- 1Department of Gastroenterology, 904 Hospital of PLA Joint Logistic Support Force, Wuxi, China
- 2Department of Gastroenterology, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou, China
Inflammatory bowel disease (IBD), comprised of Crohn’s disease (CD) and ulcerative colitis (UC), is a group of chronic inflammatory disorders. IBD is regarded as a severe healthcare problem worldwide, with high morbidity and lethality. So far, despite of numerous studies on this issue, the specific mechanisms of IBD still remain unclarified and ideal treatments are not available for IBD. The intestinal mucosal barrier is vital for maintaining the function of the intestinal self-defensive system. Among all of the components, macrophage is an important one in the intestinal self-defensive system, normally protecting the gut against exotic invasion. However, the over-activation of macrophages in pathological conditions leads to the overwhelming induction of intestinal inflammatory and immune reaction, thus damaging the intestinal functions. Autophagy is an important catabolic mechanism. It has been proven to participate the regulation of various kinds of inflammation- and immune-related disorders via the regulation of inflammation in related cells. Here in this paper, we will review the role and mechanism of intestinal macrophage autophagy in IBD. In addition, several well-studied kinds of agents taking advantage of intestinal macrophage autophagy for the treatment of IBD will also be discussed. We aim to bring novel insights in the development of therapeutic strategies against IBD.
1 Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is referred to as a group of idiopathic inflammation-related bowel diseases, which mainly affects the ileum, rectum and colon (Dun et al., 2021; Gallagher et al., 2021; Mazur et al., 2021). The clinical manifestations of IBD are atypical, including diarrhea, abdominal pain, bloody stools and so on (Li et al., 2020; Wang et al., 2020; Yeh et al., 2021). The morbidity of IBD is increasing rapidly in the recent decade, especially in the developed countries, and the diagnosed rate is relatively high in the young and middle-aged people (M’Koma, 2013; Uhlig et al., 2021). So far, the specific mechanisms for the pathogenesis and progression of IBD remain unclarified. According to recent studies, the abnormality of intestinal immune system and inflammatory responses may probably contribute to the onset and development of IBD (Cushing and Higgins, 2021; Nash et al., 2021). Certain environmental, genetic and infectious factors as well as disturbance of intestinal microbiota homeostasis may serve as vital factors to lead to the pathological state intestinal self-protective immune and inflammatory reaction (Owczarek et al., 2016; Lloyd-Price et al., 2019; Bushman et al., 2020; Ryan et al., 2020).
Currently, the intestinal tract is recognized as not only the largest digesting organ as traditionally considered, but also a critical immune organ, since it is frequently exposed and challenged with exogenous invasive microorganisms in the gut lumen (Fiorucci et al., 2021; Lu et al., 2021). As a result, a strong and complete intestinal self-defensive system have been developed in fighting against those damaged factors. In the system, the intestinal mucosal barrier has been recognized to play a vital role in protecting against invasive damage and maintaining the intestinal microbiota homeostasis (Mehandru and Colombel, 2021; Yang and Yu, 2021; You et al., 2021). It is generally revealed that the intestinal microbiota homeostasis mainly includes three layers, including the mucus layer on the surface, epithelium layer and inflammatory and immune cell layer in the submucosa (Grondin et al., 2020; Liu et al., 2020a; Shao et al., 2021). The mucus layer is a gel-like layer containing many kinds of proteins, which cover on the surface of the mucosa to get rid of the direct contacting of epithelium and intestinal lumen contents (Engevik et al., 2019; das Neves et al., 2020). The epithelium layer is referred to as a layer of gut epithelial cells, mainly including intestinal enterocytes, goblet cells, Paneth cells, enteroendocrine cells and so on, functioning in the absorption of nutrition and secretion of certain functional proteins (Shaoul et al., 2005; Haber et al., 2017; Collins et al., 2021). The immune cell layer in the submucosa is comprised of several kinds of immune- and inflammation-related cells, including macrophages, neutrophils, eosinophils, lymphocytes and so on (Perdue and McKay, 1994; Pardigol et al., 1998; Hu et al., 2021).
Among all of those immune- and inflammation-related cells of the immune cell layer in the submucosa, macrophages are one of the most studied ones. It has been shown that the gut holds the largest population of macrophages among all organs. Normally, the majority of tissue macrophages are divided into M1 and M2 subtypes, with M1 subtype displaying pro-inflammatory features and M2 subtype as anti-inflammatory and immunosuppressive characteristics (Ke et al., 2016a; Yunna et al., 2020; Wu et al., 2021a). However, for gut-resident macrophages, they have been reported to hold the characteristics of both M1 and M2 subtypes to produce both pro-inflammatory cytokines including TNF-α and anti-inflammatory cytokines such as IL-10 (Weber et al., 2011; Ke et al., 2016a). Intestinal macrophages have been revealed to play a vital role in the protection of the gut against exotic invasion and damage via the engulfment and presentation of invading antigens to other immune cells for clearance and killing (Vannella and Wynn, 2017; Muller et al., 2020; Viola and Boeckxstaens, 2020). However, the overwhelming activation of such self-protective immune and inflammatory responses induced by macrophages may lead to the disturbance of gut functions and intestinal microbiota homeostasis, thus resulting in certain gut diseases such as IBD (Baumgart and Carding, 2007; Wallace et al., 2014; Fan et al., 2017). Consequently, regulating the abnormality of inflammatory and immune reaction mediated by intestinal macrophages may serve as a potential and promising strategy for the treatment of IBD.
Autophagy is a self-eating catabolic cellular pathway, through which some long-lived proteins or cytoplasmic organelles are degraded and recycled by the integration with lysosomes to maintain cellular homeostasis and normal functions (Wang et al., 2018; Devis-Jauregui et al., 2021; Li et al., 2021). Autophagy has been reported to produce a regulatory effect in various kinds of diseases. In inflammatory and immune diseases, baseline and properly induced autophagy have been proven to effectively alleviate the onset and development of diseases via the suppression of the overwhelming inflammatory and immune reaction (Racanelli et al., 2018; Chimienti et al., 2021; McCormick et al., 2021). Autophagy has also been shown to have close connection with many forms of inflammatory and immune responses in cells, such as the inflammasomes and neutrophil extracellular traps, apoptosis and necrosis (Luo et al., 2021a; Dong et al., 2021; Qin et al., 2021). As a result, uncovering the role and underlying mechanisms of autophagy in inflammation-related disorders is necessary and vital for the development of effective therapeutic strategies. Here in this paper, the role of intestinal macrophage autophagy in IBD as well as its pharmacological applications will be reviewed and discussed based on current related studies. We hope to provide insights in the development of new strategies against IBD.
2 Autophagy in Inflammatory Bowel Disease
2.1 Biological Features of Autophagy
The word “autophagy” derives from Greet roots “auto” (self) and “phagy” (eating), which means “to eat itself” (Yoshimura et al., 2020; Joffre et al., 2021). Autophagy is a vital cellular catabolic pathway for degrading damaged proteins and organelles to recycle relying on lysosomes (Devis-Jauregui et al., 2021; Wang et al., 2021; Yao et al., 2021). After initially reported in the 1950s, the process of autophagy was systematically uncovered by Dr. Yoshinori Ohsumi, who was awarded to the 2016 Nobel Prize in Medicine or Physiology (Rubinsztein and Frake, 2016; Wollert, 2019). So far, three kinds of classic forms of autophagy have been described, namely microautophagy, macroautophagy and chaperone-mediated autophagy (Dash et al., 2019; Lin et al., 2021; Trelford and Di Guglielmo, 2021). Besides, according to the difference of degrading substrates, several kinds of selective autophagy have also been recognized and reported in various kinds of diseases, including mitophagy, pexophagy, reticulophagy, xenophagy and so on (Kuma et al., 2017; Li et al., 2021; Xu et al., 2021). Since macroautophagy is so far the most studied form of autophagy, here in this paper, the characteristics of macroautophagy and its roles in IBD will be mainly discussed (hereafter referring to “autophagy”).
