- Institut für Physikalische Biologie, Heinrich-Heine-Universität, Düsseldorf, Germany
Directed evolution leverages the principles of natural selection to engineer biomolecules with desired properties. Microbead-based approaches within water-in-oil emulsions have proven invaluable for high-throughput in vitro selections. However, highly aggregation-prone microbeads present significant challenges, including clustering, inconsistent distribution, and droplet instability. Here, we introduce a simple and cost-effective method for generating polydisperse emulsions with restored Poissonian distributions of highly aggregation-prone microbeads. This approach utilizes modified gel loader pipette tips, drawn out to create nozzles capable of disrupting microbead clusters during emulsification. Two widely utilized oil-surfactant formulations—mineral oil with Abil EM 90 and FluoSurf in HFE 7500 — were evaluated for emulsion preparation. Emulsions prepared using the modified nozzles exhibited exceptional stability, maintaining integrity during week-long incubations at 37°C, and reliably distributed microbeads into droplets in accordance with a Poissonian distribution despite the microbeads’ highly aggregation-prone property.
1 Introduction
Directed evolution is a powerful tool for engineering biomolecules with desired properties by mimicking the natural evolutionary process in a laboratory setting (Arnold and Volkov, 1999; Stucki et al., 2021; Wang et al., 2021). Among various in vitro methodologies, microbead-based approaches conducted in emulsions have gained significant attention for their ability to screen vast libraries of variants efficiently (Griffiths and Tawfik, 2003; Gan et al., 2008; Diamante et al., 2013; Price et al., 2014; Zhu et al., 2015; Mankowska et al., 2016; Siu et al., 2021; Anyaduba et al., 2022; Iizuka et al., 2022; Ito et al., 2024). These methods leverage the compartmentalization provided by water-in-oil emulsions to isolate individual genetic variants and their corresponding encoded proteins, ensuring genotype-phenotype linkage. Microbeads can be introduced to serve as a solid support for capturing and localizing genetic material or the expressed proteins, often through specific binding interactions (Diamante et al., 2013; Mankowska et al., 2016), or molecular probes to detect enzymatic activity (Griffiths and Tawfik, 2003; Zhu et al., 2015). The emulsion droplets act as microreactors, enabling high-throughput screening by creating millions of independent compartments, each containing a single microbead and the biochemical machinery for transcription and translation. The tight spatial control facilitates direct coupling between the genotype (DNA on the microbead) and the phenotype (a probe on the microbead detecting the functional activity of the protein).
There are two prominent approaches to prepare these emulsions: Highly controlled droplet-based microfluidics (Price et al., 2014; Song et al., 2003; Agresti et al., 2010; Goto et al., 2020) and preparation via stochastic one-step polydisperse emulsification methods (Gan et al., 2008; Mankowska et al., 2016; Siu et al., 2021; Tawfik and Griffiths, 1998; Griffiths and Tawfik, 2006; Wang et al., 2014; Byrnes et al., 2018).
Microfluidics-based methods utilize precisely engineered chips to manipulate small volumes of fluids at the nano-to microscale, enabling the creation of highly monodisperse droplets. Microfluidics chips, often made of PDMS (Song et al., 2003; Raj and Chakraborty, 2020) or glass (Aralekallu et al., 2023), feature intricate microchannel networks designed for controlled droplet generation and manipulation (Song et al., 2003).
Stochastic one-step polydisperse emulsification methods are used to prepare bulk emulsions using simple vortexing or mixing techniques, requiring minimal specialized equipment (Tawfik and Griffiths, 1998).
Each approach offers distinct advantages and disadvantages. Microfluidics-based systems offer precise control allowing for the generation of monodisperse droplets with highly uniform sizes, ensuring consistent reaction conditions across all compartments. On-chip manipulations of droplets such as mixing (Song et al., 2003), splitting (Link et al., 2004), merging (Song et al., 2003; Link et al., 2006), incubation (Song et al., 2003) and sorting via fluorescence or absorbance readouts (Agresti et al., 2010; Link et al., 2006; Ahn et al., 2006; Holstein et al., 2021), or droplet load and size (Nam et al., 2012; Jing et al., 2015) enable complex experimental workflows. However, microfluidics-based systems require special equipment and there are technical limits to throughput. As droplets are generated and processed in succession, the experiments are limited by flow speeds which leads to large-scale screening experiments consuming entire days of hands-on work and unfortunately needing to be closely monitored during the run, as they are highly sensitive to flow fluctuations and prone to clogging by particulate matter.
Stochastic one-step droplet generation can be performed in a single batch via mixing or vortexing of the oil- and aqueous-phases (Gan et al., 2008; Mankowska et al., 2016; Siu et al., 2021; Tawfik and Griffiths, 1998; Griffiths and Tawfik, 2006; Wang et al., 2014; Byrnes et al., 2018). This can be done in virtually any laboratory and does not require any specialized equipment. Another advantage is scalability, as larger volumes of emulsion can be generated with ease and can handle substantial library sizes without the constraints of microfluidic device throughput (Gantz et al., 2023). Furthermore, these approaches are most cost-effective, as they are inexpensive to set up and execute and consume less reagent than microfluidics-based systems. However, droplets in bulk emulsions are polydisperse. They vary in size, leading to inconsistent reaction conditions and potential bias in selection outcomes. Furthermore, the risk of cross-contamination between droplets is higher, which can compromise the fidelity of in vitro evolution processes.
