- 1Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
- 2Laboratory of Biochemistry and Enzymatic Engineering of Lipases, ENIS, University of Sfax, Sfax, Tunisia
- 3Science Department, Environmental and biotechnology research group, College of Rivière-Du-Loup, Quebec, QC, Canada
Background/objectives: The growing demand for reliable and stable biocatalysts has spurred research into microbial lipases for diverse industrial applications. This study focused on enhancing the production and purification of a lipase from Streptomyces gobitricini (LipS.g).
Methods: Maximal lipase activity (420 U/mL) was achieved during the stationary phase after 84 h of incubation at 45°C and pH 8.0, using 2% glucose and 2% yeast extract as carbon and nitrogen sources, respectively.
Results: Calcium, olive oil, and Tween, at 1%, significantly enhanced LipS.g production, highlighting the role of triglycerides and detergents in enzyme induction and substrate emulsification. The purified 50-kDa enzyme displayed maximal activity at 50°C and pH 9.0, with thermal stability between 40°C and 55°C and pH 5.0–10.0. While LipS.g efficiently hydrolyzed short and medium-chain triglycerides, it exhibited a preference for long-chain substrates, with a maximum reaction rate of 2500 μmol/min/mg and a Km value of 6.45 mM toward triolein (C18). LipS.g also demonstrated remarkable stability in detergent formulations, retaining more than 85% activity in the presence of surfactants, oxidizing agents, boron compounds, and enzyme inhibitors. Additionally, LipS.g catalyzed the esterification of oleic acid with starch and ethanol to produce starch oleate and ricinoleic acid.
Conclusion: These findings establish LipS.g as a promising biocatalyst for applications in biocatalysis and detergent formulations, with potential uses in the food, beverage, cosmetic, and pharmaceutical industries.
1 Introduction
Lipases (EC 3.1.1.3) are triacylglycerol acyl-hydrolases that hydrolyze triglycerides at the organic-aqueous interface, producing diglycerides, monoglycerides, glycerol, and free fatty acids (Adetunji and Olaniran, 2021). These enzymes are serine hydrolases with a catalytic triad of Serine, Aspartate/Glutamate, and Histidine (Gupta et al., 2004). Their molecular weight is in the range of 19–60 kDa and reported as monomeric proteins (Chandra et al., 2020; Abdelaziz et al., 2025). Lipases are classified based on their substrate specificity into non-specific, 1, 3-specific, and fatty acid-specific types (Yao et al., 2021). Non-specific lipases hydrolyze triglycerides into free fatty acids and glycerol. 1, 3-specific lipases act on the ester bonds at the C1 and C3 positions of triglycerides. Fatty acid-specific lipases preferentially hydrolyze long-chain fatty acids with cis-double bonds at the C9-C10 position, like triolein (Yao et al., 2021).
Lipases are widespread in nature and are produced by a diverse range of organisms, including plants, animals, and microorganisms (Bharathi and Rajalakshmi, 2019). The majority of microbial lipases are secreted as extracellular enzymes, making them relatively easy to isolate in high purity and ideal for large-scale production (Yao et al., 2021). Microbial lipases are preferred compared to those from animals and plants due to the rapid growth and reproduction rates of microbes facilitating the production of these enzymes with high yields (Yao et al., 2021). Among microorganisms, lipase production is especially prevalent in bacteria, fungi, and yeast. The most common bacterial sources of lipases include species from Bacillus, Pseudomonas, Staphylococcus, and Burkholderia (Bharathi and Rajalakshmi, 2019).
Bacterial lipase production is strongly affected by physicochemical conditions of medium culture, including temperature, pH, agitation speed, nitrogen and carbon sources, inorganic salts as sources of metal ions and dissolved oxygen levels (Gupta et al., 2004). Oils, triacylglycerols, fatty acids, hydrolysable esters, tweens, bile salts and glycerol are usually opadded in growth bacterial medium to induce the lipase production since they are inducible enzymes. Moreover, sugar alcohol, polysaccharides or whey as well as long-chain fatty acids, such as oleic, linoleic and linolenic acids were described to enhance lipase production (Gupta et al., 2004; Cesário et al., 2021; Al Mohaini et al., 2022). Inorganic and organic nitrogen sources, have proven effective for lipase production in some microorganisms. Likewise, divalent cations especially calcium (Ca2+) and magnesium (Mg2+) enhanced efficiency the bacterial lipase yield production (Gupta et al., 2004). Reported bacterial lipases was overexpressed generally at pH 7.0–8.0, temperature growth between 20°C and 50°C during an incubation period varying from 24 h to 96 h (Gupta et al., 2004). Only a limited number of media have been described for enhancing growth conditions of Streptomyces strains to produce extracellular lipases. For instance, Streptomyces sp. CS326 was cultivated at 28°C for 168 h in a medium containing 10 g of glucose, 10 g of soybean, and 0.1 g of Na2HPO4. Streptomyces halstedii strain ST 40 showed enhanced lipase activity when incubated with Tween 20 for 4 h. Maximal lipase production from Streptomyces griseus was achieved using sunflower oil and palm oil in an orbital shaker after 96 h (Vasconcelos, 2018).
Microbial lipases have been extensively studied due to their diverse biochemical properties and industrial applications. Typically, their maximal pH is neutral or alkaline, with maximal temperature etween 30°C and 60°C. Bacterial lipases exhibit stability across a wide pH range (pH 4.0–11.0) (Yuan et al., 2016). They are regiospecific, often 1, 3-specific, selectively hydrolyzing the ester bonds at the C1 and C3 positions of glycerol. This process breaks down triacylglycerides into free fatty acids, 1, 2(2,3)-diacylglycerides, and 2-monoacylglycerides (Gupta et al., 2004). At organic-aqueous interfaces, microbial lipases are activated, and surfactants can either enhance or inhibit their activity based on interactions and enzyme-surfactant complex formation (Delorme et al., 2011).
Due to their versatile enzymatic properties, substrate specificity, and high stability, microbial lipases are a valuable class of biotechnological enzymes used in diverse applications across industries, including food, leather, pharmaceuticals, textiles, cosmetics, and especially detergents (Adetunji and Olaniran, 2021). Furthermore, using lipases as a detergent additive is the most important industrial application (Moayd and Yan, 2017; Reyes-Reyes et al., 2022; Eskandari et al., 2024). As reported by the Brainy Insights, a company specializing in global and regional market research, the global microbial lipase market was valued at USD 518.40 million in 2021. This market is projected to grow from 2022 to 2030, reaching an estimated USD 898.40 million by 2030 (Rodríguez-Alonso et al., 2023). Detergent enzymes must be active in alkaline conditions and high temperatures, and stable in complex mixtures with Ca2+ ions, surfactants, oxidizers, and proteases. Lipases are suitable due to their broad substrate specificity, resilience in harsh washing conditions, and ability to remain active with various detergent components (Adetunji and Olaniran, 2021; Guncheva and Zhiryakova, 2011). Lipases in detergents effectively break down oily stains, reducing the need for harsh chemicals. They are also eco-friendly, leaving no toxic residues and being safe for aquatic ecosystems (Adetunji and Olaniran, 2021). Lipolase, derived from Thermomyces lanuginosus, was the first industrial lipase introduced into detergents, launched in 1988 by Novo Nordisk. Other lipases, such as Lumafast from Pseudomonas mendocina and Lipomax from Pseudomonas alcaligenes, were later commercialized by Genencor (Yao et al., 2021). The common source strains of lipase used in the detergent industry include Bacillus, Geobacillus, Pseudomonas, and Serratia genera (Chandra et al., 2020). Even though many lipases from different species are found in the literature, the enzyme market is still growing and the need for research of new enzymes remains necessary for potential biotechnological applications.
The production of low molecular weight flavor esters is crucial in food industry, where lipases serve as additives for various flavors and perfumes (Mehta et al., 2021). For instance, lipases from Bacillus aerius and Geobacillus sp., have been used to synthesize isoamyl acetate and methyl salicylate, respectively, both of which enhance the flavor of confectionery products such as chewing gums (Narwal and Gupta, 2013). Lipases were generally used as biocatalysts to enhance flavors and modify their structures through inter- or trans-esterification. Likewise, they modify flavor by synthesizing esters from short-chain fatty acids and alcohols, which are recognized as flavor and fragrance compounds (Mehta et al., 2021).
Streptomyces genus are among the most prolific producers of secondary metabolites, contributing to a significant portion of naturally derived bioactive compounds used in medicine today (Lacey and Rutledge, 2022). Historically, Streptomyces strains have been the primary source of the largest number of new antibiotic drugs as secondary metabolites compared to both bacteria and fungi. This genus represents a potent source of antibiotic including tetracycline, chloramphenicol, erythromycin, and aminoglycosides as well as quinine antibiotics (Taher et al., 2020). Additionally, Streptomyces spp. Produce a variety of extracellular enzymes especially xylanase, chitinase, and cellulase that play a crucial role in breaking down biomass into assimilable carbon units and are widely utilized in industrial and agricultural applications due to their significant large-scale fermentation capacity and proficiency in bulk manufacturing (Vasconcelos, 2018). While researchers are isolating novel Streptomyces strains from unexplored habitats to assess their secreted products potential and effectiveness, studies on Streptomyces lipases remain limited compared to those of other bacterial strains (Spasic et al., 2018). Despite the increasing number of studies in this area, further research is still needed to fully discover and understand lipase production and activity within the Streptomyces genus. As the demand for detergent-compatible enzymes and for esters production rises, ongoing efforts are being made to screen new bacteria capable of producing lipases with enhanced stability. This study focused on refining the culture conditions, purifying, and biochemically characterizing a novel thermostable lipase from Streptomyces gobitricini (S. gobitricini) strain. Additionally, the significant potential of this new lipase for industrial applications, particularly in detergent formulations and ester production, has been evaluated.
