- 1Department of Obstetrics and Gynecology, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, China
- 2Maternal-Fetal Medicine Institute, Department of Obstetrics and Gynecology, Shenzhen Baoan Women’s and Children’s Hospital, Shenzhen University, Shenzhen, China
- 3The First School of Clinical Medicine, Southern Medical University, Guangzhou, China
Recent advancements in tissue clearing and three-dimensional (3D) visualization technologies have enabled subcellular-level examination of entire organs, particularly in complex structures such as the ovary and uterus. Traditional histological approaches are limited by two-dimensional views, which restrict our understanding of female reproductive system functions. In this review, we highlight the innovations in 3D tissue clearing techniques applied to uterine and ovarian tissues, which, combined with analytical tools, facilitate comprehensive 3D visualization and image analysis. We evaluate the advantages and disadvantages of three primary categories of tissue clearing techniques: organic solvent-based, hydrogel-based, and hydrogel-embedded methods, specifically regarding the uterus and ovary. Light-sheet and multiphoton microscopy complement these techniques, providing unprecedented capabilities for high-resolution imaging of large tissue volumes. Tissue clearing technologies provide a robust strategy for early diagnosis of uterine and ovarian pathologies. Additionally, we explore the integration of tissue clearing technologies with spatial transcriptomics and AI-driven analytical tools to achieve comprehensive 3D molecular mapping. We hope this review contributes to a better understanding of tissue clearing techniques and can help researchers in navigating methodological choices for uterine and ovarian investigations.
1 Introduction
The ovaries and uterus serve as essential components of the female reproductive system. Ovarian functions include generating mature oocytes and secreting hormones such as estrogen and progesterone (Fan et al., 2019). The uterus facilitates mammalian embryo and fetal implantation, development, and growth (Mori et al., 2023). Traditional histological approaches, limited to two-dimensional sections, fail to capture the dynamic 3D spatial relationships essential for understanding complex biological processes. Comprehensive investigation of biological progress and mechanisms requires examining intact tissues in 3D spatial views rather than sections.
Recent advances in tissue clearing, including iDISCO, ScaleA2, CUBIC, and CLARITY, address the challenges of tissue opacity and light scattering (Renier et al., 2014; Malki et al., 2015; Pinheiro et al., 2021; Tomer et al., 2014). Furthermore, significant progress has occurred in imaging technologies, including light-sheet fluorescence microscopy (LSFM) and multiphoton microscopy (MPFM). These techniques enable comprehensive and high-resolution visualization of intact large samples at cellular resolution in 3D levels (Power and Huisken, 2017; Mohler et al., 2003).
Tissue clearing techniques provide valuable insights into ovarian and uterine structure and molecular mechanisms. When combined with spatial transcriptomics, these methods enable detailed visualization of ovarian follicular architecture while simultaneously mapping gene expression gradients, revealing the complex relationship between follicular development and surrounding stromal cells. Similarly, 3D imaging integrated with single-cell analysis offers comprehensive characterization of the myometrium and its molecular regulation during pregnancy and childbirth (Kagami et al., 2020). These visualization strategies have enhanced our understanding of the cellular microenvironment, providing insights into the female reproductive system. Furthermore, these approaches are crucial in examining communication between the developing embryo and maternal tissues, offering precise comprehension of processes associated with implantation and placenta formation (Kagami et al., 2017).
This review summarizes ovarian and uterine development and emphasizes the transition from traditional techniques to 3D imaging and data analysis with commercial visualization software. We also explore future directions for integration with single-cell and spatial omics technologies. Special attention is given to AI-assisted techniques for analyzing 3D imaging data, highlighting their significance in improving the accuracy and efficiency of these studies.
2 Uterus and ovary
2.1 Development of the uterus and ovary
The female reproductive system comprises external and internal genitalia. The external genitalia include the labia majora and minora, clitoris, and vestibule, whereas the internal genitalia include the ovaries, fallopian tubes, uterus, cervix, and vagina (Septadina, 2023). In vertebrates, primordial germ cells (PGCs) are gamete precursors. They originate from the epiblast and, upon migration to extraembryonic regions, undergo fate determination, representing a critical event in embryonic reproductive system establishment.
PGC specification commences during the third week of embryonic development, and by the fourth week, these cells are identifiable in the yolk sac near the allantois (Guglielmo and Albertini, 2013). They subsequently migrate into the endodermal epithelium of the hindgut, detach from the gut wall, and reach the gonadal ridge through the dorsal mesentery (Soto-Suazo et al., 2005). In XX embryos lacking SRY gene expression, factors such as WNT4, RSPO1, and FOXL2 promote the differentiation of the bipotential gonad into an ovary, with the gradual formation of cortical and medullary compartments. PGCs that settle in the cortical region initiate meiosis and arrest at prophase I, forming primary oocytes. These are surrounded by pre-granulosa cells to establish primordial follicles (Baetens et al., 2019).
Folliculogenesis advances from the primordial stage through the primary and secondary stages, the latter characterized by multiple granulosa cell layers and steroidogenic theca cells. After puberty, periodic surges of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) drive the development of a subset of follicles into the antral stage, marked by the formation of a fluid-filled cavity (Palumbo et al., 1994; Stringer et al., 2023).
The female reproductive tract primarily originates from the paramesonephric (Müllerian) ducts, which develop through cranio-caudal invagination of the coelomic epithelium adjacent to the Wolffian ducts (Orvis and Behringer, 2007). The fallopian tubes form from the upper and middle segments of the majority of the female reproductive tract, which derives from the paramesonephric (Müllerian) ducts with their ventral openings developing from the invaginated ends. During approximately the tenth week of gestation, the lower portions of both ducts merge to form the uterus and the upper portion of the vagina (Habiba et al., 2021). Considering the intricacies of the female reproductive system, we will focus on the application of tissue clearing techniques to the uterus and ovaries, aiming to reveal their underlying molecular and structural mechanisms.
2.2 Histologic methods for the analysis of the ovary and uterus
A comprehensive and high-resolution understanding of ovarian and uterine development is fundamental. The microscope’s invention transformed anatomical studies to the cellular level, necessitating organ sectioning for tissue examination. The primary method for visualizing the uterus and ovaries involves tissue sectioning and histology (Sarma et al., 2020; Pascolo et al., 2019; Pichat et al., 2018). For nearly a century, whole-organism analysis has relied on serial sectioning combined with histological staining to obtain a 2D perspective of 3D imaging. Histology remains widely utilized across biological disciplines, and modern techniques enable the reconstruction of 3D images from histological sections (Kartasalo et al., 2018; Berg et al., 2019).
Follicle count quantification methods employ age-special correction factors for representative numbers. This approach requires considerable effort, with results varying based on slice thickness precision, assessed slice statistics, and the research-specific correction factors (Berg et al., 2019). Advanced resolution methods, such as those developed by Lutton et al. (Lutton et al., 2017), enable myometrium reconstruction with approximately 50 µm per voxel side resolution. This facilitates specific imaging of smooth muscular tissue formation and fibrous microarchitecture within the uterus. These reconstructions correspond directly with original histological slides, enabling detailed examination of functional anatomical context in 3D images (Lutton et al., 2017). While stereology offers greater precision, its widespread adoption is limited by specialized equipment requirements and expertise needs. Furthermore, these methods are constrained by tissue distortion during sectioning and sectioning techniques inconsistencies (Gurcan et al., 2014).
2.3 Challenges in visualizing the intact uterus and ovary
Ovaries contain multiple tissue types, including epithelial and connective tissues, vasculature, and nerve fibers. These encompass various components, including membranes, nuclei, lipids, proteins, collagens, blood, and tissue fluids. The uterus comprises three distinct histological layers: mesometrium, myometrium, and endometrium. The mesometrium, the outermost layer, interfaces with the visceral peritoneal layer (Bates and Bowling, 2013; Taylor and Gomel, 2008). The intermediate myometrium layer contains three smooth muscle layers. Intact organ imaging presents multiple challenges: requiring long-working-distance microscopes and objectives for large samples, substantial computational resources for extensive datasets, and preservation of antigenicity, fluorophore integrity, and tissue morphology in cleared specimens (Dodt et al., 2007). Tissue clearing techniques, combined with multiphoton microscopy, light-sheet microscopy, and spinning disk confocal microscopy (SDCM), address these challenges, enabling comprehensive 3D examination of entire organs and organisms without mechanical sectioning. The non-uniform distribution of light-absorbing and scattering molecules within tissues can cause uneven light scattering, resulting in tissue opacity (Tuchin, 2015). Optical clearing techniques enhance transparency by equalizing sample RI through component modification, removal, or substitution. This adjustment reduces light scattering, improving optical transparency (Tainaka et al., 2016; Ryu et al., 2022). The application of 3D imaging for complete embryos, organs, and adult bodies has expanded through improvements in computational analysis, optical techniques, and image rendering. (Vieites-Prado and Renier, 2021; Zhu et al., 2013; Susaki et al., 2015).
3 Tissue-clearing methods optimized for uterus and ovary imaging
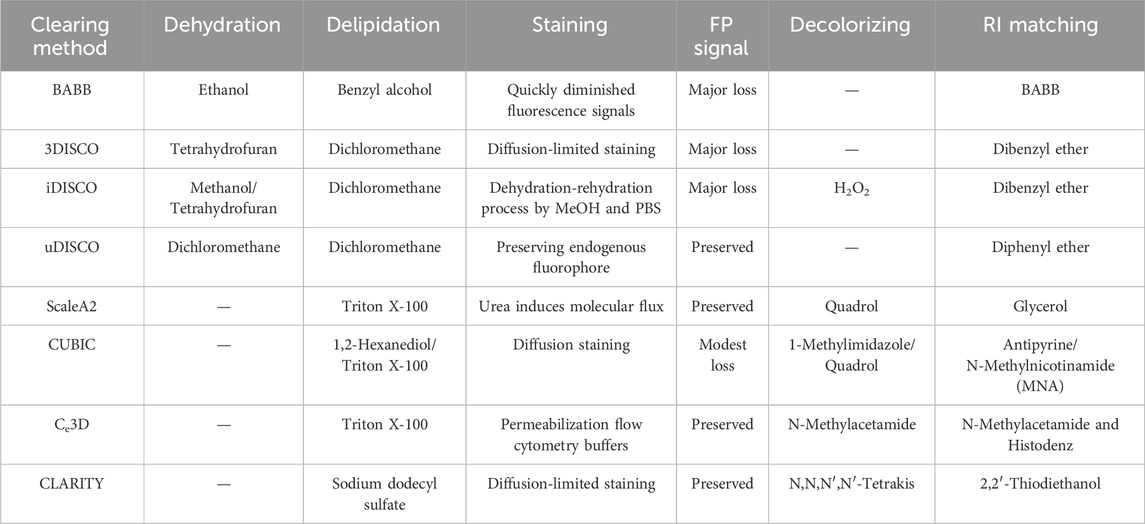
The achievement of tissue transparency in ovarian and uterine samples is crucial for reproductive biology and pathology research, enabling detailed structural and cellular dynamic analysis. Recent technological advances have significantly improved tissue transparency methods and instruments. Here, we summarize the tissue clearing methods applied to ovarian and uterine tissues (Figure 1; Table 1) and recent developments in tissue-clearing reagents (Table 2).
