- 1Medical Oncology Unit, Santa Chiara Hospital, Trento, Italy
- 2Digestive Molecular Clinical Oncology Research Unit, Università degli Studi di Verona, Verona, Italy
- 3Investigational Cancer Therapeutics Clinical Unit, Azienda Ospedaliera Universitaria Integrata, Verona, Italy
- 4Oncology Unit, A.R.N.A.S. Civico, Palermo, Italy
The mainstay treatment for patients with immediate resectable pancreatic cancer remains upfront surgery, which represents the only potentially curative strategy. Nevertheless, the majority of patients surgically resected for pancreatic cancer experiences disease relapse, even when a combination adjuvant therapy is offered. Therefore, aiming at improving disease free survival and overall survival of these patients, there is an increasing interest in evaluating the activity and efficacy of neoadjuvant and perioperative treatments. In this view, it is of utmost importance to find biomarkers able to select patients who may benefit from a preoperative therapy rather than upfront surgical resection. Defined genomic alterations and a dynamic inflammatory microenvironment are the major culprits for disease recurrence and resistance to chemotherapeutic treatments in pancreatic cancer patients. Signal transduction pathways or tumor immune microenvironment could predict early recurrence and response to chemotherapy. In the last decade, distinct molecular subtypes of pancreatic cancer have been described, laying the bases to a tailored therapeutic approach, started firstly in the treatment of advanced disease. Patients with homologous repair deficiency, in particular with mutant germline BRCA genes, represent the first subgroup demonstrating to benefit from specific therapies. A fraction of patients with pancreatic cancer could take advantage of genome sequencing with the aim of identifying possible targetable mutations. These genomic driven strategies could be even more relevant in a potentially curative setting. In this review, we outline putative predictive markers that could help in the next future in tailoring the best therapeutic strategy for pancreatic cancer patients with a potentially curable disease.
Introduction
Pancreatic adenocarcinoma has a dismal prognosis accounting for a 5-year overall survival (OS) rate lower than 10%. This proportion could exceed 30% when considering localized disease and surgical resection represents the only hope for cure. However, only a small proportion of patients has a resectable disease at diagnosis and local or distance relapse occurs after surgery in most of cases (1, 2).
Adjuvant therapy could prolong median disease-free survival (DFS) and OS after resection, reaching 21.6 and 54.4 months, respectively, with the most active treatment (3). Anyway, <50% of patients are disease free even after an adjuvant triplet regimen.
One of the main reasons of this aggressiveness is that even localized pancreatic cancer could be considered a systemic disease ab initio, since metastatic subclones exist before the clinical evidence of disease (4). Furthermore, circulating tumor cells could be isolated in the bloodstream of patients with pancreatic cancer, also in early stage, and their levels have been correlated with the probability of survival (5–7).
Growing evidence supports an anticipation of postoperative treatments to the neoadjuvant or perioperative setting. In fact, neoadjuvant therapy enables an early treatment of the micrometastatic disease (8). Furthermore, it improves R0 resection rate and decreases lymph node positivity rate, which translates in a positive prognostic impact (9, 10). Preoperative treatment has been also proposed as a tool for measuring in vivo tumor response. Moreover, after pancreatic surgery a significant fraction of patients is not fit to receive a postoperative chemotherapy due to surgical complications, poor performance status or early disease progression (3, 11, 12). Furthermore, more than 20% of patients initiating adjuvant therapy fail to complete the preplanned cycles (3). Another acknowledged advantage of preoperative therapy is the opportunity to select patients with rapid progression who would not have benefit from surgery (13).
In the last decade oncological approach has been revolutionized by a growing personalization of the treatment, even if pancreatic cancer has been characterized by slower progress. To date, the decisional algorithm in localized disease is mainly based on clinical patient features and anatomical surgical criteria (14). No predictive or prognostic markers have been recognized for driving therapeutic approach in early-stage pancreatic cancer. However, a tailored strategy for selecting patients for upfront surgery vs. preoperative therapy, and possibly which type of therapy, is of utmost importance particularly in a potentially curable setting (Figure 1).
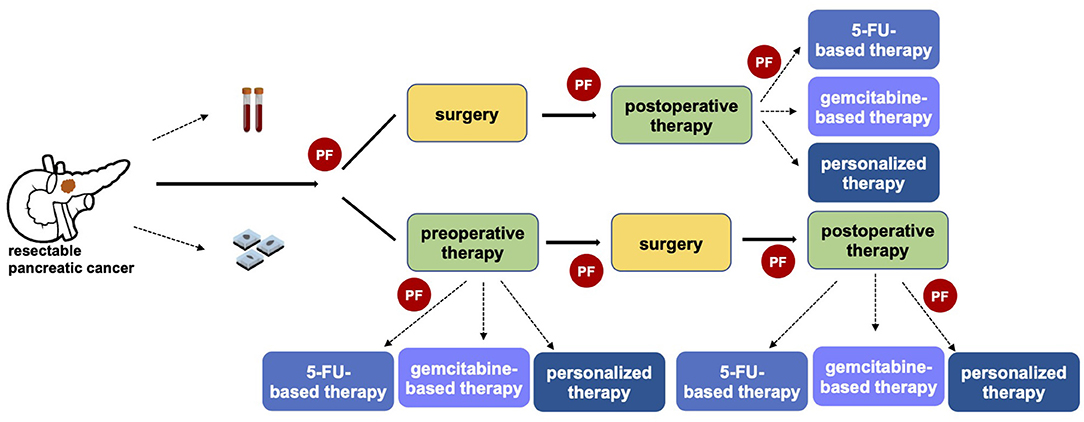
Figure 1. The importance of predictive factors in defining a personalized clinical approach in resectable pancreatic cancer. The identification of predictive factors (PF) has a central role in driving the therapeutic strategy, both in referring patients toward upfront surgery or preoperative therapy and in the choice of the optimal systemic pre- and post-operative therapy.
We performed a research on Pubmed/Medline, Cochrane library and Scopus using the keywords “predictive factors pancreatic cancer” OR “predictive pancreatic cancer” OR “predictive resected pancreatic cancer” OR “predictive resectable pancreatic cancer”. We selected the most relevant and pertinent studies, both in the preclinical and clinical setting, considering the quality of the studies and the relevance to the topic of this review. For ongoing clinical trials, we searched in the clinicatrials.gov database for recruiting and active phase II/III trials focused on resectable or resected pancreatic cancer.
Adjuvant and Neoadjuvant Treatment in Resectable Pancreatic Cancer
International guidelines recommend a preoperative treatment in borderline resectable and locally advanced disease (15, 16). Whether preoperative therapy had to be offered in resectable pancreatic cancer patients remains instead matter of debate and results in this setting are controversial. The main current standard therapeutic approach of resectable disease is upfront surgery followed by adjuvant chemotherapy.
The first trial proving a significant survival increase using adjuvant therapy was the ESPAC-1 trial, in which patients with resected pancreatic cancer received adjuvant fluorouracil or only observation (17). The phase III ESPAC-3 trial did not show a statistically significant difference between adjuvant fluorouracil plus folinic acid compared with gemcitabine after resection of pancreatic ductal adenocarcinoma (18). The combination of gemcitabine and capecitabine showed longer OS compared with gemcitabine in the phase III ESPAC-4, though lack of a postoperative imaging restaging and of CA19.9 level limits for enrollment were valuable points of weakness (19). More recently, in the phase III multicenter PRODIGE 24/CCTG PA trial the modified FOLFIRINOX regimen (comprised of oxaliplatin, irinotecan, leucovorin, and fluorouracil) prolonged survival compared to gemcitabine in patients with pancreatic cancer (3). However, even with this most active regimen, more than fifty percent of patients relapse after 2 years.
The actual trend among pancreatic cancer experts in the treatment of resectable disease is to move toward neoadjuvant or perioperative treatment.
Retrospective series have firstly proposed a survival benefit of neoadjuvant therapy, especially in patients who are not fit after upfront surgery to receive adjuvant therapy (20).
The randomized phase III PREOPANC trial investigated upfront surgery with adjuvant gemcitabine compared with perioperative gemcitabine combined with preoperative radiation in patients with resectable and borderline resectable pancreatic cancer (21). Preoperative chemoradiotherapy has improved OS, together with DFS and R0 resection rate.
SWOG S1505 is a phase II non-comparative trial randomizing patients with resectable pancreatic cancer to perioperative FOLFIRINOX or gemcitabine plus nab-paclitaxel (22). Neither of the two arms met the preplanned 2-year overall survival based on historical data from adjuvant trials. Similarly, the recent phase II NEONAX trial did not meet its primary endpoint in 18-month DFS rate neither in the perioperative arm nor in the adjuvant arm with gemcitabine plus nab-paclitaxel.
Several studies investigating preoperative or perioperative therapeutic strategies are ongoing (Table 1), including the PREOPANC-2 trial (23), the nITRO trial (24), the NorPACT-1 (25), the NEPAFOX trial (26), and the PANACHE01-PRODIGE48 trial (27).
