- Centre of Nanomaterials and Biotechnology, Faculty of Science, Jan Evangelista University in Ústí Nad Labem, Ústí Nad Labem, Czech Republic
Despite extensive study of extracellular vesicles (EVs), specifically exosomes (EXs) as biomarkers, important modulators of physiological or pathological processes, or therapeutic agents, relatively little is known about nonconventional sources of EXs, such as invertebrate or plant EXs, and their uses. Likewise, there is no clear information on the overview of storage conditions and currently used isolation methods, including new ones, such as microfluidics, which fundamentally affect the characterization of EXs and their other biomedical applications. The purpose of this review is to briefly summarize conventional and nonconventional sources of EXs, storage conditions and typical isolation methods, widely used kits and new “smart” technologies with emphasis on the influence of isolation techniques on EX content, protein detection, RNA, mRNA and others. At the same time, attention is paid to a brief overview of the direction of biomedical application of EXs, especially in diagnostics, therapy, senescence and aging and, with regard to the current situation, in issues related to Covid-19.
Introduction
Exosomes (EXs) are nano-sized extracellular vesicles (EVs) released from a subdivision of previous endosomes. EXs were first reported in the mid-1980s (Pan and Johnstone, 1983) (Pan and Johnstone, 1983). Initially, researchers assumed that EXs simply served as “garbage bags” to enable cells to get rid of unwanted components (Lin et al., 2015). However, increasing evidence has proven that EXs play an important role in many cellular processes, and provide a unique mode of cell-to-cell communication and also influence both physiological and pathological processes (Yamashita et al., 2016; Ailuno et al., 2020). EXs are released from many cell sources.
They can also be found in most body fluids (Théry et al., 2002). In recent decades, an increasing number of studies have revealed many other sources of EXs, e.g., nonconventional - animal product derived EXs, bacterial, fungal, parasitic, plant derived EXs (Schuh et al., 2019). In general, due to their biological function, EXs play an increasingly important role in diagnosis, aging studies, and as various therapeutic agents (Aheget et al., 2020).
In this review we focus mainly on a brief summary of various sources (conventional and nonconventional) of EXs, typical isolation methods, widely used kits and new “smart” technologies with emphasis on the influence of isolation techniques on EX content, protein detection, RNA, mRNA and others. At the same time, attention is paid to a brief overview of the directions of biomedical application of EXs, especially in diagnostics, therapy, senescence and aging and, with regard to the current situation, in issues related to Covid-19.
Exosomes–Biogenesis, Composition, Function and Sources
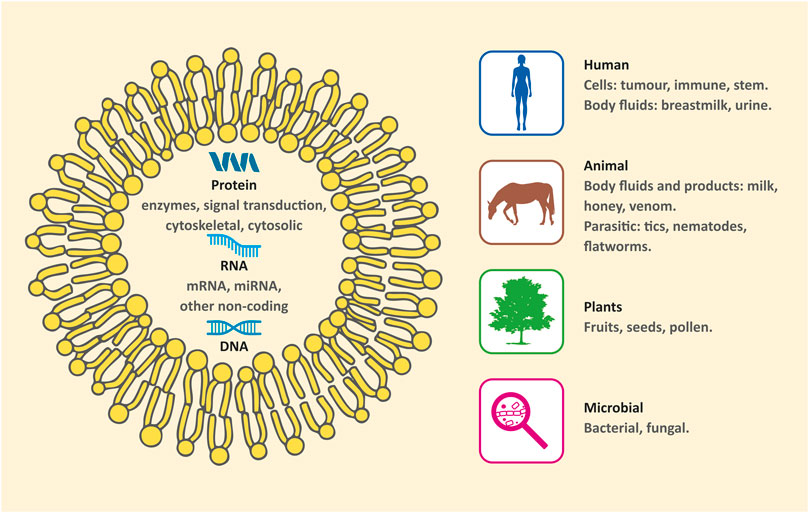
EXs as nano-sized extracellular vesicles have a diameter of about 30–150 nm (Tanziela et al., 2020) and density of 1.13–1.19 g/ml (Deb et al., 2021), dependent on their place of origin, as well as a lipid bilayer structure in the cell (Kalluri and LeBleu, 2020). EXs were first described in maturing mammalian reticulocytes (Pan and Johnstone, 1983). They release transferrin receptors (TFR) throughout small vesicles into the extracellular environment (Lin et al., 2015; Batista and Melo, 2019). Ever since Valadi et al. first stated in 2007, that EXs carry RNAs, the composition and function of these vesicles have been extensively studied (Valadi et al., 2007).
Now it is known that EXs contain various types of molecular constituents, such as nucleic acids, proteins, lipids, cytokines, transcription factors, regulatory RNAs and some other bioactive substances (Jing et al., 2018; Jeppesen et al., 2019; Zhang et al., 2020). Structurally, EX surfaces are rich in transmembrane proteins, receptors, and other functional molecules (Jafari et al., 2020). EXs have a membrane structure with a lipid bilayer in which lipids such as polyglycerol, phospholipid, ceramide, sphingomyelin, cholesterol and phosphatidylinositol are included.
The biogenesis of EXs starts with endocytosis of cell membranes, including receptor-mediated endocytosis, nuclear endocytosis of inclusion proteins and pinocytosis (Kim et al., 2021). The early endosomes are formed by the endocytosis and later matured into late endosomes. EXs carry information from the cells of origin thanks to continuous intracellular exchange of substances by invagination. The invagination in the late endosomes leads to the formation of intraluminal vesicles (ILVs), so called future EXs (Kalluri and LeBleu, 2020). ILVs are finally secreted as EXs through multivesicular bodies (MVBs) fusion to the plasma membrane and exocytosis. The exocytosis permits EXs to be released with their characteristic lipid bilayer structure (Dai et al., 2020; Jiang et al., 2020). The whole process of EX biogenesis is strictly regulated (Aheget et al., 2020).
The main elements that influence the biogenesis of EXs include certain biological factors that are essential for their formation, secretion, and yield. Studies have suggested that EX yield depends for example on the cell type, biotic and abiotic stress or that confluence of cell cultures also plays a critical role (Gurunathan et al., 2019; Aheget et al., 2020). As in all membrane vesicular processes, lipids are also important players in the biogenesis of EXs (Pegtel and Gould, 2019).
EXs are released from many cell sources, such as lymphocytes, dendritic cells, epithelial cells, endothelial cells, mast cells or neurons. They can also be found in most body fluids inclusive of blood, saliva, urine, breast milk, amniotic fluids, hydro thoracic fluids, ascitic fluid and in the culture medium of almost all cell types (Théry et al., 2002). These “conventional” sources undoubtedly provide an important basis for understanding the roles of EXs in a number of physiological processes, as well as for their subsequent possible use in biomedicine (Jing et al., 2018).
However, in the last decades, an increasing number of studies have revealed that many other sources exhibit remarkable characteristics and clarify some biological processes. Moreover, they could also be useful alternatives for medical applications. Thus, EXs can be categorized by sources as conventional (human/mammal) and nonconventional (nonhuman/nonmammal). The latter can be further subdivided into 1) animal product derived EXs, 2) bacterial, fungal, and parasitic EXs, and 3) plant derived EXs (Figure 1) (Schuh et al., 2019).
It is generally accepted that EXs play a crucial role in many physiological and pathological processes (Yamashita et al., 2016; Ailuno et al., 2020). A considerable part of the literature indicates that EXs display a wide range of functions, which depend on their cell or tissue of origin. Especially, EXs from certain types of immune cells may ensure adaptive immune responses to pathogens or tumors (Schwarzenbach and Gahan, 2021). Moreover, EXs are usually able to guard their loads against removal from the body or destruction, due to their dual layered membrane and nano-size (Tanziela et al., 2020). Therefore, they have the potential to be used to deliver therapeutic molecules (e.g., small interfering RNA, microRNAs) or small molecule drugs (e.g. doxorubicin, curcumin) to target cells for treating various types of disease. Exosomal chemical or biological modification can also increase the characteristic therapeutic competence of EXs (Tian et al., 2014).
Nonconventional Sources of EXs
Most of the exosome production and isolation techniques focus on the use of mammalian sources (cells, tissues, and body fluids). These conventional sources provided a key understanding of the role of EXs in basic (mammalian) physiological processes, and their potential role in biomedicine as therapeutic agents. Extracellular vesicles are highly evolutionary conserved across all eukaryotic organisms (Gill et al., 2019). Non-mammalian resources of EXs are gaining in popularity for their interesting characteristics and complex roles (ranging from inter-species interactions to inter-kingdom communication) providing possible alternatives for biomedical applications (Schatz and Vardi, 2018; Schuh et al., 2019).
EXs, representing a common mechanism for mediating communication in host-parasite interactions, are of particular relevance to biomedicine, since they are known to act as vectors for virulence factors and effector proteins from parasites to their hosts (Zhou et al., 2018; Nawaz et al., 2019; Wu et al., 2019; Xander et al., 2020). EXs were shown to mediate pathogenic processes through influencing gene expression and host response of the immune system (Nawaz et al., 2019). However, EXs are produced by the host cells in response to the parasite, thus a bidirectional regulatory effect on parasitic infection is present. Isolation of EXs provides novel approaches to diagnostics and possible treatment of various parasitic infections (Wu et al., 2019).
EXs and exosome-like vesicles have recently been isolated from bee glandular secretion products (honey, royal jelly, and bee pollen) and their bacteriostatic, bactericidal, and biofilm-inhibiting effects were demonstrated together with the ability to increase migration of human mesenchymal stem cells (MSCs) in vitro (inter-kingdom activity) (Schuh et al., 2019).
EXs have been isolated from lyophilized snake venom (Carregari et al., 2018) and analyzed by mass spectrometry-based proteomic approach, suggesting their role in the cytotoxicity of the venom, and providing new insight into the envenomation process (Carregari et al., 2018).
From studies on model organisms, D. melanogaster and C. Elegans (Beer and Wehman, 2017; Russell et al., 2020), it seems that extracellular vesicles (EVs) and EXs play a conserved role in developmental processes (i.e., carrying patterning morphogens) and presumably in adult mating-specific behavior. Moreover, the functional role of extracellular vesicular transport of Wnt proteins in D. melanogaster and similarly in murine model was demonstrated (Gross et al., 2012).
Other numerous non-mammalian eukaryotic organisms have been shown to use EXs/EXs-like vesicles; lower vertebrates, i.e. danio (Verweij et al., 2019), xenopus (Danilchik and Tumarkin, 2017), turtles (Zhu et al., 2019), molluscs, i.e. oysters, where EXs have been found to be involved in reparation of the outer shell (Johnstone et al., 2015), protozoa (Lin et al., 2019) and fungi (Rizzo et al., 2020). However, many possibly interesting sources of EXs with diagnostic or clinical potential remain to be discovered.
Nowadays, in addition to animal sources of EXs, plant-derived EXs come into focus. The term “EXs” has traditionally been used for animal vesicles. Similar structures have recently been discovered in plants as well, they are called plant exosome like vesicles (PELVs) and they seem to possess similar size (50–150 nm), structure and analogical function, but research is still at the beginning. Since fruits, seeds, leaves and roots of various plants are a natural part of human nutrition, PELVs, included in plant tissues, are considered to have low toxicity and immunogenicity. For research purposes, iPELVs can be isolated in large quantities at very low cost from juiced plant parts or apoplastic fluid, avoiding lengthy isolation protocols.
As an example, traditional plant model organisms, such as Arabidopsis thaliana or Nicotiana tabacum, have been used for PELVs characterization (Rutter and Innes, 2017). Research of plant vesicles is very specific, due to its potential as nanoplatforms for drug delivery (Dad et al., 2021).
Recently, PELVs have been widely studied for their therapeutic potential. For example, PELVs isolated from ginger protected mice from alcohol-induced liver damage (Zhuang et al., 2015). Ginger (Zhang et al., 2016) grape (Ju et al., 2013) and broccoli (Deng et al., 2017) PELVs enhanced intestinal repair and reduced acute colitis in murine models. Moreover, ginger PELVs mediated transport of therapeutic agent Doxorubicin, enhanced chemotherapeutic action and inhibited growth of tumors better than free drug (Zhang et al., 2016).
Significant anticancer potential of Dendropanax morbifera PELVs was also observed. Vesicles isolated from this Korean shrub reduced growth of mouse melanoma (Lee et al., 2020). Inhibition of cancer cell line proliferation of PELVs isolated from fruits like grapefruit or lemon was observed in vitro using various tumor cell lines (Raimondo et al., 2015). Lemon PELVs inhibit cancer cell proliferation in different tumor lines, but, in vivo, promotion of apoptosis in the fight against chronic myeloid leukemia has been proved (Raimondo et al., 2015). Moreover, miRNAs from grapefruit and ginger PELVs were reported to promote the growth of Lactobacillus rhamnosus in mice gut, boosting the immune response and wellbeing of the mice gut (Mu et al., 2014).
The regeneration and wound healing potential of PELVs has also been investigated. In vitro studies, using wheat-derived PELVs, the expression of collagen mRNA was detected as well as increased vascular formation, providing new possibilities for skin regeneration strategies (Şahin et al., 2019).
Use of plant derived nanovesicles is still at the beginning, concealing a large future potential for broader use. The latest trend in this field is the production of transgenic plant-derived virus-like agents for the production of vaccines (Chen and Lai, 2013), e.g. plant-derived virus-like vaccine against influenza is now an ongoing Phase III clinical trial (Pillet et al., 2019).
Exosome Isolation Methods
EXs isolation techniques are based on three basic biophysical principles; 1) Size and density; 2) Polymer precipitation; 3) Immunoaffinity capture (Supplementary Table S1). Comparisons of recent strategies and technologies for EXs isolation and purification are described in many reviews (Tang et al., 2017; Chen et al., 2019; Sidhom et al., 2020; Yang et al., 2020). Recently Chen et al. focused on comparison of isolation methods with emphasis on the EXs yield and purity (Chen et al., 2022). The very specific “smart” isolation method, which combines the aforementioned principles, are microfluidics-based methods(Yu et al., 2018; Yang et al., 2020; Zhou et al., 2020). Currently, there are many EXs isolation kits available on the market. Many studies comparing isolation methods were performed, where characteristics like exosome yield, size distribution, marker-based assessment and RNA or protein quantitation and qualification were taken under the scope (Patel et al., 2019). All isolation methods are still under progress with the goal to establish a versatile, reliable and robust isolation method that is a crucial aspect for further EXs utilization in non-invasive diagnostic and therapeutic approaches (Patel et al., 2019; Deb et al., 2021) target drug delivery systems or novel modulators for vaccination (Deb et al., 2021; Kim et al., 2021; Song et al., 2021).
Size and Density-Based Isolation Methods
In general, EXs isolation is based on removing larger constituents like cells, dead cells and cell debris followed by elimination of smaller elements like macromolecules from the sample. This can be performed by 1) ultracentrifugation (UC); 2) density gradient centrifugation; 3) ultrafiltration; or 4) size-exclusion chromatography (SEC).
Ultracentrifugation is currently signed as the gold standard for EXs isolation (Zhang et al., 2020). (Chen et al., 2019)lower speed, EXs are obtained by centrifugation at min. 100,000xg for at least 1 h. Centrifugation time, centrifugal force or rotor type can affect the yield and purity of the EXs, but also their destruction and aggregation (Cvjetkovic et al., 2014; Livshts et al., 2015). The purest EXs are obtained by density gradient centrifugation. Sucrose and Iodixanol are two commonly used media to create the gradient, when Iodixanol is more stable and less viscous than sucrose, which results in shorter centrifugation times (Ford et al., 1994; Van Veldhoven et al., 1996). Moreover, iodixanol gradient enables separation of EXs from some types of retroviruses (Cantin et al., 2008). Advantages of centrifuge-based methods are their purity and suitability for large volume isolation. Disadvantages are high instrumental costs, time consuming procedures and non-negligible loss of the EXs during the procedure (Gurunathan et al., 2019; Zhou et al., 2020).
Ultrafiltration separates EXs from the sample by membranes with different molecular weight cut-offs. By simple adjusting of the order and filter size, ultrafiltration allows to sort specific subsets of EXs with defined particle sizes with yield comparable to that of the ultracentrifugation method (Heinemann and Vykoukal, 2017). Moreover, ultrafiltration-based exosome isolation dramatically shortens processing time with minimal equipment requirement.
Advanced methods of tangential stream filtration utilize so-called tangential flow streams. The feed stream flows parallel to the membrane face. The applied pressure causes a defined fraction of the flow stream to pass through the membrane filter according to the pore size. During the tangential flow filtration procedure, several membranes are included and simultaneously several fractions of the sample can be collected (Heinemann and Vykoukal, 2017).
