- 1Department of Biomedical Engineering, National University of Singapore, Singapore, Singapore
- 2Department of Biomedical Engineering, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 3Hong Kong Center for Cerebro-Cardiovascular Health Engineering (COCHE), Hong Kong, Hong Kong SAR, China
- 4City University of Hong Kong Shenzhen Research Institute (CityUSRI), Shenzhen, China
- 5Mechanobiology Institute, National University of Singapore, Singapore, Singapore
- 6Institute for Health Innovation and Technology (iHealthtech), National University of Singapore, Singapore, Singapore
Patient-derived cancer cells (PDCCs) have emerged as a key strategy for advancing personalized cancer treatment. Unlike traditional cancer cell lines, PDCCs retain the genetic and phenotypic characteristics of the patient’s original tumor and can more accurately reflect tumor biology. This review explores recent advances in methods for culturing PDCCs, highlighting the role of these models in drug discovery and high-throughput screening of personalized therapeutic options. By establishing living models directly from patient tumors, PDCCs can more faithfully recapitulate tumor heterogeneity and microenvironmental features than traditional cell lines. These cultures bridge laboratory research and clinical reality, allowing functional testing of patients' cancer cells. Despite the promise of PDCCs, their culture remains fraught with challenges, including the extremely low number of cancer cells that can be obtained, difficulty maintaining tumor heterogeneity, low culture initiation success rates, and ethical considerations for using patient tissues. In addition, controversy remains regarding the reproducibility of results between different laboratories and patient samples. By examining the field’s current state, this review identifies gaps in the application of PDCCs, such as limited modeling capabilities for specific tumor types and the lack of comprehensive, scalable protocols for broad clinical use. This article discusses future directions, including integration with advanced microengineering and AI-driven analysis, which have the potential to overcome existing limitations and optimize PDCCs-based therapeutic strategies. PDCCs are expected to transform the future of cancer treatment as they ultimately provide more accurate drug testing and personalized medicine models.
1 Introduction
Advancing cancer research and treatment requires model systems that accurately reflect human tumors. Traditionally, researchers have relied on established 2D cancer cell lines and animal models (e.g., mouse xenografts) to study tumor biology and test drugs (Kamb, 2005; Ledur et al., 2017). While these models play an important role, they often fail to capture the full complexity of human cancer. For example, traditional 2D monolayer cell cultures lack the three-dimensional architecture, multicellular interactions, cellular diversity, and tumor microenvironment of real tumors (Seidel et al., 2015; Aboulkheyr Es et al., 2018). Animal models provide a richer microenvironment, but patient-derived tumor xenograft (PDX) generation is time-consuming and expensive, and the interaction between the immune system and tumorigenesis cannot be studied in PDX models due to their immunodeficient nature (Perez et al., 2025). As a result, many therapies that appear to be effective in 2D cancer cultures and current animal models do not translate into clinical success, and the FDA approval rate for oncology therapies is as low as 3%. These limitations highlight the need for more physiologically relevant in vitro cancer models.
The emergence of patient-derived cancer cells (PDCCs) fills this gap, allowing researchers to culture and study cancer cells obtained directly from patient tumor samples. Compared with immortalized cell lines, PDCCs better retain the genetic and phenotypic heterogeneity of the original tumor (Vlachogiannis et al., 2018). PDCCs can be obtained by surgical resection of solid parts (Dekkers et al., 2021), puncture, fine needle aspiration (Lai et al., 2020; Sachs et al., 2019) or liquid biopsy (Sachs et al., 2019; Kopper et al., 2019; Schutgens et al., 2019). PDCCs culture covers a range of techniques, from 2D cell monolayers (Ricci-Vitiani et al., 2007), 3D tumor spheroids (De Witt Hamer et al., 2008), organoids (van de Wetering et al., 2015) and advanced co-culture systems (Tsai et al., 2018). Researchers usually choose or combine these techniques according to specific research questions and clinical application requirements. Currently, PDCCs have been used to study a variety of cancers, including breast cancer, lung cancer, gastrointestinal cancer, gastroesophageal cancer, pancreatic cancer, ovarian cancer, prostate cancer, glioblastoma, liver cancer, colorectal cancer, retinoblastoma and bladder cancer (El Harane et al., 2023; Boj et al., 2015; van de Wetering et al., 2015; Broutier et al., 2017). Using PDCCs to establish in vitro models for personalized drug screening (Vlachogiannis et al., 2018), immunotherapy efficacy evaluation (Tsai et al., 2018), individualized vaccine design (Ott et al., 2017; Sahin et al., 2017) and real-time monitoring (Kumari et al., 2021; Shen et al., 2023; Shickh et al., 2022) can enable the formulation of precise treatment strategies and promote clinical translation (Schmid et al., 2018).
This review provides a comprehensive overview of PDCCs culture technologies and their application in personalized medicine, and discusses the challenges faced in technical and clinical translation. We outline future developments at the intersection of PDCCs culture and AI that are expected to improve model fidelity and clinical utility.
2 PDCCs culture techniques and development
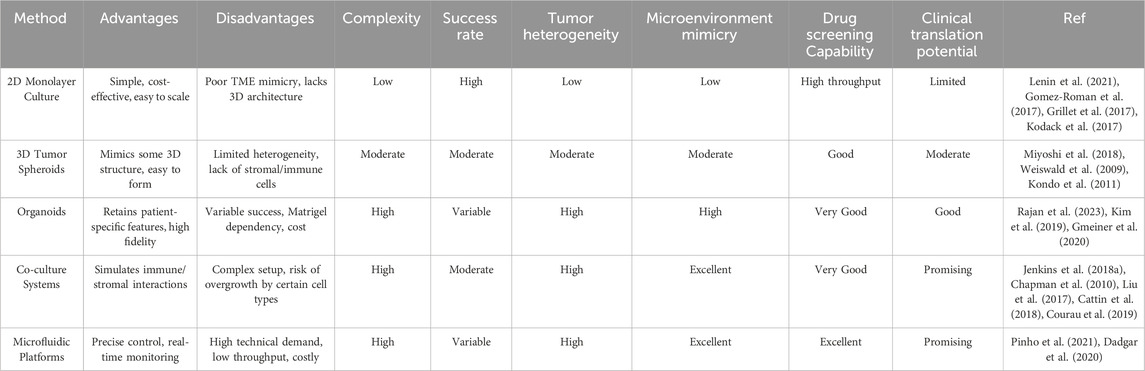
The primary methods for culturing PDCCs include traditional 2D monolayer culture, 3D spheroid models, organoid culture, co-culture systems including multiple cell types, and microfluidic chip-based culture (Figure 1). Each method has different operational complexity and tumor fidelity (Table 1).
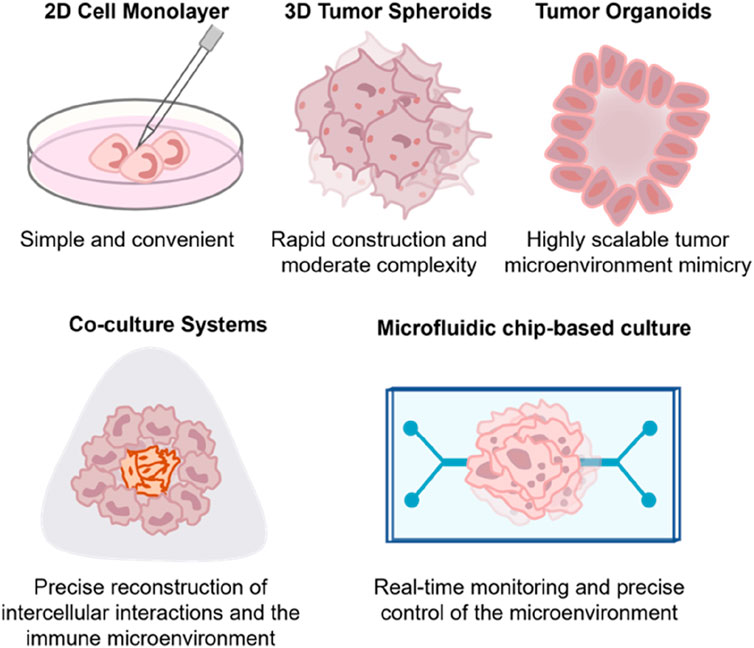
Figure 1. Comparative overview of PDCCs’ cultural typical features. This schematic illustrates five representative strategies for culturing PDCCs: 2D monolayers, 3D tumor spheroids, organoids, co-culture systems, and microfluidic platforms. Each approach differs in structural complexity and cellular interactions, offering advantages for modeling tumor biology and assessing personalized therapeutic responses.
2.1 2D cell monolayers and 3D tumor spheroids
The two-dimensional culture of PDCCs is the most straightforward approach, and it shares similarities with classical cancer cell line cultures, such as ease of manipulation and rapid cell proliferation, which have made them a workhorse in cancer research for decades (Kang et al., 2023) (Figure 2a). Large-scale drug sensitivity screening has traditionally relied on panels of two-dimensional cell lines from different patients or tumor types (Bresnahan et al., 2020; Qiu et al., 2019). However, PDCCs grown in a two-dimensional environment often fail to maintain the original tumour phenotype, with cells undergoing genetic and epigenetic drift, resulting in a loss of tumor-specific heterogeneity (Kamb, 2005; Ledur et al., 2017). In addition, cell-cell and cell-matrix interactions are absent or abnormal in 2D monolayers due to the absence of their native extracellular matrix (ECM) and tissue architecture (Kapałczyńska et al., 2018).
Figure 2. Spectrum and Evolution of PDCCs Culture Techniques. (a) Morphology of patient-derived cancer cell cultures when grown in 2D cell culture. The images are reproduced from reference (Kang et al., 2023) with permission from the Korean Cancer Association, copyright 2022. (b) Representative bright-field images of the tumour spheroids were shown and taken at ×40 total magnification. Scale bars correspond to 200 μm. The images are reproduced from the reference (Zhang et al., 2018) with permission from the Public Library of Science, copyright 2018. (c) Bright-field images depicting primary breast cancer organoid phenotypes. The top row shows cohesive organoids (left and middle: dense and solid; right: cystic and hollow), while the bottom row shows increasingly discohesive organoids (from left to right). Scale bar, 100 μm. The images are reproduced from reference (Sachs et al., 2018) with permission from Elsevier, copyright 2018. Shown in (d) are sample brightfield images of individual glioblastoma organoids over 4 weeks. Scale bar, 500 μm. The images are reproduced from reference (Jacob et al., 2020a) with permission from Elsevier, copyright 2020. (e) Images of bladder cancer organoids (BCOs) co-cultured with either MUC1 or CD19 CAR-T cells at 72 h. Scale bar, 200 μm. Immunostaining images of immunostaining for DAPI, CD8 and Granzyme B in BCOs after co-culturing with MUC1 CAR-T cells or CD19 CAR-T cells. Note the presence of activated and proliferating T cells near apoptotic BCOs. Scale bar, 100 μm. The images are reproduced from reference (Yu et al., 2021) with permission from John Wiley and Sons, copyright 2021. (f) Workflow for production of esophageal adenocarcinoma (EAC) assembloids. EAC organoids and primary CAFs are expanded before model creation, dissociated to single cells, counted, and mixed at a 2:1 ratio of CAFs to organoid cells. The cell suspension is then plated at 75,000 cells per well in an ultra-low attachment 96-well U-bottom plate. The next day, assembloids are plated in a 3:1 mixture of collagen I: BME2, and complete DMEM media are overlayed once set. Assembloids are cultured for a further 7 days before harvest. The structure formation of EAC assembloids was imaged with a ×4 objective under phase contrast microscopy every day for 7 days after matrix embedding. Scale bars: 500 μm. The images are reproduced from reference (Sharpe et al., 2024) with permission from Elsevier, copyright 2024. (g) Schematic representation of a chip filled with Pancreatic Ductal Adenocarcinoma (PDAC) organoids and pancreatic stellate cells. The cells are mixed with extracellular matrix, and these are seeded in the gel channel upon pipetting into the gel inlet and subsequently distributed along the gel channel due to capillary forces. Phaseguides separate the channels, as well as capillary pressure barriers, which keep the channels separate and allow the stratified loading of culture components. After gelation, cell culture medium was added to the medium inlet and outlet. Representative Phase Contrast image of a PDAC organoid monoculture in an OrganoPlate 2-Lane. 4× acquisition, Scale bar: 200 μm. The images are reproduced from the reference (Geyer et al., 2023) with permission from Springer Nature, copyright 2023. (h) Circulating tumor cells (CTCs) were pumped over multiple artificial organs, including lung, liver, bone, and muscle cells in the biomimetic model, followed by quantitation of organ-specific metastasis of CTCs. The images are reproduced from the reference (Kong et al., 2016) with permission from Impact Journals, copyright 2016. Bladder cancer organoids (BCOs); esophageal adenocarcinoma (EAC); Pancreatic Ductal Adenocarcinoma (PDAC); Circulating tumor cells (CTCs).
