- 1Faculty of Artificial Intelligence, Kafrelsheikh University, Kafrelsheikh, Egypt
- 2Faculty of Computer Science, King Khalid University, Abha, Saudi Arabia
- 3Computers and Systems Department, Electronics Research Institute, Cairo, Egypt
Introduction: Parasitic infections remain a major public health concern, particularly in healthcare and community settings where rapid and accurate diagnosis is essential for effective treatment and prevention. Traditional parasite detection methods rely on manual microscopic examinations, which are time-consuming, labor-intensive, and susceptible to human error. Recent advancements in automated microscopic imaging and deep learning offer promising solutions to enhance diagnostic accuracy and efficiency.
Methods: This study proposes a novel framework, the YOLO Convolutional Block Attention Module (YCBAM), to automate the detection of pinworm parasite eggs in microscopic images. The YCBAM architecture integrates YOLO with self-attention mechanisms and the Convolutional Block Attention Module (CBAM), enabling precise identification and localization of parasitic elements in challenging imaging conditions.
Results and Discussion: Experimental evaluation of the YCBAM model demonstrated a precision of 0.9971, a recall of 0.9934, and a training box loss of 1.1410, indicating efficient learning and convergence. The model achieved a mean Average Precision (mAP) of 0.9950 at an IoU threshold of 0.50 and a mAP50–95 score of 0.6531 across varying IoU thresholds, confirming its superior detection performance. The integration of YOLO with self-attention and CBAM significantly improves the automated detection of pinworm eggs, offering a highly accurate and reliable diagnostic tool for medical parasitology. This framework has the potential to reduce diagnostic errors, save time, and support healthcare professionals in making informed decisions.
1 Introduction
Pinworm parasite egg detection is a significant challenge in parasitology diagnostics due to the small size and morphological similarities of pinworm eggs with other microscopic particles. Traditional diagnostic methods, such as manual microscopic examination, are time-consuming, labor-intensive, and human error, especially in settings with high sample volumes. Moreover, these manual methods often lack sensitivity based on the examiner, leading to false negatives and delayed diagnoses, particularly in resource-constrained environments. The study aims to overcome the challenges faced by healthcare providers in accurately diagnosing pinworm infections in clinical settings. Microscopic detection of pinworm eggs faces challenges due to their small size, similarity to other microscopic particles, and the need for specialized expertise. Moreover, manual diagnostic techniques often lead to delays, misdiagnoses, and increased healthcare costs.
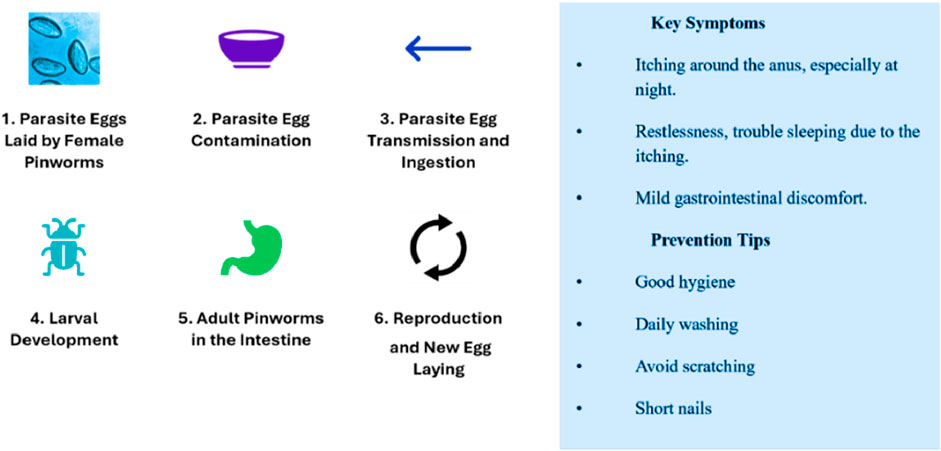
The advancement of deep learning improves diagnostic accuracy, speed, and scalability. Recent advancements in computer vision and machine learning have led to improvements in the diagnostic process, presenting a more efficient and reliable solution to parasitic egg detection. Diagnosis process of pinworm parasite eggs is difficult due to their small size and morphological similarity to other microscopic particles, measuring 50–60 μm in length and 20–30 μm in width, and the traditional examination methods are laborious and time-consuming, can lead to delayed diagnosis and increased infection rate, particularly among children (Mirzaei et al., 2022a; Mirzaei et al., 2022b).
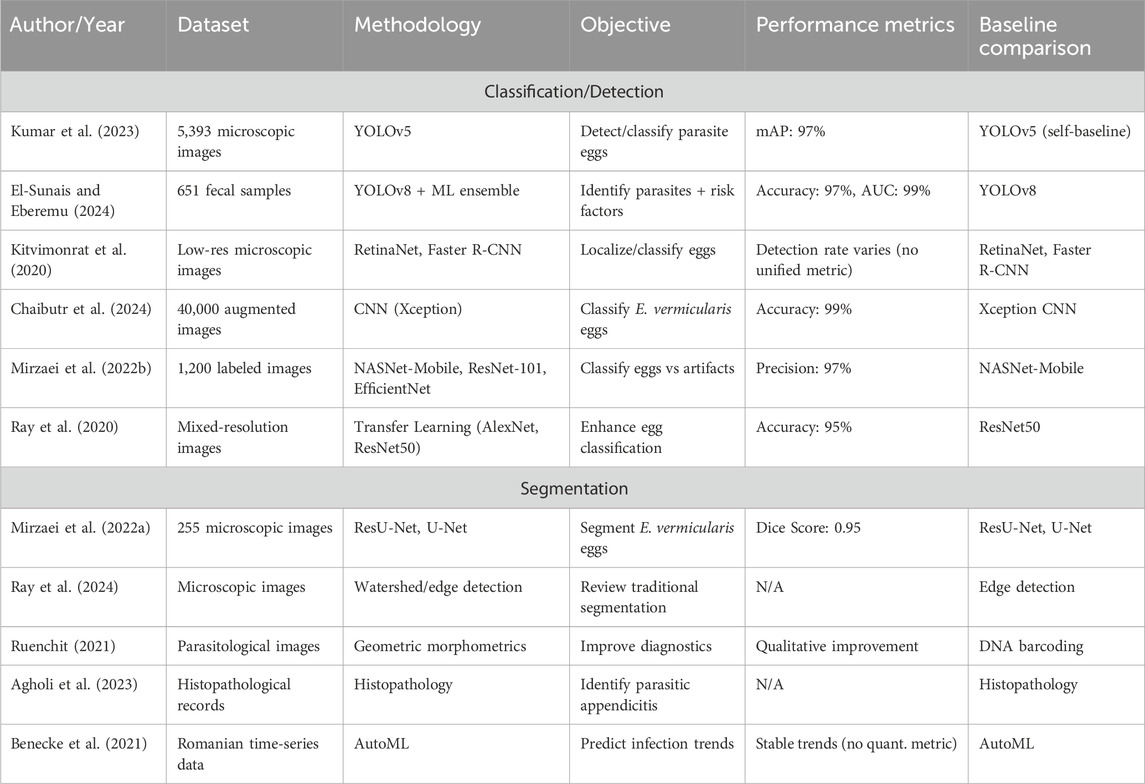
Freshly placed Pinworm eggs appear colorless or transparent, revealing the larva. Pinworm eggs have a thin, clear, bi-layered shell that protects the embryo, as shown in Figure 1 (Mirzaei et al., 2022a). The embryonated larva in the egg often curls up and moves under a microscope, showing viability (Mirzaei et al., 2022b). These eggs hatch in the small intestine of the host (Ray et al., 2024). Pinworms, also known as Enterobius, are spread through contaminated objects such as surfaces and clothing, and infected persons. Small transparent eggs can live for weeks and are transmissible, making them difficult to notice (Chaibutr et al., 2024; Agholi et al., 2023). The scotch tape test and other E. vermicularis egg identification procedures, including perianal microscopy, are based on the examiner’s ability and give false negatives due to limited sensitivity and repeated sampling (Benecke et al., 2021; Kumar et al., 2023).
Thus, an automated and accurate diagnostic workflow is needed for effective and timely early diagnosis. Recently developed Deep Learning (DL) has automated pinworm egg identification to avoid these limits; these solutions aim to save time, enhance accuracy, and reduce reliance on specialists (Kitvimonrat et al., 2020); (Elbedwehy et al., 2024). Deep learning, especially CNNs, has transformed biomedical image processing, improving E. vermicularis egg detection from microscopic images. U-Net and ResU-Net segmentation algorithms separated pinworm eggs from complex digital microscopy backgrounds, achieving high dice scores and minimal diagnostic errors (Mirzaei et al., 2022a; Mirzaei et al., 2022b). Over 97% of advanced classification models, such as NASNet-Mobile and ResNet-101, can distinguish E. vermicularis eggs from other artifacts in microscopic slides (Mirzaei et al., 2022b). The DL technique has improved parasite diagnostics detection accuracy, eliminating human error and operator training complications to learn detailed pinworm egg shape patterns from vast datasets of tagged microscopic images with performing complicated image analysis tasks faster and more consistently than manual approaches, making them ideal for large-scale screening and diagnostic applications in clinical and resource-constrained situations (Benecke et al., 2021; Kumar et al., 2023; Kitvimonrat et al., 2020; El-Sunais and Eberemu, 2024; Zhang et al., 2024; Pun et al., 2023).
