- 1The Center for Heart Development, College of Life Science, Hunan Normal University, Changsha, China
- 2Guangdong Provincial Key Laboratory of Pathogenesis, Targeted Prevention and Treatment of Heart Disease, Guangzhou Key Laboratory of Cardiac Pathogenesis and Prevention, Guangzhou, China
- 3Medical Research Center, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, Guangzhou, China
Introduction: Mutations in the DDX3X gene are the primary cause of DDX3X syndrome, with over 800 diagnosed families worldwide. DDX3X is also recognized as a single-gene driver for rare syndromes associated with epilepsy, autism, and developmental disorders. Clinical studies suggest potential links between DDX3X mutations and various cardiac comorbidities. However, there is no report on whether Ddx3xa knockout leads to cardiac phenotypes or whether a zebrafish ddx3xa gene knockout model has been used for such research. This study is based on the high genomic conservation between zebrafish and humans, utilizing a zebrafish model to investigate the potential links between DDX3X mutations and various cardiac comorbidities, as well as the underlying mechanisms.
Methods: A ddx3xa knockout model was constructed using CRISPR/Cas9 technology. To elucidate the molecular mechanisms, we performed transcriptome-wide profiling via RNA-Seq to identify differentially expressed genes and dysregulated signaling pathways. The spatiotemporal expression patterns of key genes were assessed using whole-mount in situ hybridization (WISH). Additionally, the critical role of Wnt/β-catenin signaling in the mutant phenotype was further validated using the Wnt inhibitor IWR-1.
Results: Homozygous knockout (ddx3xa−/−) embryos exhibited developmental delay, trunk malformations, and severe cardiac abnormalities, including pericardial edema, defective cardiac looping, and cardiac contractile dysfunction. Ribonucleic Acid Sequencing (RNA-seq) analysis of ddx3xa−/− zebrafish at 72 h post-fertilization (hpf) revealed significant enrichment in pathways related to actin cytoskeleton organization, calcium signaling, cardiac and vascular smooth muscle contraction, and Wnt signaling. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (QRT-PCR) and in situ hybridization confirmed dysregulated expression of key cardiac development genes (bmp4, actn2b, tbx5, nppb) and significantly impaired cardiac function. Given the role of Wnt signaling in cardiogenesis, we further analyzed this pathway and found that ddx3xa knockout upregulated the key Wnt/β-catenin transcription factor Tcf/Lef1 (T Cell Factor/Lymphoid Enhancer Factor 1) and disrupted its target genes (bmp4, tbx5) expression. Crucially, treatment of 72 hpf mutant embryos with the Wnt inhibitor IWR-1 partially rescued both the cardiac malformations and the aberrant expression of its target genes.
Discussion: This study provides the first evidence that ddx3xa regulates cardiac morphogenesis by modulating the Wnt/β-catenin signaling pathway, offering direct experimental insight into the mechanisms underlying cardiac comorbidities in DDX3X syndrome. It also highlights the unique value of the zebrafish model for dissecting conserved pathogenic pathways and exploring targeted therapeutic strategies.
1 Introduction
Proteins belonging to the DDX family are characterized by a highly conserved structural motif, the Asp-Glu-Ala-Asp (D-E-A-D) amino acid sequence (Linder and Jankowsky, 2011; Mo et al., 2021). DEAD-box proteins possess essential functions, including ATP hydrolysis, modulation of helicase activity, and participation in RNA metabolism as RNA-binding proteins (Jankowsky et al., 2011). Research indicates that DEAD-box family proteins play regulatory roles in critical cellular processes such as the cell cycle, apoptosis, and tumorigenesis, with clinical studies linking DDX protein mutations to the pathogenesis of various diseases, including cancer (Mo et al., 2021). DDX3 is a member of the DEAD-box helicase family. The human genome encodes two paralogous genes: DDX3X and its homolog DDX3Y (Kim et al., 2001). The DDX3 gene encodes a highly conserved RNA helicase that is crucial for RNA metabolism, translation initiation, and innate immune responses, and it has emerged as a significant therapeutic target in oncology (Bol et al., 2015; Devasahayam Arokia Balaya et al., 2025). Specifically, mutations in the DDX3X gene represent the primary cause of DDX3X syndrome, a neurodevelopmental disorder diagnosed in over 800 families worldwide. This syndrome accounts for approximately 3% of intellectual disability (ID) cases and exhibits a strong female predominance (Tang et al., 2021; von Mueffling et al., 2024). Studies have established clear associations between DDX3X mutations and a spectrum of neurological conditions, including epilepsy, autism spectrum disorder (ASD), and intellectual disability, alongside links to immune dysregulation and oncogenesis (Boitnott et al., 2021; von Mueffling et al., 2024). Although clinical observations have implicated DDX3X mutations in cardiac comorbidities (Dikow et al., 2017), the molecular mechanisms underpinning these cardiac contractile impairments remain largely unexplored.
The DDX3X gene is highly conserved across eukaryotes. It exhibits 98.6% amino acid identity between humans and mice, and targeted knockout of the murine Ddx3x gene results in early embryonic lethality, underscoring its critical role in early embryonic survival (Owsianka and Patel, 1999; Ditton et al., 2004; Padmanabhan et al., 2016). In hepatocytes, DDX3X protects against drug-induced acute liver injury by modulating stress granule formation and oxidative stress responses (Luo et al., 2023). Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development (Lennox et al., 2020). Clinical data from breast cancer patients indicate that high expression of the RNA helicase DDX3 is significantly associated with shortened overall survival and bone metastasis; consequently, small-molecule inhibitors targeting DDX3X are being explored as therapeutic agents for breast cancer bone metastasis (Winnard et al., 2024). Furthermore, DDX3X establishes a dynamic balance between viral propagation and host defense within the antiviral innate immune response (Ma et al., 2025). While the roles of DDX3X in liver injury, neurodevelopment, cancer, and innate immunity have been partially elucidated, its association with cardiac pathologies remains largely unexplored. Thus, elucidating the regulatory mechanisms of DDX3X within the cardiac system necessitates in vivo investigations using model organisms.
The zebrafish (Danio rerio) has emerged as a powerful model for studying developmental disorders due to its high degree of genetic and physiological conservation with humans (Dooley and Zon, 2000). Approximately 84% of human disease genes possess orthologs in zebrafish, and its transparent embryos facilitate real-time observation of organogenesis, making it particularly valuable for cardiovascular research (Lawrence, 2007; Dahme et al., 2009). The zebrafish genome harbors two paralogous ddx3x genes, ddx3xa and ddx3xb, both exhibiting high conservation with human DDX3X (Duan et al., 2024). Given their highly similar sequences and overlapping expression patterns, ddx3xa and ddx3xb display functional redundancy in certain contexts; for instance, the combined deficiency of DDX3X function leads to defects in neurogenesis, a process where these paralogs compensate for each other (Duan et al., 2024). However, studies targeting ddx3xb function specifically have revealed that the Ddx3xb helicase regulates the maternal-to-zygotic transition (MZT) in zebrafish embryogenesis through liquid-liquid phase separation (Shi et al., 2022). Thus, existing evidence indicates that while ddx3xa and ddx3xb share functional redundancy, they also possess distinct regulatory mechanisms. Previous research has demonstrated that loss of ddx3xa in zebrafish results in developmental delay and brain abnormalities (Duan et al., 2024). Nevertheless, its specific role in cardiac contractile function remains undefined and constitutes a primary focus of this investigation.
Numerous studies have established that Wnt signaling pathways are activated and play pivotal regulatory roles in both physiological and pathological processes, including cardiovascular diseases and cancer (Nusse and Clevers, 2017). Among these, the Wnt/β-catenin signaling pathway is evolutionarily highly conserved. It participates extensively in nearly all stages of cardiac development, encompassing early specification, cardiomyocyte differentiation, heart tube looping, chamber and valve formation, and conduction system patterning (Naito et al., 2006; Li et al., 2022). Notably, over half of patients diagnosed with arrhythmogenic right ventricular cardiomyopathy (ARVC) harbor mutations in genes encoding desmosomal proteins (Towbin, 2011). These mutant desmosomal proteins competitively bind to the transcription factors Tcf/Lef, displacing β-catenin. This disrupts canonical Wnt signaling and promotes the transdifferentiation of cardiomyocytes into adipocytes and fibroblasts, leading to adipogenic replacement and the development of arrhythmogenic cardiomyopathy (Garcia-Gras et al., 2006; Lombardi et al., 2011). Key downstream target genes of the canonical Wnt pathway include C-Myc, cyclin D1, C-fos, BMP4, TBX5, mesp1, and actn2b, among others (Nishimoto et al., 2015; Medina et al., 2016; Ye et al., 2019; Zhang et al., 2024). Intriguingly, studies have identified an interaction between zebrafish ddx3x and csnk2a1 (casein kinase 2 alpha 1), a known component of the Wnt signaling machinery, suggesting a potential regulatory role for ddx3x in Wnt/β-catenin signaling (Wang et al., 2023). However, it remains unclear whether ddx3x specifically modulates Wnt signaling during cardiac morphogenesis. Furthermore, there is currently no evidence regarding whether ddx3x knockout models induce cardiac phenotypes, nor have zebrafish ddx3x gene knockout models been established.
In this study, we employed CRISPR/Cas9 technology to generate a ddx3xa knockout zebrafish model (Sun et al., 2020), specifically to investigate its role in cardiac development and function. We observed that loss of ddx3xa function resulted in embryonic developmental abnormalities, including developmental delay, pericardial edema, bradycardia, and defective cardiac looping. qRT-PCR and in situ hybridization analyses demonstrated that ddx3xa knockout led to the upregulation of key Wnt/β-catenin pathway components, such as the transcription factor Tcf/Lef1. This dysregulation consequently disrupted the expression of critical downstream cardiac genes (bmp4, actn2b, tbx5, nppb), ultimately leading to severely impaired cardiac defects. Crucially, pharmacological inhibition of the Wnt/β-catenin pathway using the small-molecule inhibitor IWR-1 partially rescued both the aberrant expression of these molecular markers and the associated cardiac defects. These findings underscore a central role for ddx3xa in mediating cardiac regulation through the Wnt/β-catenin signaling pathway, demonstrating that ddx3xa governs zebrafish embryonic cardiac defects by modulating Wnt signaling activity. Collectively, our work establishes the essential role of ddx3xa in cardiac morphogenesis and provides crucial mechanistic insights into the pathogenesis of cardiac comorbidities associated with DDX3X syndrome, offering a valuable potential disease model.
