target circRNAs were PCR amplified with RNA-specific primers using the following program: denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 5 s, annealing, and extension at 60°C for 20 s (Supplementary Table S5) (Panda and Gorospe, 2018). The PCR amplicons were resolved on a 2% agarose gel stained with SYBR-Gold and visualized on an ultraviolet transilluminator. The circRNA backsplice junction sequences are confirmed by Sanger sequencing of the PCR products amplified with the divergent primers. Quantitative (q)PCR analysis of target RNAs was performed using target-specific primers and 2X PowerUP SYBR Green Master Mix (Thermo Fisher Scientific). The relative expression levels of the target RNA were calculated using the delta-CT method considering 18S rRNA or Gapdh mRNA as internal controls. Total RNA from C2C12 cells was treated with RNase R enzyme (Epicentre) followed by RT-qPCR analysis to test the circular nature of selected circRNAs (Panda and Gorospe, 2018).
Total RNA was reverse transcribed with miR-X first-strand synthesis kit following the manufacturer’s protocol (Takara). The cDNA was diluted, and RT-qPCR was performed using diluted cDNA, PowerUP SYBR Green Master Mix, and the specific forward primers for the miRNAs with the common reverse primer provided with the miR-X first-strand synthesis kit. The relative miRNA expression was measured with the delta-CT method using U6 as the endogenous loading control (Livak and Schmittgen, 2001).
Identification of circRNA-miRNA-mRNA Interactions
The spliced sequences of the selected circRNAs were provided as input in the custom prediction tool of the miRDB web tool and TargetScan software (release 7.2) to identify circRNA-associated miRNAs (Agarwal et al., 2015; Chen and Wang, 2020). The list of miRNAs expressed in mouse skeletal muscle was derived from a previous publication (GEO accession ID: GSE55164) (Kim et al., 2014). The miRNAs expressed in mouse skeletal muscle and the validated target genes in the miRTarBase release 8.0 were considered for further analysis (Huang et al., 2020) (Supplementary Table S6).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
The functions of circRNAs were determined by the genes targeted in the circRNA-miRNA-mRNA regulatory network. The STRING database (v11.5) was used for comprehensive analysis for the target genes using GO (http://www.geneontology.org/) and the KEGG (https://www.kegg.jp/) to understand the biological functions (Supplementary Table S7) (Kanehisa et al., 2016; The Gene Ontology, 2019; Szklarczyk et al., 2021). The GO enrichment analysis of the target genes found several significantly enriched (False Discovery Rate: FDR, p-value<0.05) GO terms, including Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). The enrichment score for the GO terms and KEGG pathways were calculated as Log10 of the FDR value and plotted as bubble plots.
Construction of the Competing Endogenous RNA (ceRNA) Network and Visualization
GraphPad Prism 6.0 software, R studio or Microsoft excel was used to plot the graphs for data visualization. We used Cytoscape software (v3.8.2) to construct and visualize the circRNA-miRNA-mRNA network in this study. The bubble plots were generated using ggplot2 in R studio. Statistical significance was calculated by student’s t-test and considered significant with a p-value of <0.05.
Results
Characteristics of circRNAs in Mouse Skeletal Muscle Tissue
We used gastrocnemius skeletal muscle of young and aged mice for the identification of skeletal muscle circRNAs. As shown in Figure 1A, RNA-seq libraries were prepared from circRNA enriched RNA samples followed by sequencing. The RNA-seq reads were further processed to identify circRNAs using CIRCexplorer2 (Zhang et al., 2016). Interestingly, the aged samples contained a higher percentage of chimeric reads and non-canonical splice junctions. Moreover, the total number of circRNAs identified in aged skeletal muscle was significantly higher than in young samples (Figure 1B). Consistent with previous reports, our data suggest an overall upregulation and accumulation of circRNAs in aged skeletal muscle (Gruner et al., 2016).
