- IBG-1: Biotechnology, Institute of Bio- and Geosciences, Forschungszentrum Jülich, Jülich, Germany
The oxidation of NADH with the concomitant reduction of a quinone is a crucial step in the metabolism of respiring cells. In this study, we analyzed the relevance of three different NADH oxidation systems in the actinobacterial model organism Corynebacterium glutamicum by characterizing defined mutants lacking the non-proton-pumping NADH dehydrogenase Ndh (Δndh) and/or one of the alternative NADH-oxidizing enzymes, L-lactate dehydrogenase LdhA (ΔldhA) and malate dehydrogenase Mdh (Δmdh). Together with the menaquinone-dependent L-lactate dehydrogenase LldD and malate:quinone oxidoreductase Mqo, the LdhA-LldD and Mdh-Mqo couples can functionally replace Ndh activity. In glucose minimal medium the Δndh mutant, but not the ΔldhA and Δmdh strains, showed reduced growth and a lowered NAD+/NADH ratio, in line with Ndh being the major enzyme for NADH oxidation. Growth of the double mutants ΔndhΔmdh and ΔndhΔldhA, but not of strain ΔmdhΔldhA, in glucose medium was stronger impaired than that of the Δndh mutant, supporting an active role of the alternative Mdh-Mqo and LdhA-LldD systems in NADH oxidation and menaquinone reduction. In L-lactate minimal medium the Δndh mutant grew better than the wild type, probably due to a higher activity of the menaquinone-dependent L-lactate dehydrogenase LldD. The ΔndhΔmdh mutant failed to grow in L-lactate medium and acetate medium. Growth with L-lactate could be restored by additional deletion of sugR, suggesting that ldhA repression by the transcriptional regulator SugR prevented growth on L-lactate medium. Attempts to construct a ΔndhΔmdhΔldhA triple mutant were not successful, suggesting that Ndh, Mdh and LdhA cannot be replaced by other NADH-oxidizing enzymes in C. glutamicum.
Introduction
Corynebacterium glutamicum is a Gram-positive, non-pathogenic soil bacterium that was isolated in a screen for microorganisms which excrete the flavor enhancer L-glutamate (Kinoshita et al., 1957). It is used in industry for the biotechnological production of amino acids and proteins (Eggeling and Bott, 2015; Freudl, 2017). Furthermore, strains for efficient production of many other metabolites have been constructed, such as organic acids (Wieschalka et al., 2013), diamines (Wendisch et al., 2018), and various other compounds (Becker and Wittmann, 2012). The majority of the corresponding production processes are performed under aerobic conditions and require a functional respiratory chain (Bott and Niebisch, 2003; Matsushita, 2013). Several dehydrogenases oxidize their substrates with concomitant reduction of menaquinone, the only respiratory quinone in corynebacteria. Finally, electron transfer from menaquinol to oxygen is catalyzed either by a cytochrome bc1-aa3 supercomplex (Niebisch and Bott, 2003; Graf et al., 2016; Kao et al., 2016) or by cytochrome bd oxidase (Kusumoto et al., 2000; Kabus et al., 2007).
As the majority of reducing equivalents from substrate oxidation are initially transferred to NAD+, a sufficient rate of NADH reoxidation is crucial to sustain carbon flux in central metabolism. C. glutamicum possesses a non-proton-pumping, single-subunit NADH dehydrogenase Ndh (Nantapong et al., 2005), but neither a proton-pumping complex I-type NADH dehydrogenase (Parey et al., 2020) nor a sodium-ion-pumping Nqr-type NADH dehydrogenase (Steuber et al., 2014). Analysis of a Δndh mutant of C. glutamicum revealed slightly decreased growth on glucose and acetate minimal agar plates and almost no growth defect in glucose minimal medium (Molenaar et al., 2000; Nantapong et al., 2004). Two alternative systems were proposed that can couple the oxidation of NADH to menaquinone reduction and thus compensate the lack of Ndh (Molenaar et al., 2000; Nantapong et al., 2004). These are the NAD+-dependent malate dehydrogenase Mdh (Genda et al., 2003) acting in concert with the membrane-associated malate:quinone oxidodreductase Mqo, and the NAD+-dependent L-lactate dehydrogenase LdhA acting in concert with the membrane-associated L-lactate dehydrogenase LldD (Figure 1).
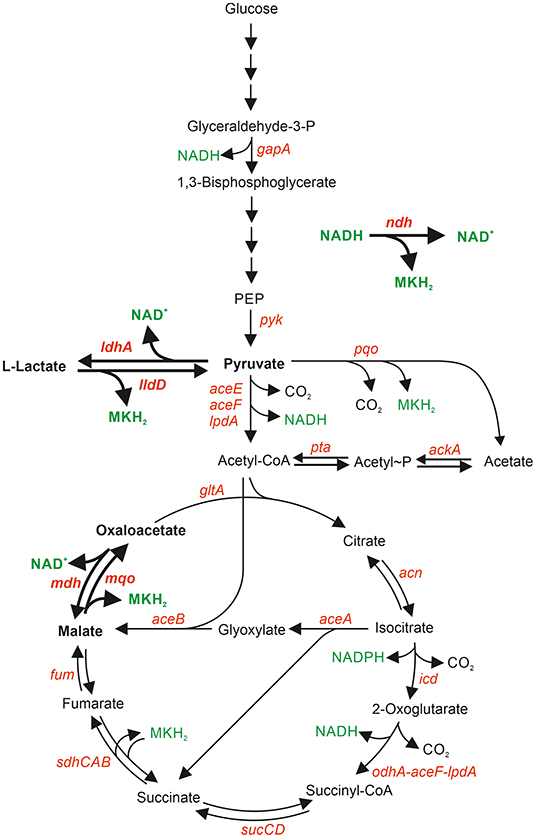
Figure 1. Central metabolic pathways relevant for NADH formation and oxidation in C. glutamicum. Genes are shown in red, NAD+, NADH, NADPH, and MKH2 in green. Reactions, metabolites, and genes relevant for NADH oxidation coupled to menaquinone (MK) reduction are shown in bold.
The net reaction of the Mdh-Mqo couple equals that of an Ndh and it could serve as an alternative “NADH dehydrogenase,” as Mdh reduces oxaloacetate with NADH to L-malate (ΔG0′ = −28.9 kJ/mol), and the membrane-associated Mqo subsequently re-oxidizes L-malate back to oxaloacetate and reduces MK (ΔG0′ = −18.5 kJ/mol). It was shown that Mqo is crucial for oxidation of malate to oxaloacetate during aerobic growth of C. glutamicum (Molenaar et al., 2000). Mdh was able to complement the absence of Mqo activity only when the Δmqo mutant was supplied with nicotinamide in order to increase the concentrations of NAD+ and NADH (Molenaar et al., 2000). Under these conditions, the NAD+/NADH ratio decreased from 4.4 in the absence of nicotinamide to 2.8 in the presence of 1 mg L[ENTX]X02212[/ENTX]1 nicotinamide, which presumably enabled the thermodynamically unfavorable oxidation of malate by Mdh (Molenaar et al., 2000). Similar to the Mdh-Mqo couple, the net reaction of the LdhA-LldD couple also corresponds to that of Ndh, as LdhA reduces pyruvate with NADH to L-lactate (ΔG0′ = −25.1 kJ/mol) and LldD re-oxidizes L-lactate to pyruvate and reduces MK (ΔG0′ = −22.4 kJ/mol) (Nantapong et al., 2004).
