- 1College of Medicine, Alfaisal University, Riyadh, Saudi Arabia
- 2International School of Medicine, Istanbul Medipol University, Istanbul, Türkiye
- 3Laboratory of Tissue/Organ Bioengineering & BioMEMS, Organ Transplant Centre of Excellence (TR&I-Dpt), King Faisal Specialist Hospital & Research Centre, Riyadh, Saudi Arabia
- 4Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 5Division of Basic and Translational Research, Department of Emergency Medicine, The Ohio State University, Columbus, OH, United States
- 6Laboratory of Molecular Immunology, Institute for Quantitative Biosciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan
- 7Department of Biological and Biomedical Sciences, Aga Khan University, Karachi, Pakistan
Cardiac organoid is a miniature and simplified three-dimensional (3D) cellular model system grown from progenitor cells or stem cells that more accurately mimic the significant biological characteristics and functions of the normal cardiac system than conventional two-dimensional (2D) models. With continued advances in 3D culture approaches, the cardiac organoid models produced through self-organization strategy following developmental induction conditions exhibit higher metabolic similarities and physiological relevance. Increasing evidence demonstrates that cardiac organoids based on the in vitro model system are useful platforms for studying human cardiac biology and pathophysiology. Despite significant advancements, the development of cardiac organoids has not progressed as far as other types of organoids due to the intricate cellular structure and microenvironment of the heart. In this review, we highlight the current classification and bioengineering strategies for establishing cardiac organoids using Matrigel and decellularized extracellular matrix derived culture platforms followed by a review of contemporary reports of their use in development biology, disease modeling, drug testing and efficacy evaluation. We also shed the light in the current limitations and future perspective of the cardiac organoid to motivate future research and accelerate the widespread adoption of organoids platforms.
1 Introduction
Cardiovascular diseases (CVD) are the leading cause of mortality and substantially contribute to healthcare and economic burdens worldwide (1). Despite enormous funding investments and implementation of various drug development strategies, ninety percent of new drugs fails in clinical trials (2). Cardiovascular disease and cancer have the lowest success rates in discovering new drug candidates, largely due to adverse effects of the therapeutic agents that result in clinical and subclinical cardiotoxicity (3–6). Because the elucidation of disease pathogenesis and high-throughput screening are of paramount importance in the development of new drugs, various 2D and 3D cell model systems have been published over the past several decades (7–9). In the traditional setting, cells are routinely cultured in 2D format for the purposes of disease research and drug screening, but 2D models inherently lack the structural complexity of their in vivo counterparts (10–12).Therefore, intensive research efforts have been made to develop next-generation culture methods that mimic important features of native tissues, such as the cellular and acellular microenvironment and cell-cell interactions when performing 3D cell culture experiments in vitro. The rapidly evolving fields of organoid engineering and biological model systems are continually providing new insights into basic experimental biology, human disease mechanisms and drug response efficacy research and are driving new therapeutic innovations (13, 14). Cardiac organoids are organized structures that self-assemble into miniaturized models composed of progenitor cells, cardiomyocytes, endothelial cells, and fibroblasts in a three-dimensional microenvironment that mimic the in vivo native organs (15, 16). Unlike traditional 2D myocardial culture systems, cardiac organoids recapitulate the human-specific aspects of heart histogenesis, physiology and developmental trajectory. Therefore, cardiac organoids have grown in popularity because they provide a unique opportunity to model cardiac diseases drug screening, and toxicity testing (17, 18). They are considered as powerful gateway to understand cardiac functions not only from a basic science perspective but also for the development of personalized therapies (19, 20). Towards that end, a wide range of cardiac organoid models have been reported over the past few years, applying a variety of experimental strategies (21, 22). Nonetheless, progress in the study of cardiac organoids is much slower than that of its other counterparts including brain, liver, kidney, and intestinal models (20–25). This represents an opportunity for active research related to the biofabrication techniques for the engineering of different models of cardioids for translational regenerative medicine applications (23–28).
In this glance article, we begin with history of the development of organoid technology and the use of organ-specific biomaterials derived from decellularized hearts in culture for cardiac organoid research. We then summarized their applications as in vitro model systems and drug screening tools. Despite the tremendous potentials of organoids models, it is essential to understand the existing hurdles and limitations of organoid technology. Therefore, we highlighted the current challenges and respective advantages of cardiac organoid research for basic and translational applications.
2 Timeline and developmental history of organoid technology
The initial efforts to generate organs in vitro began with pioneering dissociation-reaggregation experiments. In these early studies, Henry Van Peters Wilson showed that sponge cells, when mechanically separated, have the ability to come together again and self-organize, ultimately forming a complete organism (29). Several decades after this initial experiment, multiple research teams conducted dissociation-reaggregation studies, successfully creating various types of organs from separated cells of the amphibian pronephros and chicken embryos (30, 31). However, the initial observation of in vitro tissue-like colonies formation occurred through the co-culture of keratinocytes and 3T3 fibroblasts (32). An important milestone in this journey was the development of the differential adhesion hypothesis, which was prompted by the observation that cells, upon mechanical dissociation and subsequent aggregation, could autonomously reorganize into the original tissue structures from which they were derived. This phenomenon underscored the inherent capacity of cells for spatial organization and tissue reconstruction. Advancements in stem cell biology further elucidated the potential of stem cells to differentiate and organize into organ-like structures in vitro, as evidenced by the formation of teratomas and embryoid bodies. These entities demonstrated the ability of differentiated cells to form arrangements mimicking various tissue types. From the end of the 19th century (1998) to the beginning of the 20th century (2006), intensive research on stem cell technology, especially human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), triggered a wave of studies on the mechanisms and fate of stem cells in different culture environments (33, 34). In 2009, Hans Clever and team found that individual intestinal stem cells expressing leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) stem cells produce crypt-villus structures in vitro, independent of mesenchymal niche (i.e., stroma), marking the creation of the first organoids. Their findings showed that LGR5+ stem cells are involved in rapid regeneration of intestinal tissue and can self-organize the intestinal crypt-villus units from a single stem cell without requiring a surrounding cell niche (35). Since then, a variety of organoid models have been reported by different research groups, including intestinal, brain, heart, kidney, and liver, among others (35–39). These studies laid the groundwork for numerous other studies on organoids across various systems, such as the mesendoderm (including organs like the stomach, liver, pancreas, lung, and kidney) and neuroectoderm (such as the brain and retina), employing either adult stem cells (ASCs) or pluripotent stem cells (40).