Over the few decades since the initial description of autophagy, there are more than 30 autophagy-related genes (ATGs) reported to be vital in the process of autophagy (Hu, 2019; Kamel et al., 2020; Zhang et al., 2021). The process of autophagy is generally divided into two steps, including the formation of the autophagosome and integration with the lysosome to form the autolysosome (Wang et al., 2018; Yu et al., 2018; Colletti et al., 2020; Lorincz and Juhasz, 2020; Tan and Chen, 2021). Under the challenge of stressful conditions, inflammation, nutrient insufficiency or ischemia, autophagy is initiated via the triggering of the first step. In the first step, the autophagy-related proteins including ATG13, ATG101, Unc-51-like kinase 1 (ULK1) and focal adhesion kinase family interacting protein of 200 kD (FIP200) are integrated to form the ATG1 complex, which subsequently lead to the assembly of Becline-1, ATG14, VSP15 and VSP34 to form the Class III phosphatidylinositol 3-kinase (PI3K) complex. The activation of Class III PI3K signaling promote the nucleation of membrane to form the single-layer and cup-shaped phagophore. The membrane further expanded and lengthened along with the formation of ATG5-ATG12-ATG16L1 complex and participation of ATG8 [well known as “light chain 3 (LC3)”], which leads to the formation of the double-membrane and cycled autophagosome. The function of autophagosome is to facilitate cargo recruitment and engulfment. In the second step, with the participation of ATG3, ATG4 and ATG7, autophagosome fuses with the lysosome to form the autolysosome, which is regarded as the functional unit of autophagy. In the process of autophagy, Class I PI3K signaling pathway has been revealed as a classic autophagy inhibiting pathway, with the downstream proteins including Class I PI3K, Akt and mammalian target of rapamycin complex 1 (mTORC1) (Yu et al., 2015; Aoki and Fujishita, 2017; Wang et al., 2017).
Recently, the roles of autophagy in various kinds of diseases have been increasingly studied. In central nervous system, it has been demonstrated that autophagy plays a protective role in the cerebral ischemic stroke via the inhibition of neuronal apoptosis in patients and oxygen-glucose deprivation (OGD)-induced models (Han et al., 2018; Ahsan et al., 2021; Tang et al., 2021). In addition, autophagy has been shown to alleviate the severity of symptoms through the suppression of inflammatory reaction in patients with multiple sclerosis and experimental autoimmune encephalomyelitis (EAE) mice models (Hassanpour et al., 2020; Di Rita and Strappazzon, 2021; Yao et al., 2021). In cardiovascular diseases, autophagy has been revealed to alleviate the formation of macrophage-derived foam cells, thus producing a protective effect on the pathogenesis and progression of atherosclerosis and the subsequent myocardial ischemia (Shao et al., 2016; Ouimet et al., 2017; Kumar et al., 2021). So far, the role of autophagy in IBD, especially in intestinal macrophages, has been drawn increasing attention to by researchers. The effects of intestinal macrophage autophagy will be detailly described and discussed based on the reviewing of related previous studies in the following contents.
2.2 Intestinal Macrophage Autophagy in Inflammatory Bowel Disease
Autophagy has been revealed to have close crosstalk with many inflammatory and immune responses. So far, autophagy in intestinal macrophages has been demonstrated to play a vital role in the regulation of IBD. The mutation of autophagy-related genes has been shown to produce a detrimental effect on the pathogenesis and progression of IBD. In addition, autophagy has also been shown to contribute to modulating intestinal inflammation and maintaining intestinal microbiota homeostasis. Those points of contents will be discussed in this section, respectively (illustrated in Figure 1).
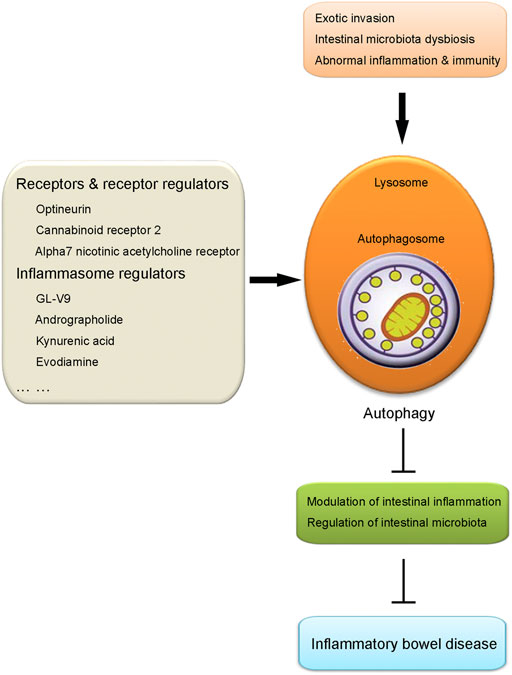
FIGURE 1. Illustration of the role of intestinal macrophage autophagy in IBD. Under the challenge of certain pathological conditions including exotic invasion, intestinal microbiota dysbiosis and abnormal inflammation and immunity, autophagy process in macrophages is induced, which results in the suppression of intestinal inflammation and regulation of intestinal microbiota. Those effects produced by the induction of macrophage autophagy contribute to the protection against IBD. Several kinds of agents have been proven to alleviate IBD taking advantage of intestinal macrophage autophagy, including receptors and receptor regulators (optineurin, cannabinoid receptor 2, alpha7 nicotinic acetylcholine receptor) and inflammasome regulators (GL-V9, andrographolide, kynurenic acid, evodiamine), etc.
2.2.1 Autophagy-Related Gene Mutation
So far, several autophagy-related genes are vital in the process of intestinal macrophage autophagy (Homer et al., 2010; Parkes, 2012; Saxena et al., 2017; Schwerd et al., 2017). The mutation of such genes, including Atg16l1, immunity-related GTPase M (Irgm), nucleotide-binding oligomerization domain-containing protein 2 (Nod2) and so on, has been reported to contribute to the pathogenesis and progression of IBD. In patients with CD, the existence of polymorphism of Atg16l1, Nod2 and Irgm in macrophages has been shown to be the etiology of CD via leading to the deficiency of autophagy (Caprilli et al., 2010). Monocyte-derived macrophages obtained from patients with CD genotyped for Nod2, Atg16l1 and Irgm mutations were reported to be unable to restrict adherent-invasive Escherichia coli (AIEC) replication and trigger the abnormality in the inflammatory responses influenced by disease activity (Lapaquette et al., 2012; Vazeille et al., 2015). In addition, defective Nod2 function in intestinal macrophages from CD patients was found to lead to a pro-inflammatory cytokine bias relying on the toll-like receptor 2 (TLR2) signaling (Netea et al., 2004), and loss of wildtype Nod2 resulted in dysregulated homeostasis of activated fibroblasts and macrophages (Nayar et al., 2021).
In mice models for IBD, it was previously revealed that mice with myeloid Atg16l1 deficiency largely exacerbated the severity of colitis with the increase of pro-inflammatory cytokine production and decrease of anti-inflammatory cytokine production (Zhang et al., 2017a). The detection of Atg16l1 T300A variant in macrophages was also proven as a risk factor for CD, acting as a dominant-negative variant (Zhang et al., 2017a; Gao et al., 2017; Samie et al., 2018). According to a study of single-nucleotide polymorphism (SNP), the SNP for Atg16l1 (rs2241880, Thr300Ala) was revealed to be strongly associated with the incidence of CD via caspase-3 activation (Luo et al., 2021b). In addition, Nod2 was reported to mediate an alleviative effect on dextran sulphate sodium (DSS)-induced colitis mice models via suppressing the gram-positive bacteria invasion and damage, while knocking out Nod2 abolished such effects (Jamontt et al., 2013; Jing et al., 2014; Luo et al., 2021b). For Irgm, its loss-of-function mutation was shown to affect the clearance by macrophages of CD-associated AIEC in mice IBD models (Parkes, 2012).
2.2.2 Modulation of Intestinal Inflammation
As discussed previously, the abnormal induction of intestinal inflammatory reaction contributes greatly to the pathogenesis and progression of IBD. Intestinal macrophages act as an important component for the triggering of inflammation and immune reaction. Deficiency of autophagy has been reported to propose an important event in IBD via the enhancement of macrophage-mediated inflammatory reaction (Schwerd et al., 2017; Samoila et al., 2020; Wu et al., 2021b). In patients with IBD, the application of cannabis, a cannabinoid receptor 2 (CB2R) agonist, was shown to alleviate the severity of IBD via the induction of autophagy and suppression of macrophage-mediated inflammation (Tartakover Matalon et al., 2020). Another study conducting on patients revealed that targeting on ATG2B for the inhibition of autophagy by microRNA-143 enhanced the production of macrophage-induced pro-inflammatory cytokines, thus playing a detrimental effect on CD (Lin et al., 2018). High-density lipoprotein (HDL) and apolipoprotein A-I (apoA-I) were reported to suppress the level of intestinal inflammation via autophagy in human intestinal epithelial cell, indicating the intestinal inflammation-suppression effect of autophagy in IBD (Gerster et al., 2015).
