Abstract
Although coral reef ecosystems across the globe are in decline due to climate change and other anthropogenic stressors, certain inshore reefs of the Upper Florida Keys reef tract have persisted, with some even thriving, under marginalized conditions. To better understand the molecular basis of the thermotolerance displayed by these corals, a laboratory-based temperature challenge experiment that also featured conspecifics from a more stress-susceptible offshore reef was conducted with the common Caribbean reef-builder Orbicella faveolata, and the proteomes of both the coral hosts and their endosymbiotic dinoflagellate communities were profiled in (1) controls, (2) corals that succumbed to high-temperature stress and bleached, and (3) those that instead acclimated to high temperatures ex situ. Proteomic signatures varied most significantly across temperatures, host genotypes, and Symbiodiniaceae assemblages, and the two eukaryotic compartments of this mutualism exhibited distinct proteomic responses to high temperatures. Both partners maintained high levels of molecular chaperones and other canonical (eukaryotic) stress response (CSR) proteins in all treatments (including controls). Instead, proteins involved in lipid trafficking, metabolism, and photosynthesis played greater roles in the holobionts’ high-temperature responses, and these energy mobilization processes may have sustained the elevated protein turnover rates associated with the constitutively active CSR.
Introduction
Corals reefs the world over are threatened by global climate change (GCC; Hoegh-Guldberg et al., 2017), and especially the rapidly increasing seawater temperatures associated with this phenomenon (Brown, 1997). South Florida’s coral reefs, in particular, have deteriorated dramatically over the past decades (Manzello, 2015) as a result of land-based pollution, GCC, and disease, yet certain inshore reefs continue to persist (even thriving in certain cases; Gintert et al., 2018). These seemingly marginalized reefs are characterized by high summer seawater temperatures, high sediment loads, extreme high or low light levels (depending on season), high nutrient levels, and otherwise poor seawater quality conditions for coral survival (Manzello et al., 2015). Nevertheless, they feature a large number of massive coral colonies that do not regularly succumb to bleaching or disease [including the now-prevalent stony coral tissue loss disease (SCTLD)]. Only 1–2 km away, the offshore reefs have suffered to a greater extent (<5% live coral cover), and their resident corals appear to have lower resilience with respect to elevated temperatures and disease (Manzello et al., 2019); this is despite the presumably more favorable seawater quality associated with these habitats.
To gain insight into the thermotolerance of massive corals of the Upper Florida Keys’ inshore reef tract, we previously exposed Orbicella faveolata genotypes from sites of differing environmental tolerances to each of two high-temperature conditions: a 5-day (d) extreme high-temperature hold at 33°C and a 31-day exposure to 32°C (a temperature these corals encounter in situ, albeit not typically over multi-week durations; Manzello et al., 2019). As hypothesized, offshore corals were less thermotolerant than inshore genotypes, which failed to bleach at 33°C. Furthermore, some corals shuffled their endosymbiotic dinoflagellate (family Symbiodiniaceae) communities over the course of the study, though switching from Breviolum-dominated assemblages to those with higher proportions of the hypothetically more thermotolerant genus Durusdinium did not demonstrate improved survival at 32°C. Although the inshore-offshore gradient in thermotolerance was confirmed, even the most resilient inshore genotypes had at least begun to bleach slightly after 31 days at 32°C.
We sought to exploit this sample set to better understand the molecular basis of thermotolerance in this Caribbean reef-builder by specifically characterizing the cellular processes underlying (1) acclimation in corals found to be relatively more high-temperature resilient and (2) the stress response in those that were beginning to bleach. A mass spectrometry (MS)-based proteomics approach was taken because, although many works have attempted to characterize the cellular biology of thermotolerant corals based on mRNA profiling alone (e.g., Bellantuono et al., 2012; Pinzón et al., 2015), the lack of correlation between gene expression and concentration of the respective protein in both scleractinian corals and Symbiodiniaceae dinoflagellates (Mayfield et al., 2016, 2018a,b) signifies that those seeking to make inferences about the cellular biology of corals and their endosymbionts must instead profile proteins; unlike mRNAs, these macromolecules directly effect behavioral changes in cells.
Materials and Methods
The Experiment
Key features of the experiment have been detailed in the Supplementary Material. Briefly, 20 O. faveolata colonies from each of three sites (Table 1; see Manzello et al., 2019 for maps and detailed site information.)-“Little Conch” (UKO2; offshore), “The Rocks” (UKI2; inshore), and “Cheeca Rocks” (UKI1; inshore)- were tagged, genotyped, and cored. After in situ recovery of the cores, they were moved to the laboratory, acclimated to aquarium conditions for several days, adhered to tile “pucks” (∼4 × 4 cm), again allowed to recover for several days, and then exposed to the treatments described above (33°C for 5 d or 32°C for 31 d, with a 7-d ramping period between the acclimation temperature of 30°C and the target temperature). The trailing two-week mean at the time of sampling (July 2017), 30°C, was used for the control treatment. Historical temperature data from Cheeca Rocks can be found at https://www.pmel.noaa.gov/co2/pco2data/cheeca/alldata/.
TABLE 1
| Sample code | Temp. (°C) (level) | Time (days) | Temp. × time | Genotype (2b-RAD) | Shelf | Health status |
| A4-1 | 30 (control) | 5 | Control-short | Skyblue | Inshore | Healthy control |
| A4-5 | 32 (high) | 31 | High-long | Skyblue | Inshore | Stressed @ high temp. |
| A4-7 | 33 (very high) | 5 | High-short | Skyblue | Inshore | Healthy @ high temp. |
| A4-8* | 30 (control) | 31 | Control-long | Skyblue | Inshore | Healthy control |
| B5-1 | 30 (control) | 31 | Control-long | Lightyellow | Inshore | Healthy control |
| B5-2* | 32 (high) | 31 | High-long | Lightyellow | Inshore | Stressed @ high temp. |
| B5-4 bu | 33 (very high) | 5 | High-short | Lightyellow | Inshore | Healthy @ high temp. |
| B5-7 bu | 30 (control) | 5 | Control-short | Lightyellow | Inshore | Healthy control |
| C5-1 | 30 (control) | 5 | Control-short | Black(a) | Offshore | Healthy control |
| C5-2 | 32 (high) | 31 | High-long | Black(a) | Offshore | Stressed @ high temp. |
| C5-7 | 30 (control) | 31 | Control-long | black(a) | Offshore | Healthy control |
| C5-8a,b | 33 (very high) | 5 | High-short | Black(a) | Offshore | Stressed @ high temp. |
| D5-2a | 30 (control) | 5 | Control-short | Grey60 | Inshore | Healthy control |
| D5-3 | 32 (high) | 31 | High-long | Grey60 | Inshore | Stressed @ high temp. |
| D5-5 | 33 (very high) | 5 | High-short | Grey60 | Inshore | Healthy @ high temp. |
| D5-8 bub | 30 (control) | 31 | Control-long | Grey60 | Inshore | Healthy control |
Orbicella faveolata samples analyzed by shotgun proteomics.
Underlined samples possessed Durusdinium spp.-dominated Symbiodiniaceae (Sym) communities; all others were instead Breviolum spp.-dominated unless noted otherwise. In the “health status” column, designations were made from Coral Watch scores (Siebeck et al., 2006); those samples colored < E3 were considered to be potentially stressed. Samples denoted by asterisks (*) were omitted from analysis due to low host coral protein yield (<30 μg). “bu” = backup (i.e., original extraction attempt failed). temp. = temperature.
aAssessed Sym proteins only.
bMix of Breviolum spp. and Durusdinium spp. dinoflagellates.
Extractions and MS
A subset of 16 samples was chosen for proteomic analyses (Table 1), and an “identical twin” design was employed in which the responses of fragments made from the same source genotype were tracked across all treatments. Genetic analyses (Manzello et al., 2019) revealed that all 20 offshore colonies from Little Conch are unique genotypes; one such clone of the black(a) genotype (2b-RAD nomenclature), colony C5, was randomly chosen for proteomics. In contrast, the inshore reef The Rocks lacks genetic diversity and is dominated by a single O. faveolata clone: skyblue (Manzello et al., 2019). One skyblue colony, A4, was chosen for proteomics. Randomly selected colonies from two genotypes of Cheeca Rocks, light-yellow (B5) and grey60 (D5), constituted the final two colonies whose fragments were analyzed in each of the four treatments. Four and two host coral and Symbiodiniaceae samples yielded few proteins upon MS (discussed below) and were omitted. This resulted in a total sample size of 12 and 14 for the host and endosymbionts, respectively (Table 1), with 3–4, 3, and 6–7 samples from Little Conch, The Rocks, and Cheeca Rocks, respectively (3–4 offshore and 9–10 inshore samples).
Coral pucks were flash-frozen in liquid nitrogen (LN2) immediately upon sampling and quickly pulverized in additional LN2 with a Baileigh Industrial hydraulic press (United States) into a wet, sand-like consistency prior to freezing (−80°C). Later, RNAs, DNAs, and proteins were extracted from ∼100 mg of partially ground tissue (including powdered skeleton) as described in the Supplementary Material. Briefly, coral and endosymbiont proteins were extracted with a modified TRIzolTM (Invitrogen, United States) protocol, and 15 μg were dissolved in ammonium bicarbonate (AB) supplemented with sodium dodecyl sulfate, quantified, electrophoresed [for protein quality control (QC)], repurified, resuspended in AB, denatured with urea, alkylated with iodoacetamide, digested overnight with trypsin, repurified, and resuspended in 20 μl of 2% acetonitrile with 0.1% formic acid (to aid in ionization) prior to nano-liquid chromatography (LC) on an Easy Nano LCTM 1000 [Thermo Fisher Scientific (TFS), United States] with a Nanospray Flex ion source (TFS) as described in Musada et al. (2020). Peptide eluates from a 2–98% acetonitrile gradient (84 min) were run on a Q Exactive Orbitrap MS (TFS) as in Musada et al. (2020). Details for all aforementioned steps can be found in the Supplementary Material.
MS Data Analysis
RAW data files from the MS were uploaded into Proteome Discoverer (ver. 2.2, TFS), and each of three fasta mRNA sequence libraries (conceptually translated to proteins) was queried: an O. faveolata transcriptome, a Symbiodiniaceae (in hospite with O. faveolata) transcriptome, and a Pocillopora acuta-Symbiodiniaceae transcriptome (Mayfield et al., 2014). All three sequence databases are described in detail in the Supplementary Material. The peak integration tolerance was 20 ppm, and the peak integration method was based on the most confident centroid. Precursor and fragment mass tolerances were 10 ppm and 0.02 Da, respectively, and up to two missed cleavages were permitted. Each of the 16 RAW files was queried separately against the three fasta libraries, and a false discovery rate (FDR)-adjusted q-value of 0.01 was set a priori. It should be noted that these stringent search parameters reduced the overall number of peptide “hits” (see “Results.”) yet ensured confidence in assignment of the peptide to the correct compartment of origin (host coral vs. dinoflagellate endosymbiont). All proteomic data (RAW MS, MZML, and MZID files), as well as the fasta libraries against which the MZML files were queried, were submitted to “MassIVE” (University of California, San Diego; accession: MSV000086098), ProteomeXchange (Vizcaíno et al., 2014; accession PXD021349), and NOAA’s Coral Reef Information System (CoRIS; accession 0227133). Finally, a tab-delineated Online Supplementary Data File (OSDF) accompanies this article and includes all data featured in analyses, figures, and tables.
