- Department of Respiratory Diseases, Qilu Hospital of Shandong University, Jinan, China
Lung adenocarcinoma (LUAD) is one of the most lethal malignancies, posing a threat to human health. However, the molecular mechanisms underlying LUAD development remain largely unknown. In this study, we found that miR-1-3p was significantly downregulated in human LUAD tissues and cell lines and played an inhibitory role in LUAD cell tumorigenesis, as evidenced by the significantly reduced viability, migration, and invasion of LUAD cells in response to miR-1-3p overexpression. Mechanistically, microRNA (miR)-1-3p physically interacted with the 3′-untranslated region (UTR) of protein regulator of cytokinesis 1 (PRC1) mRNA, leading to downregulation of PRC1. Overexpression of PRC1 reversed the inhibitory effects of miR-1-3p on LUAD cell tumorigenesis, suggesting that the miR-1-3p/PRC1 axis is majorly involved in suppressing LUAD development and progression. Consistently, PRC1 was dramatically induced in LUAD tissues and cell lines as well as associated with a poor prognosis in LUAD patients. Taken together, our study identified the miR-1-3p/PRC1 axis as an important regulatory mechanism contributing to LUAD inhibition and provided valuable clues for the future development of therapeutic strategies against LUAD.
Introduction
Lung adenocarcinoma (LUAD) is the most common subtype of non-small cell lung cancer (NSCLC), accounting for 80–85% of all lung cancers; worldwide, approximately 40% of all lung cancer patients are diagnosed with LUAD (1). Compared to other subtypes of NSCLC, LUAD has a higher incidence and a shorter survival time among patients, with a 5-year survival rate as low as 10–15% (2, 3); thus, LUAD poses a serious threat to human health. Currently, chemotherapy is a relatively effective therapeutic option for NSCLC (4). However, the existence and development of intrinsic or acquired chemoresistance greatly limit the application of chemotherapy in cancer treatment. Therefore, there is still an urgent need to develop novel therapeutic strategies against LUAD that are based on the mechanisms underlying the development and progression of LUAD.
MicroRNAs (miRNAs) are a class of small endogenous non-coding RNA molecules (~22 nucleotides) found in animals and plants that are responsible for the degradation or translation repression of mRNAs by binding to the 3′-untranslated region (UTR) of target mRNAs (5). A variety of miRNAs have been identified as novel biomarkers or promising therapeutic targets of human malignant tumors. Among them, miR-1-3p plays an antitumor role in multiple cancer types, including rhabdomyosarcoma as well as lung, thyroid, prostatic, bladder, colorectal, and hepatocellular carcinomas (6–11). However, the role of miR-1-3p in LUAD has not yet been investigated. In addition, the association between miR-1-3p and its target genes may deepen our understanding of the molecular mechanisms contributing to LUAD development, thus facilitating the discovery of improved therapies for LUAD.
The microtubule-associated protein regulator of cytokinesis 1 (PRC1) has been found to be majorly involved in the organization of antiparallel microtubules in the central spindle during cytokinesis. The human PRC1 gene, located on chromosome 15q26.1, encodes a 620-amino-acid protein with a molecular weight of 71 kDa (3). An abnormally high expression of PRC1 has been observed in breast cancer (12), bladder cancer (13), hepatocellular carcinoma (14), and pancreatic cancer (15), which suggests a promotive role of PRC1 in tumorigenesis. However, it remains largely unknown whether there is a functional association between miR-1-3p and the regulation of PRC1 in LUAD. In this study, we examined the role of miR-1-3p in LUAD growth and metastasis as well as the underlying mechanism.
Materials and Methods
Tissue Samples From LUAD Patients
Human LUAD tissues were collected from LUAD patients undergoing pulmonary resection or bronchoscopy biopsy, and normal tissues adjacent to cancer were collected from LUAD patients undergoing pulmonary resection at Qilu Hospital between May and September 2018. None of the patients had received chemotherapy or radiotherapy prior to surgery. All the fresh samples were stored in RNAlater Stabilization Solution (Ambion) at −80°C until use. This study was approved by the Ethics Committee of Shandong University, and written informed consent was obtained from all patients prior to enrollment in the present study. The clinicopathological characteristics of the patients are shown in Supplementary Table 1.
Cell Culture
Three LUAD cell lines (A549, H1299, and H1975 cells) and a human alveolar epithelial cell line (HPAEpiC) were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in RPMI 1640 medium (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified atmosphere of 5% CO2 at 37°C. Cells in the exponential phase of growth were used for the following experiments.
Construction of the miR-1-3p Overexpression Cell Lines
The pre-miR-1-3p sequences were synthesized by Biosune Biotechnology Company (Shanghai, China) and cloned into the lentiviral vector pGIPZ. Lentivirus was produced in HEK293T cells using the packaging vectors psPAX2 and pMD2.G. The cells were infected with lentivirus for 24 h and then cultured for 1 week in medium containing 2 μg/mL puromycin (Merck Millipore, USA) for screening to acquire cells with stable expression of miR-1-3p.
Transient Transfection
The miR-1-3p mimic and its negative control (NC) were chemically synthesized by GenePharma Co., Ltd. (Shanghai, China). The cells were transiently transfected with 50 nM miR-1-3p mimic or 50 nM NC (Boshang, Inc., China) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The cells were harvested at 24 or 48 h after the transfection. The NC was a scrambled oligonucleotide that does not encode any known miRNA. The transfection efficiency was confirmed by detecting the miR-1-3p expression level using the SYBR green (Takara)-based real-time quantitative polymerase chain reaction (qPCR) system.
RNA Isolation and qPCR
Total RNA was extracted from the cells using Trizol reagents (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The cDNA of miRNA was synthesized with the One Step PrimeScript miRNA cDNA Synthesis Kit (Takara Biotechnology, Co., Ltd., Dalian, China). qPCR was performed using the SYBR green Premix Ex Taq II (Takara Biotechnology, Co., Ltd.) with the Step One Plus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression of U6 was used as an internal control. The primers for miR-1-3p are indicated in Supplementary Table 2.
