- TecNM-Instituto Tecnológico El Llano Aguascalientes (ITEL), El Llano, Mexico
The field of bio-nano interfaces paves the way for a better understanding, development, and implementation of the advanced biotechnological process. Interfacing biomolecules with the nanomaterials will result in the development of new tools and techniques that, in turn, will enable to explore the fundamental process at the nano level and fabricate cost-effective portable devices. Fascinating biomolecules like DNA, RNA and proteins in the regime of nanoscale are intelligent materials that are capable of storing the information and controlling the basic structure and function of the complex biological systems. Following this concept, the current pandemic situation would be a natural selection process, where the selective pressure is on the ssRNA of Covid-19 to choose the suitable progeny for survival. Consequently, the interaction of human DNA invoking response with Covid-19 happens at the nanoscale and it could be a better candidate to provoke combat against the virus. The extent of this interaction would give us the insights at the nanotechnological level to tackle the prevention, diagnosis and treatment for Covid-19. Herein, the possible features and obstacles in Covid-19 and a probable solution from the advent of nanotechnology are discussed to address the current necessity. Moreover, the perspective sustainable green graph mask that can be prepared using green plant extract/graphene (Bio-Nano composite mask) is suggested for the possible protection of virus-like Covid-19. The composite material will not only effectively trap the virus but also inactivate the virus due to the presence of antiviral compounds in the plant extracts.
Introduction
Bio-Nano Interface deals with the convergence between the subcellular biomolecular machinery operating at the nanoscale and their interaction with nanoscale materials. The exploitation of this fundamental interface interaction leads to the emergence of a highly important area of research coined as “Nanobiotechnology.” The research on interfaces under the area of “Nanobiotechnology,” helps in understanding the fundamental biomolecular machinery and its dimensions at the nanoscale. Also, it leads to the attempts of artificial mimicking to invoke specific biological responses at the gene level. The possible interactions with nanoscale materials emerge either from man-made stimuli or through natural stimulations such as the physiochemical environmental interactions (volcanic eruption, cosmic dust, etc.,) with/without the involvement of biological systems (i.e., Carbon quantum dot presents naturally in honey, etc.) were explored. Hence, the interfacing of biological molecules using the principles and techniques in nanotechnology will aid in the understanding, development and improvement of the advanced biotechnological process essential for the product development. Implantable medical devices, nano-robots are one among the few examples of devices developed using the interface science.
Biomolecules operating at the nano level are intelligent nanomachines capable of storing the genetic information and controlling the complex biological macrosystems. The role of biomolecules in combating the diseases can be traced back to the sixth century. The deadliest plague of Justinian in the sixth century eradicated half of the global population at that time. Later on, in the fourteenth century, high mortality due to the black death was reported. In 1918 influenza pandemic infected one in every three people on the global population (Wagner et al., 2014; Rai et al., 2020). From the history of infectious diseases (might be a selective pressure), it is learned that the human race is continuously surviving with the herd immunity as a result of natural selection. By the conceptual natural evolution, the ssRNA of virus interacts with the DNA of Human species and leads to the outbreak of a pandemic disease designated as Covid-19 (SARS-CoV-2). Possibly this might be due to the selective pressure to choose the suitable progeny for the next generation. Subsequently, the interaction of engineered nanomaterials with biological functional materials at the nanoscale opens an avenue for efficient prevention, diagnostics, and treatment that can provokes the combat against the virus. Herein, the exploration of the aspects of interfacial Bio-nanotechnology would give an insight into the possible solutions for the pandemic of Covid-19.
Life Cycle of COVID-19
In the first phase, the Covid-19 Spike protein (S1 subunits contain receptor-binding domain (RBD) and S2 subunits) binds with the host cell peptidase domain (PD) of the angiotensin-converting enzyme 2 (ACE2) (Nalawansha and Samarasinghe, 2020). At the same time, S2 subunits cleavage sites are sliced by host serine protease TMPRSS2, and as a result, it leads to membrane fusion and initiates viral entry. Hence, TMPRSS2 and RBD, the interaction sites of Covid-19 are considered as the target for clinical cure by blocking the entry and designing the inhibitor for treatment. The proteolytic cleavage of Covid-19 envelope releases the genomic RNA (Plus strand RNA) in the host cytoplasm. In the next stage, the virion mRNA (Negative strand RNA) exploits the host machinery (transcription, translation, endoplasmic reticulum (ER), and Golgi apparatus), to produce the necessary proteins such as S, E, and M proteins via translation process. Finally, the budding assembly holds the functional proteins and genome RNA and completes the formation of Covid-19 (Zhang and Liu, 2020). The newly synthesized Covid-19 viral particles have to be translocated by vesicles and must be released through exocytosis. This valuable information regarding the invasion, cell multiplication and the life cycle of Covid-19 will aid researchers to design the new therapeutic strategies and repurpose the existing drugs that counteract Covid-19 (Chauhan G. et al., 2020) (Figure 1). From the chemo-proteomic Covid-19 studies, 26 proteins are cloned, tagged and expressed in Human Cells (Skariyachan et al., 2019). As a result of affinity-purification mass spectrometry, 332 protein-proteins interactions are found between human proteins and Covid-19 that are physically associated with each of Covid-19. And the results helped in the identification of 69 lead compounds for host factors/human proteins as a target (Zeng et al., 2020). At the same time, the US Food and Drug Administration has approved 29 drugs, 12 clinical trials, and 28 preclinical compounds (Gordon et al., 2020). In addition to that, multiple ongoing viral assays studies reveal the inhibitors of mRNA translation and regulators of sigma-1 & 2 receptors as the two sets of pharmacological agents that exhibited antiviral activity.
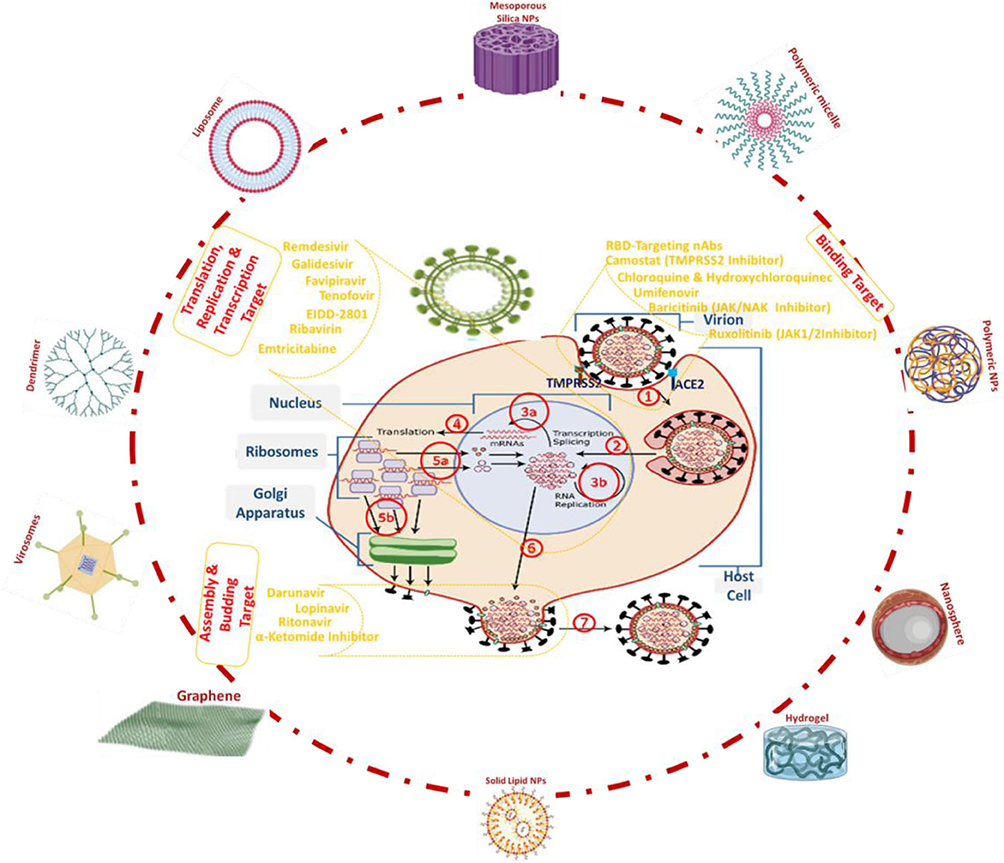
Figure 1. The life cycle of Covid-19, (1) host cell recognition, binding, fusion and endocytosis (2) viral RNA genome is released into the cytoplasm (de Queiroz et al., 2020) (3) uncoating of the RNA and replication (4) translation into two replicate polyproteins (5) translocation of virion assemble and Golgi as vesicles (6) translocation to host cell membrane (7) exocytosis and their possible nanocarrier systems to target Covid-19.
When Covid-19 transmits into the host, the median incubation period is ~5 days (Lauer et al., 2020; Omolo et al., 2020) and the active virus replicates in the tissues of the upper respiratory tract. Pharyngeal virus shedding will be very high during the first week of infections having 7.11 × 108 RNA copies per throat swab on day 4 (Wölfel et al., 2020) and after 7 days, seroconversion occurs in 50% of patients. The presence of viral replicative RNA intermediates in the throat samples, preferably by analyzing throat swab and serological samples for the identification of Covid-19 infections. The presence of viral replicative RNA intermediates in the throat samples, preferably by analyzing throat swab and serological samples for the identification of Covid-19 infections. The fatality rate of COVID-19 is substantially lower, when compared to the earlier reported Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV). Whereas, being more transmissible, the virus has spread to over 213 countries so far, infected more than 9,050,000 people and claimed over 470,000 lives to date (Worldometer, June 22, 2020)1. In the case of Covid-19, the particle diameter is around 50–200 nm (Chen N. et al., 2020) and It can be transmitted through human respiratory bioaerosols that could produce 3,000 droplets and possibly cause infections for 3 h both through airborne droplets and direct contact with infected persons. These bioaerosol droplets can spread from person to person if the people are <2 meters apart or touching surfaces upon which droplets have landed and that have not been disinfected. Recent studies demonstrated that the COVID-19 capable of surviving on plastic and stainless steel for up to 72 h, <4 h on copper and cardboard for <24 h (WHO).
Inspiration From Virology in the Realm of Nanoscience and Nanotechnology
The expansion of crystallographic studies toward biological samples like proteins and other complex organic materials got trigged in the early 1980s. Viruses have DNA or RNA as their functional genetic material and it is surrounded as well as protected by a protein coat (capsid). In some cases that the protein coat envelope is made of lipids. The wide spectrum architecture of the virus is icosahedral, and it possesses a surface antigen that restraints the geometric designs, insinuation for viral evolution, and has a binding affinity that exposes the pathological relevance. In the biological studies, two scientists contributed to the discovery of the first virus, Tobacco mosaic virus. In 1892, Ivanoski reported that the Chamberland filter-candle filtered leaf extract retains the potential infectious behavior. However, Ivanovski did not mention the virus. In 1898, Beijerinck was the first to term it as “virus,” after the incident of the tobacco mosaic. Inadequate but conclusive and complementary contributions to the discovery of viruses fetched by Ivanovski and Beijerinck (Lecoq, 2001). After the development of the first electron microscope by Max Knoll and Ersnt Ruska in 1931, it has been used to study bacteria at around 1938. And the electron microscopy has been used to elucidate the mouse Ectromelia virus, a member of the Poxviridae family. Further, the discovery of enzyme crystallization by J. B. Sumner (Nobel prize in 1946), lead to the structural elucidation of proteins (myoglobin protein, J. C. Kendrew and M. Perutz) and then the discovery of X-ray diffraction studies revealed the DNA structure (F. Crick, Jss. Watson and M. Wilkins) Later, in 1959 Richard Feynman had presented a lecture entitled “There's Plenty of Room at the Bottom” in the annual meeting of the American Physical Society, at the California Institute of Technology (Caltech). This period accelerated the importance of material science and the search toward the nanoscale dimensions. Firstly in 1974, Norio Taniguchi used the word “Nano” meaning “dwarf” in Greece and thereby Taniguchi raised the term “Nanotechnology.” The first high 2.9 Å resolution of Tomato bushy stunt virus was elucidated by Harrison and colleagues in 1978 (Harrison et al., 1978). In 1981, Wilson et al. first elucidated the surface antigen of an influenza virus and this human viral pathogen exposed two glycoproteins (haemagglutinin and neuraminidase) on its lipid membrane envelope. Haemagglutinin glycoprotein is a homotrimer that comprises two structurally distinct regions of a triple-stranded coiled-coil of α-helices and a globular region of antiparallel β-sheet (Wilson et al., 1981). These comprise the host cell surface receptor binding sites that facilitate the initial viral entry, and the variable antigenic determinants that could target and neutralize antibodies. This pioneering crystallography work firmly established the relevance to the viral structures with receptor binding for biomedical applications. The most significant technologic developments were carried out after the use of a scanning tunneling microscope by Binning and Rohrer in 1981. The technique triggered the interest toward visualization, measurement, and manipulation of materials at the nanometer scale, as well as to measure the various forces. With the aid of atomic force microscopy, the enzyme activity (Radmacher et al., 1994) and the RNA polymerase activity (Kasas et al., 1997) at the nanoscale domain could be observed directly. Eric Betzig, Stefan W. Hell and William E. Moerner in 2014, were awarded the Nobel prize for to overcome the limitation of optical microscope resolution enhancement (>0.2 micrometers). Recent advanced frontier technologies such as Microarray, MiniION—nanopore sequencing device and DNA based computation are examples of the synergic combination of materials science with the functional biomolecules. The techniques help in analyzing the functional biomolecule expression of thousands of genes at the same time and were portable to high-throughput benchtop devices for the rapid, accurate and reduced sample processing time. The new techniques with advanced technology could store larger data in memory and conduct multiple operations at once, thus solving decomposable problems much faster than conventional computers. The progress in material science and its contribution to Bio-Nano advancements is summarized in Figure 2. Since the identification of the first virus, the coherence of advanced material science toward nanoscale has contributed to every aspect of understanding the virus from their basic structure, characteristics (i.e., mode of action) and functions at a molecular level. Recently, the structure of the S-proteins of Covid-19 was revealed using cryogenic electron microscopy (Wrapp et al., 2020). To understand the structure, 3207 micrographs were taken and combined into a 3D reconstructed model with a resolution of 0.35 nm (Le Ferrand, 2020). The obtained structural information paves the way to progress the antiviral drug development. A research team led by Tiangang Liu (Wuhan University) and Yan Li (Renmin Hospital) developed a nanopore target sequencing (NTS) method for rapid identification of COVID-19 along with other respiratory viruses concurrently. As one of the prominent field of the past decade, bio-nano interface technology admits prodigious potential contributes innovative solutions to the prevention, diagnosis, and treatment of COVID-19.
