- 1Department of Systems Biology, Human Microbiology Institute, New York, NY, United States
- 2Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, United States
- 3Tetz Laboratories, New York, NY, United States
Introduction: Antimicrobial resistance remains a major global public health challenge that necessitates novel drugs with a low resistance rate.
Methods: Herein, we evaluate TGV-49, a novel broad-spectrum antimicrobial agent, against multidrug-resistant Gram-negative bacteria, including ESKAPE pathogens (Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae) and pathogens from agriculture that infect humans (Ralstonia solanacearum and Aeromonas hydrophila).
Results: TGV-49 was highly effective in overcoming resistance to conventional antibiotics. The experimental evolution of A. baumannii using a morbidostat revealed minimal development of resistance.
Conclusion: Our findings suggest TGV-49 as a potential alternative for combating MDR infections in clinical and agricultural settings.
1 Introduction
Antimicrobial resistance (AMR) remains among the most challenging global public health threats. Over 4.71 million deaths reported in 2021 were associated with bacterial AMR, including over 1.1 million deaths directly attributable to bacterial AMR (Naghavi et al., 2024). Among bacteria with the highest contribution to the AMR burden, Gram-negative pathogens, including Burkholderia cepacia complex, Serratia marcescens spp., and representatives of the ESKAPE group Enterobacter spp., Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa, pose a significant healthcare risk (Exner et al., 2017; Cristina et al., 2019; Dacco et al., 2023; Halawa et al., 2024; Miller and Arias, 2024).
ESKAPE pathogens are the primary causative agents for lower respiratory and bloodstream infections, the carbapenem-resistant mutants of which are associated with significant morbidity and mortality rates of <70% in certain cases (Tumbarello et al., 2015; Tamma et al., 2024; De Oliveira et al., 2020; Bassetti and Garau, 2021).
While ESKAPE pathogens are primarily associated with healthcare settings, another group of Gram-negative pathogens that poses an evolving threat to human health are plant and aquaculture primary pathogens (Li et al., 2024; Verweij et al., 2022; Rooney et al., 2020; Piotrowska and Popowska, 2014). These pathogens cause infections in immunocompromised individuals and act as reservoirs for antibiotic cross-resistance to related human pathogenic bacterial species within the pangenome (Reynolds and Kollef, 2021).
Effective antibiotic therapy faces several limitations, leading to a shortage of viable treatment options for human infections caused by Gram-negative bacteria (Dan and Talapan, 2024). One of the primary limitations is the intrinsic resistance of Gram-negative bacteria owing to multiple or pan-resistance mechanisms, rapid acquisition of resistance through gene transfer and mutations, and biofilm formation, spreading antibiotic resistance to healthcare pathogens from agriculture and aquaculture (Gerba, 2009; Li et al., 2024). The lack of efficient methods for antibiotic selection in healthcare settings is another major challenge (Manyi-Loh et al., 2018; Iwu et al., 2020; Tetz and Tetz, 2022a; Tetz et al., 2023b). Finally, not all antibiotic resistance mechanisms have been elucidated, making it difficult to fully understand and resolve this problem. In recent years, several previously unknown regulatory pathways contributing to bacterial resistance have been discovered, changing the paradigms on how AMR is established and maintained in the bacterial population.
Among them is the newly described Universal Receptive System formed by the extracellular DNA- and RNA-based Teazeled receptors on the outer surface of prokaryotic and eukaryotic cells, and includes the work of integrases and recombinases (Tetz and Tetz, 2022b; Tetz et al., 2025a). In bacteria, the Universal Receptive System regulates interactions between cells and the outer environment, including responses to antimicrobial agents on genetic and epigenetic levels. It also orchestrates the formation and maintenance of the cell memory (Tetz and Tetz, 2022b; Tetz et al., 2025a, 2025b).
Therefore, understanding the genetic mechanisms driving the development of AMR in Gram-negative pathogens is critical for developing novel drugs with a low resistance rate. The morbidostat device is one of the most sophisticated approaches for investigating resistance development in laboratory environments. It integrates experimental evolution through continuous culturing cycles with genome sequencing of the evolving isolates and characterization of the drug-resistant isolates produced (Leyn et al., 2021). The morbidostat can identify various resistance mechanisms based on specific genetic mutations in the presence of different concentrations of the tested antimicrobial agent (Leyn et al., 2021).
Our group previously developed Mul-1867, a novel antimicrobial agent exhibiting broad-spectrum antimicrobial activity by attacking the microbial cell wall. Mul-1867 is highly effective against resistant clinical bacterial and fungal isolates, as well as pre-formed biofilms (Tetz and Tetz, 2015; Tetz et al., 2016). Its derivative TGV-28 has high activity against plant pathogens (Kardava et al., 2023; Tetz et al., 2023a).
