- 1Discovery & Innovation Platforms, The Procter & Gamble Company, Mason, OH, United States
- 2Global Oral Care R&D, The Procter & Gamble Company, Mason, OH, United States
Objective: To determine whether outer membrane vesicles (OMVs) of the periodontal pathogen Porphyromonas gingivalis (P. gingivalis) can infect gingival keratinocytes and stimulate reactive oxygen species (ROS) production, and to assess whether stannous fluoride (SnF2), stannous chloride (SnCl2) or 0.454% SnF2 toothpaste diluents can inhibit OMV infection.
Methods: OMVs were isolated from P. gingivalis culture and their morphology was characterized using scanning electron microscopy and transmission electron microscopy. OMVs were harvested, separated from parent bacteria, labeled with fluorescent probes, and added to proliferating gingival keratinocytes. Infection was monitored by measuring uptake of fluorescence. Free radicals and ROS were quantified by adding a separate CellROX fluorescent probe following 24 h incubation with OMVs, and automated fluorescence imaging was used to assess ROS generation rates. A dose response range of SnF2 and SnCl2 concentrations as well as 0.454% SnF2 toothpaste dilutions were added to OMVs to examine their potential to neutralize OMV infectivity and protect gingival keratinocytes from development of oxidative stress. The mechanism of SnF2 inhibition of OMV infection was studied by binding SnF2 with purified lipopolysaccharides (LPS) from the bacterial culture and examining the binding of stannous to LPS using mass spectrometry.
Results: Large numbers of OMVs were formed in P. gingivalis culture medium. They were purified along with isolating soluble LPS. Fluorescence imaging revealed that OMVs infected gingival keratinocytes and promoted oxidative stress in a dose-dependent manner. SnF2, SnCl2, and SnF2 toothpaste inhibited OMV infectivity (p < 0.05) and likewise protected gingival keratinocytes from oxidative stress (p < 0.05). Stannous precipitated LPS and OMVs from solution, forming insoluble aggregates easily isolated by centrifugation. Mass spectroscopic analysis revealed that stannous was bound to LPS in a one-to-one molecular equivalent ratio.
Conclusion: SnF2 not only kills bacteria, but also inhibits bacterial virulence factors, such as LPS and OMVs. SnF2, SnCl2 and stannous-containing toothpastes can precipitate OMVs and LPS to in principle protect gingival keratinocyte cells from infection leading to inflammation and oxidative stress.
1 Introduction
Periodontal diseases are epidemic worldwide. It is estimated that up to 50% of the global population exhibits some form of periodontal disease, which is elevated in older populations (1, 2). Gingivitis, the earliest stage of periodontal disease, presents with symptoms including swelling, redness, and bleeding of the tissues along the gingival margin (3, 4). In more advanced stages, the destruction of alveolar bone and ligament structures of the teeth result in tooth mobility and, in the worst cases, tooth loss (5, 6).
Periodontal diseases are the result of microbial dysbiosis in bacterial plaque biofilms that form above the gumline and in the gingival sulcus of developing periodontal pockets (3–6). The microbial biofilms in these locations prompt inflammatory responses in the gingival tissues, and it is the effects of the pronounced chronic inflammation and compromised host response that produce the tissue damage associated with periodontal disease. The dental plaque bacteria contributing to periodontal disease include a mixture of microorganisms with anaerobic species predominating as the disease progresses (7, 8). Microbes that appear dominant in the advancement of disease include the so-called “red three” gram negative anaerobic species; Tannerella forsythia, Treponema denticola and Porphyromonas gingivalis (P. gingivalis) (9, 10). P. gingivalis is a particularly important pathogen associated with the progression of periodontal disease (10–12). The local destruction of periodontal tissues caused by infection with this organism promotes increases in the flow of gingival crevicular fluid-containing serum (including heme) compounds and collagen-degradation products into the periodontal pockets. These changes in environmental conditions favor further growth of a mixture of pathogens in the subgingival microflora producing microbial dysbiosis (7, 12–15).
One of the virulence factors generally associated with gram negative anaerobic bacteria is their production of outer membrane vesicles (OMVs) (16, 17). OMVs are double layered spherical membrane-like bodies with diameters much smaller than the native bacteria, ranging in size from 50 to 250 nanometers (18). OMVs are continuously released from gram negative bacteria during their growth and proliferation (16, 19–22). In 1985, researchers first reported that P. gingivalis can produce OMVs, although at that time their physiological actions and pathogenic effects were not completely appreciated (23). The number of OMVs relative to parent P. gingivalis bacteria can be extreme. For example, the ratio of P. gingivalis bacteria to OMVs can range up to 1–2,000, and OMVs are adherent compared to their parent bacterium (24). Bacterial OMVs are comprised of lipopolysaccharides (LPS), phospholipids, outer membrane proteins and a portion of the periplasm that is captured during the membrane formation process (16–18). OMVs may form by a variety of mechanisms (16, 17, 25); however, regardless of their origin many components of OMVs are pathogenic and include factors which contribute to host cell immune system activation, destruction, immune system escape and host cell invasion (26). For example, OMVs present surfaces containing antigens to host tissues, most notably including endotoxin (e.g., LPS) (26). It has been suggested that virulence factors present in OMVs have advantages protecting them from host deactivation. Haurat et al. showed that P. gingivalis selectively package certain membrane proteins including gingipains into OMVs (27). Gingipains contribute to barrier function degradation of gingival tissues (28) and data show that gingipain levels in OMVs of P. gingivalis are 3–5 times those in the parent bacteria (29). Contemporary research has progressed to suggest that P. gingivalis OMVs play significant roles in the pathogenesis of periodontal diseases promoted by this bacteria (26).
Our laboratories have been carrying out research to understand the magnitude, duration and mechanisms of the high bioavailable stannous fluoride (SnF2) formulated in dentifrices for the prevention of periodontal diseases. In a previous study, we evaluated the histomorphology of SnF2 reactivity with P. gingivalis bacteria using TEM and identified significant deposition of insoluble aggregates in cell membranes leading to lysis and bactericidal activity (30). We posited that the reactivity of SnF2 with P. gingivalis was most likely associated with precipitation of endotoxin and phospholipids in cell membranes (30). Notably, LPS reactivity with SnF2 has been shown to reduce the antigenicity of reacted LPS to toll receptor promotion of inflammation pathways (31–33). During the aforementioned TEM study (30) we observed significant numbers of OMVs in incubation mixtures of P. gingivalis and we likewise saw significant SnF2 reactivity with membranes of these vesicles. The purpose of this study was to expand upon those learnings and carry out a more comprehensive study, specifically to establish if OMVs of P. gingivalis can infect and stimulate reactive oxygen species (ROS) production in gingival keratinocytes and to assess whether SnF2, stannous chloride (SnCl2) or 0.454% SnF2 toothpaste diluents can inhibit OMVs. TEM analyses were performed to assess the location and tenacity of SnF2 interactions with P. gingivalis matrix vesicles. In addition, OMVs treated with SnF2 and SnCl2 were examined to see if reactivity diminished the inflammatory promotion of OMVs with gingival keratinocytes using ROS generation as a probe for inflammation promotion. Lastly, mass spectrometry analysis was carried out to confirm the nature of SnF2 reactivity with LPS.
2 Materials and methods
2.1 Bacterial growth and OMV isolation
P. gingivalis (ATCC catalog #33277, American Type Culture Collection, Manassas, VA) was cultured in 30 ml MTGE media (Anaerobic Enrichment Broth, Anaerobe Systems, 6 ml tubes- catalog #AS-778 & 500 ml bottles- catalog #AS-7785, Anaerobe System, Morgan Hill, CA) in a sterile 125 ml Erlenmeyer flask under anaerobic conditions at 37°C for 48 h as seeding bacteria. The seeding bacterial culture was inoculated with seven liters of fresh MTGE media and continued to grow for 48 h under anaerobic conditions at 37°C. The bacteria were harvested at the end of culture by centrifugation in a JA-10 rotor at 10,000 g, 4°C for 60 min in Avanti J-26 XPI High-Performance Centrifuge of Beckman Coulter (Indianapolis, IN). The supernatant was collected and filtered through 0.45 μm pore PVDF membranes to remove cell debris.
OMVs were secreted by P. gingivalis into the MTGE media. To isolate OMVs, the conditioned culture medium volume was reduced by filtration using a tangential flow filtration Minimate TFF System (PALL Life Sciences, Port Washington, NY) with filter capsules of molecular weight cutoff 100 kD at 40 Psi. The retentate of the filtration was centrifuged at 140,000 × g for 1 h at 4°C using an SW32 swinging bucket rotor on a Beckman XL-100 K Ultracentrifuge (Beckman Coulter, Atlanta, GA) to separate the OMV pellet from the supernatant. The supernatant was retained as the starting material for secreted LPS purification (see below). The pellets were resuspended in dPBS buffer [1X Dulbecco's Phosphate Buffered Saline (dPBS): Life Technologies, Grand Island, NY] and centrifuged at 200,000 × g for 1 h at 4°C (using an SW41 swinging bucket rotor) to yield a standard OMV preparation.
