- 1Department of Diagnostics and Clinical Sciences, Faculty of Veterinary Medicine, University of Agriculture in Krakow, Kraków, Poland
- 2Department of Plant Biotechnology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland
- 3Department of Infectious Diseases and Public Health, Faculty of Veterinary Medicine, University of Agriculture in Krakow, Kraków, Poland
- 4Department of Plant Physiology and Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland
Leptospirosis is a zoonotic infectious disease of growing importance in both human and veterinary medicine. Gram-negative spirochetes of Leptospira are traditionally classified into serovars based on their antigenic identity, which must be ascertained to design effective treatment procedures for humans and appropriate vaccination strategies in pets and livestock. Unfortunately, identifying Leptospira serovars is challenging and currently requires access to a wide panel of reference strains, animal-derived antisera, or monoclonal antibodies. Here, we describe a new method for the identification of Leptospira serovars that is based on monosaccharide composition analysis of the polysaccharide part of bacterial lipopolysaccharide (LPS) structures. Our approach requires no animal sacrifice and can be implemented in any laboratory equipped for chromatographic analysis. An LPS sugar fingerprint that is specific to each bacterial isolate that we studied can be generated. Importantly, sugar profiling of LPS enables distinguishing Leptospira serovars that are antigenically very similar. Using our new approach, we discover that the LPS structures of two cattle pathogens belonging to two different species: Leptospira interrogans and Leptospira borgpetersenii, and to one serovar: Hardjo, can be distinguished despite sharing major similarities. Through extensive phylogenetic analysis, we reveal which specific glycosyltransferases of the LPS biosynthesis rfb locus likely drove the emergence of these similarities and identify a single glycosyltransferase that might have contributed to the formation of saccharide differences in the LPS structure. Our findings have implications for future work on the evolution of bacterial polysaccharide synthesis and highlight the importance of preventing horizontal gene transfer between pathogenic bacteria.
Introduction
Leptospira is a genus of Gram-negative spirochetes that includes pathogens with major clinical relevance in human and veterinary medicine. Leptospira are characterized by the presence of lipopolysaccharide (LPS) molecules that are antigenic but have low endotoxic activity in animals (Sun et al., 2020). For epidemiological purposes, in addition to standard genetic species, Leptospira are classified into serovars that have different host ranges and geographical distributions (Arent et al., 2022). Correct annotation of serovars not only enhances our understanding of the epidemiology of these infections but is also crucial for the selection of strains for diagnostic purposes and for their use as components in vaccine development. The LPS is thought to be the main determinant of the serovar identity, with different genetic species frequently belonging to the same serovars and showing similar clinical importance. Previous work has demonstrated that the majority of genes implicated in LPS biosynthesis, including glycosyltransferases (GTs) responsible for polysaccharide formation, are encoded on the rfb locus of Leptospira (Medeiros et al., 2022). Importantly, the locus has been subject to horizontal gene transfer (HGT) events between Leptospira species, which likely contributed to serovar evolution (de la Peña-Moctezuma et al., 1999; Haake et al., 2004; Nieves et al., 2022; Ca Ferreira et al., 2024).
Current estimates indicate that leptospirosis affects more than one million people annually, with 60,000 deaths reported because of the condition (Centre for Disease Control and Prevention, 2024). Recent reports point to the prevalence of antibiotic resistance in specific serovars, forcing modifications of prescribed treatment protocols (Pineda et al., 2024). The disease is of particular importance in veterinary medicine. In cattle farming, leptospirosis decreases animal health, productivity, and fertility, particularly in unvaccinated herds. Chronic infections caused by serovar Hardjo are of primary concern, as they result in long-term renal and reproductive colonization (Sohm et al., 2023). Current estimates indicate up to 84% loss in gross margin for farms where outbreaks are not controlled (Carvalho et al., 2024). Importantly, vaccines for specific Leptospira serovars show only limited cross-reactivity (Barazzone et al., 2022; Rinehart et al., 2012), which necessitates designing dedicated vaccination strategies based on the prevalence of specific serovars in the area of interest.
Correct characterization of Leptospira serovars present in clinical or environmental samples is crucially important, but all currently available methods for detecting Leptospira serovars have some limitations. The gold standard is the cross-agglutinin absorption test (CAAT), which is a laborious method requiring bacterial culturing and access to a panel of sera isolated from animals exposed to specific serovars (Pinto et al., 2022; Rajapakse et al., 2015). Importantly, the identification of selected Leptospira serovars can be significantly enhanced through the use of specific monoclonal antibodies (Arent et al., 2023). However, due to the challenges in obtaining monoclonal antibodies that target serovar-specific epitopes, such antibodies are not currently available for all known serovars.
