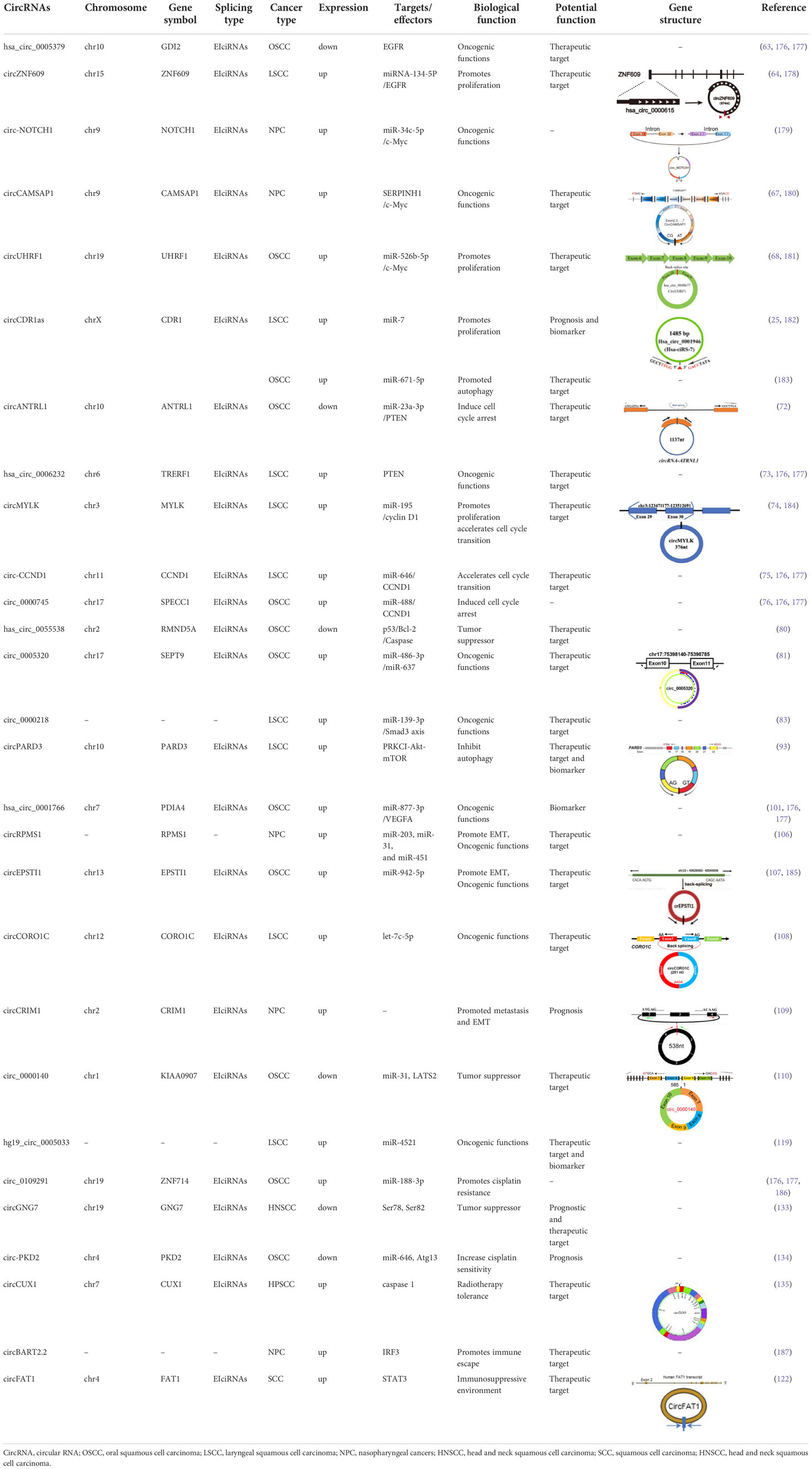
- 1Department of Otorhinolaryngology Head and Neck Surgery, Yantai Yuhuangding Hospital, Qingdao University, Yantai, Shandong, China
- 2Department of Otorhinolaryngology Head and Neck Surgery, Shandong Provincial Clinical Research Center for Otorhinolaryngologic Diseases, Yantai, Shandong, China
- 3School of Clinical Medicine, Weifang Medical University, Weifang, Shandong, China
Head and neck squamous cell carcinoma (HNSCC), the most common head and neck malignant tumor, with only monotherapy, is characterized by poor prognosis, and low 5-year survival rate. Due to the lack of therapeutic targets, the targeted drugs for HNSCC are rare. Therefore, exploring the regulation mechanism of HNSCC and identifying effective therapeutic targets will be beneficial to its treatment of. Circular RNA (CircRNA) is a class of RNA molecules with a circular structure, which is widely expressed in human body. CircRNAs regulate gene expression by exerting the function as a miRNA sponge, thereby mediating the occurrence and development of HNSCC cell proliferation, apoptosis, migration, invasion, and other processes. In addition, circRNAs are also involved in the regulation of tumor sensitivity to chemical drugs and other biological functions. In this review, we systematically listed the functions of circRNAs and explored the regulatory mechanisms of circRNAs in HNSCC from the aspects of tumor growth, cell death, angiogenesis, tumor invasion and metastasis, tumor stem cell regulation, tumor drug resistance, immune escape, and tumor microenvironment. It will assist us in discovering new diagnostic markers and therapeutic targets, while encourage new ideas for the diagnosis and treatment of HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC), originating from the mucosal epithelium of the mouth, pharynx, and larynx, is the sixth most common cancer worldwide (1). Approximately 42% of the patients with HNSCC are diagnosed as intermediate or advanced disease, and these patients often have local lymph node metastases (2). Moreover, HNSCC has a low response to treatment and high drug resistance, with a 5-year survival rate of only 40-50% and a poor prognosis (3). Therefore, the early diagnosis, intervention and seeking potential therapeutic targets for HNSCC are of great significance to improve the prognosis and survival quality of patients. In recent years, an increasing number of studies have been devoted to the identification of specific biomarkers for the clinical decision-making. Currently, some emerging biomarkers have encouraged new ideas for the clinical diagnosis and treatment of HNSCC, including some non-coding RNAs (ncRNAs), like long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), whose aberrant expression has been shown to be closely associated with the development of HNSCC (4). Particularly, circular RNAs (circRNAs), the newly discovered non-coding RNA molecules with closed-loop structure, have been proven to be a kind of new and stable biomarkers for diagnosis and treatment of HNSCC (5). CircRNAs exist widely in different species and cell lines, and are abundant, stable, conserved, and tissue-specific. In humans, they can be secreted into blood, saliva, and exosomes, playing an significant role in the tumor microenvironment. With the rapid development of next-generation sequencing technology, the basic structure and function of circRNAs have been fully confirmed, among which the functions ass HNSCC biomarkers and miRNA sponge, are extensively studied in recent years. However, the specific regulatory mechanism of circRNAs in the critical process of HNSCC development is still worth exploring. This review comprehensively described of the regulatory role of circRNAs in the development of HNSCC, interpreting the role and mechanism of circRNAs in the occurrence of this cancer, offering clues for diagnostic markers and therapeutic targets, and inspiring new ideas for the diagnosis and treatment of HNSCC.
CircRNAs
CircRNAs, first identified in viroids in 1976, are classified as non-coding RNA (6). They are characterized by extensive distribution, stability and high conservation (7). In humans, circRNAs can express in the vast majority of tissues (8–12). However, different circRNAs express with tissue specificity or developmental stage specificity (13). Based on different splicing methods, circRNAs are divided into three main categories: exon circRNAs (ecircRNAs), exon-intron circRNAs (EIciRNAs), and intron circRNAs (ciRNAs). Many studies have focused on the most common circRNAs, namely ecircRNAs (9, 14). According to recent research, the cyclization modes of circRNAs can be divided into intron cyclization and exon cyclization. There are several explanation models for the cyclization mechanism of circRNA: lariat-driven circularization (9, 15–19) (Figure 1A), intron reverse complementary sequence driven cyclization (9, 20, 21) (Figure 1B), ciRNAs model (22) (Figure 1C), and RNA binding protein driven cyclization (23, 24) (Figure 1D).