Size-exclusion chromatography allows sequential elution of different-sized extracellular vesicles fractions from one column, where small particles remain trapped in the pores of the statical phase and bigger particles are eluted along the pores with the mobile phase (Zhang et al., 2020). This method is successful in the separation of EXs from proteins, but may doped with particles of similar size like aggregates, lipoparticles or particulate matter (Böing et al., 2014; Gurunathan et al., 2019). Based on this principle, there are two commercial EXs separation kits on the market, qEV (iZON) and PURE-EVs (Hansa Biomed).
Polymer-Based Precipitation
The objective of those methods is to reduce the EXs solubility by introducing water soluble, slightly amphiphilic polymer molecules. Those molecules interact with water molecules surrounding the EXs to create a hydrophobic micro-environment, resulting in exosome precipitation. Obviously, the precipitation methods are based on 1) Polymer precipitation; or 2) Aqueous two phase systems separation (ATPSs).
Several commercially available kits are based on polymer precipitation: mExoPrep (Hansa BioMed), Total Exosome Isolation Reagent (TEI) (Invitrogen), ExoQuick (System Biosciences), Exosome Purification Kit (Norgen Biotek), miRCURY, PureExo (101Bio) and Exosome Isolation Kit (Exiqon).
In general, cells, dead cells, apoptotic bodies and debris are removed before EXs precipitation by filtration or centrifugation. Typical precipitation methods use polyethylenglycol, with molecular weights in range 6–20 kDa and overnight incubation at 4 °C. Finally, the EXs are harvested via low-speed centrifugation. Compared to ultracentrifugation methods, EXs precipitation is fast and relatively easy to provide (Zhang et al., 2020). However, water-excluding polymers can also precipitate other water-soluble materials such as nucleic acids, lipoproteins, proteins, and even viruses (Sim et al., 2012; Kimura et al., 2019). To overcome the problem with impurities, lectin agglutination can be performed. Lectins are a family of proteins that recognise carbohydrate moieties with very high specificity. Similar to PEG molecules, lectins bind to carbohydrates on the surface of the EXs, causing them to precipitate out of the solution. Therefore, only highly glycosylated impurities can be co-precipitated with EXs utilizing the lectin-induced precipitation (Samsonov et al., 2016; Doyle and Wang, 2019).
The aqueous two phase system for EXs separation (ATPSs) is a more advanced precipitation method yielding purer harvested EXs compared to basic polymer precipitation. ATPSs are based on separation of different kinds of particles in different phases. After mixing samples with dextran and PEG solutions and using low speed centrifugation, the dextran solution forms the lower phase with accumulated EXs. The upper phase, which is a relatively more hydrophobic solution of PEG, preferentially accumulates proteins and other macromolecular complexes (Shin et al., 2015; Yang et al., 2020).
Immunoaffinity Capture
All aforementioned methods allow only the nonspecific isolation of the EXs. Immunoaffinity methods remain the only method to distinguish between subgroups of vesicles or fishing of preferred subgroups from the sample. Most of the targeted moieties are proteins tetraspanins CD9, CD81 and CD63, because those markers are expressed on nearly all EXs (Clayton et al., 2001). Other markers like Rab5, CD82, annexin TGS101, or Alix have been also published for selective exosome isolation (Andreu and Yáñez-Mó, 2014; Willms et al., 2018; Liu and Su, 2019). There are two principles of EXs isolation based on immunoaffinity capture: 1) Enzyme-linked immunosorbent assay (ELISA); 2) Immunoprecipitation.
Enzyme-linked immunosorbent assay (ELISA) requires antibodies against an antigen of interest immobilized on the microplate surface. Unbound elements are washed away while immobilized EXs are detected by another antibody containing a detection tag. When the quantitative standards are available, a calibration curve can be measured and therefore ELISA can also be utilized as a quantitative method (Doyle and Wang, 2019). Commercial ELISA kits are available for the most common antigens, CD9, CD81 and CD63, e.g., ExoELISA Complete Kit (System Biosciences), Human Exosome ELISA colorimetric Kit (Novus Biologicals), Overall Exosome ELISA Kit (Abnova), ExoSense ELISA Kit (EsiBio), ExoQuant (BioVision) and many others.
For immunoprecipitation, antibodies or recombinant proteins should be fixed to solid state matrices, where the submicron-sized magnetic or polymeric beads are the most common option. This method exhibits high yield efficiency and very good sensitivity, due to its large surface. Moreover, this method enables processing of large starting volumes of samples, EXs preconcentration and also their surface immunoanalysis (Li et al., 2017). Therefore, this method is suitable for future use in diagnostics through the detection of disease-specific marker. s.
Again, there are many commercial kits on the market, such as Exosome Isolation Kit (Miltenyi Biotec), Exosome Capture beads (Abcam), ExoCap Streptavidin Kit (JSR Life Science Company), Exosome-Human Isolation/Detection Reagent (Invitrogen) as well as Exo-Flow Capture Kit (System Biosciences) and others.
The non-antibody affinity beads represent a universal immunoprecipitation based option for EXs isolation. The technology, named EVtrap (extracellular vesicle total recovery and purification) is based on beads modified with a combination of hydrophilic and aromatic groups that exhibit high affinity toward the lipid bilayer of the EXs (Iliuk et al., 2021). So-called chemical affinity EVtrap beads (Tymora analytical operations) are already available on the market.
Conclusion Remarkes to Conventional Isolation Methods
In conclusion, EXs isolation is not a completely solved process, and is still varying with respect to the source of the EXs, or the purpose for which they are isolated.
Human EXs, animal exosome-like vesicles or plant exosome like vesicles (PELVs) exhibit very similar properties on the interkingdom level. Therefore, many methods such as ultracentrifugation, ultrafiltration, size-exclusion chromatography, or precipitation are universal tools for EXs isolation.
Ultracentrifugation is a gold standard for EXs isolation and it is also universal for isolation from all known sources such as from plants (Urzì et al., 2021), animal secretions (Schuh et al., 2019), cell cultures (Théry et al., 2006) or human body fluids (Keller et al., 2011). The limit of this method is the high amount of exosomal source required. In the case that only a small volume of the exosomal source is available, precipitation based methods offer a good solution. This method is also universal for a wide spectrum of exosomal sources such as plants (Suharta et al., 2021), cell culture and body fluids. As there is a wide spectrum of commercial kits on the market, several comparative studies have been performed e.g., (Martins et al., 2018).
The most specific isolation method is based on immunoaffinity recognition. Commercial kits based on this princple are targeted to a specific marker (mostly human), therefore, cannot be applied on all EXs sources. Moreover, every immunoaffinity isolation is also optimized for the type of body fluid or the target of interest and the purpose of investigation. These procedures are mostly connected with cancer disease. Because EXs can play a very important role as the biomarkers in the cancer diagnosis and prognosis (Czystowska-Kuzmicz and Whiteside, 2021)a variety of studies have been already published e.g.(Whitesides, 2006; Hong et al., 2014).
The most advanced method for EXs isolation is microfluidics because this access for isolation also enables the EXs characterization in one microdevice (He et al., 2014).
Microfluidics
A new and rapidly evolving approach to exosome isolation, as well as detection and analysis, is utilizing microfluidic technologies. Microfluidics in general refers to the study and manipulation of fluids that are geometrically constrained in channels with dimensions of tens of micrometers. At this scale, fluid behavior differs from conventional flow theory and the impact of volumetric forces is overtaken by the surface ones (Whitesides, 2006)Miniaturization, automation and precise fluidic control achieved by implementing microfluidics leads to many benefits such as reduced sample consumption, lower reagent volume, enhanced purity or the possibility to combine multiple processes into a single step and onto a single microfluidic chip. Having effective on-chip solutions for exosome related processes would minimize cost in the long run, speed-up otherwise time consuming isolations and enable point-of-care acquisition and diagnosis. These are all crucial steps in bringing EXs to everyday laboratory and clinical praxis (Liga et al., 2015; Contreras-Naranjo et al., 2017; Jia et al., 2019).
Different authors categorize numerous microfluidic chips for exosome isolation developed in recent years into various categories and subcategories (Chen et al., 2019; Wang et al., 2021). However, at the basic level, there are currently only two possible approaches. Immunoaffinity is the common denominator of an otherwise broad range of various clever microfluidic device constructions.
The first approach, microfluidics, utilizes antibodies and immune specificity of involved reactions (Liu et al., 2018; Chen et al., 2019) to isolate EXs.
The second approach aims at the physical properties of EXs, size, surface charge and density. These microfluidic chips oriented at EXs’ physical properties utilize standard processes such as filtering through membranes (Liang et al., 2017), sieving/trapping at nanostructures (Yasui et al., 2017), and using effects of electric field (Heineck et al., 2017; Ibsen et al., 2017). They also utilize more “niche principles” such as viscoelastic separation (Liu et al., 2017; Zhou et al., 2019), deterministic lateral displacement (Liu et al., 2017)or acting on the particles via surface acoustic waves (Ku et al., 2018; Wang et al., 2021).
Both described approaches have their pros and cons. Devices using immune-affinity can be very efficient and can achieve high throughput, but they are not yet suited for all exosome types, and usually require complicated sample pre-treatment. Antibody labeling may also induce impurities. On the other hand, devices aimed at physical properties are label-free and capable of processing complex bodily fluids, but their usage generally leads to higher percentages of contaminants.
The optimal solution may lie in combining the best of both approaches and using multiple isolation principles in a single chip. Moreover, the key potential of microfluidic devices is the possibility to integrate both exosome isolation and characterization onto the same chip, thus making a huge step in enabling POC acquisition and unlocking all the advantages described earlier.
For the purpose of this all-encompassing review, we have only briefly scratched the surface of both realized and potential benefits that microfluidics could bring to the field of exosome research. However, there are several comprehensive reviews from recent years that are focused solely on microfluidics and go into great detail in their description and comparison (Liga et al., 2015; Contreras-Naranjo et al., 2017; Jia et al., 2019; Choe et al., 2021; Ding et al., 2021; Wang et al., 2021).
Storage
The following crucial topic following the EXs isolation is their storage. Their stability for extended storage is low. The technology of conservation of EXs, which protects their biological activities and makes them suitable for transport and subsequent clinical applications, is therefore a major challenge (Aheget et al., 2020). At present, applied protection techniques mostly include freezing, lyophilization and spray drying (Zhang et al., 2020).
Cryopreservation is a storage technique in which the temperature is reduced below the temperature necessary for biochemical reactions, to maintain the functional stability of biological particles. Typically, temperatures of 4°C, −80°C and −196°C are used (Zhang et al., 2020). Nevertheless, a major disadvantage of this method is the “susceptibility to frost injury”. The “frostbites” usually refer to the annexation of osmosis during the freezing process and to the formation of ice crystals inside the biological particles. To overcome this shortcoming, one or more antifreeze mixtures, so called cryoprotectants with appropriate concentrations, are frequently selectively added (Jeyaram and Jay, 2018; Kusuma et al., 2018; Bahr et al., 2020).
These cryoprotectants are usually divided into two main types, based on their permeability: 1) intracellular compounds (penetrating cryoprotectants) and 2) extracellular agents (non-penetrating cryoprotectants). The penetrating cryoprotectants (e.g., ethylene glycol and dimethyl sulfoxide) have a low molecular weight and can penetrate the membrane into the exosome and prevent the formation of ice crystals and subsequent membrane rupture (Bahr et al., 2020). Non-penetrating cryoprotectants, which do not penetrate the membrane (e.g., trehalose, sucrose and other carbohydrates), because of their high molecular mass, can form hydrogen bonds with water (Kusuma et al., 2018).
Many studies, based on obtained proofs, suggested combining both penetrating and non-penetrating cryoprotectants for higher effectivity (Ha et al., 2005; Bahr et al., 2020). It is necessary to adjust the concentrations of cryoprotectants, as it has been shown that excessively low concentrations can lead to freezing shock (the damage caused by the freezing process), while excessively high concentrations can be toxic (Best, 2015).
Freeze-drying, or lyophilization, is a method of pre-cooling materials that contain moisture. The lyophilized material freezes below the freezing point to a solid, directly sublimes the ice in vacuo and removes it as water vapor. This technology is divided into three main phases: 1) the pre-freeze phase; 2) the sublimation drying phase and 3) the analytical drying phase (Yang et al., 2020). The freeze-dried exosomal lyophilized powder can effectively reduce the complexity of storage conditions. Because the dehydration and drying of the products is completed under low temperature and vacuum conditions, the material can better retain its original activity and possible damage is significantly reduced. In addition, such a material can be easily maintained in a stable, storable state and can be easily reconstituted by simply adding water.
Similar to cryopreservation, the freeze drying and dehydration stress generated during this process can lead to destructive effects on the structure of EXs. For this reason, the use of “lyoprotectants” is proposed to protect EXs and their cargo (Nireesha et al., 2013). The most commonly used stabilizers (lyoprotectants) are disaccharides, such as lactose, sucrose and trehalose. Some studies have shown that trehalose appears to be the most effective disaccharide for preserving EXs during lyophilization, because it effectively prevents aggregation of EXs during lyophilization, increases their colloidal stability and does not alter their morphology (Bosch et al., 2016; Charoenviriyakul et al., 2018).
Spray drying is an approach that is systematically applied to the drying of various materials. After spraying the exosome solution in the drying room, the moisture evaporates rapidly on contact with hot air to give a dry powder. Factors that affect the stability of EXs during the process are mainly atomization pressure and outlet temperature. Compared to lyophilization, spray drying is faster and, since the drying method is designed to be continuous, it is also more economical (Kusuma et al., 2018).
The spray drying process is composed of five steps: 1) concentration, the material is normally concentrated before introduction into the spray dryer; 2) atomization, according to the required properties of the dried product, the atomization phase creates optimal conditions for evaporation; 3) droplet–air contact, in a special chamber, the atomized liquid then comes into contact with the hot substance of the gas, which causes evaporation of 95% of the contained water (in droplets) within a few seconds; 4) droplet drying, this takes place in two phases; during the first phase (it proceeds at a slightly constantspeed) there is enough moisture in the drop to replace the evaporated liquid on the surface. The second phase occurs when there is no longer enough moisture to maintain a saturated state on the surface of the droplets, thus, a dried shell is formed around the surface of the droplet; 5) separation, the final phase, where cyclones, bag filters, and/or electrostatic precipitators are used (Bahr et al., 2020).
At present, frozen storage at −80°C is generally the best complex storage method (Zhang et al., 2020). Nevertheless, for the right long-term storage and stability of EXs (depending on the use of different sources and different experimental techniques), a well-chosen temperature has a significant influence. Some studies have found that, in comparison with freshly extracted EXs, storage at −80°C for 4 days can change the morphology of EXs (Maroto et al., 2017) and that the biological activity of EXs can be decreased during storage at −80°C for a period longer than 28 days (Lorincz et al., 2014). Kalra H. et al. (2013) observed that exosomal recovery rate is not dependent on the viscosity of plasma or the instability of EXs after freezing at −80°C (Kalra et al., 2013). However, at the same time, some other studies have shown that milk-derived EXs are stored at −80°C for 4 weeks without any change in their physical properties (Agrawal et al., 2017). Therefore, it is necessary to study the storage stability of EXs from multiple perspectives (e.g., in terms of source, application technology or future research direction), especially for long-term storage.
Influence of EXs Isolation Method for Downstream Application
EXs can serve as indicators of normal biological processes, pathological development, or pharmacological response to a therapeutic intervention. The good biomarkers are sensitive, specific, noninvasive, and efficiently assayed using a fast and relatively inexpensive method (Alvarez et al., 2012). The isolation method strongly determines which subpopulations of EXs are captured, which in turn determines how suitable resulting samples are for various downstream applications (Buschmann et al., 2018).The isolation methods of EXs fundamentally influence EV composition (proteins, DNA, RNA or other studied compounds), size, concentration, purity and functionality (Abramowicz et al., 2016; Zhao et al., 2016). Moreover, the source of EXs can influence the choice of isolation method.
There is also a fundamental issue that is not satisfactorily addressed, i.e., the technical standardization of exosome isolation (Witwer et al., 2013). Unfortunately, no standard methods are available for exosome extraction. A few groups have conducted comparative studies on different methodologies (Witwer et al., 2013; Rekker et al., 2014; Greening et al., 2015; Royo et al., 2016). The most often used methods include ultracentrifugation, density gradient centrifugation, chromatography, filtration, polymer-based precipitation and immunoaffinity. Based on the high sensitivity of current molecular techniques, even minor components of EXs can be detected and identified. Thus, the co-isolation of contaminating non-exosomal material can generate a significant artifact (Taylor and Shah, 2015).