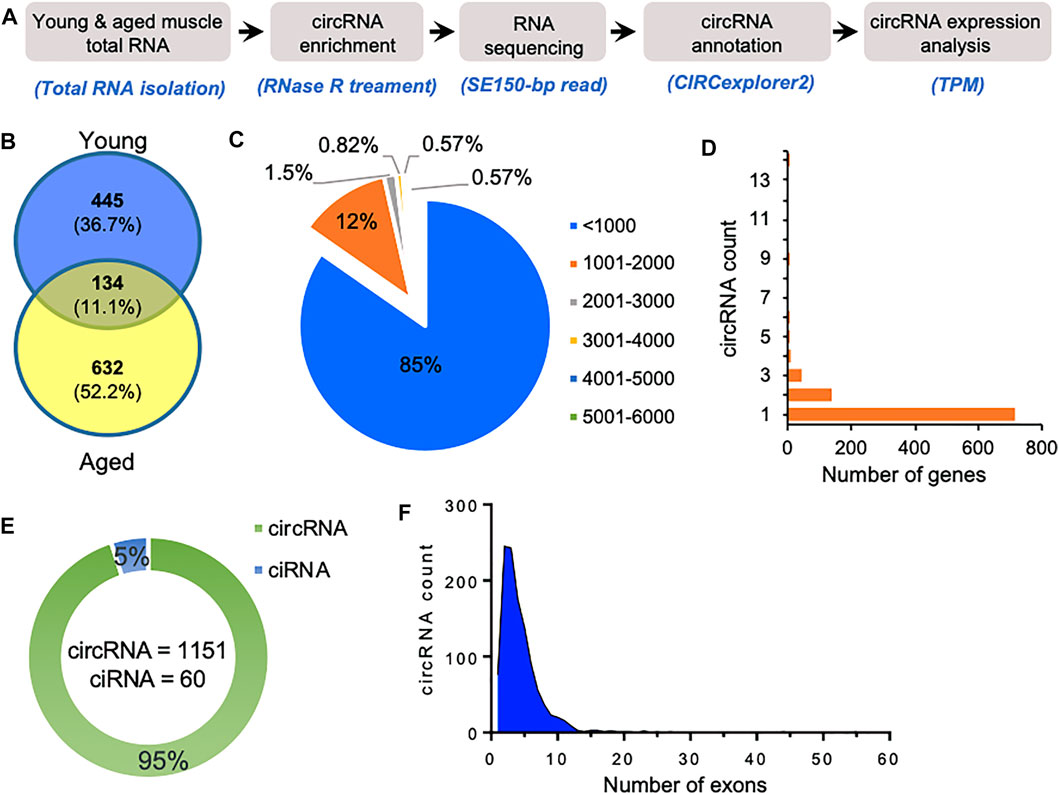
FIGURE 1. Characteristics of circRNAs expressed in aging mice skeletal muscle. (A). The experimental design and annotation of circRNAs using CIRCexplorer2 in mouse gastrocnemius muscle. (B). Venn diagram showing the number of circRNAs expressed in young and aged gastrocnemius muscle identified using CIRCexplorer2. (C). Percentage distribution of length of circRNAs in mouse skeletal muscle. (D). Number of genes corresponding to the number of circRNAs generated by them. (E). Number of exonic circRNA and intronic ciRNAs expressed in mouse skeletal muscle. (F). Distribution of the number of exons in the exonic circRNAs expressed in gastrocnemius muscle detected with CIRCexplorer2.
As shown in Figure 1C, most of the identified circRNAs were smaller than 1,000 nucleotides in length. Interestingly, most circRNA host genes generated only one or two circRNAs, while a few host genes generated multiple circRNAs (Figure 1D). Since circRNAs can be generated from the exonic and intronic sequences, our circRNA annotation suggested that ∼95% of circRNAs were exonic circRNAs while a few were intronic ciRNAs (Figure 1E). Analysis of the number of exons in the exonic circRNAs revealed that most circRNAs harbor less than ten exons (Figure 1F). The expression of circRNAs in young and aged skeletal muscle was performed using the TPM values. The complete list of the 1,211 circRNAs identified in aging mouse gastrocnemius muscle is provided (Supplementary Table S1), including circRNA annotation, chromosomal coordinates, length, and expression values. Moreover, only a few circRNAs were differentially expressed, and most of the circRNAs identified in mice skeletal muscle samples were expressed at a very low level. The low abundance and small number of circRNAs in the sequencing data could be due to fewer replicates, low depth, and smaller read-length in RNA sequencing.
Since circRNAs are known to be conserved across various species, we sought to identify circRNAs expressed in aging human skeletal muscle and muscle satellite cells. To identify circRNAs in human skeletal muscle, previously published total RNA-seq data from young (22–35 years) and aged (70–82 years) human skeletal muscle RNA-seq data were analyzed using CIRCexplorer2 pipeline (GEO ID: GSE164471) (Supplementary Table S3). We have identified more than 14,000 circRNAs, most derived from exonic sequences and less than 1,000 nucleotides in length (Supplementary Figure S1A–C). Moreover, most of the genes produced a few circRNAs, while a few genes such as TTN and NEB generate hundreds of circRNAs (Supplementary Figure S1D,E). Furthermore, we also analyzed the circRNAs expressed in young and aged muscle satellite cells (Supplementary Table S4, Supplementary Figure S2). We also identified more than 15,000 circRNAs, where 90% are exonic circRNAs, and the rest are intronic ciRNAs (Supplementary Figure S2).
Validation of circRNAs in Mice Skeletal Muscle
Since most of the circRNAs were very low abundant in the RNA-seq data, we selected a few highly expressed circRNAs for differential expression analysis using quantitative RT-PCR (Figure 2A). Using divergent primers, RT-PCR amplification of the circRNAs specifically amplified the target circRNAs without amplifying the no-RT control (Supplementary Table S5, Figure 2B). We also performed RNase R exonuclease treatment followed by RT-qPCR to analyze the circular nature of the circRNAs. The linear Gapdh mRNA was degraded to minimal levels while all tested circRNAs were resistant to RNase R digestion, confirming the circular nature of the circRNAs (Figure 2C). Furthermore, Sanger sequencing of the PCR products confirmed the amplification of the backsplice junction of the target circRNAs (Figure 2D, Supplementary Figure S3). Among the tested circRNAs, circCrebrf was moderately upregulated along with elevated expression of age-associated p53, p16, and Smad3 mRNAs in the aged muscle samples compared to the young muscle (Figures 2E,F).
