Abstract
Well-being depends on the integrated operation of biological processes at all levels of system organization, from individual cells to tissues and organ systems, collectively sustaining homeostasis and optimal bodily functions. The regulation of cooperation among these processes is mediated by information flow within networks possessing diverse structural, functional, and temporal properties. Disruption in these networks is observed in conditions such as infections, inflammatory diseases, and cancer. To advance understanding of immune system roles and to elucidate mechanisms underlying health vulnerability during disease, we utilized proteomics data related to 4,800 diseases along with protein swarm-based cause-effect analyses to identify principles governing plasticity and self-organizing capabilities of immune systems. Our findings demonstrate that the precision of immune system functions is regulated by dynamic alterations in the topologies of cooperation networks that are partially modulated by microRNAs. Additionally, our analysis indicates that investigating the underlying causes of diseases through the study of cooperative network functions and their interactions with microRNAs—rather than concentrating exclusively on individual protein targets or microRNAs—provides significant insights for devising effective treatment strategies for infections, cardiovascular conditions, Alzheimer's disease, cancer, aging, and related health concerns.
Introduction
Swarm intelligence is widely recognized as a key factor in the remarkable precision of immune systems, and this emergent property is now being simulated through computer (1, 2) Immune systems have evolved to protect organisms against a broad spectrum of challenges, including cancer, harmful substances, damaged proteins, DNA, RNA, and infections caused by viruses, bacteria, fungi, and parasites (3–5). Key functions of the innate immune system include rapid elimination of various pathogens (6, 7), development of immunological memory to prevent reinfection (8), avoiding miscarriage and maintaining pregnancy (9), supporting beneficial relationships between the host and microbiome (10), and minimizing the risk of autoimmune disease (11).
The role of the innate immune system extends beyond these core functions to regulation of tissue homeostasis (12), metabolism (13), stem cell differentiation (14), the regulation of interactions between organ systems (15), development of the central nervous system (16), and the persistence of memory and cognition (17, 18). These diverse actions rely on immune cells that display plasticity and adapt their behavior depending on injuries in a macro and micro environment-specific manner (19–21). The rules regulating this adaptability involve epigenetic regulation at all system levels (22–24). Dysregulation of these processes can occur in the context of infections (25–31), cardiovascular diseases (32, 33), dementias (34, 35), metabolic diseases (36, 37), and cancer (38, 39). The complexity and interdependence (cause-effect relationships) of these dynamic systems create significant challenges in the development of safe and effective disease treatment and prevention options (40–44).
Addressing these challenges requires an understanding of the cause-effect relationships involving interactions of multimode cooperation and communication networks with dynamic network topologies (45–55). Because variables governing dynamic network-network interaction in biological networks are often unknown, we have developed methods—such as protein swarm analysis and cause-effect analysis – to capture plasticity and emergent properties of these complex dynamic interaction systems (56). By integrating concepts from pharmacological cause-effect analysis (57), information theory (51), communication networks (47), cooperation networks (58), and particle swarm optimization (53, 59, 60), protein swarm-based cause- effect analysis determines variations in information exchange among protein swarms to identify network structures associated with discrete system perturbations (50, 57, 61–63). This analytical approach (cause-effect linkage) organizes information within large, orthogonal, decentralized data sets and sets constraints for solutions to global network architectures that generate emergent properties (64). This method aims to be robust against local data gaps or inaccuracies and provides global, unbiased insight into complex cause-effect relationships (65–67).
Given recent findings that COVID-19 may be associated with negative cardiovascular outcomes (such as heart failure and acute coronary syndromes), and that bidirectional relationships exist among atherosclerosis, Alzheimer's disease, and COVID-19 (68, 69), this study applies cause-effect analysis to investigate how COVID-19 influences regulatory rules in immune system function across 4,800 diseases—including cardiovascular diseases, dementias, 80 types of cancer, and over 350 different infectious diseases (70).
Materials
Protein swarms are generated by dividing tissue and cell-associated protein expression data published in the Human Protein Atlas (71), into information transfer modules using the String platform's gene enrichment analysis for identifying overlaps between tissue proteomes and biological processes (Gene ontology) (72). Proteins captured in this network overlap analysis containing no more than 5 proteins are collected and labeled for identifying tissue and functional associations (73). Using 7,323 of these protein swarms and a data mining tool developed by SystaMedic Inc. in collaboration with the University of Connecticut enabled the determination of co-citation frequencies of these 7,323 protein swarms with 4,800 diseases in millions of PubMed abstracts (74). Viewing collected co-citation frequency measurements as indicators of the capacity of a wide range of system perturbations to impact information exchanges between protein swarms, we collected 7,323 × 4,800 of these measurements and determined similarities between the resulting information spectra using hierarchical clustering in Spotfire (75–77). The outcome of this analysis produced an equilibrium network structure organizing 7,323 protein swarms and 4,800 diseases (56, 57, 78–84). The translation of protein swarm information into protein network structures and retrieval of pre-existing knowledge associated with protein networks used the STRING platform (85, 86).
Methods
To identify rules regulating immune system functions in over 4,800 diseases, we selected COVID-19-associated proteomics and microRNA expression data generated during the recent COVID-19 pandemic (87, 88), and accessible via PubMed (89), HMDD (90), and miRBase (91) as a starting point for this investigation (Figure 1).
Figure 1
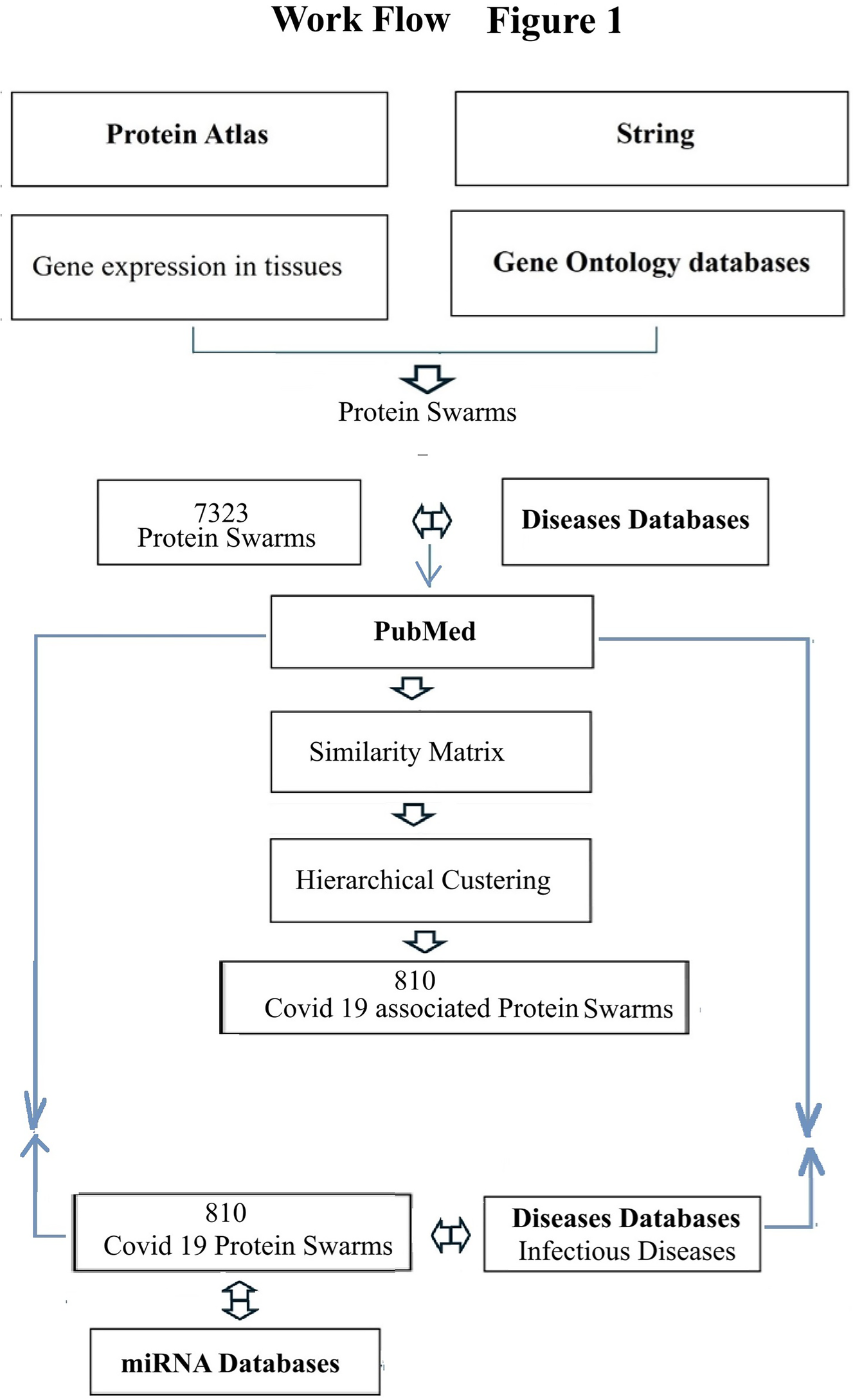
Process map of steps taken for swarm-based, cause-effect analsyis. Bolded text indicate the main databases that were used.
Terminology
The term cooperation refers to working together on accomplishing specific tasks or solving problems. The term coordination refers to the process of organizing and integrating distinct functions, making sure that distinct parts of a system are working together as needed to produce desired outcomes. The term disease-associated proteomics information refers to data describing proteins associated with a disease, how they are modified, and how they interact. The term emergent property refers to properties that arise when interactions between individual components produce new functions. The term network topology refers to the way different nodes in a network are positioned and interconnected, as well as how information flows. The term dynamic network topology refers to a network structure where nodes can join or leave the network, and information transmission can adapt to changing needs or conditions. The term network overlap analysis refers to the process of identifying shared nodes within networks. The term plasticity refers to the ability of the immune system to change its phenotype or function in response to environmental stimuli. The term protein swarm refers to a collection of proteins capable of interacting with other proteins and participating in biological functions involving these protein interactions. The term self-organization capacity of proteins refers to the emergence of an overall order in time and space resulting from the collective interactions of individual proteins. The term spectral clustering refers to a machine learning technique that groups data points into clusters by analyzing the connectivity between them using values of a similarity matrix.
Results
Spectral clustering
The hierarchical clustering of co-citation frequency measurements for 4,800 diseases and 7,323 protein swarms has been described in a previous publication (94). Building upon the observation of a notable association between COVID-19 infections and cardiovascular disease, this study utilized this published analysis to identify COVID-19-associated proteomics data, applying a cluster similarity confidence value (CCSV) threshold greater than 0.92. As outlined in Supplementary Table S1 of the Supplemental section, the analysis revealed 33 protein swarm associations (PSA), encompassing a total of 810 protein swarms and 444 host proteins. A network-based perspective demonstrates in Figure 2 that these 444 host proteins exhibit potential for physical interaction. Furthermore, network overlaps analysis indicates that COVID-19 affects host proteins integral to immune system disorders (false discovery rate: 2.14 × 10−30) (92), and respiratory disease (false discovery rate: 9.82 × 10−27) (93, 94). These results provide evidence that COVID-19 induces profound system-wide disturbances of immune system functions (95–98).
Figure 2
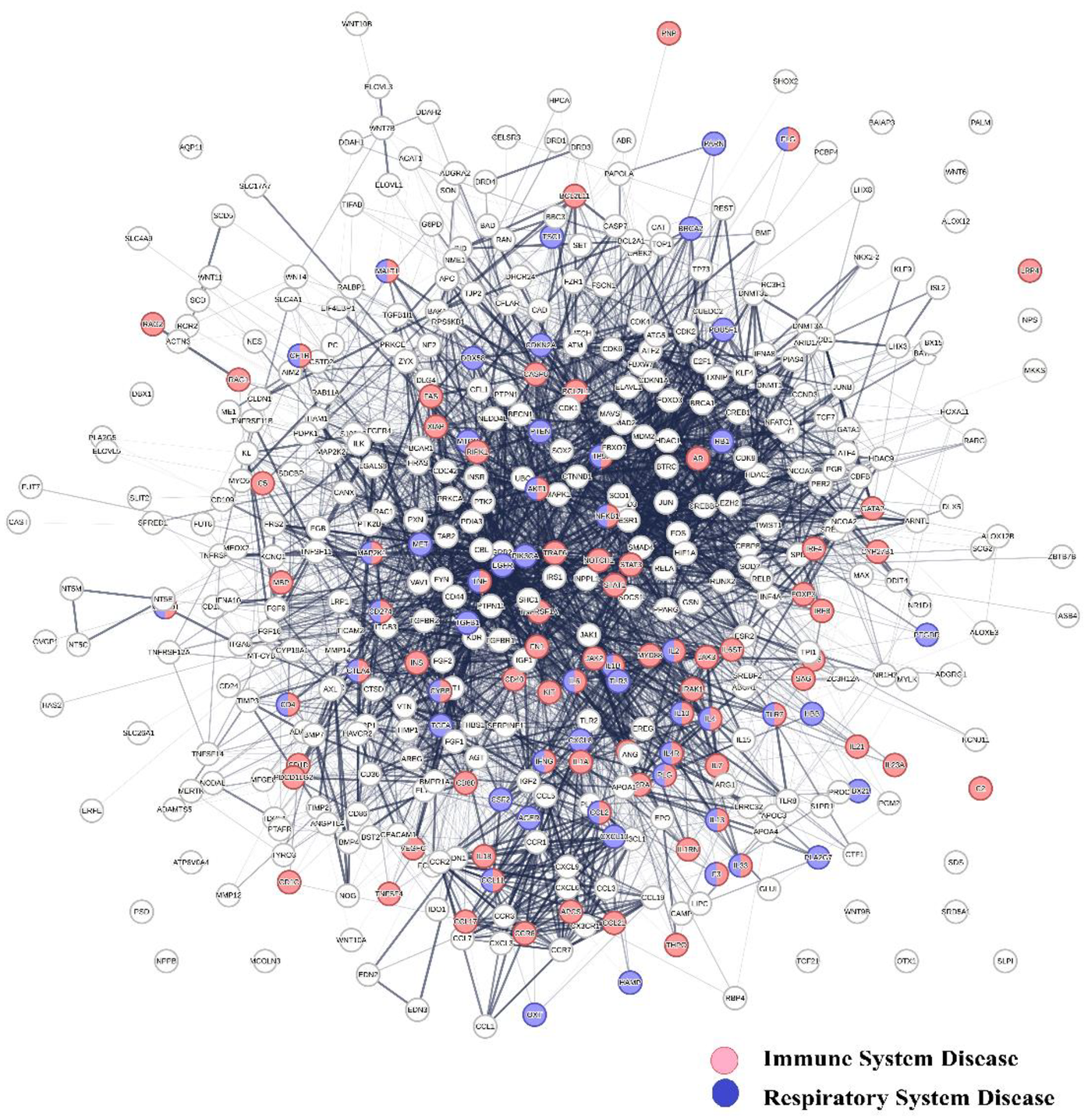
Shows the physical interaction between 444 host proteins associated with COVID 19 and identifies network overlaps between immune system diseases (red network nodes) and respiratory system diseases (blue nodes).
To evaluate the potential of COVID-19-associated proteomics data as a basis for identifying regulatory mechanisms governing system-wide immune system functions (99, 100), we analyzed co-citation frequencies of COVID-19-associated protein swarms listed in Supplementary Table S1 and 4,800 diseases described in PubMed abstracts. Hierarchical clustering of the resulting 810 × 4,800 information spectra matrix revealed alignments between COVID-19-associated proteomics profiles and those associated with these 4,800 diseases. To enable effective visual data comparison, we applied a cluster similarity confidence threshold of >0.973 for segmenting and dividing COVID-19-associated proteomics information into 20 discrete clusters. Examination of co-citation frequency measurements within these clusters indicated that 1,276 diseases—including Alzheimer's disease, 38 cancer types, 85 carcinomas, 157 inflammatory conditions, and 347 infections—demonstrated close proteomics data alignment within these sections. This finding is illustrated in Figure 3 which shows the alignment of proteomics information associated with 347 infectious diseases, vitamin D deficiency (101), Alzheimer's disease (102), and atherosclerosis (103, 104). The specific names of the infectious diseases referenced in Figure 3 can be found in Table 1 of the supplemental section.
Figure 3
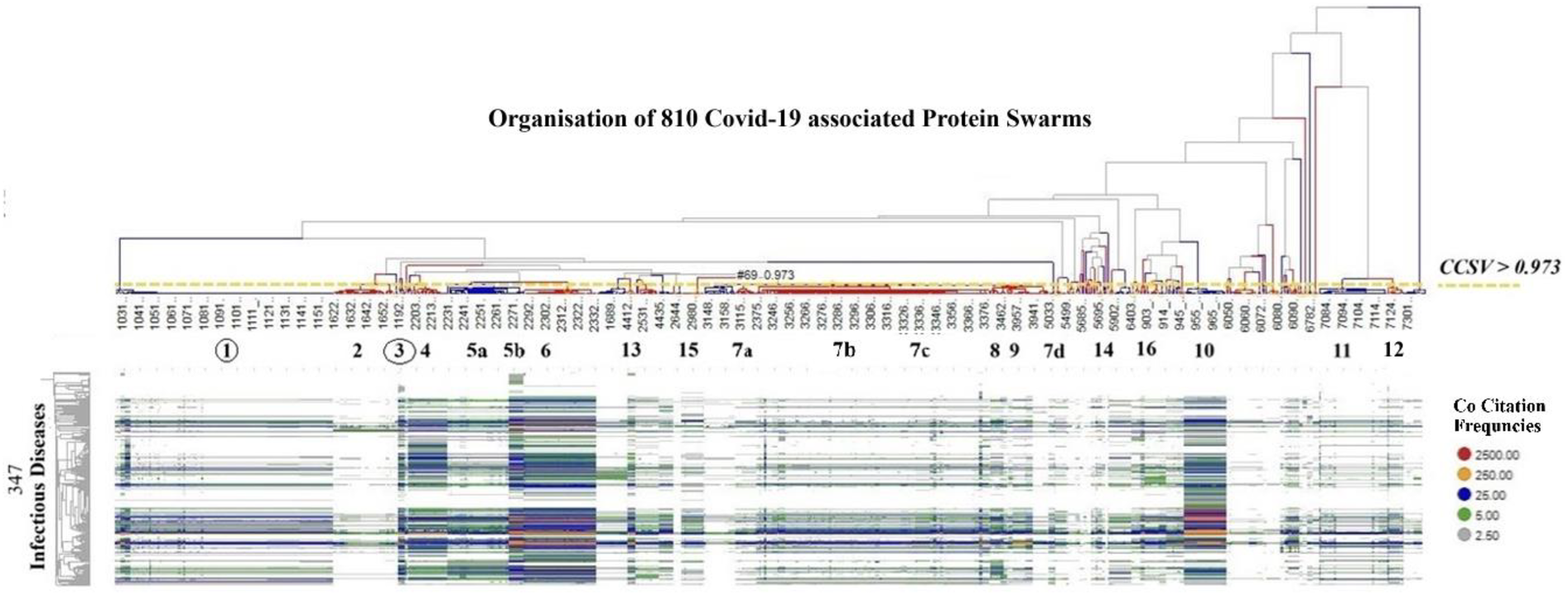
Shows the alignment of proteomics information associated with 347 infectious diseases, Vitamin D deficiency, Alzheimer's disease, and atherosclerosis, using as comparative standard 20 protein swarm clusters associated within confidence in cluster similarity values of >0.973. This high confidence in cluster similarity value shows that infectious diseases, vitamin D deficiency, Alzheimer's disease and atherosclerosis have closely aligned proteomics information in the highlighted protein swarm clusters. Red identifies the highest and grey the lowest number of co-citation frequencies.