Limitations of EXs Isolation Methods
Each of the most often used EXs isolation methods carry some limitations, in view of the EXs further characterization or application. Ultracentrifugation (UC) can reduce the amount of non-EV particles co-isolated with the EVs, however, it also results in reduced particle yield, due to lost and damaged EVs during high speed centrifugation. Polymer-based precipitation uses volume-excluding polymers to reduce the solubility of EVs and similarly sized proteins and particles, which are subsequently isolated using low speed centrifugation. However, polymer-based approaches require the use of protein removal kits to reduce the amount of co-precipitated proteins. Size-exclusion chromatography (SEC), using e.g., the IZON qEV columns, allows EVs larger than 70 nm to be separated from smaller particles and proteins that take longer to pass through the column. However this method does not completely separate EVs from non-EV material, and the presence of EVs in multiple fractions leads to dilute samples that often need to be pooled and requires an additional concentration step. Density gradient centrifugation uses a density gradient medium such as iodixanol and centrifugal force to separate and purify EVs based on their buoyant density. A second ultracentrifugation step is then required to isolate EVs from a fixed density range, which can result in the loss and damage of some EVs while co-isolating non-EV particles of a similar density.
Faster and more efficient methods to isolate EXs including ultrafiltration (UF) (Cheruvanky et al., 2007), also carry their own set of limitations. For example, highly abundant proteins present in urine tend to be retained by UF, and subsequently interfere with the identification of exosomal proteins (Rood et al., 2010).
EXs Isolation Method in View of Downstream Proteomic Analysis
The most widely used method for the isolation of EXs from biological fluids, is based on a two-step differential UC (Alvarez et al., 2012). Although this method efficiently isolates EXs, it is time-consuming and requires expensive equipment. The UC method provided the lowest exosome recovery rate, but a higher protein purity of EXs obtained by UC than e.g., with ExoQuick and TEI. Therefore, UC is more suitable for proteomic research. Centrifuging at the high speeds used for EXs isolation could also possibly lead to fusion of the particles with contaminants and other proteins, affecting the physical properties of the EXs and the sensitivity of proteomic analysis (van der Pol et al., 2012; Linares et al., 2015)). It can also cause loss of EXs from the sample, leading to lower and more variable exosome yield (Alvarez et al., 2012).
Three different techniques: UC and two commercial kits, exoEasy (EE; membrane affinity spin column method) and ExoQuick (EQ; polymer-based precipitation) for EXs isolation of patients with breast carcinoma, and further cytokine content analysis were compared (Jung et al., 2020). The dramatic enrichment of CD63 in EXs isolated by UC, but not from EE and EQ, was observed. The high protein content and the low particle-protein ratio together with the low levels of exosome markers isolated from the EQ and EE methods suggest co-isolation of contaminating serum proteins. The cytokine expressions per exosomal particle were the highest in the UC method, which seems to be the most appropriate method when isolating EXs from blood for cytokine profiling. The UC method, however, needs to be tightly controlled because the yield varies with several parameters such as rotor type, sample viscosity, and centrifugation time (Linares et al., 2015).
Kalra H et al. optimized methods to isolate EXs from blood plasma, based on three exosome isolation techniques; differential centrifugation coupled with UC, epithelial cell adhesion molecule immunoaffinity pull-down, and iodixanol density gradient separation. The iodixanol density gradient method was superior in isolating pure exosomal populations, devoid of highly abundant plasma proteins (Kalra et al., 2013). (Gemoll et al., 2020) demonstrated that EV protein expression is highly dependent on the isolation approach. On the proteomic level, UC and ExoQuick seem to be two complementary approaches allowing the detection of different proteoforms with different abundance and purity levels (Gemoll et al., 2020). Thereby, engagement of highly standardized operating procedure for EV isolation, handling and analyzing in combination with an increased transparency for data reporting are needed for implementation of non-invasive EV-based biopsies for cancer diagnostic and prognostic.
EXs Isolation Method in View of Downstream Transcriptomic Analysis
Similar exosomal small RNA profiles from iodixanol gradient and UC exosome isolation methods, suggesting that the presence of remaining cell debris does not influence downstream transcriptomic analysis, in particular, for the detection of small RNA biomarkers, was observed by Queck et al. (Quek et al., 2017). The commercial kits for EXs isolation are robust, fast, use little sample, and hence serve as ideal choices for the identification of exosomal miRNA disease biomarkers. Compared to UC as the most commonly used method to isolate EXs from plasma/serum, higher miRNA yield in serum EXs was detected by some commercial precipitation kits compared with UC (Keller et al., 2011).
Alvarez et al. (2012) also showed that the exosome isolation method based on the commercial precipitation reagent ExoQuick-TC, yields the highest quantity and quality of exosomal miRNA and mRNA from urine (Alvarez et al., 2012). Helva et al. described a disproportionate relationship between the concentration of particles and the concentration of miRNAs after EXs isolation using UC and exosome isolation kits (miRCURY, ExoQuick, and Invitrogen Total Exosome Isolation Reagent). This is probably due to the heterogeneity of exRNA distributions across the exosome populations and the potential contamination of non-exosome particles including high-density lipoproteins and RNA binding proteins aggregates, which was reported previously (Vickers et al., 2011; Mikaelian et al., 2012; Chevillet et al., 2014). However, the majority of these kits isolate or precipitate EXs and inevitably suffer from the co-isolation of other EVs and protein complexes (Helwa et al., 2017).
There are outstanding publications comparing different strategies for isolating EVs from human serum for RNA analyses that relied on RT-qPCR to profile vesicular miRNAs, (Rekker et al., 2014; Andreu et al., 2016; Crossland et al., 2016). Buschman et al. comprehensively analyzed vesicular microRNAs (miRNAs) in patient biofluids for biomarker studies (Buschmann et al., 2018). They compared five different methods of EVs isolation in combination with two RNA extraction methods regarding their suitability for biomarker discovery. Their findings reveal that isolation by precipitation and membrane affinity was highly suitable for miRNA-based biomarker discovery; whilst methods based on SEC failed to separate patients from healthy volunteers. Isolated vesicles differed in size, quantity, purity and composition, indicating that each method captured distinctive populations of EVs, as well as additional contaminants (Rekker et al., 2014).
Comparative data on methods for isolating EVs from patient biofluids are scarce, despite clear interest in utilizing EVs and their miRNA cargo for biomarker studies. Although a strong correlation of exosomal miRNA profiles was observed between the UC and ExoQuick precipitation method from blood serum of healthy individuals followed by determination of the expression profile of 375 miRNAs by real time PCR using Exiqon miRCURY LNA™ microRNA Human panel I assays, distinct clusters of miRNA levels between the used methods were identified. The detected levels of two miRNAs, miR-92a and miR-486-5p, were significantly. influenced by the exosome isolation method used (Rekker et al., 2014).
Conclusion Remarks to Influence of Isolation Methods for Downstream EX Content Analysis
The choice of EV isolation method used should depend also on the amount of starting material together with the downstream application, and will be influenced by the need to remove all, or only distinct groups, of non-EV serum components. A recent survey conducted by Gardiner et al. revealed that although UC remains the most commonly used isolation method, other approaches have gained preference when starting volume is limited (Gardiner et al., 2016). It was shown that both qEV and exoRNeasy recover more particles from sepsis sera (Buschmann et al., 2018). Studies relating to small volumes of serum or plasma have either compared different commercial EV isolation kits (Macías et al., 2019) or compared commercial EV isolation kits to size exclusion columns or UC (Andreu et al., 2016; Takov et al., 2019). Density gradients have not been compared and are normally used for large volume EV isolations, such as cell culture conditioned media (Lobb et al., 2015).
We can conclude that most evidence clearly indicates a significant failure to recover exosomal protein and RNA following UC and density gradient centrifugation (Taylor et al., 2011). SEC appears to be a better alternative for exosome isolation, since there is no loss of EXs or damage to the vesicle structure. However, like the centrifugation approaches, total EXs are isolated by this approach. Most studies support the finding that immunoaffinity capture can isolate both total EXs and pathology-specific EXs, while maintaining vesicle integrity and cargo content (Mathivanan et al., 2010; Tauro et al., 2012). Since all of these isolation approaches have limitations, new technologies are being developed, but not commonly available as field flow fractionation (FFF) (Contado, 2017). While still being developed, FFF is based on laminar flow of particles in a solution, where a mixture of particles is propelled through a channel, perpendicular to the direction of flow, resulting in separation of the particles present in the suspension. This method is unique from other separation methods, since it can separate components over a wide colloidal size range.
Biomedical Applications of EXs
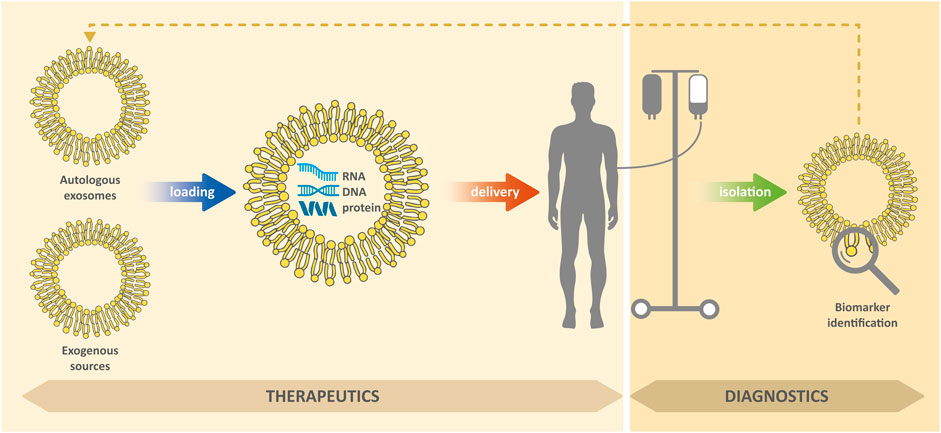
EXs in body fluids can carry markers of various diseases, including biomolecules, which are otherwise difficult to detect and which (due to lipid membrane) can be protected against degradation (Raimondo et al., 2011; Tu et al., 2011; Lin et al., 2015). In general, successful treatment of diseases usually depends, among other things, on their timely detection. Due to their presence in readily available body fluids (such as blood or urine), they are a valuable biomedical tool for non-invasive diagnostic approaches for cancer, neurodegenerative, cardiovascular and other diseases (Shi et al., 2015; Hirshman et al., 2016; Patel et al., 2019; Deb et al., 2021). In addition, EXs can be used as therapeutic agents to modulate inflammation and promote tissue regeneration, novel modulators for vaccination, or as delivery systems to deliver therapeutic molecules to target cells (Deb et al., 2021; Kim et al., 2021); see Figure 2. However, for medical applications research is necessary, in particular: 1) the ensuring the isolation of pure EXs with a minimum of contaminants; 2) the development of standardization of methods for their isolation; 3) the testing of efficacy and safety of their administration.
EXs in Diagnostics
The specific nature of EXs can be used, inter alia, for the prediction of cancer. An example is a study of lung adenocarcinoma, in which it has been reported that circulating miRNAs may act as potent biomarkers overall, helping to differentiate patients from healthy controls (Rabinowits et al., 2009). Many miRNAs (e.g., miR-17-5p, miR-10b, miR-550, miR-21) have been isolated from several different EXs obtained from the peripheral blood of pancreatic cancer patients. Subsequently, these miRNAs have been found to be an effective tool for cancer screening (Qiu et al., 2018). It has also been found that the level of miR-373 is highly prominent in the serum of breast cancer patients (Eichelser et al., 2014). Studies of gastric cancer have revealed that an exosomal load-containing long non-coding RNAs (such as INHBA-AS1, CEBPA-AS1, UCA1, AK001058, MIR4435-2HG and LINC00152) is a potent biomarker in blood samples (Ke et al., 2017).
Research into ovarian cancer has shown that EXs containing a tight linker protein claudin-4 are present in the peripheral blood. Several reports indicate that adaptation of the sensitive assay technique for determination of claudin-4 levels can subsequently be used as an effective screening criterion for this type of cancer (Li et al., 2009). Several other studies have revealed other indicators (biomarkers) of various types of cancer, such as for example the presence of excessive amounts of proteoglycan (glypican-1) in EXs obtained from patients with pancreatic cancer, or the presence of MIF indicating the occurrence of liver metastases (Costa-Silva et al., 2015).
As was mentioned before, in addition to cancer as one of the most common serious diseases today, exosomes can also be used in the diagnosis of various other illnesses, such as various neurological, renal or cardiovascular disorders.
The research of various inflammatory and signaling molecules, proteins and RNAs, which occur in the cerebral EXs may serve as a suitable tool in the detection of a different spectrum of neurological disorders (Shaimardanova et al., 2020). Because EXs can cross the blood-brain barrier (BBB) (Xu et al., 2017); they can be isolated from cerebrospinal fluid (CSF) or even blood plasma for diagnostic use (Stuendl et al., 2016).
One of the biomarkers that are present in EXs (and which are also potential players in the pathogenesis of the disease) is protein. For example, in the case of chronic traumatic encephalopathy, which is very common in athletes, so-called tau proteins are found in increased values in plasma EXs (Stern et al., 2016). Another example is the reduction in the level of functional synaptic proteins in Alzheimer’s disease as well as frontotemporal dementia. Decreased functional levels of these proteins are considered a potential biomarker, because in the early stages of the disease the synaptic dysfunction is noticed. The value of proteins in astrocyte-derived EXs is an indicator of patients’ cognitive functions and also prognostic potential (Goetzl et al., 2016).
Extracellular RNAs, which are also found in neuronal cell-derived EXs, are currently considered to be the most potential markers of disease (Rao et al., 2013). Any changes in RNA levels are considered anomalies and indicate a disease state, because circular RNA, miRNA, or even long non-coding RNAs are key to maintaining human cellular homeostasis. Specific examples are circulating fragments of U2 small nuclear RNAs, which are considered a useful biomarker in primary CNS lymphoma (Baraniskin et al., 2016) or levels of long non-coding RNAs RP11462G22.1 in Alzheimer’s and Parkinson’s disease (Gui et al., 2015; Song et al., 2021). Some studies in this area have shown the diagnostic and prognostic role of miRNAs also in trauma, where miR-212 levels decrease and, on the contrary, miR-7a, miR-21, miR-7b and miR-146 expression levels increase (Harrison et al., 2016; Meissner et al., 2016).
Cardiovascular disease is one of the main causes of mortality worldwide. This disease causes almost 20 million deaths a year. In this case, EXs play an important role in regulating the development of disease through the transport and exchange of signaling molecules (Sahoo and Losordo, 2014; Barile et al., 2017). Some studies have shown that EXs regulate processes such as apoptosis, angiogenesis, fibrosis and inflammation in cardiac tissues (Guo et al., 2020). In the cardiovascular system, the number and content of EXs vary depending on physiological conditions (Zamani et al., 2019). Injured cardiomyocytes have been found to release EXs that contain certain miRNAs (e.g., miRNA-1 and miRNA-133a) specific for the heart. Thus, myocardial infarction and any other acute coronary syndrome can be easily identified by controlling the expression of these miRNAs (Kuwabara et al., 2011).
Research has also shown that these miRNAs are upregulated in myocardial infarction and peri infarction in patients with angina pectoris (Cheng et al., 2012). A study by Matsumat et al. revealed that EXs also overexpress miRNA-34a, miRNA-192 and miRNA-194 in heart failure (Matsumoto et al., 2013). At the same time, it has been found that also miRNA-92 is upregulated in such affected patients (Goren et al., 2012). Exosomal proteins may indicate disease states. Dejong et al. described that EXs obtained from hypoxic cardiac cells contain collagen, fibronectin, and lysyl oxidase (de Jong et al., 2012). It has also been found that changes in exosomal proteomes occur after heart transplantation (Kennel et al., 2018).
All types of renal cells, such as glomerular epithelial cells, podocytes, proximal/distal epithelial cells and collecting duct cells (Miranda et al., 2010), secrete EXs. These can also be generated in very small quantities from the systemic circulation (Cheng et al., 2012). However, it can be assumed that most urinary EXs originate from the kidney. EXs secreted by the kidneys contain several biomolecules that could indicate possible abnormalities in the kidneys (Prabu et al., 2019), such as in the case of acute renal injury, where there is (with reference to control, healthy subjects) elevated GPRC5B levels, making this protein a possible diagnostic marker for this type of disease (Kwon et al., 2014). In addition to GPRC5B, ATF3 (activating transcription factor-3) and Fetuin A (Zhou et al., 2006) are also biomarkers indicating acute renal injury.
Other possible and effective biomarkers marking glomerular as well as tubular damage further include, for example, neutrophil gelatinase-associated lipocalin (NGAL) and Wilms Tumor 1 homolog (WT1). The biomarker WT1 was obtained from EXs isolated from the urine of patients suffering from focal segmental glomerulosclerosis (Zhou et al., 2013) and also from a maximum of diabetic patients (Kalani et al., 2013). An above standard level incidence of NGAL was then reported in patients with delayed graft function after kidney transplantation (Alvarez et al., 2013). In contrast, the absence of Prominin-1 (CD133) was indicated as a biomarker in the end-stage renal disease (Dimuccio et al., 2014).