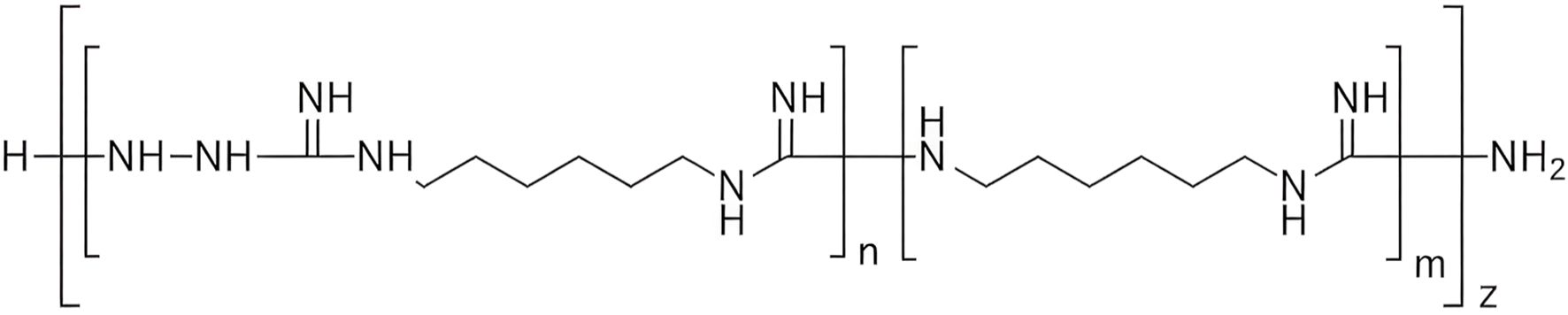
In the present study, we investigated another derivative of Mul-1867 developed by our group, TGV-49 (Tetz Group Variant-49, poly-N1-hydrazino(imino)methyl-1,6-hexanediamine) against Gram-negative pathogens, including those from the ESKAPE group (Tetz and Tetz, 2015; Tetz et al., 2016). TGV-49 shares the exact mechanism of action against bacterial cells; its positively charged hexanediamine groups bind to negatively charged bacterial membrane components (e.g., phospholipids, fatty acids), while the hydrazine groups react with carbonyl groups, thereby disrupting the microbial membrane.
This dual interaction leads to disruption of the cell wall/membrane, rapid leakage of intracellular contents (evidenced by DNA release), and eventual cell lysis. Electron microscopy confirmed that treated bacteria exhibit membrane damage and collapse (Tetz and Tetz, 2015).
TGV-49 was selected over other tested derivatives due to its higher specific activity against Gram-negative pathogens. We also used a morbidostat-based resistomics workflow to elucidate the mechanism underlying the development of resistance to TGV-49 in Acinetobacter baumannii, one of the most difficult Gram-negative pathogens to treat (Magnet et al., 2001; Zhang et al., 2024).
2 Materials and methods
2.1 Bacterial strains and growth conditions
The bacterial strains A. baumannii (ATCC BAA-1605, ATCC BAA-1710, and ATCC 17978), K. pneumoniae (ATCC BAA-1705), B. cenocepacia (ATCC-25416), and P. aeruginosa (ATCC BAA-2108) were purchased from the American Type Culture Collection (Manassas, VA, USA). Bacterial and fungal clinical isolates K. pneumoniae (VT-2646), K. oxytoca (VT-8943), B. cenocepacia (VT-2613), P. aeruginosa (VT-7530, VT-2824), P. fluorescens (VT-2535), Enterobacter cloacae (VT-1667, VT-4918), S. marcescens (VT-3904), Aeromonas hydrophila (VT-1409), Flavobacterium columnare (VT-5506), Ralstonia solanacearum (VT-5012), Xanthomonas campestris (VT-4657), Erwinia amylovora, (VT-3286), and Agrobacterium tumefaciens (VT-2679) were obtained from a private collection provided by Human Microbiology Institute (NY, USA).
{/it}All specimens were microbial suspensions derived from isolated colonies cultivated on agar plates using Columbia Blood Agar Base (Oxoid, ThermoFisher, USA). The suspensions were incubated in Mueller–Hinton broth (MHB) (Oxoid, ThermoFisher, USA) to guarantee uniform growth conditions for subsequent experimental analysis.