Table 1
| Acinetobacter baumannii pneumonia | Diphtheria | Japanese encephalitis | Paracoccidioidomycosis | Severe acute respiratory syndrome |
|---|---|---|---|---|
| Acquired immunodeficiency syndrome | Dysentery | Japanese spotted fever | Paragonimiasis | Severe combined immunodeficiency |
| Acquired thrombocytopenia | Early congenital syphilis | Kaposi's sarcoma | Paratuberculosis | Smallpox |
| Adenovirus pneumonia | Eastern equine encephalitis | Kawasaki disease | Paratyphoid fever | Sporotrichosis |
| AIDS-related Kaposi's sarcoma | Ehrlichiosis | Keratitis | Paronychia | Spotted fever |
| Alveolar echinococcosis | Encephalitis | Klebsiella pneumonia | Pasteurellosis | St. Louis encephalitis |
| Amebic colitis | Endemic typhus | Laryngeal tuberculosis | Pemphigus | Staphylococcal pneumonia |
| Avian influenza | Eosinophilic meningitis | Lassa fever | Pericarditis | Staphylococcus aureus septicemia |
| Avian malaria | Eosinophilic pneumonia | Latent syphilis | Periodontitis | Streptococcal necrotizing fasciitis |
| Babesiosis | Ephemeral fever | Legionellosis | Peritonitis | Streptococcal pharyngitis |
| Bacterial conjunctivitis | Equine encephalitis | Legionnaires' disease | Pertussis | Streptococcal pneumonia |
| Bacterial endocarditis | Erysipelas | Leishmaniasis | Pharyngitis | Streptococcal septicemia |
| Bacterial gastroenteritis | Erythema infectiosum | Lepromatous leprosy | Plague | Subacute bacterial endocarditis |
| Bacterial infectious disease | Extrapulmonary tuberculosis | Leprosy | Plasmodium falciparum malaria | Swine influenza |
| Bacterial meningitis | Fascioliasis | Leptospirosis | Plasmodium vivax malaria | Syphilis |
| Bacterial pneumonia | Feline infectious peritonitis | Listeria meningitis | Pleural tuberculosis | Systemic inflammatory response syndrome |
| Bacterial prostatitis | Female genital tuberculosis | Listeriosis | Pneumatosis cystoides intestinalis | Systemic mycosis |
| Bacterial vaginosis | Filariasis | Lobomycosis | Pneumococcal meningitis | Takayasu's arteritis |
| Botulism | Folliculitis | Louse-borne relapsing fever | Pneumococcal pneumonia | Tetanus |
| Bovine tuberculosis | Fowlpox | Lyme disease | Pneumoconiosis | Tick-borne encephalitis |
| Bubonic plague | Fungal meningitis | Lymphocytic choriomeningitis | Pneumocystosis | Tinea |
| Campylobacteriosis | Fusariosis | Malaria | Pneumonic pasteurellosis | Toxic shock syndrome |
| Central nervous system tuberculosis | Genital herpes | Mastitis | Podoconiosis | Toxoplasma myocarditis |
| Cerebral malaria | Giardiasis | Measles | Polioencephalitis | Toxoplasmosis |
| Cerebral toxoplasmosis | Glossitis | Melioidosis | Poliomyelitis | Trichinosis |
| Chagas cardiomyopathy | Gonococcal urethritis | Meningitis | Polycythemia vera | Trichomonas vaginitis |
| Chagas disease | Gonorrhea | Meningococcal meningitis | Polymyositis | Trichomoniasis |
| Chickenpox | Haemophilus influenzae meningitis | Meningoencephalitis | Pontiac fever | Trichuriasis |
| Chronic fatigue syndrome | Helicobacter pylori gastritis | Microsporidiosis | Porcine reproductive and respiratory syndrome | Trypanosomiasis |
| Chuvash polycythemia | Hemorrhagic disease | Mixed malaria | Primary amebic meningoencephalitis | Tuberculosis |
| Classic Kaposi's sarcoma | Hepatic encephalopathy | Molluscum contagiosum | Primary syphilis | Tularemia |
| Classical swine fever | Hepatitis A | Monkeypox | Primary tuberculosis | Tungiasis |
| Coccidioidomycosis | Hepatitis B | Mouth disease | Prion disease | Typhoid fever |
| Coccidiosis | Hepatitis C | Mucocutaneous leishmaniasis | Pseudorabies | Typhus |
| Colitis | Hepatitis D | Mucormycosis | Pulmonary cryptococcosis | Ulcerative colitis |
| Colorado tick fever | Hepatitis E | Multidrug-resistant tuberculosis | Pulmonary mucormycosis | Vaccinia |
| Congenital rubella | Herpes simplex | Mumps | Pulmonary sporotrichosis | Variant Creutzfeldt-Jakob disease |
| Congenital syphilis | Herpes simplex virus encephalitis | Mycoplasma pneumoniae pneumonia | Pulmonary tuberculosis | Viral encephalitis |
| Conjunctivitis | Histoplasmosis | Mycoplasmal pneumonia | Pyelonephritis | Viral gastroenteritis |
| Coronavirus infectious disease | HIV encephalopathy | Mycosis fungoides | Q fever | Viral hepatitis |
| COVID 19 | HIV-associated lipodystrophy syndrome | Myositis | Rabies | Viral infectious disease |
| Cowpox | HIV-associated nephropathy | Neuritis | Respiratory syncytial virus bronchiolitis | Viral meningitis |
| Crimean-Congo hemorrhagic fever | Human monocytic ehrlichiosis | Newcastle disease | Respiratory syncytial virus pneumonia | Viral pneumonia |
| Cryptococcal meningitis | Indian tick typhus | Norovirus gastroenteritis | Rhinocerebral mucormycosis | Vitamin D deficiency |
| Cryptococcosis | Infectious canine hepatitis | Ocular toxoplasmosis | Rhinosporidiosis | Vulvovaginal candidiasis |
| Cryptosporidiosis | Infectious mononucleosis | Ocular tuberculosis | Rickets | West Nile fever |
| Cyclosporiasis | Infective endocarditis | Omsk hemorrhagic fever | Rift Valley fever | Western equine encephalitis |
| Cystic echinococcosis | Inflammatory bowel disease | Onchocerciasis | Rocky Mountain spotted fever | Wuchereria bancrofti filariasis |
| Cystoisosporiasis | Influenza | Onychomycosis | Rotavirus gastroenteritis | Wound botulism |
| Cytomegalovirus colitis | Influenza encephalopathy | Opisthorchiasis | Rubella | Xeroderma pigmentosum |
| Cytomegalovirus pneumonia | Intermediate uveitis | Oral candidiasis | Salmonella gastroenteritis | Yellow fever |
| Dengue disease | Intestinal schistosomiasis | Oral tuberculosis | Salpingitis | Zika fever |
| Dengue hemorrhagic fever | Intestinal tuberculosis | Orchitis | Scabies | SARS-CoV-2 spike protein S1 |
| Dengue shock syndrome | Invasive aspergillosis | Oropharyngeal candidiasis | Scarlet fever | SARS-CoV-2 nucleocapsid protein |
| Dermatomycosis | Iridocyclitis | Osteomyelitis | Schistosomiasis | COVID-19 vaccine |
List of diseases having closely-aligned proteomics information with COVID-19.
Identification of protein networks involved in immune responses
To identify the roles of protein interactions within protein swarm associations 1–20 in Figure 3 for immune responses, Figure 2 was used for protein network overlap analysis (105). Using a false discovery rate of 5.42 × 10−5 in the Kegg database (106), and 3.94 × 10−9 in the Gene Ontology database (107), we identified 94 different immune system functions involving proteins in these protein swarm associations. These functions are listed in Table 2 of the supplemental section.
Table 2
| Adipocytokine signaling pathway | mTOR signaling pathway |
| AGE-RAGE signaling pathway in diabetic complications | Myeloid leukocyte activation |
| Aldosterone-regulated sodium reabsorption | Myeloid leukocyte differentiation |
| Allograft rejection | Myeloid leukocyte migration |
| AMPK signaling pathway | Natural killer cell mediated cytotoxicity |
| Apelin signaling pathway | Negative regulation of T cell activation |
| Apoptosis | Neurotrophin signaling pathway |
| Axon guidance | Neutrophil chemotaxis |
| B cell receptor signaling pathway | NOD-like receptor signaling pathway |
| Cell cycle | p53 signaling pathway |
| Cellular response to interferon-gamma | Pathways in cancer |
| Cellular response to virus | Positive regulation of angiogenesis |
| Cellular senescence | Positive regulation of ERK1 and ERK2 cascade |
| cGMP-PKG signaling pathway | Positive regulation of leukocyte chemotaxis |
| Chemokine signaling pathway | Positive regulation of MAP kinase activity |
| Chemokine-mediated signaling pathway | PPAR signaling pathway |
| Cholesterol metabolism | Protein kinase B signaling |
| C-type lectin receptor signaling pathway | Ras signaling pathway |
| Cytokine-cytokine receptor interaction | Regulation of actin cytoskeleton |
| Cytosolic DNA-sensing pathway | Regulation of chemokine production |
| Dendritic cell differentiation | Regulation of endothelial cell migration |
| EGFR tyrosine kinase inhibitor resistance | Regulation of epithelial to mesenchymal transition |
| Endoderm development | Regulation of glial cell differentiation |
| Epidermal growth factor receptor signaling pathway | Regulation of gliogenesis |
| ERBB signaling pathway | Regulation of immune system process |
| ErbB signaling pathway | Regulation of leukocyte chemotaxis |
| Estrogen signaling pathway | Regulation of osteoblast differentiation |
| Fc epsilon RI signaling pathway | Regulation of T cell differentiation |
| Fc gamma R-mediated phagocytosis | Regulation of T cell proliferation |
| FoxO signaling pathway | Relaxin signaling pathway |
| Genitalia development | Renal tubule development |
| Glial cell activation | Response to interferon-gamma |
| GnRH signaling pathway | Response to lipopolysaccharide |
| Granulocyte chemotaxis | Response to molecule of bacterial origin |
| HIF-1 signaling pathway | Response to stress |
| Hippo signaling pathway | RIG-I-like receptor signaling pathway |
| IL-17 signaling pathway | Signaling pathways regulating pluripotency of stem cells |
| Intestinal immune network for IgA production | Sphingolipid signaling pathway |
| JAK-STAT signaling pathway | T cell differentiation |
| Leukocyte chemotaxis | T cell receptor signaling pathway |
| Longevity regulating pathway | TGF-beta signaling pathway |
| Macrophage activation | Th1 and Th2 cell differentiation |
| Macrophage Polarization | Th17 cell differentiation |
| MAPK signaling pathway | Toll-like receptor signaling pathway |
| Microglia Polarization | Ubiquitin mediated proteolysis |
| Monocyte chemotaxis | Viral carcinogenesis |
| Mononuclear cell migration | Wnt signaling pathway |
List of biological processes involved in immune responses.
To determine functional relationships between protein networks among these immune functions, co-citation frequencies of COVID-19-associated protein swarms with the 94 immune functions were calculated from Medline abstracts. The result of the hierarchical clustering of these data, presented in Figure 4, shows that protein swarm associations 1–20, linked within confidence in cluster similarity values greater than 0.969, divide the 94 immune system functions into discrete clusters. Additionally, the characteristic distribution of proteomics information in these clusters indicates that the 94 immune functions and protein swarm associations 1–20 in Figure 3 coregulate immune responses in 347 infections, vitamin D deficiency, Alzheimer's disease, and atherosclerosis (108–110).
Figure 4
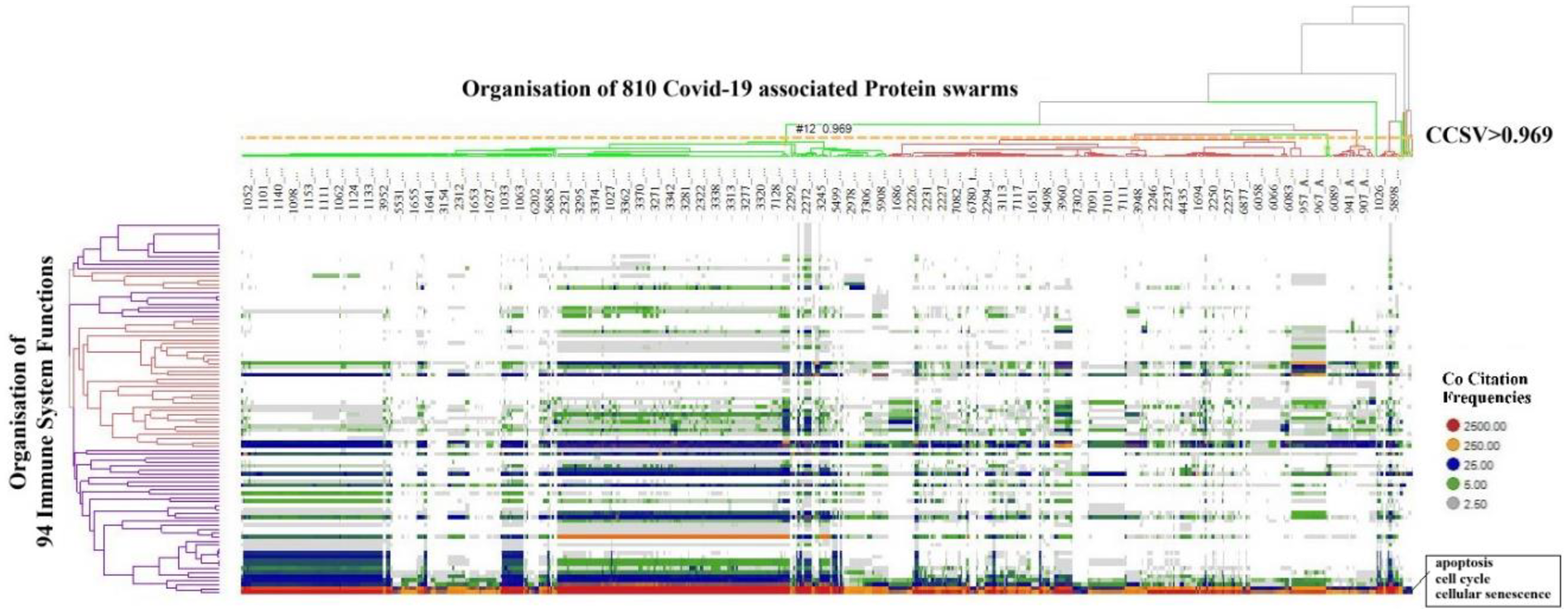
Identifies alignment of proteomics information associated with COVID 19 and proteomics information associated with 94 biological functions involved in host responses to infections.
Impact of disease-associated microRNAs on host responses to infections
To investigate the effects of disease-associated microRNAs on protein network-network interactions as depicted in Figure 4 (111), we applied two distinct methodologies. The first method aimed to identify functional relationships among proteins targeted by disease-associated microRNAs. Specifically, we examined proteins captured in protein swarm associations 1–20 and determined their co-citation frequencies with 136 mature microRNAs associated with COVID-19 and Alzheimer's Disease in Medline abstracts (112–115). Hierarchical clustering of the resulting similarity matrix visualized in the heatmap in Figure 5A indicates that disease-associated microRNAs—particularly mir-155, mir-146a, mir-34a, and mir-21, which are recognized for their role in immune system functions (116–119), Alzheimer's disease (120–123), and atherosclerosis (124–127),—regulate expression levels of all proteins captured in protein swarm associations 1–20. From a protein network perspective, as shown in Figure 5B, these microRNAs alter the expression levels of proteins located within intracellular membrane-bound organelles (128), and these proteins participate in complexes that are critical to immune responses.
Figure 5
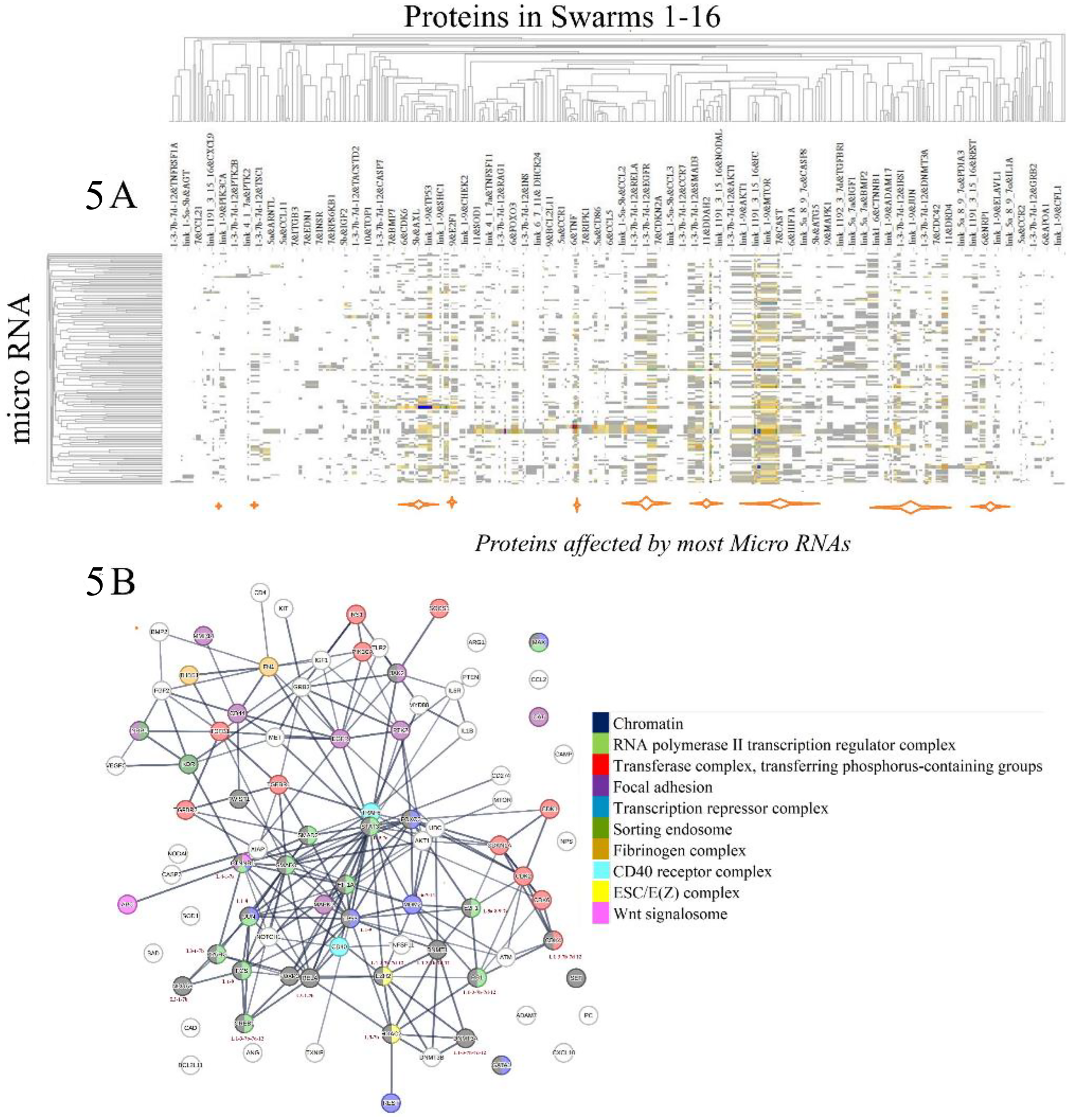
(A) identifies the effects of disease-associated microRNAs on proteins constituting protein swarm associations 1-20. Orange bars at the bottom of (A) identify the positions of proteins targeted by most disease-associated microRNAs. (B) Identifies physical interactions between proteins targeted by disease-associated microRNA. Proteins highlighted in colors are members of protein complexes located in an intracellular membrane-bounded organelle known to be targeted in infections.
For example, chromatin is implicated at the intersections of viral infections and DNA damage control (129), as well as Alzheimer's disease (130) atherosclerosis (131). The RNA polymerase II transcription regulator complex and the transferase complex transferring phosphorus-containing groups play key roles in influenza infections (132, 133), Alzheimer's disease (134), and atherosclerosis (135).
Regulation of focal adhesion is the target of pathogenic microbes (136), contributes to cell death in Alzheimer's disease (137), and is involved in the progression of atherosclerosis (138). The transcription repressor complex plays a key role in RNA virus infections (139), Alzheimer's disease (140), and atherosclerosis (141). Sorting endosome functions are hijacked in viral infections (142), Alzheimer's disease (143), and atherosclerosis (144), while the fibrinogen complex functions are targeted in bacterial infections (145), Alzheimer's disease (146), and atherosclerosis (147).
Functions of the CD40 receptor complex are central to immunity (148), Alzheimer's disease (149), and atherosclerosis (150), and the ESC/E(Z) complex governs the expression of immune-related genes (151, 152), participates in Alzheimer's disease (153), and atherosclerosis (154). Furthermore, the Wnt signalosome is pivotal in host responses to infections (155), Alzheimer's disease (156), and atherosclerosis (157). These findings indicate that protein swarm associations 1–20 coordinate functions of various protein complexes, and that microRNAs such as mir-155, mir-146a, mir-34a, and mir-21, influence the expression levels of proteins within these protein swarm associations 1–20, thereby, affecting the topology of networks that regulate functions of protein-machinery involved in immune responses to injuries and infections (158). Consequently, protein target-focused cause-effect analysis provides insight into the proximities of microRNA targets within protein networks (159), the cellular localization, and the functions of cellular machinery influenced by microRNAs (160). Additionally, proteins within swarm associations 1–20 are components of protein complexes engaged in multiple immune functions. However, due to the extensive array of proteins targeted by microRNAs, establishing unambiguous regulatory relationships between the 136 analyzed microRNAs and the proteins in swarm associations 1–20 is not feasible (161–163).
The second method for evaluating the impacts of disease-associated microRNAs on protein swarm associations 1–20 (as depicted in Figure 3) examines their influence on the organization of protein swarms. To assess this effect, we determined co-citation frequencies between protein swarms in Supplementary Table S1 and 136 disease-associated mature microRNAs in Medline abstracts. The result of the hierarchical clustering based on these collected co-citation frequency measurements are presented in Figure 6.
Figure 6

Identifies protein swarm associations (1–16) collected within a confidence in cluster similarity value of >0.897 and 23 microRNA clusters collected within a confidence in cluster similarity value of >0.67.
Using a confidence threshold in cluster similarity values greater than 0.897 to separate protein swarm associations shown in Figure 6, we identified 20 clusters (PSA 1–20 in Table 3) with swarm compositions previously described in Figure 3. Similarly, applying a confidence threshold in cluster similarity values above 0.67 for grouping microRNAs resulted in 23 clusters, as shown in Table 3.
Table 3
| Cluster identifier | microRNA within the cluster | Cluster identifier | microRNA within the cluster |
|---|---|---|---|
| A | mir-125b-5p, mir-138, mir-451a | M | mir-26a, mir-29a |
| B | mir-143, mir-17, mir-192, mir-421 | N | mir-126, mir-92a |
| C | mir-196, mir-618 | O | mir-200c, mir-203 |
| D | mir-125a-3p, mir-369 | P | mir-181b, mir-326 |
| E | mir-1207, mir-936 | Q | mir-133a, mir-423 |
| F | mir-1246, mir-183 | R | mir-133b, mir-193a, mir-200a, mir-7 |
| G | mir-425-5p, mir-212 | S | mir-34a, mir-34c |
| H | mir-129, mir-298 | T | mir-221, mir-222 |
| I | mir-29b-2, mir-590 | U | mir-107, mir-21 |
| J | mir-106a, mir-483-5p | V | mir-150, mir-223, mir-155, |
| K | mir-208a, mir-483 | mir-142-3p | |
| L | mir-520c, mir-100, mir-486, | W | mir-146a, mir-146a-5p |
| mir-5100, mir-542 |
List of microRNA clusters associated with protein swarm associations 1–20.
Bolded values represent microRNAs that play a key role in COVID-19, atherosclerosis, Alzheimer's disease.