Formerly, renal mRNA has been widely used as a diagnostic and prognostic tool for renal disorders. Current research is also focused on the possible use of miRNA obtained by non-invasive isolation from urinary EXs. One study reported that in a patient with diabetic nephropathy, levels of exosomal miR-130a and miR-145 were found to be elevated from normal and levels of miR-424 and, contrary, miR-155, were decreased (Barutta et al., 2013). Recently, several biomarkers for chronic kidney disease and renal fibrosis have also been characterized. Levels of exosomal miR-29a, miR-29c, miR-200b and miR-200c have been shown to be observed in patients with moderate disease to advanced fibrosis (Lv et al., 2013).
EXs in Therapy
EXs have several characteristics suitable for drug delivery. These include nano-size, non-toxicity, biocompatibility, low immunogenicity as well as targeting ability and organotropis (Lim and Kim, 2019). The similarities between EXs and liposomes in size and ability to carry both hydrophilic and lipophilic molecules is obvious. However, EXs’ asymmetric lipid distribution and specific protein composition of their membranes justify their organotropism and ability to target (Antimisiaris et al., 2018). This is supported by evidence that cancer-derived EXs preferentially fuse with their parent cells (Qiao et al., 2020). However, the clinical use of EXs as drug carriers is fraught with several technical problems, such as: 1) relatively low production yields; 2) considerable structural heterogeneity; 3) difficulties in drug loading 4) the development of standard, scalable and cost-effective procedures for their isolation and purification (Lu and Huang, 2020).
To overcome these problems, bioinspired exosome vesicles have been developed as an alternative to naturally derived Exs. Most of the, until now proposed, artificial exosomal mimetic systems, so-called hybrid Exs, are formed by the fusion of exosomal and liposome membranes (Sato et al., 2016) or are obtained by serial extrusion of a mother cell suspension by reducing pore size (Jang et al., 2013).
Exs are currently seen as ideal candidates for the delivery of antitumor agents, miRNAs, siRNAs, and various other biomolecules with therapeutic potential. Possible therapeutic targeting of Exs involves cell targeting, altering the native result of Exs (so that the process of tumorigenesis can be disrupted by interactions with tumor cells), stromal cells, and other immune cells (Guo et al., 2020; Deb et al., 2021).
It has been found that the removal of Exs from the circulatory system appears to be a possible functional treatment option to eliminate the metastatic effect of cancer (Tickner et al., 2014). At the same time, some studies also suggest that inhibition of exosome generation can arrest tumor formation using several different techniques, such as proton pump inhibition, microtubule assembly targeting, or endosomal sorting (Pilzer et al., 2005; Azmi et al., 2013).
Some studies suggest that the use of Exs in the treatment of cancer for the delivery of chemotherapeutic agents appears promising (Yeo et al., 2013). For example, the growth of cancer cells in breast and colon tumors has been successfully inhibited by doxorubicin-containing Exs or by mimetic exosomal nanosvesicles (Jang et al., 2013; Tian et al., 2014; Smyth et al., 2015). Based on the observation that exosomal microRNAs act effectively on the target mRNA and thus suppress gene expression in recipient cells, cancer research has also focused on delivering a specific miRNA or a small useful amount of interfering RNA (siRNA). Preclinical testing for the delivery of therapeutic miRNAs or siRNAs via Exs has focused, for example, on antitumor therapy in rodents with breast cancer (Ohno et al., 2013), glioma (Katakowski et al., 2013) or pancreatic cancer (Kamerkar et al., 2017; Mendt et al., 2018).
Further studies also indicate that Exs can be a successful alternative for the delivery of antigens which are derived from a tumor to elicit an immune response (El Andaloussi et al., 2013). Some studies based on this finding are already in the clinical trials phase (Escudier et al., 2005; Morse et al., 2005; Dai et al., 2008). It has been found that Exs play a role in exosome-mediated cell communication in cancer and that Exs derived from cancer stem cells are involved in their differentiation (Koch et al., 2014; Lin et al., 2015; Parimon et al., 2018). It has been observed that Exs participate in conferring resistance to cancer cells as well (Qu et al., 2016). All of the above studies (and many others) suggest that tumor cell-derived Exs indeed play a key role in both cancer cell proliferation and their possible elimination. Finally, they may also play a crucial role in the potential resistance to some drugs.
There are several causes that can lead to CNS tissue damage. These include injuries, hereditary diseases, infections, ischemia and many others. The fact that the BBB is only very selectively permeable is a major limitation in the treatment of CNS diseases (Timbie et al., 2015). Therefore, recent research has focused on finding various new techniques for successful BBB penetration, an example being the use of nanoparticles (Zhang and Yang, 2018).
In addition to being a very potent biomarker, EXs are potential candidates for drug delivery. EXs are highly desirable as drug carriers, especially because of their nanoscale dimensions, ability to cross the BBB, and protection against degradation - they can retain their original state (Kourembanas, 2015). However, in addition to their advantageous properties, EXs have a disadvantage as well, they were shown to assist the progression of some diseases by spreading pathogenic agents into healthy cells (Bakhshandeh et al., 2016). Despite this possible disadvantage, the potential use of EXs has been identified in several neurodegenerative diseases such as Parkinson’s disease (Zhao et al., 2014), Alzheimer’s disease (Alvarez-Erviti et al., 2011; Stuendl et al., 2016), Huntington’s disease (Didiot et al., 2016), epilepsy (Longa et al., 2017), multiple sclerosis and amyotrophic lateral sclerosis (Rajan et al., 2016).
A number of studies have been presented on the role of EXs in myocardial infarction. Some studies revealed that customized CD34 + stem cells can release EXs containing pro-angiogenic factors and a sound hedgehog (Shh) (MacKie et al., 2012). These specifically treated CD34 + cells, when injected into a mice model of acute myocardial infarction, showed a protective function against ventricular dilatation (MacKie et al., 2012). Another study showed that EXs derived from mesenchymal stem cells can increase the division of M1 macrophages into M2 macrophages, resulting in a significant enhancement of the phenomenon of inflammation in cardiac tissues and thus a reduction in infarct size.
Another study reported that exosomal miR-19a derived from the aforementioned cell type has anti-apoptotic effects caused by activation of the AKT protein, targeting SOX6, and blockade of the JNK3/caspase-3 pathway (Lu and Huang, 2020). EXs from cardiosphere-derived cells have been described as possible therapeutic agents in the treatment of cardiovascular disease. In the case of post myocardial infarction, these EXs were found to be able to enhance cardiac fibrosis. At the same time, it has been observed that, in cardiomyocyte apoptosis, the process of endothelial cell tube generation is impaired (Tseliou et al., 2015; Namazi et al., 2018).
Research revealed that another promising nanotechnology tool that can be used for treatment of cardiovascular diseases is cardiac progenitor cell (CPC) derived EXs (Guo et al., 2020). Studies in vivo models of myocardial infarction have shown that CPC-derived EXs lead to an increase in miR-210, resulting in a decrease in ephrin A3 and PTP1b, causing a decrease in apoptosis in cardiomyocytes (Barile et al., 2014). These EXs are also able to activate PAPP-A, leading to the release of IGF-1 and ultimately increasing intracellular Akt and ERK1/2 (Barile et al., 2014).
In addition to the above-mentioned uses as biomarkers, EXs also offer a number of therapeutic possibilities in kidney diseases. Examples include their use in modulating kidney transplant rejection, correcting several metabolic deficiencies, or promoting renal regeneration (Deb et al., 2021). A group of researchers, led by Grange, demonstrated that labeled MSC-Exos administered intravenously was able to target the injured kidney (Nassar et al., 2016). In another study, it was observed that, in patients with stage 3 or 4 chronic kidney disease, MSC-EXs were able to extemporize the glomerular filtration rate and also reduce albumin excretion (Dorronsoro and Robbins, 2013).
Other research has shown that repeated administration of EXs can prevent the development of chronic tubular injury. EXs isolated from bone marrow mesenchymal cells and umbilical cord mesenchymal cells have a protective effect against cisplatin-induced nephrotoxicity in both in vivo and in vitro models. The presumed reason is probably the fact that the human umbilical cord MSCs-Exo can successfully reduce the level of Bax (a protein similar to 4 bcl-2) and, conversely, increase Bcl-2 (B-cell lymphoma 2) to prevent apoptosis and activate the Erk1/2 pathway, leading to increased proliferation after cisplatin-induced renal injury (Dorronsoro and Robbins, 2013).
Diabetes is one of the main causes of chronic kidney disease, especially in developed and Western countries. Around 40% of patients with this disease develop diabetic neuropathy as the main complication (Prabu et al., 2019). Jiang et al. published the results of a 1-week study in intravenous EXs and studied their effect on renal damage and streptozotocin - induced angiogenesis in a Sprague-Dawley rat model. These results showed a reduction in urine volume and microalbuminuria as well as suppression of apoptosis of tubocytes and tubular epithelial cells induced by high glucose (Jiang et al., 2016). Some of the other remarkable work done with the EXs in the field of renal treatment include, for example, studies of Jia et al. (Jia et al., 2018), Sanches et al. (Sanchez et al., 2014) or Cantaluppi et al. (Cantaluppi et al., 2012).
Role of EXs in Senescence and Aging
Senescence is the cellular part of tissue aging due to irreversible growth arrest and other physiological changes that occur in the morphology, behavior and function of cells. Aging is also a well-known risk factor in the development of many human diseases, such as neurodegenerative disorders, cardiovascular disease, or cancer, which are common causes of death or invalidity, especially in the elderly population (Xu and Tahara, 2013). miRNAs, that regulate gene expression, play an important role in many biological processes. In terms of aging, some miRNAs were proven to be key regulators during cell aging (Weilner et al., 2013). Many studies have already shown that EXs contain miRNAs and play an important role in cell to cell communication and information transfer (Simons and Raposo, 2009; Camussi et al., 2010; Mathivanan et al., 2010; Bang and Thum, 2012). EXs with miRNAs were found to participate in the senescence of a complex cell network and contribute to aging. There is clear evidence that aging increases the number of circulating EXs in tissues. Especially from aging cells, the secretion of immunosuppressive EXs is strongly increased.
There are studies that have shown that EXs from aging cells are relatively significantly involved in the expansion of aging into neighboring cells (Alibhai et al., 2020; Salminen et al., 2020). Very interestingly, age-related EXs also contain, among other things, immunosuppressive costs that increase immunosuppression in recipient immune cells, i.e., tissue resident and accepted immune cells, e.g., M2 macrophages, myeloid-deriv ed suppressor cells (MDSC) or regulatory T cells (Salminen et al., 2020). It seems that senescent cells secrete immunosuppressive EXs in an attempt to escape the immune surveillance, and their accumulation within tissues consequently augments the aging process (Xu and Tahara, 2013; Salminen et al., 2020).
EXs and COVID-19 Virus Infection
Since December 2019, the SARS-CoV-2 (COVID-19) pandemic has become a serious public health problem worldwide. Although a number of vaccines already exist today (e.g., Pfizer and BioNTech, Moderna, Astra-Zeneca, Johnson and Johnson), finding a safe and effective prognostic, diagnostic and therapeutic tool for patients, especially those with severe COVID-19 infection, is still an actual theme. Knowledge of the host response to the new type of coronavirus SARS-CoV-2 remains limited, which prevents a detailed understanding of the pathogenesis of the disease and, consequently, the development of effective therapeutic strategies (Hassanpour et al., 2020) (Hassanpour et al., 2020). It is common for host cells to release EXs (and other extracellular vesicles) that carry viral as well as host components that can modulate the immune response during viral infection (Tan et al., 2016; Kurywchak et al., 2018; Li et al., 2019). The same seems to be the case with this disease.
In one research focused on the role of EXs in Covidem-19 infection, the scientific team asked several basic questions for this statement: 1) does COVID-19 virus use EXs and other extracellular vesicles for infection? and 2) can extracellular vesicles be targeted for the treatment of COVID-19 virus infection? The results of this study revealed that EXs from COVID-19 infected cells may contribute to spread and infection. EXs contain receptors for COVID-19 virus such as CD9 and ACE2, which may be involved in promoting COVID-19 virus infection. At the same time, the possibility of preclinical and clinical application of EXs for the treatment of COVID-19 infection was outlined, which would include stem cell-derived exosome therapy, exosome-based drug delivery, possible inhibition of exosome biogenesis and absorption, and, last but not least, a new exosome-based vaccine (Hassanpour et al., 2020).
A study by Barberis et al. investigated how SARS-CoV-2 infection affects exosome content, the involvement of EXs in disease progression, and the possible use of plasma EXs as biomarkers in determining disease severity. The results of proteomic analysis of EXs obtained from patients with this type of virus identified several molecules that participate in immune responses, inflammation, and activation of coagulation and complement pathways, which are likely to be major mechanisms leading to tissue damage and subsequent multi-organ dysfunction. Moreover, several potential biomarkers, such as fibrinogen, fibronectin, the C1r subcomponent of complement and the P-component of serum amyloid, have been shown to correlate with established disease severity. In addition, the presence of SARS-CoV-2 RNA in the exosomal load was also detected in this study, supporting the hypothesis that the virus may use the endocytosis pathway to spread infection (Barberis et al., 2021).
Another study addressed the issue of relapse of COVID-19 infection. The authors hypothesized that one of the possible mechanisms of relapse of this infection may be a cellular transport pathway associated with the release of EXs containing SARS-CoV-2 and other extracellular vesicles. The results suggest that this so-called “Trojan horse” strategy provides a possible explanation for viral RNA recurrence, even in recovered patients with COVID-19 7–14 days after discharge. It appears that viral material could be hidden in EXs or other extracellular vesicles during this time and then spread again (Elrashdy et al., 2020) (Elrashdy et al., 2020).
Several other current studies were focused on the use of EXs (e.g., from mesenchymal cells) as nanoplatforms for therapeutics and drug delivery in the fight against COVID-19 (Alzahrani et al., 2020; Sengupta et al., 2020; Pinky et al., 2021), exosome-mediated mRNA delivery for SARS-CoV-2 vaccination (Tsai et al., 2020) or their potential role on cytokine storm and treatment (Kadriyan et al., 2020).
Conclusion
EXs are now well known as important mediators of cell to cell communication, as biomarkers, and as crucial players in both physiological and pathological processes, due to their secretion from various types of cells, and detection in almost all kinds of body fluids. The exosome capacity to envelope a wide range of content, including lipids, RNAs, proteins or therapeutics, to signal specific recipient cells or tissues, makes them a promising diagnostic and therapeutic nanosize tool.
In addition to conventional (usually mammal) sources of EXs, there is a wide range of “nonconventional” (non-mammalian) sources of EXs, which are at present intensively studied, mainly for their inter-species and inter-kingdom communication, which can have important environmental, ecological or medicinal impact.
Furthermore, the isolation and purification methods of EXs is still a “hot topic”, which needs to be standardized for the feasible clinical application of EXs, or given a clearer overview of the advantages and limitations of individual isolation and purification methods, as well as storage conditions, for further downstream EXs characterization or application.
Undoubtedly, EXs represent a promising tool in the field of nanomedicine in diagnostics, therapy, and recently, with more emphasis on senescence and aging, in issues related to Covid-19.
Author Contributions
OJ contributed to the organization of the preparation, writing, reading, correction and submission of the manuscript, as well as to the writing of the Abstract and Chapters 5, 6, and 8; RH contributed to the writing of Chapters 1, 5, and 7; HM and ML contributed to the writing of Chapter 3; AS and DP contributed to the writing of Chapter 4; JM contributed to the editing and reading of all manuscripts.
Funding
The review was supported by the SGS Faculty of Science under the project with No. 53221 16 2013 01, by the Internal Grant Agency, Jan Evangelista Purkyne University in Usti nad Labem; by the Czech Science Foundation project No.20-21421S; by the European Union – European Structural and Investments Funds in the frame of the Operational Programme Research Development and Education, the ERDF/ESF project “UniQSurf - Centre of biointerfaces and hybrid functional materials” (No. CZ.02.1.01/0.0/0.0/17 _048/0007411) and Project U21 - Improving the Quality of the Grant Competition and Teaching in Doctoral Study Programmes at UJEP, CZ.02.2.69/0.0/0.0/19_073/0016947.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.846650/full#supplementary-material
Supplementary Table S1 | Overview of main Exosomes isolation methods.