The incorporation of microbeads into microfluidic systems presents significant technical challenges (Chen et al., 2023a). For successful addition of microbeads into droplets using microfluidic chips, it is crucial to ensure a uniform dispersion and dilution of microbeads in the precursor solution prior to droplet formation (Anyaduba et al., 2022). The efficiency of these approaches is usually constrainted by the Poisson distribution. Microbead encapsulation occurs randomly, with a theoretical maximum of only 37% of droplets containing a single microbead. This inefficiency results in significant waste of both microbeads in droplets containing more than one microbead and reagent in empty droplets. Recently, microfluidic strategies emerged that have overcome the limitation imposed by random Poisson distribution during microbead encapsulation into droplets (Chen et al., 2023a; Abate et al., 2009; Link et al., 2022; Yue et al., 2022; Luo and Lee, 2023). These approaches mostly rely on close packing of microbeads enabling controlled release and precise single-microbead addition into droplets. However, these approaches require the microbeads to be compressible and lubricous to prevent clogging (Chen et al., 2023a; Abate et al., 2009; Yue et al., 2022; Luo and Lee, 2023). Another approach uses surface acoustic waves (SAW) to actively encapsulate single cells into droplets. This method uses laser-assisted detection of individual cells during droplet formation and actively cutting-off droplets at the T-junction upon detection of a cell. This approach, however, results in the formation of polydisperse emulsions and requires diluted and nicely dispersed particles (Link et al., 2022).
In any of these cases, particle aggregation represents the nemesis of these microfluidics-based particle encapsulation approaches. Clumping of microbeads must be avoided, as their settling in the feed line can result in uneven encapsulation, disruption of droplet formation, or complete clogging of microfluidic channels. Certain microbead surface modifications—which might be necessary for your experimental setup—can exacerbate microbead aggregation, making it extremely difficult, if not impossible, for microfluidic systems to handle these samples effectively. When designing a selection scheme for a directed evolution campaign, predicting the effects of specific surface modifications on microbead aggregation remains challenging. Hydrophobic surface modifications, such as those introduced by certain peptides, proteins, fluorescent dyes, or polycyclic aromatic hydrocarbons, can be expected to promote microbead aggregation (Bharti et al., 2011). But also the conjugation of nucleic acids, particularly single-stranded DNA or RNA, can enhance aggregation due to complementary strand hybridization (Valignat et al., 2005; Leslie et al., 2012; Sloan et al., 2016). The exact influence of individual surface chemistries is often difficult to predict, and aggregation may unexpectedly occur and jeopardize the experimental setup. And in other cases, surface modifications that are expected to induce microbead aggregation may be unavoidable due to experimental requirements.
In such cases, polydisperse emulsification methods offer a viable alternative. These methods are less susceptible to issues related to microbead settling because precursor solutions are typically processed immediately, eliminating the idle time required in microfluidic systems. However, even polydisperse emulsification techniques face limitations when dealing with highly aggregation-prone microbeads. In case microbead clusters survive sonification and other means of dispersion or rapidly reassemble, clumps of multiple microbeads may still be encapsulated into single droplets.
