- 1State Key Laboratory of Swine and Poultry Breeding Industry, South China Agricultural University, Guangzhou, Guangdong, China
- 2Guangdong Provincial Key Lab of Agro-Animal Genomics and Molecular Breeding, and Key Lab of Chicken Genetics, Breeding and Reproduction, Ministry of Agriculture and Rural Affair, South China Agricultural University, Guangzhou, China
Current achievements in omics technologies have modernized livestock biotechnology, offering extraordinary comprehension of animal productivity, health, and reproduction. This extensive study examines the integration and implementation of the omics approaches, genomics, transcriptomics, proteomics, metabolomics, and epigenomics in livestock production systems. We reconnoitered how genomic novelties redesign breeding strategies with marker-assisted selection and CRISPR-based gene editing. Together, transcriptomic analyses indicate key insights into gene expression patterns governing economically essential traits such as muscle growth and milk production. This study also shows the role of proteomics in identifying biomarkers for health surveillance and product quality improvement along with metabolomics, which contributes to understanding feed efficiency and disease resistance. Particular attention is given to epigenomics studies exploring DNA methylation and histone modifications in reproductive efficacy, underlining their importance in fertility and embryonic development. Integrating multi-omics data through systems biology approaches is discussed, demonstrating its perspective in evolving precision livestock production. We also observed how omics technologies improve assisted reproductive technologies (ART) by better understanding of molecular mechanisms underlying fertility and embryo development. While acknowledging the potential of these technologies, we discuss critical challenges, data integration complications, and ethical respect for genetic modification. This review outlines prospect directions and potential novelties in livestock biotechnology, highlighting the crucial role of omics approaches in addressing global food security contests through better livestock productivity and reproductive efficiency. This study suggests that continuous improvement in omics technologies might be the underlying cause of the determination of the future of sustainable livestock production.
1 Introduction
Livestock production is key to global agriculture, providing significant resources such as meat, milk, wool, and leather (Krostitz, 1993; Rout et al., 2021). Livestock products are projected to increase considerably, prompting the need for more effective and supportable agricultural practices (Röös et al., 2017). Traditional livestock breeding, management, and productivity improvement approaches have relied on phenotypic selection and basic genetic approaches, contributing to substantial achievements over the past few years (van Arendonk, 2011). However, modern biotechnology has reformed this field, offering new ways to enhance livestock productivity, health, and reproductive efficiency (Sharma et al., 2021; Das et al., 2022).
Integrating omics technologies like genomics, transcriptomics, proteomics, metabolomics, and epigenomics into livestock biotechnology signifies a paradigm shift in how we procedure livestock production and reproductive management (Chakraborty et al., 2022). These omics tools assist an inclusive approach to the molecular and cellular mechanisms prevailing key traits such as growth, milk production, feed efficiency, disease resistance, and fertility (Maru and Kumar, 2024). The capability to arrest vast amounts of data at multiple biological levels provides unique insights into the complex collaborations between genes, proteins, and metabolites (McCue and McCoy, 2017). This integrated approach, often called systems biology, can optimize livestock breeding programs, enhance animal welfare, and improve the permanence of livestock farming systems (Brito et al., 2020).
Genomics has contributed to advancing livestock biotechnology (ASLAM et al., 2024). Sequencing livestock genomics has opened up new provisions for understanding the genetic basis of economically key traits. Genomic selection, a method that influences dense marker information across the genome, permits the prediction of the genome perspective of animals with better precision (Eggen, 2012; Chakraborty et al., 2022). This has contributed to further rapid genetic improvement, particularly in traits that are difficult to measure directly, such as disease resistance and reproductive productivity (Xu et al., 2017). Genomics is also instrumental in isolating genes and regulatory regions related to desirable traits, which can be directed for precision breeding through innovative gene-editing technologies such as CRISPR-Cas9 (Bohra et al., 2020).
Transcriptomics, which comprises the study of RNA transcripts, is another layer of complexity to knowing livestock biology (Lowe et al., 2017). By studying gene expression patterns in several tissues and growing stages, transcriptomics describes insights into how genes are synchronized in response to environmental, nutritional, and physiological changes (Chandhini and Rejish Kumar, 2019). This information is valuable for improving traits associated with growth, lactation, and reproductive enactment (Long, 2020). Identifying differentially expressed genes and non-coding RNAs offers new goals for genetic and nutritional involvements that can improve animal productivity and health (Jilo et al., 2024).
Proteomics is a comprehensive study of proteins and their functions, and associated genomics and transcriptomics provide information about biological effectors in cells and tissues (Satrio et al., 2024). Meanwhile, proteins are the primary molecules responsible for cellular functions. Understanding its abundance, modifications, and interactions is crucial for revealing the mechanisms underlying key traits in livestock (Wang and Ibeagha-Awemu, 2021). Proteomics has been applicable to identify biomarkers for disease resistance, meat and milk quality, and reproductive traits (Ribeiro et al., 2020). Post-translational modifications, like phosphorylation, acetylation, and glycosylation, have revealed key regulatory pathways influencing cellular processes such as muscle growth, immune response, and reproductive efficiency (Nissa et al., 2021).
Metabolomics studies identify small molecules or metabolites in biological processes, offering an exclusive approach to the metabolic processes associated with livestock growth and reproduction (Goldansaz et al., 2017). Profiling the metabolome can increase insights into animals’ nutritional and physiological status and identify biomarkers allied to feed efficiency, energy metabolism, and health (Marina et al., 2024). Metabolomics can improve livestock production by optimizing diets, finding metabolic bottlenecks, and selecting more efficient animals to convert feed into valuable products (Sun et al., 2019). Metabolomics studies can develop our understanding of metabolic adjustments during pregnancy, lactation, and growth, leading to targeted involvements that improve animal reproduction (Choudhary et al., 2024).
Epigenomics, exploring the heritable changes in gene expression that do not involve modifications in the DNA sequence, signifies an emerging frontline in livestock biotechnology (Ibeagha-Awemu and Zhao, 2015). Epigenetic modifications, such as DNA methylation and histone modifications, are key in regulating gene expression during growth and responding to environmental dynamics such as nutrition, stress, and disease (Kumar et al., 2020). Livestock epigenomics will likely improve reproductive efficiency, as epigenetic mechanisms impact fertility and embryonic development (Wang and Ibeagha-Awemu, 2021). Understanding the epigenetic role of livestock species may lead to new approaches for managing reproductive tasks, such as embryonic loss, low fertility rates, and early embryonic death (Zhu et al., 2021) (Figure 1).
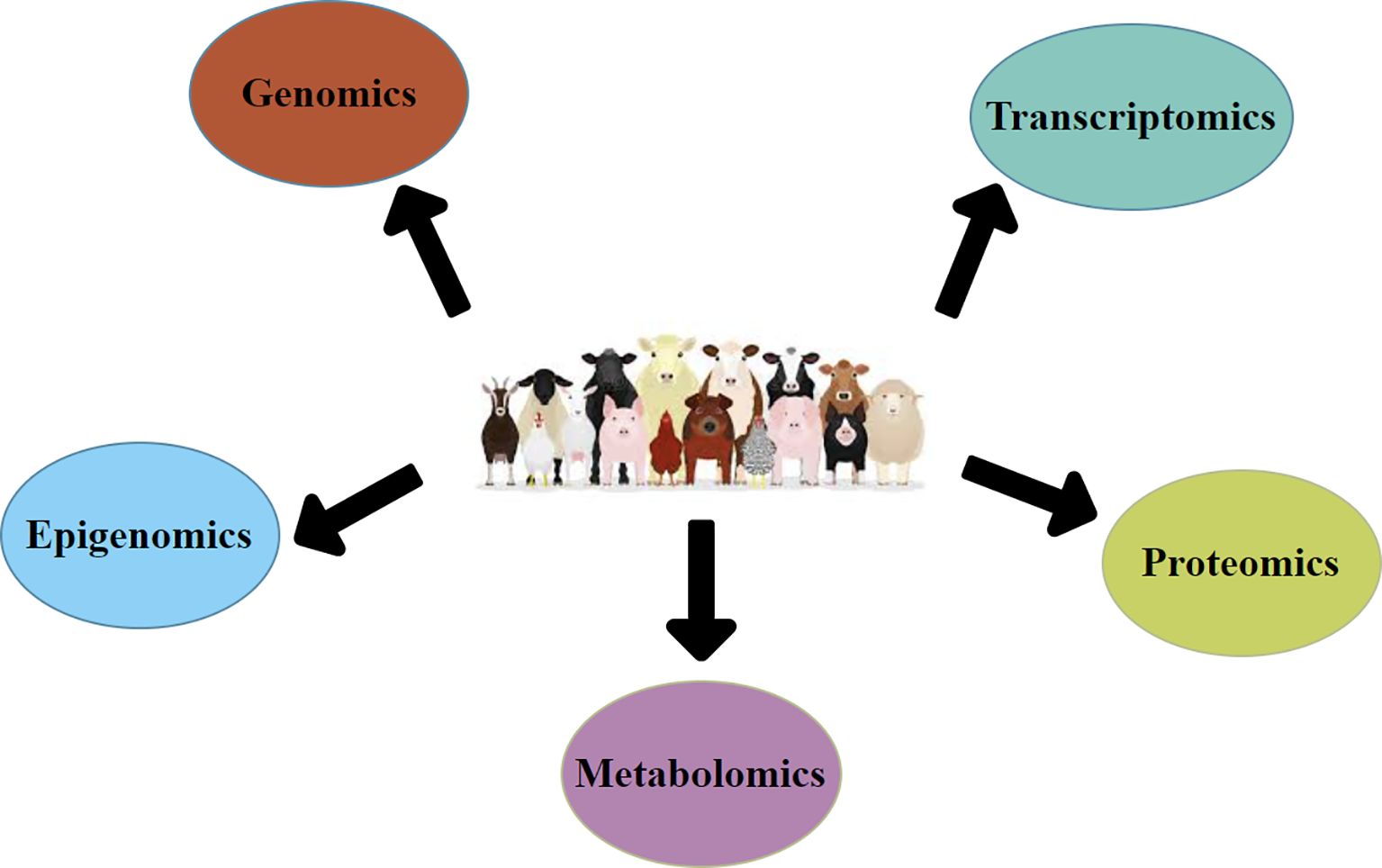
Figure 1. This graphic diagram shows multi-omics integration in livestock biotechnology to advance productivity, health, and reproduction, enabling innovative breeding and management.
Integrating these omics methodologies into livestock biotechnology presents significant challenges. A key issue is the enormous amount of data produced by omics technologies, which requires advanced bioinformatics tools for practical analysis, interpretation, and integration (Fillinger et al., 2019). Integrating data from various omics tools, including genomics, transcriptomics, proteomics, metabolomics, and epigenomics, can reveal complex regulatory networks and molecular pathways underlying key traits. However, this process demands careful consideration of data compatibility and robust statistical analysis (Canzler et al., 2020; Graw et al., 2021). Developing computational models and machine learning algorithms is key for harnessing the full potential of omics technologies in livestock biotechnology (Koltes et al., 2019; Chakraborty et al., 2022). These models must consider the dynamic interactions between genes, proteins, and metabolites across different tissues, developmental stages, and environmental stresses.
Even though integrating omics into cattle biotechnology has several advantages, many studies have not fully examined the variety of uses across various species. Other significant livestock, like sheep, goats, poultry, and even aquaculture species, have the potential to advance livestock biotechnology in addition to more conventional species like cattle and pigs (Prasad et al., 2023). Because of these differences in species-specific biological systems, customized strategies that consider each species’ distinct genetic composition and physiological traits are required (Council et al., 2002). In this regard, one example of the technological developments in the field is the function of non-coding RNAs (ncRNAs), a crucial area of genomics study. Although most ncRNA research has been on cattle, studies on the functional roles of ncRNAs in other species, such as pigs, sheep, and poultry, are gaining popularity (Gong et al., 2023). NcRNAs that regulate gene expression during essential physiological processes like immunological responses, growth, and reproduction include long non-coding RNAs, circular RNAs, and microRNAs (Yadav et al., 2024). It has been discovered that several miRNAs influence sheep follicular development, providing insight into how to improve reproductive success (Yang et al., 2019). Additionally, miRNAs have been shown to modify poultry’s immune responses, which could be utilized to enhance disease resistance and vaccine efficacy (Do et al., 2021).
Reproductive management and animal breeding are changing due to omics technologies other than ncRNAs. Cattle and pig breeding operations now rely heavily on genomic selection, which uses genomic data to forecast an animal’s genetic potential (Misztal et al., 2020). However, genomic selection also spreads to other species, such as poultry, where genomics increases meat yield and egg production, and goats, where genome-wide association studies (GWAS) are used to find loci linked to disease resistance features (Ren et al., 2024). Furthermore, proteomics and metabolomics provide fresh perspectives on the metabolic processes that control development and fertility in different species (Panner Selvam et al., 2021). Researchers are finding biomarkers that predict fertility and feed efficiency by examining the proteome profiles of dairy cows and pigs (Aranciaga et al., 2020; Mills, 2021). This could lessen the environmental effect of livestock farming (Figure 2).
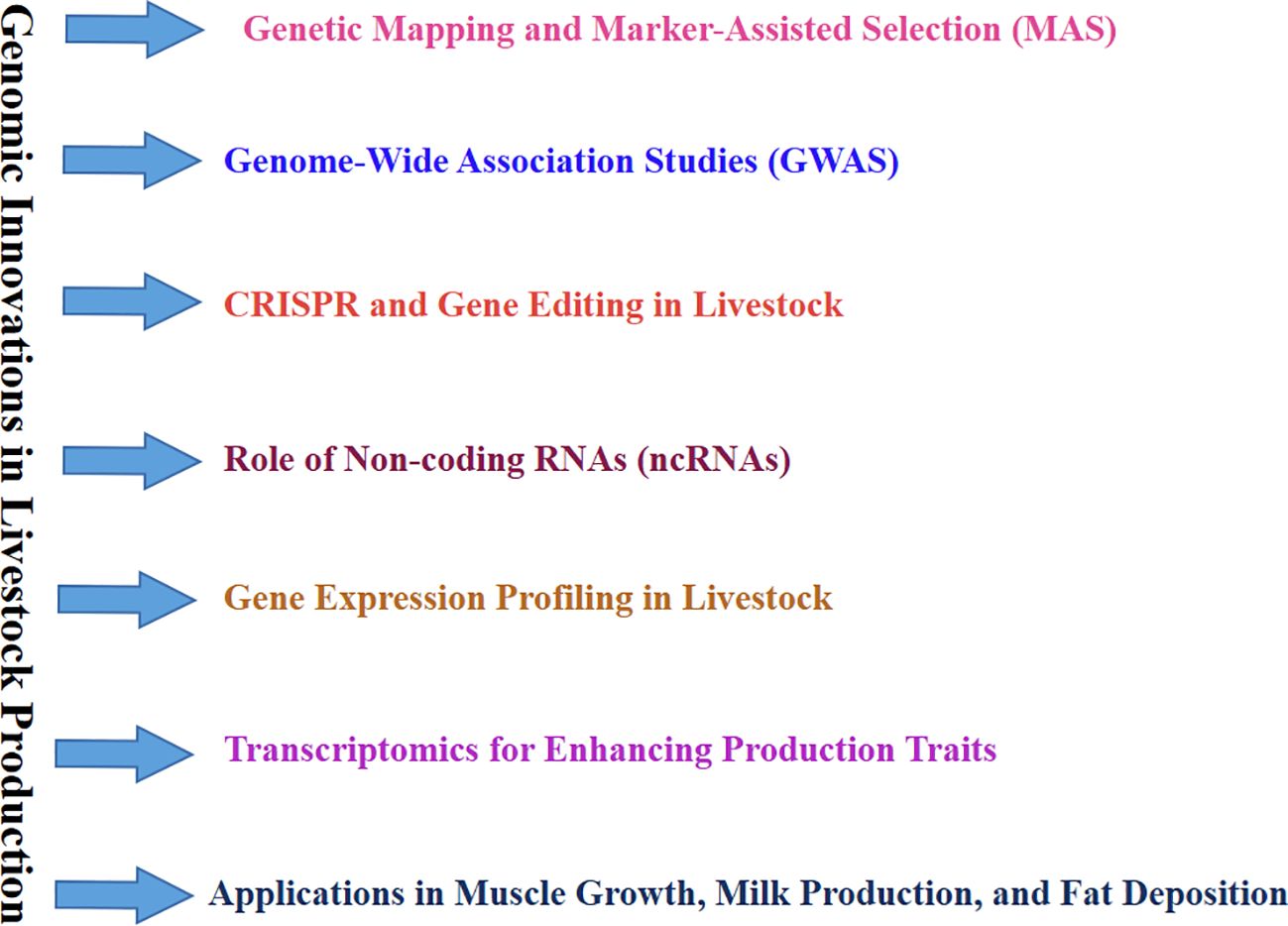
Figure 2. Overview of Genomic Innovations in Livestock Biotechnology. The flow diagram outlines key genomic approaches, including genetic mapping, marker-assisted selection (MAS), genome-wide association studies (GWAS), CRISPR and gene editing, and transcriptomics. These approaches are pivotal in improving production traits and exploring gene expression, aiming at muscle growth, milk production, and fat deposition. Integrating these technologies depicts advancements in livestock breeding, productivity, and reproductive proficiency.
The potential assistance of integrating omics approaches in livestock biotechnology is enormous. Understanding the molecular mechanisms, growth, reproduction, and health, we can develop more accurate and targeted strategies for improving livestock production (Singh et al., 2014). For instance, genomic selection with transcriptomic and proteomic data can improve the accuracy of breeding programs, leading to more practical selection for traits such as feed efficiency, disease resistance, and reproductive performance (Gutierrez-Reinoso et al., 2021). Additionally, metabolomics and epigenomics insights can apprise nutritional interventions that enhance animal growth and fertility while reducing the environmental stress of livestock farming (Loor, 2022) (Figure 3).