2.2 Antimicrobial susceptibility testing
The MICs were determined using the broth macrodilution method, as described by the Clinical and Laboratory Standards Institute M27-A3 (CLSI) (Clinical and Laboratory Standards Institute, 2019). The MIC was defined as the lowest concentration of the antimicrobial agent that inhibits bacterial growth and possesses an optical density (OD570) increase ≤ 0.05 under cultivation conditions (Dong et al., 2012). Bacteria were cultivated in Mueller-Hinton broth at 37°C, except for Flavobacterium columnare, which was cultivated at 25°C for 24 h.
Resistance or sensitivity to antibiotics was evaluated based on the breakpoints established by the CLSI and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines, and it was dependent on the antibiotic used and the pathogen tested (CLSI, 2019, 2022).
The following antimicrobial agents were used in the study: Ampicillin, Amoxicillin, Piperacillin, Ceftriaxone, Ceftazidime, Cefepime, Aztreonam, Imipenem, Meropenem, Fosfomycin, Gentamicin, Tobramycin, Amikacin, Ciprofloxacin, Levofloxacin, Polymyxin B, Colistin, Doxycycline, Tetracycline, Chloramphenicol, Trimethoprim, Sulfamethoxazole, and Rifaximin (all from Sigma-Aldrich).
TGV-49 (Human Microbiology Institute) is a derivative of Mul-1867 with more polymeric chains, resulting in a higher level of antimicrobial activity (Figure 1). For each study, the control samples were cultivated under the same conditions in antibiotic-free medium.
2.4 Experimental evolution in the morbidostat and clonal analysis
For the experimental evolution under TGV-49 pressure, we used the morbidostat device, custom-engineered based on general principles introduced by Toprak et al. (2013). Briefly, the morbidostat is a computer-controlled chemostat-like continuous culturing bioreactor where density of bacterial culture, which is constantly monitored by OD600, is controlled by varying an antibiotic concentration in media. Automated dilutions switch between addition of; (i) drug-containing media (increase of a drug concentration) when bacterial cultures are growing faster than dilution rate, and (ii) drug-free media (decrease of a drug concentration) when the growth rate slows down due to excessive drug pressure. This algorithm enables a gradually growing selective pressure driving the evolution of higher drug resistance. The detailed description of morbidostat implementation is provided on GitHub (https://github.com/sleyn/morbidostat_construction). Morbidostat-based experimental evolution of resistance to TGV-49 for A. baumannii ATCC 17978 was performed as previously described (Zlamal et al., 2021). Briefly, a morbidostat run was performed for 5 days upon inoculation of all six glass reactors with six individually prepared log-phase cultures derived from glycerol stocks of six individual colonies of A. baumannii ATCC17978 (Zlamal et al., 2021) at starting OD600 = 0.02 (20 mL in MHB/DMSO). During the run, the TGV-49 concentration in the drug-containing feed bottle was 200 mg/L (approximately 32xMIC of the unevolved parental strain). Glass reactor tubes were replaced every ~24 h (10 mL samples were taken) to minimize the impact of a biofilm accumulating on the walls of each reactor. For each reactor, glycerol stocks were prepared for all samples (taken daily) (B–F), and individual clones were randomly selected upon plating an aliquot of sample F on MHB-agar (15 clones from each reactor).
The obtained 90 clones were profiled for acquired resistance by growing them in 96-well microtiter plates in liquid MHB media without drug and supplemented with TGV-49 at 20 and 40 mg/L (corresponding to 1x and 2x MIC of unevolved strain, respectively). Clones with confirmed acquired resistance (≥ 2xMIC) were selected for the Illumina-based whole genome sequencing (WGS).
Genomic DNA was extracted using the GenElute bacterial genomic DNA kit (Sigma-Aldrich) using the manufacturer’s NA2110 protocol for Gram-negative bacteria. Libraries for sequencing were prepared using a NEBNext Ultra II FS DNA library prep kit for Illumina modules E7810L and E7595L (New England BioLabs) as per the manufacturer’s protocol (DNA input ≥100 ng); TruSeq DNA PCR-free CD index 20015949 (Illumina) adapters were used to eliminate PCR amplification. Size selection and cleanup were performed using magnetic beads AMPure XP (Beckman Coulter). Prepared libraries were quantified using a NEBNext Library Quant kit for Illumina E7630L (New England BioLabs) and pooled with volumes adjusted to normalize concentrations for ∼200-fold genomic coverage. Pooled library size and quality were analyzed with the 2100 Bioanalyzer instrument (Agilent). Pooled DNA libraries were sequenced by Novogene Co. using HiSeq 4000 (A. baumannii clones) or HiSeq X10 (all other samples) instruments (Illumina) with paired-end 150-bp read length. Our computational pipeline used for WGS data analysis and variant calling has been previously described (Zlamal et al., 2021).