In DSS-induced colitis model, it was demonstrated that macrophage-specific V-ATPase subunit ATP6V0D2 significantly alleviated the severity of DSS-induced colitis in mice via restricting the inflammasome activation in macrophages and bacterial infection of the pathogenic Salmonella typhimurium (Xia et al., 2019). Those effects were facilitated by the enhancement of autophagosome-lysosome fusion and level of autophagy flux (Xia et al., 2019). In addition, the induction of PTEN-induced putative kinase 1 (PINK1)/Parkin-driven autophagy was shown to protect against DSS-induced colitis through the inactivation of the NLR family pyrin domain containing 3 (NLRP3) inflammasome in macrophages (Mai et al., 2019). A previous study revealed that the induction of AMP-activated protein kinase (AMPK)-induced autophagy could endow the anti-inflammatory properties of intestinal macrophages, thus alleviating DSS-induced colitis (Liu et al., 2020b). In another colitis model induced by trinitro-benzene-sulfonic acid (TNBS), interleukin-33 (IL-33), a well-known anti-inflammatory cytokine, was reported to ameliorate colitis through the enhancement of intestinal macrophage autophagy in the inflammatory gut tissue via regulation of TLR4 signaling pathway (Wang et al., 2019). Furthermore, pharmacological induction of intestinal autophagy via rapamycin, a classic autophagy inducer, was demonstrated to reduce intestinal inflammation from macrophages and improved murine colitis (Macias-Ceja et al., 2017).
2.2.3 Regulation of Intestinal Microbiota
Intestinal microbiota is a vital component of the gut environment (Adak and Khan, 2019; Gomaa, 2020; Jarbrink-Sehgal and Andreasson, 2020). A good intestinal microbiota habitat is necessary for the maintenance of normal gut functions and health for organisms. In normal conditions, four phyla of microbiota are detected to be dominant in the gut of normal human, including Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria (Zhang et al., 2017b; Invernizzi et al., 2021; Mao et al., 2021). However, dysbiosis of intestinal microbiota has been investigated in patients with various digestive disorders, such as colorectal cancer and IBD (Shin et al., 2015; Tomasello et al., 2016; Sun and Shen, 2018; Fan et al., 2021; Sultan et al., 2021). Under certain pathologic conditions, some kinds of harmful microbiota are invaded and largely residue in the gut. Among those harmful microbiota, Escherichia coli (E. coli) is the most studied one, which belongs to the family of Enterobacteriaceae and is usually detected in the lower intestine (Castillo et al., 2005; Rousset et al., 2021). E. coli, especially AIEC, have been found to be enriched in the gut of both UC and CD patients (Rolhion and Darfeuille-Michaud, 2007; Mirsepasi-Lauridsen et al., 2019). The assembly and large residue of AIEC have been proven to be harmful for the gut wall via the disturbance of intestinal microbiota homeostasis and triggering of gut inflammatory reaction (Derer et al., 2020; Leccese et al., 2020).
So far, modern studies have revealed a positive role of intestinal macrophage autophagy in fighting against the invasion and damage of AIEC. Dysfunctional autophagy was found to lead to overwhelming inflammatory reaction induced by intestinal macrophages and subsequent gut microbiota dysbiosis in CD (Larabi et al., 2020). In addition, the functional autophagy in macrophages contributes greatly to maintaining the ability of macrophages in recognizing and dealing with the invasion of AIEC (Rolhion and Darfeuille-Michaud, 2007; Caprilli et al., 2010; Buisson et al., 2019). Deficiency of autophagy was shown to lead to the escape of AIEC from macrophages (Caprilli et al., 2010; Palmela et al., 2018; Buisson et al., 2019). Collectively, autophagy is vital to maintain the function of macrophages in dealing with AIEC.
3 Applications of Intestinal Macrophage Autophagy in the Treatment of Inflammatory Bowel Disease
As discussed above, intestinal macrophage autophagy plays an important role in the regulation of pathogenesis and progression of IBD. So far, with the efforts of researchers, we are lucky to have several kinds of agents which have been proven to be effective in the alleviation of IBD taking advantage of regulating intestinal macrophage autophagy. Among those agents, two kinds of autophagy regulators, including receptors and receptor regulators, and inflammasome regulators are the most popular and studied ones. As a result, in the following contents, those two kinds of autophagy regulators will be detailly described and discussed, respectively (illustrated in Figure 1 and Table 1).

TABLE 1. Pharmacological applications of intestinal macrophage autophagy regulators in the treatment of IBD.
3.1 Receptors and Receptor Regulators
In the recent few years, several kinds of receptors and receptor regulators have been demonstrated to produce an alleviative effect on IBD. For instance, optineurin, a selective autophagy receptor, has been increasing recognized as a critical factor for the maintenance of pathogen clearance and regulation of cytokine production in macrophages (Tschurtschenthaler and Adolph, 2018; Xu et al., 2020). It was reported that the activation of optineurin contributed to the alleviation of IBD, especially CD, via the induction of autophagy, thus suppressing the inflammatory responses mediated by intestinal macrophages (Tschurtschenthaler and Adolph, 2018).
Furthermore, CB2R, a member of the family of G-protein-coupled receptors (GPCR), has been increasingly investigated as an immune and inflammatory modulator (Carbone et al., 2014; Kong et al., 2014; Ke et al., 2016b). Unlike CB1R which is majorly expressed in central nervous system, it is mainly located in the immune system including macrophages and other inflammatory and immune cells in periphery tissues (Carbone et al., 2014; Kong et al., 2014; Ke et al., 2016b). It was demonstrated that the administration of CB2R agonist HU308 significantly alleviated the severity of DSS-induced colitis in mice via the induction of intestinal macrophage autophagy (Ke et al., 2016b). Those protective effects of CB2R agonist on IBD was proven to be mediated by the AMPK-mTOR-p70S6K signaling pathway, a classic autophagy pathway (Ke et al., 2016b).
In addition, alpha7 nicotinic acetylcholine receptor (α7nAChR), a member of the superfamily of cys-loop cationic ligand-gated channels, has been demonstrated to protect against several kinds of inflammation- and immune-related diseases through the triggering of the “cholinergic anti-inflammatory pathway” (Cheng and Yakel, 2015; Qin et al., 2017; Shao et al., 2017; Shao et al., 2019a). Nicotine, an α7nAChR non-selective agonist, has been shown to suppress the production of pro-inflammatory cytokines by macrophages via the microRNA-124/signal transducing activator of transcription (STAT) system in IBD (Qin et al., 2017). A previous study revealed that the administration of PNU282987, a selective α7nAChR agonist, protected against DSS-induced colitis via the induction of AMPK-mTOR-p70S6K signaling-mediated autophagy in intestinal macrophages (Shao et al., 2019a).
3.2 Inflammasome Regulators
The inflammasome, a multi-protein oligomer, is widely recognized as a member of innate immunity and also a special form of inflammatory reaction (Mariathasan et al., 2004). Inflammasome is mainly formed in inflammation- and immune-related cells like macrophages, triggering the substantial inflammatory and immune responses through the recognition of pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) (Alexandre et al., 2014; Lei-Leston et al., 2017; Shao et al., 2019b). Inflammasome has been reported to participate in the onset and development of various kinds of inflammation- and immune-related diseases via the production and secretion of certain pro-inflammatory cytokines (Ge et al., 2021; Moretti and Blander, 2021). So far, an increasing number of studies have investigated the role of macrophage inflammasome in IBD, and autophagy has been widely reported to regulate the level of inflammasome in the gut.
According to the recent related studies, several agents have been proven to be effective in the alleviation of IBD taking advantage of inducing autophagy-mediated inflammasome suppression. GL-V9, a small-molecule of AMPK activator, was shown to protect against colitis as well as colitis-associated colorectal cancer through the inhibition of the NLRP3 inflammasome, a well-studied form of inflammasome, by the induction of autophagy in intestinal macrophages (Zhao et al., 2017). Similar effect on colitis and colitis associated colorectal cancer was reported to be produced by another small molecule andrographolide via the downregulation of the PIK3CA-Akt1-mTOR-RPS6KB1 pathway (Guo et al., 2014). In addition, kynurenic acid, an endogenous regulator of social stress-exacerbated colitis, was shown to induce autophagy-dependent degradation of NLRP3 in macrophages through the kynurenic acid/G Protein-Coupled Receptor 35 (GPR35) axis (Zheng et al., 2019). Evodiamine, a chemical component extracted from Evodiae Fructus, was shown to suppress the initiation and assembly of the NLRP3 inflammasome via autophagy-mediated regulation of nuclear factor-κB (NF-κB) pathway in macrophages, thus alleviating DSS-induced colitis (Ding et al., 2020).
4 Conclusion
In this paper, we have reviewed the role of intestinal macrophage autophagy in the pathogenesis and progression of IBD. We have introduced the biological characteristics of autophagy and discussed the role of intestinal macrophage autophagy in IBD based on reviewing the related studies. In addition, two well-studied kinds of autophagy regulators, including receptors and receptor regulators as well as inflammasome regulators were described. Although the mechanisms of intestinal macrophage autophagy in IBD are increasingly investigated in the recent studies, yet the specific mechanisms remain unclarified. Furthermore, few autophagy inducers have been successfully applied in clinical practice. Consequently, to ultimately take advantage of intestinal macrophage autophagy in the treatment of IBD, further studies are demanded on this issue.
