Abstract
The goal of regenerative agriculture is to utilize technologies that build healthy soils and improve the environment. Microbial technologies could play a significant role in reducing reliance on synthetic herbicides for weed control. In the United States, the expenditure on herbicides exceeds $5 billion annually and accounts for 58% of the total pesticide use nationally. This overreliance on chemical weed control has exacerbated herbicide resistance in a multitude of weed species, leading to aggressive cultivation practices that contribute to soil erosion and depletion. The proliferation of microbiome research in agriculture has increased our understanding of the complex interactions between plant species and their microbiota. Microbial technologies offer novel weed management strategies that could reduce the need for herbicides. Some of these strategies could also help rebuild soil and improve environmental quality. Specifically, we propose three emerging areas in microbiome science that can enhance weed management: (1) identifying soil microorganisms that inhibit weed growth; (2) discovering microbial natural products that suppress weeds; and (3) developing field management approaches that promote weed suppression by enhancing soil microbiome function.
Introduction
Weed management in the United States (U.S.) has been largely reliant on synthetic herbicides since the 1970's. In some cropping systems, synthetic herbicides have reduced or eliminated the need for tillage or cultivation to manage weeds. Dependence on chemical weed control fuels a global herbicide industry that accounts for 40% of pesticide use worldwide (1). In the U.S. alone, the expenditure on herbicides exceeds $5 billion each year and accounts for 57% of the total pesticide use nationally (2). However, the continued sole reliance on chemical control has led to the evolved resistance of many weed species to an increasing number of widely used herbicides (3). Herbicide-resistant weeds, particularly weeds that are resistant to multiple herbicides, threaten agricultural productivity and sustainability.
The prevalence of single-tactic approaches to weed control stems largely from the early commercial success of glyphosate. Indeed, the development of glyphosate-resistant genetically modified (GM) crops expanded the use of glyphosate almost 15-fold by the twenty-first century (4) and encompassed multiple crops (Figure 1). Glyphosate applications in the U.S. exceed 1 billion kg/yr and now account for 67% of quantities used globally (4). As a result, severe outbreaks of glyphosate-resistant or tolerant weed populations have been reported in 54 plant species (5), resulting in an expected annual cost of over $10 billion in increased chemical costs (6). Herbicide resistance in weeds has been developing rapidly, now compromising 21 of the 31 currently known herbicide sites of action (7). Many weed populations have evolved resistance to multiple sites of action (7). The second generation of GM crops that have stacked herbicide-resistant traits is likely to accelerate the evolution of weeds resistant to multiple sites of action and further compromise the efficacy of chemical control methods (3). Pivoting toward a regenerative agricultural system that limits or delays the development of herbicide-resistant weeds will require innovative approaches.
Figure 1
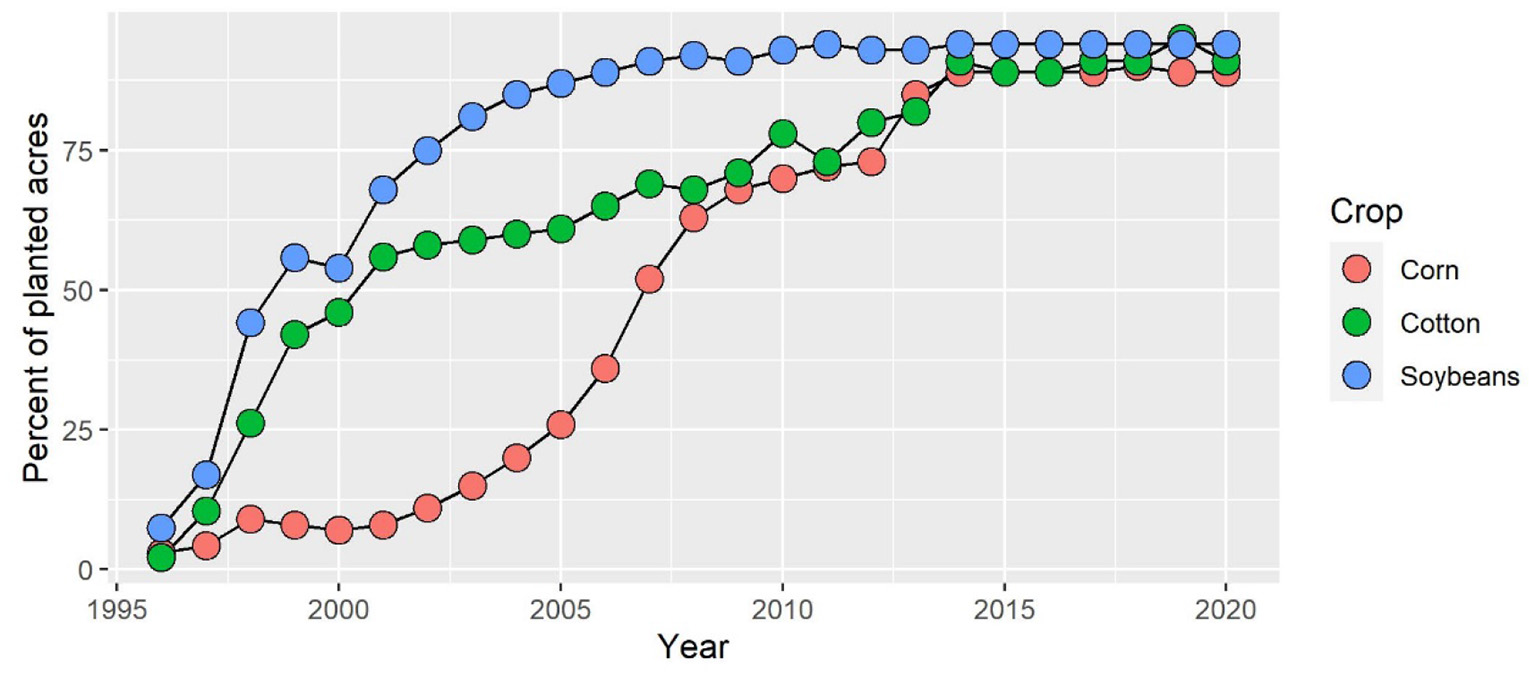
Adoption of herbicide-tolerant crops in the United States since the 1990's. The crop data indicate herbicide-tolerant varieties. Source: USDA, Economic Research Service using data from the 2002 ERS report, Adoption of Bioengineered Crops (AER-810) for the years 1996–99 and National Agricultural Statistics Service (annual) June Agricultural Survey for the years 2000–20.
While there is an urgent need to develop new herbicide compounds with novel modes of action, the pace of discovery has been slow in the early twenty-first century. In 2020, inhibition of fatty acid thioesterase (Cinmethylin, branded as Luximo by BASF), became the first new mode of action approved in the past 35 years by HRAC (Herbicide Resistance Action Committee) (Figure 2). This compound was originally discovered synthetically in the early 1980's (8). Naturally occurring microorganisms that suppress weeds are a potential source of novel herbicides. Microorganisms associated with plants (collectively, the plant microbiome) likely co-evolved strategies to contend with neighboring plant competitors, so they may be a promising reservoir for compounds that inhibit plant growth. Several microorganisms have been formulated as bioherbicides to control weedy and invasive plants in agricultural and natural areas (9). An alternative to using living microorganisms for weed control is the application of compounds produced by these organisms, which are referred to as “natural products.” Weed-suppressive and allelopathic compounds can be isolated as natural products derived from microorganisms and plants (10). Natural products isolated from microorganisms also play important roles outside of agriculture. For example, over 60% of FDA-approved anti-infective and anti-tumorigenic agents currently on the market were discovered from microorganisms found in natural environments, like soil and water (11, 12).