3 Procedural requirements for generating of cardiac organoids
The primary basis for the development of organoid culture relies on the ability of homogeneous cells to self-organize and mimic the key features of the source tissue. The key steps in self-organization rely heavily on a series of highly regulated signaling pathways that play an important role in the self-organization and cell differentiation processes in the developmental cascade of pattern formation through a variety of morphogenetic rearrangements. The traditional method of organoid culture, expansion and development involves the enzymatic digestion of tissue fragments and derivation of stem cells, progenitor cells, followed by culturing them in a 3D microenvironment using growth factor-conditioned medium (15). A variety of morphogens are used to culture of cardiac organoids, including Wnt-3a epidermal growth factor (EGF), fibroblast growth factor (FGF), R-spondin, gastrin, noggin, Human Bone Morphogenetic Protein 4, Human Activin-A, Insulin, B-27 supplement, L-ascorbic acid 2 phosphate, Aprotinin, Palmitic acid. The methods for creating in vitro cardiac organoids using stem cells, progenitor cells, cardiomyocytes (CMs), endothelial cells (ECs), and cardiac fibroblasts, fall into two categories: scaffold-based and scaffold-free techniques (Figure 1). Scaffold-based methods utilize biomaterials like hydrogels or decellularized bioscaffolds, whereas scaffold-free techniques typically involve promoting the spherical aggregation of cultured cells in an anti-adhesive setting, in addition to some newly emerged techniques, used to facilitate cardiac organoid formation, including microarray technology, 3D bioprinted models, and scaffolds based on electrospun fiber mats. Scaffold-free 3D cultures are created by the self-assembly of cells, while scaffold-based cultures use hydrogels or other scaffolds to support tissue replicas. In either case, the cells are organized into a three-dimensional structure, which is essential for preserving their morphology, phenotype, and polarity. Scaffold-free systems can create artificial 3D heart tissues that maintain mechanical integrity without the need for external support (41). This paper will focus solely on the development of cardiac models using scaffold formation methods with decellularized materials. To understand the basic concepts and operating principles of biomaterials and bioengineering strategies suitable for organoids applications, the readers can refer to more specialized reviews (42, 43).
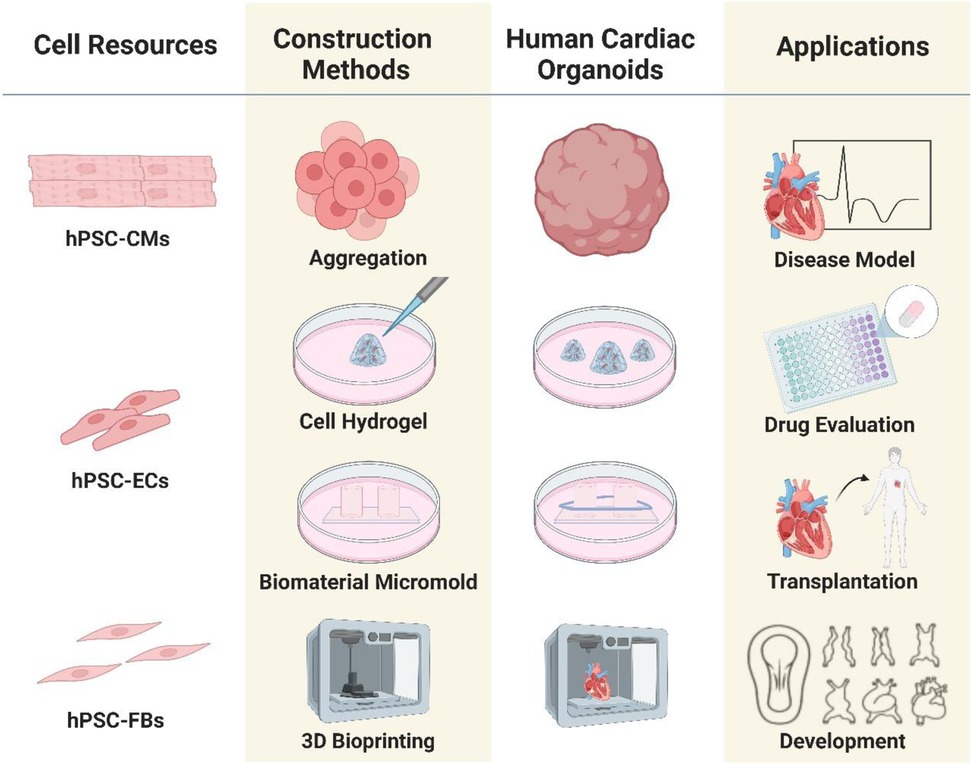
Figure 1. Outline of the process of organoids culture techniques and bioengineering strategies: organoids can be generated from different cell sources isolated from human/animal tissue samples. The construction methods play crucial role in the proper growth and development of organoid models.