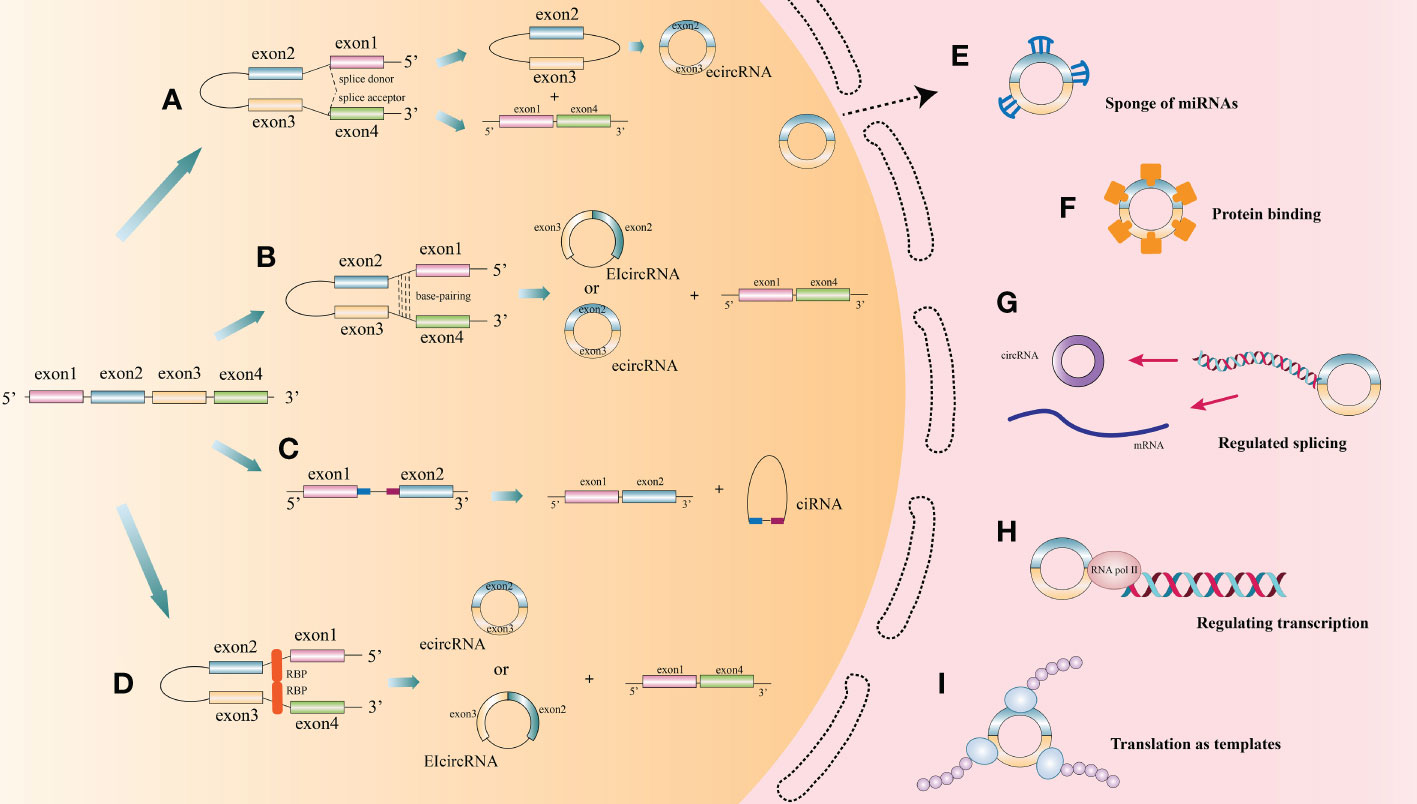
Figure 1 The biogenesis process and function of circRNA. (A) Lariat-driven circularization: During splicing of mRNA precursors, exon skipping produces a lariat intermediate containing both exons and introns. This intermediate will be reverse spliced and introns will be removed to form ecircRNAs. (B) Some introns on either side of one exon of a circRNA contain reverse complementary sequences. the RNA duplexes form two different types of circRNAs by variable splicing: circRNAs with introns and without introns, or introns within and on either side of one exon, which can compete for RNA pairing. (C) CiRNAs model. CiRNAs are generated by splicing reactions of introns. GU-rich elements near the 5’-splice site and C-rich elements near the branching site stabilize CiRNAs to avoid splicing. (D) RNA-binding proteins (RBPs) drive the cyclization of circRNAs. RBPs facilitate the formation of circRNAs by bridging the two sides of introns together. (E) CircRNAs act as endogenous competitive RNAs or miRNA sponge in cytoplasm. (F) CircRNAs act as protein sponge or protein storage library. (G) CircRNAs regulate splicing. (H) CircRNAs regulate transcription. (I) Translation function of circRNA.
At present, the functions of circRNAs as endogenous competitive RNAs or miRNA sponge in cytoplasm are wide studied (Figure 1E). Competitive endogenous RNAs (ceRNAs) contain shared miRNA response elements (MRE), such as mRNAs, pseudogenes, and LncRNAs), enabling them to competitively bind miRNAs. CircRNAs with the same specific miRNA binding site may regulate the activity of competing endogenous RNA, thereby interfering the effect of miRNA on its target through competitive miRNA binding (14). Cerebellar degeneration‐related protein 1 antisense RNA (CDR1as), also known as circular RNA sponge for miR-7 (ciRS-7), was reported to antagonize miR-7 availability (25). By targeting different miRNAs and mRNAs, circRNAs are involved in the pathogenesis of a variety of diseases, including neurological diseases (26), psychiatric diseases (27), autoimmune diseases (28), cardiovascular diseases (29, 30), cartilage degenerative diseases (31, 32), diabetes (33, 34), pulmonary fibrosis (35, 36), and various cancers (10, 37–39).
CircRNAs can also function as a protein sponge or protein storage library (Figure 1F). For example, circMBL can play a crucial role in balancing the expression levels of MBL mRNA and circMBL by isolating the excessive MBL protein (40).
CircRNAs regulate alternative splicing or transcription (Figures 1G, H). Back spliced of circRNAs may compete with linear spliced pre-mRNA for splicing sites. For example, the circMBL generated from the second exon of MBL has MBL binding sites flanking the intron (40). Thus, MBL level significantly affects circMBL biogenesis. Nuclear EIciRNAs with intronic sequences from parental genes, such as circEIF3J and circPAIP2, can interact with U1 small nuclear ribonucleotide proteins (snRNPs) and then bind to RNA polymerase II (Pol II) on the promoter of the parental gene, thereby enhancing gene expression (41). Besides, knocking down the expression of ElciRNAs has the potential to reduce the transcription of their parental genes (41).
Since the ring structure of circRNA is formed by attaching its 3’-end with 5’-end through a unique back splicing, reducing essential translation elements such as 5’-caps and poly-A tails, resulting in the blocked translation of most circRNAs. However, some circRNAs, such as circZNF609, circMb1, circ-FBXW7, circPINTexon2, and circ-SHPRH (42–47), exerted their regulatory functions as polypeptides translated by ribosomes at internal ribosome entry sites (IRESs) or by incorporating with m6A ribonucleic acid in the untranslated region (UTR) on the 5’-end (48–53), thus exerting their regulatory functions (22) (Figure 1I).
With the rapid development of bioinformatics, thousands of circRNAs in humans have been identified. CircRNAs have unique characteristics and a broad range of functions. As new important participant in the ncRNA network, circRNAs have been identified as key regulatory factors in various cancers. Based on existing knowledge of HNSCC characteristics, we assumed that circRNAs may play a possible role in various aspects of HNSCC.
CircRNAs regulate tumor cell proliferation
Cancer cells can synthesize and respond to growth factors (GFs), which in turn promote their own proliferation, forming a positive feedback growth signaling circuit (54), that is, continuous proliferation signal stimulation and infinite division. This is the basis of tumorigenesis.
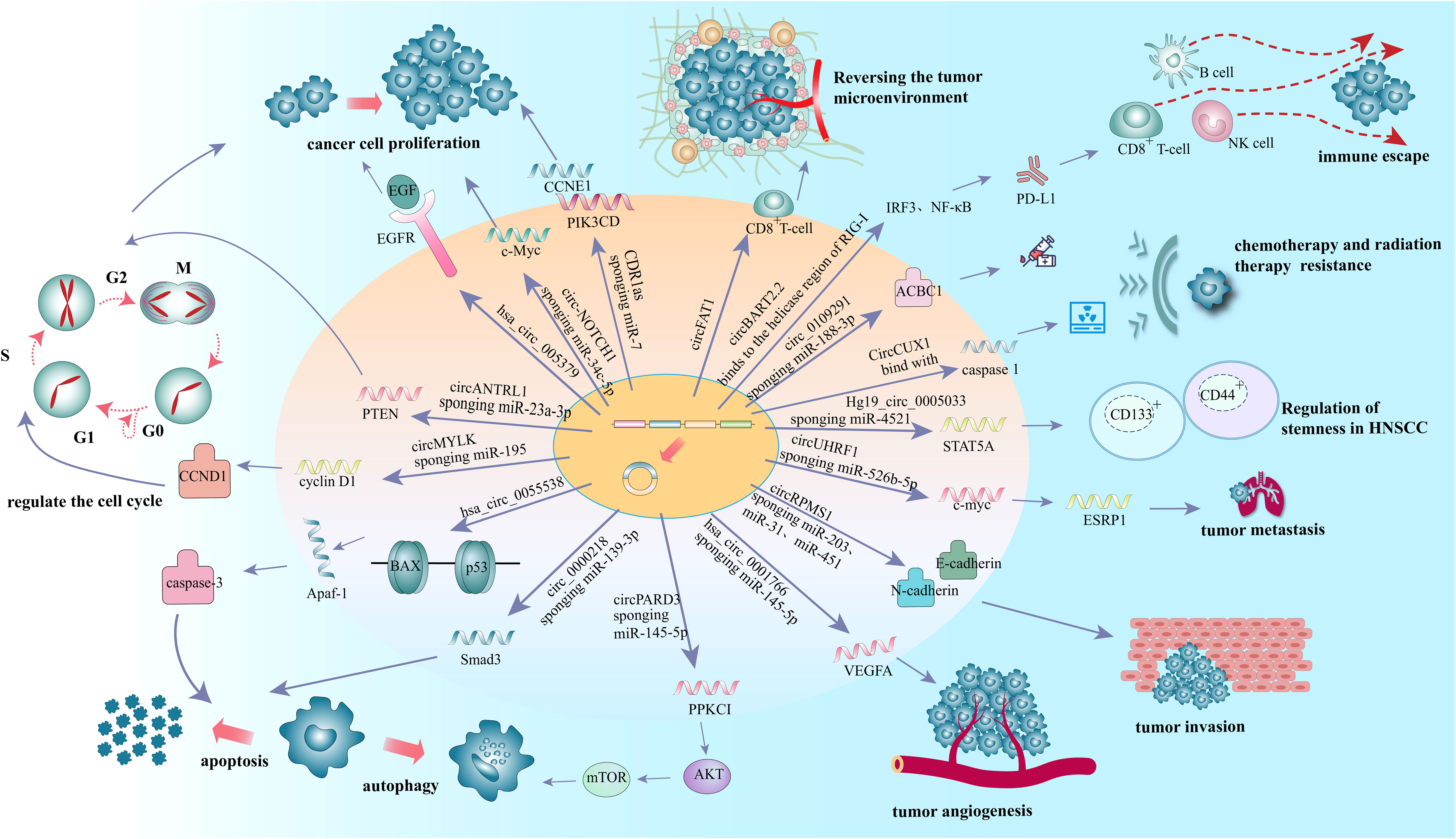
More than 90% of HNSCC patients were tested positive for epidermal growth factor receptor (EGFR) (55), which is highly expressed in a variety of solid tumors and triggers an intracellular growth factor transduction cascade to regulate cell growth (56, 57). EGFR is overexpressed in HNSCC and can activate related pathways leading to the proliferation of cancer cells (58). Previous studies have confirmed that circRNAs can affect the proliferation of tumor cells by regulating the expression of EGFR in lung cancer (59), colorectal cancer (60), glioblastoma (61), melanoma (62) and other malignant tumors. In oral squamous cell carcinoma (OSCC), hsa_circ_0005379 is located in the upstream of EGFR. Overexpression of hsa_circ_0005379 expression can inhibit the expression levels of EGFR and phosphorylated EGFR, thus regulating the proliferation of OSCC tumor cells (63) (Figure 2 and Table 1). Overexpression of the circZNF609 can stimulate miRNA-134-5P, and then up-regulate EGFR to promote the proliferation of laryngeal squamous cell carcinoma (LSCC) cells and the progression of LSCC (64) (Table 1).
