- 1 UMR 8195, Centre de Neurosciences Paris-Sud, Université Paris-Sud, Orsay, France
- 2 UMR 8195, CNRS, Orsay, France
Egr1, a member of the Egr family of transcription factors, and Arc are immediate early genes known to play major roles in synaptic plasticity and memory. Despite evidence that Egr family members can control Arc transcriptional regulation, demonstration of a selective role of Egr1 alone is lacking. We investigated the extent to which activity-dependent Arc expression is dependent on Egr1 by analyzing Arc mRNA expression using fluorescence in situ hybridization in the dorsal dentate gyrus and CA1 of wild-type (WT) and Egr1 knockout mice. Following electroconvulsive shock, we found biphasic expression of Arc in area CA1 in mice, consisting in a rapid (30 min) and transient wave followed by a second late-phase of expression (8 h), and a single but prolonged wave of expression in the dentate gyrus. Egr1 deficiency abolished the latest, but not the early wave of Arc expression in CA1, and curtailed that of the dentate gyrus. Since the early wave of Arc expression was not affected in Egr1 mutant mice, we next analyzed behaviorally induced Arc expression patterns as an index of neural ensemble activation in the dentate gyrus and area CA1 of WT and Egr1 mutant mice. Spatial exploration of novel or familiar environments induced in mice a single early and transient wave of Arc expression in the dentate gyrus and area CA1, which were not affected in Egr1 mutant mice. Analyses of Arc-expressing cells revealed that exploration recruits similar size dentate gyrus and CA1 neural ensembles in WT and Egr1 knockout mice. These findings suggest that hippocampal neural ensembles are normally activated immediately following spatial exploration in Egr1 knockout mice, indicating normal hippocampal encoding of information. They also provide evidence that in condition of strong activation Egr1 alone can control late-phases of activity-dependent Arc transcription in the dentate gyrus and area CA1 of the hippocampus.
Introduction
Current hypotheses on the molecular mechanisms of learning and memory suggest that rapid regulation of gene programs and synthesis of new proteins leading to persistent synaptic modification constitute a key mechanism for the stabilization of long-term memory (Bruel-Jungerman et al., 2007 for a review). This genomic response includes a group of immediate early genes (IEGs) that encode two classes of proteins: nuclear transcription factors that regulate late-response genes, and proteins that directly modify synaptic function (Lanahan and Worley, 1998). Among these, Egr1/Zif268 and Arc/Arg3.1 are some of the best-characterized activity-regulated IEGs for their roles in synaptic plasticity and memory.
Egr1, a member of the early growth response (Egr) gene family, encodes a nuclear transcription factor that is rapidly and transiently induced during synaptic plasticity and in defined brain structures during memory consolidation or recall (reviewed in Davis et al., 2003; Knapska and Kaczmarek, 2004). Our previous studies in mutant mice have shown that Egr1 is required for the expression of late-phase hippocampal long-term potentiation (LTP) and for the consolidation of several forms of long-term memory (Jones et al., 2001; Bozon et al., 2002, 2003; Davis et al., 2010). Like Egr1, Arc is required for synaptic plasticity and for several forms of long-term memory (Plath et al., 2006). Arc, however, is a direct effector protein at the synapse. Upon cell activation, Arc mRNA traffics to dendrites and accumulates at sites of synaptic activity, where it is locally translated (Link et al., 1995; Lyford et al., 1995; Steward et al., 1998) and plays important roles in homeostatic scaling of AMPA receptors and structural modifications at the synapse (Rial Verde et al., 2006; Shepherd et al., 2006; Messaoudi et al., 2007). The expression of Arc is regulated as an IEG (Pintchovski et al., 2009), but also as a late-response gene by a protein synthesis-dependent mechanism (Wallace et al., 1998; Li et al., 2005).
Despite the fact that Egr1 has a clear role in mediating gene expression required for learning and memory, the specific molecular mechanisms that are involved are poorly defined. Several potential Egr1 target genes have been suggested (Petersohn et al., 1995; James et al., 2005; Baumgartel et al., 2009), but few have been identified as potentially implicated in the Egr1 mutant phenotype. Recently, Li et al. (2005) identified Arc as a direct target of the Egr family of transcription factors, showing Egr members can bind the Arc promoter in vivo after kainic acid-induced seizure and transactivate Arc through an Egr response element (ERE). In this experiment however, Egr3 rather than Egr1 was found to be required for seizure-induced Arc expression in the dentate gyrus, whereas full induction of Arc after behavioral experience was suggested to depend upon both Egr1 and Egr3 (Li et al., 2005).
Here, we used Egr1 knockout mice (Egr1−/−) to investigate the extent to which activity-dependent Arc expression is dependent on Egr1. For this, we used fluorescence in situ hybridization (FISH; Guzowski et al., 1999), a sensitive method that has been extensively utilized to map neuronal networks activated by experience (reviewed in Miyashita et al., 2008). We examined Arc expression in wild-type (WT) and Egr1−/− mice following both electroconvulsive shock (ECS), a procedure known to induce robust expression of Egr1 and Arc in the hippocampus, and following behavioral exploration of novel or familiar environments (Guzowski et al., 1999; Nakamura et al., 2010). Because activity-dependent Arc induction in hippocampus can be prolonged up to 8 h (Ramirez-Amaya et al., 2005), we analyzed the temporal dynamics of its expression over long time-intervals. Furthermore, we investigated Arc expression in both the dentate gyrus and area CA1, a region where Egr1 is expressed constitutively in contrast to the dentate gyrus, to explore whether there is any structure-specificity in the ability of Egr1 to control activity-dependent Arc expression. Finally, we used Arc FISH as an index of neural ensemble activation (Guzowski et al., 1999) to assess the proportion of dentate gyrus and CA1 cells activated by spatial exploration of novel and familiar environments in WT and Egr1−/− mice.
Materials and Methods
Subjects
Egr1−/− mice were generated using 129S2 ES cells injected into C57BL/6J blastocytes (Topilko et al., 1998) and backcrossed onto C57BL/6J background for 24 generations. Targeted inactivation of the Egr1 gene involved insertion of a lacZ-neo cassette between the promoter and coding sequence and an additional frameshift mutation at the level of an Ndel restriction site that corresponds with the beginning of the DNA binding domain. As described previously (Jones et al., 2001), histochemical, physiological, and behavioral screening has shown that gross brain anatomy, basal hippocampal synaptic transmission and cell excitability, and general behavior and motor activity are normal in Egr1−/− mice. In situ hybridization studies also confirmed the complete absence of Egr1 in mutant mice, while both constitutive and LTP-inducible expression of the lacZ gene in the Egr1−/− was comparable to that of the Egr1 gene in WT mice, suggesting that signaling events upstream of Egr1 transcription are not affected in the mice. WT and Egr1−/− littermate male mice (9–14 months old) used in this study were generated by crossing heterozygous Egr1+/− mice to obtain progeny in which male siblings were either of a mutant or WT genotype, as before (Jones et al., 2001; Bozon et al., 2003). Mice were housed in a temperature and light-controlled colony room (12 h light/dark cycle) in groups of 4/5 with food and water ad libitum. The genotype was verified by PCR on tail DNA. All experiments were conducted during the light phase. All efforts were made to minimize the number of animals and their suffering throughout the experiments. Experiments were performed in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and the French National Committee (87/848).
