- Hong Kong Polytechnic University, Hong Kong, Hong Kong SAR, China
The alignment of images through deformable image registration is vital to clinical applications (e.g., atlas creation, image fusion, and tumor targeting in image-guided navigation systems) and is still a challenging problem. Recent progress in the field of deep learning has significantly advanced the performance of medical image registration. In this review, we present a comprehensive survey on deep learning-based deformable medical image registration methods. These methods are classified into five categories: Deep Iterative Methods, Supervised Methods, Unsupervised Methods, Weakly Supervised Methods, and Latest Methods. A detailed review of each category is provided with discussions about contributions, tasks, and inadequacies. We also provide statistical analysis for the selected papers from the point of view of image modality, the region of interest (ROI), evaluation metrics, and method categories. In addition, we summarize 33 publicly available datasets that are used for benchmarking the registration algorithms. Finally, the remaining challenges, future directions, and potential trends are discussed in our review.
1. Introduction
Image registration, also called image alignment, is a process of establishing spatial transformations between images (1). Image registration has wide applications in various medical image analysis and computer-assisted intervention tasks. According to the type of the transformation, it can be categorized as rigid, affine, and deformable registration (2). A rigid transformation consists of rotation and translation; an affine transformation includes translations, rotations, scaling, and sheering; the two kinds of transformations are described as a 2D single matrix. Unlike rigid and affine transformation, deformable transformation is a high-dimension problem that we need to formulate by a 3D matrix (for 2D deformable registration) or a 4D matrix (for 3D registration), i.e., deformation field. While rigid and affine registration algorithms have already achieved satisfactory performance in many applications, deformable registration is still a challenging task due to its intrinsic complexity, particularly when the deformation is large.
In clinical practice, however, deformable registration can find much more applications than rigid and affine registration. It is utilized to fuse the information that is acquired from different modalities as medical images from different modalities keep different characteristics; for example, magnetic resonance imaging (MRI) provides better contrast for soft tissues, and computed tomography (CT) has clear details of bone (3); fusing these data helps to assure better diagnosis and treatment. Even the images from one modality have distinctions when they are collected at different time points, from different views, or from various people. In this scenario, image registration is used to monitor organ or lesion variation (4).
Additionally, deformable image registration has also been utilized for various computer-assisted interventions in recent years (5–9). For transrectal ultrasound-guided (TRUS) prostate biopsy, it is the most effective way to diagnose prostate cancer, with the advantages of real-time detection, simplicity, and low cost, but for systematic sextant biopsies, its poor imaging quality and lack of sharp contrast between cancer and normal tissue results in false-negative rates of up to 30% (10). Magnetic resonance imaging (MRI), different from TRUS, is the most effective imaging technique for examining anatomical features and targeting prostate tumors. Thus, the deformable registration of pre-operative MR images and inter-operative TRUS images is utilized to fuse their information for enhancing biopsy accuracy. Moreover, registration is also of great significance in radiotherapy (11, 12); it is performed to calculate the offset of the current target from the planned position, and the offset determined from the registration is used to adjust the patient position or the radiation beam.
Deformable image registration aims to calculate the voxel-to-voxel correspondences between a moving image (i.e., source image that needs to be transformed) and a fixed image (i.e., target image used as the template). During image registration, The moving image is transformed to align with the fixed image by minimizing the dissimilarity between the fixed image and the transformed moving image.
Given the source and the target image: IS, lT∈RH×W×C , the formula is as follows:
where F represents the function space of f, Φf denotes Φ with f when the input is (IS, IT), and ℓ is the loss function to compute the discrepancy between the target image IT and the registration result T(IS|Φf). Additionally, R(Φf) is the regularization term and the hyperparameter λ is used to balance its importance on the training.
Various traditional registration methods and toolboxes have been devised over the last few decades, e.g., Elastix (13) and ANTs (14). The traditional registration algorithms are constructed as continuous optimization problems (15, 16) or discrete optimization problems (17). Their computational performance, however, is hampered by the high dimension and non-convex properties, and their capability to capture complex deformations is limited (18). Recently, deep learning-based deformable registration methods have greatly attracted researchers’ attention, as data-driven methods benefit significantly from a large number of paired/unpaired images when compared with traditional methods.
Deep learning-based models are capable of improving the deformable registration performance. Firstly, deep neural networks can prompt the iterative optimization procedure to the training stage and achieve fast inference in the test stage. Secondly, neural networks are able to work as an approximator of the similarity between the image pairs to help registration. Thirdly, the deformation field can be predicted directly through an end-to-end model without pre-defining a transformation model.
Though previous reviews about deep learning-based medical image registration literature have been published (19–22), there are still some deficiencies. On one hand, these reviews lack a summary of the public dataset for benchmarking registration algorithms. On the other hand, some details of the selected literature were missing. This review will serve as a supplement to them by adding more recent studies, by discussing the selected literature with comprehensive details, by concluding the most commonly used public dataset, and by providing some suggestions about remaining problems and further research.
Specifically, our review aims to
1. Conclude the most commonly used public datasets with details on modality, organ, dimension, quantity, disease, release time, etc.
2. Summarize literatures on deep learning-based deformable image registration, especially for recent research, and list organ, modality, dimension, model, evaluation metrics, publication source, etc. in tables.
3. Provide detailed statics on research interests (Modalities, ROIs, Evaluation metrics, and Methods).
4. Discuss the remaining issues that need to be studied and the directions for future research.
Other contents of this review are organized as follows: In Section 2, we present detailed statistics on modalities, organs, etc. Section 3 summarizes the frequently used dataset. We discuss deep learning-based deformable medical registration methods from five categories in Section 4. Then, we discuss the limitations and future potential in Section 5. Section 6 concludes the review.
2. Statistical analysis
For the purpose of this review, as completely as possible, to include relevant studies in the past decade and potentially advanced studies in the upcoming decade, this review mainly includes “deep learning”, “medical image registration”, “supervised”, “unsupervised”, “motion estimation”, “GAN”, “deep similarity”, “Transformer”, “contrastive learning”, “meta learning” and “knowledge distillation” as the search keywords. Due to the fact that medical image registration and deep learning-based methods could be involved in different conferences and journeys focusing on various specializations, the conference and journal sources of the selected papers were from but were not limited to Computer Vision and Pattern Recognition (CVPR), Medical Image Computing and Computer Assisted Interventions (MICCAI), Social Science and Management Innovation (SSMI), Medical Image Analysis (MIA), Innovative Management, Information Production (IMIP), Technology Modernization and Innovation (TMI), International Symposium on Biomedical Imaging (ISBI), Machine Learning Machine Intelligence (MLMI), and Medical Imaging With Deep Learning (MIDL). Google Scholar, arXiv, and PubMed were searched to find the targeted publications. Papers that do not notify the details about the training datasets such as not clarifying the name, the number of participants, and implementation descriptions were excluded in this review. Other papers that do not clearly state the methods and validations were also excluded in this review. After the selection and screening, 91 studies related to learning-based medical image registration were finally included. Figure 1 describes the statistical results of the studies in this review.
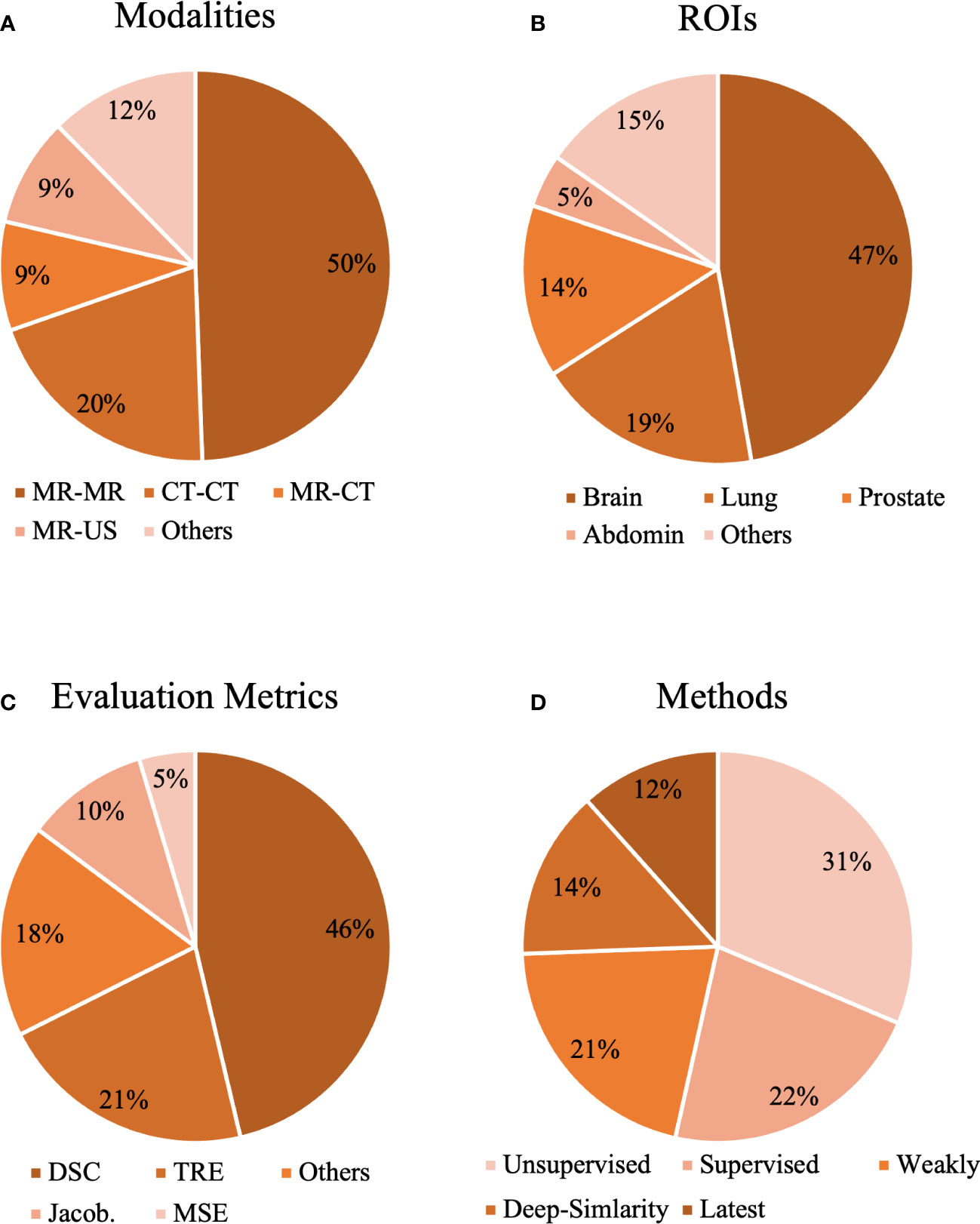
Figure 1 The statistical analysis of our selected papers from the aspects of (A) modalities, (B) organs, (C) evaluation metrics, and (D) methods.
Registration in mono-modality (MR-MR, CT-CT, etc.) and multimodality (MR-CT, MR-US, etc.) can be seen in Figure 1A. As shown in Figure 1A, MR-MR and CT-CT are the most commonly studied modalities, and each accounts for 50% and 30% of the selected studies, which might indicate that registration in mono-modality is relatively convenient and simple to implement and registration in cross-modalities encounter more challenges. Based on ROIs, we collected methods for the brain, lung, prostate, heart, liver, knee, torso, and abdomen in Figure 1B. The three most popular ROIs being investigated are brain, lung, and the prostate, and each accounts for 47%, 19%, and 14% of all the studies. The public datasets normally contain scans within these three ROIs as common diseases exist in these regions. Thus, studies utilizing public datasets could be more inclined to investigate the brain, lung, and prostate. With respect to evaluation metrics, cross-correlation (CC), mean square error (MSE), Dice coefficient (DSC), target registration error (TRE), Jacobian determinant (Jaco.Det.), and other metrics are shown in Figure 1C. The evaluation metrics such as the MSE measure the numerical difference between warped images and reference images, and other metrics such as the Jacobian determinant quantifies the smoothness of the deformation field to keep the results of the registration plausible. Based on Figure 1C, DSC is the most popular metric and accounts for 46% of all the studies. DSC would be used if the ground truth labels were provided, and this statistical analysis demonstrates the current trend for validating the accuracy of the proposed registration model that commonly requires the segmentation labels as the ground truth. This review also demonstrates the ratio of similarity-based, supervised, weakly supervised, unsupervised, and latest methods in Figure 1D. Unsupervised and supervised methods are two most popular methods and each accounts for 31% and 22% of all the studies. Although the ratio of latest methods is the smallest, this review tends to provide certain insights and suggest potential trends in developing deep learning methods of medical image registration.
3. Datasets
The absence of relevant data, as one of the main bottlenecks in current learning-based medical image analysis, resulted from several challenges including data collection with limited but subtle equipment, label annotation with human expertise, and data access under ethical considerations. Compared to commonly used datasets such as the MINST (23), qualified medical images cannot be collected from handwritten ones but need to be acquired by strict protocols and expensive scanning devices. Considering MRI method as the example, each session generally lasts 2 h under the supervision and manipulation of professional researchers, and during the scanning process, the subjects are almost fully constrained in the specialized machine in order to obtain accurate data (24). Other than sophisticated protocols to collect data mentioned above, labels and the annotation of the medical images have to be achieved by specialized professionals. Unlike datasets such as the autopilot that could be labeled by less professional annotators, in order to keep the accuracy and possibly avoid the bias of the ground truth, a single scan might be labeled by multiple experts. Finally, accessing medical data is not allowed for most researchers since the clinical data contain private information and only researchers with authorization could utilize the dataset (25).