C-Myc, a proto-oncogene and is highly expressed as a biomarker in different types of cancer (65), is also an important regulator for tumor cell proliferation. It has been shown that the circRNA circ-NOTCH1 plays an important regulatory role in the cell proliferation process of nasopharyngeal carcinoma (NPC) by targeting the miR-34c-5p/c-Myc axis (66) (Figure 2 and Table 1). Alternatively, circCAMSAP1 facilitates the expression of SERPINH1, a molecular chaperone closely associated with tumor proliferation and metastasis, by improving its mRNA stability through the 3’-untranslated region (3’UTR). High-level expression of SERPINH1 can reduce the ubiquitination-mediated degradation rate of c-Myc, boosting cell proliferation and promotion of the occurrence of NPC (67) (Table 1). Wei Zhao et al. suggested that circUHRF1 acts as a foam of mir-526b-5p in OSCC, thereby positively regulating c-Myc, promoting cell proliferation and tumorigenesis in OSCC (68) (Table 1).
Nevertheless, tumor cells can evade from the supervision of inhibitory growth signals by blocking the expression or activation of tumor suppressors. MiR-7 is a tumor suppressor that regulates a variety of biological processes. In LSCC, the expression of CDR1as is negatively correlated with that of miR-7. Induced overexpression of CDR1as can promot the proliferation of two LSCC cell lines (Hep2 and AMC-HN-8). These effects, however, can be eliminated by knocking down CDR1as or overexpressing miR-7. This suggests that CDR1as acts as the miR-7 sponges. Further in vivo experiments showed that overexpression of CDR1as can upregulat Cyclin E1 (CCNE1) and PIK3CD, the key targets of miR-7, in Hep2 and AMC-HN-8 cells, increasing tumor proliferation index and facilitating tumor growth (25) (Figure 2 and Table 1).
CircRNAs regulate tumor cell cycle
Multiple anti-proliferative signals serve to maintain cellular quiescence in normal cells (54) by blocking cell proliferation in the manner of cell cycle arrest (69). Phosphatase and tensin homolog gene (PTEN), is a tumor suppressor that induce cell cycle arrest, mostly at G1phase in tumor cells (70). While the germline mutations in PTEN substantially increase the risk of cancer (71). CircANTRL1 may act as a sponge for miR-23a-3p to promote PTEN expression and induce cell cycle arrest, ultimately contributing to improvement of radiosensitivity in OSCC (72) (Figure 2 and Table 1). Research also showed that silencing hsa_circ_0006232 in LSCC can enhance the expression of PTEN in tumor tissues and affect the process of LSCC (73) (Table 1).
In addition to regulation of anti-proliferative signals, circRNAs can regulate tumor growth by modulating cell cycle mediators. Cyclin D1, a cell cycle regulator protein encoded by CCND1, promotes the G1-S transition by activating cyclin dependent kinases 4 (CDK4) or cyclin dependent kinases 6 (CDK6). The expression level of circMYLK in LSCC carcinoma tissues was significantly higher than that in adjacent para-cancerous tissues, can increase the level of cell cycle protein D1 in Tu-177 cells (LSCC cell line) through miR-195/cyclin D1 axis, accelerate the G1-S transition of cells, and then promote the proliferation of tumor cells (74) (Figure 2 and Table 1). Yan et al. showed that circ-CCND1 can increase CCND1 levels and promote LSCC cancer cell proliferation by interacting with HuR and miR-646 (75) (Table 1). Kuang et al. suggested that in OSCC, circ_0000745 promotes OSCC progression by acting as a miR-488 sponge and by interacting with HuR proteins to regulate the expression of CCND1 (76) (Table 1).
CircRNAs are involved in resistance to tumor cell death
Apart from cell proliferation rate, cell death rate also determines the number of cells (54). Apoptosis and autophagy are the main mechanisms leading to cell depletion, and cancer cells have the ability to resist these mechanisms (69).
Tumor cells can develop resistance to apoptosis by several mechanisms. Mutations can result in the loss of pro-apoptotic regulators, most commonly in the p53 tumor suppressor gene (54). B-cell lymphoma 2 associated X-protein gene (Bax) is regulated by P53 gene and encodes B-cell lymphoma 2 (Bcl-2) homologous water-soluble protein BAX, which promotes cell apoptosis (77). Overexpression of Bax would antagonize the protective effect of Bcl-2 and lead to cell death. Bax can also regulate the expression of apoptotic protease activator 1 (Apaf-1) and affect the mitochondrial apoptosis pathway (78). Apaf-1 ultimately regulates the caspase-family related proteins such as caspase-3, the most important end cleaving enzyme in apoptosis (79). When hsa_circ_0055538 was overexpressed in OSCC cell lines SCC9 and CAL27 cells, the expression levels of p53, p21, Bax, Apaf-1, caspase-3, and cleaved caspase-3 are increased, while that of Bcl-2 is decreased. When hsa_circ_0055538 is adjusted downward, the opposite result as above is obtained, which suggests that hsa_circ_0055538 may regulate the apoptotic process of OSCC tumor cells through the p53 signaling pathway (80) (Figure 2 and Table 1). Circ_0005320 knockdown can reduce the expression level of Bcl-2 and increase the expression level of BAX by regulating mir-637/mir-486-3p, thus promoting the apoptosis of OSCC cells (81) (Table 1).
Overexpression of Smad3 can promote apoptosis of many human cancers (82). Circ_0000218 was highly expressed in LSCC cells, while miR-139-3p was less expressed in LSCC cells and negatively regulated by circ_0000218. Silencing of circ_0000218 has been shown to suppress LSCC cell viability and promote apoptosis by negatively regulating miR-139-3p expression levels, while the effects can be reversed by Smad family member 3 (Smad3) overexpression. Thus, circ_0000218 inhibition inhibits the growth of LSCC by targeting the miR-139-3p/Smad3 axis (83) (Figure 2 and Table 1).
Autophagy is a lysosome-dependent cellular degradation program that maintains energy metabolic homeostasis by eliminating toxic and potentially compromised cellular components, providing a source of nutrition and energy for cell survival in a starved state (84). Higher bioenergetic and nutritional requirements of cancer cells than normal cells. In the advanced process of malignancy, they may enable themselves to survive under low nutrient and hypoxic conditions by inducing autophagy (85). The signaling pathways involved in autophagy, including mammalian target of rapamycin (mTOR), phosphatidylinositol-3 kinase (PI3K)-protein kinase B (AKT) and mitogen-activated protein kinases (MAPK)/extracellular signal-regulated kinase (ERK), and protein kinase C iota type 1 (PRKC1) have been well established (86–88). Among them, PRKCI is a member of the kinase C protein family and is considered to be an important target for cancer therapy (89–91). It has been shown that PRKCI inhibits autophagy and promotes LSCC cell proliferation, migration, invasion, and chemoresistance. Qu et al. found that a decreasing expression of PRKCI in the U2OS cells (LSCC cell line) can enhance autophagy while suppress cell phenotype (92). In contrast, circPARD3 was able to inhibit Akt and mTOR phosphorylation and suppress autophagy in LSCC cells by activating the PRKCI-Akt-mTOR pathway, promoting malignant progression and chemoresistance in LSCC cells (93) (Figure 2 and Table 1). In OSCC, circCDR1as was significantly increased in OSCC cancer tissues (94). Transcription factor EB (TFEB), a major regulator of lysosomal and autophagic function, is enhanced under hypoxic conditions, and it can coordinate autophagosomal degradation by driving autophagy and lysosomal gene expression (95). The expression level of lysosomal-associated membrane protein 2 (LAMP2) also steadily increases under hypoxic conditions. CircCDR1as can act as a sponge for miR-671-5p to promote autophagy in cells under hypoxic conditions, thereby inducing elevated levels of TFEB and LAMP2 and regulating lysosomal function. CircCDR1as also promotes hypoxia-induced AKT and inhibits mTOR activity, suggesting that AKT/mTOR pathway and lysosomal activity contribute to circCDR1as-induced activation of autophagy under hypoxic conditions in OSCC (96) (Table 1).
CircRNAs regulate persistent tumor angiogenesis
The growth of tumors is often accompanied by angiogenesis. Vascular endothelial growth factor-A (VEGFA; also known as VEGF) is one of the main factors driving the generation and expansion of tumor vascular beds (97). Carbonic anhydrase 9 (CAIX) is involved in cancer angiogenesis under hypoxia (98). Tumor cells produce carbon dioxide intracellularly, which enters the extracellular environment by diffusion, resulting in a lower extracellular pH. Extracellular carbon dioxide undergoes a reversible hydration reaction in the presence of CAIX to produce bicarbonate and release protons. Subsequently, bicarbonate is transported into the cell by the Bt transporter protein, which binds to intracellular protons to form H2O and CO2. To overcome the lack of nutrients in tumor cells undera hypoxic state, vascular endothelial growth factor A (VEGFA) promotes angiogenesis by participating in the activation of intracellular pathways associated with angiogenesis (99), and for tumor cells, angiogenesis is an important process that promotes their metastasis (100). A recent study showed that hsa_circ_0001766 is involved in the ceRNA machinery in OSCC and plays an important role in OSCC cell progression through the hsa_circ_0001766/miR-877-3p/VEGFA axis (101) (Figure 2 and Table 1).
Activation of the janus kinase-2 (JAK2)/activator of transcription 3 (STAT3) pathway has been reported to play a key role in a variety of oncogenic processes, including angiogenesis. In OSCC, circ_0005320/miR-486-3p or circ_0005320/miR-637 axis can activate the JAK2/STAT3 pathway on OSCC cells and promote angiogenesis (81).
