Editorial on the Research Topic
Fusion proteins for the detection of pathogens or pathogen receptors
Introduction
Menaces from pathogens, such as viruses, prions, bacteria and fungi (and their toxins), are widespread. Novel tools for the detection of pathogens and their host receptors are of critical importance for improving our understanding of the molecular mechanisms underlying diseases and for developing fast, affordable, and accurate diagnostics.
Recombinant DNA technology has enabled the genetic fusion of DNA fragments coding for different proteins and linkers, to produce single-chain fusion proteins comprising multiple functionalities. The simplest engineered fusion proteins are composed of a tag or fluorescent protein, an optional linker with specific properties (e.g., flexible, rigid, or containing a protease recognition site) and a protein domain of interest. As an example, a fusion protein comprising a fluorescent protein, connected via a flexible linker to the receptor-binding domain of SARS-CoV-2 (Bierig et al., 2020), can be used as a tool to visualize the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) on cells in fluorescent microscopy. Examples of more complex engineered proteins include biosensors that can detect changes in mammalian cell lines, for example, aspartate levels (Davidsen et al., 2024), or biosensors for the detection of specific bacterial second messengers (Kaczmarczyk et al., 2024).
Engineered protein-based sensors hold promise to enable efficient detection or monitoring of a wide range of pathogens and pathogen-induced changes in host cells.
Research Topic
The aim of this Research Topic was to collect articles focused on fusion proteins that enable new possibilities for pathogen research and diagnosis. The Research Topic comprises three articles presenting original research, and one review article.
The properties of linkers in fusion proteins are often critical for achieving a desired function. While flexible linkers allow freedom of motion between two connected domains, rigid linkers (Collu et al., 2022; Jeong et al., 2016; Kwon et al., 2020) can improve accessibility of specific protein regions.
Zane et al. describe their work on genetic fusions comprising pneumococcal surface protein A (PspA) and detoxified pneumolysin (PdT) to develop improved vaccines against the bacterial pathogen Streptococcus pneumoniae. Their data show the importance of linker composition and length for the stability of fusion proteins for medical applications (Figure 1A).
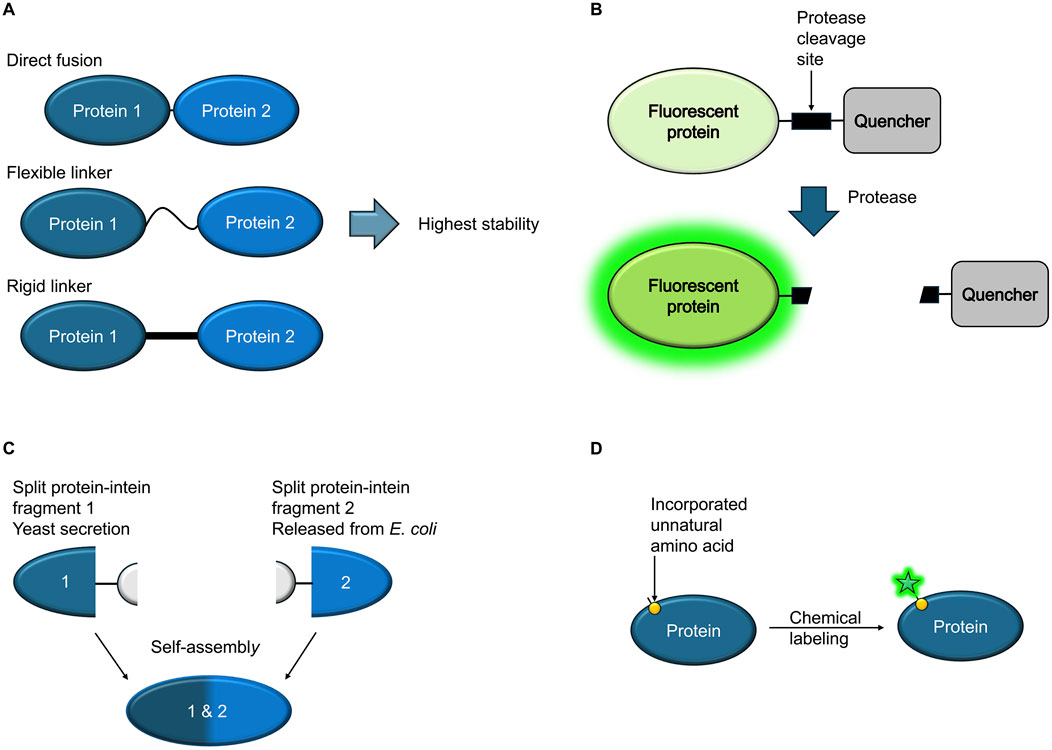
Figure 1. Schematic overview (A) Hybrid proteins consisting of two different vaccine candidate proteins, genetically fused with or without a linker, constitute a feasible alternative to available vaccines. The composition of the linker between the two proteins can affect the stability and biological activity of the fusion proteins. (B) A fluorescent protein is genetically fused to a quencher via a linker comprising a protease cleavage site that is recognized by a specific protease from a pathogen. If exposed to a solution containing the protease, the quencher is uncoupled from the fluorescent protein, resulting in increased fluorescence. (C) Inteins can be used to express two protein segments separately, followed by self-assembly into a complete, functional protein. The assembly can take place in situ in the culture medium. (D) Incorporation of unnatural amino acids displaying bioorthogonal handles on the protein surface enables subsequent selective chemical labeling with small molecule dyes that are less bulky than fluorescent proteins. Labeling can be carried out in live cells.