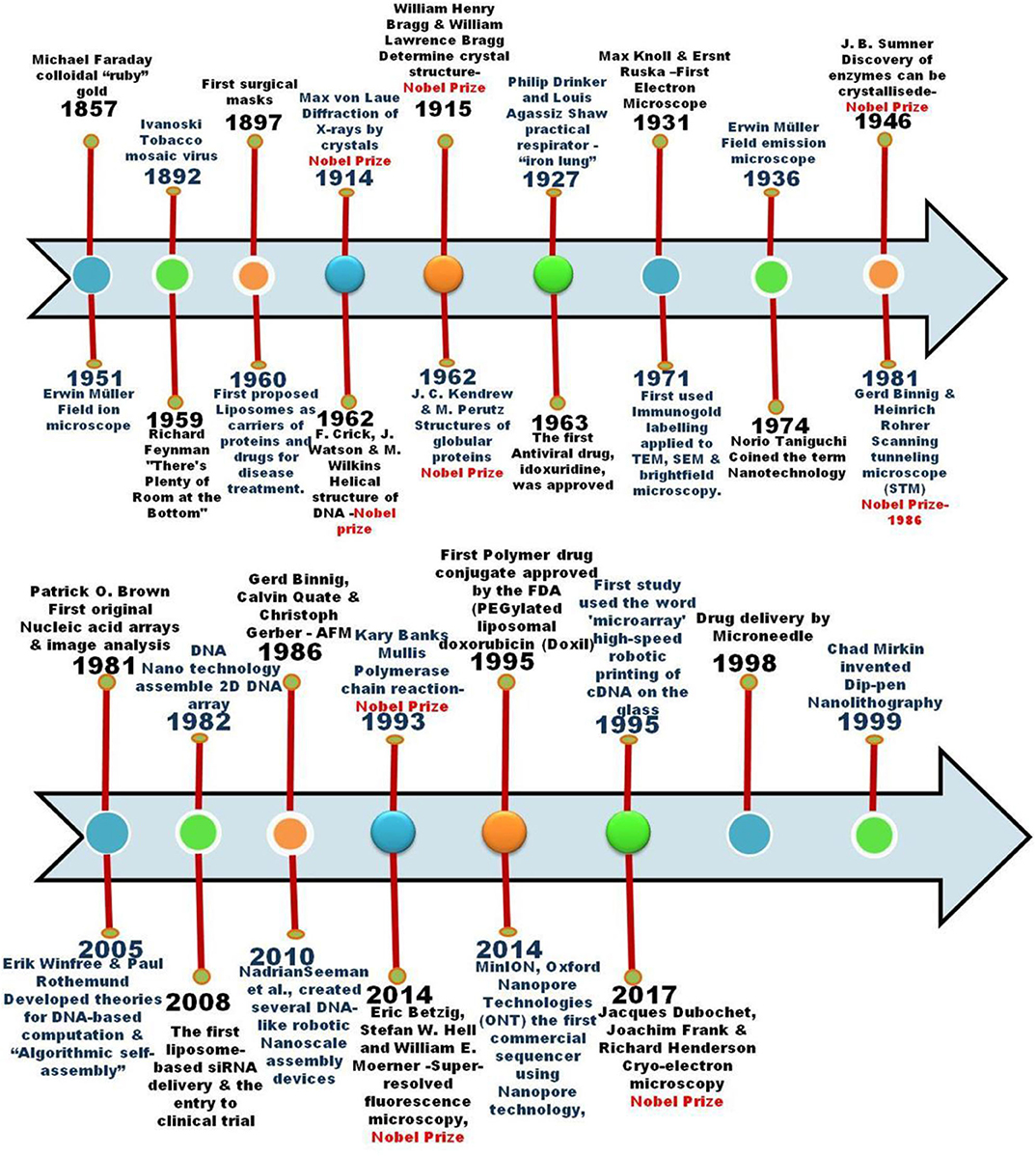
Figure 2. Timeline of progress made in material science toward the contribution of Bio-Nano advancements.
Bio-Nano Interface Technology Toward COVID-19
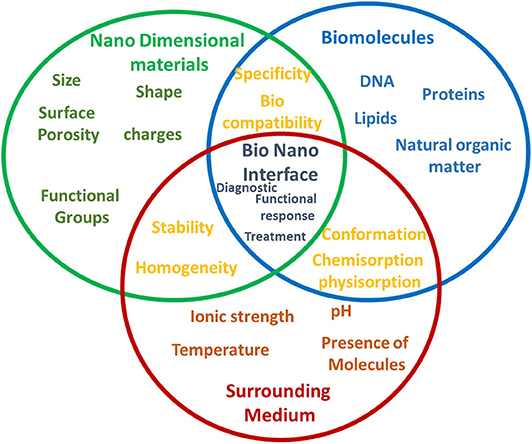
The term “interface” refers to a point where two systems, subjects, organizations, etc. meet and interact. In case of the bio-nano interface, the biological machinery is known to distinguish between natural structures differing from one another at the nanoscale based on a specific functional response. Nano dimensional materials inherits exceptional properties like mechanical, electrical, magnetic, optical, and various chemical and functional properties including the increase in the surface-to-volume ratio of the materials. When bio and nanosystems interact with each other, they could invoke a specific biological functional response and identify the target pathological conditions both at in vitro and in vivo. The physical meaning of the interaction between bio-nano systems is also influenced by the surrounding medium that could interfere and triggers the conformation, chemisorption, physisorption process along with the nanodimentional materials surface properties (Figure 3) (Lynch et al., 2015). Further, this leads to the mechanisms of specificity, cellular uptake, intracellular trafficking as the results provoke the functional response (Nel et al., 2009). Consequences of this effect could act as a key component to interface between the functional biological molecules of virus and human systems and as an advent to the understanding, development and improvement of an astonishing number of novel techniques and tools enabling the solution to Covid-19. Figure 4 shows the information about the patent registration office filing status related to Covid-19, as on July, 18 2020, from Nano-Nature database. This infers the technological insight into the Covid-19 (Gupta and Gupta, 2020). Here, we discuss the recent progress under the commercial product development stage in the field of nanoscale engineering that operates with a bio-nano interface and could potentially be employed to address significant challenges in prevention, detection and treatment of Covid-19.
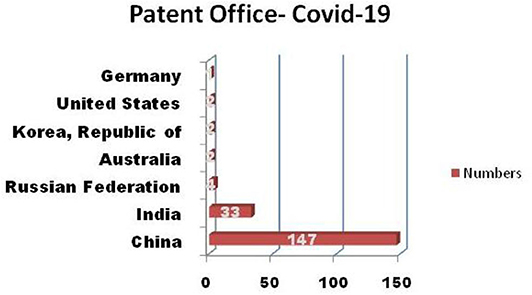
Figure 4. Patent registration office filing status of “Covid-19” from Nano-Nature database (as on 18/07/2020).
Prevention
The priority of the defense is to prevent and control the infectious respiratory disease of COVID-19, the first line of defense should be preventing exposures through the implementation of control measures such as isolation, quarantine, restriction or closure of gatherings, and/or the use of ventilation by proper local extraction. Also, during the outbreak of the pandemic, it might be useful to practice effective measures, such as respiratory hygiene/cough etiquette and hand washing. The detailed information of Covid-19 transmission is yet to come, but the droplets of around 5 μm released from coughing or sneezing are considered as the primary source of respiratory infection (van Doremalen et al., 2020; Wang and Du, 2020). Further, it is reported that the droplets smaller than 1 μm persist for longer durations up to 8 h (Morawska, 2006; Konda et al., 2020) in the environment. Covid-19 aerosol droplets have shown to remain suspended in the air for ~3 h (Huang et al., 2020; van Doremalen et al., 2020).
Self-Disinfecting Surfaces
Protecting the front line healthcare workers from Covid-19 is of prime importance to continue fighting the pandemic. Hence, enabling nanotechnology in the personal protective equipment (PPE) may introduce additional features like hydrophobicity and antimicrobial activity without altering the characteristics of the original texture of the material, breathability, durability and comfort. An example is the use of nanowhiskers in the textiles that provides hydrophobicity employing 3 fold smaller than cotton fiber and hence results in an increase of surface tension that prevents the water droplets. Also, antimicrobial activity on the different surface materials can be achieved by different coating methods using nano dimensional materials like metal NPs (zinc oxide, tin oxide nanowires, titanium dioxide, Au, Ag and Silicon), natural bioactive polymer and chemically functionalized polymers that can inhibit the growth of the microbes. These agents with specific target might intervene in the synthesis of viral RNA replication and inhibit the viral assembly. Weng et al. (2012), reported a self-disinfecting surface by depositing the nanocrystals of CuInZn4S6 (CIZS). The nanocrystals could get activated by absorbing visible light and liberate active oxidative species (H., OH., , 1O2,) which are responsible for the oxidation of the amino acid residues present in the protein envelope of the viruses. And the reported works encourages further research on the disinfection of virus through the inactivation of their protein envelopes.
Respiratory Masks
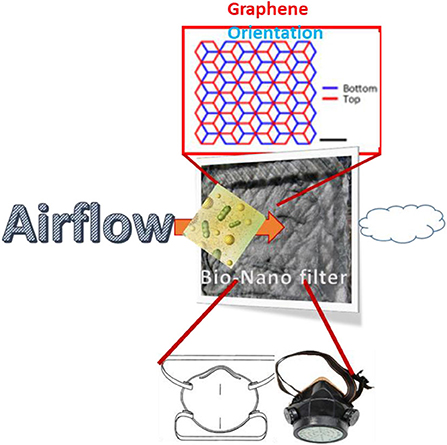
Effective use of surgical masks and N95 respirators among healthcare workers prevents influenza-like infections (Long et al., 2020; Samrat et al., 2020) and SARS (Seto et al., 2003). Hence masks play a vital in preventing the Covid-19 progression. On the other hand, the global shortage of surgical masks and N95 respirators is a major concern (Esposito et al., 2020). In line with the US (Centers for Disease Control Prevention, 2019) (CDC), healthy people are recommended to wear a cloth face mask in public (CDC)2. A report by Konda et al. (2020), summarized that cotton, natural silk, and chiffon exhibited better protection against the infection, generally above 50% in the entire 10 nm to 6.0 μm range furnished the tight weave (Konda et al., 2020). Further, they have also concluded that the higher threads per inch of cotton with tighter weaves resulted in effective filtration (for aerosol-based virus transmission) efficiencies. The use of cloth masks, in general, has several advantages such as cost-effectiveness, sustainable re-use and convenience for the common people. In addition to that, it will also reduce the usage of disposable surgical masks and N95 respirators by the general population, so that it can be reserved for the frontline workers. In this perspective, our team has come up with the idea of sustainable development of a plant extract (comprising antiviral compounds) along with graphene to produce a bio-nano composite solution to modify the commercial cloth materials, surgical mask and air filters as shown in Figure 5. Interestingly, graphene with its nano dimension (atomic thickness 0.335 nm) could trap the virus-like particles and along with the impeded antiviral compounds from plant extract, it could be able to inactivate the virus particles. Further, our method is cost-effective and scaled to the industrial process, realizing the technology readiness level (TRL) of 3. More importantly, the aim of our method is to simplify and make it adaptable to prepare in home by utilizing the available materials such as graphite pencil, ginger, aloe vera and commercial mixer (Under the submission of IMPI-México).