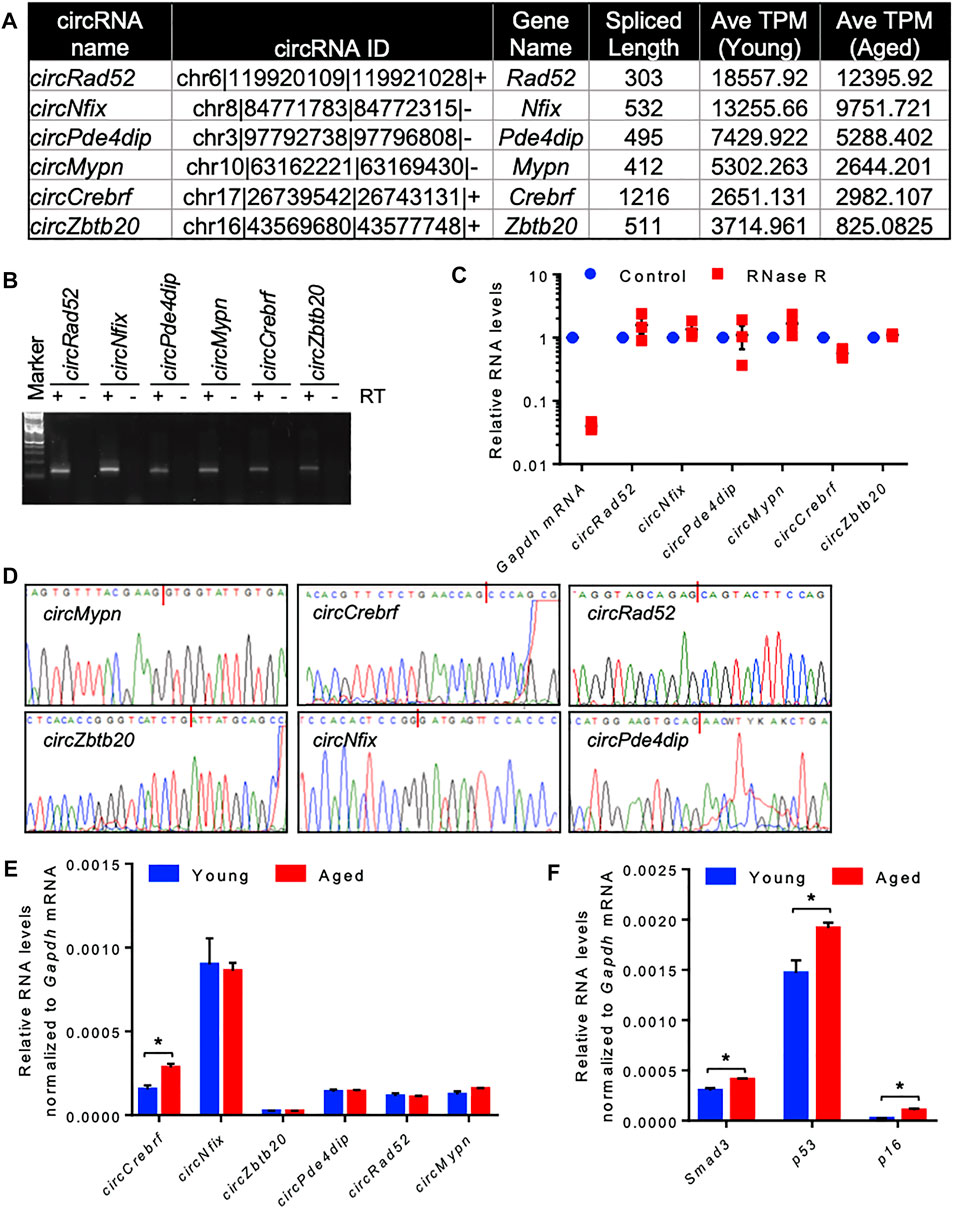
FIGURE 2. Differential expression and validation of circRNAs in aging skeletal muscle. (A). A list of abundant circRNAs was selected for further validation. (B). The RT-PCR products of circRNAs from muscle tissue were resolved on a 2% agarose gel stained with SYBR Gold. (C). RT-PCR analysis of circRNAs and linear RNAs upon RNase R treatment. (D). PCR products of circRNAs from muscle were purified and sequenced to confirm the backsplice sequences. (E,F). RT-qPCR analysis of circRNAs (E) and mRNAs (F) in young and aged skeletal muscle normalized to Gapdh mRNA. Data in (C,E,F) are the means ± SEM from three independent experiments. *, p < 0.05.
Prediction of Potential circRNA-miRNA-mRNA Regulatory Axis in Skeletal Muscle
Recent studies established that circRNAs function as inhibitors of circRNA-interacting miRNAs, thereby regulating the target gene expression. To identify the circRNA associated miRNAs, the circRNA sequences were used to predict the target miRNAs using miRDB and TargetScan software (Agarwal et al., 2015; Chen and Wang, 2020). Computational prediction identified 102 potential miRNAs targeting the six validated circRNAs using miRDB, and 234 miRNAs were identified with TargetScan (Supplementary Table S6). The previous publication reported the expression of 877 miRNAs in mouse gastrocnemius muscle (Supplementary Table S6) (Kim et al., 2014). To find the functional miRNA in skeletal muscle targeted by these circRNAs, we identified the common muscle miRNAs targeted by these six circRNAs and have experimentally validated mRNA targets reported by miRTarBase (Figure 3A; Supplementary Table S6) (Huang et al., 2020). This analysis identified fifteen functional circRNA-associated miRNAs, including mouse miR-181a-5p, miR-181b-5p, miR-181c-5p, miR-181d-5p, miR-194-5p, miR-204-5p, miR-211-5p, miR-218-5p, miR-27a-3p, miR-27b-3p, miR-299a-3p, miR-3064-5p, miR-3085-3p, miR-488-3p, and miR-668-3p (Figure 3A). Interestingly, these fifteen miRNAs were associated with five out of six circRNAs (Figures 3B,C). Analysis of the downstream genes for these miRNAs using the miRTarBase 8.0 database revealed that these fifteen miRNAs have 866 validated mRNA targets (Supplementary Table S6).