Devoy et al. engineered a modular protease sensor that enables detection of the presence of proteases using a cell-free setting. Specific protease activities can be used as biomarkers to detect viruses or cancer. Overall, the sensor consists of a green fluorescent protein (GFP), a protease cleavage site, a quencher and two different tags, one at the N-terminus, the other at the C-terminus. A specific protease, matching the chosen recognition/cleavage site in the construct, hence cleaves off the quencher, resulting in an increase in fluorescence, providing a simple readout (Figure 1B). Another possible readout is provided through the uncoupling of the two tags upon protease cleavage.
Typically, fusion proteins consist of a single continuous protein chain resulting from the expression of genetically fused elements. Inteins (protein introns) make it possible to express two protein segments separated by an intervening sequence, followed by autocatalytic splicing out of the intein part, resulting in ligation of the two flanking proteins (Lennon and Belfort, 2017). In this Research Topic, Wang et al. describe the use of intein-based splicing of proteins in situ by engineered microbial consortia. By secreting one intein-fused protein fragment from yeast cell cultures and releasing a matching complementary domain from co-cultured bacterial cells using an autolysis system, extracellular reconstitution takes place directly in the culture (Figure 1C). Possible uses include modular production of functional proteins, as well as logic computation or antibiotic resistance engineering.
While fusion protein technology often allows the engineering of valuable tools for the detection of pathogens or their receptors, there are also applications that pose technical challenges. For example, bulky fluorescent proteins can disrupt proper secretion of effectors. Singh and Kenney in their review article outline available bacterial protein labeling strategies in the context of host-pathogen interactions in detail and discuss novel approaches for the visualization of bacterial proteins and host-pathogen interactions that can overcome such problems. For example, genetic code expansion enables the incorporation of bioorthogonal handles into the protein of interest, allowing subsequent chemical labeling with dyes, even in live cells (Figure 1D).
Future directions
The study of pathogens and their interaction with host receptors is an active field of research. Novel tools, such as the strategies described in this Research Topic, play an important role towards enabling detection of specific proteins, or towards optimizing processes for the identification and characterization of pathogen-induced disorders.
Structural information can be highly useful for the design of fusion proteins, allowing a precise choice of domain boundaries. Natural proteins can furthermore be used as a guide on how to design or link proteins [molecular biomimetics, (e.g., Collu et al., 2022)].
Computational structure prediction and de novo protein design have recently come of age (e.g., Baek et al., 2021; Jumper et al., 2021; Kortemme, 2024; Pacesa et al., 2025) and their use for the design and optimization of constructs, linkers, and binders will likely become widespread.
Another current trend in structural research is the aim to solve structures of proteins and protein complexes not in their isolated form, but as much as possible in their native environments. In situ structural biology methods such as correlated light and electron microscopy (CLEM) (de Boer et al., 2015) or X-ray based methods such as holographic nano-tomography (Kuan et al., 2020) or ptychography (Bosch et al., 2024) allow new insights into cellular ultrastructure. Challenges include low contrast or the localization of proteins of interest in the crowded cellular environment. There is a need for novel tools, such as electron-dense tags like ferritin (Clarke and Royle, 2018) that can increase contrast or reveal the localization of specific cellular features. Fusion protein tools have a high potential to enable new possibilities in this field for pathogen research as well as for other areas of research.