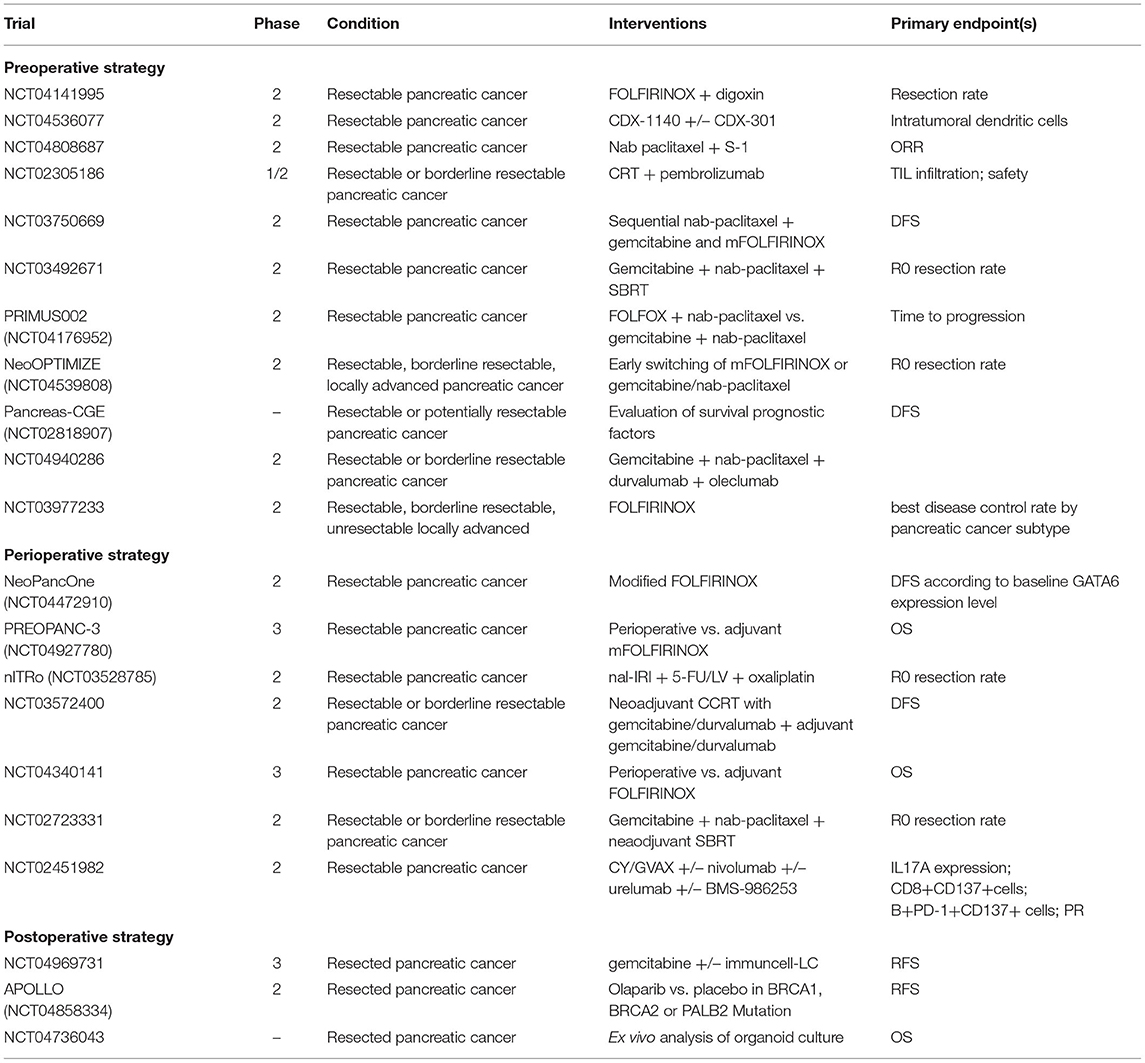
Table 1. Ongoing phase II/III trials investigating systemic treatment strategies in resectable pancreatic cancer.
Molecular Mechanisms Involved in Pancreatic Cancer Resistance to Chemotherapeutic Agents
The most promising biomarkers for supporting the benefit of a preoperative chemotherapeutic strategies in resectable PC patients emerge directly from most relevant molecular mechanisms for the intrinsic chemoresistance of pancreatic cancer cells. The resistance of solid tumors to the cytotoxic effect of cancer chemotherapy is generally accountable to the activation in key pathways involved in the regulation of cell-cycle, and, most importantly, in the suppression of programmed cell death, or apoptosis, induced by these DNA damage agents (28).
Different autocrine or paracrine pro-inflammatory factors lead to the activation of transcription factors involved in apoptosis control. Nuclear Factor κB (NF-κB) and AP-1 are among the most relevant of these transcription factors by representing a key mechanistic link between inflammation and cancer chemoresistance (29, 30). In the last decade, we contributed to demonstrate the role of the serine/threonine kinase Transforming Growth Factor-β (TGF-β)-activated kinase 1 (TAK1, also called MAP3K7) as a major determinant in the integration of different pro-inflammatory signals relevant for chemoresistance—including IL-1 (31, 32) and TGF-β (33)—to regulate, in turn, different transcription factors, including NF-κB, AP-1 (34), and YAP/TAZ (35). In this regard, TAK1 has been demonstrated as a major determinant of the resistance of cancer cells to the proapoptotic activity of chemotherapeutic agents in a number of preclinical models of solid tumors [reviewed in (36)].
More recently, with the aim of identifying circulating markers of TAK-1 pathway activation that could potentially serve as resistance biomarkers for the nanoliposomal irinotecan (nal-IRI) in patients with gemcitabine-resistant advanced pancreatic cancer, we identified CXCL8, the gene coding for IL-8, as the most significant gene regulated by TAK1 expression among those coding for secreted proteins. Consistently, circulating IL-8 was the most significant predictive marker of survival in metastatic pancreatic cancer patients treated with nal-IRI in a large panel of different cytokines, chemokines and growth factors (37). Collectively this evidence delineates a model in which autocrine or paracrine proinflammatory signaling sustain the activation of TAK-1/NF-κB cascade, and IL-8 appears to be the most significantly regulated factor by this intracellular pathway and one of the most significant candidates for selecting those patients with resectable pancreatic cancer more likely to benefit from preoperative chemotherapeutic regimens.
Transcriptomic Classification Defines Different Molecular Subtypes of Pancreatic Cancer
Molecular subtyping could lead to many advantages, including better prognostic definition and optimizing therapeutic patient management. Gene expression profiling, mainly performed in resected tumors, outlined different subgroups, which have not yet practical applications in clinical routine. The three subtypes described by Collisson include classical, quasimenchymal and exocrine-like type (38). Bailey and colleagues identified four subtypes (immunogenic, progenitor, ADEX and squamous) (39). A two-subdivision was proposed by Moffitt, distinguishing classical and basal-like subtype (40). Although they do not overlap, the quasimenchymal (Collisson), squamous (Bailey) and basal-like subtype (Moffitt) share similar features and all three have been associated with a poor prognosis. Furthermore, these have been associated with mutations in genes involved in chromatin modification, including DNA methylation and acetylation (e.g., methylation of HNF4A and GATA6 genes). The immunogenic subtype is defined by a stromal immune infiltrate and decreased tumor cellularity).
Collisson examined untreated primary resected pancreatic cancer tumors. Quasimesenchymal subtype was characterized by high tumor grade and poor survival. The classical subtype was KRAS dependent and was correlated with GATA6 expression.
GATA6 is a transcription factor involved in the normal pancreatic development (41). High GATA6 expression has been showed to correlate with the classical phenotype, also according to Moffitt classification; viceversa, the basal-like subtype showed low GATA6 RNA expression (42). Consistent with this finding, GATA6 expression has demonstrated a prognostic value.
The negative prognostic effect of low GATA6 expression has been postulated also in resected pancreatic cancer patients (42). Furthermore, it has also been proposed that patients with basal-like GATA6low tumors do not benefit from 5-fluorouracil. In an analysis of the ESPAC-3 adjuvant trial, a low GATA6 expression was correlated with lower survival in patients treated with 5-fluorouracil, while it was not associated with response to gemcitabine (43). Another group hypothesized resistance to FOLFIRINOX of the basal-like subtype (42).
The paucicellularity of pancreatic cancer makes challenging an in-depth molecular characterization. The use of patient–derived organoids has provided additional data, recapitulating the mutational landscape and transcriptional subtypes of pancreatic cancer and delineating chemotherapy signatures (44). Furthermore, they have been demonstrated to be able to predict treatment response. Interestingly, the basal-like cohort subgroup has been found to have most likely an oxaliplatin-resistant signature. Moreover, about one third of the pancreatic cancer patient–derived organoids showed no sensitivity to any of the chemotherapeutic drugs tested (gemcitabine, paclitaxel, SN-38, 5-fluorouracil, oxaliplatin). However, about half of these were sensitive to at least one of the targeted agents used.
Noteworthy, the basal-like subgroup is characterized by a hypoxia-associated gene signature, higher PD-L1 and PD-1 expression and enrichment of a T-cell-inflamed signature (42, 45).
Role of MicroRNAs as Prognostic and Predictive Factors in Early-Stage Pancreatic Cancer
MicroRNAs (miRNAs) regulate post-transcriptional gene expression affecting physiological and pathological processes (46). Hundreds of messenger RNAs (mRNAs) may be targeted by a single miRNA (47). miRNAs are classified as oncogenic or tumor suppressor, depending on the type of activity on oncogene or tumor suppressor genes (48). A prognostic value of miRNAs has been proposed since they are involved in cell survival, proliferation, invasion and metastasis (49).