Figure 2
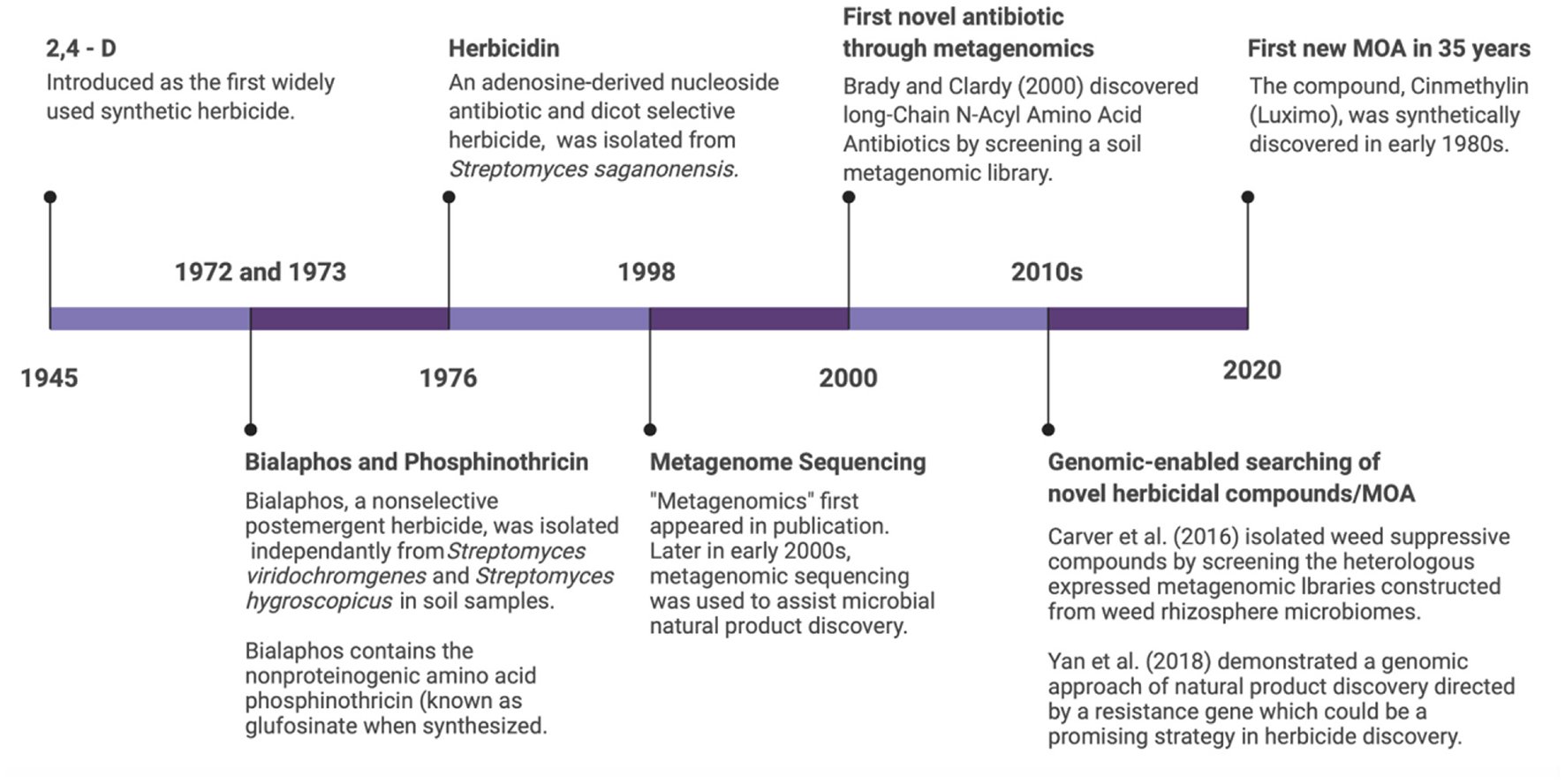
Timeline of herbicide development indicating synthetic, natural, and genomics-enabled screening tools. Chemical synthesis approaches have stagnated since the start of the twenty-first century, while microbial natural product discovery and genomics show promising applications in weed management.
Natural product discovery has historically been limited by the fact that most soil microorganisms are not cultivable in a laboratory setting. The biosynthetic potential of soil microbiomes may be underestimated because research may be biased toward the few microbial phyla that have high representation of cultivable bacteria with full reference genomes (13). In addition, research efforts have sometimes overlooked environments that could serve as potential reservoirs of natural products. For example, an analysis of park soils from New York City showed a large diversity of natural product biosynthetic gene clusters (14). Soil microbiomes producing natural products relevant to agriculture are found worldwide and in a wide variety of environments.
Innovations in sequencing and molecular biology have enabled metagenomic approaches to be developed for isolation of microbial antibiotics and enzymes, which could lead to the discovery of new modes of action for weed control (15). Notably, shotgun metagenomics facilitates de novo sequencing of microbiomes (16). Metagenomic approaches to natural product discovery have been used to identify new antibiotics for the pharmaceutical industry (17). The same methods used for the isolation of antibiotics can accelerate the discovery of weed-suppressive compounds. For example, antibiotic compounds such as herbicidin, blasticidin and 5-hydro-xylmethyl-blasticidin S exhibit herbicidal activity (15). Compounds other than antibiotics, such as glufosinate, were also discovered from soil bacteria and subsequently synthesized in large quantities as commercial herbicides (18). Most recently, genome mining of soil fungal species led to the discovery of a potent weed-suppressive compound that could be developed as an herbicide having a novel mode of action (19). Once identified, natural products must be synthesized in large quantities to be used commercially. Microbial technologies focused on product biosynthesis are based on the concept of microorganisms serving as production factories for natural products.
An alternative to the use of microbial natural products for weed control is the use of ecological management strategies that enhance microbiome function to suppress weeds. Integrated weed management (IWM), which combines various management practices based on ecological principles, can be an effective approach to manage herbicide-resistant weeds (3). An IWM approach focuses on managing herbicide-resistant weeds through mechanical and cultural practices, such as growing cover crops (20), cultivation, increased seeding rates, and reducing weed seed drop during harvest (21). However, IWM research has largely focused on aboveground processes, such as plant competition and herbivory, with more limited attention given to understanding the soil microbiome despite its significant influence on weed establishment and growth (1). The soil microbiome can be affected by land management practices. For example, agricultural practices such as tillage, crop rotation, cover crops, and fertilization can influence the diversity of arbuscular mycorrhizal fungi (22, 23), which in turn can impact a range of weed species (24, 25). This interaction suggests that field management practices could be used to create weed-suppressive soil microbiomes, supplementing current IWM strategies. Sequencing technologies could be harnessed to better understand the effects of agricultural practices on the soil microbiome and consequently on weed species and crop-weed interactions.
In this paper, we review: (1) the status and challenges of microbial biocontrol with bioherbicides; (2) current approaches of natural product discovery for novel herbicides; and (3) IWM strategies for managing the field microbiome and suppressing weeds with negative plant-soil feedbacks. Lastly, we address the need for soil microbiome research that uses emerging technologies and methodologies to discover novel weed-suppressive compounds. Our review differs from previous treatments of microbial biocontrol and natural products [e.g., (26, 27)] because we focus on the potential of emerging technologies to assist natural product discovery and the role of cropping system management in shaping the soil microbiome.
Microbial Biocontrol With Bioherbicide Agents
Decades of research have focused on bacteria and fungi for the control of undesirable plants. The microbial agents or their compounds are referred to as “bioherbicides” and suppress weeds through plant-pathogen interactions or allelopathy. For example, novel pathogens were able to accumulate and suppress a highly invasive species, Japanese stiltgrass (Microstegium vimineum), under field conditions (28). Most bioherbicides are target-specific pathogens that require large quantities of product (e.g., infective spores) to control mostly annual weeds in cropping or turfgrass systems (26). This approach is often referred to as “inundative” biological control. It is distinct from the “inoculative” or classical biological control approach, which typically uses imported insects to target non-native perennial weeds occupying extensive rangelands. Although some microbes might be candidates for the inoculative approach, this approach is challenging to implement and generally unsuitable for agricultural systems (27).
Among the most promising bioherbicides are microbial strains that can reduce the weed seedbank by promoting weed seed decay, inhibiting germination, or arresting germination. Such bioherbicides, which target the earliest stages of weed establishment, have great potential in reduced-till or no-till cropping systems (26). For example, Pseudomonas strains isolated from weed rhizospheres have been developed into effective preemergent bioherbicides. A strain of Pseudomonas fluorescens formulated as a bioherbicide caused a 90% reduction in emergence of an annual weedy grass, green foxtail (Setaria viridis) (29). Many Fusarium strains with seed-decaying potential have been evaluated for their capacity to kill weed seeds (30). In contrast with these preemergent bioherbicides, postemergent bioherbicides control the weed seed bank by reducing seed production. For example, Puccinia carduorum suppressed musk thistle (Carduus nutans) seed production by as much as 57% (31). Other research showed that pseudomonas spp. developed for biocontrol reduced seed production of downy brome (Bromus tectorum) by 64% (32).
Several commercial biocontrol agents have been developed from weed-suppressive microorganisms isolated from soil (1). A limited number of bioherbicides (products of living or dead microorganisms) are currently registered in the United States (Table 1). The first registered bioherbicide in the United States was DeVine®, introduced in 1981. The product is a facultative fungal pathogen (Phytophthora palmivora) that causes root rot in strangler vine (Morrenia odorata). Since then, the number of biopesticides has increased around the world, but the market share of bioherbicides represents <10% of all biopesticides (26). The following paragraphs discuss reasons for the limited commercial success of bioherbicides: the difficulty of studying some microbial taxa, the need to screen numerous isolates, and the unpredictable behavior of some candidate agents under field conditions.