2 Results
2.1 Effect of medium components on LipS.g production
2.1.1 Incubation time
The effect of incubation time on LipS.g production was monitored over 176 h, as shown in Figure 1. During the initial 24 h, a notable increase in absorbance was observed, indicating the exponential growth phase, where S. gobitricini focused on rapid cell division and biomass accumulation rather than lipase production, a pattern common to many Streptomyces species (Ousaadi et al., 2021). Lipase activity started to rise significantly after 48 h, peaking at 84 h with an absorbance of 1.25 ± 0.06 (Figure 1). The production of lipase, reaching 22.50 ± 2.12 U/mL corresponding to 58925,5 U/g, was closely tied to cell growth during the stationary phase, when nutrient depletion and stress signals triggered the production of secondary metabolites like extracellular lipases (Bhatt et al., 2020). This increase was also attributed to the bacteria’s need to scavenge lipids as alternative carbon sources under nutrient-limited conditions. After 84 h, enzymatic activity declined gradually, falling to 6 U/mL (28284,2 U/g) by 152 h (Figure 1). The biomass, measured by absorbance at 600 nm, gradually declined between 84 and 176 h, reaching approximately 0.97 ± 0.042. This decrease is likely due to autolysis, as cells undergo breakdown in response to nutrient depletion and waste accumulation a typical process during the bacterial decline phase. This reduction in biomass also corresponded with decreased enzymatic activity, reflecting a loss of cell viability (Ousaadi et al., 2021). Similar findings were observed with Streptomyces sp. Al-Dhabi-49, isolated from Saudi Arabian soil, where lipase activity was initially detected at 24 h, peaking at 253 ± 4.4 U/mL after 5 days, before slightly declining to 172 ± 2.1 U/mL after 6 days 23 (Al-Dhabi et al., 2020). Additionally, Streptomyces sp. A3301 produced a thermostable lipase with an enzymatic activity of 108 U/mL after 168 h of incubation in the production medium (Al-Dhabi et al., 2020). Likewise, recombinant lipase from Streptomyces bacillaris expressed in Bacillus subtilis reached its maximal production after 12H fermentation (Gao et al., 2021). A potential thermostable lipase (designated MAS1) from the marine Streptomyces sp. Strain W007 was expressed in Pichia pastoris X-33. The growth curve showed that the maximum production of MAS1 (2.1 U/mL) was achieved at 72 h, when the cell density at 600 nm reached approximately (Yuan et al., 2016).
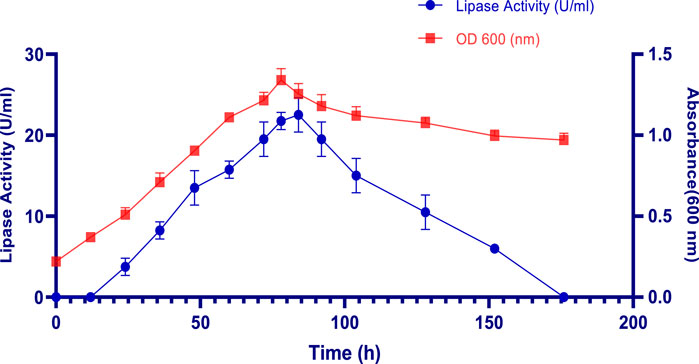
Figure 1. Effect of incubation time on S. gobitricini cell growth and lipase production. Results were presented as mean ± SD from three independent experiments.
2.1.2 Effect of physicochemical parameters on lipase production
To identify the favorable conditions for extracellular lipase production, key factors such as pH, temperature, and agitation speed were examined. These parameters are crucial for influencing enzyme yield and activity. Figure 2A illustrates the effect of temperature on lipase activity from S. gobitricini. At 30°C, activity was 21.75 ± 1.06 U/mL, increasing to 27 ± 4.24 U/mL at 40°C. The highest activity (34.5 ± 2.12 U/mL) occurred at 45°C, indicating a maximal temperature. This aligns with typical enzymatic behavior, where increased temperature enhances molecular motion and reaction rates (Iyer and Ananthanarayan, 2008; Yuan et al., 1999). However, activity declined at 50°C (28.5 ± 2.12 U/mL) and 55°C (21 ± 4.24 U/mL), suggesting enzyme denaturation. The ability of S. gobitricini to maintain extracellular lipase activity at elevated temperatures highlights its adaptation to extreme conditions.
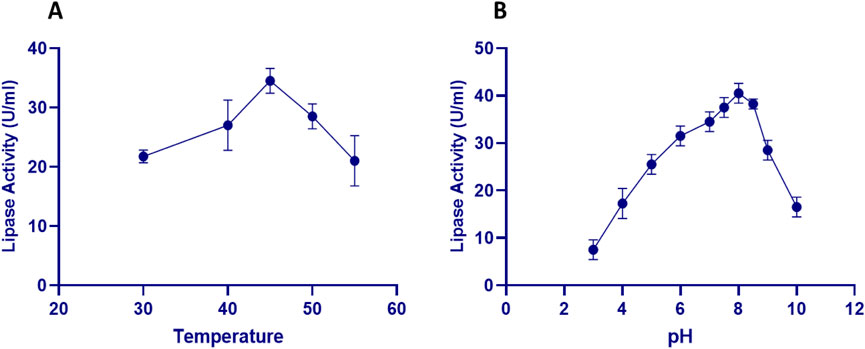
Figure 2. Variation in lipase activity produced by S. gobitricini at different temperatures (A) and pH values (B) using the basal medium containing 1% yeast extract and 1% glucose. Results were presented as mean ± SD from three independent experiments.
Lipase production by S. gobitricini varied with pH, showing a clear correlation between pH and enzyme functionality (Figure 2B). At pH 3.0, activity was low (7.5 ± 2.12 U/mL), indicating inhibition in acidic conditions. Activity increased with pH, peaking at 40.5 ± 2.12 U/mL at pH 8.0, suggesting performance in a slightly alkaline environment. Beyond this, activity declined, reaching 16.5 ± 2.12 U/mL at pH 10.0, highlighting reduced stability in highly alkaline conditions (Iyer and Ananthanarayan, 2008). Streptomyces adaptability to pH variations may be linked to physiological and biochemical mechanisms, including secondary metabolite production, intracellular pH regulation, and extracellular enzyme secretion (Sousa and Olivares, 2016). Similar trends were observed in Streptomyces strains like Streptomyces sp. Al-Dhabi-49 (pH 8.0, 168 ± 7.8 U/mL) (Al-Dhabi et al., 2020), S. clavuligerus (pH 6.8) (Dos Santos et al., 2017), and S. exfoliatus LP10 (pH 7.0) (Magda et al., 2012).
2.1.3 Carbone and nitrogen sources
Due to the significant importance of carbon catabolism in fermentation, the classical method ‘one variable at a time’ was adopted to enhance LipS.g production. The impact of carbon sources on lipase production by S. gobitricini is shown in Figure 3A. Without an added carbon source, activity was low (12.75 ± 1.06 U/mL), confirming the necessity of an external carbon source. Sorbitol moderately increased activity (26.25 ± 3.18 U/mL), while lactose and galactose further enhanced it (33 ± 4.24 and 34.5 ± 2.12 U/mL). Mannitol and xylose yielded similar levels (36 ± 4.24 and 36.75 ± 1.06 U/mL), with starch promoting higher activity (40.5 ± 2.12 U/mL). Glucose was the most effective, achieving 49.5 ± 3.53 U/mL. These findings align with studies on carbon source influence in Streptomyces strains like S. tanashiensis A2D, S. griseocarneus, and S. padanus PMS-702 (Ni et al., 2021). Glucose metabolism, regulated by glucokinase, plays a key role in enzyme production and alternative carbon utilization in Streptomyces (Diana et al., 2021). LipS.g activity peaked at 57.5 ± 3.53 U/mL at 2% glucose but declined at higher concentrations (Figure 3C), likely due to carbon catabolite repression (CCR), which regulates secondary metabolism and bioactive compound biosynthesis in Streptomyces (Lertcanawanichakul and Sahabuddeen, 2023).
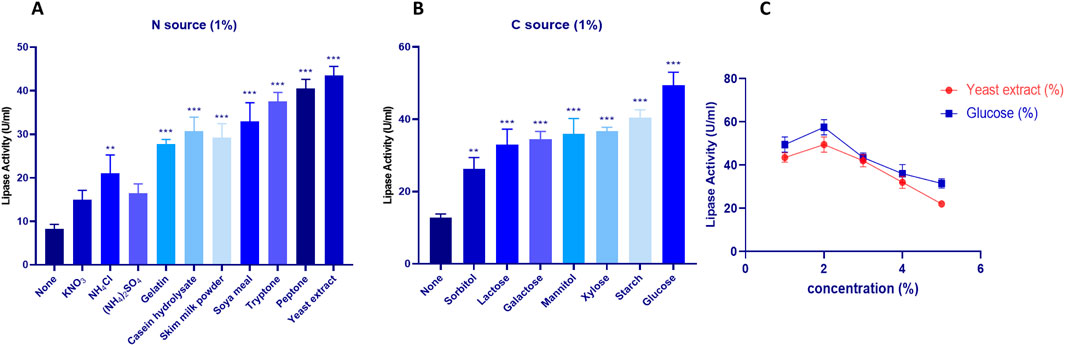
Figure 3. Impact of various Carbone sources at a concentration of 1% (A) and Nitrogen sources at a concentration 1% (B) on S. gobitricini lipase production. (C) Variation of lipase production with different concentrations of the selected Carbone source (Glucose) and Nitrogen source (Yeast extract). Results were presented as mean ± SD from three independent experiments, with asterisks indicating levels of significance (e.g., *p < 0.05; **p < 0.01; ***p < 0.001) above the bars in graphical representations.
Nitrogen is essential for metabolism and enzyme synthesis, influencing microbial growth and biochemical pathways (Blieva et al., 2021). Figure 3B shows the effect of nitrogen sources on lipase production by S. gobitricini. Without nitrogen, activity was low (8.25 ± 1.06 U/mL), confirming its necessity. Among inorganic sources, KNO3 led to moderate activity (15 ± 2.12 U/mL), while NH4Cl and (NH4)2SO4 produced 21 ± 4.24 and 16.5 ± 2.12 U/mL, respectively. Organic nitrogen sources significantly enhanced production, with gelatin (27.75 ± 1.06 U/mL) and casein hydrolysate (30.75 ± 3.18 U/mL). Higher activities were observed with skim milk (29.25 ± 3.18 U/mL) and soya meal (33 ± 4.24 U/mL), while tryptone (37.5 ± 2.12 U/mL), peptone (40.5 ± 2.12 U/mL), and yeast extract (43.5 ± 2.12 U/mL) yielded the highest activity. Yeast extract at 2% further enhanced production (Figure 3C), likely due to its rich composition of amino acids, vitamins, and growth factors that promote enzyme biosynthesis (Im et al., 2021). Similar trends were reported in Streptomyces strains, where glucose and malt extract enhanced lipase production in Streptomyces sp. Al-Dhabi-49 (Al-Dhabi et al., 2020), while NaNO3, tryptone, and peptone were the most effective for Streptomyces sp. TEM 33 (Cadirci et al., 2016).
