- 1Functional Neurosurgery Research Center, Shohada Tajrish Comprehensive Neurosurgical Center of Excellence, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Biophysics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder whose early diagnosis leads to a chance for successful treatment and decreases the side effects. Hyperphosphorylation of tau proteins is a pathological hallmark of AD that causes it to lose its attachment ability to the microtubules. Alteration of tau structure due to its hyperphosphorylation is an exciting challenge regarding AD treatments. Here, we aimed to examine the structural alterations of short helical segments of tau protein with one to three phosphorylated sites by molecular dynamics simulation. Results indicated that the interaction of two similar segments with three phosphorylated sites (P-Ser262, 285, and 289) formed a compact and more stable structure than the one phosphorylated site complex (P-Ser262). Moreover, due to the high dynamics of the P-Ser262 complex, several structures were made with different conformational dynamics, but there was only one stable cluster of the P-Ser262, 285, and 289 complex during simulation. It seems that the P-Ser262, 285, and 289 complex plays an important role in the formation of paired helical filaments (PHFs) by forming a stable dimer. Generally, it is important to identify how structural features of segments in tau protein change when the phosphorylated sites increase from one to three sites and their effects on the formation of PHFs for drug design and diagnostic biomarkers.
Introduction
Alzheimer’s disease (AD) is a neuropathological disease characterized by two lesions, including the formation of amyloid plaques and abnormally phosphorylated tau proteins. So far, more than 20 different neurological diseases have been identified in humans that are associated with the abnormal accumulation of tau protein (Williams, 2006; Iqbal et al., 2010). This protein exists in six isoforms in the human brain and is expressed by a gene on chromosome 17. They vary in length from 352 to 441 amino acids created by different pre-mRNA splicing and post-translational modifications (Nizynski et al., 2017; Dzwolak et al., 2004; Wang and Mandelkow, 2016). Moreover, each isoform comprises three main domains, including the projection domains, the proline-rich domain, and the microtubule-binding domains (MBDs). The projection domain is made of acidic residues followed by a proline-rich region, and the MBD consists of several repeats of microtubule-binding sequences that consist of 31 or 32 residues (Sottejeau et al., 2015; Barbier et al., 2019).
The interaction between filamentous actin (F-actin) and microtubules is important to stabilize the cytoskeleton, which has an essential role in cellular processes such as axonal growth, cell division, migration, and development of neurons. The interaction is mediated through the tau protein. Tau binds simultaneously to microtubules and actin filaments (Fulga et al., 2007; Mohan and John, 2015). Studies have shown that tau is bonded to F-actin with high affinity by repeated domains of tau protein and that residues 254–290 have an essential role in the interactions. The segment (254–290) has two important motifs, including KXGS and VQIINK, which exhibited high preference values for phosphorylation and aggregation (Von Bergen et al., 2000; Cabrales Fontela et al., 2017). Moreover, the VQIINK motif is important for forming paired helical filaments (PHFs). Phosphorylation at serine 262 attenuates the binding of tau protein to F-actin but alone cannot lead to the formation of PHFs (Seubert et al., 1995; Schneider et al., 1999a).
When the tau protein has malfunctioned, it causes a wide range of problems such as progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), traumatic brain injury (TBI), and frontotemporal dementia (FTD) (Graff-Radford and Woodruff, 2007; Mietelska-Porowska et al., 2014). Studies have shown that the amount of soluble tau protein in the brain of an Alzheimer’s patient is the same as in the brain of a healthy person, but the phosphorylated sites per tau molecule are about four times higher than those in a healthy person. There are naturally 2–3 moles of phosphorylated tau protein in the brain of a healthy person (Iqbal et al., 2010; Wang et al., 2016; Naseri et al., 2019).
When the phosphorylation of tau proteins increases abnormally (hyperphosphorylation), it leads to the polymerization of these proteins, which form neurofibrillary tangles (NFTs) and neuropil threads (NTs) (Omalu et al., 2010; Pradeepkiran et al., 2019). More than 85 sites in the tau protein are known to have the potential to be phosphorylated, including 5 tyrosine, 35 threonine, and 45 serine residues. Due to the expanded structure of tau, protein kinases can easily access their target sites for phosphorylation, so the amount of phosphorylation in this protein is highly variable. To date, no study has shown that all of these sites in the protein are phosphorylated simultaneously (Martin et al., 2011; Perez et al., 2011; Neddens et al., 2018).
Phosphorylation at all of these sites is not entirely independent, meaning that phosphorylation at one site affects phosphorylation at another site. Many kinases are involved in the phosphorylation of tau proteins such as PKA, CaMKII, PKC, and MAPKs (Kimura et al., 2018; Ramkumar et al., 2018). Tau phosphorylation is performed on specific motifs such as KXGS, so kinases identify those. Phosphorylated Ser262 within the KXGS motifs is an important factor in characterizing AD (Schwalbe et al., 2013; Cook et al., 2014; Kellogg et al., 2018). Increased phosphorylation of tau protein leads to the formation of PHF that hexapeptide VQIINK (PHF6*) and VQIVYK (PHF6) play an essential role in this formation. Ser285, Ser289, Ser293, Ser305, and Tyr310, located near the hexapeptides, are identified residues for phosphorylation (Hanger et al., 2007; Smit et al., 2017; Liu et al., 2019; Pradeepkiran et al., 2022). On the other hand, Ser285 and Ser289 near the C-terminus of the PHF6* and PHF6 sequences are known to be phosphorylation hotspots for casein kinase 1 (CK1), casein kinase 2 (CK2), and glycogen synthase kinase-3 (GSK-3) in the brains of Alzheimer’s disease patients. Also, phosphorylation at these sites leads to the formation of PHFs (Mendoza et al., 2013; Pradeepkiran and Reddy, 2019). However, the mechanism of its formation is not clearly understood. One of the most critical challenges in designing a biomarker for the early detection of AD is how the structure of a particular region of tau protein changes as the number of phosphorylated sites increases.
The present study aimed to identify how structural features of two docked similar segments of tau protein change when the phosphorylated sites increase from one site in Ser262 to three sites in Ser262, Ser285, and Ser289 and their effects on PHF formation. Studies have shown that phosphorylation in Ser262 decreases the binding of tau to microtubules and plays a critical role in tau accumulation and toxicity (Biernat et al., 1993; Lauckner et al., 2003; Iijima et al., 2010; Ando et al., 2016). The phosphorylated tau protein at this site is detected in preneurofibrillary tangles and is linked to an increased accumulation of seeding potency. Therefore, it is a starting point for tau abnormalities (Oba et al., 2020). On the other hand, phosphorylation in Ser262 alone does not participate in filament formation (Seubert et al., 1995; Schneider et al., 1999b). Recent studies showed that Δ280-tau and Δ187-tau mutants and phosphorylated K18Δ280 initially formed tangled dimers and tau protein oligomers (Barghorn et al., 2000; Nieznanska et al., 2021). So, we used Δ280-tau as a positive control and compared the results to one and three phosphorylation sites because the presence of P-Ser262 is important for tau protein dissociation from the microtubule, although it may not be directly involved in PHF formation.