Table 1
| Product name and time | Active microbe species | Target | Mechanism | Status | References |
|---|---|---|---|---|---|
| DeVine™, 1981 | Phytophthora palmivora | Strangler vine (Morrenia odorata) in citrus crops | Initiates a root infection in strangler vine that starts to kill the plant in 6–10 weeks | Not available | (26) |
| Collego™/LockDown™, 1982/2006 | Colletotrichum gloeosporioides f.sp. aeschynomene | Northern jointvetch (Aeschynomene virginica L. B.S.P.) in rice and soybean | Primarily infects the stems of the weed, causes disease lesions that will completely encircle the stems of the northern jointvetch plants. | Not available | (33) |
| Dr. BioSedge®, 1987 | Puccinia canaliculala | Yellow nutsedge (Cyperus esculentus L.) in soybean, potato, corn, and cotton | Inhibits yellow nutsedge flowering, reduces plant density and new tuber formation | Product failed due to mass production issue | (34) |
| Woad Warrior®, 2002 | Puccinia thlaspeos woad | Dyer's woad (Isatis tinctoria L.) | Fungal rust that reproduces and spreads using only dyer's woad as a host. | Not commercially available | (35) |
| Myco-Tech® Paste /Chontrol® Paste, 2005/2020 | Chondrostereum purpureum HQ1/ PFC2139* | Susceptible deciduous tree species in forests | Colonizes the stump and inhibits sprouting and regrowth, causes subsequent wood decomposition. The reduced stem density helps mechanical cut. | Commercially available | (34) |
| Smoulder® G/WP, 2008 | Alternaria destruens 059 | Dodders (Cuscuta spp.) in fields and ornamental nurseries | Infects live or dead dodder plant tissue, suppressing dodder at early and late stages of growth | Not commercially available | (26) |
| SolviNix™, 2009 | Tobacco mild green mosaic tobamovirus (TMGMV U2) | Tropical soda apple (Solanum viarum) in rangelands | The virus enters the plant cells through minute injuries and kills tropical soda apple by triggering a systemic lethal hypersensitive plant response. | Commercially available | (34) |
| Phoma P/H/TECH, 2012 | Phoma macrostoma 94-44B | Dicots in golf courses, agriculture and agroforestry | Produces macrocidins that cause photobleaching in dicots. | Commercially available | (26) |
| OpportuneTM/MBI-005, 2012* | Streptomyces acidiscabies | Broadleaves and sedges in turf, wheat, rice, and corn | The fermentation produces Thaxtomin A that inhibits cellulose biosynthesis in the meristem of sensitive plant species. | Undergoing further formulation refinement | EPA, Marrone Bio Innovations Inc |
| Battalion Pro, 2020 | Pseudomonas fluorescens ACK55 | Downy brome (Bromus tectorum), medusahead (Taeniatherum caput-medusae), jointed goatgrass (Aegilops cylindrica) | The bacteria affect the roots, seeds, or young seedlings and inhibit root-cell elongation. The suppressive compound inhibits lipopolysaccharide production in the cell wall and membrane and reduces root-cell wall elongation. | Absence of an industrial partner | EPA |
| Venerate /MBI-012, 2021*† | Burkholderia rinojensis A396 | Pigweed family (Amaranthaceae) | Produces herbicidal compounds including Templamide A/B and Templazole A/B | Registered as bioinsecticide. Spectrum of herbicidal activity and crop safety to be determined. WDG formulation in development. | EPA, Marrone Bio Innovations Inc |
Registered bioherbicides in the United States.
Adapted from Cordeau et al. (26), Aneja et al. (9), and Abbas et al. (34).
OpportuneTM/MBI-005 and Venerate/MBI-012 are microorganism-based products that contains non-viable microbe cells. According to Marrone Bio Innovations Inc, WDG (water dispersible granules) formulation is being developed for commercially viability.
Venerate/MBI-012 is registered as a bioinsecticide but the same strain possesses herbicidal activity. The product is sold as Venerate or Venerate XC. The herbicidal activity is from multiple metabolites (undisclosed) produced during fermentation (Marrone Bio Innovations Inc at IR-4 western regional workshop).
Bioherbicide development is impeded by challenges in accessing the pool of potential agents. Many surveys are restricted to a small subset of microbial taxa that are primarily fungi (1) and dependent on cultivation-based methods. The potential pool of biological agents for biocontrol is largely untapped, given the vast diversity of microbial taxa that have no reference genome (36). The identification and isolation of microorganisms that are elusive to standard laboratory cultivation may be facilitated by recent advances in cell capture technologies. One example is the “ichip” platform developed by Epstein et al., where cultivation in native soil habitats is achieved using diffusion chambers (37). Another example is using microfluidic techniques to simulate the chemical conditions and physical structures of native growth conditions (38). Greater focus on technologies to isolate microorganisms from their environments should expand the pool of candidate bioherbicides, increasing the likelihood that novel products for weed control will be discovered.
Another major challenge in bioherbicide discovery is the time- and resource- consuming process of isolate screening and testing. For instance, Kennedy and Stubbs (39) recovered more than 10,000 isolates using a conventional agar plating method. After several rounds of bioassay and growth chamber screening, only six strains showed promise and were field tested. Pathogenic isolates collected from diseased plants might reveal potential bioherbicides useful in downstream screening but require significant testing for off-target pathogenicity. A Bipolaris bicolor strain was isolated from severely diseased leaves of goosegrass (Eleusine indica) in a tea plantation system. Further tests on pathogenicity and host range demonstrated the potential of this strain as a biocontrol agent against Poaceae weeds in tea and broadleaf vegetable production (40). The initial field collection of 10 candidate isolates was obtained from 16 tea plantations through a time-consuming process. Soil microbiomes can serve as a pool of potential bioherbicides as well but could have similar limitations (time- and resource-intensive screening processes). For example, only one promising phytotoxic isolate was obtained after an herbicidal assay of 1,300 field-collected Streptomyces strains, even though Streptomyces are well-known for producing secondary metabolites relevant to natural product discovery (41). For bioherbicides to be successfully commercialized, additional testing and evaluation of host range, formulation, soil survival, production, and application need to be conducted to meet both consumer demand and regulatory requirements. Click or tap here to enter text.
Commercial use of biocontrol agents will be easiest if these bioherbicides, like most synthetic herbicides, are reliably effective when applied to the soil. However, the behavior of bioherbicide agents in soil can be unpredictable if the product is comprised of living organisms that are expected to perform a specific function. For example, a recent study on weed-suppressive Pseudomonas fluorescens strains showed that these strains reduced plant growth when grown on agar media but not in soil (42). Competition from other soil microorganisms was suggested as a possible reason for the loss of efficacy in the field. Indeed, microbial communities often work as a cohort and microbial functions that suppress weeds could be affected by both microbial interactions and environmental conditions. A single strain is less likely to be effective under field conditions. Recent studies have demonstrated that synthetic microbial communities (SynCom) designed with metagenomic data have tremendous potential in plant improvement (43). In fact, commercial biofertilizers or plant growth-promoting products on the market often include multiple microbial species or strains. An example is the Mammoth P consortium (Growcentia, Fort Collins, CO, USA), which is an assemblage of phosphorus-mobilizing bacteria from four taxa. The product showed higher rates of phosphate solubilization compared with single strains (44). Currently, all the registered bioherbicides in the United States are based on single species and strains. Outside of the United States, the only bioherbicide product that contains more than one strain is Organo-Sol®, registered in Canada (26). This product, which contains several species of lactic acid bacteria that produce lactic acid and citric acid, suppresses white clover (Trifolium repens) and red clover (Trifolium pratense) in lawns. Emerging technologies enabling researchers to study difficult-to-culture microorganisms may facilitate the development of novel SynCom-based bioherbicides. A more in-depth understanding of microbial interactions and their effects on microbiome function would help ensure that bioherbicides that are effective in lab and greenhouse settings can also suppress weeds in the field.
The rising cost of managing herbicide-resistant weeds and restrictions on the use of some herbicides should drive demand for innovations in weed biocontrol. The global bioherbicide market, valued at USD 1.28 billion in 2016, is expected to reach USD 4.14 billion by 2024 (45). However, challenges in bioherbicide commercialization still exist [reviewed by (27)]. Although bioherbicides have been the focus of research for decades, their modes of action are not well-understood [reviewed by (26, 46)]. Environmental factors, such as temperature and humidity, and their interactions can significantly affect bioherbicide efficacy under field conditions (47). Microbial interactions could also impact the organism's virulence through quorum sensing (47). Other factors impeding commercialization may include inconsistent product quality with scaled-up production (47) and concerns about non-target dispersal through adaptation to the new environment and host exposure over time. The potential to release a bioherbicide agent that becomes a non-target pathogen is a major limitation to wider adoption of practice. One solution to concerns about non-target dispersal is to apply active compounds produced by microorganisms (i.e., natural products) rather than releasing live microorganisms as biocontrol agents.
Discovery of Weed-Suppressive Natural Products
Microbial natural products have been a prolific source of compounds for medical and agricultural uses (12). Natural products are secondary metabolites produced by organisms that are not involved in primary growth, reproduction, and development. Some secondary metabolites increase fitness by altering interactions with other organisms. For example, secondary metabolites can be used as biological weaponry to outcompete other organisms for resources. Microorganisms in the plant rhizosphere, which is a narrow band (1–2 mm) of soil surrounding plant roots, could co-evolve with their plant hosts by generating novel allelopathic compounds that suppress competing plant species. Because these compounds suppress plants, they could be a rich reservoir for the discovery of natural products with novel herbicidal properties. Some natural products isolated from microbes can be used as herbicides without modification, while others can be modified or used to identify new herbicide targets (27).
Research on natural products developed rapidly after the discovery of penicillin, one of the world's first antibiotics (10). This discovery ushered in the “Golden Age” of natural product discovery of the 1950's and 1960's, which focused primarily on microorganisms and plants. During this early period, researchers developed systematic screening processes for soils, typically including acquisition of environmental samples, culturing and isolation of microorganisms, followed by testing of the fermentation broth or purified products against test organisms. More than 1,000 natural products with antibacterial or antifungal activities were discovered during this time (10). Most of these natural products were produced by organisms in the bacterial phylum Actinomycetes, containing the highly cultivable members of the Streptomyces genus. The many species of Streptomyces are widely distributed across nearly all ecosystems, including the microbiomes inhabited by higher eukaryotes. At present, actinobacteria produce two thirds of all known antibiotics (48). Actinobacteria also produce a vast array of anti-cancer compounds, immunosuppressants, anthelmintics, antiviral compounds, and extracellular enzymes (48). Compounds produced by several actinobacteria species have also led to novel herbicide discovery (Figure 2). These compounds include bialaphos produced by Streptomyces hygroscopicus SF1293, herbicidins produced by Streptomyces saganonensis, phosphinothricin produced by Streptomyces viridochromogenes (known as glufosinate when synthesized), and Thaxtomin A from Streptomyces acidiscabies (49) (Figure 2).
