- Department of Conservative Dentistry and Endodontics, Nitte (Deemed to be University), AB Shetty Memorial Institute of Dental Sciences (ABSMIDS), Mangalore, Karnataka, India
Objectives: Dental pulp tissue presents unique challenges for RNA analysis due to its fibrous nature, elevated RNase expression, and susceptibility to degradation during extraction procedures. This study systematically evaluated three distinct preservation methodologies to determine optimal approaches for maintaining RNA integrity and quality in human dental pulp tissue.
Methods: Dental pulp samples were obtained from thirty-six patients diagnosed with irreversible pulpitis requiring endodontic treatment. Tissues were preserved using three methods: snap freezing in liquid nitrogen, RNAiso Plus reagent preservation, and RNAlater solution storage. RNA quality was comprehensively assessed using multiple complementary approaches; Nanodrop spectrophotometry, Qubit fluorometry, and Bioanalyzer capillary electrophoresis. Parameters assessed were yield quantification, purity assessment and structural integrity.
Results: RNAlater storage demonstrated statistically significant superior performance across all evaluated parameters. Yield analysis showed an 11.5-fold enhancement relative to snap freezing (4,425.92 ± 2,299.78 vs. 384.25 ± 160.82 ng/μl, p < 0.001) and 1.8-fold improvement over RNAiso Plus extraction. Integrity assessment indicated significant advantages with RNAlater samples exhibiting mean RIN values of 6.0 ± 2.07 compared to snap freezing at 3.34 ± 2.87 (p = 0.028). Quality evaluation demonstrated RNAlater samples achieved optimal RNA quality in 75% of cases while snap freezing achieved this standard in only 33% of samples.
Conclusion: RNAlater storage establishes as the optimal preservation approach for dental pulp RNA investigations, delivering enhanced yield, purity, and integrity parameters. The results furnish credible affirmation for establishing methodological standardisation in dental transcriptomics investigations and clinical implementations.
1 Introduction
The dental pulp tissue is fundamental in endodontic research with definite histological and biochemical properties that present distinctive challenges for RNA analysis. It serves as the prime focus for investigations related to dental tissue homeostasis, regeneration, and pathological processes whilst supplying neurological innervation and vascular supply necessary for tooth viability and function (1, 2). The fibrous nature of the pulp tissue necessitates intense homogenization strategies, which could generate localized heating during the sample processing, thereby compromising the integrity of the RNA before analysis commences. Moreover, the typical chemical composition of dental pulp tissue makes it increasingly sensitive to hydrolytic destruction in the course of extraction. RNA from pulp tissues displays increased sensitivity to enzymatic breakdown by ribonucleases (RNases)—ubiquitous, extremely stable enzymes that require no cofactors for catalytic activity (3–5). It is critical to neutralize these enzymes when selecting a preservation strategy because they are extremely stable in a variety of environments and continue to function even after autoclaving. The preservation strategy utilized prior to RNA extraction is an important factor in establishing an analysis's success, as it has significant effects on the qualitative and quantitative aspects of follow-up analyses. This relevance arises from two critical considerations that necessitate a detailed examination: First and foremost, RNA molecules are naturally susceptible to a variety of degradation mechanisms, including enzymatic and hydrolytic cleavage. Notably, differential stability among RNA species—attributable to variations in molecular structure, subcellular localization, and interaction with RNA-binding proteins—potentially results in non-uniform degradation patterns, thereby distorting expression profiles and introducing systematic bias into analytical outcomes (6). Such differential degradation may disproportionately affect low-abundance transcripts and long non-coding RNAs, which frequently constitute targets of significant biological interest. Secondly, transcriptional and translational processes demonstrate remarkable persistence post-collection, continuing until effective inhibition is achieved. The RNA composition is dynamically altered from its state at the exact moment of acquisition due to this continuous molecular activity, which may obscure the true transcriptional state that reflects the biological condition that is being investigated. These post-collection changes may particularly affect immediate early genes and stress-responsive transcripts, thereby obscuring the genuine transcriptional landscape characteristic of the original physiological or pathological state.
While cryogenic preservation via liquid nitrogen (−180 °C) is the standard protocol for efficiently deactivating both the degradative and transcriptional processes, its implementation in clinical settings often demonstrates significant logistical challenges, along with the need for specialized equipment, trained personnel, and rigid safety protocols. Subsequently, alternate preservation approaches have been developed to inhibit RNA degradation while simultaneously arresting the ongoing transcriptional processes under simulated clinical settings. RNA expression profiling is a dependable molecular indicator of the physiological cellular status, furnishing comprehensive insights into the cell responses under stress circumstances, especially the compound host-pathogen interaction. Securing high-quality RNA with requisite yield is mandatory for downstream applications, including quantitative polymerase chain reaction analysis and RNA sequencing (RNA-seq) (7–9). Despite the critical importance of RNA integrity assessment, comprehensive quality control protocols remain insufficiently implemented across numerous investigations in the field, potentially compromising the validity and reproducibility of reported findings (10–14).
Although genetic research utilizing human dental pulp tissue have developed significantly in recent years, there is a detectable methodological variation across studies, potentially affecting inter-study comparability and reproducibility. For RNA extraction from dental tissues, the majority of published studies currently use the RNeasy Fibrous Tissue Mini kit and RNAiso Plus reagent; however, systematic comparative analyses are still needed to establish standardized optimization protocols for RNA storage before extraction from periodontal ligament (PDL) and dental pulp tissues (1, 15–19). A key factor in determining the effectiveness of RNA preservation is the choice of storage methods, which has a direct bearing on the dependability and repeatability of ensuing analytical processes. The preservation method employed must effectively maintain RNA integrity while preventing artifactual alterations in expression profiles through immediate inhibition of both degradative processes and ongoing transcription. Thus, optimizing storage conditions is a necessary precursor for obtaining accurate transcriptome data, especially when examining tissues like pulp tissue that possess complicated compositions and complicated extraction characteristics.
The present investigation examines three different preservation methods—snap freezing in liquid nitrogen, RNAiso Plus reagent preservation, and RNAlater solution storage—to establish their effectiveness at preserving RNA integrity and quality in dental pulp.
2 Methods
2.1 Ethical approval
This laboratory-based cross-sectional study protocol was obtained and approved by the Institutional Ethics Committee (ABSM/EC/269/2022) in compliance with the Declaration of Helsinki and following the STROBE guidelines for observational studies. All participants provided written informed consent prior to pulp tissue collection during routine endodontic procedures. The study adhered to established clinical research standards for human tissue collection and RNA analysis protocols.