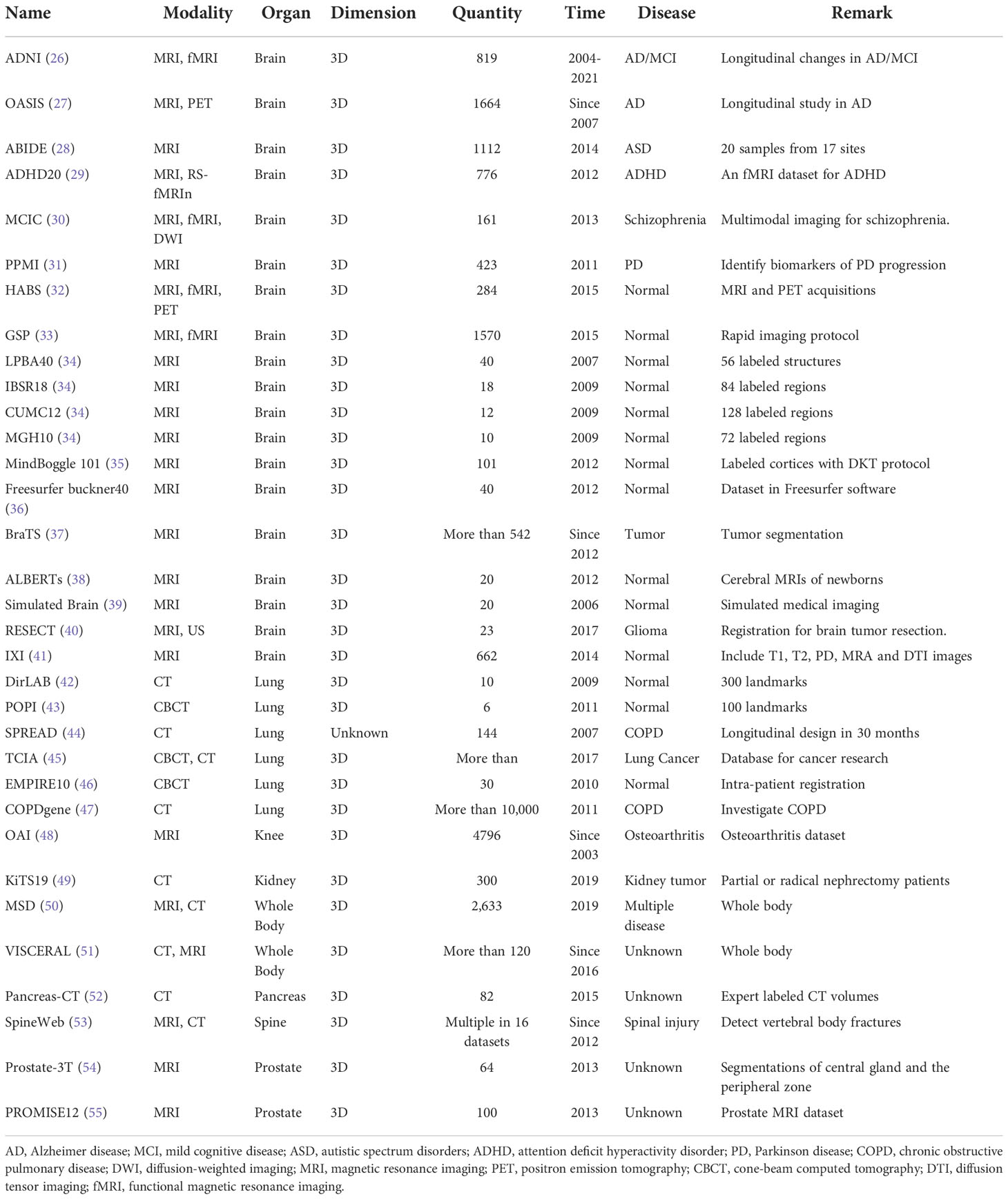
Overall, based on the characteristics of the medical dataset discussed above, prevalent and testified public databases are thus normally chosen by research teams in deep learning-based medical image analysis. Moreover, in addition to alleviating the challenges mentioned, the training and assessment of DL models on multiple public datasets could also better demonstrate the generalizability of the proposed methods and clearly compare the results on fair benchmarks, including computational time and registration accuracy. Our review summarized 30 public datasets from deep learning registration studies in Table 1. Most of the datasets consist of brain MR and lung CT images, and other modalities in different ROIs such as the abdomen and knee were also included. The ADNI dataset as one example in brain datasets contains MRI longitudinal image scans in AD patients from 63 sites across the US and Canada. ADNI develops standardized protocols for multi-center comparison and helps over 1,000 scientific publications explore the prevention and treatment of AD since 2004 and would be funded further in the future (26). COPDGene as one example in lung datasets contains chest CT scans of more than 10,000 individuals for chronic obstructive pulmonary disease (COPD). More than 375 publications have used COPDGene to explore the assessment and identify the biomarkers of COPD since 2009 (47). OAI as the example in the knee dataset contains MRI and x-ray longitudinal measurements from 4,796 subjects, and more than 400 research manuscripts have used the OAI to explore the assessments and interventions of knee osteoarthritis (48). PROMISE12 as the example in the prostate dataset is the dataset made for MICCAI prostate segmentation challenge, which was used by 11 academic teams for various segmentation algorithms and showed promising results (55).
In Table 1, the date of publication for each dataset was also listed. Some datasets are still collecting data for further imaging analysis support, such as the data collected for BraTs since 2012 (37). The quantity of each dataset demonstrates the size of each dataset, and the remarks of each dataset illustrate the significance of each dataset.
4. Methods
Figure 2 displays the chronological development of deep learning-based deformable medical image registration. In this review, deep learning-based registration methods are classified into the following four categories by evolution: Deep Iterative Methods, Supervised Methods, Unsupervised Methods, and Weakly Supervised Methods.
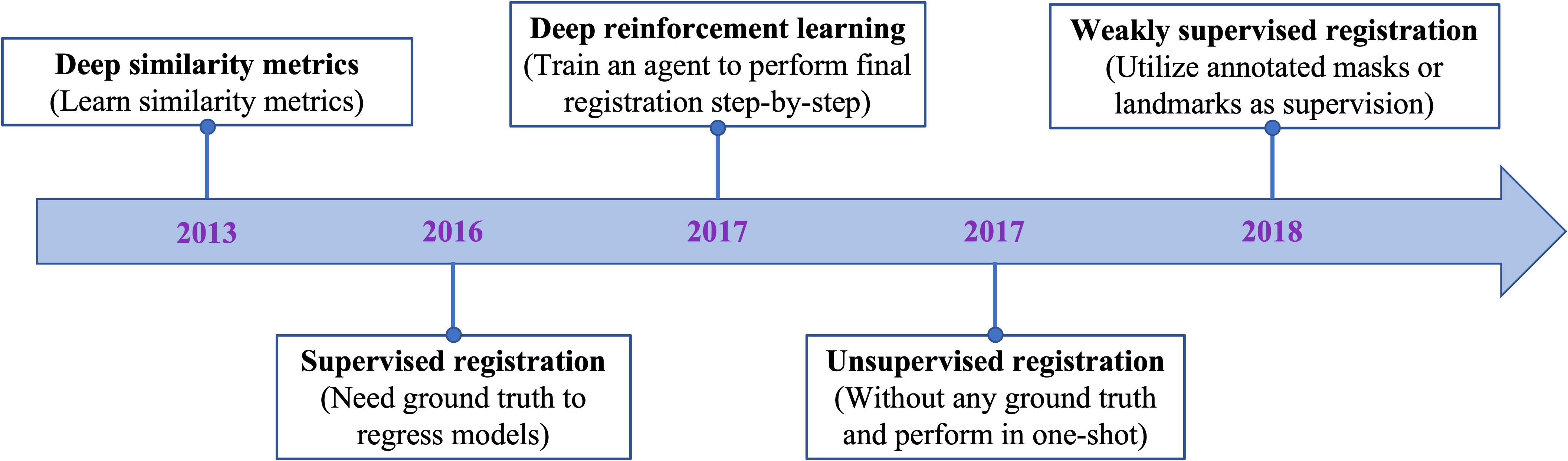
Figure 2 The evolution of deep learning-based methods on deformable medical registration from deep similarity methods to weakly supervised methods.
4.1. Deep iterative methods
Early research on deep learning-based registration directly extended the traditional registration framework by using deep neural networks as an approximator of the similarity or dissimilarity between the source image and the target image. Another early research direction is reinforcement learning. This paradigm trained an agent to perform a sequence of actions and iteratively improve image alignments by maximizing rewards. These two kinds of paradigms both adopt iterative strategy.
4.1.1. Deep similarity metrics
For traditional image registration methods, the commonly used similarity metrics are intensity-based, including mean square distance (MSD), sum-of-square distance (SSD), (normalized) mutual information (MI), and (normalized) cross-correlation (CC). Generally, these intensity-based similarity measurements work quite well for mono-modality image registration (e.g., CT-CT and MRI-MRI image registration), in which the image pair shares similar intensity distribution.
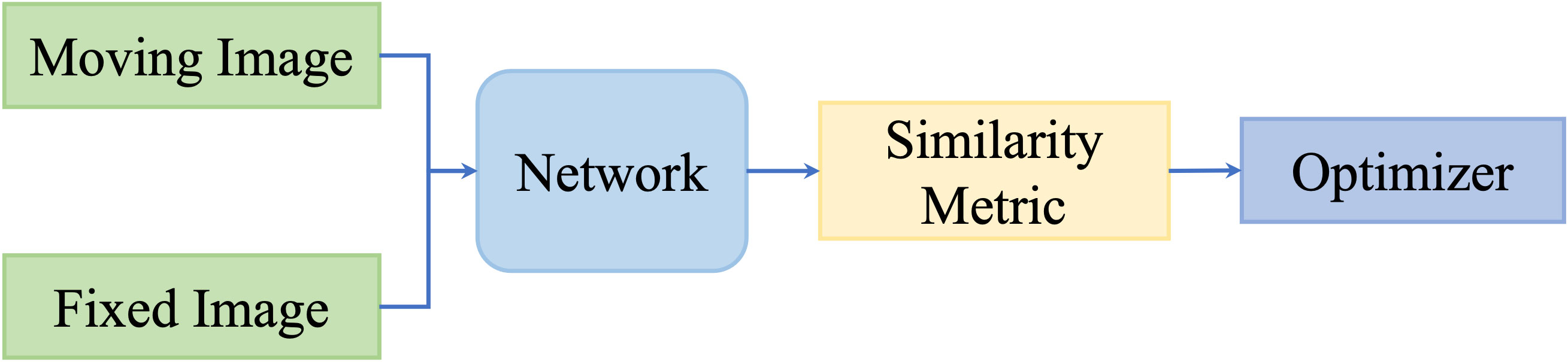
However, these metrics focus on intensity values; thus, they are not capable of measuring multimodality registration due to the diverse intensity distributions across modalities. To take advantage of deep neural networks, researchers proposed to replace the intensity-based similarity metrics with metrics that have been learned through deep networks and that achieved promising registration performance. Figure 3 shows the general pipeline of deep similarity-based registration methods.
Wu et al. (56) firstly proposed to adopt deep learning technology to obtain the similarity metric for registration. They adopted convolutional-stacked autoencoder (CAE) to extract the discriminative features for 3D deformable brain MRI registration. Then, the registration was performed by optimizing the NCC of the two features and improved registration accuracy was achieved. So et al. (57) proposed a novel learning-based metric by using Bhattacharyya Distances. The dissimilarity of the testing image pairs is calculated by incorporating the expected intensity distributions learned from the registered training image pairs. A list of research of this category is presented in Table 2.
So far, a number of registration methods for medical image based on deep similarity metrics have been studied and have shown great potential in multimodality image registration. However, a sufficient number of explicitly annotated ground truths are required for deep similarity metric training, which hinders the development of such approaches. Moreover, it is difficult to interpret the learned deep similarity metrics, and the iterative process still limits the use of these methods. Nowadays, the number of literatures for this category has decreased, and this trend is projected to continue.
4.1.2. Deep reinforcement learning
Since the initial publishing from Mnih et al. (68) and Silver et al. (69), Reinforcement Learning (RL) has gained a lot of attention and has been used for diverse applications including robotics, video games, and healthcare. In RL, an intelligent agent learns to map states to actions iteratively to maximize rewards received from a designed environment. For RL-based registration, the inputted image pairs are constructed in a given environment, and an artificial agent learns to generate the final transformation subsequently so that the rewards received from that environment can be maximized. A general pipeline of deep reinforcement learning-based deformable image registration methods is illustrated in Figure 4.