4 Necessity to develop Matrigel alternative biomaterials for organoids research
A large portion of organoids have been grown in Matrigel, a complex and poorly defined biomaterial produced from the secretions of Engelbreth-Holm-Swarm mouse sarcoma cells (44). Although affordable and versatile, Matrigel is highly complex; a proteomic study reveals that it comprises more than 1,800 distinct proteins (45). It is challenging to determine the signals required for organoid construction and function due to Matrigel's vague nature, which is made more challenging by lot-to-lot variances in Matrigel (46, 47). Additionally, Matrigel based culture environment may lack some of the constituents required for the production of healthy organoid models. For example, gut organoids generated and maintained in Matrigel lack the distinctive villous structure of mammalian intestines, which may be caused by insufficient laminin-511 and associated components (48, 49). Also, it is becoming increasingly evident that the permeabilities and mechanical characteristics of 3D environments can significantly impact the development of cells (50), organoids (51), tissues, and organs (52, 53). The mechanical behaviors-like pore size, elasticity, stress relaxation moduli, and creep compliances (54, 55) -cannot be readily distinguished from the biochemical signal outputs in the Matrigel-based culture environments. Moreover, Matrigel samples have diverse mechanical characteristics; certain parts of these hydrogels are been known to exhibit elastic moduli and stiffness related properties that are considerably higher than the average elastic modulus of the sample (56, 57). Finally, due to possible immunogenicity, Matrigel's origins in mouse cells make it unsuitable for use in clinical transplantation of humans (58). With all these shortcomings, there is growing demand to create Matrigel-free culture techniques for the growth and maintenance of organoids. Extracellular matrix (ECM) proteins function as an adhesive substrate, a signaling cue source, and a growth factor sequestration mechanism during organ development (59).
4.1 Decellularized extracellular matrix as suitable material for cardiac organoids
Because of its resemblance to the original tissue, cardiac decellularized extracellular matrix (dECM) has been studied as a potential strategy in tissue-regenerative medicine (60). In a dECM, the basic characteristics of the original tissue can be preserved (60–63). Therefore, cardiac dECM offers myocardiocytes the ideal biomimetic environment in which they can proliferate and repair damaged cardiac tissue (64). There are two types of decellularization methods: chemical and mechanical. Hydrostatic pressure or freeze-thawing are examples of mechanical techniques used to eliminate genetic materials and cellular components. Due to the partial elimination of genetic elements, these approaches are considered advantageous in terms of preserving, biomechanical and biochemical features, but may provoke immunogenicity (65). On the other hand, chemical methods lyse cells by displacing the phospholipid cell membranes with surfactants, acids, and bases. These chemicals eliminate undesirable molecules entirely but can also harm structural and signaling proteins necessary for cell regulation (66). In order to maintain the essential features of dECM, a combination of chemical and mechanical techniques can be used, taking into account the benefits and drawbacks of each method, as indicated earlier (67). The ability of various types of stem cells, including induced pluripotent stem cells (iPSCs), mesenchymal stem cells (MSCs), and embryonic stem cells (ES cells), to promote spontaneous tissue repair after seeding with dECM indicates that acellular ECM-based materials may be useful in cardiac organoids research (67–70).
Certain organoids have been cultured on dECM derived from human or animal donors, with the aim of precisely replicating the composition, structure, and vascularization of native ECM throughout organ development. The target tissue determines the decellularization techniques used, which makes them difficult to generalize (71). Even though xenogeneic ECM may trigger immunological reactions, this danger can be significantly decreased with the right preparation methods (72). FDA-approved similar animal-derived ECM scaffolds are used in clinical applications, including orthopedic implants, face reconstruction, and cardiac valve replacement (59, 73). Decellularized extracellular matrix can also offer extra signals that promote the repair of injured tissue, ultimately stabilizing the organoid transplant and enhancing its functionality (74).
4.2 Pros and cons of decellularized extracellular matrix
ECM-based techniques that have been decellularized can rapidly replicate organ function. There is no need for the surface chemistry modification of the ECM because many, if not all, of the chemical signals necessary for the engineering of a spatially defined organ (including glycoproteins that are difficult to introduce) are already present. A decellularized extracellular matrix preserves compositional variations between the basal and apical areas (62). Decellularized ECMs also have some drawbacks. Most notably, the availability of human or animal donors limits the amount of ECM that can be studied, and donor quality can impact the quality of ECM. For instance, the architecture of fibrotic or emphysematous lung tissue has changed and become harder. These modifications may result in cells that do not survive in culture for more than one week (75, 76) or in significant alterations to the phenotype of seeded cells that can survive (76). On the other hand, myocardial infarction is known to induce changes in the biophysical and biochemical properties of the ECM; changes in surface chemistry make the ECM stiffer and more susceptible to topological changes. Nevertheless, when cells are grown on infarcted tissue, they secrete an abundant amount of immunomodulatory as well as pro-survival growth factors (61). Although myocardial infarction appears to increase the survival of incorporated cells, it is essential to consider the detrimental effects of other unhealthy tissues on the formation and maintenance of organoid models (62). Furthermore, batch-to-batch variability remains a challenging problem, even for tissue derived from healthy donors. The physicochemical and mechanical properties of decellularized ECM are difficult to control and modify due to its dynamic and complex network, limiting the efficiency of the matrix and its wider adoption. In addition to being chemically ambiguous, decellularized extracellular matrices often lack established drivers of differentiation. Aggressive decellularization may eliminate surface proteoglycans required for effective organoid formation (77). Another challenge is that different decellularization techniques have different levels of success in eliminating cells or other immunogenic species, which can lead to different host immunological reactions and implant failure in clinical trials (78). Lastly, there are times when PSC differentiation into organ-specific progenitor cells that are subsequently incorporated into the decellularized matrix needs a further step.