Apparatus, Behavior, and Tissue Collection
Electroconvulsive shock was administered via ear-clip electrodes using a constant-current generator. Eighty-four mice were used in this experiment. Mice were anesthetized lightly with fluothane and a single 200 V shock was delivered for 2 s. Immediately following the shock, mice displayed tonic–clonic seizures and were placed back in their home cages before being sacrificed at delays of 5 min, 30 min, 4 h, 6 h, or 8 h after ECS (WT: n = 6, 12, 6, 6, 5; Egr1−/−: n = 6, 6, 6, 6, 7, respectively). WT and Egr1−/− cage control (CC) mice were killed at the same time of day as experimental mice (WT: n = 11; Egr1−/−: n = 7).
In behavioral experiments, mice were first handled twice daily for 2 days before training. The next day (D1), mice in the “familiar environment” condition were placed in an open-field arena (45 cm × 45 cm × 30 cm) with wood shavings on the floor, positioned on a table (120 cm high) in an experimental room containing several distal cues. They were allowed to explore the arena for 10 min before being placed back in their home cages. This was repeated twice daily for 3 days (D1–D3), and once more on D4. Mice in the “novel environment” condition were allowed to explore the open-field arena (for 10 min) only once. Behavior of the animals was monitored automatically via a camera above the arena and using the ANY-maze program (Stoelting). Subgroups of WT and Egr1−/− mice from both experimental conditions were killed 30 min or 8 h after the end of exploration. Both WT and Egr1−/− mice displayed extensive exploration of the open-field arena, as evidenced by their multiple crossings of the 25 virtual square sectors in the arena (mice of both genotypes entered each square at least eight times on average). Fifty-four mice were used in the behavioral experiment (WT and Egr1−/−, respectively: familiar 30 min n = 4 and 4; novel 30 min n = 4 and 4; familiar 8 h n = 5 and 4; novel 8 h n = 7 and 7). Undisturbed WT and Egr1−/− CC mice were killed at the same time of day as experimental mice (WT: n = 8; Egr1−/−: n = 7).
Fluorescent in situ Hybridization
After cervical dislocation, the brains were removed rapidly, quick-frozen in isopentane (−40°C), and stored at −80°C until being sectioned on a cryostat. Twenty-micrometer-thick sections were mounted on slides such that the groups were distributed on the slides in a pseudorandom manner. For normalization purposes, each slide contained a WT CC and a WT section from the 30-min delay. Slides were air dried and stored frozen at −80°C until use. A commercial transcription kit and premixed RNA labeling nucleotide mixes containing digoxigenin-labeled UTP (Roche, France) were used to generate cRNA riboprobes. Riboprobes were purified on Mini Quick Spin RNA columns (G-50, Roche). The yield and integrity of riboprobes was confirmed by gel electrophoresis. The plasmid used to generate the Arc antisense and sense riboprobes contained a nearly full-length cDNA (∼3 kbp, courtesy of Dr. D. Kuhl) of the Arc transcript (Lyford et al., 1995). Slide-mounted sections were fixed in 4% buffered paraformaldehyde, treated with 0.75% acetic anhydride/1.1% triethanolamine (Roth), then with 50% acetone/50% methanol (Roth), and equilibrated in 2× SSC (Roth). Slides were incubated in 1× hybridization buffer, consisting of 4× SSC, 50% formamide (Roth), 1× Denhardt’s solution (Sigma), 10% dextran-sulfate (Roth), 0.05% fish sperm DNA (Roche), 0.025% yeast tRNA (Roche), for 30 min at room temperature. Riboprobe (100 ng) was diluted to 100 μl in the hybridization buffer, heat denatured, chilled on ice, and then added to each slide. Hybridization was carried out at 55°C for 16 h. Slides were washed to a final stringency of 0.5× SSC at 56°C; these washes included an earlier wash step at 37°C in 2× SSC with RNase A (10 μg/ml, Sigma). Endogenous peroxidase activity was quenched with 2% H2O2 in 1× SSC, slides were blocked (blocking reagent, Perkin-Elmer), and incubated with an appropriate horseradish peroxidase (HRP)-antibody conjugate (Roche) for 2 h. Slides were washed three times in Tris-buffered saline (with 0.05% Tween-20), and the conjugate was detected using TSA-direct system (Perkin-Elmer). Nuclei were stained with Sytox green (Invitrogen). Slides were coverslipped with antifade media (Fluoromount, Southern Biotech).
Image Acquisition and Analysis
All analyses were run blind to the genotype and experimental conditions. Images were acquired with a laser confocal microscope (argon–krypton laser; Zeiss MRC1024ES; Jena, Germany). Cross-over fluorescence could be ruled out as spectra of the fluorochromes did not overlap. Each optical section was averaged three times. Photomultiplier tube assignments, pinhole size, and contrast values were kept constant for each brain region within a slide. For each experiment, images were acquired from sections on two or three slides, ∼1.7 mm posterior to bregma. The free software ImageJ (http://rsb.info.nih.gov/ij) was used to analyze captured images. Only putative neurons were included in the analyses. Putative glial-cell nuclei were identified based on their small size (∼5 μm in diameter) and bright, uniform nuclear counterstaining (Guzowski et al., 1999); these cells never expressed Arc.
Two types of analyses were performed: counting the number of cells expressing Arc mRNA signal and optical density measurements, which allowed us to measure differences in signal intensity even in conditions of unchanged number of labeled cells. In the ECS experiment, only optical density analysis could be done, as 95–100% of the neurons expressed Arc after this strong stimulus, as previously described in the rat (Guzowski et al., 1999), making it impracticable to obtain cell counts. In ECS experiments, for each mouse, two to three z-stacks were collected from the CA1 and dentate gyrus cell layers (each from a different slide). Z-stacks consisted of the three middle planes of the brain section (1 μm optical thickness/plane), collected by using a 60× oil immersion objective. The field of view using this objective was 205 μm × 136 μm. For image analysis, the three stacks were “flattened” into one image, using a projection method taking into account only the highest (maximum) pixel value of the three stacks. The area of interest was then selected by visualizing only the color channel containing Sytox labeling (cell nuclei) and using the “Lasso” tool. The mean optical density value in the region of interest was determined for the color channel containing the CY3 labeling (Arc mRNA staining). Threshold levels were determined on the basis of WT CC images, where Arc expression was limited to a few scattered cells. The threshold value was held constant for each batch of slides processed together. Finally, values for experimental mice were normalized to those of WT caged controls (0%) and to WT mice with the 30-min delay post-ECS (100%). This normalization procedure minimizes artifact caused by slide-to-slide variation in signal intensity and background.