The Mdh-Mqo and LdhA-LldD couples might function as overflow systems that come into play when Ndh activity is insufficient. For example, an F1FO-ATP synthase mutant with 25% residual ATPase activity showed enhanced glucose consumption and respiration rates (Sekine et al., 2001), requiring increased NADH oxidation rates. A proteome comparison of this ATPase mutant with the wild type revealed that the amounts of Mqo and Mdh were increased (Li et al., 2007). The authors suggested that the Mdh-Mqo couple enabled the increased NADH oxidation rates in the ATP synthase mutant (Sawada et al., 2012). A ΔF1FO mutant of C. glutamicum completely devoid of ΔF1FO-ATP synthase activity also showed increased glucose consumption and respiration rates and both the mRNA level and the protein level of Mdh were increased (Koch-Koerfges et al., 2012).
In this study, we aimed at getting a better understanding of the role of the different NADH oxidation systems for growth and metabolism of C. glutamicum. For this purpose, we constructed and characterized six deletion mutants that lack one or two of the NADH oxidation systems. The phenotype of the different mutant strains strongly depended on the carbon source. In our study, we tested glucose, acetate, and L-lactate. The results indicate that NADH oxidation is predominantly catalyzed by Ndh, but that the Mdh-Mqo and LdhA-LldD couples are also involved and increase metabolic flexibility.
Materials and Methods
Bacterial Strains and Culture Conditions
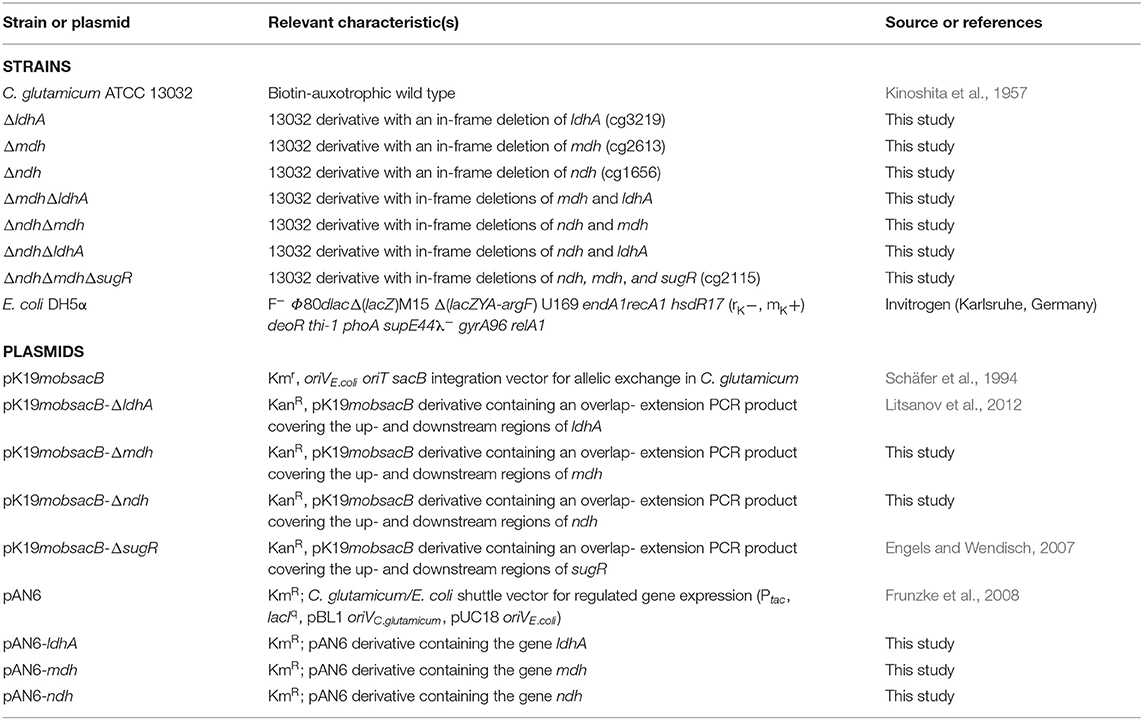
C. glutamicum strains and plasmids used in this work are listed in Table 1. For the analysis of growth, organic acid production, carbon source consumption, oxygen consumption, enzyme activities, and NAD+/NADH ratios, a 5 ml preculture in brain-heart infusion (BHI) medium was inoculated with colonies from a fresh agar plate (BHI agar) and incubated for 8 h at 30°C and 170 rpm. Cells from the preculture were transferred into 20 ml CGXII minimal medium (Keilhauer et al., 1993) containing either 4% (wt/vol) glucose (1,071 mM C), or 2–4% (wt/vol) sodium L-lactate (536–1,071 mM C), or 2–4% (wt/vol) sodium acetate (488–975 mM C) as carbon and energy source. The cultures were incubated for 16 h at 30°C and 130 rpm. After washing the cells with 0.9% (wt/vol) NaCl, the main culture with 50 ml CGXII minimal medium containing 4% (wt/vol) of the desired carbon source was inoculated to give an optical density at 600 nm (OD600) of 1. The CGXII medium was always supplemented with 30 mg/l 3,4-dihydroxybenzoic acid as iron chelator (Frunzke et al., 2008). The main cultivations were performed in baffled 500 ml Erlenmeyer flasks containing a septum for sterile sampling of the cultures at 30°C and 130 rpm. Cells were harvested during the exponential growth phase at an OD600 of 4–6 for further analysis. E. coli DH5α, which was used as host for cloning, was cultivated in LB medium or on LB agar plates at 37°C. When appropriate, kanamycin was used at a concentration of 25 μg/ml (C. glutamicum) or 50 μg/ml (E. coli).
Construction of pK19mobsacB Plasmids and Deletion Mutants
In-frame deletion mutants of C. glutamicum ATCC 13032 were constructed as described previously (Niebisch and Bott, 2001). For this purpose, the ndh or mdh upstream region, including the first seven to ten codons of each gene, and the downstream region, including the last seven codons of each gene, were amplified with the Expand High Fidelity kit (Roche Diagnostics, Mannheim, Germany) using the oligonucleotide pairs ΔXXX-1-for/ΔXXX-2-rev and ΔXXX-3-for/ΔXXX-4-rev, respectively. The “XXX” stands for the name of the gene to be deleted and is specified in Table 2, which lists the oligonucleotides used in this study. The resulting PCR products of about 500 bp were subsequently fused by overlap-extension PCR to products of ~1,000 bp. After digestion with XmaI and XbaI, these fragments were cloned into pK19mobsacB (Schäfer et al., 1994), cut with the same restriction enzymes, to yield pK19mobsacB-Δndh and pK19mobsacB-Δmdh, respectively. DNA sequence analysis revealed that the cloned PCR products contained no unwanted mutations. Subsequently, the plasmids were transferred by electroporation (van der Rest et al., 1999) into C. glutamicum strain ATCC 13032 and the transformation mixture was plated on a BHIS agar plate containing 25 μg kanamycin/ml. After selection for the first and second recombination event, kanamycin-sensitive and sucrose-resistant clones were analyzed by colony-PCR with the primer pair ΔXXX-fw/ΔXXX-rev in order to distinguish between wild type and Δndh or Δmdh clones, respectively. For the construction of the double deletion strains lacking two NADH oxidation systems, namely ΔndhΔmdh, ΔndhΔldhA, and ΔmdhΔldhA, pK19mobsacB-Δmdh or pK19mobsacB-ΔldhA were transferred by electroporation into competent Δndh or Δmdh cells and selected and analyzed as described above. For the construction of the triple deletion strain ΔndhΔmdhΔsugR, pK19mobsacB-ΔsugR was transferred by electroporation into competent ΔndhΔmdh cells and selected and analyzed as described above.
