- Library and Information Science Department, Yonsei University, Seoul, South Korea
In today’s era of information explosion, extracting entities and their relations in large-scale, unstructured collections of text to better represent knowledge has emerged as a daunting challenge in biomedical text mining. To respond to the demand to automatically extract scientific knowledge with higher precision, the public knowledge discovery tool PKDE4J (Song et al., 2015) was proposed as a flexible text-mining tool. In this study, we propose an extended version of PKDE4J to represent scientific knowledge for literature-based knowledge discovery. Specifically, we assess the performance of PKDE4J in terms of three extraction tasks: entity, relation, and event detection. We also suggest applications of PKDE4J along three lines: (1) knowledge search, (2) knowledge linking, and (3) knowledge inference. We first describe the updated features of PKDE4J and report on tests of its performance. With additional options in the processes of named entity extraction, verb expansion, and event detection, we expect that the enhanced PKDE4J can be utilized for literature-based knowledge discovery.
Introduction
Owing to the deluge of data in today’s digital world, mining useful information from large-scale, unstructured collections of text is a challenging task. The demand to discover knowledge from large amounts of data has been steadily growing over the years. Knowledge extraction requires at least two techniques, named entity recognition (NER) and relation extraction (RE). Identifying entities represented in text and the relations among them is a fundamental process of knowledge extraction. Using this process, the extracted knowledge can be utilized in a knowledge network or various systems. The US National Academy of Sciences claimed in a 2011 report that a biomedical knowledge network based on biological data and knowledge is essential for precision medicine (National Research Council, 2011). For knowledge extraction, Song et al. (2015) proposed PKDE4J, the Public Knowledge Discovery Engine for Java. The goal of PKDE4J is to extract biomedical knowledge from unstructured texts for literature-based knowledge discovery. This is a daunting goal requiring long-term research and development. In our previous study, we introduced PKDE4J as a knowledge extraction system (Song et al., 2015). In this paper, as a first step, we extend PKDE4J to make it flexible as possible, such that it can be applied to various knowledge extraction tasks. In the second step, knowledge identification, we focus on how PKDE4J can be used to represent scientific knowledge.
Several data mining-based approaches to represent biological knowledge have been proposed. Bio2RDF is a mash-up system that can be used to integrate knowledge from multiple bioinformatics databases (Belleau et al., 2008). Bell et al. (2011) integrated bio-entities and their relations into an existing database. A feature of this approach is that it utilizes structured data. However, in this paper, we introduce an extended version of PKDE4J based on text mining for users in the biomedical domain to transition from the micro-level of knowledge entities to macro-topical level by applying it to unstructured data. Swanson’s ABC model (Swanson, 1986) helped unveil knowledge discovery in terms of constructing a knowledge network and discovering new knowledge.
Prior to knowledge discovery with techniques from text mining, a knowledge extraction stage is needed. A biological entity is tagged according to its type, such as gene, disease, cell, and tissue. In PKDE4J, we extended the NER process so that it can be conducted in several modes, such as dictionary-based and machine learning-based methods combined with ontology. The RE process follows to determine the relations among the entities. This process is performed by using a set of predefined rules.
In past studies, NER has been used to extract entities and their types from text (Hanisch et al., 2005; Yang et al., 2008; Munkhdalai et al., 2015; Tang et al., 2015; Leaman and Lu, 2016). In the biomedical field, types usually include gene, disease, and chemicals.
A dictionary-based or a lexicon-based approach is widely used in biomedicine. It matches terms from prepared dictionaries to a given text. Despite its simplicity and high accuracy, there are two major problems in the dictionary-based approach. The first is its possible omission of new terminology not included in the dictionary, and the second problem is a matching problem of variants and synonyms in the dictionary. Several studies (Yang et al., 2008; Munkhdalai et al., 2015) have attempted to combine various dictionaries to solve these problems.
The rule-based approach observes general features of an entity in text and extracts entities based on heuristically acquired rules. These features include parts-of-speech tags, dependencies, and grammatical features. ProMiner (Hanisch et al., 2005) used contextual rules to achieve an accuracy of 92.9%. However, the relevant study also identified the risk of overfitting of the proposed rules.
For machine learning-based approaches, conditional random fields (CRFs), support vector machines (SVMs), and Markov models are widely used. Deep learning-based techniques are also being researched. Munkhdalai et al. (2015) proposed BANNER-CHEMDNER that uses semi-supervised learning to extract chemical entities. It recorded an F-measure score of 85.68% on the testing set Chemical Entity Mention. Tang et al. (2015) and Li et al. (2015) used CRFs with a system based on MapReduce and Hadoop to process big data. Although many studies have used CRFs to calculate the probability of the occurrence of a certain word as a biomedical/chemical entity, Tang et al. (2015) proposed an SSVM-based system (F-score: 85.05%) that outperforms CRF-based systems. Leaman and Lu (2016) used a semi-Markov, structured linear classifier that works well, especially with diseases (NCBI Disease corpus, F-score: 0.829) and chemicals (BioCreative 5 CDR corpus, F-score: 0.914). Recently, to process large amounts of bio-literature data, machine learning-based approaches have often been combined with parallel and distributed systems (Li et al., 2015; Tang et al., 2015).
Determining the relations among entities is also a fundamental task in discovering knowledge from biomedical text. Although early studies (Jelier et al., 2005) focused on extracting binary relations by using the co-occurrence approach, techniques for the extraction of complex relationships among biomedical entities have received a considerable amount of research interest because complicated and accurate relationships among entities in text can be extracted as knowledge. Extracting these complex relations involves processing using pattern and rule matching (Fundel et al., 2006) or, recently, machine learning-based techniques (Bunescu et al., 2005).
In pattern and rule matching, predefined rules based on a dependency tree and a relation trigger word are used to identify relations between entities, whereas several techniques, including SVM, Markov models, and RNN, are used in machine learning approaches. In past studies, RE for specific types of biomedical entity have been studied widely. Protein–protein interactions (PPIs) have been the subject of extensive focus (Thomas et al., 2011, Li et al., 2015). Li et al. (2015) described miRTex designed for microRNA-gene RE, and it achieved an F-score of 88%. Others like Bravo et al. (2015) focused on the relation between gene and disease.
It is challenging to find integrated systems for all types of biomedical entities. It requires sophisticated techniques and expertise that can be applied to various entity types. To overcome this limitation, Yimam et al. (2016) proposed an interactive machine learning (iML) approach to improve biomedical knowledge extraction. Holzinger (2016) defined iML as an algorithm that can optimize training data through interactions between a computer and a human. Although iML may help expedite the discovery process, we focus here on only systems for biomedical knowledge discovery.
An integrated system requires comprehensive techniques, and research on this has not been extensive thus far, despite its potential for knowledge extraction. As PKDE4J is an integrated system, multiple types of entities and relations can be extracted from various types of data sources using it.
In addition to NER and RE, event detection has gained attention for accurate knowledge extraction in recent years. Event detection refers to the task of extracting descriptions of the actions of and relations among one or more entities from the biomedical literature (Björne et al., 2010). In the expanded version of PKDE4J, we enhance the likelihood of extracting accurate relations by adding an event detection module.
The functions of PKDE4J can be applied to practical problems, such as knowledge search, knowledge network construction, and knowledge inference. For knowledge search, a system integrating PKDE4J with PubMed articles provides annotated articles. As such, users can perform more effective searches using annotated papers. Moreover, using PKDE4J, large amounts of knowledge constructed from various resources can be transformed into a network. Applying Swanson’s ABC model to the extracted knowledge, new knowledge that has not been found before can be inferred.
In this paper, we compare the extended PKDE4J with other well-known algorithms on various types of entity extraction, RE, and event detection. We also provide a detailed description of how the results are extracted by PKDE4J. To highlight its utility, we introduce examples of how it can be used for knowledge annotation, search, linking, and inference.
