- 1Department of Dermatology, UMass Chan Medical School, Worcester, MA, United States
- 2Tulane University School of Medicine, New Orleans, LA, United States
- 3UMass Chan Lamar Soutter Library, Worcester, MA, United States
- 4Department of Dermatology, Howard University Hospital, Washington, DC, United States
- 5Department of Comparative Pathobiology, Tufts University, Cummings School of Veterinary Medicine, North Grafton, MA, United States
Cutaneous lupus erythematosus (CLE) is a group of skin disorders where the immune system attacks skin cells. CLE can affect people who have systemic lupus erythematosus, or can occur independently. In prior studies, CXCL16 and its primary receptor, CXCR6, have been shown to be elevated at the RNA or protein level in different organs that are affected by lupus. In this systematic review, we sought to understand whether CXCR6 and its ligand CXCL16 could serve as biomarkers for lupus skin or other organ involvement. Our search strategy and protocol are registered on Prospero under # CRD42024583076. CXCL16 was shown to be a biomarker of lupus nephritis and disease activity in both urine and serum samples in multiple studies. CXCL16 was also elevated in cerebrospinal fluid in neuropsychiatric lupus patients as well as other autoimmune brain conditions. Last, we queried publicly available datasets and our own datasets to evaluate expression of CXCR6 and CXCL16 in lupus skin. CXCR6 but not CXCL16 was enriched in lupus skin across multiple datasets and model organisms. Taken together, our study corroborates the CXCR6 chemokine family as a potential biomarker of lupus organ involvement.
Introduction
Cutaneous lupus erythematosus (CLE) is a spectrum of autoimmune skin disorders that may occur independently or as part of systemic lupus erythematosus (SLE). CLE has manifestations of autoimmunity with complex pathophysiology involving dysregulated inflammatory signaling and aberrant immune cell trafficking. Chemokines play a critical role in organizing immune cell migration to sites of inflammation, with emerging evidence suggesting particular importance for the CXCR6-CXCL16 axis in immune-mediated skin diseases. This chemokine pathway may represent a fundamental mechanism underlying both the initiation and persistence of inflammatory skin lesions in lupus. Recent cross-species investigations have revealed intriguing patterns of conservation, suggesting evolutionary significance and potential therapeutic relevance of this pathway (1). Our research has focused on elucidating the specific contributions of the CXCR6-CXCL16 interaction in lupus pathogenesis, with particular attention to its role in tissue-resident memory T cell populations.
In juvenile SLE patients, the chemokine CXCL16, which is a ligand for CXCR6, was elevated in recent studies. CXCL16's expression in these patients was associated with clinical features such as alopecia, malar rash, and nephritis (2). CXCL16 is produced by keratinocytes (2, 3), and its expression is induced by TLR7 activation (4) which is a key signaling pathway implicated in both the CLE murine model used in our studies (5) and in human lupus (6, 7). It is also upregulated by ultraviolet (UV) light (3, 4), which suggests a potential role in photosensitivity, a hallmark of cutaneous lupus. CXCL16 has also been associated with Th1:Th2 imbalances in patient with autoimmune thrombocytopenia (8), a pattern that matches our own observations of T helper cell plasticity in murine CLE (9). Two independent studies have demonstrated that CXCR6 is expressed on CD8+ tissue-resident memory T cells (Trm) within the tumor microenvironment (10, 11), highlighting a potential role for the CXCL16-CXCR6 axis in Trm cell biology.
Building on these findings, we performed a systematic review of CXCL16-CXCR6 as potential biomarkers of lupus, which we present here. CXCL16 appears to be a promising biomarker of lupus nephritis. We also queried publicly available mouse, human, and canine CLE datasets (4–6) and found that CXCL16 upregulation in cutaneous lupus lesions is conserved across all three species. Interestingly, CXCL16 was lower in established CLE patients assessed by tape stripping, which we hypothesize may be due to sun avoidance and/or current treatment status. This cross-species conservation, combined with the role CXCL16 plays in immune signaling and Trm cell maintenance, identifies it as a promising biomarker and potential therapeutic target in lupus.
Methods
Systematic review
Search strategy
Our search strategy and protocol are registered on Prospero under # CRD42024583076. A comprehensive literature search was conducted by a medical librarian on August 22, 2023, using the following bibliographic databases from inception: Ovid MEDLINE® (ALL-1946 to Present); Cochrane Library (Wiley); PubMed (NIH) and Scopus (Elsevier). We used controlled vocabulary for both *lupus and *CXCL16/CXCR6. The entire search strategy is available in Supplementary Table S1.
Eligibility criteria
Eligibility criteria were as follows: 1. studies involving lupus erythematosus; 2. studies measuring CXCL16 and/or CXCR6 as biomarkers. Exclusion criteria included: 1. studies of diseases that were not lupus; 2. studies that did not include measurements of CXCR6 and/or CXCL16); and 3. studies in a language other than English. We allowed any article types that presented primary data to be included, including randomized control trials, non-randomized experimental studies, case-control studies, cohort studies, cross-sectional studies, comparative studies, systematic reviews, observational studies, prevalence studies, open-label trials, in vitro experiments, and ex vivo studies. We also included both human and murine studies.
Exclusion criteria for full-text review included the following: 1. duplicate article; 2. did not study lupus; 3. did not measure CXCR6/CXCL16; 4. full article not available in all libraries for interlibrary loan request; 5. full text not in English. Review articles, editorials, and text/opinion pieces were also excluded in the full-text review with the rationale that they would have summarized the same primary literature studies.
Methodological quality assessment
We assessed the risk of bias using the ROBINS-I tool with the following domains: bias due to missing data, bias due to deviations from intended interventions/studies, bias in selection of participants, bias in classes of interventions, bias in measurement of outcomes, bias in selection of reported results, risk of bias due to confounding.