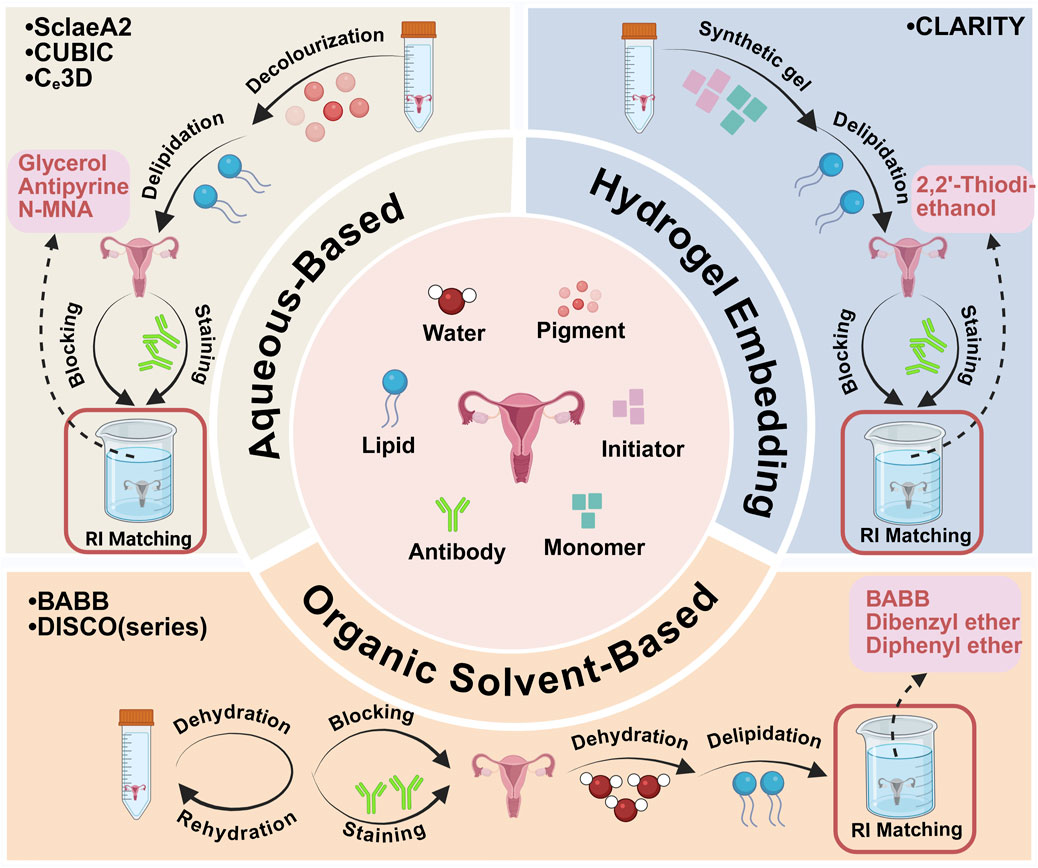
Figure 1. Major tissue clearing methods are applied to ovarian and uterus tissues. Created with BioRender.com.
3.1 Organic solvent-based tissue clearing
Organic solvent-based tissue clearing techniques provide superior tissue transparency by harmonizing components RIs via high-RI organic solvents. These protocols primarily involve tissue dehydration, lipid removal, and sample infusion with clearing solution to standardize remaining structures refractive indices. Solvent-based tissue clearing techniques compatible with immunofluorescence staining, including benzyl alcohol/benzyl benzoate (BABB) and 3D imaging of solvent-cleared organs (DISCO), represent preferred approaches for ovarian and uterine tissue clearing (Combes et al., 2009; Svingen et al., 2012; Faire et al., 2015; Dodt et al., 2007). These techniques offer cost-effectiveness and enable sample clearing within several days.
To investigate the dynamic morphogenesis of entire organs, Zucker and colleagues employed BABB alongside advanced microscopy techniques to examine whole mouse ovaries, focusing on morphology, apoptosis detection, and spectroscopy (Zucker and Jeffay, 2006). The application of LysoTracker Red to fresh, intact mouse ovaries enables identification of apoptotic cells. Tissue morphology was preserved through fixation with 4% paraformaldehyde and 1% glutaraldehyde, enhancing the background fluorescence signal and facilitating overall morphological visualization. This methodology emerged from the unexpected observation that oocyte nucleus volume increases proportionally with follicle growth, enabling differentiation between primordial and growing follicles, resulting in unprecedented quantification accuracy.
Beyond its quantification and spatial analysis capabilities, whole-mount imaging excels at detecting rare events. Whole-mount immunofluorescence staining, enhanced through BABB clearing, provides 3D visualization of the uterus and its adaptive responses to embryo implantation (Arora et al., 2016). The researchers refined the BABB protocol through meticulous uterine washing followed by incubation with fluorescently labeled secondary antibodies, specifically Alexa Fluor IgGs, to enhance protein and structural visibility within the tissue. Although BABB was initially utilized as an organic solvent for clearing mouse organs, it proved insufficient for larger samples due to rapid fluorescence signal degradation during alcohol-based dehydration. These limitations were addressed by replacing BABB with a combination of dibenzyl ether (DBE) and tetrahydrofuran (THF), known as 3DISCO solvent-based clearing, and iDISCO, which incorporates immunolabeling (Renier et al., 2014; Ertürk et al., 2012). The iDISCO method, developed by Renier et al. (Renier et al., 2014), enables whole-mount immunolabeling and volumetric imaging of mouse embryos and adult organs. In contrast to 3DISCO, uDISCO facilitates comprehensive whole-body clearing and imaging while preserving endogenous fluorescent proteins for extended periods (Pan et al., 2016; Li et al., 2018).
Within these adaptations, iDISCO application to adult ovaries achieved effective fluorescence signaling and deep antibody penetration, facilitating detailed visualization of follicular structures and the ovarian interstitial compartments in adult mice (McKey et al., 2020). McKey and colleagues examined morphogenetic events in mouse ovary development, discovering that ovary encapsulation correlates with mesonephric duct expansion and regional differentiation into the oviduct, utilizing tissue clearing and light sheet microscopy (McKey et al., 2022). Yuan and colleagues documented 3D visualization techniques revealing distinct embryo‒gland interactions within the crypt, implementing a modified 3DISCO method for tissue clearing. Through whole-mount immunostaining, 3DISCO clearing, and light-sheet imaging, they highlighted HB-EGF ’s essential role in blastocyst-gland communication (Yuan et al., 2018). Despite solvent-based tissue clearing methods demonstrating excellent clearing performance and enabling whole-body imaging at subcellular resolution, certain limitations persist, including significant sample shrinkage, organic solvent toxicity, and fluorescent protein quenching (Tian et al., 2021).
3.2 Aqueous-based tissue clearing
Aqueous-based tissue clearing methods are classified into two main categories: (a) simple immersion and (b) hyperhydration. Simple immersion involves submerging biological samples in an aqueous solution to match refractive indices, enabling gradual clearing. Common agents include sucrose (Tsai et al., 2009), fructose (Costantini et al., 2015; Ke et al., 2013), glycerol (Meglinski et al., 2004), 2,2′-thiodiethanol (TDE) (Aoyagi et al., 2015; Hou et al., 2015; Staudt et al., 2007), and formamide (Kuwajima et al., 2013). While simple immersion in aqueous clearing solutions preserves lipid structures, it inadequately clears large tissues containing connective tissues such as ovaries and uterus. An alternative method involves delipidation and reducing organ RI throughout the clearing process. Scale pioneered this mechanism, employing detergent-based lipid removal with urea-mediated hydration (supported by glycerol) for tissue clearing (Hama et al., 2011). This approach removes lipids using non-hydrophobic solvents to maintain an aqueous environment suitable for fluorescent proteins, requiring extended incubation periods of days to months with detergents such as Triton X-100, necessitating frequent solution changes (Hua et al., 2008).
When combined with sucrose preincubation, ScaleA2 enables oocyte imaging via TRA98 in fetal ovaries, particularly during meiosis and oocyte attrition (Malki et al., 2015). Between E15.5 and E18.5, total germ cell numbers decreased by approximately 27%, a smaller decline than the nearly twofold reduction observed in histological samples (Malki et al., 2014). ScaleA2 and TRA98 immunofluorescence were also employed to examine asymmetries between right and left ovaries regarding ovulation rates, confirming previous findings (Wiebold and Becker, 1987). Similar to the findings of Faire et al. (Faire et al., 2015) regarding adult ovaries, detailed analysis revealed no significant differences in germ populations between right and left ovaries at E15.5 and E18.5. This research established a framework for 3D analysis of embryonic ovaries, offering a comprehensive analysis pipeline, documenting error rates, and comparing sampling and whole-mount approaches directly (Malki et al., 2015).
The ScaleA2 technique was utilized to precisely visualize individual developing endometrial glands and their arrangement within the intact uterus, establishing a uterine gland morphology system during development comprising five stages: Bud, Teardrop, Elongated, Sinuous, and Primary Branch (Goad et al., 2017; Vue et al., 2018). Vue et al. (2018) tracked the morphological transformations of uterine glands from postnatal day 0 to day 21, revealing clear stages of gland development. Initially, at birth (P0), no glands were evident; however, by P8, exposed buds and teardrop-shaped epithelial invaginations were monitored (Filant and Spencer, 2014). This staging system uses a standardized method for reviewing and measuring prepubertal uterine gland morphology, enabling more comprehensive investigations into uterine gland growth and pathology (Vue et al., 2018). The researchers enhanced ScaleA2 for 3D imaging of perinatal mouse uterine glands, including structures such as the uterine orbit and ventral ridge. Their analysis revealed the specific temporal patterns of uterine gland formation, demonstrating that uterine adenogenesis initiates at P4, with postpartum uterine epithelial folds appearing by P5. These observations indicate that uterine morphological development exhibits greater complexity during the perinatal period than previously recognized (Vue and Behringer, 2020).
The clear, unobstructed brain/body imaging cocktails and computational analysis (CUBIC) protocol represents an innovative modification of SCALE (Susaki et al., 2014; Susaki et al., 2015; Pinheiro et al., 2021). The initial reagents used in this method consist of polyhydric alcohol, Triton-X 100, and urea, which jointly help with delipidation. The developed reagent, made up of triethanolamine, polyhydric alcohol, sucrose, and urea, aligns with the RI and enhances transparency while also protecting against endogenous fluorescent signals. CUBIC has been effectively utilized to reveal the 3D structures of ovaries, employing endogenous fluorescent reporter proteins and immunolabeled structures. Kagami et al. (Kagami et al., 2018) utilized a customized CUBIC protocol to visualize common EGFP in adult mouse ovaries as evidence of concept. Incubation with the ScaleA2-CUBIC-1 reagent alone successfully cleared fetal mouse ovaries, enabling the visualization of endogenous fluorescent signals and immunolabeling (Soygur et al., 2021). McKey and colleagues employed iDISCO+ and iDISCO + CUBIC techniques to visualize intact organ dynamic morphogenesis, revealing ovary folding, encapsulation, and integration with oviduct morphogenesis during murine development from embryonic day 14.5 to postnatal day 0 (McKey et al., 2020).
