- 1Department of Digestive Diseases, The First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, China
- 2The First Clinical Medical College, Henan University of Chinese Medicine, Zhengzhou, China
- 3Shanghai Key Laboratory of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 4Department of Pathology, The First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, China
Introduction: Taohong Siwu Decoction (THSW Decoction), a classic formula for treating blood stasis, has demonstrated significant clinical efficacy in the treatment of hepatic fibrosis. However, its primary active components and mechanisms of action remain unclear.
Methods: In this study, a carbon tetrachloride (CCl4)-induced hepatic fibrosis mouse model was established to evaluate THSW Decoction’s therapeutic effects. Ultra-high performance liquid chromatography-quadrupole/Orbitrap high-resolution mass spectrometry (UHPLC-Q-Exactive Orbitrap HRMS) was employed to identify the main prototype chemical components of THSW Decoction in the blood, while non-targeted metabolomics analysis was performed using a Waters Synapt G2-Si QTOF mass spectrometer (Synapt G2-Si QTOF HRMS system). Network pharmacology and metabolomic data were integrated to elucidate the therapeutic targets, differential metabolites, and signaling pathways of THSW Decoction. Molecular docking and binding affinity predictions between key targets and principal compounds were performed using PyMOL software. Furthermore, molecular dynamics simulations were conducted to evaluate the structural stability and binding interactions. Finally, core targets were validated in vivo.
Results: THSW Decoction effectively reduced CCl4-induced serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels, alleviating liver inflammation and collagen deposition. Forty-five blood components were detected, with 616 corresponding drug targets identified, including 419 overlapping targets for anti-hepatic fibrosis. The core protein-protein interaction (PPI) network comprised 59 nodes and 570 edges. Enrichment analysis revealed that THSW Decoction’s blood components primarily modulated biological processes such as positive regulation of response to external stimuli and oxygen content. Key signaling pathways included PI3K-Akt, estrogen, relaxin, and MAPK. Non-targeted metabolomics identified 148 differential metabolites between the model and normal groups, and 156 between the THSW Decoction and model groups. Thirty-five overlapping metabolites were enriched in cAMP, phospholipase D, and GnRH signaling pathways. Twenty intersection targets linked blood components, metabolites, and hepatic fibrosis. PPI analysis ranked JUN, PTGS2, BCL2, ESR1, and PPARG as the top five targets. A “drug-component-target-metabolite” network highlighted ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid as the top five blood components. Molecular docking and molecular dynamics simulations revealed that 3-hydroxy-4-methoxycinnamic acid binds stably to the core target PPARG, exhibiting a binding free energy of −93.68 kJ/mol. In vivo validation showed that THSW Decoction upregulated JUN and downregulated ESR1 expression in the liver.
Discussion: This study elucidates THSW Decoction’s key blood components, potential targets, and mechanisms in the treatment of hepatic fibrosis, providing a foundation for further research.
1 Introduction
Liver fibrosis is a dynamic pathological process characterized by excessive deposition of extracellular matrix (ECM), primarily driven by chronic liver injuries such as viral hepatitis, alcoholism, nonalcoholic fatty liver disease (NAFLD), or autoimmune disorders (Roehlen et al., 2020). The activation of hepatic stellate cells (HSCs) is a central event in liver fibrogenesis. However, beyond HSCs, multiple cell types—including hepatocytes, liver sinusoidal endothelial cells (LSECs), and macrophages—are also involved in the initiation and progression of liver fibrosis. Current studies suggest that liver fibrosis is not the result of a single-cell action, but rather the outcome of complex intercellular interactions. For example, hepatocyte injury and death (Roehlen et al., 2020), LSEC dedifferentiation (Gao et al., 2024), and pro-inflammatory polarization of macrophages (Matsuda and Seki, 2020) can all induce HSC activation and proliferation. In turn, activated HSCs can exacerbate hepatocyte injury, LSEC dedifferentiation, and macrophage pro-inflammatory polarization, thereby creating a vicious cycle that perpetuates fibrogenesis (Akkiz et al., 2024).
Without intervention, this condition progresses to cirrhosis, severely endangering patients’ lives. Studies indicate the annual number of cirrhosis-related deaths rose from 899,000 in 1990 to 1.32 million (GBD, 2017 Cirrhosis Collaborators, 2020). Therefore, early intervention to reverse or delay fibrosis progression helps reduce rates of decompensated cirrhosis and liver cancer, improve patients’ quality of life, and enhance disease prognosis (Ginès et al., 2022). Numerous studies show that etiological treatment (e.g., controlling or curing viral infections) can reverse hepatic fibrosis (Caligiuri et al., 2021). However, without targeted anti-fibrotic therapy, the disease frequently progresses (Schuppan et al., 2018). While many investigational drugs have undergone preclinical and clinical trials, no anti-fibrotic drugs have yet received Food and Drug Administration approval (Yoon et al., 2016).
Taohong Siwu (THSW) Decoction is a classic Chinese herbal formula in traditional Chinese medicine, composed of peach kernel, safflower, Ligusticum wallichii, Radix Rehmanniae Praeparata, Radix Paeoniae Alba, and Radix Angelicae Sinensis. THSW Decoction is primarily used to treat diseases caused by blood stasis. The pathogenesis of blood stasis is considered to be closely related to hepatic fibrosis formation (Wang et al., 2018). Recent studies have shown that THSW Decoction can alleviate liver fibrosis by improving hepatocellular lipid metabolism and mitochondrial autophagy (Wu et al., 2024). Our previous research further demonstrated that THSW Decoction significantly attenuates liver tissue damage and collagen deposition in fibrotic mice by regulating glutathione metabolism and inhibiting ferroptosis in hepatocytes (Xiao et al., 2025). However, as a multi-component traditional Chinese medicine formula, it remains unclear how the bioactive components absorbed into the bloodstream synergistically regulate glutathione metabolism and pathological processes beyond ferroptosis through multiple targets and signaling pathways. Moreover, whether THSW Decoction exerts its holistic therapeutic effects by modulating the endogenous metabolic network also warrants further investigation. Network pharmacology and non-targeted metabolomics have been widely used to investigate the components and mechanisms of traditional Chinese medicine. In this study, a CCl4-induced hepatic fibrosis mouse model was established to evaluate its efficacy. Ultra-high performance liquid chromatography-quadrupole/Orbitrap high-resolution mass spectrometry (UHPLC-Q-Exactive Orbitrap HRMS) technology was employed to analyze the prototype active components of THSW Decoction in blood. Network pharmacology, non-targeted metabolomics, molecular docking and molecular dynamics simulation were performed with a Waters Synapt G2-Si QTOF mass spectrometer (Synapt G2-Si QTOF HRMS) to explore its potential therapeutic mechanisms against hepatic fibrosis and validate these findings in vivo, providing a foundation for further research.
2 Materials and methods
2.1 Animal source
Thirty-two 6-week-old SPF-grade male C57BL/6N mice, weighing 16–18 g, were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China certificate number: 110011231102606376). The animals were housed in the Animal Experiment Center of Henan University of Chinese Medicine, with a room temperature of 25°C ± 2°C, relative humidity of 40%–60%, and a 12-h light-dark cycle, with free access to food and water. Animal experiments were approved by the Animal Welfare and Ethics Committee of Henan University of Chinese Medicine (approval number: IACUC-20230309-02).
2.2 Drugs and reagents
The experimental drugs were in granular form. Peach kernel was purchased from Guangdong Yifang Pharmaceutical Co., Ltd. (Foshan, Guangdong Province, China; batch number: 0111212); safflower, rehmannia glutinosa, white peony, and angelica were purchased from Jiangyin Tianjiang Pharmaceutical Co., Ltd. (Jiangyin, Jiangsu Province, China; batch numbers: 21016504, 21036474, 21026264, 21046824); Ligusticum wallichii was purchased from Beijing Kangrentang Pharmaceutical Co., Ltd. (Beijing, China; batch number: 20042411). THSW Decoction was prepared by mixing peach kernel, safflower, rehmannia glutinosa, white peony, angelica, and Ligusticum wallichii granules in a crude drug ratio of 3:2:4:3:3:2. Chromatographic-grade methanol, acetonitrile, and formic acid were all purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China; batch numbers: 10014108, 40064160, 40023786).
2.3 Establishment of hepatic fibrosis model and Drug Administration
Thirty-two mice were randomly divided into four groups: the blank control group (C), CCl4-induced model group (CCl4), THSW Decoction low-dose group (L), and THSW Decoction high-dose group (H). The blank control group was administered by intraperitoneal injections of olive oil at a dose of 2 mL/kg body weight three times per week for 6 weeks. The hepatic fibrosis model was established by intraperitoneal injection of a 15% CCl4-olive oil solution at a dose of 2 mL/kg body weight three times per week for 6 weeks. Starting from the 4th week, the THSW Decoction groups received granular agents (2.2 or 4.4 g/kg body weight) via oral gavage, while the CCl4 group and blank control group were administered deionized water by the same method. After 6 weeks, the mice were anesthetized with pentobarbital sodium, and blood samples were collected from the inferior vena cava along with liver tissue specimens.
2.4 Detection of serum aminotransferases
Serum ALT and AST activities were measured using commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu Province, China; catalog numbers C009-2-1 and C010-2-1).
2.5 Histopathology and immunohistochemistry
Liver tissues were fixed in neutral formalin, dehydrated with ethanol gradient, transparent with xylene, embedded in paraffin, and cut into 4 μm thick sections. Hematoxylin-eosin (H&E) staining was used to observe the inflammatory damage and Sirius red (SR) staining was used to observe the collagen deposition. The expression of Col-Ⅰ in liver tissue was detected by immunohistochemistry (1: 1,000; Servicebio, Wuhan, Hubei Province, China; catalog number: GB11022-3).
2.6 Western blot analysis
Liver tissue was lysed in RIPA buffer containing a protease inhibitor cocktail and homogenized on ice. Protein concentration was determined using the bicinchoninic acid (BCA) assay. Protein samples (25–50 μg) were denatured at 100°C for 10 min, separated by 10% SDS-PAGE, and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were incubated overnight at 4°C with primary antibodies against C-JUN, ESR1, and ACTIN (all at 1:1,000; Servicebio, Wuhan, Hubei Province, China; catalog numbers: GB111604, GB111843, and GB11001, respectively), followed by incubation with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:3,000; Servicebio, Wuhan, Hubei Province, China; catalog number: GB23303).
2.7 Preparation of drug-containing serum
Mice were fasted for 12 h before administration (with free access to water) and divided into a blank group (3 mice) and a THSW Decoction group (6 mice). The blank group received deionized water, while the THSW Decoction group received THSW Decoction by gavage at 10 mL/kg (equivalent to 15 g/kg of crude drug, ∼2× the human equivalent dose). At 1 h and 3 h post-administration, blood was collected from 3 mice in the THSW Decoction group by ocular enucleation. Serum was separated by centrifugation (8,000 rpm, 15 min, 4°C), and pooled blank serum, 1-h drug-containing serum, and 3-h drug-containing serum were prepared. Serum samples (200 μL) were protein-precipitated with 1 mL methanol, centrifuged (12,000 rpm, 10 min, 4°C), and the supernatant was dried under nitrogen at 37°C, reconstituted in 80 μL of 20% methanol, and centrifuged again for analysis.
2.8 Non-targeted metabolomics analysis of liver tissue
2.8.1 Chromatographic conditions
Chromatographic separation was performed on a Waters ACQUITY UPLC system (Waters Corporation, Milford, MA, United States) equipped with an ACQUITY HSS PFP column (100 mm × 2.1 mm, 1.8 μm). The mobile phase consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The following gradient elution program was applied: 2%–40% B from 0 to 5.0 min, 40%–98% B from 5.0 to 17.0 min, held at 98% B from 17.0 to 20.0 min, and returned to 2% B from 20.1 to 21.0 min. The column temperature was maintained at 40°C, with a flow rate of 0.3 mL/min. The injection volume was set to 1.0 μL, and the total run time was 21 min.
2.8.2 Mass spectrometry conditions
Mass spectrometric detection was conducted on a Waters Synapt G2-Si QTOF mass spectrometer (Waters Corporation, Milford, MA, United States) in both positive (ESI+) and negative (ESI−) electrospray ionization modes, scanning m/z 100–1,000. Capillary voltages were set at 3.0 kV (ESI+) and −2.5 kV (ESI−). Cone voltage, desolvation temperature (350°C), and gas flows (desolvation 600 L/h, nebulizer 50 L/h) were optimized per manufacturer’s instructions. Mass calibration was performed weekly or after maintenance using sodium formate clusters (Waters QTOF Calibration Kit), covering ions from m/z 69 to 1,350 (ESI+) and m/z 113 to 1,023 (ESI−), achieving ±2 ppm accuracy. Daily mass correction employed leucine enkephalin (1 μg/mL in 50% acetonitrile with 0.1% formic acid) as lock mass, with reference ions at m/z 556.2771 (ESI+) and 554.2615 (ESI−) to ensure ongoing mass accuracy.