Expression of miR-574-5p, miR-1244, miR-145-star, miR-328, miR-26b-star, and miR-4321 has been associated with OS and DFS in 104 advanced pancreatic cancer patients (50).
Upregulation of miR-155, miR-196a-2, miR-203, miR-210, miR-219, and miR-222 and downregulation of miR-217 have been correlated with poor prognosis and overall survival (51). MiR-155, miR-203, and miR-222 have been related to be involved in the angiogenesis pathway (51). Furthermore, miR-155 has been linked with gemcitabine resistance in pancreatic cancer by controlling exosome synthesis (52). MiR-222, miR-203, and miR-155 are also involved in cell cycle signaling pathway (51). MiR-222 targets the cell cycle inhibitors p27 and p57, whereas miR-203 and miR-155 target p53. The tumor suppressor function of miR-217 is performed by directly targeting KRAS, since miR-217 overexpression has been correlated with reduced KRAS levels (53).
High expression of miR-200c, miR-142-5p, and miR-204 has been reported to be associated with longer survival after surgical resection of pancreatic cancer (54). Indeed, up-regulation of miR-200 has been demonstrated to reverse epithelial-to-mesenchymal transition (EMT) in gemcitabine resistant pancreatic cancer cells (55). miRNA-200 family, including 200a/200b/200c/141, and miRNA-205 regulate EMT by targeting ZEB1 and ZEB2 (SIP1), that are E-cadherin suppressing factors (56).
miR-21 is an oncogenic miRNA and regulates several oncosuppressors such as CDKN1A, PTEN, PDCD4 (57–59). miR-21 expression can modulate apoptosis, Akt phosphorylation and expression of genes involved in invasive behavior, contributing to gemcitabine resistance (60). High plasma and tissue miR-21-5p levels have been associated with worse survival in patients with resectable pancreatic cancer (60, 61). In a series of 25 resectable pancreatic cancer patients, higher preoperative levels of miR-375-3p and miR-21-5p were significantly correlated with worse OS and plasma miR-21-5p concentration was independent from other clinicopathological factors (62). Low miR-21 expression has been associated with benefit from adjuvant therapy in two different cohorts of patients with pancreatic cancer and anti-miR-21 has shown to enhance anticancer drug activity in vitro (63). miR-21-5p inhibits the tumor suppressor PTEN, activating the PI3K/AKT/mTOR signaling pathway (60). In fact, elevated miR-21-5p expression has been correlated with a decreased antitumor effect of gemcitabine and 5-fluorouracil (64, 65). Moreover, miR-21-5p inhibition seems to increase sensitivity to gemcitabine (66).
High presurgical levels of miR-365a-3p, which inhibits NF-κB function inducing apoptosis, and of miR-99a-5p, which regulates mTOR, are predictors of longer survival in resected pancreatic cancer patients (67). miR-221-3p has been proposed as a marker of recurrence after resection of pancreatic cancer (68). High serum and tissue levels of miR-196a-5p, which is involved in cancer proliferation and invasiveness, have been associated with inferior median OS in patients with early-stage pancreatic cancer (69). Furthermore, patients with unresectable pancreatic cancer showed higher serum miR-196a-5p levels compared to those with resectable cancer (70). Levels of plasmatic miR-182-5p demonstrated to be negative predictors of DFS and OS in pancreatic cancer (71).
Moreover, miRNA levels could help in predicting treatment response or resistance (72). Since miRNAs could affect cell cycle, drug efflux and apoptosis, they have been proposed to be involved in chemoresistance by regulating ATP-binding cassette (ABC) membrane transporters as well as exploiting intracellular effects (73).
MiR-142-5p and miR-320c were proposed as positive predictive factors of tumor response to gemcitabine (54, 74). Similarly, downregulation of mi-R-33a, mi-R-200b, mi-R-200c, mi-R-205, let-7b, let-7c, let-7d, and let-7e has been associated with gemcitabine resistance (75). Let-7 is an oncosuppressor and reduced expression of Let-7 have been correlated with cancer progression. mRNAs target of Let-7 include KRAS, HRAS, NF2, HMGA2 and LIN28 (76). The restoration of Let-7 levels in pancreatic cancer cell lines has shown to inhibit cell proliferation (77).
Furthermore, gemcitabine-resistant pancreatic cancer cells were resensitized to gemcitabine showing decreased expression of caveolin-1 and Ki-67.
A tumor suppressing role of miR-7-5p has been proposed, as its serum levels were found decreased in patients with stage III or IV pancreatic cancer compared to normal controls. Furthermore, miR-7 was shown to be significantly lower expressed in gemcitabine-resistant pancreatic cancer patients (78).
Chemoresistant pancreatic cancer cells showed P-glycoprotein overexpression associated with miR-181a-5p and miR-218-5p dysregulation (79). Plasmatic miR-181a-5p decline has been correlated with FOLFIRINOX response in pancreatic cancer patients (80). Overexpression of miR-192-5p and miR-215-5p has shown to decrease cancer cell proliferation, affecting the S-phase of cell cycle and negatively influencing response to drugs such as 5-fluorouracil (81).
In conclusion, miRNAs could play a role in guiding the best therapeutic strategy approach in early-stage pancreatic cancer patients. The tissue- and disease-specific expression and the stability in body fluids of miRNA represent their main advantages. Furthermore, they represent a potential therapeutic target based on evidence coming from in vitro and in vivo studies (82, 83).
Molecular Profiling in Personalizing Therapeutic Strategy in Pancreatic Cancer
The molecular characterization in pancreatic cancer is not yet standard in clinical care. However, growing evidence is highlighting its relevance in the advanced setting. In a series of 71 patients whose tumor biopsies underwent whole-exome and RNA sequencing, 48% were shown to have relevant genomic alterations and 18% pathogenic or likely pathogenic germline alterations, leading to a change in clinical management in 30% of enrolled patients as a result of genomic results (84). A large US registry study showed a longer median OS in patients with actionable molecular alterations, that were about a quarter of patients screened, who received a matched therapy (85).
A recent study performing next generation sequencing (NGS) on resected pancreatic cancer specimens found likely pathogenic/pathogenic variants in 94% of samples, 18% of which were potentially actionable (86).
We recently used FoundationOne CDx or Liquid, a next-generation DNA sequencing (NGS) service to identify genomic alterations in 68 patients affected by pancreaticoduodenal cancer patients who failed standard treatments. According to ESMO Scale of Clinical Actionability for molecular Targets (ESCAT), at least one alteration ranking tier I, II, III, or IV according to ESCAT classification was detected in 8, 1, 9, and 12 patients, respectively (44.1%). Ten of them (33.3%) received a matched therapy. Patients with ESCAT tier I to IV were generally younger than the overall population (median = 54, range = 26–71 years), had an EGOG performance status score = 0 (83.3%), and an uncommon histological or clinical presentation. The most common mutations with clinical evidence of actionability (ESCAT tier I-III) involved genes of the RAF (10.3%), BRCA (5.9%) or FGFR pathways (5.9%). These results indicated that in advanced pancreaticoduodenal cancer, NGS is a feasible and valuable method for enabling precision oncology. However, this genomic profiling method might be considered only after standard treatments failure, and especially in young patients maintaining a good performance status, in order to detect potentially actionable mutations and offer molecularly targeted therapeutic approaches (87).
A clinical subgroup of patients with pancreatic cancer is identified by homologous recombination deficiency (HRD), which is caused by defects in DNA damage response (DDR) genes. The main genes that have been proposed to be involved in homologous recombination repair are BRCA1/2, PALB2, ATR, ATM, CHEK1,2, RAD51, and FANC (88). Inactivating mutations or epigenetic silencing of these genes result in HRD. The exact prevalence of HRD in pancreatic cancer has yet to be defined, partly due to the lack of a univocal definition and the variability of assays used.
If pathogenetic germline alterations have a prognostic impact remains matter of debate and available data in all stages of pancreatic cancer are conflicting (89). However, studies focusing on patients with germline mutations in DDR genes, including ATM, BRCA1/2, CDKN2A, CHEK2, ERCC4, and PALB2, have shown an improved OS (90).
Besides implications on prognosis and in terms of risk assessment and prevention, the identification of these patients has potential therapeutic repercussions (91). HRD has been identified to be a positive predictive factor of response to platinum-based therapy in patients with advanced pancreatic cancer (92–95). Furthermore, it has been speculated that the objective responsive rate of chemotherapeutic drugs containing platinum salts could reflect this subpopulation characterized by inactivation of DNA maintenance genes (96, 97). Further investigation is needed to confirm whether HRD could be a predictive factor of response to platinum therapy in early stage, since major evidence comes from advanced pancreatic cancer. However, two retrospective studies showed increased pathological complete response rates and longer OS in resectable and borderline resectable pancreatic cancer patients treated with platinum-based therapies (98, 99). Ongoing studies are investigating different therapeutic regimens in resectable and borderline resectable pancreatic cancer with HRD signature (100).
Germline BRCA mutation represents the first prospectively validated predictive factor in advanced pancreatic cancer (101). In the POLO trial the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib showed to prolong PFS compared to placebo in patients whose disease had not progressed on first-line platinum-based chemotherapy [hazard ratio (HR) of 0.53, p = 0.004], even if it lacked to improve OS (102). Various ongoing trials are investigating the efficacy of adding PARP inhibitors in the preoperative or locally advanced setting, even in combination with radiotherapy, on the basis of a possible synergy from preclinical studies (NCT04005690) (103, 104).