5 Applications of cardiac organoids
Organoid technology has recently brought a dramatic shift to biomedical research by creating 3D models that replicate cellular heterogeneity, structure, and functions of tissues (40). In recent years, organoids have been used widely in disease modeling and drug discovery in several organs, such as the brain (79), the kidney (39), the liver (80, 81), and the intestine (82). However, due to the heart's complex structure and vascularization, the progress of developing cardiac organoids has been slower than that of other organs. Regardless of these limitations, human cardiac organoids are widely considered as a novel model system for studying cardiac diseases (Table 1), drug screening and cardiotoxicity testing (84–91) (Tables 1, 2).
6 Future directions: enhancing maturation and improving fidelity to native tissue
Advancements in cardiac organoids that allow appropriate structural and functional elements of the normal heart are imperative. These include the formation of chambers, myocardium thickness, and contractibility. One recent advancement in cardiac chamber formation using organoids includes the hHO model by Lewis-Israeli et al., which involves self-organization into multiple miniature chambers via BMP4 and Activin A (37). Additionally, Lin et al. successfully established a “human-heart-in-a-jar”, demonstrating an electrically and mechanically functional miniature ventricle (83). Although these characteristics provide an obvious advantage for studying diseases and drug interactions over using simpler 2D or 3D models, they lack essential features including left-right symmetry, conductance, and valves (91). Bioreactors have been suggested in the literature to improve the maturity of cardiac organoids (92). They work by improving media circulation, which leads to elevated uptake of nutrients, and providing mechanical stretch stimuli, which promote organoid maturation (93, 94).
Lastly, poor vascularization of cardiac organoids has been observed in several models, with some even developing necrosis from limited perfusion (24). Organ-chip systems have addressed this drawback, allowing 3D microchannel scaffolds supporting millimeter-thick cardiac tissue that is optimally perfused (24, 95). A limitation of these systems is the inability to replicate the 3D configuration and multicellular composition of normal cardiac tissue. Indeed, a multicellular composition, incorporating endothelial cells and fibroblasts, enables enhanced disease modeling and cardiomyocyte maturation (87). A potential solution to this has been observed with a Heart-on-Chip (HoC) model described by Cofiño-Fabres et al., involving the addition of hiPSC-derived cardiomyocytes with other cells typically found in cardiac tissue (96). Another alternative to improve vascularization involves the development of hiPSC-derived self-assembling vascular spheres, followed by encasing with hiPSC-derived cardiomyocytes (97).
7 Conclusion
In summary, while cardiac organoids represent a significant advancement in heart disease modeling and therapeutic research, challenges remain in fully replicating the complexity of the heart's structure and function. The absence of a cardiac conduction system, functional vascularization, and the lack of standardized fabrication methods are key obstacles that hinder the development of more accurate and reliable models. However, with ongoing advancements in 3D modeling techniques, such as Heart-on-Chip systems, these limitations are gradually being addressed. As the field progresses, cardiac organoids hold great promise for improving disease modeling, drug screening, and personalized medicine, paving the way for more effective treatments and better understanding of cardiovascular diseases. Continued research into fabrication methods, tissue engineering, and functional integration will be essential for realizing the full potential of cardiac organoids in both clinical and preclinical applications.