To generate highly pure OMVs, the initial OMVs from the first ultracentrifugation were then resuspended in 800 μl HEPES buffer (50 mM HEPES, 150 mM NaCl, pH 6.8, Life Technologies, Grand Island, NY), stained with Vybrant™ Multicolor Cell-Labeling Kit with DiD Solutions (ThermoFisher Scientific, Waltham, MA, USA), and underwent another round of ultracentrifugation using OptiPrep™ (60% w/v iodixanol in water, Sigma-Aldrich, St. Louis, MO, USA) discontinuous density gradient. The initial OMV preparation was separated into four samples and each resuspended in 3 ml HEPES buffer containing 45% w/v iodixanol and placed in 4 Ultra-Clear™, 14 ml, 14 × 95 mm tubes (Beckman Coulter, Atlanta, GA). A discontinuous iodixanol gradient was achieved in each sample by layering successive 1.5 ml of HEPES buffer containing 45%, then 40%, 35%, 30%, 25% & 20% w/v iodixanol, with 45% at the bottom, in a total of 9.5 ml. Tubes were centrifuged at 173,000 × g for 72 h at 4°C using a 70.1Ti rotor installed in a Beckman XL-100 K Ultracentrifuge (Beckman-Coulter, Atlanta, GA). Eight 0.5 ml gradient fractions from each sample (1, 2, 3 & 4) were collected from top to bottom of the density gradient solution and measured at A260 and A280 for DNA/RNA and proteins, respectively, using an 8-channel NanoDrop spectrophotometer according to the manufacturer's instructions (ThermoFisher Scientific). OMVs containing fractions were identified by measuring endotoxin contents using the Pierce™ LAL Chromogenic Endotoxin Quantitation Kit, per manufacturer's instructions (ThermoFisher Scientific).
Fractions containing the purified OMVs were washed with endotoxin-free water (Sigma, St. Louis, MO) and centrifuged twice at 200,000 × g for 2 h at 4°C using a SW40 Ti rotor installed in a Beckman XL-100 K Ultracentrifuge (Beckman-Coulter, Atlanta, GA). The highly pure OMVs were resuspended in 30 ml of endotoxin-free water, and aliquoted into 0.5 ml Eppendorf tubes and stored at −80°C. OMVs were quantified using Bradford assay to estimate the amounts of proteins. OMVs were subsequently fluorescently labeled (see below) to assess their integration and infection into gingival keratinocytes.
2.2 Fluorescence-labeling of OMVs
OMVs were labelled with either green (Lipophilic Tracers DiO) or red (Lipophilic Tracers DiD) fluorescence dye following the manufacturer's instructions (Invitrogen™ Vybrant™ Multicolor Cell-Labeling Kit with DiO, DiI, DiD Solutions, 1 ml each, ThermoFisher Scientific). Briefly, the lipophilic dyes were added directly to the OMVs. The mix was incubated at 37°C for 20 min under a light proof condition. Unlabeled dyes were removed using Sephadex LH-20 resin in a Spin Columns-Snap Cap with a Collection Tube (ThermoFisher Scientific).
2.3 SnF2 and SnCl2 solution and SnF2 toothpaste supernatant reactivity
For compound treatment, SnF2 and/or SnCl2 (Sigma, St. Louis, MO) were freshly weighed and dissolved in UPW water at 10 mM as stock solutions on the day of treatment. They were then added to cell culture medium. For toothpaste diluents treatments, SnF2 0.454% W/W (Crest Pro-Health Advanced Gum Restore Deep Clean Toothpaste and Crest Pro-Health Enamel Repair and Gum Toothpaste, Procter & Gamble, Cincinnati, OH) were used to prepare diluted slurries with pure water. Toothpaste solution was freshly prepared right before the experiment in anaerobic conditions. Toothpaste was weighed and dissolved in UPW water at 10% as a stock solution. Vigorous vortexing of toothpaste stock was done for 30 min in flasks to assure complete suspension. Then 1 ml of the toothpaste suspension was transferred to a microtube and centrifuged at high speed (12,000 g) for 10 min at 4°C. The supernatant was collected into a 15 ml tube and diluted with complete cell culture medium for treatment.
2.4 Gingival keratinocyte cell culture and treatment
Immortalized Human Gingival Keratinocytes were purchased from Applied Biological Materials Inc. (Richmond, BC, V6V 2J5, Canada). Cells were maintained in a complete growth medium (proprietary to Applied Biological Materials Inc) under an atmosphere of 5% CO2 and 95% air, 95% humidity at 37°C. For OMV infections, cells were harvested from flasks at 60%–80% confluence, counted using a cell counter (Invitrogen™ Countess™ 3 Automated Cell Counter, ThermoFisher Scientific) and suspended in fresh complete growth medium. The cells were seeded at 7,000 in 100 μl of culture medium in each well of a 96-well plate. Fluorescence-labelled OMVs were subsequently added to the cell culture the next day. Both the stannous solutions and the toothpaste supernatants were diluted with complete cell culture medium and mixed with OMVs. These mixtures were incubated for 10 min at room temperature before application. The cells were then fed with the 100 μl of the mixture, incubated in the Incycyte S3 (Sartorius, Bohemia, New York), which was placed inside a regular incubator with an atmosphere of 5% CO2 and 95% air, 95% humidity at 37°C.
2.5 OMV infection of gingival keratinocytes. ROS generation following OMV infection of gingival keratinocytes
OMVs bind Toll-like receptors on the surface of, and inside, the gingival keratinocytes and modulate their metabolic pathways. Consequently, free radicals and oxidative species are generated within the cells. The internalization of OMVs into keratinocytes was recorded by taking images at preset intervals. To quantify the oxidative molecules within the cells, we applied CellROX fluorescence dye to the cells 24 h after addition of the OMVs to the cells. Both Deep Red and Green CellROX dyes (ThermoFisher Scientific) are fluorogenic probes for quantifying ROS in live cells. CellROX Green Reagent is weakly fluorescent while in a reduced state and exhibits bright green photostable fluorescence upon oxidation by ROS and survives detergent treatment. CellROX Deep Red Reagent is also a cell-permeant dye. It is non-fluorescent in a reduced state, and exhibits bright fluorescence upon oxidation by ROS. To make sure that SnF2, SnCl2 and toothpaste supernatants did not quench specific dyes, we used both Deep Red and Green CellROX reagents in our experiments. CellROX dyes (2.5 μM) were added to cells 24 h after addition of OMVs into cell culture. Images were taken at a regular interval for fluorescence quantification.
2.6 Imaging
Green and red fluorescence and phase contrast images were acquired every one to three hours for the first 24 h, every hour after adding the CellROX dyes for 10 h, and then every two hours for 3 days with an objective of 10X or 20X in the Incycyte S following the manufacturer's instruction. Three images were taken in each well at pre-selected sites in a 96-well plate, where cells were likely evenly distributed. Fluorescence-labeled OMVs were imaged in the focus plane of the objective. The CellROX™ Green or Red Detection Reagent is cell-permeable and non-fluorescent or very weakly fluorescent while in the reduced state. The fluorescent dyes bind DNA and generate strong fluorogenic signals. The images were taken and analyzed using the software from the manufacturer. Two dyes, Vybrant™ DiO Cell-Labeling and Vybrant™ DiD Cell-Labeling dyes, were used to label the OMV. Two ROS detection dyes, CellROX™ Green and CellROX™ Red, were used to visualize the oxidation conditions inside gingival keratinocytes. The fluorescence signals were different between the four fluorescence dyes. For comparisons, the fluorescence signals were normalized by dividing raw fluorescence counts with the mean fluorescence counts of 0.65 OMV μg/ml at 36 h in each plate. All images of CellROX oxidation were exported from instrument Incycyte S at 36 h. The time stamps (0 d 0 h 0 m) were default when exporting individual images.
2.7 Isolation of secreted LPS from P. gingivalis
The supernatant collected from OMV isolation was used as the source of secreted LPS from the bacterium. Secreted LPS were extracted and purified using previously described procedures (34, 35). Briefly, bacteria and bacterial fragments were removed using centrifugation and consecutive filtration with 0.45 μm and 0.22 μm membrane filters. OMVs were then concentrated from 7 liters to 100 ml using tangential flow filtration with filter capsules of a 100 kD molecular weight cutoff at 40 Psi. The concentrated OMV preparation was diluted with 500 ml of water and reconcentrated using tangential flow filtration to remove any molecules smaller than 100 kD. Individual LPS molecules, which are around 10–20 kD, would be filtered out, while only LPS aggregated as vesicles were retained. This dilution process was repeated twice to ensure that only bacterial components larger than 100 kD remained in the OMV preparation. The concentrated OMV preparation was then pelleted by ultracentrifugation, and the supernatant was saved for LPS isolation. It is likely that LPS in the supernatant existed in some form of vesicles that were not precipitated during ultracentrifugation. The supernatant was adjusted to 300 ml buffer containing 10 mM Tris-Cl buffer (pH 8), 2% Sodium Dodecyl Sulphate, 2 mM MgCl2 and 40 mg Proteinase K (all chemicals and proteinase K were purchase from Sigma, St. Louis MO). The mixture was vortexed and placed an incubator at 68°C for 24 h. Sixty ml of 3 M sodium acetate pH 5.2 and 800 ml 100% ethanol were added and kept at −20°C. The crude LPS preparation was precipitated using a JA-10 rotor at 13,000 RPM, 4°C for 60 min in Avanti J-26 XPI High-Performance Centrifuge of Beckman, and was further purified using affinity chromatography following procedures described by Hirayama et al. (36) and Sakata et al. (37) The endotoxin activities were analyzed in the fractions using the Pierce™ LAL Chromogenic Endotoxin Quantitation Kit, per manufacturer's instructions (ThermoFisher Scientific).