2.5 Statistical analyses
Data were analyzed using GraphPad Prism version 9.3.1. Two-way analysis of variance (ANOVA) was used for multiple comparisons at the 95% confidence level (p < 0.05). Data are presented as the mean ± standard deviation (SD) with three independent replicates. All experiments were performed in triplicate in three independent experiments.
3 Results
3.1 Susceptibility and broad-spectrum activity of TGV-49 against human, animal, and plant pathogens
The antimicrobial susceptibility testing demonstrated the potent broad-spectrum activity of TGV-49 against various Gram-negative bacterial strains. Notably, all tested multidrug-resistant clinical isolates (Acinetobacter baumannii, Klebsiella pneumoniae, Burkholderia cepacia, Pseudomonas aeruginosa, and Enterobacter cloacae) were resistant to a wide range of antibiotics, including beta-lactams, aminoglycosides, carbapenems, macrolides, and quinolones (Figure 2A). Some of the tested strains exhibited resistance even to last-resort treatments such as colistin. However, all these strains, including carbapenem-resistant Gram-negative bacilli from ESKAPE pathogens, were highly susceptible to TGV-49, with minimum inhibitory concentrations (MIC) ranging from 0.06 to 4.0 μg/mL.
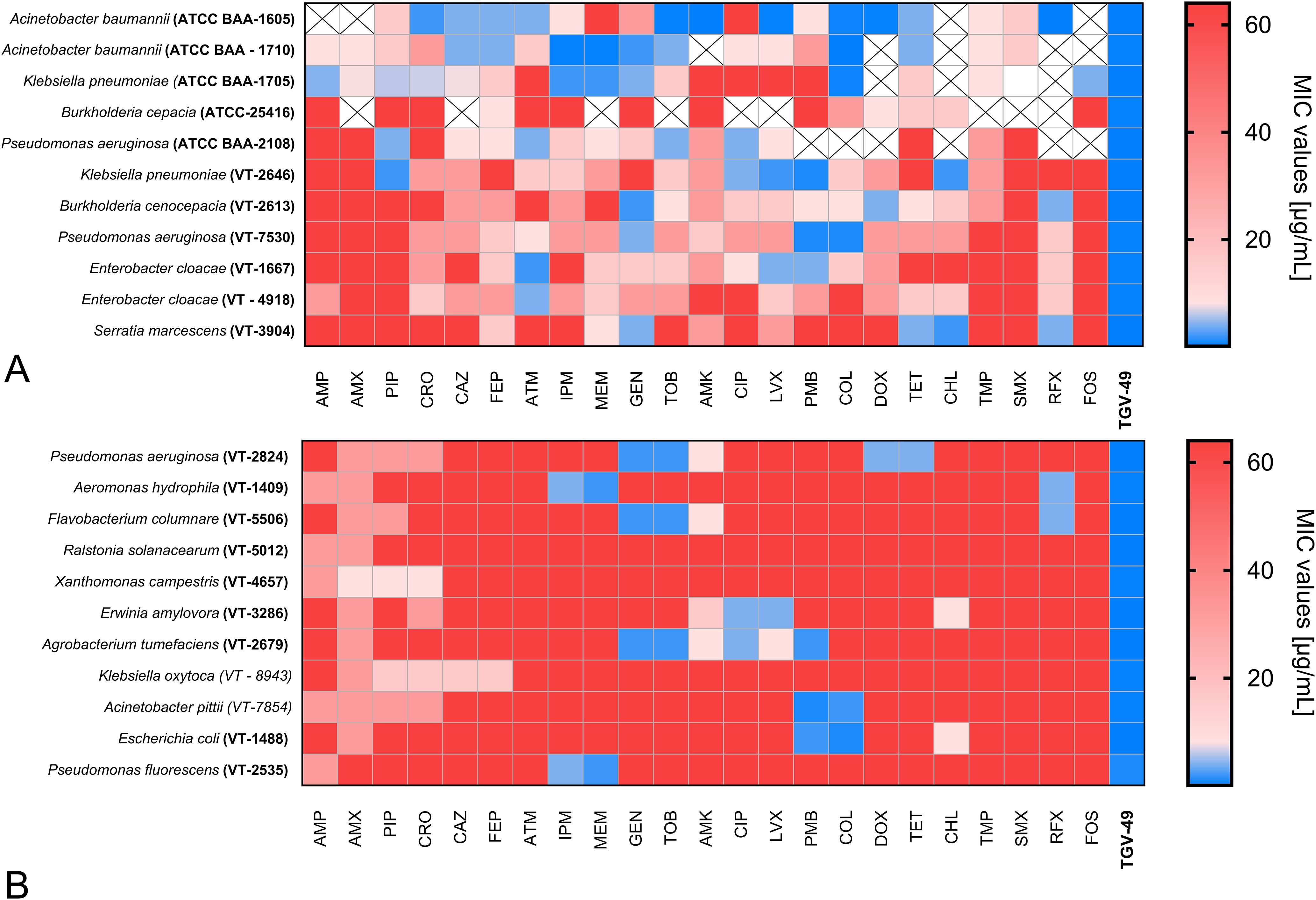
Figure 2. Antibiotic susceptibility of Gram-negative bacteria to TGV-49 and conventional antibiotics. The heatmap illustrates the antibiotic susceptibility of various Gram-negative bacterial (A) human pathogens and (B) plant and fish pathogens. The antibiotics tested against the particular bacterial strain are listed at the bottom of the heatmap. The color range reflects the MIC values (µg/mL); blue represents lower MIC values, while red represents higher MIC values (corresponding to higher resistance). The “cross” states that a particular pathogen was not tested against that particular antibiotic. Antibiotic AMP, Ampicillin; AMX, Amoxicillin; PIP, Piperacillin; CRO, Ceftriaxone; CAZ, Ceftazidime; FEP, Cefepime; ATM, Aztreonam; IPM, Imipenem; MEM, Meropenem; FOS, Fosfomycin; GEN, Gentamicin; TOB, Tobramycin; AMK, Amikacin; CIP, Ciprofloxacin; LVX, Levofloxacin; PMB, Polymyxin B; COL, Colistin; DOX, Doxycycline; TET, Tetracycline; CHL, Chloramphenicol; TMP, Trimethoprim; SMX, Sulfamethoxazole; RIF, Rifaximin.
Similarly, the antimicrobial testing of plant and fish pathogens (P. aeruginosa, P. fluorescens, A. hydrophila, A. tumerafaciens, F. columnare, R. solanacearum, X. campestris, and E. amylovora) demonstrated significant resistance to various common antibiotics used in humans (Figure 2B). However, these pathogens were susceptible to TGV-49 with low MIC values, comparable with those for human pathogens. The list of antibiotics used in this study and the results of susceptibility testing are presented in Figures 2A, B.