References
Abramowicz, A., Widlak, P., and Pietrowska, M. (2016). Proteomic Analysis of Exosomal Cargo: the challenge of High Purity Vesicle Isolation. Mol. Biosyst. 12, 1407–1419. doi:10.1039/C6MB00082G
Agrawal, A. K., Aqil, F., Jeyabalan, J., Spencer, W. A., Beck, J., Gachuki, B. W., et al. (2017). Milk-derived Exosomes for Oral Delivery of Paclitaxel. Nanomedicine: Nanotechnology, Biol. Med. 13, 1627–1636. doi:10.1016/j.nano.2017.03.001
Aheget, H., Tristán-Manzano, M., Mazini, L., Cortijo-Gutierrez, M., Galindo-Moreno, P., Herrera, C., et al. (2020). Exosome: A New Player in Translational Nanomedicine. Jcm 9, 2380. doi:10.3390/jcm9082380
Ailuno, G., Baldassari, S., Lai, F., Florio, T., and Caviglioli, G. (2020). Exosomes and Extracellular Vesicles as Emerging Theranostic Platforms in Cancer Research. Cells 9, 1–24. doi:10.3390/cells9122569
Alibhai, F. J., Lim, F., Yeganeh, A., DiStefano, P. V., Binesh‐Marvasti, T., Belfiore, A., et al. (2020). Cellular Senescence Contributes to Age‐dependent Changes in Circulating Extracellular Vesicle Cargo and Function. Aging Cell 19, 1–14. doi:10.1111/acel.13103
Alvarez, M. L., Khosroheidari, M., Kanchi Ravi, R., and Distefano, J. K. (2012). Comparison of Protein, microRNA, and mRNA Yields Using Different Methods of Urinary Exosome Isolation for the Discovery of Kidney Disease Biomarkers. Kidney Int. 82, 1024–1032. doi:10.1038/ki.2012.256
Alvarez, S., Suazo, C., Boltansky, A., Ursu, M., Carvajal, D., Innocenti, G., et al. (2013). Urinary Exosomes as a Source of Kidney Dysfunction Biomarker in Renal Transplantation. Transplant. Proc. 45, 3719–3723. doi:10.1016/j.transproceed.2013.08.079
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. A. (2011). Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 29, 341–345. doi:10.1038/nbt.1807
Alzahrani, F. A., Saadeldin, I. M., Ahmad, A., Kumar, D., Azhar, E. I., Siddiqui, A. J., et al. (20202020). The Potential Use of Mesenchymal Stem Cells and Their Derived Exosomes as Immunomodulatory Agents for COVID-19 Patients. Stem Cell Int. 2020, 1–11. doi:10.1155/2020/8835986
Andreu, Z., Rivas, E., Sanguino-Pascual, A., Lamana, A., Marazuela, M., González-Alvaro, I., et al. (2016). Comparative Analysis of EV Isolation Procedures for miRNAs Detection in Serum Samples. J. Extracellular Vesicles 5, 31655. doi:10.3402/jev.v5.31655
Andreu, Z., and Yã¡Ã±ez-Mã³, M. a. (2014). Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 5, 1–13. doi:10.3389/fimmu.2014.00442
Antimisiaris, S., Mourtas, S., and Marazioti, A. (2018). Exosomes and Exosome-Inspired Vesicles for Targeted Drug Delivery. Pharmaceutics 10, 218. doi:10.3390/pharmaceutics10040218
Azmi, A. S., Bao, B., and Sarkar, F. H. (2013). Exosomes in Cancer Development, Metastasis, and Drug Resistance: A Comprehensive Review. Cancer Metastasis Rev. 32, 623–642. doi:10.1007/s10555-013-9441-9
Bahr, M. M., Amer, M. S., Abo-El-Sooud, K., Abdallah, A. N., and El-Tookhy, O. S. (2020). Preservation Techniques of Stem Cells Extracellular Vesicles: a Gate for Manufacturing of Clinical Grade Therapeutic Extracellular Vesicles and Long-Term Clinical Trials. Int. J. Vet. Sci. Med. 8, 1–8. doi:10.1080/23144599.2019.1704992
Bakhshandeh, B., Kamaleddin, M., and Aalishah, K. (2016). A Comprehensive Review on Exosomes and Microvesicles as Epigenetic Factors. Cscr 12, 31–36. doi:10.2174/1574888x11666160709211528
Bang, C., and Thum, T. (2012). Exosomes: New Players in Cell-Cell Communication. Int. J. Biochem. Cel Biol. 44, 2060–2064. doi:10.1016/j.biocel.2012.08.007
Baraniskin, A., Zaslavska, E., Nöpel-Dünnebacke, S., Ahle, G., Seidel, S., Schlegel, U., et al. (2016). Circulating U2 Small Nuclear RNA Fragments as a Novel Diagnostic Biomarker for Primary central Nervous System Lymphoma. Neuro. Oncol. 18, 361–367. doi:10.1093/neuonc/nov144
Barberis, E., Vanella, V. V., Falasca, M., Caneapero, V., Cappellano, G., Raineri, D., et al. (2021). Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 8, 1–18. doi:10.3389/fmolb.2021.632290
Barile, L., Lionetti, V., Cervio, E., Matteucci, M., Gherghiceanu, M., Popescu, L. M., et al. (2014). Extracellular Vesicles from Human Cardiac Progenitor Cells Inhibit Cardiomyocyte Apoptosis and Improve Cardiac Function after Myocardial Infarction. Cardiovasc. Res. 103, 530–541. doi:10.1093/cvr/cvu167
Barile, L., Moccetti, T., Marbán, E., and Vassalli, G. (2017). Roles of Exosomes in Cardioprotection. Eur. Heart J. 38, ehw304–1379. doi:10.1093/eurheartj/ehw304
Barutta, F., Tricarico, M., Corbelli, A., Annaratone, L., Pinach, S., Grimaldi, S., et al. (2013). Urinary Exosomal MicroRNAs in Incipient Diabetic Nephropathy. PLoS One 8, e73798–8. doi:10.1371/journal.pone.0073798
Batista, I., and Melo, S. (2019). Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Ijms 20, 567. doi:10.3390/ijms20030567
Beer, K. B., and Wehman, A. M. (2017). Mechanisms and Functions of Extracellular Vesicle Release In Vivo-What We Can Learn from Flies and Worms. Cell Adhes. Migration 11, 135–150. doi:10.1080/19336918.2016.1236899
Best, B. P. (2015). Cryoprotectant Toxicity: Facts, Issues, and Questions. Rejuvenation Res. 18, 422–436. doi:10.1089/rej.2014.1656
Böing, A. N., van der Pol, E., Grootemaat, A. E., Coumans, F. A. W., Sturk, A., and Nieuwland, R. (2014). Single-step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J. Extracellular Vesicles 3, 23430. doi:10.3402/jev.v3.23430
Bosch, S., De Beaurepaire, L., Allard, M., Mosser, M., Heichette, C., Chrétien, D., et al. (2016). Trehalose Prevents Aggregation of Exosomes and Cryodamage. Sci. Rep. 6, 1–11. doi:10.1038/srep36162
Buschmann, D., Kirchner, B., Hermann, S., Märte, M., Wurmser, C., Brandes, F., et al. (2018). Evaluation of Serum Extracellular Vesicle Isolation Methods for Profiling miRNAs by Next-Generation Sequencing. J. Extracellular Vesicles 7, 1481321. doi:10.1080/20013078.2018.1481321
Camussi, G., Deregibus, M. C., Bruno, S., Cantaluppi, V., and Biancone, L. (2010). Exosomes/microvesicles as a Mechanism of Cell-To-Cell Communication. Kidney Int. 78, 838–848. doi:10.1038/ki.2010.278
Cantaluppi, V., Gatti, S., Medica, D., Figliolini, F., Bruno, S., Deregibus, M. C., et al. (2012). Microvesicles Derived from Endothelial Progenitor Cells Protect the Kidney from Ischemia-Reperfusion Injury by microRNA-dependent Reprogramming of Resident Renal Cells. Kidney Int. 82, 412–427. doi:10.1038/ki.2012.105
Cantin, R., Diou, J., Bélanger, D., Tremblay, A. M., and Gilbert, C. (2008). Discrimination between Exosomes and HIV-1: Purification of Both Vesicles from Cell-free Supernatants. J. Immunological Methods 338, 21–30. doi:10.1016/j.jim.2008.07.007
Carregari, V. C., Rosa-Fernandes, L., Baldasso, P., Bydlowski, S. P., Marangoni, S., Larsen, M. R., et al. (2018). Snake Venom Extracellular Vesicles (SVEVs) Reveal Wide Molecular and Functional Proteome Diversity. Sci. Rep. 8, 1–17. doi:10.1038/s41598-018-30578-4
Charoenviriyakul, C., Takahashi, Y., Nishikawa, M., and Takakura, Y. (2018). Preservation of Exosomes at Room Temperature Using Lyophilization. Int. J. Pharmaceutics 553, 1–7. doi:10.1016/j.ijpharm.2018.10.032
Chen, B.-Y., Sung, C. W.-H., Chen, C., Cheng, C.-M., Lin, D. P.-C., Huang, C.-T., et al. (2019). Advances in Exosomes Technology. Clinica Chim. Acta 493, 14–19. doi:10.1016/j.cca.2019.02.021
Chen, J., Li, P., Zhang, T., Xu, Z., Huang, X., Wang, R., et al. (2022). Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 9. doi:10.3389/fbioe.2021.811971
Chen, Q., and Lai, H. (2013). Plant-derived Virus-like Particles as Vaccines. Hum. Vaccin. Immunother. 9, 26–49. doi:10.4161/hv.22218
Cheng, Y., Wang, X., Yang, J., Duan, X., Yao, Y., Shi, X., et al. (2012). A Translational Study of Urine miRNAs in Acute Myocardial Infarction. J. Mol. Cell Cardiol. 53, 668–676. doi:10.1016/j.yjmcc.2012.08.010
Cheruvanky, A., Zhou, H., Pisitkun, T., Kopp, J. B., Knepper, M. A., Yuen, P. S. T., et al. (2007). Rapid Isolation of Urinary Exosomal Biomarkers Using a Nanomembrane Ultrafiltration Concentrator. Am. J. Physiology-Renal Physiol. 292, F1657–F1661. doi:10.1152/ajprenal.00434.2006
Chevillet, J. R., Kang, Q., Ruf, I. K., Briggs, H. A., Vojtech, L. N., Hughes, S. M., et al. (2014). Quantitative and Stoichiometric Analysis of the microRNA Content of Exosomes. Proc. Natl. Acad. Sci. U.S.A. 111, 14888–14893. doi:10.1073/pnas.1408301111
Choe, S.-w., Kim, B., and Kim, M. (2021). Progress of Microfluidic Continuous Separation Techniques for Micro-/Nanoscale Bioparticles. Biosensors 11, 464. doi:10.3390/bios11110464
Clayton, A., Court, J., Navabi, H., Adams, M., Mason, M. D., Hobot, J. A., et al. (2001). Analysis of Antigen Presenting Cell Derived Exosomes, Based on Immuno-Magnetic Isolation and Flow Cytometry. J. Immunological Methods 247, 163–174. doi:10.1016/S0022-1759(00)00321-5
Contado, C. (2017). Field Flow Fractionation Techniques to Explore the “Nano-World”. Anal. Bioanal. Chem. 409, 2501–2518. doi:10.1007/s00216-017-0180-6
Contreras-Naranjo, J. C., Wu, H.-J., and Ugaz, V. M. (2017). Microfluidics for Exosome Isolation and Analysis: Enabling Liquid Biopsy for Personalized Medicine. Lab. Chip 17, 3558–3577. doi:10.1039/c7lc00592j
Costa-Silva, B., Aiello, N. M., Ocean, A. J., Singh, S., Zhang, H., Thakur, B. K., et al. (2015). Pancreatic Cancer Exosomes Initiate Pre-metastatic Niche Formation in the Liver. Nat. Cel Biol. 17, 816–826. doi:10.1038/ncb3169
Crossland, R. E., Norden, J., Bibby, L. A., Davis, J., and Dickinson, A. M. (2016). Evaluation of Optimal Extracellular Vesicle Small RNA Isolation and qRT-PCR Normalisation for Serum and Urine. J. Immunological Methods 429, 39–49. doi:10.1016/j.jim.2015.12.011
Cvjetkovic, A., Lötvall, J., and Lässer, C. (2014). The Influence of Rotor Type and Centrifugation Time on the Yield and Purity of Extracellular Vesicles. J. Extracellular Vesicles 3, 23111. doi:10.3402/jev.v3.23111
Czystowska-Kuzmicz, M., and Whiteside, T. L. (2021). The Potential Role of Tumor-Derived Exosomes in Diagnosis, Prognosis, and Response to Therapy in Cancer. Expert Opin. Biol. Ther. 21, 241–258. doi:10.1080/14712598.2020.1813276
Dad, H. A., Gu, T.-W., Zhu, A.-Q., Huang, L.-Q., and Peng, L.-H. (2021). Plant Exosome-like Nanovesicles: Emerging Therapeutics and Drug Delivery Nanoplatforms. Mol. Ther. 29, 13–31. doi:10.1016/j.ymthe.2020.11.030
Dai, J., Su, Y., Zhong, S., Cong, L., Liu, B., Yang, J., et al. (2020). Exosomes: Key Players in Cancer and Potential Therapeutic Strategy. Sig Transduct Target. Ther. 5. doi:10.1038/s41392-020-00261-0
Dai, S., Wei, D., Wu, Z., Zhou, X., Wei, X., Huang, H., et al. (2008). Phase I Clinical Trial of Autologous Ascites-Derived Exosomes Combined with GM-CSF for Colorectal Cancer. Mol. Ther. 16, 782–790. doi:10.1038/mt.2008.1
Danilchik, M., and Tumarkin, T. (2017). Exosomal Trafficking inXenopusdevelopment. Genesis 55, e23011. doi:10.1002/dvg.23011
de Jong, O. G., Verhaar, M. C., Chen, Y., Vader, P., Gremmels, H., Posthuma, G., et al. (2012). Cellular Stress Conditions Are Reflected in the Protein and RNA Content of Endothelial Cell-Derived Exosomes. J. Extracellular Vesicles 1, 18396. doi:10.3402/jev.v1i0.18396
Deb, A., Gupta, S., and Mazumder, P. B. (2021). Exosomes: A New Horizon in Modern Medicine. Life Sci. 264, 118623. doi:10.1016/j.lfs.2020.118623
Deng, Z., Rong, Y., Teng, Y., Mu, J., Zhuang, X., Tseng, M., et al. (2017). Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase. Mol. Ther. 25, 1641–1654. doi:10.1016/j.ymthe.2017.01.025
Didiot, M.-C., Hall, L. M., Coles, A. H., Haraszti, R. A., Godinho, B. M., Chase, K., et al. (2016). Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 24, 1836–1847. doi:10.1038/mt.2016.126
Dimuccio, V., Ranghino, A., Praticò Barbato, L., Fop, F., Biancone, L., Camussi, G., et al. (2014). Urinary CD133+ Extracellular Vesicles Are Decreased in Kidney Transplanted Patients with Slow Graft Function and Vascular Damage. PLoS One 9, e104490. doi:10.1371/journal.pone.0104490
Ding, L.-N., Yang, X., Gao, Z., Effah, C. Y., Zhang, X., Wu, Y., et al. (2021). A Holistic Review of the State-Of-The-Art Microfluidics for Exosome Separation: An Overview of the Current Status, Existing Obstacles, and Future Outlook. Small 17, e2007174. doi:10.1002/smll.202007174
Dorronsoro, A., and Robbins, P. D. (2013). Regenerating the Injured Kidney with Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes. Stem Cel Res. Ther. 4, 39. doi:10.1186/scrt187
Doyle, L., and Wang, M. (2019). Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 8, 727. doi:10.3390/cells8070727
Eichelser, C., Stückrath, I., Müller, V., Milde-Langosch, K., Wikman, H., Pantel, K., et al. (2014). Increased Serum Levels of Circulating Exosomal microRNA-373 in Receptor-Negative Breast Cancer Patients. Oncotarget 5, 9650–9663. doi:10.18632/oncotarget.2520
El Andaloussi, S., Mäger, I., Breakefield, X. O., and Wood, M. J. A. (2013). Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 12, 347–357. doi:10.1038/nrd3978
Elrashdy, F., Aljaddawi, A. A., Redwan, E. M., and Uversky, V. N. (2020). On the Potential Role of Exosomes in the COVID-19 Reinfection/reactivation Opportunity. J. Biomol. Struct. Dyn. 39, 5831–5842. doi:10.1080/07391102.2020.1790426