Recently, much attention has been given to the development of strategies for serovar determination based on PCR. Some progress has been made when targeting genes of the rfb locus of the Leptospira genome with a complex combination of primer pairs (Wenderlein et al., 2024) or with in-depth analysis of RT-qPCR amplification curves (Waggoner et al., 2014; Esteves et al., 2018; Ahmed et al., 2020). However, these tools still need validation and improvement to provide confident annotation of serovars with clinical relevance.
Here, we present a new approach to the determination of Leptospira serovars that relies on the analysis of the monosaccharide composition of the bacterial LPS structures. The method utilizes high-performance liquid chromatography (HPLC) to generate an LPS sugar profile that can be considered a fingerprint of each Leptospira serovar and isolate. We apply our tool to discriminate exemplary serovars with major relevance for veterinary medicine, demonstrating that LPS composition in cattle pathogens belonging to one serovar, Hardjo, and to two species, L. interrogans and L. borgpetersenii, can be distinguished, despite high antigenic similarity, which classical serological methods cannot differentiate. Through molecular phylogenetic analysis, we demonstrate which specific GTs in the rfb locus of Leptospira are likely responsible for the biosynthesis of the similar and differentiating LPS structures in the two genetically distant cattle pathogens.
Materials and methods
LPS isolation and compositional analysis
Each bacterial strain (Supplementary Table S1) was initially assigned to serogroup and serovar by cross-agglutination using a microscopic agglutination test described previously (Dikken and Kmety, 1978). Bacterial isolates were grown in biological triplicate in a T80/40/LH culture medium (Ellis et al., 1985) at 29°C for 7–10 days, after which their LPS was isolated as previously described (Bonhomme and Werts, 2020). LPS composition was analyzed following TFA hydrolysis as detailed in Supplementary Protocol S1. A graphical summary of the LPS isolation and compositional analysis is shown in Supplementary Figure S1. Statistical analysis was performed in R.
Bioinformatics and molecular phylogenetics
Sequences of 16S rRNA were obtained from Silva (Quast et al., 2013). Sequences of GT2, GT4, and GTnc enzymes were sourced from the Carbohydrate Active Enzymes (CAZY - http://www.cazy.org/) database (Drula et al., 2022). Molecular phylogenies were reconstructed with MEGA11 (Tamura et al., 2021) using the WAG algorithm, including the gamma distribution of rates. A dendroscope was used for visualization of the GT2 phylogeny (Huson and Scornavacca, 2012).
Results
LPS monosaccharide composition analysis is a powerful tool to discriminate even closely related Leptospira serovars
We hypothesized that the variable region of the LPS molecule can act as a unique molecular signature of individual Leptospira serovars. We hoped that the simplest possible approach, the analysis of LPS monosaccharide composition, would be sufficient to discriminate between serovars. To evaluate that, we first isolated LPS structures from two distant Leptospira strains: L. borgpetersenii serovar Serjoe, strain M84, and Leptospira kirschneri serovar Grippotyphosa, strain KR100. The strains belong to two different genetic species and two different serogroups, Serjoe and Grippotyphosa, respectively, making them a perfect case study to evaluate whether analysis of the monosaccharide composition of the LPS is a suitable method for serovar annotation. Following LPS breakdown to monosaccharides and HPLC, we quantified the molar contribution of each monosaccharide to the LPS molecule (Figure 1A). The analysis revealed major differences between the two isolates, including the presence or absence of different monosaccharides (N-acetyl-glucosamine and galacturonic acid) and quantitative differences in the content of six other monosaccharides. Encouraged by our initial findings, we wanted to establish if a more closely related pair of serovars could also be discriminated. To this end, we evaluated the LPS composition in L. interrogans sv. Copenhageni M20 and L. interrogans sv. Icterohaemorrhagiae KR93. These serovars are members of one serogroup (Icterohaemorrhagiae) and belong to one species, making them a challenging pair to distinguish even with well-established serological methods (Arent et al., 2023). We were encouraged by determining that the analysis of the monosaccharide composition of the LPS in the two serovars revealed quantitative differences in the amount of xylose (Figure 1B). This observation strongly suggests that our approach is capable of distinguishing even very closely related Leptospira serovars.
