- 1Department of Mycology and Plant Pathology, Institute of Agriculture Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 2Department of Kayachikitsa, Faculty Ayurveda, Institute of Medical sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 3Bihar Agricultural University, Bhagalpur, Bihar, India
- 4ICAR-National Bureau of Agriculturally Important Microorganisms, Mau, Uttar Pradesh, India
Nanotechnology has significantly advanced the detection of plant diseases by introducing nano-inspired biosensors that offer distinct advantages over traditional diagnostic methods. These biosensors, enhanced with novel nanomaterials, exhibit increased sensitivity, catalytic activity, and faster response times, resulting in improved diagnostic efficiency. The increasing impact of climate-induced stress and emerging plant pathogens have created an urgent demand for real-time monitoring systems in agriculture. Nanobiosensors are revolutionizing plant disease management by enabling on-site detection of pests and weeds, facilitating precise pesticide applications. This article provides a comprehensive overview of the development and application of nanobiosensors in real-time plant disease diagnosis. It highlights key innovations, such as smartphone-integrated nanozyme biosensing and lab-on-a-chip technologies. Special emphasis is placed on the detection of molecular biomarkers, demonstrating the critical role of nanobiosensors in addressing the evolving challenges of plant disease management and agricultural sustainability.
1 Introduction
A biosensor is a tool used to detect biomarkers with sensitivity and selectivity, providing benefits compared to traditional diagnostic methods. Detecting a multitude of diseases with biosensors requires exceptionally precise disease-associated biomarkers, a minimally invasive or non-invasive approach, and meticulous checks to differentiate among markers linked to various health conditions. The importance of biosensors in disease detection lies in their capacity for promptly identifying disease onset, monitoring overall health, and facilitating rapid interventions for affected individuals (Mahapatra and Chandra, 2020). Fluorescence-based nanobiosensors have emerged due to recent progress, serving various medical purposes. There is research focused on employing biosensors to detect conditions such as cardiovascular diseases, cancer, and diabetes (Gouvea, 2011). Nanotechnology has been instrumental in introducing a groundbreaking type of biosensor known as the nanobiosensor. These biosensors have proven highly effective in numerous modern research fields, including environmental studies (Mahmoudpour et al., 2019), cell physiology (Shi et al., 2013), clinical detection (Mahapatra and Chandra, 2020; Mahato et al., 2018; Shetti et al., 2020), and to examine the space consequences on astronauts (Roda et al., 2018; Roda et al., 2020). Biosensors consist of three key components: (a) a biorecognition element (BRE), (b) a transducer, and (c) an amplifier and processor. A nanobiosensor is a small-scale apparatus that employs magnetic, optical, or electronic methodologies within a tiny sensor to analyze biological or biochemical occurrences (Shi et al., 2013; Roda et al., 2018; Roda et al., 2020; Di Giusto et al., 2005).
Nanobiosensors are the result of interdisciplinary research, drawing from fields like nanotechnology, biology, chemistry, and medical science (Giraldo et al., 2019; Roda et al., 2020). In most cases, nanobiosensors entail linking a biological recognition element onto a signal transducer’s surface. This initiates a heterogeneous reaction between the biorecognition element and the analyte, highlighting the pivotal importance of biosensing interface design in nanobiosensor advancement. The current progress in nanobiotechnology and advanced electronics fabrication technology has converged, giving rise to a novel category of biosensors known as nanobiosensors. These progressions represent a modern phase in nanobiotechnology, notably in the realm of diagnosing plant illnesses. Figure 1 demonstrates the utilization of nanobiosensors for detecting plant pathogens. The utilization of functionalized (bio) sensors, operating through diverse transduction processes and integrated with nanomaterial-based structures, holds the potential to establish distinct associations with a wide range of substances linked to plant diseases. These devices based on nanomaterials seem to offer promising alternatives to the conventional, more extensive methods of pathogen detection. Despite several recent articles discussing the application of nanotechnology in smart plant sensing, there remains a substantial amount of work to be undertaken in this field.
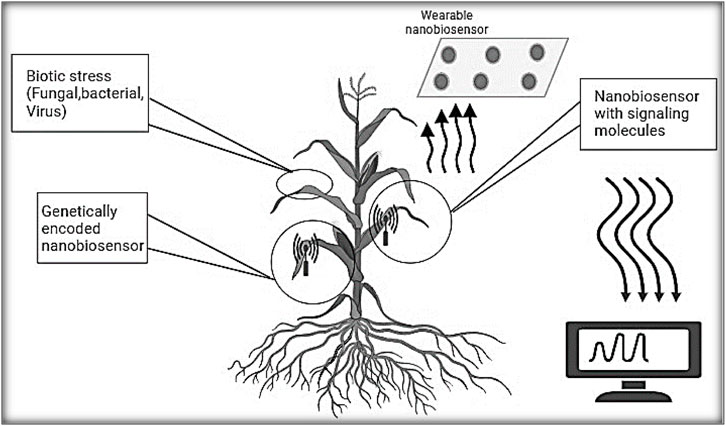
Figure 1. Nanomaterial-based sensors for detection plant diseases by monitoring bacterial, fungal and viral plant pathogens.
It's crucial to detect plant pathogens prior to the onset of disease symptoms in order to proactively monitor plant wellbeing and devise an informed disease management strategy. It is crucial to differentiate between causative species because many fungal infections elicit similar changes in plants as the disease progresses. Various direct and indirect methods are employed in the detection and diagnosis of plant diseases (Feng et al., 2015; Martinelli et al., 2015). Direct methods for examining plant pathogens and biomolecular markers, like nucleic acids, proteins, and carbohydrates, involve analyzing infected plant tissues. On the other hand, indirect methods detect plant diseases by observing changes in parameters such as emissions of volatile organic compounds, as well as alterations in physiological or histological characteristics like leaf surface temperature or humidity, spectroscopic attributes of plant tissues, morphology, and growth rate (Li et al., 2020). Both direct and indirect detection methods encompass a broad spectrum of technologies, including spectroscopic, electrochemical (Valekunja et al., 2016), and molecular approaches (Verosloff et al., 2019).
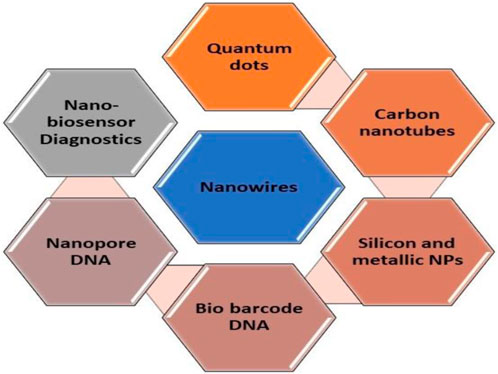
Recent endeavors have primarily focused on advancing early pathogen detection technologies to enhance sensitivity, precision, and detection speed. These efforts encompass three categories of molecular assays: enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR) testing, and loop-mediated isothermal amplification (LAMP) assay, all of which rely on protein-based or nucleic acid technologies. ELISA is recognized as a highly developed serology-based diagnostic method for fungal pathogens, allowing for pathogen identification via a colorimetric reaction visible without the aid of magnification. While the classic ELISA method has become the established approach for diagnosing various pathogens across different domains such as environmental, chemical, biotechnological, health, and agricultural analyses, it continues to exhibit certain limitations due to its limited sensitivity and accuracy. Despite improvements in targeting specific pathogens with increased accuracy and precision, these commonly employed techniques still present some drawbacks. Among these challenges are extended diagnostic timelines, complicated sample preparation steps, the necessity of transporting samples from field sites to specialized labs, and a dependence on skilled professionals (Dyussembayev et al., 2021). Recent initiatives have concentrated on merging DNA and immunological techniques while incorporating various nanomaterials like silica, metallic nanoparticles, nanowires, carbon tubes, quantum dots (QDs), bio barcode DNA, and other nanomaterials (Cardoso et al., 2021). This integration has led to the development of systems like nanoparticle-based biosensors (Figure 2), which facilitate the visual detection of disease-causing agents with economic significance (Zhan et al., 2018). Some of the primary nanoscale instruments utilized in agricultural diagnostics include microneedle patches, nanopore sequencing platforms, plant wearables, and nanoparticle or array-based sensors, which can be employed in both direct and indirect methods.
2 Detection of plant diseases using portable sensors
A multitude of sensors, crafted with versatility in mind for applications including environmental tracking and medical analysis, have been developed and made available for commercial use. These sensors can detect analytes through a variety of signal modes, including electrical, chemical, electrochemical, optical, magnetic, or vibrational, depending on the sensor’s operating mode. Incorporating nanomaterial matrices as transducers can enhance the detection limit, while the use of bio-recognition elements like DNA, antibodies, and enzymes can enhance accuracy.
2.1 Biosensor platforms based on nanomaterials
2.1.1 Quantum dots
Quantum dots, also known as QDs, are semiconductor nanocrystals distinguished by their distinctive photophysical characteristics, granting them remarkable potential as optical nanoprobes (Gao et al., 2018).