3.2 Experimental evolution of TGV-49 resistance in A. baumannii ATCC 17978
To investigate the dynamics of TGV-49 resistance development, we employed a morbidostat-based comparative resistomics workflow, as described by Zlamal et al. (2021), for clinically relevant Gram-negative bacteria, Acinetobacter baumannii ATCC 17978. Experimental evolution was performed in 6 parallel, 20 mL glass reactors of the custom-designed continuous culturing device (morbidostat), as described by Leyn et al. (2021). The procedure was performed under increasing pressure of TGV-49 drug at a concentration range of 0–200 mg/L as a function of growth rate assessed by constant monitoring of optical density (OD600). Five days of experimental evolution were sufficient for A. baumannii to acquire robust resistance against all tested clinical and experimental antibiotics (Zlamal et al., 2021; Leyn et al., 2023; Zampaloni et al., 2024). During this period, we observed no appreciable trend toward resistance against TGV-49. This observation was confirmed by MIC profiling of randomly picked clones (15 clones from each of the 6 reactors). Indeed, a mild resistance (2-4xMIC) was detected only among the clones from one (R5) of the six reactors.
We selected five clones (5F1–5F5, isolated from sample F taken from Reactor 5 close to the end of the 5-day evolutionary run) for analysis performed using Illumina-based whole-genomic sequencing (WGS). Data processing was performed by our computational pipeline (Zlamal et al., 2021), yielding and annotating Single Nucleotide Variants (SNV), short indels, Copy Number Variants (CNV), and insertion of IS-elements using the iJump algorithm (Zlamal et al., 2021). The results are listed in Supplementary Table S1A-C, and the significant variants are summarized in Supplementary Table S1D. All clones harbored essentially the same set of variants, including (i) SNV in the tRNA-Arg-CCG, (ii) IS-insert upstream of the pgsA gene, which encodes phospholipid biosynthesis enzyme (CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase), and (iii) ~5–10-fold amplification of two genomic loci (24.3 and 13.5 Kb). Of these, the most likely hypothetical candidate for driver event is the amplification of the second locus, which contains three paralogous genes adeT1, adeT2, and adeT3 encoding a putative RND-type efflux pump; this has previously been implicated in aminoglycoside resistance (Magnet et al., 2001).
While the identity of the actual drug target(s) and resistance driver gene(s) (and, thus, the mechanisms of action and resistance) remain elusive, with the hypothesized impact of AdeT-driven efflux being a subject of prospective genetic exploration, this study points to a very low propensity of A. baumannii to acquire TGV-49 resistance.