Construction of pAN6-Based Expression Plasmids
The genes ldhA, mdh, and ndh were amplified from chromosomal DNA of C. glutamicum ATCC 13032 with the gene-specific oligonucleotides XXX_F and XXX_R (Table 2). The former one introduced an NdeI restriction site including the start codon, the latter one an NheI restriction site. The resulting PCR products, with a size of 960 bp for ldhA, 1,001 bp for mdh, and 1,418 bp for ndh, were cut with NdeI and NheI and cloned separately into the expression plasmid pAN6 (Frunzke et al., 2008) cut with the same enzymes. The correctness of the cloned DNA fragments was confirmed by DNA sequencing. Subsequently, the plasmids were transferred by electroporation into C. glutamicum wild type and mutants and the transformation mixture was plated on a BHIS agar plate containing 25 μg/mL[ENTX]X02212[/ENTX]1 kanamycin. Enzymes used for DNA restriction, ligation or dephosphorylation were obtained either from Roche Diagnostics (Mannheim, Germany) or New England Biolabs (Frankfurt am Main, Germany). Plasmid DNA from E. coli or C. glutamicum was isolated with the QIAprep Spin Miniprep kit according to the manufacturer's instructions (Qiagen, Hilden, Germany).
Determination of Growth Parameters and Consumption Rates of Glucose, Organic Acids, and Oxygen
Growth was followed by measuring the optical density at 600 nm (OD600) with an Ultrospec 500-pro spectrophotometer (Amersham Biotech). The biomass concentration was calculated from OD600 values using an experimentally determined correlation factor of 0.25 gCDW L[ENTX]X02212[/ENTX]1 for OD600 = 1 (Kabus et al., 2007). Quantitative determination of glucose and organic acids in culture supernatants, and calculation of carbon source uptake rates was carried out as described (Koch-Koerfges et al., 2012).
Oxygen uptake rates (OUR) of non-growing cells were measured with a Clarke-type oxygen electrode using a thermostatically controlled, magnetically stirred 2-ml chamber at 30°C (Oxygraph, Hansatech Instruments, Germany) as described previously (Koch-Koerfges et al., 2013). The cells where harvested during exponential growth in shake flasks and oxygen consumption was followed using a cell suspension with an OD600 of 0.5–2.5 in the chamber. The protein content was calculated by assuming that 50% of the cell dry weight corresponds to protein.
Preparation of Cell Fractions for Enzymatic Assays
C. glutamicum cells were harvested by centrifugation (10,000 g, 4°C, 5 min) during the exponential growth phase and washed twice with 20 mM potassium phosphate buffer (PPB), pH 7.5. One ml of the cell suspension was mixed with 1 g zirconia/silica beads (0.1 mm diameter; Biospec, Bartlesville, USA) in a 2-ml Eppendorf tube and the cells were mechanically disrupted by three 30 s shakings in a Silamat S5 (Ivoclar Vivadent, Ellwangen, Germany). Cell debris and unbroken cells were separated by centrifugation for 5 min at 16,000 g and 4°C. The supernatant was then ultracentrifuged at 171,000 g for 60 min at 4°C. The resulting supernatant containing the soluble proteins and the resuspended sedimented membrane fraction (in 20 mM PPB pH 7.5) were used for the assays.
Enzyme Activity of Membrane-Bound Enzymes
LldD and Mqo activities were measured spectrophotometrically at 25°C by following the change in the absorbance of 2,6-dichlorophenolindophenol (DCPIP) at 600 nm (Molenaar et al., 2000; Nantapong et al., 2004). The reaction mixture contained 100 μg membrane protein, 0.2 mM DCPIP, 0.4 mM phenazine methosulfate (PMS), 1 mM NaN3, 1 mM substrate (L-lactate or L-malate for LldD and Mqo, respectively), and 50 mM PPB, pH 7.0, in a total volume of 1 ml. The amount of enzyme reducing 1 μmol/min DCPIP was defined as 1 unit, using an extinction coefficient of 22 mM−1 cm−1. Ndh activity was determined as described previously (Molenaar et al., 2000). The reaction mixture contained 100 μg membrane fraction, 0.4 mM DCPIP, 1 mM NaN3, 0.4 mM NADH, and 50 mM PPB, pH 6.5, in a total volume of 1 ml. Before adding the substrate, the reaction mixtures were incubated for 5 min at 25°C. Using a reaction mixture with buffer instead of membrane protein as a control, the decrease in absorption at 340 nm was measured and subtracted from each measurement to calculate the net value. The activity unit was defined as 1 μmol NADH oxidized per min, using an extinction coefficient of 6.3 mM−1 cm−1.
Enzyme Activity of Cytoplasmic Enzymes
LdhA and Mdh activities were determined spectrophotometrically at 25°C by following the decrease in the absorbance of NADH at 340 nm. The reaction mixture contained 100 μg soluble protein fraction, 0.2 mM NADH, 1 mM pyruvate or oxaloacetate (for Ldh or Mdh, respectively), and 50 mM PPB (pH 7.0 or pH 7.5, for the Ldh or Mdh activity assay, respectively) in a total volume of 1 ml. The activity unit was defined as 1 μmol/min NADH oxidized, using an extinction coefficient of 6.3 mM−1 cm−1. All enzyme activity results for cells grown on glucose, acetate or L-lactate minimal medium were performed in triplicates.
Determination of Intracellular NAD+/NADH Ratios
The concentrations of the pyridine nucleotides NADH and NAD+ were determined with the EnzyChrom™ NAD+/NADH assay kit (BioAssay Systems, Hayward, USA) following the manufacturer's instructions and as described previously (Siedler et al., 2011). Cells in the exponential growth phase were cooled with ice water, harvested by centrifugation (10,000 g, 4°C, 5 min), and washed twice with cold phosphate-buffered saline (PBS), pH 7.5. The sedimented cells were resuspended either in acid extraction buffer or in base extraction buffer (both part of the kit) to extract oxidized pyridine nucleotides or reduced pyridine nucleotides, respectively. The concentrations of NAD+ and NADH in each extract were then quantified by the lactate dehydrogenase cycling reaction, in which the formed NADH reduces a formazan (MTT) reagent.
Results
Growth of the C. glutamicum Strains Δndh, Δmdh, and ΔldhA on Glucose and Acetate
In order to determine the relevance of NADH dehydrogenase Ndh, malate dehydrogenase Mdh and L-lactate dehydrogenase LdhA for aerobic growth of C. glutamicum, we characterized the deletion mutants Δndh, Δmdh, and ΔldhA during cultivation in shake flasks with glucose, L-lactate, or acetate, as carbon source. These substrates were chosen as their oxidation is linked to different numbers of NADH-forming reactions. Glucose oxidation to 6 CO2 involves three NADH-forming reactions [glyceraldehyde 3-phosphate dehydrogenase, pyruvate dehydrogenase complex, and unusual 2-oxoglutarate dehydrogenase complex (Niebisch et al., 2006; Hoffelder et al., 2010; Bruch et al., 2020; Kinugawa et al., 2020)] and is coupled to the formation of 6 NADH, 2 NADPH, and 4 MQH2 if the oxidative pentose phosphate pathway is neglected. L-Lactate oxidation to 3 CO2 involves two NADH-forming reactions (pyruvate dehydrogenase complex, and 2-oxoglutarate dehydrogenase complex) and leads to the formation of 2 NADH, 1 NADPH, and 3 MQH2. Acetate oxidation to 2 CO2 involves only one NADH-forming reaction (2-oxoglutarate dehydrogenase complex) and is linked to the formation of 1 NADH, 1 NADPH, and 2 MQH2 (Figure 1).
