- 1Institute for Molecular Biotechnology, RWTH Aachen University, Aachen, Germany
- 2Electron Microscopy Facility, Institute of Pathology, RWTH Aachen University Hospital, Aachen, Germany
- 3Fraunhofer Institute for Molecular Biology and Applied Ecology IME, Aachen, Germany
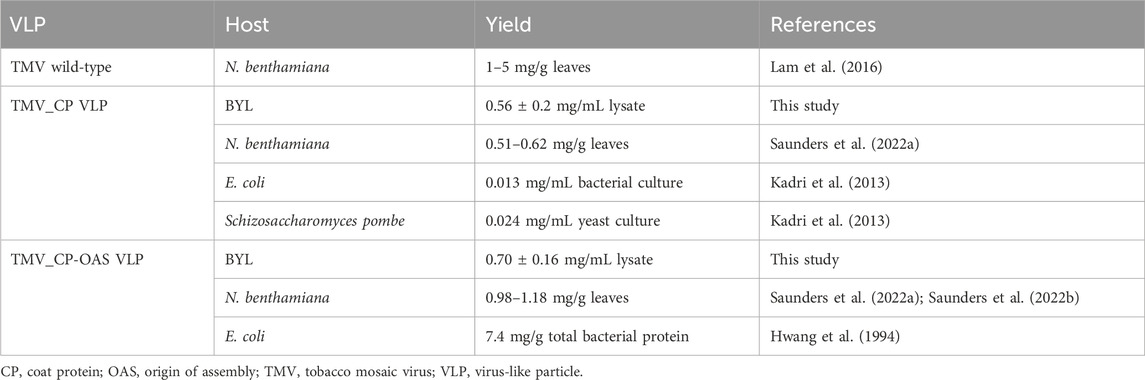
Plant virus nanoparticles (VNPs) can be used to generate versatile, functionalized nanomaterials, but they contain replicative genomic DNA whose impact on the environment must be assessed carefully. In contrast, plant virus-like particles (VLPs) lack replicative genomic information and are therefore non-infectious. However, the production of VLPs in plants is associated with challenges such as low yields and a purification process that is difficult to scale up. To address these issues, we used cell-free lysates derived from Nicotiana tabacum BY-2 cells to produce tobacco mosaic virus (TMV)-like particles. The objective of this approach was to generate VLPs with and without encapsulated RNA. SDS-PAGE and Western blotting indicated the accumulation of abundant quantities of the viral coat protein, with yields of up to 3.56 ± 0.62 mg per ml of cell-free lysate confirmed by enzyme-linked immunosorbent assay before purification. The yield of purified TMV VLP is comparable to that obtained from plants due to the low assembly efficiency, with assembled VLPs yielding up to 0.70 ± 0.16 mg per ml of lysate. Notably, this yield is approximately 23 times higher than that obtained through microbial expression systems, obviating the need for the labor-intensive processes typically associated with plant-based methods. Transmission electron microscopy revealed that VLPs assembled in the lysate at pH 7, and remained stable at up to pH 8.5. Cell-free expression therefore offers a rapid, straightforward and cost-effective method for the production of plant VLPs at high yields, and establishes a foundation for the on-demand production of functionalized VLPs under controlled and reproducible conditions.
1 Introduction
Nanomaterials have become an integral component of many industries, but synthetic nanoparticles can be toxic and persist in the environment (Malik et al., 2023). In contrast, nanoparticles derived from plant viruses – known as plant virus nanoparticles (VNPs) – provide valuable nanomaterials for many research applications. These naturally occurring structures typically range from 20 to 500 nm in size and have distinctive physical and biochemical properties that make them suitable for diverse nanoscale functions, including vaccination, diagnostics, tissue engineering, and drug delivery (Röder et al., 2019; Chung et al., 2020). VNPs are easily modified by chemical conjugation and/or genetic engineering to extend their application range (Steele et al., 2017; Wijesundara et al., 2022). They can be synthesized inexpensively in large amounts by molecular farming in plants (Röder et al., 2019), but their replicative genomic information means they remain infectious and have the potential to cause plant diseases. This can be avoided by the functionalization of virus-like particles (VLPs), which are self-assembling coat protein (CP) subunits resembling VNPs but lacking replicative genomic information. However, CP assembly without the encapsulation of RNA often results in lower yields, and for some viruses it is not possible at all (Hwang et al., 1994; Saunders et al., 2022a).
Tobacco mosaic virus (TMV) is a widely used model system in the material and analytical sciences (Lomonossoff and Wege, 2018). The 300 × 18 nm rod-shaped TMV capsid features 2130 identical CP subunits helically arranged around a 6.4 kb (+)ssRNA genome (Namba et al., 1989). The origin of assembly (OAS) is 0.9 kb from the 3′ end of the RNA and interacts with the CP subunits. Heterologous RNAs containing the OAS have been used to create diverse VLP structures in vivo and in vitro (Wege and Koch, 2020). CPs can also assemble into VLPs without RNA in defined conditions of pH and ionic strength (Butler, 1984), but a small quantity of native CPs are needed to initiate assembly (Eiben et al., 2014). The drawbacks of this approach include the cost of scale-up (CPs must be produced separately, usually from TMV-infected plants) and the loss of stability in the absence of viral RNA (Lee et al., 2021). Plant-derived TMV CPs can form nanorods without the OAS, but the assembly process is 60% less efficient (Saunders et al., 2022a). However, the resulting VLPs show variations in shape and length, making them unsuitable for applications where precise, monodisperse particles are required.
Cell-free systems circumvent many of the limitations associated with cell-based expression, including poor product yields and protein instability (Li et al., 2018; Bogart et al., 2021). Production times are shorter and on-demand production is possible because the cell-free lysate can be stored at −80 °C for more than a year before protein synthesis is initiated by the addition of the appropriate template DNA (Buntru et al., 2015). The tobacco BY-2 cell-free lysate (BYL) prepared from cell suspension cultures has been commercialized by the company LenioBio under the brand name ALiCE (LenioBio, 2024). It allows production at up to the 10-L scale, with yields of some recombinant proteins reaching grams per batch (Gupta et al., 2023). The expression of hepatitis B tandem-core VLPs in Escherichia coli lysates achieved more consistently assembled particles (Colant et al., 2021a), but the BYL platform increased the yield by 2.5-fold (Armero-Gimenez et al., 2023; Gupta et al., 2023). Given its source, BYL is more appropriate than bacterial lysates for the production of plant virus CPs.
We therefore tested the BYL for the controlled and efficient production of TMV-like particles. We developed a cell-free production method for these VLP carrier nanomaterials, addressing the issues of labor-intensive procedures, low yields, and containment inherent to cell-based production. To investigate the feasibility of assembling TMV CPs into VLPs using a cell-free lysate, we constructed two plasmids for the expression of the TMV CP. One contained the cp gene alone, whereas the other also provided the OAS, facilitating the formation of TMV VLPs with and without encapsulated RNA. We assessed the expression and assembly of TMV VLPs by SDS-PAGE, Western blot and transmission electron microscopy (TEM), and determined the quantity of particles using an enzyme-linked immunosorbent assay (ELISA). Finally, we tested the stability of the cell-free TMV VLPs in buffers spanning a range of pH values to evaluate the BYL for streamlined and scalable VLP production.