4 Discussion
This study underscores the broad-spectrum antimicrobial activity of TGV-49 against a wide range of Gram-negative pathogens, including those linked to human, animal, and plant health.
TGV-49 (poly-N1-hydrazino(imino)methyl-1,6-hexanediamine) has a non-specific action against bacterial cells by targeting negatively charged bacterial membrane components and disrupting the microbial membrane. Among human pathogens, TGV-49 was highly active against Gram-negative representatives of the ESKAPE group, including pathogens harboring resistance to beta-lactams, aminoglycosides, quinolones, and, most clinically important, carbapenem-resistant strains. These organisms are often implicated in severe hospital-acquired infections, particularly in immunocompromised patients with limited treatment options (Lehman and Grabowicz, 2019; Banneman and Rana, 2020; Reynolds and Kolef, 2021; Kunz Coyne et al., 2022). The demonstrated susceptibility of these strains to TGV-49 presents a promising alternative for managing difficult-to-treat infections. Given TGV-49’s activity against various multidrug-resistant pathogens, we focused on the development of resistance to TGV-49. For this purpose, we used a morbidostat system to study the evolution of resistance in A. baumannii to TGV-49. In these settings, A. baumannii showed no significant resistance even under continuous selective pressure, with only mild resistance emerging in a small subset of bacterial clones. This finding is particularly important as it suggests that TGV-49 has a lower tendency to promote rapid resistance evolution than many current antibiotics, which often become ineffective due to the swift development of resistant bacterial strains.
In conclusion, this study demonstrated the effectiveness of TGV-49 against human pathogens and its activity against agricultural and aquacultural pathogens. As antibiotic resistance continues to pose a global public health threat, TGV-49 provides a promising alternative for topical therapies to existing drugs, with the added benefit of minimizing the risk of resistance development. The broad applicability across diverse pathogen groups necessitates further research into the potential uses of TGV-49 and its derivatives.
Data availability statement
The DNAseq data files have been deposited into bioProject, Accession ID PRJNA1280665. Bacterial strains from the Human Microbiology Institute are available upon request from George Tetz lab (aW5mb0BobWktdXMuY29t).
Author contributions
VT: Writing – original draft, Visualization, Data curation, Methodology, Writing – review & editing, Conceptualization. KK: Investigation, Writing – original draft, Validation, Methodology, Data curation, Supervision. MV: Writing – review & editing, Methodology, Investigation. SL: Formal Analysis, Data curation, Methodology, Writing – review & editing, Investigation. ME: Methodology, Software, Data curation, Investigation, Writing – review & editing. AO: Supervision, Writing – review & editing, Conceptualization, Data curation, Validation, Investigation, Software, Formal Analysis, Project administration, Methodology. GT: Writing – review & editing, Supervision, Writing – original draft, Conceptualization, Data curation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The work of AO, SL and ME was supported by NIAID grant 5R01AI167977 to AO.
Conflict of interest
Author GT was employed by the company Tetz Laboratories.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frabi.2025.1615821/full#supplementary-material
References
Baneman E. and Rana M. M. (2020). Management of Acinetobacter infections in the immunosuppressed host. Emerg. Infect. Dis. 371–389. doi: 10.1016/B978-0-12-814458-9.00043-4
Bassetti M. and Garau J. (2021). Current and future perspectives in the treatment of multidrug-resistant Gram-negative infections. J. Antimicrob. Chemother. 76, iv23–iv37. doi: 10.1093/jac/dkab352
Clinical and Laboratory Standards Institute (2019). “Performance standards for antifungal susceptibility testing of yeasts; third edition,” in CLSI document M27-A3 (Clinical and Laboratory Standards Institute, Wayne, PA).
Clinical and Laboratory Standards Institute (2022). “Performance standards for antimicrobial susceptibility testing,” in CLSI supplement M100, 32nd (MD, USA: Clinical and Laboratory Standards Institute).