2.1.4 Triglycerides, detergents and metallic ions
The primary determinant factor for bacterial lipase expression has consistently been the Carbon source, as lipases are typically inducible enzymes activated by lipid-based substrates, such as oils or triglycerides, fatty acids, hydrolysable esters, and tweens (Gupta et al., 2004). The impact of various triglycerides and detergents on lipase production by S. gobitricini was investigated to evaluate their effect on enzyme activity and substrate availability in the culture medium. Table 1 shows that triglycerides and detergents significantly influenced lipase production. Olive oil induced the highest activity (107.5 ± 4.94 U/mL), likely due to its favorable fatty acid profile, followed by soybean and sunflower oils. Tributyrin and trioctanoin had moderate effects. Among detergents, Tween 80 significantly boosted activity (126.5 ± 3.53 U/mL), likely by enhancing substrate emulsification, while Triton X-100 had a lesser effect, and Tween 20 slightly reduced production. These findings highlight the role of both triglycerides and detergents in maximal lipase production, likely by improving substrate availability (Gupta et al., 2004). Olive oil and Tween 80 were also identified as the most effective carbon sources in other Streptomyces strains, including S. griseus (117.88 U/mL) (Vishnupriya et al., 2010) and Streptomyces sp. TEM 33 (Cadirci et al., 2016). Maximum enzyme production (486.66 U/mL) was achieved using Tween 20 as a carbon source in soil-derived Streptomyces sp. (Praveen Kumar et al., 2017).

Table 1. Effect of triglycerides, detergents and metallic ions (at 1% concentration) on S. gobitricini lipase production. Results were presented as mean ± SD from three independent experiments, Significant differences in lipase production compared to control group, with asterisks indicating levels of significance (e.g., *p < 0.05; **p < 0.01; ***p < 0.001).
The effect of metal ions on lipase production by S. gobitricini showed that CaCl2 had the most significant impact, increasing activity to 65.25 ± 3.18 U/mL, suggesting its role in stabilizing enzyme structure and enhancing catalytic efficiency (Table 1). BaCl2 and CoCl2 also slightly increased activity (48 ± 4.24 and 47 ± 2.82 U/mL, respectively), while Mg2+, Zn2+, and Fe3+ had minimal or inhibitory effects possibly due to toxicity or interference with enzyme regulation. Metal ions likely influence lipase production by interacting with the enzyme’s active site, altering fatty acid solubility, and affecting bacterial metabolism. The enhancement observed with certain metal ions could be attributed to their ability to form complexes with ionized fatty acids in the medium, modifying their solubility and interfacial behavior. Additionally, metal ions play a role in bacterial metabolism, signaling pathways, and transport. While magnesium and zinc had little effect, Fe3+ significantly inhibited lipase production, potentially due to oxidative stress or disruption of essential cofactors required for enzyme expression (Ülker et al., 2011) (Hasan et al., 2006) (Zhang et al., 2012). In Streptomyces sp. Al-Dhabi-49, 0.1% Mg2+ increased lipase production to 163.7 ± 6.2 U/mL, whereas Hg2+ drastically reduced it (Al-Dhabi et al., 2020).
Beyond improving lipase production by S. gobitricini, the choice of fermentation method is crucial for ensuring scalability and efficiency. Two key approaches for microbial lipase production-submerged fermentation (SmF) and solid-state fermentation (SSF)-utilize agro-industrial residues as substrates. SmF, widely adopted in industrial applications, operates in a liquid medium, facilitating enzyme recovery and allowing precise control over critical parameters such as pH, temperature, and oxygen levels. In contrast, SSF, which requires minimal water, offers higher productivity and environmental benefits but presents engineering challenges at larger scales, such as maintaining uniform temperature and moisture gradients (Szymczak et al., 2021). Further studies are needed to enhance the scale-up of S. gobitricini production, which typically begins in bench-scale bioreactors before transitioning to larger systems for commercial use. The primary objective of scaling up is to replicate and enhance the efficiency of small-scale findings in larger bioreactors. Successful scale-up strategies depend on key parameters, including the volumetric oxygen transfer coefficient, volumetric power consumption, impeller tip speed, and mixing time.
2.2 LipS.g purification
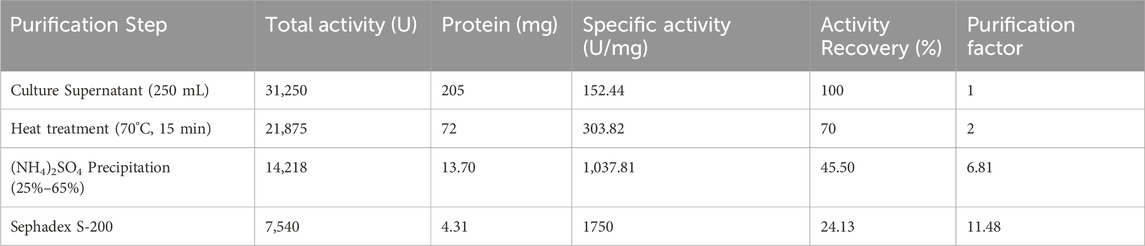
LipS.g was purified according to the procedure outlined in the materials and methods section. Initially, 250 mL of cultured supernatant was heat-treated at 70°C for 15 min. This was followed by fractionation using ammonium sulfate ((NH4)2SO4) at concentrations ranging from 25% to 65% (w/v). The resulting sample was then applied to a Sephadex S-200 column for further separation. Fractions exhibiting lipase activity, determined under standard assay conditions (Figure 4A) using olive oil as the substrate, were analyzed by SDS-PAGE. This revealed a single band with an approximate molecular weight of 50 kDa (Figure 4B). The molecular weight of the purified LipS.g was further confirmed by MALDI-TOF mass spectrometry, which determined a molecular mass of 53,262 Da (Figure 4C). Table 2 summarized the specific activity (SA) and recovery rates for LipS.g at each purification stage. The process achieved a recovery rate of 24.13% and a purification factor of 11.48. The specific activity of the purified lipase was 1750 U/mg under standard assay conditions. This result was consistent with other lipases purified from Streptomyces strains. For instance, a partially purified lipase from soil-derived Streptomyces sp. Exhibited a specific activity of 172.04 U/mg, a purification factor of 2.03, and a molecular weight of 45 kDa, as determined by SDS-PAGE analysis (Praveen Kumar et al., 2017). An organic solvent-tolerant lipase from Streptomyces sp. CS133 showed a 1.8-fold purification factor, with an estimated molecular mass of 39.8 kDa, also determined by SDS-PAGE (Zhang et al., 2012). In contrast, other studies reported lipases from Streptomyces strains with smaller molecular masses. For example, lipases from Streptomyces sp. CS326 and marine Streptomyces sp. W007 were found to migrate as single protein bands corresponding to molecular masses of 17 kDa and 29 kDa, respectively (Cho et al., 2012; Yuan et al., 1999).
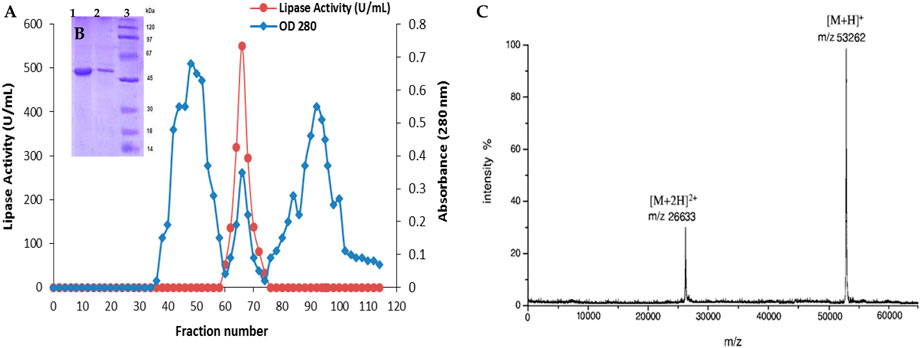
Figure 4. Chromatography profile on Sephadex S-200 column (A), SDS-PAGE analysis (B) and MALDITOF analysis (C) of the purified LipS.g. Figure 4B: Lane 1 and 2: purified LipS.g (5 and 10 μg, respectively). Line 3: molecular mass markers.
While the purification of LipS.g was successfully achieved at the laboratory scale, scaling up the process introduces several technical and economic challenges that must be carefully addressed. In chromatography-based purification, scale-up is typically achieved by increasing the column diameter and volumetric flow rate while maintaining a constant media bed height and linear flow rate. This approach ensures consistent residence time across different scales, preserving separation efficiency. However, large-scale purification presents additional complexities, including buffer selection, media packing, column engineering, and process hygiene, all of which can impact commercial biopharmaceutical manufacturing. The choice of chromatography media is particularly critical, as it directly influences purity levels and downstream processing efficiency (Milne, 2017).
2.3 LipS.g biochemical characterization
2.3.1 Effect of temperature and pH on LipS.g activity and stability
The purified LipS.g exhibited maximal activity at 50°C, with stability maintained between 40°C and 55°C but declining above 55°C due to thermal denaturation (Figure 5A). Its thermostability may be linked to structural factors such as hydrophobic clustering and salt-bridge density (Rabbani et al., 2023). LipS.g also showed highest activity at pH 9.0, with stability maintained from pH 5.0 to 10.0, but decreasing in extreme acidic or basic conditions (Figure 5B). These characteristics align with microbial lipases, such as Streptomyces sp. CS326 (pH 7.0, 40°C) (Cho et al., 2012), S. bacillaris (45°C, pH 9.0) (Gao et al., 2021), and lipase LS133 from Streptomyces sp. CS133 (stable at pH 5.0–9.0, maximal activity at pH 7.5, 40°C) (Mander et al., 2012). The purified LipS.g exhibited peak activity at 50°C, with stability maintained between 40°C and 55°C but declining above 55°C due to thermal denaturation (Figure 5A). Its thermostability may be linked to structural factors such as hydrophobic clustering and salt-bridge density (Rabbani et al., 2023). LipS.g also showed a highest activity at pH 9.0, with stability maintained from pH 5.0 to 10.0, but decreasing in extreme acidic or basic conditions (Figure 5B). These characteristics align with microbial lipases, such as Streptomyces sp. CS326 (maximal activity at pH 7.0, 40°C) (Cho et al., 2012), S. bacillaris (maximal activity at 45°C, pH 9.0) (Gao et al., 2021), and lipase LS133 from Streptomyces sp. CS133 (stable at pH 5.0–9.0, maximal activity at pH 7.5, 40°C) (Mander et al., 2012).