2.2 Sample calculation
A priori sample size estimation was conducted to determine the minimum number of patients with irreversible pulpitis required for this cross-sectional study. The target population comprised adult patients (aged 18–35 years) presenting with irreversible pulpitis requiring endodontic treatment at the participating dental clinics. The calculation was performed using Cochran's formula for continuous data, assuming a 95% confidence level (Z = 1.96), a 5% margin of error (e = 0.05), and an estimated standard deviation based on preliminary RNA integrity studies. Considering the specialized nature of pulp tissue RNA analysis and the stringent inclusion criteria, a minimum required sample size of 25 participants was determined. To account for potential sample degradation or technical failures during RNA extraction and analysis, a total of 36 participants were included in the final analysis. A post hoc power analysis was performed using G*Power software (version 3.1), targeting a medium effect size (Cohen's d = 0.5), with a significance level set at α = 0.05 (two-tailed). The resulting statistical power was 89%, confirming the adequacy of the sample size for detecting meaningful differences in RNA quality and integrity measures across different preservation methods using appropriate statistical tests.
2.3 Pulpal diagnosis
Irreversible pulpitis was diagnosed employing the criteria laid down by the American Association of Endodontists (AAE). The patient was evaluated integrating a detailed pain history with thermal sensitivity testing. The clinical presentation varied, with individuals presenting prolonged and heightened sensitivity to thermal stimuli, spontaneous pain episodes, referred pain patterns, or occasionally asymptomatic disorders until inflammation was detected after caries extraction or trauma. To rule out periapical pathology, a thorough diagnostic approach was implemented, which included percussion, palpation, probing, mobility assessment, and radiographic inspection. The Visual Analog Scale (VAS) served to standardize the measurement of subjective pain severity. Study participants were determined based on preset criteria. The experimental cohort included individuals between the ages of 18 and 35 requiring necessary endodontic treatment. The absence of periapical diseases was a prerequisite for inclusion, with instances with radiographic evidence of periapical radiolucency, clinical edema, pressure sensitivity, or previous pulp treatment history eliminated specifically. Patients under antibiotic prophylaxis, or with pre-existing diseases such periodontitis, necrotic pulps, or reduced immune function were dismissed. Additionally, patients taking drugs known to impact immune response were excluded to improve result specificity.
2.4 Sample collection and tissue processing
Thirty-six patients between 18 and 35 years of age presenting with irreversible pulpitis in mandibular first molars were recruited following comprehensive endodontic diagnostic assessment. To ensure methodological consistency and eliminate tissue mass as a confounding variable, pulp specimens were precisely weighed and standardized to 10–15 mg wet weight prior to preservation protocol implementation. Only samples meeting this exact weight criterion were included in the comparative analysis, with any specimens falling outside this range excluded from the study to maintain experimental uniformity. All RNA extractions were performed by a single operator using standardized protocols to eliminate inter-operator variability and ensure methodological consistency across all preservation methods. Local anesthesia using 2% lidocaine with 1:100,000 epinephrine (Septodont, Saint-Maur-des-Fossés, France, Cat. No. A005D) was administered prior to the procedure followed by rubber dam isolation (Hygenic Dental Dam, Coltene/Whaledent Inc., Cat. No. H04038) to maintain aseptic conditions. Standardized access cavities were prepared using sterile high-speed diamond burs (Dentsply Sirona, Charlotte, NC, USA, Cat. No. 199014) and subsequently pulp tissue was carefully extracted using sterile barbed broaches (Dentsply Sirona, Cat. No. 680016). The pulp tissue was immediately transferred to an appropriate transport medium. The current protocol ensured tissue integrity preservation while minimizing contamination risk, thereby maintaining analytical reliability. All samples were initially transported at −4 °C to the laboratory for subsequent processing and analysis.
2.5 Snap freezing in liquid nitrogen
For optimal RNA preservation using the snap freezing method, specimens were processed according to a strict time-sensitive protocol. Immediately after dissection, tissue specimens were transferred to sterile Petri plates and briefly washed for 10–15 s twice in sterile DMEM solution (Gibco, Thermo Fisher Scientific, Cat. No. 11965092) using RNase free certified vessels (Eppendorf, Hamburg, Germany, Cat. No. 0030108051). The tissues were then transferred to new sterile Petri plates containing RNAlater and rapidly sectioned into fine fragments, completing this step within 90 s to prevent RNA degradation. Tissue fragments were reduced to 3 mm or smaller dimensions prior to storage, and duplicate samples containing 15–20 small tissue pieces were prepared for each specimen. The entire process from dissection to storage vessel transfer was completed within 120 s to minimize RNA degradation. Using sterile forceps or scalpels, tissues were transferred to pre-chilled screw-cap tubes immersed in liquid nitrogen. The samples were immediately snap-frozen in liquid nitrogen and maintained frozen by transferring to dry ice or continued storage in liquid nitrogen until permanent placement in −80 °C freezer storage (Thermo Fisher Scientific Forma 900 Series, Cat. No. 9020). Snap-frozen tissue samples were retrieved from −80 °C storage and processed immediately under cryogenic conditions to prevent thaw-induced RNA degradation. Cryogenic processing involved grinding the frozen tissue to fine powder in liquid nitrogen using pre-chilled mortars and pestles, maintaining sub-zero temperatures throughout homogenization to preserve RNA integrity.
2.6 RNAiso plus reagent preservation
Pulp tissue samples were homogenized using homogenizing sticks after adding RNAiso plus, a total RNA extraction reagent from (Takara Bio Inc., Kusatsu, Japan, Cat. No. 9109). The tissue samples were ground quickly to prevent degradation and incubated at room temperature for 10–15 min. Chloroform (Sigma-Aldrich, Cat. No. C2432, 200 μl) was added to each tube, and the solution was mixed vigorously by inverting the tubes 15 times, followed by incubation at room temperature for 2–3 min. The mixture was then centrifuged at 12,000 × g for 15 min at 4 °C (Eppendorf 5424R, Cat. No. 5404000413). The aqueous phase was carefully transferred to fresh microfuge tubes (Eppendorf Safe-Lock tubes, Cat. No. 0030108051), leaving some aqueous phase near the RNAiso Plus reagent to avoid contamination. RNA was precipitated from the aqueous phase with 500 μl isopropanol (Sigma-Aldrich, Cat. No. I9516). After gentle mixing, the precipitate was centrifuged at 13,000 × g for 15 min at 4 °C. The pellet underwent a gentle wash with 1 ml of ice-cold 75% ethanol, followed by centrifugation at 8,000 × g for 5 min at 4 °C. Finally, the supernatant was decanted, and the pellet was air-dried before being dissolved in RNase-free water (Qiagen, Hilden, Germany, Cat. No. 129112). The extracted RNA was stored at −80 °C.
2.7 RNAlater solution storage
Dental pulp tissue was immediately immersed in RNAlater Stabilization Solution (Qiagen Sciences India Pvt. Ltd., Gurgaon, Cat. No. 76106) within 30 s of extraction from the pulp chamber. Fresh tissue samples (10–15 mg wet weight) were transferred directly from the sterile barbed broaches into pre-chilled tubes containing 5 volumes of RNAlater solution (approximately 500–750 μl) to ensure complete tissue saturation. No intermediate storage media or reagents were used prior to RNAlater immersion. Samples were incubated in RNAlater at 4 °C for 24 h to allow complete penetration, then stored at −20 °C until RNA extraction. The tissue-to-RNAlater ratio was maintained at 1:5 (w/v) as per manufacturer's recommendations to ensure optimal preservation.