2.8 OMV and SnF2 precipitation assay
P. gingivalis 33,277 OMV was used for the precipitation assay. SnF2 solution was freshly prepared in each experiment in water. OMV, prepared in water, was added to the wells of a 96-well plate first, and then SnF2 was added. The plate was transferred to spectrometry reader (Molecular Devices SpectraMax M5 Multi-Mode Microplate Reader, Molecular Devices, LLC, San Jose, CA) for OD600 kinetic reading in 10 min intervals.
2.9 Mass spectrometry—LPS binding measures
Matrix-assisted laser desorption ionization (MALDI) is a soft ionization technique used in mass spectrometry (MS) (38). MALDI mass spectrometry can be used for the analysis of biomolecules, including peptides, proteins, polysaccharides, and large organic molecules and polymers. In MALDI, the analyte is first co-crystallized with a UV-absorbing matrix such as alpha-cyano-4-hydroxycinnamic acid, then subjected to pulse laser radiation. This causes the desorption of the analyte/matrix crystals and produces ions which are transmitted into a mass analyzer for detection. In MALDI TOF, a time-of-flight (TOF) mass analyzer is used. MALDI TOF data can be acquired in MS mode to generate molecular weight information and in MS/MS mode to generate structural information. Typically, MALDI mass spectrometry data acquisition takes less than a minute, so the technique can be used to quickly screen for molecular species in samples of interest.
In this study, P. gingivalis secreted LPS molecular weight profiles and their interaction with SnF2 were investigated via MALDI TOF. An equal volume of soluble LPS solutions with or without added SnF2 was mixed with alpha-cyano-4-hydroxycinnamic acid (10 mg/ml in 80% acetonitrile/20% water). Next, 0.7 ul of the sample solution was spotted on a MALDI plate, air-dried, and analyzed in negative-ion mode using a MALDI TOF/TOF 4,800 Plus Analyzer (AB-SCIEX, Framingham, MA, USA). Data were acquired using a mass scan range of 500–3,000 Da and a laser power of 4,500. Data were collected in an automated fashion using random sampling over the sample spot with 250 shots per subspectrum and a total of 2,500 shots per spectrum. The mass of LPS corresponded to part of the mass of Lipid A in the LPS molecules, as shown previously (40–42).
2.10 TEM sample preparation and imaging
P. gingivalis isolated OMVs were placed in a fixative solution (2% glutaraldehyde in PBS buffer) immediately, and stored at 4°C. The fixed samples were post-fixed with 1% OsO4 in PBS buffer overnight at 4°C. The samples were then hydrated by gradually increasing the concentration (50%, 70%, 85%, 90%, 100%) with ddH2O for 3 h at room temperature and filtrated by gradually increasing the concentrate of Epon 812 with acetone (30%, 50%, 75%, 100%) at room temperature. All samples were embedded in Epon 812 Epoxy (Electron Microscopy Sciences, Hatfield, PA) and cured overnight at 65°C. Each sample block was trimmed, sectioned by Leica ultramicrotome (Leica Microsystems, Deerfield, IL) a Diatome ultracut 45° diamond knife. Roughly 70 nm thick sections were collected, placed on 200 mesh copper grid with formvar, and post-stained with uranyl acetate for 30 min and lead citrate for 10 min. All samples were examined by Hitachi S5200 STEM (Hitachi High-Tech, Hillsboro, OR) for high resolution imaging analysis at 30 KV and with Bruker EDS detector (Bruker, Madison, WI) for elemental information.
2.11 SEM sample preparation and imaging
Bacterial cultures were centrifuged to collect the bacteria after 48 h, and the bacterial pellets were placed in a fixative solution (2% glutaraldehyde in PBS buffer) immediately and stored at 40°C. The fixed samples were post-fixed with 1% OsO4 in PBS buffer overnight at 40°C. The samples were then dehydrated by gradually increasing the concentration (50%, 70%, 85%, 90%, 100%) with ddH2O for 3 h at room temperature. The dehydrated samples were deposited onto a filter paper and secured with silver paint on a sample mount. Additional sputter-coating with Au/Pd was performed in a Gatan Alto 2,500 (Ametek, Berwyn, PA) for 60 s. High-resolution imaging was performed in a Hitachi S-4700 (Hitachi High-Tech, Hillsboro, OR) SEM with an accelerating voltage of 3 kV.
2.12 Statistical analysis
Results were organized and analyzed using R. Analysis of variance (ANOVA) was performed first to analyze the whole set of data. When significant variations were found, a t-test was used to assess the differences between groups using the stat_compare_means function in the ggpubr package for bar plots. For line plots, ANOVA was performed, and differences between treatment groups were compared using pairwise t-test, with p-values adjusted using the Bonferroni method. Two fluorescent dyes, Lipophilic Tracers DiO (green) and DiD (red), were used to stain the OMVs. Additionally, two CellROX dyes (green CellROX and red CellROX) were employed to monitor ROS contents in the cells, ensuring the validity of the fluorescence readings. For statistical analysis, fluorescence readings were normalized in each experiment as relative fluorescence. The fluorescence reading was normalized with 0.65 µg/ml OMV at 36 h, where the relative fluorescence of 0.65 µg/ml OMV at 36 h was set at 100.
3 Results
3.1 OMV isolation
Figure 1A shows an SEM of P. gingivalis capturing the formation of OMVs on the surface of the bacterium. TEM images in Figure 1B show ubiquitous OMVs being created in P. gingivalis culture. Dimensions of the vesicles are shown in Figure 1C. OMVs were isolated from growth cultures of P. gingivalis bacteria using gradient ultracentrifugation. A visual example of the separated band is shown in Figure 1D. The OMVs were then purified and resuspended in media as described in the Methods. Figures 1E,F show a collection of suspended OMVs as illustrated by the TEM microscopic image.
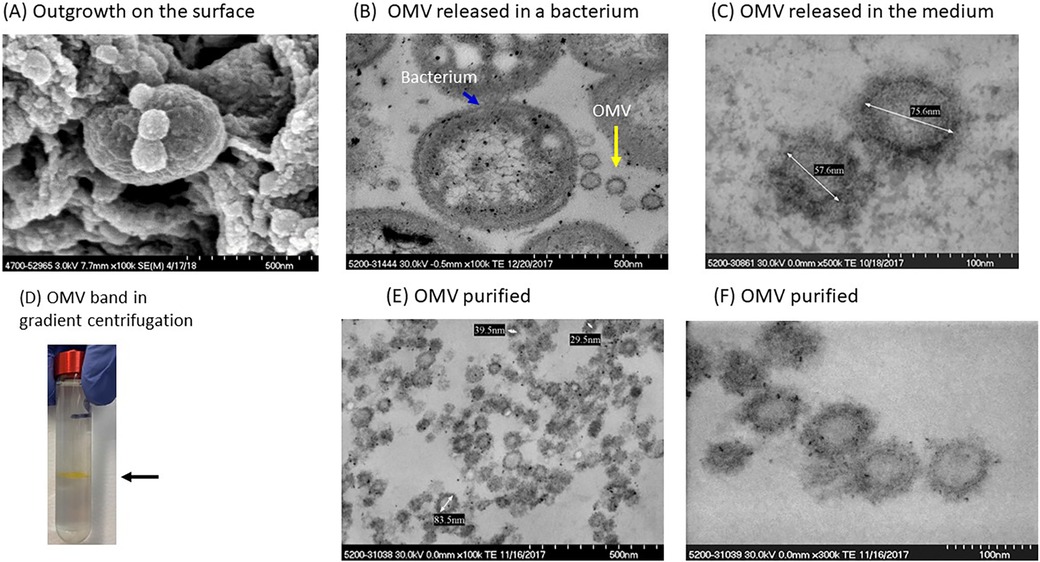
Figure 1. Isolation and characterization of P. gingivalis OMVs. (A) SEM of P. gingivalis bacteria with small outgrowths on surface. (B) Several OMVs (yellow arrow) adjacent to a P. gingivalis bacterium (blue arrow). (C) P. gingivalis OMVs in the culture medium. (D) A visible band of OMVs in OptiPrep gradient gel. (E,F) Purified OMVs of P. gingivalis.
3.2 Entry of OMVs into gingival keratinocytes
Fluorescence-stained OMVs were suspended in the culture medium. The microscopy objective captured images only in the focused plane, ensuring that only fluorescence-stained OMVs attached to or internalized by cells were visible (Figure 2A). OMVs entered gingival keratinocytes in a dose- and time-dependent manner (Figure 2B). Significant increases in OMV entry were observed (p < 0.05) at 3 h with 0.65 and 1.29 μg/ml OMVs, at 12 h with 0.32 μg/ml OMVs, and at 63 h with 0.16 μg/ml OMVs. OMVs did not have obvious deleterious effects in the first 69 h of OMV infection at the doses used in the dose curve study (Supplementary Figure S1, p < 0.05). It is worth noting that P. gingivalis OMVs are highly virulent and kill almost all gingival keratinocytes after about 100 h of infection through either pyroptosis or apoptosis (unpublished results from our labs) at concentrations of 0.75 μg/ml or higher in separate experiments.