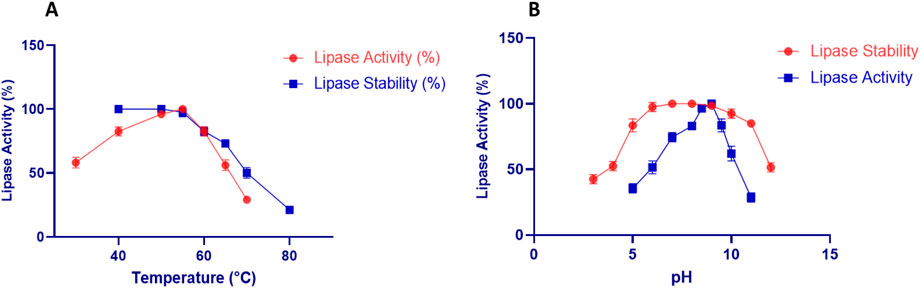
Figure 5. Impact of various temperatures (A) and pHs (B) on LipS.g activity and stability. Results were presented as mean ± SD from three independent experiments.
2.3.2 Effect of calcium on LipS.g activity
While cofactors are generally not essential for lipase activity, divalent cations, particularly calcium, often enhance catalytic activity. This enhancement is attributed to increased interaction between calcium ions and long-chain fatty acids during hydrolysis, which facilitates the enzyme’s catalytic function (Gupta et al., 2004). In this context, the effect of calcium on LipS.g activity showed that increasing calcium concentrations activated the enzyme, with maximum activity (100%) observed at 1.5–2 mM (Figure 6). At lower concentrations or in total absence of calcium (0 and 0.5 mM), enzyme activity was significantly reduced, indicating that calcium is essential for highest functional performance. Moreover, metal ions like calcium are known to interact with specific pockets on the enzyme surface, stabilizing and maintaining the conformation of flexible segments of the polypeptide chain through coordination. This structural support further enhanced LipS.g activity (Shirazi et al., 2013). However, beyond 2 mM, enzyme activity started to decline, with a steady decrease observed at higher calcium concentrations (3–8 mM) (Figure 6).
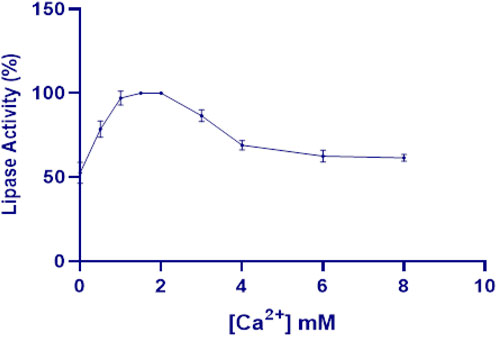
Figure 6. Effect of various concentrations of calcium on LipS.g activity. Results were presented as mean ± SD from three independent experiments.
Several Streptomyces lipases exhibit similar responses to calcium ions. For example, lipase from S. bacillaris displayed increased activity at calcium concentrations of 1 mM and 10 mM, reaching 116.2% ± 1.3% and 124.3% ± 2.7% activity, respectively (Gao et al., 2021). In contrast, lipase activities from Streptomyces sp. Strain W007 and Streptomyces sp. CS133 were unaffected by calcium at 1 mM, retaining 100% activity regardless of calcium presence (Mander et al., 2012; Yuan et al., 1999).
2.3.3 LipS.g substrate specificity and apparent kinetic parameters determination
The substrate specificities of microbial lipases, especially regarding fatty acid and positional preferences, are crucial for predicting their catalytic activity and selecting specific lipases for targeted reactions, making them highly relevant for various applications (Song et al., 2008). The substrate specificity of purified LipS.g was assessed using a range of synthetic and natural substrates (Table 3). Data in Table 3 indicated that LipS.g showed the highest activity with TC18 (100%, followed closely by TC4 (92% ± 2.82%) and TC8 (91% ± 4.24%), suggesting a preference for long chain triglycerides. Among the tested oils, coconut and sesame oils exhibited high activity (88% ± 4.24% and 87% ± 4.24%, respectively), while sunflower oil showed the lowest (77% ± 2.82%). This could be attributed to their polyunsaturated fatty acid content, particularly oleic acid (C18:1 ω9) (Bacha et al., 2019). LipS.g’s substrate preference may be due to specific enzyme-substrate interactions, influenced by the acyl-binding site’s hydrophobic residues (Albayati et al., 2020). Similar selectivity has been observed in Streptomyces lipases, such as Streptomyces sp. CS326, which preferred long-chain fatty acids (C16) (Cho et al., 2012), S. bacillaris, which showed maximal activity with C16 and significant activity with C14 and C12 (Gao et al., 2021), and Streptomyces sp. CS133, which favored C10–C18 substrates (Mander et al., 2012). Streptomyces sp. W007 hydrolyzed esters from C4 to C18, with a preference for C8 (Yuan et al., 1999).
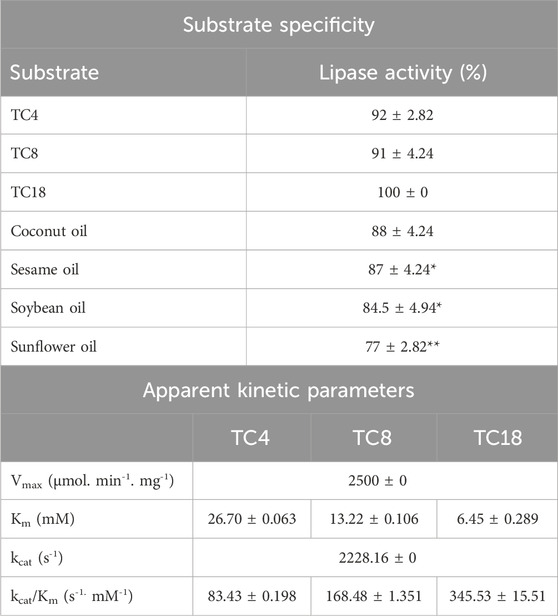
Table 3. Substrate specificity and apparent kinetic parameters of purified LipS.g. Results were presented as mean ± SD from three independent experiments Significant differences in lipase activity compared to TC18, with asterisks indicating levels of significance (e.g., *p < 0.05; **p < 0.01; ***p < 0.001).
Kinetic analysis of purified LipS.g using Lineweaver–Burk plots (Figure 7) revealed a maximum reaction rate (Vmax = 2500 μmol min-1·mg-1) and a turnover number (kcat 2228.16 s-1) across all substrates. The enzyme exhibited varying kinetic behaviors toward different triglycerides. Notably, the lowest apparent Km was observed for TC18 (6.45 mM) and the highest for TC4 (26.70 mM), while the catalytic efficiency (kcat/Km) increased with chain length-ranging from 83.43 s-1 mM-1 for TC4 to 345.53 s-1 mM-1 for TC18. While Km is sometimes interpreted as an inverse measure of enzyme-substrate affinity, this is only valid under strict Michaelis–Menten assumptions such as rapid equilibrium between Enzyme and Enzyme-Substrate and no allosteric or cooperative behavior. More precisely, Km represents the substrate concentration at which the reaction rate is half of Vmax, and its value reflects a combination of binding and catalytic rate constants. Therefore, in this study, differences in Km values likely reflect both substrate binding affinity and catalytic steps, influenced by the structural compatibility of each triglyceride with the enzyme’s active site. Kinetic parameters of various Streptomyces lipases show considerable variation, likely due to differences in substrates and methods used to measure lipase activity. For example, the lipase from Streptomyces sp. CS326, using pNPP as a substrate, displayed Km and Vmax values of 0.24 mM and 4.6 Mm min-1·mg-1, respectively (Cho et al., 2012).
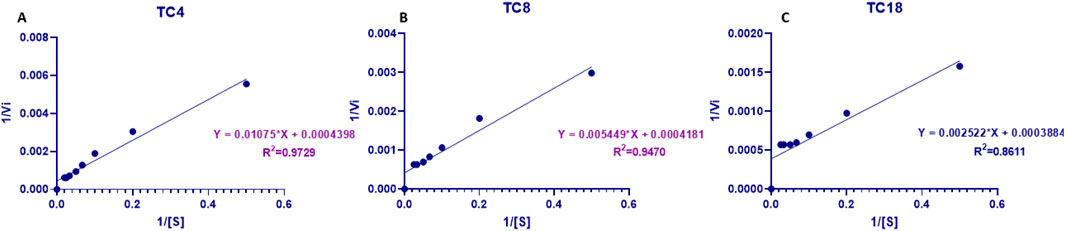
Figure 7. Lineweaver-Burk plots of LipS.g at varying substrate concentrations: (A) TC4, (B) TC8, and (C) TC18.
2.4 LipS.g biotechnological applications
2.4.1 Detergence application
Enzyme stability in the presence of surfactants and oxidizing agents is essential for applications in detergent formulations (Alonazi, 2024). For effective washing performance, a detergent lipase must be compatible with and stable in commonly used detergent components such as surfactants, bleaches, oxidizing agents, and other additives (Sarac et al., 2015). Thus, the stability of LipS.g, compared to SAL4 and Lipolase, was assessed against various surfactants, oxidizing agents, boron compounds, and enzyme inhibitors (Table 4). LipS.g exhibited the highest stability across all surfactants, maintaining or enhancing its activity, particularly in sodium cholate (110% ± 5%), likely due to enhanced electrostatic interactions with the protein (Delorme et al., 2011). It also showed strong performance in Triton X-100 (105% ± 6.24%) and Tween 80 (105% ± 5%), where hydrophobic interactions played a significant role (Delorme et al., 2011) (Table 4). Similarly, SAL4 demonstrated activity above 100% in most surfactants, with notable stability in sodium cholate (107.33% ± 2.51%) and Triton X-100 (105.66% ± 1.15%). These results were consistent with structural studies suggesting that surfactants can improve substrate availability by emulsifying, forming mixed micelles, and inducing lipase conformational changes that increase active site accessibility (Delorme et al., 2011). In contrast, Lipolase displayed a marked decline in Triton X-100 (49.66% ± 4.16%) compared to sodium taurocholate (80.16% ± 2.02%) and sodium cholate (87.9% ± 3.15%) (Table 4), likely due to surfactant interactions that reduce lipolytic efficiency by forming inactive enzyme-surfactant complexes or impairing substrate binding at the interface (Delorme et al., 2011). This highlighted the importance of carefully evaluating surfactant effects, as they could either enhance or inhibit lipase activity, depending on their type. Among purified Streptomyces lipases, only the lipase from marine Streptomyces sp. Strain W007 has been tested against various surfactants. This enzyme showed reduced activity in dioctyl sulfosuccinate and SDS but increased to 133.91% after 2 h-incubation in N-lauroyl sarcosine sodium. Zwitterionic surfactants like soy lecithin and sulfopropyl betaine also decreased activity significantly. Nonionic detergents such as Tween-20, −60, −80, Triton X-100, and nonylphenol ethoxylates inhibited this enzyme, although it retained 84.5% activity with polyethylene oxide lauryl ether (Yuan et al., 1999).

