- 1Department of Biological Sciences, Florida A&M University, Tallahassee, FL, United States
- 2College of Arts and Sciences, Florida State University, Tallahassee, FL, United States
- 3College of Liberal Arts, University of Florida, Gainesville, FL, United States
- 4Department of Biosciences, Manipal University Jaipur, Dehmi Kalan, India
- 5Bioclues.org, Hyderabad, India
Alzheimer’s disease (AD) is a growing global challenge, representing the most common neurodegenerative disorder and affecting millions of lives. As life expectancy continues to rise and populations expand, the number of individuals coping with the cognitive declines caused by AD is projected to double in the coming years. By 2050, we may see over 115 million people diagnosed with this devastating condition. Unfortunately, while we currently lack effective cures, there are preventative measures that can slow disease progression in symptomatic patients. Thus, research has shifted toward early detection and intervention for AD in recent years. With technological advances, we are now harnessing large datasets and more efficient, minimally invasive methods for diagnosis and treatment. This review highlights critical demographic insights, health conditions that increase the risk of developing AD, and lifestyle factors in midlife that can potentially trigger its onset. Additionally, we delve into the promising role of plant-based metabolites and their sources, which may help delay the disease’s progression. The innovative multi-omics research is transforming our understanding of AD. This approach enables comprehensive data analysis from diverse cell types and biological processes, offering possible biomarkers of this disease’s mechanisms. We present the latest advancements in genomics, transcriptomics, Epigenomics, proteomics, and metabolomics, including significant progress in gene editing technologies. When combined with machine learning and artificial intelligence, multi-omics analysis becomes a powerful tool for uncovering the complexities of AD pathogenesis. We also explore current trends in the application of radiomics and machine learning, emphasizing how integrating multi-omics data can transform our approach to AD research and treatment. Together, these pioneering advancements promise to develop more effective preventive and therapeutic strategies soon.
1 Background
Alzheimer’s disease (AD) is primarily characterized by the dysfunction of several brain networks responsible for maintaining homeostasis and intracellular signaling. This disease poses numerous healthcare challenges, particularly the increasing prevalence of aging populations and the lack of effective curative treatments. Current therapies mainly aim to alleviate symptoms rather than target the underlying causes. Furthermore, the lack of early detection methods complicates the diagnosis. The complex pathology of AD, marked by the accumulation of amyloid plaques and tau tangles, makes developing targeted treatments challenging. The impact of AD extends beyond medical issues, affecting caregivers both financially and emotionally, thus contributing to broader societal challenges (Passeri et al., 2022). Diagnosing AD often requires years of observed cognitive decline, and this timeline can be even longer in the absence of genetic markers. A family history of Alzheimer’s often correlates with the disease’s progression, and with no cure available, early diagnosis and preventive strategies are critical to prevent irreversible brain damage. To improve our molecular understanding of AD and enhance both treatments and early diagnoses, exploring various biological processes, including genomics, Epigenomics, transcriptomics, proteomics, lipidomics, and metabolomics, is crucial. Unlike other conditions, diagnosing AD cannot be done through brain biopsy, making advancing research on cellular structures and imaging technologies vital, given the disease’s rising prevalence. Recent studies have also utilized radiomic imaging analysis and artificial intelligence (AI) to investigate cognitive impairments linked to minor vessel diseases associated with AD (Shi et al., 2020). Efforts from prevention trials and clinicians are underway to quantify and detect AD earlier and with greater accuracy through multi-omics approaches, facilitating more comprehensive analyses of neurodegenerative disorders like Alzheimer’s (Rawat et al., 2022).
2 History and pathogenesis
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that German psychiatrist Alois Alzheimer first identified. He observed the presence of amyloid plaques and significant neuronal loss in patients experiencing memory loss and personality changes. Later, Emil Kraepelin emphasized the severity of the disease, particularly in the cerebral cortex and medial temporal lobe, contributing to cognitive decline (Braun et al., 2022). The pathophysiology of AD involves shrinkage of the cerebral cortex and hippocampus, enlargement of the ventricles, and the presence of amyloid-beta (Aβ) plaques, as well as tau neurofibrillary tangles. Neuroinflammation arises as blood vessels age, impairing the glymphatic system and leading to the buildup of Aβ plaques. Notably, AD presents features such as granulovacuolar degeneration, which is characterized by large, double membrane-bound vacuoles in neurons (Funk et al., 2011). In 1984, the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) and the Alzheimer’s Disease and Related Disorders Association (ADRDA) established diagnostic criteria based on neuropsychological testing, progressive memory loss, and impairment in daily activities. These symptoms are most found in late-onset Alzheimer’s disease (LOAD), which usually appears after the age of 65, and early-onset Alzheimer’s disease (EOAD), which can manifest as early as a person’s 40s or 50s. Both forms often begin with mild cognitive impairment (MCI), a transitional stage between normal aging and dementia characterized by subtle cognitive deficits. While individuals with MCI can typically maintain their independence in daily life, they may experience difficulties in critical thinking, memory retention, and executive functioning. Early symptoms of MCI may include forgetting recently learned information, becoming disoriented in familiar environments, and experiencing trouble with planning or problem-solving. These impairments can be overlooked or dismissed as normal aging; however, they represent the earliest clinical indicators of Alzheimer’s disease pathology (Campbell et al., 2025). Recognizing MCI is crucial as it serves as an early marker of neurodegenerative decline and reflects the onset of underlying brain changes such as synaptic dysfunction, the accumulation of beta-amyloid plaques, and tau pathology. An increasing number of patients without cognitive or behavioral symptoms are presenting with positive biomarkers, commonly referred to as “stage 1” cases. However, in the absence of active disease progression, these findings merely indicate susceptibility to the disease (Gale, 2024). Utilizing updated criteria released in 2011, biomarkers obtained from positron emission tomography (PET) scans and cerebrospinal fluid analysis, combined with machine learning, will aid in classifying these asymptomatic disease models. Nevertheless, current diagnostic methods typically identify the disease only after it has significantly progressed and caused irreversible damage (Jack et al., 2024).
3 Demographics at risk
3.1 Age and gender
Aging is the primary risk factor for AD, and sex differences significantly affect its development and progression. Research shows that women generally have lower synapse density but higher tau and amyloid-beta (Aβ) levels than men (Sundermann et al., 2020). Key contributors to these differences include gonadal hormones and sex chromosomes. Hormones such as estrogen and testosterone influence susceptibility to the disease; estrogen plays a vital role in processes involving mitochondrial function, inflammation, glucose transport and metabolism, and cholesterol homeostasis. Both testosterone and estrogen regulate apolipoprotein E (ApoE), a key biomarker for AD (Gamache et al., 2020). Additionally, the XX chromosomes in females and the XY chromosomes in males are responsible for genetic factors affecting AD risk. For example, the loss of the Y chromosome in male AD patients can increase Aβ toxicity and lead to premature cell death (Guo et al., 2022). Postmenopausal women experience increased levels of luteinizing hormone and follicle-stimulating hormone, which may contribute to AD pathology and cognitive decline (Valencia-Olvera et al., 2023). Research has shown that estrogen can lower Aβ levels by inhibiting the production of vesicles containing amyloid precursor proteins (Arjmand et al., 2024). However, after menopause, the decline in estrogen diminishes this protective mechanism, resulting in similar metabolic conditions in both sexes (Lopez-Lee et al., 2024).
3.2 Cardiovascular diseases, diabetes, and other midlife risk factors
Alzheimer’s interacts with various comorbidities, such as cardiovascular disease and diabetes, which can worsen its progression. Common health problems related to AD include high cholesterol, hypertension, and diabetes. Many cases can be traced back to midlife risk factors, including smoking, elevated blood pressure, and diabetes, accounting for up to 45% of dementia cases (Malik et al., 2021). It is essential to explore these interactions to foster the development of new treatments, especially by repurposing existing medications for Alzheimer’s management. The influence of fats and proteins on brain function and dementia risk is an area that warrants additional investigation, particularly regarding AD and vascular dementia (VD). In Type 2 diabetes mellitus (T2DM), chronic hyperglycemia exacerbates amyloid beta production and tau hyperphosphorylation, which intensifies AD pathology. Impaired insulin signaling further disrupts neuronal energy metabolism, contributing to neurodegeneration in late-onset AD. Elevated blood glucose levels in T2DM can trigger the formation of advanced glycation end-products (AGEs), which promote Aβ accumulation and tau phosphorylation, leading to increased neurodegeneration.
Genetic links between dementia and conditions like hypertension and type 2 diabetes highlight the need to understand these pathways for effective prevention and treatment. Factors such as 20-Hydroxyeicosatetraenoic acid (20-HETE), which is involved in hypertension regulation and cerebral blood flow, suggest further connections with AD and stroke risks (Gonzalez-Fernandez et al., 2021). Preventing AD relies on genetic factors, cardiovascular health, and lifestyle changes, including smoking cessation and proper nutrition (Khan et al., 2023). Therefore, focusing on cardiovascular health through lifestyle modifications and nutrition is crucial for minimizing the risk of AD and related health issues.
Chronic inflammation and oxidative stress exacerbate these health conditions, and lifestyle factors like poor diet and inactivity contribute to obesity and metabolic issues impacting cognitive and cardiovascular health. While high-density lipoprotein (HDL) cholesterol is known for lowering heart disease risk, some studies have found that elevated HDL cholesterol levels may increase risks of dementia and other health problems, highlighting the need for further exploration of these associations. In older populations, metabolic syndrome (MetS) has been linked to an increased risk of cognitive decline and cardiovascular issues, emphasizing the importance of managing lipid levels to support brain health. The brain contains substantial cholesterol, essential for nerve cell function. The transport of lipoproteins, such as low-density lipoprotein (LDL) and HDL, along with apolipoproteins like ApoE, plays a crucial role in brain fat processing. The ε4 variant of ApoE is mainly associated with a heightened susceptibility to late-onset AD. Studies surrounding obesity-related dementia implicate sedentary lifestyles, chronic stress, and genetic predisposition, particularly regarding specific ApoE alleles. The relationship between cholesterol transport and cognitive decline is an evolving research area, often yielding conflicting results. Research suggests that abdominal obesity might protect cognitive health in older adults (Pereira et al., 2023). Variations in clusterin expression may impact lipid transport in the brain, structural integrity, and cognitive function. Clusterin, or apolipoprotein J (ApoJ), is a glycoprotein associated with protein folding and linked to AD, metabolic disorders, and cardiovascular diseases. At the same time, its connection to insulin resistance and dyslipidemia implies potential as a biomarker for linking AD risk with obesity-related metabolic dysfunction.
The oral microbiota may also play a role in AD progression through various mechanisms, including oxidative stress, vascular complications, neurotoxicity, and inflammation. By causing systemic inflammation, oral bacteria could disrupt the blood-brain barrier, allowing bacteria to enter the brain. Furthermore, since amyloid-β has antibacterial properties, the interactions between oral microbiota and Aβ accumulation may be significant. Oral health can also influence dementia risk by affecting sleep, physical activity, glucose metabolism, and cardiovascular health. Moreover, midlife obesity has been linked to 7.3% of AD cases, and approximately 37% of dementia patients over 65 are also diagnosed with diabetes.
Emerging evidence suggests that obstructive sleep apnea (OSA) and other sleep disorders could increase dementia risk. The exact mechanism underlying the direct relationship between OSA and elevated dementia risk remains unknown, even as numerous epidemiological studies investigate this association with cognitive decline. These factors are often assessed in more extensive cohort studies, but the precise interplay with OSA is not yet fully understood. Clarifying these connections could enhance clinical practices and dementia prevention strategies by improving risk prediction and informing personalized treatments, particularly for individuals with mild OSA. Lifestyle and overall health delay mild cognitive impairment as individuals age. Mild cognitive impairment is often seen as a precursor to AD or dementia, and research indicates that these cognitive disorders can be influenced by ethnicity. For example, some studies have found a more significant association of symptoms in Mexican Americans compared to non-Hispanic White populations (Morgenstern et al., 2024). Conversely, research involving older Black and White Brazilian communities shows minimal racial differences in the experience of these symptoms (Wilson et al., 2021). While the influence of gender is also examined, it has been determined that brain health and neuroprotection largely depend on everyday lifestyle choices and comorbidities (Mian et al., 2024).
3.3 Early detection and long-term effects of Alzheimer’s disease
AD’s preclinical stage can span 20–50 years before noticeable symptoms, during which Aβ and oligomer formation changes begin to affect cognitive functions. However, daily life remains largely unaffected. Individuals may notice minor cognitive declines that minimally impact daily activities as the disease progresses to the symptomatic stage. Late-onset AD typically arises after age 65, while early-onset AD is less common and often linked to hereditary factors (Li et al., 2022). Key markers of AD pathogenesis include senile plaques, synaptic loss, and neurofibrillary tangles, primarily affecting memory-related areas in the brain, such as the hippocampus and cortex. Despite advancements in identifying AD biomarkers, the disease’s exact mechanisms remain poorly understood (Monteiro et al., 2023). Noteworthy symptoms of AD’s long-term effects include memory loss and impaired judgment, complicating decision-making and task management as the disease advances. The accumulation of plaques and tangles results in irreversible brain damage, impairing cognitive function, mood, and behavior (DeTure and Dickson, 2019). At the same time, memory impairments can lead to falls and challenges in maintaining adequate nutrition and hydration (Volkert et al., 2024). These enduring effects highlight the significance of early intervention and continuous supportive care before mild to moderate symptoms of cognitive impairment begin to affect a patient’s quality of life. Utilizing biomarkers to evaluate plaques and tangles while patients remain asymptomatic may help slow the progression of AD before irreversible damage occurs (Mozersky et al., 2022).
Integrating multi-omics data with machine learning and artificial intelligence (AI) offers a deeper insight into AD pathologies. AI can simultaneously analyze the relationships between various biological components of omics studies, resulting in a comprehensive dataset model. This methodology has also proven beneficial in treating and preventing other diseases, including identifying biomarkers and developing early detection strategies. In cardiovascular diseases, advancements in multi-omics and AI utilizing RNA sequencing, whole-genome sequencing, and other classification models have achieved accurate risk predictions and efficient patient classification for further treatment (DeGroat et al., 2024). In cases of leukemia, machine learning, and deep learning methods employing multi-omics have refined unclassified datasets for predicting blood cancer outcomes. Analysis techniques used in this field include gradient boosting, logistic regression, recurrent neural networks, and feedforward neural networks. These diverse datasets, which consider patient age, sex, mutation type, treatment methods, and chromosomal data, contribute to improved care and treatment (Abbasi et al., 2024). Patients undergoing dialysis have benefited from personalized medical treatments driven by AI for kidney disease. Kidney Online program utilizes deep learning and health data to offer recipes, lifestyle interventions, early health warnings, answers to inquiries, and follow-up plans. Research indicates that this intelligent online care system effectively reduces the risk of worsening kidney disease (Liu et al., 2024). As illustrated across various diseases and health conditions, integrating these multi-omics approaches into geriatric medicine and AD research can enhance patient care through comprehensive dataset assessments. Employing new technologies like AI, stem cells, and multi-omics to bridge gaps in AD research will facilitate the creation of human models to achieve improved outcomes in personalized neuropsychiatric care (Tanaka, 2025).