Author Contributions
YZ, X-FC, S-LY, and X-LT retrieved concerned literatures and wrote the manuscript. YY drew the figure and made the table. Z-GX revised the manuscript. All authors agreed to be accountable for the contents of the work.
Funding
This work was supported by General Project of Science and Technology Development Fund of Nanjing Medical University (No. NMUB2019332).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adak, A., and Khan, M. R. (2019). An Insight Into Gut Microbiota and its Functionalities. Cell Mol. Life Sci. 76 (3), 473–493. doi:10.1007/s00018-018-2943-4
Ahsan, A., Liu, M., Zheng, Y., Yan, W., Pan, L., Li, Y., et al. (2021). Natural Compounds Modulate the Autophagy with Potential Implication of Stroke. Acta Pharm. Sin. B 11 (7), 1708–1720. doi:10.1016/j.apsb.2020.10.018
Alexandre, Y. O., Cocita, C. D., Ghilas, S., and Dalod, M. (2014). Deciphering the Role of DC Subsets in MCMV Infection to Better Understand Immune Protection against Viral Infections. Front. Microbiol. 5, 378. doi:10.3389/fmicb.2014.00378
Aoki, M., and Fujishita, T. (2017). Oncogenic Roles of the PI3K/AKT/mTOR Axis. Curr. Top. Microbiol. Immunol. 407, 153–189. doi:10.1007/82_2017_6
Baumgart, D. C., and Carding, S. R. (2007). Inflammatory Bowel Disease: Cause and Immunobiology. Lancet 369 (9573), 1627–1640. doi:10.1016/S0140-6736(07)60750-8
Buisson, A., Douadi, C., Ouchchane, L., Goutte, M., Hugot, J. P., Dubois, A., et al. (2019). Macrophages Inability to Mediate Adherent-Invasive E. coli Replication is Linked to Autophagy in Crohn’s Disease Patients. Cells 8 (11), 1394. doi:10.3390/cells8111394
Bushman, F. D., Conrad, M., Ren, Y., Zhao, C., Gu, C., Petucci, C., et al. (2020). Multi-Omic Analysis of the Interaction between Clostridioides difficile Infection and Pediatric Inflammatory Bowel Disease. Cell Host Microbe 28 (3), 422–433 e427. doi:10.1016/j.chom.2020.07.020
Caprilli, R., Lapaquette, P., and Darfeuille-Michaud, A. (2010). Eating the Enemy in Crohn’s Disease: An Old Theory Revisited. J. Crohns Colitis. 4 (4), 377–383. doi:10.1016/j.crohns.2010.05.007
Carbone, F., Mach, F., Vuilleumier, N., and Montecucco, F. (2014). Cannabinoid Receptor Type 2 Activation in Atherosclerosis and Acute Cardiovascular Diseases. Curr. Med. Chem. 21 (35), 4046–4058. doi:10.2174/0929867321666140915141332
Castillo, A., Eguiarte, L. E., and Souza, V. (2005). A Genomic Population Genetics Analysis of the Pathogenic Enterocyte Effacement Island in Escherichia coli: The Search for the Unit of Selection. Proc. Natl. Acad. Sci. U.S.A. 102 (5), 1542–1547. doi:10.1073/pnas.0408633102
Cheng, Q., and Yakel, J. L. (2015). The Effect of α7 Nicotinic Receptor Activation on Glutamatergic Transmission in the hippocampus. Biochem. Pharmacol. 97 (4), 439–444. doi:10.1016/j.bcp.2015.07.015
Chimienti, G., Orlando, A., Lezza, A. M. S., D'Attoma, B., Notarnicola, M., Gigante, I., et al. (2021). The Ketogenic Diet Reduces the Harmful Effects of Stress on Gut Mitochondrial Biogenesis in a Rat Model of Irritable Bowel Syndrome. Int. J. Mol. Sci. 22 (7), 3498. doi:10.3390/ijms22073498
Colletti, M., Ceglie, D., Di Giannatale, A., and Nazio, F. (2020). Autophagy and Exosomes Relationship in Cancer: Friends or Foes? Front Cell Dev. Biol. 8, 614178. doi:10.3389/fcell.2020.614178
Collins, J. T., Nguyen, A., and Badireddy, M. (2021). Anatomy, Abdomen and Pelvis, Small Intestine. Treasure Island, FL: StatPearls.
Cushing, K., and Higgins, P. D. R. (2021). Management of Crohn Disease: A Review. JAMA 325 (1), 69–80. doi:10.1001/jama.2020.18936
das Neves, J., Sverdlov Arzi, R., and Sosnik, A. (2020). Molecular and Cellular Cues Governing Nanomaterial-Mucosae Interactions: From Nanomedicine to Nanotoxicology. Chem. Soc. Rev. 49 (14), 5058–5100. doi:10.1039/c8cs00948a
Dash, S., Aydin, Y., and Moroz, K. (2019). Chaperone-Mediated Autophagy in the Liver: Good or Bad? Cells 8 (11), 1308. doi:10.3390/cells8111308
Derer, S., Brethack, A. K., Pietsch, C., Jendrek, S. T., Nitzsche, T., Bokemeyer, A., et al. (2020). Inflammatory Bowel Disease-Associated GP2 Autoantibodies Inhibit Mucosal Immune Response to Adherent-Invasive Bacteria. Inflamm. Bowel Dis. 26 (12), 1856–1868. doi:10.1093/ibd/izaa069
Devis-Jauregui, L., Eritja, N., Davis, M. L., Matias-Guiu, X., and Llobet-Navàs, D. (2021). Autophagy in the Physiological Endometrium and Cancer. Autophagy 17 (5), 1077–1095. doi:10.1080/15548627.2020.1752548
Di Rita, A., and Strappazzon, F. (2021). A Protective Variant of the Autophagy Receptor CALCOCO2/NDP52 in Multiple Sclerosis (MS). Autophagy 17 (6), 1565–1567. doi:10.1080/15548627.2021.1924969
Ding, W., Ding, Z., Wang, Y., Zhu, Y., Gao, Q., Cao, W., et al. (2020). Evodiamine Attenuates Experimental Colitis Injury via Activating Autophagy and Inhibiting NLRP3 Inflammasome Assembly. Front. Pharmacol. 11, 573870. doi:10.3389/fphar.2020.573870
Dong, Y., Jin, C., Ding, Z., Zhu, Y., He, Q., Zhang, X., et al. (2021). TLR4 Regulates ROS and Autophagy to Control Neutrophil Extracellular Traps Formation against Streptococcus pneumoniae in Acute Otitis media. Pediatr. Res. 89 (4), 785–794. doi:10.1038/s41390-020-0964-9
Dun, S., Wang, X., Ma, Y., Hao, J., Jinshen, W., Yongqu, L., et al. (2021). Nrf2-induced miR-23a-27a-24-2 Cluster Modulates Damage Repair of Intestinal Mucosa by Targeting the Bach1/HO-1 axis in Inflammatory Bowel Diseases. Free Radic. Biol. Med. 163, 1–9. doi:10.1016/j.freeradbiomed.2020.11.006
Engevik, M. A., Luk, B., Chang-Graham, A. L., Hall, A., Herrmann, B., Ruan, W., et al. (2019). Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio 10 (3), e01087–19. doi:10.1128/mBio.01087-19
Fan, F. Y., Sang, L. X., and Jiang, M. (2017). Catechins and Their Therapeutic Benefits to Inflammatory Bowel Disease. Molecules 22 (3), 484. doi:10.3390/molecules22030484
Fan, X., Jin, Y., Chen, G., Ma, X., and Zhang, L. (2021). Gut Microbiota Dysbiosis Drives the Development of Colorectal Cancer. Digestion 102 (4), 508–515. doi:10.1159/000508328
Fiorucci, S., Carino, A., Baldoni, M., Santucci, L., Costanzi, E., Graziosi, L., et al. (2021). Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 66 (3), 674–693. doi:10.1007/s10620-020-06715-3
Gallagher, K., Catesson, A., Griffin, J. L., Holmes, E., and Williams, H. R. T. (2021). Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J. Crohns Colitis 15 (5), 813–826. doi:10.1093/ecco-jcc/jjaa227
Gao, P., Liu, H., Huang, H., Zhang, Q., Strober, W., and Zhang, F. (2017). The Inflammatory Bowel Disease-Associated Autophagy Gene Atg16L1T300A Acts as a Dominant Negative Variant in Mice. J. Immunol. 198 (6), 2457–2467. doi:10.4049/jimmunol.1502652