2 Results
2.1 Homology and evolutionary analysis of the zebrafish ddx3xa gene structure
To investigate the evolutionary conservation and functional significance of ddx3xa, we performed phylogenetic and sequence homology analyses. A phylogenetic tree was constructed using MEGA X software based on amino acid sequences of DDX3X homologous proteins from multiple species (Escherichia coli, Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Xenopus tropicalis, Pan paniscus, Homo sapiens, Mus musculus, and various teleost fishes). The results revealed close evolutionary relationships with minimal genetic divergence among these species (Figure 1A), suggesting high conservation of DDX3X structure and function throughout evolution. Further sequence alignment using DNAMAN software demonstrated >75% amino acid sequence homology between human DDX3X, mouse DDX3X, and zebrafish Ddx3xa. Notably, the RNA-binding domain exhibited exceptional conservation (Figure 1B). These findings provide further confirmation of Ddx3xa’s evolutionary conservation and support the rationale for utilizing zebrafish as a model to study its biological functions.
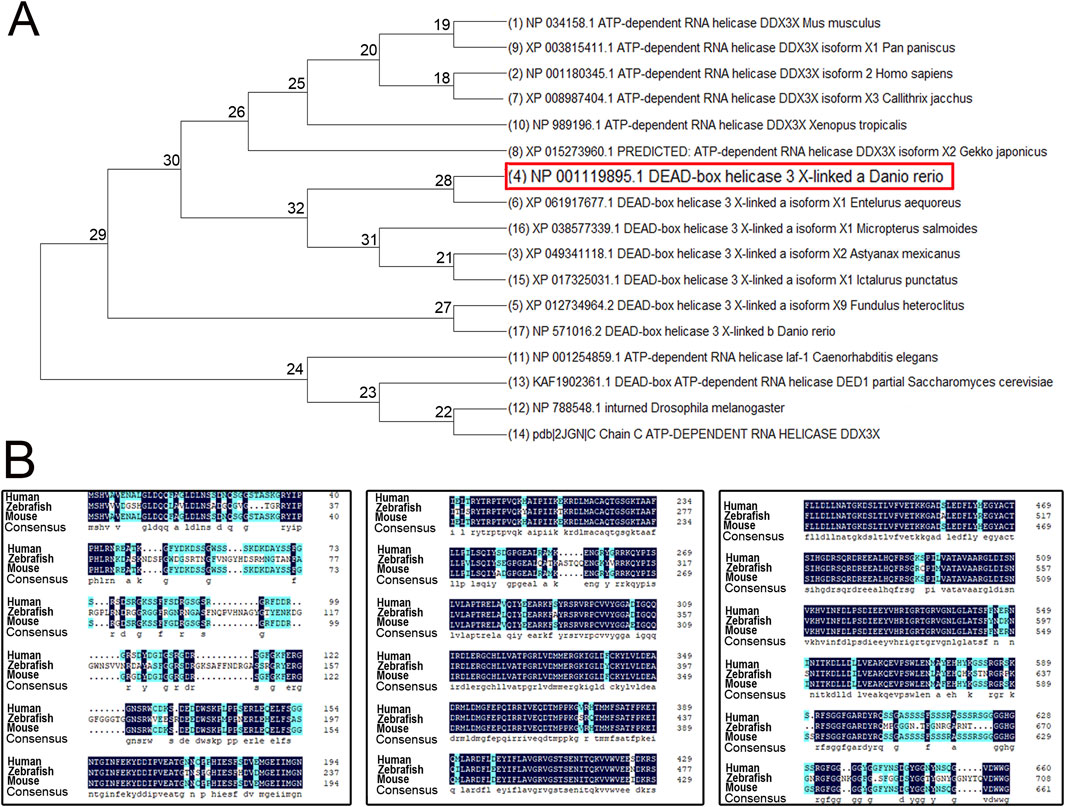
Figure 1. Phylogenetic tree and amino acid sequence analysis of DDX3X across species. (A) Phylogenetic tree of DDX3X homologs from diverse species. (B) Amino acid sequence alignment of Homo sapiens DDX3X, Mus musculus DDX3X, and Danio rerio Ddx3xa.
2.2 Expression pattern of ddx3xa in early zebrafish embryos revealed by whole-mount in situ hybridization and qRT-PCR
To delineate the spatial and temporal expression profile of ddx3xa during early development, whole-mount in situ hybridization (WISH) was performed on zebrafish embryos at six developmental stages: 0.2, 12, 24, 36, 48, and 72 h post-fertilization (hpf), using a synthesized ddx3xa-specific probe.
WISH results revealed ubiquitous ddx3xa expression at 0.2 hpf and 12 hpf (Figures 2A,B). At 24 hpf and 36 hpf, expression remained widespread throughout the embryo, with strong signals detected in the brain, skeletal muscles, and tail, exhibiting the highest intensity in the brain (Figures 2C,D,G,H). By 48 hpf, expression became more restricted, primarily localizing to the brain and pectoral fin buds, with diminished expression in the tail compared to earlier stages (Figures 2E,I). At 72 hpf, expression was confined specifically to the brain, heart, and pectoral fin buds (Figures 2F,J). These findings suggest that ddx3xa participates in various stages of zebrafish early embryogenesis, with predominant roles in brain and heart development.
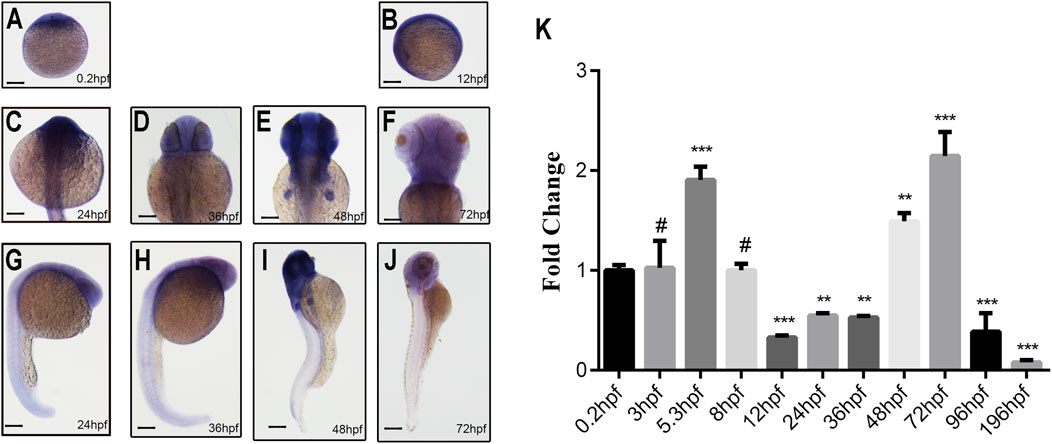
Figure 2. Spatiotemporal expression pattern of ddx3xa by whole-mount in situ hybridization and qRT-PCR analysis. (A–J) Spatiotemporal expression of ddx3xa at various developmental stages visualized by whole-mount in situ hybridization. Scale bar: 250 μm (applies to A–J). (A) 0.2 hpf; (B) 12 hpf; (C) 24 hpf; (D) 36 hpf; (E) 48 hpf; (F) 72 hpf; (G) Dorsal view, 24 hpf; (H) Dorsal view, 36 hpf; (I) Dorsal view, 48 hpf; (J) Dorsal view, 72 hpf. (K) Quantitative real-time PCR (qRT-PCR) analysis of relative ddx3xa transcript levels at various developmental time points (0.2, 3, 5.3, 8, 12, 24, 36, 48, 72, 96, 196 hpf). Expression levels were normalized to the 0.2 hpf time point. (n = 3; **P < 0.01, ***P < 0.001).
To quantitatively assess ddx3xa transcript levels across development, qRT-PCR analysis was conducted on embryos collected at 11 time points: 0.2, 3, 5.3, 8, 12, 24, 36, 48, 72, 96, and 196 hpf. Ddx3xa transcripts were detectable as early as 0.2 hpf and persisted throughout early embryonic development. Notably, transcript levels peaked at 72 hpf (Figure 2K). This expression profile indicates that ddx3xa likely functions as a widely expressed, maternally derived gene regulating early zebrafish embryogenesis, with its expression sustained across the entirety of early embryonic development.
2.3 Construction of the ddx3xa-Knockout zebrafish line
To investigate the role of ddx3xa in early zebrafish development, we established a CRISPR/Cas9-mediated knockout line using two single-guide RNAs (sgRNAs) targeting exon 4 of ddx3xa: Target 1 (5′-TGGGATGGTAGTCGTACCAA-3′) and Target 2 (5′-GGAGGACGTGGTGGCTTTCG-3′) (Figure 3A). This approach generated three stable lines (Lines 1–3) carrying frameshift mutant alleles. Sanger sequencing confirmed line-specific mutations: Line 1 exhibited a 102 bp intronic deletion with a 30 bp exonic deletion and 26 bp insertion; Line 2 showed a 169 bp exonic deletion plus 82 bp intronic deletion; and Line 3 contained a 24 bp insertion with 98-bp exonic insertion (Figure 3B). Preliminary morphological and genotypic analyses confirmed that all three mutant lines (Lines 1–3) exhibited indistinguishable core phenotypes. To ensure experimental consistency and reproducibility, all subsequent experiments were uniformly performed using Line 2.