The shift from 2D flat layers to 3D spheroid cultures has significantly improved the physiological relevance of in vitro cancer models (Tung et al., 2011; Koledova, 2024; Hagemann et al., 2017). Due to cell-cell contacts and 3D structure, tumor spheroids can more closely resemble in vivo gene expression patterns, differentiation, and therapeutic response (Ricci-Vitiani et al., 2007; El Harane et al., 2023; Pampaloni et al., 2007; Ponti et al., 2005) patterns. And as spheroids grow, they develop nutrient and oxygen gradients, with a proliferative outer layer and a more quiescent or necrotic core (Han et al., 2021). This induces physiologically relevant features such as hypoxia, acidosis, and barriers to drug penetration (Groebe and Mueller-Klieser, 1996; Grimes et al., 2014; Thakuri et al., 2019; Costa et al., 2016). However, spheroids remain simplified models of tumors (Zhang et al., 2018) (Figure 2b). They are typically composed of a single cell type (usually the cancer cell itself) and lack the supporting stromal cells, immune infiltrates, and vascular structures found in real tumors (Faria et al., 2023). Nonetheless, tumor spheroids have laid the foundation for developing more complex 3D models such as organoids with more cellular complexity.
2.2 Patient-derived tumor organoids
Patient-derived tumor organoids are miniature tumors grown from a patient’s cancer cells (usually from surgery or a biopsy sample) that reflect some characteristics of in vivo tissues and functions.
Organoids often contain multiple cell lineages present in a tumor (Dominijanni et al., 2020; Devarasetty et al., 2020). For example, patient-derived colorectal cancer organoids are composed primarily of malignant epithelial cells. Still, they may also contain cancer stem cells and embedded stromal cells from the original tissue (van de Wetering et al., 2015). Organoids often maintain tumor heterogeneity, retain key mutations and gene expression patterns, and are genetically stable even after multiple passages (Roerink et al., 2018). They can also be expanded and cryopreserved to create living biobanks of patient tumors for research or drug testing (Sachs et al., 2018; Yan et al., 2018; Lee et al., 2018) (Figure 2c). Furthermore, because organoids can be generated relatively quickly (sometimes within 1–4 weeks) (Jacob et al., 2020a; Jacob et al., 2020b) (Figure 2d), there is growing interest in using them as real-time clinical avatars to guide treatment decisions for individual patients.
Despite their promise, organoids have limitations. Most organoids lack a complete tumor microenvironment (e.g., functional blood vessels, immune cells, and nerves), making it challenging to fully assess immune checkpoint responses and angiogenesis. In addition, organoid formation efficiencies vary between tumor types; for example, non-small cell lung cancer is difficult to construct due to the overgrowth of normal airway cells into lung cancer organoids (Dijkstra et al., 2020). Furthermore, their culture requires expertise and infrastructure (Gjorevski et al., 2016), and during culture, fast-growing clones may dominate, potentially reducing heterogeneity.
Researchers are working to standardize organoid culture media, develop synthetic matrices, and increase organoid derivation rates to overcome these issues. Overall, patient-derived organoids represent a high-fidelity tumor model that has rapidly become integral to cancer research and are at the forefront of personalized medicine efforts.
2.3 Co-culture system
Co-culture systems combine patient-derived tumor organoids or cells with other cell types to fully recreate the tumor microenvironment. The goal of co-culture models is to mimic the complex cell-cell interactions within a tumor, such as cancer cells and immune cells, fibroblasts, or endothelial cells.
Co-culture models allow researchers to directly observe the interaction between patient-derived organoids and immune or stromal and endothelial cells, realistically simulating tumor immune responses and microenvironments. For example, co-culturing organoids with patient-derived autologous T cells or CAR-T cells can produce tumour-killing activity, and PD-1/PD-L1 checkpoint blockade enhances this effect (Neal et al., 2018; Yu et al., 2021; Xie et al., 2020) (Figure 2e). At the same time, co-culture with cancer-associated fibroblasts and endothelial cells can reproduce tumor-stromal interactions and angiogenesis (Sharpe et al., 2024; Lim et al., 2022) (Figure 2f), making up for the lack of a complete microenvironment in single 3D organoids.
Co-culture systems can reproduce the tumor ecosystem more fully, such as immune evasion, immune cell activation, and stromal-tumor drug interactions, which are difficult to capture in a single organoid. For example, co-culture of organoids and immune cells can not only secrete cytokine profiles, but also show cell killing dynamics similar to the patient’s actual tumor (Chakrabarti et al., 2021), providing a basis for developing personalized immunotherapy and combination therapies. However, co-culture also has challenges. The addition of multiple cell types increases complexity and variability. Immune cells are often short-lived and require specific activation. Obtaining and expanding autologous cells is difficult, and non-autologous cells may induce unreal immune responses (Wu et al., 2012). In addition, co-culture conditions (such as cell ratio, addition time, and culture format) and analysis methods still need to be further standardized.
In summary, co-culture PDCC models represent a significant advance in recreating the tumor microenvironment in vitro. By including multiple cellular players, they provide a more comprehensive understanding of tumor biology.
2.4 Microfluidic chip-based culture
Microfluidics has revolutionized in vitro modeling, including patient-derived cancer culture. Through small fluidic channels and chambers, these platforms dynamically culture 2D cells, spheroids, or organoids under tightly controlled conditions while delivering culture media, drugs, or immune cells, creating an engineered microphysiological system that recreates tissue-level structure and function (Khoo et al., 2018; Deng et al., 2021; Deng et al., 2023; Li et al., 2025; Zhang J. et al., 2025; Fu et al., 2022).
Microfluidics technology makes in vitro culture closer to the in vivo environment (Fu et al., 2023). The chip platform can precisely control flow rate, shear stress, nutrient and oxygen supply, and chemical gradients. Continuous perfusion simulates blood flow, forms nutrient gradients and waste removal similar to capillaries, and thus affects cancer cell behavior and drug sensitivity (Jung et al., 2019; Shirure et al., 2018). In addition, these systems allow for partitioned co-culture, integrating tumor organoids with endothelial, immune and other cells, and reproducing in vivo processes such as immune cell infiltration and tumor invasion of blood vessels (Aung et al., 2020; Geyer et al., 2023; Haque et al., 2022) (Figure 2g). Microfluidics platforms also support high-throughput parallel experiments, real-time imaging and sensor monitoring, providing rich data for drug screening and personalized treatment decisions (Schuster et al., 2020). The application range of PDCC chips continues to expand, including high-throughput tumor spheroid generation, tumor slice culture and multi-organ interaction simulation. It is expected to build a more biomimetic system by combining bioprinting and microfabrication technology (Skardal et al., 2010; Aleman and Skardal, 2019). The organ homing preference of cancer cells was studied in a four-organ panel, and it was demonstrated that perfused breast circulating tumour cells invaded the lung, bone, and liver, but not muscle, which is consistent with animal studies (Kong et al., 2016) (Figure 2h).
Against this backdrop, microfluidic PDCC workflows have demonstrated end-to-end feasibility from scarce patient samples to functional readouts. Chip-based workflows can directly amplify circulating tumor cells (CTCs) from liquid biopsies and enable short-term, parallel drug testing within approximately 48–72 h under controlled perfusion, demonstrating a minimal tissue approach for rapid drug profiling for precise decision-making (Khoo et al., 2018). For example, Khoo et al. reported the development of a 3D microfluidic tumour model of bladder cancer, demonstrating that the incorporation of clinically relevant microenvironmental biofilm factors modulated tumour growth and treatment response, allowing for on-chip evaluation of combination therapies at specific flow rates (Deng et al., 2021). Similarly, recent studies have combined a microfluidic PDCC platform with a deep learning classifier to automate image-based cell viability and phenotype readouts, enabling rapid and reproducible analysis of patient cells directly from liquid biopsies (Hua et al., 2023; Li et al., 2022).
Overall, microfluidics improves the accuracy and complexity of patient-derived cancer models through engineered environments, complementing the biological fidelity of organoids to build tumor chip models that more realistically reproduce the tumor microenvironment and human tumor behavior. These advanced platforms are expected to improve the predictive power of drug screening and discovery and accelerate the application of PDCCs in clinical workflows.
3 Personalized medicine based on PDCCs
The primary motivation for developing PDCC culture is that it can retain the unique characteristics of the patient’s tumour (such as gene mutations and drug sensitivity), thus providing a basis for personalised cancer treatment. As an in vitro test platform, PDCCs can be used for personalized drug screening and evaluating the effect of immunotherapy, building tumor immune models, designing personalized vaccines, and promoting clinical translational research.
3.1 Personalized drug screening
Personalized drug screening is one of the most impactful applications of PDCCs. By exposing expanded tumor organoids to different drugs (or drug combinations) in multi-well plates or microfluidic chips, cell viability, growth inhibition, and apoptosis can be measured to obtain a drug sensitivity profile of the patient’s tumor, providing a basis for selecting the most likely effective treatment. Traditionally, high-throughput screening has been challenging to achieve due to the limited number and lifespan of primary cells. Still, organoid culture and improved 3D detection technology have made this process possible (Kamb, 2005; Ledur et al., 2017).
Studies on various cancers have demonstrated the potential of PDCC-based drug screening. For example, drug responses in colorectal cancer organoids correlate with patient clinical manifestations and have been shown to successfully predict the sensitivity of metastatic colorectal cancer to targeted therapies, which were subsequently treated and clinically proven to be effective 6 (Figure 3a). Pancreatic cancer organoids have shown high consistency in drug responses and actual patient outcomes when determining chemotherapy combinations (Ponz-Sarvise et al., 2019). PDCC cultures can not only serve as therapeutic diagnostic tools in the laboratory to guide personalized medication. Still, they can also be used to screen new drugs and explore resistance mechanisms by establishing organoid biobanks of multiple tumor subtypes (Figure 3b) (Yan et al., 2018).