In glucose minimal medium the Δndh mutant showed an 18% decreased growth rate and a 12% reduced biomass formation compared to the wild type (Figure 2A, Table 3). NADH dehydrogenase activity was not detectable in extracts of the Δndh mutant, whereas it was high (2.4 U mg−1) in the wild type (Table 4). The Δndh mutant showed a 28% decreased glucose uptake rate (GUR, Table 3) and a 27% decreased oxygen uptake rate compared to the wild type (OUR, 185 ± 13 vs. 254 ± 6 nmol O2 mgProtein−1 min−1), suggesting that glycolytic flux and consequently also respiration were reduced due to an insufficient rate of NADH oxidation in the absence of Ndh. In agreement with this assumption, the NAD+/NADH ratio was reduced by more than 50% in the Δndh mutant compared to the wild type (2.0 vs. 4.7). Furthermore, the Δndh mutant secreted 22% more lactate than the wild type (Table 3). Pyruvate reduction to lactate by LdhA represents an alternative path for NADH oxidation. The enzyme activities of LdhA, Mdh, and Mqo were unchanged in the Δndh mutant, whereas LldD activity was increased 3.3-fold (Table 4).
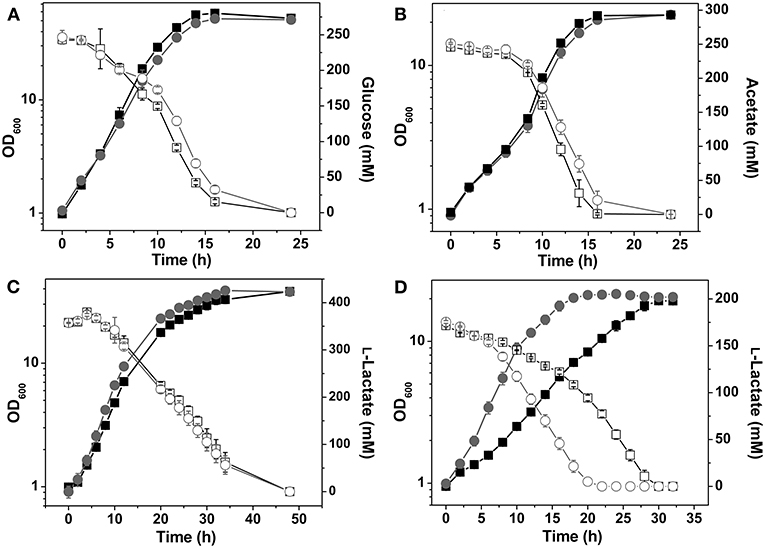
Figure 2. Growth and carbon source consumption of C. glutamicum wild type (■) and the Δndh mutant (●). Cells were grown in CGXII minimal medium containing 4% (wt/vol) glucose (A), 2% (wt/vol) acetate (B), 4% (wt/vol) L-lactate (C), or 2% (wt/vol) L-lactate (D). In (C) cells were precultivated in L-lactate minimal medium, in (D) cells were precultivated in glucose minimal medium. The concentrations of the individual carbon sources in the culture supernatant are shown by open squares (□) for the wild type and by open circles (○) for the Δndh mutant. Average values from at least three independent cultivations and standard deviations are shown.
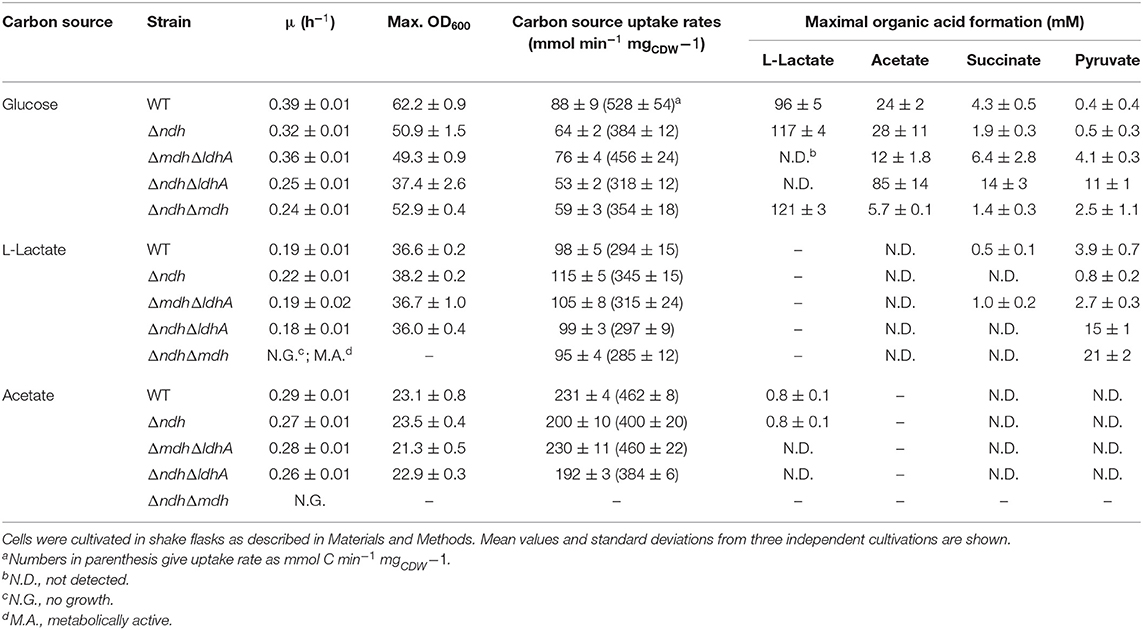
Table 3. Growth rate, biomass formation (max. OD600), carbon source uptake rates, and maximal organic acid formation of the C. glutamicum wild type, and its Δndh, ΔmdhΔldhA, ΔndhΔmdh and ΔndhΔldhA mutants.