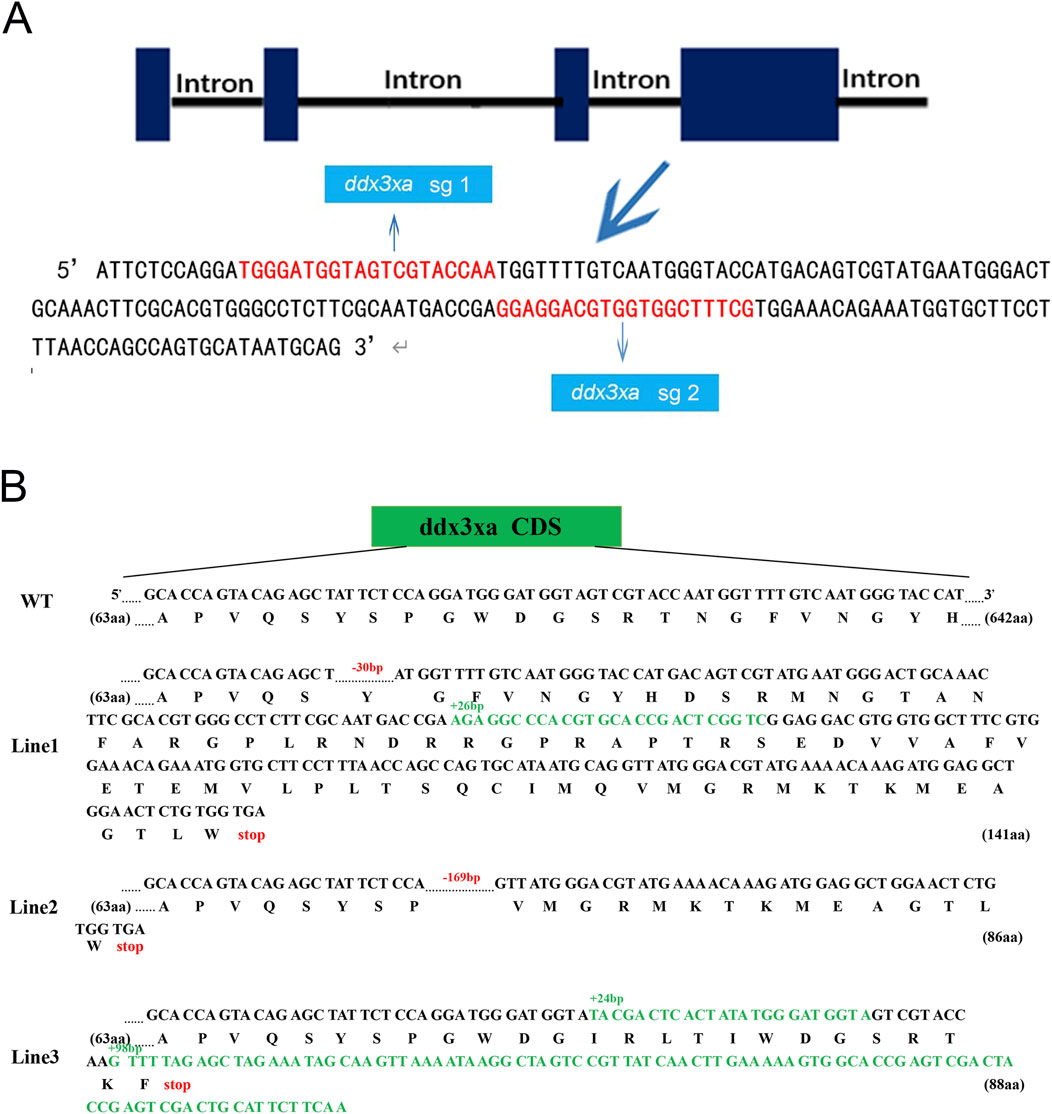
Figure 3. Schematic representation of ddx3xa knockout in zebrafish. (A) Design of sgRNAs targeting ddx3xa. Horizontal lines represent ddx3xa genomic DNA, blue rectangles denote exons, and red text indicates target site sequences. (B) Schematic of three heritable ddx3xa mutant alleles generated by CRISPR/Cas9 and their resultant protein sequences.
Stereomicroscopic observation revealed developmental delays in mutants initiating at 2 hpf and persisting through 24 hpf (Supplementary Figure S1A). By 48 hpf, ddx3xa−/− embryos displayed severe morphological abnormalities. Quantitative analysis demonstrated a 25.26% malformation rate in mutants (Figure 4A; Supplementary Figure S1B) - representing a nearly 6-fold increase compared to wild-type (WT) controls (4.4%) - indicating profound disruption of early development.
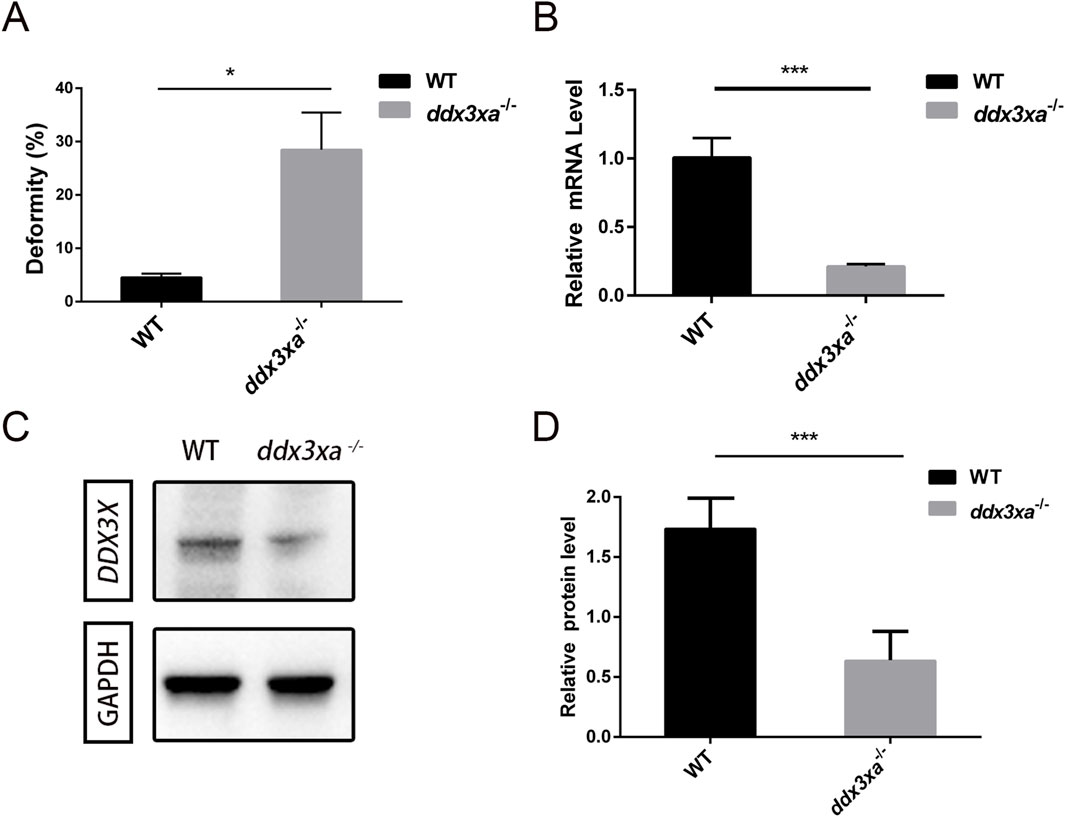
Figure 4. Malformation rate quantification and validation of ddx3xa knockout efficiency in zebrafish. (A) Malformation rate in ddx3xa−/− zebrafish at 48 hpf (**P < 0.01 vs. wild-type). (B) qRT-PCR analysis of ddx3xa mRNA levels in 72-hpf embryos showing significant downregulation in mutants (n = 3; *P < 0.05, ***P < 0.001). (C) Representative Western blot of Ddx3xa protein expression in 72-hpf larvae. Gapdh served as a loading control. (D) Quantitative densitometric analysis of Ddx3xa protein levels in wild-type and ddx3xa−/− larvae at 72 hpf.
Knockout efficiency was validated by qRT-PCR at 72 hpf, showing significantly reduced ddx3xa mRNA levels in mutants versus WT (Figure 4B). Western blot analysis further confirmed markedly decreased Ddx3xa protein expression in ddx3xa−/− embryos (Figures 4C,D), consistent with premature translational termination due to frameshift mutations. These results collectively confirm successful establishment of the ddx3xa-knockout line.
2.4 ddx3xa knockout induces cardiac malformations and functional deficiencies in zebrafish embryos
Morphological analysis of the heart revealed that a subset of mutant embryos developed pericardial edema, cardiac linearization, and impaired looping from 48 hpf. These defects were consistently observed and persisted through at least 72 hpf, in contrast to their wild-type counterparts (Figures 5A–H). Quantitative analysis of these cardiac malformations at 48 hpf revealed a significant increase in the mutant group, as measured by the percentage of embryos with pericardial edema, cardiac linearization, and impaired looping process (Supplementary Figures S2,S3; Supplementary Table S1).
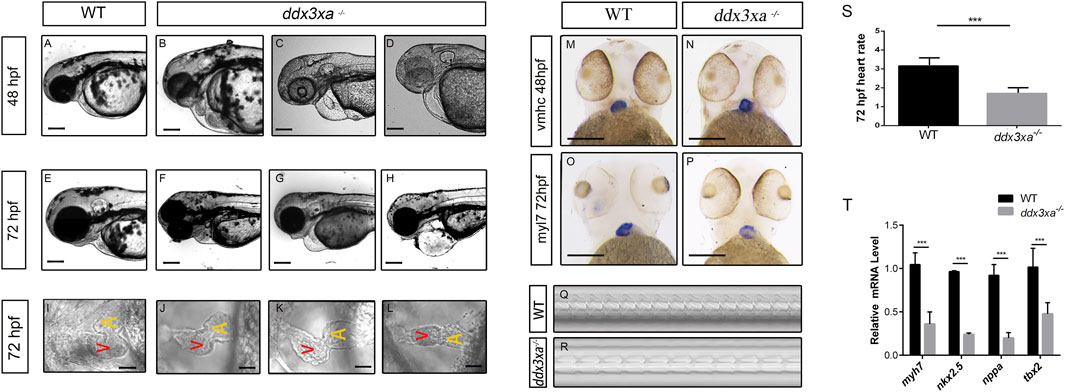
Figure 5. ddx3xa knockout induces early cardiac malformation and dysfunction in zebrafish. (A–H) Phenotypic analysis of zebrafish hearts. (A) Normal cardiac phenotype in wild-type (WT) zebrafish at 48 hpf. (B–D) Cardiac phenotypes in ddx3xa−/− zebrafish at 48 hpf. (E) Normal cardiac phenotype in WT zebrafish at 72 hpf. (F–H) Cardiac phenotypes in ddx3xa−/− zebrafish at 72 hpf. Scale bar: 500 µm (applies to A–H). (I) Dorsal view of the heart in WT zebrafish at 72 hpf. (J–L) Dorsal views of hearts in ddx3xa−/− zebrafish at 72 hpf. Scale bar: 25 µm (applies to I–L). (M–N) Whole-mount in situ hybridization (WISH) with vmhc probe at 48 hpf. ddx3xa−/− larvae display irregular ventricular contours compared to WT (scale bar: 250 μm). (M) vmhc WISH in WT zebrafish at 48 hpf. (N) vmhc WISH in ddx3xa−/− zebrafish at 48 hpf. (O–P) myl7 probe WISH at 72 hpf. ddx3xa−/− larvae exhibit defective atrioventricular looping and atrial hypoplasia compared to WT (scale bar: 250 μm). (O) myl7 WISH in WT zebrafish at 72 hpf. (P) myl7 WISH in ddx3xa−/− zebrafish at 72 hpf. (Q–T) Heartbeat analysis in WT and ddx3xa−/− zebrafish at 72 hpf. (Q) Representative heartbeat trace of WT zebrafish at 72 hpf. (R) Representative heartbeat trace of ddx3xa−/− zebrafish at 72 hpf. (S) Statistical analysis of heart rate (n = 3; ***P < 0.001). (T) qRT-PCR analysis of myh7, nkx2.5, nppa, and tbx2 transcript levels in WT and ddx3xa−/− larvae at 72 hpf (n = 3; **P < 0.01, ***P < 0.001).