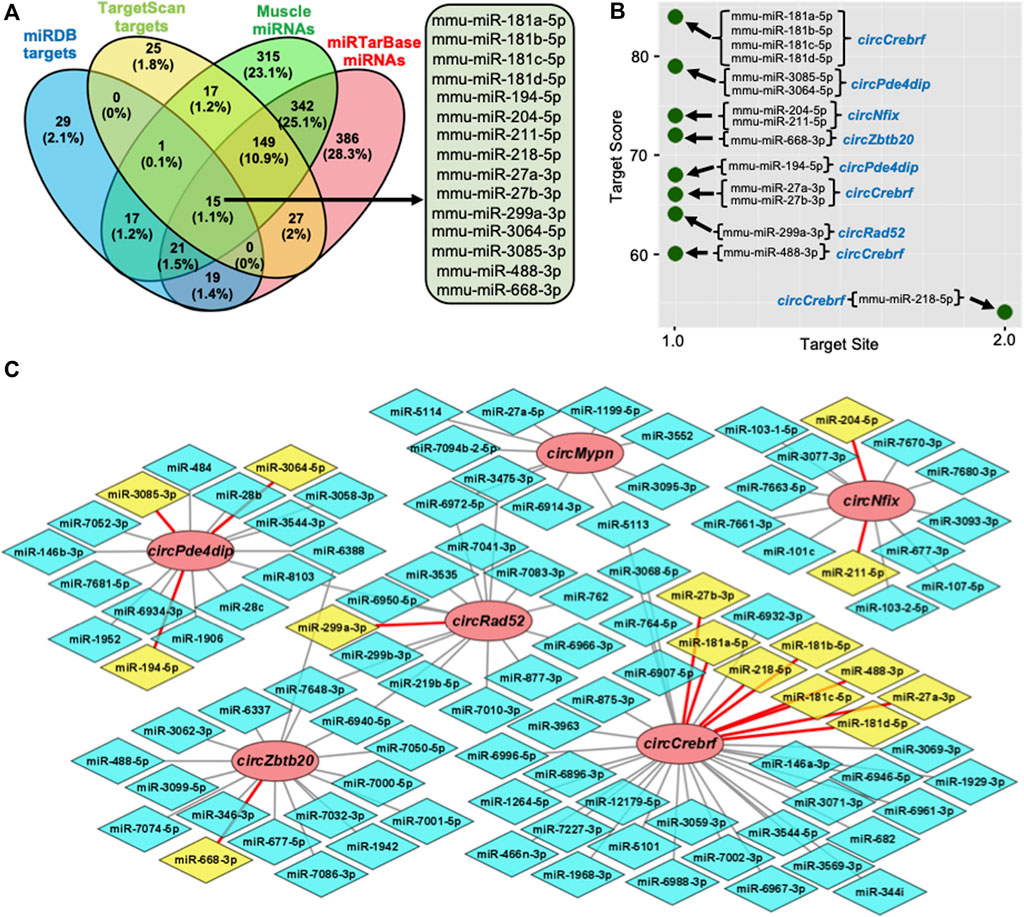
FIGURE 3. Prediction of the circRNA-miRNA-mRNA regulatory network in skeletal muscle. (A). Venn diagram depiction of the muscle miRNAs predicted to target the validated circRNAs and validated by miRTarBase. (B). Predicted miRNA targets of the six validated circRNAs using miRDB custom prediction tool. (C). The circRNA-miRNA regulatory network was constructed and visualized on Cytoscape. Circle nodes represent circRNAs, hexagon nodes represent miRNAs, and yellow miRNAs with red edges are functional miRNAs.
To find the functional significance of the downstream target genes in the circRNA-miRNA-mRNA regulatory network, we performed GO enrichment analysis for the biological process, cellular component, and molecular function (Supplementary Table S7). As shown in Supplementary Figure S4, several GO terms are enriched for the target genes of circRNAs. Briefly, multicellular organism development, cellular process, developmental process, anatomical structure development, and system development were the top GO biological processes, whereas binding, protein binding, ion binding, and organic cyclic compound binding were among the top hits in the GO molecular functions, and cell, intracellular, cytoplasm, intracellular organelle, organelle, and membrane-bounded organelle were among the top GO cellular component terms. Furthermore, KEGG pathway enrichment analysis of circRNA target genes identified several pathways critical for cell survival and aging (Supplementary Figure S4A–C; Supplementary Table S7). The common pathways with higher enrichment scores, include axon guidance, microRNAs in cancer, hippo signaling pathway, AGE-RAGE signaling pathway in diabetic complications, pathways in cancer, HIF-1 signaling pathway, and FoxO signaling pathway (Supplementary Figure S4D).
Conserved circNfix and Its Regulatory Role in Muscle Cell Differentiation
Since circRNAs are conserved across various species, we wanted to find the conserved circRNAs in mouse and human skeletal muscles (Ji et al., 2019). To identify the conserved mouse muscle circRNAs, we converted the mouse muscle circRNA coordinates to hg38 human coordinates using the LiftOver tool (Haeussler et al., 2019). Analysis of the conservation of the circRNAs expressed in mouse skeletal muscle and human muscle or satellite cells identified several conserved circRNAs (Supplementary Figure S5A). Of the six validated circRNAs, circMypn was conserved in human skeletal muscle and satellite cells, while circNfix was conserved in human satellite cells. Consistent with our mouse skeletal muscle data, circNFIX was upregulated in aged satellite cells compared to young satellite cells (Supplementary Figure S5B). The NCBI BLAST analysis suggested that the human circNFIX sequence is 96% similar to mouse circNfix, indicating conserved in humans and mice (Supplementary Figure S5C,D). In addition, the circRNA database circBase reported the expression of circNFIX (hsa_circ_0005660) in HSMM cells (Glazar et al., 2014). Since circMypn did not target any functional muscle miRNAs and the differentially expressed circCrebrf was not found to be conserved in human muscle, we wanted to analyze the role of circNfix in muscle.