2.9 Detection of blood components of THSW decoction
The UHPLC-Q Exactive HRMS procedure for the analysis of the blood components of THSW Decoction was performed according to a published protocol (Xiao et al., 2025).
Data acquisition and analysis were performed using Xcalibur 3.0 software. Molecular formulas were deduced based on precursor ions ([M + H]+, [M-H]-) and adduct ions ([M + Na]+, [M + HCOOH]-), with a mass error tolerance of ±10 ppm. Identification was achieved by comparing experimental data with reference standards, literature, and MS2 fragments from the MassBank database.
2.10 Acquisition of targets of components migrating to blood
The SMILE of the components was obtained through PuBChem, and then the relevant targets were obtained using the SEA (https://sea.bkslab.org/) database and the Tcsmp database (https://tcmspw.com/tcmsp.php). Oral bioavailability (OB) and drug-likeness (DL) were set to 30% and 0.18 (Ru et al., 2014), respectively. Finally, the targets were normalised by the Uniprot database (https://www.uniprot.org).
2.11 Acquisition of hepatic fibrosis targets
The search term “hepatic fibrosis” was uesd to Drugbank (https://www.drugbank.ca/), GeneCards (https://www.genecards.org/), TTD (http://db.idrblab.net/ttd/), OMIM (https://www.omim.org/), PharmGkb (https://www.pharmgkb.org/), and DisGeNET (https://www.disgenet.org) databases to obtain disease targets. The obtained targets were converted to standard gene names in UniProt database after removing duplicates. Potential anti-hepatic fibrosis targets of components migrating to blood were obtained based on components targets and disease targets. The bioinformatics platform (http://www.bioinformatics.com.cn/) was applied to visualize the results by outputting Venn diagram.
2.12 Screening of therapeutic targets
Venn diagrams were generated using the Bioinformatics Platform (http://www.bioinformatics.com.cn/) to identify overlapping targets between THSW Decoction blood components and hepatic fibrosis.
2.13 PPI network construction
Intersection targets were uploaded to STRING platform (https://string-db.org/) with the condition of “HOMO sapiens” and “confidence level ≥0.9”, and isolated nodes were excluded. Core targets were selected based on topological parameters (Betweenness, Closeness, Degree, Eigenvector, LAC) using Cytoscape 3.8.0.
2.14 GO and KEGG enrichment analysis
Functional enrichment analysis was performed using the DAVID database (https://davidbioinformatics.nih.gov/) for Gene Ontology (GO: biological processes, cellular components, molecular functions) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Results were visualized as bubble plots.
2.15 Non-targeted liver metabolomics
Liver tissue samples were processed by Meiji Biotech (Shanghai, China), Detailed methods can be found in the Supplementary Material.
2.16 “Drug-component-target-metabolite” network construction
The chemical structures of the common differential metabolites were retrieved from the PubChem database, and potential targets were predicted using the SwissTargetPrediction platform. Only targets with a probability score greater than 0 were included (Daina et al., 2019). Intersection targets between blood components and hepatic fibrosis targets were subjected to protein-protein interaction (PPI) analysis. A comprehensive network was subsequently constructed in Cytoscape to identify key bioactive components and metabolites.
2.17 Molecular docking
The protein structure of the targets were obtained from the PDB database (https://www.rcsb.org/), and the nuclear MOL2 files of the components were obtained from the PubChem (https://pubchem.ncbi.nlm.nih.gov). The ligand and receptor of the target protein were screened by the Pymol software. Molecular docking was performed using AutoDockTools 1.5.6 and AutoDockVina software, and the results were visualized by the Pymol software.
2.18 Molecular dynamics simulation
Molecular dynamics (MD) simulations were performed on the top-ranked target (with the highest binding free energy) and its corresponding blood-entering compound identified in Section 2.17. Simulations were conducted using the Amber 24 software package to evaluate the binding stability of the native protein and its mutant in complex with the same ligand (Case et al., 2005). System construction was carried out using the LEaP module by loading the PDB structures of both the wild-type and mutant proteins. The ff14SB force field was applied to describe the protein (Maier et al., 2015), while the General Amber Force Field (GAFF) was used to parameterize the ligand (Wang et al., 2004). Each system was solvated in a TIP3P water box with a 10.0 Å buffer from the protein surface, and neutralized by adding Na+ and Cl− ions (Zulfat et al., 2024).
To eliminate steric clashes and bad contacts in the initial structures, a two-step energy minimization was performed: first, the solvent and ions were minimized with restraints on the protein backbone, followed by full minimization of the entire system without restraints using the steepest descent and conjugate gradient methods to ensure system stability.
The equilibration phase included two steps. In the first step, the system was gradually heated to 300.0 K under constant temperature (NVT) conditions with restraints on the protein backbone. In the second step, the system was equilibrated under constant pressure (NPT, 1 bar) while maintaining the temperature at 300.0 K, to achieve proper system density and prepare a stable starting structure for production simulations.
The production phase consisted of a 1 ns MD simulation under NPT conditions without any restraints. The system was maintained at 300.0 K and 1 bar, and the trajectories were recorded for further analysis. Structural stability and dynamics were evaluated using CPPTRAJ (Roe and Cheatham, 2013), including analysis of root mean square deviation (RMSD), radius of gyration (Rg), solvent-accessible surface area (SASA), root mean square fluctuation (RMSF), and the number of hydrogen bonds. Results were visualized using Matplotlib to compare conformational differences in ligand binding between the wild-type and mutant proteins.
Furthermore, the binding free energy between protein and ligand was estimated using the MM/GBSA method (Genheden and Ryde, 2015). A total of 1,000 snapshots were extracted from the last 10 ns of a 100 ns MD trajectory. The van der Waals, electrostatic, polar, and non-polar solvation contributions were calculated, and total binding free energies were reported in kJ/mol.
2.19 Statistical analysis
Statistical analyses were performed using SPSS 21.0 software. Continuous data were analyzed by one-way ANOVA with LSD post-hoc test or Kruskal-Wallis test, as appropriate. A p-value <0.05 was considered statistically significant for all analyses.
3 Results
3.1 THSW decoction improves liver inflammation and fibrosis in mice
THSW Decoction significantly lowered serum ALT/AST levels, ameliorated hepatic collagen deposition (SR staining), and suppressed Col-1 expression in CCl4-induced mice, with the high-dose group exhibiting the most pronounced effects (Figure 1).
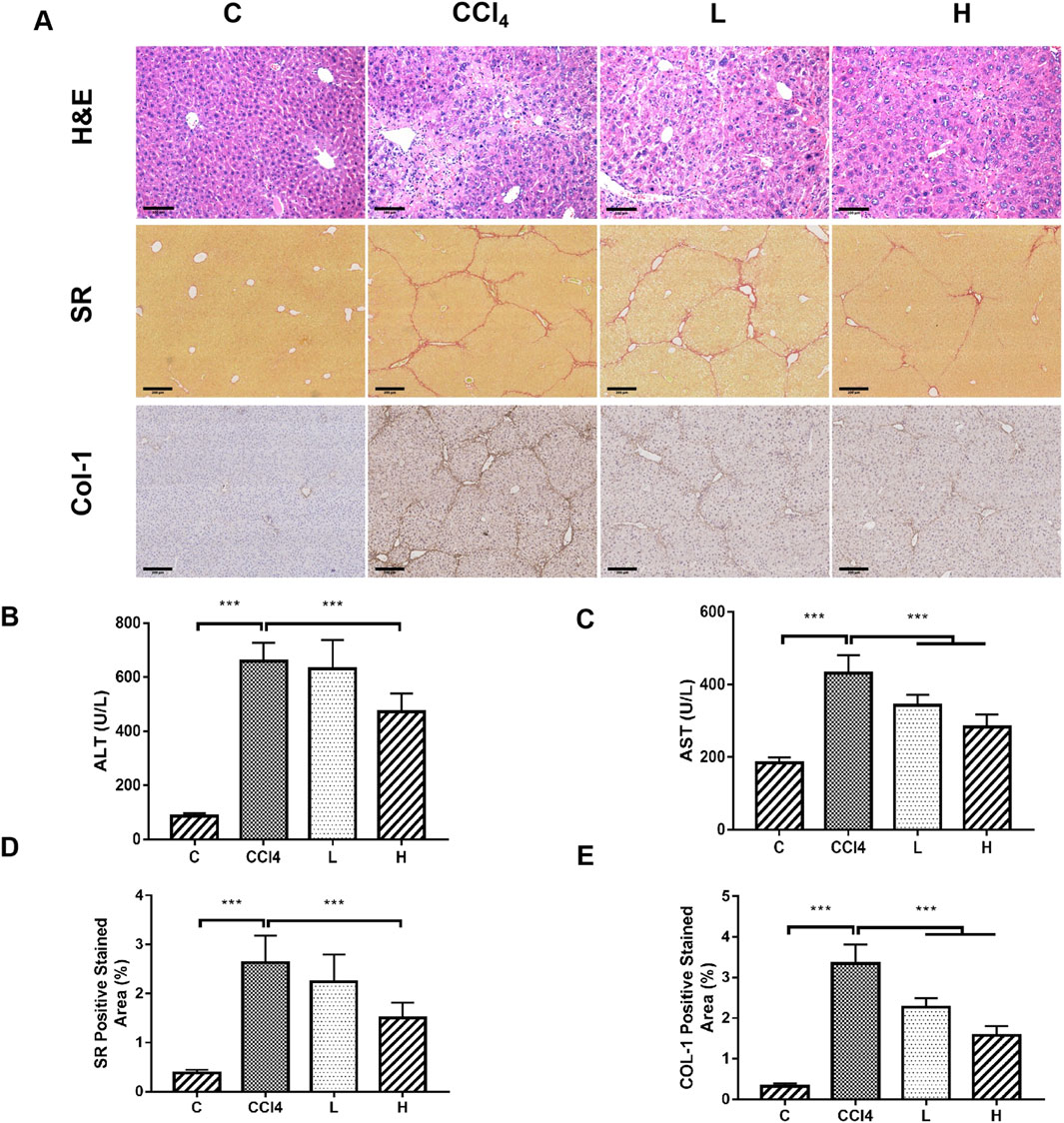
Figure 1. The ameliorative effect of THSW decoction on liver fibrosis in mice. (A): Histopathological analysis of liver tissues, including H&E staining (magnification: ×200), Sirius Red (SR) staining (magnification: ×100), and collagen type I (Col-1) immunostaining (magnification: ×100). (B): Serum alanine aminotransferase (ALT) levels. (C): Serum aspartate aminotransferase (AST) levels. (D): Semi-quantitative analysis of Sirius Red (SR) staining. (E): Semi-quantitative analysis of Col-1 staining.
3.2 Identification of components absorbed into blood from THSW decoction
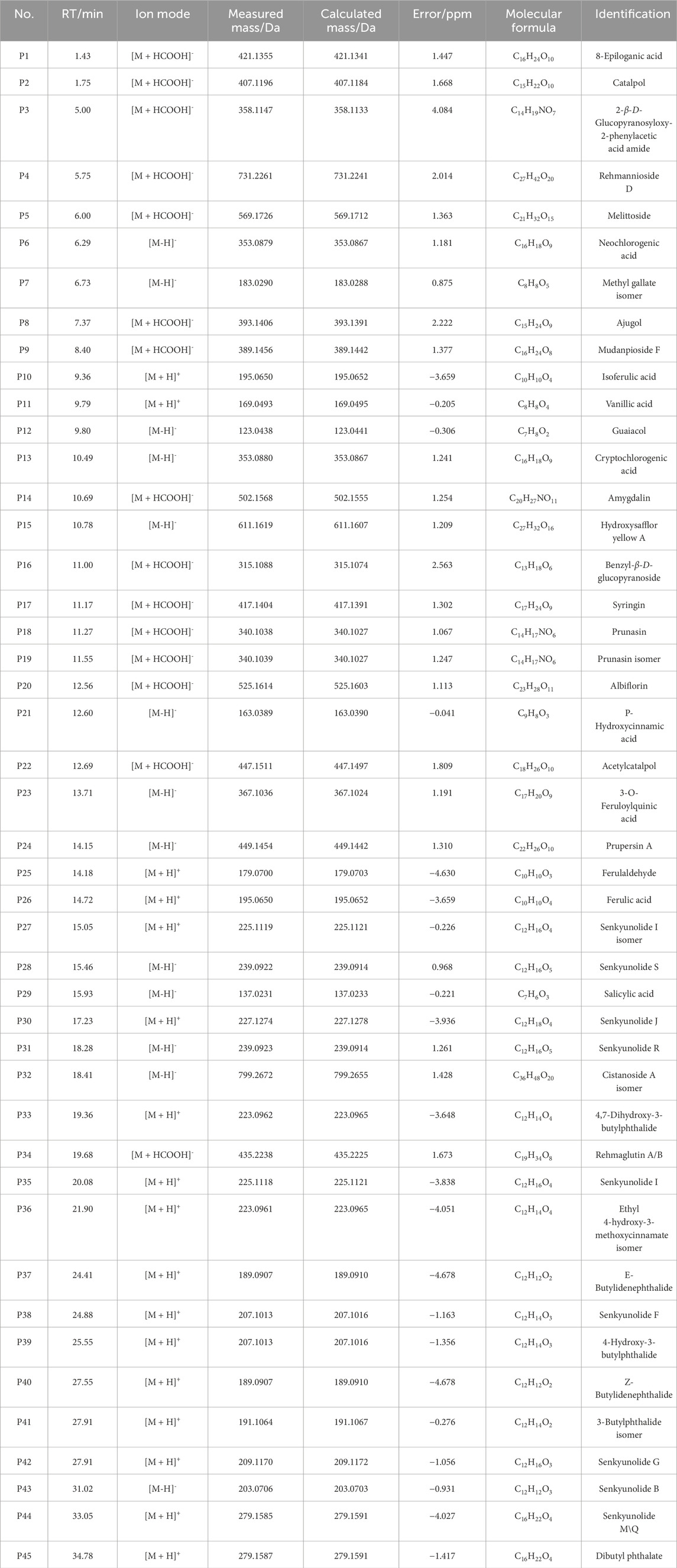
THSW Decoction and serum samples from mice after administration of THSW Decoction were collected for data collection and qualitative analyses. A total of 231 chemical components were identified from the formula, of which 45 prototype chemical components were identified in the serum of mice administered THSW Decoction, including 14 phthalides, 8 each of phenolic acids and phenylpropanoids, 6 cyclic enol ether terpenoids, 4 alkaloids, 3 terpenoids, and 1 each of flavonoids and glycosides (Table 1; Figure 2).