Data extraction
Our data extraction template included the following entries for reviewers to complete: 1. study ID; 2. title; 3. country in which the study was conducted; 4. notes; 5. aim or objective of the study; 6. study design (e.g., randomized clinical trial, case report, etc.); 7. publication date; 8. study funding sources; 9. possible conflicts of interests for study authors; 10. participants—population description (mice, humans, cell lines, etc.); 11. inclusion criteria; 12. exclusion criteria; 13. the total number of participants; 14. total number of experimental repeats; 15. plants/compounds/extracts tested; 16. active ingredients tested (if known); 17. controls used (e.g., vehicle, positive controls, negative controls, etc.); 18. primary outcome—cutaneous findings—Was the treatment efficacious (yes/no)? Statistically significant (yes/no)? 19. primary outcome—inflammation—Was the treatment efficacious (yes/no)? Statistically significant (yes/no)? 20. Primary outcome—other organ systems (e.g., kidney, brain, etc.). Was the treatment efficacious (yes/no)? Statistically significant (yes/no)? Forest plots of study outcomes were generated using GraphPad Prism software.
Primary data analysis
Patient samples
We queried our dataset of archival skin biopsies from CLE patients seen at Howard University Hospital for CXCR6/CXCL16 gene expression (1). Briefly, samples were obtained through an archival tissue IRB protocol with Memorandum of Understanding (UMass Chan & Howard University IRBs). Disease status was confirmed by chart review by a board-certified dermatologist (CF).
Microarray and statistical analysis
We used nSolver software to generate normalized gene expression counts from the NanoString myeloid v2 probeset. Samples with QC flags were excluded (1 healthy sample). Normalized gene counts were analyzed with student's t test and Receiver operator characteristic (ROC) curve functions using GraphPad Prism software v 10.
Dataset reanalysis and statistics
We queried publicly available datasets for CXCR6/CXCL16 expression using Geo2R software. These included human data from Scholtissek et al. GSE95474 (13), Yildiz-Altay et al. (1), and Seremet, Domizio et al. GSE193068 (14, 15); mouse data from Mande et al. (6); and canine data from Garelli & Wong and Amudzi et al. (16, 17). Normalized count values were plotted in GraphPad Prism software v 10. Each dataset was assessed for normality before selection of statistical tests, with normally distributed data employing t tests and non-normally distributed data employing Mann–Whitney U tests. Directional hypothesis testing for one-tailed results was to test the hypothesis that CXCR6/CXCL16 were higher in lupus than controls, which was based on our observations from the Balb/c mouse model and published studies.
Results
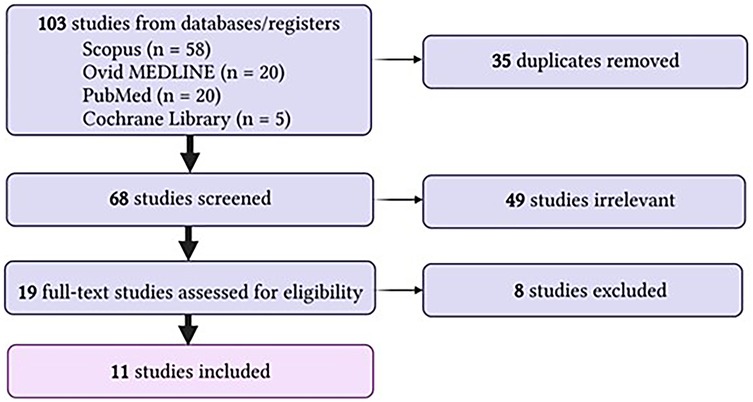
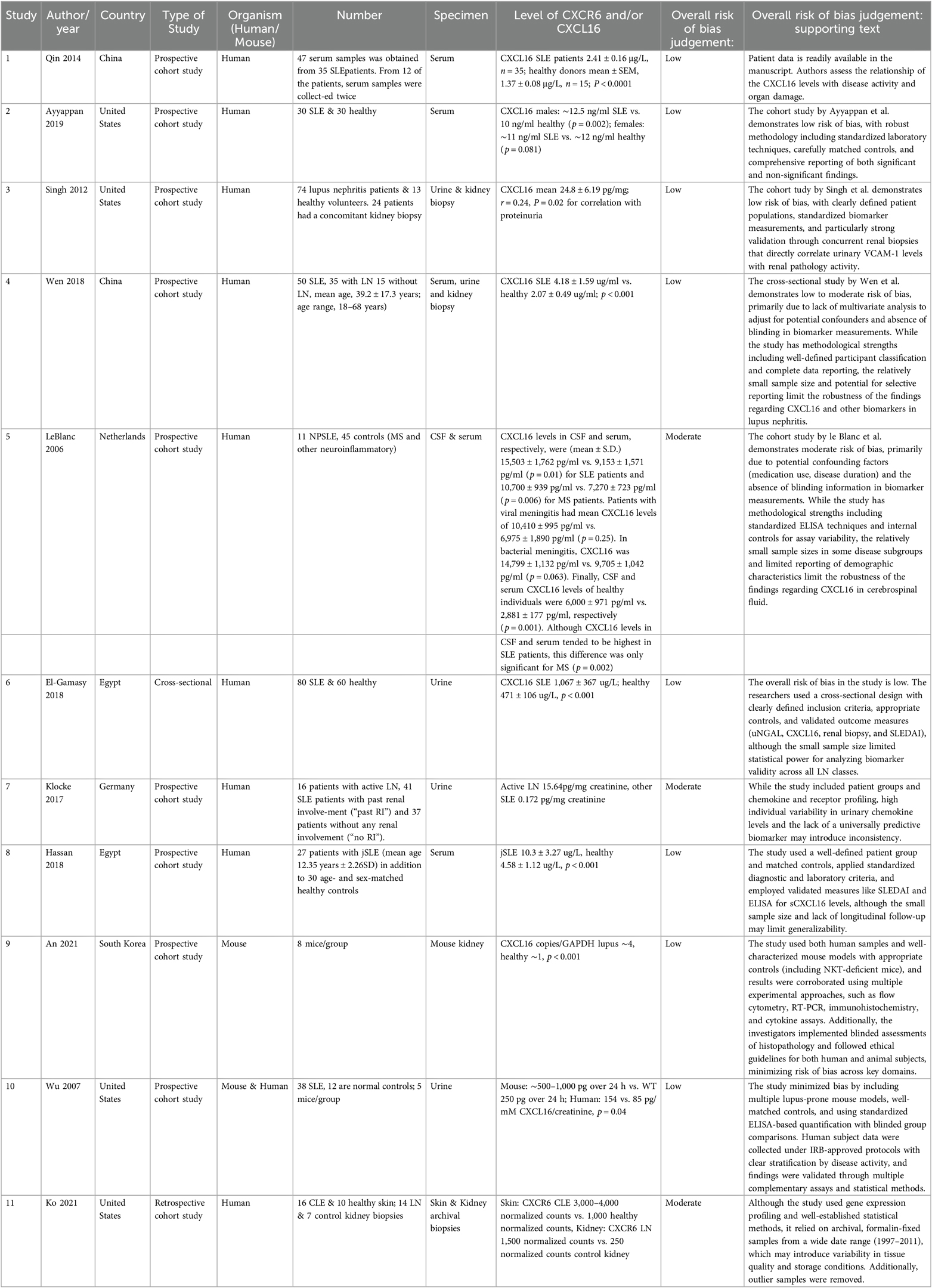
Following our search strategy, we identified 103 eligible studies from SCOPUS, Ovid MEDLINE, PubMed and Cochrane library. 35 duplicates were removed, leaving 68 studies for screening. Of these, 49 were irrelevant. 19 full-text studies were assessed, 8 of which were excluded. Reasons for exclusion included wrong outcomes (n = 1), wrong comparator (n = 2), wrong study design (n = 3) and review article with no primary data (n = 1). This search is also summarized in Figure 1. Of the 11 studies included, most used a prospective cohort design on human biological samples including serum, urine, and kidney biopsies. All of these studies noted significant increases in CXCL16 or its receptor CXCR6 (summarized in Table 1).