Western Blot Analysis
The cells were lysed in ice-cold RIPA lysis buffer, and the cell lysates were obtained by centrifugation at 12,000 rpm and 4°C for 10 min. The protein concentration was determined using the bicinchoninic acid method. The protein samples (5–10 μg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transfer to polyvinylidene difluoride membranes, and then immunoblotted with the indicated antibodies. After blocking with 5% nonfat milk, the membranes were incubated with the respective primary antibody overnight at 4°C, followed by incubation with the horseradish peroxidase-coupled secondary antibody for 1 h at room temperature. The protein bands were visualized using enhanced chemiluminescence reagents (PerkinElmer) with an ImageQuant LAS 4000 system (GE Healthcare Life Sciences). The following antibodies were used: anti-PRC1, anti-fibronectin, anti-N-cadherin, anti-vimentin, and anti-β-actin (Cell Signaling Technology).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
The cells were seeded into 96-well plates at a density of 2,000 cells/well and grown for 5 days. After the addition of 100 μL of 5 mg/mL MTT solution, the cells were incubated for an additional 4 h at 37°C, and then the supernatant was removed and dissolved in 100 μL of dimethyl sulfoxide (Sigma-Aldrich). Cell viability was assessed on the 1st, 2nd, 3rd, 4th, and 5th day. The absorbance of each well was measured in triplicate using an iMark Microplate Absorbance Reader (Bio-Rad).
Luciferase Reporter Assay
Luciferase reporter constructs containing wild-type (WT) or mutant PRC1 3′-UTR (pmirGLO-PRC1-WT or pmirGLO-PRC1-mut, respectively) were generated by GenePharma Inc. (Shanghai, China). The cells were cotransfected with 25 ng of PRC1 3′-UTR reporter constructs and 20 nM miR-1-3p mimic using Lipofectamine 2,000 (Invitrogen) in 24-well plates. At 24 h after transfection, luciferase assays were performed using the Dual-Luciferase reporter assay system (Promega). Renilla luciferase activity was used to normalize the luciferase activity of the PRC1 3′-UTR reporter constructs.
In vivo Tumorigenicity Assays
Four-week-old male BALB/c nude mice were purchased from the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China). The mice were randomly divided into two groups and injected subcutaneously with A549 cells (2 × 106 cells/mouse, n = 5 mice/group) that were infected with either control lentivirus or miR-1-3p-overexpressing lentivirus. Tumor growth was monitored by measuring the tumor diameter. Tumor volume was calculated according to the formula TV (cm3) = a × b2 × π/6, where a is the longest diameter and b is the shortest diameter. The mice were sacrificed after 3 weeks, and then the tumors were excised and weighed. All animal experiments were approved by the Shandong University Animal Care and Use Committee.
Bioinformatics Analyses
PRC1 genetic alterations and copy number variation in LUAD were retrieved from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/) (16, 17). The Cancer Genome Atlas RNA expression data of LUAD tissues were processed and analyzed by the Cancer Genomics Browser (https://xena.ucsc.edu/welcome-to-ucsc-xena/) (18). The PRC1 expression levels and copy number variation were analyzed by Proteinatlas (https://www.proteinatlas.org/), Oncomine (www.oncomine.org) (19), and Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) in LUAD and normal lung tissues via immunohistochemistry. Kaplan–Meier plots (http://kmplot.com/analysis/) (20) were used to analyze the overall survival of the LUAD patients.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Data are expressed as the mean ± standard deviation. Comparison between two groups was performed using the Student's t-test or the Mann-Whitney U-test. The correlation between the expression levels of miR-1 and PRC1 was analyzed using Pearson's correlation analysis. LUAD tissues with lower miR-1 and PRC1 expression than the median expression were assigned to the low-expression group, whereas those with higher miR-1 and PRC1 expression than the median expression were assigned to the high-expression group. Associations between the clinicopathological features and the expression levels of miR-1 and PRC1 were analyzed using the χ2 test. Overall survival curves were determined according to the Kaplan–Meier method. A p < 0.05 was considered statistically significant.
Results
MiR-1-3p Is Downregulated in Human LUAD Tissues and Cell Lines
To investigate the possible role of miR-1-3p in LUAD development, we first examined the expression levels of miR-1-3p in human LUAD tissues and cell lines. As shown in Figure 1A, miR-1-3p expression was significantly decreased in LUAD tissues, compared with the matched adjacent normal lung tissues. Similarly, marked downregulation of miR-1-3p was also observed in the human LUAD cell lines A549, H1299, and H1975, compared with the normal HPAEpiCs (Figure 1B). These in vivo and in vitro results suggest that miR-1-3p may play an inhibitory role in LUAD development.
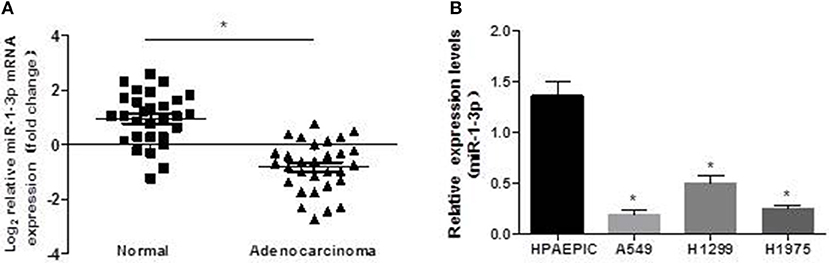
Figure 1. Expression pattern of miR-1-3p in human LUAD tissues and cell lines. qPCR was performed to determine the expression levels of miR-1-3p in human LUAD tissues (A) and cell lines (B), as indicated. *P < 0.05 in (A) (n = 30); *P < 0.05 vs. HPAEpiCs in (B) (n = 3).
Overexpression of miR-1-3p Suppresses LUAD Cell Viability in vitro
Next, we sought to investigate whether miR-1-3p indeed plays a role in suppressing LUAD development using a gain-of-function assay. As shown in Figure 2A, transfection of the miR-1-3p mimic led to a dramatic increase in miR-1-3p expression in the LUAD cell lines A549, H1299, and H1975. Importantly, overexpression of miR-1-3p significantly inhibited the viability of these three cell lines in a time-dependent manner, compared with the NC groups (Figures 2B–D). These results demonstrate that miR-1-3p is sufficient to suppress LUAD cell growth in vitro.