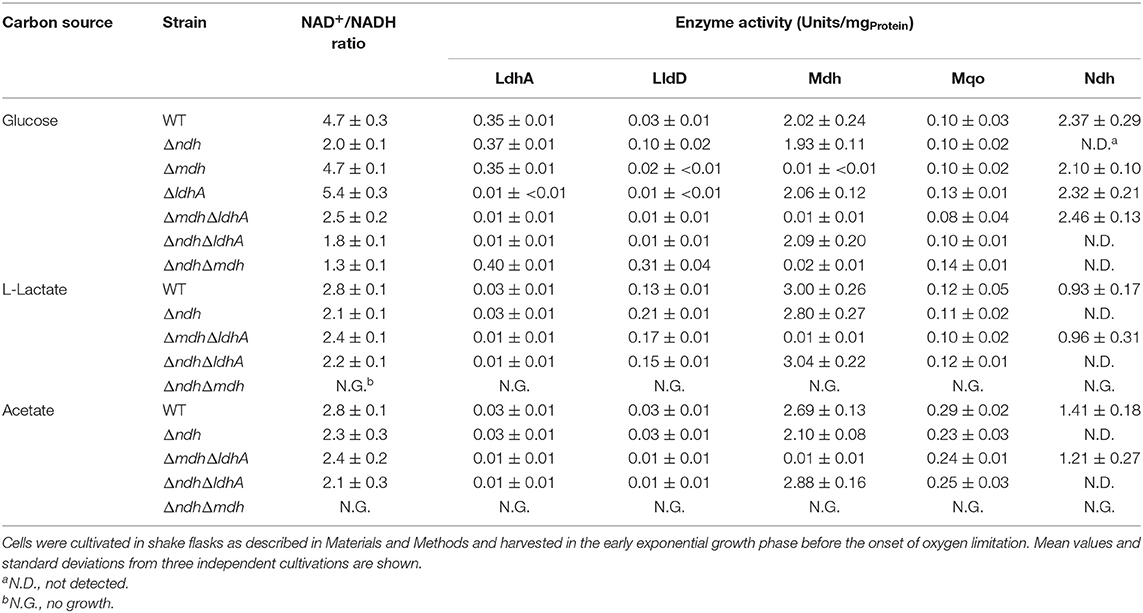
Table 4. NAD+/NADH ratios and enzyme activities in C. glutamicum wild type and its Δndh, ΔmdhΔldhA, ΔndhΔldhA, and ΔndhΔmdh mutants.
In acetate minimal medium, the growth rate of the Δndh mutant was slightly decreased by 7% (Figure 2B), the acetate uptake rate (AUR) was reduced by 13%, and the biomass yield was unchanged compared to the wild type (Table 3). The Mdh and Mqo activities were reduced by about 20% (Table 4). The NAD+/NADH ratio during growth on acetate was reduced by 18% in the Δndh mutant compared to the wild type (2.3 vs. 2.8 in the wild type, Table 4). It is noticeable that the NAD+/NADH ratio of the wild type on acetate was 40% lower than on glucose.
In contrast to the Δndh strain, growth of the Δmdh and ΔldhA mutants on glucose or acetate was comparable to that of the wild type and no changes of the NAD+/NADH ratio and of any enzyme activity tested except for the one deleted were observed (data not shown).
Growth of the Δndh Mutant on L-lactate
When cultivated in CGXII medium with L-lactate as sole carbon source, the Δndh mutant grew slightly faster than the wild type (μ = 0.22 vs. 0.19 h−1), whereas biomass formation was unchanged (Figure 2C). While the wild type required pre-cultivation in L-lactate medium to reach optimal growth rates on L-lactate, the Δndh mutant showed optimal growth irrespective of the carbon source used in the preculture (Figures 2C,D). The growth rate of non-adapted wild-type cells on L-lactate (μ = 0.13 h−1) was 34% lower compared to that of L-lactate-adapted wild-type cells (μ = 0.19 h−1). The improved growth of the Δndh mutant on L-lactate could be reversed by expression of ndh using the expression plasmid pAN6-ndh (data not shown). This indicates that the absence of Ndh has a positive influence on L-lactate utilization. The NAD+/NADH ratio on lactate was 2.8 for the wild type, comparable to acetate-grown cells, and 2.1 for the Δndh mutant. Similar to glucose-grown cells, the LldD activity was 1.6-fold increased in a Δndh mutant grown in L-lactate medium and the oxygen consumption rate of the Δndh mutant with L-lactate as substrate was increased by 17% compared to the wild type (335 ± 17 vs. 285 ± 15 nmol O2 mgProtein−1 min−1). The increased LldD activity of the Δndh mutant is presumably responsible for the increased growth rate and the increased oxygen consumption rate. The wild type secreted up to 3.9 mM pyruvate during the early exponential phase, which was metabolized during mid and late exponential phase, whereas the Δndh mutant secreted below 1 mM pyruvate (Table 3).
Characteristics of a ΔmdhΔldhA Double Mutant
To explore how the absence of more than one NADH oxidation system influences growth and other parameters, we constructed the C. glutamicum strains ΔmdhΔldhA, ΔndhΔldhA, and ΔndhΔmdh. Attempts to construct the triple mutant ΔndhΔmdhΔldhA failed, suggesting that under the conditions tested no further efficient NADH oxidation system exists in C. glutamicum that can compensate the lack of Ndh, LdhA, and Mdh.
In glucose minimal medium, the ΔmdhΔldhA mutant, which can oxidize NADH via Ndh, showed an 8% reduced growth rate, a slowed transition to the stationary phase, 21% reduced biomass formation, and a 14% reduced GUR (Figure 3A, Table 3). The slighly reduced growth rate might be due to the inability of the ΔmdhΔldhA mutant to form L-lactate, which is transiently formed by the wild type as NADH oxidation product and subsequently consumed again (Koch-Koerfges et al., 2012). The NAD+/NADH ratio (Table 4) of the ΔmdhΔldhA mutant (2.5 ± 0.2) was lower than in the wild type (4.7 ± 0.3), but higher than in the Δndh mutant (2.0 ± 2.0), suggesting that the enzyme couples LdhA-LldD and/or Mdh-Mqo contribute to NADH oxidation. In minimal medium with L-lactate or acetate as carbon sources, growth of the ΔmdhΔldhA mutant was comparable to that of the wild type (Figure 3), suggesting that both LdhA and Mdh are irrelevant for NADH oxidation on these substrates.
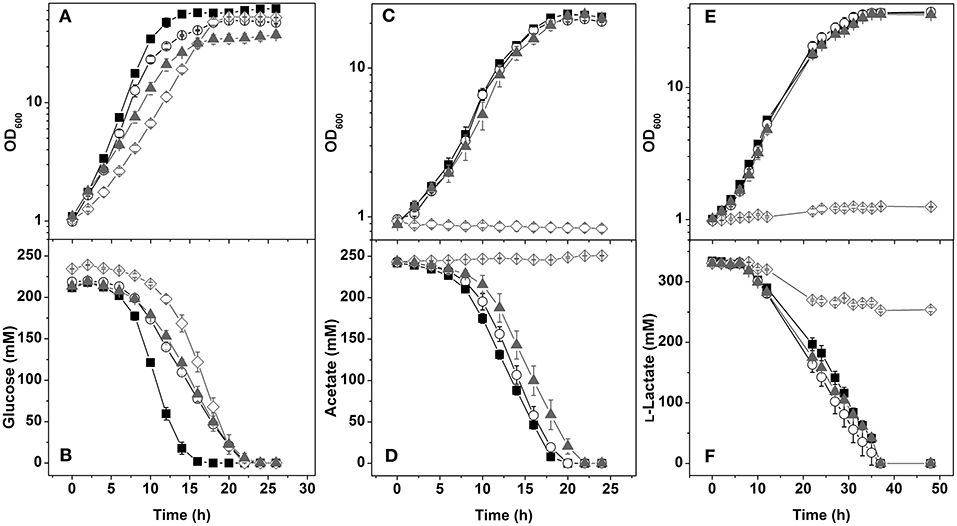
Figure 3. Growth and carbon source consumption of the C. glutamicum wild type (■) and the mutants ΔmdhΔldhA (○), ΔndhΔldhA (▴), and ΔndhΔmdh (♢). Cells were cultivated in CGXII minimal medium containing either 4% (wt/vol) glucose (A,B), 2% (wt/vol) acetate (C,D), or 4% (wt/vol) L-lactate (E,F). Growth (A,C,E) and carbon source concentration in the supernatants (B,D,F) are shown. Average values from at least three independent cultivations and the standard deviations are shown.