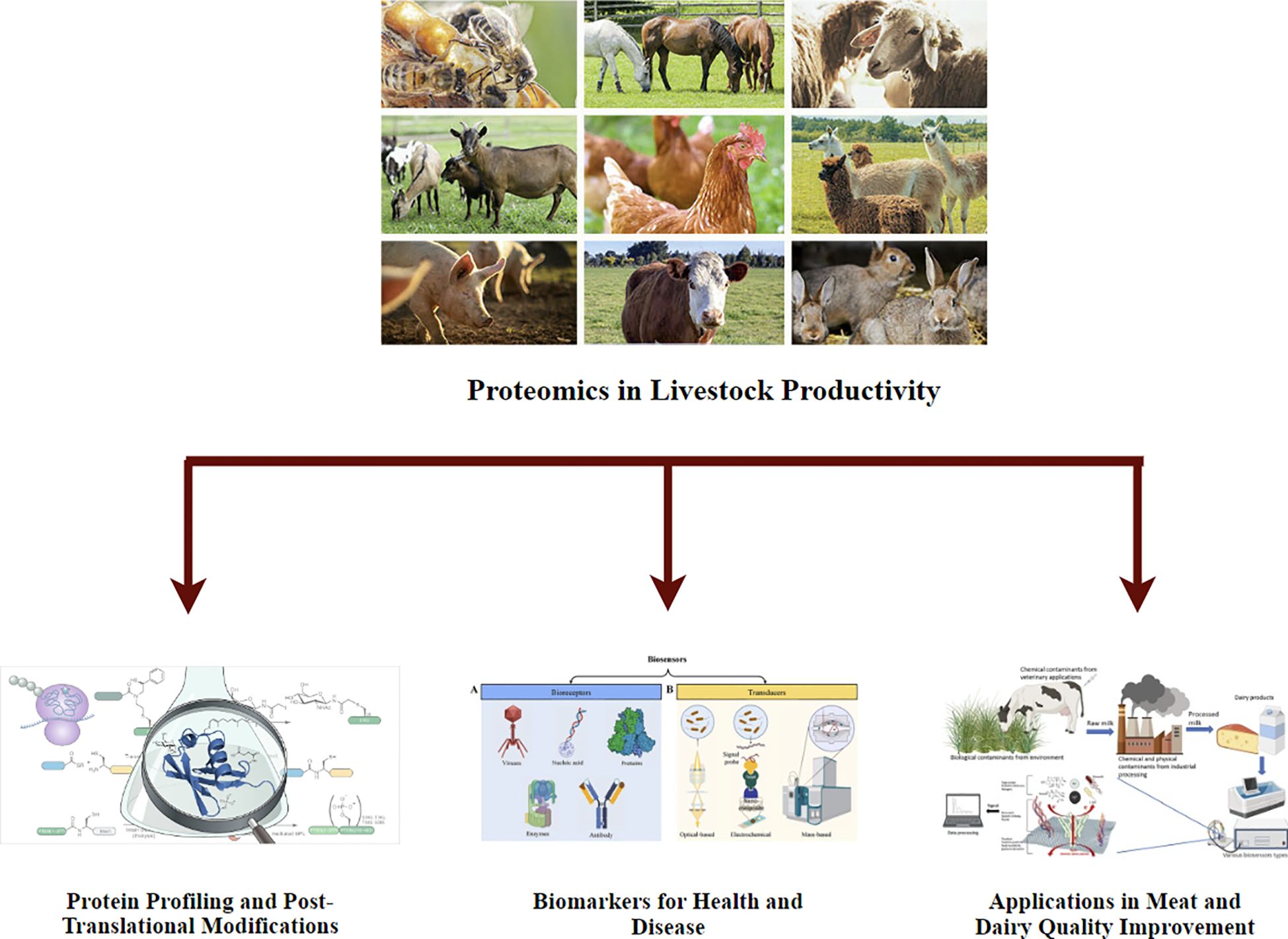
Figure 3. Proteomics analysis in livestock production emphasizes protein profiling, post-translational modifications, and biomarker detection for health and disease control. It also has essential applications in increasing meat and dairy standards. The figure outlines key proteomic approaches contributing to the advancement of livestock production.
2 Genomic innovations in livestock production
Genomic innovations have transformed livestock breeding and management, offering previously absent genetic improvement (Perisse et al., 2021). The integration of genomic technologies like genetic mapping, marker-assisted selection (MAS) (Boopathi, 2013), genome-wide association studies (GWAS) (Uffelmann et al., 2021), and CRISPR-Cas9 gene editing (Singh et al., 2017), with transcriptomics and gene expression profiling (Kumar et al., 2016) has enabled a more detailed understanding of the genetic basis of complex traits (Ribeiro et al., 2020)s. This has significantly improved growth, reproductive performance, disease resistance, and production traits such as muscle development and milk yield (Islam et al., 2020). Furthermore, the expanding repertoire of non-coding RNAs (ncRNAs) has revealed regulatory mechanisms that govern gene expression, providing deeper insights into the molecular networks that shape livestock phenotypes (Velez, 2023; Jilo et al., 2024) (Figure 4).
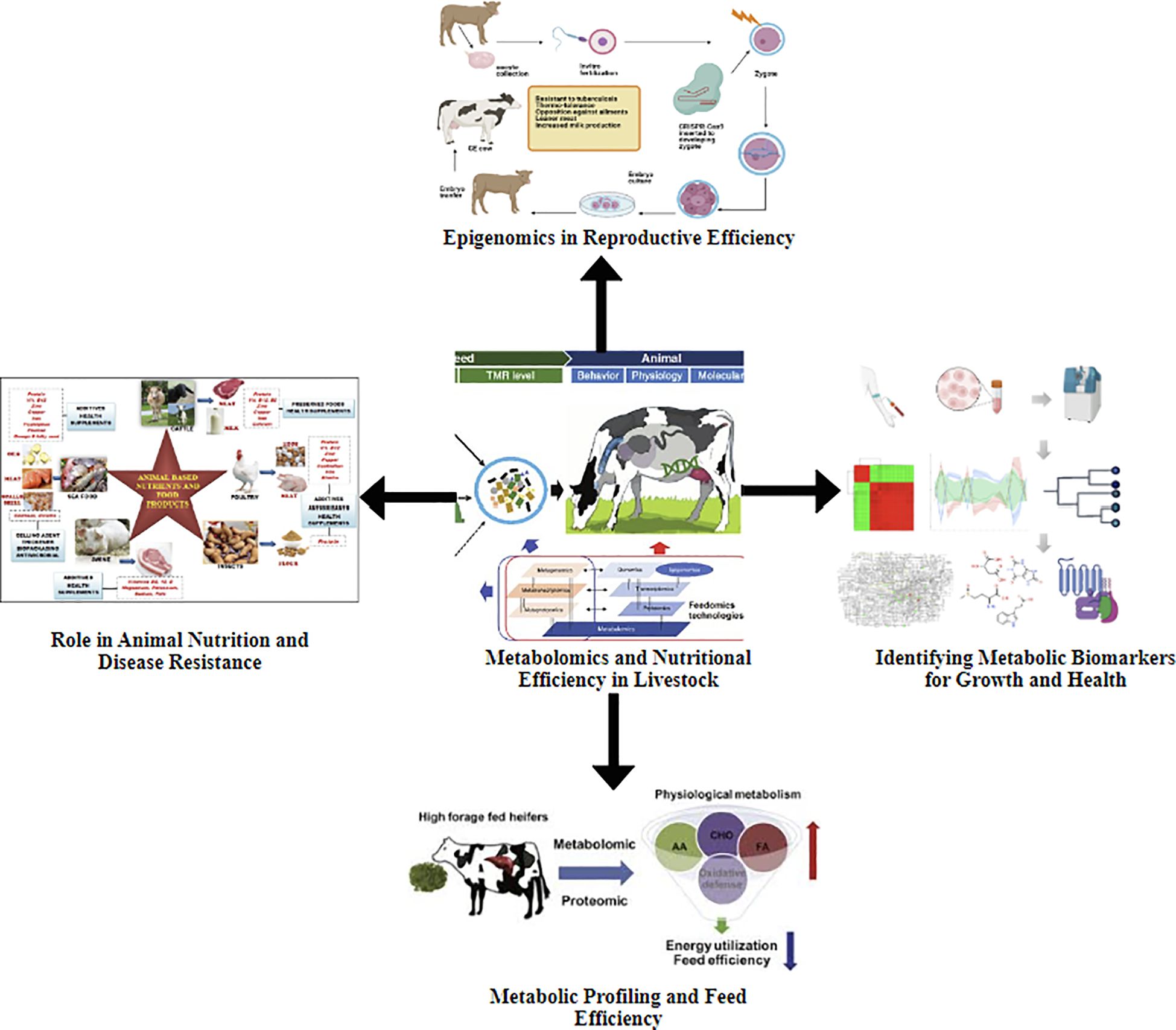
Figure 4. This schematic outlines an inclusive, integrative approach to improving livestock reproductive efficacy by leveraging epigenetic modulation. It tells the complex molecular interactions between epigenetic regulatory mechanisms, metabolomic signatures, and nutritional interventions.
2.1 Genetic mapping and marker-assisted selection
Genetic mapping discusses identifying the relative positions of genes and other genetic components within a genome (Council et al., 1988; Boopathi, 2013). High-resolution genetic maps have been recognized for livestock species, including cattle, pigs, sheep, and chickens, consenting to their recognition of quantitative trait loci (QTLs) with economically essential traits (Cockett and Kole, 2008; Stinckens et al., 2010; Hu et al., 2020). These findings of QTLs are vital for finding molecular markers for breeding, such as growth rate, milk yield, disease resistance, and reproductive productivity (Khalil, 2020). Advances in next-generation sequencing (NGS) tools have improved the accuracy of genetic maps, providing more profound attention to the genetic architecture of livestock traits (Ghosh et al., 2018). In cattle, high-density single nucleotide polymorphism (SNP) arrays have been used to identify QTLs with milk production, fertility, and meat quality (Ibeagha-Awemu et al., 2016). Genetic mapping in poultry indicates loci associated with growth rate, egg production, and resistance to infectious diseases like Marek’s disease and avian influenza (Churchil, 2023) (Figure 5).
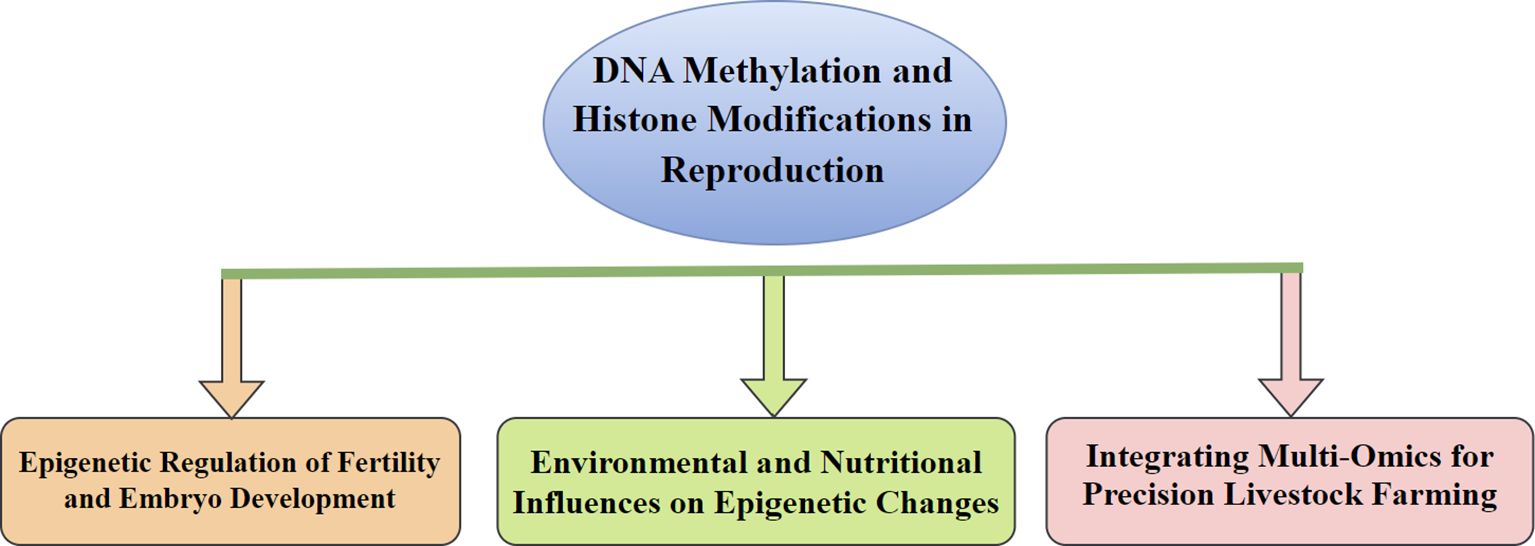
Figure 5. This indicates the key areas where epigenetic mechanisms, especially DNA methylation and histone modifications, have a potential role in reproduction. (1) Epigenetic Regulation of Fertility and Embryo Development, wherever epigenetic modifications affect gene expression patterns essential for reproductive rate; (2) Environmental and Nutritional Influences on Epigenetic Changes, underlining how external influences affect epigenetic states. (3) Integrating Multi-Omics for Precision Livestock Farming, which highlights the use of multi-omics methodologies to optimize reproductive efficacy in livestock through agriculture.
MAS leverages the association between molecular markers and desirable traits to accelerate breeding (Cobb et al., 2019). By selecting animals based on their genetic profiles rather than solely on phenotypic performance, MAS improves the accuracy and efficiency of breeding programs (Beuzen et al., 2000; Brito et al., 2020). MAS has been widely adopted in dairy cattle to enhance milk yield, fat and protein content, and somatic cell count (a measure of mastitis resistance) (Alhussien and Dang, 2018). MAS has been used in beef cattle to select traits like feed efficiency, marbling, and carcass weight (Hozáková et al., 2020). Identifying markers linked to the myostatin gene (MSTN), which regulates muscle development, has enabled the selection of animals with increased muscle mass and reduced fat deposition (Ayuti et al., 2024). Despite its success, MAS effectiveness depends on the strength of the linkage between the marker and the target trait, which can vary between populations and environments (Podlich et al., 2004). Additionally, MAS is more effective for traits controlled by a few significant genes (oligogenic traits) than polygenic traits influenced by multiple genes (Singh et al., 2015; Boopathi and Boopathi, 2020). As a result, MAS with genomic selection and GWAS capture a broader spectrum of genetic variation to develop complex traits (Tam et al., 2019) (Figure 6).
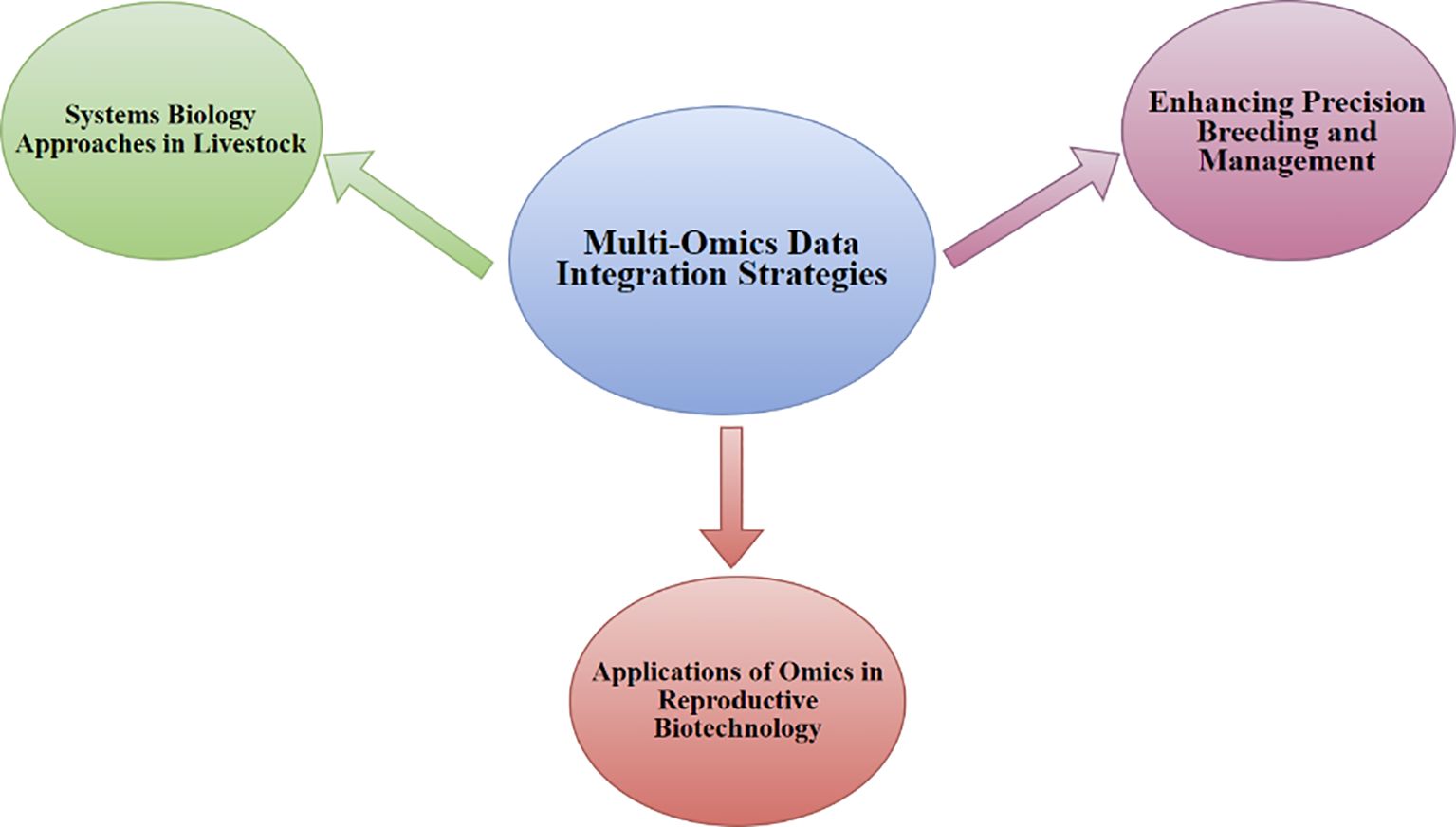
Figure 6. Signifies the integration of multi-omics data in livestock, with an emphasis on (1) Systems Biology Approaches in Livestock, which focus on understanding complex biological systems, and (2) Enhancing Precision Breeding and Management, where multi-omics data services in refining breeding practices and management decisions, and (3) Applications of Omics in Reproductive Biotechnology, illustrating how omics-based techniques contribute to advancements in reproductive technologies and genetic progresses in livestock.
2.2 Genome-wide association studies
GWAS is an effective tool for identifying the genetic variants for complex traits by scanning the entire genome for connotations (Xiao et al., 2022). Unlike MAS, which has pre-identified markers, GWAS permits the discovery of novel loci related to traits of interest (Korte and Farlow, 2013). This method has been instrumental in uncovering the genetic basis of complex traits in livestock, with growth, reproduction, disease resistance, and product quality (Te Pas et al., 2017). In cattle, GWAS has identified genetic variants with milk yield, fat composition, fertility, and resistance to mastitis (Devani et al., 2020). In pigs, GWAS has revealed loci related to feed efficiency, meat quality, and susceptibility to diseases such as porcine reproductive and respiratory syndrome (PRRS) (Lunney and Chen, 2010). In sheep and goats, GWAS has been identified in study traits like wool quality, carcass composition, and resistance to gastrointestinal parasites (Arzik et al., 2022; Carracelas et al., 2022). A critical success of GWAS in livestock is the discovery of the DGAT1 gene in dairy cattle, which shows a key role in milk fat synthesis (Khan et al., 2021). Identifying this gene has enabled the selection of animals with improved milk fat content, significantly enhancing dairy production (Barillet, 2007). Another example is identifying the FTO gene associated with fat deposition in cattle and pigs, providing opportunities to manipulate body composition for improved meat quality (Óvilo et al., 2022).