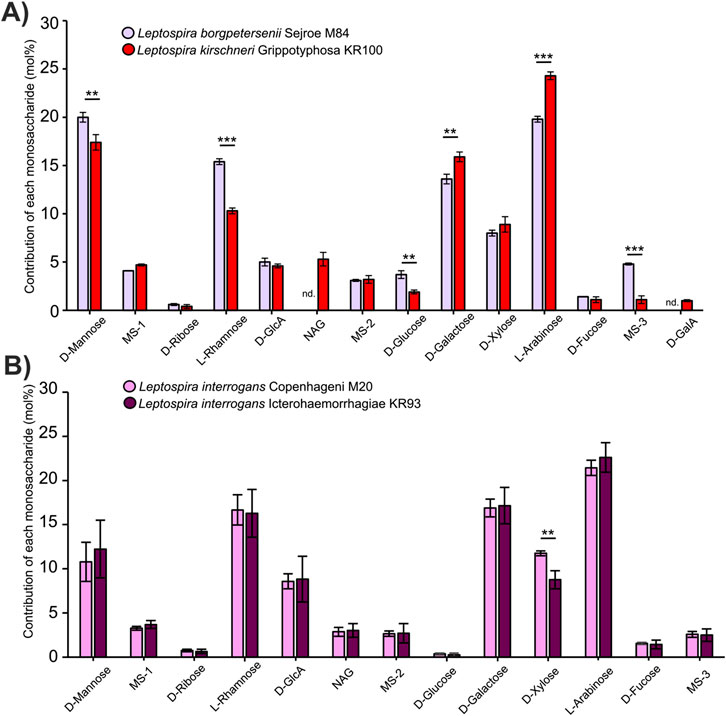
Figure 1. Monosaccharide composition of the LPS differs between Leptospira serogroups and serovars. (A) Monosaccharide composition of the LPS in Leptospira borgpetersenii serogroup Serjoe, serovar Serjoe, strain M 84, and in the Leptospira kirschneri serogroup Grippotyphosa, serovar Grippotyphosa, strain KR100. (B) Monosaccharide composition of the LPS in Leptospira interrogans serogroup Icterohaemorrhagiae, serovars: Icterohaemorrhagiae, strain KR93, and Copenhageni, strain M20. Error bars show the standard deviation of measurements performed for three biological replicates. Asterisks annotate significance in unpaired Student’s t-test with ** denoting p < 0.01 and *** denoting p < 0.001. Monosaccharide abbreviations used: NAG: N-acetylglucosamine; MS-1 to 3: unannotated monosaccharides with the same identity on all figures; D-GlcA: D-glucuronic acid; D-GalA: D-galacturonic acid. Nd. Indicates that a monosaccharide was not detected.
Having established that our method allows confident discrimination between serovars, we set out to perform an analysis of LPS from two major cattle pathogens, both belonging to the serovar Hardjo but genetically classified into two different species: L. interrogans and L. borgpetersenii. Despite belonging to the same serovar and sharing cattle as the host species, the two are clearly members of distant clades when their 16S rRNA sequences are compared (Figure 2A). In line with their belonging to the same serovar, analysis of the LPS structures from isolates of L. interrogans sv. Hardjo and L. borgpetersenii sv. Hardjo revealed that the monosaccharide compositions of the variable region of their LPS structures are very similar (Figure 2B). Importantly, we were still able to discriminate the isolates from both species, mainly by looking at the mannose, rhamnose, and xylose contents in the LPS samples. To study the differences in the LPS monosaccharide composition, we performed a principal component analysis (PCA, Figure 2C) based on all our sugar fingerprint profiles. The PCA confirmed that the compositions of LPS in L. borgpetersenii sv. Hardjo and L. interrogans sv. Hardjo are more similar to one another than to that in other members of their respective genetic species.
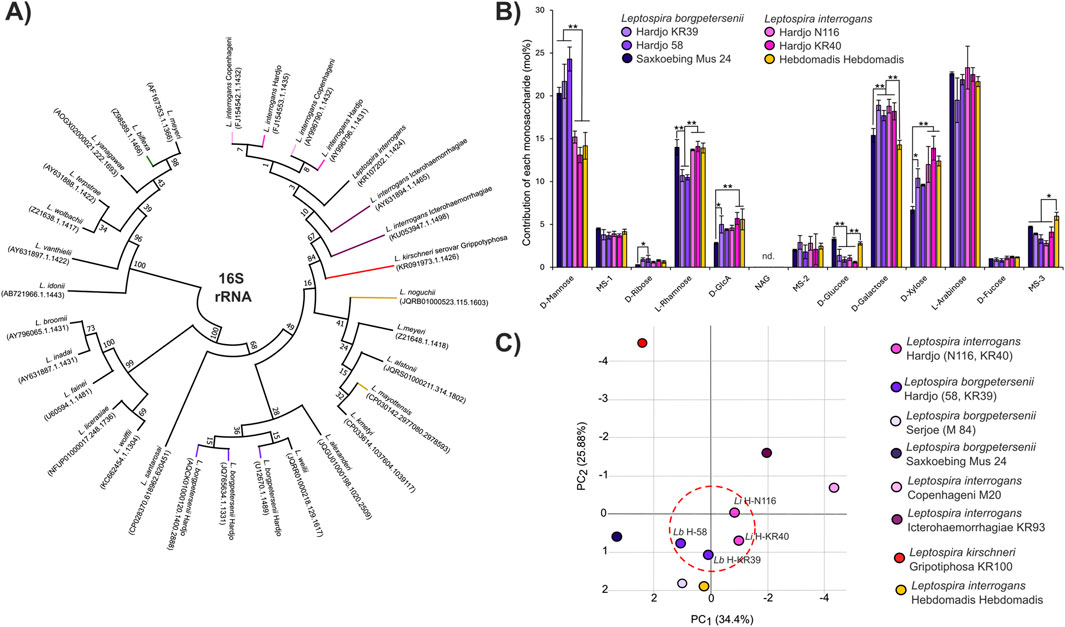
Figure 2. Composition of the LPS is similar in distant Leptospira species belonging to one serovar. (A) Result of maximum likelihood phylogenetic analysis of 16S rRNA sequence of Leptospira, including sequences from analyzed serovars. In total, 100 bootstrap replicates were used, and numerical confidence values are provided for all nodes. (B) Monosaccharide composition analysis of the LPS structures in Leptospira borgpetersenii serovar Hardjo (2 strains, KR39 and 58), Leptospira interrogans Hardjo (2 strains, N116 and KR40), Leptospira interrogans serogroup Hebdomadis serovar Hebdomadis, strain Hebdomadis, and in the Leptospira borgpetersenii serogroup Sejroe serovar Saxkoebing, strain Mus 24. Error bars show the standard deviation of measurements performed for three biological replicates. Asterisks annotate significance in ANOVA post hoc Tukey test with * denoting p < 0.05 and **p < 0.01. Nd. Indicates that a monosaccharide was not detected. (C) Principal component analysis (PCA) based on monosaccharide composition values (Supplementary Table S2) of all Leptospira serovars analyzed in this work. The LPS structures of strains belonging to the Hardjo serovar are similar and are marked with a dotted red circle.