Characteristics of a ΔndhΔldhA Double Mutant
The ΔndhΔldhA mutant, which presumably can oxidize NADH only via the Mdh-Mqo couple, showed a stronger growth defect in glucose minimal medium than the Δndh mutant (Figure 3A). The growth rate was 0.25 ± 0.1 (Δndh mutant 0.32 ± 0.1) and the final OD600 was 37.4 ± 2.6 (Δndh mutant 50.9 ± 1.5). In agreement with the reduced growth rate, the GUR was reduced by 17% compared to the Δndh mutant (53 ± 2 vs. 64 ± 2 nmol min−1 ). These results suggest that the LdhA-LldD couple plays a role in NADH oxidation on glucose besides Ndh. As expected, due to the absence of LdhA, the ΔndhΔldhA mutant did not form L-lactate. Instead, the formation of acetate, succinate, and pyruvate strongly increased, with acetate being the major by-product (Table 3). This suggests that due to limited NADH reoxidation the pyruvate dehydrogenase activity and the TCA cycle activity is limited, causing an increased overflow metabolism. The NAD+/NADH ratio of the ΔndhΔldhA mutant was 1.8 ± 0.1 and thus lower than the one of the Δndh mutant (2.0 ± 0.1).
The growth of the ΔndhΔldhA mutant in acetate minimal medium was comparable to that of the wild type (Figure 3C) and the Δndh mutant (Figure 2B) and therefore LdhA is not relevant for this substrate. Interestingly, growth of the ΔndhΔldhA mutant in L-lactate minimal medium was comparable to that of the wild type (Figure 3E). Thus, the additional deletion of ldhA abolished the positive growth effect of the ndh deletion on L-lactate utilization (see above). The positive influence of the ndh deletion on L-lactate correlated with an increased LldD activity, which was not observed in the ΔndhΔldhA mutant (Table 4). This indicates that the formation of L-lactate from pyruvate by LdhA is necessary to trigger increased lldD expression, most likely via relieve of lldD repression by the transcriptional regulator LldR (Gao et al., 2008; Georgi et al., 2008).
Characteristics of a ΔndhΔmdh Double Mutant
In glucose minimal medium, the ΔndhΔmdh mutant, which presumably can oxidize NADH only via the LdhA-LldD couple, showed a growth rate of 0.24 h−1, a GUR of 59 ± 3 nmol min−1 , and final OD600 of 52.9 ± 0.4, corresponding to reductions of 38, 40, and 33% compared to the wild type (Table 3). The ΔndhΔmdh mutant secreted 26% more L-lactate than the wild type (Table 3), which supports the importance of LdhA for NADH oxidation. With a value of 1.3 ± 0.1, the NAD+/NADH ratio of the ΔndhΔmdh mutant was the lowest one observed in our study (Table 4). This suggests that NADH oxidation by the LdhA-LldD couple is less efficient than by Ndh or Mdh-Mqo, although the activities of LdhA and LldD were highest in the ΔndhΔmdh mutant with values of 0.40 and 0.31 U mg−1, respectively (Table 4).
In contrast to the ΔmdhΔldhA and ΔndhΔldhA double mutants, the ΔndhΔmdh mutant was unable to grow with acetate or L-lactate (Figure 3). The growth defect on acetate and L-lactate could be complemented by plasmid-based expression of either ndh (pAN6-ndh) or mdh (pAN6-mdh) (data not shown). Whereas, acetate was not consumed at all by the ΔndhΔmdh mutant (Figure 3D), a decrease of the L-lactate concentration was observed within the first 20 h of incubation (Figure 3F) and pyruvate was secreted (Table 3). This suggests that the cells possessed LldD activity, allowing oxidation of L-lactate to pyruvate with concomitant reduction of menaquinone to menaquinol, which was then reoxidized by the terminal oxidases. The observation that pyruvate was excreted rather than further oxidized is probably caused by an insufficient pyruvate dehydrogenase activity due to the limited availability of the cofactor NAD+. The phenotype of the ΔndhΔmdh mutant suggests that NADH oxidation during growth on acetate and L-lactate requires either Ndh or the Mdh-Mqo couple, which cannot be replaced by the LdhA-LldD couple.
Restoration of Growth on L-lactate of the ΔndhΔmdh Mutant
It was shown that expression of ldhA in C. glutamicum is tightly repressed by the transcriptional regulator SugR in the absence of sugars (Engels et al., 2008; Toyoda et al., 2009). As LdhA is probably the only catabolic NADH oxidation enzyme left in the ΔndhΔmdh mutant, repression of ldhA by SugR might be responsible for the inhibited growth on L-lactate. To test this hypothesis we constructed a ΔndhΔmdhΔsugR mutant, which indeed was able to grow on L-lactate as sole carbon and energy source, although at a much lower growth rate (Figure 4A). The triple mutant secreted high amounts of pyruvate and later also malate (Figure 4B). Growth of the ΔndhΔmdh mutant in L-lactate minimal medium was also possible by plasmid-based expression of ldhA using plasmid pAN6-ldhA (data not shown). Thus, insufficient LdhA activity due to ldhA repression by SugR is an important reason for the growth defect of the ΔndhΔmdh mutant in L-lactate medium. Neither the ΔndhΔmdhΔsugR mutant nor the ΔndhΔmdh mutant carrying pAN6-ldhA were able to grow on acetate (data not shown), indicating that there must be additional reasons for the inability to utilize acetate.
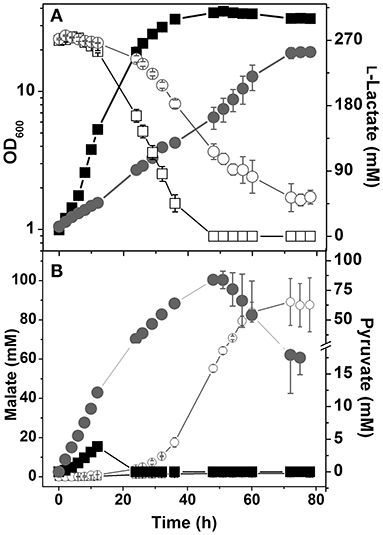
Figure 4. Growth and carbon source consumption (A) and production of organic acids (B) by C. glutamicum wild type and the mutant ΔndhΔmdhΔsugR. (A) Growth (filled symbols) and L-lactate consumption (open symbols) of the wild type (■, □) and the ΔndhΔmdhΔsugR mutant (●, ○). (B) Formation of pyruvate (closed symbols) and malate (open symbols) by the wild type (■, □) and ΔndhΔmdhΔsugR mutant (●, ○). Average values and standard deviations from at least three independent cultivations are shown.
Discussion
In the present work, we investigated a set of six mutants, namely Δndh, ΔldhA, Δmdh, ΔmdhΔldhA, ΔndhΔldhA, and ΔndhΔmdh, that lack one or two of the three known catabolic NADH oxidation enzymes in C. glutamicum to explore their role for aerobic growth and metabolism on glucose, acetate, and L-lactate. As our attempts to construct the triple mutant ΔndhΔmdhΔldhA mutant failed, it appears likely that there is no further enzyme enabling catabolic NADH oxidation in C. glutamicum besides Ndh, Mdh, and LdhA.
