Autism: A model of neurodevelopmental diversity informed by genomics
- 1Gillberg Neuropsychiatry Centre, Department of Psychiatry and Neurochemistry, Institute of Neuroscience and Physiology, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden
- 2Harvard Medical School, Boston, MA, United States
- 3Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, United States
- 4Department of Radiology, Sahlgrenska University Hospital, Gothenburg, Sweden
- 5MedTech West, Gothenburg, Sweden
A Commentary on
Autism: A model of neurodevelopmental diversity informed by genomics
by Chawner, S. J. R. A., and Owen, M. J. (2022). Front. Psychiatry 13:981691. doi: 10.3389/fpsyt.2022.981691
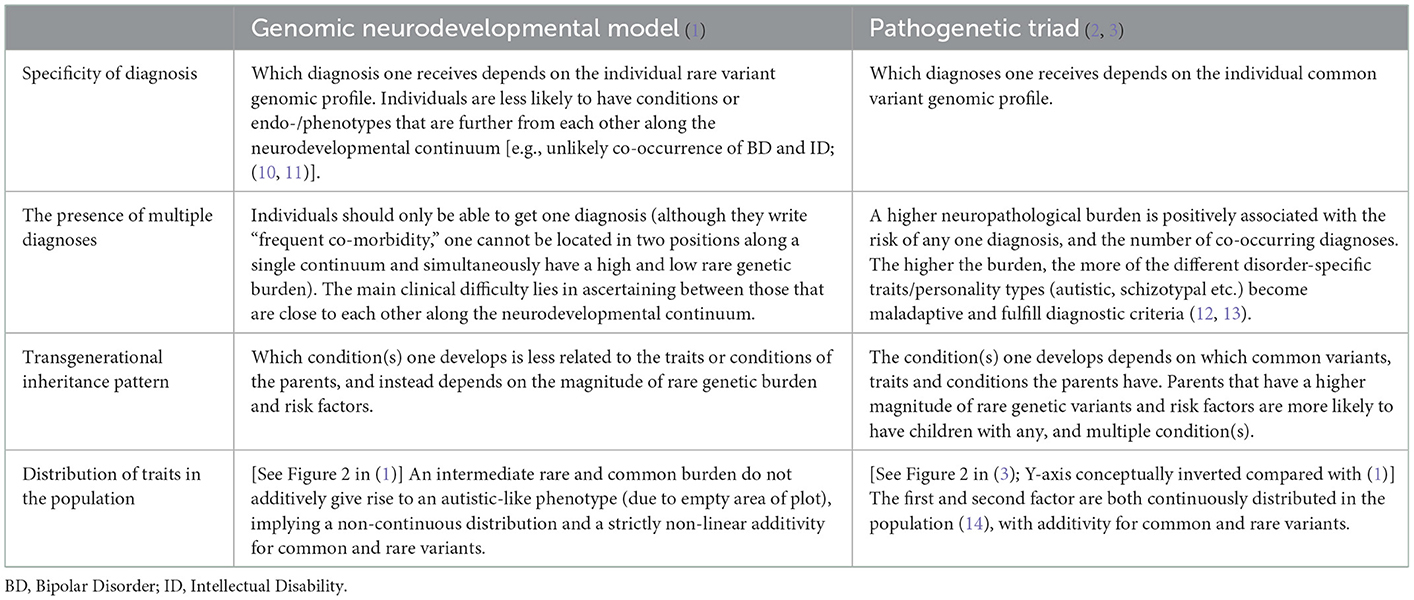
In their paper, Chawner and Owen (1) present a genetic model for autism that outlines two contributory factors: (1) a social and adaptive continuum due to common genetic variation; and (2) a neurodevelopmental continuum due to rare genetic variation that presents itself as a continuum of impairment spanning from intellectual disability, through autism and ADHD, to schizophrenia and bipolar disorder.