Neofunctionalization and horizontal transfer of specific glycosyltransferase genes in the rfb locus are the likely reasons for similarities in LPS structures
To understand the origin of the similarity in the LPS composition between the two cattle pathogens, we decided to focus our efforts on the study of GTs implicated in the formation of the glycan part of the LPS. We hypothesized that similarities in the GT toolbox of L. borgpetersenii sv. Hardjo and L. interrogans sv. Hardjo could indicate a shared biosynthetic pathway, likely originating from the previously reported horizontal gene transfer (HGT) events between the two species. The GT enzymes are subdivided into families, and the Carbohydrate Active Enzymes (CAZY) database can be used to identify families present in Leptospira genomes. The well-characterized rfb locus of L. interrogans serovar Hardjo strain Norma (Medeiros et al., 2022) comprises 12 GT enzymes that are categorized by CAZY. Of these, eight fall into the GT2 family, two into the GT4 family, and two are assigned to an as yet unknown family (GTnc). We decided to evaluate the similarities in these genes between Hardjo members from L. interrogans and L. borgpetersenii species.
To this end, we performed molecular phylogenetic analyses on the GT2 (Supplementary Figure S2), GT4 (Figure 3A), and GTnc (Supplementary Figure S3) sequences from a selection of available Leptospira genomes. For the analysis, we chose sequences from representatives of species and serovars for which we characterized the LPS composition and from species that are important for the separation of the L. interrogans and L. borgpetersenii clades (Supplementary Table S3; Figure 2A). We first evaluated GT4 enzymes (Figure 3A) and obtained a cladogram showing that Leptospiral family members separate into two major groups and a total of 12 distinct clades. Among these, most clades (10 of 12, example subtree in Figure 3B) showed topology akin to that observed in the 16S rRNA phylogeny, with L. interrogans sv. Hardjo and L. borgpetersenii sv. Hardjo sequences falling into two separate branches of a clade. Interestingly, for clades containing two GT4 enzymes from the rfb locus, the topology of the clade was different (Figures 3C,D). For one, the sequences were present only in members of the Hardjo serovar (Figure 3C), suggesting gene neofunctionalization prior to HGT. For the second one, the sequences of the Hardjo GT4 were more similar to one another than to those from other members of the same species (Figure 3D), marking the sequences as exchanged in an HGT event. A similar comparison was performed for GT2 (Supplementary Figure S2) and GTnc (Supplementary Figure S3) phylogenies, revealing neofunctionalization and HGT events for specific GT2 and GTnc enzymes.