Quantum dots have shown effectiveness as biosensors in imaging plants and detecting diseases (Wang et al., 2020). The first recorded instance of single-celled yeast generating cadmium sulfide (CdS) crystals when exposed to cadmium salt stress occurred within the framework of semiconductor nanomaterial synthesis (Dameron et al., 1989). Their miniature dimensions (1–10 nm) enable swift uptake and transportation by plants, facilitating easy detection and tracking of their fluorescent signals within biological systems. Recently, there have been reports indicating that fungal hyphae can readily assimilate CdSe–ZnS core–shell quantum dots coated with 3-mercaptopropionic acid (Rispail et al., 2014). These nanomaterials are valuable in imaging bacteria and fungi because they have low cytotoxicity and excellent biocompatibility (Kasibabu et al., 2015). The capacity of a paper sensor based on CDs-Tb to distinguish ppGpp from structurally similar nucleotides has been demonstrated. This fluorescent Tb (III)-CD paper-based sensor can detect 3′-5′-diphosphate-5′-diphosphate, even in plants or microorganisms subjected to adverse environmental conditions (Chen et al., 2018). In the creation of such QD sensors, fluorescence resonance energy transfer (FRET) is utilized in conjunction with intrinsic fluorescence amplification or quenching. In FRET sensors, QDs act as donors, and gold NPs (Shojaei et al., 2016), organic dyes, and carbon nano dots (Shojaei et al., 2016) function as acceptors. The outcome of this energy transfer process is a decrease in the emanation of fluorescence. A recent example is the FRET-based complex sensor designed for the detection of Citrus tristeza virus. Cadmium telluride (CdTe) QDs combined with CTV coat protein (CP) and CP-labeled rhodamine dye are often used as donor–acceptor pairs for this purpose. When targeted viruses are present, CP-rhodamine is replaced by free CP, leading to the restoration of QD fluorescence (Li et al., 2020). Several types of plant viruses, such as tomato ringspot virus, bean pod mottle virus, and Arabis mosaic virus, have been identified utilizing diverse methodologies. Examples include the utilization of Fe3O4/SiO2 magnetic nanoparticles, SiO2/up-conversion nanoparticles at the interface, and labeled antibodies, achieving a limit of detection (LOD) of 100 ng mL−1. In a recent study, a nanobiosensor based on quantum dots (QDs) was highly sensitive in detecting Candidatus Phytoplasma aurantifolia in damaged lime plants.
Additionally, a rapid diagnostic biosensor, utilizing CdTe QDs encapsulated with specific antibodies against the Polymyxa betae-specific glutathione S-transferase protein, was employed for efficient evaluation of plant samples, providing accurate results within 30 min (Safarpour et al., 2012). A new optical DNA biosensor utilizing quantum dots (QDs) and employing fluorescence resonance energy transfer (FRET) has been devised for discerning specific DNA sequences in Ganoderma boninense (Bakhori et al., 2013). This advancement enables the identification of analogous artificial DNA sequences of the G. boninense gene through the examination of FRET signals (Kashyap et al., 2019). The biosensor demonstrated an impressive limit of detection at 3.55 × 10−9 M, showcasing exceptional sensitivity and providing a straightforward, swift, and highly responsive approach for detecting plant diseases.
Moreover, CdS quantum dots have been biosynthesized by various microorganisms, although limited research has focused on their luminescent properties (Khiyami et al., 2014). Fungus Fusarium oxysporum, when treated with a combination of CdCl2 and SeCl4 at room temperature, produced highly luminescent CdSe QDs. Utilizing surface-modified quantum dots (QDs) as agricultural chemicals offers a potential avenue for managing plant diseases (Li et al., 2020). In another approach, the natural antibiotic Kasugamycin (KAS) was attached to the surface of ZnO QDs (Liang et al., 2019). The produced KAS-ZnO QDs exhibited exceptional pH-responsive properties and increased photostability. They were used in a greenhouse experiment to deliver KAS and Zn (II) species controllably, resulting in a significant reduction in bacterial fruit blotch incidence (Li et al., 2020).
Image analysis and a genetic algorithm using an Arduino program to distinguish between healthy and diseased plant leaves, such as those of pepper plants, potatoes, and tomatoes affected by late blight and leaf spot (Arya et al., 2018).
It’s worth noting that many quantum dots are composed of potentially toxic heavy metals like Pb, Zn, and Hg. Therefore, the potential hazards associated with their use should not be overlooked in field applications where diagnostic effectiveness is a primary concern (Pramanik et al., 2018; Tsoi et al., 2013).
2.1.2 Sequencing platform based on nanopores
Nanopore sequencing utilizes a motor protein to guide single-stranded DNA or RNA through a nanopore, which can be either protein-based or solid-state. As the nucleotides pass through the nanopore, they generate distinct electronic signals, enabling real-time sequence identification. This method, referred to as third-generation sequencing (TGS), streamlines the examination of disease-causing genetic material (Eisenstein, 2017). In recent studies, there has been significant dependence on nanopore sequencing methodologies to aid in the identification of plant pathogens (Li et al., 2020). Many investigations have utilized nanopore sequencing platforms to identify a range of plant pathogens, including bacteria, viruses, fungi, and phytoplasma, such as Penicillium digitatum affecting lemons. These inquiries make use of a portable sequencing tool created by Oxford Nanopore Technologies, named MinION. The entire process can be finalized in under 2 h, with outcomes comparable to those obtained through conventional diagnostic techniques like PCR and ELISA (Chalupowicz et al., 2019). Through the integration of nanopore sequencing with comprehensive transcriptome examination, scientists have successfully identified two viral strains, namely, Candidatus Liberibacter asiaticus and Plum Poxvirus, in peaches within a span of 24 h (Badial et al., 2018). The exceptional genome mapping capacity of MinION has facilitated the anticipation of several plant viral types within a water yam plant, encompassing Dioscorea bacilliform virus, Yam mild mosaic virus, and Yam chlorotic necrosis virus (Filloux et al., 2018).
2.1.3 Nanowire as biosensor transducer
In the twenty-first century, nanotechnology fabrication techniques have led to the creation of nanowires equipped with highly specialized, small sensors and an exceptionally miniaturized design (Ariffin et al., 2014). Surface modification of these nanowires involves treating them with an amino group solution and applying enzymes to their surfaces. The modified nanowires exhibit a high surface-area-to-volume ratio, allowing them to capture and bind to biomolecules associated with plant pathogens effectively. This specific interaction triggers measurable changes in electrical, optical, or electrochemical signals, making it possible to detect plant diseases at an early stage. Notably, these biosensors have demonstrated high efficacy in detecting viral infections such as cucumber mosaic virus (CMV) and papaya ring spot virus, both of which significantly impact crop yields. The integration of nanowire-based biosensors into portable diagnostic platforms has also expanded their applicability in real-time, on-site pathogen detection, thereby revolutionizing plant disease monitoring in precision agriculture (Ariffin et al., 2014).
2.1.4 Metallic nanoparticles-based detection
Metal nanoparticles, including Au, Ag, ZnS, PbS, and CdS nanoparticles, have gained a unique position in the field of biosensing due to their high surface-to-volume ratio, straightforward manufacturing processes, and adaptability for various surface functionalization. This has resulted in enhanced specificity and sensitivity, along with faster and more convenient detection methods such as color changes and electrochemical variations, proving to be a feasible alternative to traditional enzyme tags (Thaxton et al., 2006). For example, surface plasmon resonance was utilized to detect Karnal bunt disease in wheat (Tilletia indica) using a nanogold-based immunosensor (Singh et al., 2010). Another method involved a specific oligonucleotide labeled with fluorescein and a 2 nm gold nanoparticle at its ends, acting as a quencher, to create a nanobioreceptor for detecting phytoplasma linked to flavescence dorée in grapevines (Firrao et al., 2005). Researchers were also able to identify the pathogenic fungus Sclerotinia sclerotiorum by assessing salicylic acid levels in oilseeds using an electrochemical sensor based on a gold electrode containing copper nanoparticles (Wang et al., 2010). In a study focusing on Phytophthora species detection, a helicase-dependent isothermal amplification method combined with on-chip hybridization and subsequent deposition of silver nanoparticles (AgNPs) was utilized, enabling both visual and electrical output (facilitating ocular and electrical output). Additionally, colorimetric detection methods have been employed in the identification of Pseudomonas syringae pathovars with the assistance of AuNP-bound probes (Vaseghi et al., 2013). The colorimetric change allowed for the detection of 24–26 bacterial isolates, highlighting the suitability of the assay for the early detection of P. syringae with a sensitivity of 15 ng μL−1 of genomic DNA (Konwarh and Sharma, 2020). In a study conducted by Yüksel et al. (2015), it was demonstrated that fluorescent dyes, radioactivity, and enzyme-induced color shifts could be utilized to monitor DNA hybridization. In this approach, two distinct capture probes were attached to specific locations on an EGNP array, one for Phytophthora ramorum and the other for Phytophthora lateralis. The target DNA was hybridized with immobilized capture probes containing 2-aminopurine (2-AP) in place of adenine on the EGNP array (Yüksel et al., 2015).