I applaud the authors on relating the main mechanisms of the model to the differing views between the neurodiversity community and that of the medical model regarding the nature of autism, as they pertain to different aspects of the phenotype and both being important for explaining variability in clinical presentations. The model itself is very similar to part of a more comprehensive model I previously proposed (2, 3) and although there are many similarities between the papers, it is worth noting some empirical differences with important ramifications. I will argue that their conceptualization is not supported by the current literature and that it contains an issue that limits its practical usefulness. I will conclude by presenting testable postulates arising from the two models which will allow future studies to empirically validate them.
They write that the “neurodevelopmental continuum […] results in a diverse spectrum of outcomes,” referring to individual diagnoses under the neurodevelopmental umbrella. It does that through the effects of rare genetic mutations and environmental risk factors. As they operationalize it, the magnitude of the rare genetic burden determines which phenotype develops, and ultimately which diagnosis is received [see Figure 1 in (1)]. Conceptually, greater impairment is more closely associated with intellectual disability and autism than with schizophrenia and bipolar disorder.
Although the apparent statistical associations of these features appear in the literature, there is an issue with this operationalization that can be illustrated with an example. Consider an individual with a rare genetic burden of a given magnitude (Xinherited) and a diagnosis of bipolar disorder. If that individual were to have a child with inherited said burden, but with additional de novo variants (Xinherited + Xde novo) that child should be more likely to develop autism or ADHD than bipolar disorder. The idea that the type of condition one develops is contingent on the magnitude of rare genetic risk is not supported by empirical evidence. The conditions have partly independent genotypic (4–7), and neuroendophenotypic signatures (8, 9), suggesting that they also have partly different biological backgrounds, rather than them being part of a single continuum. A person with bipolar disorder can certainly have a lower IQ and greater “cognitive impairment” than someone with an autism diagnosis. The operationalization of the neurodevelopmental continuum alludes to a causative mechanism by which the magnitude of the rare genetic burden impacts specificity of diagnosis. This is empirically unlikely given the state of the literature, unless the continuum is a pseudo-unidimensional manifold rather than linear, and it therefore probably represents a statistical artifact.
Furthermore, following the conceptualization of a neurodevelopmental continuum, the addition of a social-adaptive factor to the model is not without issues since the autistic phenotype (which also encompasses such traits) is already conceptualized along the first factor. Clearly, the second factor is conceptualized in order to accommodate the literature on the association between autism and common genetic variation. However, within the proposed model one cannot dissociate the autistic phenotypes residing within each of the factors (whether an autistic trait belongs to the social-adaptive or the neurodevelopmental continuum), greatly limiting the practical utility of the proposed model.
Their operationalization can be contrasted with that of the pathogenetic triad (2, 3), which previously suggested that there is (1) natural variation in non-pathological traits (such as autistic or schizotypal) due to common genetic variation, and (2) a range of neurodevelopmental risk factors including, but not limited to, rare genetic variation. These risk factors negatively influence brain and cognitive development, and limit adaptive behaviors. Notably, adaptive behavior is conceptualized within a third factor that moderates the association between the first two factors in giving rise to a diagnosis. This is an important distinction since Chawner and Owen seem to conceptualize adaptive behavior within the first factor as “social-adaptive traits” (although, they do not formally operationalize it). These two factors additively influence the risk, and crucially, the first factor provides the model with disorder specificity (through common variant burden for each condition, not rare burden). Also, rather than the magnitude of neurodevelopmental risk factors affecting which condition develops (as in their model), it non-specifically determines the probability of fulfilling criteria for any one diagnosis (or multiple).
Although the models are similar, there are subtle differences that give rise to different empirical predictions, each with testable postulates. In Table 1 present a few of these predictions, and the patterns in the existing and future literature that would favor one model or the other (some of which are already supported or undermined).