Analysis of the co-citation frequency distribution in Figure 6 indicates that the microRNA clusters listed in Table 3 exhibit specific patterns within protein swarm associations (PSA) 1–20. These findings indicate that microRNAs within these clusters may modulate the expression levels of proteins associated with network topologies in cooperation network segments PSA 1–20. Data suggesting potential cause-effect relationships associated with variations in microRNA expression and network connectivity are presented in Figure 7, which outlines the connectivity and functions of protein networks captured in PSA segments (5a,13), (5b), (5a,8,13b), (7c), (9), and (10) in Figure 6. Identification of these cause-effect relationships may also identify new strategies for developing effective disease treatments.
Figure 7
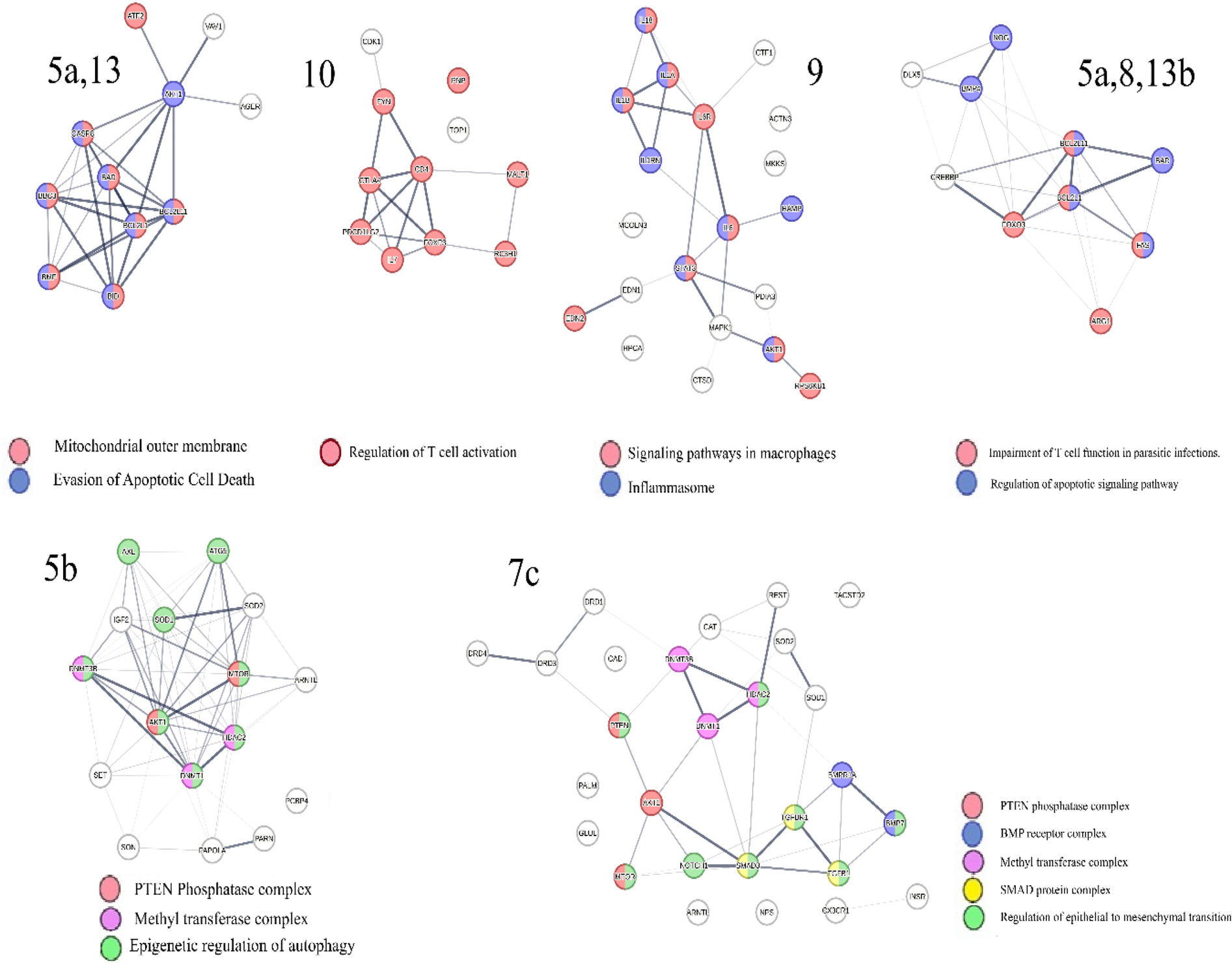
Identifies functions involving interactions between proteins in protein swarm associations (PSA) 5a,13 targeted by microRNAs clusters B, M, N, O and mir-195; PSA 7c targeted by microRNA cluster L, PSA 9 targeted by microRNA cluster O; PSA 10 targeted by microRNA cluster V and PSA 5a,8 13b targeted by microRNA cluster U; PSA 5b targeted by 61 microRNAs, PSA 5a,13: AGER, AKT1, ATF2, BAD, BBC3, BCL2L1, BCL2L11, BID, BMF, CASP8, VAV1; PSA 7c: AKT1, ARNTL, BMP7, BMPR1A, CAD, CAT, CX3CR1, DNMT1, DNMT3B, DRD1, DRD3, DRD4, GLUL, HDAC2, INSR, MTOR, NOTCH1, NPS, PALM, PTEN, REST, SMAD3, SOD1, SOD2, TACSTD2, TGFB1, TGFBR1; PSA 9: ACTN3, AKT1, CTF1, CTSD, EDN1, EDN2, HAMP, HPCA, IL18, IL1A, IL1B, IL1RN, IL6, IL6R, MAPK1, MCOLN3, MKKS, PDIA3, RPS6KB1, STAT3;PSA 5a,8,13b:ARG1, BAD, BCL2L1, BCL2L11, BMP4, CREBBP, DLX5, FAS, FOXO3, NOG; PSA 5b: DNMT3B, DNMT1, PARN, SET, HDAC2, ATG5, AXL, MTOR, PCBP4, PAPOLA, ARNTL, AKT1, SON, IGF2, SOD2, SOD1; PSA 10: CD4, CDK1, CTLA4, FOXP3, FYN, IL7, MALT1, PDCD1LG2, PNP, RC3H1, TOP1.
For example, protein network PSA (5a,13) in Figure 7 consists of proteins located in the outer mitochondrial membrane that regulate apoptosis via BH3-only proteins Bim (164). Network overlap analysis reveals that proteins in PSA (5a,13) are involved in cancer (165), infections (166–169), Alzheimer's disease (170, 171), and atherosclerosis (172). The presence of regulatory relationships between PSA (5a,13) and RNA expression levels in microRNA cluster O, which target expression levels of proteins in PSA (5a,13) is supported by observations showing that mir-195 (173–175), mir-200c (176–178), and mir-203 (179, 180) in microRNA cluster O regulate functions of BH3-only proteins and modulate activation and inhibition of apoptosis. This observation also suggests that identifying these cause-effect relationships may also identify new strategies for developing effective disease treatments for cancer, infections, Alzheimer's disease and atherosclerosis.
Similarly, proteins in cooperation network PSA (7c), which are targeted by microRNAs in Cluster L and mir-126, (associated with atherosclerosis (181, 182), and Alzheimer's disease, are members of a plasma membrane signaling receptor complex involved in the regulation of epithelial to mesenchymal transition (EMT) (183, 184). EMT is upregulated in people with Alzheimer's disease (185), and accelerates plaque growth and instability in atherosclerosis (186). Evidence for coregulation of functions within cooperation network PSA (7c) by microRNAs in cluster L, includes findings that mir-520c (187, 188), mir-100 (189), mir-486 (190), mir-5100 (191, 192), and mir-542 (193), in Cluster L, and mir 126 (189) regulate EMT and plasma levels of these microRNAs are associated with the severity of atherosclerosis (194–196), and Alzheimer's disease (197–199). These cause-effect relationships can also be used to identify new strategies for developing potentially selective disease treatments for atherosclerosis (194–196), and Alzheimer's disease.
Similarly, analysis of information related to proteins in PSA (9), which are targeted by microRNAs within cluster O, demonstrates that these proteins form part of the NF-kappa B complex and play key roles in modulating inflammation and macrophage polarization (200). The notion that microRNAs in cluster O co-regulate PSA (9) functions of is substantiated by the evidence indicating that mir-200c (201–204), and mir-203 (205–207), both in cluster O influence macrophage polarization and inflammatory processes via targeting NF-kappaB pathways. Furthermore, findings show that mir-200c (208, 209), and mir-203 (188), impact microglia polarization (210), and neuroinflammation, suggesting that the regulatory scheme captured in PSA (9) may also contribute to Alzheimer's disease (211, 212), and other dementias (213). Associations between mir-200c and mir-203 levels and the severity of coronary artery stenosis provide further evidence of the relevance of the PSA (9) regulatory scheme in atherosclerosis. These data suggest that the cause-effect relationships between PSA (9) and microRNAs within cluster O could be used for developing treatment options with a broader impact targeting inflammatory diseases, neuro-inflammation, dementia, and cardiovascular diseases.
Similarly, the retrieval of information associated with proteins in PSA (10) targeted by microRNAs in cluster V, demonstrates that these proteins play a role in regulating T cell activation, thereby bridging innate and adaptive immunity (214, 215). The hypothesis that microRNAs in cluster V co-regulate the function of PSA (10) is supported by findings indicating that mir-150, mir-223, mir-155 and mir-142 within this cluster are involved in T cell differentiation and in mediating interactions between innate and adaptive immune responses (216–220). Network overlap analysis of PSA (10) functions reveals that these mechanisms are exploited during immune evasion against parasites, cancer (221), and are also associated with the generation of pro-inflammatory T cell subsets in Alzheimer's disease (222), and atherosclerosis (223). Hence, the cause-effect relationships between PSA (10) and microRNAs within cluster V will have a substantial impact for seeking new approaches to target inflammatory disorders that are critical for immune health, protection against infections, as well as other disorders like cancer, Alzheimer's disease and atherosclerosis.
The regulation of T cell functions is further mediated by proteins in PSA (5a,8,13b), including Arginase, which is recognized as a prominent swarm member and known to be involved in the regulation of T cell activation (224). The expression levels of proteins in PSA (5a,8,13b) are influenced by microRNAs in cluster U. Evidence for co-regulatory roles of mir-21and mir-107 in cluster U is provided by experimental observations showing that these microRNAs increase Arginase 1 expression (225, 226), exhibit immunosuppressive properties (227, 228), and modulate both T-cell activation (229, 230), and T cell apoptosis (231–233). Thus, interactions between PSA (5a,8,13b) and microRNAs in cluster U could be particularly useful for treatment of multiple disorders due to aberrant regulation of T-cell functions.
Additional evidence suggesting that PSA (5a,8,13b) acts as a regulatory network in atherosclerosis is the finding that deficiency of mir-21 in cluster U induces vascular inflammation during atherogenesis (136), while mir-107, also in cluster U enhances the repair of vascular endothelial cells (234). The involvement of these protective microRNAs in Alzheimer's disease is indicated by reports demonstrating reduced levels of mir-107 and mir-21in affected individuals (235, 236). These observations broaden the value of targeting cause-effect relationships between PSA (5a,8,13b) and microRNAs in cluster U for treatment of atherosclerosis as well as Alzheimer's disease.
Retrieval of information related to proteins in PSA (5b) indicates that these proteins participate in the epigenetic regulation of autophagy, which is key for regulating immune system functions (237, 238). Levels of protein expression within the cooperation network, PSA (5b) are influenced by 61 out of 136 disease-associated microRNAs, which includes 13 microRNAs linked to Alzheimer's disease, and 6 microRNAs linked to atherosclerosis (239), as well as mir-126 and mir-92a in micro–RNA Cluster N. Evidence suggests that these microRNAs coregulate functions within cooperation network PSA (5b) as both miR-126 and miR-92a have been implicated in regulating autophagy (240, 241). Additionally, mir-let7a, which targets proteins in the cooperation network PSA (5b), has been shown to regulate autophagy in an Alzheimer's disease model (242) and plays a key role in cardiovascular disease (243). Since the topology dynamics of PSA (5b) is regulated by most of the disease-associated microRNAs, the interactions of proteins in this cooperation network represent a prime target for developing new therapies for the multiple diseases discussed in this paper.
Overall, the cause-effect relationships summarized in Figure 7 imply that cooperation networks PSA (1–20) and microRNA clusters in Table 3 are parts of an integrated regulatory system illustrated in Figure 8, which controls the functions of networks coordinating information exchanges and interactions among protein complexes shown in Figure 5.
Figure 8
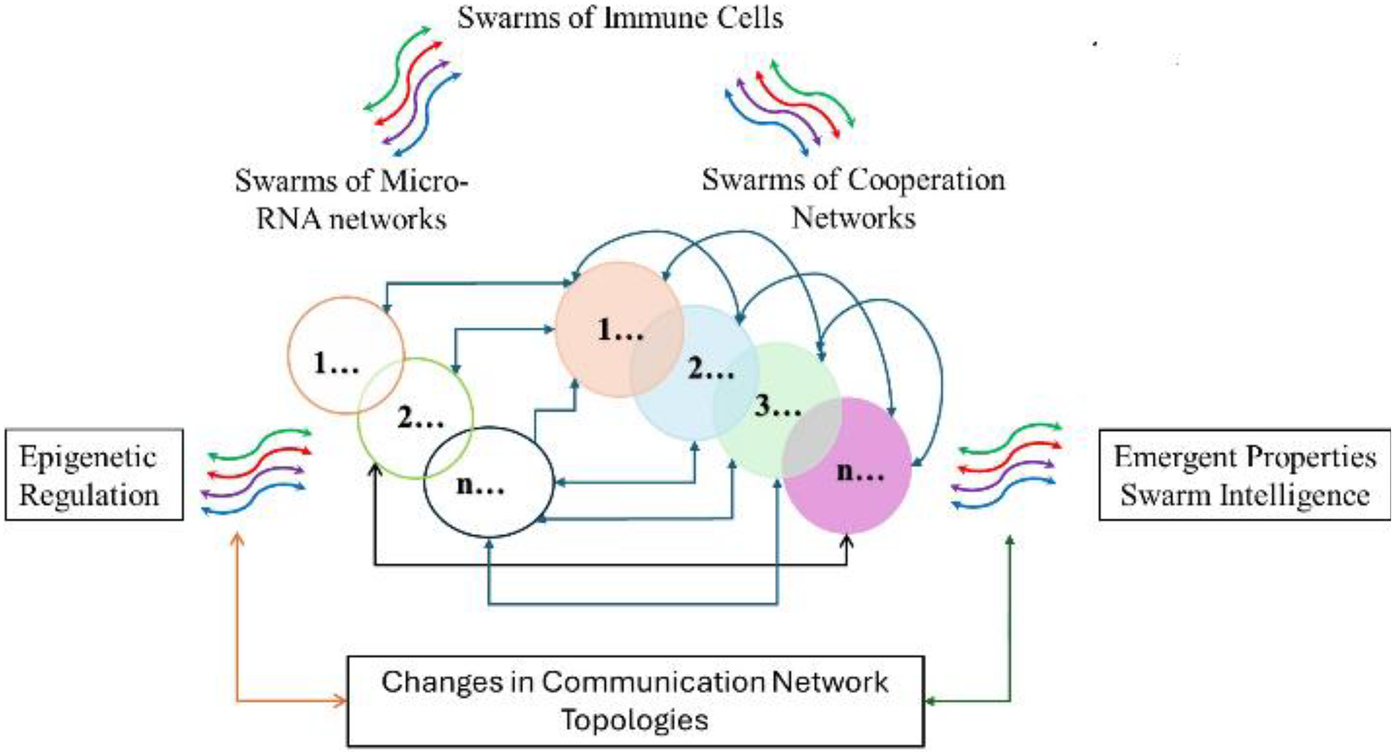
Illustrates an integrated regulatory system modulating functions of cooperation networks by modulating network topologies.
Impacts of COVID-19 on cooperation networks PSA (1–20)
Impacts of COVID-19-induced perturbations on regulatory schemes captured in PSA (1–20) are summarized in Figure 9.
Figure 9
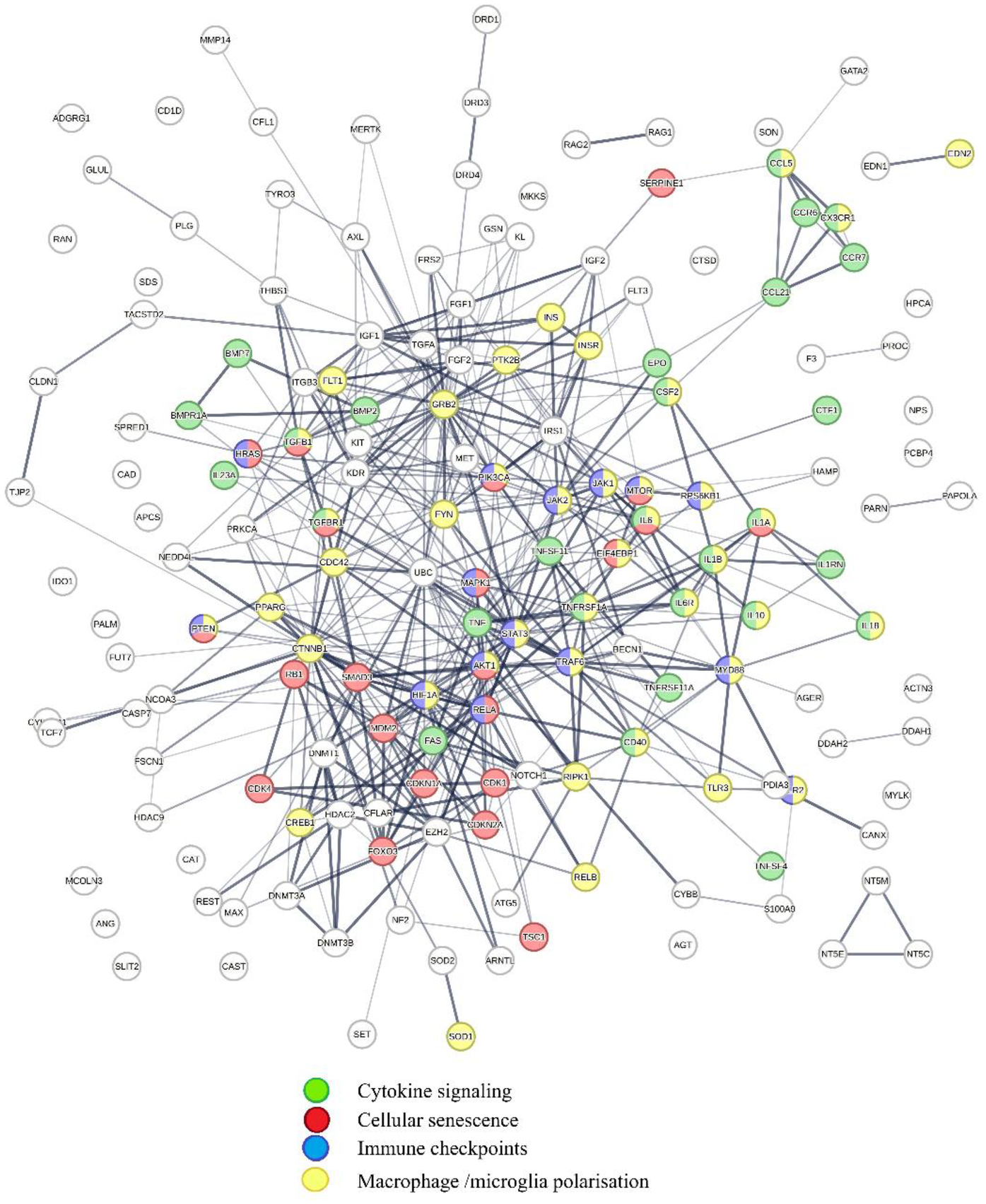
Shows physical interactions between proteins in PSA 1–20 targeted by COVID-19-associated microRNAs. Key functions affected by COVID-19 are cytokine signaling (green); cellular senescence (red); Immune checkpoints (blue); and macrophage/microglia polarization (yellow). The regulatory scheme highlighted shows that COVID-19 affects functions initiating and potentiating inflammation and regulating cellular senescence and immune checkpoints.
Retrieval of publications showing statistically significant network overlaps with protein interaction network shown in Figure 9 reveals that COVID-19-induced perturbations of cooperation networks captured in PSA (1–20) influence diseases such as cancer (244, 245), infectious diseases (246–249), metabolic diseases (250–254), cardiovascular diseases (255–257), inflammatory diseases, (258–262). Alzheimer's disease (263, 264), and aging (265, 266). Macrophages expressing ACE2 receptor and lectin CD169 can be infected by SARS-CoV-2 (267), and adopt a proinflammatory M1-like phenotype (268), which promotes both inflammation and viral replication by generating senescent cells, which secrete additional pro-inflammatory and pro-coagulation factors (269–274), and regulate immune checkpoints (275–278). Proinflammatory host responses are further amplified by SARS-CoV-2-associated spike protein, which increases M1 macrophage polarization (279).
A critical mechanism driving the amplification of inflammatory responses in COVID-19 is the dysregulation of TGFβ pathways, regulated in part by cooperation networks PSA (9) and PSA (7c) (280, 281). leading to hyperinflammation (282, 284), increased cellular senescence (283, 284), microclot formation (285), acute respiratory distress syndrome (ARDS) (286), pulmonary fibrosis (287), and life-threatening sepsis (288, 289). TGFβ signaling also lowers mir-125a-3p, mir-369 expression (cluster D; Table 3), which normally help regulate inflammation and TGFβ pathways by targeting proteins in PSA (9) and PSA (7c). Thus, mir-125a-3p, mir-369, reduce the expression of ACE2 (290), leading to loss of anti-inflammatory effects associated with ACE2 expression (291).