In this study, we present a simple and effective method for rapid generation of polydisperse emulsions containing a restored Poisson distribution of highly aggregation-prone microbeads. Our approach utilizes drawn-out 200
2 Results and discussion
As part of an ongoing project, we required the encapsulation of individual microbeads functionalized with a specifically engineered peptide into droplets. This effort was integral to a directed evolution strategy aimed at developing proteolytically active antibodies targeting the amyloid-
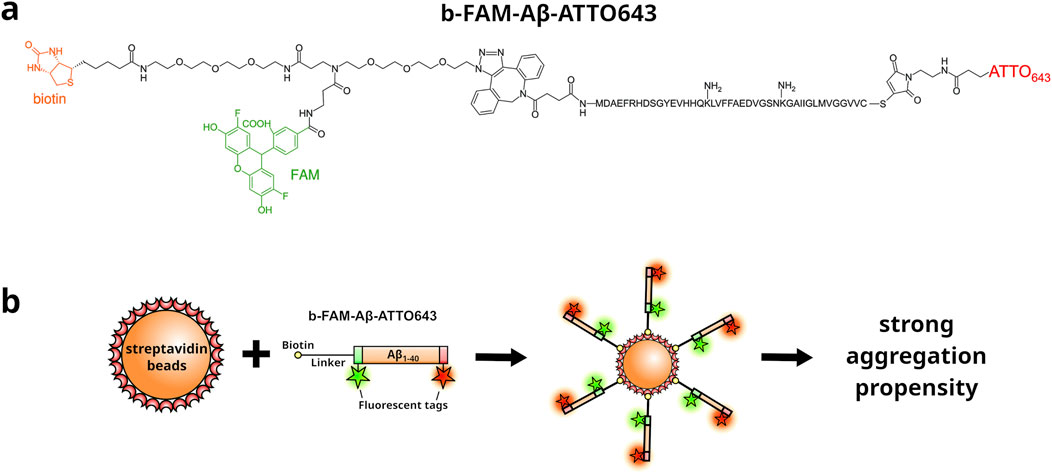
Figure 1. Schematic representation of the b-FAM-A
2.1 Homemade pipette tip-based nozzle
Low-cost and easily fabricated nozzles were created by modifying 200
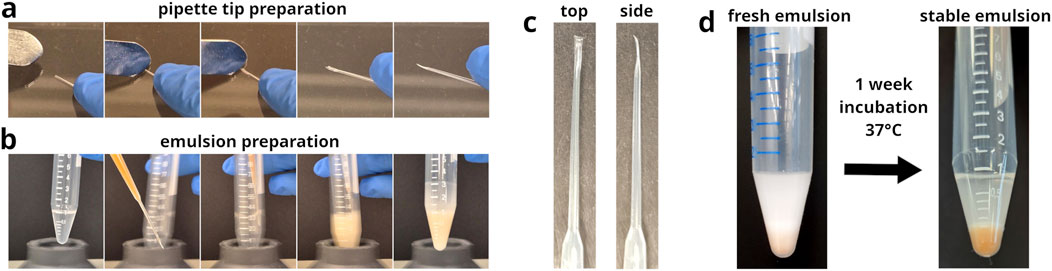
Figure 2. Preparation of modified gel loader pipette tips and their application as nozzles in emulsion generation. (a) Image series of the process used to modify 200
The morphology of the pipette tip-derived nozzles can be quite diverse (see Supplementary Figures S1, S2). However, adequate flow restriction was the important factor to achieve sufficient microbead dispersion during emulsification (see Supplementary Figure S3). Differences in nozzle morphology did not noticeably affect their functionality.
2.2 Preparation of emulsions containing highly aggregation-prone microbeads
Emulsions were prepared using two widely utilized oil-surfactant formulations for comparison: the first consisted of mineral oil with 2% (v/v) Abil EM 90% and 0.05% (v/v) Triton X-100 (Anyaduba et al., 2022; Miller et al., 2006; Courtois et al., 2008; Tubeleviciute and Skirgaila, 2010; C Hatch et al., 2011; Turchaninova et al., 2013; Takeuchi et al., 2014; Tanaka et al., 2015; Zhang et al., 2020; Tanno et al., 2020; An et al., 2020), while the second employed a commercially available formulation, 2% FluoSurf (Dolomite), comprised of the inert perfluorocarbon carrier oil HFE 7500 and a proprietary fluorous surfactant (Bussiere et al., 2019; Karamitr et al., 2020; Zhu et al., 2023).
The aqueous phase consisted of PURExpress in vitro Protein Synthesis reagent (New England Biolabs) containing 107 microbeads decorated with 106 molecules b-FAM-A
As a control, an Abil EM 90-based emulsion was prepared using an unmodified gel loader pipette tip. 100
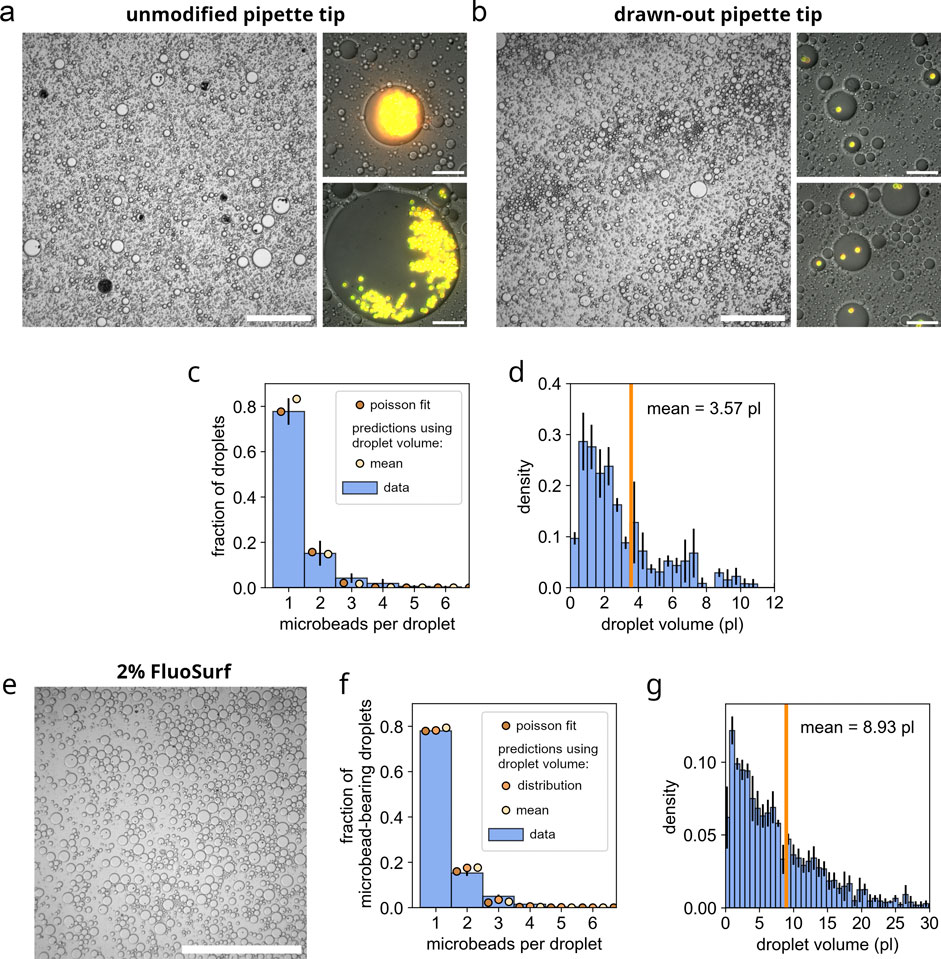
Figure 3. Restoration of Poissonian distributions of microbeads in emulsion droplets using gel loader pipette tip-derived nozzles. (a) Microscopic image of a control emulsion (Abil EM 90-based) prepared with unmodified gel loader pipette tips, showing severe clustering of microbeads and a low fraction of droplets containing microbeads. The overview image is captured using DIC microscopy at low magnification (scale bar: 250
In comparison, emulsions with the same composition were prepared using the modified, drawn-out gel loader pipette tips, as shown in Figures 2a–c. The resulting emulsions demonstrated exceptional stability, remaining intact during a 1 week incubation at 37°C without any signs of degradation (see Figure 2d; Supplementary Figure S6). Microscopic analysis revealed that microbead-containing droplets in Abil EM 90-based emulsions presented a mean volume of 3.6 pL (see Figure 3d). Wherein, approximately 80% contained a single microbead, less than 20% contained two microbeads and less than 5% contained three or more microbeads (see Figures 3b,c). Quantitative analysis of the microbead distribution within droplets closely followed a Poisson distribution fit (see Figure 3c, brown dots), yielding a fitted theoretical rate of occurence