Cristina M. L., Sartini M., and Spagnolo A. M. (2019). Serratia marcescens infections in neonatal intensive care units (NICUs). Int. J. Environ. Res. Public Health 16, 610. doi: 10.3390/ijerph16040610
Dacco V., Alicandro G., Consales A., Rosazza C., Sciarrabba C. S., Cariani L., et al. (2023). Cepacia syndrome in cystic fibrosis: A systematic review of the literature and possible new perspectives in treatment. Ped. Pulm. 58, 1337–1343. doi: 10.1002/ppul.26359
Dan M. O. and Tǎlǎpan D. (2024). Friends or foes? Novel antimicrobials tackling MDR/XDR Gram-negative bacteria: a systematic review. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1385475
De Oliveira D. M., Forde B. M., Kidd T. J., Harris P. N., Schembri M. A., Beatson S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Micr. Rev. 17, 10–128. doi: 10.1128/CMR.00181-19
Dong L., Tong Z., Linghu D., Lin Y., Tao R., Liu J., et al. (2012). Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. Int. J. Antimicrob. Agents 39, 3910–3395. doi: 10.1016/j.ijantimicag.2012.01.009
Exner M., Bhattacharya S., Christiansen B., Gebel J., Goroncy-Bermes P., Hartemann P., et al. (2017). Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg (Germany: Infect). doi: 10.3205/dgkh000290
Gerba C. P. (2009). “Environmentally transmitted pathogens,” in In environmental microbiology (AZ, USA: Acad. Press), 445–484. doi: 10.1016/B978-0-12-370519-8.00022-5
Halawa E. M., Fadel M., Al-Rabia M. W., Behairy A., Nouh N. A., Abdo M., et al. (2024). Antibiotic action and resistance: updated review of mechanisms, spread, influencing factors, and alternative approaches for combating resistance. Front. Pharmacol. 14. doi: 10.3389/fphar.2023.1305294
Iwu C. D., Korsten L., and Okoh A. I. (2020). The incidence of antibiotic resistance within and beyond the agricultural ecosystem: A concern for public health. Microbiologyopen 9, e1035. doi: 10.1002/mbo3.1035
Kardava K., Tetz V., Vecherkovskaya M., and Tetz G. (2023). Seed dressing with M451 promotes seedling growth in wheat and reduces root phytopathogenic fungi without affecting endophytes. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1176553
Kunz Coyne A. J., El Ghali A., Holger D., Rebold N., and Rybak M. J. (2022). Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 11, 661–682. doi: 10.1007/s40121-022-00601-9
Lehman K. M. and Grabowicz M. (2019). Countering Gram-negative antibiotic resistance: recent progress in disrupting the outer membrane with novel therapeutics. Antibiotics 8, 163. doi: 10.3390/antibiotics8040163
Leyn S. A., Kent J. E., Zlamal J. E., Elane M. L., Vercruysse M., and Osterman A. L. (2023). Evolution of resistance to a tricyclic pyrimidoindole DNA gyrase/topoisomerase inhibitor in Escherichia coli and Acinetobacter baumannii is driven by upregulation of efflux machinery. bioRxiv. 27, 2023–2006. doi: 10.1101/2023.06.26.546596
Leyn S. A., Zlamal J. E., Kurnasov O. V., Li X., Elane M., Myjak L. C., et al. (2021). Experimental evolution in morbidostat reveals converging genomic trajectories on the path to triclosan resistance. Microb. Genomics. doi: 10.1099/mgen.0.000575
Li T., Ou Y., Ling S., Gao M., Deng X., Liu H., et al. (2024). Suppressing Aeromonas hydrophila and bacterial antibiotic resistance genes in tomato rhizosphere soil through companion planting with basil or cilantro. Agronomy 14, 33. doi: 10.3390/agronomy14010033
Magnet S., Courvalin P., and Lambert T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001
Manyi-Loh C., Mamphweli S., Meyer E., and Okoh A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 30, 795. doi: 10.3390/molecules23040795
Miller W. R. and Arias C. A. (2024). ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Micr. 3, 1–9. doi: 10.1038/s41579-024-01054-w
Naghavi M., Vollset S. E., Ikuta K. S., Swetschinski L. R., Gray A. P., Wool E. E., et al. (2024). Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet. 28, 1199–1226. doi: 10.1016/S0140-6736(24)01867-1
Piotrowska M. and Popowska M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64, 921–934. doi: 10.1007/s13213-013-0710-3
Reynolds D. and Kollef M. H. (2021). The epidemiology, pathogenesis, and treatment of Pseudomonas aeruginosa Ralstonia solanacearum infections: an update. Drugs 81, 2117–2131. doi: 10.1007/s40265-021-01590-0
Rooney W. M., Chai R., Milner J. J., and Walker D. (2020). Bacteriocins targeting Gram-negative phytopathogenic bacteria: Plantibiotics of the future. Front. Microbiol. 18. doi: 10.3389/fmicb.2020.575981
Tamma P. D., Heil E. L., Justo J. A., Mathers A. J., Satlin M. J., Bonomo R. A., et al. (2024). Infectious diseases society of America antimicrobial-resistant treatment guidance: Gram-negative bacterial infections. Infect. Dis. Soc Am. Available online at: https://www.idsociety.org/practice-guideline/amr-guidance/.