Materials and Methods
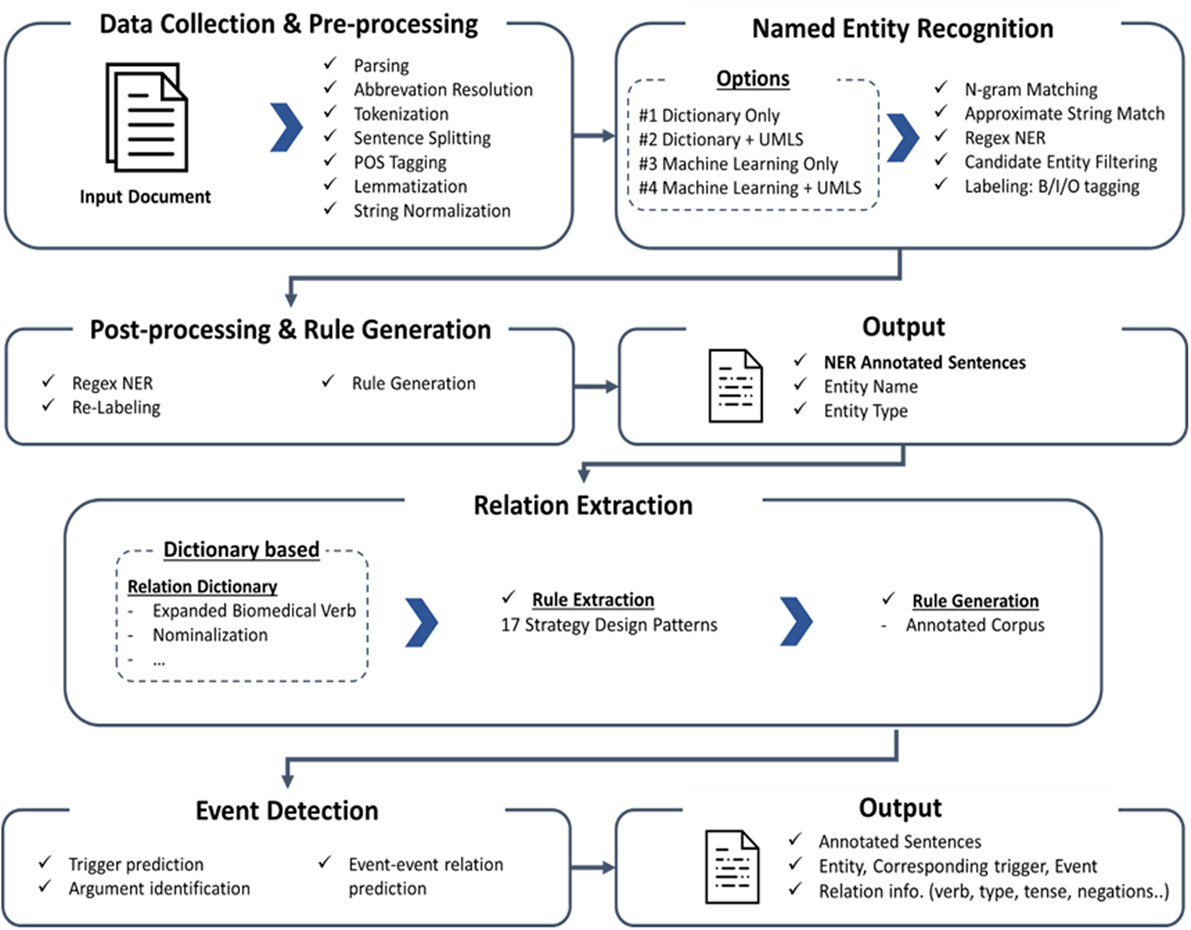
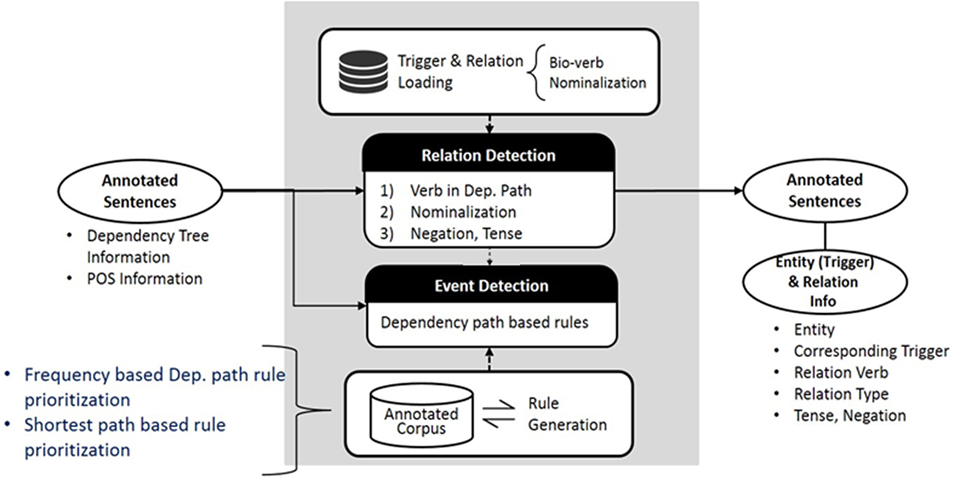
The system has been upgraded since the original version of PKDE4J was published in 2015. In this section, we introduce the updated version. The overall architecture of the system is illustrated in Figure 1. The three major modules are (1) named entity extraction, (2) RE, and (3) event detection.
Named Entity Extraction
The original PKDE4J version extracts biological entities based on a dictionary. To make the NER module more flexible, we propose three options for entity extraction in addition to the dictionary: (1) the Unified Medical Language System (UMLS) combined with the dictionary, (2) machine learning, and (3) the UMLS combined with machine learning. By adding these options, it is expected that the updated system will exhibit better performance and flexibility.
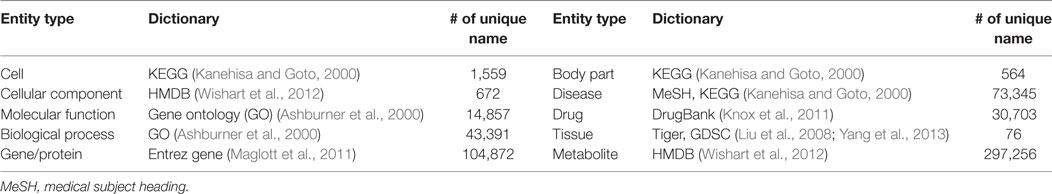
For the dictionary-based approach, we updated the previous version of PKDE4J dictionaries by integrating data from the open biomedical open database GoPubMed. GoPubMed is a search engine for biomedical literature designed to structure a large number of articles from the MEDLINE database (Doms and Schroeder, 2005). It allows users to query and explore PubMed results with controlled vocabulary, such as Gene Ontology (GO) and Medical Subject Headings (MeSH). GO aims to unify the representations of gene and gene products into structured vocabularies. Starting with three databases for organisms—FlyBase, the Saccharomyces Genome Database, and the Mouse Genome Informatics Project—GO has grown by integrating 35 major gene/protein repositories (Ashburner et al., 2000). Similarly, MeSH is the National Library of Medicine’s controlled vocabulary thesaurus used to index biomedical publications such as the PubMed database. To add GoPubMed data to various types of PKDE4J dictionaries, we retrieved articles with queries presented in Table 1. For each article, the tagged GO terms and MeSH terms were collected. Collected terms that did not represent the dictionary type were filtered by certain criteria. For example, general terms like “homo sapiens” repeatedly appeared in several queries, and thus were deleted from the list.
In addition to MeSH and GO, KEGG Disease as disease dictionary and Drugbank data as drug dictionary were added to the dictionaries for PKDE4J. KEGG Disease is a collection of disease vocabularies. It provides various information concerning diseases on perturbed molecular networks. Approximately, 2,000 items of disease information were added. Drugbank contains biochemical and pharmacological information about drugs and their targets (Wishart et al., 2006). More than 400 drugs and their 1,200 metabolites were added to the drug dictionary.
We also propose a combination of the dictionary with the UMLS-based approach, which first recognizes biological entities in text using the dictionary-based approach and then maps to UMLS terms matching the extracted entities. Many existing NER systems (Rindflesch et al., 2000; Aronson, 2001; Jimeno et al., 2008) that are lexicon based largely depend on knowledge sources such as GO (Doms and Schroeder, 2005) and the UMLS (Bodenreider, 2004). Specifically, the UMLS is a collection of multiple controlled vocabularies (e.g., NCBI and MeSH) in the biomedical domain developed by the US National Library of Medicine. It consists of over 3 million concepts, each of which is assigned to at least one of 134 semantic types from the UMLS Semantic Network, such as gene, genome, and cell. Accordingly, the integration of the UMLS into dictionary-based entity extraction can enhance the interoperability of our system and help utilize additional information concerning entities. It also allows for further analysis, such as the measurement of similarity among the extracted biological entities based on the corresponding semantic types in the Semantic Network.
Similarly, we integrated the UMLS into the machine learning-based approach. To this end, the machine learning-based entity recognizers included Abner (Settles, 2005), CheNER (Usié et al., 2013), and LingPipe (Baldwin and Carpenter, 2003). The model was chosen according to the characteristics of the experiment to be conducted. For instance, if mutation was the entity type for entity extraction, we could choose MutationFinder (Caporaso et al., 2007). After model selection, the NER process was performed using the selected model and the extracted entities were mapped to UMLS terms. Abner is a tagger for biological entities (e.g., protein, cell line, DNA, and RNA) in text and provides two models trained on the standard NLPBA (Kim et al., 2003) and BioCreative (Yeh et al., 2004) corpora. Using these models, two biological entities, gene and cell, extracted from the text are tagged and mapped into terms in the UMLS. Moreover, a named entity recognizer that performs a similar function to Abner is CheNER (Usié et al., 2013). It recognizes chemical compounds in biomedical text. The CheNER model trained on the corpora provided by Kolářik and Klinger (Klinger et al., 2008; Kolárik et al., 2008) was used to tag the drug names mentioned in text.
Relation Extraction
The core module of RE is similar to that of PKDE4J (2015). With 17 strategies, PKDE4J extracts relations between entities, where PKDE4J’s RE module focuses on the presence of verbs. Using verbs as the core of RE, we can extract more precise relations. Therefore, in the new version of PKDE4J, we expand the range of verbs in the RE module.
Expansion of Biomedical Verb List: Biomedical Verbs
The number of verbs used in the previous version of PKDE4J was 398. This means that only 398 relation types could be extracted, and the remaining relations were identified as “none” or “juxtaposed.” Therefore, a new verb list should be constructed and applied to PKDE4J to overcome this limitation.