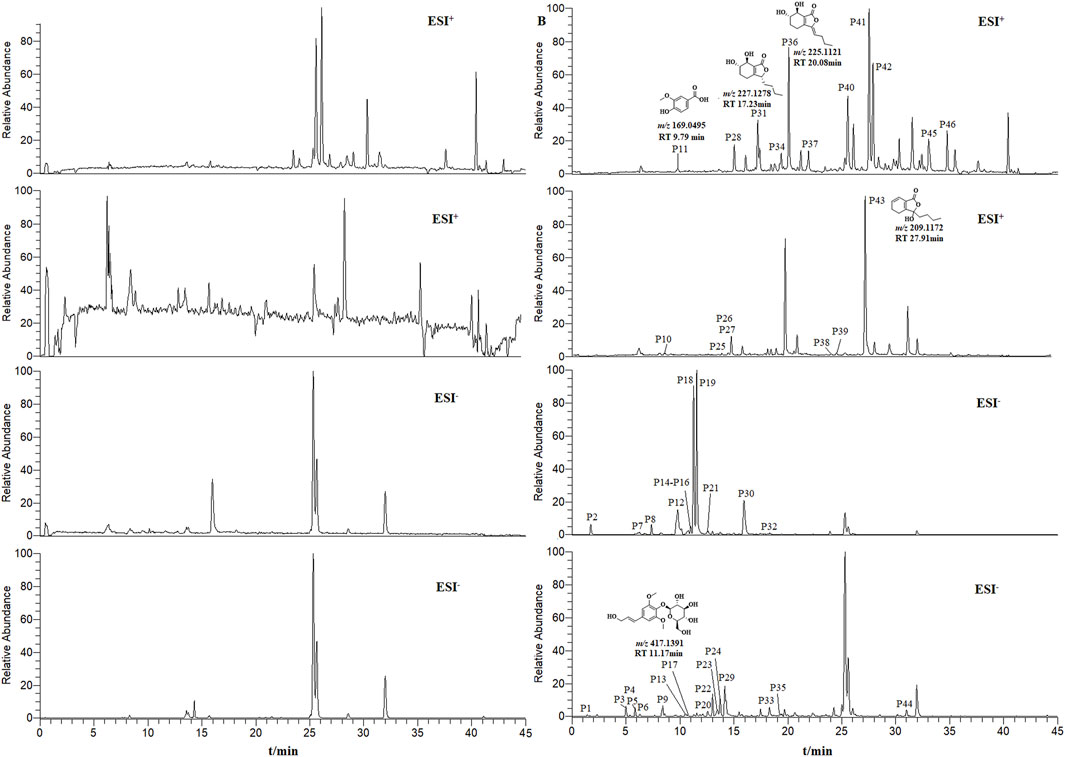
Figure 2. Extracted ion chromatograms (EICs) in positive and negative ionization modes from mouse serum samples collected before (left panels, blank serum) and after (right panels, post-administration serum) oral dosing of THSW Decoction. The displayed chromatograms correspond to the identified compounds: P11–Vanillic Acid; P17–Syringin; P31–Senkyunolide I; P36–Senkyunolide I; P43–Senkyunolide G.
3.3 Target screening for anti-hepatic fibrosis
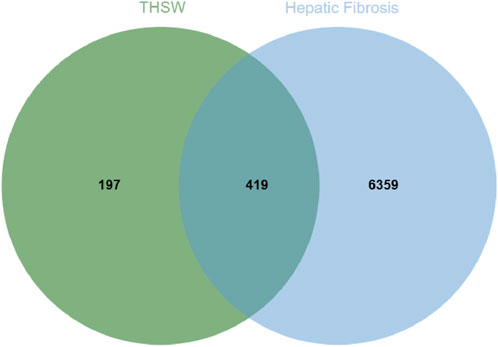
To explore the potential targets of blood-entering components in THSW Decoction, target prediction analyses were performed using the SEA and TCMSP databases. First, the SMILES structures of the identified blood-entering compounds (retrieved from PubChem) were input into the SEA database. Based on molecular structure similarity, a total of 561 candidate protein targets were predicted. Subsequently, potential targets were also retrieved from the TCMSP database under the criteria outlined in Section 2.11, yielding 221 targets. After integrating the results from both databases and removing duplicates and invalid entries, a total of 616 unique potential targets were identified (Supplementary Table S1). Meanwhile, 102, 6,512, 21, 63, 31, and 1,216 targets were obtained from Drugbank, GeneCards, TTD, OMIM, PharmGkb, and DisGeNET databases, respectively. A total of 6,778 targets were obtained after deleting duplicates (Supplementary Table S2). The 419 targets duplicated between disease targets and components targets were considered potential targets for THSW Decoction against hepatic fibrosis (Figure 3; Supplementary Table S2).
3.4 PPI network and core targets
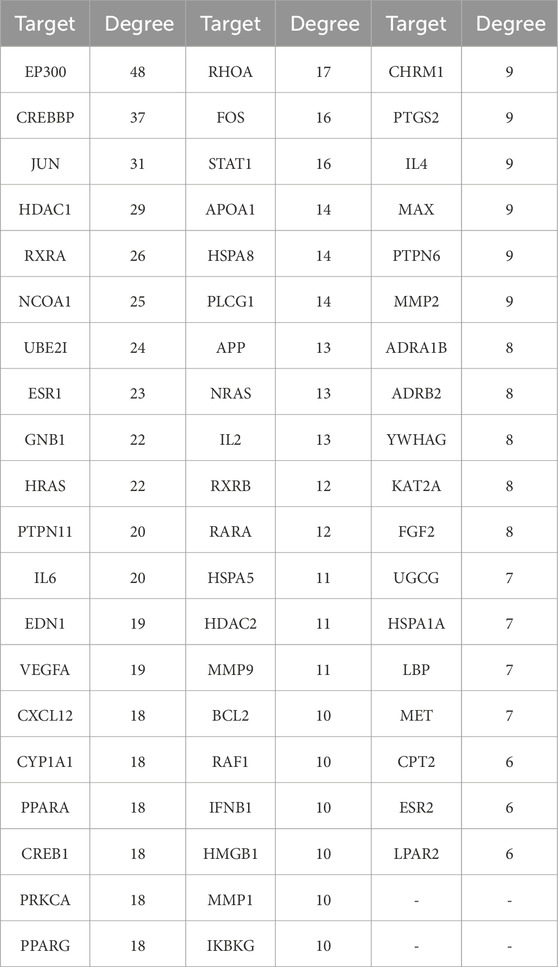
Uploaded 419 component-disease targets to the STRING database and set a minimum interaction score of 0.9. A total of 4,529 edges, 419 nodes, and 59 core targets were obtained (Figure 4; Table 2; Supplementary Table S3). Cytoscape software was used to build the PPI network and assessed the degree value of each node. The node size and color brightness reflected the degree value size. These proteins are known to play a crucial role in the PPI network, which are the key targets for the active components of THSW Decoction to regulate liver fibrosis.
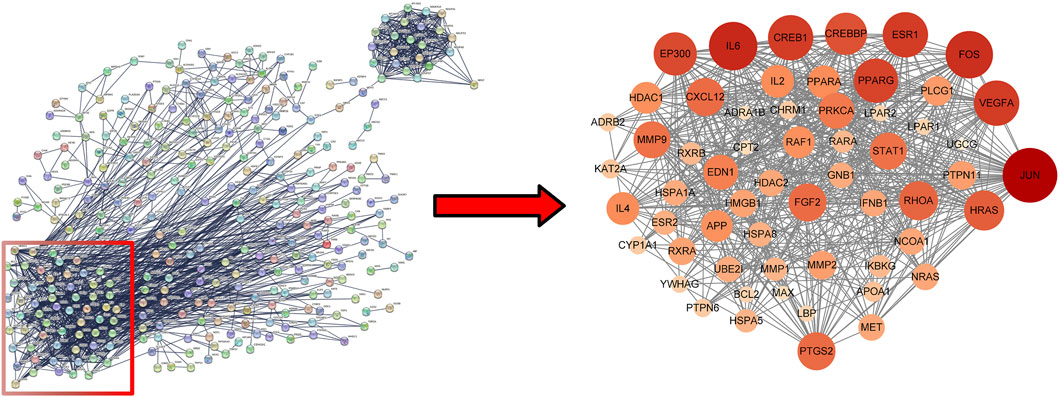
Figure 4. PPI network of core anti-hepatic fibrosis targets of components migrating to blood of THSW decoction.
3.5 GO and KEGG enrichment
GO enrichment analysis and KEGG pathway enrichment analysis were conducted on 59 core targets to further elucidate the mechanism of anti-hepatic fibrosis effect of THSW Decoction (Supplementary Table S4). A total of 1,518 BPs, 18 CCs, and 89 MFs were obtained from the GO functional annotations. BP mainly includes positive regulation of response to external stimulus, response to oxygen levels, epithelial cell migration, hormone-mediated signaling pathway, etc. CC mainly includes RNA polymerase II transcription regulator complex, transcriptional repressor complex, endoplasmic reticulum lumen, and vesicle lumen. MF mainly focuses on transcription coregulator binding, DNA-binding transcription factor binding, nuclear receptor activity, ligand-activated transcription factor activity (Figure 5A). The KEGG analysis indicated that the PI3K-Akt, estrogen, relaxin, and MAPK signaling pathways are likely the principal mechanisms through which THSW Decoction exerts its therapeutic effects on hepatic fibrosis (Figure 5B). Furthermore, the mechanism by which THSW Decoction regulates the PI3K-Akt signaling cascade in the context of anti-hepatic fibrosis was elucidated through visualization (Figure 5C).