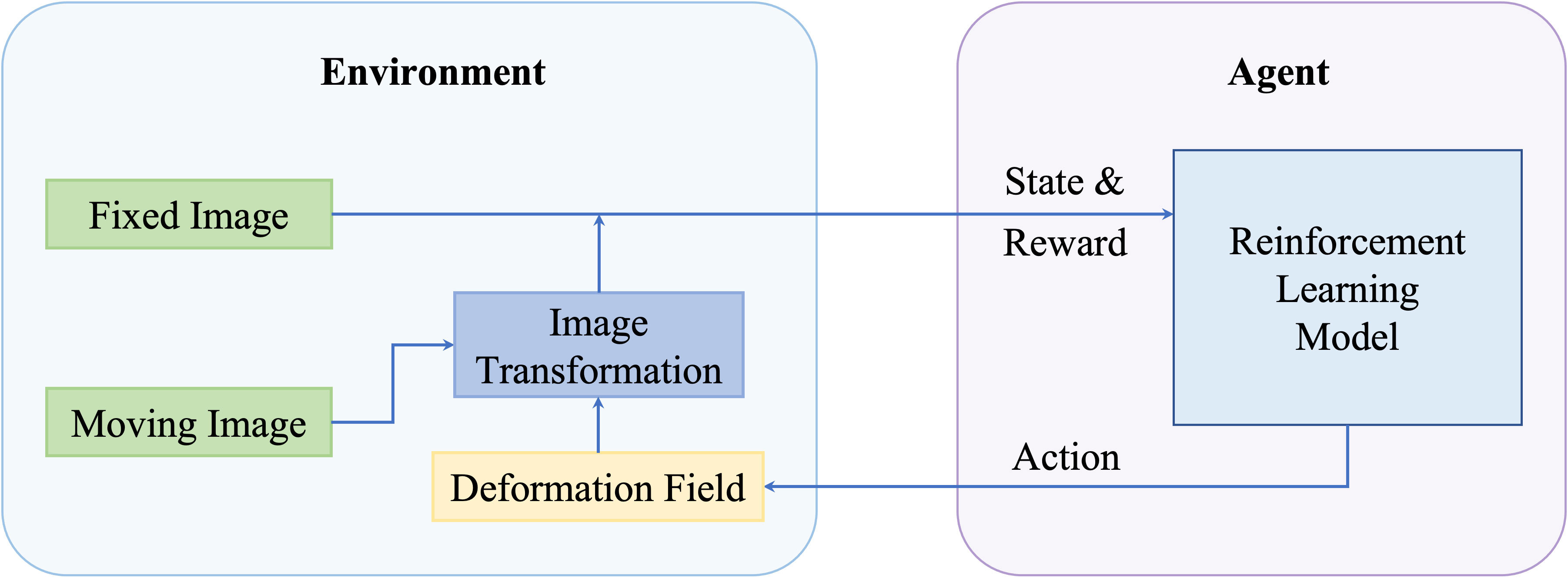
Figure 4 A general pipeline of deep reinforcement learning based deformable image registration methods.
Liao et al. (70) firstly proposed to train an artificial agent for the rigid registration of 3D CT images and cone-beam CT (CBCT) images for cardiac and abdominal images. Ma et al. (71) adopted deep reinforcement learning to extract the best feature representation to minimize the discrepancy between CT and MRI images for rigid registration. Instead, there are a few studies about RL-based algorithms on deformable registration. In 2017, Krebs et al. (72) applied RL to deformable prostate MRI registration by training an agent to investigate the parametric space of a statistical deformation model constructed with training data.
Different from deep similarity metrics-based methods, the similarity measures in these kinds of methods are routinely provided in a traditional way, e.g., MSE, NMI, or LCC. Thus, they have limited applications in multimodality registration. Moreover, RL-based methods mainly focus on rigid registration, as it is hard for agents to tackle the large state space generated from deformable vector field.
4.2. Fully supervised methods
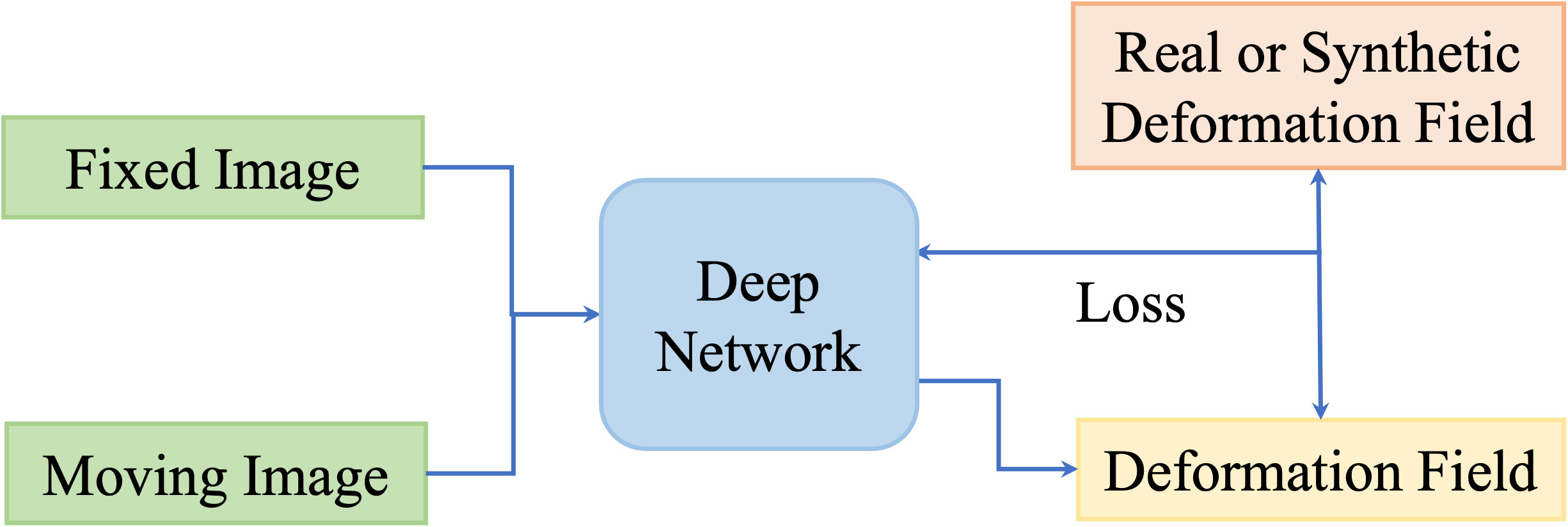
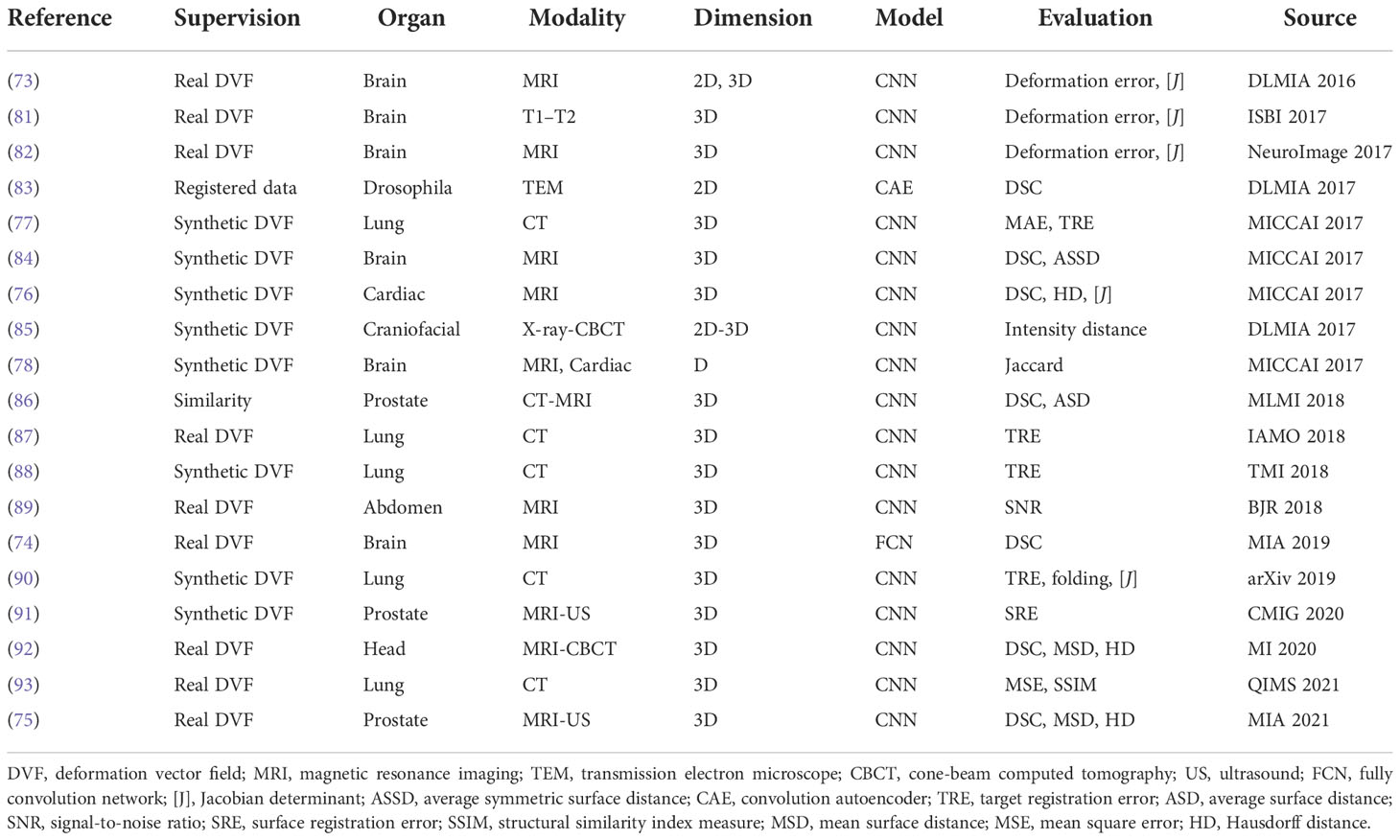
One disadvantage of deep iterative methods is that the registration process is time-consuming and iterative. Fully supervised methods help to predict transformations in one shot by the supervision of real deformation vector fields or synthetic deformation fields. The real deformation fields are generated from traditional registration, and the synthetic deformation fields are obtained from statistical models or random transformations. Figure 5 illustrates the general framework of the fully supervised methods.
At first, Yang et al. (73) proposed to register brain MRI images using a U-Net-like structure in one step. The large diffeomorphic metric mapping was utilized to generate a basis, then the original momentum values of the pixels from the images are inputted into the network, and the values are refined to predict the deformation field. Fan et al. (74) presented a Birnet for brain MRI image registration by utilizing the deformation field estimated from the traditional registration method. They also proposed gap-filling to learn more high-level characteristics and designed multi-channel inputs to learn more information. Recently, Fu et al. (75) designed an MR-TRUS registration network for prostate interventions; the supervision deformation field is generated from population-based FE models from point clouds with biomechanical constraints.
These methods have achieved notable results with real deformation fields as supervision. However, supervision by real deformation fields is limited by the size and the diversity of the dataset. Then, synthetic deformation fields are developed for the learning of deformation fields.
Rohe et al. (76) adopted a U-Net-like network to predict the deformation field for 3D cardiac MRI volume registration. The supervisions are transformations generated from mesh segmentations, and SSD between the supervision and prediction is set as the loss function. Their results are comparable with those from traditional registration. Sokooti et al. (77) presented a multi-scale network to learn a deformation field of intra-subject 3D chest CT registration. They used random DVF as supervision. Uzunova et al. (78) designed a network for the registration between 2D brain MRI and 2D cardiac MRI. Their ground truth is generated utilizing statistical appearance models (SAMs). They adapted FlowNet (79) architecture and obtained outperforming results.
The spatial transformer networks (STNs) (80) introduced in 2015 is one significant advancement that is beneficial in this era. STN is composed of three parts. The first is a localization network, whose goal is to use the input features to regress the transformation parameters. The second part is a grid generator, which generates a sampling grid that will be used to sample the input feature map. Another is a sampler that will produce the transformed feature map from the sampling grid and the input feature map by sampling and interpolation. STN can be inserted anywhere in various networks to execute a spatial transform on an input feature map because it is a completely differentiable module.
The fully supervised methods are widely studied and have notable results. A list of works about these methods are presented in Table 3. However, the generation of real or synthetic deformation fields is hard, and these deformation fields are different from the real ground truth, which will confine the accuracy and efficiency of these kinds of methods. In this situation, unsupervised methods are promising to tackle the problems.
4.3. Unsupervised methods
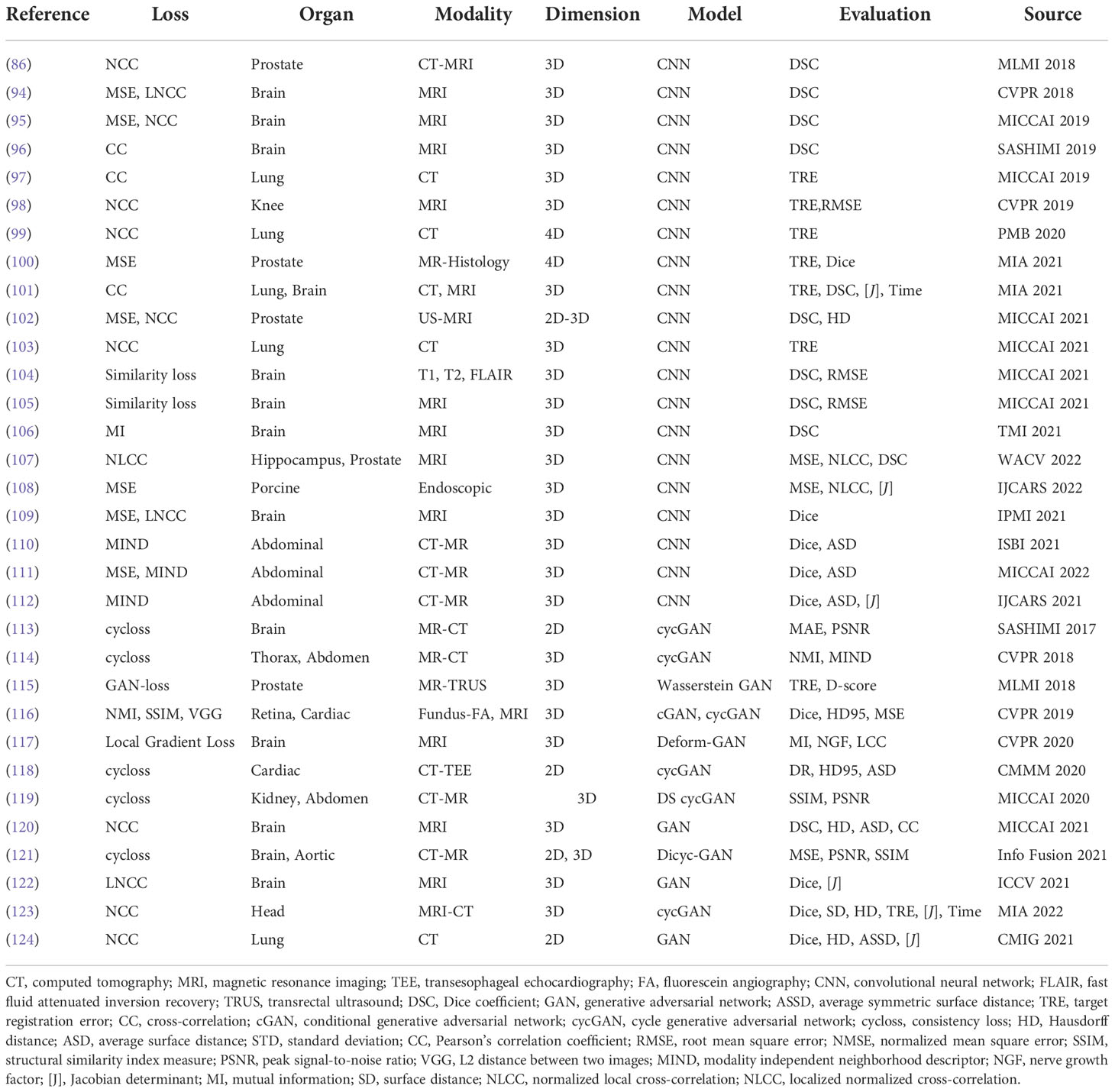
Even though different data augmentation techniques and data collection methods have been utilized in supervised learning, the preparation of the required ground truth is inconvenient and leads to the fact that the supervised framework has limitations in generalizing results in different domains and applying various registration tasks. Thus, the unsupervised registration has a more convenient training process that usually only inputs paired images without predefined DVF as the ground truth. The formulation in the loss function of the learning network constrains the output DVF to be accurate. Generally, the unsupervised learning framework consists of the similarity-based methods and GAN-based methods. Unsupervised methods are concluded in Table 4.
