- 1Indian Council of Agricultural Research-Central Citrus Research Institute, Nagpur, Maharashtra, India
- 2Soil and Water Research Institute, Agricultural Research, Education and Extension Organization, Karaj, Iran
- 3AAU-Assam Rice Research Institute, Assam Agriculture University (AAU), Titabor, Assam, India
- 4Department of Fruit Science, Faculty of Agricultural Sciences, Siksha ‘O’ Anusandhan (Deemed to be University), Bhubaneswar, Odisha, India
- 5Division of Horticulture Science, Indian Council Agricultural Research, New Delhi, India
- 6Department of Horticulture, Maharana Pratap Horticultural University, Karnal, Haryana, India
- 7ELGO-DIMITRA/Olive Tree, Subtropical Plants and Viticulture, Athens, Greece
The rhizosphere is a dynamic environment in which multiple microbial activities elicit phenotypical, physiological, and molecular crop responses. For a better understanding of the rhizosphere microbiome, researchers are utilizing next-generation sequencing to focus on microbiome regulations with an emphasis on multi-functional microbes. There are two main concepts currently being focused on: identifying microbial antagonists (between beneficial microbes and plant pathogens) from predominant stocks of plant-growth-promoting microbes, preferably with an aim towards bioprospecting soil-plant health; and secondly, developing a more microbially active rhizosphere through a process called rhizosphere hybridization (RH). The present review is focused on some recent studies on the outcome of RH in citrus cultivars, showing renewed functional corridors of the rhizosphere characterized by secondary metabolites providing a load-supporting functional dichotomy through elevated nutrient-supply, activated soil enzyme profiles, and improvements in root- shoot systems and plant defense enzymes. These response trade-offs collectively contributed to higher quality yield coupled with possibly a better shelf life of fruits. The rhizobiome of heritage trees viz., Azadirachta, Ficus, Dendrocalamus, Populus, Sasa, Acer, Alnus, Quercus, and Phyllostachys, could be effectively used in exercising RH. These observations on RH mean the concept could be expanded in other fruit crops, with an emphasis on developing a robust holobiont (climate-smart suppressive soils and engineering rhizosphere microbiomes for microbially engineered plants) as a part of regenerative agriculture.
1 Introduction
The soil region adhering to the plant root system, popularly known as rhizosphere, is said to exhibit the greatest microbiome diversity, responsible for changes in the soil bio-physico-chemical properties influenced by root growth and their elevated activity (Pinton et al., 2007). The term “Rhizosphere” was coined by the German scientist Hiltner (1904) from two Greek words i.e. rhiza (root) and sphere (field of influence). There are three broad classifications (Prashar et al., 2013) describing three different tiers of rhizosphere properties: i. root tissues enclosing endodermis and cortical layers (endorhizopshere); ii. the root surface adhering to soil particles and microbes (rhizoplane); and iii. soil immediately adjacent to the root (ectorhizosphere).
Plant roots release various organic compounds through exudation, secretion, and deposition, from seed germination through to the growth of an adult plant, thereby biologically transforming the rhizosphere (Moshiri et al., 2019; Raiesi et al., 2022; Nourgholipour et al., 2022). In this process, plants recruit active microbial communities within the rhizosphere along and within the rootzone (Mousavi et al., 2018). Organic compounds such as water soluble exudates, lysates, dead fine roots, and inorganic ions are made up of gases released by living root systems that are deposited into their surrounding environment and are collectively called rhizodeposition (Whipps and Lynch, 1985). The nature and properties of rhizodeposition increase the nutrient supply in the soil (Solanki et al., 2020). The carbon derived from plants via roots have three different routes of transformation (Cheng and Gershenson, 2007): i. root mass with living or dead root cells; ii. rhizodeposits with plant-derived materials that are effectively transformed by the rhizosphere; and iii. carbon dioxide released by roots and microbial communities. Interestingly, the microbial diversity of the rhizosphere changes in terms of its composition, structure, and function depending on plant developmental and health status, genotypes, and prevailing soil conditions (Zhang et al., 2021a). The rhizosphere microbiota have strong relationships with plant growth and health through nutrient transformation, disease resistance, and substrate metabolism (Wei et al., 2020). Studies on different plants have confirmed that rhizosphere microbiota regulated by breeding and plant domestication play pivotal roles in plant resistance to soil pathogens (Berendsen et al., 2018; Yin et al., 2021). Such complex microbial food webs (referred to as combined trophic interactions) develop in the rhizosphere, linking different microbial communities with environmental conditions and management practices (Jeffery et al., 2010).
Acknowledging the rhizosphere as a unique niche of complex microbial populations that governs soil-plant health-related issues on one hand and sustaining crop production on the other hand (Avis et al., 2008; Singh et al., 2018a) are the two major pillars of sustainable crop production. Conventional plant hybridization ensures plants inherit beneficial genes from both parents, but the influence of such hybridization on the rhizosphere microbiome has not been studied much. This area could potentially aid in identifying promising microbial communities linked to multiple stress tolerance. This kind of attempt also showcases how the plant and microbial gene pool could be integrated synergistically to obtain a microbially engineered plant with an emphasis on developing a hyper-diverse rhizosphere microbiome-driven multiple plant tolerance.
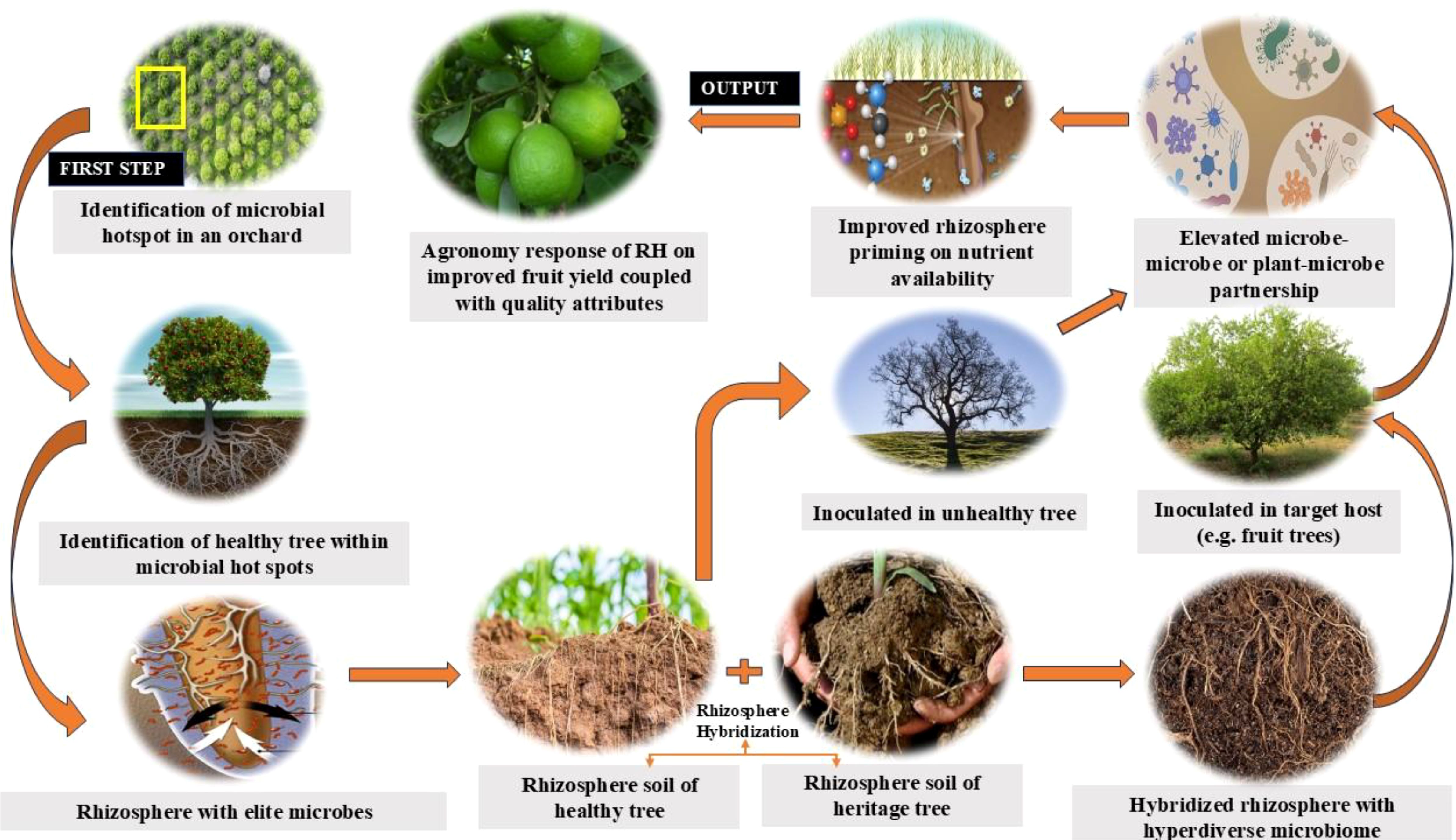
In addressing this important issue, comparative studies (Zhang et al., 2021b; Wang et al., 2023; Xie et al., 2024) on monoculture versus intercropping cultivation patterns of fruit crops have revealed significant differences in the reshaping of microbial communities and the metabolism of both beneficial microbes and microbial antagonists (Lu et al., 2019). Shen et al. (2024) observed how growing almond mushroom (Agaricus blazei Murill) intercropped with kiwifruit (Actinidia chinensis L.) aided in identifying 95 bacteria genera and 79 soil metabolites. In another study on intercropping sweet potato (Ipomoea batatas (L) Lam.) in banana (Musa nava Lour.) orchard (Li et al. 2022), soil bacterial, and fungal populations were shown to be significantly higher in intercropped banana (476.0-511.7 and 154.3-198.3 colonies) compared to monocropped banana (397.7-451.0 and 112.0-147.0 colonies). Likewise, studies on Pinto peanut (Arachis pintoi L.) intercropped in litchi (Litchi chinensis Sonn.) orchard showed improved metabolic activity of advanced bacterial communities, leading to higher potassium supply that correlated to the resistance of litchi root systems to soil borne-diseases (Zhao et al., 2022).These studies provide ample evidence of microbial communities undergoing mutational changes via different trophic interactions when conditioned by root exudates of main crops and intercrops. We propose the term “Rhizosphere Hybridization” (RH) to describe this conceptual framework (Figure 1). RH is a concept that involves combining the microbial diversity of different crop rhizospheres to improve the rhizosphere function of targeted crops for improved plant growth and development. In this paper, efforts were made to collate the work done on the rhizosphere microbiomes of different promising tree plants that could align with RH as well as put forth some future lines of research on the concept.
2 Bibliometric analysis of rhizosphere research
The authors sought to analyze various areas of rhizosphere microbiome research by reviewing publications from varied databases representing Web of Science, Google Scholar, Springer Link, and Wiley-Blackwell databases using keywords such as “rhizosphere”, “rhizosphere microbiota”, “rhizosphere interactions”, and “rhizosphere hybridization”. The results showed that 25320 publications on the rhizosphere were published from 2000-2022: 18255 research articles, 2553 review articles, and 4512 publications in the form of book chapters, encyclopedias, and conference abstracts. Of these, a majority of the 12082 publications were dedicated to agricultural and biological sciences. One notable point is that the rhizosphere studies in the subject areas of “Agricultural and Biological Sciences” and “Environmental Science” began in 2010. These results aided in understanding the process of RH and its agronomic impacts on crops. Only 1661 publications were dedicated to rhizosphere studies published from 2000-2022. Finally, based on the main objectives of the present study focusing on rhizosphere effects and RH, their impact on different crops, and rhizosphere health, 48 research and 14 review papers featuring issues such as fruit crop-based intercropping systems and rhizosphere studies of heritage trees were selected for onward discussion. Such a bibliometric analysis of rhizosphere research facilitated in identifying the gaps in rhizosphere research (Figure 2) and, at the same time, provided strong clues towards the rising popularity of rhizosphere research worldwide.
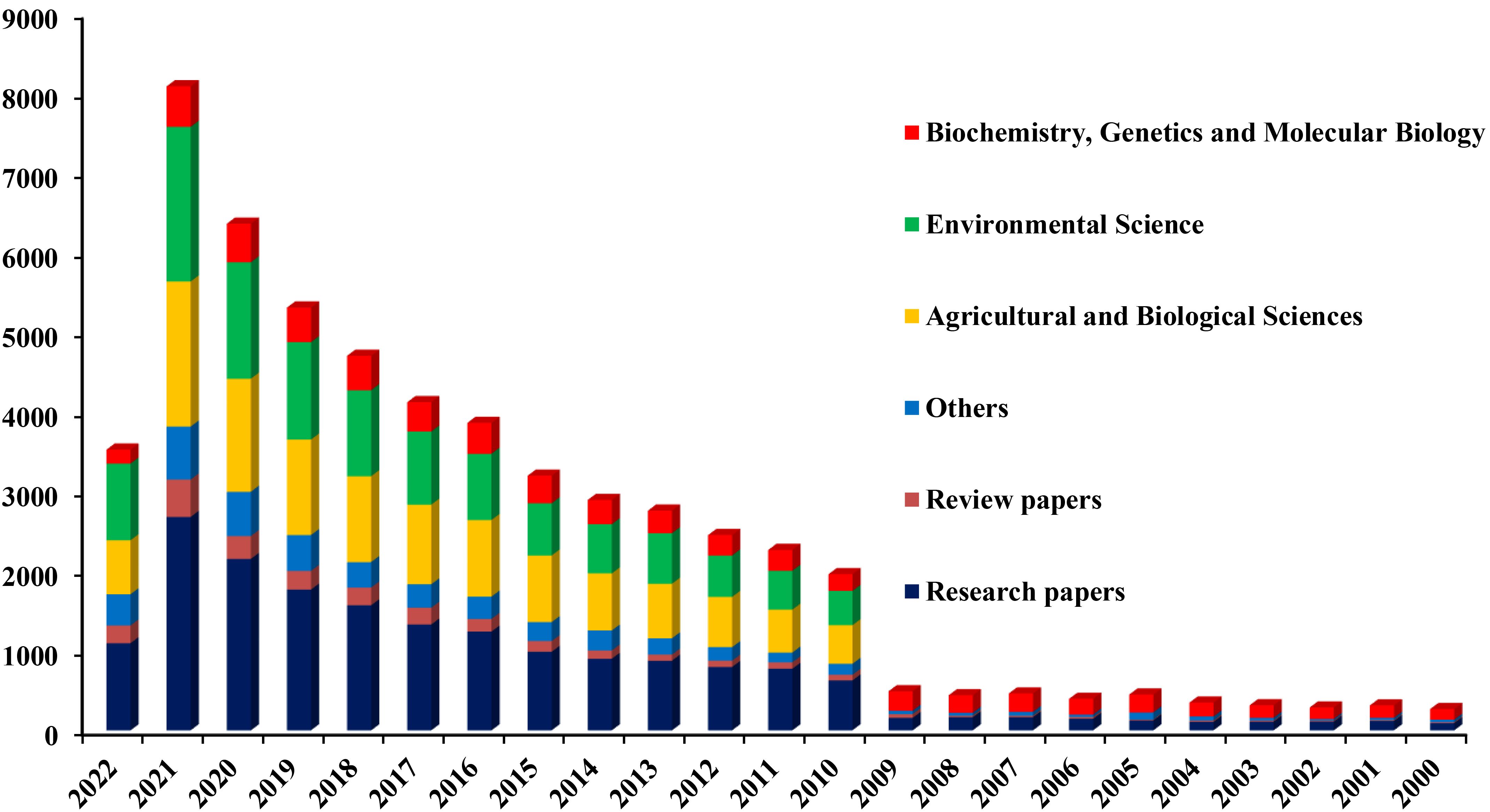
Figure 2. Bibliometric analysis of rhizosphere research highlighting the growing dynamics of rhizosphere-related issues over the past 22 years (2000-2022). Rhizosphere microbiome research has been one of the core issues among researchers worldwide.