Conserved circNfix Is Required for Efficient Myogenesis
It has been established that circRNAs act as competing endogenous RNAs (ceRNA) for target miRNAs, and circRNA expression changes positively correlate with the downstream target mRNAs. As shown in Figure 3C, circNfix is predicted to associate with many miRNAs, including miR-204-5p, that regulate myogenesis. The circNfix-associated miR-204 is known to inhibit myogenesis by suppressing the Mef2c expression in C2C12 cells (Cheng et al., 2018). Analysis of circNfix-associated miR-204 and the downstream myogenic regulator Mef2c mRNA in aged gastrocnemius muscle did not show any changes (Supplementary Figure S6A,B). To analyze the expression and function of conserved circNFIX/circNfix in human and mouse skeletal muscle, we differentiated sub-confluent HSMM and C2C12 cells for 4 days in differentiation media to induce the myotube formation. As shown in Figure 4A, the HSMM and C2C12 cells form long myotubes, and the differentiation was confirmed by upregulation of myogenic marker MYOG mRNA by qPCR analysis (Figure 4A, Supplementary Figure S7A). We further confirmed the expression of circNFIX in HSMM cells by RT-PCR followed by Sanger Sequencing (Supplementary Figure S7B). Interestingly, circNFIX expression was significantly upregulated in 4-day differentiated HSMM and C2C12 myotubes than proliferating myoblasts (Figure 4B). However, miR-204-5p levels did not alter significantly in 4-day differentiated HSMM and C2C12 myotubes compared to proliferating myoblasts (Supplementary Figure S7C,D). Notably, MEF2C mRNA, the downstream target of circNFIX, was significantly upregulated in 4-day differentiated HSMM and C2C12 myotubes supporting the circRNA ceRNA regulatory network hypothesis (Figure 4C).
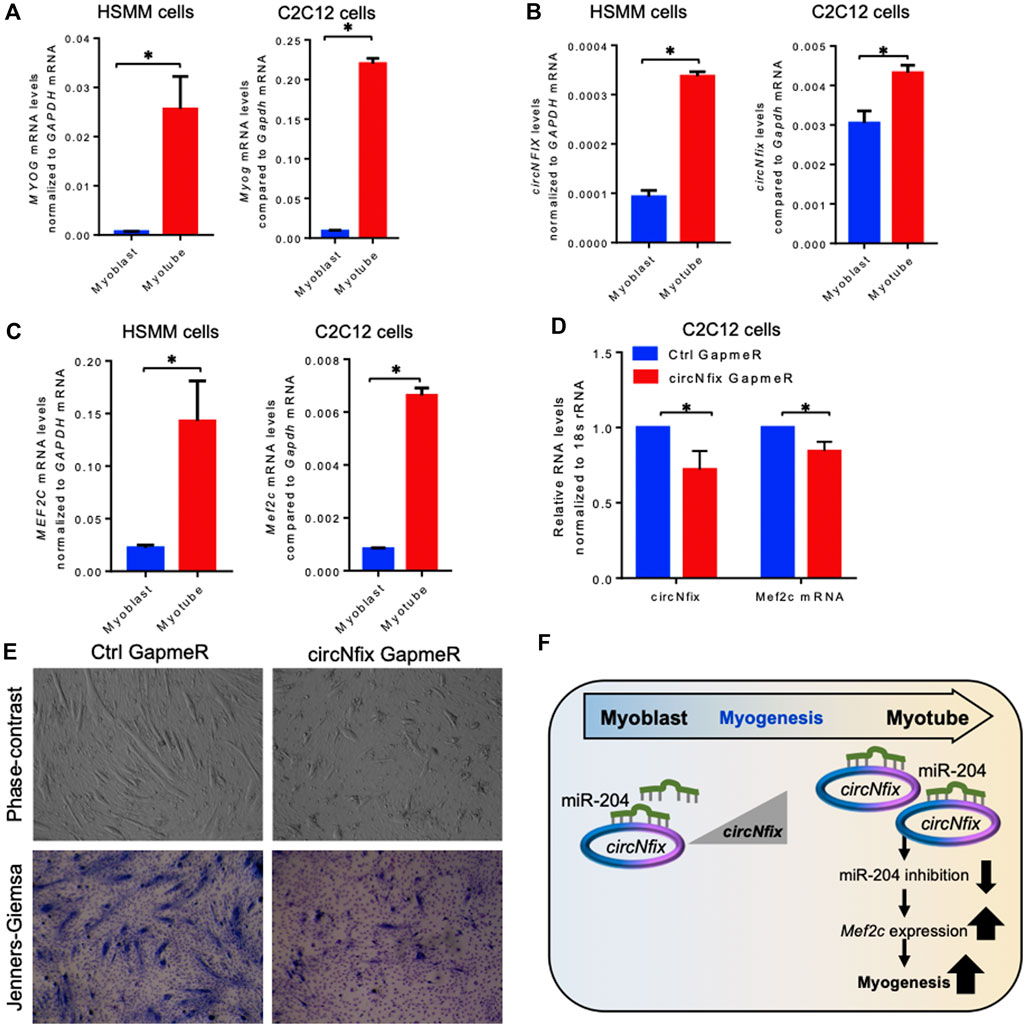
FIGURE 4. Expression analysis of circNFIX and downstream targets. (A). Relative expression of MYOG mRNAs in proliferating and 4-day differentiated myotubes of HSMM and C2C12 cells. (B). Expression levels of circNFIX measured by RT-qPCR analysis in proliferating myoblasts and 4-day differentiated myotubes of HSMM and C2C12 cells. (C). RT-qPCR analysis of MEF2C mRNA levels in proliferating myoblasts and 4-day differentiated myotubes of HSMM and C2C12 cells. (D). RT-qPCR analysis of circNfix and Mef2c expression levels in 4 day differentiated C2C12 cells transfected with Ctrl and circNfix GapmeR 24 hours before starting differentiation. (E). The phase-contrast fields and Jenners-Giemsa stained 4-day differentiated C2C12 cells transfected with Ctrl and circNfix GapmeR 24 hours before starting differentiation. (F). Proposed hypothesis of circNfix action during myoblast differentiation. Data in (A,B,C,D) are the means ± SEM from three independent experiments. *, p < 0.05.
