- Department of Neurology, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, China
Background: Ferroptosis plays a critical role in neuronal injury following cerebral infarction. However, effective therapeutic strategies targeting ferroptosis after cerebral ischemia–reperfusion injury (CI/RI) remain limited. Exosome-based therapy holds significant promise in this context. This study aims to identify key exosomal markers of ferroptosis.
Methods: By integrating and analyzing multiple GSE datasets, we identified ferroptosis-associated key genes. These findings were further validated in external databases, cellular models, and animal experiments using malondialdehyde (MDA), glutathione (GSH), iron, reactive oxygen species (ROS) assays, qRT-PCR, Western blotting. By further establishing a ferroptosis model and inhibiting DUSP1 with drugs, we further explored the potential function of DUSP1 in ferroptosis. The role of miR-101-3p was assessed in CI/RI models, while the diagnostic value of exosomal circular RNA was evaluated using receiver operating characteristic curve analysis.
Results: Combined differential analysis revealed PTGS2 and DUSP1 as ferroptosis-associated genes potentially regulated by exosomal circRNAs. In cellular and animal models, ferroptosis post-CI/RI was confirmed by elevated MDA, iron, and ROS levels, alongside reduced GSH. DUSP1 expression was significantly upregulated during ferroptosis, as demonstrated by qRT-PCR, Western blotting, and immunofluorescence. In the simple ferroptosis model, the expression of DUSP1 increases and inhibiting DUSP1 can aggravate ferroptosis. Conversely, miR-101-3p was downregulated in CI/RI, consistent with database predictions. Notably, exosomal circ_0093708 exhibited high diagnostic accuracy (Area under the curve = 0.93, sensitivity = 90%, specificity = 90%). Bioinformatics analysis suggested binding interactions among circ_0093708, miR-101-3p, and DUSP1.
Conclusion: Exosomal circ_0093708 is linked to DUSP1 and PTGS2 expression by sponging miR-101-3p, positioning it as a potential biomarker for ferroptosis in CI/RI.
1 Introduction
Stroke, a leading cause of global morbidity and mortality, poses severe threats to patient survival and often results in long-term disability among survivors (1). Furthermore, the incidence of stroke among young people has increased (2). Ischemic stroke (IS), accounting for the majority of stroke cases, is distinguished from other subtypes such as hemorrhagic stroke (3). Current clinical interventions, including intravenous thrombolysis and mechanical thrombectomy, are widely employed to restore cerebral blood flow (4). However, recanalization may paradoxically exacerbate brain damage through secondary injury mechanisms, particularly after prolonged ischemia. The cerebral ischemia–reperfusion injury (CI/RI) is driven by pathological processes such as apoptosis, the inflammatory response, oxidative stress, extracellular matrix remodeling, angiogenesis, cell hypertrophy, fibrosis, neurogenesis, blood–brain barrier damage and ferroptosis (5). Notably, ferroptosis—an iron-dependent form of regulated cell death characterized by excessive accumulation of iron ions and lipid peroxidation (6)—has been implicated as a critical contributor to CI/RI. Thus, targeting ferroptosis may represent a promising therapeutic strategy to mitigate neurodegeneration following CI/RI.
Exosomes, bilayer lipid membrane vesicles secreted by cells, facilitate intercellular communication by transporting nucleic acids, proteins, and lipids across biological barriers, including the blood–brain barrier (7). Among their cargo, circular RNAs (circRNAs) are notably abundant and stable, functioning as competitive endogenous RNAs (ceRNAs) that sequester microRNAs (miRNAs) to regulate gene expression. Recent studies reveal altered circRNAs profiles in plasma exosomes from IS patients (8), with some exhibiting diagnostic potential for large-artery atherosclerotic IS (9). However, whether exosomal circRNAs participate in ferroptosis during CI/RI, or serve as biomarkers remains unclear.
This study aims to identify ferroptosis-associated markers in plasma exosomes through integrated bioinformatics analysis, decipher the downstream miRNA-mRNA regulatory networks involved in CI/RI-induced ferroptosis, and explore novel diagnostic biomarkers for ferroptosis after CI/RI. By exploring the possible targets of plasma exosomal circRNAs in ferroptosis after CI/RI, this study may promote the understanding of the pathogenesis of CI/RI and stimulate targeted intervention measures.
2 Materials and methods
2.1 Data sources
We obtained discovery datasets GSE16561 (10) and GSE195442 (8) and validation datasets GSE110993 (11) and GSE22255 (12) from the GEO database.
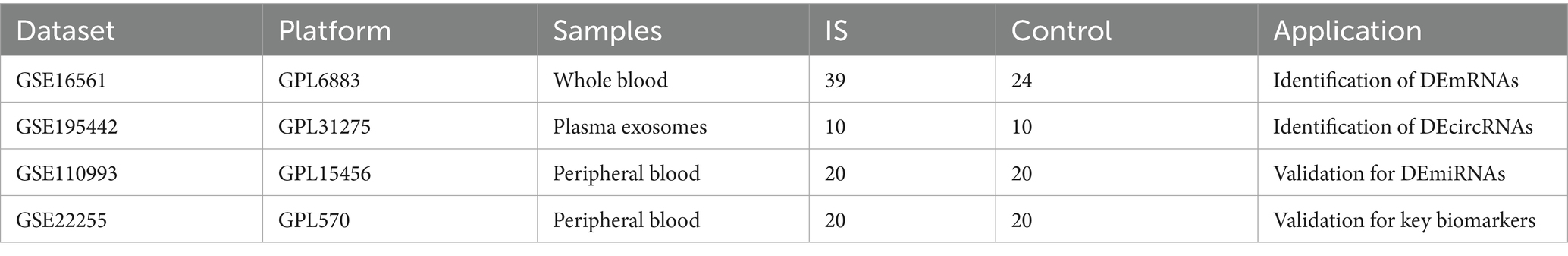
The GSE16561 dataset included 39 cerebral infarction patients and 24 healthy controls, while GSE195442 comprised 10 patients and 10 controls. For validation, GSE110993 contained 20 cerebral infarction patients and 20 healthy controls, and GSE22255 included 20 patients and 20 controls. Detailed characteristics of these datasets are presented in Table 1.
2.2 Data preprocessing and identification of DEmRNAs, DEmiRNAs, and DEcircRNAs
Following normalization of raw data, differential expression analysis was performed using the R Bioconductor limma package (v3.52.3). Genes meeting the criteria of p < 0.05 and |log2fold change| > 0.58 were identified as differentially expressed mRNAs (13), while circRNAs with p < 0.05 and |log2fold change| > 1.2 were classified as differentially expressed. Subsequently, DEcircRNAs were analyzed using the circbank database1 (14) to predict potential miRNA interactions. The resulting miRNAs were further filtered based on differential expression (p < 0.05 and |log2 fold change| > 1.2) using GSE110993 data. Heatmaps and volcano plots were generated using the R pheatmap package (v1.0.12).
2.3 Construction of the ceRNAs network
Predicted miRNAs from the circbank database (see text footnote 1) were validated against GSE110993 data. miRNA-mRNA interaction datasets were obtained from miRcode (v6.0, http://www.mircode.org/), miRTarBase (v8.0, http://mirtarbase.mbc.nctu.edu.tw/), and TargetScan (v7.2, http://www.targetscan.org/), with potential interactions identified using Perl software (v5.36.0, https://www.perl.org/). Only mRNA-miRNA pairs confirmed by all three databases were included in subsequent analyses.
2.4 Screening of key differentially expressed genes of ferroptosis
Using cerebral infarction diagnosis as the dependent variable, we analyzed 21 DEGs identified through intersection analysis of GSE16151 dataset and GSE110993 predictions. Ferroptosis-associated genes (including drivers, suppressors, markers, and unclassified genes) were retrieved from FerrDb (15).2 Key ferroptosis DEGs were visualized using the R “Venn” package (v1.11).
2.5 Cell culture and oxygen–glucose deprivation/reperfusion model
PC12 cells (rat pheochromocytoma cell line; Shanghai Institute of Biological Science, Chinese Academy of Sciences) were maintained in high-glucose DMEM (C11995500BT, Gibco, China) containing 10% fetal bovine serum (13011-8611, TIANHANG, China), along with 100 U/mL penicillin and 100 μg/mL streptomycin (C100C5, NCM Biotech, China), at 37°C under 5% CO₂ humidified conditions. To establish the OGD/R model, cells were subjected to incubation in glucose/serum-free DMEM (11,966,025, Gibco) using a tri-gas incubator (37°C, 94% N₂/5% CO₂/1% O₂) for 4 h, followed by a 24 h recovery period in complete DMEM. The experimental design comprised two groups: Control (standard culture conditions) and OGD/R.
2.6 Cell viability assay
PC12 cells (10,000 cells/well) were seeded in 96-well plates with 100 μL complete medium. After OGD/R, cells were incubated with 10 μL CCK-8 reagent (G1613-5ML, Servicebio, China) in 100 μL fresh DMEM for 1 h at 37°C. Cell viability was calculated as: [(Aexperimental − Ablank)/(Acontrol − Ablank)] × 100%.
2.7 Intracellular reactive oxygen species detection
The ROS were measured using dihydroethidium (DHE; CA1420, Solarbio, China). Cells were incubated with DHE (1:1000 dilution in DMEM) at 37°C for 30 min in the dark, washed with PBS (G4202-500, Servicebio, China), and visualized using an inverted fluorescence microscope (DMi8, Leica).
2.8 Biochemical assays
The content of intracellular iron content, malondialdehyde (MDA), and reduced glutathione (GSH) levels were quantified using commercial kits (BC5315, BC0025, and BC1175 respectively; Solarbio, China) following manufacturer protocols. Tissue samples from ischemic penumbra were processed identically. Protein content was determined using a BCA kit (BL521A, Biosharp, China), with absorbance measurements normalized to protein concentration.