While GWAS has important innovations in livestock genetics, it also has challenges. The genetic architecture of complex traits mainly includes small effect sizes, making it challenging to identify all the contributing loci (McCarthy et al., 2008; Korte and Farlow, 2013; Tam et al., 2019). The accuracy of GWAS relies on the quality and size of the datasets in some livestock species (Zhang et al., 2014). Another task is population stratification, where genetic differences between subpopulations can confuse the results (Marigorta et al., 2018). To address these tasks, scientists use multi-omics approaches integrating genomic, transcriptomic, and epigenomics data to gain an extensive understanding of trait variation (Akiyama, 2021). Advances in computational techniques and machine learning also educate the power and accuracy of GWAS, permitting the identification of subtler genetic effects (Okser et al., 2014; Prabhod, 2022). As sequencing costs continue to decrease, the whole-genome sequence data in GWAS is becoming more feasible for enhancing genetic mapping resolution (Fuentes‐Pardo and Ruzzante, 2017) (Figure 7).
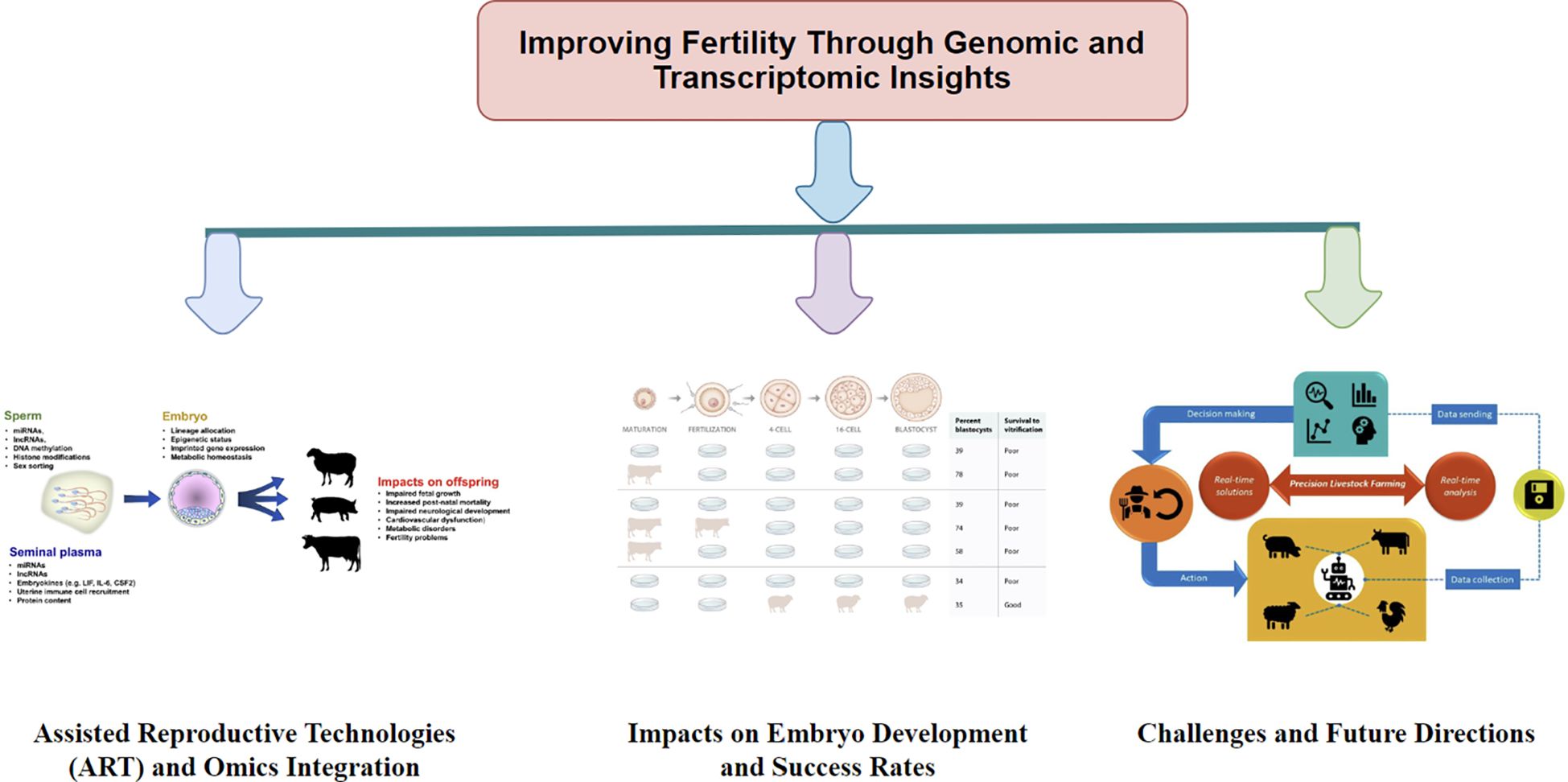
Figure 7. Demonstrates the role of genomics and transcriptomics in enhancing fertility achievement in livestock. The figure indicates three key parts: (1) Assisted Reproductive Technologies (ART) and Omics Integration, which indicates the combination of ART with genomic techniques to improve fertility; (2) Impacts on Embryo Development and Success Rates, presenting how genomic and transcriptomic influences embryo viability and developmental; and (3) Challenges and Future Directions, explaining prospective challenges and research guidance required for progressing fertility-related genetic and transcriptomic study in livestock.
2.3 CRISPR and gene editing in livestock
CRISPR-Cas9 is an innovative genome editing technique that allows precise modifications to the DNA sequence at specific loci (Gupta et al., 2019). This can potentially revolutionize livestock productivity by enabling the introduction, deletion, or alteration of genes linked with desirable traits (Quétier, 2016). Unlike traditional breeding methods, which depend on natural variation, CRISPR enables targeted genome manipulation, introducing beneficial alleles that may be rare or absent in the population (Scheben et al., 2017).
One of the most prospective implementations of CRISPR in livestock is the development of disease-resistant animals (Lamas-Toranzo et al., 2018). Scientists used CRISPR to produce pigs resistant to PRRS by editing the gene encoding the CD163 receptor, which the virus uses in the host cells (Burkard et al., 2017) and exhibits resistance to PRRS. CRISPR has improved chickens’ resistance to avian influenza and cattle’s resistance to bovine tuberculosis, providing new avenues for strengthening animals (Pal and Chakravarty, 2019; Gao et al., 2023). CRISPR has also been used to improve production in cattle by editing the myostatin gene (MSTN) to make animals with better muscle mass, resulting in higher meat yield (Kalds et al., 2023). In pigs, CRISPR has been used to modify genes linked with growth performance and feed efficiency, leading to animals that are highly productive and environmentally sustaining (Wu and Bazer, 2019). CRISPR has been used in dairy cattle to present enhanced milk production and composition alleles, offering new opportunities to strengthen dairy production (Yang, 2024).
The role of gene-edited animals in the food supply focused on strict regulatory consent and people’s perception of genetically modified organisms (GMOs) may upset the acceptance of this technology (Brookes and Smyth, 2024). We must also address ethical apprehensions regarding animal welfare, biodiversity, and food safety. Moreover, the potential for unplanned off-target effects, where CRISPR introduces changes in unintentional genome regions, requires careful checking to ensure the safety and efficacy of gene-edited animals (Cook et al., 2017; Wienert and Cromer, 2022).
2.4 Transcriptomics for enhancing production traits
Transcriptomics is a tool for understanding the gene expression patterns related to production traits in livestock (Salleh et al., 2017). In dairy cattle, transcriptomic analyses have recognized key genes involved in milk production associated with milk-protein synthesis, fat metabolism, and immune response (Zhou et al., 2019). These results have delivered valuable information for improving milk quality and productivity through selective breeding (Wang et al., 2018). In beef cattle, transcriptomics has been used to identify muscle development and meat quality. By analyzing gene expression in muscle tissues, we recognized genes linked with muscle growth, fat deposition, and meat tenderness (Wang et al., 2009). These understandings have knowledgeable breeding strategies to increase carcass quality and meat yield (Guo and Dalrymple, 2022).
2.5 Gene expression profiling in livestock
Gene expression describes accompaniments transcriptomics to allow a more focused examination of gene activity under different biological circumstances (Wolf, 2013; Kumar et al., 2016). Through techniques like RNA sequencing (RNA-Seq) and microarrays, the expression levels of thousands of genes advance a deeper understanding of the molecular networks for phenotypic traits (Singh and Singh, 2024). In livestock production, gene expression profiling has helped identify regulatory pathways for essential traits like growth, reproduction, and immune function (Loor et al., 2005). For example, gene expression profiling has elucidated key genes intricate in skeletal muscle development in cattle and pigs, enhancing meat quality and yield through targeted breeding (Karisa, 2013; Zeng and Du, 2023). The dynamic nature of the transcriptome challenges livestock production, as gene expression patterns can be affected by several environmental factors such as diet, stress, and disease (Parreira and de Sousa Araújo, 2018). While gene expression profiling offers valuable insights into trait variation, it must be taken within the broader context of other molecular data, such as epigenomics and proteomics, to recognize its full effect on livestock traits (Triantaphyllopoulos et al., 2016).
2.6 Role of non-coding RNAs
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), have emerged as crucial regulators of gene expression in livestock (Do and Suravajhala, 2023; Suravajhala, 2023). ncRNAs do not encode proteins but regulate gene expression at the transcriptional and post-transcriptional (Szymanski et al., 2003) and are associated with diverse biological processes, including development, metabolism, and immune response located ncRNAs as key players in determining complex traits in livestock (Huang et al., 2021).
New studies have implicated the ncRNAs in regulating traits like muscle growth, fat deposition, and reproductive performance (Yang et al., 2023). miRNAs have been shown to regulate muscle development by targeting genes in muscle hypertrophy (Horak et al., 2016). The lncRNAs have been linked with fat metabolism, inducing traits such as fatty tissue development (Cheng et al., 2021). ncRNAs have also been associated with milk production and composition in cattle (Lu et al., 2021). For example, specific miRNAs regulate genes related to milk protein synthesis, and others are related to lactation and immune function in the mammary gland (Hue-Beauvais et al., 2021). By modulating the expression of key genes through ncRNA-based involvements, production traits such as muscle growth, milk yield, and reproductive efficiency may be improved (Hu et al., 2022). Despite the developing interest in ncRNAs, little is known about their functions in livestock. The ncRNA-mediated regulation and the trouble of precisely measuring ncRNA expression levels present challenges for integration in the breeding programs (Ramesh, 2013). However, progress in RNA sequencing technologies and bioinformatics techniques is expected to enable the finding of new ncRNAs and their roles in livestock production.
2.7 Applications in muscle growth, milk production, and fat deposition
Muscle growth is a significant characteristic of livestock production, mainly for meat-producing animals such as cattle, pigs, and poultry (Wang et al., 2024). Genomic tools have determined numerous genes and regulatory pathways intricate in muscle development, with myogenesis, muscle hypertrophy, and protein synthesis (Yang et al., 2024). The myostatin gene (MSTN) has been specified as an essential regulator of muscle growth in cattle and pigs (Aiello et al., 2018). Animals with mutations in MSTN show better muscle mass and reduced fat deposition, enhancing carcass quantity and meat quality (Ceccobelli et al., 2022). Gene editing technologies, like CRISPR-Cas9, have been utilized to have mutations in MSTN and other muscle-related genes, causing significant muscle growth and feed efficiency in animals (Zhao et al., 2022).
Milk production is a complicated trait involving multiple genes in lactation, metabolism, and immune function (Golan and Assaraf, 2020). Genomic technologies have identified various QTLs and SNPs related to milk yield, fat and protein content, and lactation (Ma et al., 2021). The DGAT1 gene, which signifies a potential role in milk fat synthesis, has been recognized as a major contributor to milk composition in dairy cattle (Khan et al., 2021; Tian et al., 2022). MAS and genomic selection have been adopted in dairy breeding programs to improve milk production and traits like somatic cell count and mastitis resistance (Brito et al., 2021). Transcriptomics and gene expression profiling have also delivered valuable understandings of the molecular mechanisms for lactation, leading to new approaches for improving milk quality and production efficiency (Sikka et al., 2023).
Fat deposition is significant in livestock production, influencing meat quality and reproductive performance (Schumacher et al., 2022). Genomic studies have discovered several genes intricate in fat metabolism, including those related to adipogenesis, lipogenesis, and fatty acid oxidation (Ladeira et al., 2016). The FTO gene has been linked with fat deposition in cattle and pigs, giving opportunities to influence body composition for better meat quality and reproductive efficiency (Dvořáková et al., 2012; Jevsinek Skok et al., 2016). Transcriptomics and gene expression profiling have identified livestock fat deposition regulatory networks (Huang et al., 2017). Studies have shown that miRNAs and lncRNAs have critical roles in regulating adipogenesis and lipid metabolism, offering new targets for genetic progress (Lorente-Cebrián et al., 2019).
3 Proteomics in livestock productivity
Developments in molecular biology have delivered powerful techniques to attain these goals, with proteomics developing as one of the most transformative methodologies (Misra et al., 2019). Proteomics, the large-scale study of proteins and their role, offers an extraordinary prospect to comprehend the molecular tools causing important physiological and productive traits in livestock (Gao et al., 2024). It complements genomic and transcriptomic approaches by focusing on proteins that indicate cellular functions (Sobti et al., 2022). This is important because proteins are the functional products of genes, and their expression, modification, and interactions are significant to cellular processes, with metabolism, growth, reproduction, and immune responses (Udenwobele et al., 2017).
3.1 Protein profiling and post-translational modifications
In livestock, protein describing maps the proteome, or the complete set of proteins, having different tissues, phases of development, and physiological conditions (Jamwal et al., 2023). This method delivers insights into the molecular mechanisms of heavy growth, reproduction, and disease resistance (Ribeiro et al., 2020). Protein profiling has been functional extensively in livestock species, including cattle, pigs, sheep, goats, and poultry (Almeida et al., 2015). In cattle, proteomic analyses of muscle tissues have recognized proteins intricate in muscle growth, energy metabolism, and meat quality (Picard et al., 2010). In dairy cows, protein profiling of milk and mammary tissues has shown key proteins complicated in milk production, fat synthesis, and immune responses (Dai et al., 2018). Poultry protein profiling has provided insights into muscle development, egg production, and disease resistance (Kanakachari et al., 2022).
Mass spectrometry (MS) is an extensive livestock protein profiling technique (Soler et al., 2020). MS-based proteomics identifies and quantifies thousands of proteins in a single experiment (Taverna and Gaspari, 2021). MS can map protein interactions and pathways, providing a systems-level understanding of cellular processes (Richards et al., 2021). Other tools, such as two-dimensional gel electrophoresis (2D-GE) and protein microarrays, are also applied for protein profiling (Meleady, 2018), although they are less extensive than MS.
PTMs are chemical modifications that follow proteins after synthesis (Barber and Rinehart, 2018). These modifications can change protein function, stability, localization, and relations and are essential for regulating cellular processes (Karve and Cheema, 2011). PTMs regulate animal growth, reproduction, metabolism, and immune responses (Fan et al., 2024). In dairy cows, PTMs have been associated with mastitis resistance, with phosphorylated proteins identified as critical regulators of the immune response in the mammary gland (Adnane et al., 2024). The study of PTMs in livestock is still in its initial stages, but advances in proteomics technologies enable the identification of PTMs on a larger scale (Almeida et al., 2021). MS-based approaches help study PTMs, as they can identify modifications at specific amino acid residues and quantify changes in PTMs in response to different situations (Doll and Burlingame, 2015). The role of post-translational modifications (PTMs) in livestock productivity presents a promising avenue for identifying novel genetic targets. By elucidating how PTMs regulate key biological processes, researchers can enhance breeding strategies to improve traits such as growth efficiency, reproductive performance, and disease resistance.
3.2 Biomarkers for disease and health
Proteomics biomarkers are crucial as they offer real-time insights into an animal’s physiological state and response to environmental and biological challenges (Raposo de Magalhães et al., 2020). Proteomic technologies are compatible with biomarker discovery because they enable the inclusive analysis of proteins and their modifications through tissues and biofluids (Dayon et al., 2022). These biomarkers can be used to identify diseases initially, expect disease susceptibility, monitor the effectiveness of treatments, and assess animal health (Xu and Veenstra, 2008; Reinhart et al., 2012). Biomarkers have been identified for various livestock conditions, including metabolic disorders, infectious diseases and reproductive (Zachut et al., 2020).
Proteomics is a robust approach for identifying biomarkers related to infectious diseases in livestock, such as bovine tuberculosis, mastitis, foot-and-mouth disease, and avian influenza, enabling early diagnosis and optimizing disease control strategies (Suminda et al., 2022). In dairy cows, milk proteomic analyses have known key protein biomarkers related to mastitis, an inflammatory condition of the mammary gland that badly impacts milk production and quality (Giagu et al., 2022). These biomarkers contain acute-phase proteins, enzymes, and immune-related proteins upregulated due to infection (Bathla et al., 2020). Monitoring these biomarkers allows early mastitis detection, enabling timely interventions to mitigate damage to the mammary gland. Proteomics has also been used in pigs to find biomarkers for porcine reproductive and respiratory syndrome (PRRS), a viral disease associated with reproductive failure in sows and respiratory distress in piglets, aiding disease management (Genini et al., 2012). Proteomic studies have specified proteins intricate in the immune response to PRRS, which could be used to improve diagnostic tests and vaccines (Wu et al., 2022).
Besides infectious diseases, metabolic disorders like ketosis and fatty liver disease are common in high-producing livestock, particularly dairy cows (Wu, 2020). Proteomics has been used to find protein biomarkers for metabolic health in livestock, allowing primary detection and intervention (Abdelnour et al., 2019). In dairy cows, proteomic analyses of blood and milk have known biomarkers for ketosis, a metabolic disorder that occurs when cows mobilize excess fat during early lactation (Ceciliani et al., 2018). These biomarkers contain enzymes intricate in lipid metabolism, such as acyl-CoA syntheses and fatty acid-binding proteins, raised in cows with ketosis (Soares et al., 2021). Proteomics has also been used to find biomarkers for reproductive performance in livestock. For example, in cattle, proteomic studies of follicular fluid and uterine secretions have known proteins involved in ovarian function, fertilization, and early embryo development (Ferrazza et al., 2017). These biomarkers could predict fertility and develop reproductive management in dairy and beef herds (Fu et al., 2016). While proteomics has an outstanding prospective for biomarker discovery in livestock, several tasks must be addressed. One of the main tasks is the complexity of the proteome, as protein expression can differ extensively between tissues, developmental stages, and environmental conditions (Kumar et al., 2016). Another task is the need for large-scale validation of biomarkers in varied populations and environments (Trapp et al., 2014). Therefore, comprehensive validation is required before biomarkers can be implemented in routine diagnostic tests or management practices. Developing high-throughput proteomics platforms, like targeted MS and multiple reaction monitoring (MRM), facilitates the validation of biomarkers in large-scale research (Colangelo et al., 2013). As these tools develop, protein biomarkers in livestock management will likely increase, improving disease detection, animal welfare, and productivity.