Even if they represent a small proportion, mismatch repair-deficient (dMMR) tumors might benefit from immunotherapy (105, 106). The minor fraction of NTRK fusions could also represent a target for the agnostic drugs TRK inhibitors (107).
Different platforms are being used with the aim of improving precision medicine in pancreatic cancer (e.g., PRECISION-Panc in the UK, Precision Promise in the USA, EPPIC in Canada). The phase II PIONEER-Panc study is investigating novel therapeutic strategies in early-stage pancreatic cancer (NCT04481204).
Proteins involved in metabolism and mechanisms of action of chemotherapeutic drugs have been also proposed as predictive markers. Low thymidylate synthase (TYMS), excision repair cross-complementing (ERCC1) and ribonucleotide reductase M1 (RMM1) levels have been correlated with increased efficacy to fluoropirimidines, cisplatin and gemcitabine, respectively (108, 109). High levels of secreted protein acid and rich in cysteine (SPARC) have been associated with nab-paclitaxel response (110). Low expression of human equilibrative nucleoside transported 1 (hENT1) has been proposed to be involved in gemcitabine resistance (111).
Interestingly, a prospective, phase II trial enrolling patients with resectable and borderline resectable pancreatic cancer and selecting neoadjuvant therapy (fluoropyrimidine-based or gemcitabine-based) based on molecular profiling reported high resection rates (112).
Immune Signature in Predicting Treatment Response
For several years it has been observed an increased infiltration of CD8+ and CD4+ T cells together with a reduction of FOXP3+ T regs after neoadjuvant therapy in pancreatic cancer patients (113, 114).
A recent study has analyzed immune blood cells in patients with pancreatic cancer after neoadjuvant FOLFIRINOX (115). Response to FOLFIRINOX was associated with increased CD8 T cell levels, in particular CD27−Tbet+ effector/effector memory subsets, while FOLFIRINOX non-responders showed higher GATA3, CCR4 and ICOS expression in CD8 T cells. FOLFIRINOX treated patients were observed to express increased Th1 cells and decreased Th2 cells, inflammatory monocytes and regulatory T cells. Similarly, in a larger series of resectable pancreatic cancer patients high Th2 cytokines of IL-4 and IP10 and low TH1 cytokines TNFα and INFγ were significantly associated with a shorter disease-free survival (116). Low γδT cell levels have been associated to worse OS in patients with borderline resectable pancreatic cancer patients treated with neoadjuvant mFOLFIRINOX (117).
Myeloid-derived suppressor cells are also involved in tumor response to chemotherapeutic treatments (118). The negative prognostic impact of high pretreatment monocyte levels in early-stage pancreatic cancer has been extensively reported (117, 119). Aiming at targeting monocytes, the combination of FOLFIRINOX with CCR2 blockade has been explored in borderline resectable and locally advanced pancreatic cancer patients in an early phase study (120). The induction of a macrophage polarization into the “classically activated” M1 phenotype by chemotherapy is still matter of debate (121, 122).
These results indicate that neoadjuvant approaches are effective not only because of their cytotoxic effect but also for the positive impact on the immune response.
Circulating Tumor DNA as Non-invasive and Dynamic Tool
Circulating tumor DNA (ctDNA) analysis has been proposed also in pancreatic cancer as prognostic biomarker and as a tool for improving early tumor detection and monitoring tumor dynamics (123–125).
Preoperative detection of ctDNA in patients with early-stage pancreatic cancer has been correlated with decreased recurrence-free survival (RFS) and OS (126, 127). Moreover, it has been shown that a lower proportion of patients undergoing neoadjuvant chemotherapy has detectable ctDNA levels. Thus, ctDNA analysis could help in identifying patients at high risk of early recurrence after resection that might benefit from neoadjuvant chemotherapy. Some evidence has demonstrated that even high exosome DNA (exoDNA) levels are predictive of poor survival in presurgical patients (128).
Another potential application of ctDNA is the possibility of monitoring over time response to anticancer drugs. The correlation between ctDNA changes and tumor responses has been firstly demonstrated in other tumors (129–131). More recently, tumor response has been correlated with reduction of ctDNA levels also in advanced pancreatic cancer patients (132–134). Moreover, the same ctDNA dynamic trend has been observed in series of patients with resectable disease (126, 127). Therefore, serial liquid biopsies might be able to predict disease progression of on-treatment patients earlier than radiological imaging. This could be even more important in a neoadjuvant setting, where the choice of the best treatment strategy could heavily impact on patient prognosis. Furthermore, the use of liquid biopsies could find a pivotal role in patients whose tumors do not express CA19.9, that could routinely be useful in understanding clinical course.
Lastly, the mutational profile observed in ctDNA is highly overlapping compared to primary or metastatic tumor tissues (135). Thus, liquid biopsies may offer the opportunity of dynamic molecular profiling, helping in the personalization of patient treatment (136).
The most frequently used methods for measuring ctDNA levels include NGS, KRAS digital-droplet PCR assay and Safe-Sequencing System (126, 137, 138).
Organoids Recapitulating Tumor Primitive Characteristics Could Serve for Testing Chemosensitivity
Organoids are 3D cellular structures that recapitulate the identity and the cell type diversity of the organ from which they derive. Tumor organoids are novel ex vivo models that can mimic the characteristic of the original tumor in vitro, offering an additional strategy to personalizing therapeutic approaches and are being studied in pancreatic cancer (139).
Pancreatic cancer patient–derived organoid (PDO) libraries have been generated, overcoming the challenge of low neoplastic cellularity, that characterizes pancreatic cancer. PDOs have been proposed to reflect somatic mutations of primary tumor, preserving intratumoral heterogeneity (140).
Gene expression signatures of chemosensitivity have been explored in pancreatic cancer PDOs (44). PDO profiling with DNA and RNA next-generation sequencing combined with pharmacotyping, a PDO drug-testing pipeline, may predict response to conventional chemotherapeutic agents, both in the adjuvant and metastatic setting. A PDO-derived oxaliplatin sensitivity signature has been correlated with differential response to FOLFIRINOX in patients with advanced pancreatic cancer. About one third of the pancreatic cancer PDOs were not sensitive to any of the chemotherapeutic drugs tested (oxaliplatin, SN-38, 5-fluorouracil, gemcitabine, paclitaxel) but approximately half of these showed sensitivity to one or more of the targeted agents evaluated. Pharmacotyping PDO biobank has shown a chemotherapeutic efficacy that seems to recapitulate the clinical response in patients (140).
PDO pharmacotyping has a potential role in personalizing the therapeutic approach of pancreatic cancer patients, even in the early stage. PDO cultures could facilitate molecular characterization in pancreatic cancer, since its typical low cellularity could render it difficult in primary pancreatic cancer specimens. On the other side, since their high cost, the time requested and the need of specialization, they could not yet be routinely used.
Future Perspectives Involving Gut Microbiome
As in many other neoplasms, a significant field of research is focusing on analyzing the link between microbiota and pancreatic cancer, in which could play a role in cancer development, progression and therapeutic response (141).
The microbiota comprises trillions of microorganisms including bacteria, virus (virome), fungi (mycobiome) and archaea and is involved in host physiologic homeostasis.
Substantial abundance of microbiome in pancreatic tumors has been reported (142). Previous studies have demonstrated an association between gut microbial alteration and the presence of pancreatic cancer, but more recently a putative causative role has been also proposed (143–145).
The ability of gut microbiota to colonize pancreatic tumors can modify the tumor microbiome (146). Interestingly, flora from long survivors or healthy patients could shape tumor immune milieu, by recruiting and activating CD8+ T cells.
Moreover, the microbiome has been also proposed as predictor of postoperative survival in pancreatic cancer. Patients with resected pancreatic cancer showing a longer survival were characterized by higher tumor bacteria diversity (146). A signature including three tumor bacteria taxa Pseudoxanthomonas, Streptomyces and Saccharopolyspora and the species Bacillus clausii predicted patient prognosis after resection and tumor microbiome sequencing has been proposed to be used to stratify patients in adjuvant trials. Additionally, gut microbiome could impact even on postoperative complications rates after pancreatic surgery (147).
Noteworthy, recent in vitro and in vivo evidence suggests that microbiota may influence response of gastrointestinal cancers to chemotherapeutic agents. Gemcitabine metabolism has been hypothesized to be decreased by Escherichia coli through bacterial acetylation (148). Diverse bacteria from the class Gammaproteobacteria have been correlated with gemcitabine resistance in preclinical colon and pancreatic models (142). The upregulation of BIRC3, an inhibitor of apoptosis, caused by Fusobacterial nucleatum infection in colorectal cancer cells has showed decreased sensitivity to 5-FU (149). Furthermore, high levels of Fusobacterial nucleatum in resected colorectal cancer specimens have been correlated with 5-FU resistance and shorter DFS. It has been also suggested that in response to oxaliplatin treatment bacteria could mediate the infiltration of myeloid cells producing reactive oxygen species, which are responsible for the cytotoxic effect of oxaliplatin (150).