2 Materials and methods
2.1 Vector design
Two vectors (pLenEX_TMV_CP-OAS and pLenEx_TMV_CP) were prepared for the cell-free expression of TMV CP (strain U1) with and without the OAS (Figure 1; Saunders et al., 2022b). The cp gene was amplified from vector pET22b-TMV_CP (unpublished data) using primers Len-CP U1_fw/Len-CP U1_rev (Supplementary Table S1) to facilitate Gibson assembly (NEB, Frankfurt am Main, Germany). The OAS was isolated from 5 µg of wild-type TMV by RT-PCR in a reaction containing 0.2 µM primer pLen_seq_rev, 200 U M-MLV Reverse Transcriptase RNaseH Minus Point Mutant (Promega, Walldorf, Germany), 1x M-MLV reaction buffer, and 1 mM dNTPs. The reaction was incubated for 30 min at 40 °C, 20 min at 45 °C, 20 min at 50 °C, 20 min at 55 °C, and 20 min at 70 °C. The OAS was amplified using Polymerase Q5 (NEB) and primers OAS_fw/OAS_rev to generate overlaps with the cp gene, and then using primers Len-CP_fw/CPU1-OAS_rev. PCR products were resolved by 1.2% (w/v) agarose gel electrophoresis, purified using the Wizard SV Gel and PCR Clean-Up System (Promega), and the concentration was determined using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Darmstadt, Germany). The inserts were transferred to pLenEx_Strep-eYFP (Buntru et al., 2022b; Buntru et al., 2022a), pre-treated with NcoI-HF and KpnI-HF, using NEBuilder HiFi DNA Assembly Master Mix (NEB). This replaced the Strep-eYFP sequence with the TMV CP or CP-OAS sequence. Competent E. coli DH5α cells were transformed by electroporation and selected on medium containing ampicillin. Positive colonies were identified by PCR using the vector-specific primers pLen_seq_fw/pLen-seq_rev and GoTaq DNA Polymerase (Promega). The vectors were isolated using the NucleoBond Xtra Midi plasmid kit (Macherey-Nagel, Düren, Germany) and verified by sequencing (Eurofins Genomics, Ebersberg, Germany). Sequences can be found in the supplementary material.
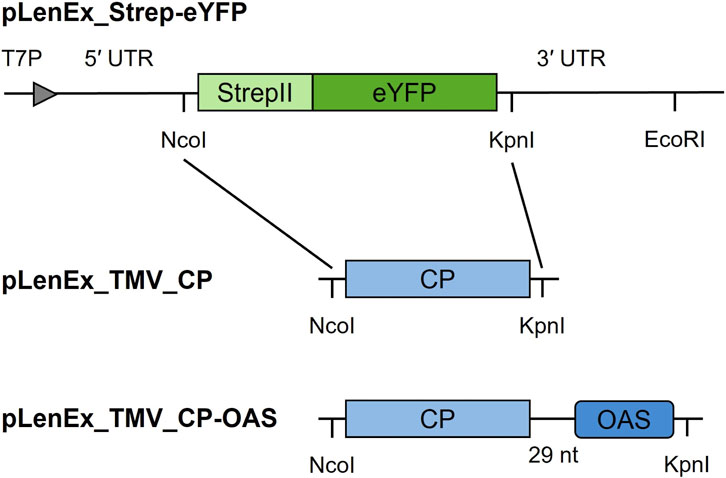
Figure 1. Schematic illustration of constructs used for the cell-free production of TMV VLPs. The TMV cp gene replaces the strepII tag and eyfp sequences. Abbreviations: 3′ UTR – TMV 3′ untranslated region, 5′ UTR – TMV omega sequence, CP – coat protein gene from TMV strain U1, eYFP – enhanced yellow fluorescent protein gene, OAS – origin of assembly, StrepII – strepII tag sequence, T7P – T7 promoter.
2.2 Cell-free production of VLPs
TMV VLPs were expressed using the ALiCE system (LenioBio, Aachen, Germany) at the 50–200-µL scale for 48 h at 25 °C, shaking at 700 rpm (Buntru et al., 2022b). Plasmid pLenEx_Strep-eYFP and water without DNA were used as controls. The eYFP signal was quantified to calculate lysate productivity by diluting reaction samples in PBS (1:10, 1:20 and 1:50) and transferring three replicate 50-µL aliquots per dilution to black microtiter plates (Greiner Bio-One, Frickenhausen, Germany). Strep-eYFP pure standards and BYL no-template reactions were used as controls. Fluorescence was measured by spectrophotometry in a plate reader (Tecan, Männedorf, Switzerland) with excitation at 485 nm (9 nm) and emission at 528 nm (20 nm).
2.3 Production of wild-type TMV particles
Wild-type TMV particles were propagated in tobacco plants growing in a phytochamber (26/20 °C day/night, 12-h photoperiod at 5,000–10,000 lux, 70% humidity). Leaves were harvested 16–18 days post-infection, and particles were extracted and purified as previously described (Schuphan et al., 2024).
2.4 Protein expression analysis
Reaction samples were boiled for 5 min in 5× reducing loading buffer (Laemmli, 1970) before SDS-PAGE using a 12% or 16% resolving gel and a 4% stacking gel (2 µL loaded per lane). Protein bands were stained with Coomassie Brilliant Blue (Thermo Fisher Scientific) and compared to P7719 pre-stained protein standards (NEB). Proteins were transferred to a Hybond-C nitrocellulose membrane (GE Healthcare, Freiburg, Germany) using a semi-dry blotting system (Bio-Rad Laboratories, Feldkirchen, Germany). The membrane was blocked for at least 30 min with 4% (w/v) skimmed milk (Roth, Karlsruhe, Germany) in PBS. Proteins were detected with a primary anti-TMV (strain U1) antibody (DSMZ, Braunschweig, Germany; diluted 1:5000 in PBS) at room temperature overnight. The membrane was washed with PBS and incubated with an alkaline phosphatase (AP)-labeled secondary goat-anti rabbit (GAR) antibody (Dianova, Hamburg, Germany; diluted 1:5000 in PBS) for at least 1.5 h. The signal was visualized with nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyphosphate p-toluidine salt (NBT/BCIP; Roth) diluted 1:100 in AP buffer (100 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, pH 9.6).
2.5 Assembly optimization and stability analysis
Reaction samples were centrifuged (15,000 x g, 4 °C, 15 min) to sediment the VLPs, which were resuspended in 30 µL PBS. We then transferred 6-µL aliquots to 30 µL of buffer that either favored (0.1 or 0.4 M sodium acetate pH 5.5) or prevented (0.4 M Tris-HCl pH 8.5) in vitro assembly and incubated the sample at 20 °C for 5–6 days.
2.6 Purification of VLPs
VLPs were purified by CaptoCore700 chromatography (Cytiva, Dreieich, Germany). The gravity flow column was prepared with 1 mL CaptoCore matrix equilibrated with 3 mL PBS. A 350-μL reaction sample was mixed with 21 mL PBS and applied to the column for 5 min. The flow-through fraction containing VLPs was concentrated using an Eppendorf Concentrator 5301 (Eppendorf, Wesseling, Germany) at room temperature to a final volume of 700 μL before analysis.
2.7 ELISA
CP-containing lysates (and non-template controls) were diluted 1:200, 1:500 and 1:1000 in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and 100-µL aliquots were coated onto high-binding microtiter plates (Greiner Bio-one) for 4 h at 37 °C. Wells were blocked with 200 µL 4% (w/v) skimmed milk in PBS for 1 h at 37 °C. CPs were detected with the antibodies described above: 100 µL of the anti-TMV antibody (diluted 1:1000 in PBS) overnight at 4 °C, then 100 µL of the AP-labeled GAR antibody (diluted 1:1000 in PBS) for at least 3 h at 37 °C. Each step was followed by three washes in PBS. The signal was visualized with 100 μL of 1 mg/mL p-nitrophenylphosphate (Roth) in substrate buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM MgCl2, pH 9.6). The absorbance of three replicate samples was recorded at 405 nm using the Tecan multiwell plate reader, with wild-type TMV as a calibration standard and PBS as the blank.