3 Rhizosphere, the seat of microbial battle
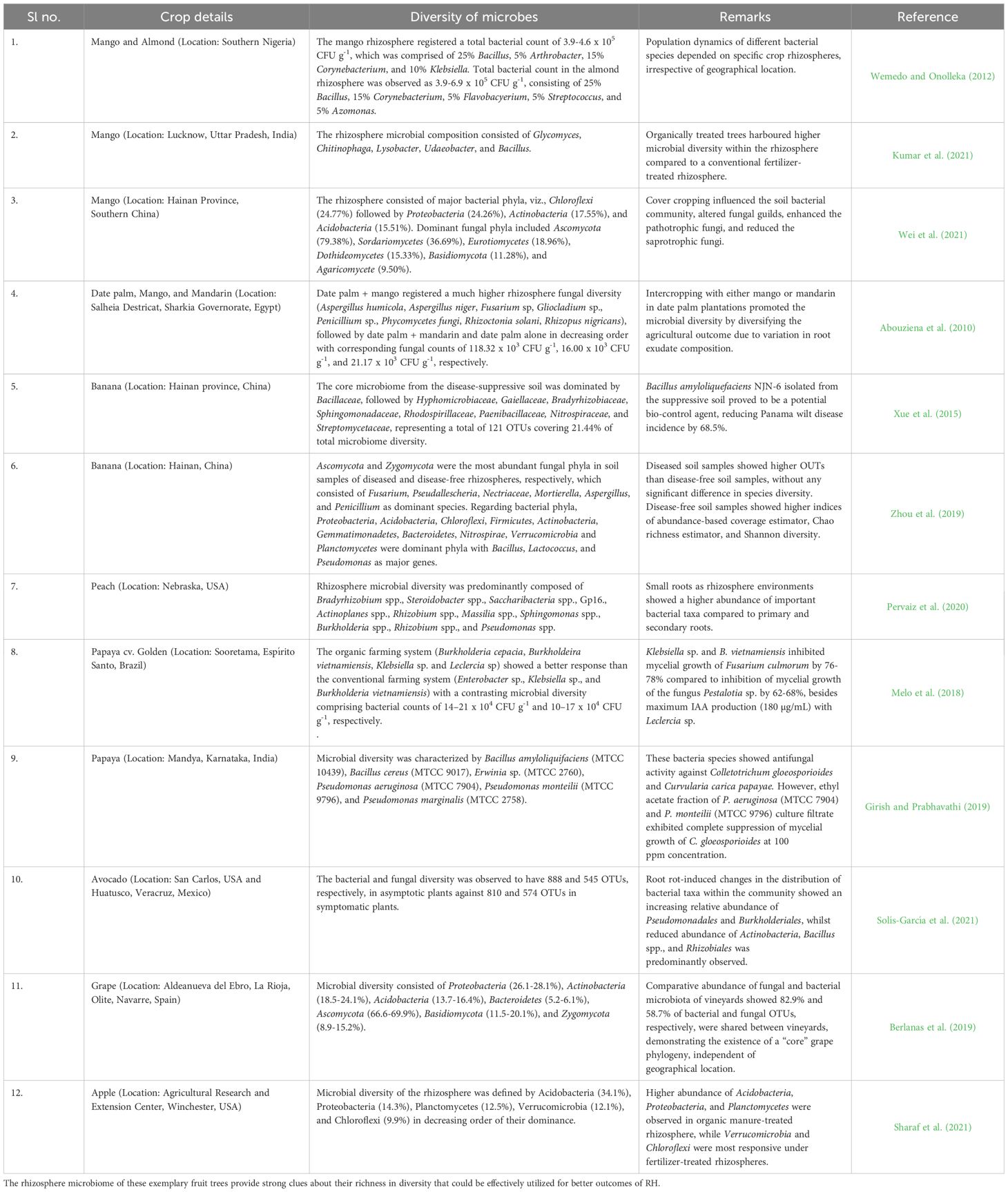
The rhizosphere is considered a dynamic battlefield between pathogenic and growth-promoting microorganisms, the outcome of which is largely determined by suitable management practices. These “battles” are where all active microorganisms interact among themselves through beneficial (mutualistic), neutral (commensalism), or detrimental (parasitic) relationships. Microbial build-up of the rhizosphere plays a vital role in mediating health and fitness against multiple stresses (Hacquard and Schadt, 2015; Zhang et al., 2021b) and, responsively, the host forms its microbial assemblage (Sasse et al., 2018). Although limited studies have highlighted the rhizosphere microbiomes of tree species (Uroz et al., 2010, 2016; Gallart et al., 2018), knowledge on the links between site factors, composition, and specific microbial niches concerning tree health is still lacking. The rhizosphere ecosystem of fruit trees is based on a fruit tree-soil-microbe relationship (representing the phytobiome) and their interactions with environmental attributes (Table 1).
Among subtropical fruit crops, citrus is one of the most extensively researched worldwide (Srivastava and Hota, 2020). In India, citrus microbiome studies have been undertaken from various angles (Srivastava et al., 2015a), with a particular focus on microbes-mediated soil fertility management (Srivastava and Ngullie, 2009; Srivastava and Malhotra, 2017) to sustain both production (Srivastava and Singh, 2008a) and fruit quality (Shamseldin et al., 2010; Srivastava et al., 2015b), as well as to invesitgate properties (Ngullie et al., 2015). Citrus is a highly nutrient-responsive crop, due to the existence of a strong nutrient sink (Srivastava and Singh, 2008b). However, regulating quality production through exploitation of the citrus rhizosphere microbiome has become a massive challenge (Srivastava and Singh, 2009). Some initial attempts have shown that citrus, like any other fruit crop, transforms the rhizosphere microenvironment according to the prevailing soil fertility gradient (Srivastava and Singh, 2009). These citrus rhizosphere-based microbes showed a strong association with citrus root either alone (Srivastava, 2010) or in combination with organic manure (Srivastava et al., 2002) and/or in combination with chemical fertilizers (Srivastava et al., 2015b). Likewise, changes in rhizosphere properties vis-à-vis fruit crops have been studied, highlighting crops such as strawberries (Kumar et al., 2020), apples (Soliman et al., 2023), citrus (Srivastava et al., 2017), fig (Abid et al., 2022), and pear (Zhang et al., 2020).
3.1 Rhizosphere Microbiome of Heritage Trees
It is common to see an unwarranted decline in the productivity of perennial crops, especially in fruit crops, after attaining peak productivity (Srivastava and Singh, 2009). This is due to the specific rhizosphere environment, popularly known as the “Negative Rhizosphere Effect” (NRE). NRE is commonly observed in crops like tea, apple, peach, and pear (Pandey et al., 2001; Somera and Mazzola, 2022). NRE is often regarded as a “Replant Disease” The underlying reasons for such a rhizosphere-driven decline in productivity is still not understood. Is it a case of the rhizosphere priming on microbial mining or nutrient mining? Sustaining the peak productive life of fruit trees is perhaps the most formidable challenge in agriculture. Researchers worldwide are still working hard to come up with well-conceptualized mechanisms associated with NRE. Modifications in the structural and functional microbial fabric of the rhizosphere due to continuous cropping is often regarded as the root cause of NRE. Sui et al. (2024), while comparing BT (perennial Poplar big trees) and CK (replanting the Poplar seedlings in soil after continuous cropping) groups of Poplar trees, reported no change in Bacillus (2.2-2.41%), while Inocybe and Geopora were 35.3 and 26.2% higher, respectively, in BT than CK groups, suggesting the rhizosphere microbiome was modified in the most effective way to ward off any NRE arising from continuous cropping. Dijkstra et al. (2013) proposed three nutrient-centered hypotheses: i. microbial mining causing a positive rhizosphere priming effect under low soil nutrient availability, ii. preferential substrate utilization inducing a shift in action from decomposing soil organic matter to utilizing rhizodeposition under high soil nutrient availability, and iii. competition causing a negative rhizosphere priming effect due to nutrient-limited microbial growth and decomposition (Weller et al., 2002).
Studies on the rhizosphere microbiome composition of f heritage trees have opened new avenues for identifying and isolating elite microbes with plant growth-promoting potential and antagonistic properties, as well as inoculation into the rhizosphere of targeted fruit crops. Due to their extended life cycle and non-deciduous nature, heritage trees are far less sensitive to biotic and abiotic stresses such as drought, salinity, herbivores, and pathogen attacks (Rodriguez et al., 2019; Zamora Ballesteros et al., 2019; Oliva et al., 2020; Pagán et al., 2022), meaning there is little threat against tree health (Reddy et al., 2013). The occurrence of a strong relationship between a rhizosphere microbiome and plant traits is a prerequisite to plant resistance against soil-borne pathogens (Bora and Bora, 2020a; b) and can be particularly helpful in heritage/forest trees. In a recent study on the rhizosphere microbiome of forest trees, Yu et al. (2022) observed considerable inhibitory effects of bacterial families (such as Propionibacteriaceae, Phycisphaeraceae, and Rokubacteria) on fungal pathogens of rhizosphere microbiota of seven forest tree species with the differential ability of recruiting key rhizosphere microbes as a function of root exudates. This strengthened the tree-microbial association, with significant differences amongst tree varieties. Hence, optimizing the microbial community of fruit tree rhizospheres with the rhizospheres of heritage trees (e.g. different species of Ficus, Azadirachta etc) could be an effective approach for improving growth and yield and developing resistance against soil-borne pathogens in fruit crops.
As many as 40% of plant photosynthates are reported to be lost by root systems, resulting in a nutrient-enriched rhizosphere that harbors a greater diversity of microbiome (Nannipieri et al., 2007) and has diversified roles in plant growth and development. Soil health with respect to physical and chemical properties depends on the biological activity of the rhizosphere (Srivastava and Singh, 2001) to be involved in the maintenance of soil health and quality. The microbiome diversity within a crop rhizosphere is, therefore, a product of crop variety or cultivar, rootstock-scion combination, age of plant, root architecture, management practices (Srivastava et al., 1994; Gupta et al., 2008), and soil properties (Srivastava and Singh, 2002). Various relationships of different microbial communities within the rhizosphere of a plant take place through the enhancement of plant growth nutrient availability and soil health and the suppression of the pathogenicity of disease-causing pathogens (Pinho et al., 2020).
4 Crops with unique rhizosphere properties
Soil microbial activities and their population dynamics are significantly affected in the presence of living roots, popularly known as the “rhizosphere effect”, which regulates the accumulation pattern of different nutrients and ecosystem functioning in terrestrial ecosystems (Yuan et al., 2020). Interestingly, the phylogenetic distance of plant hosts and associated bacterial communities are closely related (Lei et al., 2019). Sinha et al. (2009) proposed the rhizosphere properties rhizosphere microbial index, dehydrogenase, basal soil respiration/microbial biomass carbon ratio, electrical conductivity, phenol oxidase, and active microbial biomass carbon as the most critical in defending the rhizosphere of tree species such as Aegle marmelos, Azadirachta indica, Bauhinia bauhinia, Butea monosperma, Eugenia jambolana, Moringa oleifera, Dalbergia sissoo, Tamarindus indica, Morus alba, Ficus religiosa, Eucalyptus sp., and Tectona grandis.
The rhizobiome of tree species such as banyan (Ficus benghalensis L.), bamboo (Dendrocalamus strictus (Roxb.) Nees), Neem (Azadirachta indica A. Juss), which possess inherent abilities for high biomass production, higher microbial activity, and broad geographic adaptability makes them preferable choices as bio-inoculants (Table 2). But, an in-depth investigation of microbial diversity and functions to identify multi-functional microbes followed by co-evolutionary changes as a result of the introduction into the rhizosphere of target crop(s) is imperative to redefine the microbial networking of new rhizospheres, called “hybridized rhizosphere”. Evidence for such novel possibilities can be easily drawn from crop-specific rhizosphere microbiome traits and their reshaping in response to crop management inputs. Nimoni and Pongslip (2009) observed a large number of indole acetic acid (IAA) forming bacterial diversity in Ficus religiosa. Isolation and characterization of isolates showed 91% similarity with Rhizobium spp. Brevibacterium, an endophytic bacterium, isolated and characterized from the rhizosphere of F. religiosa. Studies on the efficacy of synthesizing IAA in Raphanus sativus by inoculating the bacterial species isolated from 18 soil and 10 new isolates from the roots of Brassica oleracea showed an elevated response on root and shoot development than the control. These observations showed the potential of inter-rhizosphere microbiome interactions at different trophic levels toward better crop response.
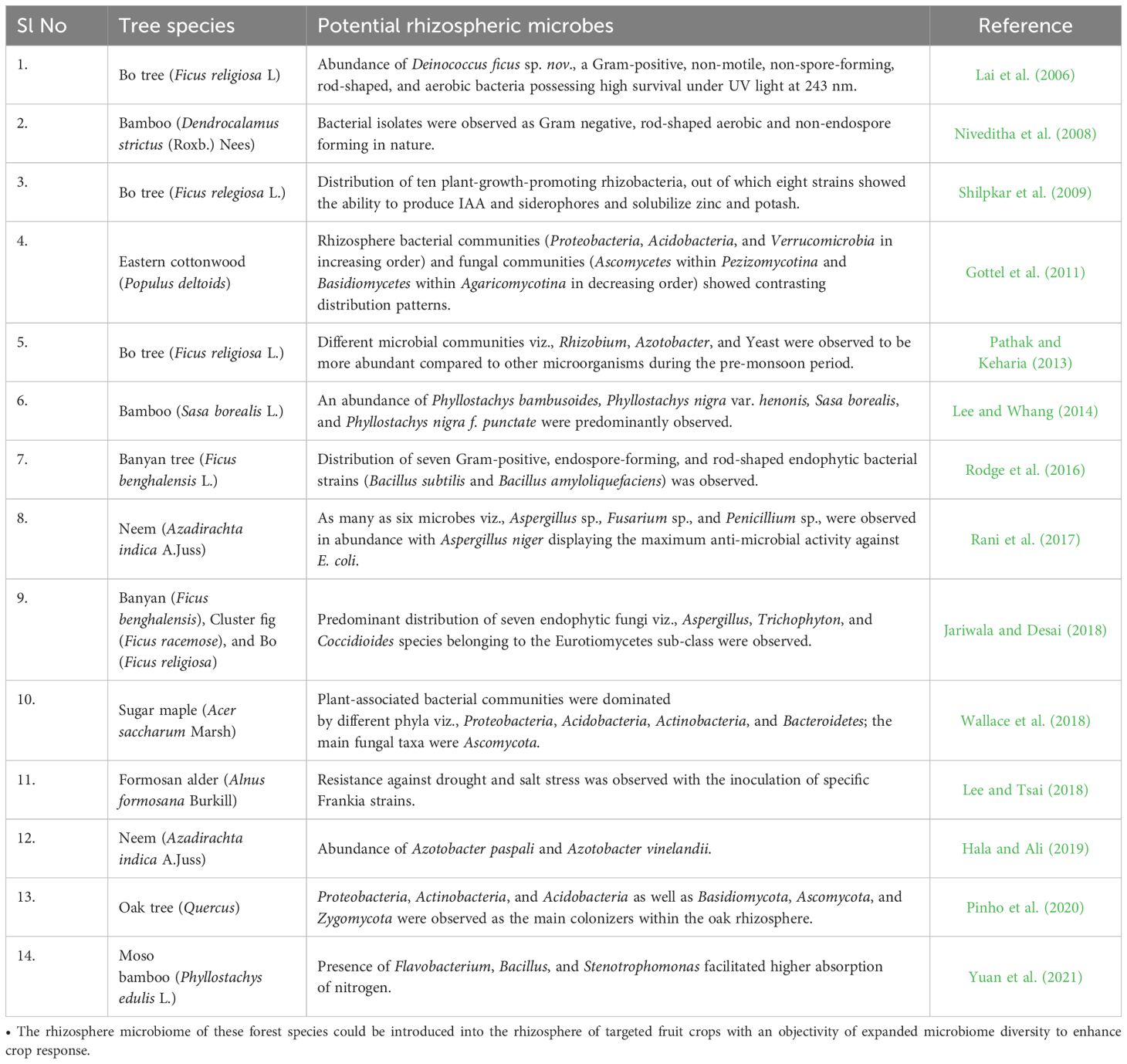
Table 2. Nature and properties of rhizosphere microbiomes of different worldwide wild forest species.