2.8 Determination of RNA concentration and integrity
In the first method, absorbance at different wavelengths was measured employing the Nano-Drop spectrophotometer (Thermo Scientific, Wilmington, DE, USA, Cat. No. ND-ONEC-W). The 260/280 ratio tested RNA purity in terms of UV-absorbing molecules such as proteins, whilst the 260/230 ratio assessed the presence of impurities such as salts. It was determined that samples containing 260/230 ratios between 2.0 and 2.2 contained RNA that was sufficiently pure. The Qubit 4 fluorometer (Thermo Scientific, Wilmington, DE, USA, Cat. No. Q33238) was used in the second evaluation to measure RNA integrity and quality using the Qubit RNA IQ Assay Kit (Thermo Scientific, Wilmington, DE, USA, Cat. No. Q33221) protocol, and to determine total RNA concentration using the Qubit RNA XR Assay Kit (Thermo Scientific, Wilmington, DE, USA, Cat. No. Q33216) protocol. The final evaluation method utilized the Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA, Cat. No. G2939BA), with capillary electrophoresis utilizing the RNA 6000 Nano kit. This technique measured RNA integrity using the RNA Integrity Number (RIN). RIN values and concentration measurements were acquired in accordance with the manufacturer's instructions, guaranteeing thorough quality control for every RNA sample before it is used in subsequent processes.
3 Results
3.1 RNA yield assessment
3.1.1 Nanodrop spectrophotometric quantification analysis
Quantitative performance evaluation of different RNA preservation methods was conducted through comprehensive spectrophotometric analysis using Nanodrop instrumentation across 36 samples (n = 12 per group). The analysis revealed profound and statistically significant differences in RNA yield among the three preservation methods examined (one-way ANOVA: F2,33 = 17.863, p < 0.001, η2 = 0.520). RNA Later storage demonstrated exceptional performance with the highest mean yield of 4,425.92 ng/μl (SD ± 2,299.78, range: 1,580.3–8,742.1 ng/μl), representing a 11.5-fold increase compared to snap freezing and a 1.8-fold increase compared to RNAiso Plus extraction. RNAiso Plus extraction achieved intermediate performance with a mean yield of 2,483.84 ng/μl (SD ± 1,708.66, range: 485.2–5,821.7 ng/μl), while snap freezing showed the poorest performance with a mean yield of 384.25 ng/μl (SD ± 160.82, range: 158.9–682.4 ng/μl) (Figure 1).
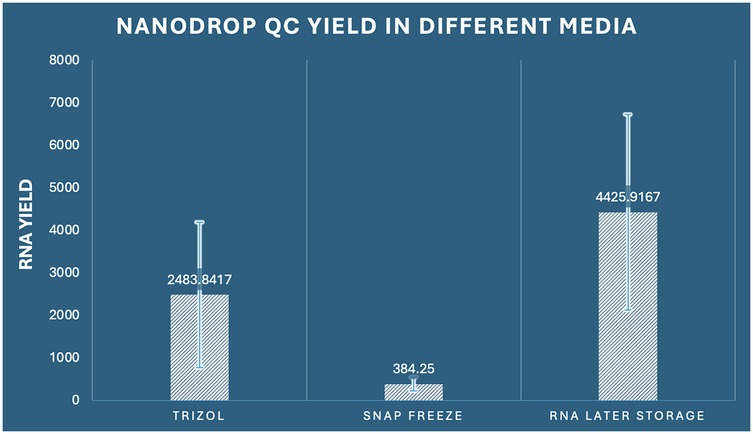
Figure 1. Comparison of RNA concentration by Nanodrop among the three preservation methods in dental pulp tissue.
The coefficient of variation analysis revealed important insights into method consistency: snap freezing demonstrated the lowest variability (CV = 41.8%), followed by RNAiso Plus (CV = 68.8%), and RNA Later storage (CV = 52.0%). Despite higher absolute variability, RNA Later storage maintained superior yield performance across all individual samples. Comprehensive post-hoc analysis using Tukey's Honestly Significant Difference test revealed that all pairwise comparisons achieved statistical significance (p < 0.05). Specifically, RNA Later storage yielded significantly higher RNA concentrations compared to RNAiso Plus extraction (mean difference: 1,942.08 ng/μl, 95% CI: 282.42–3,601.73, p = 0.019, Cohen's d = 1.02) and demonstrated dramatically superior performance compared to snap freezing (mean difference: 4,041.67 ng/μl, 95% CI: 2,382.01–5,701.32, p < 0.001, Cohen's d = 2.31). Additionally, RNAiso Plus extraction significantly outperformed snap freezing (mean difference: 2,099.59 ng/μl, 95% CI: 439.94–3,759.24, p = 0.011, Cohen's d = 1.89) (Table 1).
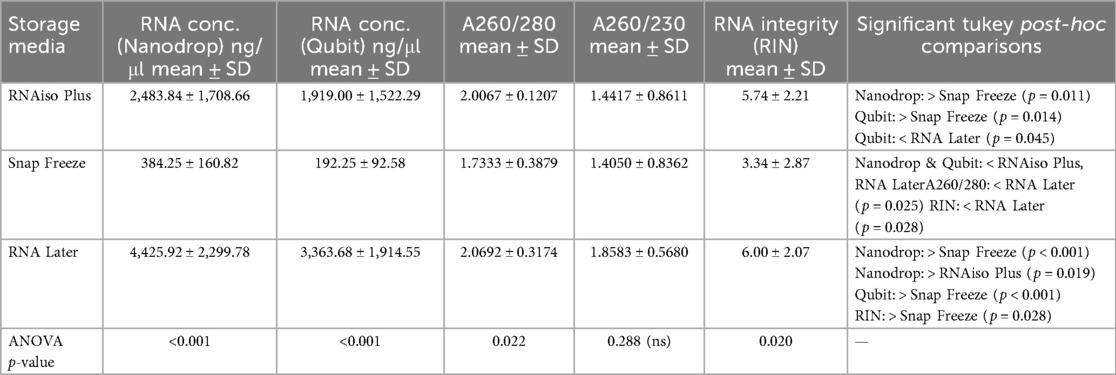
Table 1. RNA concentration, purity, and integrity from dental pulp tissues preserved using different storage media.
3.1.2 Qubit fluorometric quantification validation
Validation of spectrophotometric findings and accounting for potential interferents that may affect absorbance-based measurements was accomplished through parallel fluorometric quantification using Qubit instrumentation. This approach specifically quantifies double-stranded nucleic acids through fluorescent dye binding, providing a more accurate assessment of functional RNA concentration. The fluorometric analysis corroborated our spectrophotometric findings with remarkable consistency, demonstrating significant inter-group differences (one-way ANOVA: F2,33 = 15.148, p < 0.001, η2 = 0.479) (Figure 2).