In behavioral experiments and for Arc expression in area CA1, z-stacks consisting of 1 μm-thick optical sections were acquired with a 60× oil objective. To estimate the proportion of Arc-positive neurons, 184 neurons per mouse on average were counted (from four non-overlapping z-stacks from two slides). First, neuronal nuclei present in the median planes (representing 20% of the stack thickness) were identified and outlined. Nuclei were then characterized for the presence of Arc cytoplasmic labeling (only nuclei with labeling in very close proximity were considered as positive), and the results expressed as a percentage of total nuclei analyzed per stack. The median planes criterion reduced the likelihood of analyzing partial nuclei, which could yield false negative results (West, 1993; Guzowski et al., 1999). We also estimated the amount of Arc mRNA expression by measuring optical density in CA1 pyramidal layer, stratum radiatum and stratum oriens (containing apical and basal dendrites, respectively), following the method described above for the ECS experiment (four non-overlapping z-stacks from two slides per animal were used).
In granule cells of the dentate gyrus, Arc staining was extremely sparse, thus to avoid sampling bias we imaged the entire dentate gyrus (four to six sections/mouse, left and right side from two to three slides; Vazdarjanova et al., 2006). Each dentate gyrus image was reconstructed from overlapping 10× z-stacks by using the shape of cell groups as landmarks. Cells exhibiting Arc cytoplasmic labeling were counted. The area of each section was measured and used to estimate the total number of neurons using a correction factor that represented total neurons per square micrometer. This factor was derived from 10 z-stacks from six different mice collected at 60× magnification (granule cell counts did not vary significantly across these 10 slices). Data from the upper and lower blades of the dentate gyrus were analyzed separately. The results were expressed as percent of estimated total cell number.
Statistics
Because of small sample sizes, effects of genotype and treatment (time after ECS or behavioral testing condition and delay) on Arc mRNA expression were evaluated by non-parametric Kruskal–Wallis test. When the main effect was significant at p ≤ 0.05, additional comparisons between groups were conducted with the non-parametric Mann–Whitney U-test. Behavioral data were obtained on a larger sample, they were therefore analyzed by ANOVA, followed by Fisher’s LSD post hoc tests, when appropriate. Correlations between behavioral parameters and Arc mRNA expression in each structure were analyzed by the non-parametric Spearman’s rank correlation test.
Results
Basal Arc mRNA Expression
In caged control mice, a small percentage of CA1 pyramidal cells and dentate granule cells (DGCs) exhibited Arc expression in WT mice (9.8 and 0.32%, respectively; Figures 1–4). A similar profile was observed in Egr1−/− mice (7.5 and 0.40% in CA1 and dentate gyrus, respectively; Figures 1–4), with no significant difference between genotypes (CA1: p = 0.73; dentate gyrus: p = 0.49).
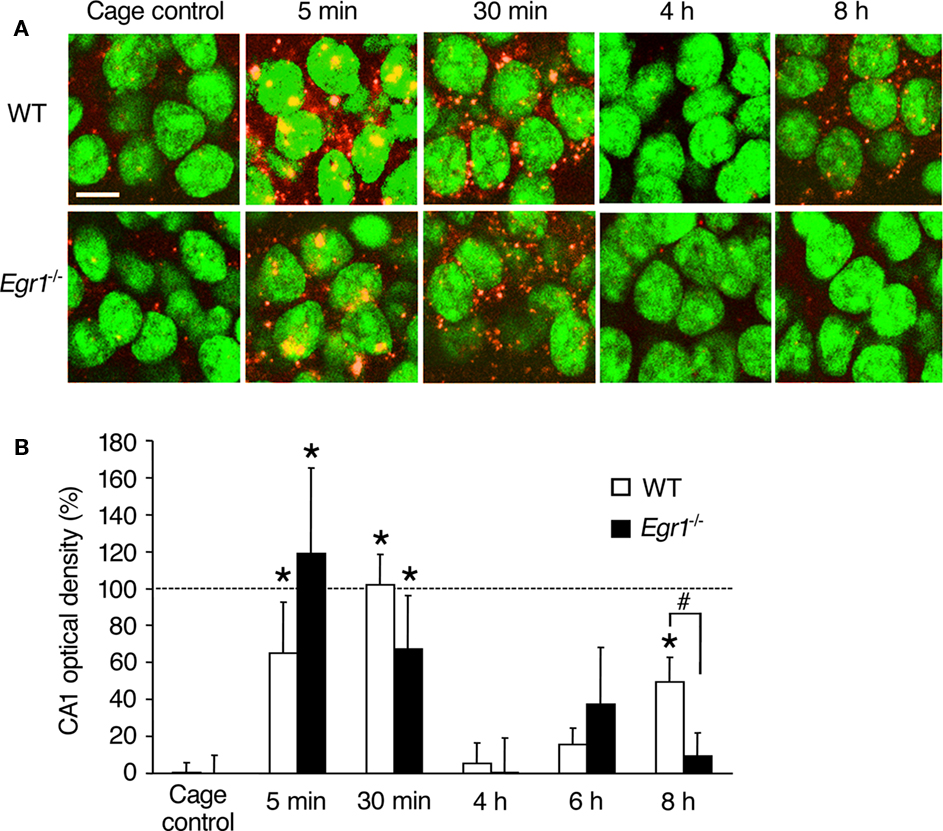
Figure 1. Arc mRNA expression after ECS in area CA1 of WT and Egr1−/− mice (cage control, CC). (A) Representative images of Arc mRNA detection using FISH in CA1 pyramidal neurons in CC mice and at different times after ECS. Cell nuclei appear in green (Sytox) and Arc mRNA signal in red (or yellow in the nucleus). Arc mRNA is intra-nuclear 5 min after ECS and mainly cytoplasmic 30 min after ECS. Nearly 100% of CA1 neurons express Arc mRNA after ECS. Scale bar, 10 μm. (B) Optical density of Arc mRNA signal in CA1. After initial Arc mRNA induction, the level of expression returns to baseline by 4–6 h, and a second wave is observed 8 h after ECS. This second wave is not observed in Egr1−/− mice. Error bars in this and subsequent figures indicate SEM. *Different from CCs of the same genotype, at least p < 0.05. #Difference between genotypes, p < 0.05. Dashed line indicates 100% (normalized to WT 30 min).