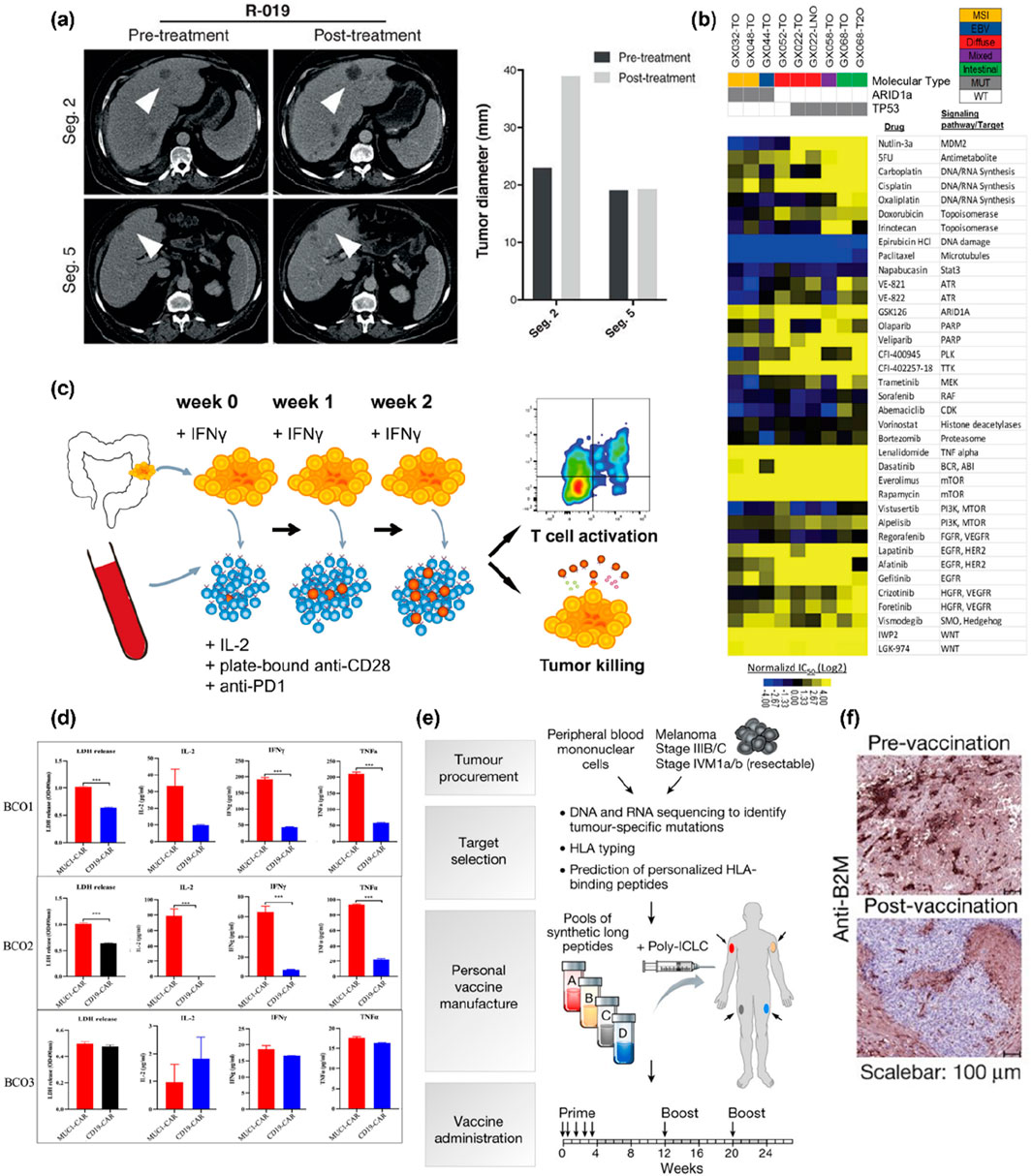
Figure 3. Applications of PDCC Cultures in Personalized Therapy. (a) Patient-derived organoids were established from a patient (R-019) with a mixed response to TAS-102. Whereas the segment two metastasis rapidly progressed, the segment five metastasis remained stable upon TAS-102 treatment (white arrowheads in the CT scan indicate metastases; bars represent pre- and posttreatment measurements of the indicated metastases). The images are reproduced from the reference (Vlachogiannis et al., 2018) with permission from AAAS, copyright 2018. (b) Large-scale drug screening in patient-derived organoid cultures. Heatmap of IC50 values for 37 compounds used to treat nine gastric cancer organoids derived from seven patients. The relative sensitivity against individual drugs is expressed as log2-transformed IC50 values and mean-centered against the therapeutic plasma steady-state concentration (Css). Therapeutic plasma concentrations of individual drugs (if available) are indicated on the x-axis of the dose-response graph. The images are reproduced from reference (Yan et al., 2018) with permission from Elsevier, copyright 2018. (c) Experimental workflow. Tumor organoids were established from dMMR CRC (resections or biopsies of primary tumors or metastases). They stimulated with IFNγ for 24 h before co-culture with peripheral blood lymphocytes (PBLs) from the same patient. PBLs were stimulated weekly with fresh tumor cells. After 2 weeks of co-culture, T cell effector functions and sensitivity of tumor organoids to T-cell-mediated killing were evaluated using flow cytometry. The images are reproduced from reference (Dijkstra et al., 2018) with permission from Elsevier, copyright 2018. (d) Quantification of lactate dehydrogenase release and cytokines products (IL-2, IFN-γ and TNF-α) from Bladder cancer organoids after co-culture with either MUC1 CAR-T cells or CD19 CAR-T cells. Values represent the mean ± SEM (n = 3; unpaired parametric t-test; ***P < 0.001). The images are reproduced from reference (Yu et al., 2021) with permission from John Wiley and Sons, copyright 2021. (e) Generation of a personal, multi-peptide neoantigen vaccine for patients with high-risk melanoma. Somatic mutations were identified by whole-exome sequencing of melanoma and germline DNA, and their expression confirmed by tumour RNA-seq. Immunizing peptides were selected based on human leukocyte antigen binding predictions. Each patient received up to 20 long peptides in four pools. The images are reproduced from the reference (Ott et al., 2017) with permission from Springer Nature, copyright 2017. (f) B2M staining of melanoma cells in pre- and post-vaccination metastases. Whereas the pre-treatment tumour sample of P04 stained almost homogenously for B2M, all tumour cells in the post-vaccine resectate were B2M-negative. The images are reproduced from the reference (Sahin et al., 2017) with permission from Springer Nature, copyright 2017. Human primary and metastatic pancreatic tumor organoid cultures (hT).
Although personalized drug screening based on PDCCs shows great potential, its widespread application still faces challenges. It is challenging to obtain results quickly to guide clinical decisions. Although most organoids can be cultured within a few weeks, this may not be timely enough for invasive cases, and growth is currently being accelerated by improving the culture medium. Second, the effect of certain drugs in organoids may not accurately predict clinical responses due to the lack of microenvironmental factors such as liver metabolism or tumour-stroma interactions, which have also promoted the integration of co-culture or microfluidics technology (Ooft et al., 2019).
3.2 Immunotherapy effect evaluation
Immunotherapy has revolutionized cancer treatment, but only a minority of patients can benefit. By adding immune components to the PDCCs co-culture system, researchers could mimic the tumor immune microenvironment, providing a new method for personalized prediction and study of immunotherapy responses.
The co-culture system of tumor organoids and immune cells can reproduce the patient’s anti-tumor immune response in vitro. By co-culturing patient-derived organoids with autologous T cells, NK cells, macrophages, etc. (Tsai et al., 2018; Chakrabarti et al., 2018; Fang et al., 2022; Chan and Ewald, 2022), researchers can observe phenomena such as immune cell infiltration, immune synapse formation, tumor cell killing and cytokine release, and use them to test the effects of immunotherapies such as PD-1/PD-L1 checkpoint blockade (Larkin et al., 2015; Garon et al., 2015; Borghaei et al., 2015; Le et al., 2015; Le et al., 2017). The PDCCs culture model reveals the mechanisms of immune escape and resistance, and by adjusting the co-culture conditions (such as the introduction of dendritic cells, specific antigen stimulation), it is observed whether the immune attack is improved. For example, adding IL-2 to the co-culture of melanoma organoids and peripheral blood lymphocytes significantly enhanced T cell proliferation and tumor killing (Dijkstra et al., 2018) (Figure 3c).
Despite the promise of co-culture systems, they also have key limitations that warrant further exploration. For example, immune cell viability and function decline rapidly in vitro, limiting the assessment of long-term efficacy (Finnberg et al., 2017; Yuki et al., 2020). Microspheres derived from human tumor suspensions containing immune cells in custom microfluidic devices have demonstrated responses to immunotherapy but lack tumor immunospecificity (Deng et al., 2018; Jenkins et al., 2018a). The static nature of co-cultures fails to recapitulate the dynamic immune cell recruitment and chemotaxis observed in vivo, reducing the predictive accuracy of therapies that rely on trafficking signals (Jenkins et al., 2018b). Comprehensive assessment of all stromal and immune components is also impeded. Furthermore, excessive immune cell proliferation can create interfering imaging features, leading to false-positive results (Stüve et al., 2023).
Advanced immune organoid platforms have emerged to overcome these challenges. Air-liquid interface organoids retain the in vivo association between native tumor-infiltrating lymphocytes and tumor cells in vitro, enabling robust assessment of the efficacy of PD-1 blockade (Neal et al., 2018; Li et al., 2016; Elbadawy et al., 2018; Li et al., 2014; Katano et al., 2013). Furthermore, a study has established an in vitro immune assessment platform based on patient-derived colorectal cancer organoids. Combined with CAR-natural killer cells, this platform can quantitatively monitor effector cell recruitment and tumor lysis in real time, and assess the killing efficacy and specificity of antigens such as EPCAM, thereby enabling personalized immunotherapy screening and off-target risk verification. (Schnalzger et al., 2019). Similarly, immune-enhancing organoids incorporating macrophages or dendritic cells can mimic antigen presentation and myeloid suppression, providing new insights into combination immunotherapy (Jiang et al., 2023; Chen et al., 2021; Subtil et al., 2023).
Furthermore, the Glioblastoma-on-a-Chip system enables real-time monitoring of T cell and tumor-associated macrophage recruitment by different glioblastoma subtypes. It analyzes the spatial expression patterns of PD-1 immune checkpoint and immunosuppressive factors such as the and TGF-β, thereby optimizing personalized immunotherapy strategies (Cui et al., 2020). PDCCs’ immune models also provide a patient-specific testbed for next-generation immunotherapies. By screening different chimeric antigen receptors in CAR-T cells co-cultured with PDCCs organoids, researchers can identify the most effective designs in vitro (Figure 3d) (Yu et al., 2021; Kumari et al., 2021; Zou et al., 2021).
In summary, PDCCs culture technology reproduces the laboratory’s interaction between tumors and the immune system, providing functional data for personalized immunotherapy research. With the help of new technologies such as microfluidic immuno-oncology chips, the culture of PDCCs is gradually becoming an important tool for predicting the efficacy of checkpoint inhibitors and customising cell therapy.
3.3 Personalized vaccine design
Personalized vaccine design is gradually becoming an important strategy for precision tumor immunotherapy. This method usually uses the patient’s tumor cells as an antigen library, identifies tumor-specific neoantigens through high-throughput sequencing and bioinformatics, and then constructs a vaccine targeting the patient’s specific mutations (Ott et al., 2017) (Figure 3e). In vitro models constructed using PDCCs, tumor spheroids or organoids can screen and verify the immunogenicity of candidate neoantigens in a short time while evaluating the effect of vaccines on activating T cell responses (Ott et al., 2017; Sahin et al., 2017) (Figure 3f). This personalized vaccine based on PDCCs can construct a personalized immune strategy for tumor-specific antigens and overcome the problem of tumor immune escape, providing patients with more precise and effective treatment options.
3.4 Real-time monitoring
In recent years, integrating microfluidic chips with machine learning has significantly improved the sensitivity and specificity of real-time patient-derived circulating tumor cells (CTCs) detection. For example, the fusion of microfluidics and deep learning in CTC detection enables automated classification and real-time monitoring, promising applications for the dynamic tracking of early-stage and metastatic cancers (Kumari et al., 2021; Shen et al., 2023; Shickh et al., 2022). Similarly, a dye-free, real-time imaging technique based on digital holographic phase microscopy and machine learning can distinguish cancer cells from blood cells at a rate of tens of cells per second, with an accuracy of 92.56% (Nissim et al., 2021). Furthermore, vapor nanobubbles-enhanced Cytophone technology has been used noninvasively to detect CTCs in melanoma patients. Accurate diagnosis was achieved in 27 of 28 patients, with a detection limit as low as 1 CTC per liter of blood, approximately 1,000-fold higher than existing detection methods (Galanzha et al., 2019).
However, the transition from research to clinical application still faces numerous obstacles. Inconsistent technical standards, high equipment costs, inadequate laboratory staff training, and incomplete regulatory policies have severely limited the adoption of liquid biopsy in routine clinical practice (Alix-Panabières and Pantel, 2021; Shickh et al., 2022; Lone et al., 2022; Martins et al., 2021; Mizutani et al., 2010). Furthermore, the inherent heterogeneity of CTCs and their extremely low concentration in blood present significant challenges for their capture and analysis (Vermesh et al., 2018; Lux et al., 2021; Chen X. et al., 2022; Kapeleris et al., 2022; Jiang W. et al., 2020; Andree et al., 2016). This leads to poor reproducibility, high false-negative/false-positive rates, and insufficient clinical confidence.
3.5 Clinical translation
The ultimate goal of PDCCs in personalized medicine is to inform and improve patient care directly. Although still in the experimental and early trial stages, significant progress has been made in the clinical translation of patient culture results.
One approach is combined clinical trials, whereby a patient receives standard care or an experimental treatment, and their tumour is studied in the laboratory in parallel using PDCC cultures. The laboratory test results are then compared with the patient’s clinical response to adjust the treatment regimen. For example, in a metastatic gastrointestinal cancer trial, organoid tests established with PDCCs were used to screen alternative therapies (Vlachogiannis et al., 2018). In another case of glioblastoma, organoids from a patient’s tumor predicted drug sensitivity, thereby guiding clinical decisions and observing tumor regression (Jacob et al., 2020a).
The main challenges facing clinical translation are scalability and turnover rate. These include ensuring that a high proportion of patient samples can quickly generate effective cultures (Centenera et al., 2018; Fu et al., 2021), especially since the success rate of some lung cancer organoids is low (Kim et al., 2019). And establishing an efficient process from hospital to laboratory to clinic. To in obtaining formal approval and widespread application. Multidisciplinary coordination and regulatory validation also need to be addressed to ensure the standardisation and predictive reliability of PDCCs testing.
Despite the challenges, PDCCs are gradually moving from the laboratory to the clinic and have played a role in trial design, such as screening out drug-sensitive patient subgroups, thereby providing information for biomarker-driven trial recruitment. PDCCs culture technology is providing solid support for the realization of truly personalized cancer treatment.
4 Current challenges
Despite the powerful capabilities of PDCC cultures for therapeutic applications, critical technical challenges, clinical translation, and ethical issues need to be addressed to ensure their reliable and responsible use.