A robust YOLO Convolutional Block Attention Module (YCBAM) architecture is presented, enhancing automatic detection of pinworm parasite eggs in microscopic images, including self-attention mechanisms and CBAM. Moreover, it is characterized by high accuracy and efficiency in object detection, such as identifying and segmenting small objects within complex backgrounds. In addition, the self-attention is used to focus on essential image regions, reducing irrelevant background features and providing dynamic feature representation for precise pinworm egg detection. CBAM enhances attention, improves feature extraction from complex backgrounds, and increases sensitivity to small, critical features such as pinworm egg boundaries, enhancing detection accuracy. The YCBAM is more effective than traditional methods and advanced detection models in detecting small objects, pinworm eggs, confirming the effectiveness of the proposed integration. The following main contributions include:
i. The YCBAM architecture, integrated into YOLOv8, enhances the performance of identifying pinworm parasite eggs in noisy and varied environments, a common challenge in microscopic imaging.
ii. Self-attention and CBAM focus on spatial and channel-wise information to improve feature extraction for achieving high detection accuracy with solid metrics: mAP of 0.995 at an IoU threshold of 0.50 and 0.6531 across many thresholds.
iii. The YCBAM architecture enhances detection accuracy and computational efficiency by integrating YOLOv8 with attention modules, enabling optimized training and inference, even with limited training data.
The successful implementation of the YCBAM architecture has several significant effects. Clinically, it could lead to faster, more reliable diagnoses, reducing the burden on healthcare professionals and improving patient outcomes by facilitating earlier detection and treatment of pinworm infections. The system was used in low-resource settings, where traditional methods lack of trained personnel or diagnostic equipment. According to healthcare and public health, this study contributes to the development of automated diagnostic systems for other parasitic infections. Additionally, the integration of attention mechanisms in the proposed model achieves similar advancements in other domains of medical image analysis, improving the accuracy of automated detection systems for a wide range of diseases.
The other section is structured as follows: Section 2 reviews related work in automated parasitic egg detection, including both traditional image processing and deep learning approaches. Section 3 explains the methodology of the YCBAM architecture, then its integration with YOLOv8, self-attention, and CBAM, with the training and experimental setup. Section 4 presents the model’s performance results, comparing it to existing models in terms of accuracy, efficiency, and robustness. Section 5 presents the findings of the proposed method, emphasizing its strengths, limitations, and suggestions for future improvements. Section 6 concludes the paper by outlining directions for future work, including expanding the model’s applicability to other diagnostic applications.
2 Related work
The identification and categorization of Enterobius vermicularis (pinworm) eggs using AI and machine learning has transformed diagnostics, improving precision and efficiency. Traditionally, pinworm egg microscopy has been the standard for diagnosing pinworm infection. The manual procedure is laborious, error-prone, and requires highly skilled professionals, making it unsuitable for high-volume clinical settings or those with limited resources (Mirzaei et al., 2022a). Researchers are using AI to achieve accuracy of diagnosis, processing time and focusing on specialized skills.
2.1 Detection and classification techniques
Deep learning automates E. vermicularis egg detection and segmentation. Mirzaei et al. (2022a) segmented pinworm eggs from microscopic images with a 0.95% dice score using ResU-Net and U-Net.
These models accurately reflect the tiny details of egg morphology. Additionally, Mirzaei et al. analyzed 255 microscopic images for segmentation and 1,200 for classification.
Pretrained models such as NASNet-Mobile, ResNet-101, and EfficientNet-b0 achieved 97% classification accuracy (Mirzaei et al., 2022b), indicating the adaptation of models to parasite eggs’ complex features, to reach accurate clinical sample detection. Ray et al. (2024) discussed parasite egg segmentation, focusing on egg size, shape, and non-egg artifacts. They achieve image improvement and noise reduction before segmentation techniques to standardize input images to reach minute egg morphological traits, and automated detection system accuracy and reliability. E. vermicularis egg classification has improved with machine learning. Chaibutr et al. (2024) developed a reliable Xception-based CNN pinworm egg classification model.
Advanced CNN architectures can improve parasite infection diagnosis, where their method attained 99% accuracy with significant data augmentation. Their study increases model generalization across visual conditions and reduces classification errors. Six pretrained models, including ResNet-101 and Inception-v3, classified E. vermicularis photos by Mirzaei et al. (2022b). These models recognized parasite eggs from other microscopic artifacts. These pretrained parasite diagnosis algorithms demonstrate how transfer learning can identify complex patterns in limited datasets or heterogeneous data sources.
2.2 Clinical applications and epidemiological insights
Medically, E. vermicularis detection is used for differential diagnoses in parasite infections, similar to other illnesses. A systematic Iranian appendectomy material examination by Agholi et al. (2023) discovered E. vermicularis in a subset of appendicitis cases, which focuses on the need for proper parasite stomach pain diagnosis. Automatic diagnostic approaches could improve clinical evaluations by presenting faster and more accurate results, enhancing patient care. Benecke et al. (2021) used machine learning to examine Romanian enterobiasis time-series data and found steady infection rates over a decade. Their study revealed that AI-based public health monitoring tools guide parasitic infection intervention efforts. AI helps doctors predict outbreaks, allocate resources, and create adapted infection control measures. For quick parasite egg detection, YOLO (You Only Look Once) object detection algorithms, particularly YOLOv5 and YOLOv8, have made significant advances.
Kumar et al. (2023) found that YOLOv5 can detect intestinal parasite eggs with 97% precision and 8.5 milliseconds per sample. YOLOv5 is more effective than Faster R-CNN and SSD in low-resource scenarios when rapid diagnostics are needed. Kitvimonrat et al. (2020) found that RetinaNet and Faster R-CNN were used to detect parasite eggs. These models performed best with huge datasets and precise annotations. Key point-based detectors CenterNet, improve detection accuracy in noisy or low-resolution images by localizing small eggs. Manual microscopic inspection of parasitic diseases is accurate but time-consuming and requires experts. Deep learning techniques such as YOLO (You Only Look Once) models automate diagnostics, AI, and machine learning (El-Sunais and Eberemu, 2024; Zhang et al., 2024). The Normalization-based Attention Module (NAM) and ODConv with YOLOv8 detect silkworm microparticle viruses and increase feature extraction and detection accuracy (Zhang et al., 2024). The technique improves agricultural virus identification by reducing detection time per image and outperforming current models (Zhang et al., 2024). Deep learning is used to detect and quantify plant-parasitic nematodes in agriculture using YOLOv5 and NemDST (Pun et al., 2023). Farmers can detect pests, eliminate laborious analysis, and improve pest control (Pun et al., 2023). AI boosts agricultural accuracy and minimizes chemical consumption (Pun et al., 2023). AI applied to cervical cancer (AlMohimeed et al., 2023; AlMohimeed et al., 2024) and lightweight deep-learning parasite detection algorithms (Xu et al., 2024). Learning-based detection is applied in human health, agriculture tasks, and other industries. The deep learning models for silkworm microparticle virus detection AI algorithms are applied in specific tasks, as it is characterized by variety and adaptability (Zhang et al., 2024).
These advances focus on intelligent diagnostic tools that use AI to improve detection in medical and agricultural pest management (Zhang et al., 2024; Pun et al., 2023). Although parasite detection using AI has improved, there are some obstacles, such as Complex parasite morphology and imaging circumstances, which make detection accuracy difficult.
Studies recommend using a group of data and robust training approaches to increase model performance across varying conditions (El-Sunais and Eberemu, 2024). While YOLOv5 and YOLOv8 have shown significant results, research is still conducted to improve these algorithms to overcome complex tasks and integrate them into diagnostic workflows (Pun et al., 2023).
2.3 Advances in data augmentation and transfer learning
Access to diverse datasets has been limited in past research. Kumar et al. (2023) modified the training dataset vertically and rotationally. The strategy makes the YOLOv5 the best model to use in a different test set of microscopic images, enhancing detection accuracy with fewer training instances. Ray et al. (2020) classified parasite eggs in feces with 95% accuracy using pre-trained deep learning models, focusing on the importance of transfer learning in data shortage and heterogeneity-challenged model training. In addition, a brain tumor (Talaat, 2024) and kidney disease (Elbedwehy et al., 2024) research shows that advanced neural networks with optimal training data have better diagnostic reliability across varied situations.
2.4 Limitations and challenges in current approaches
Despite progress, AI-based E. vermicularis detection approaches have great limitations. Kumar et al. (2023) recommended high-quality, diverse datasets. YOLOv5 model overfits, but it cannot be applied to tiny or imbalanced datasets due to the need for data augmentation, and obtaining comprehensive training data is difficult. Kitvimonrat et al. (2020) stated that the YOLOv8 model has difficulty distinguishing small, low-contrast objects in microscopic pictures. Kumar et al. noted that YOLOv8’s complexity and high processing needs make it unsuitable for resource-constrained deployment. Agholi et al. (2023) suggest that AI-based approaches may not be therapeutically useful in areas with low E. vermicularis. Automated methods can enhance diagnosis precision, but their cost-effectiveness in low-incidence areas is unclear.
According to Ruenchit, AI-driven diagnostics need expensive hardware and computing, which reduces their benefits in underdeveloped areas with high rates of parasite infection (Ruenchit, 2021). Deep learning and YOLO models improved parasite egg detection, although data quality, model complexity, and processing issues remain. These issues must be addressed to achieve reliable, scalable diagnostic systems for various clinical contexts and geographies. AI-based parasitic diagnostics could change parasitic infection management by improving speed, accuracy, and cost. The YOLOv8 silkworm microparticle virus identification model also faces challenges with data variability and model complexity. Its high computing requirements and specialized gear may limit its usage in resource-constrained settings such as small-scale agriculture or developing country labs (Zhang et al., 2024). The method improves feature extraction, but expensive hardware in resource-constrained areas (Zhang et al., 2024). The decision support tool NemDST connected to YOLOv5 can detect pests in plant-parasitic nematode management; however, it is not adaptive to different environments and crop kinds (Pun et al., 2023).
Recent advances in acute lymphoblastic leukemia detection (Hassan et al., 2024) and small object detection in controlled environments (Papadopoulos et al., 2024) show AI diagnostic model interpretability and computational cost issues (Hassan et al., 2022; Elbedwehy et al., 2024; Saber et al., 2024). Li A. et al. (2023) YOLO-SA integrates a self-attention mechanism, using the traditional backbone instead of a reparametrized module and enhancing feature fusion. This prevents detection accuracy and reduces complexity by speeding up training convergence with an anchor-based detection head.
Li Y. et al. (2023) SAE-CenterNet improves small object detection by incorporating self-attention and using Dynamic Attention Convolution (DAC) for efficient downsampling. The Attention Fusion Module (AFM) helps in multi-scale feature fusion, making it effective for detecting objects in dense environments.