Systematic review results
Wu et al. (18) examined urinary biomarkers in lupus nephritis using both murine models and human patient samples. Researchers analyzed the urine of lupus-prone mice and SLE patients for VCAM-1, P-selectin, TNFR-1, and CXCL16, finding significantly elevated levels in those with active nephritis. These markers correlated with disease severity and were primarily derived from the kidney rather than the bloodstream. Immunohistochemical analysis confirmed their presence in inflamed renal tissues, particularly in endothelial and tubular cells. In human SLE patients, urinary levels of these biomarkers distinguished those with nephritis from non-nephritic individuals with high sensitivity and specificity. VCAM-1 and P-selectin, both involved in inflammation, were particularly enriched in lupus kidneys and urine, while TNFR-1 and CXCL16 were newly identified as urinary biomarkers. The findings suggest these molecules play roles in disease pathogenesis while serving as potential noninvasive diagnostic tools. Their detection in urine provides a more accessible way to monitor disease progression without the need for invasive kidney biopsies.
Qin et al. (19) investigated serum soluble CXCL16 (sCXCL16) levels in patients with systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), and compared these levels to healthy controls. A total of 47 serum samples were collected from 35 SLE patients, with 12 patients providing samples before and after treatment with glucocorticoids and immunosuppressants. The results showed that serum sCXCL16 levels were significantly higher in SLE patients compared to RA patients and healthy controls. The study also found that sCXCL16 levels correlated with disease activity in SLE, as indicated by SLE Disease Activity Index (SLEDAI) scores, and were higher in patients with active disease flare-ups. Furthermore, the levels of sCXCL16 were linked to organ damage, specifically in patients with lupus nephritis and cutaneous involvement, suggesting that elevated sCXCL16 could serve as a marker for these conditions. Following treatment, sCXCL16 levels decreased, reflecting improvements in disease activity, although there was no significant change in chronic organ damage. The findings also suggested that sCXCL16 might help monitor disease progression and guide treatment decisions in SLE patients. The study concluded that sCXCL16 could be a useful biomarker for assessing disease activity and organ involvement, particularly in the skin and kidneys, in SLE.
Ayyappan et al. (20) analyzed changes in various immune markers in patients with systemic lupus erythematosus (SLE). The study included 30 SLE patients and 30 age- and gender-matched healthy controls, with comprehensive exclusion criteria ensuring the exclusion of those with recent infections or steroid use. It found that soluble CD14 (sCD14) levels were significantly elevated in SLE patients, with a positive correlation between sCD14 levels and disease activity, indicating a potential role for endotoxemia in SLE progression. EndoCAb IgM levels were reduced in SLE patients, suggesting that lower endotoxin neutralization might contribute to elevated sCD14. Additionally, increased levels of lysozyme were found in SLE patients, and lysozyme correlated positively with sCD14, LBP, and FABP2, suggesting a shared stimulus for these immune factors. CXCL16 levels were also elevated in SLE patients and correlated with sCD14, supporting the idea of bacterial exposure influencing these immune responses. The study noted no significant changes in galectin-3 levels in SLE patients, though a positive correlation with sCD14 and lysozyme was observed. Immunosuppressive treatments were found to reduce EndoCAb IgM levels but did not affect the ratios of EndoCAbs to total immunoglobulins.
Singh et al. (21) investigated the potential of urinary biomarkers, specifically VCAM-1, MCP-1, and CXCL16, in monitoring lupus nephritis (LN) activity and chronicity. The study involved 74 LN patients, 13 healthy volunteers, and 22 disease controls, with a focus on urinary biomarker levels before initiating any new immunosuppressive therapy. It was found that urinary VCAM-1 and MCP-1 levels were significantly elevated in patients with active renal disease, correlating strongly with disease activity scores and 24-h proteinuria. VCAM-1, in particular, was a strong discriminator of active renal disease and was most elevated in patients with class IV LN, which has a poor prognosis. The study also showed that VCAM-1 levels correlated positively with the acute inflammation index (AI), a measure of kidney inflammation, while negatively correlating with the chronicity index (CI), indicating its role in reflecting inflammation and potential reversibility of kidney damage. Interestingly, while VCAM-1 was not specific to LN and was elevated in other types of nephritis, it appeared to be a useful marker for renal injury. The results suggested that urinary VCAM-1 could serve as a valuable marker for monitoring disease activity and potentially predicting kidney damage in LN.