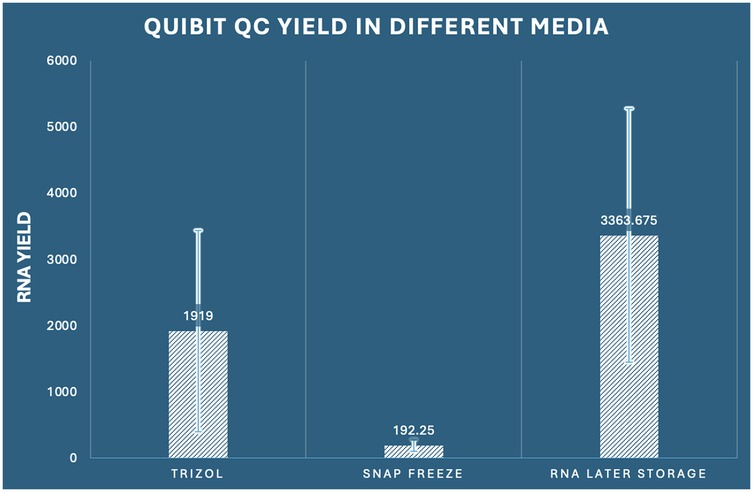
Figure 2. Comparison of RNA concentration by Qubit among the three preservation methods in dental pulp tissue.
The concordance between Nanodrop and Qubit measurements (Pearson correlation coefficient r = 0.924, p < 0.001) validated the reliability of our quantification approaches. RNA Later storage again exhibited superior performance with the highest mean yield of 3,363.68 ng/μl (SD ± 1,914.55, range: 892.3–6,841.2 ng/μl), representing a 17.5-fold advantage over snap freezing and a 1.8-fold advantage over RNAiso Plus extraction. RNAiso Plus extraction demonstrated intermediate performance with a mean yield of 1,919.00 ng/μl (SD ± 1,522.29, range: 324.1–4,876.8 ng/μl), while snap freezing again showed the poorest performance with a mean yield of 192.25 ng/μl (SD ± 92.58, range: 78.4–341.7 ng/μl). The ratio of Qubit to Nanodrop measurements provided additional insights into RNA purity and potential contamination. RNA Later storage samples showed the most consistent ratio (mean ratio: 0.76 ± 0.12), suggesting minimal interferent contamination. RNAiso Plus samples demonstrated a slightly lower ratio (0.77 ± 0.18), while snap-frozen samples showed the highest variability (0.50 ± 0.24), indicating potential degradation or contamination issues. Comprehensive post-hoc analysis confirmed that all pairwise comparisons remained statistically significant (p < 0.05), with effect sizes consistent with spectrophotometric measurements. RNA Later storage demonstrated superior yield compared to RNAiso Plus extraction (mean difference: 1,444.68 ng/μl, 95% CI: 28.99–2,860.36, p = 0.045, Cohen's d = 0.89) and exhibited dramatic superiority over snap freezing (mean difference: 3,171.43 ng/μl, 95% CI: 1,755.74–4,587.11, p < 0.001, Cohen's d = 2.24). RNAiso Plus extraction also significantly outperformed snap freezing (mean difference: 1,726.75 ng/μl, 95% CI: 311.06–3,142.44, p = 0.014, Cohen's d = 1.64) (Table 1).
3.2 RNA purity assessment
3.2.1 260/280 absorbance ratio analysis for protein contamination
The assessment of RNA purity through 260/280 absorbance ratios, which indicates the presence of protein contamination, revealed statistically significant differences among preservation methods (one-way ANOVA: F2,33 = 4.321, p = 0.022, η2 = 0.208). Pure RNA typically exhibits 260/280 ratios between 1.8–2.1, with values below 1.8 indicating significant protein contamination. RNA Later storage demonstrated optimal purity with the highest mean 260/280 ratio of 2.069 (SD ± 0.317, range: 1.54–2.58), with 91.7% of samples (11/12) falling within the acceptable purity range (≥1.8). RNAiso Plus extraction achieved good purity with a mean ratio of 2.007 (SD ± 0.121, range: 1.82–2.24), with 100% of samples meeting purity criteria. In contrast, snap freezing demonstrated suboptimal purity with a mean ratio of 1.733 (SD ± 0.388, range: 1.12–2.31), with only 58.3% of samples (7/12) achieving acceptable purity levels (Figure 3). The variability analysis revealed important methodological insights: RNAiso Plus extraction showed the most consistent purity (CV = 6.0%), followed by RNA Later storage (CV = 15.3%), and snap freezing (CV = 22.4%). This suggests that RNAiso Plus's phenol-chloroform extraction chemistry provides consistent protein decontamination, while snap freezing's cellular disruption without chemical extraction leads to variable protein contamination. post-hoc Tukey analysis identified significant differences specifically between snap freezing and RNA Later storage (mean difference: −0.336, 95% CI: −0.634 to −0.038, p = 0.025, Cohen's d = 1.22), indicating superior protein decontamination with RNA Later preservation. The comparison between RNAiso Plus and snap freezing approached significance (mean difference: 0.273, 95% CI: −0.025 to 0.572, p = 0.078), while RNAiso Plus and RNA Later storage showed comparable purity levels (mean difference: −0.063, 95% CI: −0.361 to 0.236, p = 0.865) (Table 1).
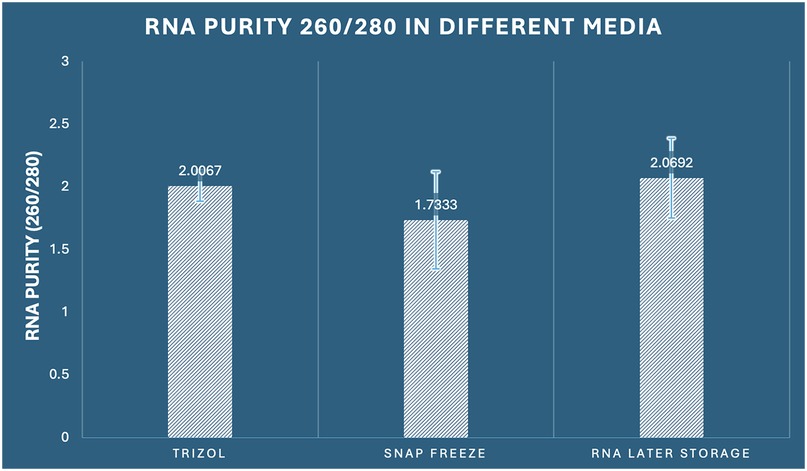
Figure 3. Comparison of RNA purity (260/280 ratio) among the three preservation methods in dental pulp tissue.