Natural product discovery continued after the Golden Age. An estimated 10–20 million microbial isolates were screened from 1950 to 2000, with efforts mostly focused on discovering antibacterial and antifungal compounds (10). There was also interest in finding natural products for the treatment of various human diseases and development of agrichemicals. Sample collection was expanded to sources beyond soils; for example, some anti-cancer compounds were successfully derived from marine samples (50). However, the pace of natural product discovery eventually slowed, partially because screening processes that require isolating organisms as pure cultures in a laboratory setting are laborious. Such cultivation-dependent screening processes are still widely used. For example, 14 phytotoxic secondary metabolites were obtained from in vitro cultures of two fungal pathogens of buffelgrass (Cenchrus ciliaris) (51). Among these compounds, radicinin was identified as a promising bioherbicide that showed target-specific toxic activities on buffelgrass. Another study conducted herbicidal assays on 1,300 field-collected Streptomyces strains and found only two herbicidal compounds from one phytotoxic isolate (41). Research in the decades after the Golden Age of natural product discovery has revealed more bioactive actinomycete species; however, most bacteria in soil remain uncultivable using standard lab culturing techniques (10, 15). Therefore, the likelihood of finding new natural products via traditional methods is decreasing. The rate of novel antibiotic discovery in the phylum Actinomycetes using traditional screening has been estimated at <1 per million (48).
Advances in genome sequencing during the twenty-first century have brought new opportunities for natural product discovery. Genome sequencing can be used to identify novel biosynthetic gene clusters (BGC) coding the production of secondary metabolites (Figure 2). Genomic sequencing data revealed that BGCs in microbial genomes are much more abundant than predictions based on expressed secondary metabolites (48). A recent study of BGC diversity and potential bioactivity in urban park soils of New York City demonstrated a higher-than-expected level of chemical novelty, suggesting that urban soils could be a valuable source of natural products (14). Genomic datasets are typically large and complex, particularly with microbiome samples or when many microbial strains are collected. To find potentially useful natural products within these datasets, bioinformatic tools such as antiSMASH6 (52) or PRISM4 (53) can be used to predict and identify novel BGCs. For example, antiSMASH was used to predict which gene clusters in actinomycetes might yield new antibiotics (54). This approach led to the discovery of corbomycin, which is an antibiotic with a novel mode of action (54). Several other bioinformatic tools have been developed to assist BCG mining. For example, with the putative gene clusters identified by antiSMASH, BiG-SCAPE can build gene cluster families (55). Additionally, MIBiG is a database of known BGCs and their products that could be used for sequence-based dereplication of the gene clusters (56). Deep learning approaches are being developed for screening of chemical libraries; these approaches can improve predictions of natural product functions based on their structures (57).
The rapid-growing datasets of BGCs discovered through high-throughput genome mining could be a challenge in natural product discovery. Because these datasets contain numerous BGCs of unknown function, many of which are irrelevant to the desired purpose or functionally redundant with known BGCs, it is difficult to determine which BGCs merit experimental study. One strategy is to cluster putative BGCs into gene cluster families (GCFs) for the purpose of dereplication and to avoid rediscovery of known compounds in the downstream experimental characterization. However, the increasingly large number of GCFs with no known functions still makes it difficult to focus downstream screening on groups that are likely to produce natural products of interest. For example, a large-scale genomic study on 3,080 bacterial genomes from the Actinomycetes phylum found nearly 18,000 GCFs, most of which have no known products (55). To increase the likelihood of obtaining chemical novelty, some researchers have tried prioritizing microbial taxa that are less well-characterized, or sampling extreme or unusual environments. For example, rare Actinomycetes bacteria of marine and wetland ecosystems were suggested to be promising sources of novel natural products (58, 59). A recent global soil survey of polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs), which are enzymes involved in the biosynthesis of numerous peptide and peptide-like natural products, found that geographic distance and biome type were associated with diversity in BGCs (11). However, knowledge about the environmental and taxonomic distributions of BGCs is still lacking (48).
Although metagenomic approaches provide great opportunities for identifying novel BGCs, it is still a challenge to link orphan (i.e., unknown corresponding metabolites) BGCs to their associated natural products (60). To address this challenge, BGCs are often cloned or transferred to heterologous expression hosts such as Streptomyces species that are known for their natural product production abilities. The heterologous expression method typically involves three steps: isolation and cloning of the DNA fragments that contain BGCs in the original host, expression in a heterologous host organism, and genetic manipulation of the cloned pathway for interrogation or activation. Zhang et al. (60) provide a comprehensive review of heterologous expression methods for microbial natural product discovery. One advantage of heterologous expression of BGCs is that it enables the expression of BGCs from uncultivable microorganisms derived from environmental samples. Another advantage is that a good heterologous expression host can provide a clean secondary metabolite background, which will make it easier to isolate and identify the compound encoded by the BGC of interest. Streptomyces coelicolor M1152/M1154, Streptomyces avermitilis SUKA17/22, and Streptomyces lividans SBT5 are good expression hosts that lack competing BGCs (61). A third advantage is that genetic tools can be used to activate cryptic BGCs in the heterologous expression host; these tools are typically not available in the original host.
Cloning is a challenging step in the heterologous expression of microbial natural products due to the large size, repetitiveness, and high GC-content of many microbial BGCs (60). Library-based cloning and heterologous expression methods provide an alternative top-down approach. The strategy is widely used to clone microbial BGCs from metagenome or environmental DNA samples, where complete genome information is lacking. Genomic DNA are randomly sheared into small fragments of ~40 kb that could contain BGCs and ligated into cosmid or fosmid vectors. The vectors are then transformed into heterologous hosts such as E. coli or Streptomyces lividans strains to be expressed. The BGCs in the random 40 kb fragments are often incomplete and need to be combined and trimmed before use (60). However, a complete BGC can be obtained using this expression method and the biosynthetic pathway can be expressed. This approach was first used by Brady and Clardy to discover novel antibiotics from uncultivated microorganisms in soil samples (62). Following the same methodology, Carver et al. (63) isolated weed-suppressive compounds by using heterologous expression to screen fosmid libraries constructed from weed rhizosphere microbiomes (Figure 2). Library vectors that hold longer DNA fragments (>55 kb) than cosmids and fosmids, such as bacterial artificial chromosomes (BACs) and P1 artificial chromosomes (PACs), could be transferred to E. coli directly by electroporation (60). Because these vectors hold longer DNA, they make it easier to screen complete biosynthetic pathways or larger BGCs. The best example is the largest heterologously expressed BGC, the quinolidomicin A1, which is over 200 kb and cloned in a BAC library (64).
Assembly and direct cloning methods provide powerful alternatives to library cloning (60). These methods, based on synthetic biology, either assemble BGCs in vitro or reconstruct larger BGCs in vivo. Examples include Gibson assembly, Golden Gate assembly, and yeast recombination (65). A recent study successfully expressed two synthesized BGCs from human metagenomic sequences in various heterologous hosts and obtained five novel antibacterial compounds (66). In addition to these cloning methods, the increased feasibility of de novo DNA synthesis is providing new opportunities for research on BGCs. De novo DNA synthesis was used to access and refactor BGCs (i.e., reorganize the cluster structure to achieve stable function) <10 kb; this approach could greatly facilitate capturing and characterizing BGCs in the future (60). Although these advances are promising, it is still challenging to express natural products in heterologous host systems, where precursors or cofactors may be lacking (67).
It is often difficult to induce BCG expression because the factors controlling activation and expression are typically not well-understood. It was estimated that <10% of BGCs are expressed under laboratory culture conditions where microorganisms are grown in artificial, simplistic environments consisting of agar petri plates and liquid broth (10). Expression of BGCs may require environmental signals and cues, such as cues from microbial interactions with hosts or other organisms (68, 69). For example, unique chemical production patterns were observed in Streptomyces coelicolor interacting with other Actinomycetes bacteria by nanospray desorption electrospray ionization (NanoDESI) and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) imaging mass spectrometry (68). A recent study showed that fungal infection of plant roots induced the production of non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) by unknown BGCs in the root endophytic microbiome (70). Computational tools such as PREDetector that predict regulatory elements of BGC expression (71), along with other meta-omics data, could be used to systematically study the triggers of BGC expression (60).
A recent paper demonstrated a genomic approach to natural product discovery directed by a resistance gene, which could be a promising strategy for herbicide discovery (19). Specifically, the authors discovered and verified the mode of action of aspterric acid (a fungal sesquiterpenoid with herbicidal activity) based on the co-clustering of its corresponding self-resistance gene in the BGC responsible for aspterric acid biosynthesis. The rationale behind this approach is that, if the fungal aspterric acid targets a plant enzyme that is also essential to fungi, the fungus might have a resistance gene in the same BGC as the genes that produce aspterric acid. Although aspterric acid was previously known as a phytotoxic compound and not suitable as a commercial herbicide due to inadequate herbicidal strength and chemical complexity, this study provides proof of concept for a resistance gene-directed approach to genome mining for novel herbicide discovery (72).