To better assess the cardiac morphological changes induced by Ddx3xa knockout, 72 hpf zebrafish larvae were immobilized in a high concentration of methylcellulose and positioned dorsally on a slide for observation. This revealed that wild-type (WT) larvae at 72 hpf displayed an atrioventricular canal and expanded heart chambers arranged side-by-side. In contrast, ddx3xa−/− larvae exhibited failure of cardiac looping, presenting a linear heart tube (Figures 5I–L).
To elucidate the molecular basis of the cardiac developmental defects, we performed spatial expression pattern analysis using whole-mount in situ hybridization (WISH) with cardiac-specific markers. At 48 hpf, staining with the ventricular marker vmhc (ventricular myosin heavy chain) showed a significantly irregular ventricular margin in mutant larvae (Figures 5M,N), suggesting impaired ventricular specification or morphogenesis. By 72 hpf, staining with the pan-cardiac marker myl7 (myosin light chain 7) revealed more profound defects: mutant hearts displayed not only abnormal atrioventricular looping but also pronounced atrial hypoplasia (Figures 5O,P). This indicates that loss of ddx3xa disrupts the normal development and patterning of both the atrium and ventricle.
To further assess whether cardiac function was compromised in the zebrafish larvae, cardiac contraction rates were measured over a 10-s interval in both WT and ddx3xa−/− larvae at 72 hpf and statistically analyzed. While rhythmic, the heart rate of ddx3xa−/− zebrafish was significantly slower than that of WT larvae.
To identify the molecular drivers underlying these morphological phenotypes, we performed qRT-PCR analysis on 72 hpf larvae to quantify the expression levels of key cardiac developmental regulatory genes. The results demonstrated significant downregulation of myh7 (myosin heavy chain 7), nkx2.5, nppa, and tbx2 in mutants (Figure 5T). These genes are involved in critical biological processes including cardiomyocyte differentiation, cardiac progenitor maintenance, morphogenesis, and contractile function regulation. Their coordinated downregulation provides a direct molecular explanation for the observed severe cardiac malformations (such as looping failure and atrial hypoplasia) and functional impairment.
In summary, this study demonstrates that ddx3xa knockout induces severe cardiac malformation and dysfunction in zebrafish embryos by disrupting the cardiac looping process, impairing cardiomyocyte differentiation, and causing abnormal heart rate. This establishes an important in vivo model for gaining deeper insights into the pathogenesis of cardiac comorbidities associated with DDX3X syndrome.
2.5 Transcriptome sequencing analysis reveals abnormal regulation of wnt signaling pathway and cardiac development-related pathways in ddx3xa-deficient zebrafish
2.5.1 Global transcriptome analysis
Principal component analysis (PCA) revealed clear separation between WT and ddx3xa−/− groups along the first principal component (PC1), which accounted for 97.6% of the total transcriptional variance (PC2: 1.5%). The distinct clustering of biological replicates within each group indicated high reproducibility, while the pronounced separation along PC1 demonstrated that the ddx3x deletion was the dominant source of transcriptional variation (Figure 6A; Supplementary Table S2). RNA-Seq analysis of wild-type (WT) and ddx3xa−/− zebrafish at 72 hpf identified 1,541 differentially expressed genes (DEGs), comprising 1,159 upregulated and 382 downregulated genes in mutants relative to WT controls (Figure 6B). Hierarchical clustering analysis of all DEGs is presented in Figure 6C. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment revealed significant alterations in actin cytoskeleton organization, neuroactive ligand-receptor interactions, calcium signaling, cardiac and vascular smooth muscle contraction, and Wnt signaling pathway (Figure 6D). Thus, this global transcriptomic profiling establishes a foundational link between ddx3xa deficiency and cardiac pathogenesis. The identified DEGs provide a valuable resource for pinpointing key cardiac regulators. Crucially, the enriched pathways directly elucidate the observed phenotypes: actin cytoskeleton and calcium signaling anomalies underlie the contractile dysfunction and bradycardia, while the significant enrichment of the Wnt pathway, alongside others, suggests a synergistic multi-pathway disruption. This evidence positions Wnt signaling as a prime candidate for the core regulatory mechanism, guiding subsequent targeted investigation.
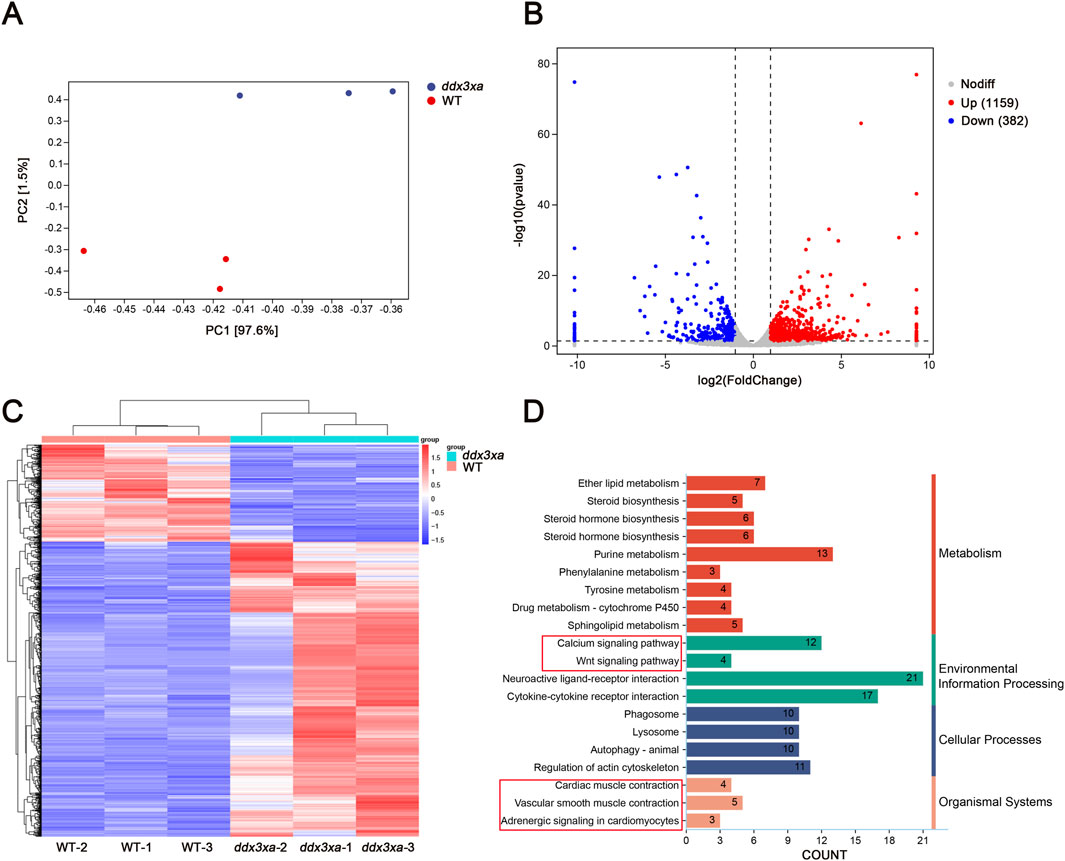
Figure 6. Transcriptomic alterations in ddx3xa−/− zebrafish at 72 hpf. (A) Principal component analysis (PCA) of the transcriptome revealed clear separation between WT and ddx3xa−/− groups along PC1, which accounted for 97.6% of the total variance (PC2: 1.5%). Sample distribution: wild-type (WT) controls (red) versus ddx3xa−/− mutants (blue). (B) Volcano plot of differentially expressed genes (DEGs): downregulated (blue), nonsignificant (gray), upregulated (red). (C) Hierarchical clustering heatmap: rows represent genes, columns represent samples. Expression levels scaled from high (red) to low (blue). (D) KEGG pathway enrichment analysis.
2.5.2 qPCR validation of key pathway genes
To validate RNA-Seq reliability, we performed qPCR analysis on candidate genes from enriched pathways: actin cytoskeleton regulation, calcium signaling, Wnt signaling, cardiac and vascular smooth muscle contraction, and cardiomyocyte adrenergic signaling. Consistent with transcriptomic data, ddx3xa−/− mutants showed significant downregulation of adcy7, itgae.1, itpka, ntrk1, ntsr1, p2rx3a, pla2g4aa, plcd1a, rac1, and chrm2b (P < 0.05), while cxcl12b, pla2g4f.1, and ptger1b were significantly upregulated (P < 0.05) (Figure 7A). Notably, aberrant expression of adrenergic signaling-related genes (adcy7, ntsr1) and myocardial contraction regulators (plcd1a, rac1) suggests ddx3xa deficiency disrupts cardiac development through impaired cardiomyocyte signaling and contractile function.
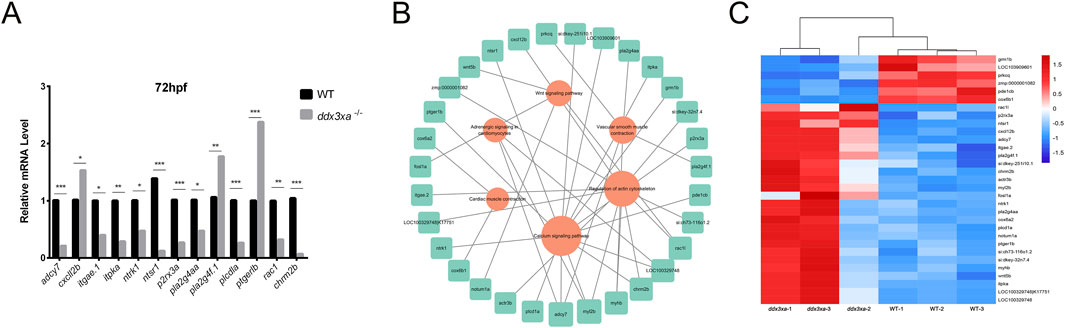
Figure 7. Validation and pathway analysis of cardiac development genes in ddx3xa−/− zebrafish. (A) qRT-PCR validation of genes related to cardiac contraction, vascular smooth muscle contraction, calcium signaling, and actin cytoskeleton regulation in ddx3xa−/− mutants versus wild-type (WT) at 72 hpf (n = 3; *P < 0.05, **P < 0.01, ***P < 0.001). (B) Pathway-gene interaction network for enriched cardiac development pathways generated using Cytoscape. (C) Heatmap of RNA-Seq expression profiles for enriched genes across key cardiac development pathways.