Statistical Analysis-Overview
Sequenced proteins were manually checked by BLASTp (against NCBI nr) to determine if any represented duplicate hits found in both the (1) O. faveolata and P. acuta holobiont transcriptomes and (2) Symbiodiniaceae and P. acuta holobiont transcriptomes; duplicate data were concatenated and scored as presence (1) vs. absence (0) for each protein. Those host and endosymbiont proteins synthesized by only 1–2 samples were excluded from differentially concentrated protein (DCP) analyses, as were those sequenced from ≧11 and ≧12 samples, respectively (“housekeeping proteins”). This difference in criteria between compartments arises from the fact that two samples that were eliminated from the host analysis due to low number of peptide hits, C5-2 and D5-2, were included in the Symbiodiniaceae analysis (Table 1). When the host and endosymbiont proteins were combined for “total holobiont” analysis, these two samples were excluded (n = 12). DCP identification was therefore carried out with only the host and endosymbiont proteins found in 3–10 and 3–11 of the 12 and 14 samples, respectively (hereafter “variable proteins”). In contrast, multivariate statistical analyses (MSA) were conducted with all host coral, endosymbiont, and total holobiont proteins.
MSA
Multi-dimensional scaling (MDS) was undertaken to depict similarity among the 5-day, 31-day, and all 12–14 samples. To assess multivariate differences across eight experimental parameters (EP)- temperature (analyzed as 1: control vs. high and 2: 30, 32, or 33°C), (3) temperature × time, (4) genotype (n = 4), (5) site (n = 3), (6) shelf (inshore vs. offshore), (7) endosymbiont assemblage (Breviolum-dominated, Durusdinium-dominated, or mixed-lineage; Table 1), and (8) pigmentation (i.e., “health status;” Table 1)- PERMANOVA was carried out with PRIMER ver. 6 (United Kingdom) after ensuring that multivariate dispersion was similar across bins with PERMDISP (alpha = 0.05 for all MSA). PRIMER’s “relate” algorithm was used to determine whether patterns in the proteomic dataset correlated with those in the experimental one after converting categorical EP data to integers (e.g., inshore = 1, offshore = 2). A Bray-Curtis (BC) similarity matrix was constructed from these recoded EP data and compared to the BC similarity matrix of the presence-absence proteomic data. These same two matrices were compared against one another with PRIMER’s “BEST” algorithm to identify the EP that accounted for the greatest degree of variation in the proteomic dataset’s similarity structure. As a second, arguably more robust approach for selecting influential EP, PRIMER’s distance-based linear modeling (DistLM) was used to generate backward selection models for which the Bayesian information criterion (BIC) was minimized.
DCPs
We sought to identify proteins sequenced in all samples of one treatment (or exclusively within one bin, e.g., genotype) and in none of the remaining. Since few fulfilled this criterion, five additional DCP identification methods were executed in JMP Pro® (United States; ver. 15): response screening (RSA; FDR-adjusted multiple comparisons), predictor screening (PSA; proteins accounting for the largest proportion of variation associated with each EP), stepwise discriminant analysis (SDA), stepwise regression (SRA), and generalized multivariate regression (GMR). Each approach is described in detail in the Supplementary Material.
Results
Overview of Proteomic Data
Across all samples, 20,185 host and 3,470 Symbiodiniaceae peptides were sequenced (1,673,323 and 495,276 amino acids, respectively); these mapped to 3,824 and 1,116 proteins, respectively, in the coral and Symbiodiniaceae sequence databases queried (FDR-adjusted q < 0.01). It should be reiterated that this represents a subset of the total number of proteins sequenced given the high degree of stringency employed; had we been less concerned with correctly elucidating the compartment of origin, host or endosymbiont, a larger number would have been mapped and included in downstream analyses. Details on protein yield per sample, host/endosymbiont protein ratios, and protein coverage can be found in the Supplementary Material (notably Supplementary Figure 1). Of the 769 presumably unique anthozoan proteins, 300 were later found to be repeated in both the O. faveolata and P. acuta databases; MSA were carried out with the remaining 469 [of which all but 44 (9.4%) could be identified using alignment-based bioinformatics approaches]. Of the 1,116 endosymbiont proteins, 315 were unique and featured in such analyses [of which all but 74 (23%) could be identified]. Of these 469 host and 315 endosymbiont proteins, 96 and 15, respectively, were housekeeping proteins (OSDF and Supplementary Figure 2), and 160 and 212, respectively, were only identified in only 1–2 samples. Univariate DCP identification was carried out with the remaining 213 and 88 “variable” host and endosymbiont proteins, respectively.
Multivariate Effects
A number of multivariate effects (Table 2) were uncovered for the holobiont (Figures 1A–C), host coral (Figures 1D–F and Supplementary Figure 2A), and Symbiodiniaceae (Figures 1G–I and Supplementary Figure 2B) proteomes. Predominant findings of interest have been included below, with details found in the Supplementary Material. The holobiont proteome was affected by temperature (BEST and DistLM), site (BEST, DistLM, and PERMANOVA), and pigmentation (DistLM), though in the MDS plot (Figure 1C) some influence of Symbiodiniaceae assemblage is also evident. For the host proteome alone (Figures 1D–F and Table 2), site (BEST and PERMANOVA) and genotype (DistLM and PERMANOVA) best distinguished samples. Unlike the host, the PERMANOVA effect of temperature was marginally significant for the Symbiodiniaceae proteome (Figures 1G–I and Table 2), though Symbiodiniaceae assemblage most strongly affected the endosymbionts’ proteomes (DistLM and PERMANOVA; i.e., the Breviolum and Durusdinium proteomes differed significantly from one another.).
TABLE 2
| Experimental parameter | df | Pseudo F | p | #Permutations | BEST model? | DistLM model? | PERMANOVA multiple comparisons |
| Holobiont (n = 784 proteins)* | |||||||
| Correlation of BEST model = 0.401. DistLM BIC = 82.9 (R2 = 0.334) | |||||||
| Temp. (control vs. high) | 1 | 1.079 | 0.306 | 406 | Yes | Yes | |
| Site (n = 3) | 2 | 1.543 | 0.048a | 958 | Yes | Yes | The Rocks≠Cheeca Rocks |
| Health status (n = 2) | 1 | 1.216 | 0.245 | 216 | Yes | ||
| Host coral (n = 469 proteins) | |||||||
| Correlation of BEST model = 0.261. DistLM BIC = 75.7 (R2 = 0.158) | |||||||
| Site | 2 | 1.738 | 0.030 | 942 | Yes | The Rocks≠Cheeca Rocks | |
| Genotype | 3 | 1.591 | 0.045b | 973 | Yes | No post-hoc differences | |
| Endosymbiont (n = 315 proteins)* | |||||||
| Correlation of BEST model = 0.366. DistLM BIC = 107 (R2 = 0.130) | |||||||
| Sym assemblage | 2 | 1.53 | 0.014c | 988 | Yes | Breviolum-dominated≠Durusdinium-dominated | |
| Health status | 1 | 1.55 | 0.053 | 630 | Yes | ||
Statistically significant multivariate effects on the Orbicella faveolata-Symbiodiniaceae (Sym) proteome.
Experimental factors that either significantly affected the holobiont, host coral, or Symbiodiniaceae proteomes (PERMANOVA p < 0.05; bold font) or were included in PRIMER’s BEST and/or DistLM models have been included in the table. For non-statistically significant model terms, please see Supplementary Table 1. Note that, although “health status” features three categories in Table 1, only two (healthy vs. bleached) were considered in the PERMANOVA. BIC = Bayesian information criterion. Temp. = temperature. *significant rho (Spearman rank-based) between experimental parameter data matrix and proteomic data matrix.
aSee Figures 1A,B. bSee Figures 1D–F. cSee Figures 1C,G–I.
FIGURE 1
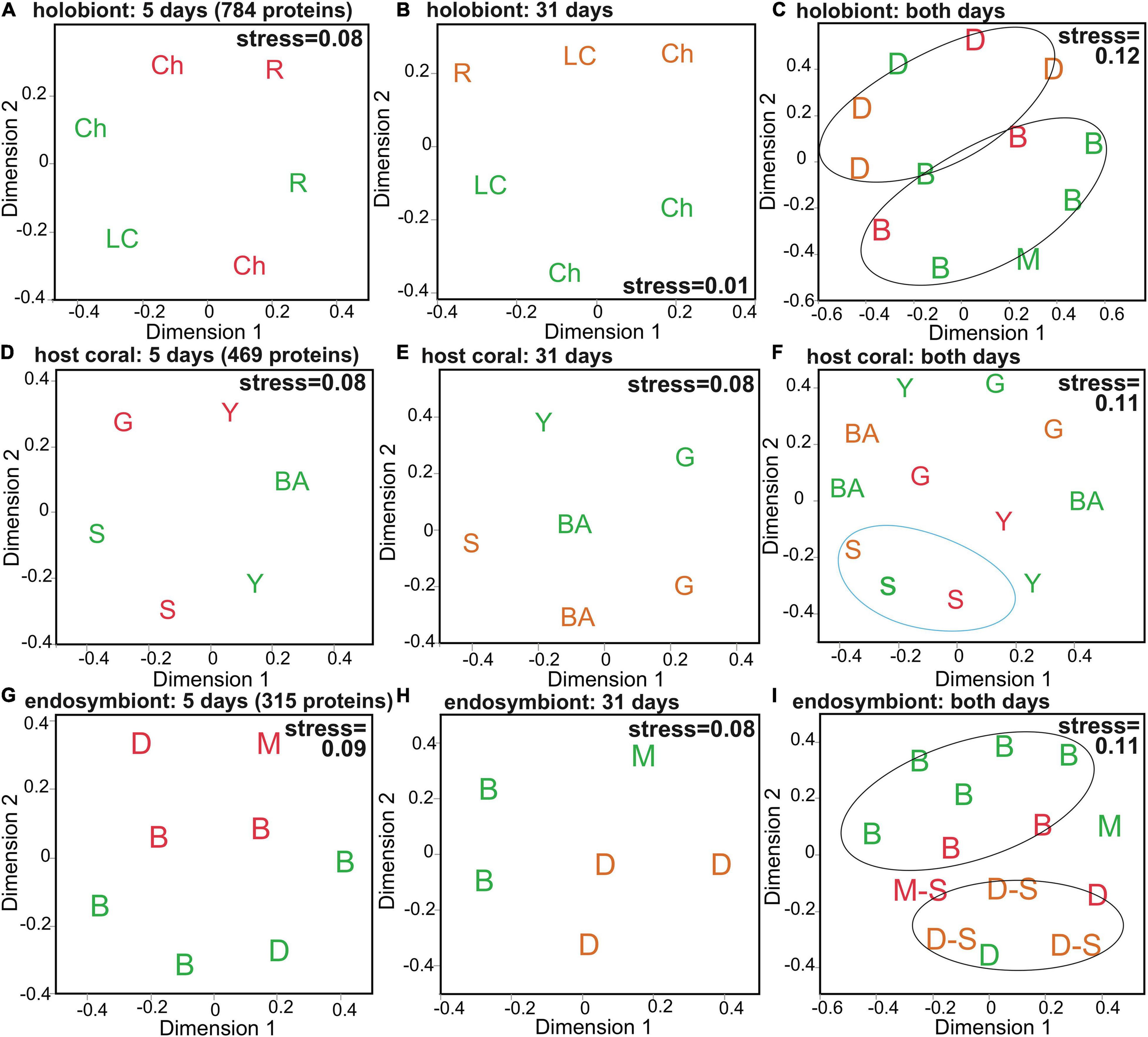
Multivariate analysis of the Orbicella faveolata-Symbiodiniaceae (Sym) proteome. Multi-dimensional scaling (MDS) depicting similarity among samples at the 5-day (A,D,G), 31-day (B,E,H), and 5+31-day (C,F,I) sampling times for all 784 holobiont proteins (A–C), the 469 host coral proteins (D–F), and the 315 Sym proteins (G–I). Icons have been coded to denote dominant PERMANOVA effects (Table 2). Site names (A,B) are abbreviated as “Ch” (Cheeca Rocks), “R” (The Rocks), and “LC” (Little Conch); Sym assemblages (C,G–I) as “B” (Breviolum-dominated), “D” (Durusdinium-dominated), and “M” (mixed-lineage; Breviolum and Durusdinium); and host genotypes (D–F) as “S” (skyblue), “G” (grey60), “Y” (lightyellow), and “BA” [black(a)]. In all panels, icons colored green, orange, and red correspond to control (30°C), high temperature (32°C), and very high temperature (33°C) samples, respectively, and samples that were Stressed at the time of sampling have been denoted by “–S” in I.