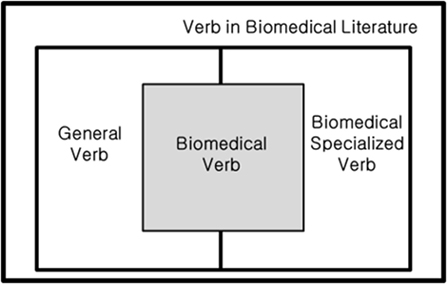
In this study, we define biomedical verbs as verbs describing the relation between biological entities used in the biomedical field. Verbs in a general field and in the biomedical field are included if they represent a relation between entities. Therefore, as shown in Figure 2, some verbs from general and specialized verbs used in biomedicine can be used as biomedical verbs.
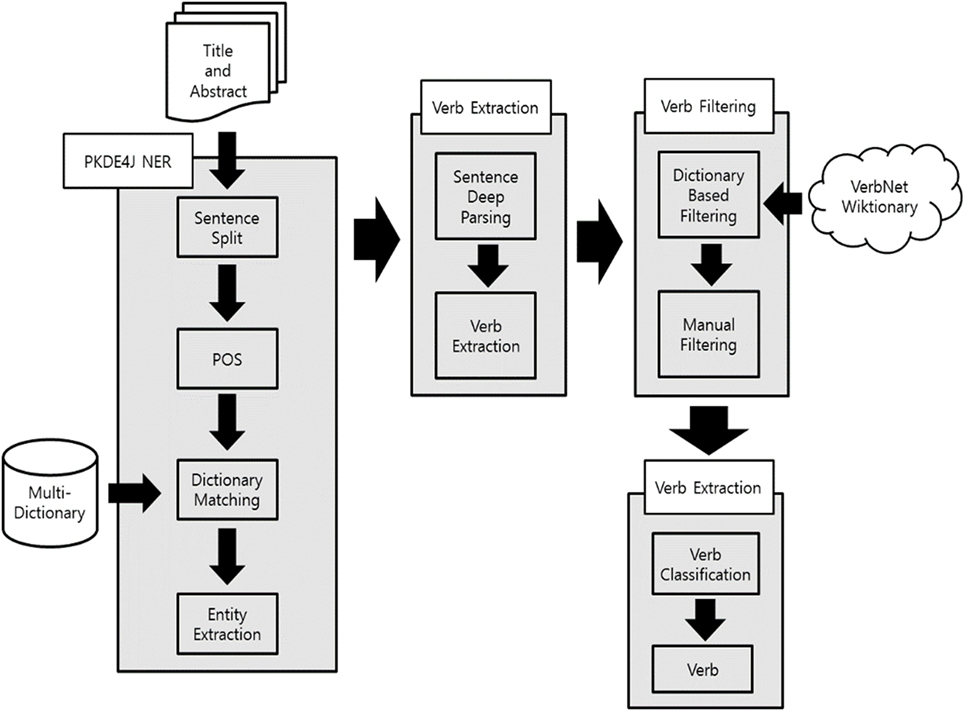
To construct biomedical verbs, we used the 2014 version of PubMed articles. We collected a total of 14,447,667 records of articles with the title and abstract of each. We then modified PKDE4J to extract verbs from sentences containing two entities. To use the dictionary-based approach for PKDE4J, we constructed dictionaries shown in Table 2. Dictionaries for each entity included KEGG, HMDB, GO, Entrez Gene, MeSH, DrugBank, Tiger, and GDSC.
Verb extraction using PKDE4J involves several processes. Figure 3 shows outline of method for biomedical verb construction. Each PubMed record consists of a title and abstract, and abstract was separated by sentence. The separated sentences were tokenized into words that were used to extract entities by mapping with the dictionaries. If more than two entities were extracted in a sentence, the related terms between them were extracted. If a certain term had a dependency relation satisfying a set of rules, with two entities on a dependency tree provided by the Stanford Core NLP (Manning et al., 2014), the term was extracted as a candidate biomedical verb. Through this process, a total of 72,844 candidate terms were extracted.
The candidate terms included verbs that could not explain biological interaction between entities, and spelling errors or incomprehensible terms. Thus, to retain only biomedical verbs, two additional filtering tasks were performed. First, to remove terms that were not verbs. WordNet (Miller, 1995) and Wiktionary (https://www.wiktionary.org/) were used. WordNet is a dictionary database that provides meaning, part of speech, and thesaurus information for each word. Until recently, various studies using WordNet had been used as an ontology for text analytics (Goikoetxea et al., 2016). Wiktionary is a Web dictionary that aims to create a multilingual dictionary as a Wiki project. At least one study recently used Wiktionary (Zesch et al., 2008). We filtered candidate terms using two reliable dictionaries. After this process, 8,855 verbs remained. The second filtering process involved the selection of verb representing meaningful relations through a manual process. In this process, a Ph.D. student in the Department of Library and Information Science and a doctor in biology conducted manual filtering. Verbs representing the relationship between entities were extracted. In the case of transitive verbs, the relationship between subject and object was indicated. For other words, the verbs were directly related to two entities. However, if the extracted transitive verbs did not provide any meaning, we deleted these verbs, such as “investigate,” “survey,” and “study.” Moreover, intransitive verbs indicating relationships between entities with preposition were added. In case of a discrepancy between the opinions of experts, a decision was made through consultation. After all the filtering processes, 4,558 verbs were obtained in a final list.
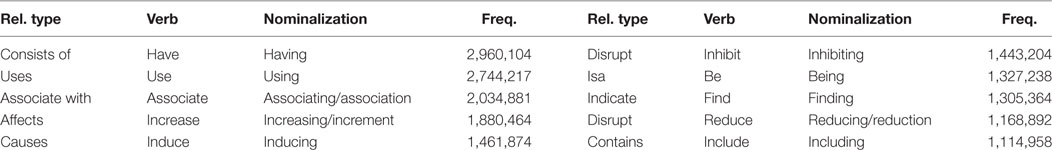
To construct a verb dictionary, two tasks needed to be performed. The 4,558 verbs were grouped into similar types depending on their meanings. If a number of verbs were classified as belonging to a similar type, the relations derived from them could represent relatively small numbers of relation types. In this study, verbs were classified using the semantic relation of UMLS consisting of hierarchical relations, and were divided into 54 categories, where the six largest categories were “ISA,” “physically_related_to,” “conceptually_related_to,” “functionally_related_to,” “temporally_related_to,” and “spatially_related_to.” To classify the verbs more accurately, manual classification process was carried out. After the classification process, the nominalized forms of the verbs were added. Specifically, a relation between entities was not identified by only verbs. For example, in the sentence “Binding A and B,” the relation between A and B was identified through “binding.” A nominalized form and a gerund can be frequently used to identify relations. Therefore, it should include not only the verb form, but also the nominalized form and gerund. This process was also performed manually.
Finally, we constructed a list of 4,558 verbs including the semantic relations from UMLS, as well as the nominalized form and gerund of each. Table 3 shows an example of a list of constructed biomedical verbs. The entire verb list can be downloaded from the following URL: http://informatics.yonsei.ac.kr/tsmm/data/Biomedical_Verb_List.xlsx.
Event Detection
By considering contextual information in the NER process, we added an event trigger detection module to the original PKDE4J process as shown in Figure 4.
