- Julius Kühn-Institute (JKI) - Federal Research Centre for Cultivated Plants, Institute for Epidemiology and Pathogen Diagnostics, Braunschweig, Germany
The use of peat alternatives in horticulture is part of an important strategy aiming at the reduction of CO2 emission and the preservation of natural wetlands. Today, different components are proposed as replacement of peat in plant growing media, very often as a heterogeneous mixture of different components. Their diverse origin is responsible for their particular physicochemical characteristics as well as the specific microbial activity. The latter brings, however, the risk of contamination with potentially pathogenic organisms, both to plants and humans. In this study, we assessed the microbial compositions of four different commercially available plant growing substrates, which ranged from 100% to 0% peat. Using amplicon sequencing of the 16S rRNA gene fragment and the ITS region, we revealed the microbial differences among those substrates and the only very minor changes in peat-reduced or peat-free substrates observable over the course of 12 weeks under greenhouse conditions. In addition, we monitored the persistence of the externally added human pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) by direct CFU enumeration on a selective medium. Results obtained in this study suggest that although S. Typhimurium was not able to persist in three of the tested substrates, in the peat-free 2 substrate its persistence may be substantially supported. Our results strengthen therefore the notion that in addition to the structural differences among the different tested substrates, the microbial community composition may add to their functional diversity. This underlines the necessity of considering microbial community compositions regarding the replacement of peat in horticultural substrates.
Introduction
Peat has been the major component of plant growing media since the mid-20th century (Bilderback et al., 2013). It is easily adaptable to the needs of different plant species due to the low pH, high water holding capacity and homogeneous inherent nutrient composition (Carlile et al., 2015; Gruda, 2012). Unfortunately, the extraction of peat for horticultural purposes significantly contributes to the emission of greenhouse gases and bears further ecological threats (Hirschler and Thrän, 2023). The partially or even complete replacement of peat in horticultural substrates was therefore a goal in many political activities and was under intensive research for the last decades. Many of these studies focused on the one hand, on optimization of the substrate composition for specific plant requirements (Nesse et al., 2019; Prisa and Caro, 2023). On the other hand, aspects of economic and ecological sustainability of peat replacers were addressed (Hirschler et al., 2022). Today, widely used peat replacers include green waste composts, wood fiber, composted bark and coconut products such as coir fiber, coir pith or coir chips (Barrett et al., 2016).
Among them, composts as products of microbe mediated oxidation of green or food waste have been found to improve plant growth promoting properties by the enrichment of substrates with nutrients, marginal nitrogen immobilization and introducing plant beneficial microorganisms (Gruda, 2019; Pot et al., 2022). Other peat replacements like wood fiber, composted bark and coconut products can remarkably increase the amount of nitrogen fertilizer required, which is a result of high nitrogen immobilization rates, occurring due to microbial decomposition of the high amounts of available carbon in these materials (Harris et al., 2020; Jackson et al., 2009).
One of the most important constraints of peat replacement is their heterogeneity. A good example for this problem is the heterogeneous composition of green composts (Hirschler et al., 2022; Vandecasteele et al., 2021). Green waste as the source of green compost can be composed of varying proportions of different plant material, leading to changing physicochemical properties. These variabilities are usually amended by organic or mineral fertilizer supply and pH adjustment, respectively (Vandecasteele et al., 2021). Apart from plant growth, substrate origin and properties strongly influence the composition of bacterial and fungal communities (Pot et al., 2022; Neher et al., 2013). Those communities are also influenced by the plant host, by the formation of the plant species-dependent rhizosphere microbiome (Trivedi et al., 2020; Berg and Smalla, 2009). Nevertheless, the composition of bacterial and fungal communities may determine the plant growth promoting function of a respective substrate to a significant extent (Pot et al., 2022). Despite that, the role of microorganisms in horticultural substrates has received little attention.
Apart from plant health, a further aspect of substrate quality is its impact on human health. Those considerations include the potential contamination of substrates with human pathogens like Salmonella enterica or pathogenic Escherichia coli from the family Enterobacteriaceae. Such contaminants originating from livestock can reach horticultural substrates via improperly sanitized irrigation water or organic fertilizers, e. g. manure or digestates (Piveteau et al., 2022; Allende and Monaghan, 2015; Jay-Russell et al., 2014). Long-term persistence of Salmonella in agricultural soils has been reported previously (Jechalke et al., 2019) and further, that its persistence is influenced by indigenous microbial communities, as presented by Schierstaedt et al. (2020) by comparing Salmonella persistence when inoculated to natural or reduced microbial communities in agricultural soils.
To address those safety concerns, the subject of the current study was to assess if the persistence of externally applied Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) depends on the composition of the respective substrate. This study was performed in four different substrates (potting soils) readily available in retail trade. The compositions of these substrates represent different peat levels and choices of peat replacers. Apart from a pure peat substrate used as a control, a peat-reduced and two peat-free alternatives containing coco fiber, wood fiber, green compost and composted bark in different proportions, were used. To gain information about the influence of substrate properties, presence of S. Typhimurium and exposure time on the dynamic of the bacterial and fungal communities, an inoculation experiment was carried out under greenhouse conditions. Further, the investigation of key taxa enabled insights into microbial processes that might account for differences in substrate functions such as potential plant growth promotion or reduction. Hence, we hypothesize that: 1) bacterial and fungal communities show strong distinctions between peat, peat-reduced and the two peat-free substrates and may be more diverse in the peat-reduced and in the peat-free substrates than in the peat-based substrate; 2) externally applied S. Typhimurium persists at different levels in the examined substrates; and that 3) bacterial and fungal taxa differentially abundant in peat, peat-reduced and peat-free substrates may be linked to possible plant growth promoting functions of the respective substrate.
Materials and methods
Experimental design and setup
Substrates
To cover the different levels of peat reduction, four different substrates were purchased. They were termed: “peat”, “peat-reduced”, “peat-free 1” and “peat-free 2”. Peat substrate used in this study consisted of fibrous peat. In the peat-reduced variant the peat fraction of fibrous and sapric peat was mixed with green compost and wood fiber, and supplemented with mineral fertilizer. Peat-free 1 contained coco fiber, wood fiber, green compost and organic fertilizer. Peat-free 2 was a mixture of bark humus, coco fiber, wood fiber, green compost and organic fertilizer.
Bacterial strain preparation
For substrate inoculation (described below) a spontaneous rifampicin-resistant mutant of Salmonella enterica subsp. enterica serovar Typhimurium strain 14028s rifR (DSM 19587; Le Minor and Popoff, 1987; Kauffmann and Edwards, 1952), in the following referred to as S. Typhimurium, was used. The strain was chosen as representing an important cause of human infections. Fresh cell material was gained by streaking LB-agar plates supplemented with 50 mg mL-1 rifampicin (Luria/Miller; Carl Roth GmbH, Karlsruhe, Germany) with stock cultures from 15% glycerol in LB broth (Luria/Miller; Carl Roth GmbH, Karlsruhe, Germany). After incubation at 37°C for 24h, cells were harvested and suspended in sterile 10mM MgCl2. The resulting cell suspension was adjusted to OD600 = 0.01 by dilution with 10mM MgCl2, representing a cell density of 107 colony forming units (CFU)/ml.
Experimental setup
All four substrates were filled into 500 mL glass jars resulting in 70 g dry matter per jar in four replicates, for both the control and Salmonella treatment (32 jars in total). The water content of substrates had been determined in duplicate by gravimetric measurement. To this end, approximately 10 g of moist substrate was weighed and dried at 105°C for 24h in a drying oven. The remaining dry matter was weighed and dry mass calculated accordingly. During the experiment, glass jars were closed by glass lids with rubber rings, which were fixed by metal clamps. The jars were placed in a greenhouse at 20°C and minimum light intensity of 1000 lx from 6 am to 10 pm.
Sampling
For the inoculation treatment, substrates in each jar were supplemented with 7 ml of S. Typhimurium cell suspension (described above) to reach an approximate number of 106 CFU/g dry matter. In the control, 7 mL of 10mM MgCl2 were added. Samples for CFU enumeration (described below), substrate moisture determination (described below) and DNA extraction (described below) were harvested at 0, 7, 14, 21, 28, 56 and 84 days post inoculation (dpi), resulting in a total number of 224 samples.
CFU enumeration and DNA extraction
Measurement of physicochemical substrate properties
The assessment of the physicochemical substrate properties such as contents of organic matter, carbon (C), nitrogen (N), phosphate, magnesium, potassium, several trace elements and density, pH and C/N ratio (Supplementary Table S1), was outsourced to an accredited laboratory (LUFA Nord-West, Oldenburg, Germany, http://www.lufa-nord-west.de), and standard methods were applied as indicated in Supplementary Table S1. It has to be noted that the results of physicochemical measurements were applied to 1 proportion per substrate (4 samples).
Substrate moisture determination
At every sampling time point, substrate moisture of each replicate was determined gravimetrically by weighing approximately 10 g of moist substrate and drying the sample at 105°C for 24h in a drying oven. After weighing the remaining dry matter, water content was calculated, accordingly (Supplementary Figure S1).
CFU enumeration
The persistence of S. Typhimurium was monitored throughout the experiment as previously performed by Jechalke et al. (2019) and Fornefeld et al. (2017). On the day of sampling, each sample was stirred thoroughly. To 1 g of substrate per replicate 9 mL of sterile 10mM MgCl2 were added and shaken using a vortex mixer for 20 seconds (10–1 dilution) and afterwards diluted in sterile 10mM MgCl2. Twenty µl of serial dilutions were dropped in duplicate on selective medium for Salmonella (XLD agar, Carl Roth GmbH, Karlsruhe, Germany) supplemented with 50 mg mL−1 rifampicin. Salmonella reduces sodium thiosulfate from the medium to H2S, which reduces ferric ammonium citrate to black colored iron sulfide. After incubation at 37°C for 24h under aerobic conditions, all colonies appearing black were considered as the inoculated S. Typhimurium and were counted. CFUs were calculated per g substrate dry weight.