DCP Overview
The five DCP identification approaches tended to yield the same suites of proteins as being responsive to each EP of interest (Supplementary Figure 3 and Supplementary Tables 2, 3), and host genotype and temperature were the EP across which the largest number of proteins differed in abundance: 18/47 DCPs (38.3%) for each (Supplementary Figure 4). Host genotype also most strongly affected the host proteome at the multivariate scale (see above.), and temperature best accounted for the structure of the holobiont proteomic dataset (Table 2). In contrast, although site significantly affected both holobiont and host proteomes (Table 2), only three site-responsive DCPs were uncovered (Supplementary Figure 4C); this is because, with the exception of Cheeca Rocks (for which two genotypes were analyzed), only one genotype was analyzed at each of the remaining sites. For this reason, we generally discuss genotype and site effects in tandem hereafter. The functional breakdowns of the 27 coral (Table 3), 20 endosymbiont (Table 4), and 47 holobiont DCPs can be seen in Figures 2A,B, and C, respectively, and the most dominant cellular processes (gene ontology) of the coral hosts and endosymbiotic dinoflagellates were metabolism (Figure 2A) and photosynthesis (Figure 2B), respectively. When analyzing all 47 DCPs together (Figure 2C), these two processes accounted for nearly 40% of all DCPs, with translation being the third most affected process.
TABLE 3
| Contig | Identity | Function | Trend(s) |
| Temperature-responsive proteins (n = 8; Figure 3) | |||
| comp111800_c2_seq1 | Ras-related protein rab11 | Vesicle fusion and trafficking | High (32°C) > all others, stressed > healthy |
| contig8136 | Large subunit ribosomal protein L18e | Translation | Control-temp. only, healthy > stressed |
| OFAVBQ_DN204682_c1_g1_i2 | 40S ribosomal protein S3 | Translation | Control temp. only |
| OFAVBQ_DN211578_c1_g2_i4 | Citrate synthase | Metabolism | Not expressed by stressed corals |
| OFAVBQ_DN212753_c2_g1_i4 | Erlin-1 | Cholesterol/lipid homeostasis | Control-5-day > all others |
| OFAVBQ_DN214053_c3_g2_i3 | Calumenin | Protein folding and sorting | High > control temp., stressed > healthy |
| OFAVBQ_DN215575_c0_g1_i3 | Glutamine synthetase-like | Metabolism | Control > high temp. |
| OFAVBQ_DN222739_c0_g1_i5 | Unknowna | Unknown | High (32°C) > all others, stressed > healthy |
| Genotype-responsive proteins (including site effects; n = 14; Figure 4) | |||
| OFAVBQ_DN221283_c3_g2 | Collagen alpha-1(I) chain-like | Structural | Offshore only |
| OFAVBQ_DN218618_c3_g1_i10 | Galaxin-like | Calcification | Not found in lightyellow |
| OFAVBQ_DN208448_c1_g1_i1 | Protein DD3-3-like | Cell-cell interactions | Not found in lightyellow |
| OFAVBQ_DN224869_c0_g1_i2 | Laccase-25-like isoform X1 | Metabolism | Grey60 > other genotypes |
| OFAVBQ_DN218284_c7_g2_i4 | Dermatopontin | Cell-cell interactions | Grey60 only |
| OFAVBQ_DN205148_c1_g2_i1 | Rhamnose-binding lectin | Cell-cell interactions | Grey60 only |
| OFAVBQ_DN103730_c0_g1_i1 | 14-3-3 protein epsilon | Signaling | Grey60 > all other genotypes |
| OFAVBQ_DN210981_c4_g1_i3 | Legumain-like | Protease | Not expressed by skyblue |
| OFAVBQ_DN213163_c3_g1_i11 | Transaldolase | Metabolism | Cheeca genotypes > others |
| OFAVBQ_DN218286_c0_g1_i1c | Glycerol kinase | Metabolism | Cheeca genotypes > others |
| OFAVBQ_DN199317_c0_g3_i1 | Collagen triple helix repeat-containing protein 1 | Structural | Lightyellow only |
| OFAVBQ_DN197100_c0_g1_i1 | Protein PRY1-like | Lipid secretion | Not found in offshore samples |
| OFAVBQ_DN203970_c3_g5_i1 | Elongation factor 1-beta-like | Translation | Lightyellow > other genotypes |
| OFAVBQ_DN197154_c0_g3_i1 | Galaxin-like | Calcification | Lightyellow only |
| Shelf-responsive proteins (n = 3; Supplementary Figure 5) | |||
| OFAVBQ_DN217467_c0_g1_i3 | 40S ribosomal protein S4-like | Translation | Offshore > inshore |
| OFAVBQ_DN204853_c0_g1_i1 | Nitrilase with biotinidase and vanin domains | Metabolism | Offshore > inshore |
| OFAVBQ_DN192542_c0_g1_i1 | Nucleoside diphosphate kinase | Metabolism | Not found in offshore samples |
| Other effect types (n = 2; Supplementary Figure 5) | |||
| OFAVBQ_DN198838_c3_g2_i3 | 40S ribosomal protein S20 | Translation | Breviolum assemblages only |
| OFAVBQ_DN218234_c4_g1_i2b | MAC/perforin domain | Immunity | Stressed > healthy |
Host coral differentially concentrated proteins (DCPs).
For details on the analytical approach(es) used to identify the 27 DCPs, please see Supplementary Table 2, and for a comparison with other studies, please see Supplementary Table 4. Sym = Symbiodiniaceae. temp. = temperature.
aalso known as sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing protein 1. bDurusdinium-dominated assemblages only.
TABLE 4
| Contig | Identity | Function | Trend(s) |
| Temperature-responsive proteins (n = 9; Figure 5) | |||
| SYMBOF_DN176524_c0_g1_i1.p1 | Rab-related GTPase family rab5 | Molecular trafficking | High-long > control long |
| SYMBOF_DN190166_c0_g1_i2.p1 | ADP-ribosylation factor | Molecular trafficking | H33 < others |
| SYMBOF_DN201098_c1_g4_i1.p1 | Photosystem II chlorophyll-binding protein CP43 | Photosynthesis | Control temp. only |
| SYMBOF_DN192802_c0_g1_i2.p1 | Unknown | Unknown | Control-short > all others |
| SYMBOF_DN196424_c4_g3_i3.p1 | Phosphoglycerate kinase | Metabolism | Control > high temp. |
| SYMBOF_DN201284_c0_g2_i5.p1 | Chaperone protein DNAJ (HSP40) | Stress response | Control > high temp., Healthy > stressed |
| SYMBOF_DN201349_c3_g4_i2.p1 | Fucoxanthin-chlorophyll a-c binding protein B | Photosynthesis | Control > high temp., Healthy > stressed |
| SYMBOF_DN163023_c0_g2_i2.p1 | 60S ribosomal protein L3 (see also Figure 6C.) | Translation | Control > high temp., Mostly D < others |
| SYMBOF_DN218526_c0_g1_i5.p1 | Chloroplast soluble PCP precursor-1 (see also Figure 6C.) | Photosynthesis | H32 < all others, Healthy > stressed, Mostly D < others |
| Sym assemblage-responsive proteins [n = 5 (excluding two that were also affected by temp. and instead found above); Figure 6] | |||
| SYMBOF_DN191788_c0_g1_i1.p1 | Peridinin chlorophyll a binding protein | Photosynthesis | Mostly B > all others, Lightyellow > others |
| SYMBOF_DN208331_c3_g1_i2.p1 | Fucoxanthin-chlorophyll a-c binding protein F-1 | Photosynthesis | Healthy > stressed, Mostly D < others |
| SYMBOF_DN211305_c3_g1_i1.p1 | Chloroplast soluble PCP precursor-2 | Photosynthesis | Mostly D only |
| SYMBOF_DN215694_c5_g7_i1.p1 | Fucoxanthin-chlorophyll a-c binding protein F-2 | Photosynthesis | Mostly B only |
| SYMBOF_DN217254_c0_g1_i1.p1 | Heat shock protein 90 (HSP90) | Stress response | Not in mix of B and D |
| Other effects (n = 6; Supplementary Figure 5) | |||
| SYMBOF_DN192258_c0_g6_i3.p1 | Unknown | Unknown | Lightyellow > all others |
| SYMBOF_DN223629_c2_g4_i1.p2 | Peridinin-chlorophyll A binding protein | Photosynthesis | Lightyellow < all others |
| SYMBOF_DN184263_c0_g2_i1.p1 | Acyl-CoA-binding protein | Metabolism | Skyblue only |
| SYMBOF_DN181028_c0_g1_i1.p1 | Luminal-binding protein 5 (BIP5) | Stress response | Grey60 only |
| SYMBOF_DN214774_c2_g1_i1.p1 | Fucoxanthin-chlorophyll a-c binding protein F | Photosynthesis | Inshore > offshore, Healthy > stressed |
| SYMBOF_DN214550_c0_g1_i1.p1 | Ubiquitin | Stress response | Cheeca Rocks < others |
Symbiodiniaceae (Sym) differentially concentrated proteins.
For details on the analytical approach(es) used to identify the 20 DCPs and a comparison with other published works, please see Supplementary Tables 3 and 5, respectively. H32 and H33 = high-temperature (temp; 32°C) treatment and very high temp. (33°C) treatments, respectively. “mostly B,” “mostly D,” and “mix of B and D” correspond to Breviolum-dominated, Durusdinium-dominated, and mixed Breviolum+Durusdinium Sym assemblages, respectively. “Short” and “long” in the “Trend(s)” column correspond to the 5- and 31-day sampling times, respectively. PCP = peridinin chlorophyll-a binding protein. QC = quality control (protein trafficking and folding).