4.1 Challenges in technology and clinical transformation
Differences in organoid culture conditions (e.g., media composition, growth factors, and ECM types) exist between laboratories (Neal et al., 2018; Maru and Hippo, 2019; Huch et al., 2017; Tuveson and Clevers, 2019), leading to inconsistent drug response results for the same tumor type, complicating comparisons between studies. For clinical application, it is urgent to develop unified standard operating procedures, including consensus culture media, chemically well-defined matrices, and unified assay readout standards (Gjorevski et al., 2016), and to verify reproducibility through inter-lab ring trials (Niepel et al., 2019).
Tumor heterogeneity is a significant challenge for PDCCs culture, as clonal selection may occur during in vitro culture, resulting in overgrowth of specific subclones and loss of representation of the original tumor (Broutier et al., 2017; Gao et al., 2014; Boretto et al., 2019). To address this issue, researchers are working to ensure that organoids retain the diversity of real tumors by optimizing culture media (van de Wetering et al., 2015; Gjorevski et al., 2016), validating key mutations (Farshadi et al., 2024) and utilizing single-cell sequencing (Wang et al., 2022). While maintaining heterogeneity is a necessary and challenging task, organoids currently perform quite well in this regard.
Not all patient samples can generate usable cultures, especially tumors with genomically unstable or extensive necrosis (e.g., the success rate for some lung cancer subtypes is less than 50%) (Dijkstra et al., 2020), which may lead to research biased towards those tumors that are easy to grow. To improve success and fairness, researchers are exploring improved culture techniques (such as air-liquid interface culture) (Esser et al., 2020), multiple sampling strategies, and in vivo support (such as short-term patient-derived xenograft passage) (Daniel et al., 2009; Rubio-Viqueira and Hidalgo, 2009; Simpson-Abelson et al., 2008) to expand the scope of application of PDCCs culture.
The time factor is critical for clinical applications because if organoid drug screening results take 2–3 months to be available, patients may have already switched to other treatments. Researchers are speeding up organoid expansion and testing to shorten this time by optimizing culture media (Georgakopoulos et al., 2020), automated processing (Jiang S. et al., 2020), and imaging readouts. In vitro tissue slice culture is faster (Centenera et al., 2013) (drug testing can be performed within a week), and the experience can provide a reference for improving PDCCs organoid co-culture methods.
4.2 Sample logistics and regulatory challenges
PDCCs are increasingly used in preclinical research and personalized medicine due to their ability to recapitulate patient-specific tumor phenotypes. However, the clinical translation of PDCC-based platforms is hampered by sample logistics and regulatory frameworks. From a logistical perspective, current tissue collection and processing workflows are fragmented across institutions. For instance, Yoko S. DeRose demonstrated that breast tumor specimens must be transported on ice immediately after surgical resection to minimize ischemic time and preserve tissue viability, which is essential for successfully establishing patient-derived models (DeRose et al., 2013). Else Driehuis further reported that placing freshly resected tumor tissue in ice-cold culture medium supplemented with Rho kinase inhibitor significantly enhances cell viability and organoid formation efficiency, with organoid generation remaining feasible after up to 72 h of cold storage at 4 °C (Driehuis et al., 2020). In addition, Michael Gock and colleagues showed that transport delays negatively affect model establishment. However, Matrigel can significantly improve tumour engraftment rates (Ding et al., 2022) during delayed cooling conditions.
From a regulatory standpoint, ensuring cross-laboratory consistency and reproducibility is essential to generating reliable, clinically actionable results. In a colorectal cancer drug screening study, the pilot study of only eight patients was expanded to a registrational clinical trial of 250 patients (NCT05189171) to verify its sensitivity and reproducibility in predicting drug responses. This move aims to meet the key regulatory requirements for data consistency and traceability of results in in vitro diagnostic models for clinical applications (Ding et al., 2022). In the application of patient-derived 3D culture systems as disease-specific drug sensitivity models, it is emphasized that quality control and cross-laboratory consistency must be met before entering clinical translation. For example, by systematically evaluating the reproducibility indicators of IC50 and maximum inhibition rate, the stability and consistency of the organoid drug sensitivity testing platform in different batches are verified to meet the reproducibility standards in the test performance regulatory guidelines (Boehnke et al., 2016).
4.3 Ethical challenges
Regarding ethics, PDCC culture involves informed consent, privacy protection, ownership, and commercialization issues. Patients are required to understand the use of their tumor samples, storage period, and sensitive genetic information that may be generated (Mollaki, 2021). There is an obligation to protect patient privacy, and whole genome sequencing of organoids may reveal sensitive information about germline mutations (Mollaki, 2021; Hendriks et al., 2020; Driehuis and Clevers, 2017). In addition, due to the high cost of personalized PDCC organoid testing, measures must be taken to ensure this technology is accessible to patients at all levels. The regulatory framework is still unclear (Munsie et al., 2017), and guidelines are urgently needed to ensure strict quality control and validation of PDCCs in clinical applications and to clarify their status in companion diagnostics.
5 Future development potential
The field of PDCC culture is rapidly evolving. Several exciting developments are expected to enhance PDCC culture further and expand its use in research and personalized medicine.
5.1 Combining advanced micro-engineering
The combination of PDCC culture with microfluidics and bioengineering methods is expected to be further deepened. Future tumour chip models will integrate multiple tissue types and achieve synergistic interactions between tumours, the immune system and normal tissues through microfluidics (Aleman and Skardal, 2019), 3D bioprinting (Chen et al., 2022b; van Pel et al., 2018) and sensors (Field 154, 155) (Figure 4a). The ultimate goal is to build personalized microphysiological systems to simulate “clinical trials on a chip” to predict tumor response, normal tissue toxicity and pharmacokinetics, formulating comprehensive personalized treatment plans.
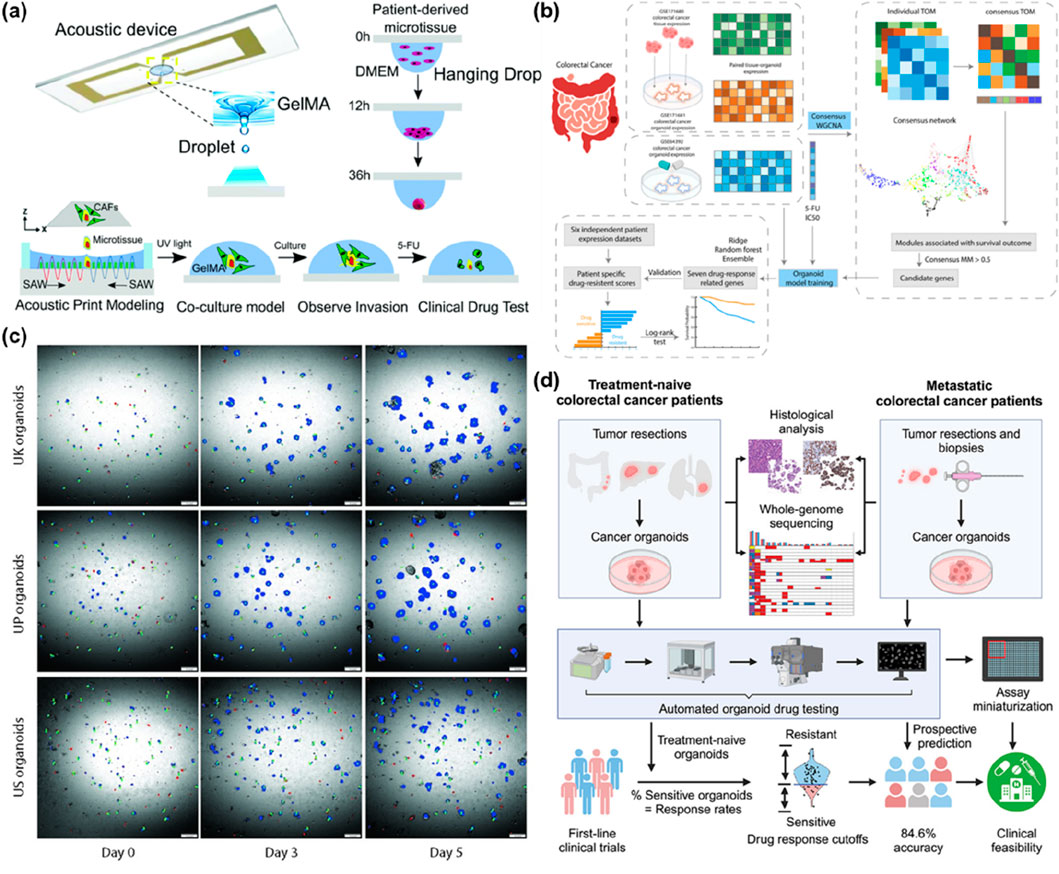
Figure 4. Future Directions for PDCC Platform Development. (a) Scheme of acoustic bioprinting for modelling cancer invasion. Acoustic device ejects GelMA acoustic droplets. Patient-derived microtissues quickly aggregate primary cells into microtissues within 36 h by hanging drop. The process of using acoustic 3D printing devices to construct a cancer invasion model. Co-cultivation of the model, observation of invasion phenomena, and clinical drug testing of the model with 5-fluorouracil (5-FU). The images are reproduced from reference (Chen H. et al., 2022) with permission from the Royal Society of Chemistry, copyright 2013. (b) Workflow for identifying gene biomarkers with Weighted Gene Co-expression Network Analysis (WGCNA) and development of an organoid model. The process begins with selecting three colorectal cancer datasets for a consensus WGCNA. Potential gene biomarkers are then chosen to train the organoid models with GSE64392 and its drug response to 5-FU. Subsequently, genes related to drug response are identified and utilized for prognosis testing on six independent colorectal cancer patient expression datasets. The images are reproduced from reference (Zhang W. et al., 2025) with permission from Elsevier, copyright 2025. (c) Organoid tracking and morphology measurement. Each patient organoid was segmented with a neural network (NN). Representative images show the changes in tracked organoids over time. Blue: live organoids, Red: dead organoids, Green: tracks. The images are reproduced from the reference (Gunnarsson et al., 2024) with permission from Public Library of Science (PLOS), copyright 2024. (d) Workflow for prospective drug testing of metastatic colorectal cancer organoids, from patient biopsy and multi-omics characterization through automated, miniaturized pharmacotyping, achieving 84.6% accuracy in predicting clinical response. The images are reproduced from reference (Tan et al., 2023) with permission from Cell, copyright 2023. 5-fluorouracil (5-FU); Weighted Gene Co-expression Network Analysis (WGCNA); neural network (NN).
5.2 AI Across Domains
As data generated from PDCCs’ culture experiments becomes increasingly complex, artificial intelligence will become a key tool for extracting actionable insights (Kong et al., 2020; Hua et al., 2024). Organoid pharmacogenomic data and network-based computational methods have successfully derived reliable drug biomarkers to treat human tumors (Kong et al., 2020). A novel deep neural network can effectively detect organoids and dynamically track them throughout the culture process (Bian et al., 2021). In addition, large databases of patient-derived culture data could be used to train predictive models (Bian et al., 2021).
Tumor spheroids, a popular preclinical model, lack reliable image segmentation. A fully convolutional network, including U-Net and HRNet, automatically segments treated and untreated multicellular tumour spheroids, achieving a Jaccard index of approximately 90% and segmentation error comparable to interobserver variability (Streller et al., 2025). This work enables more reproducible and high-throughput quantification of treatment responses in 3D tumor models, complementing software tools for high-throughput image analysis of spheroids. Using matched colorectal tumor–organoid transcriptomes, Zhang et al. developed a network-based biomarker selection strategy to predict patient-specific chemotherapy responses, achieving superior performance compared to conventional gene prioritization approaches (Zhang W. et al., 2025) (Figure 4b). Furthermore, by combining longitudinal imaging of patient-derived tumor organoids with AI-powered segmentation and mathematical modeling, the study achieved high-resolution tracking of individual organoid growth trajectories, laying the foundation for further modeling efforts aimed at predicting treatment response kinetics and the duration of drug resistance (Gunnarsson et al., 2024) (Figure 4c). Fillioux et al. developed a deep learning framework that integrates segmentation (SAM), feature extraction (DINOv2), and attention-based multiple-instance learning to analyze time-lapse videos of patient-derived organoids, enabling accurate, non-invasive prediction of chemotherapeutic efficacy over time (Fillioux et al., 2023). While these methods allow efficient and precise detection, classification, and measurement of PDCC culture platforms, they often require programming skills to create specialised code to train the networks and process the images (Spiller et al., 2021). Deep learning-based analyses can be very effective when processing large datasets, but extracting specific information requires additional data processing. Furthermore, overlapping organoids formed by PDCCs have contacts not present in individual organoids, making segmentation difficult using deep learning image processing tools (Park et al., 2023).