3.3 Applications in meat and dairy quality development
Meat quality is a key factor in livestock production, determining market value. The biochemical composition of muscle tissues initially identifies characteristics such as tenderness, flavor, juiciness, and color (Clinquart et al., 2022). Proteomics offers valuable insights into the molecular mechanisms essential to these attributes, allowing the identification of proteins and pathways that influence meat quality (Wu et al., 2015). Postmortem muscle glycolysis expressively affects meat quality by pH, water-holding capacity, and tenderness (Stajkovic et al., 2019). Proteomic studies have known key enolase and creatine kinase proteins related to glycolysis and muscle contraction, which affect meat quality traits in cattle, pigs, and poultry (Picard et al., 2010). Proteomics has also been used to study the effect of stress on meat quality (Xing et al., 2019). Stress during transport and slaughter can yield pale, soft, and exudative (PSE) meat that is less desired by consumers. Proteomic investigations of muscle tissues have identified stress-associated proteins, like heat shock proteins and enzymes involved in oxidative stress, that are upregulated in animals related to stress (Kim et al., 2010). These proteins could be utilized as stress-resistant biomarkers, allowing us to less susceptible animals to stress and produce higher-quality meat (Gagaoua et al., 2021).
Milk quality is an essential feature of livestock productivity, with proteomics having a prospect of understanding the molecular factors that affect milk composition and yield. Proteomic analyses of milk and mammary gland tissues have recognized proteins involved in milk production, secretion, and immune function, regulating milk production and quality (Bhat et al., 2020). The proteomic studies have identified caseins, whey proteins, and enzymes intricate in fat and lactose synthesis as key contributors to milk composition (Bhat et al., 2020). Proteomics has been used to study mastitis’s effects on milk quality (Reinhardt and Lippolis, 2020). Proteomic analyses of milk from mastitis cows have known proteins involved in the immune response, such as lactoferrin and serum albumin, which are raised during infection (Abdelmegid et al., 2017). These proteins may serve as biomarkers for early mastitis discovery, allowing timely treatment to inhibit impacts on milk productivity (Libera et al., 2021).
4 Metabolomics and nutritional efficiency in livestock
Metabolomics, the inclusive study of small molecules, is a powerful tool for revealing metabolic pathways that govern livestock growth, health, and productivity. Metabolic profiling contains the detailed analysis of metabolites in biological samples such as blood, urine, tissues, and ruminal fluid, providing insights into physiological and pathological states (Saleem et al., 2013). This allows the identification of key metabolites and pathways that affect feed efficiency, growth performance, and animal health (Kaur et al., 2023). In livestock, feed efficiency is a key factor of productivity and sustainability. Animals having to convert feed into body mass efficiently reduces production costs and minimizes environmental effects, thus improving the sustainability of livestock production (Taiwo, 2023). Several factors affect feed efficiency, including genetics, gut microbiota, nutrient absorption, and metabolic activity (Li et al., 2019). Metabolomics studies have helped unravel these complexities by identifying metabolic pathways that differentiate efficient from inefficient animals (Singh et al., 2023).
Studies on cattle, pigs, and poultry have confirmed that metabolic pathways associated with energy, amino acid, and lipid metabolism are key in determining feed efficiency (Zampiga et al., 2021). Healthy animals tend to have lower nitrogenous waste metabolites and higher metabolites related to better energy utilization (Ferket et al., 2002). These results deliver potential biomarkers that can be used for the genetic selection of animals with superior feed conversion ratios (FCR). Metabolomics also allows the monitoring of nutrient absorption and utilization, providing information on how different feeds affect metabolic processes (Roques et al., 2020). We can improve feed efficiency and reduce waste by optimizing feed composition based on metabolomics data.
4.1 Identifying metabolic biomarkers for growth and health
Metabolomics has allowed the identification of biomarkers correlating substantial growth and health traits in livestock (Wang and Kadarmideen, 2019). Biomarkers are signs that reveal physiological states and can be used to monitor growth rates, disease susceptibility, immune responses, and animal welfare (Staley et al., 2018). In beef cattle, metabolites such as plasma glucose, creatinine, and urea have been associated with faster growth rates and better feed efficiency (Gómez et al., 2022). These biomarkers could find animals with superior growth potential early, enabling precision breeding and management (Karisa et al., 2014). In dairy cattle, metabolites related to energy balance and lactation performance, like ketone bodies, have been known as potential biomarkers for milk production and health status (Gross and Bruckmaier, 2019).
The detection of health-related biomarkers has also progressive disease resistance research. In poultry, metabolic biomarkers associated with oxidative stress and immune function have allied to resistance against coccidiosis (Ducatelle et al., 2018). By identifying animals with better metabolic profiles, we can breed for better disease resistance, reducing antibiotic dependence and improving animal welfare (Pal and Chakravarty, 2019). Additionally, metabolic biomarkers can support the early diagnosis of subclinical diseases, which are frequently hard to detect through traditional clinical methods (Martins et al., 2021). For example, metabolomics analyses have shown distinct metabolic signatures with mastitis in dairy cows, allowing earlier intervention and more efficient treatment (Haxhiaj et al., 2022).
4.2 Role in animal nutrition and disease resistance
The application of metabolomics in livestock nutrition delivers a comprehensive understanding of how dietary mechanisms influence animal metabolism and productivity (Fontanesi, 2016). By evaluating different diets’ metabolic changes, metabolomics can help improve feed formulations to increase performance and health (Roques et al., 2020). Diets supplemented with omega-3 fatty acids have increased livestock’s metabolic profile, leading to healthier growth rates and better meat quality (Alagawany et al., 2019). Metabolomics also offers insights into how animals respond to dietary challenges, such as nutrient deficiencies (Jones et al., 2012). By understanding the metabolic variations in response to these challenges, we can modify nutritional involvements to moderate their effects (Verma et al., 2018). Metabolomics also shows that animals suffering from heat stress have altered carbohydrate and lipid metabolism (Sammad et al., 2020).
Metabolomics permits finding metabolic pathways that contribute to immune function and pathogen resistance. Elevated systemic antioxidant metabolite profiles correlate with enhanced immunological resilience, reducing susceptibility to pathogenic defies, with mastitis in bovine and respiratory infections in swine (Castro-Moretti et al., 2020). These findings show that controlling dietary antioxidant interventions may improve host immune resilience and pathogen resistance.
4.3 Epigenomics in reproductive efficiency
Epigenomics studies have revealed that environmental factors, such as nutrition, stress, and disease, can induce epigenetic alterations that affect reproductive performance (Ho et al., 2017). Maternal nutrition in gestation can alter the DNA methylation patterns in offspring, leading to modifications in growth, metabolism, and reproductive performance (Zheng et al., 2014). These results significantly affect breeding and management practices, as they advocate optimizing the maternal environment, which might develop reproductive efficiency in livestock (Greenwood and Bell, 2014). Moreover, epigenomics modifications show a crucial role in embryo development and the formation of pregnancy. Epigenetic alterations of vital reproductive genes regulate embryonic implantation, placental morphogenesis, and fetal development, contributing to reproductive success (Palini et al., 2011; Choux et al., 2015). Elucidating epigenetic modulators of reproductive initiation allows strategic interventions to optimize fertility and reproduction.
4.4 Metabolic profiling and feed efficiency
Current advances in metabolomics have developed our understanding of feed efficiency in livestock production through inclusive metabolic profiling approaches (Artegoitia et al., 2019). Incorporating high-throughput metabolomics technologies with traditional phenotypic extents has explained the complex biochemical networks underlying feed conversion efficiency (Alfaro and Young, 2018). Metabolic profiling, mainly through mass spectrometry and nuclear magnetic resonance spectroscopy, discloses significant relations between specific metabolic signs and feed efficiency phenotypes, with residual feed intake (RFI) and feed conversion ratio (FCR) (Taiwo, 2023). High-yield animals show unique metabolic features that reveal variations in energy metabolism, protein conversion, and lipid utilization pathways (Cantalapiedra-Hijar et al., 2018). These metabolic traits indicate the physiological adaptations of these proficient animals and suggest potential biomarkers for finding superior genetics (Gaughan et al., 2019; Nawaz et al., 2024). Additionally, applying metabolomics in livestock production has added to our understanding of the interaction between host metabolism, rumen microbiota, and feed efficiency, enabling the development of targeted nutritional involvements to enhance production efficiency while minimizing environmental influence (Rawal et al., 2024).
5 DNA methylation and histone modifications in reproduction
5.1 Epigenetic regulation of fertility and embryo development
Epigenetic modifications, mainly DNA methylation and histone modifications, modulate reproductive proficiency, with abnormal epigenetic signs in gametogenesis associated with low fertility and embryonic development (Jambhekar et al., 2019; Zhang et al., 2023). Histone modifications regulate key genes intricate in early embryo development, such as cell differentiation and organogenesis (Brunmeir et al., 2009). Epigenetic alterations of reproductive developments critically underpin the efficacy of assisted reproductive technologies, with in vitro fertilization and embryonic transfer strategies (Canovas et al., 2017). ART techniques can induce epigenetic changes in embryos, leading to altered gene expression and reduced developmental potential (DeAngelis et al., 2018). By understanding the epigenetic mechanisms that control fertility and embryo development, we can improve strategies to optimize ART results and increase reproductive efficiency in livestock.
5.2 Environmental and nutritional influences on epigenetic modifications
Heat stress has been revealed to change DNA methylation patterns in sperm, resulting in low fertility and reduced embryo development (Rahman et al., 2018). Maternal nutrition during gestation can increase epigenetic modifications in offspring that affect their growth, metabolism, and reproduction (Gu et al., 2015). Nutritional values can also affect epigenetic modifications in livestock. Diets rich in methyl donors, such as folate and methionine, can expand DNA methylation and enhance reproductive efficacy (Diniz et al., 2024). Likewise, diets with antioxidants can lower oxidative stress and protect against epigenetic damage, increasing fertility and embryo development (Torres-Arce et al., 2021). Understanding the environmental and nutritional influences on epigenetic modifications is key for regulating breeding and management practices in livestock.
5.3 Integrating multi-omics for precision livestock farming
Multi-omics integration allows the inclusive analysis of biological processes, providing insights into the complicated interactions between genes, proteins, metabolites, and environmental factors that influence livestock productivity (Fonseca et al., 2018). Using genomic with metabolomics data, we can identify genetic variants regulating metabolic pathways and contribute to traits like feed efficiency, growth, and disease resistance (Suravajhala et al., 2016). Integrating transcriptomic with proteomic data can deliver insights into how gene expression is transformed into protein function, allowing the identification of key regulatory networks (Kumar et al., 2016). Integrating multi-omics data also has essential applications in reproductive biotechnology, such as embryo selection and artificial insemination, by linking epigenomics with genomic data to find epigenetic markers that indicate embryo viability and reproductive values (Hernández-Vargas et al., 2020; Zhu et al., 2021; Wadood and Zhang, 2024).
6 Multi-omics data integration strategies
6.1 Systems biology approaches in livestock
Systems biology approaches have been employed to model the regulatory networks governing feed efficiency in cattle (Woelders et al., 2011). Integrating multi-omics data allows us to find the genetic and molecular aspects causing disease susceptibility and resistance (Hasin et al., 2017). These findings are used to develop targeted interventions, such as vaccines and nutritional supplements, that improve disease resistance and increase animal welfare (Colditz, 2002).
6.2 Enhancing precision breeding and management
Integrating multi-omics data can enable precision breeding and management practices in livestock production (Suravajhala et al., 2016; Gao et al., 2024). These results are applied to improve more accurate selection standards for breeding programs, leading to better productivity and sustainability. Using genomic and metabolomics data allows us to find genetic variants affecting metabolic pathways and serve traits such as feed efficiency, growth, and disease resistance (de Almeida Santana et al., 2016). Integrating the transcriptomic and proteomic data can deliver insights into how gene expression is interpreted into protein function, allowing the identification of key regulatory networks that regulate important traits (Haider and Pal, 2013). Integrating multi-omics data also has applications in reproductive biotechnology, such as embryo selection and artificial insemination (Rabaglino et al., 2021). Combining epigenomics and genomic data can reveal epigenetic markers that predict embryo viability and reproductive performance (Hernández-Vargas et al., 2020; Wu and Sirard, 2020).
6.3 Applications of omics in reproductive biotechnology
The omics techniques in reproductive biotechnology can revolutionize livestock breeding and management practices. Genomic selection assists precise identification of breeding individuals showing enhanced reproductive potential, including fertility, embryonic viability, and reproductive proficiency (Das et al., 2021). Epigenomics data can find epigenetic markers that expect reproductive values, permitting a more precise selection of animals for breeding programs (Wang et al., 2023). Transcriptomic and proteomic data can identify key genes and proteins’ roles in reproductive processes, such as oocyte maturation, embryo development, and placental function (Zhang et al., 2009b).
7 Improving fertility of livestock production through genomics and transcriptomics
7.1 Assisted reproductive technologies and omics integration
Assisted reproductive techniques have been used in livestock production to improve reproductive performance and accelerate genetics. These techniques include artificial insemination (AI), in vitro fertilization (IVF), embryo transfer (ET), and somatic cell nuclear transfer (SCNT) (Bertolini and Bertolini, 2009). With genomic and transcriptomic insights, ART can enhance fertility and reproductive rates (Ducreux et al., 2024). Genomic selection has allowed us to find animals with superior reproduction, such as early puberty, higher conception rates, and shorter calving intervals (Granleese et al., 2015). Genome-wide association studies (GWAS) have identified loci associated with fertility traits, allowing the selection of animals with required genetics (Wu et al., 2014; Ma et al., 2019). Transcriptomics delivers valued information about gene expression dynamics during key stages of reproduction, with oocyte maturation, embryo development, and implantation (Sirard, 2012). Transcriptomic profiling of reproductive tissues discloses key molecular pathways of uterine receptivity, embryo implantation, and assisted reproductive technology efficiency (He et al., 2021).
7.2 Impacts on embryo development and success rates
Embryogenesis arises from complicated molecular processes involving transcriptional regulators, involved signaling pathways, and dynamic epigenetic modulation (Chan et al., 2011). Integrative omics analyses reveal molecular mechanisms of heavy embryogenesis, optimizing assisted reproductive technology practices (Feuer et al., 2013; Ducreux et al., 2024). Preimplantation genetic screening (PGS) and preimplantation genetic diagnosis (PGD) permit the identification of embryos with the best genetics, lowering the possibility of miscarriage and improving implantation rates (Chen et al., 2014). Transcriptomic profiling tells embryonic developmental proficiency, enabling precision embryo selection (Labrecque and Sirard, 2014). The integration of epigenomics with ART has shown the significance of epigenetic regulation during embryo development (Shi and Wu, 2009). Epigenetic changes, such as DNA methylation and histone modifications, are key in controlling gene expression in early development (Lim and Maher, 2010). Abnormal epigenetic patterns can lead to developmental abnormalities and decrease fertility rates (Boissonnas et al., 2013).
7.3 Challenges and future directions
Despite considerable developments in omics technologies and their integration with Assisted Reproductive Technologies (ART), various key challenges still need to be discussed. A primary problem lies in these methodologies’ complications and the high perspective of the data generated. Genomic, transcriptomic, proteomic, and epigenomic datasets are enormous and need advanced computational and bioinformatics techniques for practical analysis, understanding, and integration (Kaur et al., 2021). Regulating the computational capacity and expertise required to manage such complex data is possible, mainly for various livestock breeding programs (Berry et al., 2011; Ahmed, 2024). Moreover, the high expenses related to omics technologies present a critical barrier to their routine application on a broad scale.
Another task is rooted in the genetic architecture of fertility traits, which are polygenic and frequently modified by complex relations between genetic and environmental influences. This polygenic nature confuses the identification of pivotal genetic variants linked with fertility, as these traits are usually affected by many small-effect loci rather than single, efficiently identifiable genes (Montgomery et al., 2014). In species with long generation intervals and low reproductive rates, such as cattle, the ability to accurately map and validate fertility-related loci is further constrained by limited generational turnover, making longitudinal studies challenging (Li, 2017). Addressing these challenges is essential for advancing the application of omics-driven insights to improve reproductive efficiency in livestock.
8 Single-cell omics: a cutting-edge approach in livestock biotechnology
The most commonly used single-cell technologies include single-cell RNA sequencing (scRNA-seq), single-cell DNA sequencing (scDNA-seq), single-cell epigenomics (such as single-cell ATAC-seq and methylation profiling), and single-cell proteomics (Evrony et al., 2021). These methods allow for the high-throughput analysis of gene expression, genetic mutations, chromatin accessibility, and protein abundance at the resolution of individual cells (Lee et al., 2020). The ability to capture molecular heterogeneity within tissues is critical in livestock, where biological systems are complex and heterogeneous at the cellular level. The application of single-cell omics in livestock biotechnology enables the identification of rare cell populations, studying dynamic cellular processes, and unravelling cellular mechanisms that underpin key traits.
8.1 Muscle biology and meat quality
Single-cell RNA sequencing has been used to explore the molecular landscape of muscle tissues in livestock species, revealing a greater cellular diversity than previously appreciated (Liu et al., 2019). Studies have identified distinct muscle fiber types and characterized gene expression patterns associated with muscle growth, fiber composition, and hypertrophy. By investigating gene expression in individual muscle cells, researchers can pinpoint molecular pathways responsible for traits like meat tenderness, marbling, and muscle size, which are crucial for improving meat quality (Wu et al., 2023). Furthermore, single-cell technologies allow examining muscle tissue responses to various environmental and genetic factors (De Micheli et al., 2020). For example, muscle cells can be studied under different stress conditions, such as disease or extreme temperatures, to identify key genes influencing stress responses and growth patterns (Potter, 2018).
8.2 Immune system and disease resistance
Immune responses in livestock, such as those to infections or vaccinations, are complex and involve various types of immune cells that interact in a highly coordinated manner. Single-cell RNA sequencing identifies and characterizes different immune cell populations in tissues like blood, spleen, and lymph nodes (Zhao et al., 2020). Single-cell sequencing has been applied to study the immune response to infectious diseases such as mastitis in dairy cattle, avian influenza in poultry, and PRRS in pigs (Taxis and Casas, 2017). Single-cell level, scientists can uncover the molecular mechanisms behind immune activation, memory, and cellular plasticity, leading to a better understanding of how livestock animals defend against pathogens (Stubbington et al., 2017). This knowledge is critical for developing more effective vaccines, improving disease resistance, and implementing precision medicine strategies in livestock populations. Additionally, single-cell technologies facilitate the discovery of rare immune cell subsets involved in pathogen recognition and immune regulation, offering new therapeutic targets for enhancing disease resistance in livestock (Yan et al., 2024).