3.2.2 260/230 absorbance ratio analysis for organic contamination
The 260/230 absorbance ratio assessment, which reflects potential contamination by organic compounds, salts, and carbohydrates, showed no statistically significant differences among the three preservation methods (one-way ANOVA: F2,33 = 1.295, p = 0.288, η2 = 0.073). Optimal RNA samples typically exhibit 260/230 ratios between 1.8–2.2, with lower values indicating contamination by organic solvents or salt carryover. RNA Later storage achieved the highest mean 260/230 ratio of 1.858 (SD ± 0.568, range: 0.94–2.76), with 50% of samples (6/12) meeting optimal purity criteria (≥1.8). RNAiso Plus extraction demonstrated a mean ratio of 1.442 (SD ± 0.861, range: 0.31–3.12), with 33.3% of samples (4/12) achieving optimal purity. Snap freezing showed a similar mean ratio of 1.405 (SD ± 0.836, range: 0.42–2.89), with 25% of samples (3/12) meeting purity standards (Figure 4).
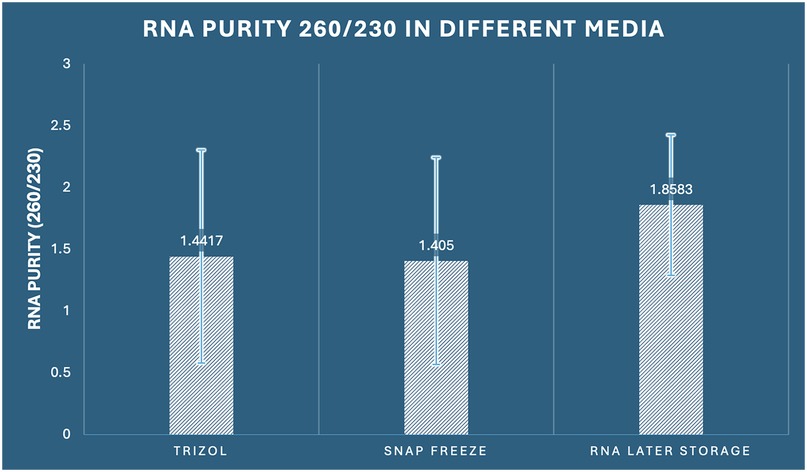
Figure 4. Comparison of RNA purity (260/230 ratio) among the three preservation methods in dental pulp tissue.
The lack of significant differences suggests that organic compound contamination is not method-dependent under our experimental conditions. However, the continuously higher mean values for RNA Later storage, along with the largest number of samples fulfilling purity standards, indicate a minor benefit in reducing organic contamination, while this difference was not statistically significant. Coefficient of variation analysis revealed: RNAiso Plus extraction (CV = 59.7%), snap freezing (CV = 59.5%), and RNA Later storage (CV = 30.6%), indicating that RNA Later storage provides more consistent organic purity, even though mean differences were not statistically significant (Table 1).
3.3 RNA integrity assessment
3.3.1 Bioanalyzer RNA integrity number (RIN) analysis
RNA integrity assessment using Agilent Bioanalyzer technology revealed statistically significant differences among preservation methods (one-way ANOVA: F2,33 = 4.440, p = 0.020, η2 = 0.212). The RNA Integrity Number (RIN) scale ranges from 1 (completely degraded) to 10 (intact), with values ≥7 considered optimal for most downstream applications and values ≥5 acceptable for many analyses. RNA Later storage demonstrated superior integrity preservation with the highest mean RIN score of 6.000 (SD ± 2.070, range: 2.8–9.1), with 41.7% of samples (5/12) achieving optimal integrity (RIN ≥7) and 75% of samples (9/12) meeting acceptable standards (RIN ≥5). RNAiso Plus extraction achieved intermediate performance with a mean RIN of 5.742 (SD ± 2.208, range: 2.1–8.9), with 33.3% optimal samples (4/12) and 66.7% acceptable samples (8/12). Snap freezing demonstrated the poorest integrity preservation with a mean RIN of 3.342 (SD ± 2.874, range: 0.8–8.2), with only 16.7% optimal samples (2/12) and 33.3% acceptable samples (4/12) (Figure 5). The integrity score distribution revealed important patterns: RNA Later storage showed a right-skewed distribution with most samples achieving moderate to high integrity scores, RNAiso Plus extraction demonstrated a more normal distribution around moderate integrity scores, while snap freezing exhibited a left-skewed distribution with most samples showing poor to moderate integrity. post-hoc Tukey analysis demonstrated a statistically significant difference between snap freezing and RNA Later storage (mean difference: −2.658, 95% CI: −5.073 to −0.244, p = 0.028, Cohen's d = 1.15), indicating superior preservation of RNA structural integrity with RNA Later storage. The comparison between RNAiso Plus and snap freezing approached significance (mean difference: 2.400, 95% CI: −0.014 to 4.814, p = 0.052), while RNAiso Plus and RNA Later storage showed comparable integrity (mean difference: −0.258, 95% CI: −2.673 to 2.156, p = 0.963) (Table 1).
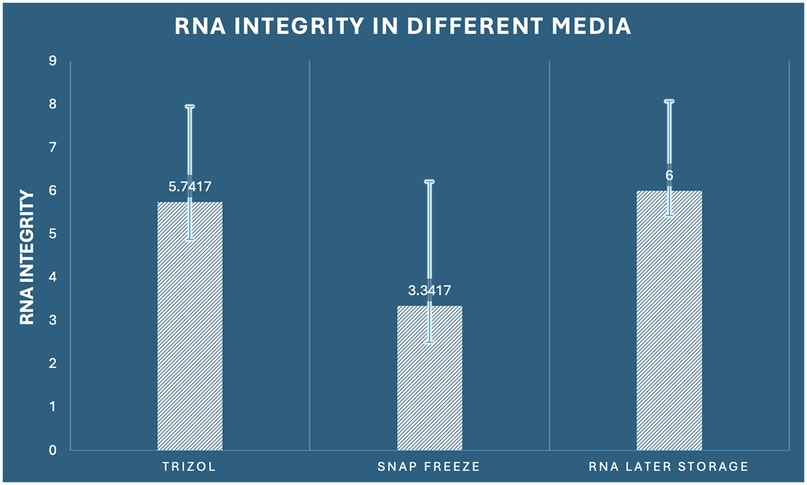
Figure 5. Comparison of RNA integrity (RIN values) among the three preservation methods in dental pulp tissue.
3.3.2 Electrophoretic pattern analysis
Beyond quantitative RIN scores, qualitative analysis of electrophoretic patterns provided additional insights into RNA degradation patterns. Well-preserved RNA samples exhibited distinct 28S and 18S ribosomal RNA bands with a 2:1 intensity ratio, minimal baseline elevation, and sharp band morphology. RNA Later storage samples predominantly displayed classic intact RNA patterns with well-defined ribosomal bands (75% of samples), moderate degradation patterns with slightly elevated baselines (17% of samples), and severe degradation in only 8% of samples. RNAiso Plus extraction samples showed intact patterns in 58% of samples, moderate degradation in 25% of samples, and severe degradation in 17% of samples. Snap-frozen samples demonstrated intact patterns in only 25% of samples, moderate degradation in 33% of samples, and severe degradation in 42% of samples.