2.1.5 Nanofabrication imaging
Nanotechnology holds great promise in addressing challenges related to toxicity, effective imaging duration, tissue selectivity, and signal intensity by allowing precise control and customization of the chemical and physical characteristics of contrast materials. Mesoscopic nanoparticles, typically ranging from 5 to 100 nm in diameter, provide extensive surface areas, making them suitable for attaching functional groups in various pathogen detection tests (Nie, 2013).
Scientists have utilized electron beam and photolithography methods to fabricate surface patterns resembling the features of plant leaves and their internal structures. Furthermore, nano-imaging tools have been deployed to examine how pathogens infiltrate and colonize leaf tissues (Mccandless, 2005), for instance, lithography was used on silicon wafers to fabricate a pillared surface. By observing the movement of Colletotrichum graminicola over this substrate, which mimicked some host plant properties, researchers identified that the fungus required slight contact (at least 4.5 μm) before forming appressoria as part of ts infection mechanism.
Nanofabrication technologies have also been employed to examine the infection mechanism and behavior of Xylella fastidiosa, the causal agent of Pierce’s disease in grapevine xylem. This research aims to develop disease-resistant grapevine cultivars (Meng et al., 2005). In a connected investigation, Szeghalmi and co-workers (2007) explored nanostructured platforms for surface-enhanced Raman scattering (SERS), aiming for precise detection with a spatial resolution of 1 micron. Their study involved employing SERS imaging to examine dried fungal hyphae cultivated on commercially accessible nanostructured gold-coated surfaces. This illustrated that the nanofabrication methods offer standardized and replicable substrates for conducting imaging studies of phytopathogen interactions, whether in situ or in vivo.
3 Plant disease detection utilizing optical sensors equipped with nanomaterials
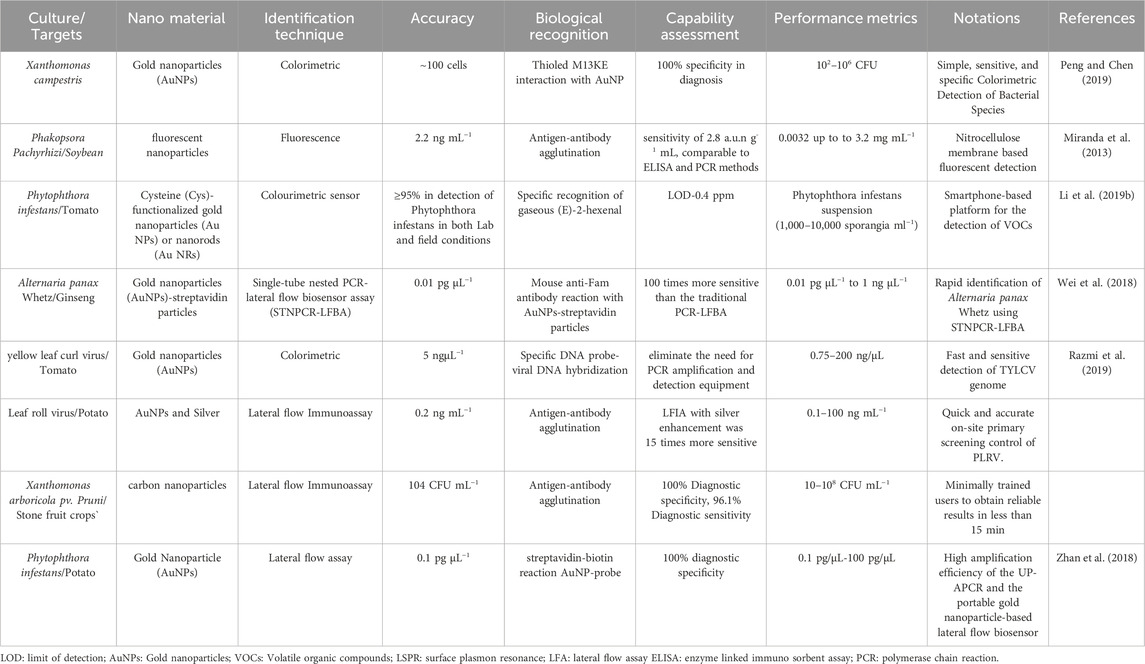
Optical sensors present numerous benefits compared to other detection techniques, such as swift identification, simplicity in operation, and comparatively affordable expenses. These sensors are devices designed to perceive and gauge alterations in the optical properties of a substance, translating them into measurable electrical signals.Common optical sensor approaches include colorimetry, fluorescence, surface plasmon resonance (SPR), flow cytometry, lateral flow assays (LFA), chemiluminescence, and bioluminescence (Yan et al., 2018). There are different types of optical sensors and nanomaterials used for the detection of plant diseases (Table 1) M13 bacteriophages were modified to exhibit a receptor-binding protein naturally inclined towards bacteria. These phages were able to attach to gold nanoparticles (AuNPs), amplifying signals and causing a visible change in color. A color-based sensor was utilized to identify pathogens, while UV-visible spectroscopy was employed for precise measurement. This method achieved a detection limit of 102 CFU mL−1, with a linear detection range spanning from 102 to 106 CFU mL−1. The primary optical techniques for detecting plant pathogen DNA typically involve color-based tests utilizing gold nanoparticles (AuNPs), such as lateral flow assays and aggregation assays. Furthermore, fluorescent and color-based microarrays, along with electrochemiluminescence analysis, are frequently employed for this objective. Leveraging lateral flow immunoassays (LFIAs) has presented exciting prospects for enhancing signals, including the integration of nanomaterials like nanoparticles and graphene, alongside straightforward adjustments to the platform structure, such as adopting a vertical flow configuration (Parolo et al., 2013a; Parolo et al., 2013b; Rivas et al., 2014; Morales-Narváez et al., 2015; Nunes-Pauli et al., 2015). LFIAs, in a sandwich format, have been utilized to detect various plant viruses such as Citrus tristeza virus (CTV) and Potato virus X (PVX), as well as plant-pathogenic bacteria like Erwinia amylovora, Xanthomonas, and Pantoea stewartii, employing AuNPs tags. For instance, in identifying Citrus tristeza virus (CTV) in citrus leaves and fruits, Salomone and colleagues (2004) formulated an LFIA using a conventional antibody-based sandwich setup with AuNPs as a marker. Qualitative findings exhibited sensitivity akin to that of an ELISA test, displaying strong correlation. The assay’s specificity also proved satisfactory, resulting in a 5% false positive rate. The inaugural LFIA for detecting a phytopathogen, specifically Tobacco mosaic virus (TMV), was documented by Tsuda (1992). Subsequently, a similar approach was developed for Potato virus X (PVX) detection (Drygin et al., 2012). The discernment of these assays was assessed against major potato seed viruses, including Potato virus Y (PVY), Potato virus M (PVM), and Potato virus A, with sensitivities reaching as low as 2 ng mL−1 for PVA (Feng et al., 2015). Employed an LFIA for swift detection of P. stewartii in corn seed samples. The LFIA was tested against three others phytopathogenic bacteria (Burkholderia glumae, Xanthomonas oryzae, and Pseudomonas syringae), and none of them showed cross-reactivity, demonstrating high selectivity. The detection limit for PSS in the test was 105 colony-forming units per milliliter (cfu mL−1) (Razo et al., 2019). Razo and co-workers in 2019 proposed enlarging gold nanoparticles (GNPs) in order to establish a sensitive lateral flow immunoassay for R. solanacearum detection, they developed lateral flow test strips containing gold nanoparticles (17.4 ± 1.0 nm) as a tag and antibodies specific to Ralstonia solanacearum conjugated to them. Tetrachloroauric (III) anion reductions on the GNP surface resulted in a signal augmentation within test zone of the test strips, and gold nanoparticles size on the test strips increased by about 100 nm, as verified by scanning electron microscopy. The method lowers R. solanacearum detection limit by 33-fold to 3 × 104 cells mL–1. Entire process, including sample prep and gold enlargement, completes in just 15 min. A novel method combines UP-APCR with AuNP-based lateral flow biosensor for direct Phytophthora infestans assessment (Zhan et al., 2018). This method offers quick, visible detection of P. infestans with high sensitivity and accuracy, achieving a low detection limit of 0.1 pg μL−1 within 1.5 h. It effectively identifies the pathogen in affected plant samples, showing great potential for field application due to its speed, simplicity, and reliability.Various studies in the literature have explored surface plasmon resonance (SPR) biosensors, including those using DNA probes, antibodies, and aptamers, for monitoring plant infections (Wang et al., 2004; Candresse et al., 2007; Lautner et al., 2010). Lin et al. (2014) developed a label-free SPR immunosensor using gold nanorods (AuNRs) to detect Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) in orchids. They coated AuNRs with antibodies against orchid viruses to enhance sensitivity and reduce color interference, achieving detection limits of 48 pg mL−1 for CymMV and 42 pg mL−1 for ORSV. Specificity was confirmed by examining target and non-target viral antigens, and performance was evaluated by detecting signal changes due to antigen-antibody binding on the AuNRs’ surface.A new surface plasmon resonance-based immunosensor was developed for detecting Pseudocerocospora fijiensis fungus in leaf extract samples. This sensor used a polyclonal antibody against P. fijiensis bacterial cell protein HF1 immobilized on a gold chip in a lateral flow assay. It showed a linear response for HF1 from 39.1 to 122 μg mL−1, with a detection limit of 11.7 μg mL−1 and a sensitivity of 0.0021 units of reflectance per ng ml−1 (Luna-Moreno et al., 2019). In various plant diseases, lateral flow (LF) test strips for DNA detection have been developed, often using gold nanoparticle (AuNP)-labeled DNA probes. Zhao et al. (2014) employed a competitive DNA hybridization method to detect Acidovorax avenae bacterial disease in melons, achieving a low detection limit of 0.48 nM. Selectivity testing against five different plant bacterial diseases showed no cross-reaction. Wei et al. (2018) used DNA hybridization on a lateral flow platform for rapid detection of Banana bunchy top virus (BBTV) using AuNPs-DNA probes, achieving a low detection limit of 0.13 nM. The BBTV-DNA lateral flow biosensor demonstrated ten times higher sensitivity than electrophoresis and showed selectivity against other viruses like Banana streak virus (BSV) and Cucumber mosaic virus (CMV).