To obtain stereoscopic whole images of the intrauterine murine embryo and placenta, Kagami and colleagues utilized transgenic mice (Kagami et al., 2017). Clear images of EGFP-positive embryos and placentas were recorded, confirming the precise 3D locations of invading trophoblasts at the feta-maternal interface (Kagami et al., 2017). Using this protocol, all placental tissues from pregnant mice (E14.5) were effectively cleared through the modified CUBIC protocol (Kagami et al., 2017). However, CUBIC employs extremely high levels of Triton (50%) to optimize lipid removal. This process, while effective, often leads to substantial protein loss (24%–41%), reducing epitope concentrations and possibly diminishing the effectiveness of immunostaining (Chung et al., 2013). Li et al. (Li et al., 2017) utilized clearing-enhanced 3D (Ce3D) techniques, whereby a blend of N-methylacetamide (22% wt/vol) with Triton X-100 and Histodenz (86% wt/vol) effectively renders tissues transparent. This approach not only maintains the fluorescence of reporter proteins but also supports the execution of direct multiplex antibody-based immunolabeling (Li et al., 2017). By utilizing the benefits of Ce3D technologies, Xu and colleagues integrated tissue transparency techniques with endogenous multicolor reporter mouse models. This integration resulted in the creation of a tissue-scale 3D imaging system with single-cell resolution. This system enables detailed imaging and tracking of angiogenesis and vascular remodeling in whole ovaries and live ovarian follicles (Xu et al., 2022).
3.3 Hydrogel embedding tissue clearing
The aqueous-based clearing methods reviewed thus far have limitations: they can clear only small samples (as in simple immersion) or operate slowly (as with hyperhydration). Moreover, techniques that employ harsh solvents or high concentrations of detergents risk significant protein loss from the tissues. The CLARITY (clear lipid-exchanged acrylamide-hybridized rigid imaging/immunostaining/in situ hybridization-compatible tissue hydrogel) technique addresses these challenges by initially embedding the tissue in a hydrogel matrix (Tomer et al., 2014; Chung et al., 2013; Yang et al., 2014). In the CLARITY method, tissues are embedded in a hydrogel using acrylamide or bisacrylamide solutions, which helps stabilize proteins and maintain the structural integrity of the samples (Malkovskiy et al., 2022). Lipids not integrated into the hydrogel are removed via electrophoretic tissue clearing or passive thermal diffusion methods (Yang et al., 2023). Feng et al. (Feng et al., 2017) developed a passive CLARITY method to reach a complete level of transparency for mouse ovaries over a period of 4–8 weeks. The quantity of follicles increased approximately 300,000-fold from the primordial follicle stage to the preovulatory stage, indicating significant follicle growth and dynamic ovarian tissue remodeling (Feng et al., 2017). Spatial evaluation revealed that follicles increasingly accumulate within the ovary as folliculogenesis advances from the primordial to the antral stages, accompanied by active ovarian remodeling in each cycle, which is consistent with prior observations (Hirshfield and Desanti, 1995). Isaacson and colleagues introduced CLARITY combined with light-sheet fluorescence microscopy for rapid volumetric imaging of both male and female reproductive systems in humans (Isaacson et al., 2020). This research paves the way for future studies focused on clarifying both normal developmental processes and the genetic underpinnings of congenital urogenital anomalies.
3.4 Designing a tissue-clearing experiment
Significant advancements have been made in various tissue clearing methods; however, several critical factors must be considered prior to initiating this experiment, including sample integrity, size, and fluorescence attenuation.
The integrity of the sample is essential during the whole process, as this experiment involves numerous steps and lasts for several weeks. Poor sample integrity undermines the retention of proteins, antigens and macromolecules, as well as overall tissue shape and stability, leading to inferior visualization of intact tissue (Weiss et al., 2021). In hydrogel embedding methods, a polymerized hydrogel or tissue gel matrix minimizes structural damage, allowing more complete removal of lipids from the tissue while reducing protein loss and distortion (Gradinaru et al., 2018). The degree of fixation, the density of the hydrogel, and the strength of the detergent can all be adapted to strike a balance between maintaining tissue integrity and increasing optical transparency.
Larger samples, with approximate dimensions of 10 × 10 × 10 mm3, present even greater challenges in staining, clearing, and imaging. Additionally, tissues with heterogeneous structures demand specialized clearing strategies, such as delipidation or depigmentation (Chi et al., 2018; Pende et al., 2020). These procedures can drastically affect factors such as transgenic signal integrity, structural morphology, and nucleotide preservation. To overcome this limitation, shrinkage-prone clearing methods, such as those utilizing organic solvents, can reduce mouse bodies to approximately one-third of their original size, fitting within the microscope’s physical constraints. This is because reductions in tissue size and improvements in resolution considerably increase the data volume. Conversely, certain hydrogel embedding and aqueous-based techniques can be adapted to expand the construct isotropically for super-resolution imaging, thereby increasing the resolution (Chen et al., 2015; Ku et al., 2016).
The retention of endogenous fluorescent (EF) protein expression, together with the use of antibodies and conjugated dyes, significantly influences the choice of protocol. Adaptations to organic solvent-based clearing protocols are continuously advancing to increase fluorescence retention, primarily with changes in dehydration processes, temperature, and pH conditions (Masselink et al., 2019). Moreover, aqueous-based methods have shown success in imaging endogenous transgenic fluorescence. In most cases, a robust signal can be achieved via the use of antibodies that amplify the signal via a stable fluorescent dye. The advantages of this approach include the ability to select wavelengths that minimize autofluorescence and absorption, increase brightness, reduce bleaching, and increase compatibility with various clearing protocols. Here, the selection of an appropriate clearing protocol is based on the integrity of the sample, the sample size, and the retention of both endogenous fluorescence and immunofluorescence (Figure 2).
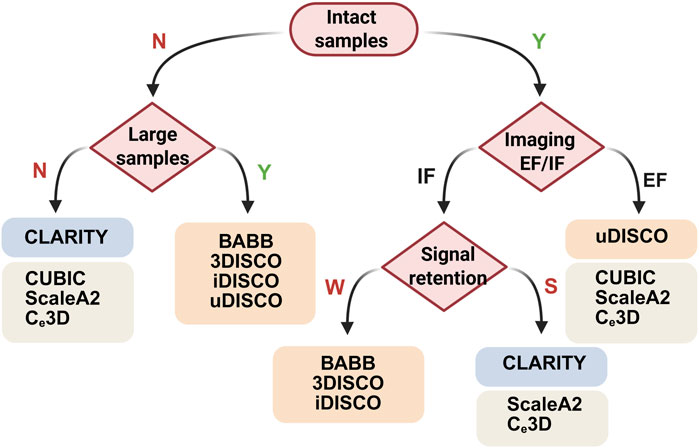
Figure 2. Decision trees for choosing clearing protocols on the basis of the integrity, size and fluorescence signals of samples. N, no; Y, yes; IF, immunofluorescence; EF, endogenous fluorescence; W, weak; S, strong. Created with BioRender.com.
3.5 Reproducibility and cost
Recent advancements have enhanced the reproducibility of tissue clearing protocols through standardized commercial solutions and equipment-assisted methodologies. Solvent-based techniques now utilize consistently formulated chemical components, while aqueous approaches benefit from commercial systems for hydrogel polymerization and electrophoretic tissue clearing (ETC), reducing inter-laboratory variability (Jensen and Berg, 2017). Nevertheless, antibody-dependent fluorescence labeling remains a critical reproducibility challenge due to batch-to-batch inconsistencies, particularly evident in large tissues where off-target binding amplifies background noise. Mitigation requires rigorous antibody validation using uncleared sections and establishment of dedicated repositories for clearing-validated reagents. Protocol harmonization ensures experimental consistency across studies by standardizing critical parameters, including sample dimensions, solution compositions, incubation conditions, tissue deformation metrics, and antibody specifications (manufacturer, lot, concentration) (Richardson et al., 2021).
The economic viability of tissue clearing techniques is primarily governed by reagent accessibility, protocol complexity, and ancillary infrastructure demands. Solvent-based methods, such as BABB and iDISCO, utilize relatively low-cost organic chemicals (Azaripour et al., 2016). For hydrogel-embedding approaches, examples like CLARITY and SHIELD incur higher reagent expenditures due to proprietary hydrogel monomers and electrophoretic instrumentation; however, these methods offset long-term costs by minimizing tissue distortion artifacts that necessitate repeat experiments (Choi et al., 2021). Aqueous techniques, including CUBIC and SeeDB, employ moderately priced reagents compatible with standard laboratory equipment but require extended processing durations, thereby increasing operational overhead (Matsumoto et al., 2019; Ke et al., 2013).
4 Tissue clearing and microscopic methods for uterus and ovary imaging
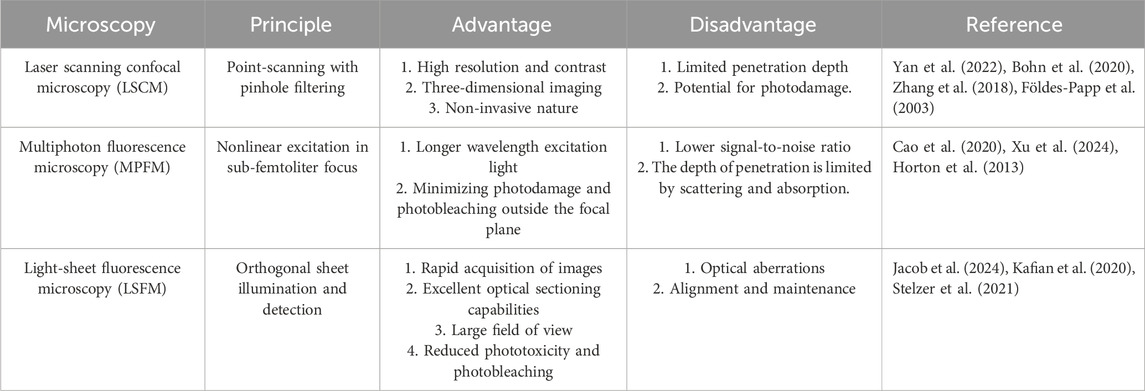
Currently, with advancements in the field of fluorescence light microscopy, numerous optical sectioning techniques are available on the market, many of which excel at imaging cleared tissues. Notable methods include confocal microscopy, multiphoton fluorescence microscopy, and light-sheet fluorescence microscopy (Table 3).