5.3 Enhancing the integration of the tumor microenvironment
Future PDCC cultures aim to fill gaps in TME mimicry by recreating a more realistic tumor microenvironment through vascularization strategies (such as co-culture with endothelial cells to form perfused capillaries (Silvestri et al., 2020)) and integrating a more complex immune system. Some studies have also explored microbiome-tumor co-cultures to evaluate the impact of the gut microbiota on cancer progression and immunotherapy response (El-Derby et al., 2024). Although there are challenges in maintaining these additional components without disrupting the core tumor culture, incremental progress brings us closer to building a “tumor ecosystem in a dish.”
5.4 Automation, standardization, and data accessibility
Automation and scale-up are essential to making the clinic’s PDCC culture model widely used. The next-generation of culture platforms will use robotic systems to automatically process each step from tissue dissociation to organoid generation and drug testing (Zhang et al., 2017) to achieve high-throughput processing, improve consistency and reduce costs. At the same time, a central facility or “living biobank” (Sachs et al., 2018) linked to the hospital will be established to send the patient’s tumor to the hospital, conduct organoid or drug testing, and quickly report to the clinician.
The lack of standardised protocols for establishing PDCC culture platforms can lead to batch-to-batch variability and an overall lack of quality control across and within institutions (Brancato et al., 2020). Furthermore, establishing PDCC culture platforms is technically challenging and requires highly trained personnel to handle and prepare patient-derived cells (Kondo and Inoue, 2019). Therefore, standard operating procedures must be adopted to govern each step, including sample handling, culture conditions, quality control measures, and result interpretation, to ensure consistent and reproducible laboratory results (Xiang et al., 2024). This is crucial for providing reliable and comparable results across institutions and developing clinical implementation guidelines. Tao Tan described a unified framework for predictive testing based on patient-derived colorectal cancer organoids, encompassing standard chemotherapy, biologics, and targeted therapy regimens (Tan et al., 2023) (Figure 4d). Interlaboratory variability and the lack of standardized protocols limit reproducibility and scalability. To overcome these obstacles, future work should focus on developing standardized culture methods and shared biobanks (Heydari et al., 2025). Training programs and collaborative platforms can also help disseminate technical expertise. The CancerModels.Org platform embodies a global open-access framework for patient-derived cancer models that standardizes, harmonizes, and integrates clinical, genomic, and functional datasets according to FAIR principles, significantly enhancing accessibility and data sharing across the research community (Perova et al., 2025).
5.5 Clinical implementation and personalized treatment algorithms
In the next 5–10 years, PDCCs data may be integrated with information such as genomic sequencing into clinical decision-making to support tumor boards, and oncologists will also use artificial intelligence to support decision-making based on these in vitro functional data (Kong et al., 2020). Personalized treatment plans based on PDCC will guide first-line treatment and provide a basis for maintenance strategies and next-line selection. At the same time, regular biopsies to generate organoids can achieve real-time monitoring, detect drug-resistant mutations early, and adjust treatment.
6 Conclusion
PDCC models, encompassing 2D monolayers, 3D spheroids, organoids, co-culture systems, and microfluidic platforms, have collectively transformed our ability to recapitulate patient-specific tumor biology and drug responses. We discuss in detail the key applications of PDCC models in personalized drug screening, immunotherapy evaluation, individualized vaccine design, real-time monitoring, and clinical translational research. While advances in advanced microengineering, AI-driven analytical techniques, and enhanced integration of the tumor microenvironment continue to refine these platforms, widespread challenges remain in clinical translation, standardized sample logistics, quality management, and navigating the evolving regulatory landscape. By overcoming these obstacles through multidisciplinary collaboration and robust validation studies, PDCCs technology has the potential to become an indispensable tool in precision oncology, enabling truly personalized treatment strategies and accelerating the translation of laboratory discoveries into patient benefit.
Author contributions
YF: Writing – original draft. BK: Writing – review and editing. CL: Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors would like to thank the funding from the Startup Grant (Grant no. A-8001301-00-00) and the Institute for Health Innovation and Technology Grant (Grant no. A-0001415-06-00) from the National University of Singapore (NUS). This work was supported by the City University of Hong Kong (7006082, 7020073, 9609332, 9609333, 9678292, 7020002), the Research Grants Council (RGC) (9048206, 8799020), the Hong Kong Center for Cerebro-Cardiovascular Health Engineering (COCHE), Innovation and Technology Commission (PRP/001/22FX), the Hetao Shenzhen-Hong Kong Science and Technology Innovation Cooperation Zone Shenzhen Park Project (HZQB-KCZYZ-2021017), and the Education Bureau Gifted Education Programme (3030780).
Acknowledgments
Figure 2a was reprinted from Lux A et al. (2021). Figure 2b was reprinted from Zhang Z et al. (2018). Figure 2c was reprinted from Sachs N et al. (2018). Figure 2d was reprinted from Jacob F et al. (2020). Figure 2e was reprinted from Yu L et al. (2021). Figure 2f was reprinted from Sharpe B P et al. (2024). Figure 2g was reprinted from Geyer M et al. (2023). Figure 2h was reprinted from Kong J et al. (2016). Figure 3a was reprinted from Vlachogiannis G et al. (2018). Figure 3b was reprinted from Yan H H N et al. (2018). Figure 3c was reprinted from Dijkstra K K et al. (2018). Figure 3d was reprinted from Yu L et al. (2021). Figure 3e was reprinted from Ott P A et al. (2017). Figure 3f was reprinted from Sahin U et al. (2017). Figure 4a was reprinted from Chen H et al. (2022). Figure 4b was reprinted from Zhang W et al. (2025). Figure 4c was reprinted from Gunnarsson E B et al. (2024). Figure 4d was reprinted from Tan T et al. (2023).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
PDCCs, patient-derived cancer cells; PDX, patient-derived tumor xenograft; ECM, extracellular matrixCTCs, circulating tumor cells.
References
Aboulkheyr Es, H., Montazeri, L., Aref, A. R., Vosough, M., and Baharvand, H. (2018). Personalized cancer medicine: an organoid approach. Trends Biotechnol. 36, 358–371. doi:10.1016/j.tibtech.2017.12.005
Aleman, J., and Skardal, A. (2019). A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol. Bioeng. 116, 936–944. doi:10.1002/bit.26871
Alix-Panabières, C., and Pantel, K. (2021). Liquid biopsy: from discovery to clinical application. Cancer Discov. 11, 858–873. doi:10.1158/2159-8290.cd-20-1311
Andree, K. C., van Dalum, G., and Terstappen, L. W. M. M. (2016). Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 10, 395–407. doi:10.1016/j.molonc.2015.12.002
Aung, A., Kumar, V., Theprungsirikul, J., Davey, S. K., and Varghese, S. (2020). An engineered tumor-on-a-chip device with breast cancer–immune cell interactions for assessing T-cell recruitment. Cancer Res. 80, 263–275. doi:10.1158/0008-5472.can-19-0342
Bian, X., Li, G., Wang, C., Liu, W., Lin, X., Chen, Z., et al. (2021). A deep learning model for detection and tracking in high-throughput images of organoid. Comput. Biol. Med. 134, 104490. doi:10.1016/j.compbiomed.2021.104490
Boehnke, K., Iversen, P. W., Schumacher, D., Lallena, M. J., Haro, R., Amat, J., et al. (2016). Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. SLAS Discov. 21, 931–941. doi:10.1177/1087057116650965
Boj, S. F., Hwang, C. I., Baker, L., Chio, I., Engle, D., Corbo, V., et al. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. doi:10.1016/j.cell.2014.12.021
Boretto, M., Maenhoudt, N., Luo, X., Hennes, A., Boeckx, B., Bui, B., et al. (2019). Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041–1051. doi:10.1038/s41556-019-0360-z
Borghaei, H., Paz-Ares, L., Horn, L., Spigel, D. R., Steins, M., Ready, N. E., et al. (2015). Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N. Engl. J. Med. 373, 1627–1639. doi:10.1056/nejmoa1507643
Brancato, V., Oliveira, J. M., Correlo, V. M., Reis, R. L., and Kundu, S. C. (2020). Could 3D models of cancer enhance drug screening? Biomaterials 232, 119744. doi:10.1016/j.biomaterials.2019.119744
Bresnahan, E., Ramadori, P., Heikenwalder, M., Zender, L., and Lujambio, A. (2020). Novel patient-derived preclinical models of liver cancer. J. hepatology 72, 239–249. doi:10.1016/j.jhep.2019.09.028
Broutier, L., Mastrogiovanni, G., Verstegen, M. M., Francies, H. E., Gavarró, L. M., Bradshaw, C. R., et al. (2017). Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435. doi:10.1038/nm.4438
Cattin, S., Ramont, L., and Rüegg, C. (2018). Characterization and in vivo validation of a three-dimensional multi-cellular culture model to study heterotypic interactions in colorectal cancer cell growth, invasion and metastasis. Front. Bioeng. Biotechnol. 6, 97. doi:10.3389/fbioe.2018.00097
Centenera, M. M., Raj, G. V., Knudsen, K. E., Tilley, W. D., and Butler, L. M. (2013). Ex vivo culture of human prostate tissue and drug development. Nat. Rev. Urol. 10, 483–487. doi:10.1038/nrurol.2013.126
Centenera, M. M., Hickey, T. E., Jindal, S., Ryan, N. K., Ravindranathan, P., Mohammed, H., et al. (2018). A patient-derived explant (PDE) model of hormone-dependent cancer. Mol. Oncol. 12, 1608–1622. doi:10.1002/1878-0261.12354
Chakrabarti, J., Holokai, L., Syu, L., Steele, N., Chang, J., Dlugosz, A., et al. (2018). Mouse-derived gastric organoid and immune cell co-culture for the study of the tumor microenvironment. Methods Mol. Biol. 1817, 157–168. doi:10.1007/978-1-4939-8600-2_16
Chakrabarti, J., Koh, V., So, J. B. Y., Yong, W. P., and Zavros, Y. (2021). A preclinical human-derived autologous gastric cancer organoid/immune cell co-culture model to predict the efficacy of targeted therapies. J. Vis. Exp. 173. doi:10.3791/61443
Chan, I., and Ewald, A. (2022). Natural killer (NK) cells: methods and protocols. Methods Mol. Biol. 2463, 235–250. doi:10.1007/978-1-0716-2160-8_17
Chapman, S., Liu, X., Meyers, C., Schlegel, R., and McBride, A. A. (2010). Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Investigation 120, 2619–2626. doi:10.1172/jci42297
Chen, J., Sun, H. W., Yang, Y. Y., Chen, H. T., Yu, X. J., Wu, W. C., et al. (2021). Reprogramming immunosuppressive myeloid cells by activated T cells promotes the response to anti-PD-1 therapy in colorectal cancer. Signal Transduct. Target. Ther. 6 (4), 4. doi:10.1038/s41392-020-00377-3
Chen, X., Xu, P., Lin, W., Jiang, J., Qu, H., Hu, X., et al. (2022a). Label-free detection of breast cancer cells using a functionalized tilted fiber grating. Biomed. Opt. Express 13, 2117–2129. doi:10.1364/boe.454645