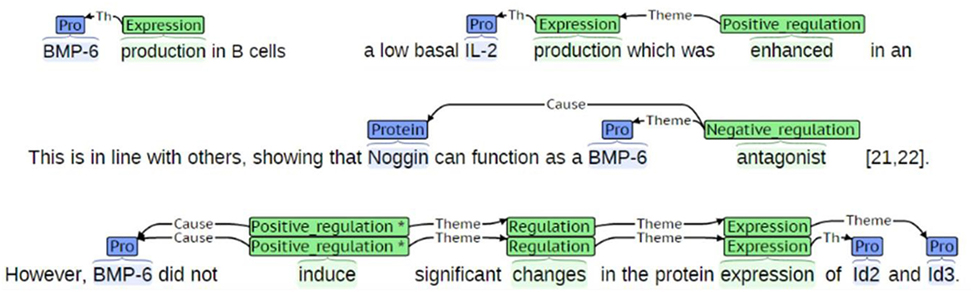
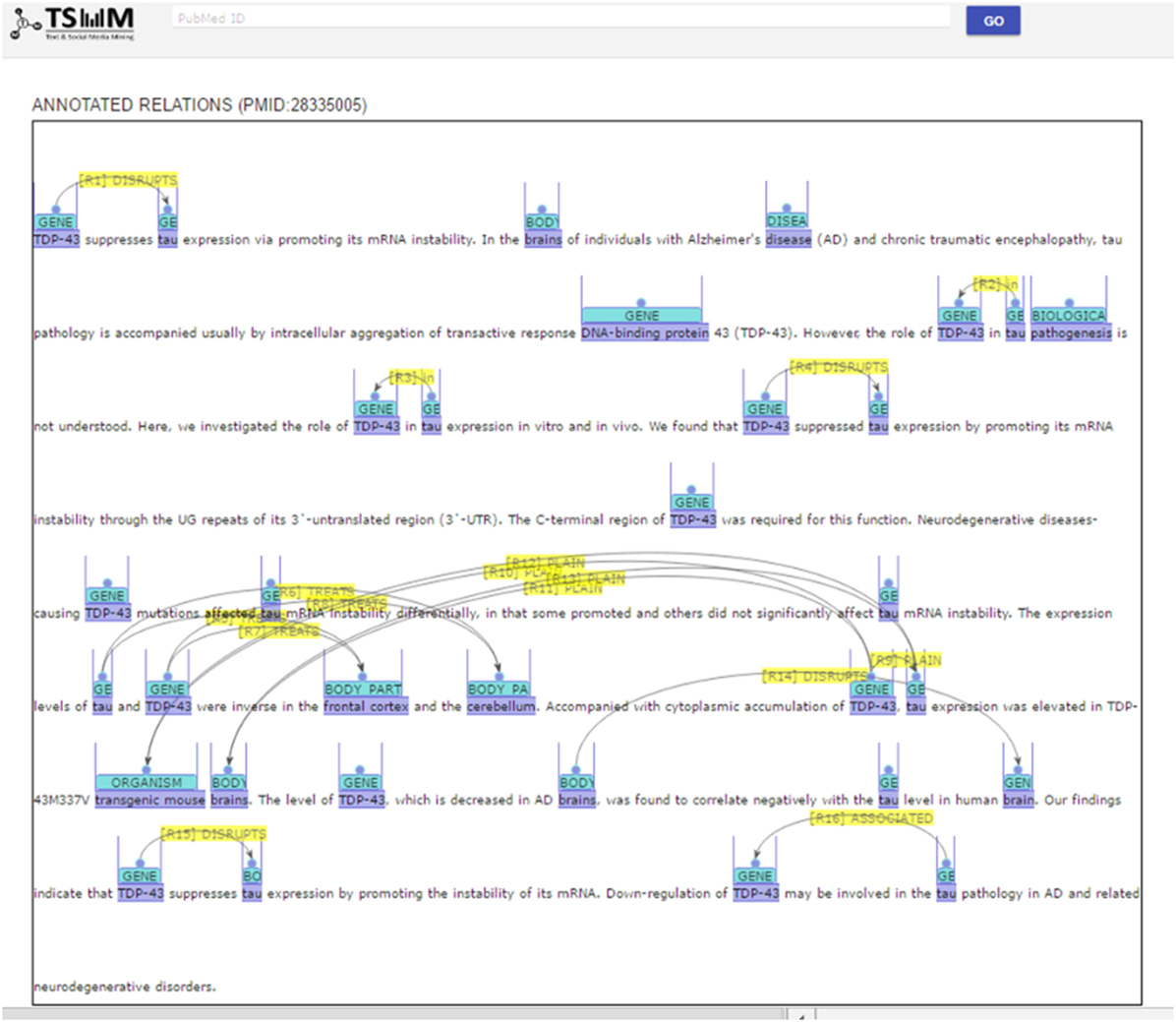
Event detection refers to the task of extracting descriptions of actions and relations from one or more entities from the biomedical literature (Björne et al., 2010). Events can function as participants in other events, thus allowing for the construction of complex conceptual networks. Events include complex interactions among biological entities, and are highly reliant on context (Miwa et al., 2012). They are usually composed of triggers that are described as words or phrases indicating the occurrence of certain events, such as “inhibition” and “expression” (Rahul et al., 2017). Figure 5 shows an example of events detected from sentences. Thus, event trigger identification is essential to extract interactions among biological entities in a more precise manner. To this end, we developed a model to recognize trigger words in text and added it to our system.
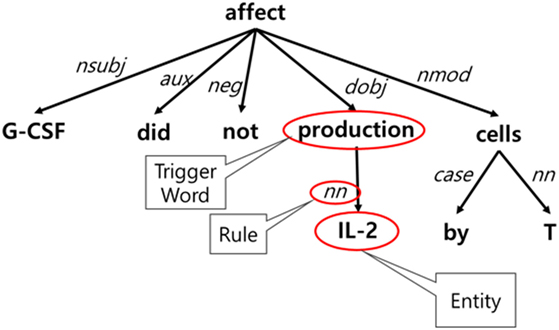
For event detection, we developed a Stanford Core NLP-based model using token-based features (lemmatization, POS tagging, and phrases) and dependency trees. The model was trained on the GENIA Event Extraction corpus (Kim et al., 2012) by selecting 653 trigger words belonging to four event types—gene expression, negative regulation, positive regulation, and regulation—and 4,132 event sets. To create rules, we applied the k-shortest path algorithm to the dependency trees. Each path from a given entity to a trigger word can be a rule, and the frequency and percentage of its occurrence were analyzed. In a given corpus, the ranking of the generated rules is based on the distance between a trigger word and an entity, and its frequency and percentage of occurrence. Figure 6 shows an example of rule generation. The event trigger detection module was added following entity extraction because events can be extracted after recognizing biological entities.
Evaluation
Named Entity Extraction
To validate the updated version of the NER module of PKDE4J, we compared the performance of the original version with that of the updated versions in terms of F-score by mapping the extracted entities to biological entities in the PubMed papers collected from GOPubMed. On average, the updated version achieved a precision of 99.9%, a recall rate of 86.6%, and an F-score of 92.8% for each biomedical entity as shown in Figure 7.
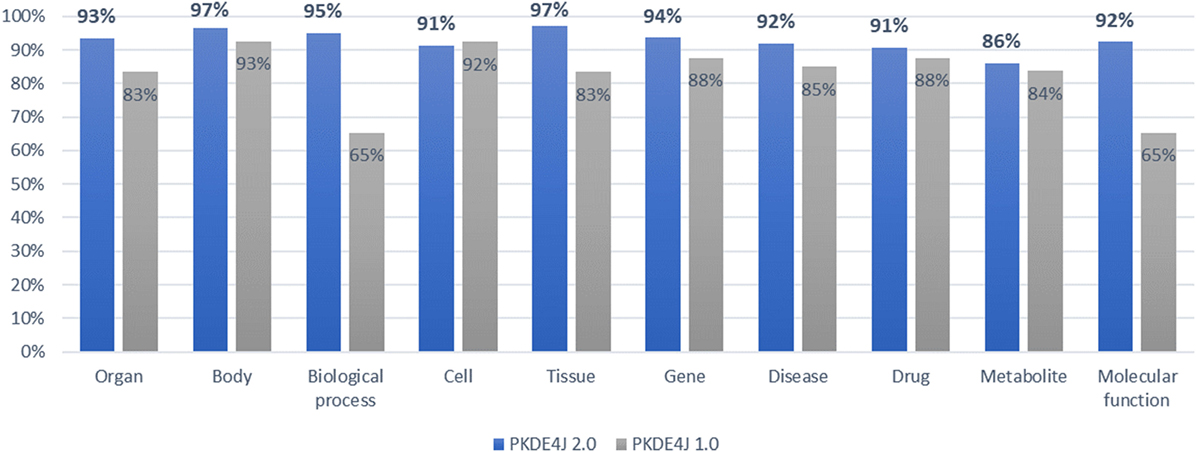
Figure 7. Comparative evaluation (F-score) of dictionary-based named entity recognition by type of biomedical entity.
When combining the UMLS with dictionary-based NER, we measured the F-score for 10 types of biomedical entity types. In Figure 8, for each entity type, the system yielded a precision of 98.1% of, a recall rate of 67.7%, and an F-score of 78.9% on average. The module combining the UMLS for entity extraction exhibited high precision and relatively low recall, as it assisted the natural language process by mapping the extracted entities to semantic entity types.
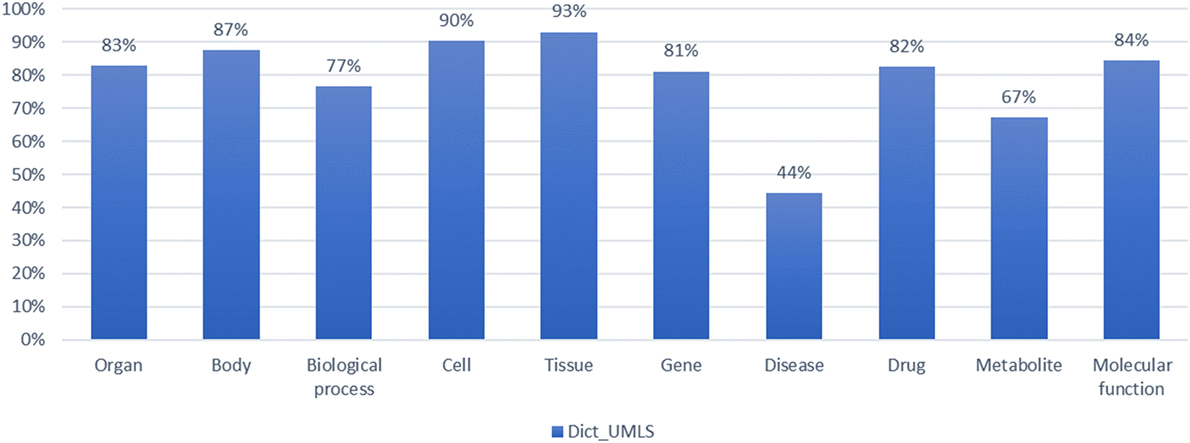
Figure 8. Evaluation of dictionary combined with Unified Medical Language System (UMLS) in the named entity recognition process by biomedical entity type.