4.1 Confocal microscopy
For decades, confocal microscopy has been a staple for imaging entire developing gonads in their entirety. Laser scanning confocal microscopy (LSCM) directs a high-energy photon to illuminate a single point and employs a pinhole to filter out-of-focus light, reducing light scattering effects. Confocal microscopy eliminates scattered light artifacts by using laser point illumination focused through an exclusionary pinhole (Figure 3A). This illuminates a single focal point in the specimen, with emitted light filtered through a conjugate detection pinhole to reject out-of-focus photons. Sequential x-y-z scanning of the focal plane builds high-resolution 3D images through optical sectioning (Zhang et al., 2018; Földes-Papp et al., 2003). This technique results in high-resolution, high-contrast optical sections of the tissue (Clendenon et al., 2011). Advances in confocal imaging and 3D analysis have enabled quantitative studies of ovarian follicles and their spatial distribution within the mouse ovary; however, these techniques have yet to be fully utilized in studying the spatiotemporal dynamics of early female meiosis. Soygur et al. developed an algorithm that utilizes confocal imaging and a novel 3D quantitative approach to investigate meiotic initiation in whole intact mouse fetal ovaries (Soygur et al., 2021). This study used 3D imaging to map meiotic initiation, revealing a novel radial pattern of meiotic onset that precedes the anterior‒posterior (A‒P) wave in mouse fetal ovaries (Soygur et al., 2021). To increase the axial resolution in a confocal point-scanning system, choosing a high numerical aperture (NA) detection lens and minimizing the pinhole size are vital. Enhanced axial resolution does not just improve lateral resolution; it likewise demands capturing more images to completely cover the tissue. This causes greater light exposure and increased bleaching of the sample; for example, a 5 mm deep tissue section imaged with 5 μm z-steps in a point-scanning system would be exposed to excitation light 1,000 times (Weiss et al., 2021).
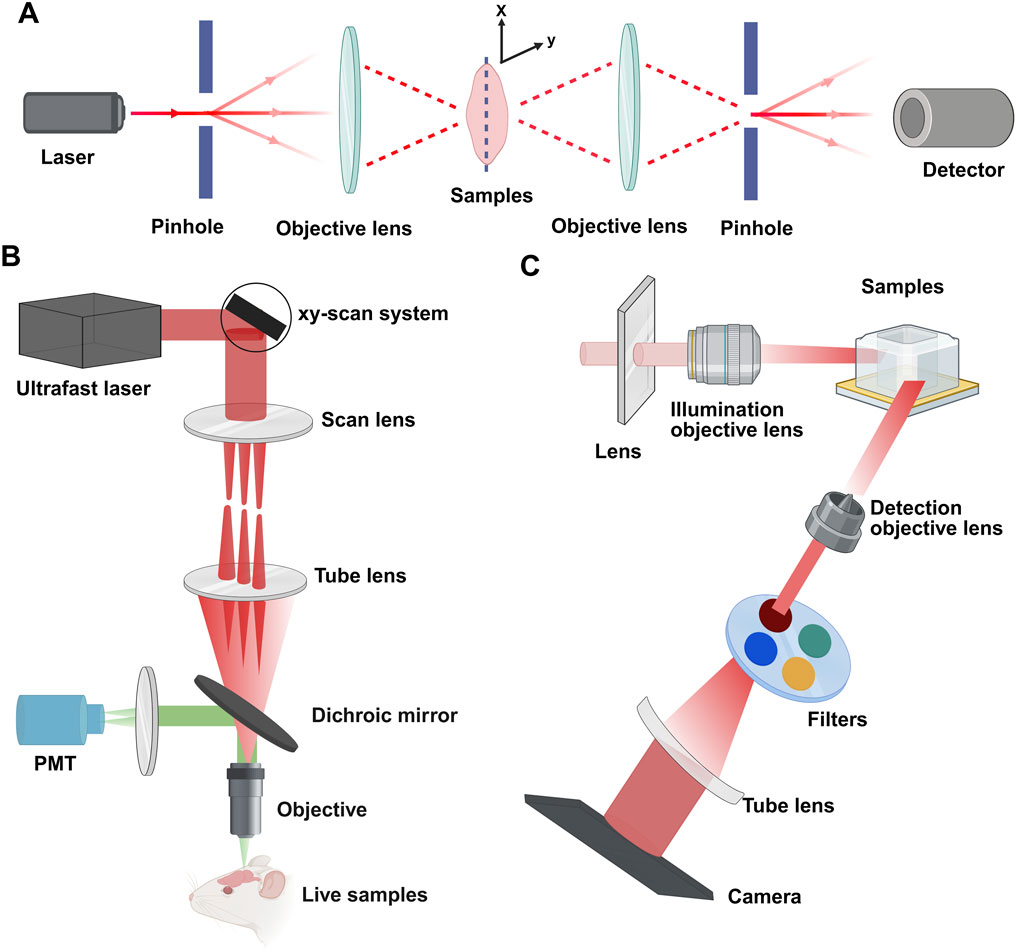
Figure 3. Principles of confocal, multiphoton, and light-sheet microscopy. (A) Confocal microscopy: a pinhole filters the illuminant to a point source focused by the objective lens. This enables optical sectioning via point-scanning in the x-y plane. Deeper scanning captures sequential images, and computer processing constructs the 3D structure. (B) Multiphoton microscopy: nonlinear excitation enables deeper tissue penetration. Illustrated with epi-fluorescence detection using one photomultiplier tube (PMT). (C) Light-sheet microscopy: a cylindrical lens forms a static light sheet illuminating the sample plane along the z-axis. Parallel illumination across the field of view minimizes photodamage. Detection occurs orthogonally (x-y plane). Created with BioRender.com.
4.2 Multiphoton microscopy
To alleviate out-of-focus photobleaching and accomplish deeper imaging, multiphoton fluorescence microscopy (MPFM) utilizes a pulsed infrared laser to excite fluorophores via nonlinear multiphoton absorption (Denk et al., 1990). In two-photon microscopy (2PFM), simultaneous absorption of two photons confines excitation to a sub-femtoliter focal volume (<1 fL) due to quadratic intensity dependence, providing intrinsic optical sectioning without confocal pinholes (Denk et al., 1990). Three-photon microscopy (3PFM) uses longer wavelengths and cubic intensity dependence to suppress background, achieving theoretical penetration depths of 3–4 mm despite marginally lower resolution (Figure 3B) (Horton et al., 2013; Xu et al., 2024). The longer excitation wavelengths of infrared light (750–1,500 nm) decrease light scattering by at least 16-fold, dramatically enhancing the possible imaging depth (Cutrale et al., 2019). MPFM is increasingly popular for deep tissue imaging across various systems because of its low toxicity, making it the preferred method for long-term intravital imaging (Mohler et al., 2003; Scheele et al., 2022). A recent study integrated ScaleA2 and CUBIC with MPFM to investigate the maturation of the ovarian reserve and analyze the patterns of follicular attrition in the perinatal mouse ovary. While MPFM yields high-resolution images and excels over confocal microscopy in deep imaging up to approximately 1.5 mm, its compatibility with standard fluorophores is limited, and the scanning of a single excitation spot across the sample by most multiphoton instruments leads to lengthy acquisition times.
4.3 Light-sheet fluorescence microscopy
Light-sheet fluorescence microscopy (LSFM) employs orthogonal illumination and detection paths (conventionally x-illumination/z-detection) to project a thin laser sheet, via cylindrical lens in selective/single plane illumination microscope (SPIM) or scanned mirror in digital scanned laser microscope (DSLM) through the sample, while a perpendicular objective collects emitted fluorescence (Figure 3C) (Stelzer et al., 2021). LSFM presents a compelling option for accelerating imaging speeds and independently optimizing lateral and axial resolution (Voie et al., 1993; Huisken et al., 2004; Dodt et al., 2007; Power and Huisken, 2017). Moreover, the technique exceeds point-scanning methods in speed, significantly reduces photobleaching, and allows for adaptable sample mounting and orientation. This flexibility is beneficial in bypassing areas that are unclear in whole-mount samples. Since Dodt et al. first applied LSFM with tissue clearing in 2007, LSFM has developed into an important method for volumetric imaging of tissues (Dodt et al., 2007).
LSFM offers a broad field of view and quick image acquisition, making it possible for researchers to swiftly and successfully capture detailed signal information of whole intact samples. This ability makes it specifically fit for imaging intricate biological structures such as ovarian follicles. Lin et al. (2017) demonstrated that LSFM can effectively visualize a range of follicular features, including diameters ranging from 70 μm to 2.5 mm, sizes of developing cumulus oophorus complexes (COCs) ranging from 40 μm to 110 μm, and follicular wall thicknesses ranging from 90 μm to 120 μm. The system excels at identifying follicles at all developmental stages, particularly the small primordial follicle clusters crucial for egg nest formation. Recent advancements have enabled the use of LSFM to characterize single-cell interactions within 3D tissues, providing insights into the cellular mechanics and interactions that are crucial for understanding physiological and pathological processes in the uterus (Fredrickson et al., 2021; Tomizawa et al., 2024). For example, optical visco-elastography combined with LSFM has been used to study the maternal–fetal interface, revealing the mechanical properties and interactions between endometrial stromal fibroblasts and placental extravillous trophoblasts (Tomizawa et al., 2024).
4.4 Strategies for bioimaging analysis
Another critical step is to process and analyze the acquired images to extract valuable biological insights. Image adjustment primarily improves visibility for presentation, whereas analysis focuses on detailed data extraction from the dataset. Handling LSFM images involves two main phases: essential image pre-processing to address imaging process and stitch image tiles into volumetric dataset, followed by information extraction through segmentation and quantitative evaluation (Richardson et al., 2021; Delgado-Rodriguez et al., 2022).
Pre-processing resolves measurement artifacts and reconstructs accurate representations, potentially with lossless compression. This often involves deskewing to ensure voxel consistency within 3D systems. Tools range from commercial software (Imaris, AMIRA) with robust 3D analysis but high costs to open-source platforms (FiJI/ImageJ, Napari) offering free customizable operations via plug-ins (Folts et al., 2024).
Object segmentation identifies regions of interest (ROIs) based on fluorescence or size. Available through both Imaris/AMIRA and open-source tools with plug-ins like Stardist/Cellpose (Lesage et al., 2023; Stringer et al., 2021). Applications range from cellular-scale oocyte studies in mouse/medaka ovaries to organ-level vasculature/morphogenesis analyses (Belle et al., 2017; Combes et al., 2009; Bunce et al., 2021). Object classification faces ovarian challenges: size-dependent segmentation limits follicle differentiation (Folts et al., 2024).