Using Cytoscape, we generated integrated pathway-gene networks for enriched cardiac development pathways (Figure 7B), with corresponding expression patterns visualized in an RNA-Seq-based heatmap (Figure 7C).
2.5.3 ddx3xa knockout disrupts wnt signaling in zebrafish
Initial experiments revealed cardiac malformations in ddx3xa−/− zebrafish larvae, suggesting that gene knockout may perturb the expression of key cardiac developmental regulators. Supporting this, prior RNA-seq data and studies in Xenopus tropicalis demonstrate that DDX3X acts as a binding partner for Casein Kinase I Isoform Epsilon (CSNK1E) within the Wnt signaling pathway, implicating it in Wnt pathway regulation (Cruciat et al., 2013). However, whether ddx3xa exerts a conserved regulatory function in zebrafish remained unknown.
To directly assess the role of ddx3xa in the zebrafish Wnt pathway, we quantified the expression of key pathway components in mutants. qRT-PCR analysis showed that at 72 hpf, transcript levels of the downstream effector tcf/lef1 and the core component ctnnb1 (β-catenin) were significantly upregulated, whereas upstream components (wnt3, wnt5b, and dvl2) were significantly downregulated (Figure 8A). This provides the first evidence that ddx3xa mediates Wnt signal transduction through regulators like tcf/lef1, and its loss creates an imbalanced state characterized by aberrant downstream activation coupled with upstream suppression. We propose that this dysregulation particularly abnormal Tcf/Lef1 activity serves as a core mechanism underlying the misregulation of critical cardiac genes (bmp4, tbx5), ultimately driving cardiac defects.
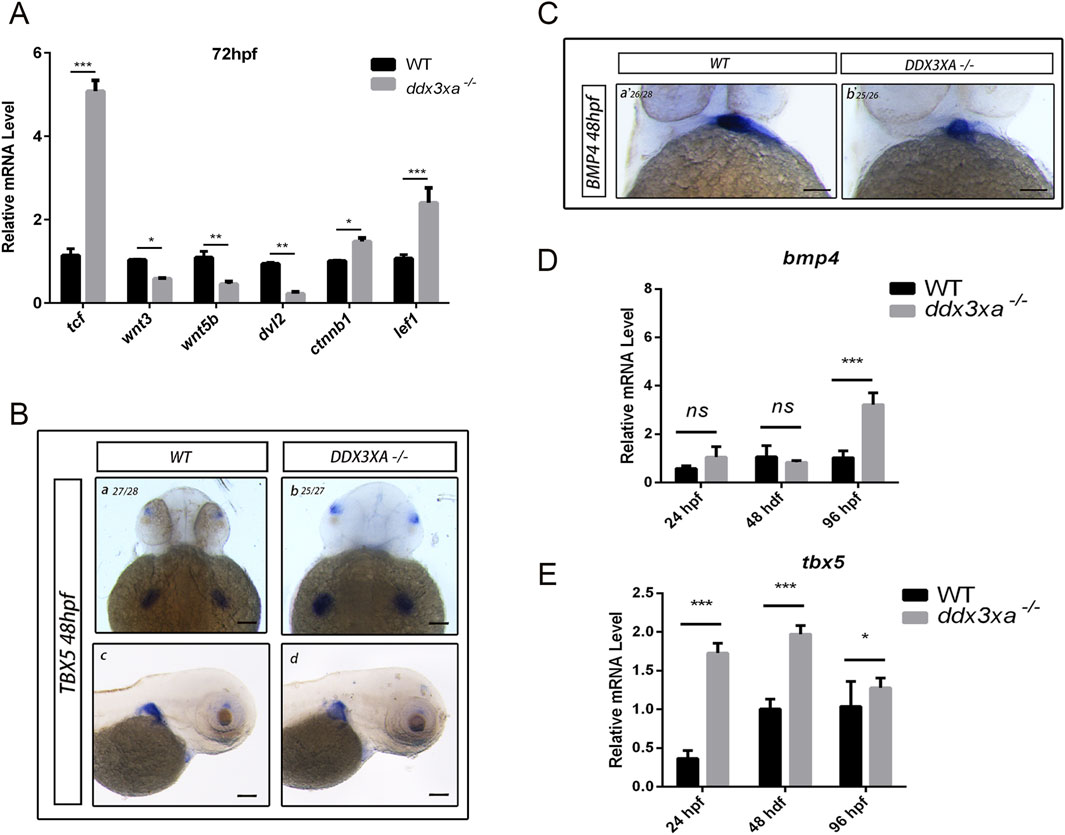
Figure 8. Wnt signaling disruption in ddx3xa−/− zebrafish. (A) Expression of key Wnt signaling components in ddx3xa−/− larvae at 72 hpf by qRT-PCR (n = 3; *p < 0.05, **p < 0.01, ***p < 0.001 vs. WT). (B) Whole-mount in situ hybridization (WISH) with tbx5 probe at 48 hpf. Numbers indicate identically stained embryos/total processed. Scale bar: 250 μm (applies to B–C). (a) WT dorsal view; (b) ddx3xa−/− dorsal view; (C) WT lateral view; (D) ddx3xa−/− lateral view. (C) WISH with bmp4 probe at 48 hpf. (a) WT ventral view; (b) ddx3xa−/− ventral view. (D) bmp4 expression levels in ddx3xa−/− embryos at 24, 48, and 96 hpf by qRT-PCR (n = 3; ***p < 0.001 vs. WT; ns: nonsignificant). (E) tbx5 expression levels in ddx3xa−/− embryos at 24, 48, and 96 hpf by qRT-PCR (n = 3; *p < 0.05, ***p < 0.001 vs. WT).
Building on these findings, we investigated whether ddx3xa modulates cardiac development via the Wnt pathway. Whole-mount in situ hybridization (WISH) analysis of Wnt-associated cardiac genes bmp4 and tbx5 revealed spatial expression abnormalities: tbx5 exhibited intensified dorsal labeling accompanied by expanded expression domains within the cardiac field (Figure 8B), while bmp4 displayed pronounced atrial-specific expression expansion at 48 hpf (Figure 8C). These spatial anomalies provide direct support for the proposed mechanism.
To resolve developmental dynamics, we analyzed the temporal expression of these genes. In ddx3xa−/− mutants, bmp4 was specifically upregulated at 96 hpf (Figure 8D), whereas tbx5 showed sustained upregulation at all stages examined (24, 48, and 96 hpf; Figure 8E). Integrated spatiotemporal analysis demonstrates that ddx3xa deficiency not only disrupts the spatial patterning of Wnt-dependent cardiac genes but, more critically, disrupts their temporally regulated expression program, leading to dysregulated coordination of key factors essential for cardiac morphogenesis.
2.6 Rescue experiments confirm ddx3xa modulates cardiac development through wnt signaling
To further confirm that ddx3xa regulates cardiac development through the Wnt signaling pathway, we performed rescue experiments using IWR-1, a specific inhibitor of β-catenin/TCF interactions in the canonical Wnt pathway. Wild-type (WT), ddx3xa−/−, myl7:EGFP transgenic line, and ddx3xa−/−; myl7:EGFP double-homozygous embryos were treated with IWR-1, and cardiac phenotypes were assessed at 72 hpf.
Untreated ddx3xa−/− embryos exhibited severe cardiac abnormalities, including pronounced pericardial edema, ventricular reduction with atrophy, atrial dilation, and overall cardiac elongation (Figure 9A; Supplementary Table S3). Notably, IWR-1 treatment significantly ameliorated cardiac malformations in ddx3xa−/− embryos at 72 hpf: pericardial edema was reduced, ventricular size was partially restored, atrial dilation was alleviated, and heart tube morphology appeared more compact and organized (Figure 9A; Supplementary Table S3). This phenotypic rescue was consistently observed in both bright-field and fluorescence views (myl7:EGFP-labeled cardiac tissue), confirming that Wnt pathway inhibition improves structural defects caused by ddx3xa deficiency.
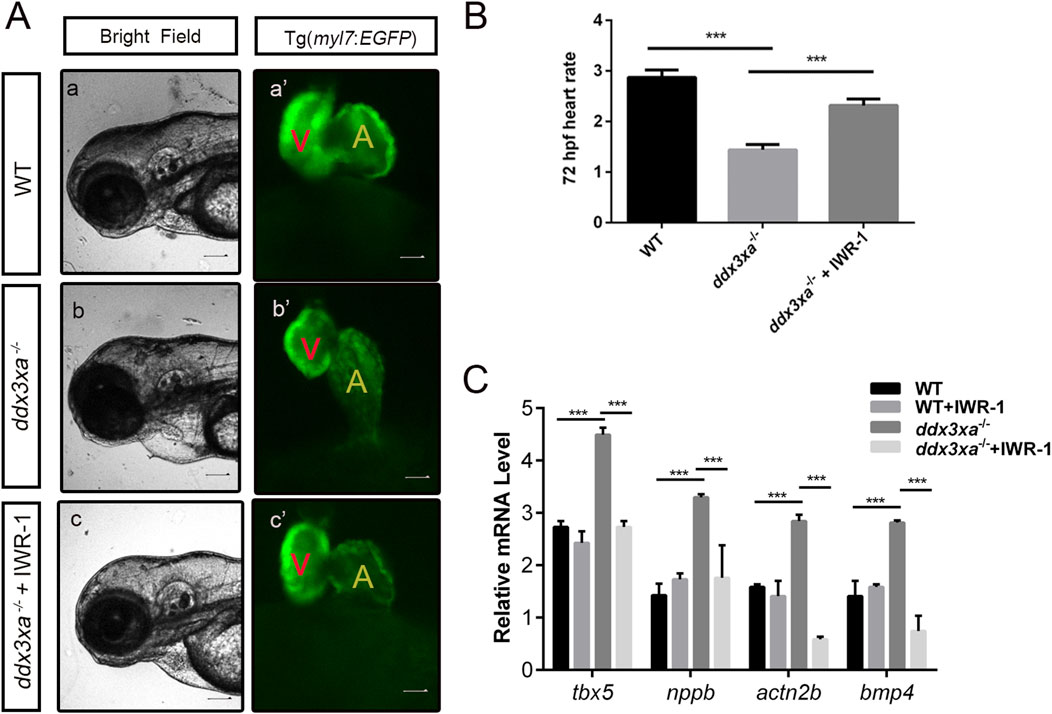
Figure 9. Pharmacological rescue of cardiac defects by Wnt inhibition. (A) Cardiac morphology at 72 hpf. Brightfield (A–C) and corresponding myl7:EGFP fluorescence (a'-c') views. (a,a') Wild-type (WT); (b,b') ddx3xa−/−; myl7:EGFP homozygous mutant; (c,c') ddx3xa−/−; myl7:EGFP mutant treated with IWR-1. Scale bar: 250 μm. (B) Heart rate quantification: ddx3xa−/− versus IWR-1-treated ddx3xa−/− mutants (n = 3; ***P < 0.001). (C) qRT-PCR analysis of Wnt-dependent cardiac genes (tbx5, nppb, actn2b, bmp4) in WT, WT + IWR-1, ddx3xa−/−, and ddx3xa−/− +IWR-1 embryos at 72 hpf (n = 3; ***P < 0.001).