Chen, H., Du, L., Li, J., Wu, Z., Gong, Z., Xia, Y., et al. (2022b). Modeling cancer metastasis using acoustically bio-printed patient-derived 3D tumor microtissues. J. Mater. Chem. B 10, 1843–1852. doi:10.1039/D1TB02789A
Costa, E. C., Moreira, A. F., de Melo-Diogo, D., Gaspar, V. M., Carvalho, M. P., and Correia, I. J. (2016). 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnol. Adv. 34, 1427–1441. doi:10.1016/j.biotechadv.2016.11.002
Courau, T., Bonnereau, J., Chicoteau, J., Bottois, H., Remark, R., Assante Miranda, L., et al. (2019). Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. cancer 7, 74. doi:10.1186/s40425-019-0553-9
Cui, X., Ma, C., Vasudevaraja, V., Serrano, J., Tong, J., Peng, Y., et al. (2020). Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. eLife 9, e52253. doi:10.7554/eLife.52253
Dadgar, N., Gonzalez-Suarez, A. M., Fattahi, P., Hou, X., Weroha, J. S., Gaspar-Maia, A., et al. (2020). A microfluidic platform for cultivating ovarian cancer spheroids and testing their responses to chemotherapies. Microsystems and Nanoeng. 6, 93. doi:10.1038/s41378-020-00201-6
Daniel, V. C., Marchionni, L., Hierman, J. S., Rhodes, J. T., Devereux, W. L., Rudin, C. M., et al. (2009). A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 69, 3364–3373. doi:10.1158/0008-5472.Can-08-4210
Dekkers, J. F., van Vliet, E. J., Sachs, N., Rosenbluth, J. M., Kopper, O., Rebel, H. G., et al. (2021). Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat. Protoc. 16, 1936–1965. doi:10.1038/s41596-020-00474-1
Deng, J., Wang, E. S., Jenkins, R. W., Li, S., Dries, R., Yates, K., et al. (2018). CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 8, 216–233. doi:10.1158/2159-8290.cd-17-0915
Deng, Y., Liu, S. Y., Chua, S. L., and Khoo, B. L. (2021). The effects of biofilms on tumor progression in a 3D cancer-biofilm microfluidic model. Biosens. Bioelectron. 180, 113113. doi:10.1016/j.bios.2021.113113
Deng, Y., Fu, Y., Chua, S. L., and Khoo, B. L. (2023). Biofilm Potentiates cancer-promoting effects of tumor-associated macrophages in a 3D multi-Faceted tumor model. Small 19, 2205904. doi:10.1002/smll.202205904
DeRose, Y. S., Gligorich, K. M., Wang, G., Georgelas, A., Bowman, P., Courdy, S. J., et al. (2013). Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr. Protoc. Pharmacol. 60, 14.23. 11–14.23. 43. doi:10.1002/0471141755.ph1423s60
Devarasetty, M., Forsythe, S. D., Shelkey, E., and Soker, S. (2020). In vitro modeling of the tumor microenvironment in tumor organoids. Tissue Eng. Regen. Med. 17, 759–771. doi:10.1007/s13770-020-00258-4
Dijkstra, K. K., Cattaneo, C. M., Weeber, F., Chalabi, M., van de Haar, J., Fanchi, L. F., et al. (2018). Generation of tumor-reactive T cells by Co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598.e12. doi:10.1016/j.cell.2018.07.009
Dijkstra, K. K., Monkhorst, K., Schipper, L. J., Hartemink, K. J., Smit, E. F., Kaing, S., et al. (2020). Challenges in establishing pure lung cancer organoids limit their utility for personalized medicine. Cell Rep. 31, 107588. doi:10.1016/j.celrep.2020.107588
Ding, S., Hsu, C., Wang, Z., Natesh, N. R., Millen, R., Negrete, M., et al. (2022). Patient-derived micro-organospheres enable clinical precision oncology. Cell Stem Cell 29, 905–917.e6. doi:10.1016/j.stem.2022.04.006
Dominijanni, A., Devarasetty, M., and Soker, S. (2020). Manipulating the tumor microenvironment in tumor organoids induces phenotypic changes and chemoresistance. Iscience 23, 101851. doi:10.1016/j.isci.2020.101851
Driehuis, E., and Clevers, H. (2017). CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiology-Gastrointestinal Liver Physiology 312, G257–g265. doi:10.1152/ajpgi.00410.2016
Driehuis, E., Kretzschmar, K., and Clevers, H. (2020). Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409. doi:10.1038/s41596-020-0379-4
El-Derby, A. M., Schaaf, C. R., Shelkey, E., Cook, K. L., Votanopoulos, K. I., and Soker, S. (2024). Immune-reactive tumor organoids system to determine the effects of microbial metabolites on cancer immunity and immunotherapies. Front. Microbiomes 3, 1411322. doi:10.3389/frmbi.2024.1411322
Elbadawy, M., Usui, T., Yamawaki, H., and Sasaki, K. (2018). Development of an experimental model for analyzing drug resistance in colorectal cancer. Cancers 10, 164. doi:10.3390/cancers10060164
Esser, L. K., Branchi, V., Leonardelli, S., Pelusi, N., Simon, A. G., Klümper, N., et al. (2020). Cultivation of Clear cell Renal cell carcinoma patient-derived organoids in an air-liquid interface system as a tool for studying individualized therapy. Front. Oncol. 10, 1775. doi:10.3389/fonc.2020.01775
Fang, H., Huang, Y., Luo, Y., Tang, J., Yu, M., Zhang, Y., et al. (2022). SIRT1 induces the accumulation of TAMs at colorectal cancer tumor sites via the CXCR4/CXCL12 axis. Cell. Immunol. 371, 104458. doi:10.1016/j.cellimm.2021.104458
Faria, C., Gava, F., Gravelle, P., Valero, J. G., Dobaño-López, C., Van Acker, N., et al. (2023). Patient-derived lymphoma spheroids integrating immune tumor microenvironment as preclinical follicular lymphoma models for personalized medicine. J. Immunother. Cancer 11, e007156. doi:10.1136/jitc-2023-007156
Farshadi, E. A., Wang, W., Mohammad, F., van der Oost, E., Doukas, M., van Eijck, C. H. J., et al. (2024). Tumor organoids improve mutation detection of pancreatic ductal adenocarcinoma. Sci. Rep. 14, 25468. doi:10.1038/s41598-024-75888-y
Fillioux, L., Gontran, E., Cartry, J., Mathieu, J. R., Bedja, S., Boilève, A., et al. (2023). “Spatio-temporal analysis of patient-derived organoid videos using deep learning for the prediction of drug efficacy.” In Proceedings of the IEEE/CVF International Conference on Computer Vision, 3930–3939. doi:10.1109/ICCVW60793.2023.00425
Finnberg, N. K., Gokare, P., Lev, A., Grivennikov, S. I., MacFarlane, A. W., Campbell, K. S., et al. (2017). Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 8, 66747–66757. doi:10.18632/oncotarget.19965
Fu, Y., Zhang, Y., and Khoo, B. L. (2021). Liquid biopsy technologies for hematological diseases. Med. Res. Rev. 41, 246–274. doi:10.1002/med.21731
Fu, Y., Zou, S., and Khoo, B. L. (2022). Label-free enrichment of human blast cells from whole blood for leukemia monitoring. Star. Protoc. 3. doi:10.1016/j.xpro.2022.101584
Fu, Y., Deng, Y., Zhang, J., Chua, S. L., and Khoo, B. L. (2023). Biofilms exacerbate atherogenesis through macrophage-induced inflammatory responses in a fibrous plaque microsystem model. Acta Biomater. 168, 333–345. doi:10.1016/j.actbio.2023.06.028
Galanzha, E. I., Menyaev, Y. A., Yadem, A. C., Sarimollaoglu, M., Juratli, M. A., Nedosekin, D. A., et al. (2019). In vivo liquid biopsy using Cytophone platform for photoacoustic detection of circulating tumor cells in patients with melanoma. Sci. Transl. Med. 11, eaat5857. doi:10.1126/scitranslmed.aat5857
Gao, D., Vela, I., Sboner, A., Iaquinta, P., Karthaus, W., Gopalan, A., et al. (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187. doi:10.1016/j.cell.2014.08.016
Garon, E. B., Rizvi, N. A., Hui, R., Leighl, N., Balmanoukian, A. S., Eder, J. P., et al. (2015). Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028. doi:10.1056/nejmoa1501824
Georgakopoulos, N., Prior, N., Angres, B., Mastrogiovanni, G., Cagan, A., Harrison, D., et al. (2020). Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 20, 4. doi:10.1186/s12861-020-0209-5
Geyer, M., Schreyer, D., Gaul, L. M., Pfeffer, S., Pilarsky, C., and Queiroz, K. (2023). A microfluidic-based PDAC organoid system reveals the impact of hypoxia in response to treatment. Cell Death Discov. 9, 20. doi:10.1038/s41420-023-01334-z
Gjorevski, N., Sachs, N., Manfrin, A., Giger, S., Bragina, M. E., Ordóñez-Morán, P., et al. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564. doi:10.1038/nature20168
Gmeiner, W. H., Miller, L. D., Chou, J. W., Dominijanni, A., Mutkus, L., Marini, F., et al. (2020). Dysregulated pyrimidine biosynthesis contributes to 5-FU resistance in SCLC patient-derived organoids but response to a novel polymeric fluoropyrimidine, CF10. Cancers 12, 788. doi:10.3390/cancers12040788
Gock, M., Kühn, F., Mullins, C. S., Krohn, M., Prall, F., Klar, E., et al. (2016). Tumor take rate optimization for colorectal carcinoma patient-derived xenograft models. BioMed Res. Int. 2016, 1–7. doi:10.1155/2016/1715053
Gomez-Roman, N., Stevenson, K., Gilmour, L., Hamilton, G., and Chalmers, A. J. (2017). A novel 3D human glioblastoma cell culture system for modeling drug and radiation responses. Neuro-Oncology 19, 229–241. doi:10.1093/neuonc/now164
Grillet, F., Bayet, E., Villeronce, O., Zappia, L., Lagerqvist, E. L., Lunke, S., et al. (2017). Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut 66, 1802–1810. doi:10.1136/gutjnl-2016-311447
Grimes, D. R., Kelly, C., Bloch, K., and Partridge, M. (2014). A method for estimating the oxygen consumption rate in multicellular tumour spheroids. J. R. Soc. Interface 11, 20131124. doi:10.1098/rsif.2013.1124
Groebe, K., and Mueller-Klieser, W. (1996). On the relation between size of necrosis and diameter of tumor spheroids. Int. J. Radiat. Oncol. Biol. Phys. 34, 395–401. doi:10.1016/0360-3016(95)02065-9
Gunnarsson, E. B., Kim, S., Choi, B., Schmid, J. K., Kaura, K., Lenz, H. J., et al. (2024). Understanding patient-derived tumor organoid growth through an integrated imaging and mathematical modeling framework. PLoS Comput. Biol. 20, e1012256. doi:10.1371/journal.pcbi.1012256
Hagemann, J., Jacobi, C., Hahn, M., Schmid, V., Welz, C., Schwenk-Zieger, S., et al. (2017). Spheroid-based 3D cell cultures enable personalized therapy testing and drug discovery in Head and Neck cancer. Anticancer Res. 37, 2201–2210. doi:10.21873/anticanres.11555
Han, S. J., Kwon, S., and Kim, K. S. (2021). Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 21, 152. doi:10.1186/s12935-021-01853-8
Haque, M. R., Wessel, C. R., Leary, D. D., Wang, C., Bhushan, A., and Bishehsari, F. (2022). Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsystems and Nanoeng. 8, 36. doi:10.1038/s41378-022-00370-6
El Harane, S., Zidi, B., El Harane, N., Krause, K. H., Matthes, T., and Preynat-Seauve, O. (2023). Cancer spheroids and organoids as novel tools for research and therapy: state of the art and challenges to guide precision medicine. Cells 12, 1001. doi:10.3390/cells12071001
Hendriks, D., Clevers, H., and Artegiani, B. (2020). CRISPR-cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 27, 705–731. doi:10.1016/j.stem.2020.10.014
Heydari, Z., Devasahayam Arokia Balaya, R., Sarkar, G., and Boardman, L. (2025). The role of organoids in advancing colorectal cancer research: insights and future directions. Cancers 17, 2129. doi:10.3390/cancers17132129
Hua, H., Zou, S., Ma, Z., Guo, W., Fong, C. Y., and Khoo, B. L. (2023). A deformability-based biochip for precise label-free stratification of metastatic subtypes using deep learning. Microsystems and Nanoeng. 9, 120. doi:10.1038/s41378-023-00577-1
Hua, H., Zhou, Y., Li, W., Zhang, J., Deng, Y., and Khoo, B. L. (2024). Microfluidics-based patient-derived disease detection tool for deep learning-assisted precision medicine. Biomicrofluidics 18, 014101. doi:10.1063/5.0172146
Huch, M., Knoblich, J. A., Lutolf, M. P., and Martinez-Arias, A. (2017). The hope and the hype of organoid research. Development 144, 938–941. doi:10.1242/dev.150201