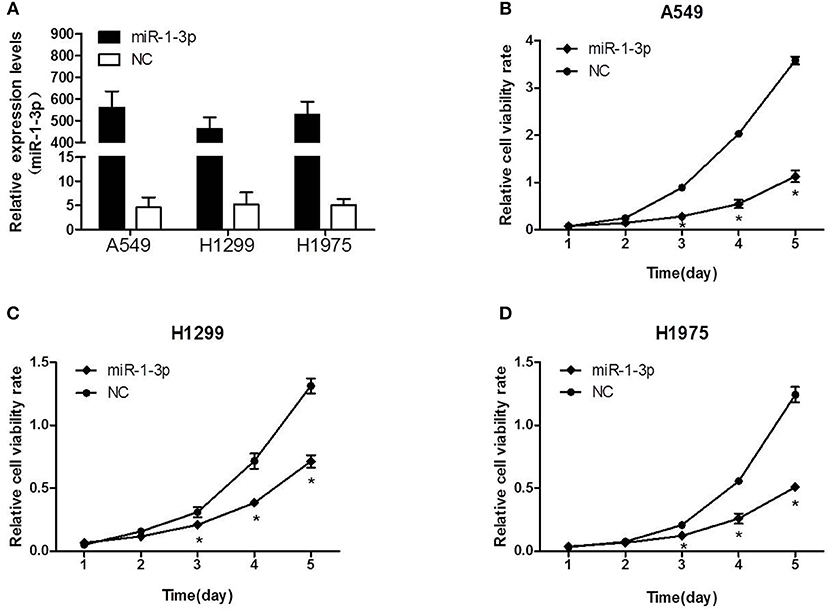
Figure 2. The effect of miR-1-3p overexpression on LUAD cell viability. (A) qPCR was performed to validate the overexpression efficiency of miR-1-3p in A549, H1299, and H1975 cells. (B–D) The MTT assay was performed to measure the viability of miR-1-3p-overexpressing A549, H1299, and H1975 cells. *P < 0.05 vs. the corresponding negative control (NC) groups (n = 3).
Overexpression of miR-1-3p Inhibits LUAD Cell Migration and Invasion in vitro
To further investigate whether miR-1-3p inhibits LUAD progression, Transwell assays were performed to examine the effects of miR-1-3p overexpression on LUAD cell migration and invasion. As shown in Figures 3A,B, overexpression of miR-1-3p resulted in a significant decrease in LUAD cell migration and invasion abilities, compared with the NC groups. Consistently, overexpression of miR-1-3p markedly suppressed epithelial-mesenchymal transition (EMT), a process contributing to tumor metastasis, as evidenced by downregulation of the mesenchymal markers fibronectin, N-cadherin, and vimentin (Figures 3C,D). These data suggest that miR-1-3p overexpression may suppress LUAD progression through reducing LUAD cell migration and invasion as well as inhibiting EMT in these cells.
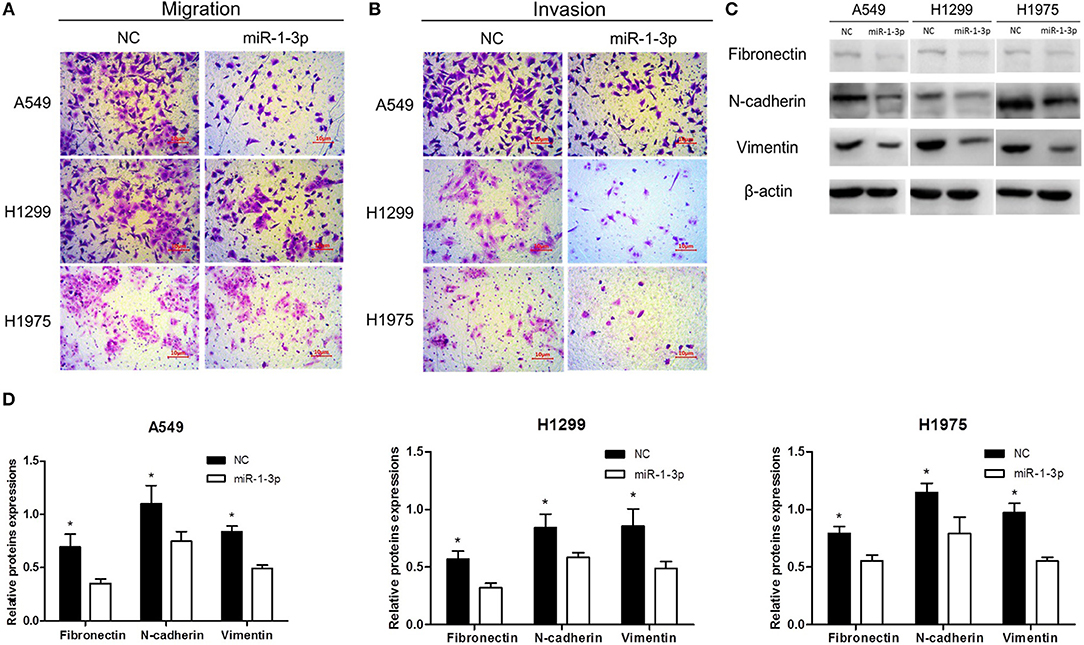
Figure 3. The effect of miR-1-3p overexpression on LUAD cell migration and invasion as well as epithelial-mesenchymal transition. Transwell assays for migration (A) and invasion (B) of miR-1-3p-overexpressing LUAD cells. (C) Western blot assay for the expression of the indicated mesenchymal markers. β-Actin was used as an internal control. (D) Quantification of the western blot assay results shown in (C). *P < 0.05 vs. miR-1-3p groups in (C) (n = 3).
miR-1-3p Inhibits Xenograft Tumor Growth of LUAD Cells
To further explore whether miR-1-3p overexpression could suppress LUAD growth in vivo, human LUAD A549 cells with and without miR-1-3p overexpression were subcutaneously inoculated into nude mice. At 1 week after inoculation, all mice had developed detectable tumors. However, at 3 weeks after inoculation, the mice bearing tumors with miR-1-3p overexpression demonstrated a dramatic decrease in the tumor size and weight (Figures 4A–C), compared to the control groups. These results show that overexpression of miR-1-3p inhibits tumorigenesis in vivo.