3.1 Electrical and electrochemical
Electrochemical and electrical approaches for plant disease detection have gained popularity, offering advantages such as straightforward procedures, sensitivity, selectivity for specific infections, and the potential for portable commercial equipment for in situ measurements (Khaled et al., 2018; Ray et al., 2017; Feng et al., 2015). These methods have been employed in various environments, including greenhouses, fields, and postharvest storage containers, allowing for the detection of infections in the air, water, and on seeds. Most of these technologies rely on identifying pathogens through the use of biosensors (Martinelli et al., 2015). Functionalized electrode interaction with the analyte causes electron transfer, enabling detection and quantification via various electrochemical assays such as amperometric, voltammetric, potentiometric, and impedimetric methods (Cesewski and Johnson, 2020; Fang, 2020), for example, Freitas and colleagues utilized gold nanoparticles (AuNPs), magnetic beads, anti-CP-CTV antibodies, and the horseradish peroxidase (HRP) enzyme to detect the capsid protein from the Citrus tristeza virus (CP-CTV). To diagnose citrus canker, Haji-Hashemi and their team designed an electrochemical immunosensor for the detection of the PthA protein. They immobilized anti-PthA antibodies on AuNPs (GNP), and the antigen was detected using the fast Fourier transform square wave voltammetry (FFT-SWV) method. The FFT-SWV peak currents decreased as the PthA concentration increased due to the formation of antigen-antibody complexes. The sensor demonstrated a linear relationship between the current response and the logarithm of PthA concentration in the range of 0.03–100 nM, with a limit of detection (LOD) of 0.01 nM. The immunosensor exhibited repeatability (a relative standard deviation of 3.9%), selectivity (as determined by studies of healthy plant sap, BSA, and myoglobin), and stability (97 percent of the original response after 7 days). It was also tested with artificially affected healthy plant sap samples. The electrochemical biosensor’s results were in agreement with those obtained using the PCR approach, suggesting its potential for early detection of citrus canker disease. Utilizing differential pulse voltammetry, Fang and co-workers detected p-ethylguaiacol using SnO2 and TiO2 nanoparticles on screen-printed carbon electrodes (SPCE) (DPV). The analyte is a volatile generated by Phytophthora cactorum fungus-infected fruits and plants. Two very different sensors had a low LOD: 35 nmol L−1 for the TiO2 electrodes and 62 nmol L−1 for the SnO2 electrodes, accordingly. An interference analysis with six chemicals revealed an increased response variation of 6.7 percent, indicating the sensors’ strong selectivity. By simulating the makeup of a genuine fruit volatile signature, the determination of p-ethylguaiacol in real infected samples was tested. Groundnut bud necrosis tospo virus (GBNV) is a disease involved for viral epidemics that needs early identification and regular monitoring to minimize rapid vector transmission. Chaudhary and co-workers employed a graphene oxide (GO) based electrochemical immunosensor to detect it (Chaudhary et al., 2021). The ITO substrates were coated with GO to enhance electrical conductivity via sp2 carbon domains, enabling modification of the sensor with anti-GBNV antibodies.The GBNV nucleocapsid (GBNV-N) protein was detected using the DPV approach in the range of 0.5–150 ng mL−1 with a LOD of 5.7 0.7 ng mL−1. The sensor’s usage was investigated, and it was discovered that throughout three and seven cycles, activity reduced by less than 3% and 10%, respectively. Some other method for detecting plant diseases is using an electronic nose (e-nose) (Cui, et al., 2018; Cellini et al., 2017). Different types of electrical, electrochemical techniques., and nanomaterials used for the detection of the plant disease are presented in Table 2.
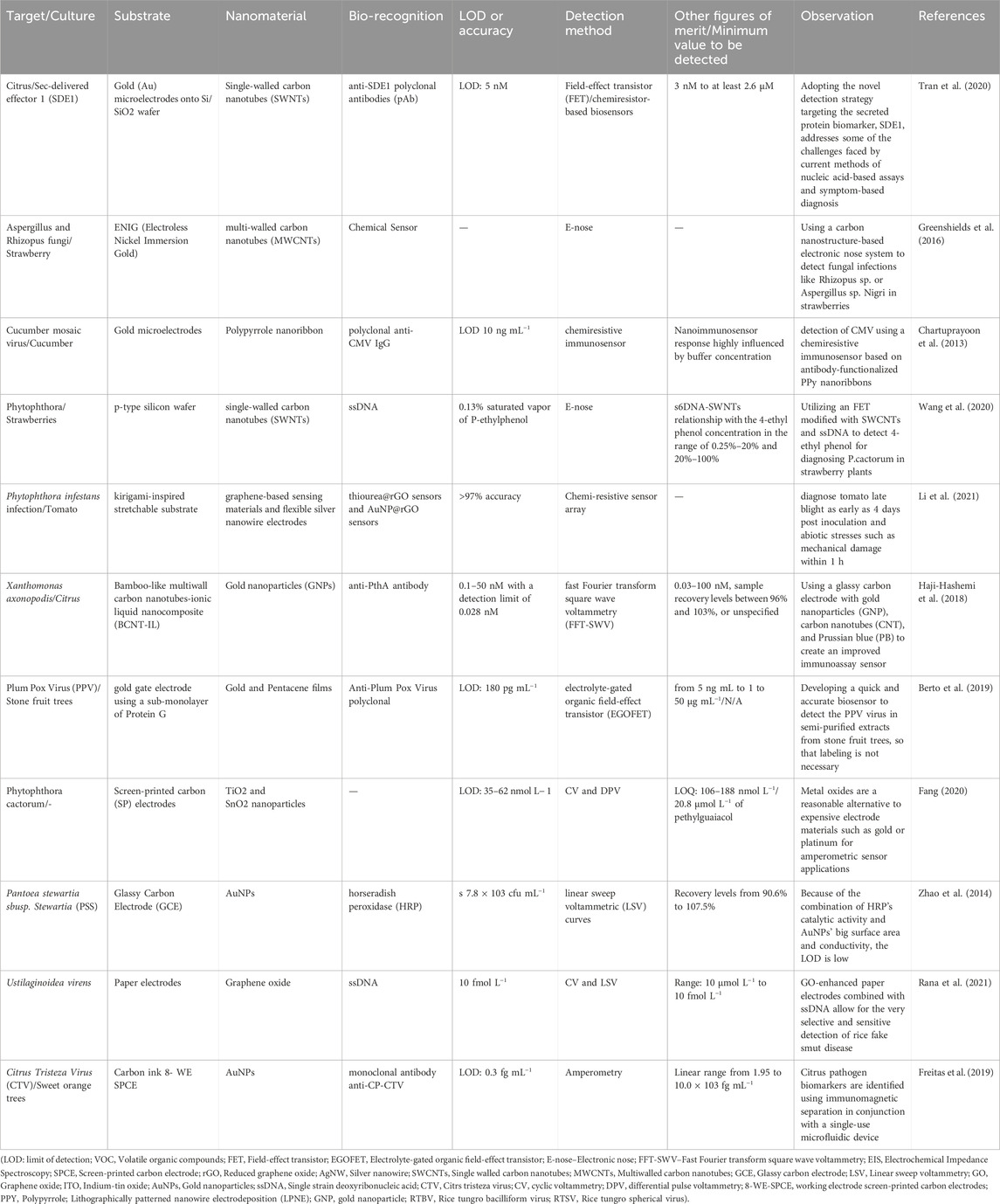
Table 2. Enhanced diagnosis of plant diseases: Combining electrical, electrochemical methods, and nanomaterials.