Peter and Pandey (2014) collected the rhizosphere samples of bamboo and isolated two strains of Pseudomonas spp. out of a total of 22 isolates. The morphological and biochemical tests confirmed the isolates as P. fluorescence (59% diversity) and P. auruginosa (40.9% diversity). The abiotic stress tolerance activity showed that these two species thrived up to 10°C at a wide pH range of 5-9, having a tolerance level of salt concentration from 2–6 ppm. P. fluorescence and P. auruginosa were also observed to inhibit Fusarium oxysporum, Rhizoctonia solani, and Alterneria solani by 59.5-64.3%, 42.0-45.9%, and 50.0-52.5%, respectively. Kanse et al. (2015) isolated six phosphate-solubilizing fungi, tentatively identified based on colony morphology as Talaromyces, Aspergillus, and Rhizobium. One isolate, SLS8, based on 8s RDNA sequence, was confirmed as T. funiculosus, showing maximum phosphate-solubilizing ability compared to others. The study also suggested T. funiculosus SLS8 as a promising inoculant (seed inoculant) for maintaining good soil phosphate levels in saline soil.
Soil respiration is an indirect measurement of the autotrophic microbial load of soil, which in turn is an indicator of ecological sustainability. In a study on in-situ measurement of soil respiration by Prasad and Baishya (2019), the highest annual soil respiration was observed under the canopy of F. religiosa (18.72 µmol CO2 m-2 s-1year) and lowest under A. indica (4.58 µmol CO2 m2/s/year) during rainy and winter seasons, respectively. Different tree species showing a decreasing order of soil respiration were observed to be F. religiosa > A. lebbeck > P. juliflora > V. leucophloea > M. pinnata > C. fistula > A. indica. The higher soil respiration under the canopy of native over non-native species further suggested the importance of the interactive effects of soil moisture and temperature in the ecosystem of heritage trees that could be used well in the process of RH. The uniqueness of rhizosphere properties in terms of microbial diversity and functions needs to be upscaled to utilize RH successfully.
4.1 Rhizosphere properties of citrus (as a case study)
Soil microorganisms in the citrus rhizosphere play a decisive role in improving soil ecology through changes in soil properties, thereby helping improve the nutrient supply chain for the betterment of citrus growth and development (Srivastava and Malhotra, 2017; Srivastava and Singh, 2002; Ortas, 2012). In response to the rhizosphere as a biological indicator for soil fertility, soil microorganisms are major contributors to defining the production sustainability of citrus orchards (Srivastava et al., 2008). Citrus trees are considered highly dependent on arbuscular mycorrhizal (AM) symbiosis as citrus roots are characterized by short and poorly distributed root hairs (Graham and Syvertsen, 1985). Van Heerden et al. (2002) reported an abundance of various fungi in the citrus rhizosphere, consisting of Aspergillus fumigatus, Absidia corymbifera, Penicillium diversum, Emericella nidulans, Rhizomucor pusillus, Paecilomyces variotii, Thermomyces lanuginosus, and Talaromyces thermophilus. Some arbuscular mycorrhizal fungi (AMF), including Glomus, Gigaspora, Entrophospora, Scutellospora, and Acaulospora species, have been reported to occur frequently in citrus orchards (Wu and Srivastava, 2012). In a fungal diversity study of Citrus unshiu Marc. trees grafted on Poncirus trifoliata, Sun et al. (2017) reported 579 and 566 operational taxonomic units (OTUs) of fungi in plant roots and rhizosphere soil, respectively. Out of these, 462 OTUs intersecting between the roots and rhizosphere soil of citrus indicated that plant roots are favorable sites for the growth and development of those fungi rather than rhizosphere soil. Considering the phylum, Ascomycota was the dominant fungal species in soil and roots. Kohli et al. (1997) reported that microbial populations in soil play a crucial role in increasing the fruit yield of Nagpur mandarin with plant-available nutrients. The correlation values of Azotobacter count (r = 0.692, p= 0.01), ammonifiers (r= 0.512, p= 0.01), and phosphate-solubilizing bacteria (r = 0.618, p= 0.01) showed a stronger connection with fruit yield than plant-available nitrogen (r = 0.489, p= 0.01), phosphorous (r= 0.316, p= 0.05), and potassium (r= 0.321, p= 0.05) in soil, signifying the potential of the population density of microbes as a bio-indicator of elevated fruit yield.
He et al. (2002) reported that soil microbial biomass nutrients viz., microbial biomass carbon, (Cmic 1.62-3.16 mg/kg), microbial biomass nitrogen, (Nmic 19.0-35.2 mg/kg), and microbial biomass phosphorous, (Pmic 20.2-42.3 mg/kg) constituted only 1.61-2.60%, 1.2-2.5% and 2.4-8.4% of total organic carbon, respectively, in citrus orchards in Zhejang province, China. The phosphorous-solubilizing capacity of Bacillus subtilis, Bacillus polymyxa, Trichoderma viridi, and Aspergillus terreus found on citrus growing belts of India is reported to be 13.30-81.68% P2O5 through insoluble tricalcium phosphate (Bhattacharya et al., 1999). Not only the bacterial species but also the mycorrhiza species like Glomus caledonium, Glomus mosseae, and Glomus clarum are abundantly observed in citrus growing belts of Italy (Palazzo et al., 1992). In eastern Spain, Glomus mosseae and Glomus intraradices are commonly observed mycorrhizal fungi in citrus rhizosphere soils, which can be inoculated back into soils through agronomic practices like crop rotation with aromatic plants viz., Thymus vulgaris, Lavandula vera, L. angustifolia, and Rosmarinus officinalis (Camprubi and Calvet, 1996a, b). In Japan, citrus orchards are reported to be richly abundant in Gigaspora, Scutellospora, and Glomus AM fungi (Ishii and Kadoya, 1996). Bhattacharya et al. (1999) reported that the Glomus population was higher in the juvenile phase (1-5-year-old orchards) as compared to the reproductive stage (5-10-year-old orchards).
Ngullie et al. (2015) evaluated 15 citrus varieties, namely four limes/lemons, three mandarins, and eight sweet oranges, at the pre-bearing stage on Alfisol. The trend in the growth of canopy volume of different citrus cultivars supported the changes in bacterial count (CFUg-1) of 7.3 x 108, 4.9 x 106, 4.5 x 106, and fungal count (CFUg-1) of 7.7 x 107, 3.1 x 105, and 2.2 x 105 in limes/lemons, sweet oranges, and mandarins, respectively. Similarly, soil microbial biomass nutrients were also observed to be higher in lime/lemons as compared to sweet oranges and mandarins, indicating the greater affinity of lime/lemon rhizosphere to reproduce microbial biomass and maintain a better nutrient pool of soil. Citrus rootstocks having greater root volume reflected their vigorousness crop phenology in terms of canopy volume, a pre-requisite for ensuring higher fruit yield. The response of different fruit crops to microbial inoculation in terms of parameters related to soil fertility changes and crop response established their strong responsive nature (Table 3).
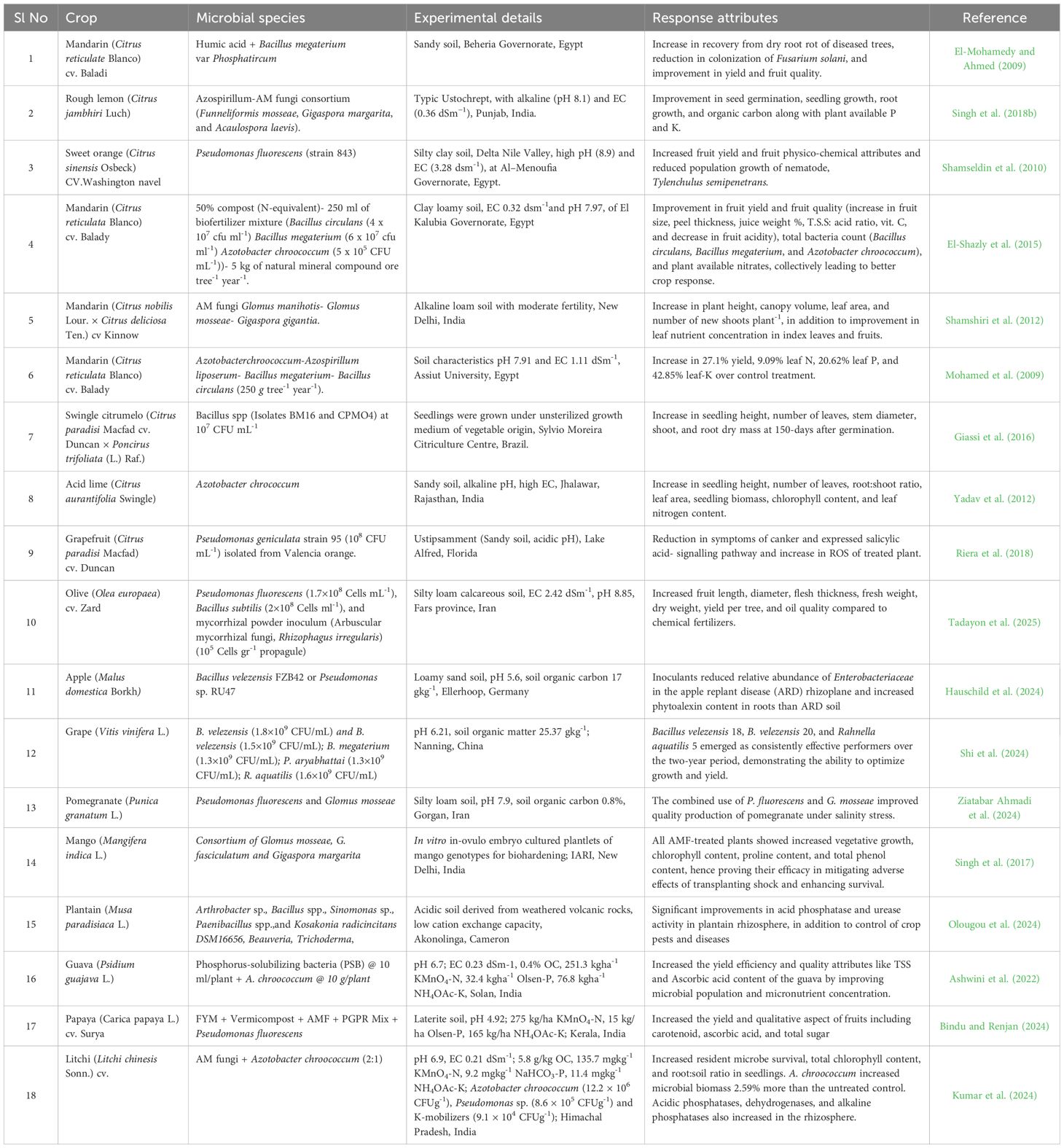
Table 3. Associative responses of fruit crops with microbial inoculants reported through worldwide literature.
Of late, culture-independent metagenomic studies aided the identification of newer microbial species and established a strong foundation for developing artificially constructed microbial communities known as artificial microbial consortia or synthetic communities (SynComs) using core microbiomes to recreate more robust structures and functions of the microbiome and representing different microbial niches. A great advantage of SynComs studies lies in the fact that members can be added, eliminated, or substituted as needed (Vorholt et al., 2017), in addition to elucidating spatial microbial interactions (Amor and Bello, 2019). Two main approaches are usually employed to artificially select the microbiomes: top-down (modifying the existing microbiome) and bottom-up (starting from individual microorganisms to build engineered microbiomes). In the top-down approach, selected environmental variables (e.g., pH, temperature, and redox potential) are used to manipulate the existing microbiome through ecological selection (Lawson et al., 2019). Although this approach is widely used for bioremediation (Atashgahi et al., 2018) and wastewater treatment (Demarche et al., 2012), it has the disadvantage of working with a complex community. In contrast, the bottom-up approach offers the biggest advantage of simplifying microbial interactions by building artificial communities from pre-selected individual organisms (Raaijmakers and Kiers, 2015).
Microbial taxa that are highly connected and more influential on the community in a pre-existing or artificially constructed microbiome are given maximum preference (Banerjee et al., 2018) for microbial screening followed by isolation and whole-genome sequencing to determine their functional capabilities (Kong et al., 2022). Consequently, their role in regulating the growth and function of other members of the microbiome can be effectively exploited to enhance specific desired functions. Such a concept of assembling SymComs is quite different from known and well-studied bacteria. Hence, it is possible to increase the number of desired bacterial strains while decreasing the number of undesired strains (Cloutier et al., 2023; Ma et al., 2022; Kugarajah et al., 2023) using the strong functional relationship between plant metabolites and the bacterial community diversity of the rhizosphere. In one of the recent incubation studies spanning 28 days, three different plant metabolites, namely benzoxazolinone, gramine, and quercetin, were added to the soil (Schutz et al., 2021) and showed that the bacterial diversity was significantly reduced by the first two metabolites only. Consequently, plants producing one or more of these metabolites are suggested to have a specific effect on the soil bacterial community.
5 Agronomic and microbial response of RH (our experiences)
While chemical or synthetic fertilization can improve fruit yield and quality, long-term use has failed to sustain the same yield expectancy due to erosion in soil carbon stock, culminating in the emergence of multiple nutrient deficiencies. Such unprecedented loss of soil fertility is likely to bring changes in microbial communities within the rhizosphere (Srivastava et al., 2008), coupled with reoriented production dynamics. Artificially, the rhizosphere could be reconstructed depending upon the need of the plant to enhance physiological efficiency through rhizosphere engineering, popularly known as RH. This is synonymous with creating an artificial environment for plant growth-promoting microorganisms to provide another protective layer against the pathogenic microbes (Rhizosphere fortification). By and large, trees react and acclimate to antagonistic soil environments, and this can happen through different strategies, including changes in root exudation and rhizo-deposition, which bring variable changes in rhizosphere soil properties (Gargallo-Garriga et al., 2018).