Figure 3. Phylogenetic analysis of Leptospira GT family 4. (A) Result of maximum likelihood phylogenetic analysis of Leptospira GT4 family protein sequences annotated by the CAZY database. One hundred bootstrap replicates were used, and numerical confidence values are provided for all nodes. Specific color codes are used for each serovar analyzed, with similar colors denoting members of one bacterial species. Letter codes are provided in brackets to indicate proteins from specific serovars. Sequences from the annotated rfb locus of L. interrogans are marked in red, and subtrees containing these sequences are presented on (B–D), with (B) showing topology expected based on genetic similarities between Leptospira species, and panels (C, D) marking topologies indicating protein neofunctionalization and HGT events between L. interrogans sv. Hardjo and L. borgpetersenii sv. Hardjo.
Discussion
In our work, we developed a new approach to the identification of Leptospira serovars that looks at the LPS monosaccharide compositions. We compared LPS structures both in distant serovars, belonging to different serogroups, such as Serjoe and Grippotyphosa (Figure 1A), and in much more similar ones, such as Copenhageni and Icterohaemorrhagiae, which belong to the same serogroup (Figure 1B). We observed clearly maintained trends in monosaccharide compositions of individual species and serovars (Supplementary Figure S4). For instance, serovar Hardjo is characterized by lower glucose and higher galactose content than other serovars. In all cases, we were able to distinguish individual Leptospira isolates based on the monosaccharide composition of their LPS structures. This confirmed that our method is suitable for discriminating Leptospira serovar pairs that are challenging to distinguish using standard serological approaches (Arent et al., 2023). Most critically, the comparative analysis of LPS structures with PCA allowed us to discriminate all studied isolates and group them according to serovar similarity. This opens the way for creating a reference library of LPS composition that can be used to assign Leptospira isolates to specific serovars.
Our method, unlike CAAT, does not require a ready collection of antisera nor any animal sacrifice (Pinto et al., 2022). Sugar fingerprinting relies on the direct analysis of the serovar-determining LPS antigen, which is advantageous when compared to PCR and RT-qPCR routes that attempt to define serovars indirectly. Moreover, our LPS compositional results are similar to those previously reported for two Leptospira isolates, which further validates our approach (Patra et al., 2015). Importantly, upon completion of the LPS compositional database, our tool will allow for rapid identification of novel Leptospira serovars, such as those isolated from unusual hosts (Grune et al., 2015). Sugar fingerprinting could be complemented by other ways of characterizing and classifying Leptospira, such as lipid A profiling (Pětrošová et al., 2023) or peptide analysis (Calderaro et al., 2014).
We applied our newly developed approach to extensively study the LPS structure in isolates belonging to two species: L. interrogans and L. borgpetersenii, and to one serovar, Hardjo, which are known cattle pathogens. We observed that their LPS structures are similar, which may be a result of the proposed HGT event between the two species (de la Peña-Moctezuma et al., 1999; Nieves et al., 2022). By combining the results from the performed phylogenies, we were able to identify six GTs that are shared between the rfb loci of L. interrogans and L. borgpetersenii sv. Hardjo (Table 1) and one GTnc (WP 000050431.1; ORF60) that is specific to L. interrogans sv. Hardjo only. We postulate that the shared enzymes, listed in Table 1, are likely responsible for the formation of a similar LPS antigen in Hardjo representatives adapted to cattle, and that the L. interrogans-specific GT may contribute to the formation of differences in LPS structure. What remains to be established is the exact activity of these enzymes and the advantage their function offers to cattle-adapted pathogens. In addition, such studies could be supplemented by linkage analysis, which would show how individual monosaccharide components of the LPS structures are connected and how LPS structure contributes to Leptospira antigenic identity. These questions are an active area of research for our group and must be addressed to fully understand the role of LPS during leptospirosis and in preventive vaccination.
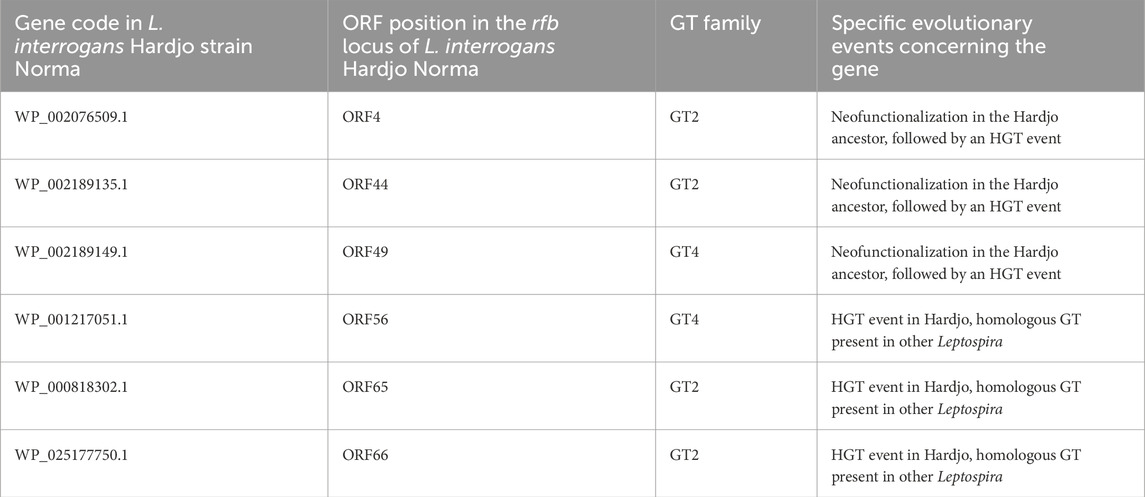
Table 1. Summary of GT enzymes proposed to be implicated in the biosynthesis of the Hardjo-specific LPS.