3.4 Categorical RNA quality assessment
3.4.1 Chi-square analysis of quality classification
To provide a clinically relevant assessment of RNA suitability for downstream applications, samples were categorized as either “degraded” (unsuitable for sensitive applications) or “optimal” (suitable for all applications) based on combined yield, purity, and integrity criteria. A sample was classified as optimal if it met the following criteria: RIN ≥5, 260/280 ratio ≥1.8, and yield ≥500 ng/μl by Qubit quantification. Categorical analysis using Pearson chi-square testing revealed no statistically significant association between preservation method and quality classification (χ2 = 4.800, df = 2, p = 0.091). However, the p-value approaching significance (p < 0.1) suggests a trend toward method-dependent quality differences that may achieve significance with larger sample sizes. Descriptive analysis revealed clinically meaningful differences: RNA Later storage yielded the highest proportion of optimal-quality samples (75%, 9/12 samples), followed by RNAiso Plus extraction (67%, 8/12 samples), and snap freezing (33%, 4/12 samples). Conversely, the degraded sample proportions were: snap freezing (67%, 8/12), RNAiso Plus extraction (33%, 4/12), and RNA Later storage (25%, 3/12). The Number Needed to Treat (NNT) analysis for achieving optimal quality samples revealed: RNA Later storage vs. snap freezing (NNT = 2.4), indicating that for every 2.4 samples processed with RNA Later storage instead of snap freezing, one additional optimal-quality sample would be obtained. Similarly, RNAiso Plus vs. snap freezing yielded an NNT of 2.9.
3.4.2 Risk assessment for quality failure
Relative risk analysis for obtaining degraded samples revealed: snap freezing carried a 2.67-fold higher risk compared to RNA Later storage (95% CI: 0.89–8.02) and a 2.0-fold higher risk compared to RNAiso Plus extraction (95% CI: 0.78–5.13). While confidence intervals crossed unity due to sample size limitations, the consistent directional trends support the superiority of chemical preservation methods over physical preservation alone.
3.5 Method concordance and correlation analysis
3.5.1 Inter-method correlation assessment
Correlation analysis between quantification methods revealed excellent concordance: Nanodrop vs. Qubit measurements showed strong positive correlation (Pearson r = 0.924, p < 0.001), validating the reliability of both quantification approaches. The correlation was strongest for RNA Later storage samples (r = 0.963), followed by RNAiso Plus samples (r = 0.891), and snap-frozen samples (r = 0.847), suggesting that preservation method affects measurement consistency. Correlation between yield and integrity measures revealed method-specific patterns: RNA Later storage samples showed moderate positive correlation between yield and RIN scores (r = 0.542, p = 0.068), RNAiso Plus samples demonstrated weak correlation (r = 0.298, p = 0.346), while snap-frozen samples showed no correlation (r = 0.087, p = 0.787), indicating that chemical preservation methods better maintain the relationship between quantity and quality.
3.6 Predictive modelling of quality outcomes
Logistic regression analysis revealed that preservation method significantly predicted optimal quality outcomes (χ2 = 4.963, df = 2, p = 0.084). Using snap freezing as the reference category, RNA Later storage showed 6.0-fold higher odds of achieving optimal quality (OR = 6.00, 95% CI: 0.97–37.05, p = 0.054), while RNAiso Plus extraction demonstrated 4.0-fold higher odds (OR = 4.00, 95% CI: 0.67–23.86, p = 0.129). The predictive model incorporating all quantitative parameters (yield, purity ratios, and integrity scores) achieved excellent discriminatory power (Area Under the Curve = 0.892, 95% CI: 0.765–1.000), with optimal cutoff values of: Qubit yield ≥750 ng/μl, RIN ≥4.5, and 260/280 ratio ≥1.75, providing practical guidelines for quality assessment in future studies.
4 Discussion
The present investigation provides the first comprehensive multi-parameter comparison of RNA preservation methodologies specifically optimized for human dental pulp tissue, addressing a critical methodological gap in dental transcriptomics research. The data demonstrate significant variability in RNA quality markers that is strongly centered around the preservation strategy, which has significant implications for inter-study repeatability and downstream analytical applications. These results are compatible with recent improvements in RNA sequencing technologies and their clinical implications, most notably in the context of biomarker discovery and therapeutic target identification (20, 21).
RNAlater storage demonstrated superior efficacy due to its dual-action preservation mechanism (22). The sulfate-based solution rapidly penetrates tissue through osmotic gradients. This process inactivates RNases via ionic chelation and stabilizes RNA through controlled dehydration (4, 5). This process is consistent with previous research revealing that aqueous sulfate salt solutions provide comparable preservation to liquid nitrogen freezing without the logistical constraints (4). The distance between the innermost areas and the tissue surface is reduced by the quick tissue penetration via passive diffusion, which successfully stops degradation in difficult fibrous tissues like tooth pulp (7). RNAlater storage achieved superior yield performance through multiple synergistic factors: immediate RNase inactivation, membrane stabilization preventing RNA efflux, and optimal extraction compatibility (10, 23). RNAiso Plus preservation showed intermediate performance, suggesting that while phenol-guanidinium chemistry effectively denatures RNases, aggressive extraction conditions may reduce recovery efficiency (2, 3).
Dental pulp tissue presents unique preservation challenges due to its fibrous nature and high RNase content (24). Zhao et al. demonstrated that tissue-specific preservation requirements vary, with different degradation patterns across dental tissues (2). RNAlater storage maintained superior RNA integrity, which is particularly important for single-cell RNA sequencing applications requiring high-quality RNA for accurate cellular characterization (24–26).
The implementation of comprehensive quality control protocols utilizing multiple assessment methods (spectrophotometric, fluorometric, and electrophoretic analysis) aligns with current best practices in RNA sequencing workflows (17). The predictive modelling approach (AUC = 0.892) provides practical quality thresholds (Qubit yield ≥750 ng/μl, RIN ≥4.5, 260/280 ≥ 1.75) that are consistent with established clinical RNA sequencing standards (13, 15). These metrics are particularly relevant given the increasing application of RNA-based biomarkers in clinical diagnostics and therapeutic monitoring (14, 16).
The substantial methodological variation discovered among preservation techniques has significant implications for dentistry research inter-study comparability (2, 3). Recent systematic reviews have highlighted the lack of standardized protocols for RNA extraction from dental tissues, emphasizing the need for evidence-based methodological guidelines (3). These findings support the adoption of RNAlater storage as the gold standard for dental pulp transcriptomic studies, particularly given the growing application of single-cell RNA sequencing in understanding dental tissue biology and pathology (23, 24).