Given that miR-204 inhibits myogenesis by suppressing MEF2C expression (Cheng et al., 2018), we hypothesized that circNfix might inhibit miR-204 activity to promote MEF2C expression and C2C12 differentiation into myotubes. Interestingly, silencing circNfix reduced Mef2c mRNA expression moderately without significant changes in miR-204 levels in the 4-day differentiated C2C12 cells (Figure 4D, Supplementary Figure S7E). These results suggested that circNfix might control the expression of Mef2c mRNA by sponging miR-204. However, the antisense oligo pulldown assay of circNfix using a biotin-labeled oligo targeting the backsplice junction could not pulldown circNfix (data not shown), which could be due to the unavailability of the circRNA junction that is either hidden by associated RBPs or due to secondary structures. Analysis of myogenesis using phase-contrast and Jenners-Giemsa staining followed by microscopy showed that silencing of circNfix decreased myogenesis and myotube formation in 4-day differentiated C2C12 cells (Figure 4E). Although Mef2c mRNA is known to be upregulated during muscle cell differentiation through transcriptional mechanisms, our results suggest that upregulation of circNfix in differentiated myotubes may partly enhance MEF2C expression by inhibiting the activity of miR-204 (Figure 4F). However, further experiments are required to establish the direct interaction and the circNfix-miR-204-MEF2C regulatory axis modulating myogenesis.
Discussions
CircRNAs are a new class of universally expressed single-stranded RNA molecules (Salzman et al., 2012; Jeck et al., 2013). Recent studies suggested that circRNAs are conserved across various species and show tissue-specific expression (Jeck et al., 2013; Ji et al., 2019). Due to their circular nature and lack of free ends, circRNAs are highly stable and resistant to cellular exonucleases (Suzuki et al., 2006; Enuka et al., 2016). It has been well established that circRNAs associate with cellular regulators and modulate cellular events by controlling transcription, splicing, mRNA stability, and translation (Vromman et al., 2021). Several studies highlighted the role of circRNAs in various diseases and normal physiological processes, including aging (Abdelmohsen et al., 2015; Cai et al., 2019). However, the role of circRNAs in skeletal muscle regeneration and myogenesis has not been well understood yet.
This study presents a comprehensive analysis of circRNAs expressed in aging mouse skeletal muscle tissues (Figure 1). Previous studies suggested an accumulation of circRNAs with advancing age (Gruner et al., 2016). Consistent with previous study, we also observed that circRNA expressions in the aged skeletal muscle were significantly higher than in the young muscle samples due to age-associated circRNA upregulation or accumulation. Since circRNAs act as miRNA sponges to regulate the downstream gene expression, we analyzed the association of circRNAs with functional miRNAs expressed in muscle (Panda, 2018). Computational analysis of the target genes of circRNA-associated miRNAs suggested their involvement in various biological processes and pathways involved in muscle cell physiology (Supplementary Figure S4).
Since circRNAs are known to be conserved, we analyzed the conservation of the six validated mouse circRNAs in the human skeletal muscle and satellite cell. Interestingly, circMypn was expressed in all three data sets, while circNfix was expressed in satellite cells and upregulated in aged muscle satellite cells. To explore the function of conserved circNfix, we used the C2C12 and HSMM myoblast differentiation models. Previous publications reported that miR-204 inhibits MEF2C expression and regulates myogenesis (Cheng et al., 2018; Tan et al., 2020). Our analysis suggested an increase in circNfix/circNFIX expression in the differentiated C2C12 and HSMM myotubes compared to myoblasts. Interestingly, the miR-204 levels in C2C12 and HSMM cells did not alter significantly after 4 days of differentiation. Moreover, silencing circNfix decreased the expression of downstream Mef2c mRNA without changes in target miR-204 supporting the ceRNA regulatory network (Figure 4). Furthermore, silencing circNfix inhibited C2C12 myotube formation suggesting that circNfix is a positive regulator of myogenesis. We propose that increased circNfix levels during myogenesis may inhibit miR-204 activity, promoting MEF2C expression in the myotubes. We acknowledge that MEF2C expression is upregulated at the transcriptional level during muscle cell differentiation (Buckingham and Rigby, 2014). Since MEF2C expression is crucial for forming functional myotubes, circNfix helps in the higher expression of MEF2C by inhibiting the activity of the miR-204 during differentiation. In sum, we propose that upregulation of circNfix during myogenesis favors myotube formation by enhancing MEF2C expression by acting as a sponge for miR-204.