Husseini-Wüsthoff, H., Riethdorf, S., Schneeweiss, A., Trumpp, A., Pantel, K., Wikman, H., et al. (2025). Cluster-based human-in-the-loop strategy for improving machine learning-based circulating tumor cell detection in liquid biopsy. Patterns 6, 101285. doi:10.1016/j.patter.2025.101285
Jacob, F., Salinas, R. D., Zhang, D. Y., Nguyen, P. T., Schnoll, J. G., Wong, S. Z. H., et al. (2020a). A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell 180, 188–204.e22. doi:10.1016/j.cell.2019.11.036
Jacob, F., Ming, G.-l., and Song, H. (2020b). Generation and biobanking of patient-derived glioblastoma organoids and their application in CAR T cell testing. Nat. Protoc. 15, 4000–4033. doi:10.1038/s41596-020-0402-9
Jenkins, R. W., Aref, A. R., Lizotte, P. H., Ivanova, E., Stinson, S., Zhou, C. W., et al. (2018a). Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 8, 196–215. doi:10.1158/2159-8290.cd-17-0833
Jenkins, R. W., Aref, A. R., Lizotte, P. H., Ivanova, E., Stinson, S., Zhou, C. W., et al. (2018b). Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 8, 196–215. doi:10.1158/2159-8290.CD-17-0833
Jiang, W., Han, L., Yang, L., Xu, T., He, J., Peng, R., et al. (2020a). Natural fish Trap-like Nanocage for label-free capture of circulating tumor cells. Adv. Sci. 7, 2002259. doi:10.1002/advs.202002259
Jiang, S., Zhao, H., Zhang, W., Wang, J., Liu, Y., Cao, Y., et al. (2020b). An automated organoid platform with inter-organoid Homogeneity and inter-patient heterogeneity. Cell Rep. Med. 1, 100161. doi:10.1016/j.xcrm.2020.100161
Jiang, S., Deng, T., Cheng, H., Liu, W., Shi, D., Yuan, J., et al. (2023). Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J. Exp. and Clin. Cancer Res. 42, 199. doi:10.1186/s13046-023-02756-4
Jung, D. J., Shin, T. H., Kim, M., Sung, C. O., Jang, S. J., and Jeong, G. S. (2019). A one-stop microfluidic-based lung cancer organoid culture platform for testing drug sensitivity. Lab a Chip 19, 2854–2865. doi:10.1039/c9lc00496c
Kamb, A. (2005). What's wrong with our cancer models? Nat. Rev. Drug Discov. 4, 161–165. doi:10.1038/nrd1635
Kang, J., Lee, J. Y., Lee, S., Kim, D., Lim, J., Jun, H. R., et al. (2023). Establishing patient-derived cancer cell cultures and xenografts in Biliary Tract cancer. Cancer Res. Treat. 55, 219–230. doi:10.4143/crt.2021.1166
Kapałczyńska, M., Kolenda, T., Przybyła, W., Zajączkowska, M., Teresiak, A., Filas, V., et al. (2018). 2D and 3D cell cultures–a comparison of different types of cancer cell cultures. Archives Med. Sci. AMS 14, 910–919. doi:10.5114/aoms.2016.63743
Kapeleris, J., Müller Bark, J., Ranjit, S., Irwin, D., Hartel, G., Warkiani, M. E., et al. (2022). Prognostic value of integrating circulating tumour cells and cell-free DNA in non-small cell lung cancer. Heliyon 8, e09971. doi:10.1016/j.heliyon.2022.e09971
Katano, T., Ootani, A., Mizoshita, T., Tanida, S., Tsukamoto, H., Ozeki, K., et al. (2013). Establishment of a long-term three-dimensional primary culture of mouse glandular stomach epithelial cells within the stem cell niche. Biochem. biophysical Res. Commun. 432, 558–563. doi:10.1016/j.bbrc.2013.02.051
Khoo, B. L., Grenci, G., Lim, Y. B., Lee, S. C., Han, J., and Lim, C. T. (2018). Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 13, 34–58. doi:10.1038/nprot.2017.125
Kim, M., Mun, H., Sung, C. O., Cho, E. J., Jeon, H. J., Chun, S. M., et al. (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10, 3991. doi:10.1038/s41467-019-11867-6
Kodack, D. P., Farago, A. F., Dastur, A., Held, M. A., Dardaei, L., Friboulet, L., et al. (2017). Primary patient-derived cancer cells and their potential for personalized cancer patient care. Cell Rep. 21, 3298–3309. doi:10.1016/j.celrep.2017.11.051
Kondo, J., and Inoue, M. (2019). Application of cancer organoid model for drug screening and personalized therapy. Cells 8, 470. doi:10.3390/cells8050470
Kondo, J., Endo, H., Okuyama, H., Ishikawa, O., Iishi, H., Tsujii, M., et al. (2011). Retaining cell–cell contact enables Prep. Cult. spheroids Compos. pure Prim. cancer cells colorectal cancer. Proceedings of the National Academy of Sciences 108, 6235–6240. doi:10.1073/pnas.1015938108
Kong, J., Luo, Y., Jin, D., An, F., Zhang, W., Liu, L., et al. (2016). A novel microfluidic model can mimic organ-specific metastasis of circulating tumor cells. Oncotarget 7, 78421–78432. doi:10.18632/oncotarget.9382
Kong, J., Lee, H., Kim, D., Han, S. K., Ha, D., Shin, K., et al. (2020). Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 11, 5485. doi:10.1038/s41467-020-19313-8
Kopper, O., de Witte, C. J., Lõhmussaar, K., Valle-Inclan, J. E., Hami, N., Kester, L., et al. (2019). An organoid platform for ovarian cancer captures intra-and interpatient heterogeneity. Nat. Med. 25, 838–849. doi:10.1038/s41591-019-0422-6
Kumari, R., Ouyang, X., Wang, J., Xu, X., Zheng, M., An, X., et al. (2021). Preclinical pharmacology modeling of chimeric antigen receptor T therapies. Curr. Opin. Pharmacol. 61, 49–61. doi:10.1016/j.coph.2021.08.008
Lai, B. F. L., Lu, R. X. Z., Hu, Y., Davenport Huyer, L., Dou, W., Wang, E. Y., et al. (2020). Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv. Funct. Mater. 30, 2000545. doi:10.1002/adfm.202000545
Larkin, J., Chiarion-Sileni, V., Gonzalez, R., Grob, J. J., Cowey, C. L., Lao, C. D., et al. (2015). Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34. doi:10.1056/nejmoa1504030
Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520. doi:10.1056/nejmoa1500596
Le, D. T., Durham, J. N., Smith, K. N., Wang, H., Bartlett, B. R., Aulakh, L. K., et al. (2017). Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413. doi:10.1126/science.aan6733
Ledur, P. F., Onzi, G. R., Zong, H., and Lenz, G. (2017). Culture conditions defining glioblastoma cells behavior: what is the impact for novel discoveries? Oncotarget 8, 69185–69197. doi:10.18632/oncotarget.20193
Lee, S. H., Hu, W., Matulay, J. T., Silva, M. V., Owczarek, T. B., Kim, K., et al. (2018). Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528.e17. doi:10.1016/j.cell.2018.03.017
Lenin, S., Ponthier, E., Scheer, K. G., Yeo, E. C. F., Tea, M. N., Ebert, L. M., et al. (2021). A drug screening pipeline using 2D and 3D patient-derived in vitro models for pre-clinical analysis of therapy response in glioblastoma. Int. J. Mol. Sci. 22, 4322. doi:10.3390/ijms22094322
Li, X., Nadauld, L., Ootani, A., Corney, D. C., Pai, R. K., Gevaert, O., et al. (2014). Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20, 769–777. doi:10.1038/nm.3585
Li, X., Ootani, A., and Kuo, C. (2016). in Gastrointestinal Physiology and diseases: methods and protocols (Springer), 33–40.
Li, W., Zhou, Y., Deng, Y., and Khoo, B. L. (2022). Early predictor tool of disease using label-free liquid biopsy-based platforms for patient-centric healthcare. Cancers 14, 818. doi:10.3390/cancers14030818
Li, W., Zhang, J., Liao, J., Zhu, M., Wang, X., Zhou, X., et al. (2025). A novel Hand-Held Spinning platform with Centrifugal microfluidics for rapid, cost-effective Urinary total protein detection at the Point of care. Anal. Chem. 97, 15049–15059. doi:10.1021/acs.analchem.5c00930
Lim, J. T. C., Kwang, L. G., Ho, N. C. W., Toh, C. C. M., Too, N. S. H., Hooi, L., et al. (2022). Hepatocellular carcinoma organoid co-cultures mimic angiocrine crosstalk to generate inflammatory tumor microenvironment. Biomaterials 284, 121527. doi:10.1016/j.biomaterials.2022.121527
Liu, X., Krawczyk, E., Suprynowicz, F. A., Palechor-Ceron, N., Yuan, H., Dakic, A., et al. (2017). Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 12, 439–451. doi:10.1038/nprot.2016.174
Lone, S. N., Nisar, S., Masoodi, T., Singh, M., Rizwan, A., Hashem, S., et al. (2022). Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. cancer 21, 79. doi:10.1186/s12943-022-01543-7
Lux, A., Bott, H., Malek, N. P., Zengerle, R., Maucher, T., and Hoffmann, J. (2021). Real-time detection of tumor cells during capture on a filter element significantly enhancing detection rate. Biosensors 11, 312. doi:10.3390/bios11090312
Martins, I., Ribeiro, I. P., Jorge, J., Gonçalves, A. C., Sarmento-Ribeiro, A. B., Melo, J. B., et al. (2021). Liquid biopsies: applications for cancer diagnosis and monitoring. Genes 12, 349. doi:10.3390/genes12030349
Maru, Y., and Hippo, Y. (2019). Current status of patient-derived ovarian cancer models. Cells 8, 505. doi:10.3390/cells8050505
Miyoshi, H., Maekawa, H., Kakizaki, F., Yamaura, T., Kawada, K., Sakai, Y., et al. (2018). An improved method for culturing patient-derived colorectal cancer spheroids. Oncotarget 9, 21950–21964. doi:10.18632/oncotarget.25134
Mizutani, T., Kondo, T., Darmanin, S., Tsuda, M., Tanaka, S., Tobiume, M., et al. (2010). A novel FRET-based Biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clin. Cancer Res. 16, 3964–3975. doi:10.1158/1078-0432.Ccr-10-0548
Mollaki, V. (2021). Ethical challenges in organoid Use. Biotech. (Basel) 10, 12. doi:10.3390/biotech10030012
Munsie, M., Hyun, I., and Sugarman, J. (2017). Ethical issues in human organoid and gastruloid research. Development 144, 942–945. doi:10.1242/dev.140111
Neal, J. T., Li, X., Zhu, J., Giangarra, V., Grzeskowiak, C. L., Ju, J., et al. (2018). Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988.e16. doi:10.1016/j.cell.2018.11.021
Niepel, M., Hafner, M., Mills, C. E., Subramanian, K., Williams, E. H., Chung, M., et al. (2019). A multi-center study on the reproducibility of drug-response assays in Mammalian cell lines. Cell Syst. 9, 35–48.e5. doi:10.1016/j.cels.2019.06.005
Nissim, N., Dudaie, M., Barnea, I., and Shaked, N. T. (2021). Real-time stain-free classification of cancer cells and blood cells using interferometric phase microscopy and machine learning. Cytom. Part A 99, 511–523. doi:10.1002/cyto.a.24227
Ooft, S. N., Weeber, F., Dijkstra, K. K., McLean, C. M., Kaing, S., van Werkhoven, E., et al. (2019). Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 11, eaay2574. doi:10.1126/scitranslmed.aay2574
Ott, P. A., Hu, Z., Keskin, D. B., Shukla, S. A., Sun, J., Bozym, D. J., et al. (2017). An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. doi:10.1038/nature22991
Pampaloni, F., Reynaud, E. G., and Stelzer, E. H. (2007). The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. cell Biol. 8, 839–845. doi:10.1038/nrm2236
Park, T., Kim, T. K., Han, Y. D., Kim, K. A., Kim, H., and Kim, H. S. (2023). Development of a deep learning based image processing tool for enhanced organoid analysis. Sci. Rep. 13, 19841. doi:10.1038/s41598-023-46485-2
van Pel, D. M., Harada, K., Song, D., Naus, C. C., and Sin, W. C. (2018). Modelling glioma invasion using 3D bioprinting and scaffold-free 3D culture. J. Cell Commun. Signal. 12, 723–730. doi:10.1007/s12079-018-0469-z
Perez, J. M., Duda, J. M., Ryu, J., Shetty, M., Mehta, S., Jagtap, P. D., et al. (2025). Investigating proteogenomic divergence in patient-derived xenograft models of ovarian cancer. Sci. Rep. 15, 813. doi:10.1038/s41598-024-84874-3
Perova, Z., Martinez, M., Mandloi, T., Gomez, F. L., Halmagyi, C., Follette, A., et al. (2025). “CancerModels,” in Org–an open global research platform for patient-derived cancer models. Nucleic acids research, 51 (1), 1360–1366. doi:10.1093/nar/gkac1021
Pinho, D., Santos, D., Vila, A., and Carvalho, S. (2021). Establishment of colorectal cancer organoids in microfluidic-based system. Micromachines 12, 497. doi:10.3390/mi12050497