4 Status of treatment
Treatments for AD are designed to address the underlying mechanisms of Aβ production and alleviate symptoms. Two main classes stand out among the approved pharmacological treatments: N-methyl aspartate receptor antagonists (NMDA) and cholinesterase inhibitors. NMDA receptor antagonists regulate glutamate activity, thereby preventing excitotoxicity due to excessive glutamate release, often linked to amyloid-induced neuronal damage. Meanwhile, cholinesterase inhibitors preserve acetylcholine levels by inhibiting the enzyme cholinesterase, which helps mitigate cognitive decline. Beyond pharmacotherapy, several herbal treatments exhibit the potential to influence the biological processes associated with AD.
Research into bioactive compounds from various plants highlights their promising applications in human health and natural remedy therapies. Phytochemicals are bioactive compounds found in plants that can be used in clinical applications when extracted. Panax ginseng is rich in compounds like gintonin, ginseng, and polysaccharides, all recognized for their health-promoting properties (Table 1). Gintonin is explicitly involved in AD management by modulating neurotransmitter levels, including acetylcholine, dopamine, and norepinephrine, promoting autophagy, and diminishing Aβ production. Natural products, including herbs and extracts, have been shown to target tau protein formation and amyloid beta effectively (Aβ) plaques due to their antioxidant properties (Lobine et al., 2021). These harmful agents arise from radical and non-radical oxygen species, reactive nitrogen species, and reactive oxygen species, which are highly chemically reactive and can contribute to oxidative damage in Alzheimer’s disease (AD), thereby impairing neuronal function. Antioxidants help mitigate this toxic stress by transforming free radicals into harmless byproducts, providing neuroprotection (Chen et al., 2021). Examples of antioxidants include Centella Asiatica, Withania somnifera, and Crocus sativus (Zieneldien et al., 2022). In addition to antioxidant therapies, anti-inflammatory treatments have also been found to support neuronal health. Studies in mice have demonstrated that mulberry extract can provide neuroprotection, reducing neuronal and astrocytic apoptosis. This effect is associated with an increase in anti-inflammatory cytokines (such as IL-4) and a decrease in pro-inflammatory cytokines (such as IL-1β, IL-6, and TNF-α), suggesting its potential as a therapeutic agent for neurodegenerative diseases like AD (Liu and Du, 2020). Similar studies have investigated on Hypericum perforatum extract and found it exhibits biological activity against Aβ-related effects (El Menuawy et al., 2024). Other anti-inflammatory herbs, such as Scutellaria baicalensis, Bacopa monnieri, and Chlorella zofingiensis, have also been shown to alleviate cognitive impairment; however, their specific neuroprotective effects in the context of AD remain unclear (Peng and Zhou, 2024). A range of additional herbal remedies with anti-inflammatory and neuroprotective properties may contribute to reducing neuronal stress and fostering repair mechanisms via the regulation of long non-coding RNAs (lncRNAs) and microRNAs (Li et al., 2024). Herbs like Ginkgo biloba and Anemone altaica offer distinctive therapeutic benefits. At the same time, ashwagandha is noted as a nerve tonic and antioxidant, potentially enhancing memory and cognitive function by raising acetylcholine levels (Mikulska et al., 2023; Supplementary Table 1).
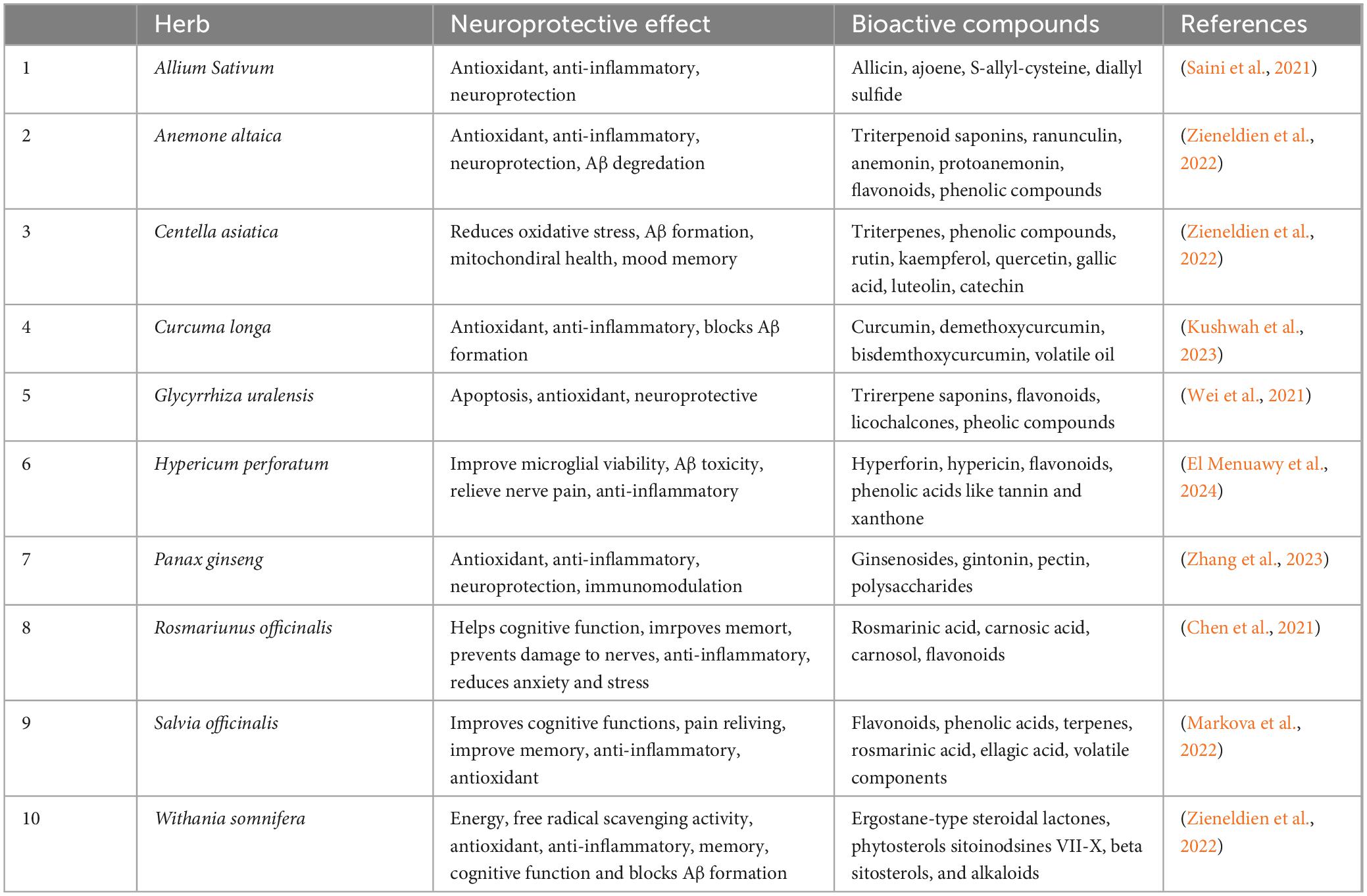
Table 1. Neuroprotective properties of selected herbs commonly used in AD treatment to restore and enhance memory and cognitive function.
The bioactive compounds identified and isolated from these medicinal plants include flavonoids, phenolic lignans, tannins, polyphenols, triterpenes, sterols, and alkaloids. Research indicates these phytochemicals exhibit antioxidant, anti-inflammatory, anti-amyloidogenic, anti-tau, and anticholinesterase activities (Chen et al., 2021).
Despite the availability of these treatments, it is important to note that there is currently no cure for AD; existing therapies broadly address symptomatic relief without stopping disease progression. However, future therapies, including immunotherapies that target amyloid plaques, are undergoing clinical trials and show promise for more specific interventions. Additionally, innovative high-throughput multi-omics approaches are making strides in identifying biomarkers that could aid in understanding AD’s pathophysiology and refining therapeutic strategies. These approaches strive to uncover reliable biomarkers associated with the characteristic features of Alzheimer’s, such as Aβ plaques and neurofibrillary tangles, highlighting the significant hurdles in biomarker discovery and disease characterization.
5 Multi-omics studies to detect early biomarkers for Alzheimer’s disease
Multi-omics studies that integrate comprehensive molecular data analysis across different stages of the disease, including preclinical, symptomatic, and advanced stages, offer valuable insights into the mechanisms of AD causes and progression while improving diagnostic accuracy. Biomarkers, measurable molecular indicators of disease presence and progression, play a crucial role in this approach. In AD, biomarkers obtained from biofluids such as urine, blood, and plasma can offer important molecular signatures. These signatures aid in early detection, targeted therapies, and ongoing disease monitoring, ultimately contributing to the development of potential treatments.
While AD and other common neurodegenerative disorders are characterized by the pathological hallmarks of tangles and plaques, identifying reliable biomarkers has been a significant challenge. Over the years, high-throughput multi-omics-based approaches have been employed to explore dependable biomarkers, creating new opportunities to understand the pathophysiology associated with different molecular states. Multi-omics studies advance our understanding of AD by examining genetic variation, regulatory mechanisms, and epigenetic modifications, exploring a broad spectrum of biological processes to identify biomarkers and elucidate disease pathology. Specifically, genomics, transcriptomics, and Epigenomics provide complementary insights into genetic predisposition, gene expression patterns, and heritable modifications that influence disease progression (Figure 1).
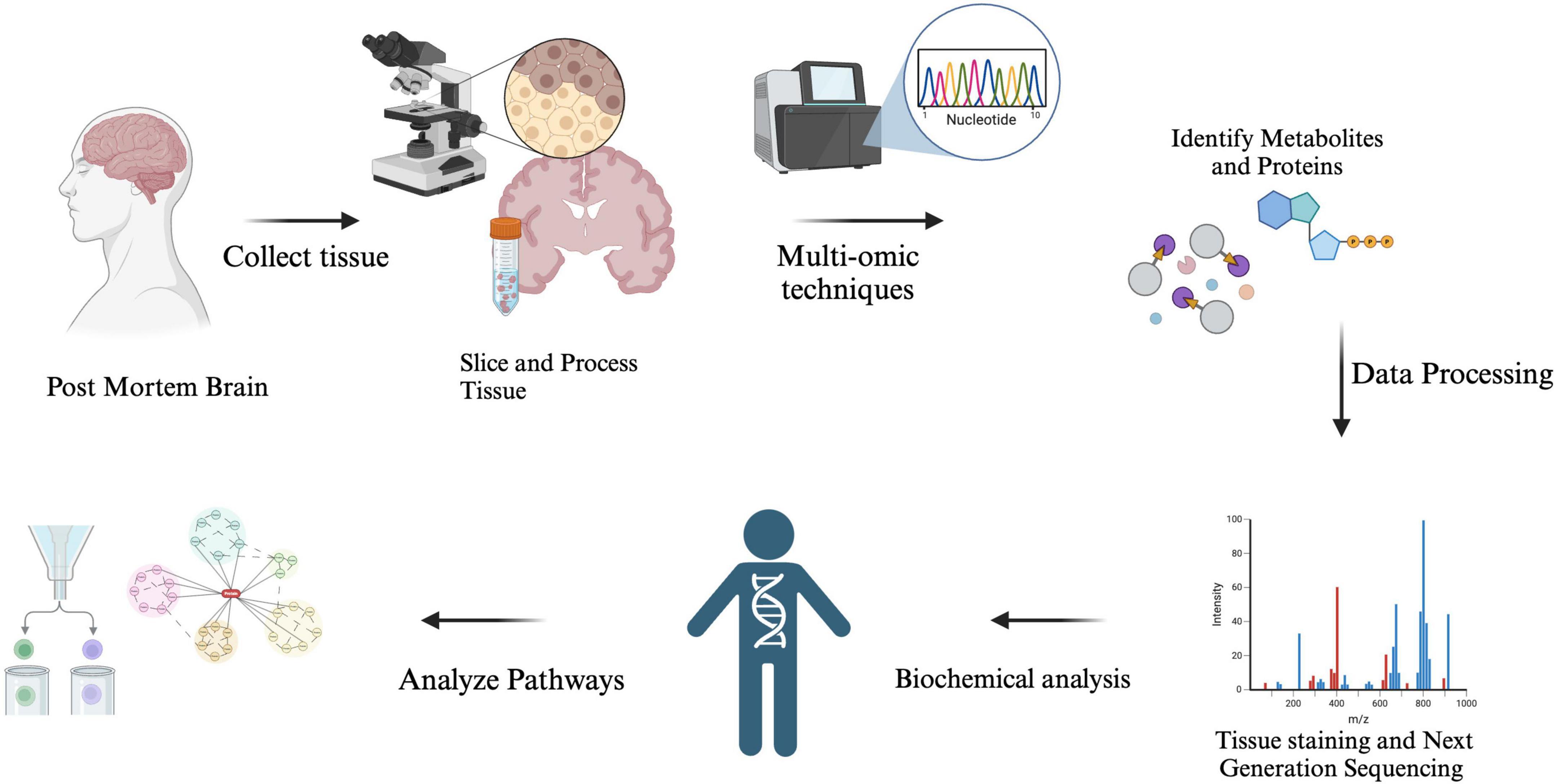
Figure 1. The analysis of Alzheimer’s disease (AD) biomarkers begins with tissue sampling, where brain tissue is collected post-mortem through biopsy. The samples are then prepared, processed, and molecular extraction is performed to isolate proteins, RNA, or metabolites. Researchers utilize techniques such as mass spectrometry and RNA sequencing, combined with computational models, to detect key biomarkers that aid in diagnosis, disease tracking, and the development of potential treatments.