2.9 RNA extraction and quantitative real-time PCR
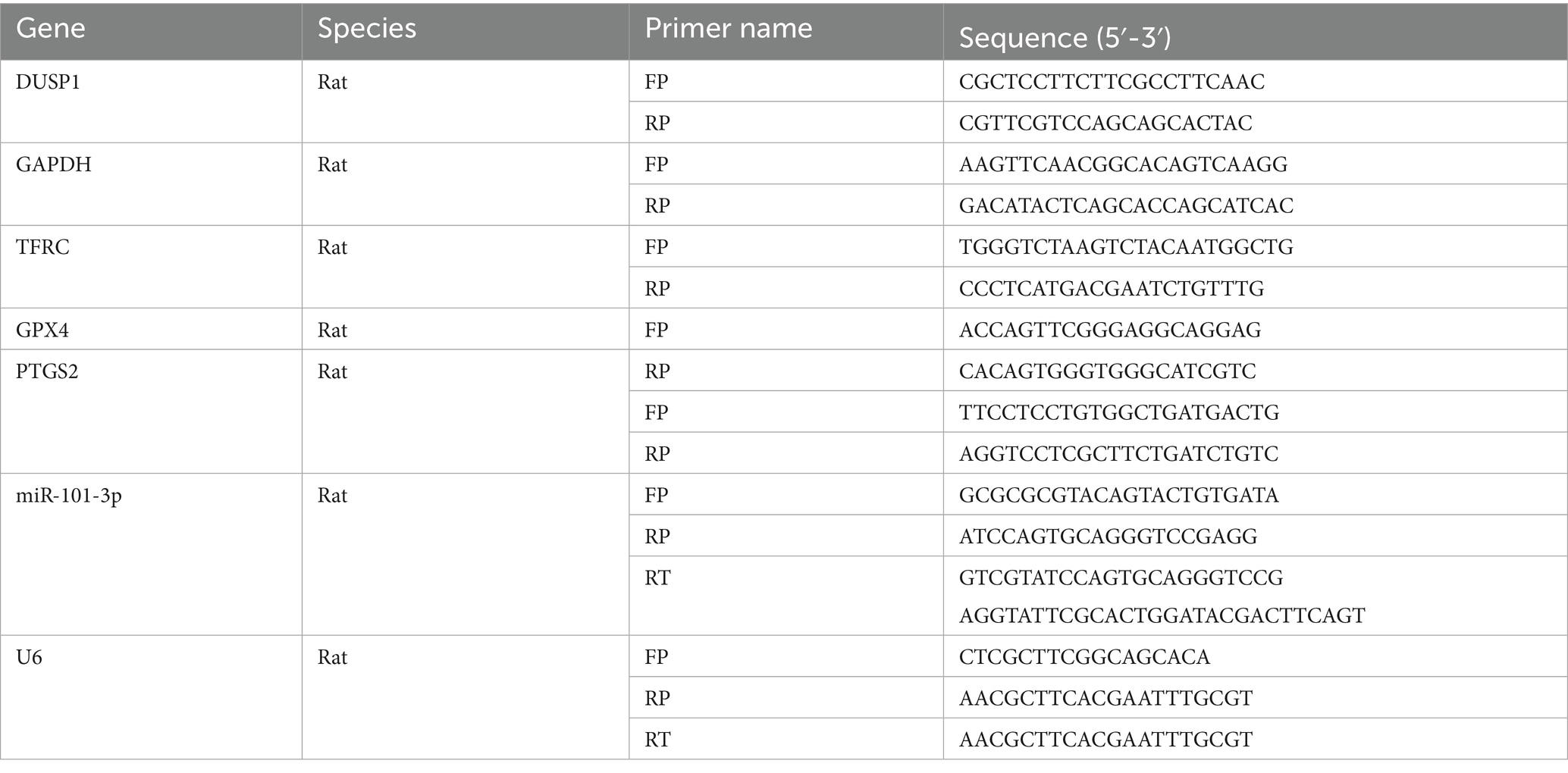
Total RNA was extracted from both cultured cells and brain tissue samples using RNAiso Plus reagent (9,109, Takara, Japan). Reverse transcription was performed using HiScript II Reverse Transcriptase (R223, Vazyme, China) following the manufacturer’s protocol. qRT-PCR amplification was carried out using ChamQ Universal SYBR qPCR Master Mix (Q311, Vazyme, China). The miRNA was reverse transcribed using the miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) (MR101-01, Vazyme, China), and quantified using the miRNA Universal SYBR qPCR Master Mix (MQ101-01, Vazyme, China). All reactions were performed on a CFX Opus Deepwell Real-Time PCR Detection System (Bio-Rad, United States). GAPDH was used as the endogenous reference gene, and relative gene expression levels were calculated using the 2−ΔΔCt method. The primer sequences used in this study are provided in Table 2.
2.10 Drug administration
CIL56 is a selective ferroptosis inducer that can induce ferroptosis by generating iron-dependent ROS. We chose CIL56 (T4309, TargetMol, America) to construct the ferroptosis model under the condition of 0.7 μM for 24 h.
(E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one (BCI) is a commonly used inhibitor of MKP-1. We used BCI (T10486, TargetMol, America) to inhibit DUSP1 in PC12 cells under the condition of 10 μM for 24 h.
2.11 Animals and middle cerebral artery occlusion/reperfusion model
Adult male Sprague–Dawley rats (250–280 g; Experimental Animal Center of Xi’an Jiaotong University, License: XJTUAE2024-076) were housed under standard conditions. The MCAO/R was performed under 2% pentobarbital sodium anesthesia (40 mg/kg, i.p.). After exposing the carotid sheath, the common carotid artery and external carotid artery were ligated. A nylon monofilament (2636A4, Beijing Cinontech, China) was inserted 18–20 mm into the internal carotid artery to occlude the middle cerebral artery for 90 min, followed by reperfusion. Sham-operated rats underwent identical procedures without filament insertion. Twelve rats were included in each group (sham and MCAO/R), with animals dying before the 24 h endpoint being excluded and replaced sequentially. Ischemic penumbra sampling followed established protocols (16), as illustrated in Figure 1A.
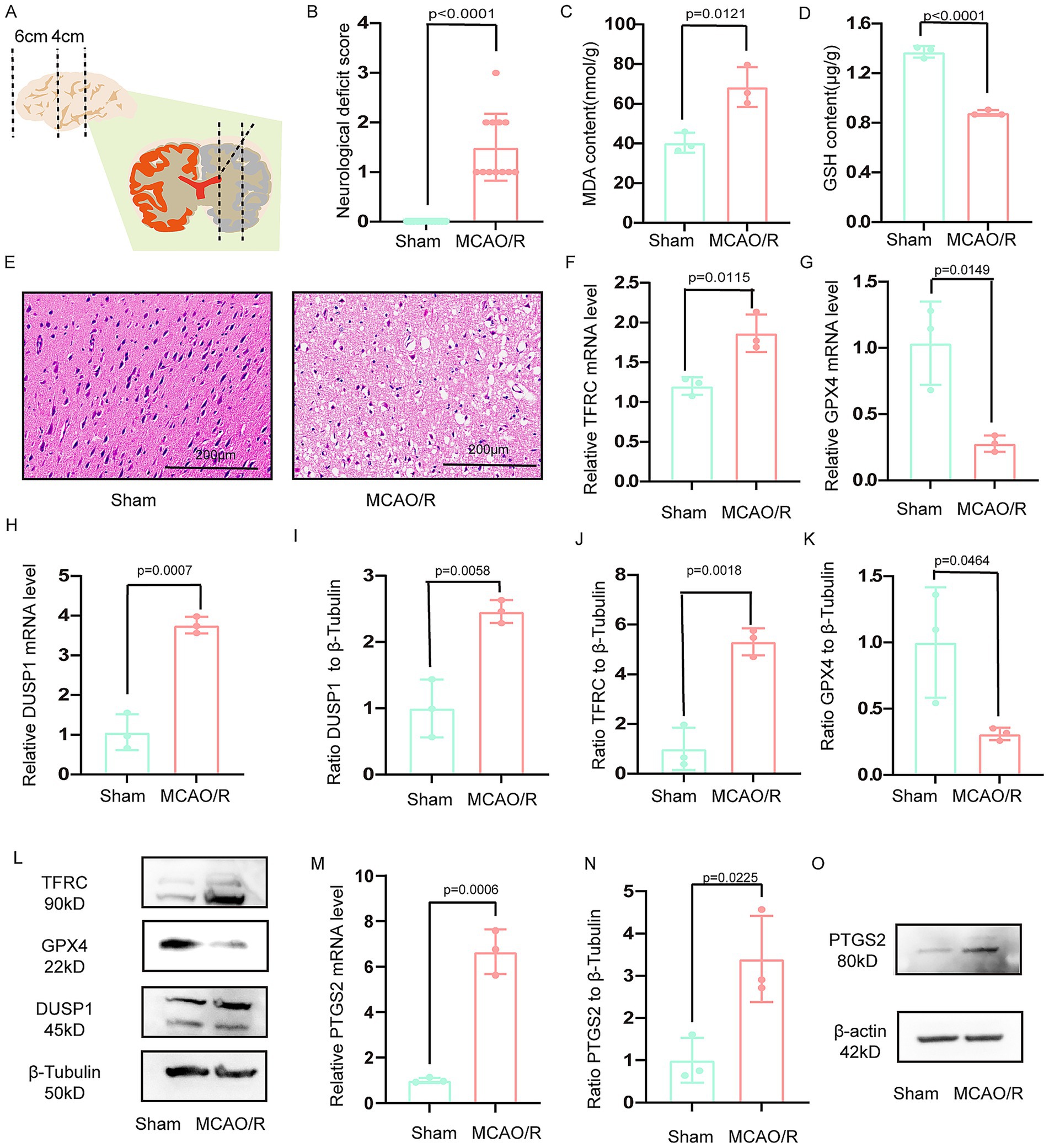
Figure 1. Verification the DUSP1 and PTGS2 in the vitro model. (A) Schematic representation of the ischemic penumbra sampling area. (B) Neurological deficit scores in MCAO/R rats. (C,D) Glutathione (GSH) and malondialdehyde (MDA) levels in the ischemic penumbra. (E) Histopathological changes in the ischemic penumbra (scale bar: 200 μm). (F,G) The mRNA expression levels of TFRC, and GPX4 in the ischemic penumbra. (H) The mRNA expression levels of DUSP1 in the ischemic penumbra. (I–L) The Western blot of DUSP1, TFRC and GPX4 in the ischemic penumbra. (M) The mRNA expression levels of PTGS2 in the ischemic penumbra. (N,O) The Western blot of PTGS2 in the ischemic penumbra. Except for the data in (B) which underwent the Mann–Whitney U test, the remaining data underwent the two independent sample t-tests after normality tests. Data are presented as mean ± SD. MCAO/R, middle cerebral artery obstruction/reperfusion.