Cardiac functional rescue was further validated by assessing heart rate. At 72 hpf, untreated ddx3xa−/− embryos showed significantly reduced cardiac contraction frequency (beats per 10 s) compared to WT. IWR-1 treatment markedly increased heart rate in ddx3xa−/− embryos, partially restoring cardiac function (Figure 9B).
To investigate the molecular basis of rescue, we analyzed expression of Wnt-downstream cardiac regulatory genes (tbx5, nppb, actn2b, bmp4) via qRT-PCR in four groups at 72 hpf: WT, WT + IWR-1, ddx3xa−/−, and ddx3xa−/−+IWR-1. Results demonstrated that IWR-1 treatment significantly downregulated tbx5, nppb, actn2b, and bmp4 expression compared to untreated ddx3xa−/− embryos (Figure 9C). These molecular changes align with observed morphological improvements, confirming that IWR-1-mediated Wnt inhibition restores dysregulated expression of key cardiac developmental genes caused by ddx3xa knockout.
Collectively, these results demonstrate that pharmacological inhibition of Wnt signaling by IWR-1 not only rescues structural abnormalities (pericardial edema, ventricular atrophy, atrial dilation) but also improves cardiac function and normalizes critical cardiac regulatory gene expression in ddx3xa−/− zebrafish. This provides direct evidence that ddx3xa governs early cardiac development and function through modulation of the Wnt signaling pathway.
3 Discussion
This study provides the first systematic in vivo evidence establishing a direct causal link between DDX3X deficiency and cardiac developmental defects. Utilizing a CRISPR/Cas9-generated ddx3xa homozygous knockout zebrafish model (Sun et al., 2020), we recapitulated potential cardiac comorbidities observed in DDX3X syndrome patients. Mutant embryos exhibited a high malformation rate (25.26%) at 48–72 h post-fertilization (hpf), characterized by pericardial edema and defective cardiac looping (Figure 5), phenotypes highly concordant with clinically reported cardiac structural abnormalities (von Mueffling et al., 2024). Notably, coupled with early-onset developmental delay (Supplementary Figure S1), underscoring the fundamental role of DDX3X in embryonic development and organogenesis. The molecular fidelity of our model is supported by the high amino acid sequence homology (>75%) between zebrafish Ddx3xa and human DDX3X, particularly within the conserved RNA-binding domains (Figure 1). This model addresses a critical gap in DDX3X syndrome research, and its highly reproducible cardiac phenotypes -including atrial hypoplasia, ventricular dysmorphology, and impaired contractile function–provide an invaluable in vivo platform for dissecting the pathological mechanisms underlying DDX3X mutation-associated cardiac disorders in humans. Considering the clinical profile of DDX3X syndrome, our findings further suggest that cardiac abnormalities may represent an under-recognized comorbid phenotype warranting increased clinical attention.
Based on our integrated findings, we propose a mechanistic model wherein ddx3xa governs cardiac morphogenesis through modulation of the Wnt/β-catenin signaling pathway (Figure 10).
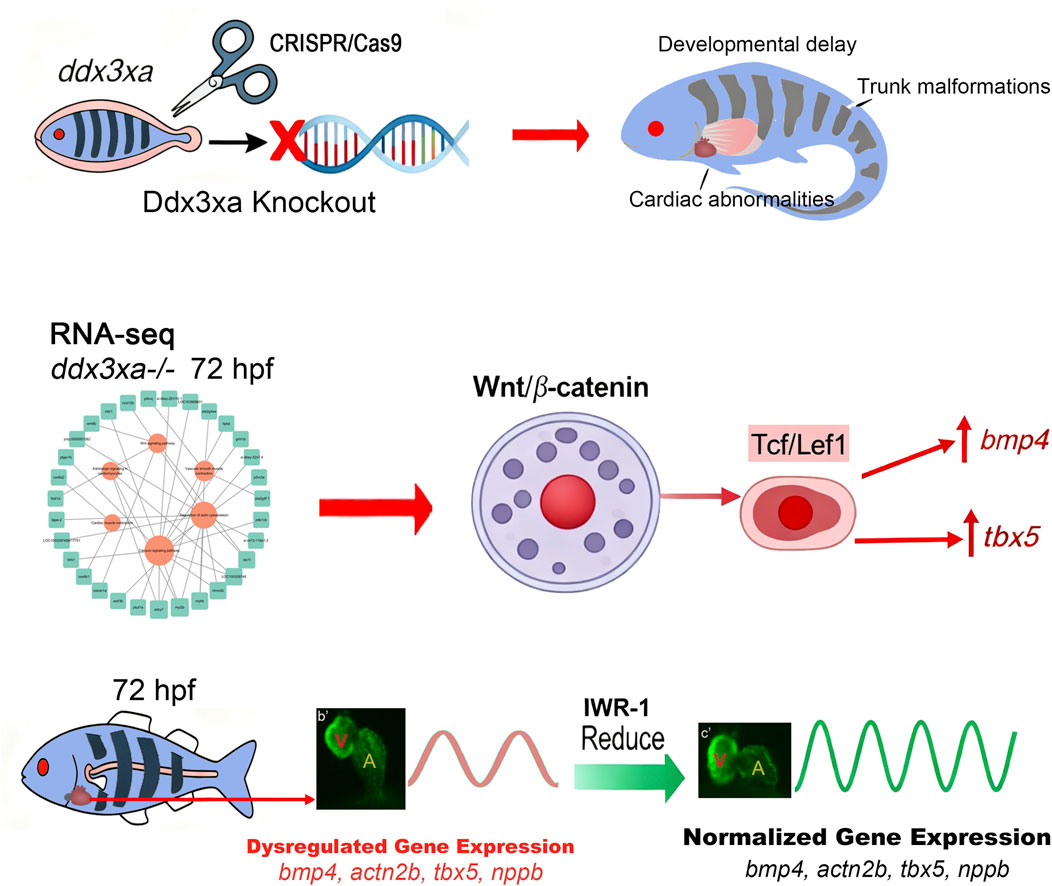
Figure 10. ddx3xa Governs Cardiac Morphogenesis Through Wnt/β-Catenin Signaling. Schematic diagram illustrating the mechanistic role of ddx3xa in zebrafish cardiac development. CRISPR/Cas9-mediated ddx3xa knockout results in developmental defects including developmental delay, trunk malformations, and cardiac abnormalities characterized by pericardial edema and impaired looping. Transcriptomic profiling of ddx3xa−/− mutants at 72 hpf identifies dysregulation of key cardiac developmental genes (bmp4, actn2b, tbx5, nppb) and reveals hyperactivation of the Wnt/β-catenin pathway. Mechanistically, ddx3xa deficiency upregulates the transcription factor Tcf/Lef1, leading to aberrant expression of its downstream targets bmp4 and tbx5. Pharmacological inhibition of Wnt signaling with IWR-1 normalizes the expression of these cardiac genes and partially rescues the morphological defects. This work establishes ddx3xa as a critical regulator of cardiac development through modulation of the Wnt/β-catenin pathway, providing novel mechanistic insights into the cardiac comorbidities of DDX3X syndrome and highlighting the therapeutic potential of Wnt pathway modulation.
Cardiac development relies on precise spatiotemporal regulation by intricate signaling networks (Liu and Foley, 2011). Our research findings approach identified the Wnt/β-catenin pathway as the central hub mediating the teratogenic effects of ddx3xa deficiency. Transcriptome sequencing revealed significant enrichment of the Wnt pathway in 72 hpf ddx3xa−/− embryos, concurrently dysregulated alongside actin cytoskeleton organization, calcium signaling, and cardiac muscle contraction pathways (Figure 6D). Mechanistically, ddx3xa knockout induced aberrant downregulation of the key Wnt transcriptional mediator Tcf/Lef1, while expression of upstream ligands (wnt3, wnt5b), cytoplasmic transducer (dvl2), and the effector β-catenin (ctnnb1) was paradoxically upregulated (Figure 8A). This “downstream activation-upstream suppression” paradox suggests that ddx3xa may regulate Tcf/Lef1 through non-canonical mechanisms. Crucially, severe spatiotemporal dysregulation was observed for Wnt-dependent cardiac target genes: bmp4 showed abnormally high expression at 96 hpf concomitant with atrial enlargement (Figures 8C,D), while tbx5 exhibited sustained upregulation and aberrant dorsal expansion within the heart field across multiple developmental stages (Figures 8B,E). Given that BMP4 drives cardiac chamber septation and TBX5 regulates atrioventricular differentiation, their dysregulated expression directly explains the observed atrial hypoplasia and looping failure in mutants (Figure 5). We propose that ddx3xa, potentially via its RNA helicase activity, could modulate the mRNA stability or translation efficiency of these target genes, or influence phase separation of Wnt pathway components (analogous to ddx3xb’s role in maternal-to-zygotic transition) (Shen et al., 2022; Shi et al., 2022), offering novel perspectives on how DDX3X fine-tunes the cardiac developmental gene network.
Cardiac contractile dysfunction, manifesting as bradycardia, was another prominent phenotype in ddx3xa mutants. Our study delineated a molecular-to-physiological cascade linking this defect to Wnt signaling dysregulation. qRT-PCR confirmed significant downregulation of key genes regulating myocardial contraction (myh7, nppa, plcd1a, rac1) and calcium signaling (itpka, plcd1a) in mutants (Figures 5T, 7A). These genes are directly involved in sarcoplasmic reticulum calcium cycling and myofilament sliding–for instance, PLCD1a hydrolyzes PIP2 to regulate calcium release, while RAC1 influences actin polymerization (Leung et al., 2014; Vachel et al., 2015). Their impaired expression resulted in significantly reduced heart rates (Figures 5Q–S), reflecting diminished contractile force. Intriguingly, KEGG analysis indicated co-enrichment of “Cardiac muscle contraction” and “Wnt signaling” pathways (Figure 6D), a connection substantiated by a pathway-gene network that integrated these enriched pathways and demonstrated strong interactions between Wnt downstream factors and contractile genes (Figure 7B). Experimentally, we demonstrated that aberrant Tcf/Lef1 upregulation suppresses the expression of nkx2.5 and myosin genes (myh7, myl7) (Figures 5O,P,T), directly disrupting contractile unit assembly. Furthermore, persistent Wnt/β-catenin activation is known to inhibit cardiomyocyte differentiation and promote fibrosis (Garcia-Gras et al., 2006). Our finding of similar phenotypes in ddx3xa mutants suggests that DDX3X deficiency, via chronic Wnt hyperactivation, may drive cardiomyocytes towards pathological remodeling, providing mechanistic insights into potential arrhythmogenic risks in DDX3X syndrome patients.