4 Detection of mycotoxins by nanobased sensor
The immune-electrode was effectively employed for the detection of AFB1 within the range of 10–100 ng dL−1, offering a sensitivity of 0.45 μAng−2, with a detection limit as low as 17.90 ng dL−1, and a rapid response time of 60 s. The primary purpose of utilizing such nanostructures is to expedite the identification of pathogens. Various nanomaterials, including carbon nanotubes, graphene, nanowires, nanocomposites, nanostructured metal oxides, and nanoparticles, are increasingly being utilized in the detection of pathogens and mycotoxins. Microfluidic systems, which can also be employed for real-time infection diagnosis with high sensitivity, represent another type of nanostructure platform (Baeummer, 2004). These systems offer the significant advantage of detecting specific target substances in small sample volumes and within a short time frame. For the detection of Aspergillus ochraceus (OTA), researchers successfully co-immobilized r-IgGs and BSA using a nanoSiO2 and chitosan-based nano-biocomposite material on an ITO substrate. Their findings indicated that the BSA/r-IgGs/CH-NanoSiO2/ITO immune-electrode exhibited optimized sensing properties for OTA recognition. In another approach, horseradish peroxidase (HRP) biosensors were employed to evaluate OTA and Penicillium viricatum in spiked beer and roasted coffee samples without the need for pretreatment, as described in the study conducted by Alonso-Lomillo and colleagues in 2010.And P. viricatum in spiked beer and roasted coffee samples with no pretreatment (Alonso-Lomilloa et al., 2010). It was improved an electrochemical immunosensor by using magnetic nanoparticles to determine ultra-trace amounts of AFM1 (up to 0.01 ppb) generated by A. flavus with foodstuffs (Paniel et al., 2010). A ‘lab-on-chip’ technique for the rapid, sensitive, and selective detection of zearalenone generated by Fusarium sp. by combining an electrokinetic magnetic bead-based electrochemical immunoassay on a microfluidic chip. For the quick and sensitive measurement of zearalenone in corn silage samples, researchers developed an immunosensor that used multi-wall carbon nanotubes and a continuous-flow technology. Ansari and co-workers (2010) showed that a sol–gel Nano-ZnO film can be used to immobilize r-IgGs and BSA can be utilized to prevent irrelevant binding affinity of r-IgGs to detect OTA with a detection range of 0.006–0.01 nM/dm. The ozonation and adsorption efficacies of altered nano-diamonds for detecting aflatoxin-B1 level were investigated. Recently it was presented an ultrasensitive approach for detecting mycotoxins using STING (signal transduction by ion nano-gating) sensing, with a detection limit of 100 fg mL−1 (Actis et al., 2010). To make a BSA/aAFB1-CAuNP/MBA/Au immune-electrode. Cysteamine functionalized gold nanoparticles (C-AuNP) and aflatoxin B1 antibodies (aAFB1) were immobilized on a 4-mercaptobenzoic acid-based self-collected monolayer on a gold electrode (MBA/Au). AFB1 in the range of 10–100 ng L−1 was detected using these electrodes. A mobile equipment that can simultaneously identify numerous bacterial, fungal toxins, and pathogens in stored food was recently created (Biswal and Misra, 2020). According to these studies, nanostructured platforms appear to be a promising alternative to traditional approaches for detecting mycotoxins and infections that damage food and agricultural products.
5 Tracking plant diseases through the identification of volatile organic compounds (VOCs)
Plants release volatile organic compounds (VOCs) as biomarkers into their environment through various parts like leaves, flowers, roots, and other organs, which can be influenced by factors such as self-healing, exposure to stress, herbivore browsing, pathogen infection, and pest repellence. These VOCs play a crucial role in plant responses to pest damage and diseases (Hoebeke et al., 2011). Among the botanical VOCs, terpenes like cis-jasmone (CJ), -pinene, limonene, and -terpinene are commonly found (Bruinsma et al., 2009; Han et al., 2016). Gas chromatography/mass spectrometry or gas chromatography electroantennographic detectors are traditionally used to detect these VOCs (Mesquita et al., 2017; Cheung et al., 2015). However, these methods are expensive, time-consuming, and impractical for real-time monitoring of plant VOCs. Therefore, there is a need for sensors that offer high accuracy, rapid response times, and immunity to interference for real-time plant VOC detection in agricultural applications. The detection of cis-jasmone vapor, a localized surface plasmon resonance (LSPR) sensor coated with a molecularly imprinted sol–gel (MISG) can be employed. The MISG coating enhances the LSPR sensor’s selectivity and reduces sensitivity. To further enhance the sensor’s sensitivity, gold nanoparticles (AuNPs) have been embedded in the MISG. According to Shang and colleagues in 2018, sensors coated with MISG containing 20 µL of 30-nm AuNPs exhibited higher sensitivity compared to sensors coated with other films. The precise detection of gaseous (E)-2-hexenal, one of the primary VOC indicators produced during P. infestans infection, researchers used cysteine (Cys)-functionalized gold nanoparticles (Au NPs) or nanorods (Au NRs) as plasmonic analytical colorants in a sensor array. This portable device incorporates a disposable colorimetric sensor array consisting of plasmonic nano-colorants and chemo-responsive organic dyes, enabling the detection of essential plant volatiles at the parts per million (ppm) level within a minute (Li Y. et al., 2019).
6 Commercially available devices
Effective plant disease detection relies on the availability of commercial bio-recognition elements, such as antibodies, DNA probes, and aptamers. Immunoassay-based technologies are commonly employed for the detection of phytopathogens, with lateral flow devices, tissue-print ELISA, and plate-ELISA kits being among the most widely used methods. Several research studies have highlighted the use of commercial kits based on immunoassays, including a portable kit designed for orchid virus detection and the Agritest lateral flow kit for identifying Erwinia amylovora, the bacterial agent responsible for pome tree disease. Furthermore, a commercial diagnostic tool has been developed for detecting xanthomonas wilt in banana plants (Hodgetts et al., 2015).
7 Challenges and future perspectives
Currently, there are three significant challenges associated with plant diagnostic tools. Concerns about the environmental impact and toxicity of synthetic nanomaterials, the urgency for faster data sharing and disease forecasting, and the durability of sensors in harsh conditions like extreme temperatures, intense sunlight, and heavy usage are paramount. Prior to field deployment, addressing safety concerns, particularly those related to hazardous nanoparticles like quantum dots (QDs), is crucial. Rigorous hazard assessment and oversight are necessary for nanosensors in contact with living plants or food to prevent toxic residues from entering the food chain and reaching consumers. The second obstacle pertains to the urgency for quicker reporting and real-time forecasting of disease outbreaks in agricultural settings. Advanced nanosensors are anticipated to be highly interconnected, facilitating nearly instantaneous monitoring. For instance, continuous tracking of plant volatile organic compound emissions offers more dynamic and precise data than sporadic measurements, thereby improving stress response monitoring. Finally, durable sensors capable of withstanding various environmental conditions, such as temperature fluctuations, humidity, and air pollution, are required before implementing these sensors in real-world agricultural settings. Further research is needed to develop innovative sensor materials, including environmentally resistant substrates with embedded nanoparticles.
8 Conclusion
One of the most significant global challenges is the substantial loss of agricultural production due to plant diseases. Plant diseases can greatly reduce crop yields, resulting in a substantial loss of resources and agricultural output. In order to develop effective strategies for disease diagnosis and mitigation, it is crucial to initially identify the prevailing plant diseases. Conventional approaches to identify plant pathogens, like scrutinizing infected tissue with a microscope or employing culture-based methods, are laborious and demand skilled personnel. However, modern techniques like biosensors offer a faster and more accessible means of disease identification. Increasing attention is being directed towards biosensors to detect phytopathogenic bacteria, with researchers striving to create portable handheld devices for swift, accurate, and site-specific detection. The convergence of biosensor technology and synthetic biology is a promising avenue of exploration for agricultural scientists.
Author contributions
JN: Writing – original draft. JC: Software, Writing – review and editing. SS: Data curation, Funding acquisition, Writing – review and editing. DP: Funding acquisition, Investigation, Visualization, Writing – review and editing. JM: Funding acquisition, Visualization, Writing – review and editing. PS: Funding acquisition, Investigation, Resources, Writing – review and editing. PB: Funding acquisition, Investigation, Writing – review and editing. SS: Formal Analysis, Funding acquisition, Writing – review and editing. P.: Data curation, Funding acquisition, Methodology, Writing – review and editing. JS: Formal Analysis, Writing – review and editing. NM: Formal Analysis, Funding acquisition, Visualization, Writing – review and editing. AK: Visualization, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Actis, P., Jejelowo, O., and Pourmand, N. (2010). Ultrasensitive mycotoxin detection by STING sensors. Biosens. Bioelectron. 26 (2), 333–337. doi:10.1016/j.bios.2010.08.016
Alonso-Lomilloa, M. A., Domınguez-Renedoa, O., Ferreira-Gonc, L., and Arcos-Martıneza, M. J. (2010). Sensitive enzyme-biosensor based on screen-printed electrodes for ochratoxin A. Biosens. Bioelectron. 25, 1333–1337. doi:10.1016/j.bios.2009.10.024
Ariffin, S. A. B., Adam, T., Hashim, U., Faridah, S., Zamri, I., and Uda, M. N. A. (2014). Plant diseases detection using nanowire as biosensor transducer. Adv. Mater Res. 832, 113–117. doi:10.4028/www.scientific.net/amr.832.113
Arya, M. S., Anjali, K., and Unni, D. (2018). Detection of unhealthy region of plant leaves using image processing and genetic algorithm with Arduino. Proc. Int. Conf. Power, Signals,Control Comput. (EPSCICON).IEEE, Thrissur, IndiaNature 550, 285–288.