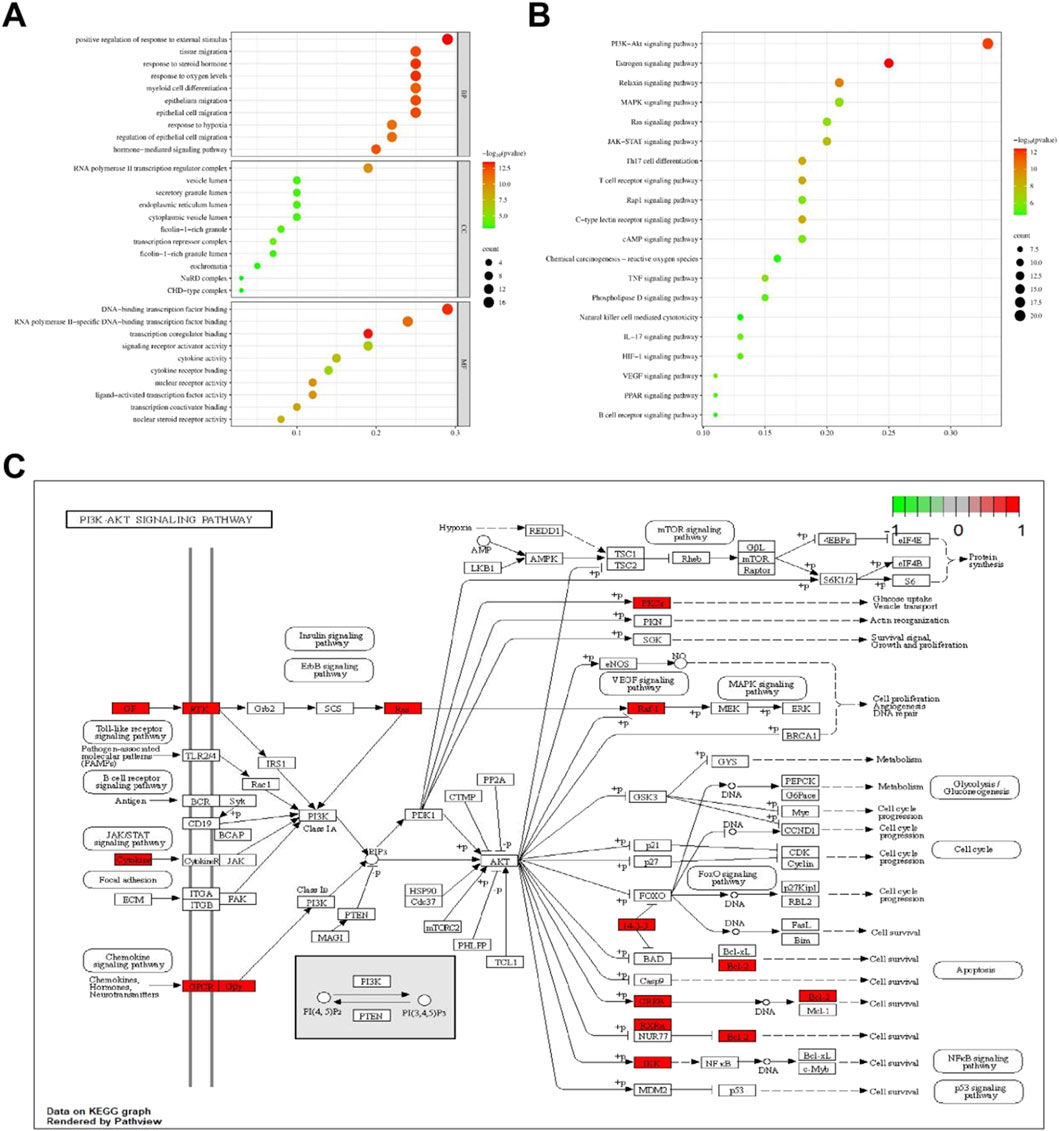
Figure 5. Enrichment analysis of core targets. (A), GO enrichment analysis. The size and color of the bubbles represent the number of targets enriched and the corrected P value, respectively. (B), KEGG signaling pathway enrichment analysis. (C), Key targets of PI3K/AKT signaling cascade. Targets in red color represents potential targets of THSW Decoction.
3.6 Non-targeted metabolomics
3.6.1 Multivariate statistical analysis
The results of 3.1 showed that the high-dose group of THSW Decoction had the most significant curative effect in regulating liver fibrosis. Therefore, in this study, the liver tissues of mice in the high-dose group were selected as samples for non-targeted metabolomics. Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were used for pattern recognition of the metabolic profile data of liver tissues, and sample distribution diagrams were obtained to reflect the similarities and differences among samples. The PCA results showed that there was an overlapping part between the normal group and the model group, while the THSW group was obviously separated from the model group. After pairwise comparison of the metabolites in the three groups of liver tissue samples by OPLS-DA, it was found that the discrimination degree among each group and the results of the permutation test were good (Figure 6).
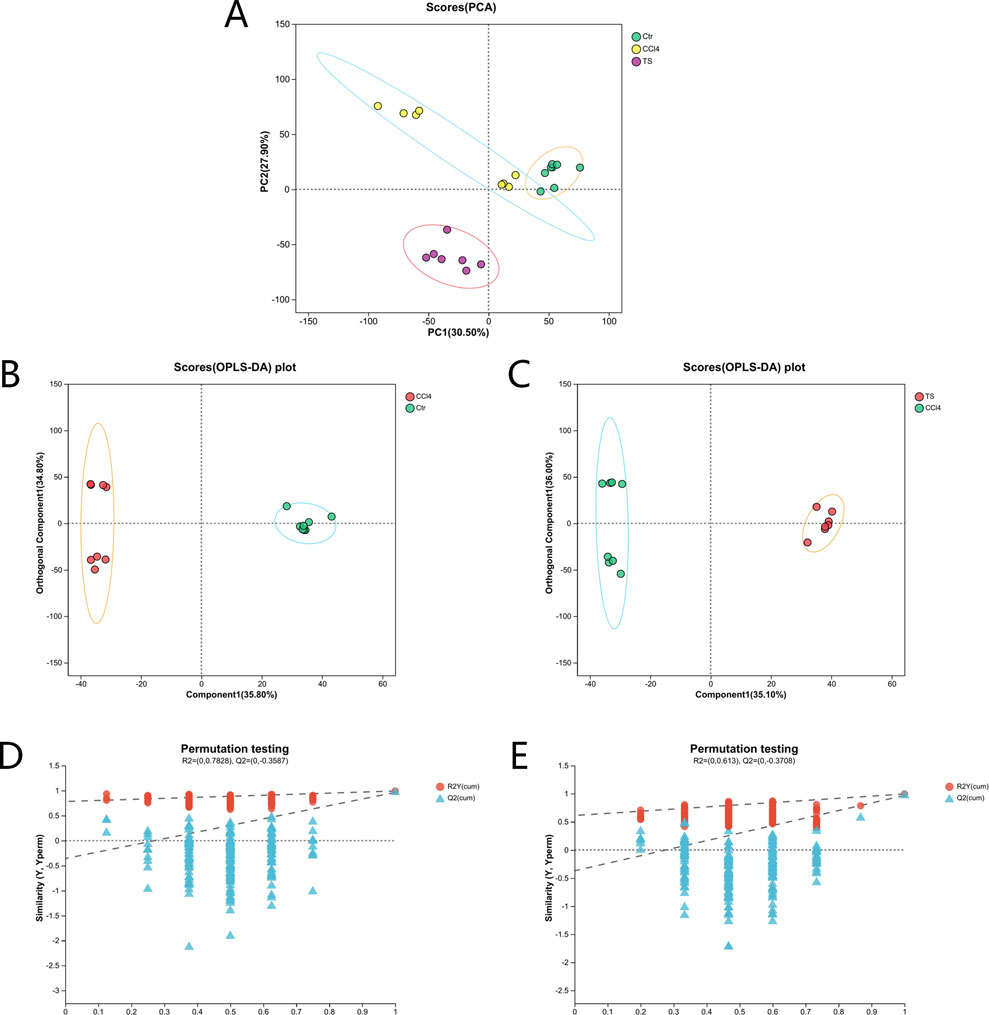
Figure 6. PCA and OPLS-DA analysis. (A) the PCA score plot. (B) the OPLS-DA score plot (normal group vs. model group). (C) the OPLS-DA score plot (model group vs. THSW group). (D) the permutation test plot of the OPLS-DA model (normal group vs. model group). (E) the permutation test plot of the OPLS-DA model (model group vs. THSW group).
3.6.2 Differential Metabolite Mining and Metabolic Pathway Analysis of Liver Tissue Samples
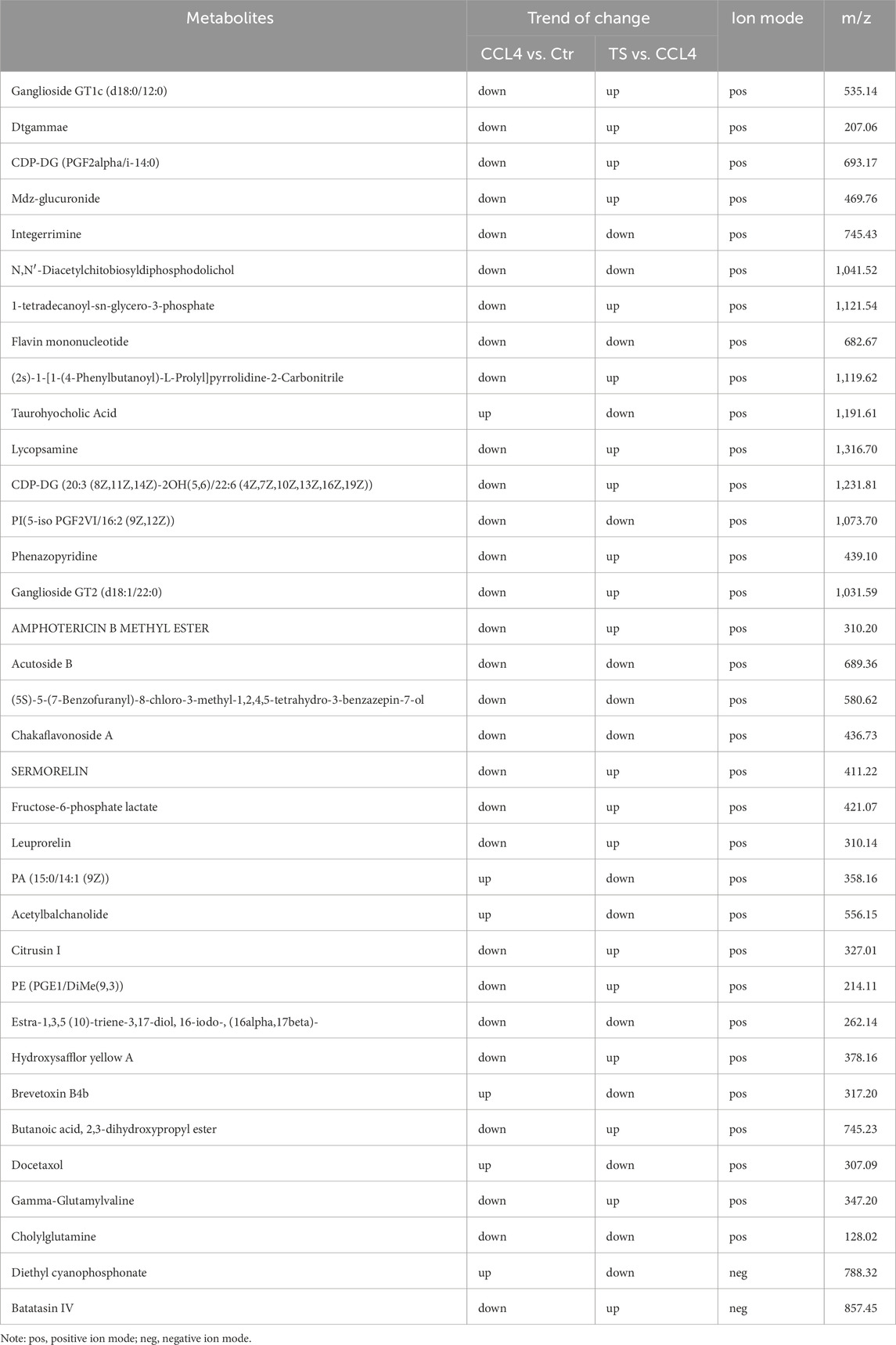
Differential metabolites among the three groups of liver tissue samples were mined through the t-test. With the screening criteria of VIP >1 and P < 0.05, the volcano plot showed the expression trends and quantities of differential metabolites in the comparisons between groups. Compared with the normal group, 148 metabolites in the mice of the model group changed, among which the levels of 9 metabolites were upregulated and those of 139 metabolites were downregulated. Compared with the model group, the levels of 56 metabolites in the THSW Decoction group were upregulated and those of 100 metabolites were downregulated (Figure 7). To clarify how THSW Decoction exerts its therapeutic effect by influencing the levels of metabolites, the common differential metabolites obtained after pairwise comparisons of the metabolites among the three groups were selected as the main biomarkers (Table 3). The screened biomarkers were analyzed by cluster heatmap, and the results indicated that the normal group, the model group, and the THSW Decoction group clustered into different categories respectively, and the change trends of the normal group and the THSW Decoction group were basically consistent, which were opposite to those of the model group (Figure 7), suggesting that THSW Decoction may exert its pharmacological effects by regulating the biomarkers related to liver fibrosis.