Agronomic Microbiome Management for Weed Suppression
Soil microorganisms play an important role in sustaining healthy soils that promote crop production and suppress pests and weeds. Management practices that enhance soil biodiversity and cause desirable changes in soil community composition are likely to increase agricultural sustainability (73). In the context of agricultural weed control, managers may be able to create weed-suppressive soils through enrichment of weed-inhibiting microorganisms (74, 75). This section focuses on identifying practices that may be useful in creating weed-suppressive soils, which include reduced tillage, reduced agrichemical inputs, and maintenance of high soil organic matter (76). In addition, we discuss the role of plant-soil feedbacks (PSF) in weed control. It is worth noting that these topics can now be studied with next-generation sequencing technology [e.g., (77, 78)], which provides a much clearer picture of the soil microbiome than the laboratory cultivation-based methods that historically limited most studies to individual bacterial species or strains.
Standard agricultural practices such as tillage and heavy use of agrichemical inputs (fertilizers and pesticides) significantly alter soil microbiomes, sometimes with undesirable results (77, 78). A recent study suggested that cropping practices of organic and conventional farming with different tillage intensities accounted for 10% of total variation in wheat rhizosphere microbial communities (79). Cropping system affected not only individual microbial groups but also microbial co-occurrences, indicating that soil microbial interactions can also be affected by common agricultural practices. A better understanding of which microbiome members influence crop and weed performance and how to optimize the microbiome could markedly improve weed management in cropping systems.
Another agronomic strategy that can be developed within a microbial weed management approach is the addition of carbon amendments or incorporation of plant residues into soil (80–82). Soil microorganisms respond rapidly to carbon additions with enhanced growth, increasing their demand for nitrogen, phosphorus, and other limiting nutrients required for primary metabolism (83). This response to increased carbon availability is referred to as “nutrient immobilization” because limiting nutrients other than carbon become immobilized in microbial cells and therefore are unavailable for biological uptake by other organisms.
Nutrient immobilization can be harnessed for weed control because plants can be poor competitors for nutrients, relative to soil microorganisms. These differences could be exploited to control weed populations that preferentially grow on highly fertile soils (84, 85). Several field studies investigating the effects of carbon addition on invasive weeds showed reduced nitrogen availability in carbon-amended soils, likely resulting from microbial nitrogen immobilization in response to carbon stimulation (86–90). These findings demonstrate the potential for soil carbon addition to promote microbial competition for nutrients during the critical period of weed control. In agronomic settings, it is important to examine which weed species are most responsive to soil carbon addition and the best timing for carbon applications and nutrient immobilization in relation to crop establishment. Future research should also describe the indirect, microorganism-mediated impacts of cover crop management on weed communities. Cover crops have been used to sequester carbon and nitrogen in soil and provide weed suppression. Several recent studies reported that soil microbial communities, including functional groups such as arbuscular mycorrhizal fungi, were positively affected by cover crop management (43). However, the role of these changes to microbial communities in influencing weed suppression by cover crops has not been examined.
Plant-soil feedback (PSF) occurs when a plant species alters biotic or abiotic conditions in soil, which in turn impact the growth of the same plant species or a different plant species (91). Negative PSF (PSF that inhibits plant growth) typically results from either allelopathic effects or accumulated host-specific pathogens in soil (92–94). Negative feedback is believed to be more common than positive feedback and plays an important role in species coexistence and the maintenance of plant biodiversity in ecosystems (91, 95).
While PSF has been primarily studied in natural environments, PSF is also important in agricultural environments and responsive to agricultural management practices. As discussed, practices such as tillage and fertilization can affect the composition and structure of the soil microbiome. These changes to the microbiome may affect PSF. van der Putten et al. (96) proposed a triangular framework consisting of symbionts, decomposers, and enemies (e.g., pathogens) to analyze the shifts in PSF due to environmental changes. The authors used this framework to predict the effects of agricultural practices (agrichemical inputs and land management in conventional and organic systems) on the soil microbiome and PSF. This study highlighted the role that PSF plays in agricultural systems but their potential for weed suppression was not discussed.
A research focus on PSF in agricultural systems could have important implications for weed management (Figure 3). There is increasing empirical evidence for a role of soil microbiomes and PSF in governing weed population dynamics. Understanding how management practices affect PSF could therefore improve ecological weed management strategies. This research is likely to be complex because PSF processes are likely to vary among weed species and cropping systems. For example, the rate at which plants accumulate species-specific pathogens and the effects of these pathogens vary among plant species (93). Plants that quickly accumulate pathogens might be more vulnerable to PSF processes limiting plant density (93). However, it is still not clear if the strength of negative PSF on a plant species corresponds with field abundance of the species, as research has provided contradictory results (97, 98). A more in-depth understanding of how weed density influences negative feedback pressures is likely to improve the efficacy of weed management strategies.
Figure 3
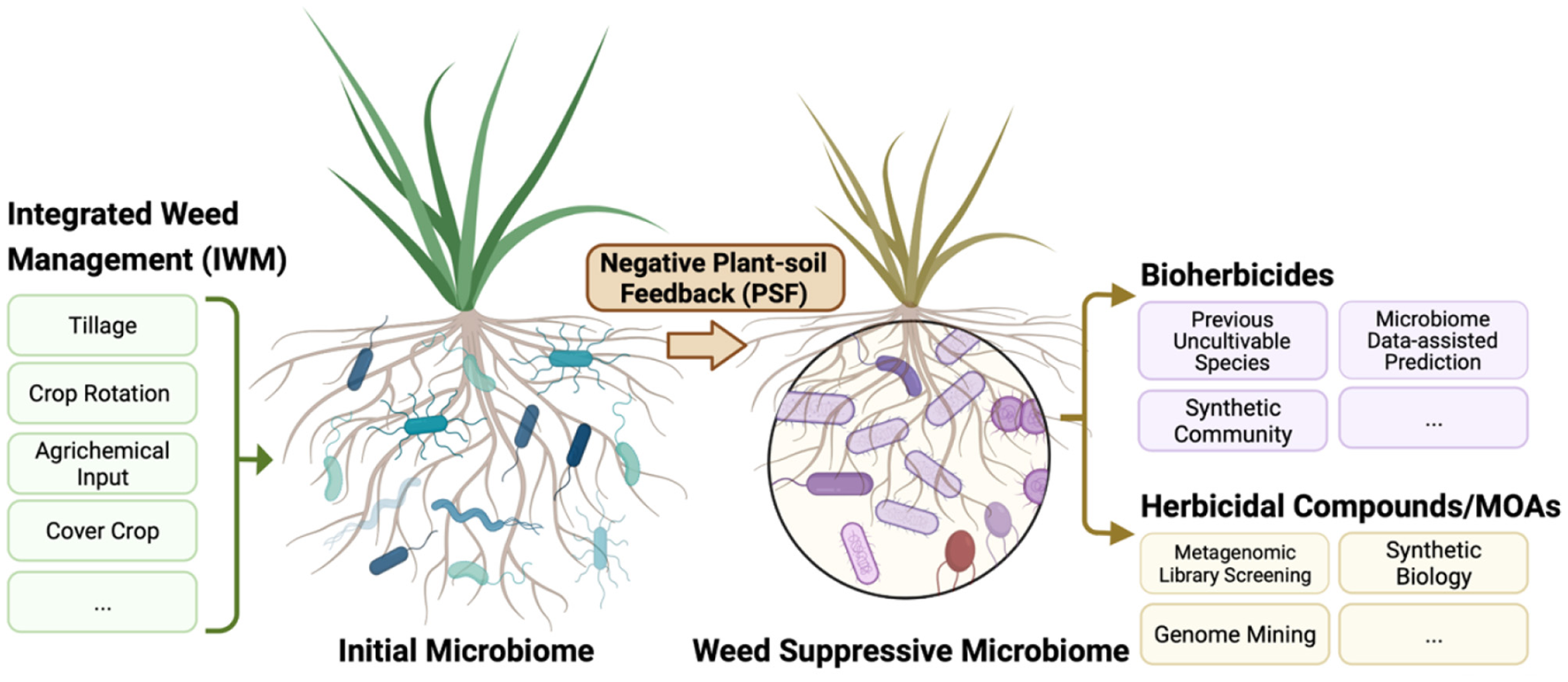
A schematic overview of approaches harnessing the soil microbiome and negative plant-soil feedback to promote weed management in regenerative agriculture. Agricultural practices alter the soil microbiome, so practices that promote weed suppression by microorganisms could be included in integrated weed management programs. Negative plant-soil feedbacks occurs when plants cause biotic and abiotic changes in the soil, which inhibit further plant growth. Research on these feedback processes may help identify potential microbial agents or compounds involved in plant growth suppression.
More generally, the direction and strength of PSF may depend on plant species or functional group (95, 99, 100). In a study of 48 grassland species, grasses and small herbs showed negative PSF, tall herbs exhibited positive PSF, and legumes showed neutral PSF, suggesting a strong correlation between PSF direction and plant functional group (99). While most weed species are small to mid-sized herbs or grasses, it is not clear if their PSF patterns are species-dependent or tend to be negative. For example, Himalayan balsam (Impatiens glandulifera), one of the most widespread weeds in the United Kingdom, showed positive PSF (101). In a study of 12 grass and forb species, early-successional species showed negative PSF while late-successional species exhibited positive PSF (102). Because PSF patterns appear to be species-specific, it is unlikely that any management program could promote negative PSF for all weed species in an agricultural field. A more realistic goal might be to promote negative PSF for most weeds or for particularly troublesome weeds, such as herbicide-resistant populations. Future research should investigate patterns in PSF strength and direction for these troublesome weeds. As sequencing technologies continue to improve in resolution and cost, the ability to characterize soil microbiomes as contributing to positive or negative PSF could improve integrated weed management strategies. For example, there may be opportunities to select cover crop species or crops that prime the soil for enrichment of weed-suppressive microbiomes and allelopathic exudates.