A similar investigation, as for the Abil EM 90-derived emulsions, was performed using emulsions prepared with a 2% FluoSurf oil-surfactant blend in perfluorocarbon carrier oil HFE 7500. Therefore, 200
These results demonstrate that this simple and rapid technique, utilizing homemade nozzles created by drawing out gel loader pipette tips, reliably produces emulsions with restored Poissonian distributions of microbeads, even for highly aggregation-prone microbeads.
Further standardization of nozzle fabrication may be feasible using the protocol published by Chen et al. (2023b), which uses a Do-It-Yourself (DIY) screwdriver-based tip modification tool to deform pipette tip orifices into an eliptical shape. To adapt this protocol for our purpose, considerably higher torques would be required to achieve sufficient flattening of the pipette tips. An adaptation of this technique would allow for standardization of nozzle production and lead to further improvement in reproducibility. On a note, however, in our experiments ultimate droplet size distributions appeared to be primarily influenced by factors such as surfactant concentrations, vortex speeds, and vortex timings, with little impact from the actual nozzle morphology.
In conclusion, we have developed a simple, cost-effective, and efficient method for the rapid generation of polydisperse emulsions, enabling the encapsulation of highly aggregation-prone microbeads in accordance with a Poissonian distribution using modified gel-loading pipette tips. This low-cost approach requires minimal equipment and materials, making it highly accessible to laboratories with limited resources. By restoring the Poissonian distribution of aggregation-prone microbeads, this method addresses a significant challenge and expands the range of applications in directed evolution systems. Microbead modifications that typically induce aggregation no longer pose a barrier to establishing directed evolution campaigns. The method’s affordability, reproducibility, and adaptability make it a compelling solution for diverse bead-based assays and droplet-based applications that necessitate the incorporation of microbeads into droplets. Beyond microbeads, this technique is likely applicable to other aggregation-prone particulate matter, provided the particle size remains within the dimensions of the nozzle’s orifice. Furthermore, modulating the applied pressure to reduce compression of the pipette tips may further accommodate the use of larger particles to some degree. In addition, our technique is expected to be compatible with microorganisms possessing robust exterior shells, such as bacteria or yeast. However, its applicability to eukaryotic cells, including animal or human cells, is likely limited due to their susceptibility to damage from the excessive shear forces generated within the pipette tip-derived nozzles.
3 Materials and methods
3.1 Materials
200
3.2 Synthesis of b-FAM-A
A precursor A
MA
20 nmol MA
3.3 Preparation of functionalized microbeads
For functionalization of microbeads, 5 × 107 ProMag 3 HP streptavidin microbeads were used. Microbeads were washed three times with 200
3.4 Preparation of gel loader pipette tip-derived nozzle
Detailed step-by-step instructions are provided in the Supplementary Material.
Modified pipette tip nozzles were prepared by manually drawing out standard 200
Step-by-Step Instructions:
1. A gel loader pipette tip was positioned on the flat surface of a sterile 96-well plate lid (Nunc, catalog number: 243656).
2. The handle side of a metal scalpel was pressed onto the frontmost 2 mm of the pipette tip, with the handle tilted at a 45°angle toward the pipette tip orifice.