Badial, A., Sherman, D., Stone, A., Gopakumar, A., Wilson, V., Schneider, W., et al. (2018). Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 102 (8), 1648–1652. doi:10.1094/pdis-04-17-0488-re
Bakhori, N. M., Yusof, N. A., Abdullah, A. H., and Hussein, M. Z. (2013). Development of a fluorescence resonance energy transfer (FRET)-based DNA biosensor for detection of synthetic oligonucleotide of Ganoderma boninense. Biosensors3 3 (4), 419–428. doi:10.3390/bios3040419
Berto, M., Vecchi, E., Baiamonte, L., Condò, C., Sensi, M., Di Lauro, M., et al. (2019). Label free detection of plant viruses with organic transistor biosensors. Sensors Actuators B Chem. 281, 150–156. doi:10.1016/j.snb.2018.10.080
Biswal, A. K., and Misra, P. K. (2020). Biosynthesis and characterization of silver nanoparticles for prospective application in food packaging and biomedical fields. Mater Chem. Phys. 250, 123014. doi:10.1016/j.matchemphys.2020.123014
Bruinsma, M., Posthumus, M. A., Mumm, R., Mueller, M. J., van Loon, J. J., and Dicke, M. (2009). Jasmonic acid-induced volatiles of Brassica oleracea attract parasitoids: effects of time and dose, and comparison with induction by herbivores. J. Exp. Bot. 60 (9), 2575–2587. doi:10.1093/jxb/erp101
Candresse, T., Lot, H., German-Retana, S., Krause-Sakate, R., Thomas, J., Souche, S., et al. (2007). Analysis of the serological variability of Lettuce mosaic virus using monoclonal antibodies and surface plasmon resonance technology. J. general virology 88 (9), 2605–2610. doi:10.1099/vir.0.82980-0
Cardoso, R. M., Pereira, T. S., Facure, M. H., dos Santos, D. M., Mercante, L. A., Mattoso, L. H., et al. (2021). Current progress in plant pathogen detection enabled by nanomaterials-based (bio) sensors. Sensors Actuators Rep. 4, 100068. doi:10.1016/j.snr.2021.100068
Cellini, A., Blasioli, S., Biondi, E., Bertaccini, A., Braschi, I., and Spinelli, F. (2017). Potential applications and limitations of electronic nose devices for plant disease diagnosis. Sensors 17 (11), 2596. doi:10.3390/s17112596
Cesewski, E., and Johnson, B. N. (2020). Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 159, 112214. doi:10.1016/j.bios.2020.112214
Chalupowicz, L., Dombrovsky, A., Gaba, V., Luria, N., Reuven, M., Beerman, A., et al. (2019). Diagnosis of plant diseases using the Nanopore sequencing platform. Plant Pathol. 68 (2), 229–238. doi:10.1111/ppa.12957
Chartuprayoon, N., Rheem, Y., Ng, J. C., Nam, J., Chen, W., and Myung, N. V. (2013). Polypyrrole nanoribbon based chemiresistive immunosensors for viral plant pathogen detection. Anal. Methods 5 (14), 3497–3502. doi:10.1039/c3ay40371h
Chaudhary, M., Verma, S., Kumar, A., Basavaraj, Y. B., Tiwari, P., Singh, S., et al. (2021). Graphene oxide based electrochemical immunosensor for rapid detection of groundnut bud necrosis orthotospovirus in agricultural crops. Talanta 235, 122717. doi:10.1016/j.talanta.2021.122717
Chen, B. B., Liu, M. L., Zhan, L., Li, C. M., and Huang, C. Z. (2018). Terbium (III) modified fluorescent carbon dots for highly selective and sensitive ratiometry of stringent. Anal. Chem. 90 (6), 4003–4009. doi:10.1021/acs.analchem.7b05149
Cheung, W. H., Pasamontes, A., Peirano, D. J., Zhao, W., Grafton-Cardwell, E. E., Kapaun, T., et al. (2015). Volatile organic compound (VOC) profiling of citrus tristeza virus infection in sweet orange citrus varietals using thermal desorption gas chromatography time of flight mass spectrometry (TD-GC/TOF-MS). Metabolomics 11 (6), 1514–1525. doi:10.1007/s11306-015-0807-6
Cui, S., Ling, P., Zhu, H., and Keener, H. (2018). Plant Pest detection using an artificial nose system: a Review. Sensors 18 (2), 378. doi:10.3390/s18020378
Dameron, C. T., Reese, R. N., Mehra, R. K., Kortan, A. R., Carroll, P. J., Steigerwald, M. L., et al. (1989). Biosynthesis of cadmium sulphide quantum semiconductor crystallites. Nature 338 (6216), 596–597. doi:10.1038/338596a0
Di Giusto, D. A., Wlassoff, W. A., Gooding, J. J., Messerle, B. A., and King, G. C. (2005). Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic acids Res. 33 (6), e64. doi:10.1093/nar/gni063
Drygin, Y. F., Blintsov, A. N., Grigorenko, V. G., Andreeva, I. P., Osipov, A. P., Varitzev, Y. A., et al. (2012). Highly sensitive field test lateral flow immunodiagnostics of PVX infection. Appl. Microbiol. Biotechnol. 93 (1), 179–189. doi:10.1007/s00253-011-3522-x
Dyussembayev, K., Sambasivam, P., Bar, I., Brownlie, J. C., Shiddiky, M. J., and Ford, R. (2021). Biosensor technologies for early detection and quantification of plant pathogens. Front. Chem. 9, 636245. doi:10.3389/fchem.2021.636245
Eisenstein, M. (2017). An ace in the hole for DNA sequencing. Nature 550 (7675), 285–288. doi:10.1038/550285a
Fang, L. (2020). “Research on plant diseases and insect pests monitoring technology under the background of internet of things technology,” in 2020 international wireless communications and mobile computing (IWCMC) (IEEE), 1999–2001.
Feng, M., Kong, D., Wang, W., Liu, L., Song, S., and Xu, C. (2015). Development of an immun–4301ochromatographic strip for rapid detection of Pantoea stewartii subsp. stewartii. Sensors 15 (2), 291.
Filloux, D., Fernandez, E., Loire, E., Claude, L., Galzi, S., Candresse, T., et al. (2018). Nanopore-based detection and characterization of yam viruses. Sci. Rep. 8 (1), 17879. doi:10.1038/s41598-018-36042-7
Firrao, G., Moretti, M., Ruiz-Rosquete, M., Gobbi, E., and Locci, R. (2005). Nanobiotransducer for detecting flavescence doree phytoplasma. J. Plant Pathol. 87, 101–107.
Freitas, T. A., Proença, C. A., Baldo, T. A., Materón, E. M., Wong, A., Magnani, R. F., et al. (2019). Ultrasensitive immunoassay for detection of Citrus tristeza virus in citrus sample using disposable microfluidic electrochemical device. Talanta 205, 120110. doi:10.1016/j.talanta.2019.07.005
Gao, G., Jiang, Y. W., Sun, W., and Wu, F. G. (2018). Fluorescent quantum dots for microbial imaging. Chin. Chem. Lett. 29 (10), 1475–1485. doi:10.1016/j.cclet.2018.07.004
Giraldo, J. P., Wu, H., Newkirk, G. M., and Kruss, S. (2019). Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol. 14 (6), 541–553. doi:10.1038/s41565-019-0470-6
Gouvea, C. (2011). Biosensors for health applications. Biosens. Health, Environ. Biosecurity (3), 16983.
Greenshields, M., Cunha, B., Coville, N., Pimentel, I., Zawadneak, M., Dobrovolski, S., et al. (2016). “Fungi active microbial metabolism detection of Rhizopus sp. and aspergillus sp,” in Section nigri on strawberry using a set of chemical sensors based on carbon nanostructures.