The proposed material made of the nanofiber membranes is attractive due to the dense nanofiber network, high surface area, biodegradability, extended durability, and re-usability. One such highly breathable, disposable, and biodegradable filter cartridge of cellulose nanofiber-based material was tested by Queensland University of technology to remove the virus size nanoparticles. Polyacrylonitrile nanofibers respiratory masks are also produced by the OxinSabzEspadan, Nano Tar Pak, Nano FanavaranKhavar, and ModiranToseeSalamat Iranian. The reusable nano-filtered face mask developed by KAIST which is washable more than 20 times has shown bactericidal properties. Likewise aligned nanofibers in orthogonal or unidirectional direction with a diameter of 100 ~ 500 nm design has a patented technology. Now, this product is awaiting the final approval from the Ministry of Food and Drug Safety. An exclusive nano-coco-carbon™ filter from the Metamasks products made of coconut shell carbon and nanofiber matrix is invented. It allows <1 mm particle and prevents toxic airborne pollution. Another innovative nanocomposite of PLACTIVE® and MDflex® from Copper3D face mask has an active three-layer filtration system of non-woven polypropylene impregnated with 5% copper oxide nanoparticles, to explore the antiviral and antibacterial activity. Silver nanoparticles functionalized graphene oxide ink synthesized by ZEN Graphene solutions Ltd, collaborated with Graphen Composites Ltd, and documented to eradicate earlier versions of coronavirus. This graphene-based composite virucidal ink could improve the protection efficiency when applied to fabrics, masks, and other personal protective equipment. Three-layer nanofiber membrane incorporated with copper oxide (CuO) nanoparticles also significantly improved the filtration efficiency (99.9%) and virucidal activity produced by the RESPILON's ReSpimask® VK. Currently, this product complies with the requirements for medical equipment of type IIR of standard EN 14683. Self-regenerating germicidal TiO2Ag nano facemask was developed by X.TiO2 Inc. They proposed that the outer TiO2 layer, when exposed to direct sunlight (UV), eradicates the accumulated germs. Also, it has a greater odor-absorption capacity and regeneration capacity by light. The same group has introduced multi-tier mask made of titanium dioxide-silver nanoparticles that can be able to destroy over 650 germs under without light conditions.
Vaccine
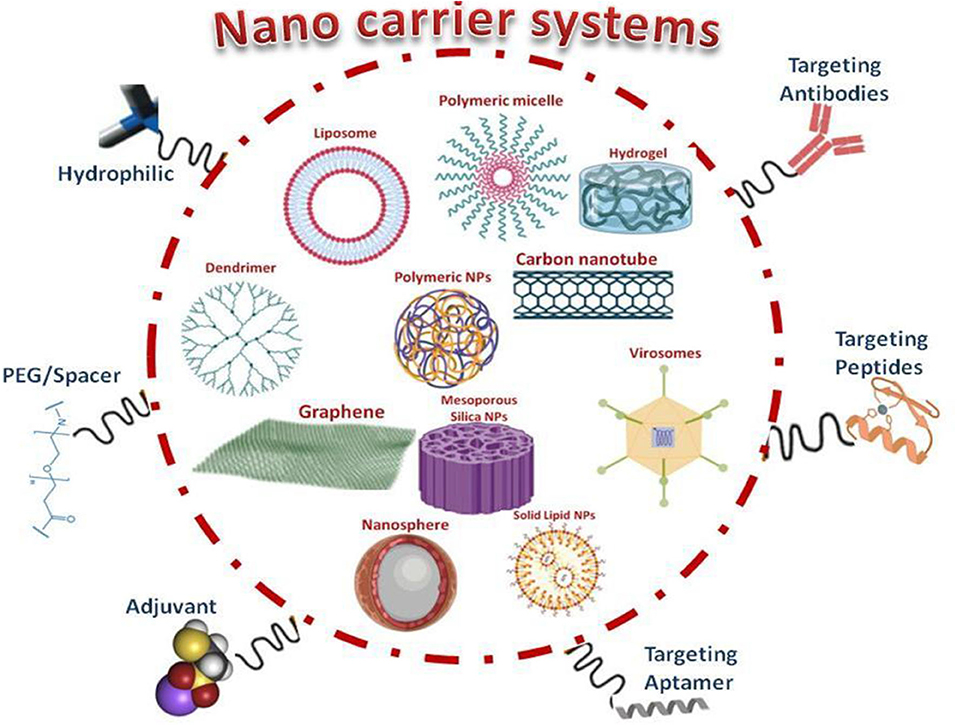
Development of a vaccine is a laborious time consuming and expensive process. Indeed, it has less probability of success and selects the better candidate from the several candidates, further it takes a long period to have a licensed vaccine (Gouglas et al., 2018). The formulations of vaccines entailed of attenuated viruses (non-virulent strain), destroyed pathogens (inactivated) or subunit of protein antigens can elicit a specific immune response (Figure 4). The efficient vaccines such as Influvac R (Daubeney et al., 1997), Vaxigrip R (Delore et al., 2006), and Fluzone R (Grohskopf et al., 2015) are used against influenza type A and type B virus which has been developed by the inactivated/fragmented trivalent or quadrivalent. Moreover, the introduction of the nasal administration of live attenuated vaccines like Nasovac R and Flumist R in young children (Carter and Curran, 2011; Dhere et al., 2011) is already in the process. Vaccines using live-attenuated viruses against influenza are shown to be risky in elderly and immune-suppressed humans (Chattopadhyay et al., 2017). However, either the use of the destroyed pathogen vaccines or virus-derived subunit vaccines induce a lower immune response, and often an adjuvant is required to boost the efficiency (Vartak and Sucheck, 2016). New vaccine modalities like the subunit vaccines (Norton et al., 2012), and antigenic proteins in pathogens that encode DNA vaccines (Luke et al., 2011) are being exploited. Interestingly, though these two new vaccines exhibit a higher safety profile, it lacks relatively lower immunogenicity. In general, researchers characteristically follow a linear sequence of steps with multiple checkpoints for data analysis or manufacturing process. But Covid-19 crisis demands an urgent need for developing a vaccine at a faster rate that requires a new pandemic paradigm of the evaluation process.A quick start and multiple steps should be executed in parallel before confirming a positive outcome of an adjacent step (Lurie et al., 2020). The exploitation of reverse genetics and next-generation sequencing technologies could be shortening the time for the development of more conventional vaccine at this pandemic. However, the development of Covid-19 vaccine has its obstacles. (1) Spike protein antigen design is critical to ensure optimal immunogen response. The most prevalent form of Covid-19 variant in the global pandemic has to carry the spike protein amino acid change at D614G (Korber et al., 2020). Additionally, there is a dispute for the best immunogen approach as to whether to target the complete protein chain or only the receptor-binding domain. (2) The exacerbating side effect of lung disease noted either directly or as a result of antibody-dependent enhancement, in the case of SARS and the Middle East respiratory syndrome (MERS). Hence, it should be concerned about the selection of a suitable animal model and laborious safety monitoring in clinical trials. (3) There is no clear evidence of protection that correlates from SARS and MERS vaccines. Further, the potential duration of immunity is unknown as with naturally acquired infection, and hence there is uncertainty in the dosage of the vaccines (Lurie et al., 2020). (4) The most important challenges of the vaccine are to stimulate both the T cell and B cell immunity against Covid-19 (Chauhan D. S. et al., 2020). (5) From the genome-wide association study on Covid-19 it reveals that the genetic variations might be associated with the severity of infection (Aiewsakun et al., 2020). Many mutations and deletions on coding and non-coding regions are also observed (Phan, 2020). Nevertheless, the high level of genetic mutations (antigenic drift) of viruses should be considered because it might have a chance to reduce the efficacy of the vaccines (Boni, 2008; Graham and Anderson, 2013). An alternative approach to the conventional vaccines is to present the relevant antigenic moieties at the particle of nanoplatforms. At present, there is a wide variety of the nanoparticles evaluated as antigen carriers, with inorganic, polymeric nanoparticles, virus-like particles (VLPs), liposomes and self-assembled protein nanoparticles (Figure 6). The main advantages of these materials are their nano dimensions as they pave the easier interaction with the biological functional molecules (DNA, RNA and proteins) at the nanoscale (Laval et al., 2000). Further, it can be administrated via subcutaneous and intramuscular injections or delivered through oral, intranasal (mucosal sites) and also through penetrating capillaries as well as mucosal surfaces (Parveen et al., 2016; Schneider et al., 2017; Smith et al., 2017). Recent signs of progress in the material science can control and tune the size, shape, solubility, surface functionalization and hydrophilicity that could allow the synthesis of nanoparticles with tailored biological properties (Angioletti-Uberti, 2017; Al-Halifa et al., 2019). Besides this, the incorporation of a wide range of antigenic molecules into the nano dimensional particles makes notable modulations of immunogenicity (Irvine et al., 2015; Szeto and Lavik, 2016). Most importantly, the available nanotechnology tools and techniques are used for the design of vaccine carriers with special focus on effectiveness, immunogenicity modulation and biosafety to tackle the current outbreak. Various institutions and bio firms have kick-started the vaccine development as soon as the first genetic information was posted. The genetic information would give us the significant genomic match of Covid-19 with the previous coronavirus and will help in elucidating the potential vaccine candidates. To get preferable vaccine target candidate nAbs are induced against S protein of Covid-19, and the ACE2- mediated host uptake must be prevented. In the circumstance of SARS/MERS, vaccine development research puts forward RBD of the S1 subunit and functional information (protein/gene) of S could act as preferred target sites (Chen W.-H. et al., 2020; Prompetchara et al., 2020; Verma et al., 2020; Zhang L. et al., 2020). Available specific antigens information on Covid-19 is limited that could obstacle the trial vaccine candidates. Covid-19 nano vaccine candidate scheme depends on (1) Antigen-Dependent Nanocarrier Selection and (2) Vaccine Adjuvant Nanoparticles.
(1) Antigen-Dependent Nanocarrier selection:
Factors such as the physicochemical characteristics, biological stability, target sites, and controlled immunomodulatory release of antigens decide the presence of antigens either inside or on the surface of the nanocarriers. The interaction of antigen on the surface of nanocarrier is based on physical adsorption of surface charge and through non-covalent hydrophobic interaction. The amphoteric nature of the antigen facilitates the adsorption or surface immobilization of antigens on nanocarriers such as chitosan, dextran sulfate-based polymeric nanoparticles, inorganic nanoparticles, and carbon nanotubes (Zhao et al., 2014; Irvine et al., 2015; Salazar-Gonzlez et al., 2015; Pati et al., 2018). The mRNA and DNA based vaccines are straggled by the degradation of extracellular RNase and nucleases, hence the delivery vehicle is vital in both cases (Geall et al., 2012; Lung P. et al., 2020). These mRNAs are entailed cell-specific receptor recognition and cross the lipid membrane. The cytosolic presence of exogenous mRNA is initiated to trigger the synthesis of functional protein (Reichmuth et al., 2016). The self-assembly of an ionizable cationic lipid resulted in lipid nanoparticles (LNPs) of size 80–200 nm (Kranz et al., 2016). It has the ability to deliver mRNA on specific target site effectively. The sustained release provokes the opting for intramuscular and intradermal routes, resulted in high antibody, B and T cells immune response (Kanasty et al., 2013). Lipid nanoparticles (LNPs) is employed as an encapsulated carrier for the mRNA-1273 that encodes for a full-length, prefusion stabilized spike (S) protein of Covid-19, under the clinical trial (NCT04405076-Phase 2 and NCT04283461-Phase 1) performed by the Moderna (Figure 6). Once the availability of the genetic sequence information, within a short period developed the mRNA based Covid-19 vaccine candidate by Moderna's that entered phase 1 clinical trial on March 16, 2020. LNPs-mRNAs vaccine is an ongoing clinical trial of phase 1/2 (2020-001038-36 and NCT04368728) that uses the four prophylactic SARS-CoV-2-RNA against Covid-19. The clinical trial of phase 1, ISRCTN17072692 used LNP-nCoVsaRNA that is encoded with the Covid-19 spike protein. The physical technologies of gene gun and electroporation also play a vital role to improve the delivery of the mRNA/DNA across the cell and nucleus membrane. Prominent elicitation of B and T cell response found in pig is based on surface electroporation of DNA coated-PLGA nanoparticles (Hutnick et al., 2012). This portable electroporation technology is now exploited as an ongoing clinical trial to insert a DNA plasmid that encodes the S-protein of Covid-19 (NCT04336410 and NCT04447781).