2.8 Semi-native gel electrophoresis
5-μL reaction samples or 10 μg of purified VLPs were supplemented with 6x gel loading purple dye (NEB) prior to separation by 1% (w/v) agarose gel electrophoresis for 60 min at 60 V in TAE buffer (40 mM Tris base, 20 mM acetic acid, 1 mM EDTA) supplemented with 0.3 μg/mL ethidium bromide (Roth). 30 μg of wild-type TMV particles were loaded in the control lane. The gel was imaged under UV light before destaining in 5.0% (v/v) methanol with 7.5% (v/v) acetic acid for 15 min, and staining with 0.25% (w/v) Coomassie Brilliant Blue G-250 in 50% (v/v) methanol +10% (v/v) acetic acid for 1 h. The gel was then destained for 3–5 days.
2.9 TEM
Pioloform-coated nickel grids (Plano, Wetzlar, Germany) were incubated with the anti-TMV antibody mAb75 (Fischer et al., 1999; diluted 1:100 in ultrapure water) for at least 30 min before blocking with 0.5% (w/v) bovine serum albumin (BBI Solutions, Freiburg, Germany) for at least 45 min. The grids were washed with three drops of ultrapure water before floating on 10–15 µL of BYL (or buffer) and 15 µL ultrapure water at room temperature overnight. After another washing step, the grids were contrasted with 1% (v/v) phosphotungstic acid. The purified VLPs were adsorbed directly onto the grids for 2 h at room temperature before washing and contrasting, and were visualized using a Hitachi HT7800 transmission electron microscope (Hitachi, Tokyo, Japan) operating at 100 kV.
2.10 Cryo-TEM
Purified VLPs were plunge-freezed in −180 °C cold ethan on lacey carbon nickel grids (Science Services, Munich, Germany) and analysed with a cryo-TEM Hitachi HT7800 transmission electron microscope (Hitachi, Tokyo, Japan) operating at an acceleration voltage of 120 kV.
2.11 Protein analysis using LC-MS/MS
To examine the acetylation, the VLPs were subjected to LC-MS/MS analysis. The protein content of the samples was reduced and separated on NuPAGE Bis-Tris 4%–12% gradient gels. Proteins were stained with Coomassie, as previously described by Kriegsheim et al. (2008). The TMV CP bands were then excised and digested using Peptidyl-Asp metalloendopeptidase (Asp-N; Promega) in accordance with the manufacturer’s protocol. The gel pieces were subsequently destained using 50% (v/v) acetonitrile. In-gel reduction of proteins was conducted using 25 mM dithiothreitol at 56 °C for 20 min, and alkylation was performed using 55 mM iodoacetamide for 20 min in the dark. Finally, digestion was carried out using 160 ng of Asp-N with a protein to enzyme ratio of 1:74 for TMV wild-type, 1:81 for TMV_CP-OAS VLPs, and 1:68 for TMV_CP VLPs over night at 37 °C. Peptides were eluted from gel matrix, concentrated for 3 h at room temperature using a vacuum concentrator (Eppendorf) and re-filled to 250 µL using 5% (v/v) acetonitrile plus 0.1% (v/v) formic acid (FA).
The acetylation of TMV VLPs was confirmed through data-dependent acquisition using a Q Exactive HF mass spectrometer (ThermoFisher Scientific) equipped with an Ultimate3000 UHPLC (ThermoFisher Scientific). The mobile phase consisted of 0.1% (v/v) formic acid (FA) in water (A) and 0.1% (v/v) formic acid (FA) in acetonitrile (B). The flow rate was set at 0.3 mL*min−1, and the autosampler was cooled to 10 °C. The separation process was performed using an Acquity aQ C18 column (2.1 × 100 mm, 80 Å) (ThermoFisher Scientific) at a temperature of 30 °C. The column was equilibrated for 2 min with 100% of mobile phase A. Thereafter, 10 μL of sample were loaded onto the column, and unbound sample components were washed out with 100% of mobile phase A for 1 min. Peptides were separated using a linear gradient from 0% to 50% of mobile phase B over 20 min. Subsequently, the column was washed with 95% of eluent B and re-equilibrated with 100% of mobile phase A for 5 min to prepare for the next run. The eluting peptides were ionized using a HESI-II ion source at a voltage of 3.5 kV and a temperature of 300 °C for the first 28 min. The sheath gas was set at a pressure of 35, while the aux gas was set at 10. The temperature of the capillary was maintained at 350 °C, and the RF level of the S-lens was configured to 50. MS scans were collected from m/z 300 to 1500 with a resolution of 60,000 at m/z 200 for MS and 15,000 for MS/MS scans. The top 5 precursors were selected at intensity thresholds above 1.6*105 for fragmentation with an isolation window of m/z 2.0. The data-dependent acquisition was executed in profile data type with an automatic gain control of 3*106 and a maximum injection time of 100 ms. Ions that were unassigned, single, and higher than 5-fold charged were excluded from the study. The dynamic exclusion parameter was set to 180 s, and the loop count was set to 5.
MS/MS spectra were searched for signals of the doubly charged deacetylated N-terminal peptide of the TMV wild-type CP at m/z 996.4875 and the acetylated N-terminal peptide at m/z 1017.4927, respectively at m/z 1032.0097 for deacetylated and m/z 1053.0113 for doubly charged acetylated N-terminal TMV_CP VLP and TMV_CP-OAS VLP CP using Freestyle 1.8 Software (ThermoFisher Scientific).
2.12 Software
NEBuilder Assembly Tool v2.10.2 was used to generate primers for Gibson assembly. Clone Manager Professional Suite 8 was used to analyze the sequences and to calculate the molecular weights and isoelectric points of proteins. ImageJ v1.51j8 (Schneider et al., 2012) was used to calculate the length of VLPs (30 per condition). Microsoft Office LTSC Professional Plus 2021 was used to generate graphs.
3 Results
3.1 TMV CPs assemble into VLPs during cell-free production
The TMV cp gene was cloned with and without the OAS in expression vector pLenEx (Buntru et al., 2022b) to ensure accumulation in the cytosolic fraction of the cell-free lysate (Figure 1).
CP monomers were produced using the commercial ALiCE BYL optimized for yields of up to 3 mg protein per ml lysate (Schillberg et al., 2019). The pLenEx_Strep-eYFP vector (Buntru et al., 2022a) was used as a positive control, and a template-free reaction was used as a negative control. The cell-free CPs were detected as an 18 kDa band by SDS-PAGE and Western blot with anti-TMV antibodies (Figure 2A). The yield determined by ELISA was 3.56 ± 0.62 mg/mL for TMV_CP and 3.23 ± 1.10 mg/mL for TMV_CP-OAS before purification. These amounts are comparable to the 3 mg/mL model protein eYFP.