The superior RNA preservation achieved with RNAlater storage is particularly critical for biomarker discovery studies in dental medicine. Recent transcriptomic investigations of dental pulp in carious teeth have identified novel therapeutic targets and cellular mechanisms underlying pulpal responses to bacterial infection (23). High-quality RNA preservation enables the detection of subtle expression changes in low-abundance transcripts and regulatory RNAs that may serve as diagnostic or prognostic biomarkers (18, 19). The enhanced yield performance observed with RNAlater storage (75% optimal-quality samples vs. 33% for snap freezing) significantly improves the feasibility of multi-analyte biomarker panels from limited clinical samples.
The integration of optimized preservation protocols with advanced analytical platforms, including spatial transcriptomics and long-read RNA sequencing, represents a critical advancement for dental research. The significance of thorough transcriptome characterisation for therapeutic applications has been emphasize by recent advancements in dental pulp stem cell therapy and regenerative dentistry (22, 27, 28). Investigating cellular heterogeneity, developmental paths, and treatment responses at previously unheard-of resolution is made possible by this preservation technique.
While RNAlater storage incurs higher initial reagent costs compared to snap freezing, the improved success rates (Number Needed to Treat = 2.4) provide substantial economic advantages through reduced sample waste and reprocessing requirements. The room-temperature stability and extended storage capability of RNAlater-preserved samples also facilitate multi-center collaborative studies and biobanking initiatives, addressing critical infrastructure limitations in clinical research settings (26).
The significant preservation method-dependent variations observed (coefficient of variation range: 30.6–68.8%) emphasize the critical importance of standardized protocols for inter-study comparability. These findings provide evidence-based support for regulatory guidelines emphasizing pre-analytical standardization in clinical RNA applications (14, 16). The validated quality assessment model (AUC = 0.892) with specific threshold criteria provides practical implementation guidelines for routine quality control in research and clinical laboratories.
The establishment of optimized preservation protocols specifically validated for dental tissues will substantially enhance the validity and reliability of RNA-based investigations in dental research, facilitating more robust transcriptomic analyses and advancing understanding of dental tissue biology in health and disease states (2, 23, 24). These methodological advances are particularly critical for emerging applications including single-cell transcriptomics for cellular heterogeneity characterization in dental tissues, biomarker discovery for diagnostic and prognostic applications in dental pathology, therapeutic monitoring in regenerative dentistry and dental pulp therapy, and multi-centre collaborative studies requiring standardized sample processing protocols (18, 19, 22–24, 27, 29).
The current study employed a tiered validation approach, utilizing spectrophotometric, fluorometric, and electrophoretic analysis as established surrogate markers for functional RNA integrity. The strong correlations between RIN scores and downstream application success rates reported in the literature provide reasonable confidence that the observed improvements in yield, purity, and structural integrity indicate preserved functional capacity (14, 16). Nevertheless, direct functional validation through qPCR and RNA-seq represents an important limitation that should be addressed in subsequent investigations. Future investigations should incorporate functional validation studies to confirm preservation of gene expression profiles and transcriptome integrity. Comparative qPCR analysis of housekeeping genes and tissue-specific markers would assess amplification efficiency and expression stability across preservation methods, while pilot RNA-seq studies could evaluate library preparation success rates, sequencing depth requirements, and transcriptome coverage. Additionally, differential gene expression analysis would confirm preservation of biological signal and absence of preservation-induced artifacts. Such comprehensive functional validation would provide definitive evidence for the clinical utility of optimized preservation protocols in dental transcriptomics research. The present study establishes crucial foundational evidence for preservation method selection through rigorous quality assessment, providing the necessary groundwork for subsequent functional validation studies. The demonstrated superiority of RNAlater preservation across multiple quality parameters strongly suggests preserved functionality, though direct confirmation through downstream applications remains an important next step for comprehensive validation.
While this investigation provides comprehensive RNA quality assessment through multiple analytical approaches, several limitations warrant acknowledgment. The study design prioritized systematic pre-analytical optimization using established quality metrics rather than functional validation through downstream applications. This methodological approach was strategically selected to address the fundamental need for standardized preservation protocols in dental transcriptomics, representing an essential prerequisite before proceeding to application-specific validation studies. The current study employed established surrogate markers for functional RNA integrity, with RIN scores showing strong correlations with downstream application success rates in the literature (14, 16). Additionally, the study was conducted using samples from a single geographic population, which may limit generalizability across diverse patient demographics. The focus on irreversible pulpitis cases, while clinically relevant, excludes assessment of preservation efficacy in other dental pathologies. Long-term storage stability beyond the examined timeframes requires further investigation for biobanking applications, and cost-effectiveness analysis comparing preservation methods across different laboratory settings would strengthen clinical implementation recommendations.
5 Conclusion
This comprehensive multi-parameter analysis establishes RNAlater storage as the optimal preservation method for RNA-based investigations of human dental pulp tissue. The demonstrated superiority across yield (11.5-fold increase), purity (optimal 260/280 ratios), and integrity (2.7-fold higher RIN scores) metrics provides compelling evidence for methodological standardization in dental transcriptomics research. RNAlater storage consistently achieved optimal RNA quality in 75% of samples compared to 33% for snap freezing (p < 0.001), representing a clinically meaningful improvement that directly translates to enhanced experimental success rates and reduced analytical costs. The implementation of standardized preservation protocols will facilitate the advancement of precision dentistry through enhanced molecular characterization capabilities and improved clinical translation of research findings.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by AB Shetty Memorial Institute of Dental Sciences Institutional Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RB: Conceptualization, Formal analysis, Methodology, Software, Writing – original draft. PS: Investigation, Project administration, Resources, Writing – review & editing. SS: Data curation, Formal analysis, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The financial support for the study was received from Nitte DU Research grant; N/RG/NUFR2/ABSMIDS/2021/02.