FIGURE 2
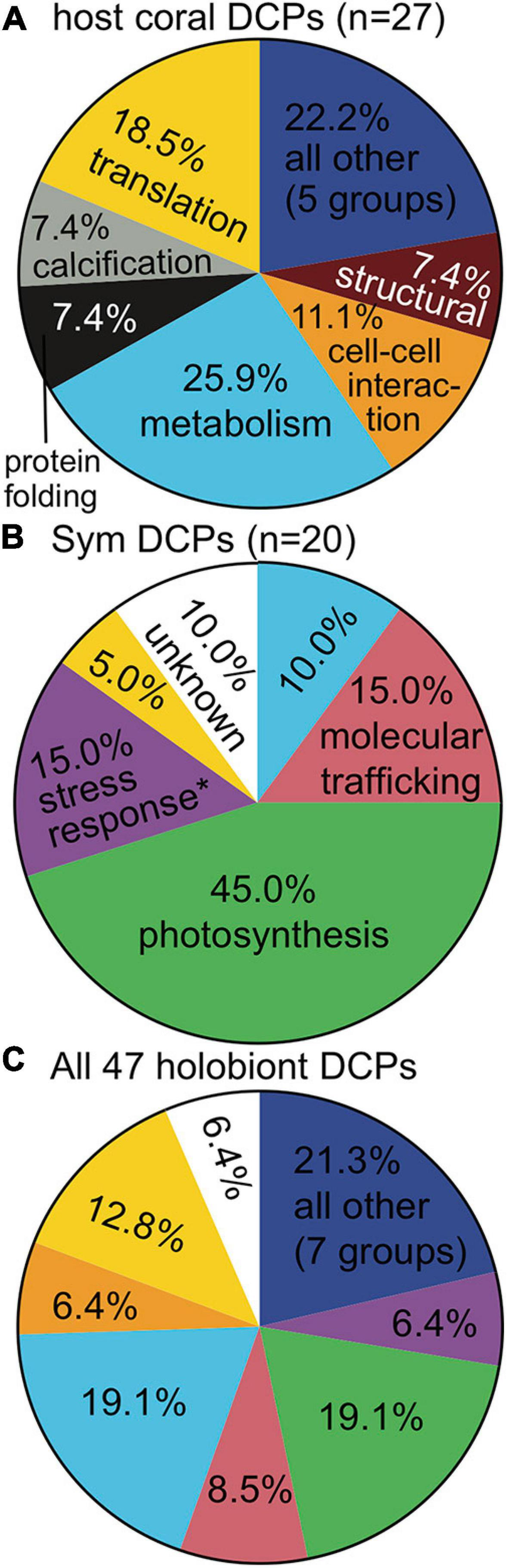
Functional breakdown of differentially concentrated proteins (DCPs). The cellular process color codes are shown only the first time they are used (the exception being the “all other” category). When proteins involved in a cellular process (e.g., stress response) were documented at a significantly higher proportion in one compartment vs. the other (Fisher’s exact test, p < 0.01), the process name in the compartment with the higher value has been denoted by an asterisk (∗). Processes specific to anthozoan hosts (A) (i.e., calcification) and dinoflagellates (B) (i.e., photosynthesis) were excluded from this analysis.
Host Coral DCPs
Of the 27 host DCPs (Table 3 and Supplementary Tables 2, 4), 8 were significantly affected by temperature (Figure 3), and a PCA with these separated samples of the control, 32, and 33°C treatments (Figure 3A). Only one DCP, ras-related protein rab11 (Figure 3B), fulfilled our optimal criterion of being synthesized only by corals of a high-temperature treatment (32°C). Three additional DCPs (Figure 3B)- large subunit ribosomal protein L18e, 40S ribosomal protein S3, and glutamine synthetase- were instead only translated by controls. Metabolism and translation were the two cellular processes featuring the largest number of temperature-sensitive host coral proteins (Figure 3C). There was also a strong effect of host genotype on 14 of the 27 DCPs (Figure 4, Table 3, and Supplementary Tables 2, 4), and 5 of these were only found in one of the four genotypes (Figure 4B). For a detailed treatise of these 14 DCPs, please consult the Supplementary Material.
FIGURE 3
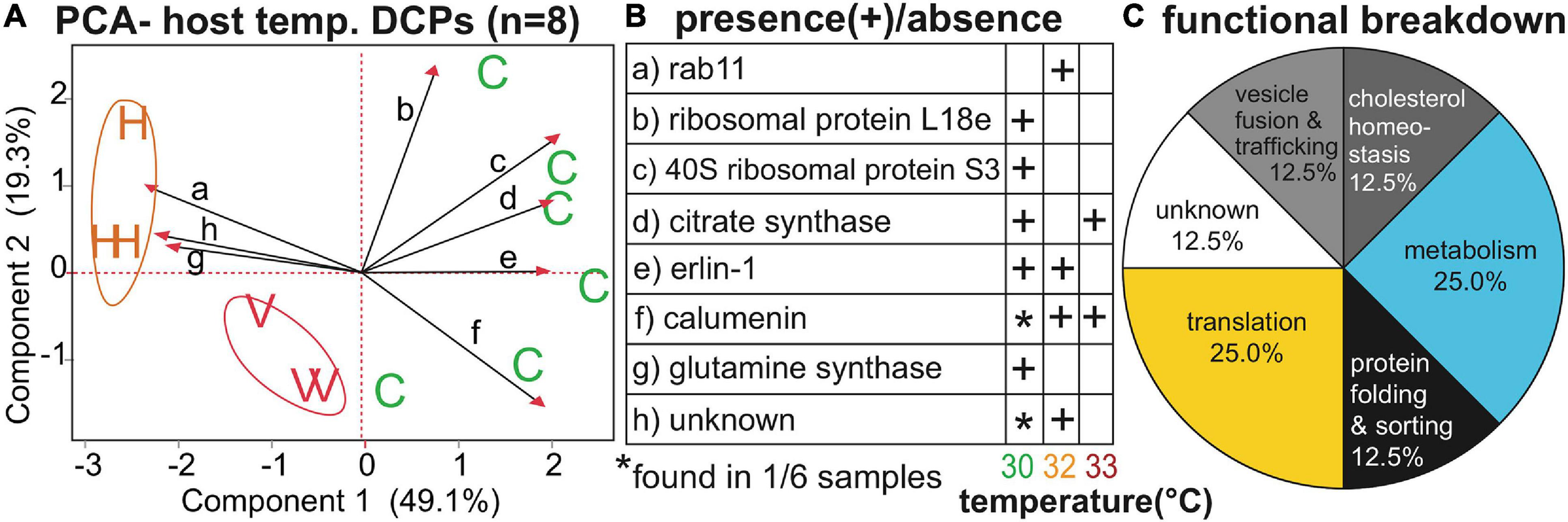
Temperature-sensitive Orbicella faveolata proteins. A PCA (A) with the eight proteins whose concentrations differed significantly across temperatures (temp.); “C,” “H,” and “V” correspond to control (30°C), high (32°C), and very high (33°C) temperatures, respectively. The black, small-font, lower-case letters over the biplot rays correspond to the row where the respective data for each protein have been displayed in (B). A functional breakdown of the proteins can be found in (C). For details on these eight proteins, please see Table 3 and Supplementary Tables 2, 4.
FIGURE 4
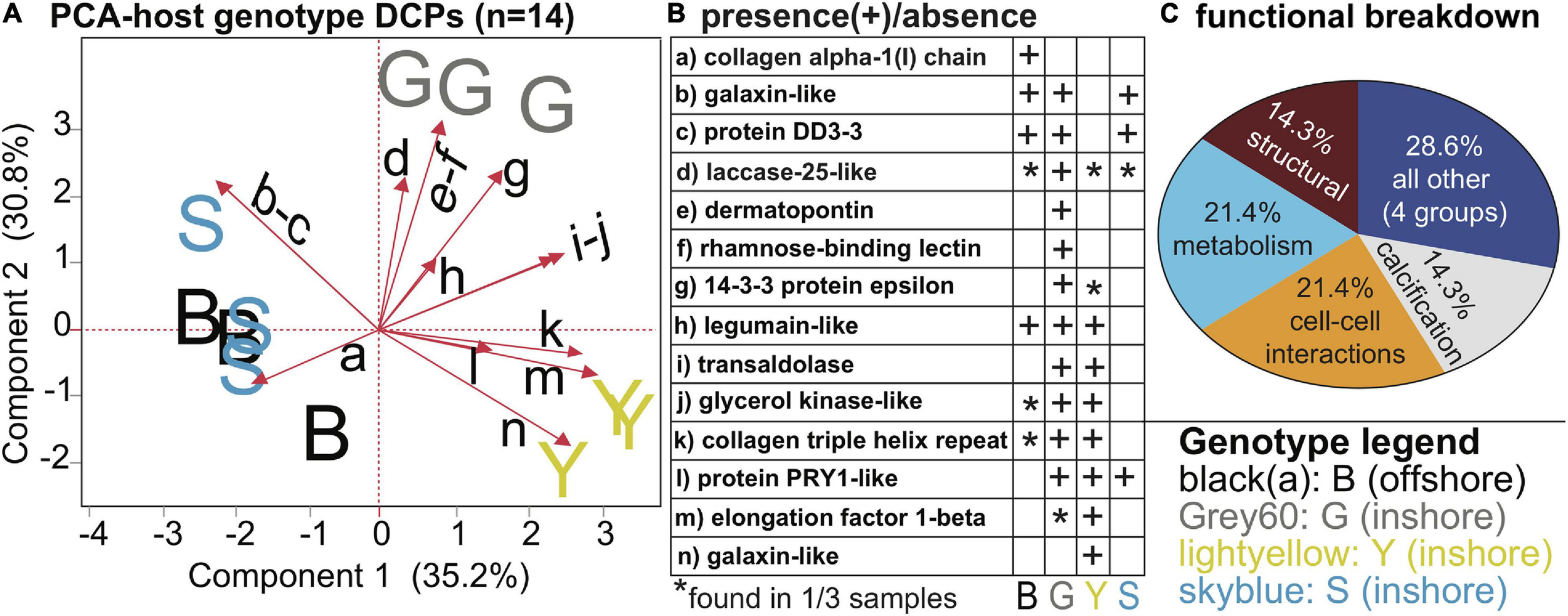
Genotypic differences in Orbicella faveolata proteins. A PCA with the 14 proteins whose concentrations differed significantly across host genotypes (A). Please note that the black, small-font, lower-case letters adjacent to the biplot rays correspond to the row where the respective data for each protein have been displayed in (B), and the legend for the sample icons can be found in the bottom right corner. A functional breakdown of these proteins is depicted in (C). For further details, please see Table 3 and Supplementary Tables 3, 5.