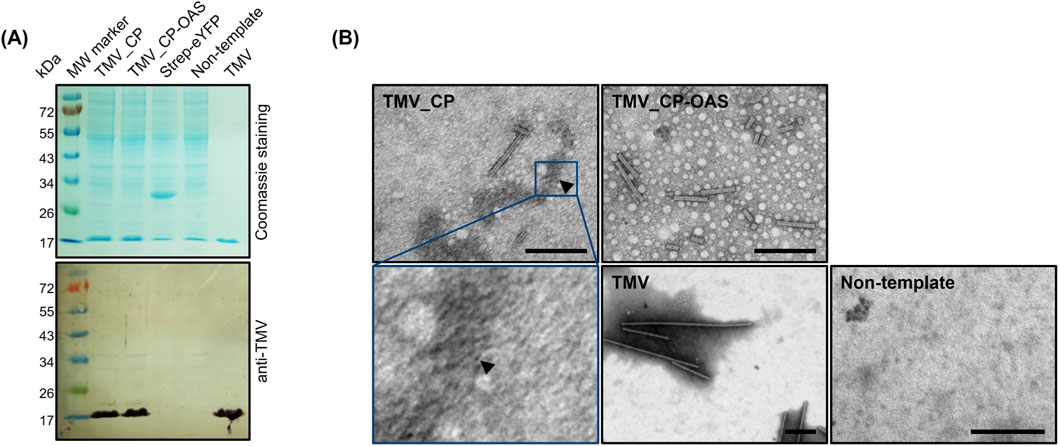
Figure 2. Ability of cell-free produced TMV CPs to assemble into VLPs. (A) Cell-free reaction samples were separated by SDS-PAGE followed by staining with Coomassie Brilliant Blue or the detection of CP (17.6 kDa) by Western blot using anti-TMV/GARAP antibodies. (B) TEM images of virus-like particles (VLPs) and disks (arrow) captured from the lysate using the anti-TMV antibody. Scale = 200 nm. H2O – non-template control, Strep-eYFP – model protein eYFP containing the StrepII tag, TMV – wild-type TMV particles, TMV_CP – cell-free expression of CP, TMV_CP-OAS – cell-free expression of CP with additional origin of assembly.
The assembly of 300-nm VLPs in the cell-free lysate, similar in morphology to native TMV particles, was confirmed by TEM (Figure 2B), although heterogeneous particles measuring 88 ± 63 nm for TMV_CP and 203 ± 91 nm for TMV_CP-OAS were also observed. More TMV_CP-OAS than TMV_CP particles were detected, as expected for RNA-guided assembly. However, the overall number of particles was lower than expected based on Western blot and ELISA data. Furthermore, the presence of numerous TMV_CP disks was suspected, although these were not clearly visible in the Figure 2B obtained due to the lower resolution of the images in comparison to the data presented in the earlier report by Finbloom et al. (2016). These results indicate either that the antibodies coated on the grids did not capture VLPs from the lysate efficiently or that assembly was inhibited by the suboptimal pH and ionic strength of the buffer (Butler, 1984).
Lysate samples were centrifuged to enrich the VLPs in the pellet, which was resuspended in PBS. The pellet and supernatant were then analyzed by SDS-PAGE, Western blot and TEM (data not shown). CP was detected in both fractions, but the majority of VLPs were found in the pellet (data not shown).
3.2 Characterization and stability of cell-free TMV VLPs
TMV_CP and TMV_CP-OAS VLPs from the resuspended pellet were transferred to different buffers and incubated for 5 days at 20 °C to test their stability. Both types of VLP formed stable rod-like structures in all tested buffers (Figure 3A), including 0.4 M Tris-HCl (pH 8.5), which usually favors monomers and A-protein structures (Butler, 1984). We compared the length of the particles to native TMV from plants (Figure 3B; Supplementary Table S2). The length of the particles varied widely in all buffers, as shown by the large standard deviations, but the average length was dependent on the buffer conditions. Native TMV particles were longest in PBS at pH 7.2 (226 ± 107 nm), were shorter but similar in 0.1 and 0.4 M sodium acetate at pH 5.5 (155 ± 63 and 139 ± 53 nm, respectively) and shortest in 0.4 M Tris-HCl at pH 8.5 (90 ± 36 nm). The TMV_CPs assembled into short VLPs (89 ± 63 nm) in the near neutral PBS but formed longer rods in 0.1 and 0.4 M acetate (194 ± 123 and 175 ± 67 nm, respectively), as expected (Butler, 1984). Surprisingly, TMV_CPs assembled into even longer rods (238 ± 124 nm) in the alkaline Tris buffer, rather than the anticipated disks or stacked disks. TMV_CP-OAS was expected to yield more particles due to the stabilizing effect of VLP assembly via interaction with RNA, and this was borne out by the longer particles observed in near neutral PBS (203 ± 91 nm). Stability was maintained in the other buffers, with VLPs of 154 ± 95 and 187 ± 84 nm in 0.1 and 0.4 M acetate, respectively, and 195 ± 66 nm in the alkaline Tris buffer. The TMV_CP-OAS VLPs also tended to aggregate end-to-end in PBS and Tris buffer. The comparison of VLPs to native TMV revealed statistically significant differences in the length of the rod-like particles in the neutral and alkaline buffers, but no significant difference at pH 5.5.
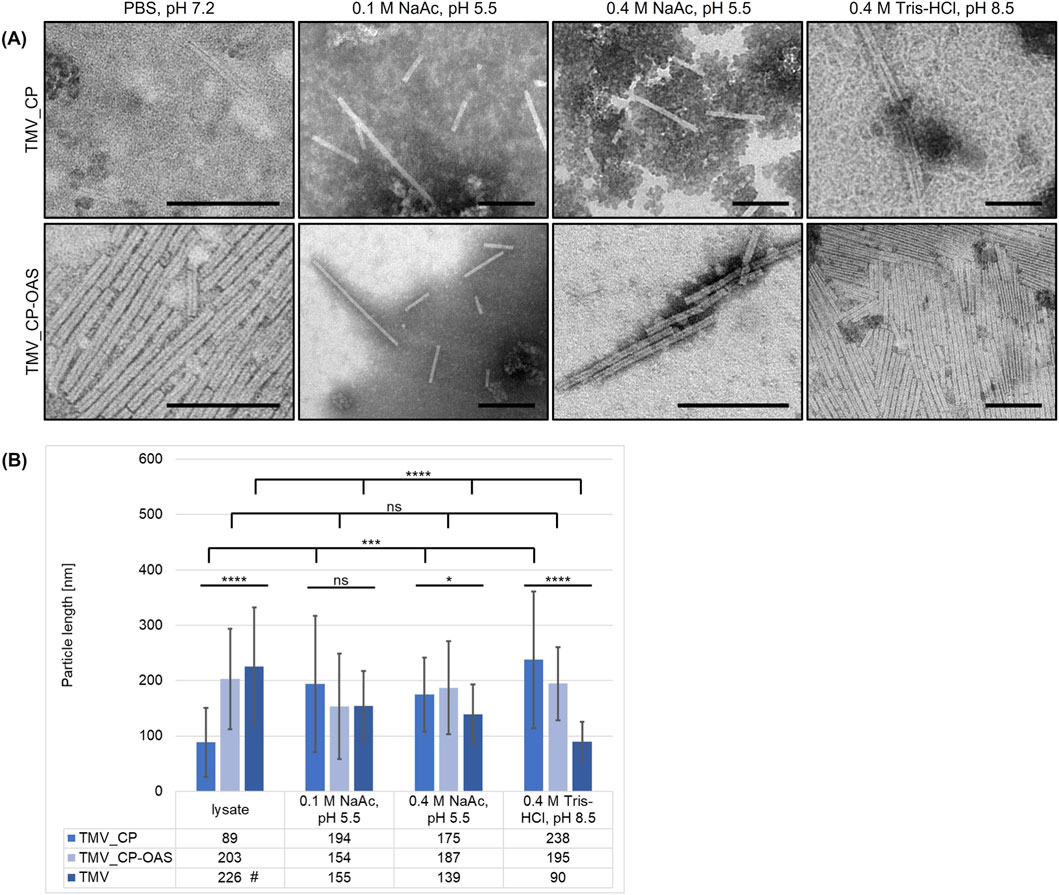
Figure 3. Length distribution and stability of cell-free produced TMV VLPs under various buffer conditions. (A) Electron micrographs and (B) length distribution of TMV_CP and TMV_CP-OAS VLPs in the crude lysate or pellet fraction (phosphate buffer, pH 7.2) incubated in different buffers (0.1 or 0.4 M sodium acetate pH 5.5, 0.4 M Tris-HCl pH 8.5). #TMV wild-type particles were dissolved in 0.01 M phosphate buffer (pH 7.2). Statistical significance was determined by one-way ANOVA (n = 30 VLPs/group; *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant). (A) Scale = 200 nm. TMV_CP – VLPs assembled from cell-free expression of CP, TMV_CP-OAS – VLPs assembled from cell-free expression of CP with additional origin of assembly.