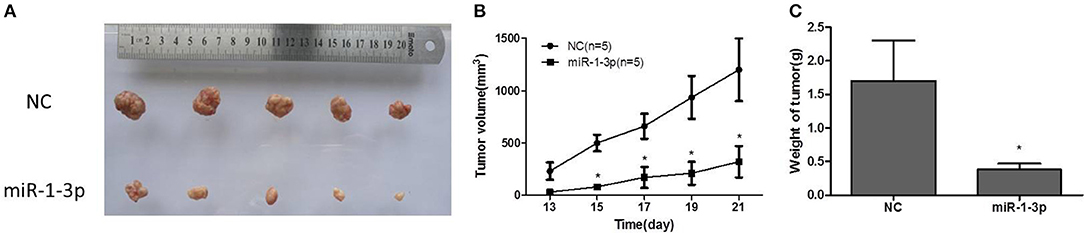
Figure 4. The effect of miR-1-3p overexpression on tumorigenesis in vivo. (A) Overexpression of miR-1-3p reduced LUAD cell-derived tumor growth in a xenograft model. (B) Summary of the tumor growth data; the error bars indicate the standard deviation. (C) miR-1-3p overexpression results in a decline of tumor weight. *P < 0.05.
PRC1 Is a Direct Target Gene of miR-1-3p
To further examine the mechanism underlying miR-1-3p-mediated suppression of LUAD development and progression, we employed the TargetScan computational algorithm to predict the target genes of miR-1-3p (21). The results indicated complementary base-pairing between miR-1-3p and the 3′-UTR of PRC1 (Figure 5A), suggesting that PRC1 may be a target gene of miR-1-3p. To verify this finding, we detected the expression of PRC1 in LUAD cells. As shown in Figures 5B,C, the protein expression of PRC1 was markedly induced in LUAD cells, compared with normal HPAEpiCs, consistent with the expression pattern of miR-1-3p in LUAD tissues and cells. Importantly, miR-1-3p overexpression led to downregulation of PRC1 in LUAD cells (Figures 5D,E), confirming that PRC1 is a downstream target of miR-1-3. To determine whether PRC1 is directly targeted by miR-1-3p, we performed a mutation assay through introducing a PRC1 3′-UTR mutation in the pmirGLO vector. The results demonstrated that the PRC1 3′-UTR mutation had no significant effect on the luciferase activity in miR-1-3p-transfected LUAD cells, compared with that in the NC-transfected cells (Figure 5F), suggesting that WT PRC1 3′-UTR is essential for the function of miR-1-3p. Taken together, our data show that PRC1 is a direct downstream target gene of miR-1-3p.
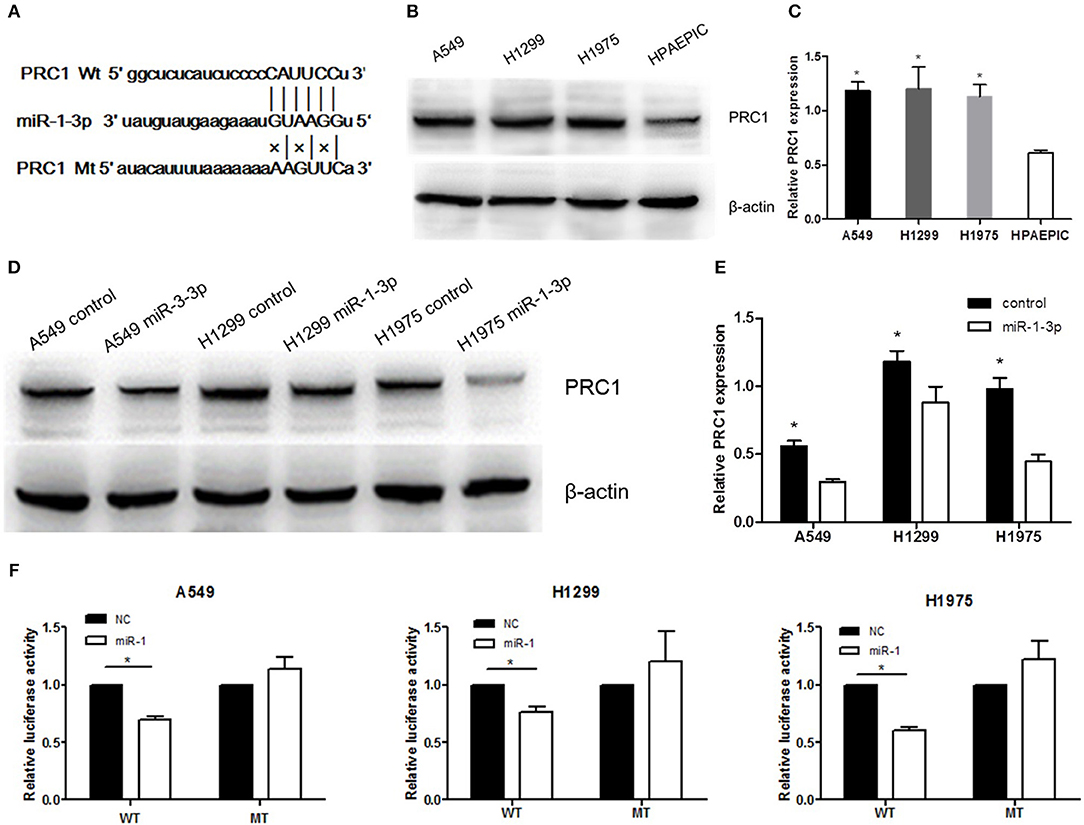
Figure 5. PRC1 is a direct target of miR-1-3p in LUAD cells. (A) The putative miR-1-3p-binding sites in PRC1 3′-UTR and the mutated binding sites are shown. (B,D) Western blot analysis of PRC1 expression in the nontransfected negative control (NC)- or miR-1-3p-transfected LUAD cells. β-Actin was used as an internal control. (C) Quantification of the western blot assay results shown in (B). *P < 0.05 vs. HPAEpiCs (n = 3). (E) Quantification of the western blot assay results shown in (D). *P < 0.05 vs. the miR-1-3p groups (n = 3). (F) Luciferase reporter assay for LUAD cells transfected with wild-type (WT) or mutated (MT) pGL3-3′-UTR. The luciferase activity was normalized to Renilla luciferase activity. *P < 0.05 (n = 3).