We used the following methods to determine the effect of this phosphorylation: first, phosphorylation was performed on Ser262 in the sequence 254–290 of tau protein and docked two phosphorylated segments. Also, Δ280-tau and non-phosphorylated tau protein were prepared as the positive control and the control, respectively. After that, MD simulation was run for the two docked segments with phosphorylation of Ser262, Ser285, and Ser289 and positive control and control during 200 ns. Results showed that two segments that were just phosphorylated in Ser262 form an unstable complex with variable structures. In comparison, two segments with three phosphorylated sites formed compact and stable structures similar to positive control during simulations.
Methods
Molecular Docking
At first, the 3D structure of microtubule-associated tau protein (seq 254–290) was obtained from RCSB with ID: 5N5A, and phosphorylation was performed by CHARMM GUI. The segment was phosphorylated in two forms, including only phosphorylation on Ser262 and another on Ser626, Ser285, and Ser289. We used the non-phosphorylated complex to control and a mutated form of tau protein (Δ280-tau leads to the formation of PHF) to positive control. Then, complexes of these segments were prepared for MD simulation. Two segments, both phosphorylated at the Ser262 position, were docked in phosphorylated complexes. The second docked complex was performed for two phosphorylated segments at Ser626, Ser285, and Ser289. The control complex was prepared by docking two non-phosphorylated segments. To make a positive control, Δ280-tau was modeled using the Modeller program available in the Chimera visualization tool, and then the two mutant segments were docked. We used Zdock, HADDOCK, Cluspro, and Hawkdock web servers for docking segments, preparing complexes, finding binding sites, and binding free energy for each complex. The best-predicted structure was then selected from each tool based on the score. Finally, the Hawkdock program selected the best prediction based on the free binding energy obtained from the web servers (Yan et al., 2020).
Molecular Dynamics Simulation
All-atom MD simulations of phosphorylated forms of tau protein were studied using GROMACS (version 2018.1) with CHARMM 36 force field. A TIP3P water model was used for the preparation of systems, and physiological concentrations of chloride and sodium (concentration of NaCl in cells is 0.15 M, which is considered a physiological concentration) were added to neutralize the systems (Callahan and Roux, 2018; Zavadlav et al., 2018; Alipour et al., 2022). The complexes are placed at least 1.0 nm in the center of the cubic box from the box edges. Furthermore, energy minimization was performed using the steepest descent algorithm with a maximum force <1,000.0 kJ/mol/nm to ensure that the system had no improper conformation. Then, equilibrium was done in a two-step, first by NVT and then NPT ensembles. A V-rescale thermostat was used to couple temperature at 310 K and a Parrinello–Rahman barostat for coupling pressure around 1 bar (Cino et al., 2012; Pachler et al., 2019). Finally, MD simulations were run for 200 ns in 2 fs and repeated three times (n = 3) for each complex (Supplementary Figs S1, S2). Periodic boundary conditions and saving energies in trajectory every 1 ps and constrain by the LINCS algorithm were set for runs too.
Binding Energy and Dissociation Constant Calculations
The complexes’ binding energy components were calculated using the molecular mechanics Poisson–Boltzmann surface area method (MM-PBSA) by g_mmpbsa in GROMACS. Moreover, the dissociation constant (Kd) was calculated with the formula (Iqbal et al., 2010) where R is the gas constant, T is the temperature in Kelvin, and ∆G is the binding energy. Kd is an important parameter to indicate the degree of affinity between two segments (Luo and Sharp, 2002).
Results and Discussion
Stability and Flexibility
MD simulation was employed for the complexes, including two docked similar segments that are phosphorylated in the Ser262 situation, and another one, the phosphorylated segments in Ser262, Ser285, and Ser289. Also, we used the non-phosphorylated and Δ280-tau complexes for the control and the positive control, respectively. As shown in Figure 1A, root mean square deviation (RMSD) fluctuated sharply in the control complex and the complex with one phosphorylated site (P-Ser262) so that the RMSD value in these complexes increased to near 3 and 2.5 nm, respectively, but the RMSD of the positive control and the complex with three phosphorylated sites (P-Ser262, 285, and 289) were stabled around 1 nm. This means more structural changes occurred in the P-Ser262 complex when compared to the P-Ser262, 285, 289 complex during 200 ns simulations. Moreover, results indicated that the root mean square fluctuation (RMSF) value of strands with one phosphorylated site and the control was higher than strands with three phosphorylated sites and positive control. Indeed, strands’ flexibility and structural changes have decreased with the increase in the number of phosphorylated sites. (Figure 1B).
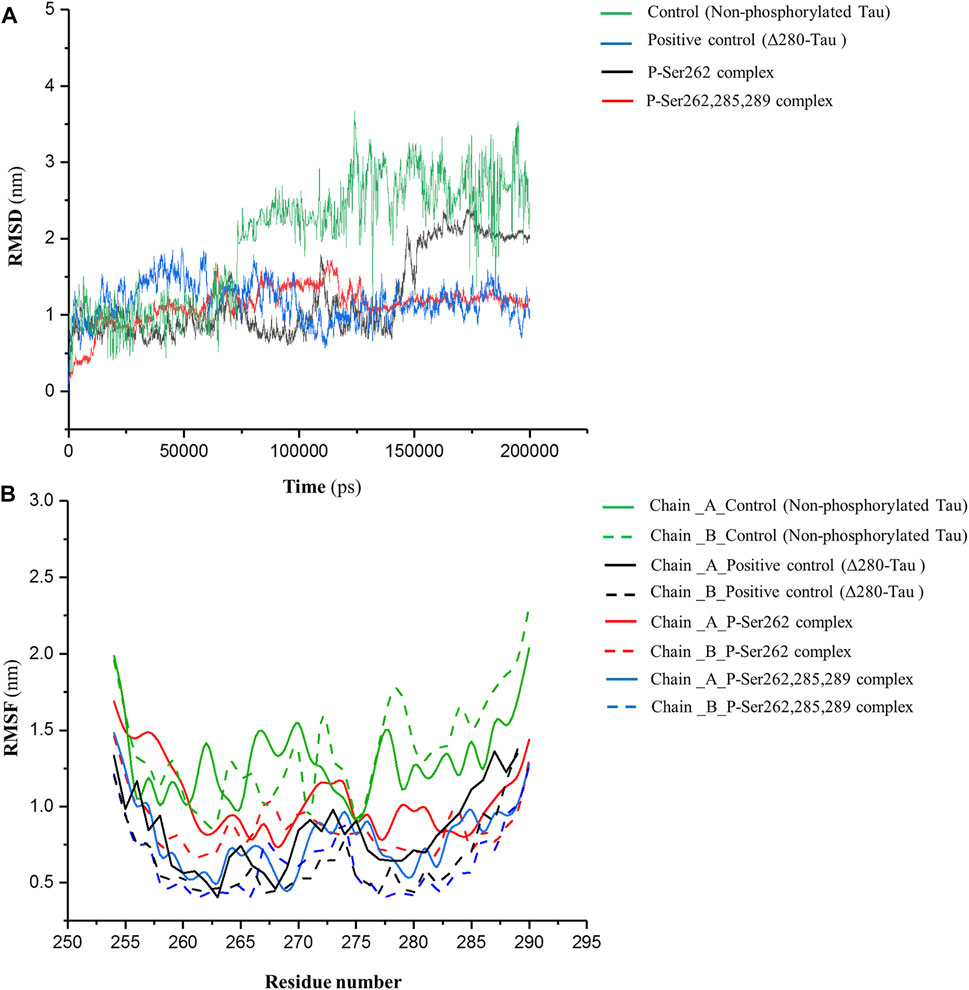
FIGURE 1. RMSD of backbone and RMSF profile for the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylation sites during 200 ns simulations. (A) RMSD plots representing the stability of the complexes and (B) RMSF of residues show each strand’s flexibility.