Extraction and purification of total community DNA
The extraction of total community DNA was performed using the FastDNA®SPIN Kit for Soil (MP Biomedicals, Santa Ana, California, USA) from 0.5 g moist substrate per sample. Total DNA was purified with the GENECLEAN®SPIN Kit (MP Biomedicals, Santa Ana, California) according to the manufacturer´s instructions. For cell lysis, a FastPrep™-24 (MP Biomedicals, Santa Ana, California, USA) homogenizer was used. Success of the DNA extraction was verified by gel electrophoresis in 0.8% agarose gel, stained with ethidium bromide (0.005%) and photographed under UV-light (Intas Gel jet Imager 2004, Intas, Göttingen, Germany). DNA concentration of samples intended for sequencing of the 16S rRNA and ITS2 gene amplicons were additionally measured photometrically using a Nanodrop™ (Thermo Fisher Scientific Inc., Waltham, Massachusetts).
Quantitative real time PCR of 16S rRNA gene amplicons
To determine the 16S rRNA gene (rrn gene) copy numbers in samples from 0, 14, 28 and 56 days after inoculation, qPCR was performed using primers and TaqMan probe specific for the domain Bacteria, as described by Suzuki et al. (2000). Briefly, the DNA standard was produced by ligation of a 16S rDNA fragment into a pGEM-T® vector (Promega GmbH, Walldorf, Germany) and transformation of competent Escherichia coli JM109 cells (Promega GmbH, Walldorf, Germany) following the manufacturer´s instructions. Plasmid DNA extraction from successfully transformed clones, that were picked from selective medium, was performed with the GeneJet Plasmid MiniPrep Kit (Thermo Fisher Scientific Inc., Waltham, Massachusetts) as indicated in the product manual. After restriction with EcoR1, the correct size of the inserts was confirmed by agarose-gel electrophoresis (described above). DNA concentration and purity were measured photometrically using a Nanodrop™ (Thermo Fisher Scientific Inc., Waltham, Massachusetts) and the concentration was adjusted to approximately 26 ng/µl. With a reaction volume of 50 µl, 40 cycles of 1 min at 95°C and 1 min at 56°C after initial non-recurring 5 min at 95°C were followed by 1 min at 60°C for detection in a QuantStudio1 Real-Time-PCR system (Thermo Fisher Scientific Inc., Waltham, Massachusetts). Log10 rrn gene copy numbers were calculated per g dry substrate.
Illumina sequencing of 16S rRNA and ITS2 gene amplicons
Amplicon sequencing of the V3-V4 region of the bacterial 16S rRNA gene and the fungal ITS2 gene on an Illumina MiSeq platform was mandated to BGI genomics (https://www.bgi.com). Processing of primer-free sequences after pre-processing of raw sequences by the sequencing provider was conducted using R version 4.4.1 (R core team, 2024). Amplicon sequence variants (ASVs) of 16S rRNA and ITS2 genes were generated with DADA2 version 1.26.0 (Callahan et al., 2016). For taxonomic affiliation, bacterial and archaeal ASVs were aligned to the SILVA SSU rel. 138 (McLaren and Callahan, 2021) database train set. Fungal ASVs were assigned to taxa using the UNITE sh-general-release-dynamic-25.07.2023 (Abarenkov et al., 2023) database train set. All raw amplicon data were deposited at NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJNA1212987 (16S, bacterial ASVs) and PRJNA1213009 (ITS, fungal ASVs). Sequences that were affiliated to Cyanobacteria/chloroplasts or mitochondria were removed from the 16S rRNA ASV data set. The bacterial data set contained a total of 9854 ASVs and exhibited an average number of 28693 reads per sample. The ITS2 data set consisted of 1179 ASVs in total, and, on average, 39818 reads were obtained per sample. No further quality filtering of the two ASV data sets was required. Respective rarefaction curves (Supplementary Figure S2) confirmed sufficient sequencing depth.
Data and statistical analyses
The software R 4.4.1 (R core team, 2024) was used to perform analyses of the 16S rRNA and ITS2 ASV data sets.
Based on cleaned ASV tables, relative abundances of ASVs were calculated. Alpha-diversity as expressed by Shannon´s index was calculated using the packages vegan (Oksanen et al., 2024), questionr (Barnier et al., 2023) and agricolae (De Mendiburu, 2023) based on 100 times subsampled data sets (samples with lowest numbers of reads, n= 24676 (16S), n= 35200 (ITS2)). Significant differences in beta-diversity induced by the factors substrate, time and inoculum were assessed by permutational analysis of variance (PERMANOVA) based on Bray-Curtis dissimilarity (10000 permutations) using the vegan package (Oksanen et al., 2024). Further, non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarity, were used to calculate and visualize differences in the compositions of bacterial and fungal communities with the vegan package (Oksanen et al., 2024). The packages phyloseq (McMurdie and Holmes, 2013) and edgeR (Robinson et al., 2010) were used to assess differential abundance of bacterial and fungal genera in the substrates, comparing two substrates at a time. According to edgeR developer recommendations, data were normalized using likelihood ratio test under negative binominal distribution and general linear models (FDR-corrected p < 0.05). To display the differences in relative abundances of the respective genera between two substrates, heatmaps based on centered and scaled log10 transformed relative abundance data were generated using the vegan (Oksanen et al., 2024) and gplots (Warnes et al., 2024) packages.
Analysis of variance (ANOVA) followed by Tukey´s HSD test using packages phyloseq (McMurdie and Holmes, 2013), multcomp (Hothorn et al., 2008), questionr (Barnier et al., 2023) and agricolae (De Mendiburu, 2023) was performed to detect significant differences between log10 CFU counts, substrate water contents, Shannon´s indices and log10 rrn gene copy numbers. Significance of differences or effects was assumed if the p-value was below 0.05.
For visualization of relative abundances, log10 CFU counts, substrate water contents, Shannon´s indices and log10 rrn gene copy numbers the package ggplot2 was used (Wickham, 2016).
Results
Physicochemical properties of peat, peat-reduced and peat-free substrates are substantially different
In order to compare the physicochemical properties of the used substrates, several abiotic parameters were assessed. Due to the low number of observations (1 result per substrate was provided, Supplementary Table S1), the results are not statistically evaluable and hence serve as approximation for the interpretation of further results (persistence of S. Typhimurium and bacterial and fungal community compositions). The four tested substrates, named: peat, peat-reduced, peat-free 1 and peat-free 2, differed remarkably (Supplementary Table S1). The peat substrate showed the highest content of organic matter and hence C amount (45.6% of fresh weight), while it had low N content, predominantly present as ammonia (Supplementary Table S1), compared to the other substrates. This resulted in a C/N ratio of 48. The peat-reduced substrate contained a rather high nitrate (NO3-) content of 115 mg/l compared to 24 mg/l in the peat substrate. Phosphate and potassium (K) contents were approximately twice as high in the peat-reduced substrate, 3.5 to 3.8 times (phosphate) and 6.8 times (K) in the two peat-free substrates compared to peat. Magnesium (Mg) content was similar in the two peat containing variants and in the two peat-free substrates. The total N content (82 mg/l) in the peat-free 1 substrate was only slightly higher than in peat, but appeared in the form of nitrate. The micronutrients Cu, Zn, Mn, B and Fe were found in clearly higher amounts in the peat-reduced and both peat-free substrates, Mn content differed the most between peat-free 1 (16 mg/l) and peat-free 2 (45 mg/l). Furthermore, peat exhibited substantially lower density than the other substrates with 120 g/l (dry weight) compared to around 200 g/l dry weight for the alternatives.
Persistence of S. Typhimurium varies in the different substrates
In a next step, we assessed the persistence of S. Typhimurium in the four substrates containing different amounts of peat or no peat, respectively. The four substrates (peat, peat-reduced and peat-free 1 as well as peat-free 2) were inoculated with Salmonella enterica subsp. enterica serovar Typhimurium strain 14028s rifR (S. Typhimurium) and its CFU numbers were enumerated over the course of 84 days at seven sampling time points. S. Typhimurium was not detected in any of the MgCl2 control samples (data not shown). Initially, all substrates showed an initial number of S. Typhimurium of approximately 7 log10 CFU of per gram dry substrate (Figure 1). In the peat and peat-free 1 substrate the values were below 2 at 56 days post inoculation (dpi) and in the peat-reduced substrate slightly above. After 84 days, S. Typhimurium was not detectable in the peat, peat-reduced and peat-free 1 substrates (Figure 1). In contrast, S. Typhimurium CFUs of log10 3.5 per gram dry substrate were still present in the peat-free 2 substrate, even at 84 dpi. In general, it seemed that the decline in log10 CFUs proceeded faster in the peat, peat-reduced and peat-free 1 substrates, even if the results from the peat substrate showed a high dispersion. S. Typhimurium in the peat-free 2 substrate seemed to persist throughout the experiment’s duration. From 7 dpi on, S. Typhimurium CFUs in the peat substrate differed significantly from those in the other substrates within the corresponding time points. Significant differences within particular substrate-types were observed between the earlier (0, 7 and 14 dpi) and the later time points (56 and 84 dpi). Taking together, the composition of the substrate had an impact on the persistence of S. Typhimurium.