CircRNAs regulate tumor invasion and metastasis
It is reported that 90% of death in cancer patients are caused by metastases rather than the primary tumor (102). During the progression of most cancers, tumor cells can migrate from the primary site, forming new colonies at distant sites. Epithelial-mesenchymal transition (EMT) has been shown to be an essential process in carcinoma cell migration and tissue metastasis (103). EMT involves a process of cellular reprogramming that convert epithelial cells into a mesenchymal-like phenotype. And this process is characterized by the loss of epithelial surface markers such as E-cadherin and the acquisition of mesenchymal markers such as vimentin and N-cadherin (104, 105). It has been demonstrated that circRPMS1 can act as a sponge for miR-203, miR-31, and miR-451 to promote the deletion of E-cadherin and the upregulation of N-cadherin and vimentin in the NPC cell line C666-1 cells, thereby regulating the EMT and invasiveness of NPC cells (106) (Figure 2 and Table 1). In OSCC, circEPSTI1 functions as a sponge for miR-942-5p to activate EMT, which in turn promotes migration and invasion of OSCC cells (107) (Table 1). In addition, circCORO1C can bind to let-7c-5p and block it to reduce the level of Pre-B-cell leukemia homeobox transcription factor 3 (PBX3), thus promoting EMT and stimulating LSCC cell invasion and migration in vitro and in vivo (108) (Table 1). CircCRIM1 is overexpressed in NPC cells and distantly metastasized NPC tissues, and it induces EMT during the progression of NPC. When circCRIM1 expression was knocked down in NPC cell lines S18 and HONE1, NPC cell morphology changed from a spindle or elongated mesenchymal morphology to an epithelial morphology. Meanwhile, the expression of the epithelial marker E-cadherin was significantly increased and the expression of the mesenchymal markers N-cadherin and Vimentin were decreased in NPC cells, suggesting that circCRIM1 has oncogenic potential and may be a marker for predicting NPC metastasis (109) (Table 1). Circ_0000140 derived from exons 7-10 of the KIAA0907 gene is downregulated in OSCC tissues and negatively correlates with the prognosis of OSCC patients. Further studies have shown that circ_0000140 can bind to miR-31 and upregulate its target gene large tumor suppressor kinase 2 (LATS2), thereby affecting the EMT of OSCC cells (110) (Table 1).
Epithelial splicing regulatory protein 1 (ESRP1), also known as RNA-binding motif protein 35A (RBM35A), is a key component in the EMT process in malignant tumors (111–114), and it has been shown to be one of the splicing factors associated with EMT in tumor metastasis (8). The circUHRF1/miR-526b-5p/c-Myc axis was found to promote ESRP1 transcript levels in OSCC, and ESRP1 was also identified to target the flanking introns of circUHRF1 and accelerate its cyclization, forming a circUHRF1/miR-526b-5p/c-Myc/TGF-β1/ESRP1 feedback loop in the EMT progression (68) (Figure 2 and Table 1).
CircRNAs regulate the stemness of HNSCC
Cancer stem cells (CSCs) are a fraction of cancer cells that have the ability to renew and initiate themselves, and are responsible for the metastasis and spread of cancer cells in the body and the failure of treatment (115–117). CircRNAs have been shown to regulate the activity of tumor stem cells. CD133+ and CD44+ CSCs, also known as TDP cells, isolated from LSCC cells and highly express the stem cell markers Sex determining Region Y (SOX2) and octamer-binding transcription factor 4 (OCT4), have enhanced proliferation, migration, and colony formation, and are more resistant to chemotherapy and radiotherapy (118). Compared with parental cells, RNA sequencing of TDP cells showed 3684 differentially expressed circRNAs (p < 0.01, log2FC > 1) (119). Among them, Hg19_circ _0005033 upregulated in TDP cells, and acted as a sponge for miR-4521 to upregulate the target STAT5A expression thus inducing stem cell-like cells (120). Therefore, Hg19_circ_0005033 may support the stem cell properties of CD133+ CD44+ laryngeal CSCs through miR4521/STAT5A axis, which needs to be verified by further studies. SOX2 has been shown to regulate self-renewal and tumorigenicity of stem cell-like cells in HNSCC (121) (Figure 2 and Table 1). In nude mouse models with HNSCC, circFat1 KD can significantly inhibit SOX2 + cells and tumor stemness (122).
CircRNAs regulate chemotherapy and radiation therapy resistance in tumors
For the treatment of locally advanced disease, radiation therapy is used as an adjunct to surgery or in conjunction with chemotherapy (2). However, acquired chemotherapy resistance is one of the main reasons for treatment failure in patients with advanced tumors (123). ATP-binding cassette B1 (ABCB1) is a multidrug resistance-associated protein that is highly expressed in resistant cell lines and can promote chemotherapy resistance by pumping intracellular drugs extracellularly (124, 125). Therefore, inhibition of ABCB1 expression is an effective way to reduce tumor drug resistance (126). Circ_0109291 promotes cisplatin resistance in oral squamous carcinoma by regulating ABCB1 expression mainly through adsorption of miR-188-3p. This provides a theoretical basis for reducing the incidence of drug resistance in OSCC (127) (Figure 2 and Table 1). Heat shock protein 27 (HSP27, also known as HSPB1), is a member of the small heat shock protein superfamily with Phospho-HSP27 (Ser15), Phospho-HSP27 (Ser78), and Phospho-HSP27 (Ser82) receptor sites (128). The function of HSP27 is regulated by post-translational phosphorylation (129, 130). HSP27 enhances multidrug resistance in tongue squamous cell carcinoma (SCCT) by activating NF-κB (131). In LSCC, HSP27 overexpression creates cellular resistance to various cellular agents, such as cisplatin and staurosporin, by inducing cell cycle arrest and remodeling actin polymerization associated with drug uptake, respectively (132). CircGNG7 can block the phosphorylation sites of Ser78 and Ser82 and inhibit the phosphorylation of HSP27 in HNSCC, thus inhibiting the phosphorylation of HSP27 in the malignant signaling cascade, which may therefore reduce the chemotherapeutic resistance of HNSCC (133) (Table 1). In addition, in OSCC, circ-PKD2 is a tumor suppressor gene that promotes autophagy related 13 (Atg13) expression by sponging miR-646, thereby accelerating cisplatin sensitivity (134) (Table 1).
Radiotherapy tolerance is also an important prognostic predictor for patients with HNSCC (135). CircCUX1, upregulated in patients with radiation-resistant hypopharyngeal squamous cell carcinoma (HPSCC), predicts poor survival outcomes. CircCUX1 binds to caspase 1 (a member of the caspase aspartate-specific protease family, also known as interleukin-1 β invertase) (136) mRNA and inhibits its expression, thus modulating the inflammatory response of tumor cells to radiation therapy and thereby producing radiation therapy tolerance (135) (Figure 2 and Table 1).
CircRNAs regulate immune escape
The programmed cell death-1/programmed cell death ligand-1 (PD-1/PD-L1) signaling pathway is an important mechanism mediating tumor immunosuppression (137, 138). PD-L1 is often expressed on the surface of tumor cells and immunosuppressive cells, and interacts with PD-1 on T cells, thus preventing tumor antigen-specific T cells from activating and killing tumor cells and leading to tumor immune escape (139–144). It has been shown that circBART2.2, which is highly expressed in NPC, can activate the transcription factors interferon regulatory Factor 3 (IRF3) and nuclear factor kappa-B (NF-κB) upon binding to the decapping enzyme region of retinoic acid inducible gene protein I (RIG-I), which promotes the transcription of PD-L1 and thus leads to tumor immune escape (122) (Figure 2 and Table 1).
Juan Sun et al. proposed the following hypothesis: hsa_circ_001859, hsa-circ_001373, and hsa_circ_002179 may regulate the expression of PD-L1 and immunosuppressive molecule IL-10 through the ceRNA network to affect the immune evasion of LSCC, and then affect the degree of malignancy of LSCC. This hypothesis is likely to represent an important mechanism of immune evasion in LSCC, however, this hypothesis needs to be tested by large-scale scientific studies in the future (145).
CircRNAs regulate tumor microenvironment
Since 1989, the association between cancer and the tumor microenvironment (TME) has received increasing attention with the “seed and soil theory” hypothesis proposed by Stephen Paget (146, 147). Growing amount of evidences have confirmed that tumor cells must recruit and reprogram surrounding normal cells in order to promote tumor progression (148). TME is mainly composed mainly of stromal cells and extracellular matrix (ECM) components. Stromal cells include immune cells, cancer-associated fibroblasts (CAFs), endothelial cells, and pericytes.
ECM is a highly dynamic structural network consisting of many stromal components bearing continuous remodeling mediated by several stromal degrading enzymes during tumorigenesis and progression (149). The protease matrix metalloproteinases (MMP) family supports the tumor cell invasion toward the basement membrane and stroma, the penetration of blood vessels, and metastasis by interacting with macromolecules on the basement membrane to degrade and stimulate ECM remodeling (150). Silencing of circZNF609 inhibited the proliferation and invasiveness of LSCC cells, and significantly increased the expression of MMP-2 protein (64), which may affect the degradation and remodeling of ECM in LSCC (Table 1). Moreover, in other tumors, like gastric cancer cells, protein expression levels of VEGF and migration-associated proteins MMP-2 and MMP-9 were significantly reduced after knockdown of circ-0000096, suggesting that circ-0000096 may affect cell growth and migration by regulating stromal remodeling and angiogenesis (151), as well as the TME. CircLMNB1, which is highly expressed in colorectal cancer (CRC), can downregulate MMP-2 and MMP-9 expression and inhibit EMT when its expression is knocked down, thereby affecting tumor dissemination and invasion (152). Silencing circDENND4C under hypoxic conditions downregulates the protein expression levels of MMP-2 and MMP-9 (153). All these provide a basis for future studies on the changes of TME in HNSCC.
Growing evidence suggests that the innate immune cells (macrophages, neutrophils, dendritic cells, innate lymphocytes, myeloid suppressor cells, and NK cells) as well as adaptive immune cells (T cells and B cells) contribute to tumor progression when present in TME (154). HNSCC is well known for having an immunosuppressive TME with low number of tumour infiltrating lymphocytes (155). In particular, the infiltration of CD8+ T cells into the TME was low. Increasing the expression of circFAT1 in HNSCC can significantly increase the infiltration of CD8+ T cells into tumors, which is an important molecular mechanism mediating the immunosuppressive environment of HNSCC (122) (Figure 2 and Table 1).