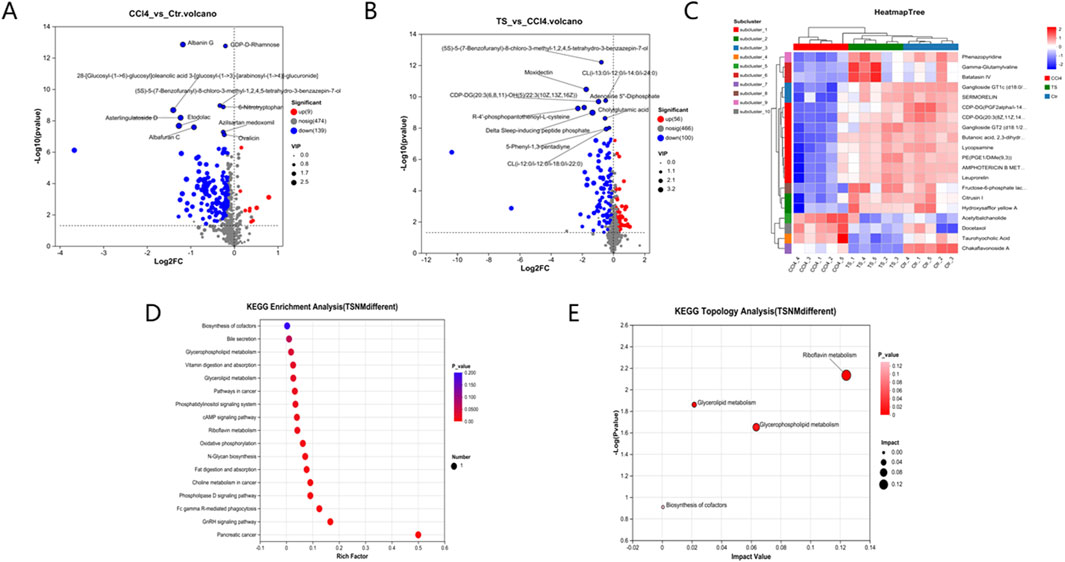
Figure 7. Differential Metabolite Mining and Metabolic Pathway Analysis of Liver Tissue Samples. (A,B), the volcano plot analyses of differential metabolites. (C), the cluster heatmap of biomarkers. (D), Kegg pathway analysis. (E), Pathway topological analysis.
Subsequently, KEGG metabolic pathway analysis and topological analysis (used to identify the principal metabolic pathways associated with the target metabolite set) were performed on the biomarkers listed in Table 3, as illustrated in Figure 7. The results demonstrated that the biomarkers were enriched in key pathways including cAMP, phospholipase D, and GnRH signaling pathways. Topological analysis further identified riboflavin metabolism as the most significant pathway.
3.7 Construction of the “drug-component-target-metabolite” network
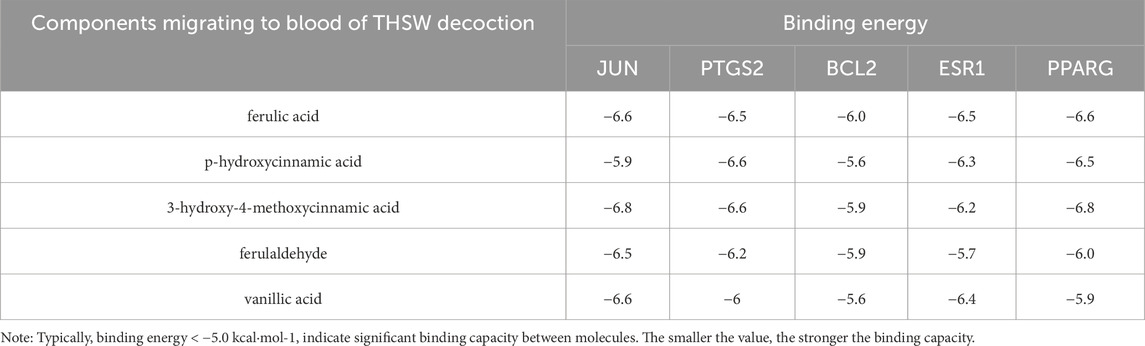
A total of 35 chemical structures of common differential metabolites were retrieved from the PubChem database. Using the SwissTargetPrediction platform, we predicted 592 potential targets (Supplementary Table S5). After intersecting these with the targets of blood-absorbed components and liver fibrosis-related targets, we identified 20 overlapping targets (Table 4). These 20 intersection targets were then uploaded to the STRING database with an interaction score threshold set at 0.4, yielding a network with 70 edges and 18 nodes. This network was subsequently imported into Cytoscape 3.8.0 for topological analysis. The top five targets identified were JUN, PTGS2, BCL2, ESR1, and PPARG. From the “Drug-Component-Target-Metabolite” network, the highest-ranking blood-absorbed components were ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid (Figure 8).
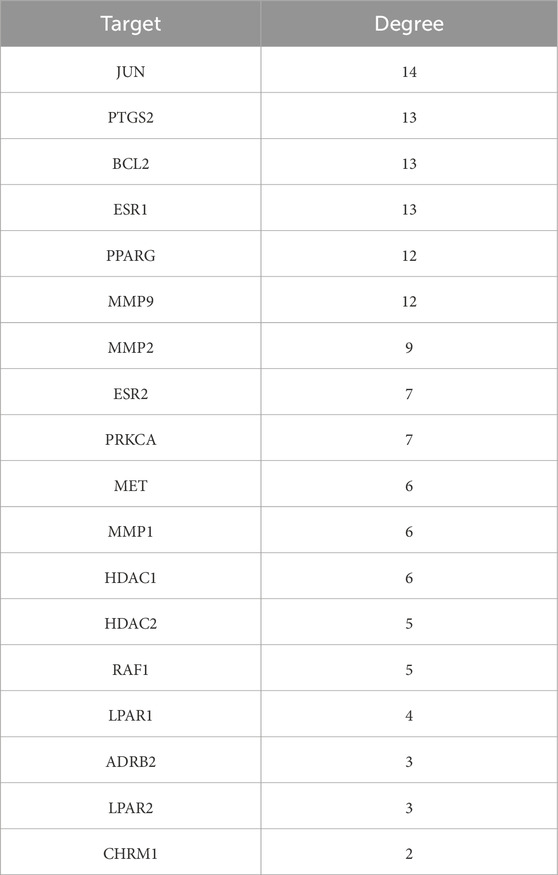
Table 4. Intersection targets of THSW decoction components in blood-liver fibrosis-differential metabolite pathways.
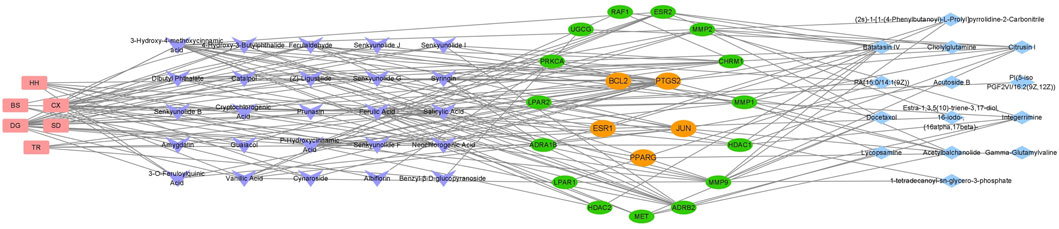
Figure 8. The “Drug-Component-Target-Metabolite” Network. Red nodes represent traditional Chinese medicine compounds, purple nodes indicate blood-absorbed components, while green and yellow nodes denote intersecting targets (with yellow highlighting core targets). Blue nodes represent differential metabolites.
3.8 Molecular docking
Molecular docking was performed between the top five target genes (JUN, PTGS2, BCL2, ESR1, and PPARG) and the top five core blood-entering components (ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid), and the results were visualized (Table 5; Figure 9) to evaluate the binding affinities between target proteins and ligand molecules. Generally, a binding energy less than–5.0 kcal mol-1 indicates a significant interaction between the molecules. The results demonstrated that the core blood-entering components of THSW Decoction exhibited favorable binding affinities with the core targets.
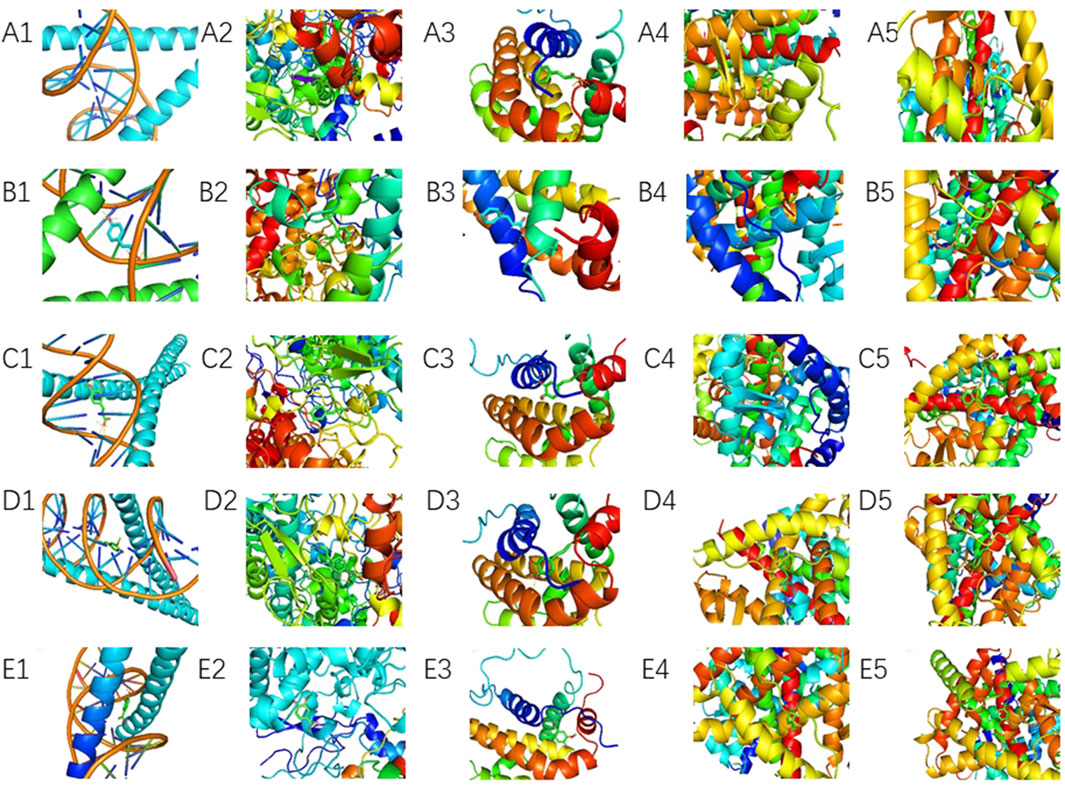
Figure 9. Molecular Docking Verification of the Core Components Entering the Blood of THSW Decoction and the Core Targets. (A1–5), ferulic acid with JUN, PTGS2, BCL2, ESR1, PPARG. (B1–5), p-hydroxycinnamic acid with JUN, PTGS2, BCL2, ESR1, PPARG. (C1–5), 3-hydroxy-4-methoxycinnamic acid with JUN, PTGS2, BCL2, ESR1, PPARG. (D1–5), ferulaldehyde with JUN, PTGS2, BCL2, ESR1, PPARG. (E1–5), vanillic acid with JUN, PTGS2, BCL2, ESR1, PPARG.
3.9 Molecular dynamics simulation
To comprehensively evaluate the binding capacity of 3-hydroxy-4-methoxycinnamic acid with PPARG and compare it with the original ligand, a systematic analysis was conducted based on hydrogen bonding interactions, conformational stability (RMSD, RMSF, Rg), surface properties (SASA), and binding free energy (MM/GBSA).
3.9.1 Hydrogen Bond Analysis
Hydrogen bonding, as a crucial non-covalent interaction, significantly stabilizes protein-ligand complexes through directional electrostatic attractions. During the 100 ns molecular dynamics simulation, the number of hydrogen bonds formed between the ligands and PPARG was monitored. The PPARG–3-hydroxy-4-methoxycinnamic acid complex consistently maintained 2–3 hydrogen bonds with minimal fluctuation, indicating relatively stable hydrophilic interactions. In comparison, the PPARG–original ligand complex formed 4–5 hydrogen bonds, which may be attributed to the presence of more polar groups in the original ligand. These findings suggest that both ligands can form stable hydrogen bond networks at the binding site (Figure 10A).