Ge, Q., Chen, X., Zhao, Y., Mu, H., and Zhang, J. (2021). Modulatory Mechanisms of NLRP3: Potential Roles in Inflammasome Activation. Life Sci. 267, 118918. doi:10.1016/j.lfs.2020.118918
Gerster, R., Eloranta, J. J., Hausmann, M., Ruiz, P. A., Cosin-Roger, J., Terhalle, A., et al. (2015). Anti-Inflammatory Function of High-Density Lipoproteins via Autophagy of IκB Kinase. Cell Mol. Gastroenterol. Hepatol. 1 (2), 171–187, e171. doi:10.1016/j.jcmgh.2014.12.006
Gomaa, E. Z. (2020). Human Gut Microbiota/Microbiome in Health and Diseases: A Review. Antonie Van Leeuwenhoek 113 (12), 2019–2040. doi:10.1007/s10482-020-01474-7
Grondin, J. A., Kwon, Y. H., Far, P. M., Haq, S., and Khan, W. I. (2020). Mucins in Intestinal Mucosal Defense and Inflammation: Learning from Clinical and Experimental Studies. Front. Immunol. 11, 2054. doi:10.3389/fimmu.2020.02054
Guo, W., Sun, Y., Liu, W., Wu, X., Guo, L., Cai, P., et al. (2014). Small Molecule-Driven Mitophagy-Mediated NLRP3 Inflammasome Inhibition Is Responsible for the Prevention of Colitis-Associated Cancer. Autophagy 10 (6), 972–985. doi:10.4161/auto.28374
Haber, A. L., Biton, M., Rogel, N., Herbst, R. H., Shekhar, K., Smillie, C., et al. (2017). A Single-Cell Survey of the Small Intestinal Epithelium. Nature 551 (7680), 333–339. doi:10.1038/nature24489
Han, B., Zhang, Y., Zhang, Y., Bai, Y., Chen, X., Huang, R., et al. (2018). Novel Insight into Circular RNA HECTD1 in Astrocyte Activation via Autophagy by Targeting MIR142-TIPARP: Implications for Cerebral Ischemic Stroke. Autophagy 14 (7), 1164–1184. doi:10.1080/15548627.2018.1458173
Hassanpour, M., Hajihassani, F., Hiradfar, A., Aghamohammadzadeh, N., Rahbarghazi, R., Safaie, N., et al. (2020). Real-State of Autophagy Signaling Pathway in Neurodegenerative Disease; Focus on Multiple Sclerosis. J. Inflamm. 17, 6. doi:10.1186/s12950-020-0237-8
Homer, C. R., Richmond, A. L., Rebert, N. A., Achkar, J. P., and McDonald, C. (2010). ATG16L1 and NOD2 Interact in an Autophagy-Dependent Antibacterial Pathway Implicated in Crohn's Disease Pathogenesis. Gastroenterology 139 (5), 1630–1641, e1–e2. doi:10.1053/j.gastro.2010.07.006
Hu, L. F. (2019). Epigenetic Regulation of Autophagy. Adv. Exp. Med. Biol. 1206, 221–236. doi:10.1007/978-981-15-0602-4_11
Hu, W., Lu, W., Li, L., Zhang, H., Lee, Y. K., Chen, W., et al. (2021). Both Living and Dead Faecalibacterium Prausnitzii Alleviate House Dust Mite-Induced Allergic Asthma through the Modulation of Gut Microbiota and Short-Chain Fatty Acid Production. J. Sci. Food Agric. 101 (13), 5563–5573. doi:10.1002/jsfa.11207
Invernizzi, R., Wu, B. G., Barnett, J., Ghai, P., Kingston, S., Hewitt, R. J., et al. (2021). The Respiratory Microbiome in Chronic Hypersensitivity Pneumonitis is Distinct from that of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 203 (3), 339–347. doi:10.1164/rccm.202002-0460OC
Jamontt, J., Petit, S., Clark, N., Parkinson, S. J., and Smith, P. (2013). Nucleotide-Binding Oligomerization Domain 2 Signaling Promotes Hyperresponsive Macrophages and Colitis in IL-10-Deficient Mice. J. Immunol. 190 (6), 2948–2958. doi:10.4049/jimmunol.1201332
Järbrink-Sehgal, E., and Andreasson, A. (2020). The Gut Microbiota and Mental Health in Adults. Curr. Opin. Neurobiol. 62, 102–114. doi:10.1016/j.conb.2020.01.016
Jing, X., Zulfiqar, F., Park, S. Y., Núñez, G., Dziarski, R., and Gupta, D. (2014). Peptidoglycan Recognition Protein 3 and Nod2 Synergistically Protect Mice from Dextran Sodium Sulfate-Induced Colitis. J. Immunol. 193 (6), 3055–3069. doi:10.4049/jimmunol.1301548
Joffre, C., Ducau, C., Poillet-Perez, L., Courdy, C., and Mansat-De Mas, V. (2021). Autophagy a Close Relative of AML Biology. Biology 10 (6), 552. doi:10.3390/biology10060552
Kamel, A. M., Badary, M. S., Mohamed, W. A., Ahmed, G. H., and El-Feky, M. A. (2020). Evaluation of Autophagy-Related Genes in Egyptian Systemic Lupus Erythematosus Patients. Int. J. Rheum. Dis. 23 (9), 1226–1232. doi:10.1111/1756-185X.13910
Ke, P., Shao, B. Z., Xu, Z. Q., Chen, X. W., and Liu, C. (2016a). Intestinal Autophagy and its Pharmacological Control in Inflammatory Bowel Disease. Front. Immunol. 7, 695. doi:10.3389/fimmu.2016.00695
Ke, P., Shao, B. Z., Xu, Z. Q., Wei, W., Han, B. Z., Chen, X. W., et al. (2016b). Activation of Cannabinoid Receptor 2 Ameliorates DSS-Induced Colitis through Inhibiting NLRP3 Inflammasome in Macrophages. PLoS One 11 (9), e0155076. doi:10.1371/journal.pone.0155076
Kong, W., Li, H., Tuma, R. F., and Ganea, D. (2014). Selective CB2 Receptor Activation Ameliorates EAE by Reducing Th17 Differentiation and Immune Cell Accumulation in the CNS. Cell Immunol. 287 (1), 1–17. doi:10.1016/j.cellimm.2013.11.002
Kuma, A., Komatsu, M., and Mizushima, N. (2017). Autophagy-Monitoring and Autophagy-Deficient Mice. Autophagy 13 (10), 1619–1628. doi:10.1080/15548627.2017.1343770
Kumar, S., Nanduri, R., Bhagyaraj, E., Kalra, R., Ahuja, N., Chacko, A. P., et al. (2021). Vitamin D3-VDR-PTPN6 Axis Mediated Autophagy Contributes to the Inhibition of Macrophage Foam Cell Formation. Autophagy 17 (9), 2273–2289. doi:10.1080/15548627.2020.1822088
Lapaquette, P., Bringer, M. A., and Darfeuille-Michaud, A. (2012). Defects in Autophagy Favour Adherent-Invasive Escherichia coli Persistence within Macrophages Leading to Increased Pro-Inflammatory Response. Cell Microbiol 14 (6), 791–807. doi:10.1111/j.1462-5822.2012.01768.x
Larabi, A., Barnich, N., and Nguyen, H. T. T. (2020). New Insights into the Interplay between Autophagy, Gut Microbiota and Inflammatory Responses in IBD. Autophagy 16 (1), 38–51. doi:10.1080/15548627.2019.1635384
Leccese, G., Bibi, A., Mazza, S., Facciotti, F., Caprioli, F., Landini, P., et al. (2020). Probiotic Lactobacillus and Bifidobacterium Strains Counteract Adherent-Invasive Escherichia coli (AIEC) Virulence and Hamper IL-23/Th17 Axis in Ulcerative Colitis, but Not in Crohn’s Disease. Cells 9 (8), 1824. doi:10.3390/cells9081824
Lei-Leston, A. C., Murphy, A. G., and Maloy, K. J. (2017). Epithelial Cell Inflammasomes in Intestinal Immunity and Inflammation. Front. Immunol. 8, 1168. doi:10.3389/fimmu.2017.01168
Li, W., He, P., Huang, Y., Li, Y. F., Lu, J., Li, M., et al. (2021). Selective Autophagy of Intracellular Organelles: Recent Research Advances. Theranostics 11 (1), 222–256. doi:10.7150/thno.49860
Li, Y., Ma, K., Zhang, L., Xu, H., and Zhang, N. (2020). Human Umbilical Cord Blood Derived-Mesenchymal Stem Cells Alleviate Dextran Sulfate Sodium-Induced Colitis by Increasing Regulatory T Cells in Mice. Front. Cell Dev. Biol. 8, 604021. doi:10.3389/fcell.2020.604021