The most pivotal breakthrough of this study lies in the phenotypic rescue achieved through targeted Wnt pathway intervention, providing direct evidence for causality. Treatment of 72 hpf mutant embryos with the specific Wnt/β-catenin inhibitor IWR-1 resulted in: (1) Morphological Rescue: Significant reduction in pericardial edema, marked improvement in ventricular hypoplasia and atrial enlargement, and partial restoration of cardiac looping (Figure 9A). (2) Functional Recovery: Substantial increase in heart contraction frequency (Figures 9B,C), indicating enhanced pumping capacity. (3) Molecular Correction: Effective suppression of aberrantly high expression of Wnt-dependent cardiac genes (tbx5, nppb, actn2b, bmp4) (Figure 9D). This multi-level rescue unequivocally confirms that the cardiac defects in ddx3xa mutants are fundamentally driven by hyperactivated Wnt/β-catenin signaling. Notably, IWR-1 acts upstream of Tcf/Lef1 by stabilizing the AXIN complex to promote β-catenin degradation (Lu et al., 2009). Its ability to downregulate Tcf/Lef1 target genes (bmp4, tbx5) demonstrates that the aberrant Tcf/Lef1 activation caused by ddx3xa deficiency is reversible via upstream intervention. This offers proof-of-concept for treating DDX3X syndrome-associated cardiac disease-small-molecule Wnt inhibitors (IWR-1) represent potential therapeutic strategies. The partial nature of the rescue (incomplete normalization of cardiac morphology) suggests that ddx3xa may also co-regulate cardiac development through additional pathways (calcium signaling, adrenergic signaling; Figure 7), necessitating combined targeted interventions for optimal efficacy.
This study is the first to integrate DDX3X deficiency, Wnt signaling dysregulation, and cardiac maldevelopment within a unified mechanistic framework, opening new dimensions for DDX3X syndrome research. The high-throughput capabilities of the zebrafish model provide a powerful platform for accelerating mechanistic dissection and therapeutic development. However, several key questions warrant further investigation: (1) Precise Molecular Interface of ddx3xa-Wnt Regulation: Does DDX3X directly bind Wnt pathway components (CSNK2A1 or β-catenin mRNA)? How does its helicase activity influence Tcf/Lef1 transcriptional complex assembly? (2) Cardiac-Specific Mechanisms: Why does ddx3xa deficiency selectively impact the heart despite expression in other organs? Tissue-specific knockout or single-cell sequencing is needed to elucidate cell-autonomous effects in cardiomyocytes. (3) Sex-Specific Considerations: Given the female predominance of DDX3X syndrome and the autosomal location of zebrafish ddx3xa, the compensatory effects of the DDX3Y homolog and the influence of sex hormone microenvironments require exploration. (4) Functional Validation of Clinical Mutations: Introducing patient-derived cardiac-associated DDX3X mutations into the zebrafish model is crucial to assess their specific impact on Wnt signaling and cardiac phenotypes.
In conclusion, this work not only establishes the central role of the ddx3xa-Wnt axis in cardiac development but also builds a bridge connecting fundamental research to clinical translation. Future integration of patient-derived induced pluripotent stem cell (iPSC)-cardiomyocyte models with in vivo zebrafish screening holds significant promise for developing precision therapeutic strategies targeting cardiac comorbidities in DDX3X syndrome.
4 Materials and methods
4.1 Zebrafish husbandry
Wild-type AB strain zebrafish (Danio rerio) were maintained at 28.5 °C ± 0.5 °C under a 15-h light/9-h dark photoperiod. Embryos were reared in E3 embryo medium (pH 7.0; conductivity ≈600 μS/cm) until 7 days post-fertilization (dpf), fed Paramecium from 1-7 dpf and Artemia nauplii thereafter, then transferred to recirculating aquaculture systems (Aisheng Technology).
4.2 Reagents and instrumentation
Key reagents included: FastPure RNA Purification Kit (Vazyme), dNTP Mix (Takara), anti-DDX3X antibody (Genecopoeia), Cas9 protein (Thermo Fisher), restriction enzymes (Promega), and T7 Transcription Kit (Promega). Major instruments comprised: PCR Thermal Cycler (Eppendorf), Microinjection System (Harvard Apparatus PLI-100A), Confocal Microscope (Leica), and qPCR System (Eppendorf) (Complete lists: Supplementary Tables S4,S5).
4.3 Primer design
sgRNA target sites for ddx3xa knockout were designed via ZIFIT (avoiding UTRs; NGG PAM), while qRT-PCR primers were designed using Primer Premier 5.0 (Supplementary Table S6).
4.4 Generation of ddx3xa mutants
sgRNAs were synthesized by PCR-amplifying T7 promoter-containing templates (Supplementary Table S3), followed by in vitro transcription (T7 Kit) and purification (FastPure RNA Kit). For microinjection, 500 pg sgRNA and 300 pg Cas9 protein were co-injected into one-cell embryos, with injected embryos incubated in E3 medium at 28 °C.
4.5 Genotyping
Genomic DNA was extracted from embryos/fin clips using alkaline lysis (50 mM NaOH, 95 °C, 10 min). Target regions were amplified by PCR (primers ddx3xa-F/R) and mutations verified via Sanger sequencing.
4.6 Transcriptome profiling via RNA-Seq
Zebrafish embryos at 72 hpf were batched into biological replicates (50 embryos per tube) and flash-frozen at −80 °C. RNA sequencing was performed commercially by Personalbio GeneCloud (Shanghai, China), with subsequent bioinformatic analyses executed on their proprietary platform (https://www.genescloud.cn/home). Pathway-gene target networks were visualized using Cytoscape software.
4.7 Imaging and quantitative analysis
Cardiac morphology in myl7:EGFP transgenics was documented by fluorescence stereomicroscopy (Leica M205FA). Whole-mount in situ hybridization specimens were imaged using a Leica digital system (v3.2.0), while larval phenotypes (24–48 hpf) were captured with a motorized Axiocam (Zeiss) and processed using AxioVision 3.0.6 and Adobe Photoshop. Cardiac function was assessed by recording ventricular contractions at 130 fps for 10-s intervals via high-speed EM-CCD camera (×48 magnification), with semi-automated analysis generating quantitative parameters including heart rate, visualized through M-mode imaging.
4.8 Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted (FastPure Kit), reverse-transcribed (Hifair III SuperMix), and amplified (SYBR Green Master Mix; Eppendorf). Using β-actin as reference gene, data were analyzed in GraphPad Prism 6.0 via Student’s t-test (P < 0.05 significance threshold).
4.9 Whole-mount in situ hybridization (WISH)
Digoxigenin-labeled antisense probes (500–1000 bp spanning ddx3xa 3′-UTR) were transcribed from PCR templates. Embryos were fixed in 4% PFA, dehydrated/rehydrated, treated with proteinase K (stage-optimized concentration/duration), pre-hybridized (65 °C, 4–6 h), and hybridized overnight (65 °C). Post-hybridization washes included sequential treatments with 50% formamide/2× SSCT, 2× SSCT, and 0.2× SSCT. After blocking, embryos were incubated with anti-DIG-AP antibody (1:3,000) and developed with NBT/BCIP, with images acquired by stereomicroscopy.
4.10 Pharmacological rescue
Adult zebrafish were sex-separated (1♀:1♂ or 1♀:2♂ ratio) in breeding tanks with transparent dividers, dark-adapted for ≥6 h, then paired under light conditions. At 6 hpf, embryos were exposed to control solution (0.05% DMSO in E3) or experimental treatment (10 nM IWR-1 in E3) in 6-well plates (20 embryos/well). Malformation and mortality rates were recorded at 24, 48, and 72 hpf, with heart rate and ejection fraction analyzed at 72 hpf via high-speed videomicroscopy.
4.11 Statistical analysis
All experiments included ≥3 biological replicates, with statistical significance determined by Student’s t-test (P < 0.05).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Biomedical Research Ethics Committee of Hunan Normal University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YC: Conceptualization, Data curation, Funding acquisition, Investigation, Writing – original draft, Writing – review and editing. ML: Investigation, Methodology, Writing – original draft. PZ: Data curation, Funding acquisition, Investigation, Writing – review and editing. HW: Formal Analysis, Writing – original draft. ZoJ: Formal Analysis, Writing – original draft. KY: Data curation, Investigation, Writing – review and editing. XY: Data curation, Investigation, Methodology, Writing – review and editing. YZ: Methodology, Writing – review and editing. XC: Investigation, Writing – review and editing. WY: Investigation, Writing – original draft. YL: Project administration, Writing – original draft. ZiJ: Conceptualization, Writing – original draft. YW: Project administration, Writing – review and editing. FL: Conceptualization, Methodology, Writing – original draft, Writing – review and editing. XW: Investigation, Writing – review and editing, Writing – original draft. XF: Conceptualization, Writing – review and editing, Writing – original draft, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China [No. 82400414 (YC); 81970324 (XF); 81800289 (FL)]. “Guangdong Special Support Plan” Provincial Health and Health Commission (Health Talent) Project (0720240240) (YC), Guangdong Major Project of Basic and Applied Basic Research (2023B0303000005) (PZ). Guangdong Provincial Special Support Program for Prominent Talents (2021JC06Y656) (PZ). Special Project of the National Natural Science Foundation of China (42542044) (XF). The Scientific Research Foundation of Hunan Provincial Educational Committee of China (No. 23A0082; 202401000473; Z2024167) (FL). the open research fund of Hunan Provincial Key Laboratory of Regional Hereditary Birth Defects Prevention and Control (Grant No. HPKL2024002) (XF).