Ding et al. (2023) developed a lightweight YOLOv4 model combined with mechanisms for security applications. The attention modules focus on key features, improving detection accuracy while maintaining efficiency, crucial for real-time security scenarios. Ji et al. (2024) YOLO-TLA, an upgraded YOLOv5, adds a detection layer for small object capture, uses the C3CrossConv module for efficiency, and applies a global attention mechanism for better feature representation. It shows a 4.6% improvement in mAP while maintaining a small model size of 9.49 million parameters. Nematode morphology and soil structures can produce false positives and negatives, impairing detection. Data processing and updates require internet connectivity, which may be problematic for farmers in remote areas with weak digital infrastructure (Pun et al., 2023). Fast real-time processing and inference are another difficulty. YOLOv5 is designed for fast detection, and high-resolution images or large datasets require processing in clinical situations when speedy diagnosis is crucial for treatment. Despite its improved accuracy, the YOLOv8 model still faces difficulty in recognizing smaller or less distinguishable targets in complicated backgrounds, such as silkworm microparticle viruses (Zhang et al., 2024). Deep learning model interpretability is a concern. The black-box structure of neural networks makes decision-making difficult to understand, which makes it hard to win medical and agricultural end-user trust (El-Sunais and Eberemu, 2024; Pun et al., 2023; Hassan, 2024) as illustrated in Table 1.
3 Proposed work
This study presents an advanced architecture, called YOLO Convolutional Block Attention Module (YCBAM), which integrates YOLOv8 with self-attention mechanisms and Convolutional Block Attention Module (CBAM) to enhance the detection and identification of pinworm parasite eggs in microscopic images.
3.1 Data preparation
Labeled pinworm egg microscopy is used. Images with different noise, magnification, and illumination settings are included in robustness. The training dataset is rotated, zoomed, and modified to prevent overfitting and increase model generalization in different images.
3.2 The proposed model architecture
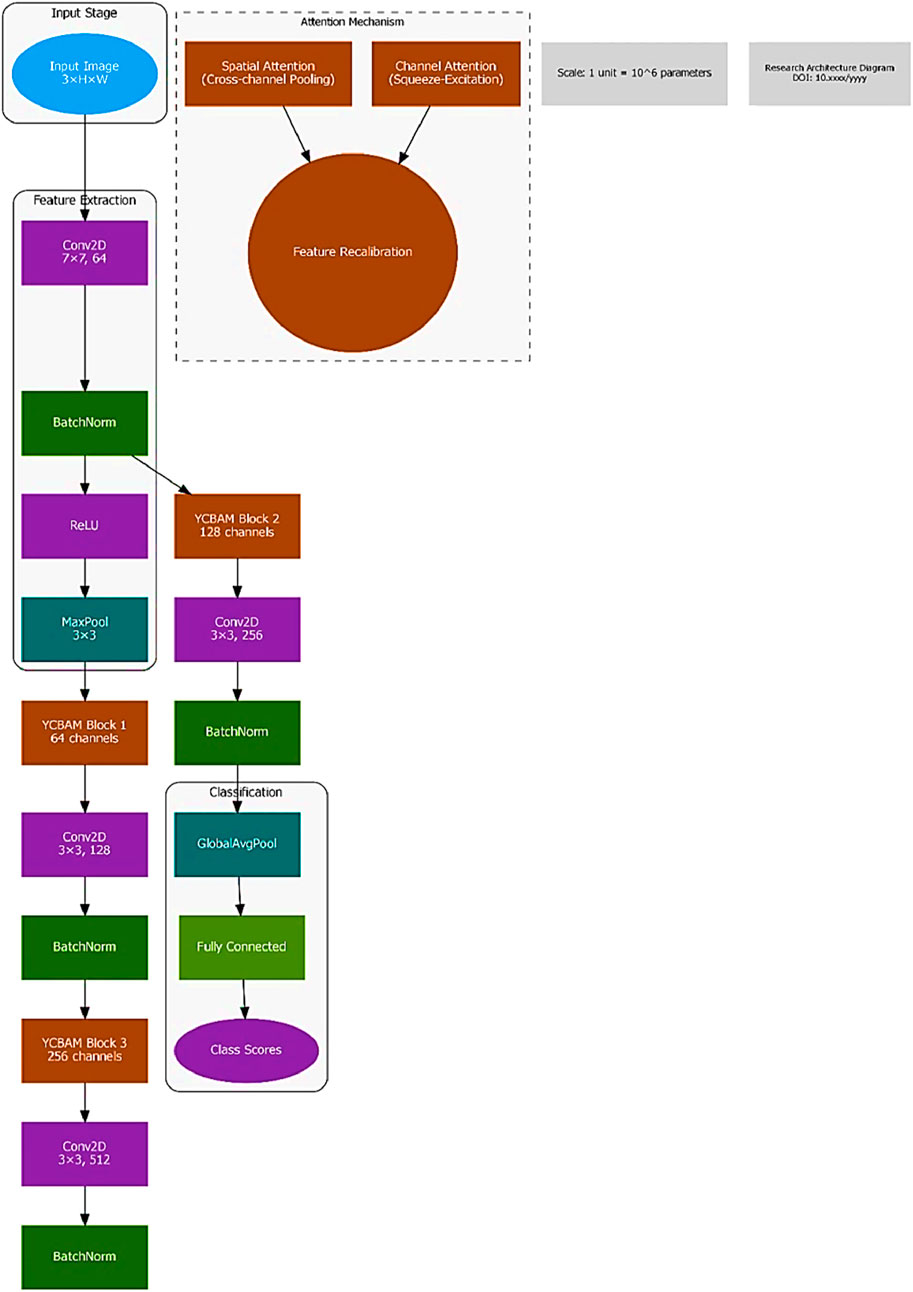
The YCBAM architecture minimizes computational cost and maximizes detection accuracy. The model integrates YOLOv8 with self-attention mechanisms and the Convolutional Block Attention Module.
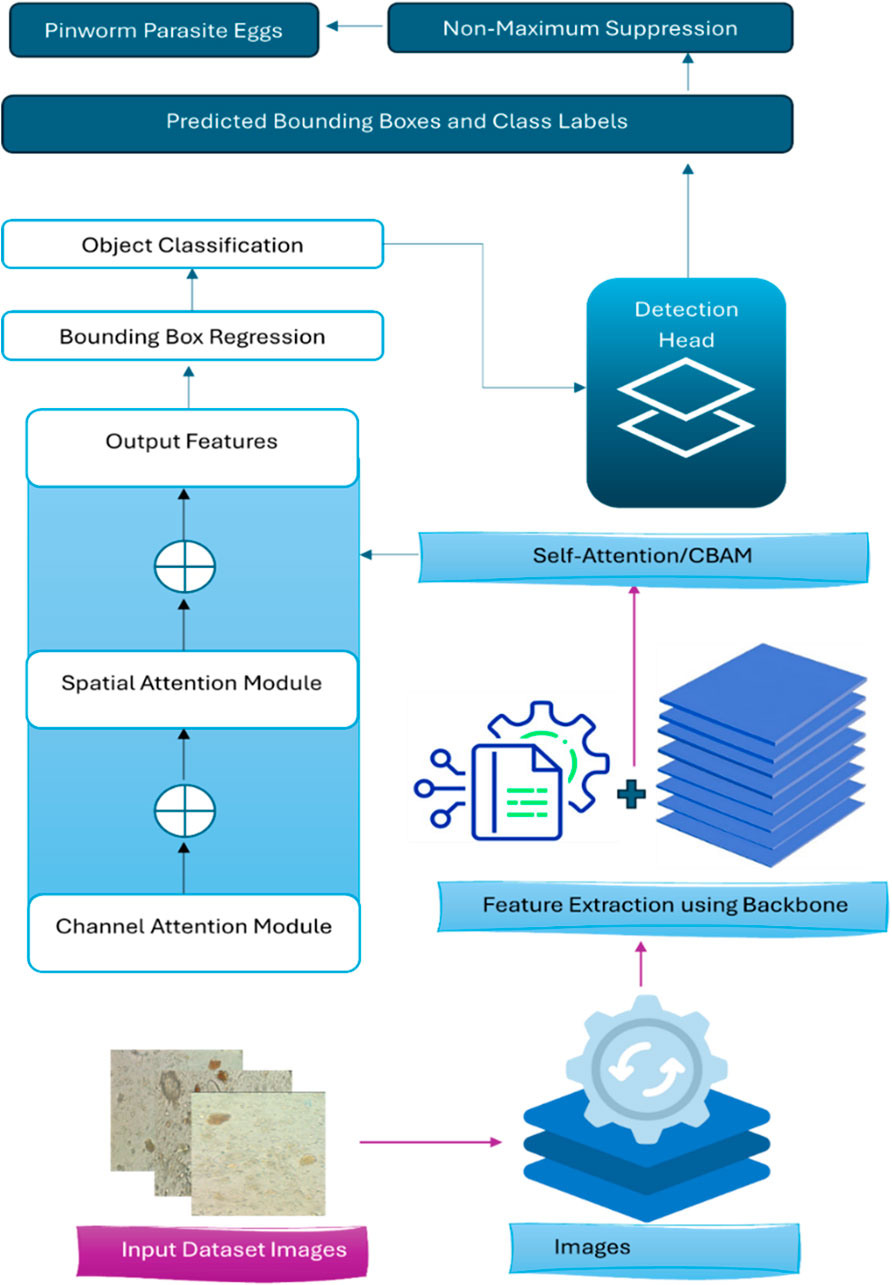
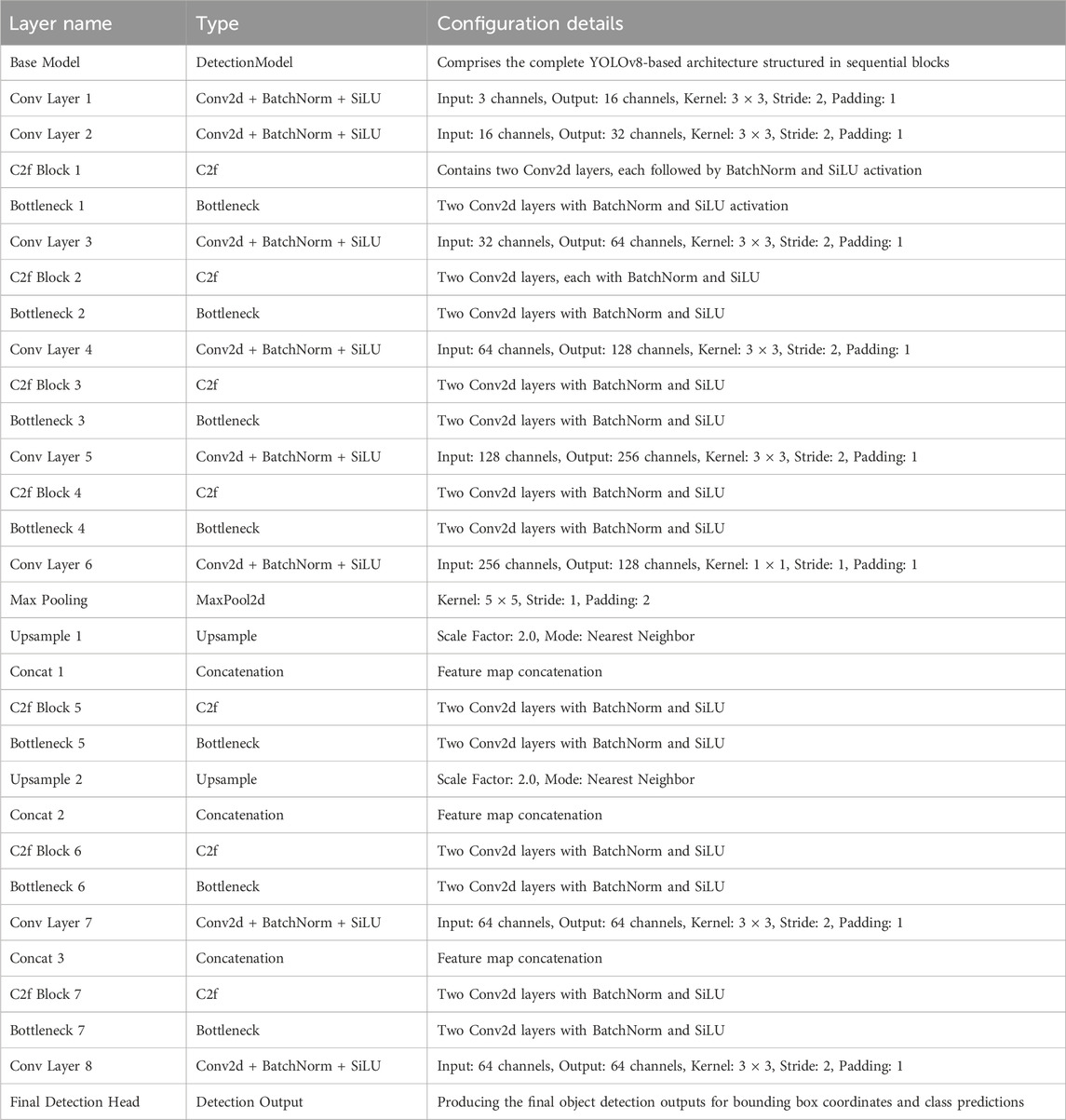
Figures 2, 3 illustrate the main components of the YOLOv8 model. The following sub-sections propose the main steps for egg image detection by YCBAM architecture. Figure 4 shows the main steps for the proposed work. Table 2 represents the layers in the YOLOv8 with CBAM model, highlighting the layer types, configurations, and activations.
3.2.1 Objectness score and bounding box prediction
For each grid cell in the feature map, YOLOv8 predicts multiple bounding boxes, each with an associated objectness score. The objectness score indicates the likelihood of an object being present in the bounding box. The total confidence score in Equation 1 for a predicted bounding box is:
Where,
3.2.2 Bounding box regression
Bounding box predictions are encoded relative to the grid cell location for object localization tasks like detecting the position of an object in an image. The bounding box coordinates in Equation 2 are computed as:
Where:
3.2.3 Loss function
A multi-task loss function is used to optimize three different components during training: objectness, classification, and localization. The total loss
Where:
The Intersection over Union (IoU) is used to evaluate the overlap between the predicted bounding box and the ground truth bounding box. IoU in Equation 4 is defined as:
Where
An enhanced IoU-based loss CIoU function that is an advanced loss function designed to improve the accuracy of bounding box regression is applied. It extends the basic IoU by incorporating additional geometric factors that affect the convergence and the performance of the object detection model in Equation 5 for more accurate bounding box regression: The CIoU loss function is defined as follows:
Where:
3.2.4 Anchor boxes
Anchor boxes, which are predefined bounding boxes of varying aspect ratios and scales are used. The network predicts adjustments to these anchor boxes to fit the objects in the image. The anchor boxes are crucial for handling objects of different sizes efficiently.
For anchor boxes and predictions, the loss function is the number of anchor boxes optimized in Equation 6:
Model Inference and Detection During inference, YOLOv8 processes the entire input image in a single pass. It predicts multiple bounding boxes and class probabilities for each grid cell. Non-Maximum Suppression (NMS) is then applied to eliminate redundant or overlapping boxes, retaining only the most confident predictions in Equation 7:
Where NMS selects boxes with the highest confidence and removes boxes with lower IoU scores.
3.2.5 Convolutional block attention module (CBAM)