Wen et al. (22) investigated the potential role of IFN-γ, CXCL16, and suPAR as biomarkers in systemic lupus erythematosus (SLE) and lupus nephritis (LN). Fifty SLE patients were divided into LN and non-LN groups, alongside 15 healthy controls, and their serum, urine, and renal tissue samples were analyzed. The results showed that serum and urine levels of IFN-γ, CXCL16, and suPAR were significantly elevated in SLE patients, particularly in those with LN, and correlated with disease activity indicators such as SLEDAI scores and 24-h proteinuria. Immunohistochemical analysis revealed increased expression of these biomarkers in renal tissues of LN patients, further linking them to kidney inflammation and damage. Strong correlations were observed between serum and urine levels of these markers, suggesting their potential as non-invasive indicators of renal involvement in SLE. Additionally, suPAR showed the strongest association with disease severity, particularly proteinuria, highlighting its potential as a key predictor of renal damage. These findings suggest that IFN-γ, CXCL16, and suPAR interact in the pathogenesis of SLE, likely contributing to local inflammation and systemic disease progression.
le Blanc et al. (23) studied the role of chemokines (CK) in T cell migration into the central nervous system (CNS) during inflammatory conditions. The researchers found that CXCL16 levels were significantly higher in cerebrospinal fluid (CSF) compared to serum, whereas CCL17 and CCL18 were absent in CSF. Elevated CXCL16 was observed in inflammatory conditions such as multiple sclerosis (MS), systemic lupus erythematosus (SLE), and meningitis. Microglia and astrocytes are likely responsible for CXCL16 production, and its increased presence may facilitate immune cell interactions and inflammation. The study also confirmed that CCL18 was elevated in SLE serum but absent in CSF, supporting its role in attracting naïve T cells outside the CNS. The findings suggest that CXCL16 contributes to neuroinflammation by promoting the influx of activated T cells, particularly when the blood-brain barrier (BBB) is compromised. The researchers highlight that CXCL16's role is not limited to neuroinflammatory diseases, as similar mechanisms were observed in rheumatoid arthritis.
El-Gamasy et al. (24) aimed to evaluate urinary neutrophil gelatinase-associated lipocalin (uNGAL) and CXCL16 levels in children and adolescents with systemic lupus erythematosus (SLE) and their role in diagnosing lupus nephritis (LN). A total of 80 SLE patients and 60 age- and sex-matched healthy controls were included, with disease activity assessed using the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) and renal biopsies performed at initial diagnosis. Urinary uNGAL and CXCL16 levels were measured using enzyme-linked immunosorbent assays (ELISA), and statistical analyses were conducted to assess their diagnostic accuracy. The study found significantly higher levels of uNGAL (8.9 ± 3.56 ng/dl) and urinary CXCL16 (1,067 ± 367 µg/L) in SLE patients compared to controls (P < 0.05). uNGAL showed a stronger correlation with 24-h urinary protein levels (r = 0.732) and SLEDAI scores (r = 0.359) than CXCL16, indicating its higher diagnostic value. The sensitivity and specificity of uNGAL in predicting LN were 95% and 90%, respectively, making it a more reliable biomarker than CXCL16. Both biomarkers were found to increase with LN severity, with the highest levels observed in Class IV LN, which is associated with worse outcomes. The findings support the use of uNGAL as a potential early predictor of LN progression, aiding in disease monitoring and treatment decisions. However, the study's limitations include the small sample size and the inability to establish specific cutoff values for different LN classes. Overall, uNGAL and CXCL16 are valuable indicators of LN activity, with uNGAL demonstrating superior diagnostic accuracy.
Klocke et al. (25) investigated the role of chemokines and their receptors in the recruitment of inflammatory cells into the kidneys of lupus nephritis (LN) patients and their potential as biomarkers. The study demonstrated that T cells and macrophages infiltrate the renal interstitium and that specific chemokines such as CCL2, CCL3-5, CXCL10, and CXCL16 play a key role in disease activity. Urine analysis revealed distinct chemokine profiles in active LN patients, with significant differences from serum levels, highlighting their potential as non-invasive biomarkers. Chemokines correlated with disease severity and urinary T-cell and macrophage counts, particularly CXCR3 ligands like CXCL10, which showed strong correlations with CD4+ T-cell levels. The study also identified that CD4+ T cells exhibit diverse chemokine receptor expressions (CCR5, CXCR3, CCR4, CCR6, CXCR6), whereas CD8+ T cells mainly rely on CCR1, CCR5, and CXCR3 for recruitment. Macrophage recruitment appeared to be driven by the CCR2 axis, with elevated CCL2, CCL7, and CCL8 levels in active LN patients. Individual variability in chemokine expression was observed, suggesting that different LN patients may have distinct chemokine recruitment pathways. Urinary chemokine concentrations did not correlate with serum levels, supporting their kidney-specific role. The study provides new insights into leukocyte migration in LN, aligning with findings from rodent models and previous human studies. These findings highlight potential therapeutic targets and reinforce urinary chemokine profiling as a promising diagnostic tool for LN.
Hassan et al. (2) investigated serum sCXCL16 levels in juvenile systemic lupus erythematosus (jSLE) patients and its correlation with disease activity, particularly lupus nephritis. It included 27 Egyptian children with jSLE diagnosed using ACR criteria and 30 healthy, age- and sex-matched controls. Clinical symptoms, laboratory tests, and renal biopsies were performed to assess disease severity, with disease activity classified using the SLEDAI score. Serum sCXCL16 levels were significantly higher in jSLE patients than in controls and correlated positively with lupus nephritis severity, SLEDAI score, anti-dsDNA levels, ESR, and blood pressure. A strong association was observed between sCXCL16 levels and specific cutaneous manifestations, particularly alopecia and malar rash. Patients with more advanced lupus nephritis (class III and IV) had significantly elevated sCXCL16 levels, suggesting its role in disease progression. The study found a negative correlation between sCXCL16 levels and C3, highlighting its potential as a biomarker for disease activity. The findings align with previous studies on the inflammatory role of CXCL16 in lupus nephritis and its association with renal damage. The study suggests that sCXCL16 could serve as a non-invasive marker for monitoring renal involvement in jSLE, reducing the need for repeated renal biopsies. Future therapeutic strategies targeting sCXCL16 could be explored to manage active disease flares and prevent renal complications in jSLE patients.