Analysis of Interactions
The number of contacts (NC) is an important parameter to analyze atomistic-resolved descriptions of the interactions occurring within and between the researched molecules. Indeed, NC can measure the binding affinity of one molecule to another (Liu et al., 2007). So, NC between two segments was calculated in control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes at a distance of less than 0.6 nm (r < 0.6 nm). This distance can be a criterion for the potential of atom-to-atom interactions such as hydrogen bonds, electrostatic interactions, and hydrophobic interactions. As shown in Figure 2A, the NC of the control and P-Ser262 complexes decreased when compared to the positive control and the P-Ser262, 285, 289 complexes during 200 ns simulations. Moreover, NC of the P-Ser262, 285, 289 complex shows an increasing trend when compared to other complexes. These results indicated that NC between two segments in the control and the P-Ser262 complex reached zero, meaning the two segments were separated at some time points.
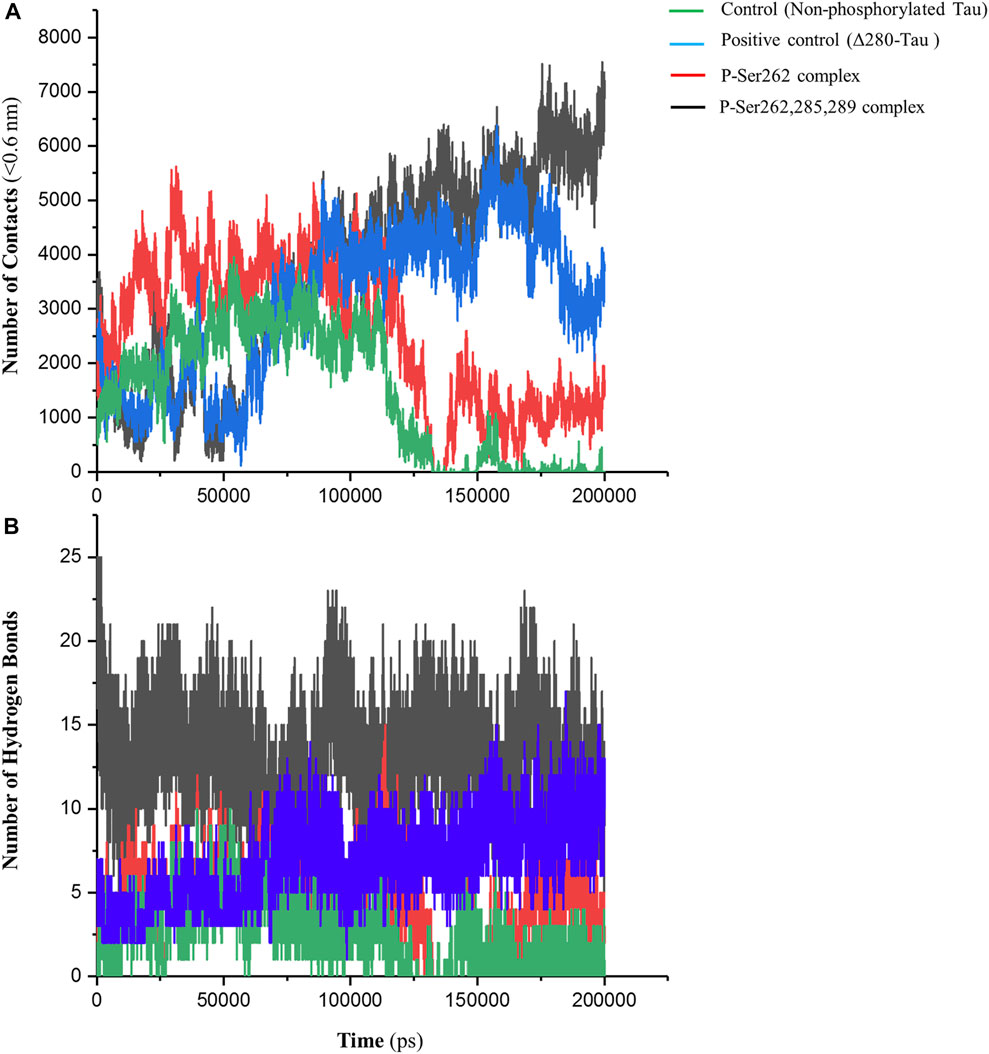
FIGURE 2. Number of contacts (NC) and number of hydrogen bonds (NHB) between two segments in the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylated sites during 200 ns simulations. (A) NC was calculated at a distance of less than 0.6 nm, which decreased in the P-Ser262 compared to P-Ser262, 285, and 289. (B) NHB in P-Ser262 is less than in P-Ser262, 285, and 289.
On the other hand, the NC of the P-Ser262 complex was more than that of control. Moreover, as another parameter to satisfy binding affinity, the number of hydrogen bonds (NHB) was calculated for each complex during simulations. Results showed that NHB in control and P-Ser262 complexes was less than that in positive control and P-Ser262, 285, 289 complex, so their average was about 4, 5, 14, and 16, respectively (Figure 2B). In general, the results showed that the interactions of the P-Ser262 complex were very similar to the control, but they increased significantly with increasing the number of phosphorylated sites to three. Indeed, as NC and NHB increase in P-Ser262, 285, 289 complex, the possibility of stable dimers increases, which is the first step in PHF formation (Luna-Viramontes et al., 2020).
Interaction Energy Analysis
Short-range Lennard-Jones and Coulomb energies describe interaction energy between two non-bonding atoms or molecules (Van Lipzig et al., 2004). So, these energies were calculated between two segments in control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes. As shown in Figure 3A, Coulomb energy of P-Ser262, 285, 289 complex becomes more negative as well as positive control than P-Ser262 during simulations, so the average value of these energies in P-Ser262, 285, and 289 and P-Ser262 was about −516.63 and −230.99 kJ/mol, respectively. Moreover, the Coulomb energy of the control and P-Ser262 complexes was zero at some time points, which means the complexes were separated into two segments during the simulations. The obtained results in Lennard-Jones energy were similar to those in Coulomb energy, and the average values of these energies in P-Ser262 and of P-Ser262, 285, 289 complexes were −156.81 and −254.63 kJ/mol, respectively. (Figure 3B). The more negative values of these energies in P-Ser262, 285, and 289 than in P-Ser262 signify that two segments were tightly bound together and made a stable dimer as an important step in PHF formation.