The Convolutional Block Attention Module (CBAM) enhances the feature extraction process by applying attention mechanisms along two dimensions: channel attention and spatial attention. The proposed mode allows the model to selectively focus on the most informative feature channels and spatial regions in the input image, improving object detection performance. CBAM consists of two sequential submodules: i) Channel Attention Module (CAM): Focuses on identifying the most important feature channels. ii)Spatial Attention Module (SAM): Identifies key spatial locations in the feature map. Both attention mechanisms are lightweight and easily integrated into existing architectures, such as YOLOv8, with minimal additional computational cost.
3.2.5.1 Channel attention module (CAM)
The Channel Attention Module focuses on which feature channels are the most important for the task. It adaptively re-weights each channel by learning a channel-wise attention map. The map emphasizes relevant channels and neglects irrelevant ones. The input feature map is
Where:
3.2.5.2 Spatial attention module (SAM)
The Spatial Attention Module focuses on identifying important spatial regions within the feature map. It produces a spatial attention map to focus on critical regions in the image with suppressing less important areas. The channel-refined feature map
Where:
3.2.5.3 Combined attention
The CBAM process can be summarized as sequentially applying channel and spatial attention in Equation 11:
Where,
Where:
Incorporating CBAM into the YOLO Model in which the pre-trained YOLO model is modified by implementing CBAM after a specific feature extraction layer. Input Image
4 Experiments and results
In this section, the YOLOv8 model was trained to enhance the accuracy and efficiency of detecting pinworm parasite eggs in microscopic images. The architecture, incorporating Self-Attention mechanisms and the Convolutional Block Attention Module (CBAM), is augmented. These enhancements improved feature extraction by enabling the model to focus on spatial and channel-wise information, leading to better detection of critical details in complex images. Key layers within the YOLOv8 architecture, including Conv, BottleneckCSP, SPPF, C2f, and the YOLO Head, have appositive effect on the performance of the model. Each layer contributed to the extraction of multi-scale features, which significantly enhanced detection accuracy.
The C2f layer provided flexibility in managing the number of channels, ensuring efficient feature extraction, and the SPPF layer’s multi-scale pooling expanded the model’s receptive field, further refining its detection capabilities. These architectural advancements contributed to improving the performance in identifying pinworm eggs with precision and reliability.
4.1 Pinworm parasite egg
The Pinworm Parasite Egg dataset comprises 2,342 high-resolution microscopic images; each annotated with precise bounding boxes around Enterobius vermicularis (pinworm) eggs, as shown in Figure 5. This dataset is organized to support the development and evaluation of deep learning models for the accurate identification of pinworm eggs, facilitating tasks such as object detection, feature extraction, and end-to-end model training. It has significant value for applications in medical diagnostics and parasitology. Each image submits a series of preprocessing steps, including automatic orientation correction based on EXIF metadata and resizing to a standardized input dimension of 640 × 640 pixels using stretch interpolation. These steps ensure uniformity across training and inference pipelines improving model performance. A data augmentation strategy was implemented to enhance model generalization and improve dataset variability. Three augmented versions of each source image were produced by applying random 90-degree rotations, each selected with equal probability. This augmentation scheme increases spatial diversity and allows the model to handle different orientations and visual contexts. All annotations were reviewed to ensure high-quality labels for supervised learning. This designed dataset presents a robust foundation for developing reliable and accurate detection models in complex microscopic environments.
4.2 Training configuration and setup
The proposed YOLO Convolutional Block Attention Module (YCBAM) architecture was implemented using Python and developed within the PyTorch deep learning framework. The model was trained and evaluated in a high-performance computing environment equipped with i) GPU: NVIDIA A100 Tensor Core GPU (40 GB), ii) Processor (Intel Xeon CPU 2.20 GHz) and iii) Memory (128 GB RAM). The model was improved using the AdamW optimizer with a learning rate of
The initial learning rate of 0.01 gradually decreased using a cosine learning rate scheduler, improving the learning process over time. Weight decay of 0.0005 was applied, and early stopping was triggered after 50 epochs if no significant improvements were observed to prevent overfitting. Data augmentation techniques, including random flips, rotations, and scaling, were used to increase the model’s robustness and generalization capability. The Intersection over Union (IoU) threshold was set to 0.2 during non-max suppression to reduce the overlap between predicted bounding boxes. While multi-scale training was not enabled by default, it was explored as a potential strategy for enhancing the model’s generalization by resizing images to various scales during training, as shown in Figure 6.