For machine learning-based NER, we exploited three prevalent models—Abner, LingPipe, and CheNER—with PKDE4J. The models were trained to recognize particular entity types as mentioned in the methodology section. Abner is a tool for cells and genes, and the entity type that can be identified by the CheNER model is limited to drugs. Therefore, we conducted an evaluation of each model with a limited number of entity types (gene, cell, and drug). As shown in Figure 9, the evaluation shows that when wrapping the Abner model with PKDE4J, in case of cells, the F-score of the model was 13% (P: 26%, R: 9%) and for genes was 31% (P: 30%, R: 33%). The CheNER model yielded an F-score of 4.8% (P: 17.3%, R: 2.8%). These results indicate that the machine learning-based approach requires a high-quality training model that represents the entire population, where an out-of-the-box train model for Abner and CheNER yields poor performance.
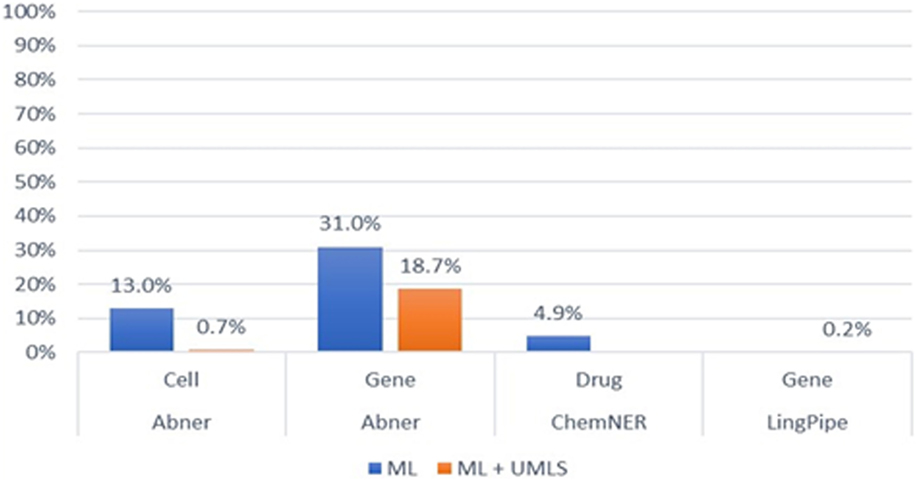
Figure 9. F-scores of the machine learning and the Unified Medical Language System (UMLS)-combined machine learning-connected systems.
When combining the UMLS with the machine learning-based system, we applied Abner and LingPipe model to PKDE4J for genes and cells. In this case, LingPipe was limited to tagging gene type. Integrating Abner and LingPipe into PKDE4J yielded low precision and recall. This can be attributed to the use of the out-of-the-box training model.
Relation Extraction
Comparison between Biomedical Verb List and Predication in SemRep and UMLS
The entity relations extracted using the biomedical verb list were compared with the entity relations’ set constructed using SemMed (Rindflesch et al., 2011) and the UMLS to confirm the agreement rate. SemMed is a database that stores a triple “subject–predicate–object” extracted by SemRep (Rindflesch and Fiszman, 2003) from Medline. It provided triples extracted from 165,670,113 sentences rom approximately 26 million PubMed abstracts. Both the subject and object were biomedical entities, and a CUI of the UMLS was assigned to them. The predicate type was provided by SemRep, and represented 61 predicate types including positive and negative distinctions. In this study, the biomedical verb list was classified using the semantic relation of the UMLS. We collected semantic relations through the UMLS using the CUI of each entity provided by SemMed, and the matching rate was computed based on it. A total of 4,406,360 sentences from approximately 20 million containing a relation verb provided by SemMed were randomly selected. We checked the agreement rate by comparing SemMed predicates with the results of applying the biomedical verb list to the same sentences. Table 4 shows the correspondence ratio between the relation “subject entity–object entity” extracted using PKDE4J based on the biomedical verbs and the relations provided by SemMed.
The “subject entity–object entity” matching rate was calculated by matching the subject and object entities, in the context of relations extracted through PKDE4J, to entities provided by SemRep. Approximately 33% matched. We discarded entities with relation type is “none” or “juxtaposed.” The former indicated that the relation between the relevant entities was likely to be positioned close to them, where a verb was located in proximity. The relation type “juxtapose” meant that two entities cooccurred without being connected to each other via a meaningful verb. In addition to filtering by these two criteria, a reason for the low match rate is the preprocessing of entities in PKDE4J. Because entities were preprocessed, a significant number of entity pairs extracted by PKDE4J did not match with those extracted by SemRep.
The agreement rate of triples was approximately 8.4%, which was relatively low. This is for the following reasons: First, Semantic Relation in UMLS determines the predefined semantic relation based on the CUI. This means that although other expressions appeared in a sentence, semantic relation was always the same if the CUIs were the same. Therefore, even if the same verb was used in several sentences, the semantic relation of the sentence varied depending on the CUI of the two entities. Moreover, it was also difficult to clearly classify the verb according to semantic relations. For example, the verb “effect” can be classified into various semantic relations such as “interacts_with,” “affects,” “causes,” “associate_with,” “result_of,” and “derive_from.” Because of the difficulty of precise classification, the concordance rate of the predicate decreased.
Although the concordance rate was low, it was useful in two respects. First, as mentioned above, if the result was extracted using the UMLS semantic type, the semantic relation was determined depending on the CUI of the extracted entity. However, if a biomedical verb was used, it had the advantage whereby the relation could be extracted through information provided in the sentence. As the information in the sentence had been used, it was possible to extract a relation more suitable for the context. Second, even if two entities did not have a semantic relation, it was possible to find an entity relation using information in a sentence in PKDE4J. SemRep or UMLS only extract entity relations with a semantic relation even if entity relations appear in the sentence structure. On the contrary, if biomedical verbs are used in RE, more relations can be extracted.
Experiments on Entity-Entity RE
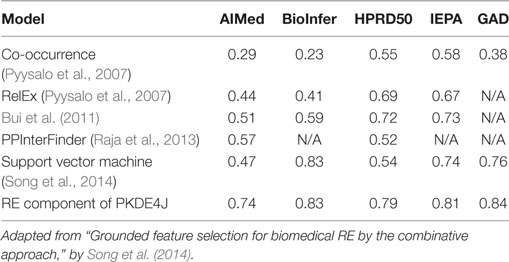
To measure the performance of the RE component of PKDE4J (PKDE4J-RE), we used five corpora with different characteristics and relation types as shown in Table 5. AIMed is among best-known corpora for PPIs (Bunescu et al., 2005). It contains 225 MEDLINE abstracts, and contains 1,955 sentences pertaining to proteins found in humans. The corpus was curated manually, and had 177 abstracts with PPI and 48 without. BioInfer and GAD are general RE corpora consisting of more than two entity types. Of the relation-type tags available in these corpora, we used only relation tagging for PPI. The BioInfer corpus is known for representing relationships among proteins, genes, and RNA (Pyysalo et al., 2007). It contains 1,100 sentences from PubMed abstracts, and the sentences contain annotations concerning entity, entity relationship, and dependency. A total of 2,662 relationship appeared in 840 sentences and the remainder had no relations. GAD (Becker et al., 2004) is a corpus that was semi-automatically annotated with three type relations: drug–disease, target–disease, and gene–disease relationships. The corpus had 5,329 sentences containing 2,800 true interactions and 2,529 false associations. The former consisted of 1,833 positive interactions and 967 negative ones. HPRD50 was created as a corpus for RelEx based on a subset of the Human Protein Reference Database (HPRD) (Fundel et al., 2006). The corpus contained (1) direct physical interactions, (2) regulatory relations, and (3) modifications (e.g., phosphorylation), which were manually annotated by two domain experts. It contained 145 sentences with a list of 433 PPI. IEPA, the Interaction Extraction Performance Assessment, is a corpus for PPIs consisting of 303 abstracts, with 486 sentences and 817 relations (Ding et al., 2002).
To evaluate RE performance, we compared the PKDE4J-RE with five approaches—SVM (Song et al., 2014), co-occurrence (Pyysalo et al., 2008), RelEx (Pyysalo et al., 2007), PPInterFinder (Raja et al., 2013), and Bui et al.’s (Bui et al., 2011) algorithm—based on results for these methods from our previous study (Song et al., 2014). The performance of each model was measured in terms of the F-score. SVM and co-occurrence are the two best-known approaches to RE. Other advanced algorithms could have been considered as well, such as CRFs, which recorded an F-score of 0.852 on 27,000 abstract from ISI in a study by Tang et al. (2015), or deep learning, which achieved an F-score of 0.613 in the SemEval-2010 Task 8 dataset in a study of Nguyen and Grishman (2015). However, our study intended to compare commonly used techniques for RE. PKDE4J-RE yielded the best performance.
RelEx is a RE technique that uses dependency trees and simple rules applied to these trees. PPInterFinder is specifically designed to extract PPIs by identifying relation keywords using a parser with Tregex and a relation keyword dictionary for 11 specific patterns based on the syntactic nature of PPI pairs. Bui et al.’s approach is tuned to PPI extraction based on dependency trees and SVM. Most of these approaches have already been evaluated using AIMed, BioInfer, HPRD50, and IEPA. As shown in Table 6, in the experiments, PKDE4J-RE outperformed the other five RE techniques over all corpora.