Although PSF processes can affect any plant species, research on PSF may be particularly important to understanding invasive plants. Many weeds are invasive species that were introduced into geographically novel ranges (with novel soil biota) and successfully competed against plants native to these novel ranges. It has long been recognized that microorganisms and PSF often play an important role in plant invasions (93, 103). Multiple hypotheses have been proposed to explain the success of plant invasions via belowground microbial effects. The “enemy release” hypothesis proposes that invasive plant species in a novel region are released from the factors limiting their population sizes in their native ranges. For example, the absence of soil-borne pathogens or growth-inhibiting soil biota in invaded regions can result in the proliferation of a plant species in these regions (104). Enemy release has been largely a theoretical concept, but there are convincing empirical studies that support this hypothesis. The establishment of Chinese tallow (Triadica sebifera), native to Asia, in the United States was influenced by soil biota: this invader was more negatively affected by soil pathogens in the native range and mycorrhizal colonization was higher in the invaded range (105). Another field study revealed that, when the invasive Lodgepole pine (Pinus contorta) is grown in soils without its native soil biota, this species can rapidly grow and spread into new regions (106). Common ragweed (Ambrosia artemisiifolia), a highly invasive annual weed native to North America, was able to escape from both aboveground and belowground enemies at a more local scale [new sites within North America; (107)]. The concept of escaping from “enemies” could become a management tool for protecting crops in situations where soil pathogen composition can be identified rapidly.
Alternatives to the enemy release hypothesis focus on soil microorganisms that are present in invaded regions. For example, invasive plant species could gain an advantage in their new ranges by stimulating the growth and abundance of pathogens that negatively affect resident competing plant species [i.e., the “accumulation of local pathogens” hypothesis; (108)]. In cropping systems, it is valuable to identify soil biota that are deleterious to weeds but not harmful to crop species. This area of study warrants further research attention. Invasive species research has also revealed that invasive plants can accumulate beneficial mutualists of their own (“enhanced mutualist” hypothesis) or suppress beneficial mutualists of resident species to gain a competitive advantage (“mutualism disruption” hypothesis). Garlic mustard (Alliaria petiolata), a noxious invasive weed, was shown to inhibit fungal mutualists of North American native plants; the authors observed stronger inhibition in its invaded range than in its native range (109). Continued research on interactions between invasive plants and soil microorganisms will improve our understanding of invasive agricultural weeds and plant-microbe interactions more broadly. Translating ecological theory into agricultural practice will require innovative approaches to identify factors influencing PSF processes, including plant, microbial, and environmental characteristics.
Conclusion
Managing weeds effectively and sustainably is essential to agricultural productivity. However, weed management in the twenty-first century is challenged by the increasing number of herbicide-resistant weeds, many of which are resistant to multiple herbicide modes of action. A key concept in regenerative agriculture is to use the most appropriate technologies to effectively manage agroecosystems. In the context of weed management, it is clear that these technologies must not be limited to synthetic herbicides. Several promising approaches are based on the soil microbiome, including bioherbicides, natural products derived from microbes, and manipulation of the existing microbiome through agricultural practices. Early research based on cultivation-dependent methods of microbial biocontrol paved the way for recent advances in genomics-enabled natural product discovery. Natural products that inhibit seed germination or arrest seedling growth can enable creative strategies for weed seedbank management. Research on microbes that contribute to weed suppression in the field can reveal additional natural products and suggest improvements to management programs (Figure 3). Continued advances in metagenomic sequencing will accelerate research on the microbial management of agricultural weeds. In addition, advances in digital agriculture will help incorporate microbiome data into predictions about crop performance and pest pressure. These technological advances are crucial to understanding how soil microbiomes affect agricultural productivity and how they might be harnessed to promote regenerative agriculture.
Funding
This work was supported by the Controlling Weedy and Invasive Plants Program (grant no. 2016-67014-24859) from the USDA National Institute of Food and Agriculture and a Cornell Institute for Digital Agriculture Student Research Grant award to LC.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
LC wrote the manuscript. JK-K and AD contributed to revisions. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Sofia Kashtelyan and Kristopher Smith for assisting with research that led to discussions on microbial natural product discovery in weed management. Thanks also to A. Sophie Westbrook for her helpful suggestions on an earlier version of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
TrognitzFHacklEWidhalmSSessitschA. The role of plant-microbiome interactions in weed establishment and control. FEMS Microbiol Ecol. (2016) 92:fiw138. 10.1093/femsec/fiw138
2.
AtwoodDPaisley-JonesC. Pesticides Industry Sales and Usage 2008–2012. Washington, DC: United States Environmental Protection Agency (2017).
3.
MortensenDAEganJFMaxwellBDRyanMRSmithRG. Navigating a critical juncture for sustainable weed management. Bioscience. (2012) 62:75–84. 10.1525/bio.2012.62.1.12
4.
BenbrookCM. Trends in glyphosate herbicide use in the United States and globally. Environ Sci Europe. (2016) 28:70. 10.1186/s12302-016-0070-0
5.
HeapI. The International Herbicide-Resistant Weed Database. Available online at: www.weedscience.org (accessed March 2, 2022).
6.
VarahAAhodoKCouttsSRHicksHLComontDCrookLet al. The costs of human-induced evolution in an agricultural system. Nat Sustainabil. (2020) 3:8. 10.1038/s41893-019-0450-8
7.
HeapI. The International Survey of Herbicide Resistant Weeds. (2022). Available online at: www.weedscience.org (accessed January 19, 2022).
8.
DukeSOBaersonSRRimandoAM. Herbicides: glyphosate. In: JR Plimmer, DW Gammon, NN Ragsdale, editors, Encyclopedia of Agrochemicals. New York, NY: John Wiley & Sons, Inc. (2003). p. 850–63. 10.1002/047126363X.agr119
9.
AnejaKRKhanSAAnejaA. Bioherbicides: strategies, challenges and prospects. In: T Satyanarayana, S Deshmukh, B Johri, editors, Developments in Fungal Biology and Applied Mycology. Singapore: Springer (2017). p. 449–70. 10.1007/978-981-10-4768-8_23
10.
KatzLBaltzRH. Natural product discovery: past, present, and future. J Indus Microbiol Biotechnol. (2016) 43:5. 10.1007/s10295-015-1723-5
11.
Charlop-PowersZOwenJGReddyBVBTerneiMGuimaraesDOde FriasUAet al. Global biogeographic sampling of bacterial secondary metabolism. eLife. (2015) 2015:e05048. 10.7554/eLife.05048
12.
PhamJvYilmaMAFelizAMajidMTMaffetoneNet al. A review of the microbial production of bioactive natural products and biologics. Front Microbiol. (2019) 10:1404. 10.3389/fmicb.2019.01404
13.
Crits-ChristophADiamondSButterfieldCNThomasBCBanfieldJF. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature. (2018) 558:440–4. 10.1038/s41586-018-0207-y
14.
Charlop-PowersZPregitzerCCLemetreCTerneiMAManikoJHoverBMet al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc Natl Acad Sci USA. (2016) 113:14811–6. 10.1073/pnas.1615581113
15.
Kao-KniffinJCarverSMDiTommasoA. Advancing weed management strategies using metagenomic techniques. Weed Sci. (2013) 61:171–84. 10.1614/WS-D-12-00114.1
16.
LibisVAntonovskyNZhangMShangZMontielDManikoJet al. Uncovering the biosynthetic potential of rare metagenomic DNA using co-occurrence network analysis of targeted sequences. Nat Commun. (2019) 10:1–9. 10.1038/s41467-019-11658-z
17.
HutchingsMTrumanAWilkinsonB. Antibiotics: past, present and future. Curr Opin Microbiol. (2019) 51:72–80. 10.1016/j.mib.2019.10.008
18.
DayanFEDukeSO. Natural compounds as next-generation herbicides. Plant Physiol. (2014) 166:1090–105. 10.1104/pp.114.239061
19.
YanYLiuQZangXYuanSBat-ErdeneUNguyenCet al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature. (2018) 559:415–8. 10.1038/s41586-018-0319-4
20.
DavisVMGibsonKDBaumanTTWellerSCJohnsonWG. Influence of weed management practices and crop rotation on glyphosate-resistant horseweed (Conyza canadensis) population dynamics and crop yield-years III and IV. Weed Sci. (2009) 57:417–26. 10.1614/WS-09-006.1
21.
WalshMJOwenMJPowlesSB. Frequency and distribution of herbicide resistance in Raphanus raphanistrum populations randomly collected across the Western Australian wheatbelt. Weed Res. (2007) 47:542–50. 10.1111/j.1365-3180.2007.00593.x
22.
AlguacilMM. The Impact of tillage practices on arbuscular mycorrhizal fungal diversity in subtropical crops. Ecological Applications. (2008) 18:527–36. 10.1890/07-0521.1
23.
van der HeydeMOhsowskiBAbbottLKHartM. Arbuscular mycorrhizal fungus responses to disturbance are context-dependent. Mycorrhiza. (2017) 27:431–40. 10.1007/s00572-016-0759-3
24.