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The author was supported by Fredrik och Ingrid Thurings stiftelse (2020-00581), the Gothenburg Society of Medicine and Kristina Stenborgs Stiftelse (GLS-960453), Stiftelsen Professor Bror Gadelius Minnesfond, Stiftelsen Systrarna Greta Johansson och Brita Anderssons minnesfond, and FoUU Department of Radiology, Sahlgrenska University Hospital.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chawner S, Owen MJ. Autism: a model of neurodevelopmental diversity informed by genomics. Front Psychiatry. (2022) 13:981691. doi: 10.3389/fpsyt.2022.981691
2. Sarovic D. A framework for neurodevelopmental disorders: operationalization of a pathogenetic triad. PsyArXiv [preprint] (2019). doi: 10.31234/osf.io/mbeqh
3. Sarovic D. A unifying theory for autism: the pathogenetic triad as a theoretical framework. Front Psychiatry. (2021) 12:767075. doi: 10.3389/fpsyt.2021.767075
4. Cross-Disorder Cross-Disorder Group of the Psychiatric Genomics C, Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. (2013) 45:984–94. doi: 10.1038/ng.2711
5. Jansen AG, Dieleman GC, Jansen PR, Verhulst FC, Posthuma D, Polderman TJC. Psychiatric polygenic risk scores as predictor for attention deficit/hyperactivity disorder and autism spectrum disorder in a clinical child and adolescent sample. Behav Genet. (2020) 50:203–12. doi: 10.1007/s10519-019-09965-8
6. Castelbaum L, Sylvester CM, Zhang Y, Yu Q, Constantino JN. On the nature of monozygotic twin concordance and discordance for autistic trait severity: a quantitative analysis. Behav Genet. (2020) 50:263–72. doi: 10.1007/s10519-019-09987-2
7. St Pourcain B, Robinson EB, Anttila V, Sullivan BB, Maller J, Golding J, et al. ASD and schizophrenia show distinct developmental profiles in common genetic overlap with population-based social communication difficulties. Mol Psychiatry. (2018) 23, 263–70. doi: 10.1038/mp.2016.198
8. Crespi B, Badcock C. Psychosis and autism as diametrical disorders of the social brain. Behav Brain Sci. (2008) 31:241–61; discussion 61–320. doi: 10.1017/S0140525X08004214
9. Opel N, Goltermann J, Hermesdorf M, Berger K, Baune BT, Dannlowski U. Cross-disorder analysis of brain structural abnormalities in six major psychiatric disorders: a secondary analysis of mega- and meta-analytical findings from the ENIGMA consortium. Biol Psychiatry. (2020) 88:678–86. doi: 10.1016/j.biopsych.2020.04.027
10. Keyes K. Psychiatric comorbidity and intellectual disability. Eur J Public Health. (2019) 29:ckz185.380. doi: 10.1093/eurpub/ckz185.380
11. Platt JM, Keyes KM, McLaughlin KA, Kaufman AS. Intellectual disability and mental disorders in a US population representative sample of adolescents. Psychol Med. (2019) 49:952–61. doi: 10.1017/S0033291718001605
12. Warrier V, Zhang X, Reed P, Havdahl A, Moore TM, Cliquet F, et al. Genetic correlates of phenotypic heterogeneity in autism. Nat Genet. (2022) 54:1293–304. doi: 10.1038/s41588-022-01072-5
13. Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. (2017) 49:978–85. doi: 10.1038/ng.3863
14. Ruzich E, Allison C, Smith P, Watson P, Auyeung B, Ring H, et al. Measuring autistic traits in the general population: a systematic review of the Autism-Spectrum Quotient (AQ) in a nonclinical population sample of 6,900 typical adult males and females. Mol Autism. (2015) 6:2. doi: 10.1186/2040-2392-6-2
Keywords: autism, theoretical framework, model, genetic architecture, neurodevelopmental disorder (NDD), rare genetic variant, common genetic variant
Citation: Sarovic D (2023) Commentary: Autism: A model of neurodevelopmental diversity informed by genomics. Front. Psychiatry 14:1113592. doi: 10.3389/fpsyt.2023.1113592
Received: 01 December 2022; Accepted: 09 January 2023;
Published: 24 January 2023.
Edited by:
Daniel Campbell, Michigan State University, United StatesReviewed by:
Leonardo Zoccante, Integrated University Hospital Verona, ItalyCopyright © 2023 Sarovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darko Sarovic,  darko.sarovic@gu.se
darko.sarovic@gu.se
 Darko Sarovic
Darko Sarovic