To confirm the assembly of VLPs in the lysate, TMV_CP and TMV_CP-OAS were purified by CaptoCore700 chromatography. The efficiency of the purification process was found to be approximately 15% for TMV_CP and 22% for TMV_CP-OAS. The presence of CPs and VLPs was confirmed by SDS-PAGE, Western blot, and TEM (Figures 4A,B). The encapsidated viral RNA was separated from the VLPs by agarose gel electrophoresis and the proteins were stained with Coomassie Brilliant Blue, with wild-type TMV as a standard (Figure 4C). The faint RNA bands and multiple protein bands representing different lengths of TMV particles were consistent with Eiben et al. (2014). Surprisingly, the TMV_CP and TMV_CP-OAS VLPs contained encapsulated nucleic acids, which could be transcribed RNA from the template DNA and/or BYL reaction. Coomassie staining showed that the mobility of the TMV_CP and TMV_CP-OAS VLPs was increased compared to wild-type TMV particles, which indicates assembly into shorter rods and is consistent with the length distribution measurement (Figure 3B). No further structural information regarding the properties of the VLPs was obtained through cryo-TEM analysis (see Supplementary Figure S1), but acetylation was verified using LC-MS/MS (see Supplementary Figure S2).

Figure 4. Characterization of purified cell-free produced TMV VLPs. (A) Purified cell-free TMV VLPs were separated by SDS-PAGE and proteins were detected with Coomassie Brilliant Blue or by Western blot using anti-TMV/GARAP antibodies to detect the 17.6-kDa CP. (B) Electron micrographs and (C) semi-native agarose gel (top imaged under UV light, bottom Coomassie Brilliant Blue). (B) Scale = 200 nm. TMV lanes contain wild-type TMV particles. Non-template – negative control using H2O in cell-free reaction, TMV_CP – VLPs purified from cell-free expression of CP, TMV_CP-OAS – VLPs purified from cell-free expression of CP with additional origin of assembly, Strep-eYFP – model protein eYFP containing the StrepII tag.
4 Discussion
Unlike synthetic nanoparticles, plant VNPs can be produced inexpensively and in large quantities by molecular farming in plants (Rybicki, 2020). They self-assemble into nanostructures with defined sizes, shapes, surface properties and functionalities, which are suitable for diverse applications (Lico et al., 2013; Eiben et al., 2019). Plant VNPs are particularly useful in medicine because they do not replicate in mammals and contain no toxic components (Manchester and Singh, 2006; Karan et al., 2024). VNPs are cleared from the body by the reticuloendothelial system within 24–96 h (Nkanga and Steinmetz, 2021). However, they retain the replicative nucleic acid of the plant virus from which they are derived and may therefore pose a risk to plants in the environment.
These environmental risks can be addressed by producing VLPs, which lack replicative nucleic acid. Not all plant viruses can form VLPs efficiently (or at all) in the absence of their genome, or at least the component that induces self-assembly, significantly reducing the yield (Rabindran and Dawson, 2001; Huang et al., 2009). VLP production is also difficult to scale up due to the inclusion of an ultracentrifugation step during purification (Hull, 2014). We used a tobacco cell-free lysate (BYL) to produce TMV_CP VLPs, and achieved yields of 0.56 ± 0.20 mg/mL lysate, up to 23 times higher than in microbial systems (Table 1; a schematic overview is represented in Supplementary Figure S3). Furthermore, it can be hypothesized that the yields of TMV_CP VLPs and TMV_CP-OAS VLPs are comparable to those observed in N. benthamiana (Table 1). It is important to note that discrepancies in experimental setup and an absence of conversion factors between biomass and culture volumes have resulted in yields being reported in their native units. Consequently, a direct comparison between these units is not possible. Cell-free systems have also been used to produce animal VLPs such as hepatitis B core particles (Lingappa et al., 1994; Spice et al., 2020; Colant et al., 2021b), and the BYL system again proved to be the most efficient (Armero-Gimenez et al., 2023). However, low purification efficiencies of up to 22% could be due to the low efficiency of VLP assembly. In order to increase the ratio of VLPs, it is recommended that lysate incubations be conducted in a lower pH range, as this has been shown to promote disk and rod formation (Durham et al., 1971). Such an approach may involve the use of a sodium acetate buffer, for example, and should be conducted over a period ranging from several days to weeks prior to purification. Moreover, mutations in the CP including R46G/E50K/E97G have been demonstrated to neutralize negatively charged residues, thereby enhancing VLP assembly (Brown et al., 2021). It is important to note that tailoring the assembly outcomes is highly dependent on the intended application. For instance, if RNA is required for silencing applications, this OAS-RNA will not only guide the assembly procedure but also enhance the stability of the uniform VLPs. An effective strategy to enhance purification efficiency involves refining the purification method utilized. Techniques such as sucrose gradient centrifugation (small scale) and Superose 6 column chromatography have shown promise in significantly improving both the purity and overall yield of correctly assembled TMV VLPs (González-Gamboa et al., 2024). These methods facilitate the separation of VLPs based on their size and density, thereby ensuring higher quality preparations. Ongoing studies are currently investigating these approaches to optimize conditions further and validate their effectiveness in different contexts.
We found that TMV CPs assembled into VLPs during the cell-free reaction at neutral pH, which is advantageous for applications under physiological conditions, whereas E. coli-derived CPs formed disk-like aggregates at neutral pH (Díaz-Avalos and Caspar, 1998; Bruckman et al., 2011). An OAS is necessary to produce rod-shaped TMV VLPs in vitro, but the process occurs at a much lower efficiency (Hwang et al., 1994; Eiben et al., 2014). This is attributed to an essential lack of N-terminal acetylation of the TMV CP in E. coli (Filner and Marcus, 1974; Shire et al., 1990). Although this limitation can be overcome by co-assembly with >20% plant-derived CP, it requires the separate infection and production of wild-type TMV, thus increasing production costs and hindering scale-up. In contrast, cell-free production can take place in a controlled environment and the TMV VLPs can be collected by size-exclusion (e.g., CaptoCore 700) chromatography in flow-through mode, eliminating the need for dialysis.