CircRNAs can interact with stromal cells, fibroblasts, and endothelial immune-related molecules in the TME of a variety of tumors, as well as various immune cells such as macrophages (156). However, the key role of more circRNAs in the TME of HNSCC is a challenge to be faced in the future.
Discussion
There are growing evidences that circRNAs have great potential in the diagnosis, treatment and prognosis of HNSCC. It is worth mentioning that regulatory effects of circRNAs described in our review are not independent of each other, but is usually closely related to various biological behaviors of cancer cells. For example, circZDBF2 promotes malignant cell behavior in OSCC, including cell proliferation, invasion, migration, and EMT processes (157). Upregulation of circRNA_100290 expression promotes proliferation, migration and invasion of LSCC cells while inhibiting apoptosis (158).
Current studies mostly focus on the clarification of mechanism of circRNAs based on full transcription sequencing of cancer tissues and normal adjacent tissues, and lack of circRNA expression data at the single-cell level of HNSCC. And these studies may be crucial for our future insight into circRNA function and advance the development of new biomarkers. However, to date, only a small number of functional circRNAs identified in HNSCC. Most of these circRNAs regulate the progression of HNSCC through miRNA sponge function. In addition, circRNAs interact with functional proteins, some of which have protein-coding potential. For example, circGNG7 mentioned in this paper can bind to serine residues 78 and 82 of HSP27 to reduce carcinogenic signal transduction.
Recent studies have also shown that circRNAs can regulate immune metabolism of HNSCC, and circ_0008068 may affect glycolysis by targeting mir-153-3p/acylglycerol kinase (AGK) axis and facilitate the CD8+ T cell response (159). It provides a better understanding of the interaction between HNSCC cellular immunity and metabolic rate, and we look forward to opening up new immune-related therapies for HNSCC. The potential network of circRNAs-RBPs and other functions of circRNAs in HNSCC have important research value and potential application value. It is expected that circRNAs can be used as important biomarker for clinical diagnosis and treatment of HNSCC.
Studies have shown that circRNA has great potential in diagnosing pathological types of lung cancer (160) and molecular subtypes of breast cancer (161). Moreover, hsa_circ_0000190 and 0001649 could be used as biomarkers to predict recurrence and treatment response in patients with OSCC (162). CircRNAs have the advantages of high abundance and good stability, and can be detected in body fluids and blood (163, 164), so it can be used as ideal diagnostic and predictive biomarkers for HNSCC diagnosis. In addition, Zhuo et al. analyzed the diagnostic value of various RNAs in hepatocellular carcinoma and with result showing that circRNAs > lncRNAs > microRNAs (165).
CircRNAs have shown great potential as therapeutic molecular tools in many cancers such as HNSCC (110, 166, 167), breast cancer (168), hepatocellular carcinoma (169), papillary thyroid cancer (170) gastric cancer (171). Compared with existing linear miRNA antagonists, circRNAs are structurally more stable, more resistant to degradation by nucleases without chemical modification, and well tolerated in vivo (172). The knockdown of oncogenic circRNAs in preclinical animal models validated circRNAs RNAi effect on tumor treatment (169, 173, 174). Some researchers have attempted to design and synthesize tumor suppressive circRNAs in vitro for delivery into disease models to explore the potential of circRNA-like drug development. In the treatment of heart disease (172) and gastric malignancies (175), engineered circRNAs were delivered to in-vivo and in-vitro disease models to reverse pathogenic mechanisms. CircRNAs will be better diagnosis and treatment targets for HNSCC patients, which will benefit for the translation of basic to clinical medicine.
In conclusion, although the technical challenges concerning circRNAs have not been fully overcome yet, with the development of biological methods, informatics techniques, and further research, we will gain more and more understanding of the physiological functions and roles of circRNAs. The new diagnostic and therapeutic strategies for HNSCC based on circRNAs will effectively serve for the clinical work in the future.
Author contributions
XH and RT conceived the paper and drafted the manuscript. XH and CW drew the figures. XS and YL edited and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Taishan Scholars Project (No. ts20190991).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Moskovitz J, Moy J, Ferris RL. Immunotherapy for head and neck squamous cell carcinoma. Curr Oncol Rep (2018) 20(2):22. doi: 10.1007/s11912-018-0654-5
2. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. Mayo Clinic Proc (2016) 91(3):386–96. doi: 10.1016/j.mayocp.2015.12.017
3. Canning M, Guo G, Yu M, Myint M, Groves MW, Byrd JK, et al. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front Cell Dev Biol (2019) 7:52. doi: 10.3389/fcell.2019.00052
4. Arantes L, De Carvalho AC, Melendez ME, Lopes Carvalho A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagnost (2018) 18(1):85–112. doi: 10.1080/14737159.2017.1404906
5. Wu S, Huang X, Tie X, Cheng Y, Xue X, Fan M. Role and mechanism of action of circular RNA and laryngeal cancer. Pathol Res Pract (2021) 223:153460. doi: 10.1016/j.prp.2021.153460
6. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U States A (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
7. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
8. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
9. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York NY) (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
10. Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C, et al. Circular RNAs and their roles in head and neck cancers. Mol Cancer (2019) 18(1):44. doi: 10.1186/s12943-019-1003-5
11. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell (2015) 58(5):870–85. doi: 10.1016/j.molcel.2015.03.027
12. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
13. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
14. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA (2015) 6(5):563–79. doi: 10.1002/wrna.1294
15. Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol (2017) 14(8):1000–6. doi: 10.1080/15476286.2016.1227904
16. Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol (2015) 427(15):2414–7. doi: 10.1016/j.jmb.2015.02.018
17. Wilusz JE. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA (2018) 9(4):e1478. doi: 10.1002/wrna.1478
18. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
19. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife (2015) 4:e07540. doi: 10.7554/eLife.07540
20. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene sry in adult mouse testis. Cell (1993) 73(5):1019–30. doi: 10.1016/0092-8674(93)90279-Y
21. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
22. Zhang Z, Yang T, Xiao J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
23. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol (2016) 17(4):205–11. doi: 10.1038/nrm.2015.32
24. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
25. Zhang J, Hu H, Zhao Y, Zhao Y. CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour's progression via miR-7 signals. Cell Prolif (2018) 51(6):e12521. doi: 10.1111/cpr.12521
26. Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy (2018) 14(7):1164–84. doi: 10.1080/15548627.2018.1458173
27. Chen Y, Li X, Meng S, Huang S, Chang S, Shi J. Identification of functional CircRNA-miRNA-mRNA regulatory network in dorsolateral prefrontal cortex neurons of patients with cocaine use disorder. Front Mol Neurosci (2022) 15:839233. doi: 10.3389/fnmol.2022.839233
28. Wen JT, Liu J, Wan L, Xin L, Guo JC, Sun YQ, et al. Triptolide inhibits cell growth and inflammatory response of fibroblast-like synoviocytes by modulating hsa-circ-0003353/microRNA-31-5p/CDK1 axis in rheumatoid arthritis. Int Immunopharmacol (2022) 106:108616. doi: 10.1016/j.intimp.2022.108616
29. Gong X, Tian M, Cao N, Yang P, Xu Z, Zheng S, et al. Circular RNA circEsyt2 regulates vascular smooth muscle cell remodeling via splicing regulation. J Clin Invest (2021) 131(24):e147031. doi: 10.1172/JCI147031
30. Wang Y, Li C, Zhao R, Qiu Z, Shen C, Wang Z, et al. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics (2021) 11(13):6315–33. doi: 10.7150/thno.52843
31. Zheng YL, Song G, Guo JB, Su X, Chen YM, Yang Z, et al. Interactions among lncRNA/circRNA, miRNA, and mRNA in musculoskeletal degenerative diseases. Front Cell Dev Biol (2021) 9:753931. doi: 10.3389/fcell.2021.753931
32. Liao H, Tu Q, Kang Y, Mao G, Li Z, Hu S, et al. CircNFIX regulates chondrogenesis and cartilage homeostasis by targeting the miR758-3p/KDM6A axis. Cell Prolif (2022) 55(11):e13302. doi: 10.1111/cpr.13302
33. Zhao W, Meng X, Liang J. Analysis of circRNA-mRNA expression profiles and functional enrichment in diabetes mellitus based on high throughput sequencing. Int Wound J (2022) 19(5):1253–62. doi: 10.1111/iwj.13838
34. Du R, Wu N, Bai Y, Tang L, Li L. circMAP3K4 regulates insulin resistance in trophoblast cells during gestational diabetes mellitus by modulating the miR-6795-5p/PTPN1 axis. J Trans Med (2022) 20(1):180. doi: 10.1186/s12967-022-03386-8
35. Cheng Z, Zhang Y, Wu S, Zhao R, Yu Y, Zhou Y, et al. Peripheral blood circular RNA hsa_circ_0058493 as a potential novel biomarker for silicosis and idiopathic pulmonary fibrosis. Ecotoxicol Environ Safety (2022) 236:113451. doi: 10.1016/j.ecoenv.2022.113451