PRC1 Is Induced in LUAD Tissues and Cell Lines
To determine whether PRC1 contributes to LUAD development and progression, we examined the expression profile of PRC1 in LUAD tissues using the publicly accessible database Oncomine. As shown in Figure 6A, the mRNA expression levels of PRC1 were significantly enhanced in the LUAD tissues, compared with the normal lung tissues. In addition, we also analyzed the mRNA expression of PRC1 in LUAD tissues using two microarray datasets from the Hou and Selamat lung cancer groups, which were downloaded from Oncomine. The results demonstrated that the mRNA expression of PRC1 was significantly induced in the LUAD tissues of these groups and positively correlated with the tumor, lymph node, metastasis (TNM) staging of LUAD (Figure 6B). These findings were further confirmed by our mRNA expression data of PRC1 and the immunohistochemical staining of PRC1 in human LUAD tissues and matched adjacent normal lung tissues (Figures 6C,E). For the in vitro study, the mRNA levels of PRC1 were dramatically increased in the LUAD cell lines A549, H1299, and H1975, compared to those in normal HPAEpiCs (Figure 6D). Collectively, these results suggest that PRC1 may be involved in LUAD development and progression.
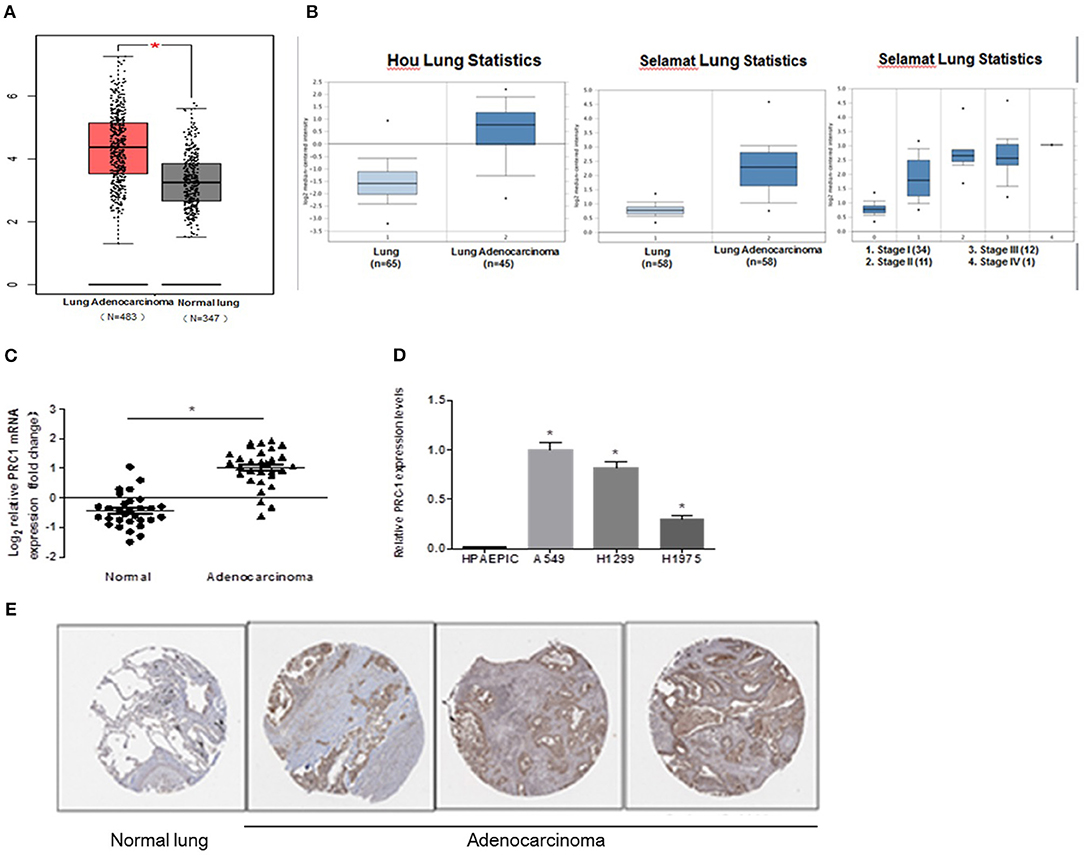
Figure 6. Expression pattern of PRC1 in LUAD tissues and cell lines. (A) The data of copy number variation in LUAD from The Cancer Genome Atlas cohort. (B) The mRNA expression of PRC1 in different TNM staging groups in Oncomine. (C) qPCR analysis of PRC1 expression in LUAD tissues (n = 30). (D) qPCR analysis of PRC1 expression in LUAD cells and HPAEpiCs. (E) Representative images of the immunohistochemical staining of PRC1 from The Human Protein Atlas in LUAD and normal lung tissues. *P < 0.05 in (C) (n = 30); *P < 0.05 vs. HPAEpiCs in (D) (n = 3).