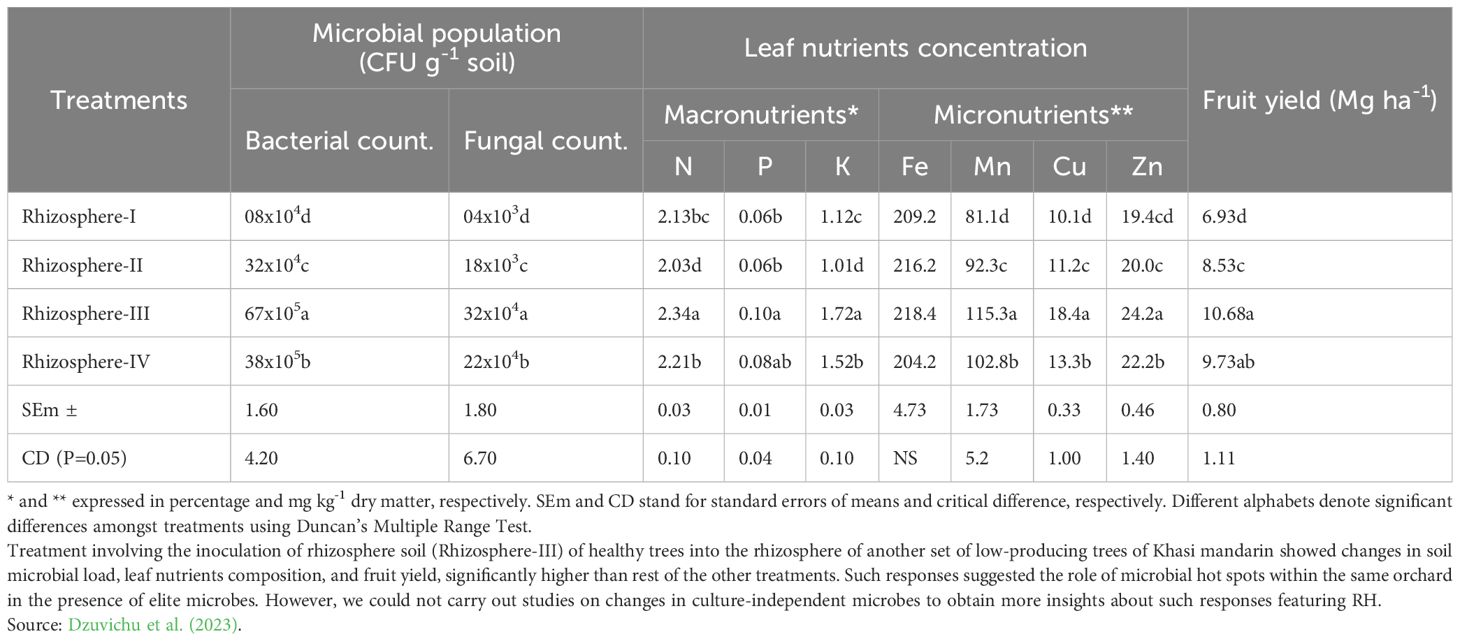
RH is a relatively recent introduction to rhizosphere research to modify rhizosphere ecology and create an environment for plant growth-promoting microbes to positively influence plant agronomy (Keditsu and Srivastava, 2014). The concept of RH is, therefore, put forward to demonstrate the value-added benefits of nutrient-microbe-plant synergy, in addition to the expected dynamism to microbial diversity in harmony with a wide range of fruit crops (Srivastava et al., 2015b, 2021). Studies were carried out in the past (Cheke et al., 2018a; b) on the inoculation response of rhizosphere soils of three perennial heritage trees, namely Ficus racemosa L. (Common name: Umber tree), Ficus benghalensis L. (Common name: Banyan tree), and Ficus religiosa L. (Common name: Pipal tree), into the rhizosphere of sweet orange trees (Citrus sinensis Osbeck). The inoculation response of F. recemosa showed the best response on rhizosphere properties of sweet orange trees, associated with a net increase of 18.0 g kg-1 Walkley-C, 62.2 kg ha-1 KMnO4-N, 19.0 kg ha-1 Olsen-P, and 95.6 kg ha-1 NH4OAc-K, in addition to a 2.34 times increase in microbial biomass nitrogen (Nmic), 4.23 times increase in microbial biomass carbon (Cmic), and 2.81 times increase in soil alkaline phosphatase over rhizosphere of sweet orange trees alone. The magnitude of these responses was, however, significantly lower when compared with the rhizosphere of F. recemosa alone, with an increase in Walkley-C by 11.2 g kg-1, KMnO4-N by 32.2 kg ha-1, Olsen-P by 11.2 kg ha-1, and NH4OAc-K by 28.2 kg ha-1, in addition to 1.34 -folds, 2.23-folds, and 1.62-folds increase in Nmic, Cmic, and soil alkaline phosphatase, respectively. Hence, microbially hybridized soil proved to be biologically highly active for better agronomic crop response (Figure 3). Later, another mode of RH (Dzuvichu et al., 2023) was studied by inoculating the rhizosphere of highly productive Khasi mandarin (Citrus reticulata Blanco) trees into lesser productive trees; after two seasons, significant improvements in fruit yield and soil fertility changes were observed (Table 4). The Rhizosphere III treatment observed significantly higher bacterial and fungal counts of 38 X 105 and 32 X 104 CFU g-1 soil, respectively, compared to other rhizosphere treatments, recording bacterial counts of 08 X 104–38 X 105 CFU g-1 soil and fungal counts of 04 X 103–22 X 104 CFU g-1 soil. These microbial changes with Rhizosphere III treatment were associated with 1.54 times (10.68 Mg ha-1) higher fruit yield compared to other rhizosphere treatments (6.93-9.73 Mg ha-1) as a function of significantly higher soil nutrients pool maintained with the Rhizosphere III treatment. Against the backdrop of such observations, how shall we identify the microbial hot spots featuring the development of elite microbial spp. within an orchard and rationalize the spatial distribution of microbes to optimize fruit yield without sacrificing the quality? These RH-based observations paved the way for the inoculation of soil rhizospheres or endosphere microbes for elevating micronutrient concentration in various plant parts viz., roots, leaves, and fruits (Ku et al., 2019), to sustain production on a long-term basis.
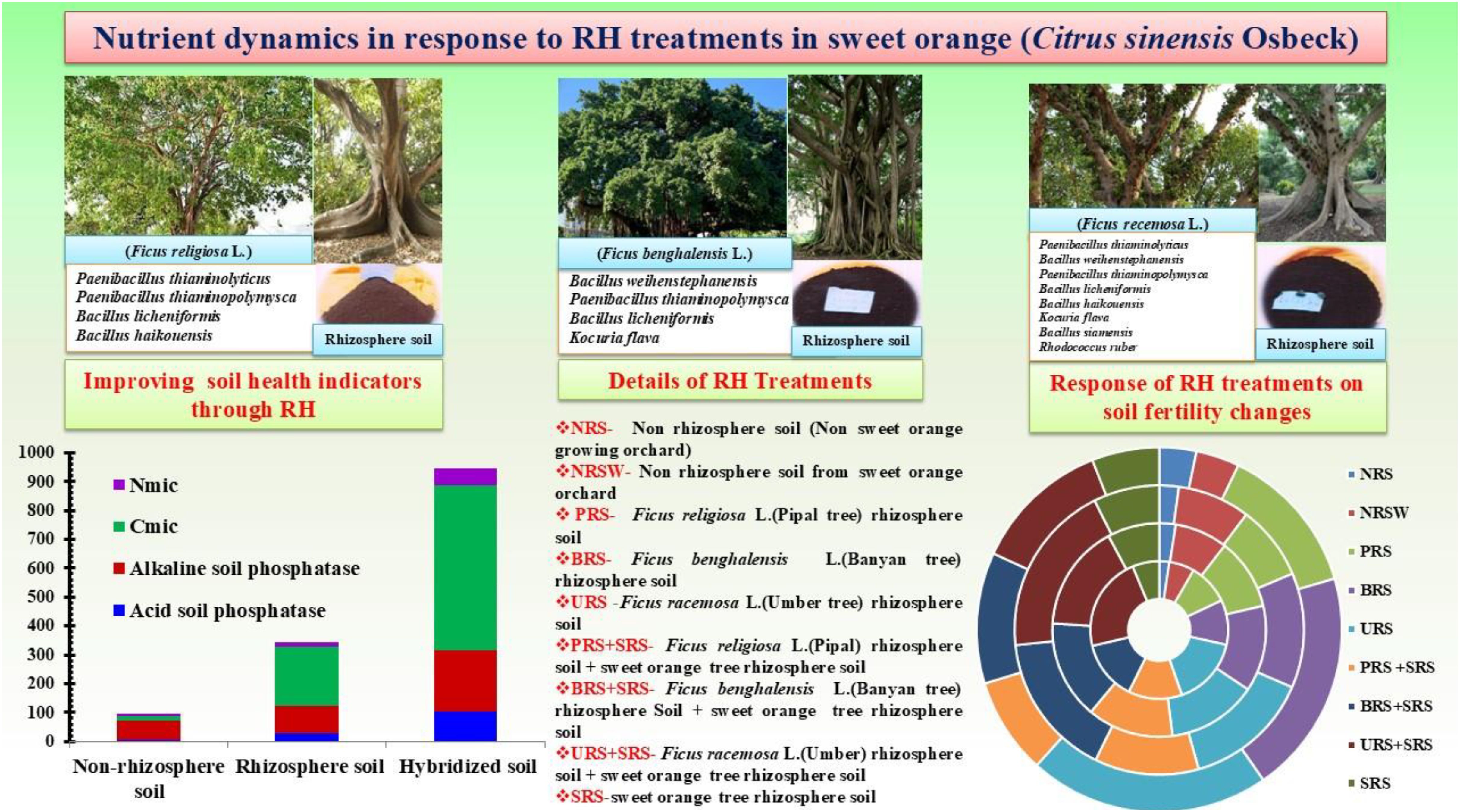
Figure 3. Diagrammatic representation on response of RH in sweet orange (Citrus sinensis Osbeck) grafted on Rangpur lime (Citrus limonia Osbeck) rootstock for improved rhizosphere health indicators (microbial biomass nitrogen, Nmic; microbial biomass carbon, Cmic; alkaline soil phosphatase; and acid soil phosphatase) and soil fertility changes (the inner most circle represents organic carbon, followed by KMnO4-N, Olson-P, and NH4OAc-K as we move on from the inner circle). Hybridized soil (URS + SRS) involving inoculation of rhizosphere of F. recemosa (URS) into the rhizosphere of sweet orange trees (SRS) proved to be far superior over either of the two alone. Source: Based on data generated through studies by Cheke et al. (2018a, b) and Srivastava et al. (2021).
Hota et al. (2021a), in their comprehensively planned study, successfully developed a hybridized soil (BbNByR) by using the rhizospheres of banyan (Ficus benghalensis L), neem (Azadirachta indica A.Juss), and bamboo (Dendrocalamus strictus (Roxb.) Nees) trees inoculated into rhizosphere of acid lime as the host fruit tree. This treatment showed significantly higher shoot growth parameters (30.4 cm seedling height, 3.0 mm seedling diameter, 5.8 branches seedling-1, and 38.9 leaves seedling -1) and root growth parameters (14.5 cm taproot length and 17.3 number of secondary roots seedling -1) over single rhizosphere effect of the control treatment involving host tree alone (Table 5). Synonymous to a hybridized rhizosphere response, the microbial consortium (Paenibacillus alvei Cheshire and Cheyne (MF113275), Micrococcus yunnanensis Cohn (MF113274), Bacillus pseudomycoides Nakamura (MF113272), Aspergillus flavus Link (MF113270), and Acinetobacter radioresistens Nishimura (MF113273)) displayed a much greater magnitude of response in mature acid lime trees through an increase in microbial biomass load of the rhizosphere, thereby adding additional rhizosphere resilience by reducing the mortality of new seedlings/buildings once planted in a new field (Hota et al., 2021b).
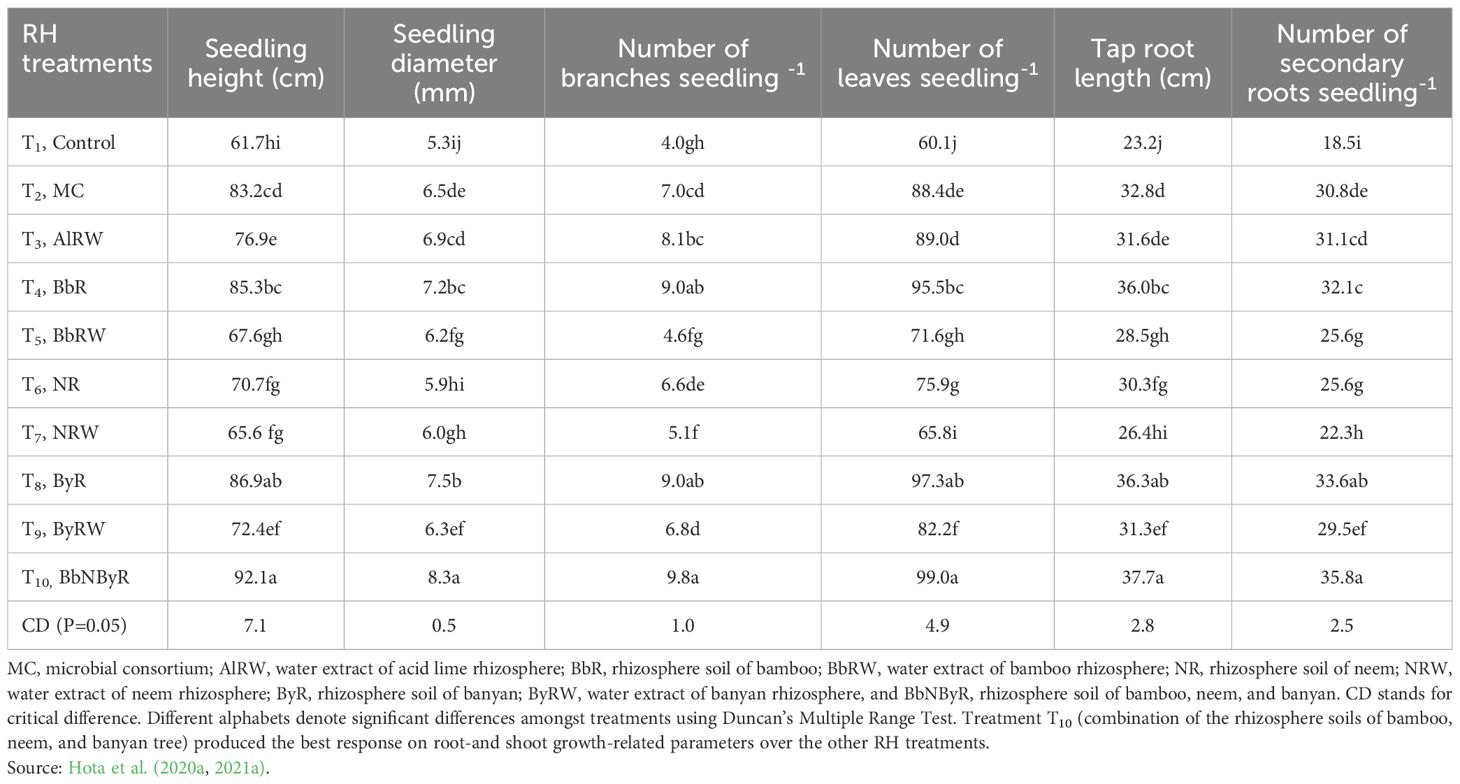
Table 5. Evaluation response of different treatments of RH on agronomic performance of acid lime (Citrus aurantifolia Swingle).