(2) Vaccine Adjuvant Nanoparticles (VANs):
The intrinsic adjuvanticity of the nanoparticles is capable of activating the complement system, inducing the autophagy and the activation of inflammasome (Hamad et al., 2010; Chauhan G. et al., 2020). Curiously the surface functional group and hydrophobicity of nanoparticles, along with other physiochemical properties, are capable of inducing the adjuvanticity mechanisms (Thomas et al., 2011) (Figure 6). As a result, VANs are conceived as amend for the efficacy and safety of the immune response. Notably, the vaccine adjuvants are capable of reducing the required dosage of antigen, and it results in making it available to a larger population (Reed et al., 2009) at this pandemic situation.
The full-length recombinant SARS CoV-2 glycoprotein nanoparticle vaccine is adjuvant with Matrix under the clinical evaluation of Phase 1/2NCT04368988) (Baviskar et al., 2020). Further, Adjuvanted recombinant protein (RBD-Dimer)NCT04445194 and recombinant spike protein with Advax™ adjuvant of NCT04453852 is under the phase 1 clinical evaluation (Table 1) (Samrat et al., 2020). Among the other COVID-19 preclinical vaccine candidates, five of the protein subunit vaccine candidates reported a combination of antigen and adjuvant. A recombinant Covid-19 glycoprotein with a matrix M (an adjuvant) used in the NVX-CoV2373 nanoparticle vaccine is expected to move into clinical trials shortly. Recently, AS03 (GlaxoSmith-Kine), MF59 (Seqirus), and CpG 1018 (Dynavax) are the licensed adjuvants developed specifically for preparing Covid-19 vaccine.
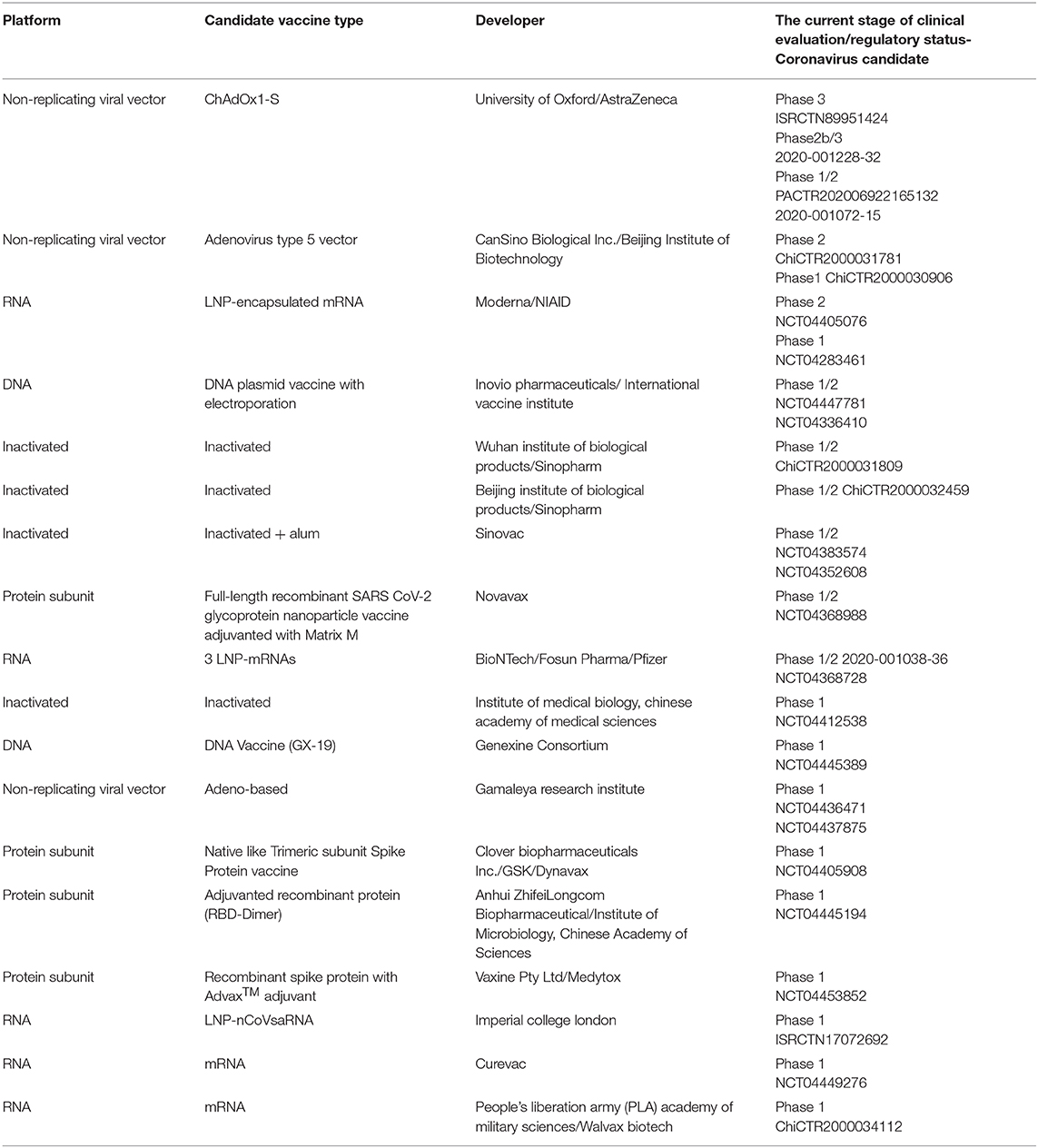
Table 1. List of vaccine candidates under the clinical evaluation (DRAFT Landscape of COVID-19 candidate vaccines– as of July 2, 2020, by WHO).
A remarkable number of nanotechnological pre-products are used in the ongoing vaccine clinical evaluation and preclinical stage as per the list from the DRAFT Landscape of COVID-19 Candidate Vaccines (as of July 2, 2020, by WHO) (Table 1). The nano dimensional materials effectively cross the cellular membrane barriers and reach its specific target site. On reaching the targeted site, it facilitates the controlled release, and stimulate the immunomodulatory activity to induce the desirable defense mechanisms (i.e., target both adaptive–T cell and B cells and innate immune systems of macrophages, monocytes, neutrophils) inside the host (Chauhan G. et al., 2020). As a result, there is eminent efficacy and safety in disease control. Further in the development of vaccine process, intrinsic adjuvant property in few nanomaterials and effective delivery of adjuvants resulted in reducing the dose-sparing which could reduce the production units and made it accessible to the larger demand at the pandemic situation. In conclusion, nanotechnology explicit its major role in the futuristic lead vaccine development.
Immunomodulation Aspect in the Context of Nanomaterial-Based Vaccine Development
Under the severe conditions, Covid-19 patients suffer from major invasion and damage impact on lung and the juxtaposed endothelial tissue by innate immune phagocytes (neutrophils and macrophages) (Pisani et al., 2020). To overcome this critical issue, it is recommended to use anti-IL-6 antibody tocilizumab and Siltuximab in the clinical trials. Nevertheless, care should be taken for the effective control and suppression of innate immunity associated with cytokine production (Pisani et al., 2020). Nano based systems plausibly counteract these critical issues and might have the potential to improve the specificity and controlled immunosuppressant delivery. And it could be attained by fine-tuning the surface charges of the nanocarrier systems. The surface charges could improve encapsulation with the required loading of the anti-immunosuppression drugs, pertinent to reduce drug dose as well as the non-target drug distribution, and possibly undesirable consequences.
Further, it can deliver targeting specific immune subpopulations to avoid the provocation of other immunological responses. Experimental results of the Octadecylamine functionalized nanodiamond with dexamethasone demonstrate the anti-inflammatory and pro-regenerative activity in human macrophages under in vitro. Additional, it also has reduced the macrophage infiltration and controlled expression of iNOS and tumor necrosis factor (TNF)-α (proinflammatory mediators) (Weiss et al., 2020). The occurrence of the ordered structure, controlled porosity with surface charges on the nano dimensional carbon materials has exhibited the effective adsorption of IL-6 and TNF-α (Weiss et al., 2020).
Diagnosis
One of the urgent needs in the Covid-19 pandemic is a multiple diagnosis test; this infers to take the strategical decision depending on the population density of all over the major cities in the world. The substantial concern in the diagnosis is sensitivity, specificity and accuracy, which further leads to the rapid implementation of proper containment of Covid-19 (i.e., identification, isolation, contact tracing, and treatment) that limit the spread.
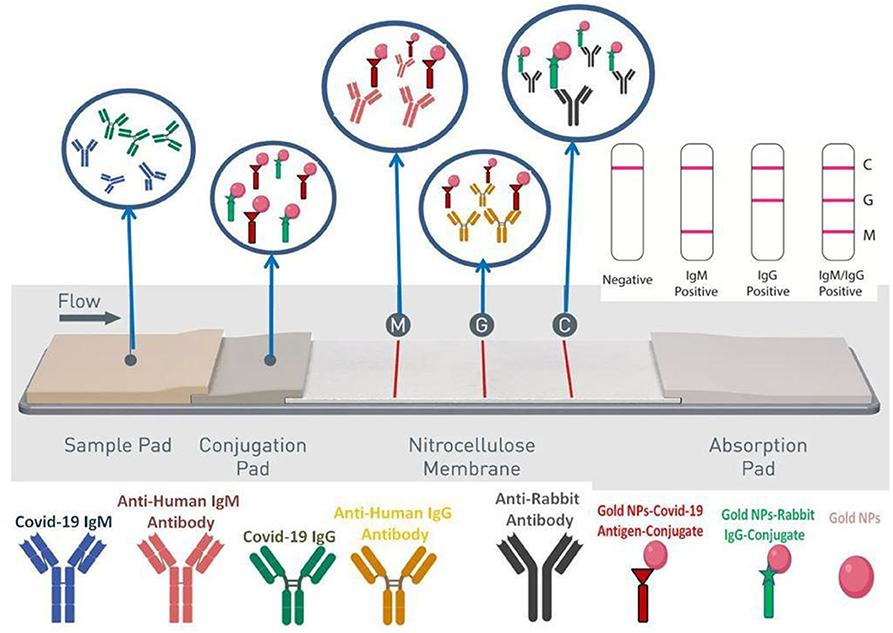
Molecular techniques are vital in diagnoses as it shows the higher target and specificity of pathogens. For the development of molecular techniques, first, we should understand the composition of the functional biomolecules (proteomic and genomic) of the pathogen, its entry into the host system and associated induction response by expression in the functional biomolecules. There are two main class of functional biomolecules where diagnosis are performed: (1) in nucleic acids (RNA-DNA) and (2) in proteins (Serological test-Induction response of viral antigen and antibodies). The priority is given to the nucleic acid-based methods by the WHO. This method is focused on direct amplification of the virus genetic material from the patient sample that offers high sensitivity (85–90%) and specificity with the aid of polymerase chain reaction (PCR) under the thermal cycler. Two strategies are used in a PCR test, (a) Reverse Transcriptase-PCR (quantitative) (b) Loop-Mediated Isothermal Amplification- PCR (LAMP-PCR) (qualitative) (Arun Krishnan et al., 2020). The RT-PCR method involves the reverse transcription of Covid-19 RNA into cDNA (Complementary DNA) strands, later by amplification of the specific gene of interest in the cDNA (Kageyama et al., 2003). This method targets the three genes of interest to detect viruses from the beta-coronavirus group (E gene- codes for envelope protein), N gene code for nucleocapsid protein and RdRP gene code RNA dependent RNA polymerase in the open reading frame ORF1. More specifically, the RdRP and E genes have high sensitivity toward Covid-19 with a technical limit of detection of 3.6 and 3.9 copies per reaction (Udugama et al., 2020). According to the WHO's recommendations, the RT-PCR test must detect three genes in a single reaction mix. This would sort out the false-positive results and guarantees double confirmation (WHO Diagnostic Detection of RT-PCT report)3. The upper respiratory (nasopharyngeal swabs, oropharyngeal swabs, nasopharyngeal washes, and nasal aspirates) samples and lower respiratory tract (sputum, BAL fluid, and tracheal aspirates) samples are used for diagnosing Covid-19. From the above, the broadly recommended sampling method is the upper respiratory sample collection (Interim Guidelines). In the RT-LAMP involves in the conversion of RNA to DNA followed by amplification of sample genome using forward inner primer and outer primer; reverse inner and reverse outer primer that targeting different regions of the DNA (Notomiet al., 2000). As a result, a higher specificity with the detection limit of ~75 copies per μL will be obtained. This method is easy to operate with a low background signal and can be visualized by a color change either by the use of pH-sensitive dye or fluorescent dye that binds to double-stranded DNA (Notomi et al., 2000; Mori et al., 2001). CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats) system have been proposed for the rapid detection of Covid-19. CRISPR is a set of DNA sequences, that results from an adaptive immune system in prokaryotic organisms (archaea and bacteria) against the previous infection of phages. Under this Cas, protein has different utilities in gene editing (Cas9) and diagnosis (Cas12a and Cas13a). Recently, CRISPR/Cas13 system has been exploited for the detection of Covid 19 in SHERLOCK [specific high sensitivity enzymatic reporter unlocking (Kilic et al., 2020)] technology by Broughton et al. (2020). In the coronavirus genome, Cas13 is targeted to the S and ORF1ab protein genes, where it binds and initiates the random cleavage of ssRNA. With the aid of a fluorescent probe as a transducer in this technology, it makes the possible way to quantifiable as well as visible confirmation of positive case (Metsky et al., 2020). Thiol functionalized Gold NPs with N-gene (nucleocapsid phosphoprotein) oligonucleotides are capable of diagnosing Covid-19 from the RNA isolated samples using colourimetric assay within 10 min. This rapid diagnosis kit progresses in the identification of infection on time, though the viral loading in the infected sample might obstruct the diagnosis. To resolve the above-mentioned issues, two-dimensional gold nanoislands are functionalized with the complementary oligonucleotide sequence of elected Covid19 gene sequence (RdRp-COVID, ORF1ab-COVID, and E genes from SARS-Cov-2) (Parihar et al., 2020; Srivastava et al., 2021). During the nucleic acid hybridization with the infected sample, two different angles of incidence at their plasmonic photothermal (PPT) and localized surface plasmon resonance (LSPR) exhibited the two different excited wavelengths (Xu et al., 2020). This unique combination feature of surface PPT and LSPR effects results in higher sensitivity (< concentration of 0.22 pM), stability and reliability in the Covid19 sensing (Talebian et al., 2020). Yet another combination of two technologies such as reverse transcription enzymatic recombinase amplification (RT-ERA) and fluorescence resonance energy transfer (FRET) was exploited in Exo FRET method with the ultrasensitive detection (0.32 × 10−18M) of Covid-19. FAM and BHQ1 were used as quenching probes and located close together along with the THF site. An exonuclease II can recognize the THF site and proceed to cleave the double-strand nature of the RNA sense sample with amplicon. As a consequence of this, the cleaved strand leaves the BHQ1 probe and restores the FAM fluorescence probe and later it helps in quantifying the FAM probe and achieve the ultrasensitive detection (Yang et al., 2020). (2) Proteins (Serological test-Induction response of viral antigen and antibodies)- The protein constitution of antigens and antibodies exploited in the serological assays can find/measure antibody response due to induction response of pathogens (Covid-19) in body fluids (blood serum or plasma). This involves the use of the different platform, including binding affinity assay (enzyme-linked immunosorbent assays-ELISAs, lateral flow assays or western blot-based assays) and functional assays such as virus neutralization, enzyme inhibition or bactericidal assays. These essential tools are involved in the management of infectious diseases, including diagnosis of infection and treatment (seroprevalence assessments and antibody titers upon vaccination) (Krammer and Simon, 2020). Once the human body encounters the Covid-19 infection, our body responds to produce IgM and IgG antibodies at the 7–10th day and 14–20th day, respectively. Even though the shorter detection of IgM can disappear earlier, however, IgG can persist over a long period after the infection. Besides antibody detection, spike glycoprotein and nucleocapsid protein antigens are under development for the early stage detection of Covid-19. Point-of-care (PoC) testing devices are used for rapid diagnosis without sending samples to a centralized well-equipped laboratory and don't require professional experts to analyze the samples experiment. This portable molecular device would significantly contribute toward cost-effective, robust and simplicity of inference results. Most widely, PoC uses one of the serological assay called as lateral flow immunoassay. This method adopted to detect Covid-19 biomarkers, IgG and IgM with impeded gold nanoparticles conjugates in the test strip. Once the patient's sample is placed on the nitrocellulose membrane strip, with the aid of capillary force the protein is drawn across the strip. The biomarkers reached with the corresponding antibodies, reveals the visible color change as a line (Figure 7). A field-effect transistor with graphene sheet coating surface functionalized with the spike protein antibody act against Covid19, and it can show the higher sensitivity against Covid-19 even with the presence of Middle East respiratory syndrome coronavirus (MERS-CoV).