An et al. (26) studied the role of IL-17-secreting invariant natural killer T (iNKT) cells in lupus nephritis (LN). They found that patients with LN had significantly higher proportions of IL-17-producing NKT cells in peripheral blood mononuclear cells (PBMCs) and increased IL-17 expression in kidney tissues compared to control groups. Using an experimental autoimmune lupus nephritis (eALN) mouse model, researchers observed that NKT cell-deficient mice had less severe disease progression, lower proteinuria, and reduced renal damage than wild-type mice. The absence of NKT cells led to decreased glomerular immune cell infiltration, lower IL-17 and proinflammatory cytokine expression, and increased levels of IL-27, which inhibits Th17 differentiation. Experiments demonstrated that NKT cells contribute to systemic inflammation and IL-17 production, promoting mesangial cell proliferation and disease progression. in vitro studies confirmed that activated NKT cells induced IL-17 secretion, which was diminished upon IL-17 blockade, reducing mesangial cell proliferation. NK1.1+ NKT cells were identified as primary regulators of IL-17 production, with their activation upregulating IL-6, STAT3, and CXCL16 expression, further linking them to LN pathogenesis. A pristane-induced lupus model reinforced these findings, showing that disease severity correlated with IL-17 and NKT cell activation. Blocking IL-17 or depleting NKT cells reduced inflammation and kidney damage, suggesting that IL-17-secreting iNKT cells are central to LN progression. These findings highlight the potential therapeutic targeting of the IL-17/NKT cell axis to mitigate LN severity.
Ko et al. (27) examined gene expression profiles in cutaneous lupus and lupus nephritis to understand their distinct clinical and histological features. Researchers analyzed 16 cutaneous lupus samples (SCLE and CCLE) and 10 normal skin samples, revealing that while lupus skin samples are distinct from normal skin, SCLE and CCLE share highly similar gene expression profiles. Despite clear clinical differences, broad gene expression analysis does not explain these distinctions, suggesting that subtle genetic variations or tissue-specific inflammation may contribute. The study identified a strong type I interferon (IFN) signature in cutaneous lupus, with upregulated genes such as IFI6, IFI44l, and CXCL10, which play key roles in inflammation and immune response. In contrast, lupus nephritis samples showed distinct gene expression patterns, including increased levels of CCL19 and TNFRSF11B, which are associated with kidney inflammation and damage. The data suggest that cutaneous lupus is characterized by strong immune activation, particularly involving T-cell chemokines and type I IFN pathways, whereas lupus nephritis exhibits a more wound-healing and fibrotic response. Using CIBERSORT analysis, the study found that cutaneous lupus had a predominance of activated CD4+ T cells, whereas lupus nephritis showed more resting memory T cells and M2 macrophages, supporting different immune mechanisms. The study proposes that lupus nephritis may result from immune complex deposition originating from inflammation in distant tissues, such as the skin. Despite similarities in early histological presentation, cutaneous lupus and lupus nephritis follow different immune response pathways, emphasizing the complexity of lupus pathogenesis. These findings provide new insights into lupus biology and may help refine diagnostic and therapeutic approaches for different lupus subtypes. A summary of the findings of these studies is presented in the forest plot in Figure 2.
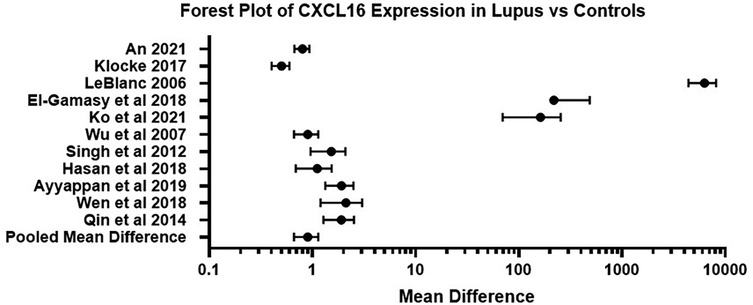
Figure 2. Forest plot of CXCL16 expression in lupus related conditions vs. control samples across studies. This forest plot summarizes the mean difference in CXCL16 levels between lupus affected individuals and healthy controls across 11 independent studies. Error bars represent 95% confidence intervals. A positive mean difference demonstrates higher CXCL16 expression in the lupus group of the study.
Evaluating CXCR6/CXCL16 expression in archival skin biopsies
We queried our mouse (1, 6), canine (16, 17) and human (1) CLE transcriptomics datasets to assess CXCR6/CXCL16 expression in skin, and compared these to other publicly available human datasets GSE95474 from Scholtissek et al. (13) and GSE193068 from Seremet, Domizio et al. (14, 15) (Figure 3). Our human dataset is presented in Yildiz-Altay et al. (1), and is comprised of n = 6 healthy and 12 biopsies from 10 DLE patients who were Black. CXCR6 was upregulated in CLE skin vs. controls across all 3 species (Figure 3A–E). CXCL16 was significantly elevated in Balb/c mouse, dog and Scholtissek et al. datasets, but did not reach significance for B6 mouse or skin of color patient samples (Figures 3F–J). To evaluate the possible utility of CXCR6 as a biomarker of cutaneous lupus, we performed Receiver Operator Characteristic curve (ROC) analysis of CXCR6 expression on our dataset, which revealed 100% sensitivity with 83.3% specificity at a normalized count value of 15.33 with an AUC of 0.8333 and P value 0.0246 (Figure 3K). Combining our dataset with Scholtissek et al, which we harmonized by using absolute RNA counts, generated an ROC with 94.4% sensitivity and 81.82% specificity with an AUC of 0.8889 and P value 0.0005 at a normalized count value of >8.071 (Figure 3L).