In long COVID, persistent proinflammatory conditions can result from reactivation of dormant viruses such as Epstein–Barr and varicella zoster (292), shifting microglia polarization in the brain to proinflammatory states and causing hyperinflammation, brain fog, and cognitive dysfunction (105, 293). Dysregulation of cooperation networks PSA (10) and PSA (5a, 8, 13b), along with sustained proinflammatory macrophage/microglia polarization and further aggravated by circulating spike proteins (92, 294–297), impairs T cell functions (298), and disrupts the regulatory system (see Figure 8) that fine-tunes inflammation (299–305).
Effects of treatments targeting the regulation of cooperation networks
The current methodology serves as an analytical tool for addressing a range of diseases. Analysis of complex relationships between microRNAs and cooperation networks identifies potential areas for the development of new treatment approaches. These strategies may complement existing drug discovery methods, as they can identify multiple disease targets (including proteins and microRNAs) and offer broader perspectives for understanding the mechanistic outcomes involved in diseases.
The summary of results presented in Table 4 introduces the concept of co-regulation of cooperation networks by microRNAs for developing therapeutic strategies relevant to the indications described in this manuscript. Additionally, it outlines approaches for examining the connections between different diseases and their mechanisms of action. For instance, the interaction between PSA (5a,8,13b) and cluster U can be used to identify specific microRNAs in cluster U that are either upregulated or downregulated in the target disease. Treatment options can then be determined through traditional screening methods by identifying substances that counteract the effects of disease-associated microRNA cooperation and protein interaction networks.
Table 4
| Protein Swarm Associations (PSA) | MicroRNA cluster | COVID-19 & other infections | Atherosclerosis & cardiometabolic diseases | Alzheimer's disease & dementias | Inflammation & immunity | Other |
|---|---|---|---|---|---|---|
| 5a.13 | O | + | + | + | Cancer, regulation of apoptosis | |
| 7c | L/mir-126 | + | + | + | Epithelial to mesenchymal transition | |
| 9 | O | + | + | + | + | Macrophage/microglial polarization |
| 10 | V | + | + | + | + | Cancer |
| 5a,8,13b | U | + | + | + | + | Aberrant regulation of T-cell functions |
| 5b | Interacts with 61/136 microRNAs | + | + | + | + | Aging (PSA5b is master coregulator of autophagy) |
| N | + | + | + | Aging/autophagy | ||
| Mir let7a | + | + | + | Aging/autophagy | ||
| I-20 | is targeted by COVID-19-associated microRNAs | + | + | + | + | Cancer, coagulopathy, aging |
Disease associations due to co-regulation of microRNAs with cooperation networks (PSAs).
Understanding these relationships supports the development of new treatment strategies, such as combinations of arginine, hesperidin, quercetin, vitamin D, and zinc for respiratory and immune disorders (see Figure 9).
Vitamin D was selected as a starting point for targeting epigenetic regulation of the regulatory scheme (see Figure 8) due to its broad effects on immune system modulation (306–308). Co-citation frequency measurements associated with Vitamin D deficiency (Figure 3) showed that vitamin D deficiency affects functions of multiple cooperation networks 1, 4, 5a, 5b, 13a, 7b, 7c, 8, 9, 7d, 14, 10, 11 and 12, many of which are regulated by microRNAs such as mir-155, mir-146a, mir-34a, and mir-21 (see Figures 3, 5B, 6, 8). SARS-CoV-2 infection increases the expression of the microRNAs mir-155, mir-146a (309), mir-34a (310), mir-21 (311), while Vitamin D supplementation reduces levels of mir-155 (312), mir-146a (313), mir-34a (314), and mir-21 (315). And has been shown in clinical trials to decrease COVID-19 severity (316). Figure 8 shows the influence of COVID-19-associated microRNAs on immune functions, suggesting that the induction of cellular senescence plays a key role in COVID-19 pathology (284, 317). The impact of cellular senescence on microRNA expression levels is provided by observations showing that the induction of cellular senescence increases the expression levels of pro-inflammatory mir-155 (318), mir-146a (319), mir-34a (320, 321), and mir-21 (322).
Counteracting effects of cellular senescence on microRNA expression levels, senolytic flavonoids such as quercetin (323) and apigenin (324), downregulate cellular senescence (325, 326), and, in doing so, decrease the expression levels of mir-155 (327, 328), mir 146a (329, 330), mir 34a (331, 340), and mir-21 (332). Benefits resulting from this senolytic action is suggested by observations showing that quercetin decreases the severity of COVID-19-induced inflammation and expedites recovery (333), and that apigenin deactivates the NLRP3 inflammasome (334).
COVID-19 is linked to arginine deprivation (335), and dysregulation of T-cell functions (335–337). Figure 7 shows that networks PSA (5a, 8, 13b) which regulate T-cell functions by regulating Arginase 1, are targeted by mir-107 and mir-21 in microRNA cluster U (Figure 6). Arginine deficiency increases mir-21 expression and inflammation by lowering endothelial nitric oxide production (338). Arginine supplementation increases mir-21 expression levels in clinical studies and shown to reduce the duration of in-hospital stays and the need for respiratory support in COVID-19 patients (339).
Zinc deficiency is associated with increased pro-inflammatory mir-21 and impaired T-cell function; supplementing zinc restores balance, lowers mortality, and shortens hospitalizations. Chronic inflammation and pulmonary fibrosis after COVID-19 are tied to dysregulation of networks 9 and 7c, leading to excessive TGFβ1 signaling and decreased mir-132. Hesperidin increases mir-132 and inhibits TGFβ1, improving immunity and controlling cytokine storms.
Zinc deficiency has also been correlated with persistent inflammation and increased mortality in COVID-19 patients. Zinc deficiency leads to overexpression of proinflammatory mir-21 (340–342). and to impair T cell functions (343, 344); zinc supplementation reverses mir-21 imbalance (345), and has been reported to lower 30-day mortality and shorten hospitalizations of COVID-19 patients (346, 347) Some of the major long-term problems associated with COVID-19 are the development of chronic inflammation (348) and pulmonary fibrosis (349). These conditions are precipitated in part by COVID-19-induced dysregulation of cooperation networks 9 and 7c, leading to excessive TGFβ1 signaling (350, 351) and decreases in mir-132 levels functioning as a suppressor of pro-inflammatory cytokine production (352, 353). Counteracting effects produced by excessive TGFβ1 signaling is the flavonoid hesperidin; it increases mir-132 levels (354, 355), and inhibits TGFβ1 activity (356). Benefits resulting from these mechanisms of action are provided by observations showing that hesperidin supplementation improves immunity against infections and controls cytokine storms (357).
Targeting the same regulatory schemes are combinations of Vitamin D and hesperidin in anti-aging studies (358), and the use of arginine and zinc combinations to improve thymic endocrine activity and peripheral immune functions in aged mice (359). The results indicate that clinical trials evaluating combinations of Vitamin D, arginine, quercetin, hesperidin, and zinc may demonstrate advantages in anti-aging and immunological research, as well as potential positive outcomes in studies related to COVID-19 and associated conditions.
Discussion
MicroRNAs are integral to various biological processes and serve as important biomarkers and potential therapeutic targets (360). To enhance the use of microRNA data for developing new avenues for disease treatment, including cardiovascular disease and cancer, bioinformatics tools have been applied to analyze microRNA expression profiles and functional roles (361–366). Nevertheless, the intricate nature of network-network interactions and the regulatory effects of microRNAs on protein networks remain largely uncharacterized, with current understanding primarily derived from theoretical bioinformatic analyses rather than direct cause-effect relationship studies (367). For instance, in efforts to elucidate the role of microRNAs in the pathogenesis of diabetic cardiomyopathy, a machine learning tool called DeepMiRBP has been used for predicting microRNA-protein network interactions (368). Additionally, extensive literature searches using multiple electronic databases such as PubMed, Scopus, Web of Science, and Google Scholar have helped examine the complex association between microRNAs and oxidative stress in cardiovascular disease (369). To identify the role of key microRNAs and mRNAs related to inflammatory bowel diseases (IBDs) and their subtypes, text mining-based approaches leveraging PubMed and PMC databases have been employed (370). Similarly, a web-based Random Walk algorithm (371) has been introduced to explore linkages between microRNA expression levels and disease-associated pathways. Furthermore, deep learning models specifically designed to predict microRNA-binding proteins have been proposed by simulating molecular interactions (372).
In contrast, the findings from our protein swarm-based cause-effect analysis, as depicted in Figures 2–9, demonstrate that interactions within the microRNA protein network modulate the precision and specificity of biological mechanisms/machinery coordinating cellular cooperation and processes across all systems levels (299, 373, 374). Examination of protein swarm organization in Figures 3, 6 provides insights into how regulatory intelligence differentiates health from disease, highlighting the influence of diseases and microRNAs on the structural regulation of cooperative functions within these networks (375).
Notably, the strong cluster similarity metrics integrating proteomics data in Figures 3, 6 suggest overlapping topologies among cooperation network segments 1–20. Figure 2 substantiates this premise by revealing that proteins within these network segments are capable of physically interacting with proteins across all 20 network segments. Additionally, Figure 5B indicates that these proteins contribute not only to information transmission but also to the execution of specific cellular functions (376, 377). These network characteristics facilitate efficient information dissemination and the coordination of cooperation among diverse functional processes. Furthermore, alterations in microRNA expression levels (378–380) can increase or decrease the number and the strength of interactions within protein networks, thereby reshaping network topology and enabling epigenetic regulation for dynamic adjustments and precise modulation of cooperation network functions.
The observed cause-effect relationships illustrated in Figures 2–9 collectively show that perturbations of microRNAs expression influence macrophage plasticity and the progression and severity of various diseases, including cardiovascular disease (381).
The section entitled “Effects of treatments targeting the regulation of cooperation networks” builds upon previous research by illustrating that integrating vitamin D's genomic and non-genomic signaling—through the coupling of nitric oxide and redox signaling—may improve the efficacy of vitamin D-based therapies for cardiovascular disease (382). Additionally, protein swarm-based, cause-effect analysis enables evaluation of treatment effects on cooperative networks via combinations such as vitamin D with quercetin, hesperidin, arginine, and zinc ascorbate, which are designed to modify disease-specific expression. This approach shows potential not only for cardiovascular diseases but also for other disorders linked to dysfunctional cooperation networks (383, 384).
Ultimately, findings from protein swarm-based cause-effect analyses support the advancement of therapies focusing on the topological architecture of cooperative networks, as opposed to targeting isolated pathways. This approach has proven effective in various contexts, including combination cancer therapies (385, 386), modulation of microbiome compositions for cardiovascular disease (387), and practices rooted in traditional medicine (388, 389).
Strengths and limitations
Understanding the influence of protein network–protein network and microRNA network–protein network interactions on health and disease is essential for developing safe and effective therapeutic strategies (390). Our findings indicate that approaches focusing solely on individual components are insufficient to capture the emergent properties resulting from dynamic network-network interactions that affect disease progression and treatment outcomes (391). To address this complexity, protein swarm-based cause–effect analysis utilizes the inherent plasticity of proteins, providing an unbiased framework to examine how emergent properties within interacting network systems modulate physiological and pathological states of organisms (392). Applying “Swarm Intelligence” as an analysis tool enables the delineation of ultra-complex cause-effect relationships that are otherwise challenging to ascertain and resolve (393).
Nevertheless, current cause–effect analyses are constrained by their predominant focus on the effects of mature microRNAs within cooperative systems, while frequently neglecting the role of post-translational modifications in proteomes. Furthermore, technical constraints restrict the resolution of studies on network–network interactions due to upper bounds on the number of protein swarms that can be sampled from tissue proteomes. Finally, the effectiveness of machine learning approaches in addressing complex biological cause–effect relationships remains constrained by insufficient data regarding concentration and temporal dependencies on a system-wide scale (413).
Moreover, methodological limitations impose constraints on the resolution of studies examining network–network interactions, as there are upper limits to the number of protein groups that can be sampled from tissue proteomes. Additionally, while machine learning approaches offer promise in elucidating complex biological cause–effect relationships, their effectiveness is currently limited by insufficient data describing concentration and temporal dependencies at the system-wide level.
Summary
Protein swarm-based cause-effect analysis was employed to investigate the principles governing immune system responses to injury, emphasizing dynamic changes in protein network topologies and the epigenetic regulation of microRNAs. These mechanisms function as computational systems that facilitate rapid adaptation to fluctuations in both micro and macro environments. Disruptions affecting the accuracy of protein network topology-mediated computations offer insights into the immune evasion strategies of pathogens (261, 394), and cancers (395–397), and parallels in pathological processes among infections (398), Alzheimer's disease (399–404), metabolic disorders (405, 406), cardiovascular diseases (407), and autoimmune diseases (408). Furthermore, deciphering complex cause-effect relationships arising from emergent computations (409–411), indicates that swarm-based cause-effect analysis may have applications not only in drug design but also in robotics and quantum computing (412, 413).
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
AF: Writing – original draft; Writing – review & editing; Formal analysis; Methodology; Conceptualization; Data curation; Investigation; Visualization. RS: Writing – review & editing. SK: Resources; Writing – review & editing; Conceptualization.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
AF, RS, SK were employed by SystaMedic Inc. AF, SK were employed by Emergent System Analytics, LLC.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2025.1577844/full#supplementary-material
References
1.
Chavali AK Gianchandani EP Tung KS Lawrence MB Peirce SM Papin JA . Characterizing emergent properties of immunological systems with multi-cellular rule-based computational modeling. Trends Immunol. (2008) 29(12):589–99. 10.1016/j.it.2008.08.006
2.
Chen ZY . Application of immunological and swarm intelligence learning-based algorithm for industrial grade computer sales prediction. Appl Artif Intell. (2024) 39(1):e2440836, 33 pages. 10.1080/08839514.2024.2440836
3.
Carpenter S O'Neill LAJ . From periphery to center stage: 50 years of advancements in innate immunity. Cell. (2024) 187(23):6780–2. 10.1016/j.cell.2024.10.013
4.
Danilova N . The evolution of immune mechanisms. J Exp Zool B Mol Dev Evol. (2006) 306(6):496–520. 10.1002/jez.b.21102
5.
InformedHealth.org. In brief: The innate and adaptive immune systems. Cologne, Germany: Institute for Quality and Efficiency in Health Care (IQWiG) (2006). Available online at:https://www.ncbi.nlm.nih.gov/books/NBK279396/(Accessed August 14, 2023).
6.
Fenn J Madon K Conibear E Derelle R Nevin S Kundu R et al An ultra-early, transient interferon-associated innate immune response associates with protection from SARS-CoV-2 infection despite exposure. EBioMedicine. (2024) 11:105475. 10.1016/j.ebiom.2024.105475
7.
Thomas P Thomma BPHJ Girardin SE Lemaitre B . The conceptual foundations of innate immunity: taking stock 30 years later. Immunity. (2024) 57(4):613–31. 10.1016/j.immuni.2024.03.007
8.
Flores-Gomez D Hobo W van Ens D Kessler EL Novakovic B Schaap NPM et al Interleukin-1β induces trained innate immunity in human hematopoietic progenitor cells in vitro. Stem Cell Rep. (2024) 19:1651–64. 10.1016/j.stemcr.2024.09.004
9.
Weng J Couture C Girard S . Innate and adaptive immune systems in physiological and pathological pregnancy. Biology (Basel). (2023) 12(3):402. 10.3390/biology12030402
10.
Zheng D Liwinski T Elinav E . Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. 10.1038/s41422-020-0332-7
11.
Waldner H . The role of innate immune responses in autoimmune disease development. Autoimmun Rev. (2009) 8(5):400–4. 10.1016/j.autrev.2008.12.019
12.
Wynn TA Chawla A Pollard JW . Macrophage biology in development, homeostasis and disease. Nature. (2013) 496(7446):445–55. 10.1038/nature12034
13.
Lackey DE Olefsky JM . Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. (2016) 12(1):15–28. 10.1038/nrendo.2015.189
14.
Naik S Larsen SB Cowley CJ Fuchs E . Two to tango: dialog between immunity and stem cells in health and disease. Cell. (2018) 175(4):908–20. 10.1016/j.cell.2018.08.071
15.
Simats A Sager HB Liesz A . Heart brain axis in health and disease: role of innate and adaptive immunity. Cardiovasc Res. (2024) 24:2325–35. 10.1093/cvr/cvae185
16.
Morimoto K Nakajima K . Role of the immune system in the development of the central nervous system. Front Neurosci. (2019) 13:916. 10.3389/fnins.2019.00916
17.
Kelvington BA Abel T . Innate immunity in neurons makes memories persist. Nature. (2024) 628(8006):40–2. 10.1038/d41586-024-00679-4
18.
Zengeler KE Lukens JR . Innate immunity at the crossroads of healthy brain maturation and neurodevelopmental disorders. Nat Rev Immunol. (2021) 21(7):454–68. 10.1038/s41577-020-00487-7
19.
Zak DE Tam VC Aderem A . Systems-level analysis of innate immunity. Annu Rev Immunol. (2014) 32:547–77. 10.1146/annurev-immunol-032713-120254
20.
Mills CD Kincaid K Alt JM Heilman MJ Hill AM . M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. (2000) 164:6166–73. 10.4049/jimmunol.164.12.6166
21.
Sumagin R . Emerging neutrophil plasticity: terminally differentiated cells no more. J Leukoc Biol. (2021) 109(3):473–5. 10.1002/JLB.1CE0720-378R
22.
Zhang Q Cao X . Epigenetic regulation of the innate immune response to infection. Nat Rev Immunol. (2019) 19:417–32. 10.1038/s41577-019-0151-6
23.
Boosani CS Agrawal DK . Epigenetic regulation of innate immunity by microRNAs. Antibodies. (2016) 5(2):8. 10.3390/antib5020008
24.
Uehata T Takeuchi O . RNA Recognition and immunity-innate immune sensing and its posttranscriptional regulation mechanisms. Cells. (2020) 9(7):1701. 10.3390/cells9071701
25.
Leseigneur C Lê-Bury P Pizarro-Cerdá J Dussurget O . Emerging evasion mechanisms of macrophage defenses by pathogenic Bacteria. Front Cell Infect Microbiol. (2020) 10:577559. 10.3389/fcimb.2020.577559
26.
Banete A Barilo J Whittaker R Basta S . The activated macrophage—a tough fortress for virus invasion: how viruses strike back. Front Microbiol. (2022) 12:803427. 10.3389/fmicb.2021.803427
27.
Yu S Ge H Li S Qiu HJ . Modulation of macrophage polarization by viruses: turning off/on host antiviral responses. Front Microbiol. (2022) 13:839585. 10.3389/fmicb.2022.839585
28.
Zhang Z Liu J Yu L Zeng R Pan W . The hijacking of HBV by small extracellular vesicles inhibits M1 macrophages to facilitate immune evasion. Sci Rep. (2024) 14:19917. 10.1038/s41598-024-70924-3
29.
Gobert AP Latour YL Asim M Finley JL Verriere TG Barry DP et al Bacterial pathogens hijack the innate immune response by activation of the reverse transsulfuration pathway. mBio. (2019) 10(5):e02174–19. 10.1128/mBio.02174-19
30.
Rojas-Pirela M Andrade-Alviárez D Quiñones W Rojas MV Castillo C Liempi A et al microRNAs: critical players during helminth infections. Microorganisms. (2022) 11(1):61. 10.3390/microorganisms11010061
31.
Thiriot JD Martinez-Martinez YB Endsley JJ Torres AG . Hacking the host: exploitation of macrophage polarization by intracellular bacterial pathogens. Pathog Dis. (2020) 78(1):ftaa009. 10.1093/femspd/ftaa009
32.
Li H Yang Y Hong W Huang M Wu M Zhao X . Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. (2020) 5(1):1. 10.1038/s41392-019-0089-y
33.
Galkina E Ley K . Immune and inflammatory mechanisms of atherosclerosis. Annu Rev Immunol. (2009) 27:165–97. 10.1146/annurev.immunol.021908.132620
34.
Castro-Gomez S Heneka MT . Innate immune activation in neurodegenerative diseases. Immunity. (2024) 57(4):790–814. 10.1016/j.immuni.2024.03.010
35.
Strauss A Swann P Kigar SL Christou R Savinykh Yarkoni N Turner L et al Peripheral innate immunophenotype in neurodegenerative disease: blood-based profiles and links to survival. Mol Psychiatry. (2024) 30:1985–94. 10.1038/s41380-024-02809-w
36.
Jin C Henao-Mejia J Flavell RA . Innate immune receptors: key regulators of metabolic disease progression. Cell Metab. (2013) 17(6):873–82. 10.1016/j.cmet.2013.05.011
37.
Paragh G Seres I Harangi M Fülöp P . Dynamic interplay between metabolic syndrome and immunity. Adv Exp Med Biol. (2014) 824:171–90. 10.1007/978-3-319-07320-0_13
38.
Shurin MR . Cancer as an immune-mediated disease. Immunotargets Ther. (2012) 1:1–6. 10.2147/ITT.S29834
39.
Huang R Kang T Chen S . The role of tumor-associated macrophages in tumor immune evasion. J Cancer Res Clin Oncol. (2024) 150(5):238. 10.1007/s00432-024-05777-4
40.
Hu A Sun L Lin H Liao Y Yang H Mao Y et al Harnessing innate immune pathways for therapeutic advancement in cancer. Sig Transduct Target Ther. (2024) 9:68. 10.1038/s41392-024-01765-9
41.
Brown KL Cosseau C Gardy JL Hancock RE . Complexities of targeting innate immunity to treat infection. Trends Immunol. (2007) 28(6):260–6. 10.1016/j.it.2007.04.005
42.
Stiel L Gaudet A Thietart S Vallet H Bastard P Voiriot G et al Innate immune response in acute critical illness: a narrative review. Ann Intensive Care. (2024) 14:137. 10.1186/s13613-024-01355-6
43.
Shen S Zhang LS . The regulation of antiviral innate immunity through non-m6A RNA modifications. Front Immunol. (2023) 14:1286820. 10.3389/fimmu.2023.1286820
44.
Holzinger A Dehmer M Jurisica I . Knowledge discovery and interactive data mining in bioinformatics—state-of-the-art, future challenges and research directions. BMC Bioinformatics. (2014) 15(Suppl 6):I1. 10.1186/1471-2105-15-S6-I1
45.