Table 4. Biotechnological applications of purified LipS.g: stability in surfactant, oxidizing agent, boron compounds and enzymes inhibitors. SAL4 and Lipolase were used as positive controls for a comparative study. Results were presented as mean ± SD from three independent experiments, with asterisks indicating levels of significance (e.g., *p < 0.05; **p < 0.01; ***p < 0.001).
When exposed to oxidizing agents, SAL4 exhibited the highest resistance, maintaining substantial activity in hydrogen peroxide (89.33% ± 4.04%) and sodium perborate (91% ± 3.06%). In contrast, Lipolase experienced significant reductions, particularly in sodium hypochlorite (51% ± 6.46%) and hydrogen peroxide (55.33% ± 5.27%) (Table 4). Among the boron compounds tested, LipS.g demonstrated the best stability, especially in BKO2 (108.33% ± 3.51%) and Na2B4O7 (106.66 %± 3.60%), while Lipolase showed the weakest performance, particularly in BKO2 (78% ± 4%) and H3BO3 (69% ± 3%) (Table 4).
The use of boron-based compounds is particularly relevant because they are commonly found in detergents and industrial formulations, where lipases are frequently applied. Testing enzyme stability in their presence helps to assess the potential compatibility and robustness of lipases under real industrial conditions. Furthermore, boron compounds such as sodium perborate and borax can act as mild oxidants or buffer agents, making them valuable tools to evaluate enzyme resilience to oxidative and ionic stress (Sarac et al., 2015).
Hydrogen peroxide is known to oxidize surface-exposed residues such as methionine, cysteine, tyrosine, and tryptophan in proteins, which can lead to a reduction in enzymatic activity. According to Törnvall et al. (2010), SAL4 and LipS.g seemed to have fewer oxidation-sensitive amino acids compared to Lipolase. Notably, all cysteine residues in SAL4 and LipS.g are likely involved in disulfide bonds, which are more resistant to oxidation than free cysteines. Oxidation can also alter the net charge and induce changes in the secondary and tertiary structures of enzymes (Törnvall et al., 2010).
In the presence of enzyme inhibitors, both LipS.g and SAL4 demonstrated strong resistance, retaining over 80% activity in SDS, PMSF, and EDTA. In contrast, Lipolase was more affected, particularly by SDS (72.33% ± 2.51%) and EDTA (64.33% ± 4.72%) (Table 4). The observed stability of LipS.g in the presence of PMSF, a well-known serine inhibitor, could be attributed to the serine residue being buried within the enzyme’s hydrophobic core, making it less accessible to PMSF (Sharma et al., 2012). Similarly, the stability of LipS.g and SAL4 in the presence of EDTA could be linked to the inaccessibility of the calcium ions, which enhance lipase activity. While EDTA likely chelates these ions, the enzyme remains unaffected due to their reduced exposure (Invernizzi et al., 2009). Additionally, the stability of LipS.g in the presence of SDS may be explained by the formation of an SDS-lipase complex, which increases the hydrophobicity of the protein surface at low concentrations (Delorme et al., 2011).
Overall, LipS.g and SAL4 consistently outperformed Lipolase in all tested conditions, making them more suitable for applications requiring high stability in detergent formulations and resistance to environmental stressors. The enhanced stability of LipS.g in the presence of oxidizing agents, boron compounds, and enzyme inhibitors makes it an ideal candidate for incorporation into detergent formulations (Guncheva and Zhiryakova, 2011).
Figure 8 illustrated the stability of LipS.g in solid and liquid detergents at concentrations of 5 mg/mL (Figure 8A) and 1/100, respectively (Figure 8B), compared to SAL4 and Lipolase. The data showed that LipS.g exhibited moderate stability in solid detergents, with residual activity ranging from 71% ± 4% to 85.3% ± 5.5% (Figure 8A). However, SAL4 consistently outperformed LipS.g, demonstrating superior stability with over 90% activity in most solid detergents, indicating its greater resistance to detergent components (Figure 8A). In contrast, Lipolase exhibited the lowest residual activity, dropping to around 50% in Nadhif and Tide, highlighting its significantly lower stability compared to both LipS.g and SAL4. In liquid detergents, SAL4 maintained the highest activity, with over 83% in all detergents, peaking at 93.33% ± 2.5% in Ariel (Figure 8B). LipS.g’s activity ranged from 64.33% ± 4.04% in Dac to 79.33% ± 4.04% in Dixan, while Lipolase was the least stable, with residual activities ranging from 46.67% ± 3.51% in Nadhif to 80.33% ± 1.52% in Fairy (Figure 8B). Thus, LipS.g and SAL4 demonstrated greater stability than Lipolase, highlighting their resistance to the denaturing effects of detergents and their potential as strong candidates for detergent formulations requiring high enzyme stability.
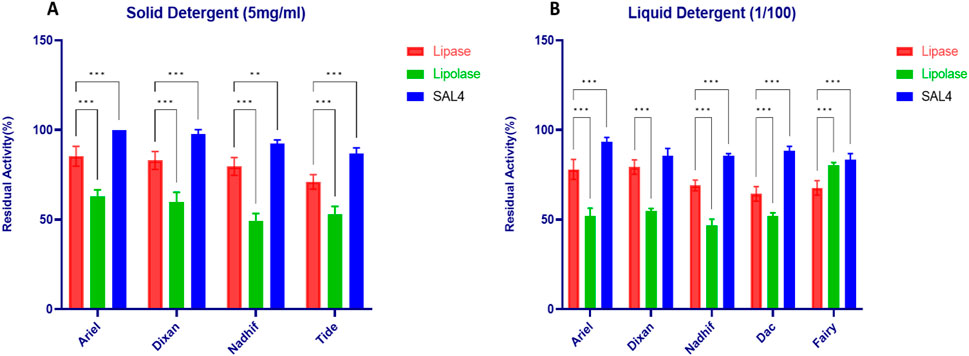
Figure 8. Stability of purified LipS.g in (A) Solid detergent at 5 mg/mL and (B) liquid detergent at 1/100 ratio. SAL4 and Lipolase were used as positive control to a comparative study. Results were presented as mean ± SD from three independent experiments, asterisks above bars indicate significant differences with the LipS.g (*p < 0.05; **p < 0.01; ***p < 0.001) (Dunnett test).
Several studies have reported bacterial lipases with high stability in liquid and solid detergents, as well as in detergent compounds such as surfactants, oxidizing agents, and enzyme inhibitors. For example, lipase produced by Pseudomonas helmanticensis HS6 strain retained 40%–80% activity after 3 h-incubation with commercial detergents, underscoring its potential for detergent industry applications (Phukon et al., 2020). A lipase from Bacillus subtilis has demonstrated resistance to surfactants, oxidizing agents, and commercial detergents (Saraswat et al., 2017). Likewise, Geobacothermophilus FMR12 exhibited high lipolytic activity at 70°C and pH 9.0, proving effective in detergent applications. Common strains used in detergent formulations include Bacillus licheniformis, Geobacillus species, Serratia marcescens DEPTK21, Bacillus flexus XJU-1, Bacillus pumilus SG2, as well as Staphylococcus arlettae, Bacillus cepacia, Pseudomonas fluorescens, and Candida species (Yao et al., 2021).
2.4.2 LipS.g-catalyzed ester formation
Lipases, a class of serine hydrolases, catalyze reactions such as esterification, interesterification, and transesterification in non-aqueous media, including organic solvents and supercritical fluids, making them valuable for producing biofuels and other esters (Stergiou et al., 2013; Martin et al., 2019; Mandari and Devarai, 2022). Among these, lipase-catalyzed esterification reactions hold significant industrial importance (Stergiou et al., 2013; Gamayurova et al., 2021). To evaluate its ester synthesis capabilities, LipS.g was tested under various conditions and compared to SAL4, a known biocatalyst in esterification (Ben Bacha et al., 2016). As shown in Table 5, LipS.g demonstrated varying conversion efficiencies across different ester substrates, similar to SAL4. Notably, LipS.g effectively synthesized antioxidants like propyl gallate and tyrosol acetate, with conversion yields of 45% ± 3.21% and 35.5% ± 4.04%, respectively (Table 4). Additionally, LipS.g catalyzed the production of butyl acetate (a pineapple flavor compound) and ethyl oleate (a biofuel) with yields of 41% ± 2.08% and 48.33% ± 2.5%, respectively, compared to SAL4’s yields of 39.34% ± 3.6% for butyl acetate and 31% ± 1.5% for ethyl oleate. Moreover, LipS.g catalyzed the esterification of oleic acid with starch, producing starch oleate, a potential fully biodegradable thermoplastic, and esterified ricinoleic acid, achieving yields of 34% ± 3.05% and 42% ± 3.05% for starch oleate and ricinoleic esters, respectively (Table 4). These esterification reactions were carried out using the ethanol as alcohol. Starch oleate, a biodegradable and non-toxic sugar fatty acid ester, is hydrolyzed in vivo by pancreatic lipase. Its synthesis through lipase-catalyzed esterification under mild conditions offers an eco-friendly alternative to conventional chemical methods that rely on acyl chlorides and toxic solvents (Rodríguez-Alonso et al., 2023). The esters synthesized serve as versatile compounds commonly used as flavors, fragrances, and antioxidants, with extensive applications in the food, beverage, cosmetic, and pharmaceutical industries (Stergiou et al., 2013). Conventional ester synthesis methods are unsustainable and generate materials with low biodegradability. In contrast, lipases offer a greener alternative for both polymer synthesis and degradation, particularly in biomedical applications. Their enzymatic approach reduces environmental impact by minimizing waste, lowering risks, and utilizing renewable resources, making them a promising tool for advancing sustainable polymer chemistry (Freije García and García Liñares, 2024).