36. Wang S, Luo W, Huang J, Chen M, Ding J, Cheng Y, et al. The combined effects of circular RNA methylation promote pulmonary fibrosis. Am J Respir Cell Mol Biol (2022) 66(5):510–23. doi: 10.1165/rcmb.2021-0379OC
37. Lei T, Zhang Y, Wang X, Liu W, Feng W, Song W. Integrated analysis of the functions and clinical implications of exosome circRNAs in colorectal cancer. Front Immunol (2022) 13:919014. doi: 10.3389/fimmu.2022.919014
38. Nie J, Yang R, Zhou R, Deng Y, Li D, Gou D, et al. Circular RNA circFARSA promotes the tumorigenesis of non-small cell lung cancer by elevating B7H3 via sponging miR-15a-5p. Cell Cycle (Georgetown Tex) (2022) 3:1–15. doi: 10.1080/15384101.2022.2105087
39. Gao F, Xu Q, Tang Z, Zhang N, Huang Y, Li Z, et al. Exosomes derived from myeloid-derived suppressor cells facilitate castration-resistant prostate cancer progression via S100A9/circMID1/miR-506-3p/MID1. J Trans Med (2022) 20(1):346. doi: 10.1186/s12967-022-03494-5
40. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
41. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct &Molecular Biol (2015) 22:256–64. doi: 10.1038/nsmb.2959
42. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
43. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
44. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
45. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Institute (2018) 110(3):304–15. doi: 10.1093/jnci/djx166
46. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene (2018) 37(13):1805–14. doi: 10.1038/s41388-017-0019-9
47. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9(1):4475. doi: 10.1038/s41467-018-02847-3
48. Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep (2015) 5:16435. doi: 10.1038/srep16435
49. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR m(6)A promotes cap-independent translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
50. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature (2015) 526(7574):591–4. doi: 10.1038/nature15377
51. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Sci (New York NY) (1995) 268(5209):415–7. doi: 10.1126/science.7536344
52. Perriman R, Ares M Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA (New York NY) (1998) 4(9):1047–54. doi: 10.1017/S135583829898061X
53. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA (New York NY) (2015) 21(2):172–9. doi: 10.1261/rna.048272.114
54. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell (2000) 100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9
55. Rubin Grandis J, Melhem MF, Barnes EL, Tweardy DJ. Quantitative immunohistochemical analysis of transforming growth factor-alpha and epidermal growth factor receptor in patients with squamous cell carcinoma of the head and neck. Cancer (1996) 78(6):1284–92. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1284::AID-CNCR17>3.0.CO;2-X
56. Huang C, Zeng X, Jiang G, Liao X, Liu C, Li J, et al. XIAP BIR domain suppresses miR-200a expression and subsequently promotes EGFR protein translation and anchorage-independent growth of bladder cancer cell. J Hematol Oncol (2017) 10(1):6. doi: 10.1186/s13045-016-0376-9
57. Sooro MA, Zhang N, Zhang P. Targeting EGFR-mediated autophagy as a potential strategy for cancer therapy. Int J Cancer (2018) 143(9):2116–25. doi: 10.1002/ijc.31398
58. Oliveira-Silva RJ, Carolina de Carvalho A, de Souza Viana L, Carvalho AL, Reis RM. Anti-EGFR therapy: Strategies in head and neck squamous cell carcinoma. Recent Patents Anti Cancer Drug Discov (2016) 11(2):170–83. doi: 10.2174/1574892811666160309121238
59. Ishola AA, Chien CS, Yang YP, Chien Y, Yarmishyn AA, Tsai PH, et al. Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res (2022) 82(1):75–89. doi: 10.1158/0008-5472.CAN-21-1473
60. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis (2018) 9(4):417. doi: 10.1038/s41419-018-0454-8
61. Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X, et al. Circular RNA-encoded oncogenic e-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol (2021) 23(3):278–91. doi: 10.1038/s41556-021-00639-4
62. Hao YL, Wang XQ. Circ-0079593 promotes proliferation and migration of melanoma cells by sponging microRNA-433 and elevating EGFR expression. Eur Rev Med Pharmacol Sci (2021) 25(2):779–86. doi: 10.26355/eurrev_202101_24639
63. Su W, Wang Y, Wang F, Sun S, Li M, Shen Y, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer (2019) 19(1):400. doi: 10.1186/s12885-019-5593-5
64. Yin X, Wang J, Shan C, Jia Q, Bian Y, Zhang H. Circular RNA ZNF609 promotes laryngeal squamous cell carcinoma progression by upregulating epidermal growth factor receptor via sponging microRNA-134-5p. Bioengineered (2022) 13(3):6929–41. doi: 10.1080/21655979.2022.2034703
65. Robaina MC, Mazzoccoli L, Klumb CE. Germinal centre b cell functions and lymphomagenesis: Circuits involving MYC and MicroRNAs. Cells (2019) 8(11):1365. doi: 10.3390/cells8111365
66. Huang W, Song W, Jiang Y, Chen L, Lu H. C-myc-induced circ-NOTCH1 promotes aggressive phenotypes of nasopharyngeal carcinoma cells by regulating the miR-34c-5p/c-Myc axis. Cell Biol Int (2021) 45(7):1436–47. doi: 10.1002/cbin.11582
67. Wang Y, Yan Q, Mo Y, Liu Y, Wang Y, Zhang S, et al. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumorigenesis via a SERPINH1/c-myc positive feedback loop. Mol Cancer (2022) 21(1):62. doi: 10.1186/s12943-022-01502-2
68. Zhao W, Cui Y, Liu L, Qi X, Liu J, Ma S, et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ (2020) 27(3):919–33. doi: 10.1038/s41418-019-0423-5
69. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol (2012) 9(6):703–19. doi: 10.4161/rna.20481
70. Radu A, Neubauer V, Akagi T, Hanafusa H, Georgescu MM. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol Cell Biol (2003) 23(17):6139–49. doi: 10.1128/MCB.23.17.6139-6149.2003
71. Worby CA, Dixon JE. PTEN. Annu Rev Biochem (2014) 83:641–69. doi: 10.1146/annurev-biochem-082411-113907
72. Chen G, Li Y, He Y, Zeng B, Yi C, Wang C, et al. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Nucleic Acids (2020) 19:961–73. doi: 10.1016/j.omtn.2019.12.031
73. Wu T, Wang G, Zeng X, Sun Z, Li S, Wang W, et al. Hsa_circ_0006232 promotes laryngeal squamous cell cancer progression through FUS-mediated EZH2 stabilization. Cell Cycle (Georgetown Tex) (2021) 20:1799 – 811. doi: 10.1080/15384101.2021.1959973
74. Duan X, Shen N, Chen J, Wang J, Zhu Q, Zhai Z. Circular RNA MYLK serves as an oncogene to promote cancer progression via microRNA-195/cyclin D1 axis in laryngeal squamous cell carcinoma. Biosci Rep (2019) 39(9):BSR20190227. doi: 10.1042/bsr20190227
75. Zang Y, Li J, Wan B, Tai Y. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J Cell Mol Med (2020) 24(4):2423–33. doi: 10.1111/jcmm.14925
76. Li K, Fan X, Yan Z, Zhan J, Cao F, Jiang Y. Circ_0000745 strengthens the expression of CCND1 by functioning as miR-488 sponge and interacting with HuR binding protein to facilitate the development of oral squamous cell carcinoma. Cancer Cell Int (2021) 21(1):271. doi: 10.1186/s12935-021-01884-1
77. Ramadan MA, Shawkey AE, Rabeh MA, Abdellatif AO. Expression of P53, BAX, and BCL-2 in human malignant melanoma and squamous cell carcinoma cells after tea tree oil treatment in vitro. Cytotechnology (2019) 71(1):461–73. doi: 10.1007/s10616-018-0287-4
78. Wang Y, Cao Y, Zhu Q, Gu X, Zhu YZ. The discovery of a novel inhibitor of apoptotic protease activating factor-1 (Apaf-1) for ischemic heart: synthesis, activity and target identification. Sci Rep (2016) 6:29820. doi: 10.1038/srep29820
79. Liu X, He Y, Li F, Huang Q, Kato TA, Hall RP, et al. Caspase-3 promotes genetic instability and carcinogenesis. Mol Cell (2015) 58(2):284–96. doi: 10.1016/j.molcel.2015.03.003
80. Su W, Sun S, Wang F, Shen Y, Yang H. Circular RNA hsa_circ_0055538 regulates the malignant biological behavior of oral squamous cell carcinoma through the p53/Bcl-2/caspase signaling pathway. J Trans Med (2019) 17(1):76. doi: 10.1186/s12967-019-1830-6
81. Zheng X, Du F, Gong X, Xu P. Circ_0005320 promotes oral squamous cell carcinoma tumorigenesis by sponging microRNA-486-3p and microRNA-637. Bioengineered (2022) 13(1):440–54. doi: 10.1080/21655979.2021.2009317
82. Millet C, Zhang YE. Roles of Smad3 in TGF-beta signaling during carcinogenesis. Crit Rev Eukaryotic Gene Expression (2007) 17(4):281–93. doi: 10.1615/CritRevEukarGeneExpr.v17.i4.30
83. Bai Y, Hou J, Wang X, Geng L, Jia X, Xiang L, et al. Circ_0000218 plays a carcinogenic role in laryngeal cancer through regulating microRNA-139-3p/Smad3 axis. Pathol Res Pract (2020) 216(9):153103. doi: 10.1016/j.prp.2020.153103
84. White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer (2012) 12(6):401–10. doi: 10.1038/nrc3262
85. Bahrami A, Khazaei M, Hassanian SM, ShahidSales S, Joudi-Mashhad M, Maftouh M, et al. Targeting the tumor microenvironment as a potential therapeutic approach in colorectal cancer: Rational and progress. J Cell Physiol (2018) 233(4):2928–36. doi: 10.1002/jcp.26041
86. Sun W, Yi Y, Xia G, Zhao Y, Yu Y, Li L, et al. Nrf2-miR-129-3p-mTOR axis controls an miRNA regulatory network involved in HDACi-induced autophagy. Mol Ther J Am Soc Gene Ther (2019) 27(5):1039–50. doi: 10.1016/j.ymthe.2019.02.010
87. Zhang Q, Meng X, Qin G, Xue X, Dang N. Lyn Kinase promotes the proliferation of malignant melanoma cells through inhibition of apoptosis and autophagy via the PI3K/Akt signaling pathway. J Cancer (2019) 10(5):1197–208. doi: 10.7150/jca.28908
88. Wang XY, Zhang XH, Peng L, Liu Z, Yang YX, He ZX, et al. Bardoxolone methyl (CDDO-me or RTA402) induces cell cycle arrest, apoptosis and autophagy via PI3K/Akt/mTOR and p38 MAPK/Erk1/2 signaling pathways in K562 cells. Am J Trans Res (2017) 9(10):4652–72.