5 Future directions in the female reproductive system and imaging
5.1 Advancing early diagnosis of uterine and ovarian pathologies
3D tissue clearing technologies overcome the fundamental limitations of traditional 2D histology and clinical imaging by enabling volumetric, subcellular-resolution visualization of intact uterine architecture. Tissue clearing technologies, such as the PEGASOS method, achieve tissue transparency through chemical treatment and integrate fluorescence labeling for subcellular-resolution 3D imaging (Jing et al., 2018). This enables spatial localization of microscopic lesions, including adenomyotic lesions, within deep tissues such as the myometrium (Martire et al., 2024). These technologies overcome the inherent limitations of 2D histopathology, which cannot reliably discern invasive growth patterns crucial for accurate tumor grading. By enabling 3D visualization of transparent tissues, such as pancreatic tumors processed using the FLASH technique, these methods reveal growth architectures and microinvasive margins that remain undetectable in traditional planar sections (Messal et al., 2019; Noë et al., 2018). This capability is particularly significant for assessing tumor margins, as 3D mapping of infiltrative borders and internal features like necrosis or abnormal vasculature provides detailed insights unattainable through 2D analysis (Almagro et al., 2021). For example, such approaches facilitate the reliable differentiation of leiomyosarcomas from benign fibroids or adenomyomas by exposing the spatial complexity of neoplastic infiltration. Unlike 2D slices, which obscure the true extent of local invasion and infiltration, 3D imaging directly captures the geometric relationship between tumor tissue and surrounding structures, offering a robust solution for precise tumor margin evaluation and enhancing differential diagnosis in clinical oncology (Guldner et al., 2016). Beyond neoplastic applications, 3D tissue clearing technologies like CUBIC have demonstrated utility in animal models, enabling whole-mount 3D imaging of murine intrauterine embryos and placentas by achieving tissue transparency (Kagami et al., 2017). This technique reveals 3D relationships between embryonic, placental, and uterine tissues—insights unattainable via conventional 2D methods—by visualizing spatial organizations of structures like embryonic membranes and placental vasculature. Such capabilities hold promise for advancing the understanding of uterine developmental anomalies, as they allow precise mapping of tissue interfaces with subcellular resolution (Arora et al., 2016). While currently validated in mice, this approach highlights the translational potential of 3D tissue clearing to improve diagnostic accuracy for human uterine malformations by exposing subtle structural deviations critical for fertility and pregnancy outcomes.
The advancement of tissue clearing methodologies and 3D ovarian cancer organoid models offers transformative potential for early and differential diagnosis of ovarian pathologies, including endometriomas, benign cysts, borderline tumors, and malignancies. In benign conditions like polycystic ovary syndrome (PCOS), a prevalent endocrine disorder impacting reproductive, metabolic, and cardiovascular health, tissue clearing enables high-resolution 3D visualization of pathological features previously inaccessible via conventional histology (Meier, 2018). For instance, Ma et al. utilized CLARITY-cleared ovaries to demonstrate that electro-acupuncture (EA) restores angiogenesis in antral follicles of PCOS-like rats, promoting folliculogenesis and ovulation (Ma et al., 2018). Subsequent CUBIC-based analysis further revealed aberrant ovarian innervation as a central pathological mechanism in PCOS, with EA exerting therapeutic effects via modulation of suprachiasmatic nucleus (SCN)-mediated neural pathways (Tong et al., 2020). In oncological diagnostics, 3D patient-derived ovarian cancer organoids replicate native tumor architecture and cellular heterogeneity, serving as physiologically relevant platforms for differential diagnosis, drug screening, and personalized therapy prediction (Clevers, 2016; Kopper et al., 2019; Maru et al., 2019). As tissue clearing methods and supporting technologies continue to progress, they are set to play an increasingly essential function in the study of ovarian diseases by enabling direct 3D validation of organoid-tumor congruence, precise mapping of cellular interactions, and identification of treatment-responsive heterogeneity (Maru et al., 2019). This synergistic integration of volumetric imaging and advanced organoid models thus provides unprecedented insights into the spatial dynamics of ovarian pathologies, accelerating biomarker discovery and refining diagnostic stratification across benign, borderline, and malignant entities.
5.2 Single-cell spatial omics in utero-ovarian tissues
Conventional single-cell transcriptomics and proteomics provide high-resolution molecular characterization but suffer from inherent spatial information loss due to tissue dissociation and sectioning (Zhou et al., 2023). This limitation impedes the identification of rare cell populations, such as ovarian micro-metastases or endometriosis-initiating cells, and obscures early pathological events at single-cell resolution. While laser-capture microdissection enables targeted single-cell analysis, its accuracy remains constrained when studying microscale pathological structures, including ovarian cancer micro-metastases (Ertürk, 2024).
Recent progress in spatial omics technologies, particularly sequential fluorescence in situ hybridization (seqFISH), combined with 3D tissue-clearing techniques has opened new avenues for resolving these challenges (Gandin et al., 2025). Tissue clearing methods like DISCO and Tris buffer–mediated retention of in situ hybridization chain reaction signal in cleared organs (TRISCO) have demonstrated remarkable potential to overcome these limitations by preserving both molecular integrity and 3D tissue architecture, enabling transcriptomic and proteomic analysis in intact millimeter-scale specimens (Bhatia et al., 2022; Kanatani et al., 2024). Achieving single-cell-resolution barcode detection in 3D-cleared tissues is essential for precise spatial mapping of molecular data, whereas implementing spatial omics technologies in volumetric formats is necessary to comprehensively analyze rare cellular events across entire organs. As these technologies mature, their integration with AI-driven data analysis pipelines will pave the way for dynamic 4D atlases of physiological processes like endometrial regeneration and ovarian aging (Ertürk, 2024; Cao et al., 2022). This transformative approach will provide fundamental new insights into the spatial regulation of gynecological pathologies, from their earliest cellular origins to tissue-scale manifestations, ultimately enabling more precise diagnostic and therapeutic strategies.
5.3 AI-based techniques for quantitative analysis
Another direction in reproductive biology will involve refining established methods for data analysis and applying AI-based approaches for quantitative analysis. The integration of AI-based analysis into image processing offers a new way to overcome bioimage analysis (Hallou et al., 2021). These AI-driven tools have overcome previous challenges by enhancing the precision of segment distinction and increasing the degree of classification contained in images. Furthermore, these methods dramatically reduce the time required for image analysis and minimize the risk of human error (Moen et al., 2019). AI-based tools are categorized into machine learning and deep learning (Moen et al., 2019; Hallou et al., 2021). Machine learning involves algorithms and statistical models that allow computers to learn from and generate predictions on the basis of data. Deep learning, a subset of machine learning, applies artificial neural networks that analyze data through a complex, layered structure, resembling the way the human brain forms connections and interprets information. All aspects of 3D bioimage analysis can be conducted manually, semi-manually, or fully automatically. Tools based on both machine learning and deep learning are instrumental in advancing our understanding of the anatomy and function of the reproductive system. The primary use of AI in image analysis of cleared reproductive system tissues involves counting ovarian follicles, a metric frequently used to assess fertility and ovarian health. Traditionally, counting follicles manually in histological sections is not only labor intensive but also prone to error. Manual counting methods often differ significantly between studies because of the absence of a universal scaling factor, leading to considerable variability in reported follicle counts (Tilly, 2003). Recently, Lesage and colleagues (Lesage et al., 2023) crafted a workflow employing open-source, cost-free, deep learning for analyzing 3D images of adult and larval medaka ovaries. They extended this approach to other typical animal models, such as zebrafish and mice. Their method segments ovarian follicles and calculates both the total number and volume of follicles. Image denoising was achieved via Noise2Void (Krull et al., 2019) within FiJI, with automated segmentation carried out via the CellPose algorithm (Stringer et al., 2021), initiated from an Anaconda command prompt. The open-source nature of these tools enhances the accessibility of this pipeline, while the adaptability of the deep learning framework allows it to be retrained for analyzing ovaries from different species.
6 Conclusion
Recent studies have laid the groundwork for understanding the functions of the mammalian ovary and mapping the 3D structure of the uterus. In the future, the creation of non-toxic clearing agents is expected to enhance the 3D visualization of samples, benefiting both laboratory analyses and clinical diagnostics. These advancements enable early detection of uterine and ovarian pathologies, significantly improving differential diagnosis and therapeutic stratification. The integration of spatial transcriptomics with cleared tissue imaging is poised to decode the molecular topography of reproductive organs, revealing previously inaccessible cell-cell communication networks during folliculogenesis and embryo implantation. AI-powered analytical platforms are emerging as essential tools for interpreting the multidimensional datasets generated by these integrated approaches, capable of identifying subtle pathological patterns in conditions like endometriosis or premature ovarian insufficiency.
As tissue clearing technologies have advanced and become more aligned with cutting-edge imaging and machine learning tools, their widespread use is anticipated to greatly increase our comprehension of the complexities of the ovary and uterus. This integration will be vital for advancing female reproductive health and deepening our understanding of reproductive aging.
Author contributions
QL: Writing – original draft, Writing – review and editing. ZS: Writing – original draft, Writing – review and editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. SL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. ZD: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review and editing. XZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. TL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review and editing. XC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Natural Science Funding of China (Grant No. 82201851), the Shenzhen Science and Technology Program (Grant No. JCYJ20210324141403009 and RCYX20210609104608036), and the Shenzhen Key Medical Discipline Construction Fund (Grant No. SZXK028).