Calderon J Berman GJ . Inferring the time-varying coupling of dynamical systems with temporal convolutional autoencoders. eLife. (2024) 13:RP100692. 10.7554/eLife.100692.1
46.
Piccolo SA Lehmann S Maier AM . Different networks for different purposes: a network science perspective on collaboration and communication in an engineering design project. Comput Ind. (2022) 142:103745. 10.1016/j.compind.2022.103745
47.
Hartle H Papadopoulos F Krioukov D . Dynamic hidden-variable network models. Phys Rev E. (2021) 103(5-1):052307. 10.1103/PhysRevE.103.052307
48.
Li S Wang L Berman M Kong YY Dorf ME . Mapping a dynamic innate immunity protein interaction network regulating type I interferon production [published correction appears in Immunity. 2011 Oct 28;35(4):647–8]. Immunity. 2011;35(3):426–40. 10.1016/j.immuni.2011.06.014
49.
Stampanoni Bassi M Iezzi E Gilio L Centonze D Buttari F . Synaptic plasticity shapes brain connectivity: implications for network topology. Int J Mol Sci. (2019) 20(24):6193. 10.3390/ijms20246193
50.
Singh MS Pasumarthy R Vaidya U Leonhardt S . On quantification and maximization of information transfer in network dynamical systems. Sci Rep. (2023) 13:5588. 10.1038/s41598-023-32762-7
51.
Janzakova K Balafrej I Kumar A Garg N Scholaert C Rouat J et al Structural plasticity for neuromorphic networks with electropolymerized dendritic PEDOT connections. Nat Commun. (2023) 14:8143. 10.1038/s41467-023-43887-8
52.
Olivença DV Davis JD Voit EO . Inference of dynamic interaction networks: a comparison between Lotka–Volterra and multivariate autoregressive models. Front Bioinform. (2022) 2:1021838. 10.3389/fbinf.2022.1021838
53.
Harush U Barzel B . Dynamic patterns of information flow in complex networks. Nat Commun. (2017) 8:2181. 10.1038/s41467-017-01916-3
54.
Gao J Barzel B Barabási A-L . Universal resilience patterns in complex networks. Nature. (2016) 530:307312. 10.1038/nature16948
55.
García Morán GA Parra-Medina R Cardona AG et al Chapter 9: Cytokines, chemokines and growth factors. In: AnayaJMShoenfeldYRojas-VillarragaALevyRACerveraR, editors. Autoimmunity: From Bench to Bedside. Bogota (Colombia): El Rosario University Press (2013). Available online at:https://www.ncbi.nlm.nih.gov/books/NBK459450/(Accessed July 18, 2013).
56.
Fliri AF Kajiji S . Functional characterization of nutraceuticals using spectral clustering: centrality of caveolae-mediated endocytosis for management of nitric oxide and vitamin D deficiencies and atherosclerosis. Front Nutr. (2022) 9:885364. 10.3389/fnut.2022.885364
57.
Fliri AF Loging WT Volkmann RA . Drug effects viewed from a signal transduction network perspective. J Med Chem. (2009) 52(24):8038–46. 10.1021/jm901001p
58.
Zhang G Li F Ren D Huang H Zhou Z Chang F . Cooperative control of self-learning traffic signal and connected automated vehicles for safety and efficiency optimization at intersections. Accid Anal Prev. (2024) 19:107890. 10.1016/j.aap.2024.107890
59.
Holzinger A Malle B Saranti A Pfeifer B . Towards multi-modal causability with graph neural networks enabling information fusion for explainable AI. Inform Fusion. (2021) 71:28–37. 10.1016/j.inffus.2021.01.008
60.
Weiel M Götz M Klein A Coquelin D Floca R Schug A . Dynamic particle swarm optimization of biomolecular simulation parameters with flexible objective functions. Nat Mach Intell. (2021) 3:727–34. 10.1038/s42256-021-00366-3
61.
Fliri AF Loging WT Volkmann RA . Analysis of information flows in interaction networks: implication for drug discovery and pharmacological research. Discov Med. (2011) 11(57):133–43.
62.
Kajihara KT Hynson NA . Networks as tools for defining emergent properties of microbiomes and their stability. Microbiome. (2024) 12(184). 10.1186/s40168-024-01868-z
63.
Anwar MS Sar GK Perc M Ghosh D . Collective dynamics of swarmalators with higher-order interactions. Commun Phys. (2024) 7(59). 10.1038/s42005-024-01556-2
64.
Mateo D Kuan YK Bouffanais R . Effect of correlations in swarms on collective response. Sci Rep. (2017) 7(1):10388. 10.1038/s41598-017-09830-w
65.
Hoogesteyn AL Rivas AL Smith SD Fasina FO Fair JM Kosoy M . Assessing complexity and dynamics in epidemics: geographical barriers and facilitators of foot-and-mouth disease dissemination. Front Vet Sci. (2023) 10:1149460. 10.3389/fvets.2023.1149460
66.
Sudakow I Reinitz J Vakulenko SA Grigoriev D . Evolution of biological cooperation: an algorithmic approach. Sci Rep. (2024) 14:1468. 10.1038/s41598-024-52028-0
67.
Muraille E . Diversity generator mechanisms are essential components of biological systems: the two queen hypothesis. Front Microbiol. (2018) 9:223. 10.3389/fmicb.2018.00223
68.
Vilaplana-Carnerero C Giner-Soriano M Dominguez À Morros R Pericas C Álamo-Junquera D et al Atherosclerosis, cardiovascular disease, and COVID-19: a narrative review. Biomedicines. (2023) 11(4):1206. 10.3390/biomedicines11041206
69.
Rudnicka-Drożak E Drożak P Mizerski G Zaborowski T Ślusarska B Nowicki G et al Links between COVID-19 and Alzheimer's Disease-What do we already know? Int J Environ Res Public Health. (2023) 20(3):2146. 10.3390/ijerph20032146
70.
Rico-Mesa JS Haloot J Anupama BK Atluri S Liu J Khalid U . The role and implications of COVID-19 in incident and prevalent heart failure. Curr Heart Fail Rep. (2024) 21(5):485–97. 10.1007/s11897-024-00677-7
71.
Uhlén M Fagerberg L Hallström BM Lindskog C Oksvold P Mardinoglu A et al Tissue-based map of the human proteome. Science. (2015) 347(6220):1260419. 10.1126/science.1260419
72.
Szklarczyk D Gable AL Nastou KC Lyon D Kirsch R Pyysalo S et al The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49(D1):D605–12. 10.1093/nar/gkaa1074
73.
Fliri AF . Method and descriptors for comparing object-induced information flows in a plurality of interaction networks; I. National Center for Biotechnology Information (2015). PubChem Patent Summary for WO-2017091822-A1. Available online at:https://pubchem.ncbi.nlm.nih.gov/patent/WO-2017091822-A1(Accessed November 25, 2015).
74.
National Library of Medicine [NLM] (2014).
75.
Fliri AF Loging WT Volkmann RA . Cause-effect relationships in medicine: a protein network perspective. Trends Pharmacol Sci. (2010) 31(11):547–55. 10.1016/j.tips.2010.07.005
76.
Girvan M Newman ME . Community structure in social and biological networks. Proc Natl Acad Sci U S A. (2002) 99(12):7821–6. 10.1073/pnas.122653799
77.
TIBCO Software Inc., Palo Alto, CA.
78.
Fierro-Monti I Wright JC Choudhary JS Vizcaíno JA . Identifying individuals using proteomics: are we there yet?Front Mol Biosci. (2022) 9:1062031. 10.3389/fmolb.2022.1062031
79.
Zhu X Shen X Qu J Straubinger RM Jusko WJ . Multi-Scale network model supported by proteomics for analysis of combined gemcitabine and birinapant effects in pancreatic cancer cells. CPT Pharmacometrics Syst Pharmacol. (2018) 7(9):549–61. 10.1002/psp4.12320
80.
Kustatscher G Collins T Gingras AC et al Understudied proteins: opportunities and challenges for functional proteomics. Nat Methods. (2022) 19(7):774–9. 10.1038/s41592-022-01454-x
81.
Ivanov PC Bartsch RP . Network physiology: mapping interactions between networks of physiologic networks. In: D'AngostinoGScalaA, editors. Networks of Networks: The Last Frontier of Complexity. Cham: Springer (2014). p. 203–22.
82.
Broido AD Clauset A . Scale-free networks are rare. Nat Commun. (2019) 10:1017. 10.1038/s41467-019-08746-5
83.
Woessmann J Kotol D Hober A Uhlén M Edfors F . Addressing the protease bias in quantitative proteomics. J Proteome Res. (2022) 21(10):2526–34. 10.1021/acs.jproteome.2c00491
84.
Bantscheff M Schirle M Sweetman G Rick J Kuster B . Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. (2007) 389(4):1017–31. 10.1007/s00216-007-1486-6
85.
Szklarczyk D Kirsch R Koutrouli M Nastou K Mehryary F Hachilif R et al The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. (2023) 51(D1):D638–46. 10.1093/nar/gkac1000
86.
Fliri AF Kajiji S . Vitamin D deficiency-associated comorbidities: a protein network dynamics perspective. Med Res Arch. (2023) 11(6). 10.18103/mra.v11i6.3996
87.
Babačić H Christ W Araújo JE Mermelekas G Sharma N Tynell J et al Comprehensive proteomics and meta-analysis of COVID-19 host response. Nat Commun. (2023) 14:5921. 10.1038/s41467-023-41159-z
88.
Harrison PW Lopez R Rahman N Allen SG Aslam R Buso N et al The COVID-19 data portal: accelerating SARS-CoV-2 and COVID-19 research through rapid open access data sharing. Nucleic Acids Res. (2021) 49(W1):W619–23. 10.1093/nar/gkab417
89.
Westergaard D Stærfeldt HH Tønsberg C Jensen LJ Brunak S . A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput Biol. (2018) 14(2):e1005962. 10.1371/journal.pcbi.1005962
90.
Cui C Zhong B Fan R Cui Q . HMDD V4.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. (2024) 52(D1):D1327–32. 10.1093/nar/gkad717
91.
Kozomara A Birgaoanu M Griffiths-Jones S . Nucleic Acids Res. (2019) 47:D155–62. 10.1093/nar/gky1141
92.
Al-Beltagi M Saeed NK Bediwy AS . COVID-19 disease and autoimmune disorders: a mutual pathway. World J Methodol. (2022) 12(4):200–23. 10.5662/wjm.v12.i4.200
93.
Singh SJ Baldwin MM Daynes E Evans RA Greening NJ Jenkins RG et al Respiratory sequelae of COVID-19: pulmonary and extrapulmonary origins, and approaches to clinical care and rehabilitation. Lancet Respir Med. (2023) 11(8):709–25. 10.1016/S2213-2600(23)00159-5
94.
Sayed S . COVID-19 and malignancy: exploration of the possible genetic and epigenetic interlinks and overview of the vaccination scenario. Cancer Treat Res Commun. (2021) 28:100425. 10.1016/j.ctarc.2021.100425
95.
Mukund K Nayak P Ashokkumar C Rao S Almeda J Betancourt-Garcia MM et al Immune response in severe and non-severe coronavirus disease 2019 (COVID-19) infection: a mechanistic landscape. Front Immunol. (2021) 12:738073. 10.3389/fimmu.2021.738073
96.
Kavanagh E . Long COVID brain fog: a neuroinflammation phenomenon?Oxf Open Immunol. (2022) 3(1):iqac007. 10.1093/oxfimm/iqac007
97.
Jit BP Qazi S Arya R Srivastava A Gupta N Sharma A . An immune epigenetic insight to COVID-19 infection. Epigenomics. (2021) 13(6):465–80. 10.2217/epi-2020-0349
98.
Anwar MM Sah R Shrestha S Ozaki A Roy N Fathah Z et al Disengaging the COVID-19 clutch as a discerning eye over the inflammatory circuit during SARS-CoV-2 infection. Inflammation. (2022) 45(5):1875–94. 10.1007/s10753-022-01674-5
99.
Sharma C Bayry J . High risk of autoimmune diseases after COVID-19. Nat Rev Rheumatol. (2023) 19(7):399–400. 10.1038/s41584-023-00964-y
100.
Bouhaddou M Reuschl AK Polacco BJ Thorne LG Ummadi MR Ye C et al SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell. (2023) 186(21):4597–614.e26. 10.1016/j.cell.2023.08.026
101.
Kearns MD Alvarez JA Seidel N Tangpricha V . Impact of vitamin D on infectious disease. Am J Med Sci. (2015) 349(3):245–62. 10.1097/MAJ.0000000000000360
102.
Piekut T Hurła M Banaszek N Szejn P Dorszewska J Kozubski W et al Infectious agents and Alzheimer's Disease. J Integr Neurosci. (2022) 21(2):73. 10.31083/j.jin2102073
103.
Leinonen M Saikku P . Evidence for infectious agents in cardiovascular disease and atherosclerosis. Lancet Infect Dis. (2002) 2(1):11–7. 10.1016/s1473-3099(01)00168-2
104.
Chidambaram V Kumar A Sadaf MI Lu E Al'Aref SJ Tarun T et al COVID-19 in the initiation and progression of atherosclerosis: pathophysiology during and beyond the acute phase. JACC Adv. (2024) 3(8):101107. 10.1016/j.jacadv.2024.101107
105.
Wang Y Chen Q Yang L Yang S He K Xie X . Corrigendum: overlapping structures detection in protein-protein interaction networks using community detection algorithm based on neighbor clustering coefficient. Front Genet. (2021) 12:799818. 10.3389/fgene.2021.799818
106.
Kanehisa M Goto S . KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28(1):27–30. 10.1093/nar/28.1.27
107.
Ashburner M Ball CA Blake JA Botstein D Butler H Cherry JM et al Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. (2000) 25(1):25–9. 10.1038/75556
108.
Thomson BJ . Viruses and apoptosis. Int J Exp Pathol. (2001) 82(2):65–76. 10.1111/j.1365-2613.2001.iep0082-0065-x
109.
Janeway CA Jr. Travers P Walport M et al The course of the adaptive response to infection. Immunobiology: The Immune System in Health and Disease. 5th ed.New York: Garland Science (2001) Available online at:https://www.ncbi.nlm.nih.gov/books/NBK27125/(Accessed June 21, 2001).
110.
Childs BG Baker DJ Kirkland JL Campisi J van Deursen JM . Senescence and apoptosis: dueling or complementary cell fates?EMBO Rep. (2014) 15(11):1139–53. 10.15252/embr.201439245
111.
Artimovič P Špaková I Macejková E Pribulová T Rabajdová M Mareková M et al The ability of microRNAs to regulate the immune response in ischemia/reperfusion inflammatory pathways. Genes Immun. (2024) 25:277–96. 10.1038/s41435-024-00283-6
112.
Liang Y Fang D Gao X Deng X Chen N Wu J et al Circulating microRNAs as emerging regulators of COVID-19. Theranostics. (2023) 13(1):125–47. 10.7150/thno.78164
113.
Arghiani N Nissan T Matin MM . Role of microRNAs in COVID-19 with implications for therapeutics. Biomed Pharmacother. (2021) 144:112247. 10.1016/j.biopha.2021.112247
114.
Abidin SZ Mat Pauzi NA Mansor NI Mohd Isa NI Hamid AA . A new perspective on Alzheimer's Disease: microRNAs and circular RNAs. Front Genet. (2023) 14:1231486. 10.3389/fgene.2023.1231486
115.
Kumar S Reddy PH . The role of synaptic microRNAs in Alzheimer's Disease. Biochim Biophys Acta Mol Basis Dis. (2020) 1866(12):165937. 10.1016/j.bbadis.2020.165937
116.
Schulte LN Westermann AJ Vogel J . Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. (2013) 41(1):542–53. 10.1093/nar/gks1030
117.
Sheedy FJ . Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. (2015) 6(19). 10.3389/fimmu.2015.00019
118.
Taheri F Ebrahimi SO Shareef S Reiisi S . Regulatory and immunomodulatory role of miR-34a in T cell immunity. Life Sci. (2020) 262:118209. 10.1016/j.lfs.2020.118209
119.
Baulina NM Kulakova OG Favorova OO . MicroRNAs: the role in autoimmune inflammation. Acta Naturae. (2016) 8(1):21–33. 10.3390/cells8020133
120.
Readhead B Haure-Mirande JV Mastroeni D Audrain M Fanutza T Kim SH et al Mir155 regulation of behavior, neuropathology, and cortical transcriptomics in Alzheimer's Disease. Acta Neuropathol. (2020) 140(3):295–315. 10.1007/s00401-020-02185-z
121.
Wang G Huang Y Wang LL Zhang YF Xu J Zhou Y et al MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer's Disease. Sci Rep. (2016) 6:26697. 10.1038/srep26697
122.
Bazrgar M Khodabakhsh P Prudencio M Mohagheghi F Ahmadiani A . The role of microRNA-34 family in Alzheimer's Disease: a potential molecular link between neurodegeneration and metabolic disorders. Pharmacol Res. (2021) 172:105805. 10.1016/j.phrs.2021.105805
123.
Garcia G Pinto S Ferreira S Lopes D Serrador MJ Fernandes A et al Emerging role of miR-21-5p in neuron-Glia dysregulation and exosome transfer using multiple models of Alzheimer's Disease. Cells. (2022) 11(21):3377. 10.3390/cells11213377
124.
Nazari-Jahantigh M Wei Y Noels H Akhtar S Zhou Z Koenen RR et al MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. (2012) 122(11):4190–202. 10.1172/JCI61716
125.
Cheng HS Besla R Li A Chen Z Shikatani EA Nazari-Jahantigh M et al Paradoxical suppression of atherosclerosis in the absence of microRNA-146a. Circ Res. (2017) 121(4):354–67. 10.1161/CIRCRESAHA.116.310529
126.
Xu Y Xu Y Zhu Y Sun H Juguilon C Li F et al Macrophage miR-34a is a key regulator of cholesterol efflux and atherosclerosis. Mol Ther. (2020) 28(1):202–16. 10.1016/j.ymthe.2019.09.008
127.
Canfrán-Duque A Rotllan N Zhang X Fernández-Fuertes M Ramírez-Hidalgo C Araldi E et al Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med. (2017) 9(9):1244–62. 10.15252/emmm.201607492
128.
Kellermann M Scharte F Hensel M . Manipulation of host cell organelles by intracellular pathogens. Int J Mol Sci. (2021) 22(12):6484. 10.3390/ijms22126484
129.
Lilley CE Chaurushiya MS Weitzman MD . Chromatin at the intersection of viral infection and DNA damage. Biochim Biophys Acta. (2010) 1799(3-4):319–27. 10.1016/j.bbagrm.2009.06.007
130.
Wang Y Zhang X Song Q Hou Y Liu J Sun Y et al Characterization of the chromatin accessibility in an Alzheimer's Disease (AD) mouse model. Alzheimers Res Ther. (2020) 12(1):29. 10.1186/s13195-020-00598-2
131.
Wang Y Gao H Wang F Ye Z Mokry M Turner AW et al Dynamic changes in chromatin accessibility are associated with the atherogenic transitioning of vascular smooth muscle cells. Cardiovasc Res. (2022) 118(13):2792–804. 10.1093/cvr/cvab347
132.
Vreede FT Chan AY Sharps J Fodor E . Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology. (2010) 396(1):125–34. 10.1016/j.virol.2009.10.003
133.
Söderholm S Kainov DE Öhman T Denisova OV Schepens B Kulesskiy E et al Phosphoproteomics to characterize host response during influenza A virus infection of human macrophages. Mol Cell Proteomics. (2016) 15(10):3203–19. 10.1074/mcp.M116.057984
134.
Dickson JR Yoon H Frosch MP Hyman BT . Cytoplasmic mislocalization of RNA polymerase II subunit RPB1 in Alzheimer disease is linked to pathologic tau. J Neuropathol Exp Neurol. (2021) 80(6):530–40. 10.1093/jnen/nlab040
135.
Mao S Song C Huang H Nie Y Ding K Cui J et al Role of transcriptional cofactors in cardiovascular diseases. Biochem Biophys Res Commun. (2024) 706:149757. 10.1016/j.bbrc.2024.149757
136.
Murphy KN Brinkworth AJ . Manipulation of focal adhesion signaling by pathogenic microbes. Int J Mol Sci. (2021) 22(3):1358. 10.3390/ijms22031358
137.
Caltagarone J Jing Z Bowser R . Focal adhesions regulate abeta signaling and cell death in Alzheimer's Disease. Biochim Biophys Acta. (2007) 1772(4):438–45. 10.1016/j.bbadis.2006.11.007
138.
Murphy JM Jeong K Tran DTK Cioffi DL Campbell PM Kim JH et al Nuclear FAK in endothelium: an intrinsic inhibitor of NF-κB activation in atherosclerosis. Atherosclerosis. (2023) 379:117189. 10.1016/j.atherosclerosis.2023.117189
139.
Li L Zhang H Chen C Huang H Tan X Wei Z et al A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding. Proc Natl Acad Sci U S A. (2021) 118(11):e2016673118. 10.1073/pnas.2016673118
140.
Aron L Qiu C Ngian ZK Liang M Drake D Choi J et al A neurodegeneration checkpoint mediated by REST protects against the onset of Alzheimer's Disease. Nat Commun. (2023) 14(1):7030. 10.1038/s41467-023-42704-6
141.
Kuznetsova T Prange KHM Glass CK de Winther MPJ . Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat Rev Cardiol. (2020) 17(4):216–28. 10.1038/s41569-019-0265-3
142.
Zeltzer S Zeltzer CA Igarashi S Wilson J Donaldson JG Goodrum F . Virus control of trafficking from sorting endosomes. mBio. (2018) 9(4):e00683–18. 10.1128/mBio.00683-18
143.
Zhang H Huang T Hong Y Hong Y Yang W Zhang X et al The retromer Complex and sorting nexins in neurodegenerative diseases. Front Aging Neurosci. (2018) 10:79. 10.3389/fnagi.2018.00079
144.