Acknowledgments
AcknowledgementsRNA sequencing was performed commercially by Personalbio.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2025.1689202/full#supplementary-material
References
Boitnott, A., Garcia-Forn, M., Ung, D. C., Niblo, K., Mendonca, D., Park, Y., et al. (2021). Developmental and behavioral phenotypes in a mouse model of DDX3X syndrome. Biol. Psychiatry 90, 742–755. doi:10.1016/j.biopsych.2021.05.027
Bol, G. M., Xie, M., and Raman, V. (2015). DDX3, a potential target for cancer treatment. Mol. Cancer 14, 188. doi:10.1186/s12943-015-0461-7
Cruciat, C. M., Dolde, C., De Groot, R. E. A., Ohkawara, B., Reinhard, C., Korswagen, H. C., et al. (2013). RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Sci. (80) 339, 1436–1441. doi:10.1126/science.1231499
Dahme, T., Katus, H. A., and Rottbauer, W. (2009). Fishing for the genetic basis of cardiovascular disease. Dis. Model. Mech. 2, 18–22. doi:10.1242/dmm.000687
Devasahayam Arokia Balaya, R., Kanekar, S., Kumar, S., and Kandasamy, R. K. (2025). Role of DEAD/DEAH-box helicases in immunity, infection and cancers. Cell Commun. Signal. 23, 292. doi:10.1186/s12964-025-02225-9
Dikow, N., Granzow, M., Graul-Neumann, L. M., Karch, S., Hinderhofer, K., Paramasivam, N., et al. (2017). DDX3X mutations in two girls with a phenotype overlapping toriello–carey syndrome. Am. J. Med. Genet. Part A 173, 1369–1373. doi:10.1002/ajmg.a.38164
Ditton, H. J., Zimmer, J., Kamp, C., Rajpert-De Meyts, E., and Vogt, P. H. (2004). The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 13, 2333–2341. doi:10.1093/hmg/ddh240
Dooley, K., and Zon, L. I. (2000). Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 10, 252–256. doi:10.1016/S0959-437X(00)00074-5
Duan, W., Huang, G., Sui, Y., Wang, K., Yu, Y., Chu, X., et al. (2024). Deficiency of DDX3X results in neurogenesis defects and abnormal behaviors via dysfunction of the notch signaling. Proc. Natl. Acad. Sci. U. S. A. 121, e2404173121. doi:10.1073/pnas.2404173121
Garcia-Gras, E., Lombardi, R., Giocondo, M. J., Willerson, J. T., Schneider, M. D., Khoury, D. S., et al. (2006). Suppression of canonical wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Invest. 116, 2012–2021. doi:10.1172/JCI27751
Jankowsky, A., Guenther, U. P., and Jankowsky, E. (2011). The RNA helicase database. Nucleic Acids Res. 39, D338–D341. doi:10.1093/nar/gkq1002
Kim, Y. S., Lee, S. G., Park, S. H., and Song, K. (2001). Gene structure of the human DDX3 and chromosome mapping of its related sequences. Mol. Cells 12, 209–214. doi:10.1016/s1016-8478(23)17085-3
Lawrence, C. (2007). The husbandry of zebrafish (danio rerio): a review. Aquaculture 269, 1–20. doi:10.1016/j.aquaculture.2007.04.077
Lennox, A. L., Hoye, M. L., Jiang, R., Johnson-Kerner, B. L., Suit, L. A., Venkataramanan, S., et al. (2020). Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron 106, 404–420. doi:10.1016/j.neuron.2020.01.042
Leung, C., Lu, X., Liu, M., and Feng, Q. (2014). Rac1 signaling is critical to cardiomyocyte polarity and embryonic heart development. J. Am. Heart Assoc. 3, e001271. doi:10.1161/JAHA.114.001271
Li, D., Sun, J., and Zhong, T. P. (2022). Wnt signaling in heart development and regeneration. Curr. Cardiol. Rep. 24, 1425–1438. doi:10.1007/s11886-022-01756-8
Linder, P., and Jankowsky, E. (2011). From unwinding to clamping — the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516. doi:10.1038/nrm3154
Liu, W., and Foley, A. C. (2011). Signaling pathways in early cardiac development. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 191–205. doi:10.1002/wsbm.112
Lombardi, R., Da Graca Cabreira-Hansen, M., Bell, A., Fromm, R. R., Willerson, J. T., and Marian, A. J. (2011). Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ. Res. 109, 1342–1353. doi:10.1161/CIRCRESAHA.111.255075
Lu, J., Ma, Z., Hsieh, J. C., Fan, C. W., Chen, B., Longgood, J. C., et al. (2009). Structure-activity relationship studies of small-molecule inhibitors of wnt response. Bioorg. Med. Chem. Lett. 19, 3825–3827. doi:10.1016/j.bmcl.2009.04.040
Luo, T., Yang, S., Zhao, T., Zhu, H., Chen, C., Shi, X., et al. (2023). Hepatocyte DDX3X protects against drug-induced acute liver injury via controlling stress granule formation and oxidative stress. Cell Death Dis. 14, 400. doi:10.1038/s41419-023-05913-x
Ma, S., Mao, Q., Weng, S., Teng, M., Luo, J., and Zhang, K. (2025). DDX3X and virus interactions: functional diversity and antiviral strategies. Front. Microbiol. 16, 1630068. doi:10.3389/fmicb.2025.1630068
Medina, M. A., Ugarte, G. D., Vargas, M. F., Avila, M. E., Necuñir, D., Elorza, A. A., et al. (2016). Alternative RUNX1 promoter regulation by Wnt/β-Catenin signaling in leukemia cells and human hematopoietic progenitors. J. Cell. Physiol. 231, 1460–1467. doi:10.1002/jcp.25258
Mo, J., Liang, H., Su, C., Li, P., Chen, J., and Zhang, B. (2021). DDX3X: structure, physiologic functions and cancer. Mol. Cancer 20, 38. doi:10.1186/s12943-021-01325-7
Naito, A. T., Shiojima, I., Akazawa, H., Hidaka, K., Morisaki, T., Kikuchi, A., et al. (2006). Developmental stage-specific biphasic roles of wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 103, 19812–19817. doi:10.1073/pnas.0605768103
Nishimoto, S., Wilde, S. M., Wood, S., and Logan, M. P. O. (2015). RA acts in a coherent feed-forward mechanism with Tbx5 to control limb bud induction and initiation. Cell Rep. 12, 879–891. doi:10.1016/j.celrep.2015.06.068
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. doi:10.1016/j.cell.2017.05.016
Owsianka, A. M., and Patel, A. H. (1999). Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257, 330–340. doi:10.1006/viro.1999.9659
Padmanabhan, P. K., Zghidi-Abouzid, O., Samant, M., Dumas, C., Aguiar, B. G., Estaquier, J., et al. (2016). DDX3 DEAD-box RNA helicase plays a central role in mitochondrial protein quality control in leishmania. Cell Death Dis. 7, e2406. doi:10.1038/cddis.2016.315
Shen, H., Yanas, A., Owens, M. C., Zhang, C., Fritsch, C., Fare, C. M., et al. (2022). Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol. Cell 82, 2588–2603.e9. doi:10.1016/j.molcel.2022.04.022
Shi, B., Heng, J., Zhou, J. Y., Yang, Y., Zhang, W. Y., Koziol, M. J., et al. (2022). Phase separation of Ddx3xb helicase regulates maternal-to-zygotic transition in zebrafish. Cell Res. 32, 715–728. doi:10.1038/s41422-022-00655-5
Sun, Y., Zhang, B., Luo, L., Shi, D. L., Wang, H., Cui, Z., et al. (2020). Systematic genome editing of the genes on zebrafish chromosome 1 by CRISPR/Cas9. Genome Res. 30, 118–126. doi:10.1101/gr.248559.119
Tang, L., Levy, T., Guillory, S., Halpern, D., Zweifach, J., Giserman-Kiss, I., et al. (2021). Prospective and detailed behavioral phenotyping in DDX3X syndrome. Mol. Autism 12, 36. doi:10.1186/s13229-021-00431-z
Towbin, J. A. (2011). Desmosomal gene variants in patients with “possible ARVC.”. Hear. Rhythm 8, 719–720. doi:10.1016/j.hrthm.2011.03.040
Vachel, L., Norez, C., Jayle, C., Becq, F., and Vandebrouck, C. (2015). The low PLC-δ1 expression in cystic fibrosis bronchial epithelial cells induces upregulation of TRPV6 channel activity. Cell Calcium 57, 38–48. doi:10.1016/j.ceca.2014.11.005
von Mueffling, A., Garcia-Forn, M., and De Rubeis, S. (2024). DDX3X syndrome: from clinical phenotypes to biological insights. J. Neurochem. 168, 2147–2154. doi:10.1111/jnc.16174
Wang, M., Yin, C., Wu, Z., Wang, X., Lin, Q., Jiang, X., et al. (2023). The long transcript of lncRNA TMPO-AS1 promotes bone metastases of prostate cancer by regulating the CSNK2A1/DDX3X complex in Wnt/β-catenin signaling. Cell Death Discov. 9, 287. doi:10.1038/s41420-023-01585-w
Winnard, P. T., Vesuna, F., Bol, G. M., Gabrielson, K. L., Chenevix-Trench, G., ter Hoeve, N. D., et al. (2024). Targeting RNA helicase DDX3X with a small molecule inhibitor for breast cancer bone metastasis treatment. Cancer Lett. 604, 217260. doi:10.1016/j.canlet.2024.217260
Ye, B., Li, L., Xu, H., Chen, Y., and Li, F. (2019). Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development. Lab. Investig. 99, 807–818. doi:10.1038/s41374-019-0204-2
Keywords: ddx3xa, zebrafish, CRISPR/Cas9, cardiac defects, wnt/β-catenin signaling, IWR-1 rescue
Citation: Chen Y, Lin M, Zhu P, Wang H, Jiao Z, Yi K, Yang X, Zhang Y, Cai X, Yuan W, Li Y, Jiang Z, Wang Y, Li F, Wu X and Fan X (2025) Ddx3xa mutations drive cardiac defects in a zebrafish model via dysregulation of wnt/β-catenin signaling. Front. Mol. Biosci. 12:1689202. doi: 10.3389/fmolb.2025.1689202
Received: 20 August 2025; Accepted: 27 October 2025;
Published: 27 November 2025.
Edited by:
Naoyuki Nishiya, Iwate Medical University, JapanReviewed by:
Kartick Patra, National Institute of Diabetes and Digestive and Kidney Diseases (NIH), United StatesSubrata Pramanik, Indian Institute of Technology Guwahati, India
Copyright © 2025 Chen, Lin, Zhu, Wang, Jiao, Yi, Yang, Zhang, Cai, Yuan, Li, Jiang, Wang, Li, Wu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Li, bGlmYW5nQGh1bm51LmVkdS5jbg==; Xiushan Wu, eGl1c2hhbnd1MjAwM0BhbGl5dW4uY29t; Xiongwei Fan, ZmFuX3hpb25nd2VpQDE2My5jb20=
†These authors have contributed equally to this work
 Yu Chen
Yu Chen Mei Lin1†
Mei Lin1† Ping Zhu
Ping Zhu Wuzhou Yuan
Wuzhou Yuan Fang Li
Fang Li Xiushan Wu
Xiushan Wu Xiongwei Fan
Xiongwei Fan