Fungal population is extremely represented in pancreatic cancer compared to normal pancreas and it migrates from the gut lumen to the pancreas retrogradely via the sphincter of Oddi (145). Fungi has been suggested to be involved in the process of pancreatic carcinogenesis. Recently, intratumoral mycobiome has been demonstrated to stimulate extracellular secretion of IL-33, that recruits TH2 cells and innate lymphoid cells 2, facilitating the type 2 immune response (151). TH2-polarized lymphoid cell tumor infiltration and circulating type 2 cytokines have been associated with poor prognosis in pancreatic cancer (116, 152). Interestingly, genetic deletion of IL-33 or anti-fungal treatment has been correlated with tumor burden decrease and survival prolongation in mice.
Increasing evidence is supporting a role of microbiota in pancreatic cancer progression and therapeutic response. Thus, therapeutic strategies targeting gut microbiome are under investigation and could highlight novel predictive factors and enhance treatment of pancreatic cancer.
Conclusions
Choosing the best treatment strategy is essential in patients affected by pancreatic cancer at early stage, when a chance of cure still exists. The optimal management of resectable pancreatic cancer is debated, and a field of research is moving toward an anticipation of the standard adjuvant therapy. To date, the main factors guiding the clinical decision making include anatomical definition of resectability and clinical and biological patient aspects. However, research is focusing on the identification of novel possible prognostic and predictive markers, able to drive a tailored treatment.
In recent years, different subgroups of pancreatic cancer have been delineated based on the molecular profile. However, this has not yet found application in clinical routine.
The tumor microenvironment and immune cell infiltration have a key role in tumor progression and in resistance to chemotherapeutic treatments in pancreatic cancer patients.
A promising role of miRNAs as prognostic and predictive markers, as well as part of a potential novel therapeutic approach, in pancreatic cancer has been proposed. In the future they could contribute to drive the treatment also before or after surgery or could also become a therapeutic target. Increasing data supports the possible benefit of molecular profiling of advanced pancreatic cancer patients and some evidence is emerging in an early setting too. CtDNA could represent a tool as predictive factors of response and for monitoring the emergence of resistance in advance compared to radiological imaging. Finally, organoids, that can recapitulate typical characteristics of pancreatic cancer, could support evaluation of chemosensitivity. The growing knowledge of the association between gut microbiome and pancreatic cancer could improve the strategic and the therapeutic management of patients affected by resectable pancreatic cancer.
Author Contributions
VM and DMe contributed to conception and design of the study. VM wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) through the Investigator Grant No: 23719 and 5x1000 Grant No: 12182, by the Italian Ministry of Health through the Ricerca Finalizzata 2016 GR-2016-02361134 grant and by the patient associations Nastro Viola and Voglio il Massimo through their donations to DMe.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Carrato A, Melisi D, Prager G, Westphalen CB, Ferreras A, D'Esquermes N, et al. Chart review of diagnostic methods, baseline characteristics and symptoms for European patients with pancreatic cancer. Future Oncol. (2021) 17:1843–54. doi: 10.2217/fon-2020-0749
2. Melisi D, Calvetti L, Frizziero M, Tortora G. Pancreatic cancer: systemic combination therapies for a heterogeneous disease. Curr Pharm Des. (2014) 20:6660–9. doi: 10.2174/1381612820666140826154327
3. Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. (2018) 379:2395–406. doi: 10.1056/NEJMoa1809775
4. Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. (2010) 467:1114–7. doi: 10.1038/nature09515
5. Kulemann B, Rosch S, Seifert S, Timme S, Bronsert P, Seifert G, et al. Pancreatic cancer: circulating tumor cells and primary tumors show heterogeneous KRAS mutations. Sci Rep. (2017) 7:4510. doi: 10.1038/s41598-017-04601-z
6. Hugenschmidt H, Labori KJ, Borgen E, Brunborg C, Schirmer CB, Seeberg LT, et al. Preoperative CTC-detection by cellsearch((R)) is associated with early distant metastasis and impaired survival in resected pancreatic cancer. Cancers. (2021) 13:485. doi: 10.3390/cancers13030485
7. Song BG, Kwon W, Kim H, Lee EM, Han YM, Kim H, et al. Detection of circulating tumor cells in resectable pancreatic ductal adenocarcinoma: a prospective evaluation as a prognostic marker. Front Oncol. (2020) 10:616440. doi: 10.3389/fonc.2020.616440
8. Hamad A, Brown ZJ, Ejaz AM, Dillhoff M, Cloyd JM. Neoadjuvant therapy for pancreatic ductal adenocarcinoma: opportunities for personalized cancer care. World J Gastroenterol. (2021) 27:4383–94. doi: 10.3748/wjg.v27.i27.4383
9. Neoptolemos JP, Stocken DD, Dunn JA, Almond J, Beger HG, Pederzoli P, et al. Influence of resection margins on survival for patients with pancreatic cancer treated by adjuvant chemoradiation and/or chemotherapy in the ESPAC-1 randomized controlled trial. Ann Surg. (2001) 234:758–68. doi: 10.1097/00000658-200112000-00007
10. Cloyd JM, Heh V, Pawlik TM, Ejaz A, Dillhoff M, Tsung A, et al. Neoadjuvant therapy for resectable and borderline resectable pancreatic cancer: a meta-analysis of randomized controlled trials. J Clin Med. (2020) 9:1129. doi: 10.3390/jcm9041129
11. Bakens MJ, van der Geest LG, van Putten M, van Laarhoven HW, Creemers GJ, Besselink MG, et al. The use of adjuvant chemotherapy for pancreatic cancer varies widely between hospitals: a nationwide population-based analysis. Cancer Med. (2016) 5:2825–31. doi: 10.1002/cam4.921
12. Altman AM, Wirth K, Marmor S, Lou E, Chang K, Hui JYC, et al. Completion of adjuvant chemotherapy after upfront surgical resection for pancreatic cancer is uncommon yet associated with improved survival. Ann Surg Oncol. (2019) 26:4108–16. doi: 10.1245/s10434-019-07602-6
13. Quiros RM, Brown KM, Hoffman JP. Neoadjuvant therapy in pancreatic cancer. Cancer Investig. (2007) 25:267–73. doi: 10.1080/07357900701206356
14. Colombo PE, Quenet F, Alric P, Mourregot A, Neron M, Portales F, et al. Distal pancreatectomy with celiac axis resection (modified appleby procedure) and arterial reconstruction for locally advanced pancreatic adenocarcinoma after FOLFIRINOX chemotherapy and chemoradiation therapy. Ann Surg Oncol. (2021) 28:1106–8. doi: 10.1245/s10434-020-08740-y
15. Tempero MA, Malafa MP, Al-Hawary M, Behrman SW, Benson AB, Cardin DB, et al. Pancreatic adenocarcinoma, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compreh Cancer Netw. (2021) 19:439–57. doi: 10.6004/jnccn.2021.0017
16. Ducreux M, Cuhna AS, Caramella C, Hollebecque A, Burtin P, Goere D, et al. Cancer of the pancreas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2015) 26 (Suppl. 5):v56–68. doi: 10.1093/annonc/mdv295
17. Neoptolemos JP, Kerr DJ, Beger H, Link K, Pederzoli P, Bassi C, et al. ESPAC-1 trial progress report: the European randomized adjuvant study comparing radiochemotherapy, 6 months chemotherapy and combination therapy versus observation in pancreatic cancer. Digestion. (1997) 58:570–7. doi: 10.1159/000201503
18. Neoptolemos JP, Stocken DD, Bassi C, Ghaneh P, Cunningham D, Goldstein D, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. (2010) 304:1073–81. doi: 10.1001/jama.2010.1275
19. Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. (2017) 389:1011–24. doi: 10.1016/S0140-6736(16)32409-6
20. Mokdad AA, Minter RM, Zhu H, Augustine MM, Porembka MR, Wang SC, et al. Neoadjuvant therapy followed by resection versus upfront resection for resectable pancreatic cancer: a propensity score matched analysis. J Clin Oncol. (2017) 35:515–22. doi: 10.1200/JCO.2016.68.5081