Vos D Kuivenhoven JA van de Sluis B . Recycling the LDL receptor to combat atherosclerosis. Aging. (2018) 10(12):3638–40. 10.18632/aging.101681
145.
Ko YP Flick MJ . Fibrinogen is at the interface of host defense and pathogen virulence in Staphylococcus aureus infection. Semin Thromb Hemost. (2016) 42(4):408–21. 10.1055/s-0036-1579635
146.
Cortes-Canteli M Zamolodchikov D Ahn HJ Strickland S Norris EH . Fibrinogen and altered hemostasis in Alzheimer's Disease. J Alzheimers Dis. (2012) 32(3):599–608. 10.3233/JAD-2012-120820
147.
Surma S Banach M . Fibrinogen and atherosclerotic cardiovascular diseases-review of the literature and clinical studies. Int J Mol Sci. (2021) 23(1):193. 10.3390/ijms23010193
148.
Elgueta R Benson MJ de Vries VC Wasiuk A Guo Y Noelle RJ . Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. (2009) 229(1):152–72. 10.1111/j.1600-065X.2009.00782.x
149.
Giunta B Rezai-Zadeh K Tan J . Impact of the CD40-CD40L dyad in Alzheimer's Disease. CNS Neurol Disord Drug Targets. (2010) 9(2):149–55. 10.2174/187152710791012099
150.
Lutgens E Lievens D Beckers L Donners M Daemen M . CD40 And its ligand in atherosclerosis. Trends Cardiovasc Med. (2007) 17(4):118–23. 10.1016/j.tcm.2007.02.004
151.
Ng J Hart CM Morgan K Simon JA . A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. (2000) 20(9):3069–78. 10.1128/MCB.20.9.3069-3078.2000
152.
Li T . The functions of polycomb group proteins in T cells. Cell Insight. (2022) 1(5):100048. 10.1016/j.cellin.2022.100048
153.
Liu T Zhu B Liu Y Zhang X Yin J Li X et al Multi-omic comparison of Alzheimer's Variants in human ESC-derived microglia reveals convergence at APOE. J Exp Med. (2020) 217(12):e20200474. 10.1084/jem.20200474
154.
Libby P Buring JE Badimon L Hansson GK Deanfield J Bittencourt MS et al Atherosclerosis. Nat Rev Dis Primers. (2019) 5(1):56. 10.1038/s41572-019-0106-z
155.
Ljungberg JK Kling JC Tran TT Blumenthal A . Functions of the WNT signaling network in shaping host responses to infection. Front Immunol. (2019) 10:2521. 10.3389/fimmu.2019.02521
156.
Palomer E Buechler J Salinas PC . Wnt signaling deregulation in the aging and Alzheimer's Brain. Front Cell Neurosci. (2019) 13:227. 10.3389/fncel.2019.00227
157.
Afroz R Goodwin JE . Wnt signaling in atherosclerosis: mechanisms to therapeutic implications. Biomedicines. (2024) 12(2):276. 10.3390/biomedicines12020276
158.
Strong M . Protein nanomachines. PLoS Biol. (2004) 2(3):E73. 10.1371/journal.pbio.0020073
159.
Xu J Li CX Li YS Lv JY Ma Y Shao TT et al MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. (2011) 39(3):825–36. 10.1093/nar/gkq832
160.
Alberts B . The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. (1998) 92(3):291–4. 10.1016/s0092-8674(00)80922-8
161.
Schira-Heinen J Czapla A Hendricks M Kloetgen A Wruck W Adjaye J et al Functional omics analyses reveal only minor effects of microRNAs on human somatic stem cell differentiation. Sci Rep. (2020) 10:3284. 10.1038/s41598-020-60065-8
162.
Ianni M Corraliza-Gomez M Costa-Coelho T Ferreira-Manso M Inteiro-Oliveira S Alemãn-Serrano N et al Spatiotemporal dysregulation of neuron–Glia related genes and pro-/anti-inflammatory miRNAs in the 5xFAD mouse model of Alzheimer's Disease. Int. J. Mol. Sci. (2024) 25:9475. 10.3390/ijms25179475
163.
Perri P Ponzoni M Corrias MV Ceccherini I Candiani S Bachetti T . A focus on regulatory networks linking MicroRNAs, transcription factors and target genes in neuroblastoma. Cancers (Basel). (2021) 13(21):5528. 10.3390/cancers13215528
164.
Woess C Tuzlak S Labi V Bertele D Schneider P Villunger A . Combined loss of the BH3-only proteins bim and bmf restores B-cell development and function in TACI-ig transgenic mice. Cell Death Differ. (2015) 22(9):1477–88. 10.1038/cdd.2015.8
165.
Sharma A Boise LH Shanmugam M . Cancer metabolism and the evasion of apoptotic cell death. Cancers (Basel). (2019) 11(8):1144. 10.3390/cancers11081144
166.
Solano-Gálvez SG Álvarez-Hernández DA Gutiérrez-Kobeh L Vázquez-López R . Leishmania: manipulation of signaling pathways to inhibit host cell apoptosis. Ther Adv Infect Dis. (2021) 8:20499361211014977. 10.1177/20499361211014977
167.
Sixt BS . Host cell death during infection with Chlamydia: a double-edged sword. FEMS Microbiol Rev. (2021) 45(1):fuaa043. 10.1093/femsre/fuaa043
168.
Campbell GR Spector SA . Current strategies to induce selective killing of HIV-1-infected cells. J Leukoc Biol. (2022) 112(5):1273–84. 10.1002/JLB.4MR0422-636R
169.
Clarke P Tyler KL . Apoptosis in animal models of virus-induced disease. Nat Rev Microbiol. (2009) 7(2):144–55. 10.1038/nrmicro2071
170.
Akhter R Sanphui P Biswas SC . The essential role of p53-up-regulated modulator of apoptosis (puma) and its regulation by FoxO3a transcription factor in β-amyloid-induced neuron death. J Biol Chem. (2014) 289(15):10812–22. 10.1074/jbc.M113.519355
171.
Biswas SC Shi Y Vonsattel JP Leung CL Troy CM Greene LA . Bim is elevated in Alzheimer's Disease neurons and is required for beta-amyloid-induced neuronal apoptosis. J Neurosci. (2007) 27(4):893–900. 10.1523/JNEUROSCI.3524-06.2007
172.
Zhaorigetu S Yang Z Toma I McCaffrey TA Hu CA . Apolipoprotein L6, induced in atherosclerotic lesions, promotes apoptosis and blocks beclin 1-dependent autophagy in atherosclerotic cells. J Biol Chem. (2011) 286(31):27389–98. 10.1074/jbc.M110.210245
173.
Delbridge ARD Strasser A . The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. (2015) 22:1071–80. 10.1038/cdd.2015.50
174.
Cao J Huang M Guo L et al MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer's Disease pathogenesis. Mol Psychiatry. (2021) 26(9):4687–701. 10.1038/s41380-020-0824-3
175.
Chen X Jiang XM Zhao LJ Sun LL Yan ML Tian Y et al MicroRNA-195 prevents dendritic degeneration and neuron death in rats following chronic brain hypoperfusion. Cell Death Dis. (2017) 8:e2850. 10.1038/cddis.2017.243
176.
Lerner M Haneklaus M Harada M Grandér D . MiR-200c regulates noxa expression and sensitivity to proteasomal inhibitors. PLoS One. (2012) 7(5):e36490. 10.1371/journal.pone.0036490
177.
Magenta A Cencioni C Fasanaro P Zaccagnini G Greco S Sarra-Ferraris G et al miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. (2011) 18(10):1628–39. 10.1038/cdd.2011.42
178.
Wu Q Ye X Xiong Y Zhu H Miao J Zhang W et al The protective role of microRNA-200c in Alzheimer's Disease pathologies is induced by Beta amyloid-triggered endoplasmic Reticulum stress. Front Mol Neurosci. (2016) 9:140. 10.3389/fnmol.2016.00140
179.
Tian L Li M Ge J Guo Y Sun Y Liu M et al MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. Tumour Biol. (2014) 35(6):5953–63. 10.1007/s13277-014-1790-7
180.
Li S Li L Li J Liang X Song C Zou Y . miR-203, fine-tunning neuroinflammation by juggling different components of NF-κB signaling. J Neuroinflammation. (2022) 19:84. 10.1186/s12974-022-02451-9
181.
Jia Z Zhang Y Xu Q Guo W Guo A . miR-126 suppresses epithelial-to-mesenchymal transition and metastasis by targeting PI3K/AKT/snail signaling of lung cancer cells. Oncol Lett. (2018) 15(5):7369–75. 10.3892/ol.2018.8207
182.
Xue B Qu Y Zhang X Xu XF . miRNA-126a-3p participates in hippocampal memory via Alzheimer's disease-related proteins. Cereb Cortex. (2022) 32(21):4763–81. 10.1093/cercor/bhab515
183.
Chockley PJ Chen J Chen G Beer DG Standiford TJ Keshamouni VG . Epithelial-mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer. J Clin Invest. (2018) 128(4):1384–96. 10.1172/JCI97611
184.
Datar I Schalper KA . Epithelial-mesenchymal transition and immune evasion during lung cancer progression: the chicken or the egg?Clin Cancer Res. (2016) 22(14):3422–4. 10.1158/1078-0432.CCR-16-0336
185.
Santos F Moreira C Nóbrega-Pereira S Bernardes de Jesus B . New insights into the role of epithelial− mesenchymal transition during aging. Int J Mol Sci. (2019) 20(4):891. 10.3390/ijms20040891
186.
Huang Q Gan Y Yu Z Wu H Zhong Z . Endothelial to mesenchymal transition: an insight in atherosclerosis. Front Cardiovasc Med. (2021) 8:734550. 10.3389/fcvm.2021.734550
187.
Wang N Wei L Huang Y Wu Y Su M Pang X et al Mir520c blocks EMT progression of human breast cancer cells by repressing STAT3. Oncol Rep. (2017) 37(3):1537–44. 10.3892/or.2017.5393
188.
Liu Y Wang J Chen J Wu S Zeng X Xiong Q et al Upregulation of miR-520c-3p via hepatitis B virus drives hepatocellular migration and invasion by the PTEN/AKT/NF-κB axis. Mol Ther Nucleic Acids. (2022) 29:47–63. 10.1016/j.omtn.2022.05.031
189.
Chen D Sun Y Yuan Y Han Z Zhang P Zhang J et al miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genet. (2014) 10(2):e1004177. 10.1371/journal.pgen.1004177
190.
Li H Mou Q Li P Yang Z Wang Z Niu J et al MiR-486-5p inhibits IL-22-induced epithelial-mesenchymal transition of breast cancer cell by repressing Dock1. J Cancer. (2019) 10(19):4695–706. 10.7150/jca.30596
191.
Li CY Wang YH Lin ZY et al MiR-5100 targets TOB2 to drive epithelial-mesenchymal transition associated with activating smad2/3 in lung epithelial cells. Am J Transl Res. (2017) 9(10):4694–706.
192.
Karere GM Glenn JP Li G Konar A VandeBerg JL Cox LA . Potential miRNA biomarkers and therapeutic targets for early atherosclerotic lesions. Sci Rep. (2023) 13(1):3467. 10.1038/s41598-023-29074-1
193.
Li C Liu HL Zhou YM Shi YC Zhang ZB Chen L et al Astrocyte elevated gene-1 serves as a target of miR542 to promote glioblastoma proliferation and invasion. Chin Med J. (2020) 133(20):2437–43. 10.1097/CM9.0000000000001072
194.
Yu B Jiang Y Wang X Wang S . An integrated hypothesis for miR-126 in vascular disease. Med Res Arch. (2020) 8(5):2133. 10.18103/mra.v8i5.2133
195.
Theofilis P Oikonomou E Vogiatzi G et al The role of MicroRNA-126 in atherosclerotic cardiovascular diseases. Curr Med Chem. (2023) 30(17):1902–21. 10.2174/0929867329666220830100530
196.
Li Z Zhao Y Suguro S Suguro R . MicroRNAs regulate function in atherosclerosis and clinical implications. Oxid Med Cell Longev. (2023) 2023:2561509. 10.1155/2023/2561509
197.
Giuliani A Gaetani S Sorgentoni G et al Circulating inflamma-miRs as potential biomarkers of cognitive impairment in patients affected by Alzheimer's Disease. Front Aging Neurosci. (2021) 13:647015. 10.3389/fnagi.2021.647015
198.
Kou X Chen D Chen N . The regulation of microRNAs in Alzheimer's Disease. Front Neurol. (2020) 11:288. 10.3389/fneur.2020.00288
199.
Peña-Bautista C Tarazona-Sánchez A Braza-Boils A Balaguer A Ferré-González L Cañada-Martínez AJ et al Plasma microRNAs as potential biomarkers in early Alzheimer disease expression. Sci Rep. (2022) 12(1):15589. 10.1038/s41598-022-19862-6
200.
Mussbacher M Derler M Basílio J Schmid JA . NF-κB in monocytes and macrophages—an inflammatory master regulator in multitalented immune cells. Front Immunol. (2023) 14:1134661. 10.3389/fimmu.2023.1134661
201.
Howe EN Cochrane DR Cittelly DM Richer JK . miR-200c targets a NF-κB up-regulated TrkB/NTF3 autocrine signaling loop to enhance anoikis sensitivity in triple negative breast cancer. PLoS One. (2012) 7(11):e49987. 10.1371/journal.pone.0049987
202.
Williams MM Christenson JL O'Neill KI Hafeez SA Ihle CL Spoelstra NS et al MicroRNA-200c restoration reveals a cytokine profile to enhance M1 macrophage polarization in breast cancer. NPJ Breast Cancer. (2021) 7(1):64. 10.1038/s41523-021-00273-1
203.
Wu G Zhang D Yang L Wu Q Yuan L . MicroRNA-200c-5p targets NIMA related kinase 7 (NEK7) to inhibit NOD-like receptor 3 (NLRP3) inflammasome activation, MODE-K cell pyroptosis, and inflammatory bowel disease in mice. Mol Immunol. (2022) 146:57–68. 10.1016/j.molimm.2022.03.121
204.
Zhao L Liu X Yang J Wang X Liu X Wu J et al MiR-200c-3p inhibits LPS-induced M1 polarization of BV2 cells by targeting RIP2. Genes Genomics. (2022) 44(4):477–86. 10.1007/s13258-021-01210-z
205.
Li S Li L Li J Liang X Song C Zou Y . miR-203, fine-tunning neuroinflammation by juggling different components of NF-κB signaling. J Neuroinflammation. (2022) 19(1):84. 10.1186/s12974-022-02451-9
206.
Chen J Liu Z Sun H Liu M Wang J Zheng C et al MiR-203a-3p attenuates apoptosis and pyroptosis of chondrocytes by regulating the MYD88/NF-κB pathway to alleviate osteoarthritis progression. Aging. (2023) 15(23):14457–72. 10.18632/aging.205373
207.
Zhang LS Chen QC Zong HT Xia Q . Exosome miRNA-203 promotes M1 macrophage polarization and inhibits prostate cancer tumor progression. Mol Cell Biochem. (2024) 479(9):2459–70. 10.1007/s11010-023-04854-5
208.
Chen T Shou L Guo X Wei M Zheng H Tao T . Magnolol attenuates the locomotor impairment, cognitive deficit, and neuroinflammation in Alzheimer's Disease mice with brain insulin resistance via up-regulating miR-200c. Bioengineered. (2022) 13(1):531–43. 10.1080/21655979.2021.2009975
209.
Park H Lee YB Chang KA . miR-200c suppression increases tau hyperphosphorylation by targeting 14-3-3γ in early stage of 5xFAD mouse model of Alzheimer's Disease. Int J Biol Sci. (2022) 18(5):2220–34. 10.7150/ijbs.66604
210.
Yang Z Zhong L Zhong S Xian R Yuan B . miR-203 protects microglia mediated brain injury by regulating inflammatory responses via feedback to MyD88 in ischemia. Mol Immunol. (2015) 65(2):293–301. 10.1016/j.molimm.2015.01.019
211.
Sun E Motolani A Campos L Lu T . The pivotal role of NF-kB in the pathogenesis and therapeutics of Alzheimer's Disease. Int J Mol Sci. (2022) 23(16):8972. 10.3390/ijms23168972
212.
Duff EP Zetterberg H Heslegrave A Dehghan A Elliott P Allen N et al Plasma proteomic evidence for increased β-amyloid pathology after SARS-CoV-2 infection. Nat Med. (2025) 30:797–806. 10.1038/s41591-024-03426-4
213.
Guo S Wang H Yin Y . Microglia polarization from M1 to M2 in neurodegenerative diseases. Front Aging Neurosci. (2022) 14:815347. 10.3389/fnagi.2022.815347
214.
Rabb H . The T cell as a bridge between innate and adaptive immune systems: implications for the kidney. Kidney Int. (2002) 61(6):1935–46. 10.1046/j.1523-1755.2002.00378.x
215.
West EE Merle NS Kamiński MM Palacios G Kumar D Wang L et al Loss of CD4+ T cell-intrinsic arginase 1 accelerates Th1 response kinetics and reduces lung pathology during influenza infection. Immunity. (2023) 56(9):2036–2053.e12. 10.1016/j.immuni.2023.07.014
216.
Jeker LT Bluestone JA . MicroRNA regulation of T-cell differentiation and function. Immunol Rev. (2013) 253(1):65–81. 10.1111/imr.12061
217.
Alivernini S Gremese E McSharry C Tolusso B Ferraccioli G McInnes IB . MicroRNA-155-at the critical interface of innate and adaptive immunity in arthritis. Front Immunol. (2018) 8:1932. 10.3389/fimmu.2017.01932
218.
Yuan X Berg N Lee JW Le TT Neudecker V Jing N et al MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol. (2018) 104(3):515–24. 10.1002/JLB.3MR0218-079R
219.
Bezman NA Chakraborty T Bender T Lanier LL . miR-150 regulates the development of NK and iNKT cells. J Exp Med. (2011) 208(13):2717–31. 10.1084/jem.20111386
220.
Berrien-Elliott MM Sun Y Neal C Ireland A Trissal MC Sullivan RP et al MicroRNA-142 is critical for the homeostasis and function of type 1 innate lymphoid cells. Immunity. (2019) 51(3):479–90.e6. 10.1016/j.immuni.2019.06.016
221.
Rodrigues V Cordeiro-da-Silva A Laforge M Ouaissi A Akharid K Silvestre R et al Impairment of T cell function in parasitic infections. PLoS Negl Trop Dis. (2014) 8(2):e2567. 10.1371/journal.pntd.0002567
222.
Guo L Li X Gould T Wang ZY Cao W . T cell aging and Alzheimer's Disease. Front Immunol. (2023) 14:1154699. 10.3389/fimmu.2023.1154699
223.
Chen J Xiang X Nie L Guo X Zhang F Wen C et al The emerging role of Th1 cells in atherosclerosis and its implications for therapy. Front Immunol. (2023) 13:1079668. 10.3389/fimmu.2022.1079668
224.
Vonwirth V Bülbül Y Werner A Echchannaoui H Windschmitt J Habermeier A et al Inhibition of arginase 1 liberates potent T cell immunostimulatory activity of human neutrophil granulocytes. Front Immunol. (2021) 11:617699. 10.3389/fimmu.2020.617699
225.
Jin J Yu G . Hypoxic lung cancer cell-derived exosomal miR-21 mediates macrophage M2 polarization and promotes cancer cell proliferation through targeting IRF1. World J Surg Onc. (2022) 20:241. 10.1186/s12957-022-02706-y
226.
Wang M Yu F Li P . Noncoding RNAs as an emerging resistance mechanism to immunotherapies in cancer: basic evidence and therapeutic implications. Front Immunol. (2023) 14:1268745. 10.3389/fimmu.2023.1268745
227.
Chi LH Cross RSN Redvers RP Davis M Hediyeh-zadeh S Mathivanan S et al MicroRNA-21 is immunosuppressive and pro-metastatic via separate mechanisms. Oncogenesis. (2022) 11(1):38. 10.1038/s41389-022-00413-7
228.
Hu GJ Jiang XY Du SY Zhang K Chen Z . miR-107-5p ameliorates neurological damage, oxidative stress, and immune responses in mice with Alzheimer's Disease by suppressing the toll-like receptor 4 (TLR4)/nuclear factor-kappaB (NF-κB) pathway. Kaohsiung J Med Sci. (2024) 40(2):119–30. 10.1002/kjm2.12797
229.
Kim C Hu B Jadhav RR Jin J Zhang H Cavanagh MM et al Activation of miR-21-regulated pathways in immune aging selects against signatures characteristic of memory T cells. Cell Rep. (2018) 25(8):2148–62.e5. 10.1016/j.celrep.2018.10.074
230.
Zhang Z Li F Tian Y Cao L Gao Q Zhang C et al Metformin enhances the antitumor activity of CD8+ T lymphocytes via the AMPK-miR-107-eomes-PD-1 pathway. J Immunol. (2020) 204(9):2575–88. 10.4049/jimmunol.1901213
231.
Ruan Q Wang P Wang T Qi J Wei M Wang S et al MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis. (2014) 5:e1095. 10.1038/cddis.2014.47
232.
Yan J Dai L Yuan J Pang M Wang Y Lin L et al miR-107 inhibits the proliferation of gastric cancer cells in vivo and in vitro by targeting TRIAP1. Front Genet. (2022) 13:855355. 10.3389/fgene.2022.855355
233.
Ma J Wang PY Zhuang J Son AY Karius AK Syed AM et al CHCHD4-TRIAP1 Regulation of innate immune signaling mediates skeletal muscle adaptation to exercise. Cell Rep. (2024) 43(1):113626. 10.1016/j.celrep.2023.113626
234.
Ma H Wang L Sun H Yu Q Yang T Wang Y et al MIR-107/HMGB1/FGF-2 axis responds to excessive mechanical stretch to promote rapid repair of vascular endothelial cells. Arch Biochem Biophys. (2023) 744:109686. 10.1016/j.abb.2023.109686
235.