Our work describes a new powerful method for the identification of Leptospira serovars. Utilizing monosaccharide profiling, we are able to perform a detailed compositional analysis of the polysaccharide part of the LPS and to generate a unique sugar fingerprint of a serovar. This allows us to distinguish antigenically similar Leptospira and to start building a reference database of serovar-specific compositional profiles. In addition, through molecular phylogenetics, we identify the exact glycosyltransferase genes implicated in the HGT events that shaped the evolution of the LPS antigen. For that work, our efforts focus on L. interrogans sv. Hardjo and L. borgpetersenii sv. Hardjo. These two pathogens prevail in different parts of the globe but their isolates have been identified in overlapping areas, such as Europe and South America (van den Brink et al., 2023; Miller et al.,1991; Hagedoorn et al., 2024), where a shared host might have been a vessel for the horizontal gene exchange between bacterial species.
Summary
Leptospirosis is a major disease of humans and animals caused by numerous distinct serovars of Leptospira bacteria. Determining which serovars are prevalent in a given area, or are causing the disease in a patient, is essential to design correct vaccination and treatment strategies. Currently we have a limited set of techniques available for identification of Leptospira serovars, with serological assays being the gold standard in the field. Unfortunately, these require the use of antisera, thus animal sacrifice, and often application of monoclonal antibodies. Our team has developed a new technique for determination of Leptospira serovar identity. By analysing the monosaccharide composition of leptospiral lipopolysaccharide (LPS) we were able to distinguish serovars showing very high degree of antigenic similarity. We further employed monosaccharide profiling to study selected serovars causing leptospirosis in cattle, and confirmed that their LPSs, though discernible with our method, share major structural similarities. Using molecular phylogenetic approaches we discover which exact glycosyltransferases may be implicated in the formation of these similar saccharide structures. Our work does not only provide a new tool for identification of Leptospira, but also sheds light on the evolution of bacterial polysaccharide biosynthesis, which may be relevant for our understanding of other host-pathogen interactions.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: Mendeley Data, doi:10.17632/zf4scz7syj.1.
Author contributions
AL: conceptualization, data curation, formal analysis, investigation, validation, visualization, writing – original draft, and writing – review and editing. JL: conceptualization, formal analysis, investigation, methodology, validation, visualization, writing – original draft, and writing – review and editing. LP: investigation, methodology, project administration, and writing – review and editing. KD: investigation and writing – review and editing. DL: conceptualization, methodology, resources, supervision, and writing – review and editing. ZA: conceptualization, funding acquisition, methodology, supervision, and writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by a grant from the National Science Centre, Poland, awarded to ZA (project number: 2019/33/B/NZ9/02159).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2025.1581587/full#supplementary-material
References
Ahmed, A. A., Goris, M. G. A., and Meijer, M. C. (2020). Development of lipL32 real-time PCR combined with an internal and extraction control for pathogenic Leptospira detection. PLoS One 15 (11), e0241584. doi:10.1371/journal.pone.0241584
Arent, Z., Gilmore, C., Pardyak, L., Dubniewicz, K., McInerney, B., and Ellis, W. (2023). The serological and genetic diversity of the Leptospira interrogans Icterohaemorrhagiae serogroup circulating in the UK. J. Vet. Res. 67 (4), 529–536. doi:10.2478/jvetres-2023-0063
Arent, Z., Pardyak, L., Dubniewicz, K., Płachno, B., and Kotula-Balak, M. (2022). Leptospira taxonomy: then and now. Med. Weter. 78 (10), 6694–2022. doi:10.21521/mw.6694
Barazzone, G. C., Teixeira, A. F., Azevedo, B. O. P., Damiano, D. K., Oliveira, M. P., Nascimento, ALTO, et al. (2022). Revisiting the development of vaccines against pathogenic Leptospira: innovative approaches, present challenges, and future perspectives. Front. Immunol. 12, 760291. doi:10.3389/fimmu.2021.760291
Bonhomme, D., and Werts, C. (2020). Purification of LPS from Leptospira. Methods Mol. Biol. 2134, 53–65. doi:10.1007/978-1-0716-0459-5_6
Ca Ferreira, L., de Fa Ferreira Filho, L., V Cosate, M. R., and Sakamoto, T. (2024). Genetic structure and diversity of the rfb locus of pathogenic species of the genus Leptospira. Life Sci. Alliance 7 (6), e202302478. doi:10.26508/lsa.202302478
Calderaro, A., Piccolo, G., Gorrini, C., Montecchini, S., Buttrini, M., Rossi, S., et al. (2014). Leptospira species and serovars identified by MALDI-TOF mass spectrometry after database implementation. BMC Res. Notes 7, 330. doi:10.1186/1756-0500-7-330
Carvalho, HGAC, Silva, D. M., Rodrigues, G. R. D., Gameiro, A. H., Dos Santos, R. F., Raineri, C., et al. (2024). Estimation of economic losses due to leptospirosis in dairy cattle. Prev. Vet. Med. 229, 106255. doi:10.1016/j.prevetmed.2024.106255
Centre for Disease Control and Prevention (2024). About leptospirosis. Available online at: https://www.cdc.gov/leptospirosis/about/index.html.