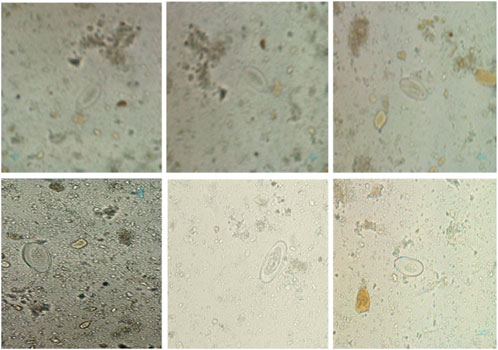
Figure 7 shows the confusion matrix, representing the model’s classification results, with predictions for “Pinworm Egg” and “Background” categories. The normalized confusion matrix provides insights into classification accuracy across both categories. The pairwise scatter plot matrix shows the distribution of bounding box parameters (x, y, width, height) used for localizing pinworm eggs, including histograms for each parameter. The heatmaps of bounding box placements and sizes indicate the spatial distribution of the detected pinworm eggs in the images. Figures 8, 9 show a sample of various circular objects in microscopic images, each labeled with a title that includes the term Enterobius vermicularis, which refers to a parasitic worm (pinworm).
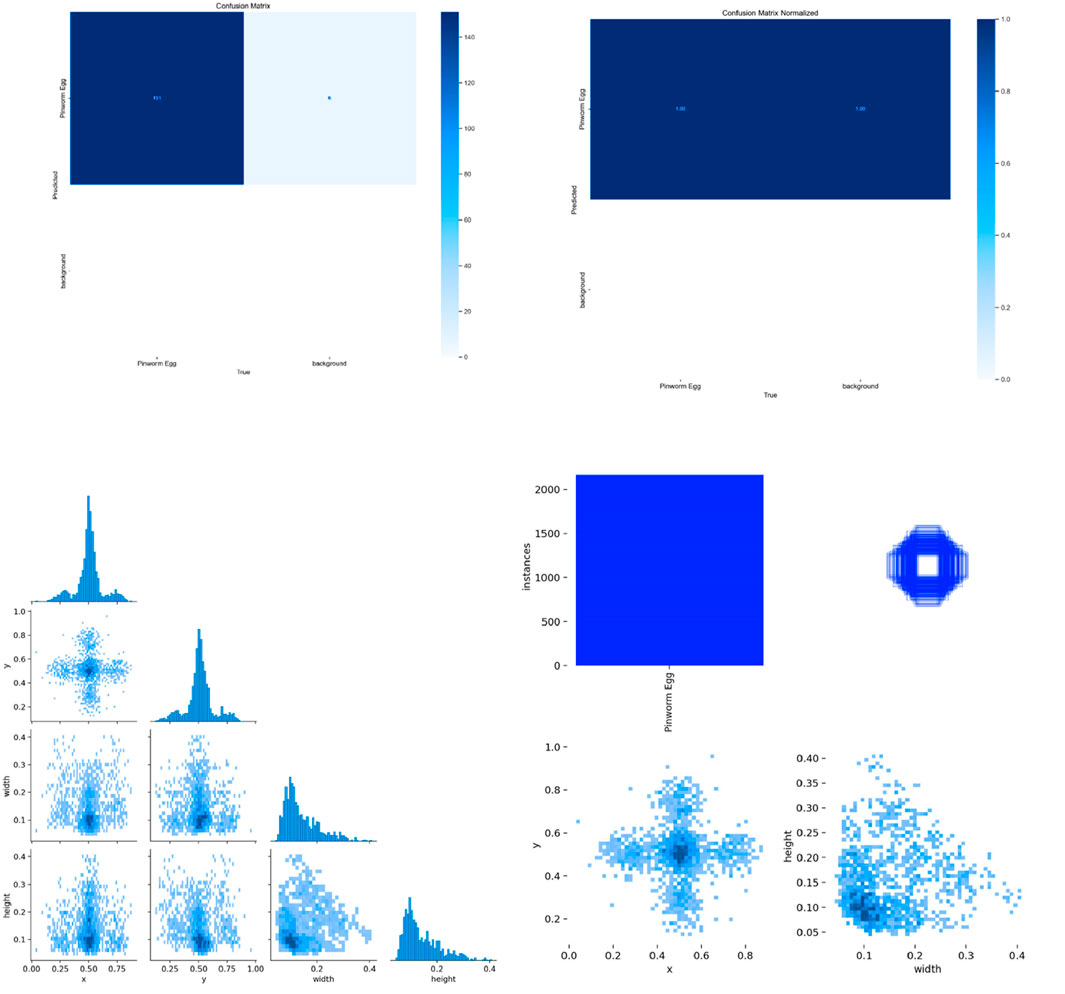
Figure 7. The various visualizations of model performance and data distribution, likely from an object detection or image classification task. It includes a confusion matrix, normalized confusion matrix, pairplots, and histograms, highlighting the accuracy of the model for each class, and the spatial distribution and sizes of detected objects.
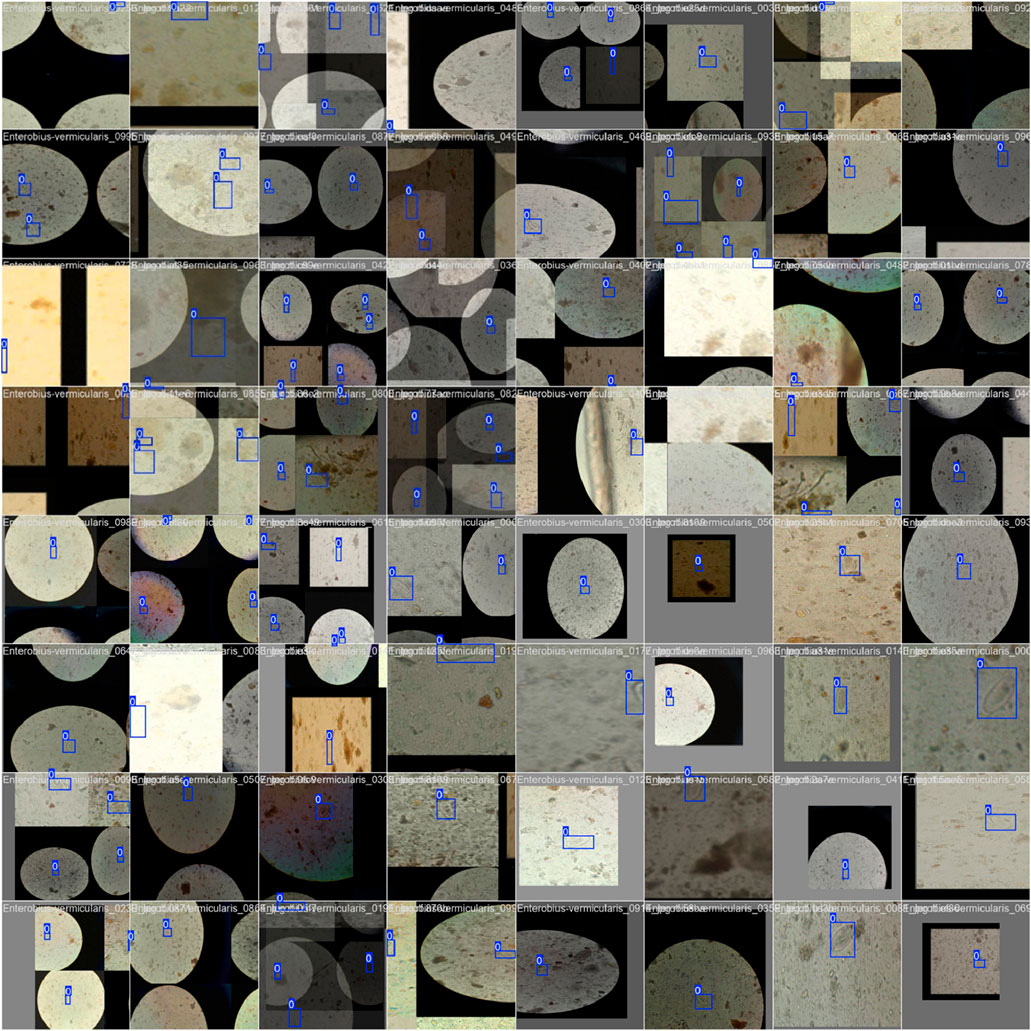
Figure 8. A sample of microscopic images showing Enterobius vermicularis samples with blue bounding boxes defining the regions of interest. The images capture various orientations and magnifications of the samples for identification or analysis purposes.