4.3.1. Similarity-based unsupervised methods
These kinds of methods update networks by minimizing the dissimilarity between the fixed images and the transformed moving image. An illustration of similarity-based unsupervised methods is presented in Figure 6.
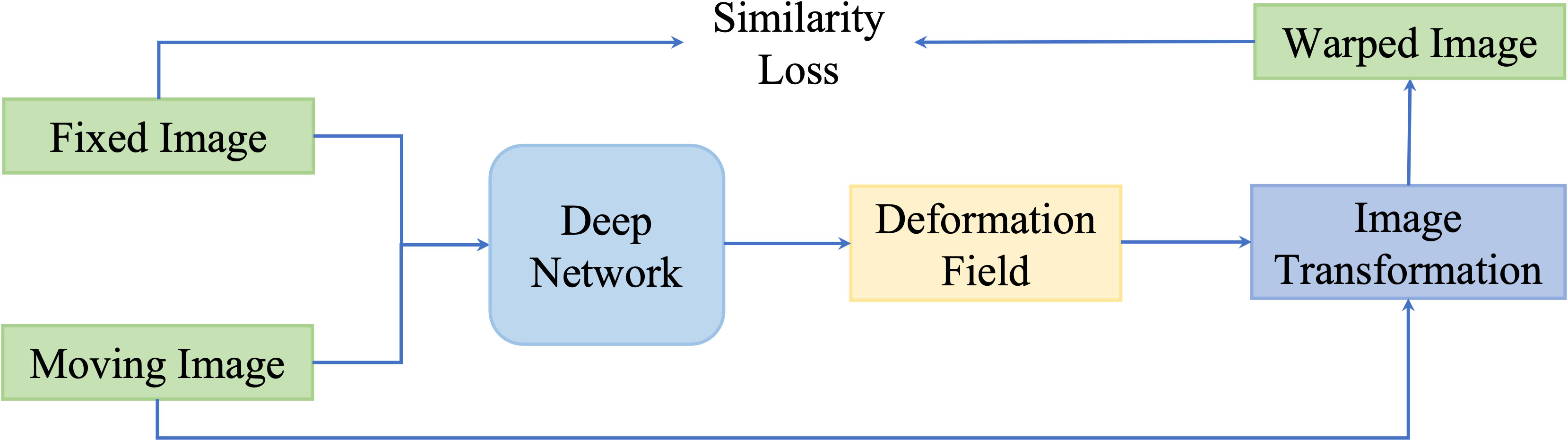
Figure 6 A general pipeline of similarity based deformable image registration methods. This network is optimized by optimizing a similarity loss function.
Balakrishnan et al. (94) designed a U-Net framework named VoxelMorph to perform DIR of brain MR images. Unlike conventional registration methods that calculate the DVF for every pair of images, they formulate the DVF as a global function that could be optimally parameterized with the trained convolutional neural network. Given the only paired images as the inputs, VoxelMorph rapidly predicts the relevant DVF and uses it to get the wrapped image. The loss function includes an unsupervised setting that minimizes the warped image and fixed image based on image intensity metrics, and an auxiliary supervised setting that minimizes the annotated segmentation errors. The proposed method performed comparably to conventional registration methods in terms of the Dice score. Estienne et al. (125) used a shared encoder with a separate decoder named U-ResNet to compute the DVF. The network inputs paired fixed and moving images and aims to output their specific segmentation maps. The registration accuracy would be optimized based on the Dice score in the segmentation results.
Not only applied in brain MR images, De et al. (126) deployed a similar unsupervised framework in cardiac MRI and chest CT. They combined the unsupervised affine and deformable registration and downsampled the input images into multiple stages, which better captures the small motions and improves the registration results. Shao et al. (127) implemented this similar coarse-to-fine registration strategy in prostate MRI images named ProsRegNet. Shen et al. (98) designed a three-phase unsupervised registration framework to calculate a transformation map for knee MRI images in a longitudinal study.
Recently, Chen et al. (105) extended this registration framework to infant tasks. They proposed an unsupervised age-conditional cerebellum atlas construction framework. Given the age input and two temporally adjacent source images, it would generate a longitudinally consistent 4D infant brain atlas with the longitudinal constraint in the loss function. Kim et al. (101) introduced the CycleMorph, which added the cycle consistency in the loss function to preserve the topology of the predicted DVF. Guo et al. (102) fused 2D TRUS image with 3D MRI volume with the frame-to-volume registration network (FVR-NET). They performed the 2D TRUS image and 3D TRUS volume registration by adopting a dual branch feature extraction module, and used the output transformation parameters to combine with the registration of 3D TRUS and 3D MRI.
Many groups have proved that their proposed similarity-based method could achieve SOTA performance compared to conventional methods; however, they mainly focus on mono-modality and the image intensity similarity metrics would be inappropriate in multimodality registration tasks.
4.3.2. GAN-based unsupervised methods
Although similarity-based unsupervised methods are not trained with existing registration transformations, analyzing transformations in terms of spatially corresponding patches could be challenging in multimodal registration and can negatively impact registration results. For overcoming issues existing in multimodality translation, generative adversarial network (GAN)-based unsupervised framework performs the training process in an adversarial setting, in which a discriminator predicts the probability that the generation of new images will match the distribution of the input training data. Figure 7 is a general framework of GAN-based deformable image registration methods.
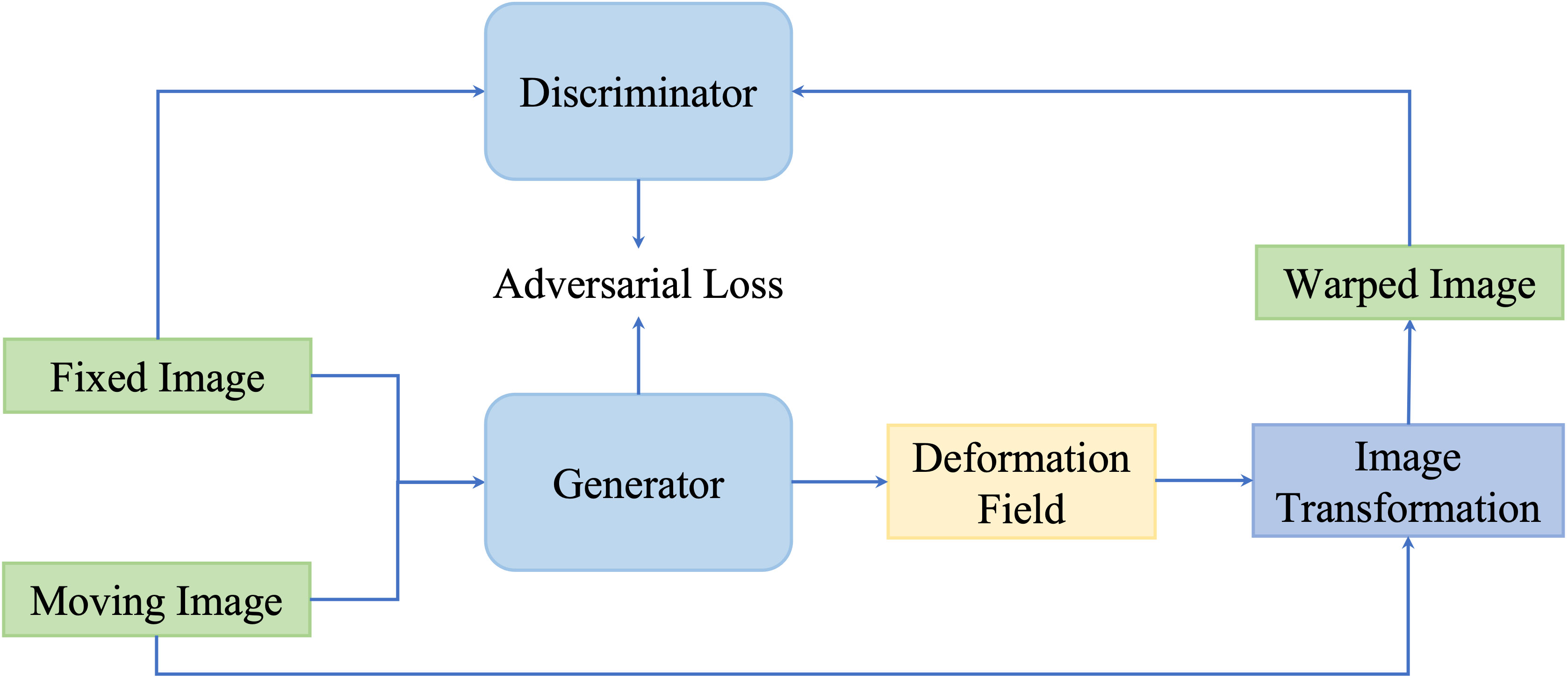
Figure 7 A general pipeline of GAN-based deformable image registration methods. This network is optimized by optimizing the adversarial loss function.
Mahapatra et al. (116) utilized both cycGAN and cGAN to predict both warped images and DVF. cGANs are used for multimodal registration so that the generated output image (the moving image after transformation) is similar to the original (based on intensity distribution) while having the same landmark locations as the reference image (from a different modality). The loss function for image generation is modified to incorporate adversarial loss and cycle consistency loss to obtain consistent and realistic deformation fields, which allow new image pairs from the untraining set to be registered. This method outperformed conventional retinal image registration with MAD, MSE, and Hausdorff distance.
The GAN framework has shown promising results in cross-modality medical image registration. Not only addressing multimodal registration, currently, more papers implement GAN in diverse registration tasks. Neel et al. claimed a GAN approach to construct the conditional deformable template across datasets from different populations. Despite GAN showing great potential in its generative and discriminative features, the validation of output warped image accuracy is still required to be investigated, especially in unpaired images with minor abnormality regions.