The IgM and IgG in the lateral flow assay have demonstrated a clinical sensitivity (57 and 81%), specificity (100 and 100%), and accuracy (69 and 86%) (Udugama et al., 2020). Clinical sensitivity of 82% realized in a test detects both IgM and IgG (Xiang et al., 2020) antibodies. Recently, Sona Nanotech, Mologic, and SureScreen Diagnostics are developed a quick response lateral-flow test to identify the COVID-19 within 5–15 min. Enzyme-Linked Immunosorbent Assay (ELISA) and Chemiluminescence Immunoassay (CLIA) are quantitative serological techniques to detect specific antigen or antibody. The ELISA test is developed to detect Covid-19 IgM and IgG antibodies by Zhang W. et al. (2020). Anti-IgG antibody conjugated with horseradish peroxidase and SARS-CoV-2 Rp3 nucleocapsid protein is adsorbed on the 96 well plate surface as biomarkers (Zhang W. et al., 2020). The manufactures of IBL international, DRG Diagnostics GmbH, Epitope and Euroimmun developed manual ELISA kits to detect the IgG and IgM antibodies (Mungroo et al., 2020). Serological detection kits are under development by the manufacturers in the worldwide. However, the major concern should be taken care of false-positive results that might be evidenced by previous virus infections other than Covid-19, such as coronavirus HKU1, NL63, OC43, or 229E. Moreover, detection limit in the “window period” (i.e., the when specific antibodies are not yet detectable range in a patient's sample) may exceed 7 days is deprived thus might prove negative results. Hence, serological tests can't be used as a predominant basic diagnosing tool in Covid-19 (Lauer et al., 2020). Concerning the current indication, WHO recommends that the new PoC immunodiagnostic test proceeds in a research lab alone. Further, it should not consider for clinical decision making until the further confirmation supporting test result is available (WHO/2019-nCoV/Sci_Brief/POC_immunodiagnostics/2020.1–as of April 8, 2020). In contrast, serology tests could help healthcare professionals to identify the development of immune response, further determining to select the donors of convalescent plasma. This would serve for the seriously ill patient of Covid-19. After the vigor's evaluation, The Food and Drug Administration (FDA)-Emergency Use Authorizations (EUA) authorized the serology test performance on their website (Supplementary Table 1)4. It should be noted that widespread testing is needed to keep the outbreaks of Covid-19 under control. Nevertheless, in many regions, there is a shortage of reagents needed to run tests and lack of infrastructure to proceed further. In that case, a serological based rapid test could be a better option.
Treatment
The fundamental studies of the Bio-Nano interactions could be revised to understand the Covid-19 infection with the human system (Chan, 2020). The coherence of material science advancements reveals the genetic information, protein structure modeling and mode of pathophysiology/immunogenicity. This information further leads to the identification of a target that would facilitate effective treatment. The possible target is the viral system—3CLpro, PLpro (viral protease) and S protein, host cell protease—transmembrane serine protease 2 (TMPRSS2) (Weiss et al., 2020), angiotensin-converting enzyme 2 (ACE2) and RNA-dependent RNA polymerase (RdRP). Based on the target sites as mentioned above, the finding of new small molecules is under development meanwhile there is search for the existing antiviral molecules (Gordon et al., 2020; Hoffmann et al., 2020; Lung J. et al., 2020; Zhang L. et al., 2020). Artificial intelligence also takes part in the identification of potential drug molecules for Covid-19 treatments (Tang et al., 2020; Zhou et al., 2020).
Nanomedicine could open a possibility for the design of a new therapeutic agent and the treatment for Covid-19 (Figure 6). A nanomedicine approach is a powerful vital tool (Petros and DeSimone, 2010), which can have the ability to transform/deliver antiviral drugs effectively and improve Covid-19 therapeutic efficacy. Nano-carriers based therapeutics would tackle the current limitations (i.e., poor aqueous solubility and low bioavailability) of antiviral therapy and offers several opportunities to improve the efficacy (Figure 6). This has a capacity of crossing the biological barriers to attain therapeutic concentrations in protected viral reservoirs (Kobayashi et al., 2014) and reaches the specific target organ/intercellular sites involved in the Covid-19 pathophysiology such as ACE2 expressing cells, viral S protein domains, cathepsin binding sites, etc. Further, the sustainable drug-releasing nanocarriers can mitigate the risk effects of deprived patient compliance and the rebound of viral during the treatment for infections. Therefore, nanocarriers based antiviral drugs delivery system could be able to modify the antiviral drug pharmacokinetics/pharmacodynamics properties and the results are reflected in the reduction of drug dosage, reduced side effects, and improved bioavailability of drugs (Lembo et al., 2018).
The respiratory disease of Covid-19 has affected lungs severely in the 14% of cases leading to lung swelling and fills it is with fluid and debris. Actively delivering a drug directly to the site may prove an effective treatment strategy. However, this task is limited by the complex mucus environment in the lungs. This problem could be overcome by the use of mucoadhesive particles (MAP) for pulmonary drug delivery. The core hydrophobic polymers have highly cationic or thiolated of decorated nanoparticles surface, which favors the interaction with negatively charged glycosylated or cysteine-rich hydrophobic domains of airway mucin fibers (Schneider et al., 2017). A consequence of this facilitates the long duration retention in the lungs by reducing the mucociliary clearance rates via rheological changes of mucus by multivalent mucus particle interactions (Schneider et al., 2017). Further, the MAP diameters (large as 200– <300 nm) smaller than mucus mesh spacings would have crossed the mucus layers easily. The repurposed Covid-19 drug composition effectively formulated with MAP system might be an effective target for the treatment.
In addition to the conventional antibodies, a single variable domain antibody without an effector domain is coined as VHH or nanobody (Nb). Heavy-chain-only antibodies (HCAbs) produced by camelids have a single variable domain that acts as the target recognition module. Nbs possess various advantages over the conventional antibodies, that includes a structure-function concert at nanoscale dimensions leads to higher solubility and homogeneity, chemical and thermal stability, docility into multiple formats, facile penetration, and reach specific target site with antigenic affinity and specificity, inferior vulnerability to steric hindrances (Konwarh, 2020b). This attractive features of nanobodies are plausible combat against Covid19. At this junction, Martinez-Delgado (2020), reported that the nanobodies derived from camels, Nb11-59 unveiled promising therapeutic against Covid-19 with ND50 of 0.55 μg/mL, exclusively via nebulization.
Regarding Off-label use of medicines for the Covid-19, the scientific brief from the WHO (update March 31, 2020) (Adhikari et al., 2020), communicate that none of the specific pharmaceutical products has yet been shown to be safe and effective for the treatment of Covid-19. Despite the promising response of remdesivir in clinical trials, it could be considered for inhibition of viral replication for the treatment of Covid-19. Prominently, to replicate the Covid-19 genome takes more than a single polymerase and ensure the pathogenicity. There are nine structural and non-structural proteins (nsps) in the viral replication-transcription complex (RTC) that are involved in the cell multiplication of Covid-19 (Chan et al., 2020; Ortiz-Prado et al., 2020). Nevertheless, a good number of medicines have been suggested as potential investigational therapies. Most of the drugs in a combined form known for closely related therapeutics have shown better response, depending on the stage and condition of the patient. Apart from this new clinical trial, the Solidarity trial is co-sponsored by WHO along with participating countries.
Solidarity trials interim results showed that the practice of the hydroxychloroquine and lopinavir/ritonavir not able to change the mortality of COVID-19 patients (“Solidarity” clinical trial for COVID-19 treatments-Latest update on treatment arms dated on July 6, 2020). Nanocarriers in the delivery of multiple drugs with diverge physicochemical properties are essentially beneficial for combination therapies (Destache et al., 2009; Shibata et al., 2013). The technological feasibility enables the fabrication of a variety of nanomaterials designed to carry the multiple drug carrier, which facilitates the sustainable control in synergistic drug compositions, improved pharmacokinetics, and reduced side effects. There have been a promising in vitro and in vivo studies of blood-brain barrier transmigration with significant anti-HIV activity evaluated in primary central nerves system cells using nanoformulation (pegylated-magneto-liposomal based nano delivery) with multidrug compounds load (antiretrovirals, latency reactivating agents, and drug abuse antagonist) (Jayant et al., 2018). To overcome the lymph node drug insufficiency, an oral combination of drugs has been formulated in a lipid nanoparticle-based delivery system accommodated with three antiretroviral drugs, lopinavir, ritonavir, and tenofovir (hydrophilic drug). The drugs have shown long-lasting plasma drug profiles and improved lymph node drug delivery levels evaluated in the macaques in vivo model (Kraft et al., 2018). These methods of multidrug based Nano delivery system could be adaptable to Covid-19. But various parameters such as analyzing the interaction between each drug compounds, controlling the release profile of each drug and balancing the antagonism/synergy/toxicity (Ma et al., 2013) should also be considered. The cholesterol modified hydroxychloroquine (CholHCQ) laden liposome can reduce the dose and toxicity of hydroxychloroquine. Further, it inhibited the proliferation of rat lung fibroblasts, and as a result, it exhibited reducing the pulmonary fibrosis (Liu et al., 2017; Chauhan G. et al., 2020). This strategy of control viral load and target site would be beneficial to the Covid-19 treatment. Hu et al. (2020) proposed a mechanism involving chloroquine-induced suppression of phosphatidylinositol binding clathrin assembly protein (PICALM), which prevents the endocytosis-mediated uptake of Covid-19 (Chauhan G. et al., 2020). The use of target-specific CRISPR-RNAs (crRNAs) can protect the host systems from the infection via CRISPR-Cas13 system (Konwarh, 2020a). This effective antiviral system specifies the target ssRNA to degrade and inhibit further proliferation of infection. In this sense, prophylactic antiviral CRISPR in the human host (PAC-MAN) approaches are to utilize Cas13d of RNA-guided RNA endonuclease activity in the host to target the Covid-19 (Nalawansha and Samarasinghe, 2020) and cleave the RNA sequences (Abbott et al., 2020). With the aid of effective nanocarrier with PAC-MAN directed toward lung epithelial cells would be repurposed treatment of Covid-19.