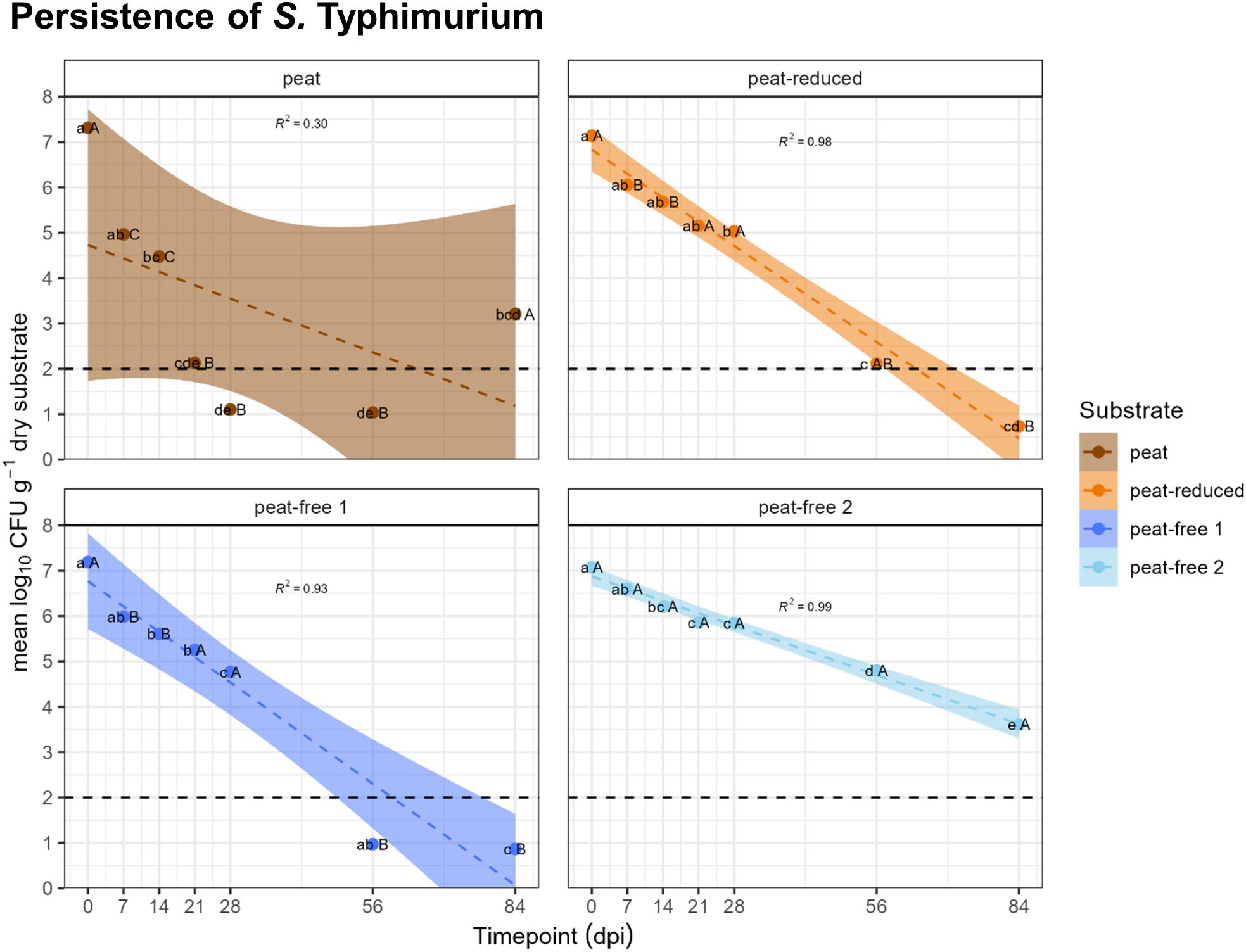
Figure 1. S. Typhimurium persists differently in peat, peat-reduced and peat-free substrates. Number of colony forming unit (CFU) of Salmonella enterica serovar Typhimurium strain 14028s (S. Typhimurium) in tested substrates termed: peat, peat-reduced, peat-free 1 and peat-free 2. Points represent mean values of four replicates ± standard deviation (SD). Lowercase letters indicate significant differences as indicated by one-way ANOVA and Tukey HSD (p < 0.05) among different time points (0, 14, 28, 56 dpi) of the same substrate variant, capital letters indicate significant differences (p < 0.05) between substrates at the same time point. The shadowed part represents the confidence interval of 95%, the black dashed lines represent the detection limit of log10 CFU/g dry substrate = 2, the colored dashed line represent the mean decrease of CFU during the experiment.
Compositions of bacterial and fungal communities revealed no impact of S. Typhimurium presence
In addition to the physicochemical differences, we assumed that the four tested substrates will differ in regard to the native microbial communities. We assessed therefore the bacterial and fungal communities in all four substrates, using the 16S rRNA and ITS gene fragment amplicon-approach. This analysis was carried out in both experimental variants, with and without prior inoculation with S. Typhimurium. The composition of bacterial and fungal communities in the four substrates over the duration of the experiment is illustrated as relative abundances of bacterial phyla and proteobacterial classes (Figure 2A) and fungal classes (Figure 2B), respectively, with a 1% threshold. Very notable was the fact that in both ASV datasets, differences between the control and S. Typhimurium-inoculated samples were marginal, in all substrates and at all time points (Figures 2A, B). In addition, the copy numbers of the rrn gene were assessed by qPCR in order to estimate the bacterial abundance. The values ranged between log 9 to log 10.5 (Supplementary Figure S3) revealing no significant differences among the substrates, except for the S. Typhimurium- inoculated peat-free 2 substrate at 14 dpi, which significantly differ from the control 7dpi, control 28dpi and S. Typhimurium 0 dpi samples from that substrate. Those results suggest that in the tested substrates, even if different, the total amount of bacteria is rather stable.
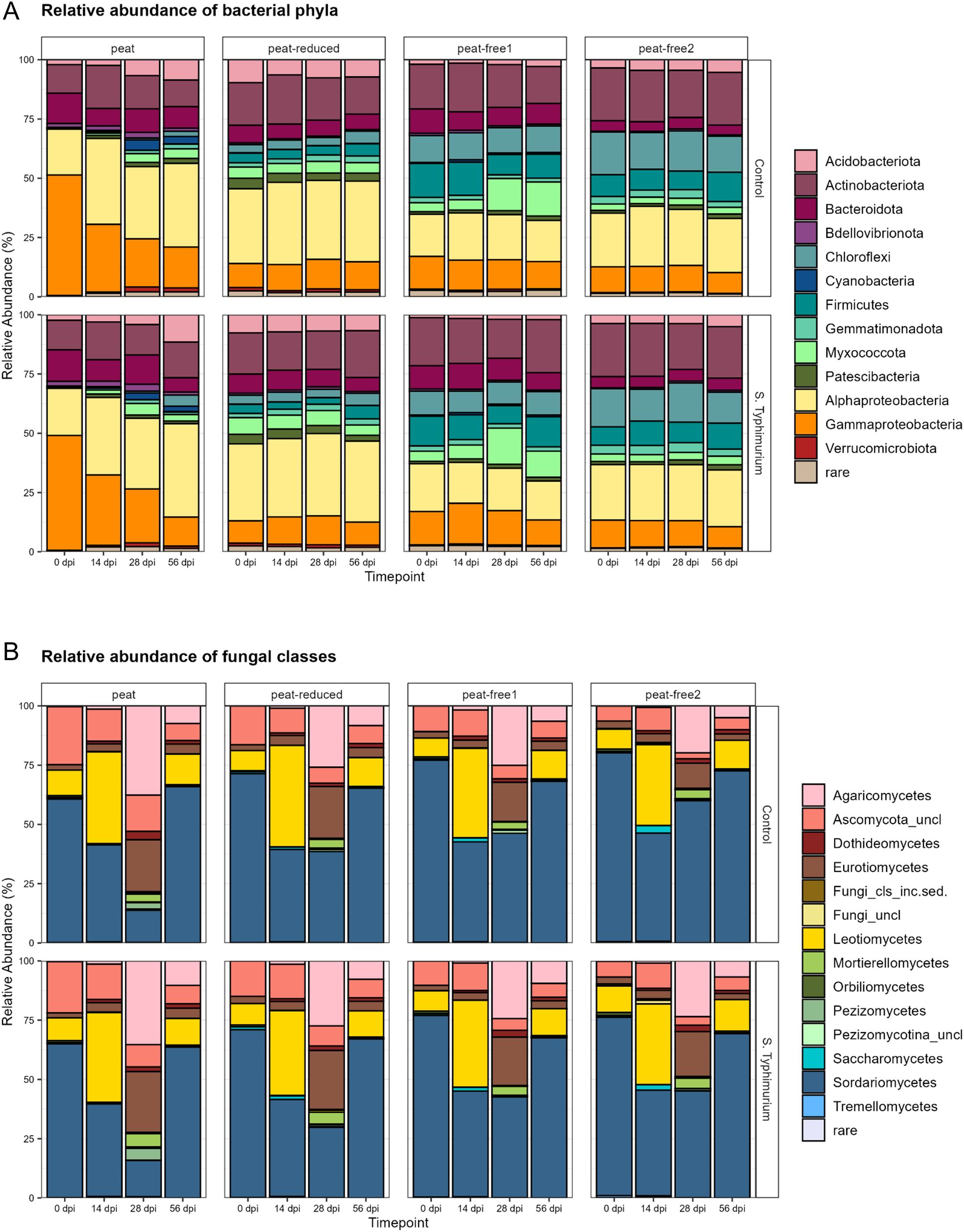
Figure 2. Relative abundance within the microbial communities differs in the tested substrates. Relative abundances (%) of bacterial phyla and proteobacterial classes (A) and fungal classes (B). The analysis was based on 16S rRNA (A) and ITS (B) gene amplicon sequence variants (ASVs). Substrates: peat, peat-reduced, peat-free 1 and peat-free 2 were assessed at time points 0, 14, 28 and 56 days post inoculation (dpi) in control and S. Typhimurium -inoculated samples, threshold >1%.
Diversified relative abundances of bacterial phyla and fungal classes
In the next step we assessed the taxonomic structure of the microbial communities.
The compositions of bacterial community, at a phylum level, differed substantially among the different substrates but not between the control and the S. Typhimurium-inoculated substrates. The differences were more pronounced between the peat and all other substrates than among the peat-reduced, peat-free 1 and peat-free 2 substrates (Figure 2A). In the peat and peat-reduced substrates, Alpha- and Gammaproteobacteria formed the biggest groups, while their proportion in the two peat-free substrates is remarkably smaller. The ratio between Alpha- and Gammaproteobacteria changed over time in peat, Gammaproteobacteria decreased in relative abundance while Alphaproteobacteria increased. In the peat-free 1 and the peat-free 2 substrates, the phyla Chloroflexi, Firmicutes and Gemmatimonadetes were present to a higher extent than in the peat and peat-reduced substrate. Myxococcota were increased in relative abundance in the peat−free 1 substrate at the time points 28 and 56 dpi. Acidobacteria, Actinobacteria and Bacteriodota were abundant in all substrates.
Similarly, the compositions of fungal communities in the four substrates also revealed differences among the substrates and also over the course of the incubation on class level (Figure 2B). Sordariomycetes formed the biggest group at 0 and 56 dpi. Leotiomycetes displayed a high proportion within the fungal community at 14 dpi in all tested substrates, while at 28 dpi an increased amount of ASVs assigned to Agaricomycetes was found. Eurotiomycetes were remarkably abundant at 14 dpi, if compared to other time points.
Bacterial and fungal diversity in the substrates displayed different patterns over time
Given the diverse composition of the studied substrates, we wondered how the diversity would change within the substrates during the incubation period. To this end, we calculated Shannon indices for both the 16S-based and the ITS-based ASV datasets (Figure 3). The obtained values are indicative of biodiversity within the given samples.