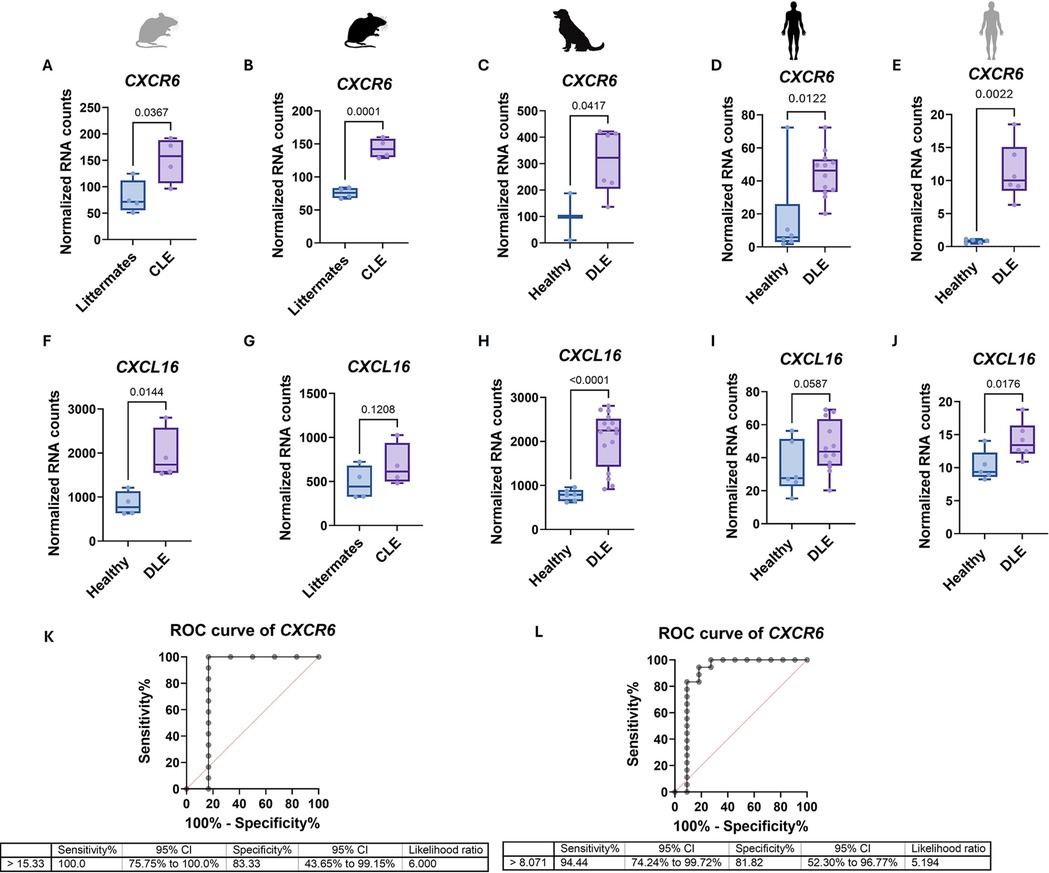
Figure 3. CXCL16 and CXCR6 are upregulated in human, mouse and dog CLE, and CXCR6 may be a useful biomarker of skin disease. CXCR6 expression in (A) Balb/c CLE mouse model (n = 4 CLE and 4 littermates, two-tailed unpaired t test p = 0.0367), (B) B6 CLE mouse model (n = 4 CLE and 4 littermates, one-tailed unpaired t test p = 0.0001), (C) canine DLE (n = 2 healthy and 6 DLE, validation cohort only had this probe, one-tailed unpaired t test p = 0.0417), (D) human DLE from a predominantly skin of color cohort (n = 6 healthy and 12 DLE, one-tailed Mann–Whitney U test p = 0.0122), and E. GSE95474 from Scholtissek et al. (2) human DLE (n = 5 healthy and 6 DLE, one-tailed Mann–Whitney U test p = 0.0022) was higher in lupus skin compared to controls. CXCL16 expression in (F) Balb/c CLE mouse model (n = 4 CLE and 4 littermates, two-tailed unpaired t test p = 0.0144), G. B6 CLE mouse model (n = 4 CLE and 4 littermates, one-tailed unpaired t test p = 0.1208), (H) canine DLE (n = 6 healthy and 16 DLE, discovery plus validation cohorts, one-tailed Mann–Whitney U test p < 0.0001), (I) human DLE from a predominantly skin of color cohort (n = 6 healthy and 12 DLE, one-tailed unpaired t test p = 0.0587), and (J) GSE95474 from Scholtissek et al. (2) human DLE (n = 5 healthy and 6 DLE, one-tailed unpaired t test p = 0.0176). While no significance was found in CXCL16 in G or I, CXCL16 was trending higher in lupus skin across all datasets. (K) Receiver operator characteristic (ROC) curve of CXCR6 from human DLE from a predominantly skin of color cohort as in panel (D) (100% sensitivity with 83.3% specificity at a normalized count value of 15.33 with an AUC of 0.8333 and p = 0.0246. (L) Combined ROC curve of human DLE from a predominantly skin of color cohort and GSE95474 from Scholtissek et al. (2) human DLE as a combination of panels (D,E) (harmonized by using absolute RNA counts; 94.4% sensitivity and 81.82% specificity with an AUC of 0.8889 and p = 0.0005 at a normalized count value of >8.071).
Discussion
CXCL16 is supported to be a biomarker of lupus nephritis and disease activity in both urine and serum samples in multiple studies from patient cohorts across the globe. Reyes-Thomas et al. also provided a nice meta analysis of the utility of CXCL16 as a protein biomarker of lupus nephritis (28).
CXCR6 but not CXCL16 appears to be enriched in lupus skin, based on our dataset, Scholtissek et al. (13) and Ko et al. (27). We hypothesize this may be due to sun avoidance behaviors by patients, as CXCL16 expression is induced by UV light (3, 4). Work from Mempel lab (11) and Huang lab (12) recently demonstrated that Trm/Tem express CXCR6 in the context of melanoma. Thus, it is possible that the enrichment of CXCR6 in CLE skin could be due to the presence of memory T cells.
The CXCR6/CXCL16 axis is likely to contribute to tissue-specific inflammation in lupus through the recruitment of CXCR6+ effector T cells to the skin and kidneys. CXCL16 binding activates downstream signaling cascades including PI3 K/AKT, MAPK/ERK and NF-kB, which promote immune cell survival, migration and cytokine release [reviewed in (29)]. Further studies should explore the therapeutic potential of targeting this axis, particularly its role in chronic tissue damage and T cell retention.
Limitations of our study include producing an exhaustive list of studies in our search strategy, such that we may have missed preclinical murine studies that identified CXCR6/CXCL16 in single cell RNA sequencing (30), or studies that may have captured CXCR6/CXCL16 in array technology without calling out individual measurements. Additionally, several studies were missing patient demographic data, and not all studies examined the impact of disease duration and/or treatment status on CXCR6/CXCL16 expression, which could contribute to bias in the interpretation of efficacy and reported results. Moreover, harmonization of measurement values would be important for establishing CXCR6/CXCL16 as a diagnostic or prognostic laboratory value for lupus disease and activity. These could include ELISA measurements of urine and serum CXCL16 protein levels, and skin CXCR6 RNA levels. Monitoring CXCR6 levels over time could also provide insight into its role and association with different pathological manifestations of CLE, including flare and non-flare conditions. This would provide knowledge of CXCR6 expression patterns that can lead to understanding disease activity and progression. Finally, for this study we excluded papers that were not in English. This may result in reduced generalizability, and possible incomplete evidence.