Haji-Hashemi, H., Norouzi, P., Safarnejad, M. R., Larijani, B., Habibi, M. M., Raeisi, H., et al. (2018). Sensitive electrochemical immunosensor for citrus bacterial canker disease detection using fast Fourier transformation square-wave voltammetry method. J. Electroanal. Chem. 820, 111–117. doi:10.1016/j.jelechem.2018.04.062
Han, Z. X., Rana, M. M., Liu, G. F., Gao, M. J., Li, D. X., Wu, F. G., et al. (2016). Green tea flavour determinants and their changes over manufacturing processes. Food Chem. 212, 739–748. doi:10.1016/j.foodchem.2016.06.049
Hodgetts, J., Karamura, G., Johnson, G., Hall, J., Perkins, K., Beed, F., et al. (2015). Development of a lateral flow device for in-field detection and evaluation of PCR-based diagnostic methods for Xanthomonas campestris pv. musacearum, the causal agent of banana xanthomonas wilt. Plant Pathol. 64 (3), 559–567. doi:10.1111/ppa.12289
Hoebeke, E. R., Smith, D. R., and Goulet, H. (2011). Athalia cornubiae benson (hymenoptera: tenthredinidae: allantinae), a sawfly genus and species new to north America. Proc. Entomological Soc. Wash. 113 (3), 309–314. doi:10.4289/0013-8797.113.3.309
Kashyap, P. L., Kumar, S., Jasrotia, P., Singh, D. P., and Singh, G. P. (2019). Nanosensors for plant disease diagnosis: current understanding and future perspectives. Nanosci. Sustain. Agric., 189–205. doi:10.1007/978-3-319-97852-9_9
Kasibabu, B. S. B., D'souza, S. L., Jha, S., Singhal, R. K., Basu, H., and Kailasa, S. K. (2015). One-step synthesis of fluorescent carbon dots for imaging bacterial and fungal cells. Anal. Methods 7 (6), 2373–2378. doi:10.1039/c4ay02737j
Khaled, A. Y., Abd Aziz, S., Bejo, S. K., Nawi, N. M., Seman, I. A., and Onwude, D. I. (2018). Early detection of diseases in plant tissue using spectroscopy–applications and limitations. Appl. Spectrosc. Rev. 53 (1), 36–64. doi:10.1080/05704928.2017.1352510
Khiyami, M. A., Almoammar, H., Awad, Y. M., Alghuthaymi, M. A., and Abd-Elsalam, K. A. (2014). Plant pathogen nanodiagnostic techniques: forthcoming changes. Biotechnol. and Biotechnol. Equip. 28 (5), 775–785. doi:10.1080/13102818.2014.960739
Konwarh, R., and Sharma, P. L. (2020). “Nanosensor platforms for surveillance of plant pathogens and phytometabolites/analytes vis-à-vis plant health status,” in Nanomaterials for agriculture and forestry applications (Elsevier), 357–385.
Lautner, G., Balogh, Z., Bardóczy, V., Mészáros, T., and Gyurcsányi, R. E. (2010). Aptamer-based biochips for label-free detection of plant virus coat proteins by SPR imaging. Analyst 135 (5), 918–926. doi:10.1039/b922829b
Li, Y., Wang, Z., Sun, L., Liu, L., Xu, C., and Kuang, H. (2019a). Nanoparticle-based sensors for food contaminants. TrAC Trends Anal. Chem. 113, 74–83. doi:10.1016/j.trac.2019.01.012
Li, Z., Liu, Y., Hossain, O., Paul, R., Yao, S., Wu, S., et al. (2021). Real-time monitoring of plant stresses via chemiresistive profiling of leaf volatiles by a wearable sensor. Matter 4 (7), 2553–2570. doi:10.1016/j.matt.2021.06.009
Li, Z., Paul, R., Ba Tis, T., Saville, A. C., Hansel, J. C., Yu, T., et al. (2019b). Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants. 5, 856–866. doi:10.1038/s41477-019-0476-y
Li, Z., Yu, T., Paul, R., Fan, J., Yang, Y., and Wei, Q. (2020). Agricultural nanodiagnostics for plant diseases: recent advances and challenges. Nanoscale Adv. 2 (8), 3083–3094. doi:10.1039/c9na00724e
Liang, Y., Duan, Y., Fan, C., Dong, H., Yang, J., Tang, J., et al. (2019). Preparation of kasugamycin conjugation based on ZnO quantum dots for improving its effective utilization. Chem. Eng. J. 361, 671–679.
Lin, H. Y., Huang, C. H., Lu, S. H., Kuo, I. T., and Chau, L. K. (2014). Direct detection of orchid viruses using nanorod-based fiber optic particle plasmon resonance immunosensor. Biosens. Bioelectron. 51, 371–378. doi:10.1016/j.bios.2013.08.009
Luna-Moreno, D., Sánchez-Álvarez, A., Islas-Flores, I., Canto-Canche, B., Carrillo-Pech, M., Villarreal-Chiu, J. F., et al. (2019). Early detection of the fungal banana black Sigatoka pathogen Pseudocercospora fijiensis by an SPR immunosensor method. Sensors 19 (3), 465. doi:10.3390/s19030465
Mahapatra, S., and Chandra, P. (2020). Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens. Bioelectron. 165, 112361. doi:10.1016/j.bios.2020.112361
Mahato, K., Kumar, S., Srivastava, A., Maurya, P. K., Singh, R., and Chandra, P. (2018). “Electrochemical immunosensors: fundamentals and applications in clinical diagnostics,” in Handbook of immunoassay technologies (Academic Press), 359–414.
Mahmoudpour, M., Dolatabadi, J. E. N., Torbati, M., and Homayouni-Rad, A. (2019). Nanomaterials based surface plasmon resonance signal enhancement for detection of environmental pollutions. Biosens. Bioelectron. 127, 72–84. doi:10.1016/j.bios.2018.12.023
Martinelli, F., Scalenghe, R., Davino, S., Panno, S., Scuderi, G., Ruisi, P., et al. (2015). Advanced methods of plant disease detection. A review. A Rev. Agron. Sustain. Dev. 35 (1), 1–25. doi:10.1007/s13593-014-0246-1
Mccandless, L. (2005). “Nanotechnology offers new insights into plant pathology,” in College of agriculture and life sciences news. Cornell University, 17–18.
Meng, Y., Li, Y., Galvani, C. D., Hao, G., Turner, J. N., Burr, T. J., et al. (2005). Upstream migration of Xylella fastidiosa via pilusdriven twitching motility. J. Bacteriol. 187 (16), 5560–5567. doi:10.1128/jb.187.16.5560-5567.2005
Mesquita, P. R., Nunes, E. C., dos Santos, F. N., Bastos, L. P., Costa, M. A., Rodrigues, F. D. M., et al. (2017). Discrimination of Eugenia uniflora L. biotypes based on volatile compounds in leaves using HS-SPME/GC–MS and chemometric analysis. Microchem. J. 130, 79–87. doi:10.1016/j.microc.2016.08.005
Miranda, B. S., Linares, E. M., Thalhammer, S., and Kubota, L. T. (2013). Development of a disposable and highly sensitive paper-based immunosensor for early diagnosis of Asian soybean rust. Biosens. Bioelectron. 45, 123–128. doi:10.1016/j.bios.2013.01.048
Morales-Narváez, E., Naghdi, T., Zor, E., and Merkoçi, A. (2015). Photoluminescent lateral-flow immunoassay revealed by graphene oxide: highly sensitive paper-based pathogen detection. Anal. Chem. 87 (16), 8573–8577. doi:10.1021/acs.analchem.5b02383
Nie, L. (2013). Biomedical nanotechnology for optical molecular imaging, diagnostics, and therapeutics. JSM Nanotechnol. Nanomed 1 (1), 1–2.
Nunes Pauli, G. E., de la Escosura-Muñiz, A., Parolo, C., Helmuth Bechtold, I., and Merkoçi, A. (2015). Lab-in-a-syringe using gold nanoparticles for rapid immunosensing of protein biomarkers. Lab a Chip 15 (2), 399–405. doi:10.1039/c4lc01123f
Paniel, N., Radoi, A., and Marty, J. L. (2010). Development of an electrochemical biosensor for the detection of afl atoxin M1 in milk. Sensors 10 (10), 9439–9448. doi:10.3390/s101009439
Parolo, C., de la Escosura-Muñiz, A., and Merkoçi, A. (2013a). Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 40 (1), 412–416. doi:10.1016/j.bios.2012.06.049
Parolo, C., Medina-Sánchez, M., De La Escosura-Muñiz, A., and Merkoçi, A. (2013b). Simple paper architecture modifications lead to enhanced sensitivity in nanoparticle based lateral flow immunoassays. Lab a Chip 13 (3), 386–390. doi:10.1039/c2lc41144j
Peng, H., and Chen, I. A. (2019). Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 13 (2), 1244–1252. doi:10.1021/acsnano.8b06395
Pramanik, S., Hill, S. K., Zhi, B., Hudson-Smith, N. V., Wu, J. J., White, J. N., et al. (2018). Comparative toxicity assessment of novel Si quantum dots and their traditional Cd-based counterparts using bacteria models Shewanella oneidensis and Bacillus subtilis. Environ. Sci. Nano 5 (8), 1890–1901. doi:10.1039/c8en00332g
Rana, K., Mittal, J., Narang, J., Mishra, A., and Pudake, R. N. (2021). Graphene based electrochemical DNA biosensor for detection of false smut of Rice (ustilaginoidea virens). plant pathology J. 37 (3), 291–298. doi:10.5423/ppj.oa.11.2020.0207
Ray, M., Ray, A., Dash, S., Mishra, A., Achary, K. G., Nayak, S., et al. (2017). Fungal disease detection in plants: traditional assays, novel diagnostic techniques and Biosensors. Biosens. Bioelectron. 87, 708–723. doi:10.1016/j.bios.2016.09.032
Razmi, A., Golestanipour, A., Nikkhah, M., Bagheri, A., Shamsbakhsh, M., and Malekzadeh-Shafaroudi, S. (2019). Localized surface plasmon resonance biosensing of Tomato Yellow Leaf Curl virus. J. Virological Methods 267, 1–7. doi:10.1016/j.jviromet.2019.02.004
Razo, S. C., Panferova, N. A., Panferov, V. G., Safenkova, I. V., Drenova, N. V., Varitsev, Y. A., et al. (2019). Enlargement of gold nanoparticles for sensitive immunochromatographic diagnostics of potato brown rot. Sensors 19 (1), 153. doi:10.3390/s19010153
Rispail, N., De Matteis, L., Santos, R., Miguel, A. S., Custardoy, L., Testillano, P. S., et al. (2014). Quantum dot and superparamagnetic nanoparticle interaction with pathogenic fungi: internalization and toxicity profile. ACS Appl. Mater. and interfaces 6 (12), 9100–9110. doi:10.1021/am501029g
Rivas, L., Medina-Sánchez, M., de la Escosura-Muñiz, A., and Merkoçi, A. (2014). Improving sensitivity of gold nanoparticle-based lateral flow assays by using wax-printed pillars as delay barriers of microfluidics. Lab. Chip 14 (22), 4406–4414. doi:10.1039/c4lc00972j
Roda, A., Mirasoli, M., Guardigli, M., Zangheri, M., Caliceti, C., Calabria, D., et al. (2018). Advanced biosensors for monitoring astronauts’ health during long-duration space missions. Biosens. Bioelectron. 111, 18–26. doi:10.1016/j.bios.2018.03.062
Roda, A., Zangheri, M., Guardigli, M., Di Nardo, F., Anfossi, L., Baggiani, C., et al. (2020). Chemiluminescence biosensor for non-invasive crew health monitoring at the international space station. Aerotecnica Missili and Spazio 99 (2), 103–109. doi:10.1007/s42496-020-00052-4
Safarpour, H., Safarnejad, M. R., Tabatabaei, M., Mohsenifar, A., Rad, F., Basirat, M., et al. (2012). Development of a quantum dots FRET-based biosensor for efficient detection of Polymyxa betae. Can. J. Plant Pathology 34 (4), 507–515. doi:10.1080/07060661.2012.709885
Salomone, A., Mongelli, M., Roggero, P., and Boscia, D. (2004). Reliability of detection of Citrus tristeza virus by an immunochromatographic lateral flow assay in comparison with ELISA. J. Plant Pathology, 43–48.