3. The scalpel handle was drawn across the pipette tip toward the orifice while applying approximately 1 kg of pressure, as determined by a precision scale. A single pass was typically sufficient; however, if compression was incomplete, a second pass could be performed to ensure proper modification.
The preparation process is demonstrated in Supplementary Video S1, and a gallery of different nozzle preparations and their QC properties is provided in to 3 Supplementary Figure S1.
3.4.1 Quality control
The proper fabrication of pipette tip-derived nozzles was usually evaluated by aspirating 200
The quality control procedure is demonstrated in Supplementary Video S2 and Supplementary Figures S2, S3.
It was observed that variations in external nozzle morphology due to variation in the preparation did not noticeably influence the emulsions and dispersion of microbeads. Proper dispersion of microbeads was rather dependent on sufficient nozzle compression.
3.5 Emulsification
Detailed step-by-step instructions are provided in the Supplementary Material.
107 microbeads decorated with 106 molecules b-FAM-A
For Abil EM 90-based emulsions 1 mL mineral oil, 2% (v/v) Abil EM 90, 0.05% (v/v) Triton X-100 was used as oil-surfactant mixture. Oil-surfactant mixture was transferred to a conical 15 mL tube and vortexed, albeit at a low speed to prevent the introduction of air. The vortexer VV3 by VWR was used on the setting 2–3 of 6, which resulted in a 1200 RPM (as determined by high speed video footage) circular path motion with a 5 mm orbital diameter. The aqueous solution with dispersed microbeads was briefly sonicated for 5 s in a sonicator bath. The 100
For FluoSurf-based emulsions, the aqueous phase was treated the exact same way, but then gradually added (over the course of 10 s) to 600
The emulsification procedure can be viewed in Supplementary Video S3.
3.6 Microscopic analysis
Microscopic imaging of emulsions was conducted using a Leica Infinity TIRF microscope operated in epifluorescence and differential interference contrast (DIC) mode using Leica LAS AF software. An 8
3.7 Data analysis
Microscopic image analysis was conducted using FIJI software (ImageJ, version 1.54f). For Abil EM 90-based emulsions, high-magnification images were utilized, as accurate differentiation of microbeads and droplet volumes was not achievable from overview images. In contrast, FluoSurf-based emulsions were analyzed using lower-magnification overview images due to the larger droplet sizes, enabling the analysis of a larger number of droplets. Diameters of droplets harboring microbeads were measured using FIJI’s ruler tool and microbeads were counted. A total of N = 4 emulsions with a total of 360 microbead-containing droplets were analyzed for Abil EM 90-based emulsions, and a total of N = 3 emulsions with a total of 1.963 microbead-containing droplets were analyzed for FluoSurf-based emulsions. Furthermore, for FluoSurf-based emulsions the droplet volume distribution (including empty droplets) was determined based on 4.874 analyzed droplets from N = 3 emulsions.
Further data analysis was performed using Python (version 3.12.8). Numpy (version 2.1.3) and Pandas (version 2.2.3) were used for data storage and handling. Matplotlib (version 3.9.3) and Seaborn (version 0.13.2) were used to render Figures. Scipy (version 1.14.1) was used to fit Poisson distributions.
Droplet volumes were determined based on the measured droplet diameters and the liquid layer height between the microscopy slide and cover slip. Liquid layer heights for Abil EM 90-based and FluoSurf-based emulsions were measured by focussing on the top of the bottom glass slide (marked by scratches with a scalpel) and then focussing on the bottom of the top glass cover slip (also marked by scratches with a scalpel) and recording the traveled z-level distance. Liquid layer heights for Abil EM 90-based and FluoSurf-based emulsions were determined to be 8.4
Poisson distributions were fitted using the probability mass function (pmf) of the scipy.stats.poisson library which utilizes the following Equation 1:
with k being the number of microbeads contained in a droplet and
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
FH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. WH: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. FH received funding from the Hans und Isle Breuer-Stiftung.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI was used to further improve the english language used in the manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2025.1568027/full#supplementary-material