4.4. Weakly supervised methods
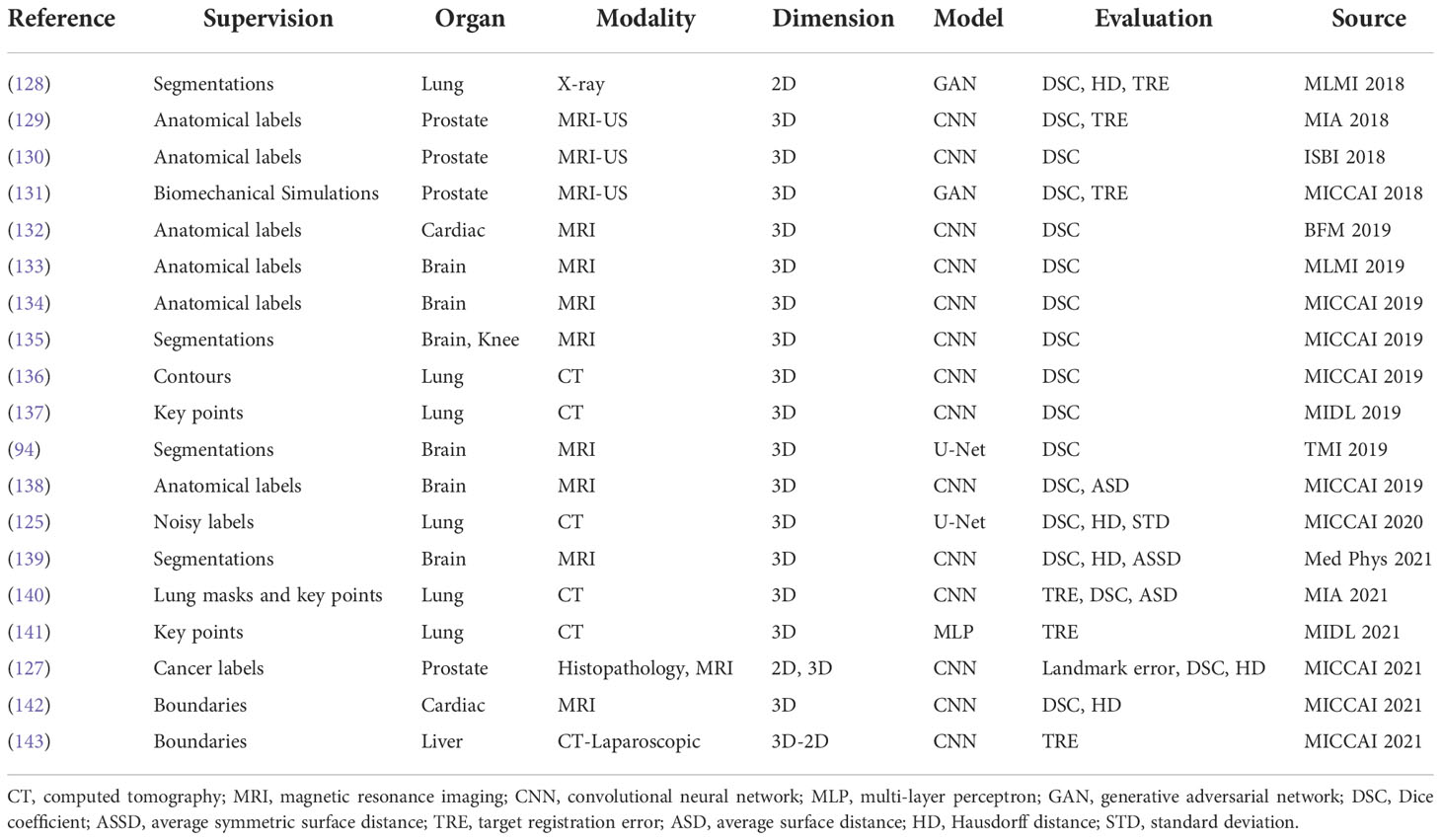
Neither using real or synthetic deformation vector field as supervision nor directly predicting deformation field without supervision, weakly supervised methods use labels as supervision, for example, segmentations and key points; such anatomical labels are feasible in practice and credible for training. Figure 8 is a general framework of weakly supervised deformable image registration methods. Generally, these networks are trained by minimizing anatomical losses, which ensures that the predicted segmentations match the anatomical labels and provide anatomy consistency monitoring. Weakly supervised methods naturally improve the performance of registration by introducing the anatomical constraints. A conclusion of weakly supervised methods is presented in Table 5.
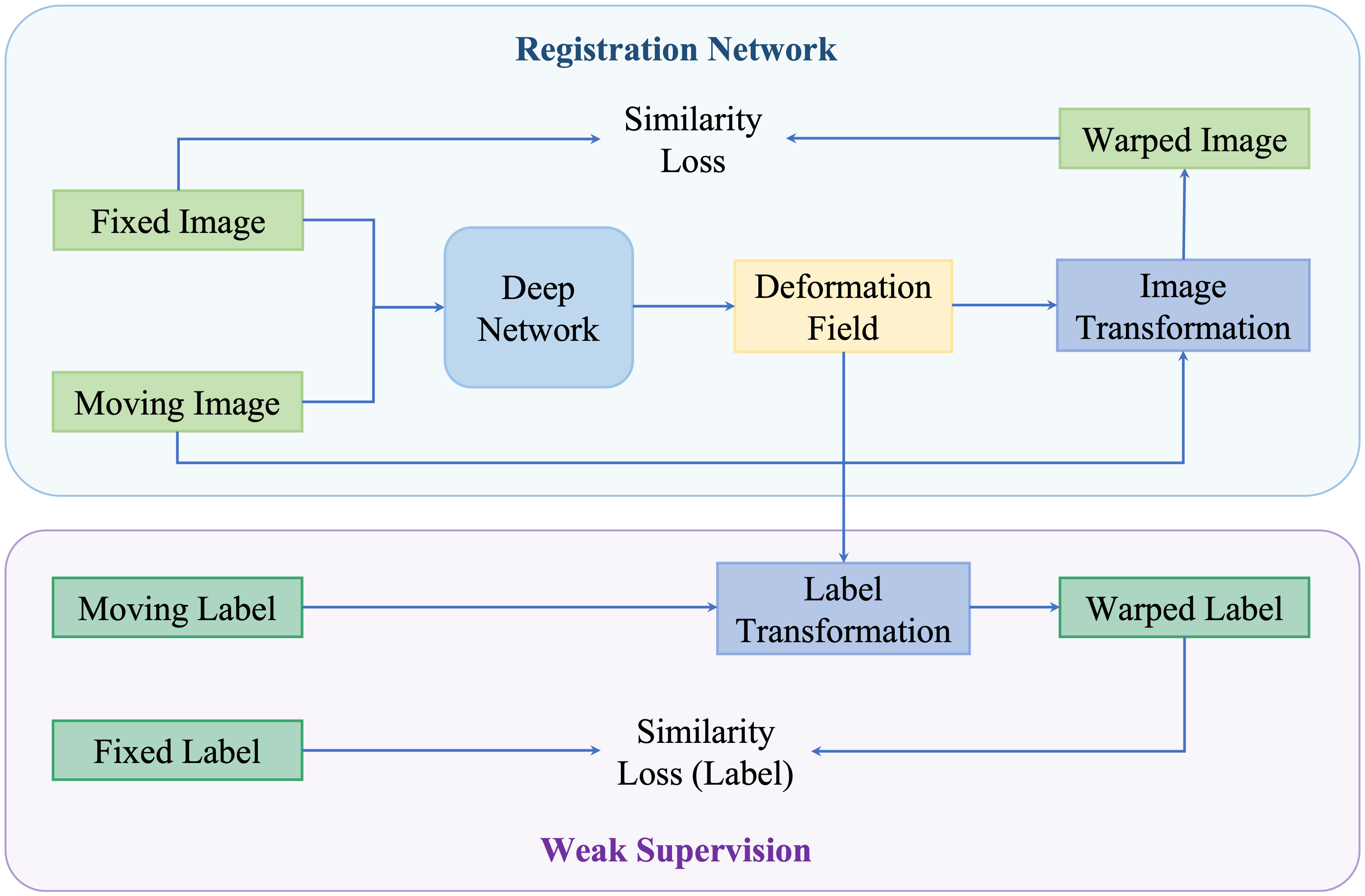
Figure 8 An illustration of weakly supervised deformable registration methods. The weak supervision is achieved by adding anatomical labels in the network.
Hu et al. (129) presented an end-to-end convolutional neural network to inference deformation field for 3D MRI and US image registration. Image pairs with multiple labeled corresponding structures were used for the training, and only unlabeled image pairs are used for testing. Xu et al. (135) proposed DeepAtlas, a network for weakly supervised registration and semi-supervised segmentation. The networks were trained jointly through an anatomy similarity loss that penalized the difference between the transformed segmentation of the moving image and the target image’s segmentation. Estienne et al. (125) developed a registration network for abdominal CT registration by applying spatial gradients and noisy segmentation labels. Recently, Hering et al. (140) proposed a multi-level framework for lung CT image registration by introducing the constraints of lung masks and key points to make the airways and arteries more aligned.
Furthermore, image registration and image segmentation are inextricably linked and can help one another. On one hand, labeled atlas images can be utilized for segmentation through image registration. On the other hand, segmentations are capable of adding extra anatomical constraints for image registration, and segmentations are also useful in evaluating the registration algorithms. Therefore, this weakly supervised paradigm is applicable in medical image segmentation, such as the DeepAtlas described in (135) and the method (144).
4.5. Latest methods/recent directions
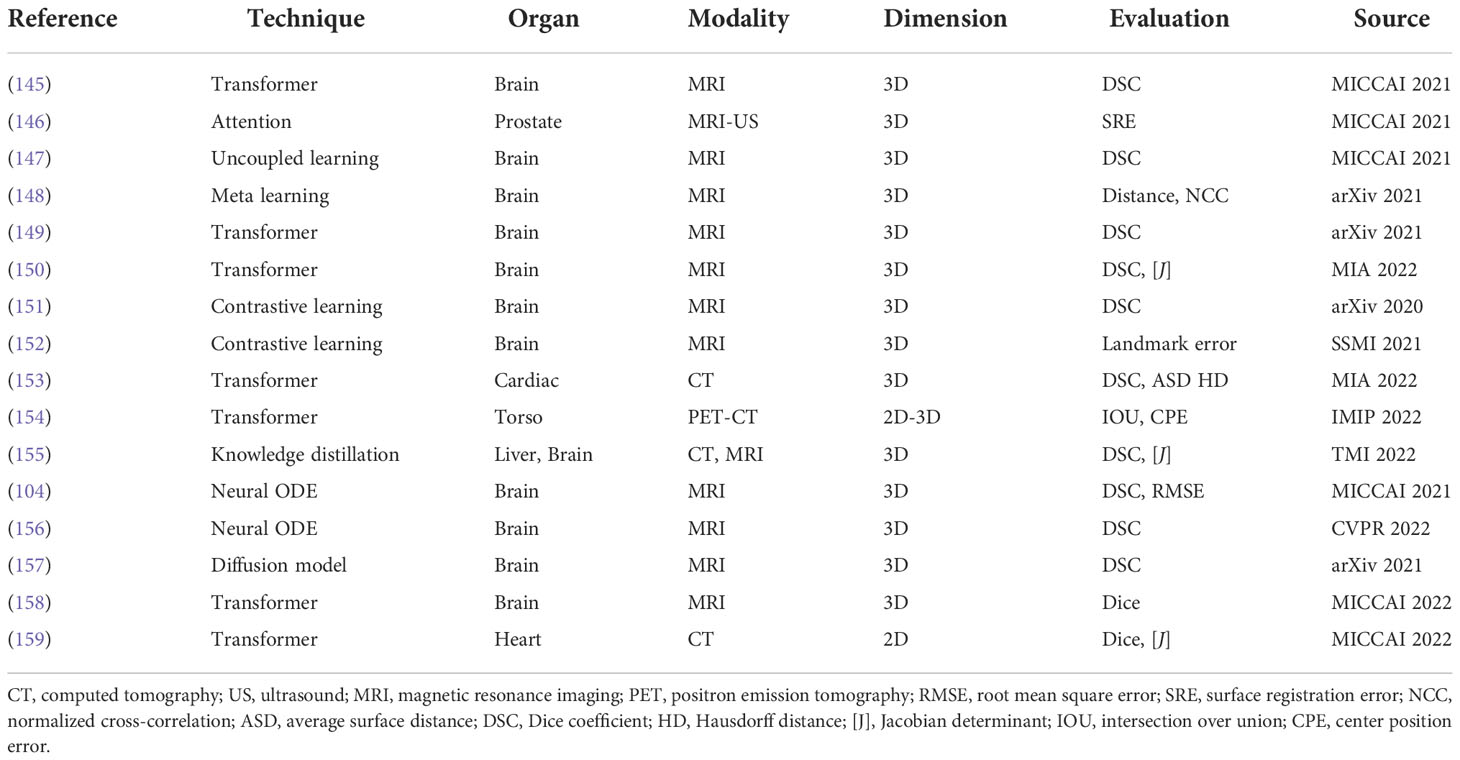
The latest deformable medical image registration algorithms are listed in Table 6; these methods adopted recent deep learning techniques, such as Transformer (160), contrastive learning (161), meta learning (162), neural ODE (163), and the diffusion model (164).
Transformer (160) is the most popular technique in medical image registration. Chen et al. (149) designed a ViT-V-Net for brain MRI image registration. They adopted a hybrid ConvNet-and-Transformer architecture to apply ViT for high-level feature learning. Their experimental results proved that simply replacing the network backbone of VoxelMorph by Vit-V-Net could improve the performance. They also extended the ViT-V-Net and presented TransMorph (150), a hybrid Transformer-ConvNet framework. In this framework, they employed the Swin Transformer (165) as the encoder to learn the spatial transformation between the input images. Then, a decoder constructed with ConvNet processed the features from the Transformer encoder and exported the dense deformation field. To provide a smooth and topology-preserving deformation field, they also presented diffeomorphic variations of TransMorph.
For contrastive learning (161), except for the application in brain MRI registration, Jiang et al. (166) developed a network to generate pseudo-CT from MRI for brain radiotherapy based on a contrastive unpaired translation network (CUT) (167). Comparing to GAN-based generation methods, their network can capture more structure and texture information that is useful to generate more realistic CT images.
Neural ODE (163) is developed to depict more complex dynamic systems. Compared to well-known deep learning models like ResNet and U-Net, neural ODE models are more efficient in terms of memory and parameters. Neural ODE models have the advantage of adaptive computing, making them potentially appropriate for application in medical applications. Additionally, the dynamics of optimization are naturally continuous. These benefits encourage researchers to study how to use neural ODEs to optimize the registration of medical images. Xu et al. (104) proposed to formulate the traditional optimization strategy in registration methods as a continuous mechanism and learn the optimizer through a multi-scale neural ODE model. Wu et al. (156) presented a novel and accurate diffeomorphic image registration framework using Neural ODES and explored the potential of combining the advantages of neural networks and flow formulations. In this work, every voxel was portrayed as a moving particle, and the entire collection of voxels was regarded as a high-dimensional dynamical system, with each voxel’s trajectory determining the appropriate deformation field.
5. Discussion
From 2013, researchers began to apply deep learning techniques to image registration, and the applications of deep models for registration flourished from 2017. Deep learning-based methods have shown high computational efficiency and comparable accuracy compared with traditional methods. Deep similarity-based models and reinforcement learning-based methods adopt iterative strategy, which is time-consuming. Supervised methods need ground truth supervision that is impractical to obtain. In contrast, unsupervised methods and weakly supervised methods are less reliant on ground truth information and become hot topics for deep learning-based registration algorithms. Recently, popular networks, such as Transformer and contrastive learning, are explored for deformable medical image registration and achieved promising results.