5.1 Genomics
The first genetic risk factor identified for AD is the dominant amyloid precursor protein (APP), a type I transmembrane protein cleaved to release Aβ. Thirty mutations in the APP gene on chromosome 21 have been discovered, with twenty-five of these mutations associated with AD and Aβ accumulation (Sirisi et al., 2024). Interestingly, a protective mutation, A673T, has been found to reduce Aβ secretion and lower the risk of developing AD. Alongside APP, other significant genes involved in AD include Presenilin-1 (PSEN1) and Presenilin-2 (PSEN2) (Kim et al., 2024). Mutations in PSEN1 account for around 80% of monogenic AD cases, while PSEN2 mutations are rarer and have a limited effect (Supplementary Table 1). It has been proposed that PSEN1 mutations may hinder neurogenesis by increasing the susceptibility of neural stem cells to amyloid toxicity, potentially leading to cognitive decline (Maksour et al., 2024). These genes influence γ-secretase activity, thereby modulating the ratios of Aβ by elevating Aβ42 levels and decreasing Aβ40 levels; however, the consequences of these alterations in AD continue to be explored (Stanciu et al., 2022). Various factors, including metabolic stress and altered transcription factors, can disrupt cellular homeostasis, further contributing to neuronal injury. Other genes implicated in AD are ATP-binding cassette transporter A1, clusterin, bridging integrator 1, evolutionarily conserved signaling intermediate in the toll pathway, estrogen receptor, and numerous vitamin D receptor gene polymorphisms (Breijyeh and Karaman, 2020). The downregulation of XRCC6, which is crucial for initiating DNA repair, has been observed in neurons characterized by AD. Additionally, age-related DNA damage that occurs during the expression of learning-related genes may accelerate the progression of AD (Lin et al., 2020).
Furthermore, genetic mutations differ based on ancestry, underscoring the importance of population-specific research on AD. Astrocytes in individuals with AD display changes in gene expression, particularly concerning glutamate receptor subunits, which can disrupt molecular pathways and ion balance (Young-Pearse et al., 2023). We present here several variants in individuals associated with AD that have been reported in ClinVar. These variants may also be suitable for application in genome editing technologies (Table 2). Furthermore, a GeneMania interaction network was deciphered between genes responsible for AD (Figure 2).
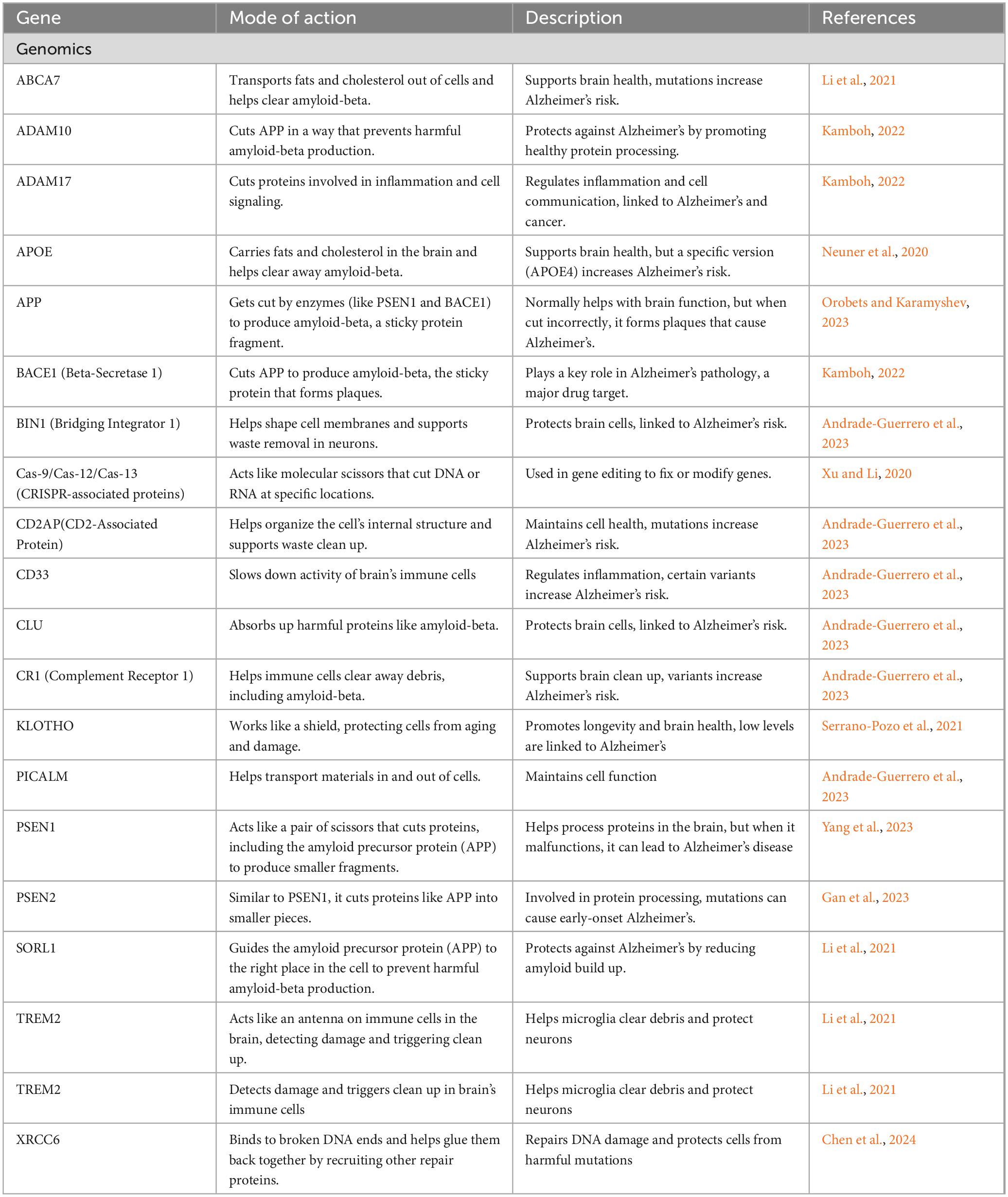
Table 2. Characterization of potential genetic markers identified via genomics research associated with AD and dementia risk.
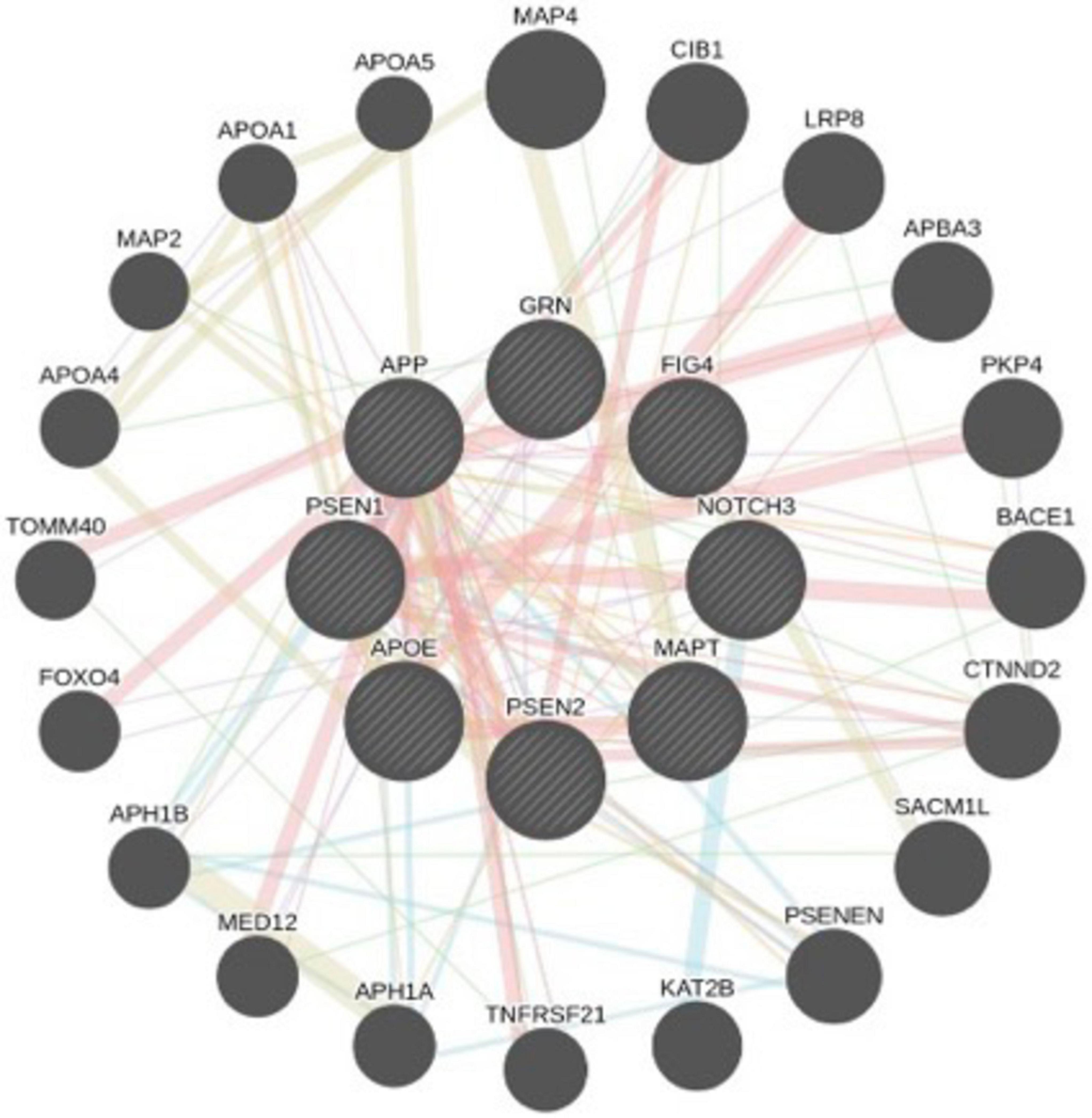
Figure 2. Systems network of AD-associated genes, with key genes highlighted in larger circles. Physical interactions and co-expression patterns are analyzed using a network prediction database. Red lines indicate physical interactions, purple lines represent co-expression, green lines denote genetic interactions, and blue lines signify pathway associations or colocalization within the same organelle. This network visualization offers insights into the complex molecular interplay underlying the pathology of Alzheimer’s disease.
Over the years, advancements in genome editing have progressed significantly, evolving from zinc finger nucleases and transcription activator-like effector nucleases (TALENs) to clustered regularly interspaced short palindromic repeats (CRISPR) methods (Katam et al., 2022). CRISPR-Cas9, a powerful genome-editing tool that can target a sizable genetic variant, aids in uncovering molecular mechanisms behind neurodegeneration, identifying potential therapeutic targets, and exploring gene-editing strategies that might ultimately prevent or treat AD. Scientists can investigate how specific genetic changes impact AD pathology by introducing or correcting mutations in these genes through cell cultures and animal models (Xu and Li, 2020). These models allow researchers to observe the effects of genetic modifications within a whole organism, which is essential for confirming physiological outcomes and understanding how genetic variations influence AD development in vivo (Blanchard et al., 2022).
The Cas system consists of two classes with six subtypes that utilize different Cas proteins depending on the genetic material and system configuration. Cas9 is a single-protein DNA cutter used in gene editing, while Cas12 targets DNA and Cas13 modifies RNA. Cas12 can detect small mutations, including those that affect DNA methylation, whereas Cas13 focuses on RNA modifications in tau proteins, characteristic of AD pathology. Researchers have applied CRISPR-Cas9 to edit genes such as APP, PSEN1, and APOE, contributing to understanding pathways related to AD. The modification of APOE alleles in vitro, particularly in neurons and glial cells derived from human stem cells, marks a significant advancement in AD research (Rahimi et al., 2024). Researchers have successfully altered the genetic sequence of the ApoE gene within cultured human neurons and glial cells using CRISPR-Cas9. This development enables the creation of more accurate cellular models of AD that mirror genetic variations found in humans, such as the ApoE4 allele, which is a significant risk factor for developing the disease. However, despite the progress made in human cell culture, the gene-editing technique has not yet been tested in ApoE knock-in mice, which is a crucial next step for advancing research (De Plano et al., 2022).
5.2 Transcriptomics
The potential for modifying AD-related gene expression presents exciting therapeutic opportunities. Transcriptomic techniques reveal alterations in gene expression that contribute to AD risk factors, including the upregulation of stress and inflammatory response genes, non-coding RNAs, alternative splicing events, and copy number variants, opening exciting therapeutic opportunities (Bagyinszky et al., 2020). Several differentially expressed genes (DEGs) across various cell types have been identified using the single cell RNA seq, enhancing our understanding of the molecular mechanisms underlying AD (Spurgat and Tang, 2022). Comparative analyses of brain cells from AD patients and age-matched controls emphasize the connection between mitochondrial dysfunction and AD. Notable genes like ZFP36L1, RERE, PURA, OGT, SPCS1, SOD1, and NDUFS5 show consistent expression alterations across 22 brain datasets (Marmolejo-Garza et al., 2022; Table 2). Transcriptomic changes reflect more widespread stress responses at later stages of AD that correlate with increasing levels of brain damage. Single-cell and single-nucleus RNA sequencing (sc/snRNA-seq) techniques have determined differentially expressed mitochondrial genes, highlighting their significant post-transcriptional roles in energy demand regulation. Expression levels of mitochondrial RNA can vary by tissue, reflecting the diverse functions of mitochondria in different cellular environments (Mei et al., 2023). Astrocytes, vital for maintaining brain homeostasis, exhibit significant transcriptomic shifts in AD. There is an upregulation of inflammatory and stress response genes, including CRYAB and GFAP, while glutamate metabolism and synaptic remodeling genes, such as SLC1A2 and SLC1A3, experience downregulation (Saura et al., 2023). Aβ proteins compromise synaptic plasticity and disrupt BDNF-TrkB retrograde signaling pathways. Treatments with Aβ1-42 have led to increased levels of axonal mRNA for AD-related genes like APP, ApoE, and CLU, suggesting their involvement in disease progression (Gao L. et al., 2022; Gao X. et al., 2022). Single-nucleus RNA sequencing has revealed cell-specific changes in gene expression during the early stages of AD, impacting processes like myelination, inflammation, and neuronal survival.