8.3 Reproductive efficiency and fertility
Single-cell omics have proven helpful in examining the molecular mechanisms regulating gametogenesis, embryonic development, and fertility. Using single-cell RNA sequencing, researchers can analyze the gene expression profiles of individual cells within the ovary and testis, identifying key regulators of oocyte maturation, sperm development, and follicular growth (Gong et al., 2022; Dong et al., 2023). Single-cell technologies have investigated how specific gene networks control fertility in livestock species like cattle, pigs, and sheep. Single-cell RNA sequencing has provided insights into regulating follicular development, luteal function, and oocyte quality, all influencing fertility outcomes (Zhao et al., 2023). These studies have led to a better understanding of how hormonal signaling and cell-to-cell interactions govern reproductive success, which can help identify genetic and molecular markers for fertility traits. Moreover, single-cell approaches have been employed to study the early stages of embryonic development, identifying cellular pathways involved in blastocyst formation and implantation (Liu et al., 2022). This information is crucial for improving reproductive technologies such as in vitro fertilization (IVF) and embryo transfer, which are integral to modern livestock breeding programs.
8.4 Milk production and lactation biology
Single-cell RNA sequencing has been applied to study the mammary gland at the single-cell level, revealing the complex cellular architecture and molecular pathways that regulate milk production. Scientists have identified specific gene sets involved in milk synthesis, secretion, and composition by profiling gene expression in lactating mammary epithelial cells (Martin Carli et al., 2020). Single-cell technologies have also been used to study the effects of environmental factors, such as diet, stress, and disease, on the mammary gland’s function. Single-cell RNA sequencing has revealed how the mammary epithelium responds to changes in nutritional intake and how metabolic stress affects milk production (Becker et al., 2021). This information is crucial for developing strategies to optimize lactation performance, improve milk yield, and enhance the nutritional quality of milk.
8.5 Adipose tissue biology and fat deposition
Single-cell omics have enabled researchers to explore the molecular underpinnings of adipogenesis, fat distribution, and metabolic regulation at the level of individual adipocytes. By analyzing gene expression in single adipose cells, researchers have uncovered molecular pathways that control the differentiation of preadipocytes into mature adipocytes and the regulation of fat storage (Audano et al., 2022). Single-cell RNA sequencing has been used in species like cattle and pigs to study adipose tissue from different depots identifying genes associated with fat deposition and metabolic rate (Ford et al., 2023). This knowledge is essential for developing breeding programs to improve feed efficiency, reduce fat accumulation, and enhance the quality of meat products, such as lean cuts and marbling.
8.6 Genetic selection and precision breeding
Single-cell RNA sequencing allows identifying gene networks and regulatory pathways associated with these traits, providing more precise targets for genetic selection (Jackson et al., 2020). Single-cell technologies enable the identification of cellular markers for desirable traits, which can be used to inform selection decisions in breeding programs (Dumitrascu et al., 2021). Integrating single-cell omics with gene editing technologies such as CRISPR/Cas9, precision breeding strategies can be developed to enhance specific traits in livestock populations with minimal unintended consequences (Sindelar, 2024). By providing high-resolution insights into cellular diversity, gene expression, and functional dynamics, single-cell technologies enable researchers to uncover novel molecular mechanisms that underpin key traits in livestock production and reproduction (Lyons et al., 2024). The applications of single-cell omics in livestock biotechnology, from muscle development and disease resistance to reproductive efficiency and milk production, pave the way for more efficient, sustainable, and resilient livestock farming systems. As these technologies evolve, their integration into breeding programs and biotechnology applications will be crucial for improving livestock productivity, health, and welfare.
9 Data analysis and integration complexities
The application of omics technologies in livestock breeding presents several ethical and regulatory challenges that require careful consideration. One prominent concern is the potential for unintended consequences, such as a reduction in genetic diversity or the inadvertent propagation of deleterious alleles (Singer et al., 2021). These possibilities highlight the need for approaches to observe and accomplish genetic variation in breeding populations to evade compromising overall herd resistance and suitability. Moreover, introducing gene-editing tools raises ethical questions about animal welfare and genetic variations’ broader ecological influences. The long-term effects of gene editing in livestock remain unclear, particularly concerning animal health, behavior, and welfare in generations.
In addition to ethical concerns, regulatory challenges present significant hurdles to integrating gene editing and Assisted Reproductive Technologies (ART) in commercial livestock breeding (Soini et al., 2006). Regulatory frameworks governing these technologies vary widely among countries, with many imposing strict guidelines or outright prohibitions on using gene-editing tools in animals. Navigating these complex regulatory landscapes can impede the approval and application of these innovations in commercial breeding programs. Furthermore, the risk of public conflict with genetically modified animals has an additional hurdle, as public opinions about genetic modification may affect consumer approval and market capability. These ethical, regulatory, and social challenges will be vital for the responsible and maintainable progression of omics-based methodologies in livestock breeding.
10 Future innovations and potential impacts on the global livestock industry
There is creative potential for the future of livestock production due to the ongoing advancements in omics tools and their use in Assisted Reproductive Technologies (ART). These novelties have the potency to considerably strengthen reproductive efficiency, intensify genetic improvement, and assist in the overall persistence of livestock systems. By application of genomic selection, we can find and transmit animals with better feed efficiency, thus reducing resource intake and minimizing the environmental factors related to livestock farming. Additionally, omics-based approaches allow a more comprehensive knowledge of the genetic aspects of health and disease resistance. This ability permits finding genetic markers associated with improved immune responses, supporting animals less vulnerable to disease. By decreasing the rate of illness in livestock populations, these policies reduce the dependency on antibiotics and other tonic procedures, thus contributing to global efforts to mitigate antimicrobial resistance.
Integrating multi-omics data, with genomics, transcriptomics, proteomics, and metabolomics, into livestock breeding programs also helps the introduction of precision livestock farming. This innovative approach allows the management of individual animals according to their exclusive genetic, physiological, and metabolic prominence, allowing for additional targeted and effective production rehearses. Precision-based management can improve key production characteristics, such as growth rates, reproductive efficiency, and product quality, alongside minimizing environmental influences. Integrating multi-omics in livestock breeding provides a data-driven, precision-oriented approach for assessment production with ecological viability, hopefully a new standard in global livestock management.
11 Summary of key findings
The application of omics technologies has significantly advanced livestock production by providing deep insights into the molecular mechanisms governing fertility, growth, and disease resistance. Genomic selection and transcriptomic profiling have played a crucial role in improving the success rates of assisted reproductive technologies (ART), allowing for more precise and data-driven breeding decisions. The integration of multi-omics approaches encompassing genomics, transcriptomics, proteomics, and metabolomics holds immense potential for further advancements in livestock reproduction. These approaches enhance reproductive efficiency, improve disease resistance, and boost overall productivity. By leveraging comprehensive molecular data, researchers can better understand the biological pathways influencing key traits, leading to more effective breeding strategies. Future advancements in livestock reproduction are expected to focus on refining precision breeding technologies. This includes the continued integration of genomic selection, transcriptomic profiling, and ART to optimize reproductive outcomes. Additionally, the development and application of gene-editing technologies, particularly CRISPR/Cas9, offer promising possibilities for directly modifying genetic traits associated with enhanced fertility, superior disease resistance, and greater resilience to environmental stressors. These innovations will contribute to more sustainable and efficient livestock production systems, ensuring long-term improvements in animal health and productivity. Furthermore, bioinformatics, data integration, and machine learning inventions will support the creation of analytical models that can improve ART protocols and guide breeding policies. These models will improve reproductive management’s precision, productivity, and sustainability of livestock systems. The extensive implementation of omics technologies in livestock production has the potential to play a significant role in global food security and the environment. By enhancing reproductive efficacy and decreasing the ecological footprint of livestock farming, these methodologies offer pathways to more resistant and sustainable livestock production systems, resolving the growing targets for food in an ecologically responsible manner.
Author contributions
AW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. FB: Data curation, Formal Analysis, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The China Agriculture Research System (Grant Number CARS-41) held the current work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelmegid S., Murugaiyan J., Abo-Ismail M., Caswell J. L., Kelton D., Kirby G. M. (2017). Identification of host defense-related proteins using label-free quantitative proteomic analysis of milk whey from cows with Staphylococcus aureus subclinical mastitis. Int. J. Mol. Sci. 19, 78. doi: 10.3390/ijms19010078
Abdelnour S. A., Abd El-Hack M. E., Khafaga A. F., Arif M., Taha A. E., Noreldin A. E. (2019). Stress biomarkers and proteomics alteration to thermal stress in ruminants: A review. J. thermal Biol. 79, 120–134. doi: 10.1016/j.jtherbio.2018.12.013
Adnane M., De Almeida A. M., Chapwanya A. (2024). Unveiling the power of proteomics in advancing tropical animal health and production. Trop. Anim. Health Production 56, 182. doi: https://doi.org/10.1007/s11250-024-04037-4
Ahmed F. (2024). Genomics and bioinformatics: integrating data for better genetic insights. Front. Biotechnol. Genet. 1, 126–146.
Aiello D., Patel K., Lasagna E. (2018). The myostatin gene: an overview of mechanisms of action and its relevance to livestock animals. Anim. Genet. 49, 505–519. doi: 10.1111/age.12696
Akiyama M. (2021). Multi-omics study for interpretation of genome-wide association study. J. Hum. Genet. 66, 3–10. doi: 10.1038/s10038-020-00842-5
Alagawany M., Elnesr S. S., Farag M. R., Abd El-Hack M. E., Khafaga A. F., Taha A. E., et al. (2019). Omega-3 and omega-6 fatty acids in poultry nutrition: effect on production performance and health. Animals 9, 573. doi: 10.3390/ani9080573
Alfaro A. C., Young T. (2018). Showcasing metabolomic applications in aquaculture: a review. Rev. Aquacult. 10, 135–152. doi: 10.1111/raq.12152
Alhussien M. N., Dang A. K. (2018). Milk somatic cells, factors influencing their release, future prospects, and practical utility in dairy animals: An overview. Veterinary World 11, 562. doi: 10.14202/vetworld.2018.562-577
Almeida A. M., Ali S. A., Ceciliani F., Eckersall P. D., Hernández-Castellano L. E., Han R., et al. (2021). Domestic animal proteomics in the 21st century: A global retrospective and viewpoint analysis. J. Proteomics 241, 104220. doi: 10.1016/j.jprot.2021.104220
Almeida A. M. D., Bassols A., Bendixen E., Bhide M., Ceciliani F., Cristobal S., et al. (2015). Animal board invited review: advances in proteomics for animal and food sciences. Animal 9, 1–17. doi: 10.1017/S1751731114002602
Aranciaga N., Morton J. D., Berg D. K., Gathercole J. L. (2020). Proteomics and metabolomics in cow fertility: a systematic review. Reproduction 160, 639–658. doi: 10.1530/REP-20-0047
Artegoitia V. M., Foote A. P., Lewis R. M., Freetly H. C. (2019). Metabolomics profile and targeted lipidomics in multiple tissues associated with feed efficiency in beef steers. ACS omega 4, 3973–3982. doi: 10.1021/acsomega.8b02494
Arzik Y., Kizilaslan M., White S. N., Piel L. M., Çınar M. U. (2022). Genomic analysis of gastrointestinal parasite resistance in akkaraman sheep. Genes 13, 2177. doi: 10.3390/genes13122177
Aslam S., Masoud M. S., Ashfaq U. A., Ansari M.-U. R., Nahid N., Imran M., et al. (2024). Genomic tools and technologies: revolutionizing livestock improvement. Anim. PRODUCTION Health 46. Available online at: https://www.researchgate.net/publication/387066695_Genomic_Tools_and_Technologies_Revolutionizing_Livestock_Improvement.
Audano M., Pedretti S., Caruso D., Crestani M., De Fabiani E., Mitro N. (2022). Regulatory mechanisms of the early phase of white adipocyte differentiation: An overview. Cell. Mol. Life Sci. 79, 139. doi: 10.1007/s00018-022-04169-6
Ayuti S. R., Lamid M., Warsito S. H., Al-Arif M. A., Lokapirnasari W. P., Rosyada Z. N. A., et al. (2024). A review of myostatin gene mutations: Enhancing meat production and potential in livestock genetic selection. Open Veterinary J. 14, 3189. doi: 10.5455/OVJ.2024.v14.i12.4
Barber K. W., Rinehart J. (2018). The abcs of ptms. Nat. Chem. Biol. 14, 188–192. doi: 10.1038/nchembio.2572
Barillet F. (2007). Genetic improvement for dairy production in sheep and goats. Small Ruminant Res. 70, 60–75. doi: 10.1016/j.smallrumres.2007.01.004
Bathla S., Sindhu A., Kumar S., Dubey S. K., Pattnaik S., Rawat P., et al. (2020). Quantitative proteomics revealed the putative biomarker for detection of early-stage intra-mammary gland infection in cow. J. Proteins Proteomics 11, 173–181. doi: 10.1007/s42485-020-00045-8
Becker D., Weikard R., Hadlich F., Kühn C. (2021). Single-cell RNA sequencing of freshly isolated bovine milk cells and cultured primary mammary epithelial cells. Sci. Data 8, 177. doi: 10.1038/s41597-021-00972-1
Berry D., Meade K. G., Mullen M. P., Butler S., Diskin M. G., Morris D., et al. (2011). The integration of ‘omic’disciplines and systems biology in cattle breeding. Animal 5, 493–505. doi: 10.1186/2046-0481-64-5
Bertolini M., Bertolini L. (2009). Advances in reproductive technologies in cattle: from artificial insemination to cloning. Rev. la Facultad Medicina Veterinaria y Zootecnia 56, 184–194. Available online at: https://revistas.unal.edu.co/index.php/remevez/article/view/13768.
Beuzen N., Stear M., Chang K. (2000). Molecular markers and their use in animal breeding. Veterinary J. 160, 42–52. doi: 10.1053/tvjl.2000.0468
Bhat S. A., Ahmad S. M., Ibeagha-Awemu E. M., Mobashir M., Dar M. A., Mumtaz P. T., et al. (2020). Comparative milk proteome analysis of Kashmiri and Jersey cattle identifies differential expression of key proteins involved in immune system regulation and milk quality. BMC Genomics 21, 1–10. doi: 10.1186/s12864-020-6574-4
Bohra A., Chand Jha U., Godwin I. D., Kumar Varshney R. (2020). Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 18, 2388–2405. doi: 10.1111/pbi.13472
Boissonnas C. C., Jouannet P., Jammes H. (2013). Epigenetic disorders and male subfertility. Fertility sterility 99, 624–631. doi: 10.1016/j.fertnstert.2013.01.124
Boopathi N. M. (2013). Genetic mapping and marker assisted selection (New Delhi: Springer). Available online at: https://link.springer.com/book/10.1007/978-981-15-2949-8.
Boopathi N. M., Boopathi N. M. (2020). Marker-assisted selection (MAS). Genetic mapping and marker assisted selection: Basics, practice and benefits, 343–388. doi: 10.1007/978-981-15-2949-8_9
Brito L., Bédère N., Douhard F., Oliveira H., Arnal M., Peñagaricano F., et al. (2021). Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 15, 100292. doi: 10.1016/j.animal.2021.100292
Brito L. F., Oliveira H. R., Mcconn B. R., Schinckel A. P., Arrazola A., Marchant-Forde J. N., et al. (2020). Large-scale phenotyping of livestock welfare in commercial production systems: a new frontier in animal breeding. Front. Genet. 11, 793. doi: 10.3389/fgene.2020.00793
Brookes G., Smyth S. J. (2024). Risk-appropriate regulations for gene-editing technologies. GM Crops Food 15, 1–14. doi: 10.1080/21645698.2023.2293510
Brunmeir R., Lagger S., Seiser C. (2009). Histone deacetylase 1 and 2-controlled embryonic development and cell differentiation. Int. J. Dev. Biol. 53. doi: 10.1387/ijdb.082649rb
Burkard C., Lillico S. G., Reid E., Jackson B., Mileham A. J., Ait-Ali T., et al. (2017). Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PloS Pathog. 13, e1006206. doi: 10.1371/journal.ppat.1006206
Canovas S., Ross P. J., Kelsey G., Coy P. (2017). DNA methylation in embryo development: epigenetic impact of ART (assisted reproductive technologies). Bioessays 39, 1700106. doi: 10.1002/bies.201700106
Cantalapiedra-Hijar G., Abo-Ismail M., Carstens G., Guan L., Hegarty R., Kenny D. A., et al. (2018). Biological determinants of between-animal variation in feed efficiency of growing beef cattle. animal 12, s321–s335. doi: 10.1017/S1751731118001489
Canzler S., Schor J., Busch W., Schubert K., Rolle-Kampczyk U. E., Seitz H., et al. (2020). Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 94, 371–388. doi: 10.1007/s00204-020-02656-y
Carracelas B., Navajas E. A., Vera B., Ciappesoni G. (2022). Genome-wide association study of parasite resistance to gastrointestinal nematodes in Corriedale sheep. Genes 13, 1548. doi: 10.3390/genes13091548
Castro-Moretti F. R., Gentzel I. N., Mackey D., Alonso A. P. (2020). Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 10, 52. doi: 10.3390/metabo10020052
Ceccobelli S., Perini F., Trombetta M. F., Tavoletti S., Lasagna E., Pasquini M. (2022). Effect of myostatin gene mutation on slaughtering performance and meat quality in marchigiana bulls. Animals 12, 518. doi: 10.3390/ani12040518
Ceciliani F., Lecchi C., Urh C., Sauerwein H. (2018). Proteomics and metabolomics characterizing the pathophysiology of adaptive reactions to the metabolic challenges during the transition from late pregnancy to early lactation in dairy cows. J. Proteomics 178, 92–106. doi: 10.1016/j.jprot.2017.10.010
Chakraborty D., Sharma N., Kour S., Sodhi S. S., Gupta M. K., Lee S. J., et al. (2022). Applications of omics technology for livestock selection and improvement. Front. Genet. 13, 774113. doi: 10.3389/fgene.2022.774113
Chan Y. S., Yang L., Ng H.-H. (2011). Transcriptional regulatory networks in embryonic stem cells. Epigenet. Disease: Pharmaceutical Opportunities, 239–252. doi: 10.1007/978-3-7643-8989-5_12
Chandhini S., Rejish Kumar V. J. (2019). Transcriptomics in aquaculture: current status and applications. Rev. Aquacult. 11, 1379–1397. doi: 10.1111/raq.12298
Chen C.-K., Yu H.-T., Soong Y.-K., Lee C.-L. (2014). New perspectives on preimplantation genetic diagnosis and preimplantation genetic screening. Taiwanese J. Obstetrics Gynecol. 53, 146–150. doi: 10.1016/j.tjog.2014.04.004
Cheng F., Liang J., Yang L., Lan G., Wang L., Wang L. (2021). Systematic identification and comparison of the expressed profiles of lncRNAs, miRNAs, circRNAs, and mRNAs with associated co-expression networks in pigs with low and high intramuscular fat. Animals 11, 3212. doi: 10.3390/ani11113212
Choudhary R. K., Kumar Bv S., Sekhar Mukhopadhyay C., Kashyap N., Sharma V., Singh N., et al. (2024). Animal wellness: the power of multiomics and integrative strategies: multiomics in improving animal health. Veterinary Med. Int. 2024, 4125118. doi: 10.1155/2024/4125118
Choux C., Carmignac V., Bruno C., Sagot P., Vaiman D., Fauque P. (2015). The placenta: phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin. Epigenet. 7, 1–20. doi: 10.1186/s13148-015-0120-2
Churchil R. R. (2023). Genetics, genomics and breeding for disease resistance in poultry. Ind. J. Vet. Anim. Sci. Res. 52, 1–7. https://agris.fao.org/search/en/providers/122648/records/6511ac7d58c30050e8a3c5f8.