2.12 Neurological deficit assessment
Neurological function was assessed 24 h after surgery using the modified Zea Longa 5-point scale (0 = no deficit; 4 = involuntary movement with consciousness disturbance). Animals were excluded if they: (1) scored 0 (model failure) or 4 (excessive injury), (2) showed subarachnoid hemorrhage at necropsy, or (3) died before the sampling endpoint.
2.13 Histopathological analysis
At 24 h post-reperfusion, rats were perfused transcardially with PBS followed by 4% paraformaldehyde. Brains were dehydrated, paraffin-embedded, and sectioned (4 μm) for hematoxylin and eosin staining and histopathological examination.
2.14 Western blotting
Protein extraction was conducted using RIPA lysis buffer (PL001, ZHHC, China) supplemented with protease inhibitors. Protein concentrations were quantified using a BCA assay (BL521A, Biosharp, China), with 30 μg of protein per sample mixed with 5 × loading buffer (ZS306-1, Zomanbio, China) at a 4:1 ratio. Following denaturation (100°C, 5 min), samples were resolved on 10% SDS-PAGE gels (PG212, Epizeme, China) and transferred to PVDF membranes (ISEQ00010, Millipore, United States) at 300 mA for 90 min. Membranes were blocked with RapidBlock solution (PS108, Epizeme, China) for 15 min at room temperature, followed by overnight incubation at 4°C with primary antibodies diluted in antibody dilution buffer (PS119, Epizeme, China). The following primary antibodies were used: DUSP1 (ET1701-82, Huabio, China; 1:1000), GPX4 (ET1706-45, Huabio, China; 1:10000), TFRC (ab269513, Abcam, United Kingdom; 1:5000), β-tubulin (66240-1-lg, Proteintech, China; 1:20000), PTGS2 (TP53818, Abmart, China,1:500), β-actin (66009-1-Ig, Proteintech, China; 1:20000). After TBST washes, membranes were incubated with HRP-conjugated secondary antibodies (SA00001-1/SA00001-2, Proteintech, China; 1:10000) for 1 h at room temperature. Protein bands were visualized using a chemiluminescence imaging system (GelView 6000Plus, BLTLUX, China). For reprobing, membranes were stripped with antibody stripping solution (WB6500, NCM Biotech, China). Band intensities were quantified using ImageJ software (v1.53t, National Institutes of Health, United States).
2.15 Statistical analysis
Statistical analyses were performed using R software (v4.3.0, https://www.r-project.org/) and GraphPad Prism (v9.0, San Diego, California, United States). Continuous variables with normal distribution were compared using Student’s t-test, while non-normally distributed data were analyzed using the Mann–Whitney U test. Brown-Forsythe tests with Dunnett’s T3 post hoc correction were used for multiple comparisons. A p-value < 0.05 was considered statistically significant. The study flowchart is presented in Figure 2.
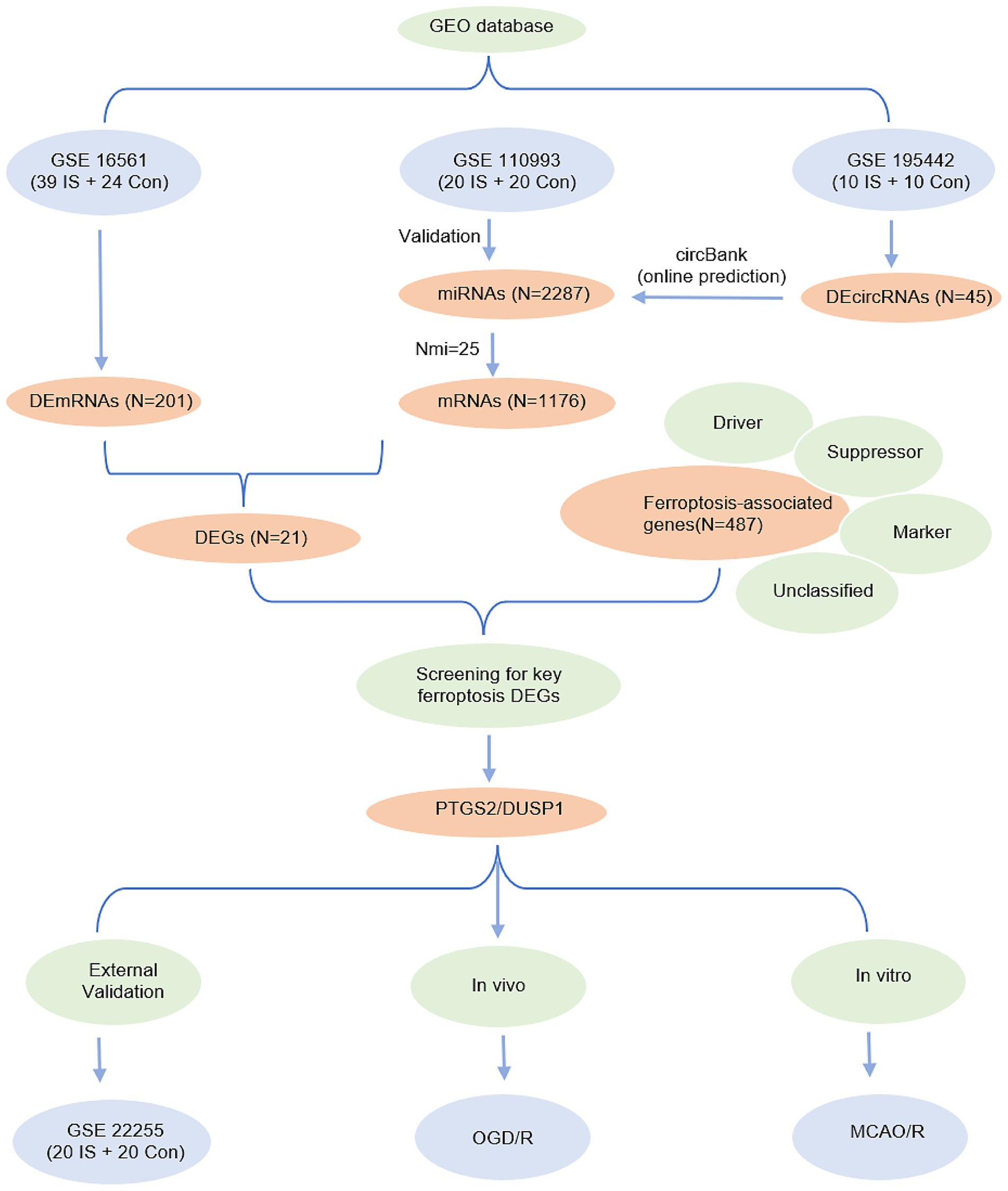
Figure 2. Entire working processes of the study. IS, ischemic stroke; Con, controls; DEGs, differently expressed genes; OGD/R, oxygen–glucose deprivation/reperfusion; MCAO/R, middle cerebral artery obstruction/reperfusion.
3 Results
3.1 Identification of differentially expressed mRNAs and circRNAs
Through comprehensive analysis of the GSE195442 and GSE16561 datasets, we identified 45 differentially expressed circRNAs (DEcircRNAs) and 201 differentially expressed mRNAs (DEmRNAs), respectively. The expression patterns of these differentially regulated circRNAs and mRNAs were clearly visualized through volcano plots (Figures 3A,B). Further intersection analysis between the DEmRNAs from GSE16561 and the potential mRNA targets predicted by miRNAs in GSE110993 revealed 21 overlapping genes. The expression profiles of these 21 DEmRNAs across different samples were subsequently illustrated using a heatmap (Figure 3C).