Agarose gel electrophoresis revealed an unexpected smear indicating the presence of RNA in both the TMV_CP and TMV_CP-OAS VLPs. Host RNAs can be encapsulated within TMV particles derived from infected plants (Siegel, 1971; Schön and Mundry, 1984), suggesting that residual RNA from BY-2 cells or in vitro transcripts are encapsulated during cell-free VLP production. Although TMV is classified as plant virus, the presence of host RNA complicates safety assessments for therapeutic or vaccine applications. This complexity arises because RNA cargo can influence immune responses, potentially leading to intended effects. Furthermore, encapsidated RNA may affect the VLP’s capacity to release its cargo or interact effectively with biological environments (McCormick and Palmer, 2008). To comprehensively validate these finding regarding encapsidated RNA in TMV_CP VLPs, a subsequent transcriptome analysis is necessary. Acetylation of the TMV CP may be essential for interaction with RNA (Hwang et al., 1994). It is reasonable to assume that the acetylation of the TMV_CP produced in the cell-free system is also likely to occur, as demonstrated in our study. The OAS serves as a nucleation center for the formation of rod-shaped VLPs, increasing their stability and yield (Gallie et al., 1987). This effect was also observed for TMV CPs produced using the BYL (Figures 3A,B).
Our results demonstrate that BYL, functioning as a cell-free analog to the natural plant host, directly stimulates the formation of VLPs during the cell-free reaction, particularly in the presence of the OAS. This agrees with Saunders et al. (2022a), who used a replicating viral vector system to produce designer-length TMV VLPs in plants. The yield decreased by ∼40% in the absence of the OAS. In contrast to wild-type TMV and TMV_CP VLPs, the assembly and stability of TMV_CP-OAS VLPs was not significantly influenced by the buffer conditions. The transcript length usually determines the particle length and can be used as a predictor (Hwang et al., 1994; Wege and Koch, 2020). In contrast, TMV_CP-OAS VLPs ranged in size from 42 to 432 nm, depending on the buffer conditions, due to the absence of a transcriptional terminator in pLenEx, resulting in unpredictable variable-length transcripts. The resulting particle length heterogeneity can lead to differences in surface area for functionalization, interactions with biological systems and payload capacity. Such batch-to-batch variability poses challenges for consistent manufacturing processes and represents a potential drawback for medical applications. It can be overcome in cell-free expression systems by using linearized PCR products as templates (personal communication, Dr. Matthias Buntru) or by inserting a terminator sequence (although this may reduce overall yields), which is currently investigated. Additionally, fine-tuning assembly conditions can minimize non-specific aggregation and promote the formation more homogeneous particles. However, the cell-free VLPs were more stable over a wider pH range than native TMV particles without affecting their morphology (Saunders et al., 2022a).
While microbial systems are generally highly scalable due to established fermentation technologies, they are not suitable for the production of TMV VLPs, as a defined content of wild-type CP is required as previously discussed. In contrast, plant-based systems suitable for VLP production demonstrate inherent scalability advantages, but may face limitations related to regulatory hurdles and spatial requirements due to the heterogeneity between individual plants and when they are grown in open fields. BYL is plant-derived, but commercially available as on-demand production system, that is stored at −80 °C and therefore offers more flexibility in usage. Although it typically requires a higher initial investment compared to microbial and traditional plant cultivation methods, it presents several advantages. Specifically, BYL operates as a controlled production system that does not require laborious extraction procedures, thereby simplifying downstream purification processes and ultimately reducing purification costs. VLP production in plants usually takes ∼4 weeks (plasmid cloning 5 days, infection 14–18 days depending on infection symptoms, purification 5 days, and analysis 2 days) whereas the BYL only requires ∼10 days (plasmid cloning 5 days, expression 2 days, purification 1–2 days, analysis 2 days). This comparison excludes the time needed for plant cultivation or cell lysate preparation, although the lysate can be produced in large batches and stored at −80 °C for convenient use. The scalability of cell-free expression (Gupta et al., 2023) aligns with sustainability goals by minimizing energy consumption and waste generation. These properties are particularly advantageous for rapid vaccine development because target epitopes can be displayed on VLPs (Lomonossoff and Wege, 2018) and RNAs can be encapsulated within them (Chan and Steinmetz, 2023). VLPs can also be used for the delivery of nucleic acids (Lam and Steinmetz, 2019; Nuñez-Rivera et al., 2020; Islam et al., 2024) or biopesticides (Chariou and Steinmetz, 2017) to plants.
The expression and assembly of TMV VLPs established in this study, provide a robust foundation for investigating one of the significant advantages of the cell-free approach: the ability to incorporate non-canonical amino acids into synthesized proteins (Wu et al., 2020). This approach could enhance the structural stability, immunogenicity and functional properties of VLPs, thereby broadening their potential applications in vaccine development and targeted delivery systems.
5 Conclusion
BYL is a versatile platform for VLP production that addresses the growing need for precise nanostructures that can be synthesized rapidly and on-demand. We used TMV as a model to show that tailored VLPs can be produced in the absence of replicative viral RNA, ensuring that the particles are non-pathogenic while enabling rapid assembly, high yields and simple purification. Our precise control over expression gives rise to VLPs that maintain structural integrity even under physiological pH conditions. The stability, on-demand production, and possibility to include non-canonical amino acids are particularly advantageous for vaccine development and targeted drug delivery, allowing the effective display of surface epitopes or therapeutic agents. In the future, cell-free produced CPs combined with the viral OAS could be used to encapsulate RNAs that silence target genes in plants or animals, allowing the development of selective biopesticides and personalized medicines.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
DHBG: Visualization, Methodology, Investigation, Writing – review and editing. EMB: Investigation, Writing – review and editing. AC: Visualization, Writing – review and editing, Investigation. SS: Supervision, Writing – review and editing. JS: Methodology, Project administration, Conceptualization, Visualization, Validation, Writing – review and editing, Funding acquisition, Writing – original draft, Supervision.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This research was funded by the ERS start-up (grant number: StUpPD_435-23), supported by the Federal Ministry of Education and Research (BMBF) and the Ministry of Culture and Science of the German State of North Rhine-Westphalia (MKW) under the Excellence Strategy of the Federal and State Governments.