Clinquart A., Ellies-Oury M., Hocquette J., Guillier L., Santé-Lhoutellier V., Prache S. (2022). On-farm and processing factors affecting bovine carcass and meat quality. Animal 16, 100426. doi: 10.1016/j.animal.2021.100426
Cobb J. N., Biswas P. S., Platten J. D. (2019). Back to the future: revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 132, 647–667. doi: 10.1007/s00122-018-3266-4
Cockett N. E., Kole C. (2008). Genome mapping and genomics in domestic animals (Springer Science & Business Media). Available online at: https://library.wur.nl/WebQuery/titel/2125464.
Colangelo C. M., Chung L., Bruce C., Cheung K.-H. (2013). Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods 61, 287–298. doi: 10.1016/j.ymeth.2013.05.004
Colditz I. (2002). Effects of the immune system on metabolism: implications for production and disease resistance in livestock. Livestock production Sci. 75, 257–268. doi: 10.1016/S0301-6226(01)00320-7
Cook N., Greenaway A., Kirk N. (2017). Annotated bibliography: Potential ethical issues and unintended effects of CRISPR gene editing and gene drives for invasive species control. Lincoln, New Zealand: Landcare Research. Available online at: https://www.landcareresearch.co.nz/assets/researchpubs/Gene-editing-annotated-bibliography.pdf.
Council, N.R, Earth, D.O, Studies, L, Sciences, C.O.L, Mapping, C.O, Genome, S.T.H (1988). Mapping and sequencing the human genome.
Council, N.R, Studies, L, Sciences, B.O.L, Biotechnology, C.O.A, Biotechnology, C.O.D.S.-B.C.a.W.P.O.A (2002). Animal biotechnology: science-based concerns.
Dai W.-T., Zou Y.-X., White R. R., Liu J.-X., Liu H.-Y. (2018). Transcriptomic profiles of the bovine mammary gland during lactation and the dry period. Funct. Integr. Genomics 18, 125–140. doi: 10.1007/s10142-017-0580-x
Das D., Karuthadurai T., Gnanasekaran S. (2021). “Genomic selection: a molecular tool for genetic improvement in livestock,” in Advances in animal genomics (Elsevier), 141–163. doi: 10.1016/B978-0-12-820595-2.00010-2
Das D., Paul D., Mondal S. (2022). “Role of biotechnology on animal breeding and genetic improvement,” in Emerging issues in climate smart livestock production (Academic Press), 317–337. doi: 10.1016/B978-0-12-822265-2.00015-6
Dayon L., Cominetti O., Affolter M. (2022). Proteomics of human biological fluids for biomarker discoveries: technical advances and recent applications. Expert Rev. Proteomics 19, 131–151. doi: 10.1080/14789450.2022.2070477
De Almeida Santana M. H., Junior G., Cesar A. S. M., Freua M. C., Da Costa Gomes R., Da Luz E Silva S., et al. (2016). Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle. J. Appl. Genet. 57, 495–504. doi: 10.1007/s13353-016-0344-7
Deangelis A. M., Martini A. E., Owen C. M. (2018). “Assisted reproductive technology and epigenetics,” in Seminars in reproductive medicine (Thieme Medical Publishers), 221–232. doi: 10.1055/s-0038-1675780
De Micheli A. J., Spector J. A., Elemento O., Cosgrove B. D. (2020). A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skeletal Muscle 10, 19. doi: 10.1186/s13395-020-00236-3
Devani K., Plastow G., Orsel K., Valente T. S. (2020). Genome-wide association study for mammary structure in Canadian Angus cows. PloS One 15, e0237818. doi: 10.1371/journal.pone.0237818
Diniz W. J., Reynolds L. P., Ward A. K., Caton J. S., Dahlen C. R., Mccarthy K. L., et al. (2024). “Epigenetics and Nutrition: Molecular Mechanisms and Tissue Adaptation in Developmental Programming,” in Molecular Mechanisms in Nutritional Epigenetics (Springer), 49–69. doi: 10.1007/978-3-031-54215-2_4
Do D. N., Dudemaine P.-L., Mathur M., Suravajhala P., Zhao X., Ibeagha-Awemu E. M. (2021). miRNA regulatory functions in farm animal diseases, and biomarker potentials for effective therapies. Int. J. Mol. Sci. 22, 3080. doi: 10.3390/ijms22063080
Do D. N., Suravajhala P. (2023). Role of Non-Coding RNAs in Animals. Animals 13 (5), 805. doi: 10.3390/ani13050805
Doll S., Burlingame A. L. (2015). Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 10, 63–71. doi: 10.1021/cb500904b
Dong F., Ping P., Ma Y., Chen X.-F. (2023). Application of single-cell RNA sequencing on human testicular samples: a comprehensive review. Int. J. Biol. Sci. 19, 2167. doi: 10.7150/ijbs.82191
Ducatelle R., Goossens E., De Meyer F., Eeckhaut V., Antonissen G., Haesebrouck F., et al. (2018). Biomarkers for monitoring intestinal health in poultry: present status and future perspectives. Veterinary Res. 49, 1–9. doi: 10.1186/s13567-018-0538-6
Ducreux B., Patrat C., Trasler J., Fauque P. (2024). Transcriptomic integrity of human oocytes used in ARTs: technical and intrinsic factor effects. Hum. Reprod. Update 30, 26–47. doi: 10.1093/humupd/dmad025
Dumitrascu B., Villar S., Mixon D. G., Engelhardt B. E. (2021). Optimal marker gene selection for cell type discrimination in single cell analyses. Nat. Commun. 12, 1186. doi: 10.1038/s41467-021-21453-4
Dvořáková V., Bartenschlager H., Stratil A., Horák P., Stupka R., Čítek J., et al. (2012). Association between polymorphism in the FTO gene and growth and carcass traits in pig crosses. Genet. Selection Evol. 44, 1–8. doi: 10.1186/1297-9686-44-13
Eggen A. (2012). The development and application of genomic selection as a new breeding paradigm. Anim. Front. 2, 10–15. doi: 10.2527/af.2011-0027
Evrony G. D., Hinch A. G., Luo C. (2021). Applications of single-cell DNA sequencing. Annu. Rev. Genomics Hum. Genet. 22, 171–197. doi: 10.1146/annurev-genom-111320-090436
Fan S., Kong C., Zhou R., Zheng X., Ren D., Yin Z. (2024). Protein post-translational modifications based on proteomics: A potential regulatory role in animal science. J. Agric. Food Chem. 72, 6077–6088. doi: 10.1021/acs.jafc.3c08332
Ferket P., Van Heugten E., Van Kempen T., Angel R. (2002). Nutritional strategies to reduce environmental emissions from nonruminants. J. Anim. Sci. 80, E168–E182. doi: 10.2527/animalsci2002.80E-Suppl_2E168x
Ferrazza R. D. A., Garcia H. D. M., Schmidt E. M. D. S., Mihm Carmichael M., Souza F. F. D., Burchmore R., et al. (2017). Quantitative proteomic profiling of bovine follicular fluid during follicle development. Biol. Reprod. 97, 835–849. doi: 10.1093/biolre/iox148
Feuer S., Camarano L., Rinaudo P. (2013). ART and health: clinical outcomes and insights on molecular mechanisms from rodent studies. Mol. Hum. Reprod. 19, 189–204. doi: 10.1093/molehr/gas066
Fillinger S., de la Garza L., Peltzer A., Kohlbacher O., Nahnsen S. (2019). Challenges of big data integration in the life sciences. Analytical Bioanalytical Chem. 411, 6791–6800. doi: 10.1007/s00216-019-02074-9
Fonseca P., Id-Lahoucine S., Reverter A., Medrano J. F., Fortes M. S., Casellas J., et al. (2018). Combining multi-OMICs information to identify key-regulator genes for pleiotropic effect on fertility and production traits in beef cattle. PloS One 13, e0205295. doi: 10.1371/journal.pone.0205295
Fontanesi L. (2016). Metabolomics and livestock genomics: Insights into a phenotyping frontier and its applications in animal breeding. Anim. Front. 6, 73–79. doi: 10.2527/af.2016-0011
Ford H., Liu Q., Fu X., Strieder-Barboza C. (2023). White adipose tissue heterogeneity in the single-cell era: From mice and humans to cattle. Biology 12, 1289. doi: 10.3390/biology12101289
Fu Q., Huang Y., Wang Z., Chen F., Huang D., Lu Y., et al. (2016). Proteome profile and quantitative proteomic analysis of buffalo (Bubalusbubalis) follicular fluid during follicle development. Int. J. Mol. Sci. 17, 618. doi: 10.3390/ijms17050618
Fuentes-Pardo A. P., Ruzzante D. E. (2017). Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol. 26, 5369–5406. doi: 10.1111/mec.14264
Gagaoua M., Terlouw E. C., Mullen A. M., Franco D., Warner R. D., Lorenzo J. M., et al. (2021). Molecular signatures of beef tenderness: Underlying mechanisms based on integromics of protein biomarkers from multi-platform proteomics studies. Meat Sci. 172, 108311. doi: 10.1016/j.meatsci.2020.108311
Gao F., Li P., Yin Y., Du X., Cao G., Wu S., et al. (2023). Molecular breeding of livestock for disease resistance. Virology 587, 109862. doi: 10.1016/j.virol.2023.109862
Gao Z., Lu Y., Li M., Chong Y., Hong J., Wu J., et al. (2024). Application of pan-omics technologies in research on important economic traits for ruminants. Int. J. Mol. Sci. 25, 9271. doi: 10.3390/ijms25179271
Gaughan J. B., Sejian V., Mader T. L., Dunshea F. R. (2019). Adaptation strategies: ruminants. Anim. Front. 9, 47–53. doi: 10.1093/af/vfy029
Genini S., Paternoster T., Costa A., Botti S., Luini M. V., Caprera A., et al. (2012). Identification of serum proteomic biomarkers for early porcine reproductive and respiratory syndrome (PRRS) infection. Proteome Sci. 10, 1–16. doi: 10.1186/1477-5956-10-48
Ghosh M., Sharma N., Singh A. K., Gera M., Pulicherla K. K., Jeong D. K. (2018). Transformation of animal genomics by next-generation sequencing technologies: a decade of challenges and their impact on genetic architecture. Crit. Rev. Biotechnol. 38, 1157–1175. doi: 10.1080/07388551.2018.1451819
Giagu A., Penati M., Traini S., Dore S., Addis M. F. (2022). Milk proteins as mastitis markers in dairy ruminants-a systematic review. Veterinary Res. Commun. 46, 329–351. doi: 10.1007/s11259-022-09901-y
Golan Y., Assaraf Y. G. (2020). Genetic and physiological factors affecting human milk production and composition. Nutrients 12, 1500. doi: 10.3390/nu12051500
Goldansaz S. A., Guo A. C., Sajed T., Steele M. A., Plastow G. S., Wishart D. S. (2017). Livestock metabolomics and the livestock metabolome: A systematic review. PloS One 12, e0177675. doi: 10.1371/journal.pone.0177675
Gómez J. F. M., Cônsolo N. R. B., Antonelo D. S., Beline M., Gagaoua M., Higuera-Padilla A., et al. (2022). Impact of cattle feeding strategy on the beef metabolome. Metabolites 12, 640. doi: 10.3390/metabo12070640
Gong Y., Lin Z., Wang Y., Liu Y. (2023). Research progress of non-coding RNAs regulation on intramuscular adipocytes in domestic animals. Gene 860, 147226. doi: 10.1016/j.gene.2023.147226
Gong X., Zhang Y., Ai J., Li K. (2022). Application of single-cell RNA sequencing in ovarian development. Biomolecules 13, 47. doi: 10.3390/biom13010047
Granleese T., Clark S. A., Swan A. A., van der Werf J. H. (2015). Increased genetic gains in sheep, beef and dairy breeding programs from using female reproductive technologies combined with optimal contribution selection and genomic breeding values. Genet. Selection Evol. 47, 1–13. doi: 10.1186/s12711-015-0151-3
Graw S., Chappell K., Washam C. L., Gies A., Bird J., Robeson M. S., et al. (2021). Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics 17, 170–185. doi: 10.1039/D0MO00041H
Greenwood P. L., Bell A. W. (2014). Consequences of nutrition during gestation, and the challenge to better understand and enhance livestock productivity and efficiency in pastoral ecosystems. Anim. Production Sci. 54, 1109–1118. doi: 10.1071/AN14480
Gross J. J., Bruckmaier R. (2019). Metabolic challenges in lactating dairy cows and their assessment via established and novel indicators in milk. Animal 13, s75–s81. doi: 10.1017/S175173111800349X
Gu L., Liu H., Gu X., Boots C., Moley K. H., Wang Q. (2015). Metabolic control of oocyte development: linking maternal nutrition and reproductive outcomes. Cell. Mol. Life Sci. 72, 251–271. doi: 10.1007/s00018-014-1739-4
Guo B, Dalrymple B. P. (2022). Transcriptomics of meat quality. In New aspects of meat quality. Woodhead Publishing. pp. 337–91. doi: 10.1016/B978-0-323-85879-3.00005-2
Gupta D., Bhattacharjee O., Mandal D., Sen M. K., Dey D., Dasgupta A., et al. (2019). CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 232, 116636. doi: 10.1016/j.lfs.2019.116636
Gutierrez-Reinoso M. A., Aponte P. M., Garcia-Herreros M. (2021). Genomic analysis, progress and future perspectives in dairy cattle selection: a review. Animals 11, 599. doi: 10.3390/ani11030599
Haider S., Pal R. (2013). Integrated analysis of transcriptomic and proteomic data. Curr. Genomics 14, 91–110. doi: 10.2174/1389202911314020003
Hasin Y., Seldin M., Lusis A. (2017). Multi-omics approaches to disease. Genome Biol. 18, 1–15. doi: 10.1186/s13059-017-1215-1
Haxhiaj K., Wishart D. S., Ametaj B. N. (2022). Mastitis: What it is, current diagnostics, and the potential of metabolomics to identify new predictive biomarkers. Dairy 3, 722–746. doi: 10.3390/dairy3040050
He A., Zou Y., Wan C., Zhao J., Zhang Q., Yao Z., et al. (2021). The role of transcriptomic biomarkers of endometrial receptivity in personalized embryo transfer for patients with repeated implantation failure. J. Trans. Med. 19, 176. doi: 10.1186/s12967-021-02837-y
Hernández-Vargas P., Muñoz M., Domínguez F. (2020). Identifying biomarkers for predicting successful embryo implantation: applying single to multi-OMICs to improve reproductive outcomes. Hum. Reprod. Update 26, 264–301. doi: 10.1093/humupd/dmz042
Ho S.-M., Cheong A., Adgent M. A., Veevers J., Suen A. A., Tam N. N., et al. (2017). Environmental factors, epigenetics, and developmental origin of reproductive disorders. Reprod. Toxicol. 68, 85–104. doi: 10.1016/j.reprotox.2016.07.011
Horak M., Novak J., Bienertova-Vasku J. (2016). Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 410, 1–13. doi: 10.1016/j.ydbio.2015.12.013
Hozáková K., Vavrišínová K., Neirurerová P., Bujko J. (2020). Growth of beef cattle as prediction for meat production: A review. Acta Fytotechnica Zootechnica 23. doi: 10.15414/afz.2020.23.02.58-69
Hu G., Do D. N., Davoudi P., Miar Y. (2022). Emerging roles of non-coding RNAs in the feed efficiency of livestock species. Genes 13, 297. doi: 10.3390/genes13020297
Hu G., Do D. N., Gray J., Miar Y. (2020). Selection for favorable health traits: a potential approach to cope with diseases in farm animals. Animals 10, 1717. doi: 10.3390/ani10091717
Huang C., Ge F., Ma X., Dai R., Dingkao R., Zhaxi Z., et al. (2021). Comprehensive analysis of mRNA, lncRNA, circRNA, and miRNA expression profiles and their ceRNA networks in the longissimus dorsi muscle of cattle-yak and yak. Front. Genet. 12, 772557. doi: 10.3389/fgene.2021.772557
Huang W., Guo Y., Du W., Zhang X., Li A., Miao X. (2017). Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci. Rep. 7, 5278. doi: 10.1038/s41598-017-05702-5
Hue-Beauvais C., Faulconnier Y., Charlier M., Leroux C. (2021). Nutritional regulation of mammary gland development and milk synthesis in animal models and dairy species. Genes 12, 523. doi: 10.3390/genes12040523
Ibeagha-Awemu E. M., Peters S. O., Akwanji K. A., Imumorin I. G., Zhao X. (2016). High density genome wide genotyping-by-sequencing and association identifies common and low frequency SNPs, and novel candidate genes influencing cow milk traits. Sci. Rep. 6, 31109. doi: 10.1038/srep31109
Ibeagha-Awemu E. M., Zhao X. (2015). Epigenetic marks: regulators of livestock phenotypes and conceivable sources of missing variation in livestock improvement programs. Front. Genet. 6, 302. doi: 10.3389/fgene.2015.00302
Islam M. A., Rony S. A., Rahman M. B., Cinar M. U., Villena J., Uddin M. J., et al. (2020). Improvement of disease resistance in livestock: application of immunogenomics and CRISPR/Cas9 technology. Animals 10, 2236. doi: 10.3390/ani10122236
Jackson C. A., Castro D. M., Saldi G.-A., Bonneau R., Gresham D. (2020). Gene regulatory network reconstruction using single-cell RNA sequencing of barcoded genotypes in diverse environments. elife 9, e51254. doi: 10.7554/eLife.51254
Jambhekar A., Dhall A., Shi Y. (2019). Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641. doi: 10.1038/s41580-019-0151-1
Jamwal S., Jena M. K., Tyagi N., Kancharla S., Kolli P., Mandadapu G., et al. (2023). Proteomic approaches to unravel the molecular dynamics of early pregnancy in farm animals: an in-depth review. J. Dev. Biol. 12, 2. doi: 10.3390/jdb12010002
Jevsinek Skok D., Kunej T., Kovac M., Malovrh S., Potocnik K., Petric N., et al. (2016). FTO gene variants are associated with growth and carcass traits in cattle. Anim. Genet. 47, 219–222. doi: 10.1111/age.2016.47.issue-2
Jilo D. D., Abebe B. K., Wang J., Guo J., Li A., Zan L. (2024). Long non-coding RNA (LncRNA) and epigenetic factors: their role in regulating the adipocytes in bovine. Front. Genet. 15, 1405588. doi: 10.3389/fgene.2024.1405588
Jones D. P., Park Y., Ziegler T. R. (2012). Nutritional metabolomics: progress in addressing complexity in diet and health. Annu. Rev. Nutr. 32, 183–202. doi: 10.1146/annurev-nutr-072610-145159
Kalds P., Zhou S., Huang S., Gao Y., Wang X., Chen Y. (2023). When less is more: targeting the Myostatin gene in livestock for augmenting meat production. J. Agric. Food Chem. 71, 4216–4227. doi: 10.1021/acs.jafc.2c08583
Kanakachari M., Ashwini R., Chatterjee R., Bhattacharya T. (2022). Embryonic transcriptome unravels mechanisms and pathways underlying embryonic development with respect to muscle growth, egg production, and plumage formation in native and broiler chickens. Front. Genet. 13, 990849. doi: 10.3389/fgene.2022.990849
Karisa B. K. (2013). Candidate genes, metabolites and biological pathways associated with residual feed intake and carcass quality in beef cattle (Canada: University of Alberta). doi: 10.7939/R3V40K93H
Karisa B., Thomson J., Wang Z., Li C., Montanholi Y., Miller S., et al. (2014). Plasma metabolites associated with residual feed intake and other productivity performance traits in beef cattle. Livestock Sci. 165, 200–211. doi: 10.1016/j.livsci.2014.03.002
Karve T. M., Cheema A. K. (2011). Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids 2011, 207691. doi: 10.4061/2011/207691
Kaur H., Kaur G., Gupta T., Mittal D., Ali S. A. (2023). Integrating omics technologies for a comprehensive understanding of the microbiome and its impact on cattle production. Biology 12, 1200. doi: 10.3390/biology12091200
Kaur P., Singh A., Chana I. (2021). Computational techniques and tools for omics data analysis: state-of-the-art, challenges, and future directions. Arch. Comput. Methods Eng. 28, 4595–4631. doi: 10.1007/s11831-021-09547-0
Khalil M. (2020). Molecular applications of candidate genes in genetic improvement programs in livestock. Egyptian J. Anim. Production 57, 1–23. doi: 10.21608/ejap.2020.97954
Khan M. Z., Ma Y., Ma J., Xiao J., Liu Y., Liu S., et al. (2021). Association of DGAT1 with cattle, buffalo, goat, and sheep milk and meat production traits. Front. Veterinary Sci. 8, 712470. doi: 10.3389/fvets.2021.712470
Kim N.-K., Park H.-R., Lee H.-C., Yoon D., Son E.-S., Kim Y.-S., et al. (2010). Comparative studies of skeletal muscle proteome and transcriptome profilings between pig breeds. Mamm. Genome 21, 307–319. doi: 10.1007/s00335-010-9264-8
Koltes J. E., Cole J. B., Clemmens R., Dilger R. N., Kramer L. M., Lunney J. K., et al. (2019). A vision for development and utilization of high-throughput phenotyping and big data analytics in livestock. Front. Genet. 10, 1197. doi: 10.3389/fgene.2019.01197
Korte A., Farlow A. (2013). The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9, 1–9. doi: 10.1186/1746-4811-9-29
Krostitz W. (1993). A world review of hides, skins, wool and hair production and marketing. PUBLICATION-EUROPEAN Assoc. FOR Anim. PRODUCTION 56, 7–7. Available online at: https://agris.fao.org/search/en/providers/122575/records/6471eabd77fd37171a70fd47.