Complexity is an important factor that needs to be considered when designing the registration networks. Parameters and floating point operations (FLOPs) are common criteria used to measure the complexity of the model. In view of the fact that the calculation of FLOPs involves the size of input images, we here discuss the model complexity in terms of parameters. Due to the large number of variants of network architecture, we provide approximate model parameters of the backbone networks that are commonly used for deformable registration in Table 7. We can see that GAN has the smallest model parameter and the parameter of Transformer expands significantly. Expect the backbone parameters, additional modules and branches also add the complexity of the registration models.
In conjunction with the previous discussion about complexity, here we discuss how to choose a registration network for a specific task from different categories. Extra label is the first factor that needs to be considered; if the label is available, then the semi-supervised methods are preferable and have the potential for high registration accuracy. The second factor for choosing a network is the data size and GPU memory; the Transformer is unavailable when the data size is large and GPU memory is limited. For multimodality registration, GAN-based methods are suitable as GAN can ensure that the generated registered image has the same characteristic (intensity distribution) as the source image while being similar to the target image in terms of structures. The supervised methods are applicable when the ground truth deformation vector fields are provided or they are easy to obtain. Lastly, the unsupervised similarity-based methods have become more popular in recent years, and they are appropriate in many deformable registration tasks as they do not require extra information.
Here, we discuss the remaining issues that need to be studied and potential directions for future research on deformable image registration:
1. Registration Models. CNNs, SAEs, GANs, DRLs, and deep RNNs make up the majority of the recently developed deep learning algorithms used for medical image registration, whereas other models also offer a significant potential for advancement. In contrast to the medical image areas, we think that the majority of future trends and contributions will originate from other subjects, such as computer vision and machine learning. Nowadays, the popular neural ODE and diffusion models have also been explored for deformable image registration. In the future, models in other areas may also have the potential to be used for deformable medical image registration.
2. Diffeomorphic Registration. Due to characteristics like topology preservation and transformation invertibility, diffeomorphic image registration using deep models has caught the attention of researchers. In deep learning-based deformable registration methods, two main strategies are proposed to guarantee the diffeomorphism of the deformation fields. The first strategy is to add an explicit constraint (regularization) for the learned deformation field. Usually, the constraints are performed by penalizing the small and large values of the Jacobian determinant. The other method is to introduce diffeomorphic integration layers. The integration is performed by scaling and squaring the stationary velocity field. However, there are still some limitations, and the guarantee for diffeomorphism in learning-based methods remains a challenging problem. Therefore, developing deformable registration networks that can guarantee the diffeomorphism of the deformation field is a prospective research topic.
3. Registration Efficiency. From the point of view of registration efficiency, the training time and network parameters still need promotion, especially for 3D image registration. For example, when training 3D lung CT registration network, a GPU card with a memory larger than 12 G is necessary. Thus, a lightweight network for 3D image registration is also a potential research direction. Considering the applications in computer-assisted surgeries, a shorter training time is preferable, so networks that can converge rapidly are appealing.
4. Large Deformation of Soft Tissues. The learning of large and irregular deformation caused by organ movement (e.g., lung respiratory motion) is still a challenging problem and needs further research. In learning-based registration methods, one strategy to address the large deformation is to adopt multi-stage coarse-to-fine architecture. This kind of method consumes large GPU memory and the training is time-consuming as the networks are trained separately. Another method for learning large deformation is to construct cascaded networks, in which each cascaded network learns an intermediate deformation field, and the source image is recursively and progressively warped by the field, finally aligned to the target image. This kind of network is still memory-consuming and is difficult to converge as all cascaded networks are trained simultaneously. Therefore, learning of large deformation remains an outstanding issue and efforts are needed.
5. Registration Constraints. Predicting the deformation field without constraints can lead to warped moving image with distorted unrealistic organ appearances. The most commonly used technique for tackling this issue is to add the L2 norm on the gradient of the deformation field to regularize the predicted deformation. The magnitude of the field might be restricted by the employment of such regularization terms. Considering this, adding anatomical constraints to deep networks can help to generate realistic deformations. More importantly, applying appropriate constraints can help the network learn a deformation field that keeps the topology of the input image pairs so that the registration is more reliable in clinical applications. Thus, exploring constraints for particular tasks is another attractive research point.
6. Conclusion
We provide a comprehensive survey for the development of deep learning-based medical image registration methods in this article. We also have a thorough analysis of publicly available datasets as well as their details in order to assist algorithm benchmarking and future studies. The evolution of learning-based image registration algorithms has followed a similar path to that of the deep learning models. Image registration neural networks are gradually moving from handling 2D images to 3D or 4D (dynamic) volumes, and converting from supervised methods to unsupervised methods and weakly supervised methods. Recent advances are also reviewed, including those methods that adopt Transformer, contrastive learning, and other latest techniques. We also present the statistical analysis of our selected papers from the aspects of modalities, organs, evaluation metrics, and supervision. Future research challenges and directions are also discussed, including how to speed up registration in higher dimensions, reduce the requirement for ground truth during training, and use anatomical constraints to produce deformation fields that are more realistic while retaining anatomical consistency.
Author contributions
JZ and BG conducted literature research and analysis. The introduction, methods, discussion, and conclusion parts of this manuscript were written by JZ. The statistical analysis, datasets, and GAN-based methods were written by BG. YS and JQ revised the paper. All authors contributed to the article and approved the submitted version.
Acknowledgments
The work described in this paper is supported by two General Research Funds of Hong Kong Research Grants Council (project no. 15205919 and 15218521).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zitova B, Flusser J. Image registration methods: A survey. Image Vision Computing (2003) 21(11) 977–1000. doi: 10.1016/S0262-8856(03)00137-9
2. Pluim JP, Fitzpatrick JM. Image registration. IEEE Trans Med Imaging (2003) 22(11):1341–3. doi: 10.1109/TMI.2003.819272
3. Huang B, Yang F, Yin M, Mo X, Zhong C. A review of multimodal medical image fusion techniques. Comput Math Methods Med (2020) 2020. doi: 10.1155/2020/8279342
4. Gao Z, Gu B, Lin J. Monomodal image registration using mutual information based methods. Image Vision Computing (2008) 26(2):164–73. doi: 10.1016/j.imavis.2006.08.002
5. Reaungamornrat S, Liu W, Wang A, Otake Y, Nithiananthan S, Uneri A, et al. Deformable image registration for cone-beam ct guided transoral robotic base-of-tongue surgery. Phys Med Biol (2013) 58(14):4951. doi: 10.1088/0031-9155/58/14/4951
6. Oh S, Kim S. Deformable image registration in radiation therapy. Radiat Oncol J (2017) 35(2):101. doi: 10.3857/roj.2017.00325
7. Rigaud B, Simon A, Castelli J, Lafond C, Acosta O, Haigron P, et al. Deformable image registration for radiation therapy: Principle, methods, applications and evaluation. Acta Oncol (2019) 58(9):1225–37. doi: 10.1080/0284186X.2019.1620331
8. Chen A, Hsu S, Lamb J, Yang Y, Agazaryan N, Steinberg M, et al. Mri-guided radiotherapy for head and neck cancer: Initial clinical experience. Clin Trans Oncol (2018) 20(2):160–8. doi: 10.1007/s12094-017-1704-4
9. Tam AL, Lim HJ, Wistuba II, Tamrazi A, Kuo MD, Ziv E, et al. Image-guided biopsy in the era of personalized cancer care: proceedings from the society of interventional radiology research consensus panel. J Vasc Interventional Radiol: JVIR (2016) 27(1):8. doi: 10.1016/j.jvir.2015.10.019
10. Norberg M, Egevad L, Holmberg L, Sparen P, Norlen B, Busch C. The sextant protocol for ultrasound-guided core biopsies of the prostate underestimates the presence of cancer. Urology (1997) 50(4):562–6. doi: 10.1016/S0090-4295(97)00306-3
11. Krilavicius T, Zliobaite I, Simonavicius H, Jaruevicius L. (2016). Predicting respiratory motion for real-time tumour tracking in radiotherapy, in: IEEE 29th International Symposium on Computer-Based Medical Systems (CBMS), (Belfast and Dublin, Ireland: IEEE). pp. 7–12.
12. Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the aapm radiation therapy committee task group no. 132. Med Phys (2017) 44(7):e43–76. doi: 10.1002/mp.12256
13. Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging (2009) 29(1):196–205. doi: 10.1109/TMI.2009.2035616
14. Avants BB, Tustison N, Song G, Avants BB, Tustison N, Johnson H. Advanced normalization tools (ants). Insight J (2009) 29(1):1–35. Available at: https://scicomp.ethz.ch/public/manual/ants/2.x/ants2.pdf
15. Thirion JP. Image matching as a diffusion process: an analogy with maxwell’s demons. Med Image Anal (1998) 2(3):243–60. doi: 10.1016/S1361-8415(98)80022-4
16. Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast mr images. IEEE Trans Med Imaging (1999) 18(8):712–21. doi: 10.1109/42.796284
17. Heinrich MP, Jenkinson M, Papież BW, Brady SM, Schnabel JA. (2013). Towards realtime multimodal fusion for image-guided interventions using self-similarities, in: International conference on medical image computing and computer-assisted intervention– MICCAI 2013. Berlin Heidelberg: Springer-Verlag 18(8):187–94.
18. Schnabel JA, Heinrich MP, Papież BW, Brady JM. Advances and challenges in deformable image registration: From image fusion to complex motion modelling. Medical Image Analysis (2016) 33:145–8. doi: 10.1016/j.media.2016.06.031
19. Boveiri HR, Khayami R, Javidan R, Mehdizadeh A. Medical image registration using deep neural networks: A comprehensive review. Medical Image Analysis (2020) 87. doi: 10.1016/j.compeleceng.2020.106767
20. Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: A review. Phys Med Biol (2020) 65(20):20TR01. doi: 10.1088/1361-6560/ab843e
21. Haskins G, Kruger U, Yan P. Deep learning in medical image registration: A survey. Mach Vision Appl (2020) 31(1):1–18. doi: 10.1007/s00138-020-01060-x
22. Xiao H, Teng X, Liu C, Li T, Ren G, Yang R, et al. A review of deep learning-based three-dimensional medical image registration methods. Quantitative Imaging Med Surg (2021) 11(12):4895. doi: 10.21037/qims-21-175
23. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE (1998) 86(11):2278–324. doi: 10.1109/5.726791
24. Young DW. What does an mri scan cost. Healthc Financ Manage (2015)69(11) 46–9. Available at: https://www.researchgate.net/profile/David-Young-8/publication/353123295_MRICost/links/60e8549cb8c0d5588ce61127/MRICost.pdf
25. Li J, Zhu G, Hua C, Feng M, Li P, Lu X, et al. A systematic collection of medical image datasets for deep learning. arXiv (2021). doi: 10.48550/arXiv.2106.12864
26. Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The alzheimer’s disease neuroimaging initiative (adni): Mri methods. J Magnetic Resonance Imaging: Off J Int Soc Magnetic Resonance Med (2008) 27(4):685–91. doi: 10.1002/jmri.21049
27. Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies (oasis): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci (2007) 19(9):1498–507. doi: 10.1162/jocn.2007.19.9.1498
28. Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry (2014) 19(6):659–67. doi: 10.1038/mp.2013.78
29. consortium A. The adhd-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci (2012) 6(62):62. doi: 10.3389/fnsys.2012.00062
30. Gollub RL, Shoemaker JM, King MD, White T, Ehrlich S, Sponheim SR, et al. The mcic collection: A shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics (2013) 11(13):367–88. doi: 10.1007/s12021-013-9184-3
31. Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, et al. The parkinson progression marker initiative (ppmi). Prog Neurobiol (2011) 95(4):629–35. doi: 10.1016/j.pneurobio.2011.09.005
32. Dagley A, LaPoint M, Huijbers W, Hedden T, McLaren DG, Chatwal JP, et al. Harvard Aging brain study: Dataset and accessibility. Neuroimage (2017) 144:255–8. doi: 10.1016/j.neuroimage.2015.03.069
33. Holmes AJ, Hollinshead MO, O’keefe TM, Petrov VI, Fariello GR, Wald LL, et al. Brain genomics superstruct project initial data release with structural, functional, and behavioral measures. Sci Data (2015) 2(1):1–16. doi: 10.1038/sdata.2015.31
34. Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain mri registration. Neuroimage (2009) 46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037
35. Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci (2012) 6:171. doi: 10.3389/fnins.2012.00171
36. FreeSurfer FB. Freesurfer. NeuroImage (2012) 62(2):774–81. doi: 10.1016/j.neuroimage.2012.01.021
37. Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv (2018). doi: 10.48550/arXiv.1811.02629
38. Gousias IS, Edwards AD, Rutherford MA, Counsell SJ, Hajnal JV, Rueckert D, et al. Magnetic resonance imaging of the newborn brain: Manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage (2012) 62(3):1499–509. doi: 10.1016/j.neuroimage.2012.05.083
39. Aubert-Broche B, Griffin M, Pike GB, Evans AC, Collins DL. Twenty new digital brain phantoms for creation of validation image data bases. IEEE Trans Med Imaging (2006) 25(11):1410–6. doi: 10.1109/TMI.2006.883453
40. Xiao Y, Fortin M, Unsgård G, Rivaz H, Reinertsen I. Re trospective evaluation of cerebral tumors (resect): A clinical database of pre-operative mri and intra-operative ultrasound in low-grade glioma surgeries. Med Phys (2017) 44(7):3875–82. doi: 10.1002/mp.12268
41. Roy AG, Conjeti S, Sheet D, Katouzian A, Navab N, Wachinger C. (2017). Error corrective boosting for learning fully convolutional networks with limited data, in: International Conference on Medical Image Computing and Computer-Assisted Intervention– MICCAI 2017, (Springer International Publishing AG):231–9.