In conclusion, our systematic review and dataset meta-analysis supports CXCR6 and its ligand CXCL16 as biomarkers for lupus organ disease, specifically in the skin and kidneys. Our systematic analysis of primary studies demonstrates consistent upregulation of CXCL16 in serum, urine and cerebrospinal fluid in active lupus, while CXCR6 is enriched in lupus skin. These findings support a model in which the CXCR6/CXCL16 pathway contributes to tissue-specific inflammation and could serve as both a diagnostic marker and therapeutic target. Future studies should further elucidate the functional roles and therapeutic modulation of this axis across different lupus phenotypes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the UMass Chan IRB and the Howard University IRB with a Memorandum of Understanding (MOU) for cross-institutional studies. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants in accordance with the national legislation and institutional requirements. The dog animal studies were approved by the Cummings School of Veterinary Medicine at Tufts University IACUC. Written informed consent was obtained from the owners for participation of their animals in the biorepository. The studies were conducted in accordance with the local legislation and institutional requirements. The mouse animal studies were approved by the UMass Chan IACUC. The studies were conducted in accordance with the local legislation and institutional requirements. All datasets (human, dog, mouse) used in this study were acquired in previous studies and were re-queried for expression of CXCR6/CXCL16.
Author contributions
FK: Investigation, Data curation, Writing – review & editing, Visualization, Writing – original draft, Validation, Funding acquisition, Formal analysis, Project administration. HD: Validation, Investigation, Writing – review & editing, Formal analysis. VR: Investigation, Writing – review & editing, Data curation, Resources, Methodology. CF: Writing – review & editing, Investigation, Supervision, Resources, Project administration, Data curation. JR: Data curation, Supervision, Investigation, Writing – review & editing, Writing – original draft, Conceptualization, Visualization, Funding acquisition, Resources, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. FK is supported by a Diversity Research Supplement Award from the Lupus Research Alliance, JR is supported by a Mechanisms & Targets Award from the Lupus Research Alliance.
Conflict of interest
JR is an inventor on use patents for human and veterinary CTCL diagnostics, and targeting CXCR3 and IL15 for the treatment of vitiligo.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/flupu.2025.1645416/full#supplementary-material
References
1. Yıldız-Altay Ü, Adhanom R, Abdi W, Romasco A, You L, Santacruz E, et al. Comparative spatial transcriptomics of hair follicle-T cell interactions in mouse, dog and human reveals conserved drivers of primary cicatricial alopecia. BioRxiv.2025.01.20.633953. (2025). doi: 10.1101/2025.01.20.633953
2. Hassan AM, Farghal NMA, Hegab DS. Serum-soluble CXCL16 in juvenile systemic lupus erythematosus: a promising predictor of disease severity and lupus nephritis. Clin Rheumatol. (2018) 37(11):3025–32. doi: 10.1007/s10067-018-4203-2
3. Tohyama M, Sayama K, Komatsuzawa H, Hanakawa Y, Shirakata Y, Dai X, et al. CXCL16 is a novel mediator of the innate immunity of epidermal keratinocytes. Int Immunol. (2007) 19:1095–102. doi: 10.1093/intimm/dxm083
4. Scholz F, Schulte A, Adamski F, Hundhausen C, Mittag J, Schwarz A, et al. Constitutive expression and regulated release of the transmembrane chemokine CXCL16 in human and murine skin. J Invest Dermatol. (2007) 127:1444–55. doi: 10.1038/sj.jid.5700751
5. Steffen S, Abraham S, Herbig M, Schmidt F, Blau K, Meisterfeld S, et al. Toll-like receptor-mediated upregulation of CXCL16 in psoriasis orchestrates neutrophil activation. J Invest Dermatol. (2018) 138(2):344-54. doi: 10.1016/j.jid.2017.08.041
6. Mande P, Zirak B, Ko W-C, Taravati K, Bride KL, Brodeur TY, et al. Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus-like inflammation. J Clin Invest. (2018) 128:2966–78. doi: 10.1172/JCI98219
7. Gies V, Schickel J-N, Jung S, Joublin A, Glauzy S, Knapp A-M, et al. Impaired TLR9 responses in B cells from patients with systemic lupus erythematosus. JCI Insight. (2018) 3(5):e96795. doi: 10.1172/jci.insight.96795
8. Brown GJ, Cañete PF, Wang H, Medhavy A, Bones J, Roco JA, et al. TLR7 gain-of-function genetic variation causes human lupus. Nature. (2022) 605:349–56. doi: 10.1038/s41586-022-04642-z
9. Hao Y, Li Y, Li H, Lyu M, Zhang D, Fu R, et al. Increased plasma sCXCL16 levels may have a relationship with Th1/Th2 imbalance in primary immune thrombocytopenia. Cytokine. (2017) 99:124-31. doi: 10.1016/j.cyto.2017.08.024
10. Haddadi N-S, Mande P, Brodeur TY, Hao K, Ryan GE, Moses S, et al. Th2 to Th1 transition is required for induction of skin lesions in an inducible and recurrent murine model of cutaneous lupus-like inflammation. Front Immunol. (2022) 13:883375. doi: 10.3389/fimmu.2022.883375
11. Di Pilato M, Kfuri-Rubens R, Pruessmann JN, Ozga AJ, Messemaker M, Cadilha BL, et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell. (2021) 184:4512–30.e22. doi: 10.1016/j.cell.2021.07.015