In two previous studies, it was reported that an ndh integration mutant grows at a slightly lower rate than the wild type ATCC 13032 on glucose and acetate agar plates (Molenaar et al., 2000) and that the growth rate of an ndh disruption mutant of the lysozyme-sensitive strain KY9714 in glucose minimal medium was not much altered compared to the parent strain (Nantapong et al., 2004). Our studies with the in-frame deletion mutant Δndh of strain ATCC 13032 revealed a slightly reduced growth rate and biomass formation in glucose minimal medium along with reduced glucose and oxygen consumption rates. A strong effect was observed for the NAD+/NADH ratio, which decreased from 4.7 in the wild type to 2.0 in the Δndh mutant. As growth and the NAD+/NADH ratio of the strains ΔldhA and Δmdh in glucose minimal medium were comparable to the wild type (Table 4), our results show that Ndh is the most important enzyme for NADH oxidation during aerobic growth of C. glutamicum on glucose.
The observation that the C. glutamicum Δndh mutant did not show a stronger growth defect on glucose was explained by the existence of alternative NADH oxidation systems formed by the couples Mdh-Mqo and LdhA-LldD, which catalyze the same net reaction as Ndh, namely the oxidation of NADH and reduction of menaquinone when acting in concert (Molenaar et al., 2000; Nantapong et al., 2004). In line with this explanation, the ΔndhΔmdh and ΔndhΔldhA double mutants showed stronger growth defects and a stronger reduction of the NAD+/NADH ratio than the Δndh mutant in glucose minimal medium. Therefore, neither the Mdh-Mqo couple alone nor the LdhA-LldD couple alone is able to compensate the lack of the other two NADH oxidation systems adequately. The ΔmdhΔldhA mutant, which still can oxidize NADH via Ndh, grew better than the ΔndhΔmdh and ΔndhΔldhA mutants, but not as good as the wild type (Table 3), suggesting that the Mdh-Mqo and LdhA-LldD couples contribute to NADH oxidation also when Ndh is present.
Growth on L-lactate was improved by the ndh deletion (Figures 2C,D). This positive effect is presumably due to a 2-fold increased activity of LldD (Table 4), which was observed also previously in the study of the ndh mutant of strain KY9714 (Nantapong et al., 2005). It has been shown that expression of the cg3226-lldD operon is regulated by the FadR-type transcriptional regulator LldR, which represses the two genes in the absence of L-lactate (Gao et al., 2008; Georgi et al., 2008). The gene cg3226 encodes a putative L-lactate permease, which is not essential for growth on L-lactate, however (Stansen et al., 2005). The reason for the increased LldD activity in the Δndh mutant is most likely the increased formation of L-lactate (Table 3), which relieves repression of the cg3226-lldD operon by LldR. In line with this explanation, the additional deletion of ldhA in the Δndh strain reversed the improved growth and the increased LldD activity (Figures 3C,D, Tables 2, 3). The reason why even L-lactate-adapted wild-type cells grew not as good as the Δndh mutant (Figure 2C) is not clear yet. The absence of Ndh might allow for an increased availability of menaquinone as electron acceptor of LldD.
Compared to glucose, growth on acetate of the strains Δndh, ΔndhΔldhA, and ΔmdhΔldhA was only minimally affected (Figures 2, 3, Table 3). This difference can be explained by the fact that glucose oxidation is coupled to the formation of up to 6 NADH/glucose, while acetate oxidation is linked to the formation of only 1 NADH/acetate (Figure 1). Thus, glucose oxidation has a much higher NADH reoxidation demand than lactate or acetate oxidation. As essentially no L-lactate is formed during growth on acetate (Table 3) and ldhA is repressed by SugR, the LdhA-LldD cycle is probably not active and NADH oxidation is accomplished via the Mdh-Mqo cycle in the mutants Δndh and ΔndhΔldhA and via Ndh in strain ΔmdhΔldhA. In agreement with this interpretation, the ΔndhΔmdh strain was unable to grow on acetate (Figure 3).
In summary, our results show that C. glutamicum uses a set of three systems for NADH oxidation coupled to menaquinone reduction. Ndh is the dominant enzyme, but the Mdh-Mqo couple and LdhA-LldD couples complement or substitute it and thereby enable NADH oxidation in situations with high catabolic fluxes or a defective Ndh enzyme. Such backup systems exist not only for NADH oxidation, but also for menaquinol oxidation, where cytochrome bd oxidase can substitute or complement the cytochrome bc1-aa3 supercomplex. This redundancy likely contributes to the metabolic flexibility and the survival of the organism in nature.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MB and AK-K designed the study. TM and AK-K performed the experiments and prepared the figures and tables. MB wrote the manuscript. All authors contributed to the interpretation of the data.
Funding
Financial support (Grant 0315598A) by the Bundesministerium für Bildung und Forschung (BMBF) is gratefully acknowledged.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Volker Wendisch (Universität Bielefeld) for providing plasmid pK19mobsacB-ΔsugR.
References
Becker, J., and Wittmann, C. (2012). Bio-based production of chemicals, materials and fuels–Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 23, 631–640. doi: 10.1016/j.copbio.2011.11.012
Bott, M., and Niebisch, A. (2003). The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104, 129–153. doi: 10.1016/s0168-1656(03)00144-5
Bruch, E., Vilela, P., Lexa-Sapart, N., Yang, L., Raynal, B., Alzari, P. M., et al. (2020). Actinobacteria challenge the paradigm: a unique protein architecture for a well-known central metabolic complex. bioRxiv. doi: 10.1101/2020.07.24.212647
Eggeling, L., and Bott, M. (2015). A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 3387–3394. doi: 10.1007/s00253-015-6508-2
Engels, V., Lindner, S. N., and Wendisch, V. F. (2008). The global repressor SugR controls expression of genes of glycolysis and of the L-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J. Bacteriol. 190, 8033–8044. doi: 10.1128/JB.00705-08
Engels, V., and Wendisch, V. F. (2007). The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 189, 2955–2966. doi: 10.1128/JB.01596-06
Freudl, R. (2017). Beyond amino acids: use of the Corynebacterium glutamicum cell factory for the secretion of heterologous proteins. J. Biotechnol. 258, 101–109. doi: 10.1016/j.jbiotec.2017.02.023
Frunzke, J., Engels, V., Hasenbein, S., Gätgens, C., and Bott, M. (2008). Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67, 305–322. doi: 10.1111/j.1365-2958.2007.06020.x
Gao, Y. G., Suzuki, H., Itou, H., Zhou, Y., Tanaka, Y., Wachi, M., et al. (2008). Structural and functional characterization of the LldR from Corynebacterium glutamicum: a transcriptional repressor involved in L-lactate and sugar utilization. Nucleic Acids Res. 36, 7110–7123. doi: 10.1093/Nar/Gkn827
Genda, T., Nakamatsu, T., and Ozaki, I. (2003). Purification and characterization of malate dehydrogenase from Corynebacterium glutamicum. J. Biosci. Bioeng. 95, 562–566. doi: 10.1016/S1389-1723(03)80162-7