42. Castillo R, Castillo E, Guerra R, Johnson VE, McPhail T, Garg AK, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol (2009) 54(7):1849. doi: 10.1088/0031-9155/54/7/001
43. Vandemeulebroucke J, Rit S, Kybic J, Clarysse P, Sarrut D. Spatiotemporal motion estimation for respiratory-correlated imaging of the lungs. Med Phys (2011) 38(1):166–78. doi: 10.1118/1.3523619
44. Stolk J, Putter H, Bakker EM, Shaker SB, Parr DG, Piitulainen E, et al. Progression parameters for emphysema: A clinical investigation. Respir Med (2007) 101(9):1924–30. doi: 10.1016/j.rmed.2007.04.016
45. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The cancer imaging archive (tcia): Maintaining and operating a public information repository. J digital Imaging (2013) 26(6):1045–57. doi: 10.1007/s10278-013-9622-7
46. Murphy K, Van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, et al. Evaluation of registration methods on thoracic ct: The empire10 challenge. IEEE Trans Med Imaging (2011) 30(11):1901–20. doi: 10.1109/TMI.2011.2158349
47. Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of copd (copdgene) study design. COPD: J Chronic Obstructive Pulmonary Dis (2011) 7(1):32–43. doi: 10.3109/15412550903499522
48. Eckstein F, Kwoh CK, Link TM. Imaging research results from the osteoarthritis initiative (oai): A review and lessons learned 10 years after start of enrolment. Ann rheumatic Dis (2014) 73(7):1289–300. doi: 10.1136/annrheumdis-2014-205310
49. Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, et al. The kits19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. arXiv (2019). doi: 10.48550/arXiv.1904.00445
50. Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, Van Ginneken B, et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv (2019). doi: 10.48550/arXiv.1904.00445
51. Jimenez-del Toro O, Müller H, Krenn M, Gruenberg K, Taha AA, Winterstein M, et al. Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: Visceral anatomy benchmarks. IEEE Trans Med Imaging (2016) 35(11)2459–75. doi: 10.1109/TMI.2016.2578680
52. Roth HR, Lu L, Farag A, Shin HC, Liu J, Turkbey EB, et al. (2015). Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2015, (Springer International Publishing Switzerland 2015) pp. 556–64.
53. Yao J, Burns JE, Munoz H, Summers RM. (2012). Detection of vertebral body fractures based on cortical shell unwrapping, in: International Conference on Medical Image Computing and Computer-Assisted Intervention- MICCAI 2012, (Berlin Heidelberg: Springer Verlag):509–16. doi: 10.1016/j.media.2013.12.002
54. Litjens G, Futterer J, Huisman H. Data from prostate-3t: the cancer imaging archive. (2015). (30 May 2022). Available at: https://wiki.cancerimagingarchive.net/display/Public/Prostate-3T
55. Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, et al. Evaluation of prostate segmentation algorithms for mri: The promise12 challenge. Med Image Anal (2014) 18(2):359–73. doi: 10.1016/j.media.2013.12.002
56. Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D. Unsupervised deep feature learning for deformable registration of mr brain images. International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer) (2013) 18(2) p. 649–56.
57. So RW, Chung AC. A novel learning-based dissimilarity metric for rigid and non-rigid medical image registration by using bhattacharyya distances. Pattern Recognition (2017) 62:161–74. doi: 10.1016/j.patcog.2016.09.004
58. Zhao L, Jia K. Deep adaptive log-demons: Diffeomorphic image registration with very large deformations. Comput Math Methods Med (2015) 2015. doi: 10.1155/2015/836202
59. Wu G, Kim M, Wang Q, Munsell BC, Shen D. Scalable high-performance image registration framework by unsupervised deep feature representations learning. IEEE Trans Biomed Eng (2015) 63(7):1505–16. doi: 10.1109/TBME.2015.2496253
60. Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N. (2016). A deep metric for multimodal registration, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2016, (Springer International Publishing AG 2016) . pp. 10–8. doi: 10.1109/TBME.2015.2496253
61. Yang X, Han X, Park E, Aylward S, Kwitt R, Niethammer M. (2016). Registration of pathological images, in: International Workshop on Simulation and Synthesis in Medical Imaging, (Springer International Publishing AG 2016) . pp. 97–107.
62. Ghosal S, Ray N. Deep deformable registration: Enhancing accuracy by fully convolutional neural net. Pattern Recognition Lett (2017) 94:81–6. doi: 10.1016/j.patrec.2017.05.022
63. Zhu X, Ding M, Huang T, Jin X, Zhang X. Pcanet-based structural representation for nonrigid multimodal medical image registration. Sensors (2018) 18(5):1477. doi: 10.3390/s18051477
64. Ferrante E, Dokania PK, Silva RM, Paragios N. Weakly supervised learning of metric aggregations for deformable image registration. IEEE J Biomed Health Inf (2018) 23(4):1374–84. doi: 10.1109/JBHI.2018.2869700
65. Eppenhof KA, Pluim JP. Error estimation of deformable image registration of pulmonary ct scans using convolutional neural networks. J Med Imaging (2018) 5(2):024003. doi: 10.1117/1.JMI.5.2.024003
66. Liu X, Jiang D, Wang M, Song Z. Image synthesis-based multi-modal image registration framework by using deep fully convolutional networks. Med Biol Eng Computing (2019) 57(5):1037–48. doi: 10.1007/s11517-018-1924-y
67. Sedghi A, O’Donnell LJ, Kapur T, Learned-Miller E, Mousavi P, Wells WM III. Image registration: Maximum likelihood, minimum entropy and deep learning. Med Image Anal (2021) 69:101939. doi: 10.1016/j.media.2020.101939
68. Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, et al. Human-level control through deep reinforcement learning. nature (2015) 518(7540):529–33. doi: 10.1038/nature14236
69. Silver D, Huang A, Maddison CJ, Guez A, Sifre L, Van Den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. nature (2016) 529(7587):484–9. doi: 10.1038/nature16961
70. Liao R, Miao S, de Tournemire P, Grbic S, Kamen A, Mansi T, et al. (2017). An artificial agent for robust image registration, in: Proceedings of the AAAI conference on artificial intelligence Palo Alto, California USA: AAAI Press, Vol. 31.
71. Ma K, Wang J, Singh V, Tamersoy B, Chang YJ, Wimmer A, et al. (2017). Multimodal image registration with deep context reinforcement learning, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2017 Springer International Publishing AG: 240–8.
72. Krebs J, Mansi T, Delingette H, Zhang L, Ghesu FC, Miao S, et al. (2017). Robust non-rigid registration through agent-based action learning, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2017 Springer International Publishing AG: 344–52.
73. Yang X, Kwitt R, Niethammer M. (2016). Fast predictive image registration, in: Deep Learning and Data Labeling for Medical Applications Springer International Publishing AG:48–57.
74. Fan J, Cao X, Yap PT, Shen D. Birnet: Brain image registration using dual-supervised fully convolutional networks. Med Image Anal (2019) 54:193–206. doi: 10.1016/j.media.2019.03.006
75. Fu Y, Lei Y, Wang T, Patel P, Jani AB, Mao H, et al. Biomechanically constrained non-rigid mr-trus prostate registration using deep learning based 3d point cloud matching. Med Image Anal (2021) 67:101845. doi: 10.1016/j.media.2020.101845
76. Rohé MM, Datar M, Heimann T, Sermesant M, Pennec X. (2017). Svf-net: learning deformable image registration using shape matching, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2017 Springer International Publishing AG:266–74.
77. Sokooti H, Vos BD, Berendsen F, Lelieveldt BP, Išgum I, Staring M. (2017). Nonrigid image registration using multi-scale 3d convolutional neural network, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2017 Springer International Publishing AG:232–9.
78. Uzunova H, Wilms M, Handels H, Ehrhardt J. (2017). Training cnns for image registration from few samples with model-based data augmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2017 Springer International Publishing AG:223–31.
79. Dosovitskiy A, Fischer P, Ilg E, Hausser P, Hazirbas C, Golkov V, et al. (2015). Flownet: Learning optical flow with convolutional networks, in: Proceedings of the IEEE international conference on computer vision The Institute of Electrical and Electronics Engineers, Inc: 2758–66.
80. Jaderberg M, Simonyan K, Zisserman A, Kavukcuoglu K. (2015). Spatial transformer networks. Adv Neural Inf Process Syst (2015). Available at: https://proceedings.neurips.cc/paper/2015/file/33ceb07bf4eeb3da587e268d663aba1a-Paper.pdf
81. Yang X, Kwitt R, Styner M, Niethammer M. (2017). Fast predictive multimodal image registration, in: IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) Melbourne, VIC, Australia:IEEE: 858–62.
82. Yang X, Kwitt R, Styner M, Niethammer M. Quicksilver: Fast predictive image registration–a deep learning approach. NeuroImage (2017) 158 378–96. doi: 10.1016/j.neuroimage.2017.07.008
83. Yoo I, Hildebrand DG, Tobin WF, Lee WCA, Jeong WK. (2017). Ssemnet: Serial-section electron microscopy image registration using a spatial transformer network with learned feature, in: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support Springer:249–57.
84. Cao X, Yang J, Zhang J, Nie D, Kim M, Wang Q, et al. (2017). Deformable image registration based on similarity-steered cnn regression, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2017 Springer International Publishing AG:300–8.
85. Pei Y, Zhang Y, Qin H, Ma G, Guo Y, Xu T, et al. (2017). Non-rigid craniofacial 2d-3d registration using cnn-based regression, in: Deep learning in medical image analysis and multimodal learning for clinical decision support Springer:117–25.
86. Cao X, Yang J, Wang L, Xue Z, Wang Q, Shen D. (2018). Deep learning based inter-modality image registration supervised by intra-modality similarity, in: International workshop on machine learning in medical imaging Springer Nature Switzerland AG:55–63.
87. Onieva J, Marti-Fuster B, Pedrero de la Puente M, San José Estépar R. (2018). Diffeomorphic lung registration using deep cnns and reinforced learning, in: Image Analysis for Moving Organ, Breast, and Thoracic Images Springer Nature Switzerland AG:284–94.
88. Eppenhof KA, Pluim JP. Pulmonary ct registration through supervised learning with convolutional neural networks. IEEE Trans Med Imaging (2018) 5 1097–105. doi: 10.1109/TMI.2018.2878316
89. Lv J, Yang M, Zhang J, Wang X. Respiratory motion correction for free-breathing 3d abdominal mri using cnn-based image registration: A feasibility study. Br J Radiol (2018) 91:20170788. doi: 10.1259/bjr.20170788
90. Sokooti H, de Vos B, Berendsen F, Ghafoorian M, Yousefi S, Lelieveldt BP, et al. 3d convolutional neural networks image registration based on efficient supervised learning from artificial deformations. arXiv (2019). doi: 10.48550/arXiv.1908.10235
91. Guo H, Kruger M, Xu S, Wood BJ, Yan P. Deep adaptive registration of multi-modal prostate images. Computerized Med Imaging Graphics (2020) 84:101769. doi: 10.1016/j.compmedimag.2020.101769
92. Fu Y, Lei Y, Zhou J, Wang T, David SY, Beitler JJ, et al. (2020). Synthetic ct-aided mri-ct image registration for head and neck radiotherapy, in: Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging International Society for Optics and Photonics 11317. p. 1131728.
93. Teng X, Chen Y, Zhang Y, Ren L. Respiratory deformation registration in 4d-ct/cone beam ct using deep learning. Quantitative Imaging Med Surg (2021) 11(2):737. doi: 10.21037/qims-19-1058
94. Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. Voxelmorph: A learning framework for deformable medical image registration. IEEE Trans Med Imaging (2019) 38(8):1788–800. doi: 10.1109/TMI.2019.2897538
95. Estienne T, Vakalopoulou M, Christodoulidis S, Battistela E, Lerousseau M, Carre A, et al. (2019). U-Resnet: Ultimate coupling of registration and segmentation with deep nets, in: International conference on medical image computing and computer-assisted intervention – MICCAI 2019 Springer Nature Switzerland AG:310–9.
96. Kuang D. (2019). Cycle-consistent training for reducing negative jacobian determinant in deep registration networks, in: International Workshop on Simulation and Synthesis in Medical Imaging Springer Nature Switzerland AG:120–9.
97. Kim B, Kim J, Lee JG, Kim DH, Park SH, Ye JC. (2019). Unsupervised deformable image registration using cycle-consistent cnn, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2019 Springer Nature Switzerland AG:166–74.
98. Shen Z, Han X, Xu Z, Niethammer M. (2019). Networks for joint affine and non-parametric image registration, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) The Institute of Electrical and Electronics Engineers, Inc :4224–33.
99. Jiang Z, Yin FF, Ge Y, Ren L. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys Med Biol (2020) 65(1) 015011. doi: 10.1088/1361-6560/ab5da0
100. Shao W, Banh L, Kunder CA, Fan RE, Soerensen SJ, Wang JB, et al. Prosregnet: A deep learning framework for registration of mri and histopathology images of the prostate. Med Image Anal (2021) 68:101919. doi: 10.1016/j.media.2020.101919
101. Kim B, Kim DH, Park SH, Kim J, Lee JG, Ye JC. Cyclemorph: Cycle consistent unsupervised deformable image registration. Med Image Anal (2021) 71 102036. doi: 10.1016/j.media.2021.102036
102. Guo H, Xu X, Xu S, Wood BJ, Yan P. (2021). End-to-end ultrasound frame to volume registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:56–65.
103. Sang Y, Ruan D. (2021). 4d-cbct registration with a fbct-derived plug-and-play feasibility regularizer, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:108–17.
104. Xu J, Chen EZ, Chen X, Chen T, Sun S. (2021). Multi-scale neural odes for 3d medical image registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:213–23.
105. Chen L, Wu Z, Hu D, Pei Y, Zhao F, Sun Y, et al. (2021). Construction of longitudinally consistent 4d infant cerebellum atlases based on deep learning, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG :139–49.