Correlation of miR-1-3p/PRC1 and Clinicopathological Characteristics of LUAD Patients
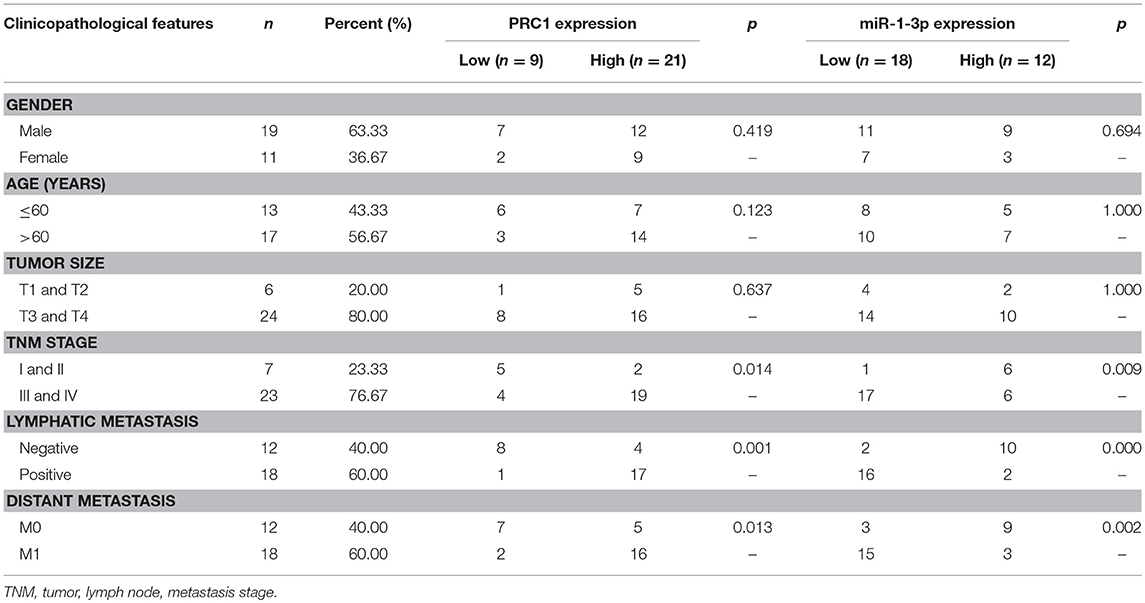
To investigate whether the miR-1-3p/PRC1 axis plays a role in LUAD development, we first analyzed the association between the miR-1-3p and PRC1 levels in LUAD tissues. The results revealed that the miR-1-3p levels were negatively correlated with the PRC1 mRNA expression (r = −0.5858; P < 0.01; Figure 7) in the LUAD tissues. Importantly, low levels of miR-1-3p and high levels of PRC1 were strongly associated with the TNM stage, lymph node metastasis, and distant metastasis (Table 1). These data suggest that LUAD development may be at least partially attributable to the miR-1-3p/PRC1 axis.
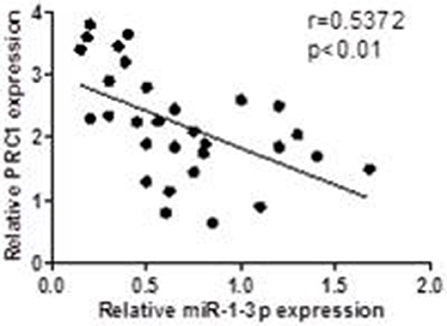
Figure 7. Association between the miR-1-3p and PRC-1 levels in LUAD patients. The PRC1 and miR-1 levels were determined by qPCR in LUAD tissues and matched adjacent normal tissues (n = 30).
miR-1-3p Inhibits LUAD Cell Metastasis via PRC1
To determine whether miR-1-3p-mediated suppression of PRC1 expression is a major mechanism inhibiting LUAD development and progression, we cotransfected LUAD cells with miR-1-3p and a PRC1-overexpression plasmid for Transwell assays. The plasmid transfection efficacy of miR-1-3p and PRC1 was validated in Figures 8A,B. We found that miR-1-3p significantly inhibited the migration and invasion of three LUAD cell lines and that the inhibitory effects of miR-1-3p were markedly reversed by PRC1 overexpression (Figures 8C–E), suggesting that miR-1-3p inhibits LUAD cell metastasis in a PRC1-depedent manner and the miR-1-3p/PRC1 axis is majorly involved in LUAD development and progression.
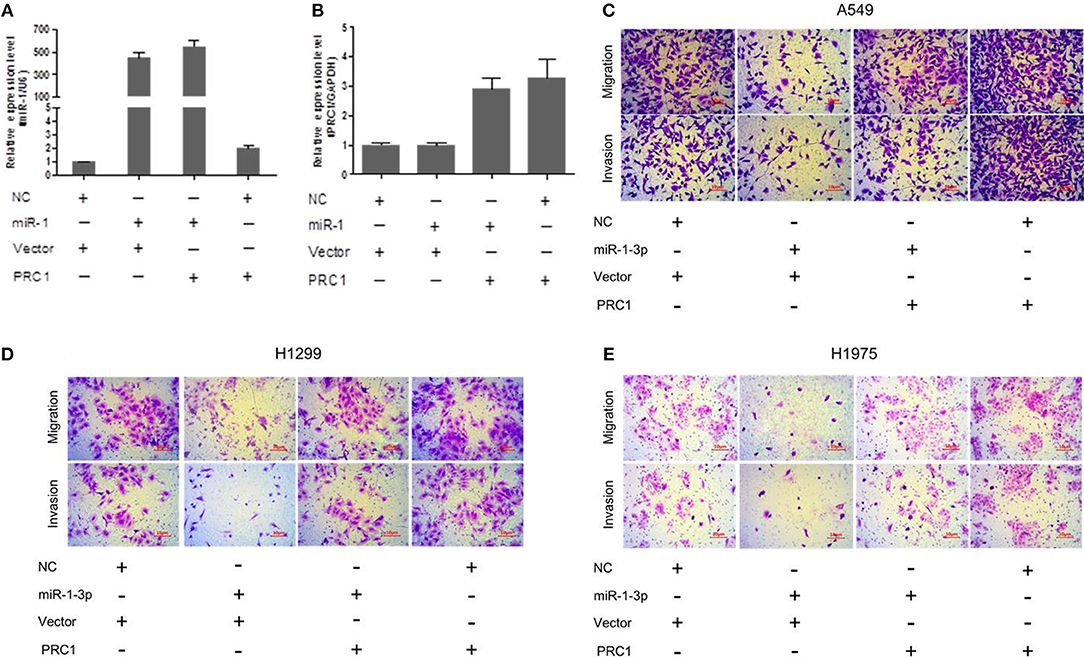
Figure 8. miR-1-3p inhibits LUAD cell migration and invasion via PRC1. (A,B) qPCR analysis of the transfection efficiency of the miR-1-3p mimic and the PRC1-overexpression plasmid in LUAD cells. (C–E) Transwell assays for migration and invasion of LUAD cells transfected with the miR-1-3p mimic and/or the PRC1-overexpression plasmid.