Although our data highlights the importance of circRNAs in myogenesis, further experiments are required to support the findings. All the circRNAs and their target miRNA or mRNAs were predicted computationally, which need to be experimentally validated. Further experiments are required to validate the differential expression of all the validated circRNAs and downstream miRNAs or mRNAs in aging skeletal muscle and myogenesis cell line models. It will be essential to perform pulldown and reporter assays to confirm the direct interaction of circNfix with miR-204. In addition, biochemical experiments are required to confirm the circNfix-miR-204-Mef2c regulatory axis that modulates skeletal muscle differentiation. Furthermore, the regulation of myogenesis by circNfix through other mechanisms such as sponging other miRNAs or RBPs remains to be studied.
In summary, we generated a comprehensive genome-wide circRNA expression profile of 1,200 circRNAs in mouse skeletal muscle. In addition, we have identified more than 14,000 circRNAs in human skeletal muscle and human muscle satellite cells. We have identified the ceRNA network for a subset of abundantly expressed circRNAs and their regulatory networks in skeletal muscle. Finally, we identified circNfix as a promoter of myogenesis by acting as a sponge for miR-204. Although our study provides new insight into the skeletal muscle gene regulation by circRNAs, further research is warranted to discover novel therapeutic targets to better manage muscle diseases and muscle regeneration.
Data Availability Statement
All data generated in this study are included in the main text or the supplementary information files. The raw reads produced in this study were deposited in the European Nucleotide Archive (ENA accession number PRJEB46548).
Ethics Statement
The animal study was reviewed and approved by Institutional animal ethics committee of the Institute of Life Sciences, Bhubaneswar, India.
Author Contributions
ACP conceived the study, acquired the funding, and supervised the work. AD, SS, SSM, and TS performed the experiments and analyzed the data. AD and ACP prepared the first draft of the manuscript. All authors reviewed, edited, and approved the manuscript for submission.
Funding
This research was supported by intramural funding from the Institute of Life Sciences and the DBT research grant (BT/PR27622/MED/30/1961/2018) awarded to Amaresh Panda. University Grant Commission of India supported AD. SS and SSM were supported by the Intermediate Fellowship of the Wellcome Trust/DBT India Alliance (Grant Number: IA/I/18/2/504017) awarded to Amaresh Panda.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Suman Singh for helpful discussions and critical proofreading of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.762185/full#supplementary-material
References
Abdelmohsen, K., Panda, A. C., De, S., Grammatikakis, I., Kim, J., Ding, J., et al. (2015). Circular RNAs in Monkey Muscle: Age-dependent Changes. Aging 7, 903–910. doi:10.18632/aging.100834
Agarwal, V., Bell, G. W., Nam, J. W., and Bartel, D. P. (2015). Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 4, e05005. doi:10.7554/eLife.05005
Bose, R., and Ain, R. (2018). Regulation of Transcription by Circular RNAs. Adv. Exp. Med. Biol. 1087, 81–94. doi:10.1007/978-981-13-1426-1_7
Buckingham, M., and Rigby, P. W. J. (2014). Gene Regulatory Networks and Transcriptional Mechanisms that Control Myogenesis. Develop. Cel. 28, 225–238. doi:10.1016/j.devcel.2013.12.020
Cai, H., Li, Y., Niringiyumukiza, J. D., Su, P., and Xiang, W. (2019). Circular RNA Involvement in Aging: An Emerging Player with Great Potential. Mech. Ageing Develop. 178, 16–24. doi:10.1016/j.mad.2018.11.002
Chen, L.-L., and Yang, L. (2015). Regulation of circRNA Biogenesis. RNA Biol. 12, 381–388. doi:10.1080/15476286.2015.1020271
Chen, Y., and Wang, X. (2020). miRDB: an Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 48, D127–D131. doi:10.1093/nar/gkz757
Cheng, X., Du, J., Shen, L., Tan, Z., Jiang, D., Jiang, A., et al. (2018). MiR-204-5p Regulates C2C12 Myoblast Differentiation by Targeting MEF2C and ERRγ. Biomed. Pharmacother. 101, 528–535. doi:10.1016/j.biopha.2018.02.096
Das, A., Das, A., Das, D., Abdelmohsen, K., and Panda, A. C. (2020). Circular RNAs in Myogenesis. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1863, 194372. doi:10.1016/j.bbagrm.2019.02.011
Das, A., Sinha, T., Shyamal, S., and Panda, A. C. (2021). Emerging Role of Circular RNA-Protein Interactions. ncRNA 7, 48. doi:10.3390/ncrna7030048
Di Timoteo, G., Rossi, F., and Bozzoni, I. (2020). Circular RNAs in Cell Differentiation and Development. Development 147, dev182725. doi:10.1242/dev.182725
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., and Yarden, Y. (2016). Circular RNAs Are Long-Lived and Display Only Minimal Early Alterations in Response to a Growth Factor. Nucleic Acids Res. 44, 1370–1383. doi:10.1093/nar/gkv1367