RinaudoVBàrberiPGiovannettiMvan der HeijdenMGA. Mycorrhizal fungi suppress aggressive agricultural weeds. Plant Soil. (2010) 333:202. 10.1007/s11104-009-0202-z
25.
el OmariBel GhachtouliN. Arbuscular mycorrhizal fungi-weeds interaction in cropping and unmanaged ecosystems: a review. Symbiosis. (2021) 83:9. 10.1007/s13199-021-00753-9
26.
CordeauSTrioletMWaymanSSteinbergCGuilleminJP. Bioherbicides: dead in the water? A review of the existing products for integrated weed management. Crop Protection. (2016) 87:44–9. 10.1016/j.cropro.2016.04.016
27.
DukeSOPanZBajsa-HirschelJDouglas BoyetteC. The potential future roles of natural compounds and microbial bioherbicides in weed management in crops. Adv Weed Sci. (2022) 40:e020210054. 10.51694/AdvWeedSci/2022
28.
StrickerKBHarmonPFGossEMClayKFlorySL. Emergence and accumulation of novel pathogens suppress an invasive species. Ecol Lett. (2016) 19:469–77. 10.1111/ele.12583
29.
DaigleDJConnickWJBoyetchkoSM. Formulating a weed-suppressive bacterium in “Pesta” 1. Weed Technol. (2002) 16:407–13. 10.1614/0890-037X(2002)0160407:FAWSBI2.0.CO;2
30.
Müller-StöverDNybroeOBaraibarBLoddoDEizenbergHFrenchKet al. Contribution of the seed microbiome to weed management. Weed Res. (2016) 56:335–9. 10.1111/wre.12218
31.
BaudoinABAMAbadRGKokLTBruckartWL. Field evaluation of Puccinia carduorum for biological control of musk thistle. Biol Control. (1993) 3:136–52. 10.1006/bcon.1993.1009
32.
KennedyACYoungFLElliottLFDouglasCL. Rhizobacteria suppressive to the weed downy brome. Soil Sci Soc Am J. (1991) 55:722–7. 10.2136/sssaj1991.03615995005500030014x
33.
BowersRC. Commercialization of Collego TM –an industrialist's view. Weed Sci. (1986) 34:24–5. 10.1017/S0043174500068326
34.
AbbasTZahirZANaveedMKremerRJ. Limitations of existing weed control practices necessitate development of alternative techniques based on biological approaches. In: DL Sparks, editor, Advances in Agronomy. Cambridge, MA: Academic Press (2018). p. 239–80. 10.1016/bs.agron.2017.10.005
35.
KroppBRHansenDRThomsonSv. Establishment and dispersal of Puccinia thlaspeos in field populations of dyer's woad. Plant Dis. (2002) 86:241–6. 10.1094/PDIS.2002.86.3.241
36.
HugLAThomasBCSharonIBrownCTSharmaRHettichRLet al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. (2016) 18:159–73. 10.1111/1462-2920.12930
37.
BerdyBSpoeringALLingLLEpsteinSS. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat Protoc. (2017) 12:2232–42. 10.1038/nprot.2017.074
38.
AleklettKKiersETOhlssonPShimizuTSCaldasVEHammerEC. Build your own soil: Exploring microfluidics to create microbial habitat structures. ISME J. (2018) 12:318–9. 10.1038/ismej.2017.184
39.
KennedyACStubbsTL. Management effects on the incidence of jointed goatgrass inhibitory rhizobacteria. Biol Control. (2007) 40:213–21. 10.1016/j.biocontrol.2006.10.006
40.
XiaoWLiJZhangYGuoYFangWValverdeBEet al. A fungal Bipolaris bicolor strain as a potential bioherbicide for goosegrass (Eleusine indica) control. Pest Manag Sci. (2021) 78:1251–64. 10.1002/ps.6742
41.
BoABKimJDKimYSSinHTKimHJKhaitovBet al. Isolation, identification and characterization of Streptomyces metabolites as a potential bioherbicide. PLoS ONE. (2019) 14:e222933. 10.1371/journal.pone.0222933
42.
LazarusBEFerisKGerminoMJ. Weed-suppressive bacteria effects differ in culture compared to in soils and with or without microbial competition and separation of active ingredient. Biol Control. (2021) 152:104422. 10.1016/j.biocontrol.2020.104422
43.
RayPLakshmananVLabbéJLCravenKD. Microbe to microbiome: a paradigm shift in the application of microorganisms for sustainable agriculture. Front Microbiol. (2020) 11:622926. 10.3389/fmicb.2020.622926
44.
BaasPBellCManciniLMLeeMNConantRTWallensteinMD. Phosphorus mobilizing consortium Mammoth PTM enhances plant growth. PeerJ. (2016) 3:e1621v1. 10.7287/peerj.preprints.1621v1
45.
Grand View Research, Inc. Bioherbicides Market Size Worth $4.14 Billion By 2024. (2018). Available online at: https://www.grandviewresearch.com/press-release/global-bioherbicides-market (accessed December 19, 2021).
46.
MastelingRLombardLde BoerWRaaijmakersJMDini-AndreoteF. Harnessing the microbiome to control plant parasitic weeds. Curr Opin Microbiol. (2019) 49:26–33. 10.1016/j.mib.2019.09.006
47.
HardingDPRaizadaMN. Controlling weeds with fungi, bacteria and viruses: a review. Front Plant Sci. (2015) 6:659. 10.3389/fpls.2015.00659
48.
van BergeijkDATerlouwBRMedemaMHvan WezelGP. Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol. (2020) 18:546–88. 10.1038/s41579-020-0379-y
49.
DukeSOAbbasHK. Natural products with potential use as herbicides. In: K Inderjit, MM Dakshini, FA Einhellig, editors, Allelopathy. Washington, DC: ACS Publications (1994). p. 348–62. 10.1021/bk-1995-0582.ch025
50.
KhalifaSAMEliasNFaragMAChenLSaeedAHegazyMEFet al. Marine natural products: a source of novel anticancer drugs. Mar Drugs. (2019) 17:491. 10.3390/md17090491
51.
MasiMFredaFSangermanoFCalabròVCimminoACristofaroMet al. Radicinin, a fungal phytotoxin as a target-specific bioherbicide for invasive buffelgrass (Cenchrus ciliaris) control. Molecules. (2019) 24:1086. 10.3390/molecules24061086
52.
BlinKShawSKloostermanAMCharlop-PowersZvan WezelGPMedemaMHet al. AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucl Acids Res. (2021) 49:gkab335. 10.1093/nar/gkab335
53.
SkinniderMAMerwinNJJohnstonCWMagarveyNA. PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucl Acids Res. (2017) 45:gkx320. 10.1093/nar/gkx320
54.
CulpEJWaglechnerNWangWFiebig-ComynAAHsuYPKotevaKet al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature. (2020) 578:582–7. 10.1038/s41586-020-1990-9
55.
Navarro-MuñozJCSelem-MojicaNMullowneyMWKautsarSATryonJHParkinsonEIet al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. (2020) 16:60–8. 10.1038/s41589-019-0400-9
56.
KautsarSABlinKShawSNavarro-MuñozJCTerlouwBRvan der HooftJJJet al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucl Acids Res. (2020) 48:gkz882. 10.1093/nar/gkz882
57.
StokesJMYangKSwansonKJinWCubillos-RuizADonghiaNMet al. A deep learning approach to antibiotic discovery. Cell. (2020) 180:688–702. 10.1016/j.cell.2020.01.021
58.
BenhadjMGacemi-KiraneDMenasriaTGueblaKAhmaneZ. Screening of rare actinomycetes isolated from natural wetland ecosystem (Fetzara Lake, northeastern Algeria) for hydrolytic enzymes and antimicrobial activities. J King Saud Univ Sci. (2019) 31:706–12. 10.1016/j.jksus.2018.03.008
59.
SubramaniRSipkemaD. Marine rare actinomycetes: a promising source of structurally diverse and unique novel natural products. Mar Drugs. (2019) 17:249. 10.3390/md17050249
60.
ZhangJJTangXMooreBS. Genetic platforms for heterologous expression of microbial natural products. Nat Prod Rep. (2019) 36:1313–32. 10.1039/C9NP00025A
61.
XuMWangYZhaoZGaoGHuangSXKangQet al. Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl Environ Microbiol. (2016) 82:5795–805. 10.1128/AEM.01383-16
62.
BradySFClardyJ. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J Am Chem Soc. (2007) 122:12903–4. 10.1128/AEM.66.6.2541-2547.2000
63.
CarverSMNikulinNKao-KniffinJ. Uncovering plant growth-mediating allelochemicals produced by soil microorganisms. Weed Sci. (2016) 64:119–28. 10.1614/WS-D-15-00095.1
64.
HashimotoTHashimotoJKozoneIAmagaiKKawaharaTTakahashiSet al. Biosynthesis of Quinolidomicin, the largest known macrolide of terrestrial origin: Identification and heterologous expression of a biosynthetic gene cluster over 200 kb. Org Lett. (2018) 20:7996–9. 10.1021/acs.orglett.8b03570
65.
HengETanLLZhangMMWongFT. CRISPR-Cas strategies for natural product discovery and engineering in actinomycetes. Process Biochem. (2021) 102:261–8. 10.1016/j.procbio.2021.01.007
66.
SugimotoYCamachoFRWangSChankhamjonPOdabasABiswasAet al. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science. (2019) 366:eaax9176. 10.1126/science.aax9176
67.
SmanskiMJBhatiaSZhaoDParkYJWoodruffLBAGiannoukosGet al. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol. (2014) 32:1241–9. 10.1038/nbt.3063
68.