Figure 3. Bacterial and fungal diversity displayed different patterns during the incubation. Shannon´s index displaying the bacterial (A) and fungal (B) alpha-diversity were based on 16S rRNA (A) and ITS (B) gene amplicon sequence variants (ASVs). Substrates: peat, peat-reduced, peat-free 1 and peat-free 2 in control and S. Typhimurium-inoculated samples were assessed. Columns represent mean ± standard deviation (SD) of four replicates. Lowercase letters indicate significant differences (p < 0.05) between time points 0, 14, 28 and 56 dpi in the same substrate variant, capital letters indicate significant differences (p<0.05) between substrates at the same time point, both according to one-way ANOVA/Tukey HSD test.
In the case of bacterial communities, the values ranged between 4 and 6.5. Interestingly, the indices seemed lower at 0 dpi, if compared to later time points. This phenomenon was observed in all four substrates, however, it was significant only in the case of peat, for both the control and the S. Typhimurium-inoculated substrates. In both experimental variants, significant differences between the peat and the other substrates were detected at respective time points (Figure 3A).
In the case of fungal communities, the calculated Shannon indices ranged between 2 and 3 (Figure 3B). Their values increased between 0 and 14 dpi and dropped until 56 dpi to a slightly higher level than at 0 dpi. The results showed a high standard deviation, especially at 28 dpi in control samples of the peat-reduced, peat-free 1 and peat-free 2 substrate. Significant differences were observed mainly between substrates at respective time points for both the control and the S. Typhimurium-inoculated substrates. Significant differences in diversity between time points in the same substrate were found only between the peat-free 1 and peat-free 2 substrate at 14 dpi in the control samples and at 28 dpi in the S. Typhimurium-inoculated samples, as well as between the peat and the peat-free 1 substrate at 56 dpi in the S. Typhimurium-inoculated samples (Figure 3B).
Those results indicate that the diversity of the bacterial community shows significant differences between the peat and the other tested substrates, most distinct at day 0. In the fungal communities, significant differences in diversity were less consistent.
Diversity among bacterial and fungal communities is governed by the original substrate
To corroborate the results even further we compared the composition of the different microbial communities in respect to the original substrate, the time of inoculation and the presence of S. Typhimurium. Using a permutational multivariate analysis of variance (PERMANOVA) we sought to identify factors influencing the composition of microbial communities, both the bacterial and the fungal. The analysis revealed that the original substrate was the factor explaining the highest proportion of variation in the bacterial (75.4%, Table 1) and the fungal (89%) ASV data set (Table 1). In both cases, the factor time had a lower (0.39 and 2.15%, respectively) but still significant influence. Nine % of variation in the composition of the bacterial community could be explained by the interaction of factors substrate and time, while this combination explained only 2.3% of the variation among fungal ASVs. The factor inoculation (S. Typhimurium) significantly influenced the composition of the fungal community, although to a rather low extent (0.2%). The interaction of the factors substrate and inoculum accounted for only 0.64% of the observed diversity.
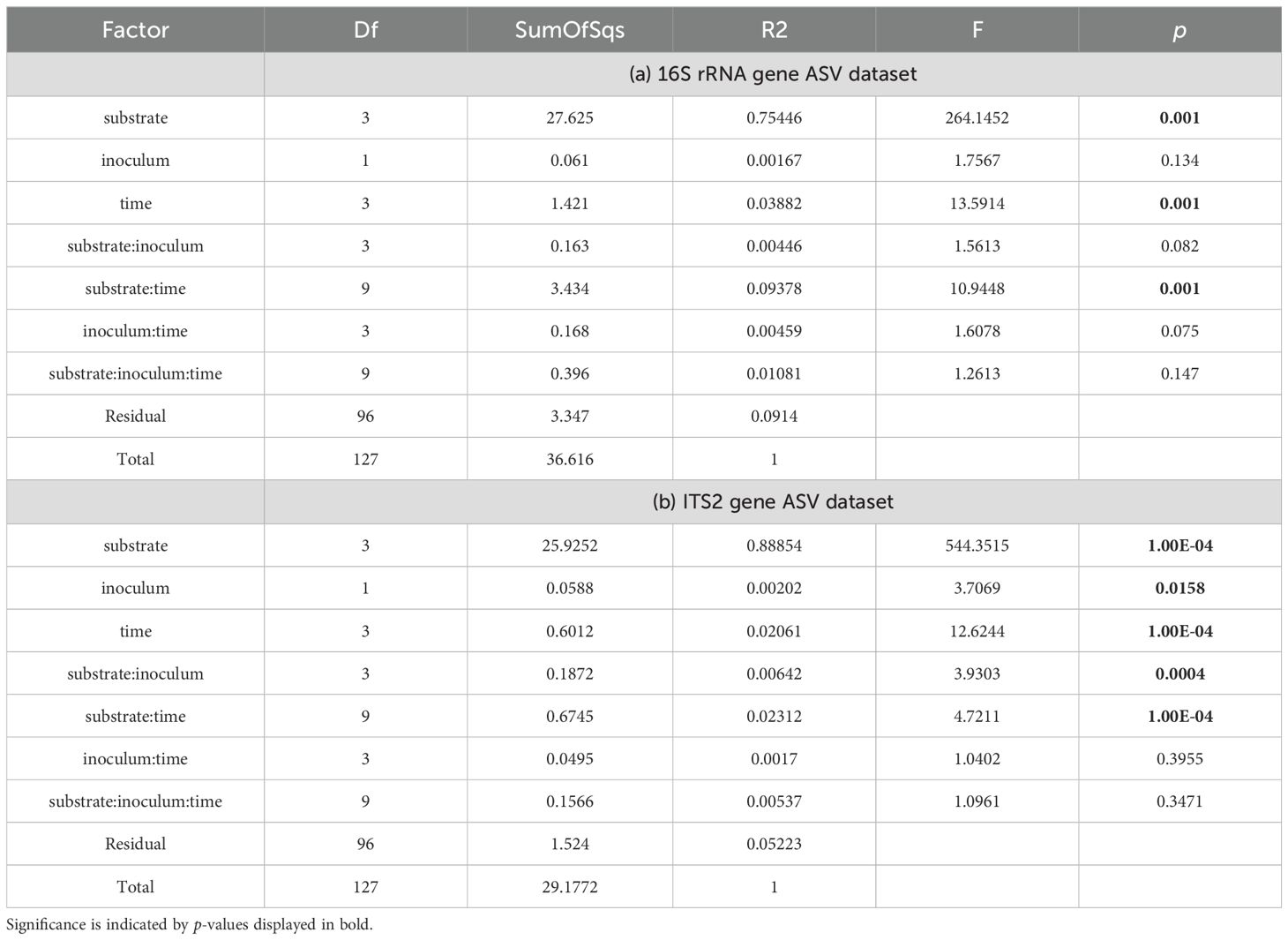
Table 1. Results of permutational analysis of variance (PERMANOVA) based on 16S rRNA (a) and ITS (b) gene amplicon sequence variant (ASV) counts.
The non-metric multidimensional scaling (NMDS) revealed a clear differentiation of both bacterial and fungal communities (Figure 4), according to the substrates. Bacterial communities in the peat and the peat-reduced substrate were separated from those of the peat-free 1 and peat-free 2 substrates along the NMDS1-axis, whereas bacterial communities of the peat-free 1 and peat-free 2 substrates seemed more similar. The 16S-based ASV data set indicated also a clearer separation of bacterial communities in the peat and peat-free 2 substrate from those in the peat-reduced and peat-free 1 substrates, along the NMDS2-axis. Bacterial communities from the peat substrate were additionally differentiated by time points (dpi) along NMDS2-axis with 0 dpi being the most distant from the later time points (Figure 4A). A time dependent separation was observed also in the case of fungal communities from the peat-free 1 substrate (Figure 4B).
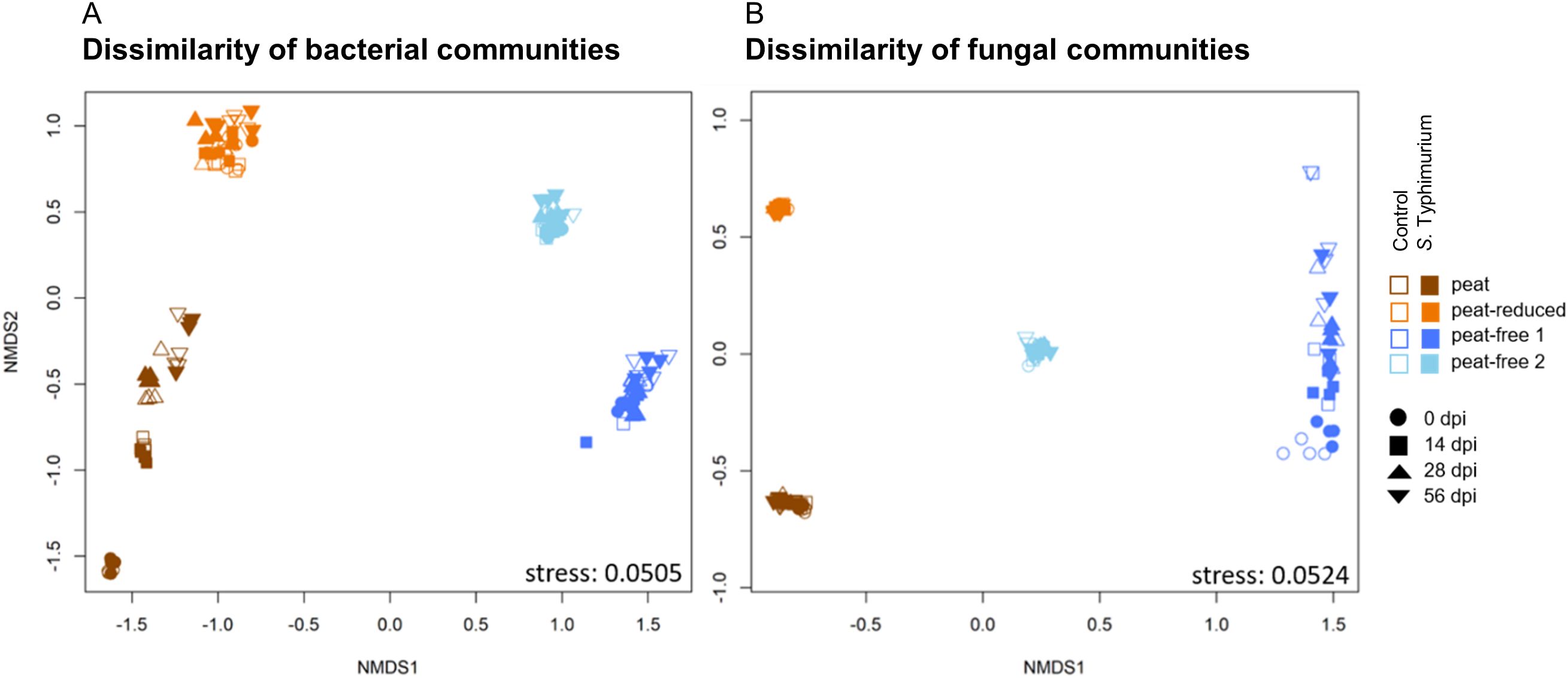
Figure 4. Original substrate dictates the composition of microbial communities. The non-metric multi-dimensional scaling (NMDS) of the bacterial (A) and fungal (B) community compositions is displayed. NMDS was based on Bray-Curtis dissimilarities calculated from 16S rRNA (A) and ITS (B) gene amplicon sequence variant (ASV) counts. The substrates peat, peat-reduced, peat-free 1 and peat-free 2 were assessed at different time points (0, 14, 28, 56 dpi) in control and S. Typhimurium-inoculated samples.