Symbiodiniaceae DCPs
Temperature and Symbiodiniaceae assemblage were the predictors that generated the greatest number of Symbiodiniaceae DCPs (Supplementary Figure 4B, Table 4, and Supplementary Tables 3, 5): 9 and 7, respectively (with 2 of these affected by both temperature and assemblage). These EP were also associated with the lowest PERMANOVA p-values (Table 2). The nine temperature-responsive DCPs led to separation among samples in a PCA (Figure 5A). Although none were maintained only in high-temperatures and not in controls, four were absent from high-temperature samples (Figure 5B): photosystem II chlorophyll-binding protein CP43, an unknown protein, a phosphoglycerate kinase, and a DNAj chaperone protein (i.e., HSP40). Two additional proteins were not synthesized by any 32°C sample (Figure 5B): 60S ribosomal protein L3 and a chloroplast-soluble peridinin chlorophyll a-binding protein (PCP). One-third of the Symbiodiniaceae temperature-sensitive DCPs were involved in photosynthesis (Figure 5C). To gain greater insight into Symbiodiniaceae assemblage effects (Figure 6), the mRNA read breakdowns for the 14 samples were plotted (Figure 6A). For concentration trends and functional breakdowns of the seven Symbiodiniaceae assemblage-responsive DCPs (Figures 6B,C), please consult the Supplementary Material (particularly Supplementary Figure 5).
FIGURE 5
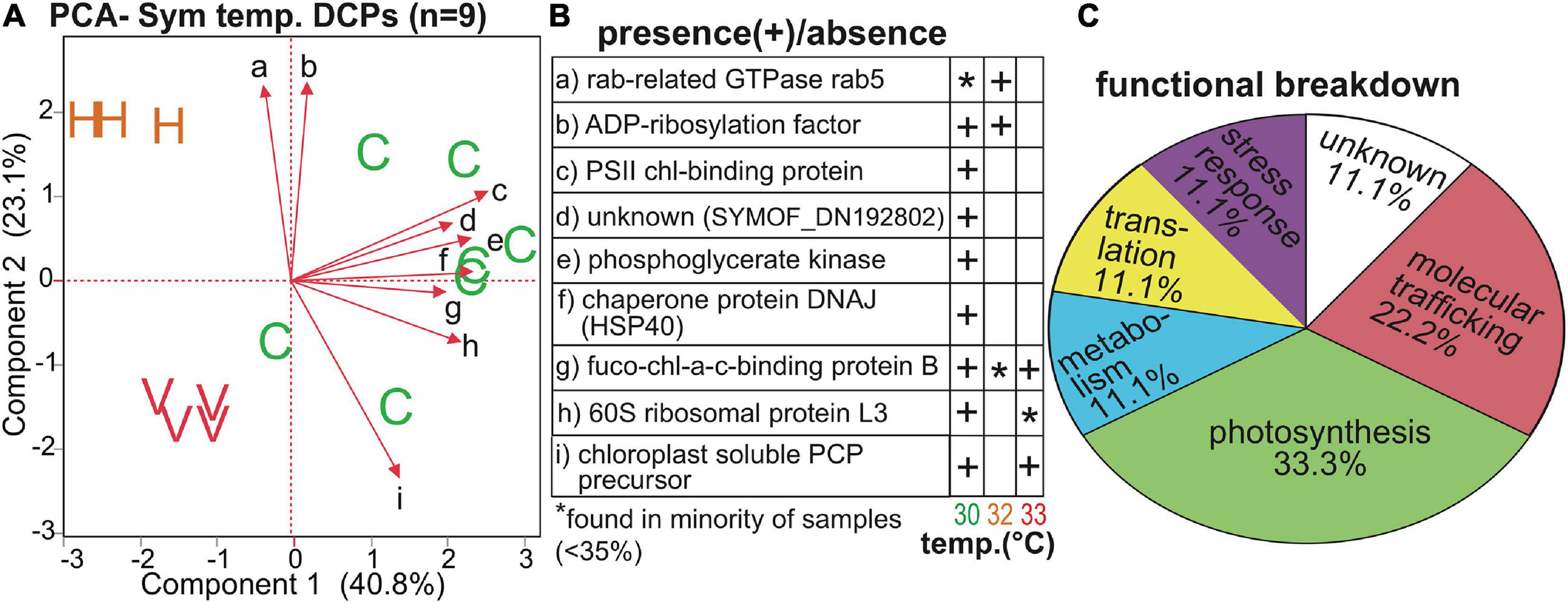
Temperature-sensitive Symbiodiniaceae proteins. A PCA with the nine proteins whose concentrations differed significantly across temperatures (A) in which “C,” “H,” and “V” correspond to control (30°C), high (32°C), and very high (33°C) temperatures, respectively. The black, small-font, lower-case letters adjacent to the biplot rays correspond to the row where the respective data for each protein have been displayed in (B). A functional breakdown of these proteins is depicted in (C). chl = chlorophyll. Fuco-chl-a-c binding = fucoxanthin-chl-a-c binding protein B. PCP = peridinin chl-a-binding protein. PS = photosystem II. For details on these nine proteins, please see Table 4 and Supplementary Tables 3, 5.
Discussion
Herein we sought to uncover high-temperature effects on cellular biology of massive coral genotypes from distinct thermal habitats that varied in their capacity to acclimate to experimentally elevated temperatures. There were clear proteomic differences among the four host genotypes and three Symbiodiniaceae assemblages, and the Symbiodiniaceae proteomes were relatively more affected by high-temperature exposure at the multivariate scale than their hosts. Symbiodiniaceae dinoflagellates also demonstrate more pronounced mRNA-level responses to high temperatures than their cnidarian hosts (Mayfield et al., 2014), and their proteomic strategy for confronting environmental change is also different from that of other corals studied to date (Mayfield et al., 2016); the latter finding was corroborated herein. Proteins involved in metabolism and translation were most likely to be affected by high-temperature exposure in the cnidarian hosts, whereas the temperature-sensitive endosymbiont DCP pool was dominated by photosynthesis proteins; this substantiates findings dating back over 20 years (e.g., Jones et al., 1998). That being said, molecular trafficking proteins (e.g., rab) were affected by thermal stress in both compartments and are discussed in greater detail below.
Of the 784 proteins profiled, only 17 (2.2%) were significantly affected by temperature. This could be due to one of at least two reasons. First, there was high proteomic fidelity to host genotype. Even after several weeks of high-temperature exposure, the host proteomes clustered as much by genotype as by their temperature treatment. Such distinct differences in protein signatures (and the associated high inter-genotype variability) may have thwarted our ability to uncover temperature-sensitive host DCPs. Instead, those eight identified represent “core” proteins exploited by all four genotypes upon high-temperature exposure. Since the low number of universal high-temperature-sensitive proteins uncovered (including nine of endosymbiont origin) could have also been due to having pooled the data from samples that were bleaching with those that had instead potentially acclimated, a separate, genotype-to-genotype analysis has been provided in the Supplementary Material. Briefly, the four host genotypes manifested distinct stress responses, as well as dissimilar strategies for acclimating to high temperatures. For instance, of the 80 and 63 proteins found to be involved in high-temperature acclimation (n = 3 controls vs. n = 3 actively acclimating corals) and bleaching (n = 3 controls vs. n = 3 bleaching corals), respectively, only 4 (5%; Supplementary Figure 6) and 5 (8%; Supplementary Figure 7) were utilized in the acclimation and bleaching responses, respectively, of multiple genotypes. The fact that coral fragments sampled from colonies that were within even several meters of each other in situ utilized entirely different means of acclimating (or succumbing) to high temperatures is puzzling, unexpected, and is under active investigation by ourselves and others working in South Florida. Perhaps, the location on the colonies from which cores were made could accommodate some of the variation (despite efforts made to be consistent with respect to the relative positioning of the drill-bit); biopsies from more shaded areas might display differing means of thermo-acclimation on account of entrained light effects, even after acclimation in a common garden prior to experimentation. Given the low overlap in the acclimation and stress responses across genotypes/sites, as well as the current inability to rigorously account for such variation, we have instead focused the remainder of our discussion on the common elements uncovered.
Of the 17 temperature-sensitive DCPs, three differed in abundance between samples that were bleaching (C5-2) or beginning to bleach (A4-5 and D5-3) at the time of sampling vs. high-temperature samples that resisted bleaching: a host ras-related protein rab11 involved in vesicle trafficking and fusion [only synthesized by visibly stressed samples (color score < E3)], an unknown host coral protein with sushi, von Willebrand factor type A, EGF, and pentraxin domains (OFAVBQ_DN222739_c0_g1_i5; only synthesized by visibly stressed samples), and an endosymbiont fucoxanthin-chlorophyll a-c binding protein F (FCPE; chloroplastic; SYMBOF_DN208331_c3_g1_i2.p1-1) involved in photosynthesis (not translated by visibly stressed samples). Given the large number of FCPE proteins even within the endosymbiont DCP pool, it is unclear whether the absence of one isoform would have significant implications for photosynthetic function. In contrast, the two host proteins were distinct, and numerous rab proteins involved in vesicle and lipid trafficking were up-regulated at high temperatures in bleaching offshore samples in the genotype-by-genotype analysis (Supplementary Figures 6, 7). Furthermore, an endosymbiont rab5 protein was up-regulated in the three bleaching samples.
Prior works have also uncovered proteins involved in vesicle, lipid, and lipid body trafficking to be affected by high-temperature exposure in host corals and their endosymbionts (Mayfield et al., 2018a,b), and the role of lipid trafficking within the anthozoan-dinoflagellate endosymbiosis has been well-studied (Chen et al., 2012, 2015, 2017). Prior work on rab11 in particular has implicated it in endocytic recycling (Lock and Stow, 2005), whereby membrane components internalized in endosomes are transported to (and then recycled at) the plasma membrane (Montealegre and van Endert, 2010). This protein has also been linked to the establishment and maintenance of the anemone-dinoflagellate endosymbiosis (Chen et al., 2003). Specifically, rab11 is normally kept away from the symbiosome, but high concentrations are documented around phagosomes containing dying endosymbionts at high temperatures (Chen et al., 2005a,b). Later, Downs et al. (2009) demonstrated that other rab proteins (e.g., rab7) are involved in symbiophagy, whereas Weston et al. (2015) took the large number of rab proteins uncovered in corals exposed to high temperatures for several hours to be indicative of dinoflagellate cell exocytosis. However, as was the case herein, their sampling regimen did not explicitly allow for the uncovering of proteins that enact bleaching, merely those associated with it. Whether up-regulation of rab11 and other rab proteins reflects the possibility of symbiophagy, dinoflagellate exocytosis, or simply an increased need to traffic membrane components to the plasma membrane during periods of elevated temperature remains to be determined. Furthermore, given that a many corals switched symbionts over the duration of the study (Figure 6), differential rab protein levels may simply be a testament to the need to resheath newly acquired dinoflagellate cells with symbiosome membranes.