Acknowledgments
AcknowledgementsWe are grateful to Prof. Hadrien Peyret (University of Nottingham) and Prof. George Lomonossoff (John Innes Centre) for providing the sequences of the CP-OAS construct and for their fruitful discussions. Our appreciation extends to Matthias Buntru (Fraunhofer IME) for his valuable discussions about the pLenEx vector, and to Karolin Richter, Michael Kupper and Katharina Schönthier (RWTH Aachen University) for their experimental help. We are indebted to Ricarda Finnern (LenioBio GmbH) for providing the ALiCE cell-free lysate. We thank Richard M. Twyman (Twyman Research Management) for critically reading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2025.1687889/full#supplementary-material
References
Armero-Gimenez, J., Wilbers, R., Schots, A., Williams, C., and Finnern, R. (2023). Rapid screening and scaled manufacture of immunogenic virus-like particles in a tobacco BY-2 cell-free protein synthesis system. Front. Immunol. 14, 1088852. doi:10.3389/fimmu.2023.1088852
Bogart, J. W., Cabezas, M. D., Vögeli, B., Wong, D. A., Karim, A. S., and Jewett, M. C. (2021). Cell-free exploration of the natural product chemical space. Chembiochem 22, 84–91. doi:10.1002/cbic.202000452
Brown, A. D., Chu, S., Kappagantu, M., Ghodssi, R., and Culver, J. N. (2021). Reprogramming virus coat protein carboxylate interactions for the patterned assembly of hierarchical nanorods. Biomacromolecules 22, 2515–2523. doi:10.1021/acs.biomac.1c00258
Bruckman, M. A., Soto, C. M., McDowell, H., Liu, J. L., Ratna, B. R., Korpany, K. V., et al. (2011). Role of hexahistidine in directed nanoassemblies of Tobacco mosaic virus coat protein. ACS Nano 5, 1606–1616. doi:10.1021/nn1025719
Buntru, M., Vogel, S., Stoff, K., Spiegel, H., and Schillberg, S. (2015). A versatile coupled cell-free transcription-translation system based on tobacco BY-2 cell lysates. Biotechnol. Bioeng. 112, 867–878. doi:10.1002/bit.25502
Buntru, M., Hahnengress, N., Croon, A., and Schillberg, S. (2022a). Plant-derived cell-free biofactories for the production of secondary metabolites. Front. Plant Sci. 12, 794999. doi:10.3389/fpls.2021.794999
Buntru, M., Vogel, S., Finnern, R., and Schillberg, S. (2022b). Plant-based cell-free transcription and translation of recombinant proteins. Methods Mol. Biol. 2480, 113–124. doi:10.1007/978-1-0716-2241-4_8
Butler, P. J. (1984). The current picture of the structure and assembly of Tobacco mosaic virus. J. Gen. Virol. 65 (Pt 2), 253–279. doi:10.1099/0022-1317-65-2-253
Chan, S. K., and Steinmetz, N. F. (2023). microRNA-181a silencing by antisense oligonucleotides delivered by virus-like particles. J. Mater Chem. B 11, 816–825. doi:10.1039/d2tb02199d
Chariou, P. L., and Steinmetz, N. F. (2017). Delivery of pesticides to plant parasitic nematodes using Tobacco mild green mosaic virus as a nanocarrier. ACS Nano 11, 4719–4730. doi:10.1021/acsnano.7b00823
Chung, Y. H., Cai, H., and Steinmetz, N. F. (2020). Viral nanoparticles for drug delivery, imaging, immunotherapy, and theranostic applications. Adv. Drug Deliv. Rev. 156, 214–235. doi:10.1016/j.addr.2020.06.024
Colant, N., Melinek, B., Frank, S., Rosenberg, W., and Bracewell, D. G. (2021a). Escherichia coli-based cell-free protein synthesis for iterative design of tandem-core virus-like particles. Vaccines (Basel) 9, 193. doi:10.3390/vaccines9030193
Colant, N., Melinek, B., Teneb, J., Goldrick, S., Rosenberg, W., Frank, S., et al. (2021b). A rational approach to improving titer in Escherichia coli -based cell-free protein synthesis reactions. Biotechnol. Prog. 37, e3062. doi:10.1002/btpr.3062
Díaz-Avalos, R., and Caspar, D. L. (1998). Structure of the stacked disk aggregate of Tobacco mosaic virus protein. Biophys. J. 74, 595–603. doi:10.1016/S0006-3495(98)77818-X
Durham, A. C., Finch, J. T., and Klug, A. (1971). States of aggregation of tobacco mosaic virus protein. Nat. New Biol. 229, 37–42. doi:10.1038/newbio229037a0
Eiben, S., Stitz, N., Eber, F., Wagner, J., Atanasova, P., Bill, J., et al. (2014). Tailoring the surface properties of Tobacco mosaic virions by the integration of bacterially expressed mutant coat protein. Virus Res. 180, 92–96. doi:10.1016/j.virusres.2013.11.019
Eiben, S., Koch, C., Altintoprak, K., Southan, A., Tovar, G., Laschat, S., et al. (2019). Plant virus-based materials for biomedical applications: trends and prospects. Adv. Drug Deliv. Rev. 145, 96–118. doi:10.1016/j.addr.2018.08.011
Filner, B., and Marcus, A. (1974). TMV coat protein synthesis in vivo: analysis of the N-terminal acetylation. Virology 61, 537–546. doi:10.1016/0042-6822(74)90288-8
Finbloom, J. A., Han, K., Aanei, I. L., Hartman, E. C., Finley, D. T., Dedeo, M. T., et al. (2016). Stable disk assemblies of a tobacco mosaic virus mutant as nanoscale scaffolds for applications in drug delivery. Bioconjug Chem. 27, 2480–2485. doi:10.1021/acs.bioconjchem.6b00424
Fischer, R., Schumann, D., Zimmermann, S., Drossard, J., Sack, M., and Schillberg, S. (1999). Expression and characterization of bispecific single-chain Fv fragments produced in transgenic plants. Eur. J. Biochem. 262, 810–816. doi:10.1046/j.1432-1327.1999.00435.x
Gallie, D. R., Sleat, D. E., Watts, J. W., Turner, P. C., and Wilson, T. M. (1987). In vivo uncoating and efficient expression of foreign mRNAs packaged in TMV-like particles. Science 236, 1122–1124. doi:10.1126/science.3472350
González-Gamboa, I., Caparco, A. A., McCaskill, J., Fuenlabrada-Velázquez, P., Hays, S. S., Jin, Z., et al. (2024). Inter-coat protein loading of active ingredients into Tobacco mild green mosaic virus through partial dissociation and reassembly of the virion. Sci. Rep. 14, 7168. doi:10.1038/s41598-024-57200-0
Gupta, M. D., Flaskamp, Y., Roentgen, R., Juergens, H., Armero-Gimenez, J., Albrecht, F., et al. (2023). Scaling eukaryotic cell-free protein synthesis achieved with the versatile and high-yielding tobacco BY-2 cell lysate. Biotechnol. Bioeng. 120, 2890–2906. doi:10.1002/bit.28461
Huang, Z., Chen, Q., Hjelm, B., Arntzen, C., and Mason, H. (2009). A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnol. Bioeng. 103, 706–714. doi:10.1002/bit.22299
Hwang, D. J., Roberts, I. M., and Wilson, T. M. (1994). Expression of Tobacco mosaic virus coat protein and assembly of pseudovirus particles in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91, 9067–9071. doi:10.1073/pnas.91.19.9067
Islam, M. R., Youngblood, M., Kim, H.-I., González-Gamboa, I., Monroy-Borrego, A. G., Caparco, A. A., et al. (2024). DNA delivery by virus-like nanocarriers in plant cells. Nano Lett. 24, 7833–7842. doi:10.1021/acs.nanolett.3c04735
Kadri, A., Wege, C., and Jeske, H. (2013). In vivo self-assembly of TMV-like particles in yeast and bacteria for nanotechnological applications. J. Virol. Methods 189, 328–340. doi:10.1016/j.jviromet.2013.02.017
Karan, S., Durán-Meza, A. L., Chapman, A., Tanimoto, C., Chan, S. K., Knobler, C. M., et al. (2024). In vivo delivery of spherical and cylindrical in vitro reconstituted virus-like particles containing the same self-amplifying mRNA. Mol. Pharm. 21, 2727–2739. doi:10.1021/acs.molpharmaceut.3c01105
Kriegsheim, A. v., Preisinger, C., and Kolch, W. (2008). Mapping of signaling pathways by functional interaction proteomics. Methods Mol. Biol. 484, 177–192. doi:10.1007/978-1-59745-398-1_12
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi:10.1038/227680a0
Lam, P., and Steinmetz, N. F. (2019). Delivery of siRNA therapeutics using Cowpea chlorotic mottle virus-like particles. Biomater. Sci. 7, 3138–3142. doi:10.1039/c9bm00785g
Lam, P., Gulati, N. M., Stewart, P. L., Keri, R. A., and Steinmetz, N. F. (2016). Bioengineering of Tobacco mosaic virus to create a non-infectious positive control for ebola diagnostic assays. Sci. Rep. 6, 23803. doi:10.1038/srep23803
Lee, K. Z., Basnayake Pussepitiyalage, V., Lee, Y.-H., Loesch-Fries, L. S., Harris, M. T., Hemmati, S., et al. (2021). Engineering Tobacco mosaic virus and its virus-like-particles for synthesis of biotemplated nanomaterials. Biotechnol. J. 16, e2000311. doi:10.1002/biot.202000311
LenioBio (2024). Efficient cell-free protein expression kits | ALiCE® by LenioBio. Available online at: https://www.leniobio.com/alice-products/ (Accessed January 10, 2025).