In a cytokine storm, the outcomes are the Hyperinflammation and macrophage activation syndrome, uncontrolled production of IL-1β, IL-6, and TNF-α (knows as proinflammatory cytokines). Under severe conditions, Covid-19 patients have the major invade and damage impact on lung and the juxtaposed endothelial tissue by innate immune phagocytes (neutrophils and macrophages) (Pisani et al., 2020). To overcome this critical issue, the recommended antibodies like anti-IL-6 antibody tocilizumab and Siltuximab under the clinical trials. Nevertheless, care should be taken for the effective control and suppression of innate immunity associated cytokine (Pisani et al., 2020). Nano based systems plausibly counteract these critical issues and might have the potential to improve the specificity and controlled immunosuppressant delivery. It could be attained by the fine-tuning of the nanocarrier systems surface charges that improve the encapsulation with the required loading of the anti-immunosuppression drugs, pertinent to reduce drug dose as well as the non-target drug distribution, and possibly undesirable consequences. Further, it can deliver targeting specific immune subpopulations to avoid the provocation of other immunological responses. Previous encouraging results of the Octadecylamine functionalized nanodiamond with dexamethasone able to demonstrate the anti-inflammatory and pro-regenerative activity in human macrophages under in vitro condition. Additional, it has also reduced the macrophage infiltration and controlled expression of iNOS and tumor necrosis factor (TNF)-α (proinflammatory mediators) (Weiss et al., 2020). The occurrence of the ordered structure, controlled porosity with surface charges on the nano dimensional carbon materials has exhibited the effective adsorption of IL-6 and TNF-α (Weiss et al., 2020). Plausible use of this kind of Nanosystem against the early stage of sepsis, where effectively complete removal of pro-inflammatory cytokines from the bloodstream, would reduce and eradicate the mortality of the critical Covid-19 patient (Weiss et al., 2020). Therefore, nano-based therapeutics could attain specific target site delivery of immunosuppressive drugs in a controlled inhibition or provoke the specific immune cell subpopulation, results in limiting the complication in a cytokine storm.
Novochizol™ along with the Bioavanta-Bosti's academic partners developed a fully biocompatible chitosan-based nanoparticle for safe and effective drug delivery technology. This could ensure sustained-release without systemic distribution when strongly adhere to lung epithelial tissues. Currently, this concept is under the extensive preclinical testing stage. Their synthesis method comprises of the two-step activation of linear chitosan, the active ingredients (small molecule or biologic) in single or combined form. Due to the intramolecular reaction, the active ingredients encapsulates and form the nano dimensional structure that targets the intra-pulmonary delivery system (Figure 8).
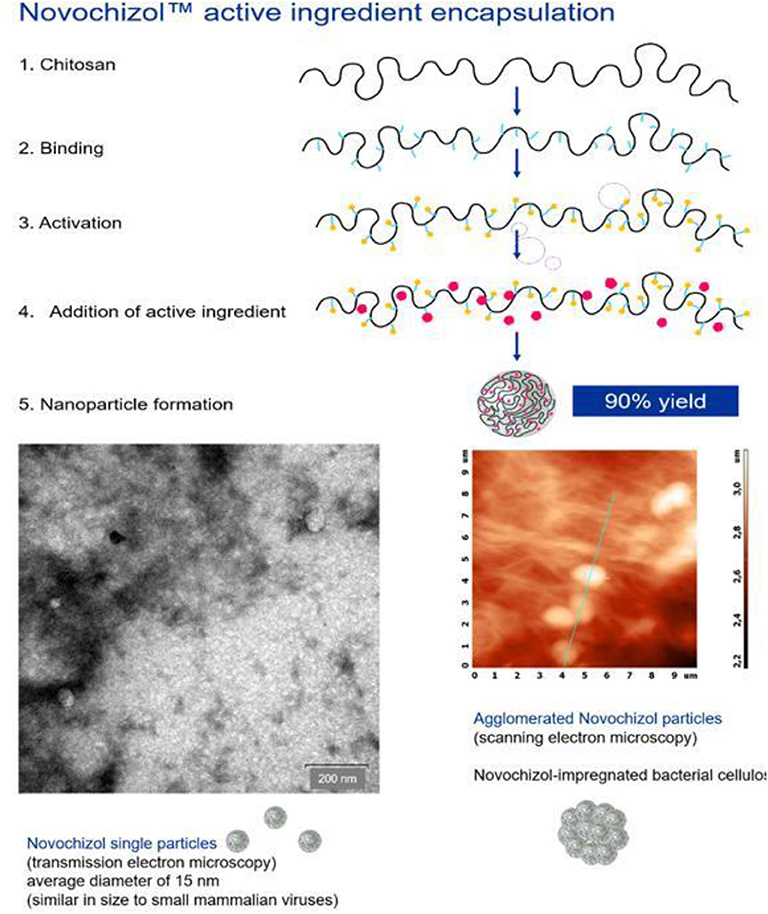
Figure 8. Biocompatible chitosan-based nanoparticle drug delivery technology from NovochizolTM (Re-produced with permission from Novochizol).
Role of in silico in Nanomedicines
Recent advancement in the computational field, artificial intelligence, and machine learning are elevating the simulation and modeling process that could predict the possible evaluation of nano theranostics and nanotoxicology response. Based on the molecular targets (3CLpro and PLpro- Viral protease, TMPRSS2-host cell produced protease, RdRp- RNA polymerase, viral S protein and host receptor ACE2) (Chauhan G. et al., 2020) and related information (either from the sequences or structural information) it is possible to predict the lead compounds (repurpose the existed antiviral molecules or new ligand molecules) and antiviral peptides, from the Zinc database, FDA approved drugs and CoronaDB-AI (Keshavarzi Arshadi et al., 2020; Lin et al., 2020). Further, the procedure is optimized by the scrutinizing of colloidal particle size determination, algorithms to establish dosimetry for inhalation toxicology, computational OMICS and nanoscale-quantitative structure-activity relationships (nano-QSAR), able to propose the refined nanomedicine with minimizing the utilization of resources. This new infesting technological era would facilitate to develop benign nanomedicine within the short span of period. In the future perspective of nanomedicine, the advantage of the futuristic technology of artificial intelligence tool can be taken for rapid screening of lead compounds, possible bioactivity screening/prediction and the rapid evaluation of these compounds in specific animal models, which enable to improve and validate the nano dimensional combination therapeutics.
Practical Snags in Bio-Nano Interface Technology Related to Covid-19
When the biological nano dimensional surfaces and synthetic nano dimensional material surfaces, are interfaced, there are numerous physicochemical interactions such as Van der Waals forces, electrostatic, solvation, solvophobic, and depletion forces. These interface forces are influenced by the surrounding environmental medium. The most probable obstacles arise in these circumstances. The precise upscale in the industrial process and durability of the stable nano dimensional products are hindering the direct application in the field of pandemic situation. In the lateral flow assay interaction of biological functional nano dimensional materials with the engineered nanomaterials convert the signal transducers (optical, electronic, magnetic, catalytic, surface plasmon, piezoelectric, and plasmonic photothermal) effectively and can be able to provide a rapid diagnosis of Covid-19. Yet, interfacing the nano dimensional materials in real-time biological complex environments are snag and could provoke protein denaturation, steric hindrance, and unspecific adsorption (Nel et al., 2009). These undesirable side effects might be revealed for estimating the durability and stability of the nano products. Nano formulated repurposing drugs and vaccines should undergo the additional clinical trials concerning the quantification of the nanomaterials in each cellular process of administrable initial concentration during the route of exposure, required concentration at the target organ and liberation or disintegration of carrier and drug (Genchi and Ciofani, 2019; Riediker, 2020). Above mentioned obstacles can be overcome with the detailed bio-nano interface technology studies. The first impressive Covid-19 vaccine candidate enters the trial is the lipid nanoparticle-based mRNA delivery. Therefore, the futuristic bio-medical field will utilize the sustainable nanotechnology to combat disease effectively.
Conclusion
The interface between the functional biological molecules with the nanotechnology principles and techniques paves the understanding, development and improvement of advanced biotechnological process and products development like implantable medical devices, nano-robots, and the use of biological molecules in nanodevices. To look for detailed biological interaction process, the observations at the nanoscale level would give a promising dimension of understanding that involves the recognition, differentiation, memory, and storage of cellular process function effectively as a collective response that can control the complex macro living systems. A similar way of understanding the nature and behavior of virus at the nanoscale level would reveal the effective control or eradication strategy. The coherence of material science provides an inevitable contribution to the biological system, which is found to be a useful tool to fight against invisible battlefield effectively. In the future perspective niche, there is plenty of room at the Bio-Nano interface.
Author Contributions
S-KK conceived the idea and wrote the manuscript. He had re-drawn the images and got permission for Figure 8 from the NovochizolTM.
Funding
S-KK would like to acknowledge the Conacyt-México: CALL 2020-1 Support for Scientific Research, Technological Development and Innovation in Health Projects in the Event of Covid-19 Contingency. Proposal number: 312032 (Under consideration, not yet approved).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
S-KK would like to acknowledge, Ernesto Lugo Ledesma, Director of Technology Institute of El Llano Aguascalientes (ITEL) and Enrique Fernández Fassnacht Director General of National Technology Institute of México (TecNM) for their constant support and facilities to promote the research activities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2020.586250/full#supplementary-material
Footnotes
1. ^https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?%22%20%5Cl%20%
2. ^Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons Under Investigation (PUIs) for Coronavirus Disease 2019 (COVID-19); Centers for Disease Control and Prevention. Available online at: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html.
3. ^Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT-PCR. Available online at: https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf.
4. ^https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance
References
Abbott, T. R., Dhamdhere, G., Liu, Y., Lin, X., Goudy, L., Zeng, L., et al. (2020). Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 181:e12. doi: 10.1016/j.cell.2020.04.020
Adhikari, S., Adhikari, U., Mishra, A., and Guragain, B. (2020). Nanomaterials for Diagnostic, Treatment and Prevention of COVID-19 Applied Science and Technology Annals 1. 1 SE-Thematic Opinions: Emerging Trends. doi: 10.3126/asta.v1i1.30295
Aiewsakun, P., Wongtrakoongate, P., Thawornwattana, Y., Hongeng, S., and Thitithanyanont, A. (2020). SARS-CoV-2 genetic variations associated with COVID-19 severity. MedRxiv 2020.05.27.20114546. doi: 10.1101/2020.05.27.20114546
Al-Halifa, S., Gauthier, L., Arpin, D., Bourgault, S., and Archambault, D. (2019). “Nanoparticle-based vaccines against respiratory viruses,” in Frontiers in Immunology Vol. 10. 22. Available online at: https://www.frontiersin.org/article/10.3389/fimmu.2019.00022
Angioletti-Uberti, S. (2017). Theory, simulations and the design of functionalized nanoparticles for biomedical applications: a soft matter perspective. NPJ Comput. Mater. 3:48. doi: 10.1038/s41524-017-0050-y
Arun Krishnan, R., Elizabeth Thomas, R., Sukumaran, A., Paul, J. K., and Vasudevan, D. M. (2020). COVID-19: current trends in in vitro diagnostics. Indian J. Clin. Biochem. 35, 285–289. doi: 10.1007/s12291-020-00906-5
Baviskar, T., Raut, D., and Bhatt, L. K. (2020). Deciphering vaccines for COVID-19: where do we stand today? Immunopharmacol. Immunotoxicol. 1–14. doi: 10.1080/08923973.2020.1837867
Boni, M. F. (2008). Vaccination and antigenic drift in influenza. Vaccine 26(Suppl. 3):C8–C14. doi: 10.1016/j.vaccine.2008.04.011
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., et al. (2020). CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874. doi: 10.1038/s41587-020-0513-4
Carter, N. J., and Curran, M. P. (2011). Live attenuated influenza vaccine (FluMist(R);Fluenz) a review of its use in the prevention of seasonalinfluenza in children and adults. Drugs 71, 1591–1622. doi: 10.2165/11206860-000000000-00000
Centers for Disease Control Prevention (2019). Coronavirus Disease (COVID-19). How to Protect Yourself and Others. Available online at: https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/prevention.html (accessed April 3, 2020).