Studies on microbe-mediated rhizosphere fortification in acid lime (Hota et al., 2020a, b, 2021b) showed that the application of vermicompost combined with a microbial consortium increased the canopy volume, improved leaf phenotypical features, and enhanced fruit yield and fruit quality parameters (such as juice content, TSS, and acidity). These observations showed that the process of RH in acid lime improved the yield and quality, as well as qualitatively improving the carrying capacity of hybridized rhizosphere with value-added efficient microbial traits. Root exudates stimulate the microbial community in the rhizosphere by providing soil microbes with the desired nutrient forms and easily degradable energy sources from root exudates and dead root cells (Kaksonen et al., 2006). The microbial diversity of heritage trees like Ficus (Rodge et al., 2016), Neem (Biswas et al., 2016), and Bamboo (Tu et al., 2014) have been studied in-depth to replicate their behavior in a conducive environment. Thus, rhizosphere properties defined in terms of the microbial pool have a strong influence on plant growth depending upon the diversity and evenness of different microbial communities within the rhizosphere of different tree species (Figure 3).
Various interactions among diverse microbial communities promote key processes associated with plant growth and soil health. Some soils are naturally suppressive to many soil-borne pathogens, although this suppression relates to agro-pedological features of the soil. In most agro-systems, the biological components are primarily important in disease suppression, paving the way for biological control of plant pathogens to be brought into the ambit of sustainable issues relating RH (Weller et al., 2023). A typical beneficial response is achieved with the ‘mycorrhiza-helper-bacteria’, a term coined by Garbaye (1994) for those bacteria widely known to stimulate mycelial proliferation of mycorrhizal fungi. Soil microorganisms are known to synthesize molecules capable of increasing the root exudation, which, in turn, stimulate mycorrhizal mycelia abundance in the rhizosphere (Wu and Srivastava, 2012). The establishment of the mycorrhizal fungus in the root cortex is known to change many key processes associated with plant physiology (Zou et al., 2016). These key functions comprise the mineral nutrient composition of plant tissues, hormonal balance, and the patterns of carbon allocation to different plant parts (Srivastava et al., 2021). Microbial communities are known to play key roles in soil carbon stabilization by incorporating organic carbon into their cellular materials and products, are stabilized by mineral associations and supply enzymes catalyzing the decomposition and transformation of plant and soil carbon (Kögel-Knabner, 2002), even though organic matter is ultimately decomposed microbially (Srivastava et al., 2015a; Srivastava, 2010).
Research on the microbiome of hybridized soil still needs to establish to what extent major microbial communities function as a collective entity, since biofilm formation in the rhizosphere is an important trait that prevents microorganisms from being detached from plant roots by various natural processes (Velmourougnane et al., 2017). Do such hybridized soils develop biofilms? Biofilms consist of syntrophic communities of microorganisms where cells stick to each other in a self-produced matrix of extracellular polymeric substances (Rana et al., 2021). Such a matrix provides the structural and functional protection through which microbes chemically link with each other by quorum sensing and function as one unit (Tan et al., 2015; Vlamakis et al., 2013). In soils, microbial communities such as bacteria and fungi develop biofilms on abiotic surfaces such as ore (minerals), water-air interfaces, and dead organisms (Rekadwad and Khobragade, 2017). In recent years, biofilmed biofertilizers (BBs) (biofertilizers containing microbial communities capable of forming biofilms) have emerged as a new inoculant strategy to improve biofertilizer efficiency and sustain soil fertility amid detrimental nutrient mining over time (Sharma et al., 2023). The idea behind BBs is that biofilm formation creates a more suitable environment for microorganisms to compete with resident organisms and negotiate with the heterogeneity of biotic and abiotic factors in soil (Unal et al., 2019).
5.1 Soil enzymes and RH
RH facilitates the development of a physiologically more active rhizosphere. Therefore, the processes and reactions taking place within the rhizosphere are influenced by the stage of root development, diversity and functionality of rhizosphere microorganisms, degree of root-microorganism association, and profile of rhizosphere enzymes— key driving factors responsible for creating rhizosphere environments conducive to positive crop responses. The enzymes are more concentrated in the rhizosphere than in bulk soil, as the rhizosphere soil is richer in organic-C substrates (Gianfreda, 2015). The balance of microbial activities, especially enzymes activities, is, therefore, responsible for the development of resilient soil health. The enzymes that are present in the rhizosphere, both through their interactions with plants and their functions in decomposition of organic compounds coupled with nutrient cycling, play a crucial role in ecological fitness and functioning of the host plant (Nannipieri et al., 2012). Furthermore, activities of rhizosphere enzymes (Table 5) are valuable indices of changes taking place in the microbial functioning within the rhizosphere soil (Gianfreda, 2015). In addition, various management practices also dictate the rhizosphere enzymatic profile of different fruit crops (Table 6).
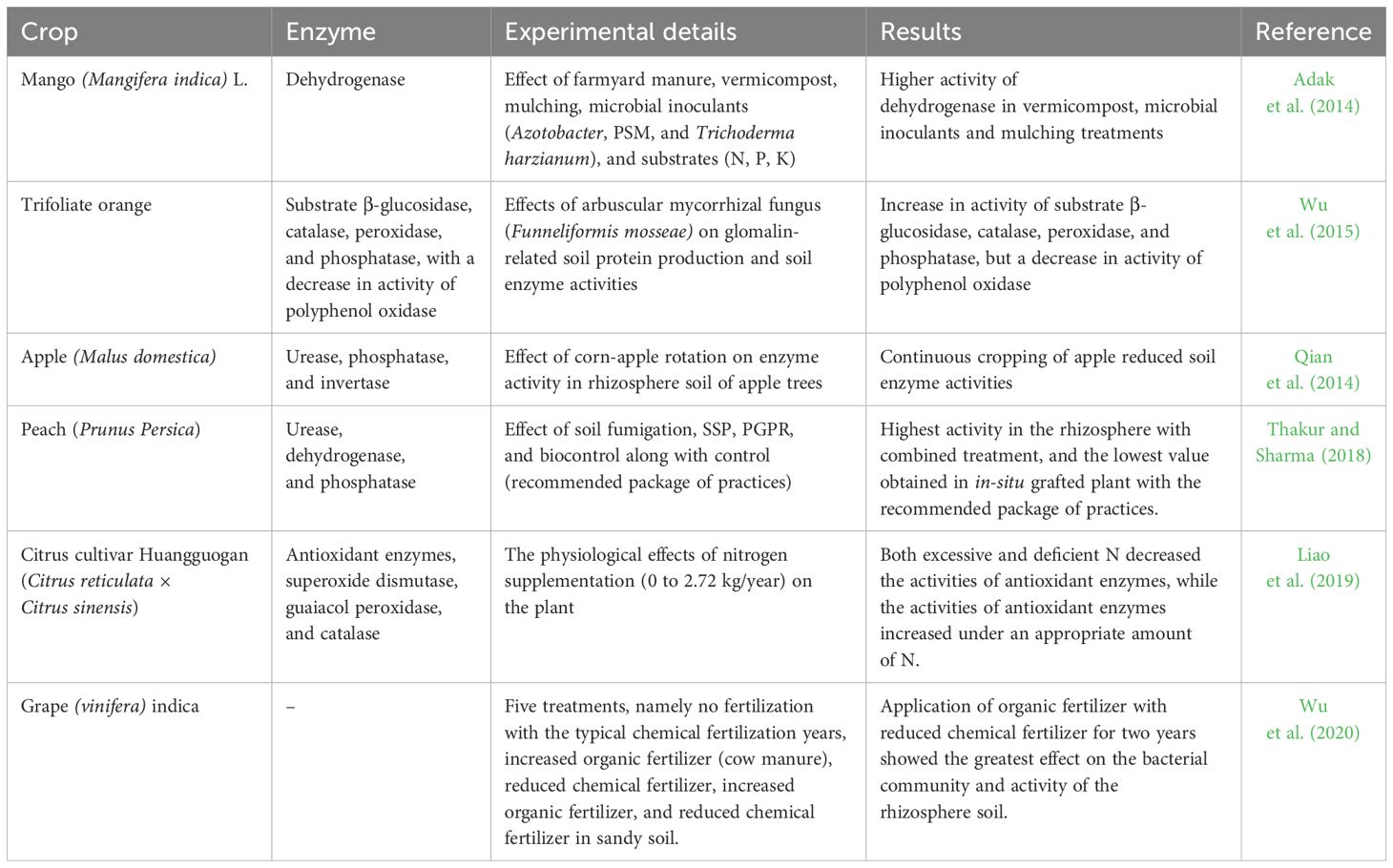
Table 6. Changes in enzymatic activities of rhizosphere soil of different fruit crops in response to soil managements practices.
5.2 RH and holobiont paradigm
Over the years, important components defining the rhizosphere function such as soil, microbes, and plants have been extensively studied in isolation, including pathogenic or symbiotic microbial interactions. Ecological approaches have only recently been developed, possibly in relation to the emergence of molecular tools and, to a lesser extent, understanding of the complex environment of the rhizosphere (Lundberg et al., 2012). In this situation, the plants and associated microbes are no longer seen as individual but rather as an association, a part of a phytobiome. Hence, the holobiont paradigm (a physiological unit of plant-microbe associative relationship) has emerged in the plant world (Zilber-Rosenberg and Rosenberg, 2008; Rosenberg et al., 2010; Vandenkoornhuyse et al., 2015), with microbes playing a key role in plant adaptation to changing environments. Crucially, the holobiont should be viewed as the unit of selection in the evolutionary process (Rosenberg et al., 2010) and, as a consequence, modification of any component of the holobiont could have a cascading effect on other components. A recent study demonstrated that the domestication of plants has affected the fabric of microbial communities, both within the rhizosphere or endosphere (Pérez-Jaramillo et al., 2016). Viewing the plant as a superorganism—representing an independent ecosystem—add a new dimension to efforts to engineer the rhizosphere microbiome, exploit the microbial gene pool, and develop microbially engineered plants. Any breakthroughs in the near future should take into account the plant-associated numerical diversity of microbial communities and their function. Hence, a better knowledge of phytobiomes is strongly advocated. This implies a better description of plant-associated microbiota associated with either one fruit cultivar or different species of the same cultivar, or both at different growth stages of development, with all these issues currently understudied. In addition, most of the physiological experiments aiming at physiological and genotypic screening are performed in the absence of the associated microbiota (Nogales, 2015).
6 Conclusion and futuristic viewpoints
The structure of indigenous microbial communities is reported to exert a multi-fold influence on the composition of plant-preferred microbiota. However, this raises another question: Is large microbial diversity or functional redundancy more important in enabling distinct microbiomes with equivalent or variable functionality? This opens up another counter question: is a rootstock-scion combination that prefers an optimal microbiome in one soil operationally effective in another soil? Do crop breeding techniques need to be highly contextual and individually tailored to specific soil types and management practices? The impacts of rhizosphere microbiomes on nutrient cycling need to be quantified through plant–soil system and relevant methods of evaluation need to be found.
The plant roots release compounds that serve as a source of energy for microorganisms. A high concentration of root exudates deposited into the rhizosphere attracts more metabolically active microorganisms to the roots than to other parts of the rhizosphere. Concerns over the excessive use of chemical/synthetic inputs have meant organic farming has become increasingly popular, promoting bio-preparations developed from rhizosphere microorganisms responsible for stimulatory responses to plant growth and development. Large-scale applications of plant growth-promoting microbes is likely to reduce the use of inorganic fertilizers to put fruit crops elevate functional corridors. A conceptual framework that exploits rhizosphere-endosphere-phyllosphere microbial diversity for bioprospecting soil-plant health to develop a revisited fruit production system has been proposed as a case study (Figure 4). The process of RH also needs to be viewed as a way to develop climate-smart soil with disease -suppressive abilities that can produce microbially engineered fruit crops. These futuristic viewpoints would lay a better foundation for understanding nutrient-microbe synergy for exploiting the productivity potential of growing, much nearer to natural farming. Rhizosphere microorganisms, apart from the activity in promoting plant growth, also offer the possibility of bioremediation of rhizospheres contaminated with pesticide residues and antagonism to different phytopathogens. Despite numerous studies on rhizosphere microorganisms, there still exists a strong need for a more elaborate understanding of the principles of rhizosphere ecology, describing microorganism function and diversity. Such candid understanding of the dynamics and composition of microbial communities developing out of RH as a newly proposed field of research could help decode microbial communication with fruit trees.
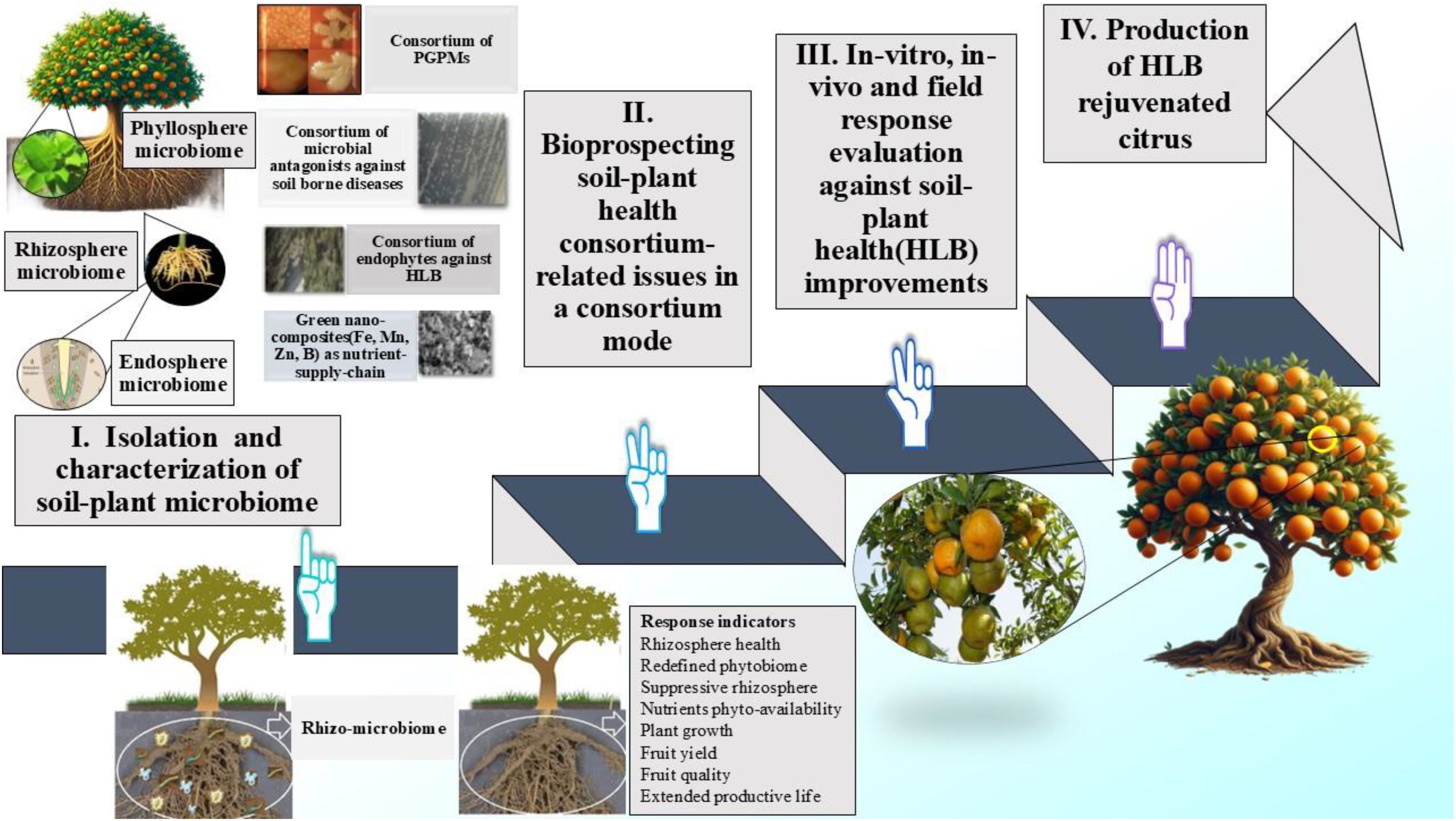
Figure 4. Conceptual framework for exploiting citrus phytobiome (citrus infected with Huanglongbing, HLB disease as a case study) by bioprospecting soil-plant health for developing a microbe-mediated citrus production system.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
AS: Conceptualization, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, Formal analysis, Project administration. SM: Conceptualization, Data curation, Formal analysis, Resources, Writing – original draft, Investigation, Methodology, Visualization, Writing – review & editing. PB: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. DH: Conceptualization, Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. VP: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Writing – review & editing. SM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. VZ: Conceptualization, Formal analysis, Methodology, Validation, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abid L., Smiri M., Federici E., Lievens B., Manai M., Yan Y., et al. (2022). Diversity of rhizospheric and endophytic bacteria isolated from dried fruit of Ficus carica. Saudi J. Biol. Sci. 29, 103398. doi: 10.1016/j.sjbs.2022.103398
Abouziena H. F., Elham Z. A., Youssef R. A., and Sahab A. F. (2010). Efficacy of intercropping mango, mandarin or Egyptian clover plants with date palm on soil properties, rhizospere microflora and quality and quantity of date fruits. J. Am. Sci. 6, 230–238.