Wang WX Rajeev BW Stromberg AJ Ren N Tang G Huang Q et al The expression of microRNA miR-107 decreases early in Alzheimer's Disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. (2008) 28(5):1213–23. 10.1523/JNEUROSCI.5065-07.2008
236.
Bai X Bian Z . MicroRNA-21 is a versatile regulator and potential treatment target in central nervous system disorders. Front Mol Neurosci. (2022) 15:842288. 10.3389/fnmol.2022.842288
237.
Kuballa P Nolte WM Castoreno AB Xavier RJ . Autophagy and the immune system. Annu Rev Immunol. (2012) 30:611–46. 10.1146/annurev-immunol-020711-074948
238.
Jiang GM Tan Y Wang H Peng L Chen HT Meng XJ et al The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol Cancer. (2019) 18(1):17. 10.1186/s12943-019-0944-z
239.
Feinberg MW Moore KJ . MicroRNA regulation of atherosclerosis. Circ Res. (2016) 118(4):703–20. 10.1161/CIRCRESAHA.115.306300
240.
Tomasetti M Monaco F Manzella N Rohlena J Rohlenova K Staffolani S et al MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget. (2016) 7(24):36338–52. 10.18632/oncotarget.8916
241.
Ma M Li H Yin S Lin T Song T . Overexpression of miR-92a attenuates kidney ischemia-reperfusion injury and improves kidney preservation by inhibiting MEK4/JNK1-related autophagy. Cell Mol Biol Lett. (2023) 28(1):20. 10.1186/s11658-023-00430-3
242.
Gu H Li L Cui C Zhao Z Song G . Overexpression of let-7a increases neurotoxicity in a PC12 cell model of Alzheimer's Disease via regulating autophagy. Exp Ther Med. (2017) 14(4):3688–98. 10.3892/etm.2017.4977
243.
Bernstein DL Jiang X Rom S . Let-7 microRNAs: their role in cerebral and cardiovascular diseases, inflammation, cancer, and their regulation. Biomedicines. (2021) 9(6):606. 10.3390/biomedicines9060606
244.
Jahankhani K Ahangari F Adcock IM Mortaz E . Possible cancer-causing capacity of COVID-19: is SARS-CoV-2 an oncogenic agent?Biochimie. (2023) 213:130–8. 10.1016/j.biochi.2023.05.014
245.
Bracken CP Scott HS Goodall GJ . A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. (2016) 17(12):719–32. 10.1038/nrg.2016.134
246.
Kuss-Duerkop SK Westrich JA Pyeon D . DNA Tumor virus regulation of host DNA methylation and its implications for immune evasion and oncogenesis. Viruses. (2018) 10(2):82. 10.3390/v10020082
247.
Du J Luo J Yu J Mao X Luo Y Zheng P et al Manipulation of intestinal antiviral innate immunity and immune evasion strategies of porcine epidemic diarrhea virus. Biomed Res Int. (2019) 2019:1862531. 10.1155/2019/1862531
248.
Liang C Lee JS Jung JU . Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin Cancer Biol. (2008) 18(6):423–36. 10.1016/j.semcancer.2008.09.003
249.
Zhao M Liu Z Shao F Zhou W Chen Z Xia P et al Communication pattern changes along with declined IGF1 of immune cells in COVID-19 patients during disease progression. Front Immunol. (2022) 12:729990. 10.3389/fimmu.2021.729990
250.
Calcaterra V Bosoni P Dilillo D Mannarino S Fiori L Fabiano V et al Impaired glucose-insulin metabolism in multisystem inflammatory syndrome related to SARS-CoV-2 in children. Children. (2021) 8(5):384. 10.3390/children8050384
251.
Milic J Barbieri S Gozzi L Brigo A Beghé B Verduri A et al Metabolic-Associated fatty liver disease is highly prevalent in the postacute COVID syndrome. Open Forum Infect Dis. (2022) 9(3):ofac003. 10.1093/ofid/ofac003
252.
Sun X Ma Z Zhao X Jin W Zhang C Ma J et al Three-dimensional bioprinting of multicell-laden scaffolds containing bone morphogenic protein-4 for promoting M2 macrophage polarization and accelerating bone defect repair in diabetes mellitus. Bioact Mater. (2020) 6(3):757–69. 10.1016/j.bioactmat.2020.08.030
253.
Xu J Lai KKY Verlinsky A Lugea A French SW Cooper MP et al Synergistic steatohepatitis by moderate obesity and alcohol in mice despite increased adiponectin and p-AMPK. J Hepatol. (2011) 55(3):673–82. 10.1016/j.jhep.2010.12.034
254.
Harris BHL Macaulay VM Harris DA Klenerman P Karpe F Lord SR et al Obesity: a perfect storm for carcinogenesis. Cancer Metastasis Rev. (2022) 41(3):491–515. 10.1007/s10555-022-10046-2
255.
Tsampasian V Bäck M Bernardi M Cavarretta E Dębski M Gati S et al Cardiovascular disease as part of long COVID: a systematic review. Eur J Prev Cardiol. (2025) 32:485–98. 10.1093/eurjpc/zwae070
256.
Khavinson V Linkova N Dyatlova A Kantemirova R Kozlov K . Senescence-associated secretory phenotype of cardiovascular system cells and inflammaging: perspectives of peptide regulation. Cells. (2022) 12(1):106. 10.3390/cells12010106
257.
Vakka A Warren JS Drosatos K . Cardiovascular aging: from cellular and molecular changes to therapeutic interventions. J Cardiovasc Aging. (2023) 3(3):23. 10.20517/jca.2023.09
258.
Silva MJA Ribeiro LR Gouveia MIM Marcelino BDR Santos CSD Lima KVB et al Hyperinflammatory response in COVID-19: a systematic review. Viruses. (2023) 15(2):553. 10.3390/v15020553
259.
Degboé Y Poupot R Poupot M . Repolarization of unbalanced macrophages: unmet medical need in chronic inflammation and cancer. Int J Mol Sci. (2022) 23(3):1496. 10.3390/ijms23031496
260.
Chung HY Kim DH Lee EK Chung KW Chung S Lee B et al Redefining chronic inflammation in aging and age-related diseases: proposal of the senoinflammation concept. Aging Dis. (2019) 10(2):367–82. 10.14336/AD.2018.0324
261.
Ferrucci L Fabbri E . Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol. (2018) 15(9):505–22. 10.1038/s41569-018-0064-2
262.
Tursi A Lopetuso LR Vetrone LM Gasbarrini A Papa A . SARS-CoV-2 infection as a potential trigger factor for de novo occurrence of inflammatory bowel disease. Eur J Gastroenterol Hepatol. (2022) 34(8):883–4. 10.1097/MEG.0000000000002379
263.
Lewis S . The long COVID haul. Nat Rev Neurosci. (2024) 25:4. 10.1038/s41583-023-00774-x
264.
He C Li Z Yang M Yu W Luo R Zhou J et al Non-Coding RNA in microglia activation and neuroinflammation in Alzheimer's Disease. J Inflamm Res. (2023) 16:4165–211. 10.2147/JIR.S422114
265.
Cao X Li W Wang T Ran D Davalos V Planas-Serra L et al Accelerated biological aging in COVID-19 patients. Nat Commun. (2022) 13(1):2135. 10.1038/s41467-022-29801-8
266.
Shin CH Kim KH Jeeva S Kang SM . Towards goals to refine prophylactic and therapeutic strategies against COVID-19 linked to aging and metabolic syndrome. Cells. (2021) 10(6):1412. 10.3390/cells10061412
267.
Jalloh S Olejnik J Berrigan J Nisa A Suder EL Akiyama H et al CD169-mediated Restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses. PLoS Pathog. (2022) 18(10):e1010479. 10.1371/journal.ppat.1010479
268.
Bain CC Lucas CD Rossi AG . Pulmonary macrophages and SARS-Cov2 infection. Int Rev Cell Mol Biol. (2022) 367:1–28. 10.1016/bs.ircmb.2022.01.001
269.
Schmitt CA Tchkonia T Niedernhofer LJ Robbins PD Kirkland JL Lee S . COVID-19 and cellular senescence. Nat Rev Immunol. (2023) 23(4):251–63. 10.1038/s41577-022-00785-2
270.
Bagheri M Nair RR Singh KK Saini DK . ATM-ROS-iNOS axis regulates nitric oxide mediated cellular senescence. Biochim Biophys Acta Mol Cell Res. (2017) 1864(1):177–90. 10.1016/j.bbamcr.2016.11.008
271.
Baz-Martínez M Da Silva-Álvarez S Rodríguez E et al Cell senescence is an antiviral defense mechanism. Sci Rep. (2016) 6:37007. 10.1038/srep37007
272.
Ren JL Pan JS Lu YP Sun P Han J . Inflammatory signaling and cellular senescence. Cell Signal. (2009) 21(3):378–83. 10.1016/j.cellsig.2008.10.011
273.
Saito Y Yamamoto S Chikenji TS . Role of cellular senescence in inflammation and regeneration. Inflamm Regen. (2024) 44(1):28. 10.1186/s41232-024-00342-5
274.
Tsioumpekou M Krijgsman D Leusen JHW Olofsen PA . The role of cytokines in neutrophil development, tissue homing, function and plasticity in health and disease. Cells. (2023) 12(15):1981. 10.3390/cells12151981
275.
Pezeshki PS Rezaei N . Immune checkpoint inhibition in COVID-19: risks and benefits. Expert Opin Biol Ther. (2021) 21(9):1173–9. 10.1080/14712598.2021.1887131
276.
Jain SS Burton Sojo G Sun H Friedland BN McNamara ME Schmidt MO et al The role of aging and senescence in immune checkpoint inhibitor response and toxicity. Int J Mol Sci. (2024) 25(13):7013. 10.3390/ijms25137013
277.
Burton DGA Stolzing A . Cellular senescence: immunosurveillance and future immunotherapy. Ageing Res Rev. (2018) 43:17–25. 10.1016/j.arr.2018.02.001
278.
Jubel JM Barbati ZR Burger C Wirtz DC Schildberg FA . The role of PD-1 in acute and chronic infection. Front Immunol. (2020) 11:487. 10.3389/fimmu.2020.00487
279.
Palestra F Poto R Ciardi R Opromolla G Secondo A Tedeschi V et al SARS-CoV-2 spike protein activates human lung macrophages. Int J Mol Sci. (2023) 24(3):3036. 10.3390/ijms24033036
280.
Mirzaei H Faghihloo E . Viruses as key modulators of the TGF-β pathway; a double-edged sword involved in cancer. Rev Med Virol. (2018) 28(2):e1967. 10.1002/rmv.1967
281.
Reece MD Taylor RR Song C Gavegnano C . Targeting macrophage dysregulation for viral infections: novel targets for immunomodulators. Front Immunol. (2021) 12:768695. 10.3389/fimmu.2021.768695
282.
Arguinchona LM Zagona-Prizio C Joyce ME Chan ED Maloney JP . Microvascular significance of TGF-β axis activation in COVID-19. Front Cardiovasc Med. (2023) 9:1054690. 10.3389/fcvm.2022.1054690
283.
Tominaga K Suzuki HI . TGF-β Signaling in cellular senescence and aging-related pathology. Int J Mol Sci. (2019) 20:5002. 10.3390/ijms20205002
284.
Onorati A Havas AP Lin B Rajagopal J Sen P Adams PD et al Upregulation of PD-L1 in senescence and aging. Mol Cell Biol. (2022) 42(10):e0017122. 10.1128/mcb.00171-22
285.
Kell DB Laubscher GJ Pretorius E . A central role for amyloid fibrin microclots in long COVID/PASC: origins and therapeutic implications. Biochem J. (2022) 479(4):537–59. 10.1042/BCJ20220016
286.
Fahy RJ Lichtenberger F McKeegan CB Nuovo GJ Marsh CB Wewers MD . The acute respiratory distress syndrome: a role for transforming growth factor-beta 1. Am J Respir Cell Mol Biol. (2003) 28(4):499–503. 10.1165/rcmb.2002-0092OC
287.
Tran S Ksajikian A Overbey J Li P Li Y . Pathophysiology of pulmonary fibrosis in the context of COVID-19 and implications for treatment: a narrative review. Cells. (2022) 11(16):2489. 10.3390/cells11162489
288.
Wu TT Travaglini KJ Rustagi A Xu D Zhang Y Andronov L et al Interstitial macrophages are a focus of viral takeover and inflammation in COVID-19 initiation in human lung. J Exp Med. (2024) 221(6):e20232192. 10.1084/jem.20232192
289.
Tao L Zhu Y Wu L Liu J . Comprehensive analysis of senescence-associated genes in sepsis based on bulk and single-cell sequencing data. Front Mol Biosci. (2024) 10:1322221. 10.3389/fmolb.2023.1322221
290.
Marchi R Sugita B Centa A Fonseca AS Bortoletto S Fiorentin K et al The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review. Infect Genet Evol. (2021) 91:104832. 10.1016/j.meegid.2021.104832
291.
Hejenkowska ED Mitash N Donovan JE Chandra A Bertrand C De Santi C et al TGF-β1 Inhibition of ACE2 mediated by miRNA uncovers novel mechanism of SARS-CoV-2 pathogenesis. J Innate Immun. (2023) 15(1):629–46. 10.1159/000533606
292.
Abdoli A Taghipour A Jahromi MAM Eftekharian F Sahraei R Sanie MS . Latent viral infections as neglected risk factors for long COVID. Lancet Glob Health. (2024) 12(2):e197. 10.1016/S2214-109X(24)00010-X
293.
Zhuo C Tian H Song X Jiang D Chen G Cai Z et al Microglia and cognitive impairment in schizophrenia: translating scientific progress into novel therapeutic interventions. Schizophrenia. (2023) 9(1):42. 10.1038/s41537-023-00370-z
294.
Low RN Low RJ Akrami A . A review of cytokine-based pathophysiology of long COVID symptoms. Front Med. (2023) 10:1011936. 10.3389/fmed.2023.1011936
295.
Dhawan M Rabaan AA Alwarthan S Alhajri M Halwani MA Alshengeti A et al Regulatory T cells (tregs) and COVID-19: unveiling the mechanisms, and therapeutic potentialities with a special focus on long COVID. Vaccines (Basel). (2023) 11(3):699. 10.3390/vaccines11030699
296.
Boretti A . mRNA vaccine boosters and impaired immune system response in immune compromised individuals: a narrative review. Clin Exp Med. (2024) 24(1):23. 10.1007/s10238-023-01264-1
297.
Briquez PS Rouhani SJ Yu J Pyzer AR Trujillo J Dugan HL et al SARS-CoV-2 infection induces cross-reactive autoantibodies against angiotensin II. Preprint. medRxiv. (2021) 2021:21265789. 10.1101/2021.11.02.21265789
298.
Yin K Peluso MJ Luo X Thomas R Shin MG Neidleman J et al Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2. Nat Immunol. (2024) 25(2):218–25. 10.1038/s41590-023-01724-6
299.
Cohen IR Efroni S . The immune system computes the state of the body: crowd wisdom, machine learning, and immune cell reference repertoires help manage inflammation. Front Immunol. (2019) 10:10. 10.3389/fimmu.2019.00010
300.
Yang J Xu J Wang W Zhang B Yu X Shi S . Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. (2023) 8(1):210. 10.1038/s41392-023-01480-x
301.
Dutta S Das N Mukherjee P . Picking up a fight: fine tuning mitochondrial innate immune defenses against RNA viruses. Front Microbiol. (2020) 11:1990. 10.3389/fmicb.2020.01990
302.
Gurtan AM Sharp PA . The role of miRNAs in regulating gene expression networks. J Mol Biol. (2013) 425(19):3582–600. 10.1016/j.jmb.2013.03.007
303.
Arif KMT Elliott EK Haupt LM Griffiths LR . Regulatory mechanisms of epigenetic miRNA relationships in human cancer and potential as therapeutic targets. Cancers (Basel). (2020) 12(10):2922. 10.3390/cancers12102922
304.
van den Berg NI Machado D Santos S Rocha I Chacón J Harcombe W et al Ecological modelling approaches for predicting emergent properties in microbial communities. Nat Ecol Evol. (2022) 6(7):855–65. 10.1038/s41559-022-01746-7
305.
Liu J Fu Z Song Y Ma R Zhao Z . How to improve the effectiveness of the cooperation networks of emergency science communication for public health emergencies. Humanit Soc Sci Commun. (2024) 11:1449. 10.1057/s41599-024-03996-1
306.
Carlberg C Velleuer E . Vitamin D and aging: central role of immunocompetence. Nutrients. (2024) 16(3):398. 10.3390/nu16030398
307.
Fetahu IS Höbaus J Kállay E . Vitamin D and the epigenome. Front Physiol. (2014) 5:164. 10.3389/fphys.2014.00164
308.
Seraphin G Rieger S Hewison M Capobianco E Lisse TS . The impact of vitamin D on cancer: a mini review. J Steroid Biochem Mol Biol. (2023) 231:106308. 10.1016/j.jsbmb.2023.106308
309.
Gaytán-Pacheco N Ibáñez-Salazar A Herrera-Van Oostdam AS Oropeza-Valdez JJ Magaña-Aquino M Adrián López J et al miR-146a, miR-221, and miR-155 are involved in inflammatory immune response in severe COVID-19 patients. Diagnostics. (2022) 13(1):133. 10.3390/diagnostics13010133
310.
Khatami A Taghizadieh M Sadri Nahand J Karimzadeh M Kiani SJ Khanaliha K et al Evaluation of MicroRNA expression pattern (miR-28, miR-181a, miR-34a, and miR-31) in patients with COVID-19 admitted to ICU and diabetic COVID-19 patients. Intervirology. (2023) 66(1):63–76. 10.1159/000529985
311.
Bautista-Becerril B Nava-Quiroz KJ Muñoz-Soria E Camarena Á Fricke-Galindo I Buendia-Roldan I et al High expression levels of miR-21-5p in younger hospitalized COVID-19 patients are associated with mortality and critical disease. Int J Mol Sci. (2023) 24(12):10112. 10.3390/ijms241210112
312.
Castillo JA Urcuqui-Inchima S . Vitamin D modulates inflammatory response of DENV-2-infected macrophages by inhibiting the expression of inflammatory-liked miRNAs. Pathog Glob Health. (2023) 117(2):167–80. 10.1080/20477724.2022.2101840
313.
Karkeni E Bonnet L Marcotorchino J Tourniaire F Astier J Ye J et al Vitamin D limits inflammation-linked microRNA expression in adipocytes in vitro and in vivo: a new mechanism for the regulation of inflammation by vitamin D. Epigenetics. (2018) 13(2):156–62. 10.1080/15592294.2016.1276681
314.
Ahmad S Ali S Mohan A Syed MA . Late breaking abstract—vitamin D suppresses LPS-induced ER stress and inflammation via modulation of mir-34a/Sirt1 axis in acute lung injury. Eur Respir J. (2019) 54(suppl 63):PA4107. 10.1183/13993003.congress-2019.PA4107
315.
Hernández-Díazcouder A Romero-Nava R Del-Río-Navarro BE Sánchez-Muñoz F Guzmán-Martín CA Reyes-Noriega N et al The roles of MicroRNAs in asthma and emerging insights into the effects of vitamin D3 supplementation. Nutrients. (2024) 16(3):341. 10.3390/nu16030341
316.
Shah K Varna VP Sharma U Mavalankar D . Does vitamin D supplementation reduce COVID-19 severity?: a systematic review. QJM. (2022) 115(10):665–72. 10.1093/qjmed/hcac040
317.
Gerdes EO W Vanichkachorn G Verdoorn BP Hanson GJ Joshi AY Murad MH et al Role of senescence in the chronic health consequences of COVID-19. Transl Res. (2022) 241:96–108. 10.1016/j.trsl.2021.10.003
318.
Yin Q Tang TT Lu XY Ni WJ Yin D Zhang YL et al Macrophage-derived exosomes promote telomere fragility and senescence in tubular epithelial cells by delivering miR-155. Cell Commun Signal. (2024) 22(1):357. 10.1186/s12964-024-01708-5
319.
Nunes ADC Weigl M Schneider A Noureddine S Yu L Lahde C et al miR-146a-5p modulates cellular senescence and apoptosis in visceral adipose tissue of long-lived Ames dwarf mice and in cultured pre-adipocytes. Geroscience. (2022) 44(1):503–18. 10.1007/s11357-021-00490-3
320.
Zuccolo E Badi I Scavello F Gambuzza I Mancinelli L Macrì F et al The microRNA-34a-induced senescence-associated secretory phenotype (SASP) favors vascular smooth muscle cells calcification. Int J Mol Sci. (2020) 21(12):4454. 10.3390/ijms21124454
321.
Zhang Y Sun Q Li X Ma X Li Y Jiao Z et al Apigenin suppresses mouse peritoneal fibrosis by down-regulating miR34a expression. Biomed Pharmacother. (2018) 106:373–80. 10.1016/j.biopha.2018.06.138
322.
Zhu S Deng S Ma Q Zhang T Jia C Zhuo D et al MicroRNA-10A* and MicroRNA-21 modulate endothelial progenitor cell senescence via suppressing high-mobility group A2. Circ Res. (2013) 112(1):152–64. 10.1161/CIRCRESAHA.112.280016
323.
Hwang HV Tran DT Rebuffatti MN Li CS Knowlton AA . Investigation of quercetin and hyperoside as senolytics in adult human endothelial cells. PLoS One. (2018) 13(1):e0190374. 10.1371/journal.pone.0190374
324.
Perrott KM Wiley CD Desprez PY Campisi J . Apigenin suppresses the senescence-associated secretory phenotype and paracrine effects on breast cancer cells. Geroscience. (2017) 39(2):161–73. 10.1007/s11357-017-9970-1
325.
Chaib S Tchkonia T Kirkland JL . Cellular senescence and senolytics: the path to the clinic. Nat Med. (2022) 28(8):1556–68. 10.1038/s41591-022-01923-y
326.