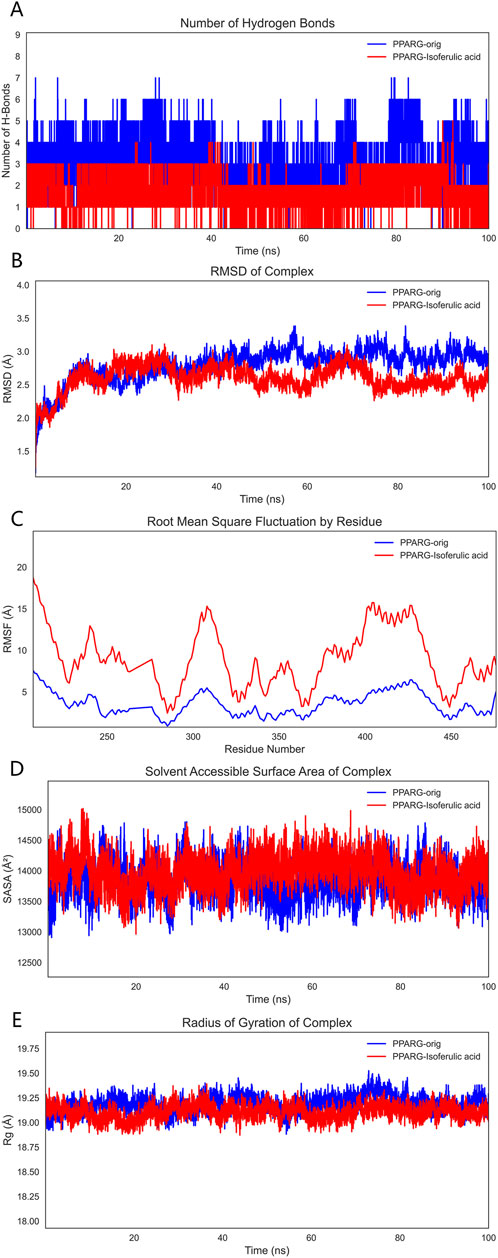
Figure 10. Molecular dynamics simulation results. Note: (A), Hydrogen bond analysis; (B), RMSD analysis; (C), RMSF analysis; (D), SASA analysis; (E), Radius of gyration (Rg) analysis.
3.9.2 RMSD analysis
RMSD (Root Mean Square Deviation) was employed to assess the overall conformational stability of the protein–ligand complexes. The RMSD trajectories indicated that both complexes maintained good stability throughout the simulation, with fluctuations remaining within 2 Å and no significant drift observed. This suggests that both systems were thermodynamically stable during the simulation period and were thus suitable for further binding energy analysis (Figure 10B).
3.9.3 RMSF analysis
RMSF (Root Mean Square Fluctuation) was used to evaluate the local flexibility of PPARG residues. The binding site residues in the PPARG–3-hydroxy-4-methoxycinnamic acid complex displayed moderate fluctuations around 10 Å, whereas the corresponding residues in the PPARG–original ligand complex exhibited lower RMSF values, indicating a stronger local stabilizing effect by the original ligand. Low flexibility at the binding site is typically associated with tighter molecular recognition, suggesting the original ligand may better stabilize the functional conformation of the protein (Figure 10C).
3.9.4 SASA analysis
SASA (Solvent Accessible Surface Area) reflects the degree of exposure of the protein surface to the solvent and is a key parameter for assessing conformational changes and hydrophilic/hydrophobic surface exposure. The SASA curve of the PPARG–3-hydroxy-4-methoxycinnamic acid complex showed a slight upward trend during the simulation, likely due to increased exposure of polar groups. In contrast, the PPARG–original ligand complex displayed more stable SASA values, suggesting a more compact structure with better shielding of hydrophobic regions. Overall, both complexes exhibited stable SASA behavior, although 3-hydroxy-4-methoxycinnamic acid may slightly increase solvent exposure, potentially affecting solubility or bioactivity (Figure 10D).
3.9.5 Rg analysis
The radius of gyration (Rg) was analyzed to reflect the compactness of the protein structure. Both PPARG–ligand complexes demonstrated stable Rg values with minimal fluctuation, indicating a tight and well-folded protein conformation that supports functional stability (Figure 10E).
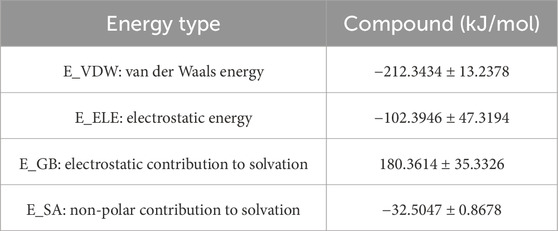
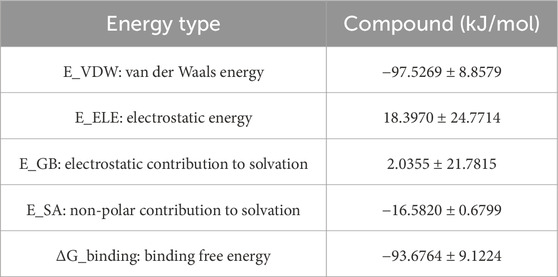
3.9.6 MM/GBSA binding free energy analysis
To quantitatively evaluate ligand–PPARG binding affinities, MM/GBSA calculations were performed using 100 frames extracted from the last 1 ns of the 100 ns MD trajectory. The binding free energy (ΔG_binding) and its components are summarized, respectively. As shown in Table 6, the PPARG–original ligand complex exhibited a binding free energy of −166.89 ± 17.86 kJ/mol, primarily driven by van der Waals interactions (−212.34 kJ/mol) and electrostatic contributions (−102.39 kJ/mol). These results indicate a strong binding affinity, largely governed by favorable non-covalent interactions. In contrast, Table 7 presents the energy components for the PPARG–3-hydroxy-4-methoxycinnamic acid complex. This complex displayed a higher ΔG_binding of −93.68 ± 9.12 kJ/mol. Notably, the electrostatic interaction term was positive (+18.40 kJ/mol), suggesting relatively unfavorable polar interactions compared to the original ligand. The reduced van der Waals contribution (−97.53 kJ/mol) also reflects a weaker overall binding affinity.
3.10 In Vivo target validation
Finally, animal experiments were conducted to verify the anti-liver fibrosis mechanism of THSW Decoction. The results further confirmed the effect of the high dose of THSW Decoction significantly increased JUN expression in the liver tissues of mice with liver fibrosis and decreased ESR1 expression (Figure 11).
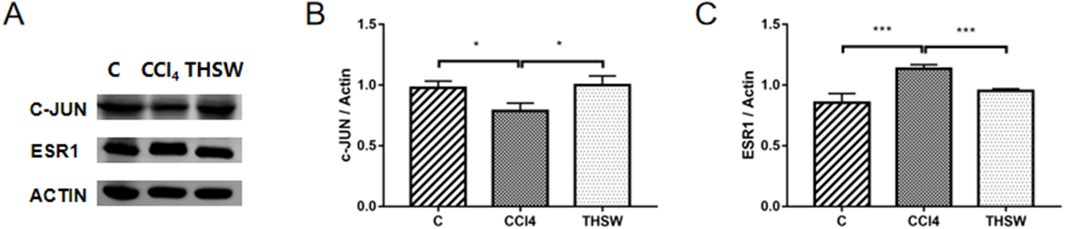
Figure 11. Effects of THSW Decoction on the Core Liver Targets (JUN, ESR1) in Mice with Liver Fibrosis. (A), Protein expressions of c-jun and ESR1 in the liver. (B,C), Semi-quantification of the protein expressions of c-jun and ESR1 in the liver. *P < 0.05, ***P < 0.001.
4 Discussion
Hepatic fibrosis is characterized by excessive deposition of collagen and other extracellular matrix (ECM) components such as fibronectin and proteoglycans. Various factors - including viral infections, alcohol consumption, obesity, and autoimmune disorders-can trigger hepatic fibrosis. These agents act through multiple pathways, including promoting liver inflammation, oxidative stress, and apoptosis (Altamirano-Barrera et al., 2017). While these pathogenic factors are well-established, therapeutic options remain limited. Interestingly, traditional Chinese medicine formulations such as THSW Decoction, have shown promise in fibrosis treatment (Xi et al., 2016), but its pharmacologically active components and precise mechanisms of action remain incompletely understood.
In this study, THSW Decoction’s efficacy in improving hepatic fibrosis was first evaluated by constructing a murine model of hepatic fibrosis induced by carbon tetrachloride. The results demonstrated that THSW Decoction significantly reduced serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in CCl4-induced hepatic fibrosis mice, ameliorated collagen deposition in liver tissues, and decreased Col-1 expression, indicating its effectiveness in alleviating liver inflammation and fibrosis. To explore its potential active components and mechanism of action, this study further employed UHPLC-Q-Exactive Orbitrap HRMS technology to analyze and identify the main prototype chemical components of THSW Decoction in the blood. A total of 45 blood components of THSW Decoction were detected. Database searches yielded 616 drug targets corresponding to 43 blood components, among which 419 were identified as potential anti-hepatic fibrosis targets. Through GO and KEGG enrichment analyses, it was revealed that the mechanism of THSW Decoction in treating liver fibrosis involves multiple aspects. In the GO enrichment analysis, the top-ranked functions include response to oxygen levels, nuclear receptor activity, ligand-activated transcription factor activity, etc. Cells need continuous oxygen supply to power energy production through mitochondrial respiration. Hypoxia can activate hypoxia - inducible factors (HIFs), which then affect the respiratory chain function and cause an increase in reactive oxygen species (ROS). Under persistent hypoxia, HIFs coordinate interactions among hepatocyte populations, promote pro - angiogenic mediator release, and drive inflammatory and fibrotic responses (Foglia et al., 2021). As ligand-regulated transcription factors, nuclear receptors control inflammation. They also mediate hepatic stellate cell (HSC) activation (Tran et al., 2018; Rudraiah et al., 2016). Building on these molecular mechanisms, our KEGG enrichment analysis reveals that THSW Decoction treats liver fibrosis mainly involves the Phosphatidylinositol-3-Kinase/Protein Kinase B(PI3K-Akt) pathway, along with estrogen, relaxin, and Mitogen-Activated Protein Kinase (MAPK) signaling pathways. The PI3K-Akt pathway promotes liver fibrosis through three key mechanisms: stimulating ECM production, activating hepatic stellate cells (HSCs), and inducing hepatic sinusoidal capillarization (Wang M. et al., 2022; Wang et al., 2019). Estrogen inhibits the activity of Kupffer cells and reduces the production of ROS via estrogen receptor α (ERα/ESR1), exerting a protective effect on various liver diseases (Kovats, 2015; Xu et al., 2022). Relaxin signals via its cognate receptor RXFP1, a G protein - coupled receptor, to: (1) inhibit collagen synthesis/deposition, (2) suppress HSC activation, and (3) enhance ECM degradation while stimulating angiogenesis (Samuel et al., 2017; Kaftanovskaya et al., 2019). Three MAPK subfamilies, namely, p38 MAPK, JNK, and ERK1/2-collectively drive HSCs activation and fibrogenesis (Jin et al., 2022; Zhang et al., 2021).
Building on network pharmacology predictions, we employed non-targeted metabolomics using a Synapt G2-Si QTOF HRMS system to correlate predicted targets and pathways with actual metabolic changes observed in vivo, systematically elucidating the overall regulatory mechanisms of traditional Chinese medicine treatment. Non-targeted metabolomics revealed 148 and 156 differential metabolites when comparing model vs. normal groups and THSW Decoction vs. model groups, respectively. Among them, 35 common differential metabolites were identified, and these were enriched in signaling pathways such as cAMP, Phospholipase D, and gonadotropin-releasing hormone (GnRH). The cAMP signaling pathway regulates liver metabolism through gene transcription and kinase activity modulation. As demonstrated previously (Cheng et al., 2008; Wahlang et al., 2018), cAMP regulates the expression of protein kinase A (PKA) and cAMP sensor family proteins (EPAC), inhibits the activation and proliferation of HSCs, regulates the synthesis of the ECM, and suppresses the inflammatory response to alleviate the degree of liver fibrosis. These findings experimentally validate the PI3K-Akt pathway predictions from our network pharmacology analysis. Based on different cellular localizations and functions, Phospholipase D can be divided into two subtypes: PLD1 and PLD2. Studies demonstrate that the expression level of PLD1 typically increases with HSCs activation, while PLD2 remains relatively unchanged (Chang et al., 2020). Targeting PLD1 can inhibit liver fibrosis. Phosphatidic acid (PA), a key PLD pathway metabolite, activates the extracellular signal-regulated kinase (ERK) pathway. This regulates DNA synthesis while influencing HSCs activation and proliferation (Benitez-Rajal et al., 2006). PA also binds to peroxisome proliferator-activated receptor alpha (PPARα), modulating lipid metabolism (Yao et al., 2024). THSW Decoction downregulates the level of PA. This suggests THSW Decoction may ameliorate liver fibrosis through a metabolism-transcription coupling mechanism. Research indicates that GnRH binds to HSCs surface receptors and regulates miR-200b expression, potentially participating in HSCs activation through metabolic reprogramming (Kyritsi et al., 2017; Xie et al., 2023).