Georgi, T., Engels, V., and Wendisch, V. F. (2008). Regulation of L-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J. Bacteriol. 190, 963–971. doi: 10.1128/JB.01147-07
Graf, S., Fedotovskaya, O., Kao, W. C., Hunte, C., Adelroth, P., Bott, M., et al. (2016). Rapid electron transfer within the III-IV supercomplex in Corynebacterium glutamicum. Sci. Rep. 6:34098. doi: 10.1038/srep34098
Hoffelder, M., Raasch, K., van Ooyen, J., and Eggeling, L. (2010). The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J. Bacteriol. 192, 5203–5211. doi: 10.1128/jb.00597-10
Kabus, A., Niebisch, A., and Bott, M. (2007). Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl. Environ. Microbiol. 73, 861–868. doi: 10.1128/AEM.01818-06
Kao, W. C., Kleinschroth, T., Nitschke, W., Baymann, F., Neehaul, Y., Hellwig, P., et al. (2016). The obligate respiratory supercomplex from actinobacteria. Biochim. Biophys. Acta 1857, 1705–1714. doi: 10.1016/j.bbabio.2016.07.009
Keilhauer, C., Eggeling, L., and Sahm, H. (1993). Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175, 5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993
Kinoshita, S., Udaka, S., and Shimono, M. (1957). Studies on amino acid fermentation. Part I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3, 193–205. doi: 10.2323/jgam.3.193
Kinugawa, H., Kondo, N., Komine-Abe, A., Tomita, T., Nishiyama, M., and Kosono, S. (2020). In vitro reconstitution and characterization of pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase hybrid complex from Corynebacterium glutamicum. MicrobiologyOpen 9:e1113. doi: 10.1002/mbo3.1113
Koch-Koerfges, A., Kabus, A., Ochrombel, I., Marin, K., and Bott, M. (2012). Physiology and global gene expression of a Corynebacterium glutamicum ΔF1FO-ATP synthase mutant devoid of oxidative phosphorylation. Biochim. Biophys. Acta 1817, 370–380. doi: 10.1016/j.bbabio.2011.10.006
Koch-Koerfges, A., Pfelzer, N., Platzen, L., Oldiges, M., and Bott, M. (2013). Conversion of Corynebacterium glutamicum from an aerobic respiring to an aerobic fermenting bacterium by inactivation of the respiratory chain. Biochim. Biophys. Acta 1827, 699–708. doi: 10.1016/j.bbabio.2013.02.004
Kusumoto, K., Sakiyama, M., Sakamoto, J., Noguchi, S., and Sone, N. (2000). Menaquinol oxidase activity and primary structure of cytochrome bd from the amino-acid fermenting bacterium Corynebacterium glutamicum. Arch. Microbiol. 173, 390–397. doi: 10.1007/s002030000161
Li, L., Wada, M., and Yokota, A. (2007). A comparative proteomic approach to understand the adaptations of an H+-ATPase-defective mutant of Corynebacterium glutamicum ATCC14067 to energy deficiencies. Proteomics 7, 3348–3357. doi: 10.1002/pmic.200700287
Litsanov, B., Brocker, M., and Bott, M. (2012). Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl. Environm. Microbiol. 78, 3325–3337. doi: 10.1128/Aem.07790-11
Matsushita, K. (2013). “Respiratory chain and energy metabolism of Corynebacterium glutamicum,” in Corynebacterium glutamicum: Biology and Biotechnology, eds H. Yukawa, M. Inui, and Steinbüchel (Berlin; Heidelberg: Springer-Verlag), 315–334.
Molenaar, D., van der Rest, M. E., Drysch, A., and Yücel, R. (2000). Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 182, 6884–6891. doi: 10.1128/jb.182.24.6884-6891.2000
Nantapong, N., Kugimiya, Y., Toyama, H., Adachi, O., and Matsushita, K. (2004). Effect of NADH dehydrogenase-disruption and over-expression on respiration-related metabolism in Corynebacterium glutamicum KY9714. Appl. Microbiol. Biotechnol. 66, 187–193. doi: 10.1007/s00253-004-1659-6
Nantapong, N., Otofuji, A., Migita, C. T., Adachi, O., Toyama, H., and Matsushita, K. (2005). Electron transfer ability from NADH to menaquinone and from NADPH to oxygen of type II NADH dehydrogenase of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 69, 149–159. doi: 10.1271/bbb.69.149
Niebisch, A., and Bott, M. (2001). Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175, 282–294. doi: 10.1007/s002030100262
Niebisch, A., and Bott, M. (2003). Purification of a cytochrome bc1-aa3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum. Identification of a fourth subunity of cytochrome aa3 oxidase and mutational analysis of diheme cytochrome c1. J. Biol. Chem. 278, 4339–4346. doi: 10.1074/jbc.M210499200
Niebisch, A., Kabus, A., Schultz, C., Weil, B., and Bott, M. (2006). Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281, 12300–12307. doi: 10.1074/jbc.M512515200
Parey, K., Wirth, C., Vonck, J., and Zickermann, V. (2020). Respiratory complex I–structure, mechanism and evolution. Curr. Opin. Struct. Biol. 63, 1–9. doi: 10.1016/j.sbi.2020.01.004
Sawada, K., Kato, Y., Imai, K., Li, L., Wada, M., Matsushita, K., et al. (2012). Mechanism of increased respiration in an H+-ATPase-defective mutant of Corynebacterium glutamicum. J. Biosci. Bioeng. 113, 467–473. doi: 10.1016/j.jbiosc.2011.11.021
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Sekine, H., Shimada, T., Hayashi, C., Ishiguro, A., Tomita, F., and Yokota, A. (2001). H+-ATPase defect in Corynebacterium glutamicum abolishes glutamic acid production with enhancement of glucose consumption rate. Appl. Microbiol. Biotechnol. 57, 534–540. doi: 10.1007/s002530100778
Siedler, S., Bringer, S., and Bott, M. (2011). Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl. Microbiol. Biotechnol. 92, 929–937. doi: 10.1007/s00253-011-3374-4
Stansen, C., Uy, D., Delaunay, S., Eggeling, L., Goergen, J. L., and Wendisch, V. F. (2005). Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71, 5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005
Steuber, J., Vohl, G., Casutt, M. S., Vorburger, T., Diederichs, K., and Fritz, G. (2014). Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase. Nature 516, 62–67. doi: 10.1038/nature14003
Toyoda, K., Teramoto, H., Inui, M., and Yukawa, H. (2009). Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding L-lactate dehydrogenase in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 83, 315–327. doi: 10.1007/s00253-009-1887-x
van der Rest, M. E., Lange, C., and Molenaar, D. (1999). A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545. doi: 10.1007/s002530051557
Wendisch, V. F., Mindt, M., and Perez-Garcia, F. (2018). Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102, 3583–3594. doi: 10.1007/s00253-018-8890-z
Keywords: NADH dehydrogenase, malate dehydrogenase, malate:quinone oxidoreductase, lactate dehydrogenase, NAD+/NADH ratio, respiratory chain, SugR
Citation: Maeda T, Koch-Koerfges A and Bott M (2021) Relevance of NADH Dehydrogenase and Alternative Two-Enzyme Systems for Growth of Corynebacterium glutamicum With Glucose, Lactate, and Acetate. Front. Bioeng. Biotechnol. 8:621213. doi: 10.3389/fbioe.2020.621213
Received: 25 October 2020; Accepted: 21 December 2020;
Published: 20 January 2021.
Edited by:
Uldis Kalnenieks, University of Latvia, LatviaCopyright © 2021 Maeda, Koch-Koerfges and Bott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Bott, bS5ib3R0QGZ6LWp1ZWxpY2guZGU=
†These authors have contributed equally to this work
‡Present address: Tomoya Maeda, RIKEN Center for Biosystems Dynamics Research, Osaka, Japan
 Tomoya Maeda†‡
Tomoya Maeda†‡ Michael Bott
Michael Bott