Abundances of bacterial and fungal genera differ significantly between substrates
Differences in the structure and diversity among the bacterial and fungal communities motivated us to assess bacterial and fungal genera associated with a particular substrate, i.e. those with significantly different relative abundances in the respective substrate. As only very few fungal or bacterial genera were detected to be enriched in response to a specific time point, we focused here on the comparisons of substrates, which matched the scope of the study. Accordingly, PERMANOVA results suggested a marginal impact of time on the bacterial and fungal community compositions. No bacterial or fungal genera were identified that were differentially abundant between control and Salmonella-inoculated samples, or within the threshold of 0.5% relative abundance. Figure 5 displays the bacterial (A) and fungal (B) genera with a relative abundance equal to or higher than 0.5% on average of all replicates in one substrate and differentially abundant between two substrates. Generally, many of the identified groups were unclassified on genus level. In total, 131 bacterial genera were differentially abundant in comparisons among the tested substrates. The majority of these were affiliated to the phyla Actinobacteriota, Gammaproteobacteria, Alphaproteobacteriota and Bacteriodota (Figure 5A, Supplementary Table S2A). More specifically, sequences affiliated to the genera Arachidicoccus (phylum Bacteriodota), Asticcacaulis (class Alphaproteobacteria), Bdellovibrio (phylum Bdellovibrionota), Burkholderia-Caballeronia-Paraburkholderia (phylum Gammaproteobacteria), Dyella (phylum Gammaproteobacteria), Pedobacter (phylum Bacteriodota), P3OB-42 (phylum Myxococcota), Streptomyces (phylum Actinobacteria) and Taibaiella (phylum Bacteriodota) were enriched in the peat substrate when compared to either the peat-reduced, peat-free 1 or peat-free 2 substrates. Chujabacter (phylum Gammaproteobacteria), Ferruginibacter (phylum Bacteriodota) and Rhodococcus (phylum Actinobacteria) were enriched in peat-reduced compared to both the peat and the peat-free 1 substrates. In the peat-free 1 substrate, the genera Nonomuraea (phylum Actinobacteriota), Allo-, Neo-, Para-Rhizobium (class Alphaproteobacteria), Ruminofilibacter (phylum Bacteroidota), Fermentimonas (phylum Bacteroidota), Sphaerobeacter (phylum Chloroflexi), Cellvibrio (class Gammaproteobacteria) and Hydrogenophaga (class Gammaproteobacteria) showed a significantly higher relative abundance than in the other tested substrates. The comparison of the peat-free 2 to either the peat or the peat-free 1 substrate revealed, that Nitrolancea (phylum Chloroflexi), Pedomicrobium (class Alphaproteobacteria) and SWB02 (class Alphaproteobacteria) were more abundant in the peat-free 2 substrate.
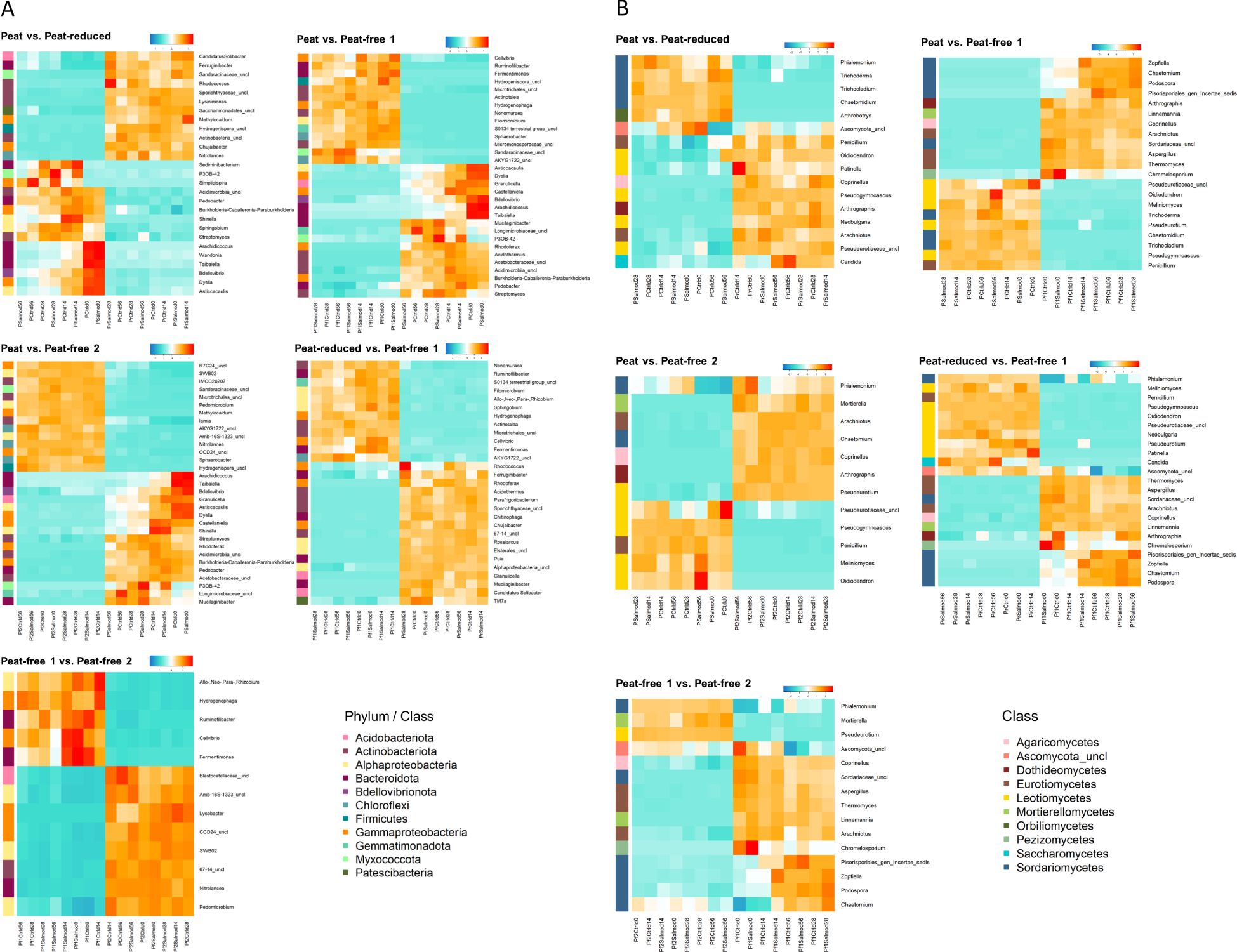
Figure 5. Abundances of genera differ significantly between substrates. Differential abundance was displayed as heatmap. The calculation was based on centered and scaled log10 transformed relative abundances of bacterial (A) and fungal (B) genera (> 0.5% relative abundance and FDR-corrected p < 0.05) as means of four replicates. Substrates peat (P); peat-reduced (Pr); peat-free 1 (Pf1) and peat-free 2 (Pf2) were assessed at different time points (d0, d14, d28, d56) in control (Ctrl) and S. Typhimurium-inoculated (Salmo) samples.
The highest proportions of fungal genera that were differentially abundant between two substrates across all comparisons belonged to the classes Leotiomycetes, Sordariomycetes and Eurotiomycetes (Figure 5B). Fungal sequences that were enriched in the peat substrate, in the comparisons between the peat and the peat-reduced or peat-free 1 substrates, were identified as Chaetomidium (class Sordariomycetes), Trichochladium (class Sordariomycetes) and Trichoderma (class Sordariomycetes). Melinomyces (class Leotiomycetes), Oidiodendron (class Leotiomycetes), Penicillium (class Eurotiomycetes) and Pseudogymnoascus (class Leotiomycetes) were enriched in the peat substrate when compared to both the peat-free 1 and peat-free 2 substrates. Candida (class Saccharomycetes), Neobulgaria (class Leotiomycetes), Oidiodendron (class Leotiomycetes), Patinella (class Ascomycetes), Penicillium (class Eurotiomycetes) and Pseudogymnoascus (class Leotiomycetes) were more abundant in peat-reduced substrate across all comparisons. The genera Coprinellus (class Agaricomycetes), Arachniotus (calss Eurotiomycetes), Chaetomium (class Sordariomycetes), Linnemannia (class Mortierellomycetes) and Podospora (class Sordariomycetes) were enriched in the peat-free 1 substrate, if compared to either the peat, peat-reduced, or peat-free 2 substrates. Groups enriched in the peat-free 2 substrate, for all comparisons, were Mortierella (class Mortierellomycetes), Phialemonium (class Sordariomycetes) and Pseudeurotium (class Leotiomycetes). Several of the differentially abundant fungal genera showed a remarkably high relative abundance, e. g. Phialemonium with over 50% in the peat and peat-free 2 substrates, ca. 40% in the peat-reduced substrate and ca. 20% in the peat-free 1 substrate (Supplementary Table S2B). Other genera with high relative abundances (between 10 and 30%) were Coprinellus (27.33% in peat-free 1), Pseudogymnoascus (25.27% in peat-reduced), Arachniotus (15.1% in peat-free 1), Chaetomidium (13.37% in peat) and Pseudeurotium (11.68% in peat-free 2), (Supplementary Table S2B).