Event Detection
Effective event detection requires in-depth analysis of sentence structure, and can benefit in particular from the use of semantic processing or deep parsing techniques that analyze both the syntactic and semantic structures of texts. Event detection is a daunting challenge, and is more complex and difficult than RE. We extended PKDE4J for event detection based on dependency trees and 17 rules applied to them. For example, in a dependency tree, if the dependency relation between a governor and a dependent contained a preposition property, such as “prep-at” + a noun phrase, the dependency relation was tagged as an event. Table 7 shows a sample sentence and the results of event detection based on the aforementioned rules.

Table 7. An example of the results of semantic parsing for event detection (Zhou and He, 2011).
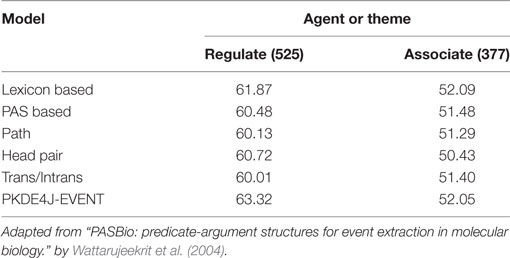
Experiments for Event Detection
To test the event detection performance of PKDE4J (PKDE4J-EVENT), we used the PASBIO corpus. It consists of 30 biomedicine-related predicates (Wattarujeekrit et al., 2004), and was built using predicate–argument structures from unstructured texts in the biomedical literature. The predicates available in PASBIO were from sentences mostly from Medline and the Embo, PNAS, NAR, and JV journals. We compared PKDE4J-EVENT with the Wattarujeekrit et al.’s approach to PASBIO. The results are shown in Table 8. The lexicon-based model consisted of six features: surface word, lemma form, head word of noun phrase, parts of speech, orthographic features, and phrase chunks. The PAS-based model consisted of features from the lexicon-based model as well as the predicate surface form, predicate lemma, voice, and surface syntactic role to represent the semantic roles of the arguments. The path model included features related to a syntactic path from the subject argument to the related predicate, and from the related predicate to the object argument. The head pair model included features of the PAS-based model and those representing a pair of subject and object heads. The trans/intrans model contained features of the PAS-based model and supplementary features indicating whether a predicate had been used in the transitive or intransitive sense. As shown in Table 8, PKDE4J-EVENT outperformed the other five models on the predicate “regulate” but achieved the second-best performance on “associate,” where the lexicon-based model achieved the best performance. This was a surprising result, in that of models with the most sophisticated feature sets, the simplest one achieved the best performance in case of two predicates. As PKDE4J is intended for entity and RE, the performance of PKDE4J-EVENT was acceptable.
Applications
The results of the evaluation show that PKDE4J is a useful and effective text-mining tool for NER, RE, and event extraction. As the results of these extraction tasks were sources for biomedical scientific knowledge representation, the results can be applied to knowledge discovery tasks such as literature-based discovery, hypothesis generation, and semantic annotation. In this section, we describe ongoing efforts to extend PKDE4J to knowledge discovery.
Knowledge Search
PKDE4J can be applied to knowledge search. Figures 10 and 11 show screenshots of its application that is publicly available at http://informatics.yonsei.ac.kr:8080/ner-re. PKDE4J can be embedded into any search engine to render the search results more meaningful for users. For example, if users enter queries, the system searches and returns the matching PubMed records with the NER results. On the results’ page, the extracted entities and relation types recognized by PKDE4J are highlighted, and the list of their relation types is also provided.
The results’ page consists of an annotated abstract and an annotated relation list. The relation list contains the types of relations between entities. The annotated abstract shows entities and relations extracted from the abstract that are highlighted in different colors by entity type, as shown in Figure 10. Moreover, the system provides a list of extracted relations of each entity and its relation type as well as a pie graph showing the ratio of the extracted types of entity relations (Figure 11).
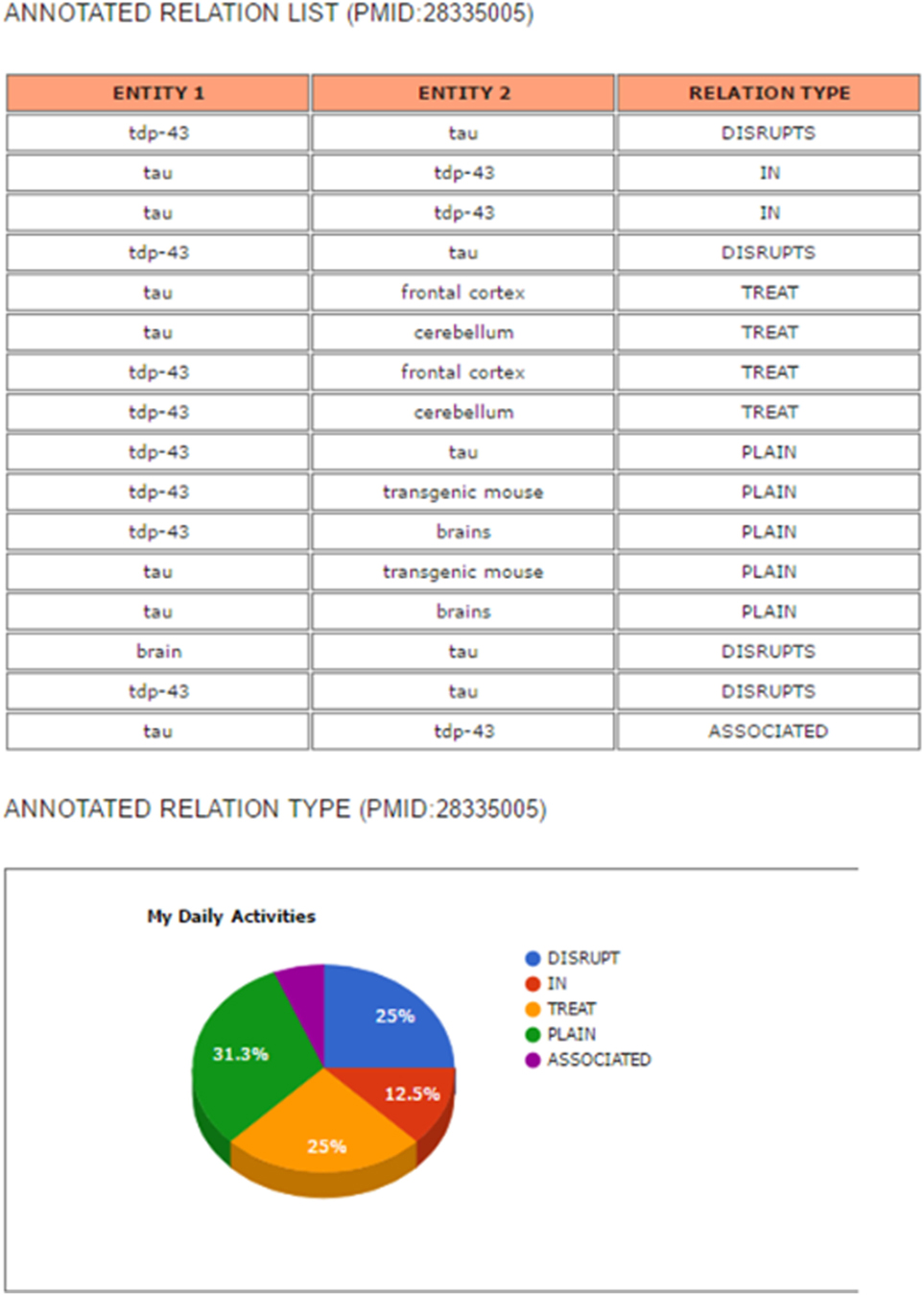
Figure 11. Example of the results’ page of search engine: an annotated relation list and a pie chart to show the ratio of annotated relation types.
Knowledge Linking
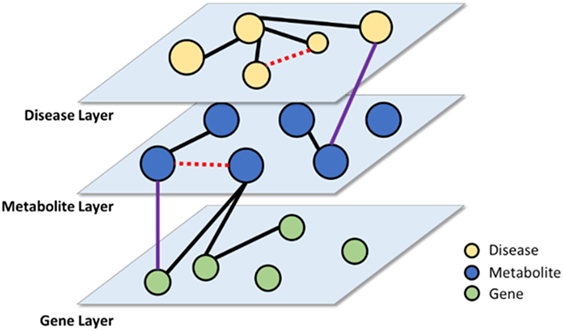
Another application of PKDE4J is knowledge linking. Biomedical data are available from various types of data sources, including research articles, clinical data, and health-care-related social media. Thus, extracting entities and relations from these heterogeneous data sources requires that they be connected to one another for knowledge discovery. These entities and their relations can be organized and linked in the form of a multi-layer network as shown in Figure 12. With the constructed network, the links among entities within the same layer as well as with those from different layers can be analyzed. If the network is used for new hypothesis generation, it can provide more sophisticated and integrated hypotheses. Moreover, the multi-layer graph can be efficiently managed using a graph database such as Neo4J (Webber, 2012).
Knowledge Inference
After searching and linking entities and their relations, we can infer knowledge as the next step of knowledge representation. The application of PKDE4J to knowledge inference can help us discover new relations or patterns based on the constructed knowledge networks. With the constructed network, PKDE4J can be applied to generate new plausible hypotheses for knowledge inference (Baek et al., 2017), as proposed by Baek et al. (2017) for literature-based discovery. This application is accessible at http://informatics.yonsei.ac.kr:8080/hypothsis_generator/index.html.