TraxlerMFWatrousJDAlexandrovTDorresteinPCKolterR. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. (2013) 4:e00459–13. 10.1128/mBio.00459-13
69.
TraxlerMFKolterR. Natural products in soil microbe interactions and evolution. Nat Prod Rep. (2015) 32:956. 10.1039/C5NP00013K
70.
CarriónVJPerez-JaramilloJCordovezVTracannaVde HollanderMRuiz-BuckDet al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. (2019) 366:606–12. 10.1126/science.aaw9285
71.
HiardSMaréeRColsonSHoskissonPATitgemeyerFvan WezelGPet al. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun. (2007) 357:861–4. 10.1016/j.bbrc.2007.03.180
72.
DukeSOStidhamMADayanFE. A novel genomic approach to herbicide and herbicide mode of action discovery. Pest Manag Sci. (2019) 75:314–7. 10.1002/ps.5228
73.
BenderSFWaggCvan der HeijdenMGA. An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol. (2016) 31:440–52. 10.1016/j.tree.2016.02.016
74.
KennedyAC. Soil microorganisms for weed management. J Crop Prod. (1999) 2:123–38. 10.1300/J144v02n01_07
75.
KremerRJLiJ. Developing weed-suppressive soils through improved soil quality management. Soil Tillage Res. (2003) 72:193–202. 10.1016/S0167-1987(03)00088-6
76.
LiJKremerRJ. Rhizobacteria associated with weed seedlings in different cropping systems. Weed Sci. (2000) 48:731–41. 10.1614/0043-1745(2000)0480734:RAWWSI2.0.CO;2
77.
CarbonettoBRascovanNÁlvarezRMentaberryAVázquezMP. Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in Argentine Pampas. PLoS ONE. (2014) 9:e99949. 10.1371/journal.pone.0099949
78.
HartmannMFreyBMayerJMäderPWidmerF. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. (2015) 9:1177–94. 10.1038/ismej.2014.210
79.
HartmanKvan der HeijdenMGAWittwerRABanerjeeSWalserJCSchlaeppiK. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome. (2018) 6:14. 10.1186/s40168-017-0389-9
80.
PerryLGGalatowitschSMRosenCJ. Competitive control of invasive vegetation: a native wetland sedge suppresses Phalaris arundinacea in carbon-enriched soil. J Appl Ecol. (2004) 41:151–62. 10.1111/j.1365-2664.2004.00871.x
81.
EschenRMortimerSRLawsonCSEdwardsARBrookAJIgualJMet al. Carbon addition alters vegetation composition on ex-arable fields. J Appl Ecol. (2007) 44:95–104. 10.1111/j.1365-2664.2006.01240.x
82.
ColeRJSoperFMLittonCMKnaufAESparksKGerowKGet al. Restoration benefits of soil nutrient manipulation and weeding in invaded dry and wet tropical ecosystems in Hawai?i. Restorat Ecol. (2021) 29:1464. 10.1111/rec.13390
83.
SinsabaughRLManzoniSMoorheadDLRichterA. Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol Lett. (2013) 16:930–9. 10.1111/ele.12113
84.
PaschkeMWMcLendonTRedenteEF. Nitrogen availability and old-field succession in a shortgrass steppe. Ecosystems. (2000) 3:144–58. 10.1007/s100210000016
85.
BlumenthalDMJordanNRRusselleMP. Soil carbon addition controls weeds and facilitates prairie restoration. Ecol Appl. (2003) 13:605–15. 10.1890/1051-0761(2003)0130605:SCACWA2.0.CO;2
86.
McLendonTRedenteEF. Effects of nitrogen limitation on species replacement dynamics during early secondary succession on a semiarid sagebrush site. Oecologia. (1992) 91:312–7. 10.1007/BF00317618
87.
Reever MorghanKJSeastedtTR. Effects of soil nitrogen reduction on nonnative plants in restored grasslands. Restorat Ecol. (1999) 7:51–5. 10.1046/j.1526-100X.1999.07106.x
88.
BaerSGBlairJMCollinsSLKnappAK. Plant community responses to resource availability and heterogeneity during restoration. Oecologia. (2004) 139:617–29. 10.1007/s00442-004-1541-3
89.
UddinMNRobinsonRWAsaedaT. Nitrogen immobilization may reduce invasibility of nutrient enriched plant community invaded by Phragmites australis. Sci Rep. (2020) 10:1601. 10.1038/s41598-020-58523-4
90.
KnaufAELittonCMColeRJSparksJPGiardinaCPGerowKGet al. Nutrient-use strategy and not competition determines native and invasive species response to changes in soil nutrient availability. Restorat Ecol. (2021) 29:e13374. 10.1111/rec.13374
91.
BeverJDDickieIAFacelliEFacelliJMKlironomosJMooraMet al. Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol. (2010) 25:468–78. 10.1016/j.tree.2010.05.004
92.
MillsKEBeverJD. Maintenance of diversity within plant communities: soil pathogens as agents of negative feedback. Ecology. (1998) 79:1595–601. 10.1890/0012-9658(1998)0791595:MODWPC2.0.CO;2
93.
KlironomosJN. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature. (2002) 417:67–70. 10.1038/417067a
94.
ManganSASchnitzerSAHerreEAMackKMLValenciaMCSanchezEIet al. Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature. (2010) 466:752–5. 10.1038/nature09273
95.
BennettJMaheraliHReinhartKLekbergYHartMKlironomosJ. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science. (2017) 355:181–4. 10.1126/science.aai8212
96.
van der PuttenWHBradfordMAPernilla BrinkmanEvan de VoordeTFJVeenGFBaileyJK. Where, when and how plant-soil feedback matters in a changing world. Funct Ecol. (2016) 30:1062–71. 10.1111/1365-2435.12657
97.
ReinhartKO. The organization of plant communities: negative plant-soil feedbacks and semiarid grasslands. Ecology. (2012) 93:2377–85. 10.1890/12-0486.1
98.
MaronJLSmithALOrtegaYKPearsonDECallawayRM. Negative plant-soil feedbacks increase with plant abundance, and are unchanged by competition. Ecology. (2016) 97:2055–63. 10.1002/ecy.1431
99.
CortoisRSchröder-GeorgiTWeigeltAvan der PuttenWHde DeynGBvan der HeijdenM. Plant-soil feedbacks: role of plant functional group and plant traits. J Ecol. (2016) 104:1608–17. 10.1111/1365-2745.12643
100.
TesteFPKardolPTurnerBLWardleDAZemunikGRentonMet al. Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science. (2017) 176:173–6. 10.1126/science.aai8291
101.
PattisonZRumbleHTannerRAJinLGangeAC. Positive plant-soil feedbacks of the invasive Impatiens glandulifera and their effects on above-ground microbial communities. Weed Res. (2016) 56:198–207. 10.1111/wre.12200
102.
KardolPMartijn BezemerTVan Der PuttenWH. Temporal variation in plant-soil feedback controls succession. Ecol Lett. (2006) 9:1080–8. 10.1111/j.1461-0248.2006.00953.x
103.
CallawayRMThelenGCRodriguezAHolbenWE. Soil biota and exotic plant invasion. Nature. (2004) 427:731–3. 10.1038/nature02322
104.
Van der PuttenWHKlironomosJNWardleDA. Microbial ecology of biological invasions. ISME J. (2007) 1:28–37. 10.1038/ismej.2007.9
105.
YangQCarrilloJJinHShangLHovickSMNijjerSet al. Plant-soil biota interactions of an invasive species in its native and introduced ranges: Implications for invasion success. Soil Biol Biochem. (2013) 65:78–85. 10.1016/j.soilbio.2013.05.004
106.
GundaleMJKardolPNilssonMCNilssonULucasRWWardleDA. Interactions with soil biota shift from negative to positive when a tree species is moved outside its native range. New Phytol. (2014) 202:415–21. 10.1111/nph.12699
107.
MacKayJKotanenPM. Local escape of an invasive plant common ragweed (Ambrosia artemisiifolia L), from above-ground and below-ground enemies in its native area. J Ecol. (2008) 96:1152–61. 10.1111/j.1365-2745.2008.01426.x
108.
EppingaMBRietkerkMDekkerSCde RuiterPCvan der PuttenWH. Accumulation of local pathogens: a new hypothesis to explain exotic plant invasions. Oikos. (2006) 114:168–76. 10.1111/j.2006.0030-1299.14625.x
109.
RatiDAPTinsonKRSLironomosJOHNK. Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology. (2008) 89:1043–55. 10.1890/07-0370.1
Summary
Keywords
bioherbicide, herbicide resistance, invasive, microbiome, natural product, weeds
Citation
Cheng L, DiTommaso A and Kao-Kniffin J (2022) Opportunities for Microbiome Suppression of Weeds Using Regenerative Agricultural Technologies. Front. Soil Sci. 2:838595. doi: 10.3389/fsoil.2022.838595
Received
18 December 2021
Accepted
15 February 2022
Published
18 March 2022
Volume
2 - 2022
Edited by
Patricia Dorr De Quadros, University of Waterloo, Canada
Reviewed by
Muneer Ahmad Malla, Dr. Hari Singh Gour University, India; Garima Singh, Pachhunga University College, India
Updates
Copyright
© 2022 Cheng, DiTommaso and Kao-Kniffin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenny Kao-Kniffin jtk57@cornell.edu
This article was submitted to Soil Biology, Ecosystems and Biodiversity, a section of the journal Frontiers in Soil Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.