Ponti, D., Costa, A., Zaffaroni, N., Pratesi, G., Petrangolini, G., Coradini, D., et al. (2005). Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511. doi:10.1158/0008-5472.can-05-0626
Ponz-Sarvise, M., Corbo, V., Tiriac, H., Engle, D. D., Frese, K. K., Oni, T. E., et al. (2019). Identification of resistance Pathways specific to malignancy using organoid models of pancreatic cancer. Clin. Cancer Res. 25, 6742–6755. doi:10.1158/1078-0432.CCR-19-1398
Qiu, Z., Li, H., Zhang, Z., Zhu, Z., He, S., Wang, X., et al. (2019). A pharmacogenomic landscape in human liver cancers. Cancer Cell 36, 179–193.e11. doi:10.1016/j.ccell.2019.07.001
Rajan, R. G., Fernandez-Vega, V., Sperry, J., Nakashima, J., Do, L. H., Andrews, W., et al. (2023). In vitro and in vivo drug-response profiling using patient-derived high-grade Glioma. Cancers 15, 3289. doi:10.3390/cancers15133289
Rejuan, R., Aulisa, E., Li, W., Thompson, T., Kumar, S., Canic, S., et al. (2025). Validation of a Microfluidic Device Prototype for Cancer Detection and Identification: Circulating Tumor Cells Classification Based on Cell Trajectory Analysis Leveraging Cell-Based Modeling and Machine Learning. Int J Numer Method Biomed Eng 41 (4), e70037. doi:10.1002/cnm.70037
Ricci-Vitiani, L., Lombardi, D. G., Pilozzi, E., Biffoni, M., Todaro, M., Peschle, C., et al. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115. doi:10.1038/nature05384
Roerink, S. F., Sasaki, N., Lee-Six, H., Young, M. D., Alexandrov, L. B., Behjati, S., et al. (2018). Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462. doi:10.1038/s41586-018-0024-3
Rubio-Viqueira, B., and Hidalgo, M. (2009). Direct in vivo xenograft tumor model for predicting chemotherapeutic drug response in cancer patients. Clin. Pharmacol. Ther. 85, 217–221. doi:10.1038/clpt.2008.200
Sachs, N., de Ligt, J., Kopper, O., Gogola, E., Bounova, G., Weeber, F., et al. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10. doi:10.1016/j.cell.2017.11.010
Sachs, N., Papaspyropoulos, A., Zomer-van Ommen, D. D., Heo, I., Böttinger, L., Klay, D., et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300. doi:10.15252/embj.2018100300
Sahin, U., Derhovanessian, E., Miller, M., Kloke, B. P., Simon, P., Löwer, M., et al. (2017). Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226. doi:10.1038/nature23003
Schmid, P., Adams, S., Rugo, H. S., Schneeweiss, A., Barrios, C. H., Iwata, H., et al. (2018). Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121. doi:10.1056/nejmoa1809615
Schnalzger, T. E., de Groot, M. H., Zhang, C., Mosa, M. H., Michels, B. E., Röder, J., et al. (2019). 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 38, e100928. doi:10.15252/embj.2018100928
Schuster, B., Junkin, M., Kashaf, S. S., Romero-Calvo, I., Kirby, K., Matthews, J., et al. (2020). Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 11, 5271. doi:10.1038/s41467-020-19058-4
Schutgens, F., Rookmaaker, M. B., Margaritis, T., Rios, A., Ammerlaan, C., Jansen, J., et al. (2019). Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313. doi:10.1038/s41587-019-0048-8
Seidel, S., Garvalov, B. K., and Acker, T. (2015). Isolation and culture of primary glioblastoma cells from human tumor specimens. Methods Mol. Biol. 1235, 263–275. doi:10.1007/978-1-4939-1785-3_19
Sharpe, B. P., Nazlamova, L. A., Tse, C., Johnston, D. A., Thomas, J., Blyth, R., et al. (2024). Patient-derived tumor organoid and fibroblast assembloid models for interrogation of the tumor microenvironment in esophageal adenocarcinoma. Cell Rep. Methods 4, 100909. doi:10.1016/j.crmeth.2024.100909
Shen, C., Rawal, S., Brown, R., Zhou, H., Agarwal, A., Watson, M. A., et al. (2023). Automatic detection of circulating tumor cells and cancer associated fibroblasts using deep learning. Sci. Rep. 13, 5708. doi:10.1038/s41598-023-32955-0
Shickh, S., Oldfield, L. E., Clausen, M., Mighton, C., Sebastian, A., Calvo, A., et al. (2022). “Game changer”: health professionals’ views on the clinical utility of circulating tumor DNA testing in hereditary cancer syndrome management. Oncol. 27, e393–e401. doi:10.1093/oncolo/oyac039
Shirure, V. S., Bi, Y., Curtis, M. B., Lezia, A., Goedegebuure, M. M., Goedegebuure, S. P., et al. (2018). Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab a Chip 18, 3687–3702. doi:10.1039/c8lc00596f
Silvestri, V. L., Henriet, E., Linville, R. M., Wong, A. D., Searson, P. C., and Ewald, A. J. (2020). A tissue-engineered 3D Microvessel model reveals the dynamics of Mosaic vessel formation in breast cancer. Cancer Res. 80, 4288–4301. doi:10.1158/0008-5472.Can-19-1564
Simpson-Abelson, M. R., Sonnenberg, G. F., Takita, H., Yokota, S. J., Conway, T. F., Kelleher, R. J., et al. (2008). Long-term engraftment and expansion of tumor-derived Memory T cells following the Implantation of non-Disrupted Pieces of human lung tumor into NOD-scid IL2Rγnull Mice. J. Immunol. 180, 7009–7018. doi:10.4049/jimmunol.180.10.7009
Skardal, A., Sarker, S. F., Crabbé, A., Nickerson, C. A., and Prestwich, G. D. (2010). The generation of 3-D tissue models based on hyaluronan hydrogel-coated microcarriers within a rotating wall vessel bioreactor. Biomaterials 31, 8426–8435. doi:10.1016/j.biomaterials.2010.07.047
Spiller, E. R., Ung, N., Kim, S., Patsch, K., Lau, R., Strelez, C., et al. (2021). Imaging-based machine learning analysis of patient-derived tumor organoid drug response. Front. Oncol. 11, 771173–772021. doi:10.3389/fonc.2021.771173
Streller, M., Michlíková, S., Ciecior, W., Lönnecke, K., Kunz-Schughart, L. A., Lange, S., et al. (2025). Image segmentation of treated and untreated tumor spheroids by fully convolutional networks. GigaScience 14, giaf027. doi:10.1093/gigascience/giaf027
Stüve, P., Nerb, B., Harrer, S., Wuttke, M., Feuerer, M., Junger, H., et al. (2023). Analysis of organoid and immune cell co-cultures by machine learning-empowered image cytometry. Front. Med. 10, 1274482–2023. doi:10.3389/fmed.2023.1274482
Subtil, B., Iyer, K. K., Poel, D., Bakkerus, L., Gorris, M. A. J., Escalona, J. C., et al. (2023). Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front. Immunol. 14, 1105244. doi:10.3389/fimmu.2023.1105244
Tan, T., Mouradov, D., Lee, M., Gard, G., Hirokawa, Y., Li, S., et al. (2023). Unified framework for patient-derived, tumor-organoid-based predictive testing of standard-of-care therapies in metastatic colorectal cancer. Cell Rep. Med. 4. doi:10.1016/j.xcrm.2023.101335
Thakuri, P. S., Gupta, M., Plaster, M., and Tavana, H. (2019). Quantitative size-based analysis of tumor spheroids and responses to therapeutics. ASSAY Drug Dev. Technol. 17, 140–149. doi:10.1089/adt.2018.895
Tsai, S., McOlash, L., Palen, K., Johnson, B., Duris, C., Yang, Q., et al. (2018). Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC cancer 18, 335–13. doi:10.1186/s12885-018-4238-4
Tung, Y.-C., Hsiao, A. Y., Allen, S. G., Torisawa, Ys, Ho, M., and Takayama, S. (2011). High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 136, 473–478. doi:10.1039/c0an00609b
Tuveson, D., and Clevers, H. (2019). Cancer modeling meets human organoid technology. Science 364, 952–955. doi:10.1126/science.aaw6985
van de Wetering, M., Francies, H., Francis, J., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi:10.1016/j.cell.2015.03.053
Vermesh, O., Aalipour, A., Ge, T. J., Saenz, Y., Guo, Y., Alam, I. S., et al. (2018). An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nat. Biomed. Eng. 2, 696–705. doi:10.1038/s41551-018-0257-3
Vlachogiannis, G., Hedayat, S., Vatsiou, A., Jamin, Y., Fernández-Mateos, J., Khan, K., et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926. doi:10.1126/science.aao2774
Wang, R., Mao, Y., Wang, W., Zhou, X., Wang, W., Gao, S., et al. (2022). Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. 23, 106. doi:10.1186/s13059-022-02673-3
Weiswald, L., Richon, S., Validire, P., Briffod, M., Lai-Kuen, R., Cordelières, F. P., et al. (2009). Newly characterised ex vivo colospheres as a three-dimensional colon cancer cell model of tumour aggressiveness. Br. J. cancer 101, 473–482. doi:10.1038/sj.bjc.6605173
De Witt Hamer, P. C., Van Tilborg, A. A. G., Eijk, P. P., Sminia, P., Troost, D., Van Noorden, C. J. F., et al. (2008). The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene 27, 2091–2096. doi:10.1038/sj.onc.1210850
Wu, R., Forget, M. A., Chacon, J., Bernatchez, C., Haymaker, C., Chen, J. Q., et al. (2012). Adoptive T-cell therapy using autologous tumor-infiltrating lymphocytes for metastatic melanoma: current status and future outlook. Cancer J. 18, 160–175. doi:10.1097/PPO.0b013e31824d4465
Xiang, D., He, A., Zhou, R., Wang, Y., Xiao, X., Gong, T., et al. (2024). Building consensus on the application of organoid-based drug sensitivity testing in cancer precision medicine and drug development. Theranostics 14, 3300–3316. doi:10.7150/thno.96027
Xie, G., Dong, H., Liang, Y., Ham, J. D., Rizwan, R., and Chen, J. (2020). CAR-NK cells: a promising cellular immunotherapy for cancer. EBioMedicine 59, 102975. doi:10.1016/j.ebiom.2020.102975
Yan, H. H., Siu, H. C., Law, S., Ho, S. L., Yue, S. S., Tsui, W. Y., et al. (2018). A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell stem cell 23, 882–897.e11. doi:10.1016/j.stem.2018.09.016
Yu, L., Li, Z., Mei, H., Li, W., Chen, D., Liu, L., et al. (2021). Patient-derived organoids of bladder cancer recapitulate antigen expression profiles and serve as a personal evaluation model for CAR-T cells in vitro. Clin. and Transl. Immunol. 10, e1248. doi:10.1002/cti2.1248
Yuki, K., Cheng, N., Nakano, M., and Kuo, C. J. (2020). Organoid models of tumor Immunology. Trends Immunol. 41, 652–664. doi:10.1016/j.it.2020.06.010
Zhang, Y. S., Aleman, J., Shin, S. R., Kilic, T., Kim, D., Mousavi Shaegh, S. A., et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. 114, E2293–E2302. doi:10.1073/pnas.1612906114
Zhang, Z., Wang, H., Ding, Q., Xing, Y., Xu, Z., Lu, C., et al. (2018). Establishment of patient-derived tumor spheroids for non-small cell lung cancer. PloS one 13, e0194016. doi:10.1371/journal.pone.0194016
Zhang, J., Fu, Y., Fong, C. Y., Hua, H., Li, W., and Khoo, B. L. (2025a). Advancements in microfluidic technology for rapid bacterial detection and inflammation-driven diseases. Lab a Chip 25, 3348–3373. doi:10.1039/d4lc00795f
Zhang, W., Wu, C., Huang, H., Bleu, P., Zambare, W., Alvarez, J., et al. (2025b). Enhancing chemotherapy response prediction via matched colorectal tumor-organoid gene expression analysis and network-based biomarker selection. Transl. Oncol. 52, 102238. doi:10.1016/j.tranon.2024.102238
Keywords: patient-derived cancer cells (PDCCs), personalized medicine, cancer therapeutics, drug discovery, 3D cell culture, tumor heterogeneity, high-throughput screening, organoids
Citation: Fu Y, Khoo BL and Lim CT (2025) Advancements and challenges in culturing patient-derived cancer cells for personalized therapeutics. Front. Lab Chip Technol. 4:1663420. doi: 10.3389/frlct.2025.1663420
Received: 10 July 2025; Accepted: 18 August 2025;
Published: 08 September 2025.
Edited by:
Duc-Huy Tran Nguyen, University of Wisconsin-Madison, United StatesReviewed by:
Surjendu Maity, Duke University, United StatesCopyright © 2025 Fu, Khoo and Lim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bee Luan Khoo, YmxraG9vQGNpdHl1LmVkdS5oaw==; Chwee Teck Lim, Y3RsaW1AbnVzLmVkdS5zZw==
 Yatian Fu1,2
Yatian Fu1,2 Bee Luan Khoo
Bee Luan Khoo Chwee Teck Lim
Chwee Teck Lim