Adak T., Singha A., Kumar K., Shukla S. K., Singh A., and Singh V. K. (2014). Soil organic carbon, dehydrogenase activity, nutrient availability and leaf nutrient content as affected by organic and inorganic source of nutrient in mango orchard soil. J. Soil Sci. Plant Nutri. 14, 394–406. doi: 10.4067/S0718-95162014005000031
Amor D. R. and Bello M. D. (2019). Bottom-up approaches to synthetic cooperation in microbial communities. Life 9, 22–28. doi: 10.3390/life9010022
Ashwini N., Kumar P., Joshi A. K., Sharma N. C., Sharma N., and Sharma N. (2022). Synergistic action of humic acid substances and bio-inoculants in guava (Psidium guajava L.): impact on growth traits, fruiting, nutrient profiling and rhizosphere stochiometry in meadow rainy season plant-soil interface. J. Pl. Nutt. 46, 1–16. doi: 10.1080/01904167.2022.2046069
Atashgahi S., Snchez-Andrea I., Heipieper H. J., Meer Jan R., Stams A. J. M., and Smidt H. (2018). Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 6390, 743–746. doi: 10.1126/science.aar3778
Avis T. J., Gravel V., Antoun H., and Tweddell R. J. (2008). Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 40, 1733–1740. doi: 10.1016/j.soilbio.2008.02.013
Banerjee S., Schlaeppi K., and van der Heijden M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Berendsen R. L., Vismans G., Yu K., Song Y., de Jonge R., Burgman W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berlanas C., Berbegal M., Elena G., Laidani M., Cibriain J. F., Sagües A., et al. (2019). The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front. Microbiol. 10, 1142. doi: 10.3389/fmicb.2019.01142
Bhattacharya P., Kumar R., and Jain R. K. (1999). “Biofertilizers use in citrus-prospects and strategies,” in Abstract International Symposium on Citriculture (National Research Centre for Citrus, Nagpur, India), 83.
Bindu B. and Renjan B. (2024). Integrated nutrient management of papaya (Carica papaya L.): Application of microbial consortium enriched organic manures for yield and fruit quality enhancement. Int. J. Plant Soil Sci. 36, 187–195. doi: 10.9734/ijpss/2024/v36i84849
Biswas K., Basu J., Ghosh A., and Giri P. (2016). Study of rhizospheric bacterial population of Azadirachta indica (Neem) of North 24 Parganas district of West Bengal for bioprospective consideration. Int. J. Exp. Res. Rev. 6, 62–66.
Bora P. and Bora L. C. (2020a). Disease management in horticulture crops through microbial interventions: An overview. Indian J. Agric. Sci. 90, 1389–1396. doi: 10.56093/ijas.v90i8.105900
Bora P. and Bora L. C. (2020b). Microbial approach for disease management in horticulture crops: an overview. Indian J. Agric. Sci. 90, 1389–1396. doi: 10.56093/ijas.v90i8.105900
Camprubi A. and Calvet C. (1996a). Isolation and screening of mycorrhizal fungi from citrus nurseries and orchards and inoculation studies. HortSci. 31, 366–369.
Camprubi A. and Calvet C. (1996b). A field inoculation system, for citrus nurseries using pre-cropping with mycorrhizal aromatic plants. Fruits 51, 133–137.
Cheke A. S., Patil V. D., Kausadikar H. K., and Srivastava A. K. (2018b). Rhizosphere hybridization: Soil nutrient availability. J. Pharmacogn. Phytochem. SP1, 3113–3117.
Cheke A. S., Patil V. D., and Srivastava A. K. (2018a). Studies on rhizosphere hybridization and nutrient dynamics in sweet orange seedling from pot culture experiment. J. Pharmacogn. Phytochem. SP1, 3077–3082.
Cheng W. and Gershenson A. (2007). “Carbon fluxes in the rhizosphere,” in The rhizosphere-an ecological perspective, 2nd ed. Eds. Cardon Z. G. and Whitbeck J. L. (Academic Press, San Diego, California, USA), 31–56.
Cloutier M., Alcaide T., Duiker S., and Bruns M. A. (2023). Tillage intensity and plant rhizosphere selection shape bacterial-archaeal assemblage diversity and nitrogen cycling genes. Soil Tillage Res. 225, 105525. doi: 10.1016/j.still.2022.105525
Demarche P., Junghanns C., Nair R. K., and Agathos S. N. (2012). Harnessing the power of enzymes in environmental stewardship. Biotchnol. Adv. 30, 933–953. doi: 10.1016/j.biotechadv.2011.05.013
Dijkstra F. A., Carillo Y., Pendall E., and Morgan E. (2013). Rhizosphere priming: a nutrient perspective. Front. Microbiol. 4. doi: 10.3389/fmicb.2013
Dzuvichu M., Alila P., Jamir S., Kanuajia S. P., and Srivastava A. K. (2023). Yiled response of rhizosphere hybridization in khasi mandrain. Intern. J. Innov. Hort. 12, 102–108. doi: 10.5958/2582-2527.2023.0011.8
El-Mohamedy R. S. R. and Ahmed M. A. (2009). Effect of biofertilizers and humic acid on control of dry root rot disease and improvement yield quality of mandarin (Citrus reticulata Blanco). Res. J. Ag. Biol. Sci. 5, 127–137.
El-Shazly S. A., El-Gazzar A. A., Soliman E. M., Abd El-Hafez A. A., Abd El-Rahman G. F., and Mohamed S. M. (2015). Effect of natural minerals compound, organic and some biofertilizers application on yield, fruit quality and leaf mineral content of Balady mandarin trees. Egyptian J. Hortic. 42, 211–230. doi: 10.21608/ejoh.2015.1288
Gallart M., Adair K. L., Love J., Meason D. F., Clinton P. W., Xue J., et al. (2018). Host genotype and nitrogen form shape the root microbiome of Pinus radiata. Microb. Ecol. 75, 419–433. doi: 10.1007/s00248-017-1055-2
Garbaye J. (1994). Helper bacteria: a new dimension to the mycorrhizal symbiosis. New Phytol. 128, 197–210. doi: 10.1111/j.1469-8137.1994.tb04003.x
Gargallo-Garriga A., Preece C., Sardans J., Oravec M., Urban O., and Peñuelas J. (2018). Root exudate metabolomes change under drought and show limited capacity for recovery. Sci. Rep. 8, 12696. doi: 10.1038/s41598-018-30150-0
Gianfreda L. (2015). Enzymes of importance to rhizosphere processes. J. Soil Sci. Plant Nutr. 15. doi: 10.4067/S0718-95162015005000022
Giassi V., Katia C. K., and Kuppe C. (2016). Bacteria as growth-promoting agents for citrus rootstocks. Microbiol. Res. 190, 46–54. doi: 10.1016/j.micres.2015.12.006
Girish K. and Prabhavathi H. R. (2019). Antifungal activity of bacteria against the phytopathogens of papaya (Carica papaya L.). EurAsian J. Biosci. 13, 83–91.
Gottel N. R., Castro H. F., Kerley M., Yang Z., Pelletier D. A., Podar M., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. doi: 10.1128/AEM.05255-11
Graham J. H. and Syvertsen J. P. (1985). Host determinants of mycorrhizal dependency of citrus rootstock seedlings. New Phytol. 101, 667–676. doi: 10.1111/j.1469-8137.1985.tb02872.x
Gupta S. G., Srivastava A. K., and Sonkar R. K. (2008). Nagpur mandarin evaluation on commercial rootstock in tropical central India. Tropic. Agric. 85, 1–5.
Hacquard S. and Schadt C. W. (2015). Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 205, 1424–1430. doi: 10.1111/nph.13133
Hala Y. and Ali A. (2019). Isolation and characterization of Azotobacter from neem rhizosphere. J. Physics: Conf. Ser. 1244, 12019. doi: 10.1088/1742-6596/1244/1/012019
Hauschild K., Orth N., Liu B., Giongo A., Gschwendtner S., and Beerhues L. (2024). Rhizosphere competent inoculants modulate the apple root–associated microbiome and plant phytoalexins. Appl. Microbiol. Biotechnol. 108, 344. doi: 10.1007/s00253-024-13181-8
He Z. L., Chen G., Yao H., Calvet D. V., Luo Y., and Huang C. Y. (2002). “Microbial biomass and its turnover: a potential diagnosing tool for soil fertility quality,” in Trans 17th World Congress on Soil Science (Queen Sirikit National Convention Centre, Bangkok, Thailand), 14–21.
Hiltner L. (1904). Über neuere erfahrungen und probleme auf dem gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arb DLG. 98, 59–78.
Hota D., Kumar V., and Singh I. P. (2020a). Response of microbial consortium to leaf phenotype of acid lime. Int. J. Adv. Sci. Res. 5, 31–33.
Hota D., Kumar V., and Singh I. P. (2021a). Pre-evaluation response of rhizosphere hybridization in acid lime. Indian J. Agric. Sci. 91, 159–163. doi: 10.56093/ijas.v91i1.110955
Hota D., Kumar V., and Singh I. P. (2021b). Agronomic performance of acid lime in response to microbial fortification of rhizosphere. Indian J. Agric. Sci. 91, 146–149. doi: 10.56093/ijas.v91i1.110951
Hota D., Srivastava A. K., Dahat S., and Dubey S. (2020b). Rhizosphere engineering through microbes enhances agronomic performance of acid lime. Prog. Hortic. 52, 120–123. doi: 10.5958/2249-5258.2020.00016.0
Ishii T. and Kadoya K. (1996). “Utilisation of vesicular-arbuscular mycorrhizal fungi in citrus orchards,” in Proceedings of the International Society of Citriculture, vol. 2 . Eds. Manicom B., Robinson J., du Plessis S. F., Joubert P., van Zyl J. L., and du Prez S. (Sun City, South Africa: International Society of Citriculture, USA), 777–780.
Jariwala B. and Desai B. (2018). Isolation and identification of endophytic fungi from various medicinal plants. BMR Microbiol. 4, 1–7. doi: 10.12691/aees-8-3-8
Jeffery S., Gardi C., Jones A., Montanarella L., Marmo L., Miko L., et al. (2010). “The soil environment,” in European atlas of soil biodiversity (European Commission, Publications office of the European Union, Luxembourg), 17–48.
Kaksonen A. H., Jussila M. M., Lindström K., and Suominen L. (2006). Rhizosphere effect of Galega orientalis in oil-contaminated soil. Soil Biol. Biochem. 38, 817–827. doi: 10.1016/j.soilbio.2005.07.011
Kanse O. S., Whitelaw-Weckert M., Kadam T. A., and Bhosale H. J. (2015). Phosphate solubilization by stress-tolerant soil fungus Talaromyces funiculosus SLS8 isolated from the Neem rhizosphere. Ann. Microbiol. 65, 85–93. doi: 10.1007/s13213-014-0839-6
Keditsu R. and Srivastava A. K. (2014). Substrate dynamics: Developments and issues. Ann. Plant Soil Res. 16, 1–8.
Kögel-Knabner I. (2002). The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 34, 139–162. doi: 10.1016/S0038-0717(01)00158-4
Kohli R. R., Srivastava A. K., Huchche A. D., Paliwal M. K., and Bhattacharya P. (1997). “Relationship of leaf nutrient status, soil available nutrients and microbial composition with fruit yield of Nagpur mandarin,” in Abstract National Symposium on Citriculture(Nagpur, Maharashtra, India: International Society of Citriculture, USA), 55.