Kumar D., Bansal G., Narang A., Basak T., Abbas T., Dash D. (2016). Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 16, 2533–2544. doi: 10.1002/pmic.201600140
Kumar H., Chaudhary A., Singh A., Sukhija N., Panwar A., Saravanan K., et al. (2020). A review on epigenetics: Manifestations, modifications, methods & challenges. J. Entomol. Zool. Stud. 8, 01–06. http://www.entomoljournal.com.
Labrecque R., Sirard M.-A. (2014). The study of mammalian oocyte competence by transcriptome analysis: progress and challenges. Mol. Hum. Reprod. 20, 103–116. doi: 10.1093/molehr/gat082
Ladeira M. M., Schoonmaker J. P., Gionbelli M. P., Dias J. C., Gionbelli T. R., Carvalho J. R. R., et al. (2016). Nutrigenomics and beef quality: a review about lipogenesis. Int. J. Mol. Sci. 17, 918. doi: 10.3390/ijms17060918
Lamas-Toranzo I., Ramos-Ibeas P., Pericuesta E., Bermejo-Álvarez P. (2018). Directions and applications of CRISPR technology in livestock research. Anim. Reprod. 15, 292. doi: 10.21451/1984-3143-AR2018-0075
Lamy E., Mau M. (2012). Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteomics 75, 4251–4258. doi: 10.1016/j.jprot.2012.05.007
Lee J., Hyeon D. Y., Hwang D. (2020). Single-cell multiomics: technologies and data analysis methods. Exp. Mol. Med. 52, 1428–1442. doi: 10.1038/s12276-020-0420-2
Li W. (2017). Studying the genetics of Mendelian and complex traits with high-throughput genotyping and sequencing in cattle (Belgium: Universite de Liege). https://hdl.handle.net/2268/203458.
Li F., Li C., Chen Y., Liu J., Zhang C., Irving B., et al. (2019). Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 7, 1–17. doi: 10.1186/s40168-019-0699-1
Libera K., Konieczny K., Grabska J., Smulski S., Szczerbal I., Szumacher-Strabel M., et al. (2021). Potential novel biomarkers for mastitis diagnosis in sheep. Animals 11, 2783. doi: 10.3390/ani11102783
Lim D. H., Maher E. R. (2010). DNA methylation: a form of epigenetic control of gene expression. Obstetrician Gynaecologist 12, 37–42. doi: 10.1576/toag.12.1.037.27556
Liu X., Chen W., Li W., Li Y., Priest J. R., Zhou B., et al. (2019). Single-cell RNA-seq of the developing cardiac outflow tract reveals convergent development of the vascular smooth muscle cells. Cell Rep. 28, 1346–1361. e1344. doi: 10.1016/j.celrep.2019.06.092
Liu D., Chen Y., Ren Y., Yuan P., Wang N., Liu Q., et al. (2022). Primary specification of blastocyst trophectoderm by scRNA-seq: New insights into embryo implantation. Sci. Adv. 8, eabj3725. doi: 10.1126/sciadv.abj3725
Long J. A. (2020). The ‘omics’ revolution: Use of genomic, transcriptomic, proteomic and metabolomic tools to predict male reproductive traits that impact fertility in livestock and poultry. Anim. Reprod. Sci. 220, 106354. doi: 10.1016/j.anireprosci.2020.106354
Loor J. J. (2022). Nutrigenomics in livestock: Potential role in physiological regulation and practical applications. Anim. Production Sci. 62, 901–912. doi: 10.1071/AN21512
Loor J. J., Dann H. M., Everts R. E., Oliveira R., Green C. A., Guretzky N., et al. (2005). Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol. Genomics 23, 217–226. doi: 10.1152/physiolgenomics.00132.2005
Lorente-Cebrián S., González-Muniesa P., Milagro F. I., Martínez J. A. (2019). MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clin. Sci. 133, 23–40. doi: 10.1042/CS20180890
Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. (2017). Transcriptomics technologies. PloS Comput. Biol. 13, e1005457. doi: 10.1371/journal.pcbi.1005457
Lu Q., Chen Z., Ji D., Mao Y., Jiang Q., Yang Z., et al. (2021). Progress on the regulation of ruminant milk fat by noncoding RNAs and ceRNAs. Front. Genet. 12, 733925. doi: 10.3389/fgene.2021.733925
Lunney J. K., Chen H. (2010). Genetic control of host resistance to porcine reproductive and respiratory syndrome virus (PRRSV) infection. Virus Res. 154, 161–169. doi: 10.1016/j.virusres.2010.08.004
Lyons A., Brown J., Davenport K. M. (2024). Single-cell sequencing technology in ruminant livestock: challenges and opportunities. Curr. Issues Mol. Biol. 46, 5291–5306. doi: 10.3390/cimb46060316
Ma L., Cole J., Da Y., Vanraden P. (2019). Symposium review: Genetics, genome-wide association study, and genetic improvement of dairy fertility traits. J. dairy Sci. 102, 3735–3743. doi: 10.3168/jds.2018-15269
Ma Y., Khan M. Z., Xiao J., Alugongo G. M., Chen X., Chen T., et al. (2021). Genetic markers associated with milk production traits in dairy cattle. Agriculture 11, 1018. doi: 10.3390/agriculture11101018
Marigorta U. M., Rodríguez J. A., Gibson G., Navarro A. (2018). Replicability and prediction: lessons and challenges from GWAS. Trends Genet. 34, 504–517. doi: 10.1016/j.tig.2018.03.005
Marina H., Arranz J. J., Suárez-Vega A., Pelayo R., Gutiérrez-Gil B., Toral P. G., et al. (2024). Assessment of milk metabolites as biomarkers for predicting feed efficiency in dairy sheep. J. Dairy Sci. doi: 10.3168/jds.2023-23984
Martin Carli J. F., Trahan G. D., Jones K. L., Hirsch N., Rolloff K. P., Dunn E. Z., et al. (2020). Single cell RNA sequencing of human milk-derived cells reveals sub-populations of mammary epithelial cells with molecular signatures of progenitor and mature states: a novel, non-invasive framework for investigating human lactation physiology. J. mammary gland Biol. neoplasia 25, 367–387. doi: 10.1007/s10911-020-09466-z
Martins A. M., Paiva M. U., Paiva D. V., De Oliveira R. M., MaChado H. L., Alves L. J., et al. (2021). Innovative approaches to assess intermediate cardiovascular risk subjects: a review from clinical to metabolomics strategies. Front. Cardiovasc. Med. 8, 788062. doi: 10.3389/fcvm.2021.788062
Maru D., Kumar A. (2024). Applications of omics technologies in livestock production, improvement and sustainability. Sustain. Agric. Reviews: Anim. Biotechnol. Livestock Production 4, 1–54. doi: 10.1007/978-3-031-54372-2_1
Mccarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., Ioannidis J. P., et al. (2008). Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369. doi: 10.1038/nrg2344
Mccue M. E., Mccoy A. M. (2017). The scope of big data in one medicine: unprecedented opportunities and challenges. Front. veterinary Sci. 4, 194. doi: 10.3389/fvets.2017.00194
Meleady P. (2018). Two-dimensional gel electrophoresis and 2D-DIGE. Diff. gel electrophoresis: Methods Protoc., 3–14. doi: 10.1007/978-1-4939-7268-5_1
Mills K. M. (2021). Identification of Protein and Lipid Biomarkers of Infertility in Young Boars and Prepubertal Gilts (Purdue University). Available online at: https://docs.lib.purdue.edu/dissertations/AAI30505196/.
Misra B. B., Langefeld C., Olivier M., Cox L. A. (2019). Integrated omics: tools, advances and future approaches. J. Mol. Endocrinol. 62, R21–R45. doi: 10.1530/JME-18-0055
Misztal I., Lourenco D., Legarra A. (2020). Current status of genomic evaluation. J. Anim. Sci. 98, skaa101. doi: 10.1093/jas/skaa101
Montgomery G. W., Zondervan K. T., Nyholt D. R. (2014). The future for genetic studies in reproduction. Mol. Hum. Reprod. 20, 1–14. doi: 10.1093/molehr/gat058
Nawaz A. H., Setthaya P., Feng C. (2024). Exploring evolutionary adaptations and genomic advancements to improve heat tolerance in chickens. Animals: an Open Access J. MDPI 14, 2215. doi: 10.3390/ani14152215
Nissa M. U., Pinto N., Mukherjee A., Reddy P. J., Ghosh B., Sun Z., et al. (2021). Organ-based proteome and post-translational modification profiling of a widely cultivated tropical water fish, Labeo rohita. J. Proteome Res. 21, 420–437. doi: 10.1021/acs.jproteome.1c00759
Okser S., Pahikkala T., Airola A., Salakoski T., Ripatti S., Aittokallio T. (2014). Regularized machine learning in the genetic prediction of complex traits. PloS Genet. 10, e1004754. doi: 10.1371/journal.pgen.1004754
Óvilo C., Trakooljul N., Núñez Y., Hadlich F., Murani E., Ayuso M., et al. (2022). SNP discovery and association study for growth, fatness and meat quality traits in Iberian crossbred pigs. Sci. Rep. 12, 16361. doi: 10.1038/s41598-022-20817-0
Pal A., Chakravarty A. (2019). Disease resistance for different livestock species. Genet. Breed. Dis. resistance livestock 271. doi: 10.1016/B978-0-12-816406-8.00019-X
Palini S., De Stefani S., Scala V., Dusi L., Bulletti C. (2011). Epigenetic regulatory mechanisms during preimplantation embryo development. Ann. New York Acad. Sci. 1221, 54–60. doi: 10.1111/j.1749-6632.2010.05937.x
Panner Selvam M. K., Finelli R., Agarwal A., Henkel R. (2021). Proteomics and metabolomics—Current and future perspectives in clinical andrology. Andrologia 53, e13711. doi: 10.1111/and.13711
Parreira J. R., De Sousa Araújo S. (2018). Studying the animal transcriptome: state of the art and challenges in the context of animal and veterinary sciences. Proteomics Domest. Animals: Farm to Syst. Biol., 421–446. doi: 10.1007/978-3-319-69682-9_20
Perisse I. V., Fan Z., Singina G. N., White K. L., Polejaeva I. A. (2021). Improvements in gene editing technology boost its applications in livestock. Front. Genet. 11, 614688. doi: 10.3389/fgene.2020.614688
Picard B., Berri C., Lefaucheur L., Molette C., Sayd T., Terlouw C. (2010). Skeletal muscle proteomics in livestock production. Briefings Funct. Genomics 9, 259–278. doi: 10.1093/bfgp/elq005
Podlich D. W., Winkler C. R., Cooper M. (2004). Mapping as you go: An effective approach for marker-assisted selection of complex traits. Crop Sci. 44, 1560–1571. doi: 10.2135/cropsci2004.1560
Potter S. S. (2018). Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492. doi: 10.1038/s41581-018-0021-7
Prabhod K. J. (2022). The role of machine learning in genomic medicine: advancements in disease prediction and treatment. J. Deep Learn. Genomic Data Anal. 2, 1–52. doi: 10.1016/j.xcrp.2022.101149
Prasad R. D., Prasad R. J., Shrivastav R. K., Charmode N., Rs P., Mamidpelliwar P., et al. (2023). A review on concept of veterinary bio-technology and livestock products in medicine. ES Food Agroforestry 14, 1004. doi: 10.30919/esfaf1004
Quétier F. (2016). The CRISPR-Cas9 technology: closer to the ultimate toolkit for targeted genome editing. Plant Sci. 242, 65–76. doi: 10.1016/j.plantsci.2015.09.003
Rabaglino M. B., O’doherty A., Bojsen-Møller Secher J., Lonergan P., Hyttel P., Fair T., et al. (2021). Application of multi-omics data integration and machine learning approaches to identify epigenetic and transcriptomic differences between in vitro and in vivo produced bovine embryos. PloS One 16, e0252096. doi: 10.1371/journal.pone.0252096
Rahman M. B., Schellander K., Luceno N. L., Van Soom A. (2018). Heat stress responses in spermatozoa: Mechanisms and consequences for cattle fertility. Theriogenology 113, 102–112. doi: 10.1016/j.theriogenology.2018.02.012
Ramesh S. (2013). Non-coding RNAs in crop genetic modification: considerations and predictable environmental risk assessments (ERA). Mol. Biotechnol. 55, 87–100. doi: 10.1007/s12033-013-9648-6
Raposo De Magalhães C. S. F., Cerqueira M., Schrama D., Moreira M. J. V., Boonanuntanasarn S., Rodrigues P. M. L. (2020). A Proteomics and other Omics approach in the context of farmed fish welfare and biomarker discovery. Rev. Aquacult. 12, 122–144. doi: 10.1111/raq.12308
Rawal S., Kaur H., Bhathan S., Mittal D., Kaur G., Ali S. A. (2024). “Ruminant Gut Microbiota: Interplay, Implications, and Innovations for Sustainable Livestock Production,” in Sustainable Agriculture Reviews: Animal Biotechnology for Livestock Production 4 (Springer), 205–228. doi: 10.1007/978-3-031-54372-2_7
Reinhardt T. A., Lippolis J. D. (2020). Characterization of bovine mammary gland dry secretions and their proteome from the end of lactation through day 21 of the dry period. J. Proteomics 223, 103831. doi: 10.1016/j.jprot.2020.103831
Reinhart K., Bauer M., Riedemann N. C., Hartog C. S. (2012). New approaches to sepsis: molecular diagnostics and biomarkers. Clin. Microbiol. Rev. 25, 609–634. doi: 10.1128/CMR.00016-12
Ren J., Gao Z., Lu Y., Li M., Hong J., Wu J., et al. (2024). Application of GWAS and mGWAS in livestock and poultry breeding. Animals: an Open Access J. MDPI 14, 2382. doi: 10.3390/ani14162382
Ribeiro D. M., Salama A. A., Vitor A. C., Argüello A., Moncau C. T., Santos E. M., et al. (2020). The application of omics in ruminant production: A review in the tropical and sub-tropical animal production context. J. Proteomics 227, 103905. doi: 10.1016/j.jprot.2020.103905
Richards A. L., Eckhardt M., Krogan N. J. (2021). Mass spectrometry-based protein–protein interaction networks for the study of human diseases. Mol. Syst. Biol. 17, e8792. doi: 10.15252/msb.20188792
Röös E., Bajželj B., Smith P., Patel M., Little D., Garnett T. (2017). Greedy or needy? Land use and climate impacts of food in 2050 under different livestock futures. Global Environ. Change 47, 1–12. doi: 10.1016/j.gloenvcha.2017.09.001
Roques S., Deborde C., Richard N., Skiba-Cassy S., Moing A., Fauconneau B. (2020). Metabolomics and fish nutrition: a review in the context of sustainable feed development. Rev. Aquacult. 12, 261–282. doi: 10.1111/raq.12316
Rout P. K., Behera B. K., Rout P., Behera B. (2021). Sustainability in Ruminant Livestock (Springer). https://link.springer.com/book/10.1007/978-981-33-4343-6.
Saleem F., Bouatra S., Guo A. C., Psychogios N., Mandal R., Dunn S. M., et al. (2013). The bovine ruminal fluid metabolome. Metabolomics 9, 360–378. doi: 10.1007/s11306-012-0458-9
Salleh M., Mazzoni G., Höglund J., Olijhoek D., Lund P., Løvendahl P., et al. (2017). RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high-and low-residual feed intake in Nordic dairy cattle. BMC Genomics 18, 1–17. doi: 10.1186/s12864-017-3622-9
Sammad A., Wang Y. J., Umer S., Lirong H., Khan I., Khan A., et al. (2020). Nutritional physiology and biochemistry of dairy cattle under the influence of heat stress: Consequences and opportunities. Animals 10, 793. doi: 10.3390/ani10050793
Satrio R. D., Fendiyanto M. H., Miftahudin M. (2024). “Tools and techniques used at global scale through genomics, transcriptomics, proteomics, and metabolomics to investigate plant stress responses at the molecular level,” in Molecular Dynamics of Plant Stress and its Management (Springer), 555–607.