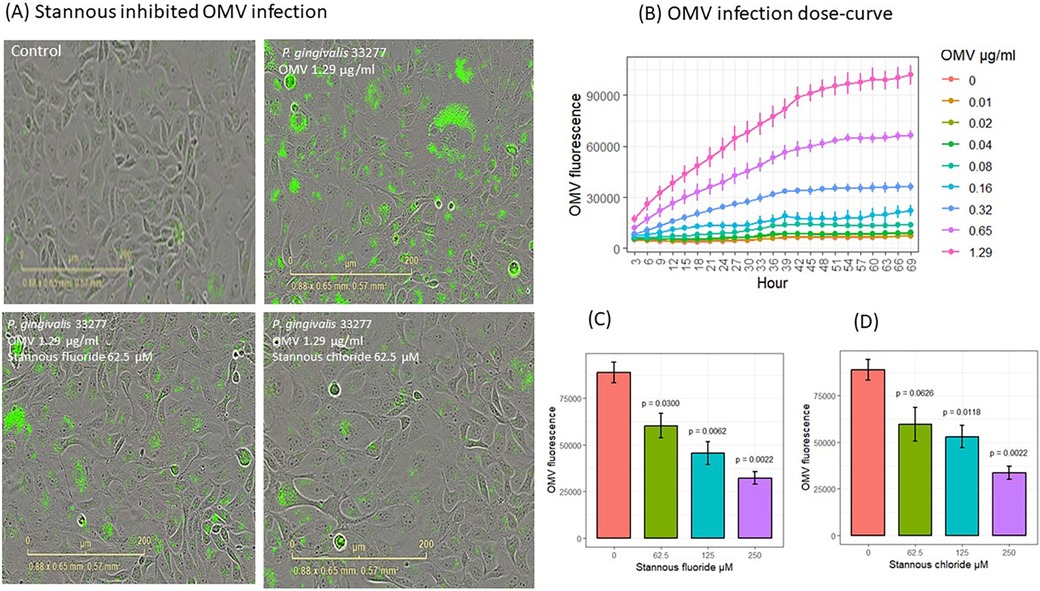
Figure 2. SnF2 and SnCl2 inhibited OMV entry into gingival keratinocytes. (A) Images of fluorescent-labeled OMVs (green) entry into gingival keratinocytes. (B) Entry of OMVs into gingival keratinocytes in a dose curve. Each time point represented the mean and SE of three separate experiments. (C) SnF2 and (D) SnCl2 inhibited entry of OMVs into gingival keratinocytes at 42 h. Each bar represented the mean and SE of three separate experiments. The plots were generated using ggpubr package in RStudio. The results were analyzed using one-way ANOVA and t-test conducted to compare the difference with OMV alone (stannous is 0 μM) as the reference group in stat_compare_means of Rstudio packages.
To understand the effect of stannous on OMV internalization into keratinocytes, we analyzed the fluorescence results at 42 h. SnF2 inhibited the entry of OMVs at 1.29 μg/ml into gingival keratinocytes at all three concentrations tested (62.5, 125, and 250 μM, p ≤ 0.030; Figures 2A,C). Similarly, SnCl2 also reduced OMV internalization at 125 and 250 μM (p ≤ 0.012; Figure 2D).
We analyzed cell proliferation and growth using phase contrast images at 42 h. Neither SnF2 nor SnCl2 reduced cell numbers at 42 h, as shown in Supplementary Figure S1B.
3.3 Oxidative stress
Following OMV exposure, keratinocytes exhibited signs of infection, as shown by images highlighting fluorescence dye-labeled OMVs (red in Figure 3A). OMVs modulated metabolic activities and increased ROS, such as hydroxyl radicals. CellROX dye was added to the cells after 24 h of incubation with OMVs. ROS oxidized the CellROX into fluorescent products (green in Figure 3A). The entry of OMVs into cells (Figures 3A,B) followed the same pattern as in the previous experiment (Figure 2B). OMV fluorescence increased inside the cells in a time- and dose-dependent manner (p < 0.05 for all concentrations from hours 14 to 42). OMVs increased CellROX fluorescence in a dose- and time-dependent manner (Figures 3A,C). OMVs increased cellular oxidation from 30 to 42 h for 1.3 μg/ml (p < 0.05) and from 32 to 40 h for 0.65 μg/ml (p < 0.05, Figure 3C).
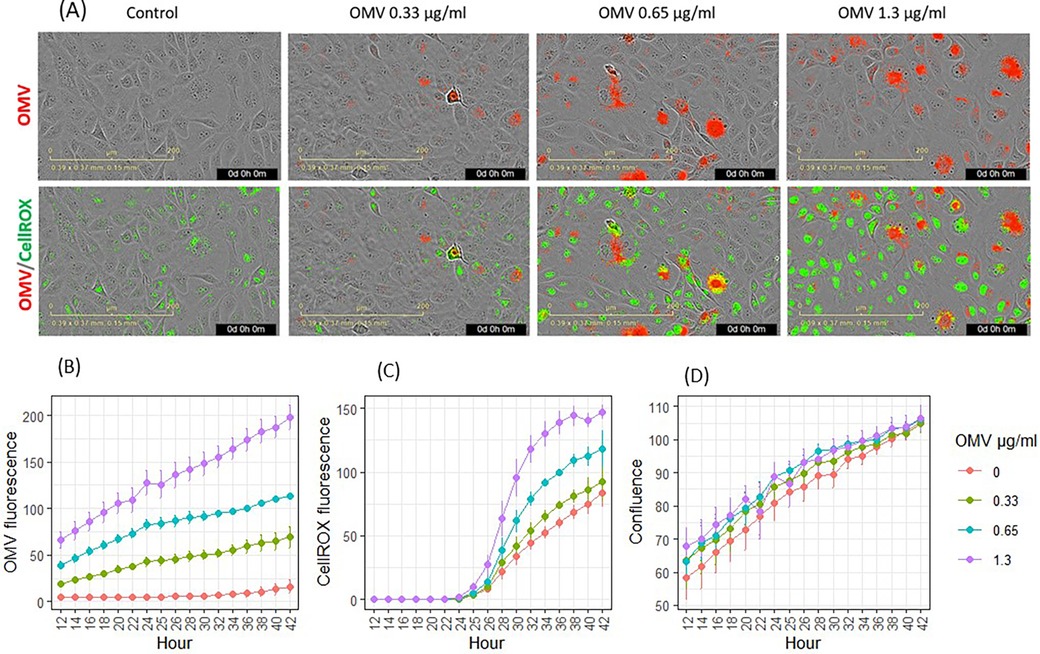
Figure 3. Effects of P. gingivalis OMVs on oxidation of CellROX dyes in gingival keratinocytes. (A) Images of OMV (red) infection and oxidation of CellROX (green). (B) Entry of OMVs into gingival keratinocytes. (C) Oxidation of CellROX dyes in gingival keratinocytes. (D) Cell confluence evaluated by phase contract images. Each time point represented the mean and SE of five separate experiments.
Both fluorescence-labeled OMVs and CellROX stain did not significantly reduce cellular coverage, as evaluated using phase contrast imaging from hours 14 to 42.
The effects of SnF2 and SnCl2 on OMV infection of keratinocytes and the subsequent generation of ROS are shown in Figure 4. Both SnF2 and SnCl2 inhibited the oxidation of CellROX in gingival keratinocytes (Figures 4B-C; p ≤ 0.008 for all three concentrations tested). Additionally, SnF2 and SnCl2 did not significantly change cell numbers (Supplementary Figure S2A). SnF2 concentrations in media containing OMVs produced substantial reductions in ROS production. These effects were observed at SnF2 concentrations as low as 31.25 μM, well within the range observed for SnF2 penetration into the subgingival sulcus following brushing with SnF2 toothpaste (42).