Glazar, P., Papavasileiou, P., and Rajewsky, N. (2014). circBase: a Database for Circular RNAs. RNA 20, 1666–1670. doi:10.1261/rna.043687
Gruner, H., Cortés-López, M., Cooper, D. A., Bauer, M., and Miura, P. (2016). CircRNA Accumulation in the Aging Mouse Brain. Sci. Rep. 6, 38907. doi:10.1038/srep38907
Haeussler, M., Zweig, A. S., Tyner, C., Speir, M. L., Rosenbloom, K. R., Raney, B. J., et al. (2019). The UCSC Genome Browser Database: 2019 Update. Nucleic Acids Res. 47, D853–D858. doi:10.1093/nar/gky1095
Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S., Hong, H. C., et al. (2020). miRTarBase 2020: Updates to the Experimentally Validated microRNA-Target Interaction Database. Nucleic Acids Res. 48, D148–D154. doi:10.1093/nar/gkz896
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 19, 141–157. doi:10.1261/rna.035667.112
Ji, P., Wu, W., Chen, S., Zheng, Y., Zhou, L., Zhang, J., et al. (2019). Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals. Cel. Rep. 26, 3444–3460. doi:10.1016/j.celrep.2019.02.078
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 44, D457–D462. doi:10.1093/nar/gkv1070
Kim, J. Y., Park, Y.-K., Lee, K.-P., Lee, S.-M., Kang, T.-W., Kim, H.-J., et al. (2014). Genome-wide Profiling of the microRNA-mRNA Regulatory Network in Skeletal Muscle with Aging. Aging 6, 524–544. doi:10.18632/aging.100677
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Okazaki, K., and Holtzer, H. (1966). Myogenesis: Fusion, Myosin Synthesis, and the Mitotic Cycle. Proc. Natl. Acad. Sci. 56, 1484–1490. doi:10.1073/pnas.56.5.1484
Panda, A. C., and Gorospe, M. (2018). Detection and Analysis of Circular RNAs by RT-PCR. Bio Protoc. 8, e2775. doi:10.21769/BioProtoc.2775
Panda, A. C. (2018). “Circular RNAs Act as miRNA Sponges,” in Circular RNAs: Biogenesis and Functions. Editor J. Xiao (Singapore: Springer Singapore), 67–79. doi:10.1007/978-981-13-1426-1_6
Pandey, P. R., Yang, J.-H., Tsitsipatis, D., Panda, A. C., Noh, J. H., Kim, K. M., et al. (2020). circSamd4 Represses Myogenic Transcriptional Activity of PUR Proteins. Nucleic Acids Res. 48, 3789–3805. doi:10.1093/nar/gkaa035
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS One 7, e30733. doi:10.1371/journal.pone.0030733
Sinha, T., Panigrahi, C., Das, D., and Chandra Panda, A. (2021). Circular RNA Translation, a Path to Hidden Proteome. Wiley. Interdiscip. Rev. RNA 2, e1685. doi:10.1002/wrna.1685
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006). Characterization of RNase R-Digested Cellular RNA Source that Consists of Lariat and Circular RNAs from Pre-mRNA Splicing. Nucleic Acids Res. 34, e63. doi:10.1093/nar/gkl151
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/measurement Sets. Nucleic Acids Res. 49, D605–D612. doi:10.1093/nar/gkaa1074
Tan, Y., Shen, L., Gan, M., Fan, Y., Cheng, X., Zheng, T., et al. (2020). Downregulated miR-204 Promotes Skeletal Muscle Regeneration. Biomed. Res. Int. 2020, 3183296. doi:10.1155/2020/3183296
The Gene Ontology, C. (2019). The Gene Ontology Resource: 20 Years and Still GOing strong. Nucleic Acids Res. 47, D330–D338. doi:10.1093/nar/gky1055
Tumasian, R. A., Harish, A., Kundu, G., Yang, J.-H., Ubaida-Mohien, C., Gonzalez-Freire, M., et al. (2021). Skeletal Muscle Transcriptome in Healthy Aging. Nat. Commun. 12, 2014. doi:10.1038/s41467-021-22168-2
Veliça, P., and Bunce, C. M. (2011). A Quick, Simple and Unbiased Method to Quantify C2C12 Myogenic Differentiation. Muscle Nerve 44, 366–370. doi:10.1002/mus.22056
Vromman, M., Vandesompele, J., and Volders, P.-J. (2021). Closing the circle: Current State and Perspectives of Circular RNA Databases. Brief. Bioinform. 22, 288–297. doi:10.1093/bib/bbz175
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., et al. (2014). Genome-wide Analysis of drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-dependent Neural Accumulation. Cel. Rep. 9, 1966–1980. doi:10.1016/j.celrep.2014.10.062
Zhang, X.-O., Dong, R., Zhang, Y., Zhang, J.-L., Luo, Z., Zhang, J., et al. (2016). Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res. 26, 1277–1287. doi:10.1101/gr.202895.115
Keywords: circular RNA, microRNA, skeletal muscle, myogenesis, ceRNA
Citation: Das A, Shyamal S, Sinha T, Mishra SS and Panda AC (2021) Identification of Potential circRNA-microRNA-mRNA Regulatory Network in Skeletal Muscle. Front. Mol. Biosci. 8:762185. doi: 10.3389/fmolb.2021.762185
Received: 21 August 2021; Accepted: 01 November 2021;
Published: 29 November 2021.
Edited by:
Pietro Laneve, Italian National Research Council, ItalyReviewed by:
Alessandra Perfetti, IRCCS Policlinico San Donato, ItalyYongjie Xu, Xinyang Normal University, China
Lei Wang, Xinyang Normal University, China
Copyright © 2021 Das, Shyamal, Sinha, Mishra and Panda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amaresh C. Panda, YW1hcmVzaC5wYW5kYUBpbHMucmVzLmlu
†These authors contributed equally to this work
 Arundhati Das1,2†
Arundhati Das1,2† Sharmishtha Shyamal
Sharmishtha Shyamal Smruti Sambhav Mishra
Smruti Sambhav Mishra Amaresh C. Panda
Amaresh C. Panda