Discussion
Different factors including temperature, pH, nutrient availability, organic matter and water content influence the composition of bacterial and fungal communities in substrates designed for plant growth. Those features determine also the potential persistence of pathogens. Such influence was previously reported for agricultural soils amended with animal manure or slurry of different origin (Jechalke et al., 2019; Shah et al., 2019; Fornefeld et al., 2017; Semenov et al., 2009). Unfortunately, little information is available for the persistence of human pathogens in soilless plant growing media. In the current study, we assessed the persistence of Salmonella enterica subsp. enterica serovar Typhimurium in four commercially available plant growing media with different levels of peat. This study used a common method for monitoring the presence of inoculants in soils (Micallef et al., 2023; Alegbeleye and Sant’Ana, 2023; Todd-Searle et al., 2020; Jechalke et al., 2019; Fornefeld et al., 2017; Gurtler et al., 2013). The direct CFU enumeration was performed from 1 g substrate extracted with 9 ml 1mM MgCl2 solution in 4 replicates to minimize potential variability. The persistence of the rifampicin resistant S. Typhimurium strain was detected on the selective XLD medium containing rifampicin. In addition, only black colonies were taken into account. Despite some methodological limitations (this method does not completely exclude the growth of bacteria different from S. Typhimurium or bacteria that have evolved a rifampicin resistance during or before the incubation) this approach is applicable in regards of the research questions raised here. The measurements of physicochemical properties of the substrates could not be used for correlation analyses with results gained in this study, as they were not statistically evaluable. Yet they serve as valuable approximation for result interpretation. The studied substrates contained in addition to or in replacement of peat, coco pulp, wood fiber, green compost or bark humus. Green compost and other composted plant materials are widely used as substrate components. The peat-free and peat-reduced variants contained higher amounts of nitrogen, phosphorus and potassium, importantly the peat substrate was not fertilized prior to this study.
Previous reports suggested that S. Typhimurium is able to survive up to 12 weeks in mature composts (Lemunier et al., 2005). On the one hand, these observations are consistent with results of this study, which revealed the longest persistence of Salmonella in the peat-free 2 substrate, a mixture of bark humus, coco fiber, wood fiber, green compost and organic fertilizer. Peat-free 2 was the only substrate in which S. Typhimurium persisted up to 84 days after inoculation. One distinctive characteristic of this substrate is the comparatively low water content of approximately 55% throughout the experiment, the other is the remarkably high manganese (Mn) content of 45 mg/L-1. Litter decomposition, and presumably aerobic bark decomposition during composting processes, are closely related to the dynamics of Mn (Peng et al., 2023). For example, Mn is essential for manganese peroxidase, which is crucial for degradation of lignin (Sun et al., 2019). For S. Typhimurium, the high Mn content might have provided an advantage. The tolerance against, for instance, oxidative or nitrosative stress could be enhanced if Mn is sufficiently available, since Mn is an important cofactor required by many enzymes involved in metabolic pathways (Ha and Lee, 2023). On the other hand, the inconsistent persistence of S. Typhimuriumin the peat substrate might be related to the relative abundance of Bdellovibrio (phylum Bdellovibrionata), which is significantly increased in peat, particularly in S. Typhimurium-inoculated samples (0.6 to 1.8%, data not shown). Bdellovibrio predates Gram-negative bacteria and was shown to reduce the number of human pathogens like Escherichia coli or Salmonella enterica in different produces, intended to be consumed fresh (Olanya et al., 2020).
Numerous studies assessed bacterial and fungal communities in natural peatlands under e. g. drained or rewetted conditions in different sampling depths (Kitson and Bell, 2020 and references therein). It is very likely that peat, once extracted from peatlands for horticultural use, exhibits rather low species diversity (Taparia et al., 2021). This phenomenon could explain the initial differences in diversity between peat and the other substrates. Those seem more heterogeneous as habitats, probably due to their original components and their inherent microorganisms. Overall, the initial nutrient content in the peat-reduced and the peat-free substrates was higher. Although the physicochemical properties were not fully assessed, it can be assumed, that due to the general microbial activity and nutrient turnover, the conditions in the respective substrate changed over the course of the experiment, probably favoring higher diversity. Furthermore, bacteria or fungi from the greenhouse environment might have colonized the substrate containers. Importantly, S. Typhimurium presence or the treatment with 10mM MgCl2 (control) did not influence the compositions of bacterial communities.
The aforementioned explanation applies most probably also to the changes in the diversity of the fungal communities. Similar to bacteria, no notable differences were observed between the control samples and those inoculated with S. Typhimurium. The fungal ASV data set revealed several genera with high relative abundances. Those genera seemed to have advantages in the competition for resources in the respective substrate. For instance, Phialemonium was present in all substrates and showed rather high relative abundances of 20 to 60%.
Since the presence of S. Typhimurium did not have a notable influence on the bacterial or fungal community compositions, other factors were noteworthy. For example, the low explanatory power of the factor “time” alone was clearly apparent, this contributed to the significant impact of the interaction with the factor “substrate” (R2 = 0.094, Table 1). This was particularly visible in the case of bacterial communities in the peat substrate, where samples of day 0 separated from the later time points. This was further confirmed by PERMANOVA analysis for samples of each substrate separately, where peat was the only substrate with a significant influence of time on the compositions of the bacterial and fungal communities. The factors “inoculum” or “time” and the interactions of all factors were less important for the fungal community composition, which was mainly shaped by the substrate, since this factor explained around 89% of the differences between the communities. The overall low importance of the factor “time” suggests only minor changes of respective substrate properties throughout the experiment.
The relative abundances of bacterial phyla (and proteobacterial classes, respectively) and fungal classes suggest overall differences in community compositions among the substrates. The majority of bacterial phyla in peat were assigned to Alpha- and Gammaproteobacteria and Bacteriodota. These can have taken advantage of the higher C/N ratio in peat, as several taxa belonging to these classes are capable of decomposition of complex organic compounds which are recalcitrant under natural (anaerobic) conditions (Mastný et al., 2021). In the current study, genera belonging to the clade Burkholderia-Caballeronia-Paraburkholderia (class Gammaproteobacteria), Dyella (phylum Gammaproteobacteria), Streptomyces (phylum Actinobacteria), Pedobacter (phylum Bacteriodota) were significantly enriched in the peat substrate, if compared to the other substrates. Theses genera were reported to have celluloytic properties and able to decompose aromatic compounds (Hetz and Horn, 2021). The family Chitinophagaceae (phylum Bacteriodota) as chitin and cellulose degraders (Rosenberg, 2014; Bailey et al., 2013) comprises several potential PGPR species (Madhaiyan et al., 2015) and was represented by Arachidococus and Taibaiella, among others, in peat. Several fungal genera in the peat substrate potentially exhibit plant beneficial traits. For example, Trichoderma species (class Sordariomycetes), ubiquitous in soils and on plant debris, can serve as biocontrol agents against plant pathogenic fungi and can form mutual endophytic symbioses (Harman et al., 2004). Meliniomyces and Oidiodendron (class Leotiomycetes) are common mycorrhizal genera (Toju and Sato, 2018). As decomposers of organic material, members of the genus Penicillium cause rots in food industry, apart from the production of antimicrobials including mycotoxins (Visagie et al., 2014).
The peat-reduced substrate did not specifically favor potential plant beneficial bacteria, but mainly those capable of decomposition of complex organic compounds like Rhodococcus (phylum Actinobacteriota), (Larkin et al., 2006) or Ferruginibacter (phylum Bacteriodota) (Busch et al., 2019; Wijaya and Oh, 2023). Fungal groups enriched in the peat-reduced substrate were overall typical saprotrophs, such as Neobulgaria (class Leotiomycetes) or Patinella (class Ascomycetes). Noteworthy were genera containing opportunistic human or animal pathogens, i.e. Candida (class Saccharomycetes) (Kumamoto et al., 2020) and Pseudogymnoascus (class Leotiomycetes) (Veselská et al., 2020).
The bacterial community of the peat-free 1 substrate seemed to be linked to the original compounds. For instance, Sphaerobacter (phylum Actinobacteriota) contains thermophilic species tolerating composting processes (Wang et al., 2022; Storey et al., 2015). Several of the enriched genera were linked to decomposition of wood components including lignocellulose degrading Ruminofilibacter (phylum Bacteroidota) (Li et al., 2020; Weiss et al., 2011), Cellvibrio (class Gammaproteobacteria) degrading lignin but also resins, gums, dyes, tannic acid, waxes, and other lipid compounds (Ranalli et al., 2019) as well as Fermentimonas, which can ferment proteinaceous compounds (Hahnke et al., 2016). The only particular plant growth promoting bacterial genus significantly enriched in the peat-free 1 substrate was identified as belonging to the clade Allo-, Neo-, Para-Rhizobium (class Alphaproteobacteria) (Trivedi et al., 2020). Examples of plant growth promoting fungal genera significantly enriched in peat-free 1 included Chaetomium (class Sordariomycetes) (Wang et al., 2016) and Linnemannia (class Mortierellomycetes) (De Tender et al., 2024; Vandepol et al., 2022). Among the other genera that were enriched in the peat-free 1 substrate, were mainly saprotrophic genera, such as Coprinellus (class Agaricomycetes) and Arachniotus (class Eurotiomycetes).
In case of the peat−free 2 substrate, the Mn-oxidizing genera Pedomicrobium (class Alphaproteobacteria), capable of biofilm formation in aqueous environments including waste water treatment, and SWB02 (class Alphaproteobacteria), seemed enriched (Sly et al., 1988; Sjöberg et al., 2020). SWB02 contributes to N-cycling by denitrification, while Nitrolancea (phylum Actinobacteriota) is able to oxidize nitrite (Sorokin et al., 2014). In case of fungal genera enriched in peat-free 2, the genera Mortierella and Pseudeurotium can be mentioned since, their members were reported to possess plant growth promoting potential (Manzotti et al., 2020). The genus Phialemonium (class Sordariomycetes) was also significantly enriched in the peat-free 2 substrate, although it was highly abundant in all substrates. Phialemonium reached an extraordinarily high relative abundance of 59.59% in the peat-free 2 substrate. It can be isolated from air and soil but also industrial waste water and sewage (Gams and McGinnis, 1983). Important to mention is the fact that this genus includes opportunistic human pathogens, which are potentially lethal for immunocompromised persons (Perdomo et al., 2011).