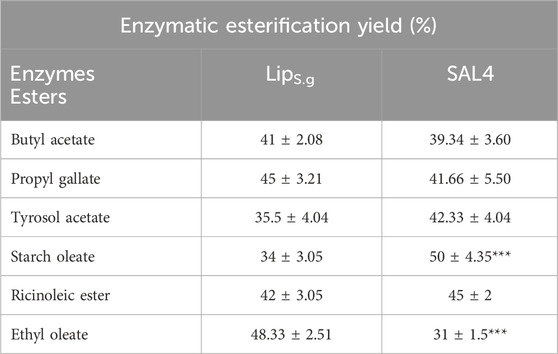
Table 5. Production of esters by LipS.g compared to SAL4. Results were presented as mean ± SD from three independent experiments, with asterisks indicating levels of significance (e.g., *p < 0.05; **p < 0.01; ***p < 0.001).
3 Materials and methods
3.1 Materials and reagents
Chromatography material (Sephadex S-200), SDS-PAGE technique, pH-stat and rotary shaker were obtained from Bio-Rad (Hercules, CA, United States). Voyager DE-RP MALDI-TOF mass spectrometer was obtained from Biosystem, Framingham, MA, United States.
Chemicals were obtained from commercial sources. Glucose, galactose, mannitol, lactose, sorbitol, starch, or xylose, casein hydrolysate, gelatin, peptone, skim milk powder, soya meal, yeast extract, tryptone, ammonium nitrate (KNO3), ammonuim chloride (NH4Cl), or ammonium dihydogen phosphate ((NH4)2SO4)), surfactants (Tween 20, Tween 80, or Triton X-100), metal ions (BaCl2, CaCl2, COCl2, MgCl2, MgSO4, FeCl3, or ZnCl2), Ethylenediaminetetraacetic acid (EDTA), Sodium Dodecyl Sulfate (SDS), or Phenylmethylsulfonyl fluoride (PMSF), surfactants (NaDC, NaTDC, Triton X-100, Tween-80, or Tween-20), boron compounds (BKO2, H3BO3, NaBO2, or Na2B4O7) and oxidants (hydrogen peroxide (H2O2), sodium perborate or sodium hypochlorite), were purchased from Bio-Rad (Hercules, CA, United States).
Lipid compounds (olive oil, coconut oil, sesame oil, soybean oil, sunflower oil, trioctanoin (TC8), and triolein (C18) were purchased from Sigma Aldrich (St. Quentin-Fallavier, France). Commercial lipolase was from Novo Nordisk, Denmark. Various laundry detergents were used including Ariel (Procter and Gamble, Switzerland), Dixan (Henkel, Spain), Nadhif (Henkel-Alki, Tunisia), and Tide (Procter and Gamble, Saudi Arabia). Liquid detergents employed were DAC (Henkel, Saudi Arabia), Dixan (Henkel, Spain), Fairy (Modern Industries, Saudi Arabia), and Nadhif (Henkel-Alki, Tunisia).
3.2 Medium conditions and components for lipase production
S. gobitricini strain used for lipase production in this study has been identified and deposited in a culture collection under the designation FA-KSU 23, with the accession number PP708563. This strain was isolated from polluted mangrove soil, identified, and preserved in the Botany and Microbiology Department, College of Science, King Saud University (Riyadh, Saudi Arabia).
To prepare the S. gobitricini culture, 50 mL of Basal Broth medium (pH 7.0) was dispensed into 500 mL Erlenmeyer flasks. After sterilization, approximately 2 mL (4 × 106 CFU/mL) of bacterial suspension, which had been grown in nutrient broth at 30°C for 2 days, was inoculated into the flask and incubated at 30°C for 7 days on a rotary shaker at 270 g.
The determination of media components, including Carbon at a concentration 1% (glucose, galactose, mannitol, lactose, sorbitol, starch, or xylose), Nitrogen at a concentration of 1% (casein hydrolysate, gelatin, peptone, skim milk powder, soya meal, yeast extract, tryptone, KNO3, NH4Cl, or (NH4)2SO4), surfactants (Tween 20, Tween 80, or Triton X-100), lipid compounds (olive oil, soybean oil, sunflower oil, TC4, or TC8), and metal ions (BaCl2, CaCl2, COCl2, MgCl2, MgSO4, FeCl3, or ZnCl2), as well as culture conditions such as incubation time (0–176 h), temperature (30°C–55°C), pH (pH 3.0–10.0), and agitation speed (67–270 g), was conducted by replacing the components in the initial production media. Samples were collected and subjected to centrifugation (10 min at 9,000 g) with the supernatant used as the crude enzyme extract for lipase assays. Growth was assessed by measuring the optical density of the culture at 600 nm. All experiments were performed in triplicate under aseptic conditions, with results reflecting the averages of these three trials.
An improved medium incorporating the best sources of carbon, nitrogen, detergent, triglycerides, and metal ions for enhanced lipase production was then developed, and the bacteria were cultivated in this refined medium.
3.3 Purification of LipS.g
Lipase from S. gobitricini was produced in the refined medium at 45°C over 84 h. Cells were removed via centrifugation (30 min at 9,000 g), and the resulting crude enzyme solution (250 mL, 31,250 Total Units (UT)) was incubated for 15 min at 70°C. Following rapid cooling and subsequent centrifugation for 30 min at 9,000 g, the supernatant (242 mL, 21,875UT) was fractionated using solid ammonium sulfate at 20%–65% saturation. The precipitate obtained post-centrifugation was resuspended in 25 mM Tris–HCl, pH 8.0, supplemented with 50 mM NaCl and 2 mM benzamidine (12 mL, 14,218UT), and loaded onto a Sephadex S-200 column (1.6 × 110 cm) pre-equilibrated with the same buffer. Every 6 min, 2.0 mL fractions were collected and analyzed for protein content and lipase activity. Fractions exhibiting high lipase activity were pooled (50 mL, 7,540 UT), concentrated, and stored at 4°C until further use.
3.4 Determination of lipase activity
Using a pH-stat at 50°C and pH 9.0, lipolytic activity was determined titrimetrically with substrates including an olive oil emulsion stabilized by 10% Gum Arabic and TC4 (Rathelot et al., 1981). Some assays were performed in the presence of different concentrations of Ca2+ (0–8 mM) at pH 9.0 and at 50°C. For kinetic studies, coconut oil, sesame oil, soybean oil, sunflower oil, trioctanoin (TC8), and triolein (C18) were also used to measure enzyme activity (Abdelkafi et al., 2009). Lipase activity was quantified in international units (U), defined as the production of 1 μmol of fatty acid per minute.
3.5 Protein analysis
Bradford protocol was used to assess the protein content (Bradford, 1976). The purified lipase was subjected to electrophoretic analysis using SDS-PAGE (15%) following the Laemmli method (Laemmli, 1970).
The molecular mass of the native protein was then accurately determined on a Voyager DE-RP MALDI-TOF mass spectrometer (Biosystem, Framingham, MA, United States). Mass spectra recorded in linear mode were externally calibrated with suitable standards and analyzed by the GRAMS/386 Software.
3.6 LipS.g characterization
3.6.1 pH and temperature effects on LipS.g activity and stability
Lipase activity was evaluated in different buffers at 0.5 M across a large range of pHs (5.0–11.0) at a temperature of 50°C. The stability of lipase at different pHs was assessed by incubating the enzyme at pH values between 3.0 and 11.0 for 1 hour at room temperature. The residual lipase activity was assessed following centrifugation under the standard assay method. The highest temperature for purified lipase activity was established by conducting enzyme assays across a range of temperatures (30°C–70°C) at pH 9.0. The thermal stability was assessed by incubating the lipase at pH 8.0 across a range of temperatures (40°C–80°C) and evaluating the residual activity after 1 h, following centrifugation, under standard titrimetric assay conditions. Each measurement was conducted on three separate assays.
3.6.2 Kinetic study
Lipase activities were assessed as a function of different concentrations (0–40 mM) of various substrates (TC18, TC8 or TC4). The maximum velocity (Vmax) and the apparent Michaelis-Menten constant (Kmapp) for each reaction were determined by Lineweaver-Burk plot.
3.6.3 Compatibility of LipS.g with oxidizing agents, surfactants, and commercial detergents
LipS.g was evaluated for its potential use in the detergent industry. 120 U sample of LipS.g (from the (NH4)2SO4 fraction) was incubated with various enzyme inhibitors (EDTA, SDS, or PMSF), surfactants (NaDC, NaTDC, Triton X-100, Tween-80, or Tween-20), boron compounds (BKO2, H3BO3, NaBO2, or Na2B4O7) and oxidants (hydrogen peroxide (H2O2), sodium perborate or sodium hypochlorite) at 1% for 60 min at 40°C.
Compatibility was also assessed with various commercial solid laundry detergents (Ariel (Procter and Gamble, Switzerland), Dixan (Henkel, Spain), Nadhif (Henkel-Alki, Tunisia), and Tide (Procter and Gamble, Saudi Arabia)) and liquid detergents (DAC (Henkel, Saudi Arabia), Dixan (Henkel, Spain), Fairy (Modern Industries, Saudi Arabia), and Nadhif (Henkel-Alki, Tunisia)) following the method of Cherif et al. (2011). Results were compared to commercial lipolase (Novo Nordisk, Denmark) and Staphylococcus aureus lipase (SAL4) from a previous study (Ben Bacha et al., 2018). All the experiments of characterization study were performed three times.
3.6.4 Enzymatic esterification
The different esterification reactions were performed according to previously described protocols (Ghamgui et al., 2007). For comparison, SAL4 and commercial lipase were used as positive controls.
3.6.5 Statistical analysis
All of the assays were done in biological triplicate with three technical replicates, and data were given as mean ± standard deviation (SD). The statistical analysis was carried out through one-way and two-way analysis of variance (ANOVA) as well as Duncan’s post hoc test using GraphPad Prism version 9 and Excel. A p-value of <0.05 was regarded as significant, and asterisks were used to indicate significance.