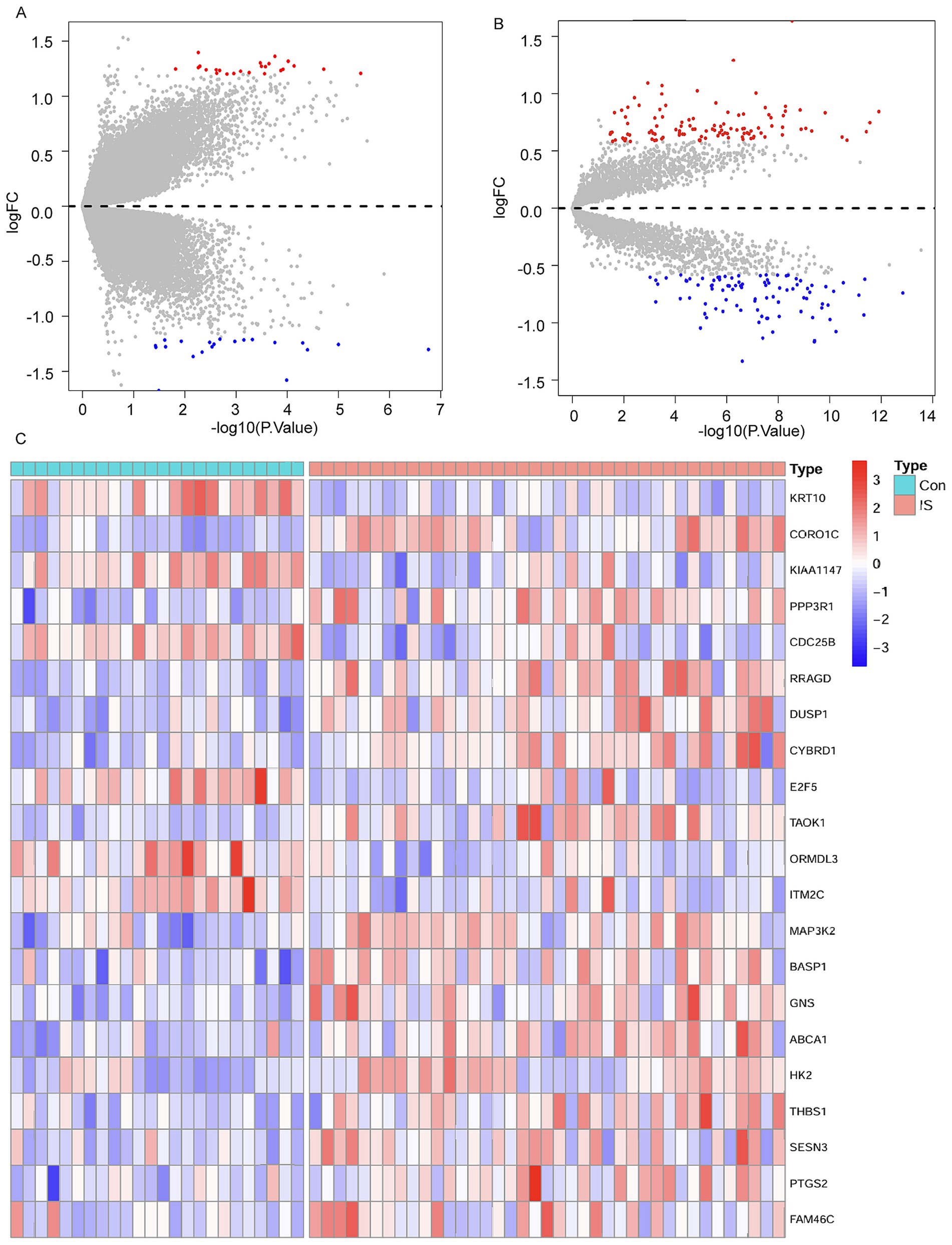
Figure 3. Differential expression analysis. (A) Volcano plot for DEcircRNAs in the GSE195442 dataset. (B) Volcano plot for DEmRNAs in the GSE16561 dataset. (C) Cluster heatmap for 21 DEmRNAs in the GSE16561 dataset.
3.2 Mining of key ferroptosis genes and external validation
From the FerrDb database, we compiled a comprehensive set of 487 ferroptosis- associated genes, consisting of 235 driver genes, 196 suppressor genes, 102 unclassified genes, and 9 marker genes. By intersecting these genes with our 21 DEmRNAs, we identified two candidate ferroptosis-associated genes: the ferroptosis marker gene PTGS2 and the unclassified gene DUSP1 (Figure 4A). In the GSE16561 dataset, DUSP1 demonstrated robust diagnostic potential, with an area under the curve (AUC) of 0.8579, sensitivity of 82.05%, and specificity of 75% (Figure 4B). PTGS2 showed moderate diagnostic performance, with an AUC of 0.7853, sensitivity of 84.62%, and specificity of 58.33% (Figure 4C). These findings were further validated in the external dataset GSE22255, where DUSP1 maintained significant diagnostic value (AUC = 0.735, sensitivity = 95%, specificity = 60%; Figure 4D), while PTGS2 exhibited slightly reduced but still notable efficacy (AUC = 0.655, sensitivity = 95%, specificity = 45%; Figure 4E).
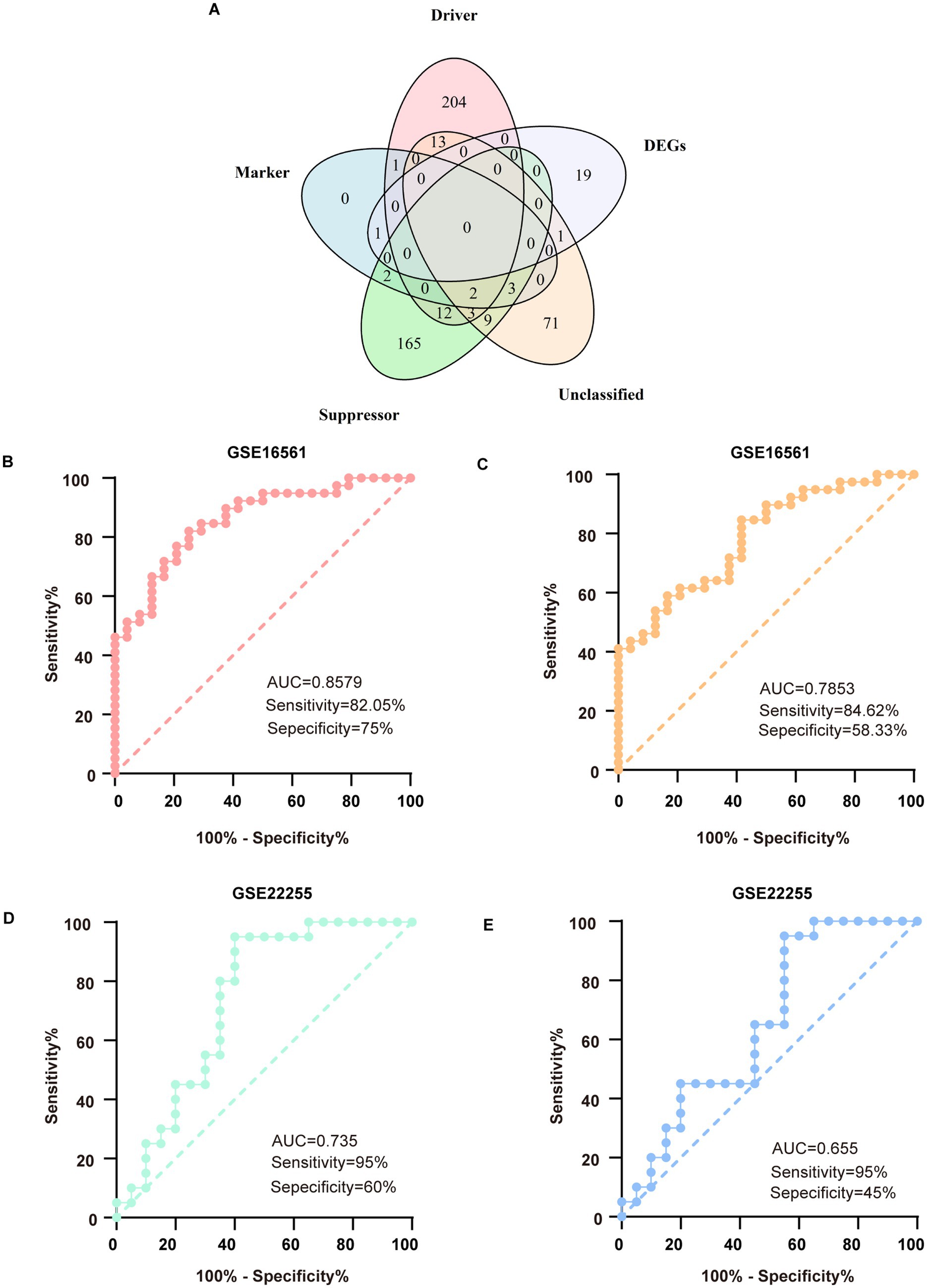
Figure 4. Mining of key genes for ferroptosis and verification in external databases. (A) The 5-set Venn diagram showing the comprehensive strategy among suppressor ferroptosis-associated genes (green circle), unclassified ferroptosis-associated genes (orange circle), marker ferroptosis-associated genes (blue circle), driver ferroptosis-associated genes (pink circle), and differentially expressed genes (DEGs) (purple circle). (B) ROC analysis for DUSP1 in the discovery dataset. (C) ROC analysis for PTGS2 in the discovery dataset. (D) ROC analysis for DUSP1 in the validation dataset. (E) ROC analysis for PTGS2 in the validation dataset. ROC, receiver operating characteristic.
3.3 In vitro validation of DUSP1 and PTGS2 in the OGD/R model
Following OGD/R treatment in PC12 cells, we observed a marked reduction in cell viability (Figure 5A). Concurrently, characteristic indicators of ferroptosis were evident, including decreased intracellular reduced GSH levels, elevated MDA and iron content, and increased ROS production (Figures 5B–E). The result of qRT-PCR further revealed upregulated TFRC and downregulated GPX4 expression (Figures 5F,G). The mRNA expression of DUSP1 was increased (Figure 5H). The result of Western blotting corroborated these findings at the protein level, demonstrating increased TFRC and decreased GPX4 levels, and significantly elevated DUSP1 protein expression (Figures 5I–L).