Conclusions
In general, the potential functional traits of the bacterial and fungal taxa identified in the current collection of 4 different commercially available horticultural substrates by the amplicon sequencing approach are not surprising when considering the original compounds. However, to our knowledge the compositions of bacterial and fungal communities in such substrates have not been assessed elsewhere. The results presented here, reveal the necessity of considering bacterial and fungal communities during the development of rather complex substrates e. g. peat-reduced or peat-free mixtures of different compounds designed for plant growth. Furthermore, secondary colonizers from the storage environment or potentially pathogenic contaminants need to be considered, in respect to user safety. Taking together, the horticultural appropriateness of peat-reduced and peat-free substrates or substrate components requires a multifaceted perspective.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1212987. https://www.ncbi.nlm.nih.gov/, PRJNA1213009.
Author contributions
AM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. JS: Investigation, Methodology, Writing – review & editing. VM: Investigation, Methodology, Writing – review & editing. OG: Investigation, Methodology, Writing – review & editing. AS: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work of Antje Müller, Oscar Gehring, Verena Maiberg and Jasmin Schmidt was financially supported by the Federal Ministry of Food and Agriculture, represented by the Fachagentur Nachwachsende Rohstoffe e.V. under grant No. 2220MT006B – “ToPGa – Entwicklung und Bewertung von torfreduzierten Produktionssystemen im Gartenbau, Mikrobiologie und Humanpathologie”.
Acknowledgments
The authors would like to thank the members of the ToPGa consortium for constructive discussions. The substrates used in this study were purchased and are freely available. As for consortium policy the producers will not be disclosed.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fhort.2025.1568055/full#supplementary-material
References
Abarenkov K., Zirk A., Piirmann T., Pöhönen R., Ivanov F., Nilsson R. H., et al. (2023). UNITE general FASTA release for Fungi. Version 18.07.2023 (UNITE Community). doi: 10.15156/BIO/2938067
Alegbeleye O. and Sant’Ana A. S. (2023). Survival of Salmonella spp. under varying temperature and soil conditions. Sci. Total Environ. 884, 163744. doi: 10.1016/j.scitotenv.2023.163744
Allende A. and Monaghan J. (2015). Irrigation water quality for leafy crops: A perspective of risks and potential solutions. Int. J. Environ. Res. Public Health 12, 7457–7477. doi: 10.3390/ijerph120707457
Bailey V. L., Fansler S. J., Stegen J. C., and McCue L. A. (2013). Linking microbial community structure to β-glucosidic function in soil aggregates. ISME J. 7, 2044–2053. doi: 10.1038/ismej.2013.87
Barnier J., Briatte F., and Larmarange J. (2023). _questionr: Functions to Make Surveys Processing Easier_. R package version 0.7.8. Available online at: https://CRAN.R-project.org/package=questionr
Barrett G. E., Alexander P. D., Robinson J. S., and Bragg N. C. (2016). Achieving environmentally sustainable growing media for soilless plant cultivation systems - A review. Scientia Horticulturae 212, 220–234. doi: 10.1016/j.scienta.2016.09.030
Berg G. and Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bilderback T. E., Riley E. D., Jackson B. E., Kraus H. T., Fonteno W. C., Owen J. J., et al. (2013). Strategies for developing sustainable substrates in nursery crop production. Acta Hortic. 1013, 43–56. doi: 10.17660/ActaHortic.2013.1013.2
Busch H., Hagedoorn P.-L., and Hanefeld U. (2019). Rhodococcus as A versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 20 (19), 4787. doi: 10.3390/ijms20194787
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carlile W. R., Cattivello C., and Zaccheo P. (2015). Organic growing media: constituents and properties. Vadose Zone J. 14, 1–13. doi: 10.2136/vzj2014.09.0125
De Mendiburu F. (2023). _agricolae: Statistical Procedures for Agricultural Research_. R package version 1.3-7. Available online at: https://CRAN.R-project.org/package=agricolae
De Tender C., Vandecasteele M., Ommeslag S., de Zutter N., Vandenbussche E., Haegeman A., et al. (2024). Linnemannia elongata: A key species in chitin-based plant growth promotion. Phytobiomes J. 8, 366–377. doi: 10.1094/PBIOMES-05-23-0031-R
Fornefeld E., Schierstaedt J., Jechalke S., Grosch R., Schikora A., and Smalla K. (2017). Persistence of salmonella typhimurium LT2 in soil enhanced after growth in lettuce medium. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00757
Gams W. and McGinnis M. R. (1983). Phialemonium, A new anamorph genus intermediate between phialophora and acremonium. Mycologia 75, 977–987. doi: 10.1080/00275514.1983.12023783
Gruda N. (2012). Current and future perspective of growing media in Europe. Acta Hortic. 960, 37–43. doi: 10.17660/actahortic.2012.960.3
Gruda N. (2019). Increasing sustainability of growing media constituents and stand-alone substrates in soilless culture systems. Agronomy 9, 298. doi: 10.3390/agronomy9060298
Gurtler J. B., Douds D. D., Dirks B. P., Quinlan J. J., Nicholson A. M., Phillips J. G., et al. (2013). Salmonella and Escherichia coli O157:H7 survival in soil and translocation into leeks (Allium porrum) as influenced by an arbuscular mycorrhizal fungus (Glomus intraradices). Appl. Environ. Microbiol. 79, 1813–1820. doi: 10.1128/AEM.02855-12
Ha N. and Lee E.-J. (2023). Manganese Transporter Proteins in Salmonella enterica serovar Typhimurium. J. Microbiol. 61, 289–296. doi: 10.1007/s12275-023-00027-7
Hahnke S., Langer T., Koeck D. E., and Klocke M. (2016). Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 66, 1466–1475. doi: 10.1099/ijsem.0.000902
Harman G. E., Howell C. R., Viterbo A., Chet I., and Lorito M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Harris C. N., Dickson R. W., Fisher P. R., Jackson B. E., and Poleatewich A. M. (2020). Evaluating peat substrates amended with pine wood fiber for nitrogen immobilization and effects on plant performance with container-grown petunia. Hortte 30, 107–116. doi: 10.21273/HORTTECH04526-19
Hetz S. A. and Horn M. A. (2021). Burkholderiaceae are key acetate assimilators during complete denitrification in acidic cryoturbated peat circles of the arctic tundra. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.628269
Hirschler O., Osterburg B., Weimar H., Glasenapp S., and Ohmes M.-F. (2022). Peat replacement in horticultural growing media: Availability of bio-based alternative materials. (Braunschweig: Johann Heinrich von Thünen-Institut), Thünen Working Paper (190) Available at: https://www.econstor.eu/handle/10419/253552.
Hirschler O. and Thrän D. (2023). Peat substitution in horticulture: interviews with german growing media producers on the transformation of the resource base. Horticulturae 9, 919. doi: 10.3390/horticulturae9080919
Jackson B. E., Wright R. D., and Alley M. M. (2009). Comparison of fertilizer nitrogen availability, nitrogen immobilization, substrate carbon dioxide efflux, and nutrient leaching in peat-lite, pine bark, and pine tree substrates. Horts 44, 781–790. doi: 10.21273/HORTSCI.44.3.781
Jay-Russell M. T., Hake A. F., Bengson Y., Thiptara A., and Nguyen T. (2014). Prevalence and characterization of Escherichia coli and Salmonella strains isolated from stray dog and coyote feces in a major leafy greens production region at the United States-Mexico border. PloS One 9, e113433. doi: 10.1371/journal.pone.0113433
Jechalke S., Schierstaedt J., Becker M., Flemer B., Grosch R., Smalla K., et al. (2019). Salmonella establishment in agricultural soil and colonization of crop plants depend on soil type and plant species. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00967
Kauffmann F. and Edwards P. R. (1952). Classification and nomenclature of enterobacteriaceae. Int. J. Syst. Bacteriol. 2, 2–8. doi: 10.1099/0096266x-2-1-2
Kitson E. and Bell N. G. A. (2020). The response of microbial communities to peatland drainage and rewetting. A review. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.582812
Kumamoto C. A., Gresnigt M. S., and Hube B. (2020). The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Opin. Microbiol. 56, 7–15. doi: 10.1016/j.mib.2020.05.006
Larkin M. I. J., Kulakov L. A., and Allen C. C. R. (2006). Biodegradation by members of the genus Rhodococcus: biochemistry, physiology, and genetic adaptation. Adv. Appl. Microbiol. 59, 1–29. doi: 10.1016/S0065-2164(06)59001-X
Le Minor L. and Popoff M. Y. (1987). Designation of Salmonella enterica sp. nov., nom. rev., as the Type and Only Species of the Genus Salmonella: Request for an Opinion. Int. J. Syst. Bacteriol. 37, 465–468. doi: 10.1099/00207713-37-4-465
Lemunier M., Francou C., Rousseaux S., Houot S., Dantigny P., Piveteau P., et al. (2005). Long-term survival of pathogenic and sanitation indicator bacteria in experimental biowaste composts. Appl. Environ. Microbiol. 71, 5779–5786. doi: 10.1128/AEM.71.10.5779-5786.2005
Li Y., Zhao J., AChinas S., Zhang Z., Krooneman J., and Euverink G. J. W. (2020). The biomethanation of cow manure in a continuous anaerobic digester can be boosted via a bioaugmentation culture containing Bathyarchaeota. Sci. Total Environ. 745, 141042. doi: 10.1016/j.scitotenv.2020.141042
Madhaiyan M., Poonguzhali S., Senthilkumar M., Pragatheswari D., Lee J.-S., and Lee K.-C. (2015). Arachidicoccus rhizosphaerae gen. nov., sp. nov., a plant-growth-promoting bacterium in the family Chitinophagaceae isolated from rhizosphere soil. Int. J. Syst. Evol. Microbiol. 65, 578–586. doi: 10.1099/ijs.0.069377-0
Manzotti A., Bergna A., Burow M., Jørgensen H. J. L., Cernava T., Berg G., et al. (2020). Insights into the community structure and lifestyle of the fungal root endophytes of tomato by combining amplicon sequencing and isolation approaches with phytohormone profiling. FEMS Microbiol. Ecol. 96 (5). doi: 10.1093/femsec/fiaa052
Mastný J., Bárta J., Kaštovská E., and Picek T. (2021). Decomposition of peatland DOC affected by root exudates is driven by specific r and K strategic bacterial taxa. Sci. Rep. 11, 18677. doi: 10.1038/s41598-021-97698-2
McLaren M. R. and Callahan B. J. (2021). Silva 138.1 prokaryotic SSU taxonomic training data formatted for DADA2. Zenodo 10, 5281.