Li, J., Zhang, L., and Liu, W. (2018). Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products. Synth. Syst. Biotechnol. 3, 83–89. doi:10.1016/j.synbio.2018.02.002
Lico, C., Schoubben, A., Baschieri, S., Blasi, P., and Santi, L. (2013). Nanoparticles in biomedicine: new insights from plant viruses. Curr. Med. Chem. 20, 3471–3487. doi:10.2174/09298673113209990035
Lingappa, J. R., Martin, R. L., Wong, M. L., Ganem, D., Welch, W. J., and Lingappa, V. R. (1994). A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of Hepatitis B virus capsid, a multimeric particle. J. Cell Biol. 125, 99–111. doi:10.1083/jcb.125.1.99
Lomonossoff, G. P., and Wege, C. (2018). TMV particles: the journey from fundamental studies to bionanotechnology applications. Adv. Virus Res. 102, 149–176. doi:10.1016/bs.aivir.2018.06.003
Malik, S., Muhammad, K., and Waheed, Y. (2023). Nanotechnology: a revolution in modern industry. Molecules 28, 661. doi:10.3390/molecules28020661
Manchester, M., and Singh, P. (2006). Virus-based nanoparticles (VNPs): platform technologies for diagnostic imaging. Adv. Drug Deliv. Rev. 58, 1505–1522. doi:10.1016/j.addr.2006.09.014
McCormick, A. A., and Palmer, K. E. (2008). Genetically engineered Tobacco mosaic virus as nanoparticle vaccines. Expert Rev. Vaccines 7, 33–41. doi:10.1586/14760584.7.1.33
Namba, K., Pattanayek, R., and Stubbs, G. (1989). Visualization of protein-nucleic acid interactions in a virus. J. Mol. Biol. 208, 307–325. doi:10.1016/0022-2836(89)90391-4
Nkanga, C. I., and Steinmetz, N. F. (2021). The pharmacology of plant virus nanoparticles. Virology 556, 39–61. doi:10.1016/j.virol.2021.01.012
Nuñez-Rivera, A., Fournier, P. G. J., Arellano, D. L., Rodriguez-Hernandez, A. G., Vazquez-Duhalt, R., and Cadena-Nava, R. D. (2020). Brome mosaic virus-like particles as siRNA nanocarriers for biomedical purposes. Beilstein J. Nanotechnol. 11, 372–382. doi:10.3762/bjnano.11.28
Rabindran, S., and Dawson, W. O. (2001). Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology 284, 182–189. doi:10.1006/viro.2001.0910
Röder, J., Dickmeis, C., and Commandeur, U. (2019). Small, smaller, nano: new applications for Potato virus X in nanotechnology. Front. Plant Sci. 10, 158. doi:10.3389/fpls.2019.00158
Rybicki, E. P. (2020). Plant molecular farming of virus-like nanoparticles as vaccines and reagents. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 12, e1587. doi:10.1002/wnan.1587
Saunders, K., Thuenemann, E. C., Peyret, H., and Lomonossoff, G. P. (2022a). The Tobacco mosaic virus origin of assembly sequence is dispensable for specific viral RNA encapsidation but necessary for initiating assembly at a single site. J. Mol. Biol. 434, 167873. doi:10.1016/j.jmb.2022.167873
Saunders, K., Thuenemann, E. C., Shah, S. N., Peyret, H., Kristianingsih, R., Lopez, S. G., et al. (2022b). The use of a replicating virus vector for in planta generation of Tobacco mosaic virus nanorods suitable for metallization. Front. Bioeng. Biotechnol. 10, 877361. doi:10.3389/fbioe.2022.877361
Schillberg, S., Raven, N., Spiegel, H., Rasche, S., and Buntru, M. (2019). Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 10, 720. doi:10.3389/fpls.2019.00720
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Schön, A., and Mundry, K. W. (1984). Coordinated two-disk nucleation, growth and properties, of virus-like particles assembled from tobacco-mosaic-virus capsid protein with poly(A) or oligo(A) of different length. Eur. J. Biochem. 140, 119–127. doi:10.1111/j.1432-1033.1984.tb08074.x
Schuphan, J., Stojanović, N., Lin, Y.-Y., Buhl, E. M., Aveic, S., Commandeur, U., et al. (2024). A combination of flexible modified plant virus nanoparticles enables additive effects resulting in improved osteogenesis. Adv. Healthc. Mater 13, e2304243. doi:10.1002/adhm.202304243
Shire, S. J., McKay, P., Leung, D. W., Cachianes, G. J., Jackson, E., Wood, W. I., et al. (1990). Preparation and properties of recombinant DNA derived Tobacco mosaic virus coat protein. Biochemistry 29, 5119–5126. doi:10.1021/bi00473a017
Siegel, A. (1971). Pseudovirions of Tobacco mosaic virus. Virology 46, 50–59. doi:10.1016/0042-6822(71)90005-5
Spice, A. J., Aw, R., Bracewell, D. G., and Polizzi, K. M. (2020). Synthesis and assembly of Hepatitis B virus-like particles in a Pichia pastoris cell-free system. Front. Bioeng. Biotechnol. 8, 72. doi:10.3389/fbioe.2020.00072
Steele, J. F. C., Peyret, H., Saunders, K., Castells-Graells, R., Marsian, J., Meshcheriakova, Y., et al. (2017). Synthetic plant virology for nanobiotechnology and nanomedicine. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 9, e1447. doi:10.1002/wnan.1447
Wege, C., and Koch, C. (2020). From stars to stripes: RNA-directed shaping of plant viral protein templates-structural synthetic virology for smart biohybrid nanostructures. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 12, e1591. doi:10.1002/wnan.1591
Wijesundara, Y. H., Herbert, F. C., Kumari, S., Howlett, T., Koirala, S., Trashi, O., et al. (2022). Rip it, stitch it, click it: a chemist's guide to VLP manipulation. Virology 577, 105–123. doi:10.1016/j.virol.2022.10.008
Keywords: BY-2 lysate, cell-free protein expression, coat protein, plant virus, virus-like particles, TMV (tobacco mosaic virus)
Citation: Große DHB, Buhl EM, Croon A, Schillberg S and Schuphan J (2025) High-yield on-demand production of tobacco mosaic virus-like particles using a plant-derived cell-free expression system. Front. Nanotechnol. 7:1687889. doi: 10.3389/fnano.2025.1687889
Received: 18 August 2025; Accepted: 03 November 2025;
Published: 19 November 2025.
Edited by:
Fu-Xing Niu, Guangxi University of Science and Technology, ChinaReviewed by:
Viswanath Buddolla, Dr. Buddolla’s Institute of Life Sciences, IndiaAshish Warghane, Mandsaur University, India
Copyright © 2025 Große, Buhl, Croon, Schillberg and Schuphan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juliane Schuphan, anVsaWFuZS5zY2h1cGhhbkBtb2xiaW90ZWNoLnJ3dGgtYWFjaGVuLmRl
†These authors have contributed equally to this work
 David H. B. Große
David H. B. Große Eva M. Buhl
Eva M. Buhl Alexander Croon
Alexander Croon Stefan Schillberg
Stefan Schillberg Juliane Schuphan
Juliane Schuphan