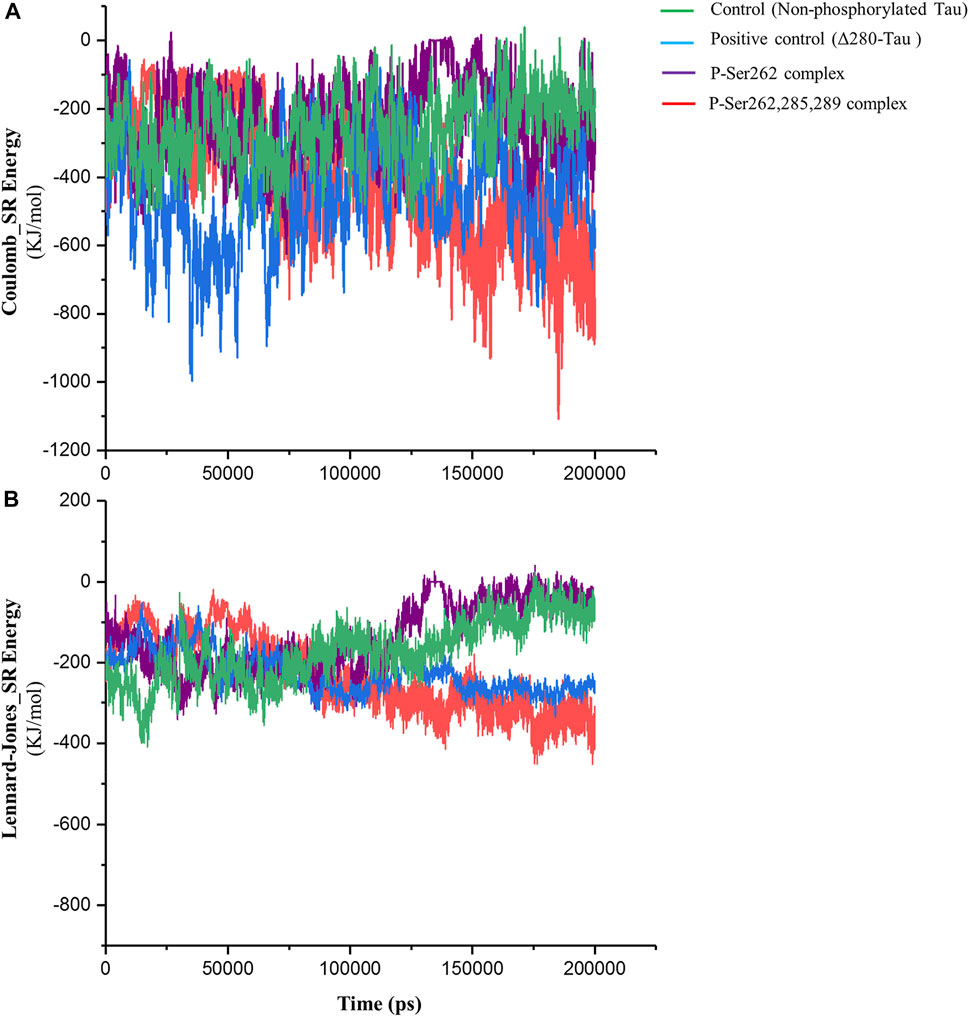
FIGURE 3. Short-range Coulomb and Lennard-Jones energies between two segments in the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylated sites during 200 ns simulations. (A) Coulomb and (B) Lennard-Jones energies are more negative in P-Ser262, 285, and 289 compared to P-Ser262.
Radius of Gyration
Radius of gyration (Rg) is a standard parameter to measure molecular size. It is the RMSD to the center of mass in the atom, molecule, peptide, protein, etc. Using this parameter, the compaction rate of one molecule can be compared with others (He and Niemeyer, 2003). So, Rg was calculated to find compaction of the complexes including the control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes during 200 ns simulations. Results indicated that Rg of the control and the P-Ser262 complex fluctuated. However, Rg values in the positive control and P-Ser262, 285, 289 complexes fluctuated up to 115 ns and stabilized around 0.75 nm. Also, Rg values were increased mainly in the control and P-Ser262 complexes when compared to the positive control and P-Ser262, 285, 289 complexes (Figure 4). Increasing the Rg value means that the structure compaction declined and two segments were separated during the simulation.
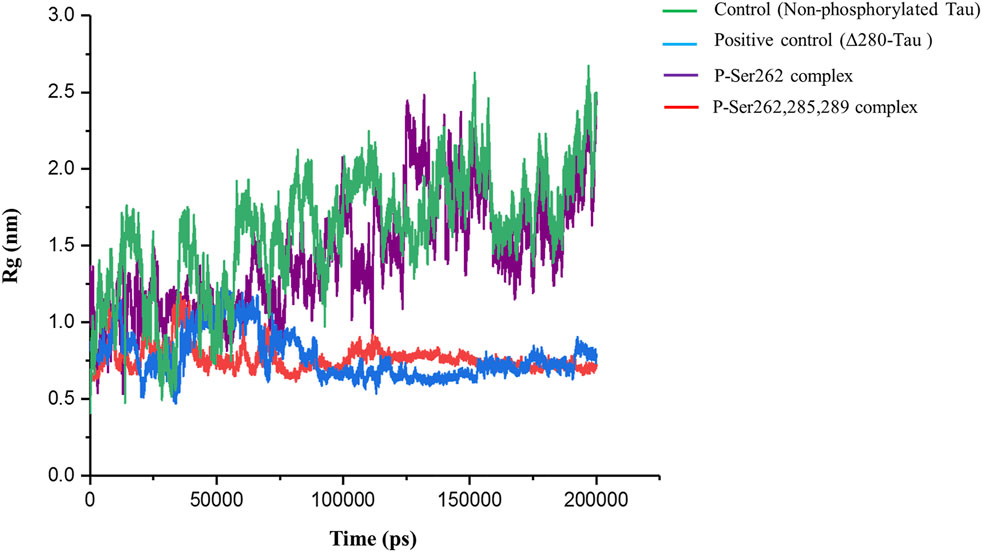
FIGURE 4. Change in the radius of gyration (Rg) of the two docked segments in the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylated sites during 200 ns simulations. Rg values were increased mainly in the P-Ser262 compared to the P-Ser262, 285, 289.