Author contributions
AY: Conceptualization, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. AJ: Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Methodology. AM: Data curation, Formal analysis, Methodology, Writing – review & editing, Validation. BT: Data curation, Formal analysis, Methodology, Writing – review & editing, Investigation. MA: Data curation, Formal analysis, Methodology, Writing – review & editing. TA: Data curation, Formal analysis, Methodology, Writing – review & editing, Validation. JK: Data curation, Formal analysis, Methodology, Validation, Writing – review & editing, Investigation. AE: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. RC: Formal analysis, Investigation, Validation, Writing – original draft. EA: Validation, Methodology, Software, Writing – review & editing. AA: Validation, Writing – review & editing, Supervision. DO: Writing – review & editing, Data curation, Methodology. IF: Writing – review & editing, Conceptualization. MK: Conceptualization, Writing – review & editing, Validation. MR: Validation, Writing – review & editing. TM: Writing – review & editing, Conceptualization, Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lindstrom M, DeCleene N, Dorsey H, Fuster V, Johnson CO, LeGrand KE, et al. Global burden of cardiovascular diseases and risks collaboration, 1990–2021. J Am Coll Cardiol. (2022) 80:2372–425. doi: 10.1016/j.jacc.2022.11.001
2. Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D, et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. (2016) 15:751–69. doi: 10.1038/nrd.2016.175
3. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. (2014) 32:40–51. doi: 10.1038/nbt.2786
4. Heinzke AL, Zdrazil B, Leeson PD, Young RJ, Pahl A, Waldmann H, et al. A compound-target pairs dataset: differences between drugs, clinical candidates and other bioactive compounds. Sci Data. (2024) 11:1160. doi: 10.1038/s41597-024-03582-9
5. Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, et al. Drug screening using a library of human induced pluripotent stem cell–derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. (2013) 127:1677–91. doi: 10.1161/CIRCULATIONAHA.113.001883
6. Frommeyer G, Eckardt L. Drug-induced proarrhythmia: risk factors and electrophysiological mechanisms. Nat Rev Cardiol. (2016) 13:36–47. doi: 10.1038/nrcardio.2015.110
7. Mohr E, Thum T, Bär C. Accelerating cardiovascular research: recent advances in translational 2D and 3D heart models. Eur J Heart Fail. (2022) 24:1778–91. doi: 10.1002/ejhf.2631
8. Le MNT, Hasegawa K. Expansion culture of human pluripotent stem cells and production of cardiomyocytes. Bioengineering. (2019) 6:48. doi: 10.3390/bioengineering6020048
9. Salem T, Frankman Z, Churko JM. Tissue engineering techniques for induced pluripotent stem cell derived three-dimensional cardiac constructs. Tissue Eng B Rev. (2022) 28:891–911. doi: 10.1089/ten.teb.2021.0088
10. Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, et al. Comparison of 2D-and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. (2015) 33:1837–43. doi: 10.3892/or.2015.3767
11. Mir TA, Nakamura M. Three-dimensional bioprinting: toward the era of manufacturing human organs as spare parts for healthcare and medicine. Tissue Eng B Rev. (2017) 23:245–56. doi: 10.1089/ten.teb.2016.0398
12. Seydel CM, Gonzaga BM de S, Coelho LL, Garzoni LR. Exploring the dimensions of Pre-clinical research: 3D cultures as an investigative model of cardiac fibrosis in Chagas disease. Biomedicines. (2024) 12:1410. doi: 10.3390/biomedicines12071410
13. Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. (2021) 6:402–20. doi: 10.1038/s41578-021-00279-y
14. Jin H, Xue Z, Liu J, Ma B, Yang J, Lei L. Advancing organoid engineering for tissue regeneration and biofunctional reconstruction. Biomater Res. (2024) 28:0016. doi: 10.34133/bmr.0016
15. Sahara M. Recent advances in generation of in vitro cardiac organoids. IJMS. (2023) 24:6244. doi: 10.3390/ijms24076244
16. Wang Y-H, Ouyang Q, Zhao S, Zhang Y, Tian R-Z, Guo Y-P, et al. Advances in cardiac organoids. Front Biosci (Landmark Ed). (2023) 28:221. doi: 10.31083/j.fbl2809221
17. Ang Y-S, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. (2016) 167:1734–49.e22. doi: 10.1016/j.cell.2016.11.033
18. Kamp TJ, Hamdan MH, January CT. Chloroquine or hydroxychloroquine for COVID-19: is cardiotoxicity a concern? J Am Heart Assoc. (2020) 9:e016887. doi: 10.1161/JAHA.120.016887
19. Miyamoto M, Nam L, Kannan S, Kwon C. Heart organoids and tissue models for modeling development and disease. Semin Cell Dev Biol. (2021) 118:119–28. doi: 10.1016/j.semcdb.2021.03.011
20. Zushin P-JH, Mukherjee S, Wu JC. FDA modernization act 2.0: transitioning beyond animal models with human cells, organoids, and AI/ML-based approaches. J Clin Invest. (2023) 133:e175824. doi: 10.1172/JCI175824
21. Li J, Li Y, Song G, Wang H, Zhang Q, Liu R, et al. Revolutionizing cardiovascular research: human organoids as a beacon of hope for understanding and treating cardiovascular diseases. Mater Today Bio. (2024) 30:101396. doi: 10.1016/j.mtbio.2024.101396