Acknowledgments
We thank BioRender for providing drawing support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almagro, J., Messal, H. A., Zaw Thin, M., Van Rheenen, J., and Behrens, A. (2021). Tissue clearing to examine tumour complexity in three dimensions. Nat. Rev. Cancer. 21, 718–730. doi:10.1038/s41568-021-00382-w
Aoyagi, Y., Kawakami, R., Osanai, H., Hibi, T., and Nemoto, T. (2015). A rapid optical clearing protocol using 2,2'-thiodiethanol for microscopic observation of fixed mouse brain. PLoS One 10, e0116280. doi:10.1371/journal.pone.0116280
Arora, R., Fries, A., Oelerich, K., Marchuk, K., Sabeur, K., Giudice, L. C., et al. (2016). Insights from imaging the implanting embryo and the uterine environment in three dimensions. Development 143, 4749–4754. doi:10.1242/dev.144386
Azaripour, A., Lagerweij, T., Scharfbillig, C., Jadczak, A. E., Willershausen, B., and Van Noorden, C. J. F. (2016). A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog. Histochem. Cytochem. 51, 9–23. doi:10.1016/j.proghi.2016.04.001
Baetens, D., Verdin, H., De Baere, E., and Cools, M. (2019). Update on the genetics of differences of sex development (DSD). Best. Pract. Res. Clin. Endocrinol. Metab. 33, 101271. doi:10.1016/j.beem.2019.04.005
Bates, G. W., and Bowling, M. (2013). Physiology of the female reproductive axis. Periodontol. 2000 61, 89–102. doi:10.1111/j.1600-0757.2011.00409.x
Belle, M., Godefroy, D., Couly, G., Malone, S. A., Collier, F., Giacobini, P., et al. (2017). Tridimensional visualization and analysis of early human development. Cell. 169, 161–173.e12. doi:10.1016/j.cell.2017.03.008
Berg, S., Kutra, D., Kroeger, T., Straehle, C. N., Kausler, B. X., Haubold, C., et al. (2019). Ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232. doi:10.1038/s41592-019-0582-9
Bhatia, H. S., Brunner, A. D., ÖztüRK, F., Kapoor, S., Rong, Z., Mai, H., et al. (2022). Spatial proteomics in three-dimensional intact specimens. Cell. 185, 5040–5058.e19. doi:10.1016/j.cell.2022.11.021
Boateng, R., Boechat, N., Henrich, P. P., and Bolcun-Filas, E. (2021). Whole ovary immunofluorescence, clearing, and multiphoton microscopy for quantitative 3D analysis of the developing ovarian reserve in mouse. J. Vis. Exp. (175), e62972. doi:10.3791/62972
Bohn, S., Sperlich, K., Stahnke, T., SchüNEMANN, M., Stolz, H., Guthoff, R. F., et al. (2020). Multiwavelength confocal laser scanning microscopy of the cornea. Biomed. Opt. Express 11, 5689–5700. doi:10.1364/boe.397615
Bunce, C., Mckey, J., and Capel, B. (2021). Concerted morphogenesis of genital ridges and nephric ducts in the mouse captured through whole-embryo imaging. Development 148, dev199208. doi:10.1242/dev.199208
Cao, R., Wallrabe, H., and Periasamy, A. (2020). Multiphoton FLIM imaging of NAD(P)H and FAD with one excitation wavelength. J. Biomed. Opt. 25, 1–16. doi:10.1117/1.Jbo.25.1.014510
Cao, Y., Chau, M. H. K., Zheng, Y., Zhao, Y., Kwan, A. H. W., Hui, S. Y. A., et al. (2022). Exploring the diagnostic utility of genome sequencing for fetal congenital heart defects. Prenat. Diagn 42, 862–872. doi:10.1002/pd.6151
Chen, F., Tillberg, P. W., and Boyden, E. S. (2015). Expansion microscopy. Science 347, 543–548. doi:10.1126/science.1260088
Chi, J., Crane, A., Wu, Z., and Cohen, P. (2018). Adipo-clear: a tissue clearing method for three-dimensional imaging of adipose tissue. J. Vis. Exp., 58271. doi:10.3791/58271
Choi, S. W., Guan, W., and Chung, K. (2021). Basic principles of hydrogel-based tissue transformation technologies and their applications. Cell. 184, 4115–4136. doi:10.1016/j.cell.2021.07.009
Chung, K., Wallace, J., Kim, S. Y., Kalyanasundaram, S., Andalman, A. S., Davidson, T. J., et al. (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. doi:10.1038/nature12107
Clendenon, S. G., Young, P. A., Ferkowicz, M., Phillips, C., and Dunn, K. W. (2011). Deep tissue fluorescent imaging in scattering specimens using confocal microscopy. Microsc. Microanal. 17, 614–617. doi:10.1017/s1431927611000535
Clevers, H. (2016). Modeling development and disease with organoids. Cell. 165, 1586–1597. doi:10.1016/j.cell.2016.05.082
Combes, A. N., Lesieur, E., Harley, V. R., Sinclair, A. H., Little, M. H., Wilhelm, D., et al. (2009). Three-dimensional visualization of testis cord morphogenesis, a novel tubulogenic mechanism in development. Dev. Dyn. 238, 1033–1041. doi:10.1002/dvdy.21925
Costantini, I., Ghobril, J. P., di Giovanna, A. P., Allegra Mascaro, A. L., Silvestri, L., Müllenbroich, M. C., et al. (2015). A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 5, 9808. doi:10.1038/srep09808
Cutrale, F., Fraser, S. E., and Trinh, L. A. (2019). Imaging, visualization, and computation in developmental biology. Annu. Rev. Biomed. Data Sci. 2, 223–251. doi:10.1146/annurev-biodatasci-072018-021305
Delgado-Rodriguez, P., Brooks, C. J., Vaquero, J. J., and MuñOZ-Barrutia, A. (2022). Innovations in ex vivo light sheet fluorescence microscopy. Prog. Biophys. Mol. Biol. 168, 37–51. doi:10.1016/j.pbiomolbio.2021.07.002
Denk, W., Strickler, J. H., and Webb, W. W. (1990). Two-photon laser scanning fluorescence microscopy. Science, 248, 73–76. doi:10.1126/science.2321027
Dodt, H.-U., Leischner, U., Schierloh, A., Jährling, N., Mauch, C. P., Deininger, K., et al. (2007). Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336. doi:10.1038/nmeth1036
Ertürk, A. (2024). Deep 3D histology powered by tissue clearing, omics and AI. Nat. Methods 21, 1153–1165. doi:10.1038/s41592-024-02327-1
ErtüRK, A., Becker, K., Jährling, N., Mauch, C. P., Hojer, C. D., Egen, J. G., et al. (2012). Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995. doi:10.1038/nprot.2012.119
Faire, M., Skillern, A., Arora, R., Nguyen, D. H., Wang, J., Chamberlain, C., et al. (2015). Follicle dynamics and global organization in the intact mouse ovary. Dev. Biol. 403, 69–79. doi:10.1016/j.ydbio.2015.04.006
Fan, X., Bialecka, M., Moustakas, I., Lam, E., Torrens-Juaneda, V., Borggreven, N. V., et al. (2019). Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat. Commun. 10, 3164. doi:10.1038/s41467-019-11036-9
Feng, Y., Cui, P., Lu, X., Hsueh, B., Moller Billig, F., Zarnescu Yanez, L., et al. (2017). CLARITY reveals dynamics of ovarian follicular architecture and vasculature in three-dimensions. Sci. Rep. 7, 44810. doi:10.1038/srep44810
Filant, J., and Spencer, T. E. (2014). Uterine glands: biological roles in conceptus implantation, uterine receptivity and decidualization. Int. J. Dev. Biol. 58, 107–116. doi:10.1387/ijdb.130344ts
Földes-Papp, Z., Demel, U., and Tilz, G. P. (2003). Laser scanning confocal fluorescence microscopy: an overview. Int. Immunopharmacol. 3, 1715–1729. doi:10.1016/S1567-5769(03)00140-1
Folts, L., Martinez, A. S., and Mckey, J. (2024). Tissue clearing and imaging approaches for in toto analysis of the reproductive system. Biol. Reprod. 110, 1041–1054. doi:10.1093/biolre/ioad182
Fredrickson, S. J., Hoog, T. G., and Udan, R. S. (2021). Visualizing blood vessel development in cultured mouse embryos using lightsheet microscopy. Methods Mol. Biol. 2319, 93–104. doi:10.1007/978-1-0716-1480-8_11
Gandin, V., Kim, J., Yang, L. Z., Lian, Y., Kawase, T., Hu, A., et al. (2025). Deep-tissue transcriptomics and subcellular imaging at high spatial resolution. Science 388, eadq2084. doi:10.1126/science.adq2084
Goad, J., Ko, Y. A., Kumar, M., Syed, S. M., and Tanwar, P. S. (2017). Differential wnt signaling activity limits epithelial gland development to the anti-mesometrial side of the mouse uterus. Dev. Biol. 423, 138–151. doi:10.1016/j.ydbio.2017.01.015
Gradinaru, V., Treweek, J., Overton, K., and Deisseroth, K. (2018). Hydrogel-tissue chemistry: principles and applications. Annu. Rev. Biophys. 47, 355–376. doi:10.1146/annurev-biophys-070317-032905
Guglielmo, M. C., and Albertini, D. F. (2013). The structural basis for coordinating oogenesis and folliculogenesis. In: G. Coticchio, D. F. Albertini, and L. De Santis, editors Oogenesis. London, UK: Springer London.
Guldner, I. H., Yang, L., Cowdrick, K. R., Wang, Q., Alvarez Barrios, W. V., Zellmer, V. R., et al. (2016). An integrative platform for three-dimensional quantitative analysis of spatially heterogeneous metastasis landscapes. Sci. Rep. 6, 24201. doi:10.1038/srep24201
Gurcan, M. N., Madabhushi, A., Lotz, J., Berger, J., MüLLER, B., Breuhahn, K., et al. (2014). Zooming in: high resolution 3D reconstruction of differently stained histological whole slide images. In: Proceedings Volume 9041, Medical Imaging 2014: Digital Pathology. San Francisco, California: SPIE, 904104. doi:10.1117/12.2043381
Habiba, M., Heyn, R., Bianchi, P., Brosens, I., and Benagiano, G. (2021). The development of the human uterus: morphogenesis to menarche. Hum. Reprod. Update 27, 1–26. doi:10.1093/humupd/dmaa036
Hallou, A., Yevick, H. G., Dumitrascu, B., and Uhlmann, V. (2021). Deep learning for bioimage analysis in developmental biology. Development 148, dev199616. doi:10.1242/dev.199616
Hama, H., Kurokawa, H., Kawano, H., Ando, R., Shimogori, T., Noda, H., et al. (2011). Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488. doi:10.1038/nn.2928
Hirshfield, A. N., and Desanti, A. M. (1995). Patterns of ovarian cell proliferation in rats during the embryonic period and the first three weeks postpartum. Biol. Reprod. 53, 1208–1221. doi:10.1095/biolreprod53.5.1208
Horton, N. G., Wang, K., Kobat, D., Clark, C. G., Wise, F. W., Schaffer, C. B., et al. (2013). In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics 7, 205–209. doi:10.1038/nphoton.2012.336
Hou, B., Zhang, D., Zhao, S., Wei, M., Yang, Z., Wang, S., et al. (2015). Scalable and DiI-compatible optical clearance of the Mammalian brain. Front. Neuroanat. 9, 19. doi:10.3389/fnana.2015.00019
Hua, L., Zhou, R., Thirumalai, D., and Berne, B. J. (2008). Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc. Natl. Acad. Sci. U. S. A. 105, 16928–16933. doi:10.1073/pnas.0808427105
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J., and Stelzer, E. H. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. doi:10.1126/science.1100035
Isaacson, D., Mccreedy, D., Calvert, M., Shen, J., Sinclair, A., Cao, M., et al. (2020). Imaging the developing human external and internal urogenital organs with light sheet fluorescence microscopy. Differentiation 111, 12–21. doi:10.1016/j.diff.2019.09.006
Jacob, A. M., Lindemann, A. F., Wagenpfeil, J., Geiger, S., Layer, Y. C., Salam, B., et al. (2024). Autofluorescence-based tissue characterization enhances clinical prospects of light-sheet-microscopy. Sci. Rep. 14, 18033. doi:10.1038/s41598-024-67366-2
Jensen, K. H. R., and Berg, R. W. (2017). Advances and perspectives in tissue clearing using CLARITY. J. Chem. Neuroanat. 86, 19–34. doi:10.1016/j.jchemneu.2017.07.005
Jing, D., Zhang, S., Luo, W., Gao, X., Men, Y., Ma, C., et al. (2018). Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell. Res. 28, 803–818. doi:10.1038/s41422-018-0049-z
Kafian, H., Lalenejad, M., Moradi-Mehr, S., Birgani, S. A., and Abdollahpour, D. (2020). Light-sheet fluorescence microscopy with scanning non-diffracting beams. Sci. Rep. 10, 8501. doi:10.1038/s41598-020-63847-2
Kagami, K., Ono, M., Iizuka, T., Matsumoto, T., Hosono, T., Sekizuka-Kagami, N., et al. (2020). A novel third mesh-like myometrial layer connects the longitudinal and circular muscle fibers -A potential stratum to coordinate uterine contractions. Sci. Rep. 10, 8274. doi:10.1038/s41598-020-65299-0
Kagami, K., Shinmyo, Y., Ono, M., Kawasaki, H., and Fujiwara, H. (2017). Three-dimensional visualization of intrauterine conceptus through the uterine wall by tissue clearing method. Sci. Rep. 7, 5964. doi:10.1038/s41598-017-06549-6
Kagami, K., Shinmyo, Y., Ono, M., Kawasaki, H., and Fujiwara, H. (2018). Three-dimensional evaluation of murine ovarian follicles using a modified CUBIC tissue clearing method. Reprod. Biol. Endocrinol. 16, 72. doi:10.1186/s12958-018-0381-7
Kanatani, S., Kreutzmann, J. C., Li, Y., West, Z., Larsen, L. L., Nikou, D. V., et al. (2024). Whole-brain spatial transcriptional analysis at cellular resolution. Science 386, 907–915. doi:10.1126/science.adn9947
Kartasalo, K., Latonen, L., Vihinen, J., Visakorpi, T., Nykter, M., and Ruusuvuori, P. (2018). Comparative analysis of tissue reconstruction algorithms for 3D histology. Bioinformatics 34, 3013–3021. doi:10.1093/bioinformatics/bty210
Ke, M. T., Fujimoto, S., and Imai, T. (2013). SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161. doi:10.1038/nn.3447
Kopper, O., De Witte, C. J., Lõhmussaar, K., Valle-Inclan, J. E., Hami, N., Kester, L., et al. (2019). An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 25, 838–849. doi:10.1038/s41591-019-0422-6
Krull, A., Vicar, T., Prakash, M., Lalit, M., and Jug, F. (2019). Probabilistic Noise2Void: unsupervised content-aware denoising. Front. Comput. Sci. 2.