21. Versteijne E, van Dam JL, Suker M, Janssen QP, Groothuis K, Akkermans-Vogelaar JM, et al. Neoadjuvant chemoradiotherapy versus upfront surgery for resectable and borderline pancreatic cancer: Long-term results of the dutch randomized PREOPANC trial. J Clin Oncol. (2022) 40:1220–30. doi: 10.1200/JCO.21.02233
22. Sohal DPS, Duong M, Ahmad SA, Gandhi NS, Beg MS, Wang-Gillam A, et al. Efficacy of perioperative chemotherapy for resectable pancreatic adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. (2021) 7:421–7. doi: 10.1001/jamaoncol.2020.7328
23. Janssen QP, van Dam JL, Bonsing BA, Bos H, Bosscha KP, Coene P, et al. Total neoadjuvant FOLFIRINOX versus neoadjuvant gemcitabine-based chemoradiotherapy and adjuvant gemcitabine for resectable and borderline resectable pancreatic cancer (PREOPANC-2 trial): study protocol for a nationwide multicenter randomized controlled trial. BMC Cancer. (2021) 21:300. doi: 10.1186/s12885-021-08031-z
24. Simionato F, Zecchetto C, Merz V, Cavaliere A, Casalino S, Gaule M, et al. A phase II study of liposomal irinotecan with 5-fluorouracil, leucovorin and oxaliplatin in patients with resectable pancreatic cancer: the nITRO trial. Ther Adv Med Oncol. (2020) 12:1758835920947969. doi: 10.1177/1758835920947969
25. Labori KJ, Lassen K, Hoem D, Gronbech JE, Soreide JA, Mortensen K, et al. Neoadjuvant chemotherapy versus surgery first for resectable pancreatic cancer (Norwegian Pancreatic Cancer Trial - 1 (NorPACT-1)) - study protocol for a national multicentre randomized controlled trial. BMC Surg. (2017) 17:94. doi: 10.1186/s12893-017-0291-1
26. Al-Batran S-E, Reichart A, Bankstahl US, Pauligk C, Kraus TW, Bechstein WO, et al. Randomized multicenter phase II/III study with adjuvant gemcitabine versus neoadjuvant/adjuvant FOLFIRINOX in resectable pancreatic cancer: the NEPAFOX trial. J Clin Oncol. (2021) 39(3_suppl):406–. doi: 10.1200/JCO.2021.39.3_suppl.406
27. Schwarz L, Vernerey D, Bachet JB, Tuech JJ, Portales F, Michel P, et al. Resectable pancreatic adenocarcinoma neo-adjuvant FOLF(IRIN)OX-based chemotherapy - a multicenter, non-comparative, randomized, phase II trial (PANACHE01-PRODIGE48 study). BMC Cancer. (2018) 18:762. doi: 10.1186/s12885-018-4663-4
28. Tamburrino A, Piro G, Carbone C, Tortora G, Melisi D. Mechanisms of resistance to chemotherapeutic and anti-angiogenic drugs as novel targets for pancreatic cancer therapy. Front Pharmacol. (2013) 4:56. doi: 10.3389/fphar.2013.00056
29. Melisi D, Chiao PJ. NF-kappa B as a target for cancer therapy. Expert Opin Ther Targets. (2007) 11:133–44. doi: 10.1517/14728222.11.2.133
30. Carbone C, Melisi D. NF-kappaB as a target for pancreatic cancer therapy. Expert Opin Ther Targets. (2012) 16 (Suppl. 2):S1–10. doi: 10.1517/14728222.2011.645806
31. Melisi D, Niu J, Chang Z, Xia Q, Peng B, Ishiyama S, et al. Secreted interleukin-1alpha induces a metastatic phenotype in pancreatic cancer by sustaining a constitutive activation of nuclear factor-kappaB. Mol Cancer Res. (2009) 7:624–33. doi: 10.1158/1541-7786.MCR-08-0201
32. Zhuang Z, Ju HQ, Aguilar M, Gocho T, Li H, Iida T, et al. IL1 receptor antagonist inhibits pancreatic cancer growth by abrogating nf-kappab activation. Clin Cancer Res. (2016) 22:1432–44. doi: 10.1158/1078-0432.CCR-14-3382
33. Melisi D, Ishiyama S, Sclabas GM, Fleming JB, Xia Q, Tortora G, et al. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol Cancer Ther. (2008) 7:829–40. doi: 10.1158/1535-7163.MCT-07-0337
34. Melisi D, Xia Q, Paradiso G, Ling J, Moccia T, Carbone C, et al. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. J Natl Cancer Inst. (2011) 103:1190–204. doi: 10.1093/jnci/djr243
35. Santoro R, Zanotto M, Simionato F, Zecchetto C, Merz V, Cavallini C, et al. Modulating TAK1 expression inhibits YAP and TAZ oncogenic functions in pancreatic cancer. Mol Cancer Ther. (2020) 19:247–57. doi: 10.1158/1535-7163.MCT-19-0270
36. Santoro R, Carbone C, Piro G, Chiao PJ, Melisi D. TAK-ing aim at chemoresistance: the emerging role of MAP3K7 as a target for cancer therapy. Drug Resist Updat. (2017) 33-35:36–42. doi: 10.1016/j.drup.2017.10.004
37. Merz V, Zecchetto C, Santoro R, Simionato F, Sabbadini F, Mangiameli D, et al. Plasma IL8 is a biomarker for TAK1 activation and predicts resistance to nanoliposomal irinotecan in patients with gemcitabine-refractory pancreatic cancer. Clin Cancer Res. (2020) 26:4661–9. doi: 10.1158/1078-0432.CCR-20-0395
38. Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. (2011) 17:500–3. doi: 10.1038/nm.2344
39. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. (2016) 531:47–52. doi: 10.1038/nature16965
40. Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. (2015) 47:1168–78. doi: 10.1038/ng.3398
41. Shi ZD, Lee K, Yang D, Amin S, Verma N, Li QV, et al. Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell stem cell. (2017) 20:675–88 e6. doi: 10.1016/j.stem.2017.01.001
42. O'Kane GM, Grunwald BT, Jang GH, Masoomian M, Picardo S, Grant RC, et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin Cancer Res. (2020) 26:4901–10. doi: 10.1158/1078-0432.CCR-19-3724
43. Martinelli P, Carrillo-de Santa Pau E, Cox T, Sainz B Jr, Dusetti N, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. (2017) 66:1665–76. doi: 10.1136/gutjnl-2015-311256
44. Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. (2018) 8:1112–29. doi: 10.1158/2159-8290.CD-18-0349
45. Connor AA, Denroche RE, Jang GH, Lemire M, Zhang A, Chan-Seng-Yue M, et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell. (2019) 35:267–82 e7. doi: 10.1016/j.ccell.2018.12.010
46. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
47. Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. (2012) 136:169–74. doi: 10.1016/j.pharmthera.2012.08.003
48. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. (2014) 24:R762–76. doi: 10.1016/j.cub.2014.06.043
49. Berindan-Neagoe I, Calin GA. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin Cancer Res. (2014) 20:6247–53. doi: 10.1158/1078-0432.CCR-13-2500
50. Namkung J, Kwon W, Choi Y, Yi SG, Han S, Kang MJ, et al. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol. (2016) 31:1160–7. doi: 10.1111/jgh.13253
51. Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. (2010) 126:73–80. doi: 10.1002/ijc.24687
52. Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, et al. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. (2017) 7:42339. doi: 10.1038/srep42339
53. Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. (2010) 31:1726–33. doi: 10.1093/carcin/bgq160
54. Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, et al. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. (2011) 18:2381–7. doi: 10.1245/s10434-011-1602-x
55. Li Y, VandenBoom TG, 2nd Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. (2009) 69:6704–12. doi: 10.1158/0008-5472.CAN-09-1298
56. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. (2008) 10:593–601. doi: 10.1038/ncb1722
57. Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. (2008) 27:2128–36. doi: 10.1038/sj.onc.1210856
58. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. (2007) 133:647–58. doi: 10.1053/j.gastro.2007.05.022
59. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. (2007) 282:14328–36. doi: 10.1074/jbc.M611393200
60. Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. (2010) 70:4528–38. doi: 10.1158/0008-5472.CAN-09-4467
61. Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. (2015) 46:539–47. doi: 10.3892/ijo.2014.2743
62. Karasek P, Gablo N, Hlavsa J, Kiss I, Vychytilova-Faltejskova P, Hermanova M, et al. Pre-operative plasma miR-21-5p is a sensitive biomarker and independent prognostic factor in patients with pancreatic ductal adenocarcinoma undergoing surgical resection. Cancer Genom Proteom. (2018) 15:321–7. doi: 10.21873/cgp.20090
63. Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE. (2010) 5:e10630. doi: 10.1371/journal.pone.0010630
64. Paik WH, Kim HR, Park JK, Song BJ, Lee SH, Hwang JH. Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. (2013) 33:1473–81.