22. Seguret M, Vermersch E, Jouve C, Hulot J-S. Cardiac organoids to model and heal heart failure and cardiomyopathies. Biomedicines. (2021) 9:563. doi: 10.3390/biomedicines9050563
23. Arai K, Murata D, Verissimo AR, Mukae Y, Itoh M, Nakamura A, et al. Fabrication of scaffold-free tubular cardiac constructs using a bio-3D printer. PLoS One. (2018) 13:e0209162. doi: 10.1371/journal.pone.0209162
24. Cho S, Lee C, Skylar-Scott MA, Heilshorn SC, Wu JC. Reconstructing the heart using iPSCs: engineering strategies and applications. J Mol Cell Cardiol. (2021) 157:56–65. doi: 10.1016/j.yjmcc.2021.04.006
25. Harrison SP, Siller R, Tanaka Y, Chollet ME, de la Morena-Barrio ME, Xiang Y, et al. Scalable production of tissue-like vascularized liver organoids from human PSCs. Exp Mol Med. (2023) 55:2005–24. doi: 10.1038/s12276-023-01074-1
26. Hofbauer P, Jahnel SM, Papai N, Giesshammer M, Deyett A, Schmidt C, et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. (2021) 184:3299–317.e22. doi: 10.1016/j.cell.2021.04.034
27. Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M, et al. In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels. Nat Commun. (2013) 4:1399. doi: 10.1038/ncomms2406
28. Thomas D, Choi S, Alamana C, Parker KK, Wu JC. Cellular and engineered organoids for cardiovascular models. Circ Res. (2022) 130:1780–802. doi: 10.1161/CIRCRESAHA.122.320305
29. Wilson H. A new method by which sponges may be artificially reared. Science. (1907) 25:912–5. doi: 10.1126/science.25.649.912
31. Weiss P, Taylor A. Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc Natl Acad Sci U S A. (1960) 46:1177–85. doi: 10.1073/pnas.46.9.1177
32. Rheinwald JG, Green H. Formation of a keratinizing epithelium in culture by a cloned cell line derived from a teratoma. Cell. (1975) 6:317–30. doi: 10.1016/0092-8674(75)90183-x
33. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. (2006) 126:663–76. doi: 10.1016/j.cell.2006.07.024
34. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. (1998) 282:1145–7. doi: 10.1126/science.282.5391.1145
35. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. (2009) 459:262–5. doi: 10.1038/nature07935
36. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. (2014) 9:2329–40. doi: 10.1038/nprot.2014.158
37. Lewis-Israeli YR, Wasserman AH, Gabalski MA, Volmert BD, Ming Y, Ball KA, et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat Commun. (2021) 12:5142. doi: 10.1038/s41467-021-25329-5
38. Liang S, Luo Y, Su Y, Zhang D, Wang S, Xu M, et al. Distinct toxicity of microplastics/TBBPA co-exposure to bioprinted liver organoids derived from hiPSCs of healthy and patient donors. Int J Bioprinting. (2024) 10:1403. doi: 10.36922/ijb.1403
39. Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. (2015) 526:564–8. doi: 10.1038/nature15695
40. Corrò C, Novellasdemunt L, Li VSW. A brief history of organoids. Am J Physiol Cell Physiol. (2020) 319:C151–65. doi: 10.1152/ajpcell.00120.2020
41. Gisone I, Cecchettini A, Ceccherini E, Persiani E, Morales MA, Vozzi F. Cardiac tissue engineering: multiple approaches and potential applications. Front Bioeng Biotechnol. (2022) 10:980393. doi: 10.3389/fbioe.2022.980393
42. Hirota A, AlMusawi S, Nateri AS, Ordóñez-Morán P, Imajo M. Biomaterials for intestinal organoid technology and personalized disease modeling. Acta Biomater. (2021) 132:272–87. doi: 10.1016/j.actbio.2021.05.010
43. Jeon EY, Sorrells L, Abaci HE. Biomaterials and bioengineering to guide tissue morphogenesis in epithelial organoids. Front Bioeng Biotechnol. (2022) 10:1038277. doi: 10.3389/fbioe.2022.1038277
44. Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. (2005) 15:378–86. doi: 10.1016/j.semcancer.2005.05.004
45. Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. (2010) 10:1886–90. doi: 10.1002/pmic.200900758
46. Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, Reiter RE, et al. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. (2011) 6:656–67. doi: 10.1038/nprot.2011.317
47. Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The hope and the hype of organoid research. Development. (2017) 144:938–41. doi: 10.1242/dev.150201
48. Gjorevski N, Ranga A, Lutolf MP. Bioengineering approaches to guide stem cell-based organogenesis. Development. (2014) 141:1794–804. doi: 10.1242/dev.101048
49. Mahoney ZX, Stappenbeck TS, Miner JH. Laminin alpha 5 influences the architecture of the mouse small intestine mucosa. J Cell Sci. (2008) 121:2493–502. doi: 10.1242/jcs.025528
50. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. (2006) 126:677–89. doi: 10.1016/j.cell.2006.06.044
51. Dahl-Jensen S, Grapin-Botton A. The physics of organoids: a biophysical approach to understanding organogenesis. Development. (2017) 144:946–51. doi: 10.1242/dev.143693
52. Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. (2014) 13:547–57. doi: 10.1038/nmat3937
53. Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol. (2017) 18:728–42. doi: 10.1038/nrm.2017.108
54. Chaudhuri O. Viscoelastic hydrogels for 3D cell culture. Biomater Sci. (2017) 5:1480–90. doi: 10.1039/c7bm00261k