Kong W. L., Wang W. Y., Xuo S. H., and Wu X. Q. (2022). Genome sequencing of Rahnella victoriana JZ-GX1 provides new insights into molecular and genetic mechanisms of plant growth promotion. Front. Microbiol. 13, 828990. doi: 10.3389/fmicb.2022.828990
Ku Y. S., Rehman H. M., and Lam H. M. (2019). Possible roles of rhizospheric and endophytic microbes to provide a safe and affordable means of crop biofortification. Agronomy 9, 764. doi: 10.3390/agronomy9110764
Kugarajah V., Nisha K. N., Jayakumar R., Sahabudeen S., Ramakrishnan P., and Mohamed S. B. (2023). Significance of microbial genome in environmental remediation. Microbiol. Res. 271, 127360. doi: 10.1016/j.micres.2023.127360
Kumar P., Joshi A. K., Sharma N., Lata S., Mehmood S., Ahlawat Y. K., et al. (2024). Integrative approaches to improve litchi (Litchi chinensis Sonn.) plant health using bio-transformations and entomopathogenic fungi. BMC Pl. Biol. 24, 902. doi: 10.1186/s12870-024-05604-5
Kumar P., Sharma N., Sharma S., and Gupta R. (2020). Rhizosphere stochiometry, fruit yield, quality attributes and growth response to PGPR transplant amendments in strawberry (Fragaria× ananassa Duch.) growing on solarized soils. Sci. Hortic. 265, 109215. doi: 10.1016/j.scienta.2020.109215
Kumar G., Suman A., Lal S., Ram R. A., Bhatt P., Pandey G., et al. (2021). Bacterial structure and dynamics in mango (Mangifera indica) orchards after long term organic and conventional treatments under subtropical ecosystem. Sci. Rep. 11, 20554. doi: 10.1038/s41598-021-00112-0
Lai W. A., Ka¨mpfer P., Arun A. B., Shen F. T., Huber B., Rekha P. D., et al. (2006). Deinococcus ficus sp. nov., isolated from the rhizosphere of Ficus religiosa L. Int. J. Sys. Evolution. Microbiol. 56, 787–791. doi: 10.1099/ijs.0.64007-0
Lawson C. E., Harcombe W. R., Hatzenpichler R., Lindemann S. R., Löffler F. E., O’Malley M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi: 10.1038/s41579-019-0255-9
Lee J. T. and Tsai S. M. (2018). The nitrogen-fixing Frankia significantly increases growth, uprooting resistance and root tensile strength of Alnus formosana. Afr. J. Biotechnol. 17, 213–225. doi: 10.5897/AJB2017.16289
Lee H. J. and Whang K. S. (2014). Streptomyces graminisoli sp. nov. and Streptomyces rhizophilus sp. nov., isolated from bamboo (Sasa borealis) rhizosphere soil. Int. J. Sys. Evolution. Microbiol. 64, 1546–1551. doi: 10.1099/ijs.0.055210-0
Lei S., Xu X., Cheng Z., Xiong J., Ma R., Zhang L., et al. (2019). Analysis of the community composition and bacterial diversity of the rhizosphere microbiome across different plant taxa. Microbiologyopen 8, e00762. doi: 10.1002/mbo3.762
Li X., Lin J., Xiao S., Feng D., Deng Y., and Yuan W. (2022). Effects of sweet potato intercropping in banana orchard on soil microbial population diversity. Annal. Microbiol. 72, 46. doi: 10.1186/s13213-022-01702-7
Liao L., Dong T., Qiu X., Rong Y., Sun G., Wang Z., et al. (2019). Antioxidant enzyme activity and growth responses of Huangguogan citrus cultivar to nitrogen supplementation. Biosci. Biotechnol. Biochem. 83, 1924–1936. doi: 10.1080/09168451.2019.1634513
Lu L.-H., Srivastava A. K., Shen Y.-L., and Wu Q.-S. (2019). A negative feedback regulation of replanted microorganisms on plant growth and soil properties of peach. Not. Bot. Horti-agrobo. 47, 255–261. doi: 10.15835/nbha47111344
Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Ma D., Liu S., Han X., Nan M., Xu Y., Qian B., et al. (2022). Complete genome sequence, metabolic model construction, and huangjiu application of Saccharopolyspora rosea A22, a thermophilic, high amylase and glucoamylase actinomycetes. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.995978
Melo J., Carvalho L., Correia P., Souza S. B., Dias T., Santana M., et al. (2018). Conventional farming disrupts cooperation among phosphate solubilising bacteria isolated from Carica papaya’s rhizosphere. App. Soil Ecol. 124, 284–288. doi: 10.1016/j.apsoil.2017.11.015
Mohamed A. K. A., Ahemed M. M. M., and El-Akkad M. E. (2009). Is it possible to compensate the annual fertilization in mandarin orchards by using the bio-fertilizers? Assiut J. Agric. Sci. 40, 37–68. doi: 10.21608/ajas.2009.268798
Moshiri F., Ebrahimi H., Ardakani M. R., Rejali F., and Mousavi S. M. (2019). Biogeochemical distribution of Pb and Zn forms in two calcareous soils affected by mycorrhizal symbiosis and alfalfa rhizosphere. Ecotoxicol. Environ. Saf. 179, 241–248. doi: 10.1016/j.ecoenv.2019.04.055
Mousavi S. M., Motesharezadeh B., Hosseini H. M., Alikhani H., and Zolfaghari A. A. (2018). Root-induced changes of Zn and Pb dynamics in the rhizosphere of sunflower with different plant growth promoting treatments in a heavily contaminated soil. Ecotoxicol. Environ. Saf. 147, 206–216. doi: 10.1016/j.ecoenv.2017.08.045
Nannipieri P., Ascher J., Ceccherini M. T., Landi L., Pietramellara G., Renella G., et al. (2007). Microbial diversity and microbial activity in the rhizosphere. Ci Suelo (Argentina) 25, 89–97. doi: 10.12691/ijebb-3-1-2
Nannipieri P., Giagnoni L., Renella G., Puglisi E., Ceccanti B., Masciandaro G., et al. (2012). Soil enzymology: classical and molecular approaches. Biol. Fertility Soils 48, 743–762. doi: 10.1007/s00374-012-0723-0
Ngullie E., Singh A. K., Sema A., and Srivastava A. K. (2015). Citrus growth and rhizosphere properties. Commun. Soil Sci. Plant Anal. 46, 1540–1550. doi: 10.1080/00103624.2015.1043460
Nimoni P. and Pongslip N. (2009). Genetic diversity and plant-growth promoting ability of the indole-3-acetic acid (IAA) synthetic bacteria isolated from agricultural soil as well as rhizosphere, rhizoplane and root tissue of Ficus religiosa L., Leucaena leucocephala and Piper sarmentosum Roxb. Res. J. Ag. Biol. Sci. 5, 29–41.
Niveditha V. R., Shewtha B., Deepa D. D., Divya N. H., and Raghavendra R. B. (2008). Plant growth promoting microorganisms (PGPMs) from bamboo rhizosphere. Advanced Biotech. 7, 33–35.
Nogales E. (2015). An electron microscopy journey in the study of microtubule structure and dynamics. Protein Sci. 24, 1912–1919. doi: 10.1002/pro.2808
Nourgholipour F., Mirseyed Hosseini H., Tehrani M. M., Motesharezadeh B., Moshiri F., and Mousavi S. M. (2022). Phosphorus fractionation affected by root induced changes of two canola cultivars. Eurasian Soil Sc. 55, 819–829. doi: 10.1134/S1064229322060102
Oliva J., Redondo M.Á., and Stenlid J. (2020). Functional ecology of forest disease. Annu. Rev. Phytopathol. 58, 343–361. doi: 10.1146/annurev-phyto-080417-050028
Olougou M. N. E., Achiri D. T., Ngone M. A., Ndzeshala S. D., Tchakounté G. V. T., Tening A. S., et al. (2024). Bio-inoculant consortia modulated plantain (Musa × paradisiaca L.) growth, rhizosphere pH, acid phosphatase and urease activity. Soil Advances. 2, 100008. doi: 10.1016/j.soilad.2024.100008
Ortas I. (2012). “Mycorrhiza in citrus: growth and nutrition,” in Advances in Citrus Nutrition. Ed. Srivastava A. K. (Springer, Dordrecht, Netherland), 333–351.
Pagán E., Robles J. M., Temnani A., Berríos P., Botía P., and Pérez-Pastor A. (2022). Effects of water deficit and salinity stress on late mandarin trees. Sci. Total Environ. 803, 150109. doi: 10.1016/j.scitotenv.2021.150109
Palazzo D., Pommerening B., and Vanadia S. (1992). “Effect of soil sterilization and vesicular arbuscular mycorrhiza on growth of sour orange (Citrus aurantium L.) seedlings,” in Proceedings of the International Society of Citriculture, vol. 2 . Eds. Tribulato E., Gentile A., and Refergiato G. (Acireale, Italy: International Society of Citriculture, USA), 621–623.
Pandey A., Palni L. M. S., and Bisht D. (2001). Dominant fungi in rhizosphere of established tea bushes and their interaction with the bacteria under in situ conditions. Microbiol. Res. 156 s, 377–382. doi: 10.1078/0944-5013-00123
Pathak K. V. and Keharia H. (2013). Characterization of fungal antagonistic bacilli isolated from aerial roots of banyan (Ficus benghalensis) using intact-cell MALDI-TOF mass spectrometry (ICMS). J. App. Microbiol. 114, 1300–1310. doi: 10.1111/jam.12161
Pérez-Jaramillo J. E., Mendes R., and Raaijmakers J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Pervaiz Z. H., Contreras J., Hupp B. M., Lindenberger J. H., Chen D., Zhang Q., et al. (2020). Root microbiome changes with root branching order and root chemistry in peach rhizosphere soil. Rhizosphere 16, 100249. doi: 10.1016/j.rhisph.2020.100249
Peter J. K. and Pandey N. (2014). Bioprospecting phosphate solubilisation and PGP activities of native strains of Pseudomonas Aeruginasa and Pseudomonas fluorescens from bamboo (Bambusa bamboo) rhizosphere. Int. J. Res. 1, 702–717.
Pinho D., Barroso C., Froufe H., Brown N., Vanguelova E., Egas C., et al. (2020). Linking tree health, rhizosphere physicochemical properties, and microbiome in acute oak decline. Forests 11, 1153. doi: 10.3390/f11111153
Pinton R., Varanini Z., and Nannipieri P. (2007). “The rhizosphere as a site of biochemical interactions among soil components, plants and microorganisms,” in The rhizosphere: biochemistry and organic substances at the soil-plant interface, 3nd ed. Eds. Pinton R., Varanini Z., and Nannipieri P. (Taylor & Francis Group, Marcel Dekker, New York, USA), 1–17.
Prasad S. and Baishya R. (2019). Interactive effects of soil moisture and temperature on soil respiration under native and non-native tree species in semi-arid forest of Delhi, India. Trop. Ecol. 60, 252–260. doi: 10.1007/s42965-019-00028-x
Prashar P., Kapoor N., and Sachdeva S. (2013). Rhizosphere: its structure, bacterial diversity and significance. Rev. Environ. Sci. Biotechnol. 13, 63–77. doi: 10.1007/s11157-013-9317-z
Qian J., Sun W., and Li Y. D. (2014). Changes in soil nutrient levels, enzyme activities, microbial community function and structure during apple orchard maturation. App. Soil Ecol. 77, 18–25. doi: 10.1016/j.apsoil.2014.01.003
Raaijmakers J. M. and Kiers E. T. (2015). Rewilding plant microbiomes. Science 378, 599–601. doi: 10.1126/science.abn6350
Raiesi T., Moradi B., and Mousavi S. M. (2022). Alterations of P fractions and some biochemical features in rhizosphere soil induced by the root activities of citrus rootstocks with different P acquisition efficiency. Eurasian Soil Sc. 20, 1–9. doi: 10.1134/S1064229322020107
Rana K., Nayak S. R., Bihary A., Sahoo A. K., Mohanty K. C., Palo S., et al. (2021). Association of quorum sensing and biofilm formation with Salmonella virulence: story beyond gathering and cross-talk. Arch. Microbiol. 203, 5887–5897. doi: 10.1007/s00203-021-02594-y
Rani N., Jain P., and Geetanjali (2017). Isolation of antimicrobial compound producing fungi from the rhizospheric soil of the medicinal plant Azadirachta Indica. J. Chem. Pharm. Res. 9, 265–270.
Reddy C. S., Sreelekshmi S., Jha C. S., and Dadhwal V. K. (2013). National assessment of forest fragmentation in India: Landscape indices as measures of the effects of fragmentation and forest cover change. Ecol. Eng. 60, 453–464. doi: 10.1016/j.ecoleng.2013.09.064
Rekadwad B. N. and Khobragade C. N. (2017). “Microbial biofilm: role in crop productivity,” in I7: Microbial applications volume 2 – biomedicine, agriculture and industry. Ed. Kalia V. C. (Springer, Cham), 107–118. doi: 10.1007/978-3-319-52669-0_5
Riera N., Wang H., Li Y., Li J., Pelz-Stelinski K., and Wang N. (2018). Induced systematic resistance against citrus canker disease by rhizobacteria. Phytopathology 108, 1038–1045. doi: 10.1094/PHYTO-07-17-0244-R
Rodge S. P., Sable S. K., Salve S. K., Sawant S. A., and Patil N. P. (2016). Isolation and characterization of PGPR from roots of Ficus religiosa growing on concrete walls and its effect on plant growth in drought condition. Int. J. Curr. Microbiol. App. Sci. 5, 583–593. doi: 10.20546/ijcmas.2016.509.066
Rodriguez P. A., Rothballer M., Chowdhury S. P., Nussbaumer T., Gutjahr C., and Falter-Braun P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Rosenberg D. E., Norman G. J., Wagner N., Patrick K., Calfas K. J., and Sallis J. F. (2010). Reliability and validity of the sedentary behavior questionnaire (SBQ) for adults. J. Phys. Activity Health 7, 697–705.101. doi: 10.1123/jpah.7.6.697
Sasse D.M., Martinoia E., and Northen T. (2018). Feed your friends: Do plants exudates shape root microbiome. Trends in Pl. Sci. 21, 25–41. doi: 10.1016/tplants.2017.09.003
Schutz V., Frindte K., Cui J., Zhag P., Hacquard S., Schulze-Lefert P., et al. (2021). Differential impact of plant secondary metabolites on the soil microbiota. Front. Microbiol. 12, 666010. doi: 10.3389/fmicb.2021.666010
Shamseldin A., El-Sheikh M. H., Hassan H. S. A., and Kabeil S. S. (2010). Microbial bio-fertilization approaches to improve yield and quality of Washington navel orange and reducing the survival of nematode in the soil. J. Am. Sci. 6, 264–271.
Shamshiri M. H., Usha K., and Singh B. (2012). Growth and nutrient uptake responses of Kinnow (Citrus nobilis × Citrus deliciosa) to vesicular arbuscular mycorrhizae. Agronomy 535846, 7. doi: 10.5402/2012/535846
Sharaf H., Thompson A. A., Williams M. A., and Peck G. M. (2021). Compost applications increase bacterial community diversity in the apple rhizosphere. Soil Sci. Soc America J. 85, 1105–1121. doi: 10.1002/saj2.20251
Sharma S., Mohler J., Mahajan S. D., Schwartz S. A., Bruggemann L., and Aalinkeel R. (2023). Microbial biofilm: A review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganism 11, 1614. doi: 10.3390/microorganisms11061614
Shen C., Li X., and Qin J. (2024). Kiwifruit-Agricus blazei intercropping effectively improved yield productivity, nutrient uptake, and rhizospheric bacterial community on the soil rhizosphere microbial community. Sci. Rep. 14, 16546. doi: 10.1038/s41598-024-66030-z
Shi H., Zhu X., Lu L., and Ye J. (2024). Effect of microbial inoculants endowed with multifarious plant growth-promoting traits on grape growth and fruit quality under organic fertilization scenarios. Agronomy 14, 491. doi: 10.3390/agronomy14030491
Shilpkar P., Mayur S., and Modi K. (2009). Assessment of microbial diversity in rhizosphere of Ficus religiosa L. tree at different moisture levels. Indian Forester. 135, 111–116.
Singh N., Sharma D. P., Kumar V., and Hota D. (2018a). Impact of different rootstocks and soil agro-techniques on rhizospheric biological activities and growth traits under apple replant sick soil. Multilogic Sci. 7, 88–92.