Tetz V., Kardava K., Krasnov K., Vecherkovskaya M., and Tetz G. (2023a). Antifungal activity of a novel synthetic polymer M451 against phytopathogens. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1176428
Tetz G., Kardava K., Vecherkovskaya M., Hahn A., Tsifansky M., Koumbourlis A., et al. (2023b). AtbFinder diagnostic test system improves optimal selection of antibiotic therapy in persons with cystic fibrosis. J. Clin. Microbiol. 61, e01558–e01522. doi: 10.1128/jcm.01558-22
Tetz G., Kardava K., Vecherkovskaya M., Khodadadi-Jamayran A., Tsirigos A., and Tetz V. (2025a). Universal receptive system as a novel regulator of transcriptomic activity of Staphylococcus aureus. Microb. Cell Fact 24, 1. doi: 10.1186/s12934-024-02637-1
Tetz V., Kardava K., Vecherkovskaya M., Khodadadi-Jamayran A., Tsirigos A., and Tetz G. (2025b). The universal receptive system: A novel regulator of antimicrobial and anticancer compound production by white blood cells. J. Leukocyte Biol. 117 (6), qiaf085. doi: 10.1093/jleuko/qiaf085
Tetz G. and Tetz V. (2015). In vitro antimicrobial activity of a novel compound, Mul-1867, against clinically important bacteria. Antimicrob. Resist. Infect. Control 4, 45. doi: 10.1186/s13756-015-0088-x
Tetz G. and Tetz V. (2022a). Overcoming antibiotic resistance with novel paradigms of antibiotic selection. Microorganisms. 10, 2383. doi: 10.3390/microorganisms10122383
Tetz G. and Tetz V. (2022b). Novel prokaryotic system employing previously unknown nucleic acids-based receptors. Microb. Cell Fact. 21, 202. doi: 10.1186/s12934-022-01923-0
Tetz G., Vikina D., and Tetz V. (2016). Antimicrobial activity of mul-1867, a novel antimicrobial compound, against multidrug-resistant Pseudomonas aeruginosa. Ann. Clin. Microbiol. Antimicrob. 15, 19. doi: 10.1186/s12941-016-0134-4
Toprak E., Veres A., Yildiz S., Pedraza J. M., Chait R., Paulsson J., et al. (2013). Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat. 8, 555–567. doi: 10.1093/jac/dkv086
Tumbarello M., Trecarichi E. M., De Rosa F. G., Giannella M., Giacobbe D. R., Bassetti M., et al. (2015). Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J. Antimicrob. Chemother. 70 (7), 2133–2143. doi: 10.1093/jac/dkv086
Verweij P. E., Arendrup M. C., Alastruey-Izquierdo A., Gold J. A., Lockhart S. R., Chiller T., et al. (2022). Dual use of antifungals in medicine and agriculture: how do we help prevent resistance developing in human pathogens? Drug Resist. Updat. 65, 100885. doi: 10.1016/j.drup.2022.100885
Zampaloni C., Mattei P., Bleicher K., Winther L., Thäte C., Bucher C., et al. (2024). A novel antibiotic class targeting the lipopolysaccharide transporter. Nature 625, 566–571. doi: 10.1038/s41586-023-06532-0
Zhang S., Di L., Qi Y., Qian X., and Wang S. (2024). Treatment of infections caused by carbapenem-resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1395260
Keywords: antibiotic resistance, TGV-49, experimental evolution, gram–negative bacteria, Acinetobacter baumannii, ESKAPE, morbidostat
Citation: Tetz VV, Kardava KM, Vecherkovskaya MF, Leyn SA, Elane ML, Osterman AL and Tetz GV (2025) Susceptibility and resistance of Gram-negative bacteria to a novel antimicrobial agent TGV-49. Front. Antibiot. 4:1615821. doi: 10.3389/frabi.2025.1615821
Received: 21 April 2025; Accepted: 20 October 2025;
Published: 03 November 2025.
Edited by:
Márió Gajdács, University of Szeged, HungaryReviewed by:
Mohammad El-Nablaway, Mansoura University, EgyptSarah Naji Aziz, Mustansiriyah University, Iraq
Copyright © 2025 Tetz, Kardava, Vecherkovskaya, Leyn, Elane, Osterman and Tetz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George V. Tetz, Zy50ZXR6QGhtaS11cy5jb20=
 Victor V. Tetz
Victor V. Tetz Kristina M. Kardava1
Kristina M. Kardava1 Maria F. Vecherkovskaya
Maria F. Vecherkovskaya Semen A. Leyn
Semen A. Leyn Andrei L. Osterman
Andrei L. Osterman George V. Tetz
George V. Tetz