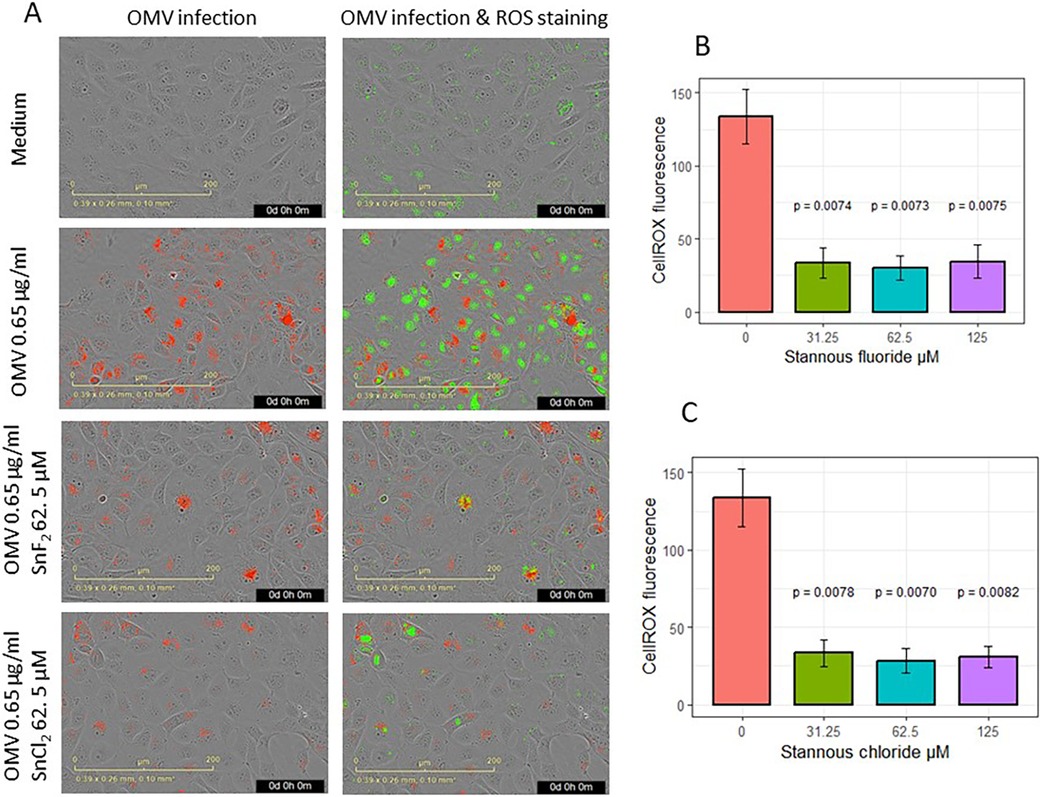
Figure 4. Effects of SnF2 and SnCl2 on the intake of fluorescence-labeled OMV (Red) and oxidized CellROX Dye (green): (A) Images of OMV intake and oxidized CellROX green in cells infected with 0.65 μg/ml OMVs without or with SnF2 or with SnCl2. (B) SnF2 and (C) SnCl2 inhibited cellular oxidation of CellROX. Each point represents the mean and SE from four separate experiments at 42 h. CellROX fluorescence was expressed as relative fluorescence. P-values listed here were from pairwise comparisons with the stannous 0 µM (OMV at 0.65 µg/ml alone) as the reference group.
Results for the effects of SnF2 toothpaste supernatants on OMV infection and inflammatory promotion are shown in Figure 5. The effects of the two toothpastes were similar, so the results were pooled as shown in Figure 5. Addition of dentifrice supernatant at 0.01% and 0.04% dilution produced significant reductions in OMV infectivity of keratinocytes (p ≤ 0.028) and suppressed subsequent generation of ROS (p ≤ 0.017), as shown in Figures 5A–C. Cell confluence was not significantly impacted (Supplementary Figure S2C) by addition of toothpaste supernatants, which is consistent with our unpublished results from other experiments.
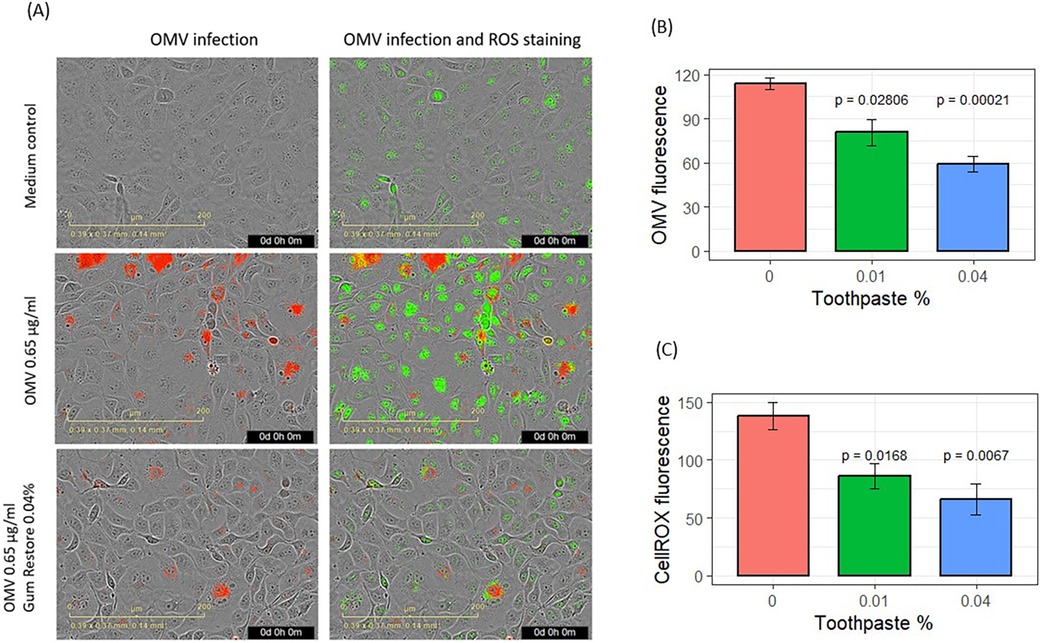
Figure 5. Effects of stannous-based toothpastes on OMV entry and oxidative stress. (A) Images showing OMV (red) entry into gingival keratinocytes and oxidized CellROX Green stains (green). (B) Entry of fluorescence-labeled OMVs into gingival keratinocytes. (C) Quantification of oxidized CellROX dyes inside the cells. CellROX fluorescence and OMV fluorescence are presented as relative fluorescence. Results from two SnF2-containing toothpastes were pooled for analysis. Each bar represents the mean and SE of four separate experiments.
During the preparation of SnF2 solutions with OMVs, some turbidity was observed. The samples were briefly centrifuged, revealing precipitation in the SnF2 and OMV tubes, but not in the OMV-alone tubes (Figure 6A). To characterize the interaction between OMVs and SnF2, we measured the turbidity (OD600) over time after mixing OMV and SnF2. The interaction was rapid, with turbidity increasing immediately upon the addition of SnF2 to the OMV solution (Figure 6B, p < 0.01). Supplementary Figure S3 shows that turbidity is dependent upon mutiple variables, including the concentrations of SnF2 and LPS. Free LPS was dramatically reduced when SnF2 increased from 0.25 to 10 mM in the reaction. Only a soluble fraction was analyzed in the mass spectrometer. Likely, the LPS was bound to SnF2 and precipitated.
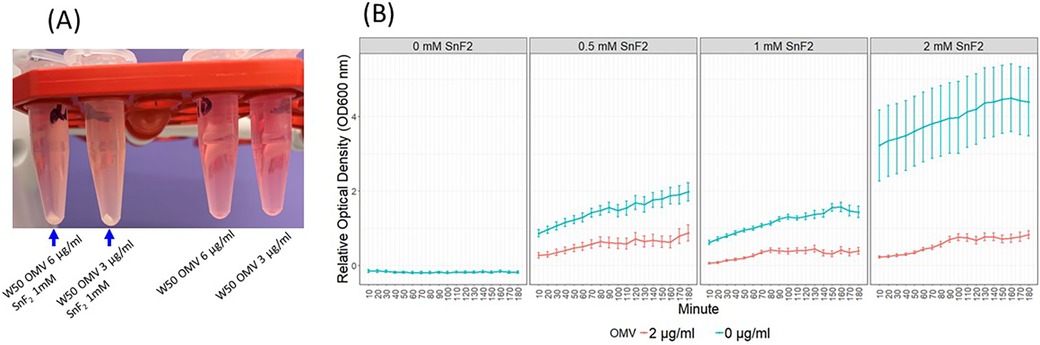
Figure 6. OMVs of P. gingivalis and SnF2 formed precipitates. (A) SnF2 binds OMVs and forms precipitates. (B) Time course of stannous binding to OMVs. Each time point represents the mean and SE of 24 independent measurements. 0 mM SnF2 was not represented in B since the OD600 was 0. Relative OD600 values were obtained by dividing OD600 by the average OD600 of the 2 µg/ml OMV + 1 mM SnF2 samples at the 60 min timepoint. Mixed effect models were fitted to the relative OD600, and all comparisons were significant at 0.01 after Bonferroni adjustment. These comparisons include SnF2 + OMV vs. SnF2, OMV + 0.5 mM SnF2 vs. OMV, and OMV + 1 mM SnF2 vs. OMV.
3.4 Stannous binds LPS on surface of OMVs directly
The precipitates formed with OMVs in the presence of SnF2 most likely result from interactions with LPS in the OMV membranes, given their high concentration in vesicles formed from P. gingivalis. We previously reported that E. coli LPS binds SnF2. In order to determine whether SnF2 bound directly to LPS on the OMV, we isolated LPS from a crude preparation and mixed it with SnF2. The direct reactivity of SnF2 with LPS from P. gingivalis was analyzed via mass spectrometry (Figure 7). Separation of P. gingivalis LPS following incubation with SnF2 produced signals associated with Sn atoms bound to the deposits. Tin has 10 stable isotopes, the highest number of any element. Three isotopes are most abundant (116 Sn, 118 Sn, 120 Sn), each about 9%. All three major tin isotopes were found to bind LPS, as the LPS peaks were shifted by 116, 118, and 120 Da (Figure 7). The molar ratio of the formed complexes appeared to be 1:1 (one stannous ion and one LPS molecule). Supplementary Figure S4 shows evaluations of both SnF2 and SnCl2 with ultrapure E. coli LPS. Results observed with P. gingivalis LPS and E. Coli LPS are similar in binding to SnF2.