89. Yin N, Liu Y, Khoor A, Wang X, Thompson EA, Leitges M, et al. Protein kinase cι and wnt/β-catenin signaling: Alternative pathways to Kras/Trp53-driven lung adenocarcinoma. Cancer Cell (2019) 36(2):156–67.e7. doi: 10.1016/j.ccell.2019.07.002
90. Sarkar S, Bristow CA, Dey P, Rai K, Perets R, Ramirez-Cardenas A, et al. PRKCI promotes immune suppression in ovarian cancer. Genes Dev (2017) 31(11):1109–21. doi: 10.1101/gad.296640.117
91. Kikuchi K, Soundararajan A, Zarzabal LA, Weems CR, Nelon LD, Hampton ST, et al. Protein kinase c iota as a therapeutic target in alveolar rhabdomyosarcoma. Oncogene (2013) 32(3):286–95. doi: 10.1038/onc.2012.46
92. Qu L, Li G, Xia D, Hongdu B, Xu C, Lin X, et al. PRKCI negatively regulates autophagy via PIK3CA/AKT-MTOR signaling. Biochem Biophys Res Commun (2016) 470(2):306–12. doi: 10.1016/j.bbrc.2016.01.059
93. Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer (2020) 19(1):166. doi: 10.1186/s12943-020-01279-2
94. Cui L, Huang C, Zhou D. Overexpression of circCDR1as drives oral squamous cell carcinoma progression. Oral Dis (2021) 00:1–11. doi: 10.1111/odi.14085
95. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Sci (New York NY) (2011) 332(6036):1429–33. doi: 10.1126/science.1204592
96. Gao L, Dou ZC, Ren WH, Li SM, Liang X, Zhi KQ. CircCDR1as upregulates autophagy under hypoxia to promote tumor cell survival via AKT/ERK(½)/mTOR signaling pathways in oral squamous cell carcinomas. Cell Death Dis (2019) 10(10):745. doi: 10.1038/s41419-019-1971-9
97. Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Internal Med (2013) 273(2):114–27. doi: 10.1111/joim.12019
98. Deron P, Vermeersch H, Mees G, Vangestel C, Pauwels P, Van de Wiele C. Expression and prognostic value of glucose transporters and hexokinases in tonsil and mobile tongue squamous cell carcinoma. Histol Histopathol (2011) 26(9):1165–72. doi: 10.14670/hh-26.1165
99. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature (2011) 473(7347):298–307. doi: 10.1038/nature10144
100. Semenza GL, Ruvolo PP. Introduction to tumor microenvironment regulation of cancer cell survival, metastasis, inflammation, and immune surveillance. Biochim Biophys Acta (2016) 1863(3):379–81. doi: 10.1016/j.bbamcr.2015.12.015
101. Shao Y, Song Y, Xu S, Li S, Zhou H. Expression profile of circular RNAs in oral squamous cell carcinoma. Front Oncol (2020) 10:533616. doi: 10.3389/fonc.2020.533616
102. Sporn MB. The war on cancer. Lancet (London England) (1996) 347(9012):1377–81. doi: 10.1016/S0140-6736(96)91015-6
103. Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol (2018) 13:395–412. doi: 10.1146/annurev-pathol-020117-043854
104. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev (2009) 28(1-2):15–33. doi: 10.1007/s10555-008-9169-0
105. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer (2009) 9(4):265–73. doi: 10.1038/nrc2620
106. Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manage Res (2019) 11:8023–31. doi: 10.2147/CMAR.S218967
107. Wang J, Jiang C, Li N, Wang F, Xu Y, Shen Z, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis (2020) 11(8):682. doi: 10.1038/s41419-020-02851-w
108. Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol Cancer (2020) 19(1):99. doi: 10.1186/s12943-020-01215-4
109. Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer (2020) 19(1):33. doi: 10.1186/s12943-020-01149-x
110. Peng QS, Cheng YN, Zhang WB, Fan H, Mao QH, Xu P. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit hippo signaling pathway. Cell Death Dis (2020) 11(2):112. doi: 10.1038/s41419-020-2273-y
111. Göttgens EL, Span PN, Zegers MM. Roles and regulation of epithelial splicing regulatory proteins 1 and 2 in epithelial-mesenchymal transition. Int Rev Cell Mol Biol (2016) 327:163–94. doi: 10.1016/bs.ircmb.2016.06.003
112. Matos ML, Lapyckyj L, Rosso M, Besso MJ, Mencucci MV, Briggiler CI, et al. Identification of a novel human e-cadherin splice variant and assessment of its effects upon EMT-related events. J Cell Physiol (2017) 232(6):1368–86. doi: 10.1002/jcp.25622
113. Lekva T, Berg JP, Lyle R, Heck A, Ringstad G, Olstad OK, et al. Epithelial splicing regulator protein 1 and alternative splicing in somatotroph adenomas. Endocrinology (2013) 154(9):3331–43. doi: 10.1210/en.2013-1051
114. Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J (2010) 29(19):3286–300. doi: 10.1038/emboj.2010.195
115. Li Y, Atkinson K, Zhang T. Combination of chemotherapy and cancer stem cell targeting agents: Preclinical and clinical studies. Cancer Lett (2017) 396:103–9. doi: 10.1016/j.canlet.2017.03.008
116. Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell (2007) 1(3):241–2. doi: 10.1016/j.stem.2007.08.012
117. Su M, Xiao Y, Ma J, Cao D, Zhou Y, Wang H, et al. Long non-coding RNAs in esophageal cancer: molecular mechanisms, functions, and potential applications. J Hematol Oncol (2018) 11(1):118. doi: 10.1186/s13045-018-0663-8
118. Wang J, Wu Y, Gao W, Li F, Bo Y, Zhu M, et al. Identification and characterization of CD133(+)CD44(+) cancer stem cells from human laryngeal squamous cell carcinoma cell lines. J Cancer (2017) 8(3):497–506. doi: 10.7150/jca.17444
119. Wu Y, Zhang Y, Niu M, Shi Y, Liu H, Yang D, et al. Whole-transcriptome analysis of CD133+CD144+ cancer stem cells derived from human laryngeal squamous cell carcinoma cells. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 47(4):1696–710. doi: 10.1159/000490992
120. Talati PG, Gu L, Ellsworth EM, Girondo MA, Trerotola M, Hoang DT, et al. Jak2-Stat5a/b signaling induces epithelial-to-Mesenchymal transition and stem-like cell properties in prostate cancer. Am J Pathol (2015) 185(9):2505–22. doi: 10.1016/j.ajpath.2015.04.026
121. Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho YS, et al. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer (2014) 111(11):2122–30. doi: 10.1038/bjc.2014.528
122. Jia L, Wang Y, Wang CY. circFAT1 promotes cancer stemness and immune evasion by promoting STAT3 activation. Adv Sci (Weinheim Baden Wurttemberg Germany) (2021) 8(13):2003376. doi: 10.1002/advs.202003376
124. Sui H, Fan ZZ, Li Q. Signal transduction pathways and transcriptional mechanisms of ABCB1/Pgp-mediated multiple drug resistance in human cancer cells. J Int Med Res (2012) 40(2):426–35. doi: 10.1177/147323001204000204
125. Bruhn O, Cascorbi I. Polymorphisms of the drug transporters ABCB1, ABCG2, ABCC2 and ABCC3 and their impact on drug bioavailability and clinical relevance. Expert Opin Drug Metab Toxicol (2014) 10(10):1337–54. doi: 10.1517/17425255.2014.952630
126. Ji N, Yang Y, Cai CY, Lei ZN, Wang JQ, Gupta P, et al. Selonsertib (GS-4997), an ASK1 inhibitor, antagonizes multidrug resistance in ABCB1- and ABCG2-overexpressing cancer cells. Cancer Lett (2019) 440–441:82–93. doi: 10.1016/j.canlet.2018.10.007
127. Gao F, Han J, Wang Y, Jia L, Luo W, Zeng Y. Circ_0109291 promotes the cisplatin resistance of oral squamous cell carcinoma by sponging miR-188-3p to increase ABCB1 expression. Cancer Biother Radiopharmaceuticals (2020) 37(4):233–45. doi: 10.1089/cbr.2020.3928
128. Kostenko S, Moens U. Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell Mol Life Sci CMLS (2009) 66(20):3289–307. doi: 10.1007/s00018-009-0086-3
129. Benjamin IJ, McMillan DR. Stress (heat shock) proteins: molecular chaperones in cardiovascular biology and disease. Circ Res (1998) 83(2):117–32. doi: 10.1161/01.RES.83.2.117
130. Lambert H, Charette SJ, Bernier AF, Guimond A, Landry J. HSP27 multimerization mediated by phosphorylation-sensitive intermolecular interactions at the amino terminus. J Biol Chem (1999) 274(14):9378–85. doi: 10.1074/jbc.274.14.9378
131. Zheng G, Zhang Z, Liu H, Xiong Y, Luo L, Jia X, et al. HSP27-mediated extracellular and intracellular signaling pathways synergistically confer chemoresistance in squamous cell carcinoma of tongue. Clin Cancer Res an Off J Am Assoc Cancer Res (2018) 24(5):1163–75. doi: 10.1158/1078-0432.CCR-17-2619
132. Lee JH, Sun D, Cho KJ, Kim MS, Hong MH, Kim IK, et al. Overexpression of human 27 kDa heat shock protein in laryngeal cancer cells confers chemoresistance associated with cell growth delay. J Cancer Res Clin Oncol (2007) 133(1):37–46. doi: 10.1007/s00432-006-0143-3
133. Ju H, Hu Z, Wei D, Huang J, Zhang X, Rui M, et al. A novel intronic circular RNA, circGNG7, inhibits head and neck squamous cell carcinoma progression by blocking the phosphorylation of heat shock protein 27 at Ser78 and Ser82. Cancer Commun (London England) (2021) 41(11):1152–72. doi: 10.1002/cac2.12213
134. Gao L, Zhang Q, Li S, Zheng J, Ren W, Zhi K. Circ-PKD2 promotes Atg13-mediated autophagy by inhibiting miR-646 to increase the sensitivity of cisplatin in oral squamous cell carcinomas. Cell Death Dis (2022) 13(2):192. doi: 10.1038/s41419-021-04497-8
135. Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis (2021) 12(4):298. doi: 10.1038/s41419-021-03558-2
136. Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. OncoTargets Ther (2018) 11:3979–87. doi: 10.2147/OTT.S158316
137. Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity (2007) 27(1):111–22. doi: 10.1016/j.immuni.2007.05.016
138. Park JJ, Omiya R, Matsumura Y, Sakoda Y, Kuramasu A, Augustine MM, et al. B7-H1/CD80 interaction is required for the induction and maintenance of peripheral T-cell tolerance. Blood (2010) 116(8):1291–8. doi: 10.1182/blood-2010-01-265975
139. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer (2019) 18(1):10. doi: 10.1186/s12943-018-0928-4
140. Ren D, Hua Y, Yu B, Ye X, He Z, Li C, et al. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol Cancer (2020) 19(1):19. doi: 10.1186s12943-020-1144-6
141. Ma BBY, Lim WT, Goh BC, Hui EP, Lo KW, Pettinger A, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: An international, multicenter study of the Mayo clinic phase 2 consortium (NCI-9742). J Clin Oncol Off J Am Soc Clin Oncol (2018) 36(14):1412–8. doi: 10.1200/JCO.2017.77.0388
142. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity (2018) 48(3):434–52. doi: 10.1016/j.immuni.2018.03.014
143. Fan C, Zhang S, Gong Z, Li X, Xiang B, Deng H, et al. Emerging role of metabolic reprogramming in tumor immune evasion and immunotherapy. Sci China Life Sci (2021) 64(4):534–47. doi: 10.1007/s11427-019-1735-4
144. Wei F, Wang D, Wei J, Tang N, Tang L, Xiong F, et al. Metabolic crosstalk in the tumor microenvironment regulates antitumor immunosuppression and immunotherapy resisitance. Cell Mol Life Sci CMLS (2021) 78(1):173–93. doi: 10.1007/s00018-020-03581-0
145. Sun J, Lian M, Ma H, Wang R, Ma Z, Wang H, et al. Competing endogenous RNA network analysis of CD274, IL−10 and FOXP3 co−expression in laryngeal squamous cell carcinoma. Mol Med Rep (2018) 17(3):3859–69. doi: 10.3892/mmr.2017.8307
146. Paget S. The distribution of secondary growths in cancer of the breast. 1889 Cancer Metastasis Rev (1989) 8(2):98–101.