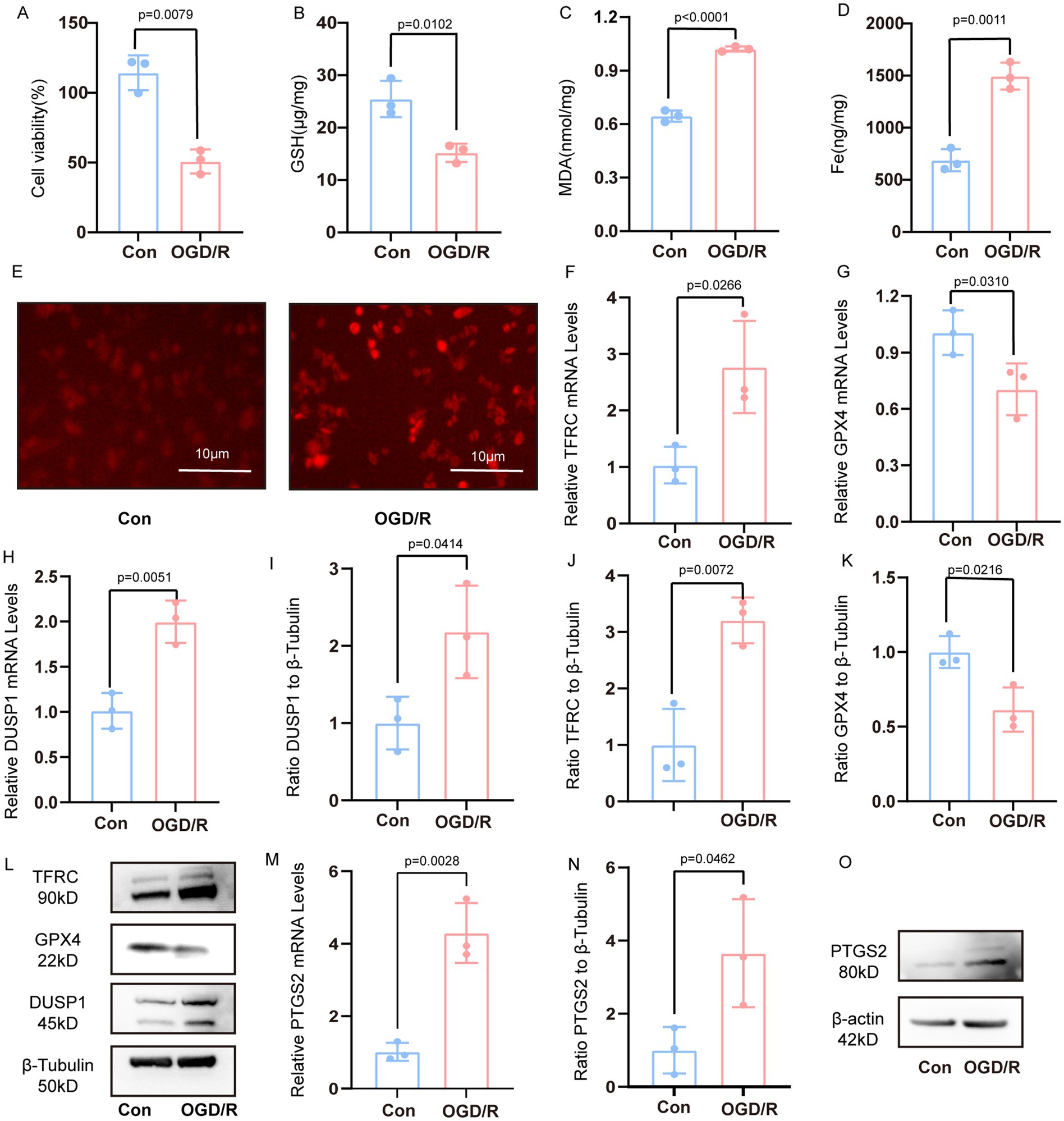
Figure 5. Verification the DUSP1 and PTGS2 in the vivo model. (A) Cell viability of PC12 cells after OGD/R treatment. (B–D) The content of GSH, MDA and Fe. (E) The content of ROS in PC12 cells after OGD/R (scale bars: 10 μm). (F,G) The mRNA expression levels of TFRC, and GPX4 in PC12 cells after OGD/R. (H) The mRNA expression levels of DUSP1 in PC12 cells after OGD/R. (I–L) The Western blot of DUSP1, TFRC, GPX4 in PC12 cells after OGD/R. (M) The mRNA expression levels of PTGS2 in PC12 cells after OGD/R. (N,O) The Western blot of PTGS2 in PC12 cells after OGD/R. All data underwent two independent sample t-tests after normality tests. Data are presented as mean ± SD. Con, controls; OGD/R, oxygen–glucose deprivation/reperfusion.
Meanwhile, the mRNA expression of PTGS2 was increased (Figure 5M). The result of Western blotting demonstrated that the protein expression of PTGS2 was increased (Figures 5N,O).
3.4 In vivo validation of DUSP1 and PTGS2 in the MCAO/R model
Successful establishment of the MCAO/R model in rats was confirmed through both behavioral and molecular assessments (Figure 1B). Consistent with the in vitro findings, the MCAO/R group showed decreased GSH and increased MDA levels in the ischemic penumbra (Figures 1C,D). The ischemic penumbra region, delineated in Figure 1A, exhibited significant pathological alterations, including neuronal shrinkage, reduced cell density, and vacuolar degeneration (Figure 1E). Molecular analyses revealed upregulated transcription of DUSP1 and TFRC, coupled with downregulated GPX4 expression (Figures 1F–H). Western blotting confirmed these trends at the protein level, with increased TFRC and decreased GPX4, and significantly elevated DUSP1 protein expression (Figures 1I–L).
Meanwhile, the mRNA expression of PTGS2 in ischemic penumbra of rat was increased (Figure 1M). The result of Western blotting demonstrated that the protein expression of PTGS2 in ischemic penumbra was increased (Figures 1N,O).
3.5 Inhibition of DUSP1 exacerbates ferroptosis in PC12 cells
Treatment of PC12 cells with CIL56 resulted in decreased cell viability. Notably, co-treatment with CIL56 and the DUSP1 inhibitor BCI led to a more pronounced reduction in cell survival compared to CIL56 treatment alone (Figure 6A). Following the induction of ferroptosis by CIL56, intracellular MDA levels were significantly elevated. Importantly, the BCI-treated group exhibited even higher MDA content than cells treated with CIL56 alone (Figure 6B). Consistent with these findings, CIL56 treatment increased intracellular ROS levels in PC12 cells. This effect was markedly enhanced when CIL56 was combined with BCI (Figure 6D).
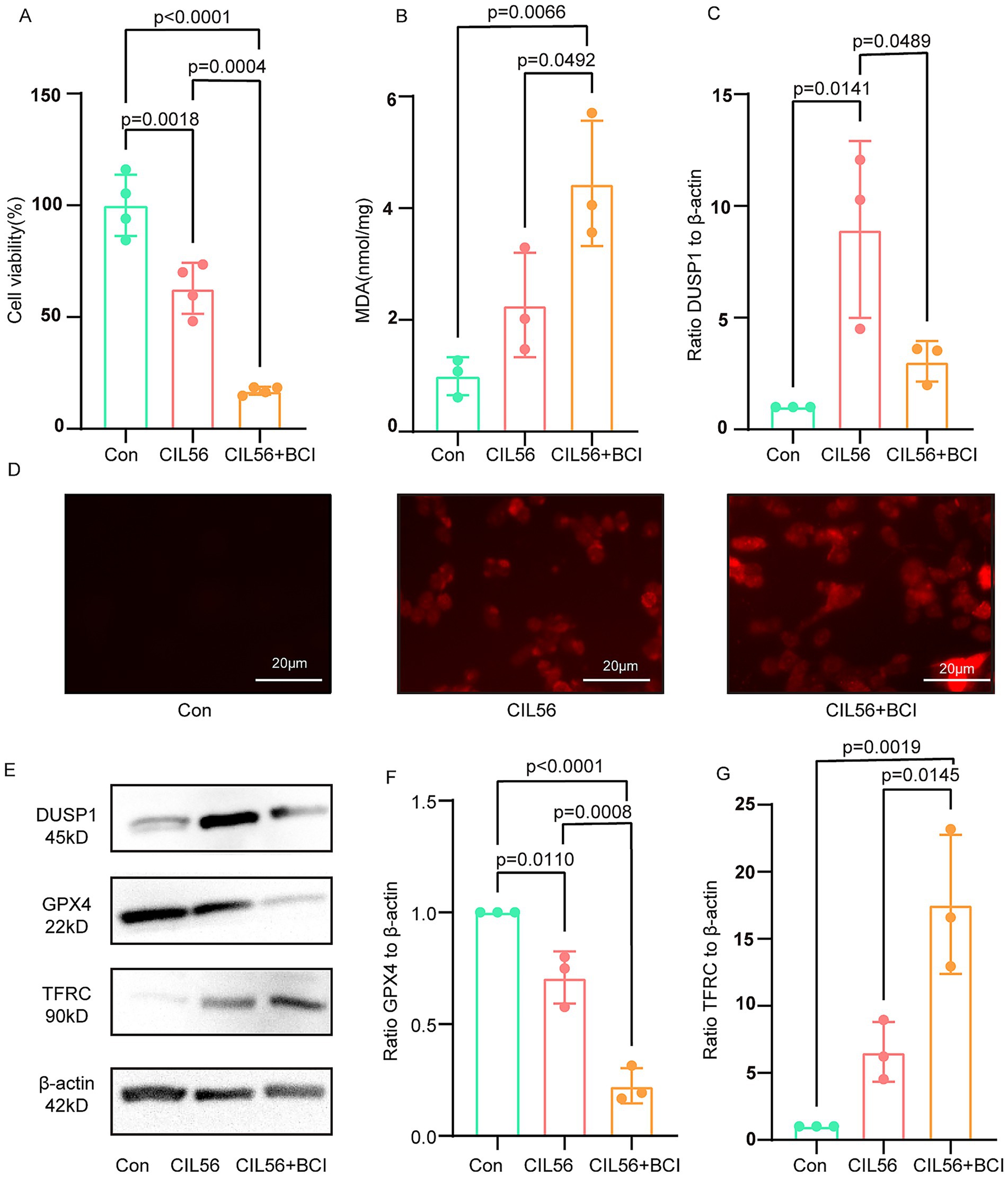
Figure 6. Inhibition of DUSP1 exacerbates ferroptosis in PC12 cells. (A) Cell viability after CIL56 treatment. (B) Intracellular MDA levels. (C) Effect of BCI on DUSP1 inhibition in the ferroptosis model. (D) Intracellular ROS levels (scale bar: 20 μm). (E) Protein expression of DUSP1, GPX4, and TFRC. (F) Quantification of GPX4 levels. (G) Quantification of TFRC levels. Data were analyzed by one-way ANOVA with Tukey’s post hoc test and presented as mean ± SD. Con, control.