5.3 Epigenomics
Advanced technologies, including proteins like CLOuD9 and light-activated dynamic looping (LADL), are used to engineer chromatin for precise gene regulation, highlighting the importance of 3D chromatin organization. CRISPR-GO and live-cell imaging enabled us to study chromatin changes in real-time (Rahman and Roussos, 2024), while epigenetic editing using CRISPR-Cas9 and pharmacological interventions shows promise as a therapeutic approach for AD (Fisher and Torrente, 2024). DNA methylation affects gene expression and chromatin accessibility, ultimately influencing the production of Aβ, calcium homeostasis, and neuronal survival, implicated in oxidative stress and synaptic plasticity (Villa and Combi, 2024). This process links environmental factors, such as homocysteine levels, to the progression of AD (Martinez-Feduchi et al., 2024). Oxidants released by immune cells, particularly from microglia, can alter DNA methylation, further exacerbating neuroinflammation and oxidative stress (Seddon et al., 2024). MicroRNAs, specifically miR-451a and miR-455-3p, play a regulatory role in neurotrophic factors like brain-derived neurotrophic factor (BDNF), neuroinflammation, and neurotransmitter balance, thereby connecting mild behavioral impairments to amyloid/tau pathology (Angelopoulou et al., 2024). Emerging research has identified novel post-translational modifications (PTMs) such as phosphorylation, acetylation, and ubiquitination as potential therapeutic targets (Qin et al., 2024; Table 2). Moreover, the epigenetic regulation of non-coding RNAs affects shared genes such as APOE, BDNF, ACE, FTO, and FNDC5, which are important for muscle mass, mobility, and cognition (Raleigh and Orchard, 2024). BRD4, a critical chromatin remodeling factor, plays a complex role in aging and disease (Sun et al., 2024). Histone acetylation, which is primarily affected in AD, is regulated by histone deacetylases. Inhibitors of these deacetylases can reverse hypoacetylation, improving cognition, memory, and neuroplasticity in preclinical models by promoting neuronal gene transcription and reducing tau and amyloid dysregulation (Pereira et al., 2024).
5.4 Proteomics
Recent advancements in mass spectrometry, dimethyl labeling, isobaric tandem mass tags (iTRAQ), and laser capture microdissection have enabled comparisons between symptomatic and asymptomatic Alzheimer’s patients’ brains against those of healthy controls. These innovative methods allow researchers to integrate all cellular proteins, leading to a comprehensive understanding of a system’s biology combined with minimally invasive diagnostic methods. Three commonly used techniques include: 1. cerebrospinal fluid (CSF) collection to assess central nervous system health and neuronal damage; 2. Plasma collection is cost-effective and contains proteins from all body tissues but is complicated by high albumin content that affects protein extraction; 3. Urine collection may contain plasma proteins and potential biomarkers like SPP1, GSN, and IGFBP7 (Jain and Sathe, 2021).
Utilizing proteins as biomarkers and studying their alterations aims to enhance early diagnosis. This breadth of analysis encompasses thousands of proteins involved in energy metabolism, glycolysis, oxidative stress, apoptosis, signal transduction, and synaptic function. However, specific issues arise, such as using polystyrene tubes, which can lead to the loss of “sticky” proteins like Aβ and introduce blood contamination that may degrade proteins, complicating biomarker analysis (Awasthi et al., 2022). Identifying specific biomarkers for Alzheimer’s is made challenging by the presence of overlapping pathologies in many patients. In neural networks, protective proteins include cytoskeleton cross-linking proteins like moesin, ezrin, and radixin, while inflammatory proteins include CAV1, COL6A1, and COL6A3 (Rayaprolu et al., 2021). Other noteworthy proteins in this context are TNF-α and miR-224, which is down-regulated in AD patients, as well as Cystatin C, angiotensin-converting enzyme (ACE), SUMO1, and Chitinase 3-like 1; Table 3). Although proteins such as β2-microglobulin and y-globulinsare associated with AD, they have not yet been validated as potential biomarkers (Mayo et al., 2021). Paraoxonase 1 (PON1) shows promise as a risk factor for AD, mainly due to its anti-inflammatory and anti-apoptotic properties, as low PON1 activity levels correlate with advanced disease.
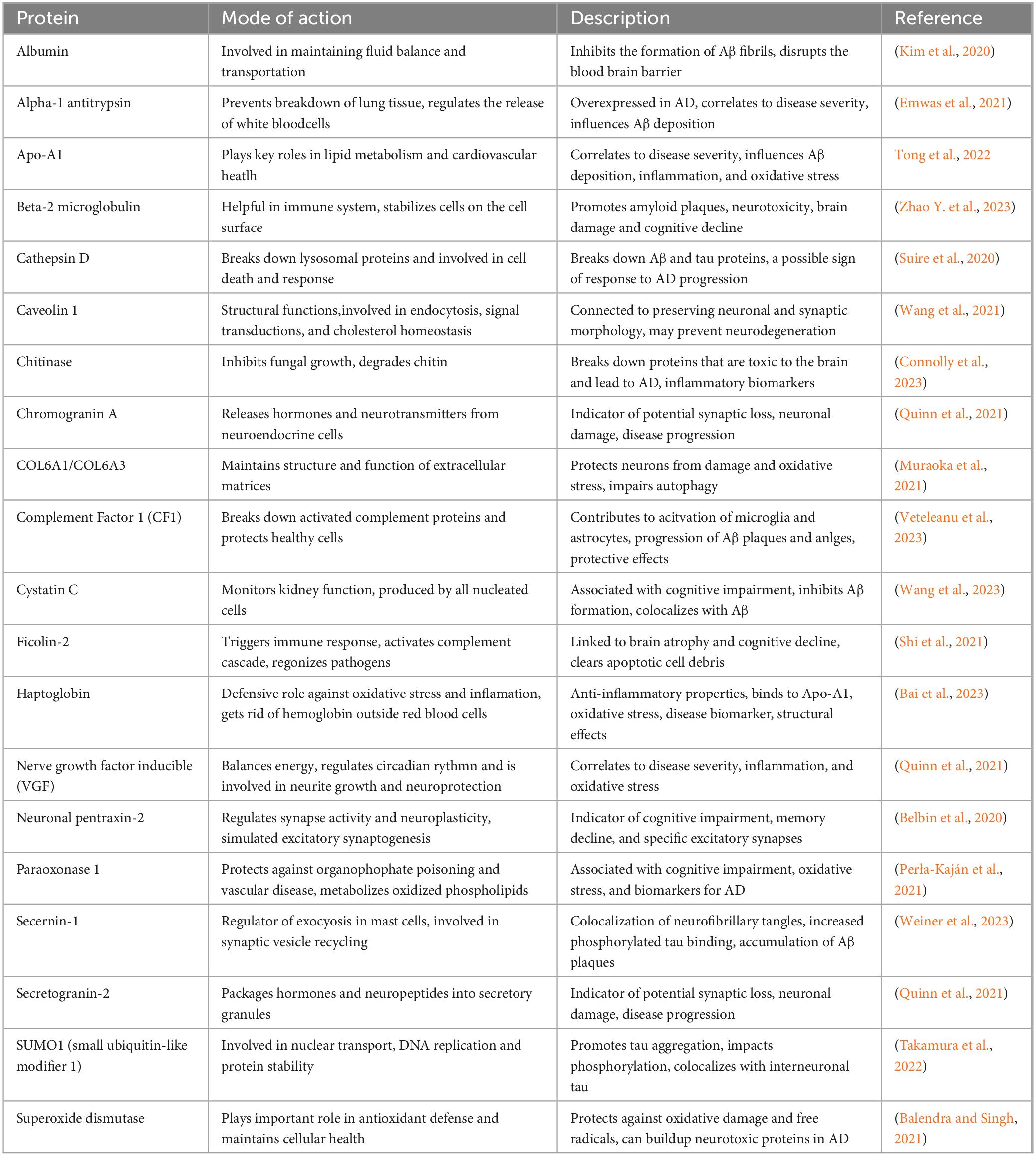
Table 3. Characterization of potential genetic markers identified via proteomics research associated with AD pathology.
In contrast, areas like the sensory cortex, motor cortex, and cerebellum show less impact (Marsillach et al., 2020). Challenges arise from small sample sizes that complicate the detection of protein differences across multiple comparisons. In contrast, larger sample sizes may yield too many variations, making it difficult to establish consistency.
5.5 Metabolomics and lipid-omics
Over the past decade, metabolomics and lipidomics have made significant advances in identifying critical changes in metabolites that affect mitochondrial function, neuroinflammation, and cognitive decline. Lipidomics focuses explicitly on the role of lipid metabolism in neuronal health and amyloid pathology. Genetic predisposition to diseases and traits can be quantified through polygenic scores derived from metabolomic data and multi-omics approaches (Joshi et al., 2023). Various metabolomic platforms are available, each with advantages and challenges, especially concerning sensitivity to external variables, reproducibility, and costs (Chen et al., 2022). Targeted metabolomics focuses on specific metabolites based on hypotheses, while untargeted methods enable broad profiling of numerous metabolites within a sample (Trifonova et al., 2023). The early applications of metabolomics in AD began in 2009 with gas chromatography-mass spectrometry and linear ion trap (LTQ) orbit trap technologies. However, challenges such as variability and small sample sizes prevented the discovery of statistically significant biomarkers (Reveglia et al., 2021).
Later research utilizing ultra-performance liquid chromatography (UPLC) with a hybrid quadrupole time-of-flight (Q-TOF mass spectrometer) and gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) platforms analyzed participants with AD, mild cognitive impairment, and healthy controls, identifying distinct metabolites like arachidonic acid, N, N-dimethylglycine, and thymine that differentiate AD patients (Yin et al., 2023). Furthermore, decreased levels of oleamide, histidine, monoglycerides, and increased phenylacetylglutamine were noted in AD patients, suggesting potential inflammatory responses (Table 4). Further analyses indicated elevated levels of cortisol and cysteine and reduced uridine in patients. Thymidine and uracil are crucial in nucleic acid metabolism, influencing mitochondrial function. Taurine, a key amino acid in the central nervous system, has been proposed to enhance cognitive function and protect against memory loss without impairing motor skills (Ramírez-Guerrero et al., 2022). Several substances with neuroprotective properties, such as kurarinone, tauroursodeoxycholic acid (TUDCA), and curcumin, have shown promise in enhancing motor behavior and reducing neuroinflammation. However, these treatments have yet to yield specific biomarkers for assessing human neuroprotection (Franco et al., 2024). Metabolomic changes linked to AD include fluctuations in phospholipids, amino acids, and other metabolites and altered kynurenine pathways. Metabolic profiling has revealed elevated levels of alanine, glutamate, and glycerophosphocholine, while decreased lactate and N-acetyl aspartate have been reported in AD patients, indicating a potential signature for the disease (Vignoli and Tenori, 2023). Research regarding insulin resistance has identified glucose and fructose as key metabolic biomarkers, while heightened ceramide levels have been associated with mitochondrial dysfunction and inflammation in AD patients (Amin et al., 2023). Notably, differences have been observed between genders, such as a lower D-serine ratio in women with AD compared to men (Yin et al., 2023). D-serine binds to receptors, activating the N-methyl-D-aspartate receptor (NMDAR) and mediating excitotoxicity. Studies indicate that D-serine contributes to excitatory neuronal damage in the hippocampus, influences neuroinflammation, and affects amino acid balance. Reducing D-serine levels in mouse models has decreased hippocampal neuronal death and neuroinflammation, presenting a viable NMDAR-based treatment strategy for AD (Ni et al., 2023). Nevertheless, a significant challenge in metabolomics is the inconsistency of results due to variability in metabolite biomarkers based on sample sources. Despite identifying numerous potential biomarkers, the statistical robustness is often limited due to challenges in controlling external influences (Batra et al., 2024).
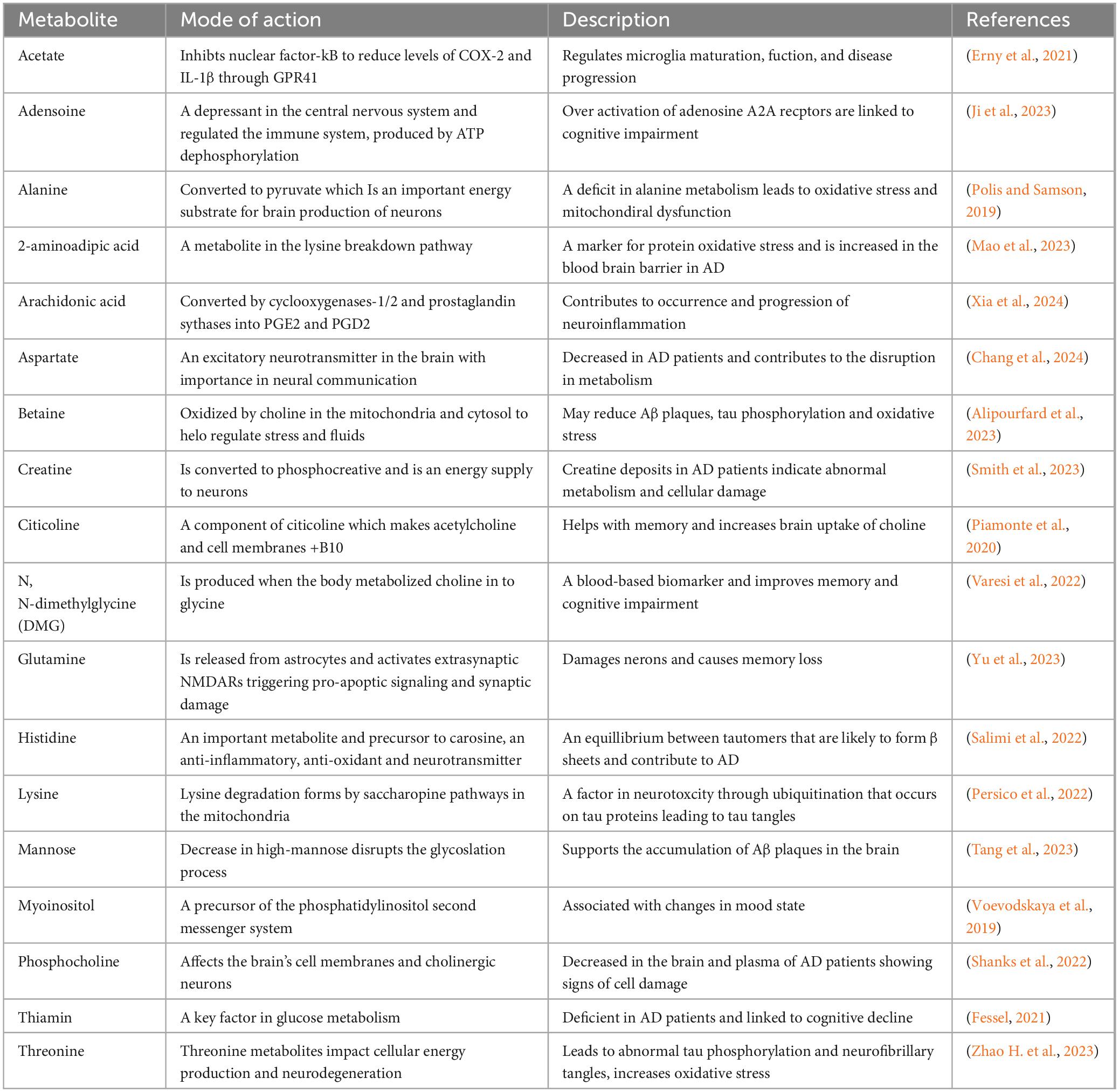
Table 4. Exploring genetic markers associated with Alzheimer’s disease and dementia risk through metabolomics and lipidomics research.