This study investigated how THSW Decoction combats liver fibrosis by combining network pharmacology and metabolomics approaches. We identified 20 overlapping targets connecting blood - absorbed components, metabolites, and liver fibrosis. PPI analysis revealed five key targets:JUN, PTGS2, BCL2, ESR1, and PPARG, indicating THSW Decoction’s multi-target anti-fibrotic mechanism. As a key downstream target of the mitogen-activated protein kinase (MAPK) pathway, JUN phosphorylation promotes HSCs proliferation and the release of inflammatory factors. THSW Decoction significantly downregulated JUN expression, potentially inhibiting HSC activation through MAPK-JUN cascade blockade (Kluwe et al., 2010; Win et al., 2018). PTGS2 drives liver fibrosis via three key mechanisms: under the induction of inflammatory factors, PTGS2 catalyzes PGE2 production, exacerbating liver inflammation (Takai et al., 2013; Rodriguez-Barbero et al., 2006). Additionally, it participates in HSCs activation by inhibiting autophagy-related proteins and upregulating α-SMA expression (Yang et al., 2020). The anti-apoptotic protein BCL2 regulates cytochrome C release from mitochondria by binding to BCL2-associated agonist of cell death (BAD), thus modulating cell apoptosis. THSW Decoction demonstrates anti-fibrotic effects by regulating BCL-2 expression to promote HSCs apoptosis, consistent with previous research (Meng et al., 2018). The functions of ESR1 were previously discussed. It inhibits liver fibrosis in both basic and clinical research (Xu et al., 2004; Cengiz et al., 2014). PPAR (peroxisome proliferator-activated receptor) serves as a potential liver-fibrosis treatment target. It alleviates fibrosis by suppressing HSCs activation, modulating inflammation/oxidative stress, improving lipid metabolism, and regulating immune responses. Clinical studies have validated PPAR agonists’ efficacy (Li et al., 2021; Liu et al., 2020).
Through network pharmacology analysis establishing a “drug-component-target-metabolite” interaction map, five components (ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid) showed high degree values among detected blood-entering components, while differential metabolites such as Batatasin IV, Citrusin I, Acetylbalchanolide, and Integerrimine also exhibited high degree values, indicating that these compounds and metabolites may serve as key bioactive substances in the therapeutic effects of THSW Decoction against liver fibrosis. ferulic acid, a derivative of cinnamic acid, has been demonstrated to possess anti-fibrotic properties in various liver fibrosis models (Shi et al., 2023). It may alleviate the intrahepatic inflammatory microenvironment by targeting PTGS2 to inhibit COX-2 expression (Gollapalli et al., 2024), and may also regulate fibrosis-related gene expression via PPARG (Cui et al., 2022; Ni et al., 2021). p-hydroxycinnamic acid, a natural phenolic compound, was reported to suppress the TLR4/NF-κB signaling pathway, reduce IL-1β precursor expression, and attenuate NLRP3 inflammasome activation in a mouse model of high-fat, high-sucrose diet-induced liver fibrosis, thereby inhibiting HSCs activation (Truong et al., 2023). 3-hydroxy-4-methoxycinnamic acid (Su et al., 2015) and ferulaldehyde (Radnai et al., 2009) also exhibit potent anti-inflammatory and antioxidant activities, although their specific roles in liver fibrosis require further investigation. In a CCl4-induced liver injury model, vanillic acid was found to significantly inhibit HSCs activation and collagen accumulation, demonstrating anti-fibrotic potential (Itoh et al., 2010), while also can regulate BCL2 expression to suppress apoptosis (Punvittayagul et al., 2021). Among the differential metabolites, Batatasin IV and Citrusin I were upregulated in the THSW Decoction group, suggesting that the THSW Decoction may promote the biosynthesis of metabolites with anti-inflammatory and antioxidant properties by modulating relevant pathways (Yang et al., 2009; Noh et al., 2015). Integerrimine, a pyrrolizidine alkaloid (PA) with known hepatotoxicity and diverse metabolic routes, was downregulated, indicating that THSW Decoction might suppress toxic PA bioactivation, helping to mitigate liver injury while maintaining anti-fibrotic efficacy (Wang Z. et al., 2022). Acetylbalchanolide, a sesquiterpene lactone, remains poorly studied and merits further investigation. Interestingly, beyond the top-ranked metabolites, PA (15:0/14:1 (9Z)) was enriched across multiple pathways and downregulated in the THSW Decoction group, potentially due to phospholipase D pathway inhibition. This suggests THSW Decoction may help restore phospholipid metabolic balance and regulate intercellular signal transduction (Sarkar and Chattopadhyay, 2021). Overall, the “drug–component–target–metabolite” network highlights that THSW Decoction may exert its anti-fibrotic effects through the synergistic action of multiple bioactive compounds. These compounds coordinately modulate key molecular targets involved in the pathogenesis of liver fibrosis, particularly pathways related to inflammation and oxidative stress. Consequently, THSW Decoction may influence critical metabolic pathways and associated metabolite levels, ultimately contributing to a multifaceted therapeutic intervention against liver fibrosis.
To validate the relationship between the main components of THSW Decoction and its core targets, molecular docking was used to predict their binding affinities. Ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid showed strong binding capacities with JUN, PTGS2, BCL2, ESR1, and PPARG. These findings suggest the primary bioactive components of THSW Decoction (ferulic acid, p-hydroxycinnamic acid, 3-hydroxy-4-methoxycinnamic acid, ferulaldehyde, and vanillic acid) may inhibit liver fibrosis by modulating JUN, PTGS2, BCL2, ESR1, and PPARG. This modulation could exert anti-inflammatory and anti-oxidative effects while regulating ECM production and HSCs activation. In the molecular dynamics simulations, the original ligand exhibited stronger binding affinity and a more stable complex structure. Nevertheless, 3-hydroxy-4-methoxycinnamic acid also maintained a stable conformation throughout the 100 ns simulation (RMSD <2 Å), established a consistent hydrogen bond network, and showed a favorable binding free energy (−93.68 kJ/mol), suggesting its ability to form a thermodynamically favorable complex with PPARG. Taken together, these results indicate that 3-hydroxy-4-methoxycinnamic acid, as a natural compound, demonstrates appreciable binding potential toward PPARG, providing theoretical support for its candidacy as a PPARG modulator. In vivo experiments showed that THSW Decoction significantly upregulated JUN expression and downregulated ESR1, which was consistent with the network prediction results. However, the direct interaction between these blood-entering components and their predicted targets requires verification, and this will be addressed in future work.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study protocol was approved by the Animal Care and Use Committee of Experimental Animal Welfare and Ethics Committee of Henan University of Chinese Medicine (No. IACUC-20230309-02). Appropriate measures were taken to minimize pain and distress during the experiment. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SL: Writing – original draft, Formal Analysis, Visualization, Methodology. SQ: Writing – review and editing, Data curation, Validation. RL: Writing – review and editing, Validation. FY: Writing – review and editing, Funding acquisition, Data curation. ZN: Methodology, Investigation, Writing – review and editing. WL: Investigation, Writing – review and editing, Writing – original draft, Methodology. SM: Funding acquisition, Writing – review and editing. ZX: Methodology, Writing – review and editing, Investigation, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work is supported by the National Natural Science Foundation of China (No. 82104772, 82205208), China Postdoctoral Science Foundation (No. 2022M721067), The Science and Technology Research Program of Henan Province (No. 222102310502), and Traditional Chinese Medicine Science Research Project of Henan Province (No. 2022ZY 2008).
Acknowledgments
The authors would like to thank Dr. Dingqi Zhang for his valuable contributions to the mass spectrometry analysis in this study. His technical support and expertise were essential for the successful completion of the metabolomics experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2025.1614341/full#supplementary-material
References
Akkiz, H., Gieseler, R. K., and Canbay, A. (2024). Liver fibrosis: from basic science towards clinical progress, focusing on the central role of hepatic stellate cells. Int. J. Mol. Sci. 25 (14), 7873. doi:10.3390/ijms25147873
Altamirano-Barrera, A., Barranco-Fragoso, B., and Mendez-Sanchez, N. (2017). Management strategies for liver fibrosis. Ann. Hepatol. 16 (1), 48–56. doi:10.5604/16652681.1226814
Benitez-Rajal, J., Lorite, M. J., Burt, A. D., Day, C. P., and Thompson, M. G. (2006). Phospholipase D and extracellular signal-regulated kinase in hepatic stellate cells: effects of platelet-derived growth factor and extracellular nucleotides. Am. J. Physiol. Gastrointest. Liver Physiol. 291 (5), G977–G986. doi:10.1152/ajpgi.00041.2006
Caligiuri, A., Gentilini, A., Pastore, M., Gitto, S., and Marra, F. (2021). Cellular and molecular mechanisms underlying liver fibrosis regression. Cells 10 (10), 2759. doi:10.3390/cells10102759
Case, D. A., Cheatham, T. R., Darden, T., Gohlke, H., Luo, R., Merz, K. J., et al. (2005). The Amber biomolecular simulation programs. J. Comput. Chem. 26 (16), 1668–1688. doi:10.1002/jcc.20290
Cengiz, M., Ozenirler, S., and Yilmaz, G. (2014). Estrogen receptor alpha expression and liver fibrosis in chronic hepatitis C virus genotype 1b: a clinicopathological study. Hepat. Mon. 14 (9), e21885. doi:10.5812/hepatmon.21885
Chang, Y., Xia, L., Song, M., Tang, M., Patpur, B. K., Li, J., et al. (2020). The in vitro effects of phospholipase D1-mTOR axis in liver fibrogenesis. Life Sci. 251, 117595. doi:10.1016/j.lfs.2020.117595
Cheng, X., Ji, Z., Tsalkova, T., and Mei, F. (2008). Epac and PKA: a tale of two intracellular cAMP receptors. Acta Biochim. Biophys. Sin. (Shanghai) 40 (7), 651–662. doi:10.1111/j.1745-7270.2008.00438.x
Cui, K., Zhang, L., La, X., Wu, H., Yang, R., Li, H., et al. (2022). Ferulic acid and p-coumaric acid synergistically attenuate non-alcoholic fatty liver disease through HDAC1/PPARG-mediated free fatty acid uptake. Int. J. Mol. Sci. 23 (23), 15297. doi:10.3390/ijms232315297
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47 (W1), W357–W364. doi:10.1093/nar/gkz382
Foglia, B., Novo, E., Protopapa, F., Maggiora, M., Bocca, C., Cannito, S., et al. (2021). Hypoxia, hypoxia-inducible factors and liver fibrosis. Cells 10 (7), 1764. doi:10.3390/cells10071764
Gao, J., Zuo, B., and He, Y. (2024). Liver sinusoidal endothelial cells as potential drivers of liver fibrosis (Review). Mol. Med. Rep. 29 (3), 40. doi:10.3892/mmr.2024.13164
GBD 2017 Cirrhosis Collaborators (2020). The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 5 (3), 245–266. doi:10.1016/S2468-1253(19)30349-8
Genheden, S., and Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10 (5), 449–461. doi:10.1517/17460441.2015.1032936
Ginès, P., Castera, L., Lammert, F., Graupera, I., Serra-Burriel, M., Allen, A. M., et al. (2022). Population screening for liver fibrosis: Toward early diagnosis and intervention for chronic liver diseases. Hepatology 75(1), 219–228. doi:10.1002/hep.32163