Shetti, N. P., Malode, S. J., Roy, S., Chandra, P., Reddy, K. R., and Chatterjee, S. (2020). “Electroanalytical techniques for investigating biofilms: applications in biosensing and biomolecular interfacing,” in Nanomaterials in diagnostic tools and devices (Elsevier), 293–329.
Shi, J., McLamore, E. S., and Porterfield, D. M. (2013). Nanomaterial based self-referencing microbiosensors for cell and tissue physiology research. Biosens. Bioelectron. 40 (1), 127–134. doi:10.1016/j.bios.2012.06.059
Shojaei, T. R., Salleh, M. A. M., Sijam, K., Rahim, R. A., Mohsenifar, A., Safarnejad, R., et al. (2016). Fluorometric immunoassay for detecting the plant virus Citrus tristeza using carbon nanoparticles acting as quenchers and antibodies labeled with CdTe quantum dots. Microchim. Acta 183 (7), 2277–2287. doi:10.1007/s00604-016-1867-7
Singh, S., Singh, M., Agrawal, V. V., and Kumar, A. (2010). An attempt to develop surface plasmon resonance based immunosensor for Karnal bunt (Tilletia indica) diagnosis based on the experience of nano-gold based lateral flow immuno-dipstick test. Thin Solid Films 519, 1156–1159. doi:10.1016/j.tsf.2010.08.061
Thaxton, C. S., Georganopoulou, D. G., and Mirkin, C. A. (2006). Gold nanoparticle probes for the detection of nucleic acid targets. Clin. ChimActa 363, 120–126. doi:10.1016/j.cccn.2005.05.042
Tran, T. T., Clark, K., Ma, W., and Mulchandani, A. (2020). Detection of a secreted protein biomarker for citrus Huanglongbing using a single-walled carbon nanotubes-based chemiresistive biosensor. Biosens. Bioelectron. 147, 111766. doi:10.1016/j.bios.2019.111766
Tsoi, K. M., Dai, Q. I. N., Alman, B. A., and Chan, W. C. (2013). Are quantum dots toxic? Exploring the discrepancy between cell culture and animal studies. Accounts Chem. Res. 46 (3), 662–671. doi:10.1021/ar300040z
Tsuda, S. (1992). A novel detection and identification technique for plant viruses: rapid Immunofilter Paper Assay (RIPA). Plant Dis. 76 (5), 466. doi:10.1094/pd-76-0466
Valekunja, R. B., Kamakoti, V., Peter, A., Phadnis, S., Prasad, S., and Nagaraj, V. J. (2016). The detection of papaya ringspot virus coat protein using an electrochemical immunosensor. Anal. Methods 8 (48), 8534–8541. doi:10.1039/c6ay02201d
Vaseghi, A., Safaie, N., Bakhshinejad, B., Mohsenifar, A., and Sadeghizadeh, M. (2013). Detection of Pseudomonas syringae pathovars by thiol-linked DNAGold nanoparticle probes. Sensors Actuators B Chem. 181, 644–651. doi:10.1016/j.snb.2013.02.018
Verosloff, M., Chappell, J., Perry, K. L., Thompson, J. R., and Lucks, J. B. (2019). PLANT-Dx: a molecular diagnostic for point-of-use detection of plant pathogens. ACS Synth. Biol. 8 (4), 902–905. doi:10.1021/acssynbio.8b00526
Wang, H., Wang, Y., Hou, X., and Xiong, B. (2020). Bioelectronic nose based on single-stranded dna and single-walled carbon nanotube to identify a major plant volatile organic compound (p-ethylphenol) released by phytophthora cactorum infected strawberries. Nanomaterials 10 (3), 479. doi:10.3390/nano10030479
Wang, R., Minunni, M., Tombelli, S., and Mascini, M. (2004). A new approach for the detection of DNA sequences in amplified nucleic acids by a surface plasmon resonance biosensor. Biosens. Bioelectron. 20 (3), 598–605. doi:10.1016/j.bios.2004.03.013
Wang, Z., Wei, F., Liu, S. Y., Xu, Q., Huang, J. Y., Dong, X. Y., et al. (2010). Electrocatalytic oxidation of phytohormone salicylic acid at copper nanoparticles-modified gold electrode and its detection in oilseed rape infected with fungal pathogen Sclerotinia sclerotiorum. Talanta80 80, 1277–1281. doi:10.1016/j.talanta.2009.09.023
Wei, S., Sun, Y., Xi, G., Zhang, H., Xiao, M., and Yin, R. (2018). Development of a single-tube nested PCR-lateral flow biosensor assay for rapid and accurate detection of Alternaria panax Whetz. PLoS One 13 (11), e0206462. doi:10.1371/journal.pone.0206462
Yan, X., Li, H., and Su, X. (2018). Review of optical sensors for pesticides. TrAC Trends Anal. Chem. 103, 1–20. doi:10.1016/j.trac.2018.03.004
Yüksel, S., Schwenkbier, L., Pollok, S., Weber, K., Cialla-May, D., and Popp, J. (2015). Label-free detection of phytophthora ramorum using surface-enhanced Raman spectroscopy. Analyst 140 (21), 7254–7262. doi:10.1039/c5an01156f
Zhan, F., Wang, T., Iradukunda, L., and Zhan, J. (2018). A gold nanoparticle-based lateral flow biosensor for sensitive visual detection of the potato late blight pathogen, Phytophthora infestans. Anal. Chim. Acta. 1036, 153–161. doi:10.1016/j.aca.2018.06.083
Keywords: nanobiosensor, nanotechnology, detection, plant pathogen, disease
Citation: Narware J, Chakma J, Singh SP, Prasad DR, Meher J, Singh P, Bhargava P, Sawant SB, Pitambara , Singh JP, Manzar N and Kashyap AS (2025) Nanomaterial-based biosensors: a new frontier in plant pathogen detection and plant disease management. Front. Bioeng. Biotechnol. 13:1570318. doi: 10.3389/fbioe.2025.1570318
Received: 03 February 2025; Accepted: 26 March 2025;
Published: 23 April 2025.
Edited by:
Damini Maithani, Dev Bhoomi Uttarakhand University, IndiaReviewed by:
Madan L. Verma, Indian Institute of Information Technology, IndiaDivya Joshi, G. B. Pant University of Agriculture and Technology, India
Copyright © 2025 Narware, Chakma, Singh, Prasad, Meher, Singh, Bhargava, Sawant, Pitambara, Singh, Manzar and Kashyap. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhijeet Shankar Kashyap, YWJoaWplZXQ0NDk3QGdtYWlsLmNvbQ==; Satyendra P. Singh, c3BzYmh1MUBnbWFpbC5jb20=
 Jeetu Narware
Jeetu Narware Jharjhari Chakma1
Jharjhari Chakma1 Nazia Manzar
Nazia Manzar Abhijeet Shankar Kashyap
Abhijeet Shankar Kashyap