Scheben A., Wolter F., Batley J., Puchta H., Edwards D. (2017). Towards CRISPR/Cas crops–bringing together genomics and genome editing. New Phytol. 216, 682–698. doi: 10.1111/nph.14702
Schumacher M., Delcurto-Wyffels H., Thomson J., Boles J. (2022). Fat deposition and fat effects on meat quality—a review. Animals 12, 1550. doi: 10.3390/ani12121550
Sharma B., Chettri D., Verma A. K. (2021). Biotechnological advancements in livestock production. Sustain. Agric. Rev. 54: Anim. Biotechnol. Livestock Production 1, 107–130. doi: 10.1007/978-3-030-76529-3_3
Shi L., Wu J. (2009). Epigenetic regulation in mammalian preimplantation embryo development. Reprod. Biol. Endocrinol. 7, 1–11. doi: 10.1186/1477-7827-7-59
Sikka P., Singh K. P., Singh I., Mishra D. C., Paul S. S., Balhara A. K., et al. (2023). Whole blood transcriptome analysis of lactating Murrah buffaloes divergent to contrasting genetic merits for milk yield. Front. Anim. Sci. 4, 1135429. doi: 10.3389/fanim.2023.1135429
Sindelar R. D. (2024). Genomics, other “OMIC” technologies, precision medicine, and additional biotechnology-related techniques. In Pharmaceutical Biotechnology: Fundamentals and Applications (pp. 209–254). (Cham: Springer International Publishing). doi: 10.1007/978-3-031-30023-3_9
Singer S. D., Laurie J. D., Bilichak A., Kumar S., Singh J. (2021). Genetic variation and unintended risk in the context of old and new breeding techniques. Crit. Rev. Plant Sci. 40, 68–108. doi: 10.1080/07352689.2021.1883826
Singh V., Braddick D., Dhar P. K. (2017). Exploring the potential of genome editing CRISPR-Cas9 technology. Gene 599, 1–18. doi: 10.1016/j.gene.2016.11.008
Singh U., Deb R., Alyethodi R. R., Alex R., Kumar S., Chakraborty S., et al. (2014). Molecular markers and their applications in cattle genetic research: A review. Biomarkers Genomic Med. 6, 49–58. doi: 10.1016/j.bgm.2014.03.001
Singh S., Sarma D. K., Verma V., Nagpal R., Kumar M. (2023). Unveiling the future of metabolic medicine: omics technologies driving personalized solutions for precision treatment of metabolic disorders. Biochem. Biophys. Res. Communications. doi: 10.1016/j.bbrc.2023.09.064
Singh V., Singh V. (2024). “Inferring interaction networks from transcriptomic data: methods and applications,” in Transcriptome Data Analysis (Springer), 11–37. doi: 10.1007/978-1-0716-3886-6_2
Singh B., Singh A., Singh B., Singh A. (2015). Marker-assisted selection. Marker-assisted Plant breeding: principles practices, 259–293. doi: 10.1007/978-81-322-2316-0
Sirard M. A. (2012). Factors affecting oocyte and embryo transcriptomes. Reprod. Domest. Anim. 47, 148–155. doi: 10.1111/j.1439-0531.2012.02069.x
Soares R., Vargas G., Muniz M., Soares M., Cánovas A., Schenkel F., et al. (2021). Differential gene expression in dairy cows under negative energy balance and ketosis: A systematic review and meta-analysis. J. Dairy Sci. 104, 602–615. doi: 10.3168/jds.2020-18883
Sobti R., Mukesh M., Sobti A. (2022). Genomic, proteomics, and biotechnology (CRC Press). doi: 10.1201/978100322083
Soini S., Ibarreta D., Anastasiadou V., Aymé S., Braga S., Cornel M., et al. (2006). The interface between assisted reproductive technologies and genetics: technical, social, ethical and legal issues. Eur. J. Hum. Genet. 14, 588–645. doi: 10.1038/sj.ejhg.5201598
Soler L., Uzbekova S., Blesbois E., Druart X., Labas V. (2020). Intact cell MALDI-TOF mass spectrometry, a promising proteomic profiling method in farm animal clinical and reproduction research. Theriogenology 150, 113–121. doi: 10.1016/j.theriogenology.2020.02.037
Stajkovic S., Vasilev D., Teodorovic V., Karabasil N. (2019). “Postmortem glycolysis and pork quality,” in IOP Conference Series: Earth and Environmental Science. 012032 (IOP Publishing). doi: 10.1088/1755-1315/333/1/012032
Staley M., Conners M. G., Hall K., Miller L. J. (2018). Linking stress and immunity: Immunoglobulin A as a non-invasive physiological biomarker in animal welfare studies. Hormones Behav. 102, 55–68. doi: 10.1016/j.yhbeh.2018.04.011
Stinckens A., Schroyen M., Peeters L., Janssens S., Buys N. (2010). Quantitative trait mutations in cattle, sheep and pigs: a review. CABI Rev., 1–13. doi: 10.1079/PAVSNNR20105026
Stubbington M. J., Rozenblatt-Rosen O., Regev A., Teichmann S. A. (2017). Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63. doi: 10.1126/science.aan6828
Suminda G. G. D., Bhandari S., Won Y., Goutam U., Pulicherla K. K., Son Y.-O., et al. (2022). High-throughput sequencing technologies in the detection of livestock pathogens, diagnosis, and zoonotic surveillance. Comput. Struct. Biotechnol. J. 20, 5378–5392. doi: 10.1016/j.csbj.2022.09.028
Sun H., Plastow G., Guan L. (2019). Invited review: Advances and challenges in application of feedomics to improve dairy cow production and health. J. dairy Sci. 102, 5853–5870. doi: 10.3168/jds.2018-16126
Suravajhala P. (2023). Role of non-coding RNAs in animals. Animals: an Open Access J. From MDPI 13. doi: 10.3390/ani13050805
Suravajhala P., Kogelman L. J., Kadarmideen H. N. (2016). Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare. Genet. Selection Evol. 48, 1–14. doi: 10.1186/s12711-016-0217-x
Szymanski M., Barciszewska M., Zywicki M., Barciszewski J. (2003). Noncoding RNA transcripts. J. Appl. Genet. 44, 1–20. https://pubmed.ncbi.nlm.nih.gov/12590177/.
Taiwo G. A. (2023). Exploring the biological basis of residual feed intake in beef cattle using multi-Omics analysis. doi: 10.33915/etd.12281
Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. (2019). Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20, 467–484. doi: 10.1038/s41576-019-0127-1
Taverna D., Gaspari M. (2021). A critical comparison of three MS-based approaches for quantitative proteomics analysis. J. Mass Spectrometry 56, e4669. doi: 10.1002/jms.v56.1
Taxis T. M., Casas E. (2017). MicroRNA expression and implications for infectious diseases in livestock. CABI Rev., 1–20. doi: 10.1079/PAVSNNR201712026
Te Pas M. F., Madsen O., Calus M. P., Smits M. A. (2017). The importance of endophenotypes to evaluate the relationship between genotype and external phenotype. Int. J. Mol. Sci. 18, 472. doi: 10.3390/ijms18020472
Tian Z., Zhang Y., Zhang H., Sun Y., Mao Y., Yang Z., et al. (2022). Transcriptional regulation of milk fat synthesis in dairy cattle. J. Funct. Foods 96, 105208. doi: 10.1016/j.jff.2022.105208
Torres-Arce E., Vizmanos B., Babio N., Marquez-Sandoval F., Salas-Huetos A. (2021). Dietary antioxidants in the treatment of male infertility: counteracting oxidative stress. Biology 10, 241. doi: 10.3390/biology10030241
Trapp J., Armengaud J., Salvador A., Chaumot A., Geffard O. (2014). Next-generation proteomics: toward customized biomarkers for environmental biomonitoring. Environ. Sci. Technol. 48, 13560–13572. doi: 10.1021/es501673s
Triantaphyllopoulos K. A., Ikonomopoulos I., Bannister A. J. (2016). Epigenetics and inheritance of phenotype variation in livestock. Epigenet. chromatin 9, 1–18. doi: 10.1186/s13072-016-0081-5
Udenwobele D. I., Su R.-C., Good S. V., Ball T. B., Varma Shrivastav S., Shrivastav A. (2017). Myristoylation: an important protein modification in the immune response. Front. Immunol. 8, 751. doi: 10.3389/fimmu.2017.00751
Uffelmann E., Huang Q. Q., Munung N. S., De Vries J., Okada Y., Martin A. R., et al. (2021). Genome-wide association studies. Nat. Rev. Methods Primers 1, 59. doi: 10.1038/s43586-021-00056-9
Van Arendonk J. A. (2011). The role of reproductive technologies in breeding schemes for livestock populations in developing countries. Livestock Sci. 136, 29–37. doi: 10.1016/j.livsci.2010.09.004
Velez C. (2023). Unraveling the Mystery of Non-Coding Genomic Content: Evolution, Regulation, and Functional Significance. http://hdl.handle.net/20.500.12648/14781.
Verma M., Hontecillas R., Tubau-Juni N., Abedi V., Bassaganya-Riera J. (2018). Challenges in personalized nutrition and health. Front. Media SA. doi: 10.3389/fnut.2018.00117
Wadood A. A., Zhang X. (2024). The omics revolution in understanding chicken reproduction: A comprehensive review. Curr. Issues Mol. Biol. 46, 6248–6266. doi: 10.3390/cimb46060373
Wang Y. H., Bower N., Reverter A., Tan S., De Jager N., Wang R., et al. (2009). Gene expression patterns during intramuscular fat development in cattle. J. Anim. Sci. 87, 119–130. doi: 10.2527/jas.2008-1082
Wang M., Ibeagha-Awemu E. M. (2021). Impacts of epigenetic processes on the health and productivity of livestock. Front. Genet. 11, 613636. doi: 10.3389/fgene.2020.613636
Wang X., Kadarmideen H. N. (2019). Metabolomics analyses in high-low feed efficient dairy cows reveal novel biochemical mechanisms and predictive biomarkers. Metabolites 9, 151. doi: 10.3390/metabo9070151
Wang X., Li W., Feng X., Li J., Liu G. E., Fang L., et al. (2023). Harnessing male germline epigenomics for the genetic improvement in cattle. J. Anim. Sci. Biotechnol. 14, 76. doi: 10.1186/s40104-023-00874-9
Wang X., Zhang L., Jin J., Xia A., Wang C., Cui Y., et al. (2018). Comparative transcriptome analysis to investigate the potential role of miRNAs in milk protein/fat quality. Sci. Rep. 8, 6250. doi: 10.1038/s41598-018-24727-y
Wang Y., Zhang D., Liu Y. (2024). Research progress on the regulating factors of muscle fiber heterogeneity in livestock: A review. Animals 14, 2225. doi: 10.3390/ani14152225
Wienert B., Cromer M. K. (2022). CRISPR nuclease off-target activity and mitigation strategies. Front. Genome Editing 4, 1050507. doi: 10.3389/fgeed.2022.1050507
Woelders H., Te Pas M., Bannink A., Veerkamp R., Smits M. (2011). Systems biology in animal sciences. Animal 5, 1036–1047. doi: 10.1017/S1751731111000036
Wolf J. B. (2013). Principles of transcriptome analysis and gene expression quantification: an RNA-seq tutorial. Mol. Ecol. Resour. 13, 559–572. doi: 10.1111/men.2013.13.issue-4
Wu G. (2020). “Management of metabolic disorders (including metabolic diseases) in ruminant and nonruminant animals,” in Animal agriculture (Elsevier), 471–491.
Wu G., Bazer F. W. (2019). Application of new biotechnologies for improvements in swine nutrition and pork production. J. Anim. Sci. Biotechnol. 10, 28. doi: 10.1186/s40104-019-0337-6
Wu W., Fu Y., Therkildsen M., Li X.-M., Dai R.-T. (2015). Molecular understanding of meat quality through application of proteomics. Food Rev. Int. 31, 13–28. doi: 10.1080/87559129.2014.961073
Wu Q., Han Y., Wu X., Wang Y., Su Q., Shen Y., et al. (2022). Integrated time-series transcriptomic and metabolomic analyses reveal different inflammatory and adaptive immune responses contributing to host resistance to PRRSV. Front. Immunol. 13, 960709. doi: 10.3389/fimmu.2022.960709
Wu C., Sirard M.-A. (2020). Parental effects on epigenetic programming in gametes and embryos of dairy cows. Front. Genet. 11, 557846. doi: 10.3389/fgene.2020.557846
Wu J., Zhang J., Teng M., Yang L., Yi J., Ha G., et al. (2014). Genome-wide association study of fertility traits in dairy cows. Adv. Anim. Biosci. 5, 256–280. doi: 10.1017/S2040470014000090
Wu J.-J., Zhu S., Tang Y.-F., Gu F., Valencak T. G., Liu J.-X., et al. (2023). Age-and microbiota-dependent cell stemness plasticity revealed by cattle cell landscape. Research 6, 0025. doi: 10.34133/research.0025
Xiao Q., Bai X., Zhang C., He Y. (2022). Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review. J. advanced Res. 35, 215–230. doi: 10.1016/j.jare.2021.05.002
Xing T., Gao F., Tume R. K., Zhou G., Xu X. (2019). Stress effects on meat quality: A mechanistic perspective. Compr. Rev. Food Sci. Food Saf. 18, 380–401. doi: 10.1111/crf3.2019.18.issue-2
Xu Y., Li P., Zou C., Lu Y., Xie C., Zhang X., et al. (2017). Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 68, 2641–2666. doi: 10.1093/jxb/erx135
Xu X., Veenstra T. D. (2008). Analysis of biofluids for biomarker research. PROTEOMICS–Clinical Appl. 2, 1403–1412. doi: 10.1002/prca.200780173
Yadav A., Mathan J., Dubey A. K., Singh A. (2024). The emerging role of non-coding RNAs (ncRNAs) in plant growth, development, and stress response signaling. Non-coding RNA 10, 13. doi: 10.3390/ncrna10010013
Yan Y., Zhu S., Jia M., Chen X., Qi W., Gu F., et al. (2024). Advances in single-cell transcriptomics in animal research. J. Anim. Sci. Biotechnol. 15, 102. doi: 10.1186/s40104-024-01063-y
Yang B. (2024). Enhancing dairy cow milk fat synthesis genes with CRISPR-cas9 technology to increase dairy product yield. Int. J. Mol. Veterinary Res. 14. doi: 10.5376/ijmvr.2024.14.0002
Yang J., Li X., Cao Y.-H., Pokharel K., Hu X.-J., Chen Z.-H., et al. (2019). Comparative mRNA and miRNA expression in European mouflon (Ovis musimon) and sheep (Ovis aries) provides novel insights into the genetic mechanisms for female reproductive success. Heredity 122, 172–186. doi: 10.1038/s41437-018-0090-1
Yang X., Li Y., Mei T., Duan J., Yan X., Mcnaughton L. R., et al. (2024). Genome-wide association study of exercise-induced skeletal muscle hypertrophy and the construction of predictive model. Physiol. Genomics 56, 578–589. doi: 10.1152/physiolgenomics.00019.2024
Yang Y., Wu J., Liu W., Zhao Y., Chen H. (2023). The function and regulation mechanism of non-coding RNAs in muscle development. Int. J. Mol. Sci. 24, 14534. doi: 10.3390/ijms241914534
Zachut M., Šperanda M., De Almeida A. M., Gabai G., Mobasheri A., Hernández-Castellano L. E. (2020). Biomarkers of fitness and welfare in dairy cattle: healthy productivity. J. Dairy Res. 87, 4–13. doi: 10.1017/S0022029920000084
Zampiga M., Calini F., Sirri F. (2021). Importance of feed efficiency for sustainable intensification of chicken meat production: implications and role for amino acids, feed enzymes and organic trace minerals. World’s Poultry Sci. J. 77, 639–659. doi: 10.1080/00439339.2021.1959277
Zeng Q., Du Z. Q. (2023). Advances in the discovery of genetic elements underlying longissimus dorsi muscle growth and development in the pig. Anim. Genet. 54, 709–720. doi: 10.1111/age.13365
Zhang P., Ni X., Guo Y., Guo X., Wang Y., Zhou Z., et al. (2009b). Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 10, 1–11. doi: 10.1186/1471-2164-10-348
Zhang Z., Ober U., Erbe M., Zhang H., Gao N., He J., et al. (2014). Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PloS One 9, e93017. doi: 10.1371/journal.pone.0093017
Zhang J., Sheng H., Hu C., Li F., Cai B., Ma Y., et al. (2023). Effects of DNA methylation on gene expression and phenotypic traits in cattle: A review. Int. J. Mol. Sci. 24, 11882. doi: 10.3390/ijms241511882
Zhao H., Wang Y., Yang Y. (2023). Follicular development and ovary aging: single-cell studies. Biol. Reprod. 109, 390–407. doi: 10.1093/biolre/ioad080
Zhao Y., Yang L., Su G., Wei Z., Liu X., Song L., et al. (2022). Growth traits and sperm proteomics analyses of myostatin gene-edited chinese yellow cattle. Life 12, 627. doi: 10.3390/life12050627
Zhao J., Zhang S., Liu Y., He X., Qu M., Xu G., et al. (2020). Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 6, 22. doi: 10.1038/s41421-020-0157-z
Zheng J., Xiao X., Zhang Q., Yu M. (2014). DNA methylation: the pivotal interaction between early-life nutrition and glucose metabolism in later life. Br. J. Nutr. 112, 1850–1857. doi: 10.1017/S0007114514002827
Zhou C., Shen D., Li C., Cai W., Liu S., Yin H., et al. (2019). Comparative transcriptomic and proteomic analyses identify key genes associated with milk fat traits in Chinese Holstein cows. Front. Genet. 10, 672. doi: 10.3389/fgene.2019.00672
Keywords: omics, biotechnology, genomics, reproduction, proteomics
Citation: Wadood AA, Bordbar F and Zhang X (2025) Integrating omics approaches in livestock biotechnology: innovations in production and reproductive efficiency. Front. Anim. Sci. 6:1551244. doi: 10.3389/fanim.2025.1551244
Received: 25 December 2024; Accepted: 31 March 2025;
Published: 28 April 2025.
Edited by:
Juliana Petrini, Clinica do Leite Ltda, BrazilReviewed by:
Dayal Nitai Das, National Dairy Research Institute (Southern Regional Station), IndiaZhe Zhang, Zhejiang University, China
S Mondal, National Institute of Animal Nutrition and Physiology (ICAR), India
Copyright © 2025 Wadood, Bordbar and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiquan Zhang, eHF6aGFuZ0BzY2F1LmVkdS5jbg==
 Armughan Ahmed Wadood
Armughan Ahmed Wadood Farhad Bordbar
Farhad Bordbar Xiquan Zhang
Xiquan Zhang