Gollapalli, P., Ashok, A. K., and S, G. T. (2024). System-level protein interaction network analysis and molecular dynamics study reveal interaction of ferulic acid with PTGS2 as a natural radioprotector. J. Biomol. Struct. Dyn. 42 (6), 2765–2781. doi:10.1080/07391102.2023.2208224
Itoh, A., Isoda, K., Kondoh, M., Kawase, M., Watari, A., Kobayashi, M., et al. (2010). Hepatoprotective effect of syringic acid and vanillic acid on CCl4-induced liver injury. Biol. Pharm. Bull. 33 (6), 983–987. doi:10.1248/bpb.33.983
Jin, L., Li, J., Yang, S., Zhang, R., Hu, C., Chen, Y., et al. (2022). MAPK p38/Ulk1 pathway inhibits autophagy and induces IL-1β expression in hepatic stellate cells. Am. J. Physiol. Gastrointest. Liver Physiol. 322 (3), G360–G367. doi:10.1152/ajpgi.00230.2021
Kaftanovskaya, E. M., Ng, H. H., Soula, M., Rivas, B., Myhr, C., Ho, B. A., et al. (2019). Therapeutic effects of a small molecule agonist of the relaxin receptor ML290 in liver fibrosis. FASEB J. 33 (11), 12435–12446. doi:10.1096/fj.201901046R
Kluwe, J., Pradere, J. P., Gwak, G. Y., Mencin, A., De Minicis, S., Osterreicher, C. H., et al. (2010). Modulation of hepatic fibrosis by c-Jun-N-terminal kinase inhibition. Gastroenterology 138 (1), 347–359. doi:10.1053/j.gastro.2009.09.015
Kovats, S. (2015). Estrogen receptors regulate innate immune cells and signaling pathways. Cell. Immunol. 294 (2), 63–69. doi:10.1016/j.cellimm.2015.01.018
Kyritsi, K., Meng, F., Zhou, T., Wu, N., Venter, J., Francis, H., et al. (2017). Knockdown of hepatic gonadotropin-releasing hormone by Vivo-Morpholino decreases liver fibrosis in multidrug resistance gene 2 knockout mice by down-regulation of miR-200b. Am. J. Pathol. 187 (7), 1551–1565. doi:10.1016/j.ajpath.2017.03.013
Li, J., Guo, C., and Wu, J. (2021). The agonists of peroxisome proliferator-activated receptor-gamma for liver fibrosis. Drug Des. devel. Ther. 15, 2619–2628. doi:10.2147/DDDT.S310163
Liu, X., Xu, J., Rosenthal, S., Zhang, L. J., McCubbin, R., Meshgin, N., et al. (2020). Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology 158 (6), 1728–1744. doi:10.1053/j.gastro.2020.01.027
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., and Simmerling, C. (2015). ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11 (8), 3696–3713. doi:10.1021/acs.jctc.5b00255
Matsuda, M., and Seki, E. (2020). Hepatic stellate cell-macrophage crosstalk in liver fibrosis and carcinogenesis. Semin. Liver Dis. 40 (3), 307–320. doi:10.1055/s-0040-1708876
Meng, D., Li, Z., Wang, G., Ling, L., Wu, Y., and Zhang, C. (2018). Carvedilol attenuates liver fibrosis by suppressing autophagy and promoting apoptosis in hepatic stellate cells. Biomed. Pharmacother. 108, 1617–1627. doi:10.1016/j.biopha.2018.10.005
Ni, X. X., Li, X. Y., Wang, Q., and Hua, J. (2021). Regulation of peroxisome proliferator-activated receptor-gamma activity affects the hepatic stellate cell activation and the progression of NASH via TGF-β1/Smad signaling pathway. J. Physiol. Biochem. 77 (1), 35–45. doi:10.1007/s13105-020-00777-7
Noh, H. J., Hwang, D., Lee, E. S., Hyun, J. W., Yi, P. H., Kim, G. S., et al. (2015). Anti-inflammatory activity of a new cyclic peptide, citrusin XI, isolated from the fruits of Citrus unshiu. J. Ethnopharmacol. 163, 106–112. doi:10.1016/j.jep.2015.01.024
Punvittayagul, C., Chariyakornkul, A., Jarukamjorn, K., and Wongpoomchai, R. (2021). Protective role of vanillic acid against diethylnitrosamine- and 1,2-dimethylhydrazine-induced hepatocarcinogenesis in rats. Molecules 26 (9), 2718. doi:10.3390/molecules26092718
Radnai, B., Tucsek, Z., Bognar, Z., Antus, C., Mark, L., Berente, Z., et al. (2009). Ferulaldehyde, a water-soluble degradation product of polyphenols, inhibits the lipopolysaccharide-induced inflammatory response in mice. J. Nutr. 139 (2), 291–297. doi:10.3945/jn.108.097386
Rodriguez-Barbero, A., Dorado, F., Velasco, S., Pandiella, A., Banas, B., and Lopez-Novoa, J. M. (2006). TGF-beta1 induces COX-2 expression and PGE2 synthesis through MAPK and PI3K pathways in human mesangial cells. Kidney Int. 70 (5), 901–909. doi:10.1038/sj.ki.5001626
Roe, D. R., and Cheatham, T. R. (2013). PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9 (7), 3084–3095. doi:10.1021/ct400341p
Roehlen, N., Crouchet, E., and Baumert, T. F. (2020). Liver fibrosis: mechanistic concepts and therapeutic perspectives. Cells 9 (4), 875. doi:10.3390/cells9040875
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. doi:10.1186/1758-2946-6-13
Rudraiah, S., Zhang, X., and Wang, L. (2016). Nuclear receptors as therapeutic targets in liver disease: are we there yet? Annu. Rev. Pharmacol. Toxicol. 56, 605–626. doi:10.1146/annurev-pharmtox-010715-103209
Samuel, C. S., Royce, S. G., Hewitson, T. D., Denton, K. M., Cooney, T. E., and Bennett, R. G. (2017). Anti-fibrotic actions of relaxin. Br. J. Pharmacol. 174 (10), 962–976. doi:10.1111/bph.13529
Sarkar, P., and Chattopadhyay, A. (2021). Insights into cellular signaling from membrane dynamics. Arch. Biochem. Biophys. 701, 108794. doi:10.1016/j.abb.2021.108794
Schuppan, D., Ashfaq-Khan, M., Yang, A. T., and Kim, Y. O. (2018). Liver fibrosis: direct antifibrotic agents and targeted therapies. Matrix Biol. 68-69, 435–451. doi:10.1016/j.matbio.2018.04.006
Shi, Y., Shi, L., Liu, Q., Wang, W., and Liu, Y. (2023). Molecular mechanism and research progress on pharmacology of ferulic acid in liver diseases. Front. Pharmacol. 14, 1207999. doi:10.3389/fphar.2023.1207999
Su, P., Shi, Y., Wang, J., Shen, X., and Zhang, J. (2015). Anticancer agents derived from natural cinnamic acids. Anticancer Agents Med. Chem. 15 (8), 980–987. doi:10.2174/1871520615666150130111120
Takai, E., Tsukimoto, M., and Kojima, S. (2013). TGF-β1 downregulates COX-2 expression leading to decrease of PGE2 production in human lung cancer A549 cells, which is involved in fibrotic response to TGF-β1. PLoS One 8 (10), e76346. doi:10.1371/journal.pone.0076346
Tran, M., Liu, Y., Huang, W., and Wang, L. (2018). Nuclear receptors and liver disease: summary of the 2017 basic research symposium. Hepatol. Commun. 2 (7), 765–777. doi:10.1002/hep4.1203
Truong, T., Seo, S. H., Chung, S., and Kang, I. (2023). Attenuation of hepatic fibrosis by p-coumaric acid via modulation of NLRP3 inflammasome activation in C57BL/6 mice. J. Nutr. Biochem. 112, 109204. doi:10.1016/j.jnutbio.2022.109204
Wahlang, B., McClain, C., Barve, S., and Gobejishvili, L. (2018). Role of cAMP and phosphodiesterase signaling in liver health and disease. Cell. Signal. 49, 105–115. doi:10.1016/j.cellsig.2018.06.005
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004). Development and testing of a general amber force field. J. Comput. Chem. 25 (9), 1157–1174. doi:10.1002/jcc.20035
Wang, J. S., Zhang, Q. Y., Cheng, J. L., Chen, L. Y., Yao, N. L., Sun, G. Z., et al. (2018). Angiogenesis and hepatic fibrosis: western and Chinese medicine therapies on the road. Chin. J. Integr. Med. 24 (9), 713–720. doi:10.1007/s11655-018-3007-1
Wang, R., Song, F., Li, S., Wu, B., Gu, Y., and Yuan, Y. (2019). Salvianolic acid A attenuates CCl4-induced liver fibrosis by regulating the PI3K/AKT/mTOR, Bcl-2/Bax and caspase-3/cleaved caspase-3 signaling pathways. Drug Des. devel. Ther. 13, 1889–1900. doi:10.2147/DDDT.S194787
Wang M., M., Zhang, J., and Gong, N. (2022). Role of the PI3K/Akt signaling pathway in liver ischemia reperfusion injury: a narrative review. Ann. Palliat. Med. 11 (2), 806–817. doi:10.21037/apm-21-3286
Wang Z., Z., Qiao, L., Zheng, Q., Han, H., Li, Z., Zhang, X., et al. (2022). Combined hepatotoxicity and toxicity mechanism of intermedine and lycopsamine. Toxins (Basel) 14 (9), 633. doi:10.3390/toxins14090633
Win, S., Than, T. A., Zhang, J., Oo, C., Min, R., and Kaplowitz, N. (2018). New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatology 67 (5), 2013–2024. doi:10.1002/hep.29689
Wu, J., Gong, L., Li, Y., Qu, J., Yang, Y., Wu, R., et al. (2024). Tao-Hong-Si-Wu-Tang improves thioacetamide-induced liver fibrosis by reversing ACSL4-mediated lipid accumulation and promoting mitophagy. J. Ethnopharmacol. 333, 118456. doi:10.1016/j.jep.2024.118456
Xi, S., Shi, M., Jiang, X., Minuk, G. Y., Cheng, Y., Peng, Y., et al. (2016). The effects of Tao-Hong-Si-Wu on hepatic necroinflammatory activity and fibrosis in a murine model of chronic liver disease. J. Ethnopharmacol. 180, 28–36. doi:10.1016/j.jep.2016.01.030
Xiao, Z., Gao, S., Li, S., Yang, F., Zhang, D., Niu, Z., et al. (2025). Taohong Siwu Decoction modulates glutathione metabolism to suppress hepatocyte ferroptosis and demonstrates anti-fibrotic effects in the liver. J. Ethnopharmacol. 350, 120025. doi:10.1016/j.jep.2025.120025
Xie, Z., Huang, J., Li, Y., Zhu, Q., Huang, X., Chen, J., et al. (2023). Single-cell RNA sequencing revealed potential targets for immunotherapy studies in hepatocellular carcinoma. Sci. Rep. 13 (1), 18799. doi:10.1038/s41598-023-46132-w
Xu, J. W., Gong, J., Chang, X. M., Luo, J. Y., Dong, L., Jia, A., et al. (2004). Effects of estradiol on liver estrogen receptor-alpha and its mRNA expression in hepatic fibrosis in rats. World J. Gastroenterol. 10 (2), 250–254. doi:10.3748/wjg.v10.i2.250
Xu, L., Yuan, Y., Che, Z., Tan, X., Wu, B., Wang, C., et al. (2022). The hepatoprotective and hepatotoxic roles of sex and sex-related hormones. Front. Immunol. 13, 939631. doi:10.3389/fimmu.2022.939631
Yang, H., Xuefeng, Y., Shandong, W., and Jianhua, X. (2020). COX-2 in liver fibrosis. Clin. Chim. Acta 506, 196–203. doi:10.1016/j.cca.2020.03.024
Yang, M. H., Yoon, K. D., Chin, Y. W., Park, J. H., and Kim, J. (2009). Phenolic compounds with radical scavenging and cyclooxygenase-2 (COX-2) inhibitory activities from Dioscorea opposita. Bioorg. Med. Chem. 17 (7), 2689–2694. doi:10.1016/j.bmc.2009.02.057
Yao, S., Kim, S. C., Li, J., Tang, S., and Wang, X. (2024). Phosphatidic acid signaling and function in nuclei. Prog. Lipid Res. 93, 101267. doi:10.1016/j.plipres.2023.101267
Yoon, Y. J., Friedman, S. L., and Lee, Y. A. (2016). Antifibrotic therapies: where are we now? Semin. Liver Dis. 36 (1), 87–98. doi:10.1055/s-0036-1571295
Zhang, J., Jiang, N., Ping, J., and Xu, L. (2021). TGF-β1-induced autophagy activates hepatic stellate cells via the ERK and JNK signaling pathways. Int. J. Mol. Med. 47 (1), 256–266. doi:10.3892/ijmm.2020.4778
Zulfat, M., Hakami, M. A., Hazazi, A., Mahmood, A., Khalid, A., Alqurashi, R. S., et al. (2024). Identification of novel NLRP3 inhibitors as therapeutic options for epilepsy by machine learning-based virtual screening, molecular docking and biomolecular simulation studies. Heliyon 10 (15), e34410. doi:10.1016/j.heliyon.2024.e34410
Keywords: hepatic fibrosis, non-targeted metabolomics, molecular docking, Taohong Siwu decoction, mice, high resolution mass spectrometry
Citation: Li S, Qi S, Li R, Yang F, Niu Z, Liu W, Ma S and Xiao Z (2025) Non-targeted metabolomics and network pharmacology of Taohong Siwu Decoction in hepatic fibrosis mouse model using high resolution mass spectrometry. Front. Mol. Biosci. 12:1614341. doi: 10.3389/fmolb.2025.1614341
Received: 18 April 2025; Accepted: 11 June 2025;
Published: 30 June 2025.
Edited by:
Andras Szeitz, University of British Columbia, CanadaReviewed by:
Yong Yang, The Affiliated Hospital of Changchun University of Traditional Chinese Medicine, ChinaMao Yongpo, Chengdu University, China
Wen Hou, Tianjin First Central Hospital, China
Copyright © 2025 Li, Qi, Li, Yang, Niu, Liu, Ma and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhun Xiao, eGlhb3podW44MThAMTYzLmNvbQ==; Suping Ma, bWFzdXBpbmdAMTYzLmNvbQ==; Wei Liu, bHdoemF5bEAxNjMuY29t
 Shengsheng Li1,2
Shengsheng Li1,2 Shenglan Qi
Shenglan Qi Wei Liu
Wei Liu Zhun Xiao
Zhun Xiao