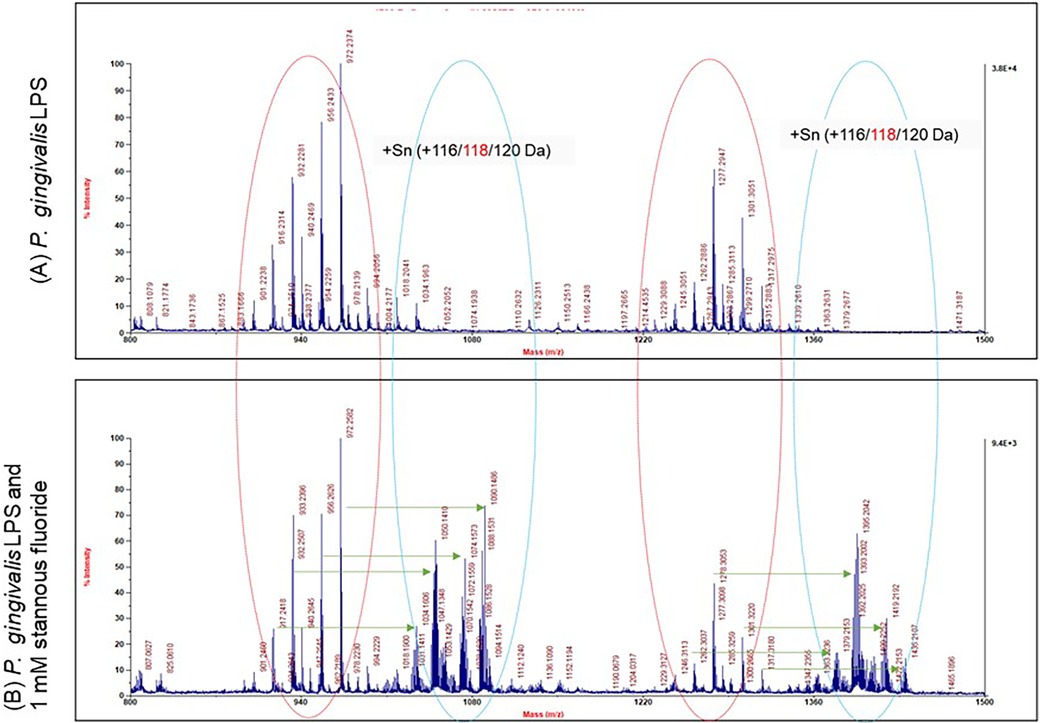
Figure 7. Mass spectrum analysis of stannous and LPS binding. P. gingivalis LPS is highly heterogeneous in size and structure. Each peak represents one molecule of LPS. There were two groups of LPS peaks in the LPS alone panel as indicated by the red circles. With SnF2, two more groups of peaks were generated as represented by the blue circles. The peaks in the blue circles were shifted by 116, 118 or 120 da to the right as indicated by the green arrows.
3.5 Aggregation of P. gingivalis OMVs
Based on the results in Figures 2–7, we propose a working model for how SnF2 inhibits the virulent effects of OMVs on gingival keratinocytes. OMVs are coated with LPS and other surface molecules. These molecules can bind to stannous ions. Stannous ions can also bind to each other, linking two OMVs together. As more OMVs aggregate, the effective concentration of OMVs is reduced. Additionally, larger particles may be less efficient at entering keratinocytes. Consequently, fewer OMVs gain entry into gingival keratinocytes (Figure 8).
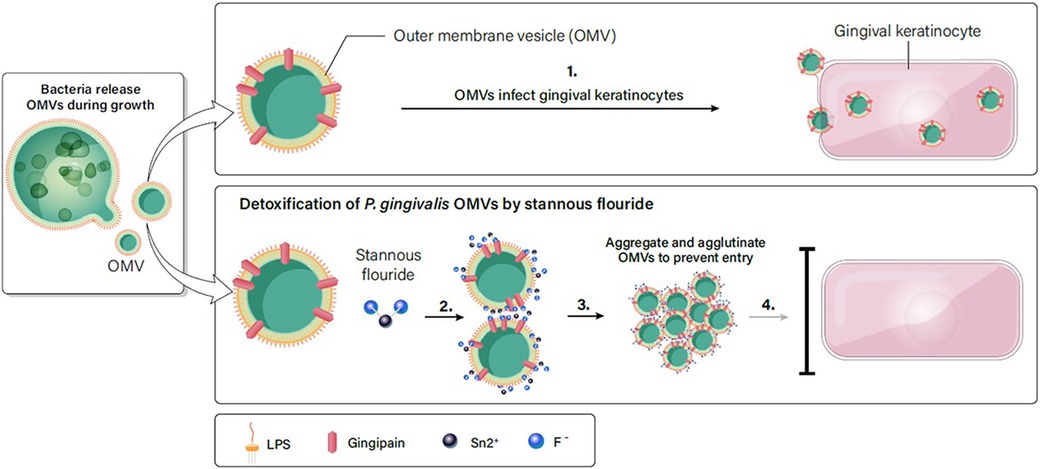
Figure 8. SnF2 interacts with OMVs to form aggregates. (1). OMVs are highly effective in entering gingival keratinocytes. (2). LPS on the surface of OMVs can interact with stannous, which is bivalent. Stannous can bind two OMV structures or bind one OMV and another stannous. (3). As more OMVs are linked together, they form a large aggregate. (4). Large aggregates of OMVs reduce the effective concentration of OMVs in the solution, and retard entry into gingival keratinocytes.
4 Discussion
In this study we examined the reactivity of SnF2 and SnCl2 both in solution and in toothpaste extracts with OMVs produced by P. gingivalis cultures. In control experiments, OMVs from P. gingivalis were observed to infect suspensions of gingival keratinocytes and, following further incubation, the exposure of the gingival keratinocyte promoted the formation of ROS associated with promotion of an inflammatory response caused by OMV infection in the cells. ROS are generated in host tissues by the innate immune system in reaction to bacterial colonization, and OMVs provoke responses as illustrated herein. Invading pathogens and the surfaces of the OMVs can be recognized by pattern recognition receptors located on the surface of host cells. In test experiments, exposure of OMVs to SnF2 produced aggregated flocs which precipitated out of solution; these could be easily separated by centrifugation. The mixtures of OMVs with SnF2 solution or with SnF2 dentifrice extract demonstrated decreased infection and diminished development of ROS in gingival keratinocytes. Precipitation of OMVs by SnF2 could decrease their ability to create inflammation and likewise might diminish their migration to contribute to systemic sequalae.
The primary rationale for the efficacy of SnF2 in inhibiting OMVs of P. gingivalis presumably stems from the strong reactivity with endotoxin (LPS) components of the OMVs (Figure 8). OMV membranes are primarily comprised of LPS. In this study, the chemical binding of SnF2 to LPS was confirmed directly with TOF-MS. This complements our previous research where we observed that SnF2 was extremely effective in binding LPS and that treatment with the compound reduced the ability of endotoxin itself to potentiate inflammatory upregulation of toll receptors TLR2 and TLR4. In clinical studies, it was confirmed that SnF2 penetrated and was retained in the gingival sulcus (subgingivally) (42). Retained concentrations of SnF2 were in the same range as those shown in this study to produce deactivation of OMV toxicity. SnF2 reactivity with parent P. gingivalis bacteria produced aggregation in cell membranes of the bacterium and lysis as evidenced by TEM.
Gingival keratinocyte cells comprise the gingival epithelium, which functions as a physical barrier to prevent the invasion of periodontal-linked bacteria (43). P. gingivalis has been shown to infect keratinocytes, impairing cellular migration and proliferation as well as passing through the epithelial barrier into underlying tissues (44). TEM from invasion assays examining P. gingivalis cultured with gingival keratinocytes demonstrated P. gingivalis internalized within the keratinocytes (45). In vitro fluorescence imaging also demonstrated P. gingivalis localization within gingival keratinocytes (46). Bacterial infection appears to be in part mediated by proteolytic enzymes, as inhibition of proteases or deletion of gingipain genes significantly inhibits the ability of P. gingivalis to invade gingival keratinocytes (47, 48).
OMVs of P. gingivalis have gained prominence in the study of virulence factors associated with periodontal disease (26). Following colonization and growth of P. gingivalis, the outer membranes of the bacterium can form discrete OMVs by swelling with the outgrowths dislodging from the bacterium itself. These OMVs present with a protected membrane structure that researchers speculate assist in their evasion of degradation (26). Their numbers and size permit their transport while escaping deactivation by myriad protective factors of the host. A large number of virulence-related molecules are enriched in OMVs, including gingipains, LPS, Mfa5, PPAD, HmuY (49, 50). For example, gingipain concentrations on OMVs are 3 to 5 fold higher than on parent P. gingivalis bacteria (29). Gingipains themselves produce important pathogenicity, for example, in compromising barrier function of periodontal tissues (28). P. gingivalis OMVs enter cells such as gingival keratinocytes more effectively than the intact P. gingivalis bacterial cells (51). P. gingivalis OMVs swiftly enter host keratinocytes via an endocytosis pathway (52). Once OMVs infect gingival keratinocytes, cellular detachment is observed in a dose dependent manner (53). OMVs impair function of keratinocytes by degrading signaling molecules required for cell migration (54). OMVs activate pattern recognition receptors (PRRs) in gingival keratinocytes leading to cell activation, cytokine secretion and apoptosis (55). Specifically, OMV infection of keratinocytes stimulates increased expression of pro-inflammatory mediators including COX-2, IL-6, IL-8, MMP-1 and MMP-3 (56).