Arc Transcription Induced by Electroconvulsive Shock
In WT mice, ECS-induced robust transcription of Arc mRNA in CA1 and dentate gyrus neurons, compared with CC mice (Figures 1 and 2). The kinetics of ECS-induced Arc mRNA expression in the cytoplasm was of different appearance in CA1 and the dentate gyrus. In area CA1 of WT mice, a clear increase in intra-nuclear Arc expression was observed 5 min after ECS, followed by cytoplasmic expression at 30 min (Figure 1). Arc expression then returned to control levels at 4 and 6 h and a second wave of expression was observed at 8 h. Overall Kruskal–Wallis analysis showed a significant effect of ECS and/or genotype on Arc mRNA expression (p = 0.0004). Subsequent Mann–Whitney U-tests showed that in CA1 of WT mice, Arc mRNA expression was elevated 5 and 30 min after ECS compared with CC mice (5 min: p = 0.005; 30 min: p < 0.00001). Optical density values were not different from that of controls at 4 and 6 h following ECS, but again a significant increase was observed during the second wave at 8 h (p = 0.002), reaching ∼50% of that observed at 30 min. In Egr1−/− mice, Arc expression was also significantly elevated at 5 and 30 min compared with CC mice (5 min: p = 0.02; 30 min: p = 0.04). In contrast to the WT, however, no second wave of elevated Arc expression at 8 h was observed in CA1 of Egr1−/− mice (p = 0.28). Moreover, between-group comparison showed that CA1 Arc mRNA expression 8 h after ECS was significantly lower in Egr1−/− than in WT mice (p = 0.04).
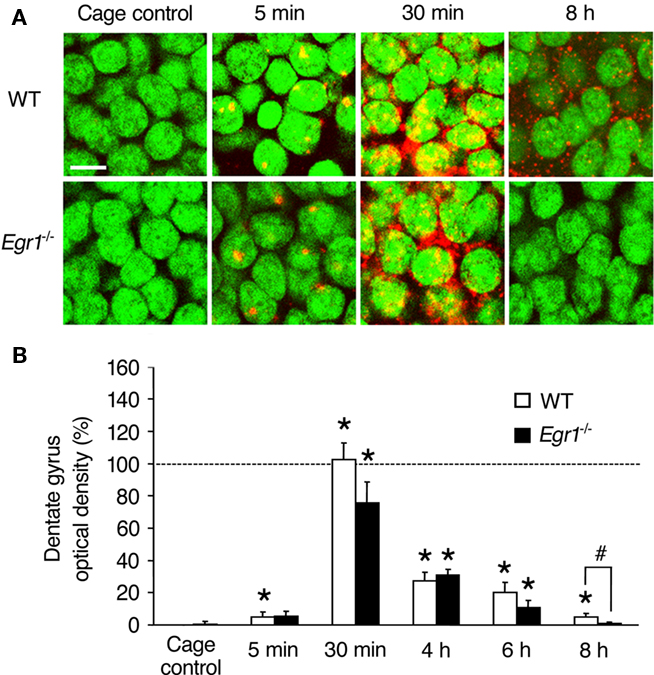
Figure 2. Arc mRNA expression after ECS in the dentate gyrus (DG) of WT and Egr1−/− mice. (A) Representative images of Arc mRNA detection in granule cells of the dentate gyrus in cage control mice and at different times after ECS. Cell nuclei appear in green (Sytox) and Arc mRNA signal in red (or yellow in the nucleus). Arc mRNA is intra-nuclear 5 min after ECS and mainly cytoplasmic 30 min after ECS. Scale bar, 10 μm. (B) Optical density of Arc mRNA signal in dentate gyrus. ECS-induced Arc mRNA expression is maintained for at least 8 h in WT mice, but decays more rapidly in Egr1−/− mice and is not significantly above the cage control baseline level after 6 h. *Different from cage controls of the same genotype, at least p < 0.05. #Difference between genotypes, p = 0.06. Dashed line indicates 100% (normalized to WT 30 min).
In the dentate gyrus of WT mice (upper blade), intra-nuclear labeling was observed in only few cells 5 min after ECS, but a large cytoplasmic increase in Arc mRNA expression was observed from 30 min up to 8 h post-ECS (Figure 2). Overall, there was a significant effect of ECS and/or genotype (Kruskal–Wallis p < 0.00001). In contrast to CA1, the increase in Arc mRNA expression in the dentate gyrus of WT mice followed a single wave and was significantly elevated compared to CC mice at all time points (5 min: p = 0.01; 30 min: p < 0.00001; 4 h: p = 0.0005; 6 h: p = 0.0005; 8 h: p = 0.004). In Egr1−/− mice, Arc mRNA was induced to a similar extent than in WT mice from 30 min to 6 h post-ECS (5 min: p = 0.10; 30 min: p = 0.001; 4 h: p = 0.001; 6 h: p = 0.01; Figure 1) with no difference between genotypes at any of these time points (all p > 0.05). However, no significant increase was detected in Egr1−/− at 8 h (p = 0.32). Moreover, comparison between groups showed a clear trend for Arc expression to be lower in Egr1−/− than in WT mice (p = 0.06), even if the level of ECS-induced expression in the dentate gyrus of WT mice represented only ∼5% of the initial increase observed at 30 min.
Arc Transcription Induced by Spatial Exploration in an Open-Field
Analysis of ECS-induced Arc mRNA expression indicated that long-term, but not short-term, expression was deficient in CA1 and the dentate gyrus of Egr1−/− mice. Thus, we next focused on the short-term (30 min) and long-term (8 h) time points to examine behaviorally induced Arc mRNA expression in WT and Egr1 mice. Two experimental conditions were used: exposure to a novel or to a familiar environment (see Materials and Methods).
Behaviorally, there was evidence for habituation in the familiar environment, as shown by the decrease in locomotor activity across the six sessions (trial 1: 30.7 m, trial 6: 20.9 m; session effect p < 0.0001). Moreover, during the session preceding sacrifice, mice in the familiar environment ambulated ∼20% less than mice exploring for the first time the novel environment (25.3 vs. 29.0 m, respectively; p = 0.03). Although Egr1−/− mice habituated similarly to the WT, a genotype effect was observed: overall locomotion in Egr1−/− mice was lower than in WT mice, with an average of ∼20% less distance moved than WT mice (genotype effect: p = 0.009). As reported previously in rats (Guzowski et al., 1999; Chawla et al., 2005; Ramirez-Amaya et al., 2005; Vazdarjanova et al., 2006), open-field exploration-induced a clear increase in Arc mRNA expression in both CA1 pyramidal cells and DGCs of WT mice, compared with the control groups (Figures 3 and 4).