Escudier, B., Dorval, T., Chaput, N., André, F., Caby, M.-P., Novault, S., et al. (2005). Vaccination of Metastatic Melanoma Patients with Autologous Dendritic Cell (DC) Derived-Exosomes: Results of the First Phase 1 Clinical Trial. J. Transl. Med. 3, 10–13. doi:10.1186/1479-5876-3-10
Ford, T., Graham, J., and Rickwood, D. (1994). Iodixanol: A Nonionic Iso-Osmotic Centrifugation Medium for the Formation of Self-Generated Gradients. Anal. Biochem. 220, 360–366. doi:10.1006/abio.1994.1350
Gardiner, C., Vizio, D. D., Sahoo, S., Théry, C., Witwer, K. W., Wauben, M., et al. (2016). Techniques Used for the Isolation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey. J. Extracellular Vesicles 5, 32945. doi:10.3402/jev.v5.32945
Gemoll, T., Strohkamp, S., Rozanova, S., Röder, C., Hartwig, S., Kalthoff, H., et al. (2020). Protein Profiling of Serum Extracellular Vesicles Reveals Qualitative and Quantitative Differences after Differential Ultracentrifugation and ExoQuick Isolation. Jcm 9, 1429. doi:10.3390/jcm9051429
Gill, S., Catchpole, R., and Forterre, P. (2019). Extracellular Membrane Vesicles in the Three Domains of Life and beyond. FEMS Microbiol. Rev. 43, 273–303. doi:10.1093/femsre/fuy042
Goetzl, E. J., Mustapic, M., Kapogiannis, D., Eitan, E., Lobach, I. V., Goetzl, L., et al. (2016). Cargo Proteins of Plasma Astrocyte‐derived Exosomes in Alzheimer's Disease. FASEB j. 30, 3853–3859. doi:10.1096/fj.201600756R
Goren, Y., Kushnir, M., Zafrir, B., Tabak, S., Lewis, B. S., and Amir, O. (2012). Serum Levels of microRNAs in Patients with Heart Failure. Eur. J. Heart Fail. 14, 147–154. doi:10.1093/eurjhf/hfr155
Greening, D. W., Xu, R., Ji, H., Tauro, B. J., and Simpson, R. J. (2015). A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. Methods Mol. Biol. 1295, 179–209. doi:10.1007/978-1-4939-2550-6_15
Gross, J. C., Chaudhary, V., Bartscherer, K., and Boutros, M. (2012). Active Wnt Proteins Are Secreted on Exosomes. Nat. Cel Biol. 14, 1036–1045. doi:10.1038/ncb2574
Gui, Y., Liu, H., Zhang, L., Lv, W., and Hu, X. (2015). Altered microRNA Profiles in Cerebrospinal Fluid Exosome in Parkinson Disease and Alzheimer Disease. Oncotarget 6, 37043–37053. doi:10.18632/oncotarget.6158
Guo, D., Xu, Y., Ding, J., Dong, J., Jia, N., Li, Y., et al. (20202020). Roles and Clinical Applications of Exosomes in Cardiovascular Disease. Biomed. Res. Int. 2020, 1–8. doi:10.1155/2020/5424281
Gurunathan, S., Kang, M.-H., Jeyaraj, M., Qasim, M., and Kim, J.-H. (2019). Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 8, 307. doi:10.3390/cells8040307
Ha, S. Y., Jee, B. C., Suh, C. S., Kim, H. S., Oh, S. K., Kim, S. H., et al. (2005). Cryopreservation of Human Embryonic Stem Cells without the Use of a Programmable Freezer. Hum. Reprod. 20, 1779–1785. doi:10.1093/humrep/deh854
Harrison, E. B., Hochfelder, C. G., Lamberty, B. G., Meays, B. M., Morsey, B. M., Kelso, M. L., et al. (2016). Traumatic Brain Injury Increases Levels of miR‐21 in Extracellular Vesicles: Implications for Neuroinflammation. FEBS Open Bio 6, 835–846. doi:10.1002/2211-5463.12092
Hassanpour, M., Rezaie, J., Nouri, M., and Panahi, Y. (2020). The Role of Extracellular Vesicles in COVID-19 Virus Infection. Infect. Genet. Evol. 85, 104422. doi:10.1016/j.meegid.2020.104422
He, M., Crow, J., Roth, M., Zeng, Y., and Godwin, A. K. (2014). Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab. Chip 14, 3773–3780. doi:10.1039/c4lc00662c
Heineck, D. P., Lewis, J. M., and Heller, M. J. (2017). Electrokinetic Device Design and Constraints for Use in High Conductance Solutions. Electrophoresis 38, 1475–1482. doi:10.1002/elps.201600563
Heinemann, M. L., and Vykoukal, J. (2017). Sequential Filtration: A Gentle Method for the Isolation of Functional Extracellular Vesicles. Methods Mol. Biol. 1660, 33–41. doi:10.1007/978-1-4939-7253-1_4
Helwa, I., Cai, J., Drewry, M. D., Zimmerman, A., Dinkins, M. B., Khaled, M. L., et al. (2017). A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One 12, e0170628–22. doi:10.1371/journal.pone.0170628
Hirshman, B. R., Kras, R. T., Akers, J. C., Carter, B. S., and Chen, C. C. (2016). Extracellular Vesicles in Molecular Diagnostics: An Overview with a Focus on CNS Diseases. Adv. Clin. Chem. 76, 37–53. doi:10.1016/bs.acc.2016.05.005
Hong, C. S., Muller, L., Boyiadzis, M., and Whiteside, T. L. (2014). Isolation and Characterization of CD34+ Blast-Derived Exosomes in Acute Myeloid Leukemia. PLoS One 9, e103310–8. doi:10.1371/journal.pone.0103310
Ibsen, S. D., Wright, J., Lewis, J. M., Kim, S., Ko, S.-Y., Ong, J., et al. (2017). Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 11, 6641–6651. doi:10.1021/acsnano.7b00549
Iliuk, A., Hadisurya, M., and Boris, R. S. (2021). through Chem. Affinity Purif. 19, 2563–2574. doi:10.1021/acs.jproteome.0c00151.Supporting
Jafari, D., Shajari, S., Jafari, R., Mardi, N., Gomari, H., Ganji, F., et al. (2020). Designer Exosomes: A New Platform for Biotechnology Therapeutics. BioDrugs 34, 567–586. doi:10.1007/s40259-020-00434-x
Jang, S. C., Kim, O. Y., Yoon, C. M., Choi, D.-S., Roh, T.-Y., Park, J., et al. (2013). Bioinspired Exosome-Mimetic Nanovesicles for Targeted Delivery of Chemotherapeutics to Malignant Tumors. ACS Nano 7, 7698–7710. doi:10.1021/nn402232g
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of Exosome Composition. Cell 177, 428–445. e18. doi:10.1016/j.cell.2019.02.029
Jeyaram, A., and Jay, S. M. (2018). Preservation and Storage Stability of Extracellular Vesicles for Therapeutic Applications. AAPS J. 20, 1–7. doi:10.1208/s12248-017-0160-y
Jia, H., Liu, W., Zhang, B., Wang, J., Wu, P., Tandra, N., et al. (2018). HucMSC Exosomes-Delivered 14-3-3ζ Enhanced Autophagy via Modulation of ATG16L in Preventing Cisplatin-Induced Acute Kidney Injury. Am. J. Transl. Res. 10, 101–113.
Jia, Y., Ni, Z., Sun, H., and Wang, C. (2019). Microfluidic Approaches toward the Isolation and Detection of Exosome Nanovesicles. IEEE Access 7, 45080–45098. doi:10.1109/ACCESS.2019.2907123
Jiang, Z.-z., Liu, Y.-m., Niu, X., Yin, J.-y., Hu, B., Guo, S.-c., et al. (2016). Exosomes Secreted by Human Urine-Derived Stem Cells Could Prevent Kidney Complications from Type I Diabetes in Rats. Stem Cel Res. Ther. 7, 1–13. doi:10.1186/s13287-016-0287-2
Jiang, Z., Liu, G., and Li, J. (2020). Recent Progress on the Isolation and Detection Methods of Exosomes. Chem. Asian J. 15, 3973–3982. doi:10.1002/asia.202000873
Jing, H., He, X., and Zheng, J. (2018). Exosomes and Regenerative Medicine: State of the Art and Perspectives. Translational Res. 196, 1–16. doi:10.1016/j.trsl.2018.01.005
Johnstone, M. B., Gohad, N. V., Falwell, E. P., Hansen, D. C., Hansen, K. M., and Mount, A. S. (2015). Cellular Orchestrated Biomineralization of Crystalline Composites on Implant Surfaces by the Eastern Oyster, Crassostrea virginica (Gmelin, 1791). J. Exp. Mar. Biol. Ecol. 463, 8–16. doi:10.1016/j.jembe.2014.10.014
Ju, S., Mu, J., Dokland, T., Zhuang, X., Wang, Q., Jiang, H., et al. (2013). Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice from DSS-Induced Colitis. Mol. Ther. 21, 1345–1357. doi:10.1038/mt.2013.64
Jung, H. H., Kim, J.-Y., Lim, J. E., and Im, Y.-H. (2020). Cytokine Profiling in Serum-Derived Exosomes Isolated by Different Methods. Sci. Rep. 10, 14069. doi:10.1038/s41598-020-70584-z
Kadriyan, H., Prasedya, E. S., Pieter, N. A. L., Gaffar, M., Punagi, A. Q., and Bukhari, A. (2020). The Potential Role of Exosome on Cytokine Storm and Treatment of Severe COVID-19 Infection. Bali Med. J. 9, 630–636. doi:10.15562/bmj.v9i3.1966
Kalani, A., Mohan, A., Godbole, M. M., Bhatia, E., Gupta, A., Sharma, R. K., et al. (2013). Wilm's Tumor-1 Protein Levels in Urinary Exosomes from Diabetic Patients with or without Proteinuria. PLoS One 8, e60177. doi:10.1371/journal.pone.0060177
Kalluri, R., and LeBleu, V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367, 367. doi:10.1126/science.aau6977
Kalra, H., Adda, C. G., Liem, M., Ang, C.-S., Mechler, A., Simpson, R. J., et al. (2013). Comparative Proteomics Evaluation of Plasma Exosome Isolation Techniques and Assessment of the Stability of Exosomes in normal Human Blood Plasma. Proteomics 13, 3354–3364. doi:10.1002/pmic.201300282
Kamerkar, S., Lebleu, V. S., Sugimoto, H., Yang, S., Ruivo, C. F., Melo, S. A., et al. (2017). Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature 546, 498–503. doi:10.1038/nature22341
Katakowski, M., Buller, B., Zheng, X., Lu, Y., Rogers, T., Osobamiro, O., et al. (2013). Exosomes from Marrow Stromal Cells Expressing miR-146b Inhibit Glioma Growth. Cancer Lett. 335, 201–204. doi:10.1016/j.canlet.2013.02.019
Ke, D., Li, H., Zhang, Y., An, Y., Fu, H., Fang, X., et al. (2017). The Combination of Circulating Long Noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 Fragments in Plasma Serve as Diagnostic Markers for Gastric Cancer. Oncotarget 8, 21516–21525. doi:10.18632/oncotarget.15628
Keller, S., Ridinger, J., Rupp, A.-K., Janssen, J. W., and Altevogt, P. (2011). Body Fluid Derived Exosomes as a Novel Template for Clinical Diagnostics. J. Transl. Med. 9, 86. doi:10.1186/1479-5876-9-86
Kennel, P. J., Saha, A., Maldonado, D. A., Givens, R., Brunjes, D. L., Castillero, E., et al. (2018). Serum Exosomal Protein Profiling for the Non-invasive Detection of Cardiac Allograft Rejection. J. Heart Lung Transplant. 37, 409–417. doi:10.1016/j.healun.2017.07.012
Kim, H., Kim, E. H., Kwak, G., Chi, S.-G., Kim, S. H., and Yang, Y. (2021). Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. Ijms 22, 14–23. doi:10.3390/ijms22010014
Kimura, T., Ferran, B., Tsukahara, Y., Shang, Q., Desai, S., Fedoce, A., et al. (2019). Production of Adeno-Associated Virus Vectors for In Vitro and In Vivo Applications. Sci. Rep. 9, 1–13. doi:10.1038/s41598-019-49624-w
Koch, R., Demant, M., Aung, T., Diering, N., Cicholas, A., Chapuy, B., et al. (2014). Populational Equilibrium through Exosome-Mediated Wnt Signaling in Tumor Progression of Diffuse Large B-Cell Lymphoma. Blood 123, 2189–2198. doi:10.1182/blood-2013-08-523886
Kourembanas, S. (2015). Exosomes: Vehicles of Intercellular Signaling, Biomarkers, and Vectors of Cell Therapy. Annu. Rev. Physiol. 77, 13–27. doi:10.1146/annurev-physiol-021014-071641
Ku, A., Lim, H. C., Evander, M., Lilja, H., Laurell, T., Scheding, S., et al. (2018). Acoustic Enrichment of Extracellular Vesicles from Biological Fluids. Anal. Chem. 90, 8011–8019. doi:10.1021/acs.analchem.8b00914
Kurywchak, P., Tavormina, J., and Kalluri, R. (2018). The Emerging Roles of Exosomes in the Modulation of Immune Responses in Cancer. Genome Med. 10, 1–4. doi:10.1186/s13073-018-0535-4
Kusuma, G. D., Barabadi, M., Tan, J. L., Morton, D. A. V., Frith, J. E., and Lim, R. (2018). To Protect and to Preserve: Novel Preservation Strategies for Extracellular Vesicles. Front. Pharmacol. 9, 1–17. doi:10.3389/fphar.2018.01199
Kuwabara, Y., Ono, K., Horie, T., Nishi, H., Nagao, K., Kinoshita, M., et al. (2011). Increased microRNA-1 and microRNA-133a Levels in Serum of Patients with Cardiovascular Disease Indicate Myocardial Damage. Circ. Cardiovasc. Genet. 4, 446–454. doi:10.1161/CIRCGENETICS.110.958975
Kwon, S.-H., Liu, K. D., and Mostov, K. E. (2014). Intercellular Transfer of GPRC5B via Exosomes Drives HGF-Mediated Outward Growth. Curr. Biol. 24, 199–204. doi:10.1016/j.cub.2013.12.010
Lee, R., Ko, H. J., Kim, K., Sohn, Y., Min, S. Y., Kim, J. A., et al. (2020). Anti‐melanogenic Effects of Extracellular Vesicles Derived from Plant Leaves and Stems in Mouse Melanoma Cells and Human Healthy Skin. J. Extracellular Vesicles 9, 1703480. doi:10.1080/20013078.2019.1703480
Li, P., Kaslan, M., Lee, S. H., Yao, J., and Gao, Z. (2017). Progress in Exosome Isolation Techniques. Theranostics 7, 789–804. doi:10.7150/thno.18133
Li, S., Li, S., Wu, S., and Chen, L. (2019). Exosomes Modulate the Viral Replication and Host Immune Responses in HBV Infection. Biomed. Res. Int. 2019, 1–9. doi:10.1155/2019/2103943
Li, Z., Bao, S., Wu, Q., Wang, H., Eyler, C., Sathornsumetee, S., et al. (2009). Hypoxia-Inducible Factors Regulate Tumorigenic Capacity of Glioma Stem Cells. Cancer Cell 15, 501–513. doi:10.1016/j.ccr.2009.03.018
Liang, L.-G., Kong, M.-Q., Zhou, S., Sheng, Y.-F., Wang, P., Yu, T., et al. (2017). An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci. Rep. 7, 46224. doi:10.1038/srep46224
Liga, A., Vliegenthart, A. D. B., Oosthuyzen, W., Dear, J. W., and Kersaudy-Kerhoas, M. (2015). Exosome Isolation: a Microfluidic Road-Map. Lab. Chip 15, 2388–2394. doi:10.1039/C5LC00240K
Lim, W., and Kim, H.-S. (2019). Exosomes as Therapeutic Vehicles for Cancer. Tissue Eng. Regen. Med. 16, 213–223. doi:10.1007/s13770-019-00190-2
Lin, J., Li, J., Huang, B., Liu, J., Chen, X., Chen, X.-M., et al. (2015). Exosomes: Novel Biomarkers for Clinical Diagnosis. Scientific World J. 2015, 1–8. doi:10.1155/2015/657086
Lin, W.-C., Tsai, C.-Y., Huang, J.-M., Wu, S.-R., Chu, L. J., and Huang, K.-Y. (2019). Quantitative Proteomic Analysis and Functional Characterization of Acanthamoeba Castellanii Exosome-like Vesicles. Parasites Vectors 12, 1–12. doi:10.1186/s13071-019-3725-z
Linares, R., Tan, S., Gounou, C., Arraud, N., and Brisson, A. R. (2015). High-speed Centrifugation Induces Aggregation of Extracellular Vesicles. J. Extracellular Vesicles 4, 29509. doi:10.3402/jev.v4.29509
Liu, C., Guo, J., Tian, F., Yang, N., Yan, F., Ding, Y., et al. (2017). Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 11, 6968–6976. doi:10.1021/acsnano.7b02277