Principal Component Analysis
Principal component analysis (PCA) is a linear transform to analyze the motion of a molecule using a covariance matrix, which is constructed from atomic displacements (Cartesian coordinates) in each conformation during simulation. Eigenvectors are obtained from diagonalization of covariance matrix used to explain molecule motions. Lower eigenvectors signify lower motion and high compactness of a molecule during simulation (David and Jacobs, 2014; Fatima et al., 2020). So PCA was calculated for the control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes from the Cα covariance matrix during 200 ns simulations. Obtained results of plotting eigenvalues against the eigenvectors indicated that 90% of the complexes’ motion was covered by the first 10 eigenvectors and reached their equilibrium in the first 20 eigenvectors (Figure 5A). Moreover, eigenvectors of the control and P-Ser262 complexes were higher than those of the positive control and P-Ser262, 285, 289 complexes, covering the areas on PC1 and PC2 that were between −60 and 80, −60 and 60 in the control and −60 and 60, −40 and 40 in the P-Ser262 complex, respectively, while in both PC1 and PC2 were between −20 and 20 in the positive control and the P-Ser262, 285, 289 complexes (Figures 5B–E). Indeed, PCA results generally suggested that various structural changes occurred in the control and P-Ser262 complexes that caused P-Ser262, 285, 289 complexes to be more stable than P-Ser262.
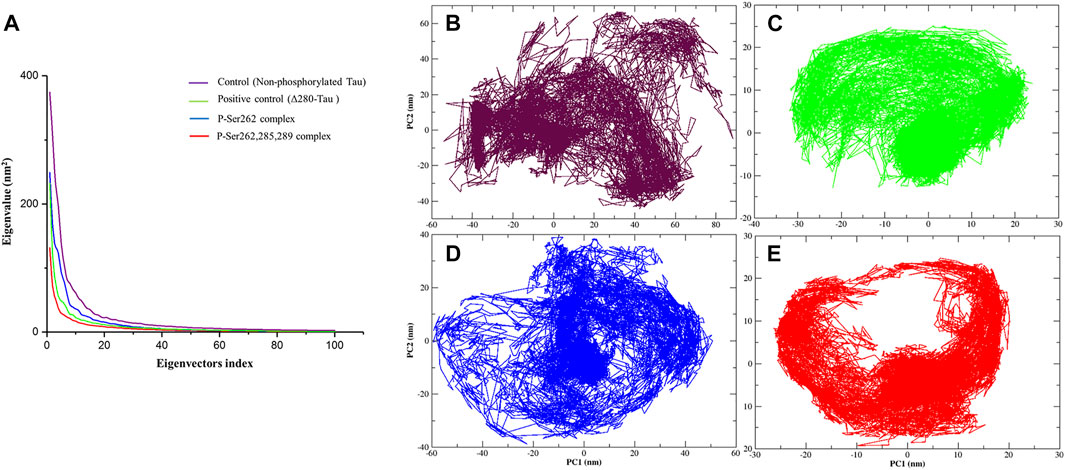
FIGURE 5. Principal component analysis (PCA) of the Cα atomic fluctuations in the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylated sites during 200 ns simulations. (A) Eigenvalues were plotted against the eigenvectors. Projections of the trajectory of (B) control and (D) P-Ser262 showed more changes compared to (C) positive control and (E) P-Ser262, 285, 289.
Gibbs Free Energy Landscape Analysis
Gibbs’s free energy landscape (FEL) is an important measure of molecule stability that allows finding the native state structures by the conformational sampling method. Native states are stable structures with minimum FEL during simulation (Borkotoky and Murali, 2018; Amir et al., 2019). Thus, FELs of the control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes were calculated to identify the conformational states using the first two principal components. Results indicated multiple minimum-energy clusters in the control and P-Ser262 complexes that depicted the structural transition from one structure to different active conformational states, while there was one minimum-energy cluster in the positive control and P-Ser262, 285, 289 complexes. Indeed, the P-Ser262, 285, 289 complex was the most stable compared to the control and P-Ser262 complex, so the two segments were wholly separated in these complexes at some time points. In contrast, a stable minimum-energy dimmer is formed in P-Ser262, 285, 289 with a similar positive control (Figure 6).
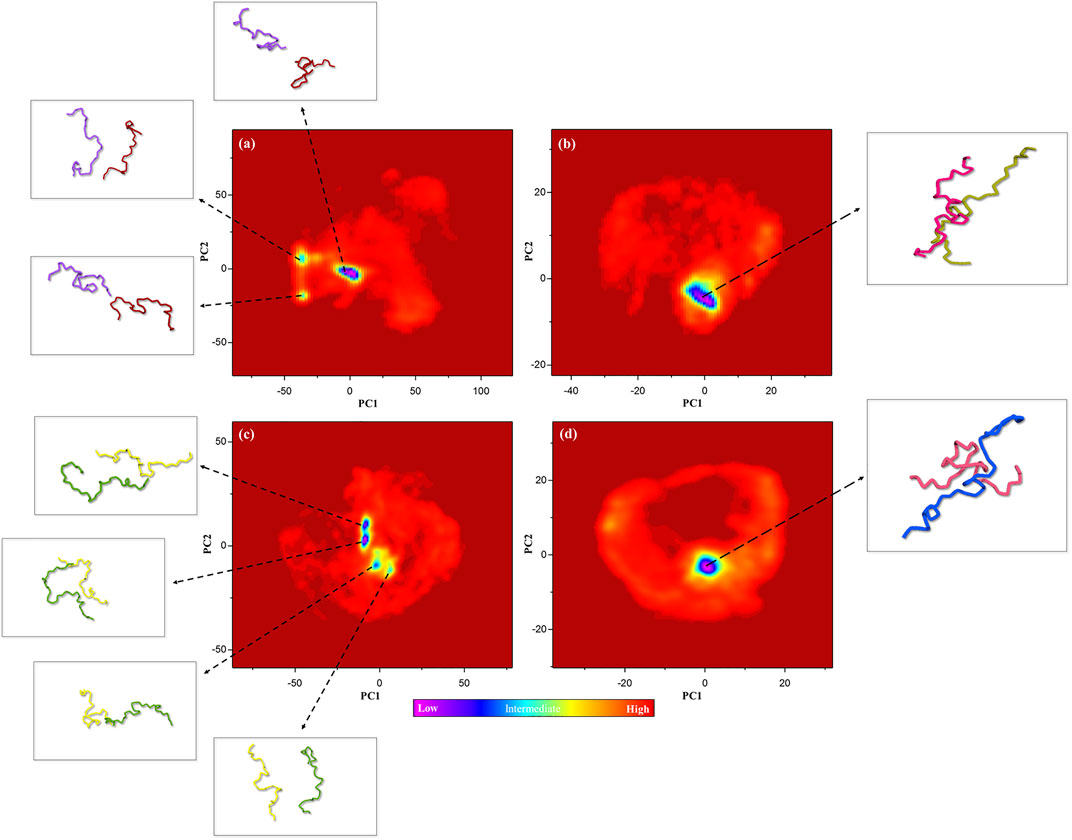
FIGURE 6. Free energy landscape (FEL) of the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylated sites on principal components 1 and 2 during 200 ns simulations. Multiple minimum-energy clusters were observed in (A) the control and (C) P-Ser262, but one cluster was observed in (B) the positive control and (D) P-Ser262, 285, 289.