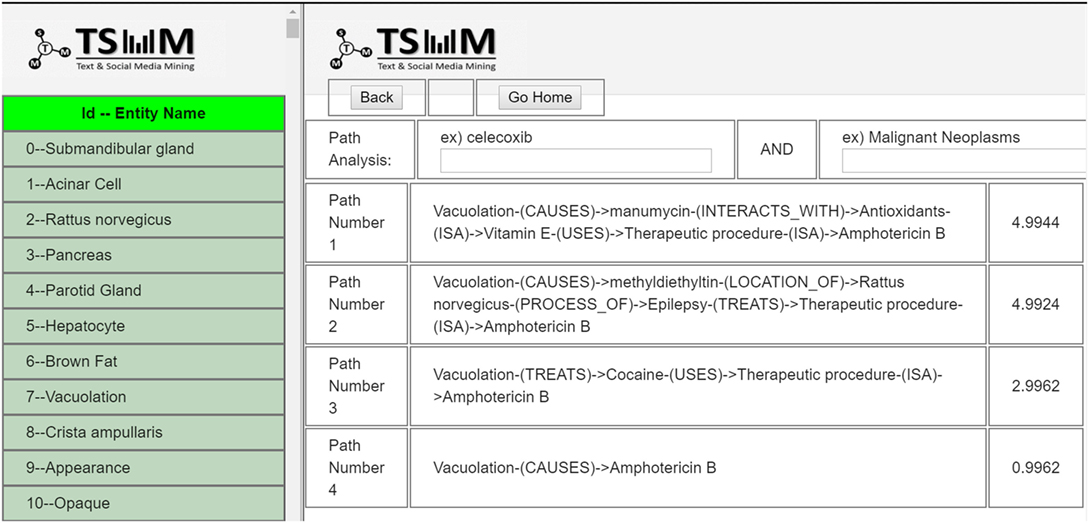
Users can search by using multiple query terms (e.g., “vacuolation”), and the system returns the matching PubMed results, including the PubMed ID, abstract, and PubMed link to the article as shown in Figure 13. Moreover, the search terms that users enter into the system are highlighted in the results. After browsing the results list, users can select the PubMed records to be included to generate new hypotheses.
When users click the “generate paths” button at the top right-hand corner of the results’ page, after choosing the number of abstracts, the results are displayed in the path analysis page as illustrated in Figure 14. In the path results’ page, entities extracted from the selected articles are listed on the left and the search bar on the right. Using the list of entities, users can generate paths by selecting two entities of interest. Based on Swanson’s ABC model, users enter two entities (A and C terms) to generate plausible hypotheses, including none, one or multiple C-terms between A and C. Moreover, these generated paths are ranked by a semantic relatedness score. If there are connected paths between the extracted entities, they are displayed in order by relatedness score. For instance, if the resulting path is “Vacuolation-(CAUSES)- > Amphotericin B,” this can be interpreted as “vacuolation” and “amphotericin B” are linked via a path that implies a causality relation.
As demonstrated by the above three applications for representing scientific knowledge, PKDE4J serves a basis for effective and automatic knowledge discovery. It is not limited to knowledge extraction either, and can be adapted to other types of knowledge representation, such as the augmentation of ontology.
Conclusion
Compared with the original PKDE4J, the upgraded version of PKDE4J was shown in this study to be a flexible system for knowledge representation. For named entity extraction, the following three options were added to it: (1) a dictionary with the UMLS, (2) machine learning, and (3) machine learning with the UMLS. For RE, verb expansion was added for more accurate detection of relations. For more precise extraction, the event trigger extraction module was attached to PKDE4J as part of the RE process based on the contextual information of sentences. The improved PKDE4J was verified to be effective compared with the original version as well as commonly used extraction techniques.
We also proposed applications of PKDE4J for knowledge representation. First, it enables knowledge search. By building a Web search system, PKDE4J helps users search for the extracted entities and their relations. Second, PKDE4J can be used to connect the extracted entities via a multi-layered network. Linking knowledge that connects parts of our knowledge can suggest new and plausible knowledge paths. Third, PKDE4J can be applied to knowledge inference. With the constructed knowledge network, PKDE4J can generate promising candidates’ hypotheses. Although we described only three applications of PKDE4J, other interesting and meaningful applications in biology as well as other domains could be developed.
Author Contributions
MS (first and corresponding author) made substantial contributions to conception and design, and evaluation and application. He also gave final approval to the version to be submitted as well as revised versions. MK, KK, YK, and SJ participated in drafting the article and revising it.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the Bio-Synergy Research Project (NRF-2013M3A9C4078138) of the Ministry of Science, ICT, and Future Planning through the National Research Foundation.
References
Aronson, A. R. (2001). “Effective mapping of biomedical text to the UMLS metathesaurus: the metamap program,” in Proceedings of the AMIA Symposium (Washington, DC: American Medical Informatics Association), 17.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25. doi: 10.1038/75556
Baek, S. H., Lee, D., Kim, M., Lee, J. H., and Song, M. (2017). Enriching plausible new hypothesis generation in PubMed. PLoS ONE 12:e0180539. doi:10.1371/journal.pone.0180539
Baldwin, B., and Carpenter, B. (2003). LingPipe. Available from World Wide Web: http://alias-i.com/lingpipe/
Becker, K. G., Barnes, K. C., Bright, T. J., and Wang, S. A. (2004). The genetic association database. Nat. Genet. 36, 431–432. doi:10.1038/ng0504-431
Bell, L., Chowdhary, R., Liu, J. S., Niu, X., and Zhang, J. (2011). Integrated bio-entity network: a system for biological knowledge discovery. PLoS ONE 6:e21474. doi:10.1371/journal.pone.0021474
Belleau, F., Nolin, M. A., Tourigny, N., Rigault, P., and Morissette, J. (2008). Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform. 41, 706–716. doi:10.1016/j.jbi.2008.03.004
Björne, J., Ginter, F., Pyysalo, S., Tsujii, J. I., and Salakoski, T. (2010). Complex event extraction at PubMed scale. Bioinformatics 26, i382–i390. doi:10.1093/bioinformatics/btq180
Bodenreider, O. (2004). The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32(Suppl._1), D267–D270. doi:10.1093/nar/gkh061
Bravo, À, Piñero, J., Queralt-Rosinach, N., Rautschka, M., and Furlong, L. I. (2015). Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinformatics 16:55. doi:10.1186/s12859-015-0472-9
Bui, Q. C., Katrenko, S., and Sloot, P. M. (2011). A hybrid approach to extract protein–protein interactions. Bioinformatics 27, 259–265. doi:10.1093/bioinformatics/btq620
Bunescu, R., Ge, R., Kate, R. J., Marcotte, E. M., Mooney, R. J., Ramani, A. K., et al. (2005). Comparative experiments on learning information extractors for proteins and their interactions. Artif. Intell. Med. 33, 139–155. doi:10.1016/j.artmed.2004.07.016
Caporaso, J. G., Baumgartner, W. A. Jr., Randolph, D. A., Cohen, K. B., and Hunter, L. (2007). MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics 23, 1862–1865. doi:10.1093/bioinformatics/btm235
Ding, J., Berleant, D., Nettleton, D., and Wurtele, E. (2002). “Mining MEDLINE: abstracts, sentences, or phrases?,” in Pacific Symposium on Biocomputing Vol. 7, (Kauai, HI), 326–337.
Doms, A., and Schroeder, M. (2005). GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res. 33(Suppl._2), W783–W786. doi:10.1093/nar/gki470
Fundel, K., Küffner, R., and Zimmer, R. (2006). RelEx—relation extraction using dependency parse trees. Bioinformatics 23, 365–371. doi:10.1093/bioinformatics/btl616
Goikoetxea, J., Agirre, E., and Soroa, A. (2016). “Single or multiple? Combining word representations independently learned from text and WordNet,” in Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, (Phoenix, AZ: AAAI Press), 2608–2614.
Hanisch, D., Fundel, K., Mevissen, H. T., Zimmer, R., and Fluck, J. (2005). ProMiner: rule-based protein and gene entity recognition. BMC Bioinformatics 6:S14. doi:10.1186/1471-2105-6-14
Holzinger, A. (2016). Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inform. 3, 119–131. doi:10.1007/s40708-016-0042-6
Jelier, R., Jenster, G., Dorssers, L. C., van der Eijk, C. C., van Mulligen, E. M., Mons, B., et al. (2005). Co-occurrence based meta-analysis of scientific texts: retrieving biological relationships between genes. Bioinformatics 21, 2049–2058. doi:10.1093/bioinformatics/bti268
Jimeno, A., Jimenez-Ruiz, E., Lee, V., Gaudan, S., Berlanga, R., and Rebholz-Schuhmann, D. (2008). Assessment of disease named entity recognition on a corpus of annotated sentences. BMC Bioinformatics 9(Suppl. 3):S3. doi:10.1186/1471-2105-9-S3-S3
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi:10.1093/nar/28.1.27
Kim, J. D., Ohta, T., Tateisi, Y., and Tsujii, J. I. (2003). GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 19(Suppl._1), i180–i182. doi:10.1093/bioinformatics/btg1023
Kim, J. D., Nguyen, N., Wang, Y., Tsujii, J. I., Takagi, T., and Yonezawa, A. (2012). The Genia event and protein coreference tasks of the BioNLP shared task 2011. BMC Bioinformatics 13(Suppl. 11):s1. doi:10.1186/1471-2105-13-S11-S1
Klinger, R., Kolářik, C., Fluck, J., Hofmann-Apitius, M., and Friedrich, C. M. (2008). Detection of IUPAC and IUPAC-like chemical names. Bioinformatics 24, i268–i276. doi:10.1093/bioinformatics/btn181
Kolárik, C., Klinger, R., Friedrich, C., Hofmann-Apitius, M., and Fluck, J. (2008). “Chemical names: terminological resources and corpora annotation,” in Workshop on Building and Evaluating Resources for Biomedical Text Mining (6th Edition of the Language Resources and Evaluation Conference) (Marrakech, Morocco), 51–58.