Altered lipid metabolism plays a significant role in aging, increasing plasma triglyceride and lipoprotein levels while decreasing triglyceride clearance and lipoprotein lipase activity. Peroxisomal disorders, which result in inherited ether lipid deficiencies, have been linked to AD (Hussain et al., 2019). These metabolic changes affect lipid transport and biochemical pathways across various organs (Chung, 2021). Mitochondria are crucial for lipid metabolism, yet their functionality declines with age, emphasizing the importance of metabolomics and lipidomics in AD research (Yin et al., 2023). The lipid metabolites and pathways strategy, or Lipid MAPS, categorizes lipids into eight groups: fatty acids, glycolipids, glycerophospholipids, sphingolipids, sterols, phenols, saccharolipids, and polyketides, each identified uniquely (Meikle et al., 2021). The brain, abundant in glycerophospholipids and sphingolipids, relies on these lipids for structural integrity and functionality. Moreover, decreased levels of plasmalogens have been observed with aging and AD; however, the results of replenishment treatments are inconsistent. Lipids are essential for numerous cellular processes, including signaling, maintaining membrane structure, and biological messaging. Although findings remain inconclusive, an upregulation of brain cholesterol synthesis in AD has been observed, and cholesterol accumulation in senile plaques suggests its potential role in the disease’s pathophysiology (Yin et al., 2023).
6 Advances in diagnostics and personalized medicine
Advancements in radiomics, neuroimaging, and machine learning are transforming the early diagnosis and management of AD. By integrating brain imaging techniques with AI-driven models, researchers can enhance the accuracy of disease prediction, monitor progression, and personalize medicine. Radiomic analyses provide insight into the structural and functional changes in the brain. Meanwhile, machine learning algorithms examine large datasets to identify biomarkers and predict disease trajectories (Figure 3).
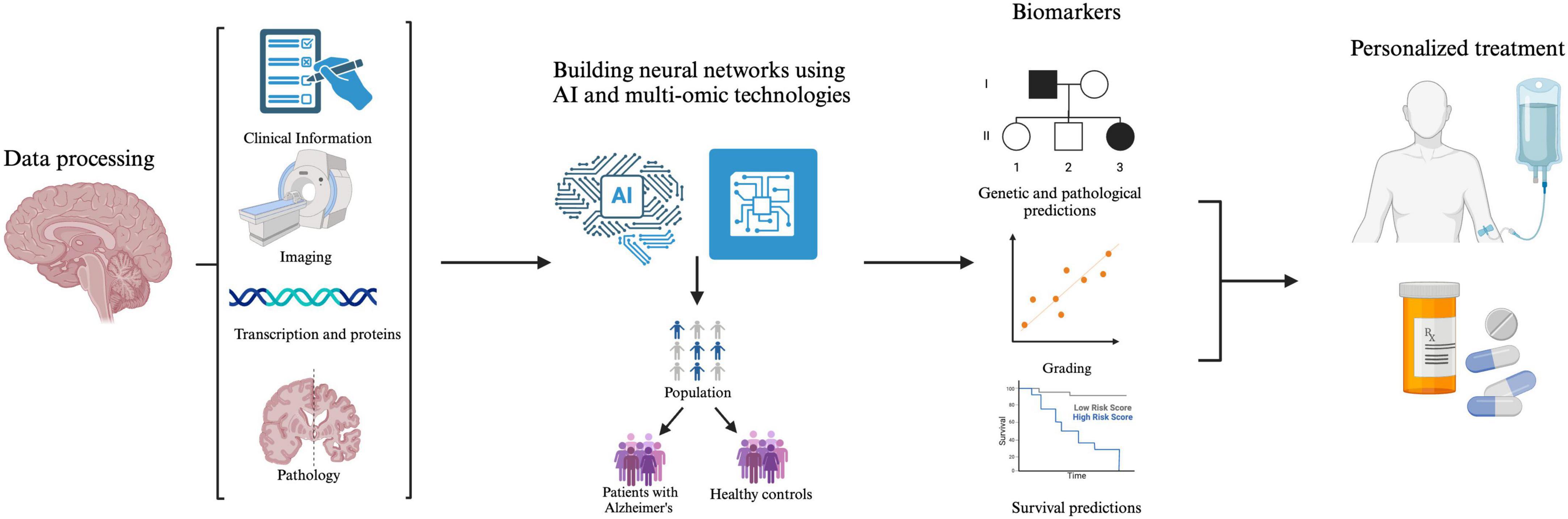
Figure 3. The integration of multi-omics data, facilitated by artificial intelligence (AI), provides a more comprehensive understanding of AD pathology. By analyzing this data, models can identify dysregulated pathways and various biomarkers, improving patients’ lives through early diagnosis, risk assessments, and targeted therapeutic interventions.
6.1 Radiomics and neuroimaging
Brain imaging and genomics are key components in systematically analyzing AD, integrating image preprocessing, region of interest identification, model building, genomic data extraction, and downstream analysis. Together, these methodologies contribute to the precision medicine approach for AD imaging biomarkers with genomic implications (Li and Luo, 2022). Radionics-based analysis and nuclear medicine tools, such as fluorodeoxyglucose (FDG), β-amyloid positron emission topography (PET), and dopamine transporter single proton emission computed tomography (SPECT), combined with advanced computer technology, can enhance classification and prediction rates for AD. The primary goal is the early diagnosis of mild to moderate cognitive impairment and monitoring its progression toward Alzheimer’s as the brain ages. Current studies utilize regions of interest-based radionics and support vector machine classifiers on PET imaging to assess decision-making accuracy in models describing AD’s stage and severity (Seo et al., 2022). Radiomic network modeling targeting the cerebellum shows promise for early identification of the preclinical stage of AD. These integrated models outperform traditional hippocampal models in patients with mild cognitive impairment, effectively predicting distinct risks associated with the progression of amyloid and tau pathologies (Chen et al., 2025). For example, models using 18F-FDG-PET images interpret deep-learning radionics to understand better and predict the pathway from mild impairment to AD. Techniques like Extreme Gradient Boosting (XGBoost) illustrate the focus areas within the model, while methods such as gradient-weighted Class Activation Mapping (Grad-CAM) and Shapley Additive exPlanations (SHAP) are crucial for identifying factors that influence predictions (Jiang et al., 2024).
6.2 Machine learning
Integrating AI with radiomics and brain imaging shows significant potential to enhance personalized medicine and clinical treatments for AD. Machine learning techniques, such as logistic regression and convolutional neural networks, are employed to develop diagnostic models that analyze various brain images for signs of AD (Bevilacqua et al., 2023). By utilizing AI to monitor and predict the formation of tangles and plaques, the approach to treating early-onset AD patients could be fundamentally transformed. GNNexplainer technology aids in identifying key variables and genetic pathways that play a crucial role in understanding treatment outcomes and success in clinical trials focused on AD (Aghakhanyan et al., 2023). Current research methodologies leverage mathematical models and computational techniques to simultaneously examine DNA and brain alterations. The amyloid-tau-neurodegeneration (ATN) framework, which encompasses amyloid, tau, and neurodegeneration, helps delineate the accumulation of these components and the resulting implications for brain function (Li et al., 2022). While a significant amount of AI research in dementia relies on the comprehensive Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset, which is praised for its size and accessibility, it has limitations, such as the underrepresentation of non-Alzheimer’s dementias and inherent biases, indicating the need for broader recruitment from memory clinics and the systematic collection of longitudinal data (Winchester et al., 2023). Emerging studies indicate that AI-driven evaluations of blood-based biomarkers and cerebrospinal fluid (CSF) markers, including plasma p-tau, the Aβ42/40 ratio, and neurofilament light (NfL), facilitate early detection of Alzheimer’s, presenting a less invasive alternative to traditional CSF tests and PET scans (Chatterjee et al., 2022). Advances in AI-powered genomic analysis, incorporating deep learning models and polygenic risk scores, enhance the identification of genetic variants linked to Alzheimer’s, allowing for more accurate risk assessments and personalized therapeutic strategies (Zhou et al., 2023). Machine learning enhances the creation of individualized treatment plans by analyzing diverse patient data, such as genetic profiles, brain scans, and medical histories, to identify patterns and predict treatment responses. These systems utilize various learning methods to analyze data, uncover new subtypes of AD, and interpret intricate datasets like brain images. By continuously tracking the patient’s condition and modifying treatment accordingly, AI aims to deliver more personalized care, increasing treatment effectiveness while minimizing side effects (Zhang et al., 2023). The rising interest in these studies among scientists underscores the need for advancements in AI models, particularly regarding how they incorporate biological networks and complex systems, including multi-omics approaches for enhanced pathway analysis. There is also a need to improve the generalizability and reproducibility of results to ensure statistical accuracy. However, despite these promising developments, these AI models are not yet ready for clinical application, as imaging alone cannot reliably diagnose AD and cognitive impairments in aging populations. The synergistic application of multi-omics and emerging brain imaging technologies holds immense potential to revolutionize the treatment landscape for patients with AD.
7 Limitations and current challenges
Current limitations on the pathology and even cure of Alzheimer’s disease (AD) are mainly due to a lack of consistent information and ethical testing. Data obtained from postmortem brains cannot provide the same educational value as insights from live brains. Although animal models aid scientists in understanding the disease, they fail to accurately replicate the symptoms of AD as they present in humans. This underlines the significance of leveraging advanced technologies to study and compare healthy aged brains with both early and late-onset Alzheimer’s patients, emphasizing the need for new research to transition into clinical trials promptly. With the genome predominantly transcribed in eukaryotes and non-coding elements playing substantial regulatory roles, there remains a limited understanding of how long non-coding RNAs (lncRNAs) influence neurodegenerative disease modulation. Recent advancements in next-generation sequencing (NGS) technology have led to the discovery of numerous lncRNAs, which warrant further investigation to clarify their regulatory functions and to enhance non-functional activity. Studies have also revealed differential methylation of microRNA (miRNA) and lncRNA genes in human hippocampal tissues affected by epilepsy, illustrating that lncRNAs participate in regulatory pathways related to inflammation and neuronal differentiation in the epileptic brain. This suggests that the differential methylation status of non-coding RNAs is crucial in the pathogenesis of neurodegenerative diseases. Given that lncRNAs affect neighboring genes, it would be intriguing to examine if mutations in these genes are associated with lncRNA modulation. If such relationships exist, they could lead to further inquiry into whether lncRNA sequences function as protospacer adjacent motifs (PAM) within the genome. A wiki-based web resource or tool may serve as an interface to predict interactions between protein-coding genes and lncRNAs. However, while the CRISPR/Cas9 system can delete lncRNA genes associated with AD, identifying functional regions and potential off-target effects from such interventions targeting the complex genomic architecture remains uncertain.
8 Conclusion
Alzheimer’s disease (AD) presents a significant global health challenge due to its rising prevalence and the absence of effective treatments. Integrating machine learning, artificial intelligence, and multi-omics technologies holds immense potential to transform AD care, allowing for minimally invasive, efficient, and more precise interventions. Nonetheless, numerous challenges persist, including the generation of vast datasets, variability among samples, divergent interpretations of biological functions, and the intricate characteristics of clinical phenotypes. Machine learning and artificial intelligence (AI) offer advanced tools to decipher complex datasets and uncover hidden patterns. However, a multidisciplinary approach and diverse cohorts characterized by detailed phenotyping are essential to improve precision medicine for dementia. Ongoing research aims to identify consistent omics signatures distinguishing between dementia subtypes, enhance biomarker development through high-dimensional data utilization, and elucidate missing heritability factors in genetic investigations. Understanding the interplay between genetics and dementia risk factors alongside associated biological processes is vital. Efficient techniques for detecting preclinical and prodromal dementia are critical for effective secondary prevention strategies. Recent advancements in blood-based biomarkers have made widespread monitoring feasible, and preventative initiatives are being tailored using digital tools for remote cognitive and behavioral tracking. Researchers could develop precision preventive methods that align risk reduction with optimal therapeutic allocation by establishing at-risk cohorts based on cardiovascular and genetic risk factors interactions.