Singh A., Srivastav M., Singh S. K., and Lal S. (2017). Effect of microbial-inoculants on growth and biochemical parameters of mango plantlets during bio-hardening. Indian J. Hortic. 74, 20–26. doi: 10.5958/0974-0112.2017.00008.1
Singh A., Thakur A., Sharma S., Gill P. P. S., and Kalia A. (2018b). Bio-inoculants enhance growth, nutrient uptake, and buddability of citrus plants under protected nursery conditions. Commun. Soil Sci. Pl. Anal. 49, 2571–2586. doi: 10.1080/00103624.2018.1526946
Sinha S., Masto R. E., Ram L. C., Selvi V. A., Srivastava N. K., Tripathi R. C., et al. (2009). Rhizosphere soil microbial index of tree species in a coal mining ecosystem. Soil Biol. Biochem. 41, 1824–1832. doi: 10.1016/j.soilbio.2008.11.022
Solanki S. P. S., Sharma N. C., Chandel J. S., and Hota D. (2020). Effect of integrated nutrient management on fruit yield and quality of peach (Prunus persica Batsch) cv July Elberta. Int. J. Pure App. Chem. 21, 152–160. doi: 10.9734/irjpac/2020/v21i1030217
Soliman S., Wang Y., Han Z., and El-Kereamy A. (2023). The attraction of apple rhizosphere microorganisms and changing leaf characteristics by Strigolactone. Hortic. 9, 528. doi: 10.3390/horticulturae9050528
Solís-García I. A., Ceballos-Luna O., Cortazar-Murillo E. M., Desgarennes D., Garay-Serrano E., Patiño-Conde V., et al. (2021). Phytophthora root rot modifies the composition of the avocado rhizosphere microbiome and increases the abundance of opportunistic fungal pathogens. Front. Microbiol. 11, 574110. doi: 10.3389/fmicb.2020.574110
Somera T. S. and Mazzola M. (2022). Toward a holistic view of orchard ecosystem dynamics: A comprehensive review of the multiple factors governing development or suppression of apple replant disease. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.949404
Srivastava A. K. (2010). Integrated nutrient management in citrus: Frontier developments. Indian J. Fert. 6, 34–44. doi: 10.1007/978-94-007-4171-3_25
Srivastava A. K. and Hota D. (2020). Fruit crops under nutrient-capped scenario: a timeless journey. Curr. Hortic. 8, 14–17. doi: 10.5958/2455-7560.2020.00014.X
Srivastava A. K., Kohli R. R., Ram L., Huchche A. D., and Das H. C. (1994). Cation exchange capacity of root as a marker for vigour of citrus species. Indian J. Agric. Sci. 64, 324–325.
Srivastava A. K. and Malhotra S. K. (2017). Nutrient –use – efficiency in perennial fruit crops- A Review. J. Plant Nutr. 40, 1928–1953. doi: 10.1080/01904167.2016.1249798
Srivastava A. K., Malhotra S. K., and Krishna Kumar N. K. (2015a). Exploiting nutrient-microbe synergy in unlocking productivity potential of perennial fruits: A review. Indian J. Agric. Sci. 85, 459–481. doi: 10.56093/ijas.v85i4.47895
Srivastava A. K. and Ngullie E. (2009). Integrated nutrient management: Theory and practice. Dynamic Soil Dynamic Plant 3, 1–30.
Srivastava A. K., Shirgure P. S., Deshmukh S. A., and Bhoyar P. A. (2017). Soil fertility and soil healthcare in citrus: A review. Ann. Plant Soil Res. 19, 127–136.
Srivastava A. K. and Singh S. (2001). Soil properties influencing yield and quality of Nagpur mandarin (Citrus reticulata Blanco). J. Indian Soc Soil Sci. 49, 226–229.
Srivastava A. K. and Singh S. (2002). Soil analysis based diagnostic norms for Indian citrus cultivar. Commun. Soil Sci. Plant Anal. 33, 1689–1706. doi: 10.1081/CSS-120004816
Srivastava A. K. and Singh S. (2008a). Analysis of citrus orchard efficiency in relation to soil properties. J. Plant Nutr. 30, 2077–2090. doi: 10.1080/01904160701700566
Srivastava A. K. and Singh S. (2008b). Citrus nutrition research in India: Problems and prospects. Indian J. Agric. Sci. 78, 3–16. doi: 10.1080/01904160701700566
Srivastava A. K. and Singh S. (2009). Citrus decline: Soil fertility and plant nutrition. J. Plant Nutr. 32, 197–145. doi: 10.1080/01904160802592706
Srivastava A. K., Singh S., and Albrigo L. G. (2008). Diagnosis and remediation of nutrient constraints in Citrus. Hortic. Rev. 34, 277–264. doi: 10.1002/9780470380147.ch5
Srivastava A. K., Singh S., and Huchche A. D. (2015b). Evaluation of INM in citrus on Vertic Ustochrept: Biometric response and soil health. J. Plant Nutr. 38, 1–15. doi: 10.56093/ijas.v85i4.47895
Srivastava A. K., Singh S., and Marathe R. A. (2002). Organic citrus: soil fertility and plant nutrition. . J. Sus. Ag 19, 5–29. doi: 10.1300/J064v19n03_03
Srivastava A. K., Wu Q. S., Mousavi S. M., and Hota D. (2021). Integrated soil fertility management in fruit crops: A overview. Int. J. Fruit Sci. 21, 413–439. doi: 10.1080/15538362.2021.1895034
Sui J., Li C., Wang Y., Li X., Liu R., Hua X., et al. (2024). Microecological shifts in the rhizosphere of perennial large trees and seedlings in continuous cropping of Poplar. Microorg 12, 58. doi: 10.3390/microorganisms12010058
Sun P., Zhang Y. C., Shen Y. L., Feng C. Q., and Wu Q. S. (2017). Identification of fungal community in citrus rhizosphere by its gene sequencing. Biotechnol. 16, 85–91. doi: 10.3923/biotech.2017.85.91
Tadayon M. S., Asgharzadeh A., Mousavi S. M., and Saghafi K. (2025). Synergistic effects of chemical and biochemical fertilization on yield enhancement and oil quality optimization in ‘Zard’ olive cultivars. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1455921
Tan C. H., Koh1 K. S., Xie C., Zhang J., Tan X. H., Lee1 G. P., et al. (2015). Community quorum sensing signalling and quenching: microbial granular biofilm assembly. Biofilms Microbes 1, 15006. doi: 10.1038/npjbiofilms
Thakur K. K. and Sharma D. P. (2018). Enzyme activities in peach rhizosphere as influenced by various soil management practices under replant situations: A pot culture study. Int. J. Chem. Study. 6, 2726–2730.
Tu Z., Chen L., Yu X., and Zheng Y. (2014). Rhizosphere soil enzymatic and microbial activities in bamboo forests in south-eastern China. Soil Sci. Plant Nutr. 60, 134–144. doi: 10.1080/00380768.2014.882219
Unal E., Erginkava Z., Korukluoglu M., and Altun G. K. (2019). “Beneficial biofilm applications in food and agricultural industry,” in Health and Safety Aspects of Food Processing Technologies. Eds. Malik A., Erginkaya Z., and Erten H. (Springer, The Netherlands), 445–469. doi: 10.1007/978-3-030-24903-8_15
Uroz S., Buée M., Murat C., Frey-Klett P., and Martin F. (2010). Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2, 281–288. doi: 10.1111/j.1758-2229.2009.00117.x
Uroz S., Oger P., Tisserand E., Cébron A., Turpault M. P., Buée M., et al. (2016). Specific impacts of beech and Norway spruce on the structure and diversity of the rhizosphere and soil microbial communities. Sci. Rep. 6, 27756. doi: 10.1038/srep27756
Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., and Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Van Heerden I., Cronje C., and Kotze J. M. (2002). Microbial, chemical and physical aspects of citrus waste composting. Bioresour. Technol. 81, 71–76. doi: 10.1016/S0960-8524(01)00058-X
Velmourougnane K., Prasanna R., and Saxena A. (2017). Agriculturally important microbial biofilms: Present status and future prospects. J. Basic Microbiol. 57. doi: 10.1002/jobm.201700046
Vlamakis H., Chai Y., Beauregard P., Losick R., and Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168. doi: 10.1038/nrmicro2960
Vorholt J. A., Vogel C., Calstrom C. I., and Mulle D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Wallace J., Laforest-Lapointe I., and Kembel S. W. (2018). Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. Peer J. 6, e5293. doi: 10.7717/peerj.5293
Wang C., Liang Q., Liu J., Zhou R., Lang R., Xu S., et al. (2023). Impact of intercropping grass on the soil rhizosphere microbial community and soil ecosystem functions in a walnut orchard. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1137590
Wei Z., Friman V. P., Pommier T., Geisen S., Jousset A., and Shen Q. R. (2020). Rhizosphere immunity: targeting the underground for sustainable plant health management. Front. Agric. Sci. Eng. 7, 317–328. doi: 10.15302/J-FASE-2020346
Wei Z., Zeng Q., and Tan W. (2021). Cover cropping impacts soil microbial communities and functions in mango orchards. Agriculture 11, 343. doi: 10.3390/agriculture11040343
Weller D. M., Raaijmakers J. M., Gardener B. B. M., and Thomashow L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Ann. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Weller D.M., Le Tourreau M., and Yang M. (2023). Classification , diversity, and microbial basis of disease suppressive soils. In: Good Microbes in Medicine, Foof Production, Biotechnology, Biorenmediation, and Agriculture. (Ed: Frans J., Brujin, Smidt H., L., Cocolin S., Sauer M, Dawling D, et al), John Wiley & sons Limited, U.K. pp. 457–465.
Wemedo S. A. and Onolleka B. (2012). Evaluation of rhizosphere bacteria of Mangifera indica (Mango) and Terminalia catappa (Almond). J. Emerg. Trends Eng. App. Sci. 3, 791–794.
Whipps J. M. and Lynch J. M. (1985). Energy losses by the plant in rhizodeposition. Annu. Proc. Phytochem. Soc. Europe. 26, 59–71.
Wu L., Jiang Y., Zhao F., He X., Liu H., and Yu K. (2020). Increased organic fertilizer application and reduced chemical fertilizer application affect the soil properties and bacterial communities of grape rhizosphere soil. Sci. Rep. 10, 1–10. doi: 10.1038/s41598-020-66648-9
Wu Q. S., Li Y., Zou Y. N., and He X. H. (2015). Arbuscular mycorrhiza mediates glomalin-related soil protein production and soil enzyme activities in the rhizosphere of trifoliate orange grown under different P levels. Mycorrhiza 25, 121–130. doi: 10.1007/s00572-014-0594-3
Wu Q. S. and Srivastava A. K. (2012). “Rhizosphere microbial communities: Isolation, characterization and value addition for substrate development,” in Advances in citrus nutrition. Ed. Srivastava A. K. (Springer, Netherland), 169–194.
Xie Q., Xu H., Wen R., Wang L., Yang Y., Zhang H., et al. (2024). Integrated management of of fruit trees and Bletilla striata implications on soil nutrient profiles and microbial community structures. Front. Microbiol. 15. doi: 10.3389/fmicb2024.1307677
Xue C., Ryan Penton C., Shen Z., Zhang R., Huang Q., Li R., et al. (2015). Manipulating the banana rhizosphere microbiome for biological control of Panama disease. Sci. Rep. 5, 11124. doi: 10.1038/srep11124
Yadav R. K., Jain M. C., and Jhakar R. P. (2012). Effect of media on growth and development of acid lime (Citrus aurantifolia Swingle) seedling with or without Azotobacter. Afr. J. Agric. Res. 7, 6421–6426. doi: 10.5897/AJAR12.1974
Yin C. T., Casa V. J. M., Schlatter D. C., Hagerty C. H., Hulbert S. H., and Paulitz T. C. (2021). Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 9, 86. doi: 10.1186/s40168-020-00997-5
Yu L., Zi H., Zhu H., Liao Y., and Li X. (2022). Rhizosphere microbiome of forest trees determines their resistance to soil-borne pathogens. Res. Square. doi: 10.21203/rs.3.rs-1234640/v1
Yuan Y., Dai X., Fu X., Kou L., Luo Y., Jiang L., et al. (2020). Differences in the rhizosphere effects among trees, shrubs and herbs in three subtropical plantations and their seasonal variations. Eur. J. Soil Biol. 100, 103218. doi: 10.1016/j.ejsobi.2020.103218
Yuan Z. S., Liu F., Liu Z. Y., Huang Q. L., Zhang G. F., and Pan H. (2021). Structural variability and differentiation of niches in the rhizosphere and endosphere bacterial microbiome of moso bamboo (Phyllostachys edulis). Sci. Rep. 11, 1–9. doi: 10.1038/s41598-021-80971-9
Zamora Ballesteros C., Diez J., Martín-García J., Witzell J., Solla A., Ahumada R., et al. (2019). Pine pitch canker (PPC): pathways of pathogen spread and preventive measures. Forests 10, 1158. doi: 10.3390/f10121158
Zhang Y., Han M., Song M., Tian J., Song B., Hu Y., et al. (2021a). Intercropping with aromatic plants increased the soil organic matter content and changed the microbial community in a pear orchard. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.616932
Zhang Q., Pang X., Chen X., Ye J., Lin S., and Jia X. (2020). Rain-shelter cultivation influence rhizosphere bacterial community structure in pear and its relationship with fruit quality of pear and soil chemical properties. Sci. Hortic. 269, 109419. doi: 10.1016/j.scienta.2020.109419
Zhang Y., Trivedi P., Xu J., Roper M. C., and Wang N. (2021b). The citrus microbiome: from structure and function to microbiome engineering and beyond. Phytobiomes J. 5, 249–262. doi: 10.1094/PBIOMES-11-20-0084-RVW
Zhao Y., Yan C., Hu F., Luo Z., Zhang S., Xiao M., et al. (2022). Intercropping into Pinto peanut in Litchi orchards effectively improved soil available potassium content, optimised soil bacterial community structure, and advanced bacterial community diversity. Front. Microbiol. 13, 868312. doi: 10.3389/fmicb.2022.868312
Zhou D., Jing T., Chen Y., Wang F., Qi D., Feng R., et al. (2019). Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC Microbiol. 19, 1–3. doi: 10.1186/s12866-019-1531-6
Ziatabar Ahmadi S. R., Seifi E., Varasteh F., and Akbarpour V. (2024). Mitigating salinity stress in pomegranate: Effects of Pseudomonas fluorescens and Glomus mosseae on stress responses of red angel and wonderful cultivars. Scien. Hortic. 330, 113036. doi: 10.1016/j.scienta.2024.113036
Zilber-Rosenberg I. and Rosenberg E. (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. doi: 10.1111/j.1574-6976.2008.00123.x
Keywords: antagonism, citrus, fruit crops, functional corridor, growth promotion, heritage trees, holobiont, microbial niche
Citation: Srivastava AK, Mousavi SM, Bora P, Hota D, Pandey V, Malhotra SK and Ziogas V (2025) Rhizosphere to rhizosphere hybridization in fruit crops: new perspectives. Front. Hortic. 4:1584807. doi: 10.3389/fhort.2025.1584807
Received: 27 February 2025; Accepted: 21 April 2025;
Published: 09 July 2025.
Edited by:
Gastón Gutiérrez Gamboa, Instituto de Investigaciones Agropecuarias, ChileReviewed by:
Kgabo Martha Pofu, University of Limpopo, South AfricaHeiplanmi Rymbai, ICAR Research Complex for NEH Region, India
Copyright © 2025 Srivastava, Mousavi, Bora, Hota, Pandey, Malhotra and Ziogas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. K. Srivastava, YWtzcml2YXMyMDA3QGdtYWlsLmNvbQ==; Popy Bora, cGJvcmEuc29uaXRwdXIxMEBnbWFpbC5jb20=; S. K. Malhotra, bWFsaG90cmFza3JhakB5YWhvby5jb20=
 A. K. Srivastava
A. K. Srivastava Seyed Majid Mousavi
Seyed Majid Mousavi Popy Bora3*
Popy Bora3* Debashish Hota
Debashish Hota S. K. Malhotra
S. K. Malhotra Vasileios Ziogas
Vasileios Ziogas