65. Wei X, Wang W, Wang L, Zhang Y, Zhang X, Chen M, et al. MicroRNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4. Cancer Med. (2016) 5:693–702. doi: 10.1002/cam4.626
66. Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or−221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. (2009) 38:e190–9. doi: 10.1097/MPA.0b013e3181ba82e1
67. Gablo N, Trachtova K, Prochazka V, Hlavsa J, Grolich T, Kiss I, et al. Identification and validation of circulating micrornas as prognostic biomarkers in pancreatic ductal adenocarcinoma patients undergoing surgical resection. J Clin Med. (2020) 9:2440. doi: 10.3390/jcm9082440
68. Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. (2013) 108:361–9. doi: 10.1038/bjc.2012.546
69. Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. (2007) 297:1901–8. doi: 10.1001/jama.297.17.1901
70. Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. (2011) 56:602–9. doi: 10.1007/s10620-010-1285-3
71. Chen Q, Yang L, Xiao Y, Zhu J, Li Z. Circulating microRNA-182 in plasma and its potential diagnostic and prognostic value for pancreatic cancer. Med Oncol. (2014) 31:225. doi: 10.1007/s12032-014-0225-z
72. Eid M, Karousi P, Kunovsky L, Tucek S, Brancikova D, Kala Z, et al. The role of circulating MicroRNAs in patients with early-stage pancreatic adenocarcinoma. Biomedicines. (2021) 9:1468. doi: 10.3390/biomedicines9101468
73. Garajova I, Le Large TY, Frampton AE, Rolfo C, Voortman J, Giovannetti E. Molecular mechanisms underlying the role of microRNAs in the chemoresistance of pancreatic cancer. BioMed Res Int. (2014) 2014:678401. doi: 10.1155/2014/678401
74. Iwagami Y, Eguchi H, Nagano H, Akita H, Hama N, Wada H, et al. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. (2013) 109:502–11. doi: 10.1038/bjc.2013.320
75. Liang C, Wang Z, Li YY, Yu BH, Zhang F, Li HY. miR-33a suppresses the nuclear translocation of beta-catenin to enhance gemcitabine sensitivity in human pancreatic cancer cells. Tumour Biol. (2015) 36:9395–403. doi: 10.1007/s13277-015-3679-5
76. Petrocca F, Lieberman J. Micromanipulating cancer: microRNA-based therapeutics? RNA Biol. (2009) 6:335–40. doi: 10.4161/rna.6.3.9013
77. Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, et al. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. (2009) 20:831–44. doi: 10.1089/hum.2008.134
78. Ye ZQ, Zou CL, Chen HB, Jiang MJ, Mei Z, Gu DN. MicroRNA-7 as a potential biomarker for prognosis in pancreatic cancer. Dis Mark. (2020) 2020:2782101. doi: 10.1155/2020/2782101
79. Gisel A, Valvano M, El Idrissi IG, Nardulli P, Azzariti A, Carrieri A, et al. miRNAs for the detection of multidrug resistance: overview and perspectives. Molecules. (2014) 19:5611–23. doi: 10.3390/molecules19055611
80. Meijer LL, Garajova I, Caparello C, Le Large TYS, Frampton AE, Vasile E, et al. Plasma miR-181a-5p downregulation predicts response and improved survival after FOLFIRINOX in pancreatic ductal adenocarcinoma. Ann Surg. (2020) 271:1137–47. doi: 10.1097/SLA.0000000000003084
81. Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. (2010) 9:2265–75. doi: 10.1158/1535-7163.MCT-10-0061
82. Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. (2005) 438:685–9. doi: 10.1038/nature04303
83. Zhao Y, Zhao L, Ischenko I, Bao Q, Schwarz B, Niess H, et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target Oncol. (2015) 10:535–48. doi: 10.1007/s11523-015-0360-2
84. Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. (2018) 8:1096–111. doi: 10.1158/2159-8290.CD-18-0275
85. Pishvaian MJ, Blais EM, Brody JR, Lyons E, DeArbeloa P, Hendifar A, et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. (2020) 21:508–18. doi: 10.1016/S1470-2045(20)30074-7
86. Krepline AN, Bliss L, Geurts J, Akinola I, Christians KK, George B, et al. Role of molecular profiling of pancreatic cancer after neoadjuvant therapy: does it change practice? J Gastrointest Surg. (2020) 24:235–42. doi: 10.1007/s11605-019-04423-6
87. Melisi D, Cavaliere A, Gobbo S, Fasoli G, Allegrini V, Simionato F, et al. Role of next-generation genomic sequencing in targeted agents repositioning for pancreaticoduodenal cancer patients. Pancreatology. (2021) doi: 10.1016/j.pan.2021.04.004. [Epub ahead of print].
88. O'Connor MJ. Targeting the DNA damage response in cancer. Molecular cell. (2015) 60:547–60. doi: 10.1016/j.molcel.2015.10.040
89. Lowery MA, Wong W, Jordan EJ, Lee JW, Kemel Y, Vijai J, et al. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms. J Natl Cancer Inst. (2018) 110:1067–74. doi: 10.1093/jnci/djy024
90. Goldstein JB, Zhao L, Wang X, Ghelman Y, Overman MJ, Javle MM, et al. Germline DNA sequencing reveals novel mutations predictive of overall survival in a cohort of patients with pancreatic cancer. Clin Cancer Res. (2020) 26:1385–94. doi: 10.1158/1078-0432.CCR-19-0224
91. Wattenberg MM, Asch D, Yu S, O'Dwyer PJ, Domchek SM, Nathanson KL, et al. Platinum response characteristics of patients with pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or PALB2 mutation. Br J Cancer. (2020) 122:333–9. doi: 10.1038/s41416-019-0582-7
92. Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. (2012) 491:399–405. doi: 10.1038/nature11547
93. Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. (2014) 111:1132–8. doi: 10.1038/bjc.2014.418
94. Sehdev A, Gbolahan O, Hancock BA, Stanley M, Shahda S, Wan J, et al. Germline and somatic DNA damage repair gene mutations and overall survival in metastatic pancreatic adenocarcinoma patients treated with FOLFIRINOX. Clin Cancer Res. (2018) 24:6204–11. doi: 10.1158/1078-0432.CCR-18-1472
95. Pokataev I, Fedyanin M, Polyanskaya E, Popova A, Agafonova J, Menshikova S, et al. Efficacy of platinum-based chemotherapy and prognosis of patients with pancreatic cancer with homologous recombination deficiency: comparative analysis of published clinical studies. ESMO Open. (2020) 5:e000578. doi: 10.1136/esmoopen-2019-000578
96. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. (2015) 518:495–501. doi: 10.1038/nature14169
97. Rombouts SJ, Walma MS, Vogel JA, van Rijssen LB, Wilmink JW, Mohammad NH, et al. Systematic review of resection rates and clinical outcomes after FOLFIRINOX-based treatment in patients with locally advanced pancreatic cancer. Ann Surg Oncol. (2016) 23:4352–60. doi: 10.1245/s10434-016-5373-2
98. Golan T, Barenboim A, Lahat G, Nachmany I, Goykhman Y, Shacham-Shmueli E, et al. Increased rate of complete pathologic response after neoadjuvant FOLFIRINOX for BRCA mutation carriers with borderline resectable pancreatic cancer. Ann Surg Oncol. (2020) 27:3963–70. doi: 10.1245/s10434-020-08469-8
99. Yu S, Agarwal P, Mamtani R, Symecko H, Spielman K, O'Hara M, et al. Retrospective survival analysis of patients with resected pancreatic ductal adenocarcinoma and a germline BRCA or PALB2 mutation. JCO Precis Oncol. (2019) 3:1–11. doi: 10.1200/PO.18.00271
100. Grose DB, McKay CJ, Cooke S, Graham JS, Duthie F, Jamieson N, et al. PRIMUS-002: a multicentre, open-label, phase II study examining FOLFOX and nab-paclitaxel (FA) and nab-paclitaxel and gemcitabine (AG) as neoadjuvant therapy for (borderline) resectable pancreatic cancer (PC), focusing on biomarker and liquid biopsy development. J Clin Oncol. (2019) 37 (15_suppl):TPS4166–TPS. doi: 10.1200/JCO.2019.37.15_suppl.TPS4166
101. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. (2019) 381:317–27. doi: 10.1056/NEJMoa1903387
102. Golan T, Hammel P, Reni M, Cutsem EV, Macarulla T, Hall MJ, et al. Overall survival from the phase 3 POLO trial: maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. J Clin Oncol. (2021) 39 (3_suppl):378–. doi: 10.1200/JCO.2021.39.3_suppl.378
103. Tuli R, Surmak AJ, Reyes J, Armour M, Hacker-Prietz A, Wong J, et al. Radiosensitization of pancreatic cancer cells in vitro and in vivo through poly (ADP-ribose) polymerase inhibition with ABT-888. Trans Oncol. (2014) 7:439–45. doi: 10.1016/j.tranon.2014.04.003
104. Tuli R, Shiao SL, Nissen N, Tighiouart M, Kim S, Osipov A, et al. A phase 1 study of veliparib, a PARP-1/2 inhibitor, with gemcitabine and radiotherapy in locally advanced pancreatic cancer. EBioMedicine. (2019) 40:375–81. doi: 10.1016/j.ebiom.2018.12.060
105. Connor AA, Denroche RE, Jang GH, Timms L, Kalimuthu SN, Selander I, et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol. (2017) 3:774–83. doi: 10.1001/jamaoncol.2016.3916
106. Rotovic B, Kulic J. Chemical structure and biochemical role of polyphosphoric acids and their derivatives. Med Pregl. (1965) 18:57–62.
107. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. (2018) 378:731–9. doi: 10.1056/NEJMoa1714448
108. Miyoshi T, Kondo K, Toba H, Yoshida M, Fujino H, Kenzaki K, et al. Predictive value of thymidylate synthase and dihydropyrimidine dehydrogenase expression in tumor tissue, regarding the efficacy of postoperatively administered UFT (tegafur+uracil) in patients with non-small cell lung cancer. Anticancer Res. (2007) 27:2641–8.