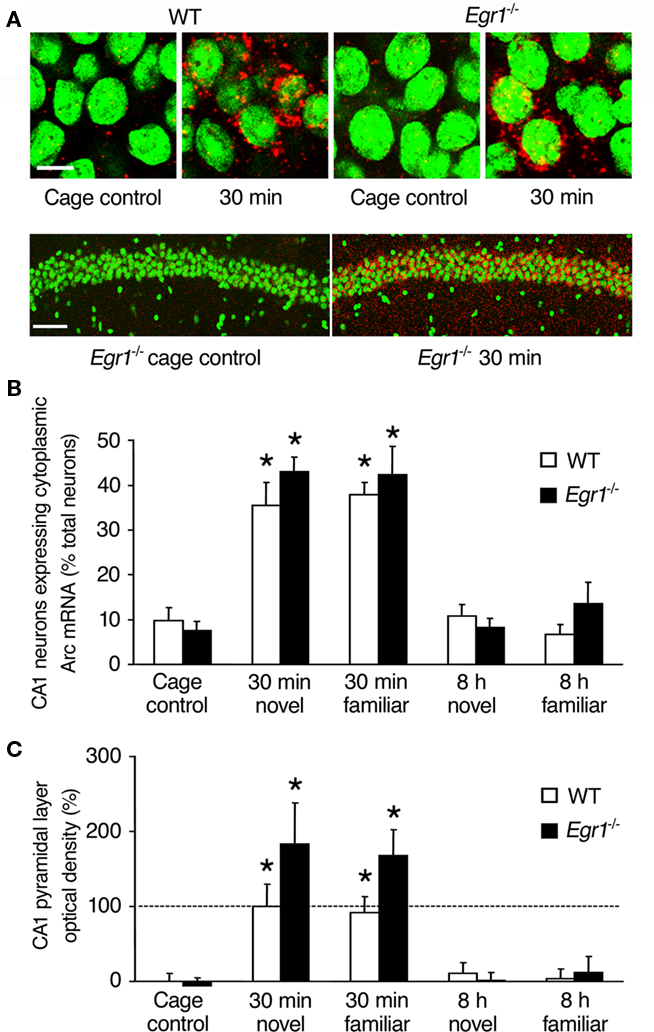
Figure 3. Arc mRNA expression after spatial exploration in CA1 of WT and Egr1−/− mice. (A) Representative images from WT and Egr1−/− cage control mice and 30 min after exploration of a novel or familiar environment. Scale bar, 10 μm. Below are sample lower definition images of Arc mRNA expression in CA1 from two Egr1−/− mice (left: cage control; right: 30 min after spatial exploration). Scale bar, 100 μm. Cell nuclei appear in green (Sytox) and Arc mRNA signal in red (or yellow in the nucleus). (B) Proportion of the neurons expressing Arc mRNA in the pyramidal layer after exploration of a novel or familiar environment. (C) Normalized optical density of Arc mRNA signal in the pyramidal layer. Arc mRNA expression was similarly induced in WT and Egr1−/− mice 30 min, but not 8 h following exploration. *Different from cage controls of the same genotype, at least p < 0.05. Dashed line indicates 100% (normalized to WT 30 min).
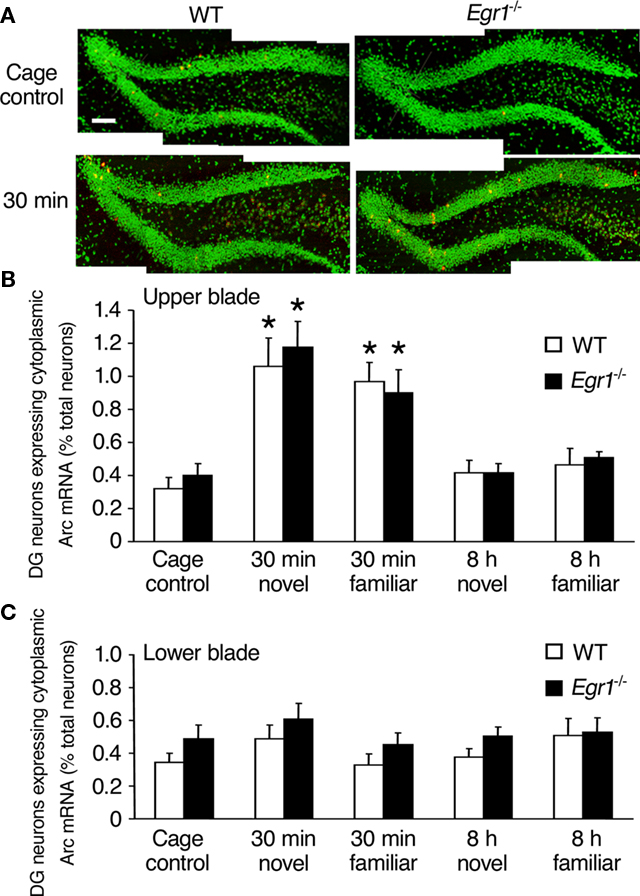
Figure 4. Arc mRNA expression after spatial exploration in the dentate gyrus (DG) of WT and Egr1−/− mice. (A) Representative images of dentate gyrus sections from WT and Egr1−/− cage control mice and 30 min after exploration of a novel or familiar environment. Scale bar, 300 μm. Cell nuclei appear in green (Sytox) and Arc mRNA signal in red (or yellow in the nucleus). (B,C) Proportion of neurons expressing Arc mRNA in the upper (B) and the lower blade (C) of the dentate gyrus after exploration of a novel or familiar environment. In the upper blade, Arc mRNA expression was similarly induced in WT and Egr1−/− mice 30 min, but not 8 h following exploration of a novel or a familiar environment. *Different from cage controls of the same genotype, at least p < 0.05.
In area CA1, we first estimated the proportion of neurons expressing cytoplasmic Arc mRNA at each time point (Figure 3). Overall Kruskal–Wallis analysis showed a significant effect of genotype, of open-field exploration and/or novelty of the environment on Arc expression (p = 0.0001; Figure 3B). Subsequent Mann–Whitney U-tests showed that Arc mRNA expression in WT mice was significantly elevated 30 min after exploration compared with CC mice, both following exposure to the novel and familiar environments (novel: p = 0.02; familiar: p = 0.006). In contrast to ECS-induced expression, there was no late wave of increased Arc expression in area CA1 of WT mice 8 h following behavioral exploration of either the novel or familiar environment. In area CA1 of Egr1−/− mice, a similar pattern of behaviorally induced Arc expression to that of WT mice was observed: Arc mRNA expression was significantly increased 30 min, but not 8 h, following exploration in both environmental conditions, compared with CC mice (30 min novel: p = 0.008; familiar: p = 0.008). Comparison of matching groups of WT and Egr1−/− mice showed that the proportion of Arc-positive cells in CA1 was similar at all time points and conditions. Moreover, novelty of the environment had no specific effect compared with familiar environment in CA1, regardless of genotype or delay.
Once established that Egr1 deficiency did not affect the proportion of neurons expressing Arc mRNA in CA1, neither at the basal condition nor after exploration, we estimated the amount of transcript expression by measuring optical densities in the pyramidal layer and in stratum radiatum and stratum oriens. Similarly to our results on the proportion of Arc-expressing neurons, optical density values 30 min after exploration were significantly increased in the pyramidal layer (Figure 3C) and stratum radiatum and oriens (data not shown) in both WT and Egr1−/− mice and in response to exposure to both the novel and familiar environments (Mann–Whitney U-test, all p < 0.05 compared with CC). The increase was slightly larger in Egr1−/− mice than in WT mice but this was not significant (all p > 0.05). Again, there was no significant increase in Arc expression at the 8-h time point in any of the different CA1 layers, genotypes or behavioral conditions (all p > 0.05). Direct comparison of matching groups of WT and Egr1−/− mice showed that Arc expression in area CA1 was similar for all time points and conditions, and there was no specific effect of spatial exploration of the novel vs. familiar environment, regardless of genotype or delay. Analyses of correlations between behavioral parameters (locomotor activity, immobility, percent time spent near the wall) and Arc mRNA expression levels (proportion of Arc-positive cells, optical density values) revealed no significant correlation at either delay (all p > 0.05).