References
Abate, A. R., Chen, C. H., Agresti, J. J., and Weitz, D. A. (2009). Beating poisson encapsulation statistics using close-packed ordering. Lab a Chip 9, 2628. doi:10.1039/b909386a
Agresti, J. J., Antipov, E., Abate, A. R., Ahn, K., Rowat, A. C., Baret, J. C., et al. (2010). Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. 107, 4004–4009. doi:10.1073/pnas.0910781107
Ahn, K., Kerbage, C., Hunt, T. P., Westervelt, R. M., Link, D. R., and Weitz, D. A. (2006). Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl. Phys. Lett. 88, 024104. doi:10.1063/1.2164911
An, X., Zuo, P., and Ye, B. C. (2020). A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 209, 120571. doi:10.1016/j.talanta.2019.120571
Anyaduba, T. D., Otoo, J. A., and Schlappi, T. S. (2022). Picoliter droplet generation and dense bead-in-droplet encapsulation via microfluidic devices fabricated via 3D printed molds. Micromachines 13, 1946. doi:10.3390/mi13111946
Aralekallu, S., Boddula, R., and Singh, V. (2023). Development of glass-based microfluidic devices: a review on its fabrication and biologic applications. Mater. and Des. 225, 111517. doi:10.1016/j.matdes.2022.111517
Arnold, F. H., and Volkov, A. A. (1999). Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 3, 54–59. doi:10.1016/S1367-5931(99)80010-6
Arya, C., Saez Cabesas, C. A., Huang, H., and Raghavan, S. R. (2017). Clustering of cyclodextrin-functionalized microbeads by an amphiphilic biopolymer: real-time observation of structures resembling blood clots. ACS Appl. Mater. and Interfaces 9, 37238–37245. doi:10.1021/acsami.7b05435
Bharti, B., Meissner, J., and Findenegg, G. H. (2011). Aggregation of silica nanoparticles directed by adsorption of lysozyme. Langmuir 27, 9823–9833. doi:10.1021/la201898v
Bussiere, V., Vigne, A., Link, A., McGrath, J., Srivastav, A., Baret, J. C., et al. (2019). High-throughput triggered merging of surfactant-stabilized droplet pairs using traveling surface acoustic waves. Anal. Chem. 91, 13978–13985. doi:10.1021/acs.analchem.9b03521
Byrnes, S. A., Chang, T. C., Huynh, T., Astashkina, A., Weigl, B. H., and Nichols, K. P. (2018). Simple polydisperse droplet emulsion polymerase chain reaction with statistical volumetric correction compared with microfluidic droplet digital polymerase chain reaction. Anal. Chem. 90, 9374–9380. doi:10.1021/acs.analchem.8b01988
Castro, D., Conchouso, D., Arevalo, A., and Foulds, I. G. (2016). “A study of the incubation of microbead agglutination assays in a microfluidic system,” in 2016 IEEE 11th annual international conference on nano/micro engineered and molecular systems (NEMS), 354–357. doi:10.1109/NEMS.2016.7758266
C Hatch, A., S Fisher, J., R Tovar, A., T Hsieh, A., Lin, R., L Pentoney, S., et al. (2011). 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab a Chip 11, 3838–3845. doi:10.1039/C1LC20561G
Chen, L., Zhang, C., Yadav, V., Wong, A., Senapati, S., and Chang, H. C. (2023b). A home-made pipette droplet microfluidics rapid prototyping and training kit for digital PCR, microorganism/cell encapsulation and controlled microgel synthesis. Sci. Rep. 13, 184. doi:10.1038/s41598-023-27470-1
Chen, L., Zhao, Y., Li, J., Xiong, C., Xu, Y., Tang, C., et al. (2023a). Exceeding 80% efficiency of single-bead encapsulation in microdroplets through hydrogel coating-assisted close-packed ordering. Anal. Chem. 95, 8889–8897. doi:10.1021/acs.analchem.3c00425
Courtois, F., Olguin, L. F., Whyte, G., Bratton, D., Huck, W. T. S., Abell, C., et al. (2008). An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem 9, 439–446. doi:10.1002/cbic.200700536
Diamante, L., Gatti-Lafranconi, P., Schaerli, Y., and Hollfelder, F. (2013). In vitro affinity screening of protein and peptide binders by megavalent bead surface display. Protein Eng. Des. Sel. 26, 713–724. doi:10.1093/protein/gzt039
Diefenbach, X. W., Farasat, I., Guetschow, E. D., Welch, C. J., Kennedy, R. T., Sun, S., et al. (2018). Enabling biocatalysis by high-throughput protein engineering using droplet microfluidics coupled to mass spectrometry. ACS Omega 3, 1498–1508. doi:10.1021/acsomega.7b01973
Gan, R., Yamanaka, Y., Kojima, T., and Nakano, H. (2008). Microbeads display of proteins using emulsion PCR and cell-free protein synthesis. Biotechnol. Prog. 24, 1107–1114. doi:10.1002/btpr.43
Gantz, M., Neun, S., Medcalf, E. J., van Vliet, L. D., and Hollfelder, F. (2023). Ultrahigh-throughput enzyme engineering and discovery in in vitro compartments. Chem. Rev. 123, 5571–5611. doi:10.1021/acs.chemrev.2c00910