109. Akita H, Zheng Z, Takeda Y, Kim C, Kittaka N, Kobayashi S, et al. Significance of RRM1 and ERCC1 expression in resectable pancreatic adenocarcinoma. Oncogene. (2009) 28:2903–9. doi: 10.1038/onc.2009.158
110. Chen G, Tian X, Liu Z, Zhou S, Schmidt B, Henne-Bruns D, et al. Inhibition of endogenous SPARC enhances pancreatic cancer cell growth: modulation by FGFR1-III isoform expression. Br J Cancer. (2010) 102:188–95. doi: 10.1038/sj.bjc.6605440
111. Spratlin J, Sangha R, Glubrecht D, Dabbagh L, Young JD, Dumontet C, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. (2004) 10:6956–61. doi: 10.1158/1078-0432.CCR-04-0224
112. Tsai S, Christians KK, George B, Ritch PS, Dua K, Khan A, et al. A phase II clinical trial of molecular profiled neoadjuvant therapy for localized pancreatic ductal adenocarcinoma. Ann Surg. (2018) 268:610–9. doi: 10.1097/SLA.0000000000002957
113. Homma Y, Taniguchi K, Murakami T, Nakagawa K, Nakazawa M, Matsuyama R, et al. Immunological impact of neoadjuvant chemoradiotherapy in patients with borderline resectable pancreatic ductal adenocarcinoma. Ann Surg Oncol. (2014) 21:670–6. doi: 10.1245/s10434-013-3390-y
114. Shibuya KC, Goel VK, Xiong W, Sham JG, Pollack SM, Leahy AM, et al. Pancreatic ductal adenocarcinoma contains an effector and regulatory immune cell infiltrate that is altered by multimodal neoadjuvant treatment. PLoS ONE. (2014) 9:e96565. doi: 10.1371/journal.pone.0096565
115. Peng H, James CA, Cullinan DR, Hogg GD, Mudd JL, Zuo C, et al. Neoadjuvant FOLFIRINOX therapy is associated with increased effector T cells and reduced suppressor cells in patients with pancreatic cancer. Clin Cancer Res. (2021) 27:6761–71. doi: 10.1158/1078-0432.CCR-21-0998
116. Piro G, Simionato F, Carbone C, Frizziero M, Malleo G, Zanini S, et al. A circulating TH2 cytokines profile predicts survival in patients with resectable pancreatic adenocarcinoma. Oncoimmunology. (2017) 6:e1322242. doi: 10.1080/2162402X.2017.1322242
117. Yoo C, Lee SS, Song KB, Jeong JH, Hyung J, Park DH, et al. Neoadjuvant modified FOLFIRINOX followed by postoperative gemcitabine in borderline resectable pancreatic adenocarcinoma: a Phase 2 study for clinical and biomarker analysis. Br J Cancer. (2020) 123:362–8. doi: 10.1038/s41416-020-0867-x
118. Liu Q, Liao Q, Zhao Y. Chemotherapy and tumor microenvironment of pancreatic cancer. Cancer Cell Int. (2017) 17:68. doi: 10.1186/s12935-017-0437-3
119. Sierzega M, Lenart M, Rutkowska M, Surman M, Mytar B, Matyja A, et al. Preoperative neutrophil-lymphocyte and lymphocyte-monocyte ratios reflect immune cell population rearrangement in resectable pancreatic cancer. Ann Surg Oncol. (2017) 24:808–15. doi: 10.1245/s10434-016-5634-0
120. Nywening TM, Wang-Gillam A, Sanford DE, Belt BA, Panni RZ, Cusworth BM, et al. Targeting tumour-associated macrophages with CCR2 inhibition in combination with FOLFIRINOX in patients with borderline resectable and locally advanced pancreatic cancer: a single-centre, open-label, dose-finding, non-randomised, phase 1b trial. Lancet Oncol. (2016) 17:651–62. doi: 10.1016/S1470-2045(16)00078-4
121. Di Caro G, Cortese N, Castino GF, Grizzi F, Gavazzi F, Ridolfi C, et al. Dual prognostic significance of tumour-associated macrophages in human pancreatic adenocarcinoma treated or untreated with chemotherapy. Gut. (2016) 65:1710–20. doi: 10.1136/gutjnl-2015-309193
122. Mota Reyes C, Teller S, Muckenhuber A, Konukiewitz B, Safak O, Weichert W, et al. Neoadjuvant therapy remodels the pancreatic cancer microenvironment via depletion of protumorigenic immune cells. Clin Cancer Res. (2020) 26:220–31. doi: 10.1158/1078-0432.CCR-19-1864
123. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. (2008) 14:985–90. doi: 10.1038/nm.1789
124. Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. (2015) 6:7686. doi: 10.1038/ncomms8686
125. Husain H, Velculescu VE. Cancer DNA in the circulation: the liquid biopsy. JAMA. (2017) 318:1272–4. doi: 10.1001/jama.2017.12131
126. Groot VP, Mosier S, Javed AA, Teinor JA, Gemenetzis G, Ding D, et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin Cancer Res. (2019) 25:4973–84. doi: 10.1158/1078-0432.CCR-19-0197
127. Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. (2016) 115:59–65. doi: 10.1038/bjc.2016.175
128. Bernard V, Kim DU, San Lucas FA, Castillo J, Allenson K, Mulu FC, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. (2019) 156:108–18.e4. doi: 10.1053/j.gastro.2018.09.022
129. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. (2013) 368:1199–209. doi: 10.1056/NEJMoa1213261
130. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. (2015) 21:3196–203. doi: 10.1158/1078-0432.CCR-14-2594
131. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. (2015) 21:795–801. doi: 10.1038/nm.3870
132. Wei T, Zhang Q, Li X, Su W, Li G, Ma T, et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol Cancer Ther. (2019) 18:196–203. doi: 10.1158/1535-7163.MCT-17-1298
133. Cheng H, Liu C, Jiang J, Luo G, Lu Y, Jin K, et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int J Cancer. (2017) 140:2344–50. doi: 10.1002/ijc.30650
134. Earl J, Garcia-Nieto S, Martinez-Avila JC, Montans J, Sanjuanbenito A, Rodriguez-Garrote M, et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. (2015) 15:797. doi: 10.1186/s12885-015-1779-7
135. Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. (2015) 5:1040–8. doi: 10.1158/2159-8290.CD-15-0274
136. Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Use of liquid biopsies in clinical oncology: pilot experience in 168 patients. Clin Cancer Res. (2016) 22:5497–505. doi: 10.1158/1078-0432.CCR-16-0318
137. Lin MT, Mosier SL, Thiess M, Beierl KF, Debeljak M, Tseng LH, et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. (2014) 141:856–66. doi: 10.1309/AJCPMWGWGO34EGOD
138. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. (2011) 108:9530–5. doi: 10.1073/pnas.1105422108
139. Yao J, Yang M, Atteh L, Liu P, Mao Y, Meng W, et al. A pancreas tumor derived organoid study: from drug screen to precision medicine. Cancer Cell Int. (2021) 21:398. doi: 10.1186/s12935-021-02044-1
140. Seppala TT, Zimmerman JW, Sereni E, Plenker D, Suri R, Rozich N, et al. Patient-derived organoid pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann Surg. (2020) 272:427–35. doi: 10.1097/SLA.0000000000004200
141. Yu Q, Jobin C, Thomas RM. Implications of the microbiome in the development and treatment of pancreatic cancer: thinking outside of the box by looking inside the gut. Neoplasia. (2021) 23:246–56. doi: 10.1016/j.neo.2020.12.008
142. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
143. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
144. Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. (2018) 39:1068–78. doi: 10.1093/carcin/bgy073
145. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
146. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806.e12. doi: 10.1016/j.cell.2019.07.008
147. Schmitt FCF, Brenner T, Uhle F, Loesch S, Hackert T, Ulrich A, et al. Gut microbiome patterns correlate with higher postoperative complication rates after pancreatic surgery. BMC Microbiol. (2019) 19:42. doi: 10.1186/s12866-019-1399-5
148. Lehouritis P, Cummins J, Stanton M, Murphy CT, McCarthy FO, Reid G, et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci Rep. (2015) 5:14554. doi: 10.1038/srep14554
149. Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, et al. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. (2019) 38:14. doi: 10.1186/s13046-018-0985-y
150. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. (2013) 342:967–70. doi: 10.1126/science.1240527
151. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. (2022) 40:153–67.e11. doi: 10.1016/j.ccell.2022.01.003
Keywords: resectable pancreatic cancer, predictive factors, neoadjuvant therapy, preoperative treatment, target therapy, molecular profiling, tumor microenvironment, microbiota
Citation: Merz V, Mangiameli D, Zecchetto C, Quinzii A, Pietrobono S, Messina C, Casalino S, Gaule M, Pesoni C, Vitale P, Trentin C, Frisinghelli M, Caffo O and Melisi D (2022) Predictive Biomarkers for a Personalized Approach in Resectable Pancreatic Cancer. Front. Surg. 9:866173. doi: 10.3389/fsurg.2022.866173
Received: 30 January 2022; Accepted: 25 March 2022;
Published: 04 May 2022.
Edited by:
Casper Van Eijck, Erasmus University Rotterdam, NetherlandsReviewed by:
Francesca Marcon, IRCCS Ca 'Granda Foundation Maggiore Policlinico Hospital, ItalyDavide Ciardiello, University of Campania Luigi Vanvitelli, Italy
Copyright © 2022 Merz, Mangiameli, Zecchetto, Quinzii, Pietrobono, Messina, Casalino, Gaule, Pesoni, Vitale, Trentin, Frisinghelli, Caffo and Melisi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valeria Merz, dmFsZXJpYW1lcnpAZ21haWwuY29t; Davide Melisi, ZGF2aWRlLm1lbGlzaUB1bml2ci5pdA==
 Valeria Merz
Valeria Merz Domenico Mangiameli
Domenico Mangiameli Camilla Zecchetto3
Camilla Zecchetto3 Alberto Quinzii
Alberto Quinzii Carlo Messina
Carlo Messina Marina Gaule
Marina Gaule Camilla Pesoni
Camilla Pesoni Orazio Caffo
Orazio Caffo Davide Melisi
Davide Melisi