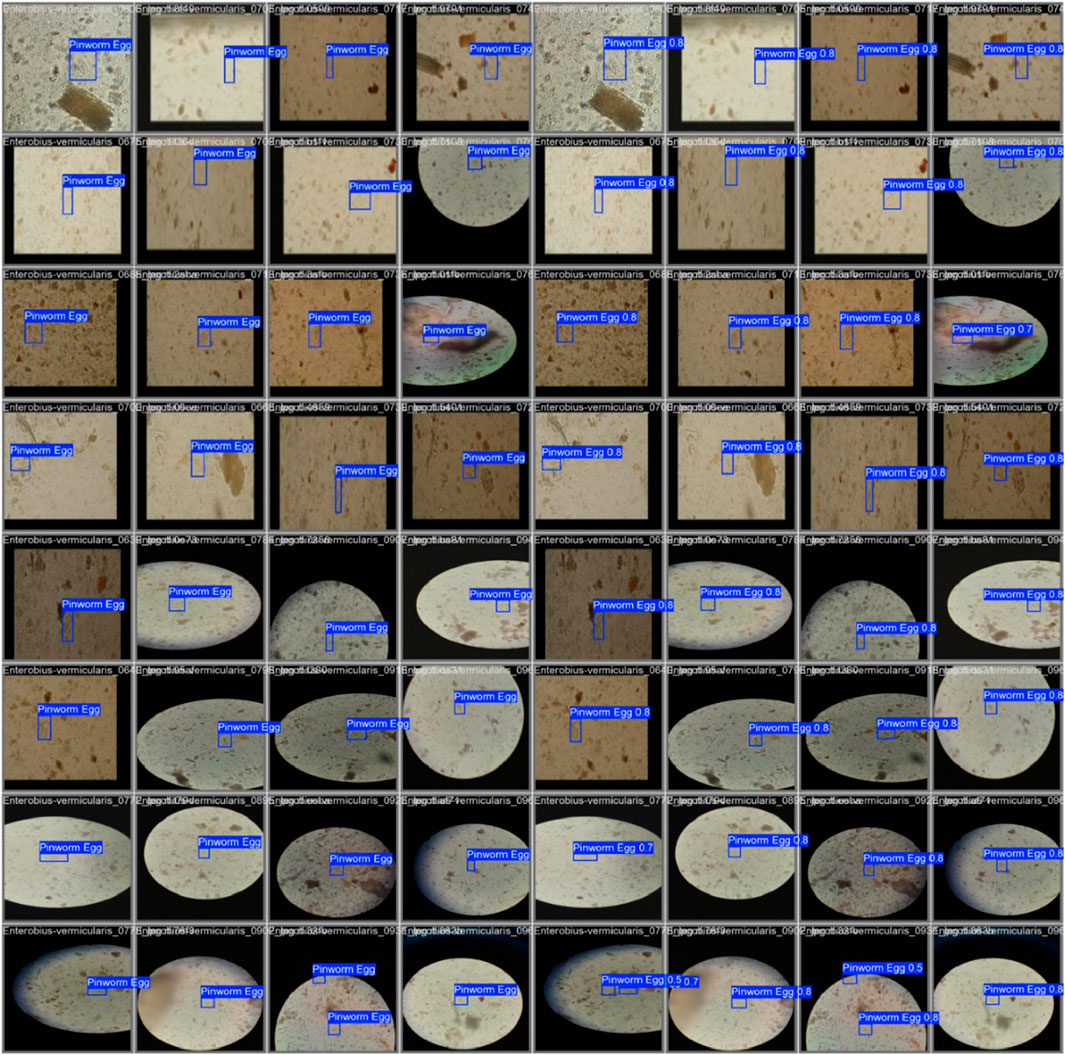
Figure 9. The grid of detection results for “Pinworm Egg” using an object detection model, showing microscope slide views, predicted classes, and confidence scores across samples, indicating successful identifications and variations in detection confidence.
The samples are captured under a microscope, and there are blue bounding boxes drawn over specific areas within each image. These boxes refer to specific regions of interest or potentially identify objects in the image, such as the parasite or some other key feature.
Table 3 shows the various layer types used in the model architecture, designed to enhance feature extraction and improve detection capabilities by using several layers for feature extraction, including Conv, BottleneckCSP, SPPF, C2f, and YoLO Head. Conv2D, BatchNorm, and SiLU activation functions help learn spatial hierarchies in input data. BottleneckCSP reduces computational complexity, while SPPF enhances the model’s receptive field. C2f residual layer keeps essential information across layers. The output layer, YoLO Head, detects objects and predicts bounding boxes, class scores, and confidence scores. Table 4 presents the hyperparameters that control the training process of the model, affecting learning dynamics by using Adam, with other optimization strategies such as Adam. The learning rate, momentum, weight decay, patience, gradient clipping, IoU threshold, data augmentation, multi-scale training, and learning rate scheduler are all crucial for effective convergence. The weight decay value helps mitigate overfitting and prevents overfitting. The model’s adaptability is further enhanced by the multi-scale training option.
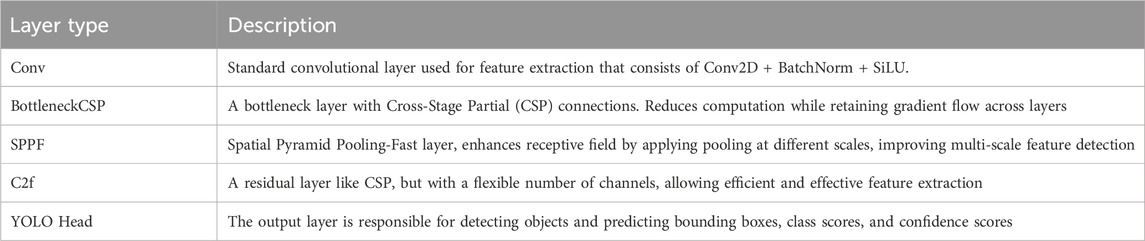
Table 3. Overview of layer types used in the YOLOv8 architecture with self-attention and CBAM integration.

Table 4. The Hyperparameters used for Training the YOLOv8 Model with Integrated Self-Attention and CBAM.
To assess the robustness and generalization capability of the proposed YOLOv8+CBAM model, it is crucial to evaluate its performance on an external dataset that was not included in the training process to ensure that the model can effectively generalize to unseen data and is not overly reliant on specific training distributions. We plan to test the model on a separate clinical dataset obtained from an independent medical facility. Performance metrics such as precision, recall, and mAP can be compared against the results from the primary dataset to determine the model’s adaptability to different imaging conditions and sample variations. Although the proposed model is computationally improved, its deployment in low-resource clinical environments such as hospitals and diagnostic labs presents certain challenges:
i. Hardware Constraints: Many clinical facilities, especially in resource-limited settings, may not have access to high-performance GPUs or cloud-based processing capabilities.
ii. Inference Speed: The real-time processing capability of the model needs to be evaluated on edge devices or embedded systems to ensure efficient deployment.
iii. User-Friendly Interface: an intuitive graphical interface and automated report generation system should be considered to facilitate adoption by healthcare professionals.
5 Discussion
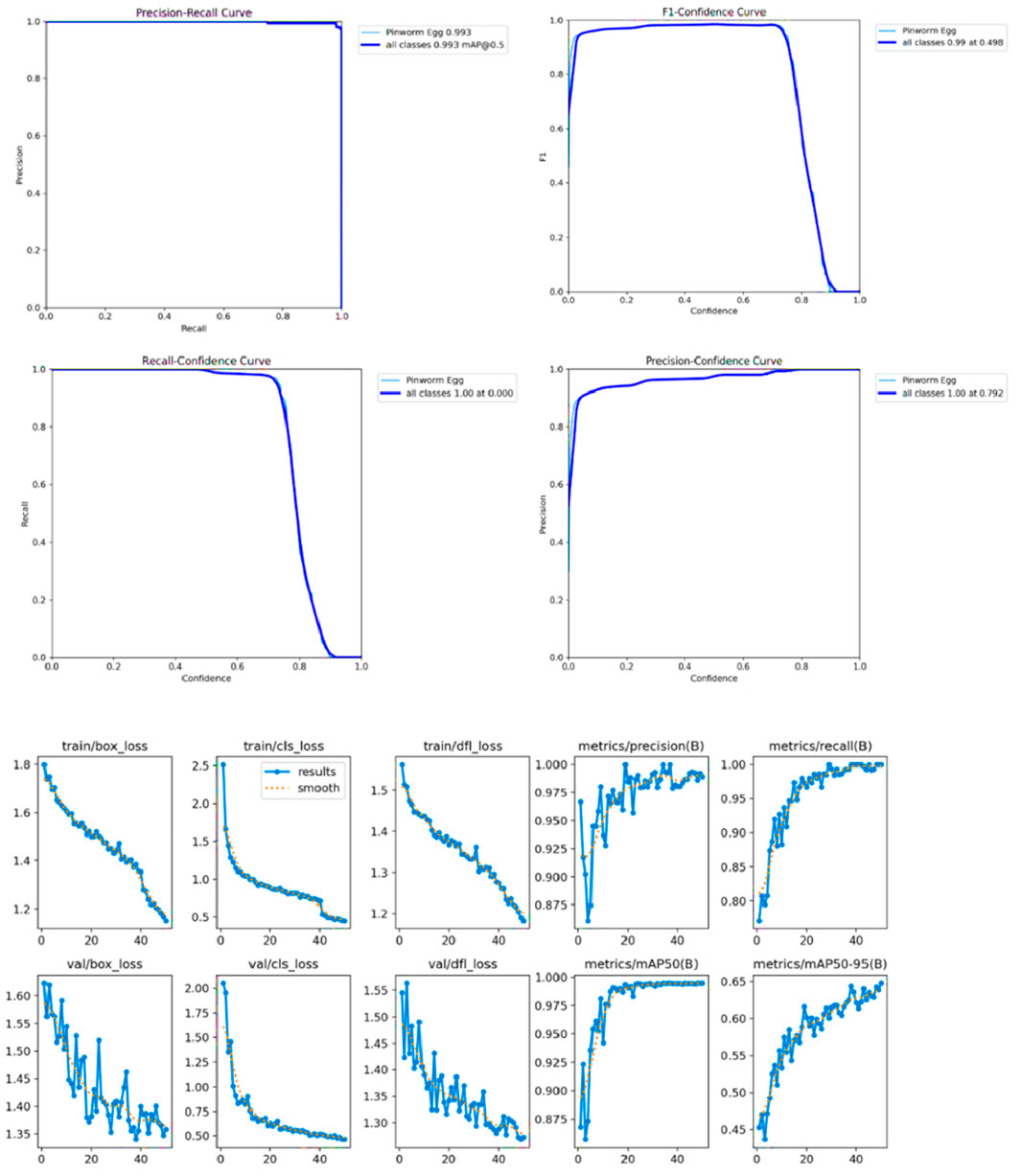
The study proposed the YCBAM model architecture for pinworm egg detection automation compared to previous studies. The model has a high mean average precision (mAP) of 0.995 at an IoU threshold of 0.50 and a mAP50-95 of 0.6531 over multiple IoU thresholds. The model’s precision of 0.99709 and recall of 0.99338 reduce false positives and negatives, which are crucial in clinical diagnostics. The training box loss is reduced to 1.141 during model optimization showing effective learning and convergence, and model robustness. The performance of model is good in the final epoch (Epoch 50), with a mean average precision (mAP@50) of 0.995, presenting its accuracy in microscopic images. The model distinguished pinworm eggs from other artifacts with 0.99709 precision, minimizing false positives.