In the dentate gyrus, Arc mRNA expression following exploration increased only in the upper blade and the apex (together referred to as upper blade hereafter), but not in the lower blade (Figure 4), as previously reported in rats (Chawla et al., 2005). This was confirmed by overall Kruskal–Wallis analysis showing statistically significant effects of genotype, open-field exploration and/or novelty on Arc mRNA expression in the upper blade (p = 0.0001), but not the lower blade (p = 0.36). In the upper blade of the dentate gyrus of WT mice, Arc mRNA expression was significantly increased compared with CC mice 30 min after exploration of both the novel and familiar environments (novel: p = 0.01; familiar: p = 0.006; Figure 4). In neither behavioral conditions was elevated Arc expression maintained at 8 h (all p > 0.05). In Egr1−/− mice, the level of Arc mRNA expression in the upper blade of the dentate gyrus was also significantly increased 30 min following exploration of both types of environments (novel: p = 0.008; familiar: p = 0.02), but not 8 h thereafter (p > 0.05 in each case). Direct comparison of matching groups of WT and Egr1−/− mice showed that Arc mRNA expression in the dentate gyrus was similar for all time points and conditions, and exploration of the novel or familiar environment had a similar effect regardless of genotype or delay (all p > 0.05). As in CA1, correlational analyses between behavioral parameters (locomotor activity, immobility, percent time spent near the wall) and Arc mRNA expression levels revealed no significant correlation at either delay (all p > 0.05).
Discussion
Two major findings of this study are that: (1) Egr1 is not necessary for Arc induction in the regions of the dorsal dentate gyrus and area CA1 analyzed, shortly (30 min) after neuronal activation; (2) Egr1 plays a significant role in activity-dependent delayed expression of Arc (8 h). Specifically, ECS-induced biphasic expression of Arc mRNA in area CA1 of WT mice, with a first wave of cytoplasmic expression 30 min after ECS and, after a decline to baseline by 4–6 h, a second, later phase of Arc expression in CA1 pyramidal cells at 8 h. We found that this late-phase of Arc expression in CA1 was completely absent in Egr1−/− mice. In DGCs, ECS rapidly (5 min) induced a single and prolonged wave of Arc mRNA expression, which decayed progressively but was still visible 8 h after ECS. With Egr1−/− mice, this wave of Arc expression in DGCs was curtailed in its longer-lasting phase (between 6 and 8 h). In the report by Li et al. (2005), Egr3, but not Egr1, was shown to be required for Arc expression in the dentate gyrus 4 h after kainic acid-induced seizures. Here, consistent with this result we did not observe a significant reduction in Arc expression 4 h after seizure in the dentate gyrus of Egr1−/− mice; however, examining longer time points following ECS, a later phase of Arc induction occurring at 8 h was not observed in hippocampal sub-regions in Egr1−/− mice, providing evidence that the Egr1 member alone can control the later phases of activity-dependent Arc gene transcription. The fact that Egr1 only affects the late induction of Arc is in agreement with the proposed role of this transcription factor in the second wave of gene expression driven by activity since Egr1 itself is induced by activity.
Short-term activity-dependent transcription of Arc is regulated as an IEG by recruitment of phosphorylated transcription factors acting at least in part via SRE and a Zeste-like response elements (Pintchovski et al., 2009). This early phase does not require protein synthesis (Wallace et al., 1998; Li et al., 2005). The early phase of Arc expression observed in WT mice is therefore probably due to post-translational modification of pre-existing transcription factors. Our results showing that short-term Arc transactivation is normal in Egr1−/− mice both after seizure and after spatial exploration suggest that Egr1 is dispensable for this early phase, even in CA1 where Egr1 is constitutively expressed. In contrast, our results strengthen the idea that the late-phase of Arc transcription requires transactivation via synthesized transcription factors, among which Egr1. Egr1 protein binds to a cognate GC-rich consensus DNA binding motif, the ERE. An ERE consensus sequence was identified proximal to the transcription start site of the Arc promoter (Li et al., 2005). In the dentate gyrus, Egr1 behaves as an inducible transcription factor with little, if any, constitutive expression (e.g., Beckmann and Wilce, 1997; French et al., 2001). It is therefore likely that activity-dependent Egr1 transcription, translation and binding to the ERE is required for initiating the late component of activity-dependent Arc transcriptional regulation. In area CA1, where basal expression of Egr1 is relatively high, its role in mediating the late wave of Arc induction could be via the same mechanisms and/or via post-translational modification of the existing Egr1 proteins leading to increased binding activity to the ERE. Further experiments would be required to determine whether the molecular mechanisms leading to Egr1-dependent transcription of Arc are similar in both hippocampal sub-regions.
Surprisingly, and in contrast to what has been reported in CA1 and dentate gyrus in rats 8 h after exploration of a novel environment (Ramirez-Amaya et al., 2005), the present experiment showed that WT mice did not express levels of Arc mRNA higher than caged controls at this late time point. This disparity between previous rat studies and our own likely reflects species differences. Notably, however, our Arc expression data in WT mice after ECS, both in CA1 and dentate gyrus, show a very similar pattern and kinetics of expression to that reported after spatial exploration in rats, with reactivation of CA1 Arc expression 8 h after ECS and prolonged expression up to 8 h in the dentate gyrus. This indicates that mouse hippocampal neurons have the intrinsic capacity for expressing activity-dependent late-phase of Arc transcriptional regulation. The reason for the absence of late expression of Arc after spatial exploration in mice compared to rats is at present unclear. One obvious possibility is that stronger initial synaptic activation is necessary in mice for the expression of the second wave of Arc in CA1 and for maintaining Arc expression over 8 h in the dentate gyrus. A stronger or more prolonged activation could possibly be achieved by using more salient behavioral paradigms such as contextual fear conditioning or spatial learning in the water maze. Alternatively, it remains possible that different molecular mechanisms are engaged in mice and rats for stabilizing neuronal ensembles activated during spatial exploration or that the process of stabilization of the activated neuronal ensembles is achieved more quickly in mice than in rats.