Overexpression of PRC1 Is Associated With a Poor Prognosis in LUAD Patients
MiR-1-3p functions through suppressing PRC1 expression, suggesting a promotive role of PRC1 in LUAD development. To confirm this, we examined the prognostic effect of PRC1 in LUAD patients from a public database by performing Kaplan–Meier analysis (http://www.kmplot.com). The results showed that the LUAD patients with a higher mRNA expression of PRC1 had shorter overall and disease-free survival times than those with a lower mRNA expression of PRC1 (Figures 9A,B). These data suggest that PRC1 overexpression may serve as a biomarker of a poor prognosis for LUAD patients, further supporting our findings that miR-1-3p plays a key role in inhibiting LUAD development through targeting PRC1.
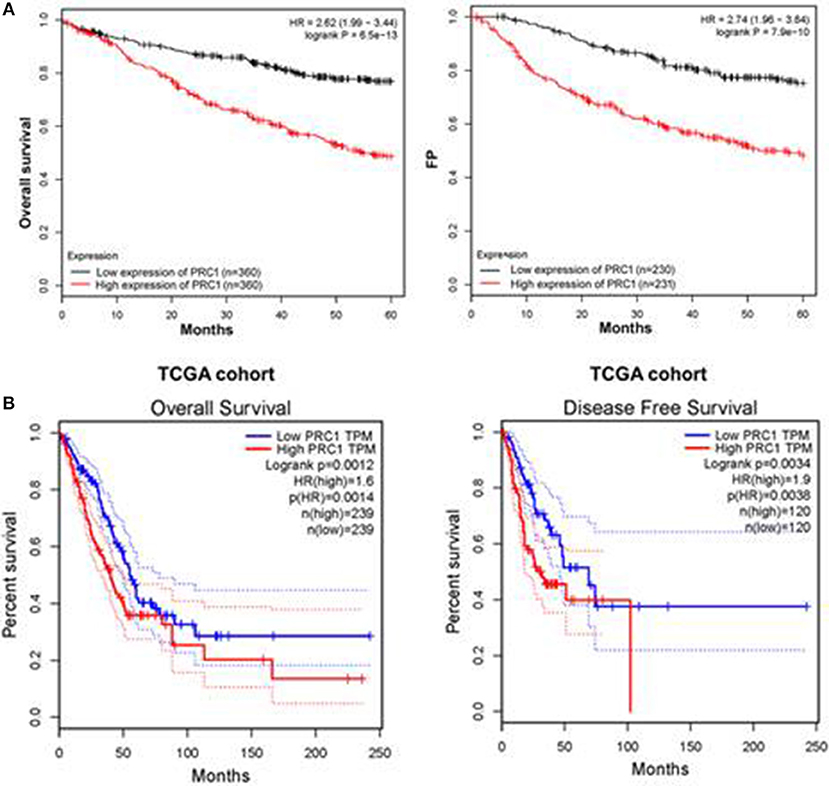
Figure 9. Overexpression of PRC1 is associated with a poor prognosis in LUAD patients. (A) The prognostic effect of PRC1 in LUAD patients was evaluated using Kaplan–Meier plots. (B) The overall and disease-free survival rates in LUAD patients with different PRC1 expression patterns were evaluated for The Cancer Genome Atlas (TCGA) cohort.
Discussion
Carcinogenesis of LUAD is a complex and multistage process involving the regulation of a wide range of genes by miRNAs (22–24). Among them, miR-1-3p, a muscle-specific miRNA, has been shown to play a key role in skeletal muscle differentiation and have inhibitory effects on the growth, migration, and invasion of LUAD (25). The present study revealed that miR-1-3p was significantly downregulated in LUAD tissues and cells. Lower levels of miR-1-3p were strongly associated with a higher TNM stage, earlier lymph node metastasis, and more distant metastasis. Therefore, miR-1-3p is suggested as a tumor suppressor in LUAD. The detection of miR-1-3p expression may be a valuable tool to evaluate the invasion and metastasis of LUAD.
There are hundreds of possible target genes of miR-1-3p, among which PRC1 is a critical protein in cytokinesis and characterized as a mitotic spindle-associated cyclin-dependent kinase substrate (26). Previous studies have provided evidence that PRC1 is involved in different types of cancer (27, 28). Loss of PRC1 leads to the accumulation of bi- and multi-nucleated cells in lung cancer, which further supports its role as the major central spindle organizer in cytokinesis (29). In view of our findings that miR-1-3p overexpression inhibits LUAD cell viability, there is a possibility that the function of PRC1 in apoptosis and senescence is due to induction of miR-1-3p. In this study, we demonstrated that the function of miR-1-3p could be suppressed by dysregulated expression of PRC1. In accordance with the above-mentioned studies, we confirmed that the overexpression of PRC1 significantly promoted the viability, invasion, and migration of LUAD cells. A higher PRC1 expression was also related to a worse outcome in patients with LUAD. Because Wnt/β-catenin signaling is dysregulated in lung cancer (30) and the overexpression of Wnt proteins (Wnt1 and Wnt5a) is significantly associated with adverse outcomes in lung cancer patients (31), we speculate that the miR-1-3p/PRC1 axis participates in dysregulation of Wnt/β-catenin signaling in LUAD development (32); however, this hypothesis requires further investigation. Although our study demonstrated that the miR-1-3p/PRC1 axis is a major mechanism underlying LUAD development, we do not exclude the possibility that other miRNAs or protein regulators besides miR-1-3p/PRC1 are also involved in LUAD pathogenesis. Therefore, more research is needed.
In summary, we identified miR-1-3p as a novel regulator of PRC1 in LUAD. A high PRC1 expression correlates with a poor prognosis in LUAD patients. Thus, targeting miR-1-3p/PRC1 may be a potential therapeutic intervention for the treatment of LUAD.
Ethics Statement
The Ethics Committee of Qilu Hospital at Shandong University approved this study [KYLL-2018 (KS)-156]. All participants in this study provided informed consent.