de la Peña-Moctezuma, A., Bulach, D. M., Kalambaheti, T., and Adler, B. (1999). Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol. Lett. 177 (2), 319–326. doi:10.1111/j.1574-6968.1999.tb13749.x
Dikken, H., and Kmety, E. (1978). “Chapter VIII: serological typing methods of leptospires,” in Methods in microbiology volume II (London: Academic Press), 259–307.
Drula, E., Garron, M. L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50 (D1), D571–D577. doi:10.1093/nar/gkab1045
Ellis, W. A., Montgomery, J., and Cassells, J. A. (1985). Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar hardjo. Res. Vet. Sci. 39 (3), 292–295. doi:10.1016/s0034-5288(18)31716-8
Esteves, L. M., Bulhões, S. M., Branco, C. C., Carreira, T., Vieira, M. L., Gomes-Solecki, M., et al. (2018). Diagnosis of human leptospirosis in a clinical setting: real-time PCR high resolution melting analysis for detection of Leptospira at the onset of disease. Sci. Rep. 8 (1), 9213. doi:10.1038/s41598-018-27555-2
Grune, L. S., Rago, V., Martínez, M., Uhart, M., Florin-Christensen, M., Romero, G., et al. (2015). Isolation of a seawater tolerant Leptospira spp. from a southern right whale (Eubalaena australis). PLoS One 10 (12), e0144974. doi:10.1371/journal.pone.0144974
Haake, D. A., Suchard, M. A., Kelley, M. M., Dundoo, M., Alt, D. P., and Zuerner, R. L. (2004). Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 186 (9), 2818–2828. doi:10.1128/JB.186.9.2818-2828.2004
Hagedoorn, N. N., Maze, M. J., Carugati, M., Cash-Goldwasser, S., Allan, K. J., Chen, K., et al. (2024). Global distribution of Leptospira serovar isolations and detections from animal host species: a systematic review and online database. Trop. Med. Int. Health 29 (3), 161–172. doi:10.1111/tmi.13965
Huson, D. H., and Scornavacca, C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61 (6), 1061–1067. doi:10.1093/sysbio/sys062
Medeiros, E. J. S., Ferreira, L. C. A., Ortega, J. M., Cosate, M. R. V., and Sakamoto, T. (2022). Genetic basis underlying the serological affinity of leptospiral serovars from serogroups Sejroe, Mini and Hebdomadis. Infect. Genet. Evol. 103, 105345. doi:10.1016/j.meegid.2022.105345
Miller, D. A., Wilson, M. A., and Beran, G. W. (1991). Relationships between prevalence of Leptospira interrogans in cattle, and regional, climatic, and seasonal factors. Am. J. Vet. Res. 52 (11), 1766–1768. doi:10.2460/ajvr.1991.52.11.1766
Nieves, C., Vincent, A. T., Zarantonelli, L., Picardeau, M., Veyrier, F. J., and Buschiazzo, A. (2022). Horizontal transfer of the rfb cluster in Leptospira is a genetic determinant of serovar identity. Life Sci. Alliance 6 (2), e202201480. doi:10.26508/lsa.202201480
Patra, K. P., Choudhury, B., Matthias, M. M., Baga, S., Bandyopadhya, K., and Vinetz, J. M. (2015). Comparative analysis of lipopolysaccharides of pathogenic and intermediately pathogenic Leptospira species. BMC Microbiol. 15, 244. doi:10.1186/s12866-015-0581-7
Pětrošová, H., Mikhael, A., Culos, S., Giraud-Gatineau, A., Gomez, A. M., Sherman, M. E., et al. (2023). Lipid A structural diversity among members of the genus Leptospira. Front. Microbiol. 14, 1181034. doi:10.3389/fmicb.2023.1181034
Pineda, S., Martínez Garro, J. M., Salazar Flórez, J. E., Agudelo-Pérez, S., Monroy, F. P., and Peláez Sánchez, R. G. (2024). Detection of genes related to antibiotic resistance in Leptospira. Trop. Med. Infect. Dis. 9 (9), 203. doi:10.3390/tropicalmed9090203