Goto, H., Kanai, Y., Yotsui, A., Shimokihara, S., Shitara, S., Oyobiki, R., et al. (2020). Microfluidic screening system based on boron-doped diamond electrodes and dielectrophoretic sorting for directed evolution of NAD(P)-Dependent oxidoreductases. Lab a Chip 20, 852–861. doi:10.1039/C9LC01263J
Griffiths, A. D., and Tawfik, D. S. (2003). Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 22, 24–35. doi:10.1093/emboj/cdg014
Griffiths, A. D., and Tawfik, D. S. (2006). Miniaturising the laboratory in emulsion droplets. Trends Biotechnol. 24, 395–402. doi:10.1016/j.tibtech.2006.06.009
Hasecke, F., Miti, T., Perez, C., Barton, J., Schölzel, D., Gremer, L., et al. (2018). Origin of metastable oligomers and their effects on amyloid fibril self-assembly. Chem. Sci. 9, 5937–5948. doi:10.1039/C8SC01479E
Holstein, J. M., Gylstorff, C., and Hollfelder, F. (2021). Cell-free directed evolution of a protease in microdroplets at ultrahigh throughput. ACS Synth. Biol. 10, 252–257. doi:10.1021/acssynbio.0c00538
Iizuka, R., Tahara, K., Matsueda, A., Tsuda, S., Yoon, D. H., Sekiguchi, T., et al. (2022). Selection of green fluorescent proteins by in vitro compartmentalization using microbead-display libraries. Biochem. Eng. J. 187, 108627. doi:10.1016/j.bej.2022.108627
Ito, K., Tayama, T., Uemura, S., and Iizuka, R. (2024). Isolation of novel fluorogenic RNA aptamers via in vitro compartmentalization using microbead-display libraries. Talanta 278, 126488. doi:10.1016/j.talanta.2024.126488
Jing, T., Ramji, R., Warkiani, M. E., Han, J., Lim, C. T., and Chen, C. H. (2015). Jetting microfluidics with size-sorting capability for single-cell protease detection. Biosens. and Bioelectron. 66, 19–23. doi:10.1016/j.bios.2014.11.001
Karamitros, C. S., Morvan, M., Vigne, A., Lim, J., Gruner, P., Beneyton, T., et al. (2020). Bacterial expression systems for enzymatic activity in droplet-based microfluidics. Anal. Chem. 92, 4908–4916. doi:10.1021/acs.analchem.9b04969
Leslie, D. C., Li, J., Strachan, B. C., Begley, M. R., Finkler, D., Bazydlo, L. A. L., et al. (2012). New detection modality for label-free quantification of DNA in biological samples via superparamagnetic bead aggregation. J. Am. Chem. Soc. 134, 5689–5696. doi:10.1021/ja300839n
Lindenburg, L., and Hollfelder, F. (2021). NAD-display: ultrahigh-throughput in vitro screening of NAD(H) dehydrogenases using bead display and flow cytometry. Angewandte Chemie Int. Ed. Engl. 60, 9015–9021. doi:10.1002/anie.202013486
Lindenburg, L., Huovinen, T., van de Wiel, K., Herger, M., Snaith, M. R., and Hollfelder, F. (2020). Split and mix assembly of DNA libraries for ultrahigh throughput on-bead screening of functional proteins. Nucleic Acids Res. 48, e63. doi:10.1093/nar/gkaa270
Link, A., McGrath, J. S., Zaimagaoglu, M., and Franke, T. (2022). Active single cell encapsulation using SAW overcoming the limitations of poisson distribution. Lab a Chip 22, 193–200. doi:10.1039/D1LC00880C
Link, D. R., Anna, S. L., Weitz, D. A., and Stone, H. A. (2004). Geometrically mediated breakup of drops in microfluidic devices. Phys. Rev. Lett. 92, 054503. doi:10.1103/PhysRevLett.92.054503
Link, D. R., Grasland-Mongrain, E., Duri, A., Sarrazin, F., Cheng, Z., Cristobal, G., et al. (2006). Electric control of droplets in microfluidic devices. Angew. Chem. Int. Ed. 45, 2556–2560. doi:10.1002/anie.200503540
Luo, X., and Lee, A. P. (2023). Overcoming double poisson limitation for co-encapsulation in droplets through hydrodynamic close packing of cells. Microfluid. Nanofluidics 27, 3. doi:10.1007/s10404-022-02600-9
Macao, B., Hoyer, W., Sandberg, A., Brorsson, A. C., Dobson, C. M., and Härd, T. (2008). Recombinant amyloid beta-peptide production by coexpression with an affibody ligand. BMC Biotechnol. 8, 82. doi:10.1186/1472-6750-8-82
Mankowska, S. A., Gatti-Lafranconi, P., Chodorge, M., Sridharan, S., Minter, R. R., and Hollfelder, F. (2016). A shorter route to antibody binders via quantitative in vitro bead-display screening and consensus analysis. Sci. Rep. 6, 36391. doi:10.1038/srep36391
Miller, O. J., Bernath, K., Agresti, J. J., Amitai, G., Kelly, B. T., Mastrobattista, E., et al. (2006). Directed evolution by in vitro compartmentalization. Nat. Methods 3, 561–570. doi:10.1038/nmeth897
Nam, J., Lim, H., Kim, C., Yoon Kang, J., and Shin, S. (2012). Density-dependent separation of encapsulated cells in a microfluidic channel by using a standing surface acoustic wave. Biomicrofluidics 6, 24120–2412010. doi:10.1063/1.4718719
Price, A. K., MacConnell, A. B., and Paegel, B. M. (2014). Microfluidic bead suspension hopper. Anal. Chem. 86, 5039–5044. doi:10.1021/ac500693r