4 Conclusion
LipS.g, a novel lipase from S. gobitricini, exhibited high enzyme activity under the best conditions that we assesed and remarkable stability in detergent formulations. Its efficiency in ester synthesis, including antioxidants and biofuels, highlighted its potential as a versatile biocatalyst. These properties suggested a wide range of industrial applications, notably in the food, beverage, cosmetics, and pharmaceutical sectors.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AA: Data curation, Formal Analysis, Resources, Software, Writing – review and editing. NK: Investigation, Methodology, Validation, Writing – original draft. MA: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Writing – review and editing. JA-G: Data curation, Investigation, Project administration, Resources, Writing – review and editing. HH: Conceptualization, Data curation, Validation, Visualization, Writing – review and editing. AB: Conceptualization, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors extend their appreciation to the researchers’ Supporting Project number (RSP2025R237), King Saud University, Riyadh, Saudi Arabia, for funding this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelaziz, A. A., Abo-Kamar, A. M., Elkotb, E. S., and Al-Madboly, L. A. (2025). Microbial lipases: advances in production, purification, biochemical characterization, and multifaceted applications in industry and medicine. Microb. Cell Factories 24 (1), 40. doi:10.1186/s12934-025-02664-6
Abdelkafi, S., Fouquet, B., Barouh, N., Durner, S., Pina, M., Scheirlinckx, F., et al. (2009). In vitro comparisons between carica papaya and pancreatic lipases during test meal lipolysis: potential use of CPL in enzyme replacement therapy. Food Chem. 115 (2), 488–494. doi:10.1016/j.foodchem.2008.12.043
Adetunji, A. I., and Olaniran, A. O. (2021). Production strategies and biotechnological relevance of microbial lipases: a review. Braz. J. Microbiol. 52 (3), 1257–1269. doi:10.1007/s42770-021-00503-5
Albayati, S. H., Masomian, M., Ishak, S. N. H., Mohamad Ali, M. S. B., Thean, A. L., Mohd Shariff, F. B., et al. (2020). Main structural targets for engineering lipase substrate specificity. Catalysts 10 (7), 747. doi:10.3390/catal10070747
Al-Dhabi, N. A., Esmail, G. A., Ghilan, A.-K. M., and Arasu, M. V. (2020). Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi J. Biol. Sci. 27 (1), 474–479. doi:10.1016/j.sjbs.2019.11.011
Al Mohaini, M., Farid, A., Muzammal, M., Ghazanfar, S., Dadrasnia, A., Alsalman, A. J., et al. (2022). Enhancing lipase production of Bacillus salmalaya strain 139SI using different carbon sources and surfactants. Appl. Microbiol. 2 (1), 237–247. doi:10.3390/applmicrobiol2010017
Alonazi, M. (2024). Staphylococcus aureus alkaline protease: a promising additive for industrial detergents. Catalysts 14 (7), 446. doi:10.3390/catal14070446
Bacha, A. B., Abid, I., Nehdi, I., and Horchani, H. (2019). Hydrolysis of oils in the wadi hanifah river in Saudi Arabia by free and immobilized Staphylococcus aureus ALA1 lipase. Environ. Prog. Sustain. Energy 38 (3), e13000. doi:10.1002/ep.13000
Ben Bacha, A., Al-Assaf, A., Moubayed, N. M. S., and Abid, I. (2018). Evaluation of a novel thermo-alkaline Staphylococcus aureus lipase for application in detergent formulations. Saudi J. Biol. Sci. 25 (3), 409–417. doi:10.1016/j.sjbs.2016.10.006
Ben Bacha, A., Moubayed, N. M., and Al-Assaf, A. (2016). An organic solvent-stable lipase from a newly isolated Staphylococcus aureus ALA1 strain with potential for use as an industrial biocatalyst. Biotechnol. Appl. Biochem. 63 (3), 378–390. doi:10.1002/bab.1381
Bharathi, D., and Rajalakshmi, G. (2019). Microbial lipases: an overview of screening, production and purification. Biocatal. Agric. Biotechnol. 22, 101368. doi:10.1016/j.bcab.2019.101368
Bhatt, K., Lal, S., R, S., and Joshi, B. (2020). Bioconversion of agriculture wastes to produce α-amylase from Bacillus velezensis KB 2216: purification and characterization. Biocatal. Agric. Biotechnol. 28, 101703. doi:10.1016/j.bcab.2020.101703
Blieva, R., Mustafin, K. G., Akhmetsadykov, N. N., Suleimenova, Z., Saduyeva, Z., Zhakipbekova, A., et al. (2021). Optimization of culture medium for enhanced protease biosynthesis in Streptomyces globisporus. Rasayan J. Chem. 14 (1), 270–275. doi:10.31788/rjc.2021.1416123
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 (1–2), 248–254. doi:10.1016/0003-2697(76)90527-3
Cadirci, B. H., Yasa, I., and Kocyigit, A. (2016). Streptomyces sp. TEM 33 possesses high lipolytic activity in solid-state fermentation in comparison with submerged fermentation. Prep. Biochem. Biotechnol. 46 (1), 23–29. doi:10.1080/10826068.2014.970693
Cesário, L. M., Pires, G. P., Pereira, R. F. S., Fantuzzi, E., Da Silva Xavier, A., Cassini, S. T. A., et al. (2021). Optimization of lipase production using fungal isolates from oily residues. BMC Biotechnol. 21 (1), 65. doi:10.1186/s12896-021-00724-4
Chandra, P., Enespa, E., Singh, R., and Arora, P. K. (2020). Microbial lipases and their industrial applications: a comprehensive review. Microb. Cell Factories 19 (1), 169. doi:10.1186/s12934-020-01428-8
Cherif, S., Mnif, S., Hadrich, F., Abdelkafi, S., and Sayadi, S. (2011). A newly high alkaline lipase: an ideal choice for application in detergent formulations. Lipids Health Dis. 10 (1), 221. doi:10.1186/1476-511X-10-221
Cho, S. S., Park, D. J., Simkhada, J. R., Hong, J. H., Sohng, J. K., Lee, O. H., et al. (2012). A neutral lipase applicable in biodiesel production from a newly isolated Streptomyces sp. CS326. Bioprocess Biosyst. Eng. 35 (1–2), 227–234. doi:10.1007/s00449-011-0598-8
Delorme, V., Dhouib, R., Canaan, S., Fotiadu, F., Carrière, F., and Cavalier, J.-F. (2011). Effects of surfactants on lipase structure, activity, and inhibition. Pharm. Res. 28 (8), 1831–1842. doi:10.1007/s11095-010-0362-9
Diana, R.-M., Monserrat, M.-R., Alba, R.-R., Beatriz, R.-V., Romina, R.-S., and Sergio, S.-E. (2021). Dissecting the role of the two Streptomyces peucetius var. Caesius glucokinases in the sensitivity to carbon catabolite repression. J. Ind. Microbiol. Biotechnol. 48 (9–10), kuab047. doi:10.1093/jimb/kuab047
Dos Santos, J. B. C., Da Silva Cruz, R. G., and Tardioli, P. W. (2017). Production of whole-cell lipase from Streptomyces clavuligerus in a bench-scale bioreactor and its first evaluation as biocatalyst for synthesis in organic medium. Appl. Biochem. Biotechnol. 183 (1), 218–240. doi:10.1007/s12010-017-2440-5
Eskandari, A., Leow, T. C., Rahman, M. B. A., and Oslan, S. N. (2024). Recent insight into the advances and prospects of microbial lipases and their potential applications in industry. Int. Microbiol. 27 (6), 1597–1631. doi:10.1007/s10123-024-00498-7
Freije García, F., and García Liñares, G. (2024). Use of lipases as a sustainable and efficient method for the synthesis and degradation of polymers. J. Polym. Environ. 32 (5), 2484–2516. doi:10.1007/s10924-023-03118-z
Gamayurova, V. S., Zinov’eva, M. E., Shnaider, K. L., and Davletshina, G. A. (2021). Lipases in esterification reactions: a review. Catal. Ind. 13 (1), 58–72. doi:10.1134/S2070050421010025
Gao, K., Wang, X., Jiang, H., Sun, J., and Mao, X. (2021). Identification of a GDSL lipase from Streptomyces bacillaris and its application in the preparation of free astaxanthin. J. Biotechnol. 325, 280–287. doi:10.1016/j.jbiotec.2020.10.009
Ghamgui, H., Miled, N., Karra-chaâbouni, M., and Gargouri, Y. (2007). Immobilization studies and biochemical properties of free and immobilized rhizopus oryzae lipase onto CaCO3: a comparative study. Biochem. Eng. J. 37 (1), 34–41. doi:10.1016/j.bej.2007.03.006
Guncheva, M., and Zhiryakova, D. (2011). Catalytic properties and potential applications of Bacillus lipases. J. Mol. Catal. B Enzym. 68 (1), 1–21. doi:10.1016/j.molcatb.2010.09.002
Gupta, R., Gupta, N., and Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64 (6), 763–781. doi:10.1007/s00253-004-1568-8
Hasan, F., Shah, A. A., and Hameed, A. (2006). Industrial applications of microbial lipases. Enzyme Microb. Technol. 39 (2), 235–251. doi:10.1016/j.enzmictec.2005.10.016
Im, H., An, T., Kwon, R., Park, S., and Kim, Y.-K. (2021). Effect of organic nitrogen supplements on syngas fermentation using Clostridium autoethanogenum. Biotechnol. Bioprocess Eng. 26 (3), 476–482. doi:10.1007/s12257-020-0221-4
Invernizzi, G., Papaleo, E., Grandori, R., De Gioia, L., and Lotti, M. (2009). Relevance of metal ions for lipase stability: structural rearrangements induced in the Burkholderia glumae lipase by calcium depletion. J. Struct. Biol. 168 (3), 562–570. doi:10.1016/j.jsb.2009.07.021
Iyer, P. V., and Ananthanarayan, L. (2008). Enzyme stability and stabilization—aqueous and non-aqueous environment. Process Biochem. 43 (10), 1019–1032. doi:10.1016/j.procbio.2008.06.004
Lacey, H. J., and Rutledge, P. J. (2022). Recently discovered secondary metabolites from Streptomyces species. Molecules 27 (3), 887. doi:10.3390/molecules27030887