Lin, F., Zhu, Y. T., and Qin, Z. H. (2021). Biomarkers of Autophagy. Adv. Exp. Med. Biol. 1208, 265–287. doi:10.1007/978-981-16-2830-6_12
Lin, X. T., Zheng, X. B., Fan, D. J., Yao, Q. Q., Hu, J. C., Lian, L., et al. (2018). MicroRNA-143 Targets ATG2B to Inhibit Autophagy and Increase Inflammatory Responses in Crohn's Disease. Inflamm. Bowel Dis. 24 (4), 781–791. doi:10.1093/ibd/izx075
Liu, L., Saitz-Rojas, W., Smith, R., Gonyar, L., In, J. G., Kovbasnjuk, O., et al. (2020a). Mucus Layer Modeling of Human Colonoids during Infection with Enteroaggragative E. coli. Sci. Rep. 10 (1), 10533. doi:10.1038/s41598-020-67104-4
Liu, R., Li, X., Ma, H., Yang, Q., Shang, Q., Song, L., et al. (2020b). Spermidine Endows Macrophages Anti-inflammatory Properties by Inducing Mitochondrial Superoxide-Dependent AMPK Activation, Hif-1α Upregulation and Autophagy. Free Radic. Biol. Med. 161, 339–350. doi:10.1016/j.freeradbiomed.2020.10.029
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 569 (7758), 655–662. doi:10.1038/s41586-019-1237-9
Lőrincz, P., and Juhász, G. (2020). Autophagosome-Lysosome Fusion. J. Mol. Biol. 432 (8), 2462–2482. doi:10.1016/j.jmb.2019.10.028
Lu, R., Zhang, Y. G., Xia, Y., Zhang, J., Kaser, A., Blumberg, R., et al. (2021). Paneth Cell Alertness to Pathogens Maintained by Vitamin D Receptors. Gastroenterology 160 (4), 1269–1283. doi:10.1053/j.gastro.2020.11.015
Luo, H., He, J., Qin, L., Chen, Y., Chen, L., Li, R., et al. (2021a). Mycoplasma Pneumoniae Lipids License TLR-4 for Activation of NLRP3 Inflammasome and Autophagy to Evoke a Proinflammatory Response. Clin. Exp. Immunol. 203 (1), 66–79. doi:10.1111/cei.13510
Luo, X., Wang, X., Huang, S., Xu, B., Luo, S., Li, Y., et al. (2021b). Paeoniflorin Ameliorates Experimental Colitis by Inhibiting Gram-Positive Bacteria-Dependent MDP-NOD2 Pathway. Int. Immunopharmacol 90, 107224. doi:10.1016/j.intimp.2020.107224
M'Koma, A. E. (2013). Inflammatory Bowel Disease: an Expanding Global Health Problem. Clin. Med. Insights Gastroenterol. 6, 33–47. doi:10.4137/CGast.S12731
Macias-Ceja, D. C., Cosín-Roger, J., Ortiz-Masiá, D., Salvador, P., Hernández, C., Esplugues, J. V., et al. (2017). Stimulation of Autophagy Prevents Intestinal Mucosal Inflammation and Ameliorates Murine Colitis. Br. J. Pharmacol. 174 (15), 2501–2511. doi:10.1111/bph.13860
Mai, C. T., Wu, M. M., Wang, C. L., Su, Z. R., Cheng, Y. Y., and Zhang, X. J. (2019). Palmatine Attenuated Dextran Sulfate Sodium (DSS)-induced Colitis via Promoting Mitophagy-Mediated NLRP3 Inflammasome Inactivation. Mol. Immunol. 105, 76–85. doi:10.1016/j.molimm.2018.10.015
Mao, L. Q., Zhou, Y. L., Wang, S. S., Chen, L., Hu, Y., Yu, L. M., et al. (2021). Impact of Helicobacter pylori Eradication on the Gastric Microbiome. Gut Pathog. 13 (1), 60. doi:10.1186/s13099-021-00460-2
Mariathasan, S., Newton, K., Monack, D. M., Vucic, D., French, D. M., Lee, W. P., et al. (2004). Differential Activation of the Inflammasome by Caspase-1 Adaptors ASC and Ipaf. Nature 430 (6996), 213–218. doi:10.1038/nature02664
Mazur, M., Zielińska, A., Grzybowski, M. M., Olczak, J., and Fichna, J. (2021). Chitinases and Chitinase-Like Proteins as Therapeutic Targets in Inflammatory Diseases, with a Special Focus on Inflammatory Bowel Diseases. Int. J. Mol. Sci. 22 (13), 6966. doi:10.3390/ijms22136966
McCormick, J. J., King, K. E., Côté, M. D., Meade, R. D., Akerman, A. P., and Kenny, G. P. (2021). Impaired Autophagy Following Ex Vivo Heating at Physiologically Relevant Temperatures in Peripheral Blood Mononuclear Cells from Elderly Adults. J. Therm. Biol. 95, 102790. doi:10.1016/j.jtherbio.2020.102790
Mehandru, S., and Colombel, J. F. (2021). The Intestinal Barrier, an Arbitrator Turned Provocateur in IBD. Nat. Rev. Gastroenterol. Hepatol. 18 (2), 83–84. doi:10.1038/s41575-020-00399-w
Mirsepasi-Lauridsen, H. C., Vallance, B. A., Krogfelt, K. A., and Petersen, A. M. (2019). Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 32 (2), e00060. doi:10.1128/CMR.00060-18
Moretti, J., and Blander, J. M. (2021). Increasing Complexity of NLRP3 Inflammasome Regulation. J. Leukoc. Biol. 109 (3), 561–571. doi:10.1002/JLB.3MR0520-104RR
Muller, P. A., Matheis, F., and Mucida, D. (2020). Gut Macrophages: Key Players in Intestinal Immunity and Tissue Physiology. Curr. Opin. Immunol. 62, 54–61. doi:10.1016/j.coi.2019.11.011
Nash, P., Kerschbaumer, A., Dörner, T., Dougados, M., Fleischmann, R. M., Geissler, K., et al. (2021). Points to Consider for the Treatment of Immune-Mediated Inflammatory Diseases with Janus Kinase Inhibitors: A Consensus Statement. Ann. Rheum. Dis. 80 (1), 71–87. doi:10.1136/annrheumdis-2020-218398
Nayar, S., Morrison, J. K., Giri, M., Gettler, K., Chuang, L. S., Walker, L. A., et al. (2021). A Myeloid-Stromal Niche and Gp130 rescue in NOD2-Driven Crohn’s Disease. Nature 593 (7858), 275–281. doi:10.1038/s41586-021-03484-5
Netea, M. G., Kullberg, B. J., de Jong, D. J., Franke, B., Sprong, T., Naber, T. H., et al. (2004). NOD2 Mediates Anti-Inflammatory Signals Induced by TLR2 Ligands: Implications for Crohn’s Disease. Eur. J. Immunol. 34 (7), 2052–2059. doi:10.1002/eji.200425229
Ouimet, M., Ediriweera, H., Afonso, M. S., Ramkhelawon, B., Singaravelu, R., Liao, X., et al. (2017). microRNA-33 Regulates Macrophage Autophagy in Atherosclerosis. Arterioscler Thromb. Vasc. Biol. 37 (6), 1058–1067. doi:10.1161/ATVBAHA.116.308916
Owczarek, D., Rodacki, T., Domagała-Rodacka, R., Cibor, D., and Mach, T. (2016). Diet and Nutritional Factors in Inflammatory Bowel Diseases. World J. Gastroenterol. 22 (3), 895–905. doi:10.3748/wjg.v22.i3.895
Palmela, C., Chevarin, C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., et al. (2018). Adherent-invasive Escherichia coli in Inflammatory Bowel Disease. Gut 67 (3), 574–587. doi:10.1136/gutjnl-2017-314903
Pardigol, A., Forssmann, U., Zucht, H. D., Loetscher, P., Schulz-Knappe, P., Baggiolini, M., et al. (1998). HCC-2, a Human Chemokine: Gene Structure, Expression Pattern, and Biological Activity. Proc. Natl. Acad. Sci. U.S.A. 95 (11), 6308–6313. doi:10.1073/pnas.95.11.6308
Parkes, M. (2012). Evidence from Genetics for a Role of Autophagy and Innate Immunity in IBD Pathogenesis. Dig. Dis. 30 (4), 330–333. doi:10.1159/000338119
Perdue, M. H., and McKay, D. M. (1994). Integrative Immunophysiology in the Intestinal Mucosa. Am. J. Physiol. 267 (2 Pt 1), G151–G165. doi:10.1152/ajpgi.1994.267.2.G151