Ku, T., Swaney, J., Park, J. Y., Albanese, A., Murray, E., Cho, J. H., et al. (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981. doi:10.1038/nbt.3641
Kuwajima, T., Sitko, A. A., Bhansali, P., Jurgens, C., Guido, W., and Mason, C. (2013). ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368. doi:10.1242/dev.091844
Lesage, M., Thomas, M., Pecot, T., Ly, T. K., Hinfray, N., Beaudouin, R., et al. (2023). An end-to-end pipeline based on open source deep learning tools for reliable analysis of complex 3D images of ovaries. Development 150, dev201185. doi:10.1242/dev.201185
Lin, H.-C. A., Dutta, R., Mandal, S., Kind, A., Schnieke, A., and Razansky, D. (2017). Light-sheet microscopy for quantitative ovarian folliculometry. In: Proceedings Volume 10043, Diagnosis and Treatment of Diseases in the Breast and Reproductive System. San Francisco, California: SPIE. doi:10.1117/12.2252242
Liu, Y., Xu, R., Zhou, Y., Wang, Y., Zhang, F., Tong, X., et al. (2023). Diane-35 and metformin therapy in rats with endometrial lesions induced by dihydrotestosterone exposure. Ann. Transl. Med. 11, 247. doi:10.21037/atm-21-2441
Li, W., Germain, R. N., and Gerner, M. Y. (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (C(e)3D). Proc. Natl. Acad. Sci. U. S. A. 114, e7321–e7330. doi:10.1073/pnas.1708981114
Li, Y., Xu, J., Wan, P., Yu, T., and Zhu, D. (2018). Optimization of GFP fluorescence preservation by a modified uDISCO clearing protocol. Front. Neuroanat. 12, 67. doi:10.3389/fnana.2018.00067
Lutton, E. J., Lammers, W. J., James, S., Van Den Berg, H. A., and Blanks, A. M. (2017). A computational method for three-dimensional reconstruction of the microarchitecture of myometrial smooth muscle from histological sections. PLoS One 12, e0173404. doi:10.1371/journal.pone.0173404
Madhavan, M. K., Demayo, F. J., Lydon, J. P., Joshi, N. R., Fazleabas, A. T., and Arora, R. (2022). Aberrant uterine folding in mice disrupts implantation chamber formation and alignment of embryo-uterine axes. Development 149, dev200300. doi:10.1242/dev.200300
Malki, S., Tharp, M. E., and Bortvin, A. (2015). A whole-mount approach for accurate quantitative and spatial assessment of fetal oocyte dynamics in mice. Biol. Reprod. 93, 113. doi:10.1095/biolreprod.115.132118
Malki, S., Van der Heijden, G. W., O’Donnell, K. A., Martin, S. L., and Bortvin, A. (2014). A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell. 29, 521–533. doi:10.1016/j.devcel.2014.04.027
Malkovskiy, A. V., Tom, A., Joubert, L. M., and Bao, Z. (2022). Visualization of the distribution of covalently cross-linked hydrogels in CLARITY brain-polymer hybrids for different monomer concentrations. Sci. Rep. 12, 13549. doi:10.1038/s41598-022-17687-x
Martire, F. G., D’Abate, C., Schettini, G., Cimino, G., Ginetti, A., Colombi, I., et al. (2024). Adenomyosis and adolescence: a challenging diagnosis and complex management. Diagnostics (Basel) 14, 2344. doi:10.3390/diagnostics14212344
Maru, Y., Tanaka, N., Itami, M., and Hippo, Y. (2019). Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol. Oncol. 154, 189–198. doi:10.1016/j.ygyno.2019.05.005
Masselink, W., Reumann, D., Murawala, P., Pasierbek, P., Taniguchi, Y., Bonnay, F., et al. (2019). Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 146, dev166884. doi:10.1242/dev.166884
Ma, T., Cui, P., Tong, X., Hu, W., Shao, L. R., Zhang, F., et al. (2018). Endogenous ovarian angiogenesis in polycystic ovary syndrome-like rats induced by low-frequency electro-acupuncture: the CLARITY three-dimensional approach. Int. J. Mol. Sci. 19, 3500. doi:10.3390/ijms19113500
Matsumoto, K., Mitani, T. T., Horiguchi, S. A., Kaneshiro, J., Murakami, T. C., Mano, T., et al. (2019). Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537. doi:10.1038/s41596-019-0240-9
Mckey, J., Anbarci, D. N., Bunce, C., Ontiveros, A. E., Behringer, R. R., and Capel, B. (2022). Integration of mouse ovary morphogenesis with developmental dynamics of the oviduct, ovarian ligaments, and rete ovarii. Elife 11, e81088. doi:10.7554/eLife.81088
Mckey, J., Cameron, L. A., Lewis, D., Batchvarov, I. S., and Capel, B. (2020). Combined iDISCO and CUBIC tissue clearing and lightsheet microscopy for in toto analysis of the adult mouse ovary†. Biol. Reprod. 102, 1080–1089. doi:10.1093/biolre/ioaa012
Meglinski, I., Bashkatov, A. N., Genina, E. A., Churmakov, D. Y., and Tuchin, V. V. (2004). The enhancement of confocal images of tissues at bulk optical immersion. Laser Phys. 13, 65–69. doi:10.48550/arXiv.physics/0401104
Meier, R. K. (2018). Polycystic ovary syndrome. Nurs. Clin. North Am. 53, 407–420. doi:10.1016/j.cnur.2018.04.008
Messal, H. A., Alt, S., Ferreira, R. M. M., Gribben, C., Wang, V. M., Cotoi, C. G., et al. (2019). Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature 566, 126–130. doi:10.1038/s41586-019-0891-2
Moen, E., Bannon, D., Kudo, T., Graf, W., Covert, M., and Van Valen, D. (2019). Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246. doi:10.1038/s41592-019-0403-1
Mohler, W., Millard, A. C., and Campagnola, P. J. (2003). Second harmonic generation imaging of endogenous structural proteins. Methods 29, 97–109. doi:10.1016/s1046-2023(02)00292-x
Mori, H., Nishida, H., Kusaba, T., Kawamura, K., Oyama, Y., and Daa, T. (2023). Clinicopathological correlations of endometrioid and clear cell carcinomas in the uterus and ovary. Med. Baltim. 102, e35301. doi:10.1097/md.0000000000035301
Noë, M., Rezaee, N., Asrani, K., Skaro, M., Groot, V. P., Wu, P. H., et al. (2018). Immunolabeling of cleared human pancreata provides insights into three-dimensional pancreatic anatomy and pathology. Am. J. Pathol. 188, 1530–1535. doi:10.1016/j.ajpath.2018.04.002
Oren, R., Fellus-Alyagor, L., Addadi, Y., Bochner, F., Gutman, H., Blumenreich, S., et al. (2018). Whole organ blood and lymphatic vessels imaging (WOBLI). Sci. Rep. 8, 1412. doi:10.1038/s41598-018-19663-w
Orvis, G. D., and Behringer, R. R. (2007). Cellular mechanisms of Mullerian duct formation in the mouse. Dev. Biol. 306, 493–504. doi:10.1016/j.ydbio.2007.03.027
Palumbo, A., and Yeh, J. (1994). In situ localization of apoptosis in the rat ovary during follicular atresia. Biol. Reprod. 51(5), 888–895. doi:10.1095/biolreprod51.5.888
Pan, C., Cai, R., Quacquarelli, F. P., Ghasemigharagoz, A., Lourbopoulos, A., Matryba, P., et al. (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867. doi:10.1038/nmeth.3964
Pascolo, L., Sena, G., Gianoncelli, A., Cernogoraz, A., Kourousias, G., Metscher, B. D., et al. (2019). Hard and soft X-ray imaging to resolve human ovarian cortical structures. J. Synchrotron Radiat. 26, 1322–1329. doi:10.1107/S1600577519003680
Pende, M., Vadiwala, K., Schmidbaur, H., Stockinger, A. W., Murawala, P., Saghafi, S., et al. (2020). A versatile depigmentation, clearing, and labeling method for exploring nervous system diversity. Sci. Adv. 6, eaba0365. doi:10.1126/sciadv.aba0365
Pichat, J., Iglesias, J. E., Yousry, T., Ourselin, S., and Modat, M. (2018). A survey of methods for 3D histology reconstruction. Med. Image Anal. 46, 73–105. doi:10.1016/j.media.2018.02.004
Pinheiro, T., Mayor, I., Edwards, S., Joven, A., Kantzer, C. G., Kirkham, M., et al. (2021). CUBIC-f: an optimized clearing method for cell tracing and evaluation of neurite density in the salamander brain. J. Neurosci. Methods 348, 109002. doi:10.1016/j.jneumeth.2020.109002
Power, R. M., and Huisken, J. (2017). A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 14, 360–373. doi:10.1038/nmeth.4224
Renier, N., Wu, Z., Simon, D. J., Yang, J., Ariel, P., and Tessier-Lavigne, M. (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 159, 896–910. doi:10.1016/j.cell.2014.10.010
Richardson, D. S., Guan, W., Matsumoto, K., Pan, C., Chung, K., ErtüRK, A., et al. (2021). TISSUE CLEARING. Nat. Rev. Methods Prim. 1, 84. doi:10.1038/s43586-021-00080-9
Ryu, Y., Kim, Y., Lim, H. R., Kim, H. J., Park, B. S., Kim, J. G., et al. (2022). Single-step fast tissue clearing of thick mouse brain tissue for multi-dimensional high-resolution imaging. Int. J. Mol. Sci. 23, 6826. doi:10.3390/ijms23126826
Sarma, U. C., Winship, A. L., and Hutt, K. J. (2020). Comparison of methods for quantifying primordial follicles in the mouse ovary. J. Ovarian Res. 13, 121. doi:10.1186/s13048-020-00724-6
Scheele, C. L. G. J., Herrmann, D., Yamashita, E., Lo Celso, C., Jenne, C. N., Oktay, M. H., et al. (2022). Multiphoton intravital microscopy of rodents. Nat. Rev. Methods Prim. 2, 89. doi:10.1038/s43586-022-00168-w
Septadina, I. S. (2023). An overview of the female reproductive system: a narrative literature review. Sriwijaya J. Obstetrics Gynecol. 1, 16–23. doi:10.59345/sjog.v1i1.25
Soto-Suazo, M., and Zorn, T. M. (2005). Primordial germ cells migration: morphological and molecular aspect. Anim. Reprod. 2, 147–160.