Liu, C., and Su, C. (2019). Design Strategies and Application Progress of Therapeutic Exosomes. Theranostics 9, 1015–1028. doi:10.7150/thno.30853
Liu, C., Xu, X., Li, B., Situ, B., Pan, W., Hu, Y., et al. (2018). Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 18, 4226–4232. doi:10.1021/acs.nanolett.8b01184
Livshits, M. A., Khomyakova, E., Evtushenko, E. G., Lazarev, V. N., Kulemin, N. A., Semina, S. E., et al. (2015). Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 5, 1–14. doi:10.1038/srep17319
Lobb, R. J., Becker, M., Wen Wen, S., Wong, C. S. F., Wiegmans, A. P., Leimgruber, A., et al. (2015). Optimized Exosome Isolation Protocol for Cell Culture Supernatant and Human Plasma. J. Extracellular Vesicles 4, 27031. doi:10.3402/jev.v4.27031
Long, Q., Upadhya, D., Hattiangady, B., Kim, D.-K., An, S. Y., Shuai, B., et al. (2017). Intranasal MSC-Derived A1-Exosomes Ease Inflammation, and Prevent Abnormal Neurogenesis and Memory Dysfunction after Status Epilepticus. Proc. Natl. Acad. Sci. U.S.A. 114, E3536–E3545. doi:10.1073/pnas.1703920114
Lőrincz, Á. M., Timár, C. I., Marosvári, K. A., Veres, D. S., Otrokocsi, L., Kittel, Á., et al. (2014). Effect of Storage on Physical and Functional Properties of Extracellular Vesicles Derived from Neutrophilic Granulocytes. J. Extracellular Vesicles 3, 25465. doi:10.3402/jev.v3.25465
Lu, M., and Huang, Y. (2020). Bioinspired Exosome-like Therapeutics and Delivery Nanoplatforms. Biomaterials 242, 119925. doi:10.1016/j.biomaterials.2020.119925
Lv, L.-L., Cao, Y.-H., Ni, H.-F., Xu, M., Liu, D., Liu, H., et al. (2013). MicroRNA-29c in Urinary Exosome/microvesicle as a Biomarker of Renal Fibrosis. Am. J. Physiology-Renal Physiol. 305, F1220–F1227. doi:10.1152/ajprenal.00148.2013
Macías, M., Rebmann, V., Mateos, B., Varo, N., Perez-Gracia, J. L., Alegre, E., et al. (2019). Comparison of Six Commercial Serum Exosome Isolation Methods Suitable for Clinical Laboratories. Effect in Cytokine Analysis. Clin. Chem. Lab. Med. 57, 1539–1545. doi:10.1515/cclm-2018-1297
MacKie, A. R., Klyachko, E., Thorne, T., Schultz, K. M., Millay, M., Ito, A., et al. (2012). Sonic Hedgehog-Modified Human CD34+ Cells Preserve Cardiac Function after Acute Myocardial Infarction. Circ. Res. 111, 312–321. doi:10.1161/CIRCRESAHA.112.266015
Maroto, R., Zhao, Y., Jamaluddin, M., Popov, V. L., Wang, H., Kalubowilage, M., et al. (2017). Effects of Storage Temperature on Airway Exosome Integrity for Diagnostic and Functional Analyses. J. Extracellular Vesicles 6, 1359478. doi:10.1080/20013078.2017.1359478
Mathivanan, S., Ji, H., and Simpson, R. J. (2010). Exosomes: Extracellular Organelles Important in Intercellular Communication. J. Proteomics 73, 1907–1920. doi:10.1016/j.jprot.2010.06.006
Matsumoto, S., Sakata, Y., Suna, S., Nakatani, D., Usami, M., Hara, M., et al. (2013). Circulating P53-Responsive MicroRNAs Are Predictive Indicators of Heart Failure after Acute Myocardial Infarction. Circ. Res. 113, 322–326. doi:10.1161/CIRCRESAHA.113.301209
Meissner, L., Gallozzi, M., Balbi, M., Schwarzmaier, S., Tiedt, S., Terpolilli, N. A., et al. (2016). Temporal Profile of MicroRNA Expression in Contused Cortex after Traumatic Brain Injury in Mice. J. Neurotrauma 33, 713–720. doi:10.1089/neu.2015.4077
Mendt, M., Kamerkar, S., Sugimoto, H., McAndrews, K. M., Wu, C.-C., Gagea, M., et al. (2018). Generation and Testing of Clinical-Grade Exosomes for Pancreatic Cancer. JCI insight 3. doi:10.1172/jci.insight.99263
Mikaelian, I., Scicchitano, M., Mendes, O., Thomas, R. A., and LeRoy, B. E. (2012). Frontiers in Preclinical Safety Biomarkers. Toxicol. Pathol. 41, 18–31. doi:10.1177/0192623312448939
Miranda, K. C., Bond, D. T., McKee, M., Skog, J., Păunescu, T. G., Da Silva, N., et al. (2010). Nucleic Acids within Urinary Exosomes/microvesicles Are Potential Biomarkers for Renal Disease. Kidney Int. 78, 191–199. doi:10.1038/ki.2010.106
Morse, M. A., Garst, J., Osada, T., Khan, S., Hobeika, A., Clay, T. M., et al. (2005). A Phase I Study of Dexosome Immunotherapy in Patients with Advanced Non-small Cell Lung Cancer. J. Transl. Med. 3, 9–8. doi:10.1186/1479-5876-3-9
Mu, J., Zhuang, X., Wang, Q., Jiang, H., Deng, Z. B., Wang, B., et al. (2014). Interspecies Communication between Plant and Mouse Gut Host Cells through Edible Plant Derived Exosome‐like Nanoparticles. Mol. Nutr. Food Res. 58, 1561–1573. doi:10.1002/mnfr.201300729
Namazi, H., Namazi, I., Ghiasi, P., Ansari, H., Rajabi, S., Hajizadeh-Saffar, E., et al. (2018). Exosomes Secreted by Normoxic and Hypoxic Cardiosphere-Derived Cells Have Anti-apoptotic Effect. Iran. J. Pharm. Res. 17, 377–385. doi:10.22037/ijpr.2018.2161
Nassar, W., El-Ansary, M., Sabry, D., Mostafa, M. A., Fayad, T., Kotb, E., et al. (2016). Umbilical Cord Mesenchymal Stem Cells Derived Extracellular Vesicles Can Safely Ameliorate the Progression of Chronic Kidney Diseases. Biomater. Res. 20, 1–11. doi:10.1186/s40824-016-0068-0
Nawaz, M., Malik, M. I., Hameed, M., and Zhou, J. (2019). Research Progress on the Composition and Function of Parasite-Derived Exosomes. Acta Tropica 196, 30–36. doi:10.1016/j.actatropica.2019.05.004
Nireesha, G. R., Divya, L., Sowmya, C., Venkateshan, N., Babu, M. N., and Lavakumar, V. (2013). Lyophilization/Freeze Drying - Rev. 3, 87–98. doi:10.9790/4200-0342733
Ohno, S.-i., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor Microrna to Breast Cancer Cells. Mol. Ther. 21, 185–191. doi:10.1038/mt.2012.180
Pan, B.-T., and Johnstone, R. M. (1983). Fate of the Transferrin Receptor during Maturation of Sheep Reticulocytes In Vitro: Selective Externalization of the Receptor. Cell 33, 967–978. doi:10.1016/0092-8674(83)90040-5
Parimon, T., Brauer, R., Schlesinger, S. Y., Xie, T., Jiang, D., Ge, L., et al. (2018). Syndecan-1 Controls Lung Tumorigenesis by Regulating miRNAs Packaged in Exosomes. Am. J. Pathol. 188, 1094–1103. doi:10.1016/j.ajpath.2017.12.009
Patel, G. K., Khan, M. A., Zubair, H., Srivastava, S. K., Khushman, M. d., Singh, S., et al. (2019). Comparative Analysis of Exosome Isolation Methods Using Culture Supernatant for Optimum Yield, Purity and Downstream Applications. Sci. Rep. 9, 1–10. doi:10.1038/s41598-019-41800-2
Pillet, S., Couillard, J., Trépanier, S., Poulin, J.-F., Yassine-Diab, B., Guy, B., et al. (2019). Immunogenicity and Safety of a Quadrivalent Plant-Derived Virus like Particle Influenza Vaccine Candidate-Two Randomized Phase II Clinical Trials in 18 to 49 and ≥50 Years Old Adults. PLoS One 14, e0216533. doi:10.1371/journal.pone.0216533
Pilzer, D., Gasser, O., Moskovich, O., Schifferli, J. A., and Fishelson, Z. (2005). Emission of Membrane Vesicles: Roles in Complement Resistance, Immunity and Cancer. Springer Semin. Immun. 27, 375–387. doi:10.1007/s00281-005-0004-1
Pinky, S., Gupta, S., Krishnakumar, V., Sharma, Y., Dinda, A. K., and Mohanty, S. (2021). Mesenchymal Stem Cell Derived Exosomes: a Nano Platform for Therapeutics and Drug Delivery in Combating COVID-19. Stem Cel Rev Rep 17, 33–43. doi:10.1007/s12015-020-10002-z
Prabu, P., Rome, S., Sathishkumar, C., Gastebois, C., Meugnier, E., Mohan, V., et al. (2019). MicroRNAs from Urinary Extracellular Vesicles Are Non-invasive Early Biomarkers of Diabetic Nephropathy in Type 2 Diabetes Patients with the 'Asian Indian Phenotype'. Diabetes Metab. 45, 276–285. doi:10.1016/j.diabet.2018.08.004
Qiao, L., Hu, S., Huang, K., Su, T., Li, Z., Vandergriff, A., et al. (2020). Tumor Cell-Derived Exosomes home to Their Cells of Origin and Can Be Used as Trojan Horses to Deliver Cancer Drugs. Theranostics 10, 3474–3487. doi:10.7150/thno.39434
Qiu, J., Yang, G., Feng, M., Zheng, S., Cao, Z., You, L., et al. (2018). Extracellular Vesicles as Mediators of the Progression and Chemoresistance of Pancreatic Cancer and Their Potential Clinical Applications. Mol. Cancer 17, 1–11. doi:10.1186/s12943-017-0755-z
Qu, L., Ding, J., Chen, C., Wu, Z.-J., Liu, B., Gao, Y., et al. (2016). Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 29, 653–668. doi:10.1016/j.ccell.2016.03.004
Quek, C., Bellingham, S. A., Jung, C.-H., Scicluna, B. J., Shambrook, M. C., Sharples, R. A., et al. (2017). Defining the Purity of Exosomes Required for Diagnostic Profiling of Small RNA Suitable for Biomarker Discovery. RNA Biol. 14, 245–258. doi:10.1080/15476286.2016.1270005
Rabinowits, G., Gerçel-Taylor, C., Day, J. M., Taylor, D. D., and Kloecker, G. H. (2009). Exosomal microRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 10, 42–46. doi:10.3816/CLC.2009.n.006
Raimondo, F., Morosi, L., Chinello, C., Magni, F., and Pitto, M. (2011). Advances in Membranous Vesicle and Exosome Proteomics Improving Biological Understanding and Biomarker Discovery. Proteomics 11, 709–720. doi:10.1002/pmic.201000422
Raimondo, S., Naselli, F., Fontana, S., Monteleone, F., Lo Dico, A., Saieva, L., et al. (2015). Citrus limon-derived Nanovesicles Inhibit Cancer Cell Proliferation and Suppress CML Xenograft Growth by Inducing TRAIL-Mediated Cell Death. Oncotarget 6, 19514–19527. doi:10.18632/oncotarget.4004
Rajan, T. S., Giacoppo, S., Diomede, F., Ballerini, P., Paolantonio, M., Marchisio, M., et al. (2016). The Secretome of Periodontal Ligament Stem Cells from MS Patients Protects against EAE. Sci. Rep. 6, 1–16. doi:10.1038/srep38743
Rao, P., Benito, E., and Fischer, A. (2013). MicroRNAs as Biomarkers for CNS Disease. Front. Mol. Neurosci. 6, 1–13. doi:10.3389/fnmol.2013.00039
Rekker, K., Saare, M., Roost, A. M., Kubo, A.-L., Zarovni, N., Chiesi, A., et al. (2014). Comparison of Serum Exosome Isolation Methods for microRNA Profiling. Clin. Biochem. 47, 135–138. doi:10.1016/j.clinbiochem.2013.10.020
Rizvanov, A., Shaimardanova, A., Solovyeva, V., Chulpanova, D., James, V., and Kitaeva, K. (2020). Extracellular Vesicles in the Diagnosis and Treatment of central Nervous System Diseases. Neural Regen. Res. 15, 586–596. doi:10.4103/1673-5374.266908
Rizzo, J., Rodrigues, M. L., and Janbon, G. (2020). Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives. Front. Cel. Infect. Microbiol. 10, 1–13. doi:10.3389/fcimb.2020.00346
Rood, I. M., Deegens, J. K. J., Merchant, M. L., Tamboer, W. P. M., Wilkey, D. W., Wetzels, J. F. M., et al. (2010). Comparison of Three Methods for Isolation of Urinary Microvesicles to Identify Biomarkers of Nephrotic Syndrome. Kidney Int. 78, 810–816. doi:10.1038/ki.2010.262
Royo, F., Diwan, I., Tackett, M., Zuñiga, P., Sanchez-Mosquera, P., Loizaga-Iriarte, A., et al. (2016). Comparative miRNA Analysis of Urine Extracellular Vesicles Isolated through Five Different Methods. Cancers 8, 112. doi:10.3390/cancers8120112
Russell, J. C., Kim, T.-K., Noori, A., Merrihew, G. E., Robbins, J. E., Golubeva, A., et al. (2020). Composition of Caenorhabditis elegans Extracellular Vesicles Suggests Roles in Metabolism, Immunity, and Aging. GeroScience 42, 1133–1145. doi:10.1007/s11357-020-00204-1
Rutter, B. D., and Innes, R. W. (2017). Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol. 173, 728–741. doi:10.1104/pp.16.01253
Şahin, F., Koçak, P., Güneş, M. Y., Özkan, İ., Yıldırım, E., and Kala, E. Y. (2019). In Vitro Wound Healing Activity of Wheat-Derived Nanovesicles. Appl. Biochem. Biotechnol. 188, 381–394. doi:10.1007/s12010-018-2913-1
Sahoo, S., and Losordo, D. W. (2014). Exosomes and Cardiac Repair after Myocardial Infarction. Circ. Res. 114, 333–344. doi:10.1161/CIRCRESAHA.114.300639
Salminen, A., Kaarniranta, K., and Kauppinen, A. (2020). Exosomal Vesicles Enhance Immunosuppression in Chronic Inflammation: Impact in Cellular Senescence and the Aging Process. Cell Signal. 75, 109771. doi:10.1016/j.cellsig.2020.109771
Samsonov, R., Shtam, T., Burdakov, V., Glotov, A., Tsyrlina, E., Berstein, L., et al. (2016). Lectin-induced Agglutination Method of Urinary Exosomes Isolation Followed by Mi-RNA Analysis: Application for Prostate Cancer Diagnostic. Prostate 76, 68–79. doi:10.1002/pros.23101
Sanchez, M. B. H., Bruno, S., Grange, C., Tapparo, M., Cantaluppi, V., Tetta, C., et al. (2014). Human Liver Stem Cells and Derived Extracellular Vesicles Improve Recovery in a Murine Model of Acute Kidney Injury. Stem Cel Res. Ther. 5, 1–11. doi:10.1186/scrt514
Sato, Y. T., Umezaki, K., Sawada, S., Mukai, S.-a., Sasaki, Y., Harada, N., et al. (2016). Engineering Hybrid Exosomes by Membrane Fusion with Liposomes. Sci. Rep. 6, 1–11. doi:10.1038/srep21933
Schatz, D., and Vardi, A. (2018). Extracellular Vesicles - New Players in Cell-Cell Communication in Aquatic Environments. Curr. Opin. Microbiol. 43, 148–154. doi:10.1016/j.mib.2018.01.014
Schuh, C. M. A. P., Cuenca, J., Alcayaga-Miranda, F., and Khoury, M. (2019). Exosomes on the Border of Species and Kingdom Intercommunication. Translational Res. 210, 80–98. doi:10.1016/j.trsl.2019.03.008
Schwarzenbach, H., and Gahan, P. B. (2021). Exosomes in Immune Regulation. ncRNA 7, 4. doi:10.3390/ncrna7010004
Sengupta, V., Sengupta, S., Lazo, A., Woods, P., Nolan, A., and Bremer, N. (2020). Exosomes Derived from Bone Marrow Mesenchymal Stem Cells as Treatment for Severe COVID-19. Stem Cell Development 29, 747–754. doi:10.1089/scd.2020.0080
Shi, R., Wang, P.-Y., Li, X.-Y., Chen, J.-X., Li, Y., Zhang, X.-Z., et al. (2015). Exosomal Levels of miRNA-21 from Cerebrospinal Fluids Associated with Poor Prognosis and Tumor Recurrence of Glioma Patients. Oncotarget 6, 26971–26981. doi:10.18632/oncotarget.4699