Qin, Y., Qiu, J., Wang, P., Liu, J., Zhao, Y., Jiang, F., et al. (2021). Impaired Autophagy in Microglia Aggravates Dopaminergic Neurodegeneration by Regulating NLRP3 Inflammasome Activation in Experimental Models of Parkinson's Disease. Brain Behav. Immun. 91, 324–338. doi:10.1016/j.bbi.2020.10.010
Qin, Z., Wan, J. J., Sun, Y., Wu, T., Wang, P. Y., Du, P., et al. (2017). Nicotine Protects against DSS Colitis through Regulating microRNA-124 and STAT3. J. Mol. Med. 95 (2), 221–233. doi:10.1007/s00109-016-1473-5
Racanelli, A. C., Kikkers, S. A., Choi, A. M. K., and Cloonan, S. M. (2018). Autophagy and Inflammation in Chronic Respiratory Disease. Autophagy 14 (2), 221–232. doi:10.1080/15548627.2017.1389823
Rolhion, N., and Darfeuille-Michaud, A. (2007). Adherent-invasive Escherichia coli in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 13 (10), 1277–1283. doi:10.1002/ibd.20176
Rousset, F., Cabezas-Caballero, J., Piastra-Facon, F., Fernández-Rodríguez, J., Clermont, O., Denamur, E., et al. (2021). The Impact of Genetic Diversity on Gene Essentiality within the Escherichia coli Species. Nat. Microbiol. 6 (3), 301–312. doi:10.1038/s41564-020-00839-y
Rubinsztein, D. C., and Frake, R. A. (2016). Yoshinori Ohsumi’s Nobel Prize for Mechanisms of Autophagy: From Basic Yeast Biology to Therapeutic Potential. J. R. Coll. Physicians Edinb. 46 (4), 228–233. doi:10.4997/JRCPE.2016.403
Ryan, F. J., Ahern, A. M., Fitzgerald, R. S., Laserna-Mendieta, E. J., Power, E. M., Clooney, A. G., et al. (2020). Colonic Microbiota is Associated With Inflammation and Host Epigenomic Alterations in Inflammatory Bowel Disease. Nat. Commun. 11 (1), 1512. doi:10.1038/s41467-020-15342-5
Samie, M., Lim, J., Verschueren, E., Baughman, J. M., Peng, I., Wong, A., et al. (2018). Selective Autophagy of the Adaptor TRIF Regulates Innate Inflammatory Signaling. Nat. Immunol. 19 (3), 246–254. doi:10.1038/s41590-017-0042-6
Samoilă, I., Dinescu, S., and Costache, M. (2020). Interplay between Cellular and Molecular Mechanisms Underlying Inflammatory Bowel Diseases Development-A Focus on Ulcerative Colitis. Cells 9 (7), 1647. doi:10.3390/cells9071647
Saxena, A., Lopes, F., Poon, K. K. H., and McKay, D. M. (2017). Absence of the NOD2 Protein Renders Epithelia More Susceptible to Barrier Dysfunction Due to Mitochondrial Dysfunction. Am. J. Physiol. Gastrointest. Liver Physiol. 313 (1), G26–G38. doi:10.1152/ajpgi.00070.2017
Schwerd, T., Pandey, S., Yang, H. T., Bagola, K., Jameson, E., Jung, J., et al. (2017). Impaired Antibacterial Autophagy Links Granulomatous Intestinal Inflammation in Niemann-Pick Disease Type C1 and XIAP Deficiency with NOD2 Variants in Crohn’s Disease. Gut 66 (6), 1060–1073. doi:10.1136/gutjnl-2015-310382
Shao, B. Z., Han, B. Z., Zeng, Y. X., Su, D. F., and Liu, C. (2016). The Roles of Macrophage Autophagy in Atherosclerosis. Acta Pharmacol. Sin 37 (2), 150–156. doi:10.1038/aps.2015.87
Shao, B. Z., Ke, P., Xu, Z. Q., Wei, W., Cheng, M. H., Han, B. Z., et al. (2017). Autophagy Plays an Important Role in Anti-Inflammatory Mechanisms Stimulated by Alpha7 Nicotinic Acetylcholine Receptor. Front. Immunol. 8, 553. doi:10.3389/fimmu.2017.00553
Shao, B. Z., Wang, S. L., Fang, J., Li, Z. S., Bai, Y., and Wu, K. (2019a). Alpha7 Nicotinic Acetylcholine Receptor Alleviates Inflammatory Bowel Disease through Induction of AMPK-mTOR-p70S6K-Mediated Autophagy. Inflammation 42 (5), 1666–1679. doi:10.1007/s10753-019-01027-9
Shao, B. Z., Wang, S. L., Pan, P., Yao, J., Wu, K., Li, Z. S., et al. (2019b). Targeting NLRP3 Inflammasome in Inflammatory Bowel Disease: Putting Out the Fire of Inflammation. Inflammation 42 (4), 1147–1159. doi:10.1007/s10753-019-01008-y
Shao, B. Z., Yao, Y., Zhai, J. S., Zhu, J. H., Li, J. P., and Wu, K. (2021). The Role of Autophagy in Inflammatory Bowel Disease. Front. Physiol. 12, 621132. doi:10.3389/fphys.2021.621132
Shaoul, R., Hong, D., Okada, Y., Cutz, E., and Marcon, M. A. (2005). Lineage Development in a Patient without Goblet, Paneth, and Enteroendocrine Cells: A Clue for Intestinal Epithelial Differentiation. Pediatr. Res. 58 (3), 492–498. doi:10.1203/01.PDR.0000179408.74781.C9
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 33 (9), 496–503. doi:10.1016/j.tibtech.2015.06.011
Sultan, S., El-Mowafy, M., Elgaml, A., Ahmed, T. A. E., Hassan, H., and Mottawea, W. (2021). Metabolic Influences of Gut Microbiota Dysbiosis on Inflammatory Bowel Disease. Front. Physiol. 12, 715506. doi:10.3389/fphys.2021.715506
Sun, M. F., and Shen, Y. Q. (2018). Dysbiosis of Gut Microbiota and Microbial Metabolites in Parkinson's Disease. Ageing Res. Rev. 45, 53–61. doi:10.1016/j.arr.2018.04.004
Tan, S., and Chen, S. (2021). Macrophage Autophagy and Silicosis: Current Perspective and Latest Insights. Int. J. Mol. Sci. 22 (1), 453. doi:10.3390/ijms22010453
Tang, Y., Liu, J., Wang, Y., Yang, L., Han, B., Zhang, Y., et al. (2021). PARP14 Inhibits Microglial Activation via LPAR5 to Promote Post-Stroke Functional Recovery. Autophagy 17 (10), 2905–2922. doi:10.1080/15548627.2020.1847799
Tartakover Matalon, S., Ringel, Y., Konikoff, F., Drucker, L., Pery, S., and Naftali, T. (2020). Cannabinoid Receptor 2 Agonist Promotes Parameters Implicated in Mucosal Healing in Patients with Inflammatory Bowel Disease. United Eur. Gastroenterol J. 8 (3), 271–283. doi:10.1177/2050640619889773
Tomasello, G., Mazzola, M., Leone, A., Sinagra, E., Zummo, G., Farina, F., et al. (2016). Nutrition, Oxidative Stress and Intestinal Dysbiosis: Influence of Diet on Gut Microbiota in Inflammatory Bowel Diseases. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub 160 (4), 461–466. doi:10.5507/bp.2016.052
Trelford, C. B., and Di Guglielmo, G. M. (2021). Molecular Mechanisms of Mammalian Autophagy. Biochem. J. 478 (18), 3395–3421. doi:10.1042/BCJ20210314
Tschurtschenthaler, M., and Adolph, T. E. (2018). The Selective Autophagy Receptor Optineurin in Crohn’s Disease. Front. Immunol. 9, 766. doi:10.3389/fimmu.2018.00766
Uhlig, H. H., Charbit-Henrion, F., Kotlarz, D., Shouval, D. S., Schwerd, T., Strisciuglio, C., et al. (2021). Clinical Genomics for the Diagnosis of Monogenic Forms of Inflammatory Bowel Disease: A Position Paper From the Paediatric IBD Porto Group of European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J. Pediatr. Gastroenterol. Nutr. 72 (3), 456–473. doi:10.1097/MPG.0000000000003017
Vannella, K. M., and Wynn, T. A. (2017). Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 79, 593–617. doi:10.1146/annurev-physiol-022516-034356