Contemporary research suggests that P. gingivalis OMVs may potentially influence a variety of systemic diseases through their transport to various distant target organs (Figure 9). P. gingivalis OMVs can migrate through the epithelial barrier and into the underlying capillary beds introducing OMVs into the circulatory system, carrying OMVs to distant tissues and organs (57). They promote increased vascular permeability and enhance vascular edema through proteolytic damage to endothelial connexins (58). P. gingivalis OMVs also have powerful platelet aggregation activity, which is an important mechanism in the formation of atherosclerotic plaque formation (59, 60), and they carry gingipain to the liver and alter glucose metabolism in the liver, promoting the development of Diabetes Mellitus (61). P. gingivalis OMV derived LPS can activate glial cells, induce brain inflammation, and correlate with the expression of Alzheimer's Disease marker proteins and neorfibrillary tangles (62). A proposed model for the potential relationship between OMV infection of gingival keratinocytes and subsequent infection of OMVs into underlying connective tissue and capillary beds with increasing systemic exposure is shown in Figure 9. Further research is needed to evaluate the role that P. gingivalis OMVs play in the initiation and progression of periodontal diseases and their relevance to putative oral disease—systemic disease linkage. Definitive evidence will require randomized, well controlled clinical research targeting the role of OMVs in disease pathogenesis. Deactivation of OMVs could present an important target for chemotherapeutics in prevention of periodontal diseases and limiting systemic exposure to OMVs of oral origin.
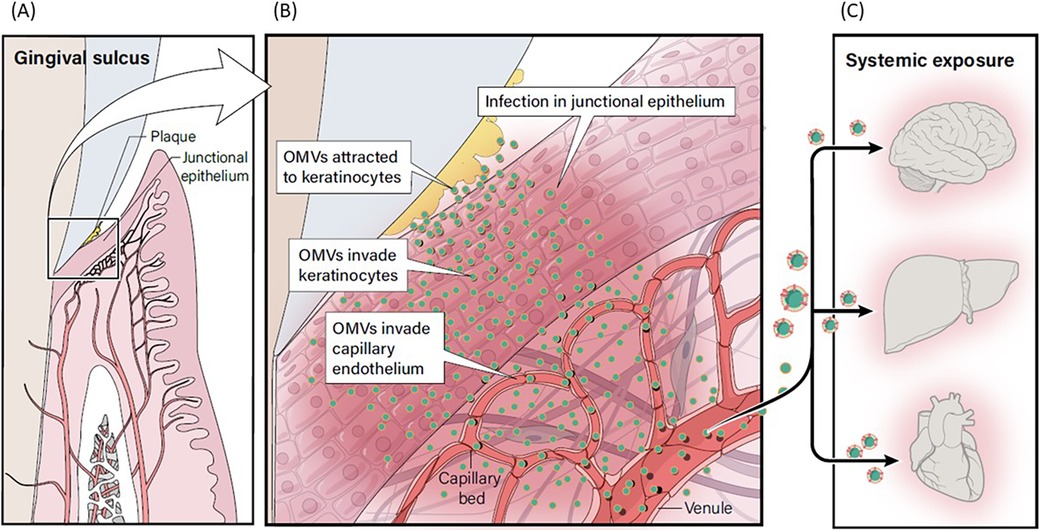
Figure 9. Proposed model for relationship between OMV infection and systemic exposure. (A) P. gingivalis within plaque in the gingival sulcus produces OMVs. (B) OMVs adhere to gingival keratinocytes, leading to invasion of keratinocytes. (C) OMVs can migrate through the disrupted epithelial barrier and into the underlying capillary beds creating a potential pathway to introduce OMVs into the circulatory system where they could be carried to distant tissues and organs, including the brain, liver and heart.
5 Conclusion
This study showed that OMVs from P. gingivalis are an infective and dose-dependent virulence factor. They are shown to invade host keratinocyte cells inducing the production of ROS by the keratinocytes. OMVs contain large concentrations of LPS and, in fact, endotoxins isolated from culture media are likely derived from OMVs rather than parent bacteria. Stannous and 0.454% SnF2 toothpaste can bind both OMVs and LPS, inhibiting the infection of keratinocytes and induction of ROS production. Reactivity of OMV LPS with stannous is likely to aggregate OMVs together, thereby precipitating out in the solution. Large aggregates of OMVs reduce infectivity.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SX: Conceptualization, Investigation, Visualization, Writing – original draft. CT: Investigation, Writing – review & editing. JA: Investigation, Writing – review & editing. FG: Conceptualization, Visualization, Writing – review & editing. NBR: Conceptualization, Supervision, Writing – review & editing. VI: Conceptualization, Investigation, Writing – review & editing. YS: Conceptualization, Investigation, Writing – review & editing. NR: Conceptualization, Writing – review & editing. AB: Conceptualization, Supervision, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by The Procter & Gamble Company.
Acknowledgments
The authors thank Paul Sagel, Ross Strand, Donald J. White, and Beth Jordan for contributions to data interpretation; Donald J. White for manuscript development; Chelsea Tan for contributions to the methodology; Xingtao Wei for help analyzing results and preparing Figure 6; and Lisa Sagel for assistance with manuscript editing and review.
Conflict of interest
All authors are employees of The Procter & Gamble Company, a manufacturer of SnF2 dentifrice.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2024.1492369/full#supplementary-material
References
1. Nazir MA. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int J Health Sci. (2017) 11(2):72–80.
2. Nazir M, Al-Ansari A, Al-Khalifa K, Alhareky M, Gaffar B, Almas K. Global prevalence of periodontal disease and lack of its surveillance. Sci World J. (2020) 2020:2146160. doi: 10.1155/2020/2146160
3. Page RC. Gingivitis. J Clin Periodontol. (1986) 13(5):345–59. doi: 10.1111/j.1600-051x.1986.tb01471.x
4. Bamashmous S, Kotsakis GA, Kerns KA, Leroux BG, Zenobia C, Chen D, et al. Human variation in gingival inflammation. Proc Natl Acad Sci U S A. (2021) 118(27):e2012578118. doi: 10.1073/pnas.2012578118
5. Slots J. Periodontitis: facts, fallacies and the future. Periodontol 2000. (2017) 75(1):7–23. doi: 10.1111/prd.12221
6. Kinane DF. Causation and pathogenesis of periodontal disease. Periodontol 2000. (2001) 25:8–20. doi: 10.1034/j.1600-0757.2001.22250102.x
7. Marsh PD. Dental diseases–are these examples of ecological catastrophes? Int J Dent Hyg. (2006) 4 Suppl 1:3–10; discussion 50–2. doi: 10.1111/j.1601-5037.2006.00195.x
8. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. (1998) 25(2):134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x
9. Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. (2010) 8(7):481–90. doi: 10.1038/nrmicro2337
10. Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. (2012) 91(9):816–20. doi: 10.1177/0022034512453589
11. Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. (2014) 15(6):768–78. doi: 10.1016/j.chom.2014.05.012
12. Hajishengallis G, Chavakis T, Lambris JD. Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontol 2000. (2020) 84(1):14–34. doi: 10.1111/prd.12331
13. Curtis MA, Diaz PI, Van Dyke TE. The role of the microbiota in periodontal disease. Periodontol 2000. (2020) 83(1):14–25. doi: 10.1111/prd.12296
14. Van Dyke TE, Bartold PM, Reynolds EC. The nexus between periodontal inflammation and dysbiosis. Front Immunol. (2020) 31(11):511. doi: 10.3389/fimmu.2020.00511
15. Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. (2012) 27(6):409–19. doi: 10.1111/j.2041-1014.2012.00663.x
16. Charpentier LA, Dolben EF, Hendricks MR, Hogan DA, Bomberger JM, Stanton BA. Bacterial outer membrane vesicles and immune modulation of the host. Membranes. (2023) 13(9):752. doi: 10.3390/membranes13090752
17. Jan AT. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front Microbiol. (2017) 8:1053. doi: 10.3389/fmicb.2017.01053
18. Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. (1999) 181(16):4725–33. doi: 10.1128/JB.181.16.4725-4733.1999
19. Kikuchi Y, Obana N, Toyofuku M, Kodera N, Soma T, Ando T, et al. Diversity of physical properties of bacterial extracellular membrane vesicles revealed through atomic force microscopy phase imaging. Nanoscale. (2020) 12(14):7950–9. doi: 10.1039/c9nr10850e
20. Sheikh A, Zechmann B, Sayes CM, Taube JH, Greathouse KL. A preparation of bacterial outer membrane with osmium tetroxide and uranyl acetate co-stain enables improved structural determination by transmission electron microscopy. Microscopy. (2023) 72(6):515–9. doi: 10.1093/jmicro/dfad027
21. Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. (1999) 65(5):1843–8. doi: 10.1128/AEM.65.5.1843-1848.1999
22. Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. (1987) 55(1):111–7. doi: 10.1128/iai.55.1.111-117.1987
23. Williams GD, Holt SC. Characteristics of the outer membrane of selected oral Bacteroides species. Can J Microbiol. (1985) 31(3):238–50. doi: 10.1139/m85-046
24. Cecil JD, O'Brien-Simpson NM, Lenzo JC, Holden JA, Chen YY, Singleton W, et al. Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS One. (2016) 11(4):e0151967. doi: 10.1371/journal.pone.0151967
25. Pérez-Cruz C, Delgado L, López-Iglesias C, Mercade E. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One. (2015) 10(1):e0116896. doi: 10.1371/journal.pone.0116896
26. Zhang Z, Liu D, Liu S, Zhang S, Pan Y. The role of Porphyromonas gingivalis outer membrane vesicles in periodontal disease and related systemic diseases. Front Cell Infect Microbiol. (2021) 10:585917. doi: 10.3389/fcimb.2020.585917
27. Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. (2011) 286(2):1269–76. doi: 10.1074/jbc.M110.185744
28. Takeuchi H, Nakamura E, Yamaga S, Amano A. Porphyromonas gingivalis infection induces lipopolysaccharide and peptidoglycan penetration through gingival epithelium. Front Oral Health. (2022) 3:845002. doi: 10.3389/froh.2022.845002
29. Mantri CK, Chen CH, Dong X, Goodwin JS, Pratap S, Paromov V, et al. Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. Microbiologyopen. (2015) 4(1):53–65. doi: 10.1002/mbo3.221
30. Xie S, Iberi V, Boissy Y, Tansky CS, Huggins T, Ramji N, et al. Stannous fluoride forms aggregates between outer and inner membranes leading to membrane rupture of Porphyromonas gingivalis and Prevotella pallens. Front Oral Health. (2024) 5:1427008. doi: 10.3389/froh.2024.1427008
31. Haught C, Xie S, Circello B, Tansky CS, Khambe D, Klukowska M, et al. Lipopolysaccharide and lipoteichoic acid virulence deactivation by stannous fluoride. J Clin Dent. (2016) 27(3):84–9.28390203
32. Haught JC, Xie S, Circello B, Tansky CS, Khambe D, Sun Y, et al. Lipopolysaccharide and lipoteichoic acid binding by antimicrobials used in oral care formulations. Am J Dent. (2016) 29(6):328–32.29178720
33. Huggins T, Haught JC, Xie S, Tansky CS, Klukowska M, Miner MC, et al. Quantitation of endotoxin and lipoteichoic acid virulence using toll receptor reporter gene. Am J Dent. (2016) 29(6):321–7.29178719
34. Westphal O, Jann K. Bacterial lipopolysaccharides extraction with phenol-water and further applications of the procedure. Meth Carb Chem. (1965) 5:83–91.