Author contributions
MC: Formal Analysis, Investigation, Software, Validation, Writing – original draft, Writing – review & editing, Methodology. KK: Formal Analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing, Conceptualization, Supervision, Funding acquisition, Resources. PS: Formal Analysis, Investigation, Methodology, Software, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Science Foundation grant # 2150087 (MC) and Title III at Florida A&M University (KK).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2025.1591796/full#supplementary-material
References
Abbasi, E. Y., Deng, Z., Ali, Q., Khan, A., Shaikh, A., Reshan, M. S. A., et al. (2024). machine learning and deep learning-based integrated multi-omics technique for leukemia prediction. Heliyon 10:e25369. doi: 10.1016/j.heliyon.2024.e25369
Aghakhanyan, G., Di Salle, G., Fanni, S. C., Francischello, R., Cioni, D., Cosottini, M., et al. (2023). Radiomics insight into the neurodegenerative “hot” brain: A narrative review from the nuclear medicine perspective. Front. Nucl. Med. 3:1143256. doi: 10.3389/fnume.2023.1143256
Alipourfard, F., Shajiee, H., Nazari-Serenjeh, F., Hojati, V., and Alirezaie, M. (2023). Betaine attenuates oxidative stress and cognitive dysfunction in an amyloid β-induced rat model of Alzheimer’s disease. Res. Pharm. Sci. 18, 270–278. doi: 10.4103/1735-5362.371583
Amin, A. M., Mostafa, H., and Khojah, H. M. J. (2023). Insulin resistance in Alzheimer’s disease: The genetics and metabolomics links. Clin. Chim. Acta 539, 215–236. doi: 10.1016/j.cca.2022.12.016
Andrade-Guerrero, J., Santiago-Balmaseda, A., Jeronimo-Aguilar, P., Vargas-Rodríguez, I., Cadena-Suárez, A. R., Sánchez-Garibay, C., et al. (2023). Alzheimer’s disease: An updated overview of its genetics. Int. J. Mol. Sci. 24:3754. doi: 10.3390/ijms24043754
Angelopoulou, E., Bougea, A., Hatzimanolis, A., Scarmeas, N., and Papageorgiou, S. G. (2024). Unraveling the potential underlying mechanisms of mild behavioral impairment: Focusing on amyloid and tau pathology. Cells 13:1164. doi: 10.3390/cells13131164
Arjmand, S., Ilaghi, M., Sisakht, A. K., Guldager, M. B., Wegener, G., Landau, A. M., et al. (2024). Regulation of mitochondrial dysfunction by estrogens and estrogen receptors in Alzheimer’s disease: A focused review. Basic Clin. Pharmacol. Toxicol. 135, 115–132. doi: 10.1111/bcpt.14035
Awasthi, S., Spellman, D. S., and Hatcher, N. G. (2022). Proteomic discovery and validation of novel fluid biomarkers for improved patient selection and prediction of clinical outcomes in Alzheimer’s disease patient cohorts. Proteomes 10:26. doi: 10.3390/proteomes10030026
Bagyinszky, E., Giau, V. V., and An, S. A. (2020). Transcriptomics in Alzheimer’s disease: Aspects and challenges. Int. J. Mol. Sci. 21:3517. doi: 10.3390/ijms21103517
Bai, H., Naj, A. C., Benchek, P., Dumitrescu, L., Hohman, T., Hamilton-Nelson, K., et al. (2023). A haptoglobin (HP) structural variant alters the effect of APOE alleles on Alzheimer’s disease. Alzheimers Dement. 19, 4886–4895. doi: 10.1002/alz.13050
Balendra, V., and Singh, S. K. (2021). Therapeutic potential of astaxanthin and superoxide dismutase in Alzheimer’s disease. Open Biol. 11:210013. doi: 10.1098/rsob.210013
Basheer, A., Agarwal, A., Mishra, B., Gupta, A., Padma Srivastava, M. V., Kirubakaran, R., et al. (2022). Use of Bacopa monnieri in the treatment of dementia due to Alzheimer disease: Systematic review of randomized controlled trials. Int. J. Med. Res. 11:e38542. doi: 10.2196/38542
Batra, R., Krumsiek, J., Wang, X., Allen, M., Blach, C., Kastenmüller, G., et al. (2024). Comparative brain metabolomics reveals shared and distinct metabolic alterations in Alzheimer’s disease and progressive supranuclear palsy. Alzheimers Dement. 20, 8294–8307. doi: 10.1002/alz.14249
Belbin, O., Xiao, M. F., Xu, D., Carmona-Iragui, M., Pegueroles, J., Benejam, B., et al. (2020). Cerebrospinal fluid profile of NPTX2 supports role of Alzheimer’s disease-related inhibitory circuit dysfunction in adults with Down syndrome. Mol. Neurodegener. 15:46. doi: 10.1186/s13024-020-00398-0
Bevilacqua, R., Barbarossa, F., Fantechi, L., Fornarelli, D., Paci, E., Bolognini, S., et al. (2023). Radiomics and artificial intelligence for the diagnosis and monitoring of Alzheimer’s disease: A systematic review of studies in the field. J. Clin. Med. 12:5432. doi: 10.3390/jcm12165432
Blanchard, J. W., Victor, M. B., and Tsai, L. H. (2022). Dissecting the complexities of Alzheimer disease with in vitro models of the human brain. Nat. Rev. Neurol. 18, 25–39. doi: 10.1038/s41582-021-00578-6
Braun, B., Demling, J., and Loew, T. H. (2022). Alzheimer’s disease: History, ethics and medical humanities in the context of assisted suicide. Philos. Ethics Hum. Med. 17:4. doi: 10.1186/s13010-021-00111-z
Breijyeh, Z., and Karaman, R. (2020). Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules 25:5789. doi: 10.3390/molecules25245789
Campbell, J., Lavoie, L., Farraia, M., Huelin, R., Zhang, Q., and Tahami Monfared, A. A. (2025). Quality of life in mild cognitive impairment and mild dementia associated with Alzheimer’s disease: A systematic review. Neurol. Ther. 14, 7–26. doi: 10.1007/s40120-024-00676-9
Chang, C. W., Hsu, J. Y., Lo, Y. T., Liu, Y. H., Mee-Inta, O., Lee, H. T., et al. (2024). Characterization of hair metabolome in 5xFAD mice and patients with Alzheimer’s disease using mass spectrometry-based metabolomics. ACS Chem. Neurosci. 15, 527–538. doi: 10.1021/acschemneuro.3c00587
Chatterjee, P., Pedrini, S., Ashton, N. J., Tegg, M., Goozee, K., Singh, A. K., et al. (2022). Diagnostic and prognostic plasma biomarkers for preclinical Alzheimer’s disease. Alzheimers Dement. 18, 1141–1154. doi: 10.1002/alz.12447
Chen, X., Drew, J., Berney, W., and Lei, W. (2021). Neuroprotective natural products for Alzheimer’s disease. Cells 10:1309. doi: 10.3390/cells10061309
Chen, Y., Li, E. M., and Xu, L. Y. (2022). Guide to metabolomics analysis: A bioinformatics workflow. Metabolites 12:357. doi: 10.3390/metabo12040357
Chen, Y., Qi, Y., Hu, Y., Qiu, X., Qiu, T., Li, S., et al. (2025). Integrated cerebellar radiomic-network model for predicting mild cognitive impairment in Alzheimer’s disease. Alzheimers Dement. 21:e14361. doi: 10.1002/alz.14361
Chen, Y., Xiao, M., Mo, Y., Ma, J., Han, Y., Li, Q., et al. (2024). Nuclear porcupine mediates XRCC6/Ku70 S-palmitoylation in the DNA damage response. Exp. Hematol. Oncol. 13:109. doi: 10.1186/s40164-024-00572-w
Chung, K. W. (2021). Advances in understanding of the role of lipid metabolism in aging. Cells 10:880. doi: 10.3390/cells10040880
Connolly, K., Lehoux, M., O’Rourke, R., Assetta, B., Erdemir, G. A., Elias, J. A., et al. (2023). Potential role of chitinase-3-like protein 1 (CHI3L1/YKL-40) in neurodegeneration and Alzheimer’s disease. Alzheimers Dement. 19, 9–24. doi: 10.1002/alz.12612
De Plano, L. M., Calabrese, G., Conoci, S., Guglielmino, S. P. P., Oddo, S., and Caccamo, A. (2022). Applications of CRISPR-Cas9 in Alzheimer’s disease and related disorders. Int. J. Mol. Sci. 23:8714. doi: 10.3390/ijms23158714
DeGroat, W., Abdelhalim, H., Peker, E., Sheth, N., Narayanan, R., Zeeshan, S., et al. (2024). Multimodal AI/ML for discovering novel biomarkers and predicting disease using multi-omics profiles of patients with cardiovascular diseases. Sci. Rep. 14:26503. doi: 10.1038/s41598-024-78553-6
DeTure, M. A., and Dickson, D. W. (2019). The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 14:32. doi: 10.1186/s13024-019-0333-5
El Menuawy, A., Brüning, T., Eiriz, I., Hähnel, U., Marthe, F., Möhle, L., et al. (2024). Apolar extracts of St. John’s wort alleviate the effects of β-amyloid toxicity in early Alzheimer’s disease. Int. J. Mol. Sci. 25:1301. doi: 10.3390/ijms25021301
Emwas, A. H., Alghrably, M., Dhahri, M., Sharfalddin, A., Alsiary, R., Jaremko, M., et al. (2021). Living with the enemy: From protein-misfolding pathologies we know, to those we want to know. Ageing Res. Rev. 70:101391. doi: 10.1016/j.arr.2021.101391
Erny, D., Dokalis, N., Mezö, C., Castoldi, A., Mossad, O., Staszewski, O., et al. (2021). Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 33, 2260–2276.e7. doi: 10.1016/j.cmet.2021.10.010
Faizan, M., Jahan, I., Ishaq, M., Alhalmi, A., Khan, R., Noman, O. M., et al. (2023). Neuroprotective effects of trigonelline in kainic acid-induced epilepsy: Behavioral, biochemical, and functional insights. Saudi Pharm. J. 31:101843. doi: 10.1016/j.jsps.2023.101843
Falchi, F. A., Pizzoccheri, R., and Briani, F. (2022). Activity and function in human cells of the evolutionary conserved exonuclease polynucleotide phosphorylase. Int. J. Mol. Sci. 23:1652. doi: 10.3390/ijms23031652
Fessel, J. (2021). Supplemental thiamine as a practical, potential way to prevent Alzheimer’s disease from commencing. Alzheimers Dement. (NY) 7:e12199. doi: 10.1002/trc2.12199
Fisher, R. M. A., and Torrente, M. P. (2024). Histone post-translational modification and heterochromatin alterations in neurodegeneration: Revealing novel disease pathways and potential therapeutics. Front. Mol. Neurosci. 17:1456052. doi: 10.3389/fnmol.2024.1456052
Franco, R., Garrigós, C., Lillo, J., and Rivas-Santisteban, R. (2024). The potential of metabolomics to find proper biomarkers for addressing the neuroprotective efficacy of drugs aimed at delaying parkinson’s and Alzheimer’s disease progression. Cells 13:1288. doi: 10.3390/cells13151288
Funk, K. E., Mrak, R. E., and Kuret, J. (2011). Granulovacuolar degeneration (GVD) bodies of Alzheimer’s disease (AD) resemble late-stage autophagic organelles. Neuropathol. Appl. Neurobiol. 37, 295–306. doi: 10.1111/j.1365-2990.2010.01135.x
Gale, S. A. (2024). Language and meaning: Asymptomatic Alzheimer’s disease in the clinic and society. J. Alzheimers Dis. 99, 489–492. doi: 10.3233/JAD-240195
Gamache, J., Yun, Y., and Chiba-Falek, O. (2020). Sex-dependent effect of APOE on Alzheimer’s disease and other age-related neurodegenerative disorders. Dis. Model Mech. 13:dmm045211. doi: 10.1242/dmm.045211
Gan, J., Zhou, H., Liu, C., and Fang, L. (2023). PSEN2 and ABCA7 variants causing early-onset preclinical pathological changes in Alzheimer’s disease: A case report and literature review. Neurol. Sci. 44, 1987–2001. doi: 10.1007/s10072-023-06602-5
Gao, L., Zhang, Y., Sterling, K., and Song, W. (2022). Brain-derived neurotrophic factor in Alzheimer’s disease and its pharmaceutical potential. Transl. Neurodegener. 11:4. doi: 10.1186/s40035-022-00279-0
Gao, X., Chen, Q., Yao, H., Tan, J., Liu, Z., Zhou, Y., et al. (2022). Epigenetics in Alzheimer’s disease. Front. Aging Neurosci. 14:911635. doi: 10.3389/fnagi.2022.911635
Gonzalez-Fernandez, E., Liu, Y., Auchus, A. P., Fan, F., and Roman, R. J. (2021). Vascular contributions to cognitive impairment and dementia: The emerging role of 20-HETE. Clin. Sci. (Lond) 135, 1929–1944. doi: 10.1042/CS20201033
Gregory, J., Vengalasetti, Y. V., Bredesen, D. E., and Rao, R. V. (2021). Neuroprotective herbs for the management of Alzheimer’s disease. Biomolecules 11:543. doi: 10.3390/biom11040543
Guo, L., Zhong, M. B., Zhang, L., Zhang, B., and Cai, D. (2022). Sex differences in Alzheimer’s disease: Insights from the multiomics landscape. Biol. Psychiatry 91, 61–71. doi: 10.1016/j.biopsych.2021.02.968
Hussain, G., Wang, J., Rasul, A., Anwar, H., Imran, A., Qasim, M., et al. (2019). Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis. 18:26. doi: 10.1186/s12944-019-0965-z
Jack, C. R., Andrews, J. S., Beach, T. G., Buracchio, T., Dunn, B., Graf, A., et al. (2024). Revised criteria for diagnosis and staging of Alzheimer’s disease: Alzheimer’s association workgroup. Alzheimers Dement. 20, 5143–5169. doi: 10.1002/alz.13859
Jain, A. P., and Sathe, G. (2021). Proteomics landscape of Alzheimer’s disease. Proteomes 9:13. doi: 10.3390/proteomes9010013
Ji, Q., Yang, Y., Xiong, Y., Zhang, Y. J., Jiang, J., Zhou, L. P., et al. (2023). Blockade of adenosine A2A receptors reverses early spatial memory defects in the APP/PS1 mouse model of Alzheimer’s disease by promoting synaptic plasticity of adult-born granule cells. Alzheimers Res. Ther. 15:187. doi: 10.1186/s13195-023-01337-z
Jiang, J., Li, C., Lu, J., Sun, J., Sun, X., Yang, J., et al. (2024). Using interpretable deep learning radiomics model to diagnose and predict progression of early AD disease spectrum: A preliminary [18F]FDG PET study. Eur. Radiol. 35, 2620–2633. doi: 10.1007/s00330-024-11158-9
Joshi, A. D., Rahnavard, A., Kachroo, P., Mendez, K. M., Lawrence, W., Julián-Serrano, S., et al. (2023). An epidemiological introduction to human metabolomic investigations. Trends Endocrinol. Metab. 34, 505–525. doi: 10.1016/j.tem.2023.06.006
Kamboh, M. I. (2022). Genomics and functional genomics of Alzheimer’s disease. Neurotherapeutics 19, 152–172. doi: 10.1007/s13311-021-01152-0