Pinto, G. V., Senthilkumar, K., Rai, P., Kabekkodu, S. P., Karunasagar, I., and Kumar, B. K. (2022). Current methods for the diagnosis of leptospirosis: issues and challenges. J. Microbiol. Methods 195, 106438. doi:10.1016/j.mimet.2022.106438
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi:10.1093/nar/gks1219
Rajapakse, S., Rodrigo, C., Handunnetti, S. M., and Fernando, S. D. (2015). Current immunological and molecular tools for leptospirosis: diagnostics, vaccine design, and biomarkers for predicting severity. Ann. Clin. Microbiol. Antimicrob. 14, 2. doi:10.1186/s12941-014-0060-2
Rinehart, C. L., Zimmerman, A. D., Buterbaugh, R. E., Jolie, R. A., and Chase, C. C. (2012). Efficacy of vaccination of cattle with the Leptospira interrogans serovar hardjo type hardjoprajitno component of a pentavalent Leptospira bacterin against experimental challenge with Leptospira borgpetersenii serovar hardjo type hardjo-bovis. Am. J. Vet. Res. 73 (5), 735–740. doi:10.2460/ajvr.73.5.735
Sohm, C., Steiner, J., Jöbstl, J., Wittek, T., Firth, C., Steinparzer, R., et al. (2023). A systematic review on leptospirosis in cattle: a European perspective. One Health 17, 100608. doi:10.1016/j.onehlt.2023.100608
Sun, A. H., Liu, X. X., and Yan, J. (2020). Leptospirosis is an invasive infectious and systemic inflammatory disease. Biomed. J. 43 (1), 24–31. doi:10.1016/j.bj.2019.12.002
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
van den Brink, KMJA, Aalberts, M., Fabri, N. D., and Santman-Berends, IMGA (2023). Effectiveness of the Leptospira hardjo control programme and detection of new infections in dairy cattle in The Netherlands. Anim. (Basel) 13 (5), 831. doi:10.3390/ani13050831
Waggoner, J. J., Balassiano, I., Abeynayake, J., Sahoo, M. K., Mohamed-Hadley, A., Liu, Y., et al. (2014). Sensitive real-time PCR detection of pathogenic Leptospira spp. and a comparison of nucleic acid amplification methods for the diagnosis of leptospirosis. PLoS One 9 (11), e112356. doi:10.1371/journal.pone.0112356
Wenderlein, J., Zitzl, T., Dufay-Simon, N., Cachet, N., Pantchev, N., Le Guyader, M., et al. (2024). “Detection and identification of pathogenic Leptospira spp,” in Serogroups in Europe between 2017 and 2020 applying a novel gene-based molecular approach. Transboundary and Emerging Diseases doi:10.1155/2024/11018411101841
Keywords: lipopolysaccharide, leptospirosis and lipopolysaccharide, serotyping, glycosyl transferase, molecular evolution
Citation: Lewicka AJ, Lyczakowski JJ, Pardyak L, Dubniewicz K, Latowski D and Arent Z (2025) Beyond serology: saccharide profiling enables identification of antigenically similar Leptospira and prompts re-evaluation of bacterial lipopolysaccharide evolution. Front. Mol. Biosci. 12:1581587. doi: 10.3389/fmolb.2025.1581587
Received: 22 February 2025; Accepted: 21 April 2025;
Published: 17 June 2025.
Edited by:
Marcelo Lima, Keele University, United KingdomReviewed by:
Pumtiwitt McCarthy, Morgan State University, United StatesRuben T. Almaraz, Sonoma State University, United States
Copyright © 2025 Lewicka, Lyczakowski, Pardyak, Dubniewicz, Latowski and Arent. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandra J. Lewicka, YWxla3NhbmRyYS5sZXdpY2thQHVyay5lZHUucGw=; Dariusz Latowski, ZGFyaXVzei5sYXRvd3NraUB1ai5lZHUucGw=
 Aleksandra J. Lewicka
Aleksandra J. Lewicka Jan J. Lyczakowski
Jan J. Lyczakowski Laura Pardyak
Laura Pardyak Klaudia Dubniewicz3
Klaudia Dubniewicz3 Dariusz Latowski
Dariusz Latowski Zbigniew Arent
Zbigniew Arent