Raj, M. K., and Chakraborty, S. (2020). PDMS microfluidics: a mini review. J. Appl. Polym. Sci. 137, 48958. doi:10.1002/app.48958
Siu, R. H. P., Liu, Y., Chan, K. H. Y., Ridzewski, C., Slaughter, L. S., and Wu, A. R. (2021). Optimization of on-bead emulsion polymerase chain reaction based on single particle analysis. Talanta 221, 121593. doi:10.1016/j.talanta.2020.121593
Sloane, H. S., Landers, J. P., and Kelly, K. A. (2016). Hybridization-induced aggregation technology for practical clinical testing. J. Mol. Diagnostics 18, 546–553. doi:10.1016/j.jmoldx.2016.02.004
Song, H., Tice, J. D., and Ismagilov, R. F. (2003). A microfluidic system for controlling reaction networks in time. Angew. Chem. Int. Ed. 42, 768–772. doi:10.1002/anie.200390203
Stucki, A., Vallapurackal, J., Ward, T. R., and Dittrich, P. S. (2021). Droplet microfluidics and directed evolution of enzymes: an intertwined journey. Angew. Chem. Int. Ed. 60, 24368–24387. doi:10.1002/anie.202016154
Takeuchi, R., Choi, M., and Stoddard, B. L. (2014). Redesign of extensive protein–DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl. Acad. Sci. 111, 4061–4066. doi:10.1073/pnas.1321030111
Tanaka, H., Yamamoto, S., Nakamura, A., Nakashoji, Y., Okura, N., Nakamoto, N., et al. (2015). Hands-off preparation of monodisperse emulsion droplets using a poly(dimethylsiloxane) microfluidic chip for droplet digital PCR. Anal. Chem. 87, 4134–4143. doi:10.1021/ac503169h
Tanno, H., McDaniel, J. R., Stevens, C. A., Voss, W. N., Li, J., Durrett, R., et al. (2020). A facile technology for the high-throughput sequencing of the paired VH:VL and TCRβ:TCRα repertoires. Sci. Adv. 6, eaay9093. doi:10.1126/sciadv.aay9093
Tawfik, D. S., and Griffiths, A. D. (1998). Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656. doi:10.1038/nbt0798-652
Tubeleviciute, A., and Skirgaila, R. (2010). Compartmentalized self-replication (CSR) selection of Thermococcus litoralis Sh1B DNA polymerase for diminished uracil binding. Protein Eng. Des. Sel. 23, 589–597. doi:10.1093/protein/gzq032
Turchaninova, M. A., Britanova, O. V., Bolotin, D. A., Shugay, M., Putintseva, E. V., Staroverov, D. B., et al. (2013). Pairing of T-cell receptor chains via emulsion PCR. Eur. J. Immunol. 43, 2507–2515. doi:10.1002/eji.201343453
Valignat, M. P., Theodoly, O., Crocker, J. C., Russel, W. B., and Chaikin, P. M. (2005). Reversible self-assembly and directed assembly of DNA-Linked micrometer-sized colloids. Proc. Natl. Acad. Sci. 102, 4225–4229. doi:10.1073/pnas.0500507102
Wang, J., Gong, Q., Maheshwari, N., Eisenstein, M., Arcila, M. L., Kosik, K. S., et al. (2014). Particle display: a quantitative screening method for generating high-affinity aptamers. Angew. Chem. Int. Ed. 53, 4796–4801. doi:10.1002/anie.201309334
Wang, Y., Xue, P., Cao, M., Yu, T., Lane, S. T., and Zhao, H. (2021). Directed evolution: methodologies and applications. Chem. Rev. 121, 12384–12444. doi:10.1021/acs.chemrev.1c00260
Yue, X., Fang, X., Sun, T., Yi, J., Kuang, X., Guo, Q., et al. (2022). Breaking through the poisson distribution: a compact high-efficiency droplet microfluidic system for single-bead encapsulation and digital immunoassay detection. Biosens. Bioelectron. 211, 114384. doi:10.1016/j.bios.2022.114384
Zeng, Y., Woolley, M., Chockalingam, K., Thomas, B., Arora, S., Hook, M., et al. (2023). Click display: a rapid and efficient in vitro protein display method for directed evolution. Nucleic Acids Res. 51, e89. doi:10.1093/nar/gkad643
Zhang, F., Liao, P., Sun, Y., Chen, Z., Pang, Y., and Huang, Y. (2020). Surfactant and oil formulations for monodisperse droplet emulsion PCR. Lab a Chip 20, 2328–2333. doi:10.1039/D0LC00052C
Zhu, B., Du, Z., Dai, Y., Kitaguchi, T., Behrens, S., and Seelig, B. (2023). Nanodroplet-based reagent delivery into water-in-fluorinated-oil droplets. Biosensors 13, 768. doi:10.3390/bios13080768
Keywords: microbeads, in vitro compartmentalization, emulsion, encapsulation, aggregation, clustering, poisson distribution
Citation: Hasecke F and Hoyer W (2025) A sticky situation – simple method for rapid poissonian encapsulation of highly aggregation-prone microbeads in polydisperse emulsions. Front. Bioeng. Biotechnol. 13:1568027. doi: 10.3389/fbioe.2025.1568027
Received: 28 January 2025; Accepted: 16 June 2025;
Published: 30 June 2025.
Edited by:
Steven R. Bischoff, NovoHelix, United StatesCopyright © 2025 Hasecke and Hoyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Filip Hasecke, ZmlsaXAuaGFzZWNrZUBoaHUuZGU=
 Filip Hasecke
Filip Hasecke Wolfgang Hoyer
Wolfgang Hoyer