Binding Energy and Dissociation Constant
Binding energy (ΔG) and dissociation constant (Kd) are important parameters to estimate the stability and affinity of two segments (Wang et al., 2017). So, these parameters were calculated for the control, positive control, and P-Ser262 and P-Ser262, 285, 289 complexes during 200 ns simulations. Results showed that ΔG of the positive control and P-Ser262, 285, 289 complexes were more negative than the control and P-Ser262 complexes, and their values were −76.36 kJ/mol, -80.26 kJ/mol, −3.69 kJ/mol, and −6.29 kJ/mol, respectively. Also, Kd in the positive control and P-Ser262, 285, 289 were lower than that in the control and P-Ser262 (Table 1). So, as a result, two segments in the P-Ser262, 285, 289 complex were tightly bound together compared to the P-Ser262 complex and did not separate easily.
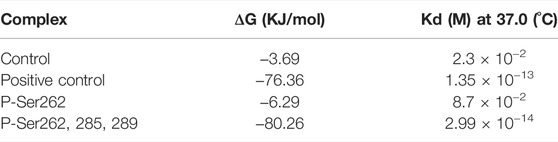
TABLE 1. Binding energy (∆G) and dissociation constant (Kd) values for the control (non-phosphorylated tau), positive control (Δ280-tau), and the complexes with one (P-Ser262) and three (P-Ser262, 285, and 289) phosphorylation sites during 200 ns simulations.
In conclusion, increasing tau phosphorylation occurs in the early stage of AD. Indeed, hyperphosphorylation of tau protein decreases its capacity to bind to microtubules, leading to microtubule destabilization and disruption of the axons and dendrites, the formation of neurofibrillary tangles, and finally, neuronal death. Studies have shown that phosphorylation of Ser262 is an important marker to distinguish AD, and also, several motifs are hotspots for phosphorylation (Majd et al., 2017). Our results show that the P-Ser262 complex is much more unstable than a P-Ser262, 285, 289 complex, and there are many structural fluctuations. Moreover, two segments in the P-Ser262 complex are separated so that the number of contacts, number of hydrogen bonds, and interaction energy become zero at some time points during simulation. So, Rg of the P-Ser262 complex dramatically increased when compared to the P-Ser262, 285, 289 complex. Indeed, the results obtained from the MD simulation of P-Ser262 are very similar to the non-phosphorylated complex (control).
Since the two segments are not tightly bonded in the P-Ser262 complex, they have a higher dynamic than the P-Ser262, 285, 289 complex. On the other hand, the high structural dynamics of the P-Ser262 complex increases the likelihood of diverse structures in it, so that there are several clusters in the P-Ser262 than in the P-Ser262, 285, 289 complex. Evidence implies that small aggregated protein intermediates, which may form soluble oligomers before forming highly structured fibrils, are the principal toxic species involved in starting neurodegenerative disorders (Vieira et al., 2007; Peterson et al., 2008; Pavlova et al., 2016). In other words, the formation of intertwined dimers and oligomers is the first step in forming PHFs and finally the fibrils. PHF structures can be assembled from a different number of these subunits (Barghorn et al., 2000; Nieznanska et al., 2021). On the other hand, our results showed that phosphorylation at these three sites leads to an intertwined dimer that may create more complex structures such as PHFs. Generally, our study shows that two segments only phosphorylated in Ser262 form an unstable complex with variable structure than two segments with three phosphorylated sites.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
MA, MM, PA, and BH-V conceived and designed the study and performed and analyzed the data. AZ and FA conceived and proposed the study. SS and BH-V, as corresponding authors, conducted research, sponsored the study, and made all the arrangements. MA wrote the original draft, and all authors reviewed and edited the final version of the manuscript.
Funding
This study was financially supported by the Functional Neurosurgery Research Center, Shahid Beheshti University of Medical Sciences (SBMU), grant number 28031.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.884705/full#supplementary-material
References
Alipour, M., Hajipour-Verdom, B., Abdolmaleki, P., and Javan, M. (2022). Molecular properties of Ca2+ transport through TRPV2 channel: A molecular dynamics simulations study. J. Biomol. Struct. Dyn., 1–8. doi:10.1080/07391102.2022.2058615
Amir, M., Mohammad, T., Kumar, V., Alajmi, M. F., Rehman, M., Hussain, A., et al. (2019). Structural analysis and conformational dynamics of STN1 gene mutations involved in coat plus syndrome. Front. Mol. Biosci. 6, 41. doi:10.3389/fmolb.2019.00041
Ando, K., Oka, M., Ohtake, Y., Hayashishita, M., Shimizu, S., Hisanaga, S., et al. (2016). Tau phosphorylation at Alzheimer's disease-related Ser356 contributes to tau stabilization when PAR-1/MARK activity is elevated. Biochem. Biophys. Res. Commun. 478 (2), 929–934. doi:10.1016/j.bbrc.2016.08.053
Barbier, P., Zejneli, O., Martinho, M., Lasorsa, A., Belle, V., Smet-Nocca, C., et al. (2019). Role of tau as a microtubule-associated protein: Structural and functional aspects. Front. Aging Neurosci. 11, 204. doi:10.3389/fnagi.2019.00204
Barghorn, S., Zheng-Fischhöfer, Q., Ackmann, M., Biernat, J., Von Bergen, M., Mandelkow, E-M., et al. (2000). Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39 (38), 11714–11721. doi:10.1021/bi000850r
Biernat, J., Gustke, N., Drewes, G., and Mandelkow, E. (1993). Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: Distinction between PHF-like immunoreactivity and microtubule binding. Neuron 11 (1), 153–163. doi:10.1016/0896-6273(93)90279-z
Borkotoky, S., and Murali, A. (2018). A computational assessment of pH-dependent differential interaction of T7 lysozyme with T7 RNA polymerase. BMC Struct. Biol. 17 (1), 7. doi:10.1186/s12900-017-0077-9
Cabrales Fontela, Y., Kadavath, H., Biernat, J., Riedel, D., Mandelkow, E., Zweckstetter, M., et al. (2017). Multivalent cross-linking of actin filaments and microtubules through the microtubule-associated protein Tau. Nat. Commun. 8 (1), 1981. doi:10.1038/s41467-017-02230-8
Callahan, K. M., and Roux, B. (2018). Molecular dynamics of ion conduction through the selectivity filter of the NaVAb sodium channel. J. Phys. Chem. B 122 (44), 10126–10142. doi:10.1021/acs.jpcb.8b09678
Cino, E. A., Choy, W-Y., and Karttunen, M. (2012). Comparison of secondary structure formation using 10 different force fields in microsecond molecular dynamics simulations. J. Chem. Theory Comput. 8 (8), 2725–2740. doi:10.1021/ct300323g
Cook, C., Carlomagno, Y., Gendron, T. F., Dunmore, J., Scheffel, K., Stetler, C., et al. (2014). Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 23 (1), 104–116. doi:10.1093/hmg/ddt402
David, C. C., and Jacobs, D. J. (2014). Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 1084, 193–226. doi:10.1007/978-1-62703-658-0_11
Dzwolak, W., Smirnovas, V., Jansen, R., and Winter, R. (2004). Insulin forms amyloid in a strain‐dependent manner: An FT‐IR spectroscopic study. Protein Sci. 13 (7), 1927–1932. doi:10.1110/ps.03607204
Fatima, S., Mohammad, T., Jairajpuri, D. S., Rehman, M. T., Hussain, A., Samim, M., et al. (2020). Identification and evaluation of glutathione conjugate gamma-l-glutamyl-l-cysteine for improved drug delivery to the brain. J. Biomol. Struct. Dyn. 38 (12), 3610–3620. doi:10.1080/07391102.2019.1664937
Fulga, T. A., Elson-Schwab, I., Khurana, V., Steinhilb, M. L., Spires, T. L., Hyman, B. T., et al. (2007). Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 9 (2), 139–148. doi:10.1038/ncb1528
Graff-Radford N. R., and Woodruff B. K. (Editors) (2007). “Frontotemporal dementia,” Seminars in neurology (New York: Medical Publishers, Inc. Seventh Avenue), 333. Copyright© 2007 by Thieme.