Knox, C., Law, V., Jewison, T., Liu, P., Ly, S., Frolkis, A., et al. (2011). DrugBank 3.0: a comprehensive resource for ‘OMICS’ research on drugs Nucleic Acids Res. 39(Suppl. 1) D1035–D1041. doi:10.1093/nar/gkq1126
Leaman, R., and Lu, Z. (2016). TaggerOne: joint named entity recognition and normalization with semi-Markov models. Bioinformatics 32, 2839–2846. doi:10.1093/bioinformatics/btw343
Li, G., Ross, K. E., Arighi, C. N., Peng, Y., Wu, C. H., and Vijay-Shanker, K. (2015). miRTex: a text mining system for miRNA-gene relation extraction. PLoS Comput. Biol. 11:e1004391. doi:10.1371/journal.pcbi.1004391
Liu, X., Yu, X., Zack, D. J., Zhu, H., and Qian, J. (2008). TiGER: a database for tissue-specific gene expression and regulation. TiGER: a database for tissue-specific gene expression and regulation, 9, 271.
Maglott, D., Ostell, J., Pruitt, K. D., and Tatusova, T. (2011). Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 39(Suppl. 1):D52–D57. doi:10.1093/nar/gkq1237
Manning, C. D., Surdeanu, M., Bauer, J., Finkel, J. R., Bethard, S., and McClosky, D. (2014). “The Stanford coreNLP natural language processing toolkit,” in Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Baltimore, Maryland: Association for Computational Linguistics), 55–60.
Miller, G. A. (1995). WordNet: a lexical database for English. Commun. ACM 38, 39–41. doi:10.1145/219717.219748
Miwa, M., Thompson, P., and Ananiadou, S. (2012). Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. Bioinformatics 28, 1759–1765. doi:10.1093/bioinformatics/bts237
Munkhdalai, T., Li, M., Batsuren, K., Park, H. A., Choi, N. H., and Ryu, K. H. (2015). Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations. J. Cheminform. 7, S9. doi:10.1186/1758-2946-7-S1-S9
National Research Council. (2011). Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press.
Nguyen, T. H., and Grishman, R. (2015). “Relation extraction: perspective from convolutional neural networks,” in Proceedings of the 1st Workshop on Vector Space Modeling for Natural Language Processing (Denver, Colorado: Association for Computational Linguistics), 39–48.
Pyysalo, S., Airola, A., Heimonen, J., Björne, J., Ginter, F., and Salakoski, T. (2008). Comparative analysis of five protein-protein interaction corpora. BMC Bioinformatics 9:S6. doi:10.1186/1471-2105-9-S3-S6
Pyysalo, S., Ginter, F., Heimonen, J., Björne, J., Boberg, J., Järvinen, J., et al. (2007). BioInfer: a corpus for information extraction in the biomedical domain. BMC Bioinformatics 8:50. doi:10.1186/1471-2105-8-50
Rahul, P. V., Sahu, S. K., and Anand, A. (2017). Biomedical Event Trigger Identification Using Bidirectional Recurrent Neural Network Based Models Vancouver, Canada: Association for Computational Linguistics, 316–321.
Raja, K., Subramani, S., and Natarajan, J. (2013). PPInterFinder—a mining tool for extracting causal relations on human proteins from literature. Database (Oxford) 2013, bas052. doi:10.1093/database/bas052
Rindflesch, T. C., and Fiszman, M. (2003). The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J. Biomed. Inform. 36, 462–477. doi:10.1016/j.jbi.2003.11.003
Rindflesch, T. C., Kilicoglu, H., Fiszman, M., Rosemblat, G., and Shin, D. (2011). Semantic MEDLINE: an advanced information management application for biomedicine. Inf. Serv. Use 31, 15–21. doi:10.3233/ISU-2011-0627
Rindflesch, T. C., Tanabe, L., Weinstein, J. N., and Hunter, L. (2000). “EDGAR: extraction of drugs, genes and relations from the biomedical literature,” in Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing (Honolulu, Hawaii: NIH Public Access), 517.
Settles, B. (2005). ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 21, 3191–3192. doi:10.1093/bioinformatics/bti475
Song, M., Kim, W. C., Lee, D., Heo, G. E., and Kang, K. Y. (2015). PKDE4J: entity and relation extraction for public knowledge discovery. J. Biomed. Inform. 57, 320–332. doi:10.1016/j.jbi.2015.08.008
Song, S. J., Heo, G. E., Kim, H. J., Jung, H. J., Kim, Y. H., and Song, M. (2014). “Grounded feature selection for biomedical relation extraction by the combinative approach,” in Proceedings of the ACM 8th International Workshop on Data and Text Mining in Bioinformatics (Shanghai, China: ACM), 29–32.
Swanson, D. R. (1986). Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect. Biol. Med. 30, 7–18. doi:10.1353/pbm.1986.0087
Tang, B., Feng, Y., Wang, X., Wu, Y., Zhang, Y., Jiang, M., et al. (2015). A comparison of conditional random fields and structured support vector machines for chemical entity recognition in biomedical literature. J. Cheminform. 7, S8. doi:10.1186/1758-2946-7-S1-S8
Thomas, P., Solt, I., Klinger, R., and Leser, U. (2012). “Learning protein protein interaction extraction using distant supervision,” in Proceedings of Robust Unsupervised and Semi-Supervised Methods in Natural Language Processing (Workshop at International Conference Recent Advances in Natural Language Processing), (Hissar, Bulgaria: INCOMA Ltd).
Usié, A., Alves, R., Solsona, F., Vázquez, M., and Valencia, A. (2013). CheNER: chemical named entity recognizer. Bioinformatics 30, 1039–1040. doi:10.1093/bioinformatics/btt639
Wattarujeekrit, T., Shah, P. K., and Collier, N. (2004). PASBio: predicate-argument structures for event extraction in molecular biology. BMC Bioinformatics 5:155. doi:10.1186/1471-2105-5-155
Webber, J. (2012). “A programmatic introduction to neo4j,” in Proceedings of the 3rd annual conference on Systems, programming, and applications: software for humanity (Tucson, Arizona: ACM), 217–218.
Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., et al. (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic. Acids Res. 34(Suppl_1), D668–D672. doi:10.1093/nar/gkj067
Wishart, D. S., Jewison, T., Guo, A. C., Wilson, M., Knox, C., Liu, Y., et al. (2012). HMDB 3.0 – the human metabolome database in 2013. Nucleic Acids Res 41, D801–D807.
Yang, W., Soares, J., Greninger, P., Edelman, E. J., Lightfoot, H., Forbes, S., et al. (2013). Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41, D955–D961. doi:10.1093/nar/gks1111
Yang, Z., Lin, H., and Li, Y. (2008). Exploiting the performance of dictionary-based bio-entity name recognition in biomedical literature. Comput. Biol. Chem. 32, 287–291. doi:10.1016/j.compbiolchem.2008.03.008
Yeh, A., Morgan, A., Colosimo, M., and Hirschman, L. (2005). BioCreAtIvE task 1A: gene mention finding evaluation in BMC Bioinformatics, 6:S2. doi:10.1186/1471-2105-6-2
Yimam, S. M., Biemann, C., Majnaric, L., Šabanović, Š, and Holzinger, A. (2016). An adaptive annotation approach for biomedical entity and relation recognition. Brain Inform. 3, 157–168. doi:10.1007/s40708-016-0036-4
Zesch, T., Müller, C., and Gurevych, I. (2008). “Using wiktionary for computing semantic relatedness,” in Proceedings of the 23rd National Conference on Artificial Intelligence, Vol. 2, (Chicago, IL: AAAI Press), 861–866.
Keywords: text mining, named entity recognition, relation extraction, scientific knowledge discovery tool, scientific knowledge representation
Citation: Song M, Kim M, Kang K, Kim YH and Jeon S (2018) Application of Public Knowledge Discovery Tool (PKDE4J) to Represent Biomedical Scientific Knowledge. Front. Res. Metr. Anal. 3:7. doi: 10.3389/frma.2018.00007
Received: 20 September 2017; Accepted: 08 February 2018;
Published: 26 February 2018
Edited by:
Xianwen Wang, Dalian University of Technology (DUT), ChinaReviewed by:
Andreas Holzinger, Medical University of Graz, AustriaSam Henry, Virginia Commonwealth University, United States
Copyright: © 2018 Song, Kim, Kang, Kim and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Song, bWluLnNvbmdAeW9uc2VpLmFjLmty
 Min Song
Min Song Munui Kim
Munui Kim Keunyoung Kang
Keunyoung Kang Yong Hwan Kim
Yong Hwan Kim