Western blot analysis revealed that CIL56 successfully induced ferroptosis in PC12 cells, as evidenced by decreased GPX4 expression and increased TFRC levels. Interestingly, DUSP1 expression was upregulated in response to CIL56 treatment (Figure 6C). The ferroptosis effects were further aggravated upon BCI co-treatment, with more pronounced changes in both GPX4 and TFRC protein levels compared to the CIL56-only group (Figures 6E–G). These results collectively suggest that pharmacological inhibition of DUSP1 potentiates ferroptosis in PC12 cells, indicating that DUSP1 may function as a negative regulator of ferroptosis.
3.6 Verification of miRNA and construction of the ceRNAs network
Through systematic analysis, we constructed a ceRNAs network. Validation using the GSE110993 and GSE195442 datasets confirmed the involvement of these regulatory molecules, as illustrated by the Sankey plots (Figure 7A). Notably, miR-101-3p emerged as a key regulator capable of simultaneously targeting both DUSP1 and PTGS2. Subsequent experiments in both the OGD/R and MCAO/R model demonstrated significant downregulation of miR-101-3p (Figures 7B,C), establishing its role in ferroptosis regulation following CI/RI.
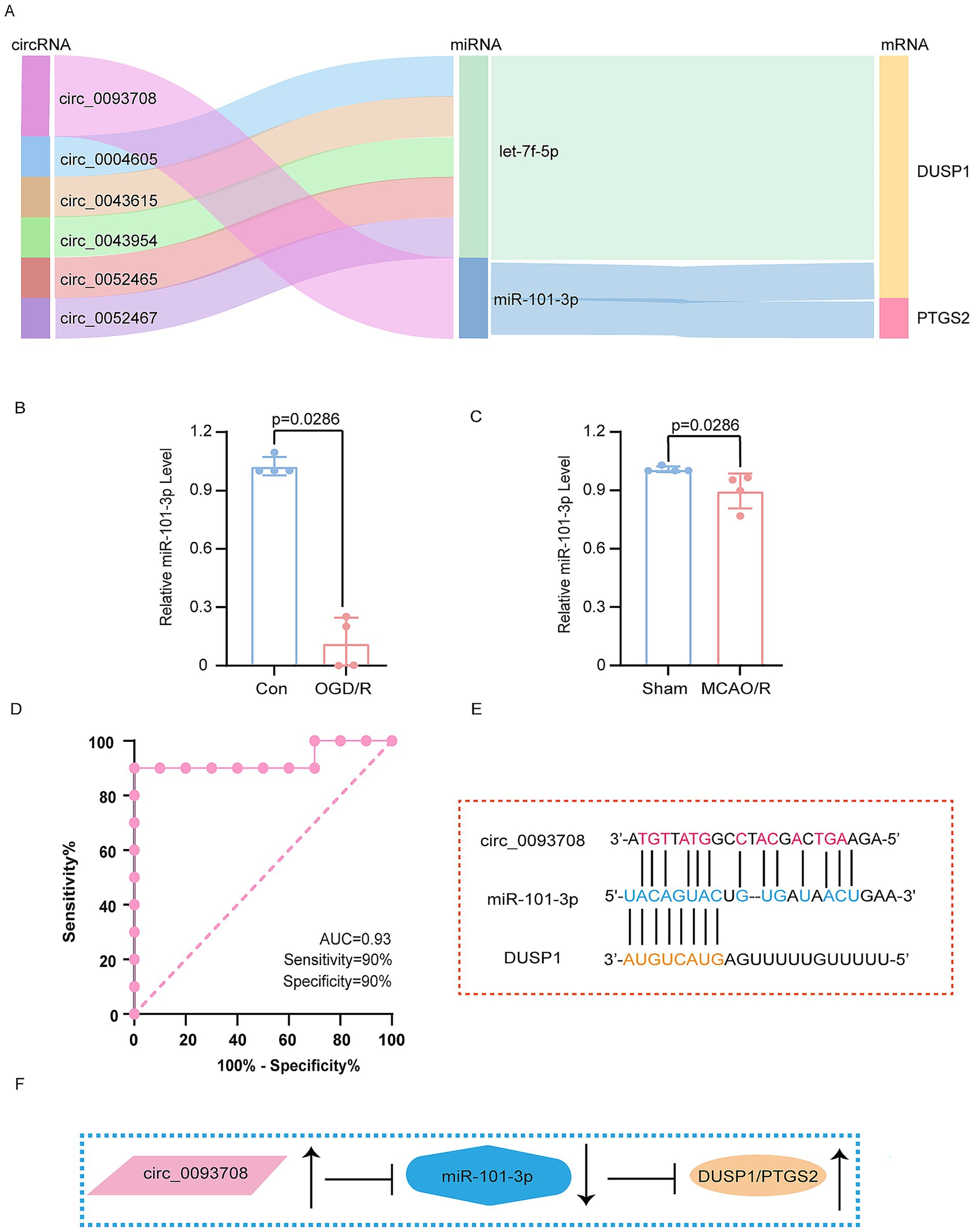
Figure 7. Verification of miRNA and construction of ceRNAs network. (A) The Sankey plots of ceRNAs networks. (B) The expression of miR-101-3p in OGD/R model. (C) The expression of miR-101-3p in MCAO/R model. (D) The ROC analysis of circ_0093708 in exosomes. (E) The schematic diagram of the binding of circ_0093708, miR-101-3p and DUSP1. (F) The schematic diagram of the regulation of circ_0093708, miR-101-3p, DUSP1 and PTGS2. The data in (B,C) underwent the Mann–Whitney U test. Con, controls; OGD/R, oxygen–glucose deprivation/reperfusion; MCAO/R, middle cerebral artery obstruction/reperfusion. ROC, receiver operating characteristic.
Further investigation revealed that circ_0093708, through competitive binding with miR-101-3p, promotes the expression of both DUSP1 and PTGS2. Remarkably, circ_0093708 exhibited exceptional diagnostic potential for cerebral infarction in the GSE195442 dataset, with an AUC of 0.93, sensitivity of 90%, and specificity of 90% (Figure 7D). The proposed regulatory mechanism of the circ_0093708/miR-101-3p/DUSP1/PTGS2 axis is comprehensively illustrated in Figures 7E,F. This integrated approach not only confirms the central role of DUSP1 in ferroptosis following CI/RI but also identifies a novel ceRNAs network with significant diagnostic and therapeutic potential.
4 Discussion
Our study employed bioinformatics approaches to integrate data from multiple databases, constructing a ceRNAs network involving exosomal circRNAs, miRNA, and mRNA. We validated the newly identified ferroptosis-associated gene DUSP1 in external databases, as well as in cellular and animal models. Furthermore, we elucidated the alterations of key miRNAs in these models and demonstrated that exosomal circRNAs could serve as novel biomarkers and potential therapeutic targets for ferroptosis following CI/RI.
Our findings revealed that PTGS2 (prostaglandin-endoperoxide synthase 2, also known as COX2) acts as a downstream target of miR-101-3p and modulates ferroptosis after CI/RI. PTGS2 is a well-established inflammatory mediator that catalyzes prostaglandin synthesis and participates in diverse pathophysiological processes. It is also a confirmed biomarker of ferroptosis, with its mRNA levels positively correlating with ferroptosis sensitivity in various tumor models (17). Post CI/RI, ferroptosis occurs alongside elevated PTGS2 expression (18). Ferroptosis inhibition has been shown to suppress the COX-2/PGE2 pathway (19), while exosomal miR-137 can inhibit COX2/PGE2 activation, conferring neuroprotection (20). Previous study corroborated that PTGS2 levels are increased in the infarcted cortex of MCAO rats. PTGS2 inhibition reduces infarct volume, improves neurological deficit scores, and enhances outcomes (21). Similarly, blocking the p53/PTGS2 pathway mitigates hippocampal neuronal ferroptosis and alleviates CI/RI (22). Collectively, these findings suggest that PTGS2 promotes ferroptosis after CI/RI.
DUSP1, a member of the dual-specificity phosphatase family, dephosphorylates tyrosine and threonine residues, thereby inhibiting mitogen-activated protein kinases (MAPK). In the central nervous system, the MAPK pathway mediates apoptosis, inflammation, autophagy, oxidative stress, and ferroptosis (23–25). The DUSP1 has emerged as an endogenous neuroprotective factor in stroke. Its inhibition exacerbates infarct volume, neurological deficits, and hemorrhagic transformation (26), while its overexpression suppresses the c-Jun N-terminal Kinase (JNK) pathway and neuronal death in OGD/R and MCAO/R models (27). In endothelial cells, DUSP1 overexpression preserves blood–brain barrier integrity by inactivating MAPK, thereby reducing brain injury and improving post-stroke outcomes (28). In ferroptosis research, DUSP1 expression increases upon ferroptosis induction in cancer cell lines (17). Intriguingly, DUSP1 has been identified as a ferroptosis inhibitor in pancreatic cancer, where it blocks lipid peroxidation-dependent ferroptosis (29). Our prior work identified DUSP1 as a reliable oxidative stress marker in ischemic stroke (30). Given its role in reducing ROS-induced cell death and preserving mitochondrial membrane potential (31), DUSP1 likely functions as a ferroptosis suppressor in CI/RI by mitigating ROS accumulation, a hallmark of ferroptosis. Our research results also preliminarily indicate that DUSP1 shows a protective effect in ferroptosis after CI/RI.