Chan, J. F. W., Kok, K. H., Zhu, Z., Chu, H., To, K. K., Yuan, S., et al. (2020). Genomic characterization of the 2019 novel human pathogenic coronavirus isolated from a patient with a typical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9, 221–236. doi: 10.1080/22221751.2020.1719902
Chan, W. C. W. (2020). Nano Research for COVID-19. ACS Nano 14, 3719–3720. doi: 10.1021/acsnano.0c02540
Chattopadhyay, S., Chen, J. Y., Chen, H. W., and Hu, C. J. (2017). Nanoparticle vaccinesadopting virus-like features for enhanced immune potentiation. Nanotheranostics 1, 244–260. doi: 10.7150/ntno.19796
Chauhan, D. S., Prasad, R., Srivastava, R., Jaggi, M., Chauhan, S. C., and Yallapu, M. M. (2020). Comprehensive review on current interventions, diagnostics, and nanotechnology perspectives against SARS-CoV-2. Bioconjugate Chem. 31, 2021–2045. doi: 10.1021/acs.bioconjchem.0c00323
Chauhan, G., Madou, M. J., Kalra, S., Chopra, V., Ghosh, D., and Martinez-Chapa, S. O. (2020). Nanotechnology for COVID-19: therapeutics and Vaccine Research. ACS Nano 14, 7760–7782. doi: 10.1021/acsnano.0c04006
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513. doi: 10.1016/S0140-6736(20)30211-7
Chen, W.-H., Strych, U., Hotez, P. J., and Bottazzi, M. E. (2020). The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 7, 61–64. doi: 10.1007/s40475-020-00201-6
Daubeney, P., Taylor, C. J., McGaw, J., Brown, E. M., Ghosal, S., Keeton, B. R., et al. (1997). Immunogenicity and tolerability of a trivalent influenza subunit vaccine (Influvac) in high-risk children aged 6 months to 4 years. Br. J. Clin. Pract. 51, 87–90.
de Queiroz, N. M. G. P., Marinho, F. V., Chagas, M. A., Leite, L. C. C., Homan, E. J., de Magalhães, M. T. Q., et al. (2020). Vaccines for COVID-19: perspectives from nucleic acid vaccines to BCG as delivery vector system. Microbes Infect. 2, 4–8. doi: 10.1016/j.micinf.2020.09.004
Delore, V., Salamand, C., Marsh, G., Arnoux, S., Pepin, S., and Saliou, P. (2006). Long-term clinical trial safety experience with the inactivated split influenza vaccine, Vaxigrip. Vaccine 24, 1586–1592. doi: 10.1016/j.vaccine.2005.10.008
Destache, C. J., Belgum, T., Christensen, K., Shibata, A., Sharma, A., and Dash, A. (2009). Combination antiretroviral drugs in PLGA nanoparticle for HIV-1. BMC Infect. Dis. 9:198. doi: 10.1186/1471-2334-9-198
Dhere, R., Yeolekar, L., Kulkarni, P., Menon, R., Vaidya, V., and Ganguly, M. (2011). A pandemic influenza vaccine in India: from strain to sale within 12 months. Vaccine 29, A16–A21. doi: 10.1016/j.vaccine.2011.04.119
Esposito, S., Principi, N., Leung, C. C., and Migliori, G. B. (2020). Universal use of face masks for success against COVID-19: evidence and implications for prevention policies. Eur. Respiratory J. 55:2001260. doi: 10.1183/13993003.01260-2020
Geall, A. J., Verma, A., Otten, G. R., Shaw, C. A., Hekele, A., Banerjee, K., et al. (2012). Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U.S.A. 109, 14604–14609. doi: 10.1073/pnas.1209367109
Genchi, G. G., and Ciofani, G. (2019). Editorial: smart tools for caring: nanotechnology meets medical challenges. Front. Bioeng. Biotech. 7:11. doi: 10.3389/978-2-88945-806-6
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Gouglas, D., Thanh Le, T., Henderson, K., Kaloudis, A., Danielsen, T., Hammersland, N. C., et al. (2018). Estimating the cost of vaccine development against epidemic infectious diseases: a cost minimisation study. Lancet Global Health 6:e1386–96. doi: 10.1016/S2214-109X(18)30346-2
Graham, B. S., and Anderson, L. J. (2013). Challenges and opportunities for respiratory syncytial virus vaccines. Curr. Top. Microbiol. Immunol. 372, 391–404. doi: 10.1007/978-3-642-38919-1_20
Grohskopf, L. A., Sokolow, L. Z., Olsen, S. J., Bresee, J. S., Broder, K. R., and Karron, A. (2015). Prevention and control of influenza with vaccines: recommendations of the advisory committee on immunization practices, United States, 2015-16 influenza season. Morb. Mortal. Wkly. Rep. 64, 818–825. doi: 10.15585/mmwr.mm6430a3
Gupta, T., and Gupta, S. K. (2020). Potential adjuvants for the development of a SARS-CoV-2 vaccine based on experimental results from similar coronaviruses. Int. Immunopharmacol. 86:106717. doi: 10.1016/j.intimp.2020.106717
Hamad, I., Al-Hanbali, O., Hunter, A. C., Rutt, K. J., Andresen, T. L., and Moghimi, S. M. (2010). Distinct polymer architecture mediates switching of complement activation pathways at the nanosphere-serum interface: implications for stealth nanoparticle engineering. ACS Nano 4, 6629–6638. doi: 10.1021/nn101990a
Harrison, S. C., Olson, A. J., Schutt, C. E., Winkler, F. K., and Bricogne, G. (1978). Tomato bushy stunt virus at 2.9 Å resolution. Nature 276, 368–373. doi: 10.1038/276368a0
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entrydepends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280. doi: 10.1016/j.cell.2020.02.052
Hu, T. Y., Frieman, M., and andWolfram, J. (2020). Insightsfromnanomedicineintochloroquineefficacyagainst COVID-19. NatureNanotechnol. 15, 247–249. doi: 10.1038/s41565-020-0674-9
Huang, H., Fan, C., Li, M., Nie, H.-L., Wang, F.-B., Wang, H., et al. (2020). COVID-19: a call forphysical scientists and engineers ACS Nano. 14, 3747–3754. doi: 10.1021/acsnano.0c02618
Hutnick, N. A., Myles, D. J., Ferraro, B., Lucke, C., Lin, F., Yan, J., et al. (2012). Intradermal DNA vaccination enhanced by low-current electroporation improves antigen expression and induces robust cellular and humoral immune responses. Hum. Gene Ther. 23, 943–950. doi: 10.1089/hum.2012.055
Irvine, D. J., Hanson, M. C., Rakhra, K., and Tokatlian, T. (2015). Synthetic nanoparticles for vaccines and immunotherapy. Chem. Rev. 115, 11109–11146. doi: 10.1021/acs.chemrev.5b00109
Jayant, R. D., Tiwari, S., Atluri, V., Kaushik, A., Tomitaka, A., Yndart, A., et al. (2018). Multifunctional nanotherapeutics for the treatment of neuro AIDS in drug abusers. Sci. Rep. 8:12991. doi: 10.1038/s41598-018-31285-w
Kageyama, T., Kojima, S., Shinohara, M., Uchida, K., Fukushi, S, Hoshino, F. B., et al. (2003). Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41, 1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003
Kanasty, R., Dorkin, J. R., Vegas, A., and Anderson, D. (2013). Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977. doi: 10.1038/nmat3765
Kasas, S., Thomson, N. H., Smith, B. L., Hansma, H. G., Zhu, X., Guthold, M., et al. (1997). Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry 36, 461–468. doi: 10.1021/bi9624402
Keshavarzi Arshadi, A., Webb, J., Salem, M., Cruz, E., Calad-Thomson, S., Ghadirian, N., et al. (2020). Artificial intelligence for COVID-19 drug discovery and vaccine development. Front. Artific. Intelligence 3:65. doi: 10.3389/frai.2020.00065
Kilic, T., Weissleder, R., and Lee, H. (2020). Molecular and immunological diagnostic tests of COVID-19: current status and challenges. Science 23:101406. doi: 10.1016/j.isci.2020.101406
Kobayashi, K., Wei, J., Iida, R., Ijiro, K., and Niikura, K. (2014). Surface engineering of nanoparticles for therapeutic applications. Polym. J. 46, 460–468. doi: 10.1038/pj.2014.40
Konda, A., Prakash, A., Moss, G. A., Schmoldt, M., Grant, G. D., and Guha, S. (2020). Aerosol filtration efficiency of common fabrics used in respiratory cloth masks. ACS Nano 14, 6339–6347. doi: 10.1021/acsnano.0c03252
Konwarh, R. (2020a). Can CRISPR/cas technology be a felicitous stratagem against the COVID-19 Fiasco? prospects and hitches. Front. Mol. Biosci. 7:244. doi: 10.3389/fmolb.2020.557377
Konwarh, R. (2020b). Nanobodies: prospects of expanding the gamut of neutralizing antibodies against the novel coronavirus, SARS-CoV-2. Front. Immunol. 11:1531. doi: 10.3389/fimmu.2020.01531
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al. (2020). Tracking changes in SARS-CoV-2 spike: evidence that d614g increases infectivity of the COVID-19 virus. Cell 182:e19. doi: 10.1016/j.cell.2020.06.043
Kraft, J. C., McConnachie, L. A., Koehn, J., Kinman, L., Sun, J., Collier, A. C., et al. (2018). Mechanism-basedpharmacokinetic (MBPK) models describe thecomplex plasma kinetics of three antiretrovirals delivered by a long-acting anti-HIV drug combination nanoparticle formulation. J. Control. Release 275, 229–241. doi: 10.1016/j.jconrel.2018.02.003
Krammer, F., and Simon, V. (2020). Serologyassays to manage COVID-19. Science 368, 1060–1061. doi: 10.1126/science.abc1227
Kranz, L. M., Diken, M., Haas, H., Kreiter, S., Loquai, C., Reuter, K. C., et al. (2016). Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401. doi: 10.1038/nature18300
Lauer, S. A., Grantz, K. H., Bi, Q., Jones, F. K., Zheng, Q., Meredith, H. R., et al. (2020). The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Annals Int. Med. 172, 577–582. doi: 10.7326/M20-0504
Laval, J. M., Mazeran, P. E., and Thomas, D (2000). Nanobiotechnology and its role in the development of new analytical devices. Analyst 125, 29–33. doi: 10.1039/a907827d
Le Ferrand, H. (2020). How can materials science contribute to fighting against the new coronavirus? MRS Bulletin 45, 327–330. doi: 10.1557/mrs.2020.129
Lecoq, H. (2001). Découverte du premier virus, le virus de la mosaïque du tabac: 1892 ou 1898? ComptesRendus de l'Académie Des Sciences. Series IIISciences de La Vie 324, 929–933. doi: 10.1016/S0764-4469(01)01368-3
Lembo, D., Donalisio, M., Civra, A., Argenziano, M., and Cavalli, R. (2018). Nanomedicine formulations for the delivery of antiviral drugs:a promising solution for thetreatment of viral infections. Exp. Opin. Drug Delivery 15, 93–114. doi: 10.1080/17425247.2017.1360863
Lin, S., Shen, R., He, J., Li, X., and Guo, X. (2020). Molecular modeling evaluation of the binding effect of ritonavir, lopinavir and darunavir to severe acute respiratory syndrome coronavirus 2 proteases. BioRxiv 2020.01.31.929695. doi: 10.1101/2020.01.31.929695
Liu, L., Ren, J., He, Z., Men, K., Mao, Y., Ye, T., et al. (2017). Cholesterol-modified hydroxychloroquine loaded nanocarriers in bleomycin-induced pulmonary fibrosis. Sci. Rep. 7:10737. doi: 10.1038/s41598-017-11450-3
Long, Y., Hu, T., Liu, L., Chen, R., Guo, Q., Yang, L., et al. (2020). Effectiveness of N95 respirators versus surgical masks against influenza: a systematic review and meta-analysis. J. Evidence Based Med. 13, 93–101. doi: 10.1111/jebm.12381
Luke, J. M., Simon, G. G., Söderholm, J., Errett, J. S., August, J. T., Gale, M., et al. (2011). Coexpressed RIG-I agonist enhances humoral immune response to influenza virus DNA vaccine. J. Virol. 85, 1370–1383. doi: 10.1128/JVI.01250-10
Lung, J., Lin, Y. S., Yang, Y. H., Chou, Y. L., Shu, L. H., Cheng, Y. C., et al. (2020). The potential chemical structure of AntiSARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 92, 693–697. doi: 10.1002/jmv.25761
Lung, P., Yang, J., and Li, Q. (2020). Nanoparticle formulated vaccines:opportunities and challenges. Nanoscale 12, 5746–5763. doi: 10.1039/C9NR08958F
Lurie, N., Saville, M., Hatchett, R., and Halton, J. (2020). Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973. doi: 10.1056/NEJMp2005630
Lynch, I., Feitshans, I. L., and Kendall, M. (2015). 'Bio-nano interactions: new tools, insights and impacts': summary of the Royal Society discussion meeting. Philosoph. Transact. R. Soc. 370:20140162. doi: 10.1098/rstb.2014.0162