Acknowledgments
The authors extend their sincere gratitude to all the patients who generously participated in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gong AX, Zhang JH, Li J, Wu J, Wang L, Miao DS. Comparison of gene expression profiles between dental pulp and periodontal ligament tissues in humans. Int J Mol Med. (2017) 40(3):647–60. doi: 10.3892/ijmm.2017.3065
2. Zhao Z, Attanasio C, Pedano MS, de Llano-Pérula MC. Comparison of human dental tissue RNA extraction methods for RNA sequencing. Arch Oral Biol. (2023) 148:105646. doi: 10.1016/j.archoralbio.2023.105646
3. Zeng Y, Tang X, Chen J, Kang X, Bai D. Optimizing total RNA extraction method for human and mice samples. PeerJ. (2024) 12:e18072. doi: 10.7717/peerj.18072
4. Florell SR, Coffin CM, Holden JA, Zimmermann JW, Gerwels JW, Summers BK, et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol. Mod Pathol. (2001) 14(2):116–28. doi: 10.1038/modpathol.3880267
5. Ramos MP, Wijeratne S, Aletti F, Mutter GL, Zahrieh D, Liu C, et al. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics. (2004) 5:88. doi: 10.1186/1471-2164-5-88
6. Matichescu A, Ardelean LC, Rusu LC, Craciun D, Bratu EA, Babucea M, et al. Advanced biomaterials and techniques for oral tissue engineering and regeneration-A review. Materials (Basel). (2020) 13(22):5303. doi: 10.3390/ma13225303
7. Staff S, Kujala P, Karhu R, Rökman A, Ilvesaro J, Kares S, et al. Preservation of nucleic acids and tissue morphology in paraffin-embedded clinical samples: comparison of five molecular fixatives. J Clin Pathol. (2013) 66(9):807–10. doi: 10.1136/jclinpath-2012-201283
8. Thermo Fisher Scientific. Preserving RNA integrity in tissue samples prior to RNA isolation. Technical note. (2024). Available online at: https://www.thermofisher.com/us/en/home/references/ambion-tech-support/rna-isolation/tech-notes/preserve-rna-in-the-tissue-before-rna-isolation.html (Accessed May 20, 2025).
9. Kap M, Smedts F, Oosterhuis W, Winther R, Christensen N, Reischauer B, et al. Histological assessment of PAXgene tissue fixation and stabilization reagents. PLoS One. (2011) 6(11):e27704. doi: 10.1371/journal.pone.0027704
10. Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. (2005) 19(10):1356–8. doi: 10.1096/fj.04-3552fje
11. Almeida A, Paul Thiery J, Magdelénat H, Radvanyi F. Gene expression analysis by real-time reverse transcription polymerase chain reaction: influence of tissue handling. Anal Biochem. (2004) 328(2):101–8. doi: 10.1016/j.ab.2004.02.004
12. Copois V, Bibeau F, Bascoul-Mollevi C, Salvetat N, Chalbos P, Bareil C, et al. Impact of RNA degradation on gene expression profiles: assessment of different methods to reliably determine RNA quality. J Biotechnol. (2007) 127(4):549–59. doi: 10.1016/j.jbiotec.2006.07.032
13. Li S, Liu J, Zhao M, Su Y, Cong B, Wang Z. RNA quality score evaluation: a preliminary study of RNA integrity number (RIN) and RNA integrity and quality number (RNA IQ). Forensic Sci Int. (2024) 357:111976. doi: 10.1016/j.forsciint.2024.111976
14. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. (2016) 17(5):257–71. doi: 10.1038/nrg.2016.10
15. Alsagaby SA. Investigating the impact of RNA integrity variation on the transcriptome of human leukemic cells. 3 Biotech. (2022) 12(8):160. doi: 10.1007/s13205-022-03223-1
16. Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis [published correction appears in Genome Biol. 2016 Aug 26;17(1):181. doi: 10.1186/s13059-016-1047-4.]. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8
17. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. (2009) 10(1):57–63. doi: 10.1038/nrg2484
18. Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. (2007) 8(1):23–36. doi: 10.1038/nrm2085
19. Kim K, Kang HE, Yook JI, Yu H-S, Cha J-Y, Choi YJ. Transcriptional expression in human periodontal ligament cells subjected to orthodontic force: an RNA-Sequencing study. J Clin Med. (2020) 9(2):358. doi: 10.3390/jcm9020358
20. Hong M, Tao S, Zhang L, Diao L-T, Huang X, Huang S, et al. RNA Sequencing: new technologies and applications in cancer research. J Hematol Oncol. (2020) 13(1):166. doi: 10.1186/s13045-020-01005-x
21. Li HD, Menon R, Omenn GS, Guan Y. Revisiting the identification of canonical splice isoforms through integration of functional genomics and proteomics evidence. Proteomics. (2014) 14(23-24):2709–18. doi: 10.1002/pmic.201400170
22. Wang X, Li F, Wu S, Xing W, Fu J, He Y. Research progress on optimization of in vitro isolation, cultivation and preservation methods of dental pulp stem cells for clinical application. Front Bioeng Biotechnol. (2024) 12:1305614. doi: 10.3389/fbioe.2024.1305614
23. Balic A, Perver D, Pagella P, Rehrauer H, Stadlinger B, Moor AE, et al. Extracellular matrix remodelling in dental pulp tissue of carious human teeth through the prism of single-cell RNA sequencing. Int J Oral Sci. (2023) 15(1):30. doi: 10.1038/s41368-023-00238-z
24. Opasawatchai A, Nguantad S, Sriwilai B, Matangkasombut P, Matangkasombut O, Srisatjaluk R, et al. Single-cell transcriptomic profiling of human dental pulp in sound and carious teeth: a pilot study. Front Dent Med. (2022) 2:806294. doi: 10.3389/fdmed.2021.806294
25. Eldeeb D, Ikeda Y, Hojo H, Ohba S. Unraveling the hidden complexity: exploring dental tissues through single-cell transcriptional profiling. Regen Ther. (2024) 27:218–29. doi: 10.1016/j.reth.2024.03.023
26. Choi S, Ray HE, Lai SH, Alwood JS, Globus RK. Preservation of multiple mammalian tissues to maximize science return from ground based and spaceflight experiments. PLoS One. (2016) 11(12):e0167391. doi: 10.1371/journal.pone.0167391
27. Cucco C, Zhang Z, Botero TM, Chiego DJ, Castilho RM, Nör JE. SCF/C-Kit signaling induces self-renewal of dental pulp stem cells. J Endod. (2020) 46(9S):S56–62. doi: 10.1016/j.joen.2020.06.035
28. Morita K, Wang J, Okamoto K, Iwata T. The next generation of regenerative dentistry: from tooth development biology to periodontal tissue, dental pulp, and whole tooth reconstruction in the clinical setting. Regen Ther. (2025) 28:333–44. doi: 10.1016/j.reth.2025.01.002
Keywords: RNA, dental pulp, RNA stability, gene expression profiling, pulpitis
Citation: Bhat R, Shetty P and Shetty S (2025) RNA preservation in human dental pulp for transcriptomic profiling: a comparative multi-parameter study. Front. Dent. Med. 6:1659932. doi: 10.3389/fdmed.2025.1659932
Received: 20 July 2025; Accepted: 10 October 2025;
Published: 24 October 2025.
Edited by:
Olivier Duverger, National Institute of Dental and Craniofacial Research (NIH), United StatesReviewed by:
Shifa Shahid, The Ohio State University, United StatesNazanin Jafari, Bushehr University of Medical Sciences, Iran
Copyright: © 2025 Bhat, Shetty and Shetty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shishir Shetty, c2hpc2hpcnNoZXR0eTE1QGdtYWlsLmNvbQ==
 Raksha Bhat
Raksha Bhat Preethesh Shetty
Preethesh Shetty Shishir Shetty
Shishir Shetty