FIGURE 6
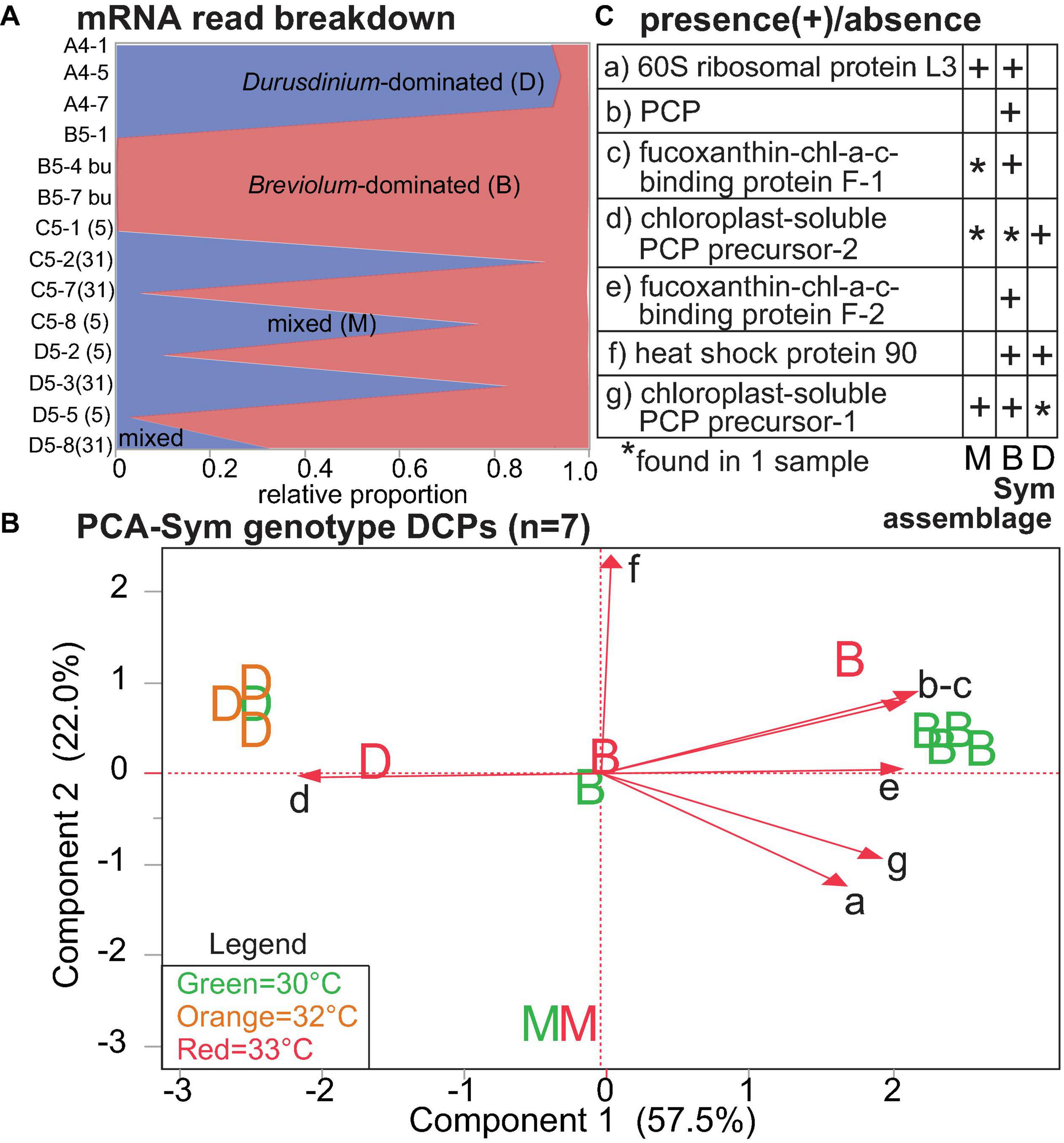
Proteomic differences across Symbiodiniaceae (Sym) lineages. In the mRNA breakdown of the 14 samples analyzed by shotgun proteomics (A), the blue and red shading reflect the proportions of Durusdinium spp. (“D”) and Breviolum spp. (“B”) dinoflagellates, respectively. Samples for which no genus comprised >80% of the mapped mRNA reads were deemed “mixed-lineage” (“M”). The numbers in parentheses following the sample codes for the two genotypes that underwent endosymbiont shuffling, C5 [black(a) from Little Conch (offshore)] and D5 [grey60 from Cheeca Rocks (inshore)], represent sampling days. A PCA with seven proteins whose concentrations differed significantly across Sym assemblages (B); letters correspond to Sym assemblages, and the colors correspond to temperatures (see inset legend.) The black, small-font, lower-case letters adjacent to the biplot rays correspond to the row where the respective data for each protein have been displayed in (C). Five, one, and one of these proteins were involved in photosynthesis (71.4%), the stress response (14.3%), and translation (14.3%), respectively. chl = chlorophyll. PCP = peridinin chl-a binding protein. For details on these nine proteins, please see Table 4 and Supplementary Tables 3, 5.
Prior works on endosymbiotic anthozoans have yielded conflicting results with respect to the relative importance of canonical eukaryotic stress response (CSR) proteins in thermo-acclimation (Table 5 and Supplementary Tables 4–6). Whereas both corals and Symbiodiniaceae from upwelling reefs constitutively express mRNAs encoding heat shock proteins (HSPs; Mayfield et al., 2011, 2012), others have observed a more traditional heat shock response at the gene (Rosic et al., 2011) and protein (Coles and Brown, 2003) levels in corals from stable temperature environments. Herein numerous CSR proteins in both the host and dinoflagellate compartments were among the most abundant housekeeping proteins found in all samples, including controls (Table 5 and OSDF). Although one HSP90 was a DCP, it only differed in concentration across endosymbiont assemblages. Furthermore, an endosymbiont HSP40 (chaperone protein DNAj) was only sequenced from controls, and another endosymbiont molecular chaperone, DNAK, was only up-regulated at high temperatures in one host genotype (grey60; Supplementary Figure 7 and OSDF). As an additional example, peroxiredoxins, which are important in the stress responses of both corals (Weston et al., 2015) and fish (Kultz et al., 2007), were instead constitutively maintained in the corals sampled herein (Table 5).
TABLE 5
| Accession | Compartment | Protein name | Prior mRNA/protein trend | mRNA/protein reference(s) | Prior trend corroborated? |
| HOUSEKEEPING CSR PROTEINS | |||||
| OFAVBQ_DN205531_c1_g1_i1 | Coral host | 78 kDa glucose-regulated protein (GRP78) | Stress-sensitive | Aranda et al. (2011) Lõhelaid et al. (2014) a | No No |
| OFAVBQ_DN217505_c2_g1_i3 | Coral host | Endoplasmin-like isoform X1 | Stress-sensitive | Ruiz-Jones and Palumbi (2017) | No |
| OFAVBQ_DN202907_c9_g3_i1 | Coral host | Glutathione S-transferase | Constitutively maintained | Whalen et al. (2010) a | Yes |
| OFAVBQ_DN215553_c1_g3_i1 | Coral host | Heat shock cognate 71 kDa protein | Constitutively maintained | Oakley et al. (2017) b Poli et al. (2017) | Yes Yesc |
| OFAVBQ_DN214823_c0_g2_i2 | Coral host | HSP60 | Stress-sensitive | Chow et al. (2009) | No |
| OFAVBQ_DN218952_c1_g1_i5 | Coral host | HSP90-beta | Constitutively maintained | Nakamura et al. (2012) | Yes |
| OFAVBQ_DN225001_c3_g1_i2 | Coral host | Peroxiredoxin-5 | Stress-sensitive | Levy et al. (2016) | No |
| OFAVBQ_DN208085_c0_g2_i1 | Coral host | Peroxiredoxin-6 | Stress-sensitive | Mayfield et al. (2018b) | No |
| SYMBOF_DN199915_c1_g4_i1.p1 | Endosymbiont | HSP70 | Constitutively maintained | Putnam et al. (2013) | Yes |
| SYMBOF_DN200180_c0_g1_i1.p1 | Endosymbiont | HSP70 | Constitutively maintained | Mayfield et al. (2011) | Yes |
| SYMBOF_DN197301_c4_g6_i3.p1 SYMBOF_DN217254_c0_g1_i1.p1 | Endosymbiont Endosymbiont | HSP90d HSP90 | Constitutively maintained Stress-sensitive | Mayfield et al. (2017a,b, c)Rosic et al. (2011)e | Yes No |
| SYMBOF_DN198813_c2_g3_i2.p1 | Endosymbiont | HSP90 | Stress-sensitive | Lin et al. (2019) | No |
| ANTHOZOAN-DINOFLAGELLATE STRESS RESPONSE AND BLEACHING-ASSOCIATED PROTEINS IDENTIFIED IN PRIOR WORKS | |||||
| OFAVBQ_DN172775_c0_g1_i1 | Coral host | Beta-gamma crystallin | Stress-sensitive | Downs et al. (2002) | No |
| comp97680_c0_seq1 | Coral host | Calmodulin | Stress-sensitive | Ricaurte et al. (2016) | No |
| SYMBOF_DN174588_c0_g1_i1.p1 | Endosymbiont | Calmodulin | Stress-sensitive | Weston et al. (2015) | No |
| OFAVBQ_DN223119_c2_g2_i5.p1 | Coral host | Carbonic anhydrase | Stress-sensitive | Weston et al. (2015) | No |
| OFAVBQ_DN214557_c1_g2_i5 | Coral host | Caspase | Stress-sensitive | Tchernov et al. (2011) | No |
| SYMBOF_DN215553_c1_g1_i2.p1 | Endosymbiont | Chaperone protein DNAK | Stress-sensitive | Weston et al. (2015) | No |
| SYMBOF_DN182054_c0_g1_i1.p1 | Endosymbiont | E3 ubiquitin-protein ligase | Stress-sensitive | Mayfield et al. (2018b) | No |
| OFAVBQ_DN194262_c1_g1_i1 | Coral host | HSP70 | Stress-sensitive | Seveso et al. (2017) | No |
| SYMBOF_DN204865_c1_g2_i1.p1 | Endosymbiont | HSP90 | Stress-sensitive | Ross (2014) | No |
| SYMBOF_DN165260_c0_g1_i2.p1 | Endosymbiont | Glutathione S-transferase | Stress-sensitive | Weston et al. (2015) | No |
| SYMBOF_DN239554_c0_g1_i1.p1 | Endosymbiont | Peptidylprolyl isomerase D | Stress-sensitive | Mayfield et al. (2018b) | Yes |
| SYMBOF_DN228106_c0_g1_i1.p1 | Endosymbiont | Peroxiredoxin-1 | Stress-sensitive | Weston et al. (2015) | No |
| SYMBOF_DN205301_c4_g1_i1.p1 | Endosymbiont | Peroxiredoxin-6e | Stress-sensitive | Mayfield et al. (2018b) | No |
| OFAVBQ_DN213774_c4_g1_i3 | Coral host | Superoxide dismutase | Constitutively maintained Stress-sensitive | Coles and Brown (2003) Oakley et al. (2017) | No No |
| OFAVBQ_DN199726_c0_g2_i1 | Coral host | Thioredoxin | Constitutively maintained Stress-sensitive | Cziesielski et al. (2018) Ricaurte et al. (2016) | No No |
| Contig14748 | Coral host | (poly)ubiquitin-B | Stress-sensitive | Coles and Brown (2003) | No |
| SYMBOF_DN214550_c0_g1_i1.p1 | Endosymbiont | Ubiquitinf | Stress-sensitive | Barshis et al. (2010) | No |
Select stress response “housekeeping” proteins, stress-sensitive proteins involved in the canonical eukaryotic stress response (CSR), and anthozoan-dinoflagellate bleaching-associated proteins identified in prior works.