35. Darveau RP, Hancock RE. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. (1983) 155(2):831–8. doi: 10.1128/jb.155.2.831-838.1983
36. Hirayama C, Sakata M, Nakamura M, Ihara H, Kunitake M, Todokoro M. Preparation of poly(epsilon-lysine) adsorbents and application to selective removal of lipopolysaccharides. J Chromatogr B Biomed Sci Appl. (1999) 721(2):187–95. doi: 10.1016/s0378-4347(98)00479-4
37. Sakata M, Todokoro M, Kai T, Kunitake M, Hirayama C. Effect of cationic polymer adsorbent pKa on the selective removal of endotoxin from an albumin solution. Chromatographia. (2001) 53:619–23. doi: 10.1007/BF02493008
38. Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. (1991) 63(24):1193A–203A. doi: 10.1021/ac00024a002
39. Mcilwaine C, Strachan A, Harrington Z, Jerreat M, Belfield LA, Sandor V, et al. Comparative analysis of total salivary lipopolysaccharide chemical and biological properties with periodontal status. Arch Oral Biol. (2020) 110:104633. doi: 10.1016/j.archoralbio.2019.104633
40. Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. (2004) 72(9):5041–51. doi: 10.1128/IAI.72.9.5041-5051.2004
41. Nativel B, Couret D, Giraud P, Meilhac O, d'Hellencourt CL, Viranaïcken W, et al. Porphyromonas gingivalis lipopolysaccharides act exclusively through TLR4 with a resilience between mouse and human. Sci Rep. (2017) 7(1):15789. doi: 10.1038/s41598-017-16190-y
42. Klukowska MA, Ramji N, Combs C, Milleman JL, Milleman KR, Ramsey DL, et al. Subgingival uptake and retention of stannous fluoride from dentifrice: gingival crevicular fluid concentrations in sulci post-brushing. Am J Dent. (2018) 31(4):184–8.30106533
43. Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front Biosci. (2007) 12:3965–74. doi: 10.2741/2363
44. Kato T, Kawai S, Nakano K, Inaba H, Kuboniwa M, Nakagawa I, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. (2007) 3:753–65. doi: 10.1111/j.1462-5822.2006.00825.x
45. Duncan MJ, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. (1993) 61(5):2260–5. doi: 10.1128/iai.61.5.2260-2265
46. Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. (1999) 1(3):215–23. doi: 10.1046/j.1462-5822.1999.00022.x
47. Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. (1995) 63(10):3878–85. doi: 10.1128/iai.63.10.3878-3885.1995
48. Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by porphrymonas gingivalis. Infect Immun. (2004) 72:4689–98. doi: 10.1128/IAI.72.8.4689-4698
49. Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O'Brien-Simpson NM, et al. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. (2014) 13(5):2420–32. doi: 10.1021/pr401227e
50. Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One. (2011) 6(6):e21372. doi: 10.1371/journal.pone.0021372; Erratum in: PLoS One. (2018) 13(8):e0203154. doi: 10.1371/journal.pone.0203154.21731719
51. Ho MH, Chen CH, Goodwin JS, Wang BY, Xie H. Functional advantages of Porphyromonas gingivalis vesicles. PLoS One. (2015) 10(4):e0123448. doi: 10.1371/journal.pone.0123448
52. Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun. (2009) 77(10):4187–96. doi: 10.1128/IAI.00009-09
53. Nakao R, Takashiba S, Kosono S, Yoshida M, Watanabe H, Ohnishi M, et al. Effect of Porphyromonas gingivalis outer membrane vesicles on gingipain-mediated detachment of cultured oral epithelial cells and immune responses. Microbes Infect. (2014) 16(1):6–16. doi: 10.1016/j.micinf.2013.10.005
54. Furuta N, Takeuchi H, Amano A. Entry of Porphrymonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. (2009) 77:4761–70. doi: 10.1128/IAI.00841-09
55. Cecil JD, Sirisaengtaksin N, O’Brien-Simpson NM, Krachler AM. Outer membrane vesicle—host cell interactions. Microbiol Spectr. (2019) 7(1):201–14. doi: 10.1128/microbiolspec.PSIB-0001-2018
56. Kou Y, Inaba H, Kato T, Tagashira M, Honma D, Kanda T, et al. Inflammatory responses of gingival epithelial cells stimulated with Porphyromonas gingivalis vesicles are inhibited by hop-associated polyphenols. J Periodontol. (2008) 79(1):174–80. doi: 10.1902/jop.2008.070364
57. Aguayo S, Schuh CMAP, Vicente B, Aguayo LG. Association between Alzheimer’s disease and oral and gut microbiota: are pore forming proteins the missing link? J Alzheimers Dis. (2018) 65(1):29–46. doi: 10.3233/JAD-180319
58. Farrugia C, Stafford GP, Murdoch C. Porphyromonas gingivalis outer membrane vesicles increase vascular permeability. J Dent Res. (2020) 99(13):1494–501. doi: 10.1177/0022034520943187
59. Okamura H, Hirota K, Yoshida K, Weng Y, He Y, Shiotsu N, et al. Outer membrane vesicles of Porphyromonas gingivalis: novel communication tool and strategy. Jpn Dent Sci Rev. (2021) 57:138–46. doi: 10.1016/j.jdsr.2021.07.003
60. Sharma A, Novak EK, Sojar HT, Swank RT, Kuramitsu HK, Genco RJ. Porphyromonas gingivalis platelet aggregation activity: outer membrane vesicles are potent activators of murine platelets. Oral Microbiol Immunol. (2000) 15(6):393–6. doi: 10.1034/j.1399-302x.2000.150610.x
61. Seyama M, Yoshida K, Yoshida K, Fujiwara N, Ono K, Eguchi T, et al. Outer membrane vesicles of Porphyromonas gingivalis attenuate insulin sensitivity by delivering gingipains to the liver. Biochim Biophys Acta Mol Basis Dis. (2020) 1866(6):165731. doi: 10.1016/j.bbadis.2020.165731
Keywords: stannous fluoride, antibacterial agents, reactive oxygen species, Porphyromonas gingivalis, keratinocyte infection, periodontal diseases, scanning electron microscopy, transmission electron microscopy
Citation: Xie S, Tansky CS, Ashe J, Gao F, Ramji NB, Iberi V, Sun Y, Ramji N and Biesbrock AR (2024) Stannous fluoride protects gingival keratinocytes against infection and oxidative stress by Porphyromonas gingivalis outer membrane vesicles. Front. Dent. Med 5:1492369. doi: 10.3389/fdmed.2024.1492369
Received: 6 September 2024; Accepted: 30 October 2024;
Published: 19 November 2024.
Edited by:
Rhythm Bains, King George's Medical University, IndiaReviewed by:
Rok Gašperšič, University of Ljubljana, SloveniaMayank Hans, ESIC Medical College, India
Copyright: © 2024 Xie, Tansky, Ashe, Gao, Ramji, Iberi, Sun, Ramji and Biesbrock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aaron R. Biesbrock, Ymllc2Jyb2NrLmFyQHBnLmNvbQ==
 Sancai Xie
Sancai Xie Cheryl S. Tansky1
Cheryl S. Tansky1 Fei Gao
Fei Gao Vighter Iberi
Vighter Iberi Yiping Sun
Yiping Sun Aaron R. Biesbrock
Aaron R. Biesbrock