Hanger, D. P., Byers, H. L., Wray, S., Leung, K-Y., Saxton, M. J., Seereeram, A., et al. (2007). Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 282 (32), 23645–23654. doi:10.1074/jbc.M703269200
He, L., and Niemeyer, B. (2003). A novel correlation for protein diffusion coefficients based on molecular weight and radius of gyration. Biotechnol. Prog. 19 (2), 544–548. doi:10.1021/bp0256059
Iijima, K., Gatt, A., and Iijima-Ando, K. (2010). Tau Ser262 phosphorylation is critical for Abeta42-induced tau toxicity in a transgenic Drosophila model of Alzheimer's disease. Hum. Mol. Genet. 19 (15), 2947–2957. doi:10.1093/hmg/ddq200
Iqbal, K., Liu, F., Gong, C-X., and Grundke-Iqbal, I. (2010). Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 7 (8), 656–664. doi:10.2174/156720510793611592
Kellogg, E. H., Hejab, N. M., Poepsel, S., Downing, K. H., DiMaio, F., Nogales, E., et al. (2018). Near-atomic model of microtubule-tau interactions. Science 360 (6394), 1242–1246. doi:10.1126/science.aat1780
Kimura, T., Sharma, G., Ishiguro, K., and Hisanaga, S. (2018). Phospho-tau bar code: Analysis of phosphoisotypes of tau and its application to tauopathy. Front. Neurosci. 12, 44. doi:10.3389/fnins.2018.00044
Lauckner, J., Frey, P., and Geula, C. (2003). Comparative distribution of tau phosphorylated at Ser262 in pre-tangles and tangles. Neurobiol. Aging 24 (6), 767–776. doi:10.1016/s0197-4580(02)00228-2
Liu, F-F., Dong, X-Y., Wang, T., and Sun, Y. (2007). Rational design of peptide ligand for affinity chromatography of tissue-type plasminogen activator by the combination of docking and molecular dynamics simulations. J. Chromatogr. A 1175 (2), 249–258. doi:10.1016/j.chroma.2007.10.074
Liu, H., Zhong, H., Liu, X., Zhou, S., Tan, S., Liu, H., et al. (2019). Disclosing the mechanism of spontaneous aggregation and template-induced misfolding of the key hexapeptide (PHF6) of tau protein based on molecular dynamics simulation. ACS Chem. Neurosci. 10 (12), 4810–4823. doi:10.1021/acschemneuro.9b00488
Luna-Viramontes, N. I., Campa-Córdoba, B. B., Ontiveros-Torres, M. Á., Harrington, C. R., Villanueva-Fierro, I., Guadarrama-Ortíz, P., et al. (2020). PHF-core tau as the potential initiating event for tau pathology in Alzheimer’s disease. Front. Cell. Neurosci. 14, 247. doi:10.3389/fncel.2020.00247
Luo, H., and Sharp, K. (2002). On the calculation of absolute macromolecular binding free energies. Proc. Natl. Acad. Sci. U. S. A. 99 (16), 10399–10404. doi:10.1073/pnas.162365999
Majd, S., Power, J. H., Koblar, S. A., and Grantham, H. J. (2017). The impact of tau hyperphosphorylation at Ser262 on memory and learning after global brain ischaemia in a rat model of reversible cardiac arrest. IBRO Rep. 2, 1–13. doi:10.1016/j.ibror.2016.12.002
Martin, L., Latypova, X., and Terro, F. (2011). Post-translational modifications of tau protein: Implications for Alzheimer's disease. Neurochem. Int. 58 (4), 458–471. doi:10.1016/j.neuint.2010.12.023
Mendoza, J., Sekiya, M., Taniguchi, T., Iijima, K. M., Wang, R., Ando, K., et al. (2013). Global analysis of phosphorylation of tau by the checkpoint kinases Chk1 and Chk2 in vitro. J. Proteome Res. 12 (6), 2654–2665. doi:10.1021/pr400008f
Mietelska-Porowska, A., Wasik, U., Goras, M., Filipek, A., and Niewiadomska, G. (2014). Tau protein modifications and interactions: Their role in function and dysfunction. Int. J. Mol. Sci. 15 (3), 4671–4713. doi:10.3390/ijms15034671
Mohan, R., and John, A. (2015). Microtubule-associated proteins as direct crosslinkers of actin filaments and microtubules. IUBMB life 67 (6), 395–403. doi:10.1002/iub.1384
Naseri, N. N., Wang, H., Guo, J., Sharma, M., and Luo, W. (2019). The complexity of tau in Alzheimer’s disease. Neurosci. Lett. 705, 183–194. doi:10.1016/j.neulet.2019.04.022
Neddens, J., Temmel, M., Flunkert, S., Kerschbaumer, B., Hoeller, C., Loeffler, T., et al. (2018). Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol. Commun. 6 (1), 52. doi:10.1186/s40478-018-0557-6
Nieznanska, H., Boyko, S., Dec, R., Redowicz, M. J., Dzwolak, W., Nieznanski, K., et al. (2021). Neurotoxicity of oligomers of phosphorylated Tau protein carrying tauopathy-associated mutation is inhibited by prion protein. Biochim. Biophys. Acta. Mol. Basis Dis. 1867 (11), 166209. doi:10.1016/j.bbadis.2021.166209
Nizynski, B., Dzwolak, W., and Nieznanski, K. (2017). Amyloidogenesis of tau protein. Protein Sci. 26 (11), 2126–2150. doi:10.1002/pro.3275
Oba, T., Saito, T., Asada, A., Shimizu, S., Iijima, K. M., Ando, K., et al. (2020). Microtubule affinity–regulating kinase 4 with an Alzheimer's disease-related mutation promotes tau accumulation and exacerbates neurodegeneration. J. Biol. Chem. 295 (50), 17138–17147. doi:10.1074/jbc.RA120.014420
Omalu, B. I., Fitzsimmons, R. P., Hammers, J., and Bailes, J. (2010). Chronic traumatic encephalopathy in a professional American wrestler. J. Forensic Nurs. 6 (3), 130–136. doi:10.1111/j.1939-3938.2010.01078.x
Pachler, M., Kabelka, I., Appavou, M-S., Lohner, K., Vácha, R., Pabst, G., et al. (2019). Magainin 2 and PGLa in bacterial membrane mimics I: Peptide-peptide and lipid-peptide interactions. Biophys. J. 117 (10), 1858–1869. doi:10.1016/j.bpj.2019.10.022
Pavlova, A., Cheng, C-Y., Kinnebrew, M., Lew, J., Dahlquist, F. W., Han, S., et al. (2016). Protein structural and surface water rearrangement constitute major events in the earliest aggregation stages of tau. Proc. Natl. Acad. Sci. U. S. A. 113 (2), E127–E136. doi:10.1073/pnas.1504415113