Please note that a more detailed version of this table featuring additional proteins can be found in the Supplementary Material (Supplementary Table 6). In the “Prior mRNA/protein trend” column, molecules have either been deemed “stress-sensitive” or “constitutively maintained” based on results of prior gene expression (reference in regular font) or protein-based (bold font) studies. In the second half of the table, we instead looked at CSR and bleaching-associated proteins identified in prior proteomic works of anthozoans to determine whether trends were congruent. Please note that this does not represent an exhaustive list of all proteins involved in the CSR (Kultz, 2005), and some heat shock proteins (HSPs) found previously at the mRNA level to be stress sensitive (e.g., hsp32 in Seveso et al., 2020) were not sequenced herein. One CSR protein uncovered herein, an endosymbiont HSP40 (SYMBOF_DN201284_c0_g2_i5.p1), was down-regulated in bleaching corals.
aOctocoral study. bSea anemone study. cSubtle depth effects noted. dNote that one Symbiodiniaceae HSP90 differed in concentration across Symbiodiniaceae assemblages. eAffected by infection status in Sproles et al. (2019), with another isoform (peroxiredoxin-1) up-regulated at high temperatures in Weston et al. (2015). fSee Table 4.
As a notable exception, one CSR protein, peptidylprolyl isomerase (Kim et al., 2017), was found only in stressed endosymbiont communities in Mayfield et al. (2018b), and, when profiling the bleaching response on a genotype-to-genotype basis herein (Supplementary Material), this antioxidant was also up-regulated at high temperatures in endosymbiont communities within two of the three inshore genotypes (skyblue and grey60). This remains, at present, the lone protein that both represents a CSR protein and has been found to be up-regulated at high temperature in more than one study (Table 5). The peptidylprolyl isomerase is also critical in the cellular response of sea squirts (Lopez et al., 2017), and even marine fish (Kultz et al., 2007), to environmental stress. With the exception of this molecule, our results obfuscate efforts to use CSR mRNA or protein concentration levels as biomarkers for coral health/resilience given their constitutively high abundance. Rab protein levels, as also recommended by Weston et al. (2015), appear to be better candidates for gauging levels of coral heat dosing.
Constitutive activation of the CSR is not a common strategy for most life forms (Hochachka and Somero, 2002) given the high metabolic burden it poses (Sokolova et al., 2012). Specifically, the energy associated with re-establishing homeostasis could detract from growth and reproduction. Perhaps the excess energy from endosymbiont photosynthesis permits sustained periods of engaged CSR. It is also possible that the constitutive maintenance of a large number of CSR and antioxidant proteins could simply be indicative of the high protein turnover and metabolic rate associated with these corals in summer; the low frequencies of oxidative stress and apoptosis markers (Table 5 and OSDF) further signify that the control corals may not have actually been exhibiting a CSR. In the future, we will analyze the proteomes of field biopsies from these same coral colonies collected during different seasons to determine whether (1) the high control coral levels of certain CSR proteins documented herein reflect acclimation to a relatively high summer mean of 30°C or (2) they are presented year-round (sensu Mayfield et al., 2019). It will also be worthwhile repeating the experiment in winter (when ambient temperatures are only 21–22°C) to determine whether high-temperature-challenged corals acclimated to colder temperatures exhibit a more traditional CSR upon heating.
It was hypothesized over 20 years ago that protein turnover is the cellular process that ultimately dictates whether corals will effectively acclimatize (Gates and Edmunds, 1999), with highest turnover rates generally associated with the more thermotolerant massive corals like O. faveolata. If high concentrations of CSR and protein turnover-associated proteins are not necessarily indicative of stress, but instead high protein turnover rates, then the observation that the majority of the temperature-responsive proteins were involved in host coral lipid trafficking and metabolism and Symbiodiniaceae lipid trafficking and photosynthesis (see Supplementary Material.) could signify that these energy mobilization processes are modulated in a way that ensures that there is sufficient cellular ATP to maintain the high metabolic rate and turnover needed to continually break down proteins, refold denatured ones, and otherwise maintain proteostasis under elevated temperatures (beyond those already high protein turnover rates characteristic of controls). This could also explain the up-regulation of a key regulator of protein turnover, calumenin, in bleaching samples (herein and in Oakley et al., 2017); indeed, this protein is up-regulated upon chronic high-temperature exposure in other marine invertebrates (Sleight et al., 2018). Although signaling molecules went largely undetected by our proteomic approach, it is possible that lipid-signaling molecules generated by the lipid trafficking proteins uncovered served as the molecular “intermediaries” through which proteostasis and lipid metabolism were linked. Whether or not constitutively high protein turnover, or even an active CSR, is an energetically sustainable strategy over longer-term timescales remains to be determined, but, at the time of writing, the inshore corals continue to persist and reproduce annually, even as those around them succumb to bleaching and/or SCTLD.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories, as well as in the Online Supplementary Data File. The names of the repository/repositories and accession numbers can be found in the article and its Supplementary Material
Ethics statement
Ethical review and approval was not required for the animal study because this study was carried out with reef corals (see Supplementary Material for permitting information).
Author contributions
AM, CA, and GK collected the samples, ran the experiments, and carried out the molecular benchwork. AM analyzed the data, created the figures, and wrote the manuscript. IE and DM secured funding and mobilized boat and laboratory support.
Funding
This project was funded by the NOAA ’OMICs Initiative (Project #NRDD18978 to DM and Kelly Goodwin).
Acknowledgments
We thank members of the AOML Coral Program (OCED/AOML/NOAA) for field and experimental support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.660153/full#supplementary-material
References
1
Aranda M. Banaszak A. T. Bayer T. Luyten J. R. Medina M. Voolstra C. R. (2011). Differential sensitivity of coral larvae to natural levels of ultraviolet radiation during the onset of larval competence.Mol. Ecol.202955–2972. 10.1111/j.1365-294x.2011.05153.x
2
Barshis D. J. Stillman J. H. Gates R. D. Toonen R. J. Smith L. W. Birkeland C. (2010). Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: does host genotype limit phenotypic plasticity?Mol. Ecol.191705–1720. 10.1111/j.1365-294x.2010.04574.x
3
Bellantuono A. J. Granados-Cifuentes C. Miller D. J. Hoegh-Guldberg O. Rodriguez-Lanetty M. (2012). Coral thermal tolerance: tuning gene expression to resist thermal stress.PLoS One7:e50685. 10.1371/journal.pone.0050685
4
Brown B. E. (1997). Coral bleaching: causes and consequences.Coral Reefs16129–138.
5
Chen H. K. Mayfield A. B. Wang L. H. Chen C. S. (2017). Coral lipid bodies as the relay center interconnecting diel-dependent lipidomic changes in different cellular compartments.Sci. Rep.7:3244. 10.1038/s41598-017-02722-z
6
Chen H. K. Song S. N. Wang L. H. Mayfield A. B. Chen Y. J. Chen W. N. U. et al (2015). A compartmental comparison of major lipid species in a coral-Symbiodinium endosymbiosis: evidence that the coral host regulates lipogenesis of its cytosolic lipid bodies.PLoS ONE10:e0132519. 10.1371/journal.pone.0132519
7
Chen M. C. Cheng Y. M. Hong M. C. Fang L. S. (2005a). Molecular cloning of Rab5 (ApRab5) in Aiptasia pulchella and its retention in phagosomes harboring live zooxanthellae.Biochem. Biophys. Res. Commun.3241024–1033. 10.1016/j.bbrc.2004.09.151
8
Chen M. C. Cheng Y. M. Sung P. J. Kuo C. E. Fang L. S. (2003). Molecular identification of Rab7 (ApRab7) in Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae.Biochem. Biophys. Res. Commun.308585–594.
9
Chen M. C. Hong M. C. Huang Y. S. Liu M. C. Cheng Y. M. Fang L. S. (2005b). ApRab11, a cnidarian homologue of the recycling regulatory protein Rab11, is involved in the establishment and maintenance of the Aiptasia-Symbiodinium endosymbiosis.Biochem. Biophys. Res. Commun.3381607–1616. 10.1016/j.bbrc.2005.10.133
10
Chen W. N. U. Kang H. J. Weis V. M. Mayfield A. B. Fang L. S. Chen C. S. (2012). Diel rhythmicity of lipid body formation in a coral-Symbiodinium endosymbiosis.Coral Reefs31521–534. 10.1007/s00338-011-0868-6
11
Chow A. M. Ferrier-Pagès C. Khalouei S. Reynaud S. Brown I. R. (2009). Increased light intensity induces heat shock protein Hsp60 in coral species.Cell Stress Chaperones14469–476. 10.1007/s12192-009-0100-6
12
Coles S. L. Brown B. E. (2003). Coral bleaching-capacity for acclimatization and adaptation. Adv. Mar. Biol. 46183–223. 10.1016/s0065-2881(03)46004-5
13
Cziesielski M. J. Liew Y. J. Cui G. Schmidt-Roach S. Campana S. Marondedze C. et al (2018). Multi-omics analysis of thermal stress response in a Zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts.Proc. R. Soc. B.28520172654. 10.1098/rspb.2017.2654
14
Downs C. A. Fauth J. E. Halas J. C. Dustan P. Bemiss J. Woodley C. M. (2002). Oxidative stress and seasonal coral bleaching.Free Radical Biol. Med.33533–543. 10.1016/s0891-5849(02)00907-3
15
Downs C. A. Kramarsky-Winter E. Martinez J. Kushmaro A. Woodley C. M. Loya Y. et al (2009). Symbiophagy as a cellular mechanism for coral bleaching.Autophagy5211–216. 10.4161/auto.5.2.7405
16
Gates R. D. Edmunds P. J. (1999). The physiological mechanisms of acclimatization in tropical reef corals.Am. Zool.3930–43. 10.1093/icb/39.1.30
17
Gintert B. E. Manzello D. P. Enochs I. C. Kolodziej G. Carlton R. D. Gleason A. C. R. et al (2018). Marked annual coral bleaching resilience of an inshore patch reef in the Florida Keys: a nugget of hope, aberrance, or last man standing?Coral Reefs37533–547. 10.1007/s00338-018-1678-x
18
Hochachka P. W. Somero G. N. (2002). Biochemical Adaptation. Oxford: Oxford University Press.