Besides this substantial difference between the two rodent species, several other features of Arc expression in hippocampal neurons following spatial exploration appear similar in mice and rats. First, similar to the findings in rats (Chawla et al., 2005; Ramirez-Amaya et al., 2005), we found that Arc-expressing neurons in CA1 were relatively homogeneously scattered along the pyramidal layer, whereas in the dentate gyrus Arc was induced in the upper blade and genu of the hilus, but virtually absent in the lower blade. Second, comparison between the present results in WT mice with previous reports in rats (Guzowski et al., 1999; Vazdarjanova et al., 2002; Ramirez-Amaya et al., 2005) suggests that spatial exploration activates neural ensembles of similar size in both species. Activity-dependent Arc expression is believed to be a reliable marker of neural ensembles activated by experience (Guzowski et al., 1999, 2006). In dorsal CA1, we found that Arc was expressed in 35.5% of the neurons 30 min after exploration, nearly identical to the proportion reported in rats (e.g., 38% in Vazdarjanova et al., 2002). In the dentate gyrus, the population of DGCs expressing Arc in response to spatial exploration is usually much smaller than in CA1 (Guzowski et al., 1999; Ramirez-Amaya et al., 2005; Vazdarjanova et al., 2006). The proportion of Arc-positive cells in the upper blade of the dentate gyrus in our experiment seemed slightly smaller (1.1%) than found in a comparable study in rats (2.4%; Chawla et al., 2005). In our mice, however, the proportion of Arc-positive neurons was also lower at the basal state than in rats (0.3 vs. 0.6% in the study of Chawla et al., 2005). Therefore, the ratio of Arc-expressing neurons after exploration to the basal state is closely similar (∼4×) in both species. In all, comparison of estimated numbers of activated cells suggests that spatial exploration recruits similar size dentate gyrus and CA1 neural ensembles in rats and mice. Moreover, our finding of similar size Arc-positive neural ensembles activated in both the dentate gyrus and CA1 between the two genotypes suggests normal neuronal encoding of information in the two hippocampal areas of Egr1−/− mice.
We also examined the contribution of relative familiarity of the environment. In WT mice, we found no difference in the proportion of Arc-positive cells after exploring novel or familiar environments, neither in area CA1 (35.5 vs. 38.0%, respectively) nor in the dentate gyrus (1.06 vs.1.17%, respectively), indicating that Arc expression is not directly linked to novelty. Of note, Arc transactivation was unaltered in Egr1−/− mice despite the fact that the mice displayed lower scores of locomotor activity compared to WT mice (by 20%). Moreover, locomotor activity also decreased by 20% in the familiar environment across successive sessions of habituation. Despite this, however, mice of both genotypes showed similar numbers of Arc-positive neurons in the familiar and novel environments. Thus, although this could be due to the fact that both genotypes displayed important locomotor activity, it points to the possibility that locomotor activity per se might not be the most critical factor determining the extent of the neural ensembles activated by experience in this paradigm. Our results are in line with those reported in rats (Guzowski et al., 2006: CA1: 37 vs. ∼39%; Chawla et al., 2005: dentate gyrus: 1.9 vs. 1.7%), showing that exploration-induced Arc transcription in CA1 and dentate gyrus does not habituate with repeated exploration of the same environment when exposures are separated by 24 h. Our study thus confirms in mice that Arc gene transcription is coupled to neural activation and does not distinguish between neural activity associated with new learning or memory retrieval (Miyashita et al., 2008).
With Egr1−/− mice, the patterns of Arc induction in the dentate gyrus and in area CA1 following exploration of a novel or familiar environment were qualitatively and quantitatively similar to those observed in WT mice. Furthermore, Egr1 deficiency had no effect in CA1 on Arc distribution into dendrites. Since there was no detectable late-phase increase in Arc transcription following exploration of the novel or familiar environments in mice, it was not possible to further assess the impact of Egr1 deficiency on Arc transcription in this experiment. However, since WT and Egr1−/− mice displayed similar patterns of Arc expression shortly after exploration, our results suggest that hippocampal neural ensembles in dentate gyrus and CA1 are normally activated during spatial exploration in Egr1−/− mice. In CA1 in particular, Guzowski et al. (1999) showed that the proportion of CA1 neurons expressing Arc immediately after spatial exploration in one or two environments is consistent with the proportion of pyramidal neurons that exhibit place cell properties in different environments, supporting the notion that Arc expression in CA1 neurons is related to the formation of a neural representation of specific environmental contexts. Hence, the normal early expression of Arc mRNA in CA1 pyramidal cells reported here after spatial exploration is consistent with our recent electrophysiological data showing that Egr1−/− mice can form new place cell representations normally in novel environments, although Egr1 deficiency impairs the ability to maintain newly formed neural representations over long delays (Renaudineau et al., 2009).
Conclusion
This study of Arc mRNA expression in mice using FISH reveals several similarities, but also substantial differences in activity-dependent transcription of Arc in hippocampal neurons between mice and rats. We observed a single early and transient wave of Arc expression in the dentate gyrus and area CA1 after spatial exploration of novel or familiar environments in mice and our results show this wave of Arc expression was not affected in Egr1 mutant mice, suggesting that spatial exploration recruits similar size dentate gyrus and CA1 neural ensembles in WT and Egr1 knockout mice immediately following spatial exploration. Our results with strong neuronal activation using ECS provide evidence that the Egr1 member of the Egr transcription factor family can alone control late-phases of activity-dependent hippocampal Arc transcription. Thus, it remains possible that a defect in the regulation of Arc may be one mechanism downstream of Egr1 deficiency which could underlie some of the long-term plasticity and long-term memory deficits that have been characterized in Egr1−/− mice. In these experiments, we specifically focused on the dorsal hippocampus, a segment primarily implicated in cognitive processes of learning and memory associated with navigation and exploration (reviewed in Fanselow and Dong, 2010). Future studies might usefully extend these observations to other regions of the hippocampus and to other brain structures, in relation to forms of memory that would be associated with longer-lasting Arc transcription patterns in neurons of mice.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research was funded by CNRS, Université Paris-Sud and by EU grant RTN-CT-2003-504231 to Serge Laroche Zsuzsa Penke was supported by EU grant RTN-CT-2003-504231. Egr1−/− mice were bred in Orsay from breeders generously provided by Patrick Charnay, Piotr Topilko, and Sonia Garel. We are grateful to Claudia Mahlke and Dietmar Kuhl for providing the Arc probes and for their help with the setting of the FISH technique, to Marion Salou for help with FISH and to Pascale Veyrac and Nathalie Devignes for providing animal care and for rearing and genotyping the mice.
References
Baumgartel, K., Tweedie-Cullen, R. Y., Grossmann, J., Gehrig, P., Livingstone-Zatchej, M., and Mansuy, I. M. (2009). Changes in the proteome after neuronal zif268 overexpression. J. Proteome Res. 8, 3298–3316.
Beckmann, A. M., and Wilce, P. A. (1997). Egr transcription factors in the nervous system. Neurochem. Int. 31, 477–510.
Bozon, B., Davis, S., and Laroche, S. (2002). Regulated transcription of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 12, 570–577.
Bozon, B., Davis, S., and Laroche, S. (2003). A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron 40, 695–701.
Bruel-Jungerman, E., Davis, S., and Laroche, S. (2007). Brain plasticity mechanisms and memory: a party of four. Neuroscientist 13, 492–505.
Chawla, M. K., Guzowski, J. F., Ramirez-Amaya, V., Lipa, P., Hoffman, K. L., Marriott, L. K., Worley, P. F., McNaughton, B. L., and Barnes, C. A. (2005). Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus 15, 579–586.