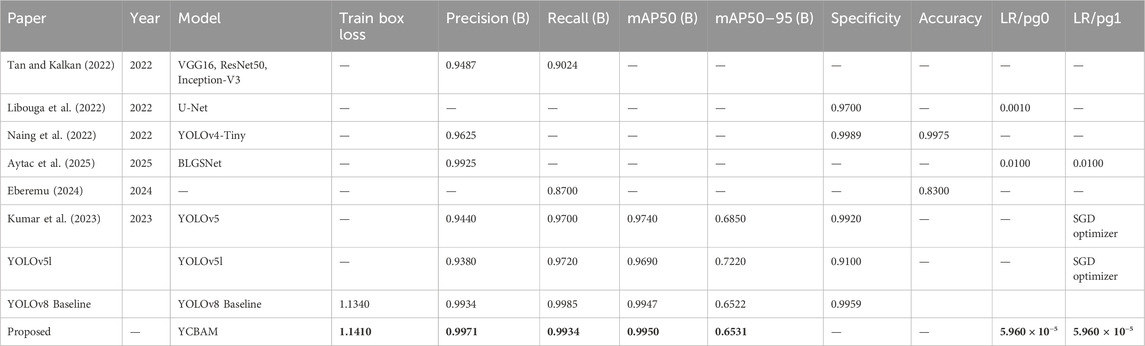
The model’s recall of 0.99338 showed that it detected nearly all pinworm eggs with few missed detections, proving its clinical diagnostic reliability. These findings improve past research. i) The model is better than YOLOv5, which showed 97% mAP. The higher accuracy of 0.995 shows better detection and recognition in complicated and noisy contexts. ii) It improves precision and recall over earlier research that averaged 97%. With 0.99709 precision and 0.99338 recalls, the model lowers diagnostic errors and false positives, enhancing clinical dependability as shown in Table 5. iii) The study uses CBAM-enhanced YOLOv8 to selectively focus on essential spatial and channel information, enabling accurate detection in low-contrast and noisy images, where earlier CNN models struggled. Despite attention modules, the model is computationally efficient, which is useful for clinical applications that need fast processing.
Unlike segmentation methods such as ResU-Net and U-Net, the model balances accuracy and efficiency, making it suitable for resource-constrained scenarios. iv) The proposed model strong training methodology, which includes data augmentation techniques like rotation, zooming, and contrast modifications and adjusted hyperparameters (learning rate of 5.96E-05, momentum of 0.937), improves its generalizability across imaging settings. Table 6 presents a comparative analysis of the performance of various state-of-the-art models for pinworm parasite egg detection. The results demonstrate that the YOLOv8-based models outperform traditional architectures such as Faster R-CNN, EfficientNet, and ResU-Net across key evaluation metrics, including precision, recall, and mean Average Precision (mAP). The baseline YOLOv8 model achieves a precision of 0.997, recall of 0.993, and mAP@50 of 0.995, significantly surpassing Faster R-CNN and ResU-Net in detection accuracy. The integration of Convolutional Block Attention Module (CBAM) and Self-Attention Mechanisms further enhances detection performance. The YOLOv8 + CBAM + Self-Attention model achieves the highest accuracy, with a precision of 0.999, recall of 0.995, and mAP@50 of 0.997, confirming the effectiveness of attention-based feature refinement in improving object localization and classification. The incremental improvements in mAP@50-95 also highlight the robustness of attention-enhanced models in handling variations in microscopic image conditions.
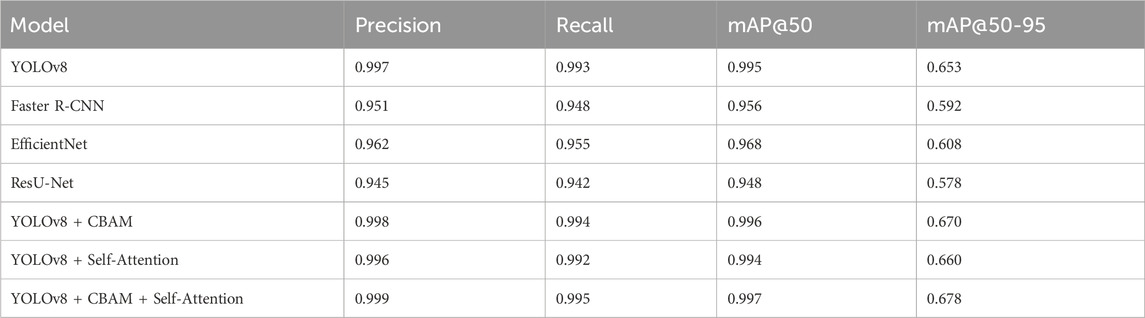
Table 6. The performance comparative analysis with other state-of-the-art Models for Pinworm Parasite Egg Detection.
Figure 9 presents a comparative analysis of different deep learning models, including YOLOv8, Faster R-CNN, EfficientNet, and ResU-Net, for pinworm parasite egg detection. The results indicate that YOLOv8 is better than other models in terms of precision (0.997), recall (0.993), and mAP@50 (0.995), highlighting its superior detection capability. ResU-Net showed the lowest performance, focusing the advantages of using advanced object detection architectures such as YOLOv8 in medical diagnostics. Figure 10 illustrates an ablation study that evaluates the effect of integrating the Convolutional Block Attention Module (CBAM) and Self-Attention into YOLOv8. The results reveal that the combined YOLOv8 + CBAM + Self-Attention model achieves the highest scores across all metrics with a precision of 0.999, a recall of 0.995, and an mAP@50 of 0.997. Figure 11 illustrates improvements in detection accuracy achieved through enhanced feature extraction and attention mechanisms, based on an ablation study assessing the effects of CBAM and Self-Attention on YOLOv8 performance.
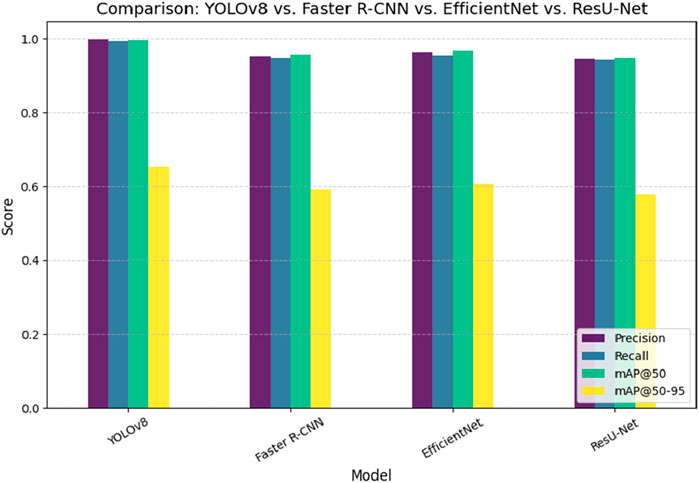
Figure 10. Performance comparison of YOLOv8, Faster R-CNN, EfficientNet, and ResU-Net in pinworm parasite egg detection.
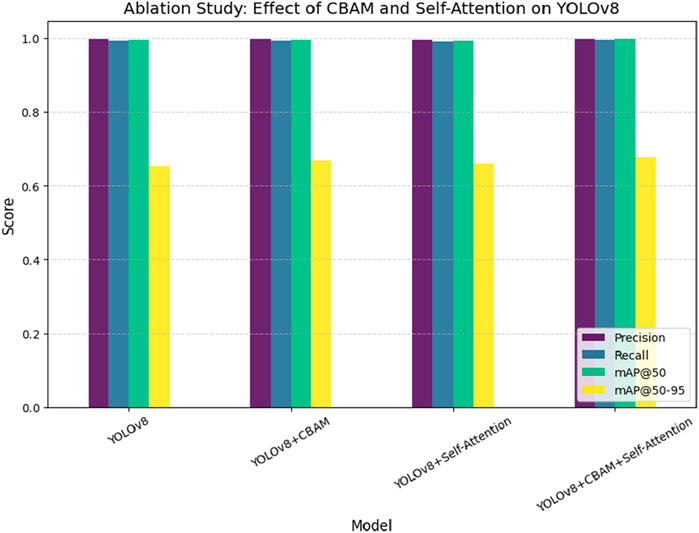
Figure 11. Ablation study assessing the effect of CBAM and Self-Attention on YOLOv8 performance, showing improvements in detection accuracy through enhanced feature extraction and attention mechanisms.
6 Conclusion and future work
According to this study, the YOLO Convolutional Block Attention Module (YCBAM) architecture is proposed to improve the detection of pinworm parasite eggs in microscopic images. The need for advancement is due to the limitations of traditional diagnostic methods, which are time-consuming and human error. With the growing need for automated, efficient, and reliable diagnostic systems in both resource-constrained and high-volume settings, the contributions of this study present a solution to improve both accuracy and scalability in parasitic egg detection. Results show the effectiveness of the YCBAM model, improving the performance in detecting pinworm eggs with high precision and recall values. The mean average precision (mAP) scores of 0.995 at an IoU threshold of 0.50 and 0.6531 across multiple thresholds further substantiate the robustness of our approach. These results focused on the competitive performance of the study’s model compared to state-of-the-art techniques.
Additionally, the integration of self-attention mechanisms and Convolutional Block Attention Module (CBAM) significantly enhances the model’s sensitivity to critical features of pinworm eggs, even in noisy and low-contrast environments. The computational efficiency of the proposed model also positions it as a suitable solution used in clinical environments with limited resources. These findings contribute to advancing automated diagnostic systems in parasitology and other medical domains. This study presents a scalable and robust solution that can be adapted to other diagnostic applications exceeding pinworm detection by determining challenges such as detection speed, small object recognition, and model efficiency. Future improvements in AI-based medical diagnostics include the integration of multi-modal data, such as genetic information, clinical records, and imaging data, to enhance diagnostic accuracy and personalized treatment. Combining microscopic images, patient history, lab test results, and genomic data can provide a more comprehensive understanding of diseases, reduce misdiagnosis risks, and improve early detection.