Our study demonstrated that miR-101-3p is downregulated in OGD/R and MCAO/R models, as well as in the GSE110993 dataset. This miRNA is a critical regulator in neurological diseases, influencing cerebrovascular and neurodegenerative pathologies (32–35). In human umbilical vein endothelial cells, miR-101-3p mimics induce ROS production and NF-κB activation, whereas its inhibition attenuates endothelial injury, highlighting its role in atherosclerosis (36). In young MCAO mice, miR-101-3p is downregulated but upregulates HDAC9; its overexpression improves neuronal morphology and apoptosis, suggesting a neuroprotective role (37). Conversely, miR-101-3p inhibition enhances DUSP1 expression, suppressing MAPK p38 and NF-κB pathways to reduce inflammation and apoptosis (38). After acute brachial plexus injury, JHDM1D-AS1 targets miR-101-3p to upregulate DUSP1, further underscoring its neuroprotective potential (39). Regarding exactly how miR-101-3p plays a role in CI/RI, more studies are needed for further exploration.
Our research implicates circ_0093708 in exosomes as a potential biomarker for ferroptosis after CI/RI. Growing evidence suggests that exosomal circRNAs can modulate ischemic stroke via ceRNAs network. Exosomal circBBS2 inhibits ferroptosis by targeting miR-494 to activate SLC7A11 signaling in ischemic stroke (40). The circOGDH is elevated in plasma exosomes of acute stroke patients and may mark the ischemic penumbra (41). Hypoxia-preconditioned adipose-derived stem cell exosomes deliver circRps5, promoting M2 microglial polarization and mitigating brain injury (42). circFUNDC1 knockdown alleviates OGD-induced endothelial injury by sponging miR-375 to inhibit PTEN (43). The circ_0000647 interference protects SK-N-SH cells by regulating the miR-126-5p/TRAF3 axis (44). Astrocyte-derived exosomal circSHOC2 reduces neuronal autophagy and apoptosis via the miR-7670-3p/SIRT1 pathway (45). The circRNAs has been increasingly investigated for its critical role in apoptosis, autophagy, angiogenesis, inflammation, oxidative stress, and blood–brain barrier after ischemic stroke by regulating target mRNAs and others, which have a promising future as diagnostic and prognostic biomarkers for ischemic stroke (46).
Our work also has several limitations. First, the binding interactions within the ceRNAs network requires further verification. We will conduct essential experimental validation (e.g., dual-luciferase reporter assay, RNA pull-down followed by qPCR for miR-101-3p, or biotinylated miRNA capture assay) to confirm the direct physical interaction between circ_0093708 and miR-101-3p. Second, circ_0093708 was analyzed bioinformatically but lacks experimental validation of its regulatory role. We plan to verified the effect of circ_0093708 knockdown or overexpression on miR-101-3p levels, DUSP1/PTGS2 expression, ferroptosis markers, and ultimately cell viability in the OGD/R model. Finally, more large-sample clinical data are needed to provide evidence for the clinical effect of circ_0093708. We will develop it in the subsequent work, fully explore the diagnostic value of circ_0093708 in ischemic stroke, and further combine it with the clinical outcomes of patients, in order to provide ideas for the treatment of ischemic stroke.
In summary, our study delineates the involvement of DUSP1 in ferroptosis after CI/RI and explores the circ_0093708/miR-101-3p/DUSP1/PTGS2 ceRNAs axis. Exosomal circ_0093708 holds promise as a diagnostic marker for ferroptosis after CI/RI.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, GSE16561; https://www.ncbi.nlm.nih.gov/, GSE195442; https://www.ncbi.nlm.nih.gov/, GSE110993; https://www.ncbi.nlm.nih.gov/, GSE22255.
Ethics statement
The animal study was approved by the Xi’an Jiaotong University Animal Ethics Review Committee (Approval number: XJTUAE2024-076). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SM: Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Visualization. XZ: Conceptualization, Data curation, Writing – original draft. JF: Investigation, Methodology, Writing – original draft. MC: Software, Writing – original draft. QY: Supervision, Writing – original draft. NZ: Formal analysis, Writing – original draft. KS: Resources, Visualization, Writing – original draft. SD: Project administration, Writing – review & editing. YC: Supervision, Validation, Writing – review & editing. HQ: Supervision, Validation, Writing – review & editing. MD: Visualization, Writing – review & editing. HY: Supervision, Validation, Writing – review & editing. TG: Visualization, Writing – review & editing. SZ: Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China [grant number 81070999], the Foundation of Shaanxi key research plan projects – General projects – Social development field [grant number 2022SF-380], the Foundation of Shaanxi health research project [grant number 2022A006], the Foundation of Shaanxi social development and technology research project [grant number 2016SF-020], the Foundation of Xi’an science and technology plan project [grant number 2019115113YX006SF039-2], the new medical technology of the Second Affiliated Hospital of Xi’an Jiaotong University [grant numbers 2019-32, 2018-16, 2010-22], the Fundamental Research Funds for the Central Universities of Xi’an Jiaotong University [grant numbers xjj2014153, 2009-95], and the Foundation of Second Affiliated Hospital of Xi’an Jiaotong University [grant number RC (GG)201109].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Katan, M, and Luft, A. Global burden of stroke. Semin Neurol. (2018) 38:208–11. doi: 10.1055/s-0038-1649503
2. Thomas, P, Jonika, T, and Farhaan, V. A contemporary review of epidemiology, risk factors, etiology, and outcomes of premature stroke. Curr Atheroscler Rep. (2022) 24:939–48. doi: 10.1007/s11883-022-01067-x
3. Benjamin, EJ, Virani, SS, Callaway, CW, Chamberlain, AM, Chang, AR, Cheng, S, et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. (2018) 137:e67–e492. doi: 10.1161/CIR.0000000000000558
4. Datta, A, Sarmah, D, Mounica, L, Kaur, H, Kesharwani, R, Verma, G, et al. Cell death pathways in ischemic stroke and targeted pharmacotherapy. Transl Stroke Res. (2020) 11:1185–202. doi: 10.1007/s12975-020-00806-z
5. Meng, Z, Qian, L, Hui, M, Hongxia, D, Liu, X, Wu, J, et al. Ischemia-reperfusion injury: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. (2024) 9:12. doi: 10.1038/s41392-023-01688-x
6. Dixon, SJ, Lemberg, KM, Lamprecht, MR, Skouta, R, Zaitsev, EM, Gleason, CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. (2012) 149:1060–72. doi: 10.1016/j.cell.2012.03.042
7. Schneider, A, and Simons, M. Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. (2013) 352:33–47. doi: 10.1007/s00441-012-1428-2
8. Yang, J, Hao, J, Lin, Y, Guo, Y, Liao, K, Yang, M, et al. Profile and functional prediction of plasma exosome-derived circRNAs from acute ischemic stroke patients. Front Genet. (2022) 13:810974. doi: 10.3389/fgene.2022.810974
9. Xiao, Q, Hou, R, Li, H, Zhang, S, Zhang, F, Zhu, X, et al. Circulating exosomal circRNAs contribute to potential diagnostic value of large artery atherosclerotic stroke. Front Immunol. (2021) 12:830018. doi: 10.3389/fimmu.2021.830018
10. Bar, TL, Conley, RY, Ding, J, Dillman, A, Warach, S, Singleton, A, et al. Genomic biomarkers and cellular pathways of ischemic stroke by RNA gene expression profiling. Neurology. (2010) 75:1009–14. doi: 10.1212/WNL.0b013e3181f2b37f
11. Tiedt, S, Prestel, M, Malik, R, Schieferdecker, N, Duering, M, Kautzky, V, et al. RNA-Seq identifies circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res. (2017) 121:970–80. doi: 10.1161/CIRCRESAHA.117.311572
12. Krug, T, Gabriel, JP, Taipa, R, Fonseca, BV, Domingues-Montanari, S, Fernandez-Cadenas, I, et al. TTC7B emerges as a novel risk factor for ischemic stroke through the convergence of several genome-wide approaches. J Cereb Blood Flow Metab. (2012) 32:1061–72. doi: 10.1038/jcbfm.2012.24
13. Tan, P, Chen, H, Huang, Z, Huang, M, Du, Y, Li, T, et al. MMP25-AS1/hsa-miR-10a-5p/SERPINE1 axis AS a novel prognostic biomarker associated with immune cell infiltration in KIRC. Mol Ther Oncolytics. (2021) 22:307–25. doi: 10.1016/j.omto.2021.07.008
14. Liu, M, Wang, Q, Shen, J, Yang, BB, and Ding, X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. (2019) 16:899–905. doi: 10.1080/15476286.2019.1600395
15. Bao, J, and Zhou, N. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. (2020) 2020:baaa021. doi: 10.1093/database/baaa021