Davis, S., Bozon, B., and Laroche, S. (2003). How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav. Brain. Res. 142, 17–30.
Davis, S., Renaudineau, S., Poirier, R., Poucet, B., Save, E., and Laroche, S. (2010). The formation and stability pf recognition memory: what happens upon recall? Front. Behav. Neurosci. 4:177. doi: 10.3389/fnbeh.2010.00177.
Fanselow, M. S., and Dong, H. W. (2010). Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65, 7–19.
French, P. J., O’Connor, V., Jones, M. W., Davis, S., Errington, M. L., Voss, K., Truchet, B., Wotjak, C., Stean, T., Doyere, V., Maroun, M., Laroche, S., and Bliss, T. V. (2001). Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo. Eur. J. Neurosci. 13, 968–976.
Guzowski, J. F., McNaughton, B. L., Barnes, C. A., and Worley, P. F. (1999). Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 2, 1120–1124.
Guzowski, J. F., Miyashita, T., Chawla, M. K., Sanderson, J., Maes, L. I., Houston, F. P., Lipa, P., McNaughton, B. L., Worley, P. F., and Barnes, C. A. (2006). Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc. Natl. Acad. Sci. U.S.A. 103, 1077–1082.
James, A. B., Conway, A. M., and Morris, B. J. (2005). Genomic profiling of the neuronal target genes of the plasticity-related transcription factor Zif268. J. Neurochem. 95, 796–810.
Jones, M. W., Errington, M. L., French, P. J., Fine, A., Bliss, T. V., Garel, S., Charnay, P., Bozon, B., Laroche, S., and Davis, S. (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4, 289–296.
Knapska, E., and Kaczmarek, L. (2004). A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog. Neurobiol. 74, 183–211.
Lanahan, A., and Worley, P. F. (1998). Immediate-early genes and synaptic function. Neurobiol. Learn. Mem. 70, 37–43.
Li, L., Carter, J., Gao, X., Whitehead, J., and Tourtellotte, W. G. (2005). The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell. Biol. 25, 10286–10300.
Link, W., Konietzko, U., Kauselmann, G., Krug, M., Schwanke, B., Frey, U., and Kuhl, D. (1995). Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc. Natl. Acad. Sci. U.S.A. 92, 5734–5738.
Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lanahan, A. A., and Worley, P. F. (1995). Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14, 433–445.
Messaoudi, E., Kanhema, T., Soule, J., Tiron, A., Dagyte, G., da Silva, B., and Bramham, C. R. (2007). Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J. Neurosci. 27, 10445–10455.
Miyashita, T., Kubik, S., Lewandowski, G., and Guzowski, J. F. (2008). Networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol. Learn. Mem. 89, 269–284.
Nakamura, N. H., Fukunaga, M., Akama, K. T., Soga, T., Ogawa, S., and Pavlides, C. (2010). Hippocampal cells encode places by forming small anatomical clusters. Neuroscience 166, 994–1007.
Petersohn, D., Schoch, S., Brinkmann, D. R., and Thiel, G. (1995). The human synapsin II gene promoter. Possible role for the transcription factor zif268/egr-1, polyoma enhancer activator 3, and AP2. J. Biol. Chem. 270, 24361–24369.
Pintchovski, S. A., Peebles, C. L., Kim, H. J., Verdin, E., and Finkbeiner, S. (2009). The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene Arc/Arg3.1 in neurons. J. Neurosci. 29, 1525–1537.
Plath, N., Ohana, O., Dammermann, B., Errington, M. L., Schmitz, D., Gross, C., Mao, X., Engelsberg, A., Mahlke, C., Welzl, H., Kobalz, U., Stawrakakis, A., Fernandez, E., Waltereit, R., Bick-Sander, A., Therstappen, E., Cooke, S. F., Blanquet, V., Wurst, W., Salmen, B., Bosl, M. R., Lipp, H. P., Grant, S. G., Bliss, T. V., Wolfer, D. P., and Kuhl, D. (2006). Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52, 437–444.
Ramirez-Amaya, V., Vazdarjanova, A., Mikhael, D., Rosi, S., Worley, P. F., and Barnes, C. A. (2005). Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J. Neurosci. 25, 1761–1768.
Renaudineau, S., Poucet, B., Laroche, S., Davis, S., and Save, E. (2009). Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc. Nat. Acad. Sci. U.S.A. 106, 11771–11775.
Rial Verde, E. M., Lee-Osbourne, J., Worley, P. F., Malinow, R., and Cline, H. T. (2006). Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron 52, 461–474.
Shepherd, J. D., Rumbaugh, G., Wu, J., Chowdhury, S., Plath, N., Kuhl, D., Huganir, R. L., and Worley, P. F. (2006). Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484.
Steward, O., Wallace, C. S., Lyford, G. L., and Worley, P. F. (1998). Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21, 741–751.
Topilko, P., Schneider-Maunoury, S., Levi, G., Trembleau, A., Gourdji, D., Driancourt, M. A., Rao, C. V., and Charnay, P. (1998). Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol. Endocrinol. 12, 107–122.
Vazdarjanova, A., McNaughton, B. L., Barnes, C. A., Worley, P. F., and Guzowski, J. F. (2002). Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J. Neurosci. 22, 10067–10071.
Vazdarjanova, A., Ramirez-Amaya, V., Insel, N., Plummer, T. K., Rosi, S., Chowdhury, S., Mikhael, D., Worley, P. F., Guzowski, J. F., and Barnes, C. A. (2006). Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. J. Comp. Neurol. 498, 317–329.
Wallace, C. S., Lyford, G. L., Worley, P. F., and Steward, O. (1998). Differential intracellular sorting of immediate early gene mRNAs depends on signals in the mRNA sequence. J. Neurosci. 18, 26–35.
Keywords: transcription factors, immediate early genes, hippocampus, spatial exploration, electroconvulsive shock, FISH, mutant mice
Citation: Penke Z, Chagneau C and Laroche S (2011) Contribution of Egr1/zif268 to activity-dependent Arc/Arg3.1 transcription in the dentate gyrus and area CA1 of the hippocampus. Front. Behav. Neurosci. 5:48. doi: 10.3389/fnbeh.2011.00048
Received: 21 April 2011; Accepted: 27 July 2011;
Published online: 17 August 2011.
Edited by:
Riccardo Brambilla, San Raffaele Scientific Institute and University, ItalyReviewed by:
Riccardo Brambilla, San Raffaele Scientific Institute and University, ItalyGilberto Fisone, Karolinska Institutet, Sweden
Copyright: © 2011 Penke, Chagneau and Laroche. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Serge Laroche, CNRS UMR 8195, Centre de Neurosciences Paris-Sud, Université Paris-Sud, Bât 446, 91405 Orsay, France. e-mail:c2VyZ2UubGFyb2NoZUB1LXBzdWQuZnI=
 Zsuzsa Penke1,2
Zsuzsa Penke1,2