Advanced deep learning models, including transformer-based architectures and graph neural networks (GNNs), can be used to efficiently process and correlate multimodal data. Additionally, federated learning can enable privacy-preserving collaboration across multiple healthcare institutions, improving model robustness and keeping data security. Further research should focus on standardizing data formats, improving interoperability between different medical systems, and determining computational challenges to ensure seamless integration of multi-modal information into AI-driven diagnostic workflows.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
EH: Writing – review and editing. FA: Data curation, Funding acquisition, Writing – original draft. SE: Data curation, Resources, Writing – original draft. AT: Investigation, Methodology, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Small Research Project under grant number RGP1/225/46.
Acknowledgments
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Small Research Project under grant number RGP1/225/46.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agholi, M., Esfandiari, F., Heidarian, H. R., Khajeh, F., Sharafi, Z., Masoudi, E., et al. (2023). The histopathological findings in appendectomy specimens in an Iranian population. GMJ 12, e2482. doi:10.31661/gmj.v12i.2482
AlMohimeed, A., Saleh, H., Mostafa, S., Saad, R. M. A., and Talaat, A. S. (2023). Cervical cancer diagnosis using stacked ensemble model and optimized feature selection: an explainable artificial intelligence approach. Computers 12 (10), 200. doi:10.3390/computers12100200
AlMohimeed, A., Shehata, M., El-Rashidy, N., Mostafa, S., Samy Talaat, A., and Saleh, H. (2024). ViT-PSO-SVM: cervical cancer predication based on integrating vision transformer with particle swarm optimization and support vector machine. Bioengineering 11 (7), 729. doi:10.3390/bioengineering11070729
Aytac, O., Senol, F. F., Tuncer, I., Dogan, S., and Tuncer, T. (2025). An innovative approach to parasite classification in biomedical imaging using neural networks. Eng. Appl. Artif. Intell. 143, 110014. doi:10.1016/j.engappai.2025.110014
Benecke, J., Benecke, C., Ciutan, M., Dosius, M., Vladescu, C., and Olsavszky, V. (2021). Retrospective analysis and time series forecasting with automated machine learning of ascariasis, enterobiasis and cystic echinococcosis in Romania. PLOS Neglected Trop. Dis. 15, e0009831. doi:10.1371/journal.pntd.0009831
Chaibutr, N., Pongpanitanont, P., Laymanivong, S., Thanchomnang, T., and Janwan, P. (2024). Development of a machine learning model for the classification of Enterobius vermicularis egg. J. Imaging 10, 212. doi:10.3390/jimaging10090212
Ding, P., Qian, H., Zhou, Y., and Chu, S. (2023). Object detection method based on lightweight YOLOv4 and attention mechanism in security scenes. J. Real-Time Image Process. 20 (2), 34. doi:10.1007/s11554-023-01263-1
Eberemu, N. C. (2024). Machine learning-based approach for diagnosing intestinal parasitic infections in northern Nigeria. Fudma J. Sci. 8 (3), 1–8. doi:10.33003/fjs-2024-0803-2404
El-Sunais, Y., and Eberemu, N. (2024). Revolutionizing parasitic infection diagnosis in northern Nigeria: an integrated machine learning approach for the identification of intestinal parasites and associated risk factors.
Elbedwehy, S., Hassan, E., Saber, A., and Elmonier, R. (2024). Integrating neural networks with advanced optimization techniques for accurate kidney disease diagnosis. Sci. Rep. 14 (1), 21740. doi:10.1038/s41598-024-71410-6
Hassan, E. (2024). Enhancing coffee bean classification: a comparative analysis of pre-trained deep learning models. Neural Comput. Applic 36, 9023–9052. doi:10.1007/s00521-024-09623-z
Hassan, E., El-Rashidy, N., and Talaa, M. (2022). Mask R-CNN models. Nile J. Commun. Comput. Sci. 3 (1), 17–27.
Hassan, E., Saber, A., and Elbedwehy, S. (2024). Knowledge distillation model for acute lymphoblastic leukemia detection: exploring the impact of nesterov-accelerated adaptive moment estimation optimizer. Biomed. Signal Process. Control 94, 106246. doi:10.1016/j.bspc.2024.106246
Ji, C. L., Yu, T., Gao, P., Wang, F., and Yuan, R. Y. (2024). YOLO-TLA: an efficient and lightweight small object detection model based on YOLOv5. J. Real-Time Image Process. 21 (4), 141. doi:10.1007/s11554-024-01519-4
Kitvimonrat, A., Hongcharoen, N., Marukatat, S., and Watchrabutsarakham, S., (2020). “Automatic detection and characterization of parasite eggs using deep learning methods,” in 2020 International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON), Phuket, Thailand, 24-27 June 2020 (IEEE).
Kumar, S., Arif, T., Ahamad, G., Chaudhary, A. A., Khan, S., and Ali, M. A. M. (2023). An efficient and effective framework for intestinal parasite egg detection using YOLOv5. Diagnostics 13, 2978. doi:10.3390/diagnostics13182978
Lee, C.-C., Huang, P. J., Yeh, Y. M., Li, P. H., Chiu, C. H., Cheng, W. H., et al. (2022). Helminth egg analysis platform (HEAP): an opened platform for microscopic helminth egg identification and quantification based on the integration of deep learning architectures. J. Microbiol. Immunol. Infect. 55 (3), 395–404. doi:10.1016/j.jmii.2021.07.014
Li, A., Song, X., Sun, S., Zhang, Z., Cai, T., and Song, H. (2023a). “YOLO-SA: an efficient object detection model based on self-attention mechanism,” in InAsia-Pacific web (APWeb) and web-age information management (WAIM) joint international conference on web and big data (Singapore: Springer Nature), 1–15.
Li, Y., Liu, L., and Lu, T. (2023b). SAE-CenterNet: self-attention enhanced CenterNet for small dense object detection. Electron. Lett. 59 (3), e212732. doi:10.1049/ell2.12732
Libouga, I. O., Bitjoka, L., Gwet David, L. L., Boukar, O., and Njan Nloga, A. M. (2022). A supervised U-Net based color image semantic segmentation for detection and classification of human intestinal parasites. e-Prime-Advances Electr. Eng. Electron. Energy 2, 100069.
Mirzaei, O., Guler, E., Akkaya, N., Bilgehan, O., and Suer, K. (2022a). Automated early-stage Enterobius vermicularis diagnosis using segmentation model applied to the deep learning method. Preprint.
Mirzaei, O., Ibrahim, A. U., Guler, E., Akkaya, N., Bilgehan, O., and Suer, K. (2022b). Artificial intelligence-assisted segmentation and classification of Enterobius vermicularis. SSRN Electron. J.
Naing, K. M., Boonsang, S., Chuwongin, S., Kittichai, V., Tongloy, T., Prommongkol, S., et al. (2022). Automatic recognition of parasitic products in stool examination using object detection approach. PeerJ Comput. Sci. 8, e1065. doi:10.7717/peerj-cs.1065
Papadopoulos, A., Melissas, P., Kastellos, A., Katranitsiotis, A., Zaparas, P., Stavridis, P., et al. (2024). “TenebrioVision: a fully annotated dataset of Tenebrio molitor larvae worms in a controlled environment for accurate small object detection and segmentation,” in Icpram.
Pun, T. B., Neupane, A., and Koech, R. (2023). A deep learning-based decision support tool for plant-parasitic nematode management. J. Imaging 9 (11), 240. doi:10.3390/jimaging9110240
Ray, K., Shil, S., Saharia, K., Sarma, N., and Karbasanavar, N. (2020). “Detection and identification of parasite eggs from microscopic images of fecal samples,” in Computational intelligence in pattern recognition: proceedings of CIPR 2019 (Springer).
Ray, K., Saharia, S., and Sarma, N. (2024). Segmentation approaches of parasite eggs in microscopic images: a survey. SN Comput. Sci. 5, 401. doi:10.1007/s42979-024-02709-4
Ruenchit, P. (2021). State-of-the-Art techniques for diagnosis of medical parasites and arthropods. Diagnostics 11, 1545. doi:10.3390/diagnostics11091545
Saber, A., Elbedwehy, S., Awad, W. A., and Hassan, E. (2024). An optimized ensemble model based on meta-heuristic algorithms for effective detection and classification of breast tumors. Neural Comput. Appl. 37, 4881–4894. doi:10.1007/s00521-024-10719-9
Talaat, A. S. (2024). “A novel ensemble model for brain tumor diagnosis,” in New mathematics and natural computation, 1–17.
Tan, H., and Kalkan, A. (2022). Using deep learning models to detect parasites early. J. Glob. Strategic Manag. 16 (2), 16. doi:10.20460/jgsm.2023.319
Xu, W., Zhai, Q., Liu, J., Xu, X., and Hua, J. (2024). A lightweight deep-learning model for parasite egg detection in microscopy images. Parasites Vectors 17 (1), 454. doi:10.1186/s13071-024-06503-2
Keywords: convolutional block attention module, microscopic image analysis, pinworm parasite, YOLO, medical parasitology
Citation: Hassan E, Alqahtani F, Elbedwehy S and Talaat AS (2025) Automated detection of pinworm parasite eggs using YOLO convolutional block attention module for enhanced microscopic image analysis. Front. Bioeng. Biotechnol. 13:1559987. doi: 10.3389/fbioe.2025.1559987
Received: 13 January 2025; Accepted: 18 August 2025;
Published: 15 October 2025.
Edited by:
Yi Zhao, The Ohio State University, United StatesReviewed by:
Yu Fenghua, Shenyang Agricultural University, ChinaGabriel Avelino Sampedro, University of the Philippines Diliman, Philippines
Copyright © 2025 Hassan, Alqahtani, Elbedwehy and Talaat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esraa Hassan, ZXNyYWEuaGFzc2FuQGFpLmtmcy5lZHUuZWc=; Felwah Alqahtani, ZmFscWh0YW5pQGtrdS5lZHUuc2E=
 Esraa Hassan
Esraa Hassan Felwah Alqahtani2*
Felwah Alqahtani2* Samar Elbedwehy
Samar Elbedwehy Amira Samy Talaat
Amira Samy Talaat