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 (5259), 680–685. doi:10.1038/227680a0
Lertcanawanichakul, M., and Sahabuddeen, T. (2023). Characterization of Streptomyces sp. KB1 and its cultural optimization for bioactive compounds production. PeerJ 11, e14909. doi:10.7717/peerj.14909
Magda, M. A., Sanaa, T., Saleh, M. A. G., and Lubna, N. (2012). Production of lipase from genetically improved Streptomyces exfoliates LP10 isolated from oil-contaminated soil. Afr. J. Microbiol. Res. 6 (6), 1125–1137. doi:10.5897/AJMR11.1123
Mandari, V., and Devarai, S. K. (2022). Biodiesel production using homogeneous, heterogeneous, and enzyme catalysts via transesterification and esterification reactions: a critical review. BioEnergy Res. 15 (2), 935–961. doi:10.1007/s12155-021-10333-w
Mander, P., Cho, S. S., Simkhada, J. R., Choi, Y. H., Park, D. J., Ha, J. W., et al. (2012). An organic solvent-tolerant alkaline lipase from Streptomyces sp. CS268 and its application in biodiesel production. Biotechnol. Bioprocess Eng. 17 (1), 67–75. doi:10.1007/s12257-011-0347-5
Martin, L. S., Cerón, A. A., Molinari, D., De Moraes, F. F., Arroyo, P. A., De Castro, H. F., et al. (2019). Enhancement of lipase transesterification activity by immobilization on β–cyclodextrin-based polymer. J. Sol-Gel Sci. Technol. 91 (1), 92–100. doi:10.1007/s10971-019-05011-5
Mehta, A., Guleria, S., Sharma, R., and Gupta, R. (2021). “The lipases and their applications with emphasis on food industry,” in Microbial biotechnology in food and health (Elsevier), 143–164. doi:10.1016/B978-0-12-819813-1.00006-2
Milne, J. J. (2017). Scale-up of protein purification: downstream processing issues. Protein Chromatogr. Methods Protoc. 1485, 71–84. doi:10.1007/978-1-4939-6412-3_5
Moayd, W., and Yan, Y. (2017). Lipases, definition, and their application. IOSR J. Pharm. Biol. Sci. 12 (03), 55–60. doi:10.9790/3008-1203025560
Narwal, S. K., and Gupta, R. (2013). Biodiesel production by transesterification using immobilized lipase. Biotechnol. Lett. 35 (4), 479–490. doi:10.1007/s10529-012-1116-z
Ni, H.-J., Lv, S.-Y., Sheng, Y.-T., Wang, H., Chu, X.-H., and Zhang, H.-W. (2021). Optimization of fermentation conditions and medium compositions for the production of chrysomycin a by a marine-derived strain Streptomyces sp. 891. Prep. Biochem. Biotechnol. 51 (10), 998–1003. doi:10.1080/10826068.2021.1885046
Ousaadi, M. I., Merouane, F., Berkani, M., Almomani, F., Vasseghian, Y., and Kitouni, M. (2021). Valorization and optimization of agro-industrial orange waste for the production of enzyme by halophilic Streptomyces sp. Environ. Res. 201, 111494. doi:10.1016/j.envres.2021.111494
Phukon, L. C., Chourasia, R., Kumari, M., Godan, T. K., Sahoo, D., Parameswaran, B., et al. (2020). Production and characterisation of lipase for application in detergent industry from a novel Pseudomonas helmanticensis HS6. Bioresour. Technol. 309, 123352. doi:10.1016/j.biortech.2020.123352
Praveen Kumar, P., Sagaya Jansi, R., Saravana Kumar, P., Nimal Christhudas, I. V. S., Preetam Raj, J. P., Vijayakumar, A., et al. (2017). Optimization of biosynthesis parameters, partial purification and characterization of extracellular lipase from soil derived Streptomyces sp. Loyola lipase-1. Biocatal. Agric. Biotechnol. 12, 241–247. doi:10.1016/j.bcab.2017.10.011
Rabbani, G., Ahmad, E., Ahmad, A., and Khan, R. H. (2023). Structural features, temperature adaptation and industrial applications of microbial lipases from psychrophilic, mesophilic and thermophilic origins. Int. J. Biol. Macromol. 225, 822–839. doi:10.1016/j.ijbiomac.2022.11.146
Rathelot, J., Julien, R., Bosc-Bierne, I., Gargouri, Y., Canioni, P., and Sarda, L. (1981). Horse pancreatic lipase. Interaction with colipase from various species. Biochimie 63 (3), 227–234. doi:10.1016/S0300-9084(81)80196-4
Reyes-Reyes, A. L., Valero Barranco, F., and Sandoval, G. (2022). Recent advances in lipases and their applications in the food and nutraceutical industry. Catalysts 12 (9), 960. doi:10.3390/catal12090960
Rodríguez-Alonso, G., Toledo-Marcos, J., Serrano-Aguirre, L., Rumayor, C., Pasero, B., Flores, A., et al. (2023). A novel lipase from Streptomyces exfoliatus DSMZ 41693 for biotechnological applications. Int. J. Mol. Sci. 24 (23), 17071. doi:10.3390/ijms242317071
Sarac, N., Ugur, A., Boran, R., and Elgin, E. S. (2015). The use of boron compounds for stabilization of lipase from Pseudomonas aeruginosa ES3 for the detergent industry. J. Surfactants Deterg. 18 (2), 275–285. doi:10.1007/s11743-014-1653-7
Saraswat, R., Verma, V., Sistla, S., and Bhushan, I. (2017). Evaluation of alkali and thermotolerant lipase from an indigenous isolated Bacillus strain for detergent formulation. Electron. J. Biotechnol. 30, 33–38. doi:10.1016/j.ejbt.2017.08.007
Sharma, P. K., Singh, K., Singh, R., Capalash, N., Ali, A., Mohammad, O., et al. (2012). Characterization of a thermostable lipase showing loss of secondary structure at ambient temperature. Mol. Biol. Rep. 39 (3), 2795–2804. doi:10.1007/s11033-011-1038-1
Shirazi, N. H., Ranjbar, B., Khajeh, K., and Moghadam, T. T. (2013). Structure–function analysis of a new bacterial lipase: effect of local structure reorganization on lipase activity. Int. J. Biol. Macromol. 54, 180–185. doi:10.1016/j.ijbiomac.2012.12.020
Song, X., Qi, X., Hao, B., and Qu, Y. (2008). Studies of substrate specificities of lipases from different sources. Eur. J. Lipid Sci. Technol. 110 (12), 1095–1101. doi:10.1002/ejlt.200800073
Sousa, J. A. D. J., and Olivares, F. L. (2016). Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications. Chem. Biol. Technol. Agric. 3 (1), 24. doi:10.1186/s40538-016-0073-5
Spasic, J., Mandic, M., Djokic, L., and Nikodinovic-Runic, J. (2018). Streptomyces spp. in the biocatalysis toolbox. Appl. Microbiol. Biotechnol. 102 (8), 3513–3536. doi:10.1007/s00253-018-8884-x
Stergiou, P.-Y., Foukis, A., Filippou, M., Koukouritaki, M., Parapouli, M., Theodorou, L. G., et al. (2013). Advances in lipase-catalyzed esterification reactions. Biotechnol. Adv. 31 (8), 1846–1859. doi:10.1016/j.biotechadv.2013.08.006
Szymczak, T., Cybulska, J., Podleśny, M., and Frąc, M. (2021). Various perspectives on microbial lipase production using agri-food waste and renewable products. Agriculture 11 (6), 540. doi:10.3390/agriculture11060540
Taher, N. A., Husen, A. S., Mahmood, Z.Sh., and Shanior, G. J. (2020). A study on actinorhodin-like substance production by Streptomyces IQ45. Al-Mustansiriyah J. Sci. 31 (3), 6–13. doi:10.23851/mjs.v31i3.93
Törnvall, U., Hedström, M., Schillén, K., and Hatti-Kaul, R. (2010). Structural, functional and chemical changes in pseudozyma Antarctica lipase B on exposure to hydrogen peroxide. Biochimie 92 (12), 1867–1875. doi:10.1016/j.biochi.2010.07.008
Ülker, S., Özel, A., Çolak, A., and Karaoğlu, Ş. A. (2011). Isolation, production, and characterization of an extracellular lipase from trichoderma harzianum isolated from soil. Turk. J. Biol. doi:10.3906/biy-1004-107
Vasconcelos, E. D. S. (2018). Production of lipase from Streptomyces. J. Bacteriol. Mycol. Open Access 6 (2). doi:10.15406/jbmoa.2018.06.00184
Vishnupriya, B., Sundaramoorthi, C., Kalaivani, M., and Selvam, K. (2010). Production of lipase from Streptomyces griseus and evaluation of bioparameters. Int. J. Chem. Tech. Res. 2 (3), 1380–1383.
Yao, W., Liu, K., Liu, H., Jiang, Y., Wang, R., Wang, W., et al. (2021). A valuable product of microbial cell factories: microbial lipase. Front. Microbiol. 12, 743377. doi:10.3389/fmicb.2021.743377
Yuan, C., Byeon, I.-J. L., Poi, M.-J., and Tsai, M.-D. (1999). Structural analysis of phospholipase A2 from functional perspective. 2. Characterization of a molten globule-like state induced by site-specific mutagenesis. Biochemistry 38 (10), 2919–2929. doi:10.1021/bi9822123
Yuan, D., Lan, D., Xin, R., Yang, B., and Wang, Y. (2016). Screening and characterization of a thermostable lipase from marine Streptomyces sp. Strain W007. Biotechnol. Appl. Biochem. 63 (1), 41–50. doi:10.1002/bab.1338
Keywords: industrial applications, biocatalysis, lipase purification, stability, detergent formulations, esterification
Citation: Alzahrani AA, Krayem N, Alonazi M, Al-Ghamdi JM, Horchani H and Ben Bacha A (2025) Versatile biocatalyst: lipase from Streptomyces gobitricini for ester synthesis and detergent innovation. Front. Bioeng. Biotechnol. 13:1589087. doi: 10.3389/fbioe.2025.1589087
Received: 06 March 2025; Accepted: 02 May 2025;
Published: 16 May 2025.
Edited by:
Noha M. Mesbah, Suez Canal University, EgyptReviewed by:
César Alonso Godoy Vargas, University of the Valley, ColombiaDiego Carballares, Complutense University of Madrid, Spain
Copyright © 2025 Alzahrani, Krayem, Alonazi, Al-Ghamdi, Horchani and Ben Bacha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abir Ben Bacha, YWFsZ2hhbm91Y2hpQGtzdS5lZHUuc2E=
 Areej Ali Alzahrani
Areej Ali Alzahrani Najeh Krayem
Najeh Krayem Mona Alonazi
Mona Alonazi Jihan M. Al-Ghamdi
Jihan M. Al-Ghamdi Habib Horchani
Habib Horchani Abir Ben Bacha
Abir Ben Bacha