Perez, D. I., Gil, C., and Martinez, A. (2011). Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med. Res. Rev. 31 (6), 924–954. doi:10.1002/med.20207
Peterson, D. W., Zhou, H., Dahlquist, F. W., and Lew, J. (2008). A soluble oligomer of tau associated with fiber formation analyzed by NMR. Biochemistry 47 (28), 7393–7404. doi:10.1021/bi702466a
Pradeepkiran, J. A., Munikumar, M., Reddy, A. P., and Reddy, P. H. (2022). Protective effects of a small molecule inhibitor ligand against hyperphosphorylated tau-induced mitochondrial and synaptic toxicities in Alzheimer disease. Hum. Mol. Genet. 31 (2), 244–261. doi:10.1093/hmg/ddab244
Pradeepkiran, J. A., Reddy, A. P., and Reddy, P. H. (2019). Pharmacophore-based models for therapeutic drugs against phosphorylated tau in Alzheimer’s disease. Drug Discov. Today 24 (2), 616–623. doi:10.1016/j.drudis.2018.11.005
Pradeepkiran, J. A., and Reddy, P. H. (2019). Structure based design and molecular docking studies for phosphorylated tau inhibitors in Alzheimer’s disease. Cells 8 (3), 260. doi:10.3390/cells8030260
Ramkumar, A., Jong, B. Y., and Ori-McKenney, K. M. (2018). ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule‐associated proteins. Dev. Dyn. 247 (1), 138–155. doi:10.1002/dvdy.24599
Schneider, A., Biernat, J., Von Bergen, M., Mandelkow, E., and Mandelkow, E-M. (1999). Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry 38 (12), 3549–3558. doi:10.1021/bi981874p
Schneider, A., Biernat Jvon, B. M., Mandelkow, E., and Mandelkow, E. M. (1999). Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry 38 (12), 3549–3558. doi:10.1021/bi981874p
Schwalbe, M., Biernat, J., Bibow, S., Ozenne, V., Jensen, M. R., Kadavath, H., et al. (2013). Phosphorylation of human Tau protein by microtubule affinity-regulating kinase 2. Biochemistry 52 (50), 9068–9079. doi:10.1021/bi401266n
Seubert, P., Mawal-Dewan, M., Barbour, R., Jakes, R., Goedert, M., Johnson, G. V., et al. (1995). Detection of phosphorylated Ser262 in fetal tau, adult tau, and paired helical filament tau (-). J. Biol. Chem. 270 (32), 18917–18922. doi:10.1074/jbc.270.32.18917
Smit, F. X., Luiken, J. A., and Bolhuis, P. G. (2017). Primary fibril nucleation of aggregation prone Tau fragments PHF6 and PHF6. J. Phys. Chem. B 121 (15), 3250–3261. doi:10.1021/acs.jpcb.6b07045
Sottejeau, Y., Bretteville, A., Cantrelle, F-X., Malmanche, N., Demiaute, F., Mendes, T., et al. (2015). Tau phosphorylation regulates the interaction between BIN1’s SH3 domain and Tau’s proline-rich domain. Acta Neuropathol. Commun. 3 (1), 58. doi:10.1186/s40478-015-0237-8
Van Lipzig, M. M., Ter Laak, A. M., Jongejan, A., Vermeulen, N. P., Wamelink, M., Geerke, D., et al. (2004). Prediction of ligand binding affinity and orientation of xenoestrogens to the estrogen receptor by molecular dynamics simulations and the linear interaction energy method. J. Med. Chem. 47 (4), 1018–1030. doi:10.1021/jm0309607
Vieira, M. N., Forny-Germano, L., Saraiva, L. M., Sebollela, A., Martinez, A. M. B., Houzel, J. C., et al. (2007). Soluble oligomers from a non-disease related protein mimic Abeta-induced tau hyperphosphorylation and neurodegeneration. J. Neurochem. 103 (2), 736–748. doi:10.1111/j.1471-4159.2007.04809.x
Von Bergen, M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E-M., Mandelkow, E., et al. (2000). Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl. Acad. Sci. U. S. A. 97 (10), 5129–5134. doi:10.1073/pnas.97.10.5129
Wang, G., Huang, Y., Wang, L-L., Zhang, Y-F., Xu, J., Zhou, Y., et al. (2016). MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep. 6 (1), 26697. doi:10.1038/srep26697
Wang, Y., and Mandelkow, E. (2016). Tau in physiology and pathology. Nat. Rev. Neurosci. 17 (1), 5–21. doi:10.1038/nrn.2015.1
Wang, Y., Martins, J. M., and Lindorff-Larsen, K. (2017). Biomolecular conformational changes and ligand binding: From kinetics to thermodynamics. Chem. Sci. 8 (9), 6466–6473. doi:10.1039/c7sc01627a
Williams, D. R. (2006). Tauopathies: Classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Intern. Med. J. 36 (10), 652–660. doi:10.1111/j.1445-5994.2006.01153.x
Yan, Y., Tao, H., He, J., and Huang, S-Y. (2020). The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15 (5), 1829–1852. doi:10.1038/s41596-020-0312-x
Keywords: Alzheimer’s disease, tau protein, hyperphosphorylation, conformational dynamics, molecular dynamics simulation
Citation: Alipour M, Motavaf M, Abdolmaleki P, Zali A, Ashrafi F, Safari S and Hajipour-Verdom B (2022) Structural Analysis and Conformational Dynamics of Short Helical Hyperphosphorylated Segments of Tau Protein (Sequence 254–290) in Alzheimer’s Disease: A Molecular Dynamics Simulation Study. Front. Mol. Biosci. 9:884705. doi: 10.3389/fmolb.2022.884705
Received: 26 February 2022; Accepted: 09 June 2022;
Published: 08 August 2022.
Edited by:
Roberta Cascella, University of Florence, ItalyCopyright © 2022 Alipour, Motavaf, Abdolmaleki, Zali, Ashrafi, Safari and Hajipour-Verdom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saeid Safari, c2FmYXJpLnNAc2JtdS5hYy5pcg==; Behnam Hajipour-Verdom, Yi5oYWppcG91ckBtb2RhcmVzLmFjLmly
 Mozhgan Alipour1,2
Mozhgan Alipour1,2 Mahsa Motavaf
Mahsa Motavaf Parviz Abdolmaleki
Parviz Abdolmaleki Saeid Safari
Saeid Safari Behnam Hajipour-Verdom
Behnam Hajipour-Verdom