Ali D Okla M Abuelreich S Vishnubalaji R Ditzel N Hamam R et al Apigenin and rutaecarpine reduce the burden of cellular senescence in bone marrow stromal stem cells. Front Endocrinol. (2024) 15:1360054. 10.3389/fendo.2024.1360054
327.
Boesch-Saadatmandi C Loboda A Wagner AE Stachurska A Jozkowicz A Dulak J et al Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. (2011) 22(3):293–9. 10.1016/j.jnutbio.2010.02.008
328.
Arango D Diosa-Toro M Rojas-Hernandez LS Cooperstone JL Schwartz SJ Mo X et al Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol Nutr Food Res. (2015) 59(4):763–72. 10.1002/mnfr.201400705
329.
Chuammitri P Srikok S Saipinta D Boonyayatra S . The effects of quercetin on microRNA and inflammatory gene expression in lipopolysaccharide-stimulated bovine neutrophils. Vet World. (2017) 10(4):403–10. 10.14202/vetworld.2017.403-410
330.
Chumsakul O Wakayama K Tsuhako A Baba Y Takai Y Kurose T et al Apigenin regulates activation of microglia and counteracts retinal degeneration. J Ocul Pharmacol Ther. (2020) 36(5):311–9. 10.1089/jop.2019.0163
331.
Kim M Jee SC Shin MK Han DH Bu KB Lee SC et al Quercetin and isorhamnetin reduce benzo[a]pyrene-induced genotoxicity by inducing RAD51 expression through downregulation of miR-34a. Int J Mol Sci. (2022) 23(21):13125. 10.3390/ijms232113125
332.
Nozari E Moradi A Samadi M . Effect of atorvastatin, curcumin, and quercetin on miR-21 and miR-122 and their correlation with TGFβ1 expression in experimental liver fibrosis. Life Sci. (2020) 259:118293. 10.1016/j.lfs.2020.118293
333.
Di Pierro F Khan A Iqtadar S Mumtaz SU Chaudhry MNA Bertuccioli A et al Quercetin as a possible complementary agent for early-stage COVID-19: concluding results of a randomized clinical trial. Front Pharmacol. (2023) 13:1096853. 10.3389/fphar.2022.1096853
334.
Tirado-Kulieva VA Hernández-Martínez E Choque-Rivera TJ . Phenolic compounds versus SARS-CoV-2: an update on the main findings against COVID-19. Heliyon. (2022) 8(9):e10702. 10.1016/j.heliyon.2022.e10702
335.
Sacchi A Grassi G Notari S Gili S Bordoni V Tartaglia E et al Expansion of myeloid derived suppressor cells contributes to platelet activation by L-arginine deprivation during SARS-CoV-2 infection. Cells. (2021) 10(8):2111. 10.3390/cells10082111
336.
Rodriguez PC Zea AH DeSalvo J Culotta KS Zabaleta J Quiceno DG et al L-arginine consumption by macrophages modulates the expression of CD3ζ chain in T lymphocytes. J Immunol. (2003) 171(3):1232–9. 10.4049/jimmunol.171.3.1232
337.
Yin K Peluso MJ Luo X Thomas R Shin MG Neidleman J et al Long COVID manifests with T cell dysregulation, inflammation, and an uncoordinated adaptive immune response to SARS-CoV-2. bioRxiv [Preprint]. (2023):2023.02.09.527892. 10.1101/2023.02.09.527892
338.
Fleissner F Jazbutyte V Fiedler J Gupta SK Yin X Xu Q et al Short communication: asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ Res. (2010) 107(1):138–43. 10.1161/CIRCRESAHA.110.216770
339.
Adebayo A Varzideh F Wilson S Gambardella J Eacobacci M Jankauskas SS et al l-Arginine and COVID-19: an update. Nutrients. (2021) 13(11):3951. 10.3390/nu13113951
340.
Fong LY Huebner K Jing R Smalley KJ Brydges CR Fiehn O et al Zinc treatment reverses and anti-zn-regulated miRs suppress esophageal carcinomas in vivo. Proc Natl Acad Sci U S A. (2023) 120(20):e2220334120. 10.1073/pnas.2220334120
341.
Hung LW Liu MY Yu T Hung KC Tsai YW Lai CC et al Zinc deficiency and post-acute outcomes in patients with COVID-19: a six-month retrospective cohort analysis of 3,726 patients. Cureus. (2024) 16(10):e71609. 10.7759/cureus.71609
342.
Chen KY Lin CK Chen NH . Effects of vitamin D and zinc deficiency in acute and long COVID syndrome. J Trace Elem Med Biol. (2023) 80:127278. 10.1016/j.jtemb.2023.127278
343.
Kulik L Maywald M Kloubert V Wessels I Rink L . Zinc deficiency drives Th17 polarization and promotes loss of treg cell function. J Nutr Biochem. (2019) 63:11–8. 10.1016/j.jnutbio.2018.09.011
344.
Hönscheid A Rink L Haase H . T-lymphocytes: a target for stimulatory and inhibitory effects of zinc ions. Endocr Metab Immune Disord Drug Targets. (2009) 9(2):132–44. 10.2174/187153009788452390
345.
Fong LY Taccioli C Jing R Smalley KJ Alder H Jiang Y et al MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency. Oncotarget. (2016) 7(10):10723–38. 10.18632/oncotarget.7561
346.
Abdallah S B Mhalla Y Trabelsi I et al Twice-Daily oral zinc in the treatment of patients with coronavirus disease 2019: a randomized double-blind controlled trial [published correction appears in Clin Infect Dis. 2023 Apr 17;76(8):1532. doi: 10.1093/cid/ciad014]. Clin Infect Dis. (2023) 76(2):185–91. 10.1093/cid/ciac807
347.
Vinceti M Filippini T Fairweather-Tait S . A potential role for zinc to enhance treatment for coronavirus disease 2019 (COVID-19)?Clin Infect Dis. (2023) 76(2):192–3. 10.1093/cid/ciac849
348.
Tandon P Abrams ND Avula LR Carrick DM Chander P Divi RL et al Unraveling links between chronic inflammation and long COVID: workshop report. J Immunol. (2024) 212(4):505–12. 10.4049/jimmunol.2300804
349.
Alrajhi NN . Post-COVID-19 pulmonary fibrosis: an ongoing concern. Ann Thorac Med. (2023) 18(4):173–81. 10.4103/atm.atm_7_23
350.
Witkowski M Tizian C Ferreira-Gomes M Niemeyer D Jones TC Heinrich F et al Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature. (2021) 600(7888):295–301. 10.1038/s41586-021-04142-6
351.
Yao C Parimon T Espindola MS et al Maladaptive TGF-β signals to the alveolar epithelium drive fibrosis after COVID-19 infection. Am J Respir Crit Care Med. (2023) 208(2):201–4. 10.1164/rccm.202302-0264LE
352.
Redenšek Trampuž S Vogrinc D Goričar K Dolžan V . Shared miRNA landscapes of COVID-19 and neurodegeneration confirm neuroinflammation as an important overlapping feature. Front Mol Neurosci. (2023) 16:1123955. 10.3389/fnmol.2023.1123955
353.
Li D Wang A Liu X Meisgen F Grünler J Botusan IR et al MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. J Clin Invest. (2015) 125(8):3008–26. 10.1172/JCI79052
354.
Tan S Dai L Tan P Liu W Mu Y Wang J et al Hesperidin administration suppresses the proliferation of lung cancer cells by promoting apoptosis via targeting the miR-132/ZEB2 signalling pathway. Int J Mol Med. (2020) 46(6):2069–77. 10.3892/ijmm.2020.4756
355.
Cannataro R Fazio A La Torre C Caroleo MC Cione E . Polyphenols in the Mediterranean diet: from dietary sources to microRNA modulation. Antioxidants. (2021) 10(2):328. 10.3390/antiox10020328
356.
Zhou Z Kandhare AD Kandhare AA Bodhankar SL . Hesperidin ameliorates bleomycin-induced experimental pulmonary fibrosis via inhibition of TGF-beta1/Smad3/AMPK and IkappaBalpha/NF-kappaB pathways. EXCLI J. (2019) 18:723–45. 10.17179/excli2019-1094
357.
Haggag YA El-Ashmawy NE Okasha KM . Is hesperidin essential for prophylaxis and treatment of COVID-19 infection?Med Hypotheses. (2020) 144:109957. 10.1016/j.mehy.2020.109957
358.
Santa K Kumazawa Y Watanabe K Nagaoka I . The potential use of vitamin D3 and phytochemicals for their anti-ageing effects. Int J Mol Sci. (2024) 25(4):2125. 10.3390/ijms25042125
359.
Mocchegiani E Muzzioli M Santarelli L Fabris N . Restoring effect of oral supplementation of zinc and arginine on thymic endocrine activity and peripheral immune functions in aged mice. Arch Gerontol Geriatr. (1992) 15(Suppl 1):267–75. 10.1016/s0167-4943(05)80026-3
360.
Chakrabortty A Patton DJ Smith BF Agarwal P . miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes (Basel). (2023) 14(7):1375. 10.3390/genes14071375
361.
Duman ET Tuna G Ak E Avsar G Pir P . Optimized network based natural language processing approach to reveal disease comorbidities in COVID-19. Sci Rep. (2024) 14:2325. 10.1038/s41598-024-52819-5
362.
Jin S Zeng X Fang J Lin J Chan SY Erzurum SC et al A network-based approach to uncover microRNA-mediated disease comorbidities and potential pathobiological implications. NPJ Syst Biol Appl. (2019) 5(41). 10.1038/s41540-019-0115-2
363.
Lozano-Velasco E Inácio JM Sousa I Guimarães AR Franco D Moura G et al miRNAs in heart development and disease. Int J Mol Sci. (2024) 25(3):1673. 10.3390/ijms25031673
364.
Lei H Liu W Si J Wang J Zhang T . Analyzing the regulation of miRNAs on protein-protein interaction network in hodgkin lymphoma. BMC Bioinformatics. (2019) 20:449. 10.1186/s12859-019-3041-9
365.
Bose B Moravec M Bozdag S . Computing microRNA-gene interaction networks in pan-cancer using miRDriver. Sci Rep. (2022) 12:3717. 10.1038/s41598-022-07628-z
366.
Phuah NH Nagoor NH . Regulation of microRNAs by natural agents: new strategies in cancer therapies. Biomed Res Int. (2014) 2014:804510. 10.1155/2014/804510
367.
Seitz H . A new perspective on microRNA-guided gene regulation specificity, and its potential generalization to transcription factors and RNA-binding proteins. Nucleic Acids Res. (2024) 52(16):9360–8. 10.1093/nar/gkae694
368.
Chatterjee B Thakur SS . Interaction networks among miRNA, protein, and metabolite fingerprints identify the regulatory networks and key players in the pathogenesis of diabetic cardiomyopathy. Front Cell Dev Biol. (2025) 13:1602320. 10.3389/fcell.2025.1602320
369.
Abolhasani S Ahmadi Y Fattahi D Rostami Y Chollou KM . microRNA-Mediated regulation of oxidative stress in cardiovascular diseases. J Clin Lab Anal. (2025) 39(9):e70017. 10.1002/jcla.70017
370.
Li Y Wang Y Chen S Liu L . The landscape of miRNA-mRNA regulatory network and cellular sources in inflammatory bowel diseases: insights from text mining and single cell RNA sequencing analysis. Front Immunol. (2024) 15:1454532. 10.3389/fimmu.2024.1454532
371.
Kim D Ko Y . MIMR: development of a web-based system for miRNA and mRNA integrated analysis. Int J Mol Sci. (2024) 25(21):11819. 10.3390/ijms252111819
372.
Azizian S Cui J . DeepMiRBP: a hybrid model for predicting microRNA-protein interactions based on transfer learning and cosine similarity. BMC Bioinformatics. (2024) 25(1):381. 10.1186/s12859-024-05985-2
373.
Altan-Bonnet G Mukherjee R . Cytokine-mediated communication: a quantitative appraisal of immune complexity. Nat Rev Immunol. (2019) 19(4):205–17. 10.1038/s41577-019-0131-x
374.
Cohen IR . The cognitive paradigm and the immunological homunculus. Immunol Today. (1992) 13(12):490–4. 10.1016/0167-5699(92)90024-2
375.
Laub MT . Keeping signals straight: how cells process information and make decisions. PLoS Biol. (2016) 14(7):e1002519. 10.1371/journal.pbio.1002519
376.
Spirin V Mirny LA . Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. (2003) 100(21):12123–8. 10.1073/pnas.2032324100
377.
Bich L Pradeu T Moreau JF . Understanding multicellularity: the functional organization of the intercellular space. Front Physiol. (2019) 10:1170. 10.3389/fphys.2019.01170
378.
Ratti M Lampis A Ghidini M Salati M Mirchev MB Valeri N et al MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. (2020) 15(3):261–78. 10.1007/s11523-020-00717-x
379.
Mann M Mehta A Zhao JL Lee K Marinov GK Garcia-Flores Y et al An NF-κB-microRNA regulatory network tunes macrophage inflammatory responses. Nat Commun. (2017) 8:851. 10.1038/s41467-017-00972-z
380.
Perry MM Moschos SA Williams AE Shepherd NJ Larner-Svensson HM Lindsay MA . Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. (2008) 180(8):5689–98. 10.4049/jimmunol.180.8.5689
381.
Baghi H B Bayat M Mehrasa P Alavi SMA Lotfalizadeh MH Memar MY et al Regulatory role of microRNAs in virus-mediated inflammation. J Inflamm. (2024) 21(1):43. 10.1186/s12950-024-00417-7
382.
Fliri A Kajiji S . Effects of vitamin D signaling in cardiovascular disease: centrality of macrophage polarization. Front Cardiovasc Med. (2024) 11:1388025. 10.3389/fcvm.2024.1388025
383.
Wang X Chen L Wei J Zheng H Zhou N Xu X et al The immune system in cardiovascular diseases: from basic mechanisms to therapeutic implications. Signal Transduct Target Ther. (2025) 10(1):166. 10.1038/s41392-025-02220-z
384.
Moore AR Zheng H Ganesan A Hasin-Brumshtein Y Maddali MV Levitt JE et al International multi-cohort analysis identifies novel framework for quantifying immune dysregulation in critical illness: results of the SUBSPACE consortium. Preprint. bioRxiv. (2024) 2024:623298. 10.1101/2024.11.12.623298
385.
Tang J Aittokallio T . Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles. Curr Pharm Des. (2014) 20(1):23–36. 10.2174/13816128113199990470
386.
Dai X Tan C . Combination of microRNA therapeutics with small-molecule anticancer drugs: mechanism of action and co-delivery nanocarriers. Adv Drug Deliv Rev. (2015) 81:184–97. 10.1016/j.addr.2014.09.010
387.
Ionescu RF Enache RM Cretoiu SM Cretoiu D . The interplay between gut Microbiota and miRNAs in cardiovascular diseases. Front Cardiovasc Med. (2022) 9:856901. 10.3389/fcvm.2022.856901
388.
Sun M Xu S Mei Y Li J Gu Y Zhang W et al MicroRNAs in medicinal plants. Int J Mol Sci. (2022) 23(18):10477. 10.3390/ijms231810477
389.
Li S Huang Q Yang Q Peng X Wu Q . MicroRNAs as promising therapeutic agents: a perspective from acupuncture. Pathol Res Pract. (2023) 248:154652. 10.1016/j.prp.2023.154652
390.
Keresztes D Kerestély M Szarka L Kovács BM Schulc K Veres DV et al Cancer drug resistance as learning of signaling networks. Biomed Pharmacother. (2025) 29:117880. 10.1016/j.biopha.2025.117880
391.
Zhang J Qu Q Chen X . Understanding collective behavior in biological systems through potential field mechanisms. Sci Rep. (2025) 15(1):3709. 10.1038/s41598-025-88440-3
392.
Fountzilas E Pearce T Baysal MA Chakraborty A Tsimberidou AM . Convergence of evolving artificial intelligence and machine learning techniques in precision oncology. NPJ Digit Med. (2025) 8(1):75. 10.1038/s41746-025-01471-y
393.
Abbass H Mostaghim S . The road forward with swarm systems. Philos Trans A Math Phys Eng Sci. (2025) 383(2289):20240135. 10.1098/rsta.2024.0135
394.
Wen Z Zhang Y Feng J Aimulajiang K Aleem MT Lu M et al Excretory/secretory proteins inhibit host immune responses by downregulating the TLR4/NF-κB/MAPKs signaling pathway: a possible mechanism of immune evasion in parasitic nematode haemonchus contortus. Front Immunol. (2022) 13:1013159. 10.3389/fimmu.2022.1013159
395.
Benharroch D Osyntsov L . Infectious diseases are analogous with cancer. Hypothesis and implications. J Cancer. (2012) 3:117–21. 10.7150/jca.3977
396.
Goldszmid RS Dzutsev A Trinchieri G . Host immune response to infection and cancer: unexpected commonalities. Cell Host Microbe. (2014) 15(3):295–305. 10.1016/j.chom.2014.02.003
397.
Howley PM . Gordon wilson lecture: infectious disease causes of cancer: opportunities for prevention and treatment. Trans Am Clin Climatol Assoc. (2015) 126:117–32.
398.
Powers CN . Diagnosis of infectious diseases: a cytopathologist's Perspective. Clin Microbiol Rev. (1998) 11(2):341–65. 10.1128/CMR.11.2.341
399.
Ahmad MA Kareem O Khushtar M Akbar Md Haque MdR Iqubal A et al Neuroinflammation: a potential risk for dementia. Int J Mol Sci. (2022) 23(2):616. 10.3390/ijms23020616
400.
Shajahan SR Kumar S Ramli MDC . Unravelling the connection between COVID-19 and Alzheimer's Disease: a comprehensive review. Front Aging Neurosci. (2024) 15:1274452. 10.3389/fnagi.2023.1274452
401.
Vojtechova I Machacek T Kristofikova Z Stuchlik A Petrasek T . Infectious origin of Alzheimer's Disease: amyloid beta as a component of brain antimicrobial immunity. PLoS Pathog. (2022) 18(11):e1010929. 10.1371/journal.ppat.1010929
402.
Honjo K van Reekum R Verhoeff NP . Alzheimer's disease and infection: do infectious agents contribute to progression of Alzheimer's Disease?Alzheimers Dement. (2009) 5(4):348–60. 10.1016/j.jalz.2008.12.001
403.
Catumbela CSG Giridharan VV Barichello T Morales R . Clinical evidence of human pathogens implicated in Alzheimer's Disease pathology and the therapeutic efficacy of antimicrobials: an overview. Transl Neurodegener. (2023) 12(1):37. 10.1186/s40035-023-00369-7
404.
Sipilä PN Heikkilä N Lindbohm JV Hakulinen C Vahtera J Elovainio M et al Hospital-treated infectious diseases and the risk of dementia: a large, multicohort, observational study with a replication cohort. Lancet Infect Dis. (2021) 21(11):1557–67. 10.1016/S1473-3099(21)00144-4
405.
Wensveen FM Šestan M Polić B . The immunology of sickness metabolism. Cell Mol Immunol. (2024) 21(9):1051–65. 10.1038/s41423-024-01192-4
406.
Du Z Ren Y Wang J Li Sx Hu Yf Wang L et al The potential association between metabolic disorders and pulmonary tuberculosis: a Mendelian randomization study. Eur J Med Res. (2024) 29:277. 10.1186/s40001-024-01845-0
407.
Napiórkowska-Baran K Schmidt O Szymczak B Lubański J Doligalska A Bartuzi Z . Molecular linkage between immune system disorders and atherosclerosis. Curr Issues Mol Biol. (2023) 45(11):8780–815. 10.3390/cimb45110552
408.
Yakoub AM Schülke S . A model for apoptotic-cell-mediated adaptive immune evasion via CD80-CTLA-4 signaling. Front Pharmacol. (2019) 10:562. 10.3389/fphar.2019.00562
409.
Chopra R Hengen KB . What if what matters is emergent?Neuron. (2025) 113(2):187–9. 10.1016/j.neuron.2024.12.027
410.
Crutchfield JP Mitchell M . The evolution of emergent computation. Proc Natl Acad Sci U S A. (1995) 92(23):10742–6. 10.1073/pnas.92.23.10742
411.
Levenson RM Singh Y Rieck B Hathaway QA Farrelly C Rozenblit J et al Advancing precision medicine: algebraic topology and differential geometry in radiology and computational pathology. Lab Invest. (2024) 104(6):102060. 10.1016/j.labinv.2024.102060
412.
Sen S Mitchell AK . Many-Body quantum interference route to the two-channel kondo effect: inverse design for molecular junctions and quantum dot devices. Phys Rev Lett. (2024) 133(7):076501. 10.1103/PhysRevLett.133.076501
413.
Timmis J Ismail AR Bjerknes JD Winfield AF . An immune-inspired swarm aggregation algorithm for self-healing swarm robotic systems. Biosystems. (2016) 146:60–76. 10.1016/j.biosystems.2016.04.001
Summary
Keywords
material protein swarms, gene ontology, proteomics, atherosclerosis, microRNAs, infections, Alzheimer's disease, cardiovascular conditions
Citation
Fliri A, Sostek R and Kajiji S (2025) Protein swarm-based cause-effect analysis: effects of microRNAs on cooperation networks linking COVID-19 infections, atherosclerosis, and Alzheimer's disease. Front. Cardiovasc. Med. 12:1577844. doi: 10.3389/fcvm.2025.1577844
Received
16 February 2025
Accepted
03 November 2025
Published
08 December 2025
Volume
12 - 2025
Edited by
Alexander Akhmedov, University of Zurich, Switzerland
Reviewed by
Sanjay Kumar Dey, University of Delhi, India
Wu Zhou, Guangzhou University of Chinese Medicine, China
Updates
Copyright
© 2025 Fliri, Sostek and Kajiji.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Anton Fliri anton.fliri@emergentsa.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.