Author contributions
RB: Investigation, Visualization, Conceptualization, Writing – review and editing, Validation, Methodology, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373 (6557), 871–876. doi:10.1126/science.abj8754
Bierig, T., Collu, G., Blanc, A., Poghosyan, E., and Benoit, R. M. (2020). Design, expression, purification, and characterization of a YFP-tagged 2019-nCoV spike receptor-binding domain construct. Front. Bioeng. Biotechnol. 8, 618615. doi:10.3389/fbioe.2020.618615
Bosch, C., Aidukas, T., Holler, M., Pacureanu, A., Müller, E., Peddie, C. J., et al. (2024). Non-destructive X-ray tomography of brain tissue ultrastructure. bioRxiv. doi:10.1101/2023.11.16.567403
Clarke, N. I., and Royle, S. J. (2018). FerriTag is a new genetically-encoded inducible tag for correlative light-electron microscopy. Nat. Commun. 9 (1), 2604. doi:10.1038/s41467-018-04993-0
Collu, G., Bierig, T., Krebs, A. S., Engilberge, S., Varma, N., Guixà-González, R., et al. (2022). Chimeric single α-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology. Structure 30 (1), 95–106.e7. doi:10.1016/j.str.2021.09.002
Davidsen, K., Marvin, J. S., Aggarwal, A., Brown, T. A., and Sullivan, L. B. (2024). An engineered biosensor enables dynamic aspartate measurements in living cells. Elife 12, RP90024. doi:10.7554/eLife.90024
de Boer, P., Hoogenboom, J. P., and Giepmans, B. N. (2015). Correlated light and electron microscopy: ultrastructure lights up. Nat. Methods 12 (6), 503–513. doi:10.1038/nmeth.3400
Jeong, W. H., Lee, H., Song, D. H., Eom, J. H., Kim, S. C., Lee, H. S., et al. (2016). Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker. Nat. Commun. 7, 11031. doi:10.1038/ncomms11031
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kaczmarczyk, A., van Vliet, S., Jakob, R. P., Teixeira, R. D., Scheidat, I., Reinders, A., et al. (2024). A genetically encoded biosensor to monitor dynamic changes of c-di-GMP with high temporal resolution. Nat. Commun. 15 (1), 3920. doi:10.1038/s41467-024-48295-0
Kortemme, T. (2024). De novo protein design-From new structures to programmable functions. Cell 187 (3), 526–544. doi:10.1016/j.cell.2023.12.028
Kuan, A. T., Phelps, J. S., Thomas, L. A., Nguyen, T. M., Han, J., Chen, C. L., et al. (2020). Dense neuronal reconstruction through X-ray holographic nano-tomography. Nat. Neurosci. 23 (12), 1637–1643. doi:10.1038/s41593-020-0704-9
Kwon, N. Y., Kim, Y., and Lee, J. O. (2020). The application of helix fusion methods in structural biology. Curr. Opin. Struct. Biol. 60, 110–116. doi:10.1016/j.sbi.2019.12.007
Lennon, C. W., and Belfort, M. (2017). Inteins. Curr. Biol. 27 (6), R204–R206. doi:10.1016/j.cub.2017.01.016
Keywords: fusion proteins, protein sensors, inteins, protein labeling, pathogen research, electron-dense protein tags, protein design, ultrastructure
Citation: Benoit RM (2025) Editorial: Fusion proteins for the detection of pathogens or pathogen receptors. Front. Bioeng. Biotechnol. 13:1660729. doi: 10.3389/fbioe.2025.1660729
Received: 06 July 2025; Accepted: 21 July 2025;
Published: 29 July 2025.
Edited and reviewed by:
Jean Marie François, Institut Biotechnologique de Toulouse (INSA), FranceCopyright © 2025 Benoit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger M. Benoit, cm9nZXIuYmVub2l0QHBzaS5jaA==
 Roger M. Benoit
Roger M. Benoit