Katam, R., Hasanvand, F., Teniyah, V., Noel, J., and Gottschalk, V. (2022). “Biosafety issue related to genome editing in plants using CRISPR-Cas9,” in Genome editing, eds S. H. Wani and G. Hensel (Cham: Springer), doi: 10.1007/978-3-031-08072-2_16
Khan, M., Jaiswal, A., and Wandile, B. A. (2023). Comprehensive review of modifiable cardiovascular risk factors and genetic influences in dementia prevention. Cureus 15:e48430. doi: 10.7759/cureus.48430
Kim, J. W., Byun, M. S., Lee, J. H., Yi, D., Jeon, S. Y., Sohn, B. K., et al. (2020). Serum albumin and beta-amyloid deposition in the human brain. Neurology 95, e815–e826. doi: 10.1212/WNL.0000000000010005
Kim, J., Yoo, I. D., Lim, J., and Moon, J. S. (2024). Pathological phenotypes of astrocytes in Alzheimer’s disease. Exp. Mol. Med. 56, 95–99. doi: 10.1038/s12276-023-01148-0
Kushwah, S., Maurya, N. S., Kushwaha, S., Scotti, L., Chawade, A., and Mani, A. (2023). Herbal therapeutics for Alzheimer’s disease: Ancient indian medicine system from the modern viewpoint. Curr. Neuropharmacol. 21, 764–776. doi: 10.2174/1570159X21666230216094353
Li, L., Jin, M., Tan, J., and Xiao, B. (2024). NcRNAs: A synergistically antiapoptosis therapeutic tool in Alzheimer’s disease. CNS Neurosci. Ther. 30:e14476. doi: 10.1111/cns.14476
Li, L., Yu, X., Sheng, C., Jiang, X., Zhang, Q., Han, Y., et al. (2022). A review of brain imaging biomarker genomics in Alzheimer’s disease: Implementation and perspectives. Transl. Neurodegener. 11:42. doi: 10.1186/s40035-022-00315-z
Li, Y., and Luo, Y. (2022). Metabolomics of aging and Alzheimer’s disease: From single-omics to multi-omics. arXiv [Preprint]. doi: 10.48550/arXiv.2212.09870
Li, Y., Laws, S. M., Miles, L. A., Wiley, J. S., Huang, X., Masters, C. L., et al. (2021). Genomics of Alzheimer’s disease implicates the innate and adaptive immune systems. Cell Mol. Life Sci. 78, 7397–7426. doi: 10.1007/s00018-021-03986-5
Lin, X., Kapoor, A., Gu, Y., Chow, M. J., Peng, J., Zhao, K., et al. (2020). Contributions of DNA damage to Alzheimer’s disease. Int. J. Mol. Sci. 21:1666. doi: 10.3390/ijms21051666
Liu, D., and Du, D. (2020). Mulberry fruit extract alleviates cognitive impairment by promoting the clearance of amyloid-β and inhibiting neuroinflammation in Alzheimer’s disease mice. Neurochem. Res. 45, 2009–2019. doi: 10.1007/s11064-020-03062-7
Liu, L. C., Liang, J. Y., Liu, Y. H., Liu, B., Dong, X. H., Cai, W. H., et al. (2024). The Intersection of cerebral cholesterol metabolism and Alzheimer’s disease: Mechanisms and therapeutic prospects. Heliyon 10:e30523. doi: 10.1016/j.heliyon.2024.e30523
Lobine, D., Sadeer, N., Jugreet, S., Suroowan, S., Keenoo, B. S., Imran, M., et al. (2021). Potential of medicinal plants as neuroprotective and therapeutic properties against Amyloid-β-Related toxicity, and glutamate-induced excitotoxicity in human neural cells. Curr. Neuropharmacol. 19, 1416–1441. doi: 10.2174/1570159X19666210412095251
Lopa, S. S., Al-Amin, M. Y., Hasan, M. K., Ahammed, M. S., Islam, K. M., Alam, A. H. M. K., et al. (2021). Phytochemical analysis and cholinesterase inhibitory and antioxidant activities of enhydra fluctuans relevant in the management of Alzheimer’s disease. Int. J. Food Sci. 2021:8862025. doi: 10.1155/2021/8862025
Lopez-Lee, C., Torres, E. R. S., Carling, G., and Gan, L. (2024). Mechanisms of sex differences in Alzheimer’s disease. Neuron 112, 1208–1221. doi: 10.1016/j.neuron.2024.01.024
Maksour, S., Finol-Urdaneta, R. K., Hulme, A. J., Cabral-da-Silva, M. E. C., Targa Dias Anastacio, H., Balez, R., et al. (2024). Alzheimer’s disease induced neurons bearing PSEN1 mutations exhibit reduced excitability. Front. Cell Neurosci. 18:1406970. doi: 10.3389/fncel.2024.1406970
Malik, R., Georgakis, M. K., Neitzel, J., Rannikmäe, K., Ewers, M., Seshadri, S., et al. (2021). Midlife vascular risk factors and risk of incident dementia: Longitudinal cohort and mendelian randomization analyses in the UK Biobank. Alzheimers Dement. 17, 1422–1431. doi: 10.1002/alz.12320
Mao, H., Huang, H., Zhou, R., Zhu, J., Yan, J., Jiang, H., et al. (2023). High preoperative blood oxaloacetate and 2-aminoadipic acid levels are associated with postoperative delayed neurocognitive recovery. Front. Endocrinol. (Lausanne) 14:1212815. doi: 10.3389/fendo.2023.1212815
Markova, E., Taneska, L., Kostovska, M., Shalabalija, D., Mihailova, L., Glavas Dodov, M., et al. (2022). Design and evaluation of nanostructured lipid carriers loaded with Salvia officinalis extract for Alzheimer’s disease treatment. J. Biomed. Mater. Res. B Appl. Biomater. 110, 1368–1390. doi: 10.1002/jbm.b.35006
Marmolejo-Garza, A., Medeiros-Furquim, T., Rao, R., Eggen, B. J. L., Boddeke, E., and Dolga, A. M. (2022). Transcriptomic and epigenomic landscapes of Alzheimer’s disease evidence mitochondrial-related pathways. Biochim. Biophys. Acta Mol. Cell Res. 1869:119326. doi: 10.1016/j.bbamcr.2022.119326
Marsillach, J., Adorni, M. P., Zimetti, F., Papotti, B., Zuliani, G., and Cervellati, C. H. D. L. (2020). Proteome and Alzheimer’s disease: Evidence of a Link. Antioxidants (Basel) 9:1224. doi: 10.3390/antiox9121224
Martinez-Feduchi, P., Jin, P., and Yao, B. (2024). Epigenetic modifications of DNA and RNA in Alzheimer’s disease. Front. Mol. Neurosci. 17:1398026. doi: 10.3389/fnmol.2024.1398026
Mayo, S., Benito-León, J., Peña-Bautista, C., Baquero, M., and Cháfer-Pericás, C. (2021). Recent evidence in epigenomics and proteomics biomarkers for early and minimally invasive diagnosis of Alzheimer’s and Parkinson’s diseases. Curr. Neuropharmacol. 19, 1273–1303. doi: 10.2174/1570159X19666201223154009
McDowall, S., Bagda, V., Hodgetts, S., Mastaglia, F., and Li, D. (2024). Controversies and insights into PTBP1-related astrocyte-neuron transdifferentiation: Neuronal regeneration strategies for Parkinson’s and Alzheimer’s disease. Transl. Neurodegener. 13:59. doi: 10.1186/s40035-024-00450-9
Mei, T., Li, Y., Orduña Dolado, A., Li, Z., Andersson, R., Berliocchi, L., et al. (2023). Pooled analysis of frontal lobe transcriptomic data identifies key mitophagy gene changes in Alzheimer’s disease brain. Front. Aging Neurosci. 15:1101216. doi: 10.3389/fnagi.2023.1101216
Meikle, T. G., Huynh, K., Giles, C., and Meikle, P. J. (2021). Clinical lipidomics: Realizing the potential of lipid profiling. J. Lipid Res. 62:100127. doi: 10.1016/j.jlr.2021.100127
Mian, M., Tahiri, J., Eldin, R., Altabaa, M., Sehar, U., and Reddy, P. H. (2024). Overlooked cases of mild cognitive impairment: Implications to early Alzheimer’s disease. Ageing Res. Rev. 98:102335. doi: 10.1016/j.arr.2024.102335
Mikulska, P., Malinowska, M., Ignacyk, M., Szustowski, P., Nowak, J., Pesta, K., et al. (2023). Ashwagandha (Withania somnifera)-current research on the health-promoting activities: A narrative review. Pharmaceutics 15:1057. doi: 10.3390/pharmaceutics15041057
Monteiro, A. R., Barbosa, D. J., Remião, F., and Silva, R. (2023). Alzheimer’s disease: Insights and new prospects in disease pathophysiology, biomarkers and disease-modifying drugs. Biochem. Pharmacol. 211:115522. doi: 10.1016/j.bcp.2023.115522
Morgenstern, L. B., Briceño, E. M., Mehdipanah, R., Chang, W., Lewandowski-Romps, L., Gonzales, X. F., et al. (2024). A community-based study of dementia in Mexican american and non-hispanic white individuals. J. Alzheimers Dis. 97, 649–658. doi: 10.3233/JAD-230729
Mozersky, J., Roberts, J. S., Rumbaugh, M., Chhatwal, J., Wijsman, E., Galasko, D., et al. (2022). Spillover: The approval of new medications for Alzheimer’s disease dementia will impact biomarker disclosure among asymptomatic research participants. J. Alzheimers Dis. 90, 1035–1043. doi: 10.3233/JAD-220113
Muraoka, S., DeLeo, A. M., Yang, Z., Tatebe, H., Yukawa-Takamatsu, K., Ikezu, S., et al. (2021). Proteomic profiling of extracellular vesicles separated from plasma of former national football league players at risk for chronic traumatic encephalopathy. Aging Dis. 12, 1363–1375. doi: 10.14336/AD.2020.0908
Neuner, S. M., Tcw, J., and Goate, A. M. (2020). Genetic architecture of Alzheimer’s disease. Neurobiol. Dis. 143:104976. doi: 10.1016/j.nbd.2020.104976
Ni, X., Inoue, R., Wu, Y., Yoshida, T., Yaku, K., Nakagawa, T., et al. (2023). Regional contributions of D-serine to Alzheimer’s disease pathology in male AppNL-G-F/NL-G-F mice. Front. Aging Neurosci. 15:1211067. doi: 10.3389/fnagi.2023.1211067
Orobets, K. S., and Karamyshev, A. L. (2023). Amyloid precursor protein and Alzheimer’s disease. Int. J. Mol. Sci. 24:14794. doi: 10.3390/ijms241914794
Passeri, E., Elkhoury, K., Morsink, M., Broersen, K., Linder, M., Tamayol, A., et al. (2022). Alzheimer’s disease: Treatment strategies and their limitations. Int. J. Mol. Sci. 23:13954. doi: 10.3390/ijms232213954
Peng, Y., and Zhou, C. (2024). Network pharmacology and molecular docking identify the potential mechanism and therapeutic role of Scutellaria baicalensis in Alzheimer’s disease. Drug Des. Dev. Ther. 18, 1199–1219. doi: 10.2147/DDDT.S450739
Pereira, J. P. D., da Silva Diniz, A., Pinho Ramiro, C. P. S., and Cabral, P. C. (2023). Abdominal obesity and hydration status as protective factors against mortality in older adults: A prospective study. Nutrition 116:112155. doi: 10.1016/j.nut.2023.112155
Pereira, M., Cruz, M. T., Fortuna, A., and Bicker, J. (2024). Restoring the epigenome in Alzheimer’s disease: Advancing HDAC inhibitors as therapeutic agents. Drug Discov. Today 29:104052. doi: 10.1016/j.drudis.2024.104052
Perła-Kaján, J., Włoczkowska, O., Zioła-Frankowska, A., Frankowski, M., Smith, A. D., de Jager, C. A., et al. (2021). Paraoxonase 1, B vitamins supplementation, and mild cognitive impairment. J. Alzheimers Dis. 81, 1211–1229. doi: 10.3233/JAD-210137
Persico, G., Casciaro, F., Amatori, S., Rusin, M., Cantatore, F., Perna, A., et al. (2022). Histone H3 Lysine 4 and 27 trimethylation landscape of human Alzheimer’s disease. Cells 11:734. doi: 10.3390/cells11040734
Piamonte, B. L. C., Espiritu, A. I., and Anlacan, V. M. M. (2020). Effects of citicoline as an adjunct treatment for Alzheimer’s disease: A systematic review. J. Alzheimers Dis. 76, 725–732. doi: 10.3233/JAD-200378
Polis, B., and Samson, A. O. (2019). “A new perspective on alzheimer’s disease as a brain expression of a complex metabolic disorder,” in Alzheimer’s disease, ed. T. Wisniewski (Brisbane (AU): Codon Publications), doi: 10.15586/alzheimersdisease.2019.ch1
Qin, Y., Chen, F., Tang, Z., Ren, H., Wang, Q., Shen, N., et al. (2022). Ligusticum chuanxiong Hort as a medicinal and edible plant foods: Antioxidant, anti-aging and neuroprotective properties in Caenorhabditis elegans. Front. Pharmacol. 13:1049890. doi: 10.3389/fphar.2022.1049890
Qin, Y., Yang, P., He, W., Li, D., Zeng, L., Li, J., et al. (2024). Novel histone post-translational modifications in Alzheimer’s disease: Current advances and implications. Clin. Epigenet. 16:39. doi: 10.1186/s13148-024-01650-w
Quinn, J. P., Kandigian, S. E., Trombetta, B. A., Arnold, S. E., and Carlyle, B. C. (2021). VGF as a biomarker and therapeutic target in neurodegenerative and psychiatric diseases. Brain Commun. 3:fcab261. doi: 10.1093/braincomms/fcab261
Rahimi, S., Balusamy, S. R., Perumalsamy, H., Ståhlberg, A., and Mijakovic, I. (2024). CRISPR-Cas target recognition for sensing viral and cancer biomarkers. Nucleic Acids Res. 52, 10040–10067. doi: 10.1093/nar/gkae736
Rahman, S., and Roussos, P. (2024). The 3D genome in brain development: An exploration of molecular mechanisms and experimental methods. Neurosci. Insights 19:26331055241293455. doi: 10.1177/26331055241293455
Raleigh, S. M., and Orchard, K. J. A. (2024). Sarcopenia as a risk factor for Alzheimer’s disease: Genetic and epigenetic perspectives. Genes (Basel) 15:561. doi: 10.3390/genes15050561
Ramírez-Guerrero, S., Guardo-Maya, S., Medina-Rincón, G. J., Orrego-González, E. E., Cabezas-Pérez, R., and González-Reyes, R. E. (2022). Taurine and astrocytes: A homeostatic and neuroprotective relationship. Front. Mol. Neurosci. 15:937789. doi: 10.3389/fnmol.2022.937789
Rawat, P., Sehar, U., Bisht, J., Selman, A., Culberson, J., and Reddy, P. H. (2022). Phosphorylated tau in Alzheimer’s disease and other tauopathies. Int. J. Mol. Sci. 23:12841. doi: 10.3390/ijms232112841
Rayaprolu, S., Higginbotham, L., Bagchi, P., Watson, C. M., Zhang, T., Levey, A. I., et al. (2021). Systems-based proteomics to resolve the biology of Alzheimer’s disease beyond amyloid and tau. Neuropsychopharmacology 46, 98–115. doi: 10.1038/s41386-020-00840-3
Reveglia, P., Paolillo, C., Ferretti, G., De Carlo, A., Angiolillo, A., Nasso, R., et al. (2021). Challenges in LC-MS-based metabolomics for Alzheimer’s disease early detection: Targeted approaches versus untargeted approaches. Metabolomics 17:78. doi: 10.1007/s11306-021-01828-w
Saini, N., Kadian, M., Khera, A., Aggarwal, A., and Kumar, A. (2021). Therapeutic potential of Allium Sativum against the Aβ(1-40)-induced oxidative stress and mitochondrial dysfunction in the Wistar rats. Am. J. Neurodegener. Dis. 10, 13–27.