55. Miroshnikova YA, Jorgens DM, Spirio L, Auer M, Sarang-Sieminski AL, Weaver VM. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys Biol. (2011) 8:026013. doi: 10.1088/1478-3975/8/2/026013
56. Reed J, Walczak WJ, Petzold ON, Gimzewski JK. In situ mechanical interferometry of matrigel films. Langmuir. (2009) 25:36–9. doi: 10.1021/la8033098
57. Soofi SS, Last JA, Liliensiek SJ, Nealey PF, Murphy CJ. The elastic modulus of matrigel as determined by atomic force microscopy. J Struct Biol. (2009) 167:216–9. doi: 10.1016/j.jsb.2009.05.005
58. Schneeberger K, Spee B, Costa P, Sachs N, Clevers H, Malda J. Converging biofabrication and organoid technologies: the next frontier in hepatic and intestinal tissue engineering? Biofabrication. (2017) 9:013001. doi: 10.1088/1758-5090/aa6121
59. Hussey GS, Dziki JL, Badylak SF. Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater. (2018) 3:159–73. doi: 10.1038/s41578-018-0023-x
60. Barbulescu GI, Bojin FM, Ordodi VL, Goje ID, Barbulescu AS, Paunescu V. Decellularized extracellular matrix scaffolds for cardiovascular tissue engineering: current techniques and challenges. Int J Mol Sci. (2022) 23:13040. doi: 10.3390/ijms232113040
61. Sullivan KE, Quinn KP, Tang KM, Georgakoudi I, Black LD. Extracellular matrix remodeling following myocardial infarction influences the therapeutic potential of mesenchymal stem cells. Stem Cell Res Ther. (2014) 5:14. doi: 10.1186/scrt403
62. Kozlowski MT, Crook CJ, Ku HT. Towards organoid culture without Matrigel. Commun Biol. (2021) 4:1387. doi: 10.1038/s42003-021-02910-8
63. Mir TA, Alzhrani A, Nakamura M, Iwanaga S, Wani SI, Altuhami A, et al. Whole liver derived acellular extracellular matrix for bioengineering of liver constructs: an updated review. Bioengineering. (2023) 10:1126. doi: 10.3390/bioengineering10101126
64. Chae S, Cho D-W. Three-dimensional bioprinting with decellularized extracellular matrix-based bioinks in translational regenerative medicine. MRS Bull. (2022) 47:70–9. doi: 10.1557/s43577-021-00260-8
65. Xing Q, Yates K, Tahtinen M, Shearier E, Qian Z, Zhao F. Decellularization of fibroblast cell sheets for natural extracellular matrix scaffold preparation. Tissue Eng Part C Methods. (2015) 21:77–87. doi: 10.1089/ten.tec.2013.0666
66. Nazari M, Kurdi M, Heerklotz H. Classifying surfactants with respect to their effect on lipid membrane order. Biophys J. (2012) 102:498–506. doi: 10.1016/j.bpj.2011.12.029
67. Guyette JP, Charest JM, Mills RW, Jank BJ, Moser PT, Gilpin SE, et al. Bioengineering human myocardium on native extracellular matrix. Circ Res. (2016) 118:56–72. doi: 10.1161/CIRCRESAHA.115.306874
68. Esmaeili Pourfarhangi K, Mashayekhan S, Asl SG, Hajebrahimi Z. Construction of scaffolds composed of acellular cardiac extracellular matrix for myocardial tissue engineering. Biologicals. (2018) 53:10–8. doi: 10.1016/j.biologicals.2018.03.005
69. Kc P, Hong Y, Zhang G. Cardiac tissue-derived extracellular matrix scaffolds for myocardial repair: advantages and challenges. Regen Biomater. (2019) 6:185–99. doi: 10.1093/rb/rbz017
70. Lu T-Y, Lin B, Kim J, Sullivan M, Tobita K, Salama G, et al. Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat Commun. (2013) 4:2307. doi: 10.1038/ncomms3307
71. Keane TJ, Swinehart IT, Badylak SF. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods. (2015) 84:25–34. doi: 10.1016/j.ymeth.2015.03.005
72. Allman AJ, McPherson TB, Badylak SF, Merrill LC, Kallakury B, Sheehan C, et al. Xenogeneic extracellular matrix grafts elicit a TH2-restricted immune response. Transplantation. (2001) 71:1631–40. doi: 10.1097/00007890-200106150-00024
73. Parmaksiz M, Dogan A, Odabas S, Elçin AE, Elçin YM. Clinical applications of decellularized extracellular matrices for tissue engineering and regenerative medicine. Biomed Mater. (2016) 11:022003. doi: 10.1088/1748-6041/11/2/022003
74. Yu Y, Alkhawaji A, Ding Y, Mei J. Decellularized scaffolds in regenerative medicine. Oncotarget. (2016) 7:58671–83. doi: 10.18632/oncotarget.10945
75. Wagner DE, Bonenfant NR, Parsons CS, Sokocevic D, Brooks EM, Borg ZD, et al. Comparative decellularization and recellularization of normal versus emphysematous human lungs. Biomaterials. (2014) 35:3281–97. doi: 10.1016/j.biomaterials.2013.12.103
76. Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. (2012) 186:866–76. doi: 10.1164/rccm.201204-0754OC
77. Shojaie S, Ermini L, Ackerley C, Wang J, Chin S, Yeganeh B, et al. Acellular lung scaffolds direct differentiation of endoderm to functional airway epithelial cells: requirement of matrix-bound HS proteoglycans. Stem Cell Rep. (2015) 4:419–30. doi: 10.1016/j.stemcr.2015.01.004
78. Keane TJ, Londono R, Turner NJ, Badylak SF. Consequences of ineffective decellularization of biologic scaffolds on the host response. Biomaterials. (2012) 33:1771–81. doi: 10.1016/j.biomaterials.2011.10.054
79. Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. (2013) 501:373–9. doi: 10.1038/nature12517