Soygur, B., Foecke, M. H., Gaylord, E. A., Fries, A., Li, J., Arora, R., et al. (2023). A roadmap for three-dimensional analysis of the intact mouse ovary. In: M. Buszczak, editor Germline stem cells: methods and protocols. New York, NY: Springer US.
Soygur, B., Jaszczak, R. G., Fries, A., Nguyen, D. H., Malki, S., Hu, G., et al. (2021). Intercellular bridges coordinate the transition from pluripotency to meiosis in mouse fetal oocytes. Sci. Adv. 7, eabc6747. doi:10.1126/sciadv.abc6747
Staudt, T., Lang, M. C., Medda, R., Engelhardt, J., and Hell, S. W. (2007). 2,2'-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 70, 1–9. doi:10.1002/jemt.20396
Stelzer, E. H. K., Strobl, F., Chang, B.-J., Preusser, F., Preibisch, S., Mcdole, K., et al. (2021). Light sheet fluorescence microscopy. Nat. Rev. Methods Prim. 1, 73. doi:10.1038/s43586-021-00069-4
Stringer, J. M., Alesi, L. R., Winship, A. L., and Hutt, K. J. (2023). Beyond apoptosis: evidence of other regulated cell death pathways in the ovary throughout development and life. Hum. Reprod. Update 29, 434–456. doi:10.1093/humupd/dmad005
Stringer, C., Wang, T., Michaelos, M., and Pachitariu, M. (2021). Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106. doi:10.1038/s41592-020-01018-x
Susaki, E. A., Tainaka, K., Perrin, D., Kishino, F., Tawara, T., Watanabe, T. M., et al. (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 157, 726–739. doi:10.1016/j.cell.2014.03.042
Susaki, E. A., Tainaka, K., Perrin, D., Yukinaga, H., Kuno, A., and Ueda, H. R. (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727. doi:10.1038/nprot.2015.085
Svingen, T., Francois, M., Wilhelm, D., and Koopman, P. (2012). Three-dimensional imaging of Prox1-EGFP transgenic mouse gonads reveals divergent modes of lymphangiogenesis in the testis and ovary. PLoS One 7, e52620. doi:10.1371/journal.pone.0052620
Tainaka, K., Kuno, A., Kubota, S. I., Murakami, T., and Ueda, H. R. (2016). Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell. Dev. Biol. 32, 713–741. doi:10.1146/annurev-cellbio-111315-125001
Taylor, E., and Gomel, V. (2008). The uterus and fertility. Fertil. Steril. 89, 1–16. doi:10.1016/j.fertnstert.2007.09.069
Tian, T., Yang, Z., and Li, X. (2021). Tissue clearing technique: recent progress and biomedical applications. J. Anat. 238, 489–507. doi:10.1111/joa.13309
Tilly, J. L. (2003). Ovarian follicle counts-not as simple as 1, 2, 3. Reprod. Biol. Endocrinol. 1, 11. doi:10.1186/1477-7827-1-11
Tomer, R., Ye, L., Hsueh, B., and Deisseroth, K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697. doi:10.1038/nprot.2014.123
Tomizawa, Y., Wali, K. H., Surti, M., Suhail, Y., and Kshitiz and Hoshino, K. (2024). Lightsheet microscopy integrates single-cell optical visco-elastography and fluorescence cytometry of 3D live tissues. bioRxiv (Preprint), 2024.04.20.590392. doi:10.1101/2024.04.20.590392
Tong, X., Liu, Y., Xu, X., Shi, J., Hu, W., Ma, T., et al. (2020). Ovarian innervation coupling with vascularity: the role of electro-acupuncture in follicular maturation in a rat model of polycystic ovary syndrome. Front. Physiol. 11, 474. doi:10.3389/fphys.2020.00474
Tsai, P. S., Kaufhold, J. P., Blinder, P., Friedman, B., Drew, P. J., Karten, H. J., et al. (2009). Correlations of neuronal and microvascular densities in Murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 29, 14553–14570. doi:10.1523/jneurosci.3287-09.2009
Tuchin, V. V. (2015). Tissue Opt. Photonics Biol. Tissue Struct. 1, 19. doi:10.18287/jbpe-2015-1-1-3
Vieites-Prado, A., and Renier, N. (2021). Tissue clearing and 3D imaging in developmental biology. Development 148, dev199369. doi:10.1242/dev.199369
Voie, A. H., Burns, D. H., and Spelman, F. A. (1993). Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236. doi:10.1111/j.1365-2818.1993.tb03346.x
Vue, Z., and Behringer, R. R. (2020). Epithelial morphogenesis in the perinatal mouse uterus. Dev. Dyn. 249, 1377–1386. doi:10.1002/dvdy.234
Vue, Z., Gonzalez, G., Stewart, C. A., Mehra, S., and Behringer, R. R. (2018). Volumetric imaging of the developing prepubertal mouse uterine epithelium using light sheet microscopy. Mol. Reprod. Dev. 85, 397–405. doi:10.1002/mrd.22973
Weiss, K. R., Voigt, F. F., Shepherd, D. P., and Huisken, J. (2021). Tutorial: practical considerations for tissue clearing and imaging. Nat. Protoc. 16, 2732–2748. doi:10.1038/s41596-021-00502-8
Wiebold, J. L., and Becker, W. C. (1987). Inequality in function of the right and left ovaries and uterine horns of the mouse. J. Reprod. Fertil. 79, 125–134. doi:10.1530/jrf.0.0790125
Xu, C., Nedergaard, M., Fowell, D. J., Friedl, P., and Ji, N. (2024). Multiphoton fluorescence microscopy for in vivo imaging. Cell. 187, 4458–4487. doi:10.1016/j.cell.2024.07.036
Xu, X., Mu, L., Li, L., Liang, J., Zhang, S., Jia, L., et al. (2022). Imaging and tracing the pattern of adult ovarian angiogenesis implies a strategy against female reproductive aging. Sci. Adv. 8, eabi8683. doi:10.1126/sciadv.abi8683
Yang, B., Treweek, J. B., Kulkarni, R. P., Deverman, B. E., Chen, C. K., Lubeck, E., et al. (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 158, 945–958. doi:10.1016/j.cell.2014.07.017
Yang, X., Ye, Z., Zhang, M., Ouyang, Y., He, D., Wang, X., et al. (2023). Tissue clearing to examine glioma complexity in 3 dimensions. J. Neuropathol. Exp. Neurol. 82, 376–389. doi:10.1093/jnen/nlad024
Yan, T., Wang, X., Liu, S., Fan, D., Xu, X., Zeng, Q., et al. (2022). Confocal laser scanning microscopy based on a silicon photomultiplier for multicolor in vivo imaging in near-infrared regions I and II. Small Methods 6, e2201105. doi:10.1002/smtd.202201105
Yuan, J., Deng, W., Cha, J., Sun, X., Borg, J. P., and Dey, S. K. (2018). Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation. Nat. Commun. 9, 603. doi:10.1038/s41467-018-03092-4
Zhang, Z., Ibrahim, M., Fu, Y., Wu, X., Ren, F., and Chen, L. (2018). Application of laser scanning confocal microscopy in the soft tissue exquisite structure for 3D scan. Int. J. Burns Trauma 8, 17–25.
Zhou, Z., Tan, C., Chau, M. H. K., Jiang, X., Ke, Z., Chen, X., et al. (2023). TEDD: a database of temporal gene expression patterns during multiple developmental periods in human and model organisms. Nucleic Acids Res. 51, D1168–d1178. doi:10.1093/nar/gkac978
Zhu, D., Larin, K. V., Luo, Q., and Tuchin, V. V. (2013). Recent progress in tissue optical clearing. Laser Phot. Rev. 7, 732–757. doi:10.1002/lpor.201200056
Keywords: tissue clearing, 3D visualization, ovary, uterus, spatial omics, AI
Citation: Liu Q, Song Z, Liu S, Dong Z, Zheng X, Leung TY and Chen X (2025) 3D visualization of uterus and ovary: tissue clearing techniques and biomedical applications. Front. Bioeng. Biotechnol. 13:1610539. doi: 10.3389/fbioe.2025.1610539
Received: 12 April 2025; Accepted: 24 June 2025;
Published: 07 July 2025.
Edited by:
Maria Angelica Miglino, Universidade de Marília, BrazilReviewed by:
Jose Amilcar Rizzo Sierra, Polytechnic University of Querétaro, MexicoFrancesco Giuseppe Martire, University of Rome Tor Vergata, Italy
Copyright © 2025 Liu, Song, Liu, Dong, Zheng, Leung and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Chen, Y2hlbnhpYW95YW5AY3Voay5lZHUuaGs=; Tak Yeung Leung, dHlsZXVuZ0BjdWhrLmVkdS5oaw==
†These authors have contributed equally to this work
 Qiqi Liu
Qiqi Liu Zhuo Song2†
Zhuo Song2† Zirui Dong
Zirui Dong Tak Yeung Leung
Tak Yeung Leung Xiaoyan Chen
Xiaoyan Chen