Author Contributions
TL designed the questionnaire and drafted the manuscript. XW did the statistical analysis. TL and LJ did the relationship analysis and collected the data. YL conceived the study, supervised and reviewed the entire study, and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00120/full#supplementary-material
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
2. Zhang W, Fan J, Chen Q, Lei C, Qiao B, Liu Q. SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma. Oncol Lett. (2018) 15:7028–36. doi: 10.3892/ol.2018.8235
3. Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, et al. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. (1998) 2:877–85. doi: 10.1016/S1097-2765(00)80302-0
4. Peters S, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2012) 23(Suppl. 7):vii56–64. doi: 10.1093/annonc/mds226
5. Leva GD, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol. (2013) 3:153. doi: 10.3389/fonc.2013.00153
6. Han C, Yu Z, Duan Z, Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res Int. (2014) 2014:428371. doi: 10.1155/2014/428371
7. Li J, Guan J, Long X, Wang Y, Xiang X. mir-1-mediated paracrine effect of cancer-associated fibroblasts on lung cancer cell proliferation and chemoresistance. Oncol Rep. (2016) 35:3523–31. doi: 10.3892/or.2016.4714
8. Li SM, Wu HL, Yu X, Tang K, Wang SG, Ye ZQ, et al. The putative tumour suppressor miR-1–3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J Exp Clin Cancer Res. (2018) 37:219. doi: 10.1186/s13046-018-0895-z
9. Diniz GP, Lino CA, Moreno CR, Senger N, Barreto-Chaves MLM. MicroRNA-1 overexpression blunts cardiomyocyte hypertrophy elicited by thyroid hormone. J Cell Physiol. (2017) 232:3360–8. doi: 10.1002/jcp.25781
10. Gao L, Yan P, Guo FF, Liu HJ, Zhao ZF. MiR-1–3p inhibits cell proliferation and invasion by regulating BDNF-TrkB signaling pathway in bladder cancer. Neoplasma. (2018) 65:89–96. doi: 10.4149/neo_2018_161128N594
11. Zhu D, Sun Y, Zhang D, Dong M, Jiang G, Zhang X, et al. miR1 inhibits the progression of colon cancer by regulating the expression of vascular endothelial growth factor. Oncol Rep. (2018) 40:589–98. doi: 10.3892/or.2018.6463
12. Shimo A, Nishidate T, Ohta T, Fukuda M, Nakamura Y, Katagiri T. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci. (2007) 98:174–81.doi: 10.1111/j.1349-7006.2006.00381.x
13. Kanehira M, Katagiri T, Shimo A, Takata R, Shuin T, Miki T, et al. Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. (2007) 67:3276–85. doi: 10.1158/0008-5472.CAN-06-3748
14. Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/beta-catenin signalling pathway. Gut. (2016) 65:1522–34. doi: 10.1136/gutjnl-2015-310625
15. Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. (2004) 23:2385–400. doi: 10.1038/sj.onc.1207392
16. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6:pl1. doi: 10.1126/scisignal.2004088
17. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
18. Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, et al. The UCSC cancer genomics browser. Nat Methods. (2009) 6:239–40. doi: 10.1038/nmeth0409-239
19. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. (2004) 6:1–6. doi: 10.1016/S1476-5586(04)80047-2
20. Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. (2013) 8:e82241. doi: 10.1371/journal.pone.0082241
21. Jiao D, Chen J, Li Y, Tang X, Wang J, Xu W, et al. miR-1–3p and miR-206 sensitizes HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signalling and EMT. J Cell Mol Med. (2018) 22:3526–36. doi: 10.1111/jcmm.13629
22. Nadal E, Chen G, Gallegos M, Lin L, Ferrer-Torres D, Truini A, et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res. (2013) 19:6842–52. doi: 10.1158/1078-0432.CCR-13-0736
23. Yu QQ, Wu H, Huang X, Shen H, Shu YQ, Zhang B, et al. MiR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells. Biomed Pharmacother. (2014) 68:155–61. doi: 10.1016/j.biopha.2014.01.005
24. Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. (2013) 123:2921–34. doi: 10.1172/JCI66353
25. Chiu KL, Lin YS, Kuo TT, Lo CC, Huang YK, Chang HF, et al. ADAM9 enhances CDCP1 by inhibiting miR-1 through EGFR signaling activation in lung cancer metastasis. Oncotarget. (2017) 8:47365–78. doi: 10.18632/oncotarget.17648
26. Zhan P, Zhang B, Xi GM, Wu Y, Liu HB, Liu YF, et al. PRC1 contributes to tumorigenesis of lung adenocarcinoma in association with the Wnt/beta-catenin signaling pathway. Mol Cancer. (2017) 16:108. doi: 10.1186/s12943-017-0682-z
27. Brynychova V, Ehrlichova M, Hlavac V, Nemcova-Furstova V, Pecha V, Leva J, et al. Genetic and functional analyses do not explain the association of high PRC1 expression with poor survival of breast carcinoma patients. Biomed Pharmacother. (2016) 83:857–64. doi: 10.1016/j.biopha.2016.07.047
28. Hanselmann S, Wolter P, Malkmus J, Gaubatz S. The microtubule-associated protein PRC1 is a potential therapeutic target for lung cancer. Oncotarget. (2018) 9:4985–97. doi: 10.18632/oncotarget.23577
29. Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. (2009) 10:9–20. doi: 10.1038/nrm2609
30. Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. (2014) 106:djt356. doi: 10.1093/jnci/djt356
31. Jin J, Zhan P, Qian H, Wang X, Katoh M, Phan K, et al. Prognostic value of wingless-type proteins in non-small cell lung cancer patients: a meta-analysis. Transl Lung Cancer Res. (2016) 5:436–42. doi: 10.21037/tlcr.2016.08.08
Keywords: lung adenocarcinoma, miR-1-3p, protein regulator of cytokinesis 1, malignant behavior, mechanism
Citation: Li T, Wang X, Jing L and Li Y (2019) MiR-1-3p Inhibits Lung Adenocarcinoma Cell Tumorigenesis via Targeting Protein Regulator of Cytokinesis 1. Front. Oncol. 9:120. doi: 10.3389/fonc.2019.00120
Received: 13 November 2018; Accepted: 11 February 2019;
Published: 01 March 2019.
Edited by:
Jian-ye Zhang, Guangzhou Medical University, ChinaReviewed by:
Frank Arfuso, Curtin University, AustraliaKuzhuvelil B. Harikumar, Rajiv Gandhi Centre for Biotechnology, India
Copyright © 2019 Li, Wang, Jing and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Li, cWxsaXl1cmVzQDE2My5jb20=
 Tao Li
Tao Li Yu Li
Yu Li