12. Vella JL, Molodtsov A, Angeles CV, Branchini BR, Turk MJ, Huang YH. Dendritic cells maintain anti-tumor immunity by positioning CD8 skin-resident memory T cells. Life Sci Alliance. (2021) 4:e202101056. doi: 10.26508/lsa.202101056
13. Scholtissek B, Zahn S, Maier J, Klaeschen S, Braegelmann C, Hoelzel M, et al. Immunostimulatory endogenous nucleic acids drive the lesional inflammation in cutaneous lupus erythematosus. J Invest Dermatol. (2017) 137:1484–92. doi: 10.1016/j.jid.2017.03.018
14. Domizio JD, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. (2022) 603:145–51. doi: 10.1038/s41586-022-04421-w
15. Seremet T, Di Domizio J, Girardin A, Yatim A, Jenelten R, Messina F, et al. Immune modules to guide diagnosis and personalized treatment of inflammatory skin diseases. Nat Commun. (2024) 15:10688. doi: 10.1038/s41467-024-54559-6
16. Amudzi AA, Piedra-Mora C, Ma DJ, Wong NB, David CN, Robinson NA, et al. Using gene expression analysis to understand complex autoimmune skin disease patients: a series of four canine cutaneous lupus erythematosus cases. Front Vet Sci. (2022) 9:778934. doi: 10.3389/fvets.2022.778934
17. Garelli CJ, Wong NB, Piedra-Mora C, Wrijil LM, Scarglia G, David CN, et al. Shared inflammatory and skin-specific gene signatures reveal common drivers of discoid lupus erythematosus in canines, humans and mice. Curr. Res. Immunol. (2021) 2:41–51. doi: 10.1016/j.crimmu.2021.03.003
18. Wu T, Xie C, Wang HW, Zhou XJ, Schwartz N, Calixto S, et al. Elevated urinary VCAM-1, P-selectin, soluble TNF receptor-1, and CXC chemokine ligand 16 in multiple murine lupus strains and human lupus nephritis. J Immunol. (2007) 179:7166–75. doi: 10.4049/jimmunol.179.10.7166
19. Qin M, Guo Y, Jiang L, Wang X. Elevated levels of serum sCXCL16 in systemic lupus erythematosus; potential involvement in cutaneous and renal manifestations. Clin Rheumatol. (2014) 33:1595–601. doi: 10.1007/s10067-014-2741-9
20. Ayyappan P, Harms RZ, Buckner JH, Sarvetnick NE. Coordinated induction of antimicrobial response factors in systemic lupus erythematosus. Front Immunol. (2019) 10:658. doi: 10.3389/fimmu.2019.00658
21. Singh S, Wu T, Xie C, Vanarsa K, Han J, Mahajan T, et al. Urine VCAM-1 as a marker of renal pathology activity index in lupus nephritis. Arthritis Res Ther. (2012) 14:R164. doi: 10.1186/ar3912
22. Wen S, He F, Zhu X, Yuan S, Liu H, Sun L. IFN-γ, CXCL16, uPAR: potential biomarkers for systemic lupus erythematosus. Clin Exp Rheumatol. (2018) 36:36–43. https://pubmed.ncbi.nlm.nih.gov/28628472/28628472
23. le Blanc LMP, van Lieshout AWT, Adema GJ, van Riel PLCM, Verbeek MM, Radstake TRDJ. CXCL16 is elevated in the cerebrospinal fluid versus serum and in inflammatory conditions with suspected and proved central nervous system involvement. Neurosci Lett. (2006) 397:145–8. doi: 10.1016/j.neulet.2005.12.029
24. El-Gamasy MA, El-Naghy W. Urinary neutrophil gelatinase-associated lipocalin and urinary soluble CXCL16 as biomarkers of activity in pediatric lupus nephritis. Indian J Nephrol. (2018) 28:427–32. doi: 10.4103/ijn.IJN_265_17
25. Klocke J, Kopetschke K, Grießbach A-S, Langhans V, Humrich JY, Biesen R, et al. Mapping urinary chemokines in human lupus nephritis: potentially redundant pathways recruit CD4+ and CD8+ T cells and macrophages. Eur J Immunol. (2017) 47:180–92. doi: 10.1002/eji.201646387
26. An JN, Ryu S, Kim YC, Yoo KD, Lee J, Kim HY, et al. NK1.1-natural killer T cells upregulate interleukin-17 expression in experimental lupus nephritis. Am J Physiol Renal Physiol. (2021) 320:F772–88. doi: 10.1152/ajprenal.00252.2020
27. Ko W-CC, Li L, Young TR, McLean-Mandell RE, Deng AC, Vanguri VK, et al. Gene expression profiling in the skin reveals strong similarities between subacute and chronic cutaneous lupus that are distinct from lupus nephritis. J Invest Dermatol. (2021) 141:2808–19. doi: 10.1016/j.jid.2021.04.030
28. Reyes-Thomas J, Blanco I, Putterman C. Urinary biomarkers in lupus nephritis. Clin Rev Allergy Immunol. (2011) 40:138–50. doi: 10.1007/s12016-010-8197-z
29. Bao N, Fu B, Zhong X, Jia S, Ren Z, Wang H, et al. Role of the CXCR6/CXCL16 axis in autoimmune diseases. Int Immunopharmacol. (2023) 121:110530. doi: 10.1016/j.intimp.2023.110530
Keywords: lupus, CXCR6, CXCL16, plasma, urine, cerebrospinal fluid (CSF), skin
Citation: Kernizan F, Dave H, Rossetti V, Frey C and Richmond JM (2025) Evaluating CXCR6 and its ligand CXCL16 as biomarkers for lupus organ involvement: a mini review and brief research report. Front. Lupus 3:1645416. doi: 10.3389/flupu.2025.1645416
Received: 11 June 2025; Accepted: 11 August 2025;
Published: 29 August 2025.
Edited by:
Amr Sawalha, University of Pittsburgh, United StatesReviewed by:
Rahul Kakalij, University of Nebraska Medical Center, United StatesMehmet Hocaoglu, University of Pittsburgh Medical Center, United States
Copyright: © 2025 Kernizan, Dave, Rossetti, Frey and Richmond. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faradia Kernizan, Zmtlcm5pemFuQHR1bGFuZS5lZHU=; Jillian M. Richmond, amlsbGlhbi5yaWNobW9uZEB0dWZ0cy5lZHU=
 Faradia Kernizan
Faradia Kernizan Himanee Dave
Himanee Dave Victoria Rossetti3
Victoria Rossetti3 Jillian M. Richmond
Jillian M. Richmond