Salimi, A., Chatterjee, S., and Lee, J. Y. (2022). Mechanistic insights into the polymorphic associations and cross-seeding of Aβ and hIAPP in the presence of histidine tautomerism: An all-atom molecular dynamic study. Int. J. Mol. Sci. 23:1930. doi: 10.3390/ijms23041930
Saura, C. A., Deprada, A., Capilla-López, M. D., and Parra-Damas, A. (2023). Revealing cell vulnerability in Alzheimer’s disease by single-cell transcriptomics. Semin. Cell Dev. Biol. 139, 73–83. doi: 10.1016/j.semcdb.2022.05.007
Seddon, A. R., MacArthur, C. P., Hampton, M. B., and Stevens, A. J. (2024). Inflammation and DNA methylation in Alzheimer’s disease: Mechanisms of epigenetic remodelling by immune cell oxidants in the ageing brain. Redox Rep. 29:2428152. doi: 10.1080/13510002.2024.2428152
Seo, Y., Jang, H., and Lee, H. (2022). Potential applications of artificial intelligence in clinical trials for Alzheimer’s disease. Life (Basel) 12:275. doi: 10.3390/life12020275
Serrano-Pozo, A., Das, S., and Hyman, B. T. (2021). APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 20, 68–80. doi: 10.1016/S1474-4422(20)30412-9
Shanks, H. R. C., Onuska, K. M., Barupal, D. K., Schmitz, T. W., Alzheimer’s Disease Neuroimaging Initiative, and Alzheimer’s Disease Metabolomics Consortium (2022). Serum unsaturated phosphatidylcholines predict longitudinal basal forebrain degeneration in Alzheimer’s disease. Brain Commun. 4:fcac318. doi: 10.1093/braincomms/fcac318
Shi, L., Buckley, N. J., Bos, I., Engelborghs, S., Sleegers, K., Frisoni, G. B., et al. (2021). Plasma proteomic biomarkers relating to Alzheimer’s disease: A meta-analysis based on our own studies. Front. Aging Neurosci. 13:712545. doi: 10.3389/fnagi.2021.712545
Shi, M. G., Feng, X. M., Zhi, H. Y., Hou, L., and Feng, D. F. (2024). Machine learning-based radiomics in neurodegenerative and cerebrovascular disease. MedComm (2020) 5:e778. doi: 10.1002/mco2.778
Sirisi, S., Sánchez-Aced, É, Belbin, O., and Lleó, A. (2024). APP dyshomeostasis in the pathogenesis of Alzheimer’s disease: Implications for current drug targets. Alzheimers Res. Ther. 16:144. doi: 10.1186/s13195-024-01504-w
Smith, A. N., Morris, J. K., Carbuhn, A. F., Herda, T. J., Keller, J. E., Sullivan, D. K., et al. (2023). Creatine as a therapeutic target in Alzheimer’s disease. Curr. Dev. Nutr. 7:102011. doi: 10.1016/j.cdnut.2023.102011
Sofela, S. O., Ibrahim, A., Ogbodo, U. C., Bodun, D. S., Nwankwo, D. O., Mafimisebi, M., et al. (2024). Computational identification of potential acetylcholinesterase (AChE) and monoamine oxidase-B inhibitors from Vitis vinifera: A case study of Alzheimer’s disease (AD). Silico Pharmacol. 12:49. doi: 10.1007/s40203-024-00214-3
Spurgat, M. S., and Tang, S. J. (2022). Single-Cell RNA-Sequencing: Astrocyte and microglial heterogeneity in health and disease. Cells 11:2021. doi: 10.3390/cells11132021
Stanciu, G. D., Ababei, D. C., Rusu, R. N., Bild, V., and Tamba, B. I. (2022). Exploring the involvement of the amyloid precursor protein A673T mutation against amyloid pathology and Alzheimer’s disease in relation to therapeutic editing tools. Pharmaceutics 14:1270. doi: 10.3390/pharmaceutics14061270
Suire, C. N., Abdul-Hay, S. O., Sahara, T., Kang, D., Brizuela, M. K., Saftig, P., et al. (2020). Cathepsin D regulates cerebral Aβ42/40 ratios via differential degradation of Aβ42 and Aβ40. Alzheimers Res. Ther. 12:80. doi: 10.1186/s13195-020-00649-8
Sun, J., Gui, Y., Zhou, S., and Zheng, X. L. (2024). Unlocking the secrets of aging: Epigenetic reader BRD4 as the target to combatting aging-related diseases. J. Adv. Res. 63, 207–218. doi: 10.1016/j.jare.2023.11.006
Sundermann, E. E., Panizzon, M. S., Chen, X., Andrews, M., Galasko, D., Banks, S. J., et al. (2020). Sex differences in Alzheimer’s-related Tau biomarkers and a mediating effect of testosterone. Biol. Sex Differ. 11:33. doi: 10.1186/s13293-020-00310-x
Takamura, H., Nakayama, Y., Ito, H., Katayama, T., Fraser, P. E., and Matsuzaki, S. (2022). SUMO1 modification of tau in progressive supranuclear palsy. Mol. Neurobiol. 59, 4419–4435. doi: 10.1007/s12035-022-02734-5
Tanaka, M. (2025). From serendipity to precision: Integrating AI, multi-omics, and human-specific models for personalized neuropsychiatric care. Biomedicines 13:167. doi: 10.3390/biomedicines13010167
Tang, X., Tena, J., Di Lucente, J., Maezawa, I., Harvey, D. J., Jin, L. W., et al. (2023). Transcriptomic and glycomic analyses highlight pathway-specific glycosylation alterations unique to Alzheimer’s disease. Sci. Rep. 13:7816. doi: 10.1038/s41598-023-34787-4
Tong, J. H., Gong, S. Q., Zhang, Y. S., Dong, J. R., Zhong, X., Wei, M. J., et al. (2022). Association of circulating apolipoprotein ai levels in patients with Alzheimer’s disease: A systematic review and meta-analysis. Front. Aging Neurosci. 14:899175. doi: 10.3389/fnagi.2022.899175
Trifonova, O. P., Maslov, D. L., Balashova, E. E., and Lokhov, P. G. (2023). Current state and future perspectives on personalized metabolomics. Metabolites 13:67. doi: 10.3390/metabo13010067
Valencia-Olvera, A. C., Maldonado Weng, J., Christensen, A., LaDu, M. J., and Pike, C. J. (2023). Role of estrogen in women’s Alzheimer’s disease risk as modified by APOE. J. Neuroendocrinol. 35:e13209. doi: 10.1111/jne.13209
Varesi, A., Carrara, A., Pires, V. G., Floris, V., Pierella, E., Savioli, G., et al. (2022). Blood-based biomarkers for Alzheimer’s disease diagnosis and progression: An overview. Cells 11:1367. doi: 10.3390/cells11081367
Veteleanu, A., Stevenson-Hoare, J., Keat, S., Daskoulidou, N., Zetterberg, H., Heslegrave, A., et al. (2023). Alzheimer’s disease-associated complement gene variants influence plasma complement protein levels. J. Neuroinflamm. 20:169. doi: 10.1186/s12974-023-02850-6
Vignoli, A., and Tenori, L. (2023). NMR-based metabolomics in Alzheimer’s disease research: A review. Front. Mol. Biosci. 10:1308500. doi: 10.3389/fmolb.2023.1308500
Villa, C., and Combi, R. (2024). Epigenetics in Alzheimer’s disease: A critical overview. Int. J. Mol. Sci. 25:5970. doi: 10.3390/ijms25115970
Voevodskaya, O., Poulakis, K., Sundgren, P., van Westen, D., Palmqvist, S., Wahlund, L. O., et al. (2019). Brain myoinositol as a potential marker of amyloid-related pathology: A longitudinal study. Neurology 92, e395–e405. doi: 10.1212/WNL.0000000000006852
Volkert, D., Beck, A. M., Faxén-Irving, G., Frühwald, T., Hooper, L., Keller, H., et al. (2024). ESPEN guideline on nutrition and hydration in dementia - update 2024. Clin. Nutr. 43, 1599–1626. doi: 10.1016/j.clnu.2024.04.039
Wang, S., Leem, J. S., Podvin, S., Hook, V., Kleschevnikov, N., Savchenko, P., et al. (2021). Synapsin-caveolin-1 gene therapy preserves neuronal and synaptic morphology and prevents neurodegeneration in a mouse model of AD. Mol. Ther. Methods Clin. Dev. 21, 434–450. doi: 10.1016/j.omtm.2021.03.021
Wang, S., Lin, X., Zhou, J., Li, M., and Song, D. (2023). Association between serum cystatin C level and cognition in older adults: A cross-sectional analysis. Front. Neurosci. 17:1200763. doi: 10.3389/fnins.2023.1200763
Wei, G., Da, H., Zhang, K., Zhang, J., Fang, J., and Yang, Z. (2021). Glycoside compounds from glycyrrhiza uralensis and their neuroprotective activities. Nat. Prod. Commun. 16. doi: 10.1177/1934578X21992988
Weiner, S., Sauer, M., Brinkmalm, G., Constantinescu, J., Constantinescu, R., Gomes, B. F., et al. (2023). SCRN1: A cerebrospinal fluid biomarker correlating with tau in Alzheimer’s disease. Alzheimers Dement. 19, 4609–4618. doi: 10.1002/alz.13042
Wilson, R. S., Capuano, A. W., Sampaio, C., Leurgans, S. E., Barnes, L. L., Farfel, J. M., et al. (2021). Neuropsychiatric symptoms in Brazilians with mild cognitive impairment and dementia. Alzheimers Dement. (Amst) 13:e12219. doi: 10.1002/dad2.12219
Winchester, L. M., Harshfield, E. L., Shi, L., Badhwar, A., Khleifat, A. A., Clarke, N., et al. (2023). Artificial intelligence for biomarker discovery in Alzheimer’s disease and dementia. Alzheimers Dement. 19, 5860–5871. doi: 10.1002/alz.13390
Xia, Q., Gao, W., Yang, J., Xing, Z., and Ji, Z. (2024). The deregulation of arachidonic acid metabolism in ovarian cancer. Front. Oncol. 14:1381894. doi: 10.3389/fonc.2024.1381894
Xu, Y., and Li, Z. (2020). CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 18, 2401–2415. doi: 10.1016/j.csbj.2020.08.031
Yang, Y., Bagyinszky, E., and An, S. S. A. (2023). Presenilin-1 (PSEN1) mutations: Clinical phenotypes beyond Alzheimer’s disease. Int. J. Mol. Sci. 24:8417. doi: 10.3390/ijms24098417
Yin, C., Harms, A. C., Hankemeier, T., Kindt, A., and de Lange, E. C. M. (2023). Status of metabolomic measurement for insights in alzheimer’s disease progression-what is missing? Int. J. Mol. Sci. 24:4960. doi: 10.3390/ijms24054960
Young-Pearse, T. L., Lee, H., Hsieh, Y. C., Chou, V., and Selkoe, D. J. (2023). Moving beyond amyloid and tau to capture the biological heterogeneity of Alzheimer’s disease. Trends Neurosci. 46, 426–444. doi: 10.1016/j.tins.2023.03.005
Yu, S. P., Jiang, M. Q., Shim, S. S., Pourkhodadad, S., and Wei, L. (2023). Extrasynaptic NMDA receptors in acute and chronic excitotoxicity: Implications for preventive treatments of ischemic stroke and late-onset Alzheimer’s disease. Mol. Neurodegener. 18:43. doi: 10.1186/s13024-023-00636-1
Zhang, M., Niu, H., Li, Q., Jiao, L., Li, H., and Wu, W. (2023). Active compounds of panax ginseng in the improvement of Alzheimer’s disease and application of spatial metabolomics. Pharmaceuticals (Basel) 17:38. doi: 10.3390/ph17010038
Zhao, H., Wang, L., Zhang, L., and Zhao, H. (2023). Phytochemicals targeting lncRNAs: A novel direction for neuroprotection in neurological disorders. Biomed. Pharmacother. 162:114692. doi: 10.1016/j.biopha.2023.114692
Zhao, Y., Zheng, Q., Hong, Y., Gao, Y., Hu, J., Lang, M., et al. (2023). β2-Microglobulin coaggregates with Aβ and contributes to amyloid pathology and cognitive deficits in Alzheimer’s disease model mice. Nat. Neurosci. 26, 1170–1184. doi: 10.1038/s41593-023-01352-1
Zhou, W., Lei, B., Yang, C., Silva, M., Xing, X., Yu, H., et al. (2023). Artemisia annua extract improves the cognitive deficits and reverses the pathological changes of Alzheimer’s disease via regulating YAP signaling. Int. J. Mol. Sci. 24:5259. doi: 10.3390/ijms24065259
Keywords: multi-omics, biomarkers, proteomics, metabolomics, genomics, CRISPR, radiomics, Alzheimer’s biomarkers, machine learning and radiomics
Citation: Cardillo M, Katam K and Suravajhala P (2025) Advancements in multi-omics research to address challenges in Alzheimer’s disease: a systems biology approach utilizing molecular biomarkers and innovative strategies. Front. Aging Neurosci. 17:1591796. doi: 10.3389/fnagi.2025.1591796
Received: 12 March 2025; Accepted: 07 May 2025;
Published: 23 July 2025.
Edited by:
Enzo Emanuele, 2E Science, ItalyReviewed by:
Mark Kon, Boston University, United StatesHatice Lüleci, Gebze Technical University, Türkiye
Copyright © 2025 Cardillo, Katam and Suravajhala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keyura Katam, a2V5dXJhMS5rYXRhbUBmYW11LmVkdQ==
†These authors have contributed equally to this work
 Madison Cardillo
Madison Cardillo Keyura Katam
Keyura Katam Prashanth Suravajhala
Prashanth Suravajhala