McMurdie P. J. and Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Micallef S. A., Callahan M. T., McEgan R., and Martinez L. (2023). Soil Microclimate and Persistence of Foodborne Pathogens Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella enterica Newport in Soil Affected by Mulch Type. J. Food Prot. 86, 100159. doi: 10.1016/j.jfp.2023.100159
Neher D. A., Weicht T. R., Bates S. T., Leff J. W., and Fierer N. (2013). Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PloS One 8, e79512. doi: 10.1371/journal.pone.0079512
Nesse A. S., Sogn T., Børresen T., and Foereid B. (2019). Peat replacement in horticultural growth media: the adequacy of coir, paper sludge and biogas digestate as growth medium constituents for tomato (Solanum lycopersicum L.) and lettuce (Lactuca sativa L.). Acta Agricult. Scandinavica Section B. — Soil Plant Sci. 69, 287–294. doi: 10.1080/09064710.2018.1556728
Oksanen J., Simpson G., Blanchet F., Kindt R., Legendre P., Minchin P., et al (2024). _vegan: Community Ecology Package_. R package version 2.6-6.1. Available online at: https://CRAN.R-project.org/package=vegan
Olanya O. M., Niemira B. A., Cassidy J. M., Boyd G., and Uknalis J. (2020). Pathogen reduction by predatory bacteria and survival of Bdellovibrio bacteriovorus and Escherichia coli on produce and buffer treated with low-dose gamma radiation. LWT 130, 109630. doi: 10.1016/j.lwt.2020.109630
Peng Y., Fornara D. A., Wu Q., Heděnec P., Yuan J., Yuan C., et al. (2023). Global patterns and driving factors of plant litter iron, manganese, zinc, and copper concentrations. Sci. Total Environ. 857, 159686. doi: 10.1016/j.scitotenv.2022.159686
Perdomo H., Sutton D. A., García D., Fothergill A. W., Gené J., Cano J., et al. (2011). Molecular and phenotypic characterization of Phialemonium and Lecythophora isolates from clinical samples. J. Clin. Microbiol. 49, 1209–1216. doi: 10.1128/JCM.01979-10
Piveteau P., Druilhe C., and Aissani L. (2022). What on earth? The impact of digestates and composts from farm effluent management on fluxes of foodborne pathogens in agricultural lands. Sci. Total Environ. 840, 156693. doi: 10.1016/j.scitotenv.2022.156693
Pot S., de Tender C., Ommeslag S., Delcour I., Ceusters J., Vandecasteele B., et al. (2022). Elucidating the microbiome of the sustainable peat replacers composts and nature management residues. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.983855
Prisa D. and Caro S. (2023). Alternative substrates in the cultivation of ornamental and vegetable plants. GSC Biol. Pharm. Sci. 24, 209–220. doi: 10.30574/gscbps.2023.24.1.0268
R Core Team (2024). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Ranalli G., Zanardini E., and Sorlini C. (2019). “Biodeterioration – Including Cultural Heritage” in Encyclopedia of Microbiology, 4th ed.. Ed. Schmidt T. M. (Academic Press), 491–509. doi: 10.1016/B978-0-12-809633-8.13016-X
Robinson M. D., McCarthy D. J., and Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinf. (Oxford England) 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rosenberg E. (2014). “The family chitinophagaceae,” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., and Thompson F. (Springer Berlin Heidelberg, Berlin, Heidelberg), 493–495.
Schierstaedt J., Jechalke S., Nesme J., Neuhaus K., Sørensen S. J., Grosch R., et al. (2020). Salmonella persistence in soil depends on reciprocal interactions with indigenous microorganisms. Environ. Microbiol. 22, 2639–2652. doi: 10.1111/1462-2920.14972
Semenov A. V., van Overbeek L., and van Bruggen A. H. C. (2009). Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75, 3206–3215. doi: 10.1128/AEM.01791-08
Shah M. K., Bradshaw R., Nyarko E., Handy E. T., East C., Millner P. D., et al. (2019). Salmonella enterica in Soils Amended with Heat-Treated Poultry Pellets Survived Longer than Bacteria in Unamended Soils and More Readily Transferred to and Persisted on Spinach. Appl. Environ. Microbiol. 85 (10). doi: 10.1128/AEM.00334-19
Sjöberg S., Stairs C. W., Allard B., Homa F., Martin T., Sjöberg V., et al. (2020). Microbiomes in a manganese oxide producing ecosystem in the Ytterby mine, Sweden: impact on metal mobility. FEMS Microbiol. Ecol. 96 (11). doi: 10.1093/femsec/fiaa169
Sly L. I., Arunpairojana V., and Hodgkinson M. C. (1988). Pedomicrobium manganicum from drinking-water distribution systems with manganese-related “Dirty water” Problems. Syst. Appl. Microbiol. 11, 75–84. doi: 10.1016/S0723-2020(88)80051-1
Sorokin D. Y., Vejmelkova D., Lücker S., Streshinskaya G. M., Rijpstra W. I. C., Sinninghe Damsté J. S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64, 1859–1865. doi: 10.1099/ijs.0.062232-0
Storey S., Chualain D. N., Doyle O., Clipson N., and Doyle E. (2015). Comparison of bacterial succession in green waste composts amended with inorganic fertiliser and wastewater treatment plant sludge. Bioresour. Technol. 179, 71–77. doi: 10.1016/j.biortech.2014.11.107
Sun T., Cui Y., Berg B., Zhang Q., Dong L., Wu Z., et al. (2019). A test of manganese effects on decomposition in forest and cropland sites. Soil Biol. Biochem. 129, 178–183. doi: 10.1016/j.soilbio.2018.11.018
Suzuki M. T., Taylor L. T., and DeLong E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5’-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Taparia T., Hendrix E., Nijhuis E., De Boer W., and van der Wolf J. (2021). Circular alternatives to peat in growing media: A microbiome perspective. J. Cleaner Production 327, 129375. doi: 10.1016/j.jclepro.2021.129375
Todd-Searle J., Friedrich L. M., Oni R. A., Shenge K., LeJeune J. T., Micallef S. A., et al. (2020). Quantification of Salmonella enterica transfer between tomatoes, soil, and plastic mulch. Int. J. Food Microbiol. 316, 108480. doi: 10.1016/j.ijfoodmicro.2019.108480
Toju H. and Sato H. (2018). Root-associated fungi shared between arbuscular mycorrhizal and ectomycorrhizal conifers in a temperate forest. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00433
Trivedi P., Leach J. E., Tringe S. G., Sa T., and Singh B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Vandecasteele B., Pot S., Maenhout K., Delcour I., Vancampenhout K., and Debode J. (2021). Acidification of composts versus woody management residues: Optimizing biological and chemical characteristics for a better fit in growing media. J. Environ. Manage 277, 111444. doi: 10.1016/j.jenvman.2020.111444
Vandepol N., Liber J., Yocca A., Matlock J., Edger P., and Bonito G. (2022). Linnemannia elongata (Mortierellaceae) stimulates Arabidopsis thaliana aerial growth and responses to auxin, ethylene, and reactive oxygen species. PloS One 17, e0261908. doi: 10.1371/journal.pone.0261908
Veselská T., Homutová K., García Fraile P., Kubátová A., Martínková N., Pikula J., et al. (2020). Comparative eco-physiology revealed extensive enzymatic curtailment, lipases production and strong conidial resilience of the bat pathogenic fungus Pseudogymnoascus destructans. Sci. Rep. 10, 16530. doi: 10.1038/s41598-020-73619-7
Visagie C. M., Houbraken J., Frisvad J. C., Hong S.-B., Klaassen C. H. W., Perrone G., et al. (2014). Identification and nomenclature of the genus Penicillium. Stud. Mycol. 78, 343–371. doi: 10.1016/j.simyco.2014.09.001
Wang X. W., Houbraken J., Groenewald J. Z., Meijer M., Andersen B., Nielsen K. F., et al. (2016). Diversity and taxonomy of Chaetomium and chaetomium-like fungi from indoor environments. Stud. Mycol. 84, 145–224. doi: 10.1016/j.simyco.2016.11.005
Wang B., Wang Y., Wei Y., Chen W., Ding G., Zhan Y., et al. (2022). Impact of inoculation and turning for full-scale composting on core bacterial community and their co-occurrence compared by network analysis. Bioresour. Technol. 345, 126417. doi: 10.1016/j.biortech.2021.126417
Warnes G., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., et al. (2024). _gplots: Various R Programming Tools for Plotting Data_. R package version 3.1.3.1. Available online at: https://CRAN.R-project.org/package=gplots.
Weiss S., Zankel A., Lebuhn M., Petrak S., Somitsch W., and Guebitz G. M. (2011). Investigation of mircroorganisms colonising activated zeolites during anaerobic biogas production from grass silage. Bioresour. Technol. 102, 4353–4359. doi: 10.1016/j.biortech.2010.12.076
Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag). Available at: https://doi.org/10.1007/978-3-319-24277-4_9.
Keywords: Salmonella enterica serovar Typhimurium, peat reduction, microbiome, bacterial community, fungal community
Citation: Müller A, Schmidt J, Maiberg V, Gehring O and Schikora A (2025) Microbial communities and substrate properties influence the fate of a human pathogen in horticultural substrates with different peat content. Front. Hortic. 4:1568055. doi: 10.3389/fhort.2025.1568055
Received: 28 January 2025; Accepted: 06 May 2025;
Published: 30 May 2025.
Edited by:
Chris Blok, Wageningen University and Research, NetherlandsReviewed by:
Jane Debode, Institute for Agricultural, Fisheries and Food Research (ILVO), BelgiumGabriele Bellotti, Catholic University of the Sacred Heart, Italy
Copyright © 2025 Müller, Schmidt, Maiberg, Gehring and Schikora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam Schikora, YWRhbS5zY2hpa29yYUBqdWxpdXMta3VlaG4uZGU=
 Antje Müller
Antje Müller Jasmin Schmidt
Jasmin Schmidt Adam Schikora
Adam Schikora