Ma, L., Kohli, M., and Smith, A. (2013). Nanoparticles for combination drug therapy. ACS Nano 7, 9518–9525. doi: 10.1021/nn405674m
Martinez-Delgado, G. (2020). Inhaled nanobodies against COVID-19. Nat. Rev. Immunol. 20:593. doi: 10.1038/s41577-020-00443-5
Metsky, H. C., Freije, C. A., Kosoko-Thoroddsen, T.-S. F., Sabeti, P. C., and Myhrvold, C. (2020). CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. BioRxiv 2020:967026. doi: 10.1101/2020.02.26.967026
Morawska, L. (2006). Droplet fate in indoor environments, or can we prevent the spread of infection? Indoor Air 16, 335–347. doi: 10.1111/j.1600-0668.2006.00432.x
Mori, Y., Nagamine, K., Tomita, N., and Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphateformation. Biochem. Biophys. Res. Commun. 289, 150–154. doi: 10.1006/bbrc.2001.5921
Mungroo, M., Khan, N., and Siddiqui, R. (2020). Novel coronavirus: currentunderstanding of clinicalfeatures, diagnosis, pathogenesis, andtreatmentoptions. Pathogens 9:297. doi: 10.3390/pathogens9040297
Nalawansha, D. A., and Samarasinghe, K. T. G. (2020). Double-barreled CRISPR technology as a novel treatment strategy for COVID-19. ACS Pharmacol. Transl. Sci. 3, 790–800. doi: 10.1021/acsptsci.0c00071
Nel, A. E., Mädler, L., Velegol, D., Xia, T., Hoek, E. M. V., Somasundaran, P., et al. (2009). Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 8, 543–557. doi: 10.1038/nmat2442
Norton, E. B., Lawson, L. B., and Mahdi, Z. (2012). The A subunit of Escherichia coli heat-labile enterotoxin functions as amucosal adjuvant and promotes IgG2a, IgA, and Th17responses to vaccine antigens. Infect Immun. 80, 2426–2435. doi: 10.1128/IAI.00181-12
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. doi: 10.1093/nar/28.12.e63
Omolo, C. A., Soni, N., Fasiku, V. O., Mackraj, I., and Govender, T. (2020). Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. Eur. J. Pharmacol. 883:173348. doi: 10.1016/j.ejphar.2020.173348
Ortiz-Prado, E., Simbaña-Rivera, K., Gómez- Barreno, L., Rubio-Neira, M., Guaman, L. P., Kyriakidis, N. C., et al. (2020). Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the Coronavirus Disease 2019 (COVID-19), a comprehensive literature review. Diagnost. Microbiol. Infect. Dis. 98:115094. doi: 10.1016/j.diagmicrobio.2020.115094
Parihar, A., Ranjan, P., Sanghi, S. K., Srivastava, A. K., and Khan, R. (2020). Point-of-care biosensor-based diagnosis of COVID-19 holds promise to combat current and future pandemics. ACS Appl. Bio Mater. 4:3458. doi: 10.1021/acsabm.0c01083
Parveen, K., Banse, V., and Ledwani, L. (2016). Green synthesis of nanoparticles: their advantages and disadvantages. AIP Conference Proc. 1724:20048. doi: 10.1063/1.4945168
Pati, R., Shevtsov, M., and Sonawane, A. (2018). Nanoparticle vaccines against infectious diseases. Front. Immunol. 9:2224. doi: 10.3389/fimmu.2018.02224
Petros, R. A., and DeSimone, J. M. (2010). Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discovery 9, 615–627. doi: 10.1038/nrd2591
Phan, T. (2020). Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evolution 81:104260. doi: 10.1016/j.meegid.2020.104260
Pisani, A., Pompa, P. P., and Bardi, G. (2020). Potential applications of nanomaterials to quench the cytokine storm in coronavirus disease 19. Front. Bioeng. Biotechnol. 8:906. doi: 10.3389/fbioe.2020.00906
Prompetchara, E., Ketloy, C., and Palaga, T. (2020). Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 38, 1–9. doi: 10.12932/AP-200220-0772
Radmacher, M., Fritz, M., Hansma, H. G., and Hansma, P. K. (1994). Direct observation of enzyme activity with the atomic force microscope. Science 265, 1577–1579. doi: 10.1126/science.8079171
Rai, P. K., Usmani, Z., Thakur, V. K., Gupta, V. K., and Mishra, Y. K. (2020). Tackling COVID-19 pandemic through nanocoatings: confront and exactitude. Curr. Res. Green Sustain. Chem. 3:100011. doi: 10.1016/j.crgsc.2020.100011
Reed, S. G., Bertholet, S., Coler, R. N., and Friede, M. (2009). New horizons in adjuvants for vaccine development. Trends Immunol. 30, 23–32. doi: 10.1016/j.it.2008.09.006
Reichmuth, A. M., Oberli, M. A., Jaklenec, A., Langer, R., and Blankschtein, D. (2016). mRNA vaccine delivery using lipid nanoparticles. Therapeut. Delivery 7, 319–334. doi: 10.4155/tde-2016-0006
Riediker, M. (2020). Nano-safety research lessons for dealing with aerosol transmissions of COVID-19. Nanotoxicology 14, 866–868. doi: 10.1080/17435390.2020.1786185
Salazar-Gonzlez, J. A., Gonzalez-Ortega, O., and Rosales- Mendoza, S. (2015). Gold nanoparticles and vaccine development. Expert Rev. Vaccines 14, 1197–1211. doi: 10.1586/14760584.2015.1064772
Samrat, S. K., Tharappel, A. M., Li, Z., and Li, H. (2020). Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 288:198141. doi: 10.1016/j.virusres.2020.198141
Schneider, C. S., Xu, Q., Boylan, N. J., Chisholm, J., Tang, B. C., Schuster, B. S., et al. (2017). Nanoparticles that do not adhere to mucus provide uniform and long-lasting drug delivery to airways following inhalation. Sci. Adv. 3:e1601556. doi: 10.1126/sciadv.1601556
Seto, W. H., Tsang, D., and Yung, R. W. (2003). Effectiveness of precautions against droplets and contact in prevention ofnosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 361, 1519–1520. doi: 10.1016/S0140-6736(03)13168-6
Shibata, A., McMullen, E., Pham, A., Belshan, M., Sanford, B., Zhou, Y., et al. (2013). Polymeric nanoparticles containing combination antiretroviral drugsfor HIV type 1 treatment. AIDS Res. Hum. Retroviruses 29, 746–754. doi: 10.1089/aid.2012.0301
Skariyachan, S., Challapilli, S. B., Packirisamy, S., Kumargowda, S. T., and Sridhar, V. S. (2019). Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for middle east respiratory syndrome coronavirus infections. Front. Microbiol. 10:569. doi: 10.3389/fmicb.2019.00569
Smith, T. R., Schultheis, K., Morrow, M. P., Kraynyak, K. A., McCoy, J. R., Yim, K. C., et al. (2017). Development of an intradermal DNA vaccine delivery strategy to achieve single-dose immunity against respiratory syncytial virus. Vaccine 35, 2840–2847. doi: 10.1016/j.vaccine.2017.04.008
Srivastava, M., Srivastava, N., Mishra, P. K., and Malhotra, B. D. (2021). Prospects of nanomaterials-enabled biosensors for COVID-19 detection. Sci. Total Environ. 754:142363. doi: 10.1016/j.scitotenv.2020.142363
Szeto, G. L., and Lavik, E. B. (2016). Materials design at the interface of nanoparticles and innateimmunity. J. Mater. Chem. B 4, 1610–1618. doi: 10.1039/C5TB01825K
Talebian, S., Wallace, G. G., Schroeder, A., Stellacci, F., and Conde, J. (2020). Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 15, 618–621. doi: 10.1038/s41565-020-0751-0
Tang, B., He, F., Liu, D., Fang, M., Wu, Z., and Xu, D. (2020). AI-aided design of novel targeted covalent inhibitors against SARS-CoV-2. BioRxiv doi: 10.1101/2020.03.03.972133
Thomas, S. N., van der Vlies, A. J., O'Neil, C. P., Reddy, S. T., Yu, S. S., Giorgio, T. D., et al. (2011). A engineering complement activation on polypropylene sulfide vaccine nanoparticles. Biomaterials 32, 2194–2203. doi: 10.1016/j.biomaterials.2010.11.037
Udugama, B., Kadhiresan, P., Kozlowski, H. N., Malekjahani, A., Osborne, M., Li, V. Y. C., et al. (2020). Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14, 3822–3835. doi: 10.1021/acsnano.0c02624
van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., et al. (2020). Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 382, 1564–1567. doi: 10.1056/NEJMc2004973
Vartak, A., and Sucheck, S. J. (2016). Recent advances in subunit vaccine carriers. Vaccines 4:12. doi: 10.3390/vaccines4020012
Verma, S., Twilley, D., Esmear, T., Oosthuizen, C. B., Reid, A.-M., Nel, M., et al. (2020). Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 11:1514. doi: 10.3389/fphar.2020.561334
Wagner, D. M., Klunk, J., Harbeck, M., Devault, A., Waglechner, N., Sahl, J. W., et al. (2014). Yersinia pestis and the Plague of Justinian 541 and #x2013;543 AD: a genomic analysis. Lancet Infect. Dis. 14, 319–326. doi: 10.1016/S1473-3099(13)70323-2
Wang, J., and Du, G. (2020). COVID-19 may transmit through aerosol.Ir. J. Med. Sci. 189, 1143–1144. doi: 10.1007/s11845-020-02218-2
Weiss, C., Carriere, M., Fusco, L., Capua, I., Regla-Nava, J. A., Pasquali, M., et al. (2020). Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 14, 6383–6406. doi: 10.1021/acsnano.0c03697
Weng, D., Qi, H., Wu, T.-T., Yan, M., Sun, R., and Lu, Y. (2012). Visible light powered self-disinfecting coatings for influenza viruses. Nanoscale 4, 2870–2874. doi: 10.1039/c2nr30388d
Wilson, I. A., Skehel, J. J., and Wiley, D. C. (1981). Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373. doi: 10.1038/289366a0
Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M. A., et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469. doi: 10.1038/s41586-020-2196-x
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507
Xiang, J., Yan, M., Li, H., Liu, T., Lin, C., Huang, S., et al. (2020). Evaluation of enzyme-link edimmunoassay and colloidal gold- immunochromatographicassay kit for detection of novel coronavirus (SARS-Cov-2) causingan outbreak of pneumonia (COVID-19). BioRxiv doi: 10.1101/2020.02.27.20028787
Xu, L., Li, D., Ramadan, S., Li, Y., and Klein, N. (2020). Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 170:112673. doi: 10.1016/j.bios.2020.112673
Yang, D.-M., Chang, T.-J., Wang, M.-L., Tsai, P.-H., Lin, T.-H., Wang, C.-T., et al. (2020). Hunting severe acute respiratory syndrome coronavirus 2 (2019 novel coronavirus): From laboratory testing back to basic research. J. Chinese Med. Associat. 83, 524–526. doi: 10.1097/JCMA.0000000000000332
Zeng, X., Song, X., Ma, T., Pan, X., Zhou, Y., Hou, Y., et al. (2020). Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 11, 4624–4636. doi: 10.1021/acs.jproteome.0c00316
Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamideinhibitors. Science 368, 409–412. doi: 10.1126/science.abb3405
Zhang, L., and Liu, Y. (2020). Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 92, 479–490. doi: 10.1002/jmv.25707
Zhang, W., Du, R., Li, B., Zheng, X., Yang, X., and Hu, B. (2020). Molecularand serologicalinvestigation of 2019-nCoV infectedpatients: implication of multiplesheddingroutes. Emerg. Microb. Infect. 9, 386–389. doi: 10.1080/22221751.2020.1729071
Zhao, L., Seth, A., Wibowo, N., Zhao, C.-X., Mitter, N., Yu, C., et al. (2014). Nanoparticle vaccines. Vaccine 32, 327–337. doi: 10.1016/j.vaccine.2013.11.069
Keywords: Covid-19, Bio-Nano, technology, interface, pandemic
Citation: Kamaraj S-K (2020) The Perspective on Bio-Nano Interface Technology for Covid-19. Front. Nanotechnol. 2:586250. doi: 10.3389/fnano.2020.586250
Received: 22 July 2020; Accepted: 04 November 2020;
Published: 30 November 2020.
Edited by:
Francis Verpoort, Wuhan University of Technology, ChinaReviewed by:
Rocktotpal Konwarh, Addis Ababa Science and Technology University, EthiopiaHao Song, University of Queensland, Australia
Copyright © 2020 Kamaraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sathish-Kumar Kamaraj, U2F0aGlzaC5rQGxsYW5vLnRlY25tLm14; c2F0aGlzaC5ib3RAZ21haWwuY29t
 Sathish-Kumar Kamaraj
Sathish-Kumar Kamaraj