80. Jabri A, Khan J, Taftafa B, Alsharif M, Mhannayeh A, Chinnappan R, et al. Bioengineered organoids offer new possibilities for liver cancer studies: a review of key milestones and challenges. Bioengineering. (2024) 11:346. doi: 10.3390/bioengineering11040346
81. Obeid DA, Mir TA, Alzhrani A, Altuhami A, Shamma T, Ahmed S, et al. Using liver organoids as models to study the pathobiology of rare liver diseases. Biomedicines. (2024) 12:446. doi: 10.3390/biomedicines12020446
82. Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. (2018) 23:787–93.e6. doi: 10.1016/j.stem.2018.11.016
83. Li RA, Keung W, Cashman TJ, Backeris PC, Johnson BV, Bardot ES, et al. Bioengineering an electro-mechanically functional miniature ventricular heart chamber from human pluripotent stem cells. Biomaterials. (2018) 163:116–27. doi: 10.1016/j.biomaterials.2018.02.024
84. Hildebrandt MR, Reuter MS, Wei W, Tayebi N, Liu J, Sharmin S, et al. Precision health resource of control iPSC lines for versatile multilineage differentiation. Stem Cell Rep. (2019) 13:1126–41. doi: 10.1016/j.stemcr.2019.11.003
85. Sebastião MJ, Gomes-Alves P, Reis I, Sanchez B, Palacios I, Serra M, et al. Bioreactor-based 3D human myocardial ischemia/reperfusion in vitro model: a novel tool to unveil key paracrine factors upon acute myocardial infarction. Transl Res. (2020) 215:57–74. doi: 10.1016/j.trsl.2019.09.001
86. Richards DJ, Coyle RC, Tan Y, Jia J, Wong K, Toomer K, et al. Inspiration from heart development: biomimetic development of functional human cardiac organoids. Biomaterials. (2017) 142:112–23. doi: 10.1016/j.biomaterials.2017.07.021
87. Voges HK, Foster SR, Reynolds L, Parker BL, Devilée L, Quaife-Ryan GA, et al. Vascular cells improve functionality of human cardiac organoids. Cell Rep. (2023) 42:112322. doi: 10.1016/j.celrep.2023.112322
88. Feng W, Schriever H, Jiang S, Bais A, Wu H, Kostka D, et al. Computational profiling of hiPSC-derived heart organoids reveals chamber defects associated with NKX2-5 deficiency. Commun Biol. (2022) 5:399. doi: 10.1038/s42003-022-03346-4
89. Shiti A, Arbil G, Shaheen N, Huber I, Setter N, Gepstein L. Utilizing human induced pluripotent stem cells to study atrial arrhythmias in the short QT syndrome. J Mol Cell Cardiol. (2023) 183:42–53. doi: 10.1016/j.yjmcc.2023.08.003
90. Goldfracht I, Protze S, Shiti A, Setter N, Gruber A, Shaheen N, et al. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat Commun. (2020) 11:75. doi: 10.1038/s41467-019-13868-x
91. Kostina A, Volmert B, Aguirre A. Human heart organoids: current applications and future perspectives. Eur Heart J. (2024) 45:751–3. doi: 10.1093/eurheartj/ehad841
92. Joddar B, Natividad-Diaz SL, Padilla AE, Esparza AA, Ramirez SP, Chambers DR, et al. Engineering approaches for cardiac organoid formation and their characterization. Transl Res. (2022) 250:46–67. doi: 10.1016/j.trsl.2022.08.009
93. Phelan MA, Lelkes PI, Swaroop A. Mini and customized low-cost bioreactors for optimized high-throughput generation of tissue organoids. Stem Cell Investig. (2018) 5:33. doi: 10.21037/sci.2018.09.06
94. Van Winkle AP, Gates ID, Kallos MS. Mass transfer limitations in embryoid bodies during human embryonic stem cell differentiation. Cells Tissues Organs. (2012) 196:34–47. doi: 10.1159/000330691
95. Osaki T, Sivathanu V, Kamm RD. Vascularized microfluidic organ-chips for drug screening, disease models and tissue engineering. Curr Opin Biotechnol. (2018) 52:116–23. doi: 10.1016/j.copbio.2018.03.011
96. Cofiño-Fabres C, Boonen T, Rivera-Arbeláez JM, Rijpkema M, Blauw L, Rensen PCN, et al. Micro-engineered heart tissues on-chip with heterotypic cell composition display self-organization and improved cardiac function. Adv Healthc Mater. (2024) 13(18):e2303664. doi: 10.1002/adhm.202303664
Keywords: cardiac organoids, 3D culture, decellularized matrix, disease model, drug screening
Citation: Yaqinuddin A, Jabri A, Mhannayeh A, Taftafa B, Alsharif M, Abbad T, Khan J, Elsalti A, Chinnappan R, Alshehri EA, Alzhrani A, Obeid DA, Fujitsuka I, Khan M, Rehman Mu and Mir TA (2025) Cardiac organoids: a new tool for disease modeling and drug screening applications. Front. Cardiovasc. Med. 12:1537730. doi: 10.3389/fcvm.2025.1537730
Received: 1 December 2024; Accepted: 1 May 2025;
Published: 20 May 2025.
Edited by:
Zhao-Jun Liu, University of Miami, United StatesReviewed by:
Francesco Canonico, Azienda Ospedaliero Universitaria Maggiore della Carità, ItalyElisa Ceccherini, National Research Council (CNR), Italy
Copyright: © 2025 Yaqinuddin, Jabri, Mhannayeh, Taftafa, Alsharif, Abbad, Khan, Elsalti, Chinnappan, Alshehri, Alzhrani, Obeid, Fujitsuka, Khan, Rehman and Mir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Yaqinuddin, YXlhcWludWRkaW5AYWxmYWlzYWwuZWR1; Tanveer Ahmad Mir, dG1pckBrZnNocmMuZWR1LnNh
 Ahmed Yaqinuddin
Ahmed Yaqinuddin Abdullah Jabri
Abdullah Jabri Abdulaziz Mhannayeh
Abdulaziz Mhannayeh Bader Taftafa1
Bader Taftafa1 Tasnim Abbad
Tasnim Abbad Abdulrahman Elsalti
Abdulrahman Elsalti Raja Chinnappan
Raja Chinnappan Dalia A. Obeid
Dalia A. Obeid Mahmood Khan
Mahmood Khan Tanveer Ahmad Mir
Tanveer Ahmad Mir