106. Huang W, Yang H, Liu X, Li C, Zhang I, Wang R, et al. A coarse-to-fine deformable transformation framework for unsupervised multi-contrast mr image registration with dual consistency constraint. IEEE Trans Med Imaging (2021) 40(10):2589–99. doi: 10.1109/TMI.2021.3059282
107. Gong X, Khaidem L, Zhu W, Zhang B, Doermann D. (2022). Uncertainty learning towards unsupervised deformable medical image registration, in: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) The Institute of Electrical and Electronics Engineers, Inc:2484–93.
108. Shao S, Pei Z, Chen W, Zhu W, Wu X, Zhang B. A multi-scale unsupervised learning for deformable image registration. Int J Comput Assisted Radiol Surg (2022) 17(1):157–66. doi: 10.1007/s11548-021-02511-0
109. Hoopes A, Hoffmann M, Fischl B, Guttag J, Dalca AV. (2021). Hypermorph: Amortized hyperparameter learning for image registration, in: International Conference on Information Processing in Medical Imaging Springer Nature Switzerland AG:3–17.
110. Xu Z, Yan J, Luo J, Wells W, Li X, Jagadeesan J. (2021). Unimodal cyclic regularization for training multimodal image registration networks, in: IEEE 18th International Symposium on Biomedical Imaging (ISBI) The Institute of Electrical and Electronics Engineers, Inc:1660–4.
111. Xu Z, Luo J, Lu D, Yan J, Frisken S, Jagadeesan J, et al. (2022). Double-uncertainty guided spatial and temporal consistency regularization weighting for learning-based abdominal registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2022 Springer Nature Switzerland AG:14–24.
112. Xu Z, Luo J, Yan J, Li X, Jayender J. F3rnet: Full-resolution residual registration network for deformable image registration. Int J Comput Assisted Radiol Surg (2021) 16(6):923–32. doi: 10.1007/s11548-021-02359-4
113. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. (2017). Deep mr to ct synthesis using unpaired data, in: International workshop on simulation and synthesis in medical imaging Springer International Publishing AG::14–23.
114. Tanner C, Ozdemir F, Profanter R, Vishnevsky V, Konukoglu E, Goksel O. Generative adversarial networks for mr-ct deformable image registration. arXiv (2018). doi: 10.48550/arXiv.1807.07349
115. Yan P, Xu S, Rastinehad AR, Wood BJ. Adversarial image registration with application for mr and trus image fusion International Workshop on Machine Learning in Medical Imaging (Springer) (2018):197–204.
116. Mahapatra D. Gan based medical image registration. arXiv (2018). doi: 10.48550/arXiv.1805.02369
117. Zhang X, Jian W, Chen Y, Yang S. (2020) Deform-gan: An unsupervised learning model for deformable registration. arXiv .
118. Lu Y, Li B, Liu N, Chen JW, Xiao L, Gou S, et al. (2020) Ct-tee image registration for surgical navigation of congenital heart disease based on a cycle adversarial network. Comput Math Methods Med (2020). doi: 10.1155/2020/4942121
119. Xu Z, Luo J, Yan J, Pulya R, Li X, Wells W, et al. (2020). Adversarial uni-and multi-modal stream networks for multimodal image registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2020 Springer Nature Switzerland AG:222–32.
120. Pei Y, Chen L, Zhao F, Wu Z, Zhong T, Wang Y, et al. (2021). Learning spatiotemporal probabilistic atlas of fetal brains with anatomically constrained registration network, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:239–48.
121. Wang C, Yang G, Papanastasiou G, Tsaftaris SA, Newby DE, Gray C, et al. Dicyc: Gan-based deformation invariant cross-domain information fusion for medical image synthesis. Inf Fusion (2021) 67 147–60. doi: 10.1016/j.inffus.2020.10.015
122. Dey N, Ren M, Dalca AV, Gerig G. (2021). Generative adversarial registration for improved conditional deformable templates, in: Proceedings of the IEEE/CVF International Conference on Computer Vision The Institute of Electrical and Electronics Engineers, Inc: 3929–41.
123. Han R, Jones C, Lee J, Wu P, Vagdargi P, Uneri A, et al. Deformable mr-ct image registration using an unsupervised, dual-channel network for neurosurgical guidance. Med Image Anal (2022) 11598:102292. doi: 10.1016/j.media.2021.102292
124. Luo Y, Cao W, He Z, Zou W, He Z. Deformable adversarial registration network with multiple loss constraints. Computerized Med Imaging Graphics (2021) 91:101931. doi: 10.1016/j.compmedimag.2021.101931
125. Estienne T, Vakalopoulou M, Battistella E, Carré A, Henry T, Lerousseau M, et al. (2020). Deep learning based registration using spatial gradients and noisy segmentation labels, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2020 Springer Nature Switzerland AG:87–93.
126. De Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal (2019) 52:128–43. doi: 10.1016/j.media.2018.11.010
127. Shao W, Bhattacharya I, Soerensen SJ, Kunder CA, Wang JB, Fan RE, et al. (2021). Weakly supervised registration of prostate mri and histopathology images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:98–107.
128. Mahapatra D, Ge Z, Sedai S, Chakravorty R. (2018). Joint registration and segmentation of xray images using generative adversarial networks, in: International Workshop on Machine Learning in Medical Imaging Springer Nature Switzerland AG:73–80.
129. Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, et al. Weakly-supervised convolutional neural networks for multimodal image registration. Med Image Anal (2018) 49:1–13. doi: 10.1016/j.media.2018.07.002
130. Hu Y, Modat M, Gibson E, Ghavami N, Bonmati E, Moore CM, et al. (2018). Label-driven weakly-supervised learning for multimodal deformable image registration, in: IEEE 15th International Symposium on Biomedical Imaging The Institute of Electrical and Electronics Engineers, Inc:1070–4.
131. Hu Y, Gibson E, Ghavami N, Bonmati E, Moore CM, Emberton M, et al. (2018). Adversarial deformation regularization for training image registration neural networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2018 Springer Nature Switzerland AG:774–82.
132. Hering A, Kuckertz S, Heldmann S, Heinrich MP. (2019). Enhancing label-driven deep deformable image registration with local distance metrics for state-of-the-art cardiac motion tracking, in: Bildverarbeitung für die Medizin 2019 Springer Vieweg Wiesbaden:309–14.
133. Hu S, Zhang L, Li G, Liu M, Fu D, Zhang W. (2019). Infant brain deformable registration using global and local label-driven deep regression learning, in: International Workshop on Machine Learning in Medical Imaging Cham: Springer:106–14.
134. Li B, Niessen WJ, Klein S, Groot MD, Ikram MA, Vernooij MW, et al. (2019). A hybrid deep learning framework for integrated segmentation and registration: evaluation on longitudinal white matter tract changes, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2019 Springer Nature Switzerland AG:645–53.
135. Xu Z, Niethammer M. (2019). Deepatlas: Joint semi-supervised learning of image registration and segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2019 Springer Nature Switzerland AG:420–9.
136. Hering A, Ginneken BV, Heldmann S. (2019). Mlvirnet: Multilevel variational image registration network, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2019 Springer Nature Switzerland AG:257–65.
137. Ha IY, Hansen L, Wilms M, Heinrich MP. (2019). Geometric deep learning and heatmap prediction for large deformation registration of abdominal and thoracic ct, in: International Conference on Medical Imaging with Deep Learning–Extended Abstract Track.arXiv.
138. Lee MC, Oktay O, Schuh A, Schaap M, Glocker B. Image-and-spatial transformer networks for structure-guided image registration. International Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 Springer Nature Switzerland AG (2019):337–45.
139. Zhu Z, Cao Y, Qin C, Rao Y, Lin D, Dou Q, et al. Joint affine and deformable three-dimensional networks for brain mri registration. Med Phys (2021) 48(3):1182–96. doi: 10.1002/mp.14674
140. Hering A, Häger S, Moltz J, Lessmann N, Heldmann S, van Ginneken B. Cnn-based lung ct registration with multiple anatomical constraints. Med Image Anal (2021) 72:102139. doi: 10.1002/mp.14674
141. Wolterink JM, Zwienenberg JC, Brune C. Implicit neural representations for deformable image registration. Medical Imaging with Deep Learning (2021).
142. Chen X, Xia Y, Ravikumar N, Frangi AF. (2021). A deep discontinuity-preserving image registration network, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:46–55.
143. Espinel Y, Calvet L, Botros K, Buc E, Tilmant C, Bartoli A. (2021). Using multiple images and contours for deformable 3d-2d registration of a preoperative ct in laparoscopic liver surgery, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:657–66.
144. Zhao F, Wu Z, Wang L, Lin W, Xia S, Li G, et al. (2021). A deep network for joint registration and parcellation of cortical surfaces, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:171–81.
145. Zhang Y, Pei Y, Zha H. (2021). Learning dual transformer network for diffeomorphic registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:129–38.
146. Song X, Guo H, Xu X, Chao H, Xu S, Turkbey B, et al. (2021). Cross-modal attention for mri and ultrasound volume registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:66–75.
147. Shu Y, Wang H, Xiao B, Bi X, Li W. (2021). Medical image registration based on uncoupled learning and accumulative enhancement, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021 Springer Nature Switzerland AG:3–13.
148. Park H, Lee GM, Kim S, Ryu GH, Jeong A, Park SH, et al. A meta-learning approach for medical image registration. arXiv (2021)
149. Chen J, He Y, Frey EC, Li Y, Du Y. Vit-v-net: Vision transformer for unsupervised volumetric medical image registration. arXiv (2021). doi: 10.1109/TMI.2022.3218147
150. Chen J, Du Y, He Y, Segars WP, Li Y, Frey EC. Transmorph: Transformer for unsupervised medical image registration. arXiv (2021). doi: 10.48550/arXiv.2104.06468
151. Liu L, Aviles-Rivero AI, Schönlieb CB. Contrastive registration for unsupervised medical image segmentation. arXiv (2020). doi: 10.48550/arXiv.2111.10480
152. Casamitjana A, Mancini M, Iglesias JE. Synth-by-reg (sbr): Contrastive learning for synthesis-based registration of paired images. International Workshop on Simulation and Synthesis in Medical Imaging (Springer):(2021):44–54. doi: 10.48550/arXiv.2011.08894
153. Sinclair M, Schuh A, Hahn K, Petersen K, Bai Y, Batten J, et al. Atlas-istn: joint segmentation, registration and atlas construction with image-and-spatial transformer networks. Med Image Anal (2022) 78:102383. doi: 10.1016/j.media.2022.102383
154. Liu L, Wang H. (2022). Multiple organ localization in dual-modality pet/ct images based on transformer network with one-to-one object query, in: 4th International Conference on Intelligent Medicine and Image Processing New York NY United States:Association for Computing Machinery: 36–42.
155. Tran MQ, Do T, Tran H, Tjiputra E, Tran QD, Nguyen A. Light-weight deformable registration using adversarial learning with distilling knowledge. IEEE Trans Med Imaging (2022) 41(6):1443–53. doi: 10.1109/TMI.2022.3141013
156. Wu Y, Jiahao TZ, Wang J, Yushkevich PA, Hsieh MA, Gee JC. Nodeo: A neural ordinary differential equation based optimization framework for deformable image registration. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2022):20804–13.
157. Kim B, Han I, Ye JC. Diffusemorph: Unsupervised deformable image registration along continuous trajectory using diffusion models. arXiv (2021). doi: 10.48550/arXiv.2112.05149
158. Chen J, Lu D, Zhang Y, Wei D, Ning M, Shi X, et al. (2022). Deformer: Towards displacement field learning for unsupervised medical image registration, in: International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer):141–51.
159. Shi J, He Y, Kong Y, Coatrieux JL, Shu H, Yang G, et al. (2022). Xmorpher: Full transformer for deformable medical image registration via cross attention, in: International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI 2022 Springer Nature Switzerland AG:217–26.
160. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst 30 (2017).
161. Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations. International conference on machine learning (PMLR) (2020):1597–607.
162. Vilalta R, Drissi Y. A perspective view and survey of meta-learning. Artif Intell Rev (2002) 18(2) 77–95. doi: 10.1023/A:1019956318069
163. Chen RT, Rubanova Y, Bettencourt J, Duvenaud DK. (2018). Neural ordinary differential equations, in: Advances in neural information processing systems Morgan Kaufmann Publishers Inc.
164. Ho J, Jain A, Abbeel P. Denoising diffusion probabilistic models. Adv Neural Inf Process Syst (2020) 33 6840–51.
165. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. (2021). Swin transformer: Hierarchical vision transformer using shifted windows, in: Proceedings of the IEEE/CVF International Conference on Computer Vision The Institute of Electrical and Electronics Engineers, Inc:10012–22.
166. Jiangtao W, Xinhong W, Xiao J, Bing Y, Lei Z, Yidong Y. (2021). Mri to ct synthesis using contrastive learning, in: IEEE International Conference on Medical Imaging Physics and Engineering and the 10th National Annual Meeting of Medical Imaging Physics The Institute of Electrical and Electronics Engineers, Inc:1–5.
Keywords: deformable image registration, medical imaging, clinical applications, deep learning, computer assisted surgery
Citation: Zou J, Gao B, Song Y and Qin J (2022) A review of deep learning-based deformable medical image registration. Front. Oncol. 12:1047215. doi: 10.3389/fonc.2022.1047215
Received: 17 September 2022; Accepted: 08 November 2022;
Published: 07 December 2022.
Edited by:
Mohamed Abdel-Nasser, Aswan University, EgyptCopyright © 2022 Zou, Gao, Song and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zou, jing9.zou@connect.polyu.hk
 Jing Zou
Jing Zou Bingchen Gao
Bingchen Gao Jing Qin
Jing Qin