Shin, H., Han, C., Labuz, J. M., Kim, J., Kim, J., Cho, S., et al. (2015). High-yield Isolation of Extracellular Vesicles Using Aqueous Two-phase System. Sci. Rep. 5, 1–11. doi:10.1038/srep13103
Sidhom, K., Obi, P. O., and Saleem, A. (2020). A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Ijms 21, 6466. doi:10.3390/ijms21186466
Sim, S.-L., He, T., Tscheliessnig, A., Mueller, M., Tan, R. B. H., and Jungbauer, A. (2012). Protein Precipitation by Polyethylene Glycol: A Generalized Model Based on Hydrodynamic Radius. J. Biotechnol. 157, 315–319. doi:10.1016/j.jbiotec.2011.09.028
Simons, M., and Raposo, G. (2009). Exosomes - Vesicular Carriers for Intercellular Communication. Curr. Opin. Cel Biol. 21, 575–581. doi:10.1016/j.ceb.2009.03.007
Smyth, T., Kullberg, M., Malik, N., Smith-Jones, P., Graner, M. W., and Anchordoquy, T. J. (2015). Biodistribution and Delivery Efficiency of Unmodified Tumor-Derived Exosomes. J. Controlled Release 199, 145–155. doi:10.1016/j.jconrel.2014.12.013
Soares Martins, T., Catita, J., Martins Rosa, I., A. B. da Cruz e Silva, O., and Henriques, A. G. (2018). Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PLoS One 13, e0198820–16. doi:10.1371/journal.pone.0198820
Song, Y., Kim, Y., Ha, S., Sheller‐Miller, S., Yoo, J., Choi, C., et al. (2021). The Emerging Role of Exosomes as Novel Therapeutics: Biology, Technologies, Clinical Applications, and the Next. Am. J. Reprod. Immunol. 85, 1–10. doi:10.1111/aji.13329
Stern, R. A., Tripodis, Y., Baugh, C. M., Fritts, N. G., Martin, B. M., Chaisson, C., et al. (2016). Preliminary Study of Plasma Exosomal Tau as a Potential Biomarker for Chronic Traumatic Encephalopathy. Jad 51, 1099–1109. doi:10.3233/JAD-151028
Stuendl, A., Kunadt, M., Kruse, N., Bartels, C., Moebius, W., Danzer, K. M., et al. (2016). Induction of α-synuclein Aggregate Formation by CSF Exosomes from Patients with Parkinson's Disease and Dementia with Lewy Bodies. Brain 139, 481–494. doi:10.1093/brain/awv346
Suharta, S., Barlian, A., Hidajah, A. C., Notobroto, H. B., Ana, I. D., Indariani, S., et al. (2021). Plant‐derived Exosome‐like Nanoparticles: A Concise Review on its Extraction Methods, Content, Bioactivities, and Potential as Functional Food Ingredient. J. Food Sci. 86, 2838–2850. doi:10.1111/1750-3841.15787
Takov, K., Yellon, D. M., and Davidson, S. M. (2019). Comparison of Small Extracellular Vesicles Isolated from Plasma by Ultracentrifugation or Size-Exclusion Chromatography: Yield, Purity and Functional Potential. J. Extracellular Vesicles 8, 1560809. doi:10.1080/20013078.2018.1560809
Tan, L., Wu, H., Liu, Y., Zhao, M., Li, D., and Lu, Q. (2016). Recent Advances of Exosomes in Immune Modulation and Autoimmune Diseases. Autoimmunity 49, 357–365. doi:10.1080/08916934.2016.1191477
Tang, Y.-T., Huang, Y.-Y., Zheng, L., Qin, S.-H., Xu, X.-P., An, T.-X., et al. (2017). Comparison of Isolation Methods of Exosomes and Exosomal RNA from Cell Culture Medium and Serum. Int. J. Mol. Med. 40, 834–844. doi:10.3892/ijmm.2017.3080
Tanziela, T., Shaikh, S., Jiang, H., Lu, Z., and Wang, X. (2020). Efficient Encapsulation of Biocompatible Nanoparticles in Exosomes for Cancer Theranostics. Nano Today 35, 100964. doi:10.1016/j.nantod.2020.100964
Tauro, B. J., Greening, D. W., Mathias, R. A., Ji, H., Mathivanan, S., Scott, A. M., et al. (2012). Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human colon Cancer Cell Line LIM1863-Derived Exosomes. Methods 56, 293–304. doi:10.1016/j.ymeth.2012.01.002
Taylor, D. D., and Shah, S. (2015). Methods of Isolating Extracellular Vesicles Impact Down-Stream Analyses of Their Cargoes. Methods 87, 3–10. doi:10.1016/j.ymeth.2015.02.019
Taylor, D. D., Zacharias, W., and Gercel-Taylor, C. (2011). Exosome Isolation for Proteomic Analyses and RNA Profiling. Methods Mol. Biol. 728, 235–246. doi:10.1007/978-1-61779-068-3_15
Théry, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cel Biol. 3, 22. doi:10.1002/0471143030.cb0322s30
Théry, C., Zitvogel, L., and Amigorena, S. (2002). Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2, 569–579. doi:10.1038/nri855
Tian, Y., Li, S., Song, J., Ji, T., Zhu, M., Anderson, G. J., et al. (2014). A Doxorubicin Delivery Platform Using Engineered Natural Membrane Vesicle Exosomes for Targeted Tumor Therapy. Biomaterials 35, 2383–2390. doi:10.1016/j.biomaterials.2013.11.083
Tickner, J. A., Urquhart, A. J., Stephenson, S.-A., Richard, D. J., and O’Byrne, K. J. (2014). Functions and Therapeutic Roles of Exosomes in Cancer. Front. Oncol. 4, 2–9. doi:10.3389/fonc.2014.00127
Timbie, K. F., Mead, B. P., and Price, R. J. (2015). Drug and Gene Delivery across the Blood-Brain Barrier with Focused Ultrasound. J. Controlled Release 219, 61–75. doi:10.1016/j.jconrel.2015.08.059
Tsai, S.-J., Guo, C., Atai, N. A., and Gould, S. J. (2020). Exosome-Mediated mRNA Delivery for SARS-CoV-2 Vaccination. bioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2020.11.06.371419v1.full.
Tseliou, E., Fouad, J., Reich, H., Slipczuk, L., De Couto, G., Aminzadeh, M., et al. (2015). Fibroblasts Rendered Antifibrotic, Antiapoptotic, and Angiogenic by Priming with Cardiosphere-Derived Extracellular Membrane Vesicles. J. Am. Coll. Cardiol. 66, 599–611. doi:10.1016/j.jacc.2015.05.068
Tu, Q., Li, L., Zhang, Y., Wang, J., Liu, R., Li, M., et al. (2011). The Effect of Acetylcholine-like Biomimetic Polymers on Neuronal Growth. Biomaterials 32, 3253–3264. doi:10.1016/j.biomaterials.2011.01.044
Urzì, O., Raimondo, S., and Alessandro, R. (2021). Extracellular Vesicles from Plants: Current Knowledge and Open Questions. Ijms 22, 5366. doi:10.3390/ijms22105366
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated Transfer of mRNAs and microRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cel Biol. 9, 654–659. doi:10.1038/ncb1596
van der Pol, E., Böing, A. N., Harrison, P., Sturk, A., and Nieuwland, R. (2012). Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 64, 676–705. doi:10.1124/pr.112.005983
Van Veldhoven, P. P., Baumgart, E., and Mannaerts, G. P. (1996). Iodixanol (Optiprep), an Improved Density Gradient Medium for the Iso-Osmotic Isolation of Rat Liver Peroxisomes. Anal. Biochem. 237, 17–23. doi:10.1006/abio.1996.0194
Verweij, F. J., Hyenne, V., Van Niel, G., and Goetz, J. G. (2019). Extracellular Vesicles: Catching the Light in Zebrafish. Trends Cel Biol. 29, 770–776. doi:10.1016/j.tcb.2019.07.007
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat. Cel Biol. 13, 423–433. doi:10.1038/ncb2210
Wang, J., Ma, P., Kim, D. H., Liu, B.-F., and Demirci, U. (2021). Towards Microfluidic-Based Exosome Isolation and Detection for Tumor Therapy. Nano Today 37, 101066. doi:10.1016/j.nantod.2020.101066
Weilner, S., Schraml, E., Redl, H., Grillari-Voglauer, R., and Grillari, J. (2013). Secretion of Microvesicular miRNAs in Cellular and Organismal Aging. Exp. Gerontol. 48, 626–633. doi:10.1016/j.exger.2012.11.017
Whitesides, G. M. (2006). The Origins and the Future of Microfluidics. Nature 442, 368–373. doi:10.1038/nature05058
Willms, E., Cabañas, C., Mäger, I., Wood, M. J. A., and Vader, P. (2018). Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 9. doi:10.3389/fimmu.2018.00738
Witwer, K. W., Buzás, E. I., Bemis, L. T., Bora, A., Lässer, C., Lötvall, J., et al. (2013). Standardization of Sample Collection, Isolation and Analysis Methods in Extracellular Vesicle Research. J. Extracellular Vesicles 2, 20360. doi:10.3402/jev.v2i0.20360
Wu, Z., Wang, L., Li, J., Wang, L., Wu, Z., and Sun, X. (2019). Extracellular Vesicle-Mediated Communication within Host-Parasite Interactions. Front. Immunol. 9, 3066. doi:10.3389/fimmu.2018.03066
Xander, P., Cronemberger-Andrade, A., and Torrecilhas, A. C. (2020). Extracellular Vesicles in Parasitic Disease. Exosomes 1, 179–198. doi:10.1016/b978-0-12-816053-4.00008-0
Xu, B., Zhang, Y., Du, X.-F., Li, J., Zi, H.-X., Bu, J.-W., et al. (2017). Neurons Secrete MIR-132-Containing Exosomes to Regulate Brain Vascular Integrity. Cell Res 27, 882–897. doi:10.1038/cr.2017.62
Xu, D., and Tahara, H. (2013). The Role of Exosomes and microRNAs in Senescence and Aging. Adv. Drug Deliv. Rev. 65, 368–375. doi:10.1016/j.addr.2012.07.010
Yamashita, T., Takahashi, Y., Nishikawa, M., and Takakura, Y. (2016). Effect of Exosome Isolation Methods on Physicochemical Properties of Exosomes and Clearance of Exosomes from the Blood Circulation. Eur. J. Pharmaceutics Biopharmaceutics 98, 1–8. doi:10.1016/j.ejpb.2015.10.017
Yang, D., Zhang, W., Zhang, H., Zhang, F., Chen, L., Ma, L., et al. (2020). Progress, Opportunity, and Perspective on Exosome Isolation - Efforts for Efficient Exosome-Based Theranostics. Theranostics 10, 3684–3707. doi:10.7150/thno.41580
Yasui, T., Yanagida, T., Ito, S., Konakade, Y., Takeshita, D., Naganawa, T., et al. (2017). Unveiling Massive Numbers of Cancer-Related Urinary-microRNA Candidates Via Nanowires. Sci. Adv. 3 (12). doi:10.1126/sciadv.1701133
Yeo, R. W. Y., Lai, R. C., Zhang, B., Tan, S. S., Yin, Y., Teh, B. J., et al. (2013). Mesenchymal Stem Cell: An Efficient Mass Producer of Exosomes for Drug Delivery. Adv. Drug Deliv. Rev. 65, 336–341. doi:10.1016/j.addr.2012.07.001
Yu, L.-L., Zhu, J., Liu, J.-X., Jiang, F., Ni, W.-K., Qu, L.-S., et al. (2018). A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed. Res. Int. 2018, 1–9. doi:10.1155/2018/3634563
Zamani, P., Fereydouni, N., Butler, A. E., Navashenaq, J. G., and Sahebkar, A. (2019). The Therapeutic and Diagnostic Role of Exosomes in Cardiovascular Diseases. Trends Cardiovasc. Med. 29, 313–323. doi:10.1016/j.tcm.2018.10.010
Zhang, G., and Yang, P. (2018). A Novel Cell-Cell Communication Mechanism in the Nervous System: Exosomes. J. Neuro Res. 96, 45–52. doi:10.1002/jnr.24113
Zhang, M., Viennois, E., Prasad, M., Zhang, Y., Wang, L., Zhang, Z., et al. (2016). Edible Ginger-Derived Nanoparticles: A Novel Therapeutic Approach for the Prevention and Treatment of Inflammatory Bowel Disease and Colitis-Associated Cancer. Biomaterials 101, 321–340. doi:10.1016/j.biomaterials.2016.06.018
Zhang, Y., Bi, J., Huang, J., Tang, Y., Du, S., and Li, P. (2020). Exosome: A Review of its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Ijn 15, 6917–6934. doi:10.2147/IJN.S264498
Zhao, Y., Haney, M. J., Gupta, R., Bohnsack, J. P., He, Z., Kabanov, A. V., et al. (2014). GDNF-Transfected Macrophages Produce Potent Neuroprotective Effects in Parkinson's Disease Mouse Model. PLoS One 9, e106867–11. doi:10.1371/journal.pone.0106867
Zhao, Z., Yang, Y., Zeng, Y., and He, M. (2016). A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection towards Blood-Based Ovarian Cancer Diagnosis. Lab. Chip 16, 489–496. doi:10.1039/C5LC01117E
Zhou, H., Kajiyama, H., Tsuji, T., Hu, X., Leelahavanichkul, A., Vento, S., et al. (2013). Urinary Exosomal Wilms' Tumor-1 as a Potential Biomarker for Podocyte Injury. Am. J. Physiology-Renal Physiol. 305, F553–F559. doi:10.1152/ajprenal.00056.2013
Zhou, H., Pisitkun, T., Aponte, A., Yuen, P. S. T., Hoffert, J. D., Yasuda, H., et al. (2006). Exosomal Fetuin-A Identified by Proteomics: A Novel Urinary Biomarker for Detecting Acute Kidney Injury. Kidney Int. 70, 1847–1857. doi:10.1038/sj.ki.5001874
Zhou, M., Weber, S. R., Zhao, Y., Chen, H., and Sundstrom, J. M. (2020). Methods for Exosome Isolation and Characterization. Exosomes 1, 23–38. doi:10.1016/b978-0-12-816053-4.00002-x
Zhou, W., Woodson, M., Neupane, B., Bai, F., Sherman, M. B., Choi, K. H., et al. (2018). Exosomes Serve as Novel Modes of Tick-Borne Flavivirus Transmission from Arthropod to Human Cells and Facilitates Dissemination of Viral RNA and Proteins to the Vertebrate Neuronal Cells. Plos Pathog. 14, e1006764. doi:10.1371/journal.ppat.1006764
Zhou, Y., Ma, Z., Tayebi, M., and Ai, Y. (2019). Submicron Particle Focusing and Exosome Sorting by Wavy Microchannel Structures within Viscoelastic Fluids. Anal. Chem. 91, 4577–4584. doi:10.1021/acs.analchem.8b05749
Zhu, X., Wang, S., Tarique, I., An, T., Yang, H., Bai, X., et al. (2019). Cellular Evidence and Source of Exosomes in the Biliary System of the Chinese Soft-Shelled Turtle, Pelodiscus Sinensis. Front. Physiol. 10, 1097. doi:10.3389/fphys.2019.01097
Keywords: exosomes, isolation methods, sources of exosomes, storage of exosomes, biomedical application of exosomes
Citation: Janouskova O, Herma R, Semeradtova A, Poustka D, Liegertova M, Malinska HA and Maly J (2022) Conventional and Nonconventional Sources of Exosomes–Isolation Methods and Influence on Their Downstream Biomedical Application. Front. Mol. Biosci. 9:846650. doi: 10.3389/fmolb.2022.846650
Received: 31 December 2021; Accepted: 28 March 2022;
Published: 02 May 2022.
Edited by:
Gabriele Candiani, Politecnico di Milano, ItalyReviewed by:
Satish Ramalingam, SRM Institute of Science and Technology, IndiaChang-Sook Hong, University of Pittsburgh, United States
Copyright © 2022 Janouskova, Herma, Semeradtova, Poustka, Liegertova, Malinska and Maly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga Janouskova, SmFub3Vza292YTMyNEBnbWFpbC5jb20=
 Olga Janouskova
Olga Janouskova Regina Herma
Regina Herma Alena Semeradtova
Alena Semeradtova David Poustka
David Poustka Hana Auer Malinska
Hana Auer Malinska Jan Maly
Jan Maly