19
Hoegh-Guldberg O. Poloczanska E. S. Skirving W. Dove S. (2017). Coral reef ecosystems under climate change and ocean acidification.Front. Mar. Sci.4:158. 10.3389/fmars.2017.00158
20
Jones R. J. Hoegh-Guldberg O. Larkum A. W. D. Schreiber U. (1998). Temperature-induced bleaching of corals begins with impairment of the CO2 fixation metabolism in Zooxanthellae.Plant Cell Environ.211219–1230. 10.1046/j.1365-3040.1998.00345.x
21
Kim E. Y. Choi Y. H. Choi C. G. Nam T. J. (2017). Effects of the cyclophilin-type peptidylprolyl cis-trans isomerase from Pyropia yezoensis against hydrogen peroxide-induced oxidative stress in HepG2 cells.Mol. Med. Rep.154132–4138. 10.3892/mmr.2017.6517
22
Kultz D. (2005). Molecular basis of the cellular stress response.Annu. Rev. Physiol.67225–257. 10.1146/annurev.physiol.67.040403.103635
23
Kultz D. Fiol D. Valkova N. Gomez-Jimenez S. Chan S. Y. Lee J. (2007). Functional genomics and proteomics of the osmotic stress response in “non-model” organisms.J. Exp. Biol2101593–1601. 10.1242/jeb.000141
24
Levy O. Karako-Lampert S. Ben-Asher H. W. Zoccola D. Pages G. Ferrier-Pages C. (2016). Molecular assessment of the effect of light and heterotrophy in the scleractinian coral Stylophora pistillata.Proc. R. Soc. B.28320153025. 10.1098/rspb.2015.3025
25
Lin S. Yu L. Zhang H. (2019). Transcriptomic responses to thermal stress and varied phosphorus conditions in Fugacium kawagutii.Microorganisms796. 10.3390/microorganisms7040096
26
Lock J. G. Stow J. L. (2005). Rab11 in recycling endosomes regulates the sorting and basolateral transport of e-cadherin.Mol. Biol. Cell161744–1755. 10.1091/mbc.e04-10-0867
27
Lõhelaid H. Teder T. Samel N. (2014). Lipoxygenase-allene oxide synthase pathway in octocoral thermal stress response.Coral Reefs34143–154. 10.1007/s00338-014-1238-y
28
Lopez C. E. Sheehan H. C. Vierra D. A. Azzinaro P. A. Meedel T. H. Howlett N. G. et al (2017). Proteomic responses to elevated ocean temperature in ovaries of the ascidian Ciona intestinalis.Biol. Open6943–955. 10.1242/bio.024786
29
Manzello D. P. (2015). Rapid recent warming of coral reefs in the Florida Keys.Sci. Rep.516762. 10.1038/srep16762
30
Manzello D. P. Enochs I. C. Kolodziej G. Carlton R. (2015). Recent decade of growth and calcification of Orbicella faveolata in the Florida Keys: an inshore−offshore comparison.Mar. Ecol. Prog. Ser.52181–89. 10.3354/meps11085
31
Manzello D. P. Matz M. V. Enochs I. C. Valentino L. Carlton R. D. Kolodziej G. et al (2019). Role of host genetics and heat−tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming.Glob. Change Biol.251016–1031. 10.1111/gcb.14545
32
Mayfield A. B. Chan P. H. Putnam H. M. Chen C. S. Fan T. Y. (2012). The effects of a variable temperature regime on the physiology of the reef-building coral Seriatopora hystrix: results from a laboratory-based reciprocal transplant. J. Exp. Biol. 2154183–4195. 10.1242/jeb.071688
33
Mayfield A. B. Chen C. S. Dempsey A. C. (2017a). Biomarker profiling in reef corals of Tonga’s Ha’apai and Vava’u Archipelagos.PLoS One.12:e0185857. 10.1371/journal.pone.0185857
34
Mayfield A. B. Chen C. S. Dempsey A. C. (2017b). Identifying corals displaying aberrant behavior in Fiji’s Lau Archipelago.PLoS One.12:e0177267. 10.1371/journal.pone.0177267
35
Mayfield A. B. Chen C. S. Dempsey A. C. (2017c). The molecular ecophysiology of closely related pocilloporid corals of New Caledonia.Platax141–45.
36
Mayfield A. B. Chen C. S. Dempsey A. C. (2019). Modeling environmentally-mediated variation in reef coral physiology.J. Sea Res.14544–54. 10.1016/j.seares.2019.01.003
37
Mayfield A. B. Chen Y. J. Lu C. Y. Chen C. S. (2018a). Exploring the environmental physiology of the Indo-Pacific reef coral Seriatopora hystrix using differential proteomics.Open J. Mar. Sci.8223–252. 10.4236/ojms.2018.82012
38
Mayfield A. B. Chen Y. J. Lu C. Y. Chen C. S. (2018b). The proteomic response of the reef coral Pocillopora acuta to experimentally elevated temperature.PLoS One13:e0192001. 10.1371/journal.pone.0192001
39
Mayfield A. B. Wang L. H. Tang P. C. Hsiao Y. Y. Fan T. Y. Tsai C. L. et al (2011). Assessing the impacts of experimentally elevated temperature on the biological composition and molecular chaperone gene expression of a reef coral.PLoS One6:e26529. 10.1371/journal.pone.0026529
40
Mayfield A. B. Wang Y. B. Chen C. S. Chen S. H. Lin C. Y. (2014). Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures.Mol. Ecol.235816–5830. 10.1111/mec.12982
41
Mayfield A. B. Wang Y. B. Chen C. S. Chen S. H. Lin C. Y. (2016). Dual-compartmental transcriptomic+ proteomic analysis of a marine endosymbiosis exposed to environmental change.Mol. Ecol.255944–5958. 10.1111/mec.13896
42
Montealegre S. van Endert P. M. (2010). Endocytic recycling of MHC class I molecules in non-professional antigen presenting and dendritic cells.Front. Immunol.9:3098. 10.3389/fimmu.2018.03098
43
Musada G. R. Dvoriantchikova G. Myer C. Ivanov D. Bhattacharya S. K. Hackam A. S. (2020). The effect of extrinsic Wnt/β-catenin signaling in Muller glia on retinal ganglion cell neurite growth.Dev. Neurobiol.8098–110. 10.1002/dneu.22741
44
Nakamura M. Morita M. Kurihara H. Mitarai S. (2012). Expression of hsp70, hsp90 and hsf1 in the reef coral Acropora digitifera under prospective acidified conditions over the next several decades.Biol. Open175–81. 10.1242/bio.2011036
45
Oakley C. A. Durand E. Wilkinson S. P. Peng L. Weis V. M. Grossman A. R. et al (2017). Thermal shock induces host proteostasis disruption and endoplasmic reticulum stress in the model symbiotic cnidarian Aiptasia.J. Proteome Res.162121–2134. 10.1021/acs.jproteome.6b00797
46
Pinzón J. H. Kamel B. Burge C. A. Harvell C. D. Medina M. Weil E. et al (2015). Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral.R. Soc. Open Sci.2:140214. 10.1098/rsos.140214
47
Poli D. Fabbri E. Goffredo S. Airi V. Franzellitti S. (2017). Physiological plasticity related to zonation affects hsp70 expression in the reef-building coral Pocillopora verrucosa.PLoS One12:e0171456. 10.1371/journal.pone.0171456
48
Putnam H. M. Mayfield A. B. Fan T. Y. Chen C. S. Gates R. D. (2013). The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2.Mar. Biol.1602157–2173. 10.1007/s00227-012-2129-9
49
Ricaurte M. Schizas N. V. Ciborowski P. Boukli N. M. (2016). Proteomic analysis of bleached and unbleached Acropora palmata, a threatened coral species of the Caribbean.Mar. Pollut. Bull.107224–232. 10.1016/j.marpolbul.2016.03.068
50
Rosic N. N. Pernice M. Dove S. Dunn S. Hoegh-Guldberg O. (2011). Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching.Cell Stress Chaperones1669–80. 10.1007/s12192-010-0222-x
51
Ross C. (2014). Nitric oxide and heat shock protein 90 co-regulate temperature-induced bleaching in the soft coral Eunicea fusca.Coral Reefs33513–522. 10.1007/s00338-014-1142-5
52
Ruiz-Jones L. J. Palumbi S. R. (2017). Tidal heat pulses on a reef trigger a fine-tuned transcriptional response in corals to maintain homeostasis.Sci. Adv.3e1601298. 10.1126/sciadv.1601298
53
Seveso D. Arrigoni R. Montano S. Maggioni D. Orlandi I. Berumen M. L. et al (2020). Investigating the heat shock protein response involved in coral bleaching across Scleractinian species in the central Red Sea.Coral Reefs3985–98. 10.1007/s00338-019-01878-6
54
Seveso D. Montano S. Reggente M. A. Maggioni D. Orlandi I. Galli P. et al (2017). The cellular stress response of the scleractinian coral Goniopora columna during the progression of the black band disease.Cell Stress Chaperones22225–236. 10.1007/s12192-016-0756-7
55
Siebeck U. Marshall N. Klüter A. Hoegh-Guldberg O. (2006). Monitoring coral bleaching using a colour reference card.Coral Reefs25453–460. 10.1007/s00338-006-0123-8
56
Sleight V. A. Peck L. S. Dyrynda E. A. Smith V. J. Clark M. S. (2018). Cellular stress responses to chronic heat shock and shell damage in temperate Mya truncata.Cell Stress Chaperones231003–1017. 10.1007/s12192-018-0910-5
57
Sokolova I. M. Frederich M. Bagwe R. Lannig G. Sukhotin A. A. (2012). Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates.Mar. Environ. Res.791–15. 10.1016/j.marenvres.2012.04.003
58
Sproles A. E. Oakley C. A. Matthews J. L. Peng L. Owen J. G. Grossman A. R. et al (2019). Proteomics quantifies protein expression changes in a model cnidarian colonised (sp.) by a thermally tolerant but suboptimal symbiont.ISME J.132334–2345. 10.1038/s41396-019-0437-5
59
Tchernov D. Kvitt H. Maramaty L. Bibby T. S. Gorbunov M. Y. Rosenfield H. et al (2011). Apoptosis and the selective survival of host animals following thermal bleaching in Zooxanthellate corals.Proc. Natl. Acad. Sci. U.S.A.1089905–9909. 10.1073/pnas.1106924108
60
Vizcaíno J. Deutsch E. Wang R. Csordas A. Reisinger F. Rios D. et al (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination.Nat. Biotechnol.32223–226. 10.1038/nbt.2839
61
Weston A. J. Dunlap W. C. Beltran V. H. Starcevic A. Hranueli D. Ward M. et al (2015). Proteomics links the redox state to calcium signaling during bleaching of the Scleractinian coral Acropora microphthalma on exposure to high solar irradiance and thermal stress.Mol. Cell Proteomics14585–595. 10.1074/mcp.m114.043125
62
Whalen K. E. Lane A. L. Kubanek J. Hahn M. E. (2010). Biochemical warfare on the reef: the role of glutathione transferases in consumer tolerance of dietary prostaglandins.PLoS One5:e8537. 10.1371/journal.pone.0008537
Summary
Keywords
coral reefs, dinoflagellates, global climate change, proteomics, symbiosis
Citation
Mayfield AB, Aguilar C, Kolodziej G, Enochs IC and Manzello DP (2021) Shotgun Proteomic Analysis of Thermally Challenged Reef Corals. Front. Mar. Sci. 8:660153. doi: 10.3389/fmars.2021.660153
Received
28 January 2021
Accepted
21 April 2021
Published
19 July 2021
Volume
8 - 2021
Edited by
Davide Seveso, University of Milano-Bicocca, Italy
Reviewed by
Brad Buckley, Portland State University, United States; Paul Yancey, Whitman College, United States
Updates
Copyright
© 2021 Mayfield, Aguilar, Kolodziej, Enochs and Manzello.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anderson B. Mayfield, abm64@miami.edu; anderson.mayfield@noaa.gov
This article was submitted to Aquatic Physiology, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.