16. Ashwal, S, Tone, B, Tian, HR, Cole, DJ, Liwnicz, BH, and Pearce, WJ. Core and penumbral nitric oxide synthase activity during cerebral ischemia and reperfusion in the rat pup. Pediatr Res. (1999) 46:390–400. doi: 10.1203/00006450-199910000-00006
17. Yang, WS, SriRamaratnam, R, Welsch, ME, Shimada, K, Skouta, R, Viswanathan, VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. (2014) 156:317–31. doi: 10.1016/j.cell.2013.12.010
18. Wu, X, Wang, B, Zhou, Y, Yang, Z, Jiang, L, Kou, Z, et al. NLRP3 inflammasome inhibitor MCC950 reduces cerebral ischemia/reperfusion induced neuronal ferroptosis. Neurosci Lett. (2023) 795:137032. doi: 10.1016/j.neulet.2022.137032
19. Xu, Y, Liu, Y, Li, K, Yuan, D, Yang, S, Zhou, L, et al. COX-2/PGE2 pathway inhibits the ferroptosis induced by cerebral ischemia reperfusion. Mol Neurobiol. (2022) 59:1619–31. doi: 10.1007/s12035-021-02706-1
20. Li, Y, Wang, J, Chen, S, Wu, P, Xu, S, Wang, C, et al. MiR-137 boosts the neuroprotective effect of endothelial progenitor cell-derived exosomes in oxyhemoglobin-treated SH-SY5Y cells partially via COX2/PGE2 pathway. Stem Cell Res Ther. (2020) 11:330. doi: 10.1186/s13287-020-01836-y
21. Yuan, K, Jin, X, Mo, X, Zeng, R, Zhang, X, Chen, Q, et al. Novel diagnostic biomarkers of oxidative stress, ferroptosis, immune infiltration characteristics and experimental validation in ischemic stroke. Aging. (2024) 16:746–61. doi: 10.18632/aging.205415
22. Hou, D, Hu, Y, Yun, T, Li, H, Yang, G, and Yu, D. The deubiquitinase OTUD3 stabilizes IRP2 expression to reduce hippocampal neuron ferroptosis via the p53/PTGS2 pathway to ameliorate cerebral ischemia–reperfusion injury. Eur J Med Res. (2024) 29:498. doi: 10.1186/s40001-024-02095-w
23. Ko, J, Jang, S, Kwon, W, Kim, S-Y, Jang, S, Kim, E, et al. Protective effect of GIP against monosodium glutamate-induced ferroptosis in mouse hippocampal HT-22 cells through the MAPK signaling pathway. Antioxidants. (2022) 11:189. doi: 10.3390/antiox11020189
24. Ruganzu, JB, Peng, X, He, Y, Wu, X, Zheng, Q, Ding, B, et al. Downregulation of TREM2 expression exacerbates neuroinflammatory responses through TLR4-mediated MAPK signaling pathway in a transgenic mouse model of Alzheimer’s disease. Mol Immunol. (2022) 142:22–36. doi: 10.1016/j.molimm.2021.12.018
25. Tabaa, MME, Aboalazm, HM, Shaalan, M, and Khedr, NF. Silymarin constrains diacetyl-prompted oxidative stress and neuroinflammation in rats: involvements of Dyn/GDNF and MAPK signaling pathway. Inflammopharmacology. (2022) 30:961–80. doi: 10.1007/s10787-022-00961-9
26. Liu, L, Doran, S, Xu, Y, Manwani, B, Ritzel, R, Benashski, S, et al. Inhibition of mitogen-activated protein kinase phosphatase-1 (MKP-1) increases experimental stroke injury. Exp Neurol. (2014) 261:404–11. doi: 10.1016/j.expneurol.2014.05.009
27. Koga, S, Kojima, S, Kishimoto, T, Kuwabara, S, and Yamaguchi, A. Over-expression of map kinase phosphatase-1 (MKP-1) suppresses neuronal death through regulating JNK signaling in hypoxia/re-oxygenation. Brain Res. (2012) 1436:137–46. doi: 10.1016/j.brainres.2011.12.004
28. Bu, F, Xu, L-S, Qin, X-D, Yang, T-Q, Zeng, J-H, Cai, H-B, et al. Overexpression of mitogen-activated protein kinase phosphatase-1 in endothelial cells reduces blood-brain barrier injury in a mouse model of ischemic stroke. Neural Regen Res. (2022) 18:1743–9. doi: 10.4103/1673-5374.363836
29. Xie, Y, Kuang, F, Liu, J, Tang, D, and Kang, R. DUSP1 blocks autophagy-dependent ferroptosis in pancreatic cancer. J Pancreatol. (2020) 3:154–60. doi: 10.1097/jp9.0000000000000054
30. Fan, J, Cao, S, Chen, M, Yao, Q, Zhang, X, Du, S, et al. Investigating the AC079305/DUSP1 axis as oxidative stress-related signatures and immune infiltration characteristics in ischemic stroke. Oxidative Med Cell Longev. (2022) 2022:8432352. doi: 10.1155/2022/8432352
31. Hocsak, E, Szabo, V, Kalman, N, Antus, C, Cseh, A, Sumegi, K, et al. PARP inhibition protects mitochondria and reduces ROS production via PARP-1-ATF4-MKP-1-MAPK retrograde pathway. Free Radic Biol Med. (2017) 108:770–84. doi: 10.1016/j.freeradbiomed.2017.04.018
32. Bhardwaj, U, and Singh, SK. Zika virus NS1 suppresses VE-cadherin and Claudin-5 via hsa-miR-101-3p in human brain microvascular endothelial cells. Mol Neurobiol. (2021) 58:6290–303. doi: 10.1007/s12035-021-02548-x
33. Tokarski, M, Cierzniak, A, and Baczynska, D. Role of hypoxia on microRNA-dependant regulation of HGFA – HGF – c-met signalling pathway in human progenitor and mature endothelial cells. Int J Biochem Cell Biol. (2022) 152:106310. doi: 10.1016/j.biocel.2022.106310
34. Yang, Z, Gao, Z, Yang, Z, Zhang, Y, Chen, H, Yang, X, et al. Lactobacillus plantarum-derived extracellular vesicles protect against ischemic brain injury via the microRNA-101a-3p/c-Fos/TGF-beta axis. Pharmacol Res. (2022) 182:106332. doi: 10.1016/j.phrs.2022.106332
35. Zhang, M, Liu, W, Zhang, Q, Hu, H, and Abdulhay, E. MiR-101-3p contributes to α-synuclein aggregation in neural cells through the miR-101-3p/SKP1/PLK2 pathway. J Healthc Eng. (2021) 2021:1–8. doi: 10.1155/2021/6147434
36. Chen, Q, Li, X, Kong, L, Xu, Q, Wang, Z, and Lv, Q. miR-101-3p induces vascular endothelial cell dysfunction by targeting tet methylcytosine dioxygenase 2. Acta Biochim Biophys Sin. (2020) 52:180–91. doi: 10.1093/abbs/gmz154
37. Zhang, M, Wang, J, Li, J, Kong, F, and Lin, S. miR-101-3p improves neuronal morphology and attenuates neuronal apoptosis in ischemic stroke in young mice by downregulating HDAC9. Transl Neurosci. (2023) 14:20220286. doi: 10.1515/tnsci-2022-0286
38. Xin, Y, Tang, L, Chen, J, Chen, D, Wen, W, and Han, F. Inhibition of miR-101-3p protects against sepsis-induced myocardial injury by inhibiting MAPK and NF-κB pathway activation via the upregulation of DUSP1. Int J Mol Med. (2021) 47:20. doi: 10.3892/ijmm.2021.4853
39. Liu, LP, Zhang, J, Pu, B, Li, WQ, and Wang, YS. Upregulation of JHDM1D-AS1 alleviates neuroinflammation and neuronal injury via targeting miR-101-3p-DUSP1 in spinal cord after brachial plexus injury. Int Immunopharmacol. (2020) 89:106962. doi: 10.1016/j.intimp.2020.106962
40. Hong, T, Zhao, T, He, W, Xia, J, Huang, Q, Yang, J, et al. Exosomal circBBS2 inhibits ferroptosis by targeting miR-494 to activate SLC7A11 signaling in ischemic stroke. FASEB J. (2023) 37:e23152. doi: 10.1096/fj.202300317RRR
41. Liu, Y, Li, Y, Zang, J, Zhang, T, Li, Y, Tan, Z, et al. CircOGDH is a penumbra biomarker and therapeutic target in acute ischemic stroke. Circ Res. (2022) 130:907–24. doi: 10.1161/circresaha.121.319412
42. Yang, H, Tu, Z, Yang, D, Hu, M, Zhou, L, Li, Q, et al. Exosomes from hypoxic pre-treated ADSCs attenuate acute ischemic stroke-induced brain injury via delivery of circ-Rps5 and promote M2 microglia/macrophage polarization. Neurosci Lett. (2022) 769:136389. doi: 10.1016/j.neulet.2021.136389
43. Bai, X, Liu, X, Wu, H, Feng, J, Chen, H, and Zhou, D. CircFUNDC1 knockdown alleviates oxygen-glucose deprivation-induced human brain microvascular endothelial cell injuries by inhibiting PTEN via miR-375. Neurosci Lett. (2022) 770:136381. doi: 10.1016/j.neulet.2021.136381
44. Dai, Y, Sheng, Y, Deng, Y, Wang, H, Zhao, Z, Yu, X, et al. Circ_0000647 promotes cell injury by modulating miR-126-5p/TRAF3 axis in oxygen-glucose deprivation and reperfusion-induced SK-N-SH cell model. Int Immunopharmacol. (2022) 104:108464. doi: 10.1016/j.intimp.2021.108464
45. Chen, W, Wang, H, Zhu, Z, Feng, J, and Chen, L. Exosome-shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 Axis. Mol Ther Nucleic Acids. (2020) 22:657–72. doi: 10.1016/j.omtn.2020.09.027
Keywords: exosomes, cerebral ischemia–reperfusion injury, ferroptosis, DUSP1, circular RNA
Citation: Ma S, Zhang X, Fan J, Chen M, Yao Q, Zhang N, Shi K, Du S, Cheng Y, Qu H, Duan M, Yang H, Gao T and Zhan S (2025) Exosomal circ_0093708 as a potential ferroptosis biomarker in cerebral ischemia–reperfusion injury. Front. Neurol. 16:1633393. doi: 10.3389/fneur.2025.1633393
Edited by:
Shuling Liu, Tianjin Medical University, ChinaReviewed by:
Sharifah Alawieyah Syed Mortadza, Putra Malaysia University, MalaysiaWeili Li, Shandong Academy of Medical Sciences (SDAMS), China
Copyright © 2025 Ma, Zhang, Fan, Chen, Yao, Zhang, Shi, Du, Cheng, Qu, Duan, Yang, Gao and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuqin Zhan, c3F6aGFuQG1haWwueGp0dS5lZHUuY24=
 Shuyin Ma
Shuyin Ma Xiaodong Zhang
Xiaodong Zhang Jiaxin Fan
Jiaxin Fan Nan Zhang
Nan Zhang Han Yang
Han Yang Shuqin Zhan
Shuqin Zhan