147. Mendoza M, Khanna C. Revisiting the seed and soil in cancer metastasis. Int J Biochem Cell Biol (2009) 41(7):1452–62. doi: 10.1016/j.biocel.2009.01.015
148. Yuan Y, Jiang YC, Sun CK, Chen QM. Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncol Rep (2016) 35(5):2499–515. doi: 10.3892/or.2016.4660
149. Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK. Extracellular matrix structure. Adv Drug Delivery Rev (2016) 97:4–27. doi: 10.1016/j.addr.2015.11.001
150. Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol Off J Am Soc Clin Oncol (2000) 18(5):1135–49. doi: 10.1200/JCO.2000.18.5.1135
151. Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer (2017) 116(5):626–33. doi: 10.1038/bjc.2016.451
152. He C, Huang C, Zhou R, Yu H. CircLMNB1 promotes colorectal cancer by regulating cell proliferation, apoptosis and epithelial-mesenchymal transition. OncoTargets Ther (2019) 12:6349–59. doi: 10.2147/OTT.S204741
153. Ren S, Liu J, Feng Y, Li Z, He L, Li L, et al. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J Exp Clin Cancer Res CR (2019) 38(1):388. doi: 10.1186/s13046-019-1398-2
154. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
155. Chen SMY, Krinsky AL, Woolaver RA, Wang X, Chen Z, Wang JH. Tumor immune microenvironment in head and neck cancers. Mol Carcinog (2020) 59(7):766–74. doi: 10.1002/mc.23162
156. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer (2020) 19(1):8. doi: 10.1186/s12943-019-1113-0
157. Rong L, Chen B, Liu K, Liu B, He X, Liu J, et al. CircZDBF2 up-regulates RNF145 by ceRNA model and recruits CEBPB to accelerate oral squamous cell carcinoma progression via NFκB signaling pathway. J Trans Med (2022) 20(1):148. doi: 10.1186/s12967-022-03347-1
158. Wang Z, Huang C, Zhang A, Lu C, Liu L. Overexpression of circRNA_100290 promotes the progression of laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C axis. Biomed Pharmacother = Biomed Parmacother (2020) 125:109874. doi: 10.1016/j.biopha.2020.109874
159. Long Y, Li C, Zhu B. Circ_0008068 facilitates the oral squamous cell carcinoma development by microRNA-153-3p/acylgycerol kinase (AGK) axis. Bioengineered (2022) 13(5):13055–69. doi: 10.1080/21655979.2022.2074106
160. Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer (2019) 18(1):134. doi: 10.1158/1535-7163.TARG-19-LB-A13
161. Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget (2016) 7(49):80967–79. doi: 10.18632/oncotarget.13134
162. Hung KF, Chen BH, Wang TY, Yang YP, Chien Y, Lo JF, et al. Identification of plasma hsa_circ_0000190 and 0001649 as biomarkers for predicting the recurrence and treatment response of patients with oral squamous cell carcinoma. J Chin Med Assoc JCMA. (2022) 85(4):431–7. doi: 10.1097/JCMA.0000000000000700
163. Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles (2018) 7(1):1424473. doi: 10.1080/20013078.2018.1424473
164. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
165. Han Z, Li K, Wu J, Wang K, Qiu C, Ye H, et al. Diagnostic value of RNA for hepatocellular carcinoma: a network meta-analysis. Biomarkers Med (2021) 15(18):1755–67. doi: 10.2217/bmm-2021-0327
166. Wang Z, Sun A, Yan A, Yao J, Huang H, Gao Z, et al. Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation. Mol Cancer (2022) 21(1):92. doi: 10.1186/s12943-022-01570-4
167. Zhao J, Li XD, Wang M, Song LN, Zhao MJ. Circular RNA ABCB10 contributes to laryngeal squamous cell carcinoma (LSCC) progression by modulating the miR-588/CXCR4 axis. Aging (2021) 13(10):14078–87. doi: 10.18632/aging.203025
168. Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis (2019) 10(2):55. doi: 10.1038/s41419-018-1287-1
169. Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating wnt/β-catenin pathway. Cell Death Dis (2020) 11(12):1065. doi: 10.1038/s41419-020-03276-1
170. Zhang W, Liu T, Li T, Zhao X. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther (2021) 28(3-4):279–93. doi: 10.1038/s41417-020-00218-z
171. Wang H, Wang N, Zheng X, Wu L, Fan C, Li X, et al. Circular RNA hsa_circ_0009172 suppresses gastric cancer by regulation of microRNA-485-3p-mediated NTRK3. Cancer Gene Ther (2021) 28(12):1312–24. doi: 10.1038/s41417-020-00280-7
172. Lavenniah A, Luu TDA, Li YP, Lim TB, Jiang J, Ackers-Johnson M, et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol Ther J Am Soc Gene Ther (2020) 28(6):1506–17. doi: 10.1016/j.ymthe.2020.04.006
173. Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res (2018) 78(11):2839–51. doi: 10.1158/0008-5472.CAN-17-2808
174. Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao W, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis (2019) 10(12):900. doi: 10.1038/s41419-019-2089-9
175. Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids (2018) 13:312–21. doi: 10.1016/j.omtn.2018.09.010
176. Zhong S, Feng J. CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinf (2022) 23(1):215. doi: 10.1186/s12859-022-04705-y
177. Zhong S, Wang J, Zhang Q, Xu H, Feng J. CircPrimer: a software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinf (2018) 19(1):292. doi: 10.1186/s12859-018-2304-1
178. He Y, Huang H, Jin L, Zhang F, Zeng M, Wei L, et al. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis (2020) 11(5):358. doi: 10.1038/s41419-020-2441-0
179. Zhao X, Zhong Q, Cheng X, Wang S, Wu R, Leng X, et al. miR-449c-5p availability is antagonized by circ-NOTCH1 for MYC-induced NOTCH1 upregulation as well as tumor metastasis and stemness in gastric cancer. J Cell Biochem (2020) 121(10):4052–63. doi: 10.1002/jcb.29575
180. Chen Z, Xu W, Zhang D, Chu J, Shen S, Ma Y, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression. Mol Ther Nucleic Acids (2021) 23:1120–35. doi: 10.1016/j.omtn.2020.12.013
181. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
182. Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q, et al. ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol Cancer (2021) 20(1):142. doi: 10.1186/s12943-021-01443-2
183. Dou Z, Gao L, Ren W, Zhang H, Wang X, Li S, et al. CiRS-7 functions as a ceRNA of RAF-1/PIK3CD to promote metastatic progression of oral squamous cell carcinoma via MAPK/AKT signaling pathways. Exp Cell Res (2020) 396(2):112290. doi: 10.1016/j.yexcr.2020.112290
184. Li J, Huang C, Zou Y, Yu J, Gui Y. Circular RNA MYLK promotes tumour growth and metastasis via modulating miR-513a-5p/VEGFC signalling in renal cell carcinoma. J Cell Mol Med (2020) 24(12):6609–21. doi: 10.1111/jcmm.15308
185. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics (2018) 8(14):4003–15. doi: 10.7150/thno.24106
186. Gao F, Han J, Wang Y, Jia L, Luo W, Zeng Y. Circ_0109291 promotes cisplatin resistance of oral squamous cell carcinoma by sponging miR-188-3p to increase ABCB1 expression. Cancer Biother Radiopharmaceuticals (2022) 37(4):233–45. doi: 10.1089/cbr.2020.3928
Keywords: head and neck squamous cell carcinoma, miRNA sponge function, tumor regulation, tumor microenvironment, circular RNAs
Citation: Han X, Tian R, Wang C, Li Y and Song X (2022) CircRNAs: Roles in regulating head and neck squamous cell carcinoma. Front. Oncol. 12:1026073. doi: 10.3389/fonc.2022.1026073
Received: 23 August 2022; Accepted: 31 October 2022;
Published: 22 November 2022.
Edited by:
Susanna Chiocca, European Institute of Oncology (IEO), ItalyReviewed by:
Zhaolei Cui, Fujian Medical University, ChinaBarbara Seliger, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2022 Han, Tian, Wang, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xicheng Song, ZHJ4Y2hzb25nQDE2My5jb20=; Yumei Li, bXloZWFydDEyNjNAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xiao Han
Xiao Han Ruxian Tian
Ruxian Tian Cai Wang
Cai Wang Yumei Li
Yumei Li Xicheng Song
Xicheng Song