Vazeille, E., Buisson, A., Bringer, M. A., Goutte, M., Ouchchane, L., Hugot, J. P., et al. (2015). Monocyte-Derived Macrophages from Crohn's Disease Patients Are Impaired in the Ability to Control Intracellular Adherent-Invasive Escherichia coli and Exhibit Disordered Cytokine Secretion Profile. J. Crohns Colitis. 9 (5), 410–420. doi:10.1093/ecco-jcc/jjv053
Viola, M. F., and Boeckxstaens, G. (2020). Intestinal Resident Macrophages: Multitaskers of the Gut. Neurogastroenterol. Motil. 32 (8), e13843. doi:10.1111/nmo.13843
Wallace, K. L., Zheng, L. B., Kanazawa, Y., and Shih, D. Q. (2014). Immunopathology of Inflammatory Bowel Disease. World J. Gastroenterol. 20 (1), 6–21. doi:10.3748/wjg.v20.i1.6
Wang, P., Shao, B. Z., Deng, Z., Chen, S., Yue, Z., and Miao, C. Y. (2018). Autophagy in Ischemic Stroke. Prog. Neurobiol. 163–164, 98–117. doi:10.1016/j.pneurobio.2018.01.001
Wang, S., Li, J., Du, Y., Xu, Y., Wang, Y., Zhang, Z., et al. (2017). The Class I PI3K Inhibitor S14161 Induces Autophagy in Malignant Blood Cells by Modulating the Beclin 1/Vps34 Complex. J. Pharmacol. Sci. 134 (4), 197–202. doi:10.1016/j.jphs.2017.07.001
Wang, W., Li, J., Tan, J., Wang, M., Yang, J., Zhang, Z. M., et al. (2021). Endonuclease G Promotes Autophagy by Suppressing mTOR Signaling and Activating the DNA Damage Response. Nat. Commun. 12 (1), 476. doi:10.1038/s41467-020-20780-2
Wang, X., Zhao, N. Q., Sun, Y. X., Bai, X., Si, J. T., Liu, J. P., et al. (2020). Acupuncture for Ulcerative Colitis: a Systematic Review and Meta-Analysis of Randomized Clinical Trials. BMC Complement. Med. Ther. 20 (1), 309. doi:10.1186/s12906-020-03101-4
Wang, Z., Shi, L., Hua, S., Qi, C., and Fang, M. (2019). IL-33 Ameliorates Experimental Colitis Involving Regulation of Autophagy of Macrophages in Mice. Cell Biosci 9, 10. doi:10.1186/s13578-019-0271-5
Weber, B., Saurer, L., Schenk, M., Dickgreber, N., and Mueller, C. (2011). CX3CR1 Defines Functionally Distinct Intestinal Mononuclear Phagocyte Subsets Which Maintain Their Respective Functions during Homeostatic and Inflammatory Conditions. Eur. J. Immunol. 41 (3), 773–779. doi:10.1002/eji.201040965
Wu, H., Zheng, J., Xu, S., Fang, Y., Wu, Y., Zeng, J., et al. (2021a). Mer Regulates Microglial/Macrophage M1/M2 Polarization and Alleviates Neuroinflammation Following Traumatic Brain Injury. J. Neuroinflammation 18 (1), 2. doi:10.1186/s12974-020-02041-7
Wu, M. Y., Liu, L., Wang, E. J., Xiao, H. T., Cai, C. Z., Wang, J., et al. (2021b). PI3KC3 Complex Subunit NRBF2 is Required for Apoptotic Cell Clearance to Restrict Intestinal Inflammation. Autophagy 17 (5), 1096–1111. doi:10.1080/15548627.2020.1741332
Xia, Y., Liu, N., Xie, X., Bi, G., Ba, H., Li, L., et al. (2019). The Macrophage-Specific V-ATPase Subunit ATP6V0D2 Restricts Inflammasome Activation and Bacterial Infection by Facilitating Autophagosome-Lysosome Fusion. Autophagy 15 (6), 960–975. doi:10.1080/15548627.2019.1569916
Xu, W., Ocak, U., Gao, L., Tu, S., Lenahan, C. J., Zhang, J., et al. (2021). Selective Autophagy as a Therapeutic Target for Neurological Diseases. Cell Mol. Life Sci. 78 (4), 1369–1392. doi:10.1007/s00018-020-03667-9
Xu, Y., Shen, J., and Ran, Z. (2020). Emerging Views of Mitophagy in Immunity and Autoimmune Diseases. Autophagy 16 (1), 3–17. doi:10.1080/15548627.2019.1603547
Yang, S., and Yu, M. (2021). Role of Goblet Cells in Intestinal Barrier and Mucosal Immunity. J. Inflamm. Res. 14, 3171–3183. doi:10.2147/JIR.S318327
Yao, R. Q., Ren, C., Xia, Z. F., and Yao, Y. M. (2021). Organelle-Specific Autophagy in Inflammatory Diseases: A Potential Therapeutic Target Underlying the Quality Control of Multiple Organelles. Autophagy 17 (2), 385–401. doi:10.1080/15548627.2020.1725377
Yeh, H., Kuo, C. J., Wu, R. C., Chen, C. M., Tsai, W. S., Su, M. Y., et al. (2021). Vedolizumab in Crohn's Disease With Rectal Fistulas and Presacral Abscess: A Case Report. World J. Gastroenterol. 27 (5), 442–448. doi:10.3748/wjg.v27.i5.442
Yoshimura, C., Nagasaka, A., Kurose, H., and Nakaya, M. (2020). Efferocytosis during Myocardial Infarction. J. Biochem. 168 (1), 1–6. doi:10.1093/jb/mvaa051
You, X. Y., Zhang, H. Y., Han, X., Wang, F., Zhuang, P. W., and Zhang, Y. J. (2021). Intestinal Mucosal Barrier is Regulated by Intestinal Tract Neuro-Immune Interplay. Front. Pharmacol. 12, 659716. doi:10.3389/fphar.2021.659716
Yu, L., Chen, Y., and Tooze, S. A. (2018). Autophagy Pathway: Cellular and Molecular Mechanisms. Autophagy 14 (2), 207–215. doi:10.1080/15548627.2017.1378838
Yu, X., Long, Y. C., and Shen, H. M. (2015). Differential Regulatory Functions of Three Classes of Phosphatidylinositol and Phosphoinositide 3-Kinases in Autophagy. Autophagy 11 (10), 1711–1728. doi:10.1080/15548627.2015.1043076
Yunna, C., Mengru, H., Lei, W., and Weidong, C. (2020). Macrophage M1/M2 Polarization. Eur. J. Pharmacol. 877, 173090. doi:10.1016/j.ejphar.2020.173090
Zhang, H., Zheng, L., McGovern, D. P., Hamill, A. M., Ichikawa, R., Kanazawa, Y., et al. (2017a). Myeloid ATG16L1 Facilitates Host-Bacteria Interactions in Maintaining Intestinal Homeostasis. J. Immunol. 198 (5), 2133–2146. doi:10.4049/jimmunol.1601293
Zhang, J., Ding, R., Wu, T., Jia, J., and Cheng, X. (2021). Autophagy-Related Genes and Long Noncoding RNAs Signatures as Predictive Biomarkers for Osteosarcoma Survival. Front. Cell Dev. Biol. 9, 705291. doi:10.3389/fcell.2021.705291
Zhang, S. L., Wang, S. N., and Miao, C. Y. (2017b). Influence of Microbiota on Intestinal Immune System in Ulcerative Colitis and Its Intervention. Front. Immunol. 8, 1674. doi:10.3389/fimmu.2017.01674
Zhao, Y., Guo, Q., Zhao, K., Zhou, Y., Li, W., Pan, C., et al. (2017). Small Molecule GL-V9 Protects Against Colitis-Associated Colorectal Cancer by Limiting NLRP3 Inflammasome through Autophagy. Oncoimmunology 7 (1), e1375640. doi:10.1080/2162402X.2017.1375640
Keywords: inflammatory bowel disease, macrophage, autophagy, intestinal, inflammation
Citation: Zheng Y, Yu Y, Chen X-F, Yang S-L, Tang X-L and Xiang Z-G (2021) Intestinal Macrophage Autophagy and its Pharmacological Application in Inflammatory Bowel Disease. Front. Pharmacol. 12:803686. doi: 10.3389/fphar.2021.803686
Received: 28 October 2021; Accepted: 09 November 2021;
Published: 24 November 2021.
Edited by:
Bo-Zong Shao, General Hospital of Chinese PLA, ChinaReviewed by:
Zhe-Qi Xu, Second Military Medical University, ChinaShu-Ling Wang, Second Military Medical University, China
Copyright © 2021 Zheng, Yu, Chen, Yang, Tang and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-Guo Xiang, xzg5851@163.com
 Yang Zheng